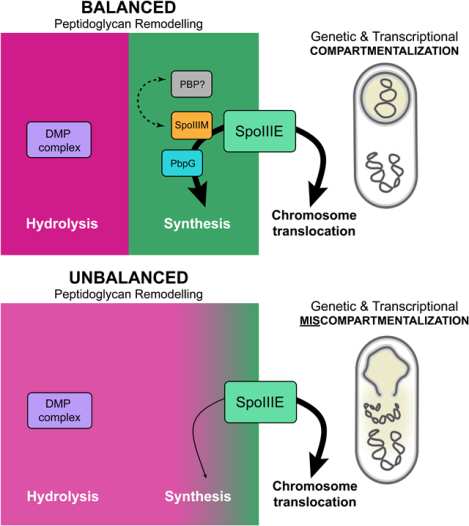
Summary
Asymmetric division, a hallmark of endospore development, generates two cells, a larger mother cell and a smaller forespore. Approximately, 75% of the forespore chromosome must be translocated across the division septum into the forespore by the DNA translocase SpoIIIE. Asymmetric division also triggers cell-specific transcription, which initiates septal peptidoglycan remodelling involving synthetic and hydrolytic enzymes. How these processes are coordinated has remained a mystery. Using Bacillus subtilis, we identified factors that revealed the link between chromosome translocation and peptidoglycan remodelling. In cells lacking these factors the asymmetric septum retracts, resulting in forespore cytoplasmic leakage and loss of DNA translocation. Importantly, these phenotypes depend on septal peptidoglycan hydrolysis. Our data support a model where SpoIIIE is anchored at the edge of a septal pore, stabilized by newly-synthesized peptidoglycan and protein-protein interactions across the septum. Together, these factors ensure coordination between chromosome translocation and septal peptidoglycan remodelling, to maintain spore development.
Keywords: Development, Sporulation, Peptidoglycan, Chromosome Translocation
Graphical Abstract
Introduction
The establishment and maintenance of genetic and transcriptional compartmentalization during cytokinesis is a key aspect of development in all organisms. In eukaryotic cells, multiple lines of evidence suggest that proteins localized at the division site coordinate chromosome segregation with cytokinesis (Fraschini, 2020). This coordination requires interplay between the reorganization of the actin and microtubule cytoskeletal elements, a variety of motor proteins and membrane trafficking (Fraschini, 2020; Seiler and Justa-Schuch, 2010). In eukaryotic cells with a cell wall (e.g. yeast), cytokinesis also requires chitin synthases and hydrolytic enzymes such as chitinases and glucanases (Lesage and Bussey, 2006). Importantly, the molecular mechanisms governing coordination between chromosome segregation with cytokinesis remain poorly understood, even in the simplest organisms, such as bacteria. Here, using the bacterium Bacillus subtilis as a model system, we investigate the mechanisms underlying the coordination between chromosome translocation and the last steps in cytokinesis during sporulation, a primordial example of cellular development.
In dividing bacteria, genetic compartmentalization is achieved by segregating most of the chromosome into each daughter cell prior to cytokinesis. Here, a membrane-anchored DNA translocase of the broadly conserved FtsK/SpoIIIE family aids in the completion of chromosome segregation (Sherratt et al., 2010; Grainge, 2010; Barre, 2007). During endospore-formation in Bacillus subtilis, a more complex situation occurs: asymmetric division generates two cellular compartments of different size and distinct developmental fates: a smaller cell called the forespore, that develops into the dormant spore, and a larger cell designated the mother cell (Stragier and Losick, 1996) (Fig. 1A). Importantly, asymmetric division precedes chromosome segregation and traps approximately 25% of the chromosome in the forespore, while the remaining 75% is translocated into the developing forespore by SpoIIIE (Wu and Errington, 1998; Sullivan et al., 2009) (Fig.1A). Remarkably, this occurs concomitantly with remodelling of the division septum and the early steps of spore envelope formation (Piggot and Hilbert, 2004). How these events are coordinated in a way that maintains transcriptional compartmentalization during development has remained unclear.
Figure 1: Identification of SpoIIIM and its requirement, together with PbpG, in maintaining compartmentalization.
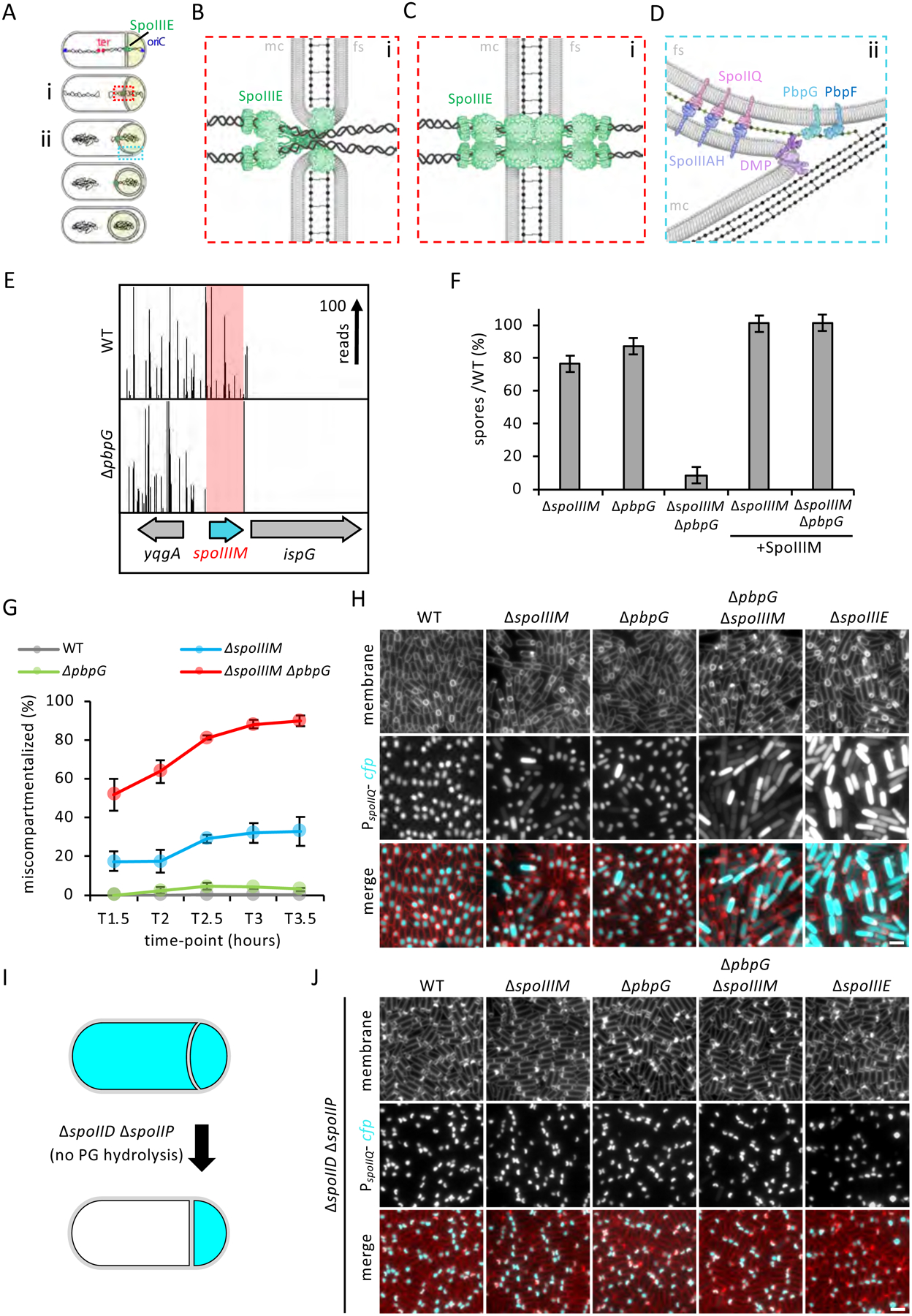
(A) Diagram of chromosome translocation and the different stages of engulfment, showing membranes (black), PG (grey), chromosome (black squiggles), SpoIIIE (green), origin (oriC, blue) and terminus (ter, red). The forespore cytoplasm is shown in beige. (B) Diagram of Aqueous Pore Model (zoomed-in from red box in Ai), showing SpoIIIE on the mother cell (mc) side and not on the forespore (fs) side, localized within unfused septal membranes that form a pore in the membrane. (C) Diagram of Channel Model (zoomed-in from red box in Ai), showing SpoIIIE on both sides of the fused septal membranes. (D) Diagram of the leading edge of the engulfing membranes (zoomed-in from blue box in Aii), showing the SpoIIIAH-SpoIIQ interaction across the engulfing membranes, PbpG (light teal) and PbpF (dark teal) in the forespore membrane, the DMP complex (purple), new PG (green dots and lines) and PG in the lateral wall of the sporangium (grey dots and lines). (E) Tn-seq profiles in WT and ΔpbpG after 24 hours of growth and sporulation in exhaustion medium. The height of each line reflects the number of sequencing reads at this position. Red box highlights the spoIIIM (yqfZ) locus, which is depleted for transposon insertions in the ΔpbpG library compared to WT. (F) Average sporulation efficiency (% ± SD, n=3) of ΔspoIIIM, ΔpbpG, ΔspoIIIM ΔpbpG and the respective spoIIIM complementation strains, ΔspoIIIM and ΔpbpG ΔspoIIIM in DSM medium. (G) Average frequency (± SD of 3 biological replicates) of miscompartmentalized cells during a sporulation time-course in WT, ΔspoIIIM, ΔpbpG and ΔspoIIIM ΔpbpG (n>900 per time-course, per strain, per replicate). (H) Representative images of miscompartmentalization in WT, ΔspoIIIM, ΔpbpG, ΔspoIIIM ΔpbpG and ΔspoIIIE at T3.5 (related to Figure 1G). Scale bar, 2 μM. (I) Diagram showing the engulfment block occurring in the ΔspoIID ΔspoIIP and its effect on compartmentalization by preventing PG hydrolysis. (J) Representative images (from one biological replicate, out of 3 examined) of miscompartmentalization in ΔspoIID ΔspoIIP at T3.5, in WT, ΔspoIIIM, ΔpbpG, ΔspoIIIM ΔpbpG and ΔspoIIIE backgrounds. Scale bar, 2 μM.
SpoIIIE is composed of three domains: an N-terminal transmembrane domain, a poorly conserved linker and a motor domain (Burton and Dubnau, 2010). The N-terminal transmembrane domain is required for SpoIIIE stability and localization to the middle of the asymmetric septum (Wu and Errington, 1997; Sharp and Pogliano, 1999; Sharp and Pogliano, 2002). The SpoIIIE motor domain contains three subdomains (α, β, and γ), being classified as a member of the RecA family of ATPases and sharing ~78% sequence similarity to FtsK (Barre, 2007). αβ assembles into a hexameric ring containing the ATPase machinery and a central channel through which double-stranded DNA is threaded (Cattoni et al., 2014). The SpoIIIE γ domain translocates DNA directionally by recognizing chromosomal DNA sequence motifs called SpoIIIE Recognition Sequences (SRS) (Ptacin et al., 2008; Cattoni et al., 2013; Besprozvannaya et al., 2013).
Two proposed models explain how SpoIIIE translocates DNA across the septum (Fig. 1B & C). In the Aqueous Pore Model, SpoIIIE is anchored at the edges of a toroidal aqueous pore surrounded by an unfused septal membrane through which the chromosome is translocated (Wu and Errington, 1997; Fiche et al., 2013). In this model, the SpoIIIE motor domains are predominantly localized on the mother-cell side of the pore and the transmembrane domain is predicted to stabilize and coordinate septal pore closure with completion of chromosome translocation (Fiche et al., 2013). In the Membrane Channel Model, SpoIIIE exists on both sides of a fused septal membrane and its transmembrane domain forms intercellular channels through which the DNA is translocated (Burton et al., 2007; Yen Shin et al., 2015; Fleming et al., 2010). A lack of clear genetic evidence in support of either model has kept the debate surrounding these models open.
Concurrent with chromosome translocation into the forespore, compartment-specific sigma factors are activated to control gene expression in the mother cell and forespore. Upon asymmetric division, σF is activated in the forespore (Fig. 1A), which controls a signal transduction pathway that triggers the activation of σE in the mother cell (Piggot and Hilbert, 2004). σE directs the synthesis of cell wall peptidoglycan (henceforth abbreviated as PG) hydrolases that assemble into the DMP complex (composed of SpoIID, SpoIIM and SpoIIP), which thin the septal PG and mediate engulfment of the forespore (Morlot et al., 2010; Chastanet and Losick, 2007) (Fig. 1D). In addition to PG degradation, engulfment is thought to involve PG synthesis by biosynthetic complexes, although these are yet to be defined (Ojkic et al., 2016). Upon completion of engulfment, membrane fission releases the forespore into the mother-cell cytoplasm as a protoplast surrounded by two membranes and a thin layer of PG in between (Khanna et al., 2019; Tocheva et al., 2013; Doan et al., 2013). Finally, during engulfment a highly-conserved SpoIIIAH-SpoIIQ zipper-like interaction holds the mother cell and forespore membranes together and functions like a ratchet, promoting efficient, forward movement of the engulfing membranes (Broder and Pogliano, 2006; Ojkic et al., 2014) (Fig. 1D).
Here, we reveal that chromosome translocation and septal PG remodelling during engulfment are coordinated through an aqueous pore stabilized by the SpoIIIE protein itself, a protein called SpoIIIM (formerly YqfZ), septal PG synthesis by the forespore-specific PG synthase PbpG and by the SpoIIIAH-SpoIIQ interaction across the septal membrane (Fig. 7A). In the absence of these factors, the asymmetric septum retracts due to septal PG hydrolysis by the DMP complex, resulting in loss of cytoplasmic and chromosomal compartmentalization, and a block to spore development (Fig. 7C). Collectively, our work defines the coordination between septal PG remodelling and chromosome segregation that ensures genetic and cytoplasmic compartmentalization during spore morphogenesis and demonstrates the primordial importance of coordinating chromosome segregation with cytokinesis during development.
Figure 7: Model illustrating coordination of PG remodelling and chromosome translocation at a highly-stabilized septal pore.
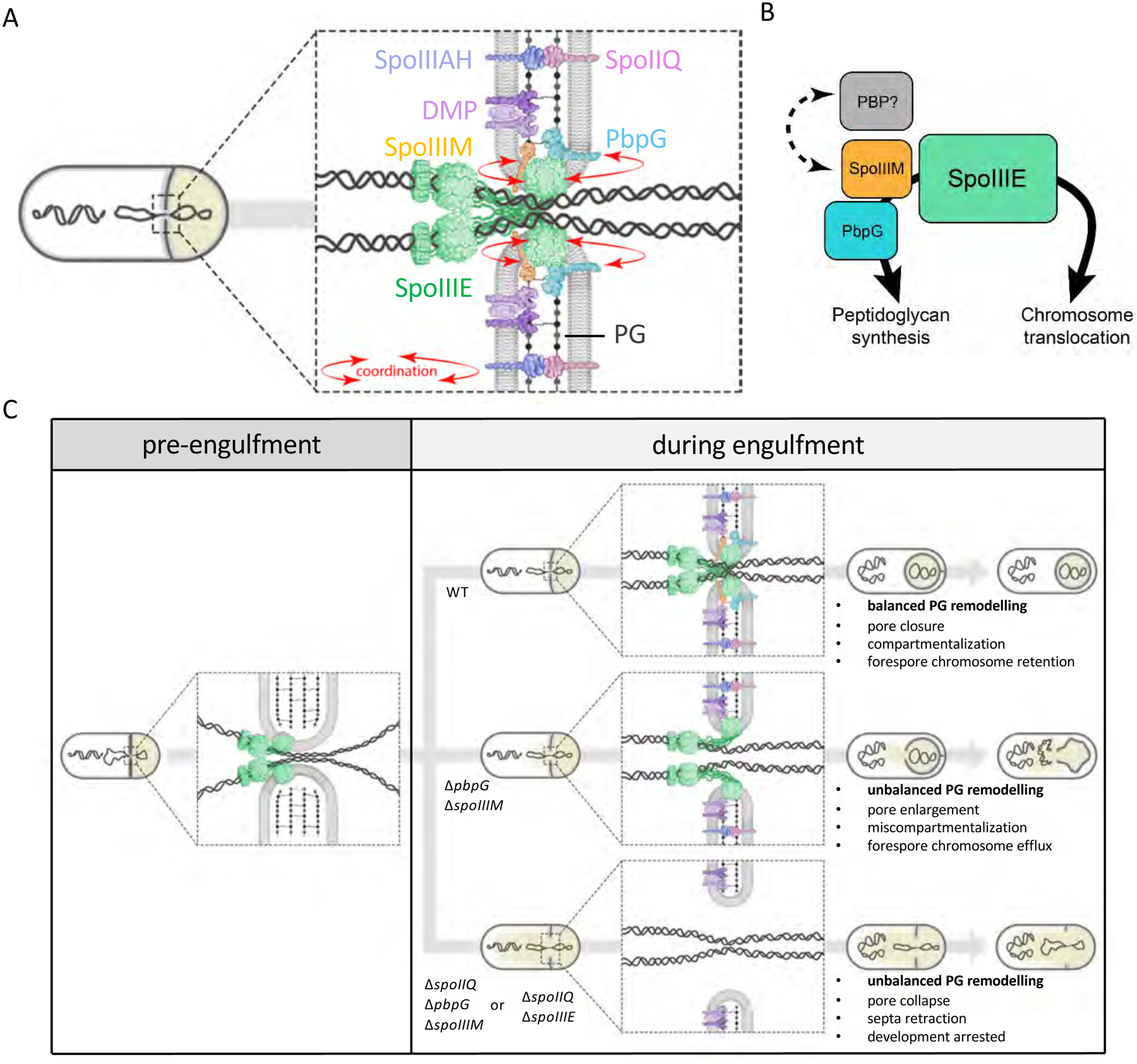
(A) Schematic illustration showing coordination (red arrows) of chromosome translocation and PG remodelling by SpoIIIM and PbpG via SpoIIIE, at the septal pore. (B) Interaction network of SpoIIIE, highlighting its role in chromosome translocation and its role in PG synthesis via interactions with SpoIIIM and PbpG. We hypothesize that a putative unidentified PG synthase (PBP) may also be involved in PG remodelling via interaction with SpoIIIM. (C) Schematic representation of the septal membrane, before and after initiation of engulfment and how different molecular components contribute to ensuring developmental compartmentalization and chromosome translocation upon initiation of engulfment. Note that in the ΔspoIIQ ΔspoIIIM ΔpbpG triple mutant, SpoIIIE is still present as focus in retracted septa (Fig. S4) but for simplicity this is not shown in the bottom panel.
Results
A screen for synthetic lethal partners with pbpG identifies spoIIIM
The forespore produces two functionally redundant class A penicillin-binding proteins (PBPs), PbpG and PbpF, that generate a thin layer of spore PG called the germ cell wall (Fig. 1D) (McPherson et al., 2001). Sporulating cells lacking one of these proteins produce close to wild-type levels of spores; however, cells lacking both are severely impaired in sporulation (McPherson et al., 2001). Wild-type (henceforth WT) and the pbpF mutant produce forespores with an oblong-shape. However, sporulating cells lacking pbpG exhibit an abnormal, jellybean-like morphology (Fig. S1A) (Rodrigues et al., 2016). Based on this morphological difference, we hypothesized that PbpG and PbpF may have specialized roles and function in separate genetic pathways. To investigate this hypothesis and identify factors that function in each pathway, we used transposon-sequencing (Tn-seq) to screen for genes that become critical for sporulation in cells lacking either pbpG or pbpF (Meeske et al., 2016).
In validation of our genetic screen, pbpF was one of the top hits in the ΔpbpG mutant transposon library and pbpG was one of the top hits in the ΔpbpF transposon library (Fig. S1D & E). One of the strongest hits that was specific to the ΔpbpG library was a gene called yqfZ that we have renamed spoIIIM (Fig. 1E and S1B). Previous work indicates that spoIIIM is transcribed in the mother cell compartment under σE control and encodes a 99 amino acid protein of unknown function (Eichenberger et al., 2003). SpoIIIM is predicted to have an N-terminal transmembrane segment and a C-terminal LysM domain (UniProt Consortium, 2018) that are implicated in binding to PG (Buist et al., 2008).
To verify that SpoIIIM becomes important for sporulation in ΔpbpG cells, we determined the sporulation efficiency of ΔspoIIIM and ΔspoIIIM ΔpbpG mutants (Fig. 1F). Consistent with previous reports, the ΔspoIIIM mutant had a mild sporulation defect (76.6% spores) and the ΔpbpG mutant sporulated to near WT levels (87.3% spores). In validation of the Tn-seq screen, the sporulation efficiency of the ΔspoIIIM ΔpbpG double mutant was 8.8% (Fig. 1F). Expression of spoIIIM from an ectopic chromosomal locus restored sporulation efficiency of ΔspoIIIM and ΔspoIIIM ΔpbpG mutants, to WT and ΔpbpG levels, respectively (Fig. 1F). Finally, consistent with the results of the Tn-seq screen and in support of the idea that SpoIIIM is specifically required in cells lacking PbpG, no synergy was observed in the ΔspoIIIM ΔpbpF double mutant (Fig. S1C).
PbpG and SpoIIIM are required for forespore morphology and compartmentalization
To begin to unravel how SpoIIIM and PbpG contribute to sporulation, we used fluorescence microscopy to determine if cells lacking these proteins exhibit morphological defects during development. Sporulating cells were imaged every 30 min after the onset of asymmetric division (1 hour and 30 minutes after the initiation of sporulation; T1.5). No obvious differences in forespore morphology were observed between the WT and spoIIIM mutant at early stages of development; however as the cells neared the completion of engulfment (T3) the spoIIIM mutant exhibited forespore morphologies that we categorized into four types: 1) WT-looking, 2) dwarf (small spherical forespores), 3) mislocalized (dwarf forespores positioned closer to the midcell of the mother cell) and 4) abnormal (forespores that lacked a spherical appearance and contained irregular membranes) (Fig. S2B & E). Consistent with the Tn-seq and sporulation efficiency data, the ΔspoIIIM ΔpbpG double mutant produced even more dwarf, mislocalized and abnormal forespores (Fig. S2E). Transmission electron microscopy (TEM) of ΔspoIIIM and ΔspoIIIM ΔpbpG confirmed the presence of dwarf forespores with wrinkled membranes (Fig. S2F). Interestingly, closer inspection of ΔpbpG cells revealed that 2.5% of the forespores have a dwarf phenotype (Fig. S2B & E), suggesting the absence of SpoIIIM enhances this defect.
The severity of the morphological defects in the ΔspoIIIM and ΔspoIIIM ΔpbpG mutants led us to wonder if their forespores retain cytoplasmic compartmentalization. We engineered strains to express a cytoplasmic, cyan fluorescent protein (CFP) in the forespore under the control of a σF-dependent promoter (PspoIIQ), and examined the distribution of the CFP signal by fluorescence microscopy during a sporulation time course (Fig. 1H). As expected, in WT cells the CFP signal was confined to the forespore throughout the entire time course (Fig. 1H). In the ΔspoIIIM mutant we observed some cells with CFP signal confined solely to the forespore, some with stronger CFP signal in the forespore and weaker CFP signal in the mother cell and others with strong CFP signal throughout the entire sporangium (Fig. 1H). The frequency of ΔspoIIIM sporulating cells with any CFP signal (weak or strong) in the mother cell increased over time and reached 33% at T3.5 (Fig. 1G). These observations suggest that ΔspoIIIM forespores leak their cytoplasmic contents into the mother cell and loose compartmentalization. We observed an even higher proportion of miscompartmentalized sporulating cells in the ΔspoIIIM ΔpbpG double mutant (90% at T3.5) (Fig. 1G). Expression of spoIIIM in trans restored compartmentalization in the double mutant (Fig. S2A). Miscompartmentalization was also observed in the ΔpbpG mutant, albeit at a low frequency (3%) (Fig. 1G & H). Highlighting the specific genetic relationship between spoIIIM and pbpG, the degree of miscompartmentalization of the ΔspoIIIM ΔpbpF double mutant was comparable to the ΔspoIIIM mutant (Fig. S2C & D). These data suggest that SpoIIIM and PbpG are also required for forespore compartmentalization.
Interestingly, miscompartmentalization and abnormal forespore morphology have both been reported for sporulating cells lacking spoIIIE (Liu et al., 2006; Hilbert et al., 2004; Doan, 2013). A side-by-side comparison of the ΔspoIIIM ΔpbpG double mutant and ΔspoIIIE revealed similar forespore morphologies and degrees of miscompartmentalization (Fig. 1H & S2B). Furthermore, combining ΔspoIIIM and/or ΔpbpG with a hypomorphic allele of spoIIIE (SpoIIIE D584A) (Burton et al., 2007), which exhibits miscompartmentalization in a small fraction of cells, resulted in a dramatic increase in miscompartmentalization (Fig. S3A & B). These observations led us to investigate if the ΔspoIIIM and ΔpbpG mutant have other phenotypes in common with ΔspoIIIE. Previous work demonstrated that ΔspoIIIE miscompartmentalization can be suppressed when cells lack the engulfment hydrolases, SpoIID and SpoIIP (Hilbert et al., 2004). Sporulating cells lacking both SpoIID and SpoIIP have flat septa due to a block in septal PG hydrolysis (Fig. 1I) (Chastanet and Losick, 2007). In a ΔspoIID ΔspoIIP mutant, ΔspoIIIE miscompartmentalization was almost completely abolished (Fig. 1J). Interestingly, this was also the case in ΔspoIIIM and ΔspoIIIM ΔpbpG mutants (Fig. 1J), suggesting that SpoIIIE, SpoIIIM and PbpG help maintain compartmentalization, by counteracting the effects of PG hydrolysis during engulfment.
SpoIIIM and PbpG are required for the dispersal of the SpoIIIE complexes
Based on the loss of compartmentalization in the ΔspoIIIE mutant, it was previously hypothesized that the chromosome spanning the septum, and/or the absence of the SpoIIIE, creates an opening in the septum through which forespore cytoplasmic contents could leak (Hilbert et al., 2004; Liu et al., 2006). We considered the possibility that SpoIIIM and PbpG are required for SpoIIIE stability and in their absence SpoIIIE becomes unstable, generating this opening. To investigate this possibility, we examined the localization of a functional SpoIIIE-GFP fluorescent fusion (Burton et al., 2007), during a sporulation time-course in WT, ΔspoIIIM, ΔpbpG and ΔspoIIIM ΔpbpG mutant cells (Fig. 2). Consistent with previous work, in WT cells, SpoIIIE-GFP localizes in a discrete focus at the centre of the asymmetric septum (Fig. 2A & B) (Burton et al., 2007; Wu and Errington, 1997; Sharp and Pogliano, 1999). As cells progress through engulfment, the focus is often located in the engulfing membranes, and as engulfment nears completion, the focus disperses, with SpoIIIE-GFP localizing as diffuse signal in the forespore membranes (Fig. 2A, B & C). SpoIIIE-GFP dispersion is thought to represent detachment of SpoIIIE from the DNA (Ben-Yehuda et al., 2003). Consistent with this idea and previous reports (Sharp and Pogliano, 1999), the number of WT sporulating cells with foci declined over time (at T3.5, 35% of the cells had a SpoIIIE-GFP focus) (Fig. 2B & D). Importantly, SpoIIIE-GFP formed a discrete focus in ΔspoIIIM and ΔspoIIIM ΔpbpG sporulating cells that resembled the focus observed in WT (Fig. 2B). Furthermore, quantification of SpoIIIE-GFP foci revealed similar signal intensities in WT and mutant strains (Fig. 2E). Thus, the miscompartmentalization in these mutants cannot be explained by instability of SpoIIIE. Interestingly, in the ΔspoIIIM and ΔspoIIIM ΔpbpG mutants, the number of cells with SpoIIIE-GFP foci persisted for a longer period of time: at T3.5, 52% and 84% of the sporulating cells, respectively, contained a SpoIIIE-GFP focus (Fig. 2B & D). Thus, SpoIIIM and PbpG are required for SpoIIIE dispersal into the forespore membranes. These results argue that forespore miscompartmentalization in cells lacking SpoIIIM and PbpG is not due to instability of SpoIIIE, suggesting these factors play a more direct role in maintaining compartmentalization at a septal pore.
Figure 2: SpoIIIE foci persist in the absence of SpoIIIM and PbpG.
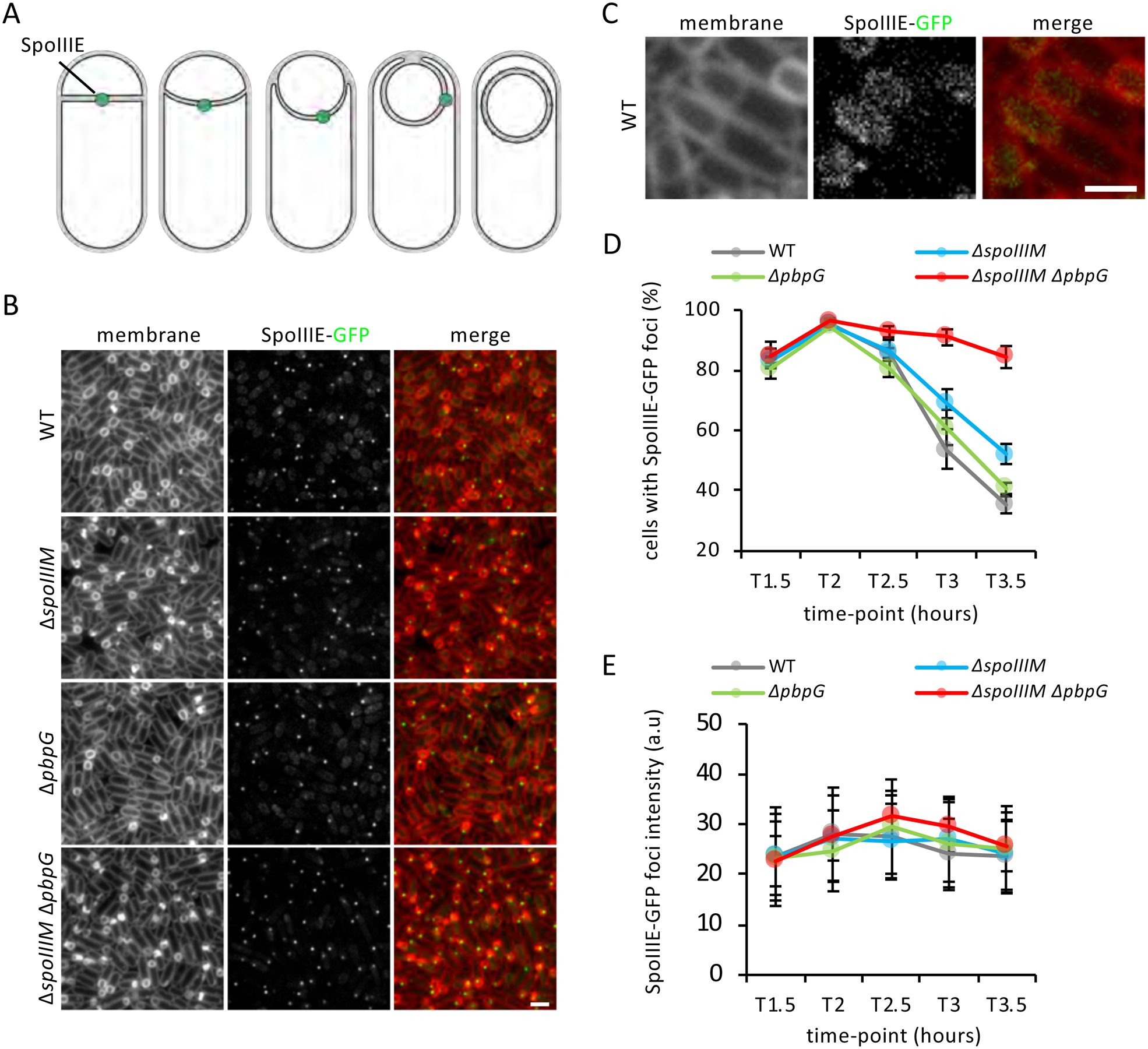
(A) Diagram showing the dynamic localization of SpoIIIE during engulfment. (B) Representative images of the localization of SpoIIIE-GFP in WT, ΔspoIIIM, ΔpbpG and ΔspoIIIM ΔpbpG at T3.5. Scale bar, 2 μM. (C) Representative image of the diffused localization of SpoIIIE in WT cells that have completed engulfment. Scale bar, 1 μM. (D) Average frequency (± SD of of 3 biological replicates) of cells with SpoIIIE-GFP foci in WT, ΔspoIIIM, ΔpbpG and ΔspoIIIM ΔpbpG during a sporulation time-course (n>1000 per time-course, per strain, per replicate). (E) Average fluorescence (± STDEVP, n>400, per time-point, per strain) intensity of SpoIIIE-GFP foci in WT, ΔspoIIIM, ΔpbpG and ΔspoIIIM ΔpbpG during a sporulation time-course.
SpoIIIM and PbpG are required for efficient chromosome translocation and retention of the chromosome in the forespore
The data above suggest that SpoIIIE can still assemble into complexes in sporulating cells lacking SpoIIIM and PbpG. To investigate if SpoIIIE is capable of DNA translocation in the mutants, we used an approach based on the LacI-lacO system (Fig. 3A) and engineered strains to produce LacI-GFP under the control of the forespore reporter (PspoIIQ-lacI-gfp) inserted at a chromosomal locus (amyE), present in the forespore at the time of asymmetric division (Wu and Errington, 1998; Marquis et al., 2008). These strains were also engineered to contain 48 lacO repeats at the pelB locus, which resides near the terminus (Fig. 3A). Using this approach, if the pelB locus is translocated into the forespore, a GFP focus will be observed in the forespore; if not, then no GFP focus will be observed (Fig. 3A). Due to miscompartmentalization in ΔspoIIIM and ΔspoIIIM ΔpbpG mutants, we would also expect to observe some sporulating cells with a GFP focus bound to the mother cell chromosome, in addition to the one in the forespore (Fig. 3A). Furthermore, if cells fail to translocate the chromosome, we would expect to observe two GFP foci in the mother cell (Fig. 3A).
Figure 3: Efficient chromosome translocation requires SpoIIIM and PbpG.
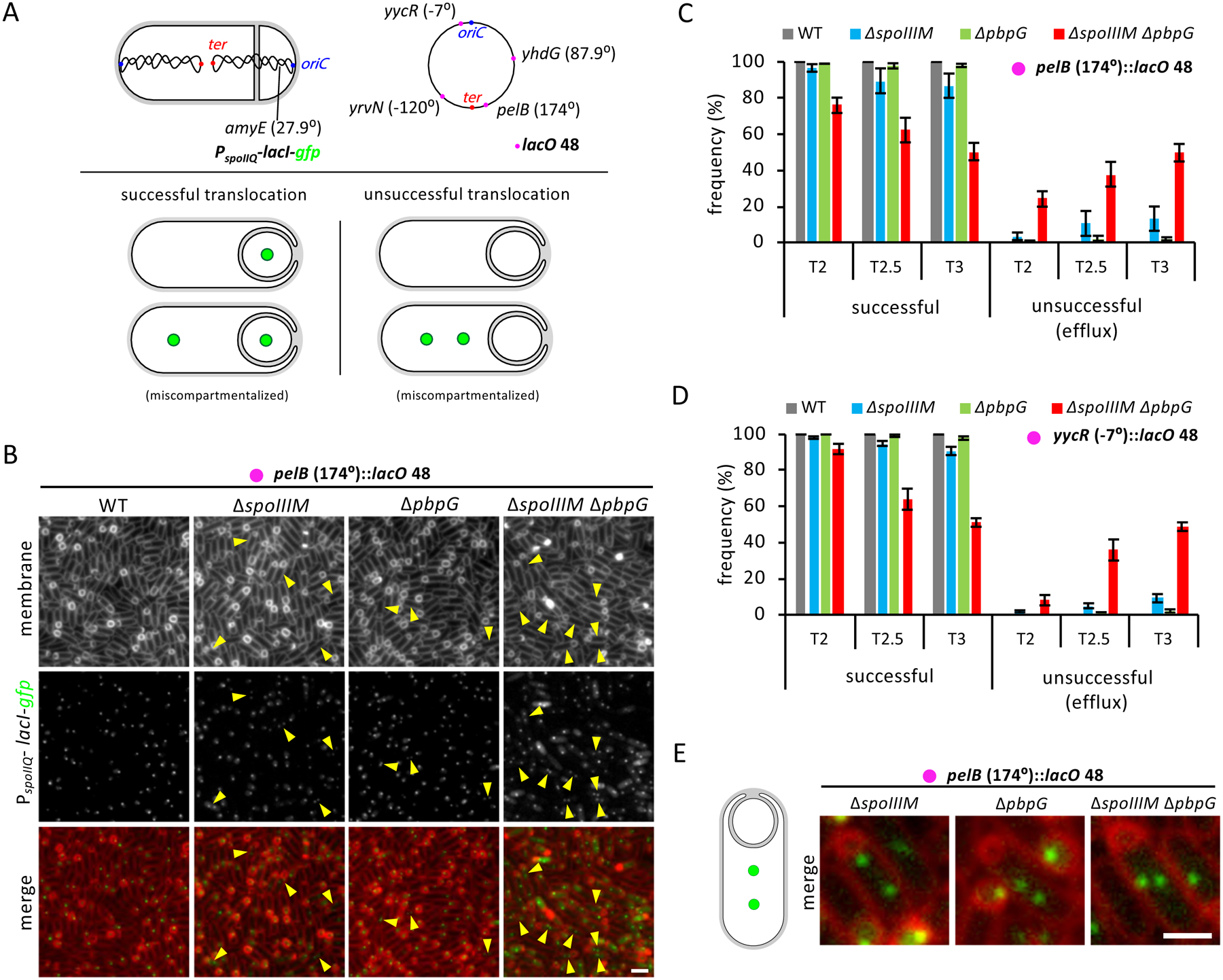
(A) Diagram explaining experimental rationale of the LacI-lacO system for visualizing chromosome translocation. LacI-GFP, expressed in the forespore (PspoIIQ) from the amyE locus, binds to lacO48 sites inserted at the yhdG, pelB, yrvN or yycR chromosomal locus. The origin (oriC) and terminus (ter) loci are shown in blue and red, respectively. Successful chromosome translocation is indicated by a LacI-GFP focus in the forespore, with an additional focus in the mother cell compartment in the event of miscompartmentalization. Unsuccessful chromosome translocation is indicated by an absence of LacI-GFP foci, or by two LacI-GFP foci in the mother cell compartment in the event of miscompartmentalization. (B) Representative images of LacI-GFP foci at T3 in WT, ΔspoIIIM, ΔpbpG and ΔspoIIIM ΔpbpG strains containing lacO48 inserted at the pelB locus (174°). Yellow arrowheads point to cells with two mother cell foci. Scale bar, 2 μM. (C) Average frequency (± SD of 3 biological replicates) of cells with a LacI-GFP focus in the forespore (successful translocation) or with no or two LacI-GFP foci in the mother cell (unsuccessful translocation, efflux) during a sporulation time-course, with lacO48 integrated at the pelB locus (174°) and (D) at the yycR locus (−7°) in WT, ΔspoIIIM, ΔpbpG and ΔspoIIIM ΔpbpG (n>650 per time-course, per strain, per replicate). (E) Representative images of cells at T3 with two LacI-GFP foci in the mother cell of ΔspoIIIM, ΔpbpG and ΔspoIIIM ΔpbpG sporulating cells containing lacO48 inserted at the pelB locus (174°).
In WT sporulating cells, DNA translocation takes ~20 minutes (Burton et al., 2007). Accordingly, virtually all forespores contained a GFP focus at all time-points examined, indicating that the chromosome terminus was efficiently translocated into the forespore (Fig. 3B & C). In ΔspoIIIM at T2, virtually all forespores (97%) contained a GFP focus, with a small fraction of cells containing two foci in the mother cell (3%) (Fig. 3C). This suggests that the majority of cells translocate the chromosome into the forespore in the absence of SpoIIIM. Interestingly, at T2.5 the number of cells with a GFP focus in the forespore decreased to 90%, while the number of cells with two GFP foci in the mother cells increased to 10%. A similar trend was observed at T3. These data suggest that in the absence of SpoIIIM the translocated chromosome is actively or passively effluxed back into the mother cell.
A similar but more dramatic trend was observed in the ΔspoIIIM ΔpbpG double mutant (Fig. 3B & C). At T2, the majority of the ΔspoIIIM ΔpbpG double mutant cells had translocated their chromosome (76% contained a GFP focus in the forespore). Remarkably, at T2.5 and T3, we observed a sharp decline in the number of cells with a GFP focus in the forespore and an increase in the number of cells with two mother cell foci (38% and 50% at T2.5 and T3, respectively) (Fig. 3B, C & E). Thus, most ΔspoIIIM ΔpbpG cells are capable of translocating a chromosome into the forespore but they fail to retain it. Finally, we note that while the vast majority of sporulating cells lacking PbpG successfully translocate a chromosome into the forespore, a small fraction of cells had two foci in the mother cell at T3 (2%) (Fig. 3B, C & E). Thus, even in the absence of pbpG, a small proportion of cells exhibit chromosome translocation defects.
Remarkably similar results were obtained when we examined chromosomes harboring the lacO array adjacent to origin at −7° (yycR) (Fig. 3D), at −120° (yrvN) or +87° (yhdG) (Fig. S3C & D). These results suggest that the entire forespore chromosome is effluxed into the mother cell in the absence of SpoIIIM and PbpG. Furthermore, analysis of origin-proximal (yycR) and terminus-proximal (pelB) loci in the ΔspoIIIM ΔpbpG cells, using different fluorescently-labeled DNA-binding proteins (LacI-YFP and TetR-CFP), revealed a pattern of YFP and CFP foci that is more consistent with passive efflux of the chromosome than active reverse translocation (Fig. S3E) (see Discussion). Collectively, these data suggest that the septal pore generated in sporulating cells lacking SpoIIIM and PbpG results in forespore chromosome loss, in addition to loss of its cytoplasmic contents.
Impairing engulfment promotes compartmentalization and efficient chromosome translocation in cells lacking SpoIIIM and PbpG
Our data indicate that blocking engulfment PG hydrolysis almost fully restores compartmentalization to ΔspoIIIM and ΔspoIIIM ΔpbpG cells. If forespore chromosome loss in these mutants is due to passive movement through a septal pore, blocking PG hydrolysis should similarly prevent chromosome loss from the forespore. This is indeed what we found. Sporulating cells lacking SpoIID and SpoIIP almost fully restored chromosome translocation, and retention, to the ΔspoIIIM and ΔspoIIIM ΔpbpG mutants (Fig.4A). Furthermore, using a mutant (ΔspoIIB) that impairs but does not abolish PG hydrolysis during engulfment (Perez et al., 2000) (Fig.4C), we found that both miscompartmentalization and forespore chromosome efflux were largely suppressed in the mutant backgrounds (Fig. 4B & C). Collectively, these data suggest that SpoIIIM and PbpG function to maintain compartmentalization of forespore cytoplasmic contents, including the chromosome, and counterbalance the activity of the PG hydrolases that remodel the septum during engulfment.
Figure 4: Blocking or impairing engulfment restores efficient chromosome translocation and compartmentalization in the absence of SpoIIIM and PbpG.
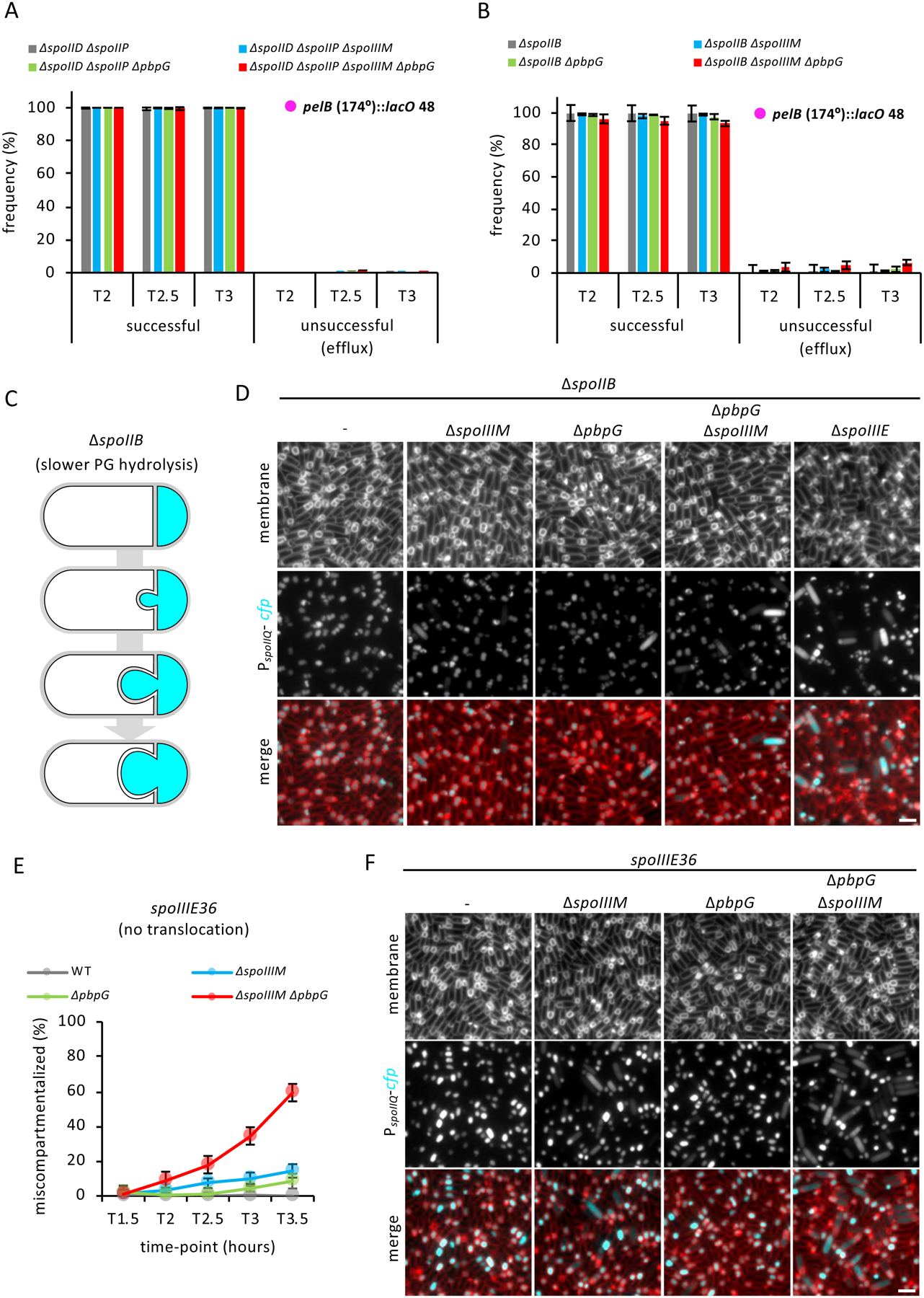
(A) Average frequency (± SD of 3 biological replicates) of cells with a LacI-GFP focus in the forespore or with two LacI-GFP foci in the mother cell during a sporulation time-course, with lacO 48 integrated at the pelB locus (174°) in the ΔspoIID ΔspoIIP alone or combined with the ΔspoIIIM, ΔpbpG and ΔpbpG ΔspoIIIM (n>600 per time-course, per strain, per replicate). (B) Average frequency (± SD of 3 biological replicates) of cells with a LacI-GFP focus in the forespore or with two LacI-GFP foci in the mother cell during a sporulation time-course, with lacO 48 integrated at the pelB locus (174°) in the ΔspoIIB alone or combined with the ΔspoIIIM, ΔpbpG and ΔspoIIIM ΔpbpG (n>600 per time-course, per strain, per replicate). (C) Schematic representation of the slower engulfment and membrane bulging phenotype observed in the ΔspoIIB. Cyan represents forespore-specific CFP. (D) Representative images of sporulating cells at T3 showing compartmentalization of forespore gene expression in ΔspoIIB mutant alone, or combined with the ΔspoIIIM, ΔpbpG, ΔspoIIIM ΔpbpG and ΔspoIIIE. Scale bar, 2 μM. (E) Average frequency (± SD of 3 biological replicates) of miscompartmentalized cells during a sporulation time-course in the spoIIIE36 mutant in otherwise WT (grey), ΔspoIIIM (blue), ΔpbpG (green) and ΔspoIIIM ΔpbpG (red) strains (n>750 per time-course, per strain). (F) Representative images of miscompartmentalization in the spoIIIE36 mutant in otherwise WT, ΔspoIIIM, ΔpbpG and ΔspoIIIM ΔpbpG strains at T3.5. Scale bar, 2 μM.
Blocking chromosome translocation partially suppresses miscompartmentalization in cells lacking SpoIIIM and PbpG
Based on recent data showing that chromosome translocation into the forespore generates turgor pressure on the septal PG (Lopez-Garrido et al., 2018), we tested if chromosome translocation into the forespore contributes to miscompartmentalization in cells lacking SpoIIIM and PbpG. If so, a SpoIIIE mutant (spoIIIE36) that produces a stable SpoIIIE focus and does not impact compartmentalization but fails to translocate the chromosome (Sharp and Pogliano, 1999; Besprozvannaya et al., 2014), should suppress miscompartmentalization in cells lacking SpoIIIM and PbpG. Indeed, spoIIIE36 partially suppressed miscompartmentalization in the ΔspoIIIM and ΔspoIIIM pbpG mutants (Fig. 4E & F). This result suggests that SpoIIIM and PbpG also function to counterbalance the effect chromosome translocation has on the septal pore.
SpoIIQ is required for septal pore stability during engulfment in cells lacking PbpG, SpoIIIM or SpoIIIE
Since slowing down engulfment by reducing the efficiency of PG hydrolysis suppressed miscompartmentalization in the ΔspoIIIM and ΔspoIIIM ΔpbpG mutants, we wondered if this would also be the case in sporulating cells lacking the SpoIIIAH-SpoIIQ zipper-like interaction. As described in the introduction, this zipper-like interaction contributes to the efficiency of engulfment (Broder and Pogliano, 2006; Ojkic et al., 2014). To this end, we introduced the ΔspoIIQ mutation into the ΔspoIIIM and ΔpbpG single mutants, and the ΔspoIIIM ΔpbpG double mutant backgrounds, and examined compartmentalization with our forespore reporter (PspoIIQ-cfp) during a sporulation time-course (Fig. 5).
Figure 5: Septa retraction and its dependency on PG hydrolysis.
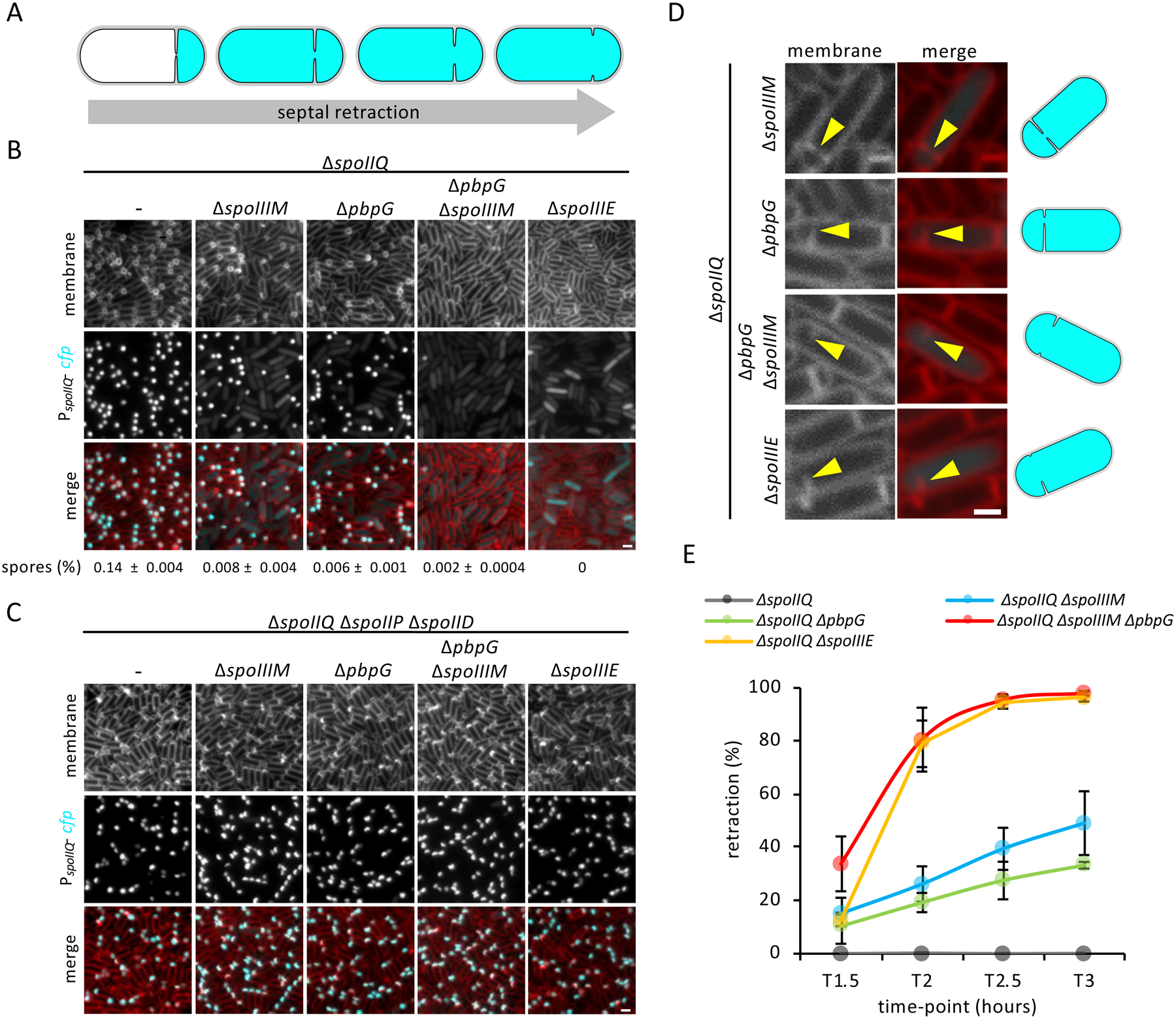
(A) Schematic representation of septal retraction, illustrating that as septal retraction progresses, CFP fluorescence (cyan) leaks from the forespore to fill the entire cell. (B) Representative images of septal retraction in WT, ΔspoIIIM, ΔpbpG, ΔspoIIIM ΔpbpG and ΔspoIIIE in a ΔspoIIQ background at T3. Scale bar,2 μM. Sporulation efficiency (%, average ± SD, n=3) is shown below the respective strains. (C) Representative images of septal retraction suppression in cells blocked for engulfment in WT, ΔspoIIIM, ΔpbpG, ΔspoIIIM ΔpbpG and ΔspoIIIE strains at T3. Scale bar, 2 μM. (D) Representative zoomed-in examples of septal retraction in ΔspoIIIM, ΔpbpG, ΔspoIIIM ΔpbpG and ΔspoIIIE at T3. Yellow arrowheads point to retracting septa. Scale bar,1 μM. Schematic representations of cells are shown on the right. (E) Average frequency (± SD of of 3 biological replicates) of cells exhibiting septal retraction in WT, ΔspoIIIM, ΔpbpG, ΔspoIIIM ΔpbpG and ΔspoIIIE during a sporulation time-course (n>600 per time-point, per strain, per replicate).
Surprisingly, instead of suppressing the compartmentalization defects, the ΔspoIIQ mutant enhanced them. Among the sporulating cells with CFP signal, a subset appeared to completely lack a forespore compartment (Fig. 5B). Since σF is required for the production of CFP and σF only becomes active after asymmetric division, sporulating cells with this phenotype must have developed an asymmetric septum, which then retracted. Indeed, closer inspection of these cells revealed vestiges of septum formation (Fig. 5D). We refer to this phenotype as septal retraction (Fig. 5A). Quantification of septal retraction during a sporulation time course revealed that the phenotype increases over time (Fig. 5E) and represented 49% and 33% of the sporulating cells at T3, in the ΔspoIIQ ΔspoIIIM and ΔspoIIQ ΔpbpG double mutant, respectively. In the ΔspoIIQ ΔspoIIIM ΔpbpG triple mutant, almost all cells (98%) displayed this phenotype by T3 (Fig. 5E).
Since the ΔspoIIIM ΔpbpG double mutant phenotypes closely resemble those in the ΔspoIIIE mutant, we examined our forespore CFP reporter in the ΔspoIIQ ΔspoIIIE double mutant. Strikingly, the ΔspoIIQ ΔspoIIIE mutant had a similar frequency of septal retraction and vestiges of septa (Fig. 5B, D & E). Transmission electron microscopy confirmed the presence of retracted septa in the ΔspoIIQ ΔspoIIIM ΔpbpG triple mutant and ΔspoIIQ ΔspoIIIE double mutant (Fig. S4A). Importantly, septal retraction in ΔspoIIQ ΔspoIIIM and ΔspoIIQ ΔpbpG double mutants or the ΔspoIIQ ΔspoIIIM ΔpbpG triple mutant was not due to destabilization of SpoIIIE, as SpoIIIE-GFP still localized as foci in cells with retracted septa (Fig. S4B). In most cells with retracted septa, regardless of the mutant, SpoIIIE-GFP was observed as a single focus (Fig. S4B, C and D). Interestingly, in a small fraction of cells, two SpoIIIE-GFP foci were observed (Fig. S4B, C & D). Three-dimensional structured illumination microscopy (3D-SIM) confirmed these observations and suggest that SpoIIIE-GFP foci reside at the edges of an enlarged septal pore in the ΔspoIIQ ΔspoIIIM ΔpbpG triple mutant (Fig. S4E & F).
Finally, we tested whether PG hydrolysis was required for septal retraction. In support of the idea that septal retraction requires PG hydrolysis, retraction was completely abolished and compartmentalization restored in all strains examined, that lack SpoIID and SpoIIP (Fig. 5C). Collectively, these data suggest that the septal membranes have not undergone fission at the time of chromosome translocation and that SpoIIIE, PbpG, SpoIIIM, and the zipper-like interaction between SpoIIIAH and SpoIIQ, all function to prevent the septal pore from enlarging during hydrolysis of the surrounding PG.
Evidence that SpoIIIM, PbpG and SpoIIIE coordinate pore closure with completion of chromosome translocation
Building on the observations we describe thus far and taking advantage of the septal retraction phenotype, we tested the hypothesis that SpoIIIM and PbpG coordinate septal pore closure with completion of chromosome translocation. We hypothesized that sporulating cells lacking SpoIIIM and/or PbpG are unable to coordinate septal pore closure with completion of chromosome translocation during engulfment PG hydrolysis, and if these cells also lack SpoIIQ, the membranes that line the pore, retract. We reasoned that if we hold back engulfment PG hydrolysis in the absence of SpoIIIM and PbpG, then the septal pore would have sufficient time to close and septal retraction will occur less frequently, unless chromosome translocation is yet to be completed and the septal pore is still open. This is indeed what we found, using the experimental system described in Figure S5A.
We performed this assay on strains that were otherwise WT, or lacked spoIIQ or both spoIIQ and pbpG. We induced the PG hydrolases at two time points after the onset of sporulation, T2 and T3, to give the cells different amounts of time to complete septal pore closure. At these time points, a similar proportion of the sporulating cells have two septa and CFP signal in the two forespore compartments (>86% for all strains) (Fig. S5A & E). Thus, after induction of the PG hydrolases, an increase in sporangia with CFP signal that have a single septum or lack septa would suggest septal retraction and failure to complete septal pore closure (i.e. membrane fission).
When the PG hydrolases were induced at T2 (Fig. S5B & C) and cells with CFP signal were examined one or 2 hours after induction (T3 & T4), most cells still had two septa and there was only a moderate increase in the number of cells with a single septum or no septa (i.e. the septa had retracted) in otherwise WT and ΔspoIIQ cells. By contrast, in the ΔspoIIQ ΔpbpG double mutant cells, the proportion of cells with a single septum or no septa was much higher (Fig. S5G).
When the PG hydrolases were induced at T3 (Fig. S5D & SE) and cells with CFP signal were examined one or 2 hours after induction (T4 & T5), although the overall pattern of septal retraction between the strains was similar to when induction occurred at T2 (more septal retraction in ΔspoIIQ ΔpbpG cells, than in otherwise WT and ΔspoIIQ cells), there was a reduction in the number of retracted septa for all strains (Fig. S5G). Thus, given sufficient time, the septal pore will undergo closure in the absence of SpoIIQ, PbpG and SpoIIIM.
Importantly, since ΔspoIIQ ΔpbpG sporulating cells have a higher frequency of retracted septa but a similar frequency of fully translocated chromosomes as otherwise WT, or ΔspoIIQ sporulating cells at both time points of PG hydrolase induction (Fig. S5F & S5G), these data suggest that septal pore closure is delayed relative to completion of chromosome translocation in sporulating cells lacking SpoIIIM and PbpG.
Finally, we performed these experiments in the ΔspoIIQ ΔspoIIIE sporulating cells that do not translocate the forespore chromosome. Interestingly, the ΔspoIIQ ΔspoIIIE mutant behaved similarly to the ΔspoIIQ ΔpbpG mutant (i.e. septal retraction decreased when the PG hydrolases are induced at T3 (Fig. S5B–E & G). Thus, given sufficient time, the septal pore will undergo closure even in the absence of SpoIIIE (and in presence of untranslocated DNA) (Fig. S5F & H). This result establishes that SpoIIIE is not absolutely required for septal membrane fission.
PG synthesis by PbpG is required for septal pore closure
Our data point to the possibility that PbpG contributes to PG synthesis around the septal pore, to maintain pore stability and drive pore closure. If this is the case, then the catalytic activity of PbpG should contribute to closure of the septal pore. To test this idea, we generated a mutant of PbpG (PbpGE94A S365A, referred to as PbpG*) harbouring alanine substitutions at predicted catalytic residues in its glycosyltransferase and transpeptidase domains (Fig. S6A), in the context of a functional N-terminal, hexahistidine-tagged version of PbpG (His6-PbpG) (Fig. S6C). We note that PbpG* may still retain some residual catalytic activity, since it partially complemented the ΔpbpG null allele in a ΔpbpG ΔpbpF (0.16%) (compared to 0.01% of the non-complemented ΔpbpG ΔpbpF double mutant) and ΔspoIIIM ΔpbpG double mutant background (57.4%) (compare Fig. S6C with Fig.1F). We examined the capacity of PbpG* to support septal pore closure by examining the different phenotypes reported thus far in the presence and absence of SpoIIIM: compartmentalization (Fig. 6A & S6D), SpoIIIE localization (Fig. 6C), chromosome translocation (Fig. 6B), and septal retraction (Fig. 6D & S6D). For all phenotypes tested, the catalytic activity of PbpG was required, although PbpG* produced phenotypes that were less penetrant than those observed in the ΔpbpG null mutant, except in the case of SpoIIIE localization and septal pore retraction, where it phenocopied the ΔpbpG null. Thus, PbpG itself, independent of its catalytic activity, is also required for the phenotypes described here. For reasons unknown, we were unable to detect His6-PbpG with His6 antibodies by immunoblot and thus confirm the levels of PbpG*. However, PbpG* in the context of a partially-functional but detectable, C-terminal, hexahistidine-tagged version of PbpG (PbpG-His6) produced similar levels of protein as the non-mutated counterpart (Fig. S6B). Thus, we conclude that PbpG catalytic mutations do not impact protein levels. Collectively, these results support the idea that PG synthesis by PbpG contributes to septal pore closure.
Figure 6: Peptidoglycan synthesis by PbpG is required for developmental characteristics and evidence that SpoIIIE, SpoIIIM and PbpG form a complex.
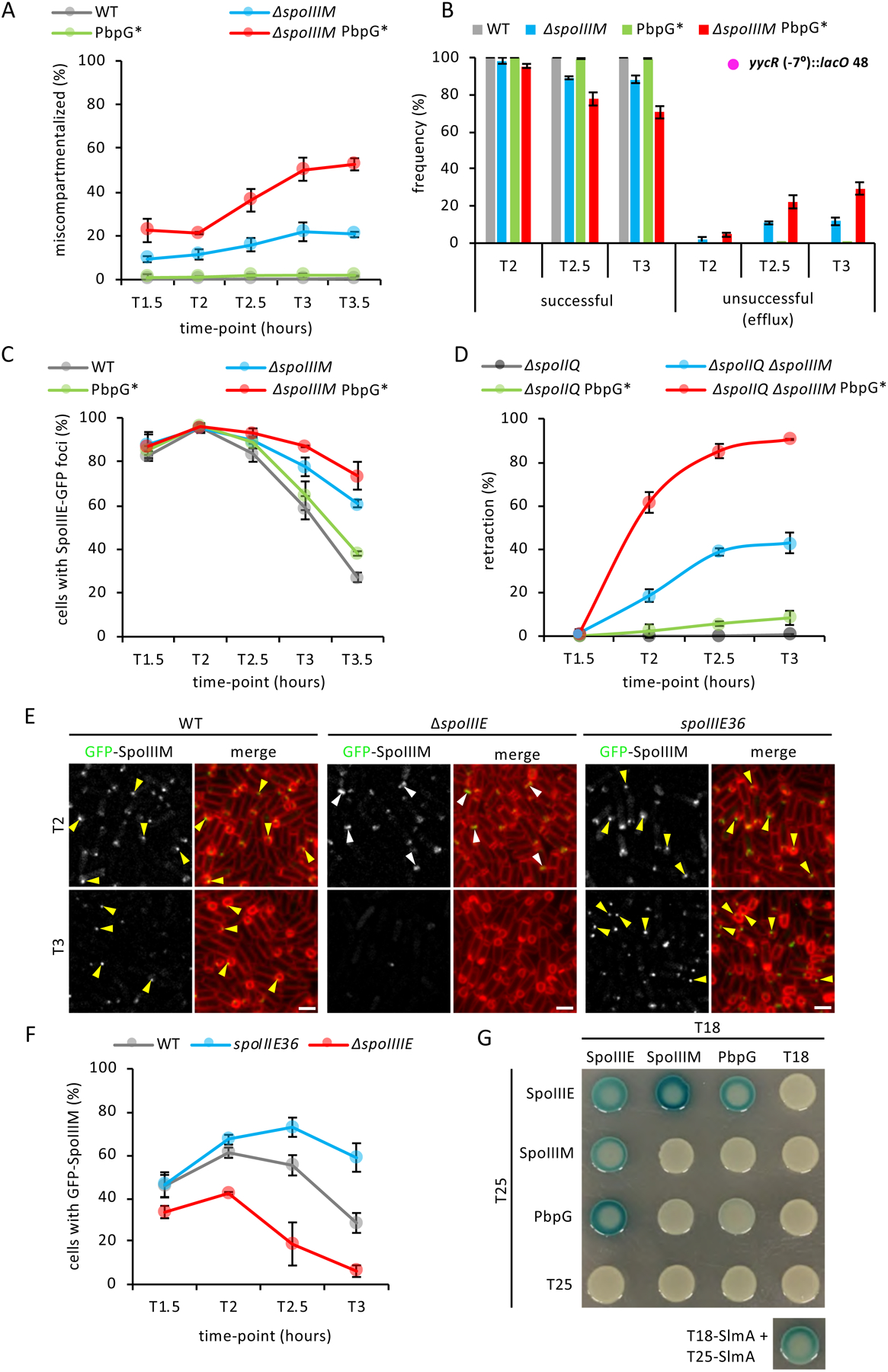
(A) Average frequency (± SD of 3 biological replicates) miscompartmentalized cells during a sporulation time-course in WT, ΔspoIIIM, pbpG* and ΔspoIIIM pbpG* strains (n>900 per time-course, per strain, per replicate). (B) Average frequency (± SD of of 3 biological replicates) of cells with a LacI-GFP focus in the forespore (successful translocation) or with no or two LacI-GFP foci in the mother cell (unsuccessful translocation, efflux) during a sporulation time-course, with lacO48 integrated at the yycR locus (−7°), in WT, ΔspoIIIM, PbpG* and ΔspoIIIM PbpG* (n>600 per time-course, per strain, per replicate). (C) Average frequency (± SD of 3 biological replicates) of cells with SpoIIIE-GFP foci during a sporulation time-course in WT (grey), ΔspoIIIM, PbpG* and ΔspoIIIM PbpG* (n>950 per time-course, per strain, per replicate). (D) Average frequency (± SD of of 3 biological replicates) of sporulating cells exhibiting septal retraction during a sporulation time-course in WT ΔspoIIIM, PbpG* and ΔspoIIIM PbpG* (n>700 per time-course, per strain, per replicate). (D) Representative images of GFP-SpoIIIM localization in WT, ΔspoIIIE and spoIIIE36 strains at T2 and T3. Yellow arrowheads indicate GFP-SpoIIIM fluorescence that appears to disperse, disappear or remain as a stable focus as engulfment nears completion in WT, ΔspoIIIE and spoIIIE36 mutant. Scale bar,2 μM. (F) Average frequency (± SD of 3 biological replicates) of cells with GFP-SpoIIIM fluorescence in WT, ΔspoIIIE and spoIIIE36 during sporulation time-course (n>750 per time-course, per strain, per replicate). (G) Bacterial two-hybrid assay of T18 and T25 fusions to SpoIIIE, SpoIIIM and PbpG. T18-SlmA and T25-SlmA interaction is included as a positive control (Cho et al., 2011).
SpoIIIM and PbpG interact with SpoIIIE
If SpoIIIM plays a role in septal pore closure, then it would likely localize to the septal membrane and exhibit a similar localization pattern as SpoIIIE. Consistent with the idea that SpoIIIM is anchored in the membrane and with its LysM domain surface-exposed, we found that a functional SpoIIIM-His6 fusion (Fig. S7C) was membrane-associated and susceptible to trypsin cleavage in a protease accessibility assay (Fig. S7A). To determine SpoIIIM’s localization, we constructed a GFP-SpoIIIM fluorescent fusion and examined its localization over a sporulation time-course (Fig. 6E & F). GFP-SpoIIIM is fully-functional as the sole source of SpoIIIM in otherwise WT cells and partially functional in the absence of PbpG (Fig. S7C). In cells that had just begun engulfment, GFP-SpoIIIM localized as a faint septal focus or septal band (Fig. 6E). A similar localization pattern was observed at intermediate stages of engulfment (Fig. 6E). As engulfment neared completion, fewer cells had GFP-SpoIIIM signal (Fig. 6E & F). GFP-SpoIIIM localization was reminiscent of the dynamic localization of GFP-SpoIIIE, suggesting that SpoIIIM may depend on SpoIIIE for its localization. To test this, we examined GFP-SpoIIIM localization in ΔspoIIIE over a sporulation time-course (Fig. 6E & F). In ΔspoIIIE, we rarely observed GFP-SpoIIIM foci; instead most cells contained GFP-SpoIIIM septal bands (Fig. 6E). Furthermore, the frequency of cells with GFP-SpoIIIM signal in ΔspoIIIE decreased earlier relative to WT cells (Fig. 6F).
To validate the localization relationship between SpoIIIE and SpoIIIM we took advantage of the spoIIIE36 mutant, which, unlike SpoIIIE, fails to disassemble as engulfment nears completion but instead persists as a stable focus (Sharp and Pogliano, 1999). If SpoIIIM localization depends on SpoIIIE, then in the spoIIIE36 mutant, GFP-SpoIIIM localization might also persist. We examined GFP-SpoIIIM localization in the spoIII36 mutant, over a sporulation time-course (Fig. 6F). Quantification of the number of cells with GFP-SpoIIIM signal (focus and bands) in the spoIIIE36 mutant, revealed that GFP-SpoIIIM foci persisted for a longer period of time (Fig. 6E & F). Similar observations, both in the ΔspoIIIE and the spoIIIE36 mutant background, were made in a merodiploid background using a stronger mother-cell promoter to drive GFP-SpoIIIM (Fig. S7D). Furthermore, immunoblot analysis of the SpoIIIM-His6 fusion confirmed that SpoIIIM stability depends on SpoIIIE (Fig. S7B). Collectively, these results suggest that SpoIIIM localization depends on SpoIIIE and these proteins form a complex in vivo.
To investigate if SpoIIIM and SpoIIIE interact directly, we used bacterial two-hybrid assays in Escherichia coli. In this assay, E. coli strain BTH101 expressed SpoIIIE and SpoIIIM fusions to domains T25 and T18 of Bordetella pertussis adenylate cyclase (Karimova et al., 1998). Positive interactions between SpoIIIE and SpoIIIM reunite T25 and T18 domains, resulting in lacZ expression, ß-galactosidase production and blue colonies on LB agar containing X-gal. We observed positive interactions, when T25 and T18 were fused to SpoIIIE and SpoIIIM (Fig. 6G). Interestingly, although PbpG appears to localize uniformly in the engulfing membranes (Ojkic et al., 2016), we also detected a positive interaction between PbpG and SpoIIIE (Fig. 6G). Collectively these results suggest that SpoIIIE, SpoIIIM and PbpG function together in maintaining PG synthesis around the septal pore during chromosome translocation, thereby ensuring compartmentalization in the face of septal PG hydrolysis.
Discussion
Critical to a better understanding of the mechanisms coordinating cytokinesis with chromosome segregation during development is a detailed appreciation of these mechanisms in simpler organisms, such as bacteria. Our data demonstrate that coordination between cytokinesis and chromosome segregation during bacterial spore development requires an intricate balance between enzymatic remodelling of cell wall material, stabilization of the division septum and chromosome segregation. Our data indicate SpoIIIE, SpoIIIM and PbpG coordinate forespore chromosome translocation with septal PG remodelling, at a highly-stabilized septal pore (Fig. 7A & C). Our observations support a model in which SpoIIIE plays two critical roles during sporulation: (1) it translocates a copy of the chromosome into the forespore compartment and (2) it functions to maintain the septal pore through which the chromosome travels during engulfment. This second role is achieved by the recruitment of SpoIIIM and PbpG. Our data further suggest that PbpG synthesizes PG around the septal pore to counteract the activity of the PG hydrolases and SpoIIIM, and SpoIIIE itself, function to reinforce the pore through protein-protein and protein-PG interactions. Finally, we reveal an important role for the highly-conserved SpoIIIAH-SpoIIQ zipper-like interaction in the stabilization of the septal membrane surrounding the pore. Together these factors maintain the septal pore and prevent its expansion due to the action of the septal PG hydrolases during engulfment (Fig.7C).
Septal PG degradation by the DMP complex plays a central role in driving engulfment (Khanna et al., 2020). However, as our data suggest, PG degradation must be counteracted to ensure the septal pore, where SpoIIIE translocates the forespore chromosome, does not expand. Our data indicate that SpoIIIE, along with PbpG and SpoIIIM, are critical for this process and suggest that they act, in part, by restoring PG surrounding the pore that is lost due to the action of the DMP complex (Fig.7B). This model is in keeping with data suggesting that remodelling of the septum during engulfment requires a balance between PG synthesis and hydrolysis (Ojkic et al., 2016) and with cryo-electron tomography data demonstrating changes in septal PG thickness before and after PG hydrolysis (Tocheva et al., 2013). Our data suggest that PbpG is responsible for localized septal PG synthesis and we hypothesize that SpoIIIE and SpoIIIM function to constrain the pore, possibly by recruiting or activating additional PG synthases in this region (Fig. 7B). Consistent with this idea, FtsW, an essential PG glycosyltransferase of the SEDS family, forms a focus in the middle of the septum at the time of asymmetric division (Burton et al., 2007).
A surprising phenotype we observed was the movement of the forespore chromosome back into the mother cell, in cells lacking SpoIIIM and PbpG (Fig. 3 and S3C & D). A similar phenotype was observed by Pogliano and co-workers in cells where SpoIIIE is targeted for degradation at the initiation of chromosome translocation (Lopez-Garrido et al., 2018). In this situation it was proposed that the chromosome is either passively expelled into the mother cell, to minimize the repulsive and hydration forces associated with packing the chromosome into the small forespore, or actively reverse translocated by SpoIIIE. Thus, it is possible that the movement out of the forespore reported here represents a similar scenario. Interestingly, we found that in cells lacking SpoIIIM and PbpG, SpoIIIE is not degraded but resides in active, stable complexes (Fig. 2). Thus, we have identified a condition where SpoIIIE is present and capable of translocating the chromosome into the forespore, but the chromosome fails to remain there. While it is tempting to hypothesize that PbpG and SpoIIIM normally function to prevent reverse translocation, we favor a simpler model in which these proteins prevent enlargement of the pore and passive efflux of forespore cytoplasmic content, and its chromosome, into the mother cell, during engulfment PG hydrolysis. In support of this model, chromosome efflux in the absence of PbpG and SpoIIIM is suppressed when engulfment PG hydrolysis is blocked or impaired (Fig. 4A and B).
An unexpected phenotype we identified in this study was that in actively engulfing cells lacking the highly-conserved SpoIIIAH-SpoIIQ zipper-like interaction and SpoIIIE, or SpoIIIM, or PbpG (or both), the asymmetric septum forms (and σF is activated), but then retracts, abolishing development (Fig.5 & S4A). The SpoIIIAH-SpoIIQ zipper-like interaction has been shown to be particularly important in sporulating cells where the PG is degraded by lysozyme (Broder and Pogliano, 2006; Blaylock et al., 2004). Similarly, we hypothesize that it also functions to prevent the septal membranes from retracting during septal PG hydrolysis. As hypothesized above, in the absence of SpoIIIE, SpoIIIM or PbpG there is reduced PG synthesis and destabilization of the septal pore. The reduced PG synthesis, combined with the thinning of septal PG at the onset of engulfment and the decreased membrane stability occurring in the absence of SpoIIIAH-SpoIIQ zipper, likely leads to progressive pore enlargement as the DMP complex makes its way outwards from the middle of the septum. Consistent with this hypothesis, we show that septal retraction can be prevented by blocking engulfment PG hydrolysis (Fig. 5C).
Overall, our results provide a more complete picture of how bacteria differentiate into spores, a morphological process that involves an asymmetric septum with a pore that must coordinate multiple developmental processes at once: the translocation of a chromosome across it, the maintenance of compartmentalization and the dramatic remodelling of the forespore envelope. More broadly, we show how coordination of different processes is critical to the establishment and maintenance of genetic and transcriptional compartmentalization during cytokinesis, key aspects of development and differentiation in all organisms. Finally, our results highlight the importance of modern approaches in genetics for distinguishing between disparate biological models.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Christopher Rodrigues (christopher.rodrigues@uts.edu.au).
Materials Availability
All materials generated in this study are available from the Lead Contact without restriction.
Data and Code Availability
The datasets generated during this study are available upon request from the Lead Contact.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Bacterial Strains and Growth Conditions
All B. subtilis strains used in this study were derived from strain 168 or PY79. B. subtilis cells were induced to sporulate via resuspension at 37°C according to the Sterlini-Mandelstam method or through exhaustion of nutrients in supplemented Difco Sporulation medium (DSM). Deletion mutant strains were either obtained from the B. subtilis Single Gene Deletion Library (Addgene) (Koo et al., 2017) or from pre-existing strains in the laboratory. All BKE mutants were back-crossed into B. subtilis 168 before assaying and prior to antibiotic cassette removal. Antibiotic cassette removal was performed using a temperature-sensitive plasmid that constitutively expresses Cre recombinase. Genetic constructs were obtained from previously existing B. subtilis strains found within the laboratory or through restriction enzyme-based cloning of PCR products into ectopic-integration, double-crossover plasmid vectors that were then transformed into B. subtilis. Tables of strains (Table S1), plasmids (Table S2), oligonucleotide primers (Table S3) can be found in the Supplemental Information.
METHOD DETAILS
Sporulation efficiency assays
Sporulation efficiency was examined using the heat-kill assay of 24–30-hour grown cultures, where the total number of heat-resistant (80°C for 20 min) colony forming units (CFUs) was compared to wild-type heat-resistant CFUs.
Transposon Insertion Sequencing
Transposon insertion sequencing (Tn-seq) was performed on WT (bDR2413) ΔpbpG (bCR1557) and ΔpbpF libraries (bCR1558), as described previously (Meeske et al., 2016). About 750,000 transformant colonies were harvested, aliquoted, and frozen. The aliquot culture was thawed on ice, resuspended in DSM, and diluted into 50 mL DSM at an OD600 of 0.05. Samples were collected at T24. The T24 samples were incubated at 80°C for 20 minutes to kill sporulation-defective cells and then plated to on LB agar allow gemination and outgrowth. Approximately, 750,000 colonies were harvested and their genomic DNA extracted and digested with MmeI enzyme to cut the MmeI restriction site inserted at the inverted repeat sequences of the magellan6 transposon. Barcode adapters were ligated and the transposon-chromosome junctions were amplified by PCR using 16 cycles. PCR products were gel-purified and sequenced on the Illumina HiSeq platform using TruSeq reagents (Tufts University TUCF Genomics facility). Reads were mapped to the B. subtilis 168 genome (NCBI NC_000964.3), tallied at each TA site.
Fluorescence Microscopy
Live-cell fluorescence imaging was performed by placing cells on a 2% (w/v) agarose pad prepared in resuspension medium and set using a gene frame (Bio-Rad). When sporulating cells reached the desired time-point, 200 μL of the culture was pelleted by centrifugation, and then resuspended in 10 μL of resuspension medium containing the membrane dye TMA-DPH (1-(4-trimethylammoniumphenyl)-6-phenyl-1,3,5-hexatriene p-toluenesulfonate) (0.05 mM) or FM 4–64 (N-(3-Triethylammoniumpropyl)-4-(6-(4-(Diethylamino) Phenyl) Hexatrienyl) Pyridinium Dibromide) (0.67 μg/μL). After gentle vortexing, 2 μL of the cell suspension was spread on the agarose pad, and a coverslip was placed on top of the gene frame. Cells were imaged by standard epifluorescence microscopy using a Zeiss Axioplan 2 Microscope equipped with 100x objective N/A 1.4. A DAPI filter to excite the TMA-DPH membrane dye with an exposure time of 400 ms. CFP, GFP, and YFP filters were used with exposure times of 600 ms, 800 ms, and 1000 ms, respectively. 3D-structured illumination microscopy was performed using the DeltaVision OMX SR microscope equipped with Olympus PlanApo N 60x objective lens N/A 1.42. The 1.515 immersion oil was selected after calculating the refractive index using softWoRx software. mCherry/A568 and GFP/A488 filters were used with exposure times of 15–20 ms and 10–15% intensity (%T).
TEM imaging
For the images in Figure S2F, sporulating cells were pelleted in 1.5-mL Eppendorf tubes at 10,000 rpm for 3 min and placed into primary fixative, consisting of 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, for 2 h at room temperature. The cells were rinsed in fresh sodium cacodylate buffer three times for 15 min each. Secondary fixation was performed using 1% osmium tetroxide and 1.5% potassium ferricyanide in cacodylate buffer for 1 h at room temperature. The cells were then rinsed in three washes of Milli-Q water for 15 min each. The fixed cell pellets were dehydrated by incubating in increasing concentrations of ethanol for 15 min, consisting of 30, 50, 70, 90 and 100% ethanol. Dehydrated cell pellets were incubated in a mixture of LR White resin and ethanol at a ratio of 1:1 for 6 h at room temperature, followed by a 2:1 LR White/ethanol mixture overnight. Cell pellets were incubated in 100% LR White resin for 6 hours, followed by another 100% resin change overnight. The cell pellets were then placed into gelatin capsules in 100% resin and the resin polymerised for 24 h in an oven at 60°C. Resin embedded tissue was sectioned with a Diatome diamond knife using a Leica UCS ultramicrotome. Sections of thickness 70 – 90 nm were collected onto formvar-coated 100 mesh copper grids and stained sequentially with 1% uranyl acetate for 10 min and lead citrate for 5 min. The sections were imaged in a JEOL 1400+ transmission electron microscope at 80kV, and images captured with a digital camera at a resolution of 2K × 2K.
For the images in Figure S4A, Sporulating cells were harvested by centrifugation at 3,220 × g for 10 min. The cell pellet (1.4 μL) was dispensed on the 200-μm side of a type A 3-mm gold platelet (Leica Microsystems), covered with the flat side of a type B 3-mm aluminum platelet (Leica Microsystems), and was vitrified by high-pressure freezing using an HPM100 system (Leica Microsystems). Following freeze-substitution at −90°C for 80 h in acetone supplemented with 1% OsO4, the samples were warmed up slowly (1°C/h) to −60°C (AFS2; Leica Microsystems). After 8 to 12 h, the temperature was raised (1°C/h) to −30°C, and the samples were kept at this temperature for another 8 to 12 h before being rinsed 4 times in pure acetone. The samples were then infiltrated with gradually increasing concentrations of resin (Embed812, EMS) in acetone (1:2, 1:1, 2:1 [v/v]) for 3 h while raising the temperature until 20°C. Pure resin was added at room temperature. After polymerization at 60°C, 80-nm-thin sections were obtained using an ultramicrotome UC7 (Leica Microsystems) and were collected on formvar-carbon-coated 100-mesh copper grids. The thin sections were post-stained for 5 min with 5% aqueous uranyl acetate, rinsed, and incubated for 2 min with lead citrate. The samples were observed using a CM12 (Philips) or Tecnai G2 Spirit BioTwin (FEI) microscope operating at 120 kV with an Orius SC1000B CCD camera (Gatan).
Immunoblot Analysis
Whole-cell lysates from sporulating cells were prepared as previously described (Rodrigues et al., 2016). Samples were heated for 15 min at 50°C prior to loading. Equivalent loading was based on OD600 at the time of harvest. Samples were separated on a 12.5% polyacrylamide gel and transferred to a PVDF membrane. Membranes were blocked in 5% non-fat milk with 0.5% Tween-20 for 1 h. Blocked membranes were probed with anti-His (1:4000) (Genscript), anti-SpoIIIAG (1:10000) (Doan et al., 2009) or anti-FtsZ (1:30000) (Hajduk et al., 2019) primary antibodies diluted into PBS with 5% non-fat milk (w/v) with 0.05% Tween- 20 at 4°C overnight. Primary antibodies were detected with horseradish-peroxidase conjugated anti-mouse or anti-rabbit antibodies (BioRad) and detected with Western Lightning ECL reagent as described by the manufacturer.
Protease accessibility assay
Protease accessibility assays were performed in sporulating cells lacking the SpoIIQ (Q) protein to ensure that the membrane proteins present in the inner and outer forespore membranes would not be artificially inaccessible because of protoplast engulfment (Broder and Pogliano, 2006). Twenty-five milliliters of sporulating cells (induced by resuspension) were harvested by centrifugation at hour 2.5 after the onset of sporulation, washed, and resuspended in 2 mL 1X SMM buffer (0.5 M sucrose, 20 mM MgCl2, 20 mM maleic acid, pH 6.5). The cells were protoplasted by lysozyme (5 mg/mL final concentration) for 10 min with slow agitation. The protoplasts were harvested by centrifugation and resuspended in 1 mL of 1X SMM. Protoplasts (100 μL) were incubated with trypsin (30 μg/mL final concentration) (Worthington), trypsin and Triton X-100 (2% final concentration), or 1X SMM for 30 min at 30°C. Reactions were terminated by the addition of 100 μL of 2X SDS-sample buffer and incubation for 5 min at 95 °C. Five microliters from each reaction were analyzed by immunoblot.
Plasmid construction
pAT001a [ycgO::spoIIIM (spec)]
pAT001a [ycgO::spoIIIM (spec)] was generated in a two-way ligation with EcoRI-BamHI PCR product containing the spoIIIM gene (oligonucleotide primers oAT001 & oAT002 and 168 genomic DNA as template) and pKM083 cut with EcoRI-BamHI. pKM083 (ycgO::spec) is an ectopic integration vector for double crossover integration at the non-essential ycgO locus.
pAT003a [ycgO::PspoIIIM-optRBS-gfp-spoIIIM (spec)]
pAT003a [ycgO::PspoIIIM-optRBS-gfp-spoIIIM (spec)] was generated in a three-way ligation with a HindIII-XhoI PCR product containing optRBS-gfp (oligonucleotide primers oAT005 & oAT006 and pCR227 DNA as template), and an XhoI-BamHI PCR product containing the spoIIIM gene (oligonucleotide primers oAT007 & oAT002) with pAT002a (ycgO::PspoIIIM) as template cut with HindIII and BamHI. pCR227 [yhdG::PgerM-gerM-gfp] (cat) contains gfp (C. Rodrigues - laboratory stock).
pAT024a [ycgO::PspoIID-optRBS-gfp-spoIIIM (spec)]
pAT024a [ycgO::PspoIID-optRBS-gfp-spoIIIM (spec)] was generated in a two-way ligation of an EcoRI-HindIII digest product from pCR113 containing PspoIID, into an HindIII-BamHI digest product of pAT003a (ycgO::PspoIIIM-optRBS-gfp-spoIIIM). pCR113 [ycgO PspoIID malE (cat)] contains the PspoIID promoter (C. Rodrigues - laboratory stock).
pAT032a [ycgO::PspoIIIM-spoIIIM-His6 (spec)]
pAT032a [ycgO::PspoIIIM-spoIIIM-His6 (spec)] was generated in a two-way ligation with EcoRI-BamHI PCR product containing the spoIIIM gene with a C-terminal hexahistidine tag (oligonucleotide primers oAT001 & oCR659 and 168 genomic DNA as template) and pKM083 cut with EcoRI-BamHI. pKM083 (ycgO::spec) is an ectopic integration vector for double crossover integration at the non-essential ycgO locus.
pAT057a [pelB::spoIIIE-GFP (cat)]
pAT057a [pelB::spoIIIE-GFP (cat)] was generated in a three-way ligation with a EcoRI-XhoI PCR product containing spoIIIE (oligonucleotide primers oCR694 & oCR695 and bBB042 DNA as template), and an XhoI-BamHI PCR product containing gfp (oligonucleotide primers oCR696 & oCR697) with pKM020 (pelB::tet) as template cut with EcoRIand BamHI.
pAT090 [ycgO::spoIIIE36 (tet)]
pAT090 [ycgO::spoIIIE36 (tet)] was generated in a two-way ligation with an EcoRI-BamHI PCR product containing spoIIIE36 (oligonucleotide primers oCR694 & oCR717 and bBB069 DNA as template) with pKM086 (ycgO::tet) cut with EcoRIand BamHI.
pCR20a [amyE::PspoIIQ-spoIVFRBS-lacI-gfp (spec)]
pCR20a [amyE::PspoIIQ-spoIVFRBS-lacI-gfp (spec)] was generated in a three-way ligation with HindIII-XhoI digest product containing spoIVFRBS-lacI (oCR416 & oCR417 and bXW1200 DNA as template), XhoI-BamHI digest product containing gfp (oCR418 and oCR353 and pCR154 as template) and pCR154 (amyE::PspoIIQ-optRBS-gfp (spec) (C. Rodrigues - laboratory stock) cut with HindIII-BamHI.
pJL013 [pelB::spoIIIE36 (tet)]
pJL013 [pelB::spoIIIE36 (tet)] was generated in a two-way ligation with an EcoRI-BamHI digest product containing spoIIIE36 from pAT090 (ycgO::spoIIIE36) with pKM033 (pelB::tet) as template cut with EcoRIand BamHI.
pHC3 [ycgO::PspoIIQ-optRBS-pbpG-His6 (tet)]
pHC3 [ycgO::PspoIIQ-optRBS-pbpG-His6 (tet)] was generated in a two-way ligation with HindIII-BamHI PCR product containing the pbpG gene [oligonucleotide primers oAT139 & oAT160 and 168 genomic DNA as template] and pCR008 cut with HindIII and BamHI. pCR008 [ycgO::PspoIIQ-optRBS-gfp-spoIIQ (tet)] contains the PspoIIQ promoter (C. Rodrigues - laboratory stock).
pHC23 [ycgO::PspoIIQ-optRBS-pbpG-His6 (E94A, S365A) (tet)]
pHC23 [ycgO::PspoIIQ-optRBS-pbpG-His6 (E94A, S365A) (tet)] was generated by sequential rounds of site-directed mutagenesis of pHC3 using oligonucleotide primers oAT96 & oAT97 to introduce the E94A mutation, followed by oAT147 & oAT148 to introduce the S365A mutation.
pHC28 [Plac::T18-spoIIIE (amp)] and pHC32 [Plac::T25-spoIIIE (kan)]
pHC28 [Plac::T18-spoIIIE (amp)] and pHC32 [Plac::T25-spoIIIE (kan)] were generated in a two-way ligation with XbaI-BamHI PCR product containing the spoIIIE gene [oligonucleotide primers oCR716 & oCR717 and 168 genomic DNA as template] and pUT18C and pKNT25 (Karimova et al., 1998) , respectively, cut with XbaI and BamHI.
pHC29 [Plac::T18-spoIIIM (amp)] and pHC33 [Plac::T25-spoIIIM (kan)]
pHC29 [Plac::T18-spoIIIM (amp)] and pHC33 [Plac::T25-spoIIIM (kan)] were generated in a two-way ligation with XbaI-BamHI PCR product containing the spoIIIM gene [oligonucleotide primers oCR718 & oCR719 and 168 genomic DNA as template] and pUT18C and pKNT25 (Karimova et al., 1998) , respectively, cut with XbaI and BamHI.
pHC30 [Plac::T18-pbpG (amp)] and pHC34 [Plac::T25-pbpG (kan)]
pHC30 [Plac::T18-pbpG (amp)] and pHC34 [Plac::T25-pbpG (kan)] were generated in a two-way ligation with XbaI-BamHI PCR product containing the pbpG gene [oligonucleotide primers oCR720 & oCR721 and 168 genomic DNA as template] and pUT18C and pKNT25 (Karimova et al., 1998), respectively, cut with XbaI and BamHI.
Other constructs
[ycgO::PpbpG-his6-pbpG (erm)] and [ycgO::PpbpG-his6-pbpGE94A,S365A (erm)]
HindIII-PpbpG was amplified by oAT137 and oHC056 using168 genomic DNA as template. The his6-pbpG-XhoI and his6-pbpGE94, S365A-XhoI DNA fragments were amplified by oHC055 and oCR704 using pHC3 and pHC23 as templates respectively. HindIII-PpbpG and his6-pbpG-XhoI and similarly HindIII-PpbpG and his6-pbpG E94,S365A-XhoI were ligated by Isothermal assembly using oHC055 and oHC056 to generate and then ligated into pKM084 (ycgO::erm) as template cut with HindIII and XhoI. Finally, after ligation into pKM084, the two constructs were transformed directly into B. subtilis.
QUANTIFICATION AND STATISTICAL ANALYSIS
Sporulation Efficiency Analysis
All the sporulation efficiency data are average (± SD) of 3 biological replicates.
Tn-seq Analysis
Mapped genes in which reads were statistically underrepresented were identified using the Mann Whitney U test. Visual inspection of transposon insertion profiles was performed with the Sanger Artemis Genome Browser and Annotation tool.
Image Analysis
Microscopy images were processed by adjusting the brightness and contrast using the Fiji software (Schindelin et al., 2012). All quantifications were performed using the manual counting tool of the Fiji software, and the raw data was then exported into Excel for data collation and graph generation. At least two fields of view (90.13 × 67.53 μM) were acquired for each biological replicate and contained hundreds of cells, each. The criteria described below, or in results and legends, was used to quantify each phenotype.
Quantification of forespore miscompartmentalization (Fig. 1G, 4E, 6A, S2D & S3B and): any cell with forespore-expressed CFP signal in the mother cell was classified as miscompartmentalized. The percentage of miscompartmentalized cells (average frequency ± SD of 3 biological replicates) was calculated based on the total number of sporulating cells that had progressed into the asymmetric division stage of development and contained CFP signal. n>750 per time-course, per strain, per replicate.
Quantification of chromosome translocation using the LacI-GFP / lacO system (Fig. 3C & D, 4A & B, 6B, S3C & D): we followed the criteria described in Figure 3A and the percentage of cells (average frequency ± SD of 3 biological replicates) was calculated based on the total number of sporulating cells that contained a GFP focus and had progressed into or beyond asymmetric division. n>600 per time-course, per strain, per replicate.
Quantification of chromosome translocation using the dual reporter system (TetR-CFP/ tetO & LacI-YFP/ lacO): we followed the criteria described in Figure S3E and the percentage of cells (average frequency ± SD of 3 biological replicates) was calculated relative to the total number of sporulating cells that contained four foci, two CFP foci and two YFP foci. Not all cells contained four visible foci within the plane of view during development: the four foci were clearly visible in approximately 25%, 30% and 47% of the sporulating cells, at T2, T2.5 and T3 respectively. n>30 per time-point, per replicate.
Quantification of septal retraction (Fig. 5E & 6D): any cell with forespore-expressed CFP signal in the mother cell and a non-continuous asymmetric septum, or vestiges of an asymmetric septum (based on the membrane signal), was classified as having a retracted septum. The percentage of cells with retracted septa (average frequency ± SD of of 3 biological replicates) was calculated based on the total number of sporulating cells that had progressed into or beyond asymmetric division. n>600 per time-point, per strain, per replicate.
Quantification of cells with a SpoIIIE-GFP focus (Fig. 2D & 6C): the percentage of cells with a SpoIIIE-GFP focus (average frequency ± SD of of 3 biological replicates) was calculated based on the number of sporulating cells that contained a GFP focus, relative to the total number of sporulating cells that contained SpoIIIE-GFP signal (foci or diffuse) in the membranes of the forespore. n>950 per time-course, per strain, per replicate.
Quantification of GFP-SpoIIIM (Fig. 6F): the percentage of cells with a GFP-SpoIIIM focus (average frequency ± SD of 3 biological replicates) was calculated based on the number of sporulating cells that contained GFP signal (focus or band), relative to the total number of sporulating cells that had progressed into or beyond asymmetric division. n>750 per time-course, per strain, per replicate.
The MicrobeJ plugin (Ducret et al., 2016) designed for the Fiji software was used to detect the signal intensity of SpoIIIE-GFP foci (Fig. 2E), on background subtracted images (Process > Subtract Background) to avoid false positive detections of the fluorescent signal. To determine the signal intensity of SpoIIIE-GFP foci, the brightfield image was set to Channel 1 and the image with the GFP signal to Channel 2. These channels were then merged into one image. For Channel 1 of the image with “Bright” background, the “Bacteria” tab was set to “Fit Shape” and “Rod-Shaped” to detect the bacteria in the brightfield image. Five parameters: “Exclude on Edges”, “Shape descriptors”, “Segmentation”, “Intensity”, and “Feature” were checked on. To refine the generated bacteria outlines, the shape descriptors (area, length, and width) were set differently for each time-point corresponding to the contours of individual cell. To resolve unprocessed cells, the manual editing tool was also used. For Channel 2 of the image with “Dark” background, the “Maxima” tab was set to “Point” and “Basic”. The tolerance was set to 15, and the intensity was set from 15-max to ensure detection of the GFP foci with a minimum signal intensity of 15. Three parameters: “Exclude on Edges”, “Shape descriptors”, and “Associations” were checked on. The raw data were displayed on a MicrobeJ results table, which was then exported into Excel for data collation and graph generation.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| THE™ His Tag Antibody, mAb, Mouse | GenScript | Cat#A00186 |
| anti-FtsZ | (Hajduk et al., 2019) | N/A |
| anti-SpoIIIAG | (Doan et al., 2009) | N/A |
| Bacterial Strains | ||
| bBB042 | (Burton et al., 2007) | N/A |
| bBB069 | (Besprozvannaya et al., 2014) | N/A |
| bDR2413 (168) | (Zeigler et al., 2008) | N/A |
| bWX1200 | (Wang et al., 2014) | N/A |
| See Table S1 for a complete list of strains used in this study. | N/A | N/A |
| Chemicals | ||
| FM4–64 | Thermo Fisher Scientific | Cat#T13320 |
| SYTOX® Orange | Thermo Fisher Scientific | Cat#S11368 |
| TMA-DPH | Thermo Fisher Scientific | Cat#T204 |
| Oligonucleotides | ||
| See Table S2 for a complete list of oligonucleotides used in this study. | Integrated DNA Technologies (IDT) | https://sg.idtdna.com/ |
| Recombinant DNA | ||
| Plac::T25-slmA (kan) | (Cho et al., 2011) | pHC535 |
| Plac::T18-slmA (amp) | (Cho et al., 2011) | pHC538 |
| See Table S3 for a complete list of plasmids constructed for this study. | N/A | N/A |
| Software and Algorithms | ||
| Fiji software | (Schindelin et al., 2012) | https://imagej.net/Welcome |
| MicrobeJ | (Ducret et al., 2016) | https://www.microbej.com/ |
Acknowledgments
We thank the members of the Duggin and Harry laboratories for their support and encouragement, the Harry laboratory for their gift of FtsZ antibody, Xindan Wang for the gift of strains, Briana Burton for the gift of strains and stimulating discussions, Leonie Herson (www.leotide.com) for scientific illustrations (Fig. 1A, B & C and 7A & C), Christian Evenhuis and Giulia Gitana from the Microbial Imaging Facility for help and advice on image data collection and analysis, particularly with 3D-SIM and Sarah Osvath for her dedicated support of our laboratory in the Bioscience Facility. This work was funded by grant DP190100793 awarded to C.M, D. Z. R and C.D.A.R, from the Australian Research Council (https://www.arc.gov.au).
Footnotes
Declaration of Interests
The authors declare no competing interests.
References
- BARRE FX 2007. FtsK and SpoIIIE: the tale of the conserved tails. Mol Microbiol, 66, 1051–1055. [DOI] [PubMed] [Google Scholar]
- BEN-YEHUDA S, RUDNER DZ & LOSICK R 2003. Assembly of the SpoIIIE DNA translocase depends on chromosome trapping in Bacillus subtilis. Curr Biol, 13, 2196–2200. [PubMed] [Google Scholar]
- BESPROZVANNAYA M, PIVORUNAS VL & BURTON BM 2014. Mechanistic study of classical translocation-dead SpoIIIE36 reveals the functional importance of the hinge within the SpoIIIE motor. Journal of Bacteriology, 196, 2481–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BESPROZVANNAYA M, PIVORUNAS VL, FELDMAN Z & BURTON BM 2013. SpoIIIE protein achieves directional DNA translocation through allosteric regulation of ATPase activity by an accessory domain. Journal of Biological Chemistry, 288, 28962–28974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BLAYLOCK B, JIANG X, RUBIO A, MORAN CP JR. & POGLIANO K 2004. Zipper-like interaction between proteins in adjacent daughter cells mediates protein localization. Genes Dev, 18, 2916–2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRODER DH & POGLIANO K 2006. Forespore Engulfment Mediated by a Ratchet-Like Mechanism. Cell, 126, 917–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUIST G, STEEN A, KOK J & KUIPERS OP 2008. LysM, a widely distributed protein motif for binding to (peptido)glycans. Molecular Microbiology, 68, 838–847. [DOI] [PubMed] [Google Scholar]
- BURTON B & DUBNAU D 2010. Membrane-associated DNA transport machines. Cold Spring Harb Perspect Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BURTON BM, MARQUIS KA, SULLIVAN NL, RAPOPORT TA & RUDNER DZ 2007. The ATPase SpoIIIE Transports DNA across Fused Septal Membranes during Sporulation in Bacillus subtilis. Cell, 131, 1301–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CATTONI DI, CHARA O, GODEFROY C, MARGEAT E, TRIGUEROS S, MILHIET PE & NÖLLMANN M 2013. SpoIIIE mechanism of directional translocation involves target search coupled to sequence-dependent motor stimulation. EMBO Reports, 14, 473–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CATTONI DI, THAKUR S, GODEFROY C, LE GALL A, LAI-KEE-HIM J, MILHIET PE, BRON P & NOLLMANN M 2014. Structure and DNA-binding properties of the Bacillus subtilis SpoIIIE DNA translocase revealed by single-molecule and electron microscopies. Nucleic Acids Res, 42, 2624–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHASTANET A & LOSICK R 2007. Engulfment during sporulation in Bacillus subtilis is governed by a multi-protein complex containing tandemly acting autolysins. Molecular Microbiology, 64, 139–152. [DOI] [PubMed] [Google Scholar]
- CHO H, MCMANUS HR, DOVE SL & BERNHARDT TG 2011. Nucleoid occlusion factor SlmA is a DNA-activated FtsZ polymerization antagonist. Proc Natl Acad Sci U S A, 108, 3773–3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOAN T 2013. FisB mediates membrane fission during sporulation in bacillus subtilis. Genes & Development, 10.1101/gad.209049.112.during, 322–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOAN T, COLEMAN J, MARQUIS KA, MEESKE AJ, BURTON BM, KARATEKIN E & RUDNER DZ 2013. FisB mediates membrane fission during sporulation in Bacillus subtilis. Genes Dev, 27, 322–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOAN T, MORLOT C, MEISNER J, SERRANO M, HENRIQUES AO, MORAN CP & RUDNER DZ 2009. Novel secretion apparatus maintains spore integrity and developmental gene expression in Bacillus subtilis. PLoS Genetics, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DUCRET A, QUARDOKUS EM & BRUN YV 2016. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat Microbiol, 1, 16077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EICHENBERGER P, JENSEN ST, CONLON EM, VAN OOIJ C, SILVAGGI J, GONZÁLEZ-PASTOR JE, FUJITA M, BEN-YEHUDA S, STRAGIER P, LIU JS & LOSICK R 2003. The σEregulon and the identification of additional sporulation genes in Bacillus subtilis. Journal of Molecular Biology, 327, 945–972. [DOI] [PubMed] [Google Scholar]
- FICHE JB, CATTONI DI, DIEKMANN N, LANGERAK JM, CLERTE C, ROYER CA, MARGEAT E, DOAN T & NÖLLMANN M 2013. Recruitment, Assembly, and Molecular Architecture of the SpoIIIE DNA Pump Revealed by Superresolution Microscopy. PLoS Biology, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FLEMING TC, SHIN JY, LEE SH, BECKER E, HUANG KC, BUSTAMANTE C & POGLIANO K 2010. Dynamic SpoIIIE assembly mediates septal membrane fission during Bacillus subtilis sporulation. Genes and Development, 24, 1160–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRASCHINI R 2020. Cytokinesis in Eukaryotic Cells: The Furrow Complexity at a Glance. Cells, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GRAINGE I 2010. FtsK - a bacterial cell division checkpoint? Molecular Microbiology, 78, 1055–1057. [DOI] [PubMed] [Google Scholar]
- HAJDUK IV, MANN R, RODRIGUES CDA & HARRY EJ 2019. The ParB homologs, Spo0J and Noc, together prevent premature midcell Z ring assembly when the early stages of replication are blocked in Bacillus subtilis. Mol Microbiol, 112, 766–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HILBERT DW, CHARY VK & PIGGOT PJ 2004. Contrasting Effects of σE on Compartmentalization of σF Activity during Sporulation of Bacillus subtilis. Journal of Bacteriology, 186, 1983–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KARIMOVA G, PIDOUX J, ULLMANN A & LADANT D 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci U S A, 95, 5752–5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KHANNA K, LOPEZ-GARRIDO J & POGLIANO K 2020. Shaping an Endospore: Architectural Transformations During Bacillus subtilis Sporulation. Annu Rev Microbiol, 10.1146/annurev-micro-022520-074650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KHANNA K, LOPEZ-GARRIDO J, ZHAO Z, WATANABE R, YUAN Y, SUGIE J, POGLIANO K & VILLA E 2019. The molecular architecture of engulfment during Bacillus subtilis sporulation. Elife, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOO BM, KRITIKOS G, FARELLI JD, TODOR H, TONG K, KIMSEY H, WAPINSKI I, GALARDINI M, CABAL A, PETERS JM, HACHMANN AB, RUDNER DZ, ALLEN KN, TYPAS A & GROSS CA 2017. Construction and Analysis of Two Genome-Scale Deletion Libraries for Bacillus subtilis. Cell Syst, 4, 291–305 e297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LESAGE G & BUSSEY H 2006. Cell wall assembly in Saccharomyces cerevisiae. Microbiol Mol Biol Rev, 70, 317–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIU NJL, DUTTON RJ & POGLIANO K 2006. Evidence that the SpoIIIE DNA translocase participates in membrane fusion during cytokinesis and engulfment. Molecular Microbiology, 59, 1097–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOPEZ-GARRIDO J, OJKIC N, KHANNA K, WAGNER FR, VILLA E, ENDRES RG & POGLIANO K 2018. Chromosome Translocation Inflates Bacillus Forespores and Impacts Cellular Morphology. Cell, 172, 758–770.e714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARQUIS KA, BURTON BM, NOLLMANN M, PTACIN JL, BUSTAMANTE C, BEN-YEHUDA S & RUDNER DZ 2008. SpoIIIE strips proteins off the DNA during chromosome translocation. Genes & development, 22, 1786–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCPHERSON DC, DRIKS A & POPHAM DL 2001. Two Class A High-Molecular-Weight Penicillin-Binding Proteins of. 183, 6046–6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MEESKE J, RODRIGUES C, BRADY J, LIM HC, BERNHARDT TG & RUDNER DZ 2016. High-Throughput Genetic Screens Identify a Large and Diverse Collection of New Sporulation Genes in Bacillus subtilis. PLoS Biology, 14, 1–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MORLOT C, UEHARA T, MARQUIS KA, BERNHARDT TG & RUDNER DZ 2010. A highly coordinated cell wall degradation machine governs spore morphogenesis in Bacillus subtilis. Genes and Development, 24, 411–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OJKIC N, LÓPEZ-GARRIDO J, POGLIANO K & ENDRES RG 2014. Bistable Forespore Engulfment in Bacillus subtilis by a Zipper Mechanism in Absence of the Cell Wall. PLoS Computational Biology, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OJKIC N, LÓPEZ-GARRIDO J, POGLIANO K & ENDRES RG 2016. Cell-wall remodeling drives engulfment during Bacillus subtilis sporulation. eLife, 5, 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PEREZ AR, ABANES-DE MELLO A & POGLIANO K 2000. SpoIIB localizes to active sites of septal biogenesis and spatially regulates septal thinning during engulfment in Bacillus subtilis. Journal of Bacteriology, 182, 1096–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PIGGOT PJ & HILBERT DW 2004. Sporulation of Bacillus subtilis. Current Opinion in Microbiology, 7, 579–586. [DOI] [PubMed] [Google Scholar]
- PTACIN JL, NOLLMANN M, BECKER EC, COZZARELLI NR, POGLIANO K & BUSTAMANTE C 2008. Sequence-directed DNA export guides chromosome translocation during sporulation in Bacillus subtilis. Nature Structural and Molecular Biology, 15, 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RODRIGUES CDA, MARQUIS KA, MEISNER J & RUDNER DZ 2013. Peptidoglycan hydrolysis is required for assembly and activity of the transenvelope secretion complex during sporulation in Bacillus subtilis. Molecular Microbiology, 89, 1039–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RODRIGUES C, HENRY X, NEUMANN E, KURAUSKAS V, BELLARD L, FICHOU Y, SCHANDA P, SCHOEHN G, RUDNER DZ & MORLOT C 2016. A ring-shaped conduit connects the mother cell and forespore during sporulation in Bacillus subtilis. Proceedings of the National Academy of Sciences, 113, 11585–11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHINDELIN J, ARGANDA-CARRERAS I, FRISE E, KAYNIG V, LONGAIR M, PIETZSCH T, PREIBISCH S, RUEDEN C, SAALFELD S, SCHMID B, TINEVEZ JY, WHITE DJ, HARTENSTEIN V, ELICEIRI K, TOMANCAK P & CARDONA A 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods, 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SEILER S & JUSTA-SCHUCH D 2010. Conserved components, but distinct mechanisms for the placement and assembly of the cell division machinery in unicellular and filamentous ascomycetes. Mol Microbiol, 78, 1058–1076. [DOI] [PubMed] [Google Scholar]
- SHARP MD & POGLIANO K 1999. An in vivo membrane fusion assay implicates SpoIIIE in the final stages of engulfment during Bacillus subtilis sporulation. Proceedings of the National Academy of Sciences, 96, 14553–14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHARP MD & POGLIANO K 2002. Role of cell-specific SpoIIIE assembly in polarity of DNA transfer. Science (New York, N.Y.), 295, 137–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHERRATT DJ, ARCISZEWSKA LK, CROZAT E, GRAHAM JE & GRAINGE I 2010. The Escherichia coli DNA translocase FtsK. [DOI] [PubMed]
- STRAGIER P & LOSICK R 1996. Molecular Genetics of Sporulation in Bacillus Subtilis. Annu. Rev. Genet, 30, 297–341. [DOI] [PubMed] [Google Scholar]
- SULLIVAN NL, MARQUIS KA & RUDNER DZ 2009. Recruitment of SMC by ParB-parS organizes the origin region and promotes efficient chromosome segregation. Cell, 137, 697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TOCHEVA EI, LÓPEZ-GARRIDO J, HUGHES HV, FREDLUND J, KURU E, VANNIEUWENHZE MS, BRUN YV, POGLIANO K & JENSEN GJ 2013. Peptidoglycan transformations during Bacillus subtilis sporulation. 88, 673–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNIPROT CONSORTIUM T 2018. UniProt: the universal protein knowledgebase. Nucleic Acids Res, 46, 2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG X, MONTERO LLOPIS P & RUDNER DZ 2014. Bacillus subtilis chromosome organization oscillates between two distinct patterns. Proc Natl Acad Sci U S A, 111, 12877–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WU LJ & ERRINGTON J 1997. Septal localization of the SpoIIIE chromosome partitioning protein in Bacillus subtilis. EMBO Journal, 16, 2161–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WU LJ & ERRINGTON J 1998. Use of asymmetric cell division and spoIIIE mutants to probe chromosome orientation and organization in Bacillus subtilis. Molecular Microbiology, 27, 777–786. [DOI] [PubMed] [Google Scholar]
- YEN SHIN J, LOPEZ-GARRIDO J, LEE S-H, DIAZ-CELIS C, FLEMING T, BUSTAMANTE C & POGLIANO K 2015. Visualization and functional dissection of coaxial paired SpoIIIE channels across the sporulation septum. 10.7554/eLife.06474.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZEIGLER DR, PRÁGAI Z, RODRIGUEZ S, CHEVREUX B, MUFFLER A, ALBERT T, BAI R, WYSS M & PERKINS JB 2008. The origins of 168, W23, and other Bacillus subtilis legacy strains. Journal of Bacteriology, 190, 6983–6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during this study are available upon request from the Lead Contact.


