Abstract
Background
High-grade gliomas are rapidly progressing tumors of the central nervous system, and are associated with poor prognosis and highly immunosuppressive microenvironments. Meanwhile, a better understanding of PD-L1, a major prognostic biomarker for checkpoint immune therapy, regulation may provide insights for developing novel immunotherapeutic strategies for treating gliomas. In the present study, we elucidate the functional significance of the orphan nuclear receptor TLX in human glioma, and its functional role in immune suppression through regulation of PD-L1/PD-1 axis.
Methods
TLX and PD-L1 expression patterns, and their association with clinicopathological parameters and immune phenotypes of glioma were analysed using CIBERSORT algorithm and single-sample gene-set enrichment analysis from The Cancer Genome Atlas (n=695) and Chinese Glioma Genome Atlas (n=1018) databases. Protein expression and cellular localization of TLX, PD-L1, and PD-1, as well as the prevalence of cytotoxic tumor-infiltrating lymphocytes (TILs), and tumor-associated macrophages (TAMs), in the glioma immune microenvironment were analyzed via tissue microarray by immunohistochemistry and multiplex immunofluorescence. Glioma allografts and xenografts with TLX manipulation (knockdown/knockout or reverse agonist) were inoculated subcutaneously, or orthotopically into the brains of immunodeficient and immunocompetent mice to assess tumor growth by imaging, and the immune microenvironment by flow cytometry. PD-L1 transcriptional regulation by TLX was analyzed by chromatin immunoprecipitation and luciferase reporter assays.
Results
TLX and PD-L1 expression was positively associated with macrophage-mediated immunosuppressive phenotypes in gliomas. TLX showed significant upregulation and positive correlation with PD-L1. Meanwhile, suppression of TLX significantly inhibited in vivo growth of glioma allografts and xenografts (p<0.05), rescued the antitumoral immune response, significantly decreased the PD-L1+, and glioma-associated macrophage population, and increased cytotoxic lymphocyte infiltration (p<0.05). Mechanistically, TLX binds directly to CD274 (PD-L1) gene promoter and activates CD274 transcription.
Conclusions
TLX contributes to glioma malignancy and immunosuppression through transcriptional activation of PD-L1 ligands that bind to PD-1 expressed on both TILs and TAMs. Thus, targeting the druggable TLX may have potential therapeutic significance in glioma immune therapy.
Keywords: immunotherapy, tumor microenvironment, brain neoplasms, programmed cell death 1 receptor, immune evation
Background
Gliomas represent the most common primary and life-threatening intracranial tumor. Nearly half of all glioma cases are diagnosed as glioblastomas (GBMs), which are regarded as the most aggressive and lethal brain tumors and are associated with dismal prognosis and poor quality of life. The median overall survival of GBM patients receiving prevalent standard therapeutic interventions (surgery, radiotherapy, and chemotherapy) is approximately 15 months with an average 5-year survival rate less than 5%.1 Although the advent of multimodal therapeutic approaches has led to a consistent, yet slow improvement in overall survival, the prognosis for patients with GBM remains poor, with most patients exhibiting early tumor progression or recurrence.2 Moreover, as this invasive tumor is located in the brain, it is difficult to achieve gross total resection through local surgery, particularly for advanced diffuse GBM. Furthermore, the blood–brain barrier limits the development of effective new drugs. Thus, novel treatment methods based on the biological attributes of GBM are urgently needed.
Although immunotherapy has proven revolutionary for the treatment of many cancers, due to their immunosuppressive tumor microenvironment, brain tumors often respond poorly to these regimens.3 Thus, understanding the unique mechanisms of the immune microenvironment of gliomas may inform the development of new effective immunotherapy options. Indeed, the poor responses of gliomas to immunotherapy, including immune checkpoint protein PD-1/PD-L1 inhibitors, may be related to the limited intratumoral infiltration of lymphocyte immune cells, the anergic nature of effector T cells to tumor-specific antigens, and low mutation rates in gliomas.4 5 However, recent studies have reported the efficacy of anti-PD-1 therapy in certain GBM patients, while indicating that neoadjuvant administration of PD-1 blockade can enhance both the local and systemic antitumor immune responses, promote survival in recurrent GBMs,6–8 and modify the tumor immune microenvironment.8 Accordingly, clinical trials are underway to identify potential patient subgroups that would benefit from immune checkpoint blockade (ICB) therapy.5
PD-L1 is considered a major prognostic biomarker for checkpoint immune therapy in many cancers. Indeed, PD-L1 expression has been immunohistochemically detectable in most GBM patients, with a significant association between PD-L1 expression with molecular GBM subtype observed.9 10 However, the relationship between PD-L1, immune cell infiltration, and cancer-associated immunosuppression remains unclear. Reports show that upregulation of PD-L1 in tumor cells is associated with an increase in tumor-infiltrating lymphocytes (TILs),11 which can trigger their own inhibition by secreting cytokines that drive PD-L1 expression. Elevated PD-L1 levels can also induce expansion and maintenance of the immunosuppressive regulatory T lymphocyte (Treg) population, which is associated with decreased survival of glioma patients.12 PD-L1 expression in cancer cells is affected by certain signaling pathways, transcription factors, and kinases. For example, loss of the tumor suppressor gene PTEN in gliomas can significantly induce PD-L1 protein expression and immuno-resistance.13 PD-L1 expression can also be attenuated by the loss of CDK5, a key enzyme associated with interferon gamma (IFN-γ) and PD-L1 upregulation.14 However, the regulation of PD-L1 expression on glioma cells, as well as its influence on tumor control for predicting treatment responses, remains unclear. Hence, elucidating the regulatory mechanism underlying PD-L1 expression in gliomas may provide insights for the development of novel immunotherapeutic strategies for glioma treatments.
The orphan nuclear receptor TLX is an essential transcriptional factor in the maintenance of stemness, and self-renewal of embryonic and adult neural stem cells (NSCs) in brain and retina development.15–20 Moreover, TLX transgenic mouse models involving genetic loss of tumor suppressors, such as PTEN or TP53, have shown that TLX overexpression can promote long-term expansion of a subgroup of NSCs, resulting in glioma development.21 22 Additionally, TLX plays an oncogenic role in promoting the development of prostate cancer through inhibition of oncogene-induced cellular senescence,23 and progression of prostate cancer towards castration-resistance via transcriptional repression of the AR gene.24 Furthermore, both natural and synthetic retinoids, such as BMS453, bind directly to TLX and regulate its transcriptional activity,25 suggesting that targeting TLX may be of potential therapeutic significance in human cancers. However, the expression of TLX in human gliomas, and its functional role in immune escape, have not yet been investigated.
In the present study, we elucidate the functional significance of TLX in immune responses via transcriptional regulation of PD-L1 in glioma.
Methods
Patients, tissue collection, and tissue microarray
Clinical glioma lesions obtained from glioma patients who underwent neurosurgical resection, and normal human brain tissue obtained from surgical trauma patients were collected from the Department of Neurosurgery, Yi Jishan Hospital, and the Shenzhen Hospital of Southern Medical University from 2016 to 2019, under the surveillance of hospital Human Ethics Committees. Each sample was confirmed by interpretation of H&E staining by two pathologists. Histological diagnosis of glioma was performed according to current WHO classification. A tumor microarray (TMA) was constructed using tissue specimens from 19 WHO grade IV GBMs, 26 WHO grade III gliomas, 25 WHO grade II gliomas, and 8 non-glioma brain tissues.
Bioinformatics analysis
Clinical information and glioma gene expression data were obtained from The Cancer Genome Atlas (TCGA) (RNA-seq, n=695) and the Chinese Glioma Genome Atlas (CGGA) (RNA-seq, n=1018) databases. Twenty-two types of TILs were identified based on CIBERSORT, a gene expression-based deconvolution algorithm that uses a set of barcoded gene expression values (a “signature matrix” of 547 genes) to characterize immune cell composition.26 27 Only cases with CIBERSORT p<0.05 were included in subsequent analyses. Enrichment levels of the 29 immune signatures in each glioma sample were quantified by single-sample gene-set enrichment analysis (ssGSEA). Hierarchical clustering of gliomas was performed based on ssGSEA scores of the 29 immune signatures.28 The ESTIMATE algorithm was used to evaluate the immune cell infiltration level (immune score) of each sample.29 TLX (NR2E1) and PD-L1 (CD274) gene expression profiles were downloaded from UCSC Xena (https://xenabrowser.net/datapages/). Survival analysis data were downloaded from the Gene Expression Profiling Interactive Analysis (http://gepia.cancer-pku.cn/) database.30
Immunohistochemical and multiplex immunofluorescence staining
The primary antibodies used for immunohistochemistry (IHC) and multiplex immunofluorescence staining are presented in the online supplemental materials and methods. IHC staining of the cancer lesions and TMA slides was performed as described previously.31 Multiplex immunofluorescence was performed using a PANO 7-plex IHC kit (Panovue, Beijing, China), according to manufacturer’s instructions. Different primary antibodies were sequentially applied, followed by horseradish peroxidase-conjugated secondary antibody incubation and tyramide signal amplification (TSA) using a TSA Fluorescence kit (Panovue, Beijing, China). After labeling with the human antigens, nuclei were stained with 4′,6-Diamidino-2-phenylindole dihydrochloride (DAPI). Stained slides were scanned using the PanoVIEW vs200 slide scanner (Panovue, Beijing, China), equipped with an Olympus 20×lens, to obtain multispectral images. Fluorescence spectra were captured at 20 nm wavelength intervals from 420 to 720 nm, with identical exposure time. Multispectral images were analyzed, and positive cells were quantified at a single-cell level by QuPath v0.2.0 image analysis software (Queen’s University Belfast, Northern Ireland, UK) on a PanoATLAS workstation.
jitc-2020-001937supp001.pdf (95.4KB, pdf)
Cell lines and cell culture
Human GBM cell line A1235, mouse GBM cell lines GL261, G422, and CT2A, human kidney HEK293 cells, and the prostate cancer cell line PC-3 were obtained from ATCC (Manassas, Virginia, USA). They were maintained in Dulbecco’s modified eagle medium supplemented with 10% fetal bovine serum, 2 mM L-glutamine, and penicillin–streptomycin (Solarbio, China). Cell lines were authenticated by regular short tandem repeat profiling.
TLX RNA interference and CRISPR/Cas9 knockdown
Stable TLX knockdown by RNA interference and CRISPR/Cas9 knockout in human and mouse glioma cells were generated as described previously.23 The sequences for shRNA and sgRNA targeting mouse TLX are presented in online supplemental materials and methods.
Quantitative reverse transcription PCR and immunoblotting
PCR and immunoblotting were performed as described previously.32
In vivo tumor growth assay
In vivo tumor growth of TLX-transduced cells was evaluated by subcutaneous or intracranial injection of cells into C57 (subcutaneous for GL261, CT2A, 5×106), ICR (intracranial for G422, 4×105), and SCID (subcutaneous for A1235, 2×106) mice. Tumor growth was monitored, and tumor volume was measured by electronic calipers and in vivo imaging. TLX inverse agonist treatment was presented in the online supplemental materials and methods. Tumors were dissected at the end of experiments, and single-cell suspensions were prepared for subsequent flow cytometric (FCM) analyses. All animal experiments were performed according to the university animal guidelines, and with prior approval from the Animal Experimentations Ethics Committee, Southern Medical University.
FCM analysis and gating
Tumor tissues from xenografted mice were dissociated into single-cell suspensions by filtering through a 40-µm cell strainer, collected by low-speed centrifugation, and resuspended in PBS before FCM analysis. Viable suspended cells (106), preblocked with Fc receptor blocking solution (BioLegend, #156603, San Diego, CA, USA), were incubated with antibodies (online supplemental materials and methods). The stained cells were subjected to FCM analysis using a Sony SA3800 analyser (Sony Biotechnology, Tokyo, Japan). Tumor cells were gated as CD45- populations; for lymphocytes, CD45+ cells were first gated by CD3+ and subsequently gated for CD4+, CD8+ and PD-1+ cells; for myeloid-associated cells, mouse macrophages were first gated by F4/80, M1 macrophages were then gated by F4/80+CD86+ and M2 macrophages by F4/80+CD206+ or F4/80+CD163+; PD-L1+ and PD-1+ cells were further characterized under the CD206+ population.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assay on the CD274 (PD-L1) gene promoter was performed in A1235 cells as previously described.23 Genomic DNA extracted from the cells was sonicated to obtain sheared fragments. DNA samples were immunoprecipitated by an antibody against TLX (Perseus Proteomics, Tokyo, JAPAN), followed by PCR analysis (30 cycles) with primer pairs specific for the PD-L1 (CD274) gene promoter.
Luciferase reporter assay
The PD-L1-luc reporter assay was performed in HEK293 cells, as described previously.33
Statistical analysis
Spearman’s correlation and Pearson’s correlation were applied to analyze the correlation between two ordinary parameters. Chi-square test was used to assess group differences. Overall survival from initial diagnosis to death or last follow-up was estimated using the Kaplan–Meier method. The log-rank test was used to assess group differences. Associations between inferred proportions of immune cell types and survival were tested using Cox regression. Quartiles of the proportion of each cell type were computed for survival analysis and modeled as continuous variables to derive interpretable HRs. Student’s t-test and one-way ANOVA were used for single comparison. A two-tailed significance level (p value) of 0.05 was applied. Data were analyzed by GraphPad Prism, SPSS V.24.0, and the statistical package of R language.
Results
Expression of TLX and PD-L1 is associated with distinct immune phenotypes in glioma
To determinate whether TLX and PD-L1 correlate with immune microenvironment regulation of glioma, we first comprehensively analyzed the diversity and landscape of immune cell tumor infiltration in glioma by CIBERSORT analysis and ssGSEA scores. The proportion of 22 types of adaptive and innate immune cells in each glioma sample was analyzed using the TCGA database (n=695); analyses were restricted to samples with a CIBERSORT p<0.05 (n=187; online supplemental figure S1A). Among the 22 tumor-infiltrating immune cells, monocytes (HR, 0.16; 95% CI, 0.45 to 0.89; p<0.001), activated NK cells (HR, 0.32; 95% CI, 0.16 to 0.63; p<0.001), and eosinophils (HR, 0.50; 95% CI, 0.30 to 0.85; p<0.001) were correlated with better prognosis, whereas macrophages, including M0 (HR, 2.25; 95% CI, 1.52 to 3.35; p<0.001), M1 (HR, 2.17; 95% CI, 1.08 to 4.37; p<0.001), and M2 (HR, 2.87; 95% CI, 1.40 to 5.89; p<0.001), as well as CD8 T cells (HR, 2.31; 95% CI, 1.43 to 3.74; p<0.001) were associated with poor prognosis (online supplemental figure S1B). Moreover, macrophages (M0, M1, and M2) were the dominant immune cells in high-grade glioma (WHO Ⅲ and Ⅳ), as evidenced by their higher percentage of infiltration (online supplemental figure S1C).
jitc-2020-001937supp002.pdf (100.6KB, pdf)
jitc-2020-001937supp003.pdf (591KB, pdf)
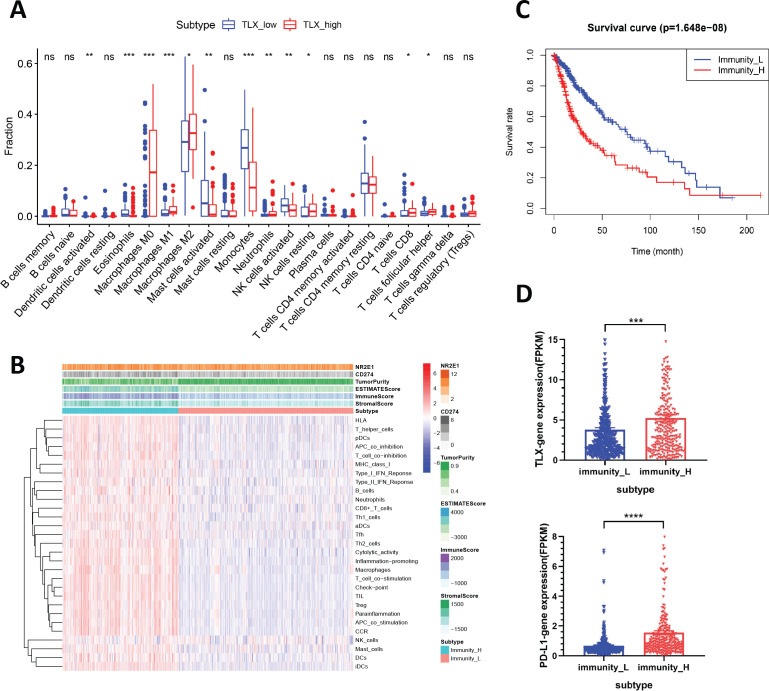
We then divided the cohort into TLX_low and TLX_high groups, and the status of immune cell infiltration was compared. Results showed that M0 and M2 macrophages were the most enriched immune cells in the TLX_high group, whereas the amount of activated mast cells and monocytes was markedly decreased (p<0.05; figure 1A), which was consistent with the status of immune cell infiltration in high-grade gliomas (Figure S1C). Furthermore, neutrophils, NK cells (activated and resting), CD8+ T cells, and follicular helper T cells also displayed significant accumulation in the TLX_high group, however, did not represent the major immune cell types in glioma lesions (p<0.05; figure 1A).
Figure 1.
Analysis of the association between TLX expression with PD-L1 and immune cell infiltration by CIBERSORT and ssGSEA. (A) The difference of immune infiltration between TLX_low and TLX_high expression group by using CIBERSORT analysis. TLX expression was segregated by median value. *P<0.05, **p<0.01, ***p<0.001 vs TLX_low patients. (B) Heatmap of 29 gene sets correlated with glioma microenvironment signatures in Immunity_H and Immunitu _L. Expression of TLX (NR2E1), PD-L1 (CD274), Tumor Purity, ESTIMATE Score, Immune Score and Stromal Score were evaluated by ESTIMATE. (C) Comparison of survival prognosis between Immunity_L and Immunity_H (log-rank test). (D) mRNA level of TLX and PD-L1 in Immunity_H and Immunity_L in TCGA. Data are mean±SEM ***p<0.001 vs Immunitu _L population. ssGSEA, single-sample gene-set enrichment analysis; TCGA, The Cancer Genome Atlas.
Hierarchical clustering of gliomas from the TCGA database revealed two distinct clusters based on the ssGSEA scores of the 29 immune signatures. ESTIMATE29 evaluation showed that the Immunity_H glioma population had elevated TLX (NR2E1) and PD-L1 (CD274) expression, as well as higher immune, stromal, and ESTIMATE scores, along with lower tumor purity than the Immunity_L population (figure 1B), and was associated with poor prognosis (p<0.001; figure 1C). Further examination of TLX and PD-L1 expression in the Immunity_L and Immunity_H subtypes revealed that both TLX and PD-L1 were significantly higher in the Immunity_H subtype than in the Immunity_L subtype (t-test, p<0.05) (figure 1D). Moreover, comparison of the ssGSEA scores for TLX_high versus TLX_low, and PD-L1_high versus PD-L1_low populations revealed higher immune cell infiltration levels and more immune-related pathways in TLX_high and PD-L1_high clusters (online supplemental figure S1E, F). Based on these results, we speculated that high expression of TLX and PD-L1 was positively associated with macrophage-related suppressive immune microenvironment and poor prognosis of gliomas.
TLX is significantly upregulated and positively correlates with PD-L1 expression in glioma
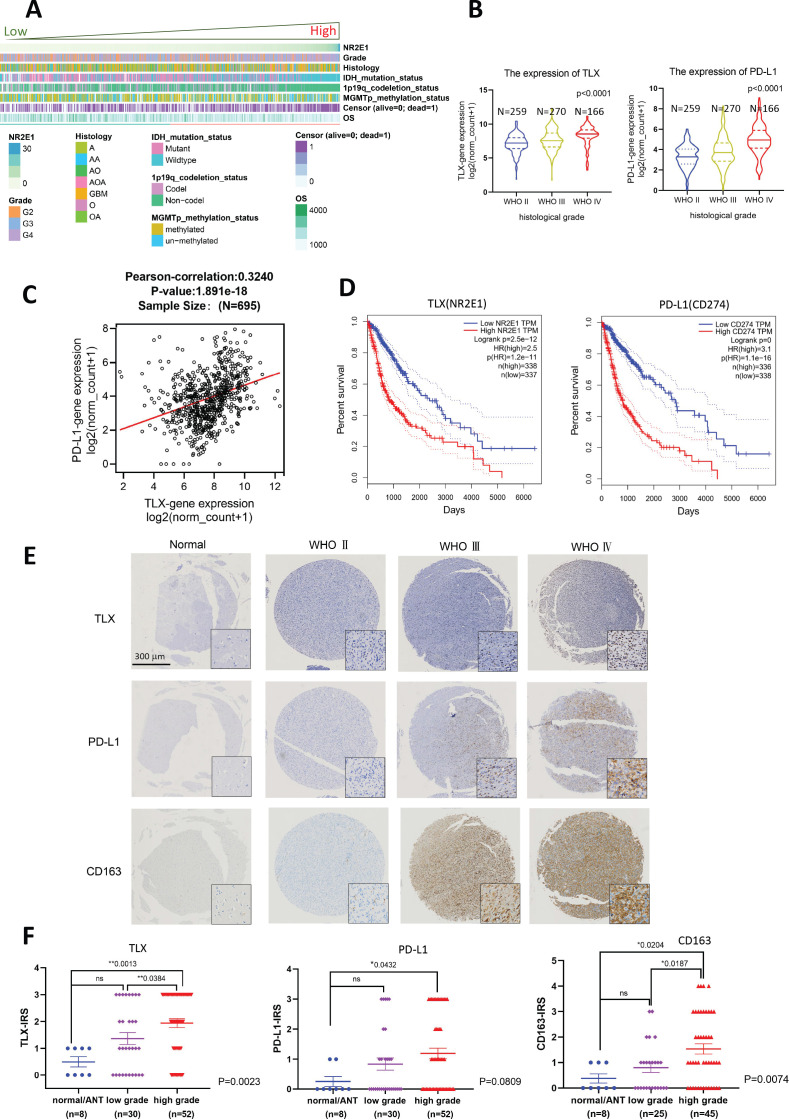
To determine whether the increase in TLX and PD-L1 expression contributed to the advanced progression of glioma, we analyzed the expression profiles of TLX and PD-L1 in clinical glioma, and their association with patient clinicopathological parameters using the cancer genomic data sets from TCGA and CGGA databases. Bioinformatics analysis of RNA-seq data showed that low TLX expression indicated lower WHO grades, IDH mutant and 1p19q deletion status. In contrast, high TLX expression was associated with an unfavorable prognosis, higher WHO grades, a wild-type IDH and 1p19q with no obvious correlation with MGMT methylation status (figure 2A and online supplemental figure S2D). Moreover, upregulation of TLX and PD-L1 in high-grade tumors (figure 2B and online supplemental figure S2A), and significant correlation between TLX and PD-L1 was confirmed in TCGA and CGGA cohorts (figure 2C and online supplemental figure S2B). Kaplan–Meier survival analysis revealed that the expression levels of TLX (775 days for TLXhigh vs 2660 days for TLXlow; p<0.001, log-rank test) and PD-L1 (828 days for PD-L1high vs 2681 days for PD-L1low; p<0.001, log-rank test) were correlated with overall survival, as per TCGA data (figure 2D), which was further verified for CGGA glioma patients (online supplemental figure S2C).
Figure 2.
Upregulation of TLX was associated with clinicopathological characteristics of aggressive glioma, PD-L1 expression and CD163+ TAMs infiltration. (A) Samples from CGGA data set (n=1018) were arranged in ascending order according to TLX expression. The relationship between TLX expression and patient’s clinicopathological parameters was depicted. (B) TLX and PD-L1 mRNA expression levels in patients from TCGA with WHO grade Ⅱ (n=259), WHO grade Ⅲ (n=270) and glioblastoma multiforme (WHO grade Ⅳ, n=166). P<0.001 vs WHO Ⅱ tumors. (C) Correlation of TLX and PD-L1 in TCGA (n=695) glioma. High expression of PD-L1 is correlated positively with TLX. Pearson-correlation=0.3240, ***p<0.001. (D) Overall survival according to expression levels of TLX (NR2E1) and PD-L1(CD274) in the tumors. (E) TLX, PD-L1, and CD163 immunohistochemistry; representative micrographs show TLX, PD-L1, and CD163 immunostaining in normal brain tissues or tumor adjacent normal tissues and glioma tissues with different grades. Magnification, ×40. Scale bar 300 µm. Insets show the enclosed areas at higher magnification. Magnification, ×400. Scale bar 40 µm. (F) Immuno-reactive score analysis of TLX, PD-L1, and CD163 performed on normal tissues or adjacent tumor tissues (n=8), low-grade gliomas (WHO Ⅱ, n=30) and high-grade gliomas (WHO Ⅲ & Ⅳ, n=52). High-grade gliomas showed significantly higher TLX, PD-L1, and CD163 expression than normal tissues and low-grade gliomas (one-way ANOVA, p=0.0013, 0.0432, and 0.0204, respectively). Data are mean±SEM *p<0.05, **<0.001 vs normal/ANT tissues. ANOVA, analysis of variance; CGGA, Chinese Glioma Genome Atlas; TAMs, tumor-associated macrophages; TCGA, The Cancer Genome Atlas.
jitc-2020-001937supp004.pdf (764.1KB, pdf)
We next performed IHC analysis using glioma TMAs and paraffin sections containing normal brain tissues and glioma lesions of different WHO grades to detect TLX and PD-L1 protein expression, as well as that of the M2 tumor-associated macrophages (TAM) marker, CD163, and cytotoxic lymphocyte marker, CD8. Patient clinical information, including baseline parameters, administered therapies, and survival, are presented in table 1. IHC analysis showed that high-grade gliomas (WHO Ⅲ and Ⅳ) displayed significantly stronger nuclear staining of TLX compared with low-grade gliomas (WHO Ⅱ, p=0.0384) and normal tissues (p=0.0013). High-grade glioma had enhanced PD-L1 expression compared with normal tissues (p=0.0432). Moreover, CD163+ cells were significantly enriched in high-grade gliomas compared with low-grade gliomas (p=0.0187) and normal tissues (p=0.0204; Figure 2D, E). Although CD8+ cells were present in glioma lesions, no significant correlation was noted between the number of CD8+ TILs and the clinical stage (online supplemental figure S3A), which was consistent with the bioinformatics results. Results of semiquantitative tumoral TLX and PD-L1 expression analysis in different histological grades (online supplemental table 1) were as follows: >50% TLX expression was observed in 38.4% of high-grade gliomas versus 20% in low-grade gliomas; >25% PD-L1 expression was in 35.6% of high-grade gliomas versus 23.3% in low-grade gliomas. Results of semiquantitative analysis of TLX+, PD-L1+, CD8+, and CD163+ cell density and distribution in glioma lesions are presented in online supplemental table 2. Correlation analysis of TLX and PD-L1 expression with CD8+ TIL and CD163+ TAM infiltration revealed significant positive correlations between PD-L1 expression and the number of CD8+ TILs (p<0.001), PD-L1 expression and CD163+ TAMs (p<0.001), and CD8+ TILs and CD163+ TAMs (p<0.001; online supplemental table 3). Together, these results indicate that upregulation of TLX and PD-L1 positively correlated with the progression of clinical glioma stage and TAM infiltration.
Table 1.
Patient characteristics
| Characteristic | Entire population (n=82) |
|
| N | % | |
| Sex | ||
| Female | 39 | 47.8 |
| Male | 43 | 52.4 |
| Age, years | ||
| ≤50 | 36 | 43.9 |
| >50 | 46 | 56.1 |
| Grade | ||
| WHO Ⅱ | 30 | 36.6 |
| WHO Ⅲ | 27 | 32.9 |
| WHO Ⅳ | 25 | 30.5 |
| Histology | ||
| Astrocytoma | 37 | 45.1 |
| Oligodendroglioma | 14 | 17.1 |
| Glioblastoma | 22 | 26.8 |
| Gliosarcoma | 4 | 4.9 |
| Mixed glioma | 5 | 6.1 |
| Extend of resection | ||
| Complete resection | 69 | 84.1 |
| Subtotal resection | 13 | 15.9 |
| Type of postoperative first-line therapy (TMZ or radiotherapy) | ||
| Yes | 54 | 65.9 |
| No | 28 | 34.1 |
| Alive at last follow-up | ||
| Yes | 21 | 25.6 |
| No | 54 | 65.9 |
| Unknown | 7 | 8.5 |
| Median overall survival in days (range) | 390 (61–2178) | |
TMZ, temozolomide.
jitc-2020-001937supp005.pdf (289.2KB, pdf)
jitc-2020-001937supp006.pdf (106.2KB, pdf)
Upregulation of TLX in glioma cells is associated with a macrophage-mediated immunosuppressive microenvironment
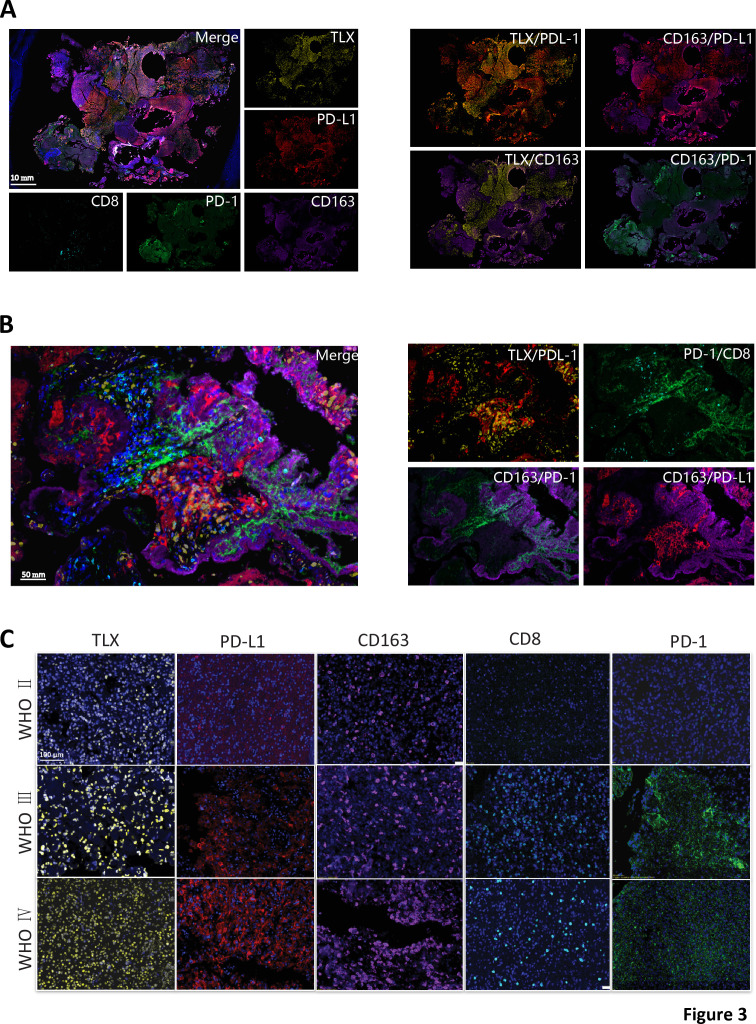
TLX, PD-L1, and markers of cytotoxic TILs (CD8, PD-1) and M2 macrophages (CD163) were detected in glioma lesions using multiplex immunofluorescence to characterize the relationship between TLX, PD-L1-positive cells, and the distribution of TILs and TAMs. A significant overlap in TLX+ (nuclear) and PD-L1+ (cytoplasm and cell membrane) cell distribution was observed in tissue sections (figure 3A, B). Meanwhile, TLX+ and CD163+ cells (cytoplasm and cell membrane) were inversely distributed. Interestingly, CD163-positive cells were effectively stained with PD-L1 and PD-1 antibodies. Furthermore, CD8+ cells were observed to unevenly infiltrate certain areas of GBM lesions, and to colocalize with PD-1+ cells (figure 3B and online supplemental figure S3B). Hence, we concluded that TLX was primarily expressed in glioma cell nuclei and was distinct from glioma infiltrated TAMs and TILs. Additionally, we confirmed that CD163+ M2 macrophages were the dominant infiltrated immune cell type in glioma.
Figure 3.
Tissue and cellular distribution of TLX, PD-L1, and TILs and TAMs. (A) Multiplex immunofluorescence staining of TLX (yellow), PD-L1 (red), PD-1 (green), CD8 (bright blue), CD163 (purple), and DAPI (blue) on GBM whole tissue section (10X), scale bar 2 mm. Left: merged and single-channel photo of the whole section. Right: combined channel of TLX/PD-L1, TLX/CD163, CD163/PD-L1, and CD163/PD-1 on whole section. (B) Representative multiplex immunofluorescence area show colocalization of TLX/PD-L1, TLX/CD163, CD163/PD-L1, and CD163/PD-1. Left: merged photo; Right: combined channels of two stainings (300X), scale bar 50 µm. (C) Representative immunofluorescence images of TLX (yellow), PD-L1 (red), PD-1 (green), CD8 (bright blue), CD163 (purple), and DAPI (blue) on TMA with different WHO grades of glioma (200X). Scale bar 100 µm. DAPI, diamidino-2-phenylindole dihydrochloride; GBM, glioblastomas; TAM, tumour-associated macrophages; TILs, tumour-infiltrating lymphocytes; TMA, tumor microarray.
To confirm these results, TLX-positive, PD-L1-positive, CD163-positive, CD8-positive, and PD-1-positive cell distribution was assessed in TMA sections stained with multicolor immunofluorescence. Results showed that TLX and PD-L1 were more highly expressed in high-grade gliomas (WHO III and IV) than low-grade lesions (WHO II), and more infiltrated TAMs were observed in advanced glioma (figure 3C). Together, these data indicated that higher expression of TLX and PD-L1 correlated with high-grade glioma and a macrophage-mediated suppressive immune microenvironment.
Inhibition of TLX significantly arrests in vivo tumor growth by rescuing antitumor immune responses
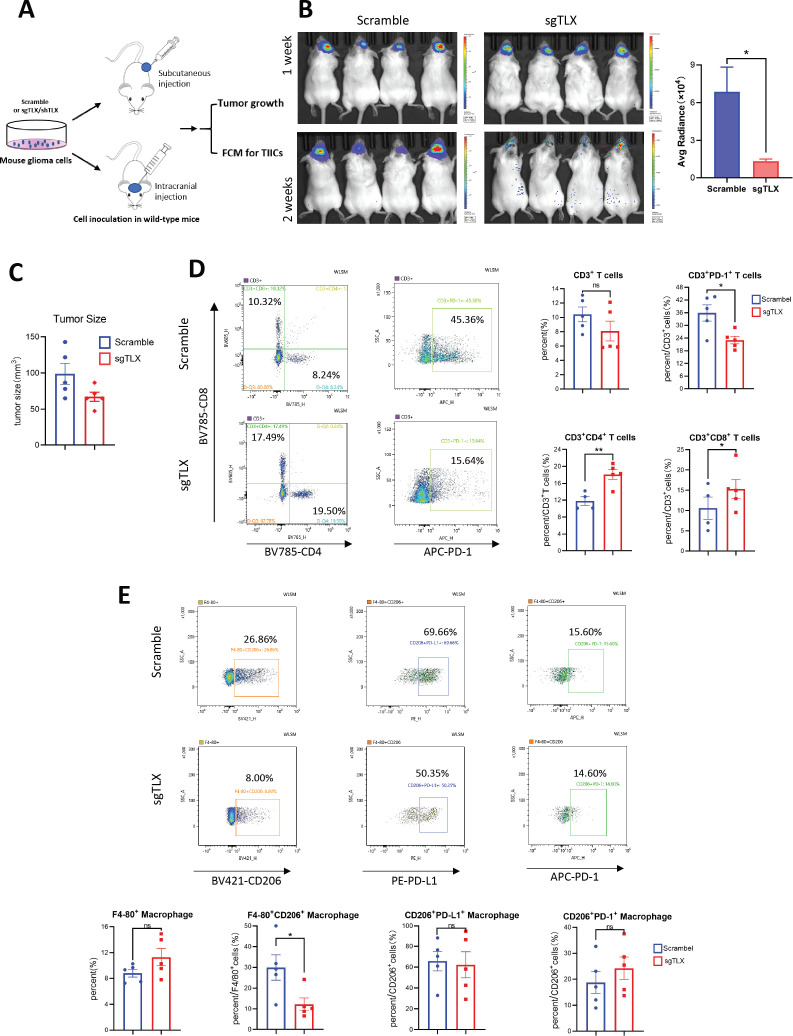
We next investigated whether TLX contributes to the antitumor immune response. In vivo glioma allograft models were established by subcutaneously implanting mouse glioma GL261 cells with shRNA-mediated TLX knockdown, and intracranial injection of G422 cells with CRISPR/Cas9-mediated TLX gene knockout into immunocompetent mice to observe tumor growth and analyze the effect of TLX inhibition on the immune microenvironment (figure 4A and online supplemental figure S4A). Results showed that TLX silencing can significantly reduce in vivo subcutaneous and brain orthotopic tumor growth in mice (figure 4B, C, and online supplemental figure S4B). We then treated CT2A allografted mice with a specific TLX inhibitor, ATRAL, at 20 mg/kg, or vehicle, to determine whether in vivo glioma growth could be suppressed by TLX inhibition (online supplemental figure S5A). ATRAL treatment markedly inhibited CT2A tumor growth, with no significant effect on body weight (online supplemental figure S5B–E. Cumulatively, these findings showed that repression of TLX could significantly inhibit glioma growth.
Figure 4.
Inhibition of TLX significantly arrested in vivo tumor growth by rescuing antitumor immune response. (A) Schematic diagram for in vivo experiment of subcutaneous and brain orthotopic tumor inoculation for tumor growth and FCM analysis. (B) Left: representative in vivo imaging show tumor growth of luciferase-labeling TLX knockout cell G422-sgTLX vs G422-scramble cells by brain orthotopical injection of at 5 µL to ICR mice. Mouse were pretreated with D-luciferin and photos were taken every week (4×105 cells per mouse, G422-sgTLX, n=9; G422-scramble, n=9). Right: quantification of luminance data from in vivo imaging. *P<0.05 vs G422-scramble tumors. (C) Subcutaneous tumors formed by G422-sgTLX vs G422-scramble infectants in ICR mice and tumor size were evaluated at the endpoint of the experiment (5×106, n=10 for each group). *P<0.05 vs G422-scramble tumors. Data are presented as mean±SD. At least two independent experiments were performed. (D, E) Flow cytometry analysis of TILs (CD3+CD4+, CD3+CD8+, CD3+PD-1+) and TAMs (F4/80+CD163+ and F4/80+CD86+) population in G422 allografts. (D) CD3+CD4+ and CD3+CD8+ populations were significantly accumulated and CD3+PD-1+ cells were remarkably decreased in G422-sgTLX tumors, *p<0.05, **p<0.001 vs scramble control. (E) M2 macrophage (F4/80+CD206+) population was remarkably decreased in G422-sgTLX tumors. *P<0.05. Although PD-1+ and PD-L1+ cells can be gated from F4/80+CD206+ cells, no difference of F4/80+CD206+/PD-1+ and F4/80+CD206+/PD-L1+ population exist between G422-sgTLX (n=5) and G422-scramble tumors (n=5). Data are mean±SEM, *p<0.05 vs scramble control. FCM, flow cytometric; TAM, tumor-associated macrophages; TILs, tumor-infiltrating lymphocytes.
jitc-2020-001937supp007.pdf (361.8KB, pdf)
jitc-2020-001937supp008.pdf (469.2KB, pdf)
We next investigated whether tumor inhibition induced by TLX repression was mediated through immune microenvironment regulation. To this end, TIL and macrophage populations in G422-sgTLX versus G422-scramble, and GL261-shTLX versus GL261-scramble allografts were examined by FCM. CD3+CD8+ cells were markedly enriched, while CDF4/80+CD206+ cells were significantly reduced in G422-sgTLX (figure 4D, E) and GL261-shTLX (online supplemental figure S4C, D) tumors (p<0.05), accompanied by a significant reduction in the CD3+PD-1+ population in subcutaneously and brain orthotopically implanted G422-sgTLX tumors (figure 4D and online supplemental figure S5F). Moreover, PD-1 expression was detected in both CD8+ T cells (CD3+CD8+PD-1+) and M2 TAMs (CDF4/80+CD206+PD-1+), although no difference in expression was observed in TAMs (figure 4E). Taken together, these results suggest that the antitumor immune response can be rescued by TLX inhibition, which subsequently contributes to in vivo tumor growth arrest in immunocompetent mice mediated through TILs and TAMs.
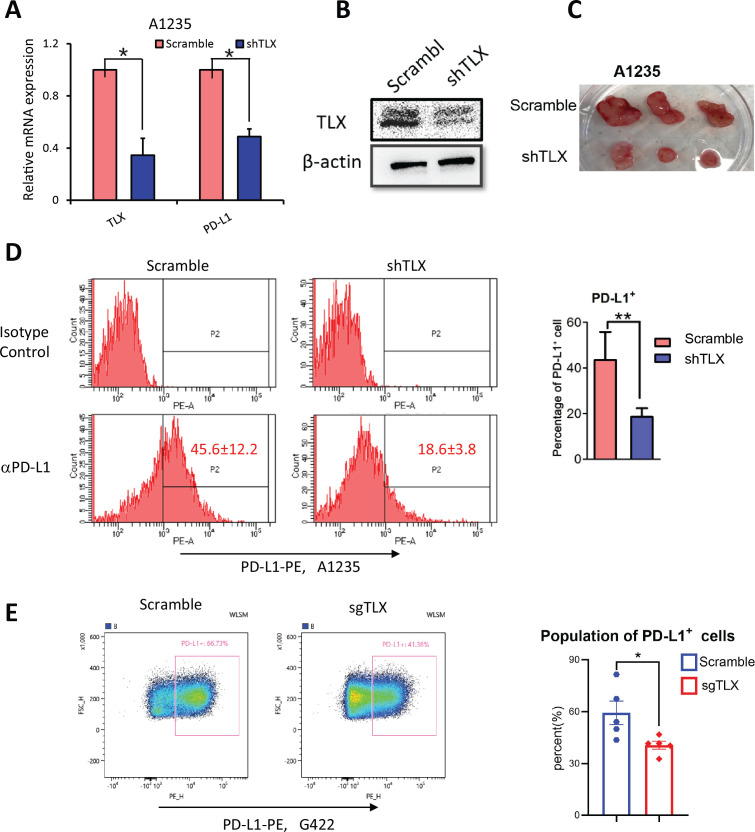
TLX regulates the expression of PD-L1 in glioma
To further illustrate the functional role of TLX in PD-L1 regulation in gliomas, we silenced TLX by shRNA in human glioma A1235 cells. TLX expression was confirmed by RT-qCR and western blotting (figure 5A, B). Mouse xenograft tumors formed by shTLX and scramble glioma A1235 cells showed that shTLX cells formed much smaller tumors in SCID mice (figure 5C). Additionally, the PD-L1+ population was markedly reduced in A1235-shTLX xenograft tumors compared with the scramble control (figure 5D). A similar reduction in PD-L1 expression was observed in mouse G422-sgTLX allografts (figure 5E, F) and GL261-shTLX allografts (online supplemental figure S4B), although no difference in PD-L1 expression was observed on M2 TAMs (CDF4/80+CD206+PD-L1+) on TLX knockout (figure 4E). To examine whether this pattern is cell-type specific, we also examined PD-L1 expression in prostate cancer cell line PC-3, for which we previously observed in vitro and in vivo promotion of tumor cell growth by TLX overexpression.23 When TLX was ectopically expressed in PC-3 cells, PD-L1 mRNA and protein were significantly upregulated (online supplemental figure S6A, B), and a significant increase in the PD-L1+ cell population was observed in PC-3-TLX compared with PC-3-pBABE tumors (online supplemental figure S6C, D). Hence, TLX can positively regulate the expression of PD-L1 in glioma, as well as in other cancer cells.
Figure 5.
TLX regulated the expression of PD-L1 in glioma. (A) mRNA level of PD-L1 in A1235-scramble and shTLX cell lines by RT-qPCR. Data are mean±SEM, *p<0.05. (B) Protein expression of TLX in A1235-scramble and shTLX by Western blot. (C) Photos of dissected tumors formed by subcutaneously injection of A1235-scramble and shTLX infectants grown in SCID mice (n=3). (D) Left: representative figures show FCM analysis of PD-L1 expression A1235-shTLX compared with scramble tumors. PD-L1 positive cells are gated according to isotype control (n=3). Right: statistic analysis of PD-L1+ population of A1235-scramble and A1235-shTLX tumors. Data are mean±SEM **p<0.01 vs scramble control. (E) FCM analysis of G422-sgTLX and G422-scramble tumors show PD-L1+ population was significantly decreased in G422-sgTLX tumors. Left: FCM gating PD-L1+ cells from tumor mass. Right: statistics analysis of PD-L1+population. *P<0.05 vs scramble control. SCID, Severe Combined Immunodeficiency; FCM, flow cytometric.
jitc-2020-001937supp009.pdf (365.1KB, pdf)
TLX binds to PD-L1 promoters, and transcriptionally activates PD-L1
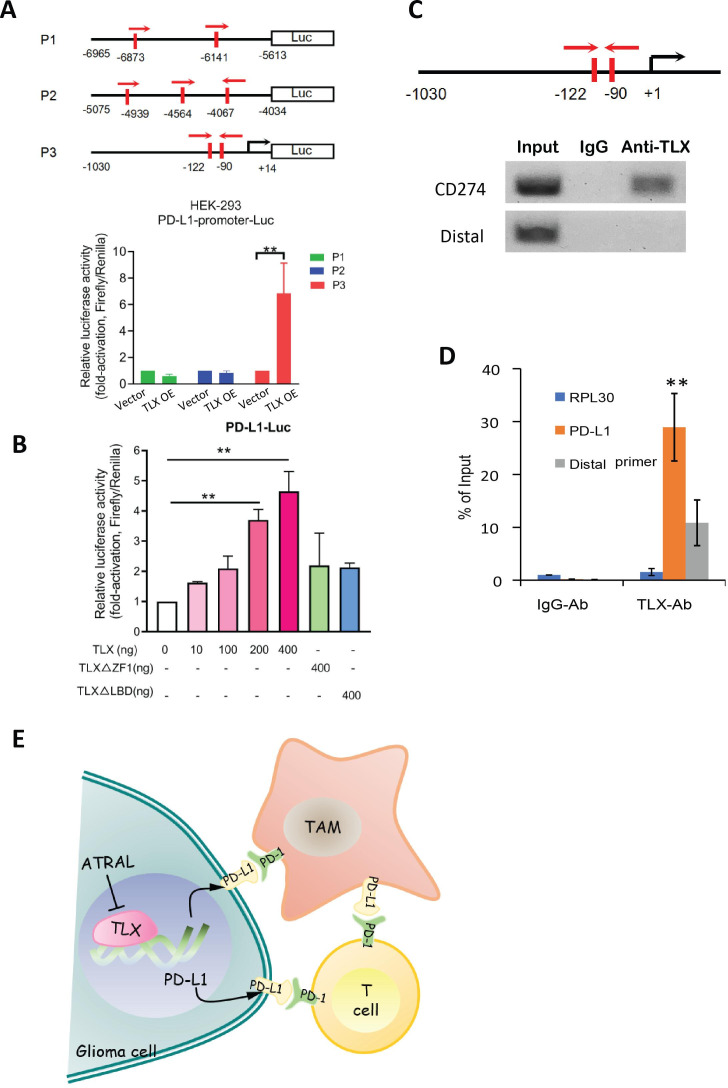
As TLX is an orphan nuclear transcriptional factor, we next investigated whether it regulates PD-L1 expression at the transcription level. In silico analyses yielded three TLX binding sites located at −6873 to −6141 bp (designated as P1), −4939 to −4067 bp (P2), and −122 to −90 bp (P3) upstream of the CD274 (PD-L1) start-site (figure 6A). To further determine whether TLX can transcriptionally activate the CD274 gene, we constructed three CD274 promoter-driven luciferase reporter plasmids. Results showed that the transfected TLX significantly boosted the activity of the P3-Luc reporter, but not the P1-Luc or P2-Luc reporters (figure 6A). Furthermore, only ectopic intact TLX, not its zine finger (ZF1) truncated mutants, increased the PD-L1-Luc reporter activity dose-dependently in transfected HEK293 cells, indicating that TLX-mediated activation of the CD274 promoter requires both an intact DNA-binding element and ligand-binding domain of TLX (figure 6B). These results suggest that TLX exerts its transcriptional activation on CD274. ChIP analysis further revealed that the proximal region that covers the human CD274 promoter with a TLX binding element (−122 to −90 bp) could be immuno-precipitated by TLX antibody in human glioma A1235 cells (figure 6C). The enrichment of CD274 promoter with TLX binding sites was confirmed by RT-qPCR (figure 6D). IgG and a distal primer of CD274 were set as negative experimental controls, wherein no accumulation of DNA was detected. Collectively, these results suggest that TLX could directly activate CD274 in glioma cells.
Figure 6.
TLX transcriptionally activates CD274 (PD-L1) gene. (A) Luciferase reporter assay of PD-L1 gene promoter transactivation in TLX-transfected HEK293 cells. Top: schematic diagram shows the three PD-L1 gene promoters driven constructs: P1-Luc, P2-Luc and P3-Luc. Bottom: only the P3-Luc reporter could be significantly transactivated by TLX. **P<0.01 vs no TLX transduction. (B) Luciferase reporter assay of PD-L1 gene promoter-driven reporters performed in HEK293 cells transfected with different dose of TLX or its truncated mutants. **P<0.01 vs blank vector transduction. (C) ChIP assay of CD274gene promoter preformed in A1235 cells. (D) qPCR analysis of precipitated CD274 promoter from ChIP assay. **P<0.01 vs IgG-Ab control. Data are presented as mean±SD of triplicate assays. (E) Schematic diagram depicts the hypothesized mechanism of TLX-mediated transcription activation of PD-L1 ligands that bind to PD-1 expressed on both TILs and TAMs, and its significance in rescue of immune response through TLX inhibition. ChIP, chromatin immunoprecipitation; TAMs, tumor-associated macrophages.
Discussion
In the present study, we demonstrated that the orphan nuclear receptor TLX, which displayed significantly upregulated expression in high-grade gliomas, and was positively associated with PD-L1 expression, could contribute to immune suppression and progression of glioma, as its inhibition caused tumor growth arrest, as well as an increase in cytotoxic immune cells and a decrease in glioma TAM infiltration. Moreover, our results suggest that TLX transcriptionally regulates PD-L1 expression, which forms an axis with TILs and TAMs expressing PD-1, contributing to the immune suppressive microenvironment of gliomas.
PD-L1 expression on tumor cells and in the tumor microenvironment (TME) has predictive value for the response to PD-L1/PD-1 blockade therapies in different tumor types. However, clinical practice shows that many PD-L1-positive tumors do not respond to PD-L1/PD-1 blockade, with only a fraction of PD-L1-negative tumors exhibiting appropriate responses. It is important to understand how the levels of PD-L1 expression at the cell surface are regulated, as well as how this regulation may vary between tumor and immune cells. PD-L1 levels in different tumors are regulated in a highly complex manner, which can be influenced by inflammatory signaling, oncogenic signaling, genetic aberrations, mRNA regulation, and protein stability.34 In glioma cells, enhanced expression of PD-L1 is reportedly induced by PTEN loss13 and CDK5 activation.14 Meanwhile, identification of novel regulatory mechanisms of the PD-L1 checkpoint remains a highly attractive research area that may offer novel opportunities for immune therapeutic interventions. Our study provides the first evidence suggesting that the druggable orphan nuclear receptor, TLX, the expression of which is remarkably enhanced in gliomas, could transcriptionally activate the expression of PD-L1 in gliomas, representing a novel regulatory mechanism of PD-L1 in gliomas. TLX regulates target genes by recruiting coregulators, such as epigenetic modifiers histone demethylase LSD1,24 35 histone deacetylases,16 24 and various newly identified coactivators. However, the involvement of potential coregulators in the regulation of TLX on PD-L1 requires further investigation. With the identification of more specific reverse agonists and antagonists of TLX, combined targeting of TLX and the PD-L1/PD-1 axis in the glioma immune microenvironment would have potential benefits for immunotherapy.
Like other solid tumors, high-grade gliomas are highly transcriptional and genetically heterogeneous. Single-cell RNA-sequencing combined with genetic, and expressional, analyses of TCGA, as well as single-cell lineage tracing, showed that malignant cells in GBMs exist in four main cellular states, which recapitulate distinct neural cell types that drive the heterogeneity of GBM malignant cells, while the frequency of cells in each state can be influenced by genetics and the TME.36 A highly immunosuppressive TME influences brain tumor outgrowth and cell plasticity. Conversely, TME complexity is also predominantly shaped by the central nervous system micromilieu or by the malignancy itself, such as IDH mutation status.37 In the present study, we also observed that TLX and PD-L1 expression correlated with IDH mutation and 1p19q deletion status. That is, low TLX and PD-L1 expression correlated with lower WHO grades, mutated IDH, and 1p19q deletion, whereas high TLX and PD-L1 expression was associated with unfavorable prognosis, higher WHO grades, wild-type IDH, and 1p19q status as per TCGA and CGGA databases. However, whether IDH mutation or 1p19q status influences the effect that TLX-induced transcriptional regulation of PD-L1 has on the TME requires further investigation.
The correlation of PD-L1 expression and immune cell infiltration in the TME has been investigated in different types of cancers, including gliomas. Indeed, PD-L1 expression and TILs are detectable in most GBMs.10 38 Moreover, apart from tumor cells, upregulation of PD-L1 expression has also been detected in glioma TAMs,39 40 myeloid-derived suppressor cells,41 and T cells.42 Upregulation of PD-L1 has a major inhibitory role in modulating the infiltration of immune cells, including TAMs, cytotoxic lymphocytes, TILs, and Tregs, and mediates tumor immune escape and ICB resistance. Herein, we found that PD-L1 was present on both glioma cells and TAMs and was positively associated with infiltration of CD163+ TAMs in high-grade gliomas. However, TLX-expressing cells and TAMs were uniquely distributed, indicating that TLX-mediated upregulation of PD-L1 occurs on glioma cells, but not on TAMs, suggesting different mechanisms responsible for driving PD-L1 upregulation in tumor and immune cells. TAMs are an attractive potential target in glioma immunotherapy, as immunosuppressive TAMs are predominantly infiltrated in gliomas and are regarded as a barrier to emerging immunotherapies.43 Our observation of significant PD-1 signaling in M2 macrophages is consistent with recent reports revealing that PD-1 ligation in macrophages subdues their phagocytic capacity,44 and promotes the differentiation of tolerogenic macrophages, which in turn suppresses T-cell activation and promotes tumor growth.42 However, the mechanism underlying PD-L1 upregulation in glioma TAMs, as well as the significance of PD-1 ligation in TAMs via PD-L1 upregulation in glioma cells requires further investigation.
It has been well established that the orphan nuclear receptor TLX plays essential roles in brain tumor initiation by regulating the self-renewal and proliferation of NSCs, as demonstrated in transgenic murine models.45–48 Our previous studies also demonstrated that TLX is important for prostate tumorigenesis23 and malignant progression,24 as well as prostate cancer stem cell proliferation.33 In the current study, in vivo tumor growth arrest was significantly induced by TLX inhibition (either by gene knockout, knockdown, or TLX reverse agonist) in both immunocompetent and immunodeficient SCID mice, which was accompanied by the rescue of antitumor immune responses, as evidenced by an increase in cytotoxic lymphocyte and decrease M2 TAM infiltration, suggesting that TLX inhibition-induced tumor growth arrest was also mediated through the immune microenvironment, especially by TAMs. Recent studies showed that GBM stem cells facilitate tumor immune evasion by secreting proteins or exosomes.49 50 Given the important role of TLX in glioma stem cell self-renewal and tumorigenesis, whether TLX-mediated immune suppression acts by regulating glioma stem cells requires further investigation.
In summary, this study provides evidence indicating that the druggable orphan nuclear receptor TLX promotes malignant growth of gliomas by suppressing the immune microenvironment via transcriptional activation of PD-L1, which binds to PD-1 expressed on both TILs and TAMs, indicating that targeting TLX signaling could be a potential therapeutic strategy for GBM treatment (figure 6E).
jitc-2020-001937supp010.pdf (717.7KB, pdf)
Footnotes
JZ, XP and YY contributed equally.
Contributors: DW, JM, and XL conceived and designed the research; JZ, YY, and ZW developed the methodology; XP and JT examined and scored clinical samples; JZ, YY, ZW, WG, ZL, RY, and WH acquired the data; JZ, XZ, and CG performed statistic analysis; FLC, JL, and XY analysed and interpreted the data; DW and XL supervised the study. All authors approved the submission of the manuscript.
Funding: This work was supported by The Science and Technology Project of Shenzhen (JCYJ20180508163203807, JCYJ20170307144115825), National natural Science Foundation of China (project number 81872283, 81572486 and 81974362), Shenzhen Key Laboratory of Viral Oncology (ZDSYS201707311140430), Sanming Project of Medicine in Shenzhen (SZSM201612023), National Key R&D Project of China (2018YFC0115301).
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available in a public, open access repository. Data are available upon reasonable request. All data relevant to the study are included in the article or uploaded as supplemental information. Yes.
References
- 1.Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. JAMA 2013;310:1842–50. 10.1001/jama.2013.280319 [DOI] [PubMed] [Google Scholar]
- 2.Tykocki T, Eltayeb M. Ten-year survival in glioblastoma. A systematic review. J Clin Neurosci 2018;54:7–13. 10.1016/j.jocn.2018.05.002 [DOI] [PubMed] [Google Scholar]
- 3.Lim M, Xia Y, Bettegowda C, et al. Current state of immunotherapy for glioblastoma. Nat Rev Clin Oncol 2018;15:422–42. 10.1038/s41571-018-0003-5 [DOI] [PubMed] [Google Scholar]
- 4.Filley AC, Henriquez M, Dey M. Recurrent glioma clinical trial, CheckMate-143: the game is not over yet. Oncotarget 2017;8:91779–94. 10.18632/oncotarget.21586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X, Guo G, Guan H, et al. Challenges and potential of PD-1/PD-L1 checkpoint blockade immunotherapy for glioblastoma. J Exp Clin Cancer Res 2019;38:87. 10.1186/s13046-019-1085-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Omuro A, Vlahovic G, Lim M, et al. Nivolumab with or without ipilimumab in patients with recurrent glioblastoma: results from exploratory phase I cohorts of CheckMate 143. Neuro Oncol 2018;20:674–86. 10.1093/neuonc/nox208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cloughesy TF, Mochizuki AY, Orpilla JR, et al. Neoadjuvant anti-PD-1 immunotherapy promotes a survival benefit with intratumoral and systemic immune responses in recurrent glioblastoma. Nat Med 2019;25:477–86. 10.1038/s41591-018-0337-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schalper KA, Rodriguez-Ruiz ME, Diez-Valle R, et al. Neoadjuvant nivolumab modifies the tumor immune microenvironment in resectable glioblastoma. Nat Med 2019;25:470–6. 10.1038/s41591-018-0339-5 [DOI] [PubMed] [Google Scholar]
- 9.Chen RQ, Liu F, Qiu XY, et al. The prognostic and therapeutic value of PD-L1 in glioma. Front Pharmacol 2018;9:1503. 10.3389/fphar.2018.01503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berghoff AS, Kiesel B, Widhalm G, et al. Programmed death ligand 1 expression and tumor-infiltrating lymphocytes in glioblastoma. Neuro Oncol 2015;17:1064–75. 10.1093/neuonc/nou307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antonios JP, Soto H, Everson RG, et al. PD-1 blockade enhances the vaccination-induced immune response in glioma. JCI Insight 2016;1. 10.1172/jci.insight.87059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DiDomenico J, Lamano JB, Oyon D, et al. The immune checkpoint protein PD-L1 induces and maintains regulatory T cells in glioblastoma. Oncoimmunology 2018;7:e1448329. 10.1080/2162402X.2018.1448329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parsa AT, Waldron JS, Panner A, et al. Loss of tumor suppressor PTEN function increases B7-H1 expression and immunoresistance in glioma. Nat Med 2007;13:84–8. 10.1038/nm1517 [DOI] [PubMed] [Google Scholar]
- 14.Dorand RD, Nthale J, Myers JT, et al. Cdk5 disruption attenuates tumor PD-L1 expression and promotes antitumor immunity. Science 2016;353:399–403. 10.1126/science.aae0477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi Y, Chichung Lie D, Taupin P, et al. Expression and function of orphan nuclear receptor TLX in adult neural stem cells. Nature 2004;427:78–83. 10.1038/nature02211 [DOI] [PubMed] [Google Scholar]
- 16.Sun G, Yu RT, Evans RM, et al. Orphan nuclear receptor TLX recruits histone deacetylases to repress transcription and regulate neural stem cell proliferation. Proc Natl Acad Sci U S A 2007;104:15282–7. 10.1073/pnas.0704089104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang C-L, Zou Y, He W, et al. A role for adult TLX-positive neural stem cells in learning and behaviour. Nature 2008;451:1004–7. 10.1038/nature06562 [DOI] [PubMed] [Google Scholar]
- 18.Sun G, Ye P, Murai K, et al. miR-137 forms a regulatory loop with nuclear receptor TLX and LSD1 in neural stem cells. Nat Commun 2011;2:529. 10.1038/ncomms1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu Y, Luo M, Ni N, et al. Reciprocal actions of microRNA-9 and TLX in the proliferation and differentiation of retinal progenitor cells. Stem Cells Dev 2014;23:2771–81. 10.1089/scd.2014.0021 [DOI] [PubMed] [Google Scholar]
- 20.Qu Q, Sun G, Li W, et al. Orphan nuclear receptor TLX activates Wnt/beta-catenin signalling to stimulate neural stem cell proliferation and self-renewal. Nat Cell Biol 2010;12:31–40. sup pp 1-9. 10.1038/ncb2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu H-K, Wang Y, Belz T, et al. The nuclear receptor tailless induces long-term neural stem cell expansion and brain tumor initiation. Genes Dev 2010;24:683–95. 10.1101/gad.560310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park H-J, Kim J-K, Jeon H-M, et al. The neural stem cell fate determinant TLX promotes tumorigenesis and genesis of cells resembling glioma stem cells. Mol Cells 2010;30:403–8. 10.1007/s10059-010-0122-z [DOI] [PubMed] [Google Scholar]
- 23.Wu D, Yu S, Jia L, et al. Orphan nuclear receptor TLX functions as a potent suppressor of oncogene-induced senescence in prostate cancer via its transcriptional co-regulation of the CDKN1A (p21(WAF1) (/) (CIP1)) and SIRT1 genes. J Pathol 2015;236:103–15. 10.1002/path.4505 [DOI] [PubMed] [Google Scholar]
- 24.Jia L, Wu D, Wang Y, et al. Orphan nuclear receptor TLX contributes to androgen insensitivity in castration-resistant prostate cancer via its repression of androgen receptor transcription. Oncogene 2018;37:3340–55. 10.1038/s41388-018-0198-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Griffett K, Bedia-Diaz G, Hegazy L, et al. The orphan nuclear receptor TLX is a receptor for synthetic and natural retinoids. Cell Chem Biol 2020;27:1272–84. 10.1016/j.chembiol.2020.07.013 [DOI] [PubMed] [Google Scholar]
- 26.Ali HR, Chlon L, Pharoah PDP, et al. Patterns of immune infiltration in breast cancer and their clinical implications: a gene-expression-based retrospective study. PLoS Med 2016;13:e1002194. 10.1371/journal.pmed.1002194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453–7. 10.1038/nmeth.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He Y, Jiang Z, Chen C, et al. Classification of triple-negative breast cancers based on Immunogenomic profiling. J Exp Clin Cancer Res 2018;37:327. 10.1186/s13046-018-1002-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoshihara K, Shahmoradgoli M, Martínez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 2013;4:2612. 10.1038/ncomms3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98–102. 10.1093/nar/gkx247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Z, Ge R, Zhou J, et al. Nuclear factor IX promotes glioblastoma development through transcriptional activation of ezrin. Oncogenesis 2020;9:39. 10.1038/s41389-020-0223-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu D, Wang Z, Lin M, et al. In Vitro and In Vivo Antitumor Activity of Cucurbitacin C, a Novel Natural Product From Cucumber. Front Pharmacol 2019;10:1287. 10.3389/fphar.2019.01287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z, Wu D, Ng C-F, et al. Nuclear receptor profiling in prostatospheroids and castration-resistant prostate cancer. Endocr Relat Cancer 2018;25:35–50. 10.1530/ERC-17-0280 [DOI] [PubMed] [Google Scholar]
- 34.Sun C, Mezzadra R, Schumacher TN. Regulation and function of the PD-L1 checkpoint. Immunity 2018;48:434–52. 10.1016/j.immuni.2018.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yokoyama A, Takezawa S, Schüle R, et al. Transrepressive function of TLX requires the histone demethylase LSD1. Mol Cell Biol 2008;28:3995–4003. 10.1128/MCB.02030-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neftel C, Laffy J, Filbin MG, et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell 2019;178:835–49. e21. 10.1016/j.cell.2019.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klemm F, Maas RR, Bowman RL, et al. Interrogation of the microenvironmental landscape in brain tumors reveals disease-specific alterations of immune cells. Cell 2020;181:1643–60. e17. 10.1016/j.cell.2020.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davidson TB, Lee A, Hsu M, et al. Expression of PD-1 by T cells in malignant glioma patients reflects exhaustion and activation. Clin Cancer Res 2019;25:1913–22. 10.1158/1078-0432.CCR-18-1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lamano JB, Lamano JB, Li YD, et al. Glioblastoma-Derived IL6 induces immunosuppressive peripheral myeloid cell PD-L1 and promotes tumor growth. Clin Cancer Res 2019;25:3643–57. 10.1158/1078-0432.CCR-18-2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aslan K, Turco V, Blobner J, et al. Heterogeneity of response to immune checkpoint blockade in hypermutated experimental gliomas. Nat Commun 2020;11:931. 10.1038/s41467-020-14642-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dubinski D, Wölfer J, Hasselblatt M, et al. CD4+ T effector memory cell dysfunction is associated with the accumulation of granulocytic myeloid-derived suppressor cells in glioblastoma patients. Neuro Oncol 2016;18:807–18. 10.1093/neuonc/nov280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Diskin B, Adam S, Cassini MF, et al. PD-L1 engagement on T cells promotes self-tolerance and suppression of neighboring macrophages and effector T cells in cancer. Nat Immunol 2020;21:442–54. 10.1038/s41590-020-0620-x [DOI] [PubMed] [Google Scholar]
- 43.Müller S, Kohanbash G, Liu SJ, et al. Single-cell profiling of human gliomas reveals macrophage ontogeny as a basis for regional differences in macrophage activation in the tumor microenvironment. Genome Biol 2017;18:234. 10.1186/s13059-017-1362-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gordon SR, Maute RL, Dulken BW, et al. PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature 2017;545:495–9. 10.1038/nature22396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zou Y, Niu W, Qin S, et al. The nuclear receptor TLX is required for gliomagenesis within the adult neurogenic niche. Mol Cell Biol 2012;32:4811–20. 10.1128/MCB.01122-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johansson E, Zhai Q, Zeng Z-J, et al. Nuclear receptor TLX inhibits TGF-β signaling in glioblastoma. Exp Cell Res 2016;343:118–25. 10.1016/j.yexcr.2016.03.028 [DOI] [PubMed] [Google Scholar]
- 47.Zhu Z, Khan MA, Weiler M, et al. Targeting self-renewal in high-grade brain tumors leads to loss of brain tumor stem cells and prolonged survival. Cell Stem Cell 2014;15:185–98. 10.1016/j.stem.2014.04.007 [DOI] [PubMed] [Google Scholar]
- 48.Cui Q, Yang S, Ye P, et al. Downregulation of TLX induces TET3 expression and inhibits glioblastoma stem cell self-renewal and tumorigenesis. Nat Commun 2016;7:10637. 10.1038/ncomms10637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Otvos B, Silver DJ, Mulkearns-Hubert EE, et al. Cancer stem cell-secreted macrophage migration inhibitory factor stimulates myeloid derived suppressor cell function and facilitates glioblastoma immune evasion. Stem Cells 2016;34:2026–39. 10.1002/stem.2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gabrusiewicz K, Li X, Wei J, et al. Glioblastoma stem cell-derived exosomes induce M2 macrophages and PD-L1 expression on human monocytes. Oncoimmunology 2018;7:e1412909. 10.1080/2162402X.2017.1412909 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2020-001937supp001.pdf (95.4KB, pdf)
jitc-2020-001937supp002.pdf (100.6KB, pdf)
jitc-2020-001937supp003.pdf (591KB, pdf)
jitc-2020-001937supp004.pdf (764.1KB, pdf)
jitc-2020-001937supp005.pdf (289.2KB, pdf)
jitc-2020-001937supp006.pdf (106.2KB, pdf)
jitc-2020-001937supp007.pdf (361.8KB, pdf)
jitc-2020-001937supp008.pdf (469.2KB, pdf)
jitc-2020-001937supp009.pdf (365.1KB, pdf)
jitc-2020-001937supp010.pdf (717.7KB, pdf)
Data Availability Statement
Data are available in a public, open access repository. Data are available upon reasonable request. All data relevant to the study are included in the article or uploaded as supplemental information. Yes.