Abstract
Coronaviruses (CoV) mature by a budding process at intracellular membranes. Here we showed that the major surface protein S of a porcine CoV (transmissible gastroenteritis virus) is not transported to the cell surface but is retained intracellularly. Site-directed mutagenesis indicated that a tyrosine-dependent signal (YXXI) in the cytoplasmic tail is essential for intracellular localization of the S protein. Surface expression of mutant proteins was evident by immunofluorescence analysis and surface biotinylation. Intracellularly retained S proteins only contained endoglycosidase H-sensitive N-glycans, whereas mutant proteins that migrated to the plasma membrane acquired N-linked oligosaccharides of the complex type. Corresponding tyrosine residues are present in the cytoplasmic tails of the S proteins of other animal CoV but not in the tail portion of the S protein of severe acute respiratory syndrome (SARS)-CoV. Changing the SEPV tetrapeptide in the cytoplasmic tail to YEPI resulted in intracellular retention of the S protein of SARS-CoV. As the S proteins of CoV have receptor binding and fusion activities and are the main target of neutralizing antibodies, the differences in the transport behavior of the S proteins suggest different strategies in the virus host interactions between SARS-CoV and other coronaviruses.
In enveloped viruses, virion morphogenesis is the result of a budding process at a cellular membrane (1). This maturation process may occur at the plasma membrane, e.g. in the case of human immunodeficiency virus or influenza viruses, or at an intracellular compartment. Coronavirus (CoV)1 maturation takes place at the cis-Golgi network also known as endoplasmic reticulum-Golgi intermediate compartment (2, 3). The surface proteins of enveloped viruses are usually transported to the membrane compartment where a budding process results in the release of virions. This has been shown for model viruses such as the influenza and vesicular stomatitis viruses (1). There are, however, examples of viruses where the sites of glycoprotein accumulation and virus maturation do not coincide. The measles virus is released from the apical plasma membrane of polarized epithelial cells, although both the H and F proteins are predominantly transported to the basolateral cell surface (4). On the other hand, the Marburg virus is released from the basolateral plasma membrane, although the viral glycoprotein gp is expressed mainly on the apical surface (5).
Coronaviruses are known human and animal pathogens that mainly affect the epithelium of the respiratory or intestinal tract. They are positive-stranded RNA viruses that contain three membrane proteins incorporated into the lipid envelope, M, S, and E. The M protein is a glycoprotein with three or four transmembrane domains (6). Its large carboxyl terminus is oriented toward the cytoplasm and most probably interacts with the nucleocapsid during budding (7). When expressed alone, the M is localized in the cis-Golgi network or cis-Golgi complex as reported for transmissible gastroenteritis virus (TGEV) (8) and for avian infectious bronchitis virus (9), or it reaches the trans-Golgi cisternae or the trans-Golgi network in the case of the murine hepatitis virus (10). The information for the intracellular localization of M resides within the first transmembrane domain (11) and additionally, in the carboxyl-terminal portion (12). The E protein is a small membrane protein with a single membrane-spanning domain. It has been reported to transiently reside in a pre-Golgi compartment (13) before it progresses to the Golgi apparatus (14, 15). The six carboxyl-terminal amino acids, RDKLYS, have been shown to be essential for the temporary retention within the pre-Golgi compartment (13). The third membrane protein, the spike (S) glycoprotein, forms the corona-like projections of the virion surface on electron micrographs (16). It has receptor-binding and membrane-fusion activities and is the main target of the immune response elicited by a coronavirus infection. Using Vaccinia virus or baculovirus expression systems, the S protein has been found to be present on the cell surface, although transport kinetics suggested a very inefficient transport (17, 18). These results together with the finding that M and E protein can induce the formation of virus-like particles (19) suggested that the S protein does not determine the site of virus maturation.
We used plasmid vectors for the expression of the S protein of porcine TGEV, and the human coronavirus associated with severe acute respiratory syndrome (SARS-CoV). We found that the TGEV S protein is intracellularly retained because of a tyrosine-based signal within the cytoplasmic tail. In contrast, the S protein of SARS-CoV lacks a tyrosine residue in the corresponding tail portion as revealed by sequence alignments, and in fact, it is transported to the cell surface. A replacement of the tetrapeptide SEPV by YPEI resulted in intracellular retention of SARS-CoV S protein.
EXPERIMENTAL PROCEDURES
Cells—BSR-T7/5 cells stably expressing T7 RNA polymerase were kindly provided by Dr. Conzelmann (Ludwig-Maximilians-Universität, Munich, Germany). The cells were propagated in minimal essential medium supplemented with 5% fetal calf serum, non-essential amino acids, and 0.5 mg/ml of Geneticin.
Construction of Plasmids—The S protein gene of TGEV, strain PUR-46-MAD, (20) was amplified from the plasmid pYATS-4 by PCR using oligonucleotides a and b (see Table I ). Primer a contained an EcoRI, primer b a PstI restriction site, which allowed to clone the PCR product into the respective sites of the pTM1 vector (21) resulting in pTM1-SSS (SSS indicates that ectodomain, membrane anchor, and cytoplasmic tail are derived from the S protein). The open reading frame of SSS was identical to the published sequence (GenBank™ accession number M94101).
Table I.
Primers used for plasmid constructions
| Oligonucleotide | 5′-Sequence-3′ |
|---|---|
| a | TTTGAATTCCACACCATGAAAAAACTATTTGTGGTTTTGG |
| b | TTTTCTGCAGTTAATGGACGTGCACTTTTTCAATTGG |
| c | CTAGATGACAAGTTCTATTTGACCC |
| d | GAGGATTTTCCGTGCTTTTTCAAGATTGACTAATGTATTGTTAATGTT |
| e | ACATTAGTCAATCTTGAAAAAGCACGGAAAATCCTCTCTGAGGTAGG |
| f | TTTTCTGCAGTCATCTTTTCTCAGCCATCGCATC |
| g | ATTTTCAAATTGTCTTCTATAAAGCACGATGACGATCACTATAATGACC |
| h | ATCGTCATCGTGCTTTATAGAAGACAATTTGAAAATTACGAACCA |
| i | CATTGACCTTTTGAGTCTACTACATATAGAGTGACAACAACTTCCTAA |
| j | TGTCACTCTATATGTAGTAGACTCAAAAGGTCAATGCTAATGGGTAAT |
| n | ATTTGAAAATGCCGAACCAATTGAA |
| o | CAATTGGTTCGGCATTTTCAAATTGT |
| p | TGAAAATTACGCCCCAATTGAAAAAG |
| q | TTTCAATTGGGGCGTAATTTTCAAAT |
| r | AAATTACGAAGCCATTGAAAAAGTG |
| s | CTTTTTCAATGGCTTCGTAATTTTC |
| t | TTACGAACCAGCCGAAAAAGTGCAC |
| u | GCACTTTTTCGGCTGGTTCGTAATT |
| v | CGAACCAATTGCCAAAGTGCACGTC |
| w | CGTGCACTTTGGCAATTGGTTCGTA |
| A | TTTTGAATTCATGTTTATTTTCTTATTATTTCTTACTCTCACTAGTGG |
| B | TTTTGGATCCTTATGTGTAATGTAATTTGACACCCTTGAG |
| C | GTTAACCAGAATGCTCAAGCATTAA |
| D | TGAGGATGACTACGAGCCAGTTCTC |
| E | GAACTGGCTCGTAGTCATCCTCATCA |
| F | TGAGGATGACTACGAGCCAATCCTCAAGGGTGTC |
| G | CACCCTTGAGGATTGGCTCGTAGTCATCCTCATC |
To construct a chimeric protein-containing parts of the TGEV S and Sendai virus F protein, the F gene region coding for the membrane anchor and carboxyl-terminal domain of the F protein of the Sendai virus (strain Fushimi) was amplified from the plasmid pcDNA3.1-F (kindly provided by Dr. Neubert, Max-Planck-Institut für Biochemie, Martinsried, Germany) by PCR using oligonucleotides e and f. In parallel, SSS was used as template for a PCR with oligonucleotides c and d. These two PCR products were purified (PCR Purification Kit, Qiagen), mixed in a molar ratio of 1:1, and heated for 2 min at 95 °C for denaturation. The mixture was incubated at 60 °C for 2 min to allow the two fragments to anneal to each other. Hybridization was mediated by overlapping complementary sequences that were introduced into the PCR fragments by 5′-overhangs of the oligonucleotides d and e. A complete double-stranded DNA hybrid was obtained after incubating the mixture with Pfu polymerase (MBI Fermentas) at 72 °C for 1 min and 30 s. The chimeric gene was subsequently amplified by PCR with oligonucleotides c and f. The PCR product was ligated into the pTM1-SSS plasmid using the PstI restriction site at the 5′-end of oligonucleotide f and the SpeI restriction site located ∼50 nucleotides upstream of the binding site of oligonucleotide c. The new part of the resulting plasmid pTM1-SFF was sequenced and found to be identical to the published sequences (GenBank™, M94101, D00152). A similar overlapping PCR technique was applied for the construction of all other chimeric and mutant protein genes. The protein domains and oligonucleotides used are indicated in Fig. 1 and Table I, respectively. For mutants E1441A, P1442A, I1443A, and E1444A the primer pairs p-q, r-s, t-u, and v-w, respectively, were used. The S protein gene of SARS-CoV (strain CUHK-W1) was amplified by PCR from the plasmid pcDNA3.1-S with oligonucleotides A and B and ligated into the pTM1 vector via restriction sites EcoRI and BamHI resulting in the plasmid pTM1-SARS-CoV-S. The sequence of the total open reading frame of SARS-CoV-S was found to be identical to the published sequence (GenBank™ accession number AY278554). Primers used for generation of mutant S1243Y are shown in Fig. 1. For mutant S1243Y/V1246I, oligonucleotides D and E were replaced by F and G. The SARS-CoV-S mutants were generated using restriction sites BamHI (5′-end of oligonucleotide B) and EcoRV (∼80 nucleotides upstream of the oligonucleotide C binding site).
Fig. 1.
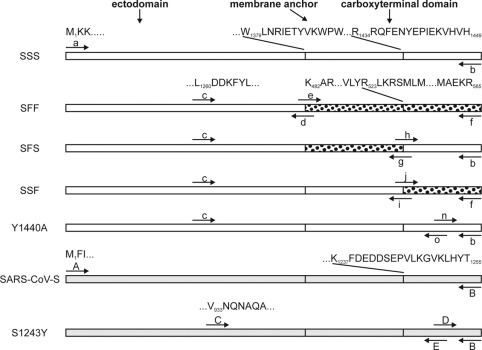
Schematic drawing of TGEV-S and SeV-F chimeras (SFF,SFS,SSF) as well as TGEV-S (SSS) and SARS-CoV-S parental and mutant proteins. TGEV-S sequences are indicated by open boxes, SeV-F sequences are indicated by dotted boxes, and SARS-CoV-S sequences are indicated by gray boxes. It should be noted that the sizes of the boxes are not proportional to the length of the amino acid chain. The carboxyl-terminal domain of TGEV-S only comprises 16 and of SARS-CoV-S 19 amino acids, whereas the tail of the F protein is 42 amino acids in length. The initial and terminal amino acids are shown once for each domain. Letters above arrows indicate the primers used for cloning (see Table I).
Immunofluorescence—BSR-T7/5 cells grown on 12-mm diameter coverslips were transfected with 1 μg of plasmid DNA, 0.5 μg of the plasmid pC-T7Pol (kindly provided by Dr. Kawaoka, University of Wisconsin-Madison, Madison, WI) coding for T7 RNA polymerase, and 2 μl of LipofectAMINE 2000 Reagent (Invitrogen) and incubated at 37 °C for 24 h. One part of the fixed cell preparations was permeabilized with 0.2% Triton/phosphate-buffered saline for 5 min. The TGEV-S ectodomain was detected with a monoclonal antibody (6A.C3) against the viral S protein (22) at a dilution of 1:200 in 1% bovine serum albumin, phosphate-buffered saline followed by incubation with a fluorescein isothiocyanate-conjugated second antibody (donkey anti-mouse, 1:200, Acris). For detection of the SARS-CoV-S ectodomain the cells were incubated with human patient serum (1:1000) and antihuman-fluorescein isothiocyanate antibody (from goat, 1:200, Sigma). Fluorescence was visualized with a Zeiss Axioplan 2 microscope.
Surface Biotinylation and Immunoprecipitation of Proteins—BSR-T7/5 cells grown in 35-mm diameter dishes were transfected with 3 μg of plasmid DNA, 1 μg of pC-T7Pol, and 10 μl of LipofectAMINE 2000 Reagent. At 24-h post-transfection cell surface proteins were labeled with a N-hydroxy-succinimide ester of biotin (0.5 mg/ml phosphate-buffered saline, Pierce). The viral antigens were immunoprecipitated from the cell lysates as described by Zimmer et al. (23, 24). For immunoprecipitation, the monoclonal anti-TGEV S protein antibody (6A.C3) was used.
Western Blot Analysis of Transfected Cells—At 24-h post-transfection BSR-T7/5 cells grown in 35-mm diameter dishes (see above) were lysed as described by Köhl et al. (25). For Western blot analysis the nitrocellulose was incubated with monoclonal antibody 6A.C3 (1:200) followed by incubation with a peroxidase-conjugated second antibody (goat anti-mouse, 1:1000, DAKO). The bound antibodies were visualized by chemoluminescence (24).
Radiolabeling and Treatment with Endoglycosidases—At 48-h post-transfection (ExGen 500, Fermentas) BSR-T7/5 cells grown in 35-mm diameter dishes (see above) were metabolically labeled with 50 μCi of [35S]methionine (Amersham Biosciences) for 4 h as described by Alfalah et al. (26). Digestion of [35S]methionine-labeled immunoprecipitates with endoglycosidase H (New England Biolabs) and N-glycosidase F (Roche Applied Science) was performed as described by Naim et al. (27). The samples were loaded on a 6% SDS-gel (6 mA, overnight, Bio-Rad). After electrophoresis, proteins were visualized using a PhosphorImager (Bio-Rad).
RESULTS
The transport of the TGEV S protein was analyzed using plasmid vectors that avoid overexpression. Expression vectors that depended on nuclear transcription turned out to be very inefficient. Therefore, all constructs (Fig. 1) were cloned into the pTM1 vector under the control of the T7 promoter and transiently expressed in BSR-T7/5 cells that stably express the T7 RNA polymerase. Fig. 2 shows that the S protein (SSS) was detectable by fluorescence microscopy after intracellular but not after surface staining. The absence of S protein from the cell surface cannot be explained by rapid internalization, because an antibody uptake assay did not provide any evidence for endocytosis of the S protein (not shown). Glycosylation analysis (see below) also indicates that the lack of surface expression is because of intracellular retention rather than endocytotic uptake. To assign a potential transport signal to the ectodomain, membrane anchor, or cytoplasmic tail, chimeric proteins were generated that contained either one of the two latter domains from the fusion protein (F) of Sendai virus. Replacement of the membrane anchor (Fig. 2, SFS) did not result in a different transport behavior. In contrast, chimeric proteins containing the tail portion (Fig. 2, SSF) or both the tail and the membrane anchor (Fig. 2, SFF) from the Sendai virus F protein were transported to the cell surface. This result strongly suggested that the information determining the intracellular retention of the TGEV S protein is present in the cytoplasmic tail.
Fig. 2.
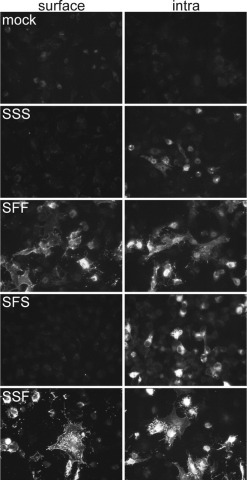
Surface and intracellular (intra) immunofluorescence analysis of parental TGEV-S (SSS) and TGEV-S/SeV-F chimeras (SFF,SFS,SSF). BSR-T7/5 cells were transfected with the pTM1 expression plasmid containing the gene indicated. Cells were analyzed for surface and intracellular expression of the indicated proteins at 24 h post-transfection by immunofluorescence microscopy.
The range of amino acids within the cytoplasmic tail of the S protein that is required for intracellular retention was narrowed down by an analysis of deletion mutants that lacked 5, 10, or 14 amino acids from the carboxyl terminus. Deletion of the carboxyl-terminal five amino acids (KVHVH) did not alter the transport characteristics of the S protein. However, further truncation of the tail by five residues resulted in a protein that was transported to the cell surface (not shown). This result indicates that the peptide 1440YEPIE1444 contains essential information for the intracellular localization of the S protein. The importance of individual amino acids within this sequence was determined by an alanine scan. Only replacement of Tyr-1440 and Ile-1443 resulted in a transport of S protein to the cell surface (Fig. 3 ). These data suggest that the intracellular localization signal of the S protein resembles the Tyr-Xxx-Xxx-Ile signal that is responsible in many proteins for endocytosis and/or transport to the basolateral surface of polarized epithelial cells.
Fig. 3.
Surface and intracellular (intra) immunofluorescence analysis of TGEV-S mutants (the amino acid exchange and position is indicated). BSR-T7/5 cells were transfected with the pTM1 expression plasmid containing the gene indicated. Cells were analyzed for surface and intracellular expression of the indicated proteins at 24 h post-transfection by immunofluorescence microscopy.
To confirm the results obtained from immunofluorescence studies, the transport of parental, mutant, and chimeric S proteins to the plasma membrane was analyzed by cell surface biotinylation. Following application of the membrane-impermeable biotinylating reagent to intact cells, only the proteins previously detected by surface immunofluorescence were labeled with biotin, as expected (Fig. 4A ). The lysates of cells expressing either of the different proteins were also analyzed by Western blot. Interestingly all proteins that were transported to the cell surface presented a double band profile, whereas intracellularly retained proteins only presented the lower band (Fig. 4B). In the case of the point mutants (Fig. 4B, lanes e and h) the upper band is less prominent than in the case of the chimeric proteins suggesting that amino acids other than the tyrosine and isoleucine residue also contribute to intracellular localization. The upper band most likely represents proteins with complex N-glycans, whereas proteins containing high mannose oligosaccharides are expected in the lower band. To verify this interpretation, radiolabeled cell lysates expressing either the SSS or the SSF protein were subjected to immunoprecipitation and treatment with either endoglycosidase H or N-glycosidase F. Both the lower and the upper band were detected with the SSF protein, whereas the SSS protein revealed only the lower band (Fig. 5 ). The band between the two forms of the S protein was present also in the mock-transfected samples and is therefore not related to the S protein. Treatment of the SSS protein with endoglycosidase H resulted in a substantial shift to a band with an apparent molecular mass of about 150 kDa. Likewise, a significant reduction in the size of the SSS protein was revealed upon N-glycosidase F treatment although not to the same level as that obtained with endoglycosidase H. This difference is because of the specificity of currently available N-glycosidase F preparations, which make it difficult to completely deglycosylate glycoproteins with a large number of N-glycans. Altogether, the combined analysis with endoglycosidase H and N-glycosidase F indicates that the glycosylation of the lower band is of the high mannose type. The effect of endoglycosidase H treatment on the lower band of the SSF protein was the same as that on the corresponding band of SSS. However, endoglycosidase H treatment of the upper band of SSF resulted only in a slight increase of the electrophoretic mobility indicating that this protein contains only a small number of oligosaccharides of the high mannose type. This band almost migrated with the nonspecific band mentioned above; however, the increased intensity compared with the mock-transfected sample indicates that the endoglycosidase H-digested protein is also located in this position. The predominant resistance of the upper band to endoglycosidase H is consistent with a substantial conversion of the oligosaccharides in this protein form to complex N-glycans in the Golgi apparatus. These results are summarized as follows. When the TGEV S protein contains the YXXI motif it is intracellularly retained, and therefore, not fully glycosylated consisting of a single low sized band. In contrast, when the C-tail of TGEV S protein is substituted by that of the F protein, the retention profile is lost being efficiently transported through the Golgi complex to the cell membrane, as demonstrated by its glycosylation profile.
Fig. 4.
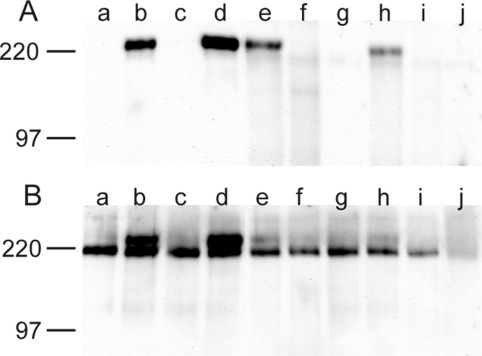
TGEV-S parental, mutant, and chimeric proteins were expressed and analyzed by surface biotinylation (A) or Western blot (B). BSR-T7/5 cells were transfected with the pTM1 expression plasmid containing either of the following TGEV-S genes, a, SSS; b, SFF; c, SFS; d, SSF; e, Y1440A; f, E1441A; g, P1442A; h, I1443A; i, E1444A; j, mock-transfected cells. At 24 h post-transfection cells were prepared for surface biotinylation or Western blot.
Fig. 5.
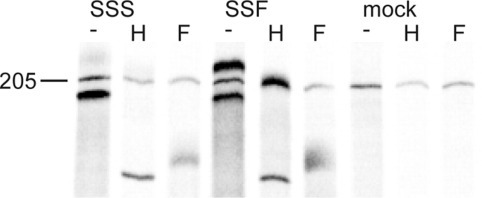
Effect of endoglycosidase treatment on parental (SSS) and chimeric (SSF) TGEV S protein. Transfected BSR-T7/5 cells were radiolabeled and prepared for treatment with endoglycosidases. Samples were incubated with endoglycosidase H (H), N-glycosidase F (F), or in the absence of enzyme (–).
Some well characterized representatives of the different taxonomic groups of coronaviruses are TGEV and feline infectious peritonitis virus (group 1), bovine coronavirus and mouse hepatitis virus (group 2), and infectious bronchitis virus (group 3). Although the S proteins of these viruses show pronounced sequence differences in the cytoplasmic tail portion, they all contain at least one tyrosine residue that might be part of a retention signal. By contrast, the tyrosine residue in the tail of the SARS coronavirus S protein is located at position –2 from the carboxyl terminus and thus cannot be part of a YXXϕ (ϕ representing a large aliphatic amino acid such as isoleucine) motif. We wondered whether the SARS-CoV S protein also contains an intracellular localization signal. Unexpectedly, in contrast to TGEV S protein the SARS-CoV S protein was efficiently transported to the cell surface as observed by surface immunofluorescence (Fig. 6 ). Interestingly, sequence comparisons showed that the tetrapeptide YEPI within the TGEV S protein that is responsible for S protein intracellular retention closely resembled the tetrapeptide SEPV present in SARS-CoV S protein tail. In both cases, the dipeptide EP is followed by a hydrophobic amino acid (V or I, respectively). Moreover, these motifs have similar locations within the respective cytoplasmic tails, amino acids 7–10 from the carboxyl terminus in the case of TGEV and amino acids 10–13 in the case of SARS-CoV. By site-directed mutagenesis a mutant protein was generated in which the serine was replaced by a tyrosine residue. The S1243Y mutant was transported to the cell surface. However, when in addition to the S → Y exchange the valine was replaced by an isoleucine residue, the resulting S1243Y/V1246I mutant was predominantly retained intracellularly. Only a few cells showed a faint surface staining. This finding demonstrates the importance of the YXXI motif for the intracellular localization of the coronavirus S protein.
Fig. 6.
Surface and intracellular (intra) immunofluorescence analysis of parental or mutant proteins of SARS-CoV-S. BSR-T7/5 cells were transfected with the pTM1 expression plasmid containing the gene indicated. Applying immunofluorescence microscopy, cells were analyzed for surface and intracellular expression of the respective protein at 24 h post-transfection.
DISCUSSION
It is well established that tyrosine residues in the cytoplasmic tail of membrane proteins play an important role in intracellular sorting events. Most of these signals conform to the minimal consensus motifs YXXϕ or NPXY. The former type of sorting signal is currently best understood. Depending on the context of a specific protein, YXXϕ signals may mediate rapid internalization from the cell surface, lysosomal targeting, localization to specialized organelles such as antigen-processing compartment or the trans-Golgi network, or delivery to the basolateral plasma membrane of polarized epithelial cells (28). These different functions require the interaction of the signal with recognition molecules associated with the different sites of protein sorting. Binding partners of YXXϕ motifs are a group of adaptor proteins. The μ2 subunit of the heterotetrameric adaptor proteins complex has been shown to interact with the tyrosine-containing sorting signal (29). The YXXϕ motif binds in an extended conformation to a region of the μ2 molecule having pockets for both the Y and ϕ residues. Variation in the recognition is achieved by different affinities of the sorting signal to the various adaptor protein complexes, which may be affected by changes within or around the YXXϕ motif.
Tyrosine-containing signals are mainly known for regulating post-Golgi transport events. Our data showed that YXXϕ motifs may also affect transport in the early secretory pathway. The CD3 ϵ chain of the T cell receptor has also been demonstrated to contain a retention signal in the cytoplasmic tail (30). Here a YXXL motif is responsible for localization within the endoplasmic reticulum. The sequence requirements of the TGEV S protein and CD3 ϵ for intracellular localization are not the same. For the latter protein it has been reported that, in addition to Tyr-177 and Leu-180, Arg-183 is involved in ER retention (31). In the corresponding position of the S protein there is a valine residue, a replacement of which does not affect intracellular retention as indicated by analysis of a deletion mutant. The two proteins appear to differ in their interaction with cellular binding partners that mediate intracellular retention. Although the tyrosine and isoleucine residues are critical for the transport behavior of the TGEV S protein, other residues are expected also to affect intracellular localization. A YTDI motif in the tail of the VSV G protein is responsible for targeting of this protein to the basolateral surface of epithelial cells. The cytoplasmic tail of G protein is known to contain a specific export signal for the transport out of the ER. A di-acidic motif around the above mentioned isoleucine residue (Asp-Ile-Glu), has been shown to efficiently recruit G and other proteins to vesicles mediating export from the ER (32). The extended sequence Tyr-Thr-Asp-Ile-Glu-Met comprising the complete YXXI motif has been reported to further increase the export efficiency and to be functional also on proteins that otherwise only inefficiently exit the ER (33).
In a recent analysis of the S protein of avian infectious bronchitis virus, the viral glycoprotein was reported to be intracellularly retained because of a dilysine motif that is present also in cellular proteins that are retained in the ER (34). These authors mainly analyzed the chimeric proteins consisting of the ectodomain and transmembrane anchor of the VSV G protein and the cytoplasmic tail of the infectious bronchitis virus. When the 11 carboxyl-terminal amino acids of this chimeric protein were replaced by the corresponding peptide of the S proteins of TGEV or SARS-CoV, the chimeric proteins were retained intracellularly. Retention was shown to be mediated by the two basic residues (lysine and histidine) among the five carboxyl-terminal amino acids (KXHXX). However, this motif appears not to be a major transport signal in the authentic S proteins, (i) a deletion mutant of the TGEV S protein lacking the five carboxyl-terminal residues was still intracellularly retained (this work), and (ii) the S protein of the SARS coronavirus is transported to the cell surface (Ref. 35 and this work). The KXHXX motif may have a modulating effect, because the endoglycosidase H-resistant form of the S protein was expressed less efficiently by the Y1440A and I1443A mutants than it was by the chimeric proteins.
Expression of S proteins from the nucleus was found to be inefficient. As TGEV replicates in the cytoplasm, the S gene may contain cryptic splice sites or other sequence elements that are detrimental for mRNA processing in the nucleus. Efficient expression of the S protein has been reported when Vaccinia virus or baculovirus was used (17, 18, 36). Although these viral vectors are very efficient expression vectors, surface transport of the S protein was inefficient. Using the Vaccinia virus for expression of the S protein of TGEV, only some protein was detected on the plasma membrane, whereas the majority of the S protein was intracellularly retained (36). In the case of feline infectious peritonitis virus, it has been reported that the Vaccinia virus expressed S protein acquired resistance to endoglycosidase H with a half-time of 3 h. This inefficient surface transport may be explained by saturation of the cellular retention machinery. Once the synthesis of an intracellularly retained protein has exceeded a threshold value, the cellular interaction partners become saturated and are not able to retain the excess amount of protein. This phenomenon has been reported for cellular proteins that are retained intracellularly, e.g. endoplasmic reticulum-Golgi intermediate compartment-53 (37).
Maturation of coronaviruses occurs by a budding process at the cis-Golgi network/endoplasmic reticulum-Golgi intermediate compartment (2). Two of the coronavirus envelope proteins, E and M, are known to be intracellularly retained (9, 10). We have shown that the S protein of TGEV is also not transported to the cell surface. For optimal virus production, it appears reasonable that the membrane proteins are enriched at the compartment where virus budding occurs. Intracellular retention of the viral membrane proteins may also delay the time point when the infected cell is recognized by the cellular defense mechanisms such as antibodies. With some coronaviruses infected cells fuse with uninfected cells forming multinucleated cells. This syncytium formation can occur when late in infection due to overproduction, the S protein cannot be retained anymore intracellularly and is transported to the cell surface. Intracellular retention in the early stage of infection may delay this cell-damaging effect and therefore contribute to optimal virus production. The absence of viral glycoprotein from the cell surface may also avoid other defense mechanisms, e.g. complement activation. Most coronaviruses contain a tyrosine residue in the cytoplasmic tail of the S proteins that may serve as a retention signal. In fact, experimental data show that the S proteins of infectious bronchitis virus (34)2 and bovine coronavirus (38) are also intracellularly retained. In this respect SARS-CoV is an exception, because the S protein is transported to the cell surface. This virus appears to have a different strategy of virus host interaction. Perhaps optimal virus production is not a point of highest priority for this virus, and interaction of the surface-expressed S protein with neighboring uninfected cells provides some advantage, e.g. cell-to-cell spread of infection. Future studies will have to show the importance of this retention signal for coronavirus infection and virulence. Additionally, identification of the cellular proteins responsible for coronavirus S protein intracellular retention will allow to study the importance of host cellular proteins in coronavirus morphogenesis and infection.
Acknowledgments
We thank Sandra Bauer for technical assistance. We thank Drs. Conzelmann, Neubert, and Kawaoka for providing cells and reagents.
Footnotes
The abbreviations used are: CoV, coronavirus; SARS, severe acute respiratory syndrome; TGEV, transmissible gastroenteritis virus; ER, endoplasmic reticulum; S, S protein.
C. Winter, C. Schwegmann-Wessels, and G. Herrler, manuscript in preparation.
References
- 1.Compans R.W., Herrler G. In: Mucosal Immunology. Ogra P.L., Mestecky J., Lamm M.E., Strober W., Bienenstock J., McGhee J.R., editors. Academic Press; New York: 1999. pp. 671–683. [Google Scholar]
- 2.Tooze J., Tooze S., Warren G. Eur. J. Cell Biol. 1984;33:281–293. [PubMed] [Google Scholar]
- 3.Krijnse-Locker J., Ericsson M., Rottier P.J., Griffiths G. J. Cell Biol. 1994;124:55–70. doi: 10.1083/jcb.124.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maisner A., Klenk H., Herrler G. J. Virol. 1998;72:5276–5278. doi: 10.1128/jvi.72.6.5276-5278.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanger C., Muhlberger E., Ryabchikova E., Kolesnikova L., Klenk H.D., Becker S. J. Virol. 2001;75:1274–1283. doi: 10.1128/JVI.75.3.1274-1283.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Escors D., Camafeita E., Ortego J., Laude H., Enjuanes L. J. Virol. 2001;75:12228–12240. doi: 10.1128/JVI.75.24.12228-12240.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Escors D., Ortego J., Laude H., Enjuanes L. J. Virol. 2001;75:1312–1324. doi: 10.1128/JVI.75.3.1312-1324.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Risco C., Muntion M., Enjuanes L., Carrascosa J.L. J. Virol. 1998;72:4022–4031. doi: 10.1128/jvi.72.5.4022-4031.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Machamer C.E., Mentone S.A., Rose J.K., Farquhar M.G. Proc. Natl. Acad. Sci. U. S. A. 1990;87:6944–6948. doi: 10.1073/pnas.87.18.6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klumperman J., Locker J.K., Meijer A., Horzinek M.C., Geuze H.J., Rottier P.J. J. Virol. 1994;68:6523–6534. doi: 10.1128/jvi.68.10.6523-6534.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swift A.M., Machamer C.E. J. Cell Biol. 1991;115:19–30. doi: 10.1083/jcb.115.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Locker J.K., Klumperman J., Oorschot V., Horzinek M.C., Geuze H.J., Rottier P.J. J. Biol. Chem. 1994;269:28263–28269. [PubMed] [Google Scholar]
- 13.Lim K.P., Liu D.X. J. Biol. Chem. 2001;276:17515–17523. doi: 10.1074/jbc.M009731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corse E., Machamer C.E. J. Virol. 2000;74:4319–4326. doi: 10.1128/jvi.74.9.4319-4326.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corse E., Machamer C.E. J. Virol. 2002;76:1273–1284. doi: 10.1128/JVI.76.3.1273-1284.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enjuanes L., Brian D., Cavanagh D., Holmes K., Lai M.M.C., Laude H., Masters P., Rottier P., Siddell S.G., Spaan W.J.M., Taguchi F., Talbot P. In: Virus Taxonomy. van Regenmortel M.H.V., Fauquet C.M., Bishop D.H.L., Carsten E.B., Estes M.K., Lemon S.M., Mayo M.A., McGeoch D.J., Pringle C.R., Wickner R.B., editors. Academic Press; New York: 2000. pp. 835–849. [Google Scholar]
- 17.Godet M., Rasschaert D., Laude H. Virology. 1991;185:732–740. doi: 10.1016/0042-6822(91)90544-L. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vennema H., Heijnen L., Zijderveld A., Horzinek M.C., Spaan W.J. J. Virol. 1990;64:339–346. doi: 10.1128/jvi.64.1.339-346.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vennema H., Godeke G.J., Rossen J.W., Voorhout W.F., Horzinek M.C., Opstelten D.J., Rottier P.J. EMBO J. 1996;15:2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Penzes Z., Gonzalez J.M., Calvo E., Izeta A., Smerdou C., Mendez A., Sanchez C.M., Sola I., Almazan F., Enjuanes L. Virus Genes. 2001;23:105–118. doi: 10.1023/A:1011147832586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moss B., Elroy-Stein O., Mizukami T., Alexander W.A., Fuerst T.R. Nature. 1990;348:91–92. doi: 10.1038/348091a0. [DOI] [PubMed] [Google Scholar]
- 22.Gebauer F., Posthumus W.P., Correa I., Sune C., Smerdou C., Sanchez C.M., Lenstra J.A., Meloen R.H., Enjuanes L. Virology. 1991;183:225–238. doi: 10.1016/0042-6822(91)90135-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zimmer G., Budz L., Herrler G. J. Biol. Chem. 2001;276:31642–31650. doi: 10.1074/jbc.M102633200. [DOI] [PubMed] [Google Scholar]
- 24.Zimmer G., Trotz I., Herrler G. J. Virol. 2001;75:4744–4751. doi: 10.1128/JVI.75.10.4744-4751.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kohl W., Zimmer G., Greiser-Wilke I., Haas L., Moennig V., Herrler G. J. Gen. Virol. 2004;85:1101–1111. doi: 10.1099/vir.0.19740-0. [DOI] [PubMed] [Google Scholar]
- 26.Alfalah M., Jacob R., Preuss U., Zimmer K.P., Naim H., Naim H.Y. Curr. Biol. 1999;9:593–596. doi: 10.1016/s0960-9822(99)80263-2. [DOI] [PubMed] [Google Scholar]
- 27.Naim H.Y., Sterchi E.E., Lentze M.J. Biochem. J. 1987;241:427–434. doi: 10.1042/bj2410427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonifacino J.S., Dell'Angelica E.C. J. Cell Biol. 1999;145:923–926. doi: 10.1083/jcb.145.5.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Owen D.J., Evans P.R. Science. 1998;282:1327–1332. doi: 10.1126/science.282.5392.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mallabiabarrena A., Jimenez M.A., Rico M., Alarcon B. EMBO J. 1995;14:2257–2268. doi: 10.1002/j.1460-2075.1995.tb07220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borroto A., Lama J., Niedergang F., Dautry-Varsat A., Alarcon B., Alcover A. J. Immunol. 1999;163:25–31. [PubMed] [Google Scholar]
- 32.Nishimura N., Balch W.E. Science. 1997;277:556–558. doi: 10.1126/science.277.5325.556. [DOI] [PubMed] [Google Scholar]
- 33.Sevier C.S., Weisz O.A., Davis M., Machamer C.E. Mol. Biol. Cell. 2000;11:13–22. doi: 10.1091/mbc.11.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lontok E., Corse E., Machamer C.E. J. Virol. 2004;78:5913–5922. doi: 10.1128/JVI.78.11.5913-5922.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simmons G., Reeves J.D., Rennekamp A.J., Amberg S.M., Piefer A.J., Bates P. Proc. Natl. Acad. Sci. U. S. A. 2004;101:4240–4245. doi: 10.1073/pnas.0306446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pulford D.J., Britton P. Virology. 1991;182:765–773. doi: 10.1016/0042-6822(91)90617-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hauri H.P., Kappeler F., Andersson H., Appenzeller C. J. Cell Sci. 2000;113:587–596. doi: 10.1242/jcs.113.4.587. [DOI] [PubMed] [Google Scholar]
- 38.Kunkel F., Herrler G. Arch. Virol. 1996;141:1123–1131. doi: 10.1007/BF01718615. [DOI] [PMC free article] [PubMed] [Google Scholar]


