Abstract
Chagas disease is a zoonotic, parasitic, vector-borne neglected tropical disease that affects the lives of over 6 million people throughout the Americas. Trypanosoma cruzi, the causative agent, presents extensive genetic diversity. Here we report the genome sequence of reference strain SC43cl1, a hybrid strain belonging to the TcV discrete typing unit (DTU). The assembled diploid genome was 79 Mbp in size, divided into 1236 contigs with an average coverage reaching 180×. There was extensive synteny of SC43cl1 genome with closely related TcV and TcVI genomes, with limited sequence rearrangements. TcVI genomes included several expansions not present in TcV strains. Comparative analysis of both nuclear and kinetoplast sequences clearly separated TcV from TcVI strains, which strongly supports the current DTU classification.
Keywords: Chagas disease, parasite, diversity, phylogeny
Résumé:
La maladie de Chagas est une zoonose parasitaire tropicale transmise par un vecteur et peu étudiée qui affecte les vies de plus de 6 millions de personnes à travers les Amériques. Le Trypanosoma cruzi, l’agent causal, présente une très grande diversité génétique. Dans ce travail, les auteurs rapportent la séquence du génome de la souche de référence SC43c11, une souche hybride appartenant au groupe (ou DTU pour « discrete typing unit ») TcV. Le génome diploïde assemblé mesure 79 Mb, formé par 1236 contigs dont la couverture moyenne était de 180×. Une grande synténie, avec peu de réarrangements, a été observée entre le génome de SC43c11 et ceux des souches apparentées des groupes TcV et TcVI. Les génomes des souches appartenant au groupe TcVI comprenaient plusieurs expansions qui sont absentes des souches du groupe TcV. Une analyse comparée des séquences nucléaires et du kinétoplaste ont permis de clairement séparer les souches des groupes TcV et TcVI, ce qui supporte fortement la classification DTU actuelle. [Traduit par la Rédaction]
Keywords: maladie de Chagas, parasite, diversité, phylogénie
Introduction
Chagas disease is a zoonotic, parasitic, vector-bome neglected tropical disease that is endemic to the Americas where it affects the lives of over 6 million people. It is caused by the protozoan parasite Trypanosoma cruzi, which belongs to the kinetoplastid group of flagellated protists that together with the euglenoids makes up the phylum Euglenozoa. While many kinetoplastids are free-living, three major groups of human pathogens have been identified, including African trypanosomes, T. cruzi, and a wide diversity of Leishmania parasites (El-Sayed et al. 2005b). A key characteristic of these organisms is the kinetoplast, which is a specialized DNA-containing organelle corresponding to the cell’s single mitochondria.
Trypanosoma cruzi itself presents extensive genetic diversity. Accordingly, it has been divided based on selected nuclear markers into seven main lineages referred to as discrete typing units (DTUs): TcI to TcVI and Tcbat (Zingales et al. 2009, 2012). These DTUs are highly stable across the American continent as well as over time. They correspond to a (near-) clade genetic structuration of the parasite as a species (Tibayrenc and Ayala 2015). The level of genetic diversity among T. cruzi DTUs is comparable to that observed among some species of Leishmania. However, this classification has been challenged based on kinetoplast markers, which yield three main clades of the parasite (Machado and Ayala 2001).
The characterization of T. cruzi parasite genetic diversity is key to understanding the dynamics of parasite transmission cycles as well as those of human infection. Indeed, the genetic diversity of T. cruzi is believed to be associated with a comparable biological diversity. It is currently hypothesized that different parasite genotypes are associated with different vertebrate hosts and transmission cycles as well as with the specific disease progression patterns in mammalian hosts (Zingales 2018).
Several T. cruzi nuclear genome sequences have been reported in recent years, but most have focused on TcI strains (Franzen et al. 2011; Grisard et al. 2014; Berry et al. 2019). Very few studies have examined strains from other DTUs (El-Sayed et al. 2005a; Baptista et al. 2018; Berna et al. 2018; Reis-Cunha et al. 2018; Diaz-Viraque et al. 2019), which is critical for a complete assessment of parasite genetic diversity and its evolution. Here we report the genome sequence of reference strain SC43, a hybrid strain belonging to the TcV DTU. It was originally isolated in 1981 from Triatoma infestans in Santa Cruz, Bolivia (Carreno et al. 1987; Barnabe et al. 2000). In mouse models, this strain was found to produce mild disease characterized by a veiy low parasitemia during the acute phase and limited cardiac damage (Rodriguez et al. 2014). Phylogenetic analysis of the SC43 genome and its kinetoplast maxicircle in comparison with closely related strains sheds light on the genetic structure of T. cruzi.
Methods
Parasite culture and sequencing
Trypanosoma cruzi strain SC43cl1 was obtained from ATCC (#50798) and cultured in liver infusion tryptose (LIT) medium at 28 °C. Parasites were first cloned by limiting dilution to ensure that a single clone would be sequenced (Ramirez et al. 2012). Briefly, an aliquot of SC43cl1 culture was taken, stained with Trypan blue, and parasites were counted in a Neubauer chamber to adjust cell suspension density to 400 parasites/mL. Parasite serial dilutions (1:2) in a 96 wells microplates were then incubated at 28 °C and single clones isolated. DNA was extracted using Qiagen Blood and Tissue DNA extraction kit following the instructions of the manufacturer.
The variable intergenic region of the mini-exon gene was used to genotype the SC43cl1 parasite culture clones, to ensure the identity of the selected clone with SC43cl1 and the TcV DTU prior to sequencing. We performed a genotyping PCR using primers reported by Souto et al. (1996), as in previous studies (Souto et al. 1996; Majeau et al. 2019). A subclone of Sc43cl1, referred to as Sc43cl1.1, was selected for DNA extraction and sequencing. About 600 ng of SC43cl1.1 genomic DNA was used to generate a sequencing library via the Illumina Nextera DNA Flex Library prep kit following the manufacturer’s instructions. The library was then sequenced on the Illumina NextSeq platform.
Genome and kinetoplast maxicircle assembly and analysis
Over 197 million pair-end 75 bp sequences were obtained from SC43cl1.1 and assembled using the T. cruzi TCC (belonging to TcVI) diploid genome as a reference (Berna et al. 2018), which represents one of the most complete genome assemblies to date based on the use of long-read sequencing technology. Assembly was performed in Geneious 11 using five iterations of the Geneious mapper at medium sensitivity. Kinetoplast maxicircle assembly was performed similarly using the TCC maxicircle as a reference (GenBank accession: MN904528), which was also obtained from long-read sequencing (Gerasimov et al. 2020).
Assembled SC43cl1.1 contigs were concatenated and first compared with the concatenated genome of the TCC strain to assess overall synteny, and a dot plot was generated in Gepard 1.4 (Krumsiek et al. 2007) using a word length of 100 bp. Additional genome sequences for strains 9280cl2 (TcV), Tula cl2 (TcVI), Esmeraldo, and Y (TcII) were downloaded from the National Center for Biotechnology Information Sequence Read Archive (NCBI-SRA) and assembled as above for farther comparison with the SC43cl1.1 genome. Whole-genome alignment of multiple T. cruzi strains was then performed using ProgressiveMauve (Darling et al. 2010) and concatenated homologous collinear sequences were used for phylogenetic analysis. Mini-exon sequences from SC43cl1.1 genome were identified by BLAST searches of the assembled genome and aligned with Muscle with sequences from reference strains obtained from NCBI GenBank database. These included sequences from Sylvio X10/1 (TcI, #CP015667), Esmeraldo (TcII, #ANOX01015751), SC43 (TcV, #AY367127), MN (TcV, #AY367128), and CL (TcVI, #U57984). Y (TcII) and Tc231 (TcIII) mini-exon sequences were obtained by Blast searches of their respective genomes. Maximum likelihood phylogenetic analysis was performed with PHYML as implemented in Geneious for all sequences. The coding region of SC43cl1.1 kinetoplast maxicircle assembly was similarly analyzed with kDNA sequences from reference strains, including Sylvio X10/1 (TcI, #FJ203996), Esmeraldo (TcII, #DQ343646), Y (TcII, #MH144198), Tc231 (TcIII, #KC987253), and CL Brener (TcVI, #DQ343645). All the SC43cl1.1 sequence data has been deposited in NCBI under Bioproject PRJNA633093, with SRA database accession SRR11802127. The Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession JACCJE000000000, and MT554701 for the kinetoplast maxicircle assembly. The version described in this paper is version JACCJE010000000.
Results
The SC43cl1.1 assembled diploid genome was 79 Mbp in size, similar to the other hybrid genomes from TcV and TcVI DTUs. It was divided into 1235 contigs, ranging from 325 to 1303015 bp in length, with an excellent average coverage reaching 180× (Table 1). Pairwise comparison of the SC43cl1.1 genome with the reference TCC genome (TcVI DTU) indicated a strong synteny over the entire assembled sequence, suggesting a rather close evolutionary relationship (Fig. 1). Nonetheless, evidence of limited sequence rearrangements between the two genomes could be detected, suggesting possible recombination.
Table 1.
Summary of genome assemblies.
| Strain | DTU | Size* (Mb) | GC (%) | N50 | Max. contig | Min. contig | Mean contig | Contigs | Coverage | Method | Reference or accession |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SC43cl1.1 | TcV | 79.91 | 51.7 | 267 750 | 1303 015 | 325 | 64 697 | 1235 | 180 | Alumina | This study |
| TCC | TcVI | 86.77 | 51.7 | 265 169 | 1305 230 | 888 | 70 380 | 1237 | 60 | PacBio | Berna et al. 2018 |
| Tula cl2 | TcVI | 83.51 | 50.6 | 7772 | 242 476 | 200 | 1826 | 45 711 | 50 | 454 | GCA_000365225 |
| Dm28c | TcI | 53.27 | 51.6 | 317 638 | 1645 565 | 911 | 83 632 | 637 | 76 | PacBio | Berna et al. 2018 |
| Esmeraldo | TcII | 38.08 | 52.1 | 66 229 | 483 664 | 200 | 2409 | 15 803 | 60 | 454 | GCA_000327425.1 |
| Berenice | TcII | 40.89 | 51.2 | 156 193 | 926 516 | 1234 | 44 205 | 923 | 69 | Ulumina and Nanopore | Diaz-Viraque et al. 2019 |
| Y | TcII | 39.34 | 51.4 | 11 782 | 304 870 | 500 | 3885 | 10 127 | 71 | Ulumina | Callejas-Hernandez et al. 2018 |
Genome size for hybrid discrete typing units (DTUs) TcV and TcVI is for diploid genome, while for TcI and TcII genomes, haploid size is shown.
Fig. 1.
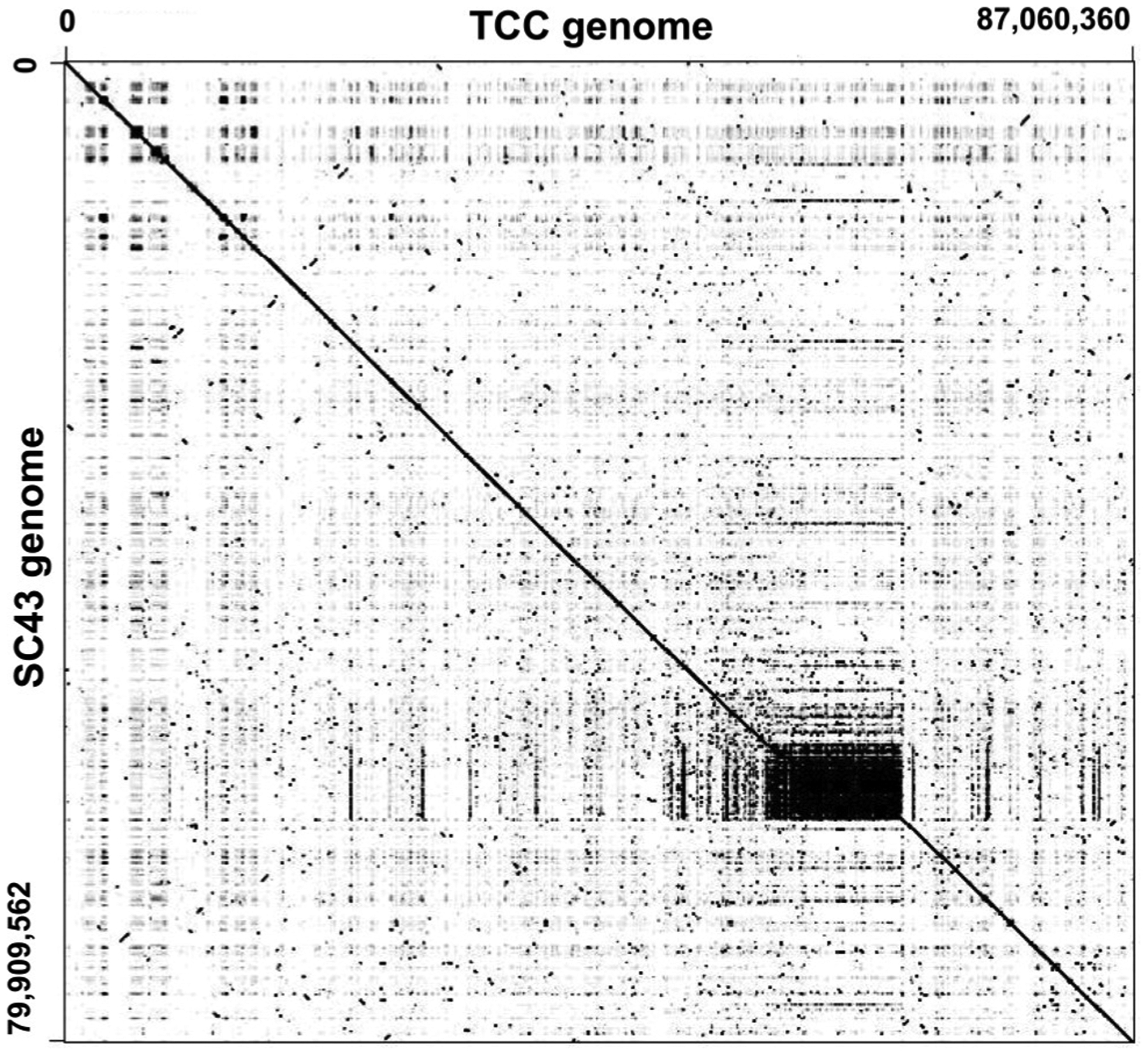
Dot plot comparison of SC43cl1.1 and TCC genomes. SC43cl1.1 contigs were concatenated (79 Mbp) and compared with the concatenated genome of the TCC strain (87 Mbp) to assess overall synteny using a dot plot. Note extensive synteny over the entire genome.
Because the distinction between TcV and TcVI DTUs has been debated due to their close relationship, genome sequences from these two DTUs were compared in more detail. Again, we detected extensive synteny among these closely related genomes, with limited sequence rearrangements (Fig. 2). However, clear differences between the two DTUs were present at the whole-genome level, with a greater similarity between SC43cl1.1 and 9280cl2 strains, and TCC and Tula cl2 strains. Also, TcVI genomes were found to be somewhat larger than TcV genomes, with an additional insertion of 4–6 assembled Mbp (Fig. 2; Table 1).
Fig. 2.
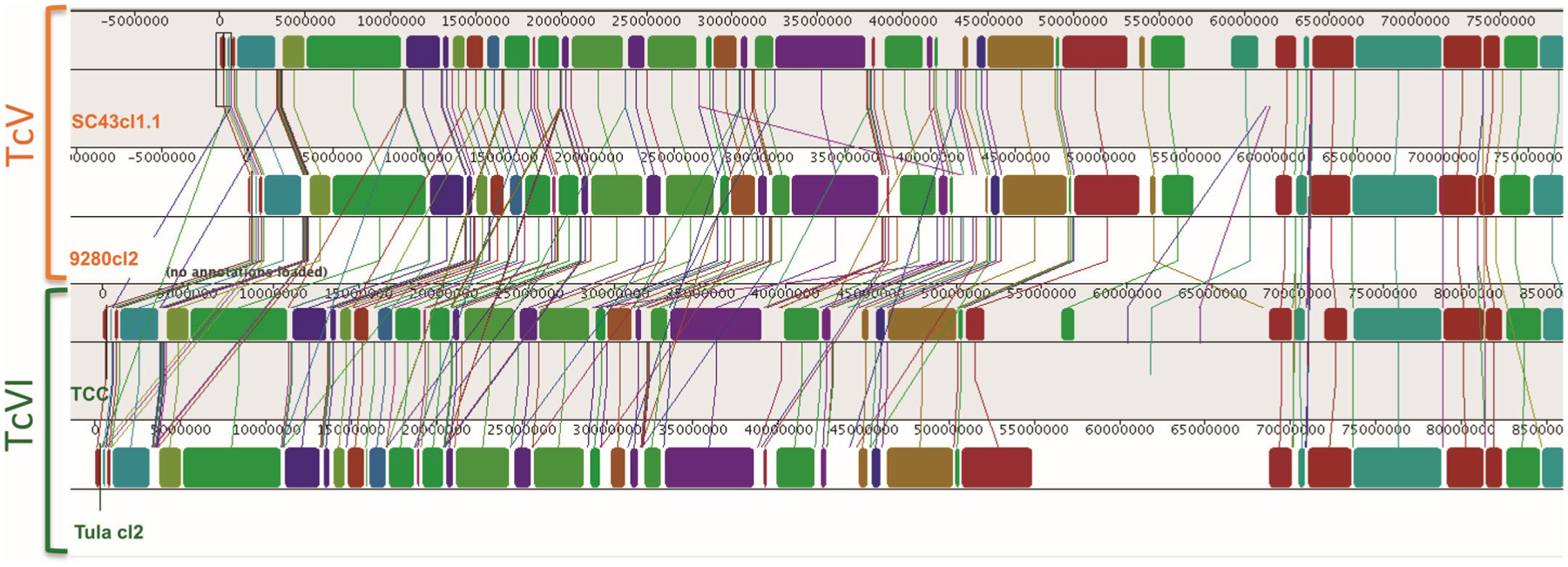
Whole-genome alignment of TcV and TcVI parasite strains. ProgressiveMauve alignment of genomes from SC43cl1.1, 9280cl2 (TcV), TCC, and Tula cl2 (TcVI) strains. Color blocks represent homologous collinear sequences that are connected among the respective genomes. There is extensive synteny, but some sequence rearrangements can be identified. TcVI genomes also have expanded regions not present in TcV.
A phylogenetic analysis of nuclear genome sequences was performed to better assess their relationship. First, we analyzed the mini-exon spliced leader, a key marker that has been extensively used for T. cruzi genotyping and shown to be able to clearly differentiate the different DTUs. This analysis clearly confirmed that SC43cl1.1 clustered with other TcV reference sequences, including the previously sequenced mini-exon marker from the SC43 strain (Fig. 3). Also, as reported before (Lewis et al. 2009; Majeau et al. 2019; Villanueva-Lizama et al. 2019), in spite of the small sequence length of this marker (about 600 bp), well-resolved clusters were identified for the closely related sequences from TcVI and TcII DTUs, while sequences from TcI and TcIII DTUs were further apart. The phylogenetic tree of the corresponding genome sequences confirmed the DTU clustering of these T. cruzi strains (Fig. 4A). In particular, the TcV and TcVI genome clusters were clearly differentiated from one another and separated from both the TcIII and the TcII parental DTUs.
Fig. 3.
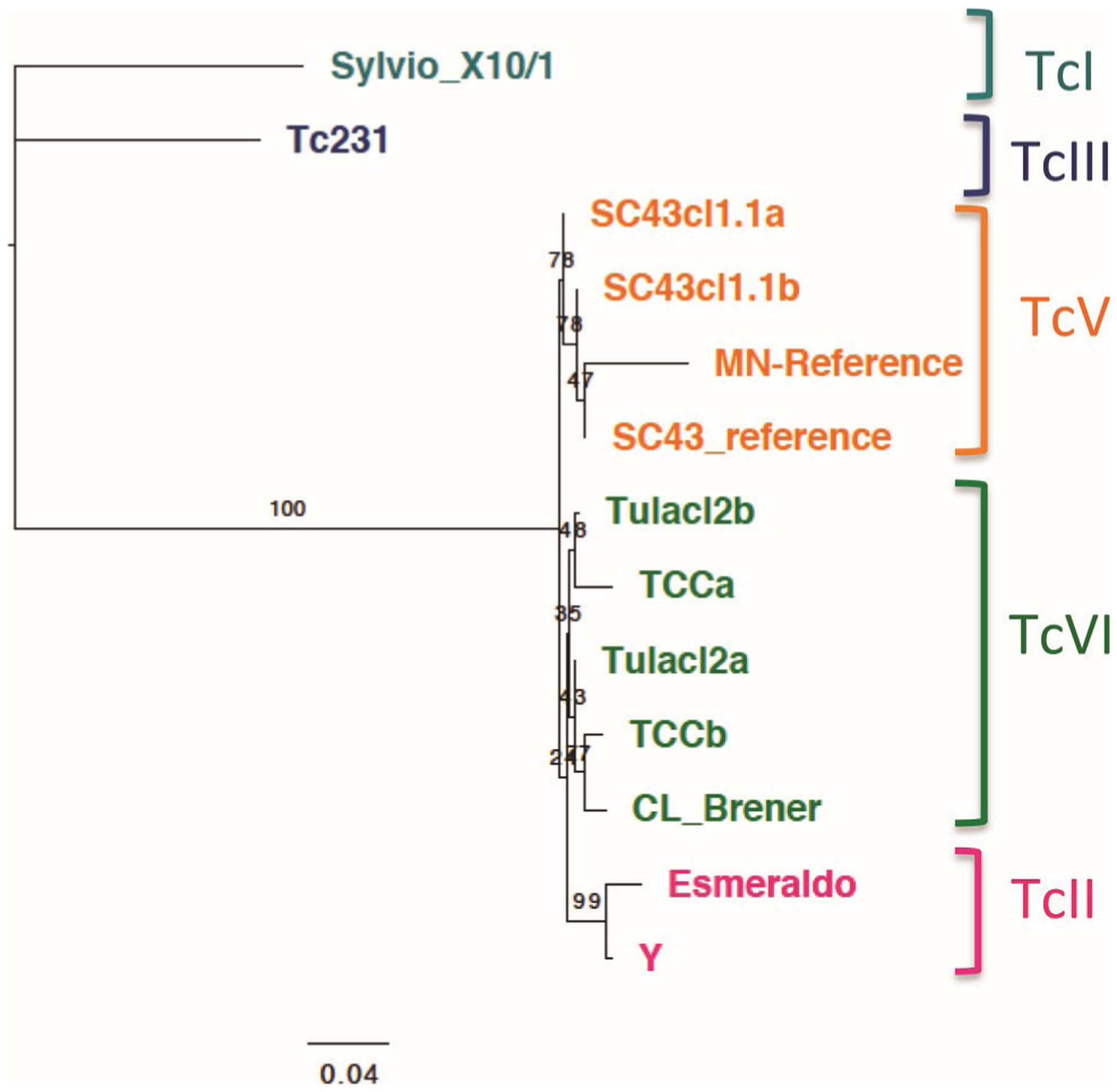
Phylogenetic analysis of the mini-exon sequence. Maximum likelihood analysis of mini-exon sequences from multiple strains. This marker allows for a good resolution of all the discrete typing units (DTUs), including the closely related TcII, TcV, and TcVI DTUs, which form well-separated clusters. Numbers on branches indicate bootstrap support.
Fig. 4.
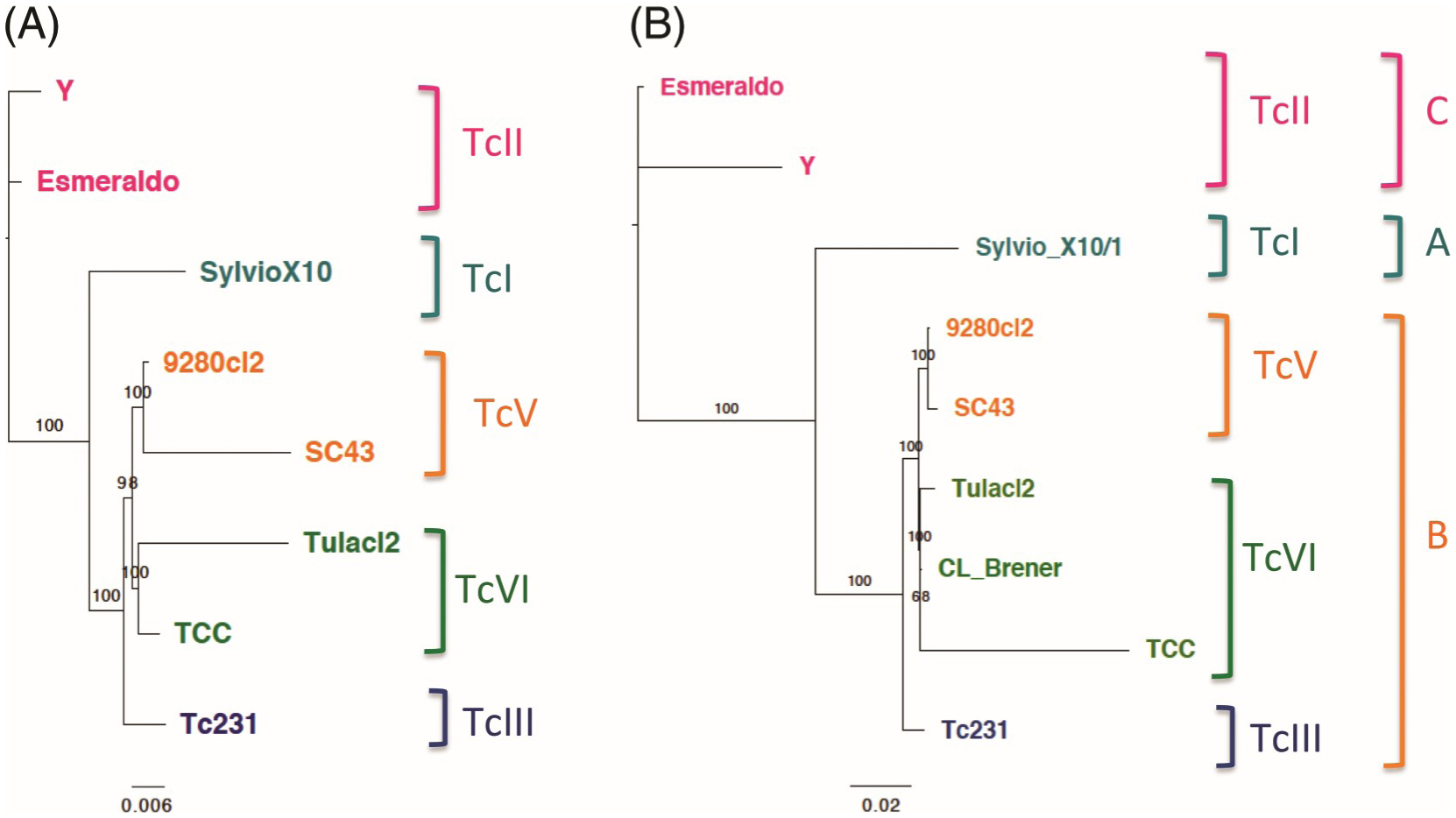
Comparison of phylogenies based on nuclear genome and kinetoplast maxicircle sequences. (A) Whole-genome alignment confirmed the clustering of strains according to discrete typing unit (DTU), and particularly resolve the closely related TcV and TcVI DTUs. (B) The corresponding ML phylogenetic tree of the kinetoplast maxicircle resulted in the previously identified three A, B, and C clades. However, there was also a clear substructuring of the maxicircles from Clade B according to the DTUs TcIII, TcV, and TcVI. Numbers on branches indicate bootstrap support.
The SC43cl1.1 kinetoplast maxicircle sequence was also analyzed. A high-quality assembly of 39 357 bp (Fig. 5) with an average coverage of 8017× was obtained. It shared 94.8% identity with the TCC maxicircle, of which there was up to 98.5% identity in the gene-coding region but only 92.4% identity in the variable/repeat region of the maxicircle. Phylogenetic analysis of the coding region of the SC43cl1.1 kinetoplast maxicircle with other available maxicircle sequences indicated three major clusters, corresponding to TcI and TcII ancestral DTUs as well as a more recent cluster that included kDNA sequences from TcIII, TcV, and TcVI DTUs (Fig. 4B). These three clusters corresponded to the previously identified maxicircle clades A, B, and C (Machado and Ayala 2001). However, a clear substructure of this diverse latter group C could also be detected, and sequences from TcIII, TcV, and TcVI DTUs were well resolved into smaller clusters with veiy high bootstrap values. Thus, the phylogeny of kDNA maxicircles matched well that of the nudear genomes of these parasite strains, strengthening the basis of current DTU classification as well as the distinction between the TcV and TcVI DTUs. These results also confirm that the TcV and TcVI DTUs inherited their kinetoplasts from a TcIII ancestor, as previously hypothesized (Westenberger et al. 2006; Zingales 2018).
Fig. 5.
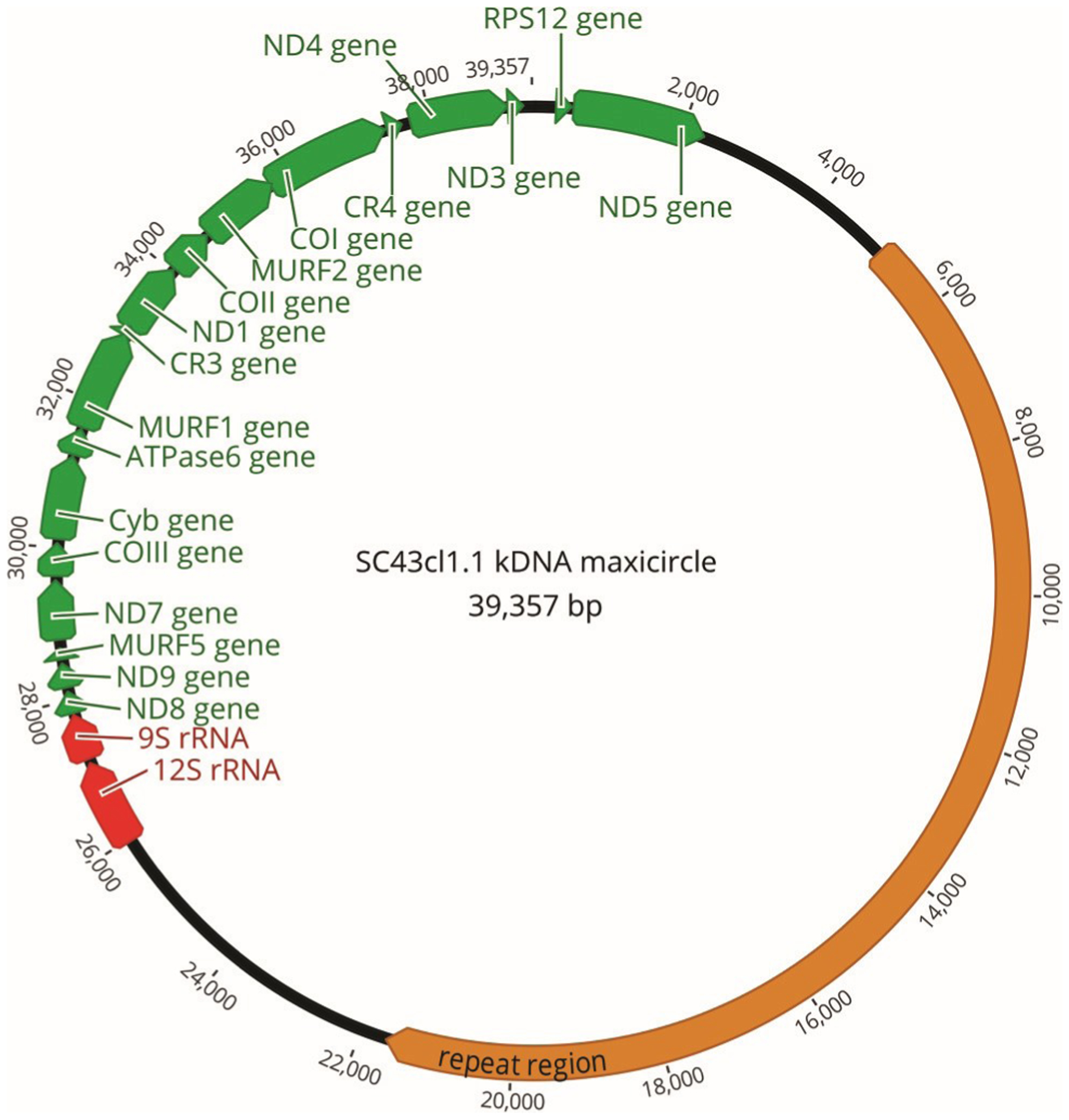
Structure and annotation of SC43cl1 kinetoplast maxicircle. Protein-coding genes are indicated in green, rRNA genes in red, and the repeat region in orange. MURF, maxicircle unidentified reading frame; ND, NADH dehydrogenase; COIII, cytochrome oxidase III; Cyb, cytochrome b; CR, cytosine-rich region; RPS12, ribosomal protein S12.
Discussion
Analysis of T. cruzi genetic diversity remains a key step to better understanding the likely associations between parasite diversity, parasite transmission cycles, and disease epidemiology. The current parasite genetic structuration into seven DTUs is based on multiple markers providing different levels of resolution (Zingales et al. 2009, 2012; Messenger et al. 2015), but there is still limited support for this classification at the genomic level due to a lack of representative genome sequences from some of these DTUs. We sequenced here the genome of strain SC43cl1, a reference strain from Bolivia belonging to the TcV DTU. This represents only the second TcV genome available, following that of strain 9280 cl2 (Reis-Cunha et al. 2018). Indeed, Bug2148 is a well characterized TcV strain, but the reported genome sequence (Callejas-Hernandez et al. 2018) is actually that from a TcI strain (E. Dumonteil et al., unpublished data).
We obtained a high-quality assembly for the SC43cl1.1 genome in only 1236 contigs with a high coverage and N50, owing mostly to the availability of the TCC genome reference. Indeed, the long-read technology used for this genome allowed for a greatly improved T. cruzi genome assembly (Berna et al. 2018), as also observed for the Berenice genome assembly (Diaz-Viraque et al. 2019). Comparison of the SC43cl1.1 genome to closely related TcV and TcVI genomes indicated extensive synteny and conservation, but a few sequence rearrangements were nonetheless detected, suggesting genetic recombination. However, further experimental validation of potential recombination is needed for confirmation. While there is growing evidence of extensive sexual reproduction in T. cruzi, particularly among TcI strains (Berry et al. 2019; Schwabl et al. 2019), is it also likely among other DTUs as well (Reis-Cunha et al. 2018). Importantly, the genome structures of TcV and TcVI parasites appeared to significantly differ, with TcVI strains presenting several expansions so far not present in TcV strains. Maximum likelihood phylogenetic analysis of genome further supported the presence of distinct genetic clusters for TcV and TcVI parasites. This is the first observation confirming DTU structure among these closely related parasites at the whole-genome level.
The concomitant analysis of the kinetoplast maxicircle provides further support for this genetic structure. Indeed, as noted before, the T. cruzi hybrid strains present uniparental inheritance of its kinetoplast DNA and bi-parental inheritance of its nuclear genome, with a nuclear genome derived from TcII and TcIII ancestors and a kinetoplast inherited from a TcIII ancestor (Machado and Ayala 2001; Westenberger et al 2006; Reis-Cunha et aL 2018). Our analysis of the large coding region of kDNA maxicircle from SC43cl1.1 and related strains agrees with the previously proposed three major kDNA clades (Machado and Ayala 2001). However, we also detected a significant sub-clustering of strains within kDNA clade C, again with a clear resolution of DTUs TcIII, TcV, and TcVI, strongly supporting the current DTU classification at the whole-genome level.
In conclusion, the genomic and kDNA sequence of T. cruzi SC43cl1.1 reported here provide further understanding of T. cruzi genetic structure and particularly indicate a significant level of genetic differentiation between the TcV and TcVI DTUs. While these observations should be further validated with additional strains from these closely related DTUs, they add to the growing number of available T. cruzi genomes and provide a key tool to associate strain biological characteristics with parasite genetic diversity.
Acknowledgements
This work was supported by the Eunice Kennedy Shriver National Institute of Child Health & Human Development (grant number 1R01HD94955-01A1) to C.H. This work received support from a grant from ByWater Institute-Louisiana to C.H., and from the Louisiana Board of Regents through the Board of Regents Support Fund [# LESASF (2018-21)-RD-A-19] to E.D.
References
- Baptista RP, Reis-Cunha JL, DeBarry JD, Chiari E, Kissinger JC, Bartholomeu DC, and Macedo AM 2018. Assembly of highly repetitive genomes using short reads: the genome of discrete typing unit III Trypanosoma cruzi strain 231. Microb. Genomics, 4: e000156. doi: 10.1099/mgen.0.000156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnabe C, Brisse S, and Tibayrenc M 2000. Population structure and genetic typing of Trypanosoma cruzi, the agent of Chagas disease: a multilocus enzyme electrophoresis approach. Parasitology, 120: 513–526. doi: 10.1017/s0031182099005661. [DOI] [PubMed] [Google Scholar]
- Berna L, Rodriguez M, Chiribao ML, Parodi-Talice A, Pita S, Rijo G, et al. 2018. Expanding an expanded genome: long-read sequencing of Trypanosoma cruzi. Microb. Genomics, 4: e000177. doi: 10.1099/mgen.0.000177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry ASF, Salazar-Sanchez R, Castillo-Neyra R, Borrini-Mayori K, Chipana-Ramos C, Vargas-Maquera M, et al. 2019. Sexual reproduction in a natural Trypanosoma cruzi population. PLoS Negl. Trop. Dis 13(5): e0007392. doi: 10.1371/journal.pntd.0007392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callejas-Hernandez F, Rastrojo A, Poveda C, Girones N, and Fresno M 2018. Genomic assemblies of newly sequenced Trypanosoma cruzi strains reveal new genomic expansion and greater complexity. Sci. Rep 8: 14631. doi: 10.1038/s41598-018-32877-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreno H, Rojas C, Aguilera X, Apt W, Miles MA, and Solari A 1987. Schizodeme analyses of Trypanosoma cruzi zymodemes from Chile. Exp. Parasitol 64: 252–260. doi: 10.1016/0014-4894(87)90150-0. [DOI] [PubMed] [Google Scholar]
- Darling AE, Mau B, and Perna NT 2010. progressive-Mauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE, 5: e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Viraque F, Pita S, Greif G, de Souza RCM, Iraola G, and Robello C 2019. Nanopore sequencing significantly improves genome assembly of the protozoan parasite Trypanosoma cruzi. Genome Biol. Evol 11: 1952–1957. doi: 10.1093/gbe/evz129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sayed NM, Myler PJ, Bartholomeu DC, Nilsson D, Aggarwal G, Tran AN, et al. 2005a. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science, 309: 409–415. doi: 10.1126/science.1112631. [DOI] [PubMed] [Google Scholar]
- El-Sayed NM, Myler PJ, Blandin G, Berriman M, Crabtree J, Aggarwal G, et al. 2005b. Comparative genomics of tiypanosomatid parasitic protozoa. Science, 309: 404–409. doi: 10.1126/science.1112181. [DOI] [PubMed] [Google Scholar]
- Franzen O, Ochaya S, Sherwood E, Lewis MD, Llewellyn MS, Miles MA, and Andersson B 2011. Shotgun sequencing analysis of Trypanosoma cruzi I Sylvio X10/1 and comparison with T. cruzi VI CL Brener. PLoS Negl. Trop. Dis 5: e984. doi: 10.1371/journal.pntd.0000984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimov ES, Zamyatnina KA, Matveeva NS, Rudenskaya YA, Kraeva N, Kolesnikov AA, and Yurchenko V 2020. Common structural patterns in the maxicircle divergent region of Trypanosomatidae. Pathogens, 9(2): 100. doi: 10.3390/pathogens9020100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisard EC, Teixeira SM, de Almeida LG, Stoco PH, Gerber AL, Talavera-Lopez C, et al. 2014. Trypanosoma cruzi clone Dm28c draft genome sequence. Genome Announc 2: e01114–13. doi: 10.1128/genomeA.01114-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumsiek J, Arnold R, and Rattei T 2007. Gepard: a rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics, 23: 1026–1028. doi: 10.1093/bioinformatics/btm039. [DOI] [PubMed] [Google Scholar]
- Lewis MD, Ma J, Yeo M, Carrasco HJ, Llewellyn MS, and Miles MA 2009. Genotyping of Trypanosoma cruzi: systematic selection of assays allowing rapid and accurate discrimination of all known lineages. Am. J. Trop. Med. Hyg 81: 1041–1049. doi: 10.4269/ajtmh.2009.09-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado CA, and Ayala FJ 2001. Nucleotide sequences provide evidence of genetic exchange among distantly related lineages of Trypanosoma cruzi. Proc. Natl. Acad. Sci. U.S.A 98: 7396–7401. doi: 10.1073/pnas.121187198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeau A, Herrera C, and Dumonteil E 2019. An improved approach to Trypanosoma cruzi molecular genotyping by next-generation sequencing of the mini-exon gene. Methods Mol. Biol 1955: 47–60. doi: 10.1007/978-1-4939-9148-8_4. [DOI] [PubMed] [Google Scholar]
- Messenger LA, Miles MA, and Bern C 2015. Between a bug and a hard place: Trypanosoma cruzi genetic diversity and the clinical outcomes of Chagas disease. Exp. Rev. Anti. Infect. Ther 13: 995–1029. doi: 10.1586/14787210.2015.1056158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez JD, Herrera C, Bogotá Y, Duqye MC, Suérez-Rivillas A, and Guhl F 2012. Validation of a Poisson-distributed limiting dilution assay (LDA) for a rapid and accurate resolution of multiclonal infections in natural Trypanosoma cruzi populations. J. Microbiol. Methods, 92: 220–225. doi: 10.1016/j.mimet.2012.11.002. [DOI] [PubMed] [Google Scholar]
- Reis-Cunha JL, Baptista RP, Rodrigues-Luiz GF, Coqueiro-Dos-Santos A, Valdivia HO, de Almeida LV, et al. 2018. Whole genome sequencing of Trypanosoma cruzi field isolates reveals extensive genomic variability and complex aneuploidy patterns within TcII DTU. BMC Genomics, 19: 816. doi: 10.1186/s12864-018-5198-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez HO, Guerrero NA, Fortes A, Santi-Rocca J, Girones N, and Fresno M 2014. Trypanosoma cruzi strains cause different myocarditis patterns in infected mice. Acta Trop 139: 57–66. doi: 10.1016/j.actatropica.2014.07.005. [DOI] [PubMed] [Google Scholar]
- Schwabl P, Imamura H, Van den Broeck F, Costales JA, Maiguashca-Sanchez J, Miles MA, et al. 2019. Meiotic sex in Chagas disease parasite Trypanosoma cruzi. Nat. Commun 10: 3972. doi: 10.1038/s41467-019-11771-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souto RP, Fernandes O, Macedo AM, Campbell DA, and Zingales B 1996. DNA markers define two major phylogenetic lineages of Trypanosoma cruzi. Mol. Biochem. Parasitol 83:141–152. doi: 10.1016/S0166-6851(96)02755-7. [DOI] [PubMed] [Google Scholar]
- Tibayrenc M, and Ayala FJ 2015. The population genetics of Trypanosoma cruzi revisited in the light of the predominant clonal evolution model. Acta Trop 151: 156–165. doi: 10.1016/j.actatropica.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva-Lizama L, Teh-Poot C, Majeau A, Herrera C, and Dumonteil E 2019. Molecular genotyping of Trypanosoma cruzi by next-generation sequencing of the mini-exon gene reveals infections with multiple parasite DTUs in Chagasic patients from Yucatan, Mexico. J. Infect. Dis 219: 1980–1988. doi: 10.1093/infdis/jiz047. [DOI] [PubMed] [Google Scholar]
- Westenberger SJ, Cerqueira GC, El-Sayed NM, Zingales B, Campbell DA, and Sturm NR 2006. Trypanosoma cruzi mitochondrial maxicircles display species- and strain-specific variation and a conserved element in the non-coding region. BMC Genomics, 7(1): 60. doi: 10.1186/1471-2164-7-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingales B 2018. Trypanosoma cruzi genetic diversity: Something new for something known about Chagas disease manifestations, serodiagnosis and drug sensitivity. Acta Trop 184: 38–52. doi: 10.1016/j.actatropica.2017.09.017. [DOI] [PubMed] [Google Scholar]
- Zingales B, Andrade SG, Briones MR, Campbell DA, Chiari E, Fernandes O, et al. 2009. A new consensus for Trypanosoma cruzi intraspecific nomenclature: second revision meeting recommends TcI to TcVI. Mem Inst. Oswaldo Cruz, 104: 1051–1054. doi: 10.1590/S0074-02762009000700021. [DOI] [PubMed] [Google Scholar]
- Zingales B, Miles MA, Campbell DA, Tibayrenc M, Macedo AM, Teixeira MM, et al. 2012. The revised Trypanosoma cruzi subspecific nomenclature: rationale, epidemiological relevance and research applications. Infect. Genet. Evol 12: 240–253. doi: 10.1016/j.meegid.2011.12.009. [DOI] [PubMed] [Google Scholar]


