Abstract
We describe a novel species isolated from walnut (Juglans regia) which comprises non-pathogenic and pathogenic strains on walnut. The isolates, obtained from a single ornamental walnut tree showing disease symptoms, grew on yeast extract–dextrose–carbonate agar as mucoid yellow colonies characteristic of Xanthomonas species. Pathogenicity assays showed that while strain CPBF 424T causes disease in walnut, strain CPBF 367 was non-pathogenic on walnut leaves. Biolog GEN III metabolic profiles disclosed some differences between strains CPBF 367 and CPBF 424T and other xanthomonads. Multilocus sequence analysis with seven housekeeping genes (fyuA, gyrB, rpoD, atpD, dnaK, efp, glnA) grouped these strains in a distinct cluster from Xanthomonas arboricola pv. juglandis and closer to Xanthomonas prunicola and Xanthomonas arboricola pv. populi. Average nucleotide identity (ANI) analysis results displayed similarity values below 93 % to X. arboricola strains. Meanwhile ANI and digital DNA–DNA hybridization similarity values were below 89 and 50 % to non-arboricola Xanthomonas strains, respectively, revealing that they do not belong to any previously described Xanthomonas species. Furthermore, the two strains show over 98 % similarity to each other. Genomic analysis shows that strain CPBF 424T harbours a complete type III secretion system and several type III effector proteins, in contrast with strain CPBF 367, shown to be non-pathogenic in plant bioassays. Taking these data altogether, we propose that strains CPBF 367 and CPBF 424T belong to a new species herein named Xanthomonas euroxanthea sp. nov., with CPBF 424T (=LMG 31037T=CCOS 1891T=NCPPB 4675T) as the type strain.
Keywords: new species, pathogenicity, walnut, Xanthomonas
Introduction
The taxonomy of the genus Xanthomonas has been extensively studied and categorized over last century, raising some disputes regarding classification and nomenclature of its members, largely due to the wide plant host diversity associated with the genus. Recorded Xanthomonas hosts include numerous economically relevant crop species, severely impacting agricultural productivity worldwide [1].
Introduction of molecular methods such as DNA–DNA hybridization allowed the first big reclassification within the genus Xanthomonas [2]. Recently, studies relying on multilocus sequence analysis (MLSA), whole-genome average nucleotide identity (ANI) and biochemical analysis have proved fruitful in the reclassification of Xanthomonas species and description amendments of several strains [3, 4]. These techniques have allowed us to reach a deeper discriminatory insight into the genetic diversity of Xanthomonas at the intrasubspecific level, consequently leading to the proposal of novel species [5–7].
Increasing reports of non-pathogenic Xanthomonas strains, such as Xanthomonas arboricola isolates recovered from asymptomatic and symptomatic host tissues, points to the uncertainty of several Xanthomonas strains regarding their host-associated lifestyle and likely host diversity [8–10]. Interestingly, X. arboricola is the species with most non-pathogenic strains within the genus Xanthomonas [11]. Characterization of several of these non-pathogenic X. arboricola strains has revealed that they are phylogenetically distant from the disease-causing strains that were isolated from their specific hosts [8]. Furthermore, plants are host to a consortium of both distant and genetically similar bacteria, displaying different host-associated phenotypes. It has been suggested that the sympatry of pathogenic and non-pathogenic strains may favour horizontal gene transfer events, likely leading to the emergence of new pathogenic lineages [8, 11].
The advent of increasingly powerful genomic tools and improved bacterial genome assemblers and accurate annotation has led to a more confident examination of bacterial evolution and phylogeny, allowing microbiologists and plant pathologists to assign specific differences to the existing taxonomy, even at the pathovar level. A pathovar of a pathogenic species refers to an infrasubspecific group that includes strains with the same host range [12, 13]. To the best of our knowledge, all Xanthomonas arboricola pv. juglandis strains described previously have been isolated from diseased walnut trees.
Furthermore, MLSA results revealed that several X. arboricola strains isolated from walnut, including both pathogenic and non-pathogenic strains, do not cluster with the members of juglandis pathovar [9, 14]. Indeed, a previous study has reported the identification of two X. arboricola strains isolated from symptomatic leaves of a pecan tree (Carya illinoinensis), which are pathogenic to both pecan and walnut trees [14]. These data raise the need to understand how the genetic makeup of plant bacterial pathogens, such as Xanthomonas , links with a diversifying range of plant hosts. Walnut (Juglans species) is an economically important tree crop worldwide, with the Persian walnut (Juglans regia L.) being the main cultivated species for walnut fruits and timber production [15–17]. X. arboricola pv. juglandis is a major threat to walnut orchards and acknowledged as the etiological agent of walnut bacterial blight (WBB) and other walnut diseases such as brown apical necrosis [18] and vertical oozing canker [19], altogether causing high yield losses [16].
In this study we characterize two novel strains, CPBF 367 and CPBF 424T, isolated from asymptomatic buds of one walnut tree showing WBB symptoms. Genotyping and comparative genomics indicate that these strains, showing a distinct pathogenicity phenotype in walnut, belong to the genus Xanthomonas , although they could not be assigned to any of the known species of this genus. This work gathers phenotypic, genotypic and genomic evidence to support that strains CPBF 367 and CPBF 424T are members of a new species of the genus Xanthomonas .
Isolation and growth of bacteria
Several isolates were obtained over three consecutive years (2014–2016) from an isolated ornamental walnut tree (Juglans regia) showing bacterial blight symptoms in leaves and fruits, located in a municipal garden in the city of Loures, Portugal (Fig. S1, available in the online version of this article). Distinct organs were sampled including symptomatic leaves (CPBF 1521 in 2014) and asymptomatic buds (CPBF 367, CPBF 424T, CPBF 426 and CPBF 427 in 2016). The procedure for preparation of plant material for bacteria isolation has been described previously [20]. Briefly, the excised plant material was disinfected by immersion in 70 % ethanol followed by washing with sterile distilled water (SDW) and then macerated in extraction bags with 5 ml SDW [21]. Suspension and correspondent dilutions were streaked on yeast extract–dextrose–carbonate (YDC) agar [22] and incubated at 26±2 °C for 4 days. Single characteristic mucoid yellow colonies of Xanthomonas were streaked on fresh nutrient agar (NA) medium to ensure purity. Selected isolates (CPBF 424T and CPBF 367) were stored at −80 °C in cryovials containing LB medium (Difco) and glycerol to a final concentration of 30 %. The strains were deposited in international bacterial collections with the accession numbers CPBF 367 (LMG 31036=CCOS 1890) and CPBF 424T (=LMG 31037T=CCOS 1891T=NCPPB 4675T). Isolates CPBF 367 and CPBF 424T were grown in YDC medium for analysis of colony morphology and pigment production for a 10 day period. The bacterial strains used throughout this study are disclosed in Table 1.
Table 1.
Strains used for pathogenicity tests and Biolog assays
|
Strain |
Species |
Host of isolation |
Geographic origin |
Year of isolation |
|---|---|---|---|---|
|
CPBF 367* |
Juglans regia |
Loures, Portugal |
2016 |
|
|
CPBF 424T* |
Juglans regia |
Loures, Portugal |
2016 |
|
|
CPBF 427* |
X. arboricola pv. juglandis |
Juglans regia |
Loures, Portugal |
2016 |
|
CPBF 1521* |
X. arboricola pv. juglandis |
Juglans regia |
Loures, Portugal |
2014 |
|
CPBF 1480 |
X. arboricola pv. juglandis |
Juglans regia |
Azeitão, Portugal |
2014 |
|
LMG 747T |
X. arboricola pv. juglandis |
Juglans regia |
New Zealand |
1956 |
|
CFBP 3123PT |
X. arboricola pv. populi |
Populus × euroamericana cv. robusta |
The Netherlands |
1979 |
*Other known names for these strains: CPBF 367=LMG 31036=CCOS 1890; CPBF 424T=LMG 31037T=CCOS 1891T=NCPPB 4675T; CPBF 427=LMG 31039=CCOS 1893; CPBF 1521=LMG 31040=CCOS 1894=NCPPB 4676. (CPBF, Portuguese Collection of Phytopathogenic Bacteria in Oeiras, Portugal; BCCM/LMG, the Belgian Coordinated Collections of Microorganisms/ LMG Bacteria Collection in Gent, Belgium; CCOS, Culture Collection of Switzerland in Wädenswil, Switzerland; NCPPB, National Collection of Plant Pathogenic Bacteria in York, UK; CFBP, French Collection of Plant associated Bacteria, France).
Pathogenicity assays
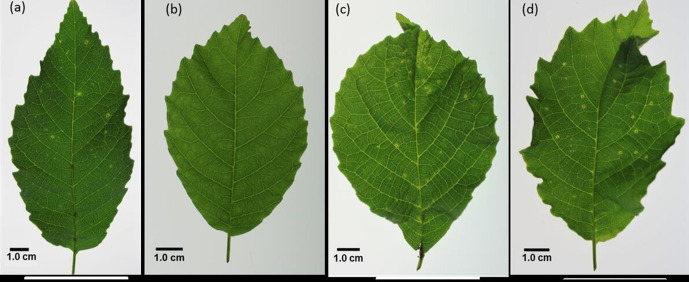
Plantlets used in the pathogenicity tests were grown from seeds collected from Juglans regia cv. Hartley on the same day. After 30 days of cold stratification treatment at 3–5 °C to break dormancy, seeds were sown in sterilized sand substrate and germinated over 60 days at alternated temperatures, 16 h day at 30 °C and 8 h night at 20 °C [23]. Walnut plantlets were then maintained in a climatic chamber under controlled environmental conditions of 16 h photoperiod (16 h of light at 24 °C and 8 h of darkness at 18 °C). Pathogenicity determination of strains CPBF 424T and CPBF 367 was carried out as previously described [20]. Briefly, inoculum suspensions of approximately 108 c.f.u. ml−1 were prepared with SDW. Plantlets with at least four young leaves expanded were inoculated by spraying with a manual atomizer until runoff. SDW was used for negative control, while X. arboricola pv. juglandis strains LMG 747T and CPBF 1480 were used as positive controls of infection. Three replicates were included for each strain tested. Symptoms were registered after 7 days and further followed for over 5 weeks. Necrotic bacterial spots were observed 7 days post inoculation on leaves of plants inoculated with walnut isolate CPBF 424T, and with positive-control strains LMG 747T and CPBF 1480 (Fig. 1). Walnut isolate CPBF 367 and negative controls did not produce any disease symptoms on walnut plantlets. In order to fulfil Koch’s postulates, typical Xanthomonas yellow mucoid bacterial colonies were re-isolated from the symptoms and confirmed by sequencing analysis of gyrB and fyuA.
Fig. 1.
Walnut plantlets leaves 7 days post-inoculation with (a) isolate CPBF 424T showing walnut bacteria blight (WBB) symptoms, (b) isolate CPBF 367 no WBB symptoms were observed, (c) Xanthomonas arboricola pv. juglandis LMG 747T and (d) X. arboricola pv. juglandis strain CPBF 1480, used as positive controls, show characteristic WBB symptoms.
Phenotypic characterization
Phenotypic characterization was carried out with Biolog GEN III MicroPlates (Biolog) according to the manufacturer’s instructions, on walnut isolates CPBF 367, CPBF 424T, CPBF 427, CPBF 1521 and reference strains X. arboricola pv. juglandis LMG 747T and X. arboricola pv. populi CFBP 3123PT. For all strains except CFBP 3123PT, three MicroPlates were assayed on different dates. Selected strains were first grown on solid NYGA medium (5 g l−1 peptone, 3 g l−1 yeast extract, 20 g l−1 glycerol and 15 g l−1 agar) for 48 h at 28 °C. The strains were subsequently grown on solid Biolog Dehydrated Growth agar for 24 h at 28 °C. Fresh colonies were then transferred into Inoculating Fluid A vials using a cotton-tipped swab. The density of the inoculum was checked and adjusted to a transmittance of 95–98 % using a turbidimeter. A total of 100 µl prepared inoculum was then dispensed into each well of the Biolog MicroPlate. MicroPlates were then incubated at 28 °C and read using a MicroStation 2 Reader (Biolog) at 24, 48, 72, 120, 192 and 240 h.
Considering compound metabolism, values 160 % superior to the negative control (water) value were considered positive, while those below 130 % the negative control were taken as negative. Values in between were considered borderline [10]. The data measured at 72 h was shown to provide the most consistent results between replicates and thus was selected for further analysis. Isolates CPBF 367 and CPBF 424T seemed to metabolize mucic acid and d-saccharic acid, in contrast to X. arboricola pv. juglandis CPBF 427, CPBF 1521 and LMG 747T. Additionally, comparisons with results from other Xanthomonas strains [4–6] revealed differences in metabolizing d-salicin, sucrose, acetic acid, melibiose, formic acid, d-arabitol, d-glucose-6-phosphate, maltose and gentiobiose (Table 2). The metabolization of some substrates was inconclusive due to the inconsistencies observed between replicates.
Table 2.
Biolog profiles of Xanthomonas euroxanthea strains in comparison with members of Xanthomonas arboricola pv. juglandis and other related Xanthomonas species
+, Positive; −, negative; BL, borderline; (+), weak positive; na, not analysed.
|
Test |
X. euroxanthea CPBF 367 |
X. euroxanthea CPBF 424T |
X. arboricola pv. juglandis CPBF 427 |
X. arboricola pv. juglandis CPBF 1521 |
X. arboricola pv. juglandis LMG 747T |
X. arboricola pv. populi CFBP 3123PT |
X. campestris pv. campestris* |
X. axonopodis pv. phaseoli* |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
d-Salicin |
− |
− |
− |
− |
− |
+ |
+ |
− |
− |
− |
n/a |
n/a |
|
Mucic acid |
+ |
+ |
− |
− |
− |
+ |
+ |
− |
+ |
+ |
n/a |
n/a |
|
d-Saccharic acid |
+ |
+ |
− |
− |
− |
+ |
+ |
− |
+ |
+ |
n/a |
− |
|
Vancomycin |
+/BL |
+/BL |
− |
− |
−/BL |
− |
− |
− |
− |
− |
n/a |
n/a |
|
Sucrose |
+ |
+ |
+ |
+ |
− |
+ |
+ |
+ |
+ |
+ |
+ |
n/a |
|
Acetic acid |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
− |
+ |
|
Melibiose |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
(+)/+ |
+ |
+ |
− |
n/a |
|
Formic acid |
+ |
+ |
+ |
+ |
+ |
+ |
(+) |
− |
− |
− |
− |
− |
|
d-Arabitol |
− |
− |
− |
− |
− |
BL |
+ |
+ |
− |
(+) |
n/a |
n/a |
|
d-Glucose-6-phosphate |
− |
− |
− |
− |
− |
+ |
− |
− |
− |
− |
− |
n/a |
|
Maltose |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
− |
− |
+ |
+ |
|
Gentiobiose |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
− |
(+) |
− |
+ |
+ |
*Vicente et al. 2017 [5] (X. floridensis WHRI 8844, WHRI 8846A, WHRI 8848T, WHRI 8851; X. nasturtii WHRI 8853T, WHRI 8930A; X. campestris pv. campestris: WHRI 1279A); X. axonopodis pv. phaseoli WHRI 1925C).
†López et al. 2018 [6] (X. prunicola CFBP 8353T, CFBP 8354, CFBP 8355).
‡Vauterin et al. 1995 [2] (X arboricola pv. corylina LMG 688, LMG 689T, LMG 8658, LMG 8660; X. arboricola pv. juglandis LMG 747T, LMG 8047; X. arboricola pv. poinsettiicola LMG 5403; X arboricola pv. populi LMG 12141PT; X. arboricola pv. pruni LMG 852T, LMG 8680).
Multilocus sequence analysis
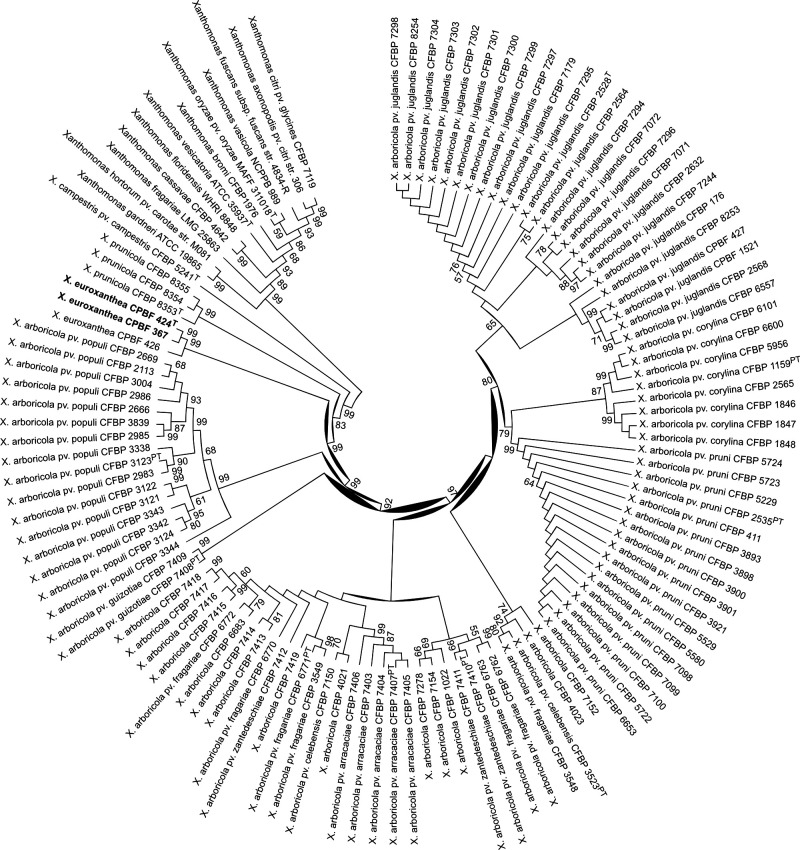
MLSA based on the concatenated partial sequences of seven genes fyuA (684 bp), gyrB (735 bp), rpoD (586 bp), atpD (750 bp), dnaK (759 bp), efp (339 bp) and glnA (675 bp) was performed. Sequences were retrieved from the NCBI database from 118 strains, including eight different X. arboricola pathovars previously used to describe the diversity of X. arboricola [24] and another 15 Xanthomonas species. The analysis was carried out using Geneious version 9.1.7 software (Biomatters) and a maximum-likelihood tree based on the General Time Reversible (GTR+G+I) model in mega 7.0 was reconstructed (Fig. 2). The novel strains CPBF 424T CPBF 367 and CPBF 426 formed a separate cluster from all other walnut-associated strains. Interestingly, although isolated from walnut trees, these strains are more closely related to X. prunicola and X. arboricola pv. pruni strains, which are known phytopathogens of Prunus trees.
Fig. 2.
Maximum-likelihood tree based on the nucleotide alignments of 118 concatenated sequences of atpD, dnaK, efp, fyuA, glnA, gyrB and rpoD partial sequences (total length of 4528 bp), emphasizing the phylogenetic relatedness of Xanthomonas euroxanthea strains CPBF 367, CPBF 424T and CPBF 426 (highlighted in bold) within the genus Xanthomonas . Bootstrap values >50 from 1000 iterations are shown over and below the branches.
Genomic characterization
The draft genome sequence of CPBF 424T was previously published [25]. Whole genome sequencing of strains CPBF 367 and CPBF 426 was carried out as described for CPBF 424T [25]. Genomes of strains CPBF 367 and CPBF 424T were approximately 4.96 and 4.90 Mbp, respectively, and comprised 22 and 10 contigs, respectively. High-quality draft genomes of strains CPBF 367 and CPBF 424T were deposited in GenBank under accession numbers UNRN00000000.1 and UIHB00000000.1, respectively.
ANI and digital DNA–DNA hybridization (dDDH) values between CPBF 367 and CPBF 424T, and genomes of X. arboricola and Xanthomonas species were calculated using the edgar version 2.0 platform [26, 27] and formula 2 of the Genome-to-Genome Distance Calculator (GGDC 2.1; http://ggdc.dsmz.de/ggdc.php) [28]. ANI analysis included the genomes of 10 X . arboricola strains, belonging to eight distinct pathovars, and 10 other Xanthomonas species (Table S1). ANI values of strains CPBF 367 and CPBF 424T were less than 93 % with X. arboricola species and less than 89 % with non-arboricola Xanthomonas species (Fig. 3). These values reside below the threshold of 95–96 % commonly considered for representatives of the same species [29]. Furthermore, these CPBF 367 and CPBF 424T strains share 98 % ANI with each other (Fig. 3). The dDDH analysis considered 20 strains representing different species of the genus Xanthomonas and revealed similarities values below the 70 % threshold (Table S2) considered for delimitation of new species [28]. Taken together, the results of ANI and dDDH analyses suggest that these strains belong to a new Xanthomonas species.
Fig. 3.
Average nucleotide identity (ANI) between novel isolates CPBF 367 and CPBF 424T (in bold) and 20 different Xanthomonas genomes using edgar. Pairwise ANI values are shown in percentages and indicated within the cells of the heat-map. Accession numbers of sequences are available in Table S1.
Type III SEcretion System (T3SS) and Type III Effectors (T3E)
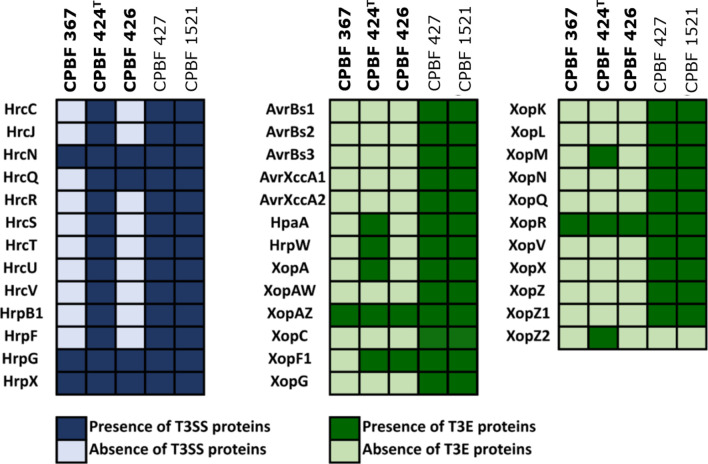
The prediction of type III secretion system (T3SS) and type III effectors (T3E) homologs was carried out with blastn analysis against previously characterized T3SS and T3Es of the Xanthomonas group [30–32]. The criteria applied as threshold was a minimal e-value of 1e−10, query length similarity cut-off ≥40 % and sequence identity with a cut-off ≥70 % (Fig. 4). The walnut pathogenic strain CPBF 424T has a T3SS gene profile similar to the strains X. arboricola pv. juglandis CPBF 427 and CPBF 1521 and possesses gene homologs described for pathogenic strains of X. arboricola [10, 33–35]. In contrast, strains CPBF 367 and CPBF 426 lack the majority of the T3SS- and T3E-encoding genes (Fig. 4), resembling the T3SS and T3E profile of atypical non-pathogenic strains of Xanthomonas species described previously [33, 35]. The absence of some of the components from the Hrp2 family and of genes that are part of the macromolecular structure of the T3SS may explain the non-pathogenic phenotype of CPBF 367, even while harbouring regulators genes of T3SS, such as hrpX and hrpG [36, 37]. Additionally, the fact that strain CPBF 426 is deficient for most T3SS genes, suggests that this strain may not be pathogenic as shown for CPBF 367. Intriguingly, CPBF 424T seems to have a functional T3SS and holds a narrow gene repertoire of T3E homologs in comparison with other X. arboricola pv. juglandis strains [9, 30, 33]. This differs markedly from X. arboricola pv. juglandis CPBF 427 and CPBF 1521 and strains CPBF 367 and CPBF 426. Although the pathogenic strain CPBF 424T possesses homologs for eight known effectors, from which only three were identified in non-pathogenic X. euroxanthea (CPBF 367 and CPBF 426), no homologs were found for most of the genes assigned as important for pathogenicity of Xanthomonas species, namely avrbs2, avrbs2, avrbs3, avrXccA1, avrXccA2, xopAW, xopC, xopG, xopK, xopL, xopN, xopQ, xopV, xopX, xopZ and xopZ1. All the aforementioned genes, with the exception of xopZ2, were present in both X. arboricola pv. juglandis CPBF 427 and CPBF 1521 strains (Fig. 4). The genomic features of this new species and the pathogen phenotype of strain CPBF 424T in walnut make these strains particularly appealing to elucidate the evolutionary hypothesis of pathogenicity in walnut, to uncover the genetic footprints of adaptation and to address speciation in Xanthomonas .
Fig. 4.
Distribution of protein homologs of the type III secretion system (T3SS) and type III effectors (T3Es) among the Xanthomonas euroxanthea strains (CPBF 367, CPBF 424T, CPBF 426) and Xanthomonas arboricola pv. juglandis (CPBF 427 and CPBF 1521) strains based on blastn analysis. Dark blue corresponds to the presence of T3SS protein homologous, and dark green corresponds to the presence of T3E homologous. The light blue and green indicate absence of T3SS and T3E homologous, respectively. The threshold criteria applied were minimal e-value of 1e−10, query length similarity cut-off ≥40 % and sequence identity cut-off ≥70 %.
Description of Xanthomonas euroxanthea sp. nov.
Xanthomonas euroxanthea (eu.ro.xan.the’a. N.L. fem. adj. euroxanthea referring to the EuroXanth COST Action CA 16107, the EU-funded network in which the isolates of this species were characterized).
Cells are Gram-stain-negative straight rods and form colonies that are yellow, circular, smooth, mucoid and slightly convex with entire margins when grown on YDC agar for 2 days. Regarding carbon source metabolism, X. euroxanthea is positive for 34 substrates (dextrin, maltose, trehalose, cellobiose, gentiobiose, sucrose, melibiose, N-acetyl-d-glucosamine, α-d-glucose, d-mannose, d-fructose, d-galactose, l-fucose, glycerol, gelatin, glycyl-l-proline, l-alanine, l-glutamic acid, l-serine, mucic acid, d-saccharic acid, methyl pyruvate, l-lactic acid, citric acid, α-keto-glutaric acid, l-malic acid, bromo-succinic acid, Tween 40, α-hydroxy-butyric acid, α-keto-butyric acid, acetoacetic acid, propionic acid, acetic acid, formic acid) and negative for 25 compounds (turanose, stachyose, raffinose, methyl β-d-glucoside, d-salicin, N-acetyl-β-d-mannosamine, N-acetyl-d-galactosamine, N-acetyl-neuraminic acid, 3-methyl glucose, l-rhamnose, d-sorbitol, d-arabitol, myo-inositol, d-glucose-6-phosphate, d-aspartic acid, d-serine, l-arginine, l-pyroglutamic acid, d-gluconic acid, quinic acid, p-hydroxy-phenylacetic acid, d-lactic acid methyl ester, d-malic acid, γ-amino-butryric acid and β-hydroxy-d,l-butyric acid).
This species includes non-pathogenic (CPBF 367) and pathogenic (CPBF 424T) strains. The type strain is CPBF 424T (LMG 31037T=CCOS 1891T=NCPPB 4675T), isolated from an asymptomatic bud of an isolated ornamental diseased walnut tree (J. regia) tree growing in a municipal garden of Loures, Portugal. The high-quality draft genome of the type strain is characterized by a size of 4.8 Mbp and a G+C content of 65.9 mol %. The GenBank accession number of the 16S rRNA gene sequence of strain CPBF 424T is MT036365 and its draft genome sequence accession number is GCA_900476395.
Supplementary Data
Funding information
This research was co-financed by the European Structural and Investment Funds (ESIFs) through COMPETE 2020, and by National Funds through FCT- Fundação para a Ciência e Tecnologia, within the framework of the project EVOXANT (PTDC/BIA-EVF/3635/2014-POCI-01–0145-FEDER-016600). C.F. and L.M. were supported by fellowships from FCT (SFRH/BD/95913/2013; SFRH/BD/137079/2018, respectively). N.C.D. was supported by grant no IZCOZ0_177064 from the Swiss National Foundation (SNF) for Scientific Research. The EDGAR platform is financially supported by the BMBF grant FKZ031A533 within the de.NBI network. This article is based upon work from COST Action CA16107 EuroXanth, supported by COST (European Cooperation in Science and Technology).
Acknowledgements
We thank Professor Dr Jean-Louis Charlet, Professor Dr Bernhard Schink and Professor Aharon Oren for advice regarding the nomenclature, and Christian Kunkel from the Culture Collection of Switzerland (CCOS AG, Wädenswil, Switzerland) for his support during the phenotypic characterization.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: ANI, average nucleotide identity; BLASTn, nucleotide Basic Local Alignment Search Tool; dDDH, digital DNA-DNA hybridization; MLSA, multilocus sequence analysis; T3E, type III effectors; T3SS, type III secretion system; WBB, walnut bacterial blight; YDC, yeast extract-dextrose-carbonate.
One supplementary figure and two supplementary tables are available with the online version of this article.
References
- 1.Vauterin L, Rademaker J, Swings J. Synopsis on the taxonomy of the genus Xanthomonas . Phytopathology. 2000;90:677–682. doi: 10.1094/PHYTO.2000.90.7.677. [DOI] [PubMed] [Google Scholar]
- 2.Vauterin L, Hoste B, Kersters K, Swings J. Reclassification of Xanthomonas . Int J Syst Bacteriol. 1995;45:472–489. doi: 10.1099/00207713-45-3-472. [DOI] [Google Scholar]
- 3.da Gama MAS, Barbosa MAG, de Farias ARG, da Silva Júnior WJ, Mariano R de LR. Taxonomic repositioning of Xanthomonas campestris pv. viticola (Nayudu 1972) Dye 1978 as Xanthomonas citri pv. viticola (Nayudu 1972) Dye 1978 comb. nov. and emendation of the description of Xanthomonas citri pv. anacardii to include pigmented isolates pathogenic to cashew plant. Phytopathology. 2018;108:1143–1153. doi: 10.1094/PHYTO-02-18-0037-R. [DOI] [PubMed] [Google Scholar]
- 4.Constantin EC, Cleenwerck I, Maes M, Baeyen S, Van Malderghem C, et al. Genetic characterization of strains named as Xanthomonas axonopodis pv. dieffenbachiae leads to a taxonomic revision of the X. axonopodis species complex. Plant Pathol. 2016;65:792–806. doi: 10.1111/ppa.12461. [DOI] [Google Scholar]
- 5.Vicente JG, Rothwell S, Holub EB, Studholme DJ. Pathogenic, phenotypic and molecular characterisation of Xanthomonas nasturtii sp. nov. and Xanthomonas floridensis sp. nov., new species of Xanthomonas associated with watercress production in Florida. Int J Syst Evol Microbiol. 2017;67:3645–3654. doi: 10.1099/ijsem.0.002189. [DOI] [PubMed] [Google Scholar]
- 6.López MM, Lopez-Soriano P, Garita-Cambronero J, Beltrán C, Taghouti G, et al. Xanthomonas prunicola sp. nov., a novel pathogen that affects nectarine (Prunus persica var. nectarina) trees. Int J Syst Evol Microbiol. 2018;68:1857–1866. doi: 10.1099/ijsem.0.002743. [DOI] [PubMed] [Google Scholar]
- 7.Triplett LR, Verdier V, Campillo T, Van Malderghem C, Cleenwerck I, et al. Characterization of a novel clade of Xanthomonas isolated from rice leaves in Mali and proposal of Xanthomonas maliensis sp. nov. Antonie van Leeuwenhoek. 2015;107:869–881. doi: 10.1007/s10482-015-0379-5. [DOI] [PubMed] [Google Scholar]
- 8.Jacques M-A, Arlat M, Boulanger A, Boureau T, Carrère S, et al. Using Ecology, Physiology, and Genomics to Understand Host Specificity in Xanthomonas . Annu Rev Phytopathol. 2016;54:163–187. doi: 10.1146/annurev-phyto-080615-100147. [DOI] [PubMed] [Google Scholar]
- 9.Essakhi S, Cesbron S, Fischer-Le Saux M, Bonneau S, Jacques M-A, et al. Phylogenetic and variable-number tandem-repeat analyses identify nonpathogenic Xanthomonas arboricola lineages lacking the canonical type III secretion system. Appl Environ Microbiol. 2015;81:5395–5410. doi: 10.1128/AEM.00835-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garita-Cambronero J, Palacio-Bielsa A, López MM, Cubero J. Comparative genomic and phenotypic characterization of pathogenic and non-pathogenic strains of Xanthomonas arboricola reveals insights into the infection process of bacterial spot disease of stone fruits. PLoS One. 2016;11:e0161977. doi: 10.1371/journal.pone.0161977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merda D, Bonneau S, Guimbaud J-F, Durand K, Brin C, et al. Recombination-prone bacterial strains form a reservoir from which epidemic clones emerge in agroecosystems. Environ Microbiol Rep. 2016;8:572–581. doi: 10.1111/1758-2229.12397. [DOI] [PubMed] [Google Scholar]
- 12.Yeates GW. A proposed nomenclature and classification for plant pathogenic bacteria. New Zeal J Agric Res. 1978;21:153–177. [Google Scholar]
- 13.Schulze-Lefert P, Panstruga R. A molecular evolutionary concept connecting nonhost resistance, pathogen host range, and pathogen speciation. Trends Plant Sci. 2011;16:117–125. doi: 10.1016/j.tplants.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 14.Fernandes C, Sousa R, Tavares F, Cruz L. First Report of Xanthomonas arboricola causing bacterial blight on pecan trees in Portugal. Plant Disease. 2018;102:2632. doi: 10.1094/PDIS-03-18-0467-PDN. [DOI] [Google Scholar]
- 15.Lamichhane JR. Xanthomonas arboricola diseases of stone fruit, almond, and walnut trees: Progress toward understanding and management. Plant Dis. 2014;98:1600–1610. doi: 10.1094/PDIS-08-14-0831-FE. [DOI] [PubMed] [Google Scholar]
- 16.Leslie C, Uratsu S, McGranahan G, Dandekar A. Agrobacterium protocols volume 2. Springer. 2006 [Google Scholar]
- 17.Belisario A, Santori A, Potente G, Fiorin A, Saphy B, et al. Brown apical necrosis (BAN): a fungal disease causing fruit drop of english walnut. In: Neil MC, editor. Vi International Walnut Symposium (Acta Horticulturae. Leuven 1: International Society of Horticultural Science. 2010. pp. 449–452. In. editor. pp. [DOI] [Google Scholar]
- 18.Moragrega C, Matias J, Aletà N, Montesinos E, Rovira M. Apical necrosis and premature drop of Persian (English) walnut fruit caused by Xanthomonas arboricola pv. juglandis . Plant Dis. 2011;95:1565–1570. doi: 10.1094/PDIS-03-11-0259. [DOI] [PubMed] [Google Scholar]
- 19.Hajri A, Meyer D, Delort F, Guillaumès J, Brin C, et al. Identification of a genetic lineage within Xanthomonas arboricola pv. juglandis as the causal agent of vertical oozing canker of Persian (English) walnut in France. Plant Pathol. 2010;59:1014–1022. doi: 10.1111/j.1365-3059.2010.02362.x. [DOI] [Google Scholar]
- 20.Fernandes C, Albuquerque P, Cruz L, Tavares F. Genotyping and epidemiological metadata provides new insights into population structure of Xanthomonas isolated from walnut trees
- 21.Fernandes C, Albuquerque P, Sousa R, Cruz L, Tavares F. Multiple DNA markers for identification of Xanthomonas arboricola pv. juglandis isolates and its direct detection in plant samples. Plant Dis. 2017;101:858–865. doi: 10.1094/PDIS-10-16-1481-RE. [DOI] [PubMed] [Google Scholar]
- 22.Stolp H, Starr MP. Bacteriophage reactions and speciation of Phytopathogenic Xanthomonads . J Phytopathol. 1964;51:442–478. doi: 10.1111/j.1439-0434.1964.tb03451.x. [DOI] [Google Scholar]
- 23.International Seed Testing Association International rules for seed testing rules. Seed Sci Technol. 1999;27 [Google Scholar]
- 24.Fischer-Le Saux M, Bonneau S, Essakhi S, Manceau C, Jacques M-A. Aggressive emerging pathovars of Xanthomonas arboricola represent widespread epidemic clones distinct from poorly pathogenic strains, as revealed by multilocus sequence typing. Appl Environ Microbiol. 2015;81:4651–4668. doi: 10.1128/AEM.00050-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernandes C, Blom J, Pothier JF, Tavares F. High-quality draft genome sequence of Xanthomonas sp. strain CPBF 424, a Walnut-Pathogenic strain with atypical features. Microbiol Resour Announc. 2018;7 doi: 10.1128/MRA.00921-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blom J, Albaum SP, Doppmeier D, Pühler A, Vorhölter F-J, et al. EDGAR: a software framework for the comparative analysis of prokaryotic genomes. BMC Bioinformatics. 2009;10:154. doi: 10.1186/1471-2105-10-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blom J, Kreis J, Spänig S, Juhre T, Bertelli C, et al. EDGAR 2.0: an enhanced software platform for comparative gene content analyses. Nucleic Acids Res. 2016;44:W22–W28. doi: 10.1093/nar/gkw255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hajri A, Pothier JF, Fischer-Le Saux M, Bonneau S, Poussier S, et al. Type three effector gene distribution and sequence analysis provide new insights into the pathogenicity of plant-pathogenic Xanthomonas arboricola . Appl Environ Microbiol. 2012;78:371–384. doi: 10.1128/AEM.06119-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryan RP, Vorhölter F-J, Potnis N, Jones JB, Van Sluys M-A, et al. Pathogenomics of Xanthomonas: understanding bacterium-plant interactions. Nat Rev Microbiol. 2011;9:344–355. doi: 10.1038/nrmicro2558. [DOI] [PubMed] [Google Scholar]
- 32.White FF, Potnis N, Jones JB, Koebnik R. The type III effectors of Xanthomonas . Mol Plant Pathol. 2009;10:749–766. doi: 10.1111/j.1364-3703.2009.00590.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cesbron S, Briand M, Essakhi S, Gironde S, Boureau T, et al. Comparative genomics of pathogenic and nonpathogenic strains of Xanthomonas arboricola unveil molecular and evolutionary events linked to pathoadaptation. Front Plant Sci. 2015;6:1126. doi: 10.3389/fpls.2015.01126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garita-Cambronero J, Palacio-Bielsa A, Cubero J. Xanthomonas arboricola pv. pruni, causal agent of bacterial spot of stone fruits and almond: its genomic and phenotypic characteristics in the X. arboricola species context. Mol Plant Pathol. 2018;19:2053–2065. doi: 10.1111/mpp.12679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garita-Cambronero J, Palacio-Bielsa A, López MM, Cubero J. Pan-genomic analysis permits differentiation of virulent and non-virulent strains of Xanthomonas arboricola that cohabit Prunus spp. and elucidate bacterial virulence factors. Front Microbiol. 2017;8:1–17. doi: 10.3389/fmicb.2017.00573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo Y, Figueiredo F, Jones J, Wang N. HrpG and HrpX play global roles in coordinating different virulence traits of Xanthomonas axonopodis pv. citri . Mol Plant Microbe Interact. 2011;24:649–661. doi: 10.1094/MPMI-09-10-0209. [DOI] [PubMed] [Google Scholar]
- 37.Jacobs JM, Pesce C, Lefeuvre P, Koebnik R. Comparative genomics of a cannabis pathogen reveals insight into the evolution of pathogenicity in Xanthomonas . Front Plant Sci. 2015;6:431. doi: 10.3389/fpls.2015.00431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.