Abstract
Background
Nucleic acids are potent stimulators of type I interferon (IFN-I) and antiviral defense, but may also promote pathological inflammation. A range of diseases are characterized by elevated IFN-I, including systemic lupus erythematosus (lupus). The DNA-activated cGAS-STING pathway is a major IFN-I-inducing pathway, and activation of signaling is dependent on trafficking of STING from the ER to the Golgi.
Methods
Here we used cell culture systems, a mouse lupus model, and material from lupus patients, to explore the mode of action of a STING antagonistic peptide, and its ability to modulate disease processes.
Findings
We report that the peptide ISD017 selectively inhibits all known down-stream activities of STING, including IFN-I, inflammatory cytokines, autophagy, and apoptosis. ISD017 blocks the essential trafficking of STING from the ER to Golgi through a mechanism dependent on the STING ER retention factor STIM1. Importantly, ISD017 blocks STING activity in vivo and ameliorates disease development in a mouse model for lupus. Finally, ISD017 treatment blocks pathological cytokine responses in cells from lupus patients with elevated IFN-I levels.
Interpretation
These data hold promise for beneficial use of STING-targeting therapy in lupus.
Funding
The Novo Nordisk Foundation, The European Research Council, The Lundbeck Foundation, European Union under the Horizon 2020 Research, Deutsche Forschungsgemeinschaft, Chulalongkorn University.
Keywords: Lupus, STING, Innate immunity, Type I interferon, Inflammation, Immunomodulatory therapy
Research in Context.
Evidence before this study
The innate immunological STING signaling pathway is activated in response to cytoplasmic DNA and contrubites to host defense and inflammatory disease. There is evidence suggesting that STING hyper-activity is involved in disease processes in a sub-group of lupus patients.
Added value of this study
This study demonstrates that the peptide ISD017 specifically blocks STING signaling, and has the capacity to inhibit disease-associated activities in lupus-prone mice and cells from lupus patients.
Implications of all the available evidence
The data presented in this study support the idea of further clinical testing of STING-targeting therapy in lupus. In addition, given the specificity of ISD017 towards STING signaling, an additional implication of the data is that the peptide may have therapeutic potential against other diseases, where STING signaling contributes to pathology.
Alt-text: Unlabelled box
1. Introduction
Nucleic acids (NA) are potent inducers of innate immune responses. There is substantial evidence for NA-activated responses playing important roles in host defense against infections in mice and humans [1], [2], [3], [4], [5], [6]. In addition, NAs also contribute to evoking protective responses against malignant cells, and defects in these responses render mice susceptible to different experimental tumors [7], [8], [9]. Several NA-based molecular structures are immunostimulatory such as double-stranded RNA (dsRNA), 5′triphosphorylated RNA, and double-stranded DNA (dsDNA) [10]. The responses evoked by NAs include induction of a broad range of antimicrobial and inflammatory cytokines, including type I interferon (IFN-I, IFNα/β), tumor necrosis factor α (TNFα), and interleukin 1β (IL-1). While the two former are induced by transcriptional programs mainly driven by the transcription factors IFN regulatory factor 3/7 and nuclear factor κB, IL1β is induced after activation of the protease complex called the inflammasome and cleavage of pro-IL1β [11]. However, in addition to the beneficial roles of NA-induced immune responses in host defense and anti-cancer immunity, there is accumulating knowledge on how inappropriate activation or regulation of NA-driven immune responses leads to immunopathology [12], [13], [14]. Interestingly, elevated levels of IFN-I seem to be common to most if not all inflammatory diseases with dysregulated signaling by NA-activated pathways and appears to contribute to pathology [15,16]. Therefore, NA-activated pathways are central players in inflammatory diseases and attractive candidates for therapeutic targeting.
Four NA-sensing pathways are known to induce IFN-I expression. First, the TLR7/9-MyD88 pathway senses RNA and DNA species in endosomes and operates predominantly in plasmacytoid dendritic cells to induce high amounts of IFNα [17,18]. Second, the TLR3-TRIF pathway senses dsRNA in endosomes [19]. Third, the cytoplasmic proteins RIG-I and MDA5 detect 5′-triphosphorylated RNA and dsRNA in the cytoplasm and signal via the mitochondria-associated adaptor protein MAVS to induce IFN-I expression [20,21]. Finally, the enzyme cGAS detects cytoplasmic DNA and is activated to produce 2′3’ cGAMP, which acts as a second messenger binding to the ER-resident protein STING [4,22]. Following cGAMP-binding, STING traffics to the ERGIC and Golgi for recruitment of the kinase TBK1 and downstream phosphorylation of STING and activation of the IFN-inducing transcription factor IRF3 [23], [24], [25], [26]. The mechanisms that govern STING trafficking are currently being elucidated, and several proteins have been demonstrated to be involved. For instance, STIM1 was reported to be important for retaining STING in the ER [27], and we showed STEEP to be essential for loading of STING into COPII bodies for trafficking to the Golgi [28]. The effector functions downstream of STING are not limited to gene expression but include apoptosis and autophagy [29], [30], [31].
Systemic lupus erythematosus (lupus) is a chronic inflammatory disease that can involve all organ systems [32] including skin, musculoskeletal system, blood vessels, and kidney. Notably, the involvement of disease activity in the kidney (lupus nephritis) is a severe manifestation [32]. The current lupus treatment is non-specific and based on broadly acting immunosuppressive drugs [33]. This includes steroids and cytotoxic compounds like cyclophosphamide. Therefore, there is an urgent need for specific and less toxic treatment options for lupus.
A key feature of lupus is antinuclear and anti-DNA antibodies, which contribute to disease development [32]. However, lupus is a heterogeneous disease, and seminal work from the last decade has revealed that patients can be subdivided into at least three categories [34,35]. Importantly, two of these categories are characterized by elevated production of IFN-I, and IFN-stimulated genes (ISGs) [35]. lupus is characterized by an overload of antigens and nucleic acid species. This leads to maturation of dendritic cells (DCs) and downstream development of antigen-specific T and B cell responses. In this respect, the development of antinuclear and anti-DNA antibody responses are pivotal [32]. Importantly, IFN-I promotes B cell responses and has been demonstrated to contribute to the autoantibody response in lupus [36,37]. Second, there is also frequently elevated neutrophil activity in lupus, and IFN-I contributes to activation and prolongation of this response [38,39]. Collectively, IFN-I stimulates two of the key drivers of lupus pathogenesis, thus promoting anti-self antibodies and excess inflammation. Interestingly, it has been reported that the levels of cGAMP are elevated in a subset of lupus patient cells, and this correlates with the clinical disease score SLEDAI [40]. This suggests that the cGAS-STING pathway is constitutively activated in lupus patients and could be a key source of the pathological IFN response.
Given the emerging knowledge on the importance of the cGAS-STING pathway in inflammatory diseases, interest has grown in developing antagonists. At present, a handful of inhibitors of the pathway has been developed [41], [42], [43], [44], [45], [46]. This includes both peptides and small molecules and inhibitory activities based on covalent and non-covalent actions. For instance, the nitrofuran derivative H-151 blocks STING palmitoylation and blocks IFN responses in Trex1−/− mice [42]. In addition, the benzimidazol-isobenzofuran derivative RU.521 binds to the active site of cGAS and prevents activation in vitro and ex vivo [45].
In this work, we report that the STING antagonistic peptide ISD017, previously reported to target murine STING [41], is also a selective inhibitor of human STING working in a STIM1-dependent manner to block STING trafficking from ER to Golgi. ISD017 inhibits all known STING downstream activities and has no overt toxic effects on cells. Importantly, ISD017 blocks STING activity in mice and improves the clinical outcome of disease in a mouse model for lupus. Finally, ISD017 blocks the pathological IFN and cytokine responses in peripheral blood mononuclear cells (PBMCs) from lupus patients with elevated IFN-I levels. Thus, ISD017 is a STING inhibitor with potential for clinical use in a subpopulation of lupus patients with elevated STING activity or other STING-driven diseases.
2. Methods
2.1. ISD017, ISD017-derivatives, H-151, RU.521, and treatment of cells
ISD017 and derived peptides were obtained from Schafer-N. H-151 and RU.521 were obtained from InvivoGen. To dissolve ISD017 a panel of solvents were tested, namely: H2O; 0.9% NaCl; TBS (pH 7.3); PBS (pH 7.4); HEPES; PBS (pH7.4) + 1 M NaOH. For the latter, which was most successful, 194 uL of PBS mixed with 6 uL 1 M NaOH was added to 1 mg ISD017 or mutant peptides. H-151 was dissolved in DMSO and diluted in PBS + 10% tween80, and RU.521 was dissolved in DMSO. For in vitro experiments, primary cells and cell lines were treated with cGAS/STING antagonists one h prior to stimulation unless otherwise stated. The antagonists were administered by direct addition to the culture medium. For stimulations, we used 60‐mer dsDNA (DNA technology) [47], 2′3′ cGAMP, and poly(I:C) (both from InvivoGen). The agonists were delivered with Lipofectamine 2000 (ThermoFischer). Liposomes were prepared as described using lipid blends containing DOTAP, DOPE and lissamine–rhodamine DOPE in the w/w/w ratio 1/1/0.1 Avanti Polar Lipids [48].
2.2. Cells
PBMCs, HEK293T and RAW264.1 cells were maintained in DMEM supplemented with 10% FBS, 100 U/ml penicillin, 100 µg/ml streptomycin, and 2 mM l-glutamine. PBMCs for experiments not involving patient material were obtained from the Aarhus University Hospital Blood Bank and isolated using Ficoll-Paque™. THP1 (RRID:CVCL_A1RT) cells and murine peritoneal cells were maintained in RPMI 1640 supplemented with 10% FBS, 100 U/ml penicillin, 100 µg/ml streptomycin, and 2 mM l-glutamine. Media containing 150 nM PMA was used for the initial 24 h to differentiate THP1 cells and later was replaced with media containing no PMA. The converted macrophage-like cells were used for the experiments after 24 h. For in vitro experiments, PBMCs, HEK293T cells, THP1, and murine peritoneal cells were seeded at a density of 5 × 105 cells/cm2, 2.5 × 105 cells/cm2, 2 × 105 cells/cm2, and 5 × 105 cells/cm2, respectively. For in vitro stimulation experiments, cells were allowed to rest for 4–6 h following seeding. For testing of inhibitory effect of ISD017 on SLE patient PCMBs ex vivo, the peptide was added to the cell culture media immediately following seeding. All cell lines used in the project were documented Mycoplasma-free.
2.3. Isolation of human material from patients and healthy donors
Patient inclusion and blood sampling have previously been described [49]. PBMCs were isolated using CPT tubes (BD Diagnostics Vacutainers). Samples were centrifuged at room temperature in a horizontal rotor for 30 min at 1800 g. Following centrifugation, the mononuclear cell layer was collected and transferred to 15-ml conical tubes. Following 2 washing steps with PBS, the cell pellets were resuspended in 10% DMSO with fetal calf serum and stored at −135 °C until use.
2.4. Isolation of extracellular vesicles from serum
Extracellular vesicles from serum of lupus patients and healthy donors were isolated using the Total Exosome Isolation Reagent (from serum) kit (InVitroGen). The isolated vesicles were enriched in exosomal markers (CD81 and CD63, data not sho3wn) and did not contain type I IFN activity (data not shown).
2.5. ΔβLUC mouse model
ΔβLUC mice have been described previously [50]. For ex-vivo quantification of the Ifnβ-luciferase reporter activity 6–12 week old mice were injected i.p. with vehicle or 30 mg/kg ISD017 in PBS (pH7.4) + 20 mM NaOH. At the indicated time points after dosing, mice were challenged i.p. with 25 mg/kg DMXAA (Invivogen). Five hours later, 107 splenocytes were lysed in 100 µl Passive Lysis Buffer (Promega) to quantify luciferase activity using the Luciferase Assay Reporter Kit (Promega) on a Clariostar Luminometer (BMG Labtech). For in vivo imaging of Ifnβ-luciferase reporter activity 6–12 week old mice were injected i.p. with vehicle or 30 mg/kg ISD017 in PBS (pH7.4) + 20 mM NaOH. At the indicated time points after dosing, mice were challenged i.v. with 20 mg/kg 2´3´-cGAMP (Invivogen). Five hours later, mice were injected with 150 mg/kg XenoLight d-luciferin (Perkin Elmer) in isotonic sodium chloride. Photon flux was quantified one minute after injection on an In-vivo Xtreme II imaging device (Bruker) with binning set to 8 × 8 pixels and an integration time of 30 s.
2.6. Fcgr2b-deficient mouse model
The Fcgr2b−/− mice on C57BL/6 background were obtained from Bolland S. (NIH, Maryland, USA). Wild type mice were purchased from the Nomura Siam International, Thailand. All animal experiments were approved by the Institutional Animal Care and Use Committees (IACUC) of the Faculty of Medicine, Chulalongkorn University (013/2563). Six-week-old mice were injected intraperitoneally (i.p.) with 10 mg/kg of ISD017 (3 times per week) until eight months of age. Handeling and evaluation of animals was done in a blinded fashion.
2.7. Histopathology
Kidney tissues were fixed in 10% Neutral buffered formalin (NBF). Tissue blocks were embedded in paraffin, 5 mm sections obtained, and then stained with hematoxylin and eosin. The pathology scores were blindly graded following the previous publication [51].
Frozen renal sections were fixed in acetone and blocked with 1% BSA in PBS. The sections were stained with Alexa 488 conjugated anti-mouse IgG antibody and Alexa 647 conjugated anti-CD45 (30-F11) (Biolegend, San Diego, CA, USA). Samples were then stained with DAPI (4′,6-Diamidino-2-Phenylindole, Dihydrochloride) (Thermo Fisher Scientific, MA USA) for 5 min in the dark at room temperature. Slides were washed 3 times and mounted with ProLongTM diamond antifade mountant (Invitrogen, CA, USA). The fluorescent signaling was visualized by ZEISS LSM 800 with Airyscan (Carl Zeiss, Germany).
2.8. Flow cytometry analysis
Splenocytes were isolated, passed through a 70-µm filter, lysed red blood cells by lysis buffer (ACK buffer: NH4Cl, KHCO3, and EDTA), and placed in staining buffer (PBS + 0.5% BSA). The splenocytes (1 × 106 cells) were stained with following antibody; anti-CD4 (GK1.5), CD8 (53–6.7), CD62l (MEL-14), CD44 (IM7), CD3ε (145–2C11), ICOS (C398.4A), CD11c (N418), B220 (RA3–6B2), CD11b (M1/70), I-Ab (AF6–120.1), PDCA-1 (129c1), CD80 (16–10A1), GL7 (GL7), and CD138 (281–2) (Biolegend, San Diego, CA, USA). The flow cytometry analysis was performed using BDTM LSR-II (BD Biosciences, USA) and FlowJo software (version 10, USA). The serum sample was measured using the LEGENDplex™ Mouse Inflammation Panel kit (IL-1α, IL-1β, IL-6, IL-10, IL-12p70, IL-17A, IL-23, IL-27, MCP-1, IFN-β, IFN-γ, TNF-α, and GM-CSF) (Biolegend, San Diego, CA, USA) and followed the manufacture protocol. The beads were analyzed using BDTM LSR-II (BD Biosciences, USA) and LEGENDplex™ Analysis Software version 8.
2.9. IFNB luciferase reporter gene assay
HEK293T cells were plated on 96 well plates and transfected with 30 ng constructs harboring IFNB-promoter firefly luciferase reporter genes and 10 ng β actin-promoter-driven Renilla luciferase together with 50 ng constructs encoding either STEEP, cGAS, TRIF, or MAVS. Eighteen h after transfection, the cells were stimulated with cGAMP or left untreated. Six hours later, the cells were treated with ISD017 (200 µg/ml) left until 24 h post transfection. The cells were lysed, and the firefly- and renilla luciferase signals were developed with Dual Glo® luciferase assay (Promega) and read on Luminoscan Ascent (Labsystems) according to manufactures instructions.
2.10. RT-qPCR
Gene expression was determined by real-time quantitative PCR, using TaqMan detection systems (Applied Biosciences). RNA was extracted using the High Pure RNA Isolation kit (Roche) and RNA quality was assessed by Nanodrop spectrometry (Thermo Fisher). RNA quantified using TaqMan assays and the RNA-to-Ct-1-Step kit according to the manufacturer's recommendations (Applied Biosciences). Taqman assays for qPCR were; ISG15 (Hs01921425_s1) and ACTB (Hs01060665) (Applied Bioscience).
2.11. ELISA
Levels of human cytokine TNFα and chemokines (CXCL10) and CCL20 in serum and culture supernatants, or murine cytokines IFN-β and IL-17A in serum, were measured by ELISA (R&D Systems), following the instructions of the manufacturer. For detection of anti-dsDNA antibodies, mitrotiter wells were coated with dsDNA (10 µg/ml) overnight. After washing (1xTBS-Tween) and blocking (PBS, 5% FBS, 3% BSA, 0.1% tween), mouse serum samples were added to the wells at a dilution of 1:100 and incubated for 1 h at 37 °C. The wells were washed and HRP conjugated goat anti-mouse antibody (Biolegend, 1:4000 in blocking solution) was added for 1 h incubation af 37 °C. After extensive washing, the ABTS Peroxidase Substrate Solution system (ThermoFisher) was used for development, and the ABTS Peroxidase Stop Solution (ThermoFisher) was use to stop the reaction. For measurements, wavelength of 450 nm was used.
2.12. Type I IFN bioassay
Bioactive human type I IFN was measured on cell supernatants using HEK‐Blue™ IFN‐α/β cells as reporter cells according to the manufacturer instructions (InvivoGen) and as described by the manufacturer.
2.13. AnnexinV staining
For assessment of cell death by flow cytometry, cells were stained using Dead Cell Apoptosis Kit with Annexin V Alexa Fluor™ 488 & Propidium Iodide (Invitrogen) according to manufacturer's protocol. Briefly, cells were centrifuged and resuspended at 1 million cells/mL in annexin binding buffer containing annexin V A488 (1:40) and PI (1:40) and stained in the dark for 15 min. Cells were then centrifuged and resuspended in annexin binding buffer and immediately analysed on a NovoCyte flow cytometer.
2.14. Peptide sequence analysis
Structural prediction of the primary sequence of ISD017 was analyzed using PEP-FOLD3.5 (https://mobyle.rpbs.univ-paris-diderot.fr/cgi-bin/portal.py#forms::PEP-FOLD3). Alphahelical wheels were generated using NetWheels (http://lbqp.unb.br/NetWheels/).
2.15. Immunoblotting
Whole‐cell extracts or immunoprecipitation samples were diluted in XT sample buffer and XT reducing agent and loaded into 4–20% SDS–PAGE gel (Bio‐Rad). The proteins were transferred from gel to PVDF membranes through Trans‐Blot Turbo™ Transfer System® (Bio‐Rad). The membrane was blocked in 5% nonfat skim milk (Sigma) for 1 hour at room temperature. The following antibodies and dilutions were used for immunoblotting anti‐STING (Cell Signaling, D2P2F/#13,647, 1:1000), sheep anti‐STING (R&D Systems, AF6516, 1:500), rabbit anti‐phosphoSTING (S366) (Cell Signaling Technology, #85,735, 1:1000), rabbit anti‐TBK1 (Cell Signaling, 3504, 1:1000), rabbit anti‐phospho-TBK1 (Ser172) (Cell Signaling, D52C2/#5483, 1:1000), IRF3 (Cell signaling, 11,904, 1:1000), phospho-IRF3 (S396) (Cell Signalling, 4947,1:1000, mouse anti‐p62 (Cell Signaling, # 88,588, 1:1000), anti-STEEP (Proteintech, 24,021–1-AP, 1:1000), anti-STIM1 (D88E10, Cell Signaling, #5668, 1:1000), rabbit anti‐LC3 (Cell Signaling, #3868, 1:1000), rabbit anti-cleaved caspase3 (Cell Signaling, #9664,1:1000), and mouse anti‐vinculin (Sigma, #V9131, 1:10,000).
2.16. Precipitation of proteins with antibodies and peptide
Cells were lysed in IP lysis buffer with 1xProtease Inhibitor Cocktail (Sigma) and 10 mM NaF, and the cell lysates were centrifuged for 10 min, 2500 g at 4 °C. The cleared cell lysates were incubated with primary antibody against sheep Anti-STING (R&D Systems, AF6516, 1:50) or with biotin-ISD017 overnight at 4 °C. Dynabeads™ Protein G or Dynabeads™ Streptavidin (Invitrogen) were added into the mixtures for extra 2 h. After 4–6 times wash with IP lysis buffer (with varying concentrations of NaCl), the precipitated complexes were eluted by glycine buffer (200 mM glycine, pH 2.5) with Protease Inhibitor Cocktail (Sigma) and 10 mM NaF, and the elutes were evaluated by Immunoblotting blotting.
2.17. Confocal microscopy
PMA‐differentiated THP1 cells were seeded on coverslips and stimulated as indicated. The cells were fixed with methanol for 5 min at −20 °C. Cells were blocked in 1xPBS with 1%BSA. Cells were stained with primary antibodies for 1 h/overnight and then stained by secondary antibody (all 1:300, Alexa Fluor; Invitrogen) for 1 h. Images were acquired on Zeiss LSM 780 confocal microscope and processed with the Zen Blue software (Zeiss). The antibodies used were sheep anti‐STING (R&D Systems,AF6516, 1:25), rabbit anti‐calreticulin (Abcam,22,683,1:50), rabbit anti-Sec24 (1:100); mouse anti‐GM130 (Cell signaling,12,238,1:1000).
2.18. Statistics and reproducibility
The data are shown as means of biological replicates ± s.d. The statistical test used was dependent on whether the data exhibited a normal distribution. Statistically significant differences between groups were determined using two-tailed Student's t-test when the data exhibited a normal distribution; two-tailed Mann–Whitney (one-way analysis of variance (ANOVA)) test when the dataset did not pass the normal distribution test. The data shown are from single experiments. The experiments were performed at least three times with similar results.
2.19. Ethical aspects
All animal experiments were approved by the Landesdirektion Sachsen (TVV 6/2019) and the Institutional Animal Care and Use Committees (IACUC) of the Faculty of Medicine, Chulalongkorn University (013/2563), and conducted in compliance with the approvals. Patient inclusion was approved by The Danish Data Protection Agency and the Central Denmark Region Committees on Health Research Ethics (#1–10–72–214–13). Patient inclusion was performed according to the Helsinki Declaration. Informed consent from all participants was obtained.
2.20. Role of funding source
The funders had no role in study design, data collection, data analyses, interpretation, or writing of the manuscript.
3. Results
3.1. ISD017 is a STING antagonist in human cells
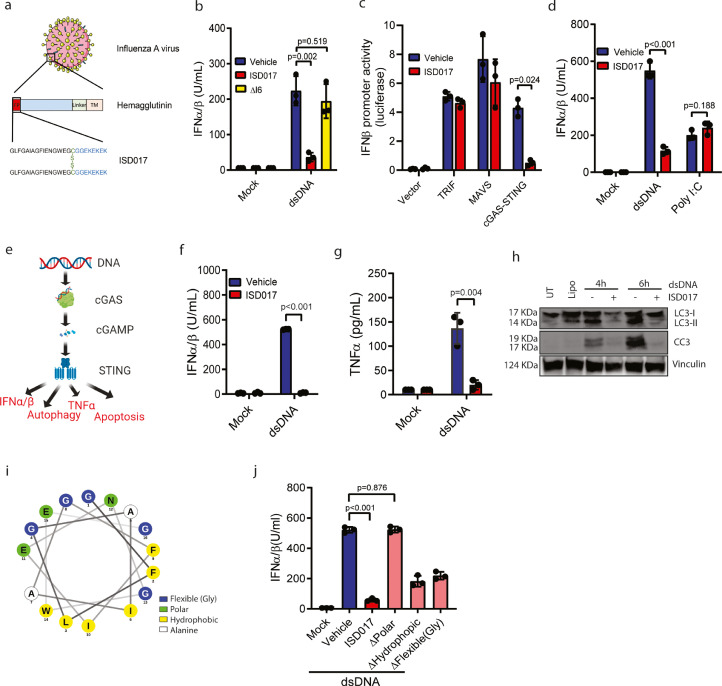
We previously reported that the fusion peptide of the influenza haemagglutinin protein possesses STING antagonistic activity in murine cells (Fig. 1a) [41]. In these studies, we found that the peptide, now called ISD017, blocked murine STING activation by fusogenic liposomes, but we did not observe any effect on cGAS-activated STING [41,48]. Fusogenic liposomes activate STING independent of the canonical cGAS pathway, but the mechanism remains unresolved [41,48]. To explore the effect of the peptide on human STING, we first improved the protocol for solubility of the peptide. A protocol using PBS and NaOH significantly elevated solubility (Supplementary Fig. 1). Under these conditions, and using higher concentrations than previously [41], ISD017 blocked human cGAS-activated STING since dsDNA-induced IFN-I expression very efficiently in PBMCs (Supplementary Fig. 1). The peptide dissolved with this protocol exhibited dose-dependent inhibition of both dsDNA-induced and liposome-induced production of IFN-I and the ISG CXCL10 in PBMCs. However, IC50 was higher in case of dsDNA induction (Supplementary Fig. 1). Likewise, ISD017 did also block both dsDNA-induced and liposome-induced production of CXCL10 in murine peritoneal cells (Supplementary Fig. 1). The inhibitory activity of ISD017 was not due to unspecific actions of the peptide since deletion of isoleucine at position 6, which significantly altered the position of residues in the predicted alphahelical structure of the peptide (see below), abolished the inhibitory activity in PMA-differentiated THP1 cells (THP1 macrophages) (Fig. 1b). To test whether ISD017 was specific for the STING pathway, we overexpressed key activating molecules from three IFN-I-inducing pathways in HEK293T cells together with an IFNβ-reporter gene and treated with ISD017. As seen in Fig. 1c, ISD017 inhibited the response in cGAS-STING-expressing cells, but not in cells expressing MAVS or TRIF. Likewise, ISD017 inhibited production of IFN-I and CXCL10 in PBMCs treated with dsDNA, but not in cells treated with the dsRNA-mimic poly(I:C) (Fig. 1d, Supplementary Fig. 1). To test whether ISD017 also inhibits non-IFN activities downstream of STING (Fig. 1e), we treated THP1 macrophage cells with dsDNA and examined for expression of TNFα, accumulation of LC3-II (a marker of autophagy), and cleavage of caspase 3 (a marker of apoptosis). Interestingly, all STING-activated functions were blocked by co-treatment with ISD017 (Fig. 1f-h, Supplementary Fig. 1).
Fig. 1.
ISD017 is a STING antagonist in human cells. (a) Illustration of the origin of the core ISD017 peptide from the influenza A fusion peptide, and its dimeric nature. (b) PMA-differentiated THP1 cells were treated with dsDNA (2 μg/ml) in the presence or absence of 1 h pre-treatment with ISD017 (200 μg/ml) or ΔI6 (mutant peptide where Isoleucine at position 6 was deleted). Supernatants were harvested 16 h post-stimulation, and IFN-I activity was determined (n = 3). (c) HEK293T cells were transfected with IFNB-luciferase reporter and the indicated expression plasmids. Six h later, the cells were treated with ISD017 (200 µg/ml), and luciferase activity was evaluated 24 h post transfection (n = 3). The bands for Vinculin and STEEP in the input lane appear “light blue”, due to saturation of the signal. (d) PBMCs were treated with ISD017 (200 µg/ml). One h later, the cells were transfected with either dsDNA or polyIC (both 2 µg/ml). Supernatants were harvested 16 h post-stimulation, and IFN-I activity was determined (n = 3). (e) Graphical illustration of the cGAS-STING pathway, and the four down-stream effector functions, IFN-I, TNFα, apoptosis, and autophagy. (f-g) PMA-differentiated macrophage-like THP1 cells were treated with ISD017 (200 µg/ml). One h later, the cells were transfected with dsDNA (2 µg/ml) or empty liposomes (mock). Levels of IFN-I and TNFα in the supernatants 16 h post-stimulation were determined by bioassay, and ELISA, respectively (n = 5). (h) PMA-differentiated macrophage-like THP1 cells were treated with ISD017 (200 µg/ml). One h later, the cells were transfected with dsDNA (2 µg/ml) and lysed 4 and 6 h later for immunoblotting with anti-LC3, anti-cleaved caspase 3 (CC3), and anti-vinculin (n = 3). (i) Illustration of the alpha-helical wheel of the ISD017 core sequence from residue 1–16. Polar residues are shown in green, hydrophobic residues in yellow, small flexible residues (Gly) are in blue, alanine residues in white. (j) PMA-differentiated macrophage-like THP1 cells were treated with parental ISD017 or one of the mutated variants (200 µg/ml). One h later, the cells were stimulated with dsDNA (2 µg/ml). Supernatants were isolated after 16 h, and level of IFN-I bioactivity in the supernatant was determined (n = 3). The data in panels b, c, d, f, g, j are presented as means of biological triplicates +/- st.dev. [Statistical analysis of the data in b-d, f, g, j were performed using two-tailed one-way ANOVA test].
ISD017 is a dimeric molecule, and we wanted to examine whether this was essential for activity. Analysis of the effect of monomeric versus dimeric ISD017 on dsDNA-induced IFN-1 expression in PBMCs showed that while both forms have antagonistic properties, the dimeric form was more potent (Supplementary Fig. 1). Next, we wanted to test the effect of timing of ISD017 treatment relative to dsDNA stimulation. THP1 macrophages were pretreated with ISD017 for up to 6 h prior to dsDNA stimulation, and IFN-I activity was measured. Pre-treatment with the peptide up to 6 h before stimulation led to a significant reduction of the IFN activity (Supplementary Fig. 1). In addition, we treated with ISD017 after stimulation with dsDNA and found that the peptide was able to block an already initiated response potently.
ISD017 is predicted to fold into an alpha helix with a polar side and a hydrophobic surface (Fig. 1i). To test which physical-chemical properties of ISD017 that were required for its activity, we made three mutants where either the polar residues, the hydrophobic residues, or the flexible residues (glycine) were mutated to alanine. All three mutants exhibited good solubility when using the protocol used for ISD017. While all mutants showed reduced inhibitory activity, the peptide lacking polar residues had entirely lost the ability to antagonize STING activities in THP1 macrophages (Fig. 1j). Thus, ISD017 is an inhibitor of human and murine STING down-stream of either fusogenic or dsDNA stimuli.
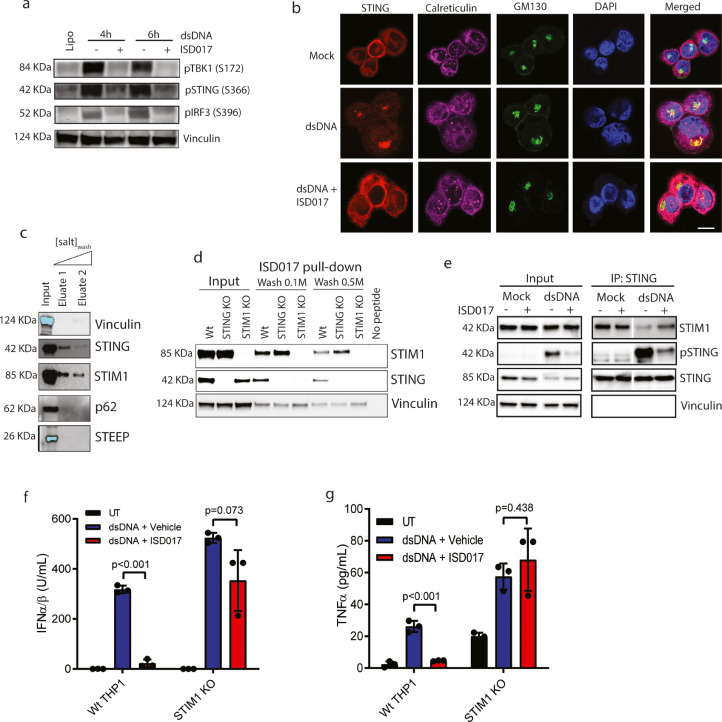
3.2. ISD017 inhibits STING ER-exit in a STIM1-dependent manner
To identify the step in the STING signaling pathway blocked by ISD017, we examined for levels of phosphorylation of TBK1 at S172, STING at S366, and IRF3 at S396 in THP1 macrophages treated with dsDNA in the presence or absence of the peptide. Treatment with ISD017 inhibited the accumulation of phosphorylated forms of both STING and TBK1 (Fig. 2a), suggesting that the inhibition occurs at a very upstream level in the pathway. Upon stimulation, STING traffics from the ER to ERGIC/Golgi where signaling occurs [23,24]. Treatment with ISD017 prevented this trafficking (Fig. 2b, Supplementary Fig. 2), demonstrating that the inhibitor acts on STING at the ER level. STING leaves the ER through COPII-mediated trafficking [28]. In agreement with this, we observed that ISD017 treatment inhibited DNA-induced elevation in Sec24 foci, a marker of COPII vesicles (Supplementary Fig. 2). We and others have found several proteins to associate with STING at the ER [27,28,[52], [53], [54], [55], [56]]. Upon precipitation of biotinylated ISD017 from THP1 macrophage lysates, we observed that STING and STIM1, but not p62 and STEEP, co-precipitated with the peptide (Fig. 2c). STIM1 has been reported to act as a STING ER retention factor [27]. Interestingly, when we compared the precipitation of STING and STIM1 with ISD017 in Wt, STING KO, and STIM1 KO THP1 macrophages, we observed that STIM1 deficiency led to an apparent reduction in the amount of STING precipitating with the peptide (Fig. 2d). By contrast, STING deficiency did not affect the interaction between STIM1 and ISD017. These data suggest that STIM1, or a STIM1-containing complex, rather than STING itself, is the target for ISD017. Moreover, we observed that the stimulation-induced dissociation of STING from STIM1, reported to enable STING activation [27], was inhibited by ISD017 (Fig. 2e). If ISD017 targets STIM1, the prediction would be that the peptide should lose STING antagonist activity in STIM1-deficient cells. Indeed, ISD017 failed to block dsDNA-induced expression of IFNB and TNFA mRNA in STIM1 deficient THP1 macrophages (Fig. 2f-g). Collectively, ISD017 blocks STING at the ER level through a mechanism dependent on STIM1.
Fig. 2.
ISD017 associates with STING and STIM1 and prevents ER-to-Golgi Trafficking. (a) PMA-differentiated THP1 cells were treated with ISD017 (200 µg/ml). One h later, the cells were transfected with dsDNA (2 µg/ml). Cells were lysed at the indicated time points post-stimulation and immunoblotted by antibodies against pTBKs (S172), pSTING (S366), pIRF3 (S396), and Vinculin. (n = 3). (b) PMA-differentiated THP1 cells were pretreated with ISD017 (200 µg/ml) for 1 h. The cells were transfected with dsDNA (6 µg/ml) and fixed 2 h later for imaging analysis after staining with anti-STING, anti-Calreticulin (ER), and anti-GM130 (Golgi) (n = 3). Scale bar, 10 µm. (c) Lysates from PMA-differentiated THP1 cells were incubated with biotinylated ISD017, washed, and precipitated with streptavidin beads. Precipitates were immunoblotted with antibodies binding the indicated proteins (n = 3). The light blue color of the bands for STEEP and vinculin in the input lane illustrates saturation of the signal. (d) Lysates from PMA-differentiated Wt, STING KO, and STIM1 KO THP1 cells were incubated with biotinylated ISD017, washed, and precipitated with streptavidin beads. Precipitates were immunoblotted with antibodies against STIM1, STING, and vinculin (n = 3). (e) PMA-differentiated THP1 cells were treated with ISD017 (200 µg/ml) for 1 h, followed by stimulation with dsDNA for 2 hs. Cell lysates were subjected to immunoprecipitation with anti-STING and immunoblotting with anti-STIM1, anti-STING, and anti-vinculin (n = 3). (f,g) Wt and STIM1 KO THP1 macrophages were treated with dsDNA in the presence or absence of ISD017 (200 µg/ml, pre-treatment). Supernatants were harvested 16 h post-stimulation, and IFN-I activity and TNFα levels were measured (n = 3). The data in panel f and g are presented as means of biological triplicates +/- st.dev. [Statistical analysis of the data in f, g were performed using two-tailed one-way ANOVA test].
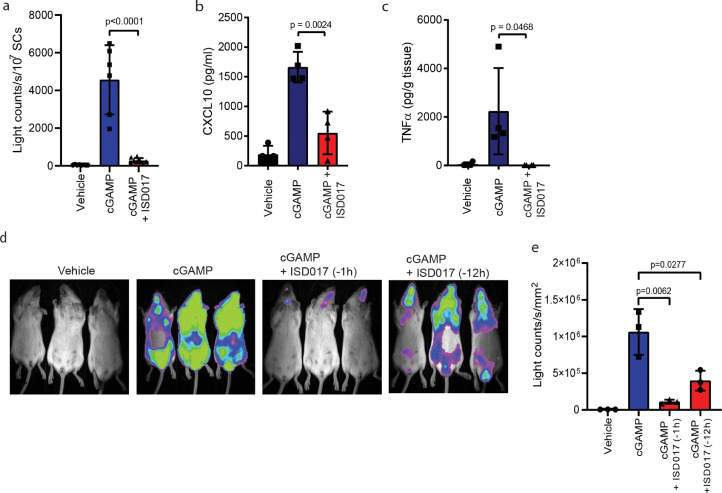
3.3. ISD017 inhibits STING activity in vivo
Next, we wanted to explore whether ISD017 exhibited STING antagonistic activity in vivo. For this purpose, we treated IFN-βΔβ-luc/Δβ-luc-reporter (ΔβLUC) mice with ISD017 followed by treatment with cGAMP or DMXAA 1 h later. STING agonist treatment lead to a strong induction of IFNβ−luciferase expression in the spleen. Importantly these responses were potently blocked by ISD017 (Fig. 3a, Supplementary Fig. 3) [42]. This was associated with parallel inhibition of expression of CXCL10 and TNFα (Fig. 3b-c). Whole-body bioimaging of the mice demonstrated that ISD017 systemically reduced IFNβ expression (Fig. 3d). To address the effect of timing of ISD017 treatment relative to the activation of the STING pathway in vivo, we pretreated mice with the peptide 1 and 12 h before cGAMP treatment. Even though we observed an apparent effect of dosing timing, the IFNβ response was still reduced by more than 60% in mice receiving ISD017 12 h before cGAMP treatment (Fig. 3d-e, Supplementary Fig. 3).
Fig. 3.
ISD017 has STING antagonistic activity in vivo. (a) IFNβΔβ-luc/Δβ-luc-reporter (ΔβLUC) mice were treated i.p. with ISD017 (30 mg/kg), and one h later injected with cGAMP (20 mg/kg) through the same route. Five h after treatment with STING agonist, spleen cells were isolated, and luciferase activity measured (n = 6). (b, c) Serum and spleen from mice treated as in a were analyzed for levels of CXCL10 and TNFα proteins by ELISA. (d) IFNβΔβ-luc/Δβ-luc-reporter (ΔβLUC) mice were treated i.p. with ISD017 (30 mg/kg), and 1 or 12 h later injected i.v. with cGAMP (20 mg/kg). Five h after treatment with STING agonist, whole animals were subjected to bioimaging (n = 3). (e) Quantification of the photon flux from the ventral view of animals shown in panel d. The data in panel a-c, e are presented as means of measurements from individual mice +/- st.dev. [Statistical analysis of the data in a, b, c, e were performed using two-tailed one-way ANOVA test].
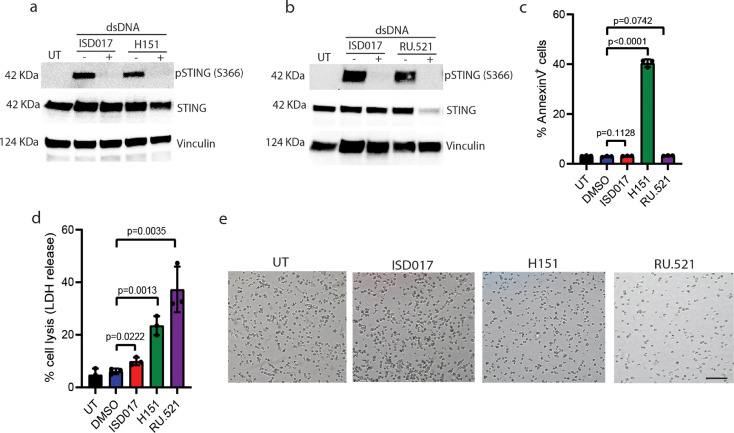
3.4. ISD017 shows no overt toxic effects on cells and mice
A series of cGAS and STING antagonists have now been described [41], [42], [43], [44], [45], [46], and we were interested in comparing ISD017 to some of these compounds. These analyses revealed that ISD017 blocks dsDNA-induced STING phosphorylation and IFN-I induction in THP1 macrophages with a potency comparable to the STING palmitoylation inhibitor H-151 and the cGAS inhibitor RU-521 (Fig. 4a-b, Supplementary Fig. 4). Although higher doses of ISD017 were required to achieve inhibition, the approximately 20 times higher molecular weight of ISD017 than, e.g., H-151 suggests comparable potency. To examine for toxic effects of ISD017 on cells and mice, we treated THP1 macrophages with the cGAS/STING inhibitors and evaluated apoptosis by annexinV staining, necrosis by LDH release, and general cell survival by microscopy. Under these experimental conditions, we observed that ISD017 induced neither apoptosis nor necrosis and was, in fact, less toxic than the compounds it was compared to (Fig. 4c-e). At lower doses of H-151, where IFN induction was still observed, this compound induced limited cell death (Supplementary Fig. 4). Examination of the cytotoxic effects of ISD017 on PBMCs from healthy donors reveated a picture simular to what was observed in THP1 macrophages (Supplementary Fig. 4). To test whether ISD017 was also well tolerated by mice, we treated mice with two doses of peptide, three times per week over seven weeks, and monitored behavior and weight development. The mice receiving ISD017 treatment exhibited no abnormalities in behavior and food intake and gained weight to the same degree as the contro4l group (Supplementary Fig. 4). Collectively, these data demonstrate that ISD017 is not toxic to cells and mice.
Fig. 4.
ISD017 shows no overt toxic effects on cells. (a, b) PMA-differentiated THP1 cells were treated with ISD017 (200 µg/ml), H-151 (4 µg/ml), or RU.521 (2 µg/ml) for 1 h followed by stimulation with dsDNA (2 µg/ml) for 2 h. Levels of pSTING, total STING, and vinculin were evaluated by immunoblotting (n = 4). (c, d) PMA-differentiated THP1 cells were treated with ISD017 (200 µg/ml), H-151 (4 µg/ml), or RU.521 (2 µg/ml) for 24 h and apoptotic and necrotic cell death was evaluated by AnnexinV staining (c) and LDH release (d), respectively (n = 3). (e) Microscopy images of cells 24 h after treatment with inhibitors of cGAS and STING. Scale bar, 100 µm. The data in panel a-d are presented as means of biological triplicates +/- st.dev. [Statistical analysis of the data in c, d were performed using two-tailed one-way ANOVA test].
3.5. ISD017 treatment improves disease outcome in a mouse model for STING-dependent lupus
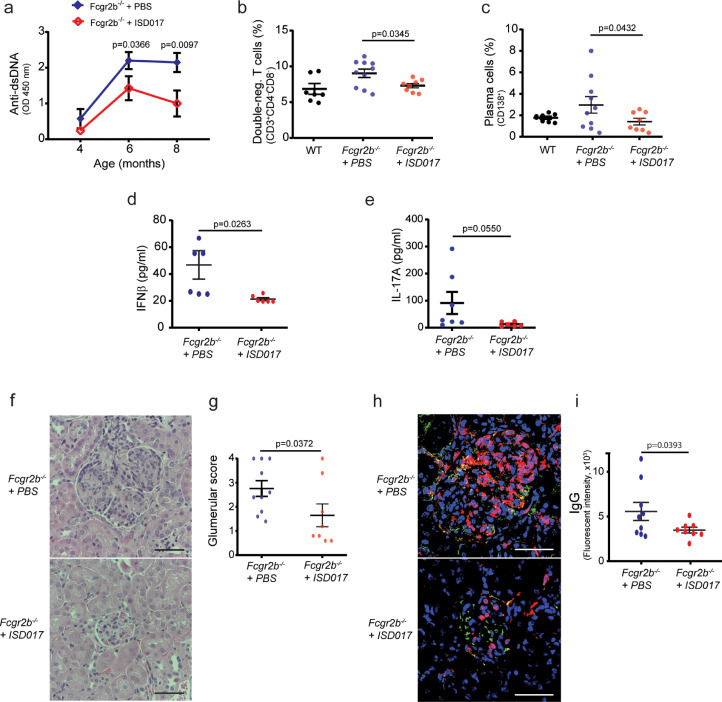
Several human diseases are now known to be associated with activation of the cGAS-STING pathway [13,40,[57], [58], [59], [60], [61]]. One of these is lupus, and we recently reported that full development of lupus-like disease in Fcgr2b−/− mice depends on STING [62]. Thus, we tested the hypothesis that ISD017 treatment could ameliorate the lupus phenotypes in the Fcgr2b-deficient mice by treating with ISD017 from 6 weeks until the age of eight months. ISD017 treatment did indeed reduce the magnitude of anti-dsDNA production in Fcgr2b−/− mice (Fig. 5a). Analysis of splenocytes from Fcgr2b−/− mice showed a reduction of double-negative T cells (Fig. 5b) and plasma cells (Fig. 5c) in the ISD017-treated group. Also, the levels of serum IFNβ was reduced in the ISD017-treated group (Fig. 5d), while the levels of IL-17A showed a clear tendency towards being lower in the ISD017-treated group (Fig. 5e). The hallmark lupus pathology of the Fcgr2b−/− mice is glomerulonephritis [63]. Significantly, treatment with ISD017 reduced kidney pathology (Fig. 5f) and the glomerular scores (Fig. 5g). The interstitial scores showed a trend to decrease by ISD017 treatment, although it did not reach statistical significance (Supplementary Fig. 5). Immunofluorescence staining of the kidney from the Fcgr2b−/− mice for IgG and CD45 showed evident IgG deposition in the glomeruli (Fig. 5h, Supplementary Fig. 5), and this was significantly lower for IgG deposition in the ISD017-treated mice (Fig. 5i). The CD45 intensity in the ISD017-treated group showed a reduction trend, but this did not reach a significant difference (Supplementary Fig. 5).
Fig. 5.
ISD017 treatment improves disease outcomes in a mouse model for lupus. (a) Fcgr2b−/- mice were treated with PBS or ISD017 (10 mg/kg). Levels of anti-dsDNA in serum (dilution 1:100) from the Fcgr2b−/− mice, harvested at age 4, 6, and 8 months, were evaluated by ELISA. (b, c) Flow cytometry analysis of splenocytes from 8-month-old Fcgr2b−/− mice treated as in panel a. The data are shown as percentage of (b) CD3+CD4−CD8− (double negative T cell) and (c) CD138+ (plasma cells). (d, e) The concentrations of serum IFNβ and IL-17A were analyzed by ELISA. (a-d, n = 8–10 per group). (f) Kidney sections of Fcgr2b−/− mice (8 months old) treated with PBS or ISD017 (10 mg/kg, 3 times per week) were stained with H&E. Data are representative of 8–10 mice per group (scale bar, 50 µm). (g) Glomerular scores of kidney sections were blindly graded (n = 8–10 per group). (h) Immunofluorescence staining of the kidneys from Fcgr2b−/− mice treated with PBS or ISD017 (10 mg/kg, 3 times per week). IgG (green), CD45 (red), and DAPI (blue). Data are representative of 8–10 mice per group (scale bar, 50 µm). (i) Quantification of immunofluorescence signals for IgG (n = 8–10 mice per group). Data in panel a-e, g, and i are shown as mean ± st.dev. [Statistical analysis of the data in a-e, g, i were performed using two-tailed one-way ANOVA test].
3.6. ISD017 blocks pathological cytokine responses in cells from lupus patients
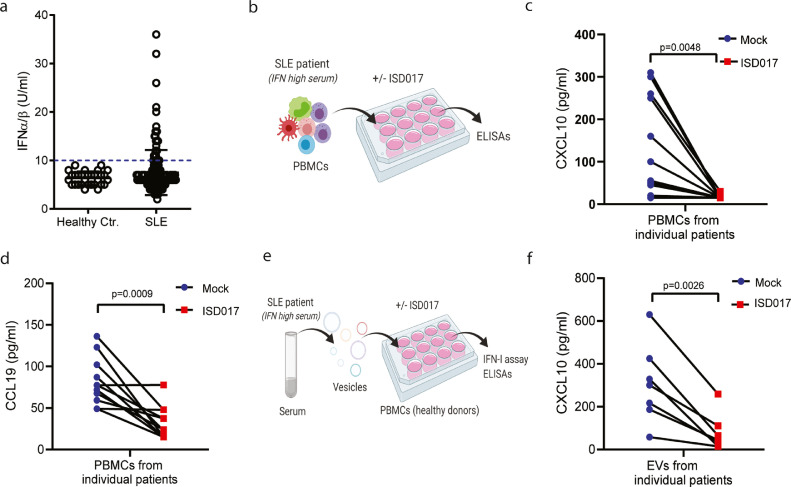
To finally explore whether ISD017 can block pathological immune activities in lupus patients, we examined a clinical cohort of 164 lupus patients (Supplementary Table 1). Evaluation of levels of IFN-I and CXCL10 in patient serum revealed that between 15 and 20% of the patients exhibited elevated levels of these cytokines (Fig. 6a, Supplementary Fig. 6). In this cohort, IFN-I serum levels were higher in the patient group with high SLEDAI disease score (Supplementary Fig. 6). Moreover, when comparing the patients showing elevated serum levels of both IFN-I and CXCL10 with those low in both cytokines, we observed that high cytokine levels were associated with high SLEDAI scores and high levels of anti-dsDNA (Supplementary Fig. 6).
Fig. 6.
ISD017 inhibits ISG responses in lupus patient cells. (a) Serum from 164 lupus patients and 21 healthy controls was analyzed for levels of IFN-I bioactivity. The data are presented as levels from individual donors. (b) Illustration of experimental set-up for the analysis of the effect of ISD017 on cytokine expression by PBMCs from lupus patients. (c) PBMCs from eleven IFN-Ihigh lupus patients were cultured for 16 h in the presence or absence of ISD017 (200 µg/ml). Supernatants were isolated and analyzed for the levels of CXCL10. (d) The same supernatants as in panel c were analyzed for CCL19. (e) Illustration of experimental set-up for analysis of the effect of ISD017 on Extracellular vesicle (EV)-induced cytokine expression in PBMCs from healthy donors. (f) PBMCs from a healthy donor were treated for 16 h with EVs isolated from seven IFN-Ihigh lupus patients in the presence or absence of ISD017 (200 µg/ml). Supernatants were isolated and analyzed for the levels of CXCL10. The data in panel a are presented as mean levels from individual donors. The data in panel c, d, and f are presented as matched values from cultures from individual patient PBMCs in the absence (blue) or presence (red) of ISD017. [Statistical analysis of the data in c, d, f i were performed using two-tailed one-way ANOVA test].
To test the effect of ISD017 treatment, we isolated and cultured PBMCs from eleven IFN-high lupus patients in the presence or absence of ISD017 (Fig. 6b) and evaluated the expression of IFN-I, CXCL10, and also the chemokine CCL19, elevated in lupus patients [64]. IFN-I could be detected in supernatants from two patient PBMCs cultures (Supplementary Fig. 6), while CXCL10 and CCL19 could be detected in all patient cell cultures (Fig. 6c-d). Importantly, the detectable levels of IFN-I and CXCL10 were reduced by ISD017 in all patient cell cultures, and CCL19 was reduced in 9 out of 11 patients (Fig. 6c-d, Supplementary Fig. 6). Gene expression analysis revealed that mRNA levels of the IFN-stimuated gene ISG15 were also reduced by ISD017 treatment (Supplementary Fig. 6). To corroborate these findings, we isolated extracellular vesicles (EV)s from serum from seven IFN-high lupus patients (Fig. 6e), since it has been reported that membrane vesicles from SLE sera has high ISG-inducing activity through the cGAS-STING pathway [65]. The EVs induced STING-dependent IFN-I expression in THP-1 macrophages (Supplementary Fig. 6). PBMCs from a healthy donor were treated with the isolated EVs in the presence or absence of ISD017, and levels of IFN-I, CXCL10, and CCL19 were measured. The EVs from all donors induced potent expression of CXCL10, while only three of the seven donors induced clear expression of IFN-I (Fig. 6f, Supplementary Fig. 6). The EVs did not induce expression of CCL19 (data not shown). All of the induced IFN-I and CXCL10 responses were inhibited by ISD017 (Fig. 6f, Supplementary Fig. 6). These data collectively suggest that ISD017 can inhibit pathological cytokine responses in cells from lupus patients with elevated IFN-I expression.
4. Discussion
Innate immune responses are important for rapid defense against infections but can also contribute to inflammatory diseases [12], [13], [14]. The cGAS-STING pathway is activated in response to cytosolic DNA, and there is now increasing evidence linking excess activation of STING to pathological inflammation. Therefore, there is a strong interest in understanding this pathway in detail and developing means to inhibit the activity of the pathway pharmacologically. Lupus is a chronic inflammatory disease, with patient groups being very heterogeneous, likely reflecting a broad panel of mechanisms as cause of the pathology. One set of patients have elevated IFN-I responses [35], and there is evidence for cGAS activity correlating with disease severity [40]. Here we report a peptide, ISD017, which effectively blocks STING signaling by specifically inhibiting STING trafficking out of the ER through a mechanism dependent on the STING ER retention factor STIM1. ISD017 displayed STING antagonistic activity in vivo and alleviated lupus symptoms and pathology in a lupus mouse model. Finally, ISD017 inhibited the expression of chemokines associated with disease in PBMCs from lupus patients.
ISD017 blocked IFNβ promoter activity induced by cGAS-STING expression, but not by TRIF or MAVS expression. In addition, the peptide inhibited IFN-I induction by dsDNA but not by poly(I:C) in PBMCs. This suggests that ISD017 specifically targets the cGAS-STING pathway. When examining for proteins associating with ISD017, we identified STING and STIM1. Interestingly, the interaction between ISD017 and STING was dependent on STIM1, and depletion of STIM1 expression abrogated the inhibitory activity of ISD017. STIM1 is reported to be a STING ER retention factor [27], and we found that treatment with ISD017 impaired the release of STING from STIM1. These data strongly suggest that ISD017 targets STIM1, or a STIM1-containing complex, to prevent dissociation of STING from STIM1, and in this way, blocks trafficking of STING from ER to Golgi. Several inhibitors of cGAS and STING have been reported at the present stage [41], [42], [43], [44], [45], [46]. These include H-151 and RU.521. H-151 blocks STING palmitoylation through irreversible covalent binding to cysteine residue at position 91, thus preventing STING oligomerization [42]. This occurs after trafficking of STING from the ER [66], and therefore inhibits activation of IRF3 and NF-κB. The effect of H-151 on STING-induced autophagy and apoptosis is not well characterized. RU.521 binds to the active site of cGAS, thus preventing the synthesis of cGAMP in response to the sensing of DNA [45]. RU.521 and other cGAS inhibitors block all STING dependent functions activated by the DNA-cGAS-cGAMP axis. Still, they will not affect STING activation by non-canonical pathways, such as ER stress and extracellular vesicles [48,67]. Thus, ISD017 is a unique STING trafficking inhibitor blocking all downstream activities irrespective of upstream stimuli.
Inflammatory diseases are characterized by ongoing inflammatory reactions, yet continuous medication is not possible. Therefore, the duration of the inhibitory activity of an inhibitory compound is important. In vitro experiments showed that the inhibitory activity of ISD017 was retained even when added to cells several hours before STING activation. Likewise, in vivo administration of ISD017 twelve hours before treatment with cGAMP led to potent inhibition of STING-mediated IFNβ production. Peptides are generally relatively unstable in free form in the extracellular environment in the body, partly due to the presence of protease and clearance by the kidney [68]. However, since ISD017 enters into cells where it exerts its activity, this may prolong half-life in the body. Also, the noticeable absence of toxic effects of ISD017 treatment in vitro and in vivo suggests that no significant secondary inflammation is induced by cellular debris from dead cells receiving ISD017. These observations may explain why treatment three times per week was sufficient to obtain a significant clinical effect in the Fcgr2b−/− lupus-like model.
In recent years it has emerged that lupus patients can be sub-categorized into different groups, with varying degrees of erythropoiesis, IFN response, neutrophils, myeloid responses, plasmablasts, and lymphoid cells [35]. The subgroup of patients with elevated IFN responses generally have severe disease, and a recent clinical trial demonstrated a beneficial effect of anti-IFNAR treatment [15]. Importantly, cGAS activity is elevated in lupus patient cells, and this correlates with disease severity [40]. Since the cGAS-STING pathway induces not only IFN production but also other inflammatory activities associated with lupus, such as Th17 responses [69], the role of the pathway in lupus may not be limited to the IFNhigh patient group. In regard to ISD017 treatment of lupus, it is also worth mentioning that STIM1-dependent calcium signaling in T cells has been reported to be a driver of Th17 pathology, thus opening up for the possibility of ISD017 dampening lupus pathology by two mechanisms. Future work should mechanistically explore the potential link between STING, STIM1 and Th17 immunity. These data suggest that the STING pathway contributes to lupus disease in a subset of patients and that ISD017 can block the associated response.
A large number of animal models have been developed to mimic lupus pathogenesis [70]. While none of them fully reflect all aspects of the disease, most are characterized by elevated levels of anti-dsDNA antibodies and significant contribution of Th17 immune to pathology [63]. The first study on the role of STING in mouse lupus models suggested a regulatory role for STING in MRL.Faslpr mice [71,72]. We recently reported that full disease development in the Fcgr2b−/− model is dependent on STING, thus revealing differences between the models, and providing a system to study the subgroup of lupus patients where STING is important for disease. In the Fcgr2b−/− model, STING mediates the activation of conventional dendritic cell maturation and plasmacytoid dendritic cell differentiation [62]. The deficiency of STING in the Fcgr2b−/− mice improved the survival of the mice and reduced the level of anti-dsDNA and the development of glomerulonephritis. Treatment of Fcgr2b−/− mice with ISD017 reduced anti-dsDNA antibody levels, improved kidney pathology, reduced serum IFNβ, and the levels of double-negative T cells. The latter is significantly associated with lupus pathology [73]. These data suggest that blocking of STING activation by ISD017 inhibits several pathological processes in lupus. Although treatment with ISD017 did not fully reach the same effect as observed upon crossing Sting-deficient and Fcgr2b−/− mice, the effect of ISD017 treatment was sufficient to reduce the pathology of glomerulonephritis. As a weakness of our study, we did not measure proteinuria in treated mice. One limitation of the current study is that we do not know how well data from Fcgr2b−/− mice translate into the human system, and which subgroups of lupus patients that this model reflect best. In addition, we still remain to test whether ISD017 treatment of Fcgr2b−/− mice has a clinical effect if initiated after on-set of symptoms. More research is needed to fully understand the mechanisms underlying the role of STING in lupus pathogenesis.
Altogether, in this work, we report that the peptide ISD017 blocks STING signaling by preventing dissociation from the ER retention factor STIM1, and thus inhibits STING signaling in vitro and in vivo. Moreover, ISD017 inhibits disease development in a lupus mouse model known to be STING dependent and also blocks pathological inflammatory responses in cells from lupus patients with elevated IFN production. Thus, ISD017 holds potential for treatment of lupus and other diseases characterized by excess activation of STING signaling.
Contributors
ThP, FSP, and SRP conceived the idea behind the project. ThP, RB, PP, and SRP designed the experiments. ThP, SK, IA, EM, RM, RN, BCZ, MC, PV, MHSM, SJW, and MBI performed the experiments. AT collected patient material and provided clinical and paraclinical information. CKH, LJØ, FSP, TrP, RB, PP, and SRP supervised experiments. SRP wrote the first draft of the manuscript, and ThP, RB, PP, and SRP finalized the manuscript. All authors contributed to revision of the work. All authors read and approved the final version of the manuscript. The underlying data used in this work have been verified by RB, PP, and SRP.
Declaration of Competing Interest
TP, CKH, LØ, and SRP own shares in ISD Immunotech, which owns the patent for ISD017 (WO2014166502A2). The patent was the patent was issued by CKH. None of the other authors have conflicts to declare.
Acknowledgments
Acknowledgments
This work was funded by NNF preseed grant (Ref number: aezh/ctoh); European Research Council (ERC-AdG ENVISION; 786602); The Novo Nordisk Foundation (NNF18OC0030274), the Lundbeck Foundation (R198-2015-171; R268-2016-3927), Chulalongkorn University GB-CU-61-25-30-15; The Ph.D. scholarship to SJW was funded by the European Union under the Horizon 2020 Research and Innovation Program, and MSCA-Innovative Training Networks Programme MSCA-ITN (EDGE, 675278); The postdoc grant for AT was funded by the Lundbeck Foundation (R264-2017-3344). RB received funding from Deutsche Forschungsgemeinschaft - Project-ID 369799452 - TRR237 and BE 5877/2-1. The Ph.D. scholarship to IA was funded by Chulalongkorn University Graduate Scholarship to Commemorate the 72nd Anniversary of His Majesty King Bhumibol Adulyadej. The Ph.D. scholarship to PV was funded by International Network for Lupus Research, International Research Network, Thailand (IRN59W0004).
Data sharing
All relevant data have been presented in the manuscript. Requests for or questions about the data can be addressed to srp@biomed.au.dk
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2021.103314.
Appendix. Supplementary materials
References
- 1.Zhang S.Y., Jouanguy E., Ugolini S., Smahi A., Elain G., Romero P. TLR3 deficiency in patients with herpes simplex encephalitis. Science. 2007;317(5844):1522–1527. doi: 10.1126/science.1139522. [DOI] [PubMed] [Google Scholar]
- 2.Ruiz-Moreno J.S., Hamann L., Shah J.A., Verbon A., Mockenhaupt F.P., Puzianowska-Kuznicka M. The common HAQ STING variant impairs cGAS-dependent antibacterial responses and is associated with susceptibility to Legionnaires' disease in humans. PLoS Pathog. 2018;14(1) doi: 10.1371/journal.ppat.1006829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441(7089):101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 4.Ishikawa H., Ma Z., Barber G.N. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461(7265):788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li X.D., Wu J., Gao D., Wang H., Sun L., Chen Z.J. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013;341(6152):1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reinert L.S., Lopusna K., Winther H., Sun C., Thomsen M.K., Nandakumar R. Sensing of HSV-1 by the cGAS-STING pathway in microglia orchestrates antiviral defense in the CNS. Nat Commun. 2016;7:13348. doi: 10.1038/ncomms13348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng L., Liang H., Xu M., Yang X., Burnette B., Arina A. STING-dependent cytosolic DNA sensing promotes radiation-induced type I interferon-dependent antitumor immunity in immunogenic tumors. Immunity. 2014;41(5):843–852. doi: 10.1016/j.immuni.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woo S.R., Fuertes M.B., Corrales L., Spranger S., Furdyna M.J., Leung M.Y. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity. 2014;41(5):830–842. doi: 10.1016/j.immuni.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomsen M.K., Skouboe M.K., Boularan C., Vernejoul F., Lioux T., Leknes S.L. The cGAS-STING pathway is a therapeutic target in a preclinical model of hepatocellular carcinoma. Oncogene. 2020;39(8):1652–1664. doi: 10.1038/s41388-019-1108-8. [DOI] [PubMed] [Google Scholar]
- 10.Schlee M., Hartmann G. Discriminating self from non-self in nucleic acid sensing. Nat Rev Immunol. 2016;16(9):566–580. doi: 10.1038/nri.2016.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paludan S.R., Bowie A.G. Immune sensing of DNA. Immunity. 2013;38:870–880. doi: 10.1016/j.immuni.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crow Y.J., Hayward B.E., Parmar R., Robins P., Leitch A., Ali M. Mutations in the gene encoding the 3 '-5 ' DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nat Genet. 2006;38(8):917–920. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y., Jesus A.A., Marrero B., Yang D., Ramsey S.E., Montealegre Sanchez G.A. Activated STING in a vascular and pulmonary syndrome. N Engl J Med. 2014;371(6):507–518. doi: 10.1056/NEJMoa1312625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu S., Wang H., Jin Y., Podolsky R., Reddy M.V., Pedersen J. IFIH1 polymorphisms are significantly associated with type 1 diabetes and IFIH1 gene expression in peripheral blood mononuclear cells. Hum Mol Genet. 2009;18(2):358–365. doi: 10.1093/hmg/ddn342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morand E.F., Furie R., Tanaka Y., Bruce I.N., Askanase A.D., Richez C. Trial of anifrolumab in active systemic lupus erythematosus. N Engl J Med. 2020;382(3):211–221. doi: 10.1056/NEJMoa1912196. [DOI] [PubMed] [Google Scholar]
- 16.Kothur K., Bandodkar S., Chu S., Wienholt L., Johnson A., Barclay P. An open-label trial of JAK 1/2 blockade in progressive IFIH1-associated neuroinflammation. Neurology. 2018;90(6):289–291. doi: 10.1212/WNL.0000000000004921. [DOI] [PubMed] [Google Scholar]
- 17.Hemmi H., Kaisho T., Takeuchi O., Sato S., Sanjo H., Hoshino K. Small anti-viral compounds activate immune cells via the TLR7 MyD88- dependent signaling pathway. Nat Immunol. 2002;3(2):196–200. doi: 10.1038/ni758. [DOI] [PubMed] [Google Scholar]
- 18.Hemmi H., Takeuchi O., Kawai T., Kaisho T., Sato S., Sanjo H. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408(6813):740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 19.Alexopoulou L., Holt A.C., Medzhitov R., Flavell R.A. Recognition of double-stranded RNA and activation of NF-kappaB by Toll- like receptor 3. Nature. 2001;413(6857):732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 20.Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5(7):730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 21.Sun Q., Sun L., Liu H.H., Chen X., Seth R.B., Forman J. The specific and essential role of MAVS in antiviral innate immune responses. Immunity. 2006;24(5):633–642. doi: 10.1016/j.immuni.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 22.Gao D., Wu J., Wu Y.T., Du F., Aroh C., Yan N. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science. 2013;341(6148):903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saitoh T., Fujita N., Hayashi T., Takahara K., Satoh T., Lee H. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci U S A. 2009;106:20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dobbs N., Burnaevskiy N., Chen D., Gonugunta V.K., Alto N.M., Yan N. STING activation by translocation from the ER is associated with infection and autoinflammatory disease. Cell Host Microbe. 2015;18(2):157–168. doi: 10.1016/j.chom.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu S., Cai X., Wu J., Cong Q., Chen X., Li T. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015 doi: 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- 26.Tanaka Y., Chen Z.J. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal. 2012;5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Srikanth S., Woo J.S., Wu B., El-Sherbiny Y.M., Leung J., Chupradit K. The Ca(2+) sensor STIM1 regulates the type I interferon response by retaining the signaling adaptor STING at the endoplasmic reticulum. Nat Immunol. 2019;20(2):152–162. doi: 10.1038/s41590-018-0287-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang B.C., Reinert L.S., Nandakumar R., Huang J., Laustsen A., Gao J. STEEP mediates STING ER exit and activation of signaling. Nat Immunol. 2020;21:868–879. doi: 10.1038/s41590-020-0730-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gulen M.F., Koch U., Haag S.M., Schuler F., Apetoh L., Villunger A. Signalling strength determines proapoptotic functions of STING. Nat Commun. 2017;8(1):427. doi: 10.1038/s41467-017-00573-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rasmussen S.B., Horan K.A., Holm C.K., Stranks A.J., Mettenleiter T.C., Simon A.K. Activation of autophagy by alpha-herpesviruses in myeloid cells is mediated by cytoplasmic viral DNA through a mechanism dependent on stimulator of IFN genes. J Immunol. 2011;187(10):5268–5276. doi: 10.4049/jimmunol.1100949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gui X., Yang H., Li T., Tan X., Shi P., Li M. Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature. 2019;567(7747):262–266. doi: 10.1038/s41586-019-1006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsokos G.C. Systemic lupus erythematosus. N Engl J Med. 2011;365(22):2110–2121. doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- 33.Gatto M., Zen M., Iaccarino L., Doria A. New therapeutic strategies in systemic lupus erythematosus management. Nat Rev Rheumatol. 2019;15(1):30–48. doi: 10.1038/s41584-018-0133-2. [DOI] [PubMed] [Google Scholar]
- 34.Bennett L., Palucka A.K., Arce E., Cantrell V., Borvak J., Banchereau J. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med. 2003;197(6):711–723. doi: 10.1084/jem.20021553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Banchereau R., Hong S., Cantarel B., Baldwin N., Baisch J., Edens M. Personalized immunomonitoring uncovers molecular networks that stratify lupus patients. Cell. 2016;165(3):551–565. doi: 10.1016/j.cell.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Domeier P.P., Chodisetti S.B., Schell S.L., Kawasawa Y.I., Fasnacht M.J., Soni C. B-cell-intrinsic type 1 interferon signaling is crucial for loss of tolerance and the development of autoreactive B cells. Cell Rep. 2018;24(2):406–418. doi: 10.1016/j.celrep.2018.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swanson C.L., Wilson T.J., Strauch P., Colonna M., Pelanda R., Torres R.M. Type I IFN enhances follicular B cell contribution to the T cell-independent antibody response. J Exp Med. 2010;207(7):1485–1500. doi: 10.1084/jem.20092695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinelli S., Urosevic M., Daryadel A., Oberholzer P.A., Baumann C., Fey M.F. Induction of genes mediating interferon-dependent extracellular trap formation during neutrophil differentiation. J Biol Chem. 2004;279(42):44123–44132. doi: 10.1074/jbc.M405883200. [DOI] [PubMed] [Google Scholar]
- 39.Aga E., Mukherjee A., Rane D., More V., Patil T., van Zandbergen G. Type-1 interferons prolong the lifespan of neutrophils by interfering with members of the apoptotic cascade. Cytokine. 2018;112:21–26. doi: 10.1016/j.cyto.2018.06.027. [DOI] [PubMed] [Google Scholar]
- 40.An J., Durcan L., Karr R.M., Briggs T.A., Rice G.I., Teal T.H. Expression of cyclic GMP-AMP synthase in patients with systemic lupus erythematosus. Arthritis Rheumatol. 2017;69(4):800–807. doi: 10.1002/art.40002. [DOI] [PubMed] [Google Scholar]
- 41.Holm C.K., Rahbek S.H., Gad H.H., Bak R.O., Jakobsen M.R., Jiang Z. Influenza A virus targets a cGAS independent STING pathway, which controls enveloped RNA viruses. Nat Commun. 2016;7:10680. doi: 10.1038/ncomms10680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haag S.M., Gulen M.F., Reymond L., Gibelin A., Abrami L., Decout A. Targeting STING with covalent small-molecule inhibitors. Nature. 2018;559(7713):269–273. doi: 10.1038/s41586-018-0287-8. [DOI] [PubMed] [Google Scholar]
- 43.Hansen A.L., Buchan G.J., Ruhl M., Mukai K., Salvatore S.R., Ogawa E. Nitro-fatty acids are formed in response to virus infection and are potent inhibitors of STING palmitoylation and signaling. Proc Natl Acad Sci U S A. 2018;115(33):E7768–E7E75. doi: 10.1073/pnas.1806239115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li S., Hong Z., Wang Z., Li F., Mei J., Huang L. The cyclopeptide astin C specifically inhibits the innate immune CDN sensor STING. Cell Rep. 2018;25(12):3405–3421. doi: 10.1016/j.celrep.2018.11.097. e7. [DOI] [PubMed] [Google Scholar]
- 45.Vincent J., Adura C., Gao P., Luz A., Lama L., Asano Y. Small molecule inhibition of cGAS reduces interferon expression in primary macrophages from autoimmune mice. Nat Commun. 2017;8(1):750. doi: 10.1038/s41467-017-00833-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siu T., Altman M.D., Baltus G.A., Childers M., Ellis J.M., Gunaydin H. Discovery of a novel cGAMP competitive ligand of the inactive form of STING. ACS Med Chem Lett. 2019;10(1):92–97. doi: 10.1021/acsmedchemlett.8b00466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Unterholzner L., Keating S.E., Baran M., Horan K.A., Jensen S.B., Sharma S. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holm C.K., Jensen S.B., Jakobsen M.R., Cheshenko N., Horan K.A., Moller H.B. Virus-cell fusion as a trigger of innate immunity dependent on the adaptor STING. Nat Immunol. 2012;13:737–743. doi: 10.1038/ni.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Troldborg A., Thiel S., Trendelenburg M., Friebus-Kardash J., Nehring J., Steffensen R. The lectin pathway of complement activation in patients with systemic lupus erythematosus. J Rheumatol. 2018;45(8):1136–1144. doi: 10.3899/jrheum.171033. [DOI] [PubMed] [Google Scholar]
- 50.Lienenklaus S., Cornitescu M., Zietara N., Lyszkiewicz M., Gekara N., Jablonska J. Novel reporter mouse reveals constitutive and inflammatory expression of IFN-beta in vivo. J Immunol. 2009;183(5):3229–3236. doi: 10.4049/jimmunol.0804277. [DOI] [PubMed] [Google Scholar]
- 51.Chan O., Madaio M.P., Shlomchik M.J. The roles of B cells in MRL/lpr murine lupus. Ann N Y Acad Sci. 1997;815:75–87. doi: 10.1111/j.1749-6632.1997.tb52046.x. [DOI] [PubMed] [Google Scholar]
- 52.Luo W.W., Li S., Li C., Lian H., Yang Q., Zhong B. iRhom2 is essential for innate immunity to DNA viruses by mediating trafficking and stability of the adaptor STING. Nat Immunol. 2016;17(9):1057–1066. doi: 10.1038/ni.3510. [DOI] [PubMed] [Google Scholar]
- 53.Wei J., Lian H., Guo W., Chen Y.D., Zhang X.N., Zang R. SNX8 modulates innate immune response to DNA virus by mediating trafficking and activation of MITA. PLoS Pathog. 2018;14(10) doi: 10.1371/journal.ppat.1007336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang L., Wang L., Ketkar H., Ma J., Yang G., Cui S. UBXN3B positively regulates STING-mediated antiviral immune responses. Nat Commun. 2018;9(1):2329. doi: 10.1038/s41467-018-04759-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun M.S., Zhang J., Jiang L.Q., Pan Y.X., Tan J.Y., Yu F. TMED2 potentiates cellular IFN responses to DNA viruses by reinforcing MITA dimerization and facilitating its trafficking. Cell Rep. 2018;25(11):3086–3098. doi: 10.1016/j.celrep.2018.11.048. e3. [DOI] [PubMed] [Google Scholar]
- 56.Prabakaran T., Bodda C., Krapp C., Zhang B., Christensen M.H., Sun C. Attenuation of cGAS-STING signaling is mediated by a p62/SQSTM1-dependent autophagy pathway activated by TBK1. EMBO J. 2018;37:e97858. doi: 10.15252/embj.201797858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gao D., Li T., Li X.D., Chen X., Li Q.Z., Wight-Carter M. Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proc Natl Acad Sci U S A. 2015;112(42):E5699–EE705. doi: 10.1073/pnas.1516465112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rodero M.P., Tesser A., Bartok E., Rice G.I., Della M.E., Depp M. Type I interferon-mediated autoinflammation due to DNase II deficiency. Nat Commun. 2017;8(1):2176. doi: 10.1038/s41467-017-01932-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lepelley A., Martin-Niclos M.J., Le Bihan M., Marsh J.A., Uggenti C., Rice G.I. Mutations in COPA lead to abnormal trafficking of STING to the Golgi and interferon signaling. J Exp Med. 2020;217(11) doi: 10.1084/jem.20200600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Konig N., Fiehn C., Wolf C., Schuster M., Cura C.E., Tungler V. Familial chilblain lupus due to a gain-of-function mutation in STING. Ann Rheum Dis. 2017;76(2):468–472. doi: 10.1136/annrheumdis-2016-209841. [DOI] [PubMed] [Google Scholar]
- 61.Jeremiah N., Neven B., Gentili M., Callebaut I., Maschalidi S., Stolzenberg M.C. Inherited STING-activating mutation underlies a familial inflammatory syndrome with lupus-like manifestations. J Clin Invest. 2014;124(12):5516–5520. doi: 10.1172/JCI79100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thim-uam A., Prabakaran T., Tansakul M., Makjaroen J., Wongkongkathep P., Chantaravisoot N. STING mediates lupus via the activation of conventional dendritic cell maturation and plasmacytoid dendritic cell differentiation. iScience. 2020:23. doi: 10.1016/j.isci.2020.101530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pisitkun P., Ha H.L., Wang H., Claudio E., Tivy C.C., Zhou H. Interleukin-17 cytokines are critical in development of fatal lupus glomerulonephritis. Immunity. 2012;37(6):1104–1115. doi: 10.1016/j.immuni.2012.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fu Q., Chen X., Cui H., Guo Y., Chen J., Shen N. Association of elevated transcript levels of interferon-inducible chemokines with disease activity and organ damage in systemic lupus erythematosus patients. Arthritis Res Ther. 2008;10(5):R112. doi: 10.1186/ar2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kato Y., Park J., Takamatsu H., Konaka H., Aoki W., Aburaya S. Apoptosis-derived membrane vesicles drive the cGAS-STING pathway and enhance type I IFN production in systemic lupus erythematosus. Ann Rheum Dis. 2018;77(10):1507–1515. doi: 10.1136/annrheumdis-2018-212988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mukai K., Konno H., Akiba T., Uemura T., Waguri S., Kobayashi T. Activation of STING requires palmitoylation at the Golgi. Nat Commun. 2016;7:11932. doi: 10.1038/ncomms11932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Petrasek J., Iracheta-Vellve A., Csak T., Satishchandran A., Kodys K., EA Kurt-Jones. STING-IRF3 pathway links endoplasmic reticulum stress with hepatocyte apoptosis in early alcoholic liver disease. Proc Natl Acad Sci U S A. 2013;110(41):16544–16549. doi: 10.1073/pnas.1308331110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu H., Huang J. Optimization of protein and peptide drugs based on the mechanisms of kidney clearance. Protein Pept Lett. 2018;25(6):514–521. doi: 10.2174/0929866525666180530122835. [DOI] [PubMed] [Google Scholar]
- 69.Van Dis E., Sogi K.M., Rae C.S., Sivick K.E., Surh N.H., Leong M.L. STING-activating adjuvants elicit a Th17 immune response and protect against mycobacterium tuberculosis infection. Cell Rep. 2018;23(5):1435–1447. doi: 10.1016/j.celrep.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Richard M.L., Gilkeson G. Mouse models of lupus: what they tell us and what they don't. Lupus Sci Med. 2018;5(1) doi: 10.1136/lupus-2016-000199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sharma S., Campbell A.M., Chan J., Schattgen S.A., Orlowski G.M., Nayar R. Suppression of systemic autoimmunity by the innate immune adaptor STING. Proc Natl Acad Sci U S A. 2015;112(7):E710–E717. doi: 10.1073/pnas.1420217112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dong G., You M., Ding L., Fan H., Liu F., Ren D. STING negatively regulates double-stranded DNA-activated JAK1-STAT1 signaling via SHP-1/2 in B cells. Mol Cells. 2015;38(5):441–451. doi: 10.14348/molcells.2015.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shivakumar S., Tsokos G.C., Datta S.K. T cell receptor alpha/beta expressing double-negative (CD4-/CD8-) and CD4+ T helper cells in humans augment the production of pathogenic anti-DNA autoantibodies associated with lupus nephritis. J Immunol. 1989;143(1):103–112. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.