![]()
Keywords: S-palmitoylation, adaptive immunity, innate immune receptors, innate immune effectors
Abstract
S-palmitoylation is a reversible posttranslational lipid modification of proteins. It controls protein activity, stability, trafficking and protein–protein interactions. Recent global profiling of immune cells and targeted analysis have identified many S-palmitoylated immunity-associated proteins. Here, we review S-palmitoylated immune receptors and effectors, and their dynamic regulation at cellular membranes to generate specific and balanced immune responses. We also highlight how this understanding can drive therapeutic advances to pharmacologically modulate immune responses.
1. Introduction
S-palmitoylation is a posttranslational modification of proteins with lipids. It typically involves the addition of a 16-carbon palmitic acid to cysteines of a protein via a thioester bond (figure 1), but other fatty acids like myristic acid and oleic acid can also be added [1]. S-palmitoylation and other lipid modifications control protein-membrane association and trafficking, thereby playing critical roles in protein function and cell signalling [2]. S-palmitoylation is unique among lipid modifications because the high energy thioester bond between the fatty acyl group and cysteine residue allows it to be a reversible modification and therefore can impart spatio-temporal control of protein function [3]. For example, this dynamic fatty acid modification of H- and N-ras enables the protein cycling between the plasma membrane and Golgi apparatus, thus maintaining subcellular compartmentalization for diversification of signal transduction (figure 1) [4]. In addition to protein targeting to different compartments and membrane microdomains, the role of S-palmitoylation is also implicated in protein stability, conformation, and homotypic and heterotypic interactions (figure 1). S-palmitoylation can exert multiple effects simultaneously to orchestrate the desired protein function. Moreover, it can act in concert with other co- and posttranslational modifications to regulate protein functions. Reported S-palmitoylated proteins include ion channels, receptors, transporters, enzymes, cell-adhesion proteins, innate immunity effectors and many others. Overall, S-palmitoylation is involved in a diverse array of physiological processes, including cellular signalling, differentiation, transcriptional regulation, metabolism and others [5–7].
Figure 1.
Protein S-palmitoylation and regulation. Palmitoylation-depalmitoylation cycle regulates protein-membrane association, lipid raft targeting, protein stability and protein–protein interactions, among others. Dynamic S-palmitoylation is mediated by DHHC palmitoyl acyl transferases (DHHC-PATs) and depalmitoylases. Image created with Biorender.com.
S-palmitoylation of proteins is mediated by palmitoyl acyltransferases (PATs) in intracellular compartments including the endoplasmic reticulum, Golgi apparatus and plasma membrane [8]. In humans, PATs are a family of 23 proteins with a characteristic Asp-His-His-Cys (DHHC) domain essential for catalysis. PATs were first discovered in yeast and are conserved across all eukaryotes, though their numbers and specificities can differ between species [9,10]. Regulation at transcriptional, translational and posttranslational levels, as well as PAT variable domains, determine localization, specific substrate profiles and functionality of individual enzymes [11]. Recent structural studies of PAT family members have provided key understanding of reaction mechanism, substrate recognition, interaction, binding and fatty acyl chain selectivity [12,13]. Genetic, cell-based and animal-based studies have implicated PATs and PAT substrates in many patho-physiological conditions including cancer and schizophrenia [14,15].
Removal of palmitate from proteins is regulated by S-depalmitoylases, which catalyse hydrolysis of thioesters [16]. Acyl-protein thioesterase 1 (APT1; also known as lysophospholipase 1, LYPLA1) and acyl-protein thioesterase 2 (APT2; also known as lysophospholipase 2, LYPLA2) were the first identified depalmitoylases for G proteins [17]. Additionally, α/β-hydrolase domain-containing protein 17 (ABHD17) and other serine hydrolase superfamily proteins were identified as depalmitoylases for PSD-95 and oncogenic protein N-Ras [18,19]. Recently, ABHD10 was identified as a depalmitoylase for peroxiredoxin (PRDX5), a key antioxidant protein, and shown to regulate its antioxidant capacity [20]. These findings provide evidence for roles of depalmitoylases in patho-physiological conditions including cancer and made them important targets for potent inhibitor development [21]. Palmitoyl-protein thioesterase 1 (PPT1), an endo-lysosomal protein, is the first known depalmitoylating enzyme to be linked to a fatal genetic lysosomal storage disorder in neurons, neuronal ceroid lipofuscinoses (NCLs) [22]. Development of inhibitors and chemical probes has advanced our understanding of individual depalmitoylase regulation and substrate specificity [23–25].
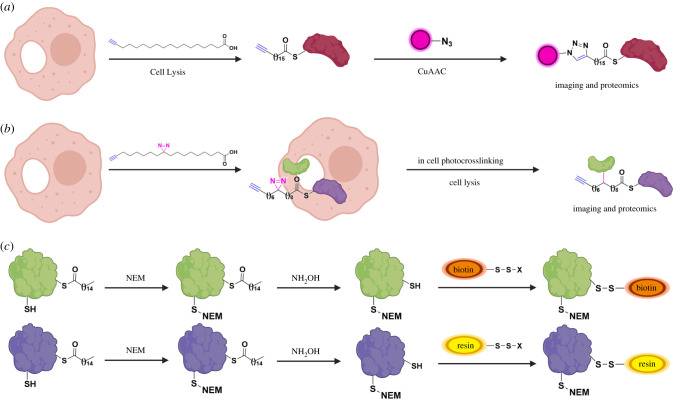
The earliest methods of detection of protein palmitoylation involved metabolic labelling of proteins with radioactive 3H, 14C or 125I containing palmitic acids. These had many limitations including low sensitivity. Development of chemical approaches to study protein S-palmitoylation have revolutionized the understanding of the field (figure 2) [26–28]. Palmitic acid chemical reporters, including palmitic acid-like molecules containing azido or alkynyl groups (e.g. alk-16/ODYA), can label cysteines of target proteins using endogenous palmitoylation machinery (figure 2a). Using bioorthogonal ligation methods, they can be reacted with fluorophores for visualization or with biotin for enrichment and identification of palmitoylated proteins [29]. Such reporters have further been incorporated in pulse-chase-like experiments to monitor palmitoylation turnover kinetics on proteins [19,30,31]. Moreover, incorporation of photocrosslinking chemical moieties in palmitic acid reporters enabled the identification of an S-palmitoylated protein interactome (figure 2b) [32]. A complementary chemical approach for the study of S-palmitoylation involves modification of thioester linked cysteines in S-palmitoylated proteins. This strategy is exploited in methods like acyl-biotin exchange (ABE) and acyl-resin-assisted capture (acyl-Rac) (figure 2c) [33,34]. In these methods, free cysteines of palmitoylated proteins are capped and thioester linkages are subsequently cleaved to generate new thiols. These thiols are then selectively labelled by biotin for ABE or thiol-reactive resin for acyl-Rac, allowing further enrichment and detection of palmitoylated peptides or proteins. A variant of this method called acyl-PEG exchange (APE) exploits PEG labelling of newly generated thiols as a mass tag for mobility-shift based assays to identify levels of protein S-palmitoylation [35]. Though the above chemical approaches each have individual limitations, they have been successfully applied for global profiling of palmitoylated proteins in yeast, protozoan and mammalian cells [36–39]. Furthermore, identification of S-palmitoylomes helped in the development of in silico predictive programmes for palmitoylation sites in proteins despite the lack of a consensus motif [40]. Other methods developed for the analysis of specific palmitoylated proteins include in-cell imaging based on bioorthogonal fatty acid labelling and in situ proximity ligation or quantification of S-palmitoylation levels by gas/liquid chromatography and mass spectrometry [41,42]. Moreover, chemical synthesis or semi-synthesis of modified peptides/proteins has been important for mechanistic understanding of S-palmitoylated proteins, as has been exemplified by the classical study on palmitoylated Ras isoforms [4,27,28]. Recent reports of site-specific chemical modifications of Ras and transmembrane protein IFITM3 with palmitate mimics in live cells and evaluation of activity afford new opportunities to study S-palmitoylation and other lipid modifications in live cells [43,44].
Figure 2.
Chemical methods for the study of S-palmitoylation. (a) Metabolic labelling of cells with palmitic acid reporter alk-16/ODYA and further copper catalysed azide-alkyne cycloaddition (CuAAC) reaction with azido-modified fluorescent or affinity tags for fluorescent visualization or affinity enrichment of S-palmitoylated proteins. (b) Metabolic labelling with a bifunctional fatty acid chemical reporter and in cell photocrosslinking allows detection and proteomic identification of S-palmitoylated protein interactome. (c) ABE and acyl-RAC methods involve capping of free cysteines with thiol-reactive reagent like N-ethylmaleimide (NEM) followed by removal of S-palmitoylation with NH2OH. Newly exposed cysteines are then reacted with a thiol-reactive biotin or resin, respectively. Further enrichment allows identification of S-palmitoylated proteins. Image created with Biorender.com.
We previously reviewed the application of these chemical tools for global profiling of palmitoylated proteins in immune cells including T cells, dendritic cells, macrophages and B cells [45]. Since the publication of this review, there has been remarkable progress in identifying new palmitoylated immunity proteins, and also a renewed emphasis on functional characterization of individual S-palmitoylated immune proteins. In this review, we will highlight mechanistic studies on the roles of S-palmitoylation in the regulation of immune protein function in immune cells (table 1). This review is organized by proteins involved in adaptive and innate immune pathways (i.e. immune sensors, signal transducers, signal regulators and immune effectors). There are excellent reviews that describe the role of protein S-palmitoylation in other physiological processes [79].
Table 1.
S-palmitoylated immunity-associated proteins.
| DHHC-PATs/ APTs | site of modification | method of study | function of modification | ref. | |
|---|---|---|---|---|---|
| T cells | |||||
| CD4 | C394, C397 | radiolabelling | lipid raft association and clustering | [46,47] | |
| CD8 | radiolabelling | CD8 association with kinase Lck | [48] | ||
| Lck | DHHC2, 21 | C3, 5 | radiolabelling | lipid raft association | [49,50] |
| Fyn | DHHC2, 3, 7, 10, 15, 20, 21 | C3, 6 | radiolabelling | plasma membrane localization | [51] |
| LAT | C26, C29 | radiolabelling | lipid raft association protein stability | [52,53] | |
| Fas | DHHC7 | C199 | ABE | lipid raft association | [54] |
| FasL | DHHC7 | C82 | ABE | lipid raft association | [55] |
| PD-1 | DHHC9 | C192 | alk-16 | protein stability | [56] |
| B cells | |||||
| CD81 | C6, C9, C80, C89, C227, C228 | radiolabelling | protein interaction | [57] | |
| HGAL | C43, C45 | radiolabelling | lipid raft association | [58] | |
| immune receptors and adapters | |||||
| PD-L1 | DHHC3 | protein stability | [59] | ||
| TLR2 | DHHC2, 3, 6, 7, 15 | C609 | alk-16 | plasma membrane localization | [60] |
| MyD88 | DHHC6 | C113, C274 | ABE | binding of IRAK4 to MyD88 | [61] |
| STING | DHHC3, 7, 15 | C88, C91 | radiolabelling | membrane clustering | [62] |
| NOD1/2 | DHHC5 | NOD1 C558, C567, C952 NOD2 C395, C1033 | ODYA, ABE | membrane association | [63] |
| FCGR2A | C208 | radiolabelling | lipid raft association | [64] | |
| ASAP2 | DHHC6 | C86 | ABE | regulation of phagocytosis | [65] |
| CD36 | DHHC 5 APT1 | C3, C7, C464, C466 | Acyl-RAC | fatty acid uptake | [66,67] |
| β1-adrenergic receptor | C392, C393, C414 | radiolabelling/alk-16 | [68] | ||
| CCR5 | C321, C323, C324 | radiolabelling | protein trafficking | [69] | |
| S1PR1 | DHHC5 | protein–protein interaction | [70] | ||
| IFNAR1 | C463 | radiolabelling | downstream STAT phosphorylation | [71] | |
| JAK1 | C541, C542 | RAC like assay | plasma membrane association | [72] | |
| STAT3 | DHHC7 APT2 | C108 | alk-14/ ABE | differentiation of TH17 | [73] |
| TNFR1 | APT2 | C248 | Acyl-RAC, ODYA | protein stability | [74] |
| immune effectors | |||||
| IFITM3 | DHHC3, 7, 15, 20 | C71, C72, C105 | alk-16 | antiviral activity | [35,75] |
| TNFα | C47 | radiolabelling | plasma membrane association | [76] | |
| DR6 | C368 | radiolabelling | [77] | ||
| DR4 | C261, C262, C263 | radiolabelling | lipid raft association and oligomerization | [78] |
2. S-palmitoylation of proteins in adaptive immunity
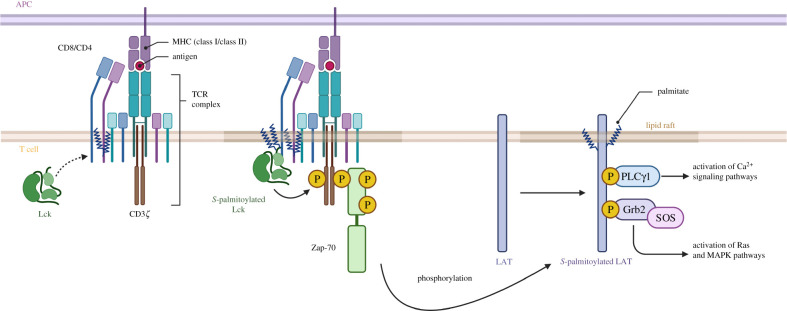
S-palmitoylation plays an important role in the regulation of host adaptive immune responses. Multiple studies investigated the S-palmitoylome of T cells using different methods and identified important factors involved in T cell activation and signal transduction (figure 3) [31,37,80–82]. The first step in T cell receptor (TCR) signal transduction involves the binding of TCR to peptide–major histocompatibility complexes (MHCs) on antigen-presenting cells (APCs). This binding step is followed by recruitment of Src family protein tyrosine kinase Lck to the cytoplasmic domains of CD4 and CD8 co-receptors. This triggers a signalling cascade leading to the formation of the LAT signalosome. Further signal propagation happens via Ca2+–calcineurin, mitogen-activated protein kinase (MAPK) and nuclear factor-κB (NF-κB) signalling pathways.
Figure 3.
S-palmitoylated proteins in T cell signalling. S-palmitoylated Src kinases (Lck) phosphorylates TCR complex leading to ZAP-70 recruitment and activation. Activated ZAP-70 phosphorylates palmitoylated LAT leading to downstream signalling pathways. Image adapted from ‘TCR Downstream Signalling’ by BioRender.com.
2.1. TCR coreceptors
S-palmitoylation of TCR subunits has not been reported, but radio labelling studies identified TCR coreceptor CD4 palmitoylation. CD4 is palmitoylated at Cys394 and Cys397, at the junction of the transmembrane and cytoplasmic domain [46]. CD4 S-palmitoylation regulates clustering with TCR/protein kinase C (PKC) θ in lipid rafts [47]. Coreceptor CD8 is a heterodimer of CD8α and CD8β. In humans, both CD8α and CD8β are S-palmitoylated, whereas in mice only CD8β is S-palmitoylated. Mouse CD8β S-palmitoylation is necessary for efficient CD8 coreceptor function as it increases CD8 association with Lck in lipid rafts [48].
2.2. Src family kinases
Src family kinase Lck is S-palmitoylated at Cys3 and Cys5 and this promotes its plasma membrane association [49,83]. Interestingly, Lck N-myristoylation at Gly2 is required for subsequent palmitoylation. Deletion of Lck S-palmitoylation reduces Lck-CD4 association and downstream signalling [84]. Further studies show that site-specific S-palmitoylation at Cys3 is important for Lck lipid raft localization [50].
S-palmitoylation of Fyn, another Src family kinase involved in T cell signal transduction, has also been shown to play an important role in its membrane association [51]. Fyn can be S-palmitoylated at Cys3 and Cys6 but Cys3 is the major site and is the most important for lipid raft association. Activation of Fyn by Lck in lipid rafts leads to TCR/CD3 activation and downstream signalling. DHHC2, 3, 7, 10, 15, 20, 21 have all been shown to mediate Fyn S-palmitoylation.
2.3. TRAPs
Activation of transmembrane adaptor proteins (TRAPs) is a crucial step in the T cell signal transduction pathway. TRAPs like LAT, phosphoprotein associated with GEMs (PAG) and Lck-interacting membrane protein (LIME) are S-palmitoylated and show lipid raft association [85]. LAT activation leads to the recruitment of key molecules for downstream signal propagation. LAT is dually S-palmitoylated at a membrane juxtaposed Cys-X-X-Cys motif. S-palmitoylation of LAT is important for its stability and plasma membrane association [52,53]. Deletion of LAT S-palmitoylation hinders TCR interaction and recruitment of PLCγ and Grb2 for downstream signal propagation as evident from impaired calcium influx and Erk activation [86].
Beyond T cell activation, LAT S-palmitoylation is also shown to regulate T cell anergy, an inability of previously responsive T cell to respond to TCR stimulation. Anergic T cells show reduced LAT palmitoylation, lipid raft association and immunological synapse recruitment [87]. This should be further explored for therapeutic interventions to inhibit or induce T cell anergy in cancer, transplantation and infectious disease.
2.4. Fas and FasL
Apoptosis of T cells that recognize self-antigens is necessary to prevent autoimmune diseases. Fas and Fas ligand (FasL) are critical regulators of T cell apoptosis. Fas binding to FasL leads to recruitment of death-inducing signalling complex (DISC) in Fas-bearing cells which activates caspase-3 mediated apoptosis. Fas is S-palmitoylated at single Cys199 in human and is required for its stability [54]. DHHC7 is identified to palmitoylate Fas. Mutation of the Fas S-palmitoylation site reduces its lipid raft association and impairs apoptosis signalling.
An ABE-based assay identified FasL S-palmitoylation [55]. FasL is palmitoylated at Cys82 located at the N-terminal end of the transmembrane domain. S-palmitoylation modulates FasL lipid raft partitioning and proteolytic cleavage by ADAM10 for efficient induction of Fas-mediated cell death.
2.5. PD-1 and PD-L1
Programmed death protein 1 (PD-1) is a T cell surface receptor that, upon activation, suppresses T cell proliferation and cytokine production. PD-1 ligands PD-L1 and PD-L2 are expressed on antigen-presenting cells and tumour cells. In silico motif-based prediction identified PD-1 S-palmitoylation at Cys192 between its transmembrane and cytosolic domain, which was confirmed by metabolic labelling studies [56]. PD-1 S-palmitoylation is catalysed by DHHC9. PD-1 palmitoylation is necessary for its stability. Recent studies show that some cancer cells also express PD-1, thus enabling them to promote tumour growth independent of adaptive immunity. S-palmitoylation of PD-1 in tumour cells can modulate downstream mammalian target of rapamycin (mTOR) signalling and proliferation.
PD-L1 expressed on tumour cells is also S-palmitoylated and this modification inhibits PD-L1 ubiquitination and trafficking to lysosomes for degradation [59]. DHHC3 catalyse PD-L1 palmitoylation. Inhibition of PD-L1 palmitoylation using 2-bromopalmitate or via DHHC3 silencing increases anti-tumour activity in cells and in mice. These discoveries of PD-1 and PD-L1 palmitoylation and targeting for modulation of T cell immune responses in cancer provide exciting opportunities for combinatorial approaches along with immune checkpoint therapy.
2.6. B cells
S-palmitoylation of critical proteins in B cells has also been identified. Indeed, an ABE-based profiling of B cells identified many candidate S-palmitoylated proteins [80]. However, there have been limited studies on functional implications of B cell protein S-palmitoylation.
B cell receptor (BCR) coreceptor CD81 is S-palmitoylated at multiple membrane juxtaposed Cys residues [57]. S-palmitoylation of CD81 is required for enhanced BCR-coreceptor complex lipid raft association [88]. It is also important for CD81 association with other BCR coreceptors CD19 and CD21 [57]. Inhibition of CD81 S-palmitoylation affects recruitment of downstream signalling molecules and activation of kinases PI3 K and PKC.
Human germinal centre-associated lymphoma (HGAL) is an adaptor protein involved in BCR signalling. Radio labelling studies identified HGAL S-palmitoylation [58]. S-palmitoylation of HGAL regulates binding and activation of Syk kinase for downstream signalling. HGAL S-palmitoylation deletion mutant localizes in the cytoplasm and significantly impairs chemoattractant-induced cell motility.
3. Palmitoylation of proteins in innate immunity
3.1. Innate immune receptors and signalling adapter proteins
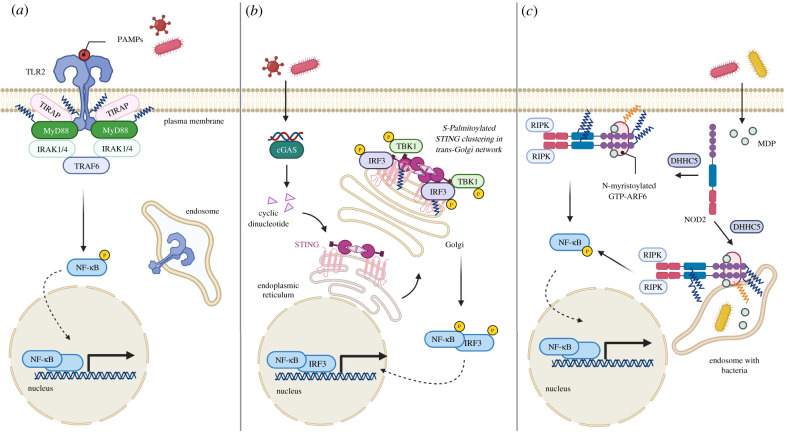
The innate immune system responds to microbial invasion by signalling through pattern recognition receptors (PRRs) that bind conserved microbial features or molecules associated with cellular damage. PRRs can be broadly classified into several families of related molecules. These families include the Toll-like receptors (TLRs), C-type lectin receptors, RIG-I-like receptors and NOD-like receptors. Additionally, a receptor known as cyclic GMP-AMP (cGAMP) synthase (cGAS) is involved in microbial DNA detection. Several of these molecules and/or their signalling adapter proteins have been demonstrated to be S-palmitoylated in recent years (figure 4). As we will describe below, S-palmitoylation, where it has been detected, is generally an activating modification for innate immune signalling through several of these microbial sensing pathways.
Figure 4.
S-palmitoylated innate immune receptors and signalling adapter proteins. (a) Activation of S-palmitoylated TLR2 by pathogen associated molecular patterns (PAMPs) leads to MyD88 signalling. S-palmitoylated MyD88 forms a complex with IL-1 receptor-associated kinase (IRAKs) for downstream signalling and subsequent translocation of nuclear factor-κB (NF-κB) to the nucleus for cytokine induction. (b) STING binds to cyclic dinucleotides at endoplasmic reticulum and translocates to Golgi apparatus where it is palmitoylated. At the trans-Golgi network, S-palmitoylated STING is clustered and recruit TBK1 and IRF3 for downstream signalling. (c) DHHC5 mediated S-palmitoylation of NOD1/2 leads to its recruitment to bacteria-containing phagosome. Further exposure to bacteria derived molecules induces NF-κB signalling. Image created with Biorender.com.
3.1.1. TLRs
Toll-like receptors (TLRs) were among the first of the microbe-sensing PRRs to be discovered. TLRs are transmembrane proteins that use leucine-rich repeat domains to detect diverse microbial products, such as proteins, nucleic acids and glycans. The human genome encodes 10 known TLRs, which localize either to the cell surface or within endosomes. Binding of a TLR to its ligand results in signalling through TLR-associated cytoplasmic adapter proteins, including MyD88. TLR signalling generally culminates in activation of the transcription factor NF-κB and production of inflammatory cytokines. S-palmitoylome profiling of murine dendritic cells using an alkynyl palmitate analogue with click chemistry-based biotin pulldowns identified TLR2 as a putative palmitoylated protein [60]. S-palmitoylation was confirmed and mapped to Cys609 via mutagenesis of candidate cysteines. C609 is located directly adjacent to the cytoplasm-facing side of the TLR2 transmembrane domain. Cys609 mutant constructs showed less responsiveness than WT TLR2 to known TLR2 microbial ligands in NF-κB reporter assays. Further, 2-BP inhibition of TLR2 palmitoylation in murine dendritic cells partially reduced TLR2 levels at the cell surface, and significantly reduced cytokine production in response to TLR2 ligands. Together, these data indicate that TLR2 palmitoylation at Cys609 is required for its proper localization and full antimicrobial activity (figure 4a) [60].
S-palmitoylation levels on TLR2 were increased by overexpression of several DHHC proteins suggesting possible enzymatic redundancy for this activating modification [60]. Cys609 is highly conserved among TLR2 proteins throughout evolution, further supporting a significant role for this modification in antimicrobial functionality. Several additional human TLRs possess cysteine residues near the cytoplasmic border of their transmembrane domains, including TLRs 1, 5, 6, 7, 8, 9 and 10. However, of the human TLRs, only TLRs 2, 5 and 10 showed robust labelling with alkynyl palmitate analogue, with TLR10 providing the strongest palmitoylation signal relative to total protein levels [60]. Whether S-palmitoylation regulates the activity of TLRs 5 and 10, or whether other TLRs are palmitoylated under specific cellular conditions, remain to be determined.
3.1.2. Myd88
MyD88 is a critical signalling adaptor protein for all TLRs, except for TLR3. The discovery of MyD88 S-palmitoylation came as a follow-up to the observation that inhibition of fatty acid synthase by the chemical inhibitor C75 reduced inflammatory responses upon TLR2, 4 or 7/8 stimulation of cells [61]. This inhibition of TLR signalling by C75 was pinpointed to MyD88 through studies examining NF-κB activation upon overexpression of various signalling molecules downstream of TLRs. Given that the product of fatty acid synthase is palmitate, the S-palmitoylation of MyD88 was investigated and confirmed using chemical reporter labelling and ABE methods [61].
Candidate S-palmitoylated cysteines, Cys113 and Cys274, were identified on MyD88 by mass spectrometry, and were further confirmed to be sites of modification via cysteine to alanine mutagenesis of MyD88 constructs and ABE analysis [61]. Interestingly, while single mutations of Cys113 or Cys274 both significantly decreased total MyD88 S-palmitoylation, a double mutant did not show a further decrease in palmitoylation. In terms of NF-κB and MAPK activation, mutation of Cys113 decreased responsiveness to the TLR4 ligand LPS, while mutation of Cys274 had no measurable effect. A Cys113 to Ala MyD88 mutant failed to recruit the IRAK4 signalling molecule upon TLR stimulation, indicating that S-palmitoylation is required for recruitment of IRAK4 to the Myddosome signalling complex downstream of TLR ligand binding (figure 4a). It will be interesting to determine whether MyD88 S-palmitoylation promotes localization with TLRs, such as TLR2, that are also palmitoylated [60]. It is also of note that MyD88 S-palmitoylation was not detected in any of the published S-palmitoyl proteome studies [40], including those done in macrophages and dendritic cells, which may suggest that robust MyD88 S-palmitoylation must be induced or that palmitoylated MyD88 exists in poorly soluble protein aggregates.
DHHC6 was identified as a leading candidate palmitoyltransferase for MyD88 based on its ability to increase MyD88 S-palmitoylation when overexpressed and based on its high expression in myeloid cell types known to have potent TLR activities [61]. As such, knockdown of DHHC6 decreased MyD88 S-palmitoylation and LPS responsiveness of macrophages. Overall, S-palmitoylation of MyD88 appears to be a required regulatory modification that can be targeted to decrease TLR-mediated inflammation either by directly inhibiting enzymatic palmitoylation of MyD88 or through limiting endogenous palmitate available for protein modification [61].
3.1.3. STING
The stimulator of interferon genes (STING) is a signalling protein involved in the response to DNA in the cytosol. STING is a multipass transmembrane protein that is activated by the cyclic dinucleotide cGAMP, which is produced by cGAS upon sensing DNA in the cytosol. Ligand-activated STING moves from the ER to the Golgi apparatus and recruits signalling molecules to activate NF-κB and interferon regulator factor 3 (IRF3), resulting in the production of proinflammatory cytokines and type I interferons (IFNs). Mukai et al. [62], hypothesized that STING is post-translationally modified at the Golgi and discovered that STING could be modified with radiolabelled palmitic acid upon cellular activation with the chemical STING agonist DMXAA. Further, overexpression of Golgi-localized DHHCs 3, 7 and 15 increased STING S-palmitoylation. Conversely, 2-BP treatment of cells eliminated STING palmitoylation and prevented cytokine production upon DMXAA stimulation, suggesting that palmitoylation is necessary for inflammatory cytokine induction by STING. The primary sites of modification on STING were mapped to Cys88 and Cys91. A STING mutant construct with serine substitutions at these positions trafficked from the ER to the Golgi similarly to WT STING upon DMXAA stimulation, but could not induce activation of NF-κB and IRF3, and thus failed to induce proinflammatory downstream gene products, including type I interferons. These results demonstrate that S-palmitoylation of STING at specific cysteines is required for its inflammatory signalling function (figure 4b) [62].
Activating STING mutations in the human population have been associated with an inflammatory autoimmune interferonopathy known as STING associated vasculopathy with onset in infancy (SAVI). Known STING mutants associated with SAVI lost their ability to spontaneously activate the IRF3- and NF-κB-dependent gene promoters when combined with S-palmitoylation-impairing mutations at Cys88 and 91 or when palmitoylation was inhibited by 2-BP treatment of cells [62]. These results suggested that palmitoylation of STING could potentially be targeted therapeutically in SAVI. Indeed, in an attempt to identify STING inhibitors by chemical screening, small molecule inhibitors that covalently react with Cys91 and block palmitoylation were discovered [89]. These inhibitors prevented clustering of STING at the Golgi and prevented STING-dependent inflammation in mouse models. Concurrently, a second group identified endogenously formed nitro-fatty acids as inhibitors of STING palmitoylation via covalent reaction with Cys88 [6]. Nitro-fatty acids were produced during DNA virus infection, suggesting that this may be a natural cellular feedback mechanism to prevent hyperactivation of the cGAS-STING pathway. Further, the addition of nitro-fatty acids to fibroblasts derived from SAVI patients prevented the constitutive type I IFN production by these cells [90]. These exciting developments in small molecule inhibitors of STING provide compelling proof-of-concept studies toward therapeutic targeting of STING palmitoylation [91].
3.1.4. NOD1/2
Nucleotide oligomerization domain (NOD) 1 and 2 proteins are cytosolic innate immune receptors that detect bacterial peptidoglycans. NOD1 and NOD2 are primarily soluble in the cytosol with a portion of these proteins associated with the cellular plasma membrane at steady state. However, they are rapidly redistributed to phagosomal membranes upon intracellular bacterial infection, where they activate NF-κB and MAPK signalling pathways. Since NOD1/2 lack transmembrane regions or traditional lipid-binding motifs, it was hypothesized that their rapid membrane association could be mediated by dynamic S-palmitoylation [63]. Indeed, treatment of cells with palmitoylation inhibitor 2-BP altered the localization of both NOD1 and NOD2 as visualized by fluorescence microscopy, a phenomenon that was confirmed by biochemical fractionation techniques to be due to loss of membrane association. 2-BP treatment of macrophages abrogated the ability of the cells to respond to NOD1/2 ligands in terms of NF-κB and MAPK pathway activation. S-palmitoylation of NOD1 and NOD2 were both subsequently confirmed by ABE and chemical reporter labelling methods. S-palmitoylation of NOD1 was mapped to three cysteine residues, Cys558, 567 and 952. A triple cysteine mutant of NOD1 lost its membrane association and ability to activate NF-κB, an effect that could be rescued by fusion of a known S-palmitoylation amino acid motif to NOD1. Similar effects were seen for NOD2, though in this case S-palmitoylation was mapped to Cys395 and 1033 (figure 4c). A BioID screen for NOD1 and NOD2 interacting proteins identified DHHC5 as a probable candidate for modification of these immune effectors [63]. Knockdown or knockout of DHHC5 resulted in decreased S-palmitoylation of NOD1 and NOD2, decreased NOD membrane association, and decreased activation of NF-κB and MAPK pathways in response to NOD ligands. Further, both DHHC5 and intact NOD S-palmitoylation sites were required for recruitment of NOD1/2 to Salmonella-containing phagosomes [63].
NOD2 variants with decreased peptidoglycan responsiveness have been associated with pathologies, such as Crohn's disease. Five of six disease variants that were tested showed major decreases in S-palmitoylation, suggesting that defective palmitoylation-dependent membrane association underlie their decreased functionality [63]. Conversely, a gain-of-function NOD2 variant showed significantly increased baseline S-palmitoylation, and 2-BP treatment eliminated its hyperactivation of NF-κB. Overall, it has emerged that S-palmitoylation is required for NOD1/2 localization with bacteria-containing membranes and activation of downstream pathways. However, it remains to be determined precisely how bacterial sensing triggers a rapid increase in NOD1/2 S-palmitoylation and how this facilitates inflammatory signalling. Recent cross-linking studies with peptidoglycan fragment muramyl-dipeptide (MDP) photoaffinity reporter showed MDP-induced NOD2 interaction with the plasma membrane or endosome resident N-myristoylated GTP-ARF6 [92]. It will therefore be interesting to understand the role of NOD2 palmitoylation in mediating interactions with membrane-bound host proteins.
3.1.5. Phagocytosis receptor
Phagocytosis for engulfing of pathogens or foreign particles by immune cells also involves initial surface receptor-mediated recognition. Phagocytosis receptor FCGR2A is S-palmitoylated and regulates its lipid raft association [64]. Furthermore, S-palmitoylation of FCGR2A associated kinase ASAP2 can regulate FCGR2A mediated phagocytosis [65].
S-palmitoylation of scavenger receptor CD36 has also been well studied. CD36 is a transmembrane glycoprotein receptor expressed on immune cells including monocytes, macrophages and other cell types, such as adipocytes and cardiac myocytes. CD36 binds to and mediates uptake of oxidized phospholipids, oxidized lipoproteins and long-chain fatty acids, thus playing a major role in lipid homeostasis in cells. In addition, CD36 plays an important role as an innate immune sensor that can recognize and bind to bacterial cell wall components, erythrocytes infected with Plasmodium falciparum and apoptotic cells. S-palmitoylation of CD36 by DHHC4 and DHHC5 was shown to play a regulatory role in adipose tissue fatty acid uptake in mice [66]. Binding of S-palmitoylated CD36 to fatty acids activates the kinase Lyn. Activated Lyn phosphorylates DHHC5 and inhibits its S-palmitoylation activity. Depalmitoylation of fatty acid bound CD36 by APT1 leads to downstream signalling to initiate endocytosis of fatty acids. Pharmacological or genetic perturbations of this dynamic palmitoylation–depalmitoylation cycle disrupts fatty acid uptake of cells [67]. Further investigations of CD36 S-palmitoylation in downstream immune signalling will be helpful to understand connections between lipid metabolism and inflammation.
Moreover, many members of the G protein-coupled receptor (GPCR) family including β1-adrenergic receptor, S1P receptor subtype 1 (S1PR1), CCR5 are known to be S-palmitoylated [68–70]. CCR5 and other S-palmitoylated cell surface receptors like EGFR are also used by viruses like human immunodeficiency virus (HIV) and influenza A virus (IAV), respectively, to enter the host cell successfully. Moreover, It should be noted that S-palmitoylation not only plays a role in the host immune system but the host S-palmitoylation machinery is also exploited by many viral or bacterial proteins for host infection [93,94]. For example, S-palmitoylation of the currently pandemic SARS-CoV-2 spike protein by DHHC5 is important for virus entry in ACE2 expressing cells [95].
3.2. Palmitoylation of innate immune effectors
Activation of innate immune receptors initiates signalling pathways resulting in the production of proinflammatory cytokines including tumour necrosis factors (TNFs), IFNs, chemokines and immune effector proteins for the elimination of microbial pathogens.
3.2.1. IFN signalling
IFNs bind to cell surface IFN receptor leading to activation of Janus tyrosine kinase (Jak)/signal-transducing activators of transcription (STAT) pathways to induce expression of IFN stimulated genes (ISGs). Type I IFNs (IFNα/β) engage the IFNAR receptor complex which is a heterodimer of IFNAR1 and IFNAR2. Radio labelling with [3H] palmitic acid identified S-palmitoylation of IFNAR1 and IFNAR2 [71]. IFNAR1 is S-palmitoylated at Cys463 present in the cytoplasmic domain proximal to the transmembrane domain. Mutagenesis of Cys463 to Ala does not affect receptor stability, endocytosis or intracellular distribution. However, loss of IFNAR1 S-palmitoylation results in decreased STAT phosphorylation and nuclear translocation, thereby affecting IFNα-stimulated gene transcription.
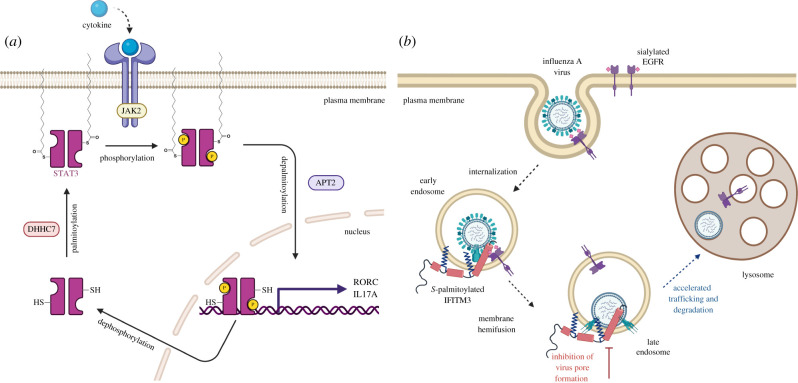
Palmitoylome profiling of adipocytes identified S-palmitoylation of four proteins in the JAK/STAT pathway JAK1, STAT1, STAT3 and STAT5A [72]. Palmitoylation of JAK1 and JAK2 was confirmed in adipocytes using an acyl-RAC-like assay. JAK1 palmitoylation was mapped to highly conserved Cys541 and Cys542. Mutation of these two Cys to Ser led to loss of S-palmitoylation and impaired JAK1 plasma membrane association. S-palmitoylation of STAT family members STAT1α, STAT1β, STAT3, STAT5B and STAT6 has been demonstrated using metabolic labelling and acyl exchange methods [73]. Canonical type I and type III IFN pathway involves both STAT1 and STAT2 activation and heterodimerization, whereas type II IFN pathway involves STAT2 activation and homodimerization. STAT2 and STAT4 S-palmitoylation has not been reported. Recently, STAT3 palmitoylation has been implicated in the regulation of TH17 cell differentiation (figure 5a) [73]. TH17 cells are proinflammatory T cells which express interlukin-17 (IL-17) and retinoic acid receptor-related orphan receptor gamma t (RORγt). STAT3 undergoes a palmitoylation–depalmitoylation cycle on Cys108 mediated by DHHC7 and APT2, respectively. This cycle accelerates TH17 cell differentiation by promoting membrane association, phosphorylation and nuclear translocation of STAT3. Interestingly, phosphorylated STAT3 is selectively depalmitoylated by APT2. Accelerated differentiation of TH17 has been connected to autoimmune diseases. DHHC7 and APT2 have been shown to be upregulated in patients with inflammatory bowel disease (IBD). Inhibition of APT2 activity or knockout of DHHC7 in a mouse model relieves IBD symptoms. Inhibition of STAT3 S-palmitoylation–depalmitoylation cycle could thus be a potential therapeutic strategy for IBD [73].
Figure 5.
S-palmitoylated innate immune effectors. (a) STAT3 dynamic S-palmitoylation in cells. STAT3 palmitoylation by DHHC7 leads to membrane association and phosphorylation. Depalmitoylation of p-STAT3 by APT2 leads to nuclear translocation of p-STAT3 and expression of STAT3 target genes. (b) S-palmitoylated IFITM3 restricts enveloped virus entry in cells through the endocytic pathway. IFITM3 colocalizes with virus-containing endosomes and prevents the release of the virus genetic material into the cytoplasm by physically restricting virus membrane pore formation and shuttling the virus for lysosomal degradation. Image created with Biorender.com.
3.2.2. IFN effectors
Palmitoylome analysis also identified several ISGs in a murine dendritic cell line and murine embryonic fibroblasts, including bone marrow stromal antigen 2 (BST2) or tetherin, immunity-related GTPase M1 (IRGM1) and interferon-induced transmembrane protein 3 (IFITM3) [60]. We have studied S-palmitoylation of IFITM3 and its regulation in our laboratories over the past 10 years. Interferon-induced transmembrane proteins (IFITM1, IFITM2 and IFITM3), which share highly conserved palmitoylation sites, are involved in host immune response to viral infections. IFITM3 is the most active isoform and restricts many viruses including influenza A virus (IAV), dengue virus (DENV), Ebola virus (EBOV), human immunodeficiency virus (HIV), hepatitis C virus (HCV) and Zika virus (ZIKV) [96,97]. IFITM3 also inhibits SARS coronavirus (SARS-CoV) infections [98]. Surprisingly, IFITM3 promotes endemic human coronavirus OC43 infection [99,100]. Recent pseudovirus infection and cell–cell fusion studies provide conflicting results regarding a role of IFITM3 in pandemic SARS-CoV-2 infection [101–104]. Infection studies with genuine SARS-CoV-2 shows IFITM3 acts primarily as an antiviral factor but an endocytosis mutant which localizes to the plasma membrane acts as a proviral factor [105].
IFITMs are present in basal levels in many different cell types. They are intrinsically expressed in embryonic stem cells for protection against virus infections [106]. IFITMs reduce the susceptibility of placental trophoblasts to viral infection, but they also inhibit trophoblast cell fusion, an essential process for placental development mediated by syncytin, which is derived from retroviral envelope [107,108]. Furthermore, the resistance of CD8+ resident memory T cells expressing IFITM3 to IAV infection extends the antiviral role of IFITMs to adaptive immune cells [109,110]. Two recent studies highlight broader roles of IFITMs in host immune pathways beyond antiviral activity. IFITM3 has been shown to be critical in phosphoinositide 3-kinase (PI3 K) signal amplification for the rapid expansion of B cells with high affinity to antigen [106]. IFITM3 has also been implicated in neuroinflammation as it modulates amyloid-β production by regulating γ-secretase activity [111].
Palmitoylome profiling of mouse dendritic cells identified IFITM3 S-palmitoylation [75]. IFITM3 is S-palmitoylated at three Cys residues (Cys71, C72 and 105) and loss of S-palmitoylation leads to abrogation of IFITM3 antiviral activity against influenza virus (figure 5b), and similar loss of antiviral activity was reported for palmitoylation-deficient IFITM1 [112]. Further APE analysis showed that Cys72 is the major site of modification of IFITM3 and mutation of this residue significantly lowers antiviral activity [35]. Also, mass spectrometric analysis of purified IFITM3 from mammalian cells revealed Cys72 as the predominant site of modification [113]. Cys72 is highly conserved across most mammals and is required for the antiviral activity of IFITM3 orthologues from mice, bats and humans [114,115]. Indeed, palmitoylation of evolutionarily ancient IFITM homologues present in mycobacteria were found to be palmitoylated when expressed in human cells [116]. Site-directed mutagenesis and live-cell imaging studies showed that Cys72 is essential for IFITM3 trafficking and colocalization with influenza virus in the endocytic pathway [117,118]. Recent NMR studies with IFITM3 chemically modified at Cys72 with maleimide-palmitate show that lipid modification at Cys72 stabilizes IFITM3 amphipathic helix membrane interaction which is important for restriction of virus infection [44,119]. These studies demonstrate the importance of site-specific palmitoylation of IFITMs in their antiviral activity. Overexpression of multiple palmitoyltransferases (DHHC 3, 7, 15 and 20) leads to increased IFITM3 S-palmitoylation but DHHC20 might be the most important for IFITM3 antiviral activity [120]. Roles for palmitoylation of IFITM3 in PI3 K signalling and regulation of γ-secretase activity remain to be investigated.
3.2.3. TNF signalling
Tumour necrosis factors (TNFs) form another important superfamily of cytokines. TNFα regulates a variety of cellular processes including inflammation, proliferation, differentiation, and can induce various forms of cell death. TNF is synthesized as a transmembrane protein (tmTNF) and presented at the plasma membrane where it is cleaved and released as soluble TNF (sTNF). Membrane-bound N-terminal fragment of TNF (NTF) is further cleaved by signal peptide peptidase-like 2b to generate intracellular domain of TNFα (ICD-TNFα). All the TNF forms show biological activities. Metabolic labelling with [3H]palmitic acid identified S-palmitoylation of tmTNF [76]. Recent studies indicate a role of palmitoylation in tmTNF lipid raft partitioning, NTF stability and efficient cleavage for ICD-TNFα formation [121]. Interaction of S-palmitoylated tmTNF with TNFR1 in lipid rafts diminishes sensitivity to sTNF, thus regulating downstream NFkB and ERK1/2 signalling pathway.
Dynamic S-palmitoylation of TNFR1 also regulates TNF signalling [74]. Plasma membrane TNFR1 activation leads to recruitment of complex 1 adapter proteins to TNFR leading to NF-κB signalling. On the other hand, K63-ubiquitylation and internalization of TNFR1 triggers apoptosis signalling. TNFR1 can be palmitoylated at multiple Cys but dynamic S-palmitoylation at transmembrane proximal Cys248 regulates its plasma membrane localization. Depalmitoylation of activated TNFR1 by APT2 is necessary for enhanced NF-κB signalling, whereas knockdown of APT2 enhances Caspase-8 mediated cell death and reduced NF-κB signalling. Other members of TNF-receptor family have also been reported to be palmitoylated. S-palmitoylation of DR4 is promotes lipid raft association and cell death signalling, whereas DR6 S-palmitoylation prevents lipid raft association [77,78,122].
4. Conclusion and perspective
Over the past decade, we have developed greater understanding of S-palmitoylation-dependent regulation of proteins in innate and adaptive immune signalling. This is a testament to the development of chemical tools to study S-palmitoylation. Use of these chemical tools and proteomics methods allowed S-palmitoylome profiling of different cell types and helped in development of in silico prediction of S-palmitoylation sites in proteins. Further use of chemical inhibitors and genetic methods helped to understand the role of S-palmitoylation in the regulation of protein function. Coupling of these tools has led to the discovery of novel protein S-palmitoylation and mechanisms of protein regulation. However, it should be noted that functional analysis remains to be performed for many putative palmitoylated proteins identified in large scale S-palmitoylome profiling studies, making this a rich area for future investigations.
Though we have learned significantly about S-palmitoylation, we still have very little understanding of the implications of a dynamic lipid modification of proteins in cell signalling and regulation in comparison to other dynamic modifications like phosphorylation or ubiquitination.
We have little understanding of the mechanism and substrate specificity of the writers and erasers of palmitoylation. Structural characterization of individual DHHCs and DHHC selective inhibitor development will be necessary future advances.
In recent years, we have recognized that the role of S-palmitoylation and modifying enzymes in immune cells extends beyond antimicrobial functions to cancer and autoimmune diseases, among others. Moreover, there has been an increasing understanding of their potential as an attractive target for therapeutic interventions, as has been discussed in this review.
Acknowledgement
We thank Hang laboratory members for helpful discussion and input into this review.
Data accessibility
This article has no additional data.
Authors' contributions
T.D., J.S.Y. and H.C.H. wrote and edited the article.
Competing interests
We declare we have no competing interests.
Funding
T.D. was supported by the Tri-Institutional Chemical Biology programme through the NIH Chemistry-Biology Training Grant T32 GM115327. This work was supported by NIH-NIGMS R01GM087544 grant (to H.C.H.).
References
- 1.Resh MD. 2013. Covalent lipid modifications of proteins. Curr. Biol. 23, R431-R435. ( 10.1016/j.cub.2013.04.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Linder ME, Deschenes RJ. 2007. Palmitoylation: policing protein stability and traffic. Nat. Rev. Mol. Cell Biol. 8, 74-84. ( 10.1038/nrm2084) [DOI] [PubMed] [Google Scholar]
- 3.Bijlmakers M-J, Marsh M. 2003. The on–off story of protein palmitoylation. Trends Cell Biol. 13, 32-42. ( 10.1016/S0962-8924(02)00008-9) [DOI] [PubMed] [Google Scholar]
- 4.Rocks O, et al. 2005. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science 307, 1746-1752. ( 10.1126/science.1105654) [DOI] [PubMed] [Google Scholar]
- 5.Zhang MM, Hang HC. 2017. Protein S-palmitoylation in cellular differentiation. Biochem. Soc. Trans. 45, 275-285. ( 10.1042/BST20160236) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen B, Sun Y, Niu J, Jarugumilli GK, Wu X. 2018. Protein lipidation in cell signaling and diseases: function, regulation, and therapeutic opportunities. Cell Chem. Biol. 25, 817-831. ( 10.1016/j.chembiol.2018.05.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chamberlain LH, Shipston MJ. 2015. The physiology of protein S-acylation. Physiol. Rev. 95, 341-376. ( 10.1152/physrev.00032.2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korycka J, et al. 2012. Human DHHC proteins: a spotlight on the hidden player of palmitoylation. Eur. J. Cell Biol. 91, 107-117. ( 10.1016/j.ejcb.2011.09.013) [DOI] [PubMed] [Google Scholar]
- 9.Roth AF, Feng Y, Chen L, Davis NG. 2002. The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase. J. Cell Biol. 159, 23-28. ( 10.1083/jcb.200206120) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lobo S, Greentree WK, Linder ME, Deschenes RJ. 2002. Identification of a Ras palmitoyltransferase in Saccharomyces cerevisiae. J. Biol. Chem. 277, 41 268-41 273. ( 10.1074/jbc.M206573200) [DOI] [PubMed] [Google Scholar]
- 11.Zmuda F, Chamberlain LH. 2020. Regulatory effects of post-translational modifications on zDHHC S-acyltransferases. J. Biol. Chem. 295, 14 640-14 652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rana MS, Kumar P, Lee C-J, Verardi R, Rajashankar KR, Banerjee A. 2018. Fatty acyl recognition and transfer by an integral membrane S-acyltransferase. Science 359, eaao6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stix R, Lee C-J, Faraldo-Gómez JD, Banerjee A. 2020. Structure and mechanism of DHHC protein acyltransferases. J. Mol. Biol. 432, 4983-4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greaves J, Chamberlain LH. 2011. DHHC palmitoyl transferases: substrate interactions and (patho)physiology. Trends Biochem. Sci. 36, 245-253. ( 10.1016/j.tibs.2011.01.003) [DOI] [PubMed] [Google Scholar]
- 15.Ko P-J, Dixon SJ. 2018. Protein palmitoylation and cancer. EMBO Rep. 19, e46666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Won SJ, Cheung See Kit M, Martin BR. 2018. Protein depalmitoylases. Crit. Rev. Biochem. Mol. Biol. 53, 83-98. ( 10.1080/10409238.2017.1409191) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duncan JA, Gilman AG. 1998. A cytoplasmic acyl-protein thioesterase that removes palmitate from G protein alpha subunits and p21(RAS). J. Biol. Chem. 273, 15 830-15 837. ( 10.1074/jbc.273.25.15830) [DOI] [PubMed] [Google Scholar]
- 18.Yokoi N, Fukata Y, Sekiya A, Murakami T, Kobayashi K, Fukata M. 2016. Identification of PSD-95 depalmitoylating enzymes. J. Neurosci. 36, 6431-6444. ( 10.1523/JNEUROSCI.0419-16.2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin DTS, Conibear E. 2015. ABHD17 proteins are novel protein depalmitoylases that regulate N-Ras palmitate turnover and subcellular localization. Elife 4, e11306. ( 10.7554/eLife.11306) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao Y, et al. 2019. ABHD10 is an S-depalmitoylase affecting redox homeostasis through peroxiredoxin-5. Nat. Chem. Biol. 15, 1232-1240. ( 10.1038/s41589-019-0399-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hernandez JL, Majmudar JD, Martin BR. 2013. Profiling and inhibiting reversible palmitoylation. Curr. Opin. Chem. Biol. 17, 20-26. ( 10.1016/j.cbpa.2012.11.023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vesa J, Hellsten E, Verkruyse LA, Camp LA, Rapola J, Santavuori P, Hofmann SL, Peltonen L. 1995. Mutations in the palmitoyl protein thioesterase gene causing infantile neuronal ceroid lipofuscinosis. Nature 376, 584-587. ( 10.1038/376584a0) [DOI] [PubMed] [Google Scholar]
- 23.Azizi S-A, Kathayat RS, Dickinson BC. 2019. Activity-based sensing of S-depalmitoylases: chemical technologies and biological discovery. Acc. Chem. Res. 52, 3029-3038. ( 10.1021/acs.accounts.9b00354) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kathayat RS, Elvira PD, Dickinson BC. 2017. A fluorescent probe for cysteine depalmitoylation reveals dynamic APT signaling. Nat. Chem. Biol. 13, 150-152. ( 10.1038/nchembio.2262) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dekker FJ, et al. 2010. Small-molecule inhibition of APT1 affects Ras localization and signaling. Nat. Chem. Biol. 6, 449-456. ( 10.1038/nchembio.362) [DOI] [PubMed] [Google Scholar]
- 26.Thinon E, Hang HC. 2015. Chemical reporters for exploring protein acylation. Biochem. Soc. Trans. 43, 253-261. ( 10.1042/BST20150004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hang HC, Linder ME. 2011. Exploring protein lipidation with chemical biology. Chem. Rev. 111, 6341-6358. ( 10.1021/cr2001977) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chuh KN, Batt AR, Pratt MR. 2016. Chemical methods for encoding and decoding of posttranslational modifications. Cell Chem. Biol. 23, 86-107. ( 10.1016/j.chembiol.2015.11.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Charron G, Zhang MM, Yount JS, Wilson J, Raghavan AS, Shamir E, Hang HC. 2009. Robust fluorescent detection of protein fatty-acylation with chemical reporters. J. Am. Chem. Soc. 131, 4967-4975. ( 10.1021/ja810122f) [DOI] [PubMed] [Google Scholar]
- 30.Zhang MM, Tsou LK, Charron G, Raghavan AS, Hang HC. 2010. Tandem fluorescence imaging of dynamic S-acylation and protein turnover. Proc. Natl Acad. Sci. USA 107, 8627-8632. ( 10.1073/pnas.0912306107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin BR, Wang C, Adibekian A, Tully SE, Cravatt BF. 2011. Global profiling of dynamic protein palmitoylation. Nat. Methods 9, 84-89. ( 10.1038/nmeth.1769) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng T, Hang HC. 2015. Bifunctional fatty acid chemical reporter for analyzing S-palmitoylated membrane protein-protein interactions in mammalian cells. J. Am. Chem. Soc. 137, 556-559. ( 10.1021/ja502109n) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forrester MT, Hess DT, Thompson JW, Hultman R, Moseley MA, Stamler JS, Casey PJ. 2011. Site-specific analysis of protein S-acylation by resin-assisted capture. J. Lipid Res. 52, 393-398. ( 10.1194/jlr.D011106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wan J, Roth AF, Bailey AO, Davis NG. 2007. Palmitoylated proteins: purification and identification. Nat. Protoc. 2, 1573-1584. ( 10.1038/nprot.2007.225) [DOI] [PubMed] [Google Scholar]
- 35.Percher A, Ramakrishnan S, Thinon E, Yuan X, Yount JS, Hang HC. 2016. Mass-tag labeling reveals site-specific and endogenous levels of protein S-fatty acylation. Proc. Natl Acad. Sci. USA 113, 4302-4307. ( 10.1073/pnas.1602244113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roth AF, Wan J, Bailey AO, Sun B, Kuchar JA, Green WN, Phinney BS, Yates JR, Davis NG. 2006. Global analysis of protein palmitoylation in yeast. Cell 125, 1003-1013. ( 10.1016/j.cell.2006.03.042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin BR, Cravatt BF. 2009. Large-scale profiling of protein palmitoylation in mammalian cells. Nat. Methods 6, 135-138. ( 10.1038/nmeth.1293) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kang R, et al. 2008. Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature 456, 904-909. ( 10.1038/nature07605) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones ML, Collins MO, Goulding D, Choudhary JS, Rayner JC. 2012. Analysis of protein palmitoylation reveals a pervasive role in Plasmodium development and pathogenesis. Cell Host Microbe 12, 246-258. ( 10.1016/j.chom.2012.06.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blanc M, David FPA, van der Goot FG. 2019. SwissPalm 2: protein S-palmitoylation database. Methods Mol. Biol. 2009, 203-214. ( 10.1007/978-1-4939-9532-5_16) [DOI] [PubMed] [Google Scholar]
- 41.Gao X, Hannoush RN. 2014. Method for cellular imaging of palmitoylated proteins with clickable probes and proximity ligation applied to Hedgehog, tubulin, and Ras. J. Am. Chem. Soc. 136, 4544-4550. ( 10.1021/ja410068g) [DOI] [PubMed] [Google Scholar]
- 42.Sorek N, Yalovsky S. 2010. Analysis of protein S-acylation by gas chromatography-coupled mass spectrometry using purified proteins. Nat. Protoc. 5, 834-840. ( 10.1038/nprot.2010.33) [DOI] [PubMed] [Google Scholar]
- 43.Li Y, Wang S, Chen Y, Li M, Dong X, Hang HC, Peng T. 2020. Site-specific chemical fatty-acylation for gain-of-function analysis of protein S-palmitoylation in live cells. Chem. Commun. (Camb) 56, 13 880-13 883. ( 10.1039/D0CC06073A) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garst E, et al. 2020. Site-specific lipidation enhances IFITM3 membrane interactions and antiviral activity. BioRxiv 293324. ( 10.1101/2020.09.11.293324) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yount JS, Zhang MM, Hang HC. 2013. Emerging roles for protein S-palmitoylation in immunity from chemical proteomics. Curr. Opin. Chem. Biol. 17, 27-33. ( 10.1016/j.cbpa.2012.11.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crise B, Rose JK. 1992. Identification of palmitoylation sites on CD4, the human immunodeficiency virus receptor. J. Biol. Chem. 267, 13 593-13 597. ( 10.1016/S0021-9258(18)42253-3) [DOI] [PubMed] [Google Scholar]
- 47.Balamuth F, Brogdon JL, Bottomly K. 2004. CD4 raft association and signaling regulate molecular clustering at the immunological synapse site. J. Immunol. 172, 5887-5892. ( 10.4049/jimmunol.172.10.5887) [DOI] [PubMed] [Google Scholar]
- 48.Arcaro A, Grégoire C, Boucheron N, Stotz S, Palmer E, Malissen B, Luescher IF. 2000. Essential role of CD8 palmitoylation in CD8 coreceptor function. J. Immunol. 165, 2068-2076. ( 10.4049/jimmunol.165.4.2068) [DOI] [PubMed] [Google Scholar]
- 49.Resh MD. 1994. Myristylation and palmitylation of Src family members: the fats of the matter. Cell 76, 411-413. ( 10.1016/0092-8674(94)90104-X) [DOI] [PubMed] [Google Scholar]
- 50.Kosugi A, Hayashi F, Liddicoat DR, Yasuda K, Saitoh S, Hamaoka T. 2001. A pivotal role of cysteine 3 of Lck tyrosine kinase for localization to glycolipid-enriched microdomains and T cell activation. Immunol. Lett. 76, 133-138. ( 10.1016/S0165-2478(01)00174-2) [DOI] [PubMed] [Google Scholar]
- 51.Wolven A, Okamura H, Rosenblatt Y, Resh MD. 1997. Palmitoylation of p59fyn is reversible and sufficient for plasma membrane association. Mol. Biol. Cell 8, 1159-1173. ( 10.1091/mbc.8.6.1159) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shogomori H, Hammond AT, Ostermeyer-Fay AG, Barr DJ, Feigenson GW, London E, Brown DA. 2005. Palmitoylation and intracellular domain interactions both contribute to raft targeting of linker for activation of T cells. J. Biol. Chem. 280, 18 931-18 942. ( 10.1074/jbc.M500247200) [DOI] [PubMed] [Google Scholar]
- 53.Tanimura N, Saitoh S, Kawano S, Kosugi A, Miyake K. 2006. Palmitoylation of LAT contributes to its subcellular localization and stability. Biochem. Biophys. Res. Commun. 341, 1177-1183. ( 10.1016/j.bbrc.2006.01.076) [DOI] [PubMed] [Google Scholar]
- 54.Rossin A, Durivault J, Chakhtoura-Feghali T, Lounnas N, Gagnoux-Palacios L, Hueber AO. 2015. Fas palmitoylation by the palmitoyl acyltransferase DHHC7 regulates Fas stability. Cell Death Differ. 22, 643-653. ( 10.1038/cdd.2014.153) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guardiola-Serrano F, Rossin A, Cahuzac N, Lückerath K, Melzer I, Mailfert S, Marguet D, Zörnig M, Hueber AO. 2010. Palmitoylation of human FasL modulates its cell death-inducing function. Cell Death Dis. 1, e88. ( 10.1038/cddis.2010.62) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yao H, et al. In press. A peptidic inhibitor for PD-1 palmitoylation targets its expression and functions. RSC Chem. Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Delandre C, Penabaz TR, Passarelli AL, Chapes SK, Clem RJ. 2009. Mutation of juxtamembrane cysteines in the tetraspanin CD81 affects palmitoylation and alters interaction with other proteins at the cell surface. Exp. Cell Res. 315, 1953-1963. ( 10.1016/j.yexcr.2009.03.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lu X, Sicard R, Jiang X, Stockus JN, McNamara G, Abdulreda M, Moy VT, Landgraf R, Lossos IS. 2015. HGAL localization to cell membrane regulates B-cell receptor signaling. Blood 125, 649-657. ( 10.1182/blood-2014-04-571331) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yao H, et al. 2019. Inhibiting PD-L1 palmitoylation enhances T-cell immune responses against tumours. Nat Biomed Eng. 3, 306-317. ( 10.1038/s41551-019-0375-6) [DOI] [PubMed] [Google Scholar]
- 60.Chesarino NM, et al. 2014. Chemoproteomics reveals Toll-like receptor fatty acylation. BMC Biol. 12, 91. ( 10.1186/s12915-014-0091-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim Y-C, Lee SE, Kim SK, Jang H-D, Hwang I, Jin S, Hong E-B, Jang K-S, Kim H-S. 2019. Toll-like receptor mediated inflammation requires FASN-dependent MYD88 palmitoylation. Nat. Chem. Biol. 15, 907-916. ( 10.1038/s41589-019-0344-0) [DOI] [PubMed] [Google Scholar]
- 62.Mukai K, Konno H, Akiba T, Uemura T, Waguri S, Kobayashi T, Barber GN, Arai H, Taguchi T. 2016. Activation of STING requires palmitoylation at the Golgi. Nat. Commun. 7, 11932. ( 10.1038/ncomms11932) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lu Y, et al. 2019. Palmitoylation of NOD1 and NOD2 is required for bacterial sensing. Science 366, 460-467. ( 10.1126/science.aau6391) [DOI] [PubMed] [Google Scholar]
- 64.Barnes NC, Powell MS, Trist HM, Gavin AL, Wines BD, Hogarth PM. 2006. Raft localisation of FcgammaRIIa and efficient signaling are dependent on palmitoylation of cysteine 208. Immunol. Lett. 104, 118-123. ( 10.1016/j.imlet.2005.11.007) [DOI] [PubMed] [Google Scholar]
- 65.Norton RL, Fredericks GJ, Huang Z, Fay JD, Hoffmann FW, Hoffmann PR. 2017. Selenoprotein K regulation of palmitoylation and calpain cleavage of ASAP2 is required for efficient FcγR-mediated phagocytosis. J. Leukoc. Biol. 101, 439-448. ( 10.1189/jlb.2A0316-156RR) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang J, et al. 2019. DHHC4 and DHHC5 facilitate fatty acid uptake by palmitoylating and targeting CD36 to the plasma membrane. Cell Rep. 26, 209-221.e5. ( 10.1016/j.celrep.2018.12.022) [DOI] [PubMed] [Google Scholar]
- 67.Hao J-W, et al. 2020. CD36 facilitates fatty acid uptake by dynamic palmitoylation-regulated endocytosis. Nat. Commun. 11, 4765. ( 10.1038/s41467-020-18565-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zuckerman DM, Hicks SW, Charron G, Hang HC, Machamer CE. 2011. Differential regulation of two palmitoylation sites in the cytoplasmic tail of the beta1-adrenergic receptor. J. Biol. Chem. 286, 19 014-19 023. ( 10.1074/jbc.M110.189977) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Blanpain C, et al. 2001. Palmitoylation of CCR5 is critical for receptor trafficking and efficient activation of intracellular signaling pathways. J. Biol. Chem. 276, 23 795-23 804. ( 10.1074/jbc.M100583200) [DOI] [PubMed] [Google Scholar]
- 70.Badawy SMM, Okada T, Kajimoto T, Ijuin T, Nakamura S-I. 2017. DHHC5-mediated palmitoylation of S1P receptor subtype 1 determines G-protein coupling. Sci. Rep. 7, 16552. ( 10.1038/s41598-017-16457-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Claudinon J, Gonnord P, Beslard E, Marchetti M, Mitchell K, Boularan C, Johannes L, Eid P, Lamaze C. 2009. Palmitoylation of interferon-alpha (IFN-alpha) receptor subunit IFNAR1 is required for the activation of Stat1 and Stat2 by IFN-alpha. J. Biol. Chem. 284, 24 328-24 340. ( 10.1074/jbc.M109.021915) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ren W, Jhala US, Du K. 2013. Proteomic analysis of protein palmitoylation in adipocytes. Adipocyte 2, 17-28. ( 10.4161/adip.22117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang M, et al. 2020. A STAT3 palmitoylation cycle promotes TH17 differentiation and colitis. Nature 586, 434-439. ( 10.1038/s41586-020-2799-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zingler P, et al. 2019. Palmitoylation is required for TNF-R1 signaling. Cell Commun. Signal. 17, 90. ( 10.1186/s12964-019-0405-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yount JS, et al. 2010. Palmitoylome profiling reveals S-palmitoylation-dependent antiviral activity of IFITM3. Nat. Chem. Biol. 6, 610-614. ( 10.1038/nchembio.405) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Utsumi T, et al. 2001. Transmembrane TNF (pro-TNF) is palmitoylated. FEBS Lett. 500, 1-6. ( 10.1016/S0014-5793(01)02576-5) [DOI] [PubMed] [Google Scholar]
- 77.Klíma M, Zájedová J, Doubravská L, Andera L. 2009. Functional analysis of the posttranslational modifications of the death receptor 6. Biochim. Biophys. Acta 1793, 1579-1587. ( 10.1016/j.bbamcr.2009.07.008) [DOI] [PubMed] [Google Scholar]
- 78.Rossin A, Derouet M, Abdel-Sater F, Hueber A-O.. 2009. Palmitoylation of the TRAIL receptor DR4 confers an efficient TRAIL-induced cell death signalling. Biochem. J. 419, 185-192. ( 10.1042/BJ20081212) [DOI] [PubMed] [Google Scholar]
- 79.Jiang H, Zhang X, Chen X, Aramsangtienchai P, Tong Z, Lin H. 2018. Protein lipidation: occurrence, mechanisms, biological functions, and enabling technologies. Chem. Rev. 118, 919-988. ( 10.1021/acs.chemrev.6b00750) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ivaldi C, Martin BR, Kieffer-Jaquinod S, Chapel A, Levade T, Garin J, Journet A. 2012. Proteomic analysis of S-acylated proteins in human B cells reveals palmitoylation of the immune regulators CD20 and CD23. PLoS ONE 7, e37187. ( 10.1371/journal.pone.0037187) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Morrison E, Kuropka B, Kliche S, Brügger B, Krause E, Freund C. 2015. Quantitative analysis of the human T cell palmitome. Sci. Rep. 5, 11598. ( 10.1038/srep11598) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wilson JP, Raghavan AS, Yang Y-Y, Charron G, Hang HC. 2011. Proteomic analysis of fatty-acylated proteins in mammalian cells with chemical reporters reveals S-acylation of histone H3 variants. Mol. Cell. Proteom. 10, M110.001198. ( 10.1074/mcp.M110.001198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yurchak LK, Sefton BM. 1995. Palmitoylation of either Cys-3 or Cys-5 is required for the biological activity of the Lck tyrosine protein kinase. Mol. Cell. Biol. 15, 6914-6922. ( 10.1128/MCB.15.12.6914) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kabouridis PS, Magee AI, Ley SC. 1997. S-acylation of LCK protein tyrosine kinase is essential for its signalling function in T lymphocytes. EMBO J. 16, 4983-4998. ( 10.1093/emboj/16.16.4983) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ladygina N, Martin BR, Altman A. 2011. Dynamic palmitoylation and the role of DHHC proteins in T cell activation and anergy. Adv. Immunol. 109, 1-44. ( 10.1016/B978-0-12-387664-5.00001-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Harder T, Kuhn M. 2000. Selective accumulation of raft-associated membrane protein LAT in T cell receptor signaling assemblies. J. Cell Biol. 151, 199-208. ( 10.1083/jcb.151.2.199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hundt M, et al. 2006. Impaired activation and localization of LAT in anergic T cells as a consequence of a selective palmitoylation defect. Immunity 24, 513-522. ( 10.1016/j.immuni.2006.03.011) [DOI] [PubMed] [Google Scholar]
- 88.Cherukuri A, Carter RH, Brooks S, Bornmann W, Finn R, Dowd CS, Pierce SK. 2004. B cell signaling is regulated by induced palmitoylation of CD81. J. Biol. Chem. 279, 31 973-31 982. ( 10.1074/jbc.M404410200) [DOI] [PubMed] [Google Scholar]
- 89.Haag SM, et al. 2018. Targeting STING with covalent small-molecule inhibitors. Nature 559, 269-273. ( 10.1038/s41586-018-0287-8) [DOI] [PubMed] [Google Scholar]
- 90.Hansen AL, et al. 2018. Nitro-fatty acids are formed in response to virus infection and are potent inhibitors of STING palmitoylation and signaling. Proc. Natl Acad. Sci. USA 115, E7768-E7775. ( 10.1073/pnas.1806239115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hansen AL, Mukai K, Schopfer FJ, Taguchi T, Holm CK. 2019. STING palmitoylation as a therapeutic target. Cell. Mol. Immunol. 16, 236-241. ( 10.1038/s41423-019-0205-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang Y-C, Westcott NP, Griffin ME, Hang HC. 2019. Peptidoglycan metabolite photoaffinity reporters reveal direct binding to intracellular pattern recognition receptors and arf gtpases. ACS Chem. Biol. 14, 405-414. ( 10.1021/acschembio.8b01038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sobocińska J, Roszczenko-Jasińska P, Ciesielska A, Kwiatkowska K. 2017. Protein palmitoylation and its role in bacterial and viral infections. Front. Immunol. 8, 2003. ( 10.3389/fimmu.2017.02003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Peng T, Hang HC. 2018. Chemical proteomic profiling of protein fatty-acylation in microbial pathogens. Curr. Top. Microbiol. Immunol. 420, 93-110. [DOI] [PubMed] [Google Scholar]
- 95.Lee M, Sugiyama M, Mekhail K, Latreille E, Negar K, Wei K, Lee WL, Antonescu C, Fairn GD. 2020. Fatty acid synthase inhibition prevents palmitoylation of SARS-CoV-2 spike protein and improves survival of mice infected with murine hepatitis virus. bioRxiv 423603. ( 10.1101/2020.12.20.423603) [DOI] [Google Scholar]
- 96.Perreira JM, Chin CR, Feeley EM, Brass AL. 2013. IFITMs restrict the replication of multiple pathogenic viruses. J. Mol. Biol. 425, 4937-4955. ( 10.1016/j.jmb.2013.09.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Savidis G, Perreira JM, Portmann JM, Meraner P, Guo Z, Green S, Brass A. 2016. The ifitms inhibit zika virus replication. Cell Rep. 15, 2323-2330. ( 10.1016/j.celrep.2016.05.074) [DOI] [PubMed] [Google Scholar]
- 98.Huang I-C, et al. 2011. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 7, e1001258. ( 10.1371/journal.ppat.1001258) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhao X, Guo F, Liu F, Cuconati A, Chang J, Block TM, Guo J-T. 2014. Interferon induction of IFITM proteins promotes infection by human coronavirus OC43. Proc. Natl Acad. Sci. USA 111, 6756-6761. ( 10.1073/pnas.1320856111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhao X, et al. 2018. Identification of residues controlling restriction versus enhancing activities of IFITM proteins on entry of human coronaviruses. J. Virol. 92, e01535-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang X, et al. 2020. Viral and host factors related to the clinical outcome of COVID-19. Nature 583, 437-440. ( 10.1038/s41586-020-2355-0) [DOI] [PubMed] [Google Scholar]
- 102.Buchrieser J, et al. 2020. Syncytia formation by SARS-CoV-2-infected cells. EMBO J. 39, e106267. ( 10.15252/embj.2020106267) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zheng M, et al. 2020. Bat SARS-Like WIV1 coronavirus uses the ACE2 of multiple animal species as receptor and evades IFITM3 restriction via TMPRSS2 activation of membrane fusion. Emerg. Microbes Infect. 9, 1567-1579. ( 10.1080/22221751.2020.1787797) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zang R, et al. 2020. Cholesterol 25-hydroxylase suppresses SARS-CoV-2 replication by blocking membrane fusion. bioRxiv 141077. ( 10.1101/2020.06.08.141077) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shi G, et al. 2021. Opposing activities of IFITM proteins in SARS-CoV-2 infection. EMBO J. 40, e106501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lee J, et al. 2020. IFITM3 functions as a PIP3 scaffold to amplify PI3 K signalling in B cells. Nature 588, 491-497. ( 10.1038/s41586-020-2884-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zani A, Zhang L, McMichael TM, Kenney AD, Chemudupati M, Kwiek JJ, Liu S-L, Yount JS. 2019. Interferon-induced transmembrane proteins inhibit cell fusion mediated by trophoblast syncytins. J. Biol. Chem. 294, 19 844-19 851. ( 10.1074/jbc.AC119.010611) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Buchrieser J, et al. 2019. IFITM proteins inhibit placental syncytiotrophoblast formation and promote fetal demise. Science 365, 176-180. [DOI] [PubMed] [Google Scholar]
- 109.Wakim LM, Gupta N, Mintern JD, Villadangos JA. 2013. Enhanced survival of lung tissue-resident memory CD8+ T cells during infection with influenza virus due to selective expression of IFITM3. Nat. Immunol. 14, 238-245. ( 10.1038/ni.2525) [DOI] [PubMed] [Google Scholar]
- 110.Yánez DC, et al. 2019. IFITM proteins drive type 2T helper cell differentiation and exacerbate allergic airway inflammation. Eur. J. Immunol. 49, 66-78. ( 10.1002/eji.201847692) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hur J-Y, et al. 2020. The innate immunity protein IFITM3 modulates γ-secretase in Alzheimer's disease. Nature 586, 735-740. ( 10.1038/s41586-020-2681-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hach JC, McMichael T, Chesarino NM, Yount JS. 2013. Palmitoylation on conserved and nonconserved cysteines of murine IFITM1 regulates its stability and anti-influenza A virus activity. J. Virol. 87, 9923-9927. ( 10.1128/JVI.00621-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Thinon E, Fernandez JP, Molina H, Hang HC. 2018. Selective enrichment and direct analysis of protein S-palmitoylation sites. J. Proteome Res. 17, 1907-1922. ( 10.1021/acs.jproteome.8b00002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Benfield, et al. In press. Bat IFITM3 restriction depends on S-palmitoylation and a polymorphic site within the CD225 domain. Life Sci. Alliance. ( 10.26508/lsa.201900542) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.John SP, et al. 2013. The CD225 domain of IFITM3 is required for both IFITM protein association and inhibition of influenza A virus and dengue virus replication. J. Virol. 87, 7837-7852. ( 10.1128/JVI.00481-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Melvin WJ, McMichael TM, Chesarino NM, Hach JC, Yount JS. 2015. IFITMs from mycobacteria confer resistance to influenza virus when expressed in human cells. Viruses 7, 3035-3052. ( 10.3390/v7062759) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Spence JS, et al. 2019. IFITM3 directly engages and shuttles incoming virus particles to lysosomes. Nat. Chem. Biol. 15, 259-268. ( 10.1038/s41589-018-0213-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Suddala KC, et al. 2019. Interferon-induced transmembrane protein 3 blocks fusion of sensitive but not resistant viruses by partitioning into virus-carrying endosomes. PLoS Pathog. 15, e1007532. ( 10.1371/journal.ppat.1007532) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chesarino NM, Compton AA, McMichael TM, Kenney AD, Zhang L, Soewarna V, et al. 2017. IFITM3 requires an amphipathic helix for antiviral activity. EMBO Rep. 18, 1740-1751. ( 10.15252/embr.201744100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.McMichael TM, et al. 2017. The palmitoyltransferase ZDHHC20 enhances interferon-induced transmembrane protein 3 (IFITM3) palmitoylation and antiviral activity. J. Biol. Chem. 292, 21 517-21 526. ( 10.1074/jbc.M117.800482) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Poggi M, et al. 2013. Palmitoylation of TNF alpha is involved in the regulation of TNF receptor 1 signalling. Biochim. Biophys. Acta 1833, 602-612. ( 10.1016/j.bbamcr.2012.11.009) [DOI] [PubMed] [Google Scholar]
- 122.Feig C, Tchikov V, Schütze S, Peter ME. 2007. Palmitoylation of CD95 facilitates formation of SDS-stable receptor aggregates that initiate apoptosis signaling. EMBO J. 26, 221-231. ( 10.1038/sj.emboj.7601460) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.