Abstract
Purpose:
Nicotinamide phosphoribosyltransferase (NAMPT) inhibitors are currently in development, but may be limited as a single agent therapy due to compound-specific toxicity and cancer metabolic plasticity allowing resistance development. To potentially lower the doses of NAMPT inhibitors required for therapeutic benefit against acute myeloid leukemia (AML), we performed a genome-wide CRISPRi screen to identify rational disease-specific partners for a novel NAMPT inhibitor, KPT-2974.
Experimental Design:
Cell lines and primary cells were analyzed for cell viability, self-renewal and responses at RNA and protein levels with loss-of-function approaches and pharmacologic treatments. In vivo efficacy of combination therapy was evaluated with a xenograft model.
Results:
We identified two histone deacetylases, HDAC8 and SIRT6, whose knockout conferred synthetic lethality with KPT-9274 in AML. Furthermore, HDAC8-specific inhibitor, PCI-34051, or clinical Class I HDAC inhibitor, AR-42, in combination with KPT-9274, synergistically decreased the survival of AML cells in a dose-dependent manner. AR-42/KPT-9274 co-treatment attenuated colony-forming potentials of patient cells while sparing healthy hematopoietic cells. Importantly, combined therapy demonstrated promising in vivo efficacy compared with KPT-9274 or AR-42 monotherapy. Mechanistically, genetic inhibition of SIRT6 potentiated the effect of KPT-9274 on PARP-1 suppression by abolishing mono-ADP ribosylation. AR-42/KPT-9274 co-treatment resulted in synergistic attenuation of homologous recombination (HR) and nonhomologous end joining (NHEJ) pathways in cell lines and leukemia initiating cells (LICs).
Conclusions:
Our findings provide evidence that HDAC8 inhibition- or shSIRT6-induced DNA repair deficiencies are potently synergistic with NAMPT targeting, with minimal toxicity towards normal cells, providing a rationale for a novel-novel combination-based treatment for AML.
Keywords: KPT-9274, HDAC8, SIRT6, HR, NHEJ, CRISPR screen
Introduction
Acute myeloid leukemia (AML) is the most commonly diagnosed leukemia in adults (1,2). The overall prognosis of the disease remains poor, with a 5-year survival rate of less than 10% for patients over 60 years old. AML cells are addicted to nicotinamide phosphoribosyltransferase (NAMPT)-mediated salvage pathway for the biosynthesis of NAD+(3,4), which serves as a co-enzyme for redox reactions and substrate for poly (ADP-ribose) polymerase (PARP) regulating DNA damage repair (DDR) gene expression and stress responses. Thus, targeting the NAMPT-dependent NAD+ generation has gained attention as a potential therapeutic strategy in AML and NAMPT inhibitors (NAMPTis) have moved to phase I trials. Our previous studies demonstrated preclinical efficacy of targeting NAMPT on eliminating AML in vitro and in vivo by employing a potent NAMPTi, KPT-9274 (5). Although our pre-clinical data are compelling, we recognize that NAMPTis will likely be ineffective as monotherapies based upon AML metabolic plasticity ultimately permitting resistance (6). In addition, dose-limiting toxicities pose another barrier for clinical success of the inhibitor (7,8). In clinical trials with the 1st generation NAMPTis, dose-limiting toxicities were observed, such as thrombocytopenia and gastrointestinal, retinal and cardiac toxicities(9). Unlike 1st generation NAMPTi, KPT-9274 was better tolerated in Phase I trials on solid tumor and non-hodgkins lymphoma, but drug-related adverse events including anemia and fatigue were reported. Therefore, strengthening the clinical efficacy of NAMPTis through rational combination therapies represents an unmet need in AML.
To address this, we have utilized an unbiased genome-wide CRISPR screen to identify genes that upon depletion confer sensitivity to KPT-9274 in AML cells. Among the top ranked hits, we identified two genes encoding NAD+-dependent histone deacetylases, namely HDAC8 and SIRT6, both with known roles in compensatory DDR pathways. We next determined the efficacy of combined NAMPTi with genetic or pharmacologic inhibition of HDAC8 or SIRT6 in AML cell lines and primary patient samples, and investigated the mechanisms that may contribute to the effect. We hypothesize that HDAC8i or SIRT6i-mediated DDR deficiencies sensitize AML cells to synthetic lethality orchestrated by NAMPTi. Therefore, concurrent inhibition of NAMPT and factors involved in compensatory DDR pathways may achieve our goals of improving the therapeutic index and effectiveness of NAMPTis in AML.
Materials and Methods
Genome-wide loss-of-function screen
The human GeCKOv2 CRISPR knockout library was obtained from Addgene. The library was amplified in bacteria and packaged into viral particles in HEK293FT cells. MOLM13 cells were transduced with lentiviral particles at a pre-determined ratio in the presence of polybrene and spinoculated at 450xg for 90 minutes. Puromycin selection was initiated after 48 hours and continued for 7 days to eliminate cells with essential gene-targeting single guide RNA (sgRNA) and non-transduced cells. The transduced cells were cultured for 3 days in the presence of 50 nM KPT-9274 or DMSO to negatively select sgRNAs that sensitize resistant cells to KPT-9274. Cells were collected on days 0 and 3 and subject to P5/P7 barcoding and deep sequencing using Illumina HiSeq4000 sequencer to detect the abundance of each sgRNA. Sequencing data were analyzed using MAGeCK VISPR algorithm to discover sgRNAs that were negatively or positively selected with KPT-9274.
Drugs
KPT-2974, AR-42, AG221, AG120, and PCI-34051 were purchased from Selleckchem. For in vivo study, AR-42 was formulated in 0.5% methylcellulose [w/v] and 0.1% Tween-80 [v/v] in sterile water.
Cell lines and shRNA transfection
MOLM13 (DSMZ Cat# ACC-554, RRID:CVCL_2119), MV4-11 (DSMZ Cat# ACC-102 RRID:CVCL_0064) and OCI-AML3 (DSMZ Cat# ACC-582, RRID:CVCL_1844) were purchased from DSMZ (Braunschweig, Germany) and cultured in RPMI1640 (Gibco) supplemented with 10% fetal bovine serum (FBS). Kasumi-1 cells (ATCC Cat# CRL-2724, RRID:CVCL_6911) were purchased from ATCC (Manassas, VA) and cultured in RPMI1640 (Gibco) supplemented with 20% FBS. Isocitrate dehydrogenase2 (IDH2)R140Q (ATCC Cat# CRL-2003IG, RRID:CVCL_UE10) and wildtype (WT) TF-1 cells (ATCC Cat# CRL-2003, RRID:CVCL_0559) were obtained from ATCC (Manassas, VA) and cultured in RPMI1640 (Gibco) supplemented with 10% FBS and 2 ng/ml recombinant human GM-CSF. MOLM13-luciferase cells were a kind gift from Dr. Ramiro Garzon (Ohio State University). HEK293FT cells were obtained from Life Technologies and cultured in DMEM (Gibco) with 10% FBS. All cell lines were used between passage three and twenty and routinely tested negative for mycoplasma contamination with Universal Mycoplasma Detection Kit (ATCC 30-1012K). Cells were authenticated with microsatellite genotyping (short tandem repeat analysis by the Ohio State University (OSU) Genomic Services Core). For shRNA transfection, shRNA oligos for knockdown of SIRT6, HDAC8, DCPS, PTGS1, HEXA and IMPA2 in lentiviral vector pLKO.1 were purchased from Sigma (Sigma-Aldrich, St. Louis, MO). shRNA sequences are listed in Supplementary Methods. All viruses were produced using the HEK293FT cells with packaging and envelope plasmids, psPax2 and VSVG. Cells were transduced by spinofection at 1,500 rpm for 90 minutes in the presence of polybrene. Seventy-two hours after transduction, puromycin was added to the culture to select shRNA-stable clones.
Patient samples
AML patient and normal donor samples were obtained from OSU Leukemia Tissue Bank. Informed written consent was obtained from each subject or each subject’s guardian under an institutional review board–approved protocol according to the Declaration of Helsinki.
MTS assay
AML cell lines or patient-derived cells were treated in a 96-well plate at 0.2-6 × 105 cells per well for 24-72 hours. Patient-derived CD34+ primary cells were cultured in 96-well plates coated with collagen in the presence of 10 ng/ml IL-3, 10 ng/ml IL-6, 10 ng/ml SCF and 10 ng/ml GM-CSF. MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] (Promega, Madison, WI) was incubated with drug-treated cells according to the manufacturer’s instructions. Plates were read by using a BioTek synergy H4 hybrid multimode microplate reader (Thermo Fisher Scientific) at 490 nM. Combenefit software was used to calculate synergy scores for pairwise combinations of drug doses.
Colony-forming unit (CFU) assays
For CFU assays, viable cells were plated at optimal densities in MethoCult H0435 (StemCell Technologies, Vancouver, Canada) in the presence of DMSO, 0.1 μM KPT-9274, 0.8 μM AR-42, or combination of inhibitors. Formed colonies were counted blindly after 7-14 days and re-plated at 10,000 cells/well. The images of colonies were captured with Echo revolving microscope. The images of Giemsa staining of cytospin preparations were acquired by CX43 Olympus inverted microscope.
EJDR reporter assay
EJDR reporter cells (10) were plated at 5X 105 cells per well in 10% FBS and RPMI1640. After drug treatments, cells were grown in 10% Tet-free FBS (100-800; Gemini) and RPMI1640. Incorporated I-Sce1 was induced with Shield1 (632189; Clontech) and triamcinolone (TA) (T6510; Sigma Aldrich) ligands for 24 hours. NHEJ and HR repair activities were assessed 48 hours post induction by quantification of DsRed− and GFP-positive cells on Cytomics FC 500 Flow Cytometer (Beckman Coulter, Brea, CA).
Limiting cell-RNA-seq (LC-RNA-seq) for primary patient LICs
LC-RNA-seq was performed as previously described (11). Briefly, patient cells were cultured in the presence of cytokines and treated with vehicle, single agents or drug combination. 300 viable LICs from treated patient samples were sorted directly into SMART-seq lysis buffer with FACSAria Fusion (BD) based on putative stem cell markers CD34+CD38−(12). Initiating/stem cells were enriched in sorted CD34+CD38− cells, as CD34+CD38− subpopulations grew significantly more colonies than CD34+CD38+ subpopulations in CFU assays (Unpublished observation). The Clontech SMARTer v4 kit (Takara Bio USA, Inc., Mountain View, CA) was used for pre-amplifying samples prior to library construction with the Nextera XT DNA Library Prep Kit (Illumina, Inc., San Diego, CA). Samples were sequenced to a depth of 15–20 million 2 × 150 bp clusters with the Illumina HiSeq 4000 platform. After quality control with CLEAR selection process and size-normalization, DEGs were called with DESeqs with (FDR) adjusted p-value <0.05. Principle component analysis (PCA) plots were created from count tables which were normalized by size and transformed. For pathway analysis, the list of DEGs, containing gene IDs and corresponding expression values, was uploaded into the IPA software (Qiagen) (Ingenuity Pathway Analysis, RRID:SCR_008653). The “core analysis” function in the software was employed to interpret the differentially expressed data and identify top enriched pathways.
In vivo MOLM13 xenograft study
All experiments were approved by the OSU Institutional Animal Care and Use Committee. Male NOD-Prkdcem26Cd52Il2rgem26Cd22/NjuCrl (NCG) mice (RRID:IMSR_CRL:572) (aged 6~8 weeks) were obtained from Charles River Laboratory. Mice were group-housed under conditions of constant photoperiod (12-hour light/12-hour dark), temperature, and humidity with ad libitum access to water and irradiated standard pelleted chow. 1X104 luciferase-tagged MOLM13 cells were injected via tail vein into NCG mice. On day 5 post-engraftment, mice were randomized to receive vehicle once daily, 100 mg/kg of KPT-9274 once daily, 20 mg/kg AR-42 every another day or the combination of KPT-9274 and AR-42 via oral gavage. Overall survival was the primary end point for the majority of the mice. A separate cohort of mice per group was used to monitor leukemic progression using IVIS imaging. Mice were monitored by animal technicians who were blinded to the treatment groups and determined when mice met early removal criteria (20% weight loss, lethargy, hunching, and poor body condition). Mice were euthanized by CO2 inhalation.
Statistical analysis
Data are presented as the mean ± SEM of independent experiments, unless otherwise specified. Statistical analyses were performed with GraphPad Prism 7.0 (GraphPad Prism, RRID:SCR_002798) or SAS/STAT software (version 9.0) with ANOVA with Tukey’s post-test correction for multigroup comparisons or a 2-tailed Student’s t test for 2-group comparisons, unless otherwise specified in the figure legends. A p-value of less than 0.01 or 0.05 was considered significant. With 8 mice/group there is 80% power to detect a difference between treated vs control group at a 5% significance level. For survival analyses, a Cox proportional hazard model was used to determine statistical significance. For CFU measurements, a negative binomial model was used to fit data.
Results
Genome-wide CRISPR screen identifies synthetic lethal partners for NAMPTi, KPT-9274, in AML cells
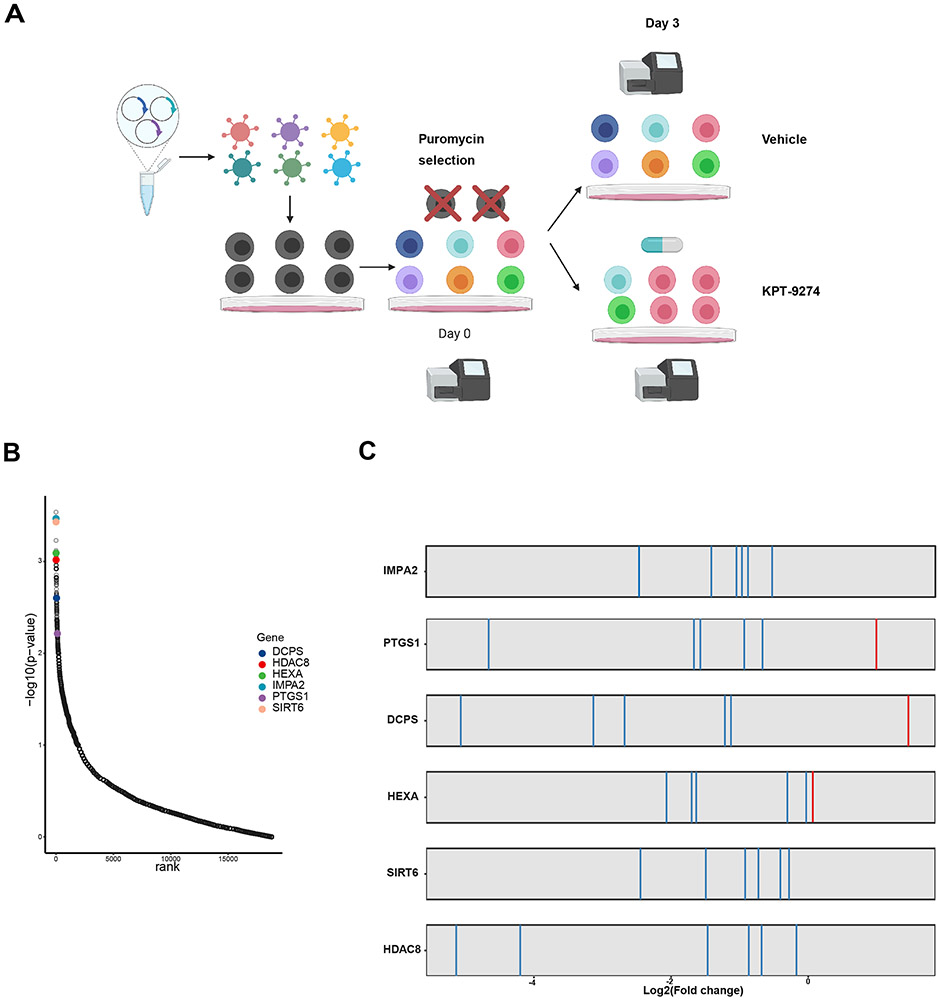
To perform the CRISPR-Cas9 screen, we used the lentivirus-based GeCKOv2 library, which contains ~130,000 sgRNAs targeting ~20,000 protein-coding genes and microRNAs(~6 sgRNAs per gene) (13). The library was packaged and transduced into Cas9-expressing MOLM13 cells (5). Then, transduced cells were selected in puromycin and received sublethal dose of 50 nM KPT-9274 or DMSO control for 3 days. After mapping sgRNA sequence reads, changes in abundance of each sgRNA were assessed using the MAGeCK program (14) (Figure 1A). 400× coverage of the library was achieved with 95% of the sgRNA sequences being retained in all samples, ensuring the sufficient sgRNA representation. By analyzing the sgRNAs that were negatively selected in the presence of KPT-9274, several genes were identified as top hits with small p-values (Figure 1B and Table 1). We prioritized top hits with available preclinical or clinical inhibitors (i.e. HDAC8, SIRT6, HEXA, IMPA2, PTGS1 and DCPS) for validation. For these hits, high proportion of sgRNAs targeting the same gene exhibited large fold depletion following KPT-9274 treatment (Figure 1C). In this unbiased manner, gene candidates whose depletions allow the broader therapeutic index and overcome NAMPTi insensitivity were nominated.
Figure 1. Genome-wide functional screening identifies two histone deacetylase genes as combination targets for NAMPTi.
A) Schematic illustration of CRISPR screen workflow. CRISPR screen was conducted by transfecting GeCKOv2 library into MOLM13 cells. Virus MOI was titrated to ensure a single sgRNA to be transduced into each cell. 50 nM KPT-9274 or DMSO vehicle was administrated to CRISPR library-transfected MOLM13 cells after puromycin selection. B) Top scored genes ranked by their −log(p-values) and displayed against each gene’s corresponding ranks of essentiality. Top hits for validation are highlighted. p-values of genes were calculated based on sgRNA efficiency. C) sgRNAs targeting selected top hits are constantly depleted following KPT-9274 treatment.
Table 1.
List of CRISPR screen top ranked hits calculated with MAGeCK VISPR
| Rank | Genes | score | p-value |
|---|---|---|---|
| 1 | CEP41 | 2.70E-05 | 0.00016 |
| 2 | C11orf52 | 3.40E-05 | 0.00019 |
| 3 | PRKAR2B | 4.60E-05 | 0.00023 |
| 4 | ARHGEF10 | 4.70E-05 | 0.00023 |
| 5 | TOMM5 | 5.90E-05 | 0.00029 |
| 6 | IMPA2 | 7.00E-05 | 0.00034 |
| 7 | SIRT6 | 7.70E-05 | 0.00037 |
| 8 | XKRY | 0.0001 | 0.00051 |
| 9 | ZNF343 | 0.00012 | 0.00058 |
| 10 | KLHL2 | 0.00012 | 0.00059 |
| 11 | KCNH2 | 0.00013 | 0.0006 |
| 12 | PRSS42 | 0.00015 | 0.00076 |
| 13 | MSN | 0.00015 | 0.00076 |
| 14 | TM6SF2 | 0.00016 | 0.0008 |
| 15 | HEXA | 0.00016 | 0.00081 |
| 16 | OR10H4 | 0.00017 | 0.00087 |
| 17 | HDAC8 | 0.00018 | 0.00096 |
| 18 | DUSP12 | 0.0002 | 0.001 |
| 19 | SEMA4C | 0.00024 | 0.0011 |
| 20 | LSMEM1 | 0.00025 | 0.0012 |
| 21 | PDIA6 | 0.00028 | 0.0012 |
| 22 | CRIP1 | 0.00029 | 0.0012 |
| 23 | HPSE | 0.0003 | 0.0012 |
| 24 | ZNF454 | 0.00032 | 0.0014 |
| 25 | CNTN6 | 0.00034 | 0.0015 |
| 26 | KMT2B | 0.00034 | 0.0015 |
| 27 | DNAJB7 | 0.00036 | 0.0016 |
| 28 | SLC17A2 | 0.00036 | 0.0016 |
| 29 | ACYP2 | 0.0004 | 0.0018 |
| 30 | OR4C16 | 0.00048 | 0.0022 |
| 31 | SLC41A2 | 0.00048 | 0.0022 |
| 32 | DCPS | 0.00053 | 0.0025 |
| 33 | CSF3R | 0.00057 | 0.0025 |
| 34 | CTSG | 0.00058 | 0.0028 |
| 35 | MOB1B | 0.00059 | 0.0028 |
| 36 | ZNF837 | 0.00065 | 0.0032 |
| 37 | FAM214A | 0.00067 | 0.0033 |
| 38 | NDUFA11 | 0.00068 | 0.0033 |
| 39 | UBE2J2 | 0.00071 | 0.0035 |
| 40 | RBAK | 0.00072 | 0.0036 |
| 41 | PDZK1IP1 | 0.00074 | 0.0037 |
| 42 | SLC25A6 | 0.00077 | 0.004 |
| 43 | MYO5A | 0.00079 | 0.0043 |
| 44 | BSN | 0.00085 | 0.0044 |
| 45 | ASF1A | 0.00085 | 0.0044 |
| 46 | MUC12 | 0.00087 | 0.0044 |
| 47 | KIAA1161 | 0.00092 | 0.0045 |
| 48 | CLPB | 0.0011 | 0.0051 |
| 49 | CXorf40B | 0.0011 | 0.0052 |
| 50 | BCL9 | 0.0011 | 0.0054 |
| 51 | GJC1 | 0.0011 | 0.0054 |
| 52 | CACNG3 | 0.0012 | 0.0057 |
| 53 | ATL3 | 0.0012 | 0.0061 |
| 54 | MTL5 | 0.0012 | 0.0061 |
| 55 | PTGS1 | 0.0012 | 0.0061 |
| 56 | ADAT2 | 0.0013 | 0.0061 |
| 57 | RASGRP2 | 0.0013 | 0.0061 |
| 58 | MRPL38 | 0.0013 | 0.0063 |
| 59 | ZNF224 | 0.0014 | 0.0067 |
| 60 | ZNF564 | 0.0015 | 0.0068 |
| 61 | HCN1 | 0.0015 | 0.0068 |
| 62 | PSEN2 | 0.0015 | 0.0071 |
| 63 | OR4D10 | 0.0015 | 0.0071 |
| 64 | ACVRL1 | 0.0015 | 0.0078 |
| 65 | STX5 | 0.0015 | 0.0081 |
| 66 | VSIG4 | 0.0015 | 0.0081 |
| 67 | ZNF778 | 0.0015 | 0.0081 |
| 68 | SLC35E3 | 0.0016 | 0.0081 |
| 69 | SLC30A9 | 0.0017 | 0.0086 |
| 70 | IL9 | 0.0017 | 0.0086 |
| 71 | IL1A | 0.0017 | 0.0087 |
| 72 | FAM19A2 | 0.0017 | 0.0087 |
| 73 | HOXD11 | 0.0018 | 0.0089 |
| 74 | KLHL18 | 0.0018 | 0.009 |
| 75 | CAP1 | 0.0018 | 0.0091 |
| 76 | GABRA3 | 0.0018 | 0.0091 |
| 77 | MTMR4 | 0.0018 | 0.0091 |
| 78 | C19orf40 | 0.0018 | 0.0092 |
| 79 | GPRC5B | 0.0018 | 0.0092 |
| 80 | CNTN2 | 0.0019 | 0.0096 |
| 81 | CHODL | 0.0021 | 0.0091 |
RRA model integrates the sgRNA-level fold change and p-values to identify interesting gene hits. RRA enrichment score and p-value for each gene are shown.
Genetic depletion of histone deacetylases, HDAC8 and SIRT6, sensitizes AML cells to NAMPT inhibition
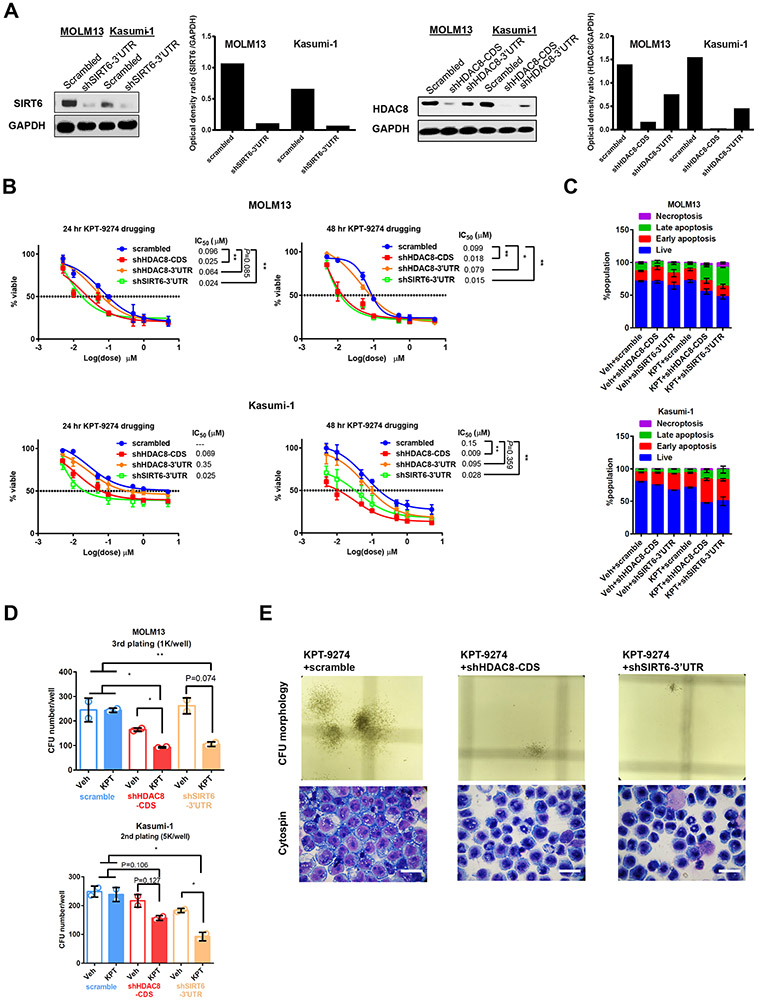
To validate the results of the CRISPR screen, we first demonstrated genetically the drug-co-essential nature of these dropout genes on KPT-9274-sensitive cell line, MOLM13, and KPT-9274-insensitive cell line, Kasumi-1. Four CDS-targeting shRNAs and one 3’UTR-targeting shRNA were designed per gene. CDS-targeting oligos were pooled and packaged into lentivirus. After puromycin selection, HDAC8-CDS targeting shRNAs effectively reduced protein expression in Kasumi-1 and MOLM13 cells, while 3'UTR-targeting oligo only marginally decreased HDAC8 abundance (Figure 2A). SIRT6 3'UTR-targeting shRNA decreased the SIRT6 level by 80%.
Figure 2. Genetic depletion of HDAC8 or SIRT6 increases vulnerability of AML cell lines to KPT-9274 treatment.
A) Knockout efficiency of CDS- and 3’UTR-targeting shRNAs in MOLM13 and Kasumi-1 cells as assessed with Western blotting. GAPDH serves as loading control. Densitometric quantification of protein band intensity is shown. n=2. B) Depletion of HDAC8 or SIRT6 sensitizes AML cell lines to KPT-9274 treatment. Dose-response curves of shRNA-stable MOLM13 and Kasumi-1 cells for 24-hour and 48-hour treatments are plotted. Cell viability was measured with MTS. Results are shown as mean±SEM of 4 technical replicates and 3~4 biological replicates. *p<0.05;**p<0.01. p-values for 24 hour KPT-9274 treatment on Kasumi-1 were not determined, as drug treatment did not achieve IC50 on scrambled control. C) Depletion of HDAC8 or SIRT6 with shRNA synergizes with KPT-9274 to enhance apoptosis of AML cell lines. Scrambled or gene-targeting shRNA-stable MOLM13 or Kasumi-1 cells were treated with IC20 dose of KPT-9274 for 48 hours and stained with Annexin/PI for flow cytometry analysis. Data are shown as mean±SEM of % population from duplicates. D) KPT-9274 attenuates AML self-renewal and induces myeloid differentiation in HDAC8 or SIRT6-depleted cells. Quantification of the number of colonies formed by shRNA-stable Kasumi-1 and MOLM13 cells in the presence or absence of KPT-9274 in CFU assays. Indicated number of cells were seeded into Methocult and colonies were counted and re-plated by a double-blind approach after 7 days. Results are shown as mean±SEM of duplicates. *p<0.05. **p<0.01. E) Representative images of colony morphologies and Giemsa staining of cytospin preparations of cells derived from KPT-9274-treated MOLM13 CFU. For each condition, images were randomly taken from 6~12 fields of view. Scale bar=50 μm.
shHDAC8-CDS or shSIRT6-3’UTR transfection reduced IC50 doses of KPT-9274 for MOLM13 and Kasumi-1, resulting in a significant reduction of cell survival (Figure 2B). The vulnerabilities of shHDAC8-3’UTR-transfected cells to KPT-9274 treatment were comparable with those of scramble-transfected cells, mirroring the low knockdown efficiency of this shRNA construct. These results imply that genetic depletion of HDAC8 decreased tumor survival in the presence of KPT-9274 proportional to the reduction in HDAC8 abundance. Knockdown of IMPA2, HEXA, PTGS1 and DCPS did not demonstrate strong synergies with KPT-9274 treatments, suggesting that they might be false positive hits from the screen (Supplementary Figure S1A). In the presence of KPT-9274 at sublethal IC20 doses, HDAC8 depletion resulted in a significant increase in the percentage of apoptotic cells in MOLM13 and Kasumi-1 cell lines (Figure 2C). Likewise, SIRT6 depletion and KPT-9274 cooperate to enhance apoptosis of AML cells. These results provided additional evidence about histone deacetylase dependency of KPT-9274-treated AML cells for survival.
Next, we examined the serial replating potentials of shRNA-stable cell lines following NAMPT inhibition. In the scramble groups, the colony numbers formed by cells treated with sublethal doses of KPT-9274 and vehicle control were comparable (Figure 2D). On the contrary, in HDAC8- or SIRT6-depleted MOLM13 and Kasumi-1 cells, KPT-2974 treatment suppressed colony formation and decreased long-term self-renewal cell populations (Figure 2D). Knockdown of HDAC8 or SIRT6 reduced the size of colonies and rendered them more compact with KPT-9274 (Fig. 2E). To evaluate the effect of treatments on cell differentiation, we stained cytospin preparations of cells derived from CFU assays. KPT-9274 treatment and depletion of HDAC8 or SIRT6 cooperatively differentiated AML cells (Figure 2E). The blasts morphologically became more mature with condensed chromatin and fragmented nuclei.
Pharmacological inhibition of HDAC8 confers vulnerability of AML subtypes to NAMPTi
To find translational relevance, we pharmacologically inhibited HDAC8 with HDAC8-specific PCI-34051 and assessed potential synergistic effects of this inhibitor with KPT-9274 on AML cells (15). PCI-34051 and KPT-9274 synergistically reduced cell survival, as assessed by the HSA independence model (Supplementary Figure S2A-C). Drug synergistic effect was most pronounced for 50 μM PCI-34051 in combination with 0.01 μM KPT-9274 in MV4-11 cells and 0.1 μM KPT-9274 in Kasumi-1 cells. MOLM13 cells exhibited synergy over a broader range, as the combination of PCI-34051 at 50 μM and KPT-9274 at doses from 0.01 μM to 0.1 μM displayed the strongest synthetic lethality. It was reported that unlike the pan-HDAC inhibitor PCI-24781, PCI-34051 at a concentration as high as 50 μM, did not induce significant tubulin acetylation, implying that it did not exert its cytotoxicity through inhibition of other HDAC isoforms (15).These data provide evidence that HDAC8 inhibition functions synergistically with NAMPT Inhibition to reduce AML viability. Nevertheless, the human equivalent dose of 50 μM of PCI-34051 is not considered to be clinically tolerable. It was reported that PCI-34051 failed to induce AML death at low micromolar doses, although it caused apoptosis on T cell leukemia in a caspase-dependent manner(15). It is possible that PCI-34051 was not metabolized effectively in AML to hit HDAC8 targets.
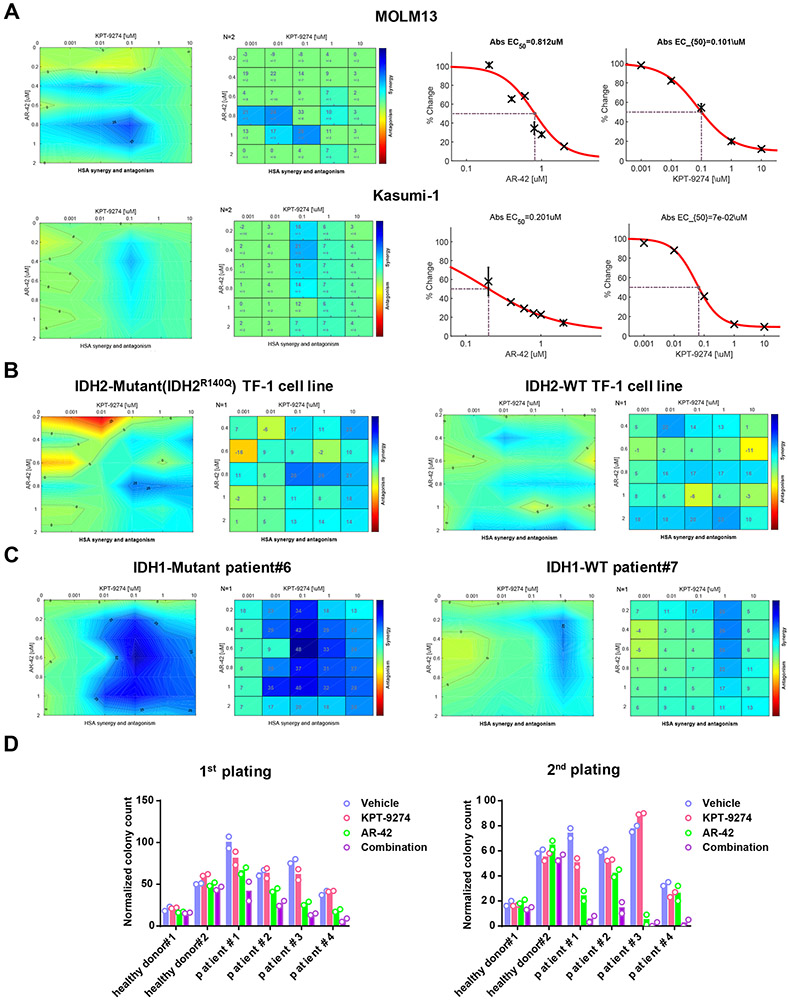
AR-42 is a potent inhibitor for class I/II HDACs, including HDAC8 (16). AR-42 entered phase I clinical trials in combination with decitabine in AML and was shown to eradicate leukemia stem cells (17-19). Depletion of HDAC1, HDAC3 or HDAC6 with CRISPR did not have significant impact on KPT-9274 sensitivity (Supplementary Figure S1B). This excluded the possibility of involvement of other HDAC isoforms in AR-42 efficacy. Therefore, we employed AR-42 as a tool compound to delineate the effect of HDAC8 inhibition on the sensitivity of AML to KPT-9274 treatment. The highest synergistic scores were observed at combination doses of 0.1 μM KPT-9274 and 0.8 μM AR-42 for MOLM13 and of 0.1 μM KPT-9274 and 0.4 μM AR-42 for Kasumi-1 (Figure 3A). Co-treatment of KPT-9274 and AR-42 also exhibited synergy on other cell lines, although to a lesser extent (Supplementary Figure S2D-E). To determine the effect of combined treatment on mitochondria respiration, we measured mitochondria membrane potential of treated cells with TMRM staining. In comparison with single agents, combined treatment attenuated TMRM fluorescence intensity and concomitantly elevated Annexin V staining in MOLM13, Kasumi-1 and MV4-11 (Supplementary Figure S3A-C). This suggests that disruption of mitochondria function may account for the observed synergism between KPT-9274 and AR-42 on AML. AR-42 and KPT-9274 co-treatment also significantly increased the percentage of apoptotic cells in a dose-dependent manner (Supplementary Figure S3D).
Figure 3. Pan-HDAC inhibitor, AR-42, and KPT-9274 synergistically reduce survival and self-renewal of AML cells.
A) Drug synergy matrix plots for the combination of AR-42 and KPT-9274 in AML cell lines, MOLM13 and Kasumi-1. MOLM13 and Kasumi-1 cells were treated with a range of dosages of AR-42 and KPT-9274 for 48 and 72 hours. Dose response curves and EC50s for single agents are shown. n=2. B) IDH2-mutant and IDH2-wildtype (WT) TF-1 erythroleukemia cell lines and C) IDH1-mutant and -WT patient cells were treated with pairwise combinations of AR-42 and KPT-9274. After treatment, MTS was added to the plate to assess mitochondria metabolism. Synergy scores were calculated with Combenefit based on HSA model. D) Quantification of CFU colonies formed by CD34+ healthy bone marrow cells and AML patient cells which were treated with vehicle, 0.1 μM KPT-9274, 0.8 μM AR-42 or drug combination.
It was reported that IDH1 and IDH2 mutations inhibit the expressions of BRCA1/2 and ATM proteins and induce HR repair defects by producing 2-hydroxyglutarate (2HG), leading to synthetic lethality triggered by PARP inhibitors (20). Here, we evaluated the effect of IDH1 or IDH2 mutation on cellular sensitivity to combined treatment of KPT-9274 and AR-42 by employing IDH2-mutant and -WT TF-1 cell lines as testing system. The most synergistic area was achieved when 0.1-1 μM KPT-9274 was combined with 0.8 μM of AR-42 in IDH2mut cells (Figure 3C). IDH2mut cells were more sensitive to the combination treatment than IDH2WT cells, suggesting that co-treatment with AR-42 and KPT-9274 resulted in a synergistic cell death in a IDH2-dependent manner (Figure 3B). We then tested the ability of the drug combination to inhibit the growth of primary AML patient cells. In accordance with findings in cell lines, exposure to AR-42 sensitized primary AML cells to KPT-9274 treatment (Figure 3C) (Table 2). Combination treatment resulted in higher maximum synergy scores in IDH1mut patient cells than in IDH1WT patient cells (max synergy score: 48 vs 26). Mutant IDH1/2 inhibitors protected IDH1/2mut AML cells against combined treatment, suggestive of a causal relationship existing between IDH1/2mut and this sensitization to AR-42/KPT-9274 combination (Supplementary Figure S4A-D). The effect of mutant IDH1/2 on drug sensitivity is 2-HG-dependent, as 2-HG treatment induces stronger synergism of AR-42/KPT-9274 combination on IDHWT cells (Supplementary Figure S4B).
Table 2.
Characterization of the AML patients selected for the study.
| Patient ID |
Age | Cytogenetics | Mutations |
|---|---|---|---|
| 1 | 74 | CN | ASXL1, PHF6, SRSF2, STAG2, TET2 |
| 2 | 74 | CN | DNMT3A, PHF6, TET2, U2AF1 |
| 3 | 72 | 46,XY, del[7][q21][13]/46, XY(6)/nonclonal[1] | IDH2, JAK3, SRSF2 |
| 4 | 65 | 47, XYY?c, del(16)(q11.2)[12]/48,idem,+8[5]/47.XYY?c[2]/nonclonal[1] | ASXL1, IDH1, JAK2, U2AF1 |
| 5 | 76 | CN | CCND2, IDH2, NRAS, STAG2 |
| 6 | 77 | CN | IDH1, DNMT3A, NPM1 |
| 7 | 75 | CN | DNMT3A, NPM1 |
| 8 | 57 | 46,XY[19]/nocloncal[1] | DNMT3A, IDH1,NPM1,PTPN11 |
| 9 | 47 | 46,XX[20] | DNMT3A, IDH2,NPM1,FLT3 |
CN, cytogenetically normal
To further characterize the synergistic effect of combined treatment on the functional subsets of AML cells, we performed CFU assays on primary cells. Compared with single agents, the drug combination significantly diminished the colony-forming and long-term replating potentials of AML blasts from patients (Figure 3D) (Table 3a-b). In contrast, the colony-forming capacities of CD34+ hematopoietic stem cells from the bone marrow of age-matched healthy donors were not substantially affected by combined treatment. Therefore, AR-42 and KPT-9274 synergistically reduced self-renewal capacities of AML cells while sparing normal human bone marrow cells.
Table 3a. Statistical analysis of CFU 1st plating of primary cells receiving different treatments.
| A negative binomial model was employed to fit the count data. | ||||||
|---|---|---|---|---|---|---|
| Model estimates: | ||||||
| Comparison | AML patients (n=4) | Healthy donors (n=2) | ||||
| Estimated ratio | 95% CI | p-value | Estimated ratio | 95% CI | p-value | |
| Combo vs. KPT-9274 | 0.35 | (0.29, 0.43) | <.001 | 0.73 | (0.56, 0.96) | 0.026 |
| Combo vs. AR-42 | 0.58 | (0.47, 0.71) | <.001 | 0.91 | (0.69, 1.21) | 0.509 |
| Combo vs. Vehicle | 0.31 | (0.26, 0.38) | <.001 | 0.85 | (0.64, 1.12) | 0.238 |
| Direct comparison between AML patients and healthy donors: | |||
|---|---|---|---|
| Comparison | Estimated ratio | 95% CI | p-value |
| Combo vs. KPT-9270 in AML vs. healthy | 0.48 | (0.35, 0.68) | <.001 |
| Combo vs. A-42 in AML vs. healthy | 0.63 | (0.45, 0.9) | 0.013 |
| Combo vs. Vehicle in AML vs. healthy | 0.37 | (0.26, 0.52) | <.001 |
Table 3b. Statistical analysis of CFU 2nd plating of primary cells receiving different treatments.
| A negative binomial model was employed to fit the count data. | ||||||
|---|---|---|---|---|---|---|
| Model estimates: | ||||||
| Comparison | AML patients (n=4) | Healthy donors (n=2) | ||||
| Estimated ratio | 95% CI | p-value | Estimated ratio | 95% CI | p-value | |
| Combo vs. KPT-9274 | 0.11 | (0.08, 0.16) | <.001 | 0.94 | (0.63, 1.4) | 0.753 |
| Combo vs. AR-42 | 0.21 | (0.14, 0.31) | <.001 | 0.8 | (0.54, 1.19) | 0.258 |
| Combo vs. Vehicle | 0.1 | (0.07, 0.14) | <.001 | 0.87 | (0.58, 1.29) | 0.468 |
| Direct comparison between AML patients and healthy donors: | |||
|---|---|---|---|
| Comparison | Estimated ratio | 95% CI | p-value |
| Combo vs. KPT-9270 in AML vs. healthy | 0.12 | (0.07, 0.2) | <.001 |
| Combo vs. A-42 in AML vs. healthy | 0.26 | (0.15, 0.45) | <.001 |
| Combo vs. Vehicle in AML vs. healthy | 0.11 | (0.06, 0.19) | <.001 |
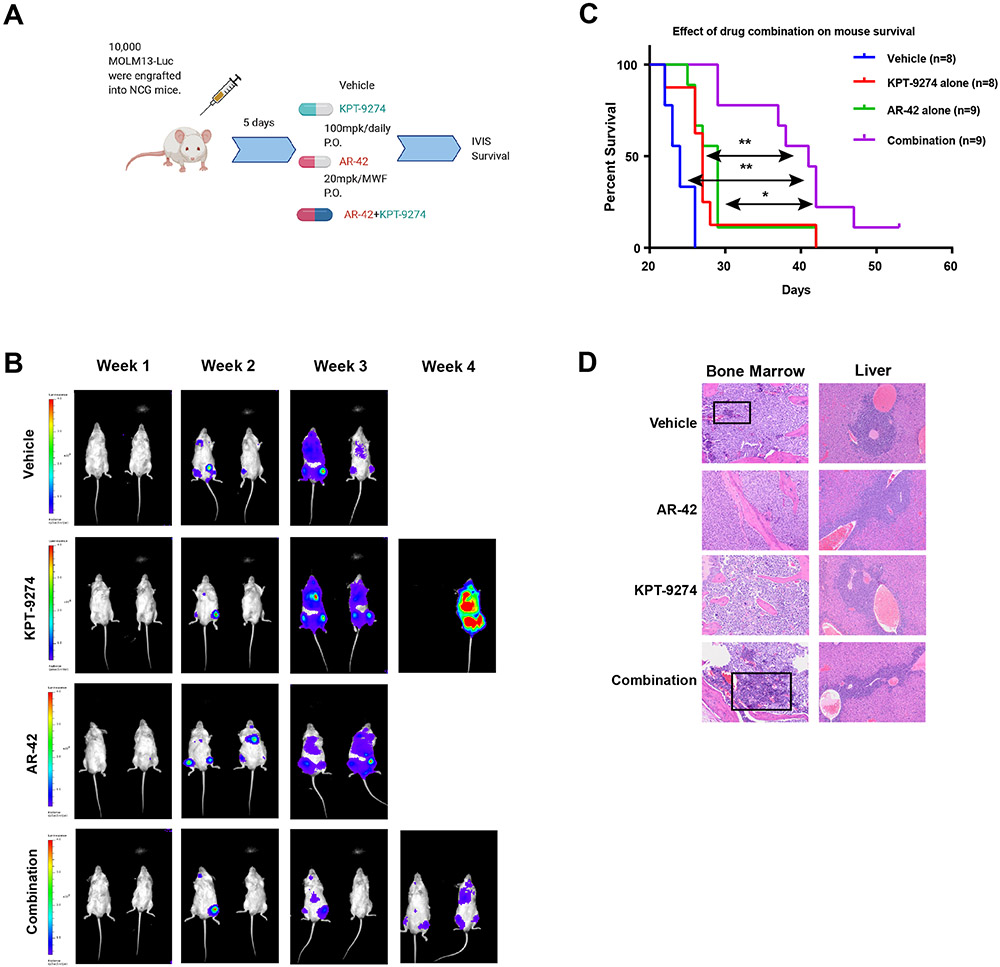
AR-42 enhances eradication of AML in combination with NAMPTi in vivo
To determine the in vivo relevance of the drug combination, we tested the efficacy of co-treatment of AR-42 and KPT-9274 in the MOLM13 xenograft model. We transplanted 1X104 luciferase-transfected MOLM13 cells by tail vein injection into NCG mice (Figure 4A). Five days after engraftment, mice were randomized to receive vehicle, KPT-9274 alone (100 mg/kg daily), AR-42 alone (20 mg/kg every another day) or combination regimen. The 20 mg/kg dose of AR-42 was tolerated according to an earlier pharmacokinetics study (21). Based on IVIS bioluminescence imaging, the disease burden was lower in the drug combination group than in either single agent or vehicle group over the course of the experiment (Figure 4B). Survival of vehicle-treated mice began to decline by week 3 (Figure 4C). KPT-9274 or AR-42 as a monotherapy added little to this effect, whereas mice in drug combination arm had a significantly longer survival. Specifically, mice in the KPT-9274 and AR-42 monotherapy groups had median survival times of 27 and 29 days, respectively (Supplementary Figure S5A). Combined therapy prolonged the lifespan, conferring survival benefits on these MOLM13-engrafted mice (median survival time: 41 days). Histopathology of a variety of organs, including bone marrow, liver, lymph node and spleen, revealed that animals in combination groups have the least infiltrating neoplastic blasts and most differentiated hematopoietic cells (myeloid, erythroid, and megakaryocytic lineages) (Figure 4D). Mice in the combination group had vacuolation of the testes with atrophy of the seminiferous tubules and a lack of spermatogenesis (Supplementary Figure S5B). No overlapping toxicities between AR-42 and KPT-9274, like cytopenia, kidney injury and liver damage, were evident (Figure 4D). In addition, we did not observe any noticeable weight loss in the AR-42 and drug combination groups over the course of the study (Supplementary Figure S5C).
Figure 4. Simultaneous inhibition of HDAC8 and NAMPT exhibits enhanced in vivo therapeutic efficacy in a mouse xenograft model of human AML.
A) Schematic illustration of experimental design. NCG mice were engrafted with luciferase-tagged MOLM13 cells and treated with vehicle, 20 mg/kg AR-42, 100 mg/kg KPT-9274 or AR-42/KPT-9274 in combination. B) IVIS imaging displays the changes of luciferase signals over weeks. C) Kaplan-Meier analysis of the mouse survival (Cox proportional hazards model; KPT-9274 vs combination p<0.005; AR-42 vs combination p<0.02; vehicle vs combination p<0.001). n=8-9 mice/group. D) Histopathology of bone marrow and liver sections from each treatment group. Black box shows the evidence of differentiated hematopoietic cells.
The synthetic lethality of AR-42 and KPT-9274 is conferred by simultaneous suppression of HR and NHEJ pathways in LICs
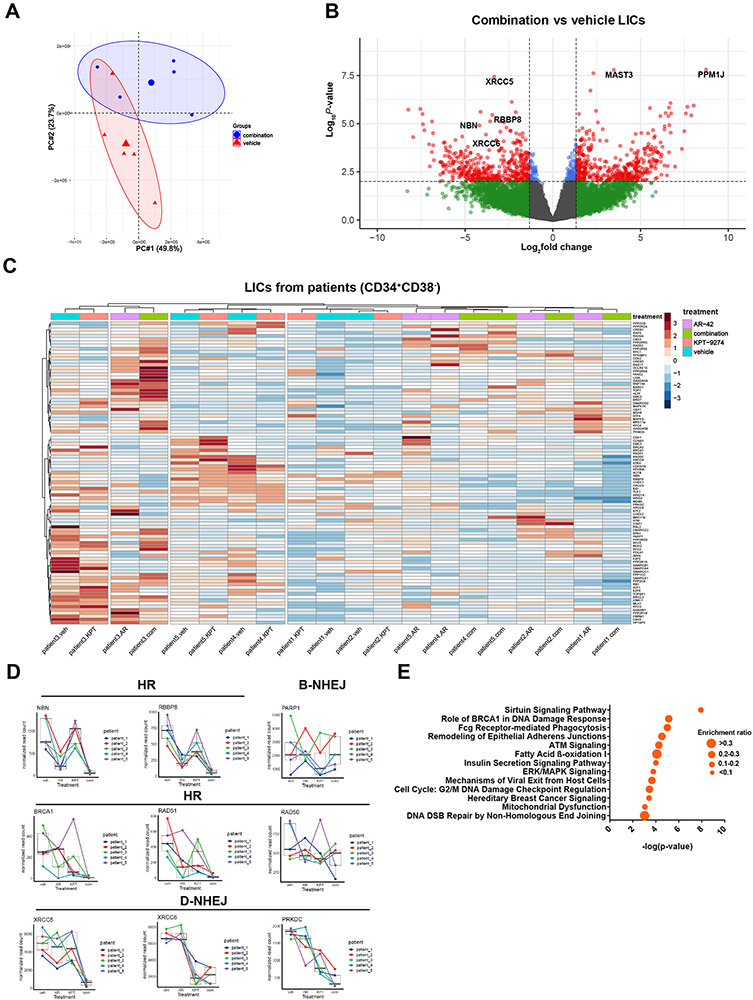
Since combined treatment of AR-42 and KPT-9274 abolishes the self-renewal potentials of patient LICs (Figure 3D), the transcriptomes of LICs could be modulated by the drug treatments. To gain insights into the mechanism by which inhibition of HDAC8 overcomes KPT-9274 insensitivity, we performed RNA-seq on patient LICs. Bone marrow cells from 5 patients were treated with vehicle, single agents or drug combination ex vivo for short time period (12 hours). Treated LICs were analyzed for transcriptional differences with DNA repair mechanisms being highlighted. Although the transcriptomes from 5 patients were quite heterogeneous, combination therapy-treated LICs were clustered separately from vehicle-treated populations in PCA analysis (Figure 5A). Consistent with PCA analysis, the divergent responses of patient LICs to drug combination treatments were observed for transcriptome patterns and differentially expressed genes (DEGs). As displayed by volcano plot, genes promoting DNA repairs, like XRCC5, XRCC6, NBN and RBBP8, were differentially downregulated, while genes involved in transferase activity (MAST3) and serine/threonine phosphatase (PPM1J) were differentially upregulated (Figure 5B). Hierarchical clustering and heatmap analysis revealed that combined treatment induced a transcriptional response profile of HR and NHEJ pathways closely related to that of AR-42 or KPT-9274 treatment but distinct from that of vehicle treatment in LIC compartments (Figure 5C).
Figure 5. AR-42 and KPT-9274 synergistically alter DNA repair gene sets of LIC transcriptomes.
Combination therapy changes the transcriptomes of patient leukemia initiating cells (LICs). Primary cells from 5 patients were treated with drug combination or vehicle for 12 hours before LICs (CD34+CD38−) were sorted for LC-RNA-seq. A) PCA plot showing that drug combination-treated LICs tend to form distinct clusters from vehicle-treated LICs. B) Volcano plot showing the significantly upregulated and downregulated DEGs in drug combination-treated LICs. Cutoff Log2FC=1.5; p-value<10e−2.C) Heatmaps showing the relative expressions of genes (normalized read counts) within DNA repair gene sets of LICs as being treated with indicated drug(s) relative to vehicle controls in 5 patients. The hierarchical clustering of genes and samples was performed using Euclidean distance matrix and Ward’s clustering method. D) Normalized read counts of selected DNA repair genes in LICs across patients under different treatment conditions. E) IPA analysis of significantly enriched pathways of differentially expressed genes in drug combination-treated LICs.
AR-42 alone or in combination with KPT-9274 downregulated the mRNA levels of HR genes, like NBN, RBBP8, RAD51 and BRCA1, D-NHEJ genes, like XRCC4 and XRCC5, and genes in ATM pathway (CHEK1 and CHEK2) (Figure 5D; Supplementary Figure S6A). Interestingly, BRCA1 and XRCC4 were upregulated by KPT-9274 alone in some patients. KPT-2974 treatment decreased the yields of D-NHEJ transcripts, like XRCC6 and PRKDC, and B-NHEJ gene, PARP1, in all patients. Based on these observations, we performed gene set enrichment analysis on drug combination-treated samples relative to control. Among the significantly enriched pathways were “The roles of BRCA1 in DNA damage response” and “ATM signaling”, suggestive of a potential impact of the combination therapy on DDR (Figure 5E). Taken together, AR-42 and KPT-9274 synergistically impair HR, NHEJ and ATM pathways by abolishing gene transcription. This suggests that synthetic lethality may be due to ineffective DNA damage response.
HDAC8 inhibition or SIRT6 knockdown causes the accumulation of DNA damage in KPT-9274-treated AML by impairing HR and D-NHEJ gene expressions and attenuating mono-ADP-ribosylation of PARP1
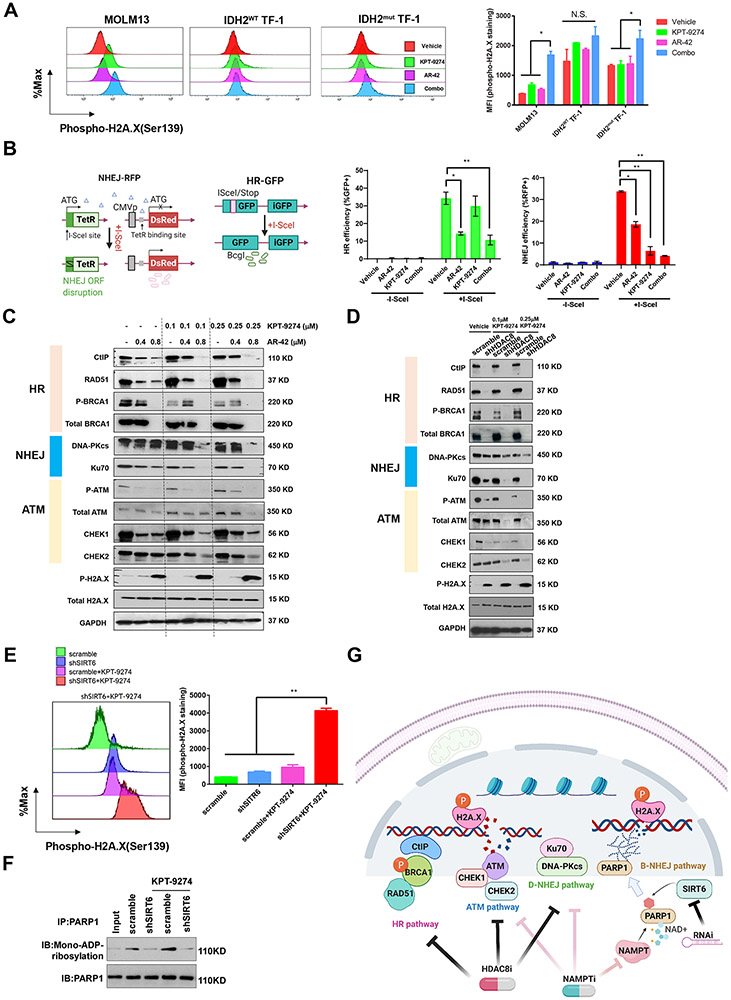
To further explore the mechanistic basis of synthetic lethality of AR-42 and KPT-9274, we detected treatment-induced changes of intracellular phospho-H2A.X levels. The drug combination created significantly more unrepaired double-strand breaks (DSBs) as indicated by an increase in phospho-H2A.X staining than either AR-42 or KPT-9274 alone in MOLM13 cells (Figure 6A). Although AR-42 or KPT-9274 slightly increased phospho-H2A.X staining in IDH2WT TF-1 cells, the drug combination failed to further upregulate phospho-H2A.X levels. In contrast, the drug combination enhanced phospho-H2A.X staining on top of the effects of single agents in IDH2mut TF-1 cells, suggesting that IDH2 mutation synergizes with HDAC8/NAMPT inhibition to impair DDR system. This effect might be attributed to HR deficiency caused by IDH2 mutation which may render cells more vulnerable to combined treatment.
Figure 6. HDAC8 inhibition or SIRT6 knockdown sensitizes AML to KPT-9274 by suppression of HR and D-NHEJ pathways or mono-ADP-ribosylation of PARP1.
A) Combined treatment of AR-42 and KPT-9274 synergistically increases unrepaired DNA damage sites as indicated by the accumulation of phosphorylated H2A.X. MOLM13, IDH2WT TF-1 and IDH2mut TF-1 cells were incubated with vehicle control, 0.8 μM AR-42 alone, 0.1 μM KPT-2974 alone or drug combination for 48 hours. Thereafter, cells were fixed, permeabilized and stained with BV421 anti-H2A.X (pS139) (Clone N1-431). Stained cells were analyzed by flow cytometry. Results are shown as mean ± SEM of duplicates. *p<0.05. N.S.: not significant. B) Left panel: schematic illustration of I-SceI-based EJDR reporter system; Right panel: Quantification of OCI-AML3-DR and -EJ reporter assays. AR-42 blocks HR and NHEJ repairs, while KPT-9274 impairs NHEJ repair. Results are shown as mean ± SEM of duplicates. *p<0.05;**p<0.01. C) Exposure of MOLM13 cells to AR-42 and KPT-9274 results in a decrease in activities of HR, NHEJ and ATM signaling, and a concomitant increase in phospho-H2A.X (pS139) (Clone 20E3), consistent with the loss of multiple pathway-mediated DSB repairs. MOLM13 cells were treated with vehicle control, 0.4 μM or 0.8 μM AR-42 alone, 0.1 μM or 0.25 μM KPT-2974 alone or drug combination for 24 hours before being subject to Western blotting analysis of HR, NHEJ and ATM markers. GAPDH serves as loading control. Results are representative of two independent experiments. D) Knockdown of HDAC8 reduces the levels of HR pathway mediators and abolishes the activities of D-NHEJ and ATM signaling in cooperation with KPT-9274. Scrambled or shHDAC8-transduced MOLM13 cells were treated with vehicle, 0.1 μM or 0.25 μM KPT-2974 before being subject to immunoblotting analysis of DNA repair targets. GAPDH serves as loading control. Results are representative of two independent experiments. E) shSIRT6 and KPT-9274 synergistically increase unrepaired DNA damage sites as indicated by the accumulation of phosphorylated H2A.X. Results are shown as mean ± SEM of duplicates. **p<0.01. F) Co-IP showing that shSIRT6 decreases mono-ADP-ribosylation of PAPR1 in response to KPT-9274 treatment. PAPR1 was immunoprecipitated and mono-ADP-ribosylation was detected by western blotting. Results are representative of two independent experiments. G) Schematic illustration of possible mechanisms of sensitizing AML cells to NAMPT inhibition by suppression of DNA repairs.
We next examined HR and NHEJ activities in AML cells exposed to these treatments using the EJDR reporter-stable U2OS cells and established OCI-AML3-DR and OCI-AML3-EJ cell lines (Figure 6B and Supplementary Figure S6B-C). KPT-9274 treatment did not impact HR activity, whereas AR-42 exposure significantly reduced I-SceI-induced HR. AR-42 treatment was also associated with NHEJ deficiency as measured by the reduction of the percentage of DsRed+ cells, but residual NHEJ activity was consistently detectable in AR-42-treated cells. KPT-9274 treatment caused a more robust reduction of NHEJ activity when applied alone or in combination with AR-42. Consistent with these findings, immunoblotting further revealed that 0.4 μM and 0.8 μM AR-42 treatment markedly reduced the levels of HR components, CtIP, Rad51, and BRCA1 as well as phosphorylated ATM as a monotherapy or in combination with KPT-9274 in MOLM13 cells (Figure 6C). The expression of D-NHEJ pathway factors, Ku70 and DNA-PKcs, and ATM pathway mediators, CHEK1 and CHEK2, were abolished by AR-42/KPT-9274 co-treatment. This effect was accompanied by induction of phospho-H2A.X. Knockdown of HDAC8 phenocopied the magnitude of reduction in the levels of HR mediators seen in AR-42-treated cells, while shHDAC8 and KPT-9274 synergistically downregulated the abundance of D-NHEJ and ATM pathway mediators (Figure 6D). Therefore, suppression of HDAC8 can attenuate HR and D-NHEJ gene expressions, resulting in NAMPTi sensitivity. To elucidate the mechanism of the synergism between shSIRT6 and KPT-9274, we measured the accumulation of phospho-H2A.X. In the presence of KPT-9274, SIRT6-depleted cells exhibited more severe DNA damage as evidenced by increased phospho-H2A.X staining compared with controls (Figure 6E). KPT-9274 enhanced mono-ADP-ribosylation of PARP1 which was eradicated by knockdown of SIRT6, suggesting that SIRT6 was responsible for PARP1 mono-ADP-ribosylation (Figure 6F). This observation provided justification for exploring the DDR-targeting mechanism of synthetic lethal effect exerted by simultaneous inhibition of NAMPT and HDAC8 or SIRT6 in AML.
Discussion
In this study, we employed a CRISPR screen to identify genes that upon depletion, increase KPT-9274 sensitivity with the ultimate goal of developing synthetic lethal therapies that allow for lowering dosage of NAMPTis. We found that depletion of the screen’s top hits, HDAC8 and SIRT6, conferred sensitivity to KPT-9274 treatment. Co-treatment with AR-42 (HDACi) and KPT-9274 resulted in a dramatic reduction of AML viability. Additionally, combination therapy showed superior efficacy in a MOLM13 xenograft. Strikingly, AR-42/KPT-9274 combination abrogated the self-renewal of patient LICs by shutting down HR and NHEJ pathways. HDAC8i, shSIRT6 and NAMPTi simultaneously suppress compensatory DSB repair processes through the regulation of transcription and post-translational modifications (Figure 6G). As inhibition of histone deacetylases and associated DNA repair machinery as a strategy to enhance NAMPTi efficacy has not been previously reported, our combination strategy represents a novel regimen to improve efficacy and tolerability of KPT-9274.
Due to the lack of potent HDAC8-specific inhibitors, AR-42 was employed in the current study for in vivo and in vitro target validation. Although AR-42 targets multiple HDAC isoforms, the observed synergistic effect with KPT-9274 is most likely attributed to its activity towards HDAC8, given that HDAC8 is the highest ranked hit among all HDACs (the only one with p< 0.01 in CRISPR screen). Additionally, knockout of other major AR-42 targets, HDAC1, HDAC3 or HDAC6, did not display any synergy with KPT-9274. Simultaneous AR-42 and KPT-9274 treatment resulted in the accumulation of phospho-H2A.X and lethal DSBs. HDAC inhibitors disable functional HR by controlling the activities and expression of HR-related genes (22,23).The roles of HDAC8 in HR were reported, as HDAC8 is associated with Rad51 and MRE11a and HDAC8 depletion leads to a decrease in Rad51 levels in multiple myeloma (24,25). Consistent with these findings, we detected substantial changes in the abundance of transcripts in HR pathway, like NBN and RBBP8(CtIP), in patient LICs upon AR-42 treatment (Figure 5C-D). We also observed a remarkable reduction of transcripts in D-NHEJ and ATM pathways with cooperative actions of AR-42 and KPT-9274. Immunoblotting results on cell lines recapitulate our patient transcriptome data by showing that AR-42 or shHDAC8 primarily abrogated the expression of HR genes, while KPT-9274 and HDAC8 inhibition synergistically downregulated the components involved in D-NHEJ and ATM pathways at protein levels, sustaining DSBs. Our findings provide evidence that HDAC8i treatment suppresses HR and D-NHEJ (“BRCAness/DNA-PKness” phenotype), which, in combination with NAMPTi, causes synthetic lethality in AML cells due to accumulation of lethal DSBs beyond the reparable threshold.
IDH1/2 mutations exacerbate the HR deficiency by upregulating 2-HG production. This “BRCAness” phenotype of IDH-mutant cells renders tumors exquisitely sensitive to PARP1 inhibitors (20). In the current study, we found that treatment of IDH1/2-deficient AML cell lines or patient cells with the drug combination resulted in elevated accumulation of γH2A.X and greater inhibition of cell growth compared with IDH-WT cells. It was not our focus to delineate the mechanism of increased sensitivity of IDH1/2-mutant cells to AR-42/KPT-9274 combination. But it is likely that IDH1/2 mutation may induce deficiency in some compensatory repair pathways, which cooperates with AR-42-induced HR and KPT-9274-induced NHEJ deficiencies to induce synergistic cell apoptosis. It was reported that 2-HG accumulation induced by IDH1(R132) mutation in AML inhibits the function of histone demethylases (KDM4A and KDM4B) that are critical for HR function and consequently TIP60 and ATM activities are also decreased (26,27). This effect is likely independent of HDAC inhibitor-mediated suppression of CtIP and BRCA1 functions. This reasoning provides a plausible explanation for why IDH1/2-mutant cells are more sensitive to NHEJ and HR inhibition than IDH1/2-WT cells. It is conceivable that inhibition of mutant IDH may attenuate 2-HG-mediated DNA repair defects and antagonize the vulnerability of AML to drug combo treatment. Therefore, co-treatment of AR-42 and KPT-9274 may benefit patients with mutant IDH1/2 in a 2-HG-depenendent manner in clinical settings and our combination strategy should not be applied concomitantly with IDH inhibitors. Therefore, IDH2 could be used as a precision medicine marker for identifying AML patients that may benefit from a therapeutic regimen combining NAMPTi and HDAC8i.
LICs are associated with relapse and drug resistance, due to their tolerance to DNA damage (28-32). Here, we report that AR-42 and KPT-9274 cooperated to shut down multiple DDR pathways and decreased self-renewal of LICs but not normal hematopoietic stem cells. AR-42 was reported to induce death of LICs by triggering caspase-dependent apoptosis (18). However, it is the first time to show that AR-42 can eradicate LICs in a combination therapy at sublethal doses. By probing LIC transcriptomes, we also found that a strong DDR-suppressive effect was achieved through combined treatments, while LICs treated with KPT-9274 alone augmented the levels of HR genes in some patients. This raises the possibility that KPT-9274-treated LIC populations could be particularly vulnerable to this anti-HR strategy. This is supported by the observation that inhibition of NAMPT downstream molecule, PARP1, induces accumulation of Rad51 and preserves HR responses (33).
Studies showed that there exists a poor-prognostic subset of AML patients relying on SIRT6 and NHEJ to compensate for DNA-replication stress (1,34). SIRT6 inhibition was shown to compromise the ability of leukemia cells to repair DSBs that, in turn, increases their sensitivity to daunorubicin and Ara-C (34). SIRT6 is known to stimulate NHEJ in the absence of DNA-PKcs, the D-NHEJ enzyme. Independent of its deacetylase function, SIRT6 mono-ADP ribosylates lysine 521 of PARP1, thereby stimulating its poly-ADP ribosylation activity (35,36). This modification is required for SIRT6-mediated stimulation of DSB repair, cooperating with NAMPT to fully unlock PARP1-mediated B-NHEJ. Our discovery provides a rationale of developing therapies targeting mono-ADP-ribosylation activity of SIRT6 while sparing deacetylase function to minimize toxicity for healthy cells.
The limitations of our study are that we only tested a limited number of AML cell lines and patient samples. Based on our previous and current studies, while KPT-9274 as a monotherapy is more efficacious towards patients with NPM1, DNMT3A and NRAS mutations, patients with NPM1, DNMT3A and IDH1 co-mutation manifest high sensitivity to combined treatment. These results advocate in favor of clinical trials of HDAC8i/NAMPTi in these patients. It was reported that NMRK1 plays a critical role in bypassing NAMPT dependence (6). AML cells vary by the degrees of NMRK1 expression. It would be of interest to study the synergistic effect of NMRK1 and NAMPT co-inhibition on the growth of NMRK1high cells in vitro and in vivo. Of note, we conducted our CRISPR screen on MOLM13 which carries MLL-AF9 translocation with truncated MLL1 (KMT2A). Future screen/validation study design may take into account KMT2A-dependency in NAMPTi sensitivity with non-MLL-AF9 cells. Other factors influencing cell sensitivity to combined treatments may include different expressions and mutations in genes encoding drug transporters and metabolic enzymes. But it is expected that our combined therapy can be extended to a broad spectrum of AML subtypes. In addition, future studies may be needed to nominate resistance dependency by using an essential screen in a resistant cell line in combination with a drug screen in a sensitive cell line. Given that HDAC8, SIRT6 and NAMPT govern different DDR pathways, it would be of great translational interest to develop a triple combination therapy of HDAC8i, SIRT6i and NAMPTi in the future. Testing drug efficacy on PDX or other spontaneous leukemia models warrants further investigations.
In summary, we validated selected histone deacetylases as attractive targets for inhibition along with NAMPTi. Based on these findings, we speculate that AML cells are particularly sensitive to concurrent silencing of multiple DDR pathways which causes cell death and lowers the risk of overdose toxicity of KPT-9274. Combined AR-42/KPT-9274 therapy may be most beneficial for patients carrying IDH mutations. Our study sheds light onto the mechanistic basis of synthetic lethality and the potential of development of HDAC8i/NAMPTi or SIRT6i/NAMPTi as a novel therapy for a broad spectrum of AML subtypes, especially those manifesting resistance to traditional therapies.
Supplementary Material
Translational Relevance.
KPT-9274 is a phase I NAMPTi which induces accumulation of DNA breaks by depleting NAD+ supply. In this study, we carried out an unbiased CRISPR screen against AML, identified two histone deacetylase members, HDAC8 and SIRT6, and validated HDAC8 pharmacologically with AR-42 and SIRT6 genetically with shRNA as novel synthetic lethal targets regulating KPT-9274 sensitivity. Our findings provide evidence that targeting DNA repair functions of HDAC8 or SIRT6 can be novel therapeutic strategies sensitizing leukemia initiating cells (LICs) to KPT-9274. Our findings support further preclinical investigation of SIRT6 as a lethal target in NAMPT-inhibited leukemia and the development of potent therapeutic agents that target its mono-ADP-ribosylase activity, while avoiding deleterious effects on its normal function. As AR-42 has been investigated in phase I trials, our discovery also provides the basis for a rational and clinically testable combination therapy with KPT-9274 at less toxic doses to treat patients with AML.
Acknowledgments:
The authors are grateful for the patients and healthy volunteers who provided blood and tissue samples for the present studies and to the OSU Comprehensive Cancer Center Leukemia Tissue Bank (supported by the National Institutes of Health [NIH], National Cancer Institute [NCI] [P30 CA016058]) for sample procurement. We also thank Dr. David Lucas and Dr. Chis Manning for their help in identification and management of primary AML samples at the Ohio State University Leukemia Tissue Bank. We thank the OSUMC genomics core facility for its help with LC-RNA-seq studies. We are grateful for Dr. Feng Zhang’s lab for CRISPR GeCKOv2 library. This work was supported by the NIH, NCI (R35 CA197734, R01 CA223165), the OSU Comprehensive Cancer Center using the Pelotonia Foundation funds, and further research support to the Byrd Laboratory from the Harry Mangurian Foundation and the D. Warren Brown Foundation.
Footnotes
Conflicts of Interest: The authors declare no potential conflicts of interest.
Data availability:
CRISPR screening and RNA-Sequencing data are available in GEO (accessions GSE162473 and GSE161397).
References:
- 1.Roboz GJ. Epigenetic targeting and personalized approaches for AML. Hematology American Society of Hematology Education Program; 2014;2014:44–51 [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA: a cancer journal for clinicians; 2018;68:7–30 [DOI] [PubMed] [Google Scholar]
- 3.Martin PR, Shea RJ, Mulks MH. Identification of a plasmid-encoded gene from Haemophilus ducreyi which confers NAD independence. J Bacteriol 2001;183:1168–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hasmann M, Schemainda I. FK866, a Highly Specific Noncompetitive Inhibitor of Nicotinamide Phosphoribosyltransferase, Represents a Novel Mechanism for Induction of Tumor Cell Apoptosis. Cancer Res 2003;63:7436–42 [PubMed] [Google Scholar]
- 5.Mitchell SR, Larkin K, Grieselhuber NR, Lai TH, Cannon M, Orwick S, et al. Selective targeting of NAMPT by KPT-9274 in acute myeloid leukemia. Blood advances 2019;3:242–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chowdhry S, Zanca C, Rajkumar U, Koga T, Diao Y, Raviram R, et al. NAD metabolic dependency in cancer is shaped by gene amplification and enhancer remodelling. Nature 2019;569:570–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holen K, Saltz LB, Hollywood E, Burk K, Hanauske AR. The pharmacokinetics, toxicities, and biologic effects of FK866, a nicotinamide adenine dinucleotide biosynthesis inhibitor. Investigational new drugs 2008;26:45–51 [DOI] [PubMed] [Google Scholar]
- 8.Zabka TS, Singh J, Dhawan P, Liederer BM, Oeh J, Kauss MA, et al. Retinal toxicity, in vivo and in vitro, associated with inhibition of nicotinamide phosphoribosyltransferase. Toxicological sciences : an official journal of the Society of Toxicology 2015;144:163–72 [DOI] [PubMed] [Google Scholar]
- 9.Galli U, Colombo G, Travelli C, Tron GC, Genazzani AA, Grolla AA. Recent Advances in NAMPT Inhibitors: A Novel Immunotherapic Strategy. Frontiers in pharmacology 2020;11:656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bindra RS, Goglia AG, Jasin M, Powell SN. Development of an assay to measure mutagenic non-homologous end-joining repair activity in mammalian cells. Nucleic acids research 2013;41:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walker LA, Sovic MG, Chiang CL, Hu E, Denninger JK, Chen X, et al. CLEAR: coverage-based limiting-cell experiment analysis for RNA-seq. Journal of translational medicine 2020;18:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costello RT, Mallet F, Gaugler B, Sainty D, Arnoulet C, Gastaut JA, et al. Human acute myeloid leukemia CD34+/CD38− progenitor cells have decreased sensitivity to chemotherapy and Fas-induced apoptosis, reduced immunogenicity, and impaired dendritic cell transformation capacities. Cancer research 2000;60:4403–11 [PubMed] [Google Scholar]
- 13.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nature methods 2014;11:783–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang BB, Wang M, Zhang WB, Xiao TF, Chen CH, Wu A, et al. Integrative analysis of pooled CRISPR genetic screens using MAGeCKFlute. Nature protocols 2019;14:756–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balasubramanian S, Ramos J, Luo W, Sirisawad M, Verner E, Buggy JJ. A novel histone deacetylase 8 (HDAC8)-specific inhibitor PCI-34051 induces apoptosis in T-cell lymphomas. Leukemia 2008;22:1026–34 [DOI] [PubMed] [Google Scholar]
- 16.Tseng YC, Kulp SK, Lai IL, Hsu EC, He WA, Frankhouser DE, et al. Preclinical Investigation of the Novel Histone Deacetylase Inhibitor AR-42 in the Treatment of Cancer-Induced Cachexia. Journal of the National Cancer Institute 2015;107:djv274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liva SG, Coss CC, Wang J, Blum W, Klisovic R, Bhatnagar B, et al. Phase I study of AR-42 and decitabine in acute myeloid leukemia. Leukemia & lymphoma 2020:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guzman ML, Yang N, Sharma KK, Balys M, Corbett CA, Jordan CT, et al. Selective activity of the histone deacetylase inhibitor AR-42 against leukemia stem cells: a novel potential strategy in acute myelogenous leukemia. Molecular cancer therapeutics 2014;13:1979–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mims A, Walker AR, Huang X, Sun J, Wang H, Santhanam R, et al. Increased anti-leukemic activity of decitabine via AR-42-induced upregulation of miR-29b: a novel epigenetic-targeting approach in acute myeloid leukemia. Leukemia 2013;27:871–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sulkowski PL, Corso CD, Robinson ND, Scanlon SE, Purshouse KR, Bai HW, et al. 2-Hydroxyglutarate produced by neomorphic IDH mutations suppresses homologous recombination and induces PARP inhibitor sensitivity. Science translational medicine 2017;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liva SG, Tseng YC, Dauki AM, Sovic MG, Vu T, Henderson SE, et al. Overcoming resistance to anabolic SARM therapy in experimental cancer cachexia with an HDAC inhibitor. EMBO molecular medicine 2020;12:e9910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JH, Choy ML, Ngo L, Foster SS, Marks PA. Histone deacetylase inhibitor induces DNA damage, which normal but not transformed cells can repair. Proceedings of the National Academy of Sciences of the United States of America 2010;107:14639–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adimoolam S, Sirisawad M, Chen J, Thiemann P, Ford JM, Buggy JJ. HDAC inhibitor PCI-24781 decreases RAD51 expression and inhibits homologous recombination. Proceedings of the National Academy of Sciences of the United States of America 2007;104:19482–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maria Gkotzamanidou MS, Sánchez Jesús Martín, Samur Mehmet Kemal, Minvielle Stephane, Magrangeas Florence, Avet-Loiseau Herve, Dimopoulos Athanasios-Meletios, Anderson Kenneth C. and Munshi Nikhil C.. HDAC8 is recruited to DNA double strand breaks sites and affects the homologous recombination efficiency in multiple myeloma. 2015; Philadelphia, PA. American Association for Cancer Research. p 30305. [Google Scholar]
- 25.Santos-Barriopedro I, Li Y, Bahl S, Seto E. HDAC8 affects MGMT levels in glioblastoma cell lines via interaction with the proteasome receptor ADRM1. Genes & cancer; 2019;10:119–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inoue S, Li WY, Tseng A, Beerman I, Elia AJ, Bendall SC, et al. Mutant IDH1 Downregulates ATM and Alters DNA Repair and Sensitivity to DNA Damage Independent of TET2. Cancer cell 2016;30:337–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sulkowski PL, Oeck S, Dow J, Economos NG, Mirfakhraie L, Liu Y, et al. Oncometabolites suppress DNA repair by disrupting local chromatin signalling. Nature 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanekamp D, Cloos J, Schuurhuis GJ. Leukemic stem cells: identification and clinical application. International journal of hematology 2017;105:549–57 [DOI] [PubMed] [Google Scholar]
- 29.De Grandis M, Mancini SJ, Aurrand-Lions M. In quest for leukemia initiating cells in AML. Oncoscience 2018;5:9–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Viale A, De Franco F, Orleth A, Cambiaghi V, Giuliani V, Bossi D, et al. Cell-cycle restriction limits DNA damage and maintains self-renewal of leukaemia stem cells. Nature 2009;457:51–6 [DOI] [PubMed] [Google Scholar]
- 31.Mohrin M, Bourke E, Alexander D, Warr MR, Barry-Holson K, Le Beau MM, et al. Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell stem cell 2010;7:174–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dai Y, Chen S, Kmieciak M, Zhou L, Lin H, Pei XY, et al. The novel Chk1 inhibitor MK-8776 sensitizes human leukemia cells to HDAC inhibitors by targeting the intra-S checkpoint and DNA replication and repair. Molecular cancer therapeutics 2013;12:878–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bellio C, DiGloria C, Foster R, James K, Konstantinopoulos PA, Growdon WB, et al. PARP Inhibition Induces Enrichment of DNA Repair-Proficient CD133 and CD117 Positive Ovarian Cancer Stem Cells. Molecular cancer research : MCR 2019;17:431–45 [DOI] [PubMed] [Google Scholar]
- 34.Cagnetta A, Soncini D, Orecchioni S, Talarico G, Minetto P, Guolo F, et al. Depletion of SIRT6 enzymatic activity increases acute myeloid leukemia cells' vulnerability to DNA-damaging agents. Haematologica 2018;103:80–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Meter M, Mao Z, Gorbunova V, Seluanov A. Repairing split ends: SIRT6, mono-ADP ribosylation and DNA repair. Aging 2011;3:829–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mao Z, Hine C, Tian X, Van Meter M, Au M, Vaidya A, et al. SIRT6 promotes DNA repair under stress by activating PARP1. Science 2011;332:1443–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
CRISPR screening and RNA-Sequencing data are available in GEO (accessions GSE162473 and GSE161397).