Abstract
Background
circular RNAs (circRNAs) are expressed abundantly in the brain and are implicated in the pathophysiology of neuropsychiatric disease. However, the potential clinical value of circRNAs in major depressive disorder (MDD) remains unclear.
Methods
RNA sequencing was conducted in whole-blood samples in a discovery set (7 highly homogeneous MDD patients and 7 matched healthy controls [HCs]). The differential expression of circRNAs was verified in an independent validation set. The interventional study was conducted to assess the potential effect of the antidepressive treatment on the circRNA expression.
Findings
in the validation set, compared with 52 HCs, significantly decreased circFKBP8 levels (Diff: -0.24; [95% CI -0.39 ~ -0.09]) and significantly elevated circMBNL1 levels (Diff: 0.37; [95% CI 0.09 ~ 0.64]) were observed in 53 MDD patients. The expression of circMBNL1 was negatively correlated with 24-item Hamilton Depression Scale (HAMD-24) scores in 53 MDD patients. A mediation model indicated that circMBNL1 affected HAMD-24 scores through a mediator, serum brain-derived neurotrophic factor. In 53 MDD patients, the amplitude of low-frequency fluctuations in the right orbital part middle frontal gyrus was positively correlated with circFKBP8 and circMBNL1 expression. Furthermore, the interventional study of 53 MDD patients demonstrated that antidepressive treatment partly increased circFKBP8 expression and the change in expression of circFKBP8 was predictive of further reduced HAMD-24 scores.
Interpretation
whole-blood circFKBP8 and circMBNL1 may be potential biomarkers for the diagnosis of MDD, respectively, and circFKBP8 may show great potential for the antidepressive treatment.
Keywords: Major depressive disorder, Biomarker, Circular RNAs, Antidepressive treatment, Brain-derived neurotrophic factor, Amplitude of low-frequency fluctuation, Neuropsychological assessment, Competing endogenous RNA
Research in context.
Evidence before this study
We searched PubMed with the terms “circRNA”, “circular RNA”, “depressive disorder”, “depression”, “blood”, “plasma”, and “serum” in English before May 31, 2020. Although previous studies revealed that circRNAs are expressed abundantly in the brain and are highly active at neuronal synapses, less study investigated the potential value of circRNAs in depressed patients, and there is no consistent findings from different laboratories. Therefore, it is unclear whether blood circRNAs can be used as the peripheral biomarker for the clinical diagnosis of depression and predicting the antidepressive treatment outcome. Considering the interplay between genetic and environmental factors may be the main cause of depression, a group of highly homogeneous depressed patients who have a history of negative life evens and suicide attempt, was used for RNA sequencing to identify potential circRNA indicators. Subsequently, the strict two-stage validation was performed to confirm the expression of circRNAs and assess the value of circRNAs as the clinical biomarkers of depression.
Added value of this study
We identified two blood circRNAs for the diagnosis of depression through the strict process of discovery and two-step validation, and these circRNAs were associated with multifaceted characteristics of depression including the severity of depressive symptom, serum brain-derived neurotrophic factor levels, and the imaging evidence of brain activity. Meanwhile, we also demonstrated that a blood circRNA had the great clinical value for the antidepressive treatment.
Implications of all the available evidence
These two blood circRNAs involved in the neuroinflammation and neuroplasticity and emotion-related brain activity alterations, may be used as promising pre-screening markers for identifying individuals with potential risk of depression. In particularly, one of them may be used as a valuable efficacy marker for guiding the individualized treatment. The present evidence further supported the potential clinical value of circRNAs in depression.
Alt-text: Unlabelled box
1. Introduction
Major depressive disorder (MDD) is the most prevalent psychiatric disease with high rates of recurrence [1]. Currently, since a diagnosis of MDD is primarily dependent on clinical manifestations, it is difficult to diagnose MDD accurately [2]. In pharmacological treatments for MDD, only about 30% of patients achieve remission with selective serotonin reuptake inhibitors (SSRIs) [3], however, the inhibitor of both norepinephrine and serotonin transporters (SNRIs), such as duloxetine, can improve efficacy in acute, adult MDD at doses of 80–120 mg/day [4]. Meanwhile, Agomelatine acting as MT1/MT2 agonist and 5-HT2C antagonist, can enhance neuroplasticity mechanisms and adult neurogenesis in brain regions (e.g. hippocampus and prefrontal cortex) and may be also a valuable option for the antidepressant treatment [5]. In addition to pharmacotherapy, psychotherapy and physical therapy [e.g. transcranial magnetic stimulation (TMS)] are considered promising treatment strategies for MDD [6]. In order to improve the accuracy of diagnosis and the efficacy of antidepressive treatment, it is important to ascertain appropriate biomarkers for determining the etiology of MDD, and development of individualized clinical treatments for MDD patients.
Circular RNAs (circRNAs), generated by a back-splicing event of specific exons of protein-coding genes, are evolutionarily conserved endogenous non-coding RNA (ncRNA) molecules [7], [8], [9]. By acting as a sponge for microRNAs (miRNAs), regulation of protein function or assisting in protein translation, circRNAs exert multiple important biological functions [10,11]. In particular, several circRNAs are expressed abundantly in the brain and are highly active at neuronal synapses involving nervous system development and differentiation, indicating that circRNAs may be crucial for understanding neuropsychiatric disorders [12,13]. Previous studies have showed that in some psychiatric disorders with the complex pathophysiology (e.g. MDD [14], [15], [16], [17], [18], schizophrenia [19], [20], [21], [22], bipolar disorder [21,23]), effective circRNA indicators may play significant roles in the diagnosis of disease and even be used for promising therapeutic targets. By reviewing the previous studies, we only found five published studies of circRNAs in depressed patients thus far, including one RNA sequencing study [14], two microarrays studies [15,16] and two real-time quantitative polymerase chain reaction (RT-qPCR) studies [17,18]. However, there is no consistency in circRNA results from different laboratories, with poor homogeneity of participants that lack multilevel assessment being the primary limitations that prevent circRNAs being considered reliable biomarkers for MDD. In addition, although we demonstrated in previous studies of circRNAs that circDYM-regulated microglial activation and the gut microbiota-circHIPK2-astrocyte axis may be involved in the etiology of depression [17,24], the precise mechanism of circRNA molecules in MDD remains unknown.
Brain-derived neurotrophic factor (BDNF) plays important roles in the modulation of neurogenesis and neuroplasticity in the brain [25]. Multiple meta-analyses have revealed that serum BDNF levels reduced in MDD patients and increased after antidepressant treatment [26,27], demonstrating a negative correlation between serum BDNF levels and severity of depressive symptoms [28]. In addition, previous study showed that circRNAs can enhance the neuronal plasticity and promote functional recovery in ischemic stroke [29]. Hence, the exploration of the relationship of BDNF (a kind of neuroplasticity-related indicator and a promising diagnostic biomarker of MDD) with circRNA indicators may contribute to revealing the possible function of circRNAs on the modulation of neuroplasticity in MDD and provide the valuable evidence to support the application of circRNA indicators in MDD.
In addition, compelling evidence has confirmed that MDD is associated with disconnection of brain function and has been characterized by abnormal brain activity [30]. Previous neuroimaging studies of MDD showed that brain regions with abnormal function activity were located mainly in emotion-related regions, such as insula, orbitofrontal cortex (OFC), and anterior cingulated cortex (ACC) [31,32], and the interaction of these brain regions can form the emotion-related brain circuit, such as the fronto-limbic system, whose functional alterations are the important characteristic of MDD [32,33]. Meanwhile, the potential mechanisms of action of antidepressant treatment response may be to change the functional connectivity between frontal and limbic brain regions [34], and the function activity in some emotion-related regions, such as ACC, may be related with treatment response to antidepressants [35]. Amplitude of low-frequency fluctuation (ALFF) is an efficient functional magnetic resonance imaging (fMRI) index of local spontaneous neuronal activity [36] and can be used to conveniently assess the abnormal activity of brain regions located in neural circuits that process emotions [37]. Furthermore, our previous study have indicated that microRNA-9 may play a crucial role in the process of brain function changes targeted prefrontal-limbic regions and induce subsequently depression [38]. Therefore, the assessment of the relationship between circRNA indicators and ALFF indicators can investigate whether circRNAs have an effect on the abnormal brain activity for leading to MDD, which contributes to understanding the underlying pathophysiology of MDD and providing neuroimaging evidence for the clinical application of circRNA indicator.
The present study aimed to identify and validate differentially expressed circRNAs in MDD patients and to evaluate their potential as disease diagnostic biomarkers and novel therapeutic targets of MDD. Meanwhile, potential clinical value and possible mechanism of circRNAs in MDD were also explored based on the neuroplasticity-related serum indicator and the brain activity-related neuroimaging indicator. Due to the interplay between genetic and environmental factors may be the main cause of MDD [39,40], we hypothesized that indicators of the epigenetics, i.e. circRNAs, can reflect the severe of depressive symptom and guide the antidepressive treatment for MDD patients.
2. Materials and methods
2.1. Study design
The present study consisted of 4 primary components. (1) Component 1 was a case-control study (MDD patient was case group and healthy subject was control group). We identified differentially expressed candidate circRNAs in a discovery set (7 MDD patients and 7 healthy controls [HCs]) using RNA high-throughput sequencing, and the expression of which was further verified using RT-qPCR in the same cases and controls from the discovery set. (2) Component 2 was also a case-control study (MDD patient was case group and healthy subject was control group). We confirmed the differential expression of candidate circRNAs in an independent validation set (53 MDD patients and 52 HCs) and primarily evaluated the potential clinical value of circRNAs in MDD through correlation and mediation analyses between these circRNA indicators and assessments of depressive symptomatology, serum BDNF indicator, and neuroimaging indicator (ALFF). (3) Component 3 was an interventional study (just for MDD patients). To explore whether the treatment can affect the expression of candidate circRNA in 53 MDD patients, we investigated the difference of circRNAs expression between actual (37 patients) and sham treatment groups (16 patients) and between before and after treatment in each treatment group to explore whether the treatment can affect the expression of candidate circRNA. Notably, the assessment of modified rTMS efficacy was not the major goal of this study, and the interventional study was not designed as a rigorous randomized control trial, although both actual and sham rTMS treatments were included. (4) Component 4 was an extension of the interventional study (Component 3), we further created a linear mixed model in 53 MDD patients to evaluate whether the expression of circRNAs can predict the severity of depressive symptomatology.
2.2. Study exposure and outcome
For study components 1 and 2, the exposure variable was the expression of candidate circRNAs, and the study outcome variable was set as binary, i.e., whether a study subject was MDD case or not.
For study component 3, the exposure variable was the targeted treatment and the study outcome variable was the expression of candidate circRNAs.
For study component 4, the exposure variable was the expression of candidate circRNAs and the study outcome variable was the severity of depressive symptomatology that can be assessed by relevant clinical scale.
2.3. Subjects
All MDD patients satisfied the following inclusion criteria: (1) age 18 to 55 years; (2) presence of a definite depressive episode in line with the Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition (DSM-IV), determined by a professional psychiatrist through the standardized structured clinical interview for DSM-IV Axis I disorders; (3) drug-naive or drug-free (i.e. free of antidepressant drug treatment longer than 2 weeks prior to the beginning of the formal study). Notably, drug-free patients were not meet criteria of treatment-resistant depression due to they had been on antidepressants for a shorter period of time and only taken one drug (e.g. selective serotonin reuptake inhibitors); (4) the 24-item Hamilton Depression Scale (HAMD-24) score of ≥ 18 was required for enrollment; (5) definite history of negative life events; (6) for women, being non-pregnant and being non-nursing; (7) with or without family history of a mental disorder. In addition, for MDD patients in the discovery set, additional inclusion criteria were added based on the above inclusion criteria: (1) more serious depressive state (i.e. HAMD-24 score ≥ 35) and definite record of attempted suicide; (2) chronic influence of negative life events; (3) for women, not in lactation or climacteric. Furthermore, for MDD patients, the following exclusion criteria were applied: (1) secondary mental disorders, such as those caused by the use of certain drugs or severe physical illness; (2) alcohol or drug abuse; (3) history of head trauma or neurological illnesses; (4) history of significant physical disorder (e.g. endocrine disease, autoimmune disease or liver or kidney dysfunction) or any type of tumor. Additionally, in the present study, HCs with no history of severe physical disorder, no personal or family history (first or second degree) of psychotic, no alcohol or drug abuse, and no gross abnormalities in brain MRI scanning, were strictly matched on the basis of age, gender, education years, and body mass index (BMI).
According to the inclusion and exclusion criteria, all HCs were randomly recruited through community health screening events and media advertisements and enrolled MDD subjects were randomly selected from an outpatient clinic or inpatient wards of the professional mental health center in the region (2017.07 – 2018.11). To obtain the scientific reliability, the following aspects were strictly controlled: (1) the diagnosis for the subject was performed collectively by two senior psychiatric physicians. If the diagnosis was inconsistent, a third professional physician would be consulted. The diagnosis of all subjects were reconfirmed by telephone follow-up prior to data analysis; (2) the evaluation of the subject was completed by two psychiatric chief physicians. Questionable terms were resolved by the thorough discussion. During the follow up of 4 weeks, clinical assessments were completed for each week by the same physicians. More details can be found in Supplementary Materials.
The discovery set, including 7 highly homogeneous MDD patients and 7 one-to-one paired HCs, was obtained from the Affiliated Zhongda Hospital of Southeast University [41], for which detailed information is presented in Supplementary Table 1. In addition, an independent validation set consisted of 53 MDD patients and 52 matched HCs recruited from the Second Affiliated Hospital at Xinxiang Medical University [18,41].
In addition, 53 MDD patients in the validation set provided consent for rTMS treatment (actually treated: 37; sham treatment: 16) as part of our previous study [42] (Chinese Clinical Trial Registry: ChiCTR1800014392), providing blood samples and clinical data to researchers. During rTMS treatment, no MDD patient received additional antidepressive therapy and all patients completed the current treatment. However, antidepressants were not limited to the 4-week follow-up observation period after rTMS treatment.
2.4. Ethics
The present study was carried out in accordance with the latest version of the Declaration of Helsinki. The ethics committee of the Affiliated Zhongda Hospital of Southeast University and the Second Affiliated Hospital of Xinxiang Medical University both approved the study (approval ID: 2019ZDSYLL055-P01). All subjects, their legal guardians or their legally authorized representatives provided informed consent.
2.5. Assessment tools
The HAMD-24 as a primary test tool, was used to assess depressive symptoms, and the Self-Rating Depression Scale (SDS) and the Beck Hopelessness Scale (BHS) were also used as second test tools to assess the severity of depression. Meanwhile, the Childhood Trauma Questionnaire-Short Form (CTQ-SF) and the Life Event Scale (LES) were used to assess the influence of negative life events, and the Beck Scale for Suicide Ideation-Chinese version-Current (BSI-CV-C) was used to assess the suicide attempt. The higher the score of these scales, the more severe of symptoms.
2.6. Sample collection
Venous blood was drawn from each subject after overnight fasting into a vacutainer tube (without anticoagulant) and a PAXgene blood RNA tube (Becton Dickinson). Detailed protocol was displayed in Supplementary Materials. Blood was sampled from the 53 MDD patients in the validation set at baseline and at the conclusion of treatment. Blood samples were collected once in all other subjects.
2.7. RNA extraction and RNA high-throughput sequencing
Total RNA was extracted using a PAXgene blood RNA kit (Qiagen) under standardized conditions using a QIAcube (Qiagen) automated processor in accordance with the manufacturer's instructions.
Sequencing libraries were generated and RNA high-throughput sequencing was performed by Genesky Biotechnologies Inc. (Shanghai, China) using standard Illumina protocols, which were described in detail in Supplementary Materials. The expression patterns of circRNAs, miRNAs, and mRNAs were obtained from the same samples of the discovery set, and subsequent bioinformatics analyses were performed.
2.8. Analysis of CircRNA sequencing data
CircRNAs were identified using CIRI software, followed by matching with data from the circBase database (http://circbase.org/). CircRNA expression was normalized using Back Spliced Reads per million mapped reads, with differential expression analysis accomplished using DEGSEQ software. The differentially expressed circRNAs between the two groups were filtered by |log2(fold change)| > 1 and p-value < 0.05 as criteria. The analyses of miRNA and mRNA sequencing data were described in Supplementary Materials.
2.9. Competing endogenous RNA (ceRNA) networks analysis and functional enrichment analysis
To investigate the ceRNA mechanism based on the differentially expressed circRNAs, miRNAs and mRNAs, a circRNA-miRNA-mRNA interaction network was constructed. Meanwhile, functional enrichment analysis was performed for Gene Ontology (GO) analysis. Supplementary Materials showed the detailed process.
2.10. RT-qPCR
Specific primers for candidate circRNAs were designed and synthesized by Shanghai Genesky Biotechnology Company, as displayed in Supplemental Table 3. cDNA was synthesized using a HiScript Q RT SuperMix for qPCR kit (Vazyme, R123-01) then analyzed by RT-qPCR using a SYBR Green Real-time PCR Master Mix (Roche, Mannheim, Germany). All reactions were performed in triplicate. The expression levels of circRNAs were calculated from the threshold cycle (Ct) value, and the relative fold change in expression (MDD vs. HC) calculated using the 2−ΔΔCt method. Glyceraldehyde-3-phosphate dehydrogenase was used as the endogenous control. Each sample was tested in triplicate.
2.11. Enzyme-linked immunosorbent assay (ELISA) analyses
The concentration of serum BDNF was measured in triplicate, using a commercial ELISA kit (R&D Systems, Minneapolis, MN, USA) in accordance with the manufacturer's protocols. The concentration in each plate was determined based on absorbance at 450 nm using a microplate reader (Thermo Scientific™, Shanghai, China). Each sample was tested in triplicate. The inter- and intra-assay coefficients of variation were < 4%.
2.12. MRI data acquisition and processing
MRI data were acquired using a Magnetom Verio (A Tim System) 3.0T superconducting magnetic resonance imaging system (Siemens, Erlangen, Germany). MRI data preprocessing was conducted using the Data Processing Assistant for Resting-State fMRI (DPARSFA 2.3) toolbox [43]. REST software (http://rest.restfmri.net) was used to calculate ALFF values. In the present study, only subjects in the validation set were analyzed by the MRI scanning, as outlined in Supplementary Materials.
2.13. rTMS treatment
The stimulation target of rTMS, the left primary visual cortex, was ascertained from the Montreal Neurological Institute coordinates [x: -1.8, y: -98.14, z: -6] [42]. The rTMS treatment was using the Magstim Rapid stimulator system with an eight-figured coil (The Magsitim Company Ltd, Whitland, UK). The resting motor threshold (RMT) was determined at each visit according to standard clinical practice [44]. The detailed parameters used for rTMS stimulation were as follows [42]: 90% RMT stimulation intensity; 10Hz frequency; 4 s on and 26 s off for 20 min; 1600 pulses per session; 2 session per days; total treatment duration: 5 days. Patients in the sham rTMS group received the same rTMS treatment protocol as the experimental rTMS group, except that the coil was turned through 90°.
2.14. Statistical analyses
2.14.1. Independent analysis of MRI data
In the validation set (Component 2), for identifying the brain region (voxels) with significantly different ALFF values between MDD and HC groups, we performed a voxel-wise one-way ANCOVA [45], [46], [47], [48] to examine the primary effect of the diagnosis (MDD vs. HC), with age [49], gender [50], and years of education [51] as covariates (dependent variable: ALFF value of each voxel; independent variable: diagnosis [MDD/HC], age, gender, years of education). The results were considered significant differences at a corrected p < 0.05 and cluster size > 4482 mm3 (166 voxels). Multiple comparison correction was performed using the AlphaSim [52,53]. Subsequently, ALFF values of these identified brain regions were extracted as neuroimaging indicators to further investigate the potential relationship of candidate circRNA expression with the neuronal activity in MDD patients.
2.14.2. Independent analysis of the composite circRNA indicator
In the validation set (component 2), the binary logistic regression [54] (dependent variable: MDD/HC; independent variable: candidate circRNAs) was used to calculate a predicted value of each subject that can be considered as the composite circRNA indicator based on these candidate circRNAs.
2.14.3. Analysis on the case-control study (components 1 and 2)
The Kolmogorov-Smirnov test was used to evaluate the normal distribution of the data and the Levene's homogeneity of variance test was also utilized. A chi-squared test was used for categorical variables (i.e. gender, smoker/nonsmoker), and continuous variables (i.e. current age, years of education, BMI, age of onset, duration of the disease, symptom assessments’ scores, serum BDNF levels, and candidate circRNA levels) were analyzed using an independent-sample t-test or a Mann-Whitney U test when appropriate between MDD and HC groups (details were displayed in Table 1).
Table 1.
Demographic characteristics and clinical informations for participants at baseline and after treatment.
| Baseline |
After treatment |
|||||
|---|---|---|---|---|---|---|
| HC (n = 52) | MDD (n = 53) | P-value | Actual rTMS (n = 37) | Sham rTMS (n = 16) | P-value | |
| Age (years) | 33.60 (10.31) | 30.66 (11.99) | 0.182* | 30.05 (12.92) | 32.06 (9.72) | 0.581* |
| Female | 28 (53.85%) | 28 (52.83%) | 0.927† | 20 (54.05%) | 8 (50.00%) | 0.997† |
| Education (years) | 13.05 (4.64) | 11.64 (3.52) | 0.400‡ | 11.68 (3.30) | 11.56 (4.10) | 0.916* |
| BMI | 23.23 (3.13) | 22.62 (3.81) | 0.367* | 22.91 (3.94) | 21.93 (3.51) | 0.396* |
| Smoking | 13 (25.00%) | 10 (18.87%) | 0.601† | 8 (21.62%) | 2 (12.50%) | 0.692† |
| Age of onset (years) | - | 26.74 (11.68) | - | 26.89 (13.04) | 26.38 (8.06) | 0.884* |
| Duration of the disease (month) | - | 49.99 (63.25) | - | 45.68 (7.51) | 75.63 (88.63) | 0.336‡ |
| HAMD-24 score (baseline) | 1.92 (2.25) | 36.09 (7.27) | < 0.001‡ | 35.81 (6.70) | 36.75 (8.65) | 0.670* |
| HAMD-24 score (5 day) | - | - | - | 18.24 (10.12) | 25.38 (10.02) | 0.022* |
| HAMD-24 score (1 week) | - | - | - | 13.89 (9.28) | 20.13 (12.22) | 0.047* |
| HAMD-24 score (2 week) | - | - | - | 11.51 (8.33) | 17.06 (12.07) | 0.058* |
| HAMD-24 score (3 week) | - | - | - | 8.65 (6.96) | 13.88 (11.29) | 0.044* |
| HAMD-24 score (4 week) | - | - | - | 6.81 (5.97) | 13.69 (12.13) | 0.008* |
| SDS score (baseline) | 34.56 (6.53) | 71.77 (10.90) | < 0.001* | 71.27 (11.09) | 72.94 (10.68) | 0.614* |
| SDS score (5 day) | - | - | - | 54.27 (17.68) | 57.06 (14.23) | 0.580* |
| BHS score (baseline) | 5.94 (1.78) | 11.17 (10.90) | < 0.001* | 11.08 (4.73) | 11.38 (5.35) | 0.843* |
| BHS score (5 day) | - | - | - | 7.19 (5.63) | 8.25 (5.67) | 0.532* |
| CTQ-SF score (baseline) | 37.85 (3.79) | 51.81 (11.82) | < 0.001‡ | 49.57 (10.79) | 57.00 (12.79) | 0.034* |
| LES score (baseline) | 5.46 (10.09) | 41.91 (28.60) | < 0.001‡ | 37.95 (25.53) | 51.06 (33.81) | 0.126* |
| BSI-CV-C score (baseline) | - | 5.92 (7.76) | - | 6.65 (8.13) | 4.25 (6.77) | 0.211‡ |
| Serum BDNF ng/ml (baseline) | 28.81 (7.29) | 21.34 (4.67) | < 0.001* | 21.53 (4.74) | 20.91 (4.61) | 0.662* |
| Serum BDNF ng/ml (5 day) | - | - | - | 25.27 (5.67) | 21.10 (4.75) | 0.013* |
Data presented as mean (standard deviation) or number of participants in each group (% of total).
MDD, major depressive disorder; HC, healthy control; BMI, body mass index; HAMD-24, the 24-item Hamilton Depression Scale; SDS, Self-Rating Depression Scale; BHS, Beck Hopelessness Scale; CTQ-SF, Childhood Trauma Questionnaire-Short Form; LES, Life Event Scale; BSI-CV-C, Beck Scale for Suicide Ideation-Chinese version-Current; rTMS, repetitive transcranial magnetic stimulation.
Independent-samples t-test.
Chi-squared test.
Mann-Whitney U-test.
In the validation set (component 2), Pearson correlation coefficient was used to assess correlations of candidate circRNA levels with assessments of symptomatology and serum and neuroimaging indicators’ levels in 53 MDD patients. Additionally, receiver operating characteristic (ROC) curves were plotted to calculate area under the curve (AUC) values so as to determine the performance of the single candidate circRNA and the composite circRNA indicator for distinguishing MDD patients from HCs. Aforementioned analyses were performed using SPSS 16.0 software (SPSS, Inc., Chicago, IL) and p < 0.05 was considered statistically significant.
Furthermore, in the validation set (component 2), a mediation effect model based on the Baron and Kenny method [55] was used to determine whether the serum BDNF or ALFF indicator could mediate the association between the candidate circRNA expression and the severity of depression in 53 patients with MDD, and covariates were controlled for clinical features (i.e. age, gender and BMI) [56], [57], [58], [59], [60]. Bootstrapping with 1000 replications was used to estimate the 95% confidence interval (CI) of the direct, indirect and total effect. β represents the regression coefficient of the linear regression model for the association between variables in the mediation analysis. The present mediation analysis was performed using SAS software version 9.4 (SAS Institute Inc., Cary, NC).
2.14.4. Analysis on the interventional study (components 3 and 4)
Component 3: The normal distribution test and homogeneity of variance test were also performed. An independent-sample t-test was used to evaluate the difference of candidate circRNA expression between actual treatment group and sham treatment group, and the paired t-test was used for the comparison of candidate circRNA expression before and after intervention in actual and sham treatment groups, respectively. For 37 MDD patients of actual treatment group, Pearson correlation analysis was performed to assess correlations of the expression of candidate circRNAs with the assessments of symptomatology and serum and neuroimaging indicators’ levels at the end of treatment and the change rate of these indexes (change rate = [baseline – the end of treatment] / baseline).
Component 4: In 53 MDD patients, we used a linear mixed model to evaluate the relationship between HAMD-24 scores and the changes of circRNA expression levels. The HAMD-24 scores at different time point was used as the outcome in the model, while the change of circRNA expression, treatment group and duration were included as the predictors. In the model, other variables including age [59], gender [16,21], and BMI [60] were regard as covariates, and some baseline data (i.e. baseline HAMD-24 score, baseline BDNF [27]) were also adjusted as covariates according to results of correlation analyses at baseline. We used the subject as the random effect in the model.
2.15. Sample-size estimation
The sample size calculation was performed using an online sample size calculators (https://sample-size.net/). The sample size used in the present study was appropriate based on the result of sample size calculation (α = 0.05, β = 0.2). Details can be found in Supplementary Materials.
2.16. Randomization
The selection of all subjects were randomized from the local population according to the inclusion and exclusion criteria. In the intervention study, MDD patients were randomly assigned to two treatment groups.
2.17. Blinding
Researchers involved in the clinical assessment, blood collection, and MRI scanning did not participate in the blood sample measurement and MRI data analysis. Additionally, researchers involved in the clinical assessment were blind to the rTMS distribution, while the rTMS physician did not participate the subsequent data analysis.
2.18. Role of the funding source
The funders had no role in the study design, data collection, data analysis, interpretation or writing of report. The corresponding author had full access to all the data and the final responsibility for the decision to submit for publication.
3. Results
3.1. Discovery and validation of differentially expressed circRNAs
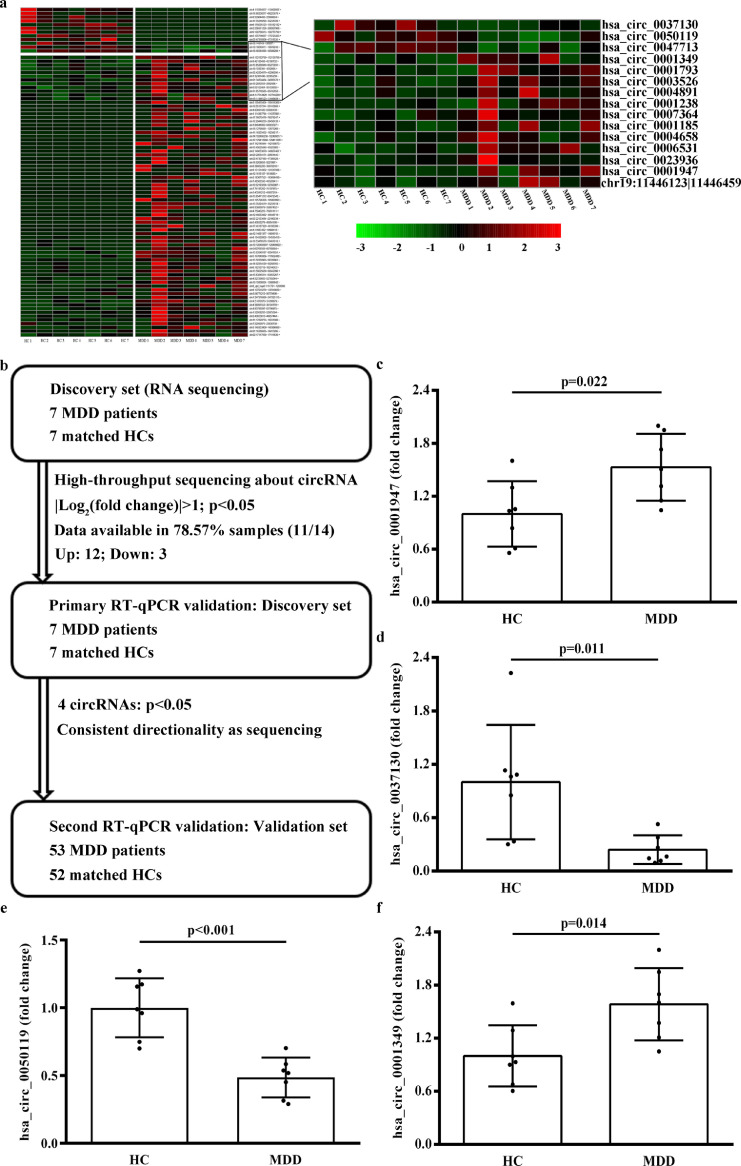
The baseline characteristics of the discovery set are listed in Supplementary Table 1. High-throughput sequencing involving 16259 circRNAs was performed in the discovery set, of which 88 were identified as being significantly different in 7 MDD patients compared with 7 HCs (SRA accession number: PRJNA606351; Fig. 1a). Subsequently, 12 upregulated circRNAs and 3 that were downregulated were detected and validated using RT-qPCR in the discovery set (Fig. 1b and Supplementary Table 2). Finally, 4 circRNAs were significantly different expression between 7 MDD patients and 7 HCs and consistent with the results of circRNA sequencing (Fig. 1c–f and Supplementary Table 4).
Fig. 1.
CircRNA expression profiles. (a) Cluster heat map showing 88 differentially expressed circRNAs with more than 2-fold change in expression and 15 candidate circRNAs in the discovery set (7 MDD patients and 7 HCs). Red represents high relative expression, green represents low relative expression. (b) Flowchart illustrating the 3-stage approach involving 2 independent cohorts for discovery and validation. (c) – (f) RT-qPCR was performed to verify the expression of the 15 candidate circRNAs in 7 MDD patients and 7 HCs. Each sample was tested in triplicate. Independent-samples t-test was used for data analysis and all data represents means ± standard deviation. circRNA, circular RNA; MDD, major depressive disorder; HC, healthy control; RT-qPCR, real-time quantitative polymerase chain reaction.
3.2. Differentially expressed circRNAs in the independent validation set
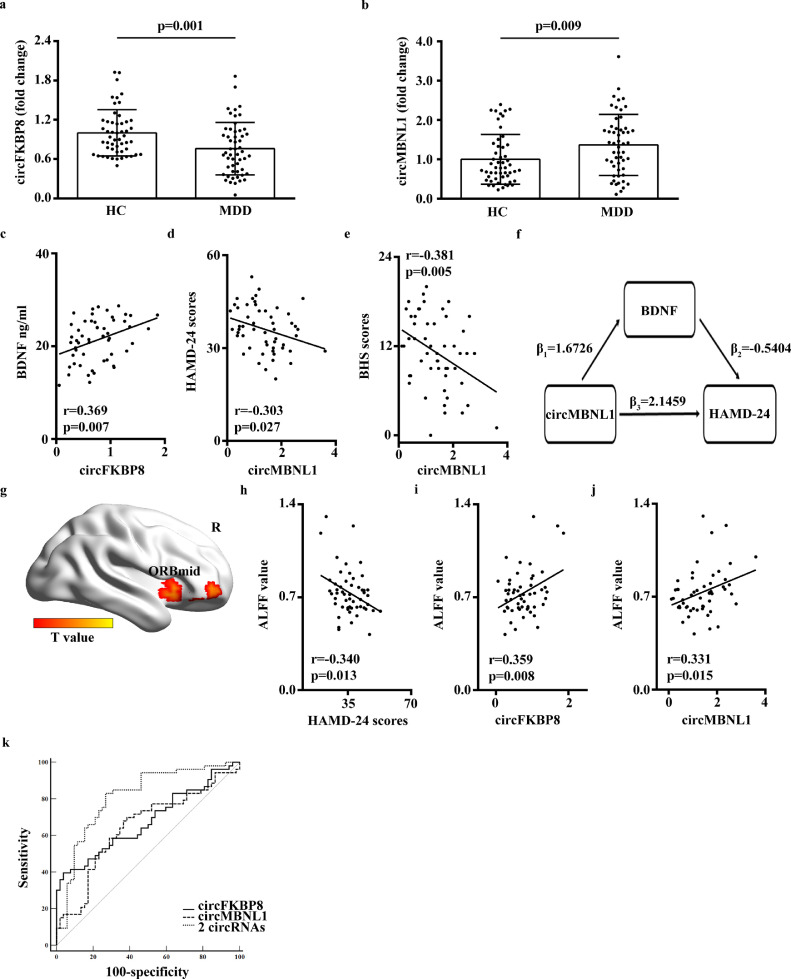
In the validation set (53 MDD patients and 52 HCs), there was no significant difference in baseline clinical features between the MDD and HC groups except for serum BDNF levels, and HAMD-24, SDS and BHS scores (Table 1). Four candidate circRNAs were further validated in the validation set, and compared with 52 HCs, hsa_circ_0050119 exhibited significantly reduced levels (p= 0.001; independent-sample t-test) and hsa_circ_0001349 had significantly increased levels (p= 0.009; independent-sample t-test) in 53 MDD patients (Fig. 2a and b). These results met statistical significance of multiple comparisons (p < 0.0125, Bonferroni correction). According to the human reference genome, hsa_circ_0050119 is assumed to be derived from the exons 5 and 6 of the FKBP8 gene and hsa_circ_0001349 from the exons 2 and 3 of the MBNL1 gene. Thus, hsa_circ_0050119 was termed circFKBP8 and hsa_circ_0001349 as circMBNL1. Besides, by searching the RNA sequencing data of the discovery set, compared with 7 HCs, the expression of the host gene of circFKBP8 was also significantly reduced in 7 MDD patients but the expression of the host gene of circMBNL1 was not significantly different.
Fig. 2.
Validation and the clinical utility of circRNAs. (a) – (b) Expression levels of circFKBP8 and circMBNL1 were determined by RT-qPCR in the independent validation set (53 MDD patients and 52 HCs). Each sample was tested in triplicate. Independent-samples t-test was used for data analysis and all data represents means ± standard deviation. (c) Correlation between circFKBP8 expression level and serum BDNF levels in 53 MDD patients. (d) –(e) Correlation between circMBNL1 expression levels and the HAMD-24 and BHS scores in 53 MDD patients. (f) Results from mediation analysis involving circMBNL1, BDNF and HAMD-24 scores in 53 MDD patients. Covariates were controlled for age, gender and BMI. β1 represents the comparable regression coefficient for association between circMBNL1 and BDNF; β2 and β3 represent comparable regression coefficients for the association between BDNF and HAMD-24 scores, and association between circMBNL1 and HAMD-24 scores with both circMBNL1 and BDNF in the linear regression model. β1 × β2 = indirect effect of circMBNL1 on HAMD-24 scores, and β3 = direct effect of circMBNL1 on HAMD-24 scores without the effect of the mediator BDNF. (g) Right ORBmid region displayed significantly increased ALFF in 53 MDD patients compared with 52 HCs (p < 0.05, Alphasim multiple comparison correction, voxel number: 166). Covariates were age, gender and years of education. (H) Correlation analysis between HAMD-24 scores and ALFF values of the right ORBmid in 53 MDD patients. (i) - (j). Correlation between expression levels of circFKBP8 and circMBNL1 and the ALFF values of the right ORBmid region in 53 MDD patients. (k) ROC curves of circFKBP8, circMBNL1 and the combination of the 2 circRNAs. circRNA, circular RNA; MDD, major depressive disorder; HC, healthy control; RT-qPCR, real-time quantitative polymerase chain reaction; BDNF, brain-derived neurotrophic factor; HAMD-24, 24-item Hamilton Depression Rating Scale; BHS, Beck Hopelessness Scale; ROC, receiver operating characteristic; ORBmid, orbital part middle frontal gyrus; R, right; ALFF, amplitude of low-frequency fluctuation.
In addition, correlation analyses in 53 MDD patients demonstrated that there was a positive correlation between circFKBP8 and BDNF levels (Fig. 2c) and the expression of circMBNL1 was negatively correlated with HAMD-24 and BHS scores (Fig. 2d and e). However, in 53 MDD patients, there was no significant correlation between the circFKBP8 or circMBNL1 expression and CTQ-SF, LES, and BSI-CV-C scores. Besides, mediation analysis in 53 MDD patients indicated that expression of circMBNL1 significantly mediated the effect of BDNF on HAMD-24 scores [indirect effect, β = -0.9038, 95% CI: (-2.35482, -0.04327), Fig. 2f] where covariates were controlled for age, gender, and BMI.
Compared with 52 HCs, 53 MDD patients exhibited decreased ALFF values in the bilateral dorsolateral superior frontal gyrus and increased ALFF values in the left middle occipital gyrus, bilateral inferior temporal gyrus, right middle temporal gyrus, right orbital part middle frontal gyrus (ORBmid), and bilateral medial prefrontal cortex and ventral ACC (Fig. 2g, Supplementary Fig. 1, AlphaSim corrected p < 0.05). Further correlation analysis revealed that the ALFFs in the right ORBmid were negatively correlated with HAMD-24 scores (Fig. 2h) and positively correlated with circFKBP8 and circMBNL1 expression in 53 MDD patients (Fig. 2i and j).
Furthermore, Fig. 2k displays the AUC values of circFKBP8 and circMBNL1, at 0.679 (95% CI: 0.576–0.781) and 0.647 (95% CI: 0.540–0.754), respectively. However, the combination of the 2 circRNAs provided greater diagnostic power with an AUC value of 0.813 (95% CI: 0.729–0.897), corresponding to a specificity of 73.1% and a sensitivity of 83.0% for the diagnosis of MDD (Fig. 2k).
3.3. rTMS intervention affected circFKBP8 expression
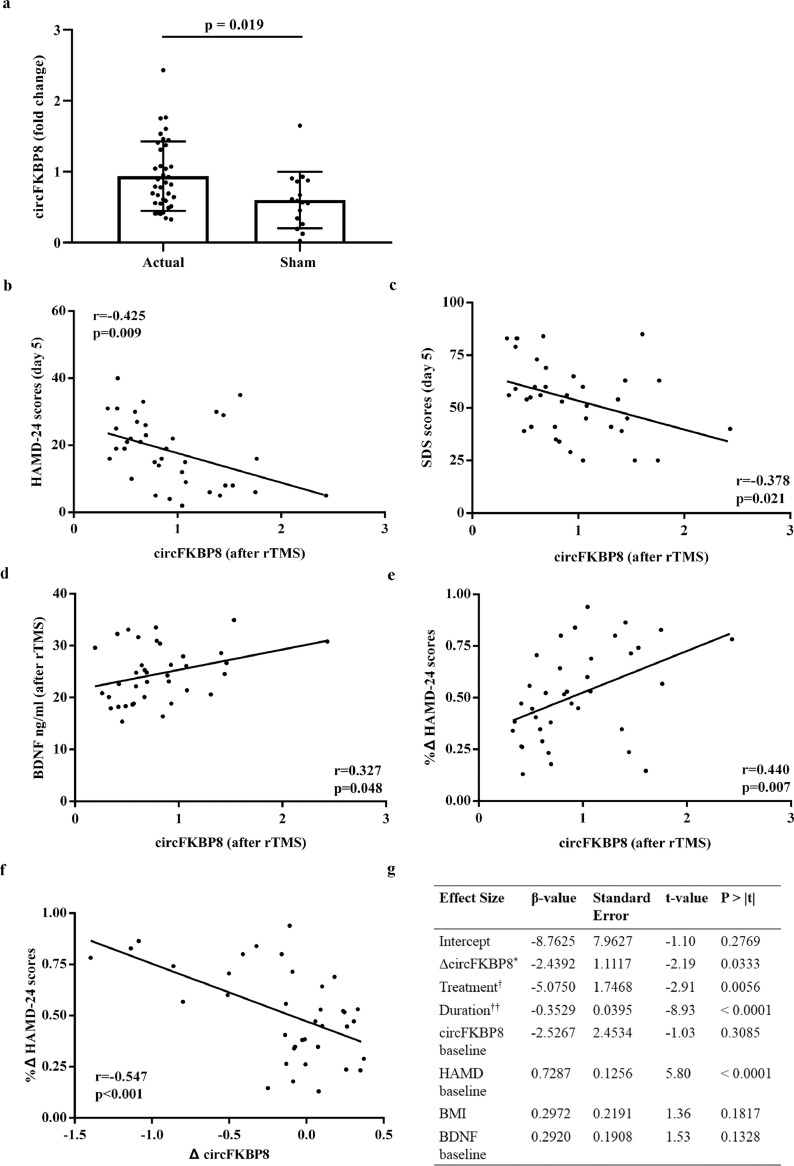
As shown in Table 1, there was no significant difference in HAMD-24 scores between rTMS treatment group and sham treatment group at baseline. However, significant differences of HAMD-24 scores were detected between two treatment groups following rTMS treatment (Table 1). Furthermore, baseline serum BDNF levels increased significantly after actual rTMS treatment, however, there was no significant difference in serum BDNF levels between before and after sham treatment (Supplementary Fig. 2a and b), and at the end of treatment, serum BDNF levels were significantly elevated in the actual treatment group as compared with the sham treatment group (Supplementary Fig. 2c). Additionally, in actual treatment group, except for the bilateral medial prefrontal cortex and ventral ACC, other brain regions had significant difference in ALFF values between before and after treatment, however, in the sham treatment group, only the left inferior temporal gyrus and dorsolateral superior frontal gyrus had significantly different ALFF values between before and after treatment (Supplementary Table 5).
CircFKBP8 exhibited differential expression in 53 MDD patients following complete rTMS intervention, with a significant increase in the expression of circFKBP8 in the actual rTMS treatment group compared with the sham treatment group (Fig. 3a), although there was only a trend in the increase in the actual rTMS treatment group before and after treatment (t = -1.990, p = 0.054, paired t-test; Supplementary Fig. 3). At the conclusion of treatment in the actual rTMS treatment, a significant negative correlation between the expression of circFKBP8 and HAMD-24 and SDS scores was observed (Fig. 3b and c), with circFKBP8 expression positively correlated with serum BDNF levels and rate of change of HAMD-24 scores (Fig. 3d and e), however, there was no significant correlation between circFKBP8 expression and the ALFFs in these identified brain regions. In addition, in the actual rTMS treatment group, the rate of change of HAMD-24 scores was negatively correlated with the change in circFKBP8 levels after treatment (Fig. 3f). But, no any correlation was detected in the sham treatment group.
Fig. 3.
Potential clinical value of circFKBP8 in the longitudinal study. (a) Expression levels of circFKBP8 in MDD patients between actual rTMS treatment group (n = 37) and sham treatment group (n = 16). The relative fold change of actual and sham groups were calculated using healthy controls as the reference, respectively. Each sample was tested in triplicate. Independent-samples t-test was used for data analysis and all data represents means ± standard deviation. (b) – (d) Correlation between circFKBP8 expression and HAMD-24 scores, SDS scores and serum BDNF levels in MDD patients at the conclusion of actual rTMS treatment (n = 37). (e) Correlation between rates of change of HAMD-24 scores and circFKBP8 expression in MDD patients at the conclusion of actual rTMS treatment (n = 37). (f) Correlation between rates of change of HAMD-24 scores and change in circFKBP8 expression in 37 MDD patients with rTMS treatment. (g) Model of change in value of circFKBP8 expression for prediction of HAMD-24 scores following rTMS treatment in 53 MDD patients. * Three sub-groups of MDD patients were obtained based on tertiles of change in circFKBP8 expression; † Influence of rTMS and sham treatments were considered in the model; †† Duration from the end of rTMS treatment to the end of the 4-week follow-up period. MDD, major depressive disorder; HC, healthy control; rTMS, repetitive transcranial magnetic stimulation; HAMD-24, 24-item Hamilton Depression Rating Scale; SDS, Self-Rating Depression Scale; BDNF, brain-derived neurotrophic factor; BMI, body mass index.
Furthermore, linear mixed effect model analysis demonstrated that a greater change in value of circFKBP8 expression (based on tertiles) was able to predict increased HAMD-24 scores after treatment in 53 MDD patients (Fig. 3g).
3.4. Possible biological mechanisms of action of circFKBP8 and circMBNL1
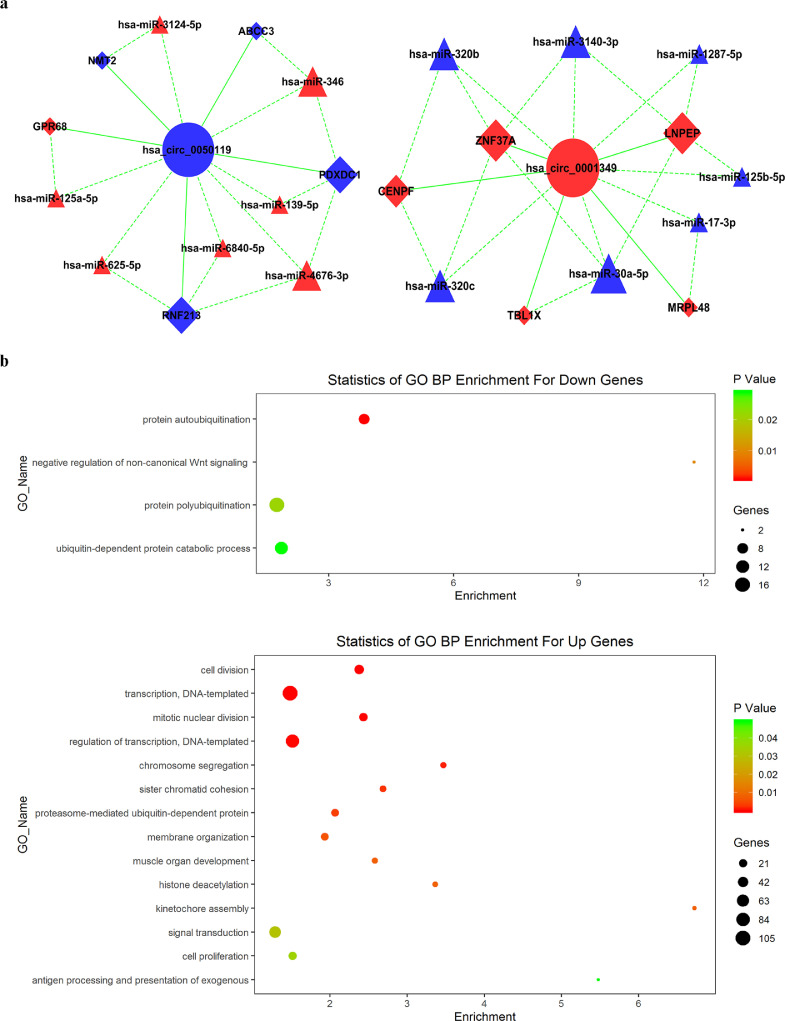
Based on the observed significantly differentially expressed circRNAs, miRNAs and mRNAs, circRNA-associated ceRNA networks were constructed in the discovery set (Supplementary Fig. 4). Subsequently, two independent ceRNA networks were obtained based on circFKBP8 and circMBNL1, including 14 miRNAs and 10 mRNAs (Fig. 4a).
Fig. 4.
Bioinformatics analysis of circFKBP8 and circMBNL1. (a) Possible binding of miRNAs and mRNAs to circFKBP8 and circMBNL1. Circles indicate circRNA, triangles indicate miRNA and squares indicate mRNA. Nodes highlighted in red and blue represent upregulation and downregulation, respectively. (b) The biological process of 10 mRNAs of ceRNA networks, identified from the Gene Ontology enrichment analysis of all significantly different mRNAs of the RNA sequencing. The above panel indicates up-regulated biological process in MDD and the bottom panel indicates down-regulated biological process in MDD. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
GO functional enrichment analysis was performed for all significantly different mRNAs, and the potential function of two ceRNA network-related mRNAs were identified (Fig. 4b). In the GO enrichment results, negative regulation of non-canonical Wnt signaling pathway (GO: 2000051), kinetochore assembly (GO: 0051382), and antigen processing and presentation of exogenous (GO: 0002480) were the mainly enriched biological process terms. In addition, GO analysis also performed for 10 mRNAs of two ceRNA networks, respectively (Supplementary Fig. 5), and the most enriched biological process terms were also located in same pathways (GO: 2000051 and GO: 0002480).
4. Discussion
The principal findings of the present study are as follows: (1) there was significantly reduced expression of circFKBP8 and significantly increased expression of circMBNL1 in whole-blood samples of MDD patients compared with HCs; (2) a mediation of the association was found between the circMBNL1 expression and HAMD-24 scores through the BDNF as mediator; (3) the expression of circFKBP8 and circMBNL1 was positively correlated with ALFFs in the right ORBmid in MDD patients; (4) the expression of circFKBP8 clearly changed following actual rTMS treatment, but not when sham treatment was performed; (5) the change in circFKBP8 expression following rTMS treatment predicted the severity of depressive symptoms following treatment. Taken together, two blood circRNAs may be meaningfully associated with MDD and/or depressive symptomatology, especially circFKBP8, which may have a potential clinical value for the antidepressive treatment.
In the present study, the method of identification and verification of potential circRNA biomarkers was well considered with multilevel assessments using serum and neuroimaging indices, significantly changed expression of circRNAs being novel in the research field of circRNAs in MDD. The specific strengths included: (1) circRNA profiles identified and validated in a highly homogeneous cohort; (2) serum BDNF and neuroimaging data used as relatively reliable evidence to evaluate the clinical utility and potential function of circRNAs; (3) the linear mixed effect model of antidepressive therapy was constructed by considering the time effect for the alteration of circRNA expression; (4) the underlying downstream mechanism was analyzed using bioinformatics analysis with ceRNA networks for guiding future research studies.
In the present study, the expression of circFKBP8 was first demonstrated to be significantly lower in MDD patients compared with HCs and show an increased change after the effective antidepressive treatment in MDD patients. Furthermore, the significant correlation between circFKBP8 expression and serum BDNF levels suggested that the circFKBP8 may regulate the neuroplasticity in MDD, which was consistent with main findings of GO analysis of circFKBP8, i.e. the enriched pathway was associated with cell differentiation and development (Supplementary Fig. 5). Importantly, circFKBP8 may be a potential biomarker for the antidepressive therapy due to the linear mixed effect model demonstrated that the tertiles of change in circFKBP8 expression accurately reflected the severity of depressive symptom at any point in time after treatment. In addition, based on the circFKBP8-associated ceRNA network, circFKBP8 may act on 5 downstream mRNAs (GPR68, PDXDC1, RNF213, NMT2 and ABCC3) in MDD. According to previous studies, GPR68 exerted pro-inflammatory function in macrophages and T cells [61], PDXDC1 involved in monoamine neurotransmitter synthesis that affects dopaminergic signaling [62,63], and RNF213 may regulate the activation of inflammatory mediators [64]. These evidence suggested that circFKBP8 may involve in the neuroinflammation of depression, but the precise mechanism (e.g. neuroplasticity) requires elucidation through additional studies. Nevertheless, based on the present findings, circFKBP8 may be a potential biomarker for the clinical practice of MDD.
Similarly, the present study also first demonstrated that the expression of circMBNL1 was significantly increased in MDD patients. However, the expression of host gene of circMNBL1 was no significantly different between MDD and HC groups, which may be caused by the independent regulatory mechanism between the circRNA and its host gene. circRNAs can act as independent gene expression regulators via various regulatory modes (e.g. miRNA sponge), therefore, the functions of circRNAs may be not affected by their host genes [65]. Meanwhile, in MDD patients, the negative correlation between circMBNL1 expression and HAMD-24 and BHS scores suggests that the compensatory increase in circMBNL1 expression may reflect the severity of depression. Interestingly, a mediator model in MDD patients showed that circMBNL1 can indirectly affect a depressive state through a change in BDNF levels, strongly indicating that BDNF-related neuronal growth and differentiation may play an important role in mediating the association of the circMBNL1 with MDD, and circMBNL1 may be implicated in the pathophysiology of MDD through regulating the neuroplasticity. Furthermore, the circMBNL1-associated ceRNA network indicated that 5 mRNAs (MRPL48, CENPF, ZNF37A, LNPEP and TBL1X) may reflect the function of circMBNL1 in MDD. Regrettably, no published study has shown any findings for these 5 mRNAs in MDD. Based on cell and animal studies, we detected that CENPF may involve in the development of neurons and glia cells [66], and LNPEP can affect the neuronal plasticity [67], which support the present findings that circMBNL1 was associated with neuroplasticity in MDD. Together, circMBNL1 may be served as a diagnostic biomarker of MDD, however, further mechanism study is necessary to determine the definite function of it in MDD although the neuroplasticity may be an important clue.
Additionally, important clinical features of the discovery set were history of negative life events and suicide attempt, which may affect identified circRNAs through some potential mechanism. Previous studies revealed that the association between the negative life event and MDD may occur via increasing inflammation or sensitivity of inflammatory responses [68,69], and MDD patients with a history of suicide attempts also significantly expressed some inflammatory response-related genetic markers [70]. Meanwhile, long-lasting consequence of negative experience occurring in life may contribute to structural modifications of neuronal plasticity to adapt to relevant environmental challenges [71,72], and there was a polygenic neurodevelopmental etiology in subjects with the suicidal behavior [73]. Likewise, the present GO analysis also indicated that mainly enriched biological processes were located in the cell proliferation and the immunity moderation, which further supported the underlying interaction between genetic and environmental factors in MDD.
There were seven associated miRNAs were identified in the respective ceRNA network. In the circFKBP8-associated ceRNA network, blood miR-139 may be a potential diagnostic biomarker for MDD [74] and the miR-346 may be useful biomarkers for schizophrenia diagnosis [75,76]. Meanwhile, in the MBNL1-associated ceRNA network, previous studies [77], [78], [79] revealed that blood miR-320b/c, miR-17, and miR-30a were associated with depression and the miR-125b was associated with Alzheimer's disease. These evidence further supported the underlying association between these circRNAs and MDD. In addition, among other 13 circRNAs that were verified again in the discovery set, 11 circRNAs acted on same downstream miRNAs of circFKBP8 or circMBNL1 according to the ceRNA network analysis and may have the similar function to circFKBP8 and circMBNL1 (Supplementary Table 2 and Fig. 3). Reviewing previous studies, hsa_circ_0007364 and hsa_circ_0001947 were associated with the cell proliferation [80,81], which suggested that the circMBNL1 may also involve in the cell development, however, no study reveals the function of the remaining circRNAs so far. Altogether, the relationship of circFKBP8 and circMBNL1 with functionally related miRNA and circRNAs in ceRNA network was a valuable direction to explore the function of circFKBP8/circMBNL1, and the constant investigation will be conducted in the subsequent study.
The present study found that ALFFs in the right ORBmid were significant higher but negatively correlated with HAMD-24 scores in MDD patients. Due to the OFC was an important emotion-related region [31], abnormal brain activity in the OFC may involve in aberrant modulation of emotional behavior in MDD patients [82]. Previous study have shown that, in comparison with HCs, MDD patients exhibited decreased ALFFs in the OFC [33,83,84], but increased ALFFs in the OFC were also detected in a cohort of treatment-naïve MDD patients [84], suggesting that a compensatory mechanism may occur in the OFC of MDD patients. Meanwhile, in the present study, significant changes in ALFF values of brain regions between before and after treatment suggested that the effective antidepressive treatment can improve significantly the function activity of most brain regions located in neural circuits that process emotions, although the present rTMS treatment as a new, fast treatment means, may not alter the abnormal activity of all brain regions. In addition, in the present study, significant correlations between the expression of circRNAs and ALFFs in the ORBmid demonstrated that these circRNAs may play a crucial role in the process of emotion-produced brain activity changes targeting the frontal region to result in MDD [85]. Consequently, these circRNAs may contribute to the pathogenesis of MDD through a neuroimaging-epigenetic interaction [38].
In the present study, two blood circRNAs for the diagnosis of MDD were proposed through the strict process of discovery and validation, which were associated with multifaceted characteristics of MDD, such as the severity of depressive symptom, the serum BDNF indicator, and the neuroimaging indicator of brain activity. As a result, these two blood circRNAs involved in the neuroinflammation and neuroplasticity and emotion-related brain activity alterations, may be used as promising pre-screening markers for identifying individuals with potential risk for MDD. On the other hand, the effective antidepressive intervention may alter the statue of neuroinflammation [86] or neuroplasticity [87] in MDD through affecting the epigenetics, e.g. the expression of circRNA. Therefore, the circFKBP8 showed the potential for guiding the individualized antidepressive treatment, especially for the rTMS treatment. Subsequently, studies of their functional role in MDD pathogenesis and further testing in various patients (e.g. schizophrenia, bipolar disorder) and various treatment approaches would be conducted to support these circRNA indicators to be used as the convenient toolkit for the clinical practice of MDD.
There are several limitations to the study. (1) The sample size of the discovery set and the validation set provided an acceptable power (> 80%) for measurements of candidate circRNAs (Supplementary Materials). However, the validation with a larger sample size may contribute to improving the power of results and supporting the use of these circRNAs for the clinical application of MDD. (2) Possible mechanisms for circFKBP8 and circMBNL1 were only predicted using bioinformatics and further research for functional verification would be performed in the subsequent study. (3) A modified and new rTMS method was used to observe the change in circRNAs dynamically. In the subsequent study, we will further verify the clinical efficacy of this method in a larger sample size and analyze the change in circRNAs in MDD patients using more therapeutic strategies, e.g. standard antidepressants and rTMS treatment of the dorsolateral prefrontal cortex. (4) Although satisfying results of two circRNAs were obtained in the present study, the bias of winner's curse may occur in the process of validation. In the future study, we will conduct the other analytical methods, such as the advanced bioinformatics analysis, machine learning analysis, to evaluate the clinical value of other circRNAs of RNA sequencing.
In conclusion, we initially illustrated the differential expression of circFKBP8 and circMBNL1 in MDD patients compared with HCs. Strong correlations between circRNAs levels and assessments of depressive symptomatology and serum BDNF and neuroimaging evidence demonstrated that the expression of circFKBP8 and circMBNL1 may serve as potential peripheral biomarkers for MDD. Furthermore, with rTMS treatment, we observed that circFKBP8 demonstrated great potential for the antidepressive treatment.
Contributors
Zhijun Zhang and Yang Zhao were responsible for the study design. Hongxing Zhang and Jianli Zhu were responsible for the recruitment of the participants, sample collection, and MRI data acquisition. Yachen Shi and Ruize Song were responsible for data validation and data collection. Yachen Shi and Zan Wang were responsible for data analysis, data interpretation, and writing the manuscript. Yang Zhao and Yuanping Yue were responsible for the quality control of the study. Zhijun Zhang, Yang Zhao, and Yachen Shi were responsible for manuscript modification and discussion of the data analysis. All the authors have critically read the manuscript and approved the submitted version.
Declaration of Competing Interest
The authors report no biomedical financial interests or potential conflicts of interest.
Acknowledgments
Acknowledgments
This study was partly supported by the National Key Research and Development Plan of China (No. 2016YFC1306700; ZJZ).
We would like to sincerely thank all participants who took part in this study, and the doctors and nurses (Mrs. Hong Zhu, Mrs. Liangliang Tan, Mrs. Dan Zhu and Mrs. Haixia Feng) for their help in recruiting them and collection of blood samples. We thank extremely Shanghai Genesky Biotechnology Company and Mr. Lingbin Sun for their help in RNA high-throughput sequencing. Meanwhile, we are grateful to Shenyang Jianjibiology Company and Mr. Haibo Chang for their help in bioinformatics analysis. Finally, we are extremely grateful to Prof. Yan Kong, Prof. Wei Du, Mr. Qingyun Wang, and Mr. Bin Han for the technical support.
Data sharing statement
All raw data were available in publicly accessible databases (SRA accession number: PRJNA606351).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2021.103337.
Contributor Information
Yang Zhao, Email: yzhao@njmu.edu.cn.
Zhijun Zhang, Email: janemengzhang@vip.163.com.
Appendix. Supplementary materials
Reference
- 1.Malhi G.S., Mann JJ. Depression. Lancet. 2018;392(10161):2299–2312. doi: 10.1016/S0140-6736(18)31948-2. [DOI] [PubMed] [Google Scholar]
- 2.Fang Y., Wu Z. Advance in diagnosis of depressive disorder. Adv Exp Med Biol. 2019;1180:179–191. doi: 10.1007/978-981-32-9271-0_9. [DOI] [PubMed] [Google Scholar]
- 3.Trivedi M.H., Rush A.J., Wisniewski S.R. Evaluation of outcomes with citalopram for depression using measurement-based care in STAR*D: implications for clinical practice. Am J Psychiatry. 2006;163(1):28–40. doi: 10.1176/appi.ajp.163.1.28. [DOI] [PubMed] [Google Scholar]
- 4.Girardi P., Pompili M., Innamorati M. Duloxetine in acute major depression: review of comparisons to placebo and standard antidepressants using dissimilar methods. Hum Psychopharmacol. 2009;24(3):177–190. doi: 10.1002/hup.1005. [DOI] [PubMed] [Google Scholar]
- 5.Pompili M., Serafini G., Innamorati M. Agomelatine, a novel intriguing antidepressant option enhancing neuroplasticity: a critical review. World J Biol Psychiatry. 2013;14(6):412–431. doi: 10.3109/15622975.2013.765593. [DOI] [PubMed] [Google Scholar]
- 6.Wang Z., Ma X., Xiao C. Standardized treatment strategy for depressive disorder. Adv Exp Med Biol. 2019;1180:193–199. doi: 10.1007/978-981-32-9271-0_10. [DOI] [PubMed] [Google Scholar]
- 7.Memczak S., Jens M., Elefsinioti A. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 8.Jeck W.R., Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X., Yang L., Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 10.Kristensen L.S., Andersen M.S., Stagsted L.V.W., Ebbesen K.K., Hansen T.B., Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 11.Abu N., Jamal R. Circular RNAs as promising biomarkers: a mini-review. Front Physiol. 2016;7:355. doi: 10.3389/fphys.2016.00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanan M., Soreq H., Kadener S. CircRNAs in the brain. RNA Biol. 2017;14(8):1028–1034. doi: 10.1080/15476286.2016.1255398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhuo C.J., Hou W.H., Jiang D.G. Circular RNAs in early brain development and their influence and clinical significance in neuropsychiatric disorders. Neural Regen Res. 2020;15(5):817–823. doi: 10.4103/1673-5374.268969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui X., Niu W., Kong L. hsa_circRNA_103636: potential novel diagnostic and therapeutic biomarker in major depressive disorder. Biomark Med. 2016;10(9):943–952. doi: 10.2217/bmm-2016-0130. [DOI] [PubMed] [Google Scholar]
- 15.Jiang G., Ma Y., An T. Relationships of circular RNA with diabetes and depression. Sci Rep. 2017;7(1):7285. doi: 10.1038/s41598-017-07931-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.An T., He Z.-.C., Zhang X.-.Q. Baduanjin exerts anti-diabetic and anti-depression effects by regulating the expression of mRNA, lncRNA, and circRNA. Chin Med. 2019;14 doi: 10.1186/s13020-019-0225-1. 3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y., Du L., Bai Y. CircDYM ameliorates depressive-like behavior by targeting miR-9 to regulate microglial activation via HSP90 ubiquitination. Mol Psychiatry. 2018 doi: 10.1038/s41380-41018-40285-41380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song R., Bai Y., Li X. Plasma circular RNA DYM related to major depressive disorder and rapid antidepressant effect treated by visual cortical repetitive transcranial magnetic stimulation. J Affect Disord. 2020;274:486–493. doi: 10.1016/j.jad.2020.05.109. [DOI] [PubMed] [Google Scholar]
- 19.Yao G., Niu W., Zhu X. hsa_circRNA_104597: a novel potential diagnostic and therapeutic biomarker for schizophrenia. Biomark Med. 2019;13(5):331–340. doi: 10.2217/bmm-2018-0447. [DOI] [PubMed] [Google Scholar]
- 20.Mahmoudi E., Fitzsimmons C., Geaghan M. Circular RNA biogenesis is decreased in postmortem cortical gray matter in schizophrenia and may alter the bioavailability of associated miRNA. Neuropsychopharmacol Off Publ Am Coll Neuropsychopharmacol. 2019;44(6):1043–1054. doi: 10.1038/s41386-019-0348-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zimmerman A., Hafez A., Amoah S. A psychiatric disease-related circular RNA controls synaptic gene expression and cognition. Mol Psychiatry. 2020;25(11):2712–2727. doi: 10.1038/s41380-020-0653-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan G., Wang L., Liu Y., Zhang H., Feng W., Liu Z. The alterations of circular RNA expression in plasma exosomes from patients with schizophrenia. J Cell Physiol. 2021;236(1):458–467. doi: 10.1002/jcp.29873. [DOI] [PubMed] [Google Scholar]
- 23.Luykx J., Giuliani F., Giuliani G., Veldink J. Coding and non-coding RNA abnormalities in bipolar disorder. Genes. 2019;10(11):946. doi: 10.3390/genes10110946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y., Huang R., Cheng M. Gut microbiota from NLRP3-deficient mice ameliorates depressive-like behaviors by regulating astrocyte dysfunction via circHIPK2. Microbiome. 2019;7(1):116. doi: 10.1186/s40168-019-0733-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewin G.R., Barde YA. Physiology of the neurotrophins. Annu Rev Neurosci. 1996;19:289–317. doi: 10.1146/annurev.ne.19.030196.001445. [DOI] [PubMed] [Google Scholar]
- 26.Polyakova M., Stuke K., Schuemberg K., Mueller K., Schoenknecht P., Schroeter ML. BDNF as a biomarker for successful treatment of mood disorders: a systematic & quantitative meta-analysis. J Affect Disord. 2015;174:432–440. doi: 10.1016/j.jad.2014.11.044. [DOI] [PubMed] [Google Scholar]
- 27.Shi Y., Luan D., Song R., Zhang Z. Value of peripheral neurotrophin levels for the diagnosis of depression and response to treatment: a systematic review and meta-analysis. Eur Neuropsychopharmacol. 2020;41:40–51. doi: 10.1016/j.euroneuro.2020.09.633. [DOI] [PubMed] [Google Scholar]
- 28.Shimizu E., Hashimoto K., Okamura N. Alterations of serum levels of brain-derived neurotrophic factor (BDNF) in depressed patients with or without antidepressants. Biol Psychiatry. 2003;54(1):70–75. doi: 10.1016/s0006-3223(03)00181-1. [DOI] [PubMed] [Google Scholar]
- 29.Yang L., Han B., Zhang Z. Extracellular vesicle-mediated delivery of circular RNA SCMH1 Promotes functional recovery in rodent and nonhuman primate ischemic stroke models. Circulation. 2020;142(6):556–574. doi: 10.1161/CIRCULATIONAHA.120.045765. [DOI] [PubMed] [Google Scholar]
- 30.Yao Z., Wang L., Lu Q., Liu H., Teng G. Regional homogeneity in depression and its relationship with separate depressive symptom clusters: a resting-state fMRI study. J Affect Disord. 2009;115(3):430–438. doi: 10.1016/j.jad.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 31.Szily E., Kéri S. Emotion-related brain regions. Ideggyogy Sz. 2008;61(3-4):77–86. [PubMed] [Google Scholar]
- 32.Zhong X., Pu W., Yao S. Functional alterations of fronto-limbic circuit and default mode network systems in first-episode, drug-naïve patients with major depressive disorder: A meta-analysis of resting-state fMRI data. J Affect Disord. 2016;206:280–286. doi: 10.1016/j.jad.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 33.Li W., Chen Z., Wu M. Characterization of brain blood flow and the amplitude of low-frequency fluctuations in major depressive disorder: a multimodal meta-analysis. J Affect Disord. 2017;210:303–311. doi: 10.1016/j.jad.2016.12.032. [DOI] [PubMed] [Google Scholar]
- 34.Dichter G.S., Gibbs D., Smoski MJ. A systematic review of relations between resting-state functional-MRI and treatment response in major depressive disorder. J Affect Disord. 2015;172:8–17. doi: 10.1016/j.jad.2014.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou M., Hu X., Lu L. Intrinsic cerebral activity at resting state in adults with major depressive disorder: a meta-analysis. Prog Neuro Psychopharmacol Biol Psychiatry. 2017;75:157–164. doi: 10.1016/j.pnpbp.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 36.Zang Y.F., He Y., Zhu C.Z. Altered baseline brain activity in children with ADHD revealed by resting-state functional MRI. Brain Dev. 2007;29(2):83–91. doi: 10.1016/j.braindev.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 37.Liu X., Li L., Li M., Ren Z., Ma P. Characterizing the subtype of anhedonia in major depressive disorder: a symptom-specific multimodal MRI study. Psychiatry Res Neuroimaging. 2021;308 doi: 10.1016/j.pscychresns.2020.111239. [DOI] [PubMed] [Google Scholar]
- 38.He C., Bai Y., Wang Z. Identification of microRNA-9 linking the effects of childhood maltreatment on depression using amygdala connectivity. Neuroimage. 2021;224 doi: 10.1016/j.neuroimage.2020.117428. [DOI] [PubMed] [Google Scholar]
- 39.Januar V., Saffery R., Ryan J. Epigenetics and depressive disorders: a review of current progress and future directions. Int J Epidemiol. 2015;44(4):1364–1387. doi: 10.1093/ije/dyu273. [DOI] [PubMed] [Google Scholar]
- 40.Mill J., Petronis A. Molecular studies of major depressive disorder: the epigenetic perspective. Mol Psychiatry. 2007;12(9):799–814. doi: 10.1038/sj.mp.4001992. [DOI] [PubMed] [Google Scholar]
- 41.Shi Y., Song R., Wang L. Identifying Plasma Biomarkers with high specificity for major depressive disorder: a multi-level proteomics study. J Affect Disord. 2020;277:620–630. doi: 10.1016/j.jad.2020.08.078. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Z., Zhang H., Xie C.M. Task-related functional magnetic resonance imaging-based neuronavigation for the treatment of depression by individualized repetitive transcranial magnetic stimulation of the visual cortex. Sci China Life Sci. 2021;64(1):96–106. doi: 10.1007/s11427-020-1730-5. [DOI] [PubMed] [Google Scholar]
- 43.Yan C.G., Zang Y.F. DPARSF: A MATLAB Toolbox for “Pipeline” Data Analysis of Resting-State fMRI. Front Syst Neurosci. 2010;4:13. doi: 10.3389/fnsys.2010.00013. 13-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McClintock S.M., Reti I.M., Carpenter L.L. Consensus Recommendations for the Clinical Application of Repetitive Transcranial Magnetic Stimulation (rTMS) in the Treatment of Depression. J Clin Psychiatry. 2018;79(1):16cs10905. doi: 10.4088/JCP.16cs10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Z., Jia X., Chen H., Feng T., Wang H. Abnormal Spontaneous brain activity in Early Parkinson's disease with mild cognitive impairment: a resting-state fMRI study. Front Physiol. 2018;9:1093. doi: 10.3389/fphys.2018.01093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhuo C., Ji F., Lin X. Global functional connectivity density alterations in patients with bipolar disorder with auditory verbal hallucinations and modest short-term effects of transcranial direct current stimulation augmentation treatment-Baseline and follow-up study. Brain Behav. 2020;10(6):e01637. doi: 10.1002/brb3.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shu H., Shi Y., Chen G. Opposite neural trajectories of apolipoprotein E ϵ4 and ϵ2 alleles with aging associated with different risks of Alzheimer's disease. Cereb Cortex. 2016;26(4):1421–1429. doi: 10.1093/cercor/bhu237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bai F., Zhang Z., Watson D.R. Abnormal functional connectivity of hippocampus during episodic memory retrieval processing network in amnestic mild cognitive impairment. Biol Psychiatry. 2009;65(11):951–958. doi: 10.1016/j.biopsych.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 49.Jacobson A., Green E., Murphy C. Age-related functional changes in gustatory and reward processing regions: An fMRI study. Neuroimage. 2010;53(2):602–610. doi: 10.1016/j.neuroimage.2010.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.AlRyalat SA. Gender similarities and differences in brain activation strategies: voxel-based meta-analysis on fMRI studies. J Integr Neurosci. 2017;16(2):227–240. doi: 10.3233/JIN-170015. [DOI] [PubMed] [Google Scholar]
- 51.Karim H.T., Tudorascu D.L., Cohen A. Relationships between executive control circuit activity, amyloid burden, and education in cognitively healthy older adults. Am J Geriatr Psychiatry. 2019;27(12):1360–1371. doi: 10.1016/j.jagp.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu Y.C., Zhang H., Zheng M.X. Local and extensive neuroplasticity in carpal tunnel syndrome: a resting-state fMRI study. Neurorehabil Neural Repair. 2017;31(10-11):898–909. doi: 10.1177/1545968317723749. [DOI] [PubMed] [Google Scholar]
- 53.Li F., Lui S., Yao L. Longitudinal changes in resting-state cerebral activity in patients with first-episode schizophrenia: a 1-year follow-up functional MR imaging study. Radiology. 2016;279(3):867–875. doi: 10.1148/radiol.2015151334. [DOI] [PubMed] [Google Scholar]
- 54.Chen S., Jiang H., Liu Y. Combined serum levels of multiple proteins in tPA-BDNF pathway may aid the diagnosis of five mental disorders. Sci Rep. 2017;7(1):6871. doi: 10.1038/s41598-017-06832-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krause M.R., Serlin R.C., Ward S.E., Rony R.Y., Ezenwa M.O., Naab F. Testing mediation in nursing research: beyond Baron and Kenny. Nurs Res. 2010;59(4):288–294. doi: 10.1097/NNR.0b013e3181dd26b3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schaakxs R., Comijs H.C., Lamers F., Kok R.M., Beekman A.T.F., Penninx B. Associations between age and the course of major depressive disorder: a 2-year longitudinal cohort study. Lancet Psychiatry. 2018;5(7):581–590. doi: 10.1016/S2215-0366(18)30166-4. [DOI] [PubMed] [Google Scholar]
- 57.Lai C.H. Major depressive disorder: gender differences in symptoms, life quality, and sexual function. J Clin Psychopharmacol. 2011;31(1):39–44. doi: 10.1097/JCP.0b013e318205a670. [DOI] [PubMed] [Google Scholar]
- 58.Dreimüller N., Lieb K., Tadić A., Engelmann J., Wollschläger D., Wagner S. Body mass index (BMI) in major depressive disorder and its effects on depressive symptomatology and antidepressant response. J Affect Disord. 2019;256:524–531. doi: 10.1016/j.jad.2019.06.067. [DOI] [PubMed] [Google Scholar]
- 59.Akhter R. Circular RNA and Alzheimer's disease. Adv Exp Med Biol. 2018;1087:239–243. doi: 10.1007/978-981-13-1426-1_19. [DOI] [PubMed] [Google Scholar]
- 60.Bao X., Zheng S., Mao S. A potential risk factor of essential hypertension in case-control study: circular RNA hsa_circ_0037911. Biochem Biophys Res Commun. 2018;498(4):789–794. doi: 10.1016/j.bbrc.2018.03.059. [DOI] [PubMed] [Google Scholar]
- 61.Alavi M.S., Karimi G., Roohbakhsh A. The role of orphan G protein-coupled receptors in the pathophysiology of multiple sclerosis: a review. Life Sci. 2019;224:33–40. doi: 10.1016/j.lfs.2019.03.045. [DOI] [PubMed] [Google Scholar]
- 62.van den Buuse M. Modeling the positive symptoms of schizophrenia in genetically modified mice: pharmacology and methodology aspects. Schizophr Bull. 2010;36(2):246–270. doi: 10.1093/schbul/sbp132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Feldcamp L.A., Boutros P.C., Raymond R., Fletcher P.J., Nobrega J.N., Wong AHC. Pdxdc1 modulates prepulse inhibition of acoustic startle in the mouse. Transl Psychiatry. 2017;7(5):e1125. doi: 10.1038/tp.2017.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vilariño-Güell C., Zimprich A., Martinelli-Boneschi F. Exome sequencing in multiple sclerosis families identifies 12 candidate genes and nominates biological pathways for the genesis of disease. PLoS genetics. 2019;15(6):e1008180. doi: 10.1371/journal.pgen.1008180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Han B., Chao J., Yao H. Circular RNA and its mechanisms in disease: from the bench to the clinic. Pharmacol Ther. 2018;187:31–44. doi: 10.1016/j.pharmthera.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 66.Hu D.J.-.K., Baffet A.D., Nayak T., Akhmanova A., Doye V., Vallee RB. Dynein recruitment to nuclear pores activates apical nuclear migration and mitotic entry in brain progenitor cells. Cell. 2013;154(6):1300–1313. doi: 10.1016/j.cell.2013.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Narwade S.C., Mallick B.N., Deobagkar DD. Transcriptome analysis reveals altered expression of memory and neurotransmission associated genes in the REM sleep deprived rat brain. Front Mol Neurosci. 2017;10:67. doi: 10.3389/fnmol.2017.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Danese A., Moffitt T.E., Pariante C.M., Ambler A., Poulton R., Caspi A. Elevated inflammation levels in depressed adults with a history of childhood maltreatment. Arch Gen Psychiatry. 2008;65(4):409–415. doi: 10.1001/archpsyc.65.4.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Danese A., Moffitt T.E., Harrington H. Adverse childhood experiences and adult risk factors for age-related disease: depression, inflammation, and clustering of metabolic risk markers. Arch Pediatr Adolesc Med. 2009;163(12):1135–1143. doi: 10.1001/archpediatrics.2009.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Galfalvy H., Haghighi F., Hodgkinson C. A genome-wide association study of suicidal behavior. Am J Med Genet B Neuropsychiatr Genet. 2015;168(7):557–563. doi: 10.1002/ajmg.b.32330. [DOI] [PubMed] [Google Scholar]
- 71.Calabrese F., Molteni R., Racagni G., Riva MA. Neuronal plasticity: a link between stress and mood disorders. Psychoneuroendocrinology. 2009;34(Suppl 1):S208–S216. doi: 10.1016/j.psyneuen.2009.05.014. [DOI] [PubMed] [Google Scholar]
- 72.Krishnan V., Nestler EJ. The molecular neurobiology of depression. Nature. 2008;455(7215):894–902. doi: 10.1038/nature07455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sokolowski M., Wasserman J., Wasserman D. Polygenic associations of neurodevelopmental genes in suicide attempt. Mol Psychiatry. 2016;21(10):1381–1390. doi: 10.1038/mp.2015.187. [DOI] [PubMed] [Google Scholar]
- 74.Liang J.Q., Liao H.R., Xu C.X. Serum exosome-derived miR-139-5p as a potential biomarker for major depressive disorder. Neuropsychiatr Dis Treat. 2020;16:2689–2693. doi: 10.2147/NDT.S277392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sun X.Y., Zhang J., Niu W. A preliminary analysis of microRNA as potential clinical biomarker for schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2015;168b(3):170–178. doi: 10.1002/ajmg.b.32292. [DOI] [PubMed] [Google Scholar]
- 76.Liu S., Zhang F., Wang X. Diagnostic value of blood-derived microRNAs for schizophrenia: results of a meta-analysis and validation. Sci Rep. 2017;7(1):15328. doi: 10.1038/s41598-017-15751-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Camkurt M.A., Acar Ş., Coşkun S. Comparison of plasma MicroRNA levels in drug naive, first episode depressed patients and healthy controls. J Psychiatr Res. 2015;69:67–71. doi: 10.1016/j.jpsychires.2015.07.023. [DOI] [PubMed] [Google Scholar]
- 78.Khandelwal N., Dey S.K., Chakravarty S., Kumar A. miR-30 family miRNAs mediate the effect of chronic social defeat stress on hippocampal neurogenesis in mouse depression model. Front Mol Neurosci. 2019;12:188. doi: 10.3389/fnmol.2019.00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Herrera-Espejo S., Santos-Zorrozua B., Álvarez-González P.E. Lopez-Lopez A systematic review of microRNA expression as biomarker of late-onset Alzheimer’s disease. Mol Neurobiol. 2019;56(12):8376–8391. doi: 10.1007/s12035-019-01676-9. [DOI] [PubMed] [Google Scholar]
- 80.Chen H., Gu B., Zhao X. Circular RNA hsa_circ_0007364 increases cervical cancer progression through activating methionine adenosyltransferase II- α(MAT2A) expression by restraining microRNA-101-5p. Bioengineered. 2020;11(1):1269–1279. doi: 10.1080/21655979.2020.1832343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Han F., Zhong C., Li W. hsa_circ_0001947 suppresses acute myeloid leukemia progression via targeting hsa-miR-329-5p/CREBRF axis. Epigenomics. 2020;12(11):935–953. doi: 10.2217/epi-2019-0352. [DOI] [PubMed] [Google Scholar]
- 82.Ongur D., Price JL. The organization of networks within the orbital and medial prefrontal cortex of rats, monkeys and humans. Cereb Cortex. 2000;10(3):206–219. doi: 10.1093/cercor/10.3.206. [DOI] [PubMed] [Google Scholar]
- 83.Zhang X., Di X., Lei H. Imbalanced spontaneous brain activity in orbitofrontal-insular circuits in individuals with cognitive vulnerability to depression. J Affect Disord. 2016;198:56–63. doi: 10.1016/j.jad.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 84.Liu J., Ren L., Womer F.Y. Alterations in amplitude of low frequency fluctuation in treatment-naïve major depressive disorder measured with resting-state fMRI. Hum Brain Mapp. 2014;35(10):4979–4988. doi: 10.1002/hbm.22526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Qin J., Wei M., Liu H. Abnormal brain anatomical topological organization of the cognitive-emotional and the frontoparietal circuitry in major depressive disorder. Magn Reson Med. 2014;72(5):1397–1407. doi: 10.1002/mrm.25036. [DOI] [PubMed] [Google Scholar]
- 86.Adzic M., Brkic Z., Mitic M. Therapeutic strategies for treatment of inflammation-related depression. Curr Neuropharmacol. 2018;16(2):176–209. doi: 10.2174/1570159X15666170828163048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gerhard D.M., Wohleb E.S., Duman RS. Emerging treatment mechanisms for depression: focus on glutamate and synaptic plasticity. Drug Discov Today. 2016;21(3):454–464. doi: 10.1016/j.drudis.2016.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.