Abstract
A progressive increase in copy number variation (CNV) characterizes the natural history of cutaneous melanoma progression toward later disease stages, but our understanding of genetic drivers underlying chromosomal arm-level CNVs remains limited. To identify candidate progression drivers, we mined the TCGA SKCM dataset and identified HDGF as a recurrently amplified gene whose high mRNA expression correlates with poor patient survival. Using melanocyte-specific overexpression in the zebrafish BRAFV600E-driven MiniCoopR melanoma model, we show that HDGF accelerates melanoma development in vivo. Transcriptional analysis of HDGF compared to control EGFP tumors showed the activation of endothelial/angiogenic pathways. We validated this observation using an endothelial kdrl:mCherry reporter line which showed HDGF to increases tumor vasculature. HDGF is frequently co-altered with the established melanoma driver SETDB1. Both genes are located on chromosome 1q, and their co-amplification is observed in up to 13% of metastatic melanoma. TCGA patients with both genes amplified and/or overexpressed have a worse melanoma specific survival. We tested co-expression of HDGF and SETDB1 in the MiniCoopR model, which resulted in faster and more aggressive melanoma development than either gene individually. Our work identifies the co-amplification of HDGF and SETDB1 as a functional driver of melanoma progression and poor patient prognosis.
1 |. INTRODUCTION
Cutaneous melanoma is the tumor type with the highest genomic mutational burden (Alexandrov et al., 2020; Consortium, 2020; Gerstung et al., 2020). While genomic analysis has identified several recurrently mutated drivers that define malignant initiation of the disease (e.g., oncogenic BRAF mutations) (Ackermann et al., 2005; Brose et al., 2002; Cancer Genome Atlas, 2015; Dhomen et al., 2009; Dovey et al., 2009; Michaloglou et al., 2005; Patton et al., 2005; Shain et al., 2015), mutational burden does not correlate with disease stage (Consortium, 2020; Gerstung et al., 2020; Priestley et al., 2019; Shain et al., 2015). Paired analyses of primary tumors and metastases have shown a surprisingly low degree of mutational heterogeneity and have not identified genes with recurrent point mutations that define metastasis (Priestley et al., 2019; Reiter et al., 2018). The same studies documented a progressive increase in copy number variation during cutaneous melanoma progression (Priestley et al., 2019; Reiter et al., 2018; Shain et al., 2015), similar to what has been observed in other solid tumors at large (Priestley et al., 2019). While some recurrent amplifications are focal, often with a clear underlying driver gene (e.g., CCND1), the genes driving selection advantages for chromosomal arm-level events, which instead often involve many genes, are not as well understood (Priestley et al., 2019). Such is the case of chromosome 1q amplification, which is observed in >20% of all metastatic pancancer samples (Priestley et al., 2019), and is the second most significant arm-level amplification observed in cutaneous melanoma (Amp q = 4.29e-9, Amp z score = 6.24) (Center, 2016). We have previously utilized mosaic transgenesis in the BRAFV600E-driven MiniCoopR (MCR) melanoma model in zebrafish to screen 17 genes located on a focally amplified region of 1q21 (chr1: 147.2–149.2 Mb), and identified SETDB1 as a driver accelerating melanoma development in vivo (Ceol et al., 2011). Whether other genes contribute to the recurrent selection of chromosome 1q amplification in cutaneous melanoma remains unknown.
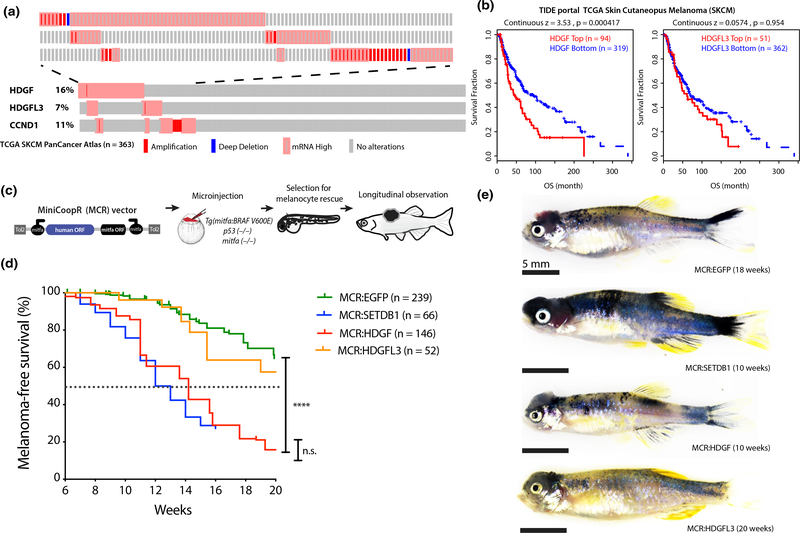
To identify additional candidate progression drivers underlying 1q arm-level amplification outside of the SETDB1 focal region, we mined the TCGA SKCM copy number, expression, and survival datasets (See online supplementary methods). We identified hepatoma-derived growth factor (HDGF) located on 1q23.1 as a recurrently amplified and/or overexpressed gene in TCGA SKCM (Figure 1a) and recurrently amplified in 3 additional independent datasets of metastatic melanoma (Figure S1a). High HDGF mRNA expression correlated with poor overall survival in melanoma patients in multiple analyses of the TCGA SKCM dataset (Figure 1b and Figure S1b). In contrast, a closely related HDGF family member HDGF Like 3 (HDGFL3, 15q25.2) is also recurrently amplified (Figure 1a), but its expression does not correlate with patient survival (Figure 1b and Figure S1c). HDGF high expression levels in TCGA bulk mRNA-seq correlate with copy number gain or amplification (Figure S1 D), and interrogation of a scRNA-seq dataset of metastatic melanoma (Tirosh et al., 2016) shows HDGF to be expressed by both MITF + cancer cells and the tumor stroma (Figure S1e).
FIGURE 1.
Frequent amplification or overexpression of HDGF correlates with poor prognosis in melanoma patients and accelerates melanoma in a zebrafish transgenic model. (a) Oncoprint of HDGF, HDGFL3 and commonly amplified gene CCND1 in TCGA skin cutaneous melanoma dataset showing copy number changes and mRNA expression (Z score > 2) compared to diploid samples. Mutations are not shown and no patient had evidence of “mRNA low.” Each individual rectangle represents a patient. The top part of the figure is a close-up of altered patients. (b) Correlation of mRNA expression of HDGF and HDGFL3, with Overall survival in TCGA skin cutaneous melanoma dataset from TIDE portal. This analysis identifies the Z expression threshold resulting in maximal curve separation (logRank Mantel-Cox p) across the two groups. For bottom versus top quartile, see Figure S1b–c. (c) Schematic of in vivo MCR experimental design (top) and (d) Kaplan–Meier survival curve (bottom) showing the tumor onset of MCR vectors microinjected in Tg(mitfa:BRAFV600E);tp53-/-;mitfa-/- to drive melanocyte-specific expression of HDGF (red) or HDGFL3 (orange), EGFP (green) and SETDB1 (blue) (****=p < .0001) (logRank Mantel-Cox p). Median onset is indicated by the dotted line. (e) Representative image of the gross anatomy of a tumor-bearing fish. Bright field image, all scale bars are 5mm. Pigmented raised melanoma tumors are visible on the head region of each fish
Given that HDGF alteration in melanoma patients co-occurs with BRAF and NRAS driver mutations (Figure S2a–b), we leveraged the zebrafish MCR model (Figure 1c) to test the functional effect of HDGF on melanoma progression in vivo. Melanoma is initiated by melanocyte-specific overexpression of human oncogenic BRAFV600E and tp53 loss, which makes it possible to test the tissue-specific effect of candidate co-operating oncogenes on the development of macroscopically visible raised melanoma tumors, thus capturing both effects on tumor initiation, and growth/early stage progression (Ceol et al., 2011) (Figure 1c schematic and online supplementary methods). Tg(mitfa:BRAFV600E); tp53-/-; mitfa-/- zebrafish lack melanocytes and do not develop melanoma. Mosaic integration of the transposon-based expression vector MiniCoopR (MCR) rescues melanocyte development by restoring mitfa, while simultaneously driving via an mitfa promoter tissue-specific expression of a gene of choice in all rescued melanocytes (Ceol et al., 2011). In this model, we tested the effect of human HDGF overexpression using EGFP and SETDB1 as established negative and positive controls, respectively (Figure 1d–e). As an additional control, we tested HDGFL3. HDGF, but not HDGFL3, accelerated the development of raised malignant melanoma tumors similarly to SETDB1, compared to the EGFP negative control (p < .0001) (Figure 1d–e).
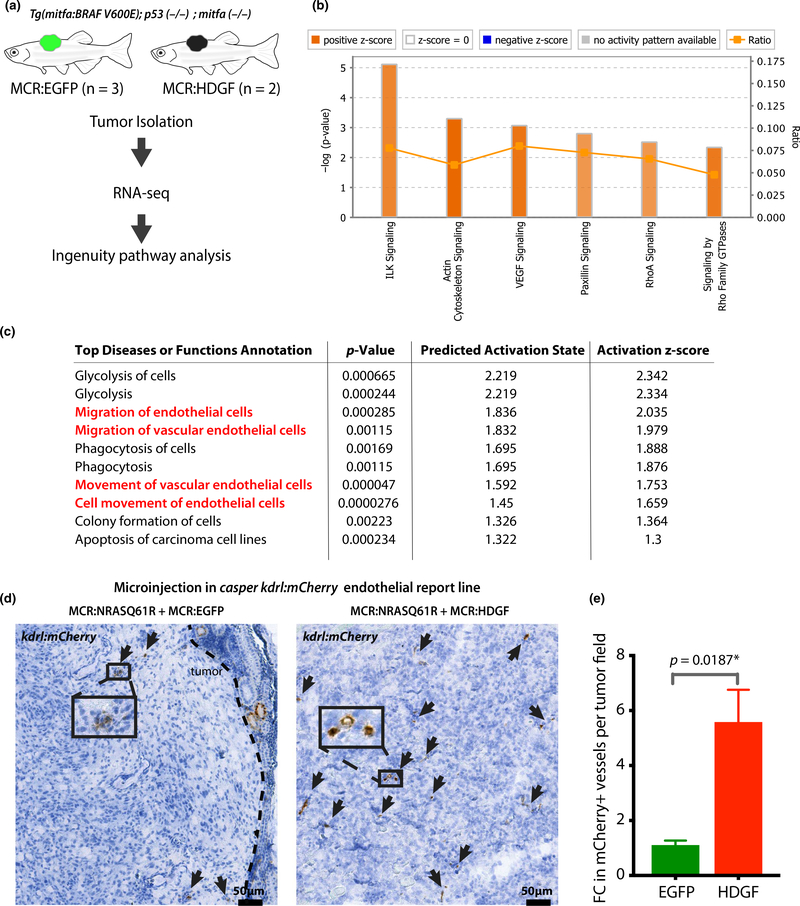
HDGF has been shown to act as a transcriptional regulator (Bao et al., 2014). To gain mechanistic insight into HDGF’s effect as a driver in the MCR model, we conducted transcriptional analysis by RNA-seq of dissected MCR:HDGF and MCR:EGFP tumors (Figure 2a, Table S1). Ingenuity Pathway Analysis (IPA) of significant differentially expressed genes between MCR:HDGF versus MCR:EGFP control tumors (Table S1) revealed the top significantly activated pathways (Z score > 2). These included VEGF signaling and other pathways related to interleukin and cytoskeletal signaling, which share several genes with VEGF-induced angiogenic responses (Figure 2b). Consistently, IPA predicted top diseases and functions contained several terms suggestive of angiogenesis and endothelial cells (Figure 2c). The top predicted upstream regulators of the transcriptional change included HIF1a and transcription factors involved in endothelial/cardiac specification (Figure S3a). Collectively, these data suggested HDGF to induce tumor vascularization, which is consistent with its known role as a regulator of angiogenesis (Bao et al., 2014; Tsai et al., 2013). GREAT pathway analysis of known HDGF targets from the ENCODE project suggests HDGF to directly bind and regulate genes involved in the VEGF, HIF1aand PDGFR signaling pathways (Figure S3b). Interestingly, TCGA SKCM patients with HDGF amplification or overexpression have a higher Buffa Hypoxia score (Buffa et al., 2010) than patients without HDGF alterations (Figure S3 C, p = 3.925e-4, q = 0.0151). HDGF transcription/copy number gain may be selected for and/or induced by an hypoxic environment during tumor progression.
FIGURE 2.
HDGF increases angiogenesis. (a) Outline of RNA isolation and transcriptional analysis in MCR:EGFP and MCR:HDGF mitfa:BRAFV600E;tp53-/--driven tumors. (b) Ingenuity pathway analysis (IPA) showing the most significant activated pathways (Z score > 2) in HDGF tumors compared to EGFP tumor controls. Color intensity corresponds to Z score value (i.e., darker orange, larger Z score). (c) IPA analysis showing the most significantly activated diseases or function annotations in MCR:HDGF tumors compared to MCR:EGFP controls. Terms relating to endothelial cell migration/angiogenesis are highlighted in red. (d) Analysis of the effect of HDGF on tumor vasculature in the MCR:NRASQ61R model. MCR:EGFP or MCR:HDGF was co-injected with MCR:NRASQ61R into one cell stage casper embryos carrying the krdl:mCherry transgenic vasculature reporter line. 8- to 10-week-old tumor-bearing zebrafish were fixed and subjected to anti-mCherry immunohistochemistry using DAB (brown). Slides were bleached to remove melanin pigment interference. A representative image per genotype is shown. The brown signal in the tumor (mCherry) represents vasculature and is highlighted with arrows. (E) Quantification of DAB mCherry + vessels in 4 MCR:EGFP and 5 MCR:HDGF tumors (multiple fields of view/tumor, at 20x). A total of 22 fields were analyzed. The number of vessels per field was averaged per animal, and fold change (FC) was calculated by normalizing over the average of EGFP controls and analyzed via unpaired t test
To validate whether HDGF tumors have increased tumor vasculature, we made use of an established fluorescent endothelial reporter line kdrl:mCherry (Proulx et al., 2010), crossed into the optically clear casper (mitfa-/-;mpv17-/-) background (White et al., 2008). We generated melanomas using microinjection of an MCR:NRASQ61R vector, which we have previously shown to induce very rapid melanoma formation in casper zebrafish in 8–12 weeks (Ablain et al., 2018; McConnell et al., 2019). We injected MCR:NRASQ61R together with equal amounts of either MCR:EGFP or MCR:HDGF (Figure S3 D) into kdrl:mCherry casper zebrafish. Immunohistochemistry analysis on fixed tumors showed a ~ 6-fold increase in mCherry + vessels in MCR:NRASQ61R + MCR:HDGF tumors compared to MCR:NRASQ61R + MCR:EGFP controls (p = .0187, Figure 2d–e) at 8–10 weeks post-injection, thus confirming HDGF to induce an increase in tumor vascularization.
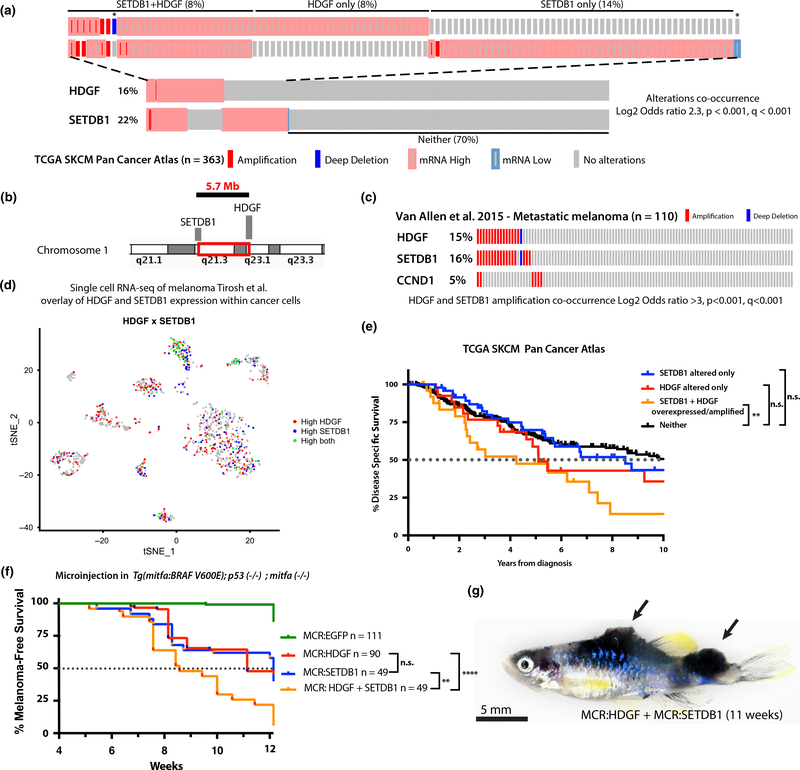
Having established HDGF as a functional driver of melanoma, we explored its genomic relationship with SETDB1 focal amplification. As expected, given their proximity (5.7 Mb distance) on chromosome 1q, SETDB1 and HDGF are significantly co-altered by amplification and/or overexpression in 8% of patients (p < .001, q < 0.001) (Figure 3a–b). In the TCGA dataset, which includes a mixture of stages and primary versus metastatic tumors as a tissues source, HDGF and SETDB1 co-amplification is observed in ~ 3% of patients (Figure S4a–b). In other genomic datasets composed exclusively of metastatic melanoma (Hugo et al., 2016; Snyder et al., 2014; Van Allen et al., 2015), their co-amplification is more pronounced and observed in up to 13% of patients (Figure 3c, Figure S4a), which is comparable to the frequencies of the most recurrently amplified melanoma oncogenes CCND1, TERT and MITF (Figure 3c and Figure S1a). Consistent with their co-variation in copy number and bulk RNA-seq, scRNA-seq of metastatic melanoma (Tirosh et al., 2016) also shows frequent high co-expression of HDGF and SETDB1 on a per cell basis (Figure 3d). Interestingly, patients in the TCGA SKCM dataset with both genes overexpressed and/or amplified have worse disease-specific survival (Figure 3a,e) (SETDB1 and HDGF co-altered versus. neither p = .0099; HDGF but not SETDB1 altered, or SETDB1 but not HDGF altered versus neither not significant). Given the two driver genes’ independent mechanistic effects (HDGF on angiogenesis and SETDB1 on senescence (Ceol et al., 2011)), we hypothesized that co-alteration of both by amplification and/or overexpression might result in a more aggressive disease, causing poor patient survival. We tested the effect of co-injection of MCR:HDGF + MCR:SETDB1 (pooled 12.5 pg of each vector for a total of 25 pg) versus MCR:HDGF, MCR:SETDB1 or MCR:EGFP control single vector injection (each at 25 pg) on tumor development in the BRAFV600E MCR melanoma model (Ceol et al., 2011) (Figure 1c). MCR:HDGF + MCR:SETDB1 co-injection resulted in a stronger acceleration in malignant melanoma formation (median onset 8 weeks) than either MCR:HDGF (median onset 11 weeks) or MCR:SETDB1 (median onset 11 weeks) vector alone (HDGF + SETDB1 versus SETDB1 p = .0013, HDGF + SETDB1 versus HDGF p < .0001, HDGF versus SETDB1 p = .25, HDGF + SETDB1, HDGF, SETDB1 versus EGFP all p < .0001) (Figure 3f). Furthermore, a higher tumor burden was observed in tumor bearing fish at 10 weeks, with 24% of individuals developing multiple (>1) distinct tumors, which is an uncommon observation in MCR:EGFP controls (4%) (Figures 3g and 1d quantified in Figure S4f). Taken together, our data suggest an additive driver effect from HDGF and SETDB1 co-amplification.
FIGURE 3.
HDGF and SETB1 frequent co-amplification or overexpression correlates with poor prognosis in melanoma patients and strongly accelerates melanoma in a zebrafish transgenic model. (a) Oncoprint of HDGF and SETDB1 in TCGA skin cutaneous melanoma dataset (Liu et al., 2018) showing significant co-occurrence of copy number changes and mRNA expression (Z score > 2). Mutations are not shown. The top part of the figure is a close-up of altered patients. Overall % prevalence in the cohort is reported for each group (SETDB1 + HDGF, HDGF only, SETDB1 only or neither). Each individual rectangle is a patient. Patients labeled with * were included in the neither group for survival analysis in 3E. (b) Illustration of the genomic localization and distance of HDGF and SETDB1 on chromosome 1. (c) Oncoprint of HDGF, SETDB1 and commonly amplified gene CCND1 in metastatic melanoma (Van Allen et al., 2015). (d) scRNA plot of metastatic melanoma (Tirosh et al., 2016) cancer cells showing co-expression of HDGF and SETDB1 at the single cell level in melanoma. Each cluster represents a patient, and only cancer cells are shown. (e) Kaplan–Meier disease-specific survival analysis of patients in Figure 1a (**p = .0099, n.s.= not significant) (logRank Mantel-Cox p). (f) Effect on tumor onset of MCR vectors microinjected into Tg(mitfa:BRAFV600E);tp53-/-;mitfa-/-. For HDGF + SETDB1, a pool of the two vectors (12.5pg + 12.5 pg of DNA) with the same total amount of plasmid DNA as the other conditions (25 pg) was injected. HDGF + SETDB1 versus SETDB1 p = .0013**, HDGF + SETDB1 versus HDGF p < .0001****, HDGF versus SETDB1 p = .25 (not significant), HDGF + SETDB1, HDGF, SETDB1 versus EGFP all p < .0001**** (logRank Mantel-Cox p). (g) Representative image of a MCR:HDGF + MCR:SETDB1 fish bearing multiple tumors. Bright field image, scale bar is 5 mm. Multiple pigmented raised melanoma tumors are visible on the dorsum and caudal peduncle regions and are marked by black arrows. See Figure S4f for quantification
A previous study reported HDGF protein expression to correlate with melanoma progression in tissue biopsies, raising the question about HDGF’s role in melanoma initiation or progression (Bernard et al., 2003). A prior perturbation study using tissue-specific expression of HDGF in an ink4a ± mouse model exposed to UV did not result in nevi or melanoma formation (Sedlmaier et al., 2011), while a study using HDGF perturbation in melanoma cell lines in vitro and in the B16-F10 mouse transplant model suggested a cancer cell autonomous role in melanoma migration (Tsai et al., 2013). Our work, in line with prior literature, solidifies HDGF’s role as a melanoma progression gene, and as a regulator of angiogenesis in melanoma. We show for the first time that recurrent co-alteration of HDGF and SETDB1 drives cutaneous melanoma progression and poor patient outcome. We speculate that recurrent amplification of chromosome 1q frequently observed in malignant melanoma may confer an evolutionary advantage in part though HDGF/SETDB1 amplification that results in its selection during disease progression. Given that this CNV event is observed in >20% of all patients in TCGA PanCancer analysis, and prior literature has established HDGF and SETDB1’s individual roles in other solid tumors (e.g., colorectal and liver cancer), it is plausible that our findings are relevant to additional tumor types beyond melanoma.
Supplementary Material
Significance.
Cutaneous melanoma has the highest genomic mutational burden of all tumor types, but neither overall mutational burden nor recurrent point mutations have been correlated with later stages of disease and progression toward metastasis. A progressive accumulation of copy number changes has been described in late disease stages, but our understanding of which specific events drive melanoma progression remains incomplete. Here, we identified HDGF and SETDB1 on chromosome 1q as additive functional drivers of melanoma progression, whose co-alteration correlates with poor patient survival. Our study pinpoints an evolutionary advantage that might underlie the recurrent amplification of chromosome 1q in malignant melanoma.
ACKNOWLEDGMENTS
MF was supported by Boehringer Ingelheim Fonds. EvR was supported by a Netherlands Organization for Scientific Research (NWO) Rubicon fellowship, and a Dutch Cancer Foundation (KWF) fellowship for Fundamental Cancer Research. LIZ is supported by: R01 CA103846, MRA (Zon, Garraway), The Starr Cancer Consortium and the Ellison Foundation. The authors would like to thank Serine Avagyan for experimental support, and Julian Ablain for providing reagents and helpful discussions.
Funding information
Boehringer Ingelheim Fonds; Ellison Medical Foundation; Starr Foundation; Melanoma Research Alliance; National Cancer Institute, Grant/Award Number: R01 CA103846
Footnotes
CONFLICT OF INTERESTS
LIZ is a founder and stockholder of Fate Therapeutics, Inc., Scholar Rock, Camp4 Therapeutics, Inc., Amagma Therapeutics, Inc., and a scientific advisor for Stemgent.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section.
REFERENCES
- Ablain J, Xu M, Rothschild H, Jordan RC, Mito JK, Daniels BH, & Yeh I (2018). Human tumor genomics and zebrafish modeling identify SPRED1 loss as a driver of mucosal melanoma. Science, 362(6418), 1055–1060. 10.1126/science.aau6509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackermann J, Frutschi M, Kaloulis K, McKee T, Trumpp A, & Beermann F (2005). Metastasizing melanoma formation caused by expression of activated N-RasQ61K on an INK4a-deficient background. Cancer Research, 65(10), 4005–4011. 10.1158/0008-5472.CAN-04-2970 [DOI] [PubMed] [Google Scholar]
- Akbani R, Akdemir KC, Aksoy BA, Albert M, Ally A, Amin SB, Arachchi H, Arora A, Auman JT, Ayala B, Baboud J, Balasundaram M, Balu S, Barnabas N, Bartlett J, Bartlett P, Bastian BC, Baylin SB, Behera M, ... Zou L. (2015). Genomic Classification of Cutaneous Melanoma. Cell, 161(7), 1681–1696. 10.1016/j.cell.2015.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, Wu Y, Boot A, Covington KR, Gordenin DA, Bergstrom EN, Islam SMA, Lopez-Bigas N, Klimczak LJ, McPherson JR, Morganella S, Sabarinathan R, Wheeler DA, Mustonen V, Getz G, ... Stratton MR. (2020). The repertoire of mutational signatures in human cancer. Nature, 578(7793), 94–101. 10.1038/s4158?6-020-1943-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao C, Wang J, Ma W, Wang X, & Cheng Y (2014). HDGF: A novel jack-of-all-trades in cancer. Future Oncology, 10(16), 2675–2685. 10.2217/fon.14.194 [DOI] [PubMed] [Google Scholar]
- Bernard K, Litman E, Fitzpatrick JL, Shellman YG, Argast G, Polvinen K, & Resing KA (2003). Functional proteomic analysis of melanoma progression. Cancer Research, 63(20), 6716–6725. [PubMed] [Google Scholar]
- Brose MS, Volpe P, Feldman M, Kumar M, Rishi I, Gerrero R, & Weber BL (2002). BRAF and RAS mutations in human lung cancer and melanoma. Cancer Research, 62(23), 6997–7000. [PubMed] [Google Scholar]
- Buffa FM, Harris AL, West CM, & Miller CJ (2010). Large meta-analysis of multiple cancers reveals a common, compact and high prognostic hypoxia metagene. Britis H Journal of Cancer, 102(2), 428–435. 10.1038/sj.bjc.6605450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Center BITGDA (2016). LowPass Copy number analysis (GISTIC2) . [Google Scholar]
- Ceol CJ, Houvras Y, Jane-Valbuena J, Bilodeau S, Orlando DA, Battisti V, Fritsch L, Lin WM, Hollmann TJ, Ferré F, Bourque C, Burke CJ, Turner L, Uong A, Johnson LA, Beroukhim R, Mermel CH, Loda M, Ait-Si-Ali S, ... Zon LI. (2011). The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature, 471(7339), 513–517. 10.1038/nature09806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium I T P C A o W G (2020). Pan-cancer analysis of whole genomes. Nature, 578(7793), 82–93. 10.1038/s41586-020-1969-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhomen N, Reis-Filho JS, da Rocha Dias S, Hayward R, Savage K, Delmas V, Larue L, Pritchard C, & Marais R (2009). Oncogenic Braf induces melanocyte senescence and melanoma in mice. Cancer Cell, 15(4), 294–303. 10.1016/j.ccr.2009.02.022 [DOI] [PubMed] [Google Scholar]
- Dovey M, White RM, & Zon LI (2009). Oncogenic NRAS cooperates with p53 loss to generate melanoma in zebrafish. Zebrafish, 6(4), 397–404. 10.1089/zeb.2009.0606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstung M, Jolly C, Leshchiner I, Dentro SC, Gonzalez S, Rosebrock D, Mitchell TJ, Rubanova Y, Anur P, Yu K, Tarabichi M, Deshwar A, Wintersinger J, Kleinheinz K, Vázquez-García I, Haase K, Jerman L, Sengupta S, Macintyre G, Van Loo P (2020). The evolutionary history of 2,658 cancers. Nature, 578(7793), 122–128. 10.1038/s41586-019-1907-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugo W, Zaretsky JM, Sun LU, Song C, Moreno BH, Hu-Lieskovan S, Berent-Maoz B, Pang J, Chmielowski B, Cherry G, Seja E, Lomeli S, Kong X, Kelley MC, Sosman JA, Johnson DB, Ribas A, & Lo RS (2016). Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell, 165(1), 35–44. 10.1016/j.cell.2016.02.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee AV, Omberg L, Wolf DM, Shriver CD, Thorsson V, Hu H, Caesar-Johnson SJ, Demchok JA, Felau I, Kasapi M, ... Mariamidze A. (2018). An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell, 173(2), 400–416 e411. 10.1016/j.cell.2018.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell AM, Mito JK, Ablain J, Dang M, Formichella L, Fisher DE, & Zon LI (2019). Neural crest state activation in NRAS driven melanoma, but not in NRAS-driven melanocyte expansion. Developmental Biology, 449(2), 107–114. 10.1016/j.ydbio.2018.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, van der Horst CM, & Peeper DS (2005). BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature, 436(7051), 720–724. [DOI] [PubMed] [Google Scholar]
- Patton EE, Widlund HR, Kutok JL, Kopani KR, Amatruda JF, Murphey RD, Berghmans S, Mayhall EA, Traver D, Fletcher CDM, Aster JC, Granter SR, Look AT, Lee C, Fisher DE, & Zon LI (2005). BRAF mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Current Biology, 15(3), 249–254. 10.1016/j.cub.2005.01.031 [DOI] [PubMed] [Google Scholar]
- Priestley P, Baber J, Lolkema MP, Steeghs N, de Bruijn E, Shale C, Duyvesteyn K, Haidari S, van Hoeck A, Onstenk W, Roepman P, Voda M, Bloemendal HJ, Tjan-Heijnen VCG, van Herpen CML, Labots M, Witteveen PO, Smit EF, Sleijfer S, ... Cuppen E. (2019). Pan-cancer whole-genome analyses of metastatic solid tumors. Nature, 10.1038/s4158?6-019-1689-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proulx K, Lu A, & Sumanas S (2010). Cranial vasculature in zebrafish forms by angioblast cluster-derived angiogenesis. Developmental Biology, 348(1), 34–46. 10.1016/j.ydbio.2010.08.036 [DOI] [PubMed] [Google Scholar]
- Reiter JG, Makohon-Moore AP, Gerold JM, Heyde A, Attiyeh MA, Kohutek ZA, Tokheim CJ, Brown A, DeBlasio RM, Niyazov J, Zucker A, Karchin R, Kinzler KW, Iacobuzio-Donahue CA, Vogelstein B, & Nowak MA (2018). Minimal functional driver gene heterogeneity among untreated metastases. Science, 361(6406), 1033–1037. 10.1126/science.aat7171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlmaier A, Wernert N, Gallitzendorfer R, Abouzied MM, Gieselmann V, & Franken S (2011). Overexpression of hepatoma-derived growth factor in melanocytes does not lead to oncogenic transformation. BMC Cancer, 11, 457. 10.1186/1471-2407-11-457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shain AH, Yeh I, Kovalyshyn I, Sriharan A, Talevich E, Gagnon A, Dummer R, North J, Pincus L, Ruben B, Rickaby W, D’Arrigo C, Robson A, & Bastian BC (2015). The Genetic Evolution of Melanoma from Precursor Lesions. New England Journal of Medicine, 373(20), 1926–1936. 10.1056/NEJMoa1502583 [DOI] [PubMed] [Google Scholar]
- Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS, Hollmann TJ, Bruggeman C, Kannan K, Li Y, Elipenahli C, Liu C, Harbison CT, Wang L, Ribas A, ... Chan T. (2014). Genetic basis for clinical response to CTLA-4 blockade in melanoma. New England Journal of Medicine, 371(23), 2189–2199. 10.1056/NEJMoa1406498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Izar B, Prakadan SM, Wadsworth MH, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, Fallahi-Sichani M, Dutton-Regester K, Lin J-R, Cohen O, Shah P, Lu D, Genshaft AS, Hughes TK, Ziegler CGK, … Garraway LA. (2016). Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science, 352(6282), 189–196. 10.1126/science.aad0501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HE, Wu JC, Kung ML, Liu LF, Kuo LH, Kuo HM, & Liu GS (2013). Up-regulation of hepatoma-derived growth factor facilitates tumor progression in malignant melanoma [corrected]. PLoS One, 8(3), e59345. 10.1371/journ?al.pone.0059345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, Sucker A, Hillen U, Geukes Foppen MH, Goldinger SM, Utikal J, Hassel JC, Weide B, Kaehler KC, Loquai C, Mohr P, Gutzmer R, Dummer R, Gabriel S, ... Garraway LA. (2015). Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science, 350(6257), 207–211. 10.1126/scien?ce.aad0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RM, Sessa A, Burke C, Bowman T, LeBlanc J, Ceol C, Bourque C, Dovey M, Goessling W, Burns CE, & Zon LI (2008). Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell, 2(2), 183–189. 10.1016/j.stem.2007.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.