Abstract
Chronic hypoxia is a major contributor to Chronic Kidney Disease (CKD) after Acute Kidney Injury (AKI). However, the temporal relation between the acute insult and maladaptive renal response to hypoxia remains unclear. In this study, we analyzed the time-course of renal hemodynamics, oxidative stress, inflammation, and fibrosis, as well as epigenetic modifications, with focus on HIF1α/VEGF signaling, in the AKI to CKD transition. Sham-operated, right nephrectomy (UNx), and UNx plus renal ischemia (IR + UNx) groups of rats were included and studied at 1, 2, 3, or 4 months. The IR + UNx group developed CKD characterized by progressive proteinuria, renal dysfunction, tubular proliferation, and fibrosis. At first month post-ischemia, there was a twofold significant increase in oxidative stress and reduction in global DNA methylation that was maintained throughout the study. Hif1α and Vegfa expression were depressed in the first and second-months post-ischemia, and then Hif1α but not Vegfa expression was recovered. Interestingly, hypermethylation of the Vegfa promoter gene at the HIF1α binding site was found, since early stages of the CKD progression. Our findings suggest that renal hypoperfusion, inefficient hypoxic response, increased oxidative stress, DNA hypomethylation, and, Vegfa promoter gene hypermethylation at HIF1α binding site, are early determinants of AKI-to-CKD transition.
Subject terms: Physiology, Nephrology, Pathogenesis
Introduction
Clinical, epidemiological, and experimental studies have shown that AKI is an independent risk factor for the development of CKD and end-stage renal disease (ESRD)1,2. In this transition, the initial insult severity and duration is proportional to the risk of CKD, besides, age is another preponderant factor3–6.
AKI is characterized by an abrupt reduction in renal blood flow with consequent hypoxia, endothelial and proximal epithelial injury, and renal dysfunction. After an AKI episode, a cascade of events occurs, such as brush border loss, cell polarity alterations, increased oxidative stress, and mitochondrial dysfunction of proximal tubular epithelial cells7,8, as a result, some of these cells undergo necrosis or apoptosis9. These processes are also accompanied by macrophage infiltration and inflammation7,10,11. Although the tubular epithelium has the capacity for regeneration12, the injured epithelium is no longer the same. A subpopulation of dedifferentiated and proliferating tubular cells that recover from the acute renal insult suffer cell cycle arrest and cannot be re-differentiated, leading to tubular atrophy; all these events contribute greatly to the tubulointerstitial fibrosis that is observed in the long term12,13.
Consequently, a maladaptive repair occurs11,14,15, where there is persistent macrophage infiltration16–18, dissociation of pericytes from the tubular capillaries19, and arrest of some tubular cells in the G2/M phase20, leading to a progressive fibrotic kidney20–22. Some mechanisms involved have been elucidated, such as trans-differentiation of pericytes into myofibroblasts19,23, uncontrolled proliferation of epithelial cells22, the emergence of an abnormal proximal-tubule phenotype24, excessive production of TGFβ by both the tubular epithelium22 and local myofibroblasts25,26, accumulation of extracellular matrix proteins27,28, vascular rarefaction29–31, chronic hypoxia30,32, and chronic stress of the endoplasmic reticulum33.
In addition, it has been recently shown that epigenetic modifications may be involved in several renal pathologies. However, the specific molecular mechanisms by which epigenetic modifications alter renal physiology are little known. The most studied epigenetic regulations in AKI are the chromatin compaction, DNA methylation and histone acetylation/deacetylation. In AKI, obstructive renal injury, and diabetic nephropathy, epigenetic modifications induced an increase in proinflammatory and profibrotic cytokines such as monocyte chemoattractant protein-1 (MCP-1), complement protein 3 (C3), transforming growth factor β (TGF-β), which in turn perpetuate inflammation and promote epithelial-to-mesenchymal transition (EMT) that contributes to renal fibrosis34–38. Although some mechanisms have been elucidated, many others remain unknown, and less is known about temporal changes during CKD progression, such as renal hemodynamics, structural injury, HIF signaling, and epigenetic modifications.
We have previously shown that an AKI episode induced by moderate or severe bilateral renal ischemia/reperfusion (IR) in male rats is sufficient to induce CKD progression after nine months22,27. Interestingly, this transition was not observed in female rats, despite a similar magnitude of AKI in both male and female rats. The unique difference in the early phase post-ischemia was that females did not exhibit oxidative stress, suggesting a pivotal role of reactive oxygen species generation in this transition21.
Thus, it is relevant to establish a temporal understanding of the pathophysiological mechanisms throughout the AKI-to-CKD transition. In this study, we used the model of unilateral renal IR plus contralateral nephrectomy, which allowed us to induce CKD after four months. We found abnormal renal hemodynamics, reduced HIF-1α signaling, increased oxidative stress, and global DNA hypomethylation in the early phase of the AKI to CKD transition. We also showed that the HIF-1α/Vegfa signaling reduction was associated with the DNA hypermethylation of the Vegfa gene promoter, beginning at an early stage post-ischemia and suggesting that reduced VEGF expression is an early contributor that triggers renal hypoxia and the consequent fibrosis.
Results
Renal injury induced by unilateral ischemia after 24 h of reperfusion
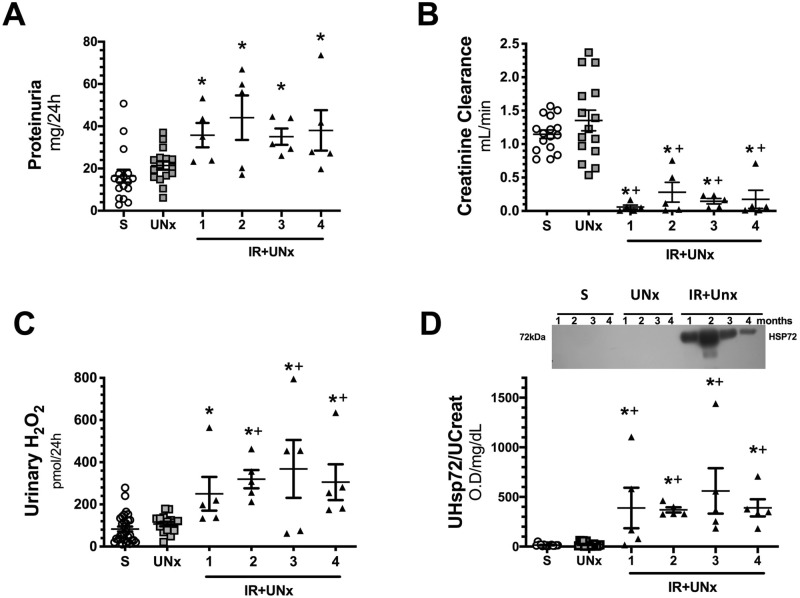
First, we corroborated that the initial insult induced by IR in the uninephrectomized rats was similar among the groups studied at 1, 2, 3, or 4 months postischemia. All IR + UNx rats were randomly assigned to the different periods of follow-up. After 24 h of inducing renal ischemia, all the IR + UNx rats exhibited significant proteinuria that was of the same magnitude among the groups assigned to 1, 2, 3 and, 4 months of follow-up (Fig. 1A), together with a similar reduction in renal function (Fig. 1B); these alterations were not observed in the S (n = 16) or UNx (n = 16) groups after 24 h of the surgery as is shown by the individual data presented in Fig. 1A, B. Consequently, the S or UNx rats were also randomly assigned to the different periods of follow-up. The urinary hydrogen peroxide levels were also evaluated and reflected significant oxidative stress in all IR + UNx groups (Fig. 1C). The urinary HSP72 levels, known to be a sensitive AKI biomarker, were also analyzed39–42. As expected, all rats that underwent IR + UNx exhibited a significant and similar increase in urinary HSP72 levels corrected by urinary creatinine (UHSP72/UCreat), (Fig. 1D). These findings show that all the IR + UNx rats exhibited a similar AKI degree. This was important to ensure that the changes observed in the long term were due to the initial insult itself rather than differences in the severity of the ischemic injury.
Figure 1.
Renal injury induced by unilateral ischemia after 24 h of reperfusion. (A) Proteinuria, (B) Creatinine clearance, (C) Urinary hydrogen peroxide, and (D) Urinary HSP72 corrected by urinary creatinine, including a representative cropped blot image. Data are represented as the mean ± SE (for S, n = 16, for UNx, n = 16 and n = 20 for IR + UNx groups). White bars represent the S, gray bars represent UNx, and black bars represent IR + UNx groups. The one-way analysis of variance (ANOVA) was used to determine statistical differences, using the Bonferroni correction for multiple comparisons. *p < 0.05 versus S group and + p < 0.05 versus UNx group. Full-length blots are presented in Supplementary Fig. 3.
Temporal progression of renal dysfunction and structural injury after an AKI episode
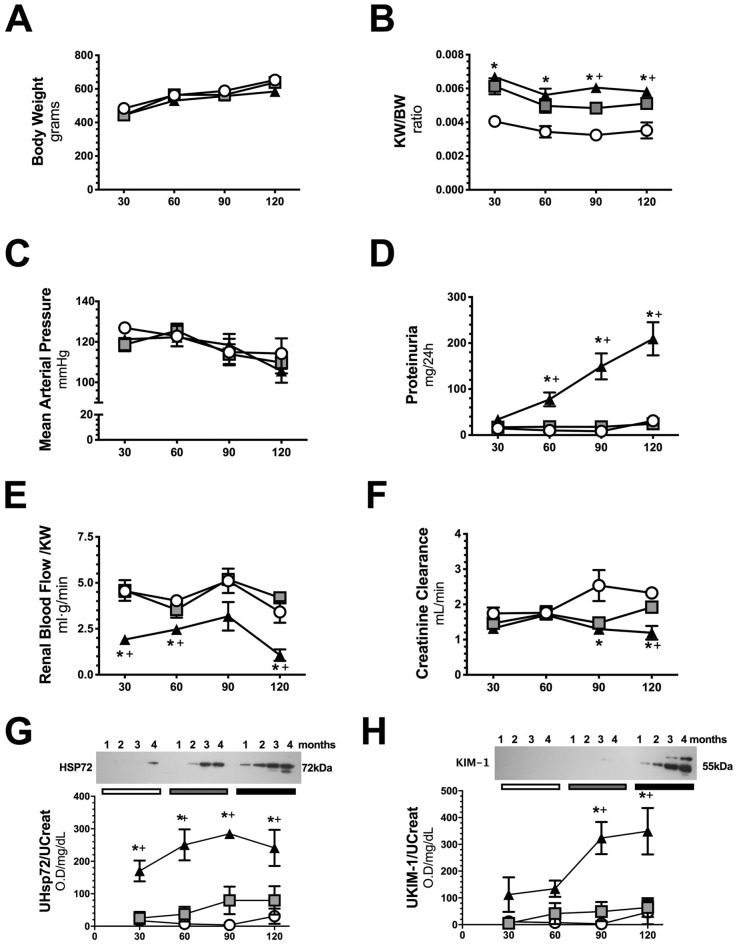
To evaluate the precise moment at which the functional, structural, and molecular alterations occur along with AKI to CKD transition, the groups were euthanized at 1, 2, 3, or 4 months after the initial ischemic insult. No differences in body weight were found among the studied groups (Fig. 2A). As expected, the UNx and IR + UNx groups showed a significant increase in kidney weight/body weight (KW/BW) starting in the first-month compared to that of the S group (Fig. 2B). All experimental groups remained normotensive at the time of evaluation (Fig. 2C). The IR + UNx group exhibited a progressive increase in proteinuria starting in the second month of follow-up that was not evident in the S and UNx groups (Fig. 2D). Due to the renal mass reduction, the UNx group exhibited renal hyperperfusion, but when it was correct to the kidney weight, renal blood flow (RBF) was similar to the S group (Fig. 2E). This finding, in the UNx group, was related to the maintenance of normal renal function (Fig. 2F). Interestingly, this compensatory response was not seen in the IR + UNx group. RBF/KW was significantly lower than that of the S and UNx groups, starting in the first-month, and renal hypoperfusion was maintaining along the study course (Fig. 2E). At the end of the study, the IR + UNx group had significant renal dysfunction (Fig. 2F). The renal urinary biomarker HSP72 and KIM1 normalized by urinary creatinine, were significantly elevated starting in the first-month and third-month respectively, and increased even more by the end of the study (Fig. 2G, H). In Table 1 appears urinary flow, fractional excretion of sodium (FENa) and osmolarity. No differences were found among the groups along the study, except that the osmolarity was lower in the IR + UNx group at fourth-month compared to S group.
Figure 2.
Follow-up of renal function in the AKI to CKD transition. (A) Body weight, (B) Ratio kidney weight/body weight, (C) Mean arterial Pressure, (D) Proteinuria, (E) Renal blood flow / kidney weight, (F) Creatinine clearance, (G) Urinary HSP72 corrected by urinary creatinine, and (H) Urinary KIM-1 corrected by urinary creatinine, including representative cropped blots, along the follow-up. Data are represented as the mean ± SE. n = 4, 4, and 5 for the S, UNx, and IR + UNx groups in each studied period: 30, 60, 90, and 120 days post-ischemia. White circles represent the S, gray squares represent UNx and black triangles represent IR + UNx groups. The one-way analysis of variance (ANOVA) was used to determine statistical differences, using the Bonferroni correction for multiple comparisons. *p < 0.05 versus S group and + p < 0.05 versus UNx group in their respective period. Full-length blots are presented in Supplementary Figs. 4 and 5.
Table 1.
Urine Chemistries along the study for all the included groups.
| Urinary flow (mL/min) | FENa (%) | Osmolarity (mOsm/L) | |
|---|---|---|---|
| Sham | |||
| 30 days | 0.031 ± 0.004 | 0.17 ± 0.03 | 350 ± 77 |
| 60 days | 0.026 ± 0.007 | 0.10 ± 0.00 | 481 ± 107 |
| 90 days | 0.023 ± 0.003 | 0.11 ± 0.02 | 729 ± 123 |
| 120 days | 0.014 ± 0.003 | 0.08 ± 0.03 | 1226 ± 178 |
| UNx | |||
| 30 days | 0.029 ± 0.005 | 0.19 ± 0.04 | 494 ± 172 |
| 60 days | 0.025 ± 0.005 | 0.15 ± 0.05 | 703 ± 119 |
| 90 days | 0.014 ± 0.007 | 0.16 ± 0.04 | 936 ± 72 |
| 120 days | 0.024 ± 0.004 | 0.09 ± 0.02 | 746 ± 129 |
| IR + UNx | |||
| 30 days | 0.026 ± 0.005 | 0.21 ± 0.07 | 448 ± 59 |
| 60 days | 0.037 ± 0.003 | 0.26 ± 0.14 | 440 ± 39 |
| 90 days | 0.030 ± 0.002 | 0.24 ± 0.07 | 501 ± 30 |
| 120 days | 0.024 ± 0.004 | 0.20 ± 0.08 | 678 ± 145* |
*p < 0.05 versus respective Sham.
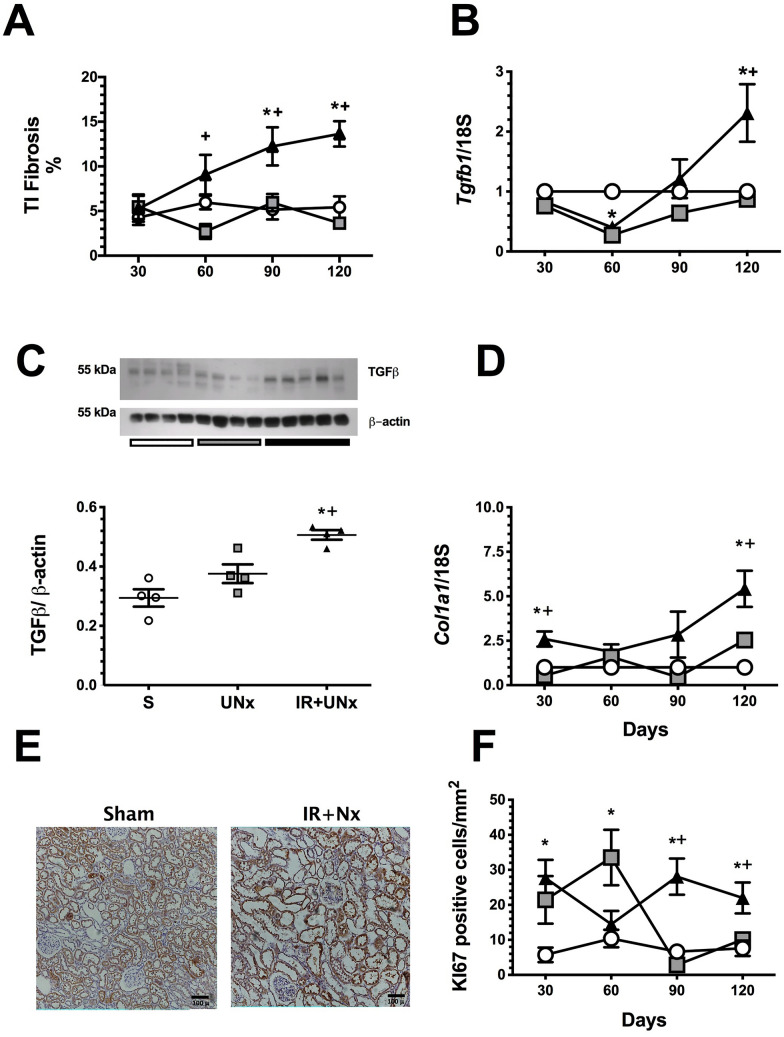
The long-term consequences of an AKI episode were also evidenced by the presence of tubulointerstitial fibrosis starting in the second-month post-ischemia, which progressively increased, whereas this injury was not detected in the UNx group (Fig. 3A). Although increased Tgfb1 mRNA levels were not observed in the early stages of the AKI to CKD transition, a significant upregulation in Tgfb1 mRNA and protein levels was evident in the fourth-month post-ischemia compared to that of the S and UNx groups (Fig. 3B, C). Accordingly with this, Col1a1 (collagen 1) mRNA levels were significantly increased in the fourth-month (Fig. 3D). We only measured TGFβ protein levels at the fourth-month, because the Tgfb1 mRNA levels and its target gene Col1a1 were only significantly increased in this point of the follow-up. Besides, the IR + UNx group exhibited higher levels of Ki67 positive tubular cells (Fig. 3E, F), similar to our previous findings22.
Figure 3.
Temporally induction of tubulointersticial fibrosis by TGFβ activation. (A) Tubulointersticial fibrosis, (B) mRNA levels of Tgfb1, (C) Protein levels of TGFβ, at fourth-month post-ischemia, including a representative cropped blots. (D) mRNA levels of Collagen1a1, (E) Representative microphotographs of Ki67 immunostaining for the S and IR + UNx groups. (F) Quantification of Ki67 positive epithelial cells (cells/mm2). Data are represented as the mean ± SE. n = 4, 4, and 5 for S, UNx, and IR + UNx groups, in each studied period: 30, 60, 90, and 120 days post-ischemia. White circles/bar represent the S, gray squares/bar represent the UNx and black triangles/bar represent the IR + UNx groups. The one-way analysis of variance (ANOVA) was used to determine statistical differences, using the Bonferroni correction for multiple comparisons. *p < 0.05 versus S group and + p < 0.05 versus UNx group in their respective period. Full-length blots are presented in Supplementary Fig. 6.
These results show that an AKI episode induced functional and structural alterations that mostly appear beginning at an early stage, highlighting the fact that there was no compensatory renal hyperperfusion expected by the renal mass lost in the IR + UNx group.
Temporal course of oxidative stress and vasoactive factors in the CKD progression induced by an AKI episode
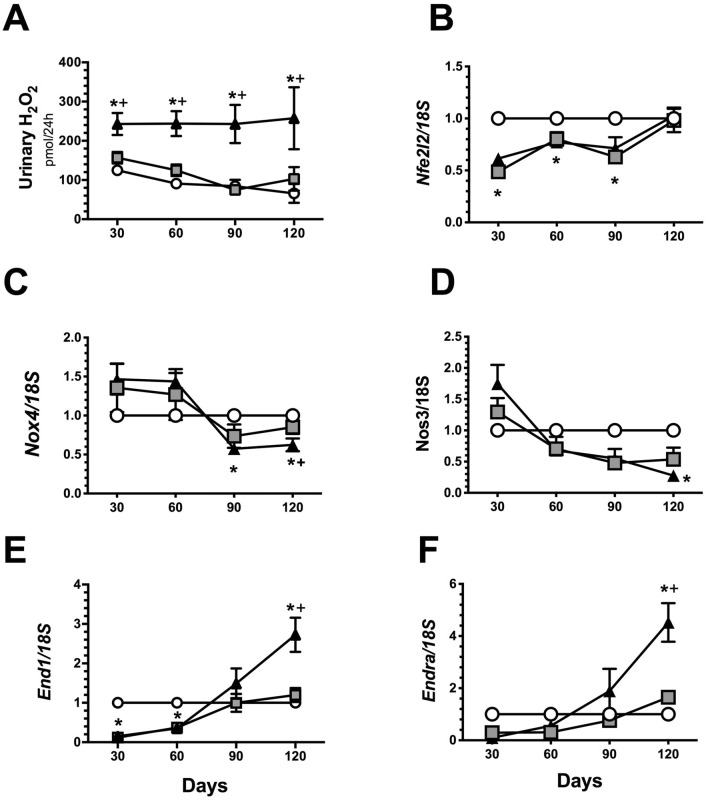
Oxidative stress and renal inflammation have a pivotal role in CKD progression; thus, the temporality of these two pathways was also analyzed. An increase in oxidative stress (Fig. 4A) and a reduction in mRNA levels of the transcription factor Nfe2l2, which stimulates the antioxidant response, was observed beginning in the initial stage of the AKI to CKD transition (Fig. 4B), even though Nox4 mRNA levels were reduced in the late stage of the disease (Fig. 4C). An imbalance in vasoactive factors was also observed at the end of the study. NOS3 mRNA levels were significantly decreased (Fig. 4D), whereas the endothelin vasoconstrictor effect was increased (Fig. 4E, F).
Figure 4.
Oxidative stress and vasoactive mediators over the course of AKI to CKD transition. (A) Urinary hydrogen peroxide, (B) mRNA levels of Nfe2l2 (C) mRNA levels of Nos3, (D) mRNA levels of NOX4, (E) mRNA levels of Edn1 and (F) mRNA levels of endothelin receptor A (Endra). Data are represented as the mean ± SE. n = 4, n = 4, and n = 5 for the S, UNx, and IR + UNx groups, in each studied period: 30, 60, 90, and 120 days post-ischemia. The one-way analysis of variance (ANOVA) was used to determine statistical differences, using the Bonferroni correction for multiple comparisons. *p < 0.05 versus S group and + p < 0.05 versus UNx group in their respective period.
Inflammatory pathways in the time course of CKD induced by AKI
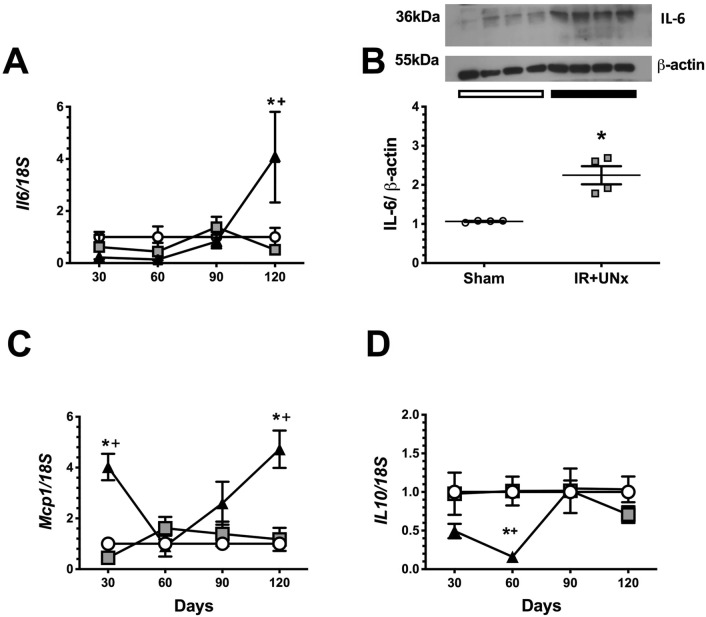
The mRNA levels of interleukin 6 (Il6), monocyte chemoattract protein (Mcp1), and interleukin 10 (Il10) were measured throughout the study. Il6 was upregulated in the IR + UNx group in the fourth-month after renal ischemia, as demonstrated by the significant elevation in interleukin 6 mRNA and protein levels (Fig. 5A, B). Because, we only observed a significant increase in Il6 mRNA levels in the IR + UNx group at fourth-month post-ischemia, the IL6 protein levels were only evaluated in the Sham and IR + UNx groups in this specific time of the study. Mcp1 mRNA levels increased in the first-month, and this elevation was observed again in the fourth month (Fig. 5C). The mRNA levels of the anti-inflammatory cytokine Il10 showed a reduction starting in the first-month that became significant in the second month and returned to normal levels by the third month compared to that of the S and UNx groups (Fig. 5D).
Figure 5.
Inflammatory mediators participation during AKI to CKD transition. (A) mRNA levels of Il6, (B) Protein levels of IL-6 at fourth-month post-ischemia, including a representative cropped blots, (C) mRNA levels of Mcp1 and (D) mRNA levels of Il10. Data are represented as the mean ± SE. n = 4, 4, and 5 for the S, UNx, and IR + UNx groups in each studied period: 30, 60, 90, and 120 days post-ischemia. White circles/bar represent the S, gray squares represent the UNx and black triangles/bar represent the IR + UNx groups. The one-way analysis of variance (ANOVA) was used to determine statistical differences, using the Bonferroni correction for multiple comparisons. *p < 0.05 versus S group and + p < 0.05 versus UNx group in their respective period. Full-length blots are presented in Supplementary Fig. 7.
These findings suggest that oxidative stress, rather than renal inflammation, is an initial trigger of the subsequent damage.
Hypoxic response in the timing of CKD progression
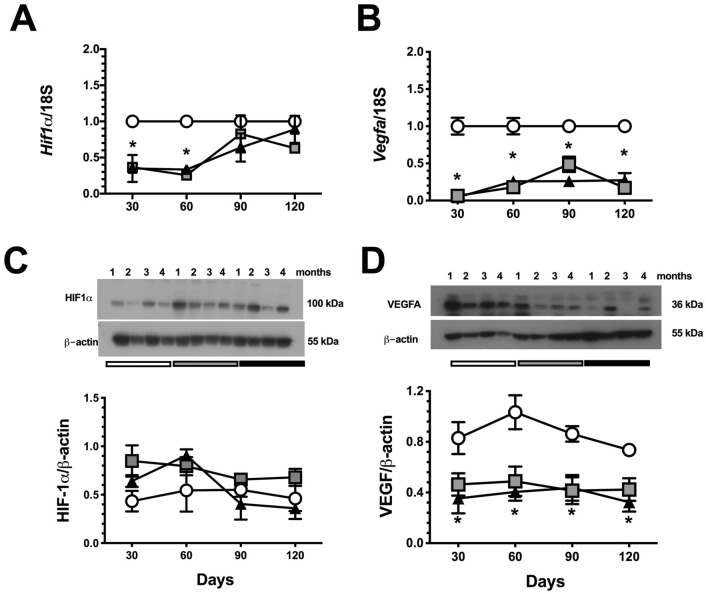
As we showed, the UNx group exhibited renal hyperperfusion starting in the first-month post ischemia (Fig. 2E). In contrast, because renal compensatory hyperperfusion was absent in the IR + UNx group, the HIF1α signaling pathway was studied. The gene expression Hif1a and one of its target genes, Vegfa, were assessed during AKI to CKD transition. In the UNx group, Hif1a mRNA levels were significantly reduced in the first and second-month post-ischemia, whereas HIF1α protein levels remained unaltered during follow-up. This response was explained and expected in part by the compensatory renal hyperperfusion seen in these rats. In contrast, there was an inefficient response to hypoxia in the IR + UNx group that exhibited renal hypoperfusion because there was a significant decrease in Hif1a mRNA levels in the first and second-month after ischemia (Fig. 6A). In support of this inefficient response, Vegfa mRNA and protein levels were significantly reduced starting in the first-month and remained so on during follow-up (Fig. 6B–D), despite Hif1a mRNA and protein levels being reestablished by the third-month, suggesting that an independent mechanism maintains Vegfa gene expression downregulation and could be related with the vascular rarefaction characteristic of the AKI to CKD transition29–31.
Figure 6.
Temporal course of HIF-1α signaling during AKI to CKD transition. (A) mRNA levels of Hif1a, (B) mRNA levels of Vegfa, (C) Protein levels of HIF-1α, including a representative cropped blot, and (D) Protein levels of VEGF, including a representative cropped blot. Data are represented as the mean ± SE. n = 4, for S, UNx, and IR + UNx groups, in each studied period: 30, 60, 90, and 120 days post-ischemia. White circles represent the S, gray squares represent the UNx and black triangles represent the IR + UNx groups. The one-way analysis of variance (ANOVA) was used to determine statistical differences, using the Bonferroni correction for multiple comparisons. *p < 0.05 versus S group and + p < 0.05 versus UNx group in their respective period. Full-length blots are presented in Supplementary Figs. 8 and 9.
Time course global DNA methylation during CKD progression induced by AKI
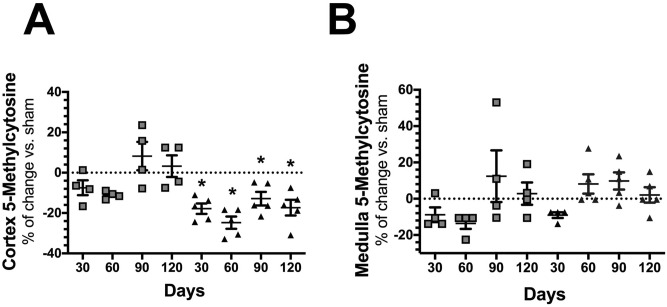
Numerous studies have demonstrated that renal injury is associated with epigenetic changes, including histone modifications, DNA methylation, and the expression of various non‐coding RNAs43. We found that rats experiencing the AKI to CKD transition exhibited mainly hypomethylation of global DNA, which started in the first-month post-ischemia and was maintained throughout follow-up (Fig. 7A). These changes were only seen in the renal cortex, whereas no differences were observed in the renal medulla (Fig. 7B). In general, the renal medulla exhibited lower levels of DNA methylation compared to that of the renal cortex.
Figure 7.
Global DNA methylation over the course of AKI to CKD transition. (A) Percentage change of global DNA methylation in renal cortex from the UNx (gray squares) and IR + UNx (black triangles) groups. (B) Percentage change of global DNA methylation in renal medulla from the UNx (gray squares) and IR + UNx (black triangles) groups, for each studied period: 30, 60, 90, and 120 days The one-way analysis of variance (ANOVA) was used to determine statistical differences, using the Bonferroni correction for multiple comparisons. *p < 0.05 versus S group.
Based on the changes observed in the global methylation of DNA and the possible independent mechanisms regulating the decreased expression of VEGF in the AKI to CKD transition, we decided to evaluate the specific methylation of the Vegfa gene promoter.
Vegfa gene promoter DNA methylation during AKI to CKD transition
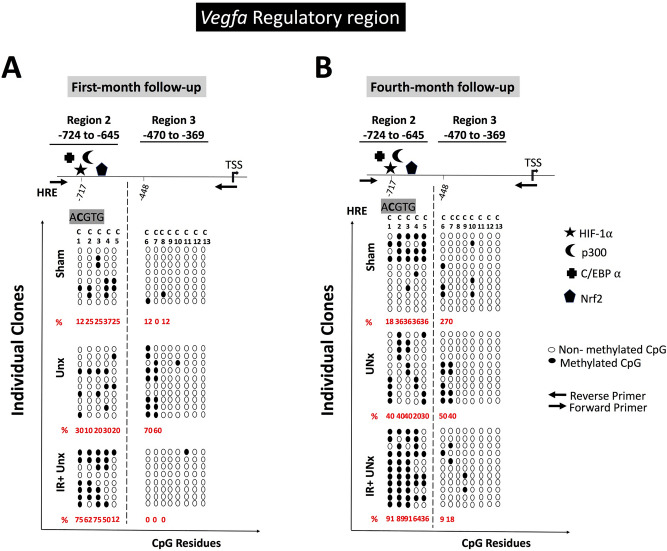
With the interest to know if DNA methylation was associated with the modulation of Vegfa and Hif1a expression in AKI to CKD transition, we assessed the methylation state on 5′-upstream promoter region by bisulfite sequencing. According to the decreased Vegfa gene expression, we localized a DNA hypermethylated region in the noncoding upstream region of this gene, since the first-month post-ischemia (Fig. 8A), which was maintained until the fourth-month of follow-up (Fig. 8B). Therefore, we discovered that in the binding site for HIF1α, located at the region 2 of the Vegfa gene promoter, contains a specific CpG that was highly methylated in the IR + UNx group since the first-month and (75%) compared to that of the S (12%) and UNx groups (30%) (Fig. 8A). Moreover, the hypermethylated region was maintained at the end of the study in the IR + UNx group (91%) compared to that of the S (18%) and UNx (40%) groups (Fig. 8B).
Figure 8.
Bisulfite sequencing of two non-codifying upstream regions of Vegfa promoter for (A) the first-month and (B) the fourth month of follow-up for the Sham, UNx and IR + UNx groups, respectively. Arrows represented the forward and reverse primers for each amplicon. HIF-1α, p300, C/EBPα, and Nrf2 binding sites as is stated. White circles represented non-methylated CpG, and black circles represented methylated CpG, each circle represented an individual clone. Each C represented a cytosine in CpG context. TSS-Transcription star site; HRE-Hypoxia response element (ACGTG).
Consistent with our transcriptional findings, we did not found any methylation in the noncoding upstream promoter region of Hif1a among the groups (Suppl. Fig. 2).
Discussion
Acute kidney injury is a public health problem and despite the advances in modern medicine, its incidence has not diminished in recent decades44. Also, the alarming increase in patients with CKD worldwide45, coupled with the recent recognition that AKI is an independent risk factor for CKD development1,2 requires joint work between biomedical and clinical researchers to avoid this complication. Therefore, it is imperative to study the temporality of the mechanisms that affect and lead to the AKI to CKD transition, which will allow for the identification of the key points and mediators in the development of this disease and propose specific therapeutic targets according to the stages of CKD. Many efforts have been made, and some pathways, such as trans-differentiation of pericytes into myofibroblasts19,23, uncontrolled proliferation of epithelial cells22, excessive production of TGFβ by both tubular epithelium22 and local myofibroblasts25,26, accumulation of extracellular matrix proteins27,28, chronic hypoxia30,32, vascular rarefaction29,31,46, and chronic stress of the endoplasmic reticulum33, have been identified, but many others remain to be elucidated.
In this study, we evaluated ischemic renal damage after 24 h of reperfusion plus right nephrectomy compared to that of the S and UNx groups. AKI induced by IR was characterized by elevated proteinuria, renal dysfunction, and oxidative stress. All IR + UNx rats exhibited the same magnitude of AKI. In the long term, the nephrectomy plus I/R model was able to accelerate the development of CKD. After four months, the IR + UNx group presented a progressive increase in proteinuria and a significant decrease in renal function. Among the alterations that occurred in the early phase of the transition, we found a significant increase in oxidative stress that was maintained throughout the study follow-up and a significant decrease in global DNA methylation, suggesting that both are early key players in the AKI to CKD transition.
As expected, the UNx group exhibited renal hyperperfusion and hyperfiltration to compensate the renal mass reduction for maintaining renal function within standard values, as the S group had. Interestingly, the IR + UNx group displayed sustained hypoperfusion that was observed beginning at an early phase and impacted the expected renal function compensation. Our results suggest that this poor renal hemodynamic response after an AKI episode could be one of the responsible mechanisms involved in the AKI to CKD transition, contributing to chronic renal hypoxia.
Previous studies have demonstrated that chronic hypoxia is a trigger mechanism in the AKI to CKD transition that coordinates the interaction between inflammation, oxidative stress, and progressive fibrosis13,16–18,20–22,47. Hif1α serves as a master regulator of adaptive responses against hypoxia, although, the levels of HIF1α can also be regulated by oxygen independent pathways48. This transcription factor induces an angiogenic pathway throughout the induction of Vegfa gene expression49,50. In this regard, we found an inefficient response against renal hypoxia in the IR + UNx group. Hif1α mRNA levels were decreased in the first and second-month post-ischemia, similar to those observed in the UNx group. Although, this response could be explained in the UNx group due to the compensatory elevation in RBF, which when normalized by kidney weight was similar to the S group. Since, HIF1α is regulated by proteasomal degradation pathways in response to oxygen levels51, it is very likely that in the UNx group, there is a greater metabolic work to maintain renal function and therefore, the state of renal oxygenation could not have changed. But this HIF1α response was unexpected in the IR + UNx group that exhibited renal hypoperfusion, a condition in which proteasomal degradation is not expected to occur. This inefficient response of HIF1α was also demonstrated by the reduction in its target gene Vegfa. Thus, Vegfa mRNA levels were reduced starting in the first-month, and interestingly, they remained reduced throughout the study, despite the restoration of HIF1α levels. The decreased Vegfa expression was also corroborated at the protein level throughout the study. Recent studies have reported that the late response of HIF1α could be related to the activation of inflammatory processes and the generation of renal fibrosis52. It is well known that the reduction in Vegfa gene expression is partly responsible for vascular rarefaction46,53–55 that accompanies the AKI to CKD transition, which also perpetuates chronic hypoxia. Although Hif1a mRNA levels were restored at the end of the study and protein levels remained unaltered, Vegfa levels continued diminishing; this opened the possibility of an alternative regulation of Vegfa gene expression through the involvement of epigenetic mechanisms.
It has been shown that the hypoxia response element (HRE) in the Vegfa promoter gene contains several cytosines in a CpG context that are potentially methylated. This methylation could reduce the HIF1α interaction with the Vegfa gene promoter56,57. In vitro studies have demonstrated that DNA hypermethylation in the Vegfa promoter region induces silencing of this gene58. To understand the reduction in Vegfa expression despite sustained renal hypoperfusion and recovery of Hif1a mRNA levels, we analyzed the methylation of the noncoding upstream region of Vegfa during the AKI to CKD transition. Interestingly, we found DNA hypermethylation in the promoter region of Vegfa. More importantly, we demonstrated that the HRE, as well as the core region for HIF1α interaction was hypermethylated since the first-month post-ischemia and it was maintained at the fourth-month of the follow-up. This site matches with the region 2, previously described to be important in Vegf gene transcription regulation59. Using the transcription factor binding site predictor tool PROMO (version 8.3 of TRANSFAC, http://alggen.lsi.upc.es), TFsitescan (http://www.ifti.org/cgi-bin/ifti/Tfsitescan.pl) and JASPAR database (http://dbcat.cgm.ntu.edu.tw), we found that this hypermethylated region contains a putative binding sequence responsible for the interaction of Hif1α, C/EBPα and the p300. This complex is essential for Vegfa transcription regulation and contributes to the pro-angiogenic pathway60,61. Our findings suggest that the reduction in Vegfa gene expression in the IR + UNx group resulted from epigenetic regulation, which could be partially responsible for inducing vascular rarefaction and chronic renal hypoxia, which are mechanisms implicated in the AKI to CKD transition19,29–31,46. In agreement with our results, it has been demonstrated that treatment with VEGF-121 was effective in suppressing the AKI to CKD transition induced by IR in rats. Although VEGF-121 did not affect AKI, the loss of peritubular capillaries in the cortex and outer stripe of the outer medulla was significantly attenuated55. Further experimental and clinical studies are required to evaluate VEGF therapeutic power in preventing the AKI to CKD transition. In this hypermethylated region, we also found a putative binding site for Nrf2, which is a master regulator of the antioxidant response62. Because hypermethylation of the Vegfa promoter region occurred from the first-month post-ischemia, it suggests that this epigenetic mechanism plays an important role in the onset of the disease, promoting chronic hypoxia and concomitant development of renal fibrosis. In a recent study, it was observed that indeed, epigenetic modifications occur early after folic acid-induced kidney damage. In particular, it was observed that de novo methylation of histone H3K4 is necessary for the differentiated cells to re-enter mitosis and regenerate the proximal tubular epithelium63. These findings together, highlighted the crucial role of the early epigenetic modifications in the long consequences after an ischemic insult. Furthermore, our results open an exciting research field to explore the mechanisms by which hypermethylation of the Vegfa promoter gene is occurring, in which cell subpopulations occur, as well as the molecular mediators of this phenomenon, such as histone modifications and the enzymes involved in this pathophysiological condition. In this context, previous reports have demonstrated that the up-regulation of DNA methyltransferase 1 (Dnmt1), DNA methylation, and transcriptional silencing are linked to fibroblast activation and kidney fibrosis37.
In addition to the mentioned changes, the IR + UNx group exhibited activation of renal inflammation. Pro-inflammatory molecules, as indicated by Mcp1 and Il6 mRNA levels, increased, while the anti-inflammatory molecule, Il10, decreased beginning at the early phase of the transition of this disease. These changes could result from the global DNA hypomethylation observed in the IR + UNx, as previous studies have shown in AKI, diabetic nephropathy, and CKD models34,37,38.
In summary, our study shows that early renal hypoperfusion, inefficient hypoxia response, increased oxidative stress, and increased inflammation play an important role in the AKI to CKD transition. Specifically, the inefficient hypoxia response results from the inadequate hypermethylation of the Vegfa promoter gene at the site of HIF1α binding that occurs in the early stage post-ischemia.
Methods
All the experimental procedures in the animals were conducted following the Guide for the Care and Use of Laboratory Animals and were approved by the animal research ethics committee of the Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán with the approval number NMM-1852. The study was carried out in compliance with the ARRIVE guidelines.
Experimental model: right nephrectomy and contralateral ischemia
Fifty-nine male Wistar rats weighing between 300 and 320 g were included. Seven rats did not meet our inclusion and exclusion criteria, two were excluded due to bleeding during the nephrectomy surgical procedure and five due to postoperative death in the first 72 h as a consequence of renal ischemia–reperfusion injury. Therefore, a total of 52 rats were included and randomly divided into three groups: the sham surgery group (S, n = 16), the right nephrectomy group (UNx, n = 16), and the group with right nephrectomy and simultaneous left renal ischemia of 45 min in the left kidney (IR + UNx, n = 20). Based on our previous experience with the IR experimental model, we calculated the sample size for comparison of two means, using the creatinine elevation after 24 h post-ischemia. Although, we did not use a method to generate the randomization sequence, each block of 3 rats was randomly assigned to each of the studied groups: S, UNx or IR + UNx and so on. The study was not blinded because no pharmacological intervention was carried out.
The animals were anesthetized with sodium pentobarbital (30 mg/kg) and kept in a thermoregulated bed to perform the surgery at 37 °C. Under anesthesia, an abdominal incision was made to expose the two kidneys, first, the nephrectomy of the right kidney was performed, dissecting the peri-renal fat, as well as, separating the adrenal gland from the kidney with delicacy to avoid damaging it. For the IR + UNx group, in addition to the nephrectomy, a clip was placed in the left kidney for 45 min, to induce the ischemic process and the reperfusion was achieved when the clip was removed, using the recovery of the coloration of the kidney, as an indicator. The animals were bred and kept in our animal facility; on a 12/12 h light/dark cycles and permitted ad libitum access to food and water. Each studied group was followed for 1, 2, 3, and 4 months, respectively. At the end of each experimental period, the following parameters were assessed: mean arterial pressure (MAP), renal blood flow (RBF), creatinine clearance, glomerular diameter, tubule-interstitial fibrosis, Ki67 positive cells, urinary H2O2 excretion urinary biomarkers of renal injury, RNA and protein levels of antioxidant enzymes and anti-inflammatory cytokines, DNA methylation and promoter VEGF methylation.
Renal functional studies
The animals were placed in metabolic cages every month, to collect urine for at least 18 h to determine urinary protein excretion and creatinine clearance. A blood sample from the retro-orbital plexus was also collected monthly. Urine collections were carried out at the same schedule in all animals, starting between 4 and 6 pm and ended 18 h later, to avoid diurnal variations. For the determination of serum and urine creatinine, the colorimetric method of picric acid was used and quantified at 510 nm in a spectrophotometer. To calculate the creatinine clearance, the formula of C = (U*V)/S was used, where U is the urinary creatinine multiplied by the urinary volume (V), and S corresponds to the serum creatinine. Urinary protein excretion was determined by the turbidimetric method of trichloroacetic acid (TCA) and quantified at 420 nm in a spectrophotometer.
By the end of each experimental period, the animals were anesthetized with sodium pentobarbital (30 mg/kg) and were placed in a thermoregulated pad. The trachea was cannulated with a PE-240 polyethylene tube and the femoral arteries were catheterized with a polyethylene tube PE-90. The mean arterial pressure was recorded by one of the catheters placed in the femoral artery, using a pressure transducer (Model p23 db, Gould, Puerto Rico). Subsequently, an abdominal incision was made to expose the left kidney, the renal artery was dissected and an ultrasound probe was placed to register the renal blood flow (1RB, Transonic, Ithaca, NY).
The right kidney from the S group was ligated and removed and the left kidney upper and lower pole for the UNx and the IR + UNx groups were excised to separate renal medulla and cortex, both sections were immediately frozen at − 70 °C for further molecular analysis. The left kidney was then perfused through the femoral artery catheter with 20 mL of saline and then fixed with 20 mL of 4% formaldehyde, and removed immediately after. The animals were euthanized with an intraperitoneal delivery of an overdose of pentobarbital (100 mg/kg) after 1, 2, 3, and 4 months of renal reperfusion.
Light microscopy and immunohistochemistry analysis
After tissue fixation, the kidneys were dehydrated and embedded in paraffin. Renal slices of 4 μm were obtained and stained with Periodic Acid Schiff (PAS) or Sirius red. In the slices stained with PAS, ten microphotographs (Magnification 400x) were obtained from different renal cortex fields of each kidney and glomerular diameter was quantified in at least 40 glomeruli per rat using a Nixon camera incorporated to the microscope. In the slices stained with Sirius red, five to eight subcortical periglomerular fields per section were randomly selected in kidneys from the groups studied (Magnification 100x). Tubulo-interstitial fibrosis consisted of extracellular matrix expansion with collagen deposition together with distortion and collapse of the tubules; fibrosis was evidenced by red coloration in Sirus red-stained slides. The affected area was delimited and the percentage of tubulointerstitial fibrosis was calculated by dividing the fibrotic by the total area, excluding the glomerular area. Researchers were blind to the experimental group.
To evaluate tubular cell proliferation, conventional immunoperoxidase assays for Ki67 (anti-Ki67 antibody, Abcam Cat. No. ab66155) were performed. For signal detection, HRP/DAB Detection System (Bio SB, Santa Barbara CA, USA Cat. No. BSB 0001) was used, slides were counterstain with hematoxylin. The number of Ki-67-positive epithelial cells on each slide was counted in at least 10-subcortical fields (100 × magnification).
Hydrogen peroxide urinary excretion
In the urine collected during the follow-up time, the determination of urine hydrogen peroxides as an oxidative stress marker was carried out, using a commercial kit (Amplex Red Hydrogen Peroxide/Peroxidase Assay, Roche, Cat. No. A22188) following the manufacturer's instructions. The determination is based on the presence of peroxidase which, when reacted with hydrogen peroxide, produces a red-fluorescent compound, which was quantified in a spectrophotometer at 560 nm and extrapolated with a standard curve.
RNA extraction and quantitative PCR
The total RNA was isolated from the kidneys using the TRIzol method (Invitrogen, Carlsbad, CA, Cat. No. 15596026) and checked for integrity using 1% agarose gel electrophoresis. To avoid DNA contamination, total RNA samples were treated with DNase (DNase I; Invitrogen, Carlsbad, CA, Cat. No. 18068015). Reverse transcription (RT) was carried out with 1 µg of total RNA and 200 U of Moloney murine leukemia virus reverse transcriptase (Invitrogen). The mRNA levels of Hif1a, Vegfa, pre-pro-endothelin (Edn1), endothelin receptor A (Ednra), endothelial NOS 3 (NOS3), transforming growth factor (Tgfb1), monocyte chemoattractant protein 1 (Mcp1), nuclear factor erythroid 2 like 2 (Nfe2l2), NADPH oxidase 4 (Nox4), collagen-1 (Col1a1) interleukin 6 (Il6), and interleukin 10 (Il10) were quantified by real-time PCR on an ABI Prism 7300 Sequence Detection System (TaqMan, ABI, Foster City, CA, Cat. No. 4331182). Primers and probes were ordered as a kit as follows: Hif1a, (Rn0057756_m1), Vegfa (Rn01511602_m1), Edn1 (Rn00561129_m1), Ednra (Rn00561137_m1), NOS3 (Rn004352204_m1), Tgfb1 (Rn00572010_m1), Mcp1 (Rn00580555_m1), Nfe2l2 (Rn00582415_m1), Nox4 (Rn00585380_m1), Col1a1 (Rn1463848_m1), Il6 (Rn01410330_m1), and Il10 (Rn99999012_m1). As an endogenous control, eukaryotic 18S rRNA (predesigned assay reagent Applied by ABI, external run, Rn03928990_g1, Cat. No. 4319413E was used. The relative quantification of each gene expression was performed with the comparative threshold cycle (Ct) method.
Western blot and antibodies
The renal cortex proteins were homogenized with a lysis buffer containing: 50 mM HEPES pH 7.4, 250 mM NaCl, 5 mM EDTA, 0.1% NP-40 and complete protease inhibitor (Roche, Cat. No.. 11,697,498,001). The proteins concentration was assessed by Lowry protein assay (Bio-Rad, Cat. No. 5000113 and 5,000,114). Renal cortex protein levels were detected by Western blot, tissue proteins (20–40 µg) were electrophoresed in a denaturing 8.5% acrylamide gel with SDS. The samples were prepared with loading buffer in a 1:1 ratio with a final volume of 20 μL. The membranes were incubated with the primary antibody TGFβ (Thermo Fisher, Cat. No. MA5-15,065, 1:1000), HIF-1alpha (Abcam, Cat. No.. ab2185, 1:5000) VEGFA (Invitrogen, MA1-16,629, 1:5000), IL-6 (Abcam, Cat. No. ab9324, 1:1000), and HRP β-Actin antibody [AC-15] (Abcam, Cat. No. ab49900, 1:1,000,000) overnight at 4 °C. Three 10-min washes were performed with TBS-1 × Tween and then incubated with the secondary antibody coupled to HRP, anti-rabbit or anti-mouse IgG (Santa Cruz, Cat. No. sc-2031 or sc-2004, respectively 1: 5000). Tissue proteins assessed by Western blot were normalized by β-Actin detection.
Detection of urinary biomarkers by western blot
Urinary HSP72 levels were detected by Western blot, each urine was diluted 1:10 in 0.9% saline solution, and 10 µL of each dilution was loaded and resolved by 8.5% SDS-PAGE, as previously described39–42,64. The membranes were incubated with mouse anti-HSP72 antibody (ENZO Life Sciences, Cat. No. ADI-SPA-819F, 1:5000 dilution) or KIM-1 (Boster, Cat. No. PA1632, 1:5000) overnight at 4 °C. Thereafter; membranes were incubated with a secondary antibody, HRP-conjugated goat anti-mouse IgG, or anti-rabbit IgG, respectively (1:5000, Cat. No. sc-2031 or sc-2004, respectively, Santa Cruz). The proteins were detected using a commercial chemiluminescence kit (Millipore, Cat. No. WBKLS0500) and were normalized by urinary creatinine (UCreat).
Genomic DNA extraction and quantification of global DNA methylation
For genomic DNA extraction, 50 mg of tissue was homogenized in 200 μL of 1X PBS (10 mM PO4, 137 mM NaCl, and 2.7 mM KCl), 60 μL of digestion buffer (Tris–HCl 1 M, EDTA 0.5 M, SDS 10%, NaCl 5 M, pH = 8), then 26 μL of Proteinase K (Sigma, Cat. No. P2308-100 mg, 10 mg/mL) was added and kept on ice for 5 min. Subsequently, the mixture was left overnight at 56 °C, then the samples were treated with 3 μL of RNase A (Qiagen, Cat. No. 19101, 10 mg/mL) and incubated for 3 h at 37 °C. By the end, 250 μL of phenol–chloroform-isoamyl alcohol (Sigma, Cat. No. 77617-100ML) was added, centrifuged for 20 min, 16.1 g at 4 °C. Only the upper phase was taken and 83 μL of ammonium acetate (7.5 M) and 250 μL of absolute ethanol were added. The samples were incubated at − 20 °C overnight, centrifuged and the supernatant was discarded. Two washes were made with 250 μL of 70% ethanol. The pellet was re-suspended in 200 μL of DNase⁄RNase-Free Distilled Water. To analyze the global DNA methylation, the commercial methylation kit of 5mC DNA (Zymo Research Cat. No. D5326) was used, 100 ng of DNA from each sample was used and it was taken to a volume of 100 μL with 5mC of coating buffer. The mix was incubated at 98 °C for 5 min and then left on ice for 10 min. Then it was added to the plate and incubated at 37 °C for 1 h and the excess was discarded and washed with 200 μL of 5mC Elisa Buffer. Thereafter, 200 μL of 5mC Elisa buffer was added to each well and incubated at 37 °C for 30 min. A mixture of 5-methyl cytosine primary antibody (1:2,000) and HRP-coupled secondary anti-rabbit antibody (1:1000) was added. Finally, 100 μL of HRP developer was added to each well, incubated 1 h, and measured in a spectrometer at 405–450 nm. The results were extrapolated with a standard curve and the correction was made for the percentage of cytosines and guanine dinucleotides (CpGs), which have been previously reported for the rat genome65.
Sequence analysis and bisulfite primer design
Three binding Hif1α sequences in the promoter Vegf gene in the rat have been previously identified and were named as region 1 (− 976 to − 857), region 2 (− 724 to − 645), and region 3 (− 470 to − 369), respectively (Suppl. Fig. 1)59. DataBase of CpG islands and Analytical Tool (DBCAT) (http://dbcat.cgm.ntu.edu.tw) software-assisted us to identify the CpG islands in the promoter and the first part of the coding regions of Vegf gene. This analysis showed that only regions 2 and 3 were enriched of CpG islands, suggesting that these regions are susceptible to methylation. Consequently, optimal primers were designed in the Methyl Primer Express software for bisulfite sequencing of Hif-1α and Vegf gene promoters. For Hif-1α the primers were: 5′-GTAGAGAGTAGAGATTGAGTT-3′ (forward) and 5′-CAAAACCTAACCAAACACTAC-3′ (reverse) that amplified the region from − 1390 to − 688 (702 bp): As was commented before there are three HIF1α binding sequences in the promoter of Vegfa, but only two are susceptible to methylation: the region 2, from − 724 to − 645, and the region 3. from − 470 to − 369, we amplified together with the following primers: 5′-GGTTTTGTTAGATTTTATAGTG-3′ (forward) and 5′-CCATAACCTAAAAATTATCTATC-3′ (reverse) yielding a product of 763 bp.
Sodium bisulfite DNA conversion and sequencing
Genomic DNA (3 μg) was processed with sodium bisulfite66, DNA fragments of interest were PCR-amplified. The amplified DNA fragments were cloned into the pGEM-T Easy system (Promega, Cat. No. A1360), and Sanger sequenced using its respective reverse primer. At least 8 clones were evaluated for each region.
Statistical analysis
The results are presented as the mean ± SE. The significance of the differences between groups was assessed by 1-way ANOVA using the Bonferroni correction for multiple comparisons. All comparisons passed the normality test. Statistical significance was defined when the p-value was < 0.05. All the graphs and statistical analyses were performed using the statistical GraphPad Prisma 8 software for Mac (GraphPad Software, San Diego, CA, USA).
Supplementary Information
Acknowledgements
We are grateful to Mariela Contreras´s staff for their aid with animal care. The results presented in this paper have not been published previously in whole or in part, except as an abstract presented at the American Society of Nephrology Kidney Week Meeting 2016 (Chicago. IL), and World Congress of Nephrology ISN 2017 (Mexico City, Mexico).
Author contributions
Conception and design A.S.N. and N.A.B. Performed the experiments and analytical techniques A.S.N., R.P.V., A.R.M.O., M.A.M.R., J.R.R.A., N.G. and M.C.B. Acquisition of data: A.S.N., and N.A.B. Analysis and interpretation of data; N.A.B., A.S.N. and F.R.T. Drafting the article and revising it critically for important intellectual content; N.A.B., A.S.N., F.R.T. and G.G.
Funding
This study was supported by Grants from the Mexican Council of Science and Technology (CONACyT) (A1-S-8715, 300151, 235855, and 235964 to NAB) and from the Universidad Nacional Autónoma de México (IN201619 to NAB). This study was performed in partial fulfillment of the requirements for the PhD degree, Andrea Sánchez-Navarro is a doctoral student from Programa de Doctorado en Ciencias Biomédicas, Universidad Nacional Autónoma de México (UNAM) and received a fellowship from CONACYT (607517).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-88000-5.
References
- 1.Chawla LS, Eggers PW, Star RA, Kimmel PL. Acute kidney injury and chronic kidney disease as interconnected syndromes. N. Engl. J. Med. 2014;371:58–66. doi: 10.1056/NEJMra1214243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hsu CY, et al. Impact of AKI on urinary protein excretion: analysis of two prospective cohorts. J. Am. Soc. Nephrol. 2019;30:1271–1281. doi: 10.1681/ASN.2018101036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bucaloiu ID, Kirchner HL, Norfolk ER, Hartle JE, Perkins RM. Increased risk of death and de novo chronic kidney disease following reversible acute kidney injury. Kidney Int. 2012;81:477–485. doi: 10.1038/ki.2011.405. [DOI] [PubMed] [Google Scholar]
- 4.Chawla LS, Amdur RL, Amodeo S, Kimmel PL, Palant CE. The severity of acute kidney injury predicts progression to chronic kidney disease. Kidney Int. 2011;79:1361–1369. doi: 10.1038/ki.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hatakeyama Y, et al. Transition from acute kidney injury to chronic kidney disease: a single-centre cohort study. Clin. Exp. Nephrol. 2018;22:1281–1293. doi: 10.1007/s10157-018-1571-5. [DOI] [PubMed] [Google Scholar]
- 6.Ishani A, et al. Acute kidney injury increases risk of ESRD among elderly. J. Am. Soc. Nephrol. 2009;20:223–228. doi: 10.1681/ASN.2007080837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Funk JA, Schnellmann RG. Persistent disruption of mitochondrial homeostasis after acute kidney injury. Am. J. Physiol. Ren. Physiol. 2012;302:F853–F864. doi: 10.1152/ajprenal.00035.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zuk A, Bonventre JV. Acute kidney injury. Annu. Rev. Med. 2016;67:293–307. doi: 10.1146/annurev-med-050214-013407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Linkermann A, et al. Two independent pathways of regulated necrosis mediate ischemia-reperfusion injury. Proc. Natl. Acad. Sci. U. S. A. 2013;110:12024–12029. doi: 10.1073/pnas.1305538110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong Y, et al. Ischemic duration and frequency determines AKI-to-CKD progression monitored by dynamic changes of tubular biomarkers in IRI mice. Front Physiol. 2019;10:153. doi: 10.3389/fphys.2019.00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fiorentino M, Grandaliano G, Gesualdo L, Castellano G. Acute kidney injury to chronic kidney disease transition. Contrib. Nephrol. 2018;193:45–54. doi: 10.1159/000484962. [DOI] [PubMed] [Google Scholar]
- 12.Bonventre JV. Dedifferentiation and proliferation of surviving epithelial cells in acute renal failure. J. Am. Soc. Nephrol. 2003;14(Suppl 1):S55–61. doi: 10.1097/01.asn.0000067652.51441.21. [DOI] [PubMed] [Google Scholar]
- 13.Bonventre JV, Yang L. Cellular pathophysiology of ischemic acute kidney injury. J. Clin. Invest. 2011;121:4210–4221. doi: 10.1172/JCI45161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonventre JV. Maladaptive proximal tubule repair: cell cycle arrest. Nephron Clin. Pract. 2014;127:61–64. doi: 10.1159/000363673. [DOI] [PubMed] [Google Scholar]
- 15.Okamura DM, Pennathur S. The balance of powers: redox regulation of fibrogenic pathways in kidney injury. Redox Biol. 2015;6:495–504. doi: 10.1016/j.redox.2015.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lech M, et al. Macrophage phenotype controls long-term AKI outcomes–kidney regeneration versus atrophy. J. Am. Soc. Nephrol. 2014;25:292–304. doi: 10.1681/ASN.2013020152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee S, et al. Distinct macrophage phenotypes contribute to kidney injury and repair. J. Am. Soc. Nephrol. 2011;22:317–326. doi: 10.1681/ASN.2009060615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang MZ, et al. CSF-1 signaling mediates recovery from acute kidney injury. J. Clin. Invest. 2012;122:4519–4532. doi: 10.1172/JCI60363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kramann R, Wongboonsin J, Chang-Panesso M, Machado FG, Humphreys BD. Gli1(+) pericyte loss induces capillary rarefaction and proximal tubular injury. J. Am. Soc. Nephrol. 2017;28:776–784. doi: 10.1681/ASN.2016030297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang L, Besschetnova TY, Brooks CR, Shah JV, Bonventre JV. Epithelial cell cycle arrest in G2/M mediates kidney fibrosis after injury. Nat. Med. 2010;16:535–543, 531p. doi: 10.1038/nm.2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lima-Posada I, et al. Gender differences in the acute kidney injury to chronic kidney disease transition. Sci. Rep. 2017;7:12270. doi: 10.1038/s41598-017-09630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodriguez-Romo R, et al. AT1 receptor antagonism before ischemia prevents the transition of acute kidney injury to chronic kidney disease. Kidney Int. 2016;89:363–373. doi: 10.1038/ki.2015.320. [DOI] [PubMed] [Google Scholar]
- 23.Humphreys BD, et al. Fate tracing reveals the pericyte and not epithelial origin of myofibroblasts in kidney fibrosis. Am. J. Pathol. 2010;176:85–97. doi: 10.2353/ajpath.2010.090517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirita Y, Wu H, Uchimura K, Wilson PC, Humphreys BD. Cell profiling of mouse acute kidney injury reveals conserved cellular responses to injury. Proc. Natl. Acad. Sci. U. S. A. 2020;117:15874–15883. doi: 10.1073/pnas.2005477117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iwano M, et al. Evidence that fibroblasts derive from epithelium during tissue fibrosis. J. Clin. Invest. 2002;110:341–350. doi: 10.1172/JCI15518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moll S, et al. Epithelial cells as active player in fibrosis: findings from an in vitro model. PLoS ONE. 2013;8:e56575. doi: 10.1371/journal.pone.0056575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barrera-Chimal J, et al. Spironolactone prevents chronic kidney disease caused by ischemic acute kidney injury. Kidney Int. 2013;83:93–103. doi: 10.1038/ki.2012.352. [DOI] [PubMed] [Google Scholar]
- 28.Horbelt M, et al. Acute and chronic microvascular alterations in a mouse model of ischemic acute kidney injury. Am. J. Physiol. Ren. Physiol. 2007;293:F688–F695. doi: 10.1152/ajprenal.00452.2006. [DOI] [PubMed] [Google Scholar]
- 29.Basile DP, Donohoe D, Roethe K, Osborn JL. Renal ischemic injury results in permanent damage to peritubular capillaries and influences long-term function. Am. J. Physiol. Ren. Physiol. 2001;281:F887–F899. doi: 10.1152/ajprenal.00050.2001. [DOI] [PubMed] [Google Scholar]
- 30.Basile DP, Donohoe DL, Roethe K, Mattson DL. Chronic renal hypoxia after acute ischemic injury: effects of L-arginine on hypoxia and secondary damage. Am. J. Physiol. Ren. Physiol. 2003;284:F338–F348. doi: 10.1152/ajprenal.00169.2002. [DOI] [PubMed] [Google Scholar]
- 31.Kramann R, Tanaka M, Humphreys BD. Fluorescence microangiography for quantitative assessment of peritubular capillary changes after AKI in mice. J. Am. Soc. 2014;25:1924–1931. doi: 10.1681/ASN.2013101121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li L, et al. FoxO3 activation in hypoxic tubules prevents chronic kidney disease. J. Clin. Invest. 2019;130:2374–2389. doi: 10.1172/JCI122256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shu S, et al. Endoplasmic reticulum stress is activated in post-ischemic kidneys to promote chronic kidney disease. EBioMedicine. 2018;37:269–280. doi: 10.1016/j.ebiom.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang N, Tan L, Xue Z, Cang J, Wang H. Reduction of DNA hydroxymethylation in the mouse kidney insulted by ischemia reperfusion. Biochem. Biophys. Res. Commun. 2012;422:697–702. doi: 10.1016/j.bbrc.2012.05.061. [DOI] [PubMed] [Google Scholar]
- 35.Pang M, et al. Inhibition of histone deacetylase activity attenuates renal fibroblast activation and interstitial fibrosis in obstructive nephropathy. Am. J. Physiol. Ren. Physiol. 2009;297:F996–F1005. doi: 10.1152/ajprenal.00282.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naito M, Zager RA, Bomsztyk K. BRG1 increases transcription of proinflammatory genes in renal ischemia. J. Am. Soc. Nephrol. 2009;20:1787–1796. doi: 10.1681/ASN.2009010118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bechtel W, et al. Methylation determines fibroblast activation and fibrogenesis in the kidney. Nat. Med. 2010;16:544–550. doi: 10.1038/nm.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodriguez-Romo R, Berman N, Gomez A, Bobadilla NA. Epigenetic regulation in the acute kidney injury (AKI) to chronic kidney disease transition (CKD) Nephrol. (Carlton) 2015;20:736–743. doi: 10.1111/nep.12521. [DOI] [PubMed] [Google Scholar]
- 39.Barrera-Chimal J, et al. Hsp72 is an early and sensitive biomarker to detect acute kidney injury. EMBO Mol. Med. 2011;3:5–20. doi: 10.1002/emmm.201000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morales-Buenrostro LE, et al. Hsp72 is a novel biomarker to predict acute kidney injury in critically ill patients. PLoS ONE. 2014;9:e109407. doi: 10.1371/journal.pone.0109407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ortega-Trejo JA, et al. Heat shock protein 72 (Hsp72) specific induction and temporal stability in urine samples as a reliable biomarker of acute kidney injury (AKI) Biomarkers. 2015;20:453–459. doi: 10.3109/1354750X.2015.1096305. [DOI] [PubMed] [Google Scholar]
- 42.Perez-Villalva R, et al. HSP72 is an early biomarker to detect cisplatin and acetaminophen nephrotoxicity. Biomarkers. 2017;22:548–556. doi: 10.1080/1354750X.2017.1315616. [DOI] [PubMed] [Google Scholar]
- 43.Reddy MA, Natarajan R. Recent developments in epigenetics of acute and chronic kidney diseases. Kidney Int. 2015;88:250–261. doi: 10.1038/ki.2015.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ronco C, Bellomo R, Kellum JA. Acute kidney injury. Lancet. 2019;394:1949–1964. doi: 10.1016/S0140-6736(19)32563-2. [DOI] [PubMed] [Google Scholar]
- 45.Mehta RL, et al. International society of nephrology's 0by25 initiative for acute kidney injury (zero preventable deaths by 2025): a human rights case for nephrology. Lancet. 2015;385:2616–2643. doi: 10.1016/S0140-6736(15)60126-X. [DOI] [PubMed] [Google Scholar]
- 46.Basile DP. Rarefaction of peritubular capillaries following ischemic acute renal failure: a potential factor predisposing to progressive nephropathy. Curr. Opin. Nephrol. Hypertens. 2004;13:1–7. doi: 10.1097/00041552-200401000-00001. [DOI] [PubMed] [Google Scholar]
- 47.Rius J, et al. NF-kappaB links innate immunity to the hypoxic response through transcriptional regulation of HIF-1alpha. Nature. 2008;453:807–811. doi: 10.1038/nature06905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iommarini L, Porcelli AM, Gasparre G, Kurelac I. Non-canonical mechanisms regulating hypoxia-inducible factor 1 alpha in cancer. Front Oncol. 2017;7:286. doi: 10.3389/fonc.2017.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Y, Cox SR, Morita T, Kourembanas S. Hypoxia regulates vascular endothelial growth factor gene expression in endothelial cells. Identification of a 5' enhancer. Circ. Res. 1995;77:638–643. doi: 10.1161/01.res.77.3.638. [DOI] [PubMed] [Google Scholar]
- 50.Pugh CW, Ratcliffe PJ. Regulation of angiogenesis by hypoxia: role of the HIF system. Nat. Med. 2003;9:677–684. doi: 10.1038/nm0603-677. [DOI] [PubMed] [Google Scholar]
- 51.Semenza GL. Hydroxylation of HIF-1: oxygen sensing at the molecular level. Physiol. Bethesda. 2004;19:176–182. doi: 10.1152/physiol.00001.2004. [DOI] [PubMed] [Google Scholar]
- 52.Yu X, et al. The balance of beneficial and deleterious effects of hypoxia-inducible factor activation by prolyl hydroxylase inhibitor in rat remnant kidney depends on the timing of administration. Nephrol. Dial. Trans. 2012;27:3110–3119. doi: 10.1093/ndt/gfr754. [DOI] [PubMed] [Google Scholar]
- 53.Basile DP, et al. Impaired endothelial proliferation and mesenchymal transition contribute to vascular rarefaction following acute kidney injury. Am. J. Physiol. Ren. Physiol. 2011;300:F721–F733. doi: 10.1152/ajprenal.00546.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Basile DP, Fredrich K, Chelladurai B, Leonard EC, Parrish AR. Renal ischemia reperfusion inhibits VEGF expression and induces ADAMTS-1, a novel VEGF inhibitor. Am. J. Physiol. Ren. Physiol. 2008;294:F928–F936. doi: 10.1152/ajprenal.00596.2007. [DOI] [PubMed] [Google Scholar]
- 55.Leonard EC, Friedrich JL, Basile DP. VEGF-121 preserves renal microvessel structure and ameliorates secondary renal disease following acute kidney injury. Am. J. Physiol. Ren. Physiol. 2008;295:F1648–F1657. doi: 10.1152/ajprenal.00099.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pisani F, et al. Potential role of the methylation of VEGF gene promoter in response to hypoxia in oxygen-induced retinopathy: beneficial effect of the absence of AQP4. J. Cell Mol. Med. 2018;22:613–627. doi: 10.1111/jcmm.13348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sundrani DP, et al. Differential placental methylation and expression of VEGF, FLT-1 and KDR genes in human term and preterm preeclampsia. Clin. Epigenet. 2013;5:6. doi: 10.1186/1868-7083-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Siddique AN, et al. Targeted methylation and gene silencing of VEGF-A in human cells by using a designed Dnmt3a-Dnmt3L single-chain fusion protein with increased DNA methylation activity. J. Mol. Biol. 2013;425:479–491. doi: 10.1016/j.jmb.2012.11.038. [DOI] [PubMed] [Google Scholar]
- 59.Shinagawa M, et al. C/EBPbeta regulates Vegf gene expression in granulosa cells undergoing luteinization during ovulation in female rats. Sci. Rep. 2019;9:714. doi: 10.1038/s41598-018-36566-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lando D, Peet DJ, Whelan DA, Gorman JJ, Whitelaw ML. Asparagine hydroxylation of the HIF transactivation domain a hypoxic switch. Science. 2002;295:858–861. doi: 10.1126/science.1068592. [DOI] [PubMed] [Google Scholar]
- 61.Mahon PC, Hirota K, Semenza GL. FIH-1: a novel protein that interacts with HIF-1alpha and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 2001;15:2675–2686. doi: 10.1101/gad.924501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schmidlin CJ, Dodson MB, Madhavan L, Zhang DD. Redox regulation by NRF2 in aging and disease. Free Radic. Biol. Med. 2019;134:702–707. doi: 10.1016/j.freeradbiomed.2019.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Soofi A, Kutschat AP, Azam M, Laszczyk AM, Dressler GR. Regeneration after acute kidney injury requires PTIP-mediated epigenetic modifications. JCI Insight. 2020 doi: 10.1172/jci.insight.130204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Institute of Laboratory Animal Resources (U.S.). Committee on care and use of laboratory animals. In NIH Publication Volumes (U.S. Dept. of Health and Human Services, Public Health Service, Bethesda, Md.).
- 65.Han L, Su B, Li WH, Zhao Z. CpG island density and its correlations with genomic features in mammalian genomes. Genome Biol. 2008;9:R79. doi: 10.1186/gb-2008-9-5-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Valdes-Quezada C, Arriaga-Canon C, Fonseca-Guzman Y, Guerrero G, Recillas-Targa F. CTCF demarcates chicken embryonic alpha-globin gene autonomous silencing and contributes to adult stage-specific gene expression. Epigenetics. 2013;8:827–838. doi: 10.4161/epi.25472. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.