Abstract
The kidney is a frequent target of autoimmune injury, including in systemic lupus erythematosus; however, how immune cells adapt to kidney’s unique environment and contribute to tissue damage is unknown. We found that renal tissue, which normally has low oxygen tension, becomes more hypoxic in lupus nephritis. In the injured mouse tissue, renal-infiltrating CD4+ and CD8+ T cells express hypoxia-inducible factor-1 (HIF-1), which alters their cellular metabolism and prevents their apoptosis in hypoxia. HIF-1-dependent gene-regulated pathways were also upregulated renal-infiltrating T cells in human lupus nephritis. Perturbation of these environmental adaptations by selective HIF-1 blockade inhibited infiltrating T cells and reversed tissue hypoxia and injury in murine models of lupus. The results suggest that targeting HIF-1 might be effective for treating renal injury in autoimmune diseases.
One Sentence Summary
HIF-1 dictates effector function of infiltrating T cells in lupus, causing tissue damage that can be abrogated by its blockade in murine models.
Introduction
The kidney is a primary target of damage in systemic lupus erythematosus (SLE, lupus), with deposition of pathogenic autoantibodies and subsequent T cell mediated tissue injury (1). Yet, little is known about how the kidney environment contributes to the adaptation of infiltrating T cells with resultant tissue damage. Changes in the local environment likely shape the phenotype and function of renal-infiltrating T cells in lupus and other autoimmune illnesses. The kidney becomes hypoxic as a common denominator following a variety of insults, with an increase in expression of transcription factor hypoxia inducible factor-1 (HIF-1) in glomerular and tubulointerstitial areas, where its incremental expression correlates with severity of injury (2). This hypoxic microenvironment could modulate effector capability of infiltrating T cells, but its effect on their phenotype with consequent contribution to tissue damage is unclear.
Aggravation of hypoxia in the renal environment may not only be the result but also the cause of tissue damage. HIF-1 enhances effector T cell function, including promotion of cytolytic activity and inflammatory cytokine production (3), while blocking terminal differentiation. These physiological stress-adaptive responses are critical for proper effector cell function in areas of pathogen invasion and in and around tumors (4). The kidney is unique in that it is physiologically more hypoxic than other organs, with oxygen gradients of ~50mm Hg (6-7%, as compared to 21% in air) in the cortex and < 10mm Hg (<1%) beneath the corticomedullary junction (5). We investigated whether the hypoxic renal environment dictates T cell function, leading to subsequent tissue damage, and if targeting these environmentally induced changes could perturb adaptive function of infiltrating cells and reverse tissue damage in lupus.
Results
Renal-infiltrating T cells are functionally active and pathogenic
We isolated from nephritic kidneys of 16-18-week-old lupus-prone MRL/MpJ-Faslpr/J (MRL/lpr) mice renal-infiltrating T cells, separating them from circulating non-resident cells that potentially lodge in the kidney at time of isolation (Fig. S1) (6), and determined their effector capacity. Consistent with published phenotypic analyses of biopsies of human lupus nephritis (7, 8), isolated CD4+ T cells expressed interferon- γ (IFN-γ) and CD8+ T cells IFN-γ and more granzyme B compared to their counterparts simultaneously isolated from spleens of mice from which kidneys were taken (Fig. S2A, B). Renal T cell populations also had increased T-bet expression, compared to those from spleens (Fig. S2C). These phenotypes were replicated in a cohort of 5-month-old lupus-prone B6.Sle1.Yaa male mice (Fig. S2D–F) (lupus in this strain is penetrant in male mice bearing a second Tlr7 locus on the Y chromosome (9)). Renal-infiltrating T cells expressed programmed cell death-1 (PD-1) (Fig. S2G), with PD-1hi CD4+ and CD8+ T cells relatively hypofunctional as determined by IFN-γ and granzyme B synthesis, respectively, as previously described (10); yet, the majority of renal-infiltrating T cells were PD-1lo (Fig. S2G–J). These data suggested that renal-infiltrating T cells are transcriptionally and functionally active, and distinct from their splenic counterparts.
To confirm pathogenicity of renal-infiltrating T cells, T-cell depleting antibody anti-Thy1.2 was injected every 3 days for 4 weeks into male MRL/lpr mice beginning at age 14 weeks, after onset of renal cell infiltration. T cell depletion effectively reduced peripheral blood T cells by >90%, yet had no effect on serum autoantibody titers and glomerular immune complex deposition (Fig. S3A–D), likely secondary to the presence of long-lived autoantibody-secreting plasma cells in lupus mice with established disease (11). In contrast, T-cell depletion reduced renal inflammation and prevented features of chronic tissue damage, including glomerulosclerosis, tubular atrophy, and interstitial fibrosis (Fig. S3E–H). These data suggested that renal-infiltrating T cells are functionally active, and contribute to tissue damage in lupus nephritis independently of autoantibody production driven by their splenic counterparts.
Adaptation to tissue hypoxia is associated with HIF-1 expression
We next sorted activated (CD44hi) CD4+ and CD8+ T cells from nephritic kidneys of 14-week-old MRL/lpr mice, again separating them from circulating cells, and subjected them to RNA-sequencing. A total of 128 shared genes were differentially upregulated (fold change > 2; p-value < 0.05) in both renal-infiltrating CD4+ and CD8+ T cells compared to splenic cells (Supplementary File 1). To determine the transcription factors (TFs) that promoted expression of these transcripts, we cross-referenced them to a database analyzing perturbations of TFs from Enrichr (12). The enriched transcripts in our dataset were preferentially downregulated upon Hif1a or Nfe2l2 (the gene expressing nuclear factor erythroid 2-related factor 2, NRF2) knockout (p = 7.8 x 10-10) and upregulated by Myc haploinsufficiency (p = 7.8 x 10-8) ((13–14). The transcriptional regulation of Hif1a (HIF-1α) was determined by referencing TRRUST database (p = 7.9 x 10-3) (15). As the transcriptional activity of Myc is suppressed upon HIF-1 activation (16, 17) and the redox balance maintained by NRF2 is important for cellular survival in hypoxia (18), we reasoned that the enriched gene sets were a consequence of cellular adaptation to hypoxia. As confirmation, our transcriptome data revealed hypoxia- and HIF-regulated gene signatures in renal-infiltrating CD4+ and CD8+ T cells (Fig. 1A,B), compared to those from the spleen. The kidney CD4+ T cells were enriched in transcripts upregulated in a gene set from procollagen-proline deoxygenase (prolyl hydroxylase, PHD)-knockout CD4+ T cells (p < 0.001) (4), whereas renal CD8+ T cells expressed genes found upon upregulated HIF-1 activity arising in virally-infected mice lacking the Von-Hippel Lindau (VHL) tumor suppressor gene (p < 0.001) (3). Similar hypoxic signatures were found in the transcriptome of renal-infiltrating CD8+ T cells (p < 0.001) taken from 23-week-old MRL/lpr mice, with renal-infiltrating CD4+ T cells showing a similar trend (p = 0.117) (Fig. S4A,B) (10). These analyses indicated that renal-infiltrating CD4+ and CD8+ T cells possess gene signatures consistent with HIF-1 mediated adaptation to hypoxia, which is known to T cell terminal differentiation and sustains T cell functionality (3).
Fig. 1. Renal-infiltrating T cells are located in areas of hypoxia.
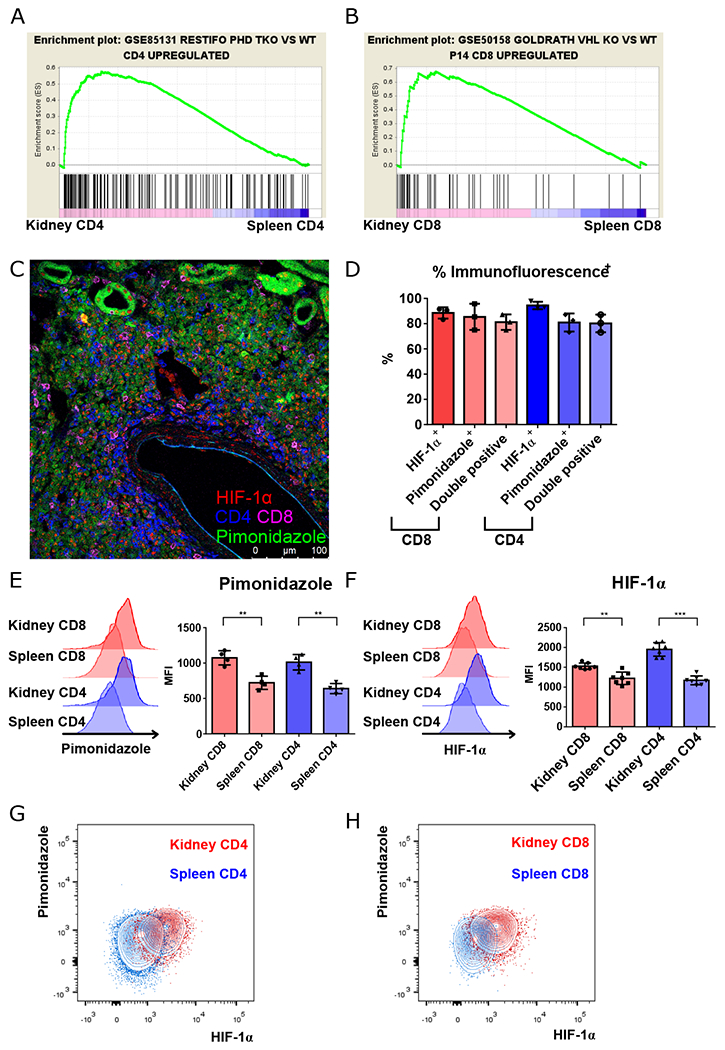
A, B. GSEA plots comparing gene signatures of renal-infiltrating CD4+ and CD8+ T cells, to splenic to CD4+ and CD8+ T cells, respectively, based upon the hypoxia signatures generated by comparing triple PHD (prolyl-4-hydroxylase domain proteins)-knockout CD4+ to wild type CD4+ T cells, and comparing VHL (Von-Hippel Lindau) tumor suppressor knockout CD8+ T cells to lymphocytic choriomenigitis virus specific P14 TCR transgenic CD8+ T cells taken from virally-infected mice. C. Representative confocal microscopy of lymphocytic aggregates of CD4+ (blue) and CD8+ T cells (magenta) with HIF-1α nuclear staining (red) located in regions of hypoxia (pimonidazole, green). D. Summary of nuclear HIF-1α and pimonidazole staining, and combined staining of CD4+ and CD8+ T cells within renal lymphocytic aggregates. E, F. Representative data and summary of pimonidazole and HIF-1α staining of activated (CD44hi) splenic vs. renal CD4+ and CD8+ T cells isolated from kidneys of 16-18-week-old MRL/lpr mice (n = 4 and 8, respectively). G, H. Representative data of pimonidazole and HIF-1α staining in kidney versus spleen CD4+ (G) and CD8+ T cells (H) as in (E) and (F). Representative of 3 experiments, n = 4 to 8 animals per group. Data shown are mean ± s.d.; statistical analysis by two-tailed paired t-test (E, F). **p < 0.01, ***p < 0.001.
To determine if this transcriptional profile resulted from changes in oxygen tension, pimonidazole (Hypoxyprobe), which binds to proteins and peptides containing thiol groups at oxygen tensions below 10 mm Hg (19), was injected into mice before sacrifice. Regions of hypoxia and renal tubular cells were then visualized by anti-pimonidazole staining (Fig. S5). By comparison to age-matched C57BL/6 (B6) control mice, in which hypoxia is limited to the renal medulla (Fig. S6A), anti-pimonidazole staining revealed extensive hypoxia involving the majority of the renal cortex of 6-month-old B6.Sle1.Yaa and 5-month-old Fas-intact MRL (MRL+/+) and MRL/lpr mice (Fig. S6B–F). Analysis of lymphocytic aggregates revealed that renal-infiltrating CD4+ and CD8+ T cells were primarily located in regions of hypoxia, with positive pimonidazole staining and nuclear staining of HIF-1α (Fig. 1D). These results were confirmed by flow cytometry, comparing T cells taken simultaneously from kidneys and spleens of MRL/lpr mice (Fig. 1E–H). Hypoxia and HIF-1α upregulation were seen in other previously described pathogenic populations, including double-negative T cells (20) and T follicular helper (Tfh)-like cells (Fig. S7A–C) (21). We also examined the expression of HIF-2α in renal T cells, demonstrating that HIF-1α was the dominant form of HIF proteins that accumulated (Fig. S8A), with the relative abundance of HIF-1α likely resulting from the differential effect of IFN-γ in regulating its transcript versus that of HIF-2α (22). Predominance of HIF-1α upregulation in renal-infiltrating T cells was also seen in 6-month-old lupus-prone B6.Sle1.Yaa mice (Fig. S8B,C), suggesting these changes are common to murine lupus. In summary, T cell adaptation to hypoxia was noted in diseased murine lupus kidneys with upregulation of HIF-1 expression.
To confirm the causal relationship between T cell infiltration and tissue hypoxia, we analyzed pimonidazole staining in male MRL/lpr mice treated with anti-mouse Thy1.2, with resultant T-cell depletion of >90%. This treatment not only prevented extensive tissue damage and fibrosis, as defined by NIH activity and chronicity indices (Fig. S3E–H), but also partially reversed hypoxia in the renal cortex (Fig. S9A–C). These findings are consistent with our previous finding that renal-infiltrating T cells are active, and that their heightened effector function contributes to tissue damage and hypoxia.
T cell survival adaptation is required in the inflamed hypoxic kidney
We next asked if renal-infiltrating T cells required modification of survival and apoptotic signals to maintain viability during hypoxia. Although Bcl-2 is overexpressed in tubulointerstitial infiltrates in NZB/W F1 lupus-prone mice and in human lupus nephritis biopsies (23), we did not find upregulation of its transcript in renal-infiltrating T cells of diseased MRL/lpr mice.
To explore expression of other pro-survival factors, we determined that of another Bcl-2 family member, BNIP3, transcriptionally controlled by HIF-1 (24). Hypoxia leads to the BH3-only Bcl2 family member-(BNIP3-) mediated opening of the mitochondrial permeability transition pore, loss of membrane potential, and increased production of reactive oxygen species (ROS), ultimately giving way to hypoxic cell death (25). Yet, certain cells, for example cancer, robustly express pyruvate dehydrogenase kinase isoform 2 (PDK2) facilitating alternative splicing of exon 3 of Bnip3 (Bnip3Δex3), the product of which stabilizes mitochondrial membrane potential and switches apoptosis to cellular survival by contrast to the full-length protein (26). The upregulation of PDK2 may be a cellular response to hypoxia, given its transcriptional control by HIF-1 in hematopoietic stem cells (27). To address the involvement of the HIF-1/PDK2/BNIP3 axis in maintaining cellular survival in hypoxia, we measured protein expression in renal-infiltrating CD4+ and CD8+ T cells, revealing that, compared to their splenic counterparts, those in the kidney expressed more PDK2 (Fig. 2A, S10A). We also found upregulation of total BNIP3 in renal-infiltrating T cells, as determined by antibody staining (Fig. 2B, S10B), with recognition of an epitope encoded by exon 2 common to both splice variants. However, quantitative PCR showed proportionate upregulation of the alternatively spliced Bnip3Δex3 transcript in addition to that of full length Bnip3FL (Fig. 2C,D), suggesting that the alternative splicing of Bnip3 by PDK2 was involved in maintaining cellular survival in the hypoxic environment.
Fig. 2. Renal T cells survive in hypoxia through Bnip3 alternative splicing mediated by HIF-1 dependent PDK2.
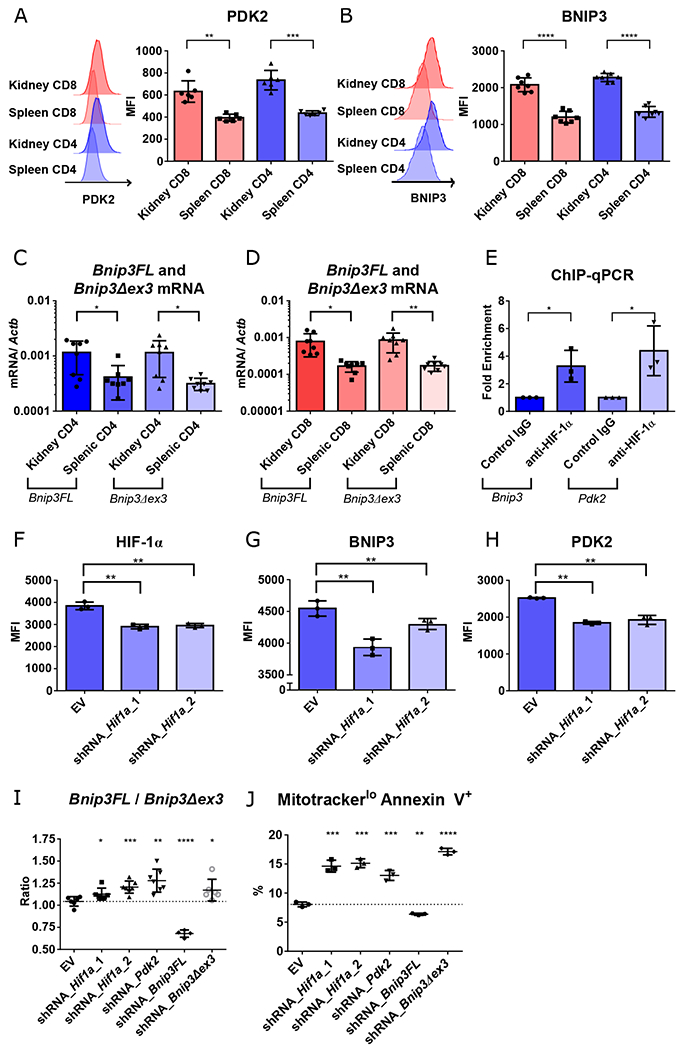
A-B. Representative data and summary of PDK2 (A) and BNIP3 (B) expression in activated (CD44hi) splenic vs. renal CD4+ and CD8+ T cells isolated from kidneys of 16-18-week-old MRL/lpr mice (n = 6, 7, respectively). C-D. Quantification of two forms of Bnip3 transcripts, full length (Bnip3FL) and exon 3 deleted (Bnip3Δex3) in CD4+ (C) and CD8+ T cells (D) (n = 8). E. Binding of HIF-1α to Bnip3 and Pdk2 promoter regions, as determined by chromatin immunoprecipitation (ChIP) and quantitative real-time PCR (qPCR). F-H. Mean fluorescence intensity of HIF-1α (F), BNIP3 (G), and PDK2 (H) in Th1-activated CD4+ T cells transduced with empty vector (EV), or two different constructs of Hif1α knockdowns after 2 days in hypoxic cultures. I. Ratio of Bnip3FL and Bnip3Δex3 mRNAs in Th1-activated CD4+ T cells transduced with either EV or knockdown constructs targeting Hif1a, Pdk2, Bnip3FL, or Bnip3Δex3 one day after adding DMOG. J. Percentage of mitotracker deep redlo annexin V+ cells of Th1-activated CD4+ T cells transduced with knockdown vectors after 3 days of hypoxic culture. Data shown are mean ± s.d.; statistical analysis by two-tailed paired t-test (A-E) and unpaired t-test (F-J). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
To further dissect the T cell response in hypoxia, we demonstrated that splenic CD4+ T cells from pre-diseased (6-8-week-old) male MRL/lpr and B6.Sle1.Yaa lupus-prone and 6-8-week-old control B6 mice had upregulation of HIF-1α and PDK2 in hypoxic (1%) conditions (Fig. S11A,B), and along with splenic CD8+ T cells, during pseudohypoxia in which the HIF protein was stabilized via addition of the PHD inhibitor dimethyloxalylglycine (DMOG) (Fig. S11C–F). These data suggested that upregulation of PDK2 in response to HIF-1 expression is a common T cell response to hypoxia.
To confirm the transcriptional control of PDK2 and BNIP3 by HIF-1, chromatin immunoprecipitation (ChIP) and qPCR was performed using CD4+ T cells from 6-8-week-old B6 mice cultured during DMOG-induced pseudohypoxia. We observed enrichment of HIF-1α binding to the promoter regions of Pdk2 and Bnip3 (Fig. 2E), with loss of protein expression observed after two days in hypoxic cultures after knockdown of Hif1a (Fig. 2F–H). Perturbation of Bnip3Δex3 relative to Bnip3FL was seen in Hif1a and Pdk2 knockdowns as well as with an shRNA targeting Bnip3Δex3, but with the reverse result upon targeting Bnip3FL (Fig. 2I). Decrease of Bnip3Δex3 expression was associated with decreased T cell survival after 3 days of hypoxia, as shown by increase of apoptotic cells with reduced active mitochondrial membrane potential (mitotracker deep red lo annexin V +) in the transduced cells (Fig. 2J, S12). These data suggested that PDK2 regulation of Bnip3 alternative splicing maintains cell survival in hypoxia, with this pathway transcriptionally controlled by HIF-1 (Fig. S13).
Proline metabolism is required for optimal T cell effector function in hypoxia
In addition to the changes in pro-survival factors, pathway analyses of the 128 commonly upregulated genes in renal-infiltrating CD4+ and CD8+ T cells revealed involvement of glutathione and amino acid metabolism pathways (Table S1). Among the upregulated genes in renal-infiltrating T cells were genes involved in the Nrf2-regulated glutathione metabolism pathway, including Idh1 (the gene encoding isocitrate dehydrogenase 1 (IDH1), and glutathione S-transferase genes (Fig. 3A). IDH1 supports redox homeostasis in reductive carboxylation (28). Alpha-ketoglutarate (αKG), a downstream metabolite of glutamine, glutamate, and proline catabolism, is converted to isocitrate through the enzymatic activity of IDH1, while reverting NADPH back to NADP+ in reductive carboxylation. Confirming our transcriptome analysis, we observed upregulation of IDH1 in renal-infiltrating T cells from both MRL/lpr and B6.Sle1.Yaa murine lupus models (Fig. S14A,B). Maintenance of redox homeostasis is critical for glycolysis to provide sufficient energy in conditions of mitochondrial dysfunction, such as hypoxia (29). Therefore, upregulation of amino acid metabolism and the downstream reductive carboxylation may support glycolysis by generating NAD+ and NADP+.
Fig. 3. T cell effector function in hypoxia is mediated by HIF-1 regulated proline metabolism facilitating glycolysis.
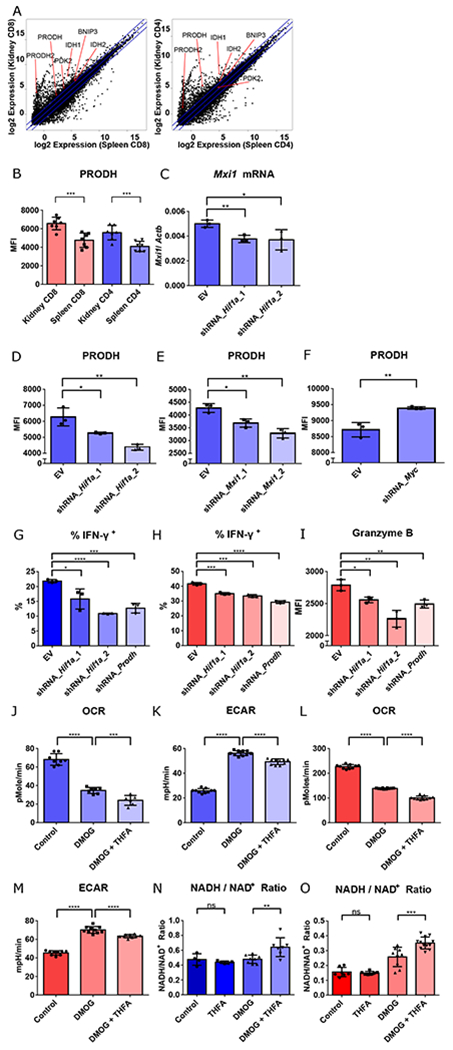
A. RNA-seq gene expression (mean) of splenic vs. renal CD8+ and CD4+ T cells isolated from kidneys of 14-week-old MRL/lpr mice. B. PRODH expression in activated (CD44hi) splenic vs. renal CD4+ and CD8+ T cells isolated from kidneys of 16-18-week-old MRL/lpr mice (n = 7). C. Mxi1 mRNA in Th1-activated CD4+ T cells transduced with either empty vector (EV), or knockdown constructs targeting Hif1a one day after adding DMOG. D-F. Mean fluorescence intensity of PRODH in Th1 activated CD4+ T cells transduced with EV, or two different Hif1a knockdown constructs (D), two different Mxi1 knockdown constructs (E), or Myc knockdown constructs (F) after 2 days of hypoxic culture. G, H. Percentage of IFN-γ+ cells of the live Th1 activated CD4+ T cells, and activated CD8+ T cells, transduced with the different knockdown vectors after 3 days (for CD4+ T cells) or 1 day (for CD8+ T cells) of hypoxia culture. I. Mean fluorescence intensity (MFI) of granzyme B in live activated CD8+ T cells transduced with the different knockdown vectors after one day of hypoxia culture. J-K. Baseline OCR (J) and extracellular acidification rate (ECAR) (K) of control-, DMOG-, DMOG- and THFA-treated Th1-activated CD4+ T cells. L-M. Baseline OCR (L) and extracellular acidification rate (ECAR) (M) of control-, DMOG-, DMOG- and THFA-treated CD8+ T cells N-O. NADH/ NAD+ ratio of control-, THFA-, DMOG-, DMOG- and THFA-treated Th1-activated CD4+ T cells (N) and CD8+ T cells (O). Data shown are mean ± s.d.; statistical analysis by two-tailed paired t-test (B) and unpaired t-test (C-O). ns = p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
To further explore amino acid metabolism, we focused on that of proline, essential for meeting cellular energy requirements and survival in hypoxia (30). Metabolomic analysis of cells with mitochondrial dysfunction have reduced proline content, suggesting a prominent role for its metabolism in hypoxia (31). By flow cytometry, we demonstrated upregulation of proline dehydrogenase (PRODH) in renal CD4+ and CD8+ T cells from MRL/lpr and B6.Sle1.Yaa mice (Fig. 3B and S14C). Using in vitro hypoxia and pseudohypoxic culture systems, we confirmed that this upregulation was controlled by hypoxia and HIF-1 in CD4+ and CD8+ T cells (Fig. S15A–C). HIF-1 is known to globally reprogram cellular metabolism, in part through transcriptional control of Max-interacting protein-1 (MXI1), which ultimately leads to suppression of c-Myc function by interfering with Myc-Max binding (16). Due to the known negative regulation of c-Myc in PRODH expression (32), we asked if upregulation of the latter by HIF-1 is through suppression of the negative regulation of c-Myc. The shRNA knockdown of Hif1a, using two different constructs, reduced Mxi1 transcript (Fig. 3C), consistent with transcriptional regulation of Mxi1 by HIF-1 (33). Knocking down either Hif1a or Mxi1 both contributed to downregulation of PRODH in CD4+ T cells after 2 days of hypoxic culture (Fig. 3D,E) indicating that HIF-1 and MXI1 are both upstream of PRODH regulation. We also found that the expression of the latter was negatively regulated by c-Myc (Fig. 3F) consistent with upstream repression of c-Myc transcriptional activity by HIF-1 and MXI1 (17). These changes during hypoxia were linked to reduced CD4+ and CD8+ T cell effector capability as evidenced by reduced IFN-γ production in both CD4+ and CD8+ T cells and reduced granzyme B production by CD8+ T cells (Fig. 3G–I).
Both CD4+ T and CD8+ T cell cells in pseudohypoxic cultures had higher extracellular acidification rate (ECAR) and lower oxygen consumption rate (OCR) (Fig. 3J–M, S16A–D), indicative of increased glycolysis and mitochondrial dysfunction, and consistent with the HIF-1 inhibition of mitochondrial biogenesis and electron transport chain function (17, 34), key features of metabolic changes in hypoxia. The addition of the selective proline dehydrogenase inhibitor, tetrahydro-2-furoic acid (THFA), reduced ECAR, suggesting that proline metabolism was critical for promotion of glycolysis upon mitochondrial dysfunction. The glycolysis-facilitating effect of proline metabolism is most likely the result of maintenance of NADH/NAD+ balance (Fig. 3N–O), as progression through glycolysis requires the oxidized form of the cofactor NAD+ (Fig. S17) (29). Taken together, these data indicate that HIF-1 controls proline metabolism through suppression of the negative regulation of c-Myc. Upregulation of proline metabolism serves as a cellular adaptation to maintain a balanced NADH/NAD+ ratio to promote glycolysis, and is critical for T cell effector functions in a hypoxic microenvironment.
Blockade of hypoxia-induced adaptation eliminates cellular infiltration, reverses tissue injury and hypoxia
As HIF-1 regulates key adaptations to T cell survival and effector function in hypoxia, it is logical that its blockade might be useful therapeutically to ameliorate tissue damage incurred by infiltrating T cells in lupus nephritis. We accordingly validated hypoxia regulated transcripts from a published dataset of kidney-infiltrating T cells (10), then confirming their phenotypic expression (Fig. S18A–F). Since EPO-producing cells are dependent on HIF-2α (35), with the latter also important in stress protection of renal tubular and endothelial cells (36), selective HIF-1 blockade may minimize perturbations in EPO production and stress adaptive renal parenchymal changes. PX-478 was selected for HIF-1α inhibition in vivo, given its role in decreasing both Hif1a transcription and translation of its protein product, alongside its enhanced ubiquitination (37).
To test the efficacy of HIF-1 blockade in vivo, five-month-old B6.Sle1.Yaa mice, early after onset of proteinuria, were treated with either gavaged PBS or PX-478 (5 mg/kg) every two days. After seven days, renal-infiltrating T cells from animals receiving PX-478 had substantial decreases of HIF-1α, PDK2, and PRODH (Fig. S19A–C). In addition to the perturbation of these adaptive pathways, we observed a reduction in T-bet expression in both CD4+ and CD8+ T cells in treated mice, while there was no difference in the percentage of Foxp3+ CD4+ T cells (Fig. S19D,E). Along with the reduction of HIF-1α and T-bet, there was reduction of IFN-γ positivity among both CD4+ and CD8+ T cells after extending therapy for 28 days (Fig. S19F). These data are consistent with the role of HIF-1 in favoring Th1 cell differentiation and controlling cytokine production (4).
After confirming the efficacy of HIF-1 blockade, an extended 28 days of treatment was performed in proteinuric 5-month-old B6.Sle1.Yaa, and 12-week-old male and 10-12-week-old female MRL/lpr mice. Four weeks after HIF-1 blockade in male MRL/lpr mice, renal-infiltrating CD4+ and CD8+ T cells decreased by more than 90% compared to control animals (Fig. 4A). Clinical efficacy was supported by substantial reduction of proteinuria in the treatment group (Fig. 4B). Similar suppression of renal inflammation and lessened proteinuria occurred in treated female MRL/lpr and male B6.Sle1.Yaa mice, compared to controls (Fig. S20A–F). Pathological scores by NIH activity index were also reduced in the PX-478 treated group (Fig. 4C), especially cellular proliferation, and glomerular leukocyte and tubulointerstitial infiltration (Fig. 4D,E, S21A–C), the latter around vessels, a feature of nephritis in the MRL/lpr strain We also observed reduction of vasculitis, a feature of nephritis in the MRL/lpr strain, in treated mice (Fig. S21D). Reduced immune complex deposition was noted in the glomeruli of the treated MRL/lpr mice (Fig. 4F, S21E). There was reversal of renal cortical hypoxia in the PX-478 treated mice to nearly the same degree observed in control Fas-intact MRL+/+ mice (Fig. 4G–I). Treated mice remained functionally active, with maintenance of hematocrits comparable to control animals (Fig. S21F), suggesting lack of off-target effects of PX-478 and that HIF-2 regulated erythropoiesis was unaffected. Extended treatment for 12 weeks with PX-478 was well tolerated in MRL/lpr mice, with preservation of renal function (Fig. S21G–H) and improvement in survival (Fig. 4J).
Fig. 4. Selective HIF-1 blockade eliminates renal injury in murine lupus nephritis.

A. Numbers of renal-infiltrating CD4+ and CD8+ T cells isolated from 16-week-old male MRL/lpr mice after four weeks of treatment with either PX-478 or PBS (n = 11, 10, respectively). B. Semi-quantitative urine dipstick analysis for proteinuria from 16-week-old male MRL/lpr mice treated with PX-478 or PBS. C. Pathological scores of the 16-week-old male MRL/lpr mice treated with PX-478 or PBS, as assessed by the NIH activity index. D-F. Representative glomerular (D), perivascular aggregates near the corticomedullary junction (E) and immunofluorescence staining of glomerular IgG2a (F) from the same mice after 4 weeks of treatment with PX-478 (D-F) and PBS (G-I). Representative of 3 experiments, n = 10 to 11 animals per group. G-I. Representative pimonidazole staining of kidney sections from 16-week-old MRL/lpr mice treated with PBS (J) or PX-478 (K), and quantification of pimonidazole positive cortical tubular cells (L). Reference line indicates pimonidazole positive renal tubular cells in control Fas-intact MRL+/+ mice. n = 5, 6 and 4 respectively. J. Kaplan Meier survival curve of MRL/lpr mice treated with PBS or PX-478. (n = 10 animals each group). Data shown are mean ± s.d.; statistical analysis by two-tailed t-test (A-C, I) and log-rank test (J). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
In addition to eliminating renal-infiltrating T cells through blockade of hypoxia adaptation, we found reduced splenic size and anti-dsDNA antibody production in MRL/lpr and B6.Sle1.Yaa mice treated with PX-478 for 4 weeks (Fig. S22A–F), coincident with the reduction in immune complex deposition in glomeruli, indicating suppression of systemic autoimmunity. These findings were likely the result of perturbation of plasma cell survival and interruption of ongoing T-B cell interactions and consequent extrafollicular (38) or germinal center (GC) B cell maturation. Consistent with the finding that hypoxia is required for bone marrow plasma cell survival (39), we found reduction of plasma cells in 16-week-old MRL/lpr mice treated with PX-478 for 4 weeks (Fig. S22G–H). The reduction in autoantibody titers, which was more pronounced in the PX-478-treated B6.Sle1.Yaa than treated MRL/lpr mice, was perhaps a consequence of less reduction in PSGL-1lo PD-1hi CXCR4hi extrafollicular helper T cells, which drive B cell maturation in the latter strain, compared to PD-1hi CXCR5hi Tfh cells which are B cell drivers in B6.Sle1.Yaa mice and were reduced upon treatment (Fig. S22I,J). There was also abolition of GC formation in PX-478-treated B6.Sle1.Yaa mice (Fig. S22K), likely related to GC hypoxia and the reliance of HIF-1 in regulating antibody class switch (40, 41).
One of the limitations of pharmacological blockade is its generalized systemic effect. To determine the role of T-cell specific HIF-1α in cellular adaptation with promotion of tissue damage, we generated Cd4cre.Hif1afl/fl.B6.Sle1.Yaa mice, finding they did not have apparent T cell developmental defects, similar to non-autoimmune Cd4cre.Hif1afl/fl mice (3), nor did we observe any effect in development of systemic autoimmunity as measured by spleen size and anti-dsDNA production, compared to Hif1afl/fl.B6.Sle1.Yaa mice (Fig. S23A–C). The lack of effect in autoantibody production was likely due to the functional redundancy of HIF-1α and HIF-2α in Tfh cells leading to generation of antibody producing cells (42). Consistent with this finding, GC sizes were similar between Cd4cre.Hif1afl/fl.B6.Sle1.Yaa and Hif1afl/fl.B6.Sle1.Yaa controls (Fig. S23D–F). However, CD4+ and CD8+ T cells isolated from Cd4cre.Hif1afl/fl.B6.Sle1.Yaa mice were less glycolytic and had higher oxidative phosphorylation (Fig. S23G–J) than controls, consistent with the known role of HIF-1 in promoting glycolysis and suppressing mitochondrial respiration (17).
Upon analysis of kidneys of Cd4cre.Hif1afl/fl.B6.Sle1.Yaa mice, we observed reduction in renal-infiltrating T cells compared to Hif1afl/fl.B6.Sle1.Yaa controls, analogous to pharmacological blockade (Fig. 5A). Even though T cell selective knockout of Hif1a had no effect on proteinuria (Fig. 5B), there was reduction in total pathological scores, with complete ablation of pathological findings as quantified by NIH activity index in 7/16 in mutant Cd4cre.Hif1afl/fl. B6.Sle1.Yaa mice (Fig. 5C). Mutant mice had similar degrees of glomerular pathology and immune complex deposition as control animals, but with reduction of tubulointerstitial inflammation (Fig. 5D–F, S23K–M). Since systemic autoimmunity was unaffected in T cell specific mutant mice, their glomerular pathology was likely the result of inflammation triggered by immune complex deposition (Fig. 23N). In contrast, blocking tissue adaptation to hypoxia largely eliminated T cells in the hypoxic tubulointerstitial compartment, which, when damaged in autoimmunity, is most correlated with poor renal function by contrast to glomerulopathy (43). We also demonstrated reduced hypoxia in the renal cortex of the genetically modified mutant mice (Fig. 5G–I), in agreement with a role of infiltrating T cells in initiation of tissue damage and resultant hypoxia. Yet, depletion of T cell tubulointerstitial infiltration did not completely reverse renal cortical hypoxia suggesting that immune complex deposition and subsequent glomerular pathology can cause tissue hypoxia. The reduction of T cell infiltration in the tissue also corresponded with increased survival of mutant Cd4cre.Hif1afl/fl B6.Sle1.Yaa mice compared to controls (Fig. 5J), further indicating the relevance of pathogenic T cell in dictating organ damage and outcome. In summary, T-cell selective HIF-1α depletion is effective in suppressing renal inflammation, implicating adaptation to hypoxia and induction of a HIF-1 transcriptional program as a feedback loop promoting tissue damage independently of humoral autoimmunity. These data also provide evidence that targeting environmental factors that regulate adaptive T cell survival and function is therapeutically beneficial in mice.
Fig. 5. Genetic ablation of HIF-1α in T cells eliminates infiltrating T cells in lupus nephritis, reverses hypoxia, and prolongs survival.
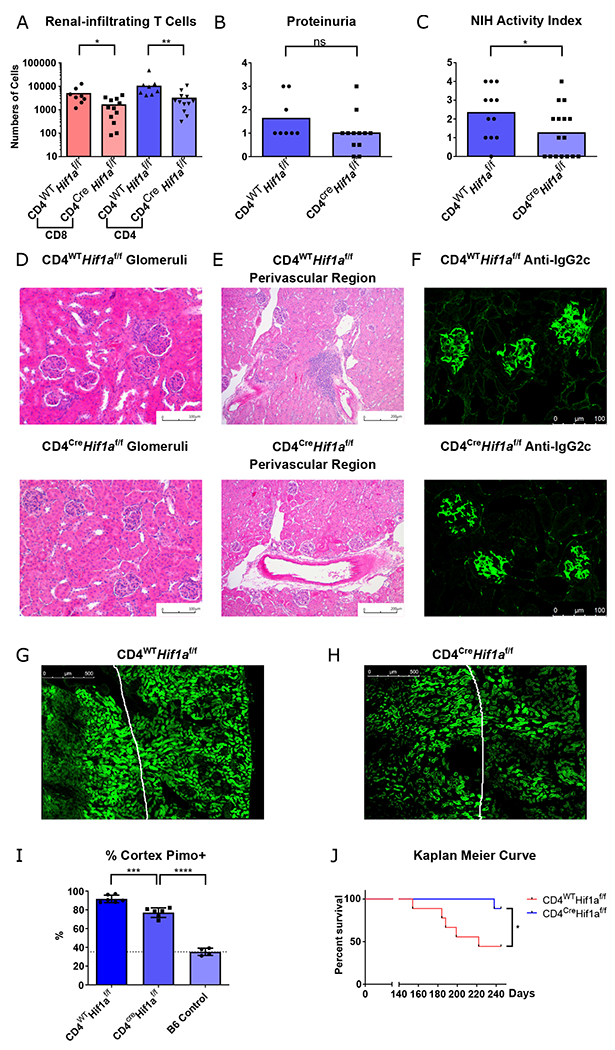
A. Numbers of renal-infiltrating CD4+ and CD8+ T cells isolated from 6-month-old Cd4cre.Hif1afl/fl and Hif1afl/fl B6.Sle1.Yaa male mice (n = 12 and 8, respectively). B. Semi-quantitative urine dipstick analysis for proteinuria the same mice as (A) C. Pathological scores of the same mice using the NIH activity index for total NIH activity index(C) (n = 16, 12, respectively). D-F. Representative glomerular (D), perivascular aggregates near the corticomedullary junction (E), and immunofluorescence staining of glomerular IgG2c (F) from the same mice. G-I Representative pimonidazole (Hypoxyprobe) staining of kidney sections and quantification of pimonidazole positive renal tubular cells in the cortex. Reference line represents the pimonidazole staining in renal cortex of control B6 mice n = 6, 6 and 4 respectively. J. Kaplan Meier survival curve of Cd4cre.Hif1afl/fl and Hif1afl/fl B6.Sle1.Yaa male mice, n = 9 animals each group. Data shown are mean ± s.d.; statistical analysis by two-tailed t-test (A-C) and log-rank test (J) ns = p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Pathways promoting tissue adaptation and effector function are upregulated in human lupus nephritis
We next asked if HIF-1 driven transcriptional pathways that are upregulated in murine lupus nephritis were similarly enhanced in renal biopsy specimens from patients with SLE. As in mice, lupus nephritis in humans is characterized by glomerular immune complex deposition and regions of dense lymphocytic aggregates, predominantly in tubulointerstitial regions, but also peri-glomerular and perivascular (21, 43). We first analyzed single cell RNA-seq data from the Accelerating Medicine Partnership (AMP) SLE project (44). Renal-infiltrating T cells from SLE patients expressed Hif1a transcripts, and other HIF-1 regulated genes identified in our work, including BNIP3 and PDK2 (Fig. S24A–C). The low PRODH signal was likely the result of its relatively low expression among T cell transcriptomes (Fig. S24D). Meanwhile, IDH1, which confers redox balance, was also expressed in renal-infiltrating T cells as in our murine lupus models (Fig. S24E).
To confirm expression of these pathways, we performed immunohistochemistry staining of lupus nephritis biopsy samples. A key enzyme for reductive carboxylation, IDH1, and HIF-1 regulated proteins, PDK2, and PRODH, were expressed in areas of dense T cell infiltration (Fig. 6A–C). By contrast, these markers were not present in T-cell dense areas (T-cell zones) of inflamed human tonsillar sections (Fig. 6D–F), although they were observed in tonsillar GCs, consistent with its hypoxia (40). The generalized diffuse staining of these markers in regions of dense lymphocytic infiltration in lupus nephritis biopsy samples was different from that of T cells in inflamed human tonsils (Fig. 6G–I). We further confirmed the expression of these hypoxia adaptive markers in T cells in biopsy samples using confocal microscopy (Fig. 6J–L), whereas these markers were only stained a portion of renal tubular cells from non-inflamed biopsies, with less fluorescence intensity compared to the inflamed region (Fig. S25). The comparison between human lymphoid tissue (tonsil) and nephritis samples was compatible with our findings that upregulation of hypoxia adaptive pathways exists in renal-infiltrating T cells in murine lupus compared to splenic T cells, and support the idea that pharmacologic HIF-1α blockade should be considered as a therapeutic option in treatment of lupus nephritis and more generally, in autoimmune tissue damage.
Fig. 6. Hypoxia-regulated T cell survival and effector pathways are upregulated in human lupus nephritis.
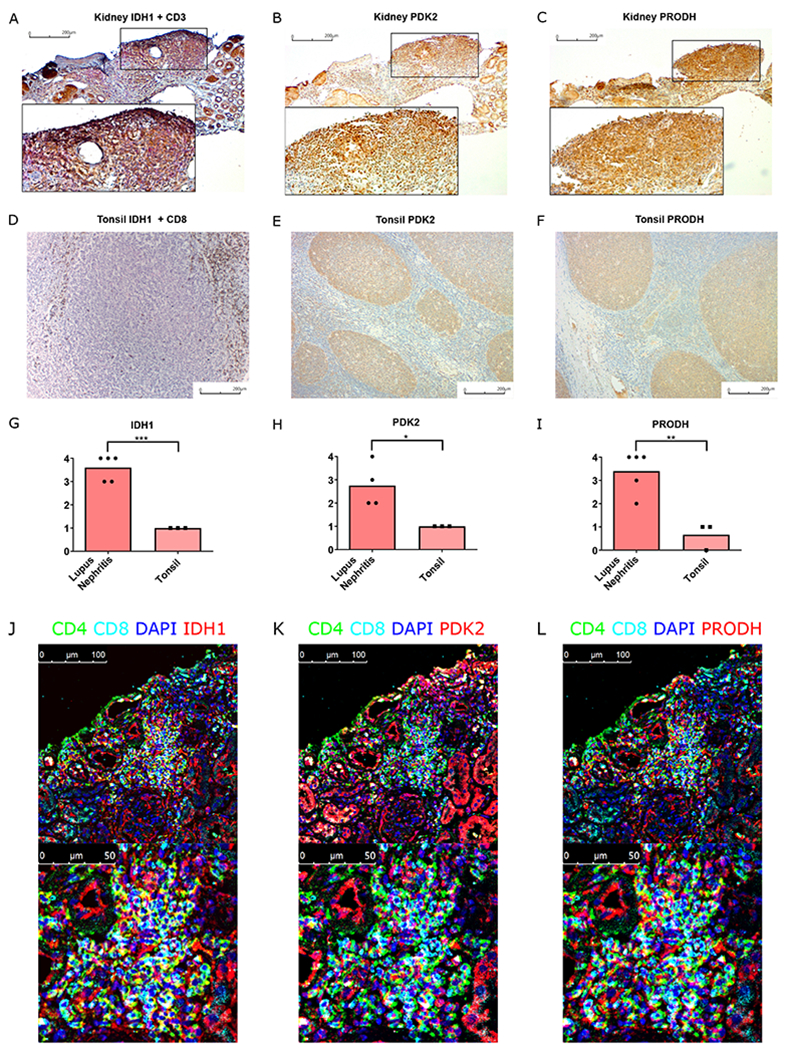
A-C. Immunohistochemistry staining of IDH1 (brown) + CD3 (red) (A), PDK2 (brown) (B), and PRODH (brown) (C) in human lupus nephritis biopsy sample. D-F. Immunohistochemistry staining of IDH1 (red) + CD8 (brown) (D), PDK2 (brown) (E), and PRODH (brown) (F) in inflamed human tonsil. G-I. Quantification of IDH1 (G), PDK2 (H), and PRODH (I) positive staining in dense lymphocytic aggregates of lupus nephritis and T cell zone in inflamed tonsils. J-L. Merged immunofluorescence staining images of CD4, CD8, DAPI with either IDH1 (J), PDK2 (K), or PRODH (L).
Discussion
We demonstrated that the hypoxic environment in the kidney dictates the phenotype of infiltrating T cells, promoting their effector functions to cause tissue damage. Immune complex deposition and destruction of glomeruli are sufficient to cause deterioration of tissue oxygen tension, which further leads to phenotypic changes of infiltrating T cells. In both T-cell depleted lupus mice and genetically modified mutant mice experiments, depletion of T cells in the kidney did not completely reverse renal cortical hypoxia. In contrast, almost complete reversal of renal cortical hypoxia was noted in the PX-478 (HIF-1 blockade) treated mice, which also had reduced autoantibody titers and immune complex deposition compared to controls. These data suggest that immune complex deposition and subsequent glomerular pathology, which are widely accepted as the initiating event of renal injury, was sufficient to cause tissue hypoxia. Tissue hypoxia phenotypically shapes infiltrating T cells, sustains their effector function with resultant tissue damage, and further aggravates tissue hypoxia.
Oligoclonal expansion of renal-infiltrating T cells in both human and murine lupus suggests antigen drive (45, 46), even though identity of the local autoantigen is unknown. The presence of PD-1hi T cells in our analysis and in previously published data (10) are most likely the result of chronic antigen stimulation with features of exhaustion (10). However, we also observed functional renal T cells that contribute to tissue damage in lupus nephritis. Hypoxia and upregulation of HIF-directed pathways sustain T cell function, despite chronic antigen stimulation and expression of surface inhibitory markers (3). These data, as observed in chronic viral infection, are consistent with our finding in the hypoxic autoimmune kidney, in that local upregulation of HIF limits T cell terminal differentiation and preserves functional capacity.
Infiltrating T cells adapt to the environment, and upregulate critical factors for cellular survival, metabolic changes, and maintained effector function dependent of HIF-1. HIF-1 plays a pro-survival role in T lymphocytes in part through PDK2-mediated alternative splicing of Bnip3, with generation of the pro-survival mRNA isoform Bnip3Δex3, which accounts for maintenance of T cell survival in hypoxic microenvironment (Fig. S11). As PDK2 is not universally expressed, we reasoned that this may be a cell-type specific escape pathway, in this case in renal-infiltrating T cells, for adaptation to hypoxia. Such regulation of apoptosis also is present in ventricular myocytes in order to regulate myocardial adaptation to ischemic stress (47).
Hypoxia driven HIF-1 activation leads to mitochondrial dysfunction, shifting cellular energy demands toward glycolysis dependence (17, 48). This alteration is partly driven by the mTORC1-HIF-1 pathway, which upregulates the glucose transporter (Glut1) to increase glucose uptake (49). Reductive carboxylation is another dominant cellular metabolic change in hypoxic cells or those with defective mitochondria (50). Reductive carboxylation maintains NADH/NAD+ redox balance via malate dehydrogenase 1 (MDH1) to facilitate glycolysis during mitochondrial dysfunction (Fig. S13) (29). Reductive carboxylation also promotes lipogenesis, which utilizes acetyl-CoA and NADPH as substrates to generate fatty acid and NADP+ (51, 52). The generation of NADP+ maintains NADH/NAD+ balance through the activity of nicotinamide nucleotide transhydrogenase (NNT) (53), and this reaction is particularly important for reductive carboxylation in mitochondrial defective cells (31). Substrates for reductive carboxylation, including oxaloacetate and acetyl-CoA, are both downstream products of citrate requiring IDH1/2 transcriptional regulation (Fig. S13) (31). This explains the cellular need for amino acid catabolism in cells with mitochondrial defects, in which αKG generated by amino acid degradation serves as a precursor for macromolecular biosynthesis (31, 54). We showed that proline metabolism is upregulated in hypoxic T cells as well as in renal-infiltrating T cells, and supports glycolysis through maintaining NADH/NAD+ redox balance, potentially through proline-derived αKG via downstream reductive carboxylation. We also demonstrated that such cellular metabolic programming in hypoxia is mainly mediated via changes in c-Myc transcription, a major metabolic regulator in activated mouse T cells (55). During hypoxia, c-Myc transcriptional activity is suppressed through HIF-1 and HIF-1 dependent regulation of MXI1 (17, 33), which interferes Myc-Max complex formation necessary for Myc-dependent gene regulation (16).
A limitation of our study is that HIF-1 likely regulates tissue injury in lupus beyond its CD4+ T cell-intrinsic effect, as we demonstrated more profound amelioration of murine nephritis using pharmacological blockade of HIF-1 compared to its genetic elimination. More detailed exploration of its role in promoting tissue injury by CD8+ T cells is warranted, as is the effect of HIF-1α blockade on other immune cells that infiltrate the lupus kidney, and its effect in renal parenchymal cells. Regulation of HIF-1 may also be independent of hypoxia (56), a facet of its regulation that merits further exploration in organ injury in lupus.
Taken together, our findings demonstrate that upon infiltration into the kidney, T cells adapt to the local environment to ensure their survival and effector capability, with remodeling of metabolic pathways. Although beneficial to the host upon local pathogen invasion, such changes in inflammatory and autoimmune diseases are maladaptive, leading to tissue damage. Understanding phenotypic changes in renal-infiltrating immune cells can lead to therapeutic targeting with disease amelioration, and offers a roadmap to further dissect mechanisms of tissue inflammation in autoimmunity. HIF-1 blockade effectively intercepts, and at least partially reverses, a cycle of tissue hypoxia, sustained T cell effector function, and kidney damage in murine lupus, that appears active in human lupus nephritis. Meanwhile, therapeutic blockade of HIF-1 effectively reduces autoantibody production in addition to prevention of T-cell mediated tissue damage, enhancing its therapeutic potential.
Materials and Methods
Study Design
We isolated and performed transcriptome analysis of renal-infiltrating and splenic T cells from two strains of lupus-prone mice, male and female MRL/lpr and male B6.Sle1.Yaa, to determine transcripts of genes that were selectively upregulated in kidneys, with the primary focus on those enhanced in hypoxia and regulated by the transcription factor HIF-1α. Differentially expressed transcripts and their protein products were confirmed as selectively upregulated in kidneys using ex vivo analysis and in situ immunofluorescence staining, and functionally interrogated using in vitro cultures to determine their roles in cell survival and metabolism. Selected proteins upregulated in renal-infiltrating T cells of lupus-prone mice were confirmed to have enhanced expression in T cells from renal biopsies of patients with lupus nephritis. We used pharmacological and genetic blockade of HIF-1α to demonstrate that this transcription factor promoted effector function of renal-infiltrating T cells, with blockade therapeutically effective in treating nephritis in male and female MRL/lpr and male B6.Sle1.Yaa mice randomly assigned to intervention or control groups of littermates. Study endpoints were death or euthanization performed because of humane concerns. All renal sections were blindingly scored by a renal pathologist (PCW). All experiments were performed at least twice, preceded by pilot experiments to determine numbers of replicates needed to reach statistical significance. The therapeutic potential of HIF-1α blockade in patients with SLE was demonstrated by analysis of these markers in human lupus nephritis biopsy samples.
Mice
Mice were housed in the pathogen-free facility in Yale Animal Resources Center (Yale University, New Haven, CT). Animal handling and experimental protocols were approved by the Yale Institutional Animal Care & Use Committee (IACUC). C57BL/6 (B6) mice were purchased from Charles River Laboratory. MRL/MpJ-Faslpr/J (MRL/lpr), CD4-Cre(B6.Cg-Tg[Cd4-cre]1Cwi/BfluJ), and Hif1afl/fl (B6.129-Hif1atm3Rsjo/J) mice were purchased from the Jackson Laboratory. B6.Sle1.Yaa mice were provided by E. Wakeland (University of Texas Southwestern Medical School). To generate Cd4cre.Hif1afl/fl.B6.Sle1.Yaa, Cd4cre mice and Hif1afl/fl mice were crossed to the B6.Sle1.Yaa background. Presence of the intact Sle1 locus was confirmed by microsatellite screening within the locus, including D1Mit15, D1Mit17, D1Mit47, D1Mit202, D1Mit113, D1Mit206, D1Mit353, D1Mit407, D1Mit105, D1Mit274, D1Mit400, and D1Mit541 (57).
Human Lupus Nephritis Biopsy Samples
Collection of archive paraffin blocks of lupus nephritis biopsy samples from Department of Pathology, Yale-New Haven Hospital, and their associated records were approved by Human Research Protection Program, Institutional Review Board, Yale University (IRB Protocol ID 1603017506). Formalin-fixed paraffin-embedded (FFPE) biopsy samples from de-identified lupus nephritis patients were obtained from the University of Chicago Human Tissue Resource Center and was used in accordance with IRB protocol # 15065B-CR009.
Isolation of Renal-infiltrating T Cells
Intravascular antibody injection of anti-Thy1.2 (Clone 53-2.1, BD, 553006), anti-CD45.1 (Clone A20, eBioscience, 11-0453-85) or anti-CD45.2 (Clone 104, eBioscience, 14-0454-85) antibodies was administered to mice 5 minutes before sacrifice, with organ harvest to distinguish intravascular and tissue-infiltrating cells performed as we have described (6). The tissue digestion protocol was modified from that used for isolation for tissue-resident T cells (58). After removal of renal capsule, renal tissues were crushed into small pieces and incubated in RPMI 1640 with 10% fetal bovine serum (FBS), 100 lU/mL collagenase D with CaCl2 (2mM) and MgCl2 (2mM) at 37°C for 45 minutes. After enzymatic digestion, gentle MACS dissociator (Program m_Spleen_01.01) was used to disrupt remaining tissue. Cell suspensions was filtered through a 70 μm strainer, then washed and resuspended in PBS. Mononuclear cells were isolated after passing the single cell suspension through Ficoll density gradient centrifugation.
In Vivo Pimonidazole Labelling
To identify regions of hypoxia in vivo, 80mg/Kg of pimonidazole (Hypoxyprobe, HP-200mg) was injected to the mice 1.5 hour prior to sacrifice as previously described (59). The mouse kidney was either freshly sampled for renal infiltrating T cells isolation or perfused with 4% paraformaldehyde for preparing tissue sections.
T Cell Depletion
To deplete T cells, 0.3mg of rat anti-mouse Thy1.2 (clone 30H12, InVivoMab, BE0066) was injected, every 3 days for a total of 4 weeks, into diseased male MRL/lpr mice. Peripheral blood was sampled 1 week after the start of treatment, and at the end of treatment to confirm T-cell depletion.
Pharmacologic HIF-1 Blockade
To block HIF-1 in vivo, PX-478 (Medchemexpress, HY-10231) treated lupus-prone MRL/lpr and B6.Sle1.Yaa lupus-prone were orally gavaged at the dose of 5mg/kg, dissolved in PBS at the concentration of 1.25mg/mL, every 2 days for the indicated duration. Control mice were gavaged with the same amount of PBS every 2 days. Treatment started at the time of proteinuria onset for these lupus mice – 12 weeks old for MRL/lpr males, 10-12 weeks for MRL/lpr females, and 5 months for B6.Sle1.Yaa mice.
RNA-sequencing
CD44hi CD4+ and CD8+ T cells from kidney and spleens of 14-week-old MRL/lpr mice were sorted, and mRNA isolated (RNeasy Plus Micro Kit, Qiagen, 74034). Library was constructed by using SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing (TaKaRa, 634893). Samples were sequenced on an Illumina HiSeq 2000 with 100-bp single reads (Yale Stem Cell Center Genomics Core facility). Sequenced reads were aligned to MM9 mouse genome by using STAR 2.5.3 and TopHat 2.1.0 through Partek Flow platform. The differentially regulated genes were analyzed using R package DEseq2 (60), and the pathway analyses were done by Enrichr (12). The human single cell RNA-sequencing data are available at ImmPort (immport.org), under study accession SDY997 AMP Lupus Network Project: Molecular Characterization of Lupus Nephritis and Correlation with Response to Therapy (44). Single cell RNA-sequencing data were analyzed by using Seurat v3.0 (61).
Statistical Analysis
Statistical analysis was performed by GraphPad Prism version 8. Bar graphs in all figures indicate the mean, and the error bars represent the standard deviation. To compare data between two groups, statistical analysis was performed by two-tailed t-test. For survival analysis, the log-rank test was used. All analyses with statistical significance were p < 0.05. Statistical results were labeled in each figure as ns = p ≥ 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Supplementary Material
Figure S1. Gating strategy for renal-infiltrating CD4+ and CD8+ T cells.
Figure S2. Renal-infiltrating CD4+ and CD8+ T cells are functionally active and phenotypically distinct.
Figure S3. Reversal of tissue damage and hypoxia after T cell depletion in lupus nephritis.
Figure S4. Validation of hypoxic signatures’ in renal-infiltrating CD4+ and CD8+ T cells using a transcriptome dataset.
Figure S5. Illustration used to define renal pimonidazole staining.
Figure S6. Regions of hypoxia extend to the renal cortex in murine lupus nephritis.
Figure S7. Effect of hypoxia on T cell populations in lupus nephritis.
Figure S8. Predominant HIF-1α but not HIF-2α expression in renal-infiltrating T cells.
Figure S9. T cell depletion reverses renal cortical hypoxia.
Figure S10. HIF-1 regulated survival pathway in renal-infiltrating T cells from B6.Sle1.Yaa lupus-prone mice.
Figure S11. HIF-1 controlled survival pathways are upregulated in hypoxia and pseudohypoxia T cell cultures.
Figure S12. Alternative splicing of BNIP3 regulated by PDK2 promotes T cell survival in hypoxia.
Figure S13. HIF-1 dependent T cell survival in renal hypoxia mediated by PDK2-driven alternative splicing of BNIP3.
Figure S14. Isocitrate dehydrogenase 1 and proline dehydrogenase are upregulated in renal-infiltrating T cells.
Figure S15. Proline dehydrogenase is upregulated in hypoxia and pseudohypoxia T cell cultures.
Figure S16. Changes in mitochondrial function and glycolysis upon blocking proline metabolism in pseudohypoxia.
Figure S17. HIF-1-controlled proline metabolism facilitates glycolysis.
Figure S18. Validation of phenotypic analyses of renal-infiltrating CD4+ and CD8+ T cells using a published transcriptome dataset.
Figure S19. Cellular adaptation in hypoxia is perturbed after selective HIF-1 blockade.
Figure S20. Selective HIF-1 blockade is therapeutic in murine lupus nephritis.
Figure S21. Phenotypic analyses of the nephritic lupus-prone mice treated with a HIF-1 antagonist.
Figure S22. HIF-1 pharmacological blockade reduces autoantibody production and abolishes germinal center formation in murine lupus.
Figure S23. Genetic ablation of HIF-1α in T cells reduces glycolytic capacity and tubulointerstitial infiltrations in lupus nephritis.
Figure S24. Hypoxia-regulated T cell survival and effector transcripts in human lupus nephritis.
Figure S25. Hypoxia-regulated proteins in non-inflamed regions of human lupus nephritis.
Tables S1 to S3
Table S1. Upregulated pathways in renal-infiltrating T cells in murine lupus nephritis.
Table S2. Targeted sequences for shRNA design.
Table S3. Primers for quantitative PCR.
Acknowledgements:
RNA sequencing was performed by the Yale Stem Cell Center Genomics Core facility, supported by the Connecticut Regenerative Medicine Research Fund and the Li Ka Shing Foundation. Murine serum renal function tests were performed by George M. O’Brien Kidney Center at Yale. We thank Amos Brooks for retrieving archive renal biopsy tissues and assisting immunohistochemistry staining.
Funding:
This research was supported by a Pilot & Feasibility Grant from the George M. O’Brien Center for Kidney Research at Yale (NIH P30 DK079310, J.C.), grants from the NIH R37 AR40072 (J.C.), R01 AR074545 (J.C.), and R21 AI142145 (J.C), NIH U19 AI082724 (M.R.C), and AbbVie (J.C.). P.M.C. was the recipient of government scholarship for graduate study from the Ministry of Education, Taiwan, and Gershon Fellowship from the Department of Immunobiology, Yale University.
Footnotes
Competing Interests: J.C. received research funding from AbbVie which was used to partially support this work.
Data and Materials Availability: Transcriptome data of renal and splenic T cells are available at Gene Expression Omnibus (GSE 139283). All the data used for the manuscript are present in the main text or supplementary material.
References
- 1.Davidson A, What is damaging the kidney in lupus nephritis? Nat Rev Rheumatol 12, 143–153 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deng W, Ren Y, Feng X, Yao G, Chen W, Sun Y, Wang H, Gao X, Sun L, Hypoxia inducible factor-1 alpha promotes mesangial cell proliferation in lupus nephritis. American journal of nephrology 40, 507–515 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Doedens AL, Phan AT, Stradner MH, Fujimoto JK, Nguyen JV, Yang E, Johnson RS, Goldrath AW, Hypoxia-inducible factors enhance the effector responses of CD8(+) T cells to persistent antigen. Nature immunology 14, 1173–1182 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clever D, Roychoudhuri R, Constantinides MG, Askenase MH, Sukumar M, Klebanoff CA, Eil RL, Hickman HD, Yu Z, Pan JH, Palmer DC, Phan AT, Goulding J, Gattinoni L, Goldrath AW, Belkaid Y, Restifo NP, Oxygen Sensing by T Cells Establishes an Immunologically Tolerant Metastatic Niche. Cell 166, 1117–1131.e1114 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whitehouse T, Stotz M, Taylor V, Stidwill R, Singer M, Tissue oxygen and hemodynamics in renal medulla, cortex, and corticomedullary junction during hemorrhage-reperfusion. American journal of physiology. Renal physiology 291, F647–653 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Laidlaw BJ, Zhang N, Marshall HD, Staron MM, Guan T, Hu Y, Cauley LS, Craft J, Kaech SM, CD4+ T cell help guides formation of CD103+ lung-resident memory CD8+ T cells during influenza viral infection. Immunity 41, 633–645 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Ito S, Chino Y, Goto D, Matsumoto I, Murata H, Tsutsumi A, Hayashi T, Uchida K, Usui J, Yamagata K, Sumida T, Laser microdissection-based analysis of cytokine balance in the kidneys of patients with lupus nephritis. Clin Exp Immunol 159, 1–10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kok HM, van den Hoogen LL, van Roon JAG, Adriaansen EJM, Fritsch-Stork RDE, Nguyen TQ, Goldschmeding R, Radstake T, Bovenschen N, Systemic and local granzyme B levels are associated with disease activity, kidney damage and interferon signature in systemic lupus erythematosus. Rheumatology (Oxford, England) 56, 2129–2134 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Mohan C, Alas E, Morel L, Yang P, Wakeland EK, Genetic dissection of SLE pathogenesis. Sle1 on murine chromosome 1 leads to a selective loss of tolerance to H2A/H2B/DNA subnucleosomes. J Clin Invest 101, 1362–1372 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tilstra JS, Avery L, Menk AV, Gordon RA, Smita S, Kane LP, Chikina M, Delgoffe GM, Shlomchik MJ, Kidney-infiltrating T cells in murine lupus nephritis are metabolically and functionally exhausted. J Clin Invest 128, 4884–4897 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoyer BF, Moser K, Hauser AE, Peddinghaus A, Voigt C, Eilat D, Radbruch A, Hiepe F, Manz RA, Short-lived plasmablasts and long-lived plasma cells contribute to chronic humoral autoimmunity in NZB/W mice. The Journal of experimental medicine 199, 1577–1584 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma’ayan A, Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic acids research 44, W90–97 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joo MS, Lee CG, Koo JH, Kim SG, miR-125b transcriptionally increased by Nrf2 inhibits AhR repressor, which protects kidney from cisplatin-induced injury. Cell death & disease 4, e899 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hofmann JW, Zhao X, De Cecco M, Peterson AL, Pagliaroli L, Manivannan J, Hubbard GB, Ikeno Y, Zhang Y, Feng B, Li X, Serre T, Qi W, Van Remmen H, Miller RA, Bath KG, de Cabo R, Xu H, Neretti N, Sedivy JM, Reduced expression of MYC increases longevity and enhances healthspan. Cell 160, 477–488 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H, Li H, Xi HS, Li S, HIF1alpha is required for survival maintenance of chronic myeloid leukemia stem cells. Blood 119, 2595–2607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zervos AS, Gyuris J, Brent R, Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell 72, 223–232 (1993). [DOI] [PubMed] [Google Scholar]
- 17.Zhang H, Gao P, Fukuda R, Kumar G, Krishnamachary B, Zeller KI, Dang CV, Semenza GL, HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer cell 11, 407–420 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Kolamunne RT, Dias IH, Vernallis AB, Grant MM, Griffiths HR, Nrf2 activation supports cell survival during hypoxia and hypoxia/reoxygenation in cardiomyoblasts; the roles of reactive oxygen and nitrogen species. Redox biology 1, 418–426 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenberger C, Rosen S, Paliege A, Heyman SN, Pimonidazole adduct immunohistochemistry in the rat kidney: detection of tissue hypoxia. Methods in molecular biology (Clifton, N.J.) 466, 161–174 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Crispin JC, Oukka M, Bayliss G, Cohen RA, Van Beek CA, Stillman IE, Kyttaris VC, Juang YT, Tsokos GC, Expanded double negative T cells in patients with systemic lupus erythematosus produce IL-17 and infiltrate the kidneys. J Immunol 181, 8761–8766 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liarski VM, Kaverina N, Chang A, Brandt D, Yanez D, Talasnik L, Carlesso G, Herbst R, Utset TO, Labno C, Peng Y, Jiang Y, Giger ML, Clark MR, Cell distance mapping identifies functional T follicular helper cells in inflamed human renal tissue. Science translational medicine 6, 230ra246 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takeda N, O’Dea EL, Doedens A, Kim JW, Weidemann A, Stockmann C, Asagiri M, Simon MC, Hoffmann A, Johnson RS, Differential activation and antagonistic function of HIF-{alpha} isoforms in macrophages are essential for NO homeostasis. Genes & development 24, 491–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ko K, Wang J, Perper S, Jiang Y, Yanez D, Kaverina N, Ai J, Liarski VM, Chang A, Peng Y, Lan L, Westmoreland S, Olson L, Giger ML, Chun Wang L, Clark MR, Bcl-2 as a Therapeutic Target in Human Tubulointerstitial Inflammation. Arthritis & rheumatology (Hoboken, N.J.) 68, 2740–2751 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo K, Searfoss G, Krolikowski D, Pagnoni M, Franks C, Clark K, Yu KT, Jaye M, Ivashchenko Y, Hypoxia induces the expression of the pro-apoptotic gene BNIP3. Cell death and differentiation 8, 367–376 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Regula KM, Ens K, Kirshenbaum LA, Inducible expression of BNIP3 provokes mitochondrial defects and hypoxia-mediated cell death of ventricular myocytes. Circ Res 91, 226–231 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Gang H, Dhingra R, Lin J, Hai Y, Aviv Y, Margulets V, Hamedani M, Thanasupawat T, Leygue E, Klonisch T, Davie JR, Kirshenbaum LA, PDK2-mediated alternative splicing switches Bnip3 from cell death to cell survival. The Journal of cell biology 210, 1101–1115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takubo K, Nagamatsu G, Kobayashi CI, Nakamura-Ishizu A, Kobayashi H, Ikeda E, Goda N, Rahimi Y, Johnson RS, Soga T, Hirao A, Suematsu M, Suda T, Regulation of glycolysis by Pdk functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. Cell stem cell 12, 49–61 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang L, Shestov AA, Swain P, Yang C, Parker SJ, Wang QA, Terada LS, Adams ND, McCabe MT, Pietrak B, Schmidt S, Metallo CM, Dranka BP, Schwartz B, DeBerardinis RJ, Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature 532, 255–258 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaude E, Schmidt C, Gammage PA, Dugourd A, Blacker T, Chew SP, Saez-Rodriguez J, O’Neill JS, Szabadkai G, Minczuk M, Frezza C, NADH Shuttling Couples Cytosolic Reductive Carboxylation of Glutamine with Glycolysis in Cells with Mitochondrial Dysfunction. Molecular cell 69, 581–593.e587 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu W, Glunde K, Bhujwalla ZM, Raman V, Sharma A, Phang JM, Proline oxidase promotes tumor cell survival in hypoxic tumor microenvironments. Cancer research 72, 3677–3686 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mullen AR, Hu Z, Shi X, Jiang L, Boroughs LK, Kovacs Z, Boriack R, Rakheja D, Sullivan LB, Linehan WM, Chandel NS, DeBerardinis RJ, Oxidation of alpha-ketoglutarate is required for reductive carboxylation in cancer cells with mitochondrial defects. Cell reports 7, 1679–1690 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu W, Le A, Hancock C, Lane AN, Dang CV, Fan TW, Phang JM, Reprogramming of proline and glutamine metabolism contributes to the proliferative and metabolic responses regulated by oncogenic transcription factor c-MYC. Proc Natl Acad Sci U S A 109, 8983–8988 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Corn PG, Ricci MS, Scata KA, Arsham AM, Simon MC, Dicker DT, El-Deiry WS, Mxi1 is induced by hypoxia in a HIF-1-dependent manner and protects cells from c-Myc-induced apoptosis. Cancer biology & therapy 4, 1285–1294 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Okamoto A, Sumi C, Tanaka H, Kusunoki M, Iwai T, Nishi K, Matsuo Y, Harada H, Takenaga K, Bono H, Hirota K, HIF-1-mediated suppression of mitochondria electron transport chain function confers resistance to lidocaine-induced cell death. Scientific reports 7, 3816 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scortegagna M, Ding K, Zhang Q, Oktay Y, Bennett MJ, Bennett M, Shelton JM, Richardson JA, Moe O, Garcia JA, HIF-2alpha regulates murine hematopoietic development in an erythropoietin-dependent manner. Blood 105, 3133–3140 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Kapitsinou PP, Sano H, Michael M, Kobayashi H, Davidoff O, Bian A, Yao B, Zhang MZ, Harris RC, Duffy KJ, Erickson-Miller CL, Sutton TA, Haase VH, Endothelial HIF-2 mediates protection and recovery from ischemic kidney injury. J Clin Invest 124, 2396–2409 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koh MY, Spivak-Kroizman T, Venturini S, Welsh S, Williams RR, Kirkpatrick DL, Powis G, Molecular mechanisms for the activity of PX-478, an antitumor inhibitor of the hypoxia-inducible factor-1alpha. Molecular cancer therapeutics 7, 90–100 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Tipton CM, Fucile CF, Darce J, Chida A, Ichikawa T, Gregoretti I, Schieferl S, Hom J, Jenks S, Feldman RJ, Mehr R, Wei C, Lee FE, Cheung WC, Rosenberg AF, Sanz I, Diversity, cellular origin and autoreactivity of antibody-secreting cell population expansions in acute systemic lupus erythematosus. Nature immunology 16, 755–765 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen DC, Garimalla S, Xiao H, Kyu S, Albizua I, Galipeau J, Chiang KY, Waller EK, Wu R, Gibson G, Roberson J, Lund FE, Randall TD, Sanz I, Lee FE, Factors of the bone marrow microniche that support human plasma cell survival and immunoglobulin secretion. Nature communications 9, 3698 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cho SH, Raybuck AL, Stengel K, Wei M, Beck TC, Volanakis E, Thomas JW, Hiebert S, Haase VH, Boothby MR, Germinal centre hypoxia and regulation of antibody qualities by a hypoxia response system. Nature 537, 234–238 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abbott RK, Thayer M, Labuda J, Silva M, Philbrook P, Cain DW, Kojima H, Hatfield S, Sethumadhavan S, Ohta A, Reinherz EL, Kelsoe G, Sitkovsky M, Germinal Center Hypoxia Potentiates Immunoglobulin Class Switch Recombination. J Immunol 197, 4014–4020 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cho SH, Raybuck AL, Blagih J, Kemboi E, Haase VH, Jones RG, Boothby MR, Hypoxia-inducible factors in CD4(+) T cells promote metabolism, switch cytokine secretion, and T cell help in humoral immunity. Proc Natl Acad Sci U S A 116, 8975–8984 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson PC, Kashgarian M, Moeckel G, Interstitial inflammation and interstitial fibrosis and tubular atrophy predict renal survival in lupus nephritis. Clinical Kidney Journal, sfx093–sfx093 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arazi A, Rao DA, Berthier CC, Davidson A, Liu Y, Hoover PJ, Chicoine A, Eisenhaure TM, Jonsson AH, Li S, Lieb DJ, Zhang F, Slowikowski K, Browne EP, Noma A, Sutherby D, Steelman S, Smilek DE, Tosta P, Apruzzese W, Massarotti E, Dall’Era M, Park M, Kamen DL, Furie RA, Payan-Schober F, Pendergraft WF 3rd, McInnis EA, Buyon JP, Petri MA, Putterman C, Kalunian KC, Woodle ES, Lederer JA, Hildeman DA, Nusbaum C, Raychaudhuri S, Kretzler M, Anolik JH, Brenner MB, Wofsy D, Hacohen N, Diamond B, The immune cell landscape in kidneys of patients with lupus nephritis. Nature immunology 20, 902–914 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winchester R, Wiesendanger M, Zhang HZ, Steshenko V, Peterson K, Geraldino-Pardilla L, Ruiz-Vazquez E, D’Agati V, Immunologic characteristics of intrarenal T cells: trafficking of expanded CD8+ T cell beta-chain clonotypes in progressive lupus nephritis. Arthritis and rheumatism 64, 1589–1600 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okamoto A, Fujio K, Tsuno NH, Takahashi K, Yamamoto K, Kidney-infiltrating CD4+ T-cell clones promote nephritis in lupus-prone mice. Kidney Int 82, 969–979 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Gang H, Hai Y, Dhingra R, Gordon JW, Yurkova N, Aviv Y, Li H, Aguilar F, Marshall A, Leygue E, Kirshenbaum LA, A novel hypoxia-inducible spliced variant of mitochondrial death gene Bnip3 promotes survival of ventricular myocytes. Circ Res 108, 1084–1092 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Lu H, Forbes RA, Verma A, Hypoxia-inducible factor 1 activation by aerobic glycolysis implicates the Warburg effect in carcinogenesis. The Journal of biological chemistry 277, 23111–23115 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Finlay DK, Rosenzweig E, Sinclair LV, Feijoo-Carnero C, Hukelmann JL, Rolf J, Panteleyev AA, Okkenhaug K, Cantrell DA, PDK1 regulation of mTOR and hypoxia-inducible factor 1 integrate metabolism and migration of CD8+ T cells. The Journal of experimental medicine 209, 2441–2453 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, Yang Y, Linehan WM, Chandel NS, DeBerardinis RJ, Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature 481, 385–388 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, Jewell CM, Johnson ZR, Irvine DJ, Guarente L, Kelleher JK, Vander Heiden MG, Iliopoulos O, Stephanopoulos G, Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 481, 380–384 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun RC, Denko NC, Hypoxic regulation of glutamine metabolism through HIF1 and SIAH2 supports lipid synthesis that is necessary for tumor growth. Cell metabolism 19, 285–292 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gameiro PA, Laviolette LA, Kelleher JK, Iliopoulos O, Stephanopoulos G, Cofactor balance by nicotinamide nucleotide transhydrogenase (NNT) coordinates reductive carboxylation and glucose catabolism in the tricarboxylic acid (TCA) cycle. The Journal of biological chemistry 288, 12967–12977 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fendt SM, Bell EL, Keibler MA, Olenchock BA, Mayers JR, Wasylenko TM, Vokes NI, Guarente L, Vander Heiden MG, Stephanopoulos G, Reductive glutamine metabolism is a function of the alpha-ketoglutarate to citrate ratio in cells. Nature communications 4, 2236 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang R, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D, McCormick LL, Fitzgerald P, Chi H, Munger J, Green DR, The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 35, 871–882 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Caielli S, Veiga DT, Balasubramanian P, Athale S, Domic B, Murat E, Banchereau R, Xu Z, Chandra M, Chung CH, Walters L, Baisch J, Wright T, Punaro M, Nassi L, Stewart K, Fuller J, Ucar D, Ueno H, Zhou J, Banchereau J, Pascual V, A CD4(+) T cell population expanded in lupus blood provides B cell help through interleukin-10 and succinate. Nature medicine 25, 75–81 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Croker BP, Gilkeson G, Morel L, Genetic interactions between susceptibility loci reveal epistatic pathogenic networks in murine lupus. Genes and immunity 4, 575–585 (2003). [DOI] [PubMed] [Google Scholar]
- 58.Steinert EM, Schenkel JM, Fraser KA, Beura LK, Manlove LS, Igyarto BZ, Southern PJ, Masopust D, Quantifying Memory CD8 T Cells Reveals Regionalization of Immunosurveillance. Cell 161, 737–749 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scharping NE, Menk AV, Whetstone RD, Zeng X, Delgoffe GM, Efficacy of PD-1 Blockade Is Potentiated by Metformin-Induced Reduction of Tumor Hypoxia. Cancer immunology research 5, 9–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Satija R, Farrell JA, Gennert D, Schier AF, Regev A, Spatial reconstruction of single-cell gene expression data. Nature biotechnology 33, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ponente M, Campanini L, Cuttano R, Piunti A, Delledonne GA, Coltella N, Valsecchi R, Villa A, Cavallaro U, Pattini L, Doglioni C, Bernardi R, PML promotes metastasis of triple-negative breast cancer through transcriptional regulation of HIF1A target genes. JCI insight 2, e87380 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dang EV, Barbi J, Yang HY, Jinasena D, Yu H, Zheng Y, Bordman Z, Fu J, Kim Y, Yen HR, Luo W, Zeller K, Shimoda L, Topalian SL, Semenza GL, Dang CV, Pardoll DM, Pan F, Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell 146, 772–784 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ray JP, Staron MM, Shyer JA, Ho PC, Marshall HD, Gray SM, Laidlaw BJ, Araki K, Ahmed R, Kaech SM, Craft J, The Interleukin-2-mTORc1 Kinase Axis Defines the Signaling, Differentiation, and Metabolism of T Helper 1 and Follicular B Helper T Cells. Immunity 43, 690–702 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP, Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Gating strategy for renal-infiltrating CD4+ and CD8+ T cells.
Figure S2. Renal-infiltrating CD4+ and CD8+ T cells are functionally active and phenotypically distinct.
Figure S3. Reversal of tissue damage and hypoxia after T cell depletion in lupus nephritis.
Figure S4. Validation of hypoxic signatures’ in renal-infiltrating CD4+ and CD8+ T cells using a transcriptome dataset.
Figure S5. Illustration used to define renal pimonidazole staining.
Figure S6. Regions of hypoxia extend to the renal cortex in murine lupus nephritis.
Figure S7. Effect of hypoxia on T cell populations in lupus nephritis.
Figure S8. Predominant HIF-1α but not HIF-2α expression in renal-infiltrating T cells.
Figure S9. T cell depletion reverses renal cortical hypoxia.
Figure S10. HIF-1 regulated survival pathway in renal-infiltrating T cells from B6.Sle1.Yaa lupus-prone mice.
Figure S11. HIF-1 controlled survival pathways are upregulated in hypoxia and pseudohypoxia T cell cultures.
Figure S12. Alternative splicing of BNIP3 regulated by PDK2 promotes T cell survival in hypoxia.
Figure S13. HIF-1 dependent T cell survival in renal hypoxia mediated by PDK2-driven alternative splicing of BNIP3.
Figure S14. Isocitrate dehydrogenase 1 and proline dehydrogenase are upregulated in renal-infiltrating T cells.
Figure S15. Proline dehydrogenase is upregulated in hypoxia and pseudohypoxia T cell cultures.
Figure S16. Changes in mitochondrial function and glycolysis upon blocking proline metabolism in pseudohypoxia.
Figure S17. HIF-1-controlled proline metabolism facilitates glycolysis.
Figure S18. Validation of phenotypic analyses of renal-infiltrating CD4+ and CD8+ T cells using a published transcriptome dataset.
Figure S19. Cellular adaptation in hypoxia is perturbed after selective HIF-1 blockade.
Figure S20. Selective HIF-1 blockade is therapeutic in murine lupus nephritis.
Figure S21. Phenotypic analyses of the nephritic lupus-prone mice treated with a HIF-1 antagonist.
Figure S22. HIF-1 pharmacological blockade reduces autoantibody production and abolishes germinal center formation in murine lupus.
Figure S23. Genetic ablation of HIF-1α in T cells reduces glycolytic capacity and tubulointerstitial infiltrations in lupus nephritis.
Figure S24. Hypoxia-regulated T cell survival and effector transcripts in human lupus nephritis.
Figure S25. Hypoxia-regulated proteins in non-inflamed regions of human lupus nephritis.
Tables S1 to S3
Table S1. Upregulated pathways in renal-infiltrating T cells in murine lupus nephritis.
Table S2. Targeted sequences for shRNA design.
Table S3. Primers for quantitative PCR.


