SUMMARY
Since their discovery, giant viruses have expanded our understanding of the principles of virology. Due to their gargantuan size and complexity, little is known about the life cycles of these viruses. To answer outstanding questions regarding giant virus infection mechanisms, we set out to determine biomolecular conditions that promote giant virus genome release. We generated four infection intermediates in Samba virus (Mimivirus genus, lineage A) as visualized by cryoelectron microscopy (cryo-EM), cryoelectron tomography (cryo-ET), and scanning electron microscopy (SEM). Each of these four intermediates reflects similar morphology to a stage that occurs in vivo. We show that these genome release stages are conserved in other mimiviruses. Finally, we identified proteins that are released from Samba and newly discovered Tupanvirus through differential mass spectrometry. Our work revealed the molecular forces that trigger infection are conserved among disparate giant viruses. This study is also the first to identify specific proteins released during the initial stages of giant virus infection.
Graphical Abstract
In Brief
An architectural and proteomic characterization of Mimivirus infection intermediates illustrates the conserved molecular forces driving infection and identifies proteins released from the giant virus early in the infection cycle.
INTRODUCTION
A hallmark of newly discovered giant viruses (GVs) is their incredibly complex biology, including gargantuan capsids and large genomes. The sheer size and complexity of these viruses, especially the inclusion of “junk” DNA in the form of introns (Azza et al., 2009; Boratto et al., 2018), challenges the canonical view of viruses as small, streamlined, and efficient killing machines. For example, most GVs are larger than 300 nm and many have genomes exceeding 1 MB, containing an estimated 1,000+ open reading frames (Colson et al., 2017). By contrast, some of the smallest viruses include the porcine circovirus (17 nm capsid; ~2,000 base genome; four proteins; Dhindwal et al., 2019) and the human rhinovirus (30 nm capsid; ~7,200 base genome; 11 proteins; Jacobs et al., 2013). ~69% of known viruses encode for less than 10 proteins (Brandes and Linial, 2019), highlighting the complexity of GVs and the true extent of our lack of knowledge concerning this new class of viruses.
GVs have been associated with a wide variety of hosts, including amoeba (Aherfi et al., 2016b), animals (Andrade et al., 2015, 2018; Dornas et al., 2014a; Khan et al., 2007), as well as human and murine cells (Ghigo et al., 2008; Lusi et al., 2017). However, amoebas also infect these creatures, casting doubt on the true GV reservoir. Although GVs have been associated with human respiratory diseases (Khan et al., 2007; La Scola et al., 2005; Saadi et al., 2013a, 2013b), inflammatory conditions (Popgeorgiev et al., 2013; Shah et al., 2014), and cancers (Aherfi et al., 2016a), no direct link between GVs and human disease has yet been established. Despite an unusually broad host range and pathogenicity, little information is available on how GVs access their hosts. Infection usually occurs via phagocytosis (Abrahão et al., 2014; Ghigo et al., 2008). Once phagocytosed, a unique capsid vertex opens, which promotes nucleocapsid release and fusion with the phagosomal membrane, ultimately releasing the genome into the host cytoplasm. A pseudo-organelle (viral factory) is then formed (Suzan-Monti et al., 2007), and host replication factors are hijacked. The endpoint of GV infections is host cell death and release of new GV progeny into the environment.
GVs are ubiquitous (Aherfi et al., 2016b; Andrade et al., 2018) and maintain infectivity in harsh environments, such as alkaline lakes (Abrahão et al., 2018), frozen permafrost (Legendre et al., 2014), 3 km deep in the ocean (Abrahão et al., 2018), and Antarctic dry valleys (Andrade et al., 2018; Kerepesi and Grolmusz, 2017). GVs have retained infectivity following exposure to harsh chemicals (Campos et al., 2012), extreme pH and salinity (Abrahão et al., 2018), and extreme temperatures (Andrade et al., 2018; Legendre et al., 2014) and are able to persist on hospital equipment (Campos et al., 2012; Dornas et al., 2014b). To survive such extremes, GVs have developed incredible capsid stability. Some GV capsids can retain infectivity for 30,000 years in permafrost (Legendre et al., 2014, 2015).
Although capsid stability is beneficial for a virus to persist in harsh environments, it also creates a thermodynamic barrier that must be overcome once a suitable host cell is encountered; all known viruses must do this to propagate. Strategies and structures used for genome translocation are generally conserved across viral families. Among the tailed double-stranded DNA (dsDNA) bacteriophages (Caudovirales), tail complexes interact with host receptor proteins to trigger conformational changes in the virion, leading to genome release (Parent et al., 2018). Similarly, many classes of eukaryotic viruses have conserved genome release mechanisms. Most enveloped viruses, including HIV, influenza, Zika virus, and herpesvirus, utilize one of three structurally conserved membrane fusion protein varieties (Harrison, 2015). Non-enveloped viruses, such as rhinovirus, poliovirus, and adenovirus, utilize conserved capsid structures to interact with host receptors to trigger genome uncoating (Suomalainen et al., 2013).
Morphologically, GV virions are either icosahedral, exemplified by Acanthamoeba polyphaga mimivirus (La Scola et al., 2003), or non-icosahedral, typified by Mollivirus and Pithovirus (Legendre et al., 2014, 2015). Like their smaller cousins, GVs also share conserved capsid structures that are used during infection. In many GVs, the unique capsid vertex provides a gateway for the infection process, but they also provide a mechanism to prevent premature loss of their precious cargo. GVs have developed at least two distinct vertex structures to seal the unique vertex until the time is right for infection: “corks” and “starfish.” Non-icosahedral GVs tend to utilize one or more cork-like structures (located flush with the capsid surface) as a seal complex (Andreani et al., 2016; Legendre et al., 2014; Philippe et al., 2013). A newly discovered class of non-icosahedral GV, consisting of members such as Pandoravirus (Legendre et al., 2014) and Orpheovirus (Andreani et al., 2018), contain an ostiole-like structure, distinct from the cork-like structure.
Mimivirus-like icosahedral GVs utilize an external proteinaceous seal complex that resembles a five-pointed starfish (Klose et al., 2010; Xiao et al., 2009). These complexes sit at the outer-most capsid layer at a unique 5-fold vertex (called the stargate vertex) and prevent it from opening (Xiao et al., 2009). Traditionally, both the unique capsid vertex and the external seal complex have been packaged together and called either the “stargate” or the “starfish.” We will refer to the unique capsid vertex as the stargate and the seal complex as the starfish. Non-mimivirus-like icosahedral GVs, such as PBCV-1 (Milrot et al., 2017), faustovirus (Klose et al., 2016), and Pacmanvirus (Andreani et al., 2017), do not utilize stargate vertices and have evolved alternative genome release strategies.
Starfish structures are found in diverse GVs, such as mimivirus (Klose et al., 2010; Xiao et al., 2009), Samba virus (SMBV) (Campos et al., 2014; Schrad et al., 2017), and the newly discovered tupanviruses (Abrahão et al., 2018; Silva et al., 2019), and are more common than the cork-like seals among GVs. Yet relatively little is known about the mechanism governing the stargate. The molecular forces and biochemical trigger(s), such as receptor proteins or phagosomal transitions, that facilitate stargate opening are unknown. Additionally, the fate of the starfish remains a mystery; is the complex removed from the capsid en masse or does the complex simply unzip?
The general steps and gross morphological changes that accompany GV infection have been visualized via thin section transmission electron microscopy (TEM) of infected cells (Abrahão et al., 2014; Silva et al., 2019). Following phagocytosis, the stargate vertex begins to open 1–3 h post-infection (Silva et al., 2019), yet little is known about the specific proteins and biomechanical forces that mediate this process. This knowledge gap is largely due to two factors: the complexity of GV virions and the lack of a robust model system for detailed biochemical and/or biophysical studies. Here, we have created an in vitro model system for studying the choreography that governs GV genome release. We were able to trap infection intermediates, identify specific proteins released during the initial stage of stargate opening, and test the efficacy of this technique on other icosahedral GVs, including SMBV, a member of Mimivirus lineage A (Campos et al., 2014); a mimivirus variant, M4 (Boyer et al., 2011); Tupanvirus soda lake (TV) (Abrahão et al., 2018); and Antarctica virus (Andrade et al., 2018). Additionally, our model reveals that members of Mimivirus lineage A unzip their starfish complexes to initiate infection.
RESULTS AND DISCUSSION
SMBV Is Resistant to the Vast Majority of Chemical Treatments
To probe the molecular forces that play a role in SMBV starfish complex stability, we exposed SMBV to treatments known to affect morphology in other viruses (Table S1). The effect of each treatment on particle stability was assessed via cryoelectron microscopy (cryo-EM). Treatments included the denaturants urea (≥9 M) and guanidinium hydrochloride (≥6 M), the detergent Triton X-100, organic solvents, such as chloroform and DMSO, as well as enzymes DNase I, bromelain, proteinase K, and lysozyme. None of these treatments resulted in disruption of the SMBV virion over the baseline of ~5% spontaneously open untreated SMBV particles (Schrad et al., 2017). Three treatments did lead to significantly increased disruption of the stargate vertex: low pH; high temperature; and high salt (see following sections). Similar treatments have been used to induce morphological changes that reflect biologically relevant structural transitions in a variety of phages and viruses (Bothner et al., 2005; Conway et al., 2001, 2007; Kant et al., 2018; Parent et al., 2010; Roos et al., 2012; Teschke et al., 2003; van de Waterbeemd et al., 2017; Yu et al., 2008; Zheng et al., 2014).
Electrostatic Interactions Are Critical for SMBV Starfish Stability
We hypothesized that pH changes occurring during and after phagocytosis may trigger SMBV stargate opening. Therefore, we dialyzed SMBV particles against different sodium phosphate buffer solutions, ranging in pH from 2 to 12 (Figure 1). Particles were visualized via cryo-EM (Figures 1A and 1B), and the percent of open particles (POP) was calculated. At and above pH 4, there was no appreciable change in the POP, compared to native (pH 7.4) levels (Figures 1A–1C). However, at and below pH 3, ~60% of the SMBV capsids had opened. Although the conditions that produced an increase in SMBV POP (lower than pH 4) are more acidic than the environment predicted within the amoebal phagosome, which is ~pH 4 (Flannagan et al., 2015; German et al., 2013; Lopez and Skaar, 2018), stargate opening is likely dependent on acidic pH, both in vivo and in vitro.
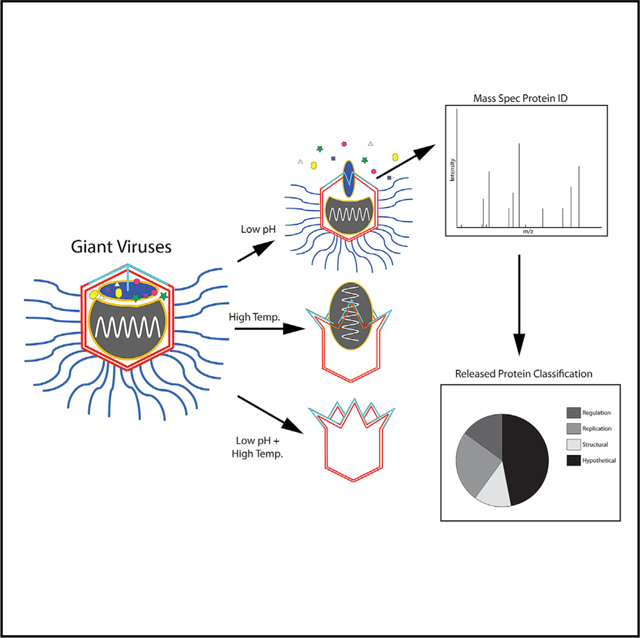
Figure 1. Low pH and High Temperature Triggered an Increase in SMBV POP.
(A and B) Representative micrographs of native (A) or low-pH-treated (B) SMBV particles. Unopened particles are denoted with stars, and those with visible cracks are marked with arrows. One particle in (B), marked with a square, is clearly empty (lacks the internal density of the nucleocapsid), but cracks in the capsid are not visible, as that vertex is not contained within the micrograph. Particles such as these were counted as open when generating the percentages of open particles (POP).
(C) POP following treatment at various pH.
(D) The POP of SMBV particles incubated at elevated temperatures.
(E–H) Bubblegram images of two different native GV particles revealing star-shaped radiation damage patterns caused by the starfish complex (highlighted in F and H). (I and J) Final micrographs in bubblegram series of two different low-pH-treated SMBV particles. Note the lack of any star-shaped radiation damage patterns. See also Figure S1.
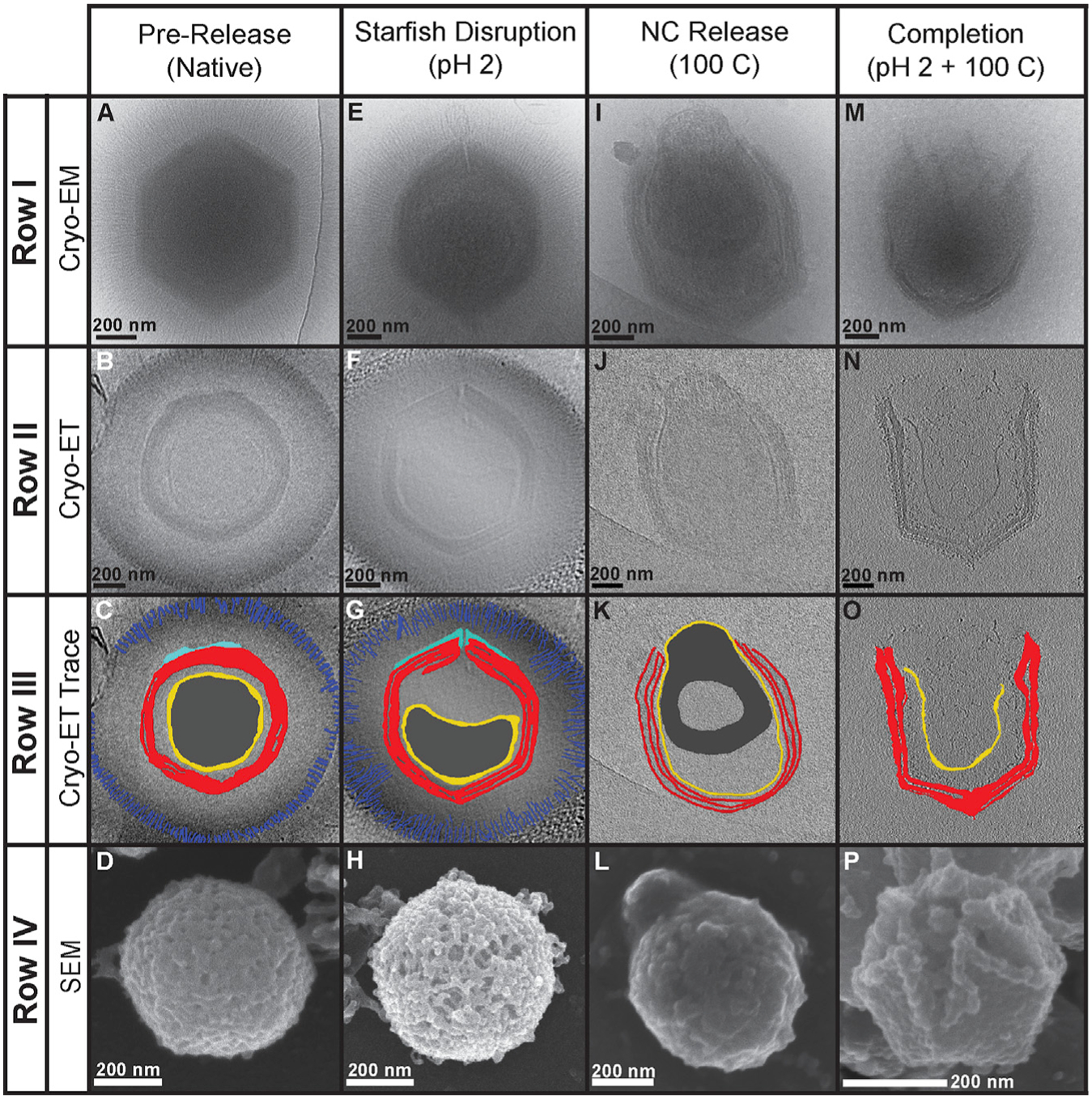
Unlike spontaneously opened GV capsids (Schrad et al., 2017; Xiao et al., 2009; Zauberman et al., 2008), these SMBV capsids were not fully open. Instead, the particles had small, noticeable cracks at one capsid vertex that assumed a star-shaped pattern. The opening of the stargate vertex at low pH is irreversible: SMBV particles returned to neutral pH still displayed star-shaped cracks in their capsids. In some particles, the extra membrane sac was caught in the process of leaving the capsid through the newly opened vertex (Figure 2E). In other particles, the sac is not visible, suggesting that it had escaped prior to imaging. Release of the extra membrane sac, also referred to as the viral seed, has been hypothesized in other GVs. The extra membrane sac is thought to contain proteins responsible for the formation of the viral factory (Mutsafi et al., 2010; Silva et al., 2019; Suzan-Monti et al., 2007), and it is distinct from the nucleocapsid membrane when visualized in 3D using cryoelectron tomography (cryo-ET) (Schrad et al., 2017; Xiao et al., 2009). To our knowledge, this is the first study to demonstrate reproducible release of the extra membrane sac and to identify some of the proteins that may be released with this complex (below).
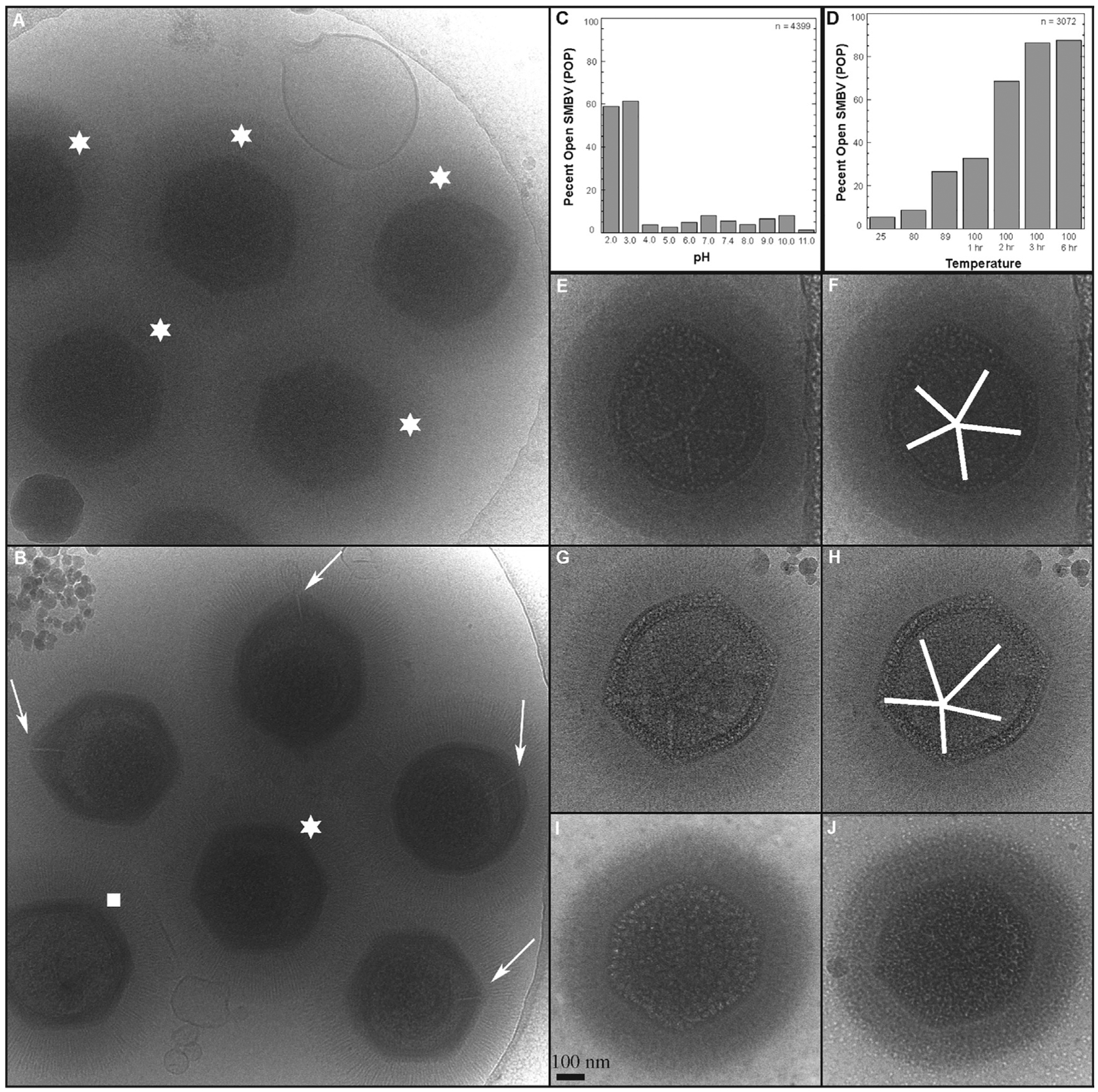
Figure 2. Electron Microscopy of SMBV Genome Release Stages.
(Row I) 2D cryoelectron micrographs of particles following either no treatment (A) or post-incubation with pH 2 (E), 100°C (I), or both pH 2 + 100°C (M). (Row II) Central slices (z = 20) of cryoelectron tomograms of particles following either no treatment (B) or post-incubation with pH 2 (F), 100°C (J), or both pH 2 + 100°C (N). (Row III) Central slices of cryo-tomograms with key features highlighted. Blue, distal tips of the external fiber layer; cyan, starfish seal complex; dark gray, dsDNA; red, capsid; yellow, lipid membranes (nucleocapsid). Slices are shown for virions following either no treatment (C) or post-incubation with pH 2 (G), 100°C (K), or both pH 2 + 100°C (O). (Row IV) Scanning electron micrographs of particles in various stages of genome release following either no treatment (D) or post-incubation with pH 2 (H), 100°C (L), or both pH 2 + 100°C (P). See Videos S2, S3, S4, S5, S6, S7, S8, and S9 for videos of the tomograms and tilt series. See EMD-20745–20748 for tomogram volumes.
We could see that the particles had indeed opened following low pH treatment. Using 2D images alone, we could not, however, determine whether the starfish complex was released en masse or whether it remained associated with the capsid. Therefore, we used scanning electron microscopy (SEM) to probe surface features. Unfortunately, SEM images of pH-2-treated SMBV particles (Figure 2H) also did not provide definitive evidence for the presence of the starfish seal, as the layer of external fibers blocked access to the capsid surface. Enzymatic removal of the fibers was needed to visualize the surface of the starfish seal in mimivirus by SEM (Xiao et al., 2009), but unfortunately, this protocol did not remove the fibers in SMBV (Table S1). We next generated 3D reconstructions of opened SMBV particles by cryo-ET (Figure 2; Video S3; EMD-20747). Tomograms confirm that the stargate vertex, and only the stargate vertex, is open in the pH-2-treated particles. Extra density corresponding to the starfish seal is clearly observed along the edges of the outer capsid layer at the stargate vertex (Video S3). Therefore, it is likely that, at least some, if not all, of the proteins that comprise the starfish seal complex remain attached to the capsid after low pH treatment. Our tomograms suggest that the SMBV starfish likely destabilizes through an “unzipping” mechanism rather than en masse release. As low pH treatment is able to trigger stargate vertex opening in vitro, we conclude that electrostatic interactions play a very important role in stabilizing this vertex prior to infection. To confirm that these interactions play a role in stargate opening, we treated GV particles with 4M NaCl and then removed the salt by dialysis for cryo-EM imaging. High salt concentrations resulted in ~40% POP (Figure S1).
We used “bubblegram” imaging, a cryo-EM technique used to localize unique features within macromolecular complexes. In this technique, samples are overexposed to produce beam-induced radiation damage. Hydrogen (H2) gas released as a result of the radiation damaging can become trapped and sometimes produces noticeable “bubbling” in the micrograph. This bubbling can be used to reveal the location and shape of the unique features in viral capsids (Parent et al., 2018), such as phage ΦKZ inner bodies (Wu et al., 2012) and also ejection proteins in phage P22 (Wu et al., 2016).
When untreated SMBV particles were exposed to excessive electron radiation, particles produced a star-shaped radiation damage pattern (Figures 1E and 1F; Video S1). By contrast, pH-2-treated SMBV particles displayed no star-shaped pattern (Figures 1I and 1J). As expected, the lack of a star-shaped radiation damage pattern is consistent with the hypothesis that the H2 gas is no longer being trapped in the SMBV virion, as the low pH treatment disrupted the stargate vertex seal.
Increased Thermal Energy Is Required for Nucleocapsid Release
Low pH or high salt alone was insufficient to fully open SMBV particles, indicating that electrostatic interactions are not solely responsible for fully sealing the stargate. Therefore, we next analyzed the effect of temperature on SMBV stability. We incubated the virions for 1 h up to 100°C, assayed the virions for morphological changes using cryo-EM, and then compared these data to images of unheated particles. After 1 h at 100°C, the POP was ~33% (Figure 1D). Following an additional incubation for up to 6 h, the POP increased to a maximum of ~88%. Additional incubation at 100°C did not increase the POP further.
Unlike low pH or high salt, which simply crack the stargate vertex, high temperatures resulted in open stargate vertices and nucleocapsids in the process of exiting the virion (Figure 2; Videos S4 and S5; EMD-20748). Within these nucleocapsids, the DNA appears to have reorganized, leaving pockets of seemingly empty space (discussed below). Much of the external fiber layer is removed (Figures 2 and S1), and the extra membrane sac is also fully released from these particles. High temperature could be an alternative GV defibering method to that proposed in Kuznetsov et al. (2010), especially as this previously described technique did not defiber SMBV particles. High temperature induces a conformational change that closely mimics a structural transition that occurs during mimivirus infection as seen in vivo (see Figure 1, panels E and F, in Abrahão et al., 2014; Figure 6), where the nucleocapsid leaves the capsid and prepares to fuse with the amoebal phagosome membrane.
Figure 6. Giant Virus Genome Release Process.
(A and B) The progression of GV genome release depicted as a cartoon (A) and through thin section TEM images (B). The thin-section micrographs represent Tupanvirus particles visualized at varying stages of the infection process in vivo. The panels for “seal disruption” and “nucleocapsid fusion” were previously published in Abrahão et al. (2018) and shown under Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, modification, and reproduction. The remaining two panels are original images.
(C) Cryoelectron micrographs of the completion of the in vitro genome release process for the four GVs studied here.
(D) Z slices of an SMBV tomogram of fully opened particles highlighting the density corresponding to the starfish seal.
Following low-pH or high-temperature treatment, there were pockets within the SMBV nucleocapsids that appear to be devoid of DNA (Figure 2). These seemingly empty pockets are not visible in the untreated SMBV particles (Figure 2) but are seen in both low-pH- and high-temperature-treated particles. Although it is possible that the void inside of SMBV nucleocapsids could be due to the extreme conditions used, it is more likely that this is biologically relevant. These pockets are only observed in SMBV particles that have begun releasing their genome, suggesting that the DNA may undergo reorganization during this process. The SMBV genome contains various chromosome condensation and histone-like proteins that could be used for this function, although their exact role in GV biology has not yet been experimentally determined. Mass spectrometry experiments (described below and shown in Table 1) suggest that many of these proteins remain with the nucleocapsid after the initial opening stage. Genome reorganization is an important stage of many virus infection processes, including HIV (Freed, 2015) and adenovirus (Mangel and San Martín, 2014). We hypothesize that genome rearrangement is also important for facilitating GV genome release into the host.
Table 1.
Identification of Proteins Released from SMBV and TV Capsids
| Presence | Ratio of Ratios | |||||
|---|---|---|---|---|---|---|
| Protein | ID | Category | Band | Shotgun | Up in Supe | Down in Pellet |
| Samba Virus | ||||||
| Actina,b | GenBank: CAA23399.1 | S | + | + | + | |
| Rpl7A, partiala | GenBank: AAY21190.1 | – | + | + | ||
| Amine oxidase | GenBank: AHJ39955.1 | M | + | + | ||
| DNA-dependent RNP subunit RPB9 | GenBank: AHJ39967.2 | Tl | + | |||
| Ubiquitin-conjugating enzyme e2 | GenBank: AHJ39993.2 | M | + | + | ||
| WD repeat-containing protein | GenBank: AHJ40002.1 | – | + | |||
| B-type lectin protein | GenBank: AHJ40019.2 | S | + | |||
| Protein phosphatase 2c | GenBank: AHJ40032.1 | Rg | + | |||
| Formamidopyrimidine-DNA glycosylase | GenBank: AHJ40038.1 | Ho | + | |||
| Hypothetical protein | GenBank: AHJ40051.1 | H/TM | + | |||
| Poly(A) polymerase catalytic subunit | GenBank: AHJ40056.1 | Tx | + | |||
| Hypothetical protein | GenBank: AHJ40060.1 | H | + | + | ||
| Hypothetical protein | GenBank: AHJ40061.1 | H | + | + | ||
| Thioredoxin domain-containing protein | GenBank: AHJ40071.1 | Ho | + | |||
| mRNA-capping enzyme | GenBank: AHJ40083.1 | Tx | + | + | ||
| Putative FtsJ-like methyltransferase | GenBank: AHJ40084.1 | Tx | + | |||
| Hypothetical protein | GenBank: AHJ40087.2 | H | + | |||
| Low-complexity protein | GenBank: AHJ40093.1 | H | + | |||
| Core protein | GenBank: AHJ40101.1 | S | + | |||
| Hypothetical protein | GenBank: AHJ40107.2 | H | + | |||
| Capsid protein 1 | GenBank: AHJ40114.2 | S | + | + | ||
| Hypothetical protein | GenBank: AHJ40128.1 | H | + | |||
| Thioredoxin domain-containing protein | GenBank: AHJ40129.2 | Ho | + | + | + | + |
| Hypothetical protein | GenBank: AHJ40139.1 | H | + | + | ||
| Hypothetical protein | GenBank: AHJ40144.1 | H | + | |||
| DNA-directed RNA polymerase subunit l | GenBank: AHJ40151.2 | Tl | + | |||
| Hypothetical protein | GenBank: AHJ40159.1 | H | + | |||
| Hypothetical protein | GenBank: AHJ40160.2 | H | + | |||
| Hypothetical protein | GenBank: AHJ40162.1 | H | + | |||
| Hypothetical protein | GenBank: AHJ40169.1 | H | + | |||
| DNA-directed RNAP subunit 1 | GenBank: AHJ40172.1 | Tl | + | + | ||
| Hypothetical protein | GenBank: AHJ40183.2 | H | + | + | ||
| Alpha beta hydrolase/esterase/lipaseb | GenBank: AHJ40190.1 | I/E | + | |||
| Hypothetical protein | GenBank: AHJ40207.1 | H | + | |||
| Hypothetical protein | GenBank: AHJ40211.1 | H | + | |||
| Hypothetical protein | GenBank: AHJ40213.2 | H | + | + | + | + |
| Hypothetical protein | GenBank: AHJ40220.1 | H | + | |||
| Hypothetical protein | GenBank: AHJ40230.1 | H | + | |||
| Hypothetical protein | GenBank: AHJ40243.1 | H | + | + | ||
| Mannose-6P isomerase | GenBank: AHJ40247.1 | M | + | + | ||
| Hypothetical protein | GenBank: AHJ40254.1 | H | + | |||
| Hypothetical protein | GenBank: AHJ40271.2 | H | + | |||
| Tat pathway signal sequence domain proteinb | GenBank: AHJ40276.1 | I | + | + | ||
| Collagen-like protein 7 | GenBank: AHJ40290.2 | S | + | + | + | |
| Hypothetical protein | GenBank: AHJ40316.2 | H | + | + | ||
| Hypothetical protein | GenBank: AHJ40318.2 | H | + | |||
| Hypothetical protein | GenBank: AHJ40319.1 | H | + | |||
| Hypothetical protein | GenBank: AHJ40326.2 | H | + | + | ||
| Low-complexity protein | GenBank: AHJ40329.1 | H | + | + | ||
| Hypothetical protein | GenBank: AHJ40333.1 | H/TM | + | + | ||
| Chemotaxis protein | GenBank: AHJ40337.1 | I/S | + | |||
| Hypothetical protein | GenBank: AHJ40339.1 | H | + | + | ||
| Hypothetical protein | GenBank: AHJ40340.1 | H | + | |||
| Ubiquitin thioesterase | GenBank: AHJ40341.2 | M | + | |||
| Hypothetical protein | GenBank: AHJ40367.2 | H/TM | + | |||
| Virion-associated membrane protein | GenBank: AHJ40371.2 | I | + | |||
| Lanosterol 14-alpha-demethylase | GenBank: AHJ40393.1 | I | + | + | ||
| Hypothetical protein | GenBank: AHJ40423.1 | H | + | + | ||
| Collagen triple helix repeat containing protein | GenBank: AMK61745.1 | S | + | + | ||
| Choline dehydrogenase-like protein | GenBank: AMK61776.1 | M | + | + | ||
| DNA topoisomerase 1b | GenBank: AMK61799.1 | Rg | + | |||
| Probable glutaredoxin | GenBank: AMK61800.1 | Rp/Ho | + | + | ||
| Hypothetical protein | GenBank: AMK61829.1 | H | + | + | + | |
| Hypothetical protein | GenBank: AMK61837.1 | H | + | + | ||
| Hypothetical protein | GenBank: AMK61849.1 | H/TM | + | |||
| Hypothetical protein | GenBank: AMK61856.1 | H | + | + | ||
| Regulator of chromosome condensationc | GenBank: AMK61866.1 | Rg/I | + | + | ||
| Thiol protease | GenBank: AMK61869.1 | E | + | |||
| Hypothetical protein | GenBank: AMK61892.1 | H | + | |||
| Hypothetical protein | GenBank: AMK61902.1 | H | + | |||
| Anaerobic nitric oxide reductase transcription regulator NorR | GenBank: AMK61903.1 | Rg | + | |||
| Ankyrin repeat protein | GenBank: AMK61918.1 | – | + | |||
| Hypothetical protein | GenBank: AMK61920.1 | H | + | |||
| Hypothetical protein | GenBank: AMK61935.1 | H | + | |||
| Hypothetical protein | GenBank: AMK61942.1 | H | + | + | ||
| N-acetyltransferase | GenBank: AMK61955.1 | M | + | |||
| Prolyl 4-hydroxylase | GenBank: AMK61959.1 | Ho | + | |||
| Proline rich protein | GenBank: AMK61968.1 | – | + | + | ||
| Hypothetical protein | GenBank: AMK61977.1 | H | + | + | ||
| NHL repeat-containing protein | GenBank: AMK61987.1 | – | + | |||
| Hypothetical protein | GenBank: AMK62013.1 | H | + | |||
| Hypothetical protein | GenBank: AMK62059.1 | H | + | + | ||
| Hypothetical protein | GenBank: AMK62082.1 | H | + | |||
| Choline dehydrogenase-like protein | GenBank: AMK62096.1 | M | + | + | ||
| Ubiquitina,d | GenBank: CAA53293.1 | M | + | + | ||
| Tupanvirus Soda Lake | ||||||
| Hypothetical protein | GenBank: AUL78681.1 | E/H | + | + | + | + |
| Hypothetical protein | GenBank: AUL77600.1 | E/H | + | |||
| Putative ORFan | GenBank: AUL77729.1 | H | + | |||
| Putative ORFan | GenBank: AUL78088.1 | H | + | + | + | + |
| Hypothetical protein | GenBank: AUL78481.1 | H | + | |||
| Hypothetical protein | GenBank: AUL78232.1 | H/Rg | + | + | ||
| Hypothetical protein | GenBank: AUL78466.1 | H | + | + | + | + |
| Hypothetical protein | GenBank: AUL77936.1 | H | + | |||
| Hypothetical protein | GenBank: AUL78214.1 | H | + | + | ||
| Hypothetical protein | GenBank: AUL77907.1 | H | + | + | + | + |
| Hypothetical protein | GenBank: AUL78468.1 | H | + | + | + | + |
| Hypothetical protein | GenBank: AUL77723.1 | H | + | + | + | + |
| Hypothetical protein | GenBank: AUL78464.1 | H | + | |||
| Hypothetical protein | GenBank: AUL78055.1 | H | + | + | + | |
| Hypothetical protein | GenBank: AUL77930.1 | H | + | + | + | |
| Putative ORFan | GenBank: AUL78635.1 | H | + | + | ||
| Hypothetical protein | GenBank: AUL77752.1 | H | + | |||
| Hypothetical protein | GenBank: AUL78219.1 | H | + | |||
| Hypothetical protein | GenBank: AUL78093.1 | H | + | + | ||
| Hypothetical protein | GenBank: AUL78067.1 | H | + | |||
| Hypothetical protein | GenBank: AUL78191.1 | H | + | + | ||
| Hypothetical protein | GenBank: AUL78287.1 | H | + | + | ||
| Hypothetical protein | GenBank: AUL77694.1 | H | + | |||
| Hypothetical protein | GenBank: AUL77820.1 | H | + | |||
| Hypothetical protein | GenBank: AUL78135.1 | H | + | + | ||
| Hypothetical protein | GenBank: AUL78143.1 | H | + | + | ||
| Hypothetical proteinc | GenBank: AUL78348.1 | H/Rg | + | + | + | |
| Hypothetical protein | GenBank: AUL77688.1 | H/TM | + | + | ||
| Hypothetical protein | GenBank: AUL78288.1 | H/TM | + | + | ||
| Hypothetical protein | GenBank: AUL77718.1 | H/TM/E | + | |||
| Cu-Zn superoxide dismutasee | GenBank: AUL78503.1 | Ho | + | |||
| mg709 proteine | GenBank: AUL77661.1 | Ho | + | + | ||
| Thioredoxin domain-containing protein | GenBank: AUL77963.1 | Ho | + | + | ||
| Catalase HPII | GenBank: AUL78097.1 | Ho | + | + | ||
| Ig family protein | GenBank: AUL78630.1 | I | + | |||
| Phosphatidylethanolamine-binding protein-like protein | GenBank: AUL77474.1 | I | + | |||
| Putative N-acetyl transferase | GenBank: AUL77680.1 | M | + | |||
| Arylsulfatase | GenBank: AUL78269.1 | M | + | |||
| Ubiquitin domain-containing proteind | GenBank: AUL78040.1 | M | + | + | + | + |
| Glyoxalase | GenBank: AUL78134.1 | M | + | |||
| Putative protein kinase | GenBank: AUL78629.1 | Rg | + | + | ||
| Glutaredoxin | GenBank: AUL78724.1 | Rp/Ho | + | + | + | |
| SNF2 family helicase | GenBank: AUL77941.1 | Rp/Rg | + | |||
| Capsid protein 1 | GenBank: AUL78147.1 | S | + | |||
| Putative fibril associated protein | GenBank: AUL78400.1 | S | + | |||
| Kinesin-like proteina | GenBank: AUL77838.1 | S | + | + | ||
| Major core protein | GenBank: AUL78082.1 | S | + | |||
| Putative pore coat assembly factor | GenBank: AUL78211.1 | S | + | + | ||
| Mimivirus elongation factor aef-2 | GenBank: AUL78714.1 | Tl | + | |||
| DNA-directed RNAP subunit | GenBank: AUL78016.1 | Tl | + | + | ||
| Intein-containing DNA-directed RNAP subunit 2 | GenBank: AUL78362.1 | Tl | + | |||
| DNA-directed RNAP subunit 6 | GenBank: AUL78368.1 | Tl | + | + | + | |
| DNA-directed RNAP subunit 1 | GenBank: AUL78302.1 | Tl | + | |||
| Putative ATP-dependent RNA helicase | GenBank: AUL77829.1 | Tx/Tl | + | |||
| Actina,b | GenBank: CAA23399.1 | S | + | + | ||
Acanthamoeba castellanii proteins.
Proteins similar to Irivovirus UPP-associated proteins.
Proteins involved in genome rearrangement.
Proteins directly involved in a putative ubiquitin-proteasome degradation pathway.
Metal-conjugating proteins.
A Combination of Low pH and High Temperature Results in Complete SMBV Genome Release
Individually, low pH and high temperature had different physical effects on SMBV. These disparate treatments are affecting both electrostatic interactions and the availability of thermal energy, respectively, and each appears to contribute to SMBV virion stability. Therefore, we hypothesized that combining low pH and high temperature might have a compound effect on stargate opening. Again, following treatment, the SMBV particle morphology was analyzed via cryo-EM (Figure 2M), cryo-ET (Figures 2N and 2O), and SEM (Figure 2P). These particles have completed the entire genome release process, as seen by the absence of the nucleocapsid. Additionally, SMBV particles were completely defibered and the internal capsid layer(s) appeared to be less rigid than the outer capsid layer (Figure 2O; Videos S6, S7, S8, and S9; EMD-20745 and EMD-20746).
SEM of dual-treated SMBV particles (Figure 2P) provides further evidence for the starfish seal. Particles treated with both low pH and high temperature clearly contain extra density around the edges of the stargate vertex, corresponding to the starfish seal. This extra density is consistent with our cryo-ET data described above, where, rather than completely dissociating from the capsid en masse, the starfish seal unzips while still retaining contacts with the capsid.
Molecular Forces that Stabilize the SMBV Stargate Vertex Are Conserved among Mimiviruses
We tested combining pH and high temperature on three other GVs: Antarctica virus (Andrade et al., 2018); Tupanvirus (Abrahão et al., 2018); and mimivirus M4 (Boyer et al., 2011). Although all four viruses analyzed here are mimivirus-like icosahedral GVs, these viruses encompass two separate GV clades belonging to the Mimiviridae family: of the genus Mimivirus (SMBV, M4, and Antarctica) and the proposed genus Tupanvirus (TV; Rodrigues et al., 2019b). Note that Tupanvirus is distinct from the others, as it has a long tail attached to the icosahedral head as seen in Abrahão et al. (2018) and Rodrigues et al. (2019a, 2019b). Following treatment, each virus was characterized via SEM and cryo-EM (Figures 3 and 6). All four GVs tested in this study opened their stargate vertices and released their nucleocapsids after being boiled in acid. Two also appeared to lose the majority of their fibers during treatment, with Tupanvirus being the exception. The fate of the fibers and their role during infection is not well known, although it has been suggested that fibers aid in viral attachment and increase phagocytosis (Rodrigues et al., 2015). These data strongly indicate that the general forces that stabilize virions and facilitate infection are conserved among distantly related amoeba-infecting members of Mimiviridae.
Figure 3. Post-genome Release Particles from Four GVs.
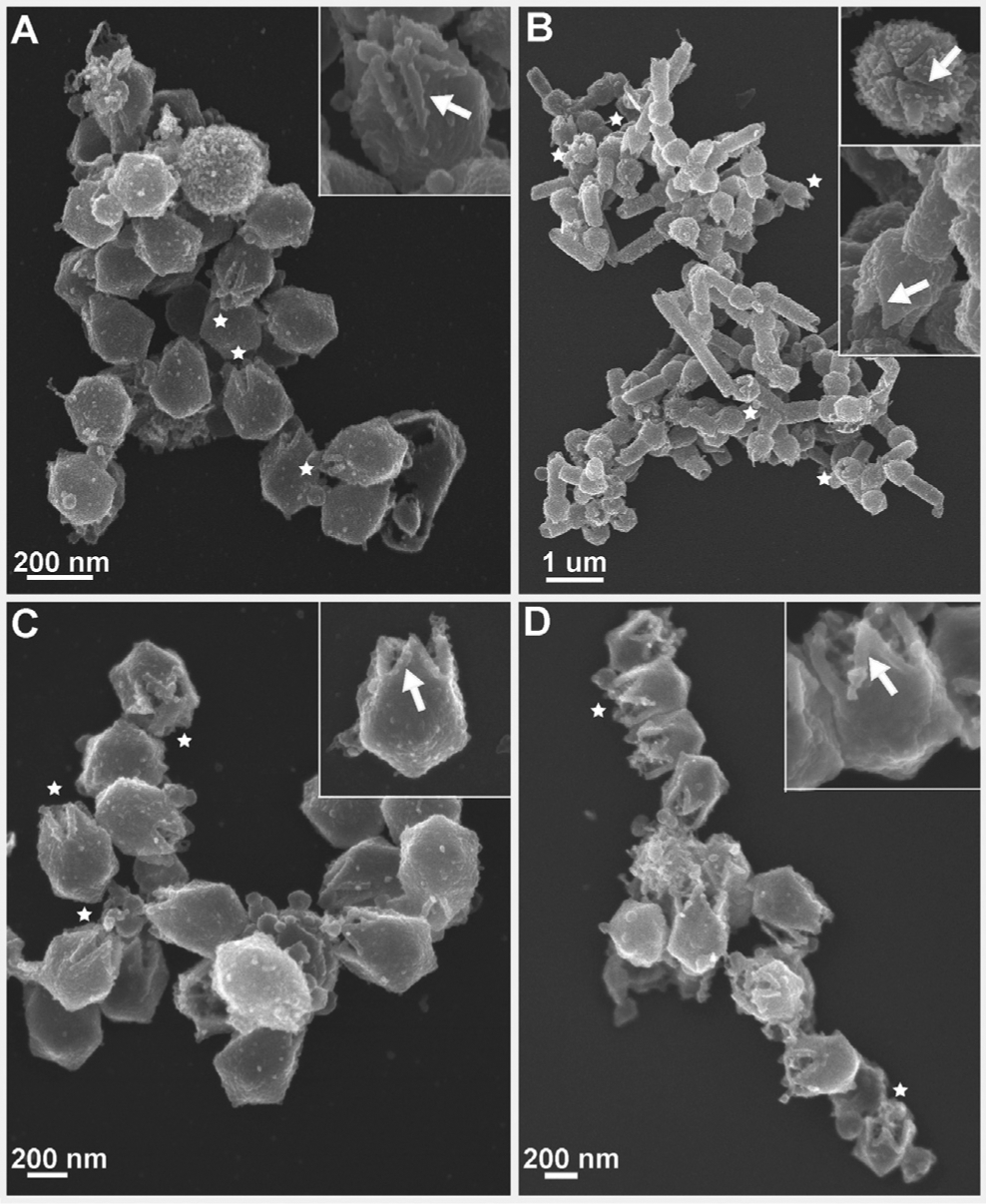
Scanning electron micrographs of low-pH- and high-temperature-treated (A) SMBV, (B) TV, (C) Antarctica virus, and (D) mimivirus particles. Insets demonstrate enlarged views highlighting capsids where clear retention of the starfish seal can be seen in SMBV, mimivirus, and Antarctica particles. Asterisks in the main panels depict selected particles with clearly visible open stargate vertices.
Although the general forces appear to be highly conserved, some specific mechanisms of starfish disruption are likely conserved only within distinct lineages. In our SEM data, Antarctica and mimivirus particles (Figures 3A and 3D, respectively) displayed density along the edges of the open stargate vertices, similar to the density seen in SMBV (Figures 2P and 3C). The presence of this extra density suggests that, like SMBV, the Antarctica and mimivirus starfish complexes unzip to facilitate stargate opening and genome release. TV, on the other hand, does not display this extra density (Figure 3B), suggesting that the TV starfish may completely dissociate from the capsid en masse during infection. TV particles also appear to fully open their stargate vertices following low-pH treatment alone. In total, our data suggest that the mechanism of seal complex unzipping may be conserved among Mimiviridae, with slight deviations present between the Mimiviruses and the proposed Tupanvirus genus.
GVs have changed our canonical view of virology, defying the previously known limits of capsid sizes and stabilities. The description of a new GV genome release strategy signifies another paradigm shift in our understanding of virology. As mentioned previously, smaller viruses tend to share conserved genome release mechanisms. This conservation can be observed within viral families, such as Flaviviridae (fusion proteins; Apellániz et al., 2014), Caudovirales (tail complexes; Parent et al., 2018), or Orthomyxoviridae and Paramyxoviridae glycoproteins (Kordyukova, 2017). This conservation also occurs across viral kingdoms. The herpesvirus portal complex shares structural similarity with many bacteriophage portal proteins (McElwee et al., 2018; Newcomb et al., 2001), and the adenovirus spike protein is homologous with the bacteriophage Sf6 tail needle knob protein (Parent et al., 2012). Mimivirus-like GVs have eschewed these known genome release structures and appear to have forged their own mechanisms, as exemplified by the common stargate mechanism.
Numerous Proteins Are Released from GV Capsids during Stargate Opening
As obvious morphological changes occurred in GVs during low-pH and high-temperature treatments, we hypothesized that proteins were likely released from the capsids at each of these stages. We analyzed proteins that remained within the SMBV and TV capsids and proteins liberated from the capsids after each treatment. We used four conditions: native virions (pH 7.4; 25°C); low pH (pH 2; 25°C); high temperature (pH 7; 100°C); and a combination (pH 2; 100°C). We then performed pellet/supernatant separations to physically separate the virions from released proteins and analyzed the contents of each via SDS-PAGE (Figure 4). A preparation scheme can be seen in Figure S2. Antarctica and mimivirus both showed a similar banding pattern as SMBV. We did not perform mass spectrometry (MS) experiments with these viruses, as there is no annotated Antarctica virus genome and mimivirus and SMBV are highly similar (Campos et al., 2012).
Figure 4. SDS-PAGE of pH-2-Treated SMBV and TV.
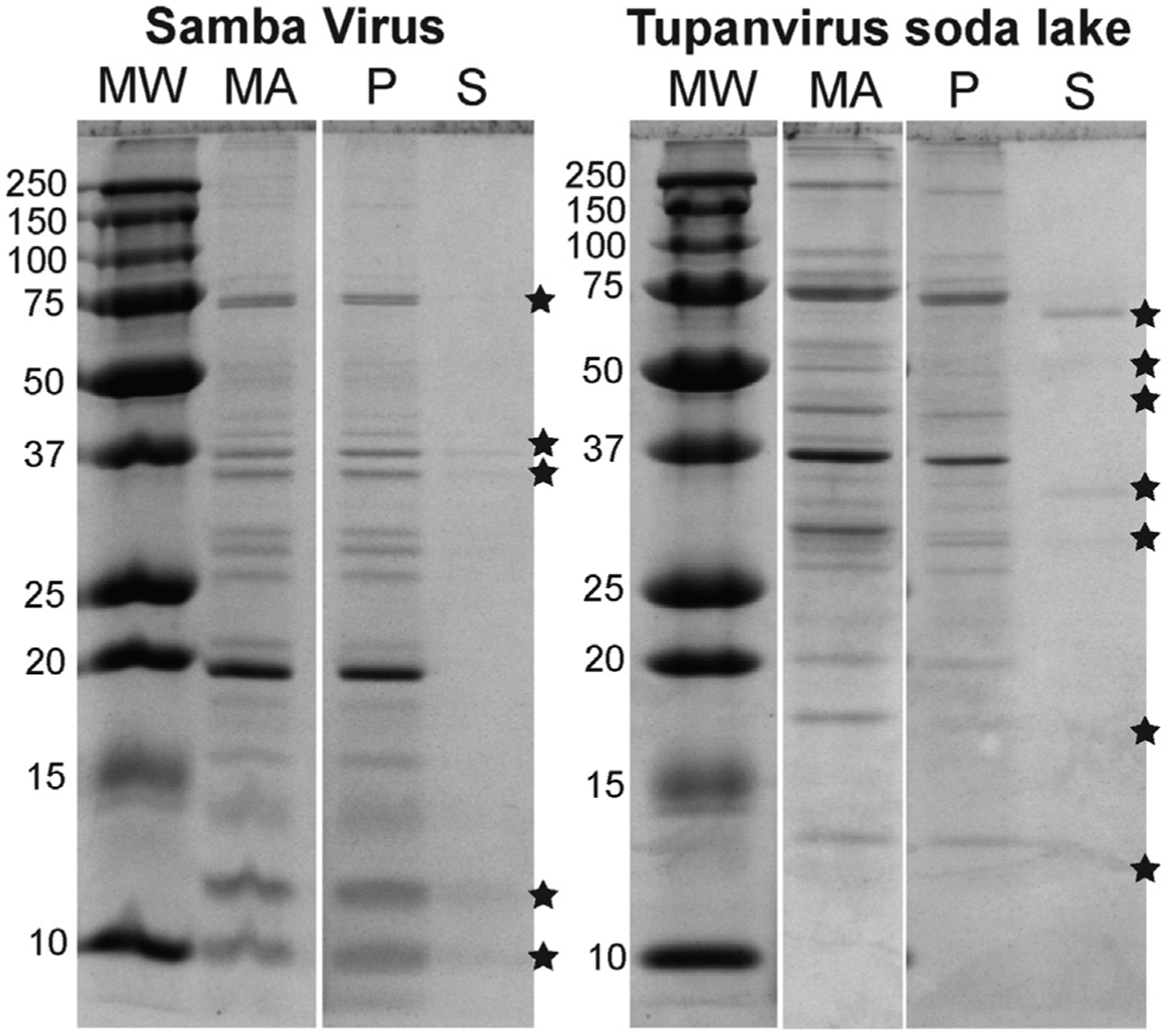
SDS-PAGE of SMBV and TV. MA, material applied (untreated viral particles); p, pellets from pH-2-treated virions; S, supernatants from pH-2-treated virions. Visible bands of proteins released into the supernatant are highlighted with asterisks. See Figure S2 for the sample preparation scheme.
For both SMBV and TV, distinct proteins were released from the capsid following low-pH treatment. Some of these proteins align with proteins in the native capsid (pellet) lane, suggesting they had been released from the capsid without significant modification/cleavage. Other proteins, especially in the TV sample, did not match proteins in the native capsid lane. These bands likely represent proteins that were cleaved during treatment. For both viruses, the native supernatant lanes did not contain any visible protein bands. When the particles were incubated at 100°C (with or without prior pH 2 treatment), it appeared that the majority of proteins were proteolytically cleaved and ran as a continuous smear on the gel, preventing detailed analysis of these samples.
Identifying the Proteins Released from SMBV and TV Virions at the Initiation of Infection
To characterize the proteins released during the initial stages of GV infection, we used MS. Initially, we used in-gel digestion of bands from the pH-2-treated SMBV and TV supernatant samples. Trypsinized fragments were analyzed via liquid chromatography-tandem mass spectrometry (LC-MS/MS), and the resultant peptides were compared to published SMBV and TV genome sequences (GenBank: KF959826.2 and GenBank: KY523104.1) as well as the A. castellanii genome (GenBank: KB007974.1) to identify host proteins. The A. castellanii actin protein was retained within these results, as this protein is known to play a role in the infection and genome release processes of Iridoviruses (Huang et al., 2018). From these initial data, we identified 48 SMBV and 26 TV proteins that are released from the virion following low-pH treatment (labeled with a [+] in the “band” column of Table 1).
Excising visible gel bands for MS analysis has the potential to miss proteins within the sample: some bands may be too faint to detect, some proteins may be too large or too small to be fully resolved or extracted, etc. Therefore, we also analyzed SMBV and TV samples using shotgun proteomics to maximize coverage in our study. We analyzed low-pH pellet and supernatant samples, as well as the untreated virus using the scheme shown in Figure S2. From this experiment, we identified 43 SMBV proteins and 37 TV proteins ([+] in the “shotgun” column of Tables 1 and S3). Of these proteins, 5 SMBV proteins and 7 TV proteins were previously identified from analysis of the gel bands.
In total, 78 SMBV proteins and 61 TV proteins were identified as having been released from the capsids at low pH. TV was isolated from an environment with high salinity and alkaline pH (9–12; Abrahão et al., 2018). SMBV, on the other hand, was isolated from a tributary of the Amazon River, a relatively neutral environment. Due to its location, TV had to evolve pH stability into its capsid to a greater extent than SMBV. Although TV was originally isolated from a basic environment, some of the strategies that the virus could have developed to stabilize its proteins, such as using a higher percentage of non-polar amino acids, could also stabilize the proteins at low pH.
187 and 169 total proteins were identified within the untreated mature virions of TV and SMBV, respectively (Figure 5). To identify proteins that had been released, we calculated the percent of the total peptide signal for each. We compared these percentages across the three samples, specifically looking at the ratios of supe:MA (material applied) and pellet:MA. Proteins where the supe:MA > 1 was enriched in the treated supernatant sample, indicating that they had been released from the capsids. These proteins are identified with a (+) in the “up in supe” column of Table 1. Conversely, proteins with pellet:MA < 1 were less abundant in the treated pellet than the native particles and likely also released. These proteins are identified with a (+) in the “down in pellet” column of Table 1. Proteins that are enriched in the supernatant samples are definitely released from the GV capsids, as no proteins were identified in the untreated supernatant samples (data not shown). Proteins that are depleted in the pellet samples are also likely released from the GV particles, although it is unlikely that any of these proteins are completely absent from the pellet samples (see POP in Figure 1A).
Figure 5. Comparison of Proteins Released by SMBV and TV Soda Lake.
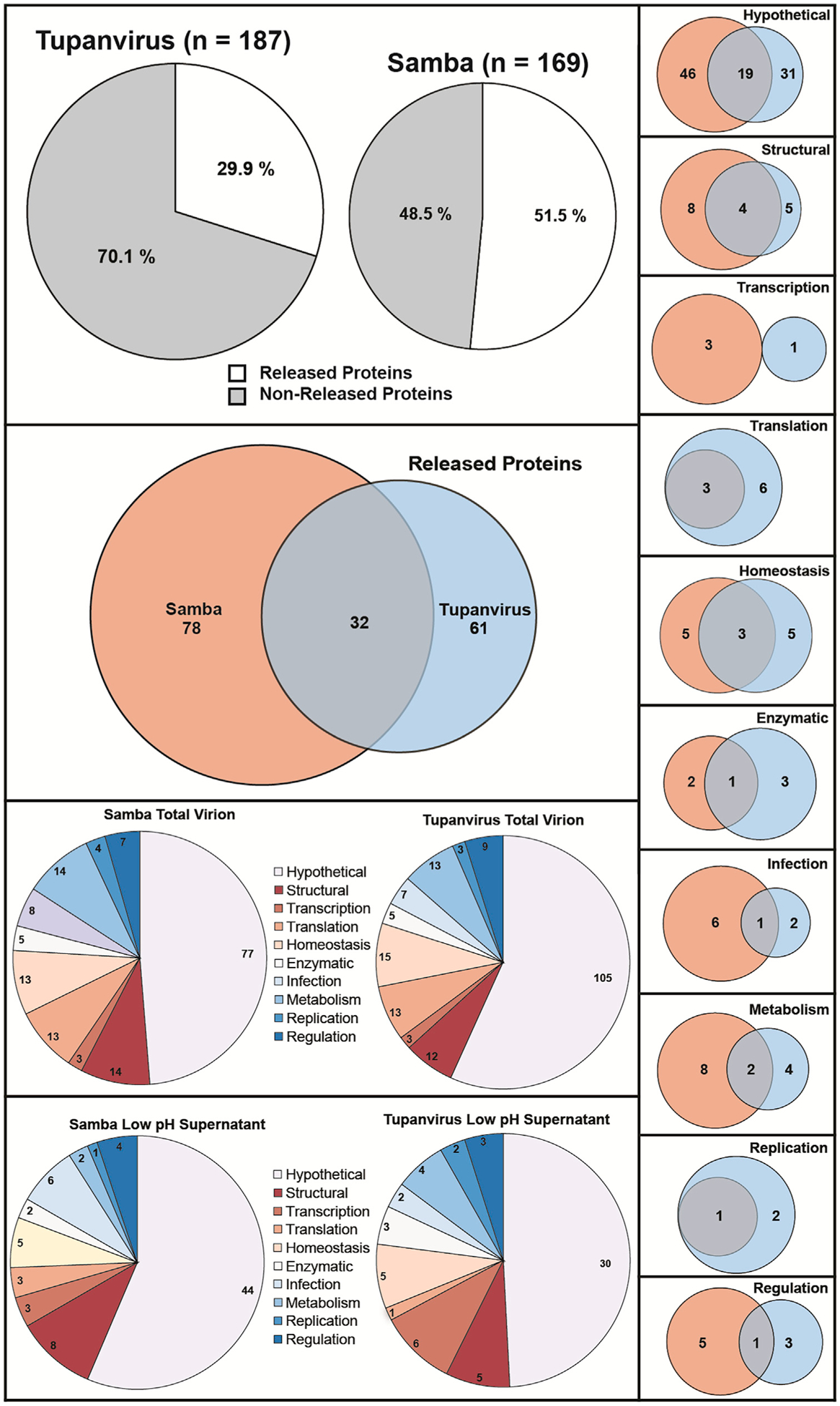
Venn diagrams comparing the total protein content and proteins released following low pH treatment of SMBV and TV particles. The homology present within these protein sets is depicted. See Table S2 for hypothetical proteins with predicted transmembrane domains and Table S3 for the relative abundance of individual proteins. See also Figure S2.
SMBV releases a higher number and percentage of these proteins (86; 51.5%) than TV particles (56; 29.9%). Putative functions for the released proteins were determined via (1) previous annotation (Abrahão et al., 2018; Campos et al., 2014), (2) NCBI BLAST analysis, (3) HHBLITS analysis (Remmert et al., 2011), (4) InterPro functional prediction (Mitchell et al., 2019), and (5) PSIPRED domain prediction using DomPred (Buchan and Jones, 2019; Jones and Cozzetto, 2015). Released proteins for each virus were separated into the following 10 categories: hypothetical; structural; transcription; translation; homeostasis; enzymatic; infection; metabolism; replication; and regulation (Figures 5B–5N).
Many of the proteins released for each virus (more than 50% for SMBV) are hypothetical proteins or proteins with unknown function. 19 of the hypothetical proteins released by the two viruses displayed obvious homology between SMBV and TV (BLAST results or functional homology prediction). All of the released SMBV proteins predicted to be involved in both translation and replication had homologs among the released TV proteins. The proteins predicted to be involved in transcription and regulation, on the other hand, did not show any readily apparent homology. The homology between the released TV and SMBV proteins in general and within each category can be found in Figure 5.
Expected Protein Types Are Released from SMBV and TV Virions during Genome Release
GVs need to carry out the same basic stages of the viral life cycle as their smaller cousins to replicate. Common stages include genome translocation into the host cell, blocking host replication, hijacking host machinery to make viral proteins, and making new viral proteins. Both SMBV and TV likely release proteins that are predicted to perform these functions, as many smaller viruses release whole proteins or peptides to facilitate this function (Manning et al., 2018; Parent et al., 2018; Wu et al., 2016). Hypothetical or unknown function proteins released from GV particles likely aid in performing these critical functions, as many of them are released during the initial phase of opening. Aside from identifying putative functions for some hypothetical proteins discussed below, determining the specific function of all of these proteins lies beyond the scope of this study.
Before the virus is able to hijack the host machinery and begin replication, it must enter the cell and translocate its genome across the phagosomal membrane into the cytoplasm. SMBV releases putative membrane proteins, such as a virion-associated membrane protein (AHJ40731.1) and as well as hypothetical proteins with predicted transmembrane domains that may play a role in membrane fusion (“H/TM” in Table 1). Both the extra membrane sac and the nucleocapsid must fuse with the phagosome to deliver the viral seed and dsDNA genome into the cytoplasm. This fusion event can clearly be seen in micrographs of infected cells (Figure 6). We will investigate the fusion capability of these newly identified proteins directly in future studies. Therefore, the results of this study help to assign putative roles to many proteins with previously unknown function, highlighting the power of this new method.
Additionally, both SMBV and TV release proteins were predicted to play a role in an ubiquitin-proteasome degradation pathway (UPP) (delineated by d in Table 1). These proteins are known to facilitate genome release in other viruses, including the large, but not quite giant, Iridoviruses (Huang et al., 2018) and herpesviruses (Greene et al., 2012). In Iridovirus infection, the UPP is coupled with metabolic, cytoskeletal, macromolecule biosynthesis, and signal transduction proteins to facilitate infection (Huang et al., 2018). Proteins predicted to carry out these functions are released from both the SMBV and TV virions along-side the UPP-related proteins (b in Table 1). Hypothetical proteins with additional functional information predicted via BLAST, HHBLIST, PSIPRED, or InterPro are listed in Table S2.
Following genome translocation, the virus forces the cell machinery to transition from making new cellular products to making viral components. Both SMBV and TV release various subunits of a DNA-dependent RNA polymerase (SMBV: AHJ39967.2, AHJ40151.2, and AHJ40172.1; TV: AUL78016.1, AUL78362.1, AUL78368.1, and AUL78302.1). This series of proteins is critical for the life cycle of the virus, as it directs the cellular machinery of the host to recognize viral DNA in lieu of cellular DNA. These proteins, especially the various DNA-dependent RNA polymerases, may play a role in transcription as hypothesized to occur following stargate opening but before nucleocapsid release (Mutsafi et al., 2014) Additional proteins in this category likely include some of the metabolic proteins released by the viruses, especially the catabolic proteins that may play a role in degrading host defenses and machinery. These proteins include a SMBV thiol protease (AMK61869.1), a SMBV amine oxidase (AHJ39955.1), and a hypothetical TV protein with a predicted inosine/uridine-favoring nucleoside hydrolase domain (AUL71835.1). Aside from these RNA polymerase subunits, both TV and SMBV release proteins that facilitate transcription. SMBV releases a poly (A) polymerase (AHJ40056.1), an mRNA-capping enzyme (AHJ40083.1), and an anaerobic transcription regulator (AMK61903.1). TV releases an SNF2 family helicase (AUL77941.1), an ATP-dependent RNA helicase (AUL77829.1), and a mimivirus-like elongation factor (AUL78714.1).
Many of the proteins we identified matched proteins that one would expect to be released during the initial stages of viral infection and greatly support our hypothesis that the in vitro stages generated in this study are reflective of those that occur in vivo. These data provide new insights into GV biology and ultimately lead to our proposed model (see next sections).
SMBV and TV Also Release Novel Proteins during Stargate Opening
SMBV and TV also release proteins that are relatively uncommon among viruses. These proteins include metal-binding homeostasis proteins as well as chemotaxis-regulating proteins. Functions for all predicted proteins are based on sequence similarity to known proteins; we did not confirm the biological function of these proteins with experimental data, and this will require follow-up studies.
Our mass spectrometry data conclusively show that both SMBV and TV release proteins predicted to play a role in maintaining homeostasis (Figure 5E). Many of these proteins likely have redox activity, protecting the virus and its cargo from reactive oxygen species (ROS) that can be found in the host phagosome (Flannagan et al., 2015). These proteins include several thioredoxin-like or thioredoxin-domain-containing proteins (SMBV: AHJ40071.1 and AHJ40129.2; TV: AUL77963.1) and glutaredoxins (SMBV: AMK618100.1; TV: AUL78724.1). TV releases a catalase protein (AUL78097.1) as well as glyoxylase (AUL78134.1) although SMBV releases a prolyl 4-hydroxylase (AMK61959.1). These proteins are also projected to protect the GV from ROS during the infection process. Here, we show that these proteins are indeed released very early in the infection process.
Redox-active proteins are also thought to play an important role in protecting the viruses from the harsh conditions present in the host phagosome. During phagocytosis, amoebal phagosomes drop to ~pH 4 (not low enough to trigger stargate opening in vitro), but they are also inundated with metals (like Cu and Zn) and ROS (German et al., 2013; Lopez and Skaar, 2018). Both viruses release metal-binding proteins (e in Table 1), including SMBV’s lanosterol demethylase (AHJ40393.1)—a cytochrome p450-like protein—and prolyl 4-hydroxylase (AMK61959.1) and TV’s mg709 (AUL77661.1)—a putative prolyl 4-hydroxylase with iron ion binding capabilities—and Cu-Zn superoxide dismutase (AUL78503.1). In conjunction with the ROS-mitigating proteins described above, these proteins likely allow GVs to survive the onslaught of low pH, high ROS, and high metal concentration found inside of the host phagosomes. We also note that the low pH of the phagosomes is similar to the low pH used in our in vitro assay, likely reflecting a physiologically relevant stage that describes GV infection mechanisms.
Although Tupanvirus infection is hypothesized to occur through phagocytosis (Abrahão et al., 2018; Silva et al., 2019), no biological data have yet been provided to substantiate said hypothesis. This proposal stems from visualization of phagocytosis of TV by Vermamoeba vermiformis and subsequent TV stargate opening via thin-section TEM (Silva et al., 2019). Thin-section TEM, embedding biological samples within epoxy resin then slicing thin, electron translucent sections off of the block, is prone to structural artifacts (Baker et al., 1999). Therefore, it is critical that any hypotheses generated from thin-section TEM imaging are supported by data from another technique. The release of proteins capable of mitigating the harsh environment of the amoebal phagosome provides biological evidence to support this hypothesis.
SMBV and TV also contain proteins that are predicted to regulate chemotaxis. SMBV releases a chemotaxis protein (AHJ40337.1) that shares homology with the putative chemotaxis protein CheD found in mimivirus (AKI80461.1) and TV (AUL78687.1). CheD proteins regulate chemotaxis via deamidation of chemotaxis receptors (InterPro). TV has been shown to shut down host chemotaxis (Oliveira et al., 2019; Rodrigues et al., 2019a), and it is likely that these CheD-like chemotaxis regulation proteins are involved in this process. Although TV does contain a CheD-like chemotaxis protein that was identified in the total virion MS data, this protein was not released following low pH treatment.
Opening the Stargate to New Avenues of GV Research
We were able to mimic four unique stages of the GV genome release process (Figure 6). GV particles that mimic these genome release stages have been seen in previous experiments (Abrahão et al., 2018; Schrad et al., 2017; Xiao et al., 2009; Zauberman et al., 2008) although relied on finding the “one-in-a-million” particle in the correct state. We are now able to mimic GV genome release stages reliably and with high frequency. Additionally, these conditions forgo the need to synchronize infection and trap GV particles in phagosomes at very specific times. Eschewing the host cell may limit specific avenues of study, such as searching for a host receptor(s), but it dramatically simplifies any studies aimed at the virus and the changes it undergoes during the genome release process.
Additionally, we have identified proteins that are released during the initial stages of infection in two GVs, SMBV and TV. Over half of the proteins released by these viruses are annotated as hypothetical, low complexity, or as an ORFan (which are new protein-coding genes restricted to taxonomically related genomes but are without homologs in other lineages). We were able to provide functional predictions for some of these proteins through homology. The release of these proteins at the initiation of stargate opening suggests that these proteins play an important role in the early stages of GV infection (phagosome survival, genome translocation, early transcription, host defense suppression, etc.). The exact functions of these proteins, as well as how their interactions mediate and orchestrate GV infection, are prime candidates for future study, and the importance is enhanced by the fact that many GVs appear to share similar strategies for genome release. All four of the GVs tested in this study responded similarly to treatment in vitro, suggesting that these GVs utilize similar molecular forces during genome release and likely similar proteins.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Kristin Parent (kparent@msu.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The full proteomics datasets generated in this study are available upon request from the Lead Contact.
Three-dimensional tomograms have been deposited to the Electron Microscopy Database (EMDB) under the ID codes EMD-20747 (pH 2, Video S3), EMD-20748 (100°C, Video S4), EMD-20746 (pH 2 + 100°C, Video S6), and EMD-20745 (pH 2 + 100°C, Video S8).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Acanthamoeba castellanii
Acanthamoeba castellanii cells were purchased from ATCC (ATCC 30010). Acanthamoeba castellanii (ATCC 30010) was cultivated in 712 PYG media w/Additives (ATCC recipe) at pH 6.5 in the presence of gentamicin (15 μg/mL) and penicillin/streptomycin (100 U/mL) at 28°C to reach a 90% confluence.
Giant Viruses
Tupanvirus soda lake (TV), Antarctica virus, and Samba virus (SMBV) were isolated previously (Abrahão et al., 2018; Andrade et al., 2018; Campos et al., 2014). M4 virus was kindly provided by Dr. Bernard La Scola and Dr. Thomas Klose (Boyer et al., 2011). Acanthamoeba castellanii (ATCC 30010) was cultivated in 712 PYG media w/Additives (ATCC recipe) at pH 6.5 in the presence of gentamicin (15 μg/mL) and penicillin/streptomycin (100 U/mL) at 28°C to reach a 90% confluence. SMBV or TV virions were diluted in phosphate buffered saline (PBS) and added to the cells to a multiplicity of infection of 5 (TV) or 10 (SMBV). An initial incubation was carried out for one hour at room temperature. After the initial incubation, additional PYG media was added to the cells and the flasks were incubated at 28°C for 48 hours. After 48 hours, more of the free amoebal cells had been lysed. Suspensions containing cell debris and cell particles were centrifuged at 900 × g to pellet residual cells. The resulting supernatant was filtered using a 2 μm filter and was immediately applied to a 22% sucrose cushion (w/w) at 15,000 × g for 30 min. Viral pellets were resuspended in PBS and stored at −80°C. Viruses were tittered using the Reed-Muench protocol (Ramakrishnan, 2016). On average, virus isolation yielded 1010 TCID50/mL (TCID = tissue culture infective dose).
METHOD DETAILS
Treatment of SMBV Particles and Image Analysis
Determining the Percentage of Open SMBV Particles
For all treatments, the percentage of open SMBV particles (POP) was determined via 2D cryo-electron microscopy. This was calculated by dividing the number of clearly opened particles by the number of total particles visualized for each condition. In general, there experiments were performed either in duplicate or triplicate in order to generate a minimum of 50 particles imaged per condition. These percentages were compared to the native (untreated) level of spontaneous SMBV particle opening, determined previously to be ~5% (Schrad et al., 2017).
Conditions That Did Not Increase POP
SMBV particles were treated with various conditions that have been shown to disrupt/destroy other viruses. These conditions include urea, guanidinium hydrochloride, DMSO, Triton X-100, chloroform, DNase I, and an enzyme cocktail (lysozyme, bromelain, proteinase K) that was previously shown to remove APMV fibers (Kuznetsov et al., 2010). Treatments were applied for 1–2 hours prior to POP determination via cryo-EM. Concentrations for the various conditions, as well as the resultant POP values, can be found in Table S1.
pH Titration of SMBV Particles
25–50 μL of SMBV particles were added to Millipore VSWP Membrane Filter dialysis discs (0.025 μm cutoff) which were then floated onto ~25 mL of 20 mM sodium phosphate buffer, adjusted to the desired pH. The samples were allowed to equilibrate for 1.5–2 hours. For conditions where low pH would interfere with additional treatment (e.g., pH 2 + DNase I or pH 2 samples submitted for mass spectrometry) the particles were dialyzed for an additional 1.5–2 hours against pH 7.0 buffer to restore neutral pH.
High Temperature Incubation
GV particles were incubated in a BioRad T100 thermal cycler at 80, 89, and 100°C for 1 hour. SMBV particles remained intact following 1 hours at 100°C, so additional incubations at 100°C were performed at 2, 3, or 6 hours. As a control, SMBV particles were also incubated for 1 hour at room temperature (25°C).
Combining High Temperature and Low pH
To determine the effect of combining low pH and high temperature, GV particles were sequentially treated with pH 2 and 100°C. First, SMBV particles were dialyzed against 20 mM sodium phosphate buffer, adjusted to pH 2, for 2 hours. Following dialysis, SMBV particles were incubated at 100°C for 3 hours.
Cryo-Electron Microscopy (Cryo-EM) and Cryo-Electron Tomography (Cryo-ET)
Sample Preparation
Samples for cryo-EM and cryo-ET were prepared as described previously (Schrad et al., 2017). Briefly, small (3–5 μL) aliquots of virus particles were applied to R2/2 (cryo-EM) or R 3.5/1 (cryo-ET) Quantifoil grids (Electron Microscopy Solutions) that had been plasma cleaned for 20 s in a Fischione model 1020 plasma cleaner. Prior to virus addition, 5–10 μL of 10 nm nanogold fiducial markers were applied to the R3.5/1 grids and were air-dried to provide markers for fiducial alignment of the tilt series. The samples were plunge frozen in liquid ethane using a manual plunge-freezing device (Michigan State University Physics Machine Shop) at room temperature using approximately 5–7 s of blotting time per grid. Frozen-hydrated samples were stored, transferred, and imaged under liquid nitrogen temperatures.
2D Cryo-Electron Microscopy
Cryo-EM experiments were performed at Michigan State University. Virus particles were imaged in either a JEOL 2200-FS TEM or an FEI Talos Arctica. The JEOL was operated operating at 200 keV, using low dose conditions controlled by SerialEM (version 3.5.0-beta, (Mastronarde, 2005)) with the use of an in-column Omega Energy Filter operating at a slit width of 35 eV. Micrographs were recorded at 25 frames per second using a Direct Electron DE-20 direct detector, cooled to −38°C. Motion correction was performed using the Direct Electron software package (Direct Electron, LLC). The Arctica was also operated at 200 keV, under low dose conditions controlled by EPU. Micrographs were recorded on a Falcon 3 direct detector operating in linear mode. Micrographs were collected between 8,000 and 10,000 × nominal magnification (6.87 and 5.30Å/pixel, respectively) on the JEOL and at 17,500 × nominal magnification (6.03Å/pixel) on the Arctica. The objective lens defocus settings ranged from 10 to 15 μm underfocus. Micrographs were collected for 5 s, resulting in a total dose of ~35 e-/Å2. For bubblegram imaging, the SMBV particles were imaged for an additional four exposures, resulting in a total does of ~140 e-/Å2.
Cryo-Electron Tomography
Cryo-ET tilt series were collected using a Titan Krios TEM operating at 300 keV with a post-column GIF (20 eV slit width) under low dose conditions controlled by SerialEM or Leginon at Purdue University. Images were collected using a Gatan K2 direct electron detector operating at 100 ms/frame. Images were collected in super resolution mode between 33,000 and 53,000 × nominal magnification (2.12 – 1.33Å/pixel). Tilt series were carried out between ± 50° with bidirectional image collection every 2°. Images were collected for 5 s (100 frames/tilt image), resulting in ~2.5 electrons/Å2 per tilt image (~125 electrons/Å2, total exposure dose).
Individual micrographs were corrected for particle motion and binned by a factor of two using MotionCor2 v1.2.0 (Zheng et al., 2017) and the corrected images were stitched back into a tilt series using the newstack functionality in IMOD (Mastronarde and Held, 2017). Tilt series alignment, using fiducial markers, and tomogram generation was carried out using IMOD v4.7.5. Final tomogram volumes were generated using ten iterations of the SIRT reconstruction method (Mastronarde, 1997) then filtered using the smooth (3×3 kernel) and median (size 3) options in IMOD. Select tomograms were annotated using Amira v2019.2 (ThermoFisher Scientific).
Scanning Electron Microscopy
SEM. Preparation and Imaging
GV particles were imaged using a JEOL JSM-7500F scanning electron microscope. Prior to imaging, virus particles were desiccated using an EM CPD300 critical point dryer, fixed with glutaraldehyde onto poly-L-Lysine treated SEM slides, and sputter coated with a ~2.7nm layer of iridium using a Q150T Turbo Pumped Coater. Particles were imaged between 8,500 × and 85,000 × nominal magnification.
Ultrathin Sections and Transmission Electron Microscopy
A. castellanii cells were infected by APMV or Tupanvirus at a multiplicity of infection of 5 and fixed at 1 hours post infection with 2.5% glutaraldehyde in a 0.1 M sodium phosphate buffer for 1 h at room temperature. The amoebas were postfixed with 2% osmium tetroxide and embedded in Epon resin. Ultrathin sections (50–100 nm) were then analyzed using transmission electron microscopy (Spirit Biotwin FEI-120 kV) at the Center of Microscopy of Universidade Federal de Minas Gerais (UFMG).
Differential Mass Spectrometry
Sample Preparation
SMBV and TV particles were dialyzed against 20 mM sodium phosphate buffer, adjusted to pH 2, for 2 hours, as described above. An aliquot of each virus was left undialyzed as a control (Material Applied, MA). Following dialysis, proteins that had been released from the viral particles were separated from the virions via centrifugation in a microcentrifuge at 8,000 × g for 15 minutes. Visible viral pellets were resuspended in the same volume as the supernatant using 20 mM sodium phosphate buffer, adjusted to pH 7.0. Two technical replicates were created for each sample. An aliquot of each sample was used for SDS-PAGE.
Each sample was TCA precipitated and submitted for LC/MS/MS analysis to the MSU Proteomics Core. Prior to submission, samples were run on a 15% polyacrylamide gel at a voltage of 200 V for 45 minutes. TV and SMBV gel bands visible by Coomassie blue stain were excised and submitted for MS analysis as well.
Proteolytic Digestion
TCA precipitated pellets were re-suspended in 270uL of 100mM ammonium bicarbonate supplemented with 10% trifluoroethanol. Samples were reduced and alkylated by adding TCEP and Iodoacetamide at 10mM and 40mM, respectively and incubating for 5min at 45C with shaking at 1400 rpm in an Eppendorf ThermoMixer. Trypsin, in 100mM ammonium bicarbonate, was added at a 1:100 ratio (wt/wt) and the mixture was incubated at 37C overnight. Final volume of each digest was ~300uL. After digestion, the samples were acidified to 2% TFA and subjected to C18 solid phase clean up using StageTips (Rappsilber et al., 2007) to remove salts.
LC/MS/MS and Data Analysis
An injection of 5uL was automatically made using a Thermo EASYnLC 1200 onto a Thermo Acclaim PepMap RSLC 0.075mm × 20mm C18 trapping column and washed for ~5min with buffer A. Bound peptides were then eluted over 95min with a gradient of 8%B to 42%B in 84min, ramping to 100%B at 85min and held at 100%B for the duration of the run (Buffer A = 99.9% Water/0.1% Formic Acid, Buffer B = 80% Acetonitrile/0.1% Formic Acid/19.9% Water) at a constant flow rate of 300nl/min. Column temperature was maintained at a constant temperature of 50°C using and integrated column oven (PRSO-V2, Sonation GmbH, Biberach, Germany). Eluted peptides were sprayed into a ThermoScientific Q-Exactive HF-X mass spectrometer using a FlexSpray spray ion source. Survey scans were taken in the Orbi trap (60,000 resolution, determined at m/z 200) and the top ten ions in each survey scan are then subjected to automatic higher energy collision induced dissociation (HCD) with fragment spectra acquired at 7,500 resolution. The resulting MS/MS spectra are converted to peak lists using MaxQuant v1.6.0.1 (Cox and Mann, 2008) and searched using the Andromeda (Cox et al., 2011) algorithm against a protein database containing sequences from SMBV or TV and Acanthamoeba castellanii (each downloaded from NCBI, https://www.ncbi.nlm.nih.gov). Common laboratory contaminants were included in the Andromeda search. Protein and peptide FDR for all searches were set to 1%.
Mass Spectrometry Data Synthesis
The percentage of the total LFQ signal each protein was responsible for in each sample was calculated by dividing the individual protein LFQ signal by the total LFQ signal for the sample, excluding contaminates. Proteins that are released from the viral particles are expected to make up a higher percentage of the supernatant sample than the whole virion (MA), so the ratios of these two percentages were calculated (Tables 1 and S3). Proteins with a supernatant:MA ratio > 1 were selected for further analysis.
Classification/Functional Annotation of Proteins Identified via MS
TV and SMBV proteins released at low pH were classified via their predicted functions and domains. Primary functional annotation had been carried out previously for both TV (Abrahão et al., 2018) and SMBV (Campos et al., 2014). Additional functional prediction, as well as homology prediction between the two viruses, was carried out through the use of the NCBI BLAST database (NCBI) as well as the HHBLITS server (Remmert et al., 2011) and the InterPro database (Mitchell et al., 2019). Domain prediction was carried out by searching the InterPro database and utilizing the PSIPRED server (Buchan and Jones, 2019) with the DISOPRED3 (Jones and Cozzetto, 2015) functionality activated.
QUANTIFICATION AND STATISTICAL ANALYSIS
Mass Spectrometry Analysis
LFQ intensities for both SMBV and TV spectra were detected in triplicate. For each virus, the initial run did not produce high quality data so these intensities were disregarded. LFQ intensities from the remaining two runs were averaged together to produce the reported intensity (Table S3).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| Samba Virus | Campos et al., 2014 | N/A |
| Tupanvirus soda lake | Abrahão et al., 2018 | N/A |
| Antarctica virus | Andrade et al., 2018 | N/A |
| Mimivirus strain M4 | Boyer et al., 2011 | N/A |
| Biological Samples | ||
| Acanthamoeba castellanii | ATCC | ATCC 30010 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Sodium phosphate, dibasic | Sigma-Aldrich | Cat # S9390 |
| Urea | VWR Life Sciences | Cat # 0568 |
| Guanidinium Hydrochloride | Amresco | Cat #0118 |
| Triton X-100 | Alfa Aesar | Cat # A16046 |
| Chloroform | EMD | Cat # CX1055 |
| Dimethyl sulfoxide | JT Baker | Cat # 9924-01 |
| Bromelain (from pineapple) | Sigma | Cat # 647-005-00-X |
| Proteinase K | Roche | Cat #03115879001 |
| DNase I | Roche | Cat # 10104159001 |
| Lysozyme | Amresco | Cat # 0663-5G |
| 712 PYG | ATCC | N/A |
| Sucrose | JT Baker | Cat # 4072-05 |
| Software and Algorithms | ||
| IMOD | Mastronarde and Held, 2017 | https://bio3d.colorado.edu/imod/download.html |
| SerialEM | Mastronarde, 2005 | https://bio3d.colorado.edu/SerialEM/download.html |
| Leginon | Suloway et al., 2005 | http://nramm.nysbc.org/downloads/ |
| Amira | ThermoFischer Scientific | https://www.thermofisher.com/us/en/home/industrial/electron-microscopy/electron-microscopy-instruments-workflow-solutions/3d-visualization-analysis-software/amira-life-sciences-biomedical.html |
| MaxQuant | Cox and Mann, 2008 | https://www.maxquant.org/ |
| Andromeda | Cox et al., 2011 | http://coxdocs.org/doku.php?id=maxquant:start/ |
| MotionCor2 | Zheng et al., 2017 | https://msg.ucsf.edu/software |
| Other | ||
| Quantifoil R2/2 Grids | Electron Microscopy Sciences | Cat # Q250-CR2 |
| 10 nm Nanogold Fiducial Markers | Aldrich (Millipore) | Cat #741957 |
| VSWP Membrane Filter Discs | Millipore | Cat # VSWP01300 |
| Quantifoil R3.5/1 Grids | Electron Microscopy Sciences | Cat # Q225CR-35 |
| Deposited Data Generated | ||
| pH 2 treated sample, Video S3 | This work | EMDB: EMD-20747 |
| 100°C treated sample, Video S4 | This work | EMDB: EMD-20748 |
| pH 2 + 100°C treated sample, Video S6 | This work | EMDB: EMD-20746 |
| pH 2 + 100°C treated sample, Video S8 | This work | EMDB: EMD-20745 |
Highlights.
Giant virus infection intermediates characterized by cryo-EM, cryo-ET, and SEM
Proteomics reveal proteins released from capsids during infection
Low pH treatment releases proteins present in the viral seed
Low pH and high temperature fully open the stargate and release the genome
ACKNOWLEDGMENTS
The authors would like to thank the MSU Proteomics and Mass Spectrometry. Facilities, especially Drs. D. Jones, C. Wilkerson, and D. Whiten, for their assistance with MS experiments. Additionally, we thank Drs. W. Jiang, T. Klose, and V. Bowman at Purdue University’s Midwest Cryo-EM Consortium (NIH Consortium no. U24GM116789-03). We would also like to thank C. Flegler at the MSU Center for Advanced Microscopy for her expertise with SEM experiments. We would like to thank the Microscopy Center of UFMG (for help with the thin-section TEM images). We would like to thank the MSU RTSF Cryo-EM Core Facility for use of the Talos Arctica. The MSU High Performance Computation Cluster (HPCC) provided computational tools and support for cryo-EM image motion correction. Dr. K. Padmanabhan and Dr. M. Feig provided additional assistance with computational resources and expert consultation in protein homology and functional predictions. Funding for this project was provided from the AAAS Marion Mason Milligan award for Women in the Chemical Sciences (K.N.P.) and the JK Billman, Jr., MD Endowed Research Professorship (K.N.P.). Funding was provided from Fundo de Amparo à Pesquisa do Estado do Rio de Janeiro e Conselho Nacional de Pesquisa and Co-ordenaç ão de Aperfeiçoamento de Pessoal de Nível superior, grant reference number 001 (J.R.C.) and the Burroughs Wellcome Fund (K.N.P.). J.R.S. has been supported by the Jack Throck Waston Fellowship and the August and Ernest Frey Research Fellowship from MSU and NIH R01 GM110185 (K.N.P.). J.S.A. has been supported by CNPq, CAPES, MSU, and FAPEMIG. Nividia provided GPU support for cryo-EM and cryo-ET image processing.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2020.04.032.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Abrahão JS, Dornas FP, Silva LC, Almeida GM, Boratto PV, Colson P, La Scola B, and Kroon EG (2014). Acanthamoeba polyphaga mimivirus and other giant viruses: an open field to outstanding discoveries. Virol. J 11, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahão J, Silva L, Silva LS, Khalil JYB, Rodrigues R, Arantes T, Assis F, Boratto P, Andrade M, Kroon EG, et al. (2018). Tailed giant Tupanvirus possesses the most complete translational apparatus of the known virosphere. Nat. Commun 9, 749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aherfi S, Colson P, Audoly G, Nappez C, Xerri L, Valensi A, Million M, Lepidi H, Costello R, and Raoult D (2016a). Marseillevirus in lymphoma: a giant in the lymph node. Lancet Infect. Dis 16, e225–e234. [DOI] [PubMed] [Google Scholar]
- Aherfi S, Colson P, La Scola B, and Raoult D (2016b). Giant viruses of amoebas: an update. Front. Microbiol 7, 349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade KR, Boratto PP, Rodrigues FP, Silva LC, Dornas FP, Pilotto MR, La Scola B, Almeida GM, Kroon EG, and Abrahão JS (2015). Oysters as hot spots for mimivirus isolation. Arch. Virol 160, 477–482. [DOI] [PubMed] [Google Scholar]
- Andrade ACDSP, Arantes TS, Rodrigues RAL, Machado TB, Dornas FP, Landell MF, Furst C, Borges LGA, Dutra LAL, Almeida G, et al. (2018). Ubiquitous giants: a plethora of giant viruses found in Brazil and Antarctica. Virol. J 15, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreani J, Aherfi S, Bou Khalil JY, Di Pinto F, Bitam I, Raoult D, Colson P, and La Scola B (2016). Cedratvirus, a double-cork structured giant virus, is a distant relative of pithoviruses. Viruses 8, E300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreani J, Khalil JYB, Sevvana M, Benamar S, Di Pinto F, Bitam I, Colson P, Klose T, Rossmann MG, Raoult D, and La Scola B (2017). Pacmanvirus, a new giant icosahedral virus at the crossroads between Asfar-viridae and Faustoviruses. J. Virol 91, e00212–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreani J, Khalil JYB, Baptiste E, Hasni I, Michelle C, Raoult D, Le-vasseur A, and La Scola B (2018). Orpheovirus IHUMI-LCC2: a new virus among the giant viruses. Front. Microbiol 8, 2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apellániz B, Huarte N, Largo E, and Nieva JL (2014). The three lives of viral fusion peptides. Chem. Phys. Lipids 181, 40–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azza S, Cambillau C, Raoult D, and Suzan-Monti M (2009). Revised Mimivirus major capsid protein sequence reveals intron-containing gene structure and extra domain. BMC Mol. Biol 10, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TS, Olson NH, and Fuller SD (1999). Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol. Mol. Biol. Rev 63, 862–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boratto PVM, Dornas FP, da Silva LCF, Rodrigues RAL, Oliveira GP, Cortines JR, Drumond BP, and Abrahão JS (2018). Analyses of the Kroon virus major capsid gene and its transcript highlight a distinct pattern of gene evolution and splicing among Mimiviruses. J. Virol 92, e01782–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothner B, Taylor D, Jun B, Lee KK, Siuzdak G, Schultz CP, and Johnson JE (2005). Maturation of a tetravirus capsid alters the dynamic properties and creates a metastable complex. Virology 334, 17–27. [DOI] [PubMed] [Google Scholar]
- Boyer M, Azza S, Barrassi L, Klose T, Campocasso A, Pagnier I, Fournous G, Borg A, Robert C, Zhang X, et al. (2011). Mimivirus shows dramatic genome reduction after intraamoebal culture. Proc. Natl. Acad. Sci. USA 108, 10296–10301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandes N, and Linial M (2019). Giant viruses-big surprises. Viruses 11, E404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan DWA, and Jones DT (2019). The PSIPRED Protein Analysis Work-bench: 20 years on. Nucleic Acids Res. 47 (W1), W402–W407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos RK, Andrade KR, Ferreira PC, Bonjardim CA, La Scola B, Kroon EG, and Abrahão JS (2012). Virucidal activity of chemical biocides against mimivirus, a putative pneumonia agent. J. Clin. Virol 55, 323–328. [DOI] [PubMed] [Google Scholar]
- Campos RK, Boratto PV, Assis FL, Aguiar ER, Silva LC, Albarnaz JD, Dornas FP, Trindade GS, Ferreira PP, Marques JT, et al. (2014). Samba virus: a novel mimivirus from a giant rain forest, the Brazilian Amazon. Virol. J 11, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson P, La Scola B, and Raoult D (2017). Giant viruses of amoebae: a journey through innovative research and paradigm changes. Annu. Rev. Virol 4, 61–85. [DOI] [PubMed] [Google Scholar]
- Conway JF, Wikoff WR, Cheng N, Duda RL, Hendrix RW, Johnson JE, and Steven AC (2001). Virus maturation involving large subunit rotations and local refolding. Science 292, 744–748. [DOI] [PubMed] [Google Scholar]
- Conway JF, Cheng N, Ross PD, Hendrix RW, Duda RL, and Steven AC (2007). A thermally induced phase transition in a viral capsid transforms the hexamers, leaving the pentamers unchanged. J. Struct. Biol 158, 224–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, and Mann M (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol 26, 1367–1372. [DOI] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, and Mann M (2011). Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res 10, 1794–1805. [DOI] [PubMed] [Google Scholar]
- Dhindwal S, Avila B, Feng S, and Khayat R (2019). Porcine Circovirus 2 uses a multitude of weak binding sites to interact with heparan sulfate, and the interactions do not follow the symmetry of the capsid. J. Virol 93, e02222–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornas FP, Rodrigues FP, Boratto PV, Silva LC, Ferreira PC, Bonjardim CA, Trindade GS, Kroon EG, La Scola B, and Abrahão JS (2014a). Mimivirus circulation among wild and domestic mammals, Amazon Region, Brazil. Emerg. Infect. Dis 20, 469–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornas FP, Silva LC, de Almeida GM, Campos RK, Boratto PV, Franco-Luiz AP, La Scola B, Ferreira PC, Kroon EG, and Abrahão JS (2014b). Acanthamoeba polyphaga mimivirus stability in environmental and clinical substrates: implications for virus detection and isolation. PLoS ONE 9, e87811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannagan RS, Heit B, and Heinrichs DE (2015). Antimicrobial mechanisms of macrophages and the immune evasion strategies of Staphylococcus aureus. Pathogens 4, 826–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed EO (2015). HIV-1 assembly, release and maturation. Nat. Rev. Microbiol 13, 484–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German N, Doyscher D, and Rensing C (2013). Bacterial killing in macrophages and amoeba: do they all use a brass dagger? Future Microbiol. 8, 1257–1264. [DOI] [PubMed] [Google Scholar]
- Ghigo E, Kartenbeck J, Lien P, Pelkmans L, Capo C, Mege JL, and Raoult D (2008). Ameobal pathogen mimivirus infects macrophages through phagocytosis. PLoS Pathog. 4, e1000087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene W, Zhang W, He M, Witt C, Ye F, and Gao SJ (2012). The ubiquitin/proteasome system mediates entry and endosomal trafficking of Kaposi’s sarcoma-associated herpesvirus in endothelial cells. PLoS Pathog. 8, e1002703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison SC (2015). Viral membrane fusion. Virology 479–480, 498–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SM, Kuo ST, Kuo HC, and Chang SK (2018). Assessment of fish iridoviruses using a novel cell line GS-1, derived from the spleen of orange-spotted grouper Epinephelus coioides (Hamilton) and susceptible to ranavirus and megalocytivirus. J. Vet. Med. Sci 80, 1766–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs SE, Lamson DM, St George K, and Walsh TJ (2013). Human rhinoviruses. Clin. Microbiol. Rev 26, 135–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, and Cozzetto D (2015). DISOPRED3: precise disordered region predictions with annotated protein-binding activity. Bioinformatics 31, 857–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant R, Llauró A, Rayaprolu V, Qazi S, de Pablo PJ, Douglas T, and Bothner B (2018). Changes in the stability and biomechanics of P22 bacteriophage capsid during maturation. Biochim. Biophys. Acta Gen. Subj 1862, 1492–1504. [DOI] [PubMed] [Google Scholar]
- Kerepesi C, and Grolmusz V (2017). The “giant virus finder” discovers an abundance of giant viruses in the Antarctic dry valleys. Arch. Virol 162, 1671–1676. [DOI] [PubMed] [Google Scholar]
- Khan M, La Scola B, Lepidi H, and Raoult D (2007). Pneumonia in mice inoculated experimentally with Acanthamoeba polyphaga mimivirus. Microb. Pathog 42, 56–61. [DOI] [PubMed] [Google Scholar]
- Klose T, Kuznetsov YG, Xiao C, Sun S, McPherson A, and Rossmann MG (2010). The three-dimensional structure of Mimivirus. Intervirology 53, 268–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose T, Reteno DG, Benamar S, Hollerbach A, Colson P, La Scola B, and Rossmann MG (2016). Structure of faustovirus, a large dsDNA virus. Proc. Natl. Acad. Sci. USA 113, 6206–6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordyukova L (2017). Structural and functional specificity of Influenza virus haemagglutinin and paramyxovirus fusion protein anchoring peptides. Virus Res. 227, 183–199. [DOI] [PubMed] [Google Scholar]
- Kuznetsov YG, Xiao C, Sun S, Raoult D, Rossmann M, and McPherson A (2010). Atomic force microscopy investigation of the giant mimivirus. Virology 404, 127–137. [DOI] [PubMed] [Google Scholar]
- La Scola B, Audic S, Robert C, Jungang L, de Lamballerie X, Drancourt M, Birtles R, Claverie JM, and Raoult D (2003). A giant virus in amoebae. Science 299, 2033. [DOI] [PubMed] [Google Scholar]
- La Scola B, Marrie TJ, Auffray JP, and Raoult D (2005). Mimivirus in pneumonia patients. Emerg. Infect. Dis 11, 449–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre M, Bartoli J, Shmakova L, Jeudy S, Labadie K, Adrait A, Lescot M, Poirot O, Bertaux L, Bruley C, et al. (2014). Thirty-thousand-year-DNA viruses with a pandoravirus morphology. Proc. Natl. Acad. Sci. USA 111, 4274–4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre M, Lartigue A, Bertaux L, Jeudy S, Bartoli J, Lescot M, Alempic JM, Ramus C, Bruley C, Labadie K, et al. (2015). In-depth study of Mollivirus sibericum, a new 30,000-y-old giant virus infecting Acanthamoeba. Proc. Natl. Acad. Sci. USA 112, E5327–E5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez CA, and Skaar EP (2018). The impact of dietary transition metals on host-bacterial interactions. Cell Host Microbe 23, 737–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusi EA, Maloney D, Caicci F, and Guarascio P (2017). Questions on unusual Mimivirus-like structures observed in human cells. F1000Res. 6, 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangel WF, and San Martín C (2014). Structure, function and dynamics in adenovirus maturation. Viruses 6, 4536–4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning KA, Quiles-Puchalt N, Penadés JR, and Dokland T (2018). A novel ejection protein from bacteriophage 80α that promotes lytic growth. Virology 525, 237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN (1997). Dual-axis tomography: an approach with alignment methods that preserve resolution. J. Struct. Biol 120, 343–352. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN (2005). Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol 152, 36–51. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN, and Held SR (2017). Automated tilt series alignment and tomographic reconstruction in IMOD. J. Struct. Biol 197, 102–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElwee M, Vijayakrishnan S, Rixon F, and Bhella D (2018). Structure of the herpes simplex virus portal-vertex. PLoS Biol. 16, e2006191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milrot E, Shimoni E, Dadosh T, Rechav K, Unger T, Van Etten JL, and Minsky A (2017). Structural studies demonstrating a bacteriophage-like replication cycle of the eukaryote-infecting Paramecium bursaria chlorella virus-1. PLoS Pathog. 13, e1006562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell AL, Attwood TK, Babbitt PC, Blum M, Bork P, Bridge A, Brown SD, Chang HY, El-Gebali S, Fraser MI, et al. (2019). InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 47 (D1), D351–D360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutsafi Y, Zauberman N, Sabanay I, and Minsky A (2010). Vaccinia-like cytoplasmic replication of the giant Mimivirus. Proc. Natl. Acad. Sci. USA 107, 5978–5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutsafi Y, Fridmann-Sirkis Y, Milrot E, Hevroni L, and Minsky A (2014). Infection cycles of large DNA viruses: emerging themes and underlying questions. Virology 466–467, 3–14. [DOI] [PubMed] [Google Scholar]
- Newcomb WW, Juhas RM, Thomsen DR, Homa FL, Burch AD, Weller SK, and Brown JC (2001). The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol 75, 10923–10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira G, Silva L, Leão T, Mougari S, da Fonseca FG, Kroon EG, La Scola B, and Abrahão JS (2019). Tupanvirus-infected amoebas are induced to aggregate with uninfected cells promoting viral dissemination. Sci. Rep 9, 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent KN, Khayat R, Tu LH, Suhanovsky MM, Cortines JR, Teschke CM, Johnson JE, and Baker TS (2010). P22 coat protein structures reveal a novel mechanism for capsid maturation: stability without auxiliary proteins or chemical crosslinks. Structure 18, 390–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent KN, Gilcrease EB, Casjens SR, and Baker TS (2012). Structural evolution of the P22-like phages: comparison of Sf6 and P22 procapsid and virion architectures. Virology 427, 177–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent KN, Schrad JR, and Cingolani G (2018). Breaking symmetry in viral icosahedral capsids as seen through the lenses of X-ray crystallography and cryo-electron microscopy. Viruses 10, E67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe N, Legendre M, Doutre G, Couté Y, Poirot O, Lescot M, Arslan D, Seltzer V, Bertaux L, Bruley C, et al. (2013). Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science 341, 281–286. [DOI] [PubMed] [Google Scholar]
- Popgeorgiev N, Michel G, Lepidi H, Raoult D, and Desnues C (2013). Marseillevirus adenitis in an 11-month-old child. J. Clin. Microbiol 51, 4102–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan MA (2016). Determination of 50% endpoint titer using a simple formula. World J. Virol 5, 85–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Mann M, and Ishihama Y (2007). Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc 2, 1896–1906. [DOI] [PubMed] [Google Scholar]
- Remmert M, Biegert A, Hauser A, and Söding J (2011). HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat. Methods 9, 173–175. [DOI] [PubMed] [Google Scholar]
- Rodrigues RA, dos Santos Silva LK, Dornas FP, de Oliveira DB, Magalhães TF, Santos DA, Costa AO, de Macêdo Farias L, Magalhães PP, Bonjardim CA, et al. (2015). Mimivirus fibrils are important for viral attachment to the microbial world by a diverse glycoside interaction repertoire. J. Virol 89, 11812–11819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues RAL, Arantes TS, Oliveira GP, Dos Santos Silva LK, and Abrahão JS (2019a). The complex nature of Tupanviruses. Adv. Virus Res 103, 135–166. [DOI] [PubMed] [Google Scholar]
- Rodrigues RAL, Mougari S, Colson P, La Scola B, and Abrahão JS (2019b). “Tupanvirus”, a new genus in the family Mimiviridae. Arch. Virol 164, 325–331. [DOI] [PubMed] [Google Scholar]
- Roos WH, Gertsman I, May ER, Brooks CL 3rd, Johnson JE, and Wuite GJ (2012). Mechanics of bacteriophage maturation. Proc. Natl. Acad. Sci. USA 109, 2342–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saadi H, Pagnier I, Colson P, Cherif JK, Beji M, Boughalmi M, Azza S, Armstrong N, Robert C, Fournous G, et al. (2013a). First isolation of Mimivirus in a patient with pneumonia. Clin. Infect. Dis 57, e127–e134. [DOI] [PubMed] [Google Scholar]
- Saadi H, Reteno DG, Colson P, Aherfi S, Minodier P, Pagnier I, Raoult D, and La Scola B (2013b). Shan virus: a new mimivirus isolated from the stool of a Tunisian patient with pneumonia. Intervirology 56, 424–429. [DOI] [PubMed] [Google Scholar]
- Schrad JR, Young EJ, Abrahão JS, Cortines JR, and Parent KN (2017). Microscopic characterization of the Brazilian giant Samba virus. Viruses 9, E30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah N, Hülsmeier AJ, Hochhold N, Neidhart M, Gay S, and Hennet T (2014). Exposure to mimivirus collagen promotes arthritis. J. Virol 88, 838–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva LCF, Rodrigues RAL, Oliveira GP, Dornas FP, La Scola B, Kroon EG, and Abrahão JS (2019). Microscopic analysis of the Tupanvirus cycle in Vermamoeba vermiformis. Front. Microbiol 10, 671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, and Carragher B (2005). Automated molecular microscopy: the new leginon system. J. Struct. Biol 151, 41–60. [DOI] [PubMed] [Google Scholar]
- Suomalainen M, Luisoni S, Boucke K, Bianchi S, Engel DA, and Greber UF (2013). A direct and versatile assay measuring membrane penetration of adenovirus in single cells. J. Virol 87, 12367–12379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzan-Monti M, La Scola B, Barrassi L, Espinosa L, and Raoult D (2007). Ultrastructural characterization of the giant volcano-like virus factory of Acanthamoeba polyphaga Mimivirus. PLoS ONE 2, e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschke CM, McGough A, and Thuman-Commike PA (2003). Penton release from P22 heat-expanded capsids suggests importance of stabilizing penton-hexon interactions during capsid maturation. Biophys. J 84, 2585–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Waterbeemd M, Llauró A, Snijder J, Valbuena A, Rodríguez-Huete A, Fuertes MA, de Pablo PJ, Mateu MG, and Heck AJR (2017). Structural analysis of a temperature-induced transition in a viral capsid probed by HDX-MS. Biophys. J 112, 1157–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Thomas JA, Cheng N, Black LW, and Steven AC (2012). Bubblegrams reveal the inner body of bacteriophage φKZ. Science 335, 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Leavitt JC, Cheng N, Gilcrease EB, Motwani T, Teschke CM, Casjens SR, and Steven AC (2016). Localization of the Houdinisome (ejection proteins) inside the bacteriophage P22 virion by bubblegram imaging. MBio 7, e01152–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Kuznetsov YG, Sun S, Hafenstein SL, Kostyuchenko VA, Chipman PR, Suzan-Monti M, Raoult D, McPherson A, and Rossmann MG (2009). Structural studies of the giant mimivirus. PLoS Biol. 7, e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu IM, Zhang W, Holdaway HA, Li L, Kostyuchenko VA, Chipman PR, Kuhn RJ, Rossmann MG, and Chen J (2008). Structure of the immature dengue virus at low pH primes proteolytic maturation. Science 319, 1834–1837. [DOI] [PubMed] [Google Scholar]
- Zauberman N, Mutsafi Y, Halevy DB, Shimoni E, Klein E, Xiao C, Sun S, and Minsky A (2008). Distinct DNA exit and packaging portals in the virus Acanthamoeba polyphaga mimivirus. PLoS Biol. 6, e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng A, Yuan F, Kleinfelter LM, and Kielian M (2014). A toggle switch controls the low pH-triggered rearrangement and maturation of the dengue virus envelope proteins. Nat. Commun 5, 3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The full proteomics datasets generated in this study are available upon request from the Lead Contact.
Three-dimensional tomograms have been deposited to the Electron Microscopy Database (EMDB) under the ID codes EMD-20747 (pH 2, Video S3), EMD-20748 (100°C, Video S4), EMD-20746 (pH 2 + 100°C, Video S6), and EMD-20745 (pH 2 + 100°C, Video S8).


