Abstract
BACKGROUND
Integrative multi-omic approaches have been increasingly applied to discovery and functional studies of complex human diseases. Short-term preoperative antibiotics have been adopted to reduce site infections in colorectal cancer (CRC) resections. We hypothesize that the antibiotics will impact analysis of multi-omic datasets generated from resection samples to investigate biological CRC risk factors.
AIM
To assess the impact of preoperative antibiotics and other variables on integrated microbiome and human transcriptomic data generated from archived CRC resection samples.
METHODS
Genomic DNA (gDNA) and RNA were extracted from prospectively collected 51 pairs of frozen sporadic CRC tumor and adjacent non-tumor mucosal samples from 50 CRC patients archived at a single medical center from 2010-2020. The 16S rRNA gene sequencing (V3V4 region, paired end, 300 bp) and confirmatory quantitative polymerase chain reaction (qPCR) assays were conducted on gDNA. RNA sequencing (IPE, 125 bp) was performed on parallel tumor and non-tumor RNA samples with RNA Integrity Numbers scores ≥ 6.
RESULTS
PERMANOVA detected significant effects of tumor vs nontumor histology (P = 0.002) and antibiotics (P = 0.001) on microbial β-diversity, but CRC tumor location (left vs right), diabetes mellitus vs not diabetic and Black/African Ancestry (AA) vs not Black/AA, did not reach significance. Linear mixed models detected significant tumor vs nontumor histology*antibiotics interaction terms for 14 genus level taxa. QPCR confirmed increased Fusobacterium abundance in tumor vs nontumor groups, and detected significantly reduced bacterial load in the (+)antibiotics group. Principal coordinate analysis of the transcriptomic data showed a clear separation between tumor and nontumor samples. Differentially expressed genes obtained from separate analyses of tumor and nontumor samples, are presented for the antibiotics, CRC location, diabetes and Black/AA race groups.
CONCLUSION
Recent adoption of additional preoperative antibiotics as standard of care, has a measurable impact on -omics analysis of resected specimens. This study still confirmed increased Fusobacterium nucleatum in tumor.
Keywords: Colorectal cancer, Antibiotics, African Continental Ancestry Group, Diabetes mellitus, 16S rRNA gene, RNA-sequencing
Core Tip: This pilot study explored the effect of five variables [tumor histology, preoperative antibiotics, laterality of colorectal cancer (CRC) location, diabetes mellitus, Black/African Ancestry (AA) race] on analysis of microbiome and host transcriptome among archived frozen CRC resection samples. The introduction of short-term preoperative antibiotics as standard of care has a measurable effect on the analysis. Despite the small sample size and variable exposure to preoperative antibiotics, it was still possible to use the data for discovery studies. Fusobacterium abundance was increased in tumor vs nontumor regions. Expression of VBP1 was decreased in expression in both Black/AA tumor and nontumor samples.
INTRODUCTION
Imbalances in mucosal associated microbiota (dysbiosis) have been reported in human colorectal cancer (CRC)[1]. The increased risk of CRC in patients with inflammatory bowel diseases and in animal models of intestinal inflammation support the concept that bacteria promoting colonic inflammation may be proto-oncogenic[2]. According to the driver and passenger hypothesis, there are key pathogens that can drive tumorigenesis and support other bacteria as passengers to proliferate and exacerbate disease in sporadic CRC[3]. Similarly, the keystone hypothesis states that certain low abundance pathogens can promote inflammation by altering a normal microbiota into a dysbiotic one[4]. There are certain pathogens that are associated with CRC including increased abundances of Peptostreptococcus, Bacteroides fragilis, Fusobacterium nucleatum (F. nucleatum), and Escherichia coli and decreased abundances of Clostridium, Bifidobacterium, Faecalibacterium and Roseburia[1-6]. The potential mechanisms at which these particular bacteria may affect the adenoma-carcinoma sequence, include gene expression alternations, promotion of chronic inflammation and release of carcinogenic metabolites[1-6]. The approach of collecting and integrating multi-omic host and microbiome data has been increasingly applied to discovery and functional studies of human gastrointestinal diseases[7,8]. In an ongoing multi-omic analysis of archived frozen mucosal samples of tumor and adjacent nontumor regions of CRC surgical resection specimens (2010-2020), we discovered that a major change was made in the preoperative antibiotic protocol as of January 2017. Prior to that time, the standard of care was to administer only intravenous antibiotics within 30 min of incision, and only a few CRC resection patients were placed on short-term oral antibiotics within a month of the surgery, for various clinical indications. However, after this time, the standard operating protocol for preoperative antibiotics was to prescribe outpatient oral neomycin and metronidazole 24 h in advance of the procedure, in order to reduce surgical site infections[9-11]. The use of antibiotics can shift the microbiome depending on the dosage and duration of the antibiotic exposure. Several studies have shown that tumorigenesis and tumor growth can be attenuated with different antibiotic cocktails and timing of antibiotic exposure with duration of inflammation. On the other hand, early exposure to antibiotics increased risk of CRC and interfered with chemotherapy efficacies due to microbial dysbiosis[5]. With these conflicting findings and this change in protocol at our institution, it allowed us to examine how differential use of antibiotics, along with other clinical/demographic factors influences integrative, multi-omic analyses of CRC.
The five following variables were included in the analysis of the microbiome and host transcriptome datasets generated in this pilot study: (1) Tumor histology (tumor vs nontumor); (2) Preoperative antibiotics (yes/no); (3) Laterality of CRC location (left vs right); (4) Diabetes mellitus (yes/no); and (5) Black/African Ancestry (AA) race (yes/no). The rationale for comparing tumor vs nontumor histology is based on multiple previous reports of differences between tumor and nontumor regions of CRC resection specimens[1,12,13]. Since antibiotics clearly have an effect on the human microbiome[14], it was crucial to assess the impact of the change in antibiotic protocol on the metagenomic and transcriptomic data, before making the commitment to expand the pilot study by analyzing more CRC resection samples. Because molecular differences have been previously reported right sided and left sided CRCs[15], laterality of CRC location was included in the analysis.
The original targets of this pilot discovery and functional study were two biological CRC risk factors: (1) Diabetes mellitus; and (2) Black/AA race. Diabetes mellitus has been associated with increased CRC risk and poorer survival[16,17], and has been implicated in CRC pathogenesis[18]. Several studies have described alterations in the fecal microbiome of patients with diabetes mellitus compared to those without diabetes mellitus in the absence of a CRC diagnosis[19-21]. Transcriptomic profiling of CRC resection tumor and adjacent nontumor samples suggested evidence of diabetic “field cancerization” of the nontumor region[22].
CRC incidence and mortality has been persistently higher in Black/AA compared to all other races in the United States[23,24]. Paredes et al[25] recently reported that transcriptomic profiling of Black/AA compared to White/European Ancestry (EA) revealed reduced immune related gene expression in tumor samples and plasma cytokine levels collected from Black/AA CRC patients. We report here the results of this pilot study to investigate the effects of the tumor histology, preoperative antibiotics, laterality of CRC location, diabetes mellitus status and Black/AA race on microbiome and human transcriptomic data of CRC patients.
MATERIALS AND METHODS
Patient samples
Sequencing analysis of 51 pairs of tumor and nontumor frozen mucosal samples collected from 50 treatment-naïve sporadic CRC subjects (excluding samples collected after neoadjuvant therapy), archived in the Stony Brook Cancer Center Tissue Analytics Shared Resource, was approved by the Stony Brook Institutional Review Board (IRB, IRB2019-00682) and by the Cold Spring Harbor Laboratory IRB (IRB 1467912). All the archived samples were collected prospectively from CRC patients scheduled between 2010-2020 for surgical resection for clinical care, who had given their informed consent for banking of surgical remnant for possible genomic sequencing, collection of longitudinal clinical metadata and access to clinical formalin-fixed paraffin embedded (FFPE) tissues for possible genomic sequencing under a protocol approved by the Stony Brook University IRB (No. 163184). CRC samples collected from individuals diagnosed with hereditary CRC syndromes or inflammatory bowel diseases and individuals who received neoadjuvant treatment, were excluded from this analysis. The clinical metadata included age at the time of tissue collection, self-identified race and ethnicity, family history of a first degree relative with CRC, tumor location (right, cecum-transverse colon; left, splenic flexure-rectum), pathologic cancer stage, smoking status (current, past, never), body mass index, diabetes mellitus status, preoperative antibiotics within a month of surgery (yes/no), cancer stage (0, 1, 2, 3, 4). Participants who enrolled in the study after 2017 were automatically assumed to be exposed to preoperative antibiotics as the new protocol prescribes oral neomycin (1 g × 3 doses) and metronidazole (500 mg × 3 doses) to the patient 24 h prior to surgery. DNA and RNA were extracted from the archived frozen tissues using the Qiagen Allprep DNA/RNA/miRNA kits (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. DNA and RNA were extracted from 5 µm sections of FFPE tumor and non-tumor samples using the Qiagen Allprep DNA/RNA FFPE (Qiagen, Hilden, Germany) kits according to the manufacturer’ protocol except that xylene was used to de-paraffinize the FFPE samples.
16S rRNA gene sequencing
The 16S rRNA libraries were constructed by generating broad-range polymerase chain reaction (PCR) amplicons using barcoded primers that target the V3V4 variable region of the 16S rRNA gene: Primers 338F (5’-ACTCCTACGGGAGGCAGCAG-3’) and 806R (5’-GGACTACHVGGGTWTCTAAT-3’), as previously described from 51 paired tumor and nontumor DNA samples[22]. Illumina paired-end sequencing was performed on the Miseq platform with versions v2.4 of the Miseq Control Software and of MiSeq Reporter, using a 600-cycle version 3 reagent kit. Illumina Miseq paired-end reads were aligned to human reference genome hg19 with bowtie2 and matching sequences were discarded as previously described[25]. All de-multiplexed, paired-end 16S rRNA gene sequence files along with associated metadata were deposited into the Gene Expression Omnibus under project accession number GSE165255. The paired end reads were assembled, aligned and classified using the SINA/SILVA classifier as previously described[26-28]. Operational taxonomic units (OTUs) were produced by binning sequences with identical taxonomic assignments.
Taqman quantitative PCR analysis of mucosal genomic DNA
Real time Taqman PCR analyses were conducted using established specific primers and probe for the F. nucleatum nusG gene[29,30] and broad range primers and probe bacterial 16S rRNA gene[31] shown in Table 1. The Taqman primers and probe for the human solute carrier family 21 or SLCO2A1 also termed prostaglandin cotransporter or PGT gene[29], a single copy autosomal gene located on human chromosome 3, were from Taqman assay ID Hs00194554_m1 (Thermo Fisher Scientific, Waltham, MA, United States). The probes and primers were custom ordered as two separate Taqman assay kits (Thermo Fisher Scientific, Waltham, MA, United States). The first kit combined the F. nucleatum NUS gene assay (FAM probe) with the total bacterial 16S rRNA gene assay (VIC probe). The second kit combined the F. nucleatum nus gene (FAM probe) with the human SLCO2A1 gene assay (VIC probe). The PCR assays were carried out in triplicate with 80 hg of sample DNA in 20 µL in each well. The thermal conditions were 10 min at 95 °C and 45 cycles of 15 s at 95 °C and 1 min at 60 °C. F. nucleatum subsp. nucleatum Knorr ATCC 25586 genomic DNA (gDNA) purchased from ATCC (Manassas, VA, United States) and a gene block was synthesized using the predicted PCR amplicon from the F. nucleatum subsp. animalis 4_8 genome reference sequence for the Human Microbiome Project (Assembly: GCA_000400875, Integrated DNA Technologies, Coralville, IA, United States) were used as positive controls. The 80 hg of DNA template was used in each 20 µL reaction and with either F. nucleatum (FAM probe)/SLCO2A1 (VIC probe) or the F. nucleatum (FAM probe) broad-range 16S rRNA gene (VIC probe) in triplicate.
Table 1.
Specific Taqman polymerase chain reaction primer and probe sequences for the Fusobacterium nucleatum nusG gene and broad range Taqman primer and probe sequences for the bacterial 16S rRNA gene
| Target gene | Sequence |
| F. nucleatum nusG gene forward primer[29] | 5’-CAACCATTACTTTAACTCTACCATGTTCA-3’ |
| F. nucleatum nusG gene reverse primer[29] | 5’-GTTGACTTTACAGAAGGAGATTATGTAAAAATC-3’ |
| F. nucleatum nusG gene probe[30] | 5’-TCAGCAACTTGTCCTTCTTGATCTTTAAATGAACC-3’ |
| Broad range 16S rRNA gene forward primer[31] | 5’-TCCTACGGGAGGCAGCAGT-3’ |
| Broad range 16S rRNA gene reverse primer[31] | 5’- GGACTACCAGGGTATCTAATCCTGTT-3’ |
| Broad range 16S rRNA gene probe[31] | 5’-CGTATTACCGCGGCTGCTGGCAC-3’ |
F. nucleatum: Fusobacterium nucleatum.
RNA sequence analysis
RNA-sequencing was conducted on a subset of tumor and nontumor RNA samples with RIN scores ≥ 6 at the CSHL Cancer Center Sequencing Technologies and Analysis Shared Resource and unpaired tumor RNA samples at the New York Genome Center. At both sequencing centers, the RNA sequence libraries were prepared with the Kapa RNA kit with RiboErase (Roche Sequencing and Life Science Kapa Biosystems, Wilmington, MA, United States) according to the manufacturer’s protocol and Illumina 125 bp PE sequencing was conducted on HiSeq2500 instruments with a targeted depth of 20 million. The sequences were aligned using Salmon in Patro et al[32]. The sequences were deposited in NCBI's Gene Expression Omnibus database with accession number GSE165255.
Statistical analysis
Statistical analysis was performed utilizing the Biostatistics and Bioinformatics Shared Resource at the Stony Brook University Cancer Center. Patient demographics were compared between the subset of samples analyzed and the total set of samples archived in the Stony Brook Cancer Center Tissue Analytics Shared Resource (2010-2020) using either Wilcoxon rank sum test for continuous variables, and chi-square test using GraphPad Prism. Alpha diversity indices [e.g., Sobs, Shannon complexity (H), Shannon Evenness (H/Hmax)] were calculated on samples yielding a total OTU count inferred through 1000 replicate resamplings using Explicet[33]. Linear mixed models were used to compare alpha-diversity (Sobs, ShannonH, ShannonE) between different groups of histology, antibiotics, location, diabetes and Black/AA race. Log-transformation was applied if the normality assumption was not satisfied. Compound symmetry (CS) covariance structure was utilized to model the correlation among measurements from the same patient. β-diversity was assessed by calculating distance matrices based on the Morisita-Horn dissimilarity index and visualized by principal coordinate analysis (PCoA). A PERMANOVA analysis[34] was performed to assess the statistical significance of differences in β-diversity with the following five variables: (1) Tumor histology (tumor/nontumor); (2) Pre-operative antibiotics (yes/no); (3) Anatomic location (left/right); (4) Diabetes mellitus (yes/no); and (5) Black/AA race (yes/no). These analyses were conducted using the vegan 2.56 package of R 3.6.1, and the default setting for number of permutations (n = 999). To compare relative abundance of each OTU between tumor vs nontumor, generalized linear mixed model (GLMM) or generalized estimating equation (GEE) models were used by using the actual counts of each OTU as the outcomes, which were assumed to follow a negative binomial distribution[26]. CS covariance structure was utilized to model the correlation among measurements from the same patient. The log overall count for each individual in each group was considered as an offset in order to normalize for differences in sequencing depth of coverage. When models had a fitting issue because of excessive 0 counts, all sample’s counts+1 were used as the outcome and log (overall count+1) was used as the offset. The two-way interaction term between tumor histology (tumor/nontumor) and preoperative antibiotics (yes/no) was first added in each model to estimate the difference between tumor vs normal within patients with or without pre-operative antibiotics. For OTUs that had model fitting difficulty and OTUs with non-significant interaction, the two-way interaction term was removed and the aforementioned models were fit again. Univariate linear mixed models were used to estimate the coefficient of correlation and the P values for the log transformed relative abundances of selected OTUs. The P values were based on the t-test for GLMM and the Z-test for GEE. The Bonferroni adjusted P values < 0.05 were considered as statistically significant. These analyses were performed using SAS 9.4 (SAS Institute Inc., Cary, NC, United States).
Differentially expressed genes (DEGs) (log2 fold change ≥ 1, adjusted P value < 0.05 were identified using DESeq2[35] for the five groups: (1) Tumor histology (tumor vs nontumor); (2) Preoperative antibiotics within a month of the surgery (yes/no); (3) Anatomic location of the CRC and adjacent nontumor region (left vs right); (4) Diabetes mellitus status (yes/no); and (5) Black/AA self-declared race (yes/no). For the latter four variables, the tumor and nontumor samples were analyzed separately. The DEG lists were also deposited in NCBI's Gene Expression Omnibus database with accession number GSE165255.
RESULTS
Patient characteristics of samples analyzed
The characteristics of the patients for the subset of samples analyzed compared to that of the total collection of archived sporadic CRC samples (2010-2020) are shown in Table 2. Since the samples in the subset were originally selected to provide Black/AA samples and “matched” White/EA samples for another transcriptomic profiling performed at a separate sequencing facility[25], there was significantly higher representation of Black/AA samples in this pilot study. Left-sided CRCs were also more highly represented in the subset compared to the overall collection. Because most of the subset samples were collected prior to January 2017, (when the protocol adding preoperative oral antibiotic prophylaxis was first introduced), there was a lower representation of samples collected from patients who had preoperative antibiotic treatment compared to the total sample collection. Of the 18 diabetic patients, only one had type 1 diabetes mellitus and the remaining 17 patients were diagnosed with type 2 diabetes mellitus.
Table 2.
Comparison of patient characteristics between the subset of sporadic colorectal cancer samples analyzed with that of the total collection of archived samples in the Stony Brook Cancer Center Tissue Analytics Shared Resource (2010-2020)
| Subset analyzed, n = 51 | Total collection, n = 190 | P value | |
| Age (yr) ± IQR | 62 ± 20 | 65 ± 20 | 0.4329 |
| Male sex (%) | 30 (58.8) | 115 (60.5) | 0.8254 |
| Race (%) | 0.0295 | ||
| White | 38 (74.5) | 163 (85.8) | |
| Black | 11 (21.6) | 14 (7.4) | |
| Asian | 1 (2.0) | 6 (3.2) | |
| Other | 1 (1.9) | 7 (3.7) | |
| Hispanic ethnicity (%) | 6 (11.8) | 22 (11.6) | 0.9707 |
| Family history 1st degree relative (%) | 7 (13.7) | 34 (17.9) | 0.4817 |
| BMI (kg/m2) | 27.8 ± 8 | 28.2 ± 8 | 0.5683 |
| Diabetes mellitus (%) | 18 (35.3) | 55 (28.9) | 0.4853 |
| Smoking (%) | 0.1103 | ||
| Current | 7 (13.7) | 10 (5.3) | |
| Past | 21 (41.2) | 88 (46.3) | |
| Never | 23 (45.1) | 92 (48.4) | |
| Anatomic location (%) | 0.0210 | ||
| Left | 31 (60.8) | 81 (42.6) | |
| Right | 20 (39.2) | 109 (57.4) | |
| Cancer stage (%) | 0.4578 | ||
| Stage 0 | 1 (2.0) | 6 (3.2) | |
| Stage 1 | 9 (17.6) | 27 (14.2) | |
| Stage 2 | 14 (27.4) | 63 (33.2) | |
| Stage 3 | 25 (49.0) | 75 (39.5) | |
| Stage 4 | 2 (3.9) | 19 (15.8) | |
| Preoperative antibiotics (%) | 16 (31.4) | 91 (47.9) | 0.0350 |
IQR: Inter-quartile range; BMI: Body mass index.
Colonic mucosal microbiome analysis
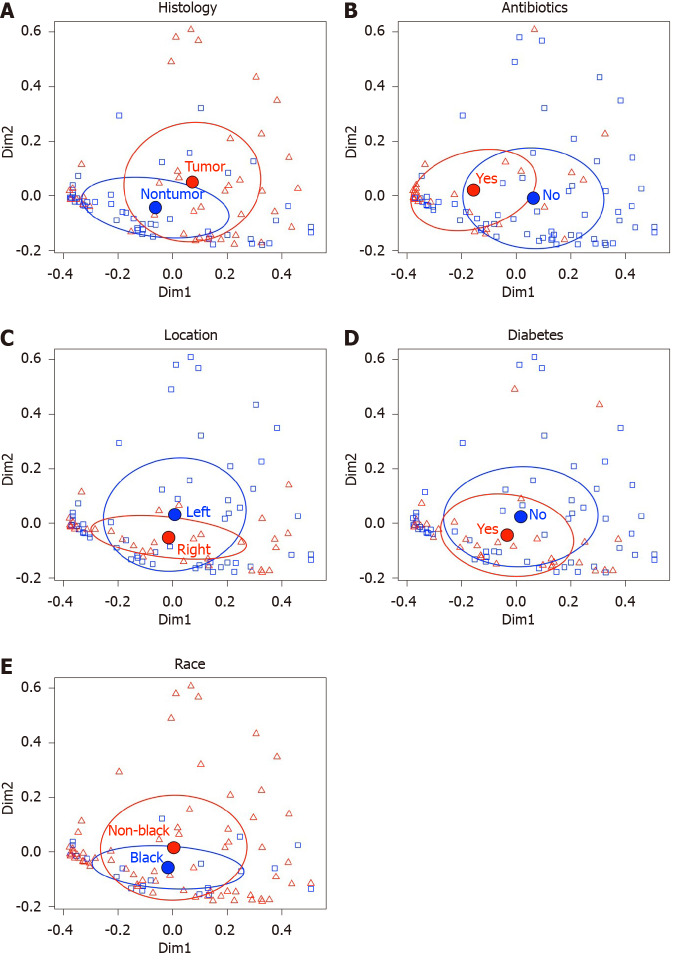
We generated 5706758 16S rRNA gene sequences from 51 pairs of CRC tumor and nontumor samples. After excluding sequence libraries with total counts < 1000, the remaining 89 sequence libraries (41 tumor, 48 nontumor, 26 preoperative antibiotics, 63 no preoperative antibiotics, 55 left, 34 right, 32 diabetic, 57 not diabetic, 21 Black/AA, 68 not Black/AA) had an average sequencing depth of 64752 (minimum 1032; maximum 248838). As shown in Table 3, no significant differences were detected in any of the three alpha diversity indices between the following five groups: (1) Preoperative antibiotics (yes/no); (2) Tumor vs nontumor histology; (3) Anatomic location of sample (left vs right); (4) Diabetes mellitus status (yes/no), and (5) Black/AA race vs not Black/AA race. Exploratory PERMANOVA using the Morasita Horn index for β-diversity was performed with the same five groups (see Table 4). Tumor histology and preoperative antibiotics demonstrated significant F statistics, but anatomic location, diabetes and Black/AA race did not reach significance. The PCoA comparisons using these indices for these five groups are displayed in Figure 1.
Table 3.
Estimated differences in alpha diversity indices between the following groups
| Variable |
Sobs |
ShannonH |
ShannonE |
|||
| Estimate (95%CI) | P value | Estimate (95%CI) | P value | Estimate (95%CI) | P value | |
| Tumor vs nontumor | -1.34 (-4.04, 1.35) | 0.32 | -0.05 (-0.25, 0.15) | 0.61 | -0.004 (-0.04, 0.03) | 0.77 |
| Pre-op antibiotics vs none | 2.93 (-3.51, 9.37) | 0.36 | 0.27 (-0.12, 0.66) | 0.17 | 0.04 (-0.02, 0.09) | 0.20 |
| Left vs right | 2.83 (-2.44, 8.10) | 0.29 | -0.10 (-0.43, 0.23) | 0.53 | -0.03 (-0.08, 0.02) | 0.26 |
| Diabetic vs nondiabetic | 3.73 (-2.30, 9.75) | 0.22 | 0.26 (-0.11, 0.62) | 0.16 | 0.03 (-0.02, 0.09) | 0.22 |
| Black/AA race vs not Black/AA race | 2.70 (-4.43, 9.83) | 0.45 | 0.09 (-0.34, 0.52) | 0.69 | 0.01 (-0.06, 0.07) | 0.80 |
Linear mixed models were used to estimate differences. The P values are based on t-tests from the linear mixed models. CI: Confidence interval; AA: African Ancestry.
Table 4.
PERMANOVA results for the following groups
| Variable | P value |
| Tumor vs nontumor | 0.002 |
| Pre-operative antibiotics vs no preoperative antibiotics | 0.001 |
| Left vs right anatomic location | 0.367 |
| Diabetes mellitus vs not diabetic | 0.061 |
| Black/AA race vs not Black/AA race | 0.258 |
AA: African Ancestry.
Figure 1.
Principal coordinate analysis plots for Morita Horn dissimilarity indices. A: Preoperative antibiotic treatment (yes/no); B: Tumor vs nontumor histology; C: Anatomic location of sample (left vs right); D: Diabetes mellitus status (yes/no); E: Black/African Ancestry race (yes/no). The filled circles and surrounding ovals indicate the mean ± SD centroid for each group.
Because PERMANOVA identified tumor histology and preoperative antibiotics as significant, fitted linear mixed models were used to measure the effect of preoperative antibiotics and tumor histology along with first order preoperative antibiotics*tumor histology interactions on the relative abundances of individual genus-level taxa. For this analysis, we included those OTUs with average relative abundances greater than 0.001% and that were observed in at least 25% of the samples. Because of model convergence issues, 4 OTUs (Firmicutes/Eubacterium, Firmicutes/Acidaminococcus, Proteobacteria/Neisseriaceae, Proteobacteria/Desulfovibrio) were also excluded because of failure of their models to converge. Of the 106 OTUs included in the relative abundance regression analysis, 14 exhibited significant first order tumor histology*preoperative antibiotics interaction terms (see Table 5). The estimated differences in tumor vs nontumor samples were analyzed separately for those 14 OTUs depending on whether the patients were prescribed additional preoperative antibiotics. One of 14 OTUs, Fusobacteria/Leptotrichia, was associated with increased relative abundance in tumor vs nontumor, regardless of preoperative antibiotic exposure (adjusted P < 0.0001 after Bonferroni correction = unadjusted P value*28 with antibiotics, and adjusted P = 0.01 after Bonferroni correction with no antibiotics). Of the 92 OTUs without significant first order preoperative antibiotic*tumor histology interaction terms, 19 OTUs, including Fusobacterium spp. demonstrated significant differences in tumor vs nontumor samples with adjusted P < 0.05 after Bonferroni correction (see Table 6).
Table 5.
Operational taxonomic units (genus level) with significant tumor histology*pre-operative antibiotics interaction terms
|
OTU (genus level)
|
| Actinobacteria/Gardnerella |
| Actinobacteria/Eggerthella |
| Firmicutes/Bacilli/unspecified |
| Firmicutes/Staphylococcus |
| Firmicutes/Parvimonas |
| Firmicutes/Anaerostipes |
| Firmicutes/Howardella |
| Firmicutes/Solobacterium |
| Fusobacteria/Fusobacteriales/unspecified |
| Fusobacteria/Leptotrichia |
| Proteobacteria/Sutterella |
| Proteobacteria/Ralstonia |
| Proteobacteria/Enterobacter |
| Proteobacteria/Haemophilus |
OTU: Operational taxonomic unit.
Table 6.
The operational taxonomic units (genus level) without significant tumor histology*preoperative antibiotics interaction terms, with significant (P < 0.05 after Bonferroni correction) estimated tumor/nontumor ratios of relative abundance and 95% confidence intervals
| OTU | Ratio (95%CI) | P value |
| Increased in tumor | ||
| Firmicutes/Carnobacteriaceae/unspecified | 22.21 (7.57, 65.21) | < 0.0001 |
| Firmicutes/ Peptoniphilus | 2.13 (1.45, 3.13) | 0.0042 |
| Firmicutes/Catenibacterium | 3.89 (1.79, 8.46) | 0.0206 |
| Firmicutes/Dialister | 2.05 (1.48, 2.83) | 0.0005 |
| Fusobacteria/Fusobacterium | 5.30 (2.82, 9.97) | < 0.0001 |
| Proteobacteria/Campylobacter | 7.40 (3.30, 16.62) | < 0.0001 |
| Proteobacteria/Citrobacter | 3.51 (2.12, 5.81) | < 0.0001 |
| Decreased in tumor | ||
| Actinobacteria/Coriobacteriaceae/unspecified | 0.45 (0.32, 0.64) | 0.0003 |
| Bacteroidetes/unspecified | 0.49 (0.32, 0.74) | 0.0240 |
| Bacteroidetes/S24-7 | 0.50 (0.36, 0.70) | 0.0016 |
| Cyanobacteria/Chloroplast | 0.44 (0.31, 0.64) | 0.0021 |
| Firmicutes/Lachnospiraceae/unspecified | 0.76 (0.68, 0.84) | < 0.0001 |
| Firmicutes/Marvinbryantia | 0.39 (0.24, 0.65) | 0.0113 |
| Firmicutes/vadinBB60 | 0.50 (0.36, 0.70) | 0.0052 |
| Firmicutes/Turicibacter | 0.44 (0.30, 0.66) | 0.0074 |
| Firmicutes/Allobaculum | 0.43 (0.29, 0.62) | 0.0002 |
| Proteobacteria/Rickettsiales/mitochondria | 0.32 (0.19, 0.53) | 0.0003 |
| Tenericutes/Anaeroplasma | 0.42 (0.28, 0.62) | 0.0004 |
| Verrucomicrobia/Akkermansia | 0.58 (0.42, 0.80) | 0.0350 |
OTU: Operational taxonomic unit; CI: Confidence interval.
Taqman quantitative PCR confirmation of increased F. nucleatum relative abundance in tumor vs non-tumor samples
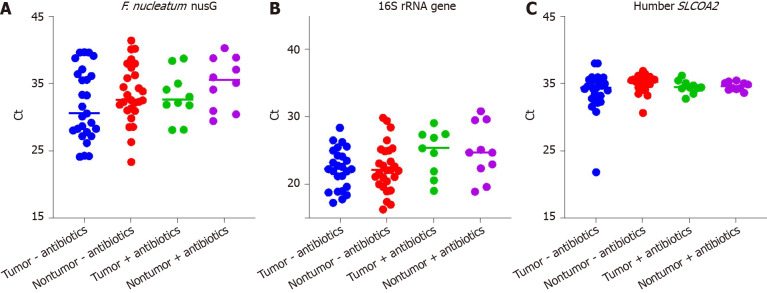
F. nucleatum (both subspecies nucleatum and subspecies animalis) were enumerated in tissues specimens by quantitative PCR (qPCR) of the nusG gene, while total bacterial load was assayed by broad-range 16S rRNA gene qPCR. The human SLCOA2 gene was assayed in parallel in order to normalize results. QPCR assays were performed on 37 paired tumor/nontumor mucosal gDNA samples, (10 pairs, +preoperative antibiotics; 27 pairs, -preoperative antibiotics). DNA samples were depleted in 7 tumor samples and 1 nontumor samples of the 51 paired tumor/nontumor samples and were therefore excluded from the analysis. F. nucleatum nusG was undetectable (undetermined) in the qPCR assays for at least one of the remaining 7 paired tumor and normal samples, and were also excluded from analysis. Both the F. nucleatum subspecies nucleatum and subspecies animalis were detected under these assay conditions. The mean threshold cycles (Ct) for the F. nucleatum nusG, broad-range 16S rRNA and human SLCOA2 assays are shown in Figure 2A-C . Pairwise comparisons of the mean tumor F. nucleatum nusG gene Ct ± SD with nontumor samples (32.2 ± 4.8 vs 34.0 ± 4.2, P = 0.0125), confirmed that there was a higher abundance of F. nucleatum in tumor samples compared with their paired adjacent nontumor samples. The mean 16S rRNA gene Ct was significantly higher in the (+)antibiotics group compared with the (-)antibiotics group (24.8 ± 3.7 vs 22.3 ± 3.4, P = 0.0085, missing 1 tumor (+)antibiotics and 1 tumor (-)antibiotics Ct value). This suggests that the preoperative antibiotics significantly reduced mucosal associated bacteria load. The difference in the mean F. nucleatum Ct between the (+)antibiotics group and the (-)antibiotics group (34.1 ± 3.7 vs 32.7 ± 4.8, P = 0.24) did not reach significance. The difference in the mean human SLCOA2 Ct between the (+)antibiotics group and the (-)antibiotics group also did not reach significance (34.5 ± 0.8 vs 34.4 ± 2.3, P = 0.74, missing 2 nontumor (-)antibiotics Ct values).
Figure 2.
Scatter plots of threshold cycle values. A: Fusobacterium nucleatum nusG gene; B: Broad range 16S rRNA gene; C: Human SLCOA2 gene quantitative polymerase chain reaction assays. F. nucleatum: Fusobacterium nucleatum.
In this current study the detection rates for the detection of F. nucleatum nusG gene were 84% and 93% respectively for the frozen tumor and nontumor samples. Of note, the Taqman probe used in the current study differed from the probe used in the previous FFPE studies, in that the probe in the current study did not overlap the reverse primer. The mean Ct values for each of the three qPCR assays performed on paired FFPE tumor/nontumor samples were compared with the Ct values obtained on the parallel pairs of frozen tumor/nontumor samples collected from the same two CRC patients (see Table 7). The broad range 16S rRNA gene Ct values were significantly higher in the FFPE samples compared to the paired frozen mucosal samples (30.6 ± 1.6 vs 22.7 ± 1.7, P = 0.0034, n = 4), despite using 80 hg of gDNA in all assays. The differences in the mean F. nucleatum nusG Ct values did not reach significance, but one of the FFPE samples bordered on being not detectable (ND). The differences in the mean human SLCOA2 Ct values did not reach significance although one of the FFPE samples was ND.
Table 7.
Comparison of the mean quantitative polymerase chain reaction mean threshold cycles ± SD between genomic DNA extracted from paired frozen and formalin-fixed paraffin embedded samples
|
F. nucleatum nusG |
16S rRNA |
Human SLCO2A1 |
||||
| Frozen | FFPE | Frozen | FFPE | Frozen | FFPE | |
| Tumor A | 25.9 ± 1.2 | 35.4 ± 0.1 | 21.8 ± 0.1 | 31.4 ± 0.1 | 32.6 ± 2.5 | 39.3 ± 0.6 |
| Nontumor A | 37.2 ± 0.6 | 39.9/ND | 22.2 ± 0.1 | 31.7 ± 0.3 | 34.8 ± 0.5 | ND |
| Tumor B | 26.0 ± 1.0 | 27.5 ± 1.0 | 21.6 ± 0.2 | 28.3 ± 0.1 | 33.2 ± 1.0 | 32.5 ± 1.3 |
| Nontumor B | 31.5 ± 1.4 | 35.1 ± 0.7 | 25.2 ± 0.1 | 31.2 ± 0.6 | 34.6 ± 0.9 | 38.2 ± 0.1 |
F. nucleatum: Fusobacterium nucleatum; FFPE: Formalin-fixed paraffin embedded; ND: Not detected.
Effect of tumor histology and preoperative antibiotics on human RNA-sequencing data
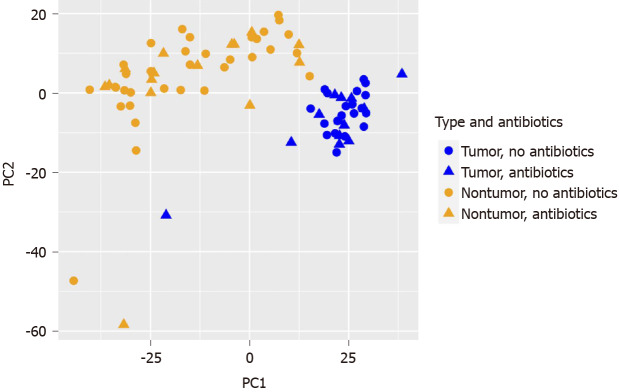
Since significant effects of tumor histology and preoperative antibiotics were noted in the microbiome analysis, we were particularly interested in the potential effect of pre-operative antibiotics on the human transcriptomic profiles generated in parallel. RNA-seq data was obtained from CSHL for 33 paired tumor/nontumor RNA samples and 4 unpaired tumor RNA samples at CSHL. RNA-sequencing was completed on an additional 11 unpaired tumor RNA samples at the NYGC sequencing center. PCoA demonstrated a clear separation between the tumor and nontumor transcriptomic profiles (see Figure 3). Out of a total of 31052 genes, 1235 DEGs were decreased in tumor (n = 48) vs nontumor (n = 33, Log2 fold change ≤ -1, adjusted P value < 0.05) and 1229 DEGs were increased.
Figure 3.
Principal coordinate analysis of human colorectal cancer RNA-seq data with respect to tumor histology (tumor vs nontumor) and preoperative antibiotics (yes/no).
No clear separation was observed on PCoA for the remaining four variables: Pre-operative antibiotics (see Figure 3), left vs right CRC location (not displayed), diabetic vs not diabetic (not displayed), or Black/AA race vs not Black/AA race (not displayed). DESeq2 analyses were conducted on the tumor samples separately from the nontumor samples for these four variables. For the (+)antibiotics (n = 15) vs the (-)antibiotics (n = 33) group of tumor samples, 30 DEGs were decreased and 188 DEGs were increased. For the (+)antibiotics (n = 13) vs (-)antibiotics (n = 20) nontumor samples, no DEGs were decreased and 32 DEGs were increased. Six upregulated DEGs were common with the same polarities in both tumor and nontumor (+)antibiotics/(-)antibiotics DEG lists (see Table 8).
Table 8.
Differentially expressed genes, (+)antibiotics vs (-)antibiotics, common to both tumor and nontumor samples
| Gene (increased) | Symbol | Tumor log2 fold change | Tumor adjusted P value | Nontumor log2 fold change | Nontumor adjusted P value |
| ENSG00000023902 | PLEKHO1 | 1.127 | 0.0461 | 1.694 | 0.0130 |
| ENSG00000253304 | TMEM200B | 1.326 | 0.0445 | 1.719 | 0.0481 |
| ENSG00000187608 | ISG15 | 1.534 | 0.0301 | 2.202 | 0.0102 |
| ENSG00000185745 | IFIT1 | 1.708 | 0.0243 | 2.245 | 0.0176 |
| ENSG00000109705 | NKX3-2 | 2.082 | 0.0476 | 2.859 | 0.0264 |
| ENSG00000145936 | KCNMB1 | 2.011 | 0.0480 | 2.874 | 0.0176 |
For the left (n = 30) vs right (n = 18) CRC location groups of tumor samples, 12 DEGs were decreased and 7 DEGs were increased, and for the left (n = 18) vs right (n = 15) CRC location groups of nontumor samples, 72 DEGs were decreased and 36 DEGs were increased. However, no DEGs were common to both tumor and nontumor left vs right DEG lists. For the diabetic (n = 18) vs nondiabetic (n = 30) groups of tumor samples, 52 DEGs and 9 DEGs were increased, and for the diabetic (n = 12) vs nondiabetic (n = 21) groups of nontumor samples, 2 DEGs were decreased and 6 DEGs were increased. No DEGs were common to both tumor and nontumor diabetic/nondiabetic DEG lists.
In the analysis of the Black/AA (n = 11) vs not Black/AA (n = 37) groups of tumor samples, 5 DEGs were decreased and 11 DEGs were increased in Black/AA samples. For the Black/AA (n = 10) vs not Black/AA (n = 33) groups of nontumor samples, 42 DEGs were decreased and 5 DEGs were increased in Black/AA samples. Three DEGs were common to both tumor and nontumor Black/AA vs not Black/AA groups (see Table 9).
Table 9.
Differentially expressed genes, Black/African Ancestry vs not Black/African Ancestry, common to both tumor and nontumor samples
|
Gene
|
Symbol
|
Tumor log2 fold change
|
Tumor adjusted P value
|
Nontumor log2 fold change
|
Nontumor adjusted P value
|
| Decreased | |||||
| ENSG00000155959 | VBP1 | -8.217 | 5.26 × 10-10 | -8.318 | 5.26 × 10-10 |
| ENSG00000102109 | PCSK1N | -5.965 | 0.0050 | -7.175 | 0.0004 |
| Increased | |||||
| ENSG00000225972 | MTND1P23 | 4.0913 | 5.67 × 10-7 | 3.801 | 0.0001 |
DISCUSSION
The results of this pilot study illustrate the need for thorough curation of the clinical metadata linked to archived gastrointestinal resection specimens for discovery and functional studies on CRC pathophysiology. Even after prospectively interviewing research subjects for antibiotic exposure prior to surgical resection and carefully reviewing the electronic medical record for prescribed medications, it was difficult to assess this potentially important confounding variable. Further discussion with the colorectal surgery service revealed that a major change in pre-operative antibiotics was established as standard of care as of January 2017 to reduce site infections. This change in protocol was made, because there was accumulating evidence that the addition of short term perioperative oral antibiotics reduced intra-abdominal surgical site infections[9-11]. The reduced mucosal associated bacterial abundance based on our 16S rRNA broad range PCR assays, may contribute to the reduced incidence of postoperative intra-abdominal infections. Our results indicate that there is a measurable effect not only on the host mucosal-associated microbial communities but also on the host colonic mucosal transcriptomic profiles. Reviewing the literature, while some studies clearly excluded CRC subjects who had received additional preoperative antibiotics[13], it is not clear in a number of published reports, whether exposure to preoperative antibiotics was taken into consideration in the analysis. Though the effect of the antibiotics is transient, and the baseline microbiome is shown to recover within 1.5 mo with the most potent antibiotics[36], the impact of preoperative antibiotics has a measurable effect on the mucosal transcriptomic profiles generated from the resection specimens.
A major limitation of this pilot study is the small number of subjects who were analyzed and potential selection bias with respect to the five variables included in the analyses. We detected significant differences in microbial β-diversity between tumor vs nontumor histology groups and (+)antibiotics vs (-)antibiotics. However, failure to detect differences with respect to CRC location, diabetes or Black/AA race could be because this study was underpowered to detect a significant effect of these three variables. Despite heterogeneity in pre-operative antibiotic protocols, qPCR analysis confirmed previous reports showing that F. nucleatum abundance was increased in tumor compared to paired nontumor samples[29,37-41]. Lower detection rates of F. nucleatum nusG gene by PCR were previously reported on archived FFPE CRC tissues, ranging from 13%-45%[37,38] while our study reports 84% and 93% detection rates for tumor and non-tumor fresh frozen regions respectively. Furthermore, F. nucleatum has been detected in advanced adenomas but not in early adenomas, which are precursor stages along the adenoma-carcinoma sequence[39,41]. F. nucleatum abundance has been associated with poor prognosis and resistance to chemotherapy[37,41,42]. In xenograft models of human CRC, antibiotic treatment for three weeks reduced Fusobacterium load, cancer cell proliferation and overall tumor growth, however treatment for only 24 h had no significant effect[38]. There has been one report that F. nucleatum abundance is higher in colonic effluents collected from Black/AA male veterans compared to White/EA male veterans undergoing screening colonoscopy[43].
Despite the small sample size and variable exposure to preoperative antibiotics, it was still possible to use the data for discovery studies. Expression of the von Hippel-Landau binding protein 1 or VBP1 in Black/AA CRC tumors (n = 64) was observed to be significantly lower compared to White/EA CRC tumors (n = 284, P = 0.026) in The Cancer Genome Atlas (TCGA) database and in a recently published transcriptomic profiling study[21]. The polarity observed for differences in proprotein convertase subtilisin/kexin type 1 inhibitor or PCSK1N Black/AA vs White/EA CRC tumors in the TCGA database however, was opposite to what was observed in this pilot study. The MTND1P23 gene was not present in the TCGA database. We are particularly intrigued by the results indicating reduced VBP1 expression in Black/AA tumor vs not Black/AA tumor and Black/AA nontumor vs not Black/AA nontumor, because of a recent report that VBP1 suppresses HIF-1α-induced epithelial‐mesenchymal transition in vitro and tumor metastasis in vivo[44].
Given that the new preoperative antibiotic protocol adopted at this medical center is 24 h of oral antibiotics in addition to current IV antibiotics administered within 30 min of incision for colorectal resections, a relevant question is how to further pursue the findings of this initial discovery and functional study. Going forward, probably the best solution is to perform targeted PCR assays on archived FFPE CRC and advanced adenoma tissues collected prior to the change in preoperative antibiotics protocol in order to sufficiently power the analysis of evaluating the effect of laterality, diabetes status and race on microbiome and transcriptomic data of CRC patients. In particular, utilizing the targeted PCR to detect Fusobacterium, it can possibly be used as a clinical prognostic biomarker for diabetic CRC patients. We also plan to shift our prospective collection of colonic tissues away from surgical resections to prospective collection of colonoscopic biopsy samples.
CONCLUSION
The recent addition of preoperative oral antibiotics 24 h to the standard administration of IV antibiotics within 30 min of incision has a measurable effect on colonic mucosal gene expression in addition to its effect on the amount and composition of mucosal associated bacteria in the resected specimen. Despite heterogeneity in the preoperative antibiotics in this study cohort, increased abundance of F. nucleatum, was observed in tumor vs nontumor regions of the resected specimen. This study identified the VBP1 gene, which may suppress CRC metastasis, as having decreased RNA expression in both tumor and nontumor regions of the resected specimen collected from Black/AA subjects.
ARTICLE HIGHLIGHTS
Research background
Imbalances in mucosal associated microbiota (dysbiosis) have been reported in human colorectal cancer (CRC). There are certain pathogens that are associated with CRC including increased abundances of Peptostreptococcus, Bacteroides fragilis, Fusobacterium nucleatum, and Escherichia coli and decreased abundances of Clostridium, Bifidobacterium, Faecalibacterium and Roseburia. The approach of collecting and integrating multi-omic host and microbiome data has been increasingly applied to discovery and functional studies of human gastrointestinal disease.
Research motivation
A major change was made in the preoperative antibiotic protocol at this hospital as of January 2017. Prior to that time, the standard of care was to administer only intravenous antibiotics within 30 min of incision, and only a few CRC resection patients were placed on short-term oral antibiotics within a month of the surgery, for various clinical indications. However, after this time, the standard operating protocol for preoperative antibiotics was to prescribe outpatient oral neomycin and metronidazole 24 h in advance of the procedure, in order to reduce surgical site infections. The use of antibiotics can shift the microbiome depending on the dosage and duration of the antibiotic exposure. Several studies have shown that tumorigenesis and tumor growth can be attenuated with different antibiotic cocktails and timing of antibiotic exposure with duration of inflammation. On the other hand, early exposure to antibiotics increased risk of CRC and interfered with chemotherapy efficacies due to microbial dysbiosis. With these conflicting findings and this change in protocol at our institution, it allowed us to examine how differential use of antibiotics, along with other clinical/demographic factors influences integrative, multi-omic analyses of CRC.
Research objectives
To examine the effect of the five following variables were included in the analysis of the microbiome and host transcriptome datasets generated in this pilot study: (1) Tumor histology (tumor vs nontumor); (2) Preoperative antibiotics (yes/no); (3) Laterality of CRC location (left vs right); (4) Diabetes mellitus (yes/no); and (5) Black/African Ancestry (AA) race (yes/no).
Research methods
Genomic DNA (gDNA) and RNA were extracted from prospectively collected 51 pairs of frozen sporadic CRC tumor and adjacent non-tumor mucosal samples from 50 CRC patients archived at a single medical center from 2010-2020. 16S rRNA gene sequencing (V3V4 region, paired end, 300 bp) and confirmatory quantitative polymerase chain reaction (PCR) assays were conducted on gDNA. RNA sequencing (IPE, 125 bp) was performed on parallel tumor and non-tumor RNA samples with RNA Integrity Numbers scores ≥ 6.
Research results
Exploratory PERMANOVA using the Morasita Horn index for β-diversity was performed with each of the five groups. Tumor vs nontumor histology (P = 0.002) and preoperative antibiotics (P = 0.001) demonstrated significant F statistics, but anatomic location, diabetes and Black/AA race did not reach significance. Differences in α-diversity did not reach significance between the five groups. Fourteen taxa at the genus level exhibited significant tumor*preoperative antibiotic interactions. Of the taxa without tumor*preoperative antibiotic interactions, 7 taxa were significantly increased in tumor vs nontumor, including Fusobacterium, and 11 taxa were significantly decreased. The increase in Fusobacterium nucleatum (F. nucleatum) abundance was confirmed by Taqman PCR assays. Additional preoperative antibiotics significantly reduced mucosa-associated total bacterial abundance, which may contribute to reduction of intra-abdominal surgical site infections. Analysis of a subset of parallel formalin-fixed paraffin embedded (FFPE) samples retained polarity of the observed trends but impaired signal strength. Principal coordinate analysis of the transcriptomic data showed a clear separation from tumor and nontumor samples. Consequently, differentially expressed genes were analyzed separately for the other four variables separately in tumor and nontumor samples. Differentially expressed genes common to the tumor and nontumor groups were identified for additional pre-operative antibiotics and Black/AA race. The VBP1 gene, which may suppress CRC metastasis, exhibits reduced expression in Black/AA subjects compared to not Black/AA subjects.
Research conclusions
The recent addition of preoperative oral antibiotics 24 h to the standard administration of IV antibiotics within 30 min of incision has a measurable effect on colonic mucosal gene expression in addition to its effect on the amount and composition of mucosal associated bacteria in the resected specimen. Despite heterogeneity in the preoperative antibiotics in this study cohort, increased abundance of F. nucleatum, was observed in tumor vs nontumor regions of the resected specimen. This study identified the VBP1 gene, which may suppress CRC metastasis, as having decreased RNA expression in both tumor and nontumor regions of the resected specimen collected from Black/AA subjects.
Research perspectives
Given that the addition of preoperative antibiotics to the standard administration of IV antibiotics within 30 min of incision has a measurable effect on colonic mucosal gene expression, in addition to its effect on the amount and composition of mucosa-associated bacteria in the resected specimen, is being widely adopted as standard of care, we plan to perform targeted PCR assays on archived CRC FFPE tissues to confirm our results collected prior to when the change in protocol was adopted (January 2017). Because patients undergoing colonoscopy are not routinely prescribed antibiotics before the procedure, we also plan to shift prospective collection of colonic neoplastic and normal tissues away from surgical resections to prospective collection of research colonoscopic biopsy samples.
ACKNOWLEDGEMENTS
We want to thank the colorectal cancer (CRC) patients who have donated their remnant surgical specimens and clinical data to the GI Clinical Resource Core and to the surgeons in the Division of Colon and Rectal Surgery, (Drs. Genua J, Lee K, Smithy W and Leiboff AR) for participating in the collection of CRC specimens for the Stony Brook Cancer Center Tissue Analytics Shared Resource. We want to thank Ji P and Dr. Williams J with assistance in extracting nucleic acids from the tissue samples. We want to thank Dr. Goodwin S for coordinating the RNA sequencing at Cold Spring Harbor Laboratory. The authors wish to acknowledge the Stony Brook Cancer Center Biostatistics and Bioinformatics Shared Resource for expert assistance on processing and analyzing the 16S rRNA sequence, the RNA-seq and clinical data. The authors also wish to acknowledge the Cold Spring Harbor Cancer Center Sequencing Technologies and Analysis Shared Resource, and the New York Genome Center for expert assistance.
Footnotes
Institutional review board statement: This study was approved by the Stony Brook University Institutional Review Board (approval No. 163184).
Informed consent statement: All study participants, or their legal guardian, provided informed written consent prior to study enrollment.
Conflict-of-interest statement: All authors declare no conflicts-of-interest related to this article.
STROBE statement: The authors have read the STROBE Statement-checklist of items, and the manuscript was prepared and revised according to the STROBE Statement-checklist of items.
Manuscript source: Unsolicited manuscript
Corresponding Author's Membership in Professional Societies: American Gastroenterology Association, No. 158070.
Peer-review started: January 21, 2021
First decision: February 9, 2021
Article in press: March 24, 2021
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B, B
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Losurdo G, Sassaki LY, Serban ED S-Editor: Gao CC L-Editor: A P-Editor: Liu JH
Contributor Information
Sarah A Malik, Department of Medicine, Renaissance School of Medicine, Stony Brook University, Stony Brook, NY 11794, United States.
Chencan Zhu, Department of Applied Mathematics and Statistics, Stony Brook University, Stony Brook, NY 11794, United States.
Jinyu Li, Stony Brook Cancer Center Biostatistics and Bioinformatics Shared Resource, Stony Brook University, Stony Brook, NY 11794, United States; Department of Pathology, Renaissance School of Medicine, Stony Brook University, Stony Brook, NY 11794, United States.
Joseph F LaComb, Department of Medicine, Renaissance School of Medicine, Stony Brook University, Stony Brook, NY 11794, United States.
Paula I Denoya, Department of Surgery, Renaissance School of Medicine, Stony Brook University, Stony Brook, NY 11794, United States.
Igor Kravets, Department of Medicine, Renaissance School of Medicine, Stony Brook University, Stony Brook, NY 11794, United States.
Joshua D Miller, Department of Medicine, Renaissance School of Medicine, Stony Brook University, Stony Brook, NY 11794, United States.
Jie Yang, Department of Applied Mathematics and Statistics, Stony Brook University, Stony Brook, NY 11794, United States; Stony Brook Cancer Center Biostatistics and Bioinformatics Shared Resource, Stony Brook University, Stony Brook, NY 11794, United States; Department of Family, Population and Preventive Medicine, Stony Brook University, Stony Brook, NY 11794, United States.
Melissa Kramer, Cold Spring Harbor Laboratory Cancer Center Sequencing Technologies and Analysis Shared Resource, Cold Spring Harbor, NY 11724, United States.
W Richard McCombie, Cold Spring Harbor Laboratory Cancer Center Sequencing Technologies and Analysis Shared Resource, Cold Spring Harbor, NY 11724, United States.
Charles E Robertson, Department of Medicine, University of Colorado Anschutz Medical Campus, Aurora, CO 80045, United States.
Daniel N Frank, Department of Medicine, University of Colorado Anschutz Medical Campus, Aurora, CO 80045, United States.
Ellen Li, Department of Medicine, Renaissance School of Medicine, Stony Brook University, Stony Brook, NY 11794, United States. ellen.li@stonybrookmedicine.edu.
Data sharing statement
All de-multiplexed, paired-end 16S rRNA gene sequence files and RNA-sequence data along with associated metadata were deposited into the Gene Expression Omnibus, which is a public repository that archives and freely distributes comprehensive sets of microarray, next-generation sequencing, and other forms of high-throughput functional genomic data submitted by the scientific community data base under project accession number GSE165255. All the tissues analyzed were collected from patients that gave informed consent for sharing results of the analysis of their anonymized tissues on public databases, where the risk of identification is very low. The statistical code used in this study is available from the corresponding author at ellen.li@stonybrookmedicine.edu.
References
- 1.Ternes D, Karta J, Tsenkova M, Wilmes P, Haan S, Letellier E. Microbiome in Colorectal Cancer: How to Get from Meta-omics to Mechanism? Trends Microbiol. 2020;28:401–423. doi: 10.1016/j.tim.2020.01.001. [DOI] [PubMed] [Google Scholar]
- 2.Brennan CA, Garrett WS. Gut Microbiota, Inflammation, and Colorectal Cancer. Annu Rev Microbiol. 2016;70:395–411. doi: 10.1146/annurev-micro-102215-095513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tjalsma H, Boleij A, Marchesi JR, Dutilh BE. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol. 2012;10:575–582. doi: 10.1038/nrmicro2819. [DOI] [PubMed] [Google Scholar]
- 4.Hajishengallis G, Darveau RP, Curtis MA. The keystone-pathogen hypothesis. Nat Rev Microbiol. 2012;10:717–725. doi: 10.1038/nrmicro2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sánchez-Alcoholado L, Ramos-Molina B, Otero A, Laborda-Illanes A, Ordóñez R, Medina JA, Gómez-Millán J, Queipo-Ortuño MI. The Role of the Gut Microbiome in Colorectal Cancer Development and Therapy Response. Cancers (Basel) 2020;12 doi: 10.3390/cancers12061406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vacante M, Ciuni R, Basile F, Biondi A. Gut Microbiota and Colorectal Cancer Development: A Closer Look to the Adenoma-Carcinoma Sequence. Biomedicines. 2020;8 doi: 10.3390/biomedicines8110489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Q, Wang K, Wu W, Giannoulatou E, Ho JWK, Li L. Host and microbiome multi-omics integration: applications and methodologies. Biophys Rev. 2019;11:55–65. doi: 10.1007/s12551-018-0491-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang T, DeSimone RA, Jiao X, Rohlf FJ, Zhu W, Gong QQ, Hunt SR, Dassopoulos T, Newberry RD, Sodergren E, Weinstock G, Robertson CE, Frank DN, Li E. Host genes related to paneth cells and xenobiotic metabolism are associated with shifts in human ileum-associated microbial composition. PLoS One. 2012;7:e30044. doi: 10.1371/journal.pone.0030044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson RL, Gladman E, Barbateskovic M. Antimicrobial prophylaxis for colorectal surgery. Cochrane Database Syst Rev. : 2014: CD001181. doi: 10.1002/14651858.CD001181.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kiran RP, Murray AC, Chiuzan C, Estrada D, Forde K. Combined preoperative mechanical bowel preparation with oral antibiotics significantly reduces surgical site infection, anastomotic leak, and ileus after colorectal surgery. Ann Surg. 2015;262:416–25; discussion 423. doi: 10.1097/SLA.0000000000001416. [DOI] [PubMed] [Google Scholar]
- 11.Espin Basany E, Solís-Peña A, Pellino G, Kreisler E, Fraccalvieri D, Muinelo-Lorenzo M, Maseda-Díaz O, García-González JM, Santamaría-Olabarrieta M, Codina-Cazador A, Biondo S. Preoperative oral antibiotics and surgical-site infections in colon surgery (ORALEV): a multicentre, single-blind, pragmatic, randomised controlled trial. Lancet Gastroenterol Hepatol. 2020;5:729–738. doi: 10.1016/S2468-1253(20)30075-3. [DOI] [PubMed] [Google Scholar]
- 12.Walther A, Johnstone E, Swanton C, Midgley R, Tomlinson I, Kerr D. Genetic prognostic and predictive markers in colorectal cancer. Nat Rev Cancer. 2009;9:489–499. doi: 10.1038/nrc2645. [DOI] [PubMed] [Google Scholar]
- 13.Flemer B, Lynch DB, Brown JM, Jeffery IB, Ryan FJ, Claesson MJ, O'Riordain M, Shanahan F, O'Toole PW. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut. 2017;66:633–643. doi: 10.1136/gutjnl-2015-309595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwartz DJ, Langdon AE, Dantas G. Understanding the impact of antibiotic perturbation on the human microbiome. Genome Med. 2020;12:82. doi: 10.1186/s13073-020-00782-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee MS, Menter DG, Kopetz S. Right Versus Left Colon Cancer Biology: Integrating the Consensus Molecular Subtypes. J Natl Compr Canc Netw. 2017;15:411–419. doi: 10.6004/jnccn.2017.0038. [DOI] [PubMed] [Google Scholar]
- 16.Larsson SC, Orsini N, Wolk A. Diabetes mellitus and risk of colorectal cancer: a meta-analysis. J Natl Cancer Inst. 2005;97:1679–1687. doi: 10.1093/jnci/dji375. [DOI] [PubMed] [Google Scholar]
- 17.El Brahimi S, Smith ML, Pinheiro PS. Role of pre-existing type 2 diabetes in colorectal cancer survival among older Americans: a SEER-Medicare population-based study 2002-2011. Int J Colorectal Dis. 2019;34:1467–1475. doi: 10.1007/s00384-019-03345-8. [DOI] [PubMed] [Google Scholar]
- 18.González N, Prieto I, Del Puerto-Nevado L, Portal-Nuñez S, Ardura JA, Corton M, Fernández-Fernández B, Aguilera O, Gomez-Guerrero C, Mas S, Moreno JA, Ruiz-Ortega M, Sanz AB, Sanchez-Niño MD, Rojo F, Vivanco F, Esbrit P, Ayuso C, Alvarez-Llamas G, Egido J, García-Foncillas J, Ortiz A DiabetesCancerConnect Consortium. 2017 update on the relationship between diabetes and colorectal cancer: epidemiology, potential molecular mechanisms and therapeutic implications. Oncotarget. 2017;8:18456–18485. doi: 10.18632/oncotarget.14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larsen N, Vogensen FK, van den Berg FW, Nielsen DS, Andreasen AS, Pedersen BK, Al-Soud WA, Sørensen SJ, Hansen LH, Jakobsen M. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One. 2010;5:e9085. doi: 10.1371/journal.pone.0009085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karlsson FH, Tremaroli V, Nookaew I, Bergström G, Behre CJ, Fagerberg B, Nielsen J, Bäckhed F. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498:99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- 21.Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, Peng Y, Zhang D, Jie Z, Wu W, Qin Y, Xue W, Li J, Han L, Lu D, Wu P, Dai Y, Sun X, Li Z, Tang A, Zhong S, Li X, Chen W, Xu R, Wang M, Feng Q, Gong M, Yu J, Zhang Y, Zhang M, Hansen T, Sanchez G, Raes J, Falony G, Okuda S, Almeida M, LeChatelier E, Renault P, Pons N, Batto JM, Zhang Z, Chen H, Yang R, Zheng W, Yang H, Wang J, Ehrlich SD, Nielsen R, Pedersen O, Kristiansen K. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 22.Del Puerto-Nevado L, Minguez P, Corton M, Solanes-Casado S, Prieto I, Mas S, Sanz AB, Gonzalez-Alonso P, Villaverde C, Portal-Nuñez S, Aguilera O, Gomez-Guerrero C, Esbrit P, Vivanco F, Gonzalez N, Ayuso C, Ortiz A, Rojo F, Egido J, Alvarez-Llamas G, Garcia-Foncillas J DiabetesCancerConnect Consortium. Molecular evidence of field cancerization initiated by diabetes in colon cancer patients. Mol Oncol. 2019;13:857–872. doi: 10.1002/1878-0261.12438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeSantis CE, Miller KD, Goding Sauer A, Jemal A, Siegel RL. Cancer statistics for African Americans, 2019. CA Cancer J Clin. 2019;69:211–233. doi: 10.3322/caac.21555. [DOI] [PubMed] [Google Scholar]
- 24.Carethers JM. Clinical and Genetic Factors to Inform Reducing Colorectal Cancer Disparitites in African Americans. Front Oncol. 2018;8:531. doi: 10.3389/fonc.2018.00531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paredes J, Zabaleta J, Garai J, Ji P, Imtiaz S, Spagnardi M, Alvarado J, Li L, Akadri M, Barrera K, Munoz-Sagastibelza M, Gupta R, Alshal M, Agaronov M, Talus H, Wang X, Carethers JM, Williams JL, Martello LA. Immune-Related Gene Expression and Cytokine Secretion Is Reduced Among African American Colon Cancer Patients. Front Oncol. 2020;10:1498. doi: 10.3389/fonc.2020.01498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mintz M, Khair S, Grewal S, LaComb JF, Park J, Channer B, Rajapakse R, Bucobo JC, Buscaglia JM, Monzur F, Chawla A, Yang J, Robertson CE, Frank DN, Li E. Longitudinal microbiome analysis of single donor fecal microbiota transplantation in patients with recurrent Clostridium difficile infection and/or ulcerative colitis. PLoS One. 2018;13:e0190997. doi: 10.1371/journal.pone.0190997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pruesse E, Peplies J, Glöckner FO. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28:1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, Holt RA. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abed J, Maalouf N, Manson AL, Earl AM, Parhi L, Emgård JEM, Klutstein M, Tayeb S, Almogy G, Atlan KA, Chaushu S, Israeli E, Mandelboim O, Garrett WS, Bachrach G. Colon Cancer-Associated Fusobacterium nucleatum May Originate From the Oral Cavity and Reach Colon Tumors via the Circulatory System. Front Cell Infect Microbiol. 2020;10:400. doi: 10.3389/fcimb.2020.00400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nadkarni MA, Martin FE, Jacques NA, Hunter N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology (Reading) 2002;148:257–266. doi: 10.1099/00221287-148-1-257. [DOI] [PubMed] [Google Scholar]
- 32.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robertson CE, Harris JK, Wagner BD, Granger D, Browne K, Tatem B, Feazel LM, Park K, Pace NR, Frank DN. Explicet: graphical user interface software for metadata-driven management, analysis and visualization of microbiome data. Bioinformatics. 2013;29:3100–3101. doi: 10.1093/bioinformatics/btt526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anderson MJ, Ellingsen KE, McArdle BH. Multivariate dispersion as a measure of beta diversity. Ecol Lett. 2006;9:683–693. doi: 10.1111/j.1461-0248.2006.00926.x. [DOI] [PubMed] [Google Scholar]
- 35.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palleja A, Mikkelsen KH, Forslund SK, Kashani A, Allin KH, Nielsen T, Hansen TH, Liang S, Feng Q, Zhang C, Pyl PT, Coelho LP, Yang H, Wang J, Typas A, Nielsen MF, Nielsen HB, Bork P, Vilsbøll T, Hansen T, Knop FK, Arumugam M, Pedersen O. Recovery of gut microbiota of healthy adults following antibiotic exposure. Nat Microbiol. 2018;3:1255–1265. doi: 10.1038/s41564-018-0257-9. [DOI] [PubMed] [Google Scholar]
- 37.Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, Yang J, Dou R, Masugi Y, Song M, Kostic AD, Giannakis M, Bullman S, Milner DA, Baba H, Giovannucci EL, Garraway LA, Freeman GJ, Dranoff G, Garrett WS, Huttenhower C, Meyerson M, Meyerhardt JA, Chan AT, Fuchs CS, Ogino S. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65:1973–1980. doi: 10.1136/gutjnl-2015-310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, Neuberg D, Huang K, Guevara F, Nelson T, Chipashvili O, Hagan T, Walker M, Ramachandran A, Diosdado B, Serna G, Mulet N, Landolfi S, Ramon Y Cajal S, Fasani R, Aguirre AJ, Ng K, Élez E, Ogino S, Tabernero J, Fuchs CS, Hahn WC, Nuciforo P, Meyerson M. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. 2017;358:1443–1448. doi: 10.1126/science.aal5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flanagan L, Schmid J, Ebert M, Soucek P, Kunicka T, Liska V, Bruha J, Neary P, Dezeeuw N, Tommasino M, Jenab M, Prehn JH, Hughes DJ. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis. 2014;33:1381–1390. doi: 10.1007/s10096-014-2081-3. [DOI] [PubMed] [Google Scholar]
- 40.Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, Baselga J, Liu C, Shivdasani RA, Ogino S, Birren BW, Huttenhower C, Garrett WS, Meyerson M. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brennan CA, Garrett WS. Fusobacterium nucleatum - symbiont, opportunist and oncobacterium. Nat Rev Microbiol. 2019;17:156–166. doi: 10.1038/s41579-018-0129-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, Qian Y, Kryczek I, Sun D, Nagarsheth N, Chen Y, Chen H, Hong J, Zou W, Fang JY. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell 2017; 170: 548-563. :e16. doi: 10.1016/j.cell.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Farhana L, Antaki F, Murshed F, Mahmud H, Judd SL, Nangia-Makker P, Levi E, Yu Y, Majumdar AP. Gut microbiome profiling and colorectal cancer in African Americans and Caucasian Americans. World J Gastrointest Pathophysiol. 2018;9:47–58. doi: 10.4291/wjgp.v9.i2.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim JA, Choi DK, Min JS, Kang I, Kim JC, Kim S, Ahn JK. VBP1 represses cancer metastasis by enhancing HIF-1α degradation induced by pVHL. FEBS J. 2018;285:115–126. doi: 10.1111/febs.14322. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All de-multiplexed, paired-end 16S rRNA gene sequence files and RNA-sequence data along with associated metadata were deposited into the Gene Expression Omnibus, which is a public repository that archives and freely distributes comprehensive sets of microarray, next-generation sequencing, and other forms of high-throughput functional genomic data submitted by the scientific community data base under project accession number GSE165255. All the tissues analyzed were collected from patients that gave informed consent for sharing results of the analysis of their anonymized tissues on public databases, where the risk of identification is very low. The statistical code used in this study is available from the corresponding author at ellen.li@stonybrookmedicine.edu.