Abstract
Aging is a complex, yet pervasive phenomenon in biology. As human cells steadily succumb to the deteriorating effects of aging, so too comes a host of age-related ailments such as neurodegenerative disorders, cardiovascular disease and cancer. Therefore, elucidation of the molecular networks that drive aging is of paramount importance to human health. Progress toward this goal has been aided by studies from simple model organisms such as Saccharomyces cerevisiae. While work in budding yeast has already revealed much about the basic biology of aging as well as a number of evolutionarily conserved pathways involved in this process, recent technological advances are poised to greatly expand our knowledge of aging in this simple eukaryote. Here, we review the latest developments in microfluidics, single-cell analysis and high-throughput technologies for studying single-cell replicative aging in S. cerevisiae. We detail the challenges each of these methods addresses as well as the unique insights into aging that each has provided. We conclude with a discussion of potential future applications of these techniques as well as the importance of single-cell dynamics and quantitative biology approaches for understanding cell aging.
1. Introduction
Rapid technological advances over the past several decades have enabled an increasingly quantitative understanding of biological systems. Cutting-edge imaging [1], sequencing [2,3], microfluidics [4] and genome editing [5,6] methods are generating increasingly complex data sets that, when combined with the requisite quantitative analysis, are producing groundbreaking biological insights. One such area benefitting from these new developments is the field of geroscience, which seeks to understand the basic biological mechanisms of aging and apply this knowledge to treat human diseases and prolong healthy lifespan [7,8]. While progress is being made by applying new experimental techniques to directly study aging in humans [9,10], conducting aging experiments in humans is difficult and in certain cases impossible. However, work from a variety of model eukaryotic organisms has revealed a number of evolutionarily conserved molecular processes that drive aging [11,12], which helps to point the way for future work in and therapies for humans. In particular, the use of the budding yeast Saccharomyces cerevisiae as a genetically tractable single-celled model system for studying aging has helped illuminate a number of important conserved pathways underlying aging [11–16] as well as potential interventions for mitigating its effects [17].
Aging in S. cerevisiae is primarily studied in two modalities, chronological aging and replicative aging [18]. For chronological aging, cells are held in stationary phase, then periodically tested for their capacity to resume division when the stationary-inducing conditions are relieved. The time in which the cells can successfully survive in stationary is defined as their chronological lifespan (CLS) [18]. In contrast, replicative aging is defined by the number of daughter cells that can be produced by a single mother cell before it stops reproducing and dies, termed as replicative lifespan (RLS) [18,19]. Because replicative aging occurs on a timescale of days rather than weeks, it has the potential of seamlessly interfacing with microfluidic devices and time-lapse fluorescence microscopy, widely used tools for studying dynamic biological processes in real-time [20,21]. However, until the last seven years, the field of yeast replicative aging relied on microdissection [19,22] as the only tool for RLS measurements. For this method, an experimenter uses a micromanipulator to separate mother and daughter cells growing on an agar pad under a microscope, thereby keeping track of and recording the number of divisions of each mother cell for RLS analysis [22]. Although microdissection has provided valuable knowledge about lifespans under various genetic and environmental conditions, its manual nature results in a number of drawbacks, including its tedium and the inability to dynamically modify the growth environment. An additional significant drawback of this technique is that the thick agar pad on which the cells grow prevents high-quality time-lapse fluorescence microscopy from being conducted [23,24]. Consequently, dynamic and quantitative single-cell measurements cannot be performed, which greatly impedes understanding of the genetic regulatory networks governing the cell aging process, which are dynamic by their very nature. To circumvent these shortcomings, new technologies and methods have been developed in recent years that offer alternatives to the microdissection approach.
Here we review the recent advances in studying yeast replicative aging using microfluidics, time-lapse fluorescent microscopy, computational modeling and high-throughput technologies. We cover recent studies utilizing each technique and discuss the unique insights provided by these approaches compared to traditional methods. Taken together, these advances provide novel insights and challenges for the field of yeast aging and suggest important and creative new avenues for applying this research to higher organisms. We conclude by discussing open questions and new directions raised by these approaches in this field that is ripe for new findings.
2. Microfluidic devices
To address the drawbacks of the traditional microdissection method, various polydimethylsiloxane (PDMS) based microfluidic devices have been developed. These devices automate mother-daughter separation during aging and allow RLS measurements to be interfaced with time-lapse fluorescence microscopy. These technologies have provided an unparalleled look at the aging process in single yeast cells by allowing the dynamics of various molecular and cellular processes to be tracked and quantified. The construction and design of such devices, however, is challenging, and microfluidic platforms capable of tracking large numbers of single mother cells throughout their lifespans require new features not necessary in the design of devices for more short-term cell tracking [25–29]. In this section, we discuss the design strategies that enable microfluidics to be applied to yeast replicative aging.
2.1. Design constraints for a yeast aging microfluidic device
One of the first considerations to have in designing microfluidics for yeast aging is the operation time of the device. An often noted feature of replicative aging is the wide distribution of lifespans one obtains from a population [19,30], with reported RLS values for long-lived wild-type (WT) cells from microdissection studies [19,22] exceeding 40 divisions. As the population doubling time for yeast is about 90 min [31], 40 divisions for single cells equates to approximately two and a half days. Considering that certain long-lived mutants can divide 60 or more times [32], it is clear that a successful microfluidic device must be able to trap mother cells for more than 4 days. Indeed, these long operation times for RLS studies result in another problem: the accumulation of a large number of daughter cells in the device. If not rapidly removed, the cells can dislodge mother cells, interfere with automated cell tracking or clog the device. Finally, the mechanisms used to trap mother cells must not stress the cells and RLS measurements should be comparable to microdissection studies. Taking these issues into account, a number of yeast aging microfluidic devices have been successfully designed (Fig. 1A), with lifespan measurements being validated in WT cells and various longevity mutants [23,24,33–39]. While each device design has its own unique features that allow it to fulfill the aforementioned criteria, the currently available microfluidic devices employ some common strategies to load and trap mother cells. Before detailing the particularities of each yeast aging microfluidic device, we describe these shared mechanisms in order to properly contextualize the motivations behind each design.
Fig. 1. Microfluidic devices for single-cell replicative aging in yeast.
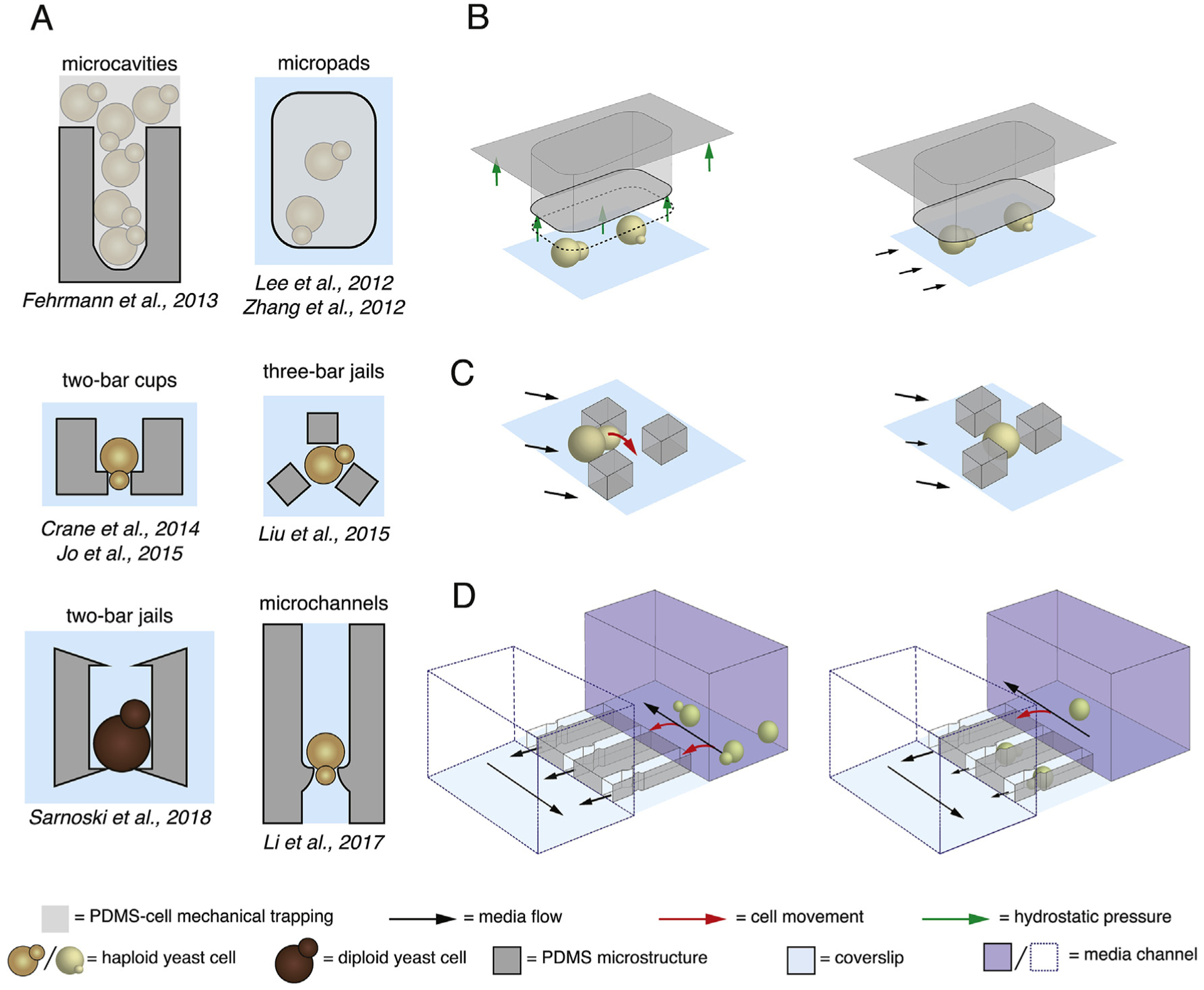
(A) The cell trap designs for currently published yeast aging microfluidic devices [23,33–39]. (B) Example of loading in a device relying on contact-based mechanical trapping, where hydrostatic pressure is used to raise the height of the micropad traps (left). Releasing pressure by allowing fluid flow to the waste port lowers the height of the micropads once more and traps cells underneath them [23,33]. (C) Devices that use hydrodynamic trapping to guide cells to the outside of the jails [37]. Cells can produce a daughter cell through the openings in the jail (left). The cell that enters the jail is geometrically confined, and is analyzed as the mother cell for RLS experiments [37,39]. (D) Loading of microchannel traps [38] using hydrodynamic trapping. Cells are guided into the traps by the flow and can produce buds through a small opening at the bottom of the trap or toward the entrance (left). Loading of a cell into the trap increases the hydraulic resistance of this path for fluid flow, facilitating the loading of adjacent traps (right). (Figure adapted from refs. 23, 33–39).
2.2. Physical mechanisms for loading and trapping mother cells
There are a plethora of methods for trapping single cells in microfluidic chambers. However, for replicative aging applications, the asymmetric nature of yeast division [40], wherein for most of their lifespans, mother cells give rise to daughters that are smaller than themselves [41], favors three primary strategies for loading cells into the trapping regions and retaining mother cells throughout their entire lifespans, as well as removing daughter cells: contact-based mechanical trapping, geometric confinement and hydrodynamic trapping. Contact-based mechanical trapping entails immobilization of cells from above, that is, between the PDMS ceiling of the microfluidic device and the glass slide. Using this approach, trapping regions smaller than or approximately the same size as a typical yeast diameter are used to push down on cells and hold them in place. Loading cells requires the ceilings or heights of these traps be raised, so that cells can actually get into the trapping regions. This is done by increasing the pressure at both the inlet and outlet of the device to generate hydrostatic pressure that raises the height of the ceiling, allowing cells to pass under the trapping areas [23,33] (Fig. 1B, left). Release of the pressure by allowing flow toward the waste port lowers the ceiling back down, immobilizing cells and trapping them [23,33] (Fig. 1B, right). Since daughter cells are smaller than their mothers, they are not as restrained by the low ceilings and can more easily escape from the trap and flow toward the waste port.
As an alternative to contact-based mechanical trapping, most recently published microfluidic devices utilize a physical mechanism known as hydrodynamic trapping. In the context of this review, hydrodynamic trapping can be defined as a technique for capturing and immobilizing cells against structures in a micro-fluidic device by exploiting the hydraulic forces that act on particles in a fluid [42–47]. Hydrodynamic trapping enables a number of desirable features for yeast aging microfluidic devices such as trapping areas with ceilings taller than the diameter of a typical yeast cell (i.e. > 4 μm). For devices of this kind, microstructures are positioned along the flow path to capture and immobilize cells. Because loading of cell traps increases the fluidic resistance along that path, this increases the flow through adjacent empty traps, which facilitates cell loading. From this point, mother cells can be obtained from daughters that enter isolated trapping regions that geometrically confines their movement (Fig. 1C) or directly from the immobilized cells that are trapped (Fig. 1D). With the physical principles used by different microfluidic devices for loading and trapping mother cells during their entire lifespans in hand, we now turn to a discussion of the design layouts of various devices.
2.3. Microfluidic design layouts
Three of the earliest published microfluidic devices for yeast aging relied on contact-based mechanical trapping of mother cells. This was accomplished via vertical micropads [23,33] or dead-end microcavities [34] (Fig. 1A, top row). The device reported by Zhang et al. [33] was an improvement on an original device developed by this group that used chemical modification of the glass coverslip and cell wall as part of its trapping mechanism [24]. The use of vertical micropads to trap cells allowed them to jettison this requirement. In the designs by Lee et al. [23] and Zhang et al. [33], the vertical micropad traps are positioned within an “open room” that is much taller than the height of the traps (for example, 15 μm in the device by Lee et al. [23]) in which media flows. The cross-sectional area of these traps is large enough to accommodate multiple mother cells under a single micropad. For the design by Fehrmann et al. [34], media channels 40 μm tall flank 3.3 μm trapping areas with dead-end cavities. Here, cells grow to fill the trapping area until a newborn cell reaches the end of the cavity where it is subsequently tracked as a mother cell for RLS analysis. While lifespan measurements of these devices produced similar results to those obtained with microdissection, the trapping methods for these devices are relatively inefficient. For designs relying on contact-based mechanical trapping, it can be difficult to retain mother cells over their entire replicative lifespans. It is also difficult to control the number of cells trapped in the case of the vertical micropad designs [23,33], or the timing at which cells reach the end of the cavities [34]. For example, in the micropad designs [23,33] larger daughter cells or cells washing down from upstream can be caught under a single micropad and dislodge mother cells from the trapping regions. Similarly, the dead-end microcavities design [34] relies on mother cells budding in a single direction for the entire course of their lifespan and a single polarity switch can remove the mother from the trap. Further, the contact-based mechanical trapping employed by these devices may not be suitable for various longevity mutants that could have altered cell size compared to wild-type cells.
With devices from Lee et al. [23], Zhang et al. [33] and Fehrmann et al. [34] paving the way for applications of microfluidics to yeast replicative aging, a new generation of devices was developed that built upon and extended these foundational technologies. As previously mentioned, these devices use hydrodynamic trapping and/or geometric confinement for loading and retaining mother cells in the traps throughout their entire lifespans. An example of a device that uses both hydrodynamic trapping and geometric confinement for operation is the three-bar “jail” design [26] that was optimized to trap single cells for replicative aging experiments by Liu et al. [37] (Fig. 1A, middle right). In this device, cells are guided toward the openings between two of the bars by the fluid flow, eventually producing a bud that enters the jail. Once there it is tracked as the mother cell. The 5.3 μm tall PDMS bars spanning the entire space between the ceiling of the device and the glass slide spatially confine each mother cell while also allowing for increases in cell size that accompany aging [48]. Buds can be produced out of three openings in the jail small enough for daughters to pass through and periodic pulses of high velocity flow are used to aid in daughter cell removal. A variation of this design was created to analyze aging in diploid cells, where a two-bar jail structure was created which functions using the same basic principles as the three-bar jail [39].
Another set of devices more fully rely on hydrodynamic trapping for proper device operation. Here, traps consist of arrays of cup shaped two-bar traps [35,36] or open-ended microchannels [38]. Devices developed by Crane et al. [35] and Jo et al. [36] are single layer devices where traps are positioned parallel to the direction of flow in an open room layout similar to devices using vertical micropads [23,33]. The device developed by Li et al. [38] consists of two layers, with 20 μm tall media channels positioned perpendicular to the 4.3 μm cell traps. In each design, traps feature small 3 μm openings that provide a docking site for mother cells, preventing their movement beyond this point while allowing passage of budding daughters. Furthermore, these openings allow for constant fluid flow through the trap, which provides the hydrodynamic force needed to secure mother cells over the course of their entire replicative lifespans. With openings on each side of the trap, mothers are able to produce buds in either direction, which are eventually washed away by the media flow. The use of hydrodynamic trapping allows the ceilings of the trapping regions to be taller than the height of the cells [35,36,38]. Along with the wider width of these traps compared to that of the cells, these designs allow room for increases in cell size during aging. Importantly, the use of hydrodynamic trapping enables very efficient loading [36,38] and also robust retention rates of mother cells [35,36,38].
2.4. Unique device features
Due to the different design layouts of each, there are a number of unique capabilities for the aforementioned microfluidic devices. A feature exclusive to the devices with dead-end microcavities [34] and jail structures [37,39] is the requirement of newborn daughter cells to load their traps. As a result, RLS analysis begins with virgin cells that have not yet divided, yielding exact lifespan measurement for all analyzed cells. Although undoubtedly useful, given that populations are disproportionally composed by young cells, with only 12.5% of cells expected to be older than two generations [49,50], this feature is not an absolute necessity for obtaining accurate RLS measurements for WT cells or longevity mutants. In regards to lifespan measurements of large numbers of cells (500 or more) or multiple strains in a single experiment, the device by Jo et al. [36] is particularly suited for such applications. Another feature that is unique to devices using microcavities [34], micropads [23,33] and microchannels [38] as traps is the ability to visualize daughter cells for some time without them being immediately washed away. For example, daughter cells, especially of old mothers, can be maintained under micropads for some time before removal. Similarly, for the microchannels used by Li et al. [38] daughters born toward the entrance of the microchannel can be retained for some time. Although removal of daughter cells is critical to prevent device clogging and for later image analysis and tracking of mothers, the daughter cells are a potential valuable source of data. Monolayer open room style devices [35–37,39] are designed to immediately remove all daughters from the field of view and therefore sacrifice the possibility of obtaining daughter cell information. In particular, the micropad design by Zhang et al. [33] and microchannel design by Li et al. [38] have been leveraged to analyze daughter cell gene expression [51] and cell morphology [38,52]. Finally, devices by Crane et al. [35] and Li et al. [38] allow media switches to be performed. This enables dynamic modifications of the cellular environment in the form of chemical perturbations or changes to the nutrient conditions. For more technical details about the design, validation and testing of many of the microfluidic devices described here, we refer the interested reader to a recent thorough review on this topic [53].
3. Quantitative single-cell analysis
The advent of microfluidic technologies as a tool for determining replicative lifespan in S. cerevisiae, combined with time-lapse fluorescence microscopy, has ushered in a new era of yeast aging research in which quantitative, real-time measurements of molecular processes during aging can be obtained at single-cell resolution (Fig. 2A). With the ability to collect data of this kind, the replicative aging process is now amenable to probing a myriad of new questions regarding the dynamics and cell-to-cell variability of gene expression, transcription factor localization, chromatin silencing, cellular morphology, organelle morphology, intracellular noise and more. Answering questions such as these is critical for developing a complete understanding of cell aging. However, the complexities of this task require sophisticated methods for analyzing and interpreting single-cell data. In this section, we consider several recent studies in which measurements of dynamic biological processes at the single-cell level obtained with microfluidic platforms have provided novel insights into yeast aging. We especially highlight those that have combined these single-cell data with computational modeling in order to form a quantitative understanding of the dynamics at play.
Fig. 2. Quantitative analysis of single-cell dynamics during replicative aging enabled from microfluidics experiments.
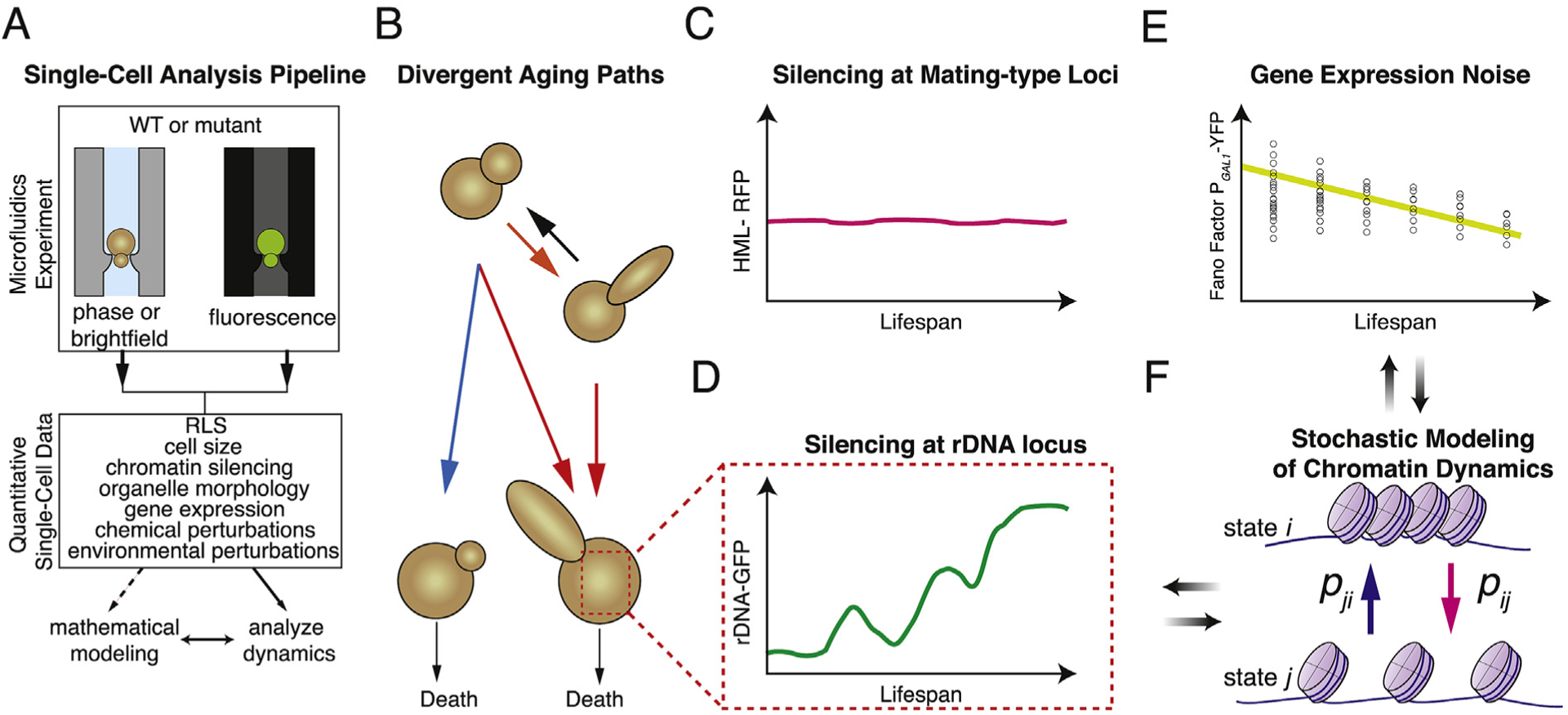
(A) Microfluidics experiments allow interfacing of yeast RLS measurements with time-lapse fluorescence microscopy. This allows dynamic and quantitative single-cell data about a number of biomolecular processes to be obtained. Mathematical modeling is a powerful too that can be utilized for such data. (B) Divergent aging trajectories uncovered by Jin et al. [52]. Cells in a population progress along two different aging paths marked by four possible phenotypic states, the dynamics of which can be described using a non-Markovian mathematical model [52]. (C) Measurement of chromatin silencing in single cells at the HML and HMR mating-type loci via a genomically integrated fluorescent reporter by Schlissel et al. [67] revealed strong silencing of this region throughout a cell’s lifespan. (D) Pulses of rDNA silencing dynamics in single cells dying with the elongated daughter phenotype were uncovered by Li et al. [38] through a GFP reporter inserted into the NTS1 region of the rDNA. (E) Liu et al. [84] found a reduction of gene expression noise during aging as determined by the declining Fano factor (variance divided by the mean) from single-cell measurements of PGAL1-YFP expression. In the cartoon figure, black circles represent single-cell values at various ages and the yellow line represents the mean Fano factor of all cells at that age [84]. As done by Liu et al. [84], this cartoon plot omits PGAL1-YFP measurements from the final four divisions of each cell, where Liu et al. [84] found that gene expression noise sharply increased. (F) Stochastic modeling of chromatin transitions between open and closed conformations at different rates or probabilities (pij and pji) at heterochromatin regions [38] and of local chromatin remodeling [84] has provided biological insight for follow-up experiments. (Figure adapted from refs. 38, 52, 67, 84). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.1. Terminal cell morphologies
A persistent finding amongst microdissection studies of yeast aging has been the existence of two different cell states at death. In one state mother cells die with a bud still attached and in the second, cells die without a bud [54–58]. Further distinctions have been made, including of cells dying with small buds and those dying with large buds that were difficult to separate from the mother [55,58]. Genetic mutations influence the percentage of cells that die in either state, suggesting that different molecular mechanisms may lie at their origin [55–57]. Subsequent studies using high-resolution, continuous monitoring of cellular morphology in microfluidic devices extended these initial observations to reveal that cells have multiple ways of aging, not a single, pre-determined pattern.
Evidence supporting this notion included the observation of distinct terminal cell morphologies, where Xie et al. [24] and Lee et al. [23] both noted two types of cell death, one with elongated and one with spherical mother morphology. Additionally, Lee et al. [23] noted that mother cells that become elongated at death usually produced ellipsoidal daughters late in life. These two death types displayed differences in cellular stress levels as well as in vacuolar and mitochondrial damage [23,24]. Interestingly, both groups found that cells of the elongated morphology had longer lifespans than cells that remained spherical [23,24]. Ensuing work from other groups using different microfluidic devices also found that some cells could produce either round or elongated buds late or at the end of their RLSs [36,38]. However the mechanisms that underlie these phenotypes are not fully known. While Xie et al. [24] noted mitochondrial damage in the cohort of cells dying with spherical morphologies, work by Fehrmann et al. [34]–which also noted two kinds of terminal states, cells that lost mitochondrial membrane potential and those that did not–suggested that loss of mitochondrial membrane potential was not linked to cell aging. Thus, future single-cell studies are needed to better understand the molecular determinants of these different terminal cell states.
Taken together, multiple, independent studies, using both microdissection and microfluidics, have documented the existence of different terminal cell morphologies in yeast. Albeit using different language and terms to describe their results, these reports ostensibly point to a common phenomenon, as work using the microdissection method [54,55,58] and different microfluidic devices [23,36,38] has detailed end state cell morphologies with small and large buds. Recent analyses using time-lapse microscopy and microfluidics have shed light on these observations and in the process have linked these terminal death types to canonical molecular processes involved in yeast aging that are discussed below.
3.2. Divergent aging trajectories
The ability to combine time-lapse microscopy with microfluidics allowed Jin et al. [52] in a recent study to quantitatively characterize cellular morphology during aging. The heterogeneity in terminal cell morphologies of yeast, as mentioned in the previous section, had been noted as early as the 1960s [54]. Using microfluidics and quantitative single-cell measurements, Jin et al. [52], discovered four different morphological states that corresponded to two divergent aging paths in single yeast cells (Fig. 2B). These cell states were determined by quantitative morphological measurements of mother-daughter pairs during aging, facilitated by using the microchannel traps developed by Li et al. [38]. The first state corresponded to a ground state in which both mothers and daughters had uniform and round shapes early during the lifespan of the mother. After their first few divisions, a second possible phenotypic state was found in which some mothers could transiently produce elongated daughters before switching back to producing daughters of the ground state. The final two states described mother-daughter morphologies toward the very end of the mothers’ lifespans, where the previously noted elongated and round but small daughters were observed [38]. These final two states were nearly mutually exclusive in WT cells [52].
These aging dynamics were investigated with a non-Markovian state transition model in which cells transition stochastically between distinct phenotypic states with different probabilities. It was found that the intermittent morphological state consisting of elongated daughters produced early in life biased cells toward the aging path that culminated with elongated buds at the very end of the mothers’ RLSs. In addition, the model revealed history-dependence in the state transitions during aging, which further reinforced the divergence of the two aging paths [52]. Subsequent experiments using genetic and environmental perturbations demonstrated that the propensity of cells to commit to either of the two divergent aging paths is tunable; Sir2p is involved in repressing the aging path with elongated daughters, whereas calorie restriction promotes a switch to this path [52].
The non-Markovian model developed by Jin et al. [52] provided crucial insights into the kinetics, history-dependence and potential divergence of single-cell aging dynamics. One advantage of this type of state transition model is that by revealing whether certain state transitions are allowed or forbidden, the inferred transition probabilities can provide insights and aid in systematically characterizing applied perturbations into the cell-state dynamics during the entire aging timescale. The model’s ability to predict the dynamics of phenotypic state changes does not require specific assumptions about the molecular mechanisms that generate these states, which is a useful feature when such mechanisms are unknown.
3.3. Chromatin silencing dynamics
Early on, studies using the microdissection method implicated the HM silent mating-type loci and the rDNA in yeast aging, two regions of the yeast genome subject to chromatin silencing via the histone-deacetylase Sir2, a major regulator of chromatin silencing in yeast [59–61]. The sterility and insensitivity of old cells to pheromone [62] was attributed to loss of chromatin silencing at the HML and HMR mating loci [49]. Similarly, the accumulation of extrachromosomal rDNA circles (ERCs), autonomously replicating units of circular DNA, formed by spontaneous excision from the highly repetitive rDNA region on chromosome XII, were linked to aging as a toxic product that accumulates in mother cells and eventually leads to cell death [63]. Further, silencing loss at the rDNA locus has been attributed to increased homologous recombination amongst the 100–200 tandem repeats within this region, with the resulting instability also seeming to negatively affect lifespan [64–66]. Given the apparent importance of chromatin maintenance at these loci, recent reports using microfluidics have focused on silencing dynamics at these regions during aging.
To investigate the silencing of HML and HMR loci and age-induced sterility at the single-cell level, Schlissel et al. [67] combined microfluidic analysis with a novel Cre-recombinase based reporter [68] which drove the expression of a red fluorescent protein (RFP) when HML was in a heterochromatic (i.e. silenced) state and a green fluorescent protein (GFP) when chromatin silencing was lost. Contrary to previous findings [49], the authors surprisingly found that loss of chromatin silencing was exceedingly rare at the HM loci, with Sir2 activity not declining with age [67]. Results from more than 1500 single cells showed that heterochromatic silencing was lost in less than 1% of cells and was seemingly unrelated to cell age (Fig. 2C) [67]. To explain the inability of old cells to respond to pheromone and mate, the authors found that aggregation of Whi3, an RNA binding protein involved in the mating response and cell division [69], was responsible for sterility in aged cells [67].
Examining the chromatin silencing dynamics at the rDNA region in single cells growing in a microfluidic device also yielded new insights into the regulation of heterochromatin during aging. By inserting a constitutively expressed GFP into non-transcribed spacer region 1 (NTS1) of the rDNA, Li et al. [38] were able to monitor changes in chromatin silencing at this locus. Unlike the strong silencing found by Schlissel et al. [67] at the HM loci, Li et al. [38] found that the rDNA region exhibited sporadic pulses of silencing loss during the early phases of aging. Further analysis revealed that about half of the cells displayed sustained silencing loss toward the end of their lifespan, which ultimately lead to cell death. The other half of cells did not display these dynamics, with the GFP silencing reporter remaining low even very late in their lifespan. In addition, silencing loss during aging was associated with production of elongated daughters (Fig. 2D). Because of the exquisite chromatin silencing dynamics of the rDNA region, the authors then developed a mathematical model to gain further insight into this process.
In order to translate their experimental findings into a computational framework, Li et al. [38] developed a phenomenological model of aging that included stochastic state transitions between silenced and unsilenced chromatin states (Fig. 2F). The model included a damage factor that accumulated in the unsilenced state, with the likelihood of cell death being proportional to this value [38]. This simple model reproduced the single-cell aging dynamics and served to drive the generation of several hypotheses about the effects of perturbations to the silencing dynamics that were subsequently experimentally verified–chemical interventions that caused prolonged silencing or loss of silencing both negatively impacted cell lifespan [38]. This combination of single-cell experiments and mathematical modeling demonstrated that chromatin silencing dynamics within the rDNA are important for controlling the aging process and raised up a conceptually interesting possibility that, in addition to specific genes or molecules per se, the temporal dynamics of molecular events themselves could also play critical roles in determining cellular longevity.
3.4. Extrachromosomal rDNA circles (ERCs)
In addition to chromatin silencing dynamics at the rDNA, studies using microfluidic devices are also uncovering new details about the detrimental effects of ERCs that originate from the rDNA region. While ERCs have long been causally linked to aging due to the fact that they accumulate in old mother cells [63,70], the exact mechanisms by which they mediated age-related cellular decline remained nebulous. Recently, great strides have been made along these lines in work by Neurohr et al. [71] and Morlot et al. [72]. Neurohr et al. [71] found that ERCs promote cell death through blocking the G1/S phase transition of the cell cycle. To piece together the events that follow rDNA silencing loss [38] and precede cell cycle arrest, Morlot et al. [72] used time-lapse microscopy to show an exponential increase in rDNA copy number, suggesting the formation of ERCs, during the late phases of aging. They further suggested that elevated pre-rRNA levels might be caused by the accumulation of ERCs in old mothers and that they in turn could lead to disruption of nuclear size and integrity [72]. Using parameter values inferred from their experimental data, follow-up mathematical modeling of a stochastic ERC excision process that leads to senescence was able to capture aspects of cell-to-cell variability in lifespan [72].
It has been shown that DNA circles are retained in mother cells through attachment to nuclear pore complexes by the chromatin regulating Spt-Ada-Gcn5 acetyltransferase (SAGA) complex [73]. Along these lines, another recent study using microfluidics by Rempel et al. [74] has detailed evidence of a buildup of defective nuclear pore complexes in aged mother cells. Findings from Morlot et al. [72], which suggest a link between ERC formation and a decline in nuclear homeostasis, nicely complement these studies.
3.5. Gene expression noise
The biological mechanisms responsible for initiating and regulating gene expression are subject to a number of stochastic effects, termed noise, that give rise to cell-to-cell variability [75–77]. Differences in molecular components between cells and the natural stochasticity of biochemical reactions themselves give rise to two forms of noise referred to as extrinsic and intrinsic noise, respectively [75]. Cells use a variety of means to either combat negative consequences of noise in gene expression or utilize the benefits of it [78–81]. In the context of aging, studies from mice [82,83] have suggested that noise increases with age in a manner that negatively impacts longevity. However, these studies were not able to follow the specific individual cells as they aged.
To gain insight into the dynamics of gene expression noise within the same cell during aging, Liu et al. [84] monitored single yeast cells growing in a microfluidic device. Quantitative measurements of yellow fluorescent protein (YFP) expression from the GAL1 promoter revealed that, surprisingly, noise decreased throughout much of cellular lifespan (Fig. 2E) before dramatically rising during the last four cell divisions [84]. To account for the observed noise reduction the authors utilized a stochastic mathematical model (Fig. 2F) of transcriptional regulation of PGAL1-YFP that incorporated chromatin remodeling and increases in cell volume during the cell cycle [84–86]. Although the model returned two ways for noise to decrease during aging, only one of these alternatives matched the experimental findings. This corresponded to the case where transition rates for open and closed chromatin states of the GAL1 promoter both increased during aging [84]. This study, together with the tightly maintained heterochromatin of the HM loci [67] and pulsatile silencing dynamics of the rDNA region [38], demonstrated that a wide range of chromatin regulation influences gene expression noise and cell-to-cell variability during aging.
The careful single-cell analysis conducted in this study [84] highlights the power of yeast as a model for aging research, where it is possible to track single cells throughout their entire lifespan. By comparison, studies in mice indicated an increase in gene expression noise with age [82,83], leading Liu et al. [84] to propose that because of methodological differences, these studies demonstrate increases in extrinsic noise whereas their study demonstrates a decrease in intrinsic noise with age. However, it is also possible that there are simply different noise trends during aging in yeast and animals. In either situation, illuminating the mechanisms behind potential similarities and differences between yeast and multicellular eukaryotes in this area represents a fascinating line of research.
4. New high-throughput technologies
Using microfluidics and time-lapse microscopy to study yeast replicative aging has the multitude of advantages previously discussed, yet this approach is necessarily limited in scale and scope by the information that can be collected. As such, systems-level questions about the transcriptome or metabolome, for example, are difficult to investigate with microfluidics-coupled microscopy alone. Furthermore, efforts to address such questions using standard approaches with liquid cultures are made prohibitively difficult due to the fact that quantities of old cells become exponentially diluted relative to newborn and younger cells as the population grows. Thus, for omics methods such as RNA-sequencing to be applied on cells of various replicative ages, a method for selectively collecting aged cells from a growing culture is required [49,50].
4.1. Devices for selective culturing of aging yeast cells
To address the problem of enrichment for aged cells, two methods [87,88] have recently been developed to specifically culture and capture mother cells at different time points along their lifespans. The procedures for both of these new technologies begin by employing previously published techniques [49,50] that allow mother cells in liquid culture to be sequestered from their progeny and toward the surface of a magnet. This is accomplished by biotinylating the cell walls of a starter culture and using magnetic streptavidin beads to bind the biotin on the surface of the cells and draw them to the magnetic surface [49,50]. Daughter cells do not inherit the biotin-coated surface from their mothers and are therefore invisible to the magnetic field [49]. As a result, the magnetic surface will collect only mother cells from the original culture (Fig. 3A). Utilizing this method, fluidically controlled columns [87] and the miniature-chemostat aging device (MAD) [88], both of which are equipped with surrounding magnets (Fig. 3B), have been designed for targeted capture of aged mother cells during cultivation. These devices operate with continuous flow of media and the replicative lifespan of mother cells is closely linked to the length of time the device is operated as a simple consequence of the relatively uniform division time of the cells [87,88]. Therefore, by taking samples at different time points during operation, mother cells of different replicative ages can be obtained at sufficient quantities for subsequent omics-level investigations (Fig. 3C and D). Although the mother enrichment program [89] has been a very fruitful tool for large-scale analysis of aging cells, these new devices provide alternate methods that do not require genetic alterations of the cells.
Fig. 3. New high-throughput technologies for systems-level insights into aging.
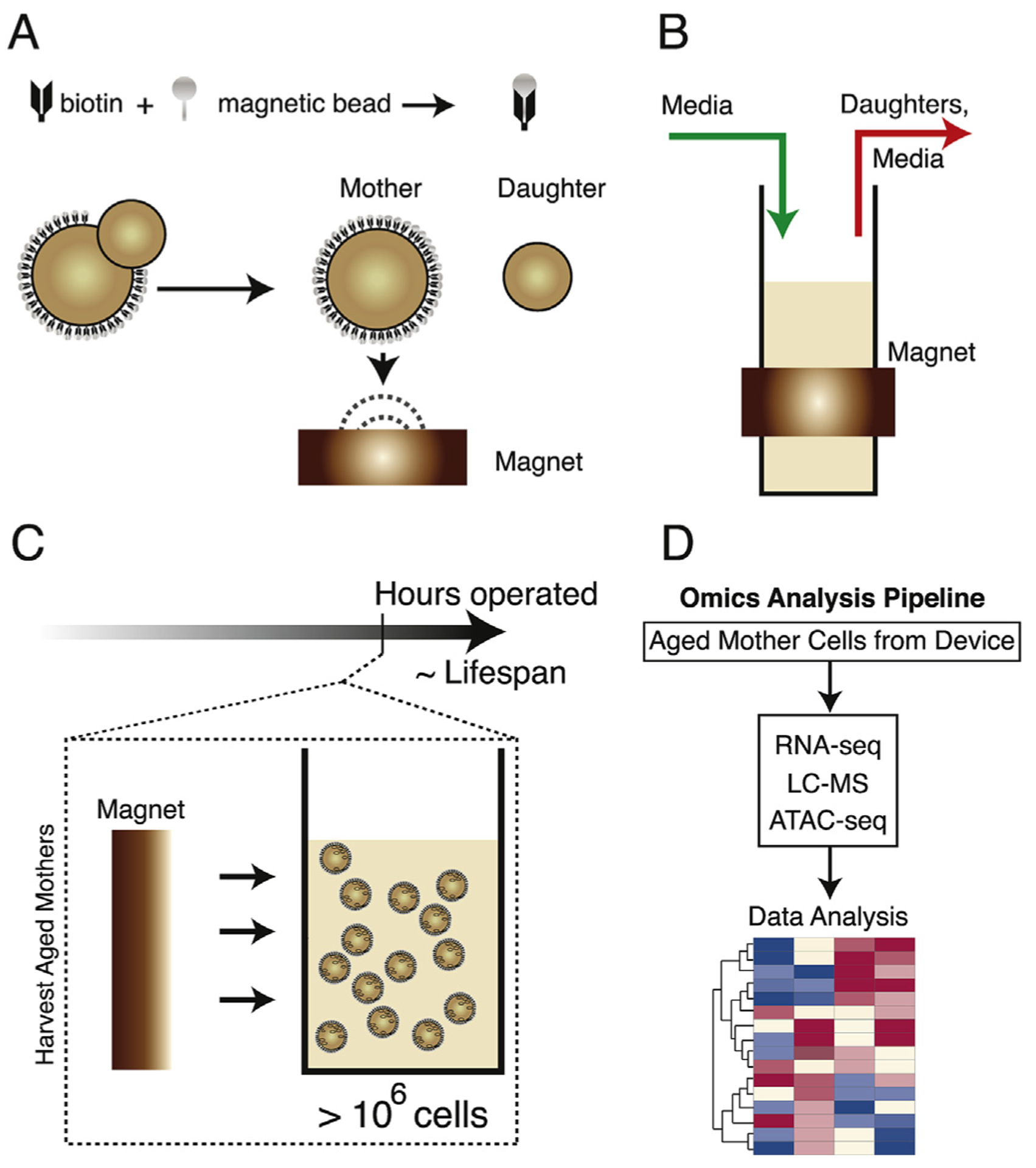
(A) Mother cells can be labeled on their surface with biotin and magnetic beads, which are not passed to their daughters. This renders mother cells able to be drawn toward magnetic surfaces [49,50]. (B) Example of MAD layout [88]. (C) Operation of these devices [87,88] allows RLS values to be highly correlated with time. Removing devices from the magnet allows release of mother cells. (D) Collecting large numbers of mother cells allows omics analysis to be done [87,88]. (Figure adapted from refs. 87, 88).
4.2. Systems-level insights
These new technologies [87,88] have already yielded fascinating insights into the changes in cell physiology that occur during replicative aging. Transcriptomics and proteomics conducted on aging cells in fluidic column devices revealed that protein abundance from genes involved in ribosome and protein production gradually increase relative to their respective mRNA levels as cells age [87]. Janssens et al. [87] found that one consequence of these dynamics is an altered stoichiometry in multiprotein complexes such as the vacuolar ATPase, which could be detrimental to cell function. A follow-up study using these columns to conduct metabolomics on aging cells identified an increased shift toward respiratory metabolism as a metabolic signature of aging [90]. To assess epigenetic changes that accompany aging, ATAC-seq was performed on wild-type cells and various longevity mutants during aging in the MAD [88]. Results from this work revealed relatively static chromatin structure in much of the genome throughout the cell’s lifespan, with notable exceptions in regions such as those regulating amino acid biosynthesis [88]. Furthermore, the authors identified increases in rDNA copy number during aging, particularly in sir2D mutants [88].
A potential drawback of these high-throughput approaches is the possibility of masking single-cell heterogeneity during aging. For this reason, an exciting prospect for the field would be to use these technologies along with microfluidics-based single-cell analysis. Such an integrated, multidisciplinary approach would offer powerful ways to investigate replicative aging, in which the systems-level insights could provide information for more targeted microfluidic studies of the regulatory networks of interest in single cells. Reciprocally, findings from microfluidic studies could be expanded to follow up with high-throughput analysis to place new results in a systems-level context.
5. Conclusion and prospects
Quantitative biology is a rapidly growing field with the potential to transform biological studies to a science with rigorous quantitative and predictive analysis. It integrates experimental technologies, such as microfluidics and advanced time-lapse imaging, with math-based theoretical frameworks, adopted from statistical mechanics and nonlinear dynamics, to address fundamental biological problems. Over the past two decades, quantitative biology approaches have been successfully exploited in the studies of gene expression [91], signal transduction [92–94], development [95,96], cell cycle and growth control [97,98], metabolism [29,99], and synthetic genetic engineering [100]. These pioneering attempts have highlighted the importance of stochasticity and spatiotemporal dynamics, two emerging concepts that are increasingly appreciated in biology, and have advanced our understanding of biological processes to a remarkable new level that is otherwise impossible with traditional methods.
For studies of aging, although quantitative biology has only been recently introduced to the field, it holds the promise to revolutionize the methodologies for studying aging as well as the ways of analyzing and thinking of the aging process. As the classic micro-dissection method has already enabled a systematic identification of a large number of genes and pathways that influence longevity [11,12,18,32,101–109], quantitative biology provides a powerful suite of new tools to elucidate how these genes interact and how these interactions change dynamically to drive the aging process, which will lead to the field’s next great leap forward.
As the aforementioned studies demonstrate, microfluidics and time-lapse imaging have started to allow direct measurements of various biological processes in individual cells across their entire lifespans. A major challenge that we are facing now, however, is to use quantitative analytical methods to interpret these newly available single-cell data and to push forward our mechanistic understanding of aging, beyond simple observations and characterization. In the section “Quantitative single-cell analysis” above, we have summarized recent progress in this direction, where computational modeling and quantitative analyses have been implemented to generate insights into the regulatory schemes of aging processes. These models and analyses are important first steps, but, at the same time, remain largely phenomenological. With resources for broader adoption of the methods described herein becoming available [110,111] and increasing amounts of single-cell dynamical data of age-related molecular processes and pathways becoming generated, the next step would be to develop dynamic mechanistic models to describe, simulate and predict how these molecular factors operate, interact and change over time to, collectively, cause aging of living cells. These efforts will generate new hypotheses to reveal new network-level dynamic properties, ultimately resulting in novel concepts, theories and principles about fundamental mechanisms of aging. As such, we envision that quantitative biology will become a powerhouse for aging research that will catalyze the emergence of conceptual breakthroughs and discoveries in the biology of aging.
Acknowledgements
This work was supported by the National Institutes of Health – National Institute on Aging grant R01-AG056440 (to N.H., J.H., L.S.T., and L.P.); Department of Defense, Air Force Office of Scientific Research, National Defense Science and Engineering graduate fellowship 32 CFR 168a (to R.O.).
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest to disclose.
References
- [1].Liu TL, Upadhyayula S, Milkie DE, Singh V, Wang K, Swin- burne IA, Mosaliganti KR, Collins ZM, Hiscock TW, Shea J, et al. , Observing the cell in its native state: imaging subcellular dynamics in multicellular organisms, Science 360 (6386) (2018), eaaq1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW, Droplet barcoding for single- cell transcriptomics applied to embryonic stem cells, Cell 161 (5) (2015) 1187–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. , Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets, Cell 161 (5) (2015) 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Zhang C, Tu H-L, Jia G, Mukhtar T, Taylor V, Rzhetsky A, Tay S, Ultra-multiplexed analysis of single-cell dynamics reveals logic rules in differentiation, Sci. Adv 5 (4) (2019), eaav7959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].McKenna A, Findlay GM, Gagnon JA, Horwitz MS, Schier AF, Shendure J, Whole-organism lineage tracing by combinatorial and cumulative genome editing, Science 353 (6298) (2016), aaf7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Frieda KL, Linton JM, Hormoz S, Choi J, Chow K-HK, Singer ZS, Budde MW, Elowitz MB, Cai L, Synthetic recording and in situ readout of lineage information in single cells, Nature 541 (7635) (2017) 107–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kennedy BK, Berger SL, Brunet A, Campisi J, Cuervo AM, Epel ES,Franceschi C, Lithgow GJ, Morimoto RI, Pessin JE, et al. , Geroscience: linking aging to chronic disease, Cell 159 (4) (2014) 709–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kaeberlein M, Translational geroscience: a new paradigm for 21st century medicine, Trans. Med. Aging 1 (2017) 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Valdes AM, Glass D, Spector TD, Omics technologies and the study of human ageing, Nat. Rev. Genet 14 (9) (2013) 601–607. [DOI] [PubMed] [Google Scholar]
- [10].Campisi J, Kapahi P, Lithgow GJ, Melov S, Newman JC, Verdin E, From discoveries in ageing research to therapeutics for healthy ageing, Nature 571 (184) (2019) 183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Fontana L, Partridge L, Longo VD, Extending healthy life span from yeast to humans, Science 328 (5976) (2010) 321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Longo VD, Kennedy BK, Sirtuins in aging and age-related disease, Cell 126 (2) (2006) 257–268. [DOI] [PubMed] [Google Scholar]
- [13].Wasko BM, Kaeberlein M, Yeast replicative aging: a paradigm for defining conserved longevity interventions, FEMS Yeast Res. 14 (1) (2014) 148–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kaeberlein M, Kennedy BK, Large-scale identification in yeast of conserved ageing genes, Mech. Ageing Dev 126 (1) (2005) 17–21. [DOI] [PubMed] [Google Scholar]
- [15].He C, Zhou C, Kennedy BK, The yeast replicative aging model, Biochim. Biophys. Acta (BBA) - Mol. Basis Dis 1864 (9) (2018) 2690–2696. [DOI] [PubMed] [Google Scholar]
- [16].Smith ED, Tsuchiya M, Fox LA, Dang N, Hu D, Kerr EO, Johnston ED, Tchao BN, Pak DN, Welton KL, et al. , Quantitative evidence for conserved longevity pathways between divergent eukaryotic species, Genome Res. 18 (4) (2008) 564–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, et al. , Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan, Nature 425 (6954) (2003) 191–196. [DOI] [PubMed] [Google Scholar]
- [18].Longo VD, Shadel GS, Kaeberlein M, Kennedy B, Replicative and chronological aging in Saccharomyces cerevisiae, Cell Metabol. 16 (1) (2012) 18–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mortimer RK, Johnston JR, Life span of individual yeast cells, Nature 183 (1959) 1751–1752. [DOI] [PubMed] [Google Scholar]
- [20].Bennett MR, Hasty J, Microfluidic devices for measuring gene network dynamics in single cells, Nat. Rev. Genet 10 (9) (2009) 628–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Duncombe TA, Tentori AM, Herr AE, Microfluidics: reframing biological enquiry, Nat. Rev. Mol. Cell Biol 16 (9) (2015) 554–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Steffen KK, Kennedy BK, Kaeberlein M, Measuring replicative life span in the budding yeast, J. Vis. Exp 28 (2009), e1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lee SS, Vizcarra IA, Huberts DH, Lee LP, Heinemann M, Whole lifespan microscopic observation of budding yeast aging through a microfluidic dissection platform, Proc. Natl. Acad. Sci. U.S.A 109 (13) (2012) 4916–4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Xie Z, Zhang Y, Zou K, Brandman O, Luo C, Ouyang Q, Li H, Molecular phenotyping of aging in single yeast cells using a novel microfluidic device, Aging Cell 11 (4) (2012) 599–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Cookson S, Ostroff N, Pang WL, Volfson D, Hasty J, Monitoring dynamics of single-cell gene expression over multiple cell cycles, Mol. Syst. Biol. 1(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ryley J, Pereira-Smith OM, Microfluidics device for single cell gene expression analysis in Saccharomyces cerevisiae, Yeast 23 (14–15) (2006) 1065–1073. [DOI] [PubMed] [Google Scholar]
- [27].Ferry MS, Razinkov IA, Hasty J, Microfluidics for synthetic biology: from design to execution, Methods Enzymol. 497 (2011) 295–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hansen AS, Hao N, O’Shea EK, High-throughput microfluidics to control and measure signaling dynamics in single yeast cells, Nat. Protoc 10 (8) (2015) 1181–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Baumgartner BL, O’Laughlin R, Jin M, Tsimring LS, Hao N, Hasty J, Flavin-based metabolic cycles are integral features of growth and division in single yeast cells, Sci. Rep 8 (1) (2018) 18045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Crane MM, Kaeberlein M, The paths of mortality: how understanding the biology of aging can help explain systems behavior of single cells, Curr. Opin. Struct. Biol 8 (2018) 25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Sherman F, Getting started with yeast, Methods Enzymol. 350 (2002) 3–41. [DOI] [PubMed] [Google Scholar]
- [32].Kaeberlein M, Powers RW, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK, Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients, Science 310 (5751) (2005) 1193–1196. [DOI] [PubMed] [Google Scholar]
- [33].Zhang Y, Luo C, Zou K, Xie Z, Brandman O, Ouyang Q, Li H, Single cell analysis of yeast replicative aging using a new generation of microfluidic device, PLoS One 7 (11) (2012), e48275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Fehrmann S, Paoletti C, Goulev Y, Ungureanu A, Aguilaniu H, Charvin G, Aging yeast cells undergo a sharp entry into senescence unrelated to the loss of mitochondrial membrane potential, Cell Rep. 5 (6) (2013) 1589–1599. [DOI] [PubMed] [Google Scholar]
- [35].Crane MM, Clark IB, Bakker E, Smith S, Swain PS, A microfluidic system for studying ageing and dynamic single-cell responses in budding yeast, PLoS One 9 (6) (2014), e100042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Jo MC, Liu W, Gu L, Dang W, Qin L, High-throughput analysis of yeast replicative aging using a microfluidic system, Proc. Natl. Acad. Sci. U. S. A 112 (30) (2015) 9364–9369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Liu P, Young TZ, Acar M, Yeast replicator: a high-throughput multiplexed microfluidics platform for automated measurements of single-cell aging, Cell Rep. 13 (3) (2015) 634–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Li Y, Jin M, O’Laughlin R, Bittihn P, Tsimring LS, Pillus L, Hasty J, Hao N, Multigenerational silencing dynamics control cell aging, Proc. Natl. Acad. Sci.U. S. A 114 (42) (2017) 11253–11258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sarnoski EA, Song R, Ertekin E, Koonce N, Acar M, Fundamental characteristics of single-cell aging in diploid yeast, iScience 7 (2018) 96–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hartwell LH, Unger MW, Unequal division in Saccharomyces cerevisiae and its implications for the control of cell division, J. Cell Biol 75 (2) (1977) 422–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kennedy BK, Austriaco NR, Guarente L, Daughter cells of Saccharomyces cerevisiae from old mothers display a reduced life span, J. Cell Biol 127 (6) (1994) 1985–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Tan WH, Takeuchi S, A trap-and-release integrated microfluidic system for dynamic microarray applications, Proc. Natl. Acad. Sci. U. S. A 104 (4) (2007) 1146–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Di Carlo D, Aghdam N, Lee LP, Single-cell enzyme concentrations, kinetics, and inhibition analysis using high-density hydrodynamic cell isolation arrays, Anal. Chem 78 (14) (2006) 4925–4930. [DOI] [PubMed] [Google Scholar]
- [44].Narayanamurthy V, Nagarajan S, Samsuri F, Sridhar T, et al. , Microfluidic hydrodynamic trapping for single cell analysis: mechanisms, methods and applications, Anal. Methods 9 (25) (2017) 3751–3772. [Google Scholar]
- [45].Yang M, Li CW, Yang J, Cell docking and on-chip monitoring of cellular reactions with a controlled concentration gradient on a microfluidic device, Anal. Chem 74 (16) (2002) 3991–4001. [DOI] [PubMed] [Google Scholar]
- [46].Ahmad Khalili A, Ahmad M, Takeuchi M, Nakajima M, Hasegawa Y,Mohamed Zulkifli R, A microfluidic device for hydrodynamic trapping and manipulation platform of a single biological cell, Appl. Sci 6 (2) (2016) 40. [Google Scholar]
- [47].Jin D, Deng B, Li J, Cai W, Tu L, Chen J, Wu Q, Wang W, A microfluidic device enabling high-efficiency single cell trapping, Biomicrofluidics 9 (1) (2015), 014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Janssens GE, Veenhoff LM, The natural variation in lifespans of single yeast cells is related to variation in cell size, ribosomal protein, and division time, PLoS One 11 (12) (2016), e0167394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Smeal T, Claus J, Kennedy B, Cole F, Guarente L, Loss of transcriptional silencing causes sterility in old mother cells of S. cerevisiae, Cell 84 (4) (1996) 633–642. [DOI] [PubMed] [Google Scholar]
- [50].Park PU, Mcvey M, Guarente L, Separation of mother and daughter cells, Methods Enzymol. (351) (2002) 468–477. [DOI] [PubMed] [Google Scholar]
- [51].Yang J, McCormick MA, Zheng J, Xie Z, Tsuchiya M, Tsuchiyama S, El-Samad H, Ouyang Q, Kaeberlein M, Kennedy BK, et al. , Systematic analysis of asymmetric partitioning of yeast proteome between mother and daughter cells reveals aging factors and mechanism of lifespan asymmetry, Proc. Natl. Acad. Sci. U.S.A 112 (38) (2015) 11977–11982.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Jin M, Li Y, O’Laughlin R, Bittihn P, Pillus L, Tsimring LS, Hasty J, Hao N, Divergent aging of isogenic yeast cells revealed through single-cell phenotypic dynamics, Cell Syst. 8 (3) (2019) 242–253 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Chen KL, Crane MM, Kaeberlein M, Microfluidic technologies for yeast replicative lifespan studies, Mech. Ageing Dev 161 (2017) 262–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Johnston J, Reproductive capacity and mode of death of yeast cells, Antonie Leeuwenhoek 32 (1) (1966) 94–98. [DOI] [PubMed] [Google Scholar]
- [55].McVey M, Kaeberlein M, Tissenbaum HA, Guarente L, The short life span of Saccharomyces cerevisiae sgs1 and srs2 mutants is a composite of normal aging processes and mitotic arrest due to defective recombination, Genetics 157 (4) (2001) 1531–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Merker RJ, Klein HL, hpr1D affects ribosomal DNA recombination and cell life span in Saccharomyces cerevisiae, Mol. Cell. Biol 22 (2) (2002) 421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Delaney JR, Chou A, Olsen B, Carr D, Murakami C, Ahmed U, Sim S, An EH, Castanza AS, Fletcher M, et al. , End-of-life cell cycle arrest contributes to stochasticity of yeast replicative aging, FEMS Yeast Res. 13 (3) (2013) 267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Falcon AA, Aris JP, Plasmid accumulation reduces life span in Saccharomyces cerevisiae, J. Biol. Chem 278 (43) (2003) 41607–41617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Moazed D, Enzymatic activities of Sir2 and chromatin silencing, Curr. Opin. Cell Biol 13 (2) (2001) 232–238. [DOI] [PubMed] [Google Scholar]
- [60].Gottschling DE, Gene silencing: two faces of Sir2, Curr. Biol 10 (19) (2000) R708–R711. [DOI] [PubMed] [Google Scholar]
- [61].Gartenberg MR, Smith JS, The nuts and bolts of transcriptionally silent chromatin in Saccharomyces cerevisiae, Genetics 203 (4) (2016) 1563–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Muller I, Parental age and the life-span of zygotes of Saccharomyces cerevisiae, Antonie Leeuwenhoek 51 (1) (1985) 1–10. [DOI] [PubMed] [Google Scholar]
- [63].Sinclair DA, Guarente L, Extrachromosomal rDNA circles: a cause of aging in yeast, Cell 91 (7) (1997) 1033–1042. [DOI] [PubMed] [Google Scholar]
- [64].Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA, Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast Sir2 and human SIRT1, J. Biol. Chem 277 (47) (2002) 45099–45107. [DOI] [PubMed] [Google Scholar]
- [65].Ganley AR, Ide S, Saka K, Kobayashi T, The effect of replication initiation on gene amplification in the rDNA and its relationship to aging, Mol. Cell 35 (5) (2009) 683–693. [DOI] [PubMed] [Google Scholar]
- [66].Ganley AR, Kobayashi T, Ribosomal DNA and cellular senescence: new evidence supporting the connection between rDNA and aging, FEMS Yeast Res. 14 (1) (2014) 49–59. [DOI] [PubMed] [Google Scholar]
- [67].Schlissel G, Krzyzanowski MK, Caudron F, Barral Y, Rine J, Aggregation of the Whi3 protein, not loss of heterochromatin, causes sterility in old yeast cells, Science 355 (6330) (2017) 1184–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Dodson AE, Rine J, Heritable capture of heterochromatin dynamics in Saccharomyces cerevisiae, eLife 4 (2015), e05007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Caudron F, Barral Y, A super-assembly of Whi3 encodes memory of deceptive encounters by single cells during yeast courtship, Cell 155 (6) (2013) 1244–1257. [DOI] [PubMed] [Google Scholar]
- [70].Shcheprova Z, Baldi S, Frei SB, Gonnet G, Barral Y, A mechanism for asymmetric segregation of age during yeast budding, Nature 454 (7205) (2008) 728–734. [DOI] [PubMed] [Google Scholar]
- [71].Neurohr GE, Terry RL, Sandikci A, Zou K, Li H, Amon A, Deregulation of the G1/S-phase transition is the proximal cause of mortality in old yeast mother cells, Genes Dev. 32 (15–16) (2018) 1075–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Morlot S, Song J, Leger-Silvestre I, Matifas A, Gadal O, Charvin G, Excessive rDNA transcription drives the disruption in nuclear homeostasis during entry into senescence in budding yeast, Cell Rep. 28 (2) (2019) 408–422. [DOI] [PubMed] [Google Scholar]
- [73].Denoth-Lippuner A, Krzyzanowski MK, Stober C, Barral Y, Role of SAGA in the asymmetric segregation of DNA circles during yeast ageing, eLife 3 (2014), e03790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Rempel IL, Crane MM, Thaller DJ, Mishra A, Jansen DP, Janssens G,Popken P, Aķsit A, Kaeberlein M, van der Giessen E, et al. , Age-dependent deterioration of nuclear pore assembly in mitotic cells decreases transport dynamics, eLife 8 (2019), e48186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Elowitz MB, Levine AJ, Siggia ED, Swain PS, Stochastic gene expression in a single cell, Science 297 (5584) (2002) 1183–1186. [DOI] [PubMed] [Google Scholar]
- [76].Blake WJ, Kærn M, Cantor CR, Collins JJ, Noise in eukaryotic gene expression, Nature 422 (6932) (2003) 633–637. [DOI] [PubMed] [Google Scholar]
- [77].Raser JM, O’Shea EK, Control of stochasticity in eukaryotic gene expression, Science 304 (5678) (2004) 1811–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Tsimring LS, Noise in biology, Rep. Prog. Phys 77 (2) (2014), 026601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Selimkhanov J, Taylor B, Yao J, Pilko A, Albeck J, Hoffmann A, Tsimring L,Wollman R, Accurate information transmission through dynamic biochemical signaling networks, Science 346 (6215) (2014) 1370–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Acar M, Mettetal JT, Van Oudenaarden A, Stochastic switching as a survival strategy in fluctuating environments, Nat. Genet 40 (4) (2008) 471–475. [DOI] [PubMed] [Google Scholar]
- [81].Raj A, van Oudenaarden A, Nature, nurture, or chance: stochastic gene expression and its consequences, Cell 135 (2) (2008) 216–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Bahar R, Hartmann CH, Rodriguez KA, Denny AD, Busuttil RA, Dolle ME, Calder RB, Chisholm GB, Pollock BH, Klein CA, et al. , Increased cell-to-cell variation in gene expression in ageing mouse heart, Nature 441 (7096) (2006) 1011–1014. [DOI] [PubMed] [Google Scholar]
- [83].Martinez-Jimenez CP, Eling N, Chen H-C, Vallejos CA, Kolodziejczyk AA,Connor F, Stojic L, Rayner TF, Stubbington MJ, Teichmann SA, et al. , Aging increases cell-to-cell transcriptional variability upon immune stimulation, Science 355 (6332) (2017) 1433–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Liu P, Song R, Elison GL, Peng W, Acar M, Noise reduction as an emergent property of single-cell aging, Nat. Commun 8 (1) (2017) 680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Peng W, Liu P, Xue Y, Acar M, Evolution of gene network activity by tuning the strength of negative-feedback regulation, Nat. Commun 6 (2015) 6226. [DOI] [PubMed] [Google Scholar]
- [86].Ferrezuelo F, Colomina N, Palmisano A, Gari E, Gallego C, Csikasz-Nagy A,Aldea M, The critical size is set at a single-cell level by growth rate to attain homeostasis and adaptation, Nat, Commun. Now 3 (2012) 1012. [DOI] [PubMed] [Google Scholar]
- [87].Janssens GE, Meinema AC, Gonzalez J, Wolters JC, Schmidt A, Guryev V,Bischoff R, Wit EC, Veenhoff LM, Heinemann M, Protein biogenesis machinery is a driver of replicative aging in yeast, eLife 4 (2015), e08527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Hendrickson DG, Soifer I, Wranik BJ, Kim G, Robles M, Gibney PA, McIsaac RS, A new experimental platform facilitates assessment of the transcriptional and chromatin landscapes of aging yeast, eLife 7 (2018), e39911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Lindstrom DL, Gottschling DE, The mother enrichment program: a genetic system for facile replicative life span analysis in Saccharomyces cerevisiae, Genetics 183 (2) (2009) 413–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Leupold S, Hubmann G, Litsios A, Meinema AC, Takhaveev V,Papagiannakis A, Niebel B, Janssens G, Siegel D, Heinemann M, Saccharomyces cerevisiae goes through distinct metabolic phases during its replicative lifespan, eLife 8 (2019), e41046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Selimkhanov J, Hasty J, Tsimring LS, Recent advances in single-cell studies of gene regulation, Curr. Opin. Biotechnol 23 (1) (2012) 34–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Hao N, O’shea EK, Signal-dependent dynamics of transcription factor translocation controls gene expression, Nat. Struct. Mol. Biol 19 (1) (2012) 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Hao N, Budnik BA, Gunawardena J, O’Shea EK, Tunable signal processing through modular control of transcription factor translocation, Science 339 (6118) (2013) 460–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Levine JH, Lin Y, Elowitz MB, Functional roles of pulsing in genetic circuits, Science 342 (6163) (2013) 1193–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Li P, Markson JS, Wang S, Chen S, Vachharajani V, Elowitz MB, Morphogen gradient reconstitution reveals hedgehog pathway design principles, Science 360 (6388) (2018) 543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Nandagopal N, Santat LA, LeBon L, Sprinzak D, Bronner ME, Elowitz MB, Dynamic ligand discrimination in the notch signaling pathway, Cell 172 (4) (2018) 869–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Feillet C, Krusche P, Tamanini F, Janssens RC, Downey MJ, Martin P,Teboul M, Saito S, Levi FA, Bretschneider T, et al. , Phase locking and multiple oscillating attractors for the coupled mammalian clock and cell cycle, Proc. Natl. Acad. Sci. U.S.A 111 (27) (2014) 9828–9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Schmoller KM, Turner J, Koivomagi M, Skotheim JM, Dilution of the cell cycle inhibitor Whi5 controls budding-yeast cell size, Nature 526 (7572) (2015) 268–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Papagiannakis A, Niebel B, Wit EC, Heinemann M, Autonomous metabolic oscillations robustly gate the early and late cell cycle, Mol. Cell 65 (2) (2017) 285–295. [DOI] [PubMed] [Google Scholar]
- [100].Hasty J, McMillen D, Collins JJ, Engineered gene circuits, Nature 420 (6912) (2002) 224–230. [DOI] [PubMed] [Google Scholar]
- [101].Veatch JR, McMurray MA, Nelson ZW, Gottschling DE, Mitochondrial dysfunction leads to nuclear genome instability via an iron-sulfur cluster defect, Cell 137 (7) (2009) 1247–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Hughes AL, Gottschling DE, An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast, Nature 492 (7428) (2012) 261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Denoth Lippuner A, Julou T, Barral Y, Budding yeast as a model organism to study the effects of age, FEMS Microbiol. Rev 38 (2) (2014) 300–325. [DOI] [PubMed] [Google Scholar]
- [104].McCormick MA, Delaney JR, Tsuchiya M, Tsuchiyama S, Shemorry A,Sim S, Chou ACZ, Ahmed U, Carr D, Murakami CJ, et al. , A comprehensive analysis of replicative lifespan in 4,698 single-gene deletion strains uncovers conserved mechanisms of aging, Cell Metabol. 22 (5) (2015) 895–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Henderson KA, Hughes AL, Gottschling DE, Mother-daughter asymmetry of pH underlies aging and rejuvenation in yeast, eLife 3 (2014), e03504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Erjavec N, Larsson L, Grantham J, Nystrom T, Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by over-producing the protein aggregation-remodeling factor Hsp104p, Genes Dev. 21 (19) (2007) 2410–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Hill SM, Hao X, Liu B, Nystrom T, Life-span extension by a meta- caspase in the yeast Saccharomyces cerevisiae, Science 344 (6190) (2014) 1389–1392. [DOI] [PubMed] [Google Scholar]
- [108].Hill SM, Hanzen S, Nystrom T, Restricted access: spatial sequestration of damaged proteins during stress and aging, EMBO Rep. 18 (3) (2017) 377–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Knorre DA, Azbarova AV, Galkina KV, Feniouk BA, Severin FF, Replicative aging as a source of cell heterogeneity in budding yeast, Mech. Ageing Dev 176 (2018) 24–31. [DOI] [PubMed] [Google Scholar]
- [110].Bakker E, Swain PS, Crane MM, Morphologically constrained and data informed cell segmentation of budding yeast, Bioinformatics 34 (1) (2017) 88–96. [DOI] [PubMed] [Google Scholar]
- [111].Chen KL, Ven TN, Crane MM, Chen DE, Feng YC, Suzuki N, Russell AE, de Moraes D, Kaeberlein M, An inexpensive microscopy system for microfluidic studies in budding yeast, Trans. Med. Aging 3 (2019) 52–56. [DOI] [PMC free article] [PubMed] [Google Scholar]


