Abstract
Diabetic kidney disease (DKD) is one of the most common microvascular complication of both type 1 (T1DM) and type 2 diabetes mellitus (T2DM), and the leading cause of end-stage renal disease (ESRD) worldwide. Persistent inflammation and subsequent chronic fibrosis are major causes of loss of renal function, which is associated with the progression of DKD to ESRD. In fact, DKD progression is affected by a combination of genetic and environmental factors. Approximately, one-third of diabetic patients progress to develop DKD despite intensive glycemic control, which propose an essential concept “metabolic memory.” Epigenetic modifications, an extensively studied mechanism of metabolic memory, have been shown to contribute to the susceptibility to develop DKD. Epigenetic modifications also play a regulatory role in the interactions between the genes and the environmental factors. The epigenetic contributions to the processes of inflammation and fibrogenesis involved in DKD occur at different regulatory levels, including DNA methylation, histone modification and non-coding RNA modulation. Compared with genetic factors, epigenetics represents a new therapeutic frontier in understanding the development DKD and may lead to therapeutic breakthroughs due to the possibility to reverse these modifications therapeutically. Early recognition of epigenetic events and biomarkers is crucial for timely diagnosis and intervention of DKD, and for the prevention of the progression of DKD to ESRD. Herein, we will review the latest epigenetic mechanisms involved in the renal pathology of both type 1 (T1DN) and type 2 diabetic nephropathy (T2DN) and highlight the emerging role and possible therapeutic strategies based on the understanding of the role of epigenetics in DKD-associated inflammation and fibrogenesis.
Keywords: Epigenetics, Metabolic memory, Diabetic kidney disease, Histone modification, Noncoding RNA
Introduction
Diabetic kidney disease (DKD) is among the most common and severe complications of diabetes mellitus (DM). Type 1 diabetes (T1DM) is mainly caused by chronic autoimmune injury to pancreatic β cells, leading to an absolute deficit of insulin [1]. In contrast, type 2 diabetes (T2DM) is a metabolic disorder characterized by chronic hyperglycemia secondary to insulin resistance. T2DM and its complications are classical diseases as a result of interactions between multiple genetic and environmental factors [2]. According to previous studies, about 30% of patients with T1DM and 20% of patients with T2DM would ultimately be followed up with diabetic nephropathy (DN). Recently, due to the increased prevalence of DKD, this complication has emerged as a primary cause of end-stage renal disease (ESRD) [3]. The pathologic features of DKD are intricate and include thickened glomerular basement membrane, increased mesangial matrix, tubular interstitial fibrosis, and loss of podocytes [4]. Intuitively, inflammation and the resulting fibrosis are two major features of DKD. Renal fibrosis is a common complication of DKD and the leading cause of ESRD.
Hyperglycemia is a major risk factor for DKD. Many clinical studies have shown that intensive glycemic control in diabetic patients can reduce the incidence and progression of DKD [5, 6]. However, many diabetic patients still develop DKD despite receiving intensified glycemic control. The phenomenon of hyperglycemic "metabolic memory" of DKD refers to the observation that diabetic patients are prone to develop diabetes-related complications due to early hyperglycemia, even if effective measures were taken to control blood glucose and maintain it within normal levels in the later stage of the disease. The underlying mechanisms of "metabolic memory" include a range of pathogenic factors associated with the diabetic complications, such as the advanced glycation end products (AGEs), oxidative stress, inflammation and epigenetic modifications [7, 8]. Among these pathogenic factors, epigenetics has emerged as "link" between metabolic memory and DKD development and progression, which has attracted the attention of the medical community.
Epigenetics refers to the changes of gene expression and function and the generation of heritable phenotype without alteration of DNA sequences [9]. Actually, the term “epigenetics” was first coined by Conrad Hal Waddington in 1942. It is believed that genotypes could determine different phenotypes through some accidental or unknown mechanisms. Based on our understanding of these relations, epigenetics and heredity can be considered close relatives [10]. Epigenetic changes primarily include DNA methylation, histone modifications, and non-coding RNA [11]. Emerging studies suggest a key role of epigenetics in the regulation of physiological and pathological processes associated with DKD development and progression including renal fibrosis, chronic inflammation, and other pathologic features [12, 13]. The present review will focus on discussing the mechanisms of metabolic memory, and its relationship with renal inflammation and fibrosis associated with DKD based on recent epigenetic perspectives, and thus highlight some possible epigenetic therapeutic strategies for DKD treatment.
Pathogenic mechanisms accounting for DKD
When progressed to DN, whether T1DM or T2DM, the histologic changes of the kidneys are similar. Multiple mechanisms are involved in the development of both T1DN and T2DN, which include metabolic disorders, such as hemodynamic changes induced by hyperglycemia, podocyte injury caused by oxidative stress, autophagy and apoptosis, renin-angiotensin system activation, infiltration of inflammatory cells and mediators, and accumulation of AGEs [14–17]. Chronic inflammation has been recognized as a promoting factor for the development of DKD to ESRD [18]. Additionally, chronic and relentless fibrosis in both glomerular and tubulointerstitial compartments is another notable and reliable indicator of the progression of DKD [19].
Role of renal inflammation in DKD
The inflammatory response can be driven by a variety of mechanisms, including activation of macrophages and accumulation of immune cells in the kidneys. Through the leukocyte adhesion molecule, including vascular cell adhesion molecule 1 (VCAM1) and intercellular adhesion molecule 1 (ICAM1), monocytes can adhere to endothelial cells and migrate through endothelial cells by secreting chemokines [20]. The chemokine (C–C motif) ligand 2 (CCL2), which represents the initial stage of glomerular and tubular inflammation, can induce the recruitment, migration, and adhesion of inflammatory cells following DKD-associated tissue injury [18]. In the kidneys of diabetic animal models and DKD patients, macrophages tend to infiltrate the kidney tissue upon the upregulation of chemokines. The accumulation of macrophages in the kidneys is a prominent feature of the progression of chronic kidney diseases [21] and is closely associated with the decline in glomerular filtration rate and poor prognosis [22]. The deletion of macrophage scavenger receptor protects DM mice from proteinuria, glomerular hypertrophy, and transforming growth factor-β (TGF-β) overproduction [23]. In turn, tumor necrosis factor-α (TNF-α) secreted by activated macrophages can stimulate the production of CCL2 in kidney cells. A recent cohort study [24] also found that multiple inflammatory proteins are involved in the terminal development of DKD and can improve the risk assessment of DKD as novel biomarkers.
The high glucose (HG) environment in the tissues of DM patients can also activate multiple cascades signaling pathways to coordinate cellular transcription, thereby induce monocyte infiltration, chemokines production and secretion, which drive inflammation, and macrophage infiltration leading to tissue damage. The Janus kinase/signal transducers and activators of transcription (JAK-STAT) signaling pathway play an important role in renal cells, including mesangial cells (MCs), podocytes, and tubular epithelial cells [25]. Zhang et al. [26] showed that the overexpression of JAK2 in podocytes can lead to increased kidney damage in T1DM mice. Additionally, the NF-E2-related factor 2/Kelch-like ECH associated protein 1 (Nrf2-keap1) pathway activation improves pathological changes in glomeruli of diabetic mice injected with streptozocin(STZ)[27]. Nrf2 inhibits the inflammatory response by directly regulating the transcription of pro-inflammatory factors such as interleukin (IL) -1 and IL-6. Moreover, downstream targets of NF-κB include adhesion molecules and pro-inflammatory cytokines (e.g., IL-6, TNF-α, CCL2), which all contribute to the development of DKD [28].
Role of renal fibrosis in DKD
The accumulation of extracellular matrix (ECM) proteins, such as type I and type III collagen, fibronectin, and laminin in the tubulointerstitial and mesangial areas is another key feature of DKD [29]. During the early stages of DKD, renal biopsy shows the accumulation of ECM proteins in the mesangial space, which gradually leads to the appearance of glomerular sclerosis [30]. The continuous protein synthesis increased deposition and decreased degradation of these ECM proteins, and their post-translationally modified isoforms contribute to the progression of fibrosis [31]. In the late stages of diabetic glomerular sclerosis, the deposition of type I and type III collagen increases dramatically [32]. Thus, the accumulation of ECM proteins plays a vital role in throughout all stages of DKD renal fibrosis. Moreover, TGF-β signaling pathway acts as a profibrotic factor, promoting renal fibrosis. TGF-β promotes the expression of downstream ECM proteins and their regulatory factor plasminogen activator inhibitor-1 (PAI-1), which is the main substance that inhibits cytoplasmic degradation, mainly by activating the transcription factors Smad2-4 [33, 34]. Connective tissue growth factor (CTGF) activation acts as a downstream mediator of fibrotic activity of TGF-β1, which increases expression of fibronectin and type IV, III and type I collagen, and promotes the deposition and assembly of ECM proteins [35, 36]. Therefore, reversing the expression of these fibrosis regulatory genes or ECM proteins may have potential renal protective effects against DKD.
Metabolic memory and epigenetic regulation in DKD
Nowadays, during the early stage of T1DM or T2DM, long-term intensive glycemic control is recognized as an effective preventive measure against diabetic complications, especially DKD development [37, 38]. The evidence was undoubtedly derived from a series of clinical trials, such as Diabetes Control and Complications Trial (DCCT) and epidemiology of diabetes interventions and complications (EDIC) where the term “metabolic memory” was first introduced. Between 1983 and 1989, 1441 patients with T1DM were recruited and randomized to receive intensive or conventional treatment. The difference of average glycated hemoglobin (HbA1c) between DCCT intensive treatment group and conventional treatment group was about 2% (7% vs. 9%). Based on a period of 20 years’ follow-up, the results of EDIC study demonstrated that despite the HbA1c of two groups were maintained within similar levels, the overall mortality rate of patients in the routine treatment group was higher than that of the general population [5]. Additionally, 3867 newly diagnosed type 2 diabetic patients were enrolled by the UK Prospective Diabetes Study (UKPDS) [6]. In this 15-year prospective study, the results also demonstrated that early intensive glycemic compliance can lead to a long-term kidney benefit. The phenomenon of metabolic memory has been termed “legacy effect” by the UKPDS investigators.
Metabolic memory, or legacy effect, refers to the "memory" effect of the body on early stage hyperglycemia, which plays an important role in the development of DKD [39]. The phenomenon of "metabolic memory" has also been confirmed in various experimental models. E1-Osta et al. [40] induced cultured aortic endothelial cells in vitro under transient hyperglycemic conditions and found that oxidative stress remained activate, and the high expression of inflammatory factors persisted even with the switch of the culture medium to normal glycemic levels. Additionally, a metabolic memory model of diabetes was successfully developed in zebrafish with 0.3% STZ solution [41]. The "metabolic memory" phenomenon has advanced our understanding of the mechanisms of diabetes complications including DKD. It has aroused people's interest in determining the potential molecular mechanisms of the metabolic memory of diabetes.
In recent years, modulation of epigenetic mechanisms has been linked to reduction of hyperglycemia and reversal of metabolic memory. Miao et al. [42] elucidated the histone modifications of peripheral blood lymphocytes and monocytes in patients with T1DM. They compared H3K9ac, H3K4me3, and H3K9me1 histone modifications in the DCCT conventional treatment group with the intensive treatment group and found a correlation between HbA1c and H3K9ac. Moreover, many genes with promoter regions recruited with high levels of H3K9ac histone modification are involved in the NF-κB pathway, which in part explains metabolic memory phenomena. Similarly, a study investigated epigenetic DNA methylation levels at specific genetic loci in patients with T1DM in the DCCT conventional and intensive treatment groups during the EDIC follow-up stage. A total of twelve annotated differentially methylated loci were discovered, including thioredoxin interacting protein (TXNIP). TXNIP has been shown to be associated with hyperglycemia and other related complications. Additionally, TXNIP-sustained hypomethylation can be induced by high glucose in cultured THP1 Monos in vitro [43]. The discovery of persistent memory embedded in these epigenetic features and the strong association with previous exposure to high glycemic conditions supports the potential role of epigenetics in metabolic memory of DKD.
Epigenetic mechanisms involved in renal inflammation and fibrosis of DKD
Recently, the role of epigenetics in the pathology of diabetic kidney disease has been established. Covalent modifications of DNA and histones, such as histone acetylation, and non-coding RNAs could interfere with gene expression, through the promoter or enhancer regions, without altering the genetic sequence of the relevant gene [44]. Next, we will review how epigenetic changes occur, accumulate, and contribute to the development and progression of inflammation and fibrosis in DKD.
DNA methylation
DNA methylation, which is considered a major transcriptional regulator, is the most widely studied epigenetic mechanism. DNA methylation is a process that occurs under normal physiological conditions, mediated by a series of DNA methyltransferases (DNMTs) [45]. This family of enzymes include DNMT1, DNMT3a, and DNMT 3b [46]. Conversely, DNA methylation can be reversed by a ten-eleven translocation (TET) protein that converts 5-methylcytosine to 5-hydroxymethylcytosine [47]. Several studies have linked high blood sugar and diabetic nephropathy to epigenetics. Although the genome-wide DNA methylation analysis surged in both T1DN and T2DN, the further studies of DNA methylation on renal inflammatory and fibrogenesis processes concentrate upon T2DN.
Genome-wide DNA methylation analysis
Recent studies attempted to identify methylation changes associated with DKD and the role of site-specific methylation editing in DKD development. Most of these studies were performed in mice or alternative types of cells, such as cells isolated from blood or saliva samples [48, 49]. A study sequenced the methylation group map of human T1DN tubule samples found that a decrease in the level of cytosine methylation in TNF-α differentially methylated region (DMR) plays an important role in controlling TNF-α transcription levels [50]. In 181 Pima Indian type 2 diabetic patients, an epigenome-wide association study examined the association of cytosine methylation levels in 397,063 genomic CpG sites with estimated glomerular filtration rate (eGFR). The result showed that methylation levels at 77 sites were significantly associated with decreased eGFR, and 3 of these sites were significantly associated with human renal tissue fibrosis [51]. To determine whether changes observed in blood samples are also related to target tissue or cell type, a recent cross-sectional study of cytosine methylation in renal tubules samples from 91 type 2 diabetic and non-diabetic patients, as well as patients with varying degrees of kidney disease, confirmed that the methylation levels of 65 probes correlated with the degree of renal fibrosis [52]. Moreover, after treatment of MCs with TGF-β1, 5140 genes with significant differences were identified by Next-Generation Sequencing (NGS) identification. Among these genes, Tnfrsf11b, Rxfp3, MMP9, Syn1, IL6, and Megf6 are known to be involved in the regulation of fibrosis and inflammatory pathway [53].
DNA methylation involved in renal inflammatory processes
DNA methylation can modulate the inflammatory processes and the expression of inflammatory-related protein in DKD. However, the current studies mainly focus on T2DN. The level of DNA methylation key enzyme DNMT1 in T2DN patients increases with the increase of inflammatory activity in peripheral blood monocytes. Inhibition of DNMT1 by 5-Aza-2′-deoxycytidine significantly increased the proportion of CD4+, CD25+ regulatory T cells in peripheral blood mononuclear cells of early-stage T2DN animals. Additionally, aberrant cytosine methylation of mTOR upstream regulators, which is induced by upregulation of DNMT1 in diabetic immune cells, induces inflammation in diabetic kidneys [54]. The decrease in the methylation level of the promoter region of the angiogenin-like protein 2 (ANGPTL2), a pro-inflammatory circulating protein, also contributes to the development and progression of albuminuria in type 2 diabetic patients [55]. These studies suggest that high glucose-induced genomic DNA methylation may be a potential cause of DKD.
DNA methylation involved in renal fibrotic processes
DNA methylation is also involved in the regulation of the fibrotic processes, which primarily revolves around T2DN as well. In db/db mice, the nuclear receptor pregnane X receptor (PXR) can play a role in fibrosis in the injured kidney by activating the response gene to complement 32 (Rgc32) [56]. Matrix metalloproteinases (MMP) are a group of peptidases involved in the degradation of ECM. A study of detecting the methylation level of the gene promoter region in peripheral blood of patients showed that the level of MMP-9 was lower in the DKD group than in the normal control group, and the simple diabetes group [57]. Moreover, the hypomethylation of TIMP-2 and AKR1B1 genes is associated with proteinuria in patients with early DKD, and TIMP-2 methylation levels are associated with increased fibronectin expression and ECM accumulation [58]. TGF-β1 stimulation also plays a regulatory role in renal fibrosis by regulating DNA methylation of various genes [53]. When TGF-β1 is stimulated, DNMT1-mediated hypermethylation of RASAL1 can lead to increased fibroblast activation and fibrosis [59]. However, in STZ-induced DKD model, TET3-mediated methylolation reverses the methylation of the RASAL1 promoter, leading to reversal of renal fibrosis [60]. Clinical studies have shown that there is a gradual decrease in DNA methylation in the TGF-β regulatory region in patients with DM and DKD. A recent study [61] also confirmed that the use of short hairpin RNA or oral inhibitors to inhibit TET2 expression can increase the methylation of CpG islands in the TGF-β regulatory region, meanwhile, significantly decrease ECM protein expression levels and mesangial cell proliferation.
Histone modification
Histone post-translational modifications (PTMs) control chromatin relaxation and gene transcription. Studies have shown that persistent changes in histone modifications caused by hyperglycemia can affect the pathways involved in DKD. The covalent PTMs of nucleosome histones occurs mainly at the tail of amino acids, including lysine acetylation (Kac), methylation (Kme), and ubiquitination, arginine methylation and serine/threonine phosphorylation [62]. Among these different types of PTMs, histone Kac and Kme role in DKD has been well studied and will be discussed here.
Common histone PTMs in DKD
Histone acetylation is primarily associated with gene activity and can promote gene transcription by neutralizing the positive charge of histone amino acid residues and attenuating the binding of histones to the negatively charged DNA [63]. In metabolic memory studies in T1DM patients, the level of histone acetylation was found to be associated with the past levels of HbA1c at earlier disease stages, which has a cumulative effect on H3K9 hyperacetylation at key genomic regions [42]. Histone acetylation plays a dynamic regulation of mutual antagonism between histone acetyltransferase (HATs) and histone deacetylase (HDACs). HKac is mediated by HATs, including P300, CBP, PCAF, and TIP60, which activate gene transcription by adding acetyl groups to conserved lysine. A study has shown that p300 is involved in upstream epigenetic mechanism of HG-induced endothelial oxidative stress, ECM protein gene expression and NF-kB signaling [64]. However, the process of deacetylation is mediated by HDAC 1–11 and Sirtuins (Sirt)1–7 [65]. Different classes of HDACs have also been shown to act as regulators in DKD pathophysiological processes such as inflammation and fibrosis.
Histone methylation, in contrast, participate in gene activation or inhibition depending on the extent of methylation and the amino acid residue modified. In general, methylation markers associated with gene transcriptional activation include K3K4me1/2/3, H3K36me2/3, and H3K79me2. Conversely, H3K9me2/3, H3K27me3, and H4K20me3 are usually associated with gene silencing or inhibition [63]. Like acetylation, the process of histone methylation is also catalyzed by enzymes. Histone methylation is mediated by histone lysine methyltransferase (HMTs) and cleared by histone lysine demethylase (HDMs). The FinnDiane Study Group has shown that the genes encoding methyltransferases SETD7, SUV39H1, and SUV39H2 are polymorphic, and the genetic variation of these genes has protective effects against diabetic microvascular complications [66].
Histone modifications involved in renal inflammation
There is accumulating evidence for mechanisms by which histone modifications control DKD inflammation (Fig. 1). In blood mononuclear cells of both T1DM and T2DM patients, HG treatment can increase the recruitment of p300/CBP and PCAF at the promoters of inflammatory genes such as TNF-α and COX-2, with increased acetylation level of histone H3K9/14 and H4K5/8/12 [67]. In STZ-induced diabetic rat model, NF-κB also can recruit p300 to the promoter region of iNOS and induce H3K9ac [68]. However, in STZ-induced diabetic mouse model, the histone deacetylase Sirt6 inhibits Notch1 and Notch4 transcription by deacetylating histone H3K9 and exerts podocyte protection through anti-inflammatory and anti-apoptotic effects [69].
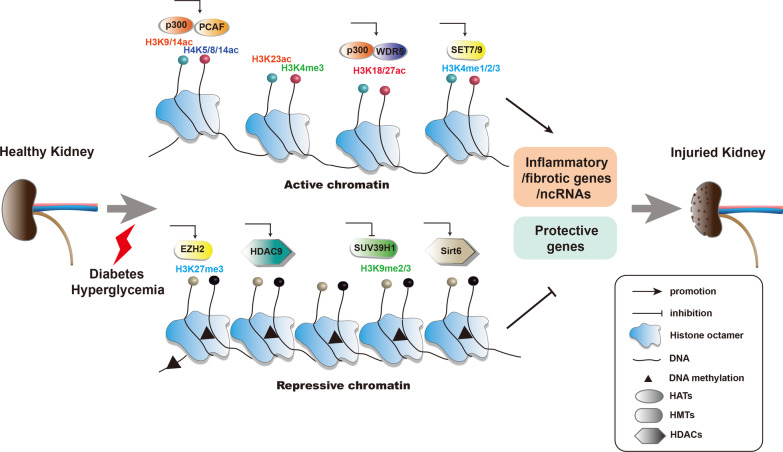
Fig. 1.
Epigenetic modifications of pathologic genes associated with DKD. In diabetic conditions, a series of epigenetic modifications occurred in protective genes, inflammatory and fibrotic genes, or even ncRNAs contribute to renal damage. In the formation of active chromatin, H3K4me1/2/3 mediated by histone methyltransferases such as SET7/9 and H3K9/14ac, H3K18/23/27ac, and H4K5/8/14ac mediated by histone acetyl transferases such as p300, WDR5, and PCAF are involved. In the contrary, H3K9me2/3 and H3K27me3 mediated by HMTs EZH2 and SUV39H1, respectively, and histone PTMs mediated by HDACs Sirt6 and HDAC9 play repressive role on the transcription of protective genes. DNA methylation mediated by DNA methyltransferases is also associated with transcriptional repression. Under disease state such as diabetes and renal injury, epigenetic alterations can also lead to the dysregulation of ncRNAs, which take part in persistent epigenetic alterations. Epigenetic modifications of pathologic genes associated with DKD play key roles in metabolic memory. HATs: histone acetyl transferases; HMTs, histone lysine methyltransferase; HDACs: histone deacetylase; PTMs, post-translational modifications
Studies of T2DN experimental models also support a role for histone modifications in renal inflammation. In diabetic db/db mice, HDAC9 silencing also reduces the release of inflammatory cytokine, podocyte apoptosis, and kidney damage [70]. Several studies have also revealed a link between chromatin histone lysine methylation and the expression of inflammation-related genes. Studies by Rama Natarajan et al. [71] and Wang et al. [72] have shown that both cultured cells and renal tissues of diabetic patients have been found to have decreased H3K9me3 concomitant with the silencing of HMT SUV39H1 at the promoters of the inflammatory genes IL-6 and MCP-1, which resulted in the upregulation of the expression of these inflammatory genes. Endoplasmic reticulum (ER) stress can trigger the expression of MCP-1 in the kidney of db/db mice induced by H3K4 methyltransferase SET7/9, which is associated with an increase in the active gene related chromatin marker, H3K4me1 [73]. The effect is also transiently present in high glucose-stimulated aortic endothelial cells. Transient or previous hyperglycemia leads to various methylation and demethylation events that allow the pro-inflammatory pathways to be continuously activated, which further confirms the role of “metabolic memory” in DKD [40]. Moreover, the administration of the H3K27 demethylase inhibitor GSK-J4 improves renal dysfunction and reduces inflammation, apoptosis, and DNA damage in diabetic db/db mice [74].
Histone modification involvement in renal fibrotic processes
Glomerulosclerosis and tubulointerstitial fibrosis are key events in the pathogenesis of DKD. Many studies have found that histone modification participates in regulating the expression of ECM-related genes in T1DN and T2DN (Fig. 1). In kidneys of STZ-induced diabetic mice, MRTF-A can recruit p300 and WDR5 to bind to the collagen I promoter, and activate the transcription of the gene by increasing the transcription of H3K18/27ac and H3K4me3 to promote renal fibrosis [75]. Furthermore, several studies have shown that the expression of profibrotic genes such as PAI-1, CTGF, and p21 can be regulated by PTMs in DKD [76]. TGF-β increased the recruitment of SET7/9 on the promoter regions of these genes, accompanied by increased level of chromatin markers associated with the active genes (H3K4me1, H3K4me2, and H3K4me3), and decline of repressed marker levels (H3K9me2 and H3K9me3) [13]. Similarly, the decrease of H3K9me2 is accompanied with the increased occupancy of H3K4me1/3 and SET7/9 on the p21 promoter, which promoted the expression of p21 gene in HG-treated rat MCs. The effect can be reversed by TGF-β1 antibody [77]. In addition, TGF-β1 can recruit HATs CBP/p300 and increase the enrichment of H3K9/14ac in the promoter region to promote the genes expression of PAI-1 and P21 genes [78]. And the increased levels of H3K9/14ac and H3K18ac in HG-induced MCs augmented the expression of profibrotic factors CTGF and TGF-β1 [79]. Moreover, emerging evidences have demonstrated that PTMs also participate in renal fibrotic processes of T2DN models. Sayyed et al. [80] have demonstrated that glomerular sclerosis is associated with increased renal H3K9ac, H3K23ac, and H3K4me2 in advanced T2DN. Under hyperglycemia/hyperinsulinemia conditions, mRNA expression of fibrin 1 is elevated in the kidneys of diabetic db/db rats due to elevated levels of histone H3 acetylation [81].
Histone PTMs-oriented therapies against DKD
Unlike methylation which is persistent and long-standing, histone acetylation is a reversible dynamic process [82]. This makes it possible to alleviate the progression of DKD by inhibiting HATs or HDACs. A series of small molecule inhibitors or drugs have been discovered in recent years in response to various mechanisms of inflammation and fibrosis regulated by histone PTMs in DKD (Table 1). However, the studies suggested that administration of histone PTMs inhibitors or drugs mainly work on T1DN. Curcumin can alleviate the increase expression of diabetic-induced ECM proteins in the kidney by inhibiting HAT p300 and its binding factor NF-κB [83]. C66, a curcumin analog, can inhibit the increase in H3K9/14ac levels and p300/CBP occupancy on gene promoters of TCGF, PAI-1, and FN-1, thus prevent diabetes-induced renal dysfunction [84]. In STZ-induced diabetic kidneys and cultured human proximal renal tubular epithelial cells, the HDACs inhibitor trichostatin A (TSA) has also been shown to block TGF-β1-induced EMT [85, 86]. Through HDAC inhibition, valproic acid cannot only significantly exert anti-inflammatory activity via reducing NF-κB [68], but it also improves kidney function by reducing damage and fibrosis [87]. In addition, some Chinese herbal active ingredient extracts, such as 2,3,5,4′-tetrahydroxysti-2-O-β-D-glucoside (TSG) and esculetin, have been found to attenuates alteration in CTGF and MMP-13 gene expression by reducing changes in the acetylation and methylation of histone 3, thus ameliorate renal fibrosis in DKD [88, 89]. In summary, these aforementioned reports provide potential therapeutic possibilities for DKD.
Table 1.
The administration and effect of histone PTMs regulators on DKD
| Inhibitors category | Experimental models | Administration stage | Duration of treatment | Mechanisms | Ref |
|---|---|---|---|---|---|
| HAT inhibitor | |||||
| Curcumin | STZ-induced SD rats | DM | 4 weeks | alleviate the increase expression of ECM proteins by inhibiting HAT p300 and its binding factor NF-κB | [83] |
| C66 | STZ-induced mice | DM | 12 weeks | inhibit the increase in H3K9/14ac levels and p300/CBP occupancy on gene promoters of TCGF, PAI-1, and FN-1 | [84] |
| HDAC inhibitor | |||||
| TSA | STZ-induced SD rats | DM | 4 weeks | Suppresses TGF-β1-induced epithelial-to-mesenchymal transition and activation of HDAC2 | [85, 86] |
| Valproic acid | STZ-induced SD rats | DM | 8 weeks | exert anti-inflammatory activity via reducing NF-κB and improve kidney function by reducing renal damage and fibrosis | [87] |
| Drugs | |||||
| TSG | STZ-induced SD rats | DM | 8 weeks |
inhibit oxidative stress, inflammatory, and expression of TGF-β1 partly mediated by activation of SIRT1 |
[88] |
| Esculetin | STZ-induced SD rats | DKD | 8 weeks |
attenuate alteration in Mmp13 and Bmp6 gene expression by involving change in acetylation and methylation of histone H3 |
[89] |
Modulation of non-coding RNAs
High-throughput sequencing technology has confirmed that less than 3% genes of the human genome can be transcribed into protein-encoding RNA, so the vast majority of RNA is non-coding RNA (ncRNAs) [90]. ncRNAs, which mainly include microRNAs (miRNAs), long non-coding RNAs (LncRNAs), and circular RNAs (CircRNAs), are once considered to be “transcriptional noise” in various diseases. However, with the in-depth research of RNA function, a growing evidence suggests that ncRNAs are involved in multiple physiological and pathological processes, including the pathogenesis of DKD [91]. As another major mechanism of epigenetic regulation, ncRNAs can influence gene expression through both transcriptional and post-transcriptional mechanisms and thereby regulate the progression of inflammation and fibrosis in DKD.
The role of microRNAs in diabetes-related renal inflammation and fibrosis
MiRNAs, a class of endogenous single-stranded RNAs with a length of 20–22 oligonucleotides, are proved to be expression-stable and tissue-specific in specific diseases [92]. Currently, various miRNAs have been reported to play vital role in the pathogenesis of DKD (Table 2) [93]. These miRNAs can be divided into T1DN-related miRNAs, T2DN-related miRNAs, and not-mentioned types. Due to the lack of in-depth research, some miRNAs, such as miR-4490[94]and miR-423-5p[95], is only limited to in vitro level. In this section, we will mainly focus on the regulatory role of miRNAs on T1DN and T2DN.
Table 2.
miRNAs involved in diabetes-related renal inflammation and fibrogenesis
| Types | Functions involved | miRNAs | Expression levels |
Target genes | Mechanisms | Ref |
|---|---|---|---|---|---|---|
| T1DN-related miRNAs | Renal Inflammation | miR-21 | Upregulation | TIMP3 | upregulation of miR-21 enhanced the excretion of pro-inflammatory factors by repressing the expression of TIMP3 | [96] |
| miR-146a | Upregulation | IRAK1/TRAF6 | upregulation of miR-146a promoted NF-kB mediated upregulation of pro-inflammatory cytokines by negative feedback to Irak1 and Traf6 | [97] | ||
| miR-146a | Downregulation | Nox4 | miR‑146a/Nox4 decreases ROS generation and inflammation and prevents DN | [98] | ||
| Renal Fibrosis | miR-192 | Upregulation | GLP1R | upregulation of miR-192 exerted its pro-fibrotic effects by directly targeting GLP1R | [99, 100] | |
| miR-214 | Upregulation | PTEN | upregulation of miR-214 contributed to renal cell hypertrophy and matrix protein expression by directly acting on PTEN | [101] | ||
| miR-22 | Upregulation | PTEN | upregulation of miR-22 promoted renal tubulointerstitial fibrosis by suppressing autophagy partially via targeting PTEN | [102] | ||
| miR-382 | Upregulation | FOXO1 | upregulation of miR-382-induced glomerular mesangial cell proliferation and ECM accumulation by targeting FOXO1 | [103] | ||
| miR-137 | Downregulation | Notch1 | miR-137 inhibition aggravated ECM protein accumulation via directly targeting Notch1 | [104] | ||
| Both inflammation and fibrosis | miR-455-3p | Downregulation | ROCK2 | downregulated miR-455-3p aggravated the progression of renal inflammation and fibrosis through promoting ROCK2 expression | [105] | |
| T2DN related miRNAs | Renal Inflammation | miR-146a | Upregulation | IRAK1/TRAF6 | upregulation of miR-146a promoted NF-kB mediated upregulation of pro-inflammatory cytokines by negative feedback to Irak1 and Traf6 | [97] |
| Renal Fibrosis | miR-133b/199b | Upregulation | Sirt 1 | upregulation of miR-133b and miR-199b enhanced TGF-β1-induced epithelial to mesenchymal transition and renal fibrosis by targeting SIRT1 in diabetic nephropathy | [107] | |
| miR-23a | Upregulation | SnoN | upregulation of miR-23a promoted high glucose-induced EMT and renal fibrogenesis by down-regulation of SnoN | [112] | ||
| miR-30e | Downregulation | GLIPR-2 | miR-30e inhibited GLIPR-2 and then promoted the proliferation of RTECs and inhibited EMT, ultimately leading to renal fibrosis in DN | [109] | ||
| miR-93 | Downregulation | Orai1 | downregulation of microRNA-93-induced TGF-b1-induced EMT and renal fibrogenesis by down-regulation of Orai1 | [110] | ||
| Both inflammation and fibrosis | miR-29b | Downregulation | SP1/Smad-3/NF-κB | miR-29b played a protective role in diabetic kidney disease by the inhibition of Sp1 expression, TGF-β/Smad3-dependent renal fibrosis, and NF-κB-driven renal inflammation | [106] | |
| Renal Inflammation | miR-423-5p | Downregulation | Nox4 | miR-423-5p suppressed high-glucose induced podocyte injury and inhibited ROS generation by targeting Nox4 | [95] | |
| Not mentioned | Renal Fibrosis | miR-4490 | Upregulation | PSMA6 | upregulation of miRNA-4490 regulated PSMA6 mRNA level post-transcriptionally | [94] |
| miR-326-3p | Downregulation | FcγRIII | miR-326-3p ameliorates high glucose and ox-LDL-IC-induced fibrotic injury in renal mesangial cells by targeting FcγRIII | [113] | ||
| Both inflammation and fibrosis | miR-199a-5p | Upregulation | Klotho | upregulated expression of miR-199a-5p decreased Klotho expression, resulting in activating the TLR4/NF-kB p65/NGAL signaling pathways and the downstream fibrosis and inflammation in HG-induced rat mesangial cells | [108] |
T1DN-related miRNAs
In STZ-induced type 1 diabetic nephropathy mice models, multiple miRNAs have emerged as important regulators in the inflammatory responses and fibrogenesis of DKD. In in vitro and in vivo studies, the upregulated expression of miR-21 is able to enhance the excretion of pro-inflammatory factors and accelerate kidney injury by repressing the expression of tissue inhibitors of metalloproteinase 3, and thereby promote inflammation [96]. Conversely, miR-146a downregulates nicotinamide adenine dinucleotide phosphate oxidase 4 (Nox4) and its downstream activities of inflammatory cytokines, such as IL-1b and IL-18 [97, 98]. miR-192 was the first miRNA which was shown to have functional role in promoting the expression of ECM and collagens in STZ-induced mice [99, 100]. In the hyperglycemic environment, the elevated miR-214 and miR-22 were found to be involved in renal fibrogenesis both by inhibiting the expression of phosphatase and tensin homolog (PTEN) [101, 102]. Many other miRNAs, such as miR-382 and miR-137, were also reported to participate glomerular and tubular fibrogenesis of T1DN [103, 104]. In addition, the overexpression of miR-455-3p in STZ-induced DN rats cannot only improve renal fibrosis but reduce inflammatory cytokines by targeting rho-associated coiled coil-containing protein kinase 2 (ROCK2) [105]. Therefore, dysregulation of miRNAs plays catalytic roles in the progression of T1DN inflammation and fibrosis.
T2DN-related miRNAs
Numerous miRNAs are now also thought to be involved in the progression of type 2 diabetic nephropathy. Among these miRNAs, miR-146a is the only one which also exert anti-inflammatory role on the pathogenesis of diabetic db/db mice[97]. However, in the T2DM experimental models, studies on the regulatory role of miRNA mainly focus on the processes of fibrogenesis. Among the miR-29 family, decreased miR-29b, not miR-29a and c, is demonstrated to ameliorate both renal inflammation and fibrogenesis. Loss of intrarenal miR-29b is able to inhibiting the progression of DKD via TGF-β/Smad3-dependent renal fibrosis and NF-κB-driven renal inflammation [106]. Mechanistically, as downstream targets of TGF-β signaling, miR-133b and miR-199b dramatically facilitate TGF-β1-induced EMT & renal fibrosis by inhibiting the expression of sirt1 in diabetic OLETF rats [107]. In addition, there is evidence that high glucose activates TLR4/NF-κB and p65/NGAL signaling pathways by upregulating miR-199a, along with the reduction of klotho, resulting in both fibrosis and inflammatory reaction [108]. Apart from miR-29b and miR-199, miR-30e [109], miR-93 [110, 111], miR-23a [112], and miR-326-3p [113] can affect the synthesis of ECM protein in MCs and the EMT by regulating their target genes, resulting in the excessive accumulation of ECM.
The role of LncRNAs in diabetes-related renal inflammation and fibrosis
Unlike miRNA, the expression of LncRNAs has poor sequence conservation among different species and is highly specific in different tissues and cell types; thus, the in-depth study of different LncRNAs could improve our understanding of the metabolic memory mechanisms of DKD. As an integral part of the epigenome, LncRNAs regulate the expression of adjacent and distal genes expression by various biological mechanisms, which include recruiting chromatin remodeling complexes, regulating gene expression as competitive endogenous RNA, and binding transcription factors or cofactors to affect the transcription of target genes as a scaffold [114, 115]. Lately, accumulating novel data have been instrumental in identifying potential biological functions of LncRNAs that contribute to renal inflammation and fibrogenesis associated with both T1DN and T2DN (Fig. 2).
Fig. 2.
Function of LncRNAs in diabetes-related renal inflammation and fibrosis. LncRNAs can regulate the expression of adjacent and distal genes by various biological mechanisms in DKD. As shown above, a LncRNAs promote the binding of H3K4me3 or H3K27me3 to the gene promoter to affect its expression via recruiting chromatin remodeling complexes such as PRC2 and MLL1. LncRNAs bind transcription factors or cofactors to affect the transcription of target genes as scaffolds: b LncRNAs regulate NLRP3 inflammasome signaling pathway by interacting with p50, the subunit of NF-κB; c LncRNA MALAT1 accelerates β-catenin nuclear accumulation through physical binding to SRSF1, and thus feedback to promote the expression of LncRNA MALAT1 and contribute to renal fibrosis; d LncRNAs also act as regulators of inflammation via directly interaction with Egr-1. In addition, e LncRNAs located in the cell cytoplasm regulate gene expression by acting as molecular sponges and competitively binding to miRNA. EED, embryonic ectoderm development; PRC2, polycomb repressive complex 2; SUZ12, PRC2 subunit; MLL1, mixed-lineage leukemia 1; Egr‐1, early growth response protein 1
Recruitment of chromatin remodeling complex
Previous studies have demonstrated that LncRNAs can mediate epigenetic changes by recruiting chromatin remodeling complexes to specific gene promoters in various diseases, including DKD [116, 117]. A 2018 study indicates that LINC00968 promotes the binding of H3K27me3 to the p21 promoter to inhibit its expression via the recruitment of EZH2, a vital subunit of PRC2 complex. The silencing of LINC00968 significantly inhibited the proliferation and fibrosis of MCs, and decreased the expression of ECM proteins such as fibronectin and collagen IV in vitro. [118]. Moreover, directed by their related LncRNAs, this model is also applicable to other chromatin modifying complexes, such as methyltransferase myeloid and lymphoid or mixed-lineage leukemia (MLL), PcG and so on. LncRNA ZEB1-AS1, which is the antisense lncRNA located in the promoter region of zinc finger E-box-binding protein 1 (ZEB1) and positively regulates its expression, displayed an anti-fibrotic role in DKD by reducing the high glucose-induced ECM accumulation. Mechanistically, LncRNA ZEB1-AS1 directly binds H3K4 methyltransferase MLL1 and induces H3K4me3 histone modification in ZEB1 promoter, which further activates ZEB1 transcription. Furthermore, the expression of lncRNA ZEB1-AS1 can be suppressed by p53 via the physical interaction with its promoter region, highlighting intricate epigenetic regulatory mechanisms in both T1DN and T2DN murine models [119].
Binding transcription factors or cofactors
Emerging studies also hint to the central role of LncRNAs in regulating transcription and post-transcription of fibrosis-associated and inflammation-related genes in DKD. Among 12 abnormally expressed LncRNAs, LincRNA-Gm4419 is verified to be the only one related to NF-κB by bioinformatic methods. To further determine its role on the pathogenesis of DKD, the deletion of LincRNA-Gm4419 by small interfering RNAs (siRNA) could significantly decrease the inflammation, fibrosis, and cell proliferation. The pro-inflammatory effect of LincRNA-Gm4419 is mediated through direct interaction with p50, the subunit of NF-κB and NLRP3 inflammasome signaling pathway [120]. LncRNA MALAT1, as a broadly expressed nuclear lncRNA in mammalian tissues, is found highly expressed not only in STZ-induced diabetic C57BL/6 mice, but also in high glucose-induced podocytes. SRSF1, also known as SF2/ASF, is one of the major pre-mRNA splicing factors that contribute to partial RNA metabolism, such as nuclear export of the mature mRNA [121]. In Lv’s study [122], LncRNA MALAT1 has been shown to accelerate β-catenin nuclear accumulation through physical binding to SRSF1, and thus promote podocyte injury and contribute to renal fibrosis. Moreover, recent studies demonstrate that LncRNA Rpph1 and LncRNA NONHSAG053901 acts as regulators of inflammation via the directly interaction with the DKD-related factor galectin-3 (Gal-3), or early growth response protein 1 (Egr‐1), respectively [123, 124]. In addition, Egr‐1 is able to augment its function in DKD inflammation by interacting with TGF‐β, and the overexpression of Egr-1 can also upregulate the lncRNA Arid2-IR level which contributes to ECM accumulation in DKD [125], in support of a complicated potential mechanism of metabolic memory in DKD.
Crosstalk between LncRNA and miRNAs
As previously mentioned, LncRNAs located in the cell cytoplasm, which act as molecular sponges, also involved in the development of renal inflammation and fibrosis through competitively binding to miRNA. Erbb4-IR, a smad3-associated profibrotic lncRNA, enhances the progression of renal fibrosis and increases albuminuria in DKD by inhibiting miR-29b in the transcriptional level. An in vitro study also revealed that the elevated expression of LncRNA Erbb4-IR was induced by AGEs rather than by high glucose [126]. A novel LncRNA, MEG3, is upregulated and promotes cell fibrosis and inflammatory in response to DKD by suppressing miR-181a and targeting the aforementioned genes, Egr-1 and TLR4. The interaction of LncRNA MEG3 with miR-181 is not only through sponging by competing with endogenous RNA mechanisms, but direct targeting by and in an Argonaute (Ago)2-dependent manner [127]. Ago2 is a major member of the RNA-induced silencing complexes (RISCs), which play a central role in post-transcriptional gene regulation. miRNAs direct complementary target mRNAs to repress and/or degrade RNA through RISCs that contains Ago family proteins [128]. The down-regulation of another lncRNA, LincRNA1700020I14Rik, promotes MCs proliferation and fibrosis in DKD through miR-34a-5p via directly targeting way and Ago2-dependent manner. Additionally, LincRNA 1700020I14Rik/miR-34a-5p/ sirt1 axis and LncRNA-NR_033515/ miR-743b-5p axis were also found to be involved in fibrogenesis of DKD [129, 130]. In any case, these findings provide a better understanding of the role of LncRNAs in inflammatory and fibrogenic progression and may lead to novel therapies for DKD.
The role of circRNAs in diabetes-related renal inflammation and fibrosis
CircRNAs are a novel class of non-coding RNAs, which are mainly generated from the back-splicing of exons, introns or a combination of both, forming a covalently closed loop [131]. In the inflammatory and fibrogenic processes of both T1DN and T2DN, circRNAs canonically function as a competing endogenous RNA to protect the target genes from miRNA‐mediated mRNA degradation, thereby achieving their biological role (Table 3).
Table 3.
circRNAs involved in diabetes-related renal inflammation and fibrogenesis
| CircRNAs | Experimental models | Functions involved | Axis | Ref |
|---|---|---|---|---|
| circRNA_15698 | T2DN, Diabetic db/db mice | ECM accumulation | circRNA_15698/miR-185/TGF‐β1 axis | [132] |
| circRNA-0080425 | T1DN, STZ-induced mice | renal fibrosis | circRNA-0080425/miR‐24‐3p/FGF11 axis | [133] |
| circ-LRP6 | Not mentioned, mesangial cells | renal inflammation and fibrosis | circLRP6/miR‐205/HMGB1 axis | [134] |
| circ-WBSCR17 | T1DN, STZ-induced mice | inflammatory responses and fibrosis | circ-WBSCR17/miR-185-5p/SOX6 axis | [135] |
| circ-RNF169 | Not mentioned, glomerular endothelial cells | cell proliferation and EMT | Unknown | [136] |
| circSTRN3 | Not mentioned, glomerular endothelial cells | cell proliferation and EMT | Unknown | [136] |
With the help of circRNA microarray analysis, circRNA_15698 was found to be upregulated in both diabetic db/db murine models and in the HG-induced MCs. The overexpression of circRNA_15698 play pivotal role in ECM accumulation via miR‐185/TGF‐β1 axis [132]. Another novel circRNA, 0080425, significantly accelerates cell proliferation and fibrosis by suppressing the expression of miR‐24‐3p. The inhibition of this miRNA releases its suppression on fibroblast growth factor 11 (FGF11), which stimulate endothelial proliferation and migration in STZ-induced T1DN [133]. Finally, the circLRP6 upregulates the expression of high mobility group box 1 (HMGB1) via the circLRP6-miR‐205-HMGB1 regulatory network, thus ameliorates HG‐induced MCs proliferation, ECM accumulation, and renal inflammation [134]. In high glucose-induced human kidney tubular cells, Circ-WBSCR17 was also reported to aggravate inflammatory responses and fibrosis by targeting miR-185-5p/SOX6 regulatory axis[135]. CircRNF169 and circSTRN3 are another two differentially expressed circRNAs that were found to be involved in HG-induced cell proliferation and EMT [136]. However, investigating their undefined mechanisms will provide novel insights for the biological role of circRNAs associated with renal inflammation and fibrosis.
Conclusion
The present review focused on the biologic role of metabolic memory, mainly in pathological processes, such as inflammation and fibrosis and potential treatment strategies, especially epigenetic changes-oriented therapies, of DKD. In short, epigenetic mechanisms that include DNA methylation, histone PTMs, and ncRNAs play critical roles in the pathogenesis of DKD, especially in the processes of inflammation and fibrogenesis. Actually, these mechanisms are not completely independent. DNA methylation can interact with histone modification, which in turn can be affected by miRNAs and LncRNAs. Once triggered by high glucose stimulation or other diabetic pathogenic factors, these intricate interactions of epigenetic changes can persistently activate pro-inflammatory and fibrotic genes in both T1DN and T2DN. Despite subsequent glycemic control, the existence of metabolic memory mediated by epigenetic mechanisms leads to the development of renal inflammation and fibrosis, and then progress to ESRD. These evidences hereinbefore demonstrate persistent epigenetic changes (DNA methylation and histone modifications) and signaling pathway mediated by non-coding RNAs may be involved in metabolic memory. In addition, the constant epigenetic changes can be transferred to offsprings, and affect the population phenotype within a very short period of time.
Therefore, in addition to intensifying blood glucose control in the early stages of diabetes, and reducing the long-term effects of hyperglycemic "metabolic memory," reasonable and effective treatment measures should be taken according to the above epigenetic mechanisms. Various epigenetic drugs, such as DNMT and HDAC inhibitors, are currently reported to play beneficial effects in preclinical studies. For ncRNAs, locked nucleic acid (LNA)-modified oligonucleotides based on antisense technology present superior sensitive and specific detecting abilities because of its high thermal stability when hybridized with their complementary mRNA. Antisense technology has been administrated successfully to targets LncRNAs in many other diseases other than DKD. In the context of diabetes, however, the administration of LNA-modified miRNA inhibitors was demonstrated to ameliorate the renal damage of DKD, especially renal fibrosis. Identifying the precise therapeutic target for antisense therapy in DKD is urgently needed, which is also the aim of this review. Unlike genetics, epigenetics is mostly reversible, which provides a new therapeutic frontier for DKD prevention and management. Thus, more researches on epigenetics and the development of epigenetic-modifying drugs are required in the future, so as to provide reliable biomarkers and novel therapeutic targets for the progression of DM to DKD by altering epigenetics.
Acknowledgements
The authors thank Dr. Yong Liu for his help in drawing figures.
Abbreviations
- AGEs
Advanced glycation end products
- ADP
Adenosine diphosphate
- ANGPTL2
Angiogenin-like protein 2
- Ago
Argonaute
- CCL2
Chemokine (C–C motif) ligand 2
- CTGF
Connective tissue growth factor
- CircRNAs
Circular RNAs
- DKD
Diabetic kidney disease
- DM
Diabetes Mellitus
- DCCT
Diabetes Control and Complications Trial
- DMR
Differentially methylated region
- DNMTs
DNA methyltransferases
- EDIC
Epidemiology of diabetes interventions and complications
- ECM
Extracellular matrix
- eGFR
Estimated glomerular filtration rate
- ESRD
End-stage renal disease
- Egr‐1
Early growth response protein 1
- ER
Endoplasmic reticulum
- FGF11
Fibroblast growth factor 11
- Gal-3
Galectin-3
- HATs
Histone acetyltransferase
- HDACs
Histone deacetylase
- HMTs
Histone lysine methyltransferase
- HDMs
Histone lysine demethylase
- HMGB1
High mobility group box 1
- HbA1c
Average glycated hemoglobin
- HG
High glucose
- ICAM1
Intercellular adhesion molecule 1
- IL
Interleukin
- JAK-STAT
Janus kinase/signal transducers and activators of transcription
- Kme1
Monomethylation
- Kme2
Demethylation
- Kme3
Trimethylation
- LNA
Locked nucleic acid
- LncRNAs
Long non-coding RNAs
- miRNAs
MicroRNAs
- MLL
Mixed-lineage leukemia
- MCs
Mesangial cells
- MMP
Matrix metalloproteinases
- 5-mC
5-Methylcytosine
- ncRNAs
Non-coding RNA
- Nox4
Nicotinamide adenine dinucleotide phosphate oxidase 4
- Nrf2-keap1
NF-E2–related factor 2/ Kelch-like ECH associated protein 1
- NAD +
Nicotinamide adenine dinucleotide
- NGS
Next-Generation Sequencing
- PTEN
Phosphatase and tensin homolog
- PAI-1
Plasminogen activator inhibitor-1
- PXR
Pregnane X receptor
- PTMs
Post-translational modifications
- ROS
Reactive oxygen species
- Rgc32
Response gene to complement 32
- RISCs
RNA-induced silencing complexes
- ROCK2
Rho-associated coiled coil-containing protein kinase 2
- Sirt
Sirtuins
- siRNA
Small interfering RNAs
- STZ
Streptozocin
- SAM
S-adenosylmethionine
- SAH
S-adenosyl homocysteine
- TSA
Trichostatin A
- TXNIP
Thioredoxin interacting protein
- TET
Ten-eleven translocation
- TGF-β
Transforming growth factor-β
- TNF-α
Tumor necrosis factor-α
- UTR
Untranslated regions
- VCAM1
Vascular cell adhesion molecule 1
- UKPDS
UK Prospective Diabetes Study
- ZEB1
Zinc finger E-box-binding protein 1
Authors' contributions
Prof. Zhang-Suo Liu devised the conceptual ideas. Dr. Wen Zheng drafted the original manuscript and drew the figures. Dr. Wen Zheng, Dr. Jia Guo, and Prof. Zhang-Suo Liu edited various versions of the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No.82070745 and No.81970633), Comprehensive and digital demonstration platform for clinical evaluation technology of new drugs for major diseases (No. 2020ZX09201-009) and the Innovation Scientists and Technicians Troop Construction Projects of Henan Province (No.182101510002).
Availability of data and materials
No additional supporting data in this article.
Declarations
Ethics approval and consent to participate
This review article is not involved in personal information and ethical approval is not required.
Consent for publication
Not applicable.
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rodriguez-Calvo T, Richardson SJ, Pugliese A. Pancreas pathology during the natural history of type 1 diabetes. Curr Diab Rep. 2018;18:124. doi: 10.1007/s11892-018-1084-3. [DOI] [PubMed] [Google Scholar]
- 2.Petersmann A, Müller-Wieland D, Müller UA, Landgraf R, Nauck M, Freckmann G, et al. Definition, classification and diagnosis of diabetes mellitus. Exp Clin Endocrinol Diabetes. 2019;127:S1–s7. doi: 10.1055/a-1018-9078. [DOI] [PubMed] [Google Scholar]
- 3.Cefalu WT, Buse JB, Tuomilehto J, Fleming GA, Ferrannini E, Gerstein HC, et al. Update and next steps for real-world translation of interventions for type 2 diabetes prevention: Reflections from a diabetes care editors' expert forum. Diabetes Care. 2016;39:1186–1201. doi: 10.2337/dc16-0873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harlan SM, Heinz-Taheny KM, Sullivan JM, Wei T, Baker HE, Jaqua DL, et al. Progressive renal disease established by renin-coding adeno-associated virus-driven hypertension in diverse diabetic models. J Am Soc Nephrol. 2018;29:477–491. doi: 10.1681/ASN.2017040385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mortality in type 1 diabetes in the dcct/edic versus the general population. Diabetes Care. 2016; 39:1378–1383. [DOI] [PMC free article] [PubMed]
- 6.Intensive blood-glucose control with sulphonylureas or insulin compared with conventional treatment and risk of complications in patients with type 2 diabetes (ukpds 33). Uk prospective diabetes study (ukpds) group. Lancet. 1998; 352:837–853. [PubMed]
- 7.Ceriello A. Hypothesis: The "metabolic memory", the new challenge of diabetes. Diabetes Res Clin Pract. 2009;86(Suppl 1):S2–6. doi: 10.1016/S0168-8227(09)70002-6. [DOI] [PubMed] [Google Scholar]
- 8.Reddy MA, Zhang E, Natarajan R. Epigenetic mechanisms in diabetic complications and metabolic memory. Diabetologia. 2015;58:443–455. doi: 10.1007/s00125-014-3462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 10.Waddington CH.The epigenotype. 1942. Int J Epidemiol. 2012; 41:10–13. [DOI] [PubMed]
- 11.Goldberg AD, Allis CD, Bernstein E. Epigenetics: A landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Keating ST, El-Osta A. Glycemic memories and the epigenetic component of diabetic nephropathy. Curr Diab Rep. 2013;13:574–581. doi: 10.1007/s11892-013-0383-y. [DOI] [PubMed] [Google Scholar]
- 13.Sun G, Reddy MA, Yuan H, Lanting L, Kato M, Natarajan R. Epigenetic histone methylation modulates fibrotic gene expression. J Am Soc Nephrol. 2010;21:2069–2080. doi: 10.1681/ASN.2010060633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Godel M, Hartleben B, Herbach N, Liu S, Zschiedrich S, Lu S, et al. Role of mtor in podocyte function and diabetic nephropathy in humans and mice. J Clin Invest. 2011;121:2197–2209. doi: 10.1172/JCI44774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reidy K, Kang HM, Hostetter T, Susztak K. Molecular mechanisms of diabetic kidney disease. J Clin Invest. 2014;124:2333–2340. doi: 10.1172/JCI72271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartleben B, Godel M, Meyer-Schwesinger C, Liu S, Ulrich T, Kobler S, et al. Autophagy influences glomerular disease susceptibility and maintains podocyte homeostasis in aging mice. J Clin Invest. 2010;120:1084–1096. doi: 10.1172/JCI39492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chilelli NC, Burlina S, Lapolla A. Ages, rather than hyperglycemia, are responsible for microvascular complications in diabetes: A "glycoxidation-centric" point of view. Nutr Metab Cardiovasc Dis. 2013;23:913–919. doi: 10.1016/j.numecd.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 18.Matoba K, Takeda Y, Nagai Y, Kawanami D, Utsunomiya K, Nishimura R.Unraveling the role of inflammation in the pathogenesis of diabetic kidney disease. Int J Mol Sci. 2019; 20: [DOI] [PMC free article] [PubMed]
- 19.Barnes JL, Glass Ii WF. Renal interstitial fibrosis: A critical evaluation of the origin of myofibroblasts. Contrib Nephrol. 2011;169:73–93. doi: 10.1159/000313946. [DOI] [PubMed] [Google Scholar]
- 20.Hojs R, Ekart R, Bevc S, Hojs N. Markers of inflammation and oxidative stress in the development and progression of renal disease in diabetic patients. Nephron. 2016;133:159–162. doi: 10.1159/000447434. [DOI] [PubMed] [Google Scholar]
- 21.Tian S, Chen SY.Macrophage polarization in kidney diseases. Macrophage. 2015; 2:10–14800/macrophage.14679. [DOI] [PMC free article] [PubMed]
- 22.Klessens CQF, Zandbergen M, Wolterbeek R, Bruijn JA, Rabelink TJ, Bajema IM, et al. Macrophages in diabetic nephropathy in patients with type 2 diabetes. Nephrol Dial Transplant. 2017;32:1322–1329. doi: 10.1093/ndt/gfw260. [DOI] [PubMed] [Google Scholar]
- 23.Usui HK, Shikata K, Sasaki M, Okada S, Matsuda M, Shikata Y, et al. Macrophage scavenger receptor-a-deficient mice are resistant against diabetic nephropathy through amelioration of microinflammation. Diabetes. 2007;56:363–372. doi: 10.2337/db06-0359. [DOI] [PubMed] [Google Scholar]
- 24.Niewczas MA, Pavkov ME, Skupien J, Smiles A, Md Dom ZI, Wilson JM, et al. A signature of circulating inflammatory proteins and development of end-stage renal disease in diabetes. Nat Med. 2019;25:805–813. doi: 10.1038/s41591-019-0415-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tuttle KR, Brosius FC, 3rd, Adler SG, Kretzler M, Mehta RL, Tumlin JA, et al. Jak1/jak2 inhibition by baricitinib in diabetic kidney disease: Results from a phase 2 randomized controlled clinical trial. Nephrol Dial Transplant. 2018;33:1950–1959. doi: 10.1093/ndt/gfx377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H, Nair V, Saha J, Atkins KB, Hodgin JB, Saunders TL, et al. Podocyte-specific jak2 overexpression worsens diabetic kidney disease in mice. Kidney Int. 2017;92:909–921. doi: 10.1016/j.kint.2017.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng H, Whitman SA, Wu W, Wondrak GT, Wong PK, Fang D, et al. Therapeutic potential of nrf2 activators in streptozotocin-induced diabetic nephropathy. Diabetes. 2011;60:3055–3066. doi: 10.2337/db11-0807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matoba K, Kawanami D, Tsukamoto M, Kinoshita J, Ito T, Ishizawa S, et al. Rho-kinase regulation of tnf-alpha-induced nuclear translocation of nf-kappab rela/p65 and m-csf expression via p38 mapk in mesangial cells. Am J Physiol Renal Physiol. 2014;307:F571–580. doi: 10.1152/ajprenal.00113.2014. [DOI] [PubMed] [Google Scholar]
- 29.Hu C, Sun L, Xiao L, Han Y, Fu X, Xiong X, et al. Insights into the mechanisms involved in the expression and regulation of extracellular matrix proteins in diabetic nephropathy. Curr Med Chem. 2015;22:2858–2870. doi: 10.2174/0929867322666150625095407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qian Y, Feldman E, Pennathur S, Kretzler M, Brosius FC, 3rd.From fibrosis to sclerosis: Mechanisms of glomerulosclerosis in diabetic nephropathy. Diabetes. 2008; 57:1439–1445. [DOI] [PMC free article] [PubMed]
- 31.Liu Y. Cellular and molecular mechanisms of renal fibrosis. Nat Rev Nephrol. 2011;7:684–696. doi: 10.1038/nrneph.2011.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stokes MB, Holler S, Cui Y, Hudkins KL, Eitner F, Fogo A, et al. Expression of decorin, biglycan, and collagen type i in human renal fibrosing disease. Kidney Int. 2000;57:487–498. doi: 10.1046/j.1523-1755.2000.00868.x. [DOI] [PubMed] [Google Scholar]
- 33.Sakharova OV, Taal MW, Brenner BM. Pathogenesis of diabetic nephropathy: Focus on transforming growth factor-beta and connective tissue growth factor. Curr Opin Nephrol Hypertens. 2001;10:727–738. doi: 10.1097/00041552-200111000-00001. [DOI] [PubMed] [Google Scholar]
- 34.Baricos WH, Reed JC, Cortez SL. Extracellular matrix degradation by cultured mesangial cells: Mediators and modulators. Exp Biol Med (Maywood) 2003;228:1018–1022. doi: 10.1177/153537020322800905. [DOI] [PubMed] [Google Scholar]
- 35.Kok HM, Falke LL, Goldschmeding R, Nguyen TQ. Targeting ctgf, egf and pdgf pathways to prevent progression of kidney disease. Nat Rev Nephrol. 2014;10:700–711. doi: 10.1038/nrneph.2014.184. [DOI] [PubMed] [Google Scholar]
- 36.Guha M, Xu ZG, Tung D, Lanting L, Natarajan R. Specific down-regulation of connective tissue growth factor attenuates progression of nephropathy in mouse models of type 1 and type 2 diabetes. Faseb j. 2007;21:3355–3368. doi: 10.1096/fj.06-6713com. [DOI] [PubMed] [Google Scholar]
- 37.Effect of intensive therapy on the development and progression of diabetic nephropathy in the diabetes control and complications trial. The diabetes control and complications (dcct) research group. Kidney Int. 1995; 47:1703–1720. [DOI] [PubMed]
- 38.Retnakaran R, Cull CA, Thorne KI, Adler AI, Holman RR.Risk factors for renal dysfunction in type 2 diabetes: U.K. Prospective diabetes study 74. Diabetes. 2006; 55:1832–1839. [DOI] [PubMed]
- 39.Kato M, Natarajan R. Diabetic nephropathy–emerging epigenetic mechanisms. Nat Rev Nephrol. 2014;10:517–530. doi: 10.1038/nrneph.2014.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.El-Osta A, Brasacchio D, Yao D, Pocai A, Jones PL, Roeder RG, et al. Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. J Exp Med. 2008;205:2409–2417. doi: 10.1084/jem.20081188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Intine RV, Olsen AS, Sarras MP, Jr.A zebrafish model of diabetes mellitus and metabolic memory. J Vis Exp. 2013; e50232. [DOI] [PMC free article] [PubMed]
- 42.Miao F, Chen Z, Genuth S, Paterson A, Zhang L, Wu X, et al. Evaluating the role of epigenetic histone modifications in the metabolic memory of type 1 diabetes. Diabetes. 2014;63:1748–1762. doi: 10.2337/db13-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Z, Miao F, Paterson AD, Lachin JM, Zhang L, Schones DE, et al. Epigenomic profiling reveals an association between persistence of DNA methylation and metabolic memory in the dcct/edic type 1 diabetes cohort. Proc Natl Acad Sci U S A. 2016;113:E3002–3011. doi: 10.1073/pnas.1603712113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dupont C, Armant DR, Brenner CA. Epigenetics: Definition, mechanisms and clinical perspective. Semin Reprod Med. 2009;27:351–357. doi: 10.1055/s-0029-1237423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang L, Zhang Q, Liu S, Chen Y, Li R, Lin T, et al. DNA methyltransferase 1 may be a therapy target for attenuating diabetic nephropathy and podocyte injury. Kidney Int. 2017;92:140–153. doi: 10.1016/j.kint.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 46.Mau T, Yung R. Potential of epigenetic therapies in non-cancerous conditions. Front Genet. 2014;5:438. doi: 10.3389/fgene.2014.00438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by mll partner tet1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bell CG, Teschendorff AE, Rakyan VK, Maxwell AP, Beck S, Savage DA. Genome-wide DNA methylation analysis for diabetic nephropathy in type 1 diabetes mellitus. BMC Med Genomics. 2010;3:33. doi: 10.1186/1755-8794-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sapienza C, Lee J, Powell J, Erinle O, Yafai F, Reichert J, et al. DNA methylation profiling identifies epigenetic differences between diabetes patients with esrd and diabetes patients without nephropathy. Epigenetics. 2011;6:20–28. doi: 10.4161/epi.6.1.13362. [DOI] [PubMed] [Google Scholar]
- 50.Park J, Guan Y, Sheng X, Gluck C, Seasock MJ, Hakimi AA, et al.Functional methylome analysis of human diabetic kidney disease. JCI Insight. 2019; 4: [DOI] [PMC free article] [PubMed]
- 51.Qiu C, Hanson RL, Fufaa G, Kobes S, Gluck C, Huang J, et al. Cytosine methylation predicts renal function decline in american indians. Kidney Int. 2018;93:1417–1431. doi: 10.1016/j.kint.2018.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gluck C, Qiu C, Han SY, Palmer M, Park J, Ko YA, et al. Kidney cytosine methylation changes improve renal function decline estimation in patients with diabetic kidney disease. Nat Commun. 2019;10:2461. doi: 10.1038/s41467-019-10378-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang B, Ji G, Naeem H, Wang J, Kantharidis P, Powell D, et al. The use of targeted next generation sequencing to explore candidate regulators of tgf-beta1's impact on kidney cells. Front Physiol. 2018;9:1755. doi: 10.3389/fphys.2018.01755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen G, Chen H, Ren S, Xia M, Zhu J, Liu Y, et al. Aberrant DNA methylation of mtor pathway genes promotes inflammatory activation of immune cells in diabetic kidney disease. Kidney Int. 2019;96:409–420. doi: 10.1016/j.kint.2019.02.020. [DOI] [PubMed] [Google Scholar]
- 55.Ishii T, Furuya F, Takahashi K, Shikata M, Takamura T, Kobayashi H, et al. Angiopoietin-like protein 2 promotes the progression of diabetic kidney disease. J Clin Endocrinol Metab. 2019;104:172–180. doi: 10.1210/jc.2017-02705. [DOI] [PubMed] [Google Scholar]
- 56.Watanabe A, Marumo T, Kawarazaki W, Nishimoto M, Ayuzawa N, Ueda K, et al. Aberrant DNA methylation of pregnane x receptor underlies metabolic gene alterations in the diabetic kidney. Am J Physiol Renal Physiol. 2018;314:F551–f560. doi: 10.1152/ajprenal.00390.2017. [DOI] [PubMed] [Google Scholar]
- 57.Yang XH, Feng SY, Yu Y, Liang Z. Study on the relationship between the methylation of the mmp-9 gene promoter region and diabetic nephropathy. Endokrynol Pol. 2018;69:269–275. doi: 10.5603/EP.a2018.0029. [DOI] [PubMed] [Google Scholar]
- 58.Aldemir O, Turgut F, Gokce C. The association between methylation levels of targeted genes and albuminuria in patients with early diabetic kidney disease. Ren Fail. 2017;39:597–601. doi: 10.1080/0886022X.2017.1358180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bechtel W, McGoohan S, Zeisberg EM, Muller GA, Kalbacher H, Salant DJ, et al. Methylation determines fibroblast activation and fibrogenesis in the kidney. Nat Med. 2010;16:544–550. doi: 10.1038/nm.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tampe B, Tampe D, Muller CA, Sugimoto H, LeBleu V, Xu X, et al. Tet3-mediated hydroxymethylation of epigenetically silenced genes contributes to bone morphogenic protein 7-induced reversal of kidney fibrosis. J Am Soc Nephrol. 2014;25:905–912. doi: 10.1681/ASN.2013070723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang L, Zhang Q, Wu Q, Wei Y, Yu J, Mu J, et al. Effect of tet2 on the pathogenesis of diabetic nephropathy through activation of transforming growth factor beta1 expression via DNA demethylation. Life Sci. 2018;207:127–137. doi: 10.1016/j.lfs.2018.04.044. [DOI] [PubMed] [Google Scholar]
- 62.Martinez-Moreno JM, Fontecha-Barriuso M, Martin-Sanchez D, Guerrero-Mauvecin J, Goma-Garces E, Fernandez-Fernandez B, et al.Epigenetic modifiers as potential therapeutic targets in diabetic kidney disease. Int J Mol Sci. 2020; 21: [DOI] [PMC free article] [PubMed]
- 63.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 64.Chen S, Feng B, George B, Chakrabarti R, Chen M, Chakrabarti S. Transcriptional coactivator p300 regulates glucose-induced gene expression in endothelial cells. Am J Physiol Endocrinol Metab. 2010;298:E127–137. doi: 10.1152/ajpendo.00432.2009. [DOI] [PubMed] [Google Scholar]
- 65.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, et al. Genome-wide mapping of hats and hdacs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Syreeni A, El-Osta A, Forsblom C, Sandholm N, Parkkonen M, Tarnow L, et al. Genetic examination of setd7 and suv39h1/h2 methyltransferases and the risk of diabetes complications in patients with type 1 diabetes. Diabetes. 2011;60:3073–3080. doi: 10.2337/db11-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miao F, Gonzalo IG, Lanting L, Natarajan R. In vivo chromatin remodeling events leading to inflammatory gene transcription under diabetic conditions. J Biol Chem. 2004;279:18091–18097. doi: 10.1074/jbc.M311786200. [DOI] [PubMed] [Google Scholar]
- 68.Khan S, Jena G, Tikoo K, Kumar V. Valproate attenuates the proteinuria, podocyte and renal injury by facilitating autophagy and inactivation of nf-kappab/inos signaling in diabetic rat. Biochimie. 2015;110:1–16. doi: 10.1016/j.biochi.2014.12.015. [DOI] [PubMed] [Google Scholar]
- 69.Liu M, Liang K, Zhen J, Zhou M, Wang X, Wang Z, et al. Sirt6 deficiency exacerbates podocyte injury and proteinuria through targeting notch signaling. Nat Commun. 2017;8:413. doi: 10.1038/s41467-017-00498-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu F, Zong M, Wen X, Li X, Wang J, Wang Y, et al. Silencing of histone deacetylase 9 expression in podocytes attenuates kidney injury in diabetic nephropathy. Sci Rep. 2016;6:33676. doi: 10.1038/srep33676. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 71.Villeneuve LM, Reddy MA, Lanting LL, Wang M, Meng L, Natarajan R. Epigenetic histone h3 lysine 9 methylation in metabolic memory and inflammatory phenotype of vascular smooth muscle cells in diabetes. Proc Natl Acad Sci U S A. 2008;105:9047–9052. doi: 10.1073/pnas.0803623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang J, Yan W, Peng X, Jiang Y, He L, Peng Y, et al. Functional role of suv39h1 in human renal tubular epithelial cells under high-glucose ambiance. Inflammation. 2018;41:1–10. doi: 10.1007/s10753-017-0657-7. [DOI] [PubMed] [Google Scholar]
- 73.Chen J, Guo Y, Zeng W, Huang L, Pang Q, Nie L, et al. Er stress triggers mcp-1 expression through set7/9-induced histone methylation in the kidneys of db/db mice. Am J Physiol Renal Physiol. 2014;306:F916–925. doi: 10.1152/ajprenal.00697.2012. [DOI] [PubMed] [Google Scholar]
- 74.Chen H, Huang Y, Zhu X, Liu C, Yuan Y, Su H, et al. Histone demethylase utx is a therapeutic target for diabetic kidney disease. J Physiol. 2019;597:1643–1660. doi: 10.1113/JP277367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu H, Wu X, Qin H, Tian W, Chen J, Sun L, et al. Myocardin-related transcription factor a epigenetically regulates renal fibrosis in diabetic nephropathy. J Am Soc Nephrol. 2015;26:1648–1660. doi: 10.1681/ASN.2014070678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yuan H, Reddy MA, Deshpande S, Jia Y, Park JT, Lanting LL, et al. Epigenetic histone modifications involved in profibrotic gene regulation by 12/15-lipoxygenase and its oxidized lipid products in diabetic nephropathy. Antioxid Redox Signal. 2016;24:361–375. doi: 10.1089/ars.2015.6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li X, Li C, Li X, Cui P, Li Q, Guo Q, et al. Involvement of histone lysine methylation in p21 gene expression in rat kidney in vivo and rat mesangial cells in vitro under diabetic conditions. J Diabetes Res. 2016;2016:3853242. doi: 10.1155/2016/3853242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yuan H, Reddy MA, Sun G, Lanting L, Wang M, Kato M, et al. Involvement of p300/cbp and epigenetic histone acetylation in tgf-beta1-mediated gene transcription in mesangial cells. Am J Physiol Renal Physiol. 2013;304:F601–613. doi: 10.1152/ajprenal.00523.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Deb DK, Chen Y, Sun J, Wang Y, Li YC. Atp-citrate lyase is essential for high glucose-induced histone hyperacetylation and fibrogenic gene upregulation in mesangial cells. Am J Physiol Renal Physiol. 2017;313:F423–f429. doi: 10.1152/ajprenal.00029.2017. [DOI] [PubMed] [Google Scholar]
- 80.Sayyed SG, Gaikwad AB, Lichtnekert J, Kulkarni O, Eulberg D, Klussmann S, et al. Progressive glomerulosclerosis in type 2 diabetes is associated with renal histone h3k9 and h3k23 acetylation, h3k4 dimethylation and phosphorylation at serine 10. Nephrol Dial Transplant. 2010;25:1811–1817. doi: 10.1093/ndt/gfp730. [DOI] [PubMed] [Google Scholar]
- 81.Gaikwad AB, Gupta J, Tikoo K. Epigenetic changes and alteration of fbn1 and col3a1 gene expression under hyperglycaemic and hyperinsulinaemic conditions. Biochem J. 2010;432:333–341. doi: 10.1042/BJ20100414. [DOI] [PubMed] [Google Scholar]
- 82.Murr R. Interplay between different epigenetic modifications and mechanisms. Adv Genet. 2010;70:101–141. doi: 10.1016/B978-0-12-380866-0.60005-8. [DOI] [PubMed] [Google Scholar]
- 83.Chiu J, Khan ZA, Farhangkhoee H, Chakrabarti S. Curcumin prevents diabetes-associated abnormalities in the kidneys by inhibiting p300 and nuclear factor-kappab. Nutrition. 2009;25:964–972. doi: 10.1016/j.nut.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 84.Wang Y, Wang Y, Luo M, Wu H, Kong L, Xin Y, et al. Novel curcumin analog c66 prevents diabetic nephropathy via jnk pathway with the involvement of p300/cbp-mediated histone acetylation. Biochim Biophys Acta. 2015;1852:34–46. doi: 10.1016/j.bbadis.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yoshikawa M, Hishikawa K, Marumo T, Fujita T. Inhibition of histone deacetylase activity suppresses epithelial-to-mesenchymal transition induced by tgf-beta1 in human renal epithelial cells. J Am Soc Nephrol. 2007;18:58–65. doi: 10.1681/ASN.2005111187. [DOI] [PubMed] [Google Scholar]
- 86.Noh H, Oh EY, Seo JY, Yu MR, Kim YO, Ha H, et al. Histone deacetylase-2 is a key regulator of diabetes- and transforming growth factor-beta1-induced renal injury. Am J Physiol Renal Physiol. 2009;297:F729–739. doi: 10.1152/ajprenal.00086.2009. [DOI] [PubMed] [Google Scholar]
- 87.Khan S, Jena G, Tikoo K. Sodium valproate ameliorates diabetes-induced fibrosis and renal damage by the inhibition of histone deacetylases in diabetic rat. Exp Mol Pathol. 2015;98:230–239. doi: 10.1016/j.yexmp.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 88.Li C, Cai F, Yang Y, Zhao X, Wang C, Li J, et al. Tetrahydroxystilbene glucoside ameliorates diabetic nephropathy in rats: Involvement of sirt1 and tgf-beta1 pathway. Eur J Pharmacol. 2010;649:382–389. doi: 10.1016/j.ejphar.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 89.Surse VM, Gupta J, Tikoo K. Esculetin induced changes in mmp13 and bmp6 gene expression and histone h3 modifications attenuate development of glomerulosclerosis in diabetic rats. J Mol Endocrinol. 2011;46:245–254. doi: 10.1530/JME-10-0154. [DOI] [PubMed] [Google Scholar]
- 90.Finishing the euchromatic sequence of the human genome Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 91.Guo J, Liu Z, Gong R. Long noncoding rna: An emerging player in diabetes and diabetic kidney disease. Clin Sci (Lond) 2019;133:1321–1339. doi: 10.1042/CS20190372. [DOI] [PubMed] [Google Scholar]
- 92.Krol J, Loedige I, Filipowicz W. The widespread regulation of microrna biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 93.Chung AC. Micrornas in diabetic kidney disease. Adv Exp Med Biol. 2015;888:253–269. doi: 10.1007/978-3-319-22671-2_13. [DOI] [PubMed] [Google Scholar]
- 94.Feng Y, Jin MY, Liu DW, Wei L.Proteasome subunit-α type-6 protein is post-transcriptionally repressed by the microrna-4490 in diabetic nephropathy. Biosci Rep. 2018; 38: [DOI] [PMC free article] [PubMed]
- 95.Xu Y, Zhang J, Fan L, He X. Mir-423-5p suppresses high-glucose-induced podocyte injury by targeting nox4. Biochem Biophys Res Commun. 2018;505:339–345. doi: 10.1016/j.bbrc.2018.09.067. [DOI] [PubMed] [Google Scholar]
- 96.Chen X, Zhao L, Xing Y, Lin B. Down-regulation of microrna-21 reduces inflammation and podocyte apoptosis in diabetic nephropathy by relieving the repression of timp3 expression. Biomed Pharmacother. 2018;108:7–14. doi: 10.1016/j.biopha.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 97.Bhatt K, Lanting LL, Jia Y, Yadav S, Reddy MA, Magilnick N, et al. Anti-inflammatory role of microrna-146a in the pathogenesis of diabetic nephropathy. J Am Soc Nephrol. 2016;27:2277–2288. doi: 10.1681/ASN.2015010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wan RJ, Li YH. Microrna-146a/napdh oxidase4 decreases reactive oxygen species generation and inflammation in a diabetic nephropathy model. Mol Med Rep. 2018;17:4759–4766. doi: 10.3892/mmr.2018.8407. [DOI] [PubMed] [Google Scholar]
- 99.Putta S, Lanting L, Sun G, Lawson G, Kato M, Natarajan R. Inhibiting microrna-192 ameliorates renal fibrosis in diabetic nephropathy. J Am Soc Nephrol. 2012;23:458–469. doi: 10.1681/ASN.2011050485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gondaliya P, Dasare A, Srivastava A, Kalia K. Mir29b regulates aberrant methylation in in-vitro diabetic nephropathy model of renal proximal tubular cells. PLoS ONE. 2018;13:e0208044. doi: 10.1371/journal.pone.0208044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bera A, Das F, Ghosh-Choudhury N, Mariappan MM, Kasinath BS, Ghosh CG. Reciprocal regulation of mir-214 and pten by high glucose regulates renal glomerular mesangial and proximal tubular epithelial cell hypertrophy and matrix expansion. Am J Physiol Cell Physiol. 2017;313:C430–c447. doi: 10.1152/ajpcell.00081.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang Y, Zhao S, Wu D, Liu X, Shi M, Wang Y, et al. Microrna-22 promotes renal tubulointerstitial fibrosis by targeting pten and suppressing autophagy in diabetic nephropathy. J Diabetes Res. 2018;2018:4728645. doi: 10.1155/2018/4728645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang S, Wen X, Han XR, Wang YJ, Shen M, Fan SH, et al. Repression of microrna-382 inhibits glomerular mesangial cell proliferation and extracellular matrix accumulation via foxo1 in mice with diabetic nephropathy. Cell Prolif. 2018;51:e12462. doi: 10.1111/cpr.12462. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 104.Han F, Wang S, Chang Y, Li C, Yang J, Han Z, et al. Triptolide prevents extracellular matrix accumulation in experimental diabetic kidney disease by targeting microrna-137/notch1 pathway. J Cell Physiol. 2018;233:2225–2237. doi: 10.1002/jcp.26092. [DOI] [PubMed] [Google Scholar]
- 105.Wu J, Liu J, Ding Y, Zhu M, Lu K, Zhou J, et al. Mir-455-3p suppresses renal fibrosis through repression of rock2 expression in diabetic nephropathy. Biochem Biophys Res Commun. 2018;503:977–983. doi: 10.1016/j.bbrc.2018.06.105. [DOI] [PubMed] [Google Scholar]
- 106.Chen HY, Zhong X, Huang XR, Meng XM, You Y, Chung AC, et al. Microrna-29b inhibits diabetic nephropathy in db/db mice. Mol Ther. 2014;22:842–853. doi: 10.1038/mt.2013.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sun Z, Ma Y, Chen F, Wang S, Chen B, Shi J. Mir-133b and mir-199b knockdown attenuate tgf-β1-induced epithelial to mesenchymal transition and renal fibrosis by targeting sirt1 in diabetic nephropathy. Eur J Pharmacol. 2018;837:96–104. doi: 10.1016/j.ejphar.2018.08.022. [DOI] [PubMed] [Google Scholar]
- 108.Wu C, Lv C, Chen F, Ma X, Shao Y, Wang Q. The function of mir-199a-5p/klotho regulating tlr4/nf-κb p65/ngal pathways in rat mesangial cells cultured with high glucose and the mechanism. Mol Cell Endocrinol. 2015;417:84–93. doi: 10.1016/j.mce.2015.09.024. [DOI] [PubMed] [Google Scholar]
- 109.Zhao D, Jia J, Shao H. Mir-30e targets glipr-2 to modulate diabetic nephropathy: In vitro and in vivo experiments. J Mol Endocrinol. 2017;59:181–190. doi: 10.1530/JME-17-0083. [DOI] [PubMed] [Google Scholar]
- 110.Badal SS, Wang Y, Long J, Corcoran DL, Chang BH, Truong LD, et al. Mir-93 regulates msk2-mediated chromatin remodelling in diabetic nephropathy. Nat Commun. 2016;7:12076. doi: 10.1038/ncomms12076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ma J, Zhang L, Hao J, Li N, Tang J, Hao L. Up-regulation of microrna-93 inhibits tgf-β1-induced emt and renal fibrogenesis by down-regulation of orai1. J Pharmacol Sci. 2018;136:218–227. doi: 10.1016/j.jphs.2017.12.010. [DOI] [PubMed] [Google Scholar]
- 112.Xu H, Sun F, Li X, Sun L. Down-regulation of mir-23a inhibits high glucose-induced emt and renal fibrogenesis by up-regulation of snon. Hum Cell. 2018;31:22–32. doi: 10.1007/s13577-017-0180-z. [DOI] [PubMed] [Google Scholar]
- 113.Wang Y, Zhang R, Zhang J, Liu F. Microrna-326-3p ameliorates high glucose and ox-ldl-ic- induced fibrotic injury in renal mesangial cells by targeting fcγriii. Nephrology (Carlton) 2018;23:1031–1038. doi: 10.1111/nep.13168. [DOI] [PubMed] [Google Scholar]
- 114.Wang KC, Chang HY. Molecular mechanisms of long noncoding rnas. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li Y, Wang Z, Shi H, Li H, Li L, Fang R, et al. Hbxip and lsd1 scaffolded by lncrna hotair mediate transcriptional activation by c-myc. Cancer Res. 2016;76:293–304. doi: 10.1158/0008-5472.CAN-14-3607. [DOI] [PubMed] [Google Scholar]
- 116.Mercer TR, Dinger ME, Mattick JS. Long non-coding rnas: Insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 117.Liu DW, Zhang JH, Liu FX, Wang XT, Pan SK, Jiang DK, et al. Silencing of long noncoding rna pvt1 inhibits podocyte damage and apoptosis in diabetic nephropathy by upregulating foxa1. Exp Mol Med. 2019;51:1–15. doi: 10.1038/s12276-019-0259-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li Z, Yu Z, Meng X, Yu P. Lncrna linc00968 accelerates the proliferation and fibrosis of diabetic nephropathy by epigenetically repressing p21 via recruiting ezh2. Biochem Biophys Res Commun. 2018;504:499–504. doi: 10.1016/j.bbrc.2018.08.048. [DOI] [PubMed] [Google Scholar]
- 119.Wang J, Pan J, Li H, Long J, Fang F, Chen J, et al. Lncrna zeb1-as1 was suppressed by p53 for renal fibrosis in diabetic nephropathy. Mol Ther Nucleic Acids. 2018;12:741–750. doi: 10.1016/j.omtn.2018.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yi H, Peng R, Zhang LY, Sun Y, Peng HM, Liu HD, et al. Lincrna-gm4419 knockdown ameliorates nf-κb/nlrp3 inflammasome-mediated inflammation in diabetic nephropathy. Cell Death Dis. 2017;8:e2583. doi: 10.1038/cddis.2016.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tripathi V, Song DY, Zong X, Shevtsov SP, Hearn S, Fu XD, et al. Srsf1 regulates the assembly of pre-mrna processing factors in nuclear speckles. Mol Biol Cell. 2012;23:3694–3706. doi: 10.1091/mbc.e12-03-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hu M, Wang R, Li X, Fan M, Lin J, Zhen J, et al. Lncrna malat1 is dysregulated in diabetic nephropathy and involved in high glucose-induced podocyte injury via its interplay with β-catenin. J Cell Mol Med. 2017;21:2732–2747. doi: 10.1111/jcmm.13189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhang P, Sun Y, Peng R, Chen W, Fu X, Zhang L, et al. Long non-coding rna rpph1 promotes inflammation and proliferation of mesangial cells in diabetic nephropathy via an interaction with gal-3. Cell Death Dis. 2019;10:526. doi: 10.1038/s41419-019-1765-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Peng W, Huang S, Shen L, Tang Y, Li H, Shi Y. Long noncoding rna nonhsag053901 promotes diabetic nephropathy via stimulating egr-1/tgf-β-mediated renal inflammation. J Cell Physiol. 2019;234:18492–18503. doi: 10.1002/jcp.28485. [DOI] [PubMed] [Google Scholar]
- 125.Yang YL, Hu F, Xue M, Jia YJ, Zheng ZJ, Li Y, et al. Early growth response protein-1 upregulates long noncoding rna arid2-ir to promote extracellular matrix production in diabetic kidney disease. Am J Physiol Cell Physiol. 2019;316:C340–c352. doi: 10.1152/ajpcell.00167.2018. [DOI] [PubMed] [Google Scholar]
- 126.Sun SF, Tang PMK, Feng M, Xiao J, Huang XR, Li P, et al. Novel lncrna erbb4-ir promotes diabetic kidney injury in db/db mice by targeting mir-29b. Diabetes. 2018;67:731–744. doi: 10.2337/db17-0816. [DOI] [PubMed] [Google Scholar]
- 127.Zha F, Qu X, Tang B, Li J, Wang Y, Zheng P, et al. Long non-coding rna meg3 promotes fibrosis and inflammatory response in diabetic nephropathy via mir-181a/egr-1/tlr4 axis. Aging (Albany NY) 2019;11:3716–3730. doi: 10.18632/aging.102011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hutvagner G, Simard MJ. Argonaute proteins: Key players in rna silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 129.Li A, Peng R, Sun Y, Liu H, Peng H, Zhang Z. Lincrna 1700020i14rik alleviates cell proliferation and fibrosis in diabetic nephropathy via mir-34a-5p/sirt1/hif-1α signaling. Cell Death Dis. 2018;9:461. doi: 10.1038/s41419-018-0527-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gao J, Wang W, Wang F, Guo C. Lncrna-nr_033515 promotes proliferation, fibrogenesis and epithelial-to-mesenchymal transition by targeting mir-743b-5p in diabetic nephropathy. Biomed Pharmacother. 2018;106:543–552. doi: 10.1016/j.biopha.2018.06.104. [DOI] [PubMed] [Google Scholar]
- 131.Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular rnas. Mol Cell. 2018;71:428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 132.Hu W, Han Q, Zhao L, Wang L. Circular rna circrna_15698 aggravates the extracellular matrix of diabetic nephropathy mesangial cells via mir-185/tgf-β1. J Cell Physiol. 2019;234:1469–1476. doi: 10.1002/jcp.26959. [DOI] [PubMed] [Google Scholar]
- 133.Liu H, Wang X, Wang ZY, Li L. Circ_0080425 inhibits cell proliferation and fibrosis in diabetic nephropathy via sponging mir-24-3p and targeting fibroblast growth factor 11. J Cell Physiol. 2020;235:4520–4529. doi: 10.1002/jcp.29329. [DOI] [PubMed] [Google Scholar]
- 134.Chen B, Li Y, Liu Y, Xu Z. Circlrp6 regulates high glucose-induced proliferation, oxidative stress, ecm accumulation, and inflammation in mesangial cells. J Cell Physiol. 2019;234:21249–21259. doi: 10.1002/jcp.28730. [DOI] [PubMed] [Google Scholar]
- 135.Li G, Qin Y, Qin S, Zhou X, Zhao W, Zhang D. Circ_wbscr17 aggravates inflammatory responses and fibrosis by targeting mir-185-5p/sox6 regulatory axis in high glucose-induced human kidney tubular cells. Life Sci. 2020;259:118269. doi: 10.1016/j.lfs.2020.118269. [DOI] [PubMed] [Google Scholar]
- 136.Ling L, Tan Z, Zhang C, Gui S, Cui Y, Hu Y, et al. Circrnas in exosomes from high glucose-treated glomerular endothelial cells activate mesangial cells. Am J Transl Res. 2019;11:4667–4682. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No additional supporting data in this article.