Summary
Genetic studies in underrepresented populations identify disproportionate numbers of novel associations. However, most genetic studies use genotyping arrays and sequenced reference panels that best capture variation most common in European ancestry populations. To compare data generation strategies best suited for underrepresented populations, we sequenced the whole genomes of 91 individuals to high coverage as part of the Neuropsychiatric Genetics of African Population-Psychosis (NeuroGAP-Psychosis) study with participants from Ethiopia, Kenya, South Africa, and Uganda. We used a downsampling approach to evaluate the quality of two cost-effective data generation strategies, GWAS arrays versus low-coverage sequencing, by calculating the concordance of imputed variants from these technologies with those from deep whole-genome sequencing data. We show that low-coverage sequencing at a depth of ≥4× captures variants of all frequencies more accurately than all commonly used GWAS arrays investigated and at a comparable cost. Lower depths of sequencing (0.5–1×) performed comparably to commonly used low-density GWAS arrays. Low-coverage sequencing is also sensitive to novel variation; 4× sequencing detects 45% of singletons and 95% of common variants identified in high-coverage African whole genomes. Low-coverage sequencing approaches surmount the problems induced by the ascertainment of common genotyping arrays, effectively identify novel variation particularly in underrepresented populations, and present opportunities to enhance variant discovery at a cost similar to traditional approaches.
Keywords: low-coverage sequencing, GWAS, GWAS arrays, whole-genome sequencing, cost comparison, study design, Africa
Introduction
Over the last decade, genome-wide association studies (GWASs) have grown rapidly, deepening biological insights into a breadth of human diseases. Data for these studies are usually generated with GWAS arrays because of their cost effectiveness and the availability of commonly used analytical pipelines. These arrays typically genotype a fixed set of hundreds of thousands to millions of common variants genome wide, and additional linked variants are then imputed with haplotype reference panels.1 The utility of this approach varies across populations, however, because most GWAS arrays consist of variants that are most common in European ancestry populations.2 Further compounding unequal genomic coverage issues, reference data for imputation are also vastly Eurocentric.3, 4, 5
Recognition of these biases in genomic infrastructure has driven concerted efforts to develop specialized, scalable arrays designed to capture variation common to different continental ancestries.6 For example, the Population Architecture using Genomics and Epidemiology (PAGE) Consortium designed the Illumina Multi-Ethnic Genotyping Array (MEGA), a dense array of ∼1.7 million variants, which aimed to improve performance for imputation across globally diverse populations.3 A significant portion of the ∼660,000 variants on the Global Screening Array (GSA)–designed to decrease costs, increase scalability, and improve imputation accuracy in European populations–consists of a subset of variants from MEGA. Additionally, the Human Heredity and Health in Africa (H3Africa) Consortium developed a dense array of ∼2.5 million variants specialized for the higher genetic diversity and smaller haplotype blocks in African genomes.7 Although these arrays all have potential benefits, an inherent weakness to their ascertained nature is that they cannot capture novel variants.
As sequencing costs have dropped, low-pass sequencing has been proposed as a similarly priced and unbiased alternative to GWAS arrays in, for example, population genetics and polygenic score analysis.8, 9, 10, 11, 12 Sequencing offers several advantages: (1) variants are unascertained, meaning that the quality of data generated is inherently unbiased toward any particular population; (2) novel, population-specific variants can be detected and used to further advance the generation of haplotype reference panels; (3) DNA strand is unambiguous given the alignment of sequencing reads to a reference genome; and (4) non-human microbiome DNA can be captured and variation analyzed with certain DNA sampling procedures. These advantages are expected to be especially beneficial in non-European populations because corresponding reference data that support arrays are often lacking.
Here, we have generated high-coverage whole-genome sequencing data from populations vastly underrepresented in genetics research to compare data quality that would be produced by sequencing at various depths versus genotyping with several commonly used arrays. We have also compared the costs and analytical approaches that are feasible from each data generation approach. To compare data generation strategies, we included whole genomes that were sequenced as part of the Neuropsychiatric Genetics of African Populations Psychosis (NeuroGAP-Psychosis) study spanning five sites across four countries in eastern and southern Africa.13 These populations are of particular interest because humans originated in Africa, resulting in high levels of genetic variation and rapid linkage disequilibrium decay, highlighting the disproportionate informativeness of African genomes for human evolutionary studies and in pinpointing causal variants. Thus, accurately capturing genetic variation in these populations in an unbiased manner is particularly important for associating, resolving, and interpreting genetic associations while ensuring equitable translation of genetic technologies. Our results highlight that low-coverage sequencing can be a more appropriate data generation strategy than GWAS arrays for assaying genetic variation across globally diverse populations.
Subjects and methods
Human subjects
Ethical and safety considerations are being taken across multiple levels, as described in greater detail previously.13 Because the subjects the study aims to recruit are deemed vulnerable populations, additional measures are taken to protect them. Potential participants are excluded if they are presenting with severe, intrusive levels of psychiatric symptoms at the time of consent. Additionally, researcher assistants use the University of California, San Diego Brief Assessment of Capacity to Consent (UBACC) system14,15 during the consent process to make sure participants understand the study, what is required of them, and that they can withdraw at any point. Participants who pass the UBACC and who want to continue are required to provide written informed consent or a fingerprint in lieu of a signature. No protected health information (PHI) or Health Insurance Portability and Accountability Act (HIPPA) identifiers are collected as part of the phenotypic or genetic dataset.
Ethical clearances to conduct this study have been obtained from all participating sites, including
-
•
Ethiopia: Addis Ababa University College of Health Sciences (#014/17/Psy) and the Ministry of Science and Technology National Research Ethics Review Committee (#3.10/14/2018);
-
•
Kenya: Moi University College of Health Sciences/Moi Teaching and Referral Hospital Institutional Research and Ethics Committee (IREC) (#IREC/2016/145, approval number: IREC 1727), Kenya National Council of Science and Technology (#NACOSTI/P/17/56302/19576) KEMRI Centre Scientific Committee (CSC# KEMRI/CGMRC/CSC/070/2016), KEMRI Scientific and Ethics Review Unit (SERU# KEMRI/SERU/CGMR-C/070/3575);
-
•
South Africa: The University of Cape Town Human Research Ethics Committee (#466/2016);
-
•
Uganda: The Makerere University School of Medicine Research and Ethics Committee (SOMREC #REC REF 2016-057) and the Uganda National Council for Science and Technology (UNCST #HS14ES);
-
•
USA: The Harvard T.H. Chan School of Public Health (#IRB17-0822).
Human whole-genome sequencing PCR-free (v.1.1–v.1.3)
Preparation of libraries for cluster amplification and sequencing
An aliquot of genomic DNA (350 ng in 50 mL) was used as the input into DNA fragmentation (also known as shearing). Shearing was performed acoustically with a Covaris focused-ultrasonicator, targeting 385 bp fragments. Following fragmentation, additional size selection was performed with a solid phase reversible immobilization (SPRI) cleanup. Library preparation was performed with a commercially available kit provided by KAPA Biosystems (KAPA Hyper Prep without amplification module, product KK8505) and with palindromic forked adapters with unique 8-base index sequences embedded within the adaptor (purchased from Roche). Following sample preparation, libraries were quantified via qPCR (kit purchased from KAPA Biosystems) with probes specific to the ends of the adapters. This assay was automated via Agilent’s Bravo liquid handling platform. On the basis of qPCR quantification, libraries were normalized to 2.2 nM and pooled into 24-plexes.
Cluster amplification and sequencing (NovaSeq 6000)
Sample pools were combined with NovaSeq Cluster Amp Reagents DPX1, DPX2, and DPX3 and loaded into single lanes of a NovaSeq 6000 S4 flow cell via the Hamilton Starlet Liquid Handling system. Cluster amplification and sequencing occurred on NovaSeq 6000 instruments utilizing sequencing-by-synthesis kits to produce 151 bp paired-end reads. We processed output from Illumina software to yield CRAM or BAM files containing demultiplexed, aggregated aligned reads. All sample information tracking was performed by automated lab information management system (LIMS) messaging.
Variant calling
We used the GATK best practices pipeline described for variant calling by using code (available via web resources). Cromwell was used to submit most jobs in parallel across the genome where possible using the Google Cloud Platform (web resources).
Depth of coverage
Depth statistics from high-coverage whole genomes were computed by the Broad Institute’s Data Science Platform team. This calculation excluded low-quality, unmapped, unpaired, and duplicate reads in depth of coverage calculations.
Downsampling sequencing reads
We downsampled reads by using the GATK DownsampleSam module, which retains a deterministically random subset of reads and their mate pairs. We calculated the probability used for downsampling on the basis of depth of coverage as described above (i.e., not simply on the basis of the total number of reads sequenced relative to the number of bases in the human genome because, for example, some reads from saliva-derived DNA may not be human).
Concordance
We computed non-reference concordance among homozygous reference, heterozygous, and homozygous non-reference calls, excluding no call and missing sites from counts, according to Table 1:
We excluded homozygous reference concordant calls (m) to avoid high concordance among rarer variants by simply imputing the most common allele.
Table 1.
Concordance among "truth" dataset of high-coverage genomes versus comparison datasets, which consist of either downsampled genomes (i.e., simulated low-coverage genomes) or filtered genomes (i.e., simulated GWAS array data)
| No call | ./. | 0/0 | 0/1 | 1/1 | |
|---|---|---|---|---|---|
| No call | a | b | c | d | e |
| ./. | f | g | h | i | j |
| 0/0 | k | l | m | n | o |
| 0/1 | p | q | r | s | t |
| 1/1 | u | v | w | x | y |
Haplotype reference
We downloaded phased 1000 Genomes haplotype reference data containing SNPs aligned to GRCh38 (web resources). We used these phased haplotypes for genotype refinement, phasing, and imputation.
Genotype refinement, phasing, and imputation
We used Beagle 4.1 for genotype refinement of variant calls in downsampled sequencing data with the 1000 Genomes Project phase 3 as reference haplotypes prior to phasing and imputation by using the genotype likelihoods (gl), ref, and map arguments with impute = false. As described in the Beagle 4.1 manual, this combination of arguments estimates the posterior genotype probability by using a reference panel with non-missing genotypes and phased data, producing as output an unphased VCF. We then used Beagle 5.1 for phasing and imputation also by using the 1000 Genomes Project phase 3 data both for low-coverage sequencing data and GWAS array data, this time with the genotype (gt), ref, map, and impute = true arguments (Figure 1D).
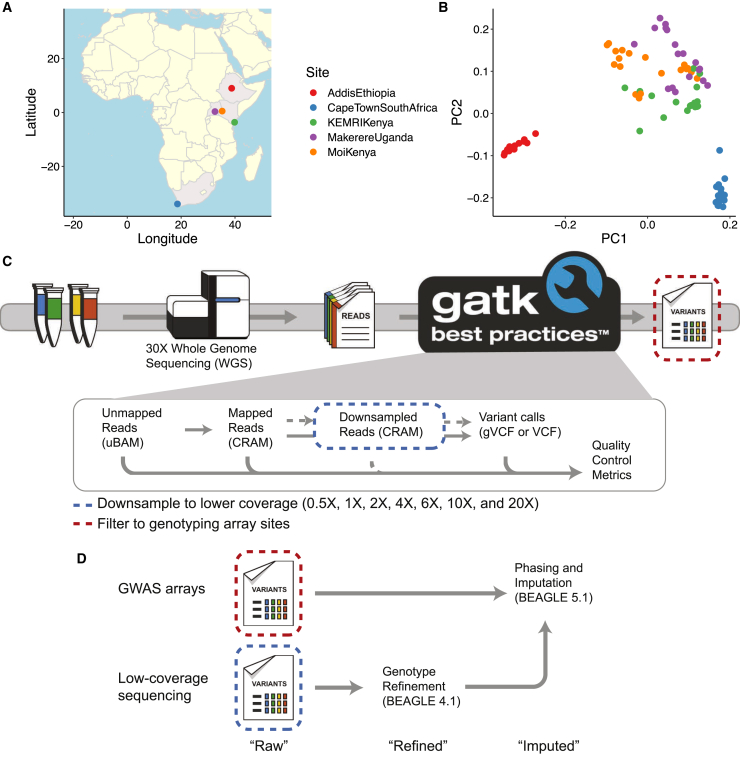
Figure 1.
Populations and sites included in high-coverage whole-genome sequence data and downsampling schema to assess the performance of lower-coverage sequencing versus GWAS arrays
(A) Map indicating where participants in the NeuroGAP-Psychosis study are enrolled in this dataset.
(B) The first two principal components (PCs) show variation within and among populations. They first distinguish the Ethiopians, and then the South Africans, from other African populations. Colors are consistent in (A) and (B).
(C) High-coverage genomes were processed with the GATK best practices pipeline. To mimic lower-coverage sequencing data, we downsampled analysis-ready CRAM files to various depths, followed by a standard implementation of the variant calling pipeline. To mimic GWAS array data, we filtered the variants called from the high-coverage sequencing data to only those sites on the arrays.
(D) After variants were filtered from high-coverage data to sites on GWAS arrays, they were phased and imputed with Beagle 5.1. After downsampling reads from high-coverage data to various depths of coverage, we refined genotypes by using Beagle 4.1 (the last version of Beagle to provide this feature), then phased and imputed them by using Beagle 5.1, as with GWAS arrays. “Raw” indicates that variant calls were produced directly from GATK with no genotype refinement or imputation, “refined” indicates variant calls from genotype refinement without imputation, and “imputed” indicates imputed variants following genotype refinement.
Gencove imputation
We generated FASTQ files from analysis-ready BAM files by using bedtools bamtofastq. We then uploaded these FASTQ files to the Gencove server, ran imputation and related analyses, and then downloaded imputation results.
Results
To compare genetic data quality from variable depths of sequencing versus commonly used GWAS arrays, we sequenced the whole genomes of participants from the NeuroGAP-Psychosis study to high coverage (target coverage of ≥30× per individual, mean coverage = 38×, all ≥20×, Figure S1). This study consists of data from five geographical sites (n = 91, with n ≥ 17 individuals per site) across eastern and southern Africa (Table 2, Figures 1A and 1B). Participants in these studies were chosen from a larger set of genotyped individuals on the basis of ancestry patterns representative of the enrollment site. They come from a range of ethnic groups, and more than five individuals per NeuroGAP-Psychosis recruitment site reported the following primary ethnicities: Amhara and Oromo from Addis Ababa, Ethiopia; Xhosa from Cape Town, South Africa; Mijikenda from Kilifi, Kenya; and the Kalenjin from Eldoret, Kenya (Table S1). There was no predominantly reported primary ethnicity among the 18 individuals from Kampala, Uganda; rather, 11 different ethnic groups were reported among these individuals.
Table 2.
Genetic samples included in these sequencing analyses
| Institution | Geographical site | Number of individuals | Number of variants (mean ± SD) | Depth of coverage (mean ± SD) |
|---|---|---|---|---|
| Addis Ababa University | Addis Ababa, Ethiopia | 17 | 3,988,434 ± 45,857 | 36.3 ± 8.03 |
| KEMRI-WT-Coast | Kilifi, Kenya | 19 | 4,284,557 ± 32,558 | 37.4 ± 6.21 |
| Makerere University | Kampala, Uganda | 18 | 4,297,527 ± 24,234 | 40.9 ± 8.56 |
| Moi University | Eldoret, Kenya | 19 | 4,246,784 ± 46,903 | 37.2 ± 7.27 |
| University of Cape Town | Cape Town, South Africa | 18 | 4,410,899 ± 14,966 | 37.6 ± 6.19 |
19 samples from Ethiopia were sequenced, but two showed significant evidence of contamination, so they were excluded from variant calling metrics and all downstream analyses. The number of variants reported are per individual non-reference variant calls.
An in silico framework for evaluating data generation strategies with high-coverage WGS data
We considered variant calls generated from all reads to be our “truth” variant calls throughout our analyses. Across all individuals and geographical sites, these high-coverage whole genomes contain 26 million variants, and there were more than 4 million non-reference variants per individual in all populations except in Ethiopia (Table 2). Consistent with our results, prior studies of Ethiopian genetics have shown reductions in genetic diversity compared with other African populations because of back-to-Africa migrations from the Middle East.16, 17, 18
We next downsampled or subset our data to simulate low-coverage and GWAS array data generation, respectively, by using two approaches (Figure 1C). First, we downsampled analysis-ready CRAM files to the number of reads corresponding to 0.5×, 1×, 2×, 4×, 6×, 10×, and 20× coverage (subjects and methods). With these downsampled data, we then generated new variant call sets corresponding to these depths (Table S2) and performed variant quality control by using standard analysis pipelines (subjects and methods). Second, we subset variants from the high-coverage “truth” data corresponding to all polymorphic sites that would have been probed by using each of the following Illumina arrays: the GSA, PsychChip, MEGA, H3Africa, and Omni2.5. For both of these datasets, we then compared the imputed data to the high-coverage variant calls to assess the number and quality of sites obtained.
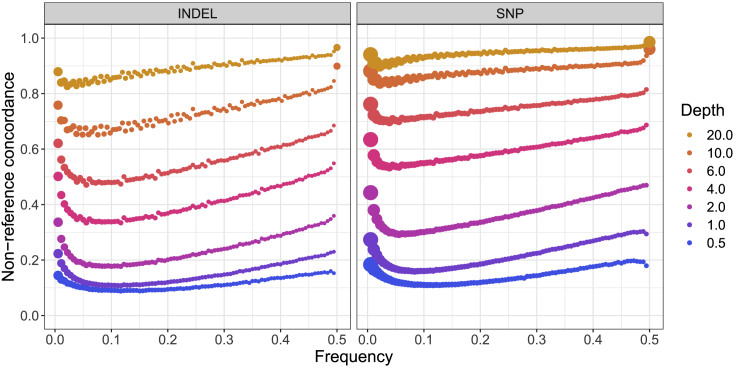
We first compared the downsampled whole-genome sequencing data (“raw”) to the highest depth “truth” prior to any genotype refinement (“refined”) or imputation (“imputed”). Compared with high-coverage sequencing data, we expect low-coverage sequencing to produce variant calls that have higher error rates and miss some genetic variants altogether because of the reduced chance of observing both alleles with high-quality reads across regions of the genome. We therefore calculated non-reference concordance (subjects and methods) between the downsampled variant call sets and the full coverage data (Figure 2, Table S3). Non-reference concordance was lower for indels than SNPs and was lowest for variants with ∼5% frequency, as has been seen previously.19 This shape reflects the need for higher genotyping quality metrics to call singleton and low-frequency variants compared with common variants; a similar shape of curve relates frequency and the mean genotype quality (GQ) metric.
Figure 2.
Pre-imputation non-reference variant concordance
We computed non-reference concordance comparing downsampled data at several depths of coverage to the highest depth sequencing call set available for all samples. The size of each dot is proportional to the number of variants in each bin. Depth summaries across samples are shown in Figure S1. Non-reference concordances averaged across variants of all allele frequencies are shown in Table S3.
After generating variant calls for low-coverage sequencing data by using GATK (“raw”), we next used Beagle, open-source software described previously,20,21 for genotype refinement and imputation of low-coverage data, an approach taken in previous studies that used low-coverage sequencing (subjects and methods, Figure 1D).9,22,23 Genotype refinement is designed to correct low-quality genotype calls via a haplotype reference panel of high-confidence genotypes and considers genotype likelihoods rather than hard calls (“refined”). Afterward, imputation uses the refined genotype calls to fill in variants from the reference panel for sites not originally called (“imputed,” Figure 1D). We performed genotype refinement and imputation on low-coverage sequencing up to 6× by using 1000 Genomes phase 3 data as a haplotype reference panel.24 We excluded the higher depths, 10× and 20×, given their already high concordance without refinement (Figure 2) and to save computational costs. To compare variant calls obtained from our whole-genome sequencing experiment with several commonly used genotyping arrays, we filtered variants from the high-coverage “truth” dataset to those on the array and then imputed genotypes by using the same methodology as in the downsampled sequencing data (Figure 1B).
Comparison of data quality from imputed GWAS array versus low-coverage sequencing data
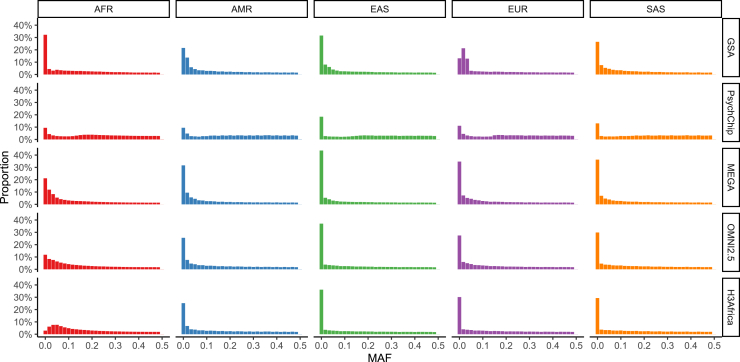
We first compared non-reference concordance in the low- versus high-coverage sequencing data by using variant calls through each step of the process, including the raw data ("raw"), after genotype refinement ("refined"), and after imputation ("imputed," subjects and methods). The total numbers of SNPs through each processing step are shown in Table 3 (imputed > raw > refined). Prior to imputation, we identify approximately 13 million variants from 1× sequencing compared to the 26 million in the high-coverage data (∼50%). This is a considerably larger number of polymorphic variants than are genotyped on any array (Table 3). A relatively low fraction of sites on some arrays are polymorphic in NeuroGAP-Psychosis (e.g., only 68.8% of sites on GSA are polymorphic). We compared this across 1000 Genomes populations by calculating the mean proportion of SNPs at various frequencies on several GWAS arrays (Figure 3). Of sites on the GSA array that were present in any individual in the 1000 Genomes Project, 3.8% versus 8.9% were monomorphic in the EUR versus AFR super populations, respectively, which were substantially better than in NeuroGAP-Psychosis. These findings reflect the fact that the 1000 Genomes Project is often used to select variants for SNP arrays and that AFR populations in the 1000 Genomes Project are poor proxies for those in NeuroGAP-Psychosis.
Table 3.
Sensitivity of various sequencing depths and GWAS arrays to detect singletons and common variants through several analytical steps
| # of SNPs | % singletons present in full set | % common variants in full set | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Call set | Raw | Refined | Imputed | Raw | Refined | Imputed | Raw | Refined | Imputed |
| 0.5× | 9,236,562 | 7,452,675 | 18,414,145 | 0.04 | 0.01 | 0.33 | 0.55 | 0.40 | 0.62 |
| 1× | 13,036,891 | 10,389,726 | 18,974,677 | 0.09 | 0.03 | 0.33 | 0.74 | 0.52 | 0.62 |
| 2× | 15,716,019 | 13,387,436 | 19,887,495 | 0.2 | 0.08 | 0.33 | 0.88 | 0.59 | 0.62 |
| 4× | 20,958,987 | 16,458,866 | 21,083,626 | 0.45 | 0.17 | 0.33 | 0.95 | 0.61 | 0.62 |
| 6× | 23,352,341 | 17,633,642 | 21,402,104 | 0.62 | 0.23 | 0.33 | 0.97 | 0.61 | 0.62 |
| 10× | 24,955,954 | N/A | N/A | 0.8 | N/A | N/A | 0.98 | N/A | N/A |
| 20× | 25,136,680 | N/A | N/A | 0.93 | N/A | N/A | 0.99 | N/A | N/A |
| All reads | 26,093,644 | N/A | N/A | 1 | N/A | N/A | 1 | N/A | N/A |
| GSA | 422,156 | N/A | 18,272,172 | N/A | N/A | 0.33 | N/A | N/A | 0.62 |
| PsychChip | 350,678 | N/A | 18,190,171 | N/A | N/A | 0.33 | N/A | N/A | 0.62 |
| MEGA | 1,152,178 | N/A | 19,219,473 | N/A | N/A | 0.33 | N/A | N/A | 0.62 |
| H3Africa | 2,151,137 | N/A | 19,709,178 | N/A | N/A | 0.33 | N/A | N/A | 0.62 |
| Omni2.5 | 2,072,034 | N/A | 19,698,788 | N/A | N/A | 0.33 | N/A | N/A | 0.62 |
All numbers reported here are from processing via Beagle. Common variants here are defined as having >5 copies (i.e., MAF > 3%). “Raw” indicates that variant calls were produced directly from GATK with no genotype refinement or imputation, “refined” indicates variant calls from genotype refinement without imputation, and “imputed” indicates imputed variants following genotype refinement.
Figure 3.
Minor allele frequency (MAF) across GWAS arrays and continental ancestries via 1000 Genomes data
AFR, Africans; AMR, admixed Americans (e.g., Hispanics/Latinos); EAS, East Asians; EUR, Europeans; SAS, South Asians. These results indicate that the GSA captures variants that are especially common in Europeans relative to elsewhere.
We also investigated the importance of the reference panel and the impact of missing population representation on sensitivity. For example, regardless of technology, we estimate that 33% of singletons in the “truth” dataset can be imputed (Table 3, i.e., 67% of singletons in the NeuroGAP-Psychosis data are absent or not tagged by the 1000 Genomes phase 3 data). This estimate is most likely optimistic given that the low sample size in this study means that many variants reported here as singletons are most likely somewhat common in the population. Additionally, 62% of common variants (allele count, AC > 5, minor allele frequency [MAF] > 3%) in the “truth” dataset can be imputed, indicating that 38% of variants in the eastern and southern African populations in NeuroGAP-Psychosis are absent or untagged in the 1000 Genomes phase 3 data. While the number of variants imputed is inherently bounded by the reference data, the raw data indicates relatively high sensitivity to variants present in the “truth” data. For example, 45% of singletons in the full dataset can be detected with 4× data (Table 3). At the same depth, 95% of common variants are detected. As expected, we observe diminishing returns in numbers of variants imputed with increasing sequencing depth. More variants can be imputed with 2× sequencing via Beagle than with any of the GWAS arrays. Our sensitivity for detecting variants common in the truth data (74%) is higher with 1× sequencing than with imputed data from any array (62%, Table 3).
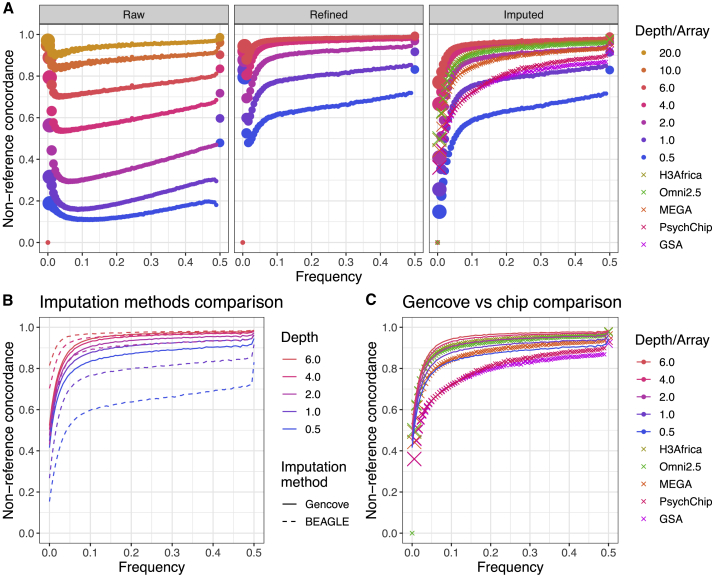
We next investigated variant call accuracy by calculating non-reference concordance across technologies. We also compared two imputation methodologies for use with low-coverage sequencing data—Beagle versus Gencove—as the latter was specifically designed for use with low-coverage data. Unlike Beagle, Gencove takes unmapped FASTQ files as an input to perform phasing and imputation, allowing consideration of genotype probabilities directly as described previously.25 Figure 4 shows non-reference concordance by allele frequency across sequencing versus array technologies and using different software for genotype refinement and imputation. Data processing steps through imputation (“refined” and “imputed” panels with results from Beagle software) are shown in Figure 4A, low-coverage sequencing imputation accuracy comparison of Beagle versus Gencove software is shown in Figure 4B, and results of low-coverage imputation with Gencove versus GWAS array data imputation with Beagle are shown in Figure 4C. Figure 4A includes different variants across panels, including fewer but more accurate variants in the “refined” panel, whereas the “imputed” panel includes more than double the number of variants but with reduced accuracy (Table 3). When using Beagle for imputing both arrays and low-coverage data, these analyses indicate that the lower-density arrays (GSA and PsychChip) perform similarly to 1× sequencing, medium-density arrays (MEGA) perform almost as well as 2× data, and high-density arrays (Omni2.5 array and H3Africa array specifically designed to capture African variation) perform between 2× and 4× sequencing (Figure 4A). We also compared the accuracy of two imputation methods, Beagle and Gencove, by using the same set of imputed sites in the low-coverage sequencing data. We find that imputation performs better with Gencove for the lowest depths (0.5×, 1×, and 2×), whereas Beagle performs better for higher depths (4× and 6×, Figure 4B, Table S4). When comparing low-coverage data imputed with Gencove versus GWAS array data imputed with Beagle, we see that 1× sequencing outperforms the low- and medium-density arrays (MEGA, GSA, and PsychChip) and that the high-density arrays (H3Africa and Omni2.5) perform comparably to 2× sequencing (Figure 4C, Table S4). Overall, these results show that GWAS arrays perform at best comparably to low-coverage sequencing.
Figure 4.
Non-reference concordance for SNPs as a function of sequencing depth or genotyping array, frequency, analysis stage, and imputation method
“Truth” dataset here is the full depth joint called sequencing dataset. All depths of sequencing data are shown for the raw data (i.e., only variant calling from GATK with no genotype refinement or imputation following). We excluded sequencing at 10× and 20× for all except the raw data because of minimal potential accuracy gains and to reduce computational costs.
(A) Non-reference concordance comparisons throughout steps of the Beagle analysis pipeline. Size of the points are proportional to the number of SNPs in each frequency bin. “Raw” indicates that variant calls were produced directly from GATK with no genotype refinement or imputation, “refined” indicates variant calls from genotype refinement without imputation, and “imputed” indicates imputed variants following genotype refinement.
(B) Non-reference concordance comparisons of Beagle versus Gencove software for imputation of low-coverage data.
(C) Non-reference concordance comparison of Gencove software for imputation of low-coverage data versus Beagle for imputation of GWAS arrays. Non-reference concordance values averaged across (B) and (C) are shown in Table S4.
In addition to imputation methods, we also compared newer imputation panels where possible. Specifically, African American and Hispanic/Latino genomes are imputed more accurately with the TOPMed imputation panel compared to the 1000 Genomes data.26 Because TOPMed neither shares harmonized individual-level data nor supports genotype refinement, we were only able to compare imputation accuracy for the GWAS arrays and not for low-coverage sequencing, which is shown in Figure S4. As shown previously, imputation accuracy is significantly higher in NeuroGAP with the TOPMed server compared with the 1000 Genomes data.
Low-coverage sequencing quality across diverse African populations
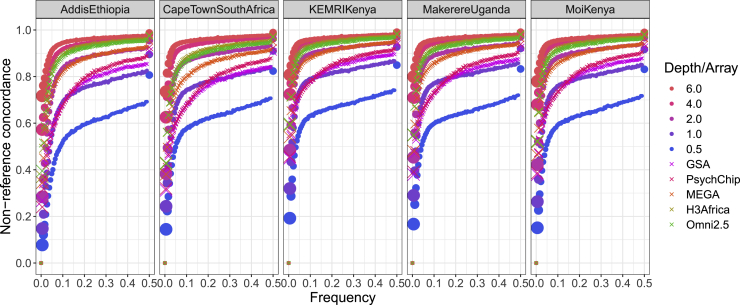
We next investigated the impact of ancestral diversity on imputation accuracy from arrays versus sequencing depth. The populations in NeuroGAP-Psychosis span a broad range of geographical, ethnolinguistic, and ancestral diversity in eastern and southern Africa. Despite this considerable diversity with a range of genetic distances from populations represented in the 1000 Genomes reference haplotypes, there is remarkable qualitative consistency in data quality from various sequencing depths and GWAS arrays (Figure 5). We quantify subtle differences across populations (Table S5). For example, imputation is least accurate among participants from Addis Ababa, Ethiopia. In contrast, imputation performs best in participants from Kilifi, Kenya, where some participants self-identify as Luhya. These differences in imputation accuracy across populations most likely reflect genetic distances between the NeuroGAP-Psychosis participants and the 1000 Genomes phase 3 reference data, which includes, for example, a Luhya population from Kenya (LWK). These findings consistently indicate that 4× sequencing data outperform all common commercial GWAS arrays for diverse African ancestry populations, including those specifically designed with African variation in mind, such as the H3Africa array.
Figure 5.
Non-reference concordance between imputed versus truth data across various populations and sites in Africa
Size of the points where applicable are proportional to the number of SNPs in each frequency bin. Quantitative comparisons across all variants and imputation methods are shown in Table S5.
Sampling and microbiome variation influence precise sequencing depth
Although this in silico framework enables us to compare two data generation strategies in a highly controlled manner with far fewer resources than data generation from many experiments, a limitation of this approach is that downsampling to an exact depth of coverage does not capture realistic variability. Important factors that can drive variation in human genome coverage here are variation arising from the sample pooling process for sequencing and variation in rates of oral microbiome contamination. Across the Broad Genomics Platform, we find that samples derived from saliva typically have bacterial contamination ranging from 5%–40% with a median around 10%, whereas blood-derived samples typically align to the human genome at 98% or higher. In these genomes specifically, contamination tends to be low: alignment rates are 93.1% ± 6.1% (mean ± SD). These alignment rates are in line with previous work.27
To better understand variability arising from these combined effects, we targeted 1× sequencing in an additional 95 non-overlapping samples sequenced from three of the sites: Addis Ababa, Ethiopia (n = 32); Eldoret, Kenya (n = 32); and Kampala, Uganda (n = 31). Similar to the high-coverage whole genomes, alignment rates were high at 93.0% ± 5.1% (mean ± SD). Coverage was close to the target at 1.13× ± 0.16× (mean ± SD): 73/95 reached 1× and the remainder were typically quite close (min = 0.72×, Figure S3). Unsurprisingly, these are correlated effects (Pearson’s r = 0.52, p = 5e−8).
A potential advantage of low-coverage sequencing over GWAS arrays is the ability to use off-target reads that do not map to Homo sapiens for further microbiome analysis. We used taxonomic profiling quantifications from the software Kraken, which were produced from 6× data input to Gencove. For each individual, we quantified relative abundances from read counts. We show the phylum-level relative abundances as a proof-of-concept (Figure S5).
Comparable list prices for low-coverage sequencing and GWAS arrays
Lastly, we list realistic pricing for low-coverage sequencing versus GWAS arrays based on current publicly available reagent costs from Illumina (Table 4). Although these do not include fixed sample and library preparation costs, we assume that these are comparable across GWAS arrays and sequencing approaches. We note that all costs can vary considerably depending on consortium pricing, sequencing facility, volume, etc. While sequencing costs list volume discounts (e.g., up to 39% discount for high volume flow cell purchasing), GWAS arrays do not; to compare these technologies as fairly as possible, we therefore list the non-discounted price but note that costs could be lower (Table S6). On the basis of these prices, we show that the high-density arrays are similar in price to 4–6× sequencing. The lowest depths of sequencing evaluated here, 0.5–1×, are cheaper than the PsychChip and GSA.
Table 4.
Costs of reagents for sequencing and genotyping options
| Depth/array | Reagent cost per sample |
|---|---|
| 30× | $1,320.83 |
| 20× | $880.55 |
| 6× | $264.17 |
| Omni2.5 | $184.43 |
| 4× | $176.11 |
| MEGA Global | $119.00 |
| 2× | $88.06 |
| PsychChip | $71.38 |
| GSA | $49.00 |
| 1× | $44.03 |
| 0.5× | $22.01 |
We aggregated the prices for reagents from Illumina’s website as of April 10, 2020. These prices notably do not include sample and library preparation costs, which we assume to be comparable between GWAS arrays and sequencing approaches. The H3Africa array is not commercially listed on Illumina’s site and is thus not included here. Sequencing reagent costs use Illumina’s list price for the NovaSeq 6000 S4 Reagent Kit. Each flow cell has a maximum output of 3,000 Gb. Prices listed assume single flow cell purchasing, which is listed at $31,700. Prices adjusting for bulk flow cell purchasing from Illumina are shown in Table S6. Sequencing costs assume 125 Gb to achieve a target depth of 30× whole-genome sequencing coverage.
Another pricing consideration regarding different depths of sequencing or GWAS arrays is the computational complexity. Genotype refinement is only necessary for low-coverage sequencing and is a more computationally complex step than imputation. Imputation is also slightly more costly with low-coverage sequencing than with GWAS arrays because more variants are called from the beginning, increasing genomic coverage. However, we find that the computational costs of genotype refinement and the slightly increased computational complexity of imputation from more variants called at the outset are negligible compared with data generation costs. For low-coverage sequencing, reagent costs alone are ≥100 times higher than the sum of refinement and imputation depending on depth of coverage (ratio increasing with higher depths), and GWAS array costs are >2,800 times higher than imputation (ratio increasing with higher array density, Table S7).
Discussion
In this study, we have compared the relative merits and costs of several genetic data generation and processing strategies in a diverse cohort of eastern and southern Africans. We conclude that 4× sequencing outperforms all GWAS arrays evaluated, including dense arrays. This outcome is in spite of the fact that the dense H3Africa array was designed to capture African variation and thus tags the most variation in the NeuroGAP-Psychosis data of all GWAS arrays analyzed here. 4× sequencing is comparable in price to high-density arrays that assay millions of SNPs and indels across the allele frequency spectrum. Among more affordable options, we find that 1× sequencing costs less than and performs similarly to or better than commonly used lower-density arrays such as the Illumina GSA. Additionally, we note that the GSA is composed of variants most common in European populations and is thus not the most appropriate technology for studies of participants with primarily non-European ancestry.
Low-coverage sequencing has several distinct advantages compared to GWAS arrays, especially the more accurate identification of genetic variation across the allele frequency spectrum particularly in underrepresented populations. In these NeuroGAP-Psychosis data, we find that 38% of common variants could not be imputed from the 1000 Genomes phase 3 data, most likely because of a dearth of eastern and southern African diversity represented in this reference panel. Among rare variants, we find that 4× sequencing detects nearly half of all singletons, an especially appealing attribute for disease studies. Previous work in psychiatric genetics has shown that while common variants explain most of the SNP heritability for schizophrenia,28,29 there are at least partially converging genetic signatures emerging from exome sequencing studies that provide new biological insights and are especially informative for severe psychiatric disorders.30 Because we are still in the early stages of gene discovery for these and many other disease areas, sequencing technologies that bridge the rare and common variant gap will be critical in fully elucidating their genetic architectures by refining causal variants, detecting variation enriched in cases and genetically clustered in particular functional domains, and identifying rare variants with large effects.31,32 Post-GWAS methodological advances with low-coverage sequencing data can facilitate these analyses; for example, pooling reads near GWAS peaks in cases separately from controls can enhance variant discovery, an important step for fine-mapping. Fully new analysis opportunities, such as using off-target reads to measure microbiome variation as demonstrated here, are also possible.
An especially valuable aspect of low-coverage sequencing in underrepresented populations is the durable opportunity to construct new haplotype reference panels used for imputation, which are mostly lacking with few exceptions.26 Recent development of genomics infrastructure, such as the TOPMed imputation server, in theory further supports data quality improvements by including more deeply sequenced and diverse haplotypes.33,34 In practice, however, because TOPMed does not currently share the harmonized individual-level data required for genotype refinement of low-coverage sequencing data, its practical utility is more limited and it is not feasible to use here. Instead, evaluation of imputation accuracy in this and other similar projects relies on existing resources that provide transparent access to requisite data, such as the 1000 Genomes Project and/or the Haplotype Reference Consortium (HRC), the latter of which aggregated low-coverage sequencing data from European ancestry populations into an imputation panel.4,35 We plan to build on the HRC’s previous work by integrating the high-coverage genomes sequenced here along with additional low-coverage whole genomes in African populations to develop a more diverse reference panel that will improve phasing and imputation for diverse African populations. New computationally efficient methods will be required to make streamlined use of low-coverage sequencing data and growing reference panels.36
GWAS arrays are currently the most commonly used data generation technology in large-scale genetic studies. Accuracy gains in European ancestry populations from low-coverage sequencing compared with GWAS arrays are more modest than in other populations because of Eurocentric SNP ascertainment on GWAS arrays.35 Yet, low-coverage sequencing still outperforms arrays in Europeans while providing several distinct advantages in populations underrepresented in genomics. These advantages are especially pronounced in African populations where overall genetic variation is higher, linkage disequilibrium is shorter, and haplotype reference data are lacking. Although African populations have the most genetic variation globally, with as much variation among individuals from different regions of Africa as between some continents, African ancestry genomes are vastly underrepresented. Further, the vast majority of African ancestry participants in genetic studies are African Americans or Afro-Caribbeans (72%–93% in the GWAS catalog and ≥90% in gnomAD) with primarily West African ancestors.37 However, large-scale efforts such as the Human, Heredity, and Health in Africa (H3Africa) Initiative and the NeuroGAP study aim to address these gaps.13,38 In addition to informing the most appropriate and cost-effective data generation strategies, this study also adds to a relatively small number of high-coverage whole genomes sequenced from Africa.
Declaration of interests
A.R.M. has consulted for 23andMe and Illumina. B.M.N. is a member of the Deep Genomics Scientific Advisory Board. He also serves as a consultant for the Camp4 Therapeutics Corporation, Takeda Pharmaceutical, and Biogen. M.J.D. is a founder of Maze Therapeutics. J.K.P. is an employee of Gencove, Inc. D.J.S. has received research grants and/or consultancy honoraria from Lundbeck and Sun. The remaining authors declare no competing interests.
Acknowledgments
We thank Juha Karjalainen for his help setting up and troubleshooting a Cromwell server for running all workflows. We thank Laura Gauthier for explanations of components of the Broad Institute Data Science Platform pipelines. We would also like to thank Moses Joloba at Makerere University College of Health Sciences in Kampala, Uganda. This study was funded by the Stanley Center for Psychiatric Research at the Broad Institute. This work was supported by funding from the National Institutes of Health (K99/R00MH117229 to A.R.M.; K01MH121659 and T32MH017119 to E.G.A.). L.B.C., B.G., K.C.K., D.J.S., S.T., and D.A. are supported, in part, by R01MH120642. A.F. is supported by the Medical Research Council and Department for International Development through the Africa Research Leader scheme.
Published: March 25, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.03.012.
Data and code availability
Data will be hosted on the Terra environment created by Broad, which contains a rich system of workspace functionalities centered on data sharing and analysis. The platform has been given a redesigned user interface under the Terra branding and extended to support a number of projects, including AnVIL (Analysis, Visualization, and Informatics Labspace). Each AnVIL data access request (DAR) is routed to the Data Access Committee (DAC) for the dataset. DAC’s are responsible for reviewing the DAR for the dataset to determine whether the research use proposed in the DAR is within the bounds of the data use limitations of the requested dataset. We are following H3Africa policies for data sharing, which are designed to enable African collaborators to make use of the data they collected before better resourced groups have access. These data will be embargoed for one year following publication. All code used is available in a GitHub repository as described in web resources.
Web resources
1000 Genomes ftp site, ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data_collections/1000_genomes_project/release/20181203_biallelic_SNV/
GATK workflows, https://github.com/gatk-workflows/gatk4-germline-snps-indels
NeuroGAP downsampling scripts, https://github.com/armartin/neurogap_downsampling
Supplemental information
References
- 1.Marchini J., Howie B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 2.Lachance J., Tishkoff S.A. SNP ascertainment bias in population genetic analyses: why it is important, and how to correct it. BioEssays. 2013;35:780–786. doi: 10.1002/bies.201300014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wojcik G.L., Fuchsberger C., Taliun D., Welch R., Martin A.R., Shringarpure S., Carlson C.S., Abecasis G., Kang H.M., Boehnke M. Imputation-Aware Tag SNP Selection To Improve Power for Large-Scale, Multi-ethnic Association Studies. G3 (Bethesda) 2018;8:3255–3267. doi: 10.1534/g3.118.200502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCarthy S., Das S., Kretzschmar W., Delaneau O., Wood A.R., Teumer A., Kang H.M., Fuchsberger C., Danecek P., Sharp K., Haplotype Reference Consortium A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang L., Li Y., Singleton A.B., Hardy J.A., Abecasis G., Rosenberg N.A., Scheet P. Genotype-imputation accuracy across worldwide human populations. Am. J. Hum. Genet. 2009;84:235–250. doi: 10.1016/j.ajhg.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmann T.J., Zhan Y., Kvale M.N., Hesselson S.E., Gollub J., Iribarren C., Lu Y., Mei G., Purdy M.M., Quesenberry C. Design and coverage of high throughput genotyping arrays optimized for individuals of East Asian, African American, and Latino race/ethnicity using imputation and a novel hybrid SNP selection algorithm. Genomics. 2011;98:422–430. doi: 10.1016/j.ygeno.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mulder N., Abimiku A., Adebamowo S.N., de Vries J., Matimba A., Olowoyo P., Ramsay M., Skelton M., Stein D.J. H3Africa: current perspectives. Pharm. Genomics Pers. Med. 2018;11:59–66. doi: 10.2147/PGPM.S141546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pasaniuc B., Rohland N., McLaren P.J., Garimella K., Zaitlen N., Li H., Gupta N., Neale B.M., Daly M.J., Sklar P. Extremely low-coverage sequencing and imputation increases power for genome-wide association studies. Nat. Genet. 2012;44:631–635. doi: 10.1038/ng.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Homburger J.R., Neben C.L., Mishne G., Zhou A.Y., Kathiresan S., Khera A.V. Low coverage whole genome sequencing enables accurate assessment of common variants and calculation of genome-wide polygenic scores. Genome Medicine. 2019;11:74. doi: 10.1186/s13073-019-0682-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pickrell J. The Gencove Blog; 2017. It is time to replace genotyping arrays with sequencing. [Google Scholar]
- 11.Alex Buerkle C., Gompert Z. Population genomics based on low coverage sequencing: how low should we go? Mol. Ecol. 2013;22:3028–3035. doi: 10.1111/mec.12105. [DOI] [PubMed] [Google Scholar]
- 12.Gilly A., Southam L., Suveges D., Kuchenbaecker K., Moore R., Melloni G.E.M., Hatzikotoulas K., Farmaki A.-E., Ritchie G., Schwartzentruber J. Very low depth whole genome sequencing in complex trait association studies. Bioinformatics. 2018;35:2555–2561. doi: 10.1093/bioinformatics/bty1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stevenson A., Akena D., Stroud R.E., Atwoli L., Campbell M.M., Chibnik L.B., Kwobah E., Kariuki S.M., Martin A.R., de Menil V. Neuropsychiatric Genetics of African Populations-Psychosis (NeuroGAP-Psychosis): a case-control study protocol and GWAS in Ethiopia, Kenya, South Africa and Uganda. BMJ Open. 2019;9:e025469. doi: 10.1136/bmjopen-2018-025469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeste D.V., Palmer B.W., Appelbaum P.S., Golshan S., Glorioso D., Dunn L.B., Kim K., Meeks T., Kraemer H.C. A new brief instrument for assessing decisional capacity for clinical research. Arch. Gen. Psychiatry. 2007;64:966–974. doi: 10.1001/archpsyc.64.8.966. [DOI] [PubMed] [Google Scholar]
- 15.Campbell M.M., Susser E., Mall S., Mqulwana S.G., Mndini M.M., Ntola O.A., Nagdee M., Zingela Z., Van Wyk S., Stein D.J. Using iterative learning to improve understanding during the informed consent process in a South African psychiatric genomics study. PLoS ONE. 2017;12:e0188466. doi: 10.1371/journal.pone.0188466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hodgson J.A., Mulligan C.J., Al-Meeri A., Raaum R.L. Early back-to-Africa migration into the Horn of Africa. PLoS Genet. 2014;10:e1004393. doi: 10.1371/journal.pgen.1004393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pagani L., Schiffels S., Gurdasani D., Danecek P., Scally A., Chen Y., Xue Y., Haber M., Ekong R., Oljira T. Tracing the route of modern humans out of Africa by using 225 human genome sequences from Ethiopians and Egyptians. Am. J. Hum. Genet. 2015;96:986–991. doi: 10.1016/j.ajhg.2015.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henn B.M., Botigué L.R., Gravel S., Wang W., Brisbin A., Byrnes J.K., Fadhlaoui-Zid K., Zalloua P.A., Moreno-Estrada A., Bertranpetit J. Genomic ancestry of North Africans supports back-to-Africa migrations. PLoS Genet. 2012;8:e1002397. doi: 10.1371/journal.pgen.1002397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crysnanto D., Pausch H. Bovine breed-specific augmented reference graphs facilitate accurate sequence read mapping and unbiased variant discovery. Genome Biol. 2020;21:184. doi: 10.1186/s13059-020-02105-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Browning S.R., Browning B.L. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Browning B.L., Zhou Y., Browning S.R. A One-Penny Imputed Genome from Next-Generation Reference Panels. Am. J. Hum. Genet. 2018;103:338–348. doi: 10.1016/j.ajhg.2018.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo Y., de Lange K.M., Jostins L., Moutsianas L., Randall J., Kennedy N.A., Lamb C.A., McCarthy S., Ahmad T., Edwards C. Exploring the genetic architecture of inflammatory bowel disease by whole-genome sequencing identifies association at ADCY7. Nat. Genet. 2017;49:186–192. doi: 10.1038/ng.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.CONVERGE consortium Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature. 2015;523:588–591. doi: 10.1038/nature14659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.1000 Genomes Project Consortium. Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wasik K., Berisa T., Pickrell J.K., Li J.H., Fraser D.J., King K., Cox C. Comparing low-pass sequencing and genotyping for trait mapping in pharmacogenetics. BioRxiv. 2019 doi: 10.1101/632141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kowalski M.H., Qian H., Hou Z., Rosen J.D., Tapia A.L., Shan Y., Jain D., Argos M., Arnett D.K., Avery C. Use of >100,000 NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium whole genome sequences improves imputation quality and detection of rare variant associations in admixed African and Hispanic/Latino populations. PLoS Genet. 2019;15:e1008500. doi: 10.1371/journal.pgen.1008500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yao R.A., Akinrinade O., Chaix M., Mital S. Quality of whole genome sequencing from blood versus saliva derived DNA in cardiac patients. BMC Med. Genomics. 2020;13:11. doi: 10.1186/s12920-020-0664-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lam M., Chen C.-Y., Li Z., Martin A.R., Bryois J., Ma X., Gaspar H., Ikeda M., Benyamin B., Brown B.C. Comparative genetic architectures of schizophrenia in East Asian and European populations. Nat. Genet. 2019;51:1670–1678. doi: 10.1038/s41588-019-0512-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh T., Poterba T., Curtis D., Akil H., Al Eissa M., Barchas J.D., Bass N., Bigdeli T.B., Breen G., Bromet E.J. Exome sequencing identifies rare coding variants in 10 genes which confer substantial risk for schizophrenia. medRxiv. 2020 doi: 10.1101/2020.09.18.20192815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., Genome Aggregation Database Consortium The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samocha K.E., Kosmicki J.A., Karczewski K.J., O’Donnell-Luria A.H., Pierce-Hoffman E., MacArthur D.G., Neale B.M., Daly M.J. Regional missense constraint improves variant deleteriousness prediction. BioRxiv. 2017 doi: 10.1101/148353. [DOI] [Google Scholar]
- 33.Das S., Forer L., Schönherr S., Sidore C., Locke A.E., Kwong A., Vrieze S.I., Chew E.Y., Levy S., McGue M. Next-generation genotype imputation service and methods. Nat. Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taliun D., Harris D.N., Kessler M.D., Carlson J., Szpiech Z.A., Torres R. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature. 2021;590:290–299. doi: 10.1038/s41586-021-03205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rubinacci S., Ribeiro D.M., Hofmeister R.J., Delaneau O. Efficient phasing and imputation of low-coverage sequencing data using large reference panels. Nat. Genet. 2021;53:120–126. doi: 10.1038/s41588-020-00756-0. [DOI] [PubMed] [Google Scholar]
- 36.Rubinacci S., Ribeiro D.M., Hofmeister R., Delaneau O. Efficient phasing and imputation of low-coverage sequencing data using large reference panels. Nat. Genet. 2021;53:120–126. doi: 10.1038/s41588-020-00756-0. [DOI] [PubMed] [Google Scholar]
- 37.Martin A.R., Teferra S., Möller M., Hoal E.G., Daly M.J. The critical needs and challenges for genetic architecture studies in Africa. Curr. Opin. Genet. Dev. 2018;53:113–120. doi: 10.1016/j.gde.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choudhury A., Aron S., Botigué L.R., Sengupta D., Botha G., Bensellak T., Wells G., Kumuthini J., Shriner D., Fakim Y.J., TrypanoGEN Research Group. H3Africa Consortium High-depth African genomes inform human migration and health. Nature. 2020;586:741–748. doi: 10.1038/s41586-020-2859-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be hosted on the Terra environment created by Broad, which contains a rich system of workspace functionalities centered on data sharing and analysis. The platform has been given a redesigned user interface under the Terra branding and extended to support a number of projects, including AnVIL (Analysis, Visualization, and Informatics Labspace). Each AnVIL data access request (DAR) is routed to the Data Access Committee (DAC) for the dataset. DAC’s are responsible for reviewing the DAR for the dataset to determine whether the research use proposed in the DAR is within the bounds of the data use limitations of the requested dataset. We are following H3Africa policies for data sharing, which are designed to enable African collaborators to make use of the data they collected before better resourced groups have access. These data will be embargoed for one year following publication. All code used is available in a GitHub repository as described in web resources.