Abstract
Background
The large airway epithelial barrier provides one of the first lines of defense against respiratory viruses, including SARS-CoV-2 that causes COVID-19. Substantial inter-individual variability in individual disease courses is hypothesized to be partially mediated by the differential regulation of the genes that interact with the SARS-CoV-2 virus or are involved in the subsequent host response. Here, we comprehensively investigated non-genetic and genetic factors influencing COVID-19-relevant bronchial epithelial gene expression.
Methods
We analyzed RNA-sequencing data from bronchial epithelial brushings obtained from uninfected individuals. We related ACE2 gene expression to host and environmental factors in the SPIROMICS cohort of smokers with and without chronic obstructive pulmonary disease (COPD) and replicated these associations in two asthma cohorts, SARP and MAST. To identify airway biology beyond ACE2 binding that may contribute to increased susceptibility, we used gene set enrichment analyses to determine if gene expression changes indicative of a suppressed airway immune response observed early in SARS-CoV-2 infection are also observed in association with host factors. To identify host genetic variants affecting COVID-19 susceptibility in SPIROMICS, we performed expression quantitative trait (eQTL) mapping and investigated the phenotypic associations of the eQTL variants.
Results
We found that ACE2 expression was higher in relation to active smoking, obesity, and hypertension that are known risk factors of COVID-19 severity, while an association with interferon-related inflammation was driven by the truncated, non-binding ACE2 isoform. We discovered that expression patterns of a suppressed airway immune response to early SARS-CoV-2 infection, compared to other viruses, are similar to patterns associated with obesity, hypertension, and cardiovascular disease, which may thus contribute to a COVID-19-susceptible airway environment. eQTL mapping identified regulatory variants for genes implicated in COVID-19, some of which had pheWAS evidence for their potential role in respiratory infections.
Conclusions
These data provide evidence that clinically relevant variation in the expression of COVID-19-related genes is associated with host factors, environmental exposures, and likely host genetic variation.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13073-021-00866-2.
Keywords: COVID-19, SARS-CoV-2, ACE2, eQTL, Bronchial epithelium
Background
Coronavirus disease 2019 (COVID-19), the clinical syndrome caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus, has led to a global crisis. As a respiratory virus, SARS-CoV-2 is hypothesized to gain entry into humans via the airway epithelium, where it initiates a host response that leads to the subsequent clinical syndrome. Despite an immense global burden of disease, the manifestations of SARS-CoV-2 infection vary enormously, from asymptomatic infection to progressive acute respiratory failure and death. The viral or host features that determine the course of disease in each individual are poorly understood. Multiple clinical risk factors for severe COVID-19 have been identified, including older age, male sex, African American race, smoking, and comorbidities such as hypertension, obesity, diabetes, cardiovascular disease, and chronic airway diseases [1–5], as well as host genetics [5–8].
The expression levels of genes that interact with the SARS-CoV-2 virus or are involved in the subsequent host response are hypothesized to be an important host factor that could partially underlie the substantial inter-individual variability in COVID-19 susceptibility and progression [9–11]. To this end, we investigate genetic and non-genetic factors influencing the expression of human genes that have been implicated in COVID-19 (study design in Fig. 1). We analyze RNA-sequencing (RNA-seq) data from bronchial brushing samples obtained from the SPIROMICS cohort (n = 163) [12], notable for the high burden of COVID-19-relevant comorbidities and rich phenotype and whole genome sequencing (WGS) data from the TOPMed Project [13]. For replication, we use two asthma RNA-seq data sets, SARP (n = 156) and MAST (n = 35) as well as expression quantitative trait loci (eQTL) data from GTEx [14]. Our analysis provides insights of the contribution of host factors and host genetics in the expression of COVID-19-related genes in the large airway epithelium for understanding inter-individual variation of COVID-19.
Fig. 1.
Study design. Graphical illustration of analyses (gray boxes) carried out to study non-genetic and genetic factors affecting the expression of COVID-19-related genes in bronchial epithelium. Input data sets for these analyses are denoted with a green box (WGS and RNA-seq) and external data sets or data resources used in these analyses are denoted with a blue box
Methods
Study population
SubPopulations and InteRmediate Outcome Measures In COPD Study (SPIROMICS)
SPIROMICS is a multi-site prospective cohort study in which the main objective is to identify subpopulations of chronic obstructive pulmonary disease (COPD) as well as markers of disease severity to enable targeted treatment and disease modification. Data were obtained from participants who underwent research bronchoscopy within SPIROMICS between February 1, 2012, and May 31, 2016. Participants ages 40–80 were enrolled across four strata (never smokers, smokers without COPD, mild/moderate COPD, and severe COPD). Full SPIROMICS study details including inclusion and exclusion criteria have been previously published [12]. Participants enrolled in SPIROMICS who consented to a research bronchoscopy and met all local requirements (e.g., any laboratory tests that are required by institutional policy to be administered prior to a bronchoscopy) were deemed eligible. Additional exclusion criteria for the SPIROMICS bronchoscopy sub-study [15] included history of cardiac disease or other comorbid condition severe enough to significantly increase risks based on investigator discretion, requirement of supplementary oxygen at rest based on arterial oxygen pressure less than 60 mmHg or arterial oxygen saturation less than 88%, severe lung function impairment defined as post-bronchodilator forced expiratory volume in 1 s (FEV1) less than 30% predicted, and use of anticoagulation or antiplatelet therapies.
Severe Asthma Research Program (SARP)
SARP is a prospective multi-center cohort study with a primary goal of improving the mechanistic and clinical understanding of severe asthma [16]. Adult and pediatric patients with and without asthma were recruited to the SARP III cohort between November 1, 2012, and October 1, 2014, by seven clinical research centers in the USA. The SARP protocol is an ongoing, six-visit, 3-year, longitudinal cohort study in which 60% of participants have severe asthma as defined by the European Respiratory Society/American Thoracic Society (ERS/ATS) criteria [17]. A subset of participants underwent research bronchoscopy. Exclusion criteria included history of smoking (> 5 pack year smoking history), co-existing lung disease, and uncontrolled comorbidities. All healthy control subjects had to have no history of asthma and normal lung function and methacholine bronchoprovocation testing. Participants with asthma had to meet ERS/ATS criteria for asthma (bronchodilator response to albuterol or positive methacholine bronchoprovocation test). Asthma had to be clinically stable at the time of bronchoscopy.
Mechanisms of ASThma study (MAST)
MAST is a single-center clinical study with a primary objective of understanding asthma biology through detailed analyses of airway secretions and tissues [18]. Mild steroid-naive asthmatics and healthy controls underwent research bronchoscopy between April 2007 and December 2011. All healthy control subjects had to have no history of asthma or allergies. Participants with asthma had to have a positive methacholine bronchoprovocation test and could not have used steroids in 6 weeks prior to enrollment. Additional exclusion criteria included respiratory infection within 4 weeks of enrollment and pregnancy.
Whole genome sequencing data
Trans-Omics for Precision Medicine (TOPMed) Project [13] data freeze 9 consist of whole genome sequences of 160,974 samples with at least 15x average coverage, including 2710 individuals from the SPIROMICS study. We obtained unphased genotypes for all individuals from the SPIROMICS study at sites with at least 10x sequencing depth (minDP10 call set) aligned to the human reference genome build GRCh38. Details regarding the DNA sample handling, quality control, library construction, clustering and sequencing, read processing, and sequence data quality control are described on the TOPMed website (https://www.nhlbiwgs.org/genetic). Variants passing all quality control (QC) filters were retained.
Derivation of airway epithelial transcriptomic data in SPIROMICS, SARP, and MAST
Cytological brushings of the airway epithelium were obtained from lower lobe bronchi at the segmental or subsegmental carina. RNA was isolated with miRNeasy extraction kits (Qiagen Inc., Valencia, CA). RNA quantity and quality were evaluated using a NanoDrop Spectrophotometer (Thermo Fisher Scientific, Wilmington, DE) and Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA), respectively. Library preparation with multiplexing was performed using Illumina TruSeq Stranded Total RNA with Ribo-zero GOLD kit (SPIROMICS, SARP) or Human/Mouse/Rat kit (MAST) per manufacturer’s protocol. Samples were sequenced using one-hundred-fifty base-pair (SPIROMICS) or one-hundred base-pair (SARP, MAST) paired end reads via the Illumina HiSeq platform at the UCSF Sandler Genomics core. FASTQ files were quality filtered and aligned to the Ensembl GRCh38 genome build using STAR [19]. Read counts were normalized using the regularized logarithm transformation function of the DESeq2 package in R [20] and batch corrected using the Combat function in the SVA package in R [21]. Outlying samples with low quality (low raw read counts, high percentage of reads mapped to multiple loci, high percentage of unmapped reads) were identified by hierarchical clustering and principal component analyses and excluded from the final data sets.
Differential expression analysis of ACE2 in relation to host/environmental factors
Linear regression models were fitted to evaluate associations between ACE2 expression (based on normalized count) and clinical variables in the SPIROMICS, SARP, and MAST cohorts with and without adjustments for covariates (see Additional file 1 for additional details).
Differential exon usage
Following alignment, we indexed and sliced the SPIROMICS BAM files to include 51.6 kb of the ACE2 genomic region (chrX:15,556,393-15,608,016 in the hg38 genome build) using samtools [22]. GTF files were manually curated to include the three exons that contribute to differential isoform expression of ACE2 [23]. Of them, the truncated ACE2 transcript (dACE2) that does not bind the SARS-CoV-2 virus but is associated with an interferon-stimulated gene response in experimental models originates from Exon 1c. The exons were counted using the ASpli package in R [24]. After correcting for overall gene counts and differences in sequence depth, linear models adjusting for batch were used to analyze differences in exon usage in association with interferon-stimulated gene signature and clinical covariates. Interpretation of differential exon usage requires consideration of the necessary adjustment for variation in total transcript count. Thus, if overall ACE2 expression is decreased in association with an outcome, a differential increase in one exon adjusts the expression of that isoform away from the overall negative association, but does not necessarily mean that the isoform is not negatively associated with the outcome to a lesser extent. Further details are provided in Additional file 1.
Gene set enrichment analysis of expression changes induced by COVID-19
We built COVID-19-relevant gene sets from publicly available differential gene expression data from participants who underwent nasal/oropharyngeal swab sampling at the time of acute respiratory illness for COVID-19 diagnosis (94 participants with COVID-19, 41 with other viral illness, 103 with no virus identified, viruses identified by metagenomic sequencing analysis) using Supplementary File 1 from Mick et al. [25]. Biological pathway gene sets were built by inputting the genes differentially downregulated between SARS-CoV-2 infection and other viral illness (P < 0.05) into the Ingenuity Pathway Analysis canonical pathway function. Gene set enrichment analyses were then performed using FGSEA [26] and the CAMERA function [27] in limma against gene lists ranked by their log fold change differential expression in association with comorbid clinical risk factors. Barcode plots were made using CAMERA. Findings were considered significant at P < 0.05 and false discovery rate (FDR) < 0.05 if multiple corrections were necessary. Additional details are provided in Additional file 1.
COVID-19-related genes
We selected 514 candidate genes implicated in COVID-19 from six different sources: Hoffmann et al. [28], Gordon et al. [29], Blanco-Melo et al. [30], COVID-19 Cell Atlas (https://www.covid19cellatlas.org/), Gassen et al. [31], and Wang et al. [32]. Of them, 496 genes were expressed in bronchial epithelium in the SPIROMICS cohort. Further details are provided in Additional file 1.
Expression quantitative trait mapping
Expression quantitative trait locus (eQTL) mapping was performed in 144 unrelated individuals from the SPIROMICS bronchoscopy sub-study with WGS genotype data from TOPMed and gene expression from bronchial epithelium profiled with RNA-seq following the analysis pipeline from the Genotype-Tissue Expression (GTEx) Consortium [14]. In short, gene expression data was normalized as follows: (1) read counts were normalized between samples using TMM [33] with edgeR [34], (2) genes with TPM ≥ 0.1 and unnormalized read count ≥6 in at least 20% of samples were retained, and (3) expression values were transformed using rank-based inverse normal transformation across samples.
Cis-eQTL mapping was performed using tensorQTL [35] across 22,738 genes and 6,605,907 variants with minor allele frequency (MAF) ≥ 0.05 and variant call rate ≥ 0.9 within ± 1 Mb from the transcription start site (TSS) of the gene. As covariates in the model, we used 15 PEER factors [36], 4 genotype principal components and sex imputed from genotype data. To control for multiple testing, 10,000 permutations were performed and FDR < 0.05 was used to identify genes with statistically significant eQTLs (eGenes). Lead cis-eQTL effect size was quantified as allelic fold change (aFC) [37], ratio of expression of the haplotype carrying the alternative allele to expression of the haplotype carrying the reference allele of an eQTL.
Additionally, cell type interacting eQTLs (ieQTLs) were mapped using an interaction model: p ~ g + i + g × i + C, where p is the expression vector (normalized as described above), g is the genotype vector, i is the normalized cell type enrichment score from xCell [38], g × i is the interaction term, and C is the covariates matrix as used in standard eQTL mapping. Only variants with MAF > 0.1 in the samples belonging to the top and bottom halves of the distribution of cell type abundance were included in the analyses. Multiple testing correction was done at the gene level using eigenMT [39], followed by Benjamini-Hochberg procedure across genes at FDR 5%. Additional details are provided in Additional file 1.
Replication of cis-eQTLs and pathway analysis
We performed replication of cis-eQTLs (gene-variant pairs) found from bronchial epithelium in 49 tissues from the GTEx project v8 release [14] based on the proportion of true positives [40], π1, and concordance rate, the proportion of gene-variant pairs with the same allelic direction for variants with nominal P value < 1 × 10−4 in the given GTEx tissue. Then, we analyzed the replication and concordance measure as a function of sample size and median cell type enrichment scores for seven cell types [41]. Further details are provided in Additional file 1.
Also, we performed gene-level lookup in GTEx v8 and eQTLGen Consortium [42] and used the functional profiling webtool g:GOSt from g:Profiler [43] to perform pathway analysis of the 492 significant eGenes in SPIROMICS not tested in GTEx v8 Lung.
pheWAS of lead COVID-19 cis-eQTLs in SPIROMICS and querying PhenoScanner
We performed a phenome-wide association study (pheWAS) in 1980 non-Hispanic White and 696 individuals from other ethnic and racial groups from SPIROMICS for the 108 lead cis-eQTLs to evaluate for phenotypic associations with spirometric measures, cell count differentials, and other variables. PheWAS regression-based models were performed using PLINK 2/0 adjusting for principal components of ancestry, sex, body mass index (BMI), age, and smoking pack-years. Significance threshold was set for the number of eQTLs tested across phenotypes (P < 4.63 × 10−4).
Additionally, PhenoScanner v2 [44, 45] was used to lookup phenotype associations of the cis-eQTL variants from large-scale genome-wide association studies (GWAS) with association P value < 10−5. The phenoscanner R package (https://github.com/phenoscanner/phenoscanner) was used to perform the queries. Additional details are provided in Additional file 1.
Colocalization analysis
To assess evidence for shared causal variant of a cis-eQTL and a GWAS trait, we used the Bayesian statistical test for colocalization, coloc [46], with conditioning and masking to overcome one single causal variant assumption. Coloc was run on a 500-kb region centered on the lead cis-eQTL with priors set to p1 = 10−4, p2 = 10−4, p3 = 5 × 10−6. We used the coloc.signals() function with mode = iterative, method = mask for GWAS traits with linkage disequilibrium (LD) data from the 1000 Genomes Project, and method = single for the eQTLs. Posterior probability for colocalization (PP4) > 0.5 was used as evidence for colocalization (see Additional file 1 for further details).
Results
Smoking, obesity, and hypertension are associated with increased airway epithelial expression of functional ACE2 isoforms
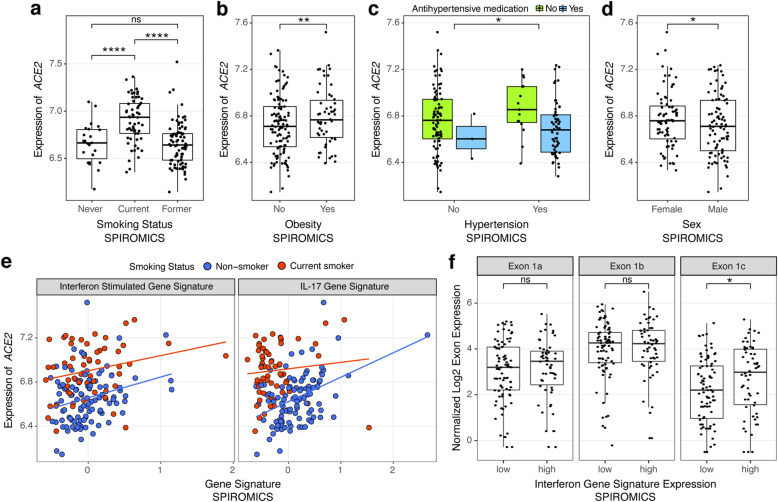
We first analyzed expression levels of ACE2, the receptor of the SARS-CoV-2 Spike protein that is the key host gene for viral entry [28, 47], in relation to non-genetic host factors (Additional file 2: Table S1). Corroborating previous reports [11, 48–50], we found that current smoking, when compared to non-smoking, had the largest overall effect on ACE2 expression of any phenotypic feature studied in SPIROMICS, before and after adjustments for covariates (log2 fold change (FC) = 0.30 ± 0.06, P = 1.7 × 10−7, Fig. 2a). This effect was absent in former smokers. In similarly adjusted models, we found no association between ACE2 levels and COPD (Additional file 3: Figure S1a), nor with asthma in MAST [50] (Additional file 3: Figure S1c). In SARP, ACE2 levels were slightly lower in asthmatics compared to healthy controls (Additional file 3: Figure S1b), which was largely driven by decreased expression of ACE2 only in asthmatics on oral steroids (Additional file 3: Figure S1d). African American race was associated with increased ACE2 expression in both SPIROMICS and SARP, but no association after adjusting for covariates suggests that this was due to a higher prevalence of comorbid conditions (Additional file 3: Figure S1e-f). However, ACE2 expression was significantly higher across data sets in association with two relevant comorbidities, obesity and hypertension (Fig. 2b-c, Additional file 3: Figure S2a-e, Additional file 3: Figure S3a-b). Of note, we further found that use of anti-hypertensives in SPIROMICS attenuates the association between ACE2 and hypertension towards levels seen in non-hypertensive participants (Fig. 2c). When stratified by anti-hypertensive class, angiotensin receptor blockers (ARBs) and diuretics, but not ACE inhibitors or calcium channel blockers, were associated with lower ACE2 levels, partially dependent on smoking status (Additional file 3: Figure S3c). Counterintuitively, modest decreases in ACE2 expression were seen in SPIROMICS in association with age (log2 FC = − 0.064 ± 0.02, P = 0.005 for every 10-year age increase, Additional file 3: Figure S4a) and male sex (log2 FC = − 0.076 ± 0.035, P = 0.03, Fig. 2d) before and after adjustments, although similar associations were not seen in SARP or MAST. Although there were no significant differences in the above reported outcomes between males and females in SPIROMICS, former smokers were older (9.1 ± 1.3 years compared to current smokers, P = 3.19 × 10−10) as were participants with hypertension (4.6 ± 1.4 years, P = 0.002, Additional file 3: Figure S5). Sex and age were, however, both adjusted for in our analyses.
Fig. 2.
ACE2 gene expression associations in SPIROMICS. a–d Box plots showing that ACE2 log2 gene expression (x-axis) was increased in association with current but not former smoking as compared to never smokers (a), obesity (b, validated in the MAST and SARP cohorts, Additional file 3: Figure S2a-b), hypertension (c, adjustments include anti-hypertensive treatment, validated in SARP, Additional file 3: Figure S3a, data not collected in MAST), and female sex (d, not replicated in either MAST or SARP, Additional file 2: Table S1A). e Scatterplots showing that ACE2 gene expression was increased in association with higher levels of our previously validated gene signatures of the airway epithelial response to interferon (left panel, replicated in SARP) and to IL-17 inflammation (right panel, replicated in MAST and SARP) after adjusting for smoking status (Additional file 2: Table S1B). f Box plots showing that ACE2 Exon 1c, which contributes to the truncated ACE2 transcript was differentially increased in association with our interferon signature while Exons 1a and 1b that contribute to the full length ACE2 transcript were not. P values indicated by: **** < 0.0001, *** < 0.001, ** < 0.01, * < 0.05, ns = not significant in linear models adjusted for covariates. In a–d and f, the boxes denote the interquartile range, the center line denotes the median, and whiskers denote the interquartile range × 1.5
As chronic airway inflammation, prevalent but heterogeneous in the airway diseases studied in the included cohorts, can influence gene expression and the host response to infections, we next studied how stereotypic adaptive airway immune responses affect ACE2 expression. We used our previously validated gene expression signatures to quantify type 2-, interferon-, and IL-17-associated inflammation [18, 51, 52]. We found that ACE2 expression was associated with increased interferon-related inflammation, as previously reported [9], as well as IL-17-related but not type 2 inflammation across data sets (Fig. 2e). Corroborating the association with IL-17 inflammation, genes highly co-expressed with ACE2 expression included genes in our IL-17 signature across data sets (Additional file 2: Table S2).
Recent reports suggested that ACE2 induction by interferon stimulation may be explained by expression of a truncated ACE2 isoform (dACE2, initiated from exon 1c instead of 1a/b) that does not bind the SARS-CoV-2 spike protein [23, 53]. We first corroborated this finding, showing that our interferon-stimulated gene signature is associated with increased exon 1c but not exons 1a or 1b usage (Fig. 2f). We also identified an increase in exon 1a usage with age. This result suggests that although overall ACE2 expression is decreased in association with age, the full length transcript initiated from exon 1a is not decreased to the same extent or is even potentially increased with age (Additional file 3: Figure S4b). Importantly, differential exon 1c usage was not associated with any other clinical/biological outcomes of interest, suggesting that the full length transcript is responsible for the observed associations.
These results overall indicate that smoking, obesity, and hypertension affect airway epithelial expression of functional ACE2 isoforms, as previously shown for smoking [11, 48–50]. The ACE2 association with interferon-related inflammation appears to be explained by the truncated version of ACE2 [23, 53]. Together, these findings suggest that smoking, obesity, and hypertension may contribute to COVID-19 severity through an association with increased ACE2 expression, while other risk factors such as male sex and airway disease likely contribute via other mechanisms, corroborating recent evidence on sex differences in the immune response to COVID-19 [54].
Obesity, hypertension, and cardiovascular disease are associated with a relative COVID-19-relevant immunosuppression at the airway epithelium
As the host’s ability to mount an appropriate response to respiratory viruses may alter susceptibility to severe infection, we next performed gene set enrichment analyses (GSEA) to determine whether clinical risk factors are associated with similar airway gene expression patterns indicative of a diminished immune response that we recently identified early in COVID-19 by nasal/oropharyngeal swab [25]. As we previously reported, the genes differentially expressed in association with SARS-CoV-2 infection compared to other viruses at diagnosis indicate a diminished innate and adaptive immune response that may allow for unabated viral infection and account for the long pre-symptomatic period associated with COVID-19 [25]. We hypothesized that clinical risk factors uniquely associated with COVID-19 severity (e.g., cardiovascular disease, hypertension) could predispose patients to develop more severe disease by contributing to this relative immunosuppression. We derived gene sets from our previously published RNA-seq data collected by nasal/oropharyngeal swab from patients at diagnosis of acute respiratory illness; 94 had COVID-19, 41 had other viral illness, and 103 had no virus identified by metagenomic sequencing analysis [25]. First, we generated gene sets derived from the 100 genes most up- and downregulated in association with infection type to use to determine if there were global similarities in gene expression changes across data sets. For pathway analyses, we then generated COVID-19-relevant gene sets specific to particular canonical pathways by inputting significantly differentially expressed genes (FDR < 0.05) between SARS-CoV-2 infection and other viral respiratory illness into the Ingenuity Pathway Analysis (IPA) canonical pathway function (Additional file 2: Table S3). GSEA was then performed using FGSEA [26] in which these gene sets were tested against gene lists ranked by their log fold change differential expression in association with comorbid clinical risk factors.
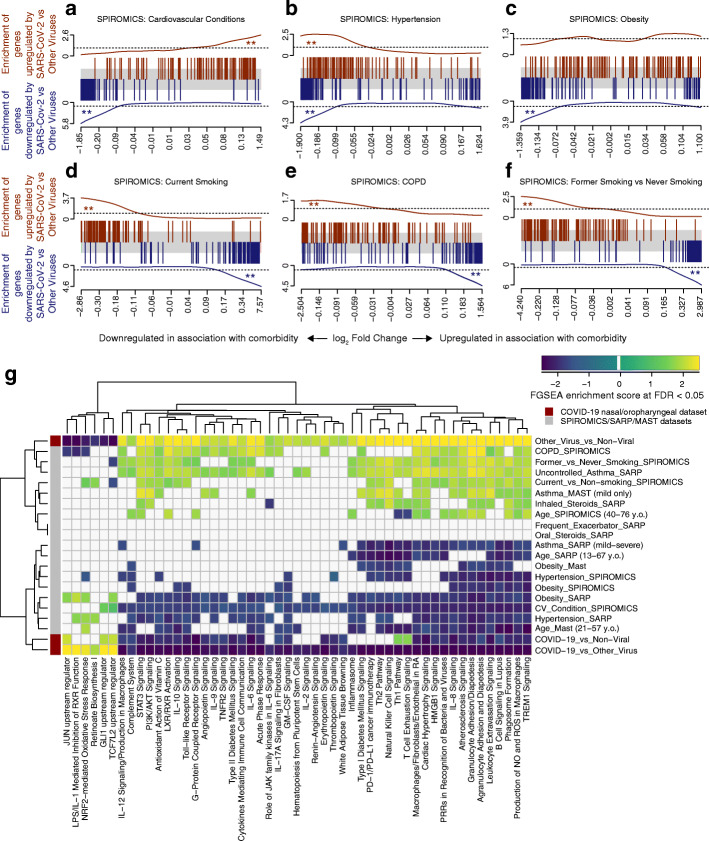
We found that the genes most downregulated in association with SARS-CoV-2 infection as compared to other viruses were significantly enriched amongst genes downregulated in association with obesity, hypertension, and cardiovascular disease in SPIROMICS (Fig. 3a–c). Findings for obesity were replicated in SARP and MAST and for hypertension in SARP (Additional file 3: Figure S6a-c, hypertension data not collected in MAST, cardiovascular disease data not collected in SARP or MAST). Conversely, genes upregulated in other viral infections (or conversely, downregulated by SARS-CoV-2) were upregulated in inflammatory airway conditions (current and former smokers, COPD) (Fig. 3d-f). Aging was associated with an enrichment in genes downregulated by SARS-CoV-2 infection only in MAST while genes upregulated with SARS-CoV-2 infection were enriched with increasing age across the data sets (Additional file 3: Figure S6d-f). Our results demonstrate a sharp contrast between SARS-CoV-2 and other viral infections, which often trigger airway disease exacerbations by potentiating the chronic airway inflammation associated with these diseases and smoking exposure. We found this same pattern in association with asthma in MAST but not when considering asthma overall in SARP, potentially due to heterogeneity of its asthma subjects. When considering just asthmatics with uncontrolled symptoms or those on inhaled compared to no steroids (a marker of severity), we did find this same enrichment of genes up and downregulated in association with non-COVID viral infections (pathway enrichment shown in Fig. 3g).
Fig. 3.
COVID-19-related gene set enrichment analyses in association with comorbidities. a–f Barcode plots in which the vertical lines represent the 100 genes most upregulated (red) or downregulated (blue) in nasal/oropharyngeal swab samples obtained from COVID-19 patients as compared to other viruses at the time of diagnosis of an acute upper respiratory infection. These gene sets are plotted against log fold gene expression changes arranged from most downregulated to most upregulated with that comorbidity (horizontal gray bar). Lines above (red) and below (blue) the bar represent the running sum statistic with a significant finding indicated when the line crosses the dashed line at either end of the plot. Genes downregulated by SARS-CoV-2 infection compared to other viruses were significantly enriched amongst genes downregulated in association with cardiovascular conditions overall (a), hypertension (b), and obesity (c), while in current (d) and former smoking (f) and in COPD (e), these downregulated genes in COVID-19 were enriched amongst upregulated genes in association with comorbidity. ** indicates FDR < 0.05. g COVID-19-related pathway gene sets were generated from an IPA analysis of the genes downregulated by SARS-CoV-2 infection compared to other viruses. Gene set enrichment scores for gene sets enriched at FDR < 0.05 (columns) are shown in the heatmap plotted against comorbidities (rows) with gene sets enriched amongst downregulated and upregulated genes indicated in blue and yellow, respectively. All pathways not enriched at FDR < 0.05 were shrunk to zero (white). Euclidean distance with average linkage was used for clustering
We used pathway gene set enrichment to determine the potential biological significance of these findings. We found across data sets that pathway gene sets derived from genes downregulated by SARS-CoV-2 infection as compared to other viruses were also enriched amongst genes downregulated in association with obesity, hypertension, cardiovascular disease, and aging (FDR < 0.05, Fig. 3g, Additional file 2: Table S4). Enriched downregulated pathways included those related to pro-inflammatory cytokines such as IL-6 and IL-17 as well as macrophage and granulocyte activation. Furthermore, pathways related to cardiovascular and metabolic disease signaling such as atherosclerosis and diabetes signaling were also enriched. We confirmed the enriched findings by separately performing IPA canonical pathway analyses on the genes differentially expressed (P < 0.05) in association with these comorbidities, finding similar results in these global/unsupervised analyses (Additional file 2: Table S5). Conversely, pro-inflammatory airway conditions such as smoking and COPD led to opposite effects. These findings suggest that obesity, hypertension, cardiovascular disease, and age are associated with a relative COVID-19-relevant immunosuppression at the airway epithelium, which, by stunting early anti-viral host responses, could contribute to increased susceptibility to SARS-CoV-2 infection and disease severity.
Host genetics has a biologically meaningful effect on the airway epithelial expression of many COVID-19-related genes
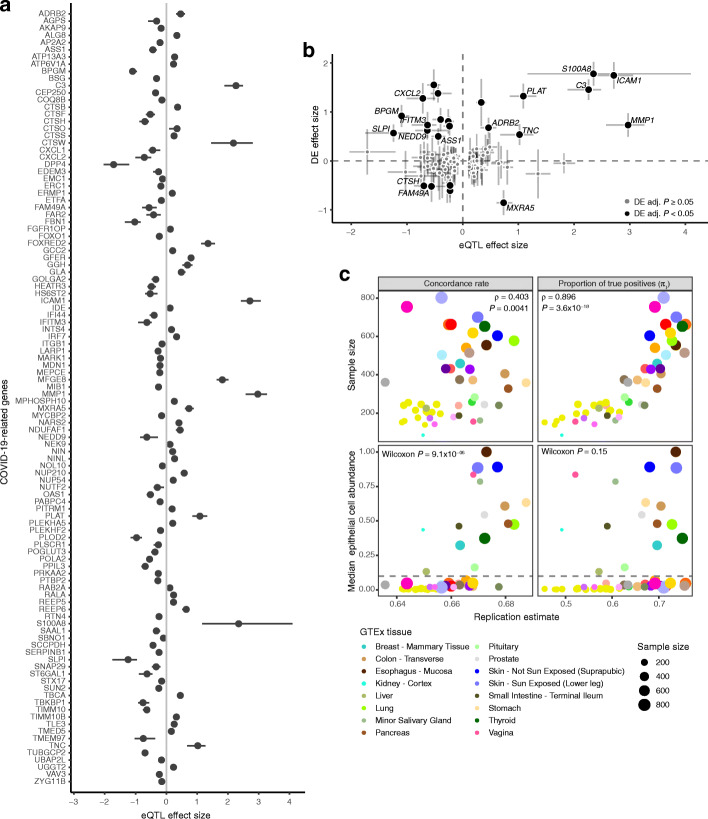
In order to map host genetic variants, we focused on 496 genes implicated in SARS-CoV-2 infection (Additional file 2: Table S6): ACE2 and TMPRSS2, key genes for viral entry [28]; CTSL, CTSB, and BSG, which may have a role as alternative routes for viral entry [28, 32]; host genes with protein-protein interactions with viral proteins [29]; differentially expressed genes as a response to the infection in cultured airway epithelial cells [30]; genes involved in autophagy that might counteract viral infection [31]; and other high interest genes from the COVID-19 Cell Atlas. Our cis-eQTL mapping in SPIROMICS (n = 144) identified significant (genome-wide FDR < 0.05) genetic regulatory variation for 108 (21.8%) of these COVID-19-related genes (Fig. 4a, Additional file 2: Table S7), with many genes also having significant eQTLs in other tissues in GTEx [14] (Additional file 2: Table S8).
Fig. 4.
Cis-eQTLs in bronchial epithelium. a Effect size measured as allelic fold change (aFC, log2) of the significant cis-eQTLs for COVID-19 candidate genes. Error bars denote 95% bootstrap confidence intervals. b Comparison of the regulatory effects and the effect of SARS-CoV-2 infection on the transcription of COVID-19 candidate genes in normal bronchial epithelial cells from Blanco-Melo et al. [30]. The graph shows regulatory effects as aFC as in a and fold change (log2) of differential expression comparing the infected with mock-treated cells with error bars denoting the 95% confidence interval. Genes with adjusted P value < 0.05 in the differential expression analysis are colored in black, genes with non-significant effect are colored in gray. Highlighted genes have eQTL effect size greater than 50% of the differential expression effect size on the absolute scale. DE—differential expression. c Replication of cis-eQTLs from bronchial epithelium in GTEx v8 using the concordance rate (proportion of gene-variant pairs with the same direction of the effect, left panel) and proportion of true positives (π1, right panel). Upper panel shows the effect of sample size on the replication and concordance measures quantified as Spearman correlation coefficient (ρ). Lower panel shows the replication and concordance measures as the function of epithelial cell enrichment of the tissues measured as median epithelial cell enrichment score from xCell. Gray dashed line denotes median enrichment score > 0.1, which classifies tissues as enriched for epithelial cells. Wilcoxon rank sum test was used to estimate the difference in replication estimates between tissues enriched or not enriched for epithelial cells. The 16 tissues enriched for epithelial cells are outlined in the figure legend, for the full legend see Additional file 3: Figure S9a
Given the sample size, we have good power to discover the vast majority of eQTLs with > 2-fold effect on gene expression [14]. Many of the genes have a substantial genetic effect on gene expression: for example, the MERS receptor DPP4 [55] has a cis-regulatory variant rs6727102 where the alternative allele decreases expression by 3.3-fold (Fig. 4a). In 16 genes, the genetic regulatory effects were > 50% of the magnitude of the differential expression induced by SARS-CoV-2 infection [30] (Fig. 4b). While the key genes ACE2 or TMPRSS2 did not have eQTLs in bronchial epithelium (Additional file 3: Figure S7a-b), as previously reported [50], TMPRSS2 has an eQTL in GTEx lung tissue. This is consistent with the lack of phenome-wide association signals [56] or COVID-19 GWAS association at these loci (round 3 meta-analyses by COVID-19 Host Genetics Initiative [8]), suggesting that genetic regulation of these two genes is unlikely to contribute to potential host genetic effects on COVID-19. Many of the genes analyzed for eQTLs had variation in expression associated to clinical factors and comorbidities, with current smoking associated with the highest number of up-and downregulated genes in association with comorbidity (Additional file 3: Figure S8a-b). Compared to ACE2, the effect of current smoking on the expression of TMPRSS2 was modest (Additional file 3: Figure S7c), and as previously reported [10], expression levels of TMPRSS2 were higher in asthmatic than healthy controls, but not in COPD, and it decreased in association with steroid use (Additional file 3: Figure S7d).
Cis-eQTLs from bronchial epithelium replicated at a high rate in those tissues from the GTEx v8 data set [14] that have a large sample size or high epithelial cell abundance (Fig. 4c, Additional file 3: Figure S9a-b), reflecting similarity in cell type composition manifesting in similarity of regulatory variant activity [14]. However, relative to GTEx lung, our bronchial epithelium eQTLs included genes enriched for sensory perception of chemical stimulus and smell (Additional file 2: Table S9). In total, 143 genes with eQTLs in SPIROMICS were not tested in GTEx nor eQTLGen Consortium [42], since bronchial epithelium is not well represented in previous eQTL catalogs. In addition to standard cis-eQTL mapping, we mapped cell type interacting eQTLs [41] but none were discovered for the COVID-19-related genes.
Regulatory variants for COVID-19-related genes as host risk factors for COVID-19 susceptibility
To study the role of these regulatory variants in COVID-19 risk, we first analyzed eQTLs in the chromosome 3 locus with a significant association with hospitalization due to COVID-19 [8] (meta-analyses round 3) and severe COVID-19 with respiratory failure [5, 7]. We found no significant eQTLs in the bronchial epithelium for any of the six genes in this locus (Additional file 3: Figure S10a), suggesting that this genetic association may be driven by other tissues or cell types with a role in COVID-19. Moreover, these genes were rather lowly expressed in bronchial epithelium (Additional file 3: Figure S10b).
Next, given that COVID-19 GWAS still have limited power, we analyzed how regulatory variants for COVID-19-relevant genes associate to other immune- or respiratory-related phenotypes in large GWAS. Indication of these variants affecting (respiratory) infections would provide hypotheses of variants that might play a role in COVID-19 risk and its comorbidities (Fig. 5a). Thus, we performed a pheWAS analysis by Phenoscanner v2 [44, 45] for the 108 lead cis-eQTLs for COVID19-related genes and diverse set of phenotypes (Additional file 2: Table S10). Furthermore, we used the SPIROMICS phenotype data to study associations for 20 phenotypes (Additional file 2: Table S11). Of these loci, 44 were associated with at least one phenotype (P < 10−5), with expected patterns—best powered GWAS traits having most associations and shared signals for highly correlated traits (Additional file 3: Figure S11).
Fig. 5.
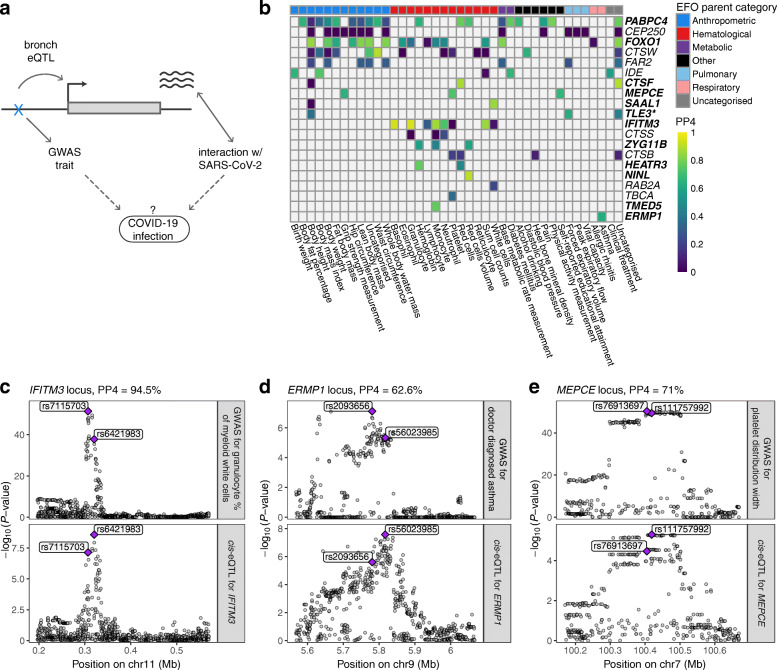
Colocalization analysis of the regulatory variants for COVID-19-related genes. a Illustration of the concept of how regulatory variants for COVID-19-related genes in bronchial epithelium can be possible candidates for genetic factors that affect infection or progression of the disease. Dotted lines denote the hypothesis we are able to create by searching for the phenotypic associations of the cis-eQTLs for COVID-19-related genes. b Heatmap of the colocalization analysis results for 20 COVID-19-related genes with eQTLs that have at least one phenotypic association belonging to the experimental factor ontology (EFO) parent categories relevant to COVID-19 (respiratory disease, hematological or pulmonary function measurement). Genes highlighted in bold indicate the loci involving COVID-19-relevant EFO categories with posterior probability for colocalization (PP4) > 0.5, suggesting evidence for shared genetic causality between eQTL and GWAS trait. In the TLE locus, the nearest genome-wide significant variant for forced expiratory volume in 1 s (FEV1) from Shrine et al. [57] is more than 1 Mb away, indicating that the association between the variant and FEV1 might be confounded by incomplete adjustment for height. c–e Regional association plot for the GWAS signal on the upper panel and cis-eQTL signal on the lower panel for IFITM3 (c), ERMP1 (d), and MEPCE (e) locus, where the eQTL for the corresponding gene colocalizes with the GWAS trait relevant to COVID-19. Genomic position of the variants is shown on the x-axis and -log10(P value) of the GWAS or eQTL association on the y-axis. The lead GWAS and eQTL variants are highlighted
We further used colocalization analysis to extract loci where the eQTL and GWAS signals are likely to share a causal variant, as opposed to spurious overlap, focusing on 20 loci with associations for hematological and respiratory system traits of which 12 colocalized (PP4 > 0.5, Fig. 5b, Additional file 2: Table S12). In Fig. 5c, we highlight IFITM3 that is upregulated by SARS-CoV-2 infection [30] and has an eQTL associated with multiple blood cell traits of the immune system [58] and neutrophil count in SPIROMICS (P < 0.002). In addition, IFITM3 has a well-characterized role in the entry of multiple viruses, including coronaviruses [59]. Another interesting gene, ERMP1 (Fig. 5d), has an eQTL colocalizing with an asthma GWAS association in the UK Biobank. ERMP1 interacts with the SARS-CoV-2 protein Orf9c [29] and ranks highly in a genome-wide CRISPR screen for genes required for SARS-CoV-2 infection [60]. Also, severe asthma is a risk factor for COVID-19 hospitalization [5] and death [61]. An eQTL for the MEPCE gene that interacts with SARS-Cov-2 protein Nsp8 [29] is associated with platelet parameters [58] (Fig. 5e). Interestingly, platelets are hyperactivated in COVID-19 [62, 63], and platelet count could be used as a prognostic biomarker in COVID-19 patients [64–66].
Discussion
Using whole genome profiling data available from biologically relevant data sets, we have generated an archive of gene expression alterations that may contribute to COVID-19 susceptibility and severity. Although we include an extensive analysis of ACE2 gene expression in bronchial epithelium and isoform usage, our findings extend beyond this, providing insight into the contribution of genetics and specific clinical risk factors in the airways’ response to the SARS-CoV-2 virus.
We demonstrate replicable associations between current smoking, obesity, hypertension, and increased bronchial epithelial ACE2 expression, potentially facilitating SARS-CoV-2 entry into host cells. Obesity and hypertension have been strongly linked with COVID-19 susceptibility and severity [1–5]. We were not well-powered to study diabetes, but in a sputum gene expression study, we did find an association between diabetes and increased ACE2 expression [67]. Together, this work suggests that one mechanism by which diseases associated with the metabolic syndrome are uniquely susceptible to COVID-19 is through increased ACE2 expression. Although ACE2 interacts with angiotensin 2 [68], we did not find that renin-angiotensin system-modifying drugs increased ACE2 expression. In fact, although our sample size was small, our data suggests that angiotensin receptor blockers are associated with lower ACE2 expression levels in smokers.
Although we observed that the largest increases in ACE2 expression were amongst current smokers, active smoking has not been identified as one of the largest risk factors for COVID-19 [1–5]. Early reports suggested a lower prevalence of smoking amongst patients with COVID-19 as compared to the general population. However, these reports have been debunked as confounded and inappropriately designed based on the flawed assumption that individuals with symptomatic COVID-19 reflect the general population when they are actually older with more comorbidities [69]. Well-adjusted studies in COVID-19 have shown that current smoking is indeed associated with increased disease severity [70, 71]. Nonetheless, current smoking does not appear to be the biggest risk factor for developing severe COVID-19 disease in large clinical studies, and thus mechanisms beyond ACE2 receptor binding of the virus must be explored.
We find evidence that the truncated dACE2 transcript is present in the bronchial epithelium and correlated with the expression of known interferon stimulated genes (ISGs). However, it does not appear to account for the observed clinical associations with overall ACE2 expression. The functional role for dACE2 is not currently known although it does not appear to bind SARS-CoV-2 [23, 53]. However, others have speculated [23] that during viral infections when ISGs are stimulated, dACE2 may act as a dummy receptor for other ACE2 ligands (e.g., microRNA-200c-3p) that if bound to ACE2 would lead to internalization of the ACE2-ligand complex and functional depletion of ACE2. Thus, dACE2 may keep ACE2 levels high during infection. Our observations suggest that it is, however, the full length transcript and not this truncated isoform that is associated with clinical risk factors.
It is likely that much of the inter-individual variation in COVID-19 is driven by a more complex molecular response to the virus in the airway than expression of ACE2 alone. This supposition is supported by our results demonstrating that obesity, hypertension, and cardiovascular comorbidities, as well as aging, are associated with a downregulation of mucosal immune response pathways similar to that seen in early SARS-CoV-2 infection in comparison to other viral infections. Together with clinical data and Mendelian randomization analyses of the causal role of smoking and BMI on severe COVID-19 [72], our result suggest that these important comorbidities increase COVID-19 susceptibility and severity by creating an airway microenvironment in which SARS-CoV-2 can gain a foothold before an effective host response is mounted. In SARS-CoV, a delayed innate immune response in tandem with early robust viral replication has been shown to lead to an enhanced late pro-inflammatory state and more severe lung injury [73]. Recent evidence suggests that SARS-CoV-2 may also impair early innate immune defenses through a host shutdown process [74]. Although diseases of the metabolic syndrome (e.g., cardiovascular conditions, obesity, and diabetes) are often associated with increased systemic inflammation, there is evidence of an associated delay in inflammatory cell recruitment to the lung during coronavirus infection in animal models [75, 76]. Further study of the lung-specific immune environment associated with these systemic diseases may be crucial to understanding susceptibility to severe SARS-CoV-2 infection.
In contrast to metabolic disorders, we find that inflammatory airway conditions increase gene expression indicative of increased innate and adaptive immune responses, potentially priming individuals for airway disease exacerbations in response to other viruses but not SARS-CoV-2. This is consistent with the large body of research showing that viruses trigger the majority of airway disease exacerbations [77]. SARS-CoV-2, however, appears to have a different immune profile and does not appear to be a major trigger for airway disease exacerbations in clinical studies [78, 79].
Furthermore, we show that host genetics has a biologically meaningful effect on the expression of many genes in the bronchial epithelium that may play an important role in COVID-19, including genes of interest as future drug targets that may not be covered by previous large eQTL catalogs from other tissue types. While we did not observe significant genetic regulatory effects for ACE2 and TMPRSS2, the effect of regulatory variants on the expression of some COVID-19-related genes can be as strong as the expression changes induced by SARS-CoV-2 infection, highlighting the possible important role of host genetics in COVID-19. Most notably, 3p21.31 locus is robustly shown to be associated with COVID-19 severity [5, 7, 8], but the functional mechanisms are unclear. The six candidate genes—SLC6A20, LZTFL1, CCR9, FYCO1, CXCR6, and XCR1—were not highly expressed in bronchial epithelium, except for LZTFL1, and did not have eQTLs in our data set, suggesting that eQTL studies from other tissues and cell types could provide more evidence for the causative gene(s) of this genetic association.
Most severe cases of SARS-CoV-2 infection progress to acute respiratory distress syndrome and respiratory failure, thus regulatory variants for COVID-19-related genes that also affect respiratory infections or immune-related outcomes of a possible host response to a virus serve as candidates for host genetic factors for COVID-19, or its severity. We pinpoint multiple COVID-19-interacting genes for which genetic regulatory variants associate with immune- or respiratory-related outcomes, including the interferon-induced transmembrane protein 3 (IFITM3), endoplasmic reticulum metallopeptidase 1 (ERMP1), and methylphosphate capping enzyme (MEPCE), making them strong candidates for host genetic risk factors.
Conclusions
Altogether, our findings of genetic and non-genetic factors affecting the expression of COVID-19-related genes in bronchial epithelium provide essential insights for understanding inter-individual variation of COVID-19 and developing therapeutic targets for COVID-19.
Supplementary Information
Additional file 1. Supplementary Methods.
Additional file 2: Table S1. Differential expression analysis of ACE2 in relation to clinical variables (A) and genomic signatures (B) in SPIROMICS, SARP, and MAST. Table S2. Top 100 genes co-expressed with ACE2 after adjustments in SPIROMICS (A), SARP (B), and MAST (C). The genes in the IL-17 signature are highlighted in yellow. Table S3. Canonical pathway gene sets based on differentially downregulated genes between SARS-CoV-2 infection and other viral illness using the Ingenuity Pathway Analysis canonical pathway function. Table S4. Association between canonical pathway gene sets from Table S3 and comorbidities in SPIROMICS (A), SARP (B), and MAST (C). Leading edge genes are enriched in association with the given comorbidity. Table S5. Canonical pathway gene sets based on genes enriched in association with each comorbidity using the Ingenuity Pathway Analysis canonical pathway function. A – cardiovascular condition in SPIROMICS, B – hypertension in SPIROMICS, C – obesity in SPIROMICS, D - hypertension in SARP, E – obesity in SARP. Table S6. COVID-19-related genes from Blanco-Melo et al. 2020, Gassen et al. 2020, Gordon et al. 2020, Hoffmann et al. 2020, Wang et al. 2020, and COVID-19 Cell Atlas. Table S7. Summary statistics of eQTL mapping in bronchial epithelium in SPIROMICS, including eQTL effect sizes, and lookup analysis from GTEx and eQTLGen Consortium. Table S8. Lookup of COVID-19-related genes with cis-eQTLs in bronchial epithelium from GTEx v8. Effect size measured as allelic fold change (log2) is given for every gene with FDR < 0.05 in GTEx v8 and its lead eQTL, or set to NA otherwise. Table S9. Pathway analysis of 492 eGenes from SPIROMICS not tested in GTEx Lung. Table S10. pheWAS of eQTLs for COVID-19-related genes in bronchial epithelium with Phenoscanner v2. Table S11. pheWAS of eQTLs for COVID-19-related genes in bronchial epithelium in (A) non-Hispanic White individuals (N = 1980) and (B) Hispanic and non-Hispanic, non-White individuals (N = 696) in SPIROMICS for 20 phenotypes. Table S12. Results of the colocalization analysis of the eQTLs in bronchial epithelium and COVID-19-relevant phenotypes.
Additional file 3: Figure S1. Associations between ACE2 gene expression and COPD, asthma, steroid use, and race. Figure S2. Associations between ACE2 gene expression and obesity. Figure S3. Associations between ACE2 gene expression and hypertension, and use of antihypertensives. Figure S4. Associations between age and ACE2 gene expression, and age and differential ACE2 exon usage. Figure S5. Associations between age and smoking status, hypertension, sex, and BMI in SPIROMICS. Figure S6. COVID-19 and other viral illness related gene set enrichment analyses in association with comorbidities in SPIROMICS, SARP, and MAST. Figure S7. Regulatory genetic effects of ACE2 and TMPRSS2, and the effect of smoking on TMPRSS2. Figure S8. Associations between COVID-19-related genes and comorbidities. Figure S9. Replication of cis-eQTLs in GTEx. Figure S10. Regulatory genetic effects of the candidate genes in the chr3 cluster associated with COVID-19. Figure S11. PheWAS associations for the 44 out of 108 lead cis-eQTLs associated with COVID-19-related genes with Phenoscanner v2.
Additional file 4: Supplementary Note. NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium Banner Authorship List.
Acknowledgements
The authors thank the SPIROMICS participants and participating physicians, investigators, and staff for making this research possible. More information about the study and how to access SPIROMICS data is available at www.spiromics.org. The authors would like to acknowledge the University of North Carolina at Chapel Hill BioSpecimen Processing Facility for sample processing, storage, and sample disbursements (http://bsp.web.unc.edu/).
We would like to acknowledge the following current and former investigators of the SPIROMICS sites and reading centers: Neil E Alexis, MD; Wayne H Anderson, PhD; Mehrdad Arjomandi, MD; Igor Barjaktarevic, MD, PhD; R Graham Barr, MD, DrPH; Patricia Basta, PhD; Lori A Bateman, MSc; Surya P Bhatt, MD; Eugene R Bleecker, MD; Richard C Boucher, MD; Russell P Bowler, MD, PhD; Stephanie A Christenson, MD; Alejandro P Comellas, MD; Christopher B Cooper, MD, PhD; David J Couper, PhD; Gerard J Criner, MD; Ronald G Crystal, MD; Jeffrey L Curtis, MD; Claire M Doerschuk, MD; Mark T Dransfield, MD; Brad Drummond, MD; Christine M Freeman, PhD; Craig Galban, PhD; MeiLan K Han, MD, MS; Nadia N Hansel, MD, MPH; Annette T Hastie, PhD; Eric A Hoffman, PhD; Yvonne Huang, MD; Robert J Kaner, MD; Richard E Kanner, MD; Eric C Kleerup, MD; Jerry A Krishnan, MD, PhD; Lisa M LaVange, PhD; Stephen C Lazarus, MD; Fernando J Martinez, MD, MS; Deborah A Meyers, PhD; Wendy C Moore, MD; John D Newell Jr, MD; Robert Paine, III, MD; Laura Paulin, MD, MHS; Stephen P Peters, MD, PhD; Cheryl Pirozzi, MD; Nirupama Putcha, MD, MHS; Elizabeth C Oelsner, MD, MPH; Wanda K O’Neal, PhD; Victor E Ortega, MD, PhD; Sanjeev Raman, MBBS, MD; Stephen I. Rennard, MD; Donald P Tashkin, MD; J Michael Wells, MD; Robert A Wise, MD; and Prescott G Woodruff, MD, MPH.
The project officers from the Lung Division of the National Heart, Lung, and Blood Institute were Lisa Postow, PhD, and Lisa Viviano, BSN; SPIROMICS was supported by contracts from the NIH/NHLBI (HHSN268200900013C, HHSN268200900014C, HHSN268200900015C, HHSN268200900016C, HHSN268200900017C, HHSN268200900018C, HHSN268200900019C, HHSN268200900020C), grants from the NIH/NHLBI (U01 HL137880 and U24 HL141762), and supplemented by contributions made through the Foundation for the NIH and the COPD Foundation from AstraZeneca/MedImmune; Bayer; Bellerophon Therapeutics; Boehringer-Ingelheim Pharmaceuticals, Inc.; Chiesi Farmaceutici S.p.A.; Forest Research Institute, Inc.; GlaxoSmithKline; Grifols Therapeutics, Inc.; Ikaria, Inc.; Novartis Pharmaceuticals Corporation; Nycomed GmbH; ProterixBio; Regeneron Pharmaceuticals, Inc.; Sanofi; Sunovion; Takeda Pharmaceutical Company; and Theravance Biopharma and Mylan.
Molecular data for the Trans-Omics in Precision Medicine (TOPMed) program was supported by the National Heart, Lung and Blood Institute (NHLBI). Genome Sequencing for “NHLBI TOPMed: SubPopulations and InteRmediate Outcome Measures In COPD Study” (phs001927) was performed at the Broad Institute Genomics Platform (HHSN268201600034I). Core support including centralized genomic read mapping and genotype calling, along with variant quality metrics and filtering were provided by the TOPMed Informatics Research Center (3R01HL-117626-02S1; contract HHSN268201800002I). Core support including phenotype harmonization, data management, sample-identity QC, and general program coordination were provided by the TOPMed Data Coordinating Center (R01HL-120393; U01HL-120393; contract HHSN268201800001I). We gratefully acknowledge the studies and participants who provided biological samples and data for TOPMed. A list of banner authors for the NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium is provided in the Additional file 4.
Abbreviations
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- COVID-19
Coronavirus disease 2019
- SPIROMICS
SubPopulations and InteRmediate Outcome Measures In COPD Study
- COPD
Chronic obstructive pulmonary disease
- SARP
Severe Asthma Research Program
- MAST
Mechanisms of ASThma Study
- eQTL
Expression quantitative trait locus
- pheWAS
Phenome-wide association study
- RNA-seq
RNA-sequencing
- WGS
Whole genome sequencing
- TOPMed
Trans-Omics for Precision Medicine
- GTEx
Genotype-Tissue Expression
- FEV1
Forced expiratory volume in 1 s
- ERS/ATS
European Respiratory Society/American Thoracic Society
- QC
Quality control
- dACE2
Truncated ACE2 transcript
- FDR
False discovery rate
- MAF
Minor allele frequency
- TSS
Transcription start site
- eGene
Gene with statistically significant eQTL
- aFC
Allelic fold change
- BMI
Body mass index
- GWAS
Genome-wide association study
- LD
Linkage disequilibrium
- PP4
Posterior support for colocalization in coloc, defined as posterior probability for observing an association with both traits driven by a shared causal variant (hypothesis four)
- FC
Fold change
- ARB
Angiotensin receptor blockers
- GSEA
Gene set enrichment analysis
- IPA
Ingenuity Pathway Analysis
- EFO
Experimental factor ontology
- ISG
Interferon stimulated genes
Authors’ contributions
S.K., T.L., S.A.C., P.G.W., and V.E.O. designed the study. S.A.C., S.G., J.N., and S.N. performed the differential expression and pathway analyses. S.K., M.M., A.P., and K.L.B. performed the eQTL analyses. V.E.O. and E.A. performed the phenome-wide association analysis. I.Z.B., R.G.B., E.R.B., R.P.B., A.P.C., C.B.C., D.J.C., G.J.C., J.L.C., M.T.D., M.K.H., N.N.H., E.A.H., R.J.K., J.A.K., F.J.M., D.A.M., R.P., S.P.P., M.C., L.C.D., S.C.E., J.V.F., E.I., N.N.J., B.D.L., X.L., W.C.M., S.E.W., J.Z., and C.L. were involved in the acquisition and processing of data. T.L. and S.A.C. supervised the work. S.K., T.L., and S.A.C. wrote the first draft. J.L.C., R.P.B., R.P., S.E.W., R.J.K., A.P.C., M.M., and K.L.B. contributed to the editing of the manuscript. All authors approved the final version of the manuscript.
Funding
This work was funded by the following funding sources: R01HL142992 (V.E.O.), R01HL137880 (V.E.O.), F30HG011194 (M.M.), T32HL144442 (K.L.B., and R.G.B.), R01MH106842 (T.L.), R01HL142028 (T.L., R.G.B., and S.K.), R01GM122924 (T.L.), UM1HG008901 (T.L.), R01GM124486 (T.L.), K23HL123778 (S.A.C.), R01HL121774 (S.A.C.), and U01HL137880 (S.A.C.). The funders had no role in study design, collection, analysis, and interpretation of data, or writing of the manuscript.
Availability of data and materials
The RNA-seq data for SPIROMICS and SARP are deposited to dbGaP at accessions phs001119.v1.p1 and phs001446, respectively. While awaiting data release via dbGaP, investigators may contact the corresponding authors or the SPIROMICS and SARP studies at https://www.spiromics.org/spiromics/ and http://www.severeasthma.org/home.html to discuss gaining access to the data via the ancillary study mechanism for these studies. MAST RNA-seq data are available at Gene Expression Omnibus (GEO) (accession number GSE67472 [80]). TOPMed WGS freeze 9 data for the SPIROMICS cohort will be available at dbGaP under accession number phs001927.
Full eQTL summary statistics for the 496 COVID-19-related genes generated during the current study can be downloaded from the GitHub repository at https://github.com/LappalainenLab/spiromics-covid19-eqtl/tree/master/eqtl/summary_stats [81]. eQTL mapping analyses code has been deposited to the GitHub repository at https://github.com/LappalainenLab/spiromics-covid19-eqtl [82].
Declarations
Ethics approval and consent to participate
The SPIROMICS study was approved by the Institutional Review Boards at each cooperating institutions: Columbia University IRB 2 (AAAE9315), University of Iowa IRB-01 (201308719), Johns Hopkins IRB-5 (NA_00035701), UCLA Medical IRB 1 (MIRB1) (10001740/18-000403), University of Michigan IRBMED B1 Board (HUM00036346/ HUM00141222), National Jewish Health IRB (HS2678), UCSF IRB Parnassus Panel (10-03169), Temple University IRB A2 (21416), U of Alabama at Birmingham IRB #2 (120906004), University of Illinois IRB #3 (2013-0939), University of Utah IRB Panel Review Board 5 (00027298/ 00108836), Wake Forest University IRB #5 (00012805/00048727), UNC Non-Biomedical IRB (10-0048), UCLA Medical IRB 1 (MIRB1) (18-000458), and University of Iowa IRB-01 (201003733). All participants provided written informed consent. The SARP and MAST studies were approved by the appropriate institutional review board at the participating sites and all participants provided written informed consent. The research conformed to the principles of the Helsinki Declaration.
Consent for publication
Not applicable
Competing interests
S.A.C. advises for AstraZeneca, GlaxoSmithKline, Glenmark Pharmaceuticals, and Amgen, gave invited lectures to Sonovion and Genentech, and writes for UpToDate. T.L. advises and has equity in Variant Bio and is a member of the scientific advisory board of Goldfinch Bio. V.E.O. has served and currently serves on Independent Data and Monitoring Committee for Regeneron and Sanofi for COVID-19 therapeutic clinical trials unrelated to the current manuscript. The remaining authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Tuuli Lappalainen and Stephanie A. Christenson contributed equally to this work.
Contributor Information
Silva Kasela, Email: skasela@nygenome.org.
Tuuli Lappalainen, Email: tlappalainen@nygenome.org.
Stephanie A. Christenson, Email: stephanie.christenson@ucsf.edu
NHLBI SubPopulations and InteRmediate Outcome Measures In COPD Study (SPIROMICS):
Neil E. Alexis, Wayne H. Anderson, Mehrdad Arjomandi, Igor Barjaktarevic, R. Graham Barr, Patricia Basta, Lori A. Bateman, Surya P. Bhatt, Eugene R. Bleecker, Richard C. Boucher, Russell P. Bowler, Stephanie A. Christenson, Alejandro P. Comellas, Christopher B. Cooper, David J. Couper, Gerard J. Criner, Ronald G. Crystal, Jeffrey L. Curtis, Claire M. Doerschuk, Mark T. Dransfield, Brad Drummond, Christine M. Freeman, Craig Galban, Mei Lan K. Han, Nadia N. Hansel, Annette T. Hastie, Eric A. Hoffman, Yvonne Huang, Robert J. Kaner, Richard E. Kanner, Eric C. Kleerup, Jerry A. Krishnan, Lisa M. LaVange, Stephen C. Lazarus, Fernando J. Martinez, Deborah A. Meyers, Wendy C. Moore, John D. Newell, Jr, Robert Paine, III, Laura Paulin, Stephen P. Peters, Cheryl Pirozzi, Nirupama Putcha, Elizabeth C. Oelsner, Wanda K. O’Neal, Victor E. Ortega, Sanjeev Raman, Stephen I. Rennard, Donald P. Tashkin, J. Michael Wells, Robert A. Wise, and Prescott G. Woodruff
NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium:
Namiko Abe, Gonçalo Abecasis, Francois Aguet, Christine Albert, Laura Almasy, Alvaro Alonso, Seth Ament, Peter Anderson, Pramod Anugu, Deborah Applebaum-Bowden, Kristin Ardlie, Dan Arking, Donna K. Arnett, Allison Ashley-Koch, Stella Aslibekyan, Tim Assimes, Paul Auer, Dimitrios Avramopoulos, John Barnard, Kathleen Barnes, R. Graham Barr, Emily Barron-Casella, Lucas Barwick, Terri Beaty, Gerald Beck, Diane Becker, Lewis Becker, Rebecca Beer, Amber Beitelshees, Emelia Benjamin, Takis Benos, Marcos Bezerra, Larry Bielak, Joshua Bis, Thomas Blackwell, John Blangero, Eric Boerwinkle, Donald W. Bowden, Russell Bowler, Jennifer Brody, Ulrich Broeckel, Jai Broome, Karen Bunting, Esteban Burchard, Carlos Bustamante, Erin Buth, Brian Cade, Jonathan Cardwell, Vincent Carey, Cara Carty, Richard Casaburi, James Casella, Peter Castaldi, Mark Chaffin, Christy Chang, Yi-Cheng Chang, Daniel Chasman, Sameer Chavan, Bo-Juen Chen, Wei-Min Chen, Yii-Der Ida Chen, Michael Cho, Seung Hoan Choi, Lee-Ming Chuang, Mina Chung, Ren-Hua Chung, Clary Clish, Suzy Comhair, Matthew Conomos, Elaine Cornell, Adolfo Correa, Carolyn Crandall, James Crapo, L. Adrienne Cupples, Joanne Curran, Jeffrey Curtis, Brian Custer, Coleen Damcott, Dawood Darbar, Sayantan Das, Sean David, Colleen Davis, Michelle Daya, Mariza de Andrade, Lisa de las Fuentes, Michael DeBaun, Ranjan Deka, Dawn DeMeo, Scott Devine, Qing Duan, Ravi Duggirala, Jon Peter Durda, Susan Dutcher, Charles Eaton, Lynette Ekunwe, Adel El Boueiz, Patrick Ellinor, Leslie Emery, Serpil Erzurum, Charles Farber, Tasha Fingerlin, Matthew Flickinger, Myriam Fornage, Nora Franceschini, Chris Frazar, Mao Fu, Stephanie M. Fullerton, Lucinda Fulton, Stacey Gabriel, Weiniu Gan, Shanshan Gao, Yan Gao, Margery Gass, Bruce Gelb, Xiaoqi ( Priscilla) Geng, Mark Geraci, Soren Germer, Robert Gerszten, Auyon Ghosh, Richard Gibbs, Chris Gignoux, Mark Gladwin, David Glahn, Stephanie Gogarten, Da-Wei Gong, Harald Goring, Sharon Graw, Daniel Grine, C. Charles Gu, Yue Guan, Xiuqing Guo, Namrata Gupta, Jeff Haessler, Michael Hall, Daniel Harris, Nicola L. Hawley, Jiang He, Ben Heavner, Susan Heckbert, Ryan Hernandez, David Herrington, Craig Hersh, Bertha Hidalgo, James Hixson, Brian Hobbs, John Hokanson, Elliott Hong, Karin Hoth, Chao ( Agnes) Hsiung, Yi-Jen Hung, Haley Huston, Chii Min Hwu, Marguerite Ryan Irvin, Rebecca Jackson, Deepti Jain, Cashell Jaquish, Min A. Jhun, Jill Johnsen, Andrew Johnson, Craig Johnson, Rich Johnston, Kimberly Jones, Hyun Min Kang, Robert Kaplan, Sharon Kardia, Sekar Kathiresan, Shannon Kelly, Eimear Kenny, Michael Kessler, Alyna Khan, Wonji Kim, Greg Kinney, Barbara Konkle, Charles Kooperberg, Holly Kramer, Christoph Lange, Ethan Lange, Leslie Lange, Cathy Laurie, Cecelia Laurie, Meryl LeBoff, Jiwon Lee, Seunggeun Shawn Lee, Wen-Jane Lee, Jonathon LeFaive, David Levine, Dan Levy, Joshua Lewis, Xiaohui Li, Yun Li, Henry Lin, Honghuang Lin, Keng Han Lin, Xihong Lin, Simin Liu, Yongmei Liu, Yu Liu, Ruth J. F. Loos, Steven Lubitz, Kathryn Lunetta, James Luo, Michael Mahaney, Barry Make, Ani Manichaikul, Jo Ann Manson, Lauren Margolin, Lisa Martin, Susan Mathai, Rasika Mathias, Susanne May, Patrick McArdle, Merry-Lynn McDonald, Sean McFarland, Stephen McGarvey, Daniel McGoldrick, Caitlin McHugh, Hao Mei, Luisa Mestroni, Deborah A. Meyers, Julie Mikulla, Nancy Min, Mollie Minear, Ryan L. Minster, Braxton D. Mitchell, Matt Moll, May E. Montasser, Courtney Montgomery, Arden Moscati, Solomon Musani, Stanford Mwasongwe, Josyf C. Mychaleckyj, Girish Nadkarni, Rakhi Naik, Take Naseri, Pradeep Natarajan, Sergei Nekhai, Sarah C. Nelson, Bonnie Neltner, Deborah Nickerson, Kari North, Jeff O’Connell, Tim O’Connor, Heather Ochs-Balcom, David Paik, Nicholette Palmer, James Pankow, George Papanicolaou, Afshin Parsa, Juan Manuel Peralta, Marco Perez, James Perry, Ulrike Peters, Patricia Peyser, Lawrence S. Phillips, Toni Pollin, Wendy Post, Julia Powers Becker, Meher Preethi Boorgula, Michael Preuss, Bruce Psaty, Pankaj Qasba, Dandi Qiao, Zhaohui Qin, Nicholas Rafaels, Laura Raffield, Vasan S. Ramachandran, D. C. Rao, Laura Rasmussen-Torvik, Aakrosh Ratan, Susan Redline, Robert Reed, Elizabeth Regan, Alex Reiner, Muagututi‘a Sefuiva Reupena, Ken Rice, Stephen Rich, Dan Roden, Carolina Roselli, Jerome Rotter, Ingo Ruczinski, Pamela Russell, Sarah Ruuska, Kathleen Ryan, Ester Cerdeira Sabino, Danish Saleheen, Shabnam Salimi, Steven Salzberg, Kevin Sandow, Vijay G. Sankaran, Christopher Scheller, Ellen Schmidt, Karen Schwander, David Schwartz, Frank Sciurba, Christine Seidman, Jonathan Seidman, Vivien Sheehan, Stephanie L. Sherman, Amol Shetty, Aniket Shetty, Wayne Hui-Heng Sheu, M. Benjamin Shoemaker, Brian Silver, Edwin Silverman, Jennifer Smith, Josh Smith, Nicholas Smith, Tanja Smith, Sylvia Smoller, Beverly Snively, Michael Snyder, Tamar Sofer, Nona Sotoodehnia, Adrienne M. Stilp, Garrett Storm, Elizabeth Streeten, Jessica Lasky Su, Yun Ju Sung, Jody Sylvia, Adam Szpiro, Carole Sztalryd, Daniel Taliun, Hua Tang, Margaret Taub, Kent D. Taylor, Matthew Taylor, Simeon Taylor, Marilyn Telen, Timothy A. Thornton, Machiko Threlkeld, Lesley Tinker, David Tirschwell, Sarah Tishkoff, Hemant Tiwari, Catherine Tong, Russell Tracy, Michael Tsai, Dhananjay Vaidya, David Van Den Berg, Peter VandeHaar, Scott Vrieze, Tarik Walker, Robert Wallace, Avram Walts, Fei Fei Wang, Heming Wang, Karol Watson, Daniel E. Weeks, Bruce Weir, Scott Weiss, Lu Chen Weng, Jennifer Wessel, Cristen Willer, Kayleen Williams, L. Keoki Williams, Carla Wilson, James Wilson, Quenna Wong, Joseph Wu, Huichun Xu, Lisa Yanek, Ivana Yang, Rongze Yang, Norann Zaghloul, Maryam Zekavat, Yingze Zhang, Snow Xueyan Zhao, Wei Zhao, Degui Zhi, Xiang Zhou, Xiaofeng Zhu, Michael Zody, and Sebastian Zoellner
References
- 1.Goyal P, Choi JJ, Pinheiro LC, Schenck EJ, Chen R, Jabri A, Satlin MJ, Campion TR, Jr, Nahid M, Ringel JB, Hoffman KL, Alshak MN, Li HA, Wehmeyer GT, Rajan M, Reshetnyak E, Hupert N, Horn EM, Martinez FJ, Gulick RM, Safford MM. Clinical characteristics of COVID-19 in New York City. N Engl J Med. 2020;382(24):2372–2374. doi: 10.1056/NEJMc2010419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta S, Hayek SS, Wang W, Chan L, Mathews KS, Melamed ML, Brenner SK, Leonberg-Yoo A, Schenck EJ, Radbel J, Reiser J, Bansal A, Srivastava A, Zhou Y, Sutherland A, Green A, Shehata AM, Goyal N, Vijayan A, Velez JCQ, Shaefi S, Parikh CR, Arunthamakun J, Athavale AM, Friedman AN, Short SAP, Kibbelaar ZA, Abu Omar S, Admon AJ, Donnelly JP, Gershengorn HB, Hernán MA, Semler MW, Leaf DE, STOP-COVID Investigators Factors associated with death in critically ill patients with coronavirus disease 2019 in the US. JAMA Intern Med. 2020;180(11):1436. doi: 10.1001/jamainternmed.2020.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Docherty AB, Harrison EM, Green CA, Hardwick HE, Pius R, Norman L, et al. Features of 20 133 UK patients in hospital with COVID-19 using the ISARIC WHO Clinical Characterisation Protocol: prospective observational cohort study. BMJ. 2020;369:m1985. doi: 10.1136/bmj.m1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petrilli CM, Jones SA, Yang J, Rajagopalan H, O’Donnell L, Chernyak Y, et al. Factors associated with hospital admission and critical illness among 5279 people with coronavirus disease 2019 in New York City: prospective cohort study. BMJ. 2020;369:m1966. doi: 10.1136/bmj.m1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shelton JF, Shastri AJ, Ye C, Weldon CH, Filshtein-Somnez T, Coker D, et al. Trans-ethnic analysis reveals genetic and non-genetic associations with COVID-19 susceptibility and severity. Preprint at medRxiv 10.1101/2020.09.04.20188318. 2020.
- 6.Williams FM, Freydin M, Mangino M, Couvreur S, Visconti A, Bowyer RC, et al. Self-reported symptoms of COVID-19 including symptoms most predictive of SARS-CoV-2 infection, are heritable. Preprint at bioRxiv 10.1101/2020.04.22.20072124. 2020. [DOI] [PubMed]
- 7.Ellinghaus D, Degenhardt F, Bujanda L, Buti M, Albillos A, Invernizzi P, et al. Genomewide association study of severe COVID-19 with respiratory failure. N Engl J Med. 2020;383(16):1522–34. 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed]
- 8.COVID-19 Host Genetics Initiative The COVID-19 Host Genetics Initiative, a global initiative to elucidate the role of host genetic factors in susceptibility and severity of the SARS-CoV-2 virus pandemic. Eur J Hum Genet. 2020;28:715–718. doi: 10.1038/s41431-020-0636-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN, et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016–1035.e19. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bradding P, Richardson M, Hinks TSC, Howarth PH, Choy DF, Arron JR, Wenzel SE, Siddiqui S. ACE2, TMPRSS2, and furin gene expression in the airways of people with asthma-implications for COVID-19. J Allergy Clin Immunol. 2020;146(1):208–211. doi: 10.1016/j.jaci.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang H, Rostami MR, Leopold PL, Mezey JG, O’Beirne SL, Strulovici-Barel Y, Crystal RG. Expression of the SARS-CoV-2 ACE2 receptor in the human airway epithelium. Am J Respir Crit Care Med. 2020;202(2):219–229. doi: 10.1164/rccm.202003-0541OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Couper D, LaVange LM, Han M, Barr RG, Bleecker E, Hoffman EA, Kanner R, Kleerup E, Martinez FJ, Woodruff PG, Rennard S, SPIROMICS Research Group Design of the Subpopulations and Intermediate Outcomes in COPD Study (SPIROMICS) Thorax. 2014;69(5):491–494. doi: 10.1136/thoraxjnl-2013-203897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taliun D, Harris DN, Kessler MD, Carlson J, Szpiech ZA, Torres R, et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Preprint at bioRxiv 10.1101/563866. 2019. [DOI] [PMC free article] [PubMed]
- 14.The GTEx Consortium The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369(6509):1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wells JM, Arenberg DA, Barjaktarevic I, Bhatt SP, Bowler RP, Christenson SA, Couper DJ, Dransfield MT, Han MLK, Hoffman EA, Kaner RJ, Kim V, Kleerup E, Martinez FJ, Moore WC, O’Beirne SL, Paine R, III, Putcha N, Raman SM, Barr RG, Rennard SI, Woodruff PG, Curtis JL. Safety and tolerability of comprehensive research bronchoscopy in chronic obstructive pulmonary disease. Results from the SPIROMICS bronchoscopy substudy. Ann Am Thorac Soc. 2019;16(4):439–446. doi: 10.1513/AnnalsATS.201807-441OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ricklefs I, Barkas I, Duvall MG, Cernadas M, Grossman NL, Israel E, et al. ALX receptor ligands define a biochemical endotype for severe asthma. JCI Insight. 2017;2(14). 10.1172/jci.insight.93534. [DOI] [PMC free article] [PubMed]
- 17.Chung KF, Wenzel SE, Brozek JL, Bush A, Castro M, Sterk PJ, Adcock IM, Bateman ED, Bel EH, Bleecker ER, Boulet LP, Brightling C, Chanez P, Dahlen SE, Djukanovic R, Frey U, Gaga M, Gibson P, Hamid Q, Jajour NN, Mauad T, Sorkness RL, Teague WG. International ERS/ATS guidelines on definition, evaluation and treatment of severe asthma. Eur Respir J. 2014;43(2):343–373. doi: 10.1183/09031936.00202013. [DOI] [PubMed] [Google Scholar]
- 18.Christenson SA, Steiling K, van den Berge M, Hijazi K, Hiemstra PS, Postma DS, Lenburg ME, Spira A, Woodruff PG. Asthma-COPD overlap. Clinical relevance of genomic signatures of type 2 inflammation in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2015;191(7):758–766. doi: 10.1164/rccm.201408-1458OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leek J, Johnson W, Parker H, Fertig E, Jaffe A, Zhang Y, et al. sva: surrogate variable analysis. 2020. [Google Scholar]
- 22.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Onabajo OO, Banday AR, Stanifer ML, Yan W, Obajemu A, Santer DM, Florez-Vargas O, Piontkivska H, Vargas JM, Ring TJ, Kee C, Doldan P, Tyrrell DL, Mendoza JL, Boulant S, Prokunina-Olsson L. Interferons and viruses induce a novel truncated ACE2 isoform and not the full-length SARS-CoV-2 receptor. Nat Genet. 2020;52(12):1283–1293. doi: 10.1038/s41588-020-00731-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mancini E, Rabinovich A, Iserte J, Yanovsky M, Chernomoretz A. ASpli: analysis of alternative splicing using RNA-Seq. 2020. [Google Scholar]
- 25.Mick E, Kamm J, Pisco AO, Ratnasiri K, Babik JM, Calfee CS, et al. Upper airway gene expression differentiates COVID-19 from other acute respiratory illnesses and reveals suppression of innate immune responses by SARS-CoV-2. Preprint at medRxiv 10.1101/2020.05.18.20105171. 2020.
- 26.Korotkevich G, Sukhov V, Sergushichev A. Fast gene set enrichment analysis. Preprint at bioRxiv 10.1101/060012. 2019.
- 27.Wu D, Smyth GK. Camera: a competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res. 2012;40(17):e133. doi: 10.1093/nar/gks461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, O’Meara MJ, Rezelj VV, Guo JZ, Swaney DL, Tummino TA, Hüttenhain R, Kaake RM, Richards AL, Tutuncuoglu B, Foussard H, Batra J, Haas K, Modak M, Kim M, Haas P, Polacco BJ, Braberg H, Fabius JM, Eckhardt M, Soucheray M, Bennett MJ, Cakir M, McGregor MJ, Li Q, Meyer B, Roesch F, Vallet T, Mac Kain A, Miorin L, Moreno E, Naing ZZC, Zhou Y, Peng S, Shi Y, Zhang Z, Shen W, Kirby IT, Melnyk JE, Chorba JS, Lou K, Dai SA, Barrio-Hernandez I, Memon D, Hernandez-Armenta C, Lyu J, Mathy CJP, Perica T, Pilla KB, Ganesan SJ, Saltzberg DJ, Rakesh R, Liu X, Rosenthal SB, Calviello L, Venkataramanan S, Liboy-Lugo J, Lin Y, Huang XP, Liu YF, Wankowicz SA, Bohn M, Safari M, Ugur FS, Koh C, Savar NS, Tran QD, Shengjuler D, Fletcher SJ, O’Neal MC, Cai Y, Chang JCJ, Broadhurst DJ, Klippsten S, Sharp PP, Wenzell NA, Kuzuoglu-Ozturk D, Wang HY, Trenker R, Young JM, Cavero DA, Hiatt J, Roth TL, Rathore U, Subramanian A, Noack J, Hubert M, Stroud RM, Frankel AD, Rosenberg OS, Verba KA, Agard DA, Ott M, Emerman M, Jura N, von Zastrow M, Verdin E, Ashworth A, Schwartz O, d’Enfert C, Mukherjee S, Jacobson M, Malik HS, Fujimori DG, Ideker T, Craik CS, Floor SN, Fraser JS, Gross JD, Sali A, Roth BL, Ruggero D, Taunton J, Kortemme T, Beltrao P, Vignuzzi M, García-Sastre A, Shokat KM, Shoichet BK, Krogan NJ. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blanco-Melo D, Nilsson-Payant BE, Liu W-C, Uhl S, Hoagland D, Møller R, et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gassen NC, Papies J, Bajaj T, Dethloff F, Emanuel J, Weckmann K, et al. Analysis of SARS-CoV-2-controlled autophagy reveals spermidine, MK-2206, and niclosamide as putative antiviral therapeutics. Preprint at bioRxiv 10.1101/2020.04.15.997254. 2020.
- 32.Wang K, Chen W, Zhou Y-S, Lian J-Q, Zhang Z, Du P, et al. SARS-CoV-2 invades host cells via a novel route: CD147-spike protein. Preprint at bioRxiv 10.1101/2020.03.14.988345. 2020.
- 33.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor-Weiner A, Aguet F, Haradhvala NJ, Gosai S, Anand S, Kim J, Ardlie K, van Allen EM, Getz G. Scaling computational genomics to millions of individuals with GPUs. Genome Biol. 2019;20(1):228. doi: 10.1186/s13059-019-1836-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stegle O, Parts L, Durbin R, Winn J. A Bayesian framework to account for complex non-genetic factors in gene expression levels greatly increases power in eQTL studies. PLoS Comput Biol. 2010;6(5):e1000770. doi: 10.1371/journal.pcbi.1000770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mohammadi P, Castel SE, Brown AA, Lappalainen T. Quantifying the regulatory effect size of cis-acting genetic variation using allelic fold change. Genome Res. 2017;27(11):1872–1884. doi: 10.1101/gr.216747.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18(1):220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davis JR, Fresard L, Knowles DA, Pala M, Bustamante CD, Battle A, Montgomery SB. An efficient multiple-testing adjustment for eQTL studies that accounts for linkage disequilibrium between variants. Am J Hum Genet. 2016;98(1):216–224. doi: 10.1016/j.ajhg.2015.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim-Hellmuth S, Aguet F, Oliva M, Muñoz-Aguirre M, Kasela S, Wucher V, Castel SE, Hamel AR, Viñuela A, Roberts AL, Mangul S, Wen X, Wang G, Barbeira AN, Garrido-Martín D, Nadel BB, Zou Y, Bonazzola R, Quan J, Brown A, Martinez-Perez A, Soria JM, GTEx Consortium§. Getz G, Dermitzakis ET, Small KS, Stephens M, Xi HS, Im HK, Guigó R, Segrè AV, Stranger BE, Ardlie KG, Lappalainen T. Cell type–specific genetic regulation of gene expression across human tissues. Science. 2020;369(6509):eaaz8528. doi: 10.1126/science.aaz8528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Võsa U, Claringbould A, Westra H-J, Bonder MJ, Deelen P, Zeng B, et al. Unraveling the polygenic architecture of complex traits using blood eQTL metaanalysis. Preprint at bioRxiv 10.1101/447367. 2018.
- 43.Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, Vilo J. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update) Nucleic Acids Res. 2019;47(W1):W191–W198. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Staley JR, Blackshaw J, Kamat MA, Ellis S, Surendran P, Sun BB, Paul DS, Freitag D, Burgess S, Danesh J, Young R, Butterworth AS. PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics. 2016;32(20):3207–3209. doi: 10.1093/bioinformatics/btw373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kamat MA, Blackshaw JA, Young R, Surendran P, Burgess S, Danesh J, Butterworth AS, Staley JR. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics. 2019;35(22):4851–4853. doi: 10.1093/bioinformatics/btz469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wallace C. Eliciting priors and relaxing the single causal variant assumption in colocalisation analyses. PLoS Genet. 2020;16(4):e1008720. doi: 10.1371/journal.pgen.1008720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lukassen S, Chua RL, Trefzer T, Kahn NC, Schneider MA, Muley T, et al. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020;39:e105114. doi: 10.15252/embj.2020105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith JC, Sausville EL, Girish V, Yuan ML, Vasudevan A, John KM, et al. Cigarette smoke exposure and inflammatory signaling increase the expression of the SARS-CoV-2 receptor ACE2 in the respiratory tract. Dev Cell. 2020;53:514–529.e3. doi: 10.1016/j.devcel.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cai G, Bossé Y, Xiao F, Kheradmand F, Amos CI. Tobacco smoking increases the lung gene expression of ACE2, the receptor of SARS-CoV-2. Am J Respir Crit Care Med. 2020;201(12):1557–1559. doi: 10.1164/rccm.202003-0693LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aliee H, Massip F, Qi C, de Biase MS, van Nijnatten JL, Kersten ETG, et al. Determinants of SARS-CoV-2 receptor gene expression in upper and lower airways. Preprint at medRxiv 10.1101/2020.08.31.20169946. 2020.
- 51.Christenson SA, van den Berge M, Faiz A, Inkamp K, Bhakta N, Bonser LR, Zlock LT, Barjaktarevic IZ, Barr RG, Bleecker ER, Boucher RC, Bowler RP, Comellas AP, Curtis JL, Han MK, Hansel NN, Hiemstra PS, Kaner RJ, Krishnanm JA, Martinez FJ, O'Neal WK, Paine R, 3rd, Timens W, Wells JM, Spira A, Erle DJ, Woodruff PG. An airway epithelial IL-17A response signature identifies a steroid-unresponsive COPD patient subgroup. J Clin Invest. 2019;129(1):169–181. doi: 10.1172/JCI121087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhakta NR, Christenson SA, Nerella S, Solberg OD, Nguyen CP, Choy DF, Jung KL, Garudadri S, Bonser LR, Pollack JL, Zlock LT, Erle DJ, Langelier C, Derisi JL, Arron JR, Fahy JV, Woodruff PG. IFN-stimulated gene expression, type 2 inflammation, and endoplasmic reticulum stress in asthma. Am J Respir Crit Care Med. 2018;197(3):313–324. doi: 10.1164/rccm.201706-1070OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ng KW, Attig J, Bolland W, Young GR, Major J, Wrobel AG, Gamblin S, Wack A, Kassiotis G. Tissue-specific and interferon-inducible expression of nonfunctional ACE2 through endogenous retroelement co-option. Nat Genet. 2020;52(12):1294–1302. doi: 10.1038/s41588-020-00732-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takahashi T, Ellingson MK, Wong P, Israelow B, Lucas C, Klein J, et al. Sex differences in immune responses that underlie COVID-19 disease outcomes. Nature. 2020;588(7837):315–320. doi: 10.1038/s41586-020-2700-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raj VS, Mou H, Smits SL, Dekkers DHW, Müller MA, Dijkman R, Muth D, Demmers JAA, Zaki A, Fouchier RAM, Thiel V, Drosten C, Rottier PJM, Osterhaus ADME, Bosch BJ, Haagmans BL. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495(7440):251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lopera Maya EA, van der Graaf A, Lanting P, van der Geest M, Fu J, Swertz M, Franke L, Wijmenga C, Deelen P, Zhernakova A, Sanna S, Lifelines Cohort Study Lack of association between genetic variants at ACE2 and TMPRSS2 genes involved in SARS-CoV-2 infection and human quantitative phenotypes. Front Genet. 2020;11:613. doi: 10.3389/fgene.2020.00613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shrine N, Guyatt AL, Erzurumluoglu AM, Jackson VE, Hobbs BD, Melbourne CA, et al. New genetic signals for lung function highlight pathways and chronic obstructive pulmonary disease associations across multiple ancestries. Nat Genet. 2019;51(3):481–493. doi: 10.1038/s41588-018-0321-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Astle WJ, Elding H, Jiang T, Allen D, Ruklisa D, Mann AL, et al. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell. 2016;167:1415–1429.e19. doi: 10.1016/j.cell.2016.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang I-C, Bailey CC, Weyer JL, Radoshitzky SR, Becker MM, Chiang JJ, Brass AL, Ahmed AA, Chi X, Dong L, Longobardi LE, Boltz D, Kuhn JH, Elledge SJ, Bavari S, Denison MR, Choe H, Farzan M. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 2011;7(1):e1001258. doi: 10.1371/journal.ppat.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Daniloski Z, Jordan TX, Wessels HH, Hoagland DA, Kasela S, Legut M, et al. Identification of required host factors for SARS-CoV-2 infection in human cells. Cell. 2021;184(1):92-105.e16. 10.1016/j.cell.2020.10.030. [DOI] [PMC free article] [PubMed]
- 61.Williamson EJ, Walker AJ, Bhaskaran K, Bacon S, Bates C, Morton CE, Curtis HJ, Mehrkar A, Evans D, Inglesby P, Cockburn J, McDonald HI, MacKenna B, Tomlinson L, Douglas IJ, Rentsch CT, Mathur R, Wong AYS, Grieve R, Harrison D, Forbes H, Schultze A, Croker R, Parry J, Hester F, Harper S, Perera R, Evans SJW, Smeeth L, Goldacre B. OpenSAFELY: factors associated with COVID-19 death in 17 million patients. Nature. 2020;584(7821):430–436. doi: 10.1038/s41586-020-2521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Manne BK, Denorme F, Middleton EA, Portier I, Rowley JW, Stubben C, Petrey AC, Tolley ND, Guo L, Cody M, Weyrich AS, Yost CC, Rondina MT, Campbell RA. Platelet gene expression and function in patients with COVID-19. Blood. 2020;136(11):1317–1329. doi: 10.1182/blood.2020007214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zaid Y, Puhm F, Allaeys I, Naya A, Oudghiri M, Khalki L, Limami Y, Zaid N, Sadki K, Ben el Haj R, Mahir W, Belayachi L, Belefquih B, Benouda A, Cheikh A, Langlois MA, Cherrah Y, Flamand L, Guessous F, Boilard E. Platelets can associate with SARS-Cov-2 RNA and are hyperactivated in COVID-19. Circ Res. 2020;127(11):1404–1418. doi: 10.1161/CIRCRESAHA.120.317703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vabret N, Britton GJ, Gruber C, Hegde S, Kim J, Kuksin M, Levantovsky R, Malle L, Moreira A, Park MD, Pia L, Risson E, Saffern M, Salomé B, Esai Selvan M, Spindler MP, Tan J, van der Heide V, Gregory JK, Alexandropoulos K, Bhardwaj N, Brown BD, Greenbaum B, Gümüş ZH, Homann D, Horowitz A, Kamphorst AO, Curotto de Lafaille MA, Mehandru S, Merad M, Samstein RM, Agrawal M, Aleynick M, Belabed M, Brown M, Casanova-Acebes M, Catalan J, Centa M, Charap A, Chan A, Chen ST, Chung J, Bozkus CC, Cody E, Cossarini F, Dalla E, Fernandez N, Grout J, Ruan DF, Hamon P, Humblin E, Jha D, Kodysh J, Leader A, Lin M, Lindblad K, Lozano-Ojalvo D, Lubitz G, Magen A, Mahmood Z, Martinez-Delgado G, Mateus-Tique J, Meritt E, Moon C, Noel J, O’Donnell T, Ota M, Plitt T, Pothula V, Redes J, Reyes Torres I, Roberto M, Sanchez-Paulete AR, Shang J, Schanoski AS, Suprun M, Tran M, Vaninov N, Wilk CM, Aguirre-Ghiso J, Bogunovic D, Cho J, Faith J, Grasset E, Heeger P, Kenigsberg E, Krammer F, Laserson U. Immunology of COVID-19: current state of the science. Immunity. 2020;52(6):910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu Y, Sun W, Guo Y, Chen L, Zhang L, Zhao S, Long D, Yu L. Association between platelet parameters and mortality in coronavirus disease 2019: retrospective cohort study. Platelets. 2020;31(4):490–496. doi: 10.1080/09537104.2020.1754383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shi S, Qin M, Shen B, Cai Y, Liu T, Yang F, et al. Association of cardiac injury with mortality in hospitalized patients with COVID-19 in Wuhan, China. JAMA Cardiol. 2020;5(7):802–10. 10.1001/jamacardio.2020.0950. [DOI] [PMC free article] [PubMed]
- 67.Peters MC, Sajuthi S, Deford P, Christenson S, Rios CL, Montgomery MT, Woodruff PG, Mauger DT, Erzurum SC, Johansson MW, Denlinger LC, Jarjour NN, Castro M, Hastie AT, Moore W, Ortega VE, Bleecker ER, Wenzel SE, Israel E, Levy BD, Seibold MA, Fahy JV. COVID-19–related genes in sputum cells in asthma. Relationship to demographic features and corticosteroids. Am J Respir Crit Care Med. 2020;202(1):83–90. doi: 10.1164/rccm.202003-0821OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vaduganathan M, Vardeny O, Michel T, McMurray JJV, Pfeffer MA, Solomon SD. Renin-angiotensin-aldosterone system inhibitors in patients with COVID-19. N Engl J Med. 2020;382(17):1653–1659. doi: 10.1056/NEJMsr2005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wenzl T. Smoking and COVID-19: did we overlook representativeness? Tob Induc Dis. 2020;18:89. doi: 10.18332/tid/129584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Patanavanich R, Glantz SA. Smoking is associated with COVID-19 progression: a meta-analysis. Nicotine Tob Res Off J Soc Res Nicotine Tob. 2020;22(9):1653–1656. doi: 10.1093/ntr/ntaa082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hopkinson NS, Rossi N, El-Sayed Moustafa J, Laverty AA, Quint JK, Freidin M, et al. Current smoking and COVID-19 risk: results from a population symptom app in over 2.4 million people. Thorax. 2021;thoraxjnl-2020-216422. 10.1136/thoraxjnl-2020-216422. [DOI] [PubMed]
- 72.Ponsford MJ, Gkatzionis A, Walker VM, Grant AJ, Wootton RE, Moore LSP, Fatumo S, Mason AM, Zuber V, Willer C, Rasheed H, Brumpton B, Hveem K, Kristian Damås J, Davies N, Olav Åsvold B, Solligård E, Jones S, Burgess S, Rogne T, Gill D. Cardiometabolic traits, sepsis and severe COVID-19: a Mendelian randomization investigation. Circulation. 2020;142(18):1791–1793. doi: 10.1161/CIRCULATIONAHA.120.050753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Channappanavar R, Fehr AR, Vijay R, Mack M, Zhao J, Meyerholz DK, Perlman S. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe. 2016;19(2):181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thoms M, Buschauer R, Ameismeier M, Koepke L, Denk T, Hirschenberger M, Kratzat H, Hayn M, Mackens-Kiani T, Cheng J, Straub JH, Stürzel CM, Fröhlich T, Berninghausen O, Becker T, Kirchhoff F, Sparrer KMJ, Beckmann R. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020;369(6508):1249–1255. doi: 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smith M, Honce R, Schultz-Cherry S. Metabolic syndrome and viral pathogenesis: lessons from influenza and coronaviruses. J Virol. 2020;94(18). 10.1128/JVI.00665-20. [DOI] [PMC free article] [PubMed]
- 76.Kulcsar KA, Coleman CM, Beck SE, Frieman MB. Comorbid diabetes results in immune dysregulation and enhanced disease severity following MERS-CoV infection. JCI Insight. 2019;4(20). 10.1172/jci.insight.131774. [DOI] [PMC free article] [PubMed]
- 77.Kurai D, Saraya T, Ishii H, Takizawa H. Virus-induced exacerbations in asthma and COPD. Front Microbiol. 2013;4:293. doi: 10.3389/fmicb.2013.00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Grandbastien M, Piotin A, Godet J, Abessolo-Amougou I, Ederlé C, Enache I, Fraisse P, Tu Hoang TC, Kassegne L, Labani A, Leyendecker P, Manien L, Marcot C, Pamart G, Renaud-Picard B, Riou M, Doyen V, Kessler R, Fafi-Kremer S, Metz-Favre C, Khayath N, de Blay F. SARS-CoV-2 pneumonia in hospitalized asthmatic patients did not induce severe exacerbation. J Allergy Clin Immunol Pract. 2020;8(8):2600–2607. doi: 10.1016/j.jaip.2020.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Beurnier A, Jutant E-M, Jevnikar M, Boucly A, Pichon J, Preda M, et al. Characteristics and outcomes of asthmatic patients with COVID-19 pneumonia who require hospitalisation. Eur Respir J. 2020;56(5):2001875. 10.1183/13993003.01875-2020. [DOI] [PMC free article] [PubMed]
- 80.Christenson SA, Arron JR, Steiling K, van den Berge M, Hijazi K, Hiemstra PS, et al. Airway epithelial gene expression in asthma versus healthy controls. GSE67472. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE67472 (2015).
- 81.Kasela S. Full eQTL summary statistics for the 496 COVID-19-related genes. Github. https://github.com/LappalainenLab/spiromics-covid19-eqtl/tree/master/eqtl/summary_stats (2020).
- 82.Kasela S. eQTL mapping analysis code. Github. https://github.com/LappalainenLab/spiromics-covid19-eqtl (2020).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Supplementary Methods.
Additional file 2: Table S1. Differential expression analysis of ACE2 in relation to clinical variables (A) and genomic signatures (B) in SPIROMICS, SARP, and MAST. Table S2. Top 100 genes co-expressed with ACE2 after adjustments in SPIROMICS (A), SARP (B), and MAST (C). The genes in the IL-17 signature are highlighted in yellow. Table S3. Canonical pathway gene sets based on differentially downregulated genes between SARS-CoV-2 infection and other viral illness using the Ingenuity Pathway Analysis canonical pathway function. Table S4. Association between canonical pathway gene sets from Table S3 and comorbidities in SPIROMICS (A), SARP (B), and MAST (C). Leading edge genes are enriched in association with the given comorbidity. Table S5. Canonical pathway gene sets based on genes enriched in association with each comorbidity using the Ingenuity Pathway Analysis canonical pathway function. A – cardiovascular condition in SPIROMICS, B – hypertension in SPIROMICS, C – obesity in SPIROMICS, D - hypertension in SARP, E – obesity in SARP. Table S6. COVID-19-related genes from Blanco-Melo et al. 2020, Gassen et al. 2020, Gordon et al. 2020, Hoffmann et al. 2020, Wang et al. 2020, and COVID-19 Cell Atlas. Table S7. Summary statistics of eQTL mapping in bronchial epithelium in SPIROMICS, including eQTL effect sizes, and lookup analysis from GTEx and eQTLGen Consortium. Table S8. Lookup of COVID-19-related genes with cis-eQTLs in bronchial epithelium from GTEx v8. Effect size measured as allelic fold change (log2) is given for every gene with FDR < 0.05 in GTEx v8 and its lead eQTL, or set to NA otherwise. Table S9. Pathway analysis of 492 eGenes from SPIROMICS not tested in GTEx Lung. Table S10. pheWAS of eQTLs for COVID-19-related genes in bronchial epithelium with Phenoscanner v2. Table S11. pheWAS of eQTLs for COVID-19-related genes in bronchial epithelium in (A) non-Hispanic White individuals (N = 1980) and (B) Hispanic and non-Hispanic, non-White individuals (N = 696) in SPIROMICS for 20 phenotypes. Table S12. Results of the colocalization analysis of the eQTLs in bronchial epithelium and COVID-19-relevant phenotypes.
Additional file 3: Figure S1. Associations between ACE2 gene expression and COPD, asthma, steroid use, and race. Figure S2. Associations between ACE2 gene expression and obesity. Figure S3. Associations between ACE2 gene expression and hypertension, and use of antihypertensives. Figure S4. Associations between age and ACE2 gene expression, and age and differential ACE2 exon usage. Figure S5. Associations between age and smoking status, hypertension, sex, and BMI in SPIROMICS. Figure S6. COVID-19 and other viral illness related gene set enrichment analyses in association with comorbidities in SPIROMICS, SARP, and MAST. Figure S7. Regulatory genetic effects of ACE2 and TMPRSS2, and the effect of smoking on TMPRSS2. Figure S8. Associations between COVID-19-related genes and comorbidities. Figure S9. Replication of cis-eQTLs in GTEx. Figure S10. Regulatory genetic effects of the candidate genes in the chr3 cluster associated with COVID-19. Figure S11. PheWAS associations for the 44 out of 108 lead cis-eQTLs associated with COVID-19-related genes with Phenoscanner v2.
Additional file 4: Supplementary Note. NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium Banner Authorship List.
Data Availability Statement
The RNA-seq data for SPIROMICS and SARP are deposited to dbGaP at accessions phs001119.v1.p1 and phs001446, respectively. While awaiting data release via dbGaP, investigators may contact the corresponding authors or the SPIROMICS and SARP studies at https://www.spiromics.org/spiromics/ and http://www.severeasthma.org/home.html to discuss gaining access to the data via the ancillary study mechanism for these studies. MAST RNA-seq data are available at Gene Expression Omnibus (GEO) (accession number GSE67472 [80]). TOPMed WGS freeze 9 data for the SPIROMICS cohort will be available at dbGaP under accession number phs001927.
Full eQTL summary statistics for the 496 COVID-19-related genes generated during the current study can be downloaded from the GitHub repository at https://github.com/LappalainenLab/spiromics-covid19-eqtl/tree/master/eqtl/summary_stats [81]. eQTL mapping analyses code has been deposited to the GitHub repository at https://github.com/LappalainenLab/spiromics-covid19-eqtl [82].