Summary
Each human genome includes de novo mutations that arose during gametogenesis. While these germline mutations represent a fundamental source of new genetic diversity, they can also create deleterious alleles that impact fitness. Whereas the rate and patterns of point mutations in the human germline are now well understood, far less is known about the frequency and features that impact de novo structural variants (dnSVs). We report a family-based study of germline mutations among 9,599 human genomes from 33 multigenerational CEPH-Utah families and 2,384 families from the Simons Foundation Autism Research Initiative. We find that de novo structural mutations detected by alignment-based, short-read WGS occur at an overall rate of at least 0.160 events per genome in unaffected individuals, and we observe a significantly higher rate (0.206 per genome) in ASD-affected individuals. In both probands and unaffected samples, nearly 73% of de novo structural mutations arose in paternal gametes, and we predict most de novo structural mutations to be caused by mutational mechanisms that do not require sequence homology. After multiple testing correction, we did not observe a statistically significant correlation between parental age and the rate of de novo structural variation in offspring. These results highlight that a spectrum of mutational mechanisms contribute to germline structural mutations and that these mechanisms most likely have markedly different rates and selective pressures than those leading to point mutations.
Keywords: copy number variation, structural variation, autism, genetic diversity, genomic structure, genomics, germline mutation, de novo mutation
Introduction
Several mechanisms, including replication infidelity,1, 2, 3 genomic damage,4, 5, 6 non-allelic recombination,7 and double-strand break repair,8 are known to create de novo mutations (DNMs) in the human germline. These mutations contribute to genomic diversity and often are primary targets in the analysis of rare, dominant genetic disorders. There is therefore a long-standing interest in understanding the frequency at which DNMs occur and the patterns that affect these rates. Numerous studies have measured the rate of germline de novo single-nucleotide variants (dnSNVs) and small insertion-deletion mutations (indels) at approximately 70 events per individual,9, 10, 11, 12, 13 and it has been established that the majority of these small point mutations arise on the paternal gamete. The frequency of single-nucleotide and insertion-deletion DNMs increases with parental age, especially paternal age.9,12,14, 15, 16, 17, 18
In contrast, precise estimates of germline mutations affecting the structure of the human genome (structural variants [SVs]) have been far more difficult to discern. De novo SVs (dnSVs) largely arise from mutational mechanisms that are distinct from those responsible for point mutations. The larger size of SVs, defined here and in many other studies as variants affecting at least 50 base pairs, increases the likelihood that any given SV will impact protein-coding genes or other critical genomic regions. Understanding the selective constraints on dnSV-specific mechanisms is essential because a broad spectrum of balanced, unbalanced, and complex structural mutations are known to underlie many developmental disorders.19, 20, 21, 22, 23, 24 However, dnSVs are predicted to occur several hundred-fold less frequently than point mutations,11 requiring a much larger sample size to achieve accurate estimates of dnSV rates.
The inherent challenge of accurately identifying SVs further complicates the measurement of dnSV rates. The short-reads that comprise most large whole-genome sequencing (WGS) datasets yield high false positive and false negative rates,25, 26, 27, 28 as paired-end short-read alignments cannot always reveal the complete structure of an SV. Most SV detection algorithms29, 30, 31, 32 screen for clusters of split alignments and paired-end reads with discordant strand orientation or insert sizes, while SVs that alter copy number are also detectable through the changes in sequence depth for the variant region.33, 34, 35
Repetitive genomic regions obfuscate SV calling by creating inconsistent or inaccurate read alignments, which can cause false negatives by attenuating the alignment signals supporting true SVs. These regions can also produce a high rate of false positive SV signals.25 Long-read sequencing technologies and de novo assembly promise to address some of these challenges and greatly improve the accuracy and sensitivity of SV detection;28 unfortunately, they remain prohibitively expensive for most large-scale analyses. Detection of SVs in repetitive regions is critical for understanding dnSV rates, as multiple homology-mediated mechanisms have been shown to drive SV formation, including those arising from non-allelic homologous recombination (NAHR)36, 37, 38 and those derived from mechanisms dependent on minimal sequence homology, such as fork stalling and template switching (FoSTeS39) and microhomology-mediated break-induced replication (MMBIR40). Identifying these variants from short-read WGS, as well as those resulting from repair mechanisms that do not require sequence homology, such as non-homologous end joining,41 requires exhaustive SV calling through the application of multiple algorithms11,42 and extensive curation of SV predictions to remove false positives.43,44 Despite these efforts, some SVs remain undetectable via short-read WGS.28 These inaccessible variants display sequence contexts distinct from those that can be captured by both short-read and long-read technologies, and estimates of dnSV rates utilizing such data are therefore lower bounds on the true SV mutation rate.45
Because of these complications, there is greater variance in estimates of dnSV rates than for single-nucleotide DNMs. Early studies from microarray technology estimated that one very large (greater than 300 kb) de novo copy number variant (CNV) occurs once in every 98 births.46 More recent studies with higher resolution short-read WGS in trio and quartet families observed one dnSV per 5–6 births,11,47 and estimates from larger population-based sequencing analyses were higher still at one dnSV per approximately 3.5 births.42 Differences in these estimates reflect variability in sample sizes, sequencing technologies, SV calling methodology, and approaches to estimating dnSV mutation rates (e.g., direct observation versus estimations via principles of population genetics), which highlights the challenges inherent to establishing precise estimates of a human dnSV rate from short-read WGS technologies.
Estimating the rate of mutations arising from mobile element insertions (MEIs) that are still active in the human genome has also been challenging. MEI mutation rates are important to understand given their ability to impact human phenotypes by creating CNVs through non-allelic homologous recombination48 or by interrupting genes through retrotransposition.49,50 A recent study in a cohort of 33 large families identified 26 de novo MEI events51 and estimated LINE1 (L1) and SVA (SINE-VNTR-Alu) retrotransposition rates at about 1/63 births, while the rate of AluY retrotranspositions was measured at about 1/40 births. Additional investigation of de novo MEIs with larger cohort sizes and more MEIs will help to refine these estimates.
dnSVs are known to play a role in the genetic etiology of sporadic autism spectrum disorder (ASD), and multiple previous studies have shown that simplex ASD-affected individuals are more likely to harbor very large dnSVs than their unaffected siblings or the general population.46,52, 53, 54 Furthermore, while parent-of-origin and parental age effects have been observed for single-nucleotide DNMs, their impact on dnSV rates remains unclear. Prior studies have indicated a large paternal contribution to dnSVs,33 while a maternal bias was reported for a subset of recurrent dnSVs in other studies.55 Efforts to identify effects of parental age on dnSV rates have generally failed to show age-based enrichment but remain inconclusive because of the small numbers of dnSVs found.33,56
In this study, we analyze the rate of de novo mutation for six major classes of SVs: deletions (DELs), duplications (DUPs), insertions (INSs, including MEIs), inversions (INVs), translocations (CTXs), and complex variants that combine more than one of the previous (CPXs). By studying the genomes of a large cohort of nuclear families, we provide an accurate, lower-bound measure of the rate of de novo SV mutation detectable with short-read WGS data. We also explore the effects of gamete of origin and parental age on dnSVs and investigate potential rate differences between ASD-affected individuals and control individuals.
Material and methods
dnSV identification
We detected SVs in the CEPH and Simons Foundation Autism Research Initiative (SFARI) cohorts with the GATK-SV discovery pipeline previously described.11,42 Briefly, GATK-SV is an ensemble approach that uses multiple established SV detection tools to maximize sensitivity while reevaluating evidence directly from BAMs (binary sequence alignment/map files) by using a random forest classifier to improve specificity in a series of variant classification modules. GATK-SV has flexibility with initial input SV algorithms, and in this case, we ran an amalgamated series of complementary algorithms that detect SVs on the basis of a variety of signatures, including discordant paired-end reads (PE), split reads (SR), and read depth (Delly [v.0.7.8], smoove [v.0.2.4], Manta [v.1.3.1], Wham [v.1.7.0] and MELT [v.2.1.4], CNVnator [v.0.3.3], and a custom version of cn.MOPS). Upon completion of the pipeline, a VCF (variant call format file) is derived containing adjudicated and integrated SVs from the raw algorithms. Furthermore, complex SVs57,58 and other SVs involving more than one breakpoint (e.g., inversions, reciprocal translocations) are fully resolved. Centromeres, telomeres, pseudo-autosomal regions, HLA genes, the mitochondrial genome, and other regions known to be highly repetitive or otherwise unmappable were excluded.
Akin to single-nucleotide variants (SNVs) and indels,59,60 careful filtering is required for precise identification of dnSVs. We have developed a set of post hoc filtering criteria that work from a GATK-SV VCF. We start with an unfiltered VCF and remove any unresolved breakpoints, SVs with split read support observed on only one side of a breakpoint, and any CNV found to have multi-allelic copy states making the determination of parental haplotypes challenging and therefore de novo calling highly inaccurate. We then exclude any variant with a parental frequency of greater than 1% for the autism cohort and 10% for the CEPH study (to account for potential F1 [second-generation] de novo transmissions). Next, we keep only variants with support in the initial raw callers, removing any potential genotyping errors, and we investigate CNV copy state overlap in parents to account for the imprecise boundaries of depth-based CNV detection that could cause an inaccurate but overlapping CNV call in a parent or child. Next, we apply a set of genotype quality (GQ) filters that were determined by an ROC (receiver operating characteristic) curve analysis with a truth set derived from molecularly validated dnSVs from a previous study on a smaller subset (n = 2,076) of the total cohort11 and false positives defined as novel de novo variants in those samples. Both child and parental GQ cutoffs were determined, the latter of which classifies variants initially predicted to be de novo that are in fact likely inherited. Optimal GQ parameters were found for an overall GQ as well as depth-based GQ and PE/SR GQ, all derived from the genotyping step in GATK-SV and present in the final VCF. Given the lack of a validated training set for CEPH and the much smaller sample size, only a simple filter of less than 30 for depth-based GQ and PE/SR GQ in parents was applied. All variants that passed these filters were manually reviewed via duphold (v.0.2.1),43 IGV,61 SV-Plaudit (v.1.0.0)44 with Samplot (v.1.0.10),62 and an internal R-based visualization script found in GATK-SV. In order to reduce the chance of missing a variant of interest, we included all private variants with a passing parental GQ in the manual investigation. In the SFARI cohort, samples found to have ten or more de novo events were excluded from manual review and classified as “outlier samples” (n = 18; 0.4% of children). No such outliers were excluded in the CEPH cohort.
We investigated potential de novo mosaic events via the following steps. Using the results of our random forest filtering, we identify CNVs greater than 10 kb from both depth- and paired-end/split-read-based algorithms that pass the random forest filtering’s p value threshold but not its separation threshold. After passing through a step where fragmented variants are stitched together, we use a cohort-based variant frequency estimate to identify variants that appear only once in the cohort. We initially set a variant frequency cutoff of 1% for SFARI, but upon manual review, we realized this was not stringent enough, so we increased the frequency cutoff in CEPH to 0.3%. We then use manual curation of the passing mosaic variants to identify true events as described above (Figure S1).
Identifying parent of origin for dnSVs
Phasing identifies the parent whose gamete underwent an error leading to a spontaneous mutation. We used SNVs, which varied in state between parents and which were inherited heterozygously (informative sites), to identify localized haplotypes that derived from either the mother or the father. We developed and applied a combination of two methods: extended read-based phasing and SNV allele-balance CNV phasing.
Extended read-based phasing
For each dnSV, we selected reads that supported the variant (split reads or discordant pairs whose gap and orientation fit the variant) and then used any nearby heterozygous SNV sites to associate other reads in the region to the variant haplotype or the reference haplotype, allowing us to extend our search for informative sites from the variant breakpoints. We then tested for informative site overlap, using any informative sites up to a maximum distance from the breakpoints of 5 kb, as long as an overlap with haplotype-assigned reads was found. If at least one informative site was found with read overlap, variant phasing was possible.
SNV allele-balance CNV phasing
CNVs have a predictable effect on the allele balance of SNVs within the region of the variant. Duplications should approximately double the number of reads that come from the duplicated region, while deletions should eliminate reads from the deleted region. Thus, where the allele balance for informative sites in the region of the variant shifts, it becomes possible to determine from which parent the de novo event was inherited. We identified all informative sites within deletions and tested for hemizygosity, where an informative site allele that should have been inherited from one parent instead disappeared and an allele not shared by both parents was inherited. We similarly identified informative sites within duplications and identified cases where allele balance was at least 2:1 rather than the null expectation of 1:1. In some cases of large CNVs, several informative sites were identified that gave contradictory phasing results. If at least 95% of sites supported one parent as the origin, we assigned the variant to that parent; otherwise, we excluded the variant from phasing.
This combination of phasing strategies allowed us to phase 268/698 deletions, duplications, inversions, and complex variants, an improvement on phasing rates for SNVs in previous work.9 Insertion variants proved the most difficult type to phase, as the split reads and discordant pairs generated by these variants (especially MEIs, which were the bulk of the insertions in our callset) often misalign or align to high-copy genomic repeats and are lost, removing the evidence needed to relate variants to a haplotype. We therefore did not analyze parent-of-origin effects on insertion variants.
Predicting causal mechanisms for dnSVs
It is impossible to confidently identify the causal mechanism for many variants, as no perfect evidence exists to confirm that a specific mechanism was responsible. However, the sequence context of a variant often provides clues that can be used to determine the most likely type of candidate mechanism that could have led to a variant’s formation. Using methodology similar to a previous analysis of SV breakpoints in mouse models,63 we analyzed the variants in our dnSV set with respect to three broad categories of mechanism that can lead to creation of a dnSV.
Microhomology-based variants
We grouped together mechanisms that lead to spontaneous rearrangement due to microhomology, including MMBIR40 and FoSTeS.39 For each variant, we collected split reads that spanned the breakpoints, requiring at least two split reads for each breakpoint. This provided strong evidence that the breakpoint coordinates were correctly identified and allowed us to test for homology of 2–100 bp between the regions upstream and downstream of each breakpoint. Variants with breakpoints that had microhomology were categorized as most likely deriving from microhomology-based mechanisms. We identified 132 microhomology SVs; 74 were in probands and 58 were in unaffected samples.
Macrohomology-based mechanisms
We used annotations of known segmental duplication pairs to identify variants that most likely resulted from NAHR. NAHR variants can be difficult to identify with short-read sequencing data, as lengthy high-identity repeats must flank the resulting variant. These repeats often mask the signals used to detect SVs by increasing the difficulty of read mapping and alignment and may be especially detrimental to the accurate identification of discordant pairs and split reads, which many SV calling tools rely on. Extremely large CNVs are the most likely NAHR-derived variants to be accurately detected, as these can be found using depth-based CNV callers, such as CNVnator and cn.MOPS. Breakpoint resolution was inexact with these variants, as they generally lacked any confident split-read support (due to flanking repeats). Thus, we grouped together CNVs whose breakpoints were flanked by segmental duplications of at least 95% sequence identity. We required that the end of the first of the pair of segmental duplication be within a distance of 20% of the CNV length from the start of the CNV and the start of the second of the segmental duplication pair be within a distance of 20% of the CNV length from the end of the CNV because of the extreme error in breakpoint calling that often arises in repeat-rich regions. 45 dnSVs were assigned to the NAHR category, including 36 in probands and 9 in unaffected samples.
Non-homology-based mechanisms
Variants which were not identified as MEIs and did not have either macrohomology or microhomology flanking the breakpoints were classified as non-homology based. These variants may arise from a number of molecular mechanisms, such as non-homologous end joining,64, in which double-strand breaks are corrected and filled in an error-prone manner. We grouped 530 variants as non-homology based, including 288 in probands and 242 in unaffected samples.
Results
Identification of dnSVs
Our analyses focused on two family-based cohorts with short-read WGS. The first cohort consisted of 572 samples in 33 large three-generation families from the CEPH-Utah cohort;65 in total, these families are comprised of 434 distinct mother, father, child trios. The second cohort consisted of 2,384 families from the SFARI Simons Simplex Collection (SSC66). The SSC cohort includes 443 ASD “trios” (consisting of one affected child and two unaffected parents) and 1,941 ASD “quartets” (consisting of one affected child, one unaffected child, and two unaffected parents). We excluded samples that failed quality control analysis (see material and methods), resulting in a final cohort of 2,363 ASD probands and 1,938 siblings, all with both parents available. Families selected had no known history of ASD, increasing the likelihood that SVs contributing to ASD arose de novo in a gamete transmitted to the affected child.
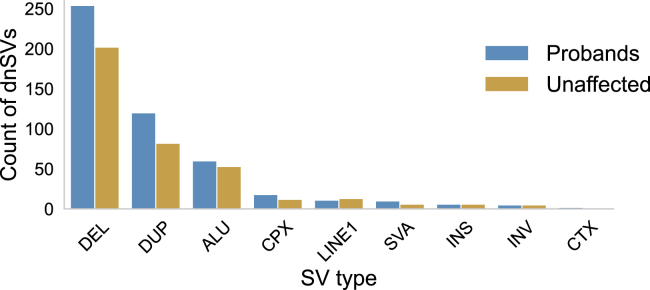
We applied a comprehensive suite of SV identification algorithms, consisting of Lumpy,31 Manta,29 Delly,32 Whamg,30 CNVnator,35 cn.MOPS,34 and MELT,67 the latter six as part of the GATK-SV framework.11,42 We then filtered putative dnSVs by using depth-of-coverage43 and visual inspection,44,61 resulting in a set of 804 high-confidence germline dnSVs, excluding trisomies and sex chromosome anomalies (Figure 1, Table S1).
Figure 1.
dnSVs identified in SFARI SSC
The count of dnSVs of each SV type found in 2,363 ASD probands and 2,372 unaffected samples.
Although a much smaller cohort, the unique, three-generation composition of the CEPH-Utah families enables direct measurement of the rate of false positive dnSV calls. Because these families have a median of eight (min = 4, max = 16) offspring in the third generation, any dnSV detected in a sample from the second generation of a CEPH-Utah family should have a 50% probability of transmission to each third-generation child. For example, the chance of at least one child inheriting a true dnSV from the second-generation parent is typically over 99% (e.g., 1 − 0.58 for a family with eight children in the third generation). Thus, any predicted dnSV observed in a second-generation individual that is absent from all third-generation offspring is considered a false positive. We identified eight second-generation dnSVs in the CEPH families, all of which were transmitted to at least one third-generation offspring.
The SV discovery and filtering methods we employed were closely based on those used in the analysis of dnSVs from a small subset of the SSC cohort containing 2,076 of the samples included in this study.11 In that analysis, 171 dnSVs were detected and 168 validated through PCR and Sanger sequencing assays, and there was a validation rate of 97% (163 dnSVs). Microarray assays were also employed to test the sensitivity via sequencing-based SV calling in that study, for dnCNVs over 40 kb (due to the relatively weaker resolution of microarrays), resulting in an estimated 2.5% false discovery rate and 99.6% sensitivity. Together, these validations suggest a low false discovery rate in CEPH as well as SFARI.
Another potential problem in dnSV detection is a false negative SV call in a parent sample, which leads to the false labeling of an inherited SV in the child as a DNM. To test for this, we also called dnSVs in the third generation of the CEPH cohort, resulting in 54 putative dnSVs. We then visually scrutinized the evidence in the parents and grandparents of each third-generation CEPH sample with a dnSV call. We examined IGV and Samplot62 images that included the offspring sample, both parents, and both sets of grandparents and carefully examined each for any missed split read, discordant pair, or coverage depth signals indicating that the putative dnSV was actually a missed transmission event. This provided an extra opportunity to detect elusive inherited variants. In 53 of the third-generation CEPH dnSVs, no evidence was detected in parents or grandparents to support the variant, while one variant presented a complex breakpoint pattern in one parent and one grandparent, which might have resulted in the third-generation dnSV call (Figure S2). We therefore estimate that ~2% of variants identified as dnSVs could be cases of missed transmission.
Many of the dnSVs we identified affect genic regions of the genome. We found that 484 dnSVs (as well as 42 somatic mosaic SVs, excluded from other analyses) overlapped gene annotations retrieved from GENCODE68 via GGD.69 Our analyses revealed that 225 of these overlaps impacted coding sequence, including 153 dnSVs in probands and 72 in unaffected samples (Table S2).
Analysis of dnSV rates and gamete of origin
We combined the SSC and CEPH dnSV sets, yielding a final set of 865 germline dnSVs in 4,735 offspring genomes (9,599 genomes with parents included), including 2,363 ASD probands and 2,372 unaffected offspring. Of the 785 samples with at least one dnSV, one contained four events, nine contained three each, and 59 contained two each.
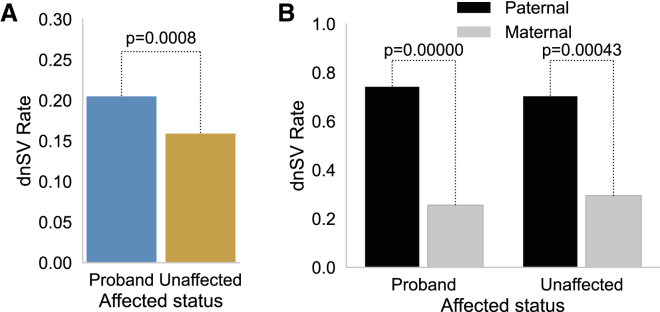
Prior studies involving fewer families have shown that ASD-affected individuals are often enriched for dnSVs compared to unaffected siblings.46,52, 53, 54 One study11 analyzed WGS in 519 SFARI families that were pre-screened for de novo loss-of-function variants or large CNVs and found no enrichment of dnSVs in probands. We leveraged this now much larger set of SSC and CEPH families to test the dnSV rate for probands and unaffected controls (Figure 2A). We found a statistically significant increase in dnSVs among ASD probands (486 dnSVs/2,363 affected individuals) compared to unaffected samples (379 dnSVs/2,372 unaffected samples; p = 0.0008, Fisher’s exact test). The rate of dnSVs in this cohort was therefore one mutation for every 0.2056 births in probands and one mutation for every 0.1598 births in control individuals. This enrichment was significant for duplications (p = 0.0212, Figure S3A) yet not significant for deletions (p = 0.0556, Figure S3B) or Alu-family MEIs (p = 0.7036, Figure S3C).
Figure 2.
Comparisons of dnSV rates
(A) Comparison by Fisher’s exact test of dnSV rates in probands versus siblings.
(B) Comparison by Fisher’s exact test of dnSV rates from variants phased to maternal or paternal gamete in probands and siblings.
We developed a method to use informative SNVs within or near dnSVs to determine the parental gamete of origin (the mutation’s haplotype phase) for de novo deletions, duplications, inversions, and complex variants. 268 dnSVs successfully phased, including 38.4% of all dnSVs of those types (see material and methods for additional details). This analysis revealed an enrichment for paternally derived SVs in both probands and unaffected samples (Figure 2B). Among unaffected samples, 66 (66%) dnSVs arose on the paternal gamete and 34 (34%) from the maternal gamete; these rates were significantly different (Fisher’s exact test; p = 0.00972). Similarly, among probands, 125 (74.4%) dnSVs had a paternal origin and 43 (25.6%) were maternally derived; the difference in these rates was also statistically significant (p < 0.0001, Fisher’s exact test).
Age effects on dnSV rates
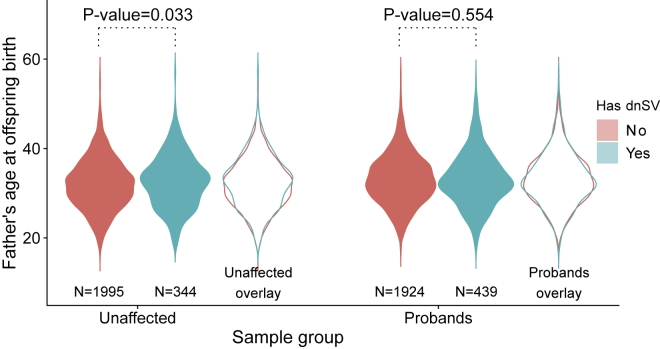
An important unanswered question is whether dnSV rates increase with parental age. Parental-age effects on the rate of de novo single-nucleotide mutations have been previously identified and are an important factor in profiling the likely causes of genetic disease in individuals with older parents. A goal of our study was therefore to determine whether there is a similar increase in the rate of dnSVs with parental age. We used the father’s age at birth of the child as a proxy for parental age and grouped samples by dnSV status (0 versus 1 or more dnSVs) and then performed a one-sided Wilcoxon rank-sum test for an increase in paternal age among samples with a dnSV versus those without (Figure 3). In probands, we found no significant difference in the distributions of father’s ages between the two groups (p = 0.554), while in unaffected samples, we found a significant increase in father’s ages (p = 0.033) that did not remain significant after Bonferroni multiple test correction for two tests (adjusted p = 0.066). We estimate that we have 80% power to detect a mean paternal age difference of 0.851 and 0.940 years between samples having a dnSV versus those without a dnSV in probands and unaffected samples, respectively (Figure S4). Thus, while undetectable within our cohort, a parental age bias may still exist, albeit with a much weaker effect than observed for some other types of mutations, such as dnSNVs; detecting a statistically significant age effect will most likely require an even larger cohort. A potential confounding variable in this comparison is the known effect of paternal age on risk for ASD,70 which could act to decrease our power to detect a parental age effect in ASD probands.
Figure 3.
Correlations of paternal age and dnSV rate
Comparison by one-sided Wilcoxon rank-sum test for increased father’s age in samples with at least one dnSV versus those without. This test is performed for unaffected samples (left) and probands (right). Overlay highlights differences in father’s age distribution between samples with and samples without a dnSV. After Bonferroni multiple testing for two tests, the adjusted marginal significance for unaffected samples is p = 0.066, and for probands, the adjusted marginal significance is p = 1.
We tested for effects of either paternal or maternal age by using the subset of dnSVs that had been phased to a parental gamete of origin (165 probands with dnSVs, 85 unaffected samples with dnSVs) and found no difference in parental ages between samples with or without a dnSV (one-sided Wilcoxon rank-sum test, Figure S5). We hypothesize that this lack of correlation speaks to the inherently different mechanisms underlying point mutations and structural changes, as the increase in point mutation rate with parental age is substantial and reproducible. As a control, we also performed a Poisson regression to test the effect of paternal age on the rate of de novo single-nucleotide mutations in this cohort and, as expected given prior findings,9,12 we found a significant correlation (p < 2e−16) between the number of dnSNVs and the age of the father for both unaffected samples and probands (Figure S6). We also tested for a correlation between the number of dnSNVs and the presence of dnSVs and found that samples with a dnSV have a significantly greater number of dnSNVs (Figure S7), yet the mechanistic basis of this correlation is unclear.
Next, we tested for parental-age effects on rates of the most common SV types: deletions (254 in probands, 202 in unaffected samples), duplications (120 in probands, 82 in unaffected samples), and MEIs (87 in probands, 78 in unaffected samples). We used father’s age as a proxy for parental age and found no enrichment with paternal age in deletions or duplications, although there was an increase in de novo MEI (dnMEI) risk to unaffected samples with increased paternal age (difference in means 2.18 years, p = 0.004, not significant after Bonferroni correction for eight tests, Figure S8). The dnMEI age effect may therefore drive the signal detected in analysis of all SV types shown in Figure 3, as neither of the other common SV types showed a significant enrichment. This may reflect the fundamentally different mechanisms underlying dnMEIs and argues for future research, especially with long-read sequencing technologies that offer greater power to characterize MEIs.
Mechanisms responsible for dnSVs
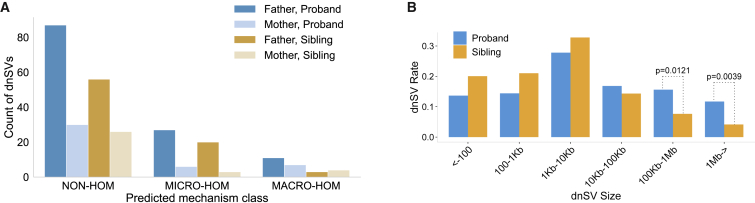
Identifying the primary mechanisms responsible for dnSVs is of fundamental interest to characterizing mutational hotspots and to understanding the mutational forces driving evolution of genome structure. However, inferring the exact mechanism underlying each SV is complicated by imprecise breakpoint mapping and low sequence complexity at SV breakpoints. We therefore grouped variants into three categories based on the degree of sequence homology observed at each dnSV breakpoint, an essential feature for several known mechanisms of structural variation, and analyzed mechanistic correlations with parental origin in the 268 phased variants (Figure 4A).
Figure 4.
A comparison of dnSV breakpoint homology and size among probands and unaffected samples
(A) Counts of phased variants grouped by predicted mechanism class, parent of origin, and affected status. Mechanism classes include those characterized by no sequence homology at breakpoints (NON-HOM), microhomology at breakpoints (MICRO-HOM), or macrohomology (matching segmental duplications) at breakpoints (MACRO-HOM).
(B) Variants binned by size and compared between probands and unaffected samples. The fraction of dnSVs assigned to each bin is statistically similar except in the largest two bins where sizes are 100 kb to 1 Mb and ≥1 Mb. The difficulty of determining the size of insertion variants, especially mobile element insertions, led to exclusion of those variants from this figure.
We found that CNVs flanked by segmental duplications, which are potential substrates for NAHR36, 37, 38, were grouped together in the macrohomology (MACRO-HOM) class. Small NAHR variants are difficult to detect, as the flanking homology at the breakpoints can decrease paired-end read signals and depth-of-coverage fluctuations that are used by Illumina-based SV-detection methods to identify variants, but large CNVs derived from NAHR are readily detected; we identified 45 NAHR-derived mutations with a median length of 1.5 Mb. Various mechanisms of SVs are identifiable by short sequences of breakpoint-flanking microhomologies, including FoSTeS39 and MMBIR.40 We therefore grouped dnSVs with a 2–10 bp homologous sequence flanking breakpoints as the microhomology (MICRO-HOM) class. Finally, we grouped variants with no breakpoint homology, most likely resulting from incorrect joining of double-strand breaks in many cases,41 as the NON-HOM (non-homology) class. Only two variants that we could capture from short-read WGS had breakpoint homology outside these size categories (18 and 22 bp).
We categorized all dnSVs except MEIs, for a total of 707 events, by the extent of breakpoint homology and inferred the mechanistic class most likely responsible (see material and methods for details). The majority of dnSVs (530, ~75%) lacked sequence homology, while fewer exhibited either macro-homology (45, ~6%) or micro-homology (132, ~19%). More dnSVs derived from the paternal gamete in each mechanism class. There were similar rates of micro-homology and non-homology dnSVs in probands and unaffected samples but a higher rate of macro-homology dnSVs in probands (n = 36 versus n = 9). This may relate to the large size of those variants (1.5 Mb) as compared to the 5.4 kb median length of all deletions and duplications we discovered. This substantially larger variant size also increases the risk of impacting genes or regulatory elements potentially involved in development of ASD. As 268, or about 38%, of these dnSVs were successfully phased, the analysis of parental effect on mechanism includes 25 macrohomology dnSVs, 54 microhomology dnSVs, and 189 non-homology dnSVs (Figure 4A). We found eight distinct macro-homology dnSVs that occurred in multiple samples, most likely via NAHR owing to the flanking segmental duplications, and appear in a total of 23 probands and two unaffected samples. These included dnSVs in regions with reported ties to ASD on chromosomes 7, 15, and 1671,72 (Table S3).
Many insertion variants were identified as MEIs. The active classes of MEIs consist of L1, Alu, and SVA retrotransposable elements (Figure 1). We identified 110 de novo Alu events, 15 de novo SVA events, and 20 de novo L1 events in probands and unaffected samples for approximate rates of 1 Alu per 42 births, 1 SVA per 309 births, and 1 L1 per 231 births. These rates of Alu-family DNMs are quite similar to a recently reported measurement of 1 Alu per 40 births in the CEPH cohort,51 while L1 and SVA rate incongruity (each reported in that study as 1/63 births) could result from the approximate 10-fold difference in cohort size, our exclusion of somatic mosaic events (which were included in the Feusier et al., 201951 rates), or from use of three independent MEI detection software tools in that study to achieve extremely high variant recovery. MEI rates were similar between probands and unaffected samples. An additional 12 insertion variants did not belong to any MEI family.
Finally, we compared the sizes of de novo deletions, duplications, and inversions found in ASD probands and unaffected samples. After Benjamini-Hochberg multiple test correction (with alpha = 0.05), we found no significant differences except for the two largest size bins, containing dnSVs impacting at least 100 kb. Such large dnSVs were observed in 112 probands compared to 38 unaffected samples (Figure 4B). This enrichment reflects the known role of large genomic alterations in ASD.46,52, 53, 54
Discussion
To our knowledge, this is the largest direct measurement of dnSVs from family-based WGS to date. Our results demonstrate that parent of origin is the predominant factor influencing the frequency of dnSV events. We also reproduce the well-established result that the de novo rates of SVs and single-nucleotide mutations are higher in ASD probands than in unaffected samples. Although there is consensus that the burden of single-nucleotide and insertion-deletion germline mutations increases with parental age, we find that any correlation between parental age and the burden of de novo structural variation in this study must be modest if present at all. This result suggests that fundamentally different endogenous and exogenous mechanisms create de novo point mutations versus structural mutations, and our results suggest an increased negative selective pressure on large chromosomal rearrangements.
We discovered large NAHR events (n = 45; median size = 1.5 Mb) identified by the presence of large, high-identity segmental duplications flanking the breakpoints of the dnSV. The size of these mutations may explain much of the imbalance between rates in ASD probands and unaffected samples of potential NAHR-derived variants, since larger variants are more likely to be deleterious and are known to be under stronger negative selection.73,74 Identification of repeat-flanked variants is also quite difficult in some cases, especially in small regions, so smaller macrohomology-mediated SVs most likely were not detected. The extensive high-identity breakpoint homologies that are required for NAHR increase the difficulty of mapping and alignment, greatly decreasing the signal of discordant pairs and split reads used by most SV detection algorithms. Smaller variants caused by NAHR are also likely to be missed by depth-based SV calling tools, as the deviations in depth of coverage are often too small for confident assessment of copy number status. Our discovery of a mechanism bias in these ASD-affected individuals is therefore most probably driven by size rather than mechanism.
We note that we identified 63 CNVs in the SSC cohort and two in the CEPH cohort that appear to be somatic in origin and mosaic in blood cells. These mutations had strong discordant and split-read alignment evidence but little to no impact on depth of coverage, most likely reflecting their low cellular prevalence. A similar number of blood-mosaic mutations were observed in probands (n = 35) and unaffected samples (n = 30). Because these mutations did not arise in the germline, they were excluded from our mutation rate analyses. However, depending on when these mutations arose in development, some may be transmittable to the next generation if they arose prior to the establishment of the individual’s germ cell lineage. None of these somatic dnSVs were shared by siblings.
Overall, our analysis of DNMs in over 2,300 families has established confident lower-bound estimates for the rate of SV mutation in the human germline and has quantified the effects of parental age on dnSV risk, as well the mechanisms underlying dnSVs. Although studying the genomes of more than 4,300 offspring and their parents provides substantial power to estimate de novo structural mutation rates, we emphasize that our estimates are lower bounds. While SV detection with short, paired-end WGS has improved dramatically over the last decade, sensitivity remains a challenge, especially for smaller SVs, insertions, and repeat-mediated SVs. Recent comparisons of SVs detected with short-read and long-read technologies, such as PacBio or Oxford Nanopore Technologies, found that thousands of SVs were missed by short-read technologies28,45 and quantified the relative impact of sequence context on detection rates. Therefore, we anticipate that future studies of dnSVs based upon long-reads will increase the detection of dnSVs, especially for smaller mutations arising in tandem-repeat sequences that are known to be hypermutable.75
Declaration of interests
The authors declare no competing interests.
Acknowledgments
We are grateful to the families participating in the Simons Foundation Autism Research Initiative (SFARI) Simplex Collection (SSC) and the Utah individuals who participated in the CEPH consortium. Research and contributing authors were supported by the National Institutes of Health (NIH) grants HG006693, HG009141, and GM124355 to A.R.Q.; MH115957, HD081256, HG008895, HD096326, and HD099547 to M.E.T.; K99DE026824 to H.B.; and GM118335 and GM059290 to L.B.J. and the Simons Foundation Autism Research Initiative (SFARI 573206 to M.E.T. and 388196 to A.R.Q.), with additional support from the Utah Genome Project and the George S. and Dolores Doré Eccles Foundation. M.E.T. was also supported by the Desmond and Ann Heathwood MGH Research Scholars Award. We would like to thank the SSC principal investigators (A.L. Beaudet; R. Bernier; J. Constantino; E.H. Cook, Jr.; E. Fombonne; D. Geschwind; D.E. Grice; A. Klin; D.H. Ledbetter; C. Lord; C.L. Martin; D.M. Martin; R. Maxim; J. Miles; O. Ousley; B. Peterson; J. Piggot; C. Saulnier; M.W. State; W. Stone; J.S. Sutcliffe; C.A. Walsh; and E. Wijsman) and the coordinators and staff at the SSC clinical sites; the SFARI staff, in particular N. Volfovsky; D.B. Goldstein for contributing to the experimental design; the Rutgers University Cell and DNA repository for accessing biomaterials; and the New York Genome Center for generating the WGS data. We also thank Ray White, Jean-Marc Lalouel, and Mark Leppert, who were instrumental in the ascertainment of the CEPH/Utah pedigrees.
Published: March 5, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.02.012.
Contributor Information
Michael E. Talkowski, Email: mtalkowski@mgh.harvard.edu.
Aaron R. Quinlan, Email: aaronquinlan@gmail.com.
Data and code availability
The data and code generated during this study are available at https://github.com/jbelyeu/dnSV-manuscript.
Supplemental information
References
- 1.Arana M.E., Kunkel T.A. Mutator phenotypes due to DNA replication infidelity. Semin. Cancer Biol. 2010;20:304–311. doi: 10.1016/j.semcancer.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Halliday J.A., Glickman B.W. Mechanisms of spontaneous mutation in DNA repair-proficient Escherichia coli. Mutat. Res. 1991;250:55–71. doi: 10.1016/0027-5107(91)90162-h. [DOI] [PubMed] [Google Scholar]
- 3.Keohavong P., Thilly W.G. Fidelity of DNA polymerases in DNA amplification. Proc. Natl. Acad. Sci. USA. 1989;86:9253–9257. doi: 10.1073/pnas.86.23.9253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friedberg E.C. DNA damage and repair. Nature. 2003;421:436–440. doi: 10.1038/nature01408. [DOI] [PubMed] [Google Scholar]
- 5.Cooke M.S., Evans M.D., Dizdaroglu M., Lunec J. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 2003;17:1195–1214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- 6.Marnett L.J. Oxyradicals and DNA damage. Carcinogenesis. 2000;21:361–370. doi: 10.1093/carcin/21.3.361. [DOI] [PubMed] [Google Scholar]
- 7.Stankiewicz P., Lupski J.R. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- 8.Kanaar R., Hoeijmakers J.H.J., van Gent D.C. Molecular mechanisms of DNA double strand break repair. Trends Cell Biol. 1998;8:483–489. doi: 10.1016/s0962-8924(98)01383-x. [DOI] [PubMed] [Google Scholar]
- 9.Sasani T.A., Pedersen B.S., Gao Z., Baird L., Przeworski M., Jorde L.B., Quinlan A.R. Large, three-generation human families reveal post-zygotic mosaicism and variability in germline mutation accumulation. eLife. 2019;8:e46922. doi: 10.7554/eLife.46922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.An J.Y., Lin K., Zhu L., Werling D.M., Dong S., Brand H., Wang H.Z., Zhao X., Schwartz G.B., Collins R.L. Genome-wide de novo risk score implicates promoter variation in autism spectrum disorder. Science. 2018;362:eaat6576. doi: 10.1126/science.aat6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Werling D.M., Brand H., An J.-Y.Y., Stone M.R., Zhu L., Glessner J.T., Collins R.L., Dong S., Layer R.M., Markenscoff-Papadimitriou E. An analytical framework for whole-genome sequence association studies and its implications for autism spectrum disorder. Nat. Genet. 2018;50:727–736. doi: 10.1038/s41588-018-0107-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jónsson H., Sulem P., Kehr B., Kristmundsdottir S., Zink F., Hjartarson E., Hardarson M.T., Hjorleifsson K.E., Eggertsson H.P., Gudjonsson S.A. Parental influence on human germline de novo mutations in 1,548 trios from Iceland. Nature. 2017;549:519–522. doi: 10.1038/nature24018. [DOI] [PubMed] [Google Scholar]
- 13.Kong A., Frigge M.L., Masson G., Besenbacher S., Sulem P., Magnusson G., Gudjonsson S.A., Sigurdsson A., Jonasdottir A., Jonasdottir A. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldmann J.M., Wong W.S.W., Pinelli M., Farrah T., Bodian D., Stittrich A.B., Glusman G., Vissers L.E.L.M., Hoischen A., Roach J.C. Parent-of-origin-specific signatures of de novo mutations. Nat. Genet. 2016;48:935–939. doi: 10.1038/ng.3597. [DOI] [PubMed] [Google Scholar]
- 15.Shendure J., Akey J.M. The origins, determinants, and consequences of human mutations. Science. 2015;349:1478–1483. doi: 10.1126/science.aaa9119. [DOI] [PubMed] [Google Scholar]
- 16.Scally A., Durbin R. Revising the human mutation rate: implications for understanding human evolution. Nat. Rev. Genet. 2012;13:745–753. doi: 10.1038/nrg3295. [DOI] [PubMed] [Google Scholar]
- 17.Roach J.C., Glusman G., Smit A.F.A., Huff C.D., Hubley R., Shannon P.T., Rowen L., Pant K.P., Goodman N., Bamshad M. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328:636–639. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turner T.N., Coe B.P., Dickel D.E., Hoekzema K., Nelson B.J., Zody M.C., Kronenberg Z.N., Hormozdiari F., Raja A., Pennacchio L.A. Genomic Patterns of De Novo Mutation in Simplex Autism. Cell. 2017;171:710–722.e12. doi: 10.1016/j.cell.2017.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma R., Deng L., Xia Y., Wei X., Cao Y., Guo R., Zhang R., Guo J., Liang D., Wu L. A clear bias in parental origin of de novo pathogenic CNVs related to intellectual disability, developmental delay and multiple congenital anomalies. Sci. Rep. 2017;7:44446. doi: 10.1038/srep44446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Redin C., Brand H., Collins R.L., Kammin T., Mitchell E., Hodge J.C., Hanscom C., Pillalamarri V., Seabra C.M., Abbott M.A. The genomic landscape of balanced cytogenetic abnormalities associated with human congenital anomalies. Nat. Genet. 2017;49:36–45. doi: 10.1038/ng.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiang C., Jacobsen J.C., Ernst C., Hanscom C., Heilbut A., Blumenthal I., Mills R.E., Kirby A., Lindgren A.M., Rudiger S.R. Complex reorganization and predominant non-homologous repair following chromosomal breakage in karyotypically balanced germline rearrangements and transgenic integration. Nat. Genet. 2012;44:390–397. doi: 10.1038/ng.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Talkowski M.E., Rosenfeld J.A., Blumenthal I., Pillalamarri V., Chiang C., Heilbut A., Ernst C., Hanscom C., Rossin E., Lindgren A.M. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell. 2012;149:525–537. doi: 10.1016/j.cell.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu B., Roos J.L., Levy S., van Rensburg E.J., Gogos J.A., Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat. Genet. 2008;40:880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- 24.Stefansson H., Rujescu D., Cichon S., Pietiläinen O.P.H., Ingason A., Steinberg S., Fossdal R., Sigurdsson E., Sigmundsson T., Buizer-Voskamp J.E., GROUP Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cameron D.L., Di Stefano L., Papenfuss A.T. Comprehensive evaluation and characterisation of short read general-purpose structural variant calling software. Nat. Commun. 2019;10:3240. doi: 10.1038/s41467-019-11146-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahmoud M., Gobet N., Cruz-Dávalos D.I., Mounier N., Dessimoz C., Sedlazeck F.J. Structural variant calling: the long and the short of it. Genome Biol. 2019;20:246. doi: 10.1186/s13059-019-1828-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kosugi S., Momozawa Y., Liu X., Terao C., Kubo M., Kamatani Y. Comprehensive evaluation of structural variation detection algorithms for whole genome sequencing. Genome Biol. 2019;20:117. doi: 10.1186/s13059-019-1720-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaisson M.J.P., Sanders A.D., Zhao X., Malhotra A., Porubsky D., Rausch T., Gardner E.J., Rodriguez O.L., Guo L., Collins R.L. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat. Commun. 2019;10:1784. doi: 10.1038/s41467-018-08148-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen X., Schulz-Trieglaff O., Shaw R., Barnes B., Schlesinger F., Källberg M., Cox A.J., Kruglyak S., Saunders C.T. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32:1220–1222. doi: 10.1093/bioinformatics/btv710. [DOI] [PubMed] [Google Scholar]
- 30.Kronenberg Z.N., Osborne E.J., Cone K.R., Kennedy B.J., Domyan E.T., Shapiro M.D., Elde N.C., Yandell M. Wham: Identifying Structural Variants of Biological Consequence. PLoS Comput. Biol. 2015;11:e1004572. doi: 10.1371/journal.pcbi.1004572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Layer R.M., Chiang C., Quinlan A.R., Hall I.M. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. 2014;15:R84. doi: 10.1186/gb-2014-15-6-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rausch T., Zichner T., Schlattl A., Stütz A.M., Benes V., Korbel J.O. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–i339. doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kloosterman W.P., Francioli L.C., Hormozdiari F., Marschall T., Hehir-Kwa J.Y., Abdellaoui A., Lameijer E.W., Moed M.H., Koval V., Renkens I., Genome of Netherlands Consortium Characteristics of de novo structural changes in the human genome. Genome Res. 2015;25:792–801. doi: 10.1101/gr.185041.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klambauer G., Schwarzbauer K., Mayr A., Clevert D.-A., Mitterecker A., Bodenhofer U., Hochreiter S. cn.MOPS: mixture of Poissons for discovering copy number variations in next-generation sequencing data with a low false discovery rate. Nucleic Acids Res. 2012;40:e69. doi: 10.1093/nar/gks003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abyzov A., Urban A.E., Snyder M., Gerstein M. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011;21:974–984. doi: 10.1101/gr.114876.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu P., Lacaria M., Zhang F., Withers M., Hastings P.J., Lupski J.R. Frequency of nonallelic homologous recombination is correlated with length of homology: evidence that ectopic synapsis precedes ectopic crossing-over. Am. J. Hum. Genet. 2011;89:580–588. doi: 10.1016/j.ajhg.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharp A.J., Locke D.P., McGrath S.D., Cheng Z., Bailey J.A., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R. Segmental duplications and copy-number variation in the human genome. Am. J. Hum. Genet. 2005;77:78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bailey J.A., Yavor A.M., Massa H.F., Trask B.J., Eichler E.E. Segmental duplications: organization and impact within the current human genome project assembly. Genome Res. 2001;11:1005–1017. doi: 10.1101/gr.187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee J.A., Carvalho C.M.B., Lupski J.R. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 40.Hastings P.J., Ira G., Lupski J.R. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lees-Miller S.P., Meek K. Repair of DNA double strand breaks by non-homologous end joining. Biochimie. 2003;85:1161–1173. doi: 10.1016/j.biochi.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 42.Collins R.L., Brand H., Karczewski K.J., Zhao X., Alföldi J., Francioli L.C., Khera A.V., Lowther C., Gauthier L.D., Wang H., Genome Aggregation Database Production Team. Genome Aggregation Database Consortium A structural variation reference for medical and population genetics. Nature. 2020;581:444–451. doi: 10.1038/s41586-020-2287-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pedersen B.S., Quinlan A.R. Duphold: scalable, depth-based annotation and curation of high-confidence structural variant calls. Gigascience. 2019;8:giz040. doi: 10.1093/gigascience/giz040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Belyeu J.R., Nicholas T.J., Pedersen B.S., Sasani T.A., Havrilla J.M., Kravitz S.N., Conway M.E., Lohman B.K., Quinlan A.R., Layer R.M. SV-plaudit: A cloud-based framework for manually curating thousands of structural variants. Gigascience. 2018;7:1–7. doi: 10.1093/gigascience/giy064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao X., Collins R.L., Lee W.-P., Weber A.M., Jun Y., Zhu Q., Weisburd B., Huang Y., Audano P.A., Wang H. Expectations and blind spots for structural variation detection from short-read alignment and long-read assembly. BioRxiv. 2020 doi: 10.1101/2020.07.03.168831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sebat J., Lakshmi B., Malhotra D., Troge J., Lese-Martin C., Walsh T., Yamrom B., Yoon S., Krasnitz A., Kendall J. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brandler W.M., Antaki D., Gujral M., Kleiber M.L., Whitney J., Maile M.S., Hong O., Chapman T.R., Tan S., Tandon P. Paternally inherited cis-regulatory structural variants are associated with autism. Science. 2018;360:327–331. doi: 10.1126/science.aan2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xing J., Watkins W.S., Witherspoon D.J., Zhang Y., Guthery S.L., Thara R., Mowry B.J., Bulayeva K., Weiss R.B., Jorde L.B. Fine-scaled human genetic structure revealed by SNP microarrays. Genome Res. 2009;19:815–825. doi: 10.1101/gr.085589.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kazazian H.H., Jr., Moran J.V. Mobile DNA in health and disease. N. Engl. J. Med. 2017;377:361–370. doi: 10.1056/NEJMra1510092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hancks D.C., Kazazian H.H., Jr. Roles for retrotransposon insertions in human disease. Mob. DNA. 2016;7:9. doi: 10.1186/s13100-016-0065-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feusier J., Watkins W.S., Thomas J., Farrell A., Witherspoon D.J., Baird L., Ha H., Xing J., Jorde L.B. Pedigree-based estimation of human mobile element retrotransposition rates. Genome Res. 2019;29:1567–1577. doi: 10.1101/gr.247965.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brandler W.M., Antaki D., Gujral M., Noor A., Rosanio G., Chapman T.R., Barrera D.J., Lin G.N., Malhotra D., Watts A.C. Frequency and Complexity of De Novo Structural Mutation in Autism. Am. J. Hum. Genet. 2016;98:667–679. doi: 10.1016/j.ajhg.2016.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sanders S.J., Ercan-Sencicek A.G., Hus V., Luo R., Murtha M.T., Moreno-De-Luca D., Chu S.H., Moreau M.P., Gupta A.R., Thomson S.A. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoon S., Xuan Z., Makarov V., Ye K., Sebat J. Sensitive and accurate detection of copy number variants using read depth of coverage. Genome Res. 2009;19:1586–1592. doi: 10.1101/gr.092981.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Duyzend M.H., Nuttle X., Coe B.P., Baker C., Nickerson D.A., Bernier R., Eichler E.E. Maternal Modifiers and Parent-of-Origin Bias of the Autism-Associated 16p11.2 CNV. Am. J. Hum. Genet. 2016;98:45–57. doi: 10.1016/j.ajhg.2015.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Girard S.L., Bourassa C.V., Lemieux Perreault L.P., Legault M.A., Barhdadi A., Ambalavanan A., Brendgen M., Vitaro F., Noreau A., Dionne G. Paternal age explains a major portion of De novo germline mutation rate variability in healthy individuals. PLoS ONE. 2016;11:e0164212. doi: 10.1371/journal.pone.0164212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brand H., Collins R.L., Hanscom C., Rosenfeld J.A., Pillalamarri V., Stone M.R., Kelley F., Mason T., Margolin L., Eggert S. Paired-Duplication Signatures Mark Cryptic Inversions and Other Complex Structural Variation. Am. J. Hum. Genet. 2015;97:170–176. doi: 10.1016/j.ajhg.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Collins R.L., Brand H., Redin C.E., Hanscom C., Antolik C., Stone M.R., Glessner J.T., Mason T., Pregno G., Dorrani N. Defining the diverse spectrum of inversions, complex structural variation, and chromothripsis in the morbid human genome. Genome Biol. 2017;18:36. doi: 10.1186/s13059-017-1158-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Francioli L.C., Cretu-Stancu M., Garimella K.V., Fromer M., Kloosterman W.P., Samocha K.E., Neale B.M., Daly M.J., Banks E., DePristo M.A., de Bakker P.I., Genome of the Netherlands consortium A framework for the detection of de novo mutations in family-based sequencing data. Eur. J. Hum. Genet. 2017;25:227–233. doi: 10.1038/ejhg.2016.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li B., Chen W., Zhan X., Busonero F., Sanna S., Sidore C., Cucca F., Kang H.M., Abecasis G.R. A likelihood-based framework for variant calling and de novo mutation detection in families. PLoS Genet. 2012;8:e1002944. doi: 10.1371/journal.pgen.1002944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Belyeu J.R., Chowdhury M., Brown J., Pedersen B.S., Cormier M.J., Quinlan A.R., Layer R.M. Samplot: A Platform for Structural Variant Visual Validation and Automated Filtering. bioRxiv. 2020 doi: 10.1101/2020.09.23.310110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Quinlan A.R., Clark R.A., Sokolova S., Leibowitz M.L., Zhang Y., Hurles M.E., Mell J.C., Hall I.M. Genome-wide mapping and assembly of structural variant breakpoints in the mouse genome. Genome Res. 2010;20:623–635. doi: 10.1101/gr.102970.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Davis A.J., Chen D.J. DNA double strand break repair via non-homologous end-joining. Transl. Cancer Res. 2013;2:130–143. doi: 10.3978/j.issn.2218-676X.2013.04.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dausset J., Cann H., Cohen D., Lathrop M., Lalouel J.M., White R. Centre d’etude du polymorphisme humain (CEPH): collaborative genetic mapping of the human genome. Genomics. 1990;6:575–577. doi: 10.1016/0888-7543(90)90491-c. [DOI] [PubMed] [Google Scholar]
- 66.Fischbach G.D., Lord C. The Simons Simplex Collection: a resource for identification of autism genetic risk factors. Neuron. 2010;68:192–195. doi: 10.1016/j.neuron.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 67.Gardner E.J., Lam V.K., Harris D.N., Chuang N.T., Scott E.C., Pittard W.S., Mills R.E., Devine S.E., 1000 Genomes Project Consortium The Mobile Element Locator Tool (MELT): population-scale mobile element discovery and biology. Genome Res. 2017;27:1916–1929. doi: 10.1101/gr.218032.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Frankish A., Diekhans M., Ferreira A.M., Johnson R., Jungreis I., Loveland J., Mudge J.M., Sisu C., Wright J., Armstrong J. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47(D1):D766–D773. doi: 10.1093/nar/gky955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cormier M.J., Belyeu J.R., Pedersen B.S., Brown J., Koster J., Quinlan A.R. Go Get Data (GGD): simple, reproducible access to scientific data. BioRxiv. 2020 doi: 10.1101/2020.09.10.291377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reichenberg A., Gross R., Weiser M., Bresnahan M., Silverman J., Harlap S., Rabinowitz J., Shulman C., Malaspina D., Lubin G. Advancing paternal age and autism. Arch. Gen. Psychiatry. 2006;63:1026–1032. doi: 10.1001/archpsyc.63.9.1026. [DOI] [PubMed] [Google Scholar]
- 71.Kumar R.A., KaraMohamed S., Sudi J., Conrad D.F., Brune C., Badner J.A., Gilliam T.C., Nowak N.J., Cook E.H., Jr., Dobyns W.B., Christian S.L. Recurrent 16p11.2 microdeletions in autism. Hum. Mol. Genet. 2008;17:628–638. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- 72.Veenstra-VanderWeele J., Cook E.H., Jr. Molecular genetics of autism spectrum disorder. Mol. Psychiatry. 2004;9:819–832. doi: 10.1038/sj.mp.4001505. [DOI] [PubMed] [Google Scholar]
- 73.Collins R.L., Brand H., Karczewski K.J., Zhao X., Alföldi J., Khera A.V., Francioli L.C., Gauthier L.D., Wang H., Watts N.A. An open resource of structural variation for medical and population genetics. bioRxiv. 2019 doi: 10.1101/578674. [DOI] [Google Scholar]
- 74.Sudmant P.H., Rausch T., Gardner E.J., Handsaker R.E., Abyzov A., Huddleston J., Zhang Y., Ye K., Jun G., Fritz M.H.-Y., 1000 Genomes Project Consortium An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chakraborty R., Kimmel M., Stivers D.N., Davison L.J., Deka R. Relative mutation rates at di-, tri-, and tetranucleotide microsatellite loci. Proc. Natl. Acad. Sci. USA. 1997;94:1041–1046. doi: 10.1073/pnas.94.3.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and code generated during this study are available at https://github.com/jbelyeu/dnSV-manuscript.