Abstract
The human genome contains multiple layers of information that extend beyond the genetic sequence. In fact, identical genetics do not necessarily yield identical phenotypes as evident for the case of two different cell types in the human body. The great variation in structure and function displayed by cells with identical genetic background is attributed to additional genomic information content. This includes large-scale genetic aberrations, as well as diverse epigenetic patterns that are crucial for regulating specific cell functions. These genetic and epigenetic patterns operate in concert in order to maintain specific cellular functions in health and disease. Single-molecule optical genome mapping is a high-throughput genome analysis method that is based on imaging long chromosomal fragments stretched in nanochannel arrays. The access to long DNA molecules coupled with fluorescent tagging of various genomic information presents a unique opportunity to study genetic and epigenetic patterns in the genome at a single-molecule level over large genomic distances. Optical mapping entwines synergistically chemical, physical, and computational advancements, to uncover invaluable biological insights, inaccessible by sequencing technologies. Here we describe the method’s basic principles of operation, and review the various available mechanisms to fluorescently tag genomic information. We present some of the recent biological and clinical impact enabled by optical mapping and present recent approaches for increasing the method’s resolution and accuracy. Finally, we discuss how multiple layers of genomic information may be mapped simultaneously on the same DNA molecule, thus paving the way for characterizing multiple genomic observables on individual DNA molecules.
Keywords: epigenetics, genetics, nanotechnology, optical mapping, single molecule, super-resolution
Introduction
Optical DNA mapping has emerged in recent years as a valuable technique for unraveling long-range information along the genome [1–3]. The method incorporates a multi-disciplinary approach (Figure 1) for extraction of long-range single-molecule genomic data, and provides an indispensable complementary perspective to DNA sequencing. The method is based on sequence-specific fluorescent labeling of up to mega-base pairs (Mbp) long chromosomal DNA molecules (Figure 1A). The labeled DNA molecules are unraveled from their entropically favored entangled state, and extended to allow orderly reading of their fluorescent markers. In nanochannel-based optical mapping, commercialized by BioNano Genomics [4,5], an electric field is applied on the inherently charged long DNA molecules to thread them into an array of nanochannels (∼36 × 36 nm² cross-section), forcing the molecules to stretch linearly due to confinement (Figure 1B.). While this extension can be achieved using various methods (thoroughly reviewed in [3,6]), confinement in nanochannels has proven to generate the most uniform DNA linearization and highest throughput and reproducibility [2,7], and will be the method focused on in this review.
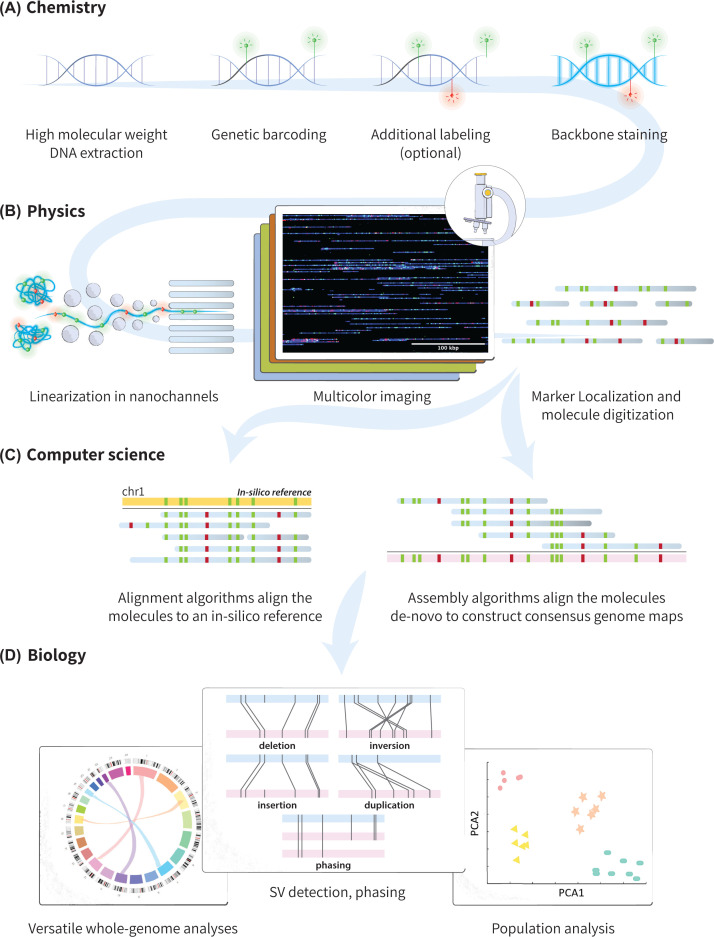
Figure 1. The multi-disciplinary process of optical mapping.
(A) High molecular weight DNA is extracted and chemically labeled at sequence-specific sites to generate a genetic barcode. Additional labeling of various information layers such as epigenetic marks can follow, and finally the DNA backbone is stained with an intercalating dye. (B) Using an electrical field, DNA molecules are unraveled from their entangled state and forced into nanochannels for linearization. Within the nanochannels, the labeled molecules are imaged in multiple colors (image obtained on BioNano Genomics Saphyr system). The molecules and their fluorescent marks are then detected and localized, and image analysis is performed for their digitization. (C) The digitized barcodes are then used to infer the molecules' genomic origin, either by their alignment to a known reference genome, or their assembly de novo. (D) The resulting genomic (and epigenetic) information obtained is used for biological analysis based on structural variants, copy number aberrations, genome phasing and population analyses.
After linearization, the confined molecules are imaged using fluorescence microscopy. The fluorescent genetic marker pattern on each DNA molecule is localized to establish a unique barcode indicating the genomic origin of the molecule. These digitized barcodes are then either aligned to a reference map, produced according to the known genome sequence, or can be assembled to create consensus contiguous maps that span many Mbps (Figure 1C) [2]. The computational methods of barcode mapping and consensus map generation have been recently discussed [2,7] and are beyond the scope of this review.
Optical mapping is a vibrant field [1,2,8], and is currently the go-to method for detecting and validating large-scale genomic rearrangements, such as structural and copy number variations (SVs, CNVs respectively) [9–13] (Figure 1D). With Mbp read lengths, optical mapping reveals such large-scale variations from the reference genome, while conventional short-read next-generation sequencing (NGS) is blind to them. In addition, as each of the mapped DNA molecules originates from a different cell, the single-molecule information extracted with optical mapping enables high-throughput characterization of cellular heterogeneity which is masked by the ensemble averages that are part of NGS analysis.
Optical mapping is commonly compared against other sequencing methods that offer long reads with single-molecule sensitivity, such as Oxford Nanopore [14] or Pacific Biosciences’ (PacBio) single-molecule real-time (SMRT) sequencing [15]. Unlike optical mapping, which is limited in its resolution (>500 bp for ensemble averaged SV detection [16], and optical resolution of ∼1500–2500 bp [17]), these methods offer single base-pair resolution with longer reads compared to NGS. Although these emerging technologies are pushing the limits of sequencing and can already capture medium-scale SVs with high precision [18], they are still limited in the fraction of ultra-long reads and price per genome coverage (see Table 1). Another important advantage of optical mapping is its ability to incorporate multiple information layers simultaneously on the same single DNA molecule. These additional layers of information include various epigenetic content, DNA damage, DNA replication sites and more, as discussed in the last section of this review, at large not accessible to other technologies in a straightforward manner.
Table 1. Long-read sequencing methods compared to optical mapping.
| BioNano Genomics Saphyr | Oxford Nanopore | PacBio SMRT sequencing, HiFi reads | ||
|---|---|---|---|---|
| MinION | PromethION | |||
| Resolution | ∼1500–2500 bp [17] | Single bp | Single bp | |
| Molecules N50* | 300 kbp** [16] | 10–60 kbp [19] | 10–20 kbp [19] | |
| Maximal throughput per cell | 0.33–5 Tbp** (∼100–1500× human genome) [2,16,20,21] | 30–50 Gbp (10–16× human) [19,22] | 180–200 Gbp (60–66×)[19,22] | 35 Gbp (11× human) [19] |
| Price per 1× human genome coverage*** | ∼$0.37–$5.50 [16] | $155–$1550 [19] | $65–$130 [19] | $133–$266 [19] |
‘Molecules N50’: half of the genetic data recorded came from reads longer or equal to this value.
Molecules under 150 kb were filtered out.
Costs include reagents and do not include instruments.
Since optical mapping in nanochannels fuses research from multiple disciplines, we chose to divide the following sections according to their primary disciplines with the intention of facilitating the reader’s orientation throughout this review. We start with a chemistry-oriented review of the main genetic labeling schemes used in optical mapping with an emphasis on those that are compatible with the BioNano Genomics systems. Next, we highlight the physical aspects of nanochannel-based optical mapping, discuss the various limitations they impose on genetic mapping accuracy and resolution, and describe methods which alleviate these restrictions. We then review major recent advances in clinical diagnosis and biological applications enabled by genetic optical mapping. Finally, we survey the emerging practices of epigenetic incorporation to optical mapping, describe the current labeling procedures and the physical limitations, and discuss the required future improvements.
Genetic labeling schemes
The labeling schemes in optical genome mapping are roughly divided into two main subgroups: sparse labeling, where enzymatic reactions are used to fluorescently tag specific sequences scattered along the long DNA molecules, creating a barcode-like pattern where each marker can be localized [4,23,24]; and continuous labeling, where frequent labeling [25], denaturation mapping [26–28] or affinity-based binding [1,27,28] generates a continuous fluorescence intensity map along the molecule [1,27,29]. The maps generated by both methods provide a unique fluorescent signature that discloses the molecule’s genomic origin [1,3–5,30,31].
Sparse labeling can be further divided into several techniques. Enzymatic labeling with nicking enzymes, for example, is a well-established scheme for sparse labeling, in which single-strand breaks are introduced at specific sequence sites by nicking enzymes [4,32,33]. The recognition sites of the enzymes span several bp and occur at distances suitable for genomic mapping. A mix of DNA polymerase and fluorescent nucleotides is then added to repair the nicked strand, fluorescently labeling the recognition sequence, followed by introduction of DNA ligase which seals the fluorescently repaired strand (Figure 2A). Multiple nicking enzymes with different recognition sites are commercially available, providing flexibility in tailoring the genetic markers distribution according to the genome or region of interest [4,32,33]. Although very popular in optical mapping, nick-translation on opposite strands of the DNA can lead to double-strand breaks, compromising the achievable molecular length.
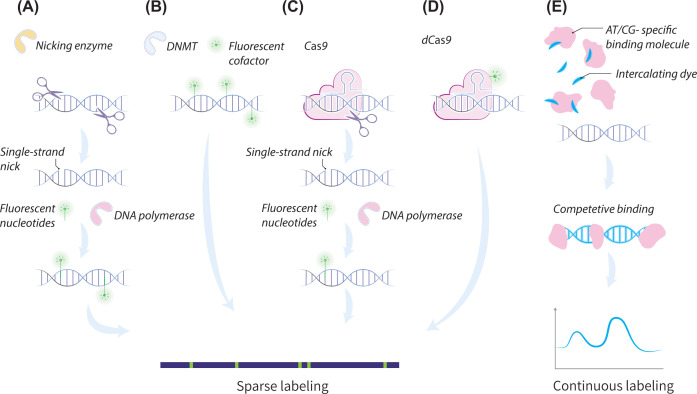
Figure 2. Genetic labeling schemes.
(A–D) sparse labeling schemes. (A) Two-step nick translation. In the first step, a nicking enzyme creates a single-strand nick in the DNA. In the second step, DNA polymerase introduces fluorescent nucleotides to the nicked site. (B) Methyltransferase-based one-step labeling. (C) Nick translation at target sequences with nicking Cas9, steps are similar to (A). (D) single-step fluorescent labeling with dCas9. (E) Continuous affinity-based labeling: competitive binding of a nonsequence-specific fluorescent intercalating dye and an AT/CG-selective molecule. This molecule blocks the dye from binding to these sites, thus keeping them dark while the rest of the genome is labeled, resulting in a sequence-specific intensity profile of the molecules.
Contrary to the two-step labeling required with nicking enzymes, methyltransferases-based labeling is a direct sequence-specific labeling method. DNA methyltransferases (DNMTs) are a family of enzymes that natively catalyze the methylation of DNA bases in biological context. By replacing the natural methyl donor cofactor with a synthetic analog that contains a fluorescent residue, the enzyme adds the modified moiety to the target sequence, creating a fluorescent spot (Figure 2B) [25,34]. The use of methyltransferases that are not sensitive to the native DNA methylation of the studied sample makes this direct labeling scheme accurate and specific while keeping the length of the molecules uncompromised. BioNano Genomics’ Direct Label and Stain (DLS) labeling assay is based on this scheme [35]. A major drawback of both of these enzymatic labeling schemes is the limited availability of enzymes and corresponding recognition sites.
Lately, Cas9 technology was adopted as a labeling scheme for optical mapping. Cas9 enables custom-designing the recognition site, thus allowing full control over the density and specificity of labels. McCaffrey et al. and Abid et al. harnessed a mutant Cas9 enzyme that can cleave single strands to perform a two-step nick labeling at customizable target sequences, such as telomere repeat sequences [36–38] (Figure 2C), while Zhang et al. presented a single-step labeling scheme that includes a mutant Cas9 with no cutting activity (dCase9) and a fluorophore embedded within the CRISPR-dCas9 complex [39] (Figure 2D). This method can improve the mapping sensitivity to structural variations, as it is able to target unique regions, not accessible by commercially available enzymes; however, up until now, Cas9 and dCas9 labeling were only implemented for whole-genome mapping together with nicking enzymes and not on their own.
Alongside the above sparse labeling schemes, dense enzymatic labeling at frequent sequence motifs can create a continuous fluorescence pattern along the DNA due to the overlap of the labels’ emission signal. These continuous patterns represent the underlying sequence and were used by Grunwald et al. to classify bacteriophage genomes in nanochannels [25]. An elegant and cost-effective alternative to such dense labeling approach uses the sequence-specific affinity of intercalating molecules. Continuous affinity-based labeling provides an alternate genome mapping method, first established by denaturation mapping [26–28], followed by affinity displacement using competitive binding. In competitive binding two agents are used for labeling: a nonsequence-specific intercalating fluorescent dye (commonly YOYO-1), and a molecule that binds selectively to either AT- or CG- rich sites (e.g., the AT-specific antibiotic Netropsin). This molecule blocks the intercalating dye from binding to these sites, otherwise free to stain the rest of the genome. In this manner, continuous profiles of AT or GC densities are generated, and can be compared to a computer-generated theoretical AT or GC content map for genomic alignment (see Figure 2E) [40]. Such profiles eliminate the need for DNA backbone staining, as the DNA molecules are already continuously marked, releasing the extra color to be used for additional observables. This method works well for small genomes, and is used for bacterial strain identification in clinical settings [29,41]. Nyberg et al. [42] used it to identify plasmids in a clinical isolate, and Müller et al. combined it with CRISPR/Cas9 to identify antibiotic resistance genes [43]. Recently, competitive binding was used by Müller et al. to map fragments of the human genome [30]. Nevertheless, the method has not yet been applied for high-throughput mapping of large genomes.
Physical aspects of optical mapping
The physical aspects of optical genome mapping in nanochannels dictate both its advantages and its limitations.
Epi-illumination fluorescence imaging, used for visualizing the barcode patterns, provides a large field of view (FOV) for high throughput data acquisition. On the other hand, as any optical imaging method, fluorescence imaging imposes a limitation on the achievable spatial resolution named the diffraction limit. Due to the diffraction of light, close-by markers could be distinguished apart only if they are separated by more than approximately half of their emitted light’s wavelength (∼250 nm that corresponds to ∼800 bp depending on the stretching factor of the DNA molecules in the nanochannels). Optical resolution sets a limit on the maximal information density that can be encoded onto the DNA molecules.
Another advantage of fluorescence imaging is the ability to use color for multiplexing and differentiating various types of genomic observables on the same DNA molecules. Yet, as discussed in a recent work by Jeffet et al. [44], introducing additional colors results in a reduction of throughput by either: (i) sequential color acquisition by emission filter switching, which increases the acquisition time linearly with each additional color; or (ii) by splitting the camera’s field of view to image multiple colors simultaneously, therefore increasing the amount of acquisitions needed to cover the sample area. Nevertheless, introducing a multi-color scheme is a crucial step in order to have a holistic understanding of the interplay between various epigenetic and genetic modifications and will be discussed in the last section of this review.
The last main physical aspect at play in this method, is the nanochannel confinement used for linearizing the DNA molecules. Since the DNA molecules are not immobilized inside the channels, they can be electrically driven in and out of the channels allowing numerous imaging cycles on the same array. This key feature enables ultra-high throughput and high coverage of the entire human genome in a single experiment [2,16]. On the other hand, this also implies that during the acquisition process the DNA molecules are free to move inside the nanochannels and accordingly experience thermal fluctuations at room temperature. These fluctuations limit two different aspects in the process of marker localization and molecule digitization (Figure 1B): (i) resolution, e.g., the ability to separate between two close-by markers and detect their location individually; and (ii) mapping accuracy, the ability to determine the correct genomic location of a marker according to its localization in the image.
As shown and calculated in some recent works [17,45–48], during the exposure time of a single frame (∼150–200 ms in the commercial system) the thermal fluctuations smear the markers' signal, which limits the resolution of two adjacent markers to ∼1.5–3 kbp separation. This value is larger than the optical diffraction limit discussed above and therefore the fluctuations are the main limit on the achievable resolution. The mapping accuracy is also limited due to the fluctuations. A single-frame-acquisition captures the DNA molecules at a nonequilibrium conformation, therefore resulting in erroneous markers location registration compared with their true genomic position (Figure 3A).
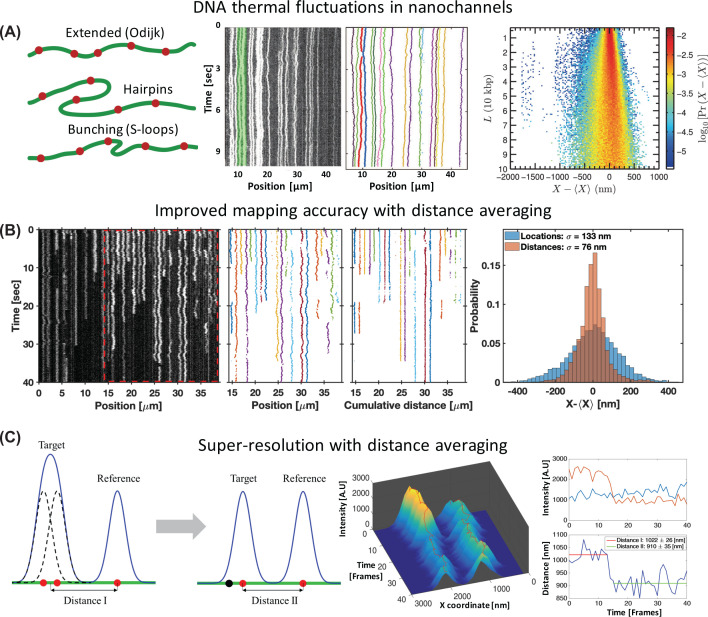
Figure 3. Physical aspects of genomic optical mapping in nanochannels.
(A) DNA fluctuations in nanochannels. Left, possible conformations of DNA molecules confined to nanochannels that contribute to the measured fluctuations. Reprinted from [47], with the permission of AIP Publishing. Middle, raw kymograph (left) and the corresponding marker localizations (right) of barcode labeled genomic DNA extracted from E. coli, showing the thermal fluctuations of confined DNA. Adapted from [48], with the permission of AIP Publishing. Right, probability distribution of observed marker separations in relation to their aligned genomic distance (calculated using 70,305 observed marker positions from single-frame images of 4557 DNA molecules confined to 43 × 43 nm² cross-section channels). The distribution gets wider as the genomic distance increases and therefore mapping accuracy reduces with genomic marker separation. Reprinted from [47], with the permission of AIP Publishing. (B) Improving mapping accuracy with pairwise distance averaging. Left, raw kymograph of the D4Z4 tandem repeat region in chromosome 4 (see optical genome mapping impact section), confined to 45 × 45 nm² cross-section channels (repetitive region marked with the red dashed frame). The repetitive region was used by Jeffet et al. [45] as a ruler to quantify the accuracy and resolution of optical mapping. Adjacent spots in the repetitive segment are distanced 3.3 kbp and each spot is composed of two unresolved fluorophores spaced 676 bp apart. Middle, comparison between localizations maps and cumulative pairwise distances maps of the repetitive region. The localizations map shows correlations in marker fluctuations at short genomic separations, while the distances map shows reduced fluctuations. Right, observed locations distribution compared to the pairwise distance distribution of the ruler molecule’s markers shown to the left. The distances standard deviation is reduced by ∼2-fold compared with locations, allowing increased mapping accuracy. (C) Enabling super-resolution with distance averaging. Left, schematic illustration of the method. A target consisting of multiple sub-diffraction-limit spaced fluorophores is imaged as a single gaussian signal on the camera. Pairwise distance recording between the target and an adjacent marker allows to remove the local collective fluctuations, and thus enable sensitivity to distance shifts originating from the target’s fluorophores blinking or bleaching behavior. Middle, 3D visualization of the intensity–time profile of two fluorescent spots presented in panel (B). Each spot is composed of two fluorophores, where a bleaching event of one of the fluorophores of the left spot is evident after 15 frames. Top right, Intensity time trace of the spots. Bottom right, pairwise distance time-trace between the two spots. The bleaching step alters the mean distance between the two spots allowing to resolve the sub-diffraction-limit distance between them with ∼30 nm resolution. Panels (B and C) were adapted with permission from [45]. Copyright (2016) American Chemical Society.
Therefore, a theoretic physical account of the thermal behavior of DNA in nanochannels, is essential for assessing and improving the limits of resolution and accuracy. When DNA is confined to nanochannels with cross-section much smaller than its statistical folding length (persistence length), it aligns with the channel and exhibits both extension and fluctuation behavior described theoretically by the classic Odijk theory [49] (leftmost panel in Figure 3A). However, optical mapping is generally done in nanochannels that have cross-sections on the same scale as the DNA persistence length, and therefore deviates from this ideal regime [45,47,48]. Adjustments to the Odijk theory have been extensively studied in recent years, both theoretically [17,46,50–54] and experimentally [17,45,47,48,55–58], and we refer the readers to two excellent reviews on the subject [6,59]. The key concepts relevant here are that deviations from the fully extended classic Odijk regime can be manifested as backfolding, hairpins, loops and bunching of the DNA molecules inside the channels (leftmost panel in Figure 3A.). These deviations were incorporated into recent theoretical models [51,60], albeit minor discrepancies still remain between theory and experimental data of molecules confined to <50 × 50 nm² cross-sections channels [17,61,62]. As a result of this research, an important reduction of nanochannels’ widths from 45 nm to 34 nm, was introduced by BioNano Genomics in their commercial Saphyr chips for optical genome mapping. This allows better DNA extension and therefore increased genomic resolution, and reduced alignment errors due to thermal fluctuations [17,45]. Despite remaining limitations [62], the current framework is well-adequate to allow the extraction of valuable genetic and epigenetic information as will be discussed in the following sections.
Although thermal fluctuations are an inherent limitation of the nanochannel-based mapping method [45,48,59,63], a practical approach to minimize their effect and to increase the mapping accuracy was recently introduced by Jeffet et al. [45]. The approach uses short exposure (40 ms) multi-frame time-lapse acquisitions instead of the longer exposure (150–200 ms) single-frame acquisitions usually used in mapping experiments. This enables recording and analysis of the thermal fluctuations of individual DNA molecules inside the nanochannels. As these fluctuations are correlated at short molecular distances (see Figure 3A,B), calculating pairwise-distances between neighboring markers removes the correlated fluctuations (middle panels in Figure 3B), and therefore provides genomic mapping that is more robust to the blinking and bleaching behavior of the markers. Averaging over these pairwise distances had shown to improve the overall mapping accuracy by ∼2-fold (see rightmost panel in Figure 3B) compared with the straightforward marker locations averaging, and more than 6-fold compared with the 450 nm (∼1.5 kbp) mapping accuracy of single-frame acquisition generally accepted in the field [4,17,45,47,48,64]. Consequently, this improved mapping accuracy enabled a 5-fold increase in mapping alignment scores compared to single-frame-acquisition mapping [45]. The core idea underlying this simple analysis is that markers along the DNA molecule can be used as ‘fluctuation reporters’, registering the local collective motion of the DNA, and thus allow fluctuation correction to the barcode registration. The method can be further improved by increasing the label density along the DNA molecule up to the resolution limit of two adjacent markers (∼1.5 kbp), enabling to characterize the DNA fluctuations with better spatial resolution and to apply better fluctuations correction.
The main caveat of this approach is the need for ∼10-fold increase in acquisition time per FOV in order to record sufficient marker positions for effective time averaging (∼40 frames are sufficient to achieve a substantial increase in accuracy [45]). This reduces the overall throughput, but can prove useful when applied to genomes that present assembling difficulties or when trying to compare molecules exhibiting small genomic differences.
Super-resolved genetic mapping
Super-resolution techniques have revolutionized microscopy and biological research since their introduction in recent years [65–68]. The evolution of the field, technical details and future directions are extensively discussed in several reviews [69–71].
In the context of optical mapping, super-resolution was implemented in recent years mainly on immobilized DNA molecules stretched on surfaces [23,72,73], where the gradual bleaching of single fluorophores was exploited [68] to produce super-resolved maps of marker locations along single DNA molecules, with down to 10 nm resolution. The fundamental assumption underlying these methods is that the molecules are fixed in space, which consequently allows recognition of the different markers according to their locations at different times. The major drawback of super-resolution imaging in the context of optical mapping, is the requirement to record many frames (typically ∼100–1000 frames [72]) of the same FOV, in order to derive the super-resolved map. Thus, the increase in resolution is achieved at the expense of a significant reduction in throughput compared with single-frame optical mapping.
Unlike immobilized DNA mapping, the dynamic movement of markers due to thermal fluctuations in nanochannel-based optical mapping, prohibits any straight-forward implementation of such super-resolution methods. Although the localization precision of markers in a single frame can be as high as ∼4 nm [47], the fluctuations completely screen out the subtle localization shifts originating from individual marker bleaching (average standard deviation of ∼140 nm, see rightmost panel in Figure 3B). This essentially limited the effective theoretical resolution of nanochannel based optical mapping to ∼1 kbp [46] and experimentally to ∼1.5–3 kbp [4,17,45,47,48]. This restriction was alleviated by applying the ‘fluctuation reporters’ concept to reduce the local collective fluctuations at the occurrence time of a bleaching event [45]. As illustrated in Figure 3C, in order to resolve sub-diffraction-limit distances in a target composed of multiple fluorophores, pairwise distance recording between the target and a close-by marker was used. Using pairwise distances, instead of marker localizations, reduces the fluctuations to a degree where they are smaller than the shifts caused by marker blinking or bleaching. This re-enables the use of super-resolution techniques [68,72] to determine the inner structure of the target by the distance step-shift at a bleaching or blinking event in the target. Jeffet et al. [45] used this principle to resolve markers separated 670 bp apart with ∼30 nm resolution (see rightmost panel in Figure 3C) [45], introducing a 15-fold resolution improvement compared to the standard ∼450 nm (∼1.5 kbp) single-frame resolution limit [4,46,47]. However, the precision and resolution of this technique depend on the proximity of the reference ‘fluctuation reporter’ marker. As the fluctuations correlation between two markers drops with increasing marker spacing, the effectiveness of the correction and therefore the super-resolution capability drops accordingly. Hence, similar to the mapping accuracy enhancement, higher label density corresponds with better super-resolution capability, permitting that the reference marker can still be resolved from the super-resolution target.
The biological impact of optical genome mapping
In the past few years, optical mapping has positioned itself as the leading technology for large-scale SVs and CNVs detection. It is considered as a high-end alternative to cytogenetic analysis and as a complement to NGS approaches. In this section, we will briefly review some of the recent progress in clinical and biological findings that were enabled by optical mapping.
Current research employs optical mapping to outline the diversity found in the human genome and thus to assemble a more comprehensive map of the genome. In several recent studies, optical mapping greatly contributed to the resolution of haplotypes, SVs and complex regions such as telomeres and sub-telomeric regions, previously inaccessible by NGS alone [11,38,74–79].
SVs are associated with a wide variety of diseases, from rare genetic disorders to cancer [80,81]. Recent works that studied cancer genomes identified new disease-relevant SVs, including highly complex rearrangements and repetitive regions, thus resolving loose ends in sequencing-based maps [12,13,82,83]. Poretti-Boltshauser syndrome and Duchenne muscular dystrophy are among the genetic disorders that have recently benefited from SV detection, enabled by optical mapping [9,20,84].
Facioscapulohumeral muscular dystrophy 1 (FSHD1) is a common type of muscular dystrophy, characterized by a CNV of the D4Z4 tandem repeat in chromosome 4. The length of each repeat unit is 3.3 kbp, and the number of repeats can reach 150 copies in healthy individuals. However, an array of less than 10 D4Z4 copies is considered pathogenic. Recently, Zhang et al. [85] and Dai et al. [21] demonstrated the clinical diagnosis of the syndrome by optical mapping and showed that it is superior to the generally used blot hybridization or FISH combing. Nevertheless, disease manifestation is not determined solely by this genetic CNV, but also by the epigenetic methylation status of the tandem repeats [86,87]. Thus, multiplexing epigenetic with genetic information enabled Sharim et al. to simultaneously detect copy number and methylation pattern of individual DNA molecules, thereby resolving different haplotypes and distinguishing an FSHD-affected patient from a healthy individual [88]. This result stresses the importance of a holistic view that combines genetic and epigenetic information in order to resolve a full genomic picture. The next section will highlight the emerging incorporation of epigenetic information into optical mapping.
Optical mapping of epigenetics
The function of the genome and the resulting gene expression profile is regulated by genetic and epigenetic mechanisms. The epigenetic information is crucial for regulating specific cell functions [89]. Major epigenetic mechanisms include covalent modifications of DNA bases [90–92] and histones [92,93], chromatin structure [89,93,94] and noncoding RNAs that can control gene expression [95]. Unlike the genetic code, which is essentially stable throughout life, the epigenetic content is dynamically changing in response to environmental cues and has great impact on cell activity and diseases [96].
The use of color to distinguish between biological entities is one of the foundations of fluorescence microscopy. In optical mapping, it allows to visualize the genetic information, together with multiple types of epigenetic contents on the same DNA molecule (Figure 4A). Once the genomic origin of the molecule is mapped according to the genetic barcode, the different epigenetic layers are instantaneously placed into their genomic context, and provide locus-specific information. Various labeling schemes of epigenetic features have been developed in recent years and will be summarized in this section together with an account of their impact on epigenetic research.
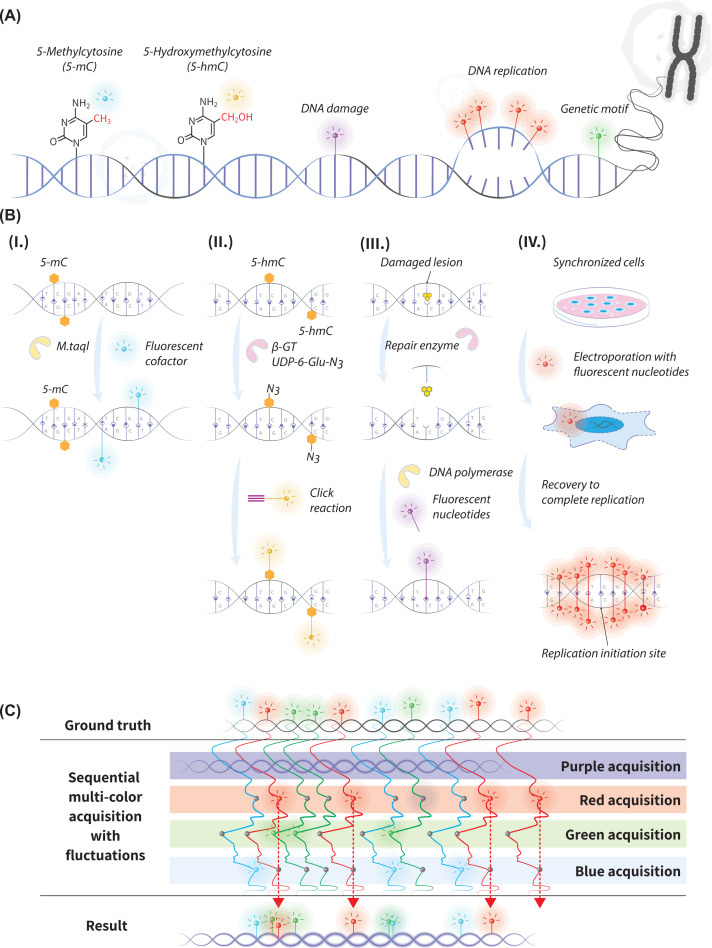
Figure 4. Genomic observables and epigenetic optical mapping in nanochannels.
(A) Example observables that can be marked and viewed in optical mapping. (B) labeling schemes of the epigenetic modifications shown in A. (i) labeling of TCGA sequences that contains an unmodified cytosine by the enzyme M.TaqI and a fluorescent cofactor; (ii) two-step labeling of 5-hmC: the enzyme β-GT attaches a modified glucose with an azide group on it from uridine-diphospho-6-azide-glucose (UDP-6-N3-Glu) to the hydroxyl group of the 5-hmC. Then the azide group is reacted with a fluorescently-labeled alkyne via click chemistry; (iii) two-step labeling of DNA damage sites: The damaged lesion is enzymatically excised, and replaced by fluorescent nucleotides; (iv) in vitro labeling of origins of DNA replication: fluorescent nucleotides enter synchronized cells following electroporation. Then, fluorescence is incorporated into the newly replicated DNA and creates symmetrical replication ‘forks’. (C) An exaggerated illustration demonstrating the impact of thermal fluctuations on multi-color marker detection. Sequential color acquisitions capture the same molecule at different off-equilibrium conformations due to fluctuations. This results in an erroneous interpretation of marker locations along the DNA molecule compared to the ground truth.
A primary epigenetic modification with direct impact on gene expression is DNA methylation of the base cytosine (5-mC) in the dinucleotide sequence CpG [90]. Unmethylated CpG sites can be specifically labeled by methyltransferase enzymes, creating a complementary map to the methylome. For example, the methyltransferase M.TaqI can directly transfer a fluorophore from a synthetic cofactor to the adenine base in the enzyme’s recognition sequence TCGA. However, if the CpG nested in this sequence context is methylated or modified, this reaction is blocked, resulting in the exclusive labeling of unmodified cytosines in every TCGA sequence context (Figure 4B,i) [25,88,97]. In 2019, Sharim et al. used this scheme to optically map a reduced representation of the human methylome for the first time. This ensemble-averaged reduced representation map (∼6% of human CpG sites are found within TCGA context) captures the majority of regulatory sites in the genome [88]. Recently, this long-read methylation data was analyzed at a single-cell level for the first time. The methylation status of promoters and their distal enhancers, simultaneously imaged on the same long DNA molecules, served to accurately deconvolve cell-type mixtures and subpopulations within a sample [98]. A recent improvement to methylation labeling, replacing the methyltransferase M.TaqI with a modified M.SssI (which recognizes any unmethylated CpG), enabled the generation of genome-wide methylation profiles of all CpG sites [99]. DNA methylation can also be labeled directly, using methyl-binding antibodies coupled with a fluorescent reporter [100]. However, the antibodies’ tendency to aggregate hampers DNA insertion into the nanochannels, therefore requiring larger channel cross-sections which reduces by ∼10-fold the achievable resolution compared to covalent base labeling [100].
Another epigenetic modification of the cytosine base, gaining increased interest in recent years due to its link to gene regulation, development, and disease, is the oxidation product of DNA methylation, 5-hydroxymethylcytosine (5-hmC) [91]. The fluorescent labeling of this modification involves two steps. First, the enzyme β-glucosyltransferase (β-GT) is used to attach an azide modified glucose moiety from a synthetic cofactor (UDP-6-N3-Glu), to the hydroxyl group of 5-hmC. In the second step, a copper-free click reaction is used to connect a fluorophore-alkyne to the azide-labeled 5-hmC (Figure 4B,ii) [101–106]. Gabrieli et al. presented a whole-genome optical 5-hmC map of human peripheral blood cells, revealing variable regions and long-range information that were not accessible by sequencing [106].
Another important information layer that could be explored by optical mapping is the DNA damage content. Cellular DNA damage lesions are continuously induced by exposure to various exogenous and endogenous agents, and are a major cause for genomic instability. A labeling scheme called RADD (repair-assisted damage detection), was developed to fluorescently label single-strand lesions in vitro by a two-step process [107–109]. First, native repair enzymes excise the damaged bases and leave a single-strand gap at the damage locus. Next, DNA polymerase and ligase are introduced and fill this gap with fluorescent nucleotides, allowing the visualization of the repaired damage sites (Figure 4B,iii). As a proof of principle, sites of DNA damage induced by the chemotherapeutic agent etoposide were recently labeled and mapped using competitive binding [30]. In the future, optical mapping can potentially be used to reveal the effect of drugs and environmental toxicants on the genome, specifically at regions that are hard to analyze by NGS, and locate genomic hotspots prone to damage and mutations.
Apart from chemical DNA modifications, multi-color optical mapping can be used to study basic biological processes. A recent example was seen in the mapping of origins of DNA replication [110–112]. Mapping these sites provides basic understanding of replication mechanisms and kinetics in normal conditions. The effect of different factors, including damaging agents, drugs, and diseases on replication, could also be studied in this manner. Lately, Wang et al. used in vivo fluorescent nucleotide pulse-labeling of human cells to mark freshly synthesized DNA and trace replication initiation sites with optical mapping, including sites active in very small percentages of cells (Figure 4B,iv) [110].
To summarize, any genomic feature that can be labeled with a fluorescent reporter and does not impair DNA linearization, has the potential to be optically mapped. Detection of multiple types of observables on the same DNA molecule remains a challenge in genomics. Optical mapping holds the potential to multiplex information on a single DNA molecule, and thus map genomic interactions, variability and context with single-molecule sensitivity.
Challenges of multi-color optical mapping and future prospects
Although dual-color optical mapping has been implemented [88,106,110], mapping more colors on the same DNA molecules imposes further constraints on the method's throughput and mapping accuracy. Meanwhile, super-resolution in two colors was only applied to immobilized optical mapping [73], and has not yet been applied to nanochannel-based methods. These limitations are the consequence of the color acquisition procedure employed in the standard fluorescence microscopy setup, where each color is imaged sequentially by inserting a suitable emission filter to the emission path. The immediate result is that each additional color acquisition requires a linear increase in acquisition time, reducing the overall throughput.
However, an additional constraint arises from the combination of sequential color acquisition and thermal fluctuations. This creates desynchronization between the barcode maps of different colors, as each color acquisition images the molecule at a different off-equilibrium conformation (see illustration in Figure 4C). Applying the multi-frame distance-averaging approach discussed previously would enable resolving this issue by allowing to better approximate the equilibrium DNA conformation. However, different color markers could not be used for accurate distance calculation, as the measurements are not synchronized. Moreover, the time delay between adjacent frames of the same color increases with every additional color, therefore multi-color acquisitions reduce the temporal resolution of the single-color fluctuations characterization. Furthermore, since each color should be corrected independently, the overall acquisition time would increase by at least 10-fold multiplied by the number of information layers, resulting in a tremendous decrease in throughput. For these reasons, and since only the genetic markers are used as anchors to map the individual molecules to the genome, additional colors would reduce the enhancement in overall genomic mapping accuracy compared with the results reported in [45]. Finally, since the different color acquisitions are not synchronized, super-resolution could only be implemented independently for each color, using only markers of the same color. This imposes the dense labeling constraint needed for super-resolution, individually to each of the colors.
Emerging possibilities of simultaneous color acquisition could alleviate these problems and allow fusing the recorded fluctuation information from the different colors. Methods such as multi-color split view [73,113], where the FOV is split between emission channels using dichroic mirrors; or spectral imaging [44,114–116], where spectral images are recorded using a dispersive element, can record marker locations of different colors at the same time. Therefore, localizations of markers with different colors can be used to create an integrated multi-color fluctuation map. This would yield a dense characterization of DNA fluctuations, greatly increasing the achievable spatial and temporal mapping resolution with reduction of overall acquisition time compared with the single-color fluctuation characterization. Moreover, simultaneous multi-color acquisition methods allow to resolve markers of different colors even when they are spaced by sub-diffraction limit distances. This enables registering the fluctuations with even higher spatial resolution that would result in unprecedented mapping accuracy and super-resolution capabilities. Finally, the constraint on label density needed for applying super-resolution, would be alleviated from dense labeling of each color independently to the total label density of all colors, allowing to apply super-resolution on all information contents even when some motifs are sparsely dispersed on the DNA.
Conclusions
In this brief review, we have introduced nanochannel-based optical genome mapping, highlighting the method’s ability to tackle outstanding problems unaddressed by available sequencing methods. We have summarized the various multidisciplinary aspects of the method including: (i) the chemically oriented mechanisms for genetic and epigenetic fluorescence labeling; (ii) the physical limitations of the method arising from both the optical acquisition and the thermodynamic properties of DNA confinement, and the ways to circumvent them; (iii) the biological impact of the method in exploration of large-scale genomic aberrations, epigenetic content and other genomic observables such as DNA damage and origins of replication.
Optical mapping’s ability to visualize multiple observables on the same single DNA molecule, will no doubt be exploited in the near future to elucidate the interactions between the various information layers encoded in our genome. We have examined the possibilities of exploring this venue and their current limitations, and pinpointed to multi-color simultaneous-acquisition techniques that would allow to remove these limitations and provide better mapping accuracy and resolution than ever.
Summary
Optical mapping in nanochannels is a single-molecule, high-throughput method that captures a high fraction of ultra-long genomic fragments and may elucidate genomic information that is otherwise inaccessible using sequencing.
Thermal fluctuations of the DNA molecules in the nanochannels pose the limit on mapping accuracy and resolution.
Fluctuation mapping can improve mapping accuracy by 6-fold and allows resolution enhancement of 15-fold by super-resolution localization.
Multi-color fluorescent labeling allows multiplexing several layers of genomic information coexisting on the same DNA molecules, potentially allowing a holistic view of the genome.
Simultaneous color acquisition, as opposed to the current sequential color acquisition, would allow to further improve the resolution and accuracy of the method.
Acknowledgments
J.J. is grateful to the Azrieli Foundation for the award of an Azrieli Fellowship.
Abbreviations
- β-GT
β-glucosyltransferase enzyme
- 3D
3-dimensional
- 5-hmC
5-hydroxymethylcytosine
- 5-mC
5-methylcytosine
- bp (kbp, Mbp, Gbp, Tbp)
(kilo-, Mega-, Giga-, Tera-) base-pairs
- Cas9
CRISPR-associated protein 9
- CNV
copy number variation
- CpG
cytosine-phosphate-guanine (cytosine base directly followed by guanine)
- CRISPR
clustered regularly interspaced short palindromic repeats
- dCase9
nuclease-deficient Cas9
- DLS
direct label and stain
- DNA
deoxyribonucleic acid
- DNMT
DNA methyltransferase
- E. coli
Escherichia coli
- FOV
field of view
- FSHD
facioscapulohumeral muscular dystrophy
- ms
millisecond
- NGS
next-generation sequencing
- nm
nanometer
- PacBio
Pacific Biosciences
- RADD
repair-assisted damage detection
- RNA
ribonucleic acid
- SMRT sequencing
single-molecule real-time sequencing
- SV
structural variation
- UDP-6-N3-Glu
uridine-diphospho-6-azide-glucose
- YOYO-1
homodimer of oxazole yellow (green fluorescent dye used for DNA staining)
Contributor Information
Jonathan Jeffet, Email: jonath7@tauex.tau.ac.il.
Yuval Ebenstein, Email: uv@tauex.tau.ac.il.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
Y.E. acknowledges support from the European Research Council Consolidator grant [grant number 817811].
Open Access
Open access for this article was enabled by the participation of Tel Aviv University in an all-inclusive Read & Publish pilot with Portland Press and the Biochemical Society under a transformative agreement with MALMAD.
Author Contribution
J.J. and S.M. wrote the manuscript. Y.M. and Y.E. reviewed and edited the manuscript.
References
- 1.Müller V. and Westerlund F. (2017) Optical DNA mapping in nanofluidic devices: principles and applications. Lab. Chip. 17, 579–590 10.1039/C6LC01439A [DOI] [PubMed] [Google Scholar]
- 2.Yuan Y., Chung C.Y.-L. and Chan T.-F. (2020) Advances in optical mapping for genomic research. Comput Struct Biotechnol. J. 18, 2051–2062 10.1016/j.csbj.2020.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levy-Sakin M. and Ebenstein Y. (2013) Beyond sequencing: optical mapping of DNA in the age of nanotechnology and nanoscopy. Curr. Opin. Biotechnol. 24, 690–698 10.1016/j.copbio.2013.01.009 [DOI] [PubMed] [Google Scholar]
- 4.Lam E.T., Hastie A., Lin C., Ehrlich D., Das S.K., Austin M.D.et al. (2012) Genome mapping on nanochannel arrays for structural variation analysis and sequence assembly. Nat. Biotechnol. 30, 771–776 10.1038/nbt.2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bocklandt S., Hastie A. and Cao H. (2019) Bionano genome mapping: high-throughput, ultra-long molecule genome analysis system for precision genome assembly and haploid-resolved structural variation discovery. In Single Mol. and Single Cell Sequencing(Suzuki Y., ed.), pp. 97–118, Springer Singapore, Singapore: 10.1007/978-981-13-6037-4_7 [DOI] [PubMed] [Google Scholar]
- 6.Dorfman K.D., King S.B., Olson D.W., Thomas J.D.P. and Tree D.R. (2013) Beyond gel electrophoresis: microfluidic separations, fluorescence burst analysis, and DNA stretching. Chem. Rev. 113, 2584–2667 10.1021/cr3002142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mendelowitz L. and Pop M. (2014) Computational methods for optical mapping. Gigascience 3, 33 10.1186/2047-217X-3-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bogas D., Nyberg L., Pacheco R., Azevedo N.F., Beech J.P., Gomila M.et al. (2017) Applications of optical DNA mapping in microbiology. BioTechniques 62, 255–267 10.2144/000114555 [DOI] [PubMed] [Google Scholar]
- 9.Barseghyan H., Tang W., Wang R.T., Almalvez M., Segura E., Bramble M.S.et al. (2017) Next-generation mapping: a novel approach for detection of pathogenic structural variants with a potential utility in clinical diagnosis. Genome Med. 9, 90 10.1186/s13073-017-0479-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balachandran P. and Beck C.R. (2020) Structural variant identification and characterization. Chromosome Res. 28, 31–47 10.1007/s10577-019-09623-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ebert P., Audano P.A., Zhu Q., Rodriguez-Martin B., Porubsky D., Bonder M.J.et al. (2021) Haplotype-resolved diverse human genomes and integrated analysis of structural variation. Science 10.1126/science.abf7117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crumbaker M., Chan E.K.F., Gong T., Corcoran N., Jaratlerdsiri W., Lyons R.J.et al. (2020) The Impact of Whole Genome Data on Therapeutic Decision-Making in Metastatic Prostate Cancer: A Retrospective Analysis. Cancers 12, 1178 10.3390/cancers12051178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peng Y., Yuan C., Tao X., Zhao Y., Yao X., Zhuge L.et al. (2020) Integrated analysis of optical mapping and whole-genome sequencing reveals intratumoral genetic heterogeneity in metastatic lung squamous cell carcinoma. Transl. Lung Cancer Res. 9, 670–681 10.21037/tlcr-19-401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jain M., Olsen H.E., Paten B. and Akeson M. (2016) The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol. 17, 239 10.1186/s13059-016-1103-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wenger A.M., Peluso P., Rowell W.J., Chang P.-C., Hall R.J., Concepcion G.T.et al. (2019) Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat. Biotechnol. 37, 1155–1162 10.1038/s41587-019-0217-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bionano Genomics Website [Internet]. [cited 2021 Jan 6]. Available from: https://bionanogenomics.com/products/bionano-data-options/
- 17.Chuang H.-M., Reifenberger J.G., Bhandari A.B. and Dorfman K.D. (2019) Extension distribution for DNA confined in a nanochannel near the Odijk regime. J. Chem. Phys. 151, 114903 10.1063/1.5121305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amarasinghe S.L., Su S., Dong X., Zappia L., Ritchie M.E. and Gouil Q. (2020) Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 21, 30 10.1186/s13059-020-1935-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Logsdon G.A., Vollger M.R. and Eichler E.E. (2020) Long-read human genome sequencing and its applications. Nat. Rev. Genet. 21, 597–614 10.1038/s41576-020-0236-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen M., Zhang M., Qian Y., Yang Y., Sun Y., Liu B.et al. (2020) Identification of a likely pathogenic structural variation in the LAMA1 gene by Bionano optical mapping. NPJ Genomic Med. 5, 31 10.1038/s41525-020-0138-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dai Y., Li P., Wang Z., Liang F., Yang F., Fang L.et al. (2020) Single-molecule optical mapping enables quantitative measurement of D4Z4 repeats in facioscapulohumeral muscular dystrophy (FSHD). J. Med. Genet. 57, 109–120 10.1136/jmedgenet-2019-106078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nanopore Products Comparison [Internet]. Oxford Nanopore Website. [cited January, 26, 2021]. Available from: https://nanoporetech.com/products/comparison?minion1b=on&minion1c=on&gridion=on&promethion=on.
- 23.Vranken C., Deen J., Dirix L., Stakenborg T., Dehaen W., Leen V.et al. (2014) Super-resolution optical DNA Mapping via DNA methyltransferase-directed click chemistry. Nucleic Acids Res. 42, e50 10.1093/nar/gkt1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das S.K., Austin M.D., Akana M.C., Deshpande P., Cao H. and Xiao M. (2010) Single molecule linear analysis of DNA in nano-channel labeled with sequence specific fluorescent probes. Nucleic Acids Res. 38, e177–e177 10.1093/nar/gkq673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grunwald A., Dahan M., Giesbertz A., Nilsson A., Nyberg L.K., Weinhold E.et al. (2015) Bacteriophage strain typing by rapid single molecule analysis. Nucleic Acids Res. 43, e117 10.1093/nar/gkv563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marie R., Pedersen J.N., Bærlocher L., Koprowska K., Pødenphant M., Sabatel C.et al. (2018) Single-molecule DNA-mapping and whole-genome sequencing of individual cells. Proc. Natl. Acad. Sci. U.S.A. 115, 11192–11197 10.1073/pnas.1804194115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reisner W., Larsen N.B., Silahtaroglu A., Kristensen A., Tommerup N., Tegenfeldt J.O.et al. (2010) Single-molecule denaturation mapping of DNA in nanofluidic channels. Proc. Natl. Acad. Sci. 107, 13294–13299 10.1073/pnas.1007081107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freitag C., Noble C., Fritzsche J., Persson F., Reiter-Schad M., Nilsson A.N.et al. (2015) Visualizing the entire DNA from a chromosome in a single frame. Biomicrofluidics 9, 044114 10.1063/1.4923262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nilsson A.N., Emilsson G., Nyberg L.K., Noble C., Stadler L.S., Fritzsche J.et al. (2014) Competitive binding-based optical DNA mapping for fast identification of bacteria–multi-ligand transfer matrix theory and experimental applications on Escherichia coli. Nucleic Acids Res. 42, e118 10.1093/nar/gku556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Müller V., Dvirnas A., Andersson J., Singh V., Sriram K.K., Johansson P.et al. (2019) Enzyme-free optical DNA mapping of the human genome using competitive binding. Nucleic Acids Res. 47, e89 10.1093/nar/gkz489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neely R.K., Deen J. and Hofkens J. (2011) Optical mapping of DNA: single-molecule-based methods for mapping genomes. Biopolymers 95, 298–311 10.1002/bip.21579 [DOI] [PubMed] [Google Scholar]
- 32.Hastie A.R., Dong L., Smith A., Finklestein J., Lam E.T., Huo N.et al. (2013) Rapid genome mapping in nanochannel arrays for highly complete and accurate de novo sequence assembly of the complex Aegilops tauschii genome. PLoS ONE 8, e55864 10.1371/journal.pone.0055864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jo K., Dhingra D.M., Odijk T., de Pablo J.J., Graham M.D., Runnheim R.et al. (2007) A single-molecule barcoding system using nanoslits for DNA analysis. Proc. Natl. Acad. Sci. 104, 2673–2678 10.1073/pnas.0611151104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goyvaerts V., Van Snick S., D'Huys L., Vitale R., Helmer Lauer M., Wang S.et al. (2020) Fluorescent SAM analogues for methyltransferase based DNA labeling. Chem. Commun. 56, 3317–3320 10.1039/C9CC08938A [DOI] [PubMed] [Google Scholar]
- 35.Deschamps S., Zhang Y., Llaca V., Ye L., Sanyal A., King M.et al. (2018) A chromosome-scale assembly of the sorghum genome using nanopore sequencing and optical mapping. Nat. Commun. 9, 4844 10.1038/s41467-018-07271-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCaffrey J., Sibert J., Zhang B., Zhang Y., Hu W., Riethman H.et al. (2016) CRISPR-CAS9 D10A nickase target-specific fluorescent labeling of double strand DNA for whole genome mapping and structural variation analysis. Nucleic Acids Res. 44, e11 10.1093/nar/gkv878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abid H.Z., Young E., McCaffrey J., Raseley K., Varapula D., Wang H.-Y.et al. (2020) Customized optical mapping by CRISPR-Cas9 mediated DNA labeling with multiple sgRNAs. Nucleic Acids Res. 49, e8 10.1093/nar/gkaa1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abid H.Z., McCaffrey J., Raseley K., Young E., Lassahn K., Varapula D.et al. (2020) Single-molecule analysis of subtelomeres and telomeres in Alternative Lengthening of Telomeres (ALT) cells. BMC Genomics 21, 485 10.1186/s12864-020-06901-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang D., Chan S., Sugerman K., Lee J., Lam E.T., Bocklandt S.et al. (2018) CRISPR-bind: a simple, custom CRISPR/dCas9-mediated labeling of genomic DNA for mapping in nanochannel arrays. bioRxiv 10.1101/371518 [DOI] [Google Scholar]
- 40.Nyberg L.K., Persson F., Berg J., Bergström J., Fransson E., Olsson L.et al. (2012) A single-step competitive binding assay for mapping of single DNA molecules. Biochem. Biophys. Res. Commun. 417, 404–408 10.1016/j.bbrc.2011.11.128 [DOI] [PubMed] [Google Scholar]
- 41.Müller V., Nyblom M., Johnning A., Wrande M., Dvirnas A., Kk S.et al. (2020) Cultivation-free typing of bacteria using optical DNA mapping. ACS Infect. Dis. 6, 1076–1084 10.1021/acsinfecdis.9b00464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nyberg L.K., Quaderi S., Emilsson G., Karami N., Lagerstedt E., Müller V.et al. (2016) Rapid identification of intact bacterial resistance plasmids via optical mapping of single DNA molecules. Sci. Rep. 6, 30410 10.1038/srep30410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Müller V., Rajer F., Frykholm K., Nyberg L.K., Quaderi S., Fritzsche J.et al. (2016) Direct identification of antibiotic resistance genes on single plasmid molecules using CRISPR/Cas9 in combination with optical DNA mapping. Sci. Rep. 6, 37938 10.1038/srep37938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jeffet J., Michaeli-Hoch Y., Torchinsky D., Israel-Elgali I., Shomron N, Craggs T.D., et al. (2020) Multi-Modal Single-Molecule Imaging with Continuously Controlled Spectral-resolution (CoCoS) Microscopy. bioRxiv 10.1101/2020.10.13.330910 [DOI] [Google Scholar]
- 45.Jeffet J., Kobo A., Su T., Grunwald A., Green O., Nilsson A.N.et al. (2016) Super-resolution genome mapping in silicon nanochannels. ACS Nano 10, 9823–9830 10.1021/acsnano.6b05398 [DOI] [PubMed] [Google Scholar]
- 46.Wang Y., Reinhart W.F., Tree D.R. and Dorfman K.D. (2012) Resolution limit for DNA barcodes in the Odijk regime. Biomicrofluidics 6, 014101 10.1063/1.3672691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reinhart W.F., Reifenberger J.G., Gupta D., Muralidhar A., Sheats J., Cao H.et al. (2015) Distribution of distances between DNA barcode labels in nanochannels close to the persistence length. J. Chem. Phys. 142, 064902 10.1063/1.4907552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sheats J., Reifenberger J.G., Cao H. and Dorfman K.D. (2015) Measurements of DNA barcode label separations in nanochannels from time-series data. Biomicrofluidics 9, 064119 10.1063/1.4938732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Odijk T. (1983) The statistics and dynamics of confined or entangled stiff polymers. Macromolecules 16, 1340–1344 10.1021/ma00242a015 [DOI] [Google Scholar]
- 50.Su T., Das S.K., Xiao M. and Purohit P.K. (2011) Transition between two regimes describing internal fluctuation of DNA in a nanochannel. Kreplak L, editor. PLoS ONE 6, e16890 10.1371/journal.pone.0016890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ödman D., Werner E., Dorfman K.D., Doering C.R. and Mehlig B. (2018) Distribution of label spacings for genome mapping in nanochannels. Biomicrofluidics 12, 034115 10.1063/1.5038417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jain A., Sheats J., Reifenberger J.G., Cao H. and Dorfman K.D. (2016) Modeling the relaxation of internal DNA segments during genome mapping in nanochannels. Biomicrofluidics 10, 054117 10.1063/1.4964927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dorfman K.D. (2017) The statistical segment length of DNA: opportunities for biomechanical modeling in polymer physics and next-generation genomics. J. Biomech. Eng. 140, 020801 10.1115/1.4037790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Odijk T. (2008) Scaling theory of DNA confined in nanochannels and nanoslits. Phys. Rev. E. 77, 1–4 10.1103/PhysRevE.77.060901 [DOI] [PubMed] [Google Scholar]
- 55.Nyberg L., Persson F., Akerman B. and Westerlund F. (2013) Heterogeneous staining: a tool for studies of how fluorescent dyes affect the physical properties of DNA. Nucleic Acids Res. 41, e184–e184 10.1093/nar/gkt755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reisner W., Morton K.J., Riehn R., Wang Y.M., Yu Z., Rosen M.et al. (2005) Statics and dynamics of single DNA molecules confined in nanochannels. Phys. Rev. Lett. 94, 196101 10.1103/PhysRevLett.94.196101 [DOI] [PubMed] [Google Scholar]
- 57.Gupta D., Miller J.J., Muralidhar A., Mahshid S., Reisner W. and Dorfman K.D. (2015) Experimental evidence of weak excluded volume effects for nanochannel confined DNA. ACS Macro Lett. 4, 759–763 10.1021/acsmacrolett.5b00340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Persson F., Utko P., Reisner W., Larsen N.B. and Kristensen A. (2009) Confinement spectroscopy: probing single DNA molecules with tapered nanochannels. Nano Lett. 9, 1382–1385 10.1021/nl803030e [DOI] [PubMed] [Google Scholar]
- 59.Dai L., Renner C.B. and Doyle P.S. (2016) The polymer physics of single DNA confined in nanochannels. Adv. Colloid Interface Sci. 232, 80–100 10.1016/j.cis.2015.12.002 [DOI] [PubMed] [Google Scholar]
- 60.Chen J.Z.Y. (2018) Self-avoiding wormlike chain confined in a cylindrical tube: scaling behavior. Phys. Rev. Lett. 121, 037801 10.1103/PhysRevLett.121.037801 [DOI] [PubMed] [Google Scholar]
- 61.Bhandari A.B. and Dorfman K.D. (2019) Simulations corroborate telegraph model predictions for the extension distributions of nanochannel confined DNA. Biomicrofluidics 13, 044110 10.1063/1.5109566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bhandari A.B. and Dorfman K.D. (2020) Limitations of the equivalent neutral polymer assumption for theories describing nanochannel-confined DNA. Phys. Rev. E. 101, 012501 10.1103/PhysRevE.101.012501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Karpusenko A., Carpenter J.H., Zhou C., Lim S.F., Pan J. and Riehn R. (2012) Fluctuation modes of nanoconfined DNA. J. Appl. Phys. 111, 24701–247018 10.1063/1.3675207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cao H., Hastie A.R., Cao D., Lam E.T., Sun Y., Huang H.et al. (2014) Rapid detection of structural variation in a human genome using nanochannel-based genome mapping technology. Gigascience 3, 34 10.1186/2047-217X-3-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rust M.J., Bates M. and Zhuang X. (2006) Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–796 10.1038/nmeth929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hell S.W. and Wichmann J. (1994) Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt. Lett. 19, 780–782 10.1364/OL.19.000780 [DOI] [PubMed] [Google Scholar]
- 67.Betzig E., Patterson G.H., Sougrat R., Lindwasser O.W., Olenych S., Bonifacino J.S.et al. (2006) Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645 10.1126/science.1127344 [DOI] [PubMed] [Google Scholar]
- 68.Gordon M.P., Ha T. and Selvin P.R. (2004) Single-molecule high-resolution imaging with photobleaching. Proc. Natl. Acad. Sci. U.S.A. 101, 6462–6465 10.1073/pnas.0401638101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moerner W.E.W.E. (2015) Single-molecule spectroscopy, imaging, and photocontrol: foundations for super-resolution microscopy (Nobel Lecture). Angew. Chem. Int. Ed. Engl. 54, 8067–8093 10.1002/anie.201501949 [DOI] [PubMed] [Google Scholar]
- 70.Schermelleh L., Ferrand A., Huser T., Eggeling C., Sauer M., Biehlmaier O.et al. (2019) Super-resolution microscopy demystified. Nat. Cell Biol. 21, 72–84 10.1038/s41556-018-0251-8 [DOI] [PubMed] [Google Scholar]
- 71.Möckl L. and Moerner W.E. (2020) Super-resolution microscopy with single molecules in biology and beyond-essentials, current trends, and future challenges. J. Am. Chem. Soc. 142, 17828–17844 10.1021/jacs.0c08178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Neely R.K., Dedecker P., Hotta J.-I., Urbanavičiūtė G., Klimašauskas S. and Hofkens J. (2010) DNA fluorocode: a single molecule, optical map of DNA with nanometre resolution. Chem. Sci. 1, 453–460 10.1039/c0sc00277a [DOI] [Google Scholar]
- 73.Baday M., Cravens A., Hastie A., Kim H., Kudeki D.E., Kwok P.-Y.et al. (2012) Multicolor super-resolution DNA imaging for genetic analysis. Nano Lett. 12, 3861–3866 10.1021/nl302069q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chaisson M.J.P., Sanders A.D., Zhao X., Malhotra A., Porubsky D., Rausch T.et al. (2019) Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat. Commun. 10, 1784 10.1038/s41467-018-08148-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Soifer L., Fong N.L., Yi N., Ireland A.T., Lam I., Sooknah M.et al. (2020) Fully Phased Sequence of a Diploid Human Genome Determined de Novo from the DNA of a Single Individual. G3: Genes|Genomes|Genetics 10, 2911–2925 10.1534/g3.119.400995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Young E., Abid H.Z., Kwok P.Y., Riethman H. and Xiao M. (2020) Comprehensive analysis of human subtelomeres by whole genome mapping. PLos Genet. 16, 1–21 10.1371/journal.pgen.1008347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wong K.H.Y., Ma W., Wei C.-Y., Yeh E.-C., Lin W.-J., Wang E.H.F.et al. (2020) Towards a reference genome that captures global genetic diversity. Nat. Commun. 11, 5482 10.1038/s41467-020-19311-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Levy-Sakin M., Pastor S., Mostovoy Y., Li L., Leung A.K.Y., McCaffrey J.et al. (2019) Genome maps across 26 human populations reveal population-specific patterns of structural variation. Nat. Commun. 10, 1025 10.1038/s41467-019-08992-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Young E., Pastor S., Rajagopalan R., McCaffrey J., Sibert J., Mak A.C.Y.et al. (2017) High-throughput single-molecule mapping links subtelomeric variants and long-range haplotypes with specific telomeres. Nucleic Acids Res. 45, e73 10.1093/nar/gkx017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li Y., Roberts N.D., Wala J.A., Shapira O., Schumacher S.E., Kumar K.et al. (2020) Patterns of somatic structural variation in human cancer genomes. Nature 578, 112–121 10.1038/s41586-019-1913-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stankiewicz P. and Lupski J.R. (2010) Structural variation in the human genome and its role in disease. Annu. Rev. Med. 61, 437–455 10.1146/annurev-med-100708-204735 [DOI] [PubMed] [Google Scholar]
- 82.Chan E.K.F., Cameron D.L., Petersen D.C., Lyons R.J., Baldi B.F., Papenfuss A.T.et al. (2018) Optical mapping reveals a higher level of genomic architecture of chained fusions in cancer. Genome Res. 28, 726–738 10.1101/gr.227975.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hadi K., Yao X., Behr J.M., Deshpande A., Xanthopoulakis C., Tian H.et al. (2020) Distinct classes of complex structural variation uncovered across thousands of cancer genome graphs. Cell 183, 197.e32–210.e32 10.1016/j.cell.2020.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shieh J.T., Penon-Portmann M., Wong K.H.Y., Levy-Sakin M., Verghese M., Slavotinek A.et al. (2020) Application of full genome analysis to diagnose rare monogenic disorders. medRxiv 10.1101/2020.10.22.20216531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang Q., Xu X., Ding L., Li H., Xu C., Gong Y.et al. (2019) Clinical application of single‐molecule optical mapping to a multigeneration FSHD1 pedigree. Mol. Genetics & Genomic Med. 7, e565 10.1002/mgg3.565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gaillard M.-C., Roche S., Dion C., Tasmadjian A., Bouget G., Salort-Campana E.et al. (2014) Differential DNA methylation of the D4Z4 repeat in patients with FSHD and asymptomatic carriers. Neurology 83, 733–742 10.1212/WNL.0000000000000708 [DOI] [PubMed] [Google Scholar]
- 87.Huichalaf C., Micheloni S., Ferri G., Caccia R. and Gabellini D. (2014) DNA methylation analysis of the macrosatellite repeat associated with FSHD muscular dystrophy at single nucleotide level. PLoS ONE 9, e115278 10.1371/journal.pone.0115278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sharim H., Grunwald A., Gabrieli T., Michaeli Y., Margalit S., Torchinsky D.et al. (2019) Long-read single-molecule maps of the functional methylome. Genome Res. 29, 646–656 10.1101/gr.240739.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Allis C.D. and Jenuwein T. (2016) The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17, 487–500 10.1038/nrg.2016.59 [DOI] [PubMed] [Google Scholar]
- 90.Dor Y. and Cedar H. (2018) Principles of DNA methylation and their implications for biology and medicine. Lancet 392, 777–786 10.1016/S0140-6736(18)31268-6 [DOI] [PubMed] [Google Scholar]
- 91.Shi D.-Q., Ali I., Tang J. and Yang W.-C. (2017) New insights into 5hmC DNA modification: generation, distribution and function. Front Genet 8, 100 10.3389/fgene.2017.00100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Heck C., Michaeli Y., Bald I. and Ebenstein Y. (2019) Analytical epigenetics: single-molecule optical detection of DNA and histone modifications. Curr. Opin. Biotechnol. 55, 151–158 10.1016/j.copbio.2018.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen Z., Li S., Subramaniam S., Shyy J.Y.-J. and Chien S. (2017) Epigenetic regulation: a new frontier for biomedical engineers. Annu. Rev. Biomed. Eng. 19, 195–219 10.1146/annurev-bioeng-071516-044720 [DOI] [PubMed] [Google Scholar]
- 94.Klein D.C. and Hainer S.J. (2020) Genomic methods in profiling DNA accessibility and factor localization. Chromosome Res. 28, 69–85 10.1007/s10577-019-09619-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Holoch D. and Moazed D. (2015) RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 16, 71–84 10.1038/nrg3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cavalli G. and Heard E. (2019) Advances in epigenetics link genetics to the environment and disease. Nature 571, 489–499 10.1038/s41586-019-1411-0 [DOI] [PubMed] [Google Scholar]
- 97.Gilboa T., Torfstein C., Juhasz M., Grunwald A., Ebenstein Y., Weinhold E.et al. (2016) Single-molecule DNA methylation quantification using electro-optical sensing in solid-state nanopores. ACS Nano 10, 8861–8870 10.1021/acsnano.6b04748 [DOI] [PubMed] [Google Scholar]
- 98.Margalit S., Abramson Y., Sharim H., Manber Z., Bhattacharya S., Chen Y.-W.et al. (2021) Long reads capture simultaneous enhancer-promoter methylation status for cell-type deconvolution. bioRxiv 10.1101/2021.01.28.428654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gabrieli T., Michaeli Y., Avraham S., Torchinsky D., Juhasz M., Coruh C.et al. (2021) Chemoenzymatic labeling of DNA methylation patterns for single-molecule epigenetic mapping.. bioRxiv 10.1101/2021.02.24.432628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lim S.F., Karpusenko A., Sakon J.J., Hook J.A., Lamar T.A. and Riehn R. (2011) DNA methylation profiling in nanochannels. Biomicrofluidics 5, 34106–341068 10.1063/1.3613671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Michaeli Y., Shahal T., Torchinsky D., Grunwald A., Hoch R. and Ebenstein Y. (2013) Optical detection of epigenetic marks: sensitive quantification and direct imaging of individual hydroxymethylcytosine bases. Chem. Commun. 49, 8599–8601 10.1039/c3cc42543f [DOI] [PubMed] [Google Scholar]
- 102.Shahal T., Gilat N., Michaeli Y., Redy-Keisar O., Shabat D. and Ebenstein Y. (2014) Spectroscopic quantification of 5-hydroxymethylcytosine in genomic DNA. Anal. Chem. 86, 8231–8237 10.1021/ac501609d [DOI] [PubMed] [Google Scholar]
- 103.Gilat N., Tabachnik T., Shwartz A., Shahal T., Torchinsky D., Michaeli Y.et al. (2017) Single-molecule quantification of 5-hydroxymethylcytosine for diagnosis of blood and colon cancers. Clin. Epigenetics 9, 70 10.1186/s13148-017-0368-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nifker G., Levy-Sakin M., Berkov-Zrihen Y., Shahal T., Gabrieli T., Fridman M.et al. (2015) One-Pot Chemoenzymatic Cascade for Labeling of the Epigenetic Marker 5-Hydroxymethylcytosine. ChemBioChem 16, 1857–1860 10.1002/cbic.201500329 [DOI] [PubMed] [Google Scholar]
- 105.Margalit S., Avraham S., Shahal T., Michaeli Y., Gilat N., Magod P.et al. (2020) 5‐Hydroxymethylcytosine as a clinical biomarker: Fluorescence‐based assay for high‐throughput epigenetic quantification in human tissues. Int. J. Cancer 146, 115–122 10.1002/ijc.32519 [DOI] [PubMed] [Google Scholar]
- 106.Gabrieli T., Sharim H., Nifker G., Jeffet J., Shahal T., Arielly R.et al. (2018) Epigenetic Optical Mapping of 5-Hydroxymethylcytosine in Nanochannel Arrays. ACS Nano 12, 7148–7158 10.1021/acsnano.8b03023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zirkin S., Fishman S., Sharim H., Michaeli Y., Don J. and Ebenstein Y. (2014) Lighting up individual DNA damage sites by in vitro repair synthesis. J. Am. Chem. Soc. 136, 7771–7776 10.1021/ja503677n [DOI] [PubMed] [Google Scholar]
- 108.Gilat N., Torchinsky D., Margalit S., Michaeli Y., Avraham S., Sharim H.et al. (2020) Rapid Quantification of Oxidation and UV Induced DNA Damage by Repair Assisted Damage Detection-(Rapid RADD). Anal. Chem. 92, 9887–9894 10.1021/acs.analchem.0c01393 [DOI] [PubMed] [Google Scholar]
- 109.Torchinsky D., Michaeli Y., Gassman N.R. and Ebenstein Y. (2019) Simultaneous detection of multiple DNA damage types by multi-colour fluorescent labelling. Chem. Commun. 55, 11414–11417 10.1039/C9CC05198H [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang W., Klein K., Proesmans K., Yang H., Marchal C., Zhu X.et al. (2020) Genome-wide mapping of human DNA replication by optical replication mapping supports a stochastic model of eukaryotic replication. bioRxiv 10.1101/2020.08.24.263459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lacroix J., Pélofy S., Blatché C., Pillaire M.-J., Huet S., Chapuis C.et al. (2016) Analysis of DNA Replication by Optical Mapping in Nanochannels. Small 12, 5963–5970 10.1002/smll.201503795 [DOI] [PubMed] [Google Scholar]
- 112.De Carli F., Menezes N., Berrabah W., Barbe V., Genovesio A. and Hyrien O. (2018) High-throughput optical mapping of replicating DNA. Small Methods 2, 1800146 10.1002/smtd.201800146 [DOI] [Google Scholar]
- 113.Ratzke C., Hellenkamp B. and Hugel T. (2014) Four-colour FRET reveals directionality in the Hsp90 multicomponent machinery. Nat. Commun. 5, 4192 10.1038/ncomms5192 [DOI] [PubMed] [Google Scholar]
- 114.Zhang Y., Song K.-H., Dong B., Davis J.L., Shao G., Sun C.et al. (2019) Multicolor super-resolution imaging using spectroscopic single-molecule localization microscopy with optimal spectral dispersion. Appl. Opt. 58, 2248 10.1364/AO.58.002248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Moon S., Yan R., Kenny S.J., Shyu Y., Xiang L., Li W.et al. (2017) Spectrally Resolved, Functional Super-Resolution Microscopy Reveals Nanoscale Compositional Heterogeneity in Live-Cell Membranes. J. Am. Chem. Soc. 139, 10944–10947 10.1021/jacs.7b03846 [DOI] [PubMed] [Google Scholar]
- 116.Mlodzianoski M.J., Curthoys N.M., Gunewardene M.S., Carter S. and Hess S.T. (2016) Super-resolution imaging of molecular emission spectra and single molecule spectral fluctuations. PLoS ONE 11, 1–12 10.1371/journal.pone.0147506 [DOI] [PMC free article] [PubMed] [Google Scholar]