Abstract
Porphyromonas gingivalis, a pathogen of chronic periodontitis, is an asaccharolytic microorganism that solely utilizes nutritional amino acids as its energy source and cellular constituents. The bacterium is considered to incorporate proteinaceous nutrients mainly as dipeptides, thus exopeptidases that produce dipeptides from polypeptides are critical for survival and proliferation. We present here an overview of dipeptide production by P. gingivalis mediated by dipeptidyl‐peptidases (DPPs), e.g., DPP4, DPP5, DPP7, and DPP11, serine exopeptidases localized in periplasm, which release dipeptides from the N‐terminus of polypeptides. Additionally, two other exopeptidases, acylpeptidyl‐oligopeptidase (AOP) and prolyl tripeptidyl‐peptidase A (PTP‐A), which liberate N‐terminal acylated di‐/tri‐peptides and tripeptides with Pro at the third position, respectively, provide polypeptides in an acceptable form for DPPs. Hence, a large fraction of dipeptides is produced from nutritional polypeptides by DPPs with differential specificities in combination with AOP and PTP‐A. The resultant dipeptides are then incorporated across the inner membrane mainly via a proton‐dependent oligopeptide transporter (POT), a member of the major facilitator superfamily. Recent studies also indicate that DPP4 and DPP7 directly link between periodontal and systemic diseases, such as type 2 diabetes mellitus and coagulation abnormality, respectively. Therefore, these dipeptide‐producing and incorporation molecules are considered to be potent targets for prevention and treatment of periodontal and related systemic diseases.
Keywords: diabetes mellitus, exopeptidase, oligopeptide transporter, periodontal disease, acylpeptidyl‐oligopeptidase, oligopeptide transporter
Abbreviations
- AOP
acylpeptidyl‐oligopeptidase
- Dcp
peptidyl‐dipeptidase
- DPP
dipeptidyl‐peptidase
- GIP
glucose‐dependent insulinotropic polypeptide
- GLP‐1
glucagon‐like peptide 1
- MCA
4‐methycoumaryl‐7‐amide
- OPT
oligopeptide transporter
- POT
proton‐dependent oligopeptide transporter
- PTP‐A
prolyl tripeptidyl‐peptidase A
- SstT
serine/threonine transporter
1. INTRODUCTION
Periodontal disease, a bacterial‐associated inflammatory condition that occurs around the gingivae, is a leading cause of tooth loss in adults, with 20%–50% of individuals affected worldwide, which results in a decrement in overall quality of life especially in elderly individuals (Nazir, 2017). A large number of epidemiological studies have shown a keen association of chronic periodontitis and type 2 diabetes mellitus (Grossi & Genco, 1998; Lalla & Papapanou, 2011; Preshaw et al., 2012), and recently much attention has been given to this oral disease because of its close relationship to systemic diseases, such as atherosclerotic cardiovascular disorder (Genco & VanDyke, 2010; Tabeta et al., 2014), decreased kidney function (Kshirsagar et al., 2007), rheumatoid arthritis (Detert et al., 2010), and Alzheimer's disease (Dominy et al., 2019; Teixeira et al., 2017).
Periodontal disease is initiated by a complex of bacterial species that form subgingival biofilm in and around the gingival crevice. When allowed to progress, inflammatory cytokines and chemokines are released from affected tissues, implying a connection of this oral inflammatory condition to other systemic diseases. In previous studies, Socransky and Haffajee and their colleagues stratified the microbiota into five major complexes, among which the first one, designated “red complex” and comprised of Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola, is strikingly associated with clinical measures of periodontal disease, and thus those members are considered to be major periodontopathic bacteria implicated in severe forms (Holt & Ebersole, 2005; Socransky & Haffajee, 2002; Socransky et al., 1998). Whole genome analysis has suggested that P. gingivalis can metabolize several sugars including melibiose and galactose (Nelson et al., 2003). Nevertheless, P. gingivalis and T. forsythia are in practical terms asaccharolytic and their growth is supported by proteinaceous substrates. Sugar utilization may be not so beneficial for periodontal bacteria, because sugar supply is quite limited in their niche. This feature of utilization of amino acids as the sole carbon and energy source is quite singular among oral microorganisms and seems to be advantageous for settlement of these pathogenic bacteria in subgingival plaque. However, until recently little has been reported regarding how nutritional amino acids are produced from extracellular proteins and become incorporated into cells of those bacteria.
Extracellular proteins in P. gingivalis are initially degraded into oligopeptides by the well‐characterized cysteine endopeptidases Arg‐gingipains (Kadowaki et al., 1994; Pavloff et al., 1995) and Lys‐gingipain (Pavloff et al., 1997; Scott et al., 1993), which are localized in the outer membrane and outer membrane vesicles. The resulting oligopeptides are transported into the periplasmic space. As they are still larger than the size incorporated across the inner membrane through the outer membrane, they must be further trimmed by exopeptidases prior to incorporation.
Collateral evidence has indicated that P. gingivalis incorporates dipeptides derived from proteinaceous nutrients. Tang‐Larsen et al. (1995) reported that additions of Met‐Met and Cys‐Cys enhanced the production of methyl mercaptan and hydrogen sulfide, respectively, whereas neither a single Met nor Cys showed increased production. The dominance of dipeptides as nutrients in P. gingivalis was also demonstrated by findings showing significant accumulation of their metabolites, such as ammonia and short‐chain fatty acids (Takahashi & Sato, 2001, 2002). These findings strongly suggest that not a single amino acid but dipeptide is the major form incorporated into P. gingivalis cells. In accordance with this notion, a series of dipeptide‐producing exopeptidases, dipeptidyl‐peptidases (DPPs), have been discovered in P. gingivalis.
In this article, we discuss the roles of DPPs and two additional exopeptidases in dipeptide production, as well as the specific distribution of dpp genes in periodontopathic bacteria. Furthermore, we demonstrate the importance of a proton‐dependent oligopeptide transporter (POT) in dipeptide incorporation in P. gingivalis. The relationship of bacterial DPPs with systemic diseases is also discussed.
2. DEGRADATION OF EXTRACELLULAR PROTEINS INITIATED BY GINGIPAINS
Gingipains are cysteine proteinases that consist of Lys‐specific (Lys‐gingipain/Kgp) (Pavloff et al., 1997; Scott et al., 1993) and two Arg‐specific (Arg‐gingipains/Rgps) proteinases (Pavloff et al., 1995). Gingipains degrade various human proteins, including complement system proteins, cytokines, and integrin (O'Brien‐Simpson et al., 2001), and their roles as virulence factors have been reviewed elsewhere (Fitzpatrick et al., 2009; Kadowaki et al., 2007; Potempa et al., 2003). The triple‐gingipain gene null mutant KDP136 was shown unable to grow in a defined medium with human albumin as the sole carbon source (Shi et al., 1999), implicating that gingipains are not only virulence factors but also essential for bacterial growth.
Following degradation of extracellular proteins by gingipains, the resultant oligopeptides are imported into periplasmic space by likely an import mechanism. Madej et al., 2020) recently reported the molecular structure of RagAB, the cell wall importer for oligopeptides, and showed that the RagAB dimer forms a large internal cavity, which seems adaptive for accepting oligopeptides from 7 to 29 residues in length.
2.1. DIPEPTIDE PRODUCTION MEDIATED BY DPPs
DPPs are exopeptidases that liberate dipeptides from the unblocked N‐terminus of oligopeptides and proteins, with eight members classified by amino acid sequence similarity and substrate specificities; DPP1–6 (or I–VI), 7, and 11. Roman numerals have been used for DPPI‐DPPVI, though for simplicity and to follow more recent usage, this study uses Arabic numbers for all DPPs. Other DPP members with various designations, i.e., QPP, FAPα (Retting et al., 1994), DPP6, DPP8, and DPP9 (Olsen & Wagtmann, 2002), DPP10 (Qi et al., 2003), and DPP‐A and DPP‐B are subtypes of eukaryotic DPP4, because of their sequence similarities (Table 1).
TABLE 1.
DPPs and two related exopeptidases expressed in P. gingivalis
| Name |
IUBMB |
MEROPS code | Gene code a | Specificity b | Synthetic and native substrate | Isozyme | Reference |
|---|---|---|---|---|---|---|---|
| DPP3 |
EC3.4.14.4 |
M49.001 |
PGN_01645 |
NH2‐RR‐|‐ |
RR‐NHMec | Ellis & Nuenke (1967); Ohara‐Nemoto et al. (2014) | |
| DPP4 |
EC3.4.14.5 |
S9.013 |
PGN_1469 |
NH2‐XP‐|‐ NH2‐XA‐|‐ |
GP‐MCA Incretin |
FAPα/seprase DPP6/DPPX DPP8/DPRP1 DPP9/DPRP2 DPP10/DPRP3 DPP‐A and B |
Banbula et al. (2000); Ohara‐Nemoto et al. (2017) |
|
DPP5 |
S9.012 |
PGN_0756 |
NH2‐AA‐|‐ NH2‐HS‐|‐ NH2‐SY‐|‐ |
KA‐MCA |
Ohara‐Nemoto et al. (2014) |
||
| DPP6 | C40.001 |
PGN_0754 |
L‐Ala‐γ‐D‐Glu‐|‐ | Guinand et al. (1979) | |||
| DPP7 | S46.001 | PGN_1479 |
NH2‐AF‐|‐ NH2‐ZZ‐|‐ |
FM‐MCA | Banbula et al. (2001); Rouf, et al. (2013); Nemoto et al. (2018) | ||
| DPP11 | S46.002 | PGN_0607 |
NH2‐ZD‐|‐ NH2‐ZE‐|‐ |
LD‐MCA LE‐MCA |
Ohara‐Nemoto et al. (2011); Rouf, et al. (2013); Nemoto et al. (2017) | ||
| AOP | S9C | PGN_1349 |
Acyl‐ZZ‐|‐ Acyl‐XZZ‐|‐ |
Benzyloxycarbonyl‐KM‐MCA | Nemoto et al. (2016); Nemoto et al. (2019) | ||
| PTP‐A |
EC3.4.14.12 |
S9.017 | PGN_1149 | NH2‐XXP‐|‐ | GAP‐β‐naphthylamide | Banbula et al. (1999); Ito et al. (2006) |
P. gingivalis ATCC 33277 (Naito et al., 2008).
X, any amino acid, Z, hydrophobic amino acid, “‐|‐”, cleavage site
The substrate specificity of DPPs is primarily defined based on the penultimate amino acid residue from the N‐terminus (P1 position), and the N‐terminal residue (P2‐position) has effects to some extent on activity towards P. gingivalis DPP7 and DPP11 (Rouf, et al., 2013). Pro at the P1’‐position, the C‐terminal side residue of the hydrolyzed peptide bond, is never accepted by any DPP, thus peptides with NH2‐Xaa1‐Xaa2‐Pro3‐ are resistant to all DPPs. P1’‐position and its C‐terminal adjacent P2’‐position residues, except for Pro, are least involved in substrate recognition. Structural analysis of human DPP4 revealed a limited interaction with P1’‐position Ser and P2’‐position Lys of YP/SKPDNPGE, first ten residues of neuropeptide (Aertgeerts et al., 2004). Similarly, P1’‐ and P2’‐position residues of Leu‐Asp/‐Val‐Trp exhibit few interactions with P. endodontalis DPP11 (Bezerra et al., 2017). These findings indicate that both P1’ and P2’ residues are not tightly associated with DPP4 and DPP11, and presumably other DPPs as well. Polypeptides with N‐terminal modification are not acceptable for DPPs. In P. gingivalis and human DPP4, this is due to the necessity of the ionic interaction between the N‐terminal α‐amino group of the substrate and carboxy groups of Glu195 and Glu196 (Rea et al., 2017).
Porphyromonas gingivalis possesses six DPPs, i.e., DPP3–7, and DPP11. A cross‐search of the oral bacterial and homology databases indicated that 43 species of 772 taxa possess DPP4, DPP5, DPP7, and DPP11 genes (Ohara‐Nemoto et al., 2018). Major bacterial species belong to the genera Bacteroides, Porphyromonas, Prevotella, Tannerella, and Capnocytophaga in the phylum Bacteroidetes, indicating that all or most of these DPPs are specifically distributed in anaerobic and facultative anaerobic oral rods, which are likely included in subgingival biofilm (Ohara‐Nemoto et al., 2018). In fact, DPP4, DPP5, DPP7, and DPP11 activities have been detected in Porphyromonas endodontalis (Nishimata et al., 2014), Tan. forsythia, and Pre. intermedia, as well as P. gingivalis and subgingival plaque specimens (Ohara‐Nemoto et al., 2018).
2.2. DPP3
DPP3 belongs to the M49.001 peptidase family and is widely distributed among eukaryotic (Ellis & Nuenke, 1967) and prokaryotic organisms, and is known to readily hydrolyze NH2‐Arg1‐Arg2‐|‐Xaa3‐. P. gingivalis DPP3 exhibits a 19.0% amino acid identity to the human entity and shows the highest activity for a synthetic substrate, Arg‐Arg‐4‐methycoumaryl‐7‐amide (MCA), followed by Leu‐Arg‐, Ala‐Arg‐, and Lys‐Ala‐MCA, the same as the eukaryotic entity (Ohara‐Nemoto et al., 2014). P. gingivalis DPP3 is composed of 906 and 886 amino acid residues in the strains ATCC 33277 and W83, respectively, which are substantially larger than 675 residues of Bacteroides thetiotaomicron DPP3 and 737 residues of human DPP3 due to the C‐terminally tagged sequence. The additional sequence is predicted at high confidence to have α‐α superhelix fold, belonging to Armadillo‐type fold family similar to the AlkD family of bacterial glycosylases (Hromić‐Jahjefendić et al., 2017). The genes encoding DPP3 with AlkD domain are specifically found within the genus Porphyromonas.
There are two lines of evidence indicating that P. gingivalis DPP3 is not responsible for dipeptide production from nutritional proteins. First, it does not contain a signal sequence, thus seems to be localized in cytoplasm, and second, P. gingivalis cells show little hydrolysis for Arg‐Arg‐MCA (Ohara‐Nemoto et al., 2014). Accordingly, we speculate that DPP3 functions for degradation of cytosolic proteins and that degradation of nutritional peptides at Arg‐Xaa bonds is already completed by Arg‐gingipains.
2.3. DPP4
DPP4 is widely distributed in a variety of species from eukaryotic (S9.003) (Misumi et al., 1992) to bacterial (S9.009) (Ahrén et al., 2000). Human DPP4/CD26 hydrolyzes the peptide bond at the carboxyl side of Pro2 most preferentially and Ala2 at a reduced rate. DPP4 is a key factor that modulates postprandial hyperglycemia by degrading incretin peptides, i.e., glucagon‐like peptide 1 (GLP‐1) and glucose‐dependent insulinotropic polypeptide (GIP), which induce secretion of insulin from pancreatic β cells. DPP4 inhibitors, a class of oral antidiabetic agents, such as sitagliptin, vildagliptin, and alogliptin, suppress degradation of incretins and improve glucose tolerance and insulin secretion (Ahrén et al., 2000; Gallwitz, 2019; Weber, 2004). Human DPP4 is a type II membrane protein, in which the N‐terminal hydrophobic sequence represents an uncleavable signal peptide, and functions as a membrane‐anchoring domain (Misumi et al., 1992), and the entity expressed in lymphocytes, and endothelial and epithelial cells as a functional receptor for human Middle East respiratory syndrome coronavirus (MERS‐CoV) (Raj et al., 2013).
Porphyromonas gingivalis DPP4 exhibits the substrate specificity similar to that of human DPP4 (Banbula et al., 2000) and a very overall structure to human entity (Rea et al., 2017), despite a limited amino acid identity (23.8%) to human DPP4 (Kiyama et al., 1998). Periodopathic bacterial DPP4 as well as human DPP4 degrades incretin and modulates blood glucose levels in mice (Ohara‐Nemoto et al., 2017). This issue will be described in detail in Section 9. P. gingivalis DPP4 also degrades substance P, fibrin inhibitory peptide, IL‐1β, IL‐2, and β‐casomorphin in vitro (Banbula et al., 2000). Mice injected with the P. gingivalis W83 strain were found to develop abscesses to a greater extent and died more frequently than those challenged with a dpp4‐deficient strain (Kumagai et al., 2000). However, this phenomenon may not represent virulence potential, but the nutritional contribution of DPP4 (Ohara‐Nemoto et al., 2014).
Human DPP4 inhibitors, i.e., P32/98, viladagliptin, and sitagliptin, also suppress the activity of periodontopathic bacterial DPP4 at 250 µM (Ohara‐Nemoto et al., 2014). A recent quantitative analysis demonstrated IC50 of vildagliptin (1.3 µM) and sitagliptin (18 µM) to human DPP4, which are 1/10.8‐ and 1/450‐fold lower IC50 concentrations of vildagliptin and sitagliptin, respectively, required for P. gingivalis DPP4 (Rea et al., 2017). These findings suggest that P. gingivalis DPP4 entering into the bloodstream is inefficiently inhibited by prescribed DPP4 inhibitors for type 2 diabetes mellitus patients (Rea et al., 2017).
2.4. DPP5
DPP5 was initially found in Aspergillus fumigatus (Beauvais et al., 1997), then later in P. gingivalis as the first bacterial species (Ohara‐Nemoto et al., 2014). Based on the discovery of P. gingivalis DPP5, its wide distribution among bacteria and archaea, as well as in eukaryotes including higher animals and plants has been revealed.
The amino acid sequence of P. gingivalis DPP5 is 28.5% identical to that of A. fumigatus DPP5. P. gingivalis DPP5 preferentially removes dipeptides with Ala and hydrophobic residues at the P1 position from synthetic dipeptidyl MCA substrates. Lys‐Ala‐MCA has been identified as the most potent substrate, though no native substrate has yet been identified.
2.5. DPP6
DPP6 removes the N‐terminal dipeptide L‐Ala‐D‐Glu by hydrolysis of the γ‐D‐glutamyl‐(L)‐diamino acid bond (Guinand et al., 1979). The substrates L‐Ala‐γ‐D‐Glu‐L‐Zaa‐Y (Zaa = a diamino acid that may be L‐lysine, meso‐diaminopimelic acid, ω‐amidated‐ or meso‐diaminopimelic acid, and Y is either D‐Ala or D‐Ala‐D‐Ala) are the peptide moieties of bacterial peptidoglycans. DPP6 belongs to C40 family, of which the representative is that of Bacillus sphaericus. B. sphaericus DPP6 is composed of 271 amino acids with no signal peptide, as expected for a cytoplasmic enzyme. The gene of DPP6 (PGN_0754) encoding a 201 amino acid protein is also present in P. gingivalis, although there is no biochemical characterization on this gene product. A biochemical analysis is needed to elucidate the role of P. gingivalis DPP6.
2.6. DPP7
DPP7, a member of S46.001, was initially discovered in P. gingivalis and does not exist in eukaryotic organisms. In this respect, it should be noted that human DPP7 is not related to bacterial DPP7, but rather is an isozyme of DPP2 (Bezerra et al., 2012). DPP7 releases the N‐terminal dipeptide NH2‐Xaa1‐Zaa2 (Zaa, hydrophobic residues) (Banbula et al., 2001; Rouf, et al., 2013), and hydrolyzes insulin B chain, type I collagen, and azocasein in vitro (Banbula et al., 2001) Since the P1‐position specificity of DPP7 overlaps with that of DPP5, findings of a specific synthetic substrate for DPP7 were eagerly anticipated and Phe‐Met‐MCA finally identified (Nemoto et al., 2018). As a result, it is now possible to distinctly measure all DPP activities expressed in bacteria and clinical specimens with Arg‐Arg‐, Gly‐Pro‐, Lys‐Ala‐, Phe‐Met‐, and Leu‐Asp‐MCA for DPP3, DPP4, DPP5, DPP7, and DPP11, respectively, as well as for determining activities in saliva and subgingival dental plaque specimens. Their activities were successfully determined in an exception of the DPP4 activity of saliva due to the presence of human DPP4 (Ohara‐Nemoto et al., 2018).
Although both DPP5 and DPP7 show a common hydrophobic P1 preference, DPP5 has no apparent amino acid preference at the P2 position (Ohara‐Nemoto et al., 2014), in contrast to the hydrophobic P2 preference of DPP7 (Rouf, et al., 2013). Thus, the difference in P2‐position preference between DPP5 and DPP7 amplifies the repertoire of peptide substrates of P. gingivalis. For example, P. gingivalis DPP7 hydrolyzes Leu‐Arg‐, Leu‐Gln‐, and Leu‐Glu‐MCA to some extent, while those are scarcely hydrolyzed by DPP5 (Ohara‐Nemoto et al., 2014). Hence, hydrolyzation of Leu‐Gly‐β‐naphthylamine, which has been observed in five P. gingivalis strains, is likely mediated by DPP7 (Suido et al., 1986).
2.7. DPP11
A metabolite analysis of P. gingivalis indicated that glutamic acid/glutamine (Glx)‐ and aspartic acid/asparagine (Asx)‐containing peptides were the most intensively consumed (Takahashi et al., 2000), and those experimental results were supported by computational analysis of the genome‐scale metabolic network using flux balance calculation (Mazumdar et al., 2009). However, how Asp‐ and Glu‐containing dipeptides are produced in the bacterium was not elucidated, until being revealed by the finding of novel Asp‐ and Glu‐specific DPP 11 (Ohara‐Nemoto et al., 2011).
A previous study noted that P. gingivalis DPP11 encoded by PGN_0607 in strain ATCC 33277 may be an isozyme of DPP7, because of its 38.4% amino acid identity to DPP7 encoded by PGN_1479 (Banbula et al., 2001). However, it was later shown that the product of PGN_0607 is a novel DPP specific for acidic amino acid residues Asp and Glu, exclusively distinct from the hydrophobic specificity of DPP7, thus DPP11 was classified as an S46.002 peptidase. S46 family peptidases are solely distributed in bacteria, and DPP11 as well as DPP7 prefer hydrophobic residues at the P2 position (Rouf, et al., 2013).
The acidic P1‐position residue is recognized by Arg673 in P. gingivalis DPP11. Biochemical studies have revealed that there are three subtypes of DPP11; (a) Porphyromonas‐type DPP11, which is Asp‐preferential, (b) Bacteroides‐type DPP11, which shows a high preference for Asp and scarcely accepts Glu, and (c) Shewanella‐type DPP11, in which S1 Ser673 instead of Arg673, primarily exhibits higher preference for Glu (Nemoto et al., 2017).
In DPP7, Arg673 of DPP11 is replaced by Gly666, which allows the recognition of bulky hydrophobic P1 residue (Ohara‐Nemoto et al., 2011). Furthermore, Gly666Arg substitution in recombinant DPP7 resulted in a partial acquisition of hydrolyzing activity against acidic residues (Rouf, et al., 2013). Also, elucidation of the three‐dimensional structures of a DPP7‐family member, DAP BII, from Pseudoxanthomonas mexicana (Sakamoto et al., 2014) and P. gingivalis DPP11 (Bezerra et al., 2017; Sakamoto et al., 2015) confirmed the essential positionings of Gly666 of DPP7 and Arg673 of DPP11 for P1‐position residue recognition. Coding sequence annotation tools that adopt sequence similarities can present misleading results regarding the classification of DPP7 and DPP11. Instead, we proposed that two S46‐family members can be readily classified by use of characteristic residues, i.e., Gly666 of DPP7 and an equivalent residue Arg673 of DPP11 (Ser673 in Shewanella‐type DPP11) (Rouf, et al., 2013) This classification was then successfully used to identify Capnocytophaga canimorsus DPP7 as it contains Gly666 (Hack et al., 2017), despite the annotation regarding a DPP11‐like protein in the MEROPS database (Rawlings et al., 2014). Besides, Bezerra et al. (2017) demonstrated a distinct thermodynamic signature in P. gingivalis DPP11 showing that protein conformational entropy is the main driving‐force for substrate binding. Since S46‐family members DPP7 and DPP11 are not present in eukaryotic organisms, they are attractive targets for drugs of periodontal disease. In this line, Sakamoto et al. (2019) developed nonpeptidyl DPP11 inhibitor SH‐5 with IC50 of 90.1 µM, which showed a dose‐dependent inhibition of the growth of P. gingivalis.
3. EXOPEPTIDASES FACILITATING DIPEPTIDE PRODUCTION
The existence of multiple DPPs should be beneficial for complete degradation of polypeptides into dipeptides. However, there appear to be two types of peptide bonds resistant to bacterial DPPs identified to date. First, N‐terminally‐acylated polypeptides are resistant to all DPPs as well as prolyl tripeptidyl‐peptidase A (PTP‐A), even though plasma proteins serving as potential nutrients for P. gingivalis are frequently N‐terminally modified. Second, polypeptides with Pro at the third position from the N‐terminus are resistant to DPPs.
The first issue is surmounted by acylpeptidyl‐oligopeptidase (AOP), which preferentially degrades polypeptides with N‐terminal modification into di‐ and tri‐peptides (Nemoto et al., 2016). The second one is also surmounted by PTP‐A, which liberates NH2‐Xaa1‐Xaa2‐Pro3 from polypeptides (Banbula et al., 1999; Ito et al., 2006). In other words, AOP and PTP‐A have possibly evolved to complete dipeptide production by DPPs in P. gingivalis.
3.1. AOP
AOP, which belongs to the S09 family, preferentially degrades N‐terminally acylated polypeptides to liberate acylated‐di‐ and tri‐peptides (Nemoto et al., 2016). The most potent synthetic substrate for P. gingivalis AOP is benzyloxycarbonyl‐Lys‐Met‐MCA. The structure of acylated compounds is not of concern, whereas concealment of the N‐terminal α‐amino group is essential for efficient degradation. Although acylaminoacyl‐peptidase (AAP), which specifically removes N‐terminal acylated amino acids from a polypeptide, has been previously reported in the human and archaea Aeropyrum pernix (Kiss et al., 2007), the amino acid sequence identity (15.8%) between P. gingivalis AOP and Aeropyrum pernix AAP is limited, while AOP scarcely removes acylated N‐terminal amino acids but dipeptides. Modeling of P. gingivalis AOP revealed the hydrophobic S1 site of AOP in accord with its hydrophobic P1 preference as well as an N‐anchor region determining the substrate specificity with a more open active site than those of DPPs accepting a broad range of N‐terminal different steric groups (Nemoto et al., 2016).
Comparisons of three AOPs from P. gingivalis, Bacteroides dorei, and Lysinibacillu sphaericus revealed that the activity of P. gingivalis AOP was most significantly enhanced by acylation of substrates, further supporting the removal of acylated N‐terminus as the primary role of that AOP (Nemoto et al., 2019).
3.2. PTP‐A
PTP‐A was first discovered in P. gingivalis and belongs to S9.017, and has been shown to hydrolyze the Pro3‐Xaa4 bond (Banbula et al., 1999). Reflecting the similarity of substrate specificities, the amino acid sequence of P. gingivalis PTP‐A is 23.5% identical to that of P. gingivalis DPP4. An unblocked N‐terminus is required for its activity and no cleavage occurs with substrates with Pro at the P1’ position. Synthetic substrates, Ala‐Ala‐Pro‐ and Gly‐Ala‐Pro‐p‐nitroanilide, are used for the assay of PTP‐A, and it has been proposed that PTP‐A is involved in the degradation of type I collagen in connection with periodontitis inflammation (Ito et al., 2006). However, because the Gly‐Xaa‐Pro repeat is located at the center of the collagen sequences, it is ambiguous whether this exopeptidase truly degrades collagens in host tissues. Hence, the primary role of PTP‐A is likely to provide oligopeptides without Pro at the third position from the N‐terminus for DPPs. Genome analysis (Naito et al., 2008) as well as biochemical studies up to date failed to find tripeptidyl‐peptidases except for PTP‐A, which strongly suggests the absence of other tripeptidyl‐peptidases in P. gingivalis.
3.3. FOUR DPPs, AOP, AND PTP‐A LOCATED IN PERIPLASM
The enzymatic activities of DPPs, AOP, and PTP‐A are scarcely detected in bacterial culture supernatant, indicating that they are cell‐associated. In addition, we previously showed the periplasmic localization of DPP5 by subcellular fractionation (Ohara‐Nemoto et al., 2014). Although the subcellular localization of other DPPs, AOP, and PTP‐A has not been biochemically verified, they possess typical signal sequences for export from the inner membrane but do not possess a conserved C‐terminal domain (CTD) of 70–80 amino acids, a prerequisite for secretion via the Type‐9 secretion system (T9SS) (Sato et al., 2010; Seers et al., 2006; Slakeski et al., 2011). In accord with signal sequence prediction of Met1‐Ala21 in P. endodontalis DPP11, the N‐terminal Asp22 was identified in an endogenous form (Ohara‐Nemoto et al., 2011). Accordingly, DPPs are transported through the inner membrane and localized in the periplasmic space. P. gingivalis AOP also possesses a potent signal sequence. Although the probability is less significant, Met1‐Ala38 in P. gingivalis PTP‐A may correspond to the signal peptide. Proteome analysis of P. gingivalis W50 also suggested the localization in the lumen of the vesicle (periplasm) of DPP4 (PG_0503), DPP5 (PG_1004), DPP7 (PG_0491), and DPP11 (PG_1283) (Veith et al., 2014). Moreover DPP4 seems to be is enriched in outer membrane vescicle under heme excess conditions (Veith et al., 2018). In contrast, DPP3 and DPP6 seem to be located in the cytoplasm, and thus, not involved in the degradation of extracellular proteins in the periplasm. Their roles should be elucidated in future studies.
Taken together, the degradation pathway of extracellular proteins in P. gingivalis, which is initiated by gingipains and finally reach dipeptides mediated by DPPs with the help of AOP and PTP‐A, is schematically illustrated in Figure 1. We currently suppose that oligopeptidyl substrates and dipeptidyl MCA are incorporated across the outer membrane via RagAB.
FIGURE 1.
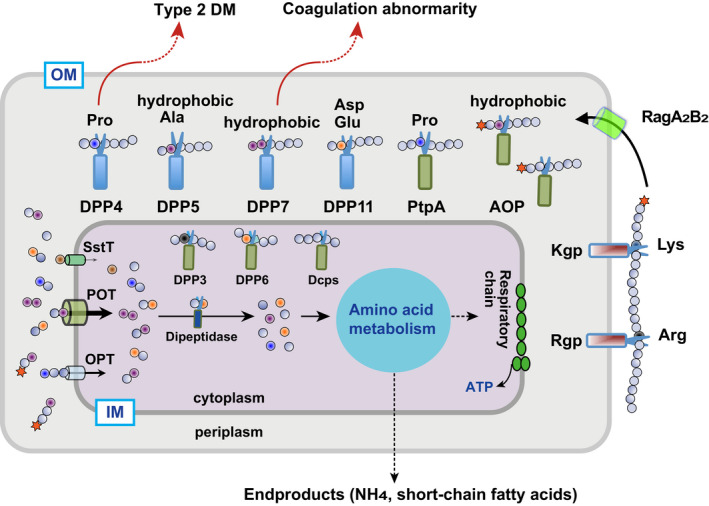
Schematic illustration on dipeptide production and incorporation in Porphyromonas gingivalis. OM and IM, outer and inner membranes, respectively [Colour figure can be viewed at wileyonlinelibrary.com]
4. DIPEPTIDE INCORPORATION BY PROTON‐DEPENDENT OLIGOPEPTIDE TRANSPORTER POT
Genome information for P. gingivalis indicates the existence of a single copy of each of three amino acid/oligopeptide transporters, i.e, serine/threonine transporter (SstT), proton‐dependent oligopeptide transporter (POT), and oligopeptide transporter (OPT) (Naito et al., 2008; Nelson et al., 2003). Among them, only P. gingivalis SstT, which incorporates amino acids with most efficiently Ser and Thr, has been reported (Dashper et al., 2001). The analysis of transporter‐gene disrupted strains of P. gingivalis indicates that a Δpot mutant of P. gingivalis had a significant defect in incorporation of dipeptides, while a Δopt strain had a moderate decrease in several dipeptides (Ohara‐Nemoto et al., 2019). As expected, a ΔsstT strain did not exhibit a decrease in dipeptide incorporation. Moreover, the growth of the Δpot strain was the most significantly retarded among the three single transporter‐deficient mutants, and the double mutant Δpot‐Δopt scarcely grew (Ohara‐Nemoto et al., 2019). These results indicated that nutritional dipeptides are primarily incorporated by POT in P. gingivalis.
It seems unusual that there is only a single copy of each of the three transporter genes in P. gingivalis, because there are multiple transporter paralogues in other bacteria. For example, E. coli possess four POT members (ydgH, ydgR, yhiP, yidL) (Ernst et al., 2009; Weitz et al., 2007), and Helicobacter pylori, limited to utilization of carbohydrates as carbon sources, has five POT (dppA, B, C, D, F) and four OPT (oppA, B, C, D) paralogs (Salama et al., 2000; Weinberg & Maier, 2007). The presence of multiple sets of transporter genes suggests that these bacteria readily respond to changes in nutritional conditions, while P. gingivalis resides in the consistently poor nutritional environment of subgingival dental plaque.
5. DIPEPTIDASES DEGRADE DIPEPTIDES INTO SINGLE AMINO ACIDS
Dipeptides incorporated into bacterial cells are cleaved by a dipeptidase to two amino acids, which enter the metabolic pathway of the cell (Figure 1). To date, little is known regarding dipeptidases in P. gingivalis and other periodontopathic bacteria. We recently identified Arg‐preferential dipeptidase A [locus tag, PIOMA14_1_1238, MEROPS code, BAU17746] in Pre. intermedia, which belongs to C69.001 (Sarwar et al., 2020). Pre. intermedia dipeptidase A most preferentially degrades Arg‐Leu and Arg‐Phe, and does other dipeptides, such as Leu‐Leu and Glu‐Glu, moderately. It never degrades tripeptides and does not show any amino peptidase ativity (Sarwar et al., 2020). Dipeptidase A/PepDA (MER0233043) from Lactobacillus helveticus, the representative enzyme of the C69.001 members, was reported to possess a hydrophobic P1 preference (Dudley et al., 1996; Vesanto et al., 1996). However, a later reexamination revealed that it also possesses an Arg preference. Pre. intermedia encodes one additional C69‐family member (BAU18827), though no hydrolyzing activity has been observed (Sarwar et al., 2020). P. gingivalis carries a single C69 family member, PGN_1103, of which the deduced sequence is 13.1% and 47.3% identical to that of BAU17746 and BAU18827 of Pre. intermedia, respectively. Therefore, PGN_1103 is likely to be an ortholog of BAU18827 and again results of our preliminary search of PGN_1103 revealed no dipeptidase activity present.
6. OTHER POTENTIAL EXOPEPTIDASES INVOLVED IN DIPEPTIDE PRODUCTION
Genome information demonstrates the presence of three additional uncharacterized S9‐family members; PGN_1542 (annotated S9 unassigned), PGN_1694 (S9 unassigned), and PGN_1878 (Ala‐DPP of S9C), of which the deduced sequences are predicted to possess signal sequences, Met1‐Ala21, Met1‐Ala20, and Met1‐Ser20, respectively, indicating their periplasm localization. They have been expressed as recombinant proteins, though their peptidase activities have yet to be identified (Nemoto et al., 2016; Ohara‐Nemoto et al., 2011). Peptidyl‐dipeptidase (Dcp) is an M3‐family exopeptidase that liberates C‐terminal dipeptides from longer polypeptides (Paschoalin et al., 2013) and when present, efficiency for producing dipeptides should be facilitated. P. gingivalis truly possesses two potential Dcp genes, PGN_1776 (MER0034589) and PGN_0788 (MER003588), though they have no signal sequences. Accordingly, these potential Dcps may be cytosolic proteins and are unlikely to be involved in dipeptide production in the periplasm.
6.1. P. GINGIVALIS DPPs RELATING TO SYSTEMIC DISEASES
Currently, the two‐way relationship between periodontal disease and type 2 diabetes mellitus is accounted for by chronic inflammation, in which an elevated level of tumor necrosis factor‐α derived from the liver by lipopolysaccharide of periodontopathic Gram‐negative bacteria, results in insulin resistance (Soorya et al., 2014; Takano et al., 2010; Teeuw et al., 2010). We recently proposed that an additional and direct link exists between the two diseases, in which periodontopathic bacterial DPP4 degrades incretins (Ohara‐Nemoto et al., 2017). Since incretins are multifunctional (Seino & Yabe, 2013) and other bioactive peptides including gastrointestinal hormones, neuropeptide, and chemokines could be potential targets, periodontopathic bacterial DPPs may modulate human homeostasis via inactivation of these molecules.
6.2. DPP4 as a modulator for type 2 diabetes mellitus
Human DPP4 cleaves the peptide bond between the second Ala and third Glu of GLP‐1 and GIP, causing rapid inactivation (Mentlein et al., 1993). Hence, DPP4 inhibitors are widely used to treat type 2 diabetes mellitus patients. Because of the common enzymatic features, the involvement of periodontopathic bacterial DPP4 in modulation of blood glucose level is reasonably inferred.
Porphyromonas gingivalis cells were found to efficiently degrade GLP‐1 and GIP, while a Δdpp4 mutant did not. In a glucose tolerance test, intravenous injection into mice of recombinant DPP4 from P. gingivalis, T. forsythia, and Pre. intermedia reduced the plasma GLP‐1 active form and insulin levels, which were accompanied by a substantial elevation in postprandial hyperglycemia together with retardation of the decrease in blood glucose levels (Ohara‐Nemoto et al., 2017). Since oral bacteremia occurs as a result of mastication, toothbrushing, and dental procedures (Tomás et al., 2012), the higher incidence of periodontopathic bacteremia in individuals suffering from severe periodontal disease seems to exacerbate type 2 diabetes mellitus through degradation of incretins by bacterial DPP4 in the bloodstream. Oral bacteria that form subgingival biofilm possess DPP4 orthologues, thus may also participate in this phenomenon. Because human DPP4 inhibitors are less efficient for bacterial DPP4 (Rea et al., 2017), development of periodontopathic DPP4‐specific inhibitors and the usage for type 2 diabetes mellitus patients may synergistically reduce blood glucose concentration.
6.3. DPP7 as a virulence factor of hemolysis
Infection by C. canimorsus caused by a severe dog or cat bite occasionally induces bleeding and coagulation abnormalities. Hack et al. (2017) reported that. C. canimorsus DPP7, which degrades and inactivates coagulation factor X at the N‐termini of its light and heavy chains, is responsible for these conditions. Their study strongly suggests that P. gingivalis and Capnocytophaga gingivalis DPP7 may retard the prothrombin time and activated partial thromboplastin time. This capability of DPP7 should be profitable for periodontopathic bacteria, because retardation of coagulation may provide hemoglobin and plasma proteins as nutrition sources for periodontopathic asaccharolytic bacteria. This issue should be further investigated in the future.
7. CONCLUDING REMARKS
Porphyromonas gingivalis, an asaccharolytic Gram‐negative rod, utilizes extracellular proteins as its sole carbon and energy sources. Since nutritional polypeptides are mainly incorporated as dipeptides in bacterial cells, they should be initially degraded into oligopeptides by endopeptidases, and Lys‐ and Arg‐gingipains located at the outer membrane, and then degraded into dipeptides by DPP4, DPP5, DPP7, and DPP11. Two exopeptidases, AOP and PTP‐A, function to transform oligopeptides into forms cleavable by DPPs. Dipeptides are incorporated in the cells mainly by the POT transporter. Bacterial DPP4 and DPP7 are closely related to human systemic diseases. Since multifunctional incretin peptides and other bioactive peptides, including gastro‐intestinal hormones, neuropeptides, and chemokines, are potent targets, periodontopathic bacterial DPPs are psossively involved in modulation of human homeostasis via degradation of these molecules. In conclusion, proteins involved in dipeptide production and incorporation have emerged as potent targets in therapy for periodontal and related systemic diseases.
CONFLICT OF INTEREST
None to declare.
Peer Review
The peer review history for this article is available at https://publons.com/publon/10.1111/omi.12317.
ACKNOWLEDGMENTS
The authors thank Drs. Yu Shimoyama (Iwate Medical University), and Toshio Ono, Tomomi T. Baba, and Takeshi Kobayakawa (Nagasaki University) for their contributions including production of experimental data, as well as Drs. Mariko Naito and Koji Nakayama (Nagasaki University) for their contributions regarding genome information for P. gingivalis and Pre. intermedia. This study was supported by JSPS KAKENHI Grants (JP19K10045 to T.K.N., JP19K10071 to Y.O.‐N.) and a Grant from the Kyushu Dental Association (to T.K.N.).
Nemoto TK, Ohara Nemoto Y. Dipeptidyl‐peptidases: Key enzymes producing entry forms of extracellular proteins in asaccharolytic periodontopathic bacterium Porphyromonas gingivalis . Mol Oral Microbiol.2021;36:145–156. 10.1111/omi.12317
REFERENCES
- Aertgeerts, K. , Ye, S. , Tennant, M. G. , Kraus, M. L. , Rogers, J. , Sang, B. C. , Skene, R. J. , Webb, D. R. , & Prasad, G. S. (2004). Crystal structure of human dipeptidyl peptidase IV in complex with a decapeptide reveals details on substrate specificity and tetrahedral intermediate formation. Protein Science, 13(2), 412–421. 10.1110/ps.03460604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrén, B. , Holst, J. J. , Mårtensson, H. , & Balkan, B. (2000). Improved glucose tolerance and insulin secretion by inhibition of dipeptidyl peptidase IV in mice. European Journal of Pharmacology, 404(1–2), 239–245. 10.1016/s0014-2999(00)00600-2 [DOI] [PubMed] [Google Scholar]
- Banbula, A. , Bugno, M. , Goldstein, J. , Yen, J. , Nelson, D. , Travis, J. , & Potempa, J. (2000). Emerging family of proline‐specific peptidases of Porphyromonas gingivalis: Purification and characterization of serine dipeptidyl peptidase, a structural and functional homologue of mammalian prolyl dipeptidyl peptidase IV. Infection and Immunity, 68(3), 1176–1182. 10.1128/iai.68.3.1176-1182.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banbula, A. , Mak, P. , Bugno, M. , Silberring, J. , Dubin, A. , Nelson, D. , Travis, J. , & Potempa, J. (1999). Prolyl tripeptidyl peptidase from Porphyromonas gingivalis. A novel enzyme with possible pathological implications for the development of periodontitis. Journal of Biological Chemistry, 274(14), 9246–9252. 10.1074/jbc.274.14.9246 [DOI] [PubMed] [Google Scholar]
- Banbula, A. , Yen, J. , Oleksy, A. , Mak, P. , Bugno, M. , Travis, J. , & Potempa, J. (2001). Porphyromonas gingivalis DPP‐7 represents a novel type of dipeptidylpeptidase. Journal of Biological Chemistry, 276(9), 6299–62305. 10.1074/jbc.M008789200 [DOI] [PubMed] [Google Scholar]
- Beauvais, A. , Monod, M. , Debeaupuis, J. P. , Diaquin, M. , Kobayashi, H. , & Latgé, J. P. (1997). Biochemical and antigenic characterization of a new dipeptidyl‐peptidase isolated from Aspergillus fumigatus . Journal of Biological Chemistry, 272(10), 6238–6244. 10.1074/jbc.272.10.6238 [DOI] [PubMed] [Google Scholar]
- Bezerra, G. A. , Dobrovetsky, E. , Dong, A. , Seitova, A. , Crombett, L. , Shewchuk, L. M. , Hassell, A. M. , Switzer, S. M. , Sweitzer, T. D. , McDevitt, P. J. , Johanson, K. O. , Kennedy‐Wilson, K. M. , Cossar, D. , Bochkarev, A. , Gruber, K. , & Dhe‐Paganon, S. (2012). Structures of human DPP7 reveal the molecular basis of specific inhibition and the architectural diversity of proline‐specific peptidases. PLoS One, 7(8), e43019. 10.1371/journal.pone.0043019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezerra, G. A. , Ohara‐Nemoto, Y. , Cornaciu, I. , Fedosyuk, S. , Hoffmann, G. , Round, A. , Márquez, J. A. , Nemoto, T. K. , & Djinović‐Carugo, K. (2017). Bacterial protease uses distinct thermodynamic signatures for substrate recognition. Scientific Reports, 7(1), 2848. 10.1038/s41598-017-03220-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashper, S. G. , Brownfield, L. , Slakeski, N. , Zilm, P. S. , Rogers, A. H. , & Reynolds, E. C. (2001). Sodium ion‐driven serine/threonine transport in Porphyromonas gingivalis . Journal of Bacteriology, 183(14), 4142–4148. 10.1128/JB.183.14.4142-4148.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detert, J. , Pischon, N. , Burmester, G. R. , & Buttgereit, F. (2010). The association between rheumatoid arthritis and periodontal disease. Arthritis Research & Therapy, 12, 218. 10.1186/ar3106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominy, S. S. , Lynch, C. , Ermini, F. , Benedyk, M. , Marczyk, A. , Konradi, A. , Nguyen, M. , Haditsch, U. , Raha, D. , Griffin, C. , Holsinger, L. J. , Arastu‐Kapur, S. , Kaba, S. , Lee, A. , Ryder, M. I. , Potempa, B. , Mydel, P. , Hellvard, A. , Adamowicz, K. , … Potempa, J. (2019). Porphyromonas gingivalis in Alzheimer's disease brains: Evidence for disease causation and treatment with small‐molecule inhibitors. Science Advances, 5(1), eaau3333. 10.1126/sciadv.aau3333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley, E. G. , Husgen, A. C. , He, W. , & Steele, J. L. (1996). Sequencing, distribution, and inactivation of the dipeptidase A gene (pepDA) from Lactobacillus helveticus CNRZ32. Journal of Bacteriology, 178(3), 701–704. 10.1128/jb.178.3.701-704.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, S. , & Nuenke, J. M. (1967). Dipeptidyl arylamidase III of the pituitary: Purification and characterization. Journal of Biological Chemistry, 242(20), 4623–4629. [PubMed] [Google Scholar]
- Ernst, H. A. , Pham, A. , Hald, H. , Kastrup, J. S. , Rahman, M. , & Mirza, O. (2009). Ligand binding analyses of the putative peptide transporter YjdL from E. coli display a significant selectivity towards dipeptides. Biochemical and Biophysical Research Communications, 389(1), 112–116. 10.1016/j.bbrc.2009.08.098 [DOI] [PubMed] [Google Scholar]
- Fitzpatrick, R. E. , Wijeyewickrema, L. C. , & Pike, R. N. (2009). The gingipains: Scissors and glue of the periodontal pathogen. Porphyromonas Gingivalis. Future Microbiology, 4(4), 471–487. 10.2217/fmb.09.18 [DOI] [PubMed] [Google Scholar]
- Gallwitz, B. (2019). Clinical use of DPP‐4 inhibitors. Frontiers in Endocrinology, 10, 389. 10.3389/fendo.2019.0089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genco, R. J. , & VanDyke, T. E. (2010). Prevention: Reducing the risk of CVD in patients with periodontitis. Nature Reviews Cardiology, 7(9), 479–480. https://10.1038/nrcardio.2010.120 . 10.1038/nrcardio.2010.120 [DOI] [PubMed] [Google Scholar]
- Grossi, S. G. , & Genco, R. J. (1998). Periodontal disease and diabetes mellitus: A two‐way relationship. Annals of Periodontology, 3, 51–61. 10.1902/annals.1998.3.1.51 [DOI] [PubMed] [Google Scholar]
- Guinand, M. , Vacheron, M. J. , Michel, G. , & Tipper, D. J. (1979). Location of peptidoglycan lytic enzymes in Bacillus sphaericus . Journal of Bacteriology, 138, 126–132. 10.1128/JB.138.1.126-132.1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hack, K. , Renzi, F. , Hess, E. , Lauber, F. , Douxfils, J. , Dogné, J. M. , & Cornelis, G. R. (2017). Inactivation of human coagulation factor X by a protease of the pathogen Capnocytophaga canimorsus . Journal of Thrombosis and Haemostasis, 15(3), 487–499. 10.1111/jth.13605 [DOI] [PubMed] [Google Scholar]
- Holt, S. C. , & Ebersole, J. L. (2005). Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: The 'red complex', a prototype polybacterial pathogenic consortium in periodontitis. Periodontology, 2000(38), 72–122. 10.1111/j.1600-0757.2005.00113.x [DOI] [PubMed] [Google Scholar]
- Hromić‐Jahjefendić, A. , Jozić, N. J. , Kazazić, S. , Branilović, M. G. , Karačić, Z. , Schrittwieser, J. H. , Das, K. M. P. , Tomin, M. , Oberer, M. , Gruber, K. , Abramić, M. , & Tomić, S. (2017). A novel Porphyromonas gingivalis enzyme: An atypical dipeptidyl peptidase III with an ARM repeat domain. PLoS One, 12(11), e0188915. 10.1371/journal.pone.0188915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, K. , Nakajima, Y. , Xu, Y. , Yamada, N. , Onohara, Y. , Ito, T. , Matsubara, F. , Kabashima, T. , Nakayama, K. , & Yoshimoto, T. (2006). Crystal structure and mechanism of tripeptidyl activity of prolyl tripeptidyl aminopeptidase from Porphyromonas gingivalis . Journal of Molecular Biology, 362(2), 228–240. 10.1016/j.jmb.2006.06.083 [DOI] [PubMed] [Google Scholar]
- Kadowaki, T. , Takii, R. , Yamatake, K. , Kawakubo, T. , Tsukuba, T. , & Yamamoto, K. (2007). A role for gingipains in cellular responses and bacterial survival in Porphyromonas gingivalis‐infected cells. Frontiers in Bioscience, 12, 4800–4809. 10.2741/2428 [DOI] [PubMed] [Google Scholar]
- Kadowaki, T. , Yoneda, M. , Okamoto, K. , Maeda, K. , & Yamamoto, K. (1994). Purification and characterization of a novel arginine‐specific cysteine proteinase (argingipain) involved in the pathogenesis of periodontal disease from the culture supernatant of Porphyromonas gingivalis . Journal of Biological Chemistry, 269(33), 21371–21378. [PubMed] [Google Scholar]
- Kiss, A. L. , Hornung, B. , Rádi, K. , Gengeliczki, Z. , Sztáray, B. , Juhász, T. , Szeltner, Z. , Harmat, V. , & Polgár, L. (2007). The acylaminoacyl peptidase from Aeropyrum pernix K1 thought to be an exopeptidase displays endopeptidase activity. Journal of Molecular Biology, 368(2), 509–520. 10.1016/j.jmb.2007.02.025 [DOI] [PubMed] [Google Scholar]
- Kiyama, M. , Hayakawa, M. , Shiroza, T. , Nakamura, S. , Takeuchi, A. , Masamoto, Y. , & Abiko, Y. (1998). Sequence analysis of the Porphyromonas gingivalis dipeptidyl peptidase IV gene. Biochimica Et Biophysica Acta, 1396(1), 39–46. 10.1016/s0167-4781(97)00225-x [DOI] [PubMed] [Google Scholar]
- Kshirsagar, A. V. , Offenbacher, S. , Moss, K. L. , Barros, S. P. , & Beck, J. D. (2007). Antibodies to periodontal organisms are associated with decreased kidney function: The dental atherosclerosis risk in communities study. Blood Purification, 25, 125–132. 10.1159/000096411 [DOI] [PubMed] [Google Scholar]
- Kumagai, Y. , Konishi, K. , Gomi, T. , Yagishita, H. , Yajima, A. , & Yoshikawa, M. (2000). Enzymatic properties of dipeptidyl aminopeptidase IV produced by the periodontal pathogen Porphyromonas gingivalis and its participation in virulence. Infection and Immunity, 68(2), 716–724. 10.1128/iai.68.2.716-724.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalla, E. , & Papapanou, P. N. (2011). Diabetes mellitus and periodontitis: A tale of two common interrelated diseases. Nature Reviews Endocrinology, 7, 738–748. 10.1038/nrendo.2011.106 [DOI] [PubMed] [Google Scholar]
- Madej, M. , White, J. B. R. , Nowakowska, Z. , Rawson, S. , Scavenius, C. , Enghild, J. J. , Bereta, G. P. , Pothula, K. , Kleinekathoefer, U. , Baslé, A. , Ranson, N. A. , Potempa, J. , & van den Berg, B. (2020). Structural and functional insights into oligopeptide acquisition by the RagAB transporter from Porphyromonas gingivalis . Nature Microbiology, 5, 1016–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumdar, V. , Snitkin, E. S. , Amar, S. , & Segre, S. (2009). Metabolic network model of a human oral pathogen. Journal of Bacteriology, 191(1), 74–90. 10.1128/JB.01123-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentlein, R. , Gallwitz, B. , & Schmidt, W. E. (1993). Dipeptidyl‐peptidase IV hydrolyses gastric inhibitory polypeptide, glucagon‐like peptide‐1(7–36)amide, peptide histidine methionine and is responsible for their degradation in human serum. European Journal of Biochemistry, 214(3), 829–835. 10.1111/j.1432-1033.1993.tb17986.x [DOI] [PubMed] [Google Scholar]
- Misumi, Y. , Hayashi, Y. , Arakawa, F. , & Ikehara, Y. (1992). Molecular cloning and sequence analysis of human dipeptidyl peptidase IV, a serine proteinase on the cell surface. Biochimica Et Biophysica Acta, 1131(3), 333–336. 10.1016/0167-4781(92)90036-y [DOI] [PubMed] [Google Scholar]
- Naito, M. , Hirakawa, H. , Yamashita, A. , Ohara, N. , Shoji, M. , Yukitake, H. , Nakayama, K. , Toh, H. , Yoshimura, F. , Kuhara, S. , Hattori, M. , Hayashi, T. , & Nakayama, K. (2008). Determination of the genome sequence of Porphyromonas gingivalis strain ATCC 33277 and genomic comparison with strain W83 revealed extensive genome rearrangements in P. gingivalis . DNA Research, 15(4), 215–225. 10.1093/dnares/dsn013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazir, M. A. (2017). Prevalence of periodontal disease, its association with 440 systemic diseases and prevention. International Journal of Health Sciences, 11(2), 72–80. [PMC free article] [PubMed] [Google Scholar]
- Nelson, K. E. , Fleischmann, R. D. , DeBoy, R. T. , Paulsen, I. T. , Fouts, D. E. , Eisen, J. A. , Daugherty, S. C. , Dodson, R. J. , Durkin, A. S. , Gwinn, M. , Haft, D. H. , Kolonay, J. F. , Nelson, W. C. , Mason, T. , Tallon, L. , Gray, J. , Granger, D. , Tettelin, Hervé , Dong, H. , … Fraser, C. M. (2003). Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. Journal of Bacteriology, 185(18), 5591–5601. 10.1128/jb.185.18.5591-5601.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto, T. K. , Bezerra, G. A. , Ono, T. , Nishimata, H. , Fujiwara, T. , & Ohara‐Nemoto, Y. (2017). Identification of a new subtype of dipeptidyl peptidase 11 and a third group of the S46‐family members specifically present in the genus Bacteroides . Biochimie, 147, 25–35. 10.1016/j.biochi.2017.10.015 [DOI] [PubMed] [Google Scholar]
- Nemoto, T. K. , Ohara‐Nemoto, Y. , Bezerra, G. A. , Shimoyama, Y. , & Kimura, S. (2016). A Porphyromonas gingivalis periplasmic novel exopeptidase, acylpeptidyl oligopeptidase, releases N‐acylated di‐ and tripeptides from oligopeptides. Journal of Biological Chemistry, 291(11), 5913–5925. 10.1074/jbc.M115.687566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto, T. K. , Ono, T. , Kobayakawa, T. , & Ohara‐Nemoto, Y. (2019). Characterization of bacterial acylpeptidyl‐oligopeptidase. Biochimie, 163, 50–57. 10.1016/j.biochi.2019.05.007 [DOI] [PubMed] [Google Scholar]
- Nemoto, T. K. , Ono, T. , & Ohara‐Nemoto, Y. (2018). Establishment of potent and specific synthetic substrate for dipeptidyl‐peptidase 7. Analytical Biochemistry, 548, 78–81. 10.1016/j.ab.2018.02.008 [DOI] [PubMed] [Google Scholar]
- Nishimata, H. , Ohara‐Nemoto, Y. , Baba, T. T. , Hoshino, T. , Fujiwara, T. , Shimoyama, Y. , Kimura, S. , & Nemoto, T. K. (2014). Identification of dipeptidyl‐peptidase (DPP)5 and DPP7 in Porphyromonas endodontalis, distinct from those in Porphyromonas gingivalis . PLoS One, 9, e114221. 10.1371/journal.pone.0114221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien‐Simpson, N. M. , Paolini, R. A. , Hoffmann, B. , Slakeski, N. , Dashper, S. G. , & Reynolds, E. C. (2001). Role of RgpA, RgpB, and Kgp proteinases in virulence of Porphyromonas gingivalis W50 in a murine lesion model. Infection and Immunity, 69(12), 7527–7534. 10.1128/IAI.69.12.7527-7534.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara‐Nemoto, Y. , Nakasato, M. , Shimoyama, Y. , Baba, T. T. , Kobayakawa, T. , Ono, T. , Yaegashi, T. , Kimura, S. , & Nemoto, T. K. (2017). Degradation of incretins and modulation of blood glucose levels by periodontopathic bacterial dipeptidyl peptidase 4. Infection and Immunity, 85(9), pii: e00277–17. 10.1128/IAI.00277-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara‐Nemoto, Y. , Rouf, S. M. A. , Naito, M. , Yanase, A. , Tetsuo, F. , Ono, T. , Kobayakawa, T. , Shimoyama, Y. , Kimura, S. , Nakayama, K. , Saiki, K. , Konishi, K. , & Nemoto, T. K. (2014). Identification and characterization of prokaryotic dipeptidyl‐peptidase 5 from Porphyromonas gingivalis . Journal of Biological Chemistry, 289(9), 5436–5448. 10.1074/jbc.M113.527333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara‐Nemoto, Y. , Sarwar, M. T. , Kobayakawa, T. , & Nemoto, T. K. (2019). Proton‐dependent dipeptide transporter in Porphyromonas gingivalis . Journal of Oral Biosciences, 286, P2–11. [Google Scholar]
- Ohara‐Nemoto, Y. , Shimoyama, Y. , Kimura, S. , Kon, A. , Haraga, H. , Ono, T. , & Nemoto, T. K. (2011). Asp‐ and Glu‐specific novel dipeptidyl peptidase 11 of Porphyromonas gingivalis ensures utilization of proteinaceous energy sources. Journal of Biological Chemistry, 286(44), 38115–38127. 10.1074/jbc.M111.278572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara‐Nemoto, Y. , Shimoyama, Y. , Nakasato, M. , Nishimata, H. , Ishikawa, T. , Sasaki, M. , Kimura, S. , & Nemoto, T. K. (2018). Distribution of dipeptidyl peptidase (DPP) 4, DPP5, DPP7 and DPP11 in human oral microbiota‐potent biomarkers indicating presence of periodontopathic bacteria. FEMS Microbiology Letters, 365(22), 10.1093/femsle/fny221 [DOI] [PubMed] [Google Scholar]
- Olsen, C. , & Wagtmann, N. (2002). Identification and characterization of human DPP9, a novel homologue of dipeptidyl peptidase IV. Gene, 299(1–2), 185–193. 10.1016/s0378-1119(02)01059-4 [DOI] [PubMed] [Google Scholar]
- Paschoalin, T. , Carmona, A. K. , & Travassos, L. R. (2013). Peptidyl‐dipeptidase Dcp. Handbook of Proteolytic Enzymes, 3rd ed. (pp. 520–524). Elsevier. [Google Scholar]
- Pavloff, N. , Pemberton, P. A. , Potempa, J. , Chen, W. C. , Pike, R. N. , Prochazka, V. , Kiefer, M. C. , Travis, J. , & Barr, P. J. (1997). Molecular cloning and characterization of Porphyromonas gingivalis lysine‐specific gingipain. A new member of an emerging family of pathogenic bacterial cysteine proteinases. Journal of Biological Chemistry, 270(3), 1595–1600. 10.1074/jbc.272.3.1595 [DOI] [PubMed] [Google Scholar]
- Pavloff, N. , Potempa, J. , Pike, R. N. , Prochazka, V. , Kiefer, M. C. , Travis, J. , & Barr, P. J. (1995). Molecular cloning and structural characterization of the Arg‐gingipain proteinase of Porphyromonas gingivalis. Biosynthesis as a proteinase‐adhesin polyprotein. Journal of Biological Chemistry, 270(3), 1007–1010. 10.1074/jbc.270.3.1007 [DOI] [PubMed] [Google Scholar]
- Potempa, J. , Sroka, A. , Imamura, T. , & Travis, J. (2003). Gingipains, the major cysteine proteinases and virulence factors of Porphyromonas gingivalis: Structure, function and assembly of multidomain protein complexes. Current Protein & Peptide Science, 4(6), 397–407. 10.2174/1389203033487036 [DOI] [PubMed] [Google Scholar]
- Preshaw, P. M. , Alba, A. L. , Herrera, D. , Jepsen, S. , Konstantinidis, A. , Makrilakis, K. , & Taylor, R. (2012). Periodontitis and diabetes: A two‐way relationship. Diabetologia, 55, 21–31. 10.1007/s00125-011-2342-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, S. Y. , Riviere, P. J. , Trojnar, J. , Junien, J. L. , & Akinsanya, K. O. (2003). Cloning and characterization of dipeptidyl peptidase 10, a new member of an emerging subgroup of serine proteases. Biochemical Journal, 373(Pt 1), 179–189. 10.1042/BJ20021914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj, V. S. , Mou, H. , Smits, S. L. , Dekkers, D. H. , Müller, M. A. , Dijkman, R. , Muth, D. , Demmers, J. A. , Zaki, A. , Fouchier, R. A. , Thiel, V. , Drosten, C. , Rottier, P. J. , Osterhaus, A. D. , Bosch, B. J. , & Haagmans, B. L. (2013). Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus‐EMC. Nature, 495(7440), 251–254. 10.1038/nature12005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings, N. D. , Waller, M. , Barrett, A. J. , & Bateman, A. (2014). MEROPS: The database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Research, 42(Database issue), D503–D509. 10.1093/nar/gkt953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea, D. , Van Elzen, R. , De Winter, H. , Van Goethem, S. , Landuyt, B. , Luyten, W. , Schoofs, L. , Van Der Veken, P. , Augustyns, K. , De Meester, I. , Fülöp, V. , & Lambeir, A. M. (2017). Crystal structure of Porphyromonas gingivalis dipeptidyl peptidase 4 and structure‐activity relationships based on inhibitor profiling. European Journal Medicinal Chemistry, 139, 482–491. 10.1016/j.ejmech.2017.08.024 [DOI] [PubMed] [Google Scholar]
- Retting, V. V. J. , Su, S. L. , Fortunato, S. R. , Scanlan, M. J. , Raj, B. K. , Garin‐Chesa, P. , Healey, J. H. , & Old, L. J. (1994). Fibroblast activation protein: Purification, epitope mapping and induction by growth factors. International Journal of Cancer, 58(3), 385–395. 10.1002/ijc.2910580314 [DOI] [PubMed] [Google Scholar]
- Rouf, S. M. A. , Ohara‐Nemoto, Y. , Hoshino, T. , Fujiwara, T. , Ono, T. , & Nemoto, T. K. (2013). Discrimination based on Gly and Arg/Ser at Position 673 between dipeptidyl‐peptidase (DPP) 7 and DPP11, widely distributed DPPs in pathogenic and environmental Gram‐negative bacteria. Biochimie, 95(4), 824–832. 10.1016/j.biochi.2012.11.019 [DOI] [PubMed] [Google Scholar]
- Rouf, S. M. A. , Ohara‐Nemoto, Y. , Ono, T. , Shimoyama, Y. , Kimura, S. , & Nemoto, T. K. (2013). Phenylalanine664 of dipeptidyl peptidase (DPP) 7 and phenylalanine671 of DPP11 mediate preference for P2‐position hydrophobic residues of a substrate. FEBS Open Bio, 3, 177–181. 10.1016/j.fob.2013.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, Y. , Suzuki, Y. , Iizuka, I. , Iizuka, I. , Tateoka, C. , Roppongi, S. , Fujimoto, M. , Inaka, K. , Tanaka, H. , Masaki, M. , Ohta, K. , Okada, H. , Nonaka, T. , Morikawa, Y. , Nakamura, K. T. , Ogasawara, W. , & Tanaka, N. (2014). S46 peptidases are the first exopeptidases to be members of clan PA. Scientific Reports, 15, 4977. 10.1038/srep04977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, Y. , Suzuki, Y. , Iizuka, Y. , Tateoka, C. , Roppongi, S. , Fujimoto, M. , Inaka, K. , Tanaka, H. , Yamada, M. , Ohta, K. , Gouda, H. , Nonaka, T. , Ogasawara, W. , & Tanaka, N. (2015). Structural and mutational analyses of dipeptidyl peptidase 11 from Porphyromonas gingivalis reveal the molecular basis for strict substrate specificity. Scientific Reports, 5, 11151. 10.1038/srep11151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, Y. , Suzuki, Y. , Nakamura, A. , Watanabe, Y. , Sekiya, M. , Roppongi, S. , Kushibiki, C. , Iizuka, I. , Tani, O. , Sakashita, H. , Inaka, K. , Tanaka, H. , Yamada, M. , Ohta, K. , Honma, N. , Shida, Y. , Ogasawara, W. , Nakanishi‐Matsui, M. , Nonaka, T. , … Tanaka, N. (2019). Fragment‐based discovery of the first nonpeptidyl inhibitor of an S46 family peptidase. Scientific Reports., 9(1), 13587. 10.1038/s41598-019-49984-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama, N. , Guillemin, K. , McDaniel, T. K. , Sherlock, G. , Tompkins, L. , & Falkow, S. (2000). A whole‐genome microarray reveals genetic diversity among Helicobacter pylori strains. Proceedings of the National Academy of Sciences USA, 97(26), 14668–14673. 10.1073/pnas.97.26.14668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarwar, M. T. , Ohara‐Nemoto, Y. , Kobayakawa, T. , Naito, M. , & Nemoto, T. K. (2020). Characterization of substrate specificity and novel autoprocessing mechanism of dipeptidase A from Prevotella intermedia . Biological Chemistry, 401(5), 629–642. 10.1515/hsz-2019-0387 [DOI] [PubMed] [Google Scholar]
- Sato, K. , Naito, M. , Yukitake, H. , Hirakawa, H. , Shoji, M. , McBride, M. J. , Rhodes, R. G. , & Nakayama, K. (2010). A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proceedings of the National Academy of Sciences of the United States of America, 107(1), 276–281. 10.1073/pnas.0912010107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, C. F. , Whitaker, E. J. , Hammond, B. F. , & Colman, R. J. (1993). Purification and characterization of a potent 70‐kDa thiol lysyl‐proteinase (Lys‐gingipain) from Porphyromonas gingivalis that cleaves kininogens and fibrinogen. Journal of Biological Chemistry, 268(11), 7935–7942. [PubMed] [Google Scholar]
- Seers, C. A. , Slakeski, N. , Veith, P. D. , Nikolof, N. , Chen, Y.‐Y. , Dashper, S. G. , & Reynolds, E. C. (2006). The RgpB C‐terminal domain has a role in attachment of RgpB to the outer membrane and belongs to a novel C‐terminal‐domain family found in Porphyromonas gingivalis . Journal of Bacteriology, 188(17), 6376–6386. 10.1128/JB.00731-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seino, Y. , & Yabe, D. (2013). Glucose‐dependent insulinotropic polypeptide and glucagon‐like peptide‐1: Incretin actions beyond the pancreas. Journal of Diabetes Investigation, 4(2), 108–130. 10.1111/jdi.12065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Y. , Ratnayake, D. B. , Okamoto, K. , Abe, N. , Yamamoto, K. , & Nakayama, K. (1999). Genetic analyses of proteolysis, hemoglobin binding, and hemagglutination of Porphyromonas gingivalis: Construction of mutants with a combination of rgpA, rgpB, kgp, and hagA . Journal of Biological Chemistry, 274(25), 17955–17960. 10.1074/jbc.274.25.17955 [DOI] [PubMed] [Google Scholar]
- Slakeski, N. , Seers, C. A. , Ng, K. , Moore, C. , Cleal, S. M. , Veith, P. D. , Lo, A. W. , & Reynolds, E. C. (2011). C‐terminal domain residues important for secretion and attachment of RgpB in Porphyromonas gingivalis . Journal of Bacteriology, 193(1), 132–142. 10.1128/JB.00773-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socransky, S. S. , & Haffajee, A. D. (2002). Periodontal microbial ecology. Periodontology 2000, 38(1), 135–187. 10.1111/j.1600-0757.2005.00107.x [DOI] [PubMed] [Google Scholar]
- Socransky, S. S. , Haffajee, A. D. , Cugini, M. A. , Smith, C. , & Kent, R. L. Jr (1998). Microbial complexes in subgingival plaque. Journal of Clinical Periodontology, 25(2), 134–144. 10.1111/j.1600-051x.1998.tb02419.x [DOI] [PubMed] [Google Scholar]
- Soorya, K. V. , Suchetha, A. , Lakshmi, P. , Sapna, N. , Apoorva, S. M. , Bhat, D. , & Mundinamane, D. B. (2014). The effect of scaling and root planing on glycaemic control, periodontal status and gingival crevicular fluid TNF‐α levels in an Indian population‐ to reveal the ambivalent link. Journal of Clinical and Diagnostic Research, 8(11), ZC22–ZC26. 10.7860/JCDR/2014/9490.5115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suido, H. , Nakamura, M. , Mashimo, P. A. , Zambon, J. J. , & Genco, R. J. (1986). Arylaminopeptidase activities of oral bacteria. Journal of Dental Research, 65(11), 1335–1340. 10.1177/00220345860650111101 [DOI] [PubMed] [Google Scholar]
- Tabeta, K. , Yoshie, H. , & Yamazaki, K. (2014). Current evidence and biological plausibility linking periodontitis to atherosclerotic cardiovascular disease. Japanese Dental Science Review, 50(3), 55–62. 10.1016/j.jdsr.2014.03.001 [DOI] [Google Scholar]
- Takahashi, N. , & Sato, T. (2001). Preferential utilization of dipeptides by Porphyromonas gingivalis . Journal of Dental Research, 80(5), 1425–1429. 10.1177/00220345010800050801 [DOI] [PubMed] [Google Scholar]
- Takahashi, N. , & Sato, T. (2002). Dipeptide utilization by the periodontal pathogens Porphyromonas gingivalis, Prevotella intermedia, Prevotella nigrescens and Fusobacterium nucleatum . Oral Microbiology and Immunology, 17(1), 50–54. 10.1046/j.0902-0055.2001.00089.x [DOI] [PubMed] [Google Scholar]
- Takahashi, N. , Sato, T. , & Yamada, T. (2000). Metabolic pathways for cytotoxic end product formation from glutamate‐ and aspartate‐containing peptides by Porphyromonas gingivalis . Journal of Bacteriology, 182(17), 4704–4710. 10.1128/jb.182.17.4704-4710.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano, M. , Nishihara, R. , Sugano, N. , Matsumoto, K. , Yamada, Y. , Takane, M. , Fujisaki, Y. , & Ito, K. (2010). The effect of systemic anti‐tumor necrosis factor‐α treatment on Porphyromonas gingivalis infection in type 2 diabetic mice. Archives of Oral Biology, 55(5), 379–384. 10.1016/j.archoralbio.2010.03.004 [DOI] [PubMed] [Google Scholar]
- Tang‐Larsen, J. , Claesson, R. , Edlund, M. B. , & Carlsson, J. (1995). Competition for peptides and amino acids among periodontal bacteria. Journal of Periodontal Research, 30(6), 390–395. 10.1111/j.1600-0765.1995.tb01292.x [DOI] [PubMed] [Google Scholar]
- Teeuw, W. J. , Gerdes, V. E. , & Loos, B. G. (2010). Effect of periodontal treatment on glycemic control of diabetic patients: A systematic review and meta‐analysis. Diabetes Care, 33(2), 421–427. 10.2337/dc09-1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira, F. B. , Saito, M. T. , Matheus, F. C. , Prediger, R. D. , Yamada, E. S. , Maia, C. S. F. , & Lima, R. R. (2017). Periodontitis and Alzheimer’s disease: A possible comorbidity between oral 495 chronic inflammatory condition and neuroinflammation. Frontiers in Aging Neuroscience, 9, 327. 10.3389/fnagi.2017.00327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomás, I. , Diz, P. , Tobías, A. , Scully, C. , & Donos, N. (2012). Periodontal health status and bacteraemia from daily oral activities: Systematic review/meta‐analysis. Journal of Clinical Periodontology, 39(3), 213–228. 10.1111/j.1600-051X.2011.01784.x [DOI] [PubMed] [Google Scholar]
- Veith, P. D. , Chen, Y.‐Y. , Gorasia, D. G. , Chen, D. , Glew, M. D. , O'Brien‐Simpson, N. M. , Cecil, J. D. , Holden, J. A. , & Reynolds, E. C. (2014). Porphyromonas gingivalis outer membrane vesicles exclusively contain outer membrane and periplasmicproteins and carry a cargo enriched with virulence factors. Journal of Proteome Research, 13(5), 2420–2432. 10.1021/pr401227e [DOI] [PubMed] [Google Scholar]
- Veith, P. D. , Luong, C. , Tan, K. H. , Dashper, S. G. , & Eric C. Reynolds, E. C. (2018). Outer membrane vesicle proteome of Porphyromonas gingivalis is differentially modulated relative to the outer membrane in response to heme availability. Journal Proteome Research, 17, 2377–2389. [DOI] [PubMed] [Google Scholar]
- Vesanto, E. , Peltoniemi, K. , Purtsi, T. , Steele, J. L. , & Palva, A. (1996). Molecular characterization, over‐expression and purification of a novel dipeptidase from Lactobacillus helveticus . Applied Microbiology and Biotechnology, 45(5), 638–645. 10.1007/s002530050741 [DOI] [PubMed] [Google Scholar]
- Weber, A. E. (2004). Dipeptidyl peptidase IV inhibitors for the treatment of diabetes. Journal of Medical Chemistry, 47(17), 4135–4141. 10.1021/jm030628v [DOI] [PubMed] [Google Scholar]
- Weinberg, M. V. , & Maier, R. J. (2007). Peptide transport in Helicobacter pylori: Roles of dpp and opp systems and evidence for additional peptide transporters. Journal of Bacteriology, 189(9), 3392–3402. 10.1128/JB.01636-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitz, D. , Harder, D. , Casagrande, F. , Fotiadis, D. , Obrdlik, P. , Kelety, B. , & Daniel, H. (2007). Functional and structural characterization of a prokaryotic peptide transporter with features similar to mammalian PEPT1. Journal of Biological Chemistry, 282(2), 2832–2839. 10.1074/jbc.M604866200 [DOI] [PubMed] [Google Scholar]


