Abstract
The intestinal epithelium represents a natural barrier against harmful xenobiotics, while facilitating the uptake of nutrients and other substances. Understanding the interaction of chemicals with constituents of the intestinal epithelium and their fate in the body requires quantitative measurement of relevant proteins in in vitro systems and intestinal epithelium. Recent studies have highlighted the mismatch between messenger RNA (mRNA) and protein abundance for several drug‐metabolizing enzymes and transporters in the highly dynamic environment of the intestinal epithelium; mRNA abundances cannot therefore be used as a proxy for protein abundances in the gut, necessitating direct measurements. The objective was to determine the expression of a wide range proteins pertinent to metabolism and disposition of chemicals and nutrients in the intestinal epithelium. Ileum and jejunum biopsy specimens were obtained from 16 patients undergoing gastrointestinal elective surgery. Mucosal fractions were prepared and analyzed using targeted and global proteomic approaches. A total of 29 enzymes, 32 transporters, 6 tight junction proteins, 2 adhesion proteins, 1 alkaline phosphatase, 1 thioredoxin, 5 markers, and 1 regulatory protein were quantified—60 for the first time. The global proteomic method identified a further 5,222 proteins, which are retained as an open database for interested parties to explore. This study significantly expands our knowledge of a wide array of proteins important for xenobiotic handling in the intestinal epithelium. Quantitative systems biology models will benefit from the novel systems data generated in the present study and the translation path offered for in vitro to in vivo translation.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
☑ The intestine expresses a variety of proteins interacting with ingested material. Some of these (e.g., enzymes and transporters) have been quantified in a limited number of cases, but, for many key proteins, there is a complete lack of quantitative data.
WHAT QUESTION DID THIS STUDY ADDRESS?
☑ We used global proteomics to quantify protein expression in two intestinal regions with the aim of understanding patterns of expression, interindividual variability, and of generating data to feed systems pharmacology models of drug distribution.
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
☑ The unique data set generated here (> 5,000 proteins) includes quantitative measurements of proteins in intestinal epithelium for the first time. It provides protein expression levels of 77 proteins involved in the metabolism and traffic of drugs across the intestinal epithelium.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
☑ The data presented here are an essential resource for systems biology models to improve our understanding of the pathogenesis of intestinal disease, the physiology of nutrient absorption, and the pharmacological basis of therapy in various patient populations.
The intestinal epithelium contains many proteins capable of modulating the influx and efflux of chemicals and even of metabolizing xenobiotics before they gain access to the portal venous circulation and, ultimately, the systemic blood. 1 Predicting the fate of xenobiotics (e.g., environmental toxins or therapeutic drugs) after ingestion is challenging, not least because animal studies are poorly predictive of human intestinal metabolism; 2 the nature, quantity, and even topology of enzymes and transporters in the human intestinal epithelium appear to be significantly different from those in experimental animals. 3 The possibility of quantitative extrapolation from in vitro to in vivo (IVIVE) systems has driven the recent surge in the use of various microphysiological systems and organ‐on‐chip methodologies. 4 However, these require quantitative knowledge of relevant proteins in both the in vitro systems and the target tissue. 5
Messenger RNA (mRNA) assays are popular surrogates for measuring proteins in human tissues, but mRNA expression is a poor indicator of protein abundance in dynamic (non–steady‐state) conditions. 6 The intestinal epithelium is a highly dynamic environment because of continuous exposure to high concentrations of inducers (ingested compounds) and the short life span of enterocytes. 7 There is no good substitute for quantitative protein measurements.
To our knowledge there have been only 11 previous publications on quantitative proteomics of human intestinal epithelium 6 , 8 , 9 , 10 , 11 , 12 , 13 , 14 , 15 and a further two exploring colon cancer. 16 , 17 Most of these involved targeted proteomics of specific proteins. Global methods are generally considered to be complementary, sacrificing some precision in quantification in return for measurement of a vast number of proteins simultaneously (> 5,000)—economical when the aim is to characterize a wide range of often interrelated proteins. Different liquid chromatography–mass spectrometry methodologies have been reviewed extensively to assess the choice of the most appropriate mass spectrometry–based approach for the quantification of proteins relevant to IVIVE. 18
Robust estimates of physiologically relevant scaling factors, such as drug‐metabolizing enzymes (DMEs) and drug transporters, are required for IVIVE integrated in physiologically‐based pharmacokinetic (PBPK) models for prediction of appropriate drug dosing for different populations. 19 However, it is often difficult to harness protein abundance values typically reported as “pmol/milligram of a protein matrix” into a mechanistic IVIVE‐PBPK strategy. 12 An often overlooked yet critical “tissue‐based” scaling factor, essentially a tissue‐matrix protein yield (mg), is needed to translate the reported abundance data to estimates of whole organ protein abundances required for scaling the kinetic activity data to characterize their impact on disposition in a whole body model.
In this novel study, global and targeted (accurate mass and retention time, AMRT) 20 proteomics were applied to determine the levels of proteins in mucosal protein preparations from the human jejunum and ileum. More than 5,000 intestinal epithelial proteins were identified and quantified. We specifically investigated cytochrome P450 (CYP), uridine diphosphate (UDP)‐glucuronosyltransferase (UGT), transporters, transferases, oxygenases, tight junction proteins, regulatory proteins, and cell markers with the aim of understanding patterns of expression, interindividual variation, and differences in abundance between jejunum and ileum. The same set of samples was analyzed in one of our previous reports, 6 which focused on a limited number of DMEs and drug transporters and addressed experimental optimization. To facilitate understanding the impact of intestinal epithelial proteins on the fate of xenobiotics, the incorporation of a translational pathway for the transporter protein abundances reported herein into an IVIVE‐PBPK strategy is addressed.
Materials and Methods
Detailed methods are described in the Supplementary Materials .
Human intestinal samples
After informed consent, human small intestinal tissues were obtained from 16 patients undergoing elective surgery at Salford Royal NHS Foundation Trust, United Kingdom. Prior ethics committee approval was granted by the North West Research Ethics Committee, United Kingdom (06/1410/126), and all procedures were carried out in accordance with the Declaration of Helsinki guidelines. The human intestinal samples were taken from macroscopically normal regions, at least 5 cm from obviously diseased bowel. However, donors were undergoing elective surgery for an array of conditions, and impact of disease on the proteome cannot be completely ruled out. Following sample removal, mucosal tissue was rapidly separated from muscle and serosal layers by blunt dissection. Samples of mucosa (~1 cm2) were snap frozen in liquid nitrogen and stored at −80°C for subsequent analysis. Demographic information is provided in Supplementary Table S1 .
Tissue preparation and isolation of human intestine mucosal tissue fractions
Snap‐frozen tissue samples were ground to powder using a cooled mortar and pestle, and resuspended in buffer containing 150 mM sodium chloride, 1% Triton X‐100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 50 mM Tris base pH 8.0, with protease inhibitors (0.5 mM phenylmethanesulfonylfluoride and 50 µL/mL Protease Inhibitor Cocktail (Sigma Aldrich, Poole, UK). Following incubation and mixing at 4°C for 30 minutes, the extract was centrifuged at 14,000 g for 5 minutes at 4°C. The supernatant was removed and stored in aliquots at −80°C. Protein concentrations were determined using a Bradford assay (Bio‐Rad, Paisley, UK).
Sample preparation and processing
Briefly, two technical replicates for each of the 16 human small intestinal samples were prepared, spiked with quantification concatemers (QconCATs), 21 and subjected to the filter‐aided sample preparation protocol. 6 For global and AMRT liquid chromatography‐tandem mass spectrometry analysis, a Q Exactive HF Hybrid Quadrupole‐Orbitrap mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) was used.
Data analysis and protein quantification
Quantitative analysis of global proteomic data was carried out using the “Hi3” method on Progenesis software version 4.0 (Nonlinear Dynamics, Newcastle upon Tyne, UK) as previously described. 20 The reference protein (ATP1A1; subunit alpha‐1 of sodium–potassium adenosine triphosphatase) was quantified previously using multiple reaction monitoring (MRM). 6 The (targeted) AMRT method was also performed ( Supplementary Material ). Percentage identical peptides and percentage identical proteins were estimated as previously described to assess the integrity of replicate analyses and the overall similarity of samples (Tables S2B and S3 ). 20
Statistical analysis
Statistical data analysis was performed using Microsoft Excel 2010 (Redmond, Washington), GraphPad Prism version 7.03 (La Jolla, California), and R version 3.4.3 (Vienna, Austria). The Spearman rank‐order correlation (Rs) test, with t distribution of the P values, was used to assess intercorrelations between protein abundance levels. Differences between expression levels in jejunum and ileum were assessed using Mann‐Whitney U test.
Results
Characteristics of human intestinal mucosa samples
Four jejunum and 12 ileum samples were analyzed from donors with an age range of 17–80 years. The identified proteins (4,442 from jejunum and 4,386 from ileum) were annotated for subcellular location and protein class using GO (Gene Ontology Consortium), UniProt (UniProt Consortium), HPA (Stockholm, Sweden), and PANTHER (Bethesda, Maryland) databases (Figure 1a–c ). Proteins were assigned as follows: plasma membrane: 18%; endoplasmic reticulum: 17%; cytoplasm: 42%; nucleus: 22%; Golgi body: 5%; mitochondria: 14%. Some proteins were expressed in more than one location. An overlap of 3,797 proteins (76%) were expressed in both intestinal segments, with only 528 and 645 proteins unique to the ileum and jejunum, respectively (Figure 1b ). Functional classes were distributed similarly in the two regions: drug‐metabolizing enzymes 955 and 959 (jejunum and ileum), oxidoreductases: 238; transporters: 134; membrane traffic proteins: 118; receptors: 83; cell junction proteins: 28. Enzymes responsible for xenobiotic detoxification included hydrolases (439) and transferases (282).
Figure 1.
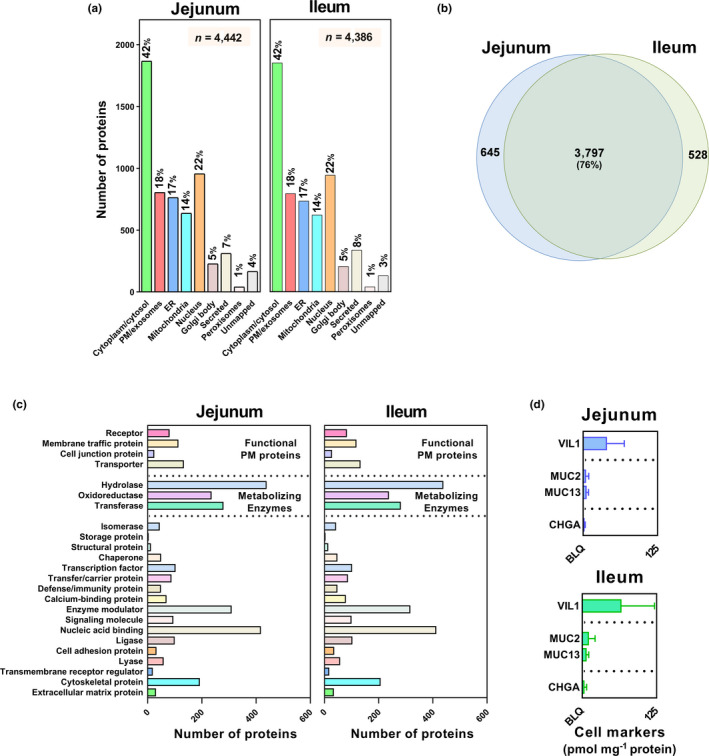
Global protein expression in human jejunal and ileal mucosa: (a) localization, (b) overlap, and (c) functional assignment of proteins identified in human jejunum and ileum; (d) markers of different cell types in the intestinal mucosa, including VIL1 (enterocytes), MUC2/MUC13 (goblet cells), and CHGA (enteroendocrine cells); no statistically significant differential protein expression was noted for markers between jejunum and ileum (Mann‐Whitney test). Protein localization was based on GO (Gene Ontology), Uniprot, and HPA (Human Protein Atlas) databases. Functional classes were assigned using the PANTHER database. (c) proteins relevant to membrane traffic and metabolizing enzymes are highlighted. BLQ, below the limit of quantification; ER, endoplasmic reticulum; n, number of proteins; PM, plasma membrane.
Cellular composition of human intestinal samples
Nearly 90% of the intestinal epithelium or villi consists of enterocytes, with 10% goblet cells (mucus cells) and 1% enteroendocrine cells. 22 To confirm the quality of mucosal preparation, cell‐specific markers of enterocytes (villin, VIL1), goblet cells (MUC2 and MUC13) and enteroendocrine cells (chromogranin A, CHGA) were quantified for the first time using global proteomics (Figure 1d ). 20 In both fractions, VIL1>>MUC2 and MUC13>CHGA, as expected.
Novel enzymes, transporters, and cell junction proteins quantified using the global approach
This is the first global proteomic study aimed at analyzing proteins involved in drug metabolism and disposition in human jejunum and ileum. Of approximately 5,700 quantifiable proteins, 77 (six CYPs, six UGTs, two flavin‐containing monooxygenases (FMOs), two auxiliary proteins, thirteen transferases, one phosphatase, one redox protein, eight ATP‐binding cassette (ABC) transporters, twenty four solute carriers (SLCs), six tight junction, two adhesion proteins, one regulatory protein and five markers) were of especial interest (Supplementary Table S1 ), 60 were measured for the first time in this study as shown in Tables 1, 2, 3, 4.
Table 1.
Quantification of six CYP enzymes, two flavin‐containing monooxygenases, CYB5A, and POR in human intestine
| Enzyme | Median | Mean ± SD | CV | Range | n a |
|---|---|---|---|---|---|
| pmol/mg | pmol/mg | % | (min–max) | ||
| pmol/mg | |||||
| Cytochrome P450 enzymes | |||||
| CYP2C9 | 0.96 | 0.87 ± 0.48 | 54.4 | 0.21–1.90 | 9 |
| CYP2C19 | 0.48 | 0.48 ± 0.01 | 2.08 | 0.47–0.49 | 2 |
| CYP2S1 | 0.48 | 0.57 ± 0.28 | 48.9 | 0.26–1.27 | 16 |
| CYP3A4 | 1.74 | 2.47 ± 2.68 | 109 | 0.13–11.4 | 14 |
| CYP4F2 | 0.28 | 0.62 ± 0.72 | 116 | 0.05–2.35 | 16 |
| CYP27A1 | 1.30 | 1.31 ± 0.57 | 43.2 | 0.53–2.42 | 16 |
| Flavin‐containing monooxygenases | |||||
| FMO1 | 0.55 | 0.58 ± 0.36 | 61.5 | 0.05–1.38 | 13 |
| FMO5 | 0.36 | 0.56 ± 0.38 | 67.1 | 0.20–1.43 | 14 |
| Auxiliary proteins | |||||
| CYB5A b | 10.5 | 12.8 ± 5.84 | 45.8 | 4.96–22.9 | 16 |
| POR b | 2.29 | 2.51 ± 1.59 | 63.6 | 0.33–6.85 | 16 |
Protein expression is represented as a median, mean ± SD (standard deviation), % CV (% coefficient of variation) and range (min–max). Abundance of enzymes is expressed in pmol/mg of intestine mucosal protein.
CYP, cytochrome P450; CYB5A, cytochrome‐b 5; max, maximum; min, minimum; POR, NADPH‐cytochrome P450 reductase.
Number of human intestine samples.
Cytochrome P450 auxiliary proteins.
Table 2.
Quantification of several phase II biotransformation enzymes
| Enzyme | Median | Mean ± SD | CV | Range | n a |
|---|---|---|---|---|---|
| pmol/mg | pmol/mg | % | (min–max) | ||
| pmol/mg | |||||
| Uridine‐5′‐diphospho (UDP)‐glucuronosyltransferase | |||||
| UGT1A1 | 0.63 | 1.30 ± 1.71 | 131 | 0.14–6.49 | 12 |
| UGT1A6 | 0.79 | 0.94 ± 0.59 | 62.9 | 0.30–1.72 | 3 |
| UGT1A10 | 0.87 | 0.98 ± 0.60 | 61.6 | 0.37–2.46 | 15 |
| UGT2A3 | 1.89 | 2.23 ± 1.60 | 71.6 | 0.64–6.92 | 16 |
| UGT2B7 | 0.60 | 1.39 ± 1.82 | 131 | 0.01–5.70 | 14 |
| UGT2B17 | 6.74 | 7.91 ± 6.71 | 84.9 | 1.45–29.8 | 16 |
| Transferase, thioredoxin, and phosphatase enzymes | |||||
| GSTA2 | 14.4 | 15.7 ± 7.49 | 47.9 | 5.27–30.6 | 15 |
| GSTK1 | 6.55 | 7.23 ± 3.31 | 45.7 | 3.38–15.6 | 16 |
| GSTM2 | 0.98 | 1.59 ± 1.59 | 100 | 0.38–6.76 | 16 |
| GSTM4 | 0.24 | 0.74 ± 1.11 | 150 | 0.01–3.58 | 13 |
| GSTO1 | 6.76 | 7.31 ± 2.98 | 40.7 | 3.25–13.8 | 16 |
| GSTP1 | 30.6 | 34.6 ± 15.6 | 45.1 | 17.7–67.3 | 16 |
| SULT1A2 | 1.49 | 1.69 ± 1.21 | 71.5 | 0.16–4.27 | 16 |
| SULT1A3 | 2.99 | 3.04 ± 2.07 | 68.1 | 0.23–6.25 | 8 |
| SULT1B1 | 5.47 | 6.25 ± 2.72 | 43.5 | 2.90–10.7 | 16 |
| SULT1E1 | 0.21 | 0.29 ± 0.20 | 70.1 | 0.03–0.61 | 13 |
| SULT2A1 | 3.95 | 3.95 ± 3.02 | 76.5 | 0.33–11.0 | 16 |
| MGST2 | 4.87 | 5.98 ± 3.19 | 53.3 | 1.18–12.2 | 16 |
| MGST3 | 5.40 | 5.69 ± 3.62 | 63.6 | 0.40–15.9 | 16 |
| TXN | 34.5 | 31.6 ± 12.2 | 39.0 | 13.9–60.3 | 16 |
| ALPI | 3.07 | 3.89 ± 3.52 | 90.5 | 0.49–13.7 | 16 |
Six UGTs (uridine‐5′‐diphospho (UDP)‐glucuronosyltransferases), six GSTs (glutathione S‐transferases), two MGSTs (microsomal glutathione S‐transferases), five SULTs (sulfotransferases), one redox enzyme (TXN, thioredoxin) and one intestinal alkaline phosphatase (ALPI) in human intestine mucosal fractions. Protein expression is represented as a median, mean ± SD (standard deviation), % CV (% coefficient of variation) and range (min–max). Abundance of enzymes is expressed in pmol/mg of intestine mucosal protein.
max, maximum; min, minimum.
Number of human intestine samples.
Table 3.
Abundance levels of 32 transporters quantified in human intestinal fractions
| Transporter | Median | Mean ± SD | CV | Range | n a |
|---|---|---|---|---|---|
| pmol/mg | pmol/mg | % | (min–max) | ||
| pmol/mg | |||||
| ATP‐binding cassette transporters | |||||
| ABCB1 | 0.86 | 1.02 ± 0.75 | 73.4 | 0.12–3.29 | 16 |
| ABCB7 | 0.09 | 0.16 ± 0.19 | 125 | 0.02–0.83 | 15 |
| ABCD1 | 0.32 | 0.48 ± 0.37 | 76.7 | 0.07–1.35 | 15 |
| ABCD3 | 0.89 | 1.00 ± 0.63 | 63.1 | 0.15–2.16 | 16 |
| ABCE1 | 0.52 | 0.57 ± 0.27 | 46.7 | 0.26–1.20 | 16 |
| ABCF1 | 0.20 | 0.28 ± 0.22 | 78.9 | 0.08–0.83 | 13 |
| ABCF3 | 0.13 | 0.15 ± 0.05 | 35.9 | 0.07–0.24 | 9 |
| ABCG2 | 0.38 | 0.37 ± 0.29 | 76.8 | 0.04–1.12 | 10 |
| Solute carriers | |||||
| SLC1A5 | 0.15 | 0.17 ± 0.07 | 40.8 | 0.06–0.32 | 15 |
| SLC2A2 | 0.47 | 0.66 ± 0.43 | 66.1 | 0.05–1.37 | 14 |
| SLC3A1 | 1.06 | 1.61 ± 1.37 | 85.2 | 0.23–4.85 | 16 |
| SLC3A2 | 0.81 | 0.84 ± 0.46 | 54.3 | 0.14–1.69 | 16 |
| SLC5A1 | 4.35 | 5.97 ± 4.57 | 76.5 | 0.91–19.7 | 16 |
| SLC8A1 | 0.18 | 0.21 ± 0.10 | 46.1 | 0.07–0.40 | 12 |
| SLC12A2 | 0.45 | 0.54 ± 0.34 | 63.1 | 0.12–1.46 | 16 |
| SLC15A1 | 1.03 | 1.08 ± 0.44 | 40.7 | 0.46–1.89 | 8 |
| SLC25A1 | 1.15 | 1.38 ± 0.75 | 54.3 | 0.17–3.09 | 16 |
| SLC25A3 | 9.17 | 9.94 ± 5.03 | 50.6 | 1.35–22.6 | 16 |
| SLC25A4 | 0.68 | 0.90 ± 0.89 | 98.7 | 0.11–3.97 | 15 |
| SLC25A5 | 12.9 | 13.9 ± 6.11 | 44.0 | 4.19–27.1 | 16 |
| SLC25A6 | 4.08 | 4.14 ± 2.22 | 53.6 | 0.51–9.50 | 16 |
| SLC25A11 | 0.86 | 1.02 ± 0.57 | 56.1 | 0.19–2.52 | 16 |
| SLC25A13 | 0.93 | 1.15 ± 0.59 | 51.8 | 0.29–2.82 | 16 |
| SLC25A15 | 0.30 | 0.46 ± 0.32 | 70.3 | 0.17–1.26 | 13 |
| SLC25A20 | 0.90 | 1.63 ± 1.96 | 121 | 0.30–8.47 | 16 |
| SLC25A24 | 3.68 | 3.23 ± 1.15 | 35.6 | 0.96–4.96 | 16 |
| SLC27A2 | 0.20 | 0.27 ± 0.24 | 87.2 | 0.02–0.92 | 16 |
| SLC27A4 | 0.68 | 0.80 ± 0.49 | 61.6 | 0.20–2.00 | 13 |
| SLC30A7 | 0.36 | 0.38 ± 0.24 | 62.8 | 0.11–1.10 | 14 |
| SLC39A5 | 0.07 | 0.08 ± 0.06 | 74.4 | 0.02–0.24 | 13 |
| SLC39A14 | 0.51 | 0.55 ± 0.32 | 59.0 | 0.08–1.14 | 14 |
| SLC44A1 | 0.29 | 0.33 ± 0.11 | 34.3 | 0.21–0.55 | 14 |
Protein expression is represented as a median, mean ± SD (standard deviation), % CV (% coefficient of variation) and range (min–max). Abundance of transporters is expressed in pmol/mg of intestine mucosal protein.
max, maximum; min, minimum.
Number of human intestine samples.
Table 4.
Expression levels of six junction proteins, two adhesion proteins, one regulatory protein, and five markers
| Protein | Median | Mean ± SD | CV | Range | n a |
|---|---|---|---|---|---|
| pmol/mg | pmol/mg | % | (min–max) | ||
| pmol/mg | |||||
| Junction Proteins | |||||
| TJP1 | 0.15 | 0.18 ± 0.07 | 41.4 | 0.05–0.29 | 12 |
| TJP2 | 0.27 | 0.31 ± 0.12 | 40.2 | 0.15–0.58 | 16 |
| TJP3 | 0.12 | 0.94 ± 1.40 | 148 | 0.04–4.49 | 10 |
| F11R | 0.78 | 0.84 ± 0.46 | 55.0 | 0.15–1.76 | 15 |
| CLDN3 | 0.25 | 0.27 ± 0.09 | 32.1 | 0.17–0.41 | 7 |
| CLDN7 | 1.40 | 1.41 ± 0.43 | 30.1 | 0.94–2.34 | 9 |
| Adhesion Proteins | |||||
| JUP | 0.79 | 0.79 ± 0.31 | 40.0 | 0.23–1.59 | 16 |
| CDH17 | 19.6 | 21.3 ± 10.8 | 50.7 | 3.99–50.3 | 16 |
| Regulatory protein | |||||
| HSP90B1 | 24.4 | 29.7 ± 17.1 | 57.6 | 8.46–71.9 | 16 |
| Markers | |||||
| MUC2 | 2.84 | 4.30 ± 4.10 | 95.5 | 1.35–18.0 | 16 |
| MUC13 | 2.95 | 4.37 ± 2.94 | 67.3 | 0.99–10.7 | 16 |
| CHGA | 1.74 | 1.89 ± 0.75 | 39.8 | 0.94–3.41 | 14 |
| VIL1 | 26.9 | 26.8 ± 11.2 | 41.6 | 11.7–47.6 | 16 |
| ATP1A1 | 16.6 | 17.3 ± 7.05 | 40.9 | 4.64–31.9 | 16 |
Protein expression is represented as a median, mean ± SD (standard deviation), % CV (% coefficient of variation) and range (min–max). Abundance of proteins is expressed in pmol/mg of intestine mucosal protein.
max, maximum; min, minimum.
Number of human intestine samples.
CYP3A4 was the highest expressed drug metabolizing CYP enzyme followed by CYP27A1 and CYP2C9 (Table 1 ). UGT2B17 showed the highest expression among quantified UGT enzymes followed by UGT2A3, UGT2B7, and UGT1A1 (Table 2 ). Eight glutathione S‐transferases (GSTs), including two microsomal glutathione S‐transferases (MGSTs), were quantified and their variability across samples ranged from 3.8‐fold to 256‐fold. The order of abundance of GSTs in the small intestine was GSTP1>GSTA2>GSTO1>GSTK1>MGST2>MGST3> GSTM2>GSTM4.
Although sulfotransferases (SULTs) were, on average, less abundant, five were detected in the order SULT1B1>SULT2A1>SULT1A3>SULT1A2>SULT1E1 (Table 2 ).
Comparison of CYP, UGT, and transporter abundance measured by label‐free and targeted proteomics
The same human samples have previously been analyzed using a QconCAT targeted strategy (MRM). 6 Intramethod quantification across technical replicates was shown to be high (see Table S2 and Table S4 in the Supplement). The overlap between targets quantified in previous report 6 and current study included only three CYPs (3A4, 2C9, and 2C19), three UGTs (1A1,1A6, and 2B7), two ABC transporters (ABCB1 and ABCG2), one tight junction protein (CDH17), and one membrane marker protein (ATP1A1). Where both measurements were available, the values were within twofold, with the exception of CYP enzymes, which were underestimated by the label‐free approach (twofold to 10‐fold). The label‐free methodology allowed quantification of many additional relevant proteins (21 enzymes, 30 transporters, 7 tight junction proteins, and 4 cell marker proteins). A summary of the results of the three methods is shown in Supplementary Table S5 . Label‐free methods are generally less sensitive than targeted approaches, and CYP2C19 and UGT1A6 were detected in very few samples and ABCC2 in none at all, prohibiting comparison.
Comparison of enzyme abundance between jejunum and ileum
Expression of CYP3A4, CYP27A1, UGT1A1, UGT2A3, GSTA2, MGST3, SULT1B1, and SULT2A1 was found to be significantly higher in the jejunum than the ileum but all other phase I and phase II enzyme abundances were similar (Figure 2a–c and Table S4 ). These differences were not related to sample preparation as the differences in CYP and UGT enzymes (CYP3A4, CYP27A1, UGT1A1, and UGT2A3) remained significant after normalization to Villin 1, a resident enterocyte marker (see Table S3 in the Supplement).
Figure 2.
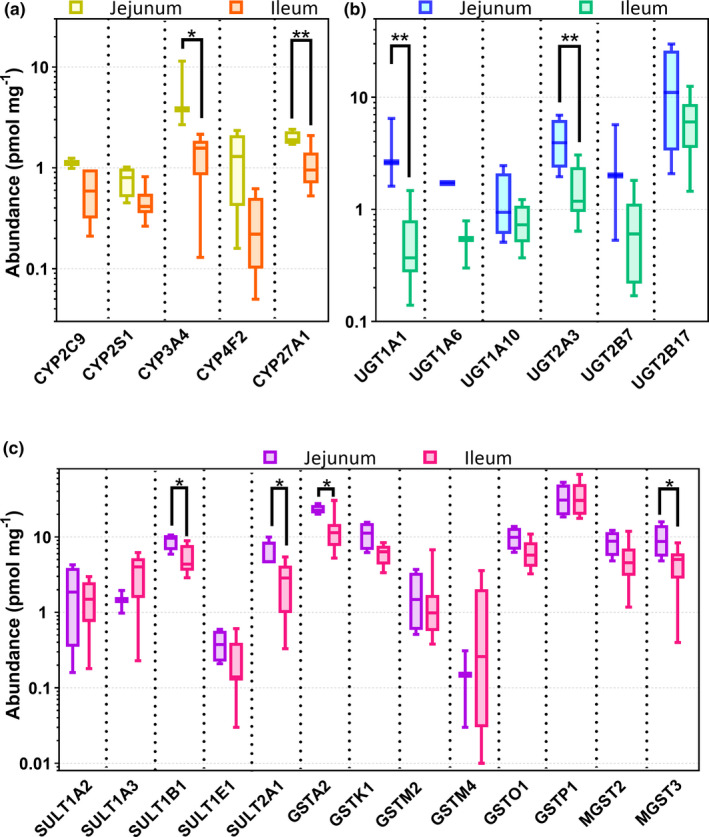
Protein expression of (a) phase I and (b and c) II enzymes in the jejunum and ileum of 16 donors. The * symbol indicates differences in expression data between jejunum and ileum (Mann‐Whitney test). *, P < 0.05; **, P < 0.01.
Protein abundance levels of ABC and SLC transporters in human jejunum and ileum
ABC and SLC transporters were present at low abundance with ABCB1, ABCD3, SLC25A3, and SLC25A5 showing the highest expression levels (Table 3 ). ABCG2 was detected in 10 samples with average expression level of only 0.37 ± 0.29 pmol/mg protein. SLC3A2, SLC12A2, SLC25A3, SLC25A6, SLC25A11, SLC25A13, SLC25A20, SLC39A14, and SLC44A1 showed higher expression in the jejunum than ileum but other transporters were found at similar levels (Table S4 ). These differences persisted for several SLC proteins (SLC3A2, SLC12A2, SLC25A13, SLC25A20, and SLC39A14) after normalization to Villin 1 (see Table S3 Supplement).
Tight junction and adhesion proteins
Six tight junction (TJ) associated proteins (TJP1, TJP2, TJP3, F11R, CLDN3, and CLDN7) and one adhesion protein (JUP) were quantified for the first time in this report. The cadherin‐like transporter (CDH17/HPT1) was confirmed to be highly abundant (Figure 3b ), as indicated in a previous study. 5 As shown in Table 4 , the average abundance of CDH17 was 21.3 pmol/mg protein, with the other seven junction proteins expressed at 1–2 orders of magnitude lower.
Figure 3.
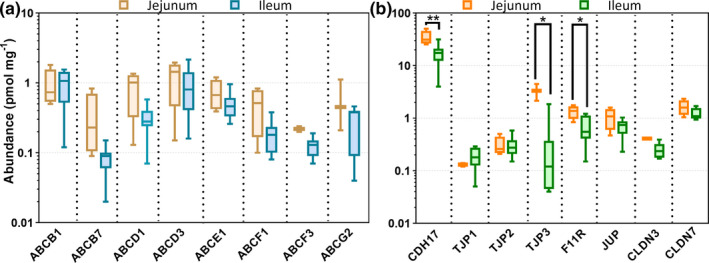
Protein expression of (a) ABC transporters and (b) tight junction and adhesion proteins in the jejunum and ileum of 16 donors. The * symbol indicates differences of expression data between jejunum and ileum for CDH17, TJP3 and F11Rusing Mann‐Whitney test (*, P < 0.05; **, P = 0.01).
AMRT approach
The AMRT targeted method was applied with two QconCATs (TransCAT and MetCAT) 21 as standards to measure, eight CYPs, four UGTs, four ABC transporters, eight SLC transporters, ATP1A1, and CDH17. The individual abundance values of proteins measured using AMRT and MRM 6 were correlated whenever the data were available from both methods. We showed a strong and statistically significant correlation (Figure S1 ) between AMRT and MRM approaches with (Rs = 0.92, P < 0.0001, R 2 = 0.70). Table S5 and Supplementary Table S2 show a summary and individual AMRT abundance values, respectively.
Protein–protein correlation of human intestine protein expression
Intercorrelation analysis was performed on protein abundance data generated by global proteomics. The enzyme pairs CYP3A4/CYP27A1 and CYP2F4/CYP27A1 showed very strong positive correlations (Figure S3 A). We found no correlations between UGT enzymes. Cross‐family correlations between CYPs and UGTs were weak overall, except for the pairs CYP4F2/UGT2B17 (Rs = 0.78, P < 0.0001, R 2 = 0.73) (Figure S3 A). ABC and SLC transporters showed very strong and significant correlations; for example, ABCB1/SLC3A1 (Rs = 0.86, P < 0.0001, R 2 = 0.60). Enzyme and transporter correlation matrices are shown in Figure S3 A,B.
Translational path for using the generated abundance data
Translational applications of PBPK models require IVIVE scaling factors 23 , 24 to translate the rates of activity (expressed based on protein abundances) to expected clearance in specific segments of small intestine, either based on the mass of tissue or its surface area. Unfortunately, there is insufficient information, specifically from these samples, to enable the total mucosal protein to be calculated for a given mass or surface area. This was due to the fact that sample processing was not originally intended for the generation of this scalar. However, surrogate values for a “total mucosal protein scalar” per mass of tissue can be derived from publications employing a mucosal scraping approach, 25 (16,015 mg total mucosal (scraped) protein per small intestine, Table S6 ). Alternatively, an enterocyte chelation method, 5 where enterocyte homogenate protein was measured (see Table S7 ), results in an average enterocyte total protein scalar of approximately 5,242 mg for the whole small intestine. It is likely that a mucosal scraping method 25 aligns more closely to a total mucosal protein yield in using our methodology.
The abundance of ABCB1 (pmol) in the entire small intestine (1000 virtual healthy White individuals; Simcyp Simulator, version 19, Certara, Sheffield, UK) was simulated by incorporating the mean absolute levels of ABCB1 from this study (pmol/mg total mucosal protein; Table 3 ) and total mucosal protein scalar (16,015 mg). A comparison was made with a case when incorporating ABCB1 total membrane protein abundances (meta‐analysis) and the total membrane protein scalar (2,737 mg) per intestine (Simcyp Simulator, version 19). 12 The total mucosal protein scaling approach provided a 15.8‐fold higher small intestinal P‐gp expression than the total membrane approach (23,393 vs. 1,481 pmol/small intestine), highlighting considerable higher abundances in vivo using these methods compared with those previously reported.
Discussion
Over the last two decades, mathematical models of human physiology and biology have greatly improved our understanding of how different organs react to endogenous and exogenous molecular interventions, and paved the way for recognizing the complex cellular processes which modify the function of organs under disease conditions. 26 However, such models are data‐hungry and in many cases have been unable to deliver translation from in vitro studies, because of a lack of information on key variables. While computational models of epithelial tissues at cellular or subcellular levels are not uncommon, 27 models that contain quantitative proteomic data to characterize responses to chemicals (including xenobiotics) are rare. Microphysiological (organ‐on‐chip) systems have facilitated in vitro rapid testing of interactions of given tissues with specific chemicals, 28 but the full potential of such systems are only realized within a quantitative framework for extrapolation from in vitro bench studies to in vivo expectations (IVIVE). 29 The importance of the oral route of drug administration ensures that many drugs have to interact with a range of intestinal proteins before exerting their effects. 30 This is the first global proteomic study aimed at analyzing these human intestinal proteins: CYPs, UGTs, FMOs, transferases, phosphatases, transporters, tight junction, regulatory proteins, and enterocyte markers. For most of these proteins, no human small intestinal expression profiles have previously been reported, and the abundances of 60 such proteins (of a total 77) are reported for the first time. The targeted analysis of the same set of samples reported previously 6 covered a limited number of intestinal enzymes and transporters, because of the requirement for internal standards. The current global analysis confirmed our previous findings and quantified a large number of additional targets, providing a more comprehensive picture of protein expression in the ileum and jejunum, while ensuring the quality of the analysis using various quality controls (replicate integrity, cross‐method, and cross‐platform confirmation). CYP2S1 and CYP27A1 are not normally detected in human liver and are reported in small intestine for the first time. CYP27A1 is key to bile acid biosynthesis, and therefore to cholesterol homeostasis; its inhibition decreases cancer metastasis in animal models. 31 CYP2S1 metabolizes exogenous and endogenous compounds including naphthalene, all‐trans retinoic acid, and various toxins. 32 , 33 Previous mRNA analysis has demonstrated low expression of CYP2S1 in liver relative to lung, stomach, small intestine, skin, and spleen, suggesting an important role in extrahepatic metabolism. 34 Our results support the literature finding that CYP3A4 and CYP2C9 are abundant in the small intestine 6 , 14 , 35 compared with other quantified CYPs. High interindividual variability was found for the expression of the most CYP and UGT enzymes; 54% coefficient of variation for CYP2C9 and 85% for UGT2B17. There is agreement among the limited published studies that UGT2B17 is the most abundant UGT in the intestine, with UGT1A1 and UGT2B7 abundance consistently highly ranked. 9 , 13 , 35 In the present study, UGT2A3 (quantified for the first time) was found be the second most abundant UGT, followed by UGT12B7, UGT1A1, and UGT1A10. Apart from UGT2B17, these enzymes are all present at low levels and differences in order were not unexpected. Differences between reported abundances of DMEs can be associated with interlaboratory differences in sample preparation. For example, intestinal microsomal fractions from enterocytes, total membrane‐enriched fractions, and entire crude mucosa containing other types of intestinal cells have all been used. These interlaboratory differences in the proteomic workflow can contribute to technical variability in reported values. 9 , 10 , 11 We have also quantified the low abundance proteins UGT1A3 and UGT1A6 (Table S5 ) using the AMRT approach. This high level of first‐time quantification reflects two important factors. A global approach allowed us to quantify unexpected proteins, whereas the targeted approach permits greater sensitivity of quantification of proteins of known interest. 20 Secondly, the use of surgical tissue allows rapid freezing, so samples are of high quality. Drozdzik et al. used postmortem tissue, allowing direct comparison of intestinal segments but with more uncertain sample harvest. 14 Relatively little attention has been paid to SULTs, GSTs, and FMOs in human intestine, although the impact of age on several SULT protein abundances in liver has been reported. 36 Sulfotransferases metabolize and regulate the levels of several hormones 37 and drugs, including oxamniquine (antischistosomiasis) and cyproterone acetate (anti–prostate cancer). In contrast to SULTs, GSTs are highly polymorphic, a factor implicated in resistance to chemotherapeutic drugs, such as busulfan. 38 We quantified five SULTs, the most abundant being SULT1B1, and eight GSTs, GSTP1 and GSTA2 being abundant (Table 2 ), as well as two FMOs, also involved in xenobiotic detoxification and possibly in aging. 39 Consistent with data from the liver and brain, no correlations were observed between protein abundance and common covariates such as age and gender. 20 , 35 , 40 , 41 ABC transporters were expressed (Supplementary Table S1 , Table 3 ) in order ABCB1≈ABCD3>ABCG2. Both ABCB1 and ABCG2 act as efflux transporters for several xenobiotics, limiting their oral bioavailability. ABCD3 is required for bile reabsorption which takes place primarily in the ileum. Disease‐related transporters quantified for the first time include ABCD1 which has about half the abundance of ABCD3. Mutation of ABCD1 has been linked to adrenoleukodystrophy, characterized by accumulation of very long fatty acids in tissues and plasma. 42 ABCE1 (expressed at similar levels to ABCD1), as well as ABCF1, ABCB7, and ABCF3 were all detected for the first time, albeit at very low levels. Sample preparation and interindividual variability can have a profound effect on quantification of low abundance transporters, leading to some conflict in the literature about rank order. 15 , 35 , 43 Assessment of interindividual variability requires the separation of the uncertainties associated with the processing and measurement of samples from the true variations between the individuals, only possible when there are replicate samples. The common practice of using single values from a given individual (whether as a mean from replicates or a single measure) may hamper obtaining reliable interindividual variability. Reproducibility of the label‐free global quantification across two replicates and confirmation of the abundances using two targeted methods were reassuring and provided more confidence in the measurements. Of the 24 SLC transporters quantified (Supplementary Table S1 ), the most abundant were SLC5A1, SLC25A3, SLC25A5, SLC25A6, and SLC25A24. SLC3A2, SLC12A2, SLC25A1, SLC25A3, SLC25A6, SLC25A11, SLC25A13, SLC25A20, SLC39A14, and SLC44A1 were expressed more highly in jejunum than ileum, whereas SLC5A1, SLC25A5, and SLC25A24 were expressed equally in the two small intestinal regions. SLC15A1, the oligopeptide transporter 1 (PEPT1), primarily transports di‐peptides but also facilitates the absorption of drugs, such as valacyclovir. 44 Disease‐related transporters quantified for the first time include choline transporter‐like proteins 1(SLC44A1) reported to be involved in abnormal choline disposition. SLC3A2 is a potential biomarker in gastric cancer. 45 SLC5A1 has a role in glycemic control in type 2 diabetes mellitus. SLC12A2 deficiency causes Kilquist syndrome, a form of hearing loss. 46 Tight junction proteins, together with adherens junctions, underpin the integrity of the gut epithelium as a selective barrier. Changes in the expression of these proteins result in increased intestinal permeability, which has been linked to the etiopathogenesis of some types of inflammatory bowel disease. 47 A range of tightjunctionproteins and two adhesion proteins (Table 4 ) were quantified in the present study, although these low abundance proteins will certainly benefit from targeted proteomic approaches to yield more sensitive measurements. Both endoplasmin (HSP90B1) and alkaline phosphatase (ALPI) are believed to be markers for intestinal inflammation and were quantified for the first time in the present study. Reduced levels of ALP mRNA have been associated with Crohn’s disease and ulcerative colitis. Regional differences in the abundance of some DMEs and drug transporters have been reported in some but not in other studies. 14 , 15 We would expect the matched postmortem tissue used by Drozdzik et al. 14 to afford more sensitive intraintestinal results than the present study; nevertheless, our data showed segment‐dependent expression levels of CYP3A4, CYP27A1, UGT1A1, UGT2A3, SULT1B1, SULT2A1, GSTA2, MGST3, F11R, CDH17, SLC3A2, SLC12A2, SLC25A1, SLC25A3, SLC25A6, SLC25A11, SLC25A13, SLC25A20, SLC39A14, and SLC44A1. CYP3A4 expression was greater in the jejunum than in the ileum, consistent with the observation that the permeability of ropivacaine (CYP3A4 substrate) and its metabolite 2′,6′‐pipecoloxylidide is higher in jejunum than in ileum. 48 The auxiliary proteins CYB5A and POR were also more highly expressed in the jejunum than ileum. A targeted approach is generally more sensitive to intercorrelations between proteins, but global proteomics can be used to scan for unexpected correlations. We observed very strong correlations between CYP3A4/CYP27A1 (Rs = 0.76, P = 0.002, R 2 = 0.56) and ABCB1/SLC3A1 (Rs = 0.86, P < 0.0001, R 2 = 0.37) (Figure S3 A,B) for the first time, but the physiological basis behind these correlations is unclear. The enzyme pairs GSTK1/GSTO1 and GSTO1/MGST2 also showed very strong correlation (Figure S3 A). These correlations are expected only if the pairs of proteins are regulated by the same nuclear receptors and/or if they share a common regulatory pathway. 49 A rich abundance data set of proteins related to absorption, distribution, metabolism and excretion can facilitate further mechanistic explorations using appropriate translational approaches described here. A common challenge, however, is that these precious human samples were initially procured for assessing protein expression by immuno‐blotting, rather than being purposed for quantitative proteomic assessment and incorporation in translational IVIVE‐PBPK strategies. A chaotropic‐detergent based sample preparation with medium speed centrifugation was sufficient for the semiquantitative assays primarily proposed. The repurposing of these samples to investigate human mucosal protein abundances by liquid chromatography‐tandem mass spectrometry was therefore a significant achievement. To apply the reported abundances for translational purposes, e.g., PBPK modeling, requires scaling of the data from total mucosal protein abundances (e.g., Table 3 ) to intestinal segmental or total small intestinal abundances (in pmol). The primary limitation is that there is insufficient information to enable the total mucosal protein yield (mg) to be calculated because the sample processing was not designed for the generation of this scalar. However, mucosal scraping homogenate protein yields from the literature and previously unpublished data from our group 5 on homogenate protein from chelated enterocytes provide surrogate options to scale the abundance data to segmental, or total small intestinal abundance data. As expected, the scraping method (Table S6 ) gave an approximate threefold higher yield compared with the more refined enterocyte elution method (Table S7 ), although it should be acknowledged these are not the same samples. It was considered that the mucosal scraping method 25 better captured the tissue processing approaches in this study, and the total mucosal protein was used to scale ABCB1 abundances (Table 3 ) by simulation. An approach to scaling transporter abundances in total membrane protein matrices to segmental organ abundances (pmol) in a PBPK model has been described. 12 The total membrane protein yield based on three different fractionation methods is approximately 2,700 mg for the entire small intestine. 12 Given the higher abundances in the total mucosal protein in these samples compared with the collated total membrane protein abundances from several sources, with the higher mucosal protein yields, it is no surprise that higher simulated abundances are obtained using the data set and approaches described here relative to those in Harwood et al., 2019. 12 Human surgical samples are precious, and going forward, recommendations are in place so that they can be collected in a way to maximize the translational value of proteomic measurements. 24 Particularly relevant to IVIVE is aligning proteomic methodologies used to define human tissue abundances with those required to define in vitro system activity‐abundance relations.
Conclusions
The information provided in the current report adds considerably to the extremely limited information on human small intestinal protein abundance data. It characterizes several enzyme, transporter, and tight junctional proteins in human intestine, reporting expression levels of 60 proteins for the first time. These provide a resource for input parameters needed in systems biology models of human small intestinal epithelium. In addition, over 5,000 proteins were quantified using a global approach, so the data also provide a rich source of information for wider investigations.
Funding
This study was supported by the Innovative Medicines Initiative Joint Undertaking (http://www.imi.europa.eu) under grant agreement no. 115369, resources of which are composed of financial contribution from the European Union's Seventh Framework Programme (FP7/2007‐2013) and European Federation of Pharmaceutical Industries and Associations (EFPIA) companies’ in kind contribution.
Conflict of Interest
M.H. and A.R.‐H. are employees of Certara UK Limited. Certara is one of the providers of PBPK modeling platforms and services to pharmaceutical companies. All other authors declared no competing interests for this work.
Author Contributions
Z.M.A.‐M., B.A., M.D.H., G.C., G.W., J.B., and A.R.‐H. wrote the manuscript. Z.M.A.‐M., N.C., B.A., J.B., and A.R.‐H. designed the research. Z.M.A.‐M. and N.C. performed the research. Z.M.A.‐M., B.A., and J.B. analyzed the data. Z.M.A.‐M. and N.C. contributed new reagents/analytical tools.
Supporting information
Supplementary Material
Table S1–S6
Acknowledgments
The authors thank the Royal Commission for the Exhibition of 1851 for their award to the University of Manchester on the basis of M.D. Harwood successfully completing an Industrial Fellowship grant in 2015 which facilitated the initial aspects of this work. The authors also thank the Bio‐MS core facility, University of Manchester, for providing access to liquid chromatography–mass spectrometry instrumentation. The authors also thank Jessica Waite for helping to format and submit the manuscript.
References
- 1. Hunt, R.H. et al. The stomach in health and disease. Gut. 64, 1650–1668 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Musther, H. , Olivares‐Morales, A. , Hatley, O.J.D. , Liu, B. & Rostami Hodjegan, A. Animal versus human oral drug bioavailability: Do they correlate? Eur. J. Pharm. Sci. 57, 280–291 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Akabane, T. , et al. A comparison of pharmacokinetics between humans and monkeys. Drug Metab. Dispos. 38, 308–316 (2010). [DOI] [PubMed] [Google Scholar]
- 4. Cirit, M. & Stokes, C.L. Maximizing the impact of microphysiological systems with in vitro–in vivo translation. Lab Chip 18, 1831–1837 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Harwood, M.D. , et al. Application of an LC‐MS/MS method for the simultaneous quantification of human intestinal transporter proteins absolute abundance using a QconCAT technique. J. Pharm. Biomed. Anal. 110, 27–33 (2015). [DOI] [PubMed] [Google Scholar]
- 6. Couto, N. et al. Quantitative proteomics of clinically relevant drug‐metabolizing enzymes and drug transporters and their intercorrelations in the human small intestine. Drug Metab. Dispos. 48, 245–254 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Darwich, A.S. , Aslam, U. , Ashcroft, D.M. & Rostami‐Hodjegan, A. Meta‐analysis of the turnover of intestinal epithelia in preclinical animal species and humans. Drug Metab. Dispos. 42, 2016–2022 (2014). [DOI] [PubMed] [Google Scholar]
- 8. Drozdzik, M. et al. Protein Abundance of Clinically Relevant Drug Transporters in the Human Liver and Intestine: A Comparative Analysis in Paired Tissue Specimens. Clin. Pharmacol. Ther. 105, 1204–1212 (2019). [DOI] [PubMed] [Google Scholar]
- 9. Sato, Y. et al. Optimized methods for targeted peptide‐based quantification of human uridine 5'‐diphosphate‐glucuronosyltransferases in biological specimens using liquid chromatography‐tandem mass spectrometry. Drug Metab. Dispos. 42, 885–889 (2014). [DOI] [PubMed] [Google Scholar]
- 10. Akazawa, T. , et al. High expression of UGT1A1/1A6 in monkey small intestine: comparison of protein expression levels of cytochromes P450, UDP‐glucuronosyltransferases, and transporters in small intestine of cynomolgus monkey and human. Mol. Pharm. 15, 127–140 (2018). [DOI] [PubMed] [Google Scholar]
- 11. Gröer, C. et al. Absolute protein quantification of clinically relevant cytochrome P450 enzymes and UDP‐glucuronosyltransferases by mass spectrometry‐based targeted proteomics. J. Pharm. Biomed. Anal. 100, 393–401 (2014). [DOI] [PubMed] [Google Scholar]
- 12. Harwood, M.D. , Zhang, M. , Pathak, S.M. & Neuhoff, S. The regional‐specific relative and absolute expression of gut transporters in adult caucasians: a meta‐analysis. Drug Metab. Dispos. 47, 854–864 (2019). [DOI] [PubMed] [Google Scholar]
- 13. Zhang, H. et al. Regional proteomic quantification of clinically relevant non‐cytochrome P450 enzymes along the human small intestine. Drug Metab. Dispos. 48, 528–536 (2020). [DOI] [PubMed] [Google Scholar]
- 14. Drozdzik, M. et al. Protein Abundance of Clinically Relevant Drug‐Metabolizing Enzymes in the Human Liver and intestine: A Comparative Analysis in PairedTissue Specimens. Clin. Pharmacol. Ther. 104, 515–524 (2018). [DOI] [PubMed] [Google Scholar]
- 15. Drozdzik, M. et al. Protein abundance of clinically relevant multidrug transporters along the entire length of the human intestine. Mol. Pharm. 11, 3547–3555 (2014). [DOI] [PubMed] [Google Scholar]
- 16. Wiśniewski, J.R. , et al. Extensive quantitative remodeling of the proteome between normal colon tissue and adenocarcinoma. Mol. Syst. Biol. 8, 611 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wiśniewski, J.R. , et al. Absolute proteome analysis of colorectal mucosa, adenoma, and cancer reveals drastic changes in fatty acid metabolism and plasma membrane transporters. J. Proteome Res. 14, 4005–4018 (2015). [DOI] [PubMed] [Google Scholar]
- 18. Al Feteisi, H. , Achour, B. , Barber, J. & Rostami‐Hodjegan, A. Choice of LC‐MS methods for the absolute quantification of drug‐metabolizing enzymes and transporters in human tissue: a comparative cost analysis. AAPS J. 17, 438–446 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rostami‐Hodjegan, A. Physiologically based pharmacokinetics joined with in vitro–in vivo extrapolation of ADME: A marriage under the arch of systems pharmacology. Clin. Pharmacol. Ther. 92, 50–61 (2012). [DOI] [PubMed] [Google Scholar]
- 20. Al‐Majdoub, Z.M. et al. Proteomic quantification of human blood‐brain barrier SLC and ABC transporters in healthy individuals and dementia patients. Mol. Pharm. 16, 1220–1233 (2019). [DOI] [PubMed] [Google Scholar]
- 21. Russell, M.R. et al. Alternative fusion protein strategies to express recalcitrant QconCAT proteins for quantitative proteomics of human drug metabolizing enzymes and transporters. J. Proteome Res. 12, 5934–5942 (2013). [DOI] [PubMed] [Google Scholar]
- 22. Cheng, H. & Leblond, C.P. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine I. Columnar cell. Am. J. Anat. 141, 461–479 (1974). [DOI] [PubMed] [Google Scholar]
- 23. Harwood, M.D. et al. In vitro–in vivo extrapolation scaling factors for intestinal P‐glycoprotein and breast cancer resistance protein: part II. the impact of cross‐laboratory variations of intestinal transporter relative expression factors on predicted drug disposition. Drug Metab. Dispos. 44, 476–480 (2016). [DOI] [PubMed] [Google Scholar]
- 24. Prasad, B. et al. Toward a consensus on applying quantitative liquid chromatography–tandem mass spectrometry proteomics in translational pharmacology research: A White Paper. Clin. Pharmacol. Ther. 106, 525–543 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Paine, M.F. et al. Characterization of interintestinal and intraintestinal variations in human CYP3A‐dependent metabolism. J. Pharmacol. Exp. Ther. 283, 1552–1562 (1997). [PubMed] [Google Scholar]
- 26. Foy, B.H. , Gonçalves, B.P. & Higgins, J.M. Unraveling disease pathophysiology with mathematical modeling. Annu. Rev. Pathol. 15, 371–394 (2020). [DOI] [PubMed] [Google Scholar]
- 27. Smallwood, R. Computational modeling of epithelial tissues. Wiley Interdiscip. Rev. Syst. Biol. Med. 1, 191–201 (2009). [DOI] [PubMed] [Google Scholar]
- 28. Mittal, R. et al. Organ‐on‐chip models: implications in drug discovery and clinical applications. J. Cell. Physiol. 234, 8352–8380 (2019). [DOI] [PubMed] [Google Scholar]
- 29. Korcsmaros, T. , Schneider, M.V. & Superti‐Furga, G. Next generation of network medicine: interdisciplinary signaling approaches. Integr. Biol. 9, 97–108 (2017). [DOI] [PubMed] [Google Scholar]
- 30. Kostewicz, E.S. et al. PBPK models for the prediction of in vivo performance of oral dosage forms. Eur. J. Pharm. Sci. 57, 300–321 (2014). [DOI] [PubMed] [Google Scholar]
- 31. Baek, A.E. et al. The cholesterol metabolite 27 hydroxycholesterol facilitates breast cancer metastasis through its actions on immune cells. Nat. Commun. 8, 864 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ingelman‐Sundberg, M. Human drug metabolising cytochrome P450 enzymes: properties and polymorphisms. Naunyn. Schmiedebergs Arch. Pharmacol. 369, 89–104 (2004). [DOI] [PubMed] [Google Scholar]
- 33. Rivera, S.P. , Saarikoski, S.T. & Hankinson, O. Identification of a novel dioxin‐inducible cytochrome P450. Mol. Pharmacol. 61, 255–259 (2002). [DOI] [PubMed] [Google Scholar]
- 34. Saarikoski, S.T. , Wikman, H.A.‐L. , Smith, G. , Wolff, C.H.J. & Husgafvel‐Pursiainen, K. Localization of cytochrome P450 CYP2S1 expression in human tissues by in situ hybridization and immunohistochemistry. J. Histochem. Cytochem. 53, 549–556 (2005). [DOI] [PubMed] [Google Scholar]
- 35. Miyauchi, E. et al. Quantitative atlas of cytochrome P450, UDP‐glucuronosyltransferase, and transporter proteins in jejunum of morbidly obese subjects. Mol. Pharm. 13, 2631–2640 (2016). [DOI] [PubMed] [Google Scholar]
- 36. Ladumor, M.K. et al. Ontogeny of hepatic sulfotransferases and prediction of age‐dependent fractional contribution of sulfation in acetaminophen metabolism. Drug Metab. Dispos. 47, 818–831 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. van der Spek, A.H. , Fliers, E. & Boelen, A. The classic pathways of thyroid hormone metabolism. Mol. Cell. Endocrinol. 458, 29–38 (2017). [DOI] [PubMed] [Google Scholar]
- 38. Gibbs, J.P. , Czerwinski, M. & Slattery, J.T. Busulfan‐glutathione conjugation catalyzed by human liver cytosolic glutathione S‐transferases. Cancer Res. 56, 3678–3681 (1996). [PubMed] [Google Scholar]
- 39. Rossner, R. , Kaeberlein, M. & Leiser, S.F. Flavin‐containing monooxygenases in aging and disease: emerging roles for ancient enzymes. J. Biol. Chem. 292, 11138–11146 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Couto, N. , et al. Quantification of proteins involved in drug metabolism and disposition in the human liver using label‐free global proteomics. Mol. Pharm. 16, 632–647 (2019). [DOI] [PubMed] [Google Scholar]
- 41. Achour, B. , Russell, M.R. , Barber, J. & Rostami‐Hodjegan, A. Simultaneous quantification of the abundance of several cytochrome P450 and uridine 5’‐diphospho‐glucuronosyltransferase enzymes in human liver microsomes using multiplexed targeted proteomics. Drug Metab. Dispos. 42, 500–510 (2014). [DOI] [PubMed] [Google Scholar]
- 42. Amorosi, C.A. et al. X‐linked adrenoleukodystrophy: molecular and functional analysis of the ABCD1 gene in Argentinean patients. PLoS One 7, e52635 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gröer, C. et al. LC‐MS/MS‐based quantification of clinically relevant intestinal uptake and efflux transporter proteins. J. Pharm. Biomed. Anal. 85, 253–261 (2013). [DOI] [PubMed] [Google Scholar]
- 44. Ganapathy, M.E. , Huang, W. , Wang, H. , Ganapathy, V. & Leibach, F.H. Valacyclovir: a substrate for the intestinal and renal peptide transporters PEPT1 and PEPT2. Biochem. Biophys. Res. Commun. 246, 470–475 (1998). [DOI] [PubMed] [Google Scholar]
- 45. Yang, Y. et al. Discovery of SLC3A2 cell membrane protein as a potential gastric cancer biomarker: implications in molecular imaging. J. Proteome Res. 11, 5736–5747 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Macnamara, E.F. et al. Kilquist syndrome: A novel syndromic hearing loss disorder caused by homozygous deletion of SLC12A2. Hum. Mutat. 40, 532–538 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ulluwishewa, D. , et al. Regulation of tight junction permeability by intestinal bacteria and dietary components. J. Nutr. 141, 769–776 (2011). [DOI] [PubMed] [Google Scholar]
- 48. Berggren, S. , et al. Regional transport and metabolism of ropivacaine and its CYP3A4 metabolite PPX in human intestine. J. Pharm. Pharmacol. 55, 963–972 (2003). [DOI] [PubMed] [Google Scholar]
- 49. Urquhart, B.L. , Tirona, R.G. & Kim, R.B. Nuclear receptors and the regulation of drug‐metabolizing enzymes and drug transporters: implications for interindividual variability in response to drugs. J. Clin. Pharmacol. 47, 566–578 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Table S1–S6


