Summary
Progressive myoclonus epilepsies (PMEs) comprise a group of clinically and genetically heterogeneous rare diseases. Over 70% of PME cases can now be molecularly solved. Known PME genes encode a variety of proteins, many involved in lysosomal and endosomal function. We performed whole-exome sequencing (WES) in 84 (78 unrelated) unsolved PME-affected individuals, with or without additional family members, to discover novel causes. We identified likely disease-causing variants in 24 out of 78 (31%) unrelated individuals, despite previous genetic analyses. The diagnostic yield was significantly higher for individuals studied as trios or families (14/28) versus singletons (10/50) (OR = 3.9, p value = 0.01, Fisher’s exact test). The 24 likely solved cases of PME involved 18 genes. First, we found and functionally validated five heterozygous variants in NUS1 and DHDDS and a homozygous variant in ALG10, with no previous disease associations. All three genes are involved in dolichol-dependent protein glycosylation, a pathway not previously implicated in PME. Second, we independently validate SEMA6B as a dominant PME gene in two unrelated individuals. Third, in five families, we identified variants in established PME genes; three with intronic or copy-number changes (CLN6, GBA, NEU1) and two very rare causes (ASAH1, CERS1). Fourth, we found a group of genes usually associated with developmental and epileptic encephalopathies, but here, remarkably, presenting as PME, with or without prior developmental delay. Our systematic analysis of these cases suggests that the small residuum of unsolved cases will most likely be a collection of very rare, genetically heterogeneous etiologies.
Keywords: progressive myoclonus epilepsy, dolichol-dependent glycosylation, whole-exome sequencing, epilepsy genetics
Introduction
The progressive myoclonus epilepsies (PMEs) are a group of rare clinically and genetically heterogeneous disorders that typically present in childhood or adolescence with action myoclonus, generalized tonic-clonic seizures, and progressive neurological decline.1 The majority of PMEs follow autosomal-recessive inheritance, with rare mitochondrial causes and a small but increasing number of autosomal-dominant genes.2,3
Clinically, the PMEs can be categorized into two broad groups. In one group, cognition is largely preserved with clinical features dominated by severe, treatment-resistant, and physically disabling myoclonus, tonic-clonic seizures, and ataxia.1 The most common and paradigmatic form is Unverricht-Lundborg disease (ULD, EPM1 [MIM: 254800]), which is caused by recessive mutation, most commonly a dodecamer repeat expansion, of cystatin B (CSTB [MIM: 601145]). The second clinical group is associated with significant cognitive impairment and decline, with the major forms including Lafora disease (EPM2A/B [MIM: 254780]) and the neuronal ceroid lipofuscinoses (NCLs) which involve a number of mostly recessive genes.
Known PME genes encode a variety of proteins, many of which have an endosomal and lysosomal function (Table S1). Despite this, there is no apparent unifying pathway leading to the phenotype.4,5 Importantly a molecular genetic diagnosis will currently be made with an established PME gene in approximately 70% of all individuals diagnosed with PME.2
We previously performed a whole-exome sequencing (WES) study on a cohort of 84 molecularly unsolved and unrelated singleton cases of PME. We identified a recurrent pathogenic variant in KCNC1 (MIM: 176258) (p.Arg320His [c.959G>A]) as a cause of PME now known as MEAK (myoclonus epilepsy and ataxia due to K+ channel mutation, EPM7 [MIM: 616187]).3,6 This heterozygous variant not only added another autosomal-dominant gene to the list of known PME genes, but also highlighted a role for de novo pathogenic variants in PME.
In this study, we aimed to identify further causative genes for the unsolved PMEs by expanding our WES data analysis to include additional unsolved cases and using, where possible, a trio-design approach to enhance the detection of de novo pathogenic variants.
Subjects and methods
Subjects
We studied a total of 84 (78 unrelated) molecularly unsolved individuals (45 males) who had been clinically diagnosed with PME. Individuals were referred for genetic research from centers in Europe, Australia, and the USA over 25 years. Informed consent for DNA analysis was obtained from individuals in line with local institutional review board requirements at the time of collection.
The majority of the cohort had previously had extensive genetic investigations, including clinical microarray and gene panel analyses (including mitochondrial gene testing where suspected), or singleton WES as part of our earlier research (n = 57).3 Specifically, all individuals had been screened and tested negative for recessive variants in CSTB, including the dodecamer repeat expansion, and for the KCNC1 recurrent pathogenic p.Arg320His variant.
A trio-design approach was used for 22/78 unrelated individuals (28%) where DNA was available for both unaffected parents for WES. Six unrelated subjects were exome sequenced with an affected first-degree relative (parent-child, n = 2; or sibling pairs, n = 4); two of the four sibling pairs had both unaffected parents available for WES and were analyzed as a quartet. The remaining 50 affected individuals were analyzed as WES singletons (Figure S1).
Exome sequencing
This study included two sequencing cohorts (Figure S1). The first cohort comprised 57 singleton individuals with PME that remained unsolved after our initial study;3 of these, 44 did not have parental DNA available for trio- or quartet- WES re-analysis. This cohort was exome sequenced previously at the Wellcome Sanger Institute, Cambridge, UK in 2011-2012 (details of sequencing described in Muona et al.3).
The second cohort comprised a total of 40 individuals with PME and 48 unaffected parents (contributing to 22 trios and 2 quartets). 27 PME-affected individuals in cohort 2 were newly referred; 13 were subjects from cohort 1 that were re-sequenced with their parents. Exome sequencing for this cohort was performed at the Broad Institute of MIT and Harvard, Cambridge, MA, USA in 2015-2016. In detail, genomic DNA (approximately 1 μg) extracted from peripheral blood for each sample was enzymatically sheared in whole-exome library preparation. In-solution hybrid exome capture was performed using the Illumina Rapid Capture Exome enrichment kit with 38 Mb target region (29 Mb baited), which includes 98.3% of the intervals in the RefSeq exome database. Sequencing was performed on either Illumina HiSeq 4000 or HiSeq X instrument with the use of 151bp paired-end reads. The mean average sequencing depth for each sample was 78-fold, with more than 80% of target bases having at least 30-fold coverage. Mitochondrial DNA (mtDNA) was not targeted in either sequencing cohort.
Variant calling
Sequence reads were processed and aligned to human genome hg19/GRCh37 as described previously.3 Variant calling of single-nucleotide variants and indels was done by GATK (v.3.7-0) HaplotypeCaller using joint calling approach. Thirteen individuals underwent WES twice (in the previous study and here), so their sequence data were merged to maximize coverage. Variant quality scores were recalibrated jointly with GATK VariantRecalibrator. A truth sensitivity cutoff of 99.8% was used for both SNVs and indels. De novo variants were called by GATK GenotypeRefinement and GATK PossibleDeNovo tools.
Sex and pedigree checks
Sex and ancestry checks for all samples and relatedness checks between all sample pairs were estimated using Peddy.7 Inbreeding coefficients for all samples were estimated using FEstim.8
Variant annotation
Variant consequences were annotated using Variant Effect Predictor tool.9 In silico prediction of deleterious variants was carried out by CADD,10 SIFT,11 PolyPhen2,12 and, in the case of splicing variants, Transcript inferred Pathogenicity (TraP) Score.13 Variant allele frequencies were obtained primarily from the Genome Aggregation Database (gnomADv2.1.1).15 Gene-phenotype associations were annotated based on OMIM database and Clinical Genomics Database.
Variant filtering
To identify potentially pathogenic variants from the annotated data, all variants within 8 bp of exonic regions were filtered based on the potential modes of inheritance: X-linked, autosomal recessive, dominant, and de novo using a similar approach to previously.3 In recessive filtering, the exome data were analyzed for rare (<150 heterozygous counts and no homozygotes in the gnomADv2.1.1 database)14 homozygous or compound heterozygous variants including missense, nonsense, splice site, inframe insertion and deletion, and frameshift variants based on Variant Effect Predictor annotations in CCDS genes (Ensembl release 88). In the dominant filtering strategy (applied to both singleton cases, affected parent-child pairs, and the de novo variant analysis), we included heterozygous variants with <5 counts in gnomADv2.1.1.
Variant prioritization
Variants surviving the filtering steps were manually assessed and prioritized. All prioritized variants were classified according to ACMG standards and guidelines.15 As these guidelines are not designed for novel research findings, and because they do not always capture the phenotypic subtleties, we also used a study-specific method of classification. We combined three lines of evidence: (1) at the variant level (e.g., using in silico prediction tools), (2) at the pedigree level (e.g., variant segregation data within families), and (3) at the gene level (e.g., prior disease phenotype associations). Each variant was given a score between 0 and 2 for the three lines of evidence making the maximum score 6 (Table S2).
We deemed variants as causative with “high confidence” if a score ≥5 was achieved and “moderate confidence” for variants with scores ≥4. Variants scoring <4 were not prioritized or reported without the support of functional data.
Variant validation and segregation
Candidate variants in known and potentially novel disease genes were confirmed by bi-directional Sanger sequencing (ABI BigDye 3.1, Applied Biosystems) on ABI3730xl DNA Analyzer. Primers were designed with Primer-BLAST.16 The sequences were analyzed using Sequencher v.5.3 (Gene Codes Corporation).
Specific splicing in silico predictions were made using Human Splicing Finder v3.1.17 Confirmation of CLN6 (GenBank: NM_017882.3; MIM: 606725) splicing effect was performed by RT-PCR from total RNA extracted from fibroblast cells followed by sequencing of the abnormal amplicon (Figure S4).
Deletion confirmation of NEU1 (GenBank: NM_000434.3; MIM: 608272) was performed by quantitative PCR (qPCR) (Figure S5).
Primers for Sanger sequencing and PCR are available upon request.
Copy number variant analysis from WES data
Copy number variants were called from the WES data based on relative sequencing depth. CNVkit was used to call the variants.18 This analysis was performed separately for WES data generated in the original study3 and in the current one owing to the different exome capture kits used. CNV analysis focused on known disease genes (annotated against Clinical Genomics Database), in particular those associated with PMEs.
Analysis of short tandem repeats
We additionally examined whether any of the probands had short tandem repeats (STRs) that were expanded at 27 known pathogenic loci (Table S3). The WES samples were examined separately using two STR detection tools, Expansion Hunter v.2.5.519 and exSTRa.20 For each locus we looked for evidence of outlying samples in terms of STR length by inspecting plots of estimated STR size (ExpansionHunter), and empirical cumulative distribution function (eCDF; exSTRa) plots of the number of repeated bases observed for each sample.
Human fibroblast culture
Fibroblast cultures were established from skin biopsy samples of PME1, PME2, PME71, and PME27 as well as control subjects. Cells were cultured in DMEM (GIBCO, Thermo Scientific) plus 10% FBS, 100 units/mL penicillin, 100 μg/mL streptomycin, and 2 mM glutamine in a 37°C and 5% CO2 humidified incubator.
Microsomal cis-prenyltransferase activity measurement
Crude microsomes were prepared as described21 with minor modifications. cis-prenyltransferase (cisPTase) assays and activity measurements in human dermal fibroblasts were performed as described22,23 with minor modifications. In brief, microsomal fractions from cells were prepared by centrifugation at 100,000 × g for 40 min at 4°C. 50 mg microsomal protein was used for cisPTase activity measurement with reaction mixture containing 45 mM FPP, 50 mM [1- 14C]- isopentenyl pyrophosphate (IPP) (55 mCi/mmol; 138,000 cpm/reaction), 25 mM Tris-HCl (pH 7.4), 5 mM MgCl2, 1.25 mM DTT, 2.5 mM sodium orthovanadate, 10 mM Zaragozic acid A, and 0.35% Triton X-100. Reactions were performed at 37°C for 1 h and stopped by the addition of 4 mL of chloroform:methanol (3:2 ratio). The protein pellet was removed by centrifugation and the supernatant was washed three times with 1/5 volume of 10 mM EDTA in 0.9% NaCl. The incorporation of radioactive IPP into organic fraction containing polyprenyl pyrophosphate was measured by scintillation counting.
Western blot analysis
Cells were washed twice with ice-cold PBS and lysed in lysis buffer (50 mM Tris-HCl, 1% NP-40, 0.1% SDS, 0.1% Deoxycholic Acid, 0.1 mM EDTA, 0.1 mM EGTA, protease and phosphatase inhibitors). Protein extracts were separated by SDS-PAGE and then transferred to nitrocellulose membrane. Primary antibodies against NUS1 (Abcam, ab168351), DHDDS (Sigma, HPA026727), ICAM1 (Santa Cruz, Sc8439), LAMP1 (BD Transduction Laboratories, 611402), and HSP90 (Cell Signaling Technology, 4877) were used. The appropriate LI-COR secondary IRDye antibodies and LI-COR Odyssey Infrared Imaging System (LI-COR, Lincoln, NE) were used for antibody detection.
Filipin staining
Cells were grown on glass cover glasses, fixed in 4% PFA for 10 min, and permeablized in 0.1% Triton X-100 for 5 min. Cells were then incubated with 50 mg/mL filipin (Sigma, F4767) for 1 h. As a positive control for induction of cholesterol accumulation, cells were treated for 16 h with 1 mM U18666A (EMD Biosciences). Relative intensity of filipin staining was quantified by calculating average pixel intensity using Adobe Photoshop according to the equation: average filipin intensity = total intensity above low threshold/number of pixels above low threshold.24
Yeast strains and culture methods
S. cerevisiae strain alg10D (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YGR227W::kanX4, Dharmacon) and its derivatives were used. Cultures were grown at 30°C in YPD or Synthetic minimal medium made of 0.67% (wt/vol) yeast nitrogen base and 2% (wt/vol) glucose supplemented with auxotrophic requirements. For solid media, agar (Becton Dickinson) was added at a 2% (wt/vol) final concentration. Yeast transformations were performed by standard yeast genetic methods.
Functional characterization of ALG10/ALG10B variants in yeast
To examine the functionality of hALG10 proteins, the N-glycosylation status of carboxypeptidase Y (CPY) was tested in S. cerevisiae alg10D strain transformed with empty pKG-GW1 plasmid (2 μ, LEU2 marker22) (negative control) or pKG-GW1 carrying yeast (y) ALG10 ORF (positive control), human (h) ALG10, hALG10 p.Lys391Valfs∗35, hALG10B or hALG10B p.Leu253Trp. Yeast transformants were inoculated from single colony and grown overnight at 30°C in synthetic medium lacking leucine. Cells from saturated overnight cultures were harvested and lysed by alkaline method. Whole-cell lysate (WCL) was subjected to SDS-PAGE (7.5% gel) and immunoblotting. Yeast CPY was detected with anti-CPY monoclonal antibodies (Fisher Scientific, clone 10A5B5).
Statistical and data analysis
Statistical analyses and graphical representation were performed with the GraphPad Prism v.7.0 software (GraphPad Software) or the R statistical programming language (v.3.6.1). Figure legends indicate the statistical test used in functional experiments. p values < 0.05 were considered significant.
Gene co-expression networks
For gene co-expression analyses, normalized brain expression values from the BrainSpan Developmental transcriptome dataset were downloaded (see web resources). Genes were removed if they had expression values missing from >50% of the 524 samples available from 42 individuals.
Using the log2 transformed expression values, a matrix of weighted correlations was generated, with weights determined as 1/√n, where n is the number of samples contributed by the respective individual. Correlation plots were visualized using the corrplot R package, with genes ordered by hierarchical clustering, using the median linkage method.
To determine whether the established and candidate PME genes were more highly co-expressed than expected by chance we randomly sampled 5,000 sets of genes. We calculated the median for each random gene set and compared this to the observed median of the PME gene set.
Results
PME cohort
The study cohort included 84 individuals with PME from 78 families who did not have a known molecular basis. Genomic ancestry checks suggested 74 families (95%) were of European descent, with more than half (n = 46) referred from hospital centers in Italy. The other four families were admixed American (n = 2) and East Asian (n = 2). Inbreeding estimates using FEstim suggested 24% of families were consanguineous (19/78). This was consistent with clinical descriptions of parental relatedness in ten families; detailed pedigree histories were not available for the other nine.
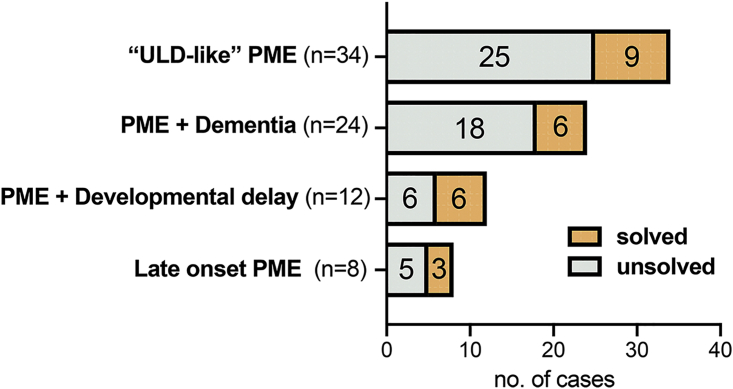
Clinically, the majority of the 78 unrelated affected individuals were classified into two well-established groups: 43% (n = 34) had “ULD-like” PME (i.e., classical childhood/adolescent onset of PME; no dementia) and 31% (n = 24) had PME + dementia. Two smaller groups comprised developmental delay predating PME onset (n = 12) and a group of late-onset (>20 years) PME (n = 8) (Figure 1). Age of disease onset across the cohort ranged from late infancy to 45 years (mean 12 years) (Figure S2).
Figure 1.
Proportion of all 78 unrelated individuals with PME with (solved) and without (unsolved) likely pathogenic variants by clinical group
In total, we identified variants in 24 out of 78 (31%) unrelated affected individuals that we regarded with moderate-to-high confidence (see subjects and methods) as causative. Interestingly, the diagnostic success was highest in one of the two newly recognized, rarer clinical groups (PME with prior developmental delay), although the numbers were small (Figure 1).
We had the most success with individuals in whom we had sequenced additional family members (14/28); we identified a likely causal variant in 45% of trios and in 67% of subjects where another affected 1st-degree relative was sequenced. The proportion of singletons with likely causative variants was significantly less, with just 10 out of 50 cases (OR = 3.9, p value = 0.01, Fisher’s exact test).
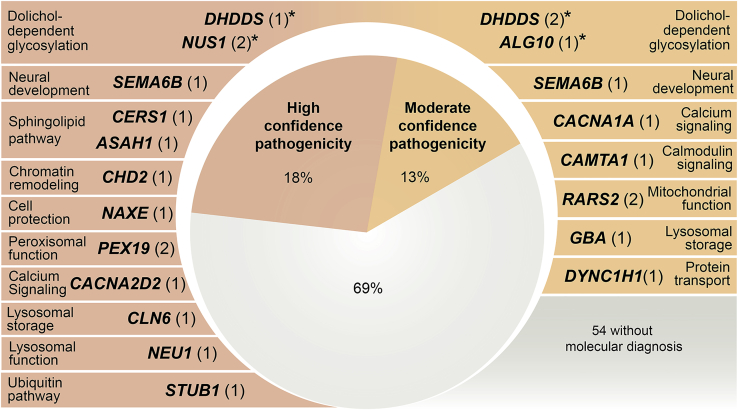
The 24 likely solved cases involved 18 genes: 1 (ALG10 [GenBank: NM_032834.4; MIM: 618355]) has no known disease associations, 6 were known PME genes, including the very recently described SEMA6B (GenBank: NM_032108.4; MIM: 608873),25 and 11 have been reported in other neurological diseases, but not previously in PME (Figure 2, Tables 1, 2, 3, S4, and S5).
Figure 2.
PME-associated genes (n = 18) with high and moderate confidence variants detected in our cohort of 78 unrelated individuals with PME that had previous extensive genetic investigations
The number of unrelated individuals with variants in each gene is shown in parentheses with the known primary function/pathway of each gene also listed. See subjects and methods for criteria followed when classifying variants as high versus moderate confidence. ∗Functionally validated genes in this study.
Table 1.
Dolichol-dependent glycosylation genes with variants identified in the PME cohort
| Patient ID country of origin | Sex | Gene (GenBank) | Variant(s) (LOVD ID) | gnomAD MAF | Inheritance | Clinical summary (onset age) | WES study design | Confidence |
|---|---|---|---|---|---|---|---|---|
| PME1 Italy | M | NUS1 (NM_138459.3) | c.740dupT (p.Asp248Glyfs∗15) (het) (#00334867) | 0 | de novo | myoclonus (13y), no seizures; no ataxia, normal cognition | trio | high |
| PME2 Italy | F | NUS1 (NM_138459.3) | c.310delG (p.Val104∗) (het) (#00334873) | 0 | de novo | absence with eyelid myoclonia (4y); myoclonus (8y); ataxia, moderate cognitive impairment; febrile seizures (4y) with developmental regression | trio | high |
| PME3 Italy | M | DHDDS (NM_024887.3) | c.632G>A (p.Arg211Gln) (het) (#00334875) | 0 | de novo | myoclonus (7y); absences with eyelid myoclonia (9y); mild ataxia, moderate cognitive impairment; developmental delay | trio | high |
| PME71 Italy | F | DHDDS (NM_024887.3) | c.614G>A (p.Arg205Gln) (het) (#00334877) | 0 | unknown | ataxia (late infancy); rare TCS (17y), mild action myoclonus (29y); normal cognition | singleton | moderate |
| PME27 Italy | F | DHDDS (NM_024887.3) | c.283G>A (p.Asp95Asn) (het) (#00334878) | 0 | unknown | tremor (21y); myoclonus (35y), single TCS (36y); ataxia, normal cognition; bilateral deafness | singleton | moderate |
| PME50 Turkey | F | ALG10 (NM_032834.4) | c.1170_1171delAA (p.Lys391Valfs∗35)(hom) (#00334880) | 0 | AR | frequent myoclonus (13y), rare TCS; ataxia, mild cognitive dysfunction (16y); scoliosis | trio | moderate |
Abbreviations: MAF, minor allele frequency; AR, autosomal recessive; het, heterozygous; hom, homozygous; gnomAD, The Genome Aggregation Database; TCS, tonic-clonic seizure. Detailed clinical summaries can be found in Table S5. See subjects and methods for criteria for classifying variants as high versus moderate confidence.
Table 2.
High and moderate confidence variants identified in established PME genes
| Patient ID country of origin | Sex | Gene (GenBank) | Variant(s) (LOVD ID) | gnomAD MAF | Inheritance | Clinical summary (onset age) | WES study design | Confidence |
|---|---|---|---|---|---|---|---|---|
| PME83 Australia | M | SEMA6B (NM_032108.4) | c.1993delC (p.Arg665Glyfs∗20) (het) (#00334899) | 0 | AD | developmental delay and regression; ataxia, tremor (2.5y); drop attacks and absence seizures (4y), TCS (11y), wheelchair (11y), multifocal myoclonus (15 y); severe ID | singletona | high |
| PME25 Canada | F | SEMA6B (NM_032108.4) | c.2032delG (p.Glu678Argfs∗7) (het) (#00334902) | 0 | AD | developmental delay; ataxia (2.5y); TCS (5y), resting and action myoclonus (10y), possible absence and focal seizures, tremor, wheelchair (14y); moderate ID | singleton | moderate |
| PME15 Italy | F | CLN6 (NM_017882.3) | c.486+28T>C (splicing) (hom)b (#00334904) | 0 | AR | ataxia (14y); severe myoclonus (32y), TCS, dementia, pyramidal signs, psychiatric co-morbidities | singleton | high |
| PME26 (dec.) Germany | M | GBA (NM_001005742.2) | c.761+4A>G (splicing) (hom) (#00334906) | 0 | AR | myoclonus (8y); ataxia, ophthalmoplegia, mild cognitive impairment, splenomegaly | singleton | moderate |
| PME10 Malaysia | M | NEU1 (NM_000434.3) | c.544A>G (p.Ser182Gly); deletion of NEU1 (comp het) (#00334907) | 0.001; 0 | AR | occasional TCS (12y); frequent myoclonus (14y), ataxia, normal cognition, visual deterioration (20y), cherry-red spots (21y) | trio | high |
| PME7 Israel | F | CERS1 (NM_021267.4) | c.210G>A (p.Trp70∗); c.202C>A (p.Leu68Met) (both hom) (#00334908), (#00334910) | 0; 0 | AR | action myoclonus (11/16yr); ataxia, occasional TCS, mild cognitive impairment | sibling pair; quartet | high |
| PME8 Israel | F | |||||||
| PME9 (dec.) Australia | M | ASAH1 (NM_004315.4) | c.966−2A>G, splicing; c.504A>C (p.Lys168Asn) (comp het) (#00334911) | 0.000004; 0.00006 | AR | multifocal myoclonus (10y). TCS, progressive limb and bulbar weakness (16y); hearing impairment (4y); deceased (19y) | trio | high |
Abbreviations: MAF, minor allele frequency; comp het, compound heterozygous; hom, homozygous; AR, autosomal recessive; gnomAD, The Genome Aggregation Database; dec., deceased; TCS, tonic-clonic seizure. Detailed clinical summaries can be found in Table S5. See subjects and methods for criteria followed when classifying variants as high versus moderate confidence.
Variant subsequently confirmed de novo by Sanger sequencing; maternal DNA did not meet quality control requirements for WES.
Splicing effect of intronic variant confirmed by RT-PCR (see Figure S4).
Table 3.
High and moderate confidence variants identified in established disease genes (not PME)
| Patient ID country of origin | Sex | Gene (GenBank) | Disease previously associated with gene (MIM ID) | Variant(s) (LOVD ID) | gnomAD MAF | Inheritance | Clinical presentation | WES study design | Confidence |
|---|---|---|---|---|---|---|---|---|---|
| PME11 Italy | M | CHD2 (NM_001271.3) | epileptic encephalopathy, childhood-onset (MIM: 615369) | c.532A>T (p.Arg178∗) (het) (#00334913) | 0 | de novo | frequent absence seizures and rare TCS (6y), severe myoclonus (14y); ataxia, dementia; developmental delay | trio | high |
| PME19 Italy | M | CACNA2D2 (NM_001174051.2) | cerebellar atrophy with seizures and variable developmental delay (MIM: 618501) | c.1260G>A (p.Thr420=) (het, de novo); c.1112A>G (p.Tyr371Cys) (het, pat inherited) (#00334914) | 0; 0 | AR | myoclonus, absence and tonic seizures (4y); dementia, no ataxia; developmental delay | trio | high |
| PME4 (dec.) Italy | F | STUB1 (NM_005861.4) | autosomal-recessive spinocerebellar ataxia 16 (MIM: 615768); spinocerebellar ataxia 48 (MIM: 618093) | c.169C>T (p.Pro57Ser) (hom) (#00334915) | 0 | AR | ataxia (12y); myoclonus, TCS (30y); dementia; tetraparesis | trio | high |
| PME16 Italy | F | CACNA1A (NM_001127222.2) | early infantile epileptic encephalopathy (MIM: 617106); spinocerebellar ataxia 6 (MIM: 183086); episodic ataxia type 2 (MIM: 108500); familial heiplegic migraine 1 (MIM: 141500); familial hemiplegic migraine 1 with progressive cerebellar ataxia (MIM: 141500) | c.4897G>A (p.Asp1633Asn) (het)a (#00334917) | 0 | unknown | ataxia, myoclonus (30y); cognitive impairment; sensorineural hearing impairment | singleton | moderate |
| PME17 Italy | F | CAMTA1 (NM_015215.4) | non-progressive cerebellar ataxia with mental retardation (MIM: 614756) | c.4418G>C (p.Ser1473Thr) (het) (#00334922), (#00334925) | 0.000004 | AD | myoclonus (18y), no TCS; no ataxia or dementia | parent-child | moderate |
| PME18 Italy | M | myoclonus, rare TCS (25y); no ataxia or dementia | |||||||
| PME21 Malta | M | PEX19 (NM_001193644.1) | peroxisome biogenesis disorder 12A (Zellweger) (MIM: 614886) | c.254C>T (p.Ala85Val) (hom) (#00334927), (#00334928) | 0.0009 | AR | progressive ataxia (7yr); myoclonus, TCS (9y), dementia (10y); limb spasticity | sibling pair | high |
| PME22 Malta | M | progressive ataxia (8y); myoclonus, TCS (9y); dementia (10y); limb spasticity | |||||||
| PME60 (dec.) Malta | F | PEX19 (NM_001193644.1) | Peroxisome biogenesis disorder 12A (Zellweger) (MIM: 614886) | c.254C>T (p.Ala85Val) (hom) (#00334930) | 0.0009 | AR | progressive severe ataxia (8y); TCS (12y); hypertonia | singleton | high |
| PME5 (dec.) Italy | F | NAXE (NM_144772.2) | encephalopathy, progressive early-onset, with brain edema and/or leukoencephalopathy (MIM: 617186) | c.128C>A (p.Ser43∗) (hom) (#00334932) | 0.00003 | AR | versive motor seizures (12y), daily absence (13y) and myoclonus (15y), rare TCS (21y); slowly progressive ataxia (19y) dementia and pyramidal signs; developmental delay | singleton | high |
| PME12 Italy | M | RARS2 (NM_020320.3) | pontocerebellar hypolasia type 6 (MIM: 611523) | c.943C>T (p.Arg315∗); c.425T>C (p.Val142Ala) (comp het) (#00334933), (#00334935) | 0.00004; 0.00005 | AR | mild ataxia (childhood), moderate cognitive impairment; rare TCS and absence seizures (9y), mild myoclonus (11y) | sibling pair; quartet | moderate |
| PME13 Italy | F | ataxia (childhood), moderate cognitive impairment; rare TCS and absence seizures (9), myoclonus (11y) | |||||||
| PME14 Italy | F | RARS2 (NM_020320.3) | pontocerebellar hypolasia type 6 (MIM: 611523) | c.1026G>A (p.Met342Ile); c.3G>A (p.Met1Ile) (comp het) (#00334936) | 0.0002; 0 | AR | prominent progressive action myoclonus (25y); no TCS, no ataxia, no dementia | trio | moderate |
| PME64 Italy | M | DYNC1H1 (NM_001376.4) | Charcot-Marie-Tooth disease axonal type 20 (MIM: 614228); mental retardation, autosomal-dominant 13 (MIM: 614563); spinal muscular atrophy lower extremity-predominant (MIM: 158600) | c.7828delC (p.Arg2610Glyfs∗23) (het) (#00334938) | 0 | de novo | myoclonus (12y), refractory TCS and absence seizures (22y); no ataxia or dementia | trio | moderate |
Abbreviations: MAF, minor allele frequency; AR, autosomal recessive; comp het, compound heterozygous; het, heterozygous; hom, homozygous; AD, autosomal dominant; gnomAD, The Genome Aggregation Database; dec., deceased; TCS, tonic-clonic seizure. Detailed clinical summaries can be found in Table S5. Please see subjects and methods for criteria followed when classifying variants as high versus moderate confidence.
See Figure S7 for molecular modeling that supports a loss-of-function effect for this CACNA1A variant.
Dolichol-dependent glycosylation identified as a PME pathway
In discovering variants in NUS1 (GenBank: NM_138459.3; MIM: 610463), DHDDS (GenBank: NM_024887.3; MIM: 608172), and ALG10 in a total of 6 unrelated subjects, we identified dolichol-dependent glycosylation as a disease pathway for PME (Figures 2 and S3). The age of onset and clinical features were heterogeneous (Table 1).
We subsequently functionally characterized the variants in these three related genes. NUS1 and DHDDS encode two subunits of cisPTase (also known as dehydrodolichyl diphosphate synthase), the first enzyme committed to dolichol synthesis (Figure S3D).22,23,26,27 CisPTase is located at a critical branchpoint of farnesyl diphosphate metabolism, with an alternate branch responsible for cholesterol synthesis. ALG10 is more distal in the dolichol pathway; it is a glucosyltransferase that transfers the terminal glucose residue from dolichyl phosphate glucose (Dol-P-Glc) onto the lipid-linked oligosaccharide precursor Glc2Man9GlcNAc(2)-PP-Dol. The terminal glucose residue added is a key element required for efficient N-linked glycosylation of proteins.28
NUS1
Two individuals with PME had variants in NUS1 (also termed NgBR), encoding the accessory subunit of cisPTase. PME1 carried a de novo frameshift variant c.740dupT (p.Asp248Glyfs∗15) and PME2 a de novo nonsense variant c.310delG (p.Val104∗) (Tables 1, S4, and S5, Figure S3A).
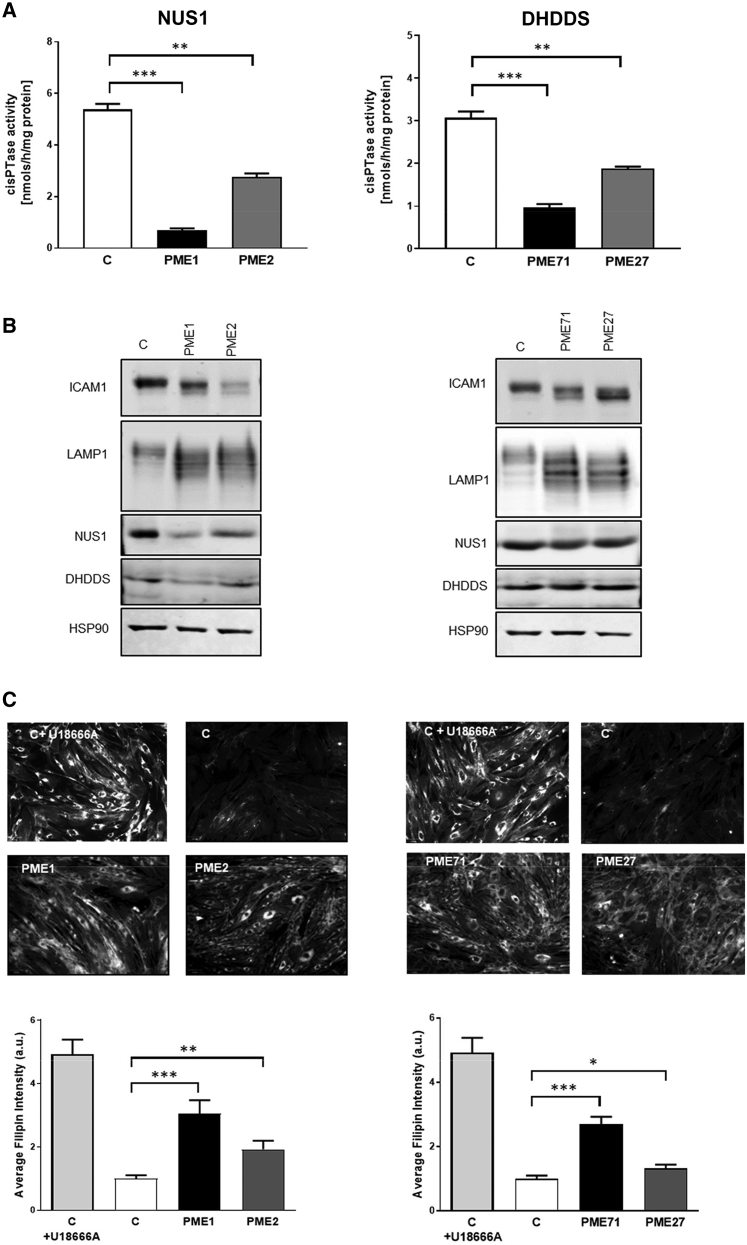
Initial analysis of fibroblast cells by western blotting revealed decreased amount of NUS1 in cells from individuals with PME compared to control subjects, implying the presence of nonsense-mediated mRNA decay and/or instability of the truncated proteins (Figure 3B). In PME1, also the amount of DHDDS appeared to be decreased, in line with the predicted truncated NUS1 product that is missing the interface region for heterodimerization with DHDDS and consequently DHDDS instability.23,29,30 CisPTase activity in cells was drastically decreased, demonstrating that lower protein levels directly affect enzymatic turnover rates (Figure 3A). In order to evaluate the consequence of the reduced cisPTase activity in fibroblast cells, the glycosylation status of ICAM1 and LAMP1, established markers for N-glycosylation defects,31,32 was examined. Altered ICAM1 and LAMP1 expression and migration were detected by western blot analysis (Figure 3B). Finally, we examined free cholesterol levels, an additional consequence of NUS1 dysfunction in cells.33 Fibroblasts were stained with filipin and free cholesterol pools were examined. Both case fibroblasts exhibited increased accumulation of free cholesterol compared to controls (Figure 3C).
Figure 3.
The NUS1 and DHDDS variants cause defects in protein glycosylation due to reduced cisPTase activity in patient-derived fibroblast cell lines
(A) Reduced microsomal cisPTase activity in isolated membranes from PME (NUS1: PME1, PME2 and DHDDS: PME71, PME27) compared to control (C) fibroblasts. ∗∗p < 0.005, ∗∗∗p < 0.001. Data presented as mean ± SEM of at least three independent measurements.
(B) Affected protein glycosylation in PME fibroblasts. Western blot analysis of NUS1, DHDDS, LAMP1, and ICAM1 levels. HSP90 was used as loading control.
(C) Increased cholesterol accumulation in PME fibroblasts. Filipin staining and quantitative representation from PME and control cells. U18666A was used as a positive control for inhibition of cholesterol trafficking. ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.001, a.u., arbitrary units. Data are representative of at least three experiments.
DHDDS
Three individuals with PME were identified with rare missense variants in DHDDS (also termed hCIT), encoding the catalytic subunit of cisPTase. PME3 was found to have a de novo missense variant c.632G>A (p.Arg211Gln) previously described in three individuals with developmental and epileptic encephalopathy (DEE).34,35 PME71 and PME27 carried heterozygous missense variants c.614G>A (p.Arg205Gln) and c.283G>A (p.Asp95Asn), respectively. No parental samples were available for PME71 for segregation analysis (Tables 1, S4, and S5, Figure S3B). For PME27, it was possible to exclude the c.283G>A variant only in the father as maternal DNA was unavailable. PME27 was also heterozygous for a rare variant in DNMT1 (c.1619A>G [p.Tyr540Cys]), but without functional support this variant did not meet our criteria for prioritization (Table S6).
Functional studies in fibroblasts from PME71 and PME27 showed apparently normal amounts of both DHDDS and NUS1 (Figure 3B) in line with the preserved capacity for heterodimerization, decreased cisPTase activity in isolated membranes (Figure 3A), and altered levels and migration of ICAM1 and LAMP1 proteins indicating protein N-glycosylation defect (Figure 3B). Furthermore, consistent with reduced cisPTase activity and protein glycosylation defect, increased cholesterol accumulation was detected in both fibroblast cells (Figure 3C). Fibroblasts were not available from PME3 but, because the variant was de novo and previously reported, we regarded it as disease causing with high confidence.
ALG10
PME50 was included in the first exome study and identified to carry the homozygous frameshift variant c.1170_1171delAA (p.Lys391Valfs∗35) (Tables 1, S4, and S5, Figure S3C) in ALG10,3 encoding a putative alpha-1,2-glucosyltransferase. At that time, with no prior disease association for ALG10 and with no functional studies performed, the variant was regarded as of uncertain significance. Here, we now provide evidence for its pathogenicity.
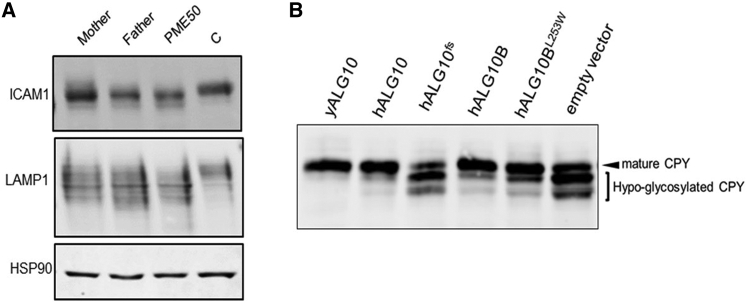
Hypo-glycosylation of reporter proteins ICAM1 and LAMP1, identified in western blot analysis of fibroblasts from PME50 (Figure 4A), predicts a defect in alpha-1,2-glucosyltransferase activity. The heterozygous carrier parents of PME50 also showed abnormal glycoslation pattern (Figure 4A). In the sixth decade of their life, both parents were neurologically normal. The mother, like PME50 (Table S5), was morbidly obese, while the father was of normal weight.
Figure 4.
The ALG10 frameshift mutation causes defects in protein N-glycosylation due to a predicted defect in alpha-1,2-glucosyltransferase activity
(A) Affected protein glycosylation in fibroblasts carrying the ALG10 and ALG10B variants. Western blot analysis of ICAM1 and LAMP1 expression. HSP90 was used as loading control.
(B) Protein N-glycosylation of CPY shows multiple hypo-glycosylated bands in a yeast alg10 deletion strain transformed with mutated human (h) ALG10 (hALG10fs) or empty vector. N-glycosylation deficiency is rescued when transformed with either wild-type yeast ALG10 (yALG10), hALG10, or hALG10B.
To confirm the predicted function of ALG10 as an alpha-1,2-glucosyltransferase and to model the ALG10 variant, we used a yeast alg10 deletion strain to re-express human wild-type and mutant ALG10 proteins for functional complementation. In the absence of dolichyl-phosphoglucose-dependent alpha-1,2-glucosyltransferase activity, the lipid-linked oligosaccharide (N-glycan precursor) lacking terminal glucose is less efficiently transferred to glycoprotein,28 resulting in the reporter protein for N-glycosylation, CPY, being hypo-glycosylated and running more quickly on SDS-polyacrylamide gel electrophoresis. Both yeast and human ALG10 did complement the yeast deletion strain (Figure 4B), as judged by the presence of mainly the mature form of CPY. Alg10 deletion strains transformed either with empty vector or with mutated ALG10 (Figure 4B) showed multiple bands of CPY corresponding to hypo-glycosylated forms of the protein, thus supporting the pathogenicity of the ALG10 p.Lys391Valfs∗35 variant. Given that the ALG10 variant was only detected in one individual with PME and has no established disease association, we classified it as disease causing with moderate confidence, despite the functional evidence for its pathogenicity.
PME50, however, is also homozygous for a missense variant c.758T>G (p.Leu253Trp) in the highly homologous ALG10B (MIM: 603313) gene encoding alpha-1,2-glucosyltransferase B.36 The missense variant is reported in gnomAD with an allele frequency of 0.004 with 5 homozygous individuals so it is unlikely to be pathogenic on its own. However, while human ALG10B complemented glycosylation in the yeast assay, the variant ALG10B was not quite as effective (Figure 4B), so we could not rule out a contribution of the homozygous ALG10B variant to the phenotype.
Likely causative variants in established PME genes
In seven families, we identified likely causative variants in six established PME genes (SEMA6B, CLN6, GBA [GenBank: NM_001005742.2; MIM: 606463], NEU1, CERS1 [GenBank: NM_021267.4; MIM: 606919], ASAH1 [GenBank: NM_004315.4; MIM: 613468]) (Table 2). These cases all defied diagnosis earlier because of unusual genetic mechanisms or very rare or newly recognized causes.
SEMA6B was recently published as a dominant PME gene with de novo variants in four individuals.26 We independently validate this finding with an additional two affected individuals (PME83, PME25). Both of our subjects had frameshift variants in the last exon of SEMA6B (Table 2) within very close proximity to the published series.26 Low coverage, due to high GC content, of this exon meant that only one of the two variants were initially called by our bioinformatics pipeline and thus both variants escaped detection until targeted SEMA6B reanalysis. Clinically, PME83 and PME25 were classified as PME with developmental delay, consistent with the published cases (Table S5). We confirmed the de novo status for PME83 by subsequent Sanger sequencing of the parents, but parental DNA was unavailable for PME25.
In the case of CLN6 and GBA, the putative causative variants (Table 2) are both intronic and were not prioritized by initial filtering strategies. Prior to genetic testing, PME15 and PME26 were clinically suspected of having Kufs Type A (MIM: 601780) and Gaucher disease type III (MIM: 231000), respectively.37,38 Both variants are homozygous and inbreeding coefficient estimates were consistent with parental consanguinity for the two families. Predictions for the GBA splice-site variant having an effect on mRNA splicing was consistent across all splicing in silico tools; however, without the ability to confirm this experimentally (PME26 deceased), we classified the variant as likely causative with moderate confidence. The deep intronic CLN6 variant was predicted in silico to create an intronic exonic splicing enhancer (ESE) site39 and RT-PCR from PME15 fibroblast cells confirmed aberrant mRNA splicing (Figure S4A). Sanger sequencing of the aberrant product revealed inclusion of 119 nucleotides of intronic sequence downstream of the 3′ end of exon 4 (Figure S4B). These data are compatible with the homozygous variant in PME15 causing activation of a non-canonical splice site through creation of an intronic ESE site (Figure S4C). The intronic inclusion creates a premature stop codon after 60 nucleotides of open reading frame in the intronic sequence. This is predicted to result in nonsense-mediated decay with partial loss-of-functional protein, compatible with the late-onset CLN6 disease in PME15. As such we classified this variant as likely causative with high confidence (Figure 2; Tables S4 and S5).
Our single CNV finding was at the NEU1 locus. In PME10, WES data initially suggested a homozygous c.544A>G (p.Ser182Gly) NEU1 variant (Table 2). Validation by Sanger sequencing showed that only the mother was a heterozygous carrier of the missense variant. Reanalysis of the WES data for a potential CNV in the region indicated the presence of a deletion on the paternal allele, confirmed by quantitative PCR (Figure S5). Subsequently, PME10’s younger brother developed symptoms and genetic analysis confirmed his compound heterozygous status for the same NEU1 variants. Clinically the presentation for both brothers was consistent with sialidosis (MIM: 256550)40 (Tables S4 and S5).
Recessive variants in very rare PME genes involved in the sphingolipid pathway, CERS141 and ASAH1,42 were identified in one family each. Siblings PME7 and PME8 were homozygous for two variants in CERS1, a nonsense and a missense variant (Table 2). Segregation analysis confirmed heterozygosity for both variants in one of the parents, respectively. The parents were known to be related, with consistent inbreeding F estimates. In PME9, WES revealed compound heterozygous variants in ASAH1, one splice-site and one missense variant (Tables 2, S4, and S5); at diagnosis the individual had PME but not spinal muscular atrophy although this subsequently developed.
Likely causative variants in other known disease genes
An additional 11 likely causative variants were identified in genes not previously associated with PME, but recognized in neurological phenotypes including seizures or ataxia (Table 3). CHD2 (GenBank: NM_001271.3; MIM: 602119), CACNA2D2 (GenBank: NM_001174051.2; MIM: 607082), and CACNA1A (GenBank: NM_001127222.2; MIM: 601011) are established DEE genes, as are NUS1 and DHDDS involved in dolichol metabolism (see above). CACNA1A is also associated with ataxia syndromes as are STUB1 (GenBank: NM_005861.4; MIM: 607207) and CAMTA1 (GenBank: NM_015215.4; MIM: 611501).
PEX19 (GenBank: NM_001193644.1; MIM: 600279), NAXE (GenBank: NM_144772.2; MIM: 608862), RARS2 (GenBank: NM_020320.3; MIM: 611524), and DYNC1H1 (GenBank: NM_001376.4; MIM: 600112) are currently associated with more complex neurological phenotypes (Table 3). These variants all met our criteria for moderate to high confidence in causation based on both the genetic data and phenotypic overlap (Tables S4 and S5) (see subjects and methods).
In the case of PEX19, this is a well-established gene for peroxisome biogenesis disorders (MIM: 614886). We identified three individuals (PME21, PME22, PME60) from two unrelated families of Maltese origin, with the same homozygous missense variant c.254C>T (p.Ala85Val). All three individuals shared a similar phenotype with onset around age 9 years involving myoclonus, tonic-clonic seizures, ataxia, cognitive decline, and marked photosensitivity (Tables 3 and S5). PME60 had a clinically similarly affected brother who was deceased and not tested. This variant is not present in the Maltese Genome project (J. Vella et al., 2013, European Society of Human Genetics, conference) with 400 individuals; however, haplotype analysis results were consistent with a distant founder effect (Figure S6). Further, independent studies have identified two additional Maltese individuals with the same homozygous variant and similar clinical phenotype (data not shown).
Filtered variants that did not meet our criteria for prioritization can be found in Table S6. Our short tandem repeat analyses did not detect any expansions at the known pathogenic loci (Table S3).
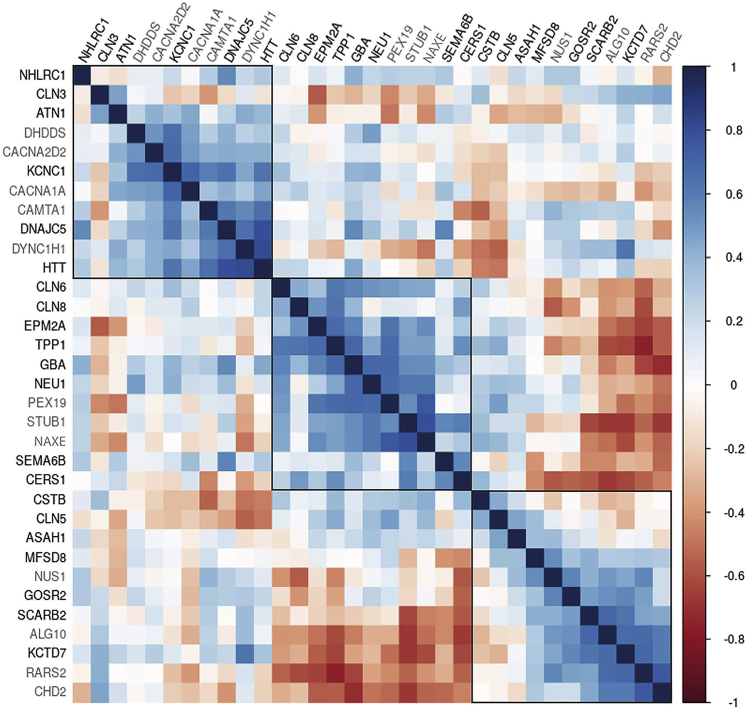
PME gene brain co-expression networks
Using brain expression data from BrainSpan, we examined the co-expression between the major established PME genes (Table S1) and all genes we report here with likely causative variants (Tables 1, 2, and 3). Expression data were not available for MT-TK (MIM: 590060) responsible for myoclonus epilepsy associated with ragged-red fibers (MERRF [MIM: 545000]); this mitochondrial gene was therefore excluded from the analysis.
The ordered correlation matrices revealed some striking patterns (Figure 5). We observed 3 large clusters of 11 positively correlated gene sets that accounted for all candidate and established (bold) PME genes. Cluster one contains NHLRC1 (MIM: 608072), CLN3 (MIM: 607042), ATN1 (MIM: 607462), DHDDS, CACNA2D2, KCNC1, CACNA1A, CAMTA1, DNAJC5 (MIM: 611203), DYNC1H1, HTT (MIM: 613004). Cluster two contains CLN6, CLN8 (MIM: 607837), EPM2A (MIM: 607566), TPP1 (MIM: 607998), GBA, NEU1, PEX19, STUB1, NAXE, SEMA6B, CERS1. Cluster three contains CSTB, CLN5 (MIM: 608102), ASAH1, MFSD8 (MIM: 611124), NUS1, GOSR2 (MIM: 604027), SCARB2 (MIM: 602257), ALG10, KCTD7 (MIM: 611725), RARS2, CHD2.
Figure 5.
Gene co-expression matrix for 33 known (black) and candidate (gray) PME genes
Pairwise Spearman correlations between genes shown, based on 524 samples from 42 individuals from the BrainSpan resource. Genes are ordered and grouped with hierarchical clustering, using the median linkage method.
Using a Monte Carlo sampling approach, we found evidence that the established and candidate PME genes were more highly co-expressed than would be expected by chance (p < 0.05). These results suggest that overall these genes have similar brain gene expression signatures. Shared biological networks are further supported by the observation that clusters 2 and 3 are negatively correlated.
Discussion
Our data uncovered dolichol-dependent protein glycosylation as an important pathway underlying PME. Additional findings were the confirmation of SEMA6B as a cause of PME and that PME can sometimes be a rare manifestation of variants in genes associated with developmental and epileptic encephalopathy or ataxia syndromes. Finally, our results suggest that there is unlikely to be a major shared genetic basis to the remaining unsolved cases, but rather the answer will most likely be a heterogenous mix of rare disorders. However, rare variants in a novel gene, particularly in the introns, and regions of low coverage cannot be excluded.
Overall, we identified plausible pathogenic variants in 24 out of 78 (31%) unrelated affected individuals. This cohort had been extensively studied for known genetic causes previously, so, it is notable that our diagnostic yield was this high. As de novo dominant mutations were recently established as an important alternative cause of PME,3 we pursued a trio-design WES analysis where possible. Overall, we had significantly greater success identifying plausible pathogenic variants in subjects that had been sequenced with other family members (i.e., as an affected trio- or quartet- with unaffected parents or part of an affected sibling or parent-offspring pair). This was driven in part by the importance of de novo variants in dominant genes, that has previously been under-appreciated for this disease group, but also the ability to confirm compound heterozygosity and/or homozygosity for variants under a recessive model. Clinically, the two primary categories of PME have historically been separated according to the presence (PME with dementia) or absence (“ULD-like”) of cognitive decline. In this analysis of cases defying molecular diagnosis, two additional clinical groups were apparent: PME with prior developmental delay and a late-onset group. Our success rate in diagnosis was highest for one of the newly recognized, albeit smaller, clinical groups: 50% for PME with prior developmental delay (Figure 1).
We associate dolichol-dependent glycosylation with the PME phenotype through the identification of variants in NUS1, DHDDS, and ALG10, supported by demonstrating glycosylation defects in fibroblast cell lines and/or in yeast assays. Protein glycosylation is a ubiquitous post-translational modification that contributes to several crucial biological and physiological processes within cells. Given that variants in NUS1, DHDDS, and ALG10 were associated with altered expression and migration of ICAM1 and LAMP1, and since ALG10 is specifically linked to N-glycosylation, it is plausible that variants in these genes result in N-glycosylation defects in cells.23,29, 30, 31, 32 N-glycosylation followed by oligomannose phosphorylation of the N-glycated protein is pivotal for lysosomal targeting of enzymes.43 Given that defects in many lysosomal enzymes have been associated with PME, hypoglycosylation caused by impaired N-glycosylation of such proteins may be contributing to the phenotype in individuals with mutations in NUS1, DHDDS, and ALG10. However, the exact mechanisms would need to be explored in further functional studies. Of note, dolichol metabolism was first associated with PME more than 30 years ago with the observation that dolichol content was significantly increased in the brains and urinary sediment of individuals with NCL.44,45 The reason for this observation remained unknown but was postulated to be caused by a possible defect in dolichol recycling or metabolism. PME now joins the expanding list of phenotypes included under the rubric of congenital disorders of glycosylation, which are quite clinically heterogeneous (see GeneReviews in web resources). Unlike most of the established PME genes where the clinical presentation is somewhat characteristic for each gene, the clinical picture of the dolichol pathway genes (Table 1) is more reminiscent of TBC1D24 (MIM: 613577) where the clinical spectrum is much wider.46
A handful of pathogenic variants in NUS1 and DHDDS have previously been associated with various phenotypes. Bi-allelic mutations in both genes have been reported in single families with congenital disorders of glycosylation showing severe, multiorgan manifestations,22,47 and in DHDDS additionally with retinitis pigmentosa.48 More recently, heterozygous de novo variants in both NUS1 and DHDDS were reported in individuals with DEE.34,35 Interestingly, one of these DHDDS variants was identified in PME3 in our cohort and recently the recurrent p.Arg37His variant was reported in a case with mild intellectual disability, rare generalized seizures, and a stable myoclonic tremor.49 NUS1 variants have also been associated with early-onset Parkinson disease with an increase in rare variant burden in PD-affected individuals versus control subjects.50 Remarkably, variants in two established recessive PME genes, GBA and SCARB2, are also risk alleles for Parkinson disease.51, 52, 53 Finally, the recent NUS1 reports of a recurrent heterozygous de novo variant in two unrelated individuals with epilepsy, myoclonus, and ataxia54 and an autosomal-dominant family with epilepsy, ataxia, and tremor segregating a heterozygous frameshift variant,55 support our conclusion that NUS1 is a PME gene. However, it is clear that the phenotypic spectrum for both NUS1 and DHDDS is broad.
The majority of proteins involved in the N-glycosylation pathway (like NUS1 and DHDDS) have been associated with mainly autosomal-recessive congenital disorders of glycosylation.56 ALG10 is a rare exception as it has previously not been associated with any clinical phenotype, the only exception being our report of it as a candidate gene for PME based on the identification of a homozygous frameshift variant in PME50.3 Here, through functional characterization of this variant, we give further support for ALG10 being a PME gene. However, despite functional evidence implying pathogenicity of the reported ALG10 variant, further individuals with PME should be identified to establish ALG10 as a disease gene. Interestingly, PME50 was also homozygous for a missense variant in the highly homologous ALG10B gene (also known as KCR1),36 that has not been previously associated with any human recessive disorder. Our yeast complementation data imply that the ALG10B variant may be a hypomorph with attenuated ability for transferring the glucose residue to the lipid-linked oligosaccharide precursor. In the absence of ALG10 activity, this may not be enough to maintain a proper level of cellular transferase activity. It is therefore possible that compromised function of both genes is required for an ALG10-related disease to manifest.
Individuals with PME are typically cognitively normal prior to epilepsy onset. Here we highlight a rare group with prior developmental delay (n = 12); six have plausible genetic findings. Importantly, two of the six had heterozygous frameshift variants in SEMA6B. SEMA6B was recently discovered as a rare PME gene, with frameshift variants all occurring in the GC-rich last exon of this gene26 in four subjects. They had mild initial developmental delay, seizure onset between 11 months and 6 years with subsequent cognitive and motor regression, needing assistance with ambulation by the early second decade. Microcephaly and spasticity were present in some. All were regarded as having severe intellectual disability and they were all alive at ages 12–28 years. Our cases had a similar course (Table S5), but did not have microcephaly or significant spasticity and the level of intellectual disability was moderate-severe. To date they have survived until 38 and 39 years without further deterioration, unlike the pattern seen in some PMEs due to storage disorders and those with mutations in SCARB2 or GOSR2 with prominent early-adult deterioration and often early death.
Traditionally, PME and DEE are regarded as distinct syndromic groups; this distinction continues to be practically useful. However, it is now clear that the boundary between these groups is blurred, both from a genetic and phenotypic view point. Three of the other four developmentally delayed PME-affected individuals with molecular findings had variants in established DEE genes. This included de novo dominant variants in DHDDS and CHD2 and a recessive CACNA2D2 mutation. Here, we associate these three DEE genes with PME, building on our initial study where we expanded the TBC1D24 phenotypic spectrum to PME.3 Similarly, KCNA2 (MIM: 176262), another established DEE gene, was recently reported in a single case of PME.57 In the reverse direction, after we discovered KCNC1 as a causative de novo dominant PME gene, it has now also been established as an important DEE gene, although the causative mutations differ.58
We also report putative pathogenic variants in a handful of known ataxia genes (Table 3), both recessive and dominant. These genes join AFG3L2 (MIM: 604581) and SACS (MIM: 604490) reported in our initial study3 as known ataxia genes with pathogenic variants in individuals with PME. We had a small number of individuals in our PME cohort of 84 who had no reported tonic-clonic seizures making their clinical presentation more consistent with progressive myoclonic ataxia (PMA [MIM: 159700]). This clinical overlap, with both PME and PMA presentations, is well established for genes such as GOSR2 and KCNC1.6,59 We also identified interesting variants in other known neurological disease genes (Table 3) that not only significantly broaden the genetic basis to the PMEs, but also highlight the need for further functional studies and larger patient numbers to fully understand genotype-phenotype correlations.
The brain gene co-expression analysis uncovered some potentially important relationships between established PME and newly reported PME genes. The advantage of using a brain-specific resource for this analysis, such as BrainSpan, is the detection of brain-specific signatures. An additional advantage of the brain gene co-expression approach is that it is not biased against genes with little known about their function or limited by published material as can be the case for other network generating data sources (e.g., protein-protein interactions or text-mining). The observation that three PME genes associated with the sphingolipid pathway (i.e., CERS1, NEU1, GBA) cluster together in gene set 2 (Figure 5) is proof of principle for the unbiased gene co-expression approach. As such, the clustering of genes that have not previously been biologically associated may indeed be highlighting biologically relevant pathways.
Future perspectives
The PMEs are the genetically best-characterized group of epilepsies. They are highly genetically heterogeneous and there are founder effects, resulting in a different distribution of particular types of PME in various populations. The most comprehensive study of ∼200 affected individuals from Italy reached a diagnosis in ∼70% of cases although not all were fully investigated.2 A number of the residual cases have been diagnosed subsequently, including via this study. Future whole-genome sequencing approaches such as long read sequencing, as well as improved bioinformatic software (e.g., for structural variant calling, repeat expansion detection) likely hold the key to uncovering the elusive genetic basis to these remaining rare genetic disorders.
We only report one pathogenic CNV in this study, but we cannot rule out CNVs as a more important genetic factor due again to exome-sequencing data being limited in its ability to detect such genetic variants. The same argument is true for the detection of repeat expansions. Over half of the known disease-causing repeat expansions are located in intronic and UTR gene regions that are not well captured by exome-sequencing data, so it is not perhaps surprising we had no positive results from this analysis. The recent discovery of pathogenic intronic pentanucleotide expansions in familial adult myoclonic epilepsy (FAME), a dominant disorder that is on the mild end of severity of the PME spectrum, reinforces the relevance and importance of searching for known and novel repeat expansions in genetically unsolved individuals with PME.60, 61, 62, 63 Finally, it is also possible that there are important mtDNA pathogenic variants that remain undetected as we did not systematically study the mitochondrial genome.
Our experience with this cohort has highlighted just how genetically heterogeneous the residuum of unsolved individuals with PME are. Of the remaining unsolved cases, they are unlikely to include another gene affecting a large proportion of cases like KCNC1,3 but rather they are probably a collection of multiple rare genetic causes. Collectively, we estimate that it is now less than 20% of individuals with PME that cannot be attributed to known disease genes with intronic variants possibly going undetected in previous analyses as was the case for our CLN6- and GBA-positive cases. The detection and interpretation of such variants will only improve as the field transitions from exome sequencing to whole-genome sequencing.
Declaration of interests
M.M. is employed by Blueprint Genetics. All other authors declare no competing interests.
Acknowledgments
The authors are indebted to the families participating in this study. We thank Paula Hakala, Katri Aksentjeff, Saara Tegelberg, Simona Allievi, and Marta Bayly for technical support and Michael Hildebrand for molecular analysis. The following funding bodies are acknowledged: Swiss National Foundation (Early Postdoc Mobility Grant [to C. Courage]), Folkhälsan Research Foundation (to A.-E.L.), NIH grant R35 HL139945 (to W.C.S.), Australian National Health and Medical Research Council (NHMRC) Program Grants GNT1054618 (to M.B.) and GNT1091593 (to S.F.B. and I.E.S.), NHMRC Senior Research Fellowship (GNT1102971) and Independent Research Institute Infrastructure Support Scheme (IRIISS) (to M.B.), Victorian Government’s Operational Infrastructure Support Program (to M.B.), Istanbul University Scientific Research Fund-BAP-2019K12-149071 (to B.B.), NHMRC Senior Research Fellowship (GNT1104718) (to L.M.D.); and NHMRC Practitioner Fellowship (GNT1104831) (to I.E.S.). A.-E.L. is a HiLIFE Fellow at the University of Helsinki.
Published: April 1, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.03.013.
Contributor Information
Samuel F. Berkovic, Email: s.berkovic@unimelb.edu.au.
Anna-Elina Lehesjoki, Email: anna-elina.lehesjoki@helsinki.fi.
Data and code availability
All variants reported in this study have been deposited to the LOVD database; individual LOVD identifiers are provided in Tables 1, 2, 3, and S4. WES data have not been deposited in a public repository due to privacy and ethical restrictions. R code used in brain co-expression analysis is available as supplemental methods.
Web resources
BrainSpan – Atlas of the Developing Human Brain, http://www.brainspan.org/
GeneReviews, Sparks, S.E., and Krasnewich, D.M. (1993). Congenital Disorders of N-Linked Glycosylation and Multiple Pathway Overview, https://www.ncbi.nlm.nih.gov/books/NBK1332/
gnomAD Browser v.2.1.1, https://gnomad.broadinstitute.org/
Human Splicing Finder, http://www.umd.be/HSF3/
OMIM, https://www.omim.org/
TraP, http://trap-score.org
Supplemental information
References
- 1.Berkovic S.F., Andermann F., Carpenter S., Wolfe L.S. Progressive myoclonus epilepsies: specific causes and diagnosis. N. Engl. J. Med. 1986;315:296–305. doi: 10.1056/NEJM198607313150506. [DOI] [PubMed] [Google Scholar]
- 2.Franceschetti S., Michelucci R., Canafoglia L., Striano P., Gambardella A., Magaudda A., Tinuper P., La Neve A., Ferlazzo E., Gobbi G., Collaborative LICE study group on PMEs Progressive myoclonic epilepsies: definitive and still undetermined causes. Neurology. 2014;82:405–411. doi: 10.1212/WNL.0000000000000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muona M., Berkovic S.F., Dibbens L.M., Oliver K.L., Maljevic S., Bayly M.A., Joensuu T., Canafoglia L., Franceschetti S., Michelucci R. A recurrent de novo mutation in KCNC1 causes progressive myoclonus epilepsy. Nat. Genet. 2015;47:39–46. doi: 10.1038/ng.3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramachandran N., Girard J.M., Turnbull J., Minassian B.A. The autosomal recessively inherited progressive myoclonus epilepsies and their genes. Epilepsia. 2009;50(Suppl 5):29–36. doi: 10.1111/j.1528-1167.2009.02117.x. [DOI] [PubMed] [Google Scholar]
- 5.Kollmann K., Uusi-Rauva K., Scifo E., Tyynelä J., Jalanko A., Braulke T. Cell biology and function of neuronal ceroid lipofuscinosis-related proteins. Biochim. Biophys. Acta. 2013;1832:1866–1881. doi: 10.1016/j.bbadis.2013.01.019. [DOI] [PubMed] [Google Scholar]
- 6.Oliver K.L., Franceschetti S., Milligan C.J., Muona M., Mandelstam S.A., Canafoglia L., Boguszewska-Chachulska A.M., Korczyn A.D., Bisulli F., Di Bonaventura C. Myoclonus epilepsy and ataxia due to KCNC1 mutation: Analysis of 20 cases and K+ channel properties. Ann. Neurol. 2017;81:677–689. doi: 10.1002/ana.24929. [DOI] [PubMed] [Google Scholar]
- 7.Pedersen B.S., Quinlan A.R. Who’s Who? Detecting and Resolving Sample Anomalies in Human DNA Sequencing Studies with Peddy. Am. J. Hum. Genet. 2017;100:406–413. doi: 10.1016/j.ajhg.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leutenegger A.L., Labalme A., Genin E., Toutain A., Steichen E., Clerget-Darpoux F., Edery P. Using genomic inbreeding coefficient estimates for homozygosity mapping of rare recessive traits: application to Taybi-Linder syndrome. Am. J. Hum. Genet. 2006;79:62–66. doi: 10.1086/504640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R., Thormann A., Flicek P., Cunningham F. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rentzsch P., Witten D., Cooper G.M., Shendure J., Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47(D1):D886–D894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sim N.L., Kumar P., Hu J., Henikoff S., Schneider G., Ng P.C. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40:W452-7. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gelfman S., Wang Q., McSweeney K.M., Ren Z., La Carpia F., Halvorsen M., Schoch K., Ratzon F., Heinzen E.L., Boland M.J. Annotating pathogenic non-coding variants in genic regions. Nat. Commun. 2017;8:236. doi: 10.1038/s41467-017-00141-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., Genome Aggregation Database Consortium The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T.L. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desmet F.O., Hamroun D., Lalande M., Collod-Béroud G., Claustres M., Béroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Research. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Talevich E., Shain A.H., Botton T., Bastian B.C. CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing. PLoS Comput. Biol. 2016;12:e1004873. doi: 10.1371/journal.pcbi.1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dolzhenko E., van Vugt J.J.F.A., Shaw R.J., Bekritsky M.A., van Blitterswijk M., Narzisi G., Ajay S.S., Rajan V., Lajoie B.R., Johnson N.H., US–Venezuela Collaborative Research Group Detection of long repeat expansions from PCR-free whole-genome sequence data. Genome Res. 2017;27:1895–1903. doi: 10.1101/gr.225672.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tankard R.M., Bennett M.F., Degorski P., Delatycki M.B., Lockhart P.J., Bahlo M. Detecting Expansions of Tandem Repeats in Cohorts Sequenced with Short-Read Sequencing Data. Am. J. Hum. Genet. 2018;103:858–873. doi: 10.1016/j.ajhg.2018.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rush J.S., Matveev S., Guan Z., Raetz C.R., Waechter C.J. Expression of functional bacterial undecaprenyl pyrophosphate synthase in the yeast rer2Delta mutant and CHO cells. Glycobiology. 2010;20:1585–1593. doi: 10.1093/glycob/cwq107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park E.J., Grabińska K.A., Guan Z., Stránecký V., Hartmannová H., Hodaňová K., Barešová V., Sovová J., Jozsef L., Ondrušková N. Mutation of Nogo-B receptor, a subunit of cis-prenyltransferase, causes a congenital disorder of glycosylation. Cell Metab. 2014;20:448–457. doi: 10.1016/j.cmet.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harrison K.D., Park E.J., Gao N., Kuo A., Rush J.S., Waechter C.J., Lehrman M.A., Sessa W.C. Nogo-B receptor is necessary for cellular dolichol biosynthesis and protein N-glycosylation. EMBO J. 2011;30:2490–2500. doi: 10.1038/emboj.2011.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pipalia N.H., Huang A., Ralph H., Rujoi M., Maxfield F.R. Automated microscopy screening for compounds that partially revert cholesterol accumulation in Niemann-Pick C cells. J. Lipid Res. 2006;47:284–301. doi: 10.1194/jlr.M500388-JLR200. [DOI] [PubMed] [Google Scholar]
- 25.Hamanaka K., Imagawa E., Koshimizu E., Miyatake S., Tohyama J., Yamagata T. De Novo Truncating Variants in the Last Exon of SEMA6B Cause Progressive Myoclonic Epilepsy. Amer. J. Hum. Genet. 2020;106:549–558. doi: 10.1016/j.ajhg.2020.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grabińska K.A., Edani B.H., Park E.J., Kraehling J.R., Sessa W.C. A conserved C-terminal RXG motif in the NgBR subunit of cis-prenyltransferase is critical for prenyltransferase activity. J. Biol. Chem. 2017;292:17351–17361. doi: 10.1074/jbc.M117.806034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grabińska K.A., Park E.J., Sessa W.C. cis-Prenyltransferase: New Insights into Protein Glycosylation, Rubber Synthesis, and Human Diseases. J. Biol. Chem. 2016;291:18582–18590. doi: 10.1074/jbc.R116.739490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burda P., Aebi M. The ALG10 locus of Saccharomyces cerevisiae encodes the α-1,2 glucosyltransferase of the endoplasmic reticulum: the terminal glucose of the lipid-linked oligosaccharide is required for efficient N-linked glycosylation. Glycobiology. 1998;8:455–462. doi: 10.1093/glycob/8.5.455. [DOI] [PubMed] [Google Scholar]
- 29.Edani B.H., Grabińska K.A., Zhang R., Park E.J., Siciliano B., Surmacz L., Ha Y., Sessa W.C. Structural elucidation of the cis-prenyltransferase NgBR/DHDDS complex reveals insights in regulation of protein glycosylation. Proc. Natl. Acad. Sci. USA. 2020;117:20794–20802. doi: 10.1073/pnas.2008381117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bar-El M.L., Vaňková P., Yeheskel A., Simhaev L., Engel H., Man P., Haitin Y., Giladi M. Structural basis of heterotetrameric assembly and disease mutations in the human cis-prenyltransferase complex. Nat. Commun. 2020;11:5273. doi: 10.1038/s41467-020-18970-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Medina-Cano D., Ucuncu E., Nguyen L.S., Nicouleau M., Lipecka J., Bizot J.C., Thiel C., Foulquier F., Lefort N., Faivre-Sarrailh C. High N-glycan multiplicity is critical for neuronal adhesion and sensitizes the developing cerebellum to N-glycosylation defect. eLife. 2018;7:7. doi: 10.7554/eLife.38309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He P., Ng B.G., Losfeld M.E., Zhu W., Freeze H.H. Identification of intercellular cell adhesion molecule 1 (ICAM-1) as a hypoglycosylation marker in congenital disorders of glycosylation cells. J. Biol. Chem. 2012;287:18210–18217. doi: 10.1074/jbc.M112.355677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harrison K.D., Miao R.Q., Fernandez-Hernándo C., Suárez Y., Dávalos A., Sessa W.C. Nogo-B receptor stabilizes Niemann-Pick type C2 protein and regulates intracellular cholesterol trafficking. Cell Metab. 2009;10:208–218. doi: 10.1016/j.cmet.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamdan F.F., Myers C.T., Cossette P., Lemay P., Spiegelman D., Laporte A.D., Nassif C., Diallo O., Monlong J., Cadieux-Dion M., Deciphering Developmental Disorders Study High Rate of Recurrent De Novo Mutations in Developmental and Epileptic Encephalopathies. Am. J. Hum. Genet. 2017;101:664–685. doi: 10.1016/j.ajhg.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lelieveld S.H., Reijnders M.R., Pfundt R., Yntema H.G., Kamsteeg E.J., de Vries P., de Vries B.B., Willemsen M.H., Kleefstra T., Löhner K. Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat. Neurosci. 2016;19:1194–1196. doi: 10.1038/nn.4352. [DOI] [PubMed] [Google Scholar]
- 36.Nakajima T., Hayashi K., Viswanathan P.C., Kim M.-Y., Anghelescu M., Barksdale K.A., Shuai W., Balser J.R., Kupershmidt S. HERG is protected from pharmacological block by α-1,2-glucosyltransferase function. J. Biol. Chem. 2007;282:5506–5513. doi: 10.1074/jbc.M605976200. [DOI] [PubMed] [Google Scholar]
- 37.Arsov T., Smith K.R., Damiano J., Franceschetti S., Canafoglia L., Bromhead C.J., Andermann E., Vears D.F., Cossette P., Rajagopalan S. Kufs disease, the major adult form of neuronal ceroid lipofuscinosis, caused by mutations in CLN6. Am. J. Hum. Genet. 2011;88:566–573. doi: 10.1016/j.ajhg.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park J.K., Orvisky E., Tayebi N., Kaneski C., Lamarca M.E., Stubblefield B.K., Martin B.M., Schiffmann R., Sidransky E. Myoclonic epilepsy in Gaucher disease: genotype-phenotype insights from a rare patient subgroup. Pediatr. Res. 2003;53:387–395. doi: 10.1203/01.PDR.0000049515.79882.94. [DOI] [PubMed] [Google Scholar]
- 39.Vaz-Drago R., Custódio N., Carmo-Fonseca M. Deep intronic mutations and human disease. Hum. Genet. 2017;136:1093–1111. doi: 10.1007/s00439-017-1809-4. [DOI] [PubMed] [Google Scholar]
- 40.Canafoglia L., Robbiano A., Pareyson D., Panzica F., Nanetti L., Giovagnoli A.R., Venerando A., Gellera C., Franceschetti S., Zara F. Expanding sialidosis spectrum by genome-wide screening: NEU1 mutations in adult-onset myoclonus. Neurology. 2014;82:2003–2006. doi: 10.1212/WNL.0000000000000482. [DOI] [PubMed] [Google Scholar]
- 41.Vanni N., Fruscione F., Ferlazzo E., Striano P., Robbiano A., Traverso M., Sander T., Falace A., Gazzerro E., Bramanti P. Impairment of ceramide synthesis causes a novel progressive myoclonus epilepsy. Ann. Neurol. 2014;76:206–212. doi: 10.1002/ana.24170. [DOI] [PubMed] [Google Scholar]
- 42.Zhou J., Tawk M., Tiziano F.D., Veillet J., Bayes M., Nolent F., Garcia V., Servidei S., Bertini E., Castro-Giner F. Spinal muscular atrophy associated with progressive myoclonic epilepsy is caused by mutations in ASAH1. Am. J. Hum. Genet. 2012;91:5–14. doi: 10.1016/j.ajhg.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim J.-J.P., Olson L.J., Dahms N.M. Carbohydrate recognition by the mannose-6-phosphate receptors. Curr. Opin. Struct. Biol. 2009;19:534–542. doi: 10.1016/j.sbi.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wolfe L.S., Ng Ying Kin N.M., Palo J., Haltia M. Dolichols in brain and urinary sediment in neuronal ceroid lipofuscinosis. Neurology. 1983;33:103–106. doi: 10.1212/wnl.33.1.103. [DOI] [PubMed] [Google Scholar]
- 45.Wolfe L.S., Palo J., Santavuori P., Andermann F., Andermann E., Jacob J.C., Kolodny E. Urinary sediment dolichols in the diagnosis of neuronal ceroid-lipofuscinosis. Ann. Neurol. 1986;19:270–274. doi: 10.1002/ana.410190308. [DOI] [PubMed] [Google Scholar]
- 46.Balestrini S., Milh M., Castiglioni C., Lüthy K., Finelli M.J., Verstreken P., Cardon A., Stražišar B.G., Holder J.L., Jr., Lesca G. TBC1D24 genotype-phenotype correlation: Epilepsies and other neurologic features. Neurology. 2016;87:77–85. doi: 10.1212/WNL.0000000000002807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sabry S., Vuillaumier-Barrot S., Mintet E., Fasseu M., Valayannopoulos V., Héron D., Dorison N., Mignot C., Seta N., Chantret I. A case of fatal Type I congenital disorders of glycosylation (CDG I) associated with low dehydrodolichol diphosphate synthase (DHDDS) activity. Orphanet J. Rare Dis. 2016;11:84. doi: 10.1186/s13023-016-0468-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Züchner S., Dallman J., Wen R., Beecham G., Naj A., Farooq A., Kohli M.A., Whitehead P.L., Hulme W., Konidari I. Whole-exome sequencing links a variant in DHDDS to retinitis pigmentosa. Am. J. Hum. Genet. 2011;88:201–206. doi: 10.1016/j.ajhg.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Togashi N., Fujita A., Shibuya M., Uneoka S., Miyabayashi T., Sato R., Okubo Y., Endo W., Inui T., Jin K. Fifteen-year follow-up of a patient with a DHDDS variant with non-progressive early onset myoclonic tremor and rare generalized epilepsy. Brain Dev. 2020;42:696–699. doi: 10.1016/j.braindev.2020.06.011. [DOI] [PubMed] [Google Scholar]
- 50.Guo J.F., Zhang L., Li K., Mei J.P., Xue J., Chen J., Tang X., Shen L., Jiang H., Chen C. Coding mutations in NUS1 contribute to Parkinson’s disease. Proc. Natl. Acad. Sci. USA. 2018;115:11567–11572. doi: 10.1073/pnas.1809969115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gan-Or Z., Dion P.A., Rouleau G.A. Genetic perspective on the role of the autophagy-lysosome pathway in Parkinson disease. Autophagy. 2015;11:1443–1457. doi: 10.1080/15548627.2015.1067364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Michelakakis H., Xiromerisiou G., Dardiotis E., Bozi M., Vassilatis D., Kountra P.M., Patramani G., Moraitou M., Papadimitriou D., Stamboulis E. Evidence of an association between the scavenger receptor class B member 2 gene and Parkinson’s disease. Mov. Disord. 2012;27:400–405. doi: 10.1002/mds.24886. [DOI] [PubMed] [Google Scholar]
- 53.Aharon-Peretz J., Rosenbaum H., Gershoni-Baruch R. Mutations in the glucocerebrosidase gene and Parkinson’s disease in Ashkenazi Jews. N. Engl. J. Med. 2004;351:1972–1977. doi: 10.1056/NEJMoa033277. [DOI] [PubMed] [Google Scholar]
- 54.Den K., Kudo Y., Kato M., Watanabe K., Doi H., Tanaka F., Oguni H., Miyatake S., Mizuguchi T., Takata A. Recurrent NUS1 canonical splice donor site mutation in two unrelated individuals with epilepsy, myoclonus, ataxia and scoliosis - a case report. BMC Neurol. 2019;19:253. doi: 10.1186/s12883-019-1489-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Araki K., Nakamura R., Ito D., Kato K., Iguchi Y., Sahashi K., Toyama M., Hamada K., Okamoto N., Wada Y. NUS1 mutation in a family with epilepsy, cerebellar ataxia, and tremor. Epilepsy Res. 2020;164:106371. doi: 10.1016/j.eplepsyres.2020.106371. [DOI] [PubMed] [Google Scholar]
- 56.Ng B.G., Freeze H.H. Perspectives on glycosylation and its congenital disorders. Trends Genet. 2018;34:466–476. doi: 10.1016/j.tig.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Canafoglia L., Castellotti B., Ragona F., Freri E., Granata T., Chiapparini L., Gellera C., Scaioli V., Franceschetti S., DiFrancesco J.C. Progressive myoclonus epilepsy caused by a gain-of-function KCNA2 mutation. Seizure. 2019;65:106–108. doi: 10.1016/j.seizure.2019.01.005. [DOI] [PubMed] [Google Scholar]
- 58.Cameron J.M., Maljevic S., Nair U., Aung Y.H., Cogné B., Bézieau S., Blair E., Isidor B., Zweier C., Reis A. Encephalopathies with KCNC1 variants: genotype-phenotype-functional correlations. Ann. Clin. Transl. Neurol. 2019;6:1263–1272. doi: 10.1002/acn3.50822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boissé Lomax L., Bayly M.A., Hjalgrim H., Møller R.S., Vlaar A.M., Aaberg K.M., Marquardt I., Gandolfo L.C., Willemsen M., Kamsteeg E.J. ‘North Sea’ progressive myoclonus epilepsy: phenotype of subjects with GOSR2 mutation. Brain. 2013;136:1146–1154. doi: 10.1093/brain/awt021. [DOI] [PubMed] [Google Scholar]
- 60.Ishiura H., Doi K., Mitsui J., Yoshimura J., Matsukawa M.K., Fujiyama A., Toyoshima Y., Kakita A., Takahashi H., Suzuki Y. Expansions of intronic TTTCA and TTTTA repeats in benign adult familial myoclonic epilepsy. Nat. Genet. 2018;50:581–590. doi: 10.1038/s41588-018-0067-2. [DOI] [PubMed] [Google Scholar]
- 61.Corbett M.A., Kroes T., Veneziano L., Bennett M.F., Florian R., Schneider A.L., Coppola A., Licchetta L., Franceschetti S., Suppa A. Intronic ATTTC repeat expansions in STARD7 in familial adult myoclonic epilepsy linked to chromosome 2. Nat. Commun. 2019;10:4920. doi: 10.1038/s41467-019-12671-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Florian R.T., Kraft F., Leitão E., Kaya S., Klebe S., Magnin E., van Rootselaar A.F., Buratti J., Kühnel T., Schröder C., FAME consortium Unstable TTTTA/TTTCA expansions in MARCH6 are associated with Familial Adult Myoclonic Epilepsy type 3. Nat. Commun. 2019;10:4919. doi: 10.1038/s41467-019-12763-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yeetong P., Pongpanich M., Srichomthong C., Assawapitaksakul A., Shotelersuk V., Tantirukdham N., Chunharas C., Suphapeetiporn K., Shotelersuk V. TTTCA repeat insertions in an intron of YEATS2 in benign adult familial myoclonic epilepsy type 4. Brain. 2019;142:3360–3366. doi: 10.1093/brain/awz267. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All variants reported in this study have been deposited to the LOVD database; individual LOVD identifiers are provided in Tables 1, 2, 3, and S4. WES data have not been deposited in a public repository due to privacy and ethical restrictions. R code used in brain co-expression analysis is available as supplemental methods.