Polygenic scores have emerged as a new tool to quantify inherited risk for a given disease based on the cumulative impact of many common sites of DNA variation.1–3 Although several programs plan to return polygenic scores to patients, the implications of a high polygenic score for a given individual’s family members have not been fully characterized. A recent analysis focused on height and cognitive ability suggested polygenic scores are largely similar between siblings and thus have limited utility for embryo selection seeking to maximize these traits.4
Here, we build upon this prior work by examining the correlation of polygenic scores and concordance of a high polygenic score according to relatedness across 4 cardiometabolic diseases: coronary artery disease, atrial fibrillation, diabetes, and severe obesity (body mass index, ≥40 kg/m2). We studied 179 monozygotic twin pairs, 22 644 sibling pairs, and 11 123 second-degree relative pairs derived from the UK Biobank study.5 Among the studied individuals, median age was 58 years, 57.1% were women, and 88.1% were of European ancestry. Relatedness was computed according to Kinshipbased Inference for Genome-wide association studies (KING) kinship coefficients as described previously.5 For each condition, we used previously published polygenic scores, adjusted for genetic ancestry using the residual of a linear regression model predicting the raw polygenic score using the first 10 principal components as described previously.1–3
We first measured the correlation of the polygenic score for coronary artery disease between related individuals. As expected, a perfect correlation (Pearson R=1.0) was observed between monozygotic twins (Figure A). This correlation was attenuated among sibling pairs—R=0.51—and decreased to 0.25 among pairs of second-degree relatives (Figure B and C). Nearly identical results were observed across the 3 additional diseases.
Figure.
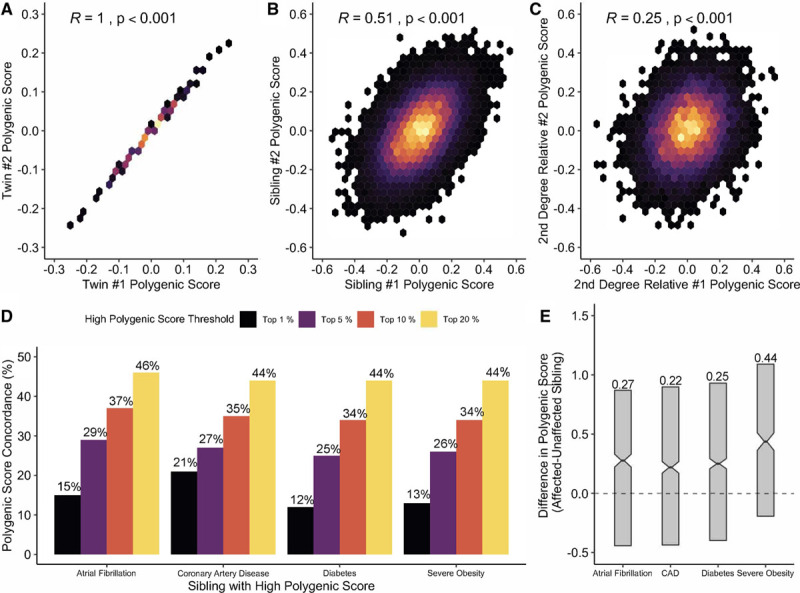
Concordance of polygenic scores among related individuals. Two-dimensional correlation heat maps of polygenic scores for coronary artery disease (CAD) between pairs of monozygotic twins (A), siblings (B), and second-degree relatives (C). D, The percentage of individuals with a sibling’s polygenic score above a certain threshold percentage who also share a similarly increased polygenic score for 4 cardiometabolic diseases. Polygenic scores were normalized to have a mean of 0 and SD of 1 as performed previously.1–3 Among sibling pairs with discordant disease status, median polygenic score was higher in the afflicted siblings for each of the 4 diseases, with effect ranging from 0.22 to 0.44 SD units (P<0.001 by paired Wilcoxon rank-sum test for each; E).
Among individuals presenting with a high polygenic score for a given disease, we asked: how common is a similarly high score among the individuals’ siblings? Because polygenic scores are normally distributed quantitative traits—there is no standard threshold for high—we explored multiple thresholds: top 1%, 5%, 10%, and 20% of the disease-specific distributions. Taking coronary artery disease as an example, for individuals with a polygenic score in the top 1% of the distribution, 21% of siblings’ polygenic scores met this threshold, corresponding to a >20-fold enrichment (Figure D). If a high polygenic score was defined as the top 20% of the distribution, 44% of siblings’ polygenic scores similarly met this threshold. The positive predictive value of a high polygenic score was largely independent of family history—for coronary artery disease, the positive predictive value of a sibling with a polygenic score in the top 20% of the distribution for a similarly elevated score was 47% for those who reported family history versus 40% for those who did not.
We next studied whether the variability in polygenic scores might contribute to disease discordance between sibling pairs, as was observed for coronary artery disease in 1844 pairs, atrial fibrillation in 1430 pairs, severe obesity in 749 pairs, and diabetes in 2376 pairs. Among these pairs, the higher polygenic score was observed in the affected sibling in 1071 of 1844 pairs with discordant coronary artery disease status (58%)—significantly higher than the 50% that would be observed by chance (P<0.001). A similar pattern was noted across the three additional diseases, with the afflicted sibling having the higher polygenic score in 60% of pairs for atrial fibrillation, 60% for diabetes, and 67% for severe obesity. We next conducted a matched-pair analysis, assessing the difference in polygenic score between affected and unaffected siblings. For each of the 4 diseases, the median difference was significantly >0, with effect size ranging from 0.22 to 0.44 SDs (Figure E). Taken together, these results suggest that chance differences in polygenic scores among siblings play a statistically significant, but quantitatively modest, role in differential disease manifestation.
These results may have important implications for clinical practice and genetic counseling. For monogenic risk variants, cascade screening is recommended to identify a binary phenotype, about 50% of first-degree relatives will harbor the same variant. By contrast, polygenic scores are continuous traits that correlate strongly across sibling pairs. Nonetheless, cascade testing to identify additional family members with a high polygenic score may warrant consideration. Indeed, when a high polygenic score is defined as the top 20% of the distribution—corresponding to a more than doubling of risk for each of the 4 diseases1,2—about 45% of siblings similarly met this threshold. Genetic counseling that contextualizes polygenic score results within the context of personal and family health history and accounts for implications of an increased score for family members is warranted.
Additional research is needed to assess the optimal approach to polygenic score disclosure and operationalize cascade screening within this context, with the ultimate goal of identifying additional individuals with high inherited risk who may benefit from targeted screening or prevention.
Acknowledgments
This research has been conducted using the UK Biobank Resource under application number 7089 and approved by the Mass General Brigham Institutional Review Board. The data that support the findings of this study are available from the corresponding author upon reasonable request and from the UK Biobank Resource.
Sources of Funding
Funding support was provided by grant 1K08HG010155 from the National Human Genome Research Institute, a Hassenfeld Scholar Award from the Massachusetts General Hospital, a Merkin Institute Fellowship from the Broad Institute of MIT and Harvard, and a sponsored research agreement from IBM Research (all to Dr Khera).
Disclosures
Dr Khera has served as a scientific advisor to Sanofi, Medicines Company, Maze Therapeutics, Navitor Pharmaceuticals, Verve Therapeutics, Amgen, Color, and Columbia University (National Institutes of Health); received speaking fees from Illumina, MedGenome, Amgen, and the Novartis Institute for Biomedical Research; received sponsored research agreements from the Novartis Institute for Biomedical Research and IBM Research; and reports a patent related to a genetic risk predictor (20190017119). The other authors report no conflicts.
Footnotes
For Sources of Funding and Disclosures, see page 245.
Contributor Information
Nicholas J. Reid, Email: nicholas_reid@hms.harvard.edu.
Deanna G. Brockman, Email: deanna.brockman@mgh.harvard.edu.
Courtney Elisabeth Leonard, Email: cleonard0@mgh.harvard.edu.
Renee Pelletier, Email: renee.pelletier@mgh.harvard.edu.
References
- 1.Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, Natarajan P, Lander ES, Lubitz SA, Ellinor PT, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018; 50:1219–1224. doi: 10.1038/s41588-018-0183-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khera AV, Chaffin M, Wade KH, Zahid S, Brancale J, Xia R, Distefano M, Senol-Cosar O, Haas ME, Bick A, et al. Polygenic prediction of weight and obesity trajectories from birth to adulthood. Cell. 2019; 177:587–596.e9. doi: 10.1016/j.cell.2019.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khera AV, Chaffin M, Zekavat SM, Collins RL, Roselli C, Natarajan P, Lichtman JH, D’Onofrio G, Mattera J, Dreyer R, et al. Whole-genome sequencing to characterize monogenic and polygenic contributions in patients hospitalized with early-onset myocardial infarction. Circulation. 2019; 139:1593–1602. doi: 10.1161/CIRCULATIONAHA.118.035658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karavani E, Zuk O, Zeevi D, Barzilai N, Stefanis NC, Hatzimanolis A, Smyrnis N, Avramopoulos D, Kruglyak L, Atzmon G, et al. Screening human embryos for polygenic traits has limited utility. Cell. 2019; 179:1424–1435.e8. doi: 10.1016/j.cell.2019.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O’Connell J, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018; 562:203–209. doi: 10.1038/s41586-018-0579-z [DOI] [PMC free article] [PubMed] [Google Scholar]


