SUMMARY
CD8+ T cell exhaustion impedes control of chronic viral infection; yet how new T cell responses are mounted during chronic infection is unclear. Unlike T cells primed at the onset of infection that rapidly differentiate into effectors and exhaust, we demonstrate that virus-specific CD8+ T cells primed after establishment of chronic LCMV infection preferentially generate memory-like transcription factor TCF1+ cells that were transcriptionally and proteomically distinct, less exhausted, and more responsive to immunotherapy. Mechanistically, adaptations of antigen presenting cells and diminished T cell signaling intensity promoted differentiation of the memory-like subset at the expense of rapid effector cell differentiation, which was now highly dependent on IL-21-mediated CD4+ T cell help for its functional generation. Chronic viral infection similarly redirected de novo differentiation of tumor-specific CD8+ T cells, ultimately preventing cancer control. Thus, targeting these T cell stimulatory pathways could enable strategies to control chronic infection, tumors and enhance immunotherapeutic efficacy.
Keywords: LCMV, CD8+ T cells, chronic infection, immunotherapy, PDL1, T cell exhaustion, cancer, tumor immunology, CD4 T cell help, IL-21, dendritic cells
Graphical Abstract
eTOC Blurb
How new T cell responses are mounted during chronic infection is unclear. Snell et al. demonstrate that unlike T cells primed at the onset of infection, CD8+ T cells primed after chronic viral infection is established are differentially programmed into less exhausted, TCF1+ memory-like cells that respond better to immunotherapy but diminish immunity to cancers that arise in chronic infection.
INTRODUCTION
CD8+ cytotoxic T lymphocytes (CTL) control viral infections by directly killing infected cells, secreting proinflammatory cytokines, and forming memory that protects against re-exposure. Although most viral infections trigger a potent CTL response that resolves infection, certain viruses are capable of outpacing the immune response and establishing a chronic infection. In response to prolonged viral replication and antigen stimulation the host initiates an immunosuppressive program that actively suppresses antiviral T cell function leading to CTL exhaustion and preventing subsequent control of infection (Wherry and Kurachi, 2015). Our understanding of T cell activation and exhaustion in chronic infection comes largely from studies characterizing T cells primed at the onset of infection, yet this represents only a fraction of the antiviral T cell response generated during chronic infection (Allen et al., 2005; Henn et al., 2012; Le et al., 2017; McGranahan et al., 2016; Vezys et al., 2006). On the other hand, little is known about how new T cell responses are mounted throughout chronic infection, despite their importance to balance attrition of exhausted cells, to target virus-escape mutants and neo-antigens, to control secondary infections and tumors that arise in the chronic infection, and for immunotherapy to rebuild the antiviral response.
At the onset of chronic infection, CD8+ T cells are activated in a setting of increasing virus titers and shifting from a pro-inflammatory to immunosuppressive environment that progressively leads to exhaustion (Angelosanto et al., 2012; Brooks et al., 2006; Wherry et al., 2003a). However, in an established chronic viral infection, this immunosuppressive environment is already entrenched and naïve T cells being primed would immediately encounter established chronic virus and inflammation, altered antigen presenting cell (APC) populations (and functions), and a long-since established suppressive environment (Cunningham et al., 2016; Snell et al., 2017). These environmental differences between the onset and the established chronic infection could all potentially affect CD8+ T cell activation, function and differentiation; having consequences toward long-term viral control and for immune restorative therapies, including hematopoietic stem cell engineering approaches that are inherently dependent on the activation of de novo T cell responses within established chronic viral infection (Kitchen et al., 2012).
Activated CD8+ T cells in chronic viral infection differentiate into two subsets: TCF1+ (T cell factor1) and TCF1− CD8+ T cells (He et al., 2016; Im et al., 2016; Leong et al., 2016; Utzschneider et al., 2016; Wu et al., 2016). The vast majority of antiviral CD8+ T cells are composed of granzyme and perforin expressing terminally differentiated TCF1− effector cells. TCF1+ cells possess decreased effector function, but maintain proliferative capacity and an ability to reseed the TCF1− cells throughout chronic infection. Consequently, the TCF1+ subset is essential for long-term virus control. Yet, the molecular and cellular mechanisms that drive the fate choices of these CD8+ T cell subsets, how they are modulated through the course of chronic infection, or how to therapeutically induce these subsets are not understood.
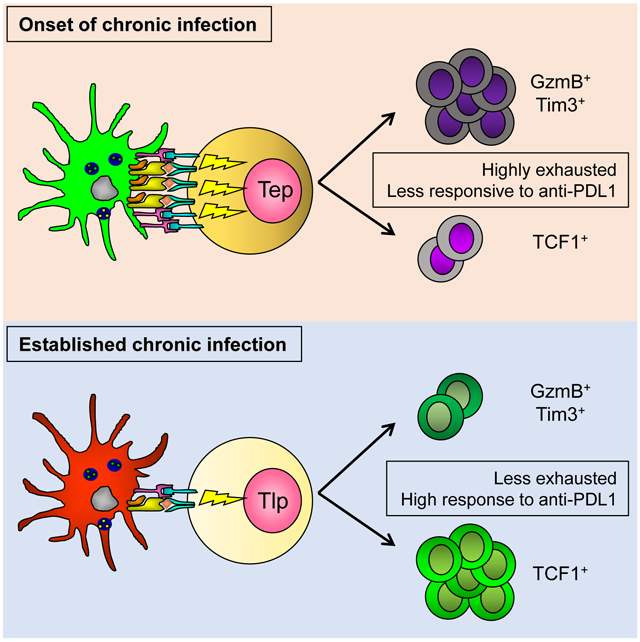
Herein, we demonstrate that unlike CD8+ T cells primed at the onset of chronic LCMV infection that rapidly differentiated into effectors and exhausted, CD8+ T cells primed after chronic infection was established preferentially generated TCF1+ memory-like T cells that resisted contraction, were less exhausted, had increased functionality and responded better to anti-PDL1 immunotherapy. Changes to the priming APC diminished the strength of activating TCR and costimulatory signals inducing TCF1+ memory-like CD8 T cell programming. Thus, by modulating APC and T cell signal strength, CD8+ T cells primed in the established infection are skewed away from rapid effector differentiation and exhaustion and are enabled for the sustained battle against a long-term chronic infection.
RESULTS
Virus-specific CD8+ T cells primed in an established persistent infection undergo an alternative pathway of transcriptional and effector differentiation.
To investigate how naïve CD8+ T cells (Tn) differentiate in the environment of an established persistent infection compared to at the onset of infection, we adoptively transferred LCMV-Glycoprotein (GP)33-41 specific CD8+ TCR transgenic (P14) T cells into mice that had been chronically-infected 21 days earlier with LCMV-Clone 13 (Cl13), or into naïve mice that were then immediately infected with Cl13, enabling comparison of the same cells primed at different times of infection. Importantly, transgenic P14 CD8+ T cells recapitulate the host-derived GP33-41 tetramer response to Cl13 (Brooks et al., 2006). In the LCMV system, chronic virus replication peaks at ~8 days after infection and then decreases to set-point and is maintained systemically for 60-80 days (Figure S1A). By 21 days, the viral titers were ~2 logs lower than at their peak from the onset of infection and remained similar at continual levels in the spleen throughout the established chronic infection (Figure S1A), consistent with the physiologic evolution of chronic viral infections. The rapid virus expansion was also observed following acute LCMV-Armstrong infection, despite the containment of the acute infection at 8 days (Figure S1A), indicating that it is not the initial high spike of virus itself, but rather many factors that cause T cell exhaustion and viral persistence.
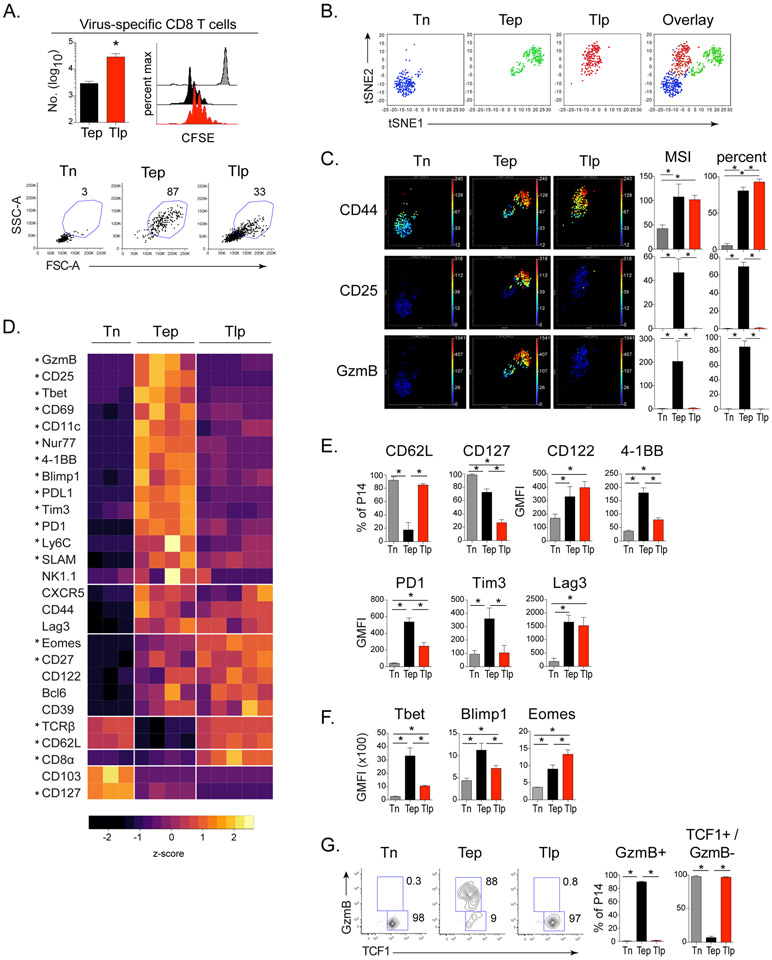
Sixty hours after transfer, CD8+ T cells primed at the onset of chronic infection (termed early prime; Tep) and CD8+ T cells primed in the established persistent infection (termed late prime; Tlp) had undergone similar proliferation, despite that Tlp cells failed to efficiently blast (Figure 1A and S1B). To more comprehensively assess phenotypic and functional differences between the Tep and Tlp populations we performed high dimensional time-of-flight mass cytometry (CyTOF) analysis. Cells belonging to the Tep, Tlp and Tn cell subsets did not overlap on the t-Distributed Stochastic Neighbor Embedding (t-SNE) maps, indicating that they had distinct high dimensional phenotypes (Figure 1B). Both Tep and Tlp cells clustered separately from Tn cells, however Tlp cells mapped closer to the naïve cells than Tep cells, suggesting more similarity between Tn and Tlp cells (Figure 1B). Consistent with activation, CD44 was upregulated by both Tep and Tlp populations compared to Tn cells (Figure 1C and 1D). However, despite their activation and proliferation, Tlp cells maintained high surface expression of the lymph node retention molecule CD62L, and failed to upregulate key effector and survival molecules including the high affinity IL-2 receptor alpha chain (CD25) and Granzyme B (GzmB) (Figure 1C-E and S1C). Although moderately increased over Tn cells, the Tlp cells had lower expression of many activation markers compared to Tep cells, including CD69, CD11c, 4-1BB, Ly6C, SLAM, PD1, Tim3 and PDL1 (Figure 1D, E). Further, Tep cells had increased expression of the transcription factors Tbet and Blimp1 (Figure 1D, F, S1D), which drive CD8+ T cell effector function and terminal differentiation (Joshi et al., 2007; Xin et al., 2016). Conversely, Tlp cells exhibited few characteristics of CTL effector differentiation, having low expression of Tbet and Blimp1 and failing to produce GzmB (Figure 1D-G, S1D). Instead, Tlp cells had higher expression of the transcriptional regulator Eomesodermin (Eomes) and sustained high TCF1 expression (Figure 1D-G and S1D), transcription factors that endow self-renewal capacity and are associated with long-term memory T cell survival. Thus, Tep cells exhibited a CTL effector signature characterized by heightened expression of multiple activation and inhibitory molecules, while Tlp cells exhibited a signature associated with memory and longevity.
Figure 1. CD8+ T cells primed in an established chronic infection have a distinct differentiation profile 60 hours post-priming.
P14 T cells were transferred into naïve mice immediately infected with Cl13 (Tep), mice that had been infected 21 days earlier with Cl13 (Tlp) or into naïve mice that were not infected (Tn). T cell responses were analyzed 60hrs after transfer.
(A) Bar graph indicates the number of CFSE-labeled P14 Tep and Tlp cells in the spleen. Histograms depict proliferation of CFSE-labeled P14s: gray (Tn), black (Tep) and red (Tlp). Flow plots depict proportion of blasting cells in the spleen.
(B) t-SNE plots of naïve, early and late primed P14 T cells analyzed by CyTOF (For complete CyTOF panel see Table S1).
(C) t-SNE plots colored by intensity of indicated markers individually scaled. Bar graphs indicate median signal intensity (MSI) and percent marker positive of P14 cells.
(D) Heat map depicts median signal intensity (MSI) or geometric mean fluorescence intensity (GMFI) of indicated extracellular markers and transcription factors derived from CyTOF and flow cytometry. Rows are scaled by z-score. * represents p<0.05 between Tep and Tlp cells. Significance was calculated by t-test.
(E) Frequency or GMFI of indicated extracellular markers on P14 cells.
(F) GMFI of indicated transcription factors in P14 T cells.
(G) Proportion of GzmB+/TCF1+ GzmB− cell subsets of P14 T cells.
Data represent 3 or more independent experiments with 3-5 mice per group. Error bars indicate standard deviation (SD). Significance was determined by one-way ANOVA unless otherwise indicated. *, p<0.05 See also Figure S1 and Table S1.
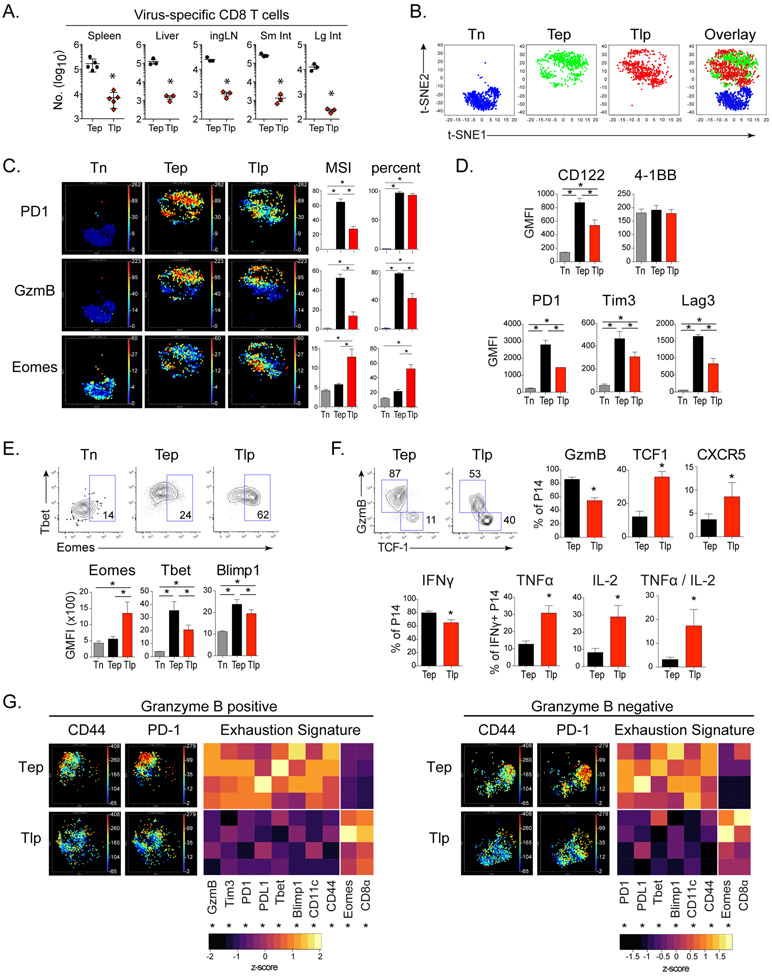
By 8 days after priming, CD8+ Tep cell numbers were increased by 20-100 fold in multiple tissues (Figure 2A). CyTOF analyses demonstrated Tep and Tlp were largely exclusive in high dimensional space, indicating a continued divergence between their differentiation states (Figure 2B, C). By day 8, CD62L, CD25 and CD127 were down-regulated on both Tep and Tlp cells (Figure S2A), whereas Tep cells exhibited higher expression of the activation and/or inhibitory proteins CD122, CD11c, PD1, PDL1, Tim3 and Lag3, as well as increased expression of Tbet, Blimp1 and GzmB (Figure 2C-F, S2B, S2C), suggesting increased stimulation, terminal differentiation and exhaustion of this subset. In contrast, Tlp cells had increased expression of chemokine receptor CXCR5 (both as a percentage of cells and single cell expression levels); and a large population of Tlp cells remained TCF1+, despite slightly decreased TCF1 expression on a per cell basis, likely due to lower activation levels (Figure 2F, S2B). This increased proportion of Tlp TCF1+ cells corresponded with a decreased percentage of GzmB producing Tlp effector cells, as well as less GzmB expression on a per cell basis (Figure 2F, S2B, S2C). Comparison of the effector GzmB+ subset of Tep versus Tlp cells (Fig S2E for backgating) demonstrated that the Tlp effectors that were present expressed a less activated and exhausted signature, characterized by decreased expression of many inhibitory and/or terminal differentiation proteins (Figure 2G). This pattern was similar when we examined GzmB− TCF1+ subsets as well (Figure 2G), suggesting that regardless of effector cell or memory-like cell differentiation, Tlp cells were less activated and exhausted overall. In line with this, the percentage of IFNγ+ Tlp cells that co-produced TNFα and IL-2 was also increased compared to Tep cells, demonstrating that at the single cell level the Tlp cells had increased polyfunctionality (Figure 2F). Finally, Tlp and Tep populations possessed similar CTL killing capacity despite decreased proportions of GzmB+ CTL in the total Tlp population (Figure 2F, S1D), thus indicating that CD8+ Tlp cells sustained a multi-parameter signature of diminished immune activation and exhaustion in both the memory-like and effector populations.
Figure 2. Virus-specific CD8+ T cells primed in an established persistent infection undergo an alternative pathway of transcriptional and effector differentiation.
P14 T cells were transferred into naïve mice immediately infected with Cl13 (Tep), into mice that had been infected 21 days earlier with Cl13 (Tlp), or into naïve mice that were not infected (Tn). T cell responses were analyzed 8 days after transfer.
(A) Total number of Tep and Tlp cells in indicated organs. * p<0.05 by t-test.
(B) t-SNE plots of naïve, early and late primed P14 T cells analyzed by CyTOF.
(C) t-SNE plots colored by intensity of indicated markers individually scaled from 0 to maximum. Bar graphs indicate median signal intensity (MSI) and percent positive of P14 cells.
(D) GMFI of indicated activation and inhibitory markers on virus-specific P14 T cells.
(E) Flow plots show proportion of Eomes positive P14 cells and bar graphs depict GMFI of indicated transcription factors in P14 T cells.
(F) Flow plots and bar graphs depict proportion of GzmB+/TCF1+ GzmB− / CXCR5+ cell subsets of P14 T cells (top). Bar graphs depict the proportion of IFNg producing P14 T cells and the proportion of TNFα, IL-2 and dual TNFα/IL-2 producing cells of IFNγ+ Tep and Tlp P14 cells (bottom). * p<0.05 by t-test.
(G) t-SNE plots of GzmB+ and GzmB− Tep and Tlp cells colored by CD44 and PD1 expression assessed by mass cytometry. Heat maps depict MSI of indicated extracellular markers and transcription factors by GzmB+ and GzmB− Tep and Tlp cells. Columns are scaled by z-score. * p<0.05 by t-test.
Data represent 3 or more independent experiments with 3-5 mice per group. Error bars indicate SD. Significance was determined by one-way ANOVA unless otherwise indicated. *, p<0.05.
See also Figure S2.
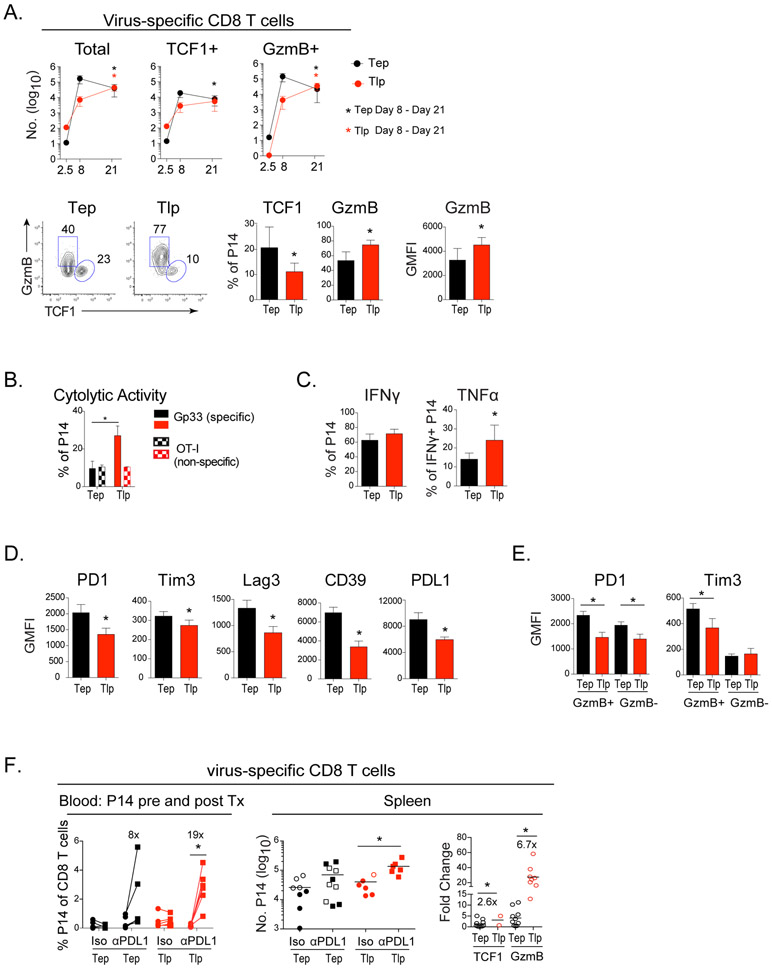
The programming of Tlp cells suggested that this population may possess long-term survival potential (He et al., 2016; Im et al., 2016; Leong et al., 2016; Utzschneider et al., 2016; Wu et al., 2016). While both TCF1+ and GzmB+ Tep cells contracted from their peak numbers after day 8 post-priming, these populations were sustained or increased in CD8+ Tlp cells between day 8 and 21 after priming (Figure 3A). GzmB expression by Tlp cells was also higher three weeks after priming compared to Tep cells on a per cell basis (Figure 3A), and a higher proportion of Tlp cells maintained cytolytic activity (Figure 3B). Further, while similar proportions of Tep and Tlp cells produced IFNγ, an increased proportion of IFNγ+ Tlp cells produced TNFα at this time point, indicating their sustained polyfunctionality (Figure 3C). Consistent with decreased CTL exhaustion and increased survival, expression of PD1, Tim3, Lag3, CD39 and PDL1 remained lower in Tlp than Tep cells, despite sustained splenic viral titers in late primed mice (Figure 3D and S1A). When Tlp and Tep cells were subdivided into GzmB+ effector versus GzmB− memory-like cells, Tlp cells expressed less PD1 on both populations, as well as decreased Tim3 on GzmB+ cells (Tim3 is not expressed on TCF1+ memory-like cells) (Figure 3E). A similar differentiation was observed in liver-infiltrating Tlp cells with an overall decrease in the number of GzmB+ Tlp effector cells remaining at day 21 (Figure S3A). Further, like the splenic Tlp cells, the single cell expression levels of GzmB were increased and PD1 decreased in liver Tlp cells (Figure S3A), indicating that the altered differentiation and decreased expression of inhibitory receptors occurred in both lymphoid and nonlymphoid tissues.
Figure 3. Late primed cells are sustained, resist exhaustion and respond better to PDL1 Blockade.
P14 T cells were transferred into naïve mice immediately infected with Cl13 (Tep) or into mice that had been infected 21 days earlier with Cl13 (Tlp).
(A) Kinetics of total, TCF1+ and GzmB+ Tep (black) and Tlp (red) P14 T cell responses. Bottom flow plots and bar graphs depict the GMFI of GzmB and the proportion of GzmB+ or TCF1+ P14 T cells at day 21 after infection.
(B) Percentage of cytolytic Tep or Tlp P14 cells to targets labeled with LCMV-Gp33 (solid) or non-specific OT-I peptide (patterned).
(C) Percentage of IFNγ+ P14s cells and percentage of TNFα producing cells of IFNγ+ P14s at day 21 post-priming.
(D & E) GMFI of various inhibitory factors on (D) total or (E) GzmB+ and GzmB− Tep and Tlp P14 T cells.
(F) Mice were CD4+ depleted prior to Cl13 infection. Tep and Tlp P14 CD8+ T cells were primed as described previously and 21 days post-priming mice were treated with anti-PDL1 or isotype control antibody over 14 days. Left graph depicts proportion of P14 T cells in the blood before (day 20) and after (day 35) antibody therapy (*, p<0.05 by paired t-test). The middle graph indicates the total number of Tep and Tlp in the spleen after isotype or anti-PDL1 treatment (2 experiments combined: open shapes and filled shapes). Right graph represents the relative increase (fold change) in the number of TCF1+ and GzmB+ Tep and Tlp cells following anti-PDL1 blockade in relation to their number of TCF1+ cells from their respective isotype treatment group. To generate these values, the number of TCF1+ and GzmB+ cells from each mouse in the anti-PDL1 group was divided by the average number of TCF1+ cells from their isotype group. As in the middle graph, 2 experiments were combined.
Data represent 2-4 independent experiments with 4-5 mice per group. Error bars indicate SD. Numbers inside the graphs show the fold change between the indicated groups. Significance was determined by t-test unless otherwise indicated. *, p<0.05.
See also Figure S3.
Virus-specific CD8+ T cells primed in the midst of infection are more responsive to PDL1 blockade
The preferential generation of TCF1+ cells coupled with their decreased expression of inhibitory receptors suggested that Tlp cells might respond better than Tep cells to anti-PDL1 therapy (He et al., 2016; Im et al., 2016; Utzschneider et al., 2016). To test this, Tep and Tlp cells were primed for 21 days and then mice were treated with anti-PDL1 blocking antibody. For this experiment, the mice were CD4+ depleted prior to infection to determine intrinsically how the Tep and Tlp cells responded to anti-PDL1 without the secondary effects of decreasing virus loads. Following anti-PDL1 blockade, an average 19-fold expansion of Tlp cells in the blood was observed with robust responses in all treated mice, whereas Tep cells expanded with only an average 8-fold increase in anti-PDL1 treated mice (Figure 3F). A similar enhanced responsiveness was observed in the spleen, with Tlp cells responding to anti-PDL1 therapy in ~91% of mice, whereas Tep responsiveness was only observed in 50% of treated mice (Figure 3F). When the fold expansion was quantified using the TCF1+ cells from isotype treated mice as baseline (the cells that give rise to the anti-PDL1 induced CD8+ T cells), the fold change in Tlp TCF1+ and GzmB+ cells was substantially higher than observed for Tep cells (Figure 3F). The enhanced expansion of Tlp cells was independent of changes in viral titers, since the therapy purposefully did not lower viral titers in the late prime condition and only minimally in the early prime condition (Figure S3B). The slightly lowered viral titers following PDL1 blockade in the early prime condition likely arose from effects on the endogenous T cell response (not the transferred P14 cells), which become less responsive to immunotherapy with time in chronic infection (early prime blockade initiated 21 days after infection whereas late prime blockade was initiated 42 days after infection). Thus, the Tlp cells responded better than the Tep cells despite the small decrease in virus titers in the early prime condition, indicating that due to their differentiation state, the Tlp cells intrinsically possessed enhanced responsiveness to anti-PDL1 therapy.
Diminished TCR and costimulatory signaling inhibit differentiation of effector CD8+ Tlp cells while preserving the TCF1+ subset
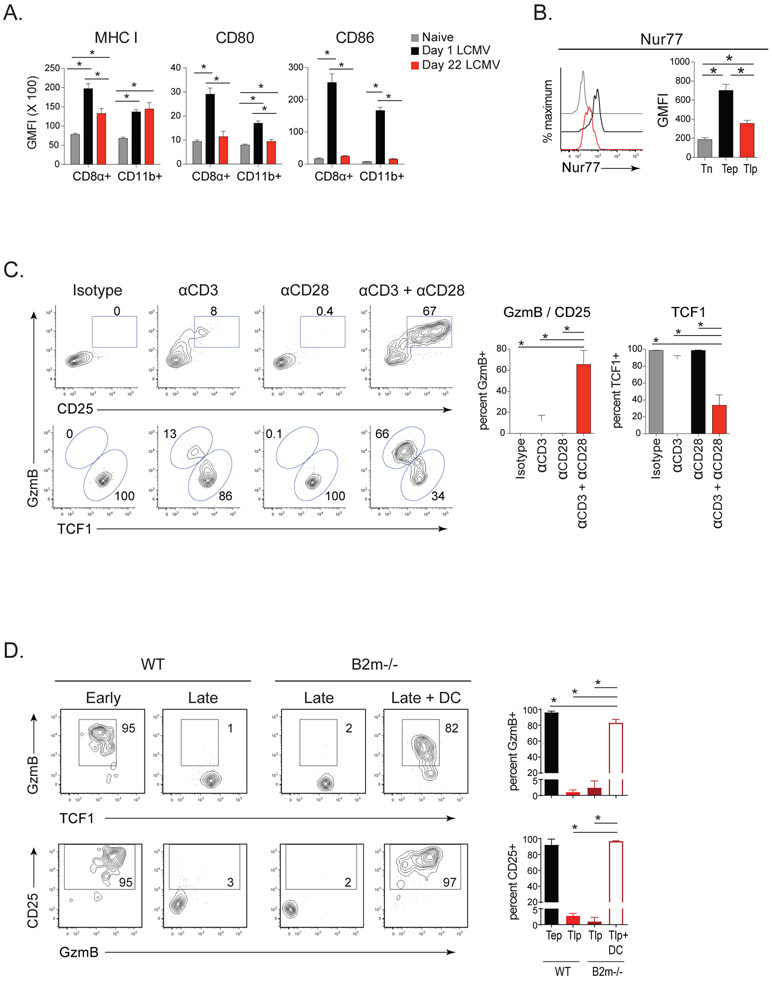
Tlp and Tep cell differentiation pathways diverged early following priming (Figure 1), suggesting that APCs may alter the induction of these subsets. Although there were fewer dendritic cells (DCs) in the midst of infection compared with 1 day after infection, the proportions of CD8α+ and CD11b+ DC subsets were similar (Figure S4A), indicating that a DC subset skewing was likely not driving differential priming. CD8α+ DCs and macrophages in the established infection expressed less MHC I than those 1 day after infection; and CD80 and CD86 expression was also dramatically reduced on DCs and macrophages during infection, similar to levels seen in naïve mice (Figure 4A, S4B). The diminished APC activation state during infection corresponded with reduced Nur77 expression (a surrogate marker for TCR signaling) on Tlp compared to Tep cells at priming (Figure 4B). Together, this suggested that the distinct differentiation of Tlp cells may result from decreased TCR and/or costimulatory signals from priming APCs. To test the need for strong stimulation to generate rapid effector cells, we supplemented TCR and costimulatory signals to Tlp cells in vivo using agonistic anti-CD3 and/or anti-CD28 antibodies. While anti-CD3 or anti-CD28 alone failed to robustly generate GzmB+ effector cell differentiation, simultaneous enhancement of TCR and costimulatory signaling at the time of Tlp priming induced the majority of TCF1+ cells to become GzmB+ CD25+ cells (Figure 4C). Increasing the amount of TCR and costimulation also drove Tlp GzmB+ effector cell differentiation in CD4−/− mice (Figure S4C), indicating that the decreased TCR and costimulatory signal strength intrinsically inhibited CD8+ T cell effector differentiation in the established chronic infection. The antibody treatments did not significantly alter the number of virus-specific CD8+ Tlp cells at this time point (Figure S4D), demonstrating that changes in the proportions of Tlp subsets reflected conversion between TCF1+ and GzmB lineages. Thus, decreases in signaling strength maintained memory-like TCF1+ differentiation and limited rapid effector cell differentiation in the established chronic infection.
Figure 4. Diminished dendritic cell responsiveness and lower T cell stimulation/costimulation suppress differentiation of CD8+ effector Tlp cells while sustaining the TCF1+ subset.
(A) GMFI of MHC I, CD80 and CD86 on CD8α+ and CD11b+ DCs at 1 day (black) and 22 days post-Cl13 infection (red) (i.e., 1 day after early and late priming, respectively) and in naïve mice (gray).
(B) Nur77 expression by Tn, Tep, and Tlp P14 cells 60 hrs after priming.
C) P14 T cells were transferred into mice that had been infected 21 days earlier with Cl13. Four hours post-priming mice were treated i.p. with anti-CD3 and/or anti-CD28 or isotype control. Flow plots and bar graphs depict the frequency of GzmB+ CD25+ effector and TCF1+ memory-like P14 cells 60hrs after transfer.
(D) P14 T cells were transferred at the onset (Early) or at day 21 (Late) after Cl13 infection of wildtype or B2microglobulin (B2m)−/− mice. At the time of priming, a group of B2m−/− received GP33 peptide loaded bmDC. GzmB, TCF1 and CD25 expression was quantified on the P14 cells 60hrs after transfer.
Data represent 2-3 independent experiments with 4-5 mice per group. Error bars indicate SD. Significance was determined by one-way ANOVA. *, p<0.05.
See also Figure S4.
As DCs are fundamental for the priming of CD8+ T cells at the onset of LCMV infection (Probst and van den Broek, 2005), we used CD11c-DTR mice to test their role in priming of Tlp.cells. DT treatment just prior to late priming resulted in ~90-95 percent depletion of DCs, and diminished the proliferation and number of Tlp cells (Figure S4E). The residual priming of Tlp cells likely resulted from incomplete DC depletion or the ability of other APC populations, such as macrophages to prime CD8+ T cells in the absence of DCs. As DCs do significantly contribute to Tlp cell priming, we chose this cell type to test whether effector Tlp cell differentiation could be restored in the established infection if the quality of APC were enhanced. We transferred GP33-41 peptide-pulsed bone marrow derived DCs (bmDC) into Cl13 infected beta 2-microglobulin (B2m)−/− mice that lack the ability to endogenously present antigen to CD8+ T cells, thus restricting priming to the transferred DC while still maintaining the early and late priming environments and viral titers. Transfer of bmDC effectively induced high level Tlp GzmB+ effector cell differentiation to the same level as Tep cells (Figure 4D). The role of priming DC was further corroborated using infection of mice with a variant of Cl13 that lacks the GP33-41 epitope recognized by P14 T cells (Cl13V35A), but contains all other LCMV-derived CD8+ T cell epitopes and has analogous replication kinetics to wildtype Cl13 (Puglielli et al., 2001). DC transfer in this system also generated Tlp GzmB+ effector differentiation (Figure S4F), demonstrating that rapid de novo effector T cell differentiation is possible in the chronic infection when strong T cell stimulation is provided. Thus, the decreased T cell stimulatory capacity during the established chronic infection preferentially sustained TCF1+ memory-like cells and limited effector differentiation.
Transcriptional profiling reveals enriched signatures of TLP function, memory generation and maintenance
To gain further insight into the mechanisms that program Tep and Tlp cells, we performed RNA-seq analysis of these subsets 60 hours after priming. Transcriptional profiling demonstrated that Tep and Tlp cells rapidly diverge, with 7970 genes differentially expressed (3730 upregulated in Tep; 4240 upregulated in Tlp cells) (Figure 5A). Gene Set Enrichment Analysis (GSEA) identified upregulation of gene signatures of DNA replication, translation, ribosome biogenesis and glycolysis in Tep cells (Figure 5B and Table S2), in line with enhanced Tep cell activation and blasting (Figure 1, S1). The myc pathway (which promotes glycolysis) (Verbist et al., 2016) and the mTORC1 signaling pathway were also enriched in Tep cells (Figure 5B), consistent with the need for glycolytic metabolism to provide the energetic needs for CD8+ effector T cell differentiation, and mTORC1’s role in driving terminal differentiated effector-like T cells (Buck et al., 2017; Pollizzi et al., 2015). Tlp cells on the other hand, showed enrichment in pathways involved in cell adhesion, integrin interactions, cell motility and the Focal Adhesion Kinase (FAK) pathway (Figure 5B and Table S2). A role of the FAK pathway is to modulate the T cell:APC interaction, and the increase in this pathway which promotes movement from interaction, suggests that the decreased TCR and costimulatory signals in Tlp cells arise from decreased interaction time with APC. Consistent with the effector vs memory cell dichotomy between Tep and Tlp cells, the effector CD8+ T cell gene signature was enriched in Tep cells while the memory CD8+ T cell pathway was enriched in Tlp cells (Figure 5C and Table S3). While Tep cells had increased expression of genes associated with T cells in chronic LCMV, Tlp cells showed upregulation of genes associated with CD8+ T cells in acute LCMV (Figure 5C and Table S3), indicating the early divergence in exhaustion programming. Tep cells expressed high levels of multiple genes associated with enhanced effector activation and/or exhaustion including the costimulatory receptors and ligands Tnfrsf8 (CD30), Tnfrsf4 (OX40), Tnfrsf9, Tnfsf9 (4-1BBL and 4-1BB, respectively), cytokines IFNg, IL21 and IL10, exhaustion factors Tigit, Entpd1 (CD39), Pdcd1 (PD1), CD276 (B7-H3), Pdcdlg2 (PDL2), Havcr2 (Tim3), and the Metallothioneins Mt1, Mt2 and Mt3 (Figure 5D) that have been identified to promote CD8+ T cell dysfunction (Singer et al., 2016). Further, multiple cytolytic factors: Prf1, Gzmb, Gzma, Fasl and Gzmc, were increased in Tep cells, whereas Tlp cells instead had decreased expression of inhibitory factors (except Lag3) (Figure 5D). Tlp cells were enriched in a variety of genes involved in programming long-lived immunity and T cell memory, including IL21R (Allard et al., 2007), Tnfsf8 (CD30L) (Nishimura et al., 2005) and CD27 (Wherry et al., 2003b) and increased expression of many transcription factors present in CD8+ memory T cells and self-renewal, including Pou6f1, Bach2, Eomes, Id3, Tcf7 (TCF1), Bcl6, Foxo1, and Lef1 (Figure 5D). Metabolically, sterol and fatty acid metabolism pathways were increased in Tlp cells, including increased SREBF1 and 2 (SREBP1 and 2), as well as increased expression of Rictor, which is involved in the mTORC2 pathway that guides CD8+ T cell memory differentiation (Figure 5D) (Buck et al., 2017; Pollizzi et al., 2015). In contrast, Tep cells exhibit a terminally differentiated transcriptome, including increased expression of Prdm1 (Blimp1), Batf, Maf1, Zbtb32, tbx21 (Tbet), and Myc (Figure 5D), which drive CTL effector differentiation and function (Giordano et al., 2015; Kuroda et al., 2011; Shin et al., 2017; Xin et al., 2016). Thus, Tep cells very rapidly exhibited a transcriptional profile of robust activation, effector differentiation and dysfunction, while Tlp cells had increased expression of genes involved in memory differentiation and maintenance.
Figure 5. Transcriptional profiling 60 hours after priming reveals TLP cells have enrichment of genes involved in CD8+ T cell memory generation and maintenance.
(A) Scatter plot of average log gene expression log2 (CPM+1) of Tep (black) vs Tlp (red). Differentially expressed genes have a Q value <0.05 and a log2 fold change >0.5.
(B) Gene Set Enrichment Analysis (GSEA) identifies select differentially expressed pathways in Tep (black) and Tlp (red) from Enrichment Map Gene Set ranked by normalized enrichment score (NES).
(C) GSEA plots of differentially expressed effector vs memory CD8+ T cell pathway (top) and acute vs chronic LCMV CD8+ T cell pathway acquired from ImmuneSigDB. ES, enrichment score.
(D) Heat maps depict expression of selected differentially expressed genes in counts per million (CPM). Rows are scaled by z-score.
(E) GSEA plots of IL-2 treated and IL-21 treated CD8+ T cell pathways acquired from ImmuneSigDB.
Data represent 2 independent experiments. Each replicate consists of groups of 5 mice pooled prior to sorting of P14 T cells.
CD4+ T cell help in conjunction with differential IL-2 and IL-21 usage promote differentiation of TCF1+ into GzmB+ effector CD8+ T cell subsets
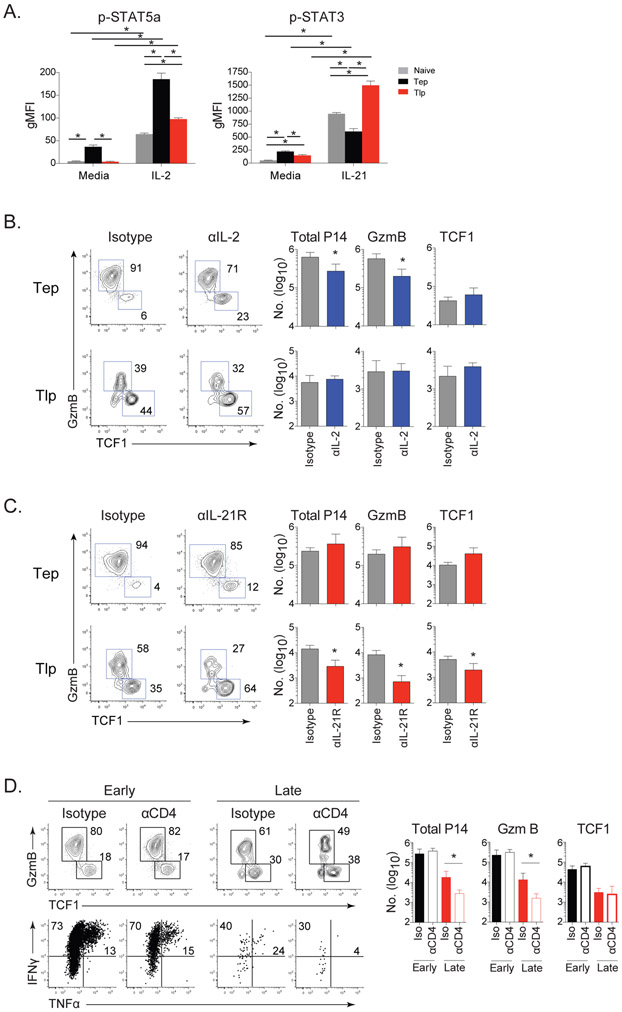
Consistent with the expression of CD25 (Figure 1C, D) and increased expression of STAT5a RNA (Figure 5D), an IL-2 signaling gene signature was elevated in Tep cells (Figure 5E). IL-2 is a strong inducer of STAT5a phosphorylation and basal pSTAT5a levels were ~10-fold higher in Tep cells (Figure 6A). Upon stimulation with IL-2, Tep cells robustly phosphorylated STAT5a, while Tlp cells phosphorylated STAT5a to a much lesser extent that was similar to basal IL-2 responsiveness in Tn cells (Figure 6A). In vivo antibody blockade of IL-2 at the onset of infection significantly reduced GzmB producing Tep cells both in proportion and number, whereas it had a minimal effect on Tlp differentiation (Figure 6B), indicating that IL-2 promoted effector CD8+ T cell differentiation at the onset of chronic infection, but that as infection progresses, other factors must take over this role.
Figure 6. CD4+ T cell help and differential IL-2 and IL-21 usage drive differentiation of TCF1+ into GzmB+ effector antiviral CD8+ T cell subsets at distinct times after infection.
(A) STAT5a and STAT3 phosphorylation by P14 Tn, (gray), Tep (black) or Tlp (red) cells isolated from mice 60hrs after priming and then cultured without stimulation (media) or following IL-2 or IL-21 stimulation for 30 minutes.
(B & C) Naïve mice immediately infected with Cl13 or mice infected 21 days prior with Cl13 received P14 T cells and were treated with (B) isotype (gray) or anti-IL-2 (blue); (C) isotype (gray) or anti-IL-21R (red) antibodies. Flow plots depict the proportion and bar graphs the number of total, GzmB+ and TCF1+ virus-specific CD8+ Tep and Tlp P14 cells 8 days after priming.
(D) Mice were CD4+ depleted or isotype treated (undepleted) prior to Cl13 infection. (Top) Flow plots depict the proportion and bar graphs show the numbers of total, GzmB+ and TCF1+ subsets of Tep or Tlp cells 8 days after priming. (Bottom) Proportion of IFNγ and TNFα producing Tep or Tlp cells upon peptide restimulation 8 days after priming.
Data represent 2 independent experiments with 4-5 mice per group. Error bars indicate SD. Significance was determined by t-test. *, p<0.05.
See also Figure S5.
Instead of IL-2, Tlp cells had enrichment of an IL-21 signaling gene signature and Tlp cells expressed significantly higher levels of IL-21R compared to Tep cells (Figure 5D, 5E). This was unexpected since the only previously identified role of IL-21 in chronic infection was to sustain long-since generated CD8+ T cells and no role of IL-21 in CD8+ T cell priming at the onset of infection was identified (Elsaesser et al., 2009; Frohlich et al., 2009; Yi et al., 2009). IL-21 signals primarily through STAT3, and IL-21 stimulation induced STAT3 phosphorylation by both Tep and Tlp cells, although much more potently in Tlp cells, with Tep cells responding less then Tn cells (Figure 6A). IL-2 did not induce Stat3 phosphorylation and IL-21 did not cause Stat5a phosphorylation in either Tep or Tlp cells, showing phosphorylation of these signaling molecules was specific to their inducing cytokine (Figure S5A). Inhibition of IL-21R signaling in vivo with an anti-IL21R blocking antibody (Zhang et al., 2015) did not affect Tep cell differentiation, whereas IL-21R blockade in the midst of infection reduced the proportion of effector GzmB producing Tlp cells by over 50% and almost doubled the frequency of TCF1+ Tlp cells (Figure 6C). The blockade of IL-21R signaling in the established chronic infection led to a decline in the overall number of Tlp cells, largely due to the decrease in GzmB+ effector Tlp cells (Figure 6C). Thus, IL-21 signaling is critical for CD8+ effector T cell differentiation in established, but not at the onset of chronic viral infection.
The distinct need of Tlp cells for IL-21, coupled with their decreased TCR signaling and costimulation, suggested that CD4+ T cell help may be particularly important when priming CD8+ T cells in the midst of infection. Depletion of CD4+ T cells prior to infection did not affect initial Tep cell expansion, subset differentiation or function (Figure 6D). However, in the absence of virus-specific CD4+ T cell help, CD8+ Tlp cells were decreased ~5-10 fold numerically, had even further decreased GzmB+ effector differentiation, and had dramatically reduced cytokine producing capacity (Figure 6D and S5B). The decreased number of total Tlp cells in the absence of CD4+ help was largely due to the decreased ability to generate GzmB+ effector cells, while the number of TCF1+ Tlp cells was either not or only minimally decreased by lack of CD4+ help (Figure 6D). The diminished Tlp effector generation over the 8 day priming and expansion period in the absence of CD4+ help was not due to changes in virus levels since no significant difference in viral titers was observed between undepleted or CD4+ depleted mice in this time (day 8 or day 29 after infection for Tep and Tlp cell priming, respectively; Figure S5C). Further, blockade of IL-21R did not additionally affect CD8+ Tlp cells in the absence of CD4+ T cell help (Figure S5D). Thus, in the established chronic virus infection (and distinct from what is observed at the onset of infection), IL-21 mediated CD4+ T cell help is critical for initially generating Tlp effector cell priming and function.
Chronic viral infection skews de novo primed anti-tumor CD8+ T cells
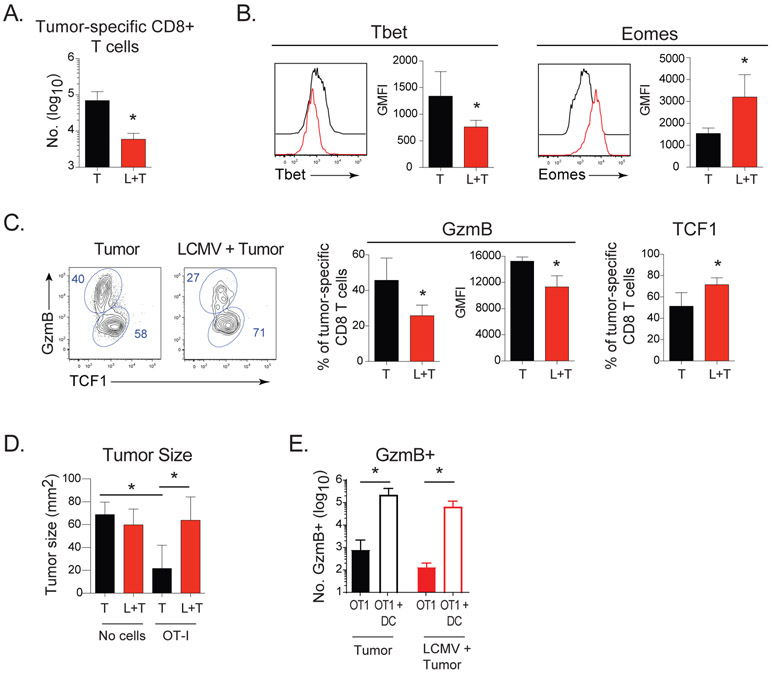
Chronic viral infections are associated with enhanced tumor formation, yet the underlying mechanisms for why this occurs are unclear. We hypothesized that like the virus-specific Tlp cells, the changes in the APC would similarly alter de novo priming of CD8+ T cells encountering a tumor in the presence of the chronic viral infection. To determine whether the altered effector differentiation observed by virus-specific Tlp cells spreads to secondary responses, we administered ovalbumin (OVA) expressing EG7 tumors to naïve or chronically infected mice. Twelve days after tumor injection there were decreased tumor-specific OT1 tumor infiltrating lymphocytes (TILs) in chronically-infected mice compared with mice that received tumor alone (Figure 7A). Like virus-specific CD8+ TLP cells, tumor-specific CD8+ TILs in chronically-infected mice were all activated (based on proliferation dye dilution), but expressed less Tbet, increased Eomes and had a differentiation pattern skewed toward TCF1+ cells and decreased GzmB+ effector cells (Figure 7B, 7C). The tumor-specific CD8+ TILs in the chronically-infected mice also expressed less GzmB on a per cell basis, analogous to virus-specific Tlp cells (Figure 7C). Although the goal of the experiment was to determine whether CD8+ T cell differentiation was similarly altered against a secondary (non-related) challenge in the chronic infection, it was interesting to observe that wherein the naïve mice were able to ultimately control the tumor at later time points at the dose given, the chronically infected mice were not (Figure S6A). When naïve OT1 cells were transferred prior to tumor, the naïve mice more rapidly controlled the tumor compared to no OT1 transfer controls (Figure 7D and S6A). On the other hand, OT1 transfer prior to tumor administration in the chronic infection did not control the tumor, although it did significantly enhance tumor control at later time points (albeit with a slower kinetic and to a lesser extent then in naïve mice; Figures S6A). These kinetics indicated that the tumor-specific OT1 cells were functioning in the naïve and chronically infected conditions, and that the addition of OT1 re-enforced the observed tumor kinetic in the given condition (likely due to their increased precursor frequency), but did not change the ultimate outcome otherwise observed when no cells were transferred. The decreased tumor-specific effector cell priming and the inability to control tumor growth in the chronic infection was overcome by providing OVA-peptide labeled bmDC that strongly activated the tumor-specific CD8+ T cells (Figure 7E and S6B), indicating that the changes in the APC spread to alter differentiation of T cells targeting tumors that arise in chronic infection, but that with the proper priming APC and CD8+ T cell activation, the tumor can be rapidly controlled during the chronic infection despite the pervasive immunosuppressive environment.
Figure 7. Chronic viral infection redirects anti-tumor CD8+ T cell differentiation and control of tumor growth.
OT-I cells were injected into naïve mice or into mice infected for 21 days with Cl13. One day later EG7 tumors were injected and tumor-specific responses quantified on day 12.
(A) Total number of tumor-specific OT-I CD8+ T cells in tumors of naïve mice (T) or in mice infected with Cl13 (T+L).
(B) Expression of Tbet and Eomes in tumor-infiltrating OT-I T cells.
(C) Proportion and GMFI of GzmB+ cells and proportion of TCF1+ OT-I cells in tumors.
(D) Tumor size on day 12 after tumor injection in naïve and chronically infected mice that received OT-I cells or no cells.
(E) OT-I cells were injected into naïve mice (black) or mice that had been infected for 21 days with Cl13 (red). One day later EG7 tumors were injected into all mice either alone (shaded bars) or in combination with OVA-peptide pulsed DCs (open bars). Graph depicts the number of tumor-specific GzmB+ effector cells 14 days later.
Data represent 2 independent experiments with 4-5 mice per group. Error bars indicate SD. Significance was determined by t-test * p<0.05.
See also Figure S6.
DISCUSSION
Although previous work has extensively characterized the differentiation and exhaustion of antiviral CD8+ T cells primed at the onset of chronic viral infection, little is known about how virus-specific or secondary antigen specific de novo CD8+ T cell responses are mounted once chronic infection is established. We demonstrate that whereas CD8+ T cells primed at the onset of chronic infection almost entirely became short-term effectors and rapidly exhausted, CD8+ T cells primed after the infection is established acquired a distinct transcriptional profile that allowed them to resist exhaustion and to retain long-term killing in the face of ongoing viral replication. Tep cells received increased TCR and costimulatory signaling, blasted more, and rapidly differentiated within the first 60 hours post-priming into effector cells with high expression of multiple activation and inhibitory markers. In contrast, Tlp cells received decreased TCR and costimulatory signals and although were equally proliferative, remained in a less differentiated state after priming, failing to produce GzmB, retaining TCF1 and expressing Eomes and multiple other transcription factors and surface proteins associated with CD8+ memory. By day 8, Tlp cells did generate a proportion of GzmB+ effector cells (albeit to a much lesser extent and expressing less GzmB at the single cell level then Tep cells), while preserving a high proportion of the memory-like subset. Both at the population level, and in particular in the GzmB+ effector subset, Tlp cells expressed a broad signature of decreased exhaustion and long-term survival potential. As chronic infection progressed Tlp cells minimally contracted, had elevated GzmB expression and killing ability, and continued to express decreased levels of inhibitory and activation receptors. The definition of T cell exhaustion is evolving from solely a description of cytokine levels to the integration of multiple criteria including functional parameters (e.g., cytokines, granzymes), distinct transcriptional and metabolic profiles, the combinatorial and quantitative expression of inhibitory receptors, and the ability to respond to immunotherapy (Bengsch et al., 2018), By all these criteria, the Tlp cells are less exhausted than their Tep cell counterparts. Due to the low numbers of Tlp cells generated, we were unable to perform transfer experiments to identify the ability of these cells to control infection compared to Tep cells. However, based on the ability of the TCF1+ population to seed the GzmB effector pool over time (He et al., 2016; Im et al., 2016; Leong et al., 2016; Utzschneider et al., 2016; Wu et al., 2016) and Tlp cells lower level of inhibitory receptors, the Tlp cells could likely lead to enhanced long-term control of chronic viruses. Overall, the change in stimulation, differentiation and function of Tlp cells may be an adaptation of the immune system to the ‘realization’ that it is no longer engaged in a short battle, but rather fighting a protracted war and needs to promote responses that are better suited for this environment.
Although many changes in the immune environment can suppress T cell responses in chronic infection, the decreased Tlp effector cell priming was initiated by intrinsic changes in the APC that led to decreased T cell stimulation. While dynamic modulation of APC subsets and function has been previously reported in Cl13 (Cunningham et al., 2016; Sevilla et al., 2004), its impact on CD8+ T cell priming, differentiation and function in vivo has not been determined. We now demonstrate that this decreased T cell stimulation specifically drove memory-like TCF1+ differentiation at the expense of rapid effector differentiation. The DC transfer experiments and the in vivo anti-CD3 and anti-CD28 treatments demonstrated that rapid Tlp cell effector differentiation is possible in the environment of the established chronic infection. Neither anti-CD3 nor anti-CD28 alone could drive effector differentiation, thus, it is not just that one strong stimulus can overcome these signaling defects but that enhancement of both TCR and CD28 signaling is needed to drive effector differentiation from the TCF1+ populations in the chronic infection. Further, in the presence of strong T cell activation signals, the need of Tlp for CD4+ T cell help was overcome, analogous to Tep priming at the onset of infection. Thus, with strongly stimulatory APCs, effector cells can be induced in the established chronic infection, whereas lower stimulation maintains TCF1 differentiation and guides the observed differentiation program. It should be noted that we are not suggesting that the immune environment itself does not also modulate Tlp cell responses. In addition to alterations in APC, the environment and the reduction in virus titers in established chronic infection likely also contribute to modulate Tlp cells, potentially augmenting the differentiation into TCF1+ cells, and likely further enabling the decreased levels of exhaustion.
Strong antigenic signaling at the onset of chronic infection increased CD25 expression that allowed IL-2 signaling to help differentiate TCF1+ CD8+ T cells into GzmB producing effectors, with minimal involvement from IL-21 signaling. In the midst of infection low antigenic signaling failed to induce CD25 expression, and coupled with the progressive switch from IL-2 to IL-21 production with viral persistence (Elsaesser et al., 2009), eventually programmed a fraction of the TCF1+ cells to differentiate into less exhausted effector cells, while still maintaining the memory-like reservoir. This requirement for IL-21 signaling at the time of priming is distinct from previous observations in which IL-21 had no notable effect on the priming or differentiation of virus-specific CD8+ Tep cells at the onset of infection (Elsaesser et al., 2009; Frohlich et al., 2009; Yi et al., 2009), but functioned exclusively to sustain the previously activated cells at later time points in chronic infection. Consistent with the strong requirement for IL-21 signaling, CD4+ T cell help was critical to promote the eventual Tlp effector cell differentiation and function in the late priming condition where APC activation is diminished, whereas CD4+ T cell help was dispensable for Tep cell priming. This data suggests that in situations wherein CD4+ T cell help is progressively limiting such as with HIV infection, diminished CD8+ T cell responses may be further compounded by lack of de novo effector cell priming to sustain the ongoing response, control virus escape mutants or to fight opportunistic cancers that arise. Thus it is interesting to speculate that the progressive switch from IL-2 toward IL-21 producing CD4+ T cells evolved not only to sustain the CD8+ T cells generated at the onset of infection, but also to promote effector cell differentiation of newly primed CD8+ T cells in the less stimulatory environment of the established chronic infection.
There is a great deal of interest in understanding what cells respond to immunotherapy and how to specifically induce these to best control infections and cancer. CD8+ Tep cells responded to anti-PDL1 therapy, however only ~50% of the mice exhibited robust responses. Such variety is similar to the reported diversity in responses seen in patient populations treated with checkpoint blockade immunotherapy, where typically only a fraction effectively responds. In contrast, Tlp cells had much higher response rates and displayed increased fold expansion, consistent with their enhanced TCF1+ memory-like cell generation and decreased levels of inhibitory receptors (Blackburn et al., 2009; He et al., 2016; Im et al., 2016; Leong et al., 2016; Utzschneider et al., 2016; Wu et al., 2016), suggesting that even among the total TCF1+ pool, distinct TCF1+ populations could be preferentially enhanced by anti-PDL1 immunotherapy. Interestingly, recent data has shown that tumors with mismatch repair defects that generate increased amounts of neo-antigens have enhanced responsiveness to checkpoint blockade (Le et al., 2017; Nebot-Bral et al., 2017). Why these cells are specifically sensitive to checkpoint blockade is unclear, but it is interesting to speculate that the priming of de novo CD8+ T cell responses in the established tumor environment similarly skews CD8+ T cell fate commitment to generate cells that are better able to respond to checkpoint blockade. Thus, CD8+ T cells primed at different times in the chronic infection may disproportionately contribute to the immunotherapeutic response, with important ramifications for virus control and the design of vaccines and immunotherapies aimed at de novo inducing distinct antiviral CD8 T cell subsets.
Many chronic viral infections are associated with increased incidence and poorer outcome to cancers. Although the mechanisms driving these relationships are likely many, mounting an initial CD8+ T cell response not well suited for immediate effector activity could allow tumors to develop that would otherwise be controlled by effector anti-tumor CD8+ T cells. Indeed, the environment in chronic infection similarly decreased rapid tumor-specific CD8+ T cell effector differentiation and in line with this, the tumor-specific CD8+ T cells primed in the chronic infection had diminished capacity to control tumor growth. Thus, while beneficial to sustain long-term immunity, this Tlp adaptation may in some instances come at a cost when immediate strong effector responses are desired to control pathogen or tumor. Transfer of tumor-peptide labeled DC at the time of tumor cell priming in chronically infected mice efficiently induced rapid and robust effector cell generation and efficient control of tumor growth, indicating that rapid effector cell differentiation against tumors and effective tumor control is possible in the environment of the chronic viral infection if strongly stimulatory APCs are present. Ultimately, targeting APC and these T cell fate determining pathways could lead to new therapies that actively control CD8+ T cell subsets and guide subsequent effector or memory responses to better control infection, tumors and enhance responses to immunotherapy.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Requests for resources and reagents should be directed to David Brooks (dbrooks@uhnresearch.ca) The anti-IL-21R blocking antibody is under MTA #2015643237 from Amgen.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
C57BL/6 mice were purchased from The Jackson Laboratory, the rodent breeding colony at the University of California, Los Angeles or at the Princess Margaret Cancer Center. LCMV-GP33-specific CD8+ TCR transgenic (P14) mice have been described previously (Brooks et al., 2006) and OVA257-64-specific CD8+ TCR transgenic (OT-I) mice, B2m−/− mice and CD11c-DTR mice were purchased from the Jackson Laboratory. Experiments were primarily performed with male mice (6-10 weeks old), although experiments were also performed with female mice for confirmation of results. Mice were housed under specific pathogen–free conditions. Mouse handling conformed to the experimental protocols approved by the University of California, Los Angeles Animal Research Committee (ARC) and the OCI Animal Care Committee at the Princess Margaret Cancer Center/University Health Network.
METHOD DETAILS
LCMV infection and P14 T cell adoptive transfer
Mice were infected i.v. via the retroorbital sinus with 2×106 PFU of LCMV-Clone13 (Cl13) or 2×106 PFU of LCMV-Cl13 variant V35A (Puglielli et al., 2001) generously provided by Dr. Dorian McGavern (NIH). Virus stocks were prepared and viral titers were quantified as described previously (Brooks et al., 2006). LCMV-specific CD8+ P14 T cells were isolated from the spleens of transgenic mice by negative selection (StemCell Technologies) and transferred i.v. in the retroorbital sinus. To assess priming and differentiation of virus-specific CD8+ T cells, 1,000 P14 cells were transferred into (1) naive mice that were infected with Cl13 immediately (early priming) or (2) into mice that had been infected 21-25 days earlier with Cl13 (late priming). For experiments in which the mice were sacrificed at 60 hours after transfer, 250,000 P14 cells were transferred to enable detection at this early time-point.
DC adoptive transfer
Bone marrow-derived dendritic cells (BMDCs) were generated as previously described (Roney, 2013). Briefly bone marrow cells from C57BL/6 mice were cultured in 20 ng/mL GM-CSF (BioLegend) with media changes at day 3, 6 and 8. 20 ng/mL IL-4 (BioLegend) was added to the culture at day 6 and day 8. Loosely adherent cells were harvested on day 10 and cultured with 100 ng/mL LPS (Sigma-Aldrich) for 20 hours. Cells were then pulsed with 1 μg/ml of LCMV-GP33-41 (for P14 T cells) or OVA257-264 (SIINFEKL; for OT-I T cells) peptide for 2 hours. Two million peptide pulsed BMDCs were injected per mouse.
EG7 tumor model
1x106 OT-I cells were injected into either naïve or mice that had been persistently infected 21 days prior. One day later, 1x106 EG7 tumor cells (generously provided by Dr. Robert Prins, University of California, Los Angeles) were injected subcutaneously into both groups of mice. In certain experiments OVA257-264 peptide-pulsed DCs were also injected on the same day as the tumor. Mice were sacrificed 12 days post-tumor injection. Tumors were digested using the Gentlemax tumor dissociation kit (Miltenyi). Tumor and single cell suspensions were acquired and analyzed by flow cytometry.
Time-of-Flight mass cytometry (CyTOF)
Up to 4×106 splenocytes were pulsed with 12.5μM Cisplatin (BioVision) in PBS for 1 min prior to quenching with CyTOF staining media (Mg+/Ca+ HBSS containing 2% FBS (Multicell), 10mM HEPES (Cornning), and FBS underlay. Cells were then resuspended in staining media containing metal-tagged surface antibodies (Table S1) and Fc block (CD16/32; in house) for 30 min at 4°C. Cells were fixed, permeablized and stained with metal tagged intracellular antibodies (Table S1) using the eBioscience™ Foxp3 / Transcription Factor Staining Buffer Set according to manufacturer’s instructions. All antibody concentrations were used at saturating concentrations previously determined by titration. Cells were then incubated overnight in PBS (Multicell) containing 0.3% (ws/v) saponin, 1.6% (v/v) paraformaldehyde (diluted from 16%; Polysciences Inc) and 50 nM Iridium (Fluidigm). Cells were analyzed on a Helios or CyTOF-2 mass cytometer (Fluidigm). EQ Four Element Calibration Beads (Fluidigm) were used to normalize signal intensity over time and data analysis was performed. P14 T cells were gated on (DNA/Iridium+, single event length, cisplatin−, B220lo NK1.1lo, TCRβ+, CD8a+ Thy1.1+). t-SNE analyses were performed on the P14 cells (perplexity = 30, theta = 0.5, iterations = 1000, equal sampling).
Flow cytometry and intracellular cytokine stimulation
Single cell suspensions were prepared from organs and were stained ex vivo using antibodies to CD8 (53-6.7), Thy1.1 (H1S51), CD62L (MEL-14), CD25 (PC61), CD127 (A7R34), CD122 (TM-β1), 4-1BB (1AH2), CD27 (LG.3A10), PD1 (29F.1A12), Tim3 (215008), Lag3 (C9B7W), MHC I (AF6-88.5), CD80 (16-10A1), CD86 (GL-1), CD11c (3.9), CD11b (M1/70). All were from Biolegend with the exception of Thy1.1 (eBiosciences), 4-1BB (BD Biosciences), and Tim3 (R&D Systems). Staining for Tbet (4B10), Blimp1 (5E7), Granzyme B (GB11), (Biolegend), TCF1 (S33-966), (BD Biosciences), EOMES (Dan11mag), and Nur77 (12.14) (eBiosciences) was performed as directed using the Foxp3 / Transcription Factor Staining kit (eBiosciences). Samples were run on a FACS Verse (BD Biosciences) and data analyzed using Flow Jo software (Treestar).
For cytokine quantification, splenocytes were restimulated for 5 hours at 37°C with 2 μg/ml of MHC class I-restricted LCMV peptide GP33-40 in the presence of 50 U/ml recombinant murine IL-2 and 1 mg/ml brefeldin A (Sigma). Following the 5 hours in vitro restimulation, cells were stained with a fixable viability stain, zombie aqua (Biolegend), extracellularly stained as above with CD8, Thy1.1, and fixed, permeabilized (Biolegend cytokine staining kit) and stained with IFNγ (XMG1.2), TNFα (MP6-XT22) and IL-2 (JES6-5H4) (Biolegend).
For phospho STAT3 and STAT5 staining, splenocytes were stimulated with 105 Units of IL-2 (Thermoscientific) or 200 ng/mL of IL-21 (R&D Systems) for 15 minutes at 37°C. Cells were then fixed immediately with 1% paraformaldehyde, then permeabilized with ice-cold 100% MeOH and stained with CD8 (53-6.7), Thy1.1 (H1S51) (Biolegend), pSTAT3 (pY705, clone 4/P-STAT3) and pSTAT5 (pY694, clone 47/Stat5) (BD Biosciences).
CTL Trogocytosis Assay
We used the trogocytosis assay to specifically measure the small population of transferred cells among the larger mixed group of endogenous virus-specific cells that would also lyse in a standard CTL assay. The trogocytosis assay was performed as described in (Daubeuf et al., 2006). EG7 target cells were labeled with biotin at 1 mg/mL and then peptide labeled for 1 hour with 1 μM LCMV-specific GP33-41 or ovalbumin-specific OVA257-264 as a non-specific control at 37°C. Target cells were mixed with effector splenoctyes from Cl13 infected mice at a 1:1 ratio and incubated for 4 hours at 37°C. Following the incubation cells were spun down, supernatants removed and resuspended in cold PBS + 2 mM EDTA to dissociate cell conjugates. Cells were stained for CD8, Thy1.1, streptavidin-APC and subsequently analyzed by flow cytometry to quantify the level of trogocytosis by P14 cells from GP33 pulsed or OVA-pulsed EG7 cells.
RNA-seq
P14 T cells were transferred into mice that had been infected 21 days earlier with Cl13 or into naïve mice immediately infected with Cl13. Sixty hours post-infection mice were sacrificed and splenocytes from 4-5 mice were pooled and B cell depleted with anti-CD19 beads (Miltenyi). Single cell suspensions were then stained for virus-specific P14 cells using CD8 and Thy1.1 and FACSorted on a Moflo Astrios (Beckman Coulter) directly into RLT buffer (Qiagen). RNA was isolated using a single cell RNA purification kit (Norgen Biotech Corp.) according to manufacturer’s instructions. SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing (Clontech) was used per manufacturer’s instructions for amplification of RNA and subsequent cDNA synthesis. All samples proceeded through NexteraXT DNA Library Preparation (Illumina) using NexteraXT Index Kit V1 or V2 Set A (Illumina) following manufacturer’s instructions. A portion of this library pool was sent for sequencing on an Illumina NextSeq HighOutput, single read at the Princess Margaret Genomics Core Facility. An average of 400M reads were obtained per pool, with an average of 40M reads/sample across the entire data set.
In vivo antibody treatments, antibody blockade and DT treatment
For the PDL1 blockade experiments and the analysis of CD4 help dependence, CD4 T cells were depleted prior to LCMV infection by administering 125 μg of anti-CD4 antibody (GK1.5) to mice 3 days and then 1 day prior to infection. CD4 depletion was confirmed with flow cytometry using a non-blocking CD4 clone (RM4.4). For in vivo blocking experiments 250μg of anti-PDL1 (10F.9G2) or isotype control (LTF-2) was administered i.p. starting 21 days post-cell transfer and then every 3 days for a total of 5 treatments. For IL-2 and IL-21R blockade experiments at the onset of Cl13 infection mice were treated with 500μg of anti-IL-2 (S4B6-1), 300 μg of anti-IL-21R (mouse IgG1; Amgen) or appropriate RatIgG or Mouse IgG isotype controls one day following P14 injection and 8 hours prior to Cl13 infection, and every 2 days following for another 3 treatments. For IL-2 and IL-21R blockade in the midst of Cl13 infection, mice were treated with the same doses described above 4 hours prior to P14 injection (day 25 post LCMV-infection), then again 1 day later, and every subsequent 2 days until sacrifice 8 days post-P14 transfer. For in vivo agonistic antibody treatments persistently infected mice were treated i.p. with 200mg anti-CD3 (1452C11) and/or 100μg anti-CD28 (PV11) or an appropriate isotype control 4 hrs after P14 transfer. All antibodies were obtained from BioXcell, except anti-IL-21R which was obtained from Amgen. For diphtheria toxin (DT)-mediated depletion of CD11c+ DCs we administered either DT at 4 ng/g or control PBS to CD11c-DTR mice 18 hours prior to late P14 priming (D20 of Cl13 infection) and then again at 30 hrs post-P14 transfer (48 hrs after the initial DT treatment).
QUANTIFICATION AND STATISTICAL ANALYSIS
Mass Cytometry and Flow Cytometry Analysis
Heatmaps were plotted in R using the viridis color package and the gplots package.
All statistical parameters are described in the figure legends. Student’s t tests (two-tailed, unpaired, or where indicated paired) and One-way ANOVA, multiple comparisons or two-way ANOVA, multiple comparisons were performed using GraphPad Prism 6 software (GraphPad Software, Inc.). In all figures, error bars indicate standard deviation.
RNAseq Analysis
Illumina reads were aligned to the Mus Musculus GRCm38 genome build 88 using HISAT2. Alignments were compressed and sorted using SAMtools. The alignments were quantified using HTSeq to obtain gene counts. Differential analysis was conducted using edgeR using modified code from the rnaseq.wiki protocol. Low count genes were excluded from analysis if at least 3 samples did not have at least 1 CPM reads for that gene. Gene counts were normalized using Trimmed Mean of M-values (TMM) normalization. Figures were plotted in R (R Core Team (2017) and the gplots package.
We performed a Gene Set Enrichment Analysis on the Enrichment Map gene set “Mouse_GOBP_AllPathways_no_GO_iea_August_01_2017_symbol.gmt” and ImmuneSigDB using genes pre-ranked by edgeR p value. Immunoglobulin genes were excluded from the GSEA analysis. ImmuneSigDB gene lists were converted from human genes to orthologoues mouse genes using Ensembl BioMart.
DATA AND SOFTWARE AVAILABILITY
The RNA seq data generated in this paper has been deposited in the Gene Expression Omnibus (GEO) under accession number GEO: GSE105044.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-CD45 (30-F11) | Fluidigm | Cat # 3089005B |
| Anti-Ly6c (HK14) | BioLegend | Cat # 128002 |
| Anti-CD44 (IM7) | BioLegend | Cat # 103002 |
| Anti-CXCR5 (145502) | BioLegend | Cat # L138D7 |
| Anti-GzmB (GB11) | Thermofischer | Cat # MA1-80734 |
| Anti-Eomes (Dan11mag) | eBioscience | Cat # 14-4875-80 |
| Anti-Thy1.1 (HIS51) | eBioscience | Cat # 14-0900-85 |
| Anti-CD11b (M1/70) | Fluidigm | Cat # 3148003B |
| Anti-CD69 (H1.273) | BioLegend | Cat # 104502 |
| Anti-Ly6G (1A8) | BioLegend | Cat # 127602 |
| Anti-CD25 (3C7) | Fluidigm | Cat # 3151007B |
| Anti-Bcl6 (K112.91) | BD Bioscence | Cat # 561520 |
| Anti-CD8a (53-6.7) | Fluidigm | Cat # 3153012B |
| Anti-CD103 (M290) | BD Bioscience | Cat # 553699 |
| Anti-PDL1 (M1H5) | eBioscience | Cat # 14-5982-85 |
| Anti-Thy1.2 (53-21) | Thermofischer | Cat # 14-0902-82 |
| Anti-CD39 (24DMS1) | eBioscience | Cat # 14-0391-82 |
| Anti-CD4 (rm4-5) | BioLegend | Cat # 100506 |
| Anti-Tbet (4B10) | Fluidigm | Cat # 3161014B |
| Anti-TCRβ (109202) | BioLegend | Cat # H57-597 |
| Anti-B220 (RA3-6B2) | Thermofischer | Cat # 14-0452-85 |
| Anti-SLAM (TC15-12F12.2) | BioLegend | Cat # 115933 |
| Anti-NK1.1 (PK136) | Fluidigm | Cat # 3170002B |
| Anti-CD95 (15A7) | eBioscience | Cat # 14-0951-85 |
| Anti-CD11c (N418) | BioLegend | Cat # 117302 |
| Anti-PD1 (RMP-30) | BioLegend | Cat # 109104 |
| Anti-MHC-II (M5/114.15.2) | Fluidigm | Cat # 3209006B |
| Anti-CD8a (53-6.7) | BioLegend | Cat # 100714 |
| Anti-Thy1.1 (H1S51) | eBiosciences | Cat # 45-0900-80 |
| Anti-CD62L (MEL-14) | BioLegend | Cat # 104418 |
| Anti-CD25 (PC61) | BioLegend | Cat # 101908 |
| Anti-CD127 (A7R34) | BioLegend | Cat # 135023 |
| Anti-CD122 (TM-β1) | BioLegend | Cat # 105906 |
| Anti-4-1BB (1AH2) | BD Biosciences | Cat # 558975 |
| Anti-CD27 (LG.3A10) | BioLegend | Cat # 124223 |
| Anti-PD1 (29F.1A12) | BioLegend | Cat # 135224 |
| Anti-Tim3 (215008) | R&D Systems | Cat # FAB1529A |
| Anti-Lag3 (C9B7W) | BioLegend | Cat # 125210 |
| Anti-MHC I (AF6-88.5) | BioLegend | Cat # 116506 |
| Anti-CD80 (16-10A1) | BioLegend | Cat # 104706 |
| Anti-CD86 (GL-1) | BioLegend | Cat # 200308 |
| Anti-CD11c (3.9) | BioLegend | Cat # 301628 |
| Anti-CD11b (M1/70) | BioLegend | Cat # 101216 |
| Anti-Tbet (4B10) | BioLegend | Cat # 644814 |
| Anti-Blimp1 (5E7) | BioLegend | Cat # 149902 |
| Anti-Granzyme B (GB11) | BioLegend | Cat # 515408 |
| Anti-TCF1 (S33-966) | BD Biosciences | Cat # 564217 |
| Anti-Eomes (Dan11mag) | eBiosciences | Cat # 12-4875-82 |
| Anti-Nur77 (12.14) | eBiosciences | Cat # 12-5965-82 |
| Anti-IFNγ (XMG1.2) | Biolegend | Cat # 505810 |
| Anti-TNFα (MP6-XT22) | Biolegend | Cat # 506327 |
| Anti-pSTAT3 pY705 (4/P-STAT3) | BD Biosciences | Cat # 562072 |
| Anti-pSTAT5 pY694 (47/Stat5(pY694) | BD Biosciences | Cat # 5601188 |
| Anti-CD4 (GK1.5) | BioXcell | Cat # BE0003-1 |
| Anti-IL-2 (S4B6-1) | BioXcell | Cat # BE0043-1 |
| Anti-IL-21R | Amgen | N/A |
| Anti-CD3 (1452C11) | BioXcell | Cat # BE0001-1 |
| Anti-CD28 (PV11) | BioXcell | Cat # BE0015-5 |
| Anti-PDL1 (10F.9G2) | BioXcell | Cat # BE0101 |
| Anti-Rat IgG2a isotype ctrl (2A3) | BioXcell | Cat # BE0089 |
| Anti-Rat IgG2b isotype ctrl (LTF-2) | BioXcell | Cat # BE0090 |
| Polyclonal Armenian hamster isotype ctrl | BioXcell | Cat # BE0091 |
| Anti-Mouse IgG1 isotype control | Leinco | Cat # I-102 I-102 |
| Bacterial and Virus Strains | ||
| LCMV Clone 13 | Michael Oldstone, Scripps | Grew up in house |
| LCMV Clone 13 variant V35A | Dorian McGavern, NIH | Grew up in house |
| Chemicals, Peptides, and Recombinant Proteins | ||
| LCMV peptide GP33-40 | New England Peptide | Custom order |
| Ovalbumin peptide OVA257-264 | New England Peptide | Custom order |
| Cisplatin | BioVision | Cat # 1550-1000 |
| Iridium | Fluidigm | Cat # 201192b |
| zombie aqua fixable viability stain | Biolegend | Cat # 423102 |
| Murine IL-2 | Thermoscientific | Cat # PMC0025 |
| Recombinant mouse IL-21 | R & D Systems | Cat # 594-ML |
| Murine GM-CSF | Biolegend | Cat # 576306 |
| Murine IL-4 | Biolegend | Cat # 574306 |
| LPS | Sigma | Cat # L4391-1mg |
| Diptheria toxin | Sigma | Cat # D0564-1mg |
| Brefeldin A | Sigma | Cat # B7651-5MG |
| CFSE | Thermoscientific | Cat # C1157 |
| Biotin | Thermoscientific | Cat # 21335 |
| Critical Commercial Assays | ||
| Single cell RNA Purification Kit | Norgen Biotek Corp. | Cat # 51800 |
| SMART-Seq v4 Ultra Low Input RNA Kit | Clontech | Cat # 634894 |
| NexteraXT DNA Library Preparation | Illumina | Cat # FC-131-1096 |
| NexteraXT Index Kit V1 | Illumina | Cat # FC-131-1002 |
| NexteraXT Index Kit V2 Set A | Illumina | Cat # FC-131-2001 |
| Foxp3 / Transcription Factor Staining kit | eBiosciences | Cat # A25866A |
| Tumor dissociation kit | Miltenyi | Cat # 130-096-730 |
| Deposited Data | ||
| RNA-seq data | NCBI - GEO | GSE105044 |
| Experimental Models: Cell Lines | ||
| EG7 | Robert Prins lab, UCLA | N/A |
| Experimental Models: Organisms/Strains | ||
| P14 mice | In house | N/A |
| OT-I mice | Jackson laboratory | Cat # 003831 |
| C57BL/6 mice | Jackson laboratory | Cat # 000664 |
| CD11c-DTR mice | Jackson laboratory | Cat # 004509 |
| B2m KO mice | Jackson laboratory | Cat # 002087 |
| Software and Algorithms | ||
| Prism 6 | Graphpad Software, Inc | https://www.graphpad.com/ |
| Flow Jo Version 9.8.5 | Treestar | https://www.flowjo.com |
| Cytobank | Cytobank, Inc. | https://www.cytobank.org/ |
| HISAT2 | Kim et al., 2015 | https://ccb.jhu.edu/software/hisat2/index.shtml |
| HTSeq | Anders et al., 2015 | https://htseq.readthedocs.io/en/release_0.9.1/ |
| SAMtools | Li et al., 2009 | http://samtools.sourceforge.net |
| edgeR | Robinson et al., 2010 McCarthy et al., 2012 |
http://bioconductor.org/packages/release/bioc/html/edgeR.html |
| GSEA | Subramanian et al., 2005 Mootha et al., 2003 |
http://www.gsea-msigdb.org/gsea/index.jsp |
| ImmuneSigDB | Subramanian et al., 2005 Godec et al., 2016 |
http://software.broadinstitute.org/gsea/msigdb/genesets.jsp?collection=C7 |
| Enrichment Map | Merico et al., 2010 | http://baderlab.org/GeneSets |
| BioMart | Kinsella et al., 2011 | https://www.ensembl.org/biomart/ |
| GRCm38 Genome Build 88 | Yates et al., 2016 | http://www.ensembl.org/Mus_musculus/Info/Annotation |
| R | R Core Team, 2017 | https://www.r-project.org |
| gplots | Warnes et al., 2016 | https://cran.r-project.org/web/packages/gplots/index.html |
| viridis | Garnier, 2017 | https://cran.r-project.org/web/packages/viridis/ |
Highlights.
CD8+ T cell priming in chronic infection (Tlp) programs TCF1+ differentiation
Tlp are less exhausted and have superior responsiveness to PDL1 blockade
Decreased APC-mediated T cell stimulation primes TCF1+ cell differentiation
Chronic infection redirects tumor-specific CD8+ T cell differentiation
ACKNOWLEDGEMENTS
We thank past and present members of the Brooks laboratory for technical help and discussion. This work was supported by the Canadian Institutes of Health Research (CIHR) Foundation Grant FDN148386, the National Institutes of Health (NIH) grant AI085043, the Scotiabank Research Chair to D.G.B, a training grant from the Fonds de la Recherche en Santé du Québec to L.M.S., and the Medicine by Design Award#C1TPA-2016-20 (D.G.B and T.L.M).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Allard EL, Hardy MP, Leignadier J, Marquis M, Rooney J, Lehoux D, and Labrecque N (2007). Overexpression of IL-21 promotes massive CD8+ memory T cell accumulation. Eur J Immunol 37, 3069–3077. [DOI] [PubMed] [Google Scholar]
- Allen TM, Altfeld M, Geer SC, Kalife ET, Moore C, O'Sullivan K M, Desouza I, Feeney ME, Eldridge RL, Maier EL, et al. (2005). Selective escape from CD8+ T-cell responses represents a major driving force of human immunodeficiency virus type 1 (HIV-1) sequence diversity and reveals constraints on HIV-1 evolution. J Virol 79, 13239–13249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelosanto JM, Blackburn SD, Crawford A, and Wherry EJ (2012). Progressive loss of memory T cell potential and commitment to exhaustion during chronic viral infection. J Virol 86, 8161–8170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengsch B, Ohtani T, Khan O, Setty M, Manne S, O'Brien S, Gherardini PF, Herati RS, Huang AC, Chang KM, et al. (2018). Epigenomic-Guided Mass Cytometry Profiling Reveals Disease-Specific Features of Exhausted CD8 T Cells. Immunity 48, 1029–1045 e1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn SD, Shin H, Haining WN, Zou T, Workman CJ, Polley A, Betts MR, Freeman GJ, Vignali DA, and Wherry EJ (2009). Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat Immunol 10, 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks DG, McGavern DB, and Oldstone MB (2006). Reprogramming of antiviral T cells prevents inactivation and restores T cell activity during persistent viral infection. J Clin Invest 116, 1675–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck MD, Sowell RT, Kaech SM, and Pearce EL (2017). Metabolic Instruction of Immunity. Cell 169, 570–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham CR, Champhekar A, Tullius MV, Dillon BJ, Zhen A, de la Fuente JR, Herskovitz J, Elsaesser H, Snell LM, Wilson EB, et al. (2016). Type I and Type II Interferon Coordinately Regulate Suppressive Dendritic Cell Fate and Function during Viral Persistence. PLoS Pathog 12, e1005356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubeuf S, Puaux AL, Joly E, and Hudrisier D (2006). A simple trogocytosis-based method to detect, quantify, characterize and purify antigen-specific live lymphocytes by flow cytometry, via their capture of membrane fragments from antigen-presenting cells. Nat Protoc 1, 2536–2542. [DOI] [PubMed] [Google Scholar]
- Elsaesser H, Sauer K, and Brooks DG (2009). IL-21 is required to control chronic viral infection. Science 324, 1569–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohlich A, Kisielow J, Schmitz I, Freigang S, Shamshiev AT, Weber J, Marsland BJ, Oxenius A, and Kopf M (2009). IL-21R on T cells is critical for sustained functionality and control of chronic viral infection. Science 324, 1576–1580. [DOI] [PubMed] [Google Scholar]
- Giordano M, Henin C, Maurizio J, Imbratta C, Bourdely P, Buferne M, Baitsch L, Vanhille L, Sieweke MH, Speiser DE, et al. (2015). Molecular profiling of CD8 T cells in autochthonous melanoma identifies Maf as driver of exhaustion. Embo J 34, 2042–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R, Hou S, Liu C, Zhang A, Bai Q, Han M, Yang Y, Wei G, Shen T, Yang X, et al. (2016). Follicular CXCR5-expressing CD8(+) T cells curtail chronic viral infection. Nature 537, 412–428. [DOI] [PubMed] [Google Scholar]
- Henn MR, Boutwell CL, Charlebois P, Lennon NJ, Power KA, Macalalad AR, Berlin AM, Malboeuf CM, Ryan EM, Gnerre S, et al. (2012). Whole genome deep sequencing of HIV-1 reveals the impact of early minor variants upon immune recognition during acute infection. PLoS Pathog 8, e1002529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im SJ, Hashimoto M, Gerner MY, Lee J, Kissick HT, Burger MC, Shan Q, Hale JS, Lee J, Nasti TH, et al. (2016). Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537, 417–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi NS, Cui W, Chandele A, Lee HK, Urso DR, Hagman J, Gapin L, and Kaech SM (2007). Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity 27, 281–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitchen SG, Levin BR, Bristol G, Rezek V, Kim S, Aguilera-Sandoval C, Balamurugan A, Yang OO, and Zack JA (2012). In vivo suppression of HIV by antigen specific T cells derived from engineered hematopoietic stem cells. PLoS Pathog 8, e1002649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda S, Yamazaki M, Abe M, Sakimura K, Takayanagi H, and Iwai Y (2011). Basic leucine zipper transcription factor, ATF-like (BATF) regulates epigenetically and energetically effector CD8 T-cell differentiation via Sirt1 expression. Proc Natl Acad Sci U S A 108, 14885–14889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, Lu S, Kemberling H, Wilt C, Luber BS, et al. (2017). Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357, 409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong YA, Chen Y, Ong HS, Wu D, Man K, Deleage C, Minnich M, Meckiff BJ, Wei Y, Hou Z, et al. (2016). CXCR5(+) follicular cytotoxic T cells control viral infection in B cell follicles. Nat Immunol 17, 1187–1196. [DOI] [PubMed] [Google Scholar]
- McGranahan N, Furness AJ, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, Jamal-Hanjani M, Wilson GA, Birkbak NJ, Hiley CT, et al. (2016). Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 351, 1463–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebot-Bral L, Brandao D, Verlingue L, Rouleau E, Caron O, Despras E, El-Dakdouki Y, Champiat S, Aoufouchi S, Leary A, et al. (2017). Hypermutated tumours in the era of immunotherapy: The paradigm of personalised medicine. Eur J Cancer 84, 290–303. [DOI] [PubMed] [Google Scholar]
- Nishimura H, Yajima T, Muta H, Podack ER, Tani K, and Yoshikai Y (2005). A novel role of CD30/CD30 ligand signaling in the generation of long-lived memory CD8+ T cells. J Immunol 175, 4627–4634. [DOI] [PubMed] [Google Scholar]
- Pollizzi KN, Patel CH, Sun IH, Oh MH, Waickman AT, Wen J, Delgoffe GM, and Powell JD (2015). mTORC1 and mTORC2 selectively regulate CD8(+) T cell differentiation. J Clin Invest 125, 2090–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst HC, and van den Broek M (2005). Priming of CTLs by lymphocytic choriomeningitis virus depends on dendritic cells. J Immunol 174, 3920–3924. [DOI] [PubMed] [Google Scholar]
- Puglielli MT, Zajac AJ, van der Most RG, Dzuris JL, Sette A, Altman JD, and Ahmed R (2001). In vivo selection of a lymphocytic choriomeningitis virus variant that affects recognition of the GP33-43 epitope by H-2Db but not H-2Kb. J Virol 75, 5099–5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roney K (2013). Bone marrow-derived dendritic cells. Methods Mol Biol 1031, 71–76. [DOI] [PubMed] [Google Scholar]
- Sevilla N, McGavern DB, Teng C, Kunz S, and Oldstone MB (2004). Viral targeting of hematopoietic progenitors and inhibition of DC maturation as a dual strategy for immune subversion. J Clin Invest 113, 737–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin HM, Kapoor VN, Kim G, Li P, Kim HR, Suresh M, Kaech SM, Wherry EJ, Selin LK, Leonard WJ, et al. (2017). Transient expression of ZBTB32 in anti-viral CD8+ T cells limits the magnitude of the effector response and the generation of memory. PLoS Pathog 13, e1006544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M, Wang C, Cong L, Marjanovic ND, Kowalczyk MS, Zhang H, Nyman J, Sakuishi K, Kurtulus S, Gennert D, et al. (2016). A Distinct Gene Module for Dysfunction Uncoupled from Activation in Tumor-Infiltrating T Cells. Cell 166, 1500–1511 e1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell LM, McGaha TL, and Brooks DG (2017). Type I Interferon in Chronic Virus Infection and Cancer. Trends Immunol 38, 542–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utzschneider DT, Charmoy M, Chennupati V, Pousse L, Ferreira DP, Calderon-Copete S, Danilo M, Alfei F, Hofmann M, Wieland D, et al. (2016). T Cell Factor 1-Expressing Memory-like CD8(+) T Cells Sustain the Immune Response to Chronic Viral Infections. Immunity 45, 415–427. [DOI] [PubMed] [Google Scholar]
- Verbist KC, Guy CS, Milasta S, Liedmann S, Kaminski MM, Wang R, and Green DR (2016). Metabolic maintenance of cell asymmetry following division in activated T lymphocytes. Nature 532, 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezys V, Masopust D, Kemball CC, Barber DL, O'Mara LA, Larsen CP, Pearson TC, Ahmed R, and Lukacher AE (2006). Continuous recruitment of naive T cells contributes to heterogeneity of antiviral CD8 T cells during persistent infection. J Exp Med 203, 2263–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry EJ, Blattman JN, Murali-Krishna K, van der Most R, and Ahmed R (2003a). Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J Virol 77, 4911–4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry EJ, and Kurachi M (2015). Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol 15, 486–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry EJ, Teichgraber V, Becker TC, Masopust D, Kaech SM, Antia R, von Andrian UH, and Ahmed R (2003b). Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat Immunol 4, 225–234. [DOI] [PubMed] [Google Scholar]
- Wu T, Ji Y, Moseman EA, Xu HC, Manglani M, Kirby M, Anderson SM, Handon R, Kenyon E, Elkahloun A, et al. (2016). The TCF1-Bcl6 axis counteracts type I interferon to repress exhaustion and maintain T cell stemness. Sci Immunol 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin A, Masson F, Liao Y, Preston S, Guan T, Gloury R, Olshansky M, Lin JX, Li P, Speed TP, et al. (2016). A molecular threshold for effector CD8(+) T cell differentiation controlled by transcription factors Blimp-1 and T-bet. Nat Immunol 17, 422–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi JS, Du M, and Zajac AJ (2009). A vital role for interleukin-21 in the control of a chronic viral infection. Science 324, 1572–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Yu G, Chan B, Pearson JT, Rathanaswami P, Delaney J, Ching Lim A, Babcook J, Hsu H, and Gavin MA (2015). Interleukin-21 receptor blockade inhibits secondary humoral responses and halts the progression of preestablished disease in the (NZB x NZW)F1 systemic lupus erythematosus model. Arthritis Rheumatol 67, 2723–2731. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA seq data generated in this paper has been deposited in the Gene Expression Omnibus (GEO) under accession number GEO: GSE105044.