Significance
The axolotl is an important model organism because it is a tetrapod with a similar body plan to humans. Unlike humans, the axolotl regenerates limbs and other complex tissues. Therefore, the axolotl contributes to understanding evolution, development, and regeneration. With sophisticated tools for gene modification and tissue labeling, a fully assembled genome sequence was a sorely missing resource. Assembly was difficult because the genome size is 10× that of humans. Here, we use a cross-linking strategy called Hi-C to link together fragmented genome sequences to chromosome scale. We show that gene regulation occurs over very large genomic distances and that mitotic chromosomes are packaged efficiently.
Keywords: genome assembly, axolotl, regeneration, Topological Associating Domains
Abstract
Vertebrates harbor recognizably orthologous gene complements but vary 100-fold in genome size. How chromosomal organization scales with genome expansion is unclear, and how acute changes in gene regulation, as during axolotl limb regeneration, occur in the context of a vast genome has remained a riddle. Here, we describe the chromosome-scale assembly of the giant, 32 Gb axolotl genome. Hi-C contact data revealed the scaling properties of interphase and mitotic chromosome organization. Analysis of the assembly yielded understanding of the evolution of large, syntenic multigene clusters, including the Major Histocompatibility Complex (MHC) and the functional regulatory landscape of the Fibroblast Growth Factor 8 (Axfgf8) region. The axolotl serves as a primary model for studying successful regeneration.
Several phylogenetically important animals such as salamanders and lungfish have exceptionally large genomes reaching more than 10-fold the size of the human genome (1). These giant genomes provide extreme examples that allow us to analyze scalable and limiting features of chromosome organization and gene regulation but also pose technical challenges for chromosome-scale genome assembly and annotation. The 32 Gb axolotl genome was previously assembled using a long-read assembly to a contig N50 of 219 Kb and optical mapping scaffolding to a scaffold N50 of 2 Mb. Contig assembly was limited by the presence of large, repetitive retroviral elements. Indeed, the massive presence of repetitive sequences both intergenically and within introns appears to be the primary basis of the expanded genome size (2).
Given the difficulty of obtaining long reads spanning these long, repetitive regions, a different approach seemed necessary to obtain a chromosome scale assembly. All-versus-all chromosome conformation capture methods (Hi-C) quantify the short- and long-distance contacts of any given part of a genome to other parts of the genome and have been instrumental in reconstructing organizational principles of chromatin (3). The frequency of short-distance contacts is used to reconstruct the linear order of genomic sequences from fragmented genome assemblies to obtain chromosome-scale contiguity of sequence information (4, 5). This has elicited a sea change in the availability of high-quality genome sequence information enabling genomic access to important organisms and the phylogenetic analysis of evolutionary history.
Analysis of longer-range Hi-C contacts in interphase cells has revealed the existence of Topologically Associating Domains (TADs) that frequently correspond to domains of long-range transcriptional regulatory interactions (6). In both vertebrates and invertebrates, arrays of conserved noncoding elements (CNEs) called gene regulatory blocks (GRBs) coincide with TADs, arguing for TADs as ancient organizational features related to gene regulation in metazoan genomes (7). Interestingly, comparison of GRBs and their associated TADs among various animal species shows a correspondence between GRB and TAD size with genome size.
An outstanding biological setting where the relationship between long-distance gene regulation and genomic architecture has been analyzed is mammalian limb development (8–10). For example, in the mouse limb, Ihh and Epha4 are adjacent genes with distinct limb expression patterns, each with their own regulatory domains spanning over 100 Kb. A large deletion encompassing the TAD boundary between these results in Ihh being expressed under the influence of EphA regulatory elements acting over hundreds of kilobases, yielding a polydactyly phenotype (10). Salamanders as basal tetrapods and the axolotl (Ambystoma mexicanum), as an experimentally accessible representative species, have been critical for evolutionary studies of limb development (11, 12). It is currently unknown whether and how such long-distance regulation and TAD organization may occur in the axolotl in the face of a roughly 13-fold larger genome. It is also an open question whether adaptations related to these large genomic distances could contribute to characteristic aspects of their limb development (13–15). For example, the conserved, long-range regulatory elements controlling Fgf8 gene expression in the apical ectodermal ridge of the mouse limb bud have been extensively characterized and act over 200 Kb ranges (8). It has not been determined whether these regulatory elements exist in the axolotl and whether their spacing scales with genome size. This is a particularly interesting question, as the axolotl displays a divergent expression domain of Fgf8 in the anterior mesenchyme instead of the distal ectoderm (16). It will be important to determine whether conserved regulatory elements exist and whether they confer mesenchymal or ectodermal expression.
The axolotl and other salamanders are extreme not only in their genome size but also in their ability to regenerate functional body parts, such as the limb, after removal. The process of limb regeneration involves the reactivation of the gene regulatory network governing limb development in the adult blastema. How this gene regulatory network is suppressed in the adult limb but reawakened upon limb amputation and blastema formation has been a key question of regeneration biology that has lain long unanswered, largely due to unavailability of a chromosome-scale genome assembly and of genome-wide chromatin profiling to analyze the genetic basis of this remarkable trait. On the other hand, the extensive availability of cell lines and transgenic animals makes the axolotl an accessible and important model to experimentally probe the genetic basis of regeneration and to understand the relationship between genome size, chromosome architectural features, and transcription. Here we describe a chromosome-scale assembly of the 32 Gb axolotl genome, an important cornerstone to study the mechanisms and evolution of gene regulation during limb development and regeneration. We use Hi-C contact information and the annotated genome to uncover the topological organization of long-distance transcriptional regulatory units and of mitotic chromosomes in this giant genome.
Results
Chromosome-Level Assembly of the Axolotl Genome.
Previous work using meiotic mapping had assigned and ordered the majority of Bionano scaffolds from a long read–based axolotl genome assembly (scaffold N50 2 Mb) (2) to chromosome-scale linkage groups, but the resolution was not sufficient to determine the orientation and local ordering of most scaffolds (17) (SI Appendix, Fig. S1A). We set out to use Hi-C data (3) to produce a high-resolution chromosome-scale assembly. Using the four-cutter DpnII restriction enzyme, we generated Hi-C libraries from d/d strain larval tissues (432 million contacts) as well as from the cultured AL1 (limb mesenchyme) cell line in interphase (766 million contacts) or synchronized in mitosis (72 million contacts) and one HindIII-based library from interphase cells (160 million contacts) in order to obtain high-resolution contiguity. We have focused on assembling the genome of the Vienna d/d strain of axolotl which is a line carrying a homozygous mutation in the Endothelin 3 locus that causes a lack of skin pigmentation (18); this strain is commonly used experimentally for molecular and live imaging experiments due to lack of pigmentation (19).
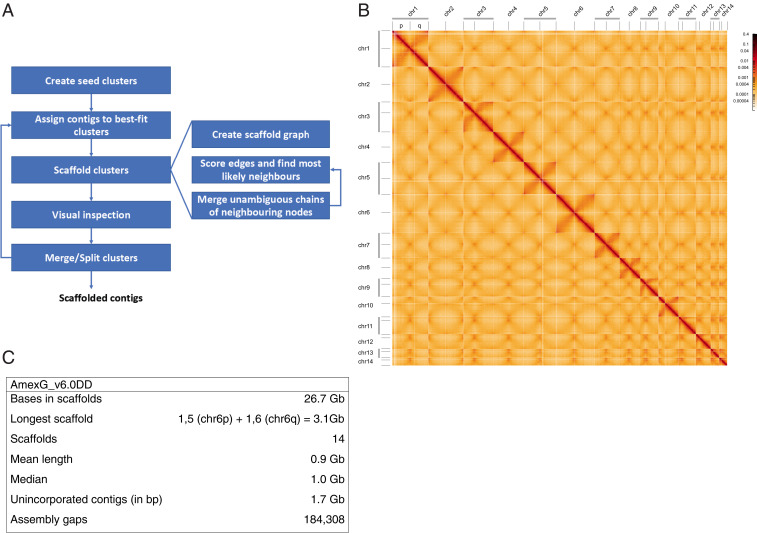
A Hi-C scaffolding algorithm was developed taking into account the depletion of contacts in the abundant repetitive regions that affects the comparison of contact frequencies between contigs (Fig. 1A and Materials and Methods), subsequently influencing their relative ordering and orientation in the scaffolds. Contact data from in vivo tissue samples and cultured cells were implemented in two parallel assembly processes that also took into account the previous meiotic mapping data to initiate the clustering (Materials and Methods). The data from the two assembly processes were compared and merged using the d/d tissue data as a base to arrive at the final chromosome-scale assembly consisting of 14 chromosomes [split into 14 shorter p and 14 longer q chromosome arms for technical reasons (20)] to generate a working dataset that includes 26.7 Gb, out of 28.4 Gb, of contigs from the original assembly (Fig. 1 B and C and SI Appendix, Fig. S1 A and B).
Fig. 1.
Hi-C–mediated scaffolding of the giant axolotl genome. (A) Procedure for scaffolding the large and repeat-rich genome. Seed clusters were created based on the linkage map data from ref. 17. Unassigned contigs were assigned to the clusters based on contact frequency. Subsequent scaffolding of the clusters and visual inspection of contact maps allowed their splitting and merging. This was repeated until no more changes could be made to the clusters. The scaffolding itself creates a graph structure from the contact data and contigs contained in a cluster turning contigs into nodes and contacts into edges. Based on the edges, the most likely left and right neighbor of each node are computed, and continuous chains of nodes having each other as their most likely neighbor merged into paths (effectively a new node) subsuming their constituent nodes. This process is repeated until no more paths can be constructed. (B) Hi-C heatmap of d/d contact data of all 28 scaffolded chromosome arms. Denser areas of red signal off-diagonal represent interactions between the arms of the same chromosome. (C) Summary of assembly characteristics. For chromosome arm lengths, see SI Appendix, Fig. S1B.
We also compared the d/d Hi-C data to that from two datasets that were generated from a separate wild-type (WT) animal to annotate potential structural polymorphisms between d/d and WT but did not detect large-scale (megabase) structural variation between the datasets (SI Appendix, Fig. S1C). Finally, we generated full-length messenger RNA (mRNA) sequences from several tissues through the Iso-Seq platform (SRR13825941) and aligned these data, along with several available RNA sequencing (RNA-Seq) datasets (PRJNA300706, PRJNA378982) (2, 21), to the genome to produce gene models representing 35,529 annotated genomic loci with a Benchmarking Universal Single-Copy Orthologs (BUSCO) C-score of 88.4% (vertebrate database) (22).
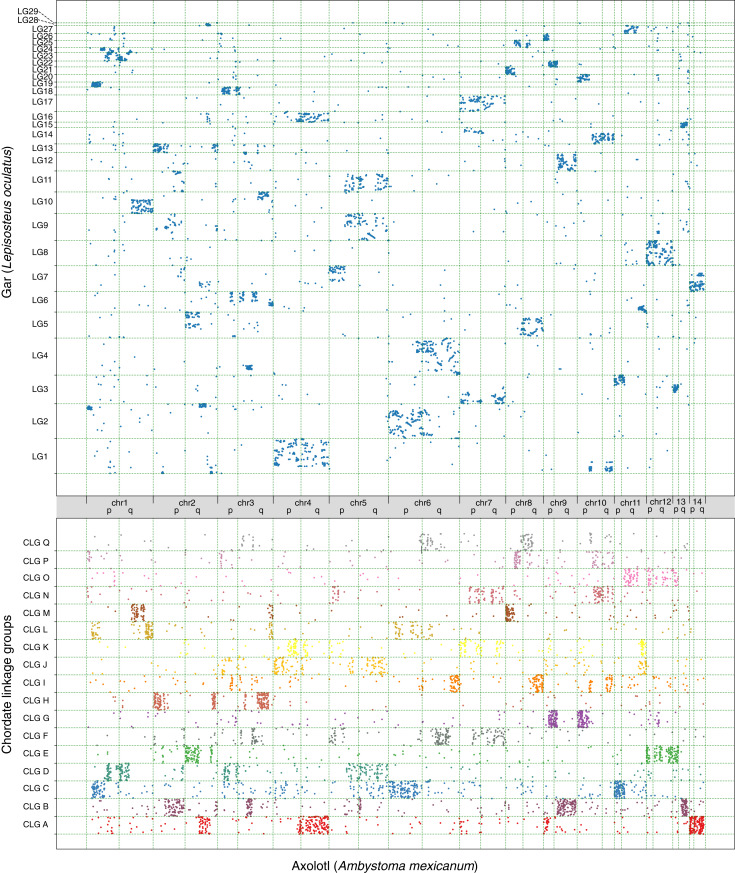
To gain an overview of gene ortholog organization in these axolotl chromosomal scaffolds, we examined syntenic relationships to chromosomal scaffolds from Lepisosteus oculatus (gar) as another representative vertebrate and to the inferred Chordate Linkage Group models (CLGs) (23) (Fig. 2). To highlight patterns of conservation, we generated Oxford plots, in which each dot represents a 1:1 homolog between two species and its respective position within their genomes. These plots reveal that the arms of the same chromosome often correspond to different CLG identities, but syntenic boundaries between the ancestral CLGs are sharp in many cases, which is indicative of recent fusion events. For instance, chr1q is a clear fusion of CLGD, CLGM, and CLGL, while there is only little intermixing between CLGM and CLGL. Similarly, chr2q most likely arose through the initial fusion of CLGA and CLGE, followed by an internal inversion and subsequently fusion with CLGH. In contrast, the entire chr12 is an old fusion of CLGE and CLGO, a feature that is shared among all jawed vertebrates (23). These data together with the Hi-C contact maps and the concurrence of our scaffolding with the meiotic mapping (17) give strong validation to our assembly.
Fig. 2.
Macrosynteny of axolotl assembly in comparison to gar genome and CLGs. (Upper) Oxford plots of 11,677 1:1 axolotl–gar orthologs showing relative arrangement on chromosomes. (Lower) Oxford plots of 4,938 axolotl–CLG orthologs showing relative arrangement on chromosomes. Clear syntenic clustering of orthologs is visible.
Evolution of the Major Histocompatibility Complex.
The chromosomal contiguity of our data allowed us to examine how large, multigene loci are organized in this giant genome. We focused on the Major Histocompatibility Complex (MHC), an attribute of jawed vertebrates, whose function was resolved over 70 y ago as a genomic region that affects tissue graft versus host compatibility (24, 25). In humans, the main locus on chromosome 6 contains over 100 protein-coding genes, many of which encode components of the adaptive immune system, namely, the machinery for antigen presentation in MHC class I and class II. In eutherian mammals, a cluster of class III genes containing complement and Tumor necrosis factor signaling component genes can be found between the classical class I and II clusters (Fig. 3A) (24, 25). Spread over 3 Mb in humans, the locus is highly complex and has been extremely dynamic over evolution. Its orthologous location in other vertebrates varies widely in composition and order of genes even between eutherian, marsupial, and monotreme mammals, while chicken shows a highly minimalized MHC (25). For these reasons, an understanding of the ancestral MHC locus has been elusive. With a well-annotated Xenopus laevis MHC locus available (26), the knowledge of the salamander MHC locus architecture contributes significantly to theories on ancestral states of the MHC.
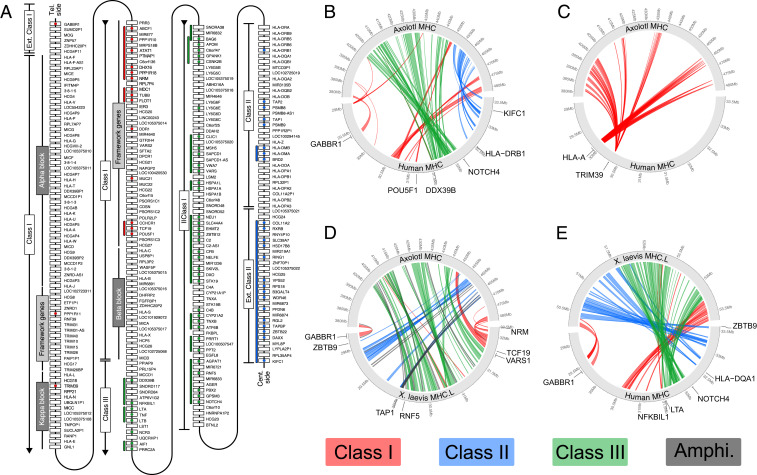
Fig. 3.
Evolution and expansion of AxMHC. (A) Representation of the human MHC, with colored circles indicating when an ortholog was found in the axolotl MHC locus. The colored lines next to the genes represent groups of genes that are neighboring genes in axolotl. Genes coming from MHC class I, II, and III in human are colored in red, blue, and green, respectively. Adapted from ref. 27. (B–E) Pairwise comparison of gene placement between. (B) axolotl and human without the highly amplified gene families HLA and TRIM. (C) Axolotl and human only showing highly amplified gene families HLA and TRIM. (D) Axolotl and X. laevis. (E) X. laevis and human.
To identify the axolotl MHC locus, we aligned the set of human MHC protein sequences against the axolotl genome using the Basic Local Alignment Search Tool (BLAST), which revealed a ∼100 Mb region of axolotl chromosome arm 13q that consisted of 126 MHC locus–related orthologs excluding the highly amplified gene sequences that include Human Leukocyte Antigen (HLA) and Tripartite Motif (TRIM) family members (SI Appendix, Fig. S2 A and B). The axolotl locus starts with the extended Class I gene Gamma-Aminobutyric Acid Type B Receptor Subunit 1 (Axgabbr1) and ends with the class I gene Discoidin Domain Receptor Tyrosine Kinase 1 (Axddr1), which is found centrally in the MHC cluster in humans suggesting substantial differences in gene order. However, examination of local blocks of genes showed conserved syntenic arrangements including those considered framework genes (Fig. 3A and Datasets S1–S3). To gain an overview of MHC gene arrangement, we compared the positions of axolotl, X. laevis, and human orthologs (Fig. 3 B–E). The presence of class I genes at the two ends of the cluster is shared between axolotl and Xenopus, indicating that this split class I arrangement is either ancestral to the tetrapods or evolved within the common ancestral lineage of extant amphibians. Interestingly, the axolotl and Xenopus class II and III gene clusters have exchanged positions with respect to the class I genes, with the axolotl having a relative arrangement of class II and III more similar to that in humans. These data suggest that the presence of the class III region between class I and II is a shared ancient arrangement in the common ancestor of modern amphibians and mammals. The MHC locus hosts several gene families that are known to evolve rapidly, including the HLAs and the Trim family (27–29). The axolotl locus contains a large number of HLA-related sequences that have translocated to other regions of the Xenopus genome, away from the main cluster (Fig. 3D and SI Appendix, Fig. S2C). Phylogenetic analysis of the HLA sequences from several fish, frogs, mouse, and human led us to conclude that all the axolotl HLA sequences clustered in a manner consistent with a salamander lineage–specific amplification (SI Appendix, Fig. S2A). Similarly, we observed an extraordinary amplification of Trim family members. Phylogenetic analysis revealed that most of these sequences fall in a branch assigned to Trim39, which is an E3 ubiquitin ligase associated with regulation of cell death and response to DNA damage (SI Appendix, Fig. S2B). In summary, the arrangement of the axolotl MHC has many shared features with Xenopus and humans, suggesting this could reflect a relatively conservative topology of the MHC cluster. An intriguing question is whether the massive expansion of the Trim39 sequences is functionally related to salamander-associated physiology.
Scaling of the Axfgf8 Regulatory Locus and TADs.
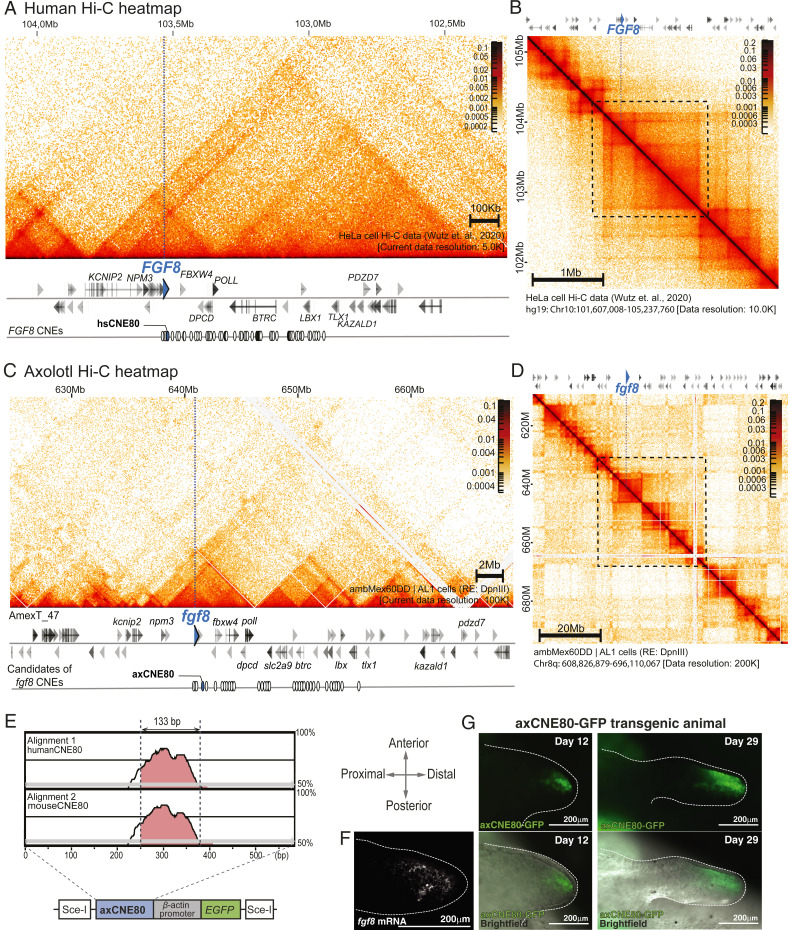
Beyond conserved gene clusters, the Hi-C data enabled us to examine the relationship of TADs with gene organization. We wondered, given the vast genomic distances in axolotl, whether TAD organization scaled with genome/gene distances or whether TAD size is universal between vertebrates and unrelated to genome expansion. To address this question, we first focused our analysis on a region containing a gene under the control of distant regulatory elements. For this purpose, we chose the regulatory landscape of the Fgf8 locus, which has been extensively characterized in mouse and human using functional annotation of Cis-Regulatory Elements (CREs) that are conserved among vertebrates (8). In human, there are 84 CNEs extending 0.57 Mb away from the hsFGF8 transcriptional start site (TSS) among which 27 drove mmFgf8-like tissue–specific expression when tested in a mouse transgenic reporter assay. Of these, five CNEs drove expression in the limb bud (CNE58, CNE59, CNE61, CNE66, and CNE80). This redundant dispersal of enhancers over large distances has been termed a holo-enhancer (8). The known hsFGF8 regulatory landscape is interspersed among eight genes extending to the hsTLX1 gene, with CNEs present in intergenic as well as intronic sequences (Fig. 4A).
Fig. 4.
Expanded regulatory architecture of Axfgf8. (A and B) Human FGF8 regulatory locus. FGF8 is marked with a blue arrowhead and a dotted line. (A) High magnification at 5 Kb resolution. White circles display CNEs surrounding the hsFGF8, 27 of which show FGF8 enhancer function. (B) Human Hi-C map at 10 Kb resolution (chr10: 101 Mb to 105 Mb). (C and D) Axolotl Fgf8 regulatory unit with Hi-C contact map. Axfgf8 is marked with a blue arrowhead and a line dotted. (C) High magnification at 100 Kb resolution. The linear sequence of genes is given below the heatmap. White circles display 43 CNEs surrounding Axfgf8. (D) Hi-C map at 200 Kb resolution (chr8q: 608 Mb to 696 Mb). (E) Characterization of the axolotl CNE80. (Top) mVISTA (40) plot showing the aligment of a 589 bp AxCNE80 candidate region against the human and mouse CNE80 sequences (mVISTA SLAGAN alignment program: Criteria 70%, 100 bp). The track is shown with a 100 bp window. The candidate region contains a highly conserved 133 bp sequence. (Bottom) Schema of transgenesis construct used to test AxCNE80 function. (F) Expression of Axfgf8 mRNA in stage 44 (St. 44) (41) axolotl limb bud revealed using fluorescent in situ hybridization chain reaction (HCR). Axfgf8 mRNA is expressed in the distal–anterior mesenchyme. The limb bud is outlined by dotted lines. (G) Axolotl CNE80 drives expression in the Axfgf8 expression domain. GFP expression is in the limb bud of an axCNE80 > EGFP transgenic animal at day 12 and 29 after injection (St. 44 to 47). (Top) GFP images. (Bottom) Overlay with widefield. Strong GFP expression is detected in the distal–anterior mesenchyme, similar to Axfgf8 mRNA.
The Axfgf8 region harbors a syntenic arrangement of these eight orthologs. In humans, the neighboring gene, hsFBXW4, sits at a distance of 81 Kb from hsFGF8, while in the axolotl, Axfbxw4 sits at a distance of 1.74 Mb from Axfgf8, a 21-fold expansion (SI Appendix, Fig. S3). The most distant gene in the regulatory landscape, TLX1 sits 0.65 and 15.3 Mb away in human and axolotl, respectively, showing that the >20-fold expansion occurs throughout the region. Through homology searches, axolotl CNEs corresponding to 43 out of 84 previously defined hsFGF8 CNEs (8) were identifiable and were found at a distance up to 14 Mb away from the Axfgf8 TSS, an expansion in distance of 25-fold (Dataset S5). When examining these expanded intergenic distances, we noted large arrays of repetitive sequences present in axolotl that are not present in the human locus (SI Appendix, Fig. S4), similar to the highly repeat-rich landscape previously described for the axolotl HoxA locus (2).
When we overlapped this gene sequence information with the Hi-C contact matrices, we observed highly similar patterns of TADs and sub-TADs between human and axolotl in relation to the genes and CNEs, albeit with the axolotl TADs being at least 20 times expanded in linear distance along the genome (Fig. 4 A–D). Whereas in humans, the TAD encompassing hsFGF8 (extending from the hsNPM3 gene to hsPOLL) is 210 Kb large, the equivalent TAD in the axolotl spanned 10.3 Mb, a 49-fold expansion. Notably, the TADs are visible in the human dataset at 5 Kb resolution (30), whereas in the axolotl, the TADs were visible at 100 Kb resolution. Examination of the human dataset at 100 Kb did not reveal any multimegabase TAD structures. These results showed that the organizational dynamics related to long-distance contacts in regulatory landscapes scale 20- to 50-fold within the Axfgf8 locus and are essentially preserved in the axolotl.
To determine whether the CNEs are functional CREs, we tested one of the CNEs, CNE80, that drove expression in the limb bud in mouse reporter assays (Fig. 4E) (8). Transgenic axolotls expressing the green fluorescent protein (GFP) under the control of the axolotl CNE80 (AxCNE80) sequence juxtaposed to a minimal actin promoter showed expression corresponding to the expected Axfgf8 expression domain in the anterior mesenchyme as revealed by in situ hybridization (14) (Fig. 4 F and G). GFP expression in the transgenic animal maintained a faithful restriction to the anterior mesenchyme at multiple timepoints in development. It should be noted that hsCNE80 faithfully drives gene expression in the apical ectodermal ridge of the mouse limb bud, and AxCNE80 faithfully drives the uniquely divergent expression in the anterior mesenchyme in axolotl (14). We infer from these data that the axolotl CNEs are indeed functioning across the larger distances of the axolotl genome. This data also suggests that the genomic changes that result in the divergent mesenchymal expression of axolotl Fgf8 lie in transregulators of the locus. This conclusion would be consistent with the observation that several other genes expressed in the mouse apical ectoderm are also expressed in axolotl limb mesenchyme (14, 31).
Interphase and Mitotic Chromosome Structure in the Giant Genome.
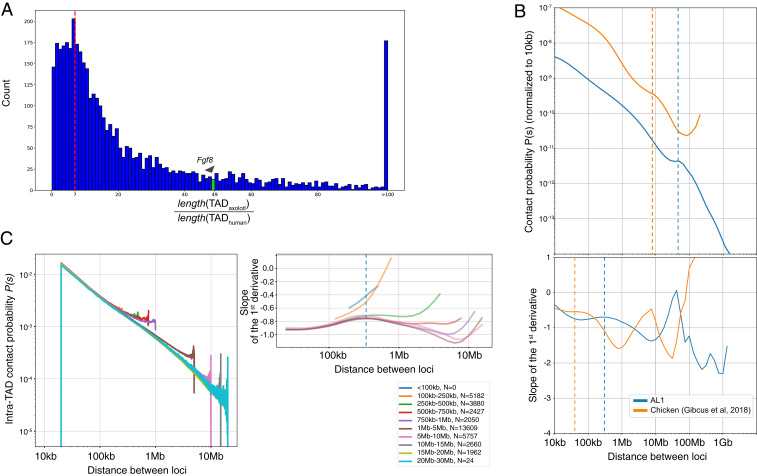
To gain a genome-wide comparison of axolotl interphase TAD organization compared to human, we used automated TAD detection in both datasets at multiple resolutions. We identified 4,086 TADs, which were homologous with respect to the 9,408 genes they contained, and calculated a TAD-by-TAD axolotl/human TAD length ratio. This procedure yielded an axolotl/human TAD length ratio distribution with a peak between sevenfold and the tail of the distribution ranging up to more than 100-fold (Fig. 5A and SI Appendix, Fig. S5).
Fig. 5.
Expanded features of axolotl interphase chromosomes. (A) TAD length comparison. Ratio of orthologous TADs between axolotl and human. TADs surrounding gene orthologs were identified genome-wide. On average, axolotl TADs are 7× longer than their human counterparts. The TAD that encompasses Axfgf8 (green bar and arrowhead) is 49× longer than its orthologous interval in human. (B) Contact probability plots for axolotl (blue) and chicken (orange). Genome-wide contact probability plots are presented versus genomic distance between loci normalized to the contact probability at 10 Kb. In the interphase data, the inflection point (dashed lines) in chicken is at 40 Kb, while in axolotl, it is at 300 Kb, suggesting that large loops accumulate in axolotl interphase chromosomes. (C) Axolotl intra-TAD contact probability as a function of distance in conserved TADs, classified into different size categories. Graphs show the same slope indicating same contact frequencies within TADs of different sizes. Upturn at end of each line reflects increased contact frequency found near TAD boundaries.
Another analytical means to gain insight into chromosome structure from Hi-C data has been through contract probability maps (32). To assess the structural features of axolotl chromosomes, we analyzed the average Hi-C contact frequency as a function of genomic separation (P[s]) (Fig. 5B). This Hi-C metric has been previously used to infer the loop sizes and helical condensation state of human and chicken chromosomes (32, 33). We therefore sought to determine how chromatin features scale by comparing the axolotl Hi-C interphase and mitotic datasets to the recently published haploid chicken cell datasets (33). In the interphase data, the inflection point revealed that in chicken, expected loop sizes are 40 Kb, while in axolotl, the predicted loop size is 300 Kb, suggesting that large loops accumulate in axolotl chromosomes (Fig. 5B). Given the read depth of our data, we are unable to conclude whether loops at the 40 Kb range also exist in the axolotl. Given the giant TADs present in the axolotl that scale on average seven to 10 times compared to human (depending on whether we compare homologous or all TADs, respectively), we asked whether the contact frequency in large TADs scale with TAD size (Fig. 5C). Categorization of TADs into size classes and showed that the contact probability with distance relationship did not change based on TAD size. Therefore, in large TADs, contact probabilities are not changed to facilitate contacts across large distances.
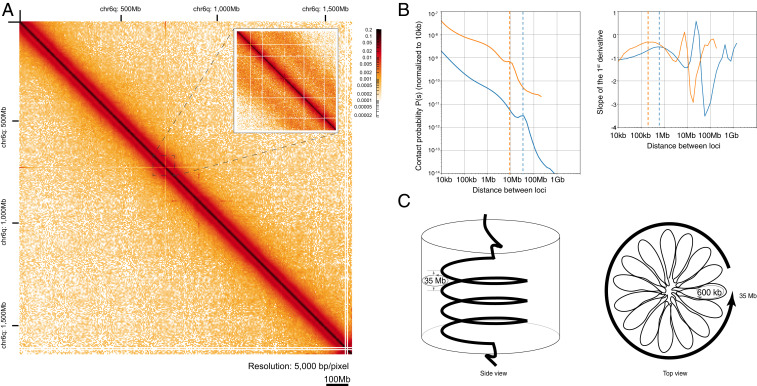
Throughout history, salamander cells have been used to study the mechanics of mitosis due to their unusually large spindles and mitotic chromosomes (34). During mitotic chromosome condensation, TADs are dissolved, and chromosomes become uniformly condensed into arrays of randomly positioned loops (32). Our mitotic Hi-C dataset provided the opportunity to predict the loop structure of axolotl mitotic chromosomes (Fig. 6A). Overall, the shape of the axolotl mitotic P(s) graph looked similar to those observed in chicken (Fig. 6B) with overall downward slope, showing 1) a shallow inflection point, 2) a nonmonotonic increase at longer genomic separations, and 3) a sharp decline at the end. However, the features in axolotl were spread over longer distances which would be consistent with larger-scale features. Taking as a reference a spiral staircase model of mitotic chromosomes (33), we predicted the main loop and helical turn sizes in axolotl mitotic chromosomes. The model predicts that condensed chicken chromosomes are organized into nested loops, with small-scale loop formation (80 Kb scale) which are nested into larger-scale loops that form a spiral winding around a central core at 12 Mb per turn in late prometaphase.
Fig. 6.
Expanded features of axolotl mitotic chromosomes. (A) Mitotic chromosome heatmap. Hi-C map of chromosome arm 6q during mitosis. The inset demonstrates the presence of the secondary diagonals, which is characteristic for mitotic Hi-C heatmaps (scale bar 100 Mb). (B) Contact probability plots. Genome-wide contact probability plots are presented versus genomic distance between loci normalized to the contact probability at 10 Kb. In mitotic chromosomes (Bottom), the peak at 35 Mb (dashed line) corresponds to one turn of the helix. The peak in the first derivative (right) at 600 Kb corresponds to the length of a single loop within that turn. (C) Proposed model of helical organization of mitotic chromosomes. During mitosis, the DNA is packaged in ∼600 Kb loops, which themselves are arranged into helical turns of the polymer. A single turn comprises ∼35 Mb DNA, which agrees well with the values observed in the contact probability plot.
From the P(s) graphs, it is possible to predict the small-scale loop and helical turn frequencies. In the axolotl data, the P(s) showed a characteristic flattening at 400 to 600 Kb, meaning that the small-scale loops in axolotl are expected to be five to 7.5 times larger than those in chicken. The other analyzable feature of the contact probability map is the peak just before the steep drop-off, which has been modeled as the length scale at which one turn of the helical winding results in peak of long-distance contacts. In chicken, this is found at 12 Mb, whereas in axolotl, a 35 Mb helical turn is predicted, which is threefold longer length scale than chicken (Fig. 6C). These data indicate that axolotl mitotic chromosomes are more condensed per base pair along the chromosomal axis than in other vertebrate genomes.
Discussion
Our work here highlights the significance of an accurate chromosome-level assembly of the axolotl genome for the understanding of the evolution of chromosome organization including structural features, gene complexes, and long-distance regulatory regions. The overall organization of gene order by comparison to gar and the CLGs revealed, despite the enormous expansion of the genome, the retention of substantial blocks of syntenic organization in this expanded genome, confirming the conclusions of Smith et al. based on meiotic linkage map data (17). Indeed, our scrutiny of the multigene Fgf8 regulatory locus showed a precise maintenance of gene order compared to the human gene locus but interestingly with an inversion of the whole locus with respect to the chromosome. These data suggest that functional long-range regulatory regions are preserved among diverse tetrapods species to keep core features of the body plan. Remarkably, the distances between genes and regulatory elements were highly expanded in axolotl, with repetitive elements riddling these expanded domains. These observations reveal that it is possible to retain functional gene regulatory relationships between CREs and genes in an expanded landscape. Correspondingly, TADs and other features such as loops show seven- to 10-fold larger features in the axolotl compared to other vertebrates. This suggests that large numbers of transposable elements have inserted into the genome, including presumably large numbers of ancient insertions in the stem salamander lineage, without negatively impacting the effectiveness of long-range gene regulation. Anecdotally, this gene regulation can apparently occur over >3 Mb distances typically thought of as potential distance limit in mammals, since the conserved and functional axolotl Shh ZRS element lies 7 Mb from the Shh TSS. How this occurs is still unknown.
Interestingly, with the expanded genome, the axolotl cells have a greatly protracted cell cycle speed of 72 to 96 h compared to a 24 h cell cycle of a typical vertebrate cell (35). This correspondence between genome size and cell cycle length may allow the decelerated cellular lifestyle in axolotl to provide enough time for these large loops to form. Alternatively, the axolotl genome, even in interphase, may form loops within loops to facilitate long-range interactions, or repetitive sequences may have special packaging properties. Unfortunately, due to the large size of the genome and the read depths achievable in these Hi-C data, it was not possible to determine whether smaller-scale loop structures are embedded within the large loops. An interesting future direction will be to determine whether the signals for large loop formation are intrinsically encoded in the axolotl genomic sequences, perhaps linked to repetitive elements or whether they are due to divergent implementation of the protein complexes that execute loop formation, for example, the processivity of cohesion complexes based on the levels of components promoting turnover such as WAPL (36).
Materials and Methods
Detailed information is provided in SI Appendix, Materials and Methods.
Animals and Cultured Cell Lines.
All animal experiments were performed in accordance with an approved license under the Austrian laws.
In Situ Hi-C Library Preparation and Hi-C Sequencing Data Preprocessing.
For genome scaffolding, 16 Hi-C datasets were prepared from interphase and mitotic-arrested AL1 cells and d/d embryos (3). Hi‐C libraries were generated from 1 × 106 cells that were fixed with 1% formaldehyde and in situ chromatin digested using HindIII or DpnII. Sequencing and read mapping details are described in SI Appendix.
Genome Scaffolding.
The high level of repetitiveness of large genomes poses a challenge for Hi-C based scaffolding. Contact depletion in repetitive regions coupled with sequence biases disturbing the assumed uniform spread of digest sites throughout the genome results in biases that lead to suboptimal scaffolding. We developed an agglomerative hierarchical clustering-based scaffolding approach utilizing various normalization techniques to overcome these hurdles while also achieving high performance. Clusters were initially populated using the contig to linkage group assignment from Smith et al. (17) Normalization and details on the workings of the scaffolder are described in SI Appendix, Materials and Methods.
Genome Error Correction.
To correct the residual errors in the contigs, we generated five datasets of Illumina reads using the DNA from the same individual as for the genome assembly was used (2).
To improve the error correction in coding regions, RNA-seq reads were mapped to the corrected reference. Since the reads originated from multiple individuals, only indels and small gaps were fixed, but not polymorphisms.
Annotation.
RNA-seq data were aligned to the genome to generate gene models. Iso-Seq reads were separately aligned, and the gene models from both sets were combined using StringTie–merge.
The final gene models were annotated using a custom annotation pipeline (SI Appendix). Additionally, open reading frames were predicted by extending the homologous alignment to the left until the left-most start codon (if possible) and to the right until the first in-frame terminal codon (if present).
Synteny Analysis.
The set of gar proteins and axolotl proteins were used to run a two-way blastp with default settings. In total, 11,677 mutually best hits in both runs were plotted on the Oxford plots. The same principle was used to visualize the synteny between the axolotl and the CLGs (23).
TAD Size Comparisons.
TADs were predicted in human and axolotl using Homer as described in SI Appendix, Materials and Methods.
Homologous TADs were identified by taking the smallest TAD for each human gene and identifying all genes that belonged to the same TAD. Homologous axolotl genes based on the gene symbol and the smallest axolotl TAD that contained all or most of those genes were then identified.
Axolotl MHC Cluster Annotation and Synteny Analysis.
The axolotl MHC was identified by using BLAST, the human MHC homologous genes (24), and the Xenopus tropicalis MHC homologs. To classify the protein families, the datasets of eight other vertebrates (zebrafish, gar, chicken, Xenopus, Nanorana parkeri, Lithobates catesbeianus mouse, and human).
AxFgf8 CNEs and Transcript Analysis.
mVISTA was used to identify the axolotl Fgf8 CNEs (8). AxCNE80-GFP transgenic axolotls were generated by I-SceI meganuclease-mediated transgenesis (37).
Whole-mount HCR fluorescent in situ hybridization with AxFgf8 probes was performed for axolotl limb buds according to previously published methods (38).
Supplementary Material
Acknowledgments
We thank the Institute of Molecular Pathology (IMP) axolotl caretakers for their dedicated work. We are grateful to Alex Schleiffer, Roman Stocsits, Oleg Simakov, Jim Kaufman, and Anton Goloborodko for sharing the data, help with the analyses and advice, and Gordana Wutz, Yuka Taniguchi-Sugiura, Alex Vogt, and the VBCF NGS as well as the IMP IT department for advice and assistance. We thank Meinrad Busslinger for reading part of the manuscript. E.M.T. was supported by core funding from the IMP and ERC AdG 742046, RegGeneMems. A.K. was supported by a JSPS Postdoctoral Fellowship for Overseas Researchers. L.O. was supported by a fellowship from the HFSP. N.T., M.C.K., J.J.S., and S.R.V. were supported by NIH R24OD010435 and P40OD019794, and ARO W911NF1810168.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2017176118/-/DCSupplemental.
Data Availability
The genome can be accessed through a genome browser: https://genome.axolotl-omics.org/ (39). The assembly, annotation, and the track hubs can be downloaded from https://www.axolotl-omics.org/assemblies (the user must expand the AmexG_v6.0-DD block) (39). This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession PGSH00000000. The version described in this paper is version PGSH00000000.2. Sequencing data have been deposited in NCBI BioProject database (PRJNA520877, PRJNA644663, and PRJNA645452). Transcriptome and genome assemblies are available at https://www.axolotl-omics.org/assemblies (39).
References
- 1.Gregory T. R., Animal Genome Size Database, http://www.genomesize.com (2021). Accessed 8 December 2020.
- 2.Nowoshilow S., et al., The axolotl genome and the evolution of key tissue formation regulators. Nature 554, 50–55 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Lieberman-Aiden E., et al., Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burton J. N., et al., Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat. Biotechnol. 31, 1119–1125 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaplan N., Dekker J., High-throughput genome scaffolding from in vivo DNA interaction frequency. Nat. Biotechnol. 31, 1143–1147 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Symmons O., et al., Functional and topological characteristics of mammalian regulatory domains. Genome Res. 24, 390–400 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harmston N., et al., Topologically associating domains are ancient features that coincide with Metazoan clusters of extreme noncoding conservation. Nat. Commun. 8, 441 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marinić M., Aktas T., Ruf S., Spitz F., An integrated holo-enhancer unit defines tissue and gene specificity of the Fgf8 regulatory landscape. Dev. Cell 24, 530–542 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Andrey G., et al., A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science 340, 1234167 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Lupiáñez D. G., et al., Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161, 1012–1025 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stopper G. F., Wagner G. P., Inhibition of sonic hedgehog signaling leads to posterior digit loss in Ambystoma mexicanum: Parallels to natural digit reduction in urodeles. Dev. Dyn. 236, 321–331 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Boisvert C. A., Joss J. M., Ahlberg P. E., Comparative pelvic development of the axolotl (Ambystoma mexicanum) and the Australian lungfish (Neoceratodus forsteri): Conservation and innovation across the fish-tetrapod transition. Evodevo 4, 3 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fröbisch N. B., Shubin N. H., Salamander limb development: Integrating genes, morphology, and fossils. Dev. Dyn. 240, 1087–1099 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Nacu E., Gromberg E., Oliveira C. R., Drechsel D., Tanaka E. M., FGF8 and SHH substitute for anterior-posterior tissue interactions to induce limb regeneration. Nature 533, 407–410 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Purushothaman S., Elewa A., Seifert A. W., Fgf-signaling is compartmentalized within the mesenchyme and controls proliferation during salamander limb development. eLife 8, e48507 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christensen R. N., Weinstein M., Tassava R. A., Expression of fibroblast growth factors 4, 8, and 10 in limbs, flanks, and blastemas of Ambystoma. Dev. Dyn. 223, 193–203 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Smith J. J., et al., A chromosome-scale assembly of the axolotl genome. Genome Res. 29, 317–324 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woodcock M. R., et al., Identification of mutant genes and introgressed tiger salamander DNA in the laboratory axolotl, Ambystoma mexicanum. Sci. Rep. 7, 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Currie J. D., et al., Live imaging of axolotl digit regeneration reveals spatiotemporal choreography of diverse connective tissue progenitor pools. Dev. Cell 39, 411–423 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.An international system for human cytogenetic nomenclature (1978), ISCN (1978). Report of the Standing Committee on Human Cytogenetic Nomenclature. Birth Defects Orig Article Ser. 14, 313–404 (1978). [PubMed]
- 21.Bryant D. M., et al., A tissue-mapped axolotl de novo transcriptome enables identification of limb regeneration factors. Cell Rep. 18, 762–776 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seppey M., Manni M., Zdobnov E. M., BUSCO: Assessing genome assembly and annotation completeness. Methods Mol. Biol. 1962, 227–245 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Simakov O., et al., Deeply conserved synteny resolves early events in vertebrate evolution. Nat. Ecol. Evol. 4, 820–830 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shiina T., Blancher A., Inoko H., Kulski J. K., Comparative genomics of the human, macaque and mouse major histocompatibility complex. Immunology 150, 127–138 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaufman J., Unfinished business: Evolution of the MHC and the adaptive immune system of jawed vertebrates. Annu. Rev. Immunol. 36, 383–409 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Session A. M., et al., Genome evolution in the allotetraploid frog Xenopus laevis. Nature 538, 336–343 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shiina T., et al., Rapid evolution of major histocompatibility complex class I genes in primates generates new disease alleles in humans via hitchhiking diversity. Genetics 173, 1555–1570 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lobkovsky A. E., et al., Multiplicative fitness, rapid haplotype discovery, and fitness decay explain evolution of human MHC. Proc. Natl. Acad. Sci. U.S.A. 116, 14098–14104 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abduriyim S., Zou D. H., Zhao H., Origin and evolution of the major histocompatibility complex class I region in eutherian mammals. Ecol. Evol. 9, 7861–7874 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wutz G., et al., ESCO1 and CTCF enable formation of long chromatin loops by protecting cohesinSTAG1 from WAPL. eLife 9, e52091 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimokawa T., Yasutaka S., Kominami R., Shinohara H., Lmx-1b and Wnt-7a expression in axolotl limb during development and regeneration. Okajimas Folia Anat. Jpn. 89, 119–124 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Naumova N., et al., Organization of the mitotic chromosome. Science 342, 948–953 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gibcus J. H., et al., A pathway for mitotic chromosome formation. Science 359, eaao6135 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Callan H. G., Chromosomes and nucleoli of the axolotl, Ambystoma mexicanum. J. Cell Sci. 1, 85–108 (1966). [DOI] [PubMed] [Google Scholar]
- 35.Maden M., Blastemal kinetics and pattern formation during amphibian limb regeneration. J. Embryol. Exp. Morphol. 36, 561–574 (1976). [PubMed] [Google Scholar]
- 36.Hill L., et al., Wapl repression by Pax5 promotes V gene recombination by Igh loop extrusion. Nature 584, 142–147 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sobkow L., Epperlein H. H., Herklotz S., Straube W. L., Tanaka E. M., A germline GFP transgenic axolotl and its use to track cell fate: Dual origin of the fin mesenchyme during development and the fate of blood cells during regeneration. Dev. Biol. 290, 386–397 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Choi H. M. T., et al., Third-generation in situ hybridization chain reaction: Multiplexed, quantitative, sensitive, versatile, robust. Development 145, dev165753 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nowoshilow S., Tanaka E. M., Introducing–An integrated -omics data portal for the axolotl research community. Exp. Cell Res. 394, 112143 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Frazer K. A., Pachter L., Poliakov A., Rubin E. M., Dubchak I., VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 32, W273–W279 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bordzilovskaya N. P., Dettlaff T. A., Table of stages of the normal development of axolotl embryos. Axolotl Newsletter 7, 2–22 (1979). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome can be accessed through a genome browser: https://genome.axolotl-omics.org/ (39). The assembly, annotation, and the track hubs can be downloaded from https://www.axolotl-omics.org/assemblies (the user must expand the AmexG_v6.0-DD block) (39). This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession PGSH00000000. The version described in this paper is version PGSH00000000.2. Sequencing data have been deposited in NCBI BioProject database (PRJNA520877, PRJNA644663, and PRJNA645452). Transcriptome and genome assemblies are available at https://www.axolotl-omics.org/assemblies (39).