Abstract
Dexamethasone, a synthetic glucocorticoid, has previously shown mortality benefit in severe coronavirus disease 2019 (COVID-19) in a randomized controlled trial. As the illness is considered to reflect a hyperinflammatory state, this therapeutic effectiveness is presumably ascribed to broad anti-inflammatory activities of glucocorticoids. Here, an unbiased analysis of available transcriptomic data on lung and blood immune cells from severe COVID-19 patients and matching cellular models of dexamethasone treatment is presented that supports this presumption. Comparison of differentially expressed genes in severe COVID-19 with that in dexamethasone treated cells reveals a small set of genes that are regulated in opposite direction between the disease and the drug, and are enriched for genes and processes related to glucocorticoid pathway and receptor binding. This expression signature differentiates as a whole various cytokines from a set of anti-cytokine/anti-inflammatory agents, with the former resembling COVID-19 and the latter dexamethasone in gene regulation. The signature apparently relates to TNF- α, IL-1α, IL-1β, IFN-α, IFN-β, and IFN-γ signaling, but not IL-6 signaling, suggesting that therapeutic effect of dexamethasone in COVID-19 does not involve IL-6 pathway. However, as all these observations are purely based on bioinformatic analysis, experimental evidence will be required to validate the inferences drawn. In conclusion, the present analysis seems to provide a proof of concept for therapeutic mechanisms of dexamethasone in COVID-19.
Abbreviations: COVID-19, Coronavirus Disease 2019; DEGs, differentially expressed genes; MSigDB, molecular signatures database; GEO, Gene Expression Omnibus; GREIN, GEO RNA-seq Experiments Interactive Navigator; FC, fold change
Keywords: COVID-19, Dexamethasone, Glucocorticoids, Inflammation, Transcriptome, Gene expression
1. Introduction
The classic synthetic glucocorticoid dexamethasone has previously shown in a randomized clinical trial mortality benefit in severely sick hospitalized patients with coronavirus disease 2019 (COVID-19) (RECOVERY Collaborative Group et al., 2020). As severe COVID-19 is seemingly characterized by a feedforward inflammatory circuit involving systemic elevations in cytokine and chemokine levels, often referred to as cytokine storm, the observed benefit is presumably ascribed to broad anti-inflammatory and immunosuppressive effects of glucocorticoids (Cain and Cidlowski, 2020, Macauley et al., 2020). The latter, a class of steroid hormones, act by binding ubiquitously expressed glucocorticoid receptor, a ligand-activated transcription factor encoded by NR3C1 gene in humans, which, upon translocation to the nucleus, binds to canonical glucocorticoid response elements and leads to transcriptional activation of its targets including anti-inflammatory genes such as TSC22D3, DUSP1, and SGK1 (Cain and Cidlowski, 2020, Hudson et al., 2018). The ligand-bound receptor can also cause transcriptional inhibition of numerous genes including pro-inflammatory genes by antagonizing activities of corresponding transcription factor like NF-κB and AP1 through protein-protein interactions (Cain and Cidlowski, 2020, Hudson et al., 2018). With context-dependent mechanisms of action, glucocorticoids may produce therapeutic effects in an inflammatory condition by acting on both immune and nonimmune cells (Quatrini and Ugolini, 2020).
Given its potential for knowledge discovery (Wang et al., 2019), the approach of integrative analysis of available transcriptomic data is applied here towards gaining insights into mechanisms underlying dexamethasone action in severe COVID-19. Transcriptomic perturbations induced by drugs across cell types are known to show high conservation, with cell type specific changes reflecting a more selective effect (Iskar et al., 2013). A combined approach may therefore be used to draw inference about mechanism of action of drugs. The present analysis is mainly focused on transcriptomic data associated with lung and blood immune cells from severe COVID-19 patients and matching cellular models of dexamethasone treatment. In particular, the COVID-19 samples used pertain to post-mortem lung samples (Blanco-Melo et al., 2020) and to peripheral blood immune cells including CD14+ monocytes, NK cells, CD8+ T cells, CD4+ T cells, and B cells (Wilk et al., 2020). On the other hand, the dexamethasone treated cells include A549 adenocarcinomic human alveolar basal epithelial cells (McDowell et al., 2018) and CD14+ monocytes isolated from peripheral blood donated by healthy human volunteers and differentiated in vitro into macrophages (Jubb et al., 2016). Although the extent to which the cancer cell line A549 would be relevant in viral infection may appear as a limitation of the analysis, it is notable that cellular assays of drug induced transcriptomic perturbations in human cancer cell lines and human primary cells are considered to show significant similarities and hence assumed to be interchangeable (Liu et al., 2020). Another limitation could be the absence of samples representing dexamethasone treatment in COVID-19. However, in the absence of such samples, it is an accepted bioinformatic approach to predict effectiveness of a drug in a given disease if gene expression signature associated with the drug show inverse correlation to that associated with the disease (Iwata et al., 2019). The underlying assumption is that the drug would suppress the disease related transcriptomic alterations. With these assumptions in place, the present analysis supports the hypothesis that broad anti-inflammatory activities of dexamethasone underlie its effectiveness in COVID-19.
2. Results
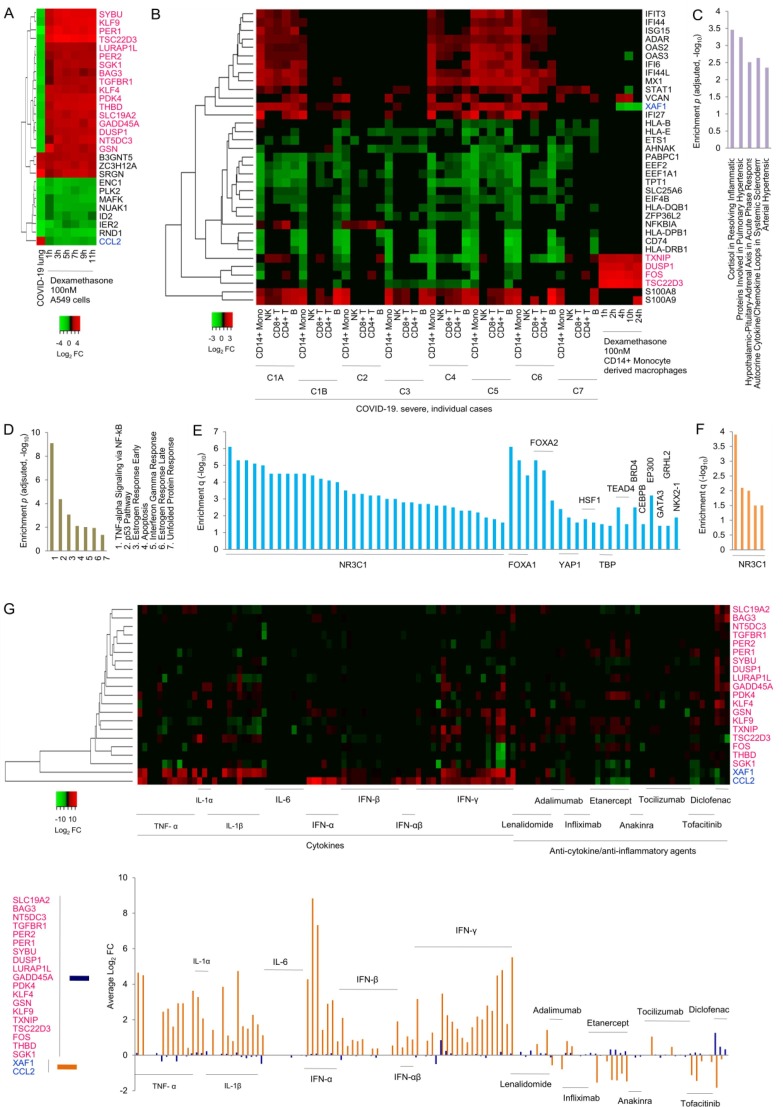
First, genes that are differentially expressed in lung samples from deceased COVID-19 patients compared to biopsied lung tissue from age-matched uninfected healthy subjects (Blanco-Melo et al., 2020) as well as across time series of dexamethasone treated A549 human lung alveolar epithelial cells compared to vehicle control (McDowell et al., 2018) were clustered together. Strikingly, the cluster showed that a majority of genes are regulated in opposite direction between disease and drug samples (Fig. 1 A). Eighteen of the total 28 differentially expressed genes (DEGs) which were common across samples showed opposite regulation, with 17 (SYBU, KLF9, PER1, TSC22D3, LURAP1L, PER2, SGK1, BAG3, TGFBR1, KLF4, PDK4, THBD, SLC19A2, GADD45A, DUSP1, NT5DC3, GSN) downregulated in the disease and upregulated in the drug group, and one (CCL2) upregulated in disease and downregulated in drug treatment. Given that TSC22D3, DUSP1, and SGK1, as mentioned above, are well known anti-inflammatory targets of NR3C1 mediated transcriptional activation, and CCL2 a well known pro-inflammatory target for NR3C1 mediated inhibition, the present analysis provided an unbiased support to the hypothesis that broad anti-inflammatory activities of dexamethasone underlie the latter's therapeutic effects in severe COVID-19.
Fig. 1.
Integrative transcriptome analysis. (A) Heatmap clustering of lung DEGs. Gene symbols shown in pink and blue indicate opposite regulation between COVID-19 and dexamethasone. COVID-19 sample represents post-mortem lung tissue of deceased patients compared to biopsied lung samples from age-matched uninfected healthy controls. A549 lung cancer cell line samples represent indicated drug treatment conditions in comparison to vehicle control. FC, fold change. Curated and annotated data along with log2 FC values are provided in Supplementary Table 1A. (B) Heatmap clustering of DEGs associated with COVID-19 immune cells (CD14+ monocytes, NK cells, CD8+ T cells, CD4+ T cells, and B cells) and CD14+ monocyte derived macrophages treated with dexamethasone under indicated conditions. C1A and C1B, and C2-C7 represent originally used patient symbols; for two disease stages in case 1 (C1A, C1B), and case 2 to case 7 (C2-C7). Curated and annotated data along with log2 FC values are presented in Supplementary Table 1B. Other details as mentioned in A. (C) Disease pathway enrichment. Top 5 terms are shown. Full results are given in Supplementary Table 1C. (D) MSigDB gene set enrichment. All enriched gene sets are shown. Detailed results are presented in Supplementary Table 1D. (E) Enrichment of transcription factor binding sites in lung. All enriched transcription factors are indicated. Each bar represents an experiment ID in NCBI's Sequence Read Archive. Detailed results are shown in Supplementary Table 1E. (F) Enrichment of transcription factor binding sites in blood. Complete set is shown. Each bar represents an experiment ID in NCBI's Sequence Read Archive. Detailed results are given in Supplementary Table 1F. (G) Clustering of DEGs associated with various cytokines, and anti-cytokine/anti-inflammatory agents in diverse cells and tissues. Heatmap is shown in the upper panel, and average log2 FC values for the two subsets of 19 and 2 genes in the lower panel. Curated and annotated data along with log2 FC values are provided in Supplementary Table 1G. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Next, a comparison of 35 most frequent DEGs among severe COVID-19 patient-specific transcriptomes of blood immune cells, namely, CD14+ monocytes, NK cells, CD8+ T cells, CD4+ T cells, and B cells, relative to healthy individuals (Wilk et al., 2020), and a time series of dexamethasone treated CD14+ monocyte derived macrophages relative to vehicle control (Jubb et al., 2016) remarkably showed that COVID-19 monocytes specifically cluster with dexamethasone treated monocyte derived macrophages (Fig. 1 B). Of a total of 9 DEGs that were associated with one or more dexamethasone samples, 4 (TXNIP, DUSP1, FOS, TSC22D3) showed upregulation in drug samples and downregulation in disease samples in general. One gene (XAF1) that was downregulated in dexamethasone samples showed upregulation in disease irrespective of cell types. Notably, like in lung samples, TSC22D3 and DUSP1 showed opposite regulation between disease related monocytes and drug related monocyte derived macrophages, supporting involvement of NR3C1 mediated transcriptional activation of anti-inflammatory genes in dexamethasone action in severe COVID-19.
The 21 genes identified above as regulated in opposite direction between disease and drug were next examined for enrichment of human disease pathways. Notably, these genes showed highest enrichment for cortisol in resolving inflammation, followed by other inflammation related pathways including hypothalamic–pituitary–adrenal axis in acute phase response and autocrine cytokine/chemokine loops in systemic scleroderma (Fig. 1 C). Similar analysis with Molecular Signatures Database (MSigDB) hallmark genes, which signify specific biological states or processes, showed enrichment for NF-kB mediated TNF-α signaling and IFN-γ response, besides others (Fig. 1 D). Further, analysis of transcription factor binding sites showed enrichment for NR3C1 binding sites in both lung (Fig. 1 E) and blood (Fig. 1 F). These findings supported involvement of dexamethasone's genomic mechanisms in COVID-19 treatment.
Finally, to independently validate the above gene set as predictive of therapeutic mechanisms, DEGs in diverse cells and tissues treated with different cytokines, and anti-cytokine or anti-inflammatory agents, compared to controls, were considered. The cytokines included TNF- α, IL-1α, IL-1β, IL-6, IFN-α, IFN-β, universal IFN-αβ, and IFN-γ. The anti-cytokine/anti-inflammatory agents included anti-TNF agents lenalidomide, adalimumab, infliximab and etanercept, IL-1α and IL-1β blocker anakinra, IL-6 antagonist tocilizumab, JAK-STAT signaling inhibitor tofacitinib, and non-steroidal anti-inflammatory drug diclofenac. Clustering of these DEGs revealed, as expected, an overall trend for opposite gene regulation between cytokines as a whole and anti-cytokine/anti-inflammatory agents as a whole, with the former group mimicking COVID-19 associated gene regulation and the latter dexamethasone associated (Fig. 1 G). Though the present analysis is based on highly diverse samples and hence may not be sufficient for separating individual cytokines and drugs in terms of their transcriptomic effects, it was notable that IL-6 among cytokines, and etanercept, tofacitinib and diclofenac among anti-cytokine/anti-inflammatory agents stand out from all others in their respective group (Fig. 1 G). IL-6 was almost devoid of any gene regulatory activity, whereas the etanercept, tofacitinib and diclofenac matched dexamethasone to certain extent in terms of effect on gene regulation.
3. Discussion
Results presented here support the hypothesis that broad anti-inflammatory effects of dexamethasone underlie its beneficial effects in severe COVID-19. The findings suggest involvement of both the major mechanisms by which glucocorticoids produce these effects, namely, transcriptional activation of anti-inflammatory genes and transcriptional suppression of pro-inflammatory genes. Conversely, immune dysregulation in COVID-19 involves both downregulation of anti-inflammatory genes and upregulation of pro-inflammatory genes. These mechanistic characteristics of disease and drug is consistent with the latter's effectiveness in the illness. As a corollary, agents that counter this dual dysregulation may possess therapeutic properties overlapping with dexamethasone in treatment of severe COVID-19. Given this, etanercept, tofacitinib, and diclofenac that showed this desirable activity may be considered promising. Etanercept, a fusion protein that acts as a soluble TNF receptor to prevent the pro-inflammatory cytokine TNF-α from activating the downstream inflammatory cascade, is one of the biologics, like the anti-TNF monoclonal antibodies adalimumab and infliximab, long in use for the treatment of rheumatoid arthritis (Mpofu et al., 2005). Given the association between elevated TNF levels and increased COVID-19 mortality, and as TNF blockade in rheumatoid arthritis is known to overall suppress pro-inflammatory cytokines implicated in COVID-19 related hyperinflammation, the biological plausibility for effectiveness of anti-TNF therapy in COVID-19 treatment is considered promising (Robinson et al., 2020aa, Robinson et al., 2020bb). Patient registry based studies also support the potential of anti-TNF agents in COVID-19 treatment (Robinson et al., 2020aa, Robinson et al., 2020bb). Clinical trials of adalimumab and infliximab in COVID-19 are currently at enrolment or pre-enrolment stages (Robinson et al., 2020). It may prove valuable to similarly assess etanercept for effectiveness in the illness. Regarding tofacitinib, it is a synthetic small molecule inhibitor of JAK family of kinases that can inhibit multiple JAK-dependent cytokine signaling pathways, and modulate, indirectly through autocrine and paracrine feedback mechanisms, production of other pro-inflammatory cytokines (Hodge et al., 2016). Multiple clinical trials of tofacitinib in COVID-19 have been undertaken (Luo et al., 2020), with findings yet to be reported. As regards diclofenac, though it is currently not a candidate treatment for severe COVID-19, a recent review of literature has supported the usefulness of non-steroidal anti-inflammatory drugs as an adjunct therapy in the illness (Zhao et al., 2020).
The observation of cytokine and anti-cytokine pair of IL-6 and tocilizumab not mimicking pathological and therapeutic gene expression signature identified here, in that order, likely suggests that dexamethasone mechanism in severe COVID-19 does not involve IL-6 signaling. Based on coexpression of NR3C1 and IL-6 in immune and nonimmune cells, it was previously suggested that therapeutic benefit of dexamethasone in severe COVID-19 possibly stems from inhibition of IL-6 production at systemic level as well as at local level in the lungs, supporting IL-6 antagonists like tocilizumab as potential treatments in the illness (Awasthi et al., 2020). Notably, in a recently reported randomized trial in hospitalized patients of COVID-19 with hypoxia and systemic inflammation, tocilizumab has been shown to improve survival and other clinical outcomes, over and above the benefits provided by systemic corticosteroids (RECOVERY Collaborative Group et al., 2021). The present analysis however does not support the above hypothesis that dexamethasone acts through IL-6 inhibition in COVID-19. Finally, given that synthetic glucocorticoids comprise of one of the most widely prescribed medicine in the world and are used in the treatment of a variety of inflammatory conditions (Freishtat et al., 2010), the proof-of-concept transcriptome analysis provided here in the specific context of dexamethasone and COVID-19 may serve as model for replication in other drug-disease combinations to infer underlying therapeutic mechanisms.
4. Methods
NCBI's PubMed and Gene Expression Omnibus (GEO) were extensively searched to identify relevant human datasets. Large and comparative studies were preferred, if available. The datasets were manually curated and annotated. Original author-identified DEGs were used, if provided in full. Otherwise, DEGs with adjusted p value significance, irrespective of fold change, were extracted using GEO2R (Barrett et al., 2013) for GEO microarray data, and GEO RNA-seq Experiments Interactive Navigator (GREIN) for RNA-seq data (Mahi et al., 2019). COVID-19 lung data represented altered gene expression in post-mortem lung tissue of deceased patients in comparison to that in biopsy samples of lung tissue obtained from age-matched uninfected healthy individuals (Blanco-Melo et al., 2020). The lung cancer cell line A549 represented transcriptomic alterations induced by treatment with 100 nM dexamethasone for 1 h, 3 h, 5 h, 9 h and 11 h, in comparison to vehicle control (McDowell et al., 2018). COVID-19 immune cells represented single-cell RNA-seq profiles of CD14+ monocytes, NK cells, CD8+ T cells, CD4+ T cells, and B cells in peripheral blood, with severe patients compared to healthy controls (Wilk et al., 2020). CD14+ monocyte derived macrophages represented gene expression changes induced by treatment with 100 nM dexamethasone for 1 h, 2 h, 4 h, 10 h and 24 h, compared to vehicle control (Jubb et al., 2016). Cytokine and cytokine inhibitor related transcriptional profiles involved diverse cell/tissue types and treatment conditions. Heatmap clustering was performed using Heatmapper (Babicki et al., 2016), with genes clustered using log2 fold change value 0 for non-DEGs, and the criteria scale type none, average linkage, and Euclidean distance. Average log2 fold change was calculated accordingly. Disease pathway enrichment analysis and MSigDB enrichment analysis were performed using Enrichr (Kuleshov et al., 2016), identifying significantly enriched terms based on adjusted p value. Transcription factor binding site enrichment was performed using Chip-Atlas (Oki et al., 2018), by selecting lung or blood as target tissue and other settings as default, and identifying significant enrichment on the basis of q value.
Declaration of Competing Interest
The author declares that he has no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gene.2021.145665.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Awasthi, S., Wagner, T., Venkatakrishnan, A.J., Puranik, A., Hurchik, M., Agarwal, V., et al. Plasma IL-6 Levels following corticosteroid therapy as an indicator of ICU length of stay in critically ill COVID-19 Patients. medRxiv 2020, 20144733. [DOI] [PMC free article] [PubMed]
- Babicki S., Arndt D., Marcu A., Liang Y., Grant J.R., Maciejewski A., Wishart D.S. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 2016;44(W1):W147–W153. doi: 10.1093/nar/gkw419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41(Database issue) doi: 10.1093/nar/gks1193. D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.-C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., Wang T.T., Schwartz R.E., Lim J.K., Albrecht R.A., tenOever B.R. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181(5):1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain D.W., Cidlowski J.A. After 62 years of regulating immunity, dexamethasone meets COVID-19. Nat. Rev. Immunol. 2020;20:587–588. doi: 10.1038/s41577-020-00421-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freishtat R.J., Nagaraju K., Jusko W., Hoffman E.P. Glucocorticoid efficacy in asthma: is improved tissue remodeling upstream of anti-inflammation. J. Investig. Med. 2010;58:19–22. doi: 10.231/JIM.0b013e3181b91654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge J.A., Kawabata T.T., Krishnaswami S., Clark J.D., Telliez J.B., Dowty M.E., et al. The mechanism of action of tofacitinib – an oral Janus kinase inhibitor for the treatment of rheumatoid arthritis. Clin. Exp. Rheumatol. 2016;34:318–328. [PubMed] [Google Scholar]
- Hudson W.H., de Vera I.M.S., Nwachukwu J.C., Weikum E.R., Herbst A.G., Yang Q., et al. Cryptic glucocorticoid receptor-binding sites pervade genomic NF-κB response elements. Nat. Commun. 2018;9:1337. doi: 10.1038/s41467-018-03780-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskar M., Zeller G., Blattmann P., Campillos M., Kuhn M., Kaminska K.H., et al. Characterization of drug-induced transcriptional modules: towards drug repositioning and functional understanding. Mol. Syst. Biol. 2013;9:662. doi: 10.1038/msb.2013.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata M., Yuan L., Zhao Q., Tabei Y., Berenger F., Sawada R., et al. Predicting drug-induced transcriptome responses of a wide range of human cell lines by a novel tensor-train decomposition algorithm. Bioinformatics. 2019;35:i191–i199. doi: 10.1093/bioinformatics/btz313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jubb A.W., Young R.S., Hume D.A., Bickmore W.A. Enhancer turnover is associated with a divergent transcriptional response to glucocorticoid in mouse and human macrophages. J. Immunol. 2016;196(2):813–822. doi: 10.4049/jimmunol.1502009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., McDermott M.G., Monteiro C.D., Gundersen G.W., Ma'ayan A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Zhu L., Thakkar S., Roberts R., Tong W. Can transcriptomic profiles from cancer cell lines be used for toxicity assessment? Chem. Res. Toxicol. 2020;33:271–280. doi: 10.1021/acs.chemrestox.9b00288. [DOI] [PubMed] [Google Scholar]
- Luo W., Li Y.X., Jiang L.J., Chen Q., Wang T., Ye D.W. Targeting JAK-STAT signaling to control cytokine release syndrome in COVID-19. Trends Pharmacol. Sci. 2020;41:531–543. doi: 10.1016/j.tips.2020.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macauley P., Martin A., Epelbaum O. Corticosteroids in the treatment of severe COVID-19 lung disease: the pulmonology perspective from the first United States epicentre. Int. J. Infect. Dis. 2020;100:309–313. doi: 10.1016/j.ijid.2020.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahi N.A., Najafabadi M.F., Pilarczyk M., Kouril M., Medvedovic M. GREIN: An Interactive Web Platform for Re-analyzing GEO RNA-seq Data. Sci. Rep. 2019;9:7580. doi: 10.1038/s41598-019-43935-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell I.C., Manandhar D., Vockley C.M., Schmid A.K., Reddy T.E., Engelhardt B.E. Clustering gene expression time series data using an infinite Gaussian process mixture model. PLoS Comput. Biol. 2018;14 doi: 10.1371/journal.pcbi.1005896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mpofu S., Fatima F., Moots R.J. Anti-TNF-alpha therapies: they are all the same (aren't they?) Rheumatology (Oxford) 2005;44:271–273. doi: 10.1093/rheumatology/keh483. [DOI] [PubMed] [Google Scholar]
- Oki S., Ohta T., Shioi G., Hatanaka H., Ogasawara O., Okuda Y., et al. ChIP-Atlas: a data-mining suite powered by full integration of public ChIP-seq data. EMBO Rep. 2018;19 doi: 10.15252/embr.201846255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quatrini L., Ugolini S. New insights into the cell- and tissue-specificity of glucocorticoid actions. Cell. Mol. Immunol. 2020;31:1–10. doi: 10.1038/s41423-020-00526-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RECOVERY Collaborative Group, Horby PW, Pessoa-Amorim G, Peto L, Brightling CE, Sarkar R et al. Tocilizumab in patients admitted to hospital with COVID-19 (RECOVERY): preliminary results of a randomised, controlled, open-label, platform trial. medRxiv 2021.02.11.21249258.
- RECOVERY Collaborative Group, Horby P, Lim WS, Emberson JR, Mafham M, Bell JL et al. Dexamethasone in hospitalized patients with Covid-19 – preliminary report. N. Engl. J. Med. 2020; NEJMoa2021436.
- Robinson P.C., Liew D.F.L., Liew J.W., Monaco C., Richards D., Shivakumar S., et al. The potential for repurposing anti-TNF as a therapy for the treatment of COVID-19. Med (N Y) 2020;1:90–102. doi: 10.1016/j.medj.2020.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson P.C., Richards D., Tanner H.L., Feldmann M. Accumulating evidence suggests anti-TNF therapy needs to be given trial priority in COVID-19 treatment. Lancet Rheumatol. 2020;2(11):e653–e655. doi: 10.1016/S2665-9913(20)30309-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Lachmann A., Ma'ayan A. Mining data and metadata from the gene expression omnibus. Biophys. Rev. 2019;11:103–110. doi: 10.1007/s12551-018-0490-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk A.J., Rustagi A., Zhao N.Q., Roque J., Martínez-Colón G.J., McKechnie J.L., et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D., Zhang S., Igawa T., Frishman W. Use of nonsteroidal anti-inflammatory drugs for COVID-19 infection: adjunct therapy? Cardiol. Rev. 2020;28:303–307. doi: 10.1097/CRD.0000000000000340. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.