Abstract
Serine palmitoyltransferase complex (SPT) mediates the first and rate-limiting step in the de novo sphingolipid biosynthetic pathway. The larger subunits SPTLC1 and SPTLC2/SPTLC3 together form the catalytic core while a smaller third subunit either SSSPTA or SSSPTB has been shown to increase the catalytic efficiency and provide substrate specificity for the fatty acyl-CoA substrates. The in vivo biological significance of these smaller subunits in mammals is still unknown. Here, using two null mutants, a conditional null for ssSPTa and a null mutant for ssSPTb, we show that SSSPTA is essential for embryogenesis and mediates much of the known functions of the SPT complex in mammalian hematopoiesis. The ssSPTa null mutants are embryonic lethal at E6.5 much like the Sptlc1 and Sptlc2 null alleles. Mx1-Cre induced deletion of ssSPTa leads to lethality and myelopoietic defect. Chimeric and competitive bone marrow transplantation experiments show that the defect in myelopoiesis is accompanied by an expansion of the Lin−Sca1+c-Kit+ stem and progenitor compartment. Progenitor cells that fail to differentiate along the myeloid lineage display evidence of endoplasmic reticulum stress. On the other hand, ssSPTb null mice are homozygous viable, and analyses of the bone marrow cells show no significant difference in the proliferation and differentiation of the adult hematopoietic compartment. SPTLC1 is an obligatory subunit for the SPT function, and because Sptlc1−/− and ssSPTa−/− mice display similar defects during development and hematopoiesis, we conclude that an SPT complex that includes SSSPTA mediates much of its developmental and hematopoietic functions in a mammalian model.
Keywords: ssSPTa, ssSPTb, serine palmitoyltransferase, sphingolipid, hematopoiesis, hematopoietic stem cells, myelopoiesis, granulopoiesis, bone marrow cells, ER stress
Abbreviations: BMCs, bone marrow cells; CMP, common myeloid precursor; EBs, embryoid bodies; GM-CSF, granulocyte macrophage colony stimulating factor; GMP, granulocyte-macrophage precursor; HSCs, hematopoietic stem cells; IL-3, interleukin; LCB, long-chain base; LK, Lin− c-Kit+ Sca1−; LSK, Lin−c-Kit+, Sca1+; LT-HSCs, long-term hematopoietic stem cells; MEP, megakaryocyte-erythrocyte precursor; MPP, multipotent progenitor cell; PLP, pyridoxal phosphate; SPT, serine palmitoyltransferase; ST-HSCs, short-term hematopoietic stem cells; UPR, unfolded protein response
Sphingolipids are important structural components of all eukaryotic and many sphingolipid-containing prokaryotic membranes (1, 2, 3). They contain a long-chain base (LCB) commonly referred to as sphingoid base as core structural element. They are 2-amino-1,3-dihydroxyalkanes-or-enes (4). The alkyl chain length could be variable, have double bonds, and further modified with N-acylation and other modifications to generate a diverse array of sphingolipids in eukaryotes and some specific sphingolipid containing prokaryotes. Sphingolipids are a minor but very important class of lipids with important roles in the biology of eukaryotes. They are amphipathic in nature providing barrier functions and influence membrane fluidity (5, 6). They form specialized structures within membrane environments playing important role in the interactions of the cell with other cells or its environment (7). While on-demand generation of a particular sphingolipid metabolite could be channeled via different metabolic routes of the sphingolipid metabolic pathway, the de novo sphingolipid biosynthetic pathway remains essential for embryonic development and functioning of several adult organs (8, 9, 10, 11, 12, 13, 14, 15). It is a well conserved biosynthetic pathway among eukaryotes and generates numerous bioactive sphingolipids (1, 16, 17, 18, 19, 20, 21, 22, 23). Because many of the intermediates are bioactive, several of the enzymes of the pathway are regulated.
The de novo pathway is essential for survival of all eukaryotes and is initiated by the condensation of serine with a fatty acyl-CoA (usually palmitoyl-CoA) to form 3-ketosphinganine (11, 13, 24). This is the first, committed, and rate limiting step of the pathway. It is catalyzed by a heterotrimeric enzyme complex called serine palmitoyltransferase (SPT) (25, 26). In mammals, the complex consists of two large subunits called SPTLC1 and SPTLC2 (or SPTLC3) and one of the two small subunits SSSPTA or SSSPTB (27, 28). The two large subunits are homologous and contain several structural elements essential for binding to the cofactor pyridoxal phosphate (PLP), although a critical residue for binding is mutated in the SPTLC1 subunit. Hence, the PLP binding is mediated by the large SPTLC2 or three subunits. The active site of the enzyme is believed to be created by an interface generated by SPTLC1-SPTLC2 interaction. Amongst the small subunits, SSSPTA primary sequence is about 99% conserved between human and mouse (1 homologous substitution in 71 amino acids). Although there are about 11 amino acid changes between the human and mouse SSSPTB sequences, most of them are conservative substitutions indicating a high degree of conservation. In mammals, SSSPTA (about 71 amino acids) and SSSPTB (about 76 amino acids) share about 63% sequence similarity (Fig. 1A). While SPTLC1-SPTLC2 dimers show basal enzymatic activity, the binding of the third subunit increases the catalytic efficiency by as much as 100-fold and also provide some fatty acyl-CoA preference to the complex (27, 29). The incorporation of SSSPTA into the complex allows for preferential utilization of palmitoyl-CoA as substrate, whereas the presence of SSSPTB has been shown in vitro to allow the incorporation of both palmitoyl-CoA or longer chain (C18-C20) fatty acyl-CoAs as substrates for the condensation reaction (2, 27, 29). The relative contributions of the two small subunits in the SPT complex during development and its function in a mammalian model system is yet to be evaluated.
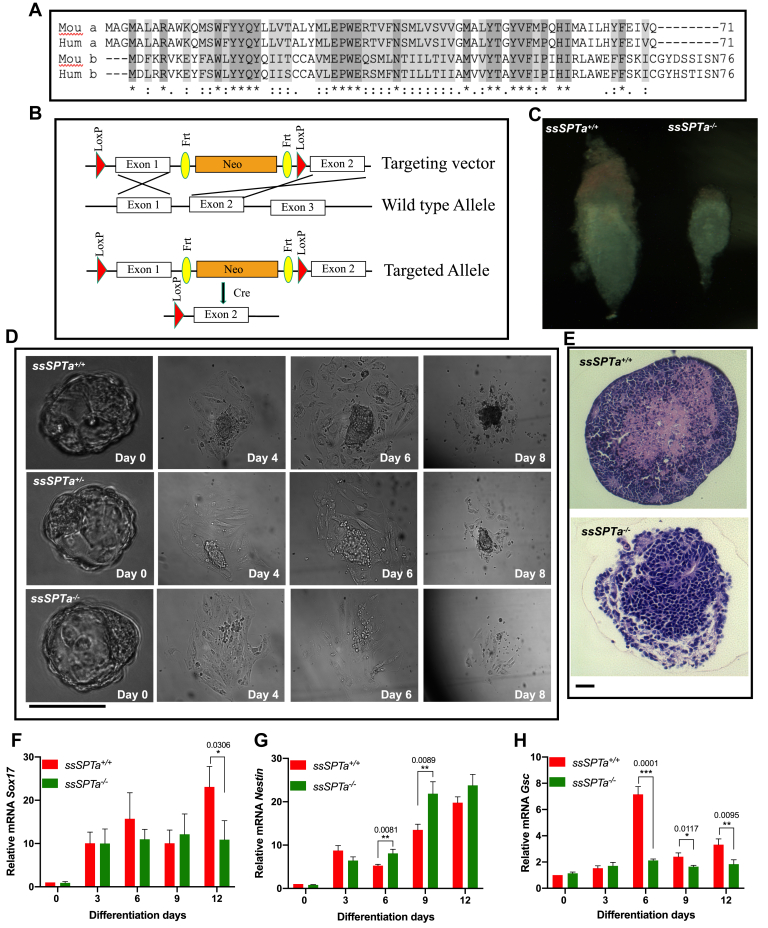
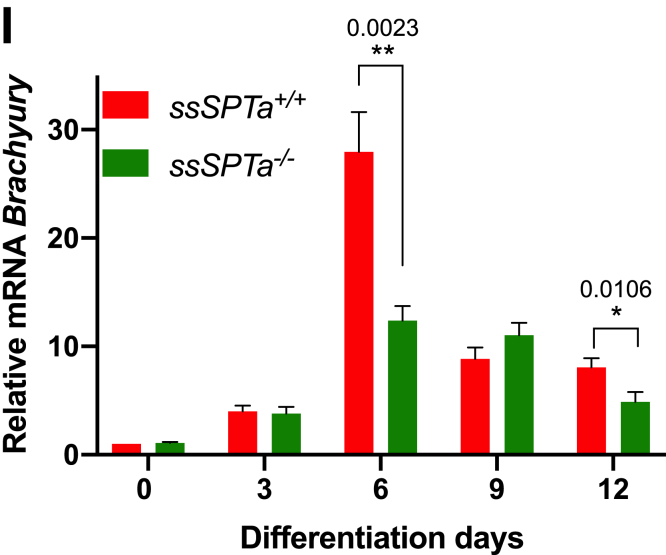
Figure 1.
ssSPTa deletion causes embryonic lethality at E6.5. A, amino acid sequence alignment of SSSPTA/B from mouse and Homo sapiens using Clustal W. B, generation of ssSPTa conditional knockout mice. Strategy to delete exon 1 and generate ssSPTa−/−. C, ssSPTa+/+ and ssSPTa−/− mouse embryos harvested at E6.5. D, microscopic images of ssSPTa+/+ and ssSPTa+/− and ssSPTa−/− embryonic inner cells mass proliferation at various stages. E, ssSPTa+/+ and ssSPTa−/− embryoid differentiation from embryonic stem cells. F–I, real time qPCR analysis of ectodermal marker—Nestin, endodermal marker—Sox17, and mesodermal marker—Gsc and mesodermal marker—Brachyury in ssSPTa+/+and ssSPTa−/− embryonic stem cells differentiation to embryoid body. The gene expressions were normalized to Gapdh. The qPCR experiments were averaged from three biological samples, and for each sample, the qPCR was performed in technical triplicates. All graphs are represented as mean ± SEM. p Value < 0.05 is significant, calculated from unpaired t test. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. Scale bar 100 μm.
Here, we report in vivo studies from conditional null mutants of the mouse ssSPTa and ssSPTb. We show that germline deletion of ssSPTa results in embryonic lethality around E6.5, and embryoid bodies (EBs) derived from ES cells show differentiation defects. We demonstrate that loss of ssSPTa in the adult hematopoietic stem cells (HSCs) results in defective myelopoietic differentiation while erythropoiesis is relatively spared. Chimeric and competitive bone marrow transplantation studies showed that defects in myelopoiesis are accompanied by relative expansion of the stem and progenitor compartment. Molecular analysis indicates that aberrant myelopoiesis is accompanied by endoplasmic reticulum stress. These phenotypes are similar to the defects observed in Sptlc1 null mutants (30). On the other hand, germline null mutants of ssSPTb are homozygous viable and do not show defects in adult hematopoiesis. Our results indicate that SSSPTA mediates much of the developmental and hematopoietic functions of the SPT complex in mammals.
Results
ssSPTa null mutants are homozygous lethal
To understand the in vivo functional significance of the SSSPTA subunit, we generated a floxed allele for the gene. The ssSPTa gene is localized to chromosome 12 (Chromosome 12, 23.46 cM), and the ORF of the gene is encoded by two exons. Using the standard Cre-lox system and Bac recombineering, we targeted exon 1 (aa 1–39) for conditional deletion of the gene (Fig. 1B) (31). To generate a zygotic null allele of ssSPTa, we employed Actin-Cre (32). Genotyping of about 289 live-born progenies from heterozygous intercross indicated that about 64% were heterozygous and about 36% were wild type, and no homozygous mutants were recovered (Table S1). To determine the embryonic stage at which the fetuses were dying, we set up timed mating of heterozygous intercross and genotyped the embryos at different times of gestation. We noticed that ssSPTa−/− embryos begin to degenerate and get absorbed between E6.5 and E7.5. Sptlc1−/− and Sptlc2−/− null have also been reported to be early embryonic lethal (24, 30). E6.5 embryos were smaller than the corresponding ssSPTa+/+ harvested from the same uterus, indicating disruption of the normal developmental process in the mutant embryos (Figs. 1C and S1A). Mutant embryos displayed significant lysotracker dye and TUNEL staining implying increased apoptosis compared with the wild-type embryos (Fig. S1, B and C). Mutant embryos also showed reduced staining with bromodeoxyuridine staining indicating reduced proliferation (Fig. S1D). Blastocysts were obtained from ssSPTa+/+, sSPTa+/−, and ssSPTa−/− conceptus, and the blastocyst inner cell mass outgrowth was assessed for 8 days. For the first 2 days, no changes were observed between control and mutant samples. The ssSPTa+/+, ssSPTa+/−, and ssSPTa−/− blastocysts successfully hatched from zona pellucida and initiated growth (Fig. 1D). However, ssSPTa−/− inner mass cells gradually stopped proliferating and were lost around day 6. The impaired growth of inner mass cells of ssSPTa−/− blastocysts suggests that ssSPTa deficiency causes a developmental arrest and eventually leads to the death of the embryos. EBs were derived from ssSPTa+/+ and ssSPTa−/− embryonic stem cells (Fig. 1E). Because EBs have been considered to be the developmental equivalent of the egg cylinder–staged mouse embryos, we compared the SSSPTA-depleted EBs with wild-type EBs. After 14 days of culture in suspension, the wild-type EBs had a distinct outer layer surrounding differentiated cells and EB cavity. In contrast, although ssSPTa null ESCs were able to form putative EBs, these EBs appeared blocked in the differentiation process, forming undifferentiated masses of cells that lacked cavity formation and differentiated structures. The differentiation of germ layers in the EBs was examined by quantitative PCR (qPCR) using markers for ectoderm (Nestin), endoderm (Sox17), and mesoderm (Gsc). Despite morphological differences in the appearance of the EBs, there were no major differences in expression of ectodermal and endodermal markers Nestin and Sox17, respectively (Fig. 1, F and G). On the other hand, Gsc and Brachyury, markers for mesoderm, showed a consistent decrease in the mutant EBs especially around day 6 (Fig. 1, H and I). Collectively, the experiments described above indicated that ssSPTa null mutants have differentiation defects and are embryonic lethal between E6.5 and E7.5 like Sptlc1 and Sptlc2 null mutants. Our observations support the notion that ssSPTa is a functional third subunit of the SPT complex necessary for mouse embryonic development.
Adult ssSPTa−/− hematopoietic stem cells fail to differentiate along myeloid lineage
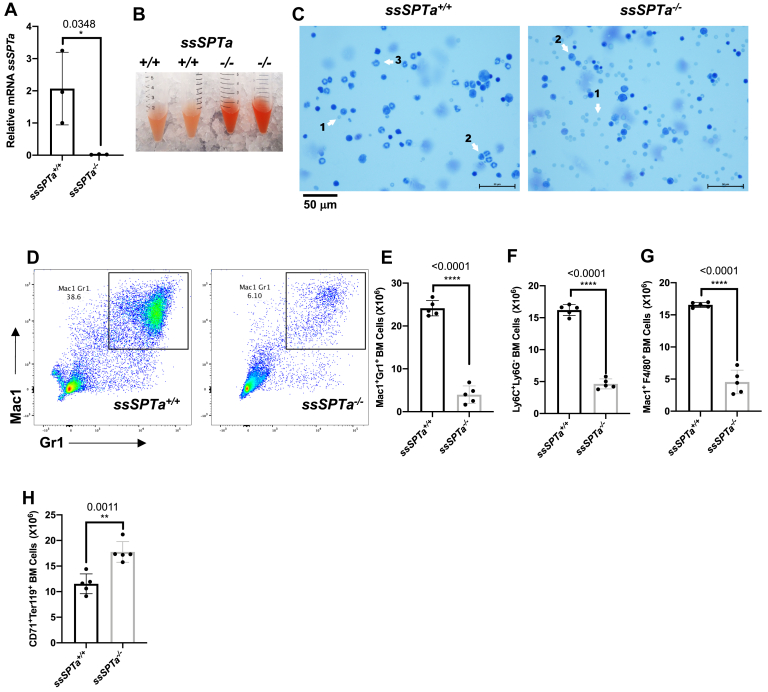
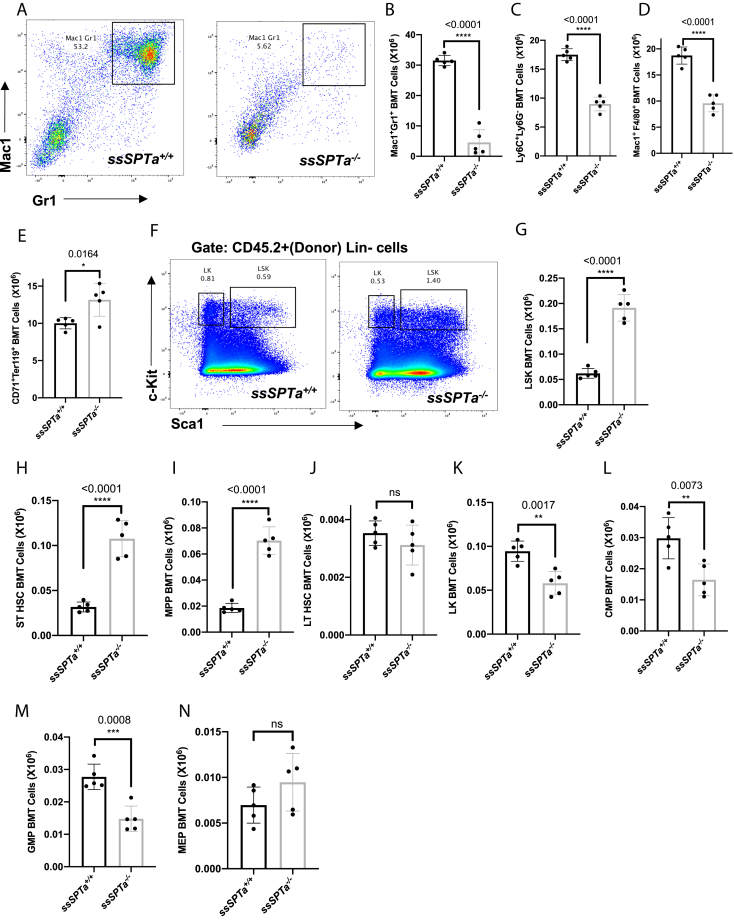
The adult hematopoietic system is a tissue of mainly mesodermal origin (33). To evaluate the contribution of SSSPTA subunit to SPT function in adult hematopoiesis, we deleted ssSPTa in the adult hematopoietic stem cells using Mx1-Cre (34). For these experiments, to activate the Mx1-Cre promoter, ssSPTaflox/flox, and control ssSPTa+/+, Mx1-Cre mice were subjected to poly(I:C) treatment. Treated animals begin to die after 8 days after poly(I:C) treatment while control animals remain relatively healthy. Therefore, for these experiments, the bone marrow cells (BMCs) were harvested at day 8 and the genotype ascertained by PCR (Fig. S2A). The disruption of the gene was confirmed by qPCR analysis of the ssSPTa transcript (Fig. 2A). Similar to Sptlc1−/− mice, ssSPTa−/− mutants also showed intestinal fluid accumulation. Hematoxylin and eosin staining of the intestine revealed loss of villi and altered appearance of the mucosal and submucosal layers (Fig. S2, B and C). Bone marrow suspensions isolated from the ssSPTa−/− were red in color, and cytocentrifuged preparations of the BMCs contained more RBCs and fewer differentiated myeloid cells (Fig. 2, B and C). The total BM cellularity of the ssSPTa−/− mice was reduced compared with similarly treated ssSPTa+/+ mice (Fig. S2D). To define how loss of ssSPTa influences lineage development in hematopoiesis, we analyzed BMCs, thymocytes, and splenocytes for lineage markers expressed on myeloid cells (Mac1;Gr1); erythroid cells (CD71;Ter119); B lymphoid cells (IgM), and T lymphocytes (CD4;CD8) using flow cytometry. Mac1+Gr1+ that mark cells of granulocytic lineage are reduced 5-fold in the ssSPTa−/− BMCs (Fig. 2, D and E). The monocyte (Ly6C+;Ly6G−) and macrophage (Mac1+;F4/80+) lineages were substantially decreased (Fig. 2, F and G). On the other hand, cells of the erythrocytic lineage were increased (Fig. 2H, Fig. S2, E–G). The cellularity of the spleen was slightly decreased, and there was a modest but consistent decrease in the number of Mac1+Gr1+ (Fig. S2, H and I). There were no significant changes in thymus except for a slight reduction in CD8+ thymocytes (Fig. S2, J–M). Also, the number of IgM+ BMCs were not affected (Fig. S2N). These data indicate that loss of ssSPTa in the adult hematopoietic system primarily compromises myeloid differentiation while relatively sparing erythropoiesis and lymphopoiesis.
Figure 2.
Mx1-Cre deletion of ssSPTa affects bone marrow myeloid cells. A, real time qPCR analysis of ssSPTa mRNA expression in BM cells isolated from ssSPTa+/+ and ssSPTa−/− mice on day eight after poly(I:C) injection. Gene expression was normalized to β-actin. qPCR results were from three independent experiments. B, bone marrow tissue isolated from ssSPTa+/+ and ssSPTa−/− mice. C, Wright-Giemsa staining of BMCs from the ssSPTa+/+ and ssSPTa−/− mice. 1 - RBC, and 2-metamyelocyte, and 3-segmented band cell. D, fluorescence activated cell sorting plots of myeloid bone marrow cells analyzed by Mac-1 and Gr-1 staining 8 days after poly(I:C) injection (n = 5). E, the total numbers of Mac-1+Gr-1+, F, Ly6C+Ly6G-. G, Mac-1+ F4/80+ cells were for ssSPTa+/+ and ssSPTa−/− mice (n = 5). H, the total numbers of CD71+Ter119+ BMCs were plotted for the ssSPTa+/+ and ssSPTa−/− mice (n = 5). All graphs are represented as mean ± SEM. p Value < 0.05 is significant, calculated from unpaired t test. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗∗p < 0.0001. BMCs, bone marrow cells.
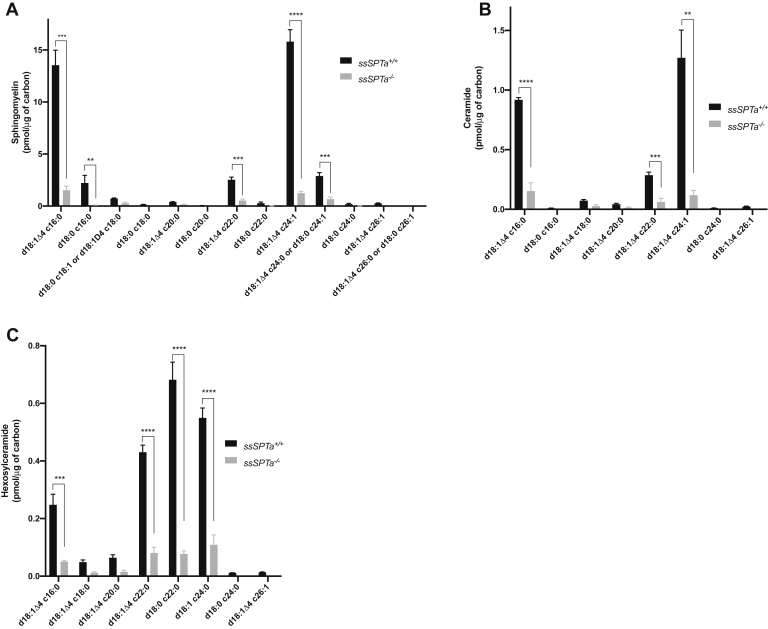
Mass spectrometric quantitative analysis of sphingolipids in the BMCs of the ssSPTa−/− indicated a severe reduction in the levels of almost all species of the major sphingomyelins, ceramides, and hexosylceramides compared with that of ssSPTa+/+ (Fig. 3, A–C). Thus, the hematopoietic defects observed in the ssSPTa null BMCs is accompanied by a severe deficiency of majority of the sphingolipids in these cells. The comparison of the sphingolipid profile between ssSPTa+/+ and ssSPTa−/− described above suggests that SSSPTA plays a major role in the de novo biosynthesis of sphingolipids in the adult hematopoietic system. As with Sptlc1−/− BMCs, we see a correlation between loss of SSSPTA function, significant decrease in major sphingolipids, and defective myelopoiesis in the BMCs (30). It is likely that SSSPTA is a critical component of the SPT complex in mammalian adult hematopoietic system.
Figure 3.
Sphingolipids measurement by mass spectrometry of ssSPTa+/+ and ssSPTa−/− BMCs. A, sphingomyelins, (B) ceramides, and (C) hexosylceramides were analyzed by mass spectrometry and normalized to carbon content and are plotted for the ssSPTa+/+ and ssSPTa−/− (n = 3) 8 days after poly(I:C) injection. The results are from three independent BM preparations from each genotype. All graphs are represented as mean ± SEM. p Value < 0.05 is significant, calculated from unpaired t test. ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. BMCs, bone marrow cells.
To assess the cell intrinsic requirement of SSSPTA in adult hematopoiesis, we performed noncompetitive and competitive bone marrow repopulation assays using ssSPTa+/+ Mx1-Cre and ssSPTaflox/flox Mx1-Cre BMCs. These assays gauge reconstitution of different lineages of BMCs in irradiated mice transplanted with donor cells and is routinely used to determine hematopoietic stem and progenitor cell (HSPC) function in vivo. For noncompetitive experiments, the transplanted mice were treated with poly(I:C) (15 μg per gram of body weight, followed by a second dose 2 days later) 6 weeks after transplantation of ssSPTa+/+ Mx1-Cre and ssSPTaflox/flox Mx1-Cre BMCs. The chimeric ssSPTa−/− mice did not survive beyond 4 to 5 weeks, and therefore, we analyzed mice 3 weeks after the last injection (Fig. S3A). The BM cellularity was reduced in the mutant compared with the control (Fig. S3B). In the chimeric mice, the total donor Mac1+Gr1+ neutrophils were decreased 6-fold, suggesting that SSSPTA is intrinsically required for the development of granulocytes (Fig. 4, A and B). Macrophage and monocytic lineages were also compromised (Fig. 4, C and D). Because CD45 is expressed in the early stages of erythroid development, we examined BMCs for donor erythroid progenitor cells and found that these cells were slightly increased in the BM of ssSPTa−/− mice, indicating erythroid development was not compromised (Fig. 4E and Fig. S3, C–E). The total number of ssSPTa−/− thymocytes were decreased accompanied by a decrease in the number of the immature CD4+CD8+ double positive cells (Fig. S3, F and G). However, we did not see a significant difference in the number of mature single positive CD4+ or CD8+ cells (Fig. S3, H and I). Finally, we observed no significant difference in B-lymphoid cells in the BM of ssSPTa+/+ and ssSPTa−/− chimeric mice (Fig. S3J). These experiments reveal that loss of SSSPTA causes defects in myeloid differentiation in the adult hematopoietic system.
Figure 4.
ssSPTa deletion impairs myeloid differentiation and spares erythroid differentiation in chimeric mice. A, fluorescence activated cell sorting plot of myeloid differentiation in BMCs analyzed by Mac-1 and Gr-1 staining. B, the total numbers of Mac-1+Gr-1+, (C) Ly6C+Ly6G-, (D) Mac-1+F4/80+ cells were plotted for the ssSPTa+/+ and ssSPTa−/− (n = 5). E, the total numbers of CD71+Ter119+ cells were plotted for the transplanted ssSPTa+/+ and ssSPTa−/− mice (n = 5). F, the LSK and LK populations of HSPCs from transplanted bone marrow were analyzed by Sca-1 and c-Kit staining 21 days after poly(I:C) injection. G–J, the total numbers of LSK, ST-HSCs, MPPs, and LT-HSCs were plotted for the ssSPTa+/+ and ssSPTa−/− mice (n = 5). K–N, the total numbers of LK, CMPs, GMPs, and MEPs were plotted for the ssSPTa+/+ and ssSPTa−/− mice (n = 5). All graphs are represented as mean ± SEM. p Value < 0.05 is significant, calculated from unpaired t test. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. CMP, common myeloid precursor; GMP, granulocyte-macrophage precursor; LK, Lin− c-Kit+ Sca1−; LSK, Lin−c-Kit+, Sca1+; LT-HSCs, long-term hematopoietic stem cells; MEP, megakaryocyte-erythrocyte precursor; MPPs, multipotent progenitor cells; ST-HSCs, short-term hematopoietic stem cells.
To understand how loss of ssSPTa leads to defects in myelopoiesis, we evaluated HSPC development in the chimeric mice 21 days after loss of ssSPTa. HSCs were gated by forward and side scatter for live cells, CD45.2 to distinguish donor cells and then for lineage-negative cells (Lin−). The Lin− cells were analyzed for the expression of c-Kit and Sca-1 which identifies Lin−c-Kit+, Sca1+ (LSK), and Lin− c-Kit+ Sca1− (LK) cells (Fig. 4F). The LSK cells were subgated using FLT3 and CD34 to isolate long-term hematopoietic stem cells (LT-HSCs), short-term hematopoietic stem cells (ST-HSCs), and multipotent progenitor cells (MPPs) (Fig. S4A). We found a trend toward expansion of the stem and progenitor cell LSK compartment (Fig. 4G) with significant increase in the ST-HSC and MPP compartments (Fig. 4, H and I), whereas LT-HSC was not significantly changed (Fig. 4J). We also subgated the LK cells for FCR and CD34 to identify common myeloid precursor (CMP, Lin−, Sca-1− c-Kit+ CD34+, FCR−), granulocyte-macrophage precursor (GMP, Lin−, Sca-1− c-Kit+ CD34+, FCR+), and megakaryocyte-erythrocyte precursor (MEP, Lin−, Sca-1− c-Kit+ CD34−, FCR−) (Fig. S4B). There was about a 2-fold decrease in the levels of LK cells (Fig. 4K). Among the progenitor cell population, there was a significant decrease in the numbers of CMP and GMP while the MEPs were not affected (Fig. 4, L–N).
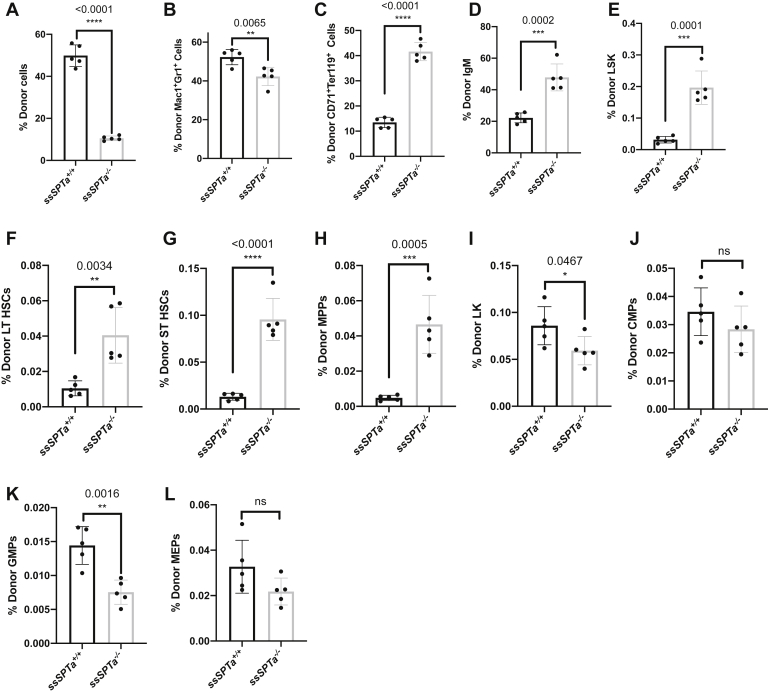
Competitive bone marrow transplant experiments were performed to evaluate HSC development. For these experiments, a 1:1 ratio of wild-type (ssSPTa+/+ Mx1-Cre) to mutant (ssSPTa−/− Mx1-Cre) BMCs were used. ssSPTa−/− cells showed a reduced donor reconstitution (10.5%) 3 weeks after ssSPTa deletion compared with about 50% for control BMCs in this model (Fig. 5A). The hematopoietic phenotypes were similar to that observed in the chimeric environment. There was a small but significant decrease in Mac1+Gr1+ cells and a slight increase in the donor CD71+ Ter119+ and the donor IgM+ cells (Fig. 5, B–D). The LSK compartment was expanded including the LT-HSC, ST-HSC, and the MPPs (Fig. 5, E–H). There was a decrease in the LK compartment with significant decrease in the GMPs (Fig. 5, I–L). These features are very similar to those observed in the Sptlc1 null mutant. Thus, as in the case of Sptlc1, ssSPTa deletion of the gene has cell autonomous effects both in the HSCs and the myeloid lineages downstream.
Figure 5.
ssSPTa deletion impairs LK differentiation and compensatory changes in HSCs in competitive repopulation studies analyzed 3 weeks after poly(I:C) injection in chimeric mice. A, proportion of donor CD45.2 cells. B, proportion of donor myeloid cells. C, proportion of donor erythroid cells. D, proportion of donor B-lymphoid cells. E, proportion of donor LSK. F, proportion of donor LT-HSCs. G, proportion of donor ST-HSCs. H, proportion of donor MPP cells. I, proportion of donor LK. J, proportion of donor CMPs. K, Proportion of donor GMPs. L, proportion of donor MEP cells, of ssSPTa+/+ and ssSPTa−/− in competitive transplant studies were plotted (n = 5). All graphs are represented as mean ± SEM. p Value < 0.05 is significant, calculated from unpaired t test. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. CMP, common myeloid precursor; GMP, granulocyte-macrophage precursor; HSCs, hematopoietic stem cells; LK, Lin− c-Kit+ Sca1−; LSK, Lin−c-Kit+, Sca1+; MEP, megakaryocyte-erythrocyte precursor; MPP, multipotent progenitor cell; ST-HSCs, short-term hematopoietic stem cells.
ER stress in ssSPTa−/− BMCs
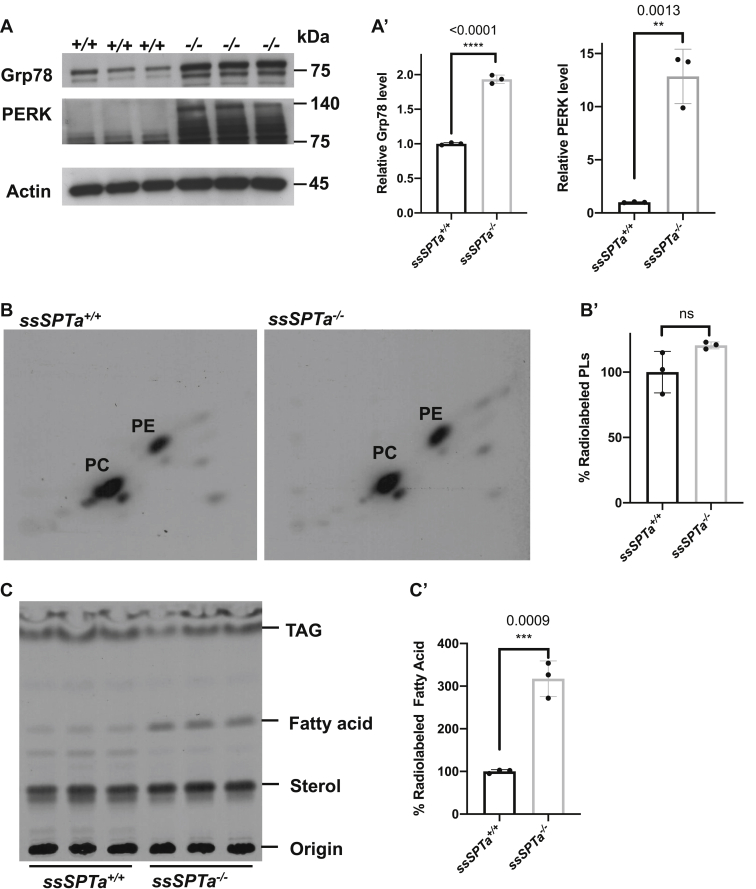
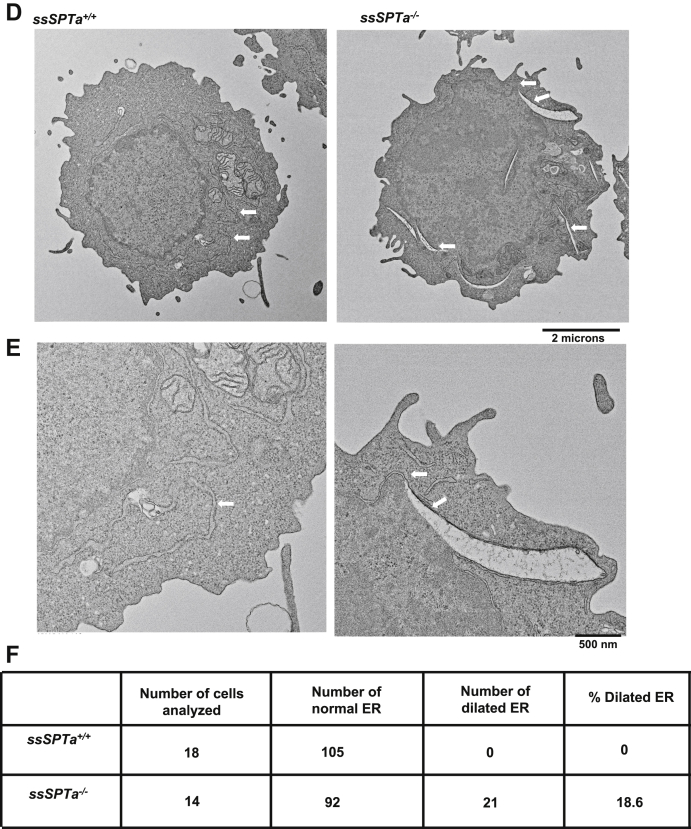
Extensive analyses in Sptlc1 revealed that loss of SPTLC1 causes myeloid differentiation defects in part because of ER stress and the differentiating BMCs show evidence of unfolded protein response (UPR) (30). Further, we showed that ER stress in general disrupts myelopoiesis while sparing erythropoiesis. To test if loss of ssSPTa also results in ER stress and activation of the UPR, we harvested BMCs from ssSPTa+/+ and ssSPTa−/− and cultured cells for 4 days in the presence of interleukin 3 (IL-3) and granulocyte macrophage colony stimulating factor (GM-CSF) to induce myeloid differentiation. Western blot analysis of extracts prepared from these differentiating BMCs showed Bip/Grp78 and PERK were elevated in these cells indicating ER stress and activation of the UPR (Fig. 6, A and A′). Metabolic labeling experiments with [32]P and [14]C and TLC analyses under these conditions showed no significant changes in total phospholipid content (Fig. 6, B and B′), however, there was a 3-fold increase in fatty acid labeling in the ssSPTa−/− extracts (Fig. 6, C and C′). The accumulation of fatty acids observed here is similar to those observed in Sptlc1−/− mutant under these conditions (30). Electron microscopic images of the differentiating cells showed grossly enlarged endoplasmic reticulum in the mutants but not in ssSPTa+/+ cells (Fig. 6, D–F). Additionally, Western analysis on bone marrow cells obtained from ssSPTa+/+ and ssSPTa−/− showed that markers of UPR such as PERK and p-IRE1α are increased in the ssSPTa−/− samples (Fig. S4, C and C′). These data together support the conclusion that lack of SSSPTA causes defects in myeloid differentiation in adult BMCs because of the accompanied ER stress in the differentiating cells.
Figure 6.
Deletion of ssSPTa shows ER stress. A, western blot analysis of cell extracts of ssSPTa+/+ and ssSPTa−/− BMCs cultured for 4 days in the presence of GM-CSF and IL-3. Lysates were probed for Grp78 and PERK. A′, quantitative data representative of three independent experiments. B, [32P] orthophosphoric acid labeling of the ssSPTa+/+ and ssSPTa−/− BMCs cultured in medium containing GM-CSF and IL-3. Lipids were normalized to equal counts per minute. B′, the percentage of radiolabeled phospholipids was plotted for the ssSPTa+/+ and ssSPTa−/−. C, [14C] acetate labeling of the ssSPTa+/+ and ssSPTa−/− BMCs cultured in medium containing GM-CSF and IL-3. Lipids were normalized to equal counts per minute. C′, the percentage of radiolabeled fatty acids was plotted for the ssSPTa+/+ and ssSPTa−/−. D, The electron micrographs of BMCs cultured in the presence of GM-CSF and IL-3, magnification 2 μs. E, expanded image of represented EM picture to show ER tubules of the cells, magnification 500 nm. The arrow represents ER tubules. In ssSPTa−/−, engorgement of ER tubules were seen. F, quantitative analysis of ER from cells described in D. All graphs are represented as mean ± SEM. p Value < 0.05 is significant, calculated from unpaired t test. ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. BMCs, bone marrow cells; GM-CSF, granulocyte macrophage colony stimulating factor; IL-3, interleukin 3.
SSSPTA is critical for SPT function
RNAseq and microarray analysis indicate that ssSPTa is more widely and abundantly expressed than ssSPTb in mammals (URL: http://www.informatics.jax.org) (35). Our qPCR analyses for the two transcripts in control mice were in line with the above reports (Fig. S5, A and B). Mass spectrometric measurements in BMCs of ssSPTa−/− showed a significant reduction in major sphingolipids compared with ssSPTa+/+ suggesting that this subunit is crucial for the functioning of the SPT complex in the synthesis of sphingolipids. It is worth noting that deletion of SSSPTA in the liver causes loss of SPT enzymatic activity similar to that observed after the deletion of SPTLC1 (Fig. S5C). Also, deletion of ssSPTa in the BMCs compromises the state of the SPT complex including decrease in the steady-state levels of SPTLC1 and SPTLC2 (Fig. S5D). These observations highlight the importance of SSSPTA in the stability and activity of the SPT complex in these organs.
ssSPTb−/− mice are viable and are not compromised in adult hematopoiesis
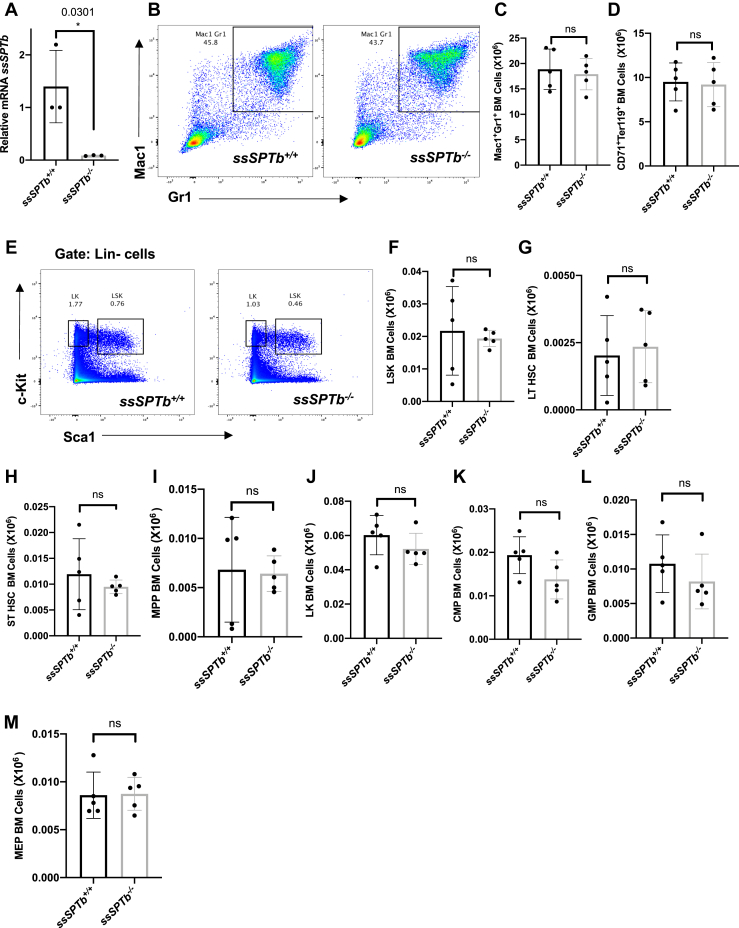
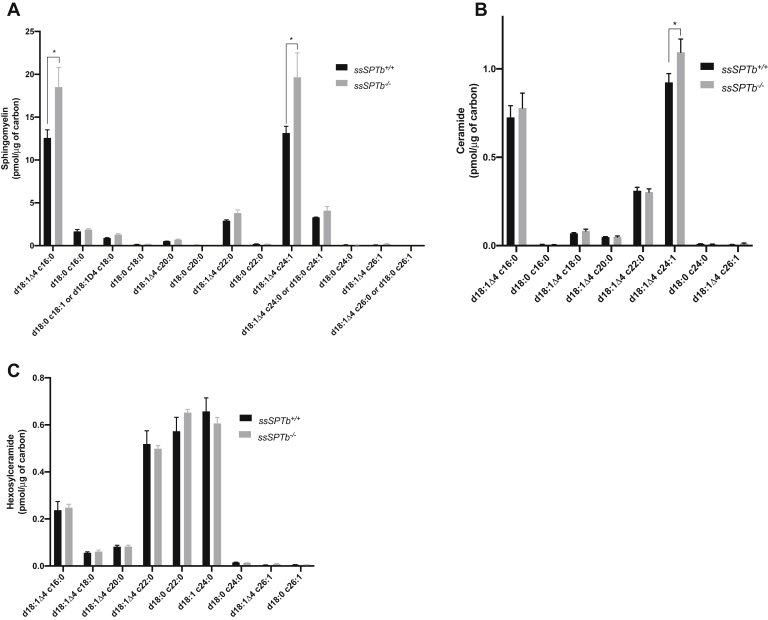
SSSPTB is the second and a well conserved small subunit of the SPT complex. The gene encoding for this subunit is localized to chromosome 3 and is 84% identical to the human homolog. We targeted exon 3 that encodes for all of the translated region of the gene (Fig. S6, A and B). A zygotic null allele was generated by mating the floxed allele to Actin-Cre (Fig. 7A). The ssSPTb−/− mice were homozygous viable. To delineate the role of SSSPTB in hematopoiesis we examined the bone marrow of the ssSPTb+/+ and ssSPTb−/− from heterozygous intercross and evaluated the components of the hematopoietic system. Visual examination appeared to show straw colored bone marrow preparations that were indistinguishable between ssSPTb+/+ and ssSPTb−/− (Fig. S6C) Wright-Giemsa-stained bone marrow preparations were also comparable between the two genotypes (Fig. S6D). The bone marrow cellularity was not compromised in the ssSPTb−/− mice (Fig. S6E). We examined the BMCs for lineage markers expressed on myeloid cells (Mac-1;Gr-1); erythroid cells (CD71;Ter119); B lymphoid cells (IgM); and T lymphocytes (CD4;CD8) using flow cytometry. There was no difference in the abundance of Mac-1+ Gr-1+ between ssSPTb+/+ and ssSPTb−/− (Fig. 7, B and C). The monocyte (Ly6C+;Ly6G−) and macrophage (Mac1+;F4/80+) lineages were also comparable between the two genotypes (Fig. S6, F and G), and there were no differences in the cells of the erythroid lineage (CD71+;Ter119+) and (Ter119+) (Figs. 7D and S6H). Examination of the LSK compartment for stem and MPPs showed no significant differences between the ssSPTb+/+ and ssSPTb−/− BMCs (Fig. 7, E–I and Fig. S6I). Similarly, subgating of the LK cells using FCR and CD34 showed no differences in the CMPs, GMPs, and MEPs (Fig. 7, J–M and Fig. S6J). The total cellularity and the number of Mac1+Gr+ cells were similar between the control and the mutant spleen (Fig. S6, K and L). We analyzed thymus for T cell differentiation and did not find significant difference in cellularity, CD4, CD8 double positive immature cells, or single positive mature cells (Fig. S5, M–P). The IgM+ B lymphocytic lineage was also not compromised in the BMCs (Fig. S6Q). Collectively, the above results indicate that that SSSPTB does not play a significant role in adult hematopoiesis. We also found that loss of SSSPTB did not compromise the levels of d-18 series of sphingomyelins, ceramides, or hexosylceramides in the bone marrow cells of the adult hematopoietic system (Fig. 8, A–C). These results indicate that SSSPTB subunit does not play a significant role in the embryonic development of mice and is not critical for the adult hematopoietic system.
Figure 7.
ssSPTb knockout shows normal hematopoiesis. A, real-time qPCR analysis of ssSPTb mRNA expression in BM cells isolated from ssSPTb+/+ and ssSPTb−/− mice. Gene expression was normalized to β-actin. qPCR results from three independent experiments. B, fluorescence activated cell sorting plots of myeloid bone marrow cells analyzed by Mac-1 and Gr-1 staining (n = 5). C, the total numbers of Mac-1+Gr-1+ were plotted for the ssSPTb+/+ and ssSPTb−/− mice (n = 5). D, the total numbers of CD71+Ter119+ cells, were plotted for the ssSPTb+/+ and ssSPTb−/− mice (n = 5). E, fluorescence activated cell sorting plots of LSK and LK populations of HSPCs from bone marrow of ssSPTb+/+ and ssSPTb−/− were analyzed by Sca-1 and c-Kit staining. F–I, the total numbers of LSK, LT-HSCs, ST-HSCs, and MPPs were plotted for the ssSPTb+/+ and ssSPTb−/− (n = 5). J–M, The total numbers of LK, CMPs, GMPs, and MEPs cells were plotted for the ssSPTb+/+ and ssSPTb−/− (n = 5). All graphs are represented as mean ± SEM. ∗p Value < 0.05 is significant, calculated from unpaired t test. CMP, common myeloid precursor; GMP, granulocyte-macrophage precursor; LK, Lin− c-Kit+ Sca1−; LSK, Lin−c-Kit+, Sca1+; LT-HSCs, long-term hematopoietic stem cells; MEP, megakaryocyte-erythrocyte precursor; MPPs, multipotent progenitor cells; ST-HSCs, short-term hematopoietic stem cells.
Figure 8.
Sphingolipids measurement by mass spectrometry of ssSPTb+/+ and ssSPTb−/− BMCs. A, sphingomyelin, (B) ceramide, and (C) hexosylceramide were analyzed by mass spectrometry and normalized to carbon content are plotted for the ssSPTb+/+ and ssSPTb−/− (n = 3). The results are from three independent BM cells of each genotype. All graphs are represented as mean ± SEM. ∗p Value < 0.05 is significant, calculated from unpaired t test. BMCs, bone marrow cells.
Discussion
In this study, we have evaluated the importance of the small subunits of the mammalian SPT complex. We show that SSSPTA subunit is essential for development and adult hematopoiesis while SSSPTB subunit does not play major roles in these programs.
SPT, that catalyzes the first and rate-limiting step of the pathway is a heterotrimeric complex comprised of the large subunits, SPTLC1 and SPTLC2 or SPTLC3 and small subunit, SSSPTA or SSSPTB. It produces 3-ketodihydrosphingosine by a Claisen-like condensation/decarboxylation reaction of serine and acyl-CoA. SPT has been shown to be critical for the development of several eukaryotic organisms. Deletion of Sptlc1 homologs in Saccharomyces cerevisiae, Arabidopsis, Drosophila, and mouse are lethal. The S. cerevisiae null mutants of lcb1 (Sptlc1 homolog) are long-chain base auxotrophs (13). Arabidopsis null mutants of Sptlc1 homolog are embryonic lethal and die before developing into a fully mature globular stage (36). Drosophila Sptlc1 and Sptlc2 null mutants are larval lethal (8, 37). Null mutants of Sptlc1 that encodes for the obligate subunit of SPT complex and null mutants of Sptlc2 are embryonic lethal indicating that the SPT complexes containing these two subunits serve important role during embryonic development in the mice (24, 30).
Pioneering studies by the Dunn laboratory have established the roles of the small subunits of the SPT complex. TSC3P null mutant was identified as temperature-sensitive lethal that had the ability to augment the basal SPT activity of the LCB1/LCB2 heterodimer (38). ssSPTa and ssSPTb were identified as mammalian functional homologs of the yeast Tsc3 in a screen using yeast lcb1 mutants expressing Sptlc1 and Sptlc2 (27). Utilization of SSSPTA allows the SPT complex to incorporate palmitoyl-CoA as a substrate for the synthesis of d-18 series of sphingolipids. Utilization of SSSPTB allows the SPT complex the ability to incorporate either palmitoyl-CoA and longer fatty acyl-CoAs (up to C20) as a substrate to generate either the d-18 or higher series of sphingolipids (27). Relatively few studies have evaluated the in vivo functions of the small subunits of the SPT complex. Arabidopsis thaliana encodes for homologs of ssSPTs, called “ssSPTa” and “ssSPTb”, which share about 88% amino acid identity between them. Both of them share more homology to human SSSPTA (25–30% shared amino acid sequence) than SSSPTB (18–25%) (39). The two proteins were equally competent to enhance SPT activity, although “ssSPTa” was more widely expressed. Homozygous loss of “ssSPTa” resulted in loss of pollen viability. A gain of function mutation in the ssSPTb gene was shown to have a 2-fold increase in d-20 long chain bases in the brain and eye of the stellar mutant mice. These mice exhibited neurodegenerative effects (2).
We set about to examine the broad importance of SSSPTA and SSSPTB in the functioning of SPT during development and in adult hematopoiesis in a mammalian model organism. Adult hematopoietic system is a mesenchymal tissue, and because embryoid body differentiation defects pointed to an aberration in the mesenchymal germ layer, it provided us with a strong impetus to evaluate the hematopoietic system. Also, studies have suggested important roles for the sphingolipid metabolic pathway in development and function of the adult hematopoietic system (22, 40, 41). Our studies with the Sptlc1 mutant demonstrated that SPT was essential for myelopoiesis, and it was logical to ask if either SSSPTA or SSSPTB was essential for this function of SPT (30). We show that loss of ssSPTa resulted in embryonic lethality at E6.5 and compromised myelopoiesis in the adult hematopoietic system.
The data from ssSPTa−/− mice have consolidated our finding that SPT function and by extension the de novo biosynthetic pathway has an important role in hematopoiesis. Loss of SPT function in these cells causes ER stress and cell death with a drastic reduction in the production of myeloid cells. Chemically induced ER stress in differentiating BMCs selectively compromises myelopoiesis while sparing erythropoiesis indicating that robust ER function in general is critical for myelopoiesis (30). While ER stress and upregulation of the UPR was initially discovered and described as an ancient, conserved mechanism to maintain proteostasis, with discoveries of integrated stress response, recent discussions have included the concept that ER stress and UPR are integrated broadly into the overall function of the ER (42, 43, 44). ER is the site of biosynthesis of not only proteins but also a host of lipids that include neutral lipid, phospholipid, sphingolipid, and cholesterol. Therefore, relative proportions and concentrations of the different intermediates of the lipid pathways and their flux will impact the structure and function of ER. They will also influence the mechanisms that control the quality of the ER. Therefore, it is not surprising that any perturbations in these pathways disrupts ER homeostasis and induces ER stress. We have previously shown that differentiating LK cell compartments of BMCs show an enrichment of ER-associated genes likely in the myeloid compartment underscoring the importance of the secretory pathway and ER function in differentiating myeloid cells (30). Thus, any disruption that leads to compromised ER function compromises myelopoiesis. Mass spectrometric measurements in BMCs of ssSPTa−/− showed a significant reduction in major sphingolipids compared with ssSPTa+/+ suggesting that this subunit is crucial for the functioning of the SPT complex in the synthesis of sphingolipids. Our observations highlight the importance of SSSPTA in the stability and activity of the SPT complex in these organs. We do not see significant contributions from the SSSPTB during embryogenesis or adult hematopoiesis. Our results imply that SSSPTA plays a role that is of equal significance as the obligate SPTLC1 subunit of the SPT complex during development and adult hematopoiesis suggesting that SSSPTA is indispensable for these functions of the SPT complex.
Experimental procedures
Generation of ssSPTa conditional knockout mice
The National Cancer Institute at Frederick animal care and user committee has approved the protocol ASP20 to 073 used in this manuscript for animal studies. For targeting the ssSPTa gene, loxP sites were inserted into the introns upstream of exon 1 and downstream of exon 1. The targeting vector pLJM containing a 10.2 Kb fragment was retrieved from BAC clones using recombineering technology (31). The targeting vector contained 4.5 Kb 5′ and 4.7 Kb 3′ homology arms. The first loxP insertion site was 327 bp upstream of exon 1, and the second loxP-Neo-loxP site was 244 bp downstream of exon 1. Upon induction of Cre, exon 1 was deleted. This results in deletion of 39 amino acids of the protein. The ssSPTa targeting vectors were electroporated into murine C57BL6 ES cells as described (45). Targeted ssSPTa transfected clones were identified by PCR analysis and infected with Adeno-Cre to remove the NeoR gene and exon 1. Independent ES cell clones were injected into C57BL6 blastocysts. Founder chimeras were bred to B6 females, and the F1 heterozygotes were intercrossed. Recombinant clones were identified by PCR.
Mouse genotypes were identified by PCR performed using DNA isolated from tail clippings.
The sequences for the flox primers are as follows: forward, 5-GATCCTTCAGTATTCTTTGAGGCA-3; mutant reverse, 5-AGCTAAACCACAAGAAGCTGAGCG-3; and wild type reverse, 5-TGTCCATCTGCACGAGACTAGTGA-3.
PCR reactions began with a denaturing step at 94 °C for 5 min followed by 30 cycles of 94 °C for 30s, 53 °C for 30s, and 68 °C for 30s. Amplification with these primers produces a band of 397 bp for the floxed allele.
For the wild type and null, the primer sequences used were as follows: forward WT, 5-CGACTCTTTAGTAGCATTCTCACC-3; reverse WT, 5-TGACCAGCAGGTACTGGTAGTAGA-3; and forward Null, 5-GGAATCCTTCTGGGCTGGTCT-3; reverse Null, 5- GCCTTTCAGTAACTTACAAAC-3.
Amplification with these primers produced a band of 600 bp for the wild type. PCR reactions began with a denaturing step at 94 °C for 4 min followed by 34 cycles of 94 °C for 30s, 55 °C for 30s, and 68 °C for 30s. Amplification with these primers produced a band of 250 bp for the mutant alleles. PCR reactions began with a denaturing step at 94 °C for 5 min followed by 30 cycles of 94 °C for 30s, 53 °C for 30s, and 68 °C for 30s.
Mice carrying the floxed ssSPTa allele were crossed with mice expressing Cre recombinase under the control of the Mx1 promoter (34). Upon injection of poly(I:C) (HMW), tlrl-pic-5 from InvivoGen, the Mx1 promoter controls expression of Cre, causing deletion of exon 1 (ssSPTa−/−). Intraperitoneal injections of 500 μg of poly(I:C) resulted in deletion of the gene in hematopoietic cells. The ssSPTa+/+ Mx1-Cre mice were used as controls for all experiments unless otherwise mentioned. Eight to 10 weeks old mice were used for all experiments.
Generation of ssSPTb null mouse
A 11.28 Kb targeting vector was generated with 4.75 Kb (loxP-Neo-loxP) upstream homology and 4.45 Kb (loxP) downstream homology.
Deletion of 2091 nucleotides included about 1.5 Kb of exon 3 encoding for all of the coding sequence of ssSPTb gene.
Heterozygote mouse (ssSPTb+/−) mating was performed to generate null mouse. Mice carrying the null ssSPTb allele were crossed with the same genotype to generate null allele. The ssSPTb+/+ mice were used as controls for all experiments unless otherwise mentioned. Eight to 10 weeks old mice were used for all experiments.
For the wild type and null, the primer sequences used were as follows: forward WT, 5-GGAATACCCATGATACAACTC-3; Reverse WT, 5-CAA CAg gTA TCC ATg TCA gTA -3; forward Null, 5-GGAATACCCATGATACAACTC-3; and reverse Null, 5-TCTCTGAGTATGTCTATAAGG-3.
Amplification with these primers produced bands of 250 bp and 350 bp for the wild-type and mutant alleles, respectively. PCR reactions began with a denaturing step at 94 °C for 5 min followed by 30 cycles of 94 °C for 30s, 53 °C for 30s, and 68 °C for 30s.
Blastocyst outgrowth
Four- to six-weeks-old ssSPTa+/− female mice were mated with ssSPTa+/− male mice. At 3.5 days after coitus, the plugged females were euthanized, and the uterus was dissected and flushed with ES cell medium without leukemia inhibitory factor. Single E3.5 blastocyst was seeded onto each well of gelatinized 96-well plates containing 200 μl of the medium and incubated at 37 °C and 5% CO2. Photographs of the cultured embryos were taken every day. After 9 days, the preparations were washed with PBS, and blastocyst DNA was isolated using lysis buffer with proteinase K and the blastocysts genotyped (14).
Embryo dissection and embryoid body formation
Timed mating was performed for ssSPTa+/− mice on a mixed genetic background (C57BL/J129). Females with copulation plugs were considered to be at day 0.5 of gestation, and embryos present in this pregnant female were designated at E0.5. Pregnant females were dissected at E6.5, and the dissected embryos were photographed under a normal inverted light microscope (Axioskop; Carl Zeiss, Inc) connected to a digital camera (Powershot S50; Canon). DNA for genotyping was isolated from small pieces of yolk sac by alkaline lysis buffer (25 mM NaOH and 0.2 mM EDTA) and neutralization buffer (40 mM Tris-HCl). For histological analysis, embryos were fixed with 4% paraformaldehyde (HCHO), embedded in paraffin, sectioned, and stained with hematoxylin and eosin. Photomicrographs were taken using a microscope (Axiophot; Carl Zeiss, Inc) and a digital camera (CoolSNAP HQ; Photometrics).
E3.5 blastocysts from the uteri of pregnant ssSPTa+/− females of heterozygous intercross were harvested and cultured on Matrigel coated plates in ES culture medium containing LIF (1000 units/ml), PD0325901 (1 μM), and CHIR99021 (3 μM). Inner cell mass outgrowths were trypsinized and passaged sequentially until ES cell lines were established.
Embryonic stem cells were placed onto bacteriological plates (100 mm-Petri dishes) and cultured in suspension in ES cell medium without LIF, PD0325901, and CHIR99021. The medium was changed every day. The partial EBs were harvested at 0, 3, 6, 9, 12 days to perform real time PCR, and EB structure was detected at 14 days (46).
EM analyses
For EM analysis, the fluorescence activated cell sorting sorted BMCs were fixed in 4% HCHO and 2% glutaraldehyde in 0.1 M sodium cacodylate buffer. The thin-sectioned cells were viewed and imaged on an electron microscope (EM300; Hitachi High-Tech).
Immunoblotting
Bone marrow cultured cells were lysed, and proteins were quantified by an absorbance assay (Bio-Rad Laboratories). The quantified lysates were mixed with 2× Laemmli buffer (Bio-Rad Laboratories) and denatured at 100 °C for 5 min. Equal amounts of denatured protein were resolved on 10% SDS-PAGE gels. The resolved proteins were subsequently transferred to nitrocellulose membranes (Pall Corporation) or PVDF membranes (Millipore). They were blocked with 5% nonfat dry milk (Bio-Rad Laboratories) or 5% BSA in PBST. After blocking, membranes were probed with primary and HRP-conjugated secondary antibodies, detected by chemiluminescence agents (Denville). In between changes, the membranes were washed with 0.05% Tween 20 in PBS. The primary antibodies BiP (9C50B12) (3177) and PERK (C33E10) (3192) were purchased from Cell Signaling Technologies. SPTLC1 antibodies were generated from our laboratory. SPTLC2 (CAT#: TA319780) antibodies were purchased from ORIGENE. The phospho-IRE1α (Ser724) from Novus biologicals (NB100–2323). HRP-linked anti-rabbit and anti-mouse IgGs were from the Jackson Laboratory. Prestained protein standards were from Bio-Rad Laboratories, SMOBIO and Santa Cruz Biotechnology. Immunoblots were quantitated by Fiji ImageJ software.
Myeloid differentiation in liquid culture
Nucleated cells were isolated from ssSPTa+/+ and ssSPTa−/− BMCs by lymphocyte separation medium (MP Biomedical). They were plated (2 × 106 cells/ml of media) and cultured for 4 days in the presence of GM-CSF and IL-3, (both at 50 ng/ml concentrations). Harvested cells were used for western blots as described above (47).
[14C]acetate labeling of BMCs
The protocol is as described before (30). ssSPTa+/+ and ssSPTa−/− BMCs were plated for myeloid differentiation with GM-CSF and IL-3 in liquid culture. After 48 h of culture, 5μci of [14C] acetate (58 mCi/mmol) (ARC 0173) were added to the medium. The cells were harvested after an additional 36 h of culture, and lipids were extracted according to Bligh and Dyer. Chloroform extracts were subjected to TLC with silica gel 60F 254 TLC plates. The solvents for the neutral lipids were resolved by petroleum ether/diethyl ether/acetic acid (65:25:5, v/v). The results are from biological triplicate experiments of cultured BMCs from three ssSPTa+/+ and three ssSPTa−/− mice, with lipids normalized to equal counts per minute. The fatty acid was identified by comparing with known standard (30).
[32P]orthophosphoric acid labeling of BMCs
The methods used were essentially as described before (30). ssSPTa+/+ and ssSPTa−/− BMCs were plated for myeloid differentiation with GM-CSF and IL-3 in liquid culture. After 48 h of culture, 50μci of [32P] orthophosphoric acid (100 mCi/ml) (ARC 0103A) were added to the medium. The cells were harvested after an additional 24 h of culture, and lipids were extracted according to Bligh and Dyer. Chloroform extracts were subjected to TLC with silica gel 60F 254 TLC plates. The solvents for the phospholipid first dimension, chloroform/methanol/ammonia (65:25:5, v/v) followed by second dimension chloroform/methanol/acetone/acetic acid/water (50:10:20:15:5, v/v). The results are from biological triplicate experiments of cultured BMCs from three ssSPTa+/+ and three ssSPTa−/− mice, with lipids normalized to equal counts per minute.
SPT activity
SPT activity was measured based on the method described previously and using microsomal preparations of the enzyme complex (48). The activity was measured with 150 μg of liver microsomal proteins in a 200 μl volume containing 50 μm PLP, 2 μl of [L-3H(G)]serine (1 mCi/ml) (NET248005MC), 100 μm palmitoyl CoA (Sigma P9716), 5 mM EDTA, and 5 mM DTT per reaction. The reaction was performed in glass tube, incubated for 60 min at 37 °C, and terminated by the addition of 200 μl of 0.5 N NH4OH and 500 μl of CHCl3 and 1 ml methanol. After vortex, an additional 0.75 ml of CHCl3 and 0.75 ml of 0.5 N NH4OH were added and vortexed. After centrifugation at 2500 rpm at room temperature for 5 min to separate the phases, the top layer was aspirated off. The organic phase was washed twice with 1 ml of 0.1 M KCl and transfer to scintillation vial and was dried under N2 gas. The pellet was resuspended in scintillation fluid for counting. The results are from biological triplicate experiments of three wild-type, three ssSPTa−/− and three SPTLC1−/− mice liver microsomal proteins.
Immunophenotyping
The methods used were as described before (30). Bone marrow cells were flushed out from mouse femurs and tibias and filtered through 40 μm cell strainers (Sysmex Celltrics). For bone marrow, nucleated cells were isolated by lymphocyte separation medium. The cells were stained for lineage-specific markers Mac-1, Gr-1, Ly6C, Ly6G, Ter119, CD71, B220, and IgM using the corresponding anti-mouse cognate antibodies (eBioscience). For staining of HSPCs, lineage-positive cells were removed by magnetic depletion with biotin-conjugated Ter119, CD71, Mac-1, Gr-1, and IgM antibodies (eBioscience) and streptavidin T1 Dynabeads (# 65601, 65602) (Invitrogen). Hematopoietic cell subsets were identified using antibodies for c-Kit, Sca-1, CD34, Flt3, FcR, IL-7R, and CD45.2 (eBioscience). All bone marrow transplant mice were analyzed using CD45.2 for identification of the donor cells. LSRII flow cytometer (BD Biosciences) was used to acquire fluorescence activated cell sorting data, which were analyzed with FlowJo software (47, 49).
BM transplantation
Female mice (aged 8–12 weeks) were used as recipients for all transplantation experiments. For competitive reconstitution assays, lethally irradiated (10 Gy, 137Cs source) congenic CD45.1 recipient mice were transplanted with 2 × 106 BMCs from ssSPTa+/+ Mx1-Cre or ssSPTaflox/flox Mx1-Cre (donor CD45.2) and CD45.1-C57BL/6 in the ratio of 1:1. For noncompetitive reconstitution assays, lethally irradiated (10 Gy, 137Cs source) congenic CD45.1 recipient mice were transplanted with 2 × 106 BMCs from ssSPTa+/+ Mx1-Cre or ssSPTaflox/flox Mx1-Cre (donor CD45.2). Each recipient mouse received 2 × 106 donor BMCs through their lateral vein. Six weeks later, after the re-establishment of the donor cells in the recipient bone marrow, poly(I:C) was injected on alternate days at a concentration of 15 μg/gm body weight (50). The bone marrow cells were harvested on day 21 after the first injection, and CD45.2 donor cells were gated and analyzed.
BMC sphingolipid analysis by MS
ssSPTa+/+ and ssSPTa−/− bone marrow cells were harvested 8 days after poly(I:C) injection for the whole bone marrow analysis. ssSPTb+/+ and ssSPTb−/− bone marrow cells also were analyzed. Lipids were prepared from the respective cells by chloroform and methanol extraction, normalized for carbon content, and analyzed by MS for sphingolipids (14).
Real-time PCR
Total RNA was extracted from wild-type and mutants (ssSPTa, ssSPTb) bone marrow cells by trizol RNA isolation protocol.
RNA samples were reverse transcribed by transcriptor first strand cDNA synthesis kit (Roche), and expression of respective gene was analyzed by real-time qPCR using the iTaq universal SYBR green supermix and step one plus (applied biosystem). For the ssSPTa, ssSPTb analyses, Actin was for used normalizing gene expression profile, whereas Gapdh was used for normalizing embryoid body differentiation markers. The primer sequences used were as follows:
| Primer sequences for real time RT-PCR | |
|---|---|
| ssSPTa | Reverse: 5′-AGGTGGGCTGGCGTTAT-3′ |
| Reverse: 5′-CTGGGGCATGAAGACGTAGC-3′ | |
| ssSPTb | Forward: 5′-CGTGAAGGAGTATTTTGCCTGG -3′ |
| Reverse: 5′-GCCACAATGGTCAGTATGATGGT-3′ | |
| Nestin | Forward: 5′-TGAGGGTCAGGTGGTTCTG-3′ |
| Reverse: 5′-AGAGCAGGGAGGGACATTC-3′ | |
| Sox 17 | Forward: 5′-AAGAAACCCTAAACACAAACAGCG-3′ |
| Reverse: 5′-TTTGTGGGAAGTGGGATCAAGAC-3′ | |
| Gsc | Forward: 5′-AAACGCCGAGAAGTGGAACAAG-3′ |
| Reverse: 5′-AAGGCAGGGTGTGTGCAAGTAG-3′ | |
| GAPDH | Forward: 5′-ACCATCTTCCAGGAGCGAG-3′ |
| Reverse: 5′-TAAGCAGTTGGTGGTGCAG-3′ | |
| Actin | Forward: 5′-TTCTTTGCAGCTCCTTCGTT-3′ |
| Reverse: 5′-ATGGAGGGGAATACAGCCC-3′ | |
| Brachyury | Forward: 5′-CGGACAATTCATCTGCTTG-3′ |
| Reverse: 5′-AGGTGGGCTGGCGTTAT-3′ | |
Statistical analysis
Our data were analyzed by two-tailed unpaired t test with Welch's correction. Results were presented as the mean and standard deviations of groups, with p-values less than 0.05 were considered to be statistically significant.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article (and/or) its supporting information.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
This study was funded by the intramural division of the National Cancer Institute, National Institutes of Health (Division of Health and Human Services). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Author contributions
V. P., J. L., J. R. K., U. A., and J. K. A. conceived the study and designed the experiments. V. P., J. L., D. A., S. M. S., T. A., performed the experiments, K.K. provided expertise during standardization of hematopoietic experiments, A. Y. provided expertise to derive ES cells, F. S. and K. N. performed electron microscopy, S. D. F. and T. A. performed mass spectrometric analysis of sphingolipids, L. T. assisted in the generation of the knockout mice, V. P., J. L., U. A., and J. K. A. wrote and edited the manuscript with assistance from all the authors.
Edited by Phyllis Hanson
Footnotes
Present address for Jing Lin: US Food and Drug Administration, Silver Spring, MD.
Present address for Diwash Acharya: MaxCyte, Inc, Gaithersburg, MD.
Present address for Sargur Madabushi Srideshikan: Department of Radiation Oncology, City of Hope, National Medical Center, Duarte, CA.
Present address for Kimberly Klarmann: Basic Science Program, Leidos Inc, Frederick National Laboratory for Cancer Research, Frederick, MD.
Contributor Information
Usha Acharya, Email: acharyaur@nih.gov.
Jairaj K. Acharya, Email: acharyaj@mail.nih.gov.
Supporting information
Table S1 and Figures S1 to S6
References
- 1.Hannun Y.A., Obeid L.M. Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018;19:175–191. doi: 10.1038/nrm.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao L., Spassieva S., Gable K., Gupta S.D., Shi L.Y., Wang J., Bielawski J., Hicks W.L., Krebs M.P., Naggert J., Hannun Y.A., Dunn T.M., Nishina P.M. Elevation of 20-carbon long chain bases due to a mutation in serine palmitoyltransferase small subunit b results in neurodegeneration. Proc. Natl. Acad. Sci. U. S. A. 2015;112:12962–12967. doi: 10.1073/pnas.1516733112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mota Fernandes C., Del Poeta M. Fungal sphingolipids: Role in the regulation of virulence and potential as targets for future antifungal therapies. Expert Rev. Anti Infect Ther. 2020;8:1083–1092. doi: 10.1080/14787210.2020.1792288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Merrill A.H., Jr. De novo sphingolipid biosynthesis: A necessary, but dangerous, pathway. J. Biol. Chem. 2002;277:25843–25846. doi: 10.1074/jbc.R200009200. [DOI] [PubMed] [Google Scholar]
- 5.Wigger D., Gulbins E., Kleuser B., Schumacher F. Monitoring the sphingolipid de novo synthesis by stable-isotope labeling and liquid chromatography-mass spectrometry. Front. Cell Dev. Biol. 2019;7:210. doi: 10.3389/fcell.2019.00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bieberich E. Sphingolipids and lipid rafts: Novel concepts and methods of analysis. Chem. Phys. Lipids. 2018;216:114–131. doi: 10.1016/j.chemphyslip.2018.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hakomori S. Structure and function of sphingoglycolipids in transmembrane signalling and cell-cell interactions. Biochem. Soc. Trans. 1993;21(Pt 3):583–595. doi: 10.1042/bst0210583. [DOI] [PubMed] [Google Scholar]
- 8.Adachi-Yamada T., Gotoh T., Sugimura I., Tateno M., Nishida Y., Onuki T., Date H. De novo synthesis of sphingolipids is required for cell survival by down-regulating c-Jun N-terminal kinase in Drosophila imaginal discs. Mol. Cell Biol. 1999;19:7276–7286. doi: 10.1128/mcb.19.10.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alexaki A., Clarke B.A., Gavrilova O., Ma Y., Zhu H., Ma X., Xu L., Tuymetova G., Larman B.C., Allende M.L., Dunn T.M., Proia R.L. De novo sphingolipid biosynthesis is required for adipocyte survival and metabolic homeostasis. J. Biol. Chem. 2017;292:3929–3939. doi: 10.1074/jbc.M116.756460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mousley C.J., Trettin K.D., Tyeryar K., Ile K.E., Schaaf G., Bankaitis V.A. Sphingolipid metabolism in trans-golgi/endosomal membranes and the regulation of intracellular homeostatic processes in eukaryotic cells. Adv. Enzyme Regul. 2010;50:339–348. doi: 10.1016/j.advenzreg.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 11.Pinto W.J., Srinivasan B., Shepherd S., Schmidt A., Dickson R.C., Lester R.L. Sphingolipid long-chain-base auxotrophs of Saccharomyces cerevisiae: Genetics, physiology, and a method for their selection. J. Bacteriol. 1992;174:2565–2574. doi: 10.1128/jb.174.8.2565-2574.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ross J.S., Russo S.B., Chavis G.C., Cowart L.A. Sphingolipid regulators of cellular dysfunction in type 2 diabetes mellitus: A systems overview. Clin. Lipidol. 2014;9:553–569. [PMC free article] [PubMed] [Google Scholar]
- 13.Wells G.B., Lester R.L. The isolation and characterization of a mutant strain of Saccharomyces cerevisiae that requires a long chain base for growth and for synthesis of phosphosphingolipids. J. Biol. Chem. 1983;258:10200–10203. [PubMed] [Google Scholar]
- 14.Wang X., Rao R.P., Kosakowska-Cholody T., Masood M.A., Southon E., Zhang H., Berthet C., Nagashim K., Veenstra T.K., Tessarollo L., Acharya U., Acharya J.K. Mitochondrial degeneration and not apoptosis is the primary cause of embryonic lethality in ceramide transfer protein mutant mice. J. Cell Biol. 2009;184:143–158. doi: 10.1083/jcb.200807176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yonamine I., Bamba T., Nirala N.K., Jesmin N., Kosakowska-Cholody T., Nagashima K., Fukusaki E., Acharya J.K., Acharya U. Sphingosine kinases and their metabolites modulate endolysosomal trafficking in photoreceptors. J. Cell Biol. 2011;192:557–567. doi: 10.1083/jcb.201004098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hannun Y.A., Obeid L.M. The ceramide-centric universe of lipid-mediated cell regulation: Stress encounters of the lipid kind. J. Biol. Chem. 2002;277:25847–25850. doi: 10.1074/jbc.R200008200. [DOI] [PubMed] [Google Scholar]
- 17.Montefusco D.J., Allegood J.C., Spiegel S., Cowart L.A. Non-alcoholic fatty liver disease: Insights from sphingolipidomics. Biochem. Biophys. Res. Commun. 2018;504:608–616. doi: 10.1016/j.bbrc.2018.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith W.L., Merrill A.H., Jr. Sphingolipid metabolism and signaling minireview series. J. Biol. Chem. 2002;277:25841–25842. doi: 10.1074/jbc.R200011200. [DOI] [PubMed] [Google Scholar]
- 19.Merrill A.H., Jr., Schmelz E.M., Dillehay D.L., Spiegel S., Shayman J.A., Schroeder J.J., Riley R.T., Voss K.A., Wang E. Sphingolipids--the enigmatic lipid class: Biochemistry, physiology, and pathophysiology. Toxicol. Appl. Pharmacol. 1997;142:208–225. doi: 10.1006/taap.1996.8029. [DOI] [PubMed] [Google Scholar]
- 20.Acharya U., Acharya J.K. Enzymes of sphingolipid metabolism in Drosophila melanogaster. Cell Mol. Life Sci. 2005;62:128–142. doi: 10.1007/s00018-004-4254-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dadsena S., Bockelmann S., Mina J.G.M., Hassan D.G., Korneev S., Razzera G., Jahn H., Niekamp P., Muller D., Schneider M., Tafesse F.G., Marrink S.J., Melo M.N., Holthuis J.C.M. Ceramides bind VDAC2 to trigger mitochondrial apoptosis. Nat. Commun. 2019;10:1832. doi: 10.1038/s41467-019-09654-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cartier A., Hla T. Sphingosine 1-phosphate: Lipid signaling in pathology and therapy. Science. 2019;366 doi: 10.1126/science.aar5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogretmen B. Sphingolipid metabolism in cancer signalling and therapy. Nat. Rev. Cancer. 2018;18:33–50. doi: 10.1038/nrc.2017.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hojjati M.R., Li Z., Jiang X.C. Serine palmitoyl-CoA transferase (SPT) deficiency and sphingolipid levels in mice. Biochim. Biophys. Acta. 2005;1737:44–51. doi: 10.1016/j.bbalip.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 25.Hanada K. Serine palmitoyltransferase, a key enzyme of sphingolipid metabolism. Biochim. Biophys. Acta. 2003;1632:16–30. doi: 10.1016/s1388-1981(03)00059-3. [DOI] [PubMed] [Google Scholar]
- 26.Hanada K., Hara T., Nishijima M. Purification of the serine palmitoyltransferase complex responsible for sphingoid base synthesis by using affinity peptide chromatography techniques. J. Biol. Chem. 2000;275:8409–8415. doi: 10.1074/jbc.275.12.8409. [DOI] [PubMed] [Google Scholar]
- 27.Han G., Gupta S.D., Gable K., Niranjanakumari S., Moitra P., Eichler F., Brown R.H., Jr., Harmon J.M., Dunn T.M. Identification of small subunits of mammalian serine palmitoyltransferase that confer distinct acyl-CoA substrate specificities. Proc. Natl. Acad. Sci. U. S. A. 2009;106:8186–8191. doi: 10.1073/pnas.0811269106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harrison P.J., Dunn T.M., Campopiano D.J. Sphingolipid biosynthesis in man and microbes. Nat. Prod. Rep. 2018;35:921–954. doi: 10.1039/c8np00019k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harmon J.M., Bacikova D., Gable K., Gupta S.D., Han G., Sengupta N., Somashekarappa N., Dunn T.M. Topological and functional characterization of the ssSPTs, small activating subunits of serine palmitoyltransferase. J. Biol. Chem. 2013;288:10144–10153. doi: 10.1074/jbc.M113.451526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parthibane V., Acharya D., Srideshikan S.M., Lin J., Myerscough D.G., Abimannan T., Vijaykrishna N., Blankenberg D., Bondada L., Klarmann K.D., Fox S.D., Andresson T., Tessarollo L., Acharya U., Keller J.R. Sptlc1 is essential for myeloid differentiation and hematopoietic homeostasis. Blood Adv. 2019;3:3635–3649. doi: 10.1182/bloodadvances.2019000729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharan S.K., Thomason L.C., Kuznetsov S.G., Court D.L. Recombineering: A homologous recombination-based method of genetic engineering. Nat. Protoc. 2009;4:206–223. doi: 10.1038/nprot.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewandoski M., Meyers E.N., Martin G.R. Analysis of Fgf8 gene function in vertebrate development. Cold Spring Harb. Symp. Quant. Biol. 1997;62:159–168. [PubMed] [Google Scholar]
- 33.Dzierzak E., Bigas A. Blood development: Hematopoietic stem cell dependence and independence. Cell Stem Cell. 2018;22:639–651. doi: 10.1016/j.stem.2018.04.015. [DOI] [PubMed] [Google Scholar]
- 34.Kuhn R., Schwenk F., Aguet M., Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 35.Smith C.M., Hayamizu T.F., Finger J.H., Bello S.M., McCright I.J., Xu J., Baldarelli R.M., Beal J.S., Campbell J., Corbani L.E., Frost P.J., Lewis J.R., Giannatto S.C., Miers D., Shaw D.R. The mouse gene expression database (GXD): 2019 update. Nucleic Acids Res. 2019;47:D774–D779. doi: 10.1093/nar/gky922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen M., Han G., Dietrich C.R., Dunn T.M., Cahoon E.B. The essential nature of sphingolipids in plants as revealed by the functional identification and characterization of the Arabidopsis LCB1 subunit of serine palmitoyltransferase. Plant Cell. 2006;18:3576–3593. doi: 10.1105/tpc.105.040774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oswald M.C., West R.J., Lloyd-Evans E., Sweeney S.T. Identification of dietary alanine toxicity and trafficking dysfunction in a Drosophila model of hereditary sensory and autonomic neuropathy type 1. Hum. Mol. Genet. 2015;24:6899–6909. doi: 10.1093/hmg/ddv390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gable K., Slife H., Bacikova D., Monaghan E., Dunn T.M. Tsc3p is an 80-amino acid protein associated with serine palmitoyltransferase and required for optimal enzyme activity. J. Biol. Chem. 2000;275:7597–7603. doi: 10.1074/jbc.275.11.7597. [DOI] [PubMed] [Google Scholar]
- 39.Kimberlin A.N., Majumder S., Han G., Chen M., Cahoon R.E., Stone J.M., Dunn T.M., Cahoon E.B. Arabidopsis 56-amino Acid serine palmitoyltransferase-interacting proteins stimulate sphingolipid synthesis, are essential, and affect mycotoxin sensitivity. Plant Cell. 2013;25:4627–4639. doi: 10.1105/tpc.113.116145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xie S.Z., Garcia-Prat L., Voisin V., Ferrari R., Gan O.I., Wagenblast E., Kaufmann K.B., Zeng A.G.X., Takayanagi S.I., Patel I., Lee E.K., Jargstorf J., Holmes G., Romm G., Pan K. Sphingolipid modulation activates proteostasis programs to govern human hematopoietic stem cell self-renewal. Cell Stem Cell. 2019;25:639–653.e637. doi: 10.1016/j.stem.2019.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loh K.C., Baldwin D., Saba J.D. Sphingolipid signaling and hematopoietic malignancies: To the rheostat and beyond. Anticancer Agents Med. Chem. 2011;11:782–793. doi: 10.2174/187152011797655159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kozutsumi Y., Segal M., Normington K., Gething M.J., Sambrook J. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature. 1988;332:462–464. doi: 10.1038/332462a0. [DOI] [PubMed] [Google Scholar]
- 43.Shamu C.E., Cox J.S., Walter P. The unfolded-protein-response pathway in yeast. Trends Cell Biol. 1994;4:56–60. doi: 10.1016/0962-8924(94)90011-6. [DOI] [PubMed] [Google Scholar]
- 44.Ron D., Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 45.Tessarollo L., Palko M.E., Akagi K., Coppola V. Gene targeting in mouse embryonic stem cells. Methods Mol. Biol. 2009;530:141–164. doi: 10.1007/978-1-59745-471-1_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fagundez C.B., Loresi M.A., Ojea Quintana M.E., Delcourt S.M., Testa R., Gogorza S.J., Argibay P.F. A simple approach for mouse embryonic stem cells isolation and differentiation inducing embryoid body formation. Cell Biol. Int. 2009;33:1196–1200. doi: 10.1016/j.cellbi.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 47.Heath V., Suh H.C., Holman M., Renn K., Gooya J.M., Parkin S., Klarmann K.D., Ortiz M., Johnson P., Keller J. C/EBPalpha deficiency results in hyperproliferation of hematopoietic progenitor cells and disrupts macrophage development in vitro and in vivo. Blood. 2004;104:1639–1647. doi: 10.1182/blood-2003-11-3963. [DOI] [PubMed] [Google Scholar]
- 48.Rutti M.F., Richard S., Penno A., von Eckardstein A., Hornemann T. An improved method to determine serine palmitoyltransferase activity. J. Lipid Res. 2009;50:1237–1244. doi: 10.1194/jlr.D900001-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh S.K., Williams C.A., Klarmann K., Burkett S.S., Keller J.R., Oberdoerffer P. Sirt1 ablation promotes stress-induced loss of epigenetic and genomic hematopoietic stem and progenitor cell maintenance. J. Exp. Med. 2013;210:987–1001. doi: 10.1084/jem.20121608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu F., Cheng G., Hamard P.J., Greenblatt S., Wang L., Man N., Perna F., Xu H., Tadi M., Luciani L., Nimer S.D. Arginine methyltransferase PRMT5 is essential for sustaining normal adult hematopoiesis. J. Clin. Invest. 2015;125:3532–3544. doi: 10.1172/JCI81749. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 and Figures S1 to S6
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article (and/or) its supporting information.