Abstract
Obstructive sleep apnea-hypopnea (OSAH) is correlated with an increased incidence of lung cancer. In our study, we explored the functional roles of microRNAs (miRNAs) in lung cancer patients that were complicated with OSAH involving the deubiquitination enzyme. The miR-320b expression pattern in lung cancer tissues and cells was determined. The interactions between ubiquitin-specific peptidase 37 (USP37) and miR-320b were evaluated by a dual-luciferase reporter gene assay, whereas USP37 and Cdc10-dependent transcript 1 (CDT1) was assessed by co-immunoprecipitation and immunofluorescence. After the induction of intermittent hypoxia (IH), a gain-of function approach was performed to investigate roles of miR-320b, USP37, and CDT1 in lung cancer cell proliferation and invasion. In addition, nude mouse xenograft models were used to study their effects on tumor growth in vivo. miR-320b was poorly expressed in lung cancer patients with OSAH. IH treatment downregulated the expression of miR-320b but promoted the proliferation and invasion capabilities of lung cancer cells, both of which were suppressed by the overexpression of miR-320b through decreasing USP37. USP37 interacted with and deubiquitinated CDT1 to protect it from proteasomal degradation. Our study uncovered that IH-induced downregulation of miR-320b promoted the tumorigenesis of lung cancer by the USP37-mediated deubiquitination of CDT1.
Keywords: obstructive sleep apnea-hypopnea, lung cancer, microRNA-320b, USP37, CDT1, deubiquitinase
Graphical Abstract
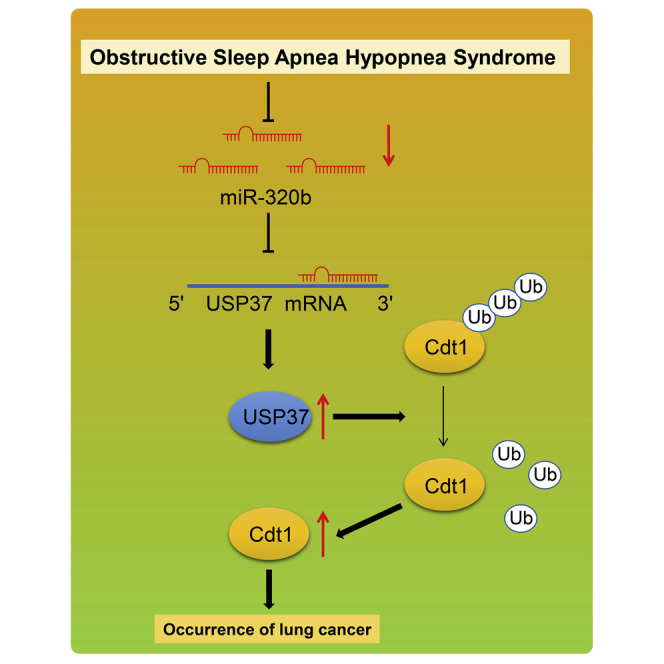
A series of in vivo and in vitro experiments were performed to investigate the functional roles of miR-320b in lung cancer complicated with OSAH. We identified poorly expressed miR-320b in lung cancer with OSAH. It uncovered that intermittent hypoxia-induced miR-320b downregulation promoted lung cancer tumorigenesis via the USP37-mediated deubiquitination of CDT1.
Introduction
Lung cancer, one of the most frequently diagnosed cancers in the world, still remains the number one cause of cancer-related deaths in men and women, accounting for approximately 1.6 million deaths each year.1 Long-term tobacco smoking is responsible for the majority of the incidence of lung cancer; however, approximately 15 percent of lung cancer cases develop in individuals with no prior history of smoking.2 More than two-thirds of patients are diagnosed with locally advanced or metastatic lung cancer, thus rendering therapeutic approaches with only limited effects on survival. Moreover, the 5-year survival rate is less than 20% across most countries in the world.3 These findings have highlighted the inefficacy of current therapeutic regimens. Therefore, a better understanding of lung cancer pathogenesis will be critical in improving the diagnosis and treatment of lung cancer.
Hypoxia, a condition where tissues in the whole body do not receive adequate levels of oxygenation, has been shown to promote genomic instability, metastasis, and aggressiveness of multiple solid tumors, including lung cancer.4 The phenomenon is restricted to cancer cells, shedding light on the therapeutic potential of hypoxia-regulated factors.5 Recent studies have associated the tumorigenesis of lung cancer with obstructive sleep apnea-hypopnea (OSAH), having a predilection of targeting the upper lobes of the lungs,6 leading to intermittent hypoxia (IH).7,8 It has been suggested that lung cancer cells exposed to IH display increased aggressiveness, as reflected by enhanced cell survival and improved metastasis.9 However, the underlying mechanisms by which IH promotes lung cancer tumorigenesis and progression is not yet fully understood.
Recent evidence suggests that the expression of miR-320b is downregulated in patients with OSAH.10 Low expressions of miR-320b have been shown to favor the initiation and progression of different cancers, most notably lung cancer.11,12 Furthermore, recent studies have suggested that the expression levels of miR-320b can be used as a promising diagnostic and prognostic biomarker of lung cancer.11, 12, 13 Therefore, it is possible that OSAH-associated IH promotes lung cancer initiation and progression by regulating miR-320b. In this study, we explored the role of miR-320b in IH-induced lung cancer tumorigenesis and progression in vitro and in vivo. Our data illustrated that upon the treatment of IH, miR-320b was subsequently downregulated, which had led to increased cancer cell proliferation and invasion. We also identified the underlying mechanism by which downregulated miR-320b promoted the initiation and development of lung cancer.
Results
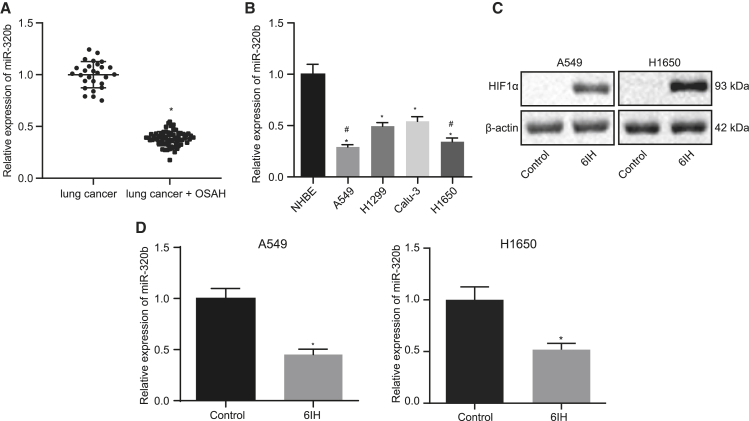
miR-320b is poorly expressed in lung cancer patients with OSAH and lung cancer cells exposed to IH
Recent studies have associated OSAH to the development of hypoxic-induced lung cancer.7,14 Mounting evidence has revealed that miR-320b, which has been shown to inhibit the tumorigenesis of lung cancer,15 is poorly expressed in patients with OSAH.10 These findings indicate that miR-320b may be involved in the tumorigenesis of lung cancer patients with OSAH. To identify the role of miR-320b in regulating tumor progression in lung cancer patients with OSAH, we evaluated the expression of miR-320b in lung cancer patients with or without OSAH. The results indicated that lung cancer patients with OSAH expressed lower levels of miR-320b compared with patients without OSAH (Figure 1A). We also compared the expressions of miR-320b in normal bronchial epithelial cell line (NHBEs) and multiple lung cancer cell lines (A549, H1299, Calu-3, and H1650) and discovered that the expression of miR-320b was significantly lower in lung cancer cells than that in NHBEs (Figure 1B). We also observed that among the lung cancer cell lines, A549 and H1650 expressed significantly lower levels of miR-320b (Figure 1B). Therefore, A549 and H1650 were selected for further studies. We then evaluated the effects of IH on the expression of miR-320b and hypoxia inducible factor 1α (HIF1α) and found that 6IH had significantly decreased the expression of miR-320b and increased the expression of HIF1α in lung cancer cells (Figures 1C and 1D), suggesting that cells were successfully treated with IH. Therefore, our data demonstrated that the expression of miR-320b was downregulated in lung cancer patients with OSAH and treatment with IH resulted in a decrease of miR-320b in lung cancer cells.
Figure 1.
miR-320b is poorly expressed in lung cancer patients with OSAH- and IH-treated lung cancer cells
(A) qRT-PCR detecting the expression of miR-320b in lung cancer patients with OSAH (n = 27) or without OSAH (n = 68) (∗p < 0.05; lung cancer + OSAH versus lung cancer). (B) qRT-PCR detecting miR-320b levels in normal bronchial epithelial cells and lung cancer cells (∗p < 0.05, H1299 and Calu-3 versus NHBE; #p < 0.05, A549 and H1650 versus H1299 and Calu-3). (C) Western blot analysis for determining the expression level of HIF1α normalized to β-actin in lung cancer cells (∗p < 0.05, 6IH versus control). (D) qRT-PCR detecting the expression of miR-320b in lung cancer cells treated with or without 6IH (∗p < 0.05, 6IH versus control). Data are presented as the mean ± standard deviation. Data between two groups were compared using an unpaired t test. Comparisons among multiple groups were performed using one-way ANOVA with Tukey’s post hoc test.
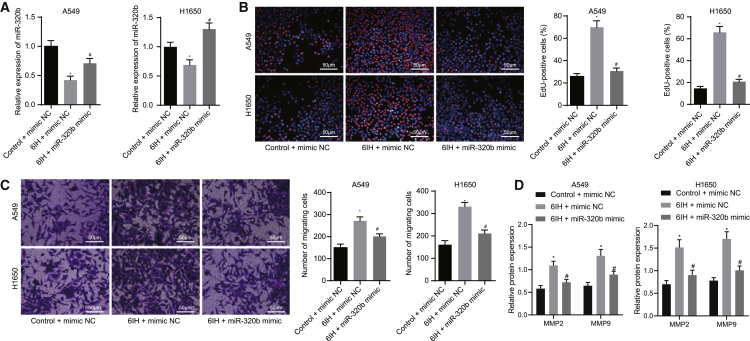
Overexpression of miR-320b inhibits proliferation and invasion in lung cancer cells exposed to IH
Subsequent experiments were performed to investigate the action of miR-320b in lung cancer cells. miR-320b has been reported to inhibit the tumorigenesis of lung cancer.15 Meanwhile, IH treatment has been shown to decrease the expression of miR-320b in lung cancer cells. Thus, it is reasonable to speculate that aberrantly expressed miR-320b is involved in the development of hypoxic-induced lung cancer. To test this hypothesis, we modified the expression of miR-320b in A549 and H1650 cells and validated the modification by quantitative reverse transcription polymerase chain reaction (qRT-PCR) (Figure 2A). The results revealed significantly downregulated expression of miR-320b in cells treated with 6IH, whereas miR-320b expression was increased in the presence of miR-320b mimic. Furthermore, we explored the role of miR-320b in IH-mediated lung cancer cell growth by performing a 5-ethynyl-2′-deoxyuridine (EdU) proliferation assay. The results suggested that 6IH treatment significantly promoted the proliferation of A549 and H1650 cells, as reflected by increased EdU-labeled nuclei (red fluorescence). However, the promotive effect of 6IH was impaired by the overexpression of miR-320b, as reflected by reduced EdU-labeled nuclei (Figure 2B). We then investigated the role of miR-320b in IH-induced lung cancer cell invasive potency by using Transwell invasion assay. We noticed that 6IH treatment increased the number of invaded cells while the promotive effect was neutralized by miR-320b mimic in lung cancer cells (A549 and H1650) (Figure 2C). We further evaluated the effect of miR-320b on the expression of tumor invasion and metastasis-related proteins, matrix metallopeptidase 2 (MMP-2) and MMP-9, in lung cancer cells exposed to 6IH treatment by western blot analysis. The results revealed that 6IH elevated the protein levels of MMP-2 and MMP-9, while the overexpression of miR-320b compromised the action of 6IH on lung cancer cells (Figure 2D). These findings suggested that miR-320b inhibited IH-mediated lung cancer progression by inhibiting cancer cell proliferation and invasion.
Figure 2.
Overexpression of miR-320b inhibits IH-induced lung cancer cell proliferation and invasion
(A) qRT-PCR detecting levels of miR-320b in A549 and H1650 cells. (B) Representative images and results of EdU proliferation evaluating the growth of lung cancer cell (A549 and H1650) with altered miR-320b expression levels (magnification 200×). (C) Representative images and results of Transwell invasion assay measuring the effect of miR-320b on lung cancer cell (A549 and H1650) invasion potency (magnification 200×). (D) Representative images and results of immunoblotting analysis detecting MMP-2 and MMP-9 proteins in lung cancer cells with altered levels of miR-320b. ∗p < 0.05, 6IH + mimic NC versus control + mimic NC; #p < 0.05, 6IH + mimic NC versus 6IH + miR-320b mimic. Data are presented as the mean ± standard deviation. Data between two groups were compared using an unpaired t test. Comparisons among multiple groups were performed using one-way ANOVA with Tukey’s post hoc test.
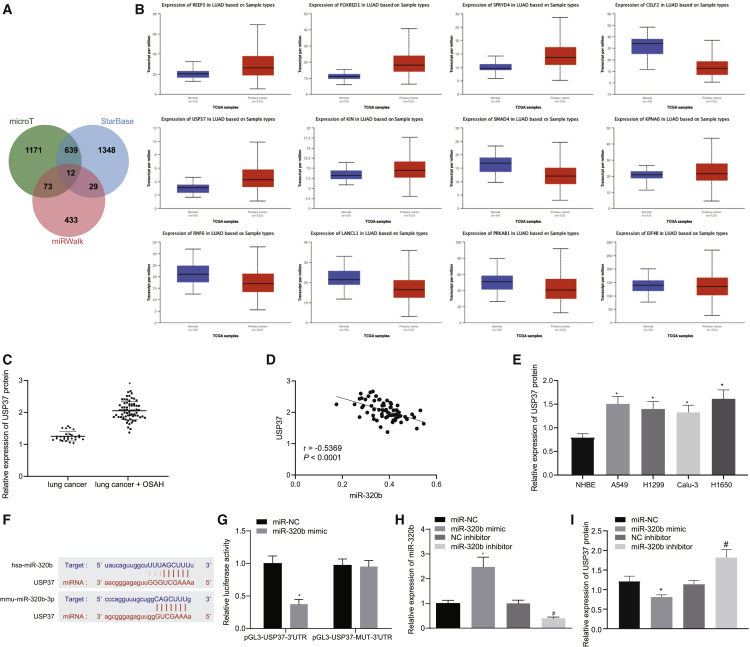
miR-320b targets ubiquitin-specific peptidase 37 (USP37)
We further pondered how miR-320b is involved in the tumorigenesis of lung cancer. Thus, we aimed to identify the targets of miR-320b in lung cancer by conducting a bioinformatics analysis. Using online prediction tools (microT, StarBase, and miRWalk), 12 potential targets of miR-320b were predicted in the intersection (Figure 3A). We then compared the expressions of those candidate genes in lung cancer and normal lung tissues by using datasets obtained from The Cancer Genome Atlas (TCGA) database from a web-based analyzing site (UALCAN) (Figure 3B). The results presented that 6 genes were significantly upregulated in lung cancer (Table 1). Among all of the candidate genes, USP37 was selected because of its ability to help promote the initiation and progression of lung cancer.16 The expression of USP37 was then evaluated in cancer tissues from patients with or without OSAH by western blot analysis, and the results revealed that lung cancer patients with OSAH expressed higher levels of USP37 than those without OSAH (Figure 3C). The results of correlation analysis between miR-320b and USP37 indicated an inverse relationship (Figure 3D). Our immunoblotting analysis results also indicated that USP37 protein levels were significantly lower in normal bronchial epithelial cells (NHBEs) than that in cancer cells (A549, H1650, Calu-3, and H1299) (Figure 3E). Therefore, it is possible that USP37 may be involved in OSAH-associated lung cancer tumorigenesis. Using online prediction tools, we predicted the targeting site of miR-320b on USP37 (Figure 3F). To validate the results from the predictive target sites, we performed dual-luciferase reporter gene assay. The 3′ untranslated region (3′ UTR) of USP37 containing wild-type (WT) targeting sequence (pGL3-USP37-3′UTR) or mutated (MUT) targeting sequence (pGL3-USP37-MUT-3′ UTR) were cloned into reporter plasmid and transfected into HEK293T cells. It was observed that the luciferase activity was significantly reduced in cells that have been transfected with miR-320b mimic and pGL3-USP37-3′ UTR, but not in those transfected with miR-320b mimic and pGL3-USP37-MUT-3′ UTR (Figure 3G). We also modified the expression levels of miR-320b by transfecting miR-320b mimic or inhibitor, followed by evaluating the expression level of USP37. The overexpression of miR-320b significantly reduced the protein level of USP37 while inhibiting miR-320b resulted in an increased expression of USP37 (Figures 3H and 3I). These findings suggested that miR-320b targeted USP37 and mediated its downregulation.
Figure 3.
miR-320b targets USP37 in lung cancer cells
(A) Different online tools (microT, StarBase, and miRWalk) predicted miR-320b targets visualized by a Venn diagram. (B) Expression of the 12 predicted targets of miR-320b in lung cancer tissues and normal lung tissues analyzed by UALCAN website. (C) Representative images and results of immunoblotting analysis detecting the expression of USP37 in lung tissues obtained from lung cancer patients with OSAH (n = 27) or without OSAH (n = 68) (∗p < 0.05, lung cancer + OSAH versus lung cancer). (D) Correlation analysis between miR-320b and USP37 in patients with lung cancer and OSAH. (E) Representative images and results of western blot analysis detecting the expression of USP37 in lung cancer cells and NHBE cells (∗p < 0.05, A549, H1299, Calu-3, and H1650 versus NHBE). (F) Predicted miR-320b targeting sequences on USP37 3′ UTR. (G) Dual-luciferase reporter gene assay evaluating the interactions between miR-320b and predicted sequences on USP37 3′ UTR (∗p < 0.05, miR-320b mimic versus miR-NC). (H) qRT-PCR evaluating the expression of miR-320b in lung cancer cells transfected with miR-320b mimic, miR-320b inhibitor, and NCs (∗p < 0.05, miR-320b mimic versus miR-NC; #p < 0.05, miR-320b inhibitor versus NC inhibitor). (I) Representative images and results of western blot analysis measuring the expression of USP37 in lung cancer cells transfected with miR-320b mimic, miR-320b inhibitor, and NCs (∗p < 0.05, miR-320b mimic versus miR-NC; #p < 0.05, miR-320b inhibitor versus NC inhibitor). Data are presented as the mean ± standard deviation. Data between two groups were compared using an unpaired t test. Comparisons among multiple groups were performed using one-way ANOVA with Tukey’s post hoc test. The correlation between miR-320b and USP37 was analyzed by Pearson correlation coefficient.
Table 1.
Comparison of candidate genes in TCGA database between lung cancer samples and normal samples
| Gene | Comparison | Statistical significance | Expression |
|---|---|---|---|
| REEP3 | normal versus primary | 1.6243673 × 10–12 | up |
| FOXRED1 | normal-versus-primary | 1.6243673 × 10-12 | up |
| SPRYD4 | normal versus primary | 1.6243673 × 10-12 | up |
| CELF2 | normal versus primary | 1.6244783 × 10-12 | down |
| USP37 | normal versus primary | 1.6244783 × 10-12 | up |
| KIN | normal versus primary | 1.2927004 × 10-10 | up |
| SMAD4 | normal versus primary | 1.2367000 × 10-9 | down |
| KPNA6 | normal versus primary | 1.0374000 × 10-6 | up |
| RNF6 | normal versus primary | 6.2391000 × 10-5 | down |
| LANCL1 | normal versus primary | 3.5555000 × 10-4 | down |
| PRKAB1 | normal versus primary | 1.7902200 × 10-1 | down |
| EIF4B | normal versus primary | 2.1964000 × 10-1 | down |
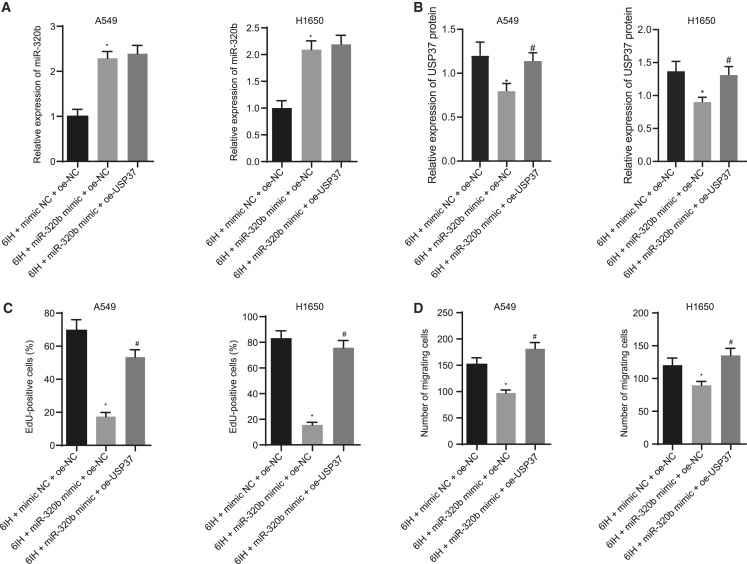
miR-320b inhibits IH-mediated lung cancer progression by targeting USP37
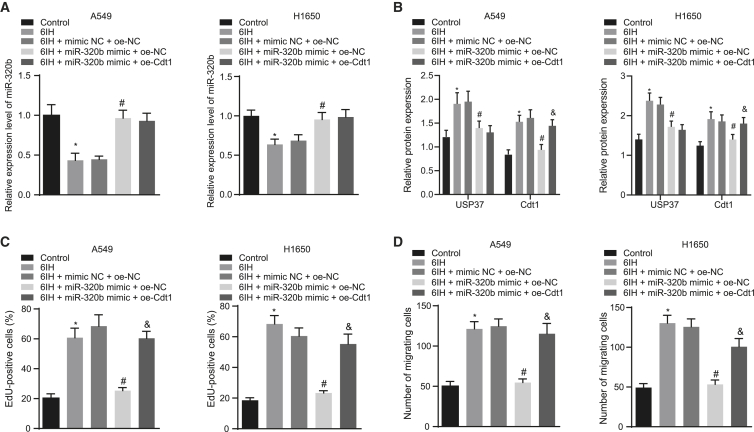
Our findings have suggested that USP37 may be involved in the tumorigenesis of IH-induced lung cancer. miR-320b, which was downregulated in lung cancer patients with OSAH, can target USP37 and mediate its downregulation. We further investigated the role of miR-320b in USP37 involved in IH-mediated lung cancer progression using lung cancer cell lines (A549 and H1650). Subsequently, we altered the expressions of miR-320b and/or USP37 and validated the modulation (Figures 4A and 4B). First, we performed qRT-PCR assay to determine the resultant expression levels of miR-320b and USP37. Results showed that following 6IH, miR-320b expression increased and USP37 expression decreased in response to the miR-320b mimic. However, when USP37 and miR-320b were overexpressed at the same time, miR-320b expression did not differ significantly, and USP37 expression was elevated. Then, EdU proliferation assay was conducted to study the role of miR-320b in USP37 involved in 6IH-induced lung cancer growth. The results suggested that miR-320b overexpression reduced 6IH-induced lung cancer cell proliferation; however, this inhibitory effect was impaired by further overexpressing USP37 in those cells (Figures 4C and S1A). This suggests that miR-320b inhibited 6IH-promoted lung cancer proliferation by targeting USP37. We then performed a Transwell invasion assay to evaluate the role of miR-320b in USP37-associated IH-induced lung cancer metastasis and discovered that 6IH-induced lung cancer invasion was inhibited by miR-320b mimic. However, co-transfecting miR-320b mimic and USP37 failed to inhibit IH-induced lung cancer cell invasion (Figures 4D and S1B), indicating that miR-320b inhibited 6IH-induced invasion in a USP37-dependent manner. Taken together, our data suggested that miR-320b inhibited IH-mediated lung cancer progression by targeting USP37.
Figure 4.
miR-320b inhibits IH-induced lung cancer cell proliferation and invasion by targeting USP37
(A) qRT-PCR evaluating the expression of miR-320b in lung cancer cells under different conditions. (B) Immunoblotting analysis detecting the expression of USP37 in lung cancer cells. (C) Results of EdU proliferation assay measuring lung cancer cell growth. (D) Results of Transwell invasion assay investigating lung cancer cell invasion potency (∗p < 0.05, 6IH + miR-320b mimic + oe-NC versus 6IH + mimic NC + oe-NC; #p < 0.05, 6IH + miR-320b + oe-USP37 versus 6IH + miR-320b + oe-NC). Data are presented as the mean ± standard deviation. Data between two groups were compared using an unpaired t test. Comparisons among multiple groups were performed using one-way ANOVA with Tukey’s post hoc test.
USP37 promoted the expression of Cdc10-dependent transcript 1 (CDT1) in a deubiquitinase activity-dependent manner
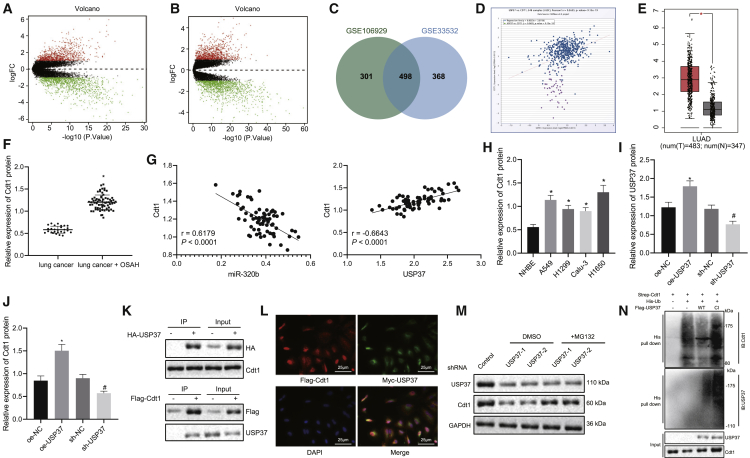
The differential expressed genes (DEGs) were further analyzed from the datasets (Gene Expression Omnibus [GEO]: GSE106929 and GSE33532) obtained from the GEO database. We found 1,968 DEGs, including 799 upregulated genes and 1,169 downregulated genes in the GEO: GSE106929 dataset (Figure 5A). We also found 2,128 DEGs consisting of 866 upregulated genes and 1,262 downregulated genes in the GEO: GSE33532 dataset (Figure 5B). We then identified 498 overlapped DEGs in the intersection of both datasets (Figure 5C). The genes were rendered functional, as classified in the DAVID bioinformatics resource (https://david.ncifcrf.gov/home.jsp) and are comprised of 6 apoptosis-associated pathways (Table 2). Among all, cyclin-dependent kinase (CDK) regulation of DNA replication pathway was analyzed with the highest level of significance (lowest p value). Therefore, we further examined the 6 enriched genes in this pathway using an online tool (Chipbase) and found that USP37 and CDT1 were closely associated with lung cancer (Figure 5D). Moreover, previous evidence has shown that USP37 promotes the expression of CDT1.17 The upregulation of CDT1 in lung cancer was also supported by data obtained from the TCGA database (Figure 5E). Thus, we hypothesized that USP37 may contribute to the tumorigenesis of lung cancer by regulating CDT1. To test this hypothesis, we evaluated the expression of CDT1 by performing western blot analysis in patients with lung cancer and OSAH, who exhibited significantly upregulated CDT1 than those only with lung cancer (Figure 5F). Further correlation analyses between miR-320b and CDT1 as well as between USP37 and CDT1 showed that miR-320b was negatively correlated with CDT1, while USP37 was positively correlated with CDT1 (Figure 5G). CDT1 expression was measured in lung cancer cells (A549, H1299, Calu-3, and H1650) and NHBEs, and it was found that lung cancer cells expressed higher levels of CDT1 than that in NHBEs (Figure 5H). We then altered the expression of USP37 in lung cancer cells and measured the expression of CDT1. The results revealed that the overexpression of USP37 increased CDT1 protein levels and USP37 silencing decreased CDT1 expressions (Figures 5I and 5J). Interestingly, accumulating evidence has shown that USP37 stabilizes proteins by its deubiquitinating activity.18,19 Therefore, it is possible that USP37 utilizes the same approach to regulate CDT1 protein levels. In order to confirm this hypothesis, we performed a co-immunoprecipitation assay followed by an immunoblotting analysis, detecting CDT1 and USP37, respectively, in order to evaluate the interactions between USP37 and CDT1. We observed that CDT1 could be co-immunoprecipitated by USP37 and vice versa (Figure 5K). Moreover, immunofluorescence staining was conducted to detect the subcellular localization of these proteins. The results showed that USP37 and CDT1 were co-localized in nuclei (Figure 5L). These findings highlighted the possibility that USP37 could stabilize CDT1 by interacting with and deubiquitinating it. To explore whether USP37 mediates the proteasomal degradation of CDT1, we treated USP37-silencing cells with MG132 or dimethyl sulfoxide (DMSO) and detected the expression of CDT1 afterward. The results illustrated that USP37 knockdown significantly reduced CDT1 protein levels while lysosomal inhibitor (MG132) preserved the CDT1 protein (Figure 5M). Last, ubiquitination assay was carried out and results revealed that, relative to CDT1 alone expression, the co-expression of WT USP37 and CDT1 resulted in decreased ubiquitination level of CDT1, while the co-expression of the MUT type USP37 without deubiquitination activity (CI USP37) and CDT1 showed no obviously changed ubiquitination level of CDT1. Besides, the overexpression of CI USP37 significantly increased the ubiquitination level of USP37 (Figure 5N). This suggested that USP37 downregulation-induced CDT1 reduction was mediated by the ubiquitin-proteasomal pathway.
Figure 5.
USP37 promotes the expression of CDT1 by deubiquitination
(A) Differential expressed genes in the dataset obtained from the GEO database (GEO: GSE106929) represented in a volcano plot (red dots indicate upregulated genes and green dots indicate downregulated genes in lung tumor samples). (B) Differentially expressed genes in the dataset obtained from the GEO database (GEO: GSE33532) represented in a volcano plot (red dots indicate upregulated genes and green dots indicate downregulated genes in lung tumor samples). (C) Overlapped upregulated genes in datasets GEO: GSE106929 and GSE33532 visualized by a Venn diagram. (D) Expression levels of USP37 and CDT1 were co-related by an online tool (Chipbase). (E) Relative expression levels of CDT1 in lung tumor (red box) and normal lung tissue (gray box) analyzed by TCGA database. (F) Representative images and results of immunoblotting analysis evaluating CDT1 expression in lung tissues obtained from lung cancer patients with OSAH (n = 27) or without OSAH (n = 68) (∗p < 0.05, lung cancer + OSAH versus lung cancer). (G) Correlation analysis between miR-320b and CDT1 as well as between USP37 and CDT1 in lung cancer patients with OSAH. (H) Representative images and results of immunoblotting analysis evaluating CDT1 expression in normal bronchial epithelial cells and lung cancer cells (∗p < 0.05, lung cancer cells versus NHBE cells). (I) Representative images and results of immunoblotting analysis validating the overexpression or knockdown of USP37 (∗p < 0.05, oe-USP37 versus oe-NC; #p < 0.05, sh-USP37 versus sh-NC). (J) Representative images and results of immunoblotting analysis evaluating CDT1 expression in lung cancer cells with the overexpression or knockdown of USP37 (∗p < 0.05, oe-USP37 versus oe-NC; #p < 0.05, sh-USP37 versus sh-NC). (K) Representative images of co-immunoprecipitation assay investigating the interactions between USP37 and CDT1. (L) Representative images of immunofluorescent staining showing co-localization of USP37 and CDT1 (magnification 400×). (M) Representative images of immunoblotting analysis evaluating the expressions of USP37 and CDT1. (N) Ubiquitination assay detecting ubiquitination levels of USP37 and CDT1. Data are presented as the mean ± standard deviation. Data between two groups were compared using an unpaired t test. Comparisons among multiple groups were performed using one-way ANOVA with Tukey’s post hoc test. The correlation between two indicators was analyzed by Pearson correlation coefficient.
Table 2.
Apoptosis-associated pathways
| Term | Count | p value | Gene |
|---|---|---|---|
| CDK regulation of DNA replication | 6 | 1.30 × 10-4 | CCNE1, CDC6, ORC6, MCM2, MCM4, CDT1 |
| Cell cycle | 4 | 3.70 × 10-2 | CCNB1, CDK1, PLK1, CHEK1 |
| Cyclins and cell cycle regulation | 4 | 3.70 × 10-2 | CCNB1, CCNE1, CDK1, CDKN2A |
| Role of Ran in mitotic spindle regulation | 3 | 4.10 × 10-2 | KIF15, TPX2, AURKA |
| Cell cycle | 4 | 5.90 × 10-2 | CCNE1, CDK1, CDKN2A, DHFR |
| Estrogen-responsive protein Efp controls cell cycle and breast tumors growth | 3 | 8.20 × 10-2 | CCNB1, CDK1, CCNB2 |
miR-320b impaired IH-mediated lung cancer progression by inhibiting CDT1 expression through targeting USP37
To understand whether the regulation of CDT1 expression by USP37 plays a role in lung cancer cell growth and invasion, the expressions of miR-320b and/or CDT1 in lung cancer cells (A549 and H1650) were altered. Cell proliferation and invasion were evaluated after cells were treated with or without 6IH. The expression of miR-320b was evaluated by qRT-PCR (Figure 6A), and the expression of USP37 and CDT1 was measured by western blot analysis (Figure 6B). The results revealed that 6IH treatment could decrease the expression of miR-320b yet increase that of USP37 and CDT1, and the overexpression of miR-320b could downregulate the expressions of USP37 and CDT1, while the effect of miR-320b mimic on the expressions of miR-320b, USP37, and CDT1 could be counteracted by overexpressed CDT1 (oe-CDT1). We found that the promoting effect of 6IH treatment on lung cancer cell proliferation was impaired by the overexpression of miR-320b, which was reflected by reduced numbers of EdU-labeled (red fluorescence) lung cancer cells in the 6IH-miR-320b mimic + oe-NC group. However, further overexpressing CDT1 compromised the inhibitory effect achieved by overexpressed miR-320b, which was reflected by increased EdU-labeled (red fluorescence) lung cancer cells in 6IH-miR-320b mimic + oe-CDT1 group (Figures 6C and S1C). We also noted that 6IH- promoted cancer cell invasion was inhibited by the overexpression of miR-320b, as reflected by a decreased number of invaded cells observed in 6IH-miR-320b mimic + oe-NC group. On the other hand, cells co-transfected with miR-320b mimic and oe-CDT1 failed to inhibit 6IH-induced cell invasion (Figures 6D and S1D). Taken together, our study suggested that miR-320b suppressed CDT1 by targeting USP37 to inhibit the progression of lung cancer.
Figure 6.
miR-320b inhibits lung cancer cell proliferation and invasion by inhibiting CDT1 through targeting USP37
(A) qRT-PCR detecting the expression of miR-320b in lung cancer cells (A549 and H1650). (B) Immunoblotting analysis detecting the expressions of USP37 and CDT1 in lung cancer cells. (C) Results of EdU proliferation evaluating lung cancer cell growth. (D) Results of Transwell invasion assay measuring the invasion potency of lung cancer cells (∗p < 0.05, 6IH versus control; #p < 0.05, 6IH + miR-320b mimic + oe-NC versus 6IH + mimic NC + oe-NC; &p < 0.05, 6IH + miR-320b mimic + oe-CDT1 versus 6IH + miR-320b + oe-NC). Data are presented as the mean ± standard deviation. Data between two groups were compared using an unpaired t test. Comparisons among multiple groups were performed using one-way analysis of variance (ANOVA) with Tukey’s post hoc test.
miR-320b inhibited tumorigenesis in vivo by inhibiting CDT1 expression through targeting USP37
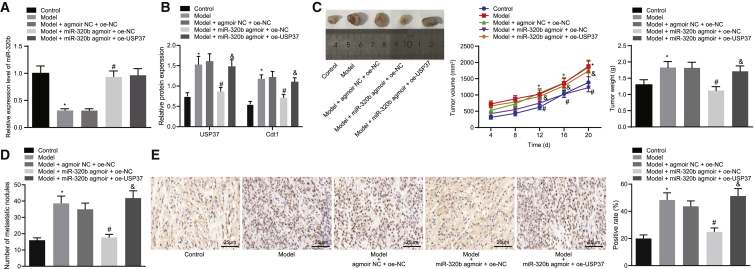
We also confirmed our findings in a mouse model of human lung cancer xenograft. Lung cancer cells that have undergone different treatments were delivered into nude mice, and tumor growth was monitored and compared. The expression of miR-320b, USP37, and CDT1 was measured by qRT-PCR and western blot analysis, respectively (Figures 7A and 7B). We found that the expression of miR-320b was decreased while that of USP37 and CDT1 were increased in mice bearing IH-treated cells, and miR-320b agmoir significantly increased the expression of miR-320b and reduced the expressions of USP37 and CDT1. We further evaluated the tumor progression by comparing the tumor size, tumor weight, growth curve, and the number of tumor nodules on the lung surface among different groups. The results indicated that IH treatment led to a promoted tumorigenesis, as reflected by increased tumor size and tumor weight with more tumor nodules observed on the lung surface in the model group. Moreover, the overexpression of miR-320b significantly impaired IH-promoted tumor progression, as reflected by decreased tumor size and tumor weight with fewer tumor nodules detected on the lung surface in the model + miR-320b agmoir + oe-negative control (oe-NC) group. We also observed that the inhibitory effect of miR-320b agmoir on tumor growth was neutralized by the overexpression of USP37, as reflected by the increased tumor size and weight, with more tumor nodules present on the lung surface (Figures 7C and 7D). Additionally, Ki67 expression in tumor tissues was identified by immunohistochemistry, results of which revealed significantly lower Ki67 expression levels in the model group, while higher expression of Ki67 was observed following HI-treatment and delivery of miR-320b agmoir. Further addition of oe-USP37 significantly increased the expression of Ki67 (Figure 7E). Taken together, our findings supported the hypothesis that miR-320b inhibited the tumorigenesis and progression of lung cancer by inhibiting CDT1 expression through targeting USP37.
Figure 7.
miR-320b inhibits tumorigenesis and development of lung cancer in vivo by regulating USP37/CDT1
(A) qRT-PCR evaluating the expression of miR-320b in lung cancer cells. (B) Representative images and results of immunoblotting analysis measuring the protein levels of USP37 and CDT1 in lung cancer cells. (C) Images of xenograft tumors at endpoint (left panel), tumor growth curve (middle panel), and final tumor weight (right panel) reflecting the development of lung tumor in vivo. (D) Representative images of tumor nodules on the lung surface. (E) Immunohistochemistry detecting the expression of Ki67 in tumor tissues from mice in each group (scale bar = 25 μm) (∗p < 0.05, model versus control; #p < 0.05, model + miR-320b agmoir + oe-NC versus model + agmoir NC + oe-USP37; &p < 0.05, model + miR-320b agmoir + oe-USP37 versus model + miR-320b + oe-NC). Data are presented as the mean ± standard deviation. Data between two groups were compared using an unpaired t test. Comparisons among multiple groups were performed using one-way ANOVA with Tukey’s post hoc test. Comparisons over time were performed using repeated-measurements of ANOVA with Bonferroni post hoc test. n = 8.
Discussion
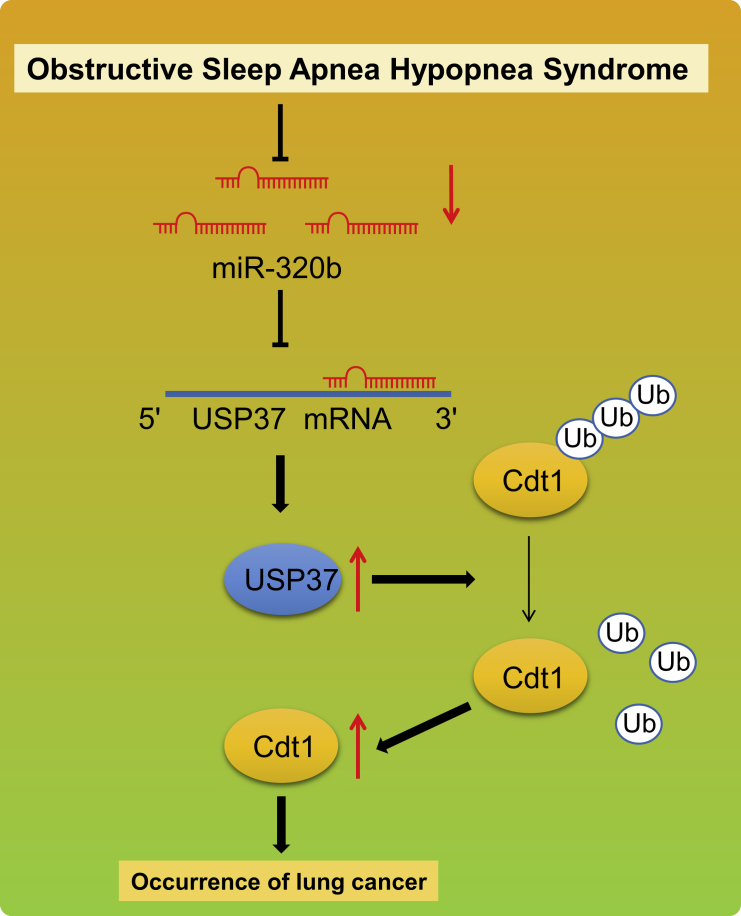
Lung cancer, the world’s leading cause of cancer death, has multiple kinds of diagnostic modalities as well as treatment approaches, depending on the subtypes and stage of cancer.20 Despite the progress achieved regarding cancer diagnosis and treatment in the past decades, the 5-year survival still remains below 20% in most countries.3 Thus, a better understanding of the initiation and progression of lung cancer is critical to improving the clinical outcome of patients with lung cancer. Accumulating evidence has demonstrated that OSAH is highly prevalent in lung cancer patients,7,21,22 suggesting a potential association between OSAH and the subsequent development of lung cancer. In our study, we tried to explore the significant roles of OSAH in lung cancer concerning the following factors: miR-320b, USP37, and CDT1. Finally, the findings of our study concluded that miR-320b inhibited the tumorigenesis and progression of lung cancer by regulating the USP37/CDT1 axis (Figure 8).
Figure 8.
The mechanism diagram depicting that IH-induced downregulation of miR-320b promoted the tumorigenesis of lung cancer by the USP37-mediated deubiquitination of CDT1
After sample collection and cell treatment, we compared the expression of miR-320b in lung cancer patients with or without OSAH and noticed that patients with OSAH expressed lower levels of miR-320b. More importantly, IH, a hallmark of OSAH, has been shown to promote lung cancer aggressiveness.9 Moreover, previous studies have shown that miR-320b, downregulated in OSAH patients,10 functions as a tumor suppressor of lung cancer,11,12 which was in line with our study.
We then explored the role of miR-320b in the progression of lung cancer in vitro. Our findings have demonstrated that overexpressing miR-320b significantly inhibited IH-induced lung cancer cell proliferation and invasion. Consistent with our results, previous studies have demonstrated the inhibitory effects of miR-320b on cancer cell proliferation, invasion, and survival.12,23,24 We further explored the molecular mechanisms to identify how miR-320b inhibited the progression of IH-induced lung cancer. Thus, we conducted bioinformatics analysis to identify potential targets of miR-320b. Among the predicted targets, USP37 has particularly caught our attention. Accumulating evidence has demonstrated that USP37 promotes lung cancer cell proliferation, migration, and invasion16 but inhibits lung cancer cell apoptosis in a deubiquitination-dependent manner.19 Therefore, it is possible that IH-mediated miR-320b reduction in lung cancer cells resulted in the upregulation of USP37, which further promoted cancer cell proliferation, invasion, and survival. Thus, we evaluated the expression of USP37 in lung cancer patients with or without OSAH and noticed that lung cancer patients with OSAH expressed higher levels of USP37. We also observed the inhibitory effects expressed by miR-320b on IH-induced lung cancer cell progression being compromised by the overexpression of USP37. These findings have confirmed that miR-320b inhibits IH-mediated lung cancer progression by targeting USP37. Moreover, USP37 has been shown to deubiquitinate and stabilize CDT1.17 Consequently, we investigated whether CDT1 is involved in miR-320b-inhibited IH-induced lung cancer progression. In alignment with previous studies,25,26 CDT1 increased lung cancer aggressiveness by promoting cell proliferation, invasion, and survival. CDT1 was also observed to compromise the anti-tumorigenic effects of miR-320b on IH-treated lung cancer cells. We have also proven that miR-320b inhibited lung cancer tumorigenesis and progression by regulating USP37/CDT1 axis, by using in vivo models of human lung cancer xenograft.
Taken together, our data have demonstrated, for the first time, a novel molecular mechanism by which miR-320b inhibits IH-induced lung cancer initiation and progression. These findings have furthered our understanding on the pathogenesis of lung cancer complicated with OSAH and will provide new insights into more promising developments of therapeutic approaches against lung cancer complicated with OSAH in the future.
Materials and methods
Bioinformatics analysis
Target genes of miR-320b were predicted by three online databases (microT [http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=microT_CDS/], StarBase [http://starbase.sysu.edu.cn/], and miRWalk [http://mirwalk.umm.uni-heidelberg.de/]) based on different algorithms. Commonly predicted targets were visualized using an online Venn diagram (http://jvenn.toulouse.inra.fr/app/index.html). The expression of target genes in lung cancer tissues and normal lung tissues, obtained in the TCGA database, was compared using the UALCAN interactive web resource (http://ualcan.path.uab.edu/). Lung-cancer-associated gene expression datasets (GEO: GSE106929 and GSE33532) were downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo/) as shown in Table 3. DEGs in lung cancer tissues were analyzed using “limma” package of R language. The correction of false discovery rates was associated with p values, while p <0.05 and |log2 (fold change)| >2 were set as the cut-off values to screen DEGs. Upregulated DEGs in both datasets were visualized using a Venn diagram and analyzed for pathways. Co-expressed genes were analyzed by accessing an online site (Chipbase, http://rna.sysu.edu.cn/chipbase/) to identify potential mechanisms.
Table 3.
Gene expression datasets from GEO database
Patient sample collection
Lung cancer tissues were collected from 68 lung cancer patients that were complicated with OSAH (male = 50; female = 18; age 35–75 years old; average age 56.09 ± 12.49 years old). The patients were selected from the First Affiliated Hospital of Zhengzhou University between January 2016 and January 2018. Recruitment criteria consist of the following: (1) patients with complete clinical records; (2) patients diagnosed with lung cancer by surgical pathological evaluation; (3) tumor characteristics meeting the tumor node metastasis classification system (TI, n = 5; TII–III, n = 63); and (4) patients diagnosed with OSAH, characterized by irregular snoring patterns and shortness of breath along with nocturia, morning headache, dizziness, parched mouth and scorched tongue. Patients that were met with the following criteria were excluded from this study: (1) surgical pathological evaluation not supporting the diagnosis of lung cancer; (2) patients taking immunosuppressive drugs or cytotoxic drugs; (3) patients with chronic inflammatory diseases; (4) patients with acute respiratory distress syndrome or other diseases that cause sleep-related hypoxia; (5) patients with tumor recurrence or distant metastasis; and (6) patients with a history of mental illness or patients coping poorly during the questionnaire. A total of 27 patients pathologically diagnosed with lung cancer were recruited (n = 27, including 2 cases of T1 and 25 cases of TII–III). All recruited patients in this study have signed informed consent documents. The study was conducted with the approval of the Ethics Committee of the First Affiliated Hospital of Zhengzhou University.
qRT-PCR
The total RNA content was extracted from tissues and cells using Trizol (15596026, Invitrogen, Carlsbad, CA, USA) reagent and was reversely transcribed into complementary DNA (cDNA) using the miRNA First Strand cDNA Synthesis kit (B532451-0200, Sangon, Shanghai, China). Amplification and quantification were performed using the Fast SYBR Green PCR kit (Applied Biosystems, Foster City, CA, USA) on an ABI7500 system. The relative expression level of miRNAs was normalized with U6. The relative expression of genes was calculated using the 2−ΔΔCt method. Primers for miRNA and U6 are shown in Table 4.
Table 4.
Primer sequences for qRT-PCR
| Sequences | |
|---|---|
| miR-320b | F, 5′-GTCGTATCCAGTGCAGGG TCCGAGGTATTCGCACTGGA TACGACTTTTCGAC-3′ |
| R, 5′-TCCGAAACGGGAGAGTTGG-3′ | |
| U6 | F, 5′-GTCGTATCCAGTGCAGGGTCC GAGGTATTCGCACTGGATACG ACAAAATA-3′ |
| R, 5′-TCCGATCGTGAAGCGTTC-3′ |
qRT-PCR, reverse transcription quantitative polymerase chain reaction; miR, microRNA; F, forward; R, reverse.
Cell culture and transfection
All lung cancer cell lines (A549, H1299, Calu-3, and H1650) and the NHBE cell line, purchased from American Type Culture Collection, were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (Gibco, Carlsbad, CA, USA), 100 μg/mL streptomycin, and 100 U/mL penicillin at 37°C in a humidified 5% CO2 environment.
An oxygen chamber with adjustable oxygen concentrations was established to mimic IH according to previous reported studies.27 Cells cultured in the oxygen chamber were subject to intermittent hypoxic conditions (60 min of hypoxia [5% oxygen] followed by 30 min of normoxia [20% oxygen]) for 6 h in total.
Different plasmids expressing miRNAs or proteins (miR-320b mimic, oe-USP37, and oe-CDT1 as well as their corresponding controls; all plasmids were purchased from GenePharma [Shanghai, China]) were delivered into cells using the Lipofectamine 2000 transfection reagent (11668030, Invitrogen) according to the manufacturer’s instructions.
EdU proliferation assay
EdU proliferation assay was performed as described previously, only with minor adjustments.28 Cells were first seeded in 24-well plates, labeled with 10 μM EdU (C10341-1, RiboBio, Guangzhou, Guangdong, China) and left in an incubator for 2 h. The cells were then fixed with 4% paraformaldehyde (PFA) for 15 min at room temperature, followed by washing with phosphate-buffered saline (PBS) containing 3% bovine serum albumin (BSA) twice. Afterward, cells were permeabilized with 0.5% Triton X-100 at room temperature for 20 min and washed with PBS containing 3% BSA twice. The cells were later incubated with Apollo 567 (RiboBio) in complete darkness for 30 min. After cells were washed twice with PBS containing 3% BSA, their nuclei were stained with Hoechst 33342 in complete darkness for 30 min. The results were monitored and recorded from 3 randomly selected views of each group of cells under the guidance of a fluorescent microscope (FM-600, PUDA, Shanghai, China). The number of red-fluorescent cells (Apollo stained) was divided by the number of total cells (Apollo stained and Hoechst 33342 stained) in order to determine the percentage of EdU-positive cells.
Transwell invasion assay
Transwell invasion assay was performed by following the same methods as previously described.29 Cells were left to culture in a serum-free medium for 12 h and seeded in Transwell inserts (Corning, Corning, NY, USA) coated with matrix gel (BD Biosciences, San Jose, CA, USA) in a serum-free medium at a density of 1 × 104 cells/well. The Transwell inserts were then placed into the wells of a 24-well plate containing complete culture medium. After 24 h, Transwell inserts were washed with PBS buffer solution and the un-invaded cells were removed with cottons. Cells were then fixed with methanol and stained with 1% toluidine blue (Sigma-Aldrich, St. Louis, MO, USA). Five randomly selected views under an inverted microscope (CarlZeiss, Germany) were photographed and analyzed.
Western blot analysis
Cells were lysed using a radio-immunoprecipitation assay buffer (BOSTER Biological Technology, Wuhan, Hubei, China). The protein concentration was measured using a bicinchoninic acid protein quantification kit (BOSTER). The same amount of total cell lysate was separated using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene fluoride membrane. The membrane was then left to be blocked for 1 h and incubated with diluted primary rabbit anti-human antibodies overnight at 4°C: USP37 (ab190814, 1:2,000, Abcam, Cambridge, MA, USA), CDT1 (ab202067, 1:500, Abcam), MMP-2 (1:1,000, ab92536, Abcam), MMP-9 (1:1,000, ab76003, Abcam), HIF1α (ab51608, 1:1,000, Abcam). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (ab181602, 1:5,000, Abcam) or β-actin (ab8227, 1:1,000, Abcam) was used as loading control. On the following day, membranes were washed with PBS buffer solution 3 times (10 min each), followed by being incubated with horseradish peroxidase-conjugated secondary antibodies (goat anti-rabbit immunoglobulin G [IgG], ab205718, 1:10,000, Abcam) at 37°C for 1 h. The membranes were then washed with PBS buffer solution 3 times (5 min each) and allowed to develop in enhanced chemiluminescence (ECL) reagent. Images were taken by a SmartView Pro 2000 imager system (UVCI-2100, Major Science, USA). The relative expression level of proteins was represented by the gray value ratio of protein bands evaluated by Quantity One software.
Dual-luciferase reporter gene assay
WT and MUT sequences of USP37 3′ UTR binding to miR-320b were cloned and constructed into target sequences of luciferase reporter gene downstream psiCheck2 plasmid (pGL3-USP37-3′ UTR and pGL3-USP37-MUT-3′ UTR). NC mimic and miR-320b mimic were respectively co-transfected with luciferase reporter vectors (pGL3-USP37-3′ UTR and pGL3-USP37-MUT-3′ UTR) into HEK293T cells. HEK293T cells of each group were seeded into 6-well plates with 2 × 105 cells per well. After the cells were adhered, transfection was performed by following the method described above. After the successful transfection of cells, they were subsequently cultured for 48 h and were collected. Target effects were expressed as the relative luciferase activity of report vector containing target sequences. The changes in the luciferase activity of miR-320b and USP37 in cells were detected by following the instructions provided by Genecopoeia’s dual-luciferase detection kit (D0010, Beijing Solibao Technology, Beijing, China). The Glomax 20/20 luminometer fluorescence detector (E5311, Shaanxi Zhongmei Biotechnology, Shaanxi, China) was used to detect the firefly luciferase activity, with Renilla luciferase activity serving as the internal reference.
Co-immunoprecipitation (coIP) assay
Cells were lysed in coIP buffer (10 mM 4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid [pH = 8.0], 300 mM NaCl, 0.1 mM ethylenediaminetetraacetic acid, 20% glycerol, and 0.2% Nonidet P-40) supplemented with protease inhibitors and phosphatase inhibitors. Cellular debris was pelleted by centrifugation, and cell lysate was pre-cleared by incubation with 25 μL protein A/G agarose for 1.5 h at 4°C. Cleared total cell lysates from each group were incubated with antibodies against USP37 or CDT1 overnight at 4°C. On the following day, 30 μL of protein A/G agarose was added into the lysate and incubated for 2 h at 4°C to immobilize antibodies and the bound proteins. Western blot analysis was used to detect the specific protein of interest.
Fluorescence staining
Cells seeded onto coverslips were fixed with 4% PFA for 15 min at room temperature and washed with PBS buffer solution three times. The cells were subsequently left to permeabilize with 0.5% Triton X-100 for 15 min at room temperature and were blocked with 10% goat serum for 1 h at room temperature. The cells were then incubated with rabbit anti-human USP37 antibody (ab190184, 1:1,000, Abcam) or mouse anti-human CDT1 antibody (sc-365305, 1:1,000, Santa Cruz, Santa Cruz, CA, USA), respectively, overnight at 4°C. On the following day, the cells were washed and incubated with tetramethylrhodamine B isothiocyanate (TRITC)-conjugated goat anti-rabbit IgG heavy and light chains (H&L) antibody (ab6718, 1:1,000, Abcam) or fluorescein isothiocyanate-conjugated goat anti-mouse IgG H&L antibody (ab6785, 1:1,000, Abcam) for 45 min at room temperature, followed by a PBS wash. The coverslips were then mounted with a Vectashield Antifade Mounting Medium (Vector Laboratories, Burlingame, CA, USA). The subcellular localizations of USP37 and CDT1 were observed and photographed under the guidance of confocal fluorescent microscope (Olympus, Tokyo, Japan).
Ubiquitination assay
The plasmids expressing Strep-CDT1, his-ubiquitin, control plasmids, WT USP37, or CI USP37 (Flag-USP37) were used to transfect the 293T cells for 48 h, which were subsequently treated with 20 μM MG132 for 4 h. After that, the cells were lysed on ice for 30 min by immunoprecipitation (IP) lysis buffer containing protease inhibitor, phosphatase inhibitor (Roche, Basel, Switzerland), and 10 μM N-ethylmaleimide (Sigma). Protein A/G agarose (Sigma) was used to pre-bind to the designated antibodies. The resin beads were washed with a lysis buffer and the sample was further eluted by SDS-PAGE. Immunoblotting was followed by an enhanced chemiluminescence (ECL) detection system (Bio-Rad, Hercules, CA, USA).
Human lung cancer xenograft in nude mice
A total of 40 specific-pathogen-free (SPF) male athymic BALB/c nude mice (4–5 weeks old, 18–20 g) purchased from Hunan SJA Laboratory Animal (Hunan, China) were raised in standard cages exposed to artificial 12/12-h light-dark cycles at 26°C ± 1°C with free access to food and water. The mice were randomly assigned into 5 groups (n = 8): control group, model group, model + agmoir-NC + oe-NC group, model + miR-320b agmoir + oe-NC group, and model + miR-320b agmoir + oe-USP37 group. In the control group, mice received A549 cells that were exposed to normoxic conditions. In the model group, mice received A549 cells that were exposed to 6IH daily. In the model + agmoir-NC + oe-NC group, mice received A549 cells with overexpressed agmoir NC and oe-NC exposed to 6IH daily. In the model + miR-320b + oe-NC group, mice were transplanted with A549 cells overexpressing miR-320b and oe-NC exposed to 6IH daily. In the model + miR-320b agmoir + oe-USP37 group, mice were treated with A549 cells overexpressing agmoir NC and USP37 exposed to 6IH daily. Under exposure to normal oxygen or IH, 6 × 106 A549 cells from each treatment group suspended in 200 μL PBS were mixed with the same amount of matrigel. A total of 0.2 mL (3 × 106) of the single-cell suspension was injected subcutaneously into the right axillary region of nude mice. The tumor growth was monitored and recorded every other day after cellular transplantation. The tumor volume (V) was calculated with the following formulae: V (mm3) = (L × D2)/2, where L and D represents the tumor length and width, respectively (in mm). Three weeks after transplantation, the mice were anesthetized by pentobarbital sodium and euthanized; the final tumor volume and weight were subsequently measured. Tumor volume in nude mice was plotted against time. All animal experiments were conducted under the protocols approved by the Institutional Animal Care and Use Committee of the First Affiliated Hospital of Zhengzhou University. Great efforts were made to minimize the pain and reduce the number of animals used.
Immunohistochemistry
The freshly resected tumors were fixed with PFA and left overnight, embedded with paraffin, and sliced into 4 x axis serial sections. Following dewaxing and dehydration, sections were heated in a citric acid buffer solution to extract antigen, incubated with primary antibodies to Ki67 (ab15580, 1:1,000, Abcam), secondary antibodies, and finally were sealed with neutral gum, by following the instructions indicated in the manual. The percentage of Ki67-positive proliferative cells was calculated.
Pulmonary metastasis model in nude mice
A total of 40 SPF male athymic BALB/c nude mice (4–5 weeks old, 18 – 20 g) were treated with the same methods as previously mentioned. A549 cells (6 × 106) were mixed with Matrigel at a ratio of 1:1. The mixture (0.1 mL) was injected into nude mice via tail vein. Five weeks after the administration of the mixture, mice were then euthanized with the lung resected and photographed. Only visible tumors that could be observed in the lungs were counted.
Statistical analysis
All data from this study were analyzed using SPSS 21.0 statistical software (IBM SPSS Statistics, Armonk, NY, USA). Data were presented as mean ± standard deviation. Data between two groups were compared using an unpaired t test. Comparisons among multiple groups were performed using one-way analysis of variance (ANOVA) with Tukey’s post hoc test. Comparisons over time were performed using repeated-measures ANOVA with Bonferroni post hoc test. The correlation between two indicators was analyzed by Pearson correlation coefficient. A value of p <0.05 was considered as statistically significant. All experiments were repeated three times independently.
Acknowledgments
The authors sincerely appreciate all members who participated in this work.
Author contributions
W.L., K.H., and F.W. wrote the paper and designed the study. G.C. and H.G. conceived the study and participated in the design of the study. Z.H. and S.Z. performed the experiments. W.L. and S.Z. coordinated the work and modified the final manuscript. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.12.023.
Supplemental information
References
- 1.Salem A., Asselin M.C., Reymen B., Jackson A., Lambin P., West C.M.L., O’Connor J.P.B., Faivre-Finn C. Targeting Hypoxia to Improve Non-Small Cell Lung Cancer Outcome. J. Natl. Cancer Inst. 2018;110 doi: 10.1093/jnci/djx160. [DOI] [PubMed] [Google Scholar]
- 2.Thun M.J., Hannan L.M., Adams-Campbell L.L., Boffetta P., Buring J.E., Feskanich D., Flanders W.D., Jee S.H., Katanoda K., Kolonel L.N. Lung cancer occurrence in never-smokers: an analysis of 13 cohorts and 22 cancer registry studies. PLoS Med. 2008;5:e185. doi: 10.1371/journal.pmed.0050185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hirsch F.R., Scagliotti G.V., Mulshine J.L., Kwon R., Curran W.J., Jr., Wu Y.L., Paz-Ares L. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389:299–311. doi: 10.1016/S0140-6736(16)30958-8. [DOI] [PubMed] [Google Scholar]
- 4.Ren W., Mi D., Yang K., Cao N., Tian J., Li Z., Ma B. The expression of hypoxia-inducible factor-1α and its clinical significance in lung cancer: a systematic review and meta-analysis. Swiss Med. Wkly. 2013;143:w13855. doi: 10.4414/smw.2013.13855. [DOI] [PubMed] [Google Scholar]
- 5.Wilson W.R., Hay M.P. Targeting hypoxia in cancer therapy. Nat. Rev. Cancer. 2011;11:393–410. doi: 10.1038/nrc3064. [DOI] [PubMed] [Google Scholar]
- 6.Byers T.E., Vena J.E., Rzepka T.F. Predilection of lung cancer for the upper lobes: an epidemiologic inquiry. J. Natl. Cancer Inst. 1984;72:1271–1275. [PubMed] [Google Scholar]
- 7.Cabezas E., Pérez-Warnisher M.T., Troncoso M.F., Gómez T., Melchor R., Pinillos E.J., El Hachem A., Gotera C., Rodríguez P., Mahillo Fernández I. Sleep Disordered Breathing Is Highly Prevalent in Patients with Lung Cancer: Results of the Sleep Apnea in Lung Cancer Study. Respiration. 2019;97:119–124. doi: 10.1159/000492273. [DOI] [PubMed] [Google Scholar]
- 8.Kang H.S., Kwon H.Y., Kim I.K., Ban W.H., Kim S.W., Kang H.H., Yeo C.D., Lee S.H. Intermittent hypoxia exacerbates tumor progression in a mouse model of lung cancer. Sci. Rep. 2020;10:1854. doi: 10.1038/s41598-020-58906-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y., Song X., Wang X., Wei L., Liu X., Yuan S., Lv L. Effect of chronic intermittent hypoxia on biological behavior and hypoxia-associated gene expression in lung cancer cells. J. Cell. Biochem. 2010;111:554–563. doi: 10.1002/jcb.22739. [DOI] [PubMed] [Google Scholar]
- 10.Yang X., Niu X., Xiao Y., Lin K., Chen X. MiRNA expression profiles in healthy OSAHS and OSAHS with arterial hypertension: potential diagnostic and early warning markers. Respir. Res. 2018;19:194. doi: 10.1186/s12931-018-0894-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lieb V., Weigelt K., Scheinost L., Fischer K., Greither T., Marcou M., Theil G., Klocker H., Holzhausen H.J., Lai X. Serum levels of miR-320 family members are associated with clinical parameters and diagnosis in prostate cancer patients. Oncotarget. 2017;9:10402–10416. doi: 10.18632/oncotarget.23781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lv Q.L., Du H., Liu Y.L., Huang Y.T., Wang G.H., Zhang X., Chen S.H., Zhou H.H. Low expression of microRNA-320b correlates with tumorigenesis and unfavorable prognosis in glioma. Oncol. Rep. 2017;38:959–966. doi: 10.3892/or.2017.5762. [DOI] [PubMed] [Google Scholar]
- 13.Jin X., Chen Y., Chen H., Fei S., Chen D., Cai X., Liu L., Lin B., Su H., Zhao L. Evaluation of Tumor-Derived Exosomal miRNA as Potential Diagnostic Biomarkers for Early-Stage Non-Small Cell Lung Cancer Using Next-Generation Sequencing. Clin. Cancer Res. 2017;23:5311–5319. doi: 10.1158/1078-0432.CCR-17-0577. [DOI] [PubMed] [Google Scholar]
- 14.Li L., Lu J., Xue W., Wang L., Zhai Y., Fan Z., Wu G., Fan F., Li J., Zhang C. Target of obstructive sleep apnea syndrome merge lung cancer: based on big data platform. Oncotarget. 2017;8:21567–21578. doi: 10.18632/oncotarget.15372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang S., Zhang X., Sun Q., Zhuang C., Li G., Sun L., Wang H. LncRNA NR2F2-AS1 promotes tumourigenesis through modulating BMI1 expression by targeting miR-320b in non-small cell lung cancer. J. Cell. Mol. Med. 2019;23:2001–2011. doi: 10.1111/jcmm.14102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan J., Deng Q., Jiang C., Wang X., Niu T., Li H., Chen T., Jin J., Pan W., Cai X. USP37 directly deubiquitinates and stabilizes c-Myc in lung cancer. Oncogene. 2015;34:3957–3967. doi: 10.1038/onc.2014.327. [DOI] [PubMed] [Google Scholar]
- 17.Hernández-Pérez S., Cabrera E., Amoedo H., Rodríguez-Acebes S., Koundrioukoff S., Debatisse M., Méndez J., Freire R. USP37 deubiquitinates Cdt1 and contributes to regulate DNA replication. Mol. Oncol. 2016;10:1196–1206. doi: 10.1016/j.molonc.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang W., Kim S.U., Ahn S.H. Non-invasive prediction of forthcoming cirrhosis-related complications. World J. Gastroenterol. 2014;20:2613–2623. doi: 10.3748/wjg.v20.i10.2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai J., Li M., Wang X., Li L., Li Q., Hou Z., Jia H., Liu S. USP37 Promotes Lung Cancer Cell Migration by Stabilizing Snail Protein via Deubiquitination. Front. Genet. 2020;10:1324. doi: 10.3389/fgene.2019.01324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nasim F., Sabath B.F., Eapen G.A. Lung Cancer. Med. Clin. North Am. 2019;103:463–473. doi: 10.1016/j.mcna.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 21.Siegel R., Naishadham D., Jemal A. Cancer statistics, 2013. CA Cancer J. Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 22.Wolner Z., Pulitzer M.P., Marchetti M.A. Isolated pink papule on the chest. Dermatol. Pract. Concept. 2017;7:19–22. doi: 10.5826/dpc.0701a03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang H., Cao F., Li X., Miao H., e J., Xing J., Fu C.G. miR-320b suppresses cell proliferation by targeting c-Myc in human colorectal cancer cells. BMC Cancer. 2015;15:748. doi: 10.1186/s12885-015-1728-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lv G.Y., Miao J., Zhang X.L. Long Noncoding RNA XIST Promotes Osteosarcoma Progression by Targeting Ras-Related Protein RAP2B via miR-320b. Oncol. Res. 2018;26:837–846. doi: 10.3727/096504017X14920318811721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liontos M., Koutsami M., Sideridou M., Evangelou K., Kletsas D., Levy B., Kotsinas A., Nahum O., Zoumpourlis V., Kouloukoussa M. Deregulated overexpression of hCdt1 and hCdc6 promotes malignant behavior. Cancer Res. 2007;67:10899–10909. doi: 10.1158/0008-5472.CAN-07-2837. [DOI] [PubMed] [Google Scholar]
- 26.Turhan A.H., Atıcı A., Muşlu N., Polat A., Helvacı İ. The effects of pentoxifylline on lung inflammation in a rat model of meconium aspiration syndrome. Exp. Lung Res. 2012;38:250–255. doi: 10.3109/01902148.2012.676704. [DOI] [PubMed] [Google Scholar]
- 27.Li R.C., Haribabu B., Mathis S.P., Kim J., Gozal D. Leukotriene B4 receptor-1 mediates intermittent hypoxia-induced atherogenesis. Am. J. Respir. Crit. Care Med. 2011;184:124–131. doi: 10.1164/rccm.201012-2039OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ning H., Albersen M., Lin G., Lue T.F., Lin C.S. Effects of EdU labeling on mesenchymal stem cells. Cytotherapy. 2013;15:57–63. doi: 10.1016/j.jcyt.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang S., Wu Y., Hou Y., Guan X., Castelvetere M.P., Oblak J.J., Banerjee S., Filtz T.M., Sarkar F.H., Chen X. CXCR2 macromolecular complex in pancreatic cancer: a potential therapeutic target in tumor growth. Transl. Oncol. 2013;6:216–225. doi: 10.1593/tlo.13133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.