In this article, Chavez et al. show that the myeloid transcription factor PU.1 plays an essential role in maintaining hematopoietic stem cell (HSC) quiescence and homeostatic protein synthesis rates, thereby regulating HSC pool size and cell cycle activity during chronic inflammatory stress.
Abstract
Hematopoietic stem cells (HSCs) are capable of entering the cell cycle to replenish the blood system in response to inflammatory cues; however, excessive proliferation in response to chronic inflammation can lead to either HSC attrition or expansion. The mechanism(s) that limit HSC proliferation and expansion triggered by inflammatory signals are poorly defined. Here, we show that long-term HSCs (HSCLT) rapidly repress protein synthesis and cell cycle genes following treatment with the proinflammatory cytokine interleukin (IL)-1. This gene program is associated with activation of the transcription factor PU.1 and direct PU.1 binding at repressed target genes. Notably, PU.1 is required to repress cell cycle and protein synthesis genes, and IL-1 exposure triggers aberrant protein synthesis and cell cycle activity in PU.1-deficient HSCs. These features are associated with expansion of phenotypic PU.1-deficient HSCs. Thus, we identify a PU.1-dependent mechanism triggered by innate immune stimulation that limits HSC proliferation and pool size. These findings provide insight into how HSCs maintain homeostasis during inflammatory stress.
Introduction
Hematopoietic stem cell (HSC) quiescence promotes lifelong blood regeneration and is controlled by a complex regulatory network, including cell-intrinsic transcription factors and epigenetic modifiers (Pietras et al., 2011), organelle homeostasis mechanisms (Hinge et al., 2020; Liang et al., 2020), and signals generated from the bone marrow (BM) niche (Morrison and Scadden, 2014). At least some portion of the HSC pool can be briefly forced out of quiescence to facilitate blood system regeneration by stressors such as infection (Prendergast and Essers, 2014), chronic stress (Heidt et al., 2014), and myeloablative injury (Harrison and Lerner, 1991), demonstrating that HSCs can replenish cells lost to disruptions in BM homeostasis (King and Goodell, 2011). However, tight regulation of cell cycle activity is crucial for maintaining the long-term functional integrity of the HSC pool (Matsumoto et al., 2011; Pietras et al., 2011). This is particularly true under inflammatory stress conditions, where increased proliferative activity can lead to functional attrition and/or aberrant expansion of the HSC compartment, including in the contexts of chronic infection and/or genetic mutation associated with BM failure and myeloid oncogenesis (Essers et al., 2009; King et al., 2011; Matatall et al., 2016; Pietras, 2017; Pietras et al., 2014; Rodríguez et al., 2021; Takizawa et al., 2017; Walter et al., 2015; Zambetti et al., 2016; Zhang et al., 2016). Highly enriched HSC fractions can limit proliferative activity and maintain long-term engraftment capacity even under a variety of inflammatory stress conditions (Bujanover et al., 2018; Hernandez et al., 2020; Pietras et al., 2014; Pietras et al., 2016; Rabe et al., 2020; Wilson et al., 2008; Zhao et al., 2019), implying the existence of mechanism(s) that prevent excessive HSC cell cycle entry.
HSC can directly respond to pathogens and physiological danger signals via direct sensing (Takizawa et al., 2017) and/or paracrine proinflammatory cytokines produced by damaged and/or infected cells, such as IFNs (Ehninger et al., 2014; Essers et al., 2009; Matatall et al., 2016), G-CSF (Schuettpelz et al., 2014), TNF (Etzrodt et al., 2019; Yamashita and Passegué, 2019), and IL-1 (Hemmati et al., 2019; Weisser et al., 2016). IL-1 consists of two cytokines (IL-1α and IL-1β) with different expression patterns that share a common receptor complex and elicit similar responses (Dinarello, 2018). IL-1 is produced in response to a wide variety of physiological stress conditions, including aging; chronic inflammatory disease; myeloablative treatments, such as radiation and/or chemotherapy; obesity; and cellular senescence (Dinarello, 2018; Laberge et al., 2015). It may also contribute to hematological malignancy and is highly expressed in cells from patients with myelodysplastic syndrome (MDS), myeloproliferative neoplasia, and acute myelogenous leukemia (Ågerstam et al., 2016; Barreyro et al., 2018; Carey et al., 2017; Ezaki et al., 1995; Zhang et al., 2016). Previous work from our group demonstrates that acute IL-1 exposure drives myeloid cell overproduction in vivo via precocious activation of the master myeloid transcription factor PU.1 (Pietras et al., 2016) in HSC. Interestingly, this effect is transient, as HSCs reenter quiescence following chronic exposure to IL-1. These findings imply the existence of a braking mechanism that limits HSC cell cycle entry in response to chronic inflammatory stress (Pietras et al., 2016; Rabe et al., 2020).
We recently found that quiescent HSCs downregulate numerous protein synthesis and cell proliferation genes in a mouse model of chronic rheumatoid arthritis (Hernandez et al., 2020). Interestingly, activation of this gene program relies, at least in part, on IL-1 signaling (Hernandez et al., 2020). In the present study, we further explore the relevance of this gene program to HSC function during chronic inflammation using a mouse model of chronic IL-1 stimulation (Pietras et al., 2016). We show that IL-1 signaling is sufficient to rapidly induce repression of a broad set of cell cycle and protein synthesis genes in HSCs, in turn limiting cell cycle activity. We find that repression of these genes in HSCs is associated with PU.1 expression, and chromatin immunoprecipitation sequencing (ChIP-seq) analysis shows that PU.1 directly binds these target genes. Strikingly, PU.1 deficiency leads to derepression of cell cycle and protein synthesis genes, licensing HSCs to increase protein synthesis activity and enter the cell cycle in response to IL-1 stimulation. Together, these data support a model in which PU.1 enforces HSC quiescence in response to inflammatory stress by limiting protein synthesis and cell cycle activity, thereby preventing aberrant expansion of the HSC pool.
Results
Chronic IL-1 induces repression of cell cycle and protein synthesis genes in long-term HSCs (HSCLT)
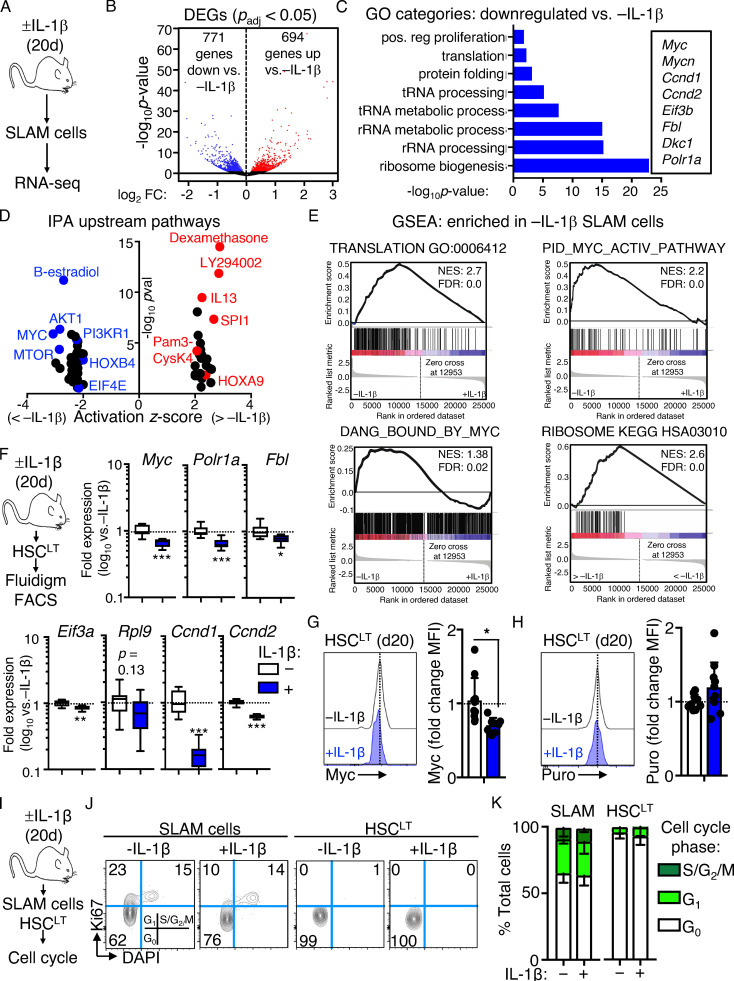
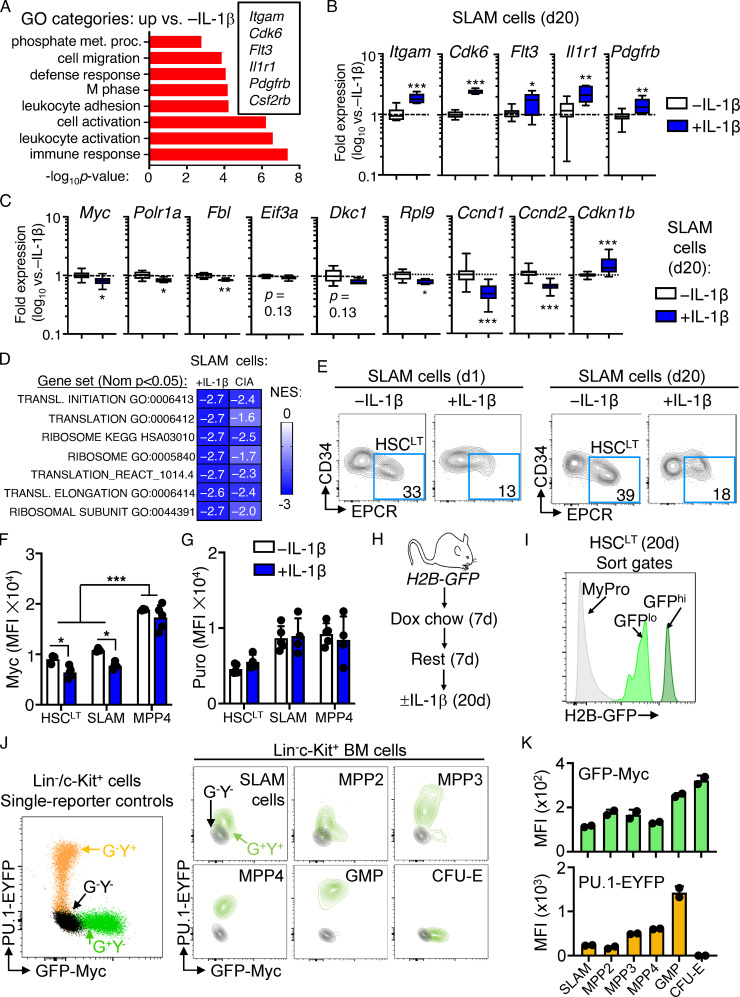
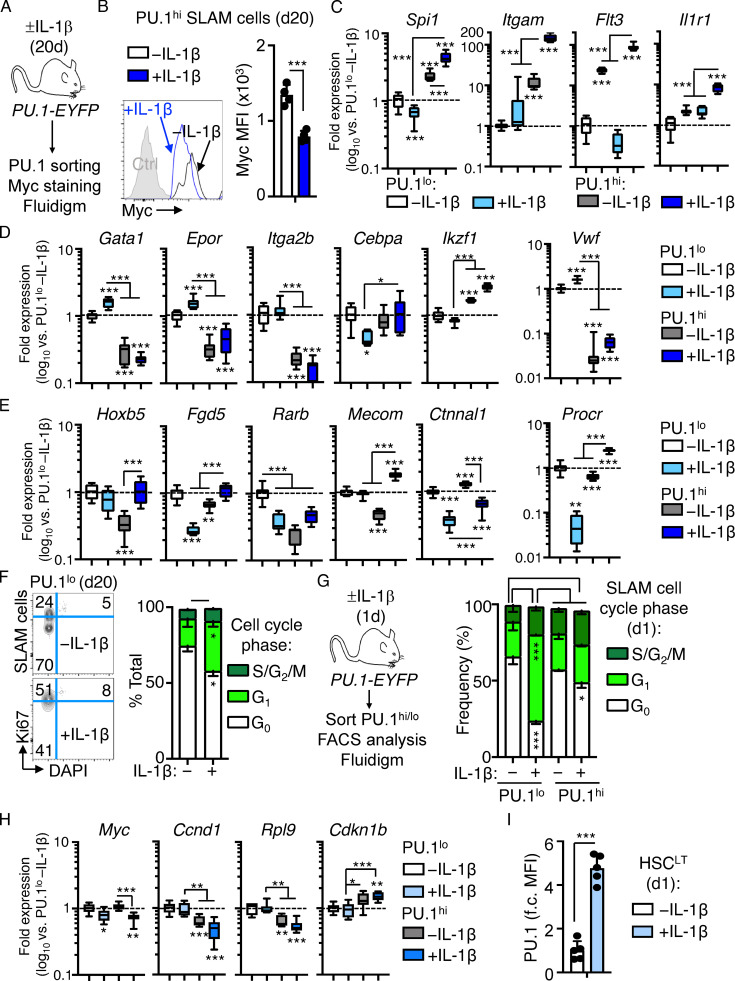
We previously found that HSCs maintain a largely quiescent state during chronic inflammatory stress driven by IL-1 and chronic inflammatory arthritis (Hernandez et al., 2020; Pietras, 2017; Pietras et al., 2014; Rabe et al., 2020). To address the mechanism(s), we analyzed gene expression by RNA sequencing (RNA-seq) in the HSC-enriched SLAM (signaling lymphocytic activation molecule) fraction (SLAM cells; LSK/Flk2−CD48−CD150+) isolated from the BM of mice treated with or without IL-1β for 20 d to model chronic inflammatory stress (Fig. 1 A; Pietras et al., 2016; Rabe et al., 2020). We found >1,400 differentially expressed genes (DEGs; Padj < 0.05; Fig. 1 B and Table S1). Gene ontology (GO) analysis identified cell activation, immune response, leukocyte adhesion, and defense response gene programs among upregulated DEGs (Fig. S1 A and Table S2). Using a custom Fluidigm quantitative RT-PCR (qRT-PCR) array, we validated increased expression of key genes in these pathways, including Itgam (Mac-1), Cdk6, Flt3, Il1r1, and Pdgfrb (Fig. S1 B). Strikingly, downregulated DEGs were enriched for ribosome biogenesis, rRNA processing, transfer RNA processing, and translation GO categories (Fig. 1 C and Table S2), while Ingenuity Pathway Analysis (IPA) and Gene Set Enrichment Analysis (GSEA) likewise identified multiple cell proliferation and mRNA translation pathways inhibited by IL-1 (Fig. 1, D and E; and Table S2), which were likewise validated by qRT-PCR (Fig. S1 C). These data were strikingly reminiscent of Myc pathway downregulation we observed in SLAM cells from mice with collagen-induced arthritis (CIA; Hernandez et al., 2020), and comparison with our prior GSEA analyses of SLAM cells from CIA mice revealed significant overlap in translation and ribosome gene signature enrichment between datasets (Fig. S1 D), suggesting repression of translation pathways in HSCs is not exclusively a property of in vivo IL-1 stimulation and can be observed in physiological models of chronic inflammatory disease.
Figure 1.
Chronic IL-1 induces repression of cell cycle and protein synthesis genes. (A) Experimental design for RNA-seq studies (n = 4–7 pools of SLAM cells from mice treated for 20 d ± IL-1β). Pools were generated from three independent cohorts of mice. (B) Volcano plot of DEGs (Padj ≤ 0.05) in IL-1–exposed SLAM cells (LSK/Flk2−/CD48−/CD150+) from A showing log2 fold change (FC) versus −log10 P value significance. See also Table S1. (C) GO category enrichment of downregulated DEGs in IL-1–exposed SLAM cells from A, expressed as −log10 P value. See also Table S2. (D) IPA showing enriched upstream regulators of DEGs in IL-1–exposed SLAM cells from A. See also Table S3. (E) GSEA analysis of significantly downregulated DEGs. GSEA plots show negative enrichment of translation and Myc pathway genes in IL-1–exposed SLAM cells from A. See also Table S4. (F) Experimental design for Fluidigm qRT-PCR analyses and intracellular FACS staining of HSCLT (LSK/Flk2−/CD48−/CD150+/CD34−/EPCR+) from mice treated for 20 d with or without IL-1β (left), and quantification by Fluidigm qRT-PCR array of cell cycle and protein synthesis gene expression in HSCLT (n = 8/group). Data are expressed as log10 fold expression versus −IL-1β. Box represents upper and lower quartiles with line representing median value. Whiskers represent minimum and maximum values. Data are representative of two independent experiments. (G) Intracellular flow cytometry analysis of Myc protein levels in HSCLT (n = 10 −IL-1β; 8 +IL-1β). Data are expressed as fold change of MFI versus −IL-1β. Individual values are shown with bars representing mean values. Data are compiled from three independent experiments. (H) Intracellular flow cytometry analysis of puro incorporation in HSCLT (n = 9 −IL-1β; 10 +IL-1β). Data are expressed as fold change of MFI versus −IL-1β. Individual values are shown with bars representing mean values. Data are compiled from three independent experiments. (I) Experimental design for cell cycle analyses of SLAM cells and HSCLT from mice treated for 20 d with or without IL-1. (J) Representative flow cytometry plots showing cell cycle distribution in SLAM cells and HSCLT from I. (K) Quantification of cell cycle phase distribution in SLAM cells and HSCLT from I (n = 5/group). Data are representative of three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by Mann-Whitney U test or ANOVA with Tukey’s test in K. Error bars represent SD. See also Fig. S1.
Figure S1.
Characterization of SLAM cells and reporter mice. (A) GO category enrichment of significantly upregulated DEGs in SLAM cells from mice treated for 20 d with or without IL-1β versus 20 d −IL-1β, expressed as −log10 P value. See Table S2 for complete list of GO categories. (B and C) Quantification by Fluidigm qRT-PCR array of gene expression in SLAM cells from mice treated with or without IL-1β for 20 d (n = 8/group). Data are expressed as log10 fold expression versus −IL-1β. Box represents upper and lower quartiles with line representing median value. Whiskers represent minimum and maximum values. Data are representative of two independent experiments. (D) Comparison of normalized enrichment scores (NESs) for indicated gene signatures from IL-1–exposed SLAM cells and SLAM cells from CIA mice (GSE129511). (E) Representative FACS plots showing frequencies of phenotypic HSCLT fraction within the SLAM gate from mice treated with IL-1β versus without IL-1β for either 1 or 20 d. Data are representative of multiple (more than three) experiments. (F) Geometric MFI of Myc from one of three experiments (n = 5/group). Individual values are shown with bars representing mean values. (G) Geometric MFI of puro from one of three experiments (n = 5/group). Individual values are shown with bars representing mean values. (H) Experimental design for H2B-GFP in vivo labeling experiments. Dox, doxycycline. (I) Representative sort gates showing identification of GFPhi and GFPlo populations within the HSCLT gate. Myeloid progenitors (MyPro), which rapidly dilute the GFP label, are shown in gray. (J) Representative FACS plot showing GFP and YFP profiles of single-color control Lin−/c-Kit+ HSPC from Myc-GFP (G+Y−) or PU1-EYFP (G−Y+) mice relative to G−Y− controls (left) and representative FACS plots of defined HSPC populations from Myc-GFP::PU.1-EYFP (G+Y+) mice versus G−Y− controls (right). (K) Quantification of GFP and YFP levels in each HSPC population from mice in J (n = 2/group). Individual values are shown with bars representing mean values. Data in J and K are representative of at least two independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by Mann-Whitney U test or ANOVA with Tukey's post-test in F and G. Error bars represent SD.
We previously showed that HSCLT-enriched SLAM cells (defined as LSK/Flk2−CD48−CD150+CD34−EPCR+; Fig. S1 E) overlap phenotypically with deeply quiescent CD49blo reserve HSCs (Zhao et al., 2019) and remain almost exclusively in a quiescent (G0) cell cycle state, despite chronic IL-1 exposure (Rabe et al., 2020). Cell cycle and protein synthesis genes were likewise repressed in chronic IL-1–exposed HSCLT (Fig. 1 F), and we confirmed reduced Myc expression in HSCs by intracellular flow cytometry staining (Fig. 1 G and Fig. S1 F). Myc levels in phenotypic MPP4 (LSK/Flk2+) were not significantly impacted by chronic IL-1, suggesting that not all progenitor cells repress Myc to the same extent in response to IL-1 (Fig. S1 F). Since chronic IL-1 exposure reduced expression of protein synthesis genes, we also measured the impact of chronic IL-1 exposure on protein synthesis rates in HSCLT by in vivo puromycin (puro) incorporation assays (Fig. 1 H). Consistent with prior work (Signer et al., 2014), puro incorporation rates in HSCLT, SLAM cells, and MPP4 were all similar (Fig. S1 G), and chronic IL-1 exposure did not impact the HSCLT protein synthesis rate (Fig. 1 H and Fig. S1 G). Lastly, we confirmed our prior published findings that HSCLT were quiescent after chronic IL-1 exposure (Fig. 1, I–K; Rabe et al., 2020). Taken together, these data suggest that repression of cell cycle and protein synthesis genes is associated with homeostatic protein synthesis and cell cycle activity in HSCLT during chronic inflammatory stress.
IL-1–induced gene repression is rapid and independent of HSCLT divisional history
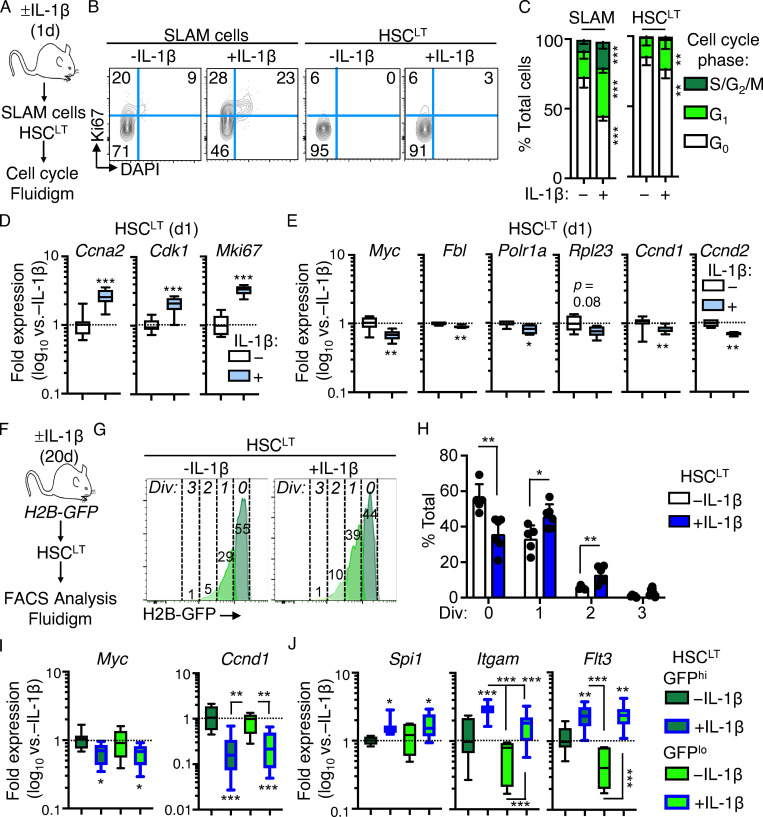
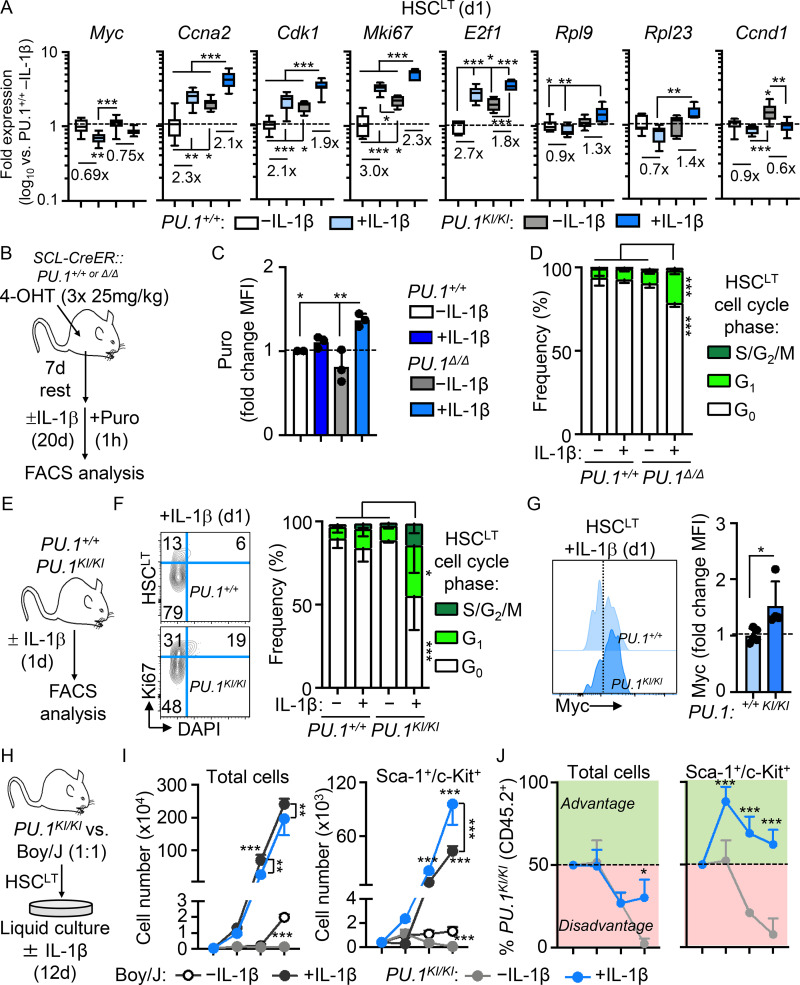
Since IL-1–repressed genes primarily appear to regulate mechanisms associated with cell proliferation, we first assessed whether this gene program was also activated in response to acute IL-1 exposure (a single IL-1 injection; Fig. 2 A). Consistent with prior investigations, we found that a single injection of IL-1 rapidly induced robust cell cycle activity in the SLAM compartment (Fig. 2, B and C). Concurrently, we observed a limited, though significant, induction of cell cycle activity in phenotypic HSCLT after acute IL-1 exposure (Fig. 2, B and C). Strikingly, Fluidigm analysis of HSCLT revealed that, while genes such as Ccna2 and Mki67 were upregulated, consistent with increased cell cycle activity in this compartment (Fig. 2 D), Myc and other protein synthesis genes downregulated under chronic IL-1 conditions were rapidly repressed even under acute IL-1 conditions (Fig. 2 E). Given this finding, we next asked whether downregulation of these genes was triggered exclusively in HSCLT that had entered the cell cycle following IL-1 exposure. Hence, we used Col1a-TetO::H2B-GFP mice (Foudi et al., 2009) to distinguish undivided phenotypic HSCLT from phenotypic HSCLT with accumulated divisional history following chronic IL-1 exposure (Fig. 2 F). We induced H2B-GFP transgene for 2 wk with doxycycline chow (Säwén et al., 2016) followed by a 1-wk rest and subsequent chase period of 20 d with or without IL-1 injection. Consistent with our cell cycle analysis showing acute induction of HSCLT cell cycle activity, we observed an increase in H2B-GFP dilution in phenotypic HSCLT from IL-1–treated mice (Fig. 2, G and H). These data were indicative of roughly a single extra division in some, but not all, HSCLT. We therefore assessed whether IL-1 repressed Myc and Ccnd1 exclusively in HSCLT that had divided or vice versa. Notably, qRT-PCR analysis of GFPhi (undivided) versus GFPlo (divided) HSCLT (Fig. S1, H and I) showed that Myc and Ccnd1 were repressed equally in HSCLT regardless of divisional history (Fig. 2 I). Likewise, both GFPhi and GFPlo HSCLT subsets also upregulated IL-1 target genes, including Spi1 (PU.1), Itgam, and Flt3 equally, suggesting proliferative history did not impact sensitivity to IL-1 (Fig. 2 I). Taken together, our data indicate that IL-1 rapidly induces repression of Myc and other cell cycle and protein synthesis genes in a manner independent of divisional history.
Figure 2.
IL-1–induced gene repression is rapid and independent of HSCLT divisional history. (A) Experimental design for cell cycle analyses of SLAM cells and HSCLT from mice treated for 1 d with or without IL-1β. (B) Representative flow cytometry plots showing cell cycle distribution in SLAM cells and HSCLT from A. (C) Quantification of cell cycle phase distribution in SLAM cells and HSCLT from A (n = 10/group). Data are compiled from three independent experiments. (D) Quantification by Fluidigm qRT-PCR array of cell cycle gene expression in HSCLT from mice treated for 1 d with or without IL-1β (n = 8/group). Data are expressed as log10 fold expression versus −IL-1β. Box represents upper and lower quartiles with line representing median value. Whiskers represent minimum and maximum values. Data are representative of two independent experiments. (E) Quantification of IL-1–repressed protein synthesis and cell cycle genes from Fluidigm qRT-PCR array in D. Data are representative of two independent experiments. (F) Experimental design for analysis of divisional history in HSCLT using H2B-GFP mice treated for 20 d with or without IL-1β. (G) Representative FACS plots showing analysis of HSCLT divisional history via GFP dilution from H2B-GFP mice treated for 20 d with or without IL-1β. (H) Quantification of divisional history of mice in F based on GFP dilution (n = 5–6/group). Data are compiled from two independent experiments. (I) Quantification by Fluidigm qRT-PCR array of IL-1–repressed genes in GFPhi and GFPlo HSCLT from H2B-GFP mice in F (n = 8/group). Data are expressed as log10 fold expression versus −IL-1β. Box represents upper and lower quartiles with line representing median value. Whiskers represent minimum and maximum values. Data are representative of two independent experiments. (J) Quantification of IL-1 target genes from Fluidigm qRT-PCR array in I. Data are representative of two independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by Mann-Whitney U test or ANOVA with Tukey’s test in C, H, I, and J. Error bars represent SD. See also Fig. S1.
IL-1–induced gene repression is associated with increased PU.1 levels
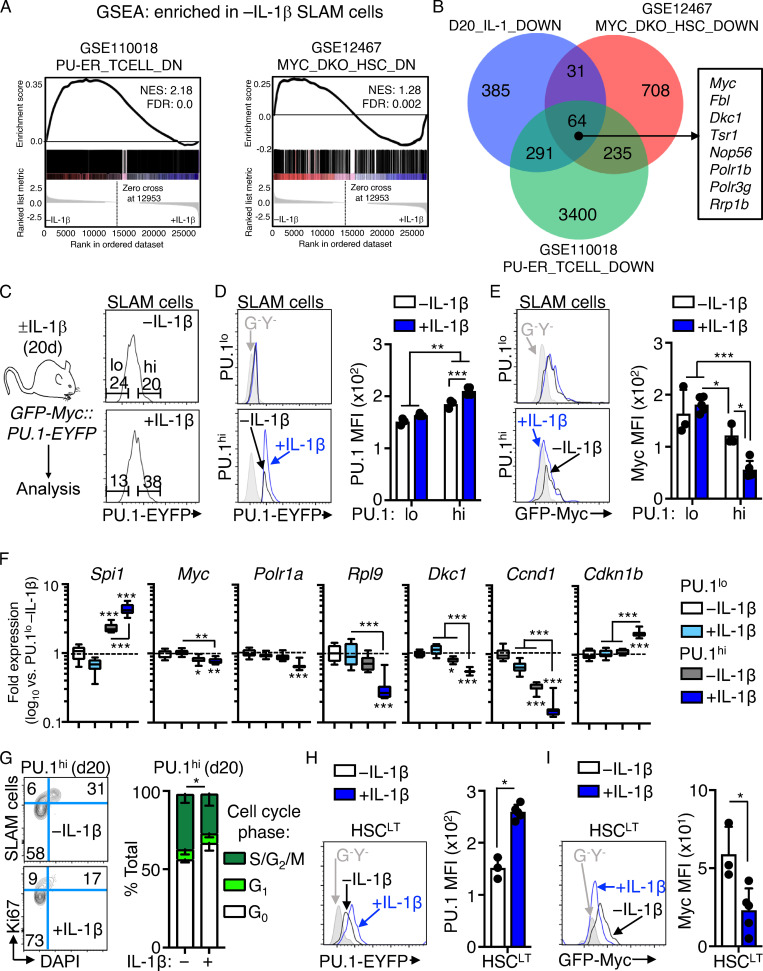
To investigate how IL-1 represses cell cycle and protein synthesis genes in HSCLT, we reanalyzed our IPA data and noticed that it had identified SPI1 (PU.1) pathway activation in IL-1–exposed SLAM cells (Fig. 1 D), consistent with our prior published findings (Etzrodt et al., 2019; Pietras et al., 2016; Rabe et al., 2020). Interestingly, PU.1 can also restrict proliferation in hematopoietic stem and progenitor cells (HSPCs), likely a mechanism to promote PU.1 accumulation during myeloid differentiation (Fukuchi et al., 2008; Kueh et al., 2013). We reasoned this mechanism could also serve to restrict HSC activation by IL-1. To establish a relationship between PU.1 and IL-1–mediated repression of cell cycle and protein synthesis genes, we first compared our RNA-seq dataset with publicly available datasets in which PU.1 was overexpressed in thymocytes (Hosokawa et al., 2018; Gene Expression Omnibus [GEO] accession no. GSE93755) and in which Myc and Mycn were conditionally deleted in HSCs, leading to reduced proliferative activity (Laurenti et al., 2008; GEO accession no. GSE12467). GSEA analysis revealed that cell cycle and protein synthesis genes repressed in these datasets were likewise repressed in SLAM cells from IL-1–exposed mice (i.e., enriched in −IL-1 SLAM cells; Fig. 3 A). We also identified a common signature of cell cycle and protein synthesis genes, including Myc itself, repressed in all three RNA-seq datasets (Fig. 3 B and Table S3).
Figure 3.
IL-1–induced gene repression is associated with high PU.1 levels. (A) GSEA enrichment of significantly downregulated genes in publicly available datasets versus RNA-seq analysis of SLAM cells from mice treated with or without IL-1β for 20 d. Data show downregulated genes as negatively enriched in SLAM cells from mice treated for 20 d with or without IL-1β. See also Table S5. (B) Venn diagram showing intersections between gene sets in A. A partial list of common genes is depicted at the right of the diagram. See also Table S6 for complete list of genes. (C) Left: Experimental design for analysis of PU.1-EYFP::GFP-Myc mice treated with or without IL-1β for 20 d. Right: Representative FACS plots showing gating strategy to identify PU.1lo and PU.1hi SLAM cells based on PU.1-EYFP expression levels in these mice. (D) Representative FACS plots (left) and quantification (right) showing PU.1-EYFP expression levels in PU.1lo and PU.1hi SLAM cell fractions from C (n = 3 −IL-1β; 5 +IL-1β). PU.1-EYFP negative control is shown in gray. Individual values are shown with bars representing mean values. Data are representative of two independent experiments. (E) Representative FACS plots (left) and quantification (right) showing GFP-Myc expression levels in PU.1lo and PU.1hi SLAM cell fractions from C (n = 3 −IL-1β; 5 +IL-1β). GFP-Myc negative control is shown in gray. Individual values are shown with bars representing mean values. Data are representative of two independent experiments. (F) Quantification by Fluidigm qRT-PCR array of cell cycle and protein synthesis gene expression in PU.1hi and PU.1lo SLAM cells from mice treated with or without IL-1β for 20 d (n = 8/group). Data are expressed as log10 fold expression versus −IL-1β. Box represents upper and lower quartiles with line representing median value. Whiskers represent minimum and maximum values. Data are representative of two independent experiments. (G) Representative FACS plots (left) and quantification (right) of cell cycle distribution in PU.1hi SLAM cells from mice treated with or without IL-1β for 20 d (n = 3/group) using Ki-67 and DAPI. Data are compiled from two independent experiments. (H) Representative FACS plots (left) and quantification (right) showing PU.1-EYFP expression levels in HSCLT from mice in C (n = 3 −IL-1β; 5 +IL-1β). PU.1-EYFP negative control is shown in gray in FACS plots. Individual values are shown with bars representing mean values. Data are representative of two independent experiments. (I) Representative FACS plots (left) and quantification (right) showing GFP-Myc expression levels in PU.1lo and PU.1hi SLAM cell fractions from mice in C. GFP-Myc negative control is shown in gray in FACS plots. Individual values are shown with bars representing mean values. Data are representative of two independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by Mann-Whitney U test or ANOVA with Tukey’s test in D–G. Error bars represent SD. See also Figs. S1 and S2.
We then generated PU.1-EYFP::GFP-Myc dual-reporter mice using previously published PU.1-EYFP and GFP-Myc reporter mouse strains (Fig. 3 C; Hoppe et al., 2016; Huang et al., 2008; Kirstetter et al., 2006). As these reporters consist of fluorescent fusion proteins knocked into the endogenous loci, we could directly read out in the same cells the correlation between PU.1 and Myc expression. We observed lineage-specific expression patterns of PU.1 and Myc in HSPCs from these mice, with granulocyte-macrophage progenitors (LK/CD41−/FcgR+; Pronk et al., 2007) expressing high levels of PU.1, whereas phenotypic CFU-erythroid (LK/CD41−/FcgR−/CD150−/CD105+) expressed high Myc levels but low PU.1 levels (Fig. S1, J and K). Consistent with our prior work, chronic IL-1 increased PU.1 expression and the frequency of PU.1hi SLAM cells (Fig. 3, C and D; Pietras et al., 2016). Notably, GFP-Myc levels decreased significantly in PU.1hi SLAM cells following chronic IL-1 exposure (Fig. 3 E), which we independently validated by staining for Myc protein in PU.1hi HSCLT (Fig. S2, A and B). These data support a model in which IL-1–induced PU.1 represses Myc. To assess whether increased PU.1 expression was associated with repression of other genes identified by our RNA-seq study, we performed Fluidigm qRT-PCR analysis on PU.1hi and PU.1lo SLAM cells isolated from mice treated with or without IL-1 for 20 d (Fig. S2 A). Expectedly, Spi1(PU.1) was further upregulated in PU.1hi SLAM cells by IL-1, whereas Myc and several cell cycle and protein synthesis genes repressed by IL-1 were specifically downregulated in PU.1hi SLAM cells (Fig. S2 C). Likewise, known IL-1/PU.1 target genes, such as Itgam, Flt3, and Il1r1, were elevated in PU.1hi SLAM cells and further upregulated by IL-1 treatment (Fig. S2 C). Our data also showed that PU.1hi SLAM cells expressed lower levels of Mk/E genes like Gata1 than PU.1lo SLAM cells, consistent with lineage-related anticorrelations between PU.1 and Gata-1 (Fig. S2 D). Furthermore, IL-1 exposure enriched for HSC genes in PU.1hi SLAM cells (Fig. S2 E), suggesting that the frequency of HSCLT in the PU.1hi phenotypic gate increases following IL-1 exposure. Consistent with PU.1-mediated repression of cell cycle and protein synthesis genes, in vivo chronic IL-1 exposure decreased cell cycle activity in PU.1hi SLAM cells, whereas IL-1 triggered increased cell cycle activity in the PU.1lo SLAM fraction (Fig. 3 G and Fig. S2 F). To link all of these findings back to phenotypic HSCLT under chronic IL-1 conditions, we assessed PU.1-EYFP and Myc-GFP levels after 20 d of IL-1 treatment. As predicted by our data, phenotypic HSCLT expressed elevated levels of PU.1-EYFP, whereas GFP-Myc levels were reduced (Fig. 3, H and I). We observed a similar pattern of cell cycle activity and gene expression after acute (1 d) in vivo IL-1 stimulation, with relatively limited induction of cell cycle activity in PU.1hi SLAM cells that coincided with repression of genes, including Myc, Ccnd1, and Rpl9 (Fig. S2, G and H), and increased expression of PU.1-EYFP in HSCLT (Fig. S2 I). Altogether, our data show that elevated PU.1 expression is associated with repression of cell cycle and protein synthesis genes in SLAM cells and HSCLT.
Figure S2.
Characterization of SLAM cell fractions based on PU.1 level. (A) Experimental design for analysis of Myc levels in PU.1hi SLAM cells. (B) Representative FACS plot (left) and quantification (right) of Myc levels in PU.1hi SLAM HSCs from PU.1-EYFP mice treated with or without IL-1β for 20 d (n = 4/group). Individual values are shown with bars representing means. Data are compiled from two independent experiments. (C–E) Quantification by Fluidigm qRT-PCR array of IL-1 target gene expression (C), lineage gene expression (D), and HSC gene expression (E) in PU.1lo and PU.1hi SLAM cells from PU.1-EYFP mice treated with or without IL-1β for 20 d (n = 8/group). Data are expressed as log10 fold expression versus −IL-1β. Box represents upper and lower quartiles with line representing median value. Whiskers represent minimum and maximum values. Data are representative of two independent experiments. (F) Representative FACS plots (left) and quantification (right) of cell cycle distribution in PU.1lo SLAM cells from mice treated with or without IL-1β for 20 d (n = 3/group) using Ki-67 and DAPI. Data are compiled from two independent experiments. (G) Experimental design (left) and quantification (right) of cell cycle distribution in PU.1lo and PU.1hi SLAM cells from mice treated with or without IL-1β for 1 d (n = 4/group). Data are compiled from two independent experiments. (H) Quantification by Fluidigm qRT-PCR array of cell cycle and protein synthesis genes in PU.1lo and PU.1hi SLAM cells from PU.1-EYFP mice treated with or without IL-1β for 1 d (n = 8/group). Data are expressed as log10 fold expression versus −IL-1β. Box represents upper and lower quartiles with line representing median value. Whiskers represent minimum and maximum values. Data are representative of two independent experiments. (I) Quantification of fold change (f.c.) in geometric MFI of HSCLT from PU.1-EYFP mice treated with or without IL-1β for 1 d (n = 5/group). Individual values are shown with bars representing mean values. Data are representative of two independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by Mann-Whitney U test or ANOVA with Tukey’s test in C–E, and H. Error bars represent SD.
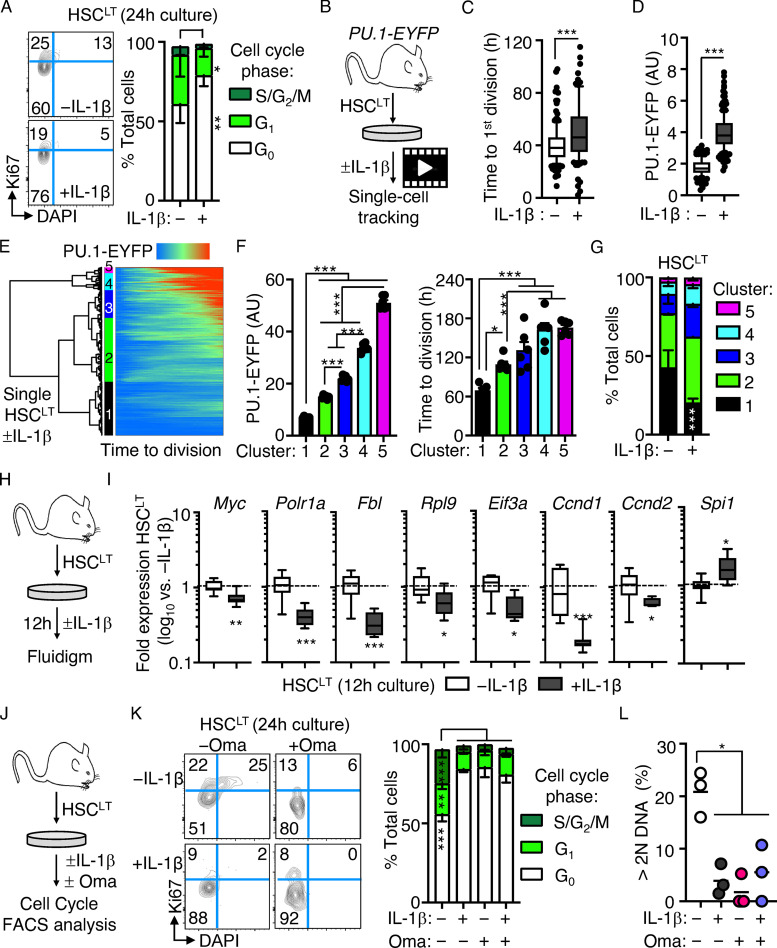
Direct IL-1 stimulation in vitro induces PU.1 and restricts HSCLT cell cycle entry
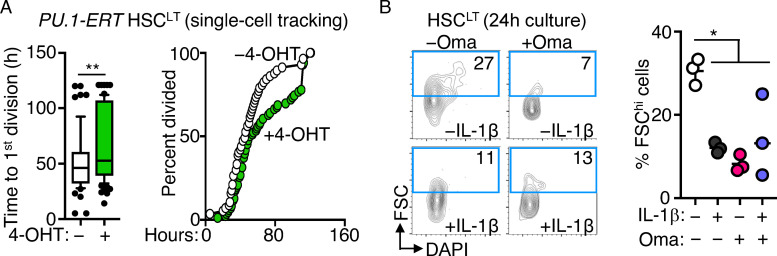
We next assessed whether IL-1 could directly restrict cell cycle activity following IL-1 stimulation of HSCLT in vitro. Notably, cell cycle entry was delayed in HSCLT cultured for 24 h with IL-1 (Fig. 4 A). To address the link with PU.1, we next analyzed HSCLT division kinetics with or without IL-1 using live single-cell imaging of cultured PU.1-EYFP HSCLT (Fig. 4 B). Consistent with our cell cycle analyses, IL-1 significantly delayed—but did not halt—the initial cell division of cultured HSCLT (Fig. 4 C), consistent with our prior single-cell tracking studies in SLAM cells (Pietras et al., 2016). PU.1-EYFP levels were also rapidly and significantly increased in HSCLT cultured with IL-1 before the first cell division, also consistent with previous observations for TNF (Fig. 4 D; Etzrodt et al., 2019). To more clearly delineate the relationship between PU.1 level and division kinetics, we performed unsupervised hierarchical clustering analysis of PU.1-EYFP expression patterns across time from first observation until cell division for all our tracked HSCLT with or without IL-1 (Fig. 4 E). Hierarchical clustering analysis identified five clusters based on PU.1-EYFP reporter expression dynamics in individual HSCLT (Fig. 4, E and F), with cluster 1 representing cells with low or nearly absent PU.1-EYFP reporter activity. Notably, HSCLT with low PU.1 expression levels (cluster 1) had significantly shorter division kinetics than HSCLT with higher PU.1 expression levels (clusters 2–5). To address the impact of IL-1, we next separated cells by treatment condition and analyzed the distribution of clusters in each. Strikingly, IL-1 treatment significantly reduced the number of rapidly dividing PU.1lo HSCs in cluster 1, with these cells instead distributing into clusters 2–5 (Fig. 4 G). We also assessed whether increased PU.1 expression was sufficient to delay HSCLT cell division by analyzing the division kinetics of transgenic PU.1-ERT HSCLT, which activate exogenous PU.1 upon induction with tamoxifen (4-OHT; Fukuchi et al., 2008). As expected, PU.1 activation enforced a delay in HSCLT division kinetics similar to the effects of IL-1 (Fig. S3 A). Taken together, these data indicate a positive correlation between PU.1 levels and cell cycle progression.
Figure 4.
Direct IL-1 stimulation in vitro induces PU.1 and restricts HSCLT cell cycle entry. (A) Representative FACS plots (left) and quantification (right) of cell cycle distribution in HSCLT cultured for 24 h with or without IL-1β (n = 6/group). Data are compiled from three independent experiments. (B) Experimental design for single-cell tracking studies of PU.1-EYFP HSCLT cultured with or without IL-1β. Time to first cell division was tracked via microscopy. (C) Graph showing kinetics of first cell division in HSCLT from B (n = 194 −IL-1β; 139 IL-1β). Data are compiled from three independent experiments. (D) PU.1-EYFP levels in HSCLT before first cell division (n = 137 −IL-1β; 192 +IL-1β). Data are compiled from three independent experiments. Box shows upper and lower quartiles with line showing median value, and whiskers upper and lower 10th percentile and individual dots represent outliers. (E) Hierarchical clustering analysis of PU.1-EYFP expression over time from the start of observation until the first division in PU.1-EYFP HSCLT cultured with or without IL-1 (Euclidean distance with Ward linkage; n = 557 −IL-1β; 489 +IL-1β). Data are compiled from three independent experiments. (F) Quantification of PU.1-EYFP level (as arbitrary units [AU]; left) and time to division (right) of HSCLT cultured with or without IL-1β in E. Individual values (representing means from cells cultured either with or without IL-1β in an experiment) are shown with bars representing mean values (n = 6). Data are compiled from three independent experiments. (G) Distribution of HSCLT from E in different clusters based on culture with or without IL-1β. Data are compiled from three independent experiments. (H) Experimental design for Fluidigm qRT-PCR array analysis of HSCLT cultured with or without IL-1β for 12 h. (I) Quantification by Fluidigm qRT-PCR array of cell cycle and protein synthesis gene expression in HSCLT from F (n = 8/group). Data are expressed as log10 fold expression versus −IL-1β. Box represents upper and lower quartiles with line representing median value. Whiskers represent minimum and maximum values. Data are representative of two independent experiments. (J) Experimental design for cell cycle analysis of HSCLT cultured with or without IL-1β and with or without 100 nM Oma for 24 h. (K) Representative FACS plots (left) and quantification (right) of cell cycle distribution in HSCLT from H (n = 3/group). Data are from one experiment. (L) Proportion of HSCLT with >2N DAPI signal from H. Individual values are shown with means (horizontal line). *, P < 0.05; **, P < 0.01; ***, P < 0.001 by Mann-Whitney U test or one-way ANOVA with Tukey’s test in F, G, K, and L. Error bars represent SD. See also Fig. S3.
Figure S3.
Impact of PU.1 expression and Oma on HSCLT. (A) Single-cell tracking studies of PU-ERT HSCLT cultured with or without 4-OHT. Quantification (left) of time to first cell division (n = 57 −4-OHT; 86 +4-OHT) and graph showing kinetics of first cell division in HSCLT (right). Data are representative of two independent experiments. Box shows upper and lower quartiles with line showing median value, and whiskers upper and lower 10th percentile and individual dots represent outliers. (B) Representative FACS plots (left) and quantification (right) of FSChi HSCs based on FSC/DAPI in HSCLT treated in vitro with or without IL-1β or with or without Oma for 24 h. Individual values are shown with lines representing mean values. Data are representative of two independent experiments. *, P < 0.05; **, P < 0.01 by Mann-Whitney U test or ANOVA with Tukey’s test in B. Error bars represent SD.
IL-1 treatment also rapidly repressed cell cycle and protein synthesis genes in HSCLT after 12 h in culture (Fig. 4, H and I), with a corresponding increase in Spi1. Given the broad downregulation of protein synthesis genes, we assessed whether reduced protein synthesis is sufficient to limit HSCLT cell cycle entry. We thus compared the impact of IL-1 versus omacetaxine (Oma), which binds the ribosomal A-site and inhibits protein synthesis (Gandhi et al., 2014), on HSCLT cell cycle entry (Fig. 4 J). Like IL-1, Oma effectively limited HSC cell cycle entry and reduced total Ki-67 protein expression (Fig. 4 K). Since reduced Ki-67 levels may be related to translation inhibition and not cell cycle changes, we independently read out cell cycle progression based on the frequency of cells with >2N DNA content (i.e., S/G2/M phases) based on DAPI alone. We observed a significant reduction in IL-1 and/or Oma-treated HSCLT in S phase or beyond, consistent with slowed cell cycle activity (Fig. 4 L). Oma and IL-1 also reduced the forward scatter (FSC) profile of cultured HSCs, consistent with previous observations correlating cell size and biosynthetic activity (Fig. S3 B; Iritani and Eisenman, 1999; Scognamiglio et al., 2016). Collectively, these data indicate that IL-1 directly and rapidly represses cell cycle and protein synthesis genes, thereby limiting HSCLT cell cycle entry in vitro.
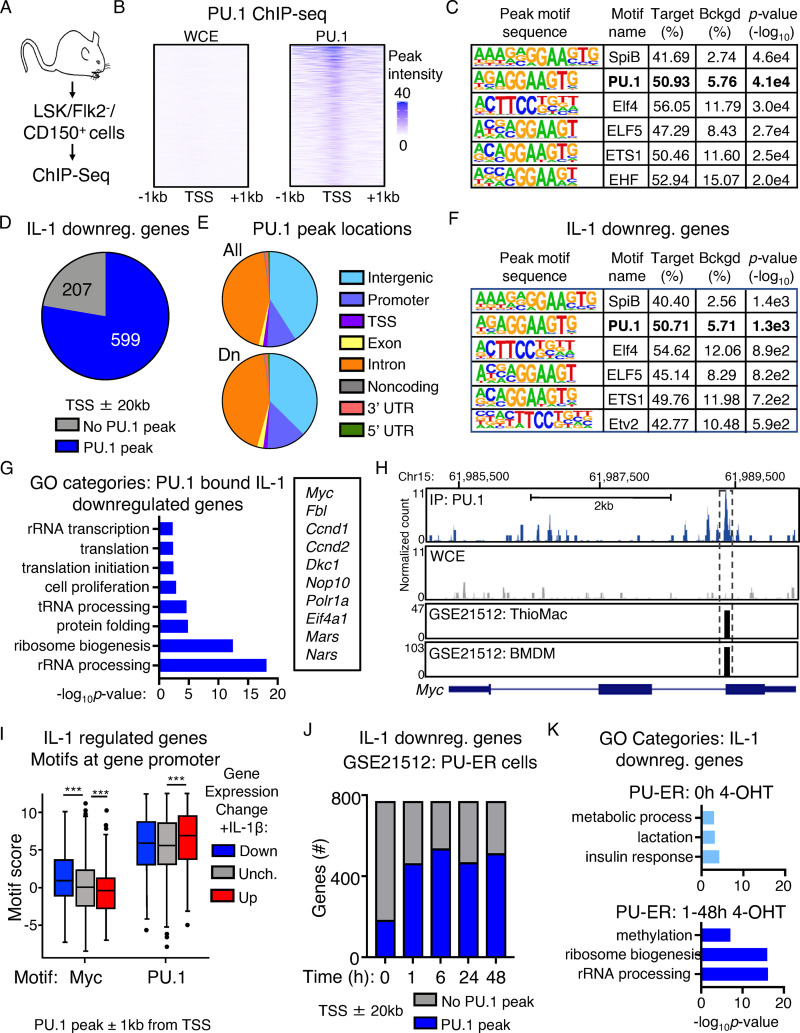
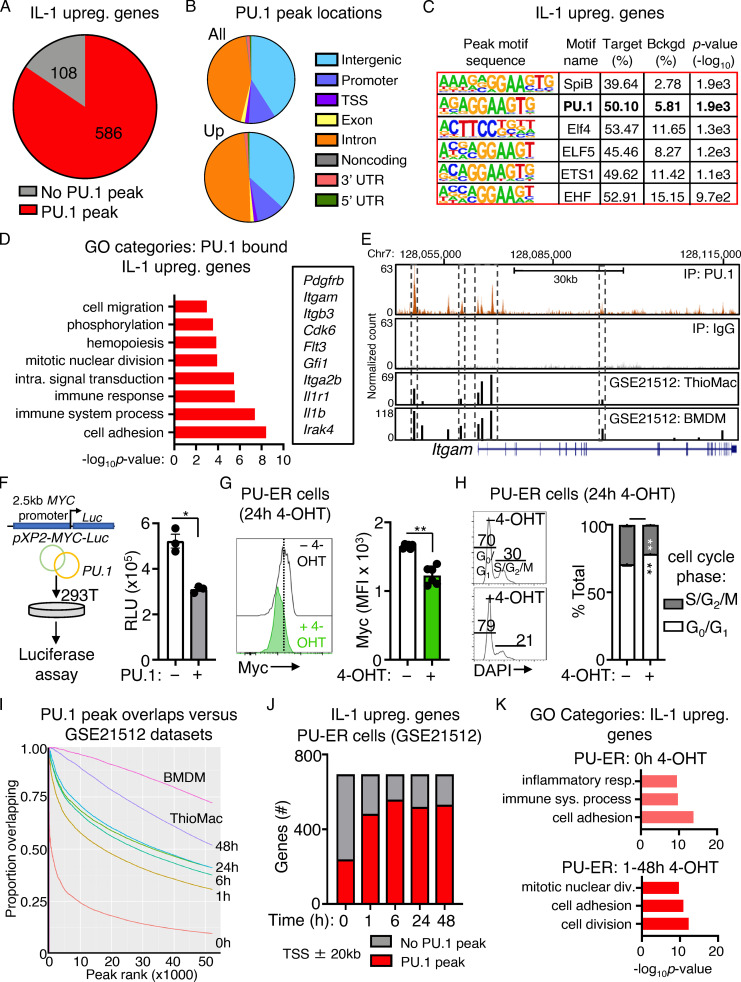
PU.1 directly binds cell cycle and protein synthesis genes repressed by IL-1
Given the association between high PU.1 levels and IL-1–induced repression of translation and cell cycle genes, we asked whether PU.1 directly interacts with these genes. We therefore analyzed genome-wide PU.1 binding using ChIP-seq analysis of HSC-enriched LSK/Flk2−/CD150+ cells (Fig. 5 A). Relative to whole-cell extract (WCE) controls, we identified 52,127 unique and specific PU.1 peaks located throughout the genome, with significant enrichment at 5′-GGAA-3′–containing consensus motifs within 200 bp of the peak sites (Fig. 5, B and C; and Table S4). Notably, the vast majority of IL-1 DEGs (586/694 IL-1 up; 599/771 IL-1 down) had PU.1 peaks located predominantly in intronic or intergenic regions within ± 20 kb of the gene transcription start site (TSS), likewise within 200 bp of 5′-GGAA-3′–containing consensus motifs (Fig. 5, D–F; Fig. S4, A–C; and Table S4). GO analysis of IL-1–repressed genes associated with PU.1 ChIP-seq peaks revealed expected enrichment for rRNA processing, cell proliferation, and translation categories (Fig. 5 G and Table S5). We next analyzed PU.1 peaks associated with the Myc gene and found several located within ±20 kb of the TSS itself, including a peak located within the gene body near the junction of intron 2/exon 3 (Fig. 5 H) and several located in the 3′ intergenic space (Table S4). The intronic Myc peak was also present in two independent published macrophage datasets (Heinz et al., 2010; GEO accession no. GSE21512). Indeed, PU.1 peaks identified in our ChIP-seq dataset corresponded closely with peaks in these two datasets (Table S4). Conversely, IL-1–induced genes with PU.1 peaks were enriched for cell adhesion, immune response, and other expected gene categories (Fig. S4 D and Table S5), including the PU.1 target gene Itgam, which contained several peaks located both in the gene body as well as in the 5′ intergenic region, including a site near the TSS/promoter as previously characterized and a major peak that may represent an enhancer site (Fig. S4 E). Interestingly, we also noted that a subset of IL-1–downregulated genes with PU.1 were also significantly enriched for Myc motifs within 1 kb of their TSS relative to upregulated or unchanged genes (Fig. 5 I and Table S4), further underscoring the negative regulatory relationship between PU.1 and Myc target genes. We also used an in vitro luciferase reporter assay to demonstrate that PU.1 can directly repress activity of the human Myc promoter (Fig. S4 F), in line with prior studies indicating that PU.1 can be recruited to the Myc promoter in complex with histone deacetylase proteins (Kihara-Negishi et al., 2001). To further characterize the dynamics of PU.1 interactions with IL-1 target genes, we reanalyzed published ChIP-seq data (Heinz et al., 2010; GEO accession no. GSE21512) generated in PU.1−/− fetal liver hematopoietic progenitor cells expressing a 4-OHT–inducible PU.1 transgene (PU-ER cells) at different time points postinduction (Heinz et al., 2010). These cells constitutively express a low level of PU.1 from the transgene, with 4-OHT rapidly inducing PU.1 protein activation (Walsh et al., 2002). We found that activation of the PU.1 transgene leads to reduced Myc expression and a small but significant cell cycle delay in PU-ER cells, suggesting that, despite the difference in cell type and origin, the response to PU.1 expression in this system resembles key features of IL-1–treated HSCLT (Fig. S4, G and H). In the PU-ER cell ChIP-seq dataset, a small fraction (177 total) of IL-1–repressed genes were constitutively bound by low levels of PU.1 present in the absence of 4-OHT (0 h). Interestingly, those genes were unrelated to cell cycle or protein synthesis (Fig. 5, J and K). The number of IL-1–repressed genes bound by PU.1 increased significantly and rapidly with 4-OHT treatment, with top GO categories of these genes now including ribosome biogenesis and rRNA processing (Fig. 5, J and K). We observed a similar pattern of expression for IL-1–upregulated genes, though in this case inflammatory response genes were already bound by PU.1 at 0 h (Fig. S4, J and K; and Table S5) and transgene-inducible peaks centered around cell adhesion and mitotic cell division gene categories (Fig. S4 K), with the latter centered around genes required for late cell cycle stages rather than quiescence exit. Taken together, these data support a model in which PU.1 represses gene expression by directly binding a broad set of cell cycle and protein synthesis genes. They also suggest that, at reduced levels, PU.1 does not bind the majority of IL-1–repressed cell cycle and protein synthesis genes.
Figure 5.
PU.1 directly binds cell cycle and protein synthesis genes repressed by IL-1. (A) Experimental design for ChIP-seq experiment (n = 2/group). (B) Heatmap showing PU.1 ChIP-seq peak intensities versus WCE control at TSS ± 1 kb. (C) Transcription factor binding site motif enrichment at ChIP-seq peak sites. (D) Pie chart comparing IL-1–downregulated genes identified in SLAM cells by RNA-seq analysis in Fig. 1 and presence of PU.1 peaks at or near these genes (TSS ± 20 kb). (E) Pie chart showing PU.1 peak locations in IL-1–downregulated genes. (F) Transcription factor binding site motif enrichment at PU.1 ChIP-seq peak sites in IL-1–downregulated genes (TSS ± 20 kb). (G) GO category enrichment of IL-1–downregulated DEGs containing PU.1 peaks in C. Representative genes from the indicated categories are shown to the right. Data are expressed as −log10 P value. (H) UCSC genome browser rendering of a PU.1 peak location in Myc gene body. Tracks show PU.1 ChIP-Seq and WCE control, with corresponding peak locations and intensities in thioglycollate-elicited primary mouse macrophage (ThioMac) and BM-derived macrophage (BMDM) PU.1 ChIP-seq datasets from GSE21512. (I) Myc and PU.1 motif enrichment (motif score) at TSS ± 1 kb in IL-1–downregulated, –upregulated, or unchanged genes containing PU.1 peaks. Box shows upper and lower quartiles with line showing median value, and whiskers upper and lower 10th percentile and individual dots represent outliers. ***, P ≤ 0.001 based on Wilcoxon rank sum test. (J) Comparison of IL-1–downregulated genes in SLAM HSCs with genes containing PU.1 peaks within TSS ± 20 kb in PU-ER cells with or without 4-OHT. Based on PU.1 ChIP-seq datasets in GSE21512. (K) GO category enrichment of IL-1–downregulated DEGs containing PU.1 peaks in PU-ER cell ChIP-seq dataset at 0 h +4-OHT versus combined 1–48 h +4-OHT. Top three GO categories are shown. Data are expressed as −log10 P value. See also Fig. S4.
Figure S4.
PU.1 binding to IL-1–upregulated genes and analysis of PU-ER cells. (A) Pie chart comparing IL-1–upregulated genes identified in SLAM cells by RNA-seq analysis in Fig. 1 D and presence of PU.1 peaks at or near these genes (TSS ± 20 kb) in ChIP-seq data. See also Table S4. (B) Pie chart showing proportion of PU.1 peak locations in all genes versus IL-1–upregulated genes. (C) Transcription factor binding site motif enrichment at PU.1 ChIP-seq peak sites located at TSS ± 20 kb in IL-1–downregulated genes. (D) GO category enrichment of IL-1–upregulated DEGs containing PU.1 peaks. Representative genes in the indicated categories are shown to the right. Data are expressed as −log10 P value. See also Table S5. (E) UCSC genome browser rendering of PU.1 peak location in Itgam gene body. Tracks show PU.1 ChIP-seq, WCE control, and peak locations and intensities in thioglycollate-elicited primary mouse macrophage (ThioMac) and BM-derived macrophage (BMDM) PU.1 ChIP-seq datasets from GSE21512. (F) Luciferase reporter assay measuring MYC promoter activity in 293T cells with or without PU.1 (n = 3/group). Data are representative of two independent experiments. (G) Quantification of Myc protein expression in PU-ER cells after 24 h culture with or without 4-OHT (n = 6/group). Data are representative of two independent experiments. Individual values are shown with bars representing mean values. (H) Cell cycle activity in PU-ER cells after 24 h culture with or without 4-OHT (n = 6/group). Data are representative of two independent experiments. (I) Comparison of PU.1 peak overlaps between PU.1 ChIP-seq data in A and datasets from GSE21512. (J) Comparison of IL-1–upregulated genes in SLAM HSCs with genes containing PU.1 peaks within TSS ± 20 kb in PU-ER cells with or without 4-OHT. Based on PU.1 ChIP-seq datasets in GSE21512. (K) GO category enrichment of IL-1–downregulated DEGs containing PU.1 peaks in PU-ER cell ChIP-seq dataset at 0 h with 4-OHT versus combined 1–48 h with 4-OHT. The top three GO categories are shown. Data are expressed as −log10 P value. *, P < 0.05; **, P < 0.01 by Mann-Whitney U test or ANOVA with Tukey’s test in H. Error bars represent SD.
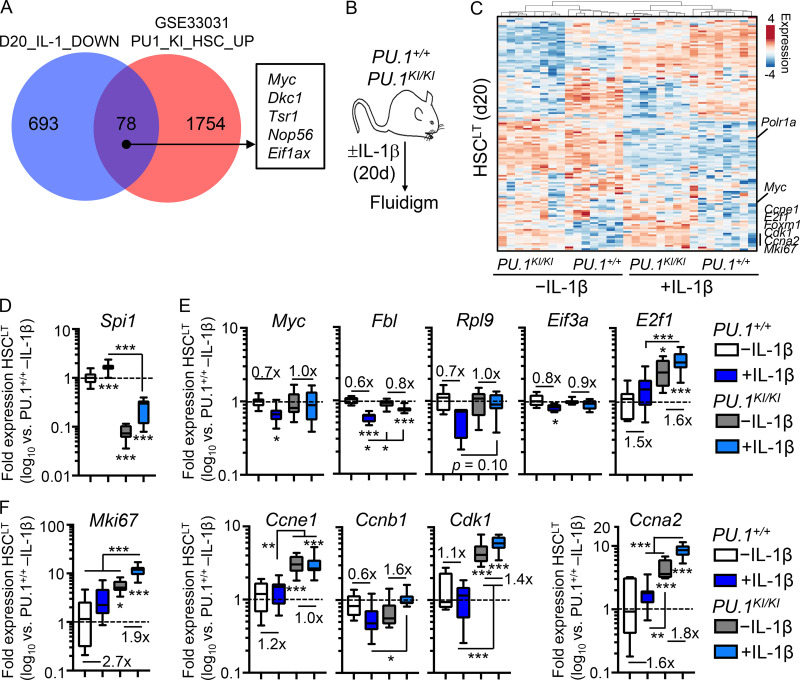
PU.1-deficient HSCLT overexpress cell cycle and protein synthesis genes
To further explore the impact of reduced PU.1 expression on HSCLT function, we analyzed PU.1KI/KI mice, which express ∼30% of normal PU.1 levels in SLAM cells due to a deactivating point mutation knocked into the 14-kb upstream Spi1 autoregulatory binding motif (Staber et al., 2013). As SLAM cells from these mice exhibit derepression of cell cycle genes, including Ccnd1, E2f, and Cdk1, we first compared our RNA-seq data to published gene expression microarray data from PU.1+/+ and PU.1KI/KI SLAM cells (Staber et al., 2013; GEO accession no. GSE33031). Expectedly, several cell cycle and protein synthesis genes repressed by IL-1, including Myc, were significantly upregulated in PU.1KI/KI SLAM cells (Fig. 6 A). We therefore assessed gene expression in HSCLT from PU.1KI/KI mice and PU.1+/+ littermate controls treated with or without IL-1 for 20 d by Fluidigm qRT-PCR array (Fig. 6 B). We confirmed a significant reduction of Spi1 gene expression in PU.1KI/KI HSCLT (Fig. 6 D). Cdk1, E2f1, and Myc were overexpressed in PU.1KI/KI HSCLT, consistent with prior characterizations of PU.1-deficient HSCs under homeostatic conditions (Fig. 6, E and F; Rosenbauer et al., 2004; Staber et al., 2013). We observed increased expression of several cell cycle and protein synthesis genes, including Ccne1, Ccna2, and Mki67 in PU.1KI/KI HSCLT (Fig. 6, E and F). Chronic IL-1 exposure further increased expression of these genes in PU.1KI/KI HSCLT (Fig. 6, E and F). On the other hand, IL-1–mediated repression of other target genes, such as Myc, Rpl9, Eif3a, and Fbl, was attenuated or absent in PU.1KI/KI HSCLT (Fig. 6, E and F). We observed broadly similar patterns of aberrant gene expression in PU.1KI/KI HSCLT after acute IL-1 treatment in vivo (Fig. S5 A). These data suggest that PU.1 is required to limit the expression of cell cycle and protein synthesis genes both at steady state and under inflammatory stress.
Figure 6.
Aberrant cell cycle and protein synthesis gene expression in PU.1-deficient HSCLT. (A) Venn diagram comparing genes downregulated by IL-1 in SLAM cells (Fig. 1) and genes upregulated in PU.1KI/KI SLAM cells. (B) Experimental design of Fluidigm qRT-PCR array analyses of HSCLT from PU.1+/+ and PU.1KI/KI mice treated with or without IL-1β for 20 d. (C) Heatmap with hierarchical clustering analysis (Pearson correlation with average linkage) of gene expression data from HSCLT in B. (D) Quantification by Fluidigm qRT-PCR array of Spi1 gene expression in HSCLT from B. Data are expressed as log10 fold expression versus −IL-1β. Box represents upper and lower quartiles with line representing median value. Whiskers represent minimum and maximum values. Data are representative of two independent experiments. (E) Quantification of cell cycle and protein synthesis gene expression by Fluidigm qRT-PCR array analysis in D. (F) Quantification of cell cycle gene expression by Fluidigm qRT-PCR array analysis in D. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by ANOVA with Tukey’s test. See also Fig. S5.
Figure S5.
PU.1 conditional knockout analysis and culture of PU.1KI/KI HSCLT. (A) Quantification by Fluidigm qRT-PCR array of cell cycle and protein synthesis gene expression in HSCLT from mice treated with or without IL-1β for 1 d (n = 8/group). Data are expressed as log10 fold expression versus −IL-1β. Box represents upper and lower quartiles with line representing median value. Whiskers represent minimum and maximum values. Data are from one experiment. (B) Study design for Cre induction with 4-OHT and analysis of HSCLT from SCL-CreERT PU.1+/+ and PU.1Δ/Δ mice treated with or without IL-1β for 20 d (n = 2–3/group). (C) Intracellular flow cytometry analysis of puro incorporation in HSCLT from mice in E. Puro was injected i.p. 1 h before BM harvest. Data are expressed as fold change of MFI versus −IL-1β. Individual values are shown with bars representing mean values. Data are representative of two independent experiments. (D) Quantification of cell cycle phase distribution in HSCLT from PU.1+/+ and PU.1Δ/Δ mice in B based on Ki-67 and DAPI. Data are representative of two independent experiments. (E) Experimental design for analysis of HSCLT from PU.1+/+ and PU.1KI/KI mice treated with or without IL-1β for 1 d. (F) Representative FACS plots (left) and quantification (right) of cell cycle phase distribution in HSCLT from mice in H based on Ki-67 and DAPI (n = 7–8/group). Data are compiled from two experiments. (G) Myc levels from PU.1+/+ and PU.1KI/KI mice treated with IL-1β for 1 d (n = 4–5/group). Individual values are shown with bars representing mean values. Data are compiled from two experiments. (H) Experimental design for competitive in vitro assays on Boy/J and PU.1KI/KI HSCLT cultured in a 1:1 ratio with or without IL-1β for 12 d. (I) Quantification of total cells (left) and immature Sca-1+/cKit+ progenitors at the indicated time points (n = 3/group). (J) Quantification of frequency of total (left) and immature (right) CD45.2+ cells derived from PU.1KI/KI HSCLT cultured with or without IL-1β at the indicated time points. Percentages >50 (color coded green in the graphs) are indicative of a competitive advantage for PU.1KI/KI cells. Data are representative of two independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by Mann-Whitney U test in G or ANOVA with Tukey’s test. Error bars represent SD.
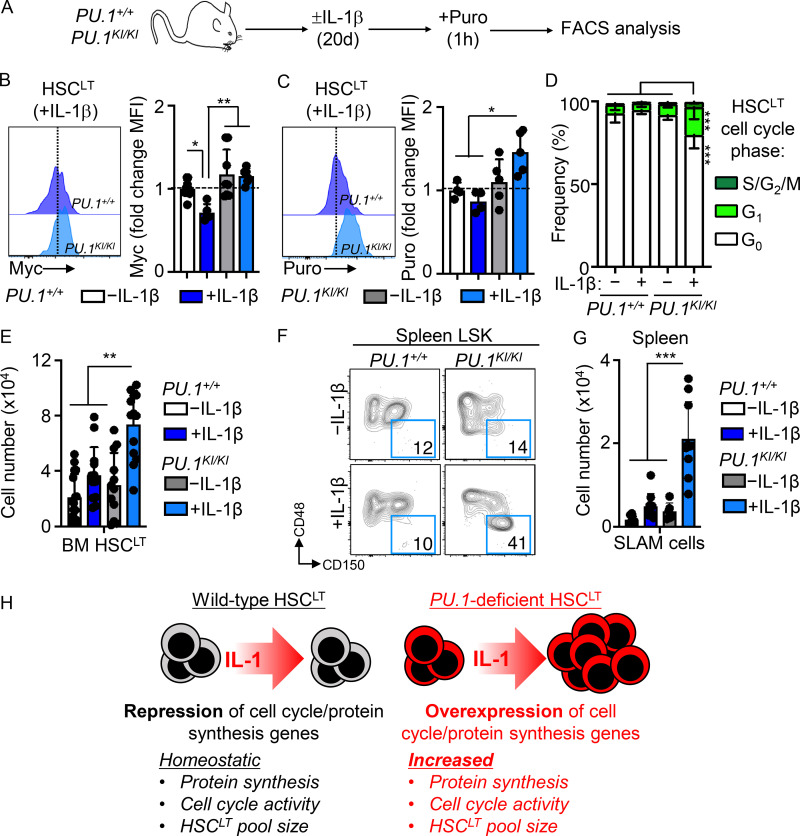
Chronic IL-1 induces aberrant cell cycle activity and expansion of PU.1KI/KI HSCLT
We next assessed how the molecular deregulations in PU.1KI/KI HSCLT impacted the properties of these cells under inflammatory stress (Fig. 7 A). Consistent with our qRT-PCR data, IL-1–exposed PU.1KI/KI HSCLT expressed higher relative levels of Myc than PU.1+/+ HSCLT (Fig. 7 B). Chronic IL-1 also triggered exuberant protein synthesis activity in PU.1KI/KI HSCLT (Fig. 7 C). To assess the functional consequences, we assessed cell cycle activity in PU.1+/+ and PU.1KI/KI HSCLT. Strikingly, IL-1 triggered increased cell cycle activity in PU.1KI/KI HSCLT (Fig. 7 D). We independently confirmed IL-1–dependent activation of aberrant protein synthesis and cell cycle activity in HSCLT from conditional SCL-Cre-ER::PU.1Δ/Δ mice treated with or without IL-1 for 20 d (Fig. S5, B–D). In addition, consistent with our qRT-PCR analyses after acute IL-1 stimulation (Fig. S5 A), we found abnormally high cell cycle activity and Myc expression in PU.1KI/KI HSCLT after single IL-1 injection (Fig. S5, E–G). Given that IL-1 triggered increased protein synthesis and cell cycle activity in PU.1-deficient HSCLT, we reasoned this could lead to aberrant expansion of the phenotypic HSCLT pool. We first addressed this question by monitoring HSCLT expansion using an expedient system in which PU.1KI/KI HSCLT were cultured 1:1 with CD45.1+ Boy/J HSCLT. Notably, PU.1KI/KI HSCLT grew poorly in culture (Fig. S5 I); however, addition of IL-1 significantly potentiated the expansion of these cells (Fig. S5 I). In fact, immature c-Kit+/Sca-1+ progenitors derived from PU.1KI/KI HSCLT rapidly dominated the cultures relative to Boy/J competitor cells (Fig. S5, I and J). These data suggested that IL-1 might trigger expansion of PU.1KI/KI HSCLT in vivo. Hence, we assessed the number of HSCLT in the BM of PU.1+/+ and PU.1KI/KI mice treated with or without IL-1 for 20 d. As anticipated, IL-1 triggered aberrant expansion of phenotypic HSCLT exclusively in the BM of PU.1KI/KI mice (Fig. 7 E). Notably, this phenotype was not confined to the BM, as we also observed significant expansion of phenotypic SLAM cells in the spleens of IL-1–treated PU.1KI/KI mice (Fig. 7, F and G). Taken together, these data demonstrate that aberrant protein synthesis and cell cycle activity are emergent properties of PU.1-deficient HSCLT that can be triggered by IL-1 and result in the expansion of phenotypic HSCLT in the BM and extramedullary sites (Fig. 7 H). Altogether, these data show that PU.1 is required to limit HSCLT cell cycle activity, thereby restricting HSCLT expansion during chronic inflammatory conditions.
Figure 7.
Chronic IL-1 induces aberrant cell cycle activity and expansion of PU.1-deficient HSCLT. (A) Experimental design for analyses of HSCLT from PU.1+/+ and PU.1KI/KI mice treated for 20 d with or without IL-1β. (B) Intracellular flow cytometry analysis of Myc protein levels in HSCLT from PU.1+/+ and PU.1KI/KI mice treated with or without IL-1β for 20 d (n = 4–9/group) Data are expressed as fold change of MFI versus −IL-1β. Individual values are shown with bars representing mean values. Data are compiled from two independent experiments. (C) Intracellular flow cytometry analysis of puro incorporation in HSCLT from PU.1+/+ and PU.1KI/KI mice treated with or without IL-1β for 20 d (n = 4 PU.1+/+; 5 PU.1KI/KI). Data are expressed as fold change of MFI versus −IL-1β. Individual values are shown with bars representing mean values. Data are from one experiment. (D) Quantification of cell cycle phase distribution in HSCLT from mice in A. Data are compiled from two independent experiments. (E) Quantification of BM HSCLT from mice in A. Individual values are shown with bars representing mean values. Data are compiled from three independent experiments. (F) Representative FACS plots showing SLAM cells in the spleens of PU.1+/+ and PU.1KI/KI mice treated for 20 d with or without IL-1β. (G) Quantification of SLAM cells in the spleens of mice in A. Individual values are shown with bars representing mean values. Data are compiled from two independent experiments. (H) Cartoon showing key features of WT and PU.1-deficient HSCLT. WT HSCLT (left) engage a cell cycle and protein synthesis repression gene program that limits protein synthesis, cell cycle activity, and HSCLT pool size following challenge with IL-1. On the other hand, PU.1-deficient HSCLT overexpress cell cycle and protein synthesis genes, priming them for increased protein synthesis and cell cycle activity that is associated with aberrant expansion of the HSCLT pool following IL-1 challenge. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by ANOVA with Tukey’s test. Error bars represent SD. See also Fig. S5.
Discussion
Here, we show that IL-1 exposure represses a broad set of genes regulating HSCLT cell cycle and protein synthesis activity. We show that this gene program is linked to restricted HSCLT cell cycle entry and thus underwrites the quiescent phenotype of HSCs under chronic inflammatory stress. Notably, PU.1 is a crucial driver of this program, with PU.1 itself binding the majority of target genes repressed by IL-1. Strikingly, our data demonstrate this molecular state leaves PU.1-deficient HSCLT poised to exit quiescence and support aberrant expansion of the phenotypic HSCLT pool when triggered by IL-1. Our data thus identify a PU.1-driven molecular mechanism enforcing HSC quiescence during inflammatory stress and support a model in which PU.1 serves as a critical limiting mechanism that regulates HSC cell cycle activity and pool size in this context.
We find that the IL-1/PU.1 axis represses cell cycle and protein synthesis gene expression and limits HSCLT proliferative activity under all conditions studied, including in vitro culture and in vivo treatment of mice under both acute and chronic conditions. The activating versus suppressive functions of the IL-1/PU.1 axis may at first appear paradoxical. Indeed, previous reports indicate that acute IL-1 administration in vivo triggers cell cycle activity in the SLAM compartment (Hemmati et al., 2019; Pietras et al., 2016; Weisser et al., 2016), which we recapitulate here. On the other hand, our H2B-GFP and cell cycle analyses show that only a limited fraction of phenotypic HSCLT enter the cell cycle following IL-1 challenge in vivo, particularly relative to the larger SLAM compartment, which is known to contain HSC-like CD41hi SLAM cells that can rapidly enter the cell cycle in response to acute inflammatory stress (Haas et al., 2015). Likewise, BrdU label-retention experiments show that, while stressors such as LPS can trigger proliferation in HSC-enriched cells, a sizeable fraction of these cells nonetheless retains the BrdU label (Takizawa et al., 2017; Wilson et al., 2008), and that a subset of CD49blo HSCs that overlap phenotypically with the EPCR+/CD34− HSCLT fraction used here (Rabe et al., 2020) can remain dormant even following 5-fluorouracil myeloablation (Zhao et al., 2019). These data all support a model in which HSCLT engage mechanisms to limit cell cycling in response to inflammatory stress. Our findings that PU.1-deficient HSCLT cycle excessively following IL-1 stimulation indicate that PU.1 is indeed one of these mechanisms. In this context, it is worth noting that published BM chimera studies show that in vivo HSC proliferation in response to IL-1 can be triggered independently of IL-1 receptor (IL-1R) on HSCs themselves (Cain et al., 2011; Ueda et al., 2009) and may rely on IL-1R–induced G-CSF production in the BM niche. Hence, the proliferative effect of IL-1 on HSCLT may in fact be the result of indirect mechanisms related to rapid mobilization of granulocytes from the BM, whereas direct IL-1 signaling triggers PU.1-dependent cell cycle restriction. Detailed studies using BM chimera models can further clarify the direct versus indirect effects of IL-1 on HSCLT proliferation in vivo.
We also previously reported that IL-1 activates precocious myeloid differentiation in HSCs (Pietras et al., 2016), and in the present study we see induction of myeloid genes in HSCLT; however, we demonstrate that IL-1 also activates a PU.1 program that restricts HSC cell cycle activity and expansion by repressing a broad set of cell cycle and protein synthesis genes. We made similar observations in HSCs from mice with CIA (Hernandez et al., 2020). Repression of cell cycle and protein synthesis genes by PU.1 may therefore be a common mechanism triggered in HSCs by at least a subset of inflammatory stimuli, including some TLRs, which share downstream signaling pathways with IL-1. On the other hand, analysis of HSCs from PU.1-EYFP mice treated acutely with TLR3 and -4 ligands polyriboinosinic: polyryibocyticylic acid and LPS, respectively, show that PU.1 is induced via a TNF-dependent mechanism (Etzrodt et al., 2019; Yamashita and Passegué, 2019). Furthermore, cytokines such as IFN do not robustly activate PU.1 expression in a direct fashion (Etzrodt et al., 2019), suggesting that other mechanism(s) may enforce HSC quiescence under chronic IFN stimulation, such as activation of p53 and/or the mRNA translation-blocking activity of several IFN-stimulated genes (Li et al., 2015; Pietras et al., 2014).
It is important to note that, while PU.1 is commonly characterized as a lineage differentiation factor, its role as a regulator of cell proliferation has also been extensively studied (Delestré et al., 2017; Fukuchi et al., 2008; Oikawa et al., 1999; Solomon et al., 2017; Ziliotto et al., 2014). The connection between cell cycle inhibition and myeloid differentiation likely centers on cell cycle lengthening as a mechanism to promote PU.1 accumulation and myeloid differentiation in hematopoietic progenitors (Kueh et al., 2013). In agreement with this model, our data show that increased PU.1 activity serves as more of a rheostat than a bona fide proliferation block in HSCLT, erecting enough of an activation barrier to prevent excessive proliferation during inflammatory challenge while still priming HSCs for myeloid differentiation as evidenced by simultaneous upregulation of myeloid determinant genes in IL-1–exposed HSCLT. This is a function common to numerous myeloid transcription factors, including Cebpa family members and Gfi1, which is itself a PU.1 target (Hock et al., 2004; Porse et al., 2005; Pulikkan et al., 2017; Staber et al., 2013; Umek et al., 1991; Zeng et al., 2004). Thus, induction of a PU.1-dependent cell cycle restriction gene program in HSCLT under inflammatory stress conditions is consistent with a model in which PU.1 promotes efficient generation of myeloid progenitors to reconstitute hematopoiesis, while protecting the HSC compartment from damage and/or depletion. Absence of this program likely underlies the failure of PU.1KI/KI mice to reconstitute hematopoiesis following 5-fluorouracil treatment (Staber et al., 2013), which we interpret as a coupled effect of increased HSC proliferation leading to apoptosis, and inefficient generation of new myeloid progenitors due to reduced PU.1 accumulation. Altogether, this mechanism may be a remarkable example of parsimony in a biological system, wherein a single molecular program can mediate distinct functional outcomes based on cell type and context.
Our data agree with prior studies showing that PU.1 deficiency leads to derepression of Myc and numerous cell cycle genes, including Cdk1 and E2f1 (Rosenbauer et al., 2004; Staber et al., 2013; Will et al., 2015), which is exacerbated by IL-1 exposure. Indeed, we find that PU.1 directly binds a wide array of genes that regulate cell cycle and protein synthesis activity. In that setting, it is unlikely that repression of a single factor, such as Myc, is the sole mechanism limiting HSCLT cell cycle activity. Indeed, we find that inhibition of protein synthesis in cultured HSCLT using Oma phenocopies the delayed cell cycle entry observed with IL-1 stimulation, which is consistent with our model that PU.1 inhibition of multiple protein synthesis genes contributes to enforced HSC quiescence. Notably, Oma is a U.S. Food and Drug Administration–approved therapy for chronic myelogenous leukemia and can directly ablate MDS stem cells with aberrant levels of protein synthesis (Stevens et al., 2018). Protein synthesis and cell cycle activity are closely linked, as cell cycle entry and mitosis require a significant amount of new protein. Hence, in eukaryotic cells, deletion and/or knockdown of key genes regulating protein synthesis, such as translation elongation initiation factors (eIFs), several of which are repressed by IL-1 in our dataset, results in delayed cell cycle entry (Polymenis and Aramayo, 2015). The importance of eIFs in initiating HSC cell cycle activity has been illustrated by elegant work in which dual deletion of 4E-BP1/2, which negatively regulates translation by inhibiting eIF4E, leads to increased protein synthesis, aberrant cell cycle activity, and HSC expansion (Signer et al., 2016). Likewise, deletion of Pten, which dephosphorylates and inhibits the eIF activator mammalian target of rapamycin, leads to similar aberrant HSC activity and hypersensitivity to inflammatory factors, such as G-CSF and IFN (Porter et al., 2016; Signer et al., 2014), indicating that protein synthesis levels must be carefully regulated for normal HSC function. The phenotype of Pten- and 4E-BP1/2–deficient HSCs is strikingly reminiscent of that of IL-1–exposed PU.1-deficient HSCLT in our model. In a similar line, inflammatory initiation of a cell cycle–primed Galert state in HSCs and other cells requires activation of mammalian target of rapamycin (Rodgers et al., 2014). We also identified decreased expression of ribosomal RNA genes, as well as genes such as Fbl required to process them for assembly into ribosomes. While cells synthesize ribosomes independently of cell cycle phase, inhibition of rRNA production can halt cell cycle activity (Polymenis and Aramayo, 2015) with accompanying decreases in cell size, similar to our findings in HSCLT exposed to IL-1 and/or Oma in culture. Along these lines, HSCs carrying a hypomorphic allele of the ribosomal protein Rpl24 exhibit decreased proliferative activity and can rescue the phenotype of Pten-deficient HSCs (Signer et al., 2014). Likewise, IL-1 downregulates expression of Myc, a crucial upstream regulator of protein synthesis and cell cycle genes that leads to defective cell cycle entry if deleted in stem cells (Laurenti et al., 2008; Scognamiglio et al., 2016; Wilson et al., 2004). Together, these data suggest that fine tuning of protein synthesis is essential for maintaining HSC quiescence and function (Signer et al., 2014). Interestingly, despite IL-1–mediated downregulation of protein synthesis genes and slowed cell cycle progression in our in vitro cultures, we do not observe decreased protein synthesis rates in IL-1–exposed HSCLT in vivo. Repression of protein synthesis genes may therefore dampen the effects of IL-1–driven mitogenic signaling and maintain homeostatic protein synthesis activity rather than block it altogether, which in vitro is read out as slowed cell cycle progression in response to the stress of being placed in culture. IL-1 activates numerous signaling pathways that directly impact protein synthesis and cell cycle activity, including PI3K/Akt, p38 MAPK, MAPK/ERK kinase (MEK)/ERK, and RAS (Dinarello, 2018). Thus, in future studies, we hope to identify which pathway(s) are potential triggers that activate aberrant protein synthesis and cell cycle activity in PU.1-deficient HSCs, as well as the extent to which inhibiting them can rescue this phenotype.
Dysregulated inflammatory signaling has emerged as a key partner alongside oncogenic mutations in the development and/or progression of hematological malignancy (Barreyro et al., 2018; Pietras, 2017). Aberrant signaling via IL-1 and the downstream IL-1 receptor associated kinase/TNF receptor associated factor pathway shared by IL-1R and TLRs has been implicated as a driver of survival and/or expansion of MDS and myeloid leukemia stem cells (Ågerstam et al., 2016; Askmyr et al., 2013; Barreyro et al., 2012; Carey et al., 2017; Muto et al., 2020; Smith et al., 2019; Zhang et al., 2016). Indeed, blockade of these pathways can limit and/or reverse disease progression (Carey et al., 2017; Mitchell et al., 2018; Rhyasen et al., 2013; Zhang et al., 2016); however, the mechanism(s) by which inflammatory signaling initiates expansion of oncogenically mutated HSCs remain obscure. Chronic inflammation, often associated with increased IL-1 activity, is a common phenotype in individuals at risk for hematological malignancy, which can be related to several preexisting factors, including aging/tissue decline, genotoxin exposure, metabolic dysfunction, and/or autoimmune disease (Barreyro et al., 2018; Pietras, 2017). Our data suggest that, while chronic IL-1 exposure can induce expansion of myeloid-biased hematopoietic progenitors and mature myeloid cells, this phenotype is essentially self-limiting and does not deterministically progress to malignancy. These data are consistent with the overall rarity of hematological malignancy among individuals with chronic inflammatory phenotypes, despite the relative increase in risk (Anderson et al., 2009; Gañán-Gómez et al., 2015). On the other hand, loss-of function mutations in PU.1 itself are rarely observed in hematological malignancy, although a wide array of myeloid leukemia–associated oncogenic lesions can interfere with PU.1 expression or function, including PML/RARA, AML1-ETO, NPM1c, CEBPA, and mitogenic kinase mutations in BCR/ABL and Flt3ITD (Gerloff et al., 2015; McKenzie et al., 2019; Mueller et al., 2006; Noguera et al., 2016; Vangala et al., 2003; Yang et al., 2012). In addition, TET2 and/or DNMT3A mutations associated with early oncogenesis may interfere with PU.1 function due to aberrant methylation of PU.1 binding sites (Kaasinen et al., 2019). Hence, deficiency in key myeloid regulators, like PU.1, related to indirect effects of mutations in other genes may serve as a crucial nexus that primes HSCs to activate aberrant protein synthesis and cell cycle activity when triggered by inflammatory signals associated with BM pathogenesis, like IL-1 and TNF-α. This model is illustrated by a parallel study by our group in which we found that IL-1 can trigger the selective expansion of Cebpa-deficient HSPCs in a mouse model of oncogenic BM competition (Higa et al., 2021). These findings are consistent with the theory of adaptive oncogenesis, which stipulates that environmental factors associated with tissue decline, such as inflammation, are crucial drivers of oncogenesis by selecting for emergent phenotypes, such as increased proliferation, self-renewal, or survival, that are linked cancer-associated mutations (Henry et al., 2015; Laconi et al., 2020). Hence, oncogenic mutations and inflammatory signals may collaborate to activate emergent proliferative phenotypes in stem cells that, in turn, lead to aberrant expansion of downstream progenitors and myeloid cells that can initiate disease. Such a model provides a rationale for exploring the use of anti-inflammatory therapies as a means of preventing and/or delaying leukemogenesis in at-risk individuals.
Materials and methods
Mice
WT C57BL/6, congenic B6.SJL-PtprcaPepcb/BoyJ (Boy/J) mice, GFP-Myc mice (Huang et al., 2008), and Col1a1-TetO::rtTA-H2B-GFP mice (Foudi et al., 2009) were obtained from The Jackson Laboratory. PU.1KI/KI mice and PU.1flox mice (Staber et al., 2013) were a kind gift of Dan Tenen (Harvard Stem Cell Institute, Cambridge, MA). PU.1flox mice were crossed onto a 4-OHT–inducible SCL-CreERT driver strain (Göthert et al., 2005) for experiments. PU.1-EYFP mice (Hoppe et al., 2016; Kirstetter et al., 2006) were a kind gift of Dr. Claus Nerlov (MRC Weatherall Institute, Oxford, UK). PU.1ERT2 mice (Fukuchi et al., 2008) were kindly provided by Dr. Hideyaki Nakajima (Department of Stem Cell and Immune Regulation, Yokohama City University School of Medicine, Yokohama, Japan). WT littermate controls were used for experiments involving PU.1KI/KI mice and PU.1flox mice. 6–12-wk-old animals of both sexes were used for experiments. All animal experiments were conducted in accordance with a protocol approved by the Institutional Animal Care and Use Committee at University of Colorado Anschutz Medical Campus (protocol no. 00091).
In vivo procedures
For in vivo IL-1 stimulation, IL-1β (Peprotech) was resuspended in sterile Dulbecco’s PBS/0.2% BSA. 0.5 µg IL-1 or PBS/BSA alone was injected i.p. in a 100-µl bolus once per day for either 1 or 20 d, as previously described (Pietras et al., 2016; Rabe et al., 2020). In vivo puro labeling assays were performed as previously described (Chapple et al., 2018), with 500 µg puro (Life Technologies) injected i.p. 1 h before euthanasia. In vivo H2B-GFP labeling was performed by feeding mice with doxycycline chow (2 g/kg; Teklad) ad libitum for 2 wk (Säwén et al., 2016). Animals were allowed to rest for 1 wk before IL-1 treatment. Induction of the SCL-CreERT transgene in PU.1 conditional KO mice was performed by injecting mice i.p. with 25 mg/kg 4-OHT resuspended in corn oil daily for 3 d. Mice were allowed to rest for 1 wk before the start of IL-1 treatment.
Flow cytometry
Analysis of BM cell populations was performed using a similar protocol as previously described (Pietras et al., 2016; Rabe et al., 2020). BM was flushed from femurs and tibiae of mice using staining media (SM; Hanks’ Buffered Saline Solution + 2% FBS) and a syringe equipped with a 21-G needle. Cells were subsequently resuspended in 1× ACK (150 mM NH4Cl/10 mM KHCO3) to remove erythrocytes, washed with SM, filtered through 70-µm mesh to remove debris, resuspended in SM, and counted on a Vicell automated counter (Beckman Coulter). For quantification of immature HSPCs, 107 BM cells were blocked with purified rat IgG (Sigma-Aldrich) and stained for 30 min on ice with SM containing the following antibodies: PE-Cy5–conjugated anti-CD3, CD4, CD5, CD8, Gr-1, and Ter119 as a lineage exclusion stain; Flk2-biotin; CD34-FITC; EPCR (endothelial protein C receptor)-PE; Mac-1–PE/Cy7; CD16/32-APC; CD48-A700; and cKit-APC/Cy7. Cells were subsequently washed, resuspended in a 1:4 dilution of Brilliant Staining Buffer (Becton Dickinson) in SM containing Sca-1-BV421, CD41-BV510, CD105-BV711, CD150-BV785, and streptavidin (SA)-BV605 and incubated for 30 min on ice. For analysis of GFP-Myc::PU.1-EYFP cells, a custom 510/20-nm (GFP) and 550/30-nm (EYFP) bandpass filter setup (Becton Dickinson) was used to discriminate the two fluorescent proteins. BM cells from single-positive mice were used for compensation controls and fluorescence-minus-one controls, and GFP−/YFP− BM cells were surface stained alongside and used as negative controls. For analysis of mature BM cell populations, cells were blocked as above and stained for 30 min on ice with the following antibodies: Gr-1–Pacific Blue, Ly6C-BV605, B220-BV786, CD4-FITC, CD8-PE, Mac-1–PE/Cy7, IgM-APC, CD3-A700, and CD19-APC/Cy7. Following surface staining, cells were washed with SM, resuspended in SM containing 1 µg/ml propidium iodide, and immediately analyzed on a three-laser, 12-channel FACSCelesta analyzer (Becton Dickinson) or a five-laser, 18-channel BD Fortessa. For analysis of spleen cells, spleens were removed and minced through a 70-µm filter basket (VWR) to make a single-cell suspension. Splenocytes were treated with ACK buffer as above and subsequently processed for staining of mature and immature hematopoietic populations, as described above.
Cell cycle analysis was performed using protocols similar to previous publications (Hernandez et al., 2020; Jalbert and Pietras, 2018). 107 BM cells were stained for 30 min on ice with the following antibodies: PE-Cy5–conjugated anti-CD3, CD4, CD5, CD8, Gr-1 and Ter119 as a lineage exclusion stain, Flk2-biotin, CD34-FITC, EPCR-PE, Sca-1–PE/Cy7, CD48-A700, and c-Kit–APC/Cy7. Cells were washed with SM and resuspended in 1:4 Brilliant Buffer:SM containing SA-BV605 and CD150-BV785, incubated for 30 min on ice, washed in SM, and fixed for 20 min at room temperature (RT) with Cytofix/Cytoperm (Becton Dickinson). Cells were subsequently washed with 1× PermWash buffer (Becton Dickinson), permeabilized for 10 min at RT with Perm Buffer Plus (Becton Dickinson), washed in PermWash, and refixed for 5 min at RT with CytoFix/Cytoperm. Cells were subsequently washed in PermWash and incubated with anti–Ki-67 antibody in PermWash buffer for 30 min at RT. Cells were then washed and either stored at 4°C or resuspended in Dulbecco’s PBS containing 1 µg/ml DAPI and analyzed on a BD LSRII analyzer equipped with a UV laser (Becton Dickinson).
Myc staining was performed as previously described (Freire and Conneely, 2018). 107 BM cells were stained for 30 min on ice with SM containing the following antibodies: PE-Cy5–conjugated anti-CD3, CD4, CD5, CD8, Gr-1 and Ter119 as a lineage exclusion stain, Flk2-biotin, CD34-FITC, EPCR-PE, Mac-1–PE/Cy7, CD48-A700, and cKit-APC/Cy7. Cells were washed in SM and then stained for 30 min in 1:4 Brilliant Buffer:SM containing Sca-1–BV421, SA-BV605, and CD150-BV785. Cells were subsequently washed in SM and fixed for 20 min with CytoFix/CytoPerm. Cells were washed with PermWash and blocked in PermWash for 1 h at RT, followed by washing and a 1-h RT incubation with anti-Myc purified antibody. Cells were subsequently stained with an A647-conjugated anti-rabbit Fab for 30 min at RT, washed in PermWash, resuspended in SM, and analyzed on an LSRII.
Puro stainings were performed as previously described (Chapple et al., 2018). Cells were surface stained and fixed using the same approach as for Myc staining. Following fixation, cells were stained with anti-puro antibody diluted in PermWash buffer for 1 h at RT, washed and stained with an anti-mouse IgG2a antibody for 30 min, washed once more, and resuspended in SM for analysis on an LSRII.
For H2B-GFP dilution analysis, we used splenocytes from Ubc-GFP mice (Schaefer et al., 2001) as a GFP compensation control to ensure comparable brightness to the GFP signal. BM cells were stained as above and analyzed on an LSRII or sorted on a FACSAria Fusion sorter. GFP dilution analyses were performed by using the proliferation analysis function of FlowJo using default settings. All samples from an individual experiment were concatenated before running the algorithm, and GFP expression bins were subsequently assigned to individual samples.
All fluorescence intensity data throughout the manuscript are based on the geometric mean fluorescence intensity (MFI) of that parameter. A complete list of FACS antibodies and dilutions used can be found in Table S6.
Cell sorting
For HSC isolation by cell sorting, arm, leg, pelvic bones, and spines were isolated from mice as previously described (Pietras et al., 2016; Rabe et al., 2020). Bones were crushed in SM using a mortar and pestle and depleted of erythrocytes with 1× ACK. Cells were subsequently placed atop a Histopaque 1119 gradient (Sigma-Aldrich) and centrifuged. To enrich c-Kit+ cells, BM cells were incubated on ice for 20 min with c-Kit microbeads (5 µl/100 µl SM per mouse; 130–091-224; Miltenyi Biotec), washed with SM, and enriched on an AutoMACS Promagnetic cell separator (Miltenyi Biotec). Enriched cells were subsequently washed, blocked with rat IgG, and stained for 30 min on ice with the following antibodies: PE-Cy5–conjugated anti-CD3, CD4, CD5, CD8, Gr-1, and Ter119 as a lineage exclusion stain, Flk2-biotin, CD34-FITC, ECPR-PE, Mac-1–PE/Cy7, CD48-A700, and c-Kit–APC/Cy7. Cells were subsequently washed with SM and resuspended in a 1:4 dilution of Brilliant Staining Buffer in SM containing Sca-1–BV421, SA-BV605, and CD150-BV785. For B220+ cell isolation by cell sorting, spleens were harvested, pressed over a 70-μm filter, and depleted of erythrocytes. To enrich for B220+ cells, splenocytes were incubated on ice for 20 min with B220 microbeads (20 µl/1,000 µl SM per mouse; Miltenyi Biotec), washed with SM, and enriched on an AutoMACS Pro. Enriched cells were washed, blocked with rat IgG, and stained with B220-APC for 15 min on ice. Purified B220+ cells were immediately fixed for 20 min at RT (100 µl; BD Cytofix/Cytoperm), washed with SM, and stored at 4°C.
In vitro culture assays
Purified HSCLT were cultured using a similar protocol as that previously published (Pietras et al., 2016). Double-sorted cells were grown in StemPro34 containing Stempro supplement, antibacterial-antimycotic (100×; Gibco), L-glutamine (100×; Gibco), stem cell factor (SCF; 25 ng/ml), thrombopoietin (TPO; 25 ng/ml), IL-3 (10 ng/ml), GM-CSF (20 ng/ml), Flt3L (50 ng/ml), IL-11 (50 ng/ml), erythropoietin (4 U/ml), and IL-1β (25 ng/ml) and were incubated at 37°C, 5% O2, 5% CO2 for either 12 or 24 h. Cell culture grade puro (1 nM; Gibco) was added 1 h before harvesting cells to monitor protein synthesis rates. In experiments containing Oma (100 nM; Teva Pharmaceuticals), cells were cultured in the presence of Oma for 1 h before IL-1β addition and remained in cultures for the duration of the experiment. Following indicated times in culture, cells were harvested and either resorted based on viability for Fluidigm gene expression analysis or were immediately fixed for 20 min at RT (100 µl; BD Cytofix/Cytoperm) for Ki-67/DAPI analysis or puro incorporation assays. Following fix/perm, 5 × 105 sorted, fixed B220+ spleen cells were added to each sample as carrier cells and excluded during analysis based on B220 expression (Matatall et al., 2018). PU.1-ERT HSCLT were cultured as above but with or without 100 nM 4-OHT. PU-ER cells were cultured in Iscove's Modification of Dulbecco’s medium (IMDM) + 10% heat-inactivated serum and 5 mg/ml IL-3 and GM-CSF. Transgene was induced by resuspending cells in media containing 100 nM 4-OHT. Cells were harvested after 24 h and analyzed for cell cycle activity or Myc expression levels as described above.
Luciferase reporter assays
pcDNA3-PU.1 (human PU.1) and pXP2-MYC luciferase reporter constructs were kind gifts of Dan Tenen (Harvard Stem Cell Institute). pXP2-MYC contains the 2.5-kb Xmn1 fragment of the human MYC promoter driving a luciferase reporter. 293T cells were transfected with pXP2-Myc and either pcDNA3-PU.1 or empty pcDNA3 using PolyJet in vitro DNA transfection reagent (Signagen) according to the manufacturer’s instructions. 24 h later, cells were lysed and analyzed for luciferase activity using the Pierce Firefly Luciferase Glow Assay (Invitrogen). Luciferase activity was read out on a microplate reader (Promega).
Single-cell tracking analysis
Time-lapse experiments were conducted at 37°C, 5% O2, 5% CO2 on μ-slide VI0,4 channels slides (IBIDI) coated for 1 h at RT with 5 or 10 µg/ml anti-CD43-biotin, as previously described (Loeffler et al., 2018). Cells were cultured in phenol red–free IMDM supplemented with 20% bovine serum albumin/human insulin/human transferrin, 2 mM L-glutamine, 50 µM 2-mercaptoethanol and 50 U/ml penicillin, 50 µg/ml streptomycin, 25 ng/ml SCF, 25 ng/ml TPO, 10 ng/ml IL-3, 20 ng/ml GM-CSF, 50 ng/ml Flt3L, 50 ng/ml IL-11, and 4 U/ml EPO. 25 ng/ml IL-1b and/or 500 nM 4-OHT (Sigma-Aldrich) was added as indicated. Images were acquired using a Nikon-Ti Eclipse equipped with a linear encoded motorized stage, Orca Flash 4.0 V2 (Hamamatsu), Spectra X fluorescent light source (Lumencor), and The Cube (Life Imaging Service) temperature control system. White light emitted by Spectra X was collimated and used as a transmitted light for brightfield illumination via a custom-made motorized mirror controlled by Arduino UNO Rev3 (Arduino). Fluorescent images were acquired using optimized filter sets eGFP (470/40 nm; 495LP; 525/50 nm), YFP (500/20 nm; 515LP; 535/30 nm), mKO2 (546/10 nm; 560LP; 577/25 nm), mCherry (550/32 nm; 585LP; 605/15 nm), and Cy5 (620/60 nm; 660LP; 700/75 nm; all AHF) to detect GFP-cMYC, PU.1-YFP, CD71-PE, tetramethylrhodamine, and CD71-APC, respectively. Time intervals of brightfield and fluorescent image acquisition were chosen to minimize phototoxicity. Images were acquired using a 10× CFI Plan Apochromat λ objective (NA 0.45). Single-cell tracking and image quantification were performed using self-written software as described (Hilsenbeck et al., 2016; Loeffler et al., 2018). Software used for data acquisition of time-lapse imaging data is published and open sourced (YouScope v.2.1; http://langmo.github.io/youscope/). Software for single-cell tracking and fluorescence quantification used in this study is published and open sourced (Hilsenbeck et al., 2016). Segmentation software is published and open sourced (Hilsenbeck et al., 2017). Acquired 16-bit images with 2048 × 2048–pixel resolution were saved as .png and linearly transformed to 8-bit using channel-optimized white points and corrected for background and shading (Peng et al., 2017) before analysis. Brightfield images were used for segmentation using fastER (Hilsenbeck et al., 2016), eroded to reduce segmentation artifacts caused by close cell proximity (settings: morphological transformation x = 3, y = 3, op = 2, shape: 2), and subsequently dilated (settings: simple dilation 6) to ensure proper segmentation and quantification results. Tracking and quantification of fluorescence channels were done as previously described (Hilsenbeck et al., 2016) and analyzed using Matlab 2018b (MathWorks). For hierarchical clustering analyses, PU.1YFP expression levels of in vitro–cultured HSCs were quantified from the start of culture until cell division. PU.1.YFP fluorescence intensities were z-normalized across replicates, with each single-cell time series normalized to 40 time points using Matlab’s spline function and clustered using hierarchical clustering with Euclidean distance and ward linkage using Matlab 2019b. The number of clusters was chosen based on (1) the reproducibility of cluster frequencies across replicates and (2) to minimize intracluster variance.
RNA-seq
RNA was isolated from individual pools of 104 to 2 × 104 double-sorted SLAM cells using an RNEasy Micro kit (Qiagen). RNA was quantified and quality checked using an Agilent Bioanalyzer 2100 (Agilent Technologies). 1 ng of total RNA was preamplified using the SMARTer Ultra-Low Input kit v4 (Clontech) according to the manufacturer’s instructions, and cDNA quality was assessed using the Qubit Fluorometer (Life Technologies) and the Bioanalyzer. 150 pg cDNA was used to generate Illumina sequencing libraries using the NexteraXT kit (Illumina) according to the manufacturer’s instructions. Libraries were subsequently hybridized to an Illumina single-end flow cell and amplified using cBot (Illumina). Sequencing of libraries was performed on Illumina HiSeq 2500v4 high-throughput sequencer. Single-ended reads of 100 nt were generated for each sample and demultiplexed using bcl2fastq v1.8.4. Trimmomatic v0.32 was used for quality filtering and adapter removal. Processed reads were mapped to the mouse genome GRCm38.p4 (mm10/mg38) using STAR_2.4.2a. Raw read counts were obtained via htseq-count v0.6.1 and Gencode-M12 gene annotations. Differential expression analyses were performed using deSeq2 in Bioconductor. Genes with Padj < 0.05 were considered significant DEGs. Upstream regulator analysis of DEGs was performed using IPA software on default settings. GSEA analyses were performed on DEGs using default settings. GO analyses were performed using DAVID v6.8 (http://david.ncifcrf.gov; Huang et al., 2009). Venn diagrams were generated from DEG lists using IteractiVenn (http://www.interactivenn.net; Heberle et al., 2015).
ChIP-seq
ChIP-seq analysis was performed on LSK/Flk2−/CD150+ cells sorted from the BM of eight C57BL/six mice per ChIP. Cells were cross-linked in 0.75% formaldehyde, pelleted, and frozen until the ChIP procedure. Pellets were thawed on ice, resuspended in lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCL [pH 8.1] plus protease inhibitor) for 10 min. Volume was brought up to 1 ml with ChIP dilution buffer (16.7 mM Tris-HCl [pH 8.1], 167 mM NaCl, 0.01% SDS, 1.1% Triton X-100, and 1.2 mM EDTA plus protease inhibitor) and samples were sonicated with a Branson sonicator to shear chromatin. Chromatin was subsequently incubated with 1 µg PU.1 antibody (sc-390405; Santa Cruz Biotechnology) and rotated overnight at 4°C. Next, ChIPs were added to a 50:50 mix of prewashed Protein A/G Dynabeads (50 µl; 10001D/10009D; Thermo Fisher Scientific) and rotated overnight at 4°C. The next day, 175 µl RIPA/140 mM NaCl buffer (0.1% sodium deoxycholate, 0.1% SDS, 1% Triton X-100,140 mM NaCl, 1 mM EDTA, 20 mM Tris-HCl [pH 8.1]) was added to each tube and beads were transferred to a 96-well plate on a magnet, washed twice with cold RIPA/500 ml NaCl buffer (0.1% sodium deoxycholate, 0.1% SDS, 1% Triton X-100, 140 mM NaCl, 1 mM EDTA, and 20 mM Tris-HCl [pH 8.1]), twice with cold LiCl buffer (0.25 M LiCl, 1% NP-40, 1% Na deoxycholate, 1 mM EDTA, and 10 mM Tris-HCl [pH 8.1]), and twice with RT TE buffer (10 mM Tris-HCl [pH 8.0] and 1 mM EDTA [pH 8.0]). Chromatin was eluted by adding 50 µl of elution buffer (10 mM Tris-Cl [pH 8.0], 5 mM EDTA, 300 mM NaCl, 0.1% SDS, and 5 mM dithiothreitol) and 8 µl of reverse cross-linking buffer (250 mM Tris-HCl [pH 6.5], 1.25 M NaCl, 62.5 mM EDTA, 5 mg/ml Proteinase K, and 62.5 μg/ml RNase A) and incubated at 65°C overnight. ChIP material was quantified on a Qubit spectrometer (Invitrogen) according to manufacturer’s instructions. Library construction was performed first using an End-it DNA End-Repair Kit (ER0720; Epicenter) according to manufacturer’s instructions, followed by DNA ligase-based adapter ligation (M2200S; New England Biolabs) and enrichment of adapter-modified DNA fragments by PCR. PCR fragments were gel purified using a Qiagen Gel Purification Kit (Qiagen) and sequenced on an Illumina single-end flow cell. Sequencing of libraries was performed on Illumina HiSeq 2500v4 high-throughput sequencer. Fastq read files were trimmed using Cutadapt (Martin, 2011). The trimmed reads were mapped to mm10 mouse genome using HISAT2 (Kim et al., 2019). Peak calling was conducted by HOMER program run in factor mode (false discovery rate < 0.001). The intergenic peaks nearest a TSS were annotated as the corresponding gene, and peaks within a gene were further categorized into exonic, intronic, 3′ or 5′ untranslated regions using HOMER mm10 annotation. Venn diagrams were generated from DEG lists using IteractiVenn (http://www.interactivenn.net; Heberle et al., 2015).
Fluidigm qRT-PCR
Fluidigm qRT-PCR analysis was performed using previously published protocols (Hernandez et al., 2020; Pietras et al., 2016; Rabe et al., 2020). Pools of 100 cells were sorted directly into a 96-well PCR plate (AB2396; Thermo Fisher Scientific) containing 5 µl of 2× CellsDirect Reaction Mix (Invitrogen). Immediately following sort, the plate was sealed, centrifuged at 500 ×g for 5 min, snap frozen, and stored at −80°C. cDNA was generated from RNA by reverse transcription using Superscript III Taq polymerase (Invitrogen) and preamplified with a custom 96-target DeltaGene (Fluidigm) primer set for 18 cycles on a thermocycler (Eppendorf). Any excess primers from the preamplification reaction were removed by Exonuclease-I (New England Biolabs) incubation, and samples were diluted in DNA suspension buffer (Teknova) before chip loading. A Fludigm 96.96 Dynamic Array integrated fluidics circuit was loaded with cDNA, primers, and SsoFast Sybr Green Master Mix (Bio-Rad) and analyzed on a BioMark HD system (Fluidigm). Subsequently, data were analyzed using Fluidigm Gene Expression Software and all values were normalized to Gusb. The ΔΔCt approach was used to identify relative changes in gene expression. Hierarchical clustering and principal component analyses of normalized expression data were performed using ClustVis (https://biit.cs.ut.ee/clustvis; Metsalu and Vilo, 2015). A complete list of qRT-PCR primers included in the custom primer sets as well as their sequences can be found in Table S6. Primer sets for Hes1, Hoxa2, Hmga1, Ms4a3, Lcn2, and Sod2 performed poorly due to low expression levels in HSCLT, and resulting data were not included in downstream analyses.
Statistical analysis
Statistical analyses were performed using Prism software (GraphPad). For RNA-seq, deSeq2 was used. Means and SD are reported except where noted. In figure legends, n refers to the number of biological replicates. The number of independent experiments from which the replicates derive is also reported. Except for RNA-seq and ChIP-seq analyses, statistical significance was determined by Student’s t test or Mann-Whitney U test (bivariate comparisons) or one-way ANOVA with Tukey's test (multivariate comparisons).
Data availability
Sequencing data from this study are publicly available from the National Center for Biotechnology Information GEO accession no. GSE165810. Other datasets generated during the current study are available from the corresponding author upon reasonable request. Any data supporting the findings of this study are available from the corresponding author upon reasonable request.
Online supplemental material
Fig. S1, related to Figs. 1, 2, and 3, shows additional analyses and Fluidigm qRT-PCR array validation of RNA-seq data, representative FACS gating for HSCLT and characterization of Myc and puro incorporation in HSCs and MPP, and representative FACS gating and controls for H2B-GFP and GFP-Myc::PU.1-EYFP mice. Fig. S2, related to Fig. 3, contains independent validation of Myc expression changes in PU.1hi SLAM cells, as well as additional characterization of PU.1hi and PU.1lo SLAM cells after acute (1 d) and chronic (20 d) IL-1 treatment. Fig. S3, related to Fig. 4, shows division kinetics of PU.1-ERT HSCLT and further characterization of HSCLT cultured with IL-1. Fig. S4, related to Fig. 5, shows further characterization of PU.1 ChIP-seq datasets, MYC luciferase reporter assay, and characterization of Myc and cell cycle activity in PU-ER cells. Fig. S5, related to Figs. 6 and 7, shows additional characterization of PU.1KI/KI HSCLT after acute IL-1 challenge, independent in vivo validation of PU.1KI/KI results using SCL-CreERT–driven PU.1 conditional knockout mice, and competitive cell culture analysis of PU.1KI/KI HSCLT. Table S1 provides the list of DEGs in SLAM cells from mice treated with or without IL-1β for 20 d. Table S2 provides GO categories, IPA upstream regulator categories, and GSEA enrichment analysis of DEGs in SLAM cells from mice treated with or without IL-1β for 20 d. Table S3 provides overlaps between IL-1 downregulated genes and public datasets. Table S4 provides PU.1 ChIP-seq peak locations, scores, and PU.1/Myc motif scores. Table S5 provides GO categories of genes enriched in PU.1 ChIP-seq peak locations. Table S6 provides a list of antibodies and primers used in this study.
Supplementary Material
displays DEGs in SLAM cells from mice treated with or without IL-1β for 20 d.
provides an analysis of DEGs, with GO categories, IPA upstream regulator categories, and GSEA enrichment analysis of DEGs in SLAM cells from mice treated with or without IL-1β for 20 d.
shows a comparison of DEGs with public data, with overlap between IL-1–downregulated genes and public datasets.
displays PU.1 ChIP-seq peak locations, scores, and PU.1/Myc motif scores.
provides an analysis of PU.1 target genes, with GO categories enriched in IL-1 DEGs containing PU.1 ChIP-seq peaks.
lists the antibodies, antibody dilutions, and Fluidigm qRT-PCR primers used in this study.
Acknowledgments
We thank Garrett Hedlund for expert assistance with flow cytometry resources. We also thank the University of Colorado Office of Laboratory Animal Resources for expert vivarium administration and assistance with animal procedures.
This work was supported by National Institutes of Health grants R01 DK119394 and K01 DK098315, the Boettcher Webb-Waring Early Career Investigator Award, and the Cleo Meador and George Ryland Scott Endowed Chair in Hematology (E.M. Pietras); National Institutes of Health grant R01 AG067584 (J. DeGregori); National Institutes of Health grant F31 HL138754 (J.L. Rabe); National Institutes of Health grants F30 CA210383 and T32 AG000279 (K.C. Higa); Swiss National Science Foundation grant 179490 (T. Schroeder); and the National Science Foundation Graduate Research Fellowship Program (T.S. Mills). This work was supported in part by the University of Colorado Cancer Center Flow Cytometry Shared Resource, funded by National Cancer Institute grant P30 CA046934.
Author contributions: Conceptualization: E.M. Pietras; methodology: E.M. Pietras, J. DeGregori, D. Loeffler, K.C. Higa, and T. Schroeder; formal analysis: D. Loeffler, N. Ahmed, H. Kim, J.R. Myers, and B.M. Stevens; investigation: J.S. Chavez, J.L. Rabe, D. Loeffler, G. Hernandez, R.L. Gessner, K.E. Niño, Z. Ke, T.S. Mills, K.C. Higa, N. Ahmed, B.M. Idler, and E.M. Pietras; resources: H. Nakajima, T. Schroeder, P. Davizon-Castillo, and R.S. Welner; writing – original draft: E.M. Pietras and J.S. Chavez; writing – review and editing: E.M. Pietras, J.S. Chavez, and J. DeGregori; supervision: E.M. Pietras, J. DeGregori, C.T. Jordan, J. Ashton, and T. Schroeder; funding acquisition: E.M. Pietras and J. DeGregori.
References
- Ågerstam, H., Hansen N., von Palffy S., Sandén C., Reckzeh K., Karlsson C., Lilljebjörn H., Landberg N., Askmyr M., Högberg C., et al. 2016. IL1RAP antibodies block IL-1-induced expansion of candidate CML stem cells and mediate cell killing in xenograft models. Blood. 128:2683–2693. 10.1182/blood-2015-11-679985 [DOI] [PubMed] [Google Scholar]
- Anderson, L.A., Pfeiffer R.M., Landgren O., Gadalla S., Berndt S.I., and Engels E.A.. 2009. Risks of myeloid malignancies in patients with autoimmune conditions. Br. J. Cancer. 100:822–828. 10.1038/sj.bjc.6604935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askmyr, M., Ågerstam H., Hansen N., Gordon S., Arvanitakis A., Rissler M., Juliusson G., Richter J., Järås M., and Fioretos T.. 2013. Selective killing of candidate AML stem cells by antibody targeting of IL1RAP. Blood. 121:3709–3713. 10.1182/blood-2012-09-458935 [DOI] [PubMed] [Google Scholar]
- Barreyro, L., Will B., Bartholdy B., Zhou L., Todorova T.I., Stanley R.F., Ben-Neriah S., Montagna C., Parekh S., Pellagatti A., et al. 2012. Overexpression of IL-1 receptor accessory protein in stem and progenitor cells and outcome correlation in AML and MDS. Blood. 120:1290–1298. 10.1182/blood-2012-01-404699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreyro, L., Chlon T.M., and Starczynowski D.T.. 2018. Chronic immune response dysregulation in MDS pathogenesis. Blood. 132:1553–1560. 10.1182/blood-2018-03-784116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujanover, N., Goldstein O., Greenshpan Y., Turgeman H., Klainberger A., Scharff Y., and Gazit R.. 2018. Identification of immune-activated hematopoietic stem cells. Leukemia. 32:2016–2020. 10.1038/s41375-018-0220-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain, D.W., Snowden P.B., Sempowski G.D., and Kelsoe G.. 2011. Inflammation triggers emergency granulopoiesis through a density-dependent feedback mechanism. PLoS One. 6:e19957. 10.1371/journal.pone.0019957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey, A., Edwards D.K. V, Eide C.A., Newell L., Traer E., Medeiros B.C., Pollyea D.A., Deininger M.W., Collins R.H., Tyner J.W., et al. 2017. Identification of Interleukin-1 by Functional Screening as a Key Mediator of Cellular Expansion and Disease Progression in Acute Myeloid Leukemia. Cell Rep. 18:3204–3218. 10.1016/j.celrep.2017.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapple, R.H., Hu T., Tseng Y.J., Liu L., Kitano A., Luu V., Hoegenauer K.A., Iwawaki T., Li Q., and Nakada D.. 2018. ERα promotes murine hematopoietic regeneration through the Ire1α-mediated unfolded protein response. eLife. 7:e31159. 10.7554/eLife.31159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delestré, L., Cui H., Esposito M., Quiveron C., Mylonas E., Penard-Lacronique V., Bischof O., and Guillouf C.. 2017. Senescence is a Spi1-induced anti-proliferative mechanism in primary hematopoietic cells. Haematologica. 102:1850–1860. 10.3324/haematol.2016.157636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinarello, C.A. 2018. Overview of the IL-1 family in innate inflammation and acquired immunity. Immunol. Rev. 281:8–27. 10.1111/imr.12621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehninger, A., Boch T., Uckelmann H., Essers M.A., Müdder K., Sleckman B.P., and Trumpp A.. 2014. Posttranscriptional regulation of c-Myc expression in adult murine HSCs during homeostasis and interferon-α-induced stress response. Blood. 123:3909–3913. 10.1182/blood-2013-10-531038 [DOI] [PubMed] [Google Scholar]
- Essers, M.A., Offner S., Blanco-Bose W.E., Waibler Z., Kalinke U., Duchosal M.A., and Trumpp A.. 2009. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature. 458:904–908. 10.1038/nature07815 [DOI] [PubMed] [Google Scholar]
- Etzrodt, M., Ahmed N., Hoppe P.S., Loeffler D., Skylaki S., Hilsenbeck O., Kokkaliaris K.D., Kaltenbach H.M., Stelling J., Nerlov C., and Schroeder T.. 2019. Inflammatory signals directly instruct PU.1 in HSCs via TNF. Blood. 133:816–819. 10.1182/blood-2018-02-832998 [DOI] [PubMed] [Google Scholar]
- Ezaki, K., Tsuzuki M., Katsuta I., Maruyama F., Kojima H., Okamoto M., Nomura T., Wakita M., Miyazaki H., Sobue R., et al. 1995. Interleukin-1 beta (IL-1 beta) and acute leukemia: in vitro proliferative response to IL-1 beta, IL-1 beta content of leukemic cells and treatment outcome. Leuk. Res. 19:35–41. 10.1016/0145-2126(94)00064-H [DOI] [PubMed] [Google Scholar]
- Foudi, A., Hochedlinger K., Van Buren D., Schindler J.W., Jaenisch R., Carey V., and Hock H.. 2009. Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat. Biotechnol. 27:84–90. 10.1038/nbt.1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire, P.R., and Conneely O.M.. 2018. NR4A1 and NR4A3 restrict HSC proliferation via reciprocal regulation of C/EBPα and inflammatory signaling. Blood. 131:1081–1093. 10.1182/blood-2017-07-795757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuchi, Y., Ito M., Shibata F., Kitamura T., and Nakajima H.. 2008. Activation of CCAAT/enhancer-binding protein alpha or PU.1 in hematopoietic stem cells leads to their reduced self-renewal and proliferation. Stem Cells. 26:3172–3181. 10.1634/stemcells.2008-0320 [DOI] [PubMed] [Google Scholar]
- Gañán-Gómez, I., Wei Y., Starczynowski D.T., Colla S., Yang H., Cabrero-Calvo M., Bohannan Z.S., Verma A., Steidl U., and Garcia-Manero G.. 2015. Deregulation of innate immune and inflammatory signaling in myelodysplastic syndromes. Leukemia. 29:1458–1469. 10.1038/leu.2015.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi, V., Plunkett W., and Cortes J.E.. 2014. Omacetaxine: a protein translation inhibitor for treatment of chronic myelogenous leukemia. Clin. Cancer Res. 20:1735–1740. 10.1158/1078-0432.CCR-13-1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerloff, D., Grundler R., Wurm A.A., Bräuer-Hartmann D., Katzerke C., Hartmann J.U., Madan V., Müller-Tidow C., Duyster J., Tenen D.G., et al. 2015. NF-κB/STAT5/miR-155 network targets PU.1 in FLT3-ITD-driven acute myeloid leukemia. Leukemia. 29:535–547. 10.1038/leu.2014.231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göthert, J.R., Gustin S.E., Hall M.A., Green A.R., Göttgens B., Izon D.J., and Begley C.G.. 2005. In vivo fate-tracing studies using the Scl stem cell enhancer: embryonic hematopoietic stem cells significantly contribute to adult hematopoiesis. Blood. 105:2724–2732. 10.1182/blood-2004-08-3037 [DOI] [PubMed] [Google Scholar]
- Haas, S., Hansson J., Klimmeck D., Loeffler D., Velten L., Uckelmann H., Wurzer S., Prendergast A.M., Schnell A., Hexel K., et al. 2015. Inflammation-Induced Emergency Megakaryopoiesis Driven by Hematopoietic Stem Cell-like Megakaryocyte Progenitors. Cell Stem Cell. 17:422–434. 10.1016/j.stem.2015.07.007 [DOI] [PubMed] [Google Scholar]
- Harrison, D.E., and Lerner C.P.. 1991. Most primitive hematopoietic stem cells are stimulated to cycle rapidly after treatment with 5-fluorouracil. Blood. 78:1237–1240. 10.1182/blood.V78.5.1237.1237 [DOI] [PubMed] [Google Scholar]
- Heberle, H., Meirelles G.V., da Silva F.R., Telles G.P., and Minghim R.. 2015. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics. 16:169. 10.1186/s12859-015-0611-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidt, T., Sager H.B., Courties G., Dutta P., Iwamoto Y., Zaltsman A., von Zur Muhlen C., Bode C., Fricchione G.L., Denninger J., et al. 2014. Chronic variable stress activates hematopoietic stem cells. Nat. Med. 20:754–758. 10.1038/nm.3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz, S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., and Glass C.K.. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 38:576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmati, S., Sinclair T., Tong M., Bartholdy B., Okabe R.O., Ames K., Ostrodka L., Haque T., Kaur I., Mills T.S., et al. 2019. PI3 kinase alpha and delta promote hematopoietic stem cell activation. JCI Insight. 4:e125832. 10.1172/jci.insight.125832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry, C.J., Casás-Selves M., Kim J., Zaberezhnyy V., Aghili L., Daniel A.E., Jimenez L., Azam T., McNamee E.N., Clambey E.T., et al. 2015. Aging-associated inflammation promotes selection for adaptive oncogenic events in B cell progenitors. J. Clin. Invest. 125:4666–4680. 10.1172/JCI83024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez, G., Mills T.S., Rabe J.L., Chavez J.S., Kuldanek S., Kirkpatrick G., Noetzli L., Jubair W.K., Zanche M., Myers J.R., et al. 2020. Pro-inflammatory cytokine blockade attenuates myeloid expansion in a murine model of rheumatoid arthritis. Haematologica. 105:585–597. 10.3324/haematol.2018.197210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilsenbeck, O., Schwarzfischer M., Skylaki S., Schauberger B., Hoppe P.S., Loeffler D., Kokkaliaris K.D., Hastreiter S., Skylaki E., Filipczyk A., et al. 2016. Software tools for single-cell tracking and quantification of cellular and molecular properties. Nat. Biotechnol. 34:703–706. 10.1038/nbt.3626 [DOI] [PubMed] [Google Scholar]
- Higa, K.C., Goodspeed A., Chavez J.S., De Dominici M., Danis E.P., Zaberezhnyy V., Rabe J.L., Tenen D.G., Pietras E.M., and DeGregori J.. 2021. Chronic Interleukin-1 triggers selection for Cebpa-knockout multipotent hematopoietic progenitors. J. Exp. Med. In press. 10.1084/jem.20200560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilsenbeck, O., Schwarzfischer M., Loeffler D., Dimopoulos S., Hastreiter S., Marr C., Theis F.J., and Schroeder T.. 2017. fastER: a user-friendly tool for ultrafast and robust cell segmentation in large-scale microscopy. Bioinformatics. 33:2020–2028. 10.1093/bioinformatics/btx107 [DOI] [PubMed] [Google Scholar]
- Hinge, A., He J., Bartram J., Javier J., Xu J., Fjellman E., Sesaki H., Li T., Yu J., Wunderlich M., et al. 2020. Asymmetrically Segregated Mitochondria Provide Cellular Memory of Hematopoietic Stem Cell Replicative History and Drive HSC Attrition. Cell Stem Cell. 26:420–430.e6. 10.1016/j.stem.2020.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock, H., Hamblen M.J., Rooke H.M., Schindler J.W., Saleque S., Fujiwara Y., and Orkin S.H.. 2004. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature. 431:1002–1007. 10.1038/nature02994 [DOI] [PubMed] [Google Scholar]
- Hoppe, P.S., Schwarzfischer M., Loeffler D., Kokkaliaris K.D., Hilsenbeck O., Moritz N., Endele M., Filipczyk A., Gambardella A., Ahmed N., et al. 2016. Early myeloid lineage choice is not initiated by random PU.1 to GATA1 protein ratios. Nature. 535:299–302. 10.1038/nature18320 [DOI] [PubMed] [Google Scholar]
- Hosokawa, H., Ungerbäck J., Wang X., Matsumoto M., Nakayama K.I., Cohen S.M., Tanaka T., and Rothenberg E.V.. 2018. Transcription Factor PU.1 Represses and Activates Gene Expression in Early T Cells by Redirecting Partner Transcription Factor Binding. Immunity. 48:1119–1134.e7. 10.1016/j.immuni.2018.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C.Y., Bredemeyer A.L., Walker L.M., Bassing C.H., and Sleckman B.P.. 2008. Dynamic regulation of c-Myc proto-oncogene expression during lymphocyte development revealed by a GFP-c-Myc knock-in mouse. Eur. J. Immunol. 38:342–349. 10.1002/eji.200737972 [DOI] [PubMed] [Google Scholar]
- Huang, W., Sherman B.T., and Lempicki R.A.. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Iritani, B.M., and Eisenman R.N.. 1999. c-Myc enhances protein synthesis and cell size during B lymphocyte development. Proc. Natl. Acad. Sci. USA. 96:13180–13185. 10.1073/pnas.96.23.13180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalbert, E., and Pietras E.M.. 2018. Analysis of Murine Hematopoietic Stem Cell Proliferation During Inflammation. Methods Mol. Biol. 1686:183–200. 10.1007/978-1-4939-7371-2_14 [DOI] [PubMed] [Google Scholar]
- Kaasinen, E., Kuismin O., Rajamäki K., Ristolainen H., Aavikko M., Kondelin J., Saarinen S., Berta D.G., Katainen R., Hirvonen E.A.M., et al. 2019. Impact of constitutional TET2 haploinsufficiency on molecular and clinical phenotype in humans. Nat. Commun. 10:1252. 10.1038/s41467-019-09198-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara-Negishi, F., Yamamoto H., Suzuki M., Yamada T., Sakurai T., Tamura T., and Oikawa T.. 2001. In vivo complex formation of PU.1 with HDAC1 associated with PU.1-mediated transcriptional repression. Oncogene. 20:6039–6047. 10.1038/sj.onc.1204756 [DOI] [PubMed] [Google Scholar]
- Kim, D., Paggi J.M., Park C., Bennett C., and Salzberg S.L.. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37:907–915. 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, K.Y., and Goodell M.A.. 2011. Inflammatory modulation of HSCs: viewing the HSC as a foundation for the immune response. Nat. Rev. Immunol. 11:685–692. 10.1038/nri3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, K.Y., Baldridge M.T., Weksberg D.C., Chambers S.M., Lukov G.L., Wu S., Boles N.C., Jung S.Y., Qin J., Liu D., et al. 2011. Irgm1 protects hematopoietic stem cells by negative regulation of IFN signaling. Blood. 118:1525–1533. 10.1182/blood-2011-01-328682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirstetter, P., Anderson K., Porse B.T., Jacobsen S.E., and Nerlov C.. 2006. Activation of the canonical Wnt pathway leads to loss of hematopoietic stem cell repopulation and multilineage differentiation block. Nat. Immunol. 7:1048–1056. 10.1038/ni1381 [DOI] [PubMed] [Google Scholar]
- Kueh, H.Y., Champhekar A., Nutt S.L., Elowitz M.B., and Rothenberg E.V.. 2013. Positive feedback between PU.1 and the cell cycle controls myeloid differentiation. Science. 341:670–673. 10.1126/science.1240831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laberge, R.M., Sun Y., Orjalo A.V., Patil C.K., Freund A., Zhou L., Curran S.C., Davalos A.R., Wilson-Edell K.A., Liu S., et al. 2015. MTOR regulates the pro-tumorigenic senescence-associated secretory phenotype by promoting IL1A translation. Nat. Cell Biol. 17:1049–1061. 10.1038/ncb3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laconi, E., Marongiu F., and DeGregori J.. 2020. Cancer as a disease of old age: changing mutational and microenvironmental landscapes. Br. J. Cancer. 122:943–952. 10.1038/s41416-019-0721-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurenti, E., Varnum-Finney B., Wilson A., Ferrero I., Blanco-Bose W.E., Ehninger A., Knoepfler P.S., Cheng P.F., MacDonald H.R., Eisenman R.N., et al. 2008. Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell. 3:611–624. 10.1016/j.stem.2008.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M.M., MacDonald M.R., and Rice C.M.. 2015. To translate, or not to translate: viral and host mRNA regulation by interferon-stimulated genes. Trends Cell Biol. 25:320–329. 10.1016/j.tcb.2015.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, R., Arif T., Kalmykova S., Kasianov A., Lin M., Menon V., Qiu J., Bernitz J.M., Moore K., Lin F., et al. 2020. Restraining Lysosomal Activity Preserves Hematopoietic Stem Cell Quiescence and Potency. Cell Stem Cell. 26:359–376.e7. 10.1016/j.stem.2020.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeffler, D., Wang W., Hopf A., Hilsenbeck O., Bourgine P.E., Rudolf F., Martin I., and Schroeder T.. 2018. Mouse and human HSPC immobilization in liquid culture by CD43- or CD44-antibody coating. Blood. 131:1425–1429. 10.1182/blood-2017-07-794131 [DOI] [PubMed] [Google Scholar]
- Martin, M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 17:10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Matatall, K.A., Jeong M., Chen S., Sun D., Chen F., Mo Q., Kimmel M., and King K.Y.. 2016. Chronic Infection Depletes Hematopoietic Stem Cells through Stress-Induced Terminal Differentiation. Cell Rep. 17:2584–2595. 10.1016/j.celrep.2016.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matatall, K.A., Kadmon C.S., and King K.Y.. 2018. Detecting Hematopoietic Stem Cell Proliferation Using BrdU Incorporation. Methods Mol. Biol. 1686:91–103. 10.1007/978-1-4939-7371-2_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto, A., Takeishi S., Kanie T., Susaki E., Onoyama I., Tateishi Y., Nakayama K., and Nakayama K.I.. 2011. p57 is required for quiescence and maintenance of adult hematopoietic stem cells. Cell Stem Cell. 9:262–271. 10.1016/j.stem.2011.06.014 [DOI] [PubMed] [Google Scholar]
- McKenzie, M.D., Ghisi M., Oxley E.P., Ngo S., Cimmino L., Esnault C., Liu R., Salmon J.M., Bell C.C., Ahmed N., et al. 2019. Interconversion between Tumorigenic and Differentiated States in Acute Myeloid Leukemia. Cell Stem Cell. 25:258–272.e9. 10.1016/j.stem.2019.07.001 [DOI] [PubMed] [Google Scholar]
- Metsalu, T., and Vilo J.. 2015. ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 43(W1):W566-W570. 10.1093/nar/gkv468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, K., Barreyro L., Todorova T.I., Taylor S.J., Antony-Debré I., Narayanagari S.R., Carvajal L.A., Leite J., Piperdi Z., Pendurti G., et al. 2018. IL1RAP potentiates multiple oncogenic signaling pathways in AML. J. Exp. Med. 215:1709–1727. 10.1084/jem.20180147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison, S.J., and Scadden D.T.. 2014. The bone marrow niche for haematopoietic stem cells. Nature. 505:327–334. 10.1038/nature12984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, B.U., Pabst T., Fos J., Petkovic V., Fey M.F., Asou N., Buergi U., and Tenen D.G.. 2006. ATRA resolves the differentiation block in t(15;17) acute myeloid leukemia by restoring PU.1 expression. Blood. 107:3330–3338. 10.1182/blood-2005-07-3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto, T., Walker C.S., Choi K., Hueneman K., Smith M.A., Gul Z., Garcia-Manero G., Ma A., Zheng Y., and Starczynowski D.T.. 2020. Adaptive response to inflammation contributes to sustained myelopoiesis and confers a competitive advantage in myelodysplastic syndrome HSCs. Nat. Immunol. 21:535–545. 10.1038/s41590-020-0663-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguera, N.I., Piredda M.L., Taulli R., Catalano G., Angelini G., Gaur G., Nervi C., Voso M.T., Lunardi A., Pandolfi P.P., and Lo-Coco F.. 2016. PML/RARa inhibits PTEN expression in hematopoietic cells by competing with PU.1 transcriptional activity. Oncotarget. 7:66386–66397. 10.18632/oncotarget.11964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa, T., Yamada T., Kihara-Negishi F., Yamamoto H., Kondoh N., Hitomi Y., and Hashimoto Y.. 1999. The role of Ets family transcription factor PU.1 in hematopoietic cell differentiation, proliferation and apoptosis. Cell Death Differ. 6:599–608. 10.1038/sj.cdd.4400534 [DOI] [PubMed] [Google Scholar]
- Peng, T., Thorn K., Schroeder T., Wang L., Theis F.J., Marr C., and Navab N.. 2017. A BaSiC tool for background and shading correction of optical microscopy images. Nat. Commun. 8:14836. 10.1038/ncomms14836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras, E.M. 2017. Inflammation: a key regulator of hematopoietic stem cell fate in health and disease. Blood. 130:1693–1698. 10.1182/blood-2017-06-780882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras, E.M., Warr M.R., and Passegué E.. 2011. Cell cycle regulation in hematopoietic stem cells. J. Cell Biol. 195:709–720. 10.1083/jcb.201102131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras, E.M., Lakshminarasimhan R., Techner J.M., Fong S., Flach J., Binnewies M., and Passegué E.. 2014. Re-entry into quiescence protects hematopoietic stem cells from the killing effect of chronic exposure to type I interferons. J. Exp. Med. 211:245–262. 10.1084/jem.20131043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras, E.M., Mirantes-Barbeito C., Fong S., Loeffler D., Kovtonyuk L.V., Zhang S., Lakshminarasimhan R., Chin C.P., Techner J.M., Will B., et al. 2016. Chronic interleukin-1 exposure drives haematopoietic stem cells towards precocious myeloid differentiation at the expense of self-renewal. Nat. Cell Biol. 18:607–618. 10.1038/ncb3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenis, M., and Aramayo R.. 2015. Translate to divide: сontrol of the cell cycle by protein synthesis. Microb. Cell. 2:94–104. 10.15698/mic2015.04.198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porse, B.T., Bryder D., Theilgaard-Mönch K., Hasemann M.S., Anderson K., Damgaard I., Jacobsen S.E., and Nerlov C.. 2005. Loss of C/EBP alpha cell cycle control increases myeloid progenitor proliferation and transforms the neutrophil granulocyte lineage. J. Exp. Med. 202:85–96. 10.1084/jem.20050067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter, S.N., Cluster A.S., Signer R.A.J., Voigtmann J., Monlish D.A., Schuettpelz L.G., and Magee J.A.. 2016. Pten Cell Autonomously Modulates the Hematopoietic Stem Cell Response to Inflammatory Cytokines. Stem Cell Reports. 6:806–814. 10.1016/j.stemcr.2016.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast, A.M., and Essers M.A.. 2014. Hematopoietic stem cells, infection, and the niche. Ann. N. Y. Acad. Sci. 1310:51–57. 10.1111/nyas.12400 [DOI] [PubMed] [Google Scholar]
- Pronk, C.J., Rossi D.J., Månsson R., Attema J.L., Norddahl G.L., Chan C.K., Sigvardsson M., Weissman I.L., and Bryder D.. 2007. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell. 1:428–442. 10.1016/j.stem.2007.07.005 [DOI] [PubMed] [Google Scholar]
- Pulikkan, J.A., Tenen D.G., and Behre G.. 2017. C/EBPα deregulation as a paradigm for leukemogenesis. Leukemia. 31:2279–2285. 10.1038/leu.2017.229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabe, J.L., Hernandez G., Chavez J.S., Mills T.S., Nerlov C., and Pietras E.M.. 2020. CD34 and EPCR coordinately enrich functional murine hematopoietic stem cells under normal and inflammatory conditions. Exp. Hematol. 81:1–15.e6. 10.1016/j.exphem.2019.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhyasen, G.W., Bolanos L., Fang J., Jerez A., Wunderlich M., Rigolino C., Mathews L., Ferrer M., Southall N., Guha R., et al. 2013. Targeting IRAK1 as a therapeutic approach for myelodysplastic syndrome. Cancer Cell. 24:90–104. 10.1016/j.ccr.2013.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers, J.T., King K.Y., Brett J.O., Cromie M.J., Charville G.W., Maguire K.K., Brunson C., Mastey N., Liu L., Tsai C.R., et al. 2014. mTORC1 controls the adaptive transition of quiescent stem cells from G0 to G(Alert). Nature. 510:393–396. 10.1038/nature13255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez, A., Zhang K., Färkkilä A., Filiatrault J., Yang C., Velázquez M., Furutani E., Goldman D.C., García de Teresa B., Garza-Mayén G., et al. 2021. MYC Promotes Bone Marrow Stem Cell Dysfunction in Fanconi Anemia. Cell Stem Cell. 28:33–47.e8. 10.1016/j.stem.2020.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbauer, F., Wagner K., Kutok J.L., Iwasaki H., Le Beau M.M., Okuno Y., Akashi K., Fiering S., and Tenen D.G.. 2004. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat. Genet. 36:624–630. 10.1038/ng1361 [DOI] [PubMed] [Google Scholar]
- Säwén, P., Lang S., Mandal P., Rossi D.J., Soneji S., and Bryder D.. 2016. Mitotic History Reveals Distinct Stem Cell Populations and Their Contributions to Hematopoiesis. Cell Rep. 14:2809–2818. 10.1016/j.celrep.2016.02.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer, B.C., Schaefer M.L., Kappler J.W., Marrack P., and Kedl R.M.. 2001. Observation of antigen-dependent CD8+ T-cell/ dendritic cell interactions in vivo. Cell. Immunol. 214:110–122. 10.1006/cimm.2001.1895 [DOI] [PubMed] [Google Scholar]
- Schuettpelz, L.G., Borgerding J.N., Christopher M.J., Gopalan P.K., Romine M.P., Herman A.C., Woloszynek J.R., Greenbaum A.M., and Link D.C.. 2014. G-CSF regulates hematopoietic stem cell activity, in part, through activation of Toll-like receptor signaling. Leukemia. 28:1851–1860. 10.1038/leu.2014.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scognamiglio, R., Cabezas-Wallscheid N., Thier M.C., Altamura S., Reyes A., Prendergast A.M., Baumgärtner D., Carnevalli L.S., Atzberger A., Haas S., et al. 2016. Myc Depletion Induces a Pluripotent Dormant State Mimicking Diapause. Cell. 164:668–680. 10.1016/j.cell.2015.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signer, R.A., Magee J.A., Salic A., and Morrison S.J.. 2014. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature. 509:49–54. 10.1038/nature13035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signer, R.A., Qi L., Zhao Z., Thompson D., Sigova A.A., Fan Z.P., DeMartino G.N., Young R.A., Sonenberg N., and Morrison S.J.. 2016. The rate of protein synthesis in hematopoietic stem cells is limited partly by 4E-BPs. Genes Dev. 30:1698–1703. 10.1101/gad.282756.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, M.A., Choudhary G.S., Pellagatti A., Choi K., Bolanos L.C., Bhagat T.D., Gordon-Mitchell S., Von Ahrens D., Pradhan K., Steeples V., et al. 2019. U2AF1 mutations induce oncogenic IRAK4 isoforms and activate innate immune pathways in myeloid malignancies. Nat. Cell Biol. 21:640–650. 10.1038/s41556-019-0314-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon, L.A., Podder S., He J., Jackson-Chornenki N.L., Gibson K., Ziliotto R.G., Rhee J., and DeKoter R.P.. 2017. Coordination of Myeloid Differentiation with Reduced Cell Cycle Progression by PU.1 Induction of MicroRNAs Targeting Cell Cycle Regulators and Lipid Anabolism. Mol. Cell. Biol. 37:e00013-17. 10.1128/MCB.00013-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staber, P.B., Zhang P., Ye M., Welner R.S., Nombela-Arrieta C., Bach C., Kerenyi M., Bartholdy B.A., Zhang H., Alberich-Jordà M., et al. 2013. Sustained PU.1 levels balance cell-cycle regulators to prevent exhaustion of adult hematopoietic stem cells. Mol. Cell. 49:934–946. 10.1016/j.molcel.2013.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, B.M., Khan N., D’Alessandro A., Nemkov T., Winters A., Jones C.L., Zhang W., Pollyea D.A., and Jordan C.T.. 2018. Characterization and targeting of malignant stem cells in patients with advanced myelodysplastic syndromes. Nat. Commun. 9:3694. 10.1038/s41467-018-05984-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa, H., Fritsch K., Kovtonyuk L.V., Saito Y., Yakkala C., Jacobs K., Ahuja A.K., Lopes M., Hausmann A., Hardt W.D., et al. 2017. Pathogen-Induced TLR4-TRIF Innate Immune Signaling in Hematopoietic Stem Cells Promotes Proliferation but Reduces Competitive Fitness. Cell Stem Cell. 21:225–240.e5. 10.1016/j.stem.2017.06.013 [DOI] [PubMed] [Google Scholar]
- Ueda, Y., Cain D.W., Kuraoka M., Kondo M., and Kelsoe G.. 2009. IL-1R type I-dependent hemopoietic stem cell proliferation is necessary for inflammatory granulopoiesis and reactive neutrophilia. J. Immunol. 182:6477–6484. 10.4049/jimmunol.0803961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umek, R.M., Friedman A.D., and McKnight S.L.. 1991. CCAAT-enhancer binding protein: a component of a differentiation switch. Science. 251:288–292. 10.1126/science.1987644 [DOI] [PubMed] [Google Scholar]
- Vangala, R.K., Heiss-Neumann M.S., Rangatia J.S., Singh S.M., Schoch C., Tenen D.G., Hiddemann W., and Behre G.. 2003. The myeloid master regulator transcription factor PU.1 is inactivated by AML1-ETO in t(8;21) myeloid leukemia. Blood. 101:270–277. 10.1182/blood-2002-04-1288 [DOI] [PubMed] [Google Scholar]
- Walsh, J.C., DeKoter R.P., Lee H.J., Smith E.D., Lancki D.W., Gurish M.F., Friend D.S., Stevens R.L., Anastasi J., and Singh H.. 2002. Cooperative and antagonistic interplay between PU.1 and GATA-2 in the specification of myeloid cell fates. Immunity. 17:665–676. 10.1016/S1074-7613(02)00452-1 [DOI] [PubMed] [Google Scholar]
- Walter, D., Lier A., Geiselhart A., Thalheimer F.B., Huntscha S., Sobotta M.C., Moehrle B., Brocks D., Bayindir I., Kaschutnig P., et al. 2015. Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature. 520:549–552. 10.1038/nature14131 [DOI] [PubMed] [Google Scholar]
- Weisser, M., Demel U.M., Stein S., Chen-Wichmann L., Touzot F., Santilli G., Sujer S., Brendel C., Siler U., Cavazzana M., et al. 2016. Hyperinflammation in patients with chronic granulomatous disease leads to impairment of hematopoietic stem cell functions. J. Allergy Clin. Immunol. 138:219–228.e9. 10.1016/j.jaci.2015.11.028 [DOI] [PubMed] [Google Scholar]
- Will, B., Vogler T.O., Narayanagari S., Bartholdy B., Todorova T.I., da Silva Ferreira M., Chen J., Yu Y., Mayer J., Barreyro L., et al. 2015. Minimal PU.1 reduction induces a preleukemic state and promotes development of acute myeloid leukemia. Nat. Med. 21:1172–1181. 10.1038/nm.3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, A., Murphy M.J., Oskarsson T., Kaloulis K., Bettess M.D., Oser G.M., Pasche A.C., Knabenhans C., Macdonald H.R., and Trumpp A.. 2004. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev. 18:2747–2763. 10.1101/gad.313104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, A., Laurenti E., Oser G., van der Wath R.C., Blanco-Bose W., Jaworski M., Offner S., Dunant C.F., Eshkind L., Bockamp E., et al. 2008. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 135:1118–1129. 10.1016/j.cell.2008.10.048 [DOI] [PubMed] [Google Scholar]
- Yamashita, M., and Passegué E.. 2019. TNF-α Coordinates Hematopoietic Stem Cell Survival and Myeloid Regeneration. Cell Stem Cell. 25:357–372.e7. 10.1016/j.stem.2019.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H., Liang H., Yan J.S., Tao R., Hao S.G., and Ma L.Y.. 2012. Down-regulation of hematopoiesis master regulator PU.1 via aberrant methylation in chronic myeloid leukemia. Int. J. Hematol. 96:65–73. 10.1007/s12185-012-1106-x [DOI] [PubMed] [Google Scholar]
- Zambetti, N.A., Ping Z., Chen S., Kenswil K.J.G., Mylona M.A., Sanders M.A., Hoogenboezem R.M., Bindels E.M.J., Adisty M.N., Van Strien P.M.H., et al. 2016. Mesenchymal Inflammation Drives Genotoxic Stress in Hematopoietic Stem Cells and Predicts Disease Evolution in Human Pre-leukemia. Cell Stem Cell. 19:613–627. 10.1016/j.stem.2016.08.021 [DOI] [PubMed] [Google Scholar]
- Zeng, H., Yücel R., Kosan C., Klein-Hitpass L., and Möröy T.. 2004. Transcription factor Gfi1 regulates self-renewal and engraftment of hematopoietic stem cells. EMBO J. 23:4116–4125. 10.1038/sj.emboj.7600419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, B., Chu S., Agarwal P., Campbell V.L., Hopcroft L., Jørgensen H.G., Lin A., Gaal K., Holyoake T.L., and Bhatia R.. 2016. Inhibition of interleukin-1 signaling enhances elimination of tyrosine kinase inhibitor-treated CML stem cells. Blood. 128:2671–2682. 10.1182/blood-2015-11-679928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, M., Tao F., Venkatraman A., Li Z., Smith S.E., Unruh J., Chen S., Ward C., Qian P., Perry J.M., et al. 2019. N-Cadherin-Expressing Bone and Marrow Stromal Progenitor Cells Maintain Reserve Hematopoietic Stem Cells. Cell Rep. 26:652–669.e6. 10.1016/j.celrep.2018.12.093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziliotto, R., Gruca M.R., Podder S., Noel G., Ogle C.K., Hess D.A., and DeKoter R.P.. 2014. PU.1 promotes cell cycle exit in the murine myeloid lineage associated with downregulation of E2F1. Exp. Hematol. 42:204–217.e1. 10.1016/j.exphem.2013.11.011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
displays DEGs in SLAM cells from mice treated with or without IL-1β for 20 d.
provides an analysis of DEGs, with GO categories, IPA upstream regulator categories, and GSEA enrichment analysis of DEGs in SLAM cells from mice treated with or without IL-1β for 20 d.
shows a comparison of DEGs with public data, with overlap between IL-1–downregulated genes and public datasets.
displays PU.1 ChIP-seq peak locations, scores, and PU.1/Myc motif scores.
provides an analysis of PU.1 target genes, with GO categories enriched in IL-1 DEGs containing PU.1 ChIP-seq peaks.
lists the antibodies, antibody dilutions, and Fluidigm qRT-PCR primers used in this study.
Data Availability Statement
Sequencing data from this study are publicly available from the National Center for Biotechnology Information GEO accession no. GSE165810. Other datasets generated during the current study are available from the corresponding author upon reasonable request. Any data supporting the findings of this study are available from the corresponding author upon reasonable request.