Abstract
Background
Sequencing of patient-derived xenograft (PDX) mouse models allows investigation of the molecular mechanisms of human tumor samples engrafted in a mouse host. Thus, both human and mouse genetic material is sequenced. Several methods have been developed to remove mouse sequencing reads from RNA-seq or exome sequencing PDX data and improve the downstream signal. However, for more recent chromatin conformation capture technologies (Hi-C), the effect of mouse reads remains undefined.
Results
We evaluated the effect of mouse read removal on the quality of Hi-C data using in silico created PDX Hi-C data with 10% and 30% mouse reads. Additionally, we generated 2 experimental PDX Hi-C datasets using different library preparation strategies. We evaluated 3 alignment strategies (Direct, Xenome, Combined) and 3 pipelines (Juicer, HiC-Pro, HiCExplorer) on Hi-C data quality.
Conclusions
Removal of mouse reads had little-to-no effect on data quality as compared with the results obtained with the Direct alignment strategy. Juicer extracted more valid chromatin interactions for Hi-C matrices, regardless of the mouse read removal strategy. However, the pipeline effect was minimal, while the library preparation strategy had the largest effect on all quality metrics. Together, our study presents comprehensive guidelines on PDX Hi-C data processing.
Keywords: Hi-C, chromatin conformation capture, xenografts, PDX, xenome
Introduction
Patient-derived tumor xenograft (PDX) mouse models are indispensable in preclinical and translational cancer research. Previous studies have demonstrated that human tumors engrafted in immunocompromised mouse models preserve each patient's genetic heterogeneity [1] and response to treatment [2,3]. Consequently, the main application of PDX systems is to elucidate the molecular mechanisms of human cancers within controlled in vivo conditions. With the wide adoption of sequencing technologies, sequencing of PDX samples is now a standard [4–7].
High-throughput sequencing of PDX samples faces challenges not present in sequencing of cell lines and homogeneous tissues. Engraftment of human cancer tissue fragments into mice leads to the rapid loss of human stroma and invasion of mouse stromal cells [1,3]. Consequently, sequencing of PDX tumor samples produces reads derived from both human and mouse genomes, with mouse read contamination ranging from 4–7% up to 20% for RNA-seq and exome data [8], and even 47% on average for whole-genome sequencing data [9]. Metastases are even more variable, and we previously identified up to 99% mouse reads in PDX RNA-seq data from lung, liver, or brain metastases [4]. Given the high similarity of human and mouse genomes, with orthologous gene products on average 85% identical [10], the presence of mouse reads introduces uncertainty in the alignment of PDX sequencing data.
Three strategies have been developed to address the removal of mouse reads from PDX sequencing data. The first strategy, referred to hereafter as “Direct,” is the direct alignment of PDX sequencing data to the human genome. The second, filtering strategy includes separation of human and mouse reads and using only human data for downstream analysis. Xenome was among the first tools implementing filtering strategy. It classifies reads into the human, mouse, both, neither, or ambiguous categories using a 25-mer matching algorithm [11]. Despite being relatively old and lacking maintenance, Xenome remains widely used in bioinformatics pipelines [12]. We refer to this strategy as “Xenome” throughout. The third strategy involves the alignment of reads to human and mouse genomes simultaneously and then filtering reads by best alignment match [8]. This approach has been implemented in Disambiguate [13], bamcmp [14], and XenoCP [15] tools. This strategy, referred to hereafter as “Combined,” includes alignment to the in silico combined human-mouse reference genome to disambiguate human and mouse reads at the alignment step [4,16].
Each strategy for mouse read removal from PDX sequencing data has its own advantages and disadvantages. The Xenome and Combined strategies require extra effort, more processing time, and in some cases doubling requirements for computational resources. Several studies investigated the benefits of removal of contaminating mouse reads from PDX sequencing data. In DNA-seq PDX data, the removal of mouse reads reduced the false-positive rate of somatic mutation detection, especially when matching normal samples are not available [8,12,13,15,17]. In RNA-seq data, the removal of mouse reads improved gene expression quantification [15], correlation with pure human gene expression [8], and enrichment in relevant pathways [14]. Benchmarking of all 3 strategies using DNA-seq convincingly demonstrated that the Xenome and Combined strategies are necessary to minimize false discovery rates in detecting genomic variants, with exome sequencing data benefiting the most [17]. The general consensus is that the removal of mouse reads from PDX sequencing data improves the extraction of human-specific signal from RNA-seq and DNA-seq PDX sequencing data [8,11–16].
Chromatin conformation capture technology and its high-throughput derivatives, such as Hi-C [18], have recently emerged as tools to assess the 3D structure of the genome. Changes in the three-dimensional (3D) structure of the genome are an established hallmark of cancer [19–21]. However, the majority of the 3D cancer genomics studies have been performed in vitro using cell lines [22–24]. Hi-C sequencing of PDX samples opens novel ways for understanding mechanisms of human cancers under controlled in vivo conditions. However, the effect of contaminating mouse reads on the quality of PDX Hi-C data, and the choice of pipeline, remains undefined.
Hi-C sequencing data possess unique qualities that need to be considered when evaluating the effect of mouse reads in Hi-C PDX data. First, Hi-C paired-end reads are processed individually, as single-end data. Second, Hi-C data undergo extensive filtering to extract “valid pairs,” i.e., reads representative of two ligated DNA fragments with proper orientation and distance between them [25,26]. Furthermore, in contrast to typical sequencing experiments, processing of Hi-C data requires high-performance computational resources because one Hi-C experiment produces more than 20x the number of reads of a typical RNA-seq experiment [27]. It remains uncertain whether efforts to remove mouse reads from PDX Hi-C data are justified and meaningfully improve the quality of human Hi-C data.
To address the effect of mouse read removal in PDX sequencing data, we evaluated 3 strategies for preprocessing PDX Hi-C data: Direct, Xenome, and Combined. Using different library preparation strategies, we generated 2 deeply sequenced Hi-C datasets of a carboplatin-resistant UCD52 breast cancer cell line [4,5]. We further created 3 in silico PDX Hi-C datasets with either 10% or 30% of mouse read contamination, mirroring the percent of mouse reads observed in our experimental Hi-C data. In particular, we used Hi-C data from normal and cancer cells to investigate whether the biological properties, such as copy number variations inherent to cancer genomes, affect the quality of Hi-C data. Human Hi-C data without mouse read contamination were used as a baseline. This design allowed us to comprehensively quantify the effect of contaminating mouse reads on the quality of Hi-C data and the downstream results.
Although several studies discuss how to process Hi-C data and what pipeline to use [25,28,29], they have not evaluated the effect of mouse read contamination. We evaluated 3 leading pipelines, Juicer [30], HiC-Pro [31], and HiCExplorer [32], in terms of Hi-C data quality, their ability to extract biological information, and computational runtime.
In total, we tested 9 combinations of strategies—all pairwise combinations of 3 strategies for mouse read handling (Direct, Xenome, and Combined) and 3 pipelines (Juicer, HiC-Pro, and HiCExplorer)—to generate contact matrices from 9 in silico and 2 experimental PDX Hi-C datasets. Furthermore, we assessed the effect of library preparation strategies on the quality of downstream results from Hi-C data. We found that removing mouse reads using the Xenome or Combined strategies minimally affects the quality of Hi-C matrices and information extracted from them, while the Direct alignment yielded comparable-quality results without the additional computational overhead. The choice of processing pipeline had negligible impact on data quality and the downstream results. Ultimately, the choice of library preparation was the single variable with largest effect on data quality. From these studies, we recommend using the Direct alignment of PDX Hi-C data to the human genome. The choice of the library preparation strategy should be given priority.
Results
A comprehensive workflow for assessing the impact of mouse read contamination in PDX Hi-C data
Sequencing of biological samples from patient-derived xenograft (PDX) mouse models faces a challenge of mixed genomic context derived from host (mouse) and graft (human) cells. Naturally, the goal is to sequence human-specific genomic information; however, highly homologous mouse reads may hinder the identification of human genomic information. We investigated whether the presence of mouse reads in human Hi-C data negatively affects Hi-C data quality and whether the removal of mouse reads improves the detection of topologically associating domains (TADs) and chromatin loops. We created in silico PDX Hi-C data and generated two experimental PDX Hi-C datasets (Table 1, Additional File 1: Table). We assessed 3 alignment strategies for mouse read removal and 3 common pipelines to generate Hi-C matrices (Fig. 1).
Table 1:
Summary of in silico and experimental PDX Hi-C data
| Hi-C data | Description | Total reads | Proportion of mouse reads (%)a | Optimal resolution (kb)b |
|---|---|---|---|---|
| Baseline | ||||
| GM12878 | Human B-lymphoblastoids | 486,848,169 | 0 | 7.0 |
| HMEC | Human mammary epithelial | 456,577,383 | 0 | 7.9 |
| KBM7 | Human myelogenous leukemia | 431,368,621 | 0 | 8.3 |
| CH12-LX (rep 1) | Mouse lymphoma cell line | 45,594,869 | 100 | N/A |
| CH12-LX (rep 2) | Mouse lymphoma cell line | 175,930,719 | 100 | N/A |
| in silico PDX | ||||
| GM12878 (10%) | GM12878 + CH12-LX (rep 1) | 532,443,038 | 8.56 | 7.0/7.1/7.0 |
| GM12878 (30%) | GM12878 + CH12-LX (rep 2) | 662,778,888 | 26.54 | 7.0/7.1/7.0 |
| HMEC (10%) | HMEC + CH12-LX (rep 1) | 502,172,252 | 9.08 | 7.9/7.9/7.9 |
| HMEC (30%) | HMEC + CH12-LX (rep 2) | 632,508,102 | 27.81 | 7.9/7.9/7.9 |
| KBM7 (10%) | KBM7 + CH12-LX (rep 1) | 476,963,490 | 9.56 | 8.3/8.3/8.3 |
| KBM7 (30%) | KBM7 + CH12-LX (rep 2) | 607,299,340 | 28.97 | 8.3/8.3/8.3 |
| Experimental PDX | ||||
| UCD52 Library 1 | Basal-like BRCA cell line | 873,892,191 | 12.16/12.38 | 11.5/11.9/11.7 |
| UCD52 Library 2 | Basal-like BRCA cell line | 708,069,622 | 25.78/29.14 | 8.9/9.1/9.0 |
Estimated using Xenome/Combined alignment strategy, respectively.
Estimated following Direct/Xenome/Combined alignment strategy, respectively.
Figure 1:
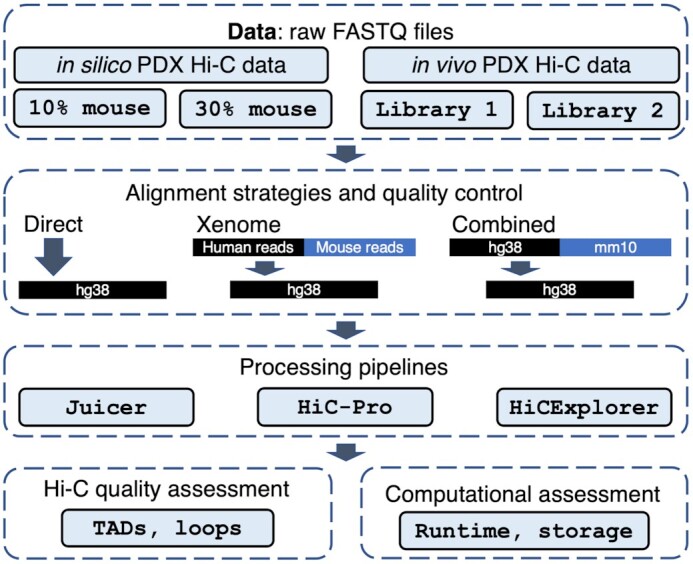
PDX Hi-C data analysis workflow. In silico (controlled mixture of human and 10/30% mouse Hi-C reads) and experimental PDX Hi-C data (2 library preparation strategies) were processed using 3 read alignment strategies (Direct: read alignment directly to the human genome, Xenome: human reads retrieved with Xenome tool, and Combined: all reads aligned to the combined human-mouse genome, respectively). Three pipelines (Juicer, HiC-Pro, HiCExplorer) were used to obtain Hi-C matrices. Hi-C data quality and runtime metrics were assessed following each processing step.
The in silico PDX Hi-C data were created by concatenating FASTA reads from previously published mouse and human Hi-C data [27] (see Methods). Human Hi-C data from GM12878 B-lymphoblastoid cells (nearly normal karyotype) and KBM7 myelogenous leukemia (near-haploid karyotype) were selected to assess the effect of mouse read contamination in normal and cancer Hi-C data, respectively. HMEC human mammary epithelial cells were selected to parallel the breast cancer origin of our experimental PDX Hi-C data. Mouse Hi-C data from B-lymphoblast CH12-LX cells were used to create the in silico PDX Hi-C data with  and
and 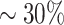 level of mouse read contamination. Human Hi-C data for the corresponding cell lines without mouse reads were used as a baseline.
level of mouse read contamination. Human Hi-C data for the corresponding cell lines without mouse reads were used as a baseline.
The main limitation of in silico PDX Hi-C data is that human and mouse reads originate from different libraries. Although in silico PDX Hi-C data may be sufficient to test the performance of aligners on a mixture of highly homologous human and mouse reads, it is unknown whether this mixture can recapitulate the complexity of experimental PDX Hi-C data, where, theoretically, crosslinking and ligation of human and mouse DNA can occur. To investigate whether the removal of mouse reads from experimental PDX Hi-C data improves the quality of Hi-C matrices, we generated replicates of Hi-C data from a triple-negative breast cancer PDX (UCD52 cells), obtained with 2 different library preparation strategies (Library 1 and Library 2; see Methods). As expected, human-specific replicates of experimental PDX Hi-C data prepared with the same library preparation strategy showed high correlation, in contrast to those prepared with a different strategy (mean Pearson correlation coefficient (PCC) = 0.9963 and 0.9547, respectively). Mouse matrices were uniformly correlated irrespective of the library preparation strategy (mean PCC = 0.9870; Additional File 2: Figure). Therefore, replicates of Hi-C data were merged for downstream processing. In total, we processed 11 PDX Hi-C datasets (Table 1).
We applied 3 alignment strategies to remove mouse read contamination: the Direct alignment of PDX Hi-C reads to the human reference genome (“Direct”), the alignment of data cleaned of mouse read data using the Xenome tool [11] (“Xenome”), or using pre-alignment to a combined human and mouse genome (“Combined”; see Methods, Fig. 1). We then applied 3 pipelines for processing of Hi-C data: Juicer [30], HiC-Pro [31], and HiCExplorer [32] (Fig. 1). The use of different methods for mouse read removal and pipelines allowed us to establish the optimal strategy for analyzing Hi-C data derived from PDX mouse models.
Experimental PDX Hi-C data have a higher proportion of ambiguously mapped reads
Xenome accurately estimated the 10%/30% proportion of mouse reads in our in silico PDX Hi-C data (Fig. 2, Additional File 3: Table). We observed a similar proportion of mouse reads in our experimental PDX data (∼12% and 30%; Table 1). Less than 1% of reads were mapped to both or neither human nor mouse genomes, and these results were consistent in the in silico and experimental PDX Hi-C data. Compared with in silico PDX data, the number of “ambiguous” reads in the experimental data was higher (4–5% vs. 1%; Additional File 3). This higher intra-population heterogeneity is expected because, in contrast to cell lines, experimental PDX samples contain a mixture of different cell types and cell states. This will introduce background noise interactions and should be considered when comparing experimental and in silico PDX Hi-C analysis results. Overall, our results indicate that in silico PDX Hi-C data reflect the level of mouse read contamination observed in experimental settings. However, the higher level of ambiguously mapped reads suggests unique biological properties in experimental PDX Hi-C data and justifies the need for their analysis.
Figure 2:
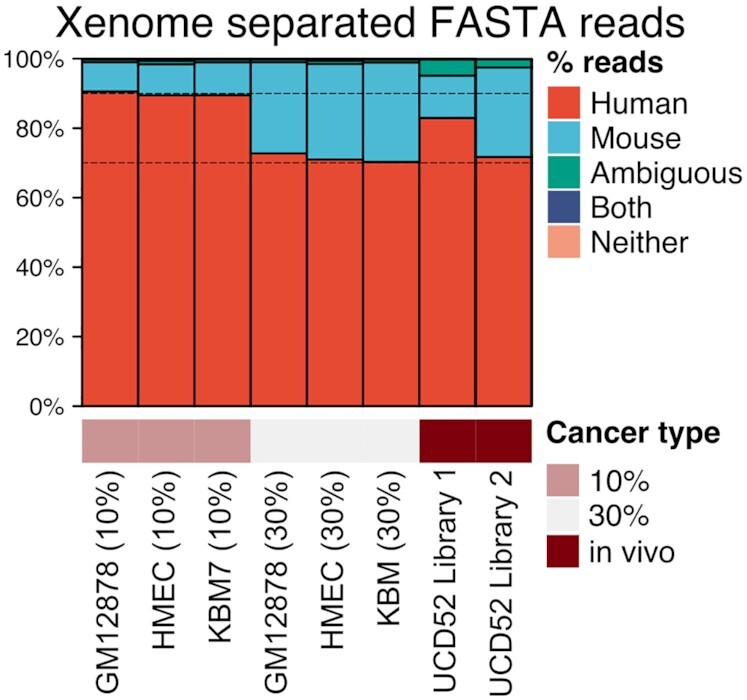
Proportions of human and mouse reads in experimental and in silico PDX Hi-C data. Dashed lines indicate the 10% and 30% mouse read contamination thresholds. Details of Xenome read separation statistics are collected in Additional File 3: Table.
Removal of mouse reads has negligible impact on the retrieval rate and quality of Hi-C contacts
Following data processing using all combinations of alignment strategies and pipelines, we investigated the level of residual mouse reads mismapped to the human genome. For that, we used in silico PDX Hi-C data where the identity of human and mouse reads can be tracked. As expected, following Xenome and Combined mouse read removal strategies, the data processed by any pipeline had, on average, 0.0064% of mouse reads, and these results were independent of the initial level of mouse read contamination (range, 0.0002–0.0125%; Additional File 4: Table). Furthermore, using the Direct alignment strategy resulted in a higher level of residual mouse reads (average, 0.0625%; range, 0.0037–0.2402%). Juicer retained the largest proportion of mouse reads with, on average, 0.1032%/0.2250% of the initial 10% and 30% mouse read contamination, respectively, while HiC-Pro retained the smallest proportion of mouse reads (Additional File 4: Table). Thus, both HiC-Pro and HiCExplorer pipelines effectively eliminated contaminating mouse reads with direct alignment of Hi-C reads to the human genome.
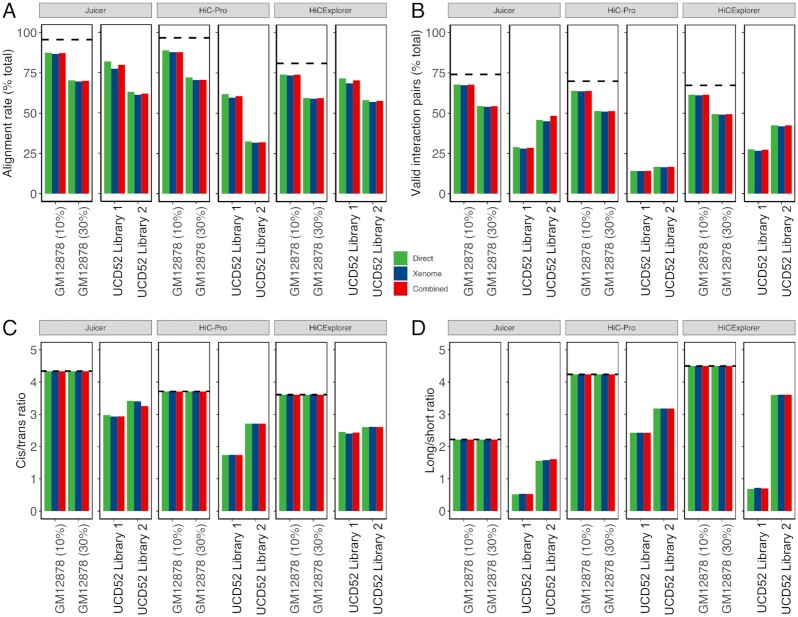
We extracted 4 Hi-C quality metrics from the log files produced by each pipeline (all QC metrics are given in Additional File 5: Table). “Alignment rate" is the proportion of reads aligned to the human genome. “Valid interaction pairs" is the proportion of reads marked as Hi-C contacts by each pipeline considering the valid restriction site within a reasonable distance. Higher values of those metrics indicate better data quality. “Cis/trans ratio" is the ratio of intra- vs. inter-chromosomal interacting reads. A higher cis/trans ratio indicates enrichment for within-chromosomal reads, expected in the Hi-C experiments. “Long/short ratio" is the ratio of cis interactions >20 kb away vs those <20 kb away. The expectation is to capture more long-distance chromatin interactions, i.e., a long/short ratio with a value >1, while a long/short ratio <1 indicates that long interactions were lost, prompting a cautious interpretation of the results. These Hi-C quality metrics allow for the comprehensive definition of optimal alignment strategy and the effect of mouse read removal.
The removal of mouse reads had minimal-to-no effect on the alignment quality metrics of in silico and experimental PDX Hi-C data (Fig. 3, Additional File 6: Figure). Expectedly, the alignment rate and the proportion of valid interaction pairs in in silico PDX Hi-C data were diminished proportionally to the percent of mouse read contamination (10% or 30%), as compared with those in pure human Hi-C data for the corresponding cell lines (dashed lines in Fig. 3A and B). The removal of mouse reads from in silico PDX Hi-C data did not markedly affect the cis/trans ratio and long/short ratio (Fig. 3C and D). These results were consistent across cell lines (Additional File 6: Figure) and suggest that, while the Direct alignment strategy retains more mismapped reads, the downstream Hi-C quality metrics perform similarly to those from data with explicitly removed mouse reads.
Figure 3:
Quality metrics for selecting the optimal pipeline for processing PDX Hi-C data. All metrics are stratified by the pipeline (Juicer, HiC-Pro, and HiCExplorer) and color-coded by the alignment strategy (green: Direct, blue: Xenome, red: Combined). (A) Alignment rate representing the proportion of all aligned reads. (B) The proportion of valid interaction pairs as determined by each pipeline. (C) The ratio of cis interacting pairs (i.e., occurring on the same chromosome) vs. trans interacting pairs (i.e., between chromosome interactions). (D) The ratio of long- vs short-interacting Hi-C contacts. Dashed lines correspond to the baseline alignment quality metrics for human Hi-C data without mouse reads.
Similar to the results obtained with in silico PDX Hi-C data, the removal of mouse reads from experimental PDX Hi-C data did not markedly affect quality metrics (Fig. 3), although more variability was observed (∼2–4%). Interestingly, although the alignment rate of data prepared with the Library 2 strategy was lower than that of Library 1–prepared data (Fig. 3A), the proportion of valid interaction pairs, cis/trans ratio, and, in particular, long/short ratio were higher (Fig. 3B–D). These results suggest that the Library 2–prepared data contain more information about intra-chromosomal long- and short-distance chromatin interactions. In summary, these results indicate that the removal of mouse reads does not substantially improve or change the alignment quality of PDX Hi-C data, but the library preparation strategy has a significant effect.
Evaluation of pipelines in terms of their ability to recover information from PDX Hi-C data
Although removing mouse reads using either strategy did not substantially affect the alignment quality of PDX Hi-C data (Fig. 3A), we noted pipeline-specific differences (Fig. 3, Additional File 7: Figure), referred to by their names for brevity. Specifically, Juicer produced a similar alignment rate as HiC-Pro in in silico PDX Hi-C data. However, it recovered 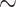 15% more alignable reads in experimental PDX Hi-C data compared to HiC-Pro. On the other hand, HiCExplorer yielded ∼20% lower alignment rate for in silico PDX Hi-C data. Yet, HiCExplorer performed nearly as well as Juicer in the alignment of experimental PDX Hi-C data (Additional File 7: Fig. A). Similarly, Juicer recovered up to 10% more valid interaction pairs in in silico PDX data as compared to HiC-Pro and HiCExplorer (Additional File 7: Fig. B). However, in experimental PDX Hi-C data, Juicer recovered nearly twice as many valid interaction pairs as the HiC-Pro and outperformed HiCExplorer by a ∼2% margin (Additional File 7: Fig. B). These results indicate that Juicer can recover more alignable reads and recover a higher proportion of valid interaction pairs. These improvements were particularly pronounced when processing experimental PDX Hi-C data.
15% more alignable reads in experimental PDX Hi-C data compared to HiC-Pro. On the other hand, HiCExplorer yielded ∼20% lower alignment rate for in silico PDX Hi-C data. Yet, HiCExplorer performed nearly as well as Juicer in the alignment of experimental PDX Hi-C data (Additional File 7: Fig. A). Similarly, Juicer recovered up to 10% more valid interaction pairs in in silico PDX data as compared to HiC-Pro and HiCExplorer (Additional File 7: Fig. B). However, in experimental PDX Hi-C data, Juicer recovered nearly twice as many valid interaction pairs as the HiC-Pro and outperformed HiCExplorer by a ∼2% margin (Additional File 7: Fig. B). These results indicate that Juicer can recover more alignable reads and recover a higher proportion of valid interaction pairs. These improvements were particularly pronounced when processing experimental PDX Hi-C data.
A typical Hi-C experiment is expected to detect the majority of interactions within chromosomes (cisinteractions) as compared with between-chromosome (trans) interactions. This should be reflected by a high cis/trans ratio. Juicer produced Hi-C data with a higher cis/transratio than the HiC-Pro and HiCExplorer pipelines. These results were consistent between in silico and experimental PDX Hi-C data (Fig. 3, Additional File 7: Fig. C). Juicer yielded lower long/short ratios compared to the other 2 pipelines (Fig. 3D), which reflects the fact that Juicer captured overwhelmingly more and most probably unwanted short-distance cis interaction (Fig. 3C). These results were consistent in in silico and experimental PDX Hi-C data (Additional File 7: Fig. D). Interestingly, HiCExplorer gave the highest long/short ratios in all in silico PDXs and in the experimental PDX using the Library 2 preparation strategy. Notably, all quality metrics were superior in Hi-C data obtained using the Library 2 preparation strategy. These results suggest that, altogether, HiCExplorer may offer the most reliable information from PDX Hi-C data, and highlight the importance of library preparation strategy.
The presence of mouse reads has a negligible effect on the detection of TADs and chromatin loops
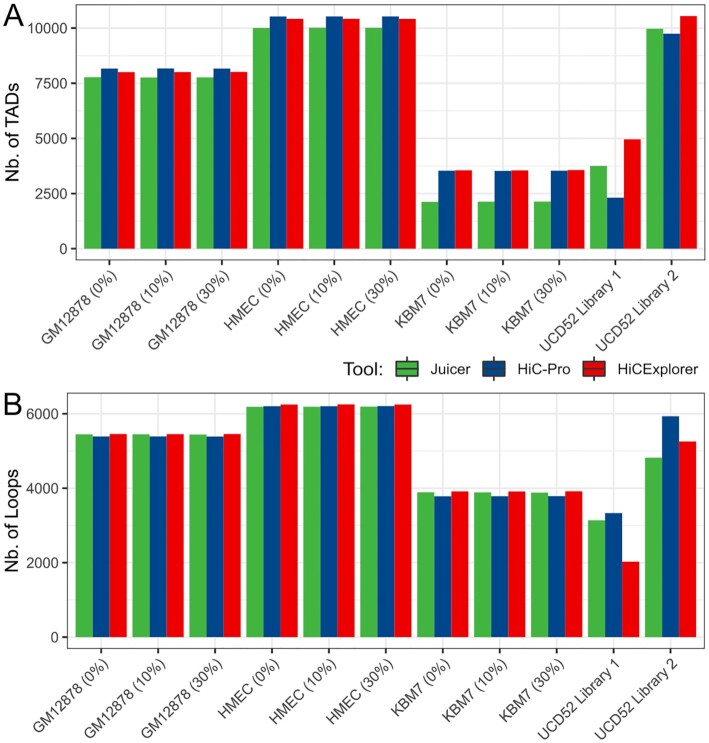
The most typical use of Hi-C data is to detect chromatin 3D structures, such as Topologically Associating Domains (TADs) and chromatin loops. Given that mouse read removal strategies had negligible impact on Hi-C data quality, we used the Direct alignment strategy for the following tests. We evaluated the number of TADs and loops detected from data processed by the 3 pipelines. To focus on the data- and pipeline-specific differences, we used the same TAD/loop calling algorithms throughout our work (see Methods). The number of TADs and loops should be considered as a suggestive indicator of data quality under the hypothesis that a deeper-sequenced high-complexity Hi-C experiment would produce Hi-C matrices where more TADs/loops can be detected.
Compared to baseline (pure human Hi-C data), the number of cell-type–specific TADs and loops was nearly identical at the 10% or 30% level of in silico mouse read contamination (Fig. 4, Additional File 8: Table). We also observed that TAD and loop boundaries detected from in silico PDX Hi-C data were highly overlapping in a condition-specific manner, and this overlap was unaffected by mouse read contamination (Additional File 9: Fig. A–C, Additional File 10: Fig. A–C). These results were consistent irrespective of the pipeline and support the notion that mouse reads do not markedly affect TAD and loop boundary detection.
Figure 4:
The library preparation strategy has the largest effect on TAD/loop detection. The number of TADs (A) and loops (B) are similar at different levels of mouse reads and across pipelines in all in silico PDX Hi-C data, whereas experimental PDX Hi-C data produced variable results. Results for the Direct alignment strategy are shown.
Library preparation strategy has the largest effect on TAD and loop detection
We observed nearly twice as many TADs and loops detected in Library 2–prepared data than in Library 1–prepared data (Fig. 4), paralleling our observation that Library 2–prepared data have better quality metrics (Fig. 3). Using experimental Hi-C data, HiC-Pro detected the fewest TADs but the most loops, while HiCExplorer detected the most TADs. Notably, the pipeline-specific differences in the numbers of TADs and loops were most pronounced for Library 1–prepared data (Fig. 4). These results suggest that, with the optimal library preparation strategy, the differences in pipelines are negligible, further emphasizing the importance of library preparation strategy.
Similar to the analysis we did on in silico PDX Hi-C data, we investigated the agreement between TAD and loop boundaries detected from experimental PDX Hi-C data, processed with different pipelines. Given the same biological origin of experimental PDX Hi-C data, we expected a high overlap of boundaries also between the 2 libraries. We found boundaries detected from data prepared with the Library 2 strategy to be highly consistent irrespective of the pipelines (Additional File 9: Fig. D, Additional File 10: Fig. D). In contrast, boundaries detected from Hi-C data prepared with the Library 1 strategy were most distinct and more variable. Notably, Juicer and HiCExplorer boundaries were most similar, while HiC-Pro boundaries were distinct from them (Additional File 9: Fig. D, Additional File 10: Fig. D). These results suggest that pipeline selection is less critical when working with high-quality data (Library 2). Of note, Juicer and HiCExplorer appear to detect concordant boundaries irrespective of data quality.
Finally, we investigated the enrichment of CTCF, a known boundary mark, at TAD and loop boundaries. As expected, co-localization enrichment of CTCF was highly significant (χ2 P-value < 2.225E−308) and similar irrespective of the initial mouse read contamination level. However, cell-line– and library-specific differences were more pronounced (Additional File 11: Figure). Similarly, enrichment of CTCF signal was highly similar (Additional File 12–13: Figure). We observed slightly higher variability in undersequenced KBM7 data and Library 1–prepared experimental PDX data, with less significant CTCF co-localization and signal enrichment in those samples (Additional File 12–13: Figure). These results suggest that boundaries supported by biological evidence can be detected irrespective of mouse read contamination and pipeline, and the library preparation strategy is essential for improved TAD/loop boundary detection.
Technical and runtime considerations
We compared the runtime and storage requirements for each alignment strategy and pipeline. Removal of mouse reads with either Xenome or Combined strategy resulted in smaller files and, consequently, faster processing time (Fig. 5A). However, when considering the additional time needed to remove mouse reads (longest for the Combined strategy), processing of the original data (Direct) was the fastest. Together with previous observations of the minimal effect of mouse read removal on Hi-C data quality, these results indicate that extra computational time used to remove mouse reads does not appear to be beneficial for the quality of downstream results.
Figure 5:
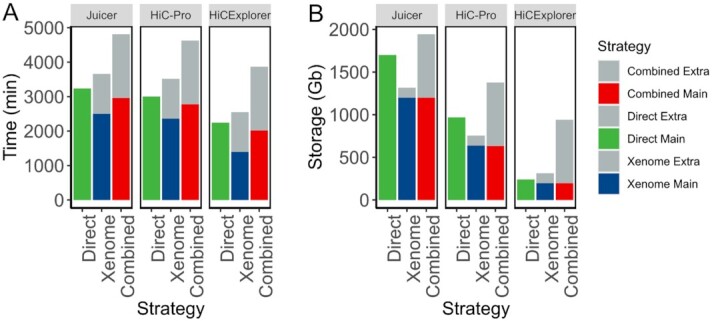
Removal of mouse reads carries a significant computational overhead. An example of runtime (A) and storage (B) resources required for processing PDX Hi-C data to obtain Hi-C matrices. Only within-pipeline runtime comparisons are valid because each pipeline used different computational resources (see Methods). Results for processing Library 2–prepared PDX Hi-C data are presented. Extra: accounting for time and storage space required to filter mouse reads. Main: time and storage determined for processing human reads.
The removal of mouse reads requires considerable extra storage space, with the Combined strategy requiring the most additional storage (Fig. 5B). Interestingly, the Juicer pipeline required the largest storage space even when processing the original data (Direct); however, it can be minimized by compressing text files produced by it. Together with additional time requirements, extra space for removing mouse reads creates a significant computational overhead with negligible benefits as compared with the Direct alignment strategy.
The choice of tools for mouse read removal is an important technical consideration requiring significant human time. Xenome, a part of the Gossamer bioinformatics suite, has not been updated since 5 January 2017 (as of 15 October 2020). It requires dependencies that can only be installed using administrative privileges, which are rarely available for bioinformaticians working in a high-performance computing environment. Furthermore, Xenome requires creating its own genome index, which also contributes to the storage and processing time, and was not included in Fig. 5. The Combined strategy can be implemented ad hoc, and the combined genomes and indexes can be downloaded using Refgenie [33] (see Methods). However, the extra hard drive space and time required for mouse read removal create an unnecessary human and computational burden and can contribute to delays in a project. We recommend using the Direct alignment strategy for the most optimal computational processing of experimental PDX Hi-C data.
Discussion
We have assessed the effect of mouse read contamination on the performance of 3 leading pipelines for Hi-C data processing. Using quality control (QC) metrics at the alignment stage, we showed that, unlike whole-exome and RNA-seq data from PDX models, Hi-C PDX data are largely unaffected by mouse read contamination. This is not unexpected because Hi-C data processing pipelines include a series of filters to select valid pairs [25]. It is highly unlikely for experimental PDX Hi-C data to contain human-mouse chimeric reads, and even if such a read pair occurs, the probability that it would be recognized as a valid Hi-C contact (e.g., mapped in the proper orientation, within a certain distance from the nearest restriction site) is negligible. Our study confirms this reasoning and recommends the Direct alignment of PDX Hi-C data to the graft (human) genome.
Our results indicate that the Juicer pipeline may recover more alignable reads and valid interaction pairs and achieves better cis/trans but worse long/short interaction ratios. Given that Juicer retains more misaligned mouse reads within in silico PDX Hi-C data (Additional File 4: Table), it remains unclear whether these reads represent true human chromatin interactions in experimental PDX Hi-C data. This performance of Juicer can be attributed to the use of the BWA-MEM aligner, which can efficiently handle split-read alignment. In contrast, HiC-Pro uses the bowtie2 aligner with the default “–end-to-end” mapping settings. The documentation for the HiCExplorer pipeline discourages end-to-end alignment of Hi-C reads because the alignment needs to accommodate for potential ligation junctions. Consequently, we used the BWA-MEM aligner with HiCExplorer. Given that Juicer and HiCExplorer both detected similar TAD/loop boundaries even in the poorer-quality Library 1–prepared data (Additional File 9–10: Fig. D), both emerge as leading tools in our study. More generally, our results suggest the use of BWA-MEM–based pipelines when processing experimental PDX Hi-C data.
Even though Juicer initially produced poor results in terms of long/short ratio metric (Fig. 3D), this did not seem to affect the final number of TADs and loops detected, as well as their boundaries. Between Juicer and HiCExplorer, we find Juicer the easiest to set up for running. On the other hand, HiCExplorer comes with a comprehensive suite of tools for downstream analysis of the Hi-C matrices with no need to change the Hi-C matrix format. Both tools perform well and we leave the choice to the user on the basis of his/her experience to install and run the tools, as well as the ability to change between different Hi-C matrix data formats.
We identified library preparation strategy as a major determinant of the downstream data quality. While differences in quality metrics between in silico PDX Hi-C datasets can be attributed to the differences in sequencing depth (Additional File 1: Table), differences in our experimental PDX Hi-C data can be directly attributed to the library preparation strategies. Although our experimental PDX Hi-C data had nearly twice as many reads as the in silico PDX Hi-C data (Table 1), their quality metrics were inferior compared to in silico–constructed Hi-C data (Fig. 3). This was most pronounced for Library 1–prepared data, which we speculate is due to the presence of nearly 40% read duplicates, as compared to 12–15% duplicates in other datasets (Additional File 1: Table). However, the higher proportion of dangling ends, self-circles, dumped reads, singletons, and so forth may have contributed to the inferior quality of Library 1–prepared data (Additional File 5: Table). Similar to the ENCODE guidelines [34], our observations suggest the importance of controlling duplicates in Hi-C data.
Despite the lower number of sequencing reads and alignment rate, data obtained with the Library 2 preparation strategy recovered more cis-interacting Hi-C contacts spanning longer distances (cis/trans ratio and long/short ratio metrics in Fig. 3C and Fig. 3D, respectively). Furthermore, the number and size of TADs detected from the Library 2–prepared data was similar to that detected in in silico PDX Hi-C data (Fig. 4). This can be attributed to multiple enzymes cutting the human genome in more than 16M sites. In contrast, the single-enzyme Library 1 preparation strategy digests the genome in ∼7.2M sites. Given that Hi-C data quality significantly affects downstream results, we suggest careful inspection of the shallow sequenced library before the deep-sequencing experiment, giving particular weight to the metrics presented in Fig. 3. The choice of restriction enzymes should be given primary consideration in designing PDX Hi-C experiments.
According to the ENCODE guidelines [34], we expected to recover ∼58% of sequenced reads as valid Hi-C interactions. While our in silico PDX Hi-C data [27] almost always achieved this threshold, our experimental PDXs did not meet these criteria (∼28 and ∼45 for Library 1 and Library 2 preparation strategies, respectively; Additional File 5: Table). Of note, other studies report a much lower rate of valid Hi-C interactions. For instance, the mean number of valid interactions across 93 Hi-C datasets was 17.72±13.04 [35]. The overall lower percentage of valid interactions in our experimental Hi-C data can be partially explained by the fact that the genome of carboplatin-resistant UCD52 cells may be affected by genome rearrangements. The presence of duplications, deletions, and inversions is known to affect the genome's 3D organization [36] and may have negatively affected the performance of our experimental PDX Hi-C data. Our results suggest the need to consider the effect of large-scale genome variation in the processing of PDX Hi-C data, in addition to the standard Hi-C data quality metrics.
Methods
Generation of experimental PDX Hi-C data
UCD52 tumors were implanted in mice and once palpable treated with a single dose of 40 mg/kg carboplatin, as previously described [4,5]. Once the tumors began growing again, they were treated with another dose of carboplatin. This was repeated until the tumor was no longer responsive to carboplatin. Xenograft tissue samples were processed by Phase Genomics (Seattle, WA) and Arima Genomics (San Diego, CA). Data generated using Phase Genomics/Arima Genomics library preparation strategy are referred to as “Library 1”/“Library 2,” respectively. The following protocols detail each strategy, as provided by the respective service providers.
Phase Genomics (Library 1) preparation strategy
Approximately 200 mg of xenograft tissue was finely chopped and then crosslinked for 20 min at room temperature (RT) with end-over-end mixing in 1 mL of Proximo Crosslinking solution. The crosslinking reaction was terminated with a quenching solution for 20 min at RT with end-over-end mixing. Quenched tissue was rinsed once with 1× Chromatin Rinse Buffer (CRB), resuspended in Proximo Animal Lysis Buffer 1, and then transferred to Dounce Homogenizer (Kontes) and homogenized with 12 strokes using the “A” homogenizer. Disrupted tissue in lysis buffer was incubated 20 min at RT. Large debris was removed following a 1-min 500g spin. Lysate was recovered and transferred to a clean tube and pelleted by spinning at 17,000g for 5 min. The supernatant was removed and pellet washed once with 1× CRB. After removing 1× CRB wash, the pellet was resuspended in 100 µL Proximo Lysis Buffer 2 and incubated at 65°C for 10 min. Chromatin was irreversibly bound to SPRI beads by adding 100 µL SPRI beads to lysate and incubating for 10 min at RT. Beads were then washed once with 1× CRB. Beads were resuspended in 150 µL of Proximo fragmentation buffer and 5 µL of Proximo fragmentation enzyme (PN LS0027; 5,000 U/m Sau3AI cutting at “GATC”) was added and incubated for 1 hour at 37°C. The sample was cooled to 12°C, and 2.5 µL of Phase Genomics Finishing Enzyme was added (PN LS0030). Sample was incubated 30 min at 12°C, adding 6 µL of Stop Solution (PN LS0004) at the completion of the incubation. The beads were then washed with 1× CRB and resuspended in 100 µL of Proximo Ligation Buffer supplemented with 5 µL of Proximity ligation enzyme. The proximity ligation reaction was incubated at RT for 4 hours with occasional gentle mixing. After the ligation step, 5 µL of Reverse Crosslinks enzyme (PN BR0012) was added and the reaction incubated at 65°C for 1 hour. After reversing crosslinks, the free DNA was recovered by adding 100 µL of SPRI buffer to the reaction. Beads were washed twice with 80% ethanol, air dried, and proximity ligation products were eluted (Elution Buffer, PN BR0014). DNA fragments containing proximity ligation junctions were enriched with streptavidin beads (PN LS0020). After washing streptavidin beads twice with PG Wash Buffer 2 (PN BR0004), once with PG Wash Buffer 1 (PN BR0016), and once with molecular biology grade water, library was constructed using Proximo library reagents (PNs LS0034, LS0035, and BR0017) amplified with high-fidelity polymerase (PN BR0018), and size selected using SPRI enriching for fragments between 300 and 700 bp. Pooled libraries were sequenced on an Illumina NovaSeq 6000 instrument using an S4 flow cell. Libraries were de-multiplexed using unique dual indexes following standard Illumina methods.
Arima Genomics (Library 2) preparation strategy
Hi-C experiments were performed by Arima Genomics (San Diego, CA) according to the Arima-HiC protocols described in the Arima-HiC kit (P/N: A510008). After the Arima-HiC protocol, Illumina-compatible sequencing libraries were prepared by first shearing purified Arima-HiC proximally ligated DNA and then size-selecting DNA fragments from ∼200 to 600 bp using SPRI beads. The size-selected fragments were then enriched for biotin and converted into Illumina-compatible sequencing libraries using the KAPA Hyper Prep kit (P/N: KK8504). After adapter ligation, DNA was PCR amplified and purified using SPRI beads. The purified DNA underwent standard QC (qPCR and Bioanalyzer) and was sequenced on the HiSeq X following the manufacturer's protocols.
Construction of in silico PDX Hi-C data
Publicly available Hi-C data from the study by Rao et al. 2014 [27] (GSE63525) were used to construct in silico PDX Hi-C data. Three human and 1 mouse cell line Hi-C datasets were selected (Table S1). To construct in silico PDX data containing a mixture of human and mouse reads, FASTA files from human and mouse cell lines were concatenated to form Hi-C datasets containing ∼10% and 30% mouse reads (Table 1). If read length differed between human and mouse datasets, reads were trimmed from the 3′ end to smallest read length using cutadapt (v2.7 [37]) before concatenation.
Removal of mouse reads from PDX Hi-C data
Three mouse read removal strategies were evaluated: Direct, Xenome, and Combined (Fig. 1). In the Direct alignment strategy, all reads were mapped to the human reference genome version GRCh38/hg38 using only autosomal and sex chromosomes. In the Xenome approach, PDX Hi-C reads were processed with the Xenome tool [11] from the gossamer GitHub repository [38], and human-only FASTA reads were kept. In the Combined strategy, the combined human-mouse genome was created by concatenating autosomal and sex chromosomes from hg38 and mm10 genomes. Chromosome names were renamed with “hg38_” or “mm10_” prefixes. Both species-specific and combined genomes, as well as the corresponding bowtie2 and BWA indexes, are available for download using refgenie v.0.9.3 [33]. Scripts to download and organize refgenie's assets are provided (see “Data Availability” section).
Raw reads were first mapped with BWA-MEM -SP5 (v.0.7.17 [39]) to the combined genome, and the resulting BAM files were then subsetted with samtools (v.1.3.1 [40]) to keep reads mapping to the hg38 chromosomes. bedtools bamtofastq (v.v2.17.0 [41]) was then applied to convert the hg38-BAM files back to FASTQ format.
Processing human Hi-C data and PDX Hi-C data
All Hi-C data were processed with three pipelines with default settings: (i) Juicer (v.1.6 [30]), (ii) HiC-Pro (v.3.0.0 [31]), and (iii) HiCExplorer (v. 3.5.1 [32]). Sites for Phase Genomics cutting enzyme (GATC) were detected using the (i) generate_site_positions.py, (ii) digest_genome.py, and (iii) findRestSite scripts that come with each tool, respectively. Sites for Arima Genomics cutting enzyme (^GATC, G^ANTC) were obtained from [42] (used for HiC-Pro and HiCExplorer) and generated with the generate_site_positions.py for Juicer pipeline. The optimal data resolution was identified using Juicer's script calculate_map_resolution.sh and set to 10 kb to analyze 3D genome structures for all Hi-C data.
Switching between Hi-C file formats and matrix normalization
Each pipeline adapts its own format for storing the data. Juicer saves the contact matrices into a binary .hic format. HiC-Pro stores results as a text file in the sparse data matrix .matrix and genomic coordinate .bed formats. HiCExplorer uses an HDF5-based binary .h5 file format. To compare data produced by each pipeline, the data at 10-kb resolution were converted to the HiCExplorer-compatible .h5 format. HiC-Pro raw text-based contact matrices were directly converted to h5 format with the HiCExplorer's hicConvertFormat tool with the default settings. Juicer's toolbox was used to extract raw text-based contact matrices with the following command: “juicer_tools_1.13.02.jar dump observed NONE file.hic chrom chrom BP 10000 outputName.txt". The text files were then converted to HiC-Pro format using a customized R script and converted to h5 format with the HiCExplorer's hicConvertFormat tool. All h5 files were then normalized using the HiCExplorer's hicCorrectMatrix tool on a per chromosome basis using the Knight and Ruiz (KR) method.
Analysis of TADs and chromatin loops
HiCExplorer's hicFindTADs tool was applied on the KR-normalized matrices to calculate a genome-wide TAD separation score with “minDepth,” “maxDepth,” and “step” set to 30, 100, and 10 kb, respectively. “thresholdComparisons” and “delta” were set to 0.05 and 0.01, and “fdr” method was chosen for “correctForMultipleTesting.”
Similarly, HiCExplorer's hicDetectLoops tool was used to detect chromatin loops with the following settings: “maxLoopDistance” set to 2,000,000, “windowSize” set to 10, “peakWidth” set to 6, “peakInteractionsThreshold” set to 10, “pValuePreselection” and “pValue” both set to 0.05.
CTCF co-localization (or overlap) enrichment was assessed using GenomeRunner [43,44]. Briefly, genomic coordinates of TAD and loop boundaries were converted to the hg19 genome assembly and tested for enrichment in the consolidated Transcription Factor ChIP-seq data from ENCODE (wgEncodeRegTfbsClusteredV2 table in the UCSC genome browser). The χ2 test was used to assess co-localization enrichment and enrichment odds ratios were presented for across-condition comparisons.
CTCF signal was plotted using HiCExplorer's computeMatrix and plotProfile tools with the default settings. The ENCFF414WYX.bigWig CTCF track was downloaded from [47] on 14 December 2020.
Technical considerations
All jobs were run on a high-performance computer cluster under the CentOS v.6.7 operating system and the PBS job submission system PBSPro_12.2.1.140 292. The Juicer pipeline was run on 1 CPU; the other pipelines were run on 16 CPUs. Owing to administrative restrictions, only time and storage space were captured. The processing scripts are available at the project home page [45].
Data Availability
Accession numbers to download the publicly available Hi-C data used in this study are listed in Table S1. Experimental PDX Hi-C data generated in this study are available at the SRA via bioproject number PRJNA668904. All codes necessary to reproduce the analyses are available at the project home page [45]. Snapshots of our code and other supporting data are openly available in the GigaScience repository, GigaDB [46].
Availability of Source Code and Requirements
Project name: PDX Hi-C processing
Project home page: https://github.com/dozmorovlab/PDX-HiC_processingScripts
Operating systems(s): Mac/Linux
Programming language: Shell, R (≥4.0)
Other requirements: None
License: MIT
Any restrictions to use by non-academics: None
Additional Files
Additional File 1: Table.Datasets used in the present study. Selected quality metrics were obtained using FastQC v.0.11.8.
Additional File 2: Figure.Correlation between Hi-C matrices obtained from each replicate of experimental PDX samples. Experimental PDX Hi-C data were processed through Xenome to separate human and mouse reads. Human Hi-C matrices showed very high correlation, most pronounced for Library 2 preparation strategy (A). As expected, mouse Hi-C matrices were similar irrespective of library preparation strategy. Pearson correlation coefficients were calculated for 1-Mb matrices (non-zero elements only) and averaged across all chromosomes.
Additional File 3: Table.Xenome filtering statistics. The values represent the proportions of total reads in each PDX as indicated.
Additional File 4: Table.The proportion of mouse reads mismapped to the human genome in insilico PDX Hi-C data. There was 10% and 30% initial mouse read contamination, processed with Direct, Xenome, and Combined strategies, and Juicer, HiC-Pro, and HiCExplorer pipelines, 0–100% range. The % values are calculated with respect to the total number of reads that define each PDX.
Additional File 5: Table.Summary statistics used to compare the efficacy of the 3 Hi-C pipelines. Pipeline-specific alignment statistics are shown in the corresponding worksheets. Statistics shown in Fig. 3 are highlighted in red.
Additional File 6: Figure.Quality metrics assessed to select the optimal pipeline for processing PDX Hi-C data. Observations using HMEC and KBM7 cell lines confirm the results shown in Fig. 3. All metrics are stratified by the pipeline (Juicer, HiC-Pro, and HiCExplorer) and color-coded by the alignment strategy (green: Direct alignment; blue: Xenome selected alignment of human reads; red: Combined human-mouse genome alignment strategy). (A) Alignment rate representing the proportion of all aligned reads. (B) The proportion of valid interaction pairs as determined by each pipeline. (C) The ratio of cis interacting pairs (i.e., occurring on the same chromosome) vs trans interacting pairs (i.e., between chromosome interactions). (D) The ratio of long- vs. short-interacting Hi-C contacts. Dashed lines correspond to the baseline alignment quality metrics for Hi-C data without mouse reads.
Additional File 7: Figure.Comparison of information extracted from in silico and experimental PDX Hi-C data by the alignment strategy. The same data as shown in Fig. 3 and Additional File 6: Figure grouped by the mouse read removal strategy (green: Juicer; blue: HiC-Pro; red: HiCExplorer). Dashed line: threshold marking the ratios equal to 1.
Additional File 8: Table.The number of TADs and loops detected in each PDX Hi-C sample by each pipeline. Results for the Direct alignment strategy are shown.
Additional File 9: Figure.Overlap between TAD boundaries detected from PDX data processed by Juicer, HiC-Pro, and HiCExplorer. Multi-dimensional scaling (MDS) plots of the (1 − Jaccard overlap) distance matrices are shown. Pipeline-specific data are shown in panels A–C. Panel D shows the overlap between TAD boundaries detected in experimental PDX Hi-C data. Results for the Direct alignment strategy are shown.
Additional File 10: Figure.Overlap between loop boundaries detected from PDX data processed by Juicer, HiC-Pro, and HiCExplorer. Multi-dimensional scaling (MDS) plots of the (1 − Jaccard overlap) distance matrices are shown. Pipeline-specific data are shown in panels A–C. Panel D shows the overlap between TAD boundaries detected in experimental PDX Hi-C data. Results for the Direct alignment strategy are shown.
Additional File 11: Figure.CTCF overlap enrichment odds ratio. The CTCF enrichment odds ratios are shown at TAD (A) and loop (B) boundaries detected from the in silico and experimental PDX Hi-C data. The pipelines (X-axis) are color-coded as follows: green: Juicer; blue: HiC-Pro; red: HiCExplorer. Results for the Direct alignment strategy are shown.
Additional File 12: Figure.CTCF signal enrichment at TAD boundaries. CTCF signal was calculated up to 25 kb upstream and downstream from the TAD boundary (referred to as “center”) and the mean values across all TAD boundaries are plotted for each PDX tested. The pipeline-specific mean signals are color-coded as follows: dark blue: HiC-Pro; light blue: HiCExplorer; yellow: Juicer. Results for the Direct alignment strategy are shown.
Additional File 13: Figure.CTCF signal enrichment at loop boundaries. CTCF signal was calculated up to 25 kb upstream and downstream from the TAD boundary (referred to as “center”) and the mean values across all TAD boundaries are plotted for each PDX tested. The pipeline-specific mean signals are color-coded as follows: dark blue: HiC-Pro; light blue: HiCExplorer; yellow: Juicer. Results for the Direct alignment strategy are shown.
Abbreviations
bp: base pairs; BWA: Burrows-Wheeler Aligner; ChIP-seq: chromatin immunoprecipitation followed by sequencing; CPU: central processing unit; CRB: Chromatin Rinse Buffer; kb: kilobase pairs; KR: Knight and Ruiz; Mb: megabase pairs; PCC: Pearson correlation coefficient; PDX: patient-derived xenograft; QC: quality control; RT: room temperature; SRA: Sequence Read Archive; TAD: topologically associating domain.
Competing interests
The authors declare that they have no competing interests.
Funding
This work was supported in part by the PhRMA Foundation Research Informatics Award and the George and Lavinia Blick Research Scholarship to M.D., the NIH/NCI (1R01CA246182–01A1) grant, and the Susan G. Komen Foundation (CCR19608826) award to J.C.H.
Author contributions
M.G.D. and J.C.H. conceived the project. J.C.H., D.C.B., and J.R. collected samples. N.C.S. created all genomic references. M.G.D., K.M.T., and A.L.O. analyzed the data. M.G.D. and K.M.T. wrote the manuscript. All authors commented on the manuscript.
Supplementary Material
Joanna Achinger-Kawecka -- 11/30/2020 Reviewed
Joanna Achinger-Kawecka -- 1/31/2021 Reviewed
Noam Kaplan -- 12/11/2020 Reviewed
Noam Kaplan -- 2/7/2021 Reviewed
Contributor Information
Mikhail G Dozmorov, Department of Biostatistics, Virginia Commonwealth University, Richmond, VA 23298, USA; Department of Pathology, Virginia Commonwealth University, Richmond, VA 23284, USA.
Katarzyna M Tyc, Department of Biostatistics, Virginia Commonwealth University, Richmond, VA 23298, USA; Department of Pharmacology and Toxicology, Virginia Commonwealth University, Richmond, VA, 23298, USA.
Nathan C Sheffield, Center for Public Health Genomics, University of Virginia, Charlottesville, VA 22908, USA.
David C Boyd, Department of Pathology, Virginia Commonwealth University, Richmond, VA 23284, USA; Integrative Life Sciences Doctoral Program, Virginia Commonwealth University, Richmond, VA 23298, USA.
Amy L Olex, C. Kenneth and Dianne Wright Center for Clinical and Translational Research, Virginia Commonwealth University, Richmond, VA 23298, USA.
Jason Reed, Virginia Commonwealth University, Massey Cancer Center, Richmond, VA, 23298, USA; Department of Physics, Virginia Commonwealth University, Richmond, VA 23220, USA.
J Chuck Harrell, Department of Pathology, Virginia Commonwealth University, Richmond, VA 23284, USA.
References
- 1. Bruna A, Rueda OM, Greenwood W, et al. A biobank of breast cancer explants with preserved intra-tumor heterogeneity to screen anticancer compounds. Cell. 2016;167(1):260–74.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Izumchenko E, Paz K, Ciznadija D, et al. Patient-derived xenografts effectively capture responses to oncology therapy in a heterogeneous cohort of patients with solid tumors. Ann Oncol. 2017;28(10):2595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. DeRose YS, Wang G, Lin Y-C, et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat Med. 2011;17(11):1514–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Turner TH, Alzubi MA, Sohal SS, et al. Characterizing the efficacy of cancer therapeutics in patient-derived xenograft models of metastatic breast cancer. Breast Cancer Res Treat. 2018;170(2):221–34. [DOI] [PubMed] [Google Scholar]
- 5. Alzubi MA, Turner TH, Olex AL, et al. Separation of breast cancer and organ microenvironment transcriptomes in metastases. Breast Cancer Res. 2019;21(1):36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Girotti MR, Gremel G, Lee R, et al. Application of sequencing, liquid biopsies, and patient-derived xenografts for personalized medicine in melanoma. Cancer Discov. 2016;6(3):286–99. [DOI] [PubMed] [Google Scholar]
- 7. Li S, Shen D, Shao J, et al. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Rep. 2013;4(6):1116–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rossello FJ, Tothill RW, Britt K, et al. Next-generation sequence analysis of cancer xenograft models. PLoS One. 2013;8:e74432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lin M-T, Tseng L-H, Kamiyama H, et al. Quantifying the relative amount of mouse and human DNA in cancer xenografts using species-specific variation in gene length. BioTechniques. 2010;48(3):211–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Makałowski W, Zhang J, Boguski MS. Comparative analysis of 1196 orthologous mouse and human full-length mRNA and protein sequences. Genome Res. 1996;6(9):846–57. [DOI] [PubMed] [Google Scholar]
- 11. Conway T, Wazny J, Bromage A, et al. Xenome–a tool for classifying reads from xenograft samples. Bioinformatics. 2012;28(12):i172–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Woo XY, Srivastava A, Graber JH, et al. Genomic data analysis workflows for tumors from patient-derived xenografts (PDXs): challenges and guidelines. BMC Med Genomics. 2019;12:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ahdesmäki MJ, Gray SR, Johnson JH, et al. Disambiguate: An open-source application for disambiguating two species in next generation sequencing data from grafted samples. F1000Res. 2016;5:2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Khandelwal G, Girotti MR, Smowton C, et al. Next-generation sequencing analysis and algorithms for PDX and CDX models. Mol Cancer Res. 2017;15(8):1012–6. [DOI] [PubMed] [Google Scholar]
- 15. Rusch M, Ding L, Arunachalam S, et al. XenoCP: Cloud-based BAM cleansing tool for RNA and DNA from xenograft. bioRxiv. 2020; doi: 10.1101/843250. [DOI] [Google Scholar]
- 16. Callari M, Batra AS, Batra RN, et al. Computational approach to discriminate human and mouse sequences in patient-derived tumour xenografts. BMC Genomics. 2018;19:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tso KY, Lee SD, Lo KW, et al. Are special read alignment strategies necessary and cost-effective when handling sequencing reads from patient-derived tumor xenografts?. BMC Genomics. 2014;15:1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lieberman-Aiden E, Berkum NL, Williams L, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326(5950):289–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rickman DS, Soong TD, Moss B, et al. Oncogene-mediated alterations in chromatin conformation. Proc Natl Acad Sci U S A. 2012;109(23):9083–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hnisz D, Weintraub AS, Day DS, et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science. 2016;351(6280):1454–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Valton A-L, Dekker J. TAD disruption as oncogenic driver. Curr Opin Genet Dev. 2016;36:34–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fritz AJ, Ghule PN, Boyd JR, et al. Intranuclear and higher-order chromatin organization of the major histone gene cluster in breast cancer. J Cell Physiol. 2018;233(2):1278–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Johnston MJ, Nikolic A, Ninkovic N, et al. High-resolution structural genomics reveals new therapeutic vulnerabilities in glioblastoma. Genome Res. 2019;29(8):1211–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kantidze OL, Gurova KV, Studitsky VM, et al. The 3D genome as a target for anticancer therapy. Trends Mol Med. 2020;26(2):141–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lajoie BR, Dekker J, Kaplan N. The hitchhiker's guide to Hi-C analysis: Practical guidelines. Methods. 2015;72:65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zheng Y, Ay F, Keles S. Generative modeling of multi-mapping reads with mHi-C advances analysis of Hi-C studies. Elife. 2019;8, doi: 10.7554/eLife.38070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rao SSP, Huntley MH, Durand NC, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159(7):1665–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pal K, Forcato M, Ferrari F. Hi-C analysis: From data generation to integration. Biophys Rev. 2019;11(1):67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Forcato M, Nicoletti C, Pal K, et al. Comparison of computational methods for Hi-C data analysis. Nat Methods. 2017;14(7):679–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Durand NC, Shamim MS, Machol I, et al. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 2016;3(1):95–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Servant N, Varoquaux N, Lajoie BR, et al. HiC-Pro: An optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015;16:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ramirez F, Bhardwaj V, Arrigoni L, et al. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat Commun. 2018;9:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stolarczyk M, Reuter VP, Smith JP, et al. Refgenie: A reference genome resource manager. Gigascience. 2020;9(2), doi: 10.1093/gigascience/giz149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. ENCODE project. Data Production and Processing Standard of the Hi-C Mapping Center.https://www.encodeproject.org/documents/75926e4b-77aa-4959-8ca7-87efcba39d79/@@download/attachment/comp_doc_7july2018_final.pdf. [Google Scholar]
- 35. Yang D, Jang I, Choi J, et al. 3DIV: A 3D-genome Interaction Viewer and database. Nucleic Acids Res. 2018;46(D1):D52–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chakraborty A, Ay F. Identification of copy number variations and translocations in cancer cells from Hi-C data. Bioinformatics. 2018;34(2):338–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetJ. 2011;17(1):10–2. [Google Scholar]
- 38. Gossamer. https://githubcom/data61/gossamer. Accessed 11 December 2019. [Google Scholar]
- 39. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6)841–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Arima restriction enzyme files. Accessed 9 April 2020.ftp://ftp-arimagenomics.sdsc.edu/pub/HiCPro_GENOME_FRAGMENT_FILES. [Google Scholar]
- 43. Dozmorov MG, Cara LR, Giles CB, et al. GenomeRunner: Automating genome exploration. Bioinformatics. 2012;28(3):419–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dozmorov MG, Cara LR, Giles CB, et al. GenomeRunner web server: Regulatory similarity and differences define the functional impact of SNP sets. Bioinformatics. 2016;32(15):2256–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. PDX-HiC project homepage. https://github.com/dozmorovlab/PDX-HiC_processingScripts. [Google Scholar]
- 46. Dozmorov M, Tyc KM, Sheffield NC, et al. Supporting data for “Hi-C sequencing of patient-derived xenografts: Analysis guidelines.”. GigaScience Database. 2021. 10.5524/100870. [DOI] [PMC free article] [PubMed]
- 47.Experiment summary for ENCSR000DZN. https://www.encodeproject.org/experiments/ENCSR000DZN/. 1st Feb 2021. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Dozmorov M, Tyc KM, Sheffield NC, et al. Supporting data for “Hi-C sequencing of patient-derived xenografts: Analysis guidelines.”. GigaScience Database. 2021. 10.5524/100870. [DOI] [PMC free article] [PubMed]
Supplementary Materials
Joanna Achinger-Kawecka -- 11/30/2020 Reviewed
Joanna Achinger-Kawecka -- 1/31/2021 Reviewed
Noam Kaplan -- 12/11/2020 Reviewed
Noam Kaplan -- 2/7/2021 Reviewed
Data Availability Statement
Accession numbers to download the publicly available Hi-C data used in this study are listed in Table S1. Experimental PDX Hi-C data generated in this study are available at the SRA via bioproject number PRJNA668904. All codes necessary to reproduce the analyses are available at the project home page [45]. Snapshots of our code and other supporting data are openly available in the GigaScience repository, GigaDB [46].