Abstract
In eukaryotes, DNA mismatch recognition is accomplished by the highly conserved MutSα (Msh2/Msh6) and MutSβ (Msh2/Msh3) complexes. Previously, in the yeast Saccharomyces cerevisiae, we determined that deleting MSH6 caused wild-type Msh2 levels to drop by ∼50%. In this work, we determined that Msh6 steady-state levels are coupled to increasing or decreasing levels of Msh2. Although Msh6 and Msh2 are reciprocally regulated, Msh3 and Msh2 are not. Msh2 missense variants that are able to interact with Msh6 were destabilized when Msh6 was deleted; in contrast, variants that fail to dimerize were not further destabilized in cells lacking Msh6. In the absence of Msh6, Msh2 is turned over at a faster rate and degradation is mediated by the ubiquitin-proteasome pathway. Mutagenesis of certain conserved lysines near the dimer interface restored the levels of Msh2 in the absence of Msh6, further supporting a dimer stabilization mechanism. We identified two alternative forms of regulation both with the potential to act via lysine residues, including acetylation by Gcn5 and ubiquitination by the Not4 ligase. In the absence of Gcn5, Msh2 levels were significantly decreased; in contrast, deleting Not4 stabilized Msh2 and Msh2 missense variants with partial function. The stabilizing effect on Msh2 by either the presence of Msh6 or the absence of Not4 are dependent on Gcn5. Taken together, the results suggest that the wild-type MutSα mismatch repair protein stability is governed by subunit interaction, acetylation, and ubiquitination.
Keywords: mismatch repair, ubiquitination, acetylation, dimer stabilization, MutS
Introduction
Colorectal cancer is the third leading cause of cancer death in both men and women in the United States and approximately 147,900 new cases and 53,200 deaths are predicted to occur in 2020 alone (American Cancer Society 2020). Of these, Hereditary Non-polyposis Colorectal Cancer, also known as Lynch Syndrome, accounts for roughly one in every 35 cases of colorectal cancer (Lynch et al. 2018). Lynch Syndrome confers a broad spectrum of cancer susceptibility including colorectal, endometrial, ovarian, stomach, small intestine, liver, gallbladder ducts, upper urinary tract, brain, skin, and prostate and is caused by mutations that impair DNA Mismatch Repair (Lynch et al. 2018; Gupta and Heinen 2019; Ryan et al. 2019).
Eukaryotic DNA mismatch repair is initiated when MutS homologs detect mismatches in the helix during DNA replication, recombination or upon exposure to DNA damaging agents (Kunkel and Erie 2005; Reyes et al. 2015). Single base-pair mismatches are recognized by MutSα (Msh2/Msh6), larger insertion/deletion loops resulting from DNA polymerase slippage at microsatellites are detected by MutSβ (Msh2/Msh3), and single nucleotide insertion/deletion loops are detectable by both MutSα and MutSβ (Acharya et al. 1996; Habraken et al. 1996; Iaccarino et al. 1996; Marsischky et al. 1996; Palombo et al. 1996). Once MutS complexes bind mispaired DNA, they recruit additional factors to excise and repair the error-containing strand (Hsieh and Yamane 2008).
Mismatch repair enhances the fidelity of DNA replication 100- to 1000-fold by eliminating single base mismatches and insertion–deletion loops (Kunkel and Erie 2005). Without proper mismatch repair, DNA accumulates mutations that eventually lead to compromised genomic integrity. Accordingly, DNA mismatch repair is an ancient and well-conserved mechanism. To date, considerable research has delineated components and mechanisms of mismatch; however, only limited information is available on how this process is regulated and the degree to which that regulation is conserved.
Previously, we engineered and characterized a large collection of MSH2 missense mutations, mostly of clinical origin, using yeast as a model system and found that half of the defective Msh2 proteins displayed reduced cellular levels (Gammie et al. 2007). We proposed two potential factors that were likely to influence Msh2 stability—dimerization (Hayes et al. 2009) and misfolding caused by certain missense substitutions (Arlow et al. 2013). We hypothesized that the low levels of the Msh2 missense variants could be due to reduced interaction with Msh6, exposed hydrophobicity, or a combination of both mechanisms. For the low-level Msh2 variants, the exposed hydrophobicity appears to cause a more pronounced instability because the variants are typically found at levels in the cell that are lower than would be expected from the loss of Msh6 alone (Hayes et al. 2009; Arlow et al. 2013). However, dimer stabilization was speculated to be the major regulatory control for the wild-type protein. In this work, we confirmed the dimerization stabilization hypothesis for MutSα and identified regulatory pathways involved in stabilization and degradation.
Materials and methods
Microbial and molecular techniques
Yeast strains (Table 1) and plasmids (Table 2) were manipulated using standard microbial and molecular techniques (Ausubel et al. 1989; Amberg et al. 2005). Primers were synthesized by Integrated DNA Technologies Inc. (Coralville, IA, USA). Restriction endonuclease digestions and polymerase chain reactions (PCR) were performed using manufacturer recommended reaction conditions (New England Biolabs, Beverly, MA, USA). The bacterial strain used for the propagation of plasmids was XL2-Blue (Stratagene, La Jolla, CA, USA). Plasmid DNA extractions were accomplished using the Qiagen procedure (Qiagen, Valencia, CA, USA). Nucleotide sequencing was performed by Genewiz (South Plainfield, NJ, USA).9
Table 1.
Strains used in this study
| Name | Genotype | Source |
|---|---|---|
| MY14904a | MAT a MSH2-myc::KanMX MSH3-myc::KanMX MSH6-myc::KanMX ura3-1 his3-11,15 leu2-3,112 | Gammie Laboratory |
| MY14082a | MAT a MSH6-myc::KanMX MSH3-myc::KanMX ura3-1 leu2-3,112 his3-11,15 | Gammie Laboratory |
| MY10733 | MAT a MSH2-myc::KanMX ura3-1 leu2-3,112 can1-100 his3-11,15 rad5-5 bar1 POL2-3xHA::LEU2 | Gammie Laboratory |
| MY14176a | MAT a MSH6-myc::KanMX msh2Δ::URA3 his3-11,15 trp1-1 | Gammie Laboratory |
| MY13569a | MAT a MSH6-myc::KanMX msh3Δ::HphMX ade2-1 trp1-1 ura3-1 leu2-3,112 can1-100 his3-11,15 | Gammie Laboratory |
| MY12073a | MAT α MSH6-myc::KanMX ura3-1 leu2-3,112 can1-100 his3-11,15 | Gammie Laboratory |
| MY14146a | MAT a MSH3-myc::KanMX msh2Δ:: URA3 leu2-3,112 his3-11,15 trp1-1 | Gammie Laboratory |
| MY13797a | MAT a MSH3-myc::KanMX msh6Δ::HphMX ura3-1 leu2-3,112 his3-11,15 trp1-1 hom3-10 POL2-3xHA::LEU2 | Gammie Laboratory |
| MY12328a | MAT a MSH3-myc::NatMX ade2-1 trp1-1 ura3-1 his3-11,15 hom3-10 | Gammie Laboratory |
| MY12490a | MATα MSH6-MYC::KanMX, msh2Δ::NatMX, his3-11,15, leu2-3,112 | Gammie Laboratory |
| MY9691a | MATα msh2Δ::URA3 his3-11,15 ade2-1 ura3-1 trp1-1 RAD5 CAN1 | Gammie Laboratory |
| MY9682a | MATα msh2Δ::URA3 msh6Δ::KanMX his3-11,15 ade2-1 ura3-1 trp1-1 RAD5 CAN1 | Gammie Laboratory |
| PJ69-4A | MAT a trp1-901 leu2-3,112 ura3-52 his3Δ200 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ | (James et al. 1996) |
| PJ69-4α | MATα trp1-901 leu2-3,112 ura3-52 his3Δ200 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ | (James et al. 1996) |
| MY12487 | MATα pre1-1 pre2-2 ura3Δ5 his3-11,15 leu2-3,112 msh6Δ::KanMX | Gammie Laboratory |
| MY11625 | MATα ura3Δ0 his3Δ1 leu2Δ0 lys2Δ0 msh6Δ::KanMX | Gammie Laboratory |
| AGY1057 | MAT a pol3-01 msh2Δ::LEU2 leu2-3,112 his3Δ200 trp1-1 ura3-1 [MSH2-MYC:: KanMX6 URA3 CEN/ARS] | Arlow et al. (2013) |
| BY4742 | MATα ura3Δ0 leu2Δ0 his3Δ1 lys2Δ0 | Brachmann et al. (1998) |
| 124-F-8 | MATα hpa2Δ::KanMX lys2Δ0 ura3Δ0 leu2Δ0 his3Δ1 | Brachmann et al. (1998) |
| 124-A-3 | MATα hat1Δ::KanMX lys2Δ0 ura3Δ0 leu2Δ0 his3Δ1 | Brachmann et al. (1998) |
| 133-A-5 | MATα sas2Δ::KanMX lys2Δ0 ura3Δ0 leu2Δ0 his3Δ1 | Brachmann et al. (1998) |
| 125-H-9 | MATα sas3Δ::KanMX lys2Δ0 ura3Δ0 leu2Δ0 his3Δ1 | Brachmann et al. (1998) |
| 123-C-8 | MATα elp3Δ::KanMX lys2Δ0 ura3Δ0 leu2Δ0 his3Δ1 | Brachmann et al. (1998) |
| 101-F-10 | MATα rtt109Δ::KanMX lys2Δ0 ura3Δ0 leu2Δ0 his3Δ1 | Brachmann et al. (1998) |
| 150-A-11 | MATα gcn5Δ::KanMX lys2Δ0 ura3Δ0 leu2Δ0 his3Δ1 | Brachmann et al. (1998) |
| 140-G-7 | MATα hda1Δ::KanMX lys2Δ0 ura3Δ0 leu2Δ0 his3Δ1 | Brachmann et al. (1998) |
| 133-D-12 | MATα hos1Δ::KanMX lys2Δ0 ura3Δ0 leu2Δ0 his3Δ1 | Brachmann et al. (1998) |
| 109-C-10 | MATα hos3Δ::KanMX lys2Δ0 ura3Δ0 leu2Δ0 his3Δ1 | Brachmann et al. (1998) |
| 143-F-10 | MATα set3Δ::KanMX lys2Δ0 ura3Δ0 leu2Δ0 his3Δ1 | Brachmann et al. (1998) |
| 122-E-3 | MATα hos2Δ::KanMX lys2Δ0 ura3Δ0 leu2Δ0 his3Δ1 | Brachmann et al. (1998) |
| 121-D-1 | MATα rpd3Δ::KanMX lys2Δ0 ura3Δ0 leu2Δ0 his3Δ1 | Brachmann et al. (1998) |
| 108-A-2 | MATα hst1Δ::KanMX lys2Δ0 ura3Δ0 leu2Δ0 his3Δ1 | Brachmann et al. (1998) |
| 123-H-6 | MATα hst2Δ::KanMX lys2Δ0 ura3Δ0 leu2Δ0 his3Δ1 | Brachmann et al. (1998) |
| 106-A-7 | MATα hst3Δ::KanMX lys2Δ0 ura3Δ0 leu2Δ0 his3Δ1 | Brachmann et al. (1998) |
| 120-H-12 | MATα hst4Δ::KanMX lys2Δ0 ura3Δ0 leu2Δ0 his3Δ1 | Brachmann et al. (1998) |
| 146-F-3 | MATα sir2Δ::KanMX lys2Δ0 ura3Δ0 leu2Δ0 his3Δ1 | Brachmann et al. (1998) |
| MY12064a | MAT a msh2Δ::URA3 leu2-3,112 ade2-1 trp1-1 ura3-1 his3-11,15 hom3-10 | Gammie Laboratory |
| MY12336a | MAT a msh2Δ::URA3 san1Δ::NatMX leu2-3,112 ade2-1 trp1-1 ura3-1 his3-11,15 hom3-10 | Gammie Laboratory |
| AGY227 | MATα his3-11,15 ade2-1 ura3-1 trp1-1 leu2-3,112 RAD5 CAN1 | Gammie Laboratory |
| MY11719b | MATα pre1-1 pre2-2 ura3Δ5 his3-11,15 leu2-3,112 | Heinemeyer et al. (1993) |
| 171-E-12 | MATα not3Δ::KanMX lys2Δ0 ura3Δ0 leu2Δ0 his3Δ1 | Brachmann et al. (1998) |
| 139-F-6 | MATα not4Δ::KanMX lys2Δ0 ura3Δ0 leu2Δ0 his3Δ1 | Brachmann et al. (1998) |
| 171-D-7 | MATα not5Δ::KanMX lys2Δ0 ura3Δ0 leu2Δ0 his3Δ1 | Brachmann et al. (1998) |
| MY12379a | MATα msh2Δ::URA3 msh6Δ::KanMX san1Δ::NatMX his3-11,15 ura3-1 ade2-1 trp1-1 leu2-3,112 | Gammie Laboratory |
| MY14638a | MATα msh2Δ::URA3 msh6Δ::KanMX not4Δ::NatMX his3-11,15 ade2-1 ura3-1 trp1-1 RAD5 CAN1 | Gammie Laboratory |
| MY14976a | MATα msh2Δ::URA3 msh6Δ::KanMX gcn5Δ::HphMX his3-11,15 ade2-1 ura3-1 trp1-1 RAD5 CAN1 | Gammie Laboratory |
| MY15204a | MATα msh2Δ::URA3 msh6Δ::KanMX gcn5Δ::HphMX not4Δ::NatMX his3-11,15 ade2-1 ura3-1 trp1-1 RAD5 CAN1 | Gammie Laboratory |
W303 strains. Confirmed to be wild-type at the RAD5 locus by PCR and at the CAN1 locus by canavanine resistance assays.
Strain obtained from Kiran Madura, UMDNJ-Robert Wood Johnson Medical School.
Table 2.
Plasmids used in this study
| Laboratory strain number | Plasmid name | Relevant markers | Source |
|---|---|---|---|
| AG17 | pMSH2 | MSH2-HA HIS3 CEN6 ARSH4 ampr | Gammie et al. (2007) |
| pRS413 | HIS3 CEN6 ARSH4 ampr | Sikorski and Hieter (1989) | |
| AG122 | pGAL-MSH2 | PGAL10-MSH2-HA 2µ HIS3 ampr | Gammie et al. (2007) |
| pRS423 | 2µ HIS3 ampr | Sikorski and Hieter (1989) | |
| AG86 | pMSH2-D524Y | msh2-D524Y-HA HIS3 CEN6 ARSH4 ampr | Gammie et al. (2007) |
| AG375 | pMSH2-E194G | msh2-E194G-HA HIS3 CEN6 ARSH4 ampr | Gammie et al. (2007) |
| AG207 | pMSH2-C195R | msh2-C195R-HA HIS3 CEN6 ARSH4 ampr | Gammie et al. (2007) |
| AG495 | pMSH2-C195Y | msh2-C195Y-HA HIS3 CEN6 ARSH4 ampr | Arlow et al. (2013) |
| AG486 | pMSH2-L183P | msh2-L183P-HA HIS3 CEN6 ARSH4 ampr | Gammie et al. (2007) |
| AG507 | pMSH2-C345F | msh2-C345F-HA HIS3 CEN6 ARSH4 ampr | Arlow et al. (2013) |
| AG208 | pMSH2-C345R | msh2-C345R-HA HIS3 CEN6 ARSH4 ampr | Gammie et al. (2007) |
| AG209 | pMSH2-C345Y | msh2-C345Y-HA HIS3 CEN6 ARSH4 ampr | Gammie et al. (2007) |
| AG418 | pMSH2-G350R | msh2-G350R-HA HIS3 CEN6 ARSH4 ampr | Gammie et al. (2007) |
| AG417 | pMSH2-T347I | msh2-T347I-HA HIS3 CEN6 ARSH4 ampr | Gammie et al. (2007) |
| AG421 | pMSH2-A618V | msh2-A618V-HA HIS3 CEN6 ARSH4 ampr | Gammie et al. (2007) |
| AG497 | pMSH2-D621G | msh2-D621G-HA HIS3 CEN6 ARSH4 ampr | Arlow et al. (2013) |
| AG422 | pMSH2-D621N | msh2-D621N-HA HIS3 CEN6 ARSH4 ampr | Gammie et al. (2007) |
| AG419 | pMSH2-R371S | msh2-R371S-HA HIS3 CEN6 ARSH4 ampr | Gammie et al. (2007) |
| AG461 | pMSH2-G711D | msh2-G711D-HA HIS3 CEN6 ARSH4 ampr | Gammie et al. (2007) |
| AG239 | pMSH2-G711R | msh2-G711R-HA HIS3 CEN6 ARSH4 ampr | Gammie et al. (2007) |
| AG31 | pMSH2-P640L | msh2-P640L-HA HIS3 CEN6 ARSH4 ampr | Gammie et al. (2007) |
| AG487 | pMSH2-P640T | msh2-P640T-HA HIS3 CEN6 ARSH4 ampr | Arlow et al. (2013) |
| AG93 | pMSH2-C716F | msh2-C716F-HA HIS3 CEN6 ARSH4 ampr | Gammie et al. (2007) |
| AG211 | pMSH2-C716R | msh2-C716R-HA HIS3 CEN6 ARSH4 ampr | Gammie et al. (2007) |
| AG420 | pMSH2-L521P | msh2-L521P-HA HIS3 CEN6 ARSH4 ampr | Gammie et al. (2007) |
| AG496 | pMSH2-R542L | msh2-R542L-HA HIS3 CEN6 ARSH4 ampr | Arlow et al. (2013) |
| AG29 | pMSH2-R542P | msh2-R542P-HA HIS3 CEN6 ARSH4 ampr | Gammie et al. (2007) |
| AG124 | pGBD-MSH2 | GBD-MSH2-HA TRP1 2µ ampr | Gammie et al. (2007) |
| pGBD-C2 | GBD TRP1 2µ ampr | James et al. (1996) | |
| AG113 | pGAD-MSH6 | GAD-MSH6 LEU2 2µ ampr | Gammie et al. (2007) |
| AG378 | pGBD-MSH2-E194G | GBD-msh2-E194G-HA TRP1 2µ ampr | Gammie et al. (2007) |
| AG234 | pGBD-MSH2-C195R | GBD-msh2-C195R-HA TRP1 2µ ampr | Gammie et al. (2007) |
| AG504 | pGBD-MSH2-C195Y | GBD-msh2-C195Y-HA TRP1 2µ ampr | Arlow et al. (2013) |
| AG491 | pGBD-MSH2-L183P | GBD-msh2-L183P-HA TRP1 2µ ampr | Arlow et al. (2013) |
| AG518 | pGBD-MSH2-C345F | GBD-msh2-C345F-HA TRP1 2µ ampr | Arlow et al. (2013) |
| AG235 | pGBD-MSH2-C345R | GBD-msh2-C345R-HA TRP1 2µ ampr | Gammie et al. (2007) |
| AG236 | pGBD-MSH2-C345Y | GBD-msh2-C345Y-HA TRP1 2µ ampr | Gammie et al. (2007) |
| AG433 | pGBD-MSH2-G350R | GBD-msh2-G350R-HA TRP1 2µ ampr | Gammie et al. (2007) |
| AG432 | pGBD-MSH2-T347I | GBD-msh2-T347I-HA TRP1 2µ ampr | Gammie et al. (2007) |
| AG448 | pGBD-MSH2-A618V | GBD-msh2-A618V-HA TRP1 2µ ampr | Gammie et al. (2007) |
| AG501 | pGBD-MSH2-D621G | GBD-msh2-D621G-HA TRP1 2µ ampr | Arlow et al. (2013) |
| AG449 | pGBD-MSH2-D621N | GBD-msh2-D621N-HA TRP1 2µ ampr | Gammie et al. (2007) |
| AG446 | pGBD-MSH2-R371S | GBD-msh2-R371S-HA TRP1 2µ ampr | Gammie et al. (2007) |
| AG456 | pGBD-MSH2-G711D | GBD-msh2-G711D-HA TRP1 2µ ampr | Gammie et al. (2007) |
| AG265 | pGBD-MSH2-G711R | GBD-msh2-G711R-HA TRP1 2µ ampr | Gammie et al. (2007) |
| AG135 | pGBD-MSH2-P640L | GBD-msh2-P640L-HA TRP1 2µ ampr | Gammie et al. (2007) |
| AG492 | pGBD-MSH2-P640T | GBD-msh2-P640T-HA TRP1 2µ ampr | Arlow et al. (2013) |
| AG141 | pGBD-MSH2-C716F | GBD-msh2-C716F-HA TRP1 2µ ampr | Gammie et al. (2007) |
| AG215 | pGBD-MSH2-C716R | GBD-msh2-C716R-HA TRP1 2µ ampr | Gammie et al. (2007) |
| AG447 | pGBD-MSH2-L521P | GBD-msh2-L521P-HA TRP1 2µ ampr | Gammie et al. (2007) |
| AG130 | pGBD-MSH2-D524Y | GBD-msh2-D524Y-HA TRP1 2µ ampr | Gammie et al. (2007) |
| AG500 | pGBD-MSH2-R542L | GBD-msh2-R542L-HA TRP1 2µ ampr | Arlow et al. (2013) |
| AG132 | pGBD-MSH2-R542P | GBD-msh2-R542P-HA TRP1 2µ ampr | Gammie et al. (2007) |
| MR5893 | pMSH2-K65A | MSH2-K65A-HA HIS3 CEN6 ARSH4 ampr | Gammie Laboratory |
| MR5904 | pMSH2-K75A | MSH2-K75A-HA HIS3 CEN6 ARSH4 ampr | Gammie Laboratory |
| MR5894 | pMSH2-K96A | MSH2-K96A-HA HIS3 CEN6 ARSH4 ampr | Gammie Laboratory |
| MR5905 | pMSH2-K193A | MSH2-K193A-HA HIS3 CEN6 ARSH4 ampr | Gammie Laboratory |
| MR5895 | pMSH2-K404A | MSH2-K404A-HA HIS3 CEN6 ARSH4 ampr | Gammie Laboratory |
| MR5896 | pMSH2-K405A | MSH2-K405A-HA HIS3 CEN6 ARSH4 ampr | Gammie Laboratory |
| MR5897 | pMSH2-K549A | MSH2-K549A-HA HIS3 CEN6 ARSH4 ampr | Gammie Laboratory |
| MR5898 | pMSH2-K555A | MSH2-K555A-HA HIS3 CEN6 ARSH4 ampr | Gammie Laboratory |
| MR5899 | pMSH2-K564K | msh2-K564K-HA HIS3 CEN6 ARSH4 ampr | Gammie Laboratory |
| MR5900 | pMSH2-K873A | MSH2-K873A-HA HIS3 CEN6 ARSH4 ampr | Gammie Laboratory |
| MR5901 | pMSH2-K875A | MSH2-K875A-HA HIS3 CEN6 ARSH4 ampr | Gammie Laboratory |
| AG569 | pGAD-SAN1-C279S | GAD- san1-C279S LEU2 2µ ampr | (Arlow et al. 2013) |
| AG342 | pGAL-MSH2-R542P | PGAL10-msh2-R542P-HA 2µ HIS3 ampr | (Gammie et al. 2007) |
| pRS415 | LEU2 CEN6 ARSH4 ampr | (Sikorski and Hieter 1989) | |
| MR5919 | pMSH6-MYC | MSH6-myc CEN6 ARSH4 LEU2 ampr | Gammie Laboratory |
Marker swaps replacing KanMX with HphMX or NatMX were accomplished by amplifying HphMX4, or NatMX3 and using single-step PCR-mediated gene replacement methods (Lorenz et al. 1995; Goldstein and McCusker 1999). Dominant drug-resistant marked gene deletions were amplified from strains obtained from the Yeast Deletion Consortium (Brachmann et al. 1998) and engineered into W303 strains using single-step PCR-mediated gene disruption (Baudin et al. 1993; Lorenz et al. 1995). Molecular confirmation of proper gene replacement was achieved by PCR of the replacement junctions. Yeast strains were transformed using the lithium acetate protocol (Amberg et al. 2005).
The creation of MSH2 and the msh2 missense variants into a GAL10 promoter (PGAL10) high-copy, construct, and the yeast 2 hybrid system was described previously (Gammie et al. 2007). Site-directed mutagenesis of lysine residues was accomplished as described previously (Gammie et al. 2007).
Diploids of the yeast 2-hybrid reporter strains expressing GAD-Msh6 or GAD-San1C279S and the GBD-Msh2 or GBD-Msh2 variants were plated onto selective media lacking histidine, leucine, and tryptophan (–HIS –LEU –TRP) and nonselective media (–LEU–TRP) to assess for interactions and spotting efficiency, respectively (James et al. 1996).
Mismatch repair assays exploiting the synthetic-lethal interaction of pol3-01 and defective alleles of MSH2 were conducted a plasmid shuffle assay as described previously (Tennen et al. 2013). In the strain background used in this analysis, the combination of the pol3-01 allele and a deletion of MSH2 (msh2Δ pol3-01) is lethal at all temperatures and therefore allows for the detection of mild mismatch repair defects. However, it should be noted that complete mismatch repair function is not required to suppress the lethality.
Immunoblot analysis
Approximately 3 × 107 cells were used to prepare protein extracts (Ohashi et al. 1982; Amberg et al. 2005). Protein preparations to preserve ubiquitinylated species were conducted as described previously (Laney and Hochstrasser 2002). Samples were fractionated on a 7% resolving gel using standard discontinuous SDS-PAGE and immunoblotting techniques (Ausubel et al. 1989). Immunoblotting was conducted according to the Amersham ECL™ Western Blotting System (GE Healthcare Life Science, Piscataway, NJ). The primary antibodies, monoclonal α-hemagglutinin, α-HA (12CA5, Santa Cruz Biotechnology), monoclonal α-myc (Myc.A7, Thermo Scientific), monoclonal α-PGK (Invitrogen), and polyclonal α-Kar2 (Rose Laboratory, Georgetown University) were used at a 1:200, 1:1000, 1:5000, and 1:5000 dilution, respectively. The α-mouse or α-rabbit IgG horseradish peroxidase (HRP) conjugated secondary antibodies (GE Healthcare Life Sciences) were used at a 1:2500 dilution. Band intensities of the target protein and the loading controls were directly quantified with a G: BOX imaging system (Syngene) or with the densitometry function of the open-source program ImageJ (Schneider et al. 2012).
Turnover assays using the GAL10 inducible/repressible promoter (PGAL10)
Cells expressing the PGAL10 fusions were grown to logarithmic phase in synthetic medium with 2% raffinose (a carbon source that neither represses nor induces expression). Cultures were diluted back to early logarithmic phase in the presence of synthetic medium with 2% galactose and incubated for 3 h to induce synthesis of MSH2 or the msh2 missense alleles. The cells were pelleted and resuspended in synthetic medium containing 2% glucose to repress synthesis and maintained in logarithmic phase with dilutions of fresh medium. At the indicated time points (ranging from 0 to 6 h) 3 × 107 cells were pelleted, flash frozen in liquid nitrogen, and stored at -80°C until processing for immunoblotting. When calculating turnover rates, corrections for cell division were conducted as described previously (Arlow et al. 2013).
Data and reagent availability
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Results
The levels of Msh6 and Msh2 are reciprocally regulated, but Msh3 is not subject to the same regulatory mechanism
We previously showed that the steady-state levels of Msh2 decrease in the absence of its heterodimer partner Msh6 and that the stabilizing effect of Msh3 is minor compared to Msh6 (Hayes et al. 2009). In that same study, imaging of fluorescently tagged Msh6 and Msh3 suggested that Msh2 stabilized Msh6, but the Msh2 stabilizing effect was less pronounced for Msh3 (Hayes et al. 2009). Our previous work mostly used epitope-tagged genes on plasmids harbored in strains with deletions in the relevant mismatch repair genes. In this work, all three genes coding for MutSα/β subunits were tagged with an identical myc epitope repeat at the endogenous chromosomal positions using the native promoters. The fusions were determined to be functional for mismatch repair (Haye and Gammie 2015). The relevant mismatch repair loci were deleted within the tagged strains and protein levels were measured by immunoblotting.
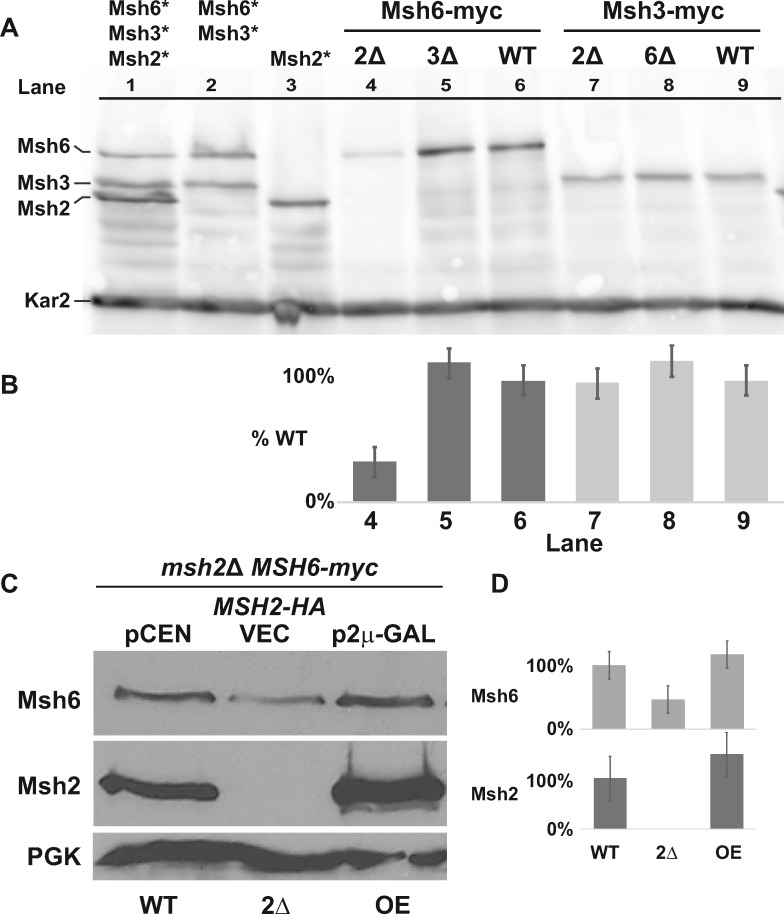
Strains with combinations of tagged MutS proteins are shown in Figure 1A (lanes 1–3) to indicate the differences in molecular weights of each subunit. In alignment with our previous results (Haye and Gammie 2015), Figure 1A (lane 1) shows a Msh2: Msh6: Msh3 ratio of 2:1:1, consistent with there being equal levels MutSα (Msh6/Msh2) and MutSβ (Msh3/Msh2) assuming all of the proteins are in a complex. We also confirmed our previous results based on fluorescence microscopy (Hayes et al. 2009) that the absence of Msh2 decreases Msh6 levels (Figure 1A, lanes 4 and 6), but does not significantly influence Msh3 levels (Figure 1A, lanes 7 and 9).
Figure 1.
Levels of Msh6, but not Msh3, are influenced by Msh2. (A) Chromosomally tagged Msh6 and Msh3 proteins display differential regulation in the absence of Msh2. Cultures grown to mid-exponential phase were processed and immunoblotted with α-myc antibodies for Msh2-myc, Msh3-myc, and Msh6-myc, and α-Kar2 for the loading control. Lanes 1-3 show the relative migration position and levels of Msh6, Msh3, or Msh2 expressed from stains where the proteins are identically tagged with the myc epitope, singly or in combinations (lanes 1-3, tagged protein indicated above the lanes with asterisks). Msh6-myc (lanes 4-6) and Msh3-myc (lanes 7-8) proteins were expressed in the absence of Msh2 (2Δ), Msh3 (3Δ) or Msh6 (6Δ), or in wild-type (WT) strain backgrounds. (B) Band intensities from Panel A of Msh6-myc and Msh3-myc were normalized to the loading controls using ImageJ and graphed as the percentage of each protein expressed in the WT strain (%WT). Lane numbers from Panel A are shown for reference. Error bars represent the stand error. (C) Msh6 levels are controlled by the abundance of Msh2. A strain with a chromosomally myc tagged MSH6 (MSH6-myc) and with a deletion in MSH2 (msh2Δ MSH6-myc) harbored plasmids expressing wild-type MSH2-HA from the endogenous promoter on a low copy, centromere-based plasmid (pCEN or WT), or overexpressed from an inducible GAL10 promoter on a high-copy, 2 μ plasmid (p2μ-GAL or OE). As a comparison, no Msh2 was expressed in the msh2Δ MSH6-myc strain with a plasmid vector (VEC, or 2Δ). The cells were grown to exponential phase in 2% galactose. Msh6-myc, Msh2-HA, and the PGK loading control were detected by α-myc, α-HA, and α-PGK, respectively. (D) Band intensities from Panel C of Msh6-myc and Msh2-HA were normalized to the loading controls using ImageJ and graphed as the percentage of each protein expressed in the WT strain described in Panel C. Error bars represent the stand error.
It is of interest that deleting Msh6 did not significantly stabilize Msh3 (Figure 1A, lanes 7 and 8) and deleting Msh3 did not significantly stabilize Msh6 (Figure 1A, lanes 4 and 5) as one might expect if more monomeric Msh2 was available for dimerization in the absence of the other competing subunit. At most, the increase is ∼15% in the absence of the competing subunit (Figure 1, A and B, compare lane 5 to 6 and lane 8 to 9); however, we reasoned that the effects may be too small to capture under wild-type conditions. Alternatively, Msh6 levels might be the limiting factor for MutSα dimerization, or other regulatory factors may dictate the pool of subunits available for dimerization.
To test whether overexpression of Msh2 might allow for better detection of the effect, we used a strain that has a deletion of the MSH2 gene as well as a chromosomally encoded, myc-epitope-tagged version of Msh6. The strain also expressed wild-type Msh2 from the endogenous promoter on a low copy centromeric plasmid or from an inducible promoter on a high-copy plasmid. Finally, the strain with an empty vector was analyzed as a control for a strain lacking Msh2. We confirmed that without Msh2, Msh6 levels dropped to roughly 50% (Figure 1, C and D, compare WT and msh2Δ) and overexpression of wild-type Msh2 resulted in a modest ∼20% increase of Msh6 over endogenous levels (Figure 1, C and D, compare WT and overexpressed, OE). This data supports the model that levels of Msh2 and Msh6 are stabilized by dimerization and that overexpression of Msh2 can elevate the levels of Msh6 to a certain extent (∼20%).
Unstable missense variants of Msh2 that fail to interact with Msh6 are not further destabilized when Msh6 is absent
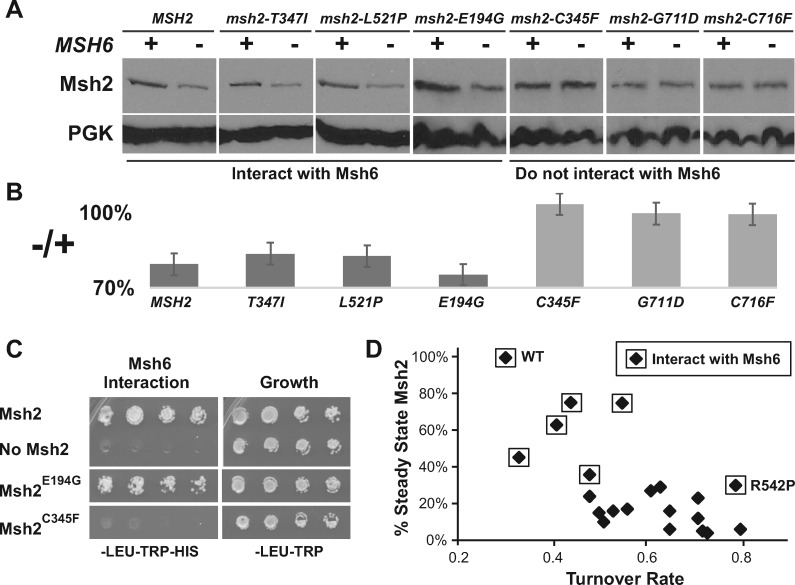
To further support the dimerization stabilization hypothesis, we examined unstable Msh2 missense variants’ steady-state levels with and without Msh6 present. We reasoned that if a Msh2 missense variant can interact with Msh6 and be stabilized, then we would expect lower protein levels in a strain lacking MSH6. Conversely, if the Msh2 missense variant cannot interact with Msh6, then the presence or absence of Msh6 should have no effect on the variant’s protein level. The results revealed that Msh2 variants capable of interacting with Msh6 show a reduction in levels in the absence of Msh6; whereas, the Msh2 variants that fail to interact with Msh6 did not display a difference in levels in the presence or absence of Msh6 (Table 3, examples in Figures 2, A–C).
Table 3.
Missense substitutions causing increased turnover of Msh2 and their ability to interact with Msh6
| Cluster location (domains)a | Yeast position | Mismatch repairb | Interaction with Msh6b | Levels lowered in absence of Msh6 |
|---|---|---|---|---|
| Wild-type | + | ++ | ++ | |
| Connector/levers (2/3) | E194G | ++/– | ++ | ++ |
| C195R | – | – | – | |
| C195Y | – | – | – | |
| L183P | – | – | – | |
| C345F | – | – | – | |
| C345R | – | – | – | |
| C345Y | – | – | – | |
| Levers/ATPase (3/5) | G350R | – | – | – |
| T347I | ++/– | ++ | ++ | |
| A618V | – | + | ++ | |
| D621G | – | – | – | |
| D621N | – | – | – | |
| R371S | – | – | – | |
| G711D | – | – | – | |
| G711R | – | – | – | |
| Levers/ATPase (3/5) | P640L | – | – | – |
| P640T | – | – | – | |
| C716F | – | – | – | |
| C716R | – | – | – | |
| Clamp/levers (3/4) | L521P | – | ++ | ++ |
| D524Y | – | + | ++ | |
| R542L | – | ++ | ++ | |
| R542P | – | ++ | ++ |
Domains correspond to the human MutSα structure (Warren et al. 2007).
The mismatch repair defects and interactions with Msh6 for most of the variants were first described in our previous publications (Gammie et al. 2007; Arlow et al. 2013).
Figure 2.
Unstable missense variants of Msh2 that fail to interact with Msh6 are not further destabilized in its absence. (A) Representative examples of the steady-state levels of Msh2 missense variants in the presence and absence of Msh6. Indicated HA-tagged missense variants (msh2-T743I, msh2-L521P, msh2-E194G, msh2-C345F, msh2-G711D, and msh2-C716F shown) were expressed in msh2Δ (+) or msh2Δmsh6Δ (–) and examined by immunoblotting. The variants that interact or do not interact with Msh6 are indicated. (B) Band intensities of Msh2 and the Msh2 variant proteins were normalized to the loading controls from Panel A using the densitometry function of the open source program ImageJ (Schneider et al. 2012) and graphed as the percentage of the Msh2 or variant Msh2 proteins expressed in the absence of Msh6 to the level expressed in the presence of Msh6 (–/+). Error bars represent the standard error. (C) Representative examples of Msh6 interactions with wild-type Msh2 and the unstable Msh2 variants. Yeast 2-hybrid strain PJ69-4A was transformed with pGBD-MSH2, pGBD-C2 (no Msh2), and all the pGBD-MSH2 low level variant plasmids including pGBD-MSH2-E194G and pGBD-MSH2-C345F shown in the figure. Yeast 2-hybrid strain PJ69-4α was transformed with pGAD-MSH6. Diploids were formed and selected for growth on medium lacking leucine and tryptophan (–LEU–TRP) and on selective medium also lacking histidine (–LEU–TRP–HIS) for the yeast 2-hybrid interaction. (C) Low level variants that do not interact with Msh6 are less stable than those that do interact with Msh6. The plot shows the steady-state levels of wild-type Msh2 (WT) and the unstable Msh2 variants as a percentage of wild-type verses the turnover rate. The turnover rates were calculated by normalizing densitometry scanned values to the zero-time point, plotting the normalized values on a log scale, and taking the average of the negative log slope. The steady-state data for most of the Msh2 variants and turnover rates were reported previously (Arlow et al. 2013). Msh2 and Msh2 missense variants that interact with Msh6 are indicated with a box around the data point. The Msh2R542P (R542P) outlier is indicated.
Because Msh6 improves the stability of Msh2, we predicted that the variants that could interact with Msh6 would have both a higher steady-state level and a slower rate of turnover if Msh6 was present in the cell. The graph in Figure 2D shows that the ability of Msh2 missense variants to interact with Msh6 is correlated with slower turnover rates (Arlow et al. 2013) and higher steady-state levels (Gammie et al. 2007), with one exception Msh2R542P, a variant we will address later in this manuscript. In summary, in the presence of Msh6, unstable Msh2 missense variants capable of forming a heterodimer are stabilized and those that fail to interact are not stabilized, further strengthening the dimer stabilization hypothesis.
Wild-type Msh2 is turned over at a faster rate in the absence of Msh6 via the ubiquitin-proteasome pathway
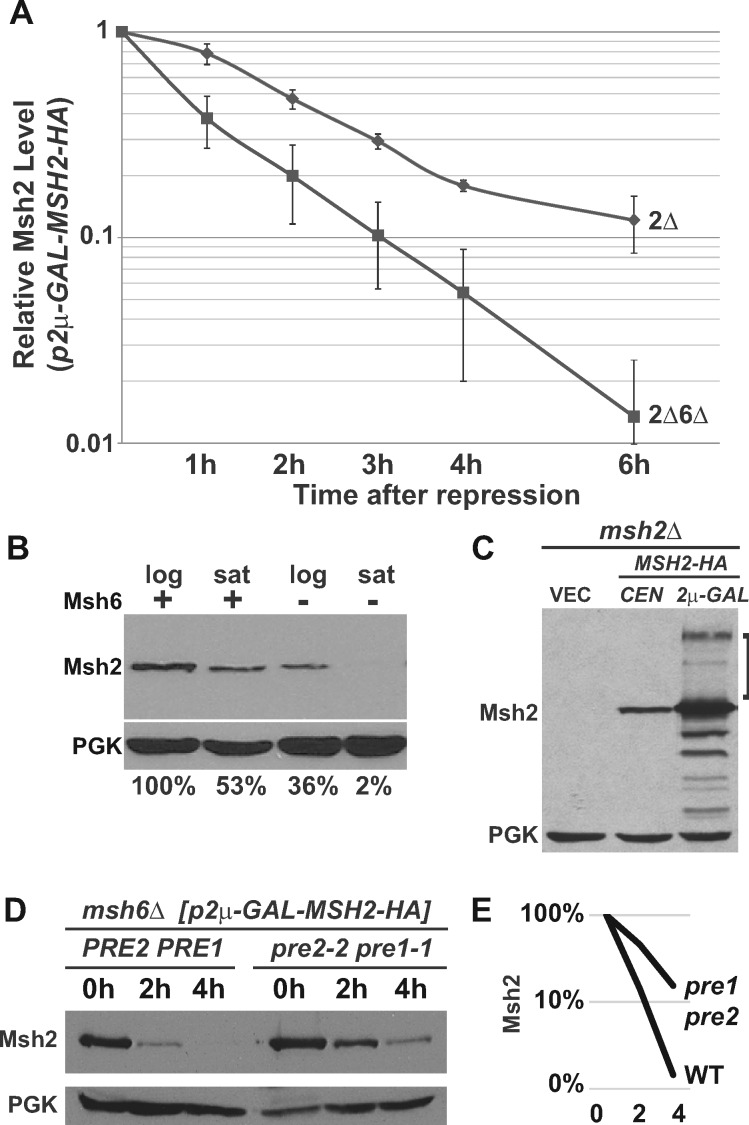
To quantify the degree to which the absence of Msh6 destabilizes Msh2, we measured the turnover rate of Msh2 in the presence and absence of Msh6 using the GAL10 inducible/repressible promoter. Exponentially growing glucose starved cells were exposed to galactose to express wild-type Msh2 in the presence and absence of MSH6. The cells were then grown in glucose to repress synthesis and kept in exponential phase throughout the experiments by diluting the cultures with fresh medium. Samples were taken over time after repression and protein immunoblotting experiments were performed. Because the turnover kinetics might be altered because of the overexpression conditions, we do not draw any conclusions about the absolute rates of turnover, rather we use this system to determine whether there is a difference in the presence or absence of Msh6. Figure 3A shows that Msh2 is degraded significantly faster in the absence of Msh6.
Figure 3.
Wild-type Msh2 is degraded more rapidly in the absence of Msh6 via the ubiquitin-proteasome pathway. Indicated msh2Δ (2Δ) or msh2Δmsh6Δ (2Δ6Δ) cells bearing the vector pRS413 (VEC), a centromere-based plasmid encoding MSH2-HA (pCEN-MSH2-HA), or a high-copy plasmid expressing MSH2 under GAL promoter (p2μ-GAL-MSH2-HA) were grown to exponential phase. Protein extracts of ∼3 × 107 cells were subjected to chemiluminescence immunoblotting methods to detect Msh2 with α-HA and a loading control (PGK) with α-PGK. (A) Msh2 has an increased turnover rate in the absence of Msh6. Indicated cells carrying p2μ-GAL-MSH2-HA were grown to exponential phase in medium containing 2% raffinose. MSH2 expression was induced with 2% galactose and repressed with 2% glucose (zero-time point) and time points were taken as described in Materials & Methods. The graph represents the average of three experiments. The data were normalized to the zero-time point. The error bars are the standard error of the mean. (B) The stabilizing effect of Msh6 on Msh2 is more pronounced when cultures become saturated. Strains that were wild-type (+) or lacking Msh6 (–) were grown in synthetic medium to an optical density at 600 nm (OD600) of 0.6 representing logarithmic phase (log) or until the cultures saturated in stationary phase (sat) at an OD600 of 1.6. Samples were prepared for immunoblotting to detect Msh2 and the PGK loading control. Band intensities of Msh2 were normalized to the loading controls using ImageJ and shown below the immunoblot images as the percentage Msh2 expressed in the presence of Msh6 during logarithmic phase. (C) High molecular weight species of wild-type Msh2 are observed when Msh2 is overexpressed. Indicated cells were grown to exponential phase in 2% galactose to overexpress MSH2. High molecular weight Msh2-HA species are indicated with a square bracket. (D) Genetic inhibition of the proteasome stabilizes monomeric Msh2. A strain with an MSH6 deletion (msh6Δ) and temperature sensitive mutations in genes for the 20S proteasome (pre1-1 pre2-2) and a msh6Δ strain with a wild-type proteasome (PRE2 PRE1) harbored p2μ-GAL-MSH2-HA and were grown to early exponential phase at 30°C in galactose containing medium. The cells were shifted to 37°C for additional 30 min to deactivate the proteasome in the proteasome mutant strain, and then 2% Glucose was added to repress MSH2 (zero-time point, 0 h). Time points at 2 and 4 h were taken at the indicated time after repression. (E) Band intensities of Msh2 from Panel D were normalized to the loading controls using ImageJ and graphed on a log scale as the percentage Msh2 expressed at 0 h for the strains with a wild-type proteasome (WT) or a defective proteasome (pre1 pre2) over the 0, 2, and 4 h time points.
Our previous work with unstable Msh2 variants showed that the ubiquitin-proteasome pathway was involved in the accelerated post-translational turnover (Arlow et al. 2013). In this work, we tested whether wild-type Msh2, in the absence of Msh6, is also degraded by the ubiquitin-proteasome pathway. Given that the ubiquitin-proteasome pathway is upregulated in stationary phase (De Virgilio 2012), we examined whether the destabilization of Msh2 in the absence of Msh6 is more pronounced in stationary phase (Figure 3B). The data are consistent with the ubiquitin-proteasome pathway playing a role in the regulation.
We sought to determine whether Msh2 is ubiquitinated by detecting high molecular weight forms of the protein, a hallmark of poly-ubiquitination. High molecular weight poly-ubiquitinated proteins are often difficult to detect because of their instability. To favor the conditions for observing these species, we overexpressed MSH2 using the inducible GAL10 promoter and prepared protein extracts in the presence of N-Ethylmaleimide to decrease de-ubiquitination (Laney and Hochstrasser 2002) and under these conditions, we observed high molecular weight species of Msh2 (Figure 3C), consistent with polyubiquitination.
To verify that the ubiquitin-proteasome pathway controls wild-type Msh2 levels in the absence of Msh6, Msh2 turnover rates were measured in a strain lacking MSH6 and with mutations in the proteasome (pre2-2 pre1-1). The turnover was compared to a strain lacking MSH6 with the proteasome intact. The turnover experiments revealed that the levels of wild-type Msh2 in the absence of Msh6 were stabilized in proteasome-defective mutant cells compared to conditions where the proteasome was functional (Figure 3, D and E). Taken together, the data provide evidence that in the absence of Msh6, wild-type Msh2 is degraded at a higher rate and the degradation is mediated by the ubiquitin-proteasome pathway.
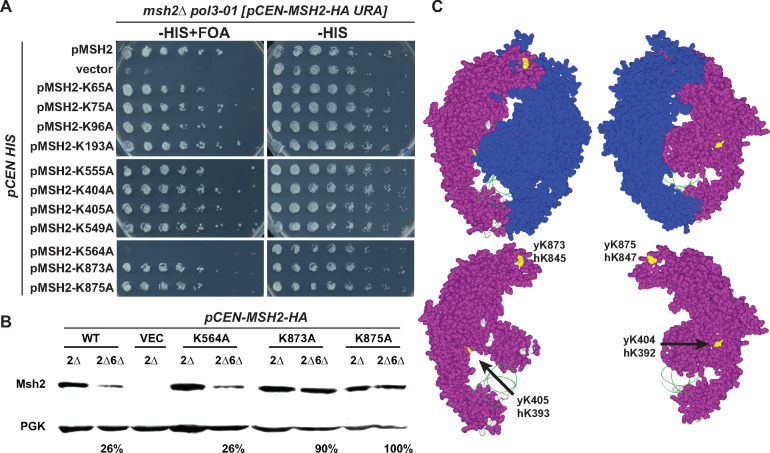
Mutagenesis of certain lysines on the surface of the dimer interface partially stabilize Msh2 in the absence of Msh6
Given that ubiquitin moieties are covalently ligated to lysine residues (Jariel-Encontre et al. 2008), we hypothesized the monomeric Msh2 protein is susceptible to degradation by exposing lysine residues or a ubiquitin ligase docking site otherwise concealed within the heterodimer interface as has been observed in other systems (Keppler and Archer 2010). We tested whether amino acid substitutions of lysine residues with alanine would inhibit ubiquitin-mediated degradation and increase Msh2 levels in the absence of Msh6. We used the Msh2 sequences from yeast and humans in conjunction with the MutSα structure bound to mismatched DNA (Warren et al. 2007) to identify conserved, surface-exposed lysines that are likely to be masked upon formation of the heterodimer. We substituted 11 potential single target lysine residues with alanines (K > A) and confirmed by DNA mismatch repair assays that the K > A variants suppressed the synthetic lethality to the same extent as the wild-type protein except for Msh2K564A (Table 4 and Figure 4A). We examined the single K > A mutant protein levels in strains in the presence and absence of MSH6 (Table 4) and identified four K > A substitutions that reproducibly increase the steady-state levels of Msh2 in the absence of Msh6 (see Figure 4B as an example). The stabilizing substitutions were in domain 5 at position K873 and K875 as well as in domain 3 at positions K404 and K405 (Table 4 and Figure 4C). Interestingly, domains 3 and 5 remain folded in a stable conformation in the MutS dimer in bacteria even in the absence of DNA binding; whereas domains 1, 2, and 4 are not sufficiently structured to analyze by crystallography (Obmolova et al. 2000). These data are consistent with the conclusion that certain lysines on Msh2 found within the stable portion of the dimer interface are targets for modification in the absence of Msh6.
Table 4.
Phenotypes of lysine substitutions in the dimer interface
| yMsh2 lysine substitution | hMsh2 lysine position | Domain in hMutSα a | Rescues msh2 Δ pol3-01 lethality | Stabilized in msh6 Δ (n = 4) |
|---|---|---|---|---|
| None | + | – | ||
| K65A | K65 | 1 | + | – |
| K75A | K73 | 1 | + | – |
| K96A | K90 | 1 | + | – |
| K193A | K197 | 2 | + | – |
| K404A | K392 | 3 | + | –/+ |
| K405A | K393 | 3 | + | –/+ |
| K549A | K531 | 4 | + | – |
| K555A | K537 | 4 | + | – |
| K564A | K546 | 4 | – | – |
| K873A | K845 | 5 | + | + |
| K875A | K847 | 5 | + | + |
Domains correspond to the human MutSα structure (Warren et al. 2007).
Figure 4.
Conserved lysines on the MutSα stabilize Msh2 in the absence of Msh6. (A) Functional assays of lysine substitution mutants. An msh2Δ pol3-01 strain kept alive by a URA3-based plasmid expressing wild-type MSH2 (pCEN-MSH2-HA URA) was transformed with HIS3 encoding plasmids (pCEN HIS) expressing endogenously expressed wild-type MSH2 (pMSH2), no MSH2 (vector), and lysine substitution variants (pMSH2-KcodonA). Stains were grown overnight in medium lacking histidine allowing for the loss of the covering wild-type MSH2 URA3 plasmid. Fivefold serial dilutions were delivered to plates lacking histidine with no drug (–HIS) or supplement with 5-FOA (–HIS+ FOA). 5-FOA selects for cells that were able to lose the MSH2 URA3 plasmid during growth, indicating full suppression of the mismatch repair defect. (B) Steady-state levels of Msh2 and its lysine substitution variants in the presence (2Δ) and absence of Msh6 (2Δ6Δ). The proteins were detected as described in Figure 1A. Band intensities were normalized to the loading controls using ImageJ and shown below the immunoblot images as the percentage of the protein level expressed in the presence of Msh6. (C) Conserved lysines highlighted on the human MutSα structure. The Msh2 protein is shown in purple with the conserved lysines that when mutated had no effect on levels (gray) or stabilized the levels of Msh2 (yellow) in the absence of Msh6. The ribbon backbone of the DNA molecule (green) is shown for orientation purposes. Two views of the heterodimer with and without Msh6 (blue) are shown. Images created without Msh6 are to reveal the concealed lysines. The potential lysine targets (yellow) are labeled with the yeast and human codon numbers in the images with just Msh2. Images were made with Swiss PDB Viewer (Guex et al. 1999) and Persistence of Vision Raytracer (Version 3.6) retrieved from http://www.povray.org/download/.
Although ubiquitination is one potential form of modification, other regulatory modulators remained a possibility, including acetylation. In fact, human Msh2 lysines K845 and K847 corresponding to yeast Msh2 K873 and K875 identified above as mediating the stabilizing effect of Msh6 are targeted by HDAC6 for deacetylation and ubiquitination (Zhang et al. 2014).
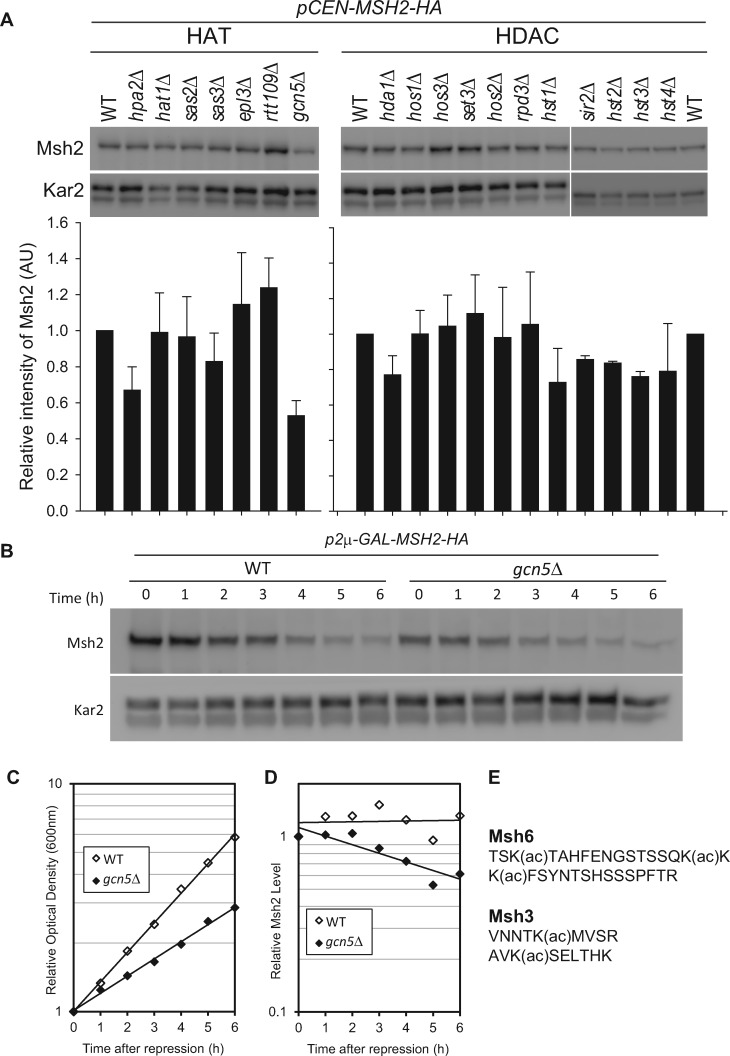
The Gcn5 acetyltransferase mediates Msh2 turnover potentially through acetylation of Msh6
To address whether acetylation might be mediating the regulation, we analyzed a panel of strains lacking acetylases and deacetylases for their ability to control Msh2 levels (Figure 5A). A deletion of the histone acetyltransferase GCN5 (gcn5Δ) had the largest effect on Msh2 steady-state levels. Using a promoter shut-off assay described above, we determined the turnover rate of Msh2 in the presence and absence of Gcn5 (Figure 5B). Because gcn5Δ cells have a growth defect (Figure 5C), the Msh2 levels were normalized to cell growth (Figure 5D). Taken together, we established that gcn5Δ enhanced the turnover of Msh2. Proteomic acetylating studies determined that Msh6 is acetylated at the N-terminal region (Figure 5E) and that deletion of GCN5 results in at least a 10-fold decrease in the acetylation of Msh6 (Downey et al. 2015). It is of interest that Msh3 was also identified as an acetylated protein in that same proteomic analysis in strains lacking several deacetylases; however, it was not identified as a significant target of Gcn5 (Downey et al. 2015). Finally, Msh2 was not identified in the analysis (Downey et al. 2015). Taken together, the data suggest that Gcn5 acetylation plays a role in the turnover of Msh2 potentially through Msh6. As GCN5 is an acetyltransferase and in other processes functions in a higher complex with ubiquitin ligase (Mao et al. 2009), we sought to identify the potential ubiquitin ligase that controls wild-type MutSα levels.
Figure 5.
Gcn5 mediates MutSα turnover through acetylation of Msh6. (A) Msh2 steady-state levels in histone acetyl transferase (HAT) mutants or histone deacetylase (HDAC) mutants. Cells were grown to exponential phase and processed for immunoblotting as described in above. The Msh2 levels from duplicate experiments were quantified, normalized by a loading control Kar2 and graphed as the relative intensity of Msh2 (AU, arbitrary units). Error bars are standard error of the mean. (B-D) Gcn5 regulates the turnover of Msh2. Turnover experiments of Msh2 were conducted in wild-type (WT) or GCN5 deletion strains (gcn5Δ). After MSH2 repression by 2% glucose, cells were harvested and assayed for Msh2 protein levels via immunoblotting (B) and growth via optical density readings (C). Msh2 levels were quantified and normalized to the Kar2 loading control (D). (E) Msh6 and Msh3 acetylated fragments. The acetylation sites were mapped previously (Downey et al. 2015).
The Not4 ubiquitin ligase controls wild-type and certain Msh2 variant levels depending on partial mismatch repair function and dimerization and the control is dependent upon the presence of Gcn5
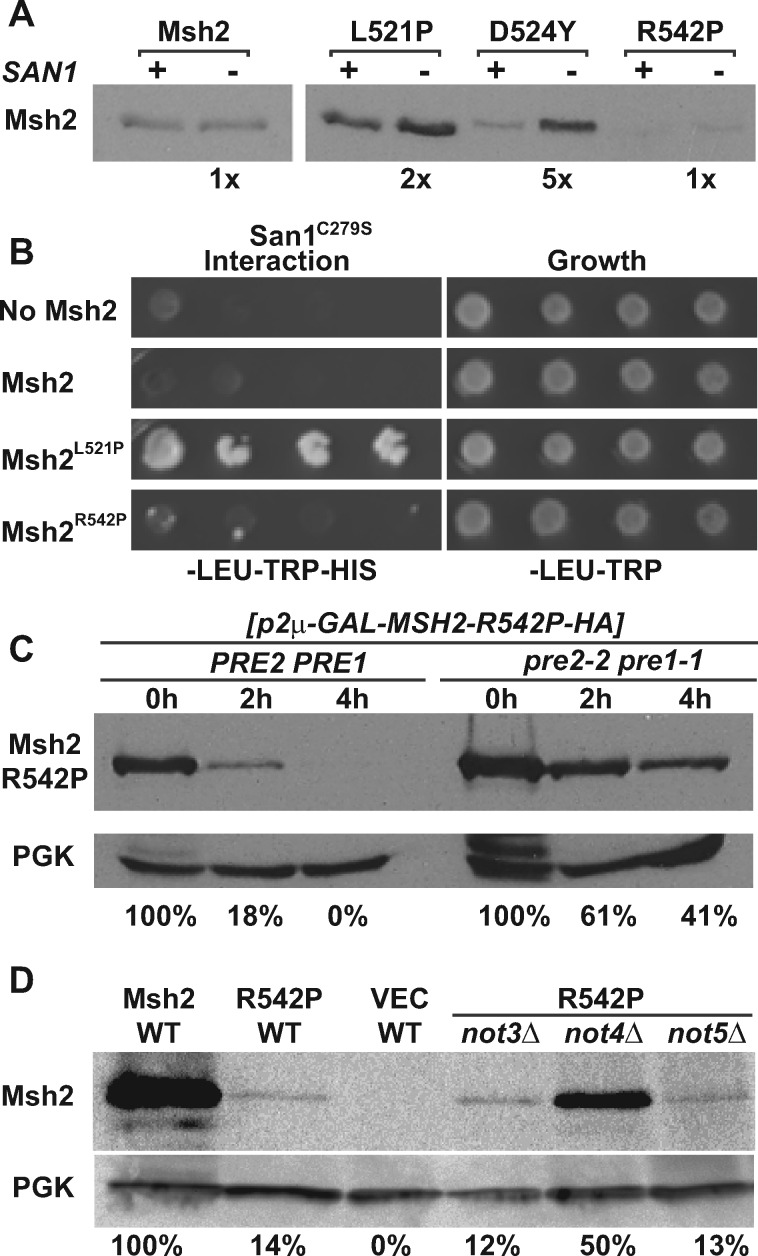
Given that wild-type Msh2 is targeted by the ubiquitin-proteasome pathway in the absence of Msh6, we sought to identify potential ligases. Previously, we showed that clinically significant low-level Msh2 missense variants are degraded by the proteasome pathway via the San1 ubiquitin ligase, a protein that targets misfolded nuclear proteins. However, as would be expected, properly folded wild-type Msh2 is not a target of San1 (Arlow et al. 2013). To find the ligase for wild-type Msh2, we deployed a missense variant Msh2R542P, the only Msh2 low-level variant that is also not regulated by San1 and therefore more likely to be folded properly. Data shown in Figure 6 illustrates our previous findings (Arlow et al. 2013) that unlike other low-level variants (e.g., Msh2D524Y), Msh2R542P is similar to wild-type Msh2. Msh2R542P failed to be stabilized in the absence of SAN1 (Figure 6A) and did not interact with a San1 variant (San1C279S) known to prolong substrate-ligase interactions (Figure 6B). We reasoned that Msh2R542P might be targeted by the same ligase as wild-type Msh2 and could be useful to identify the potential ubiquitin ligase in a screen of ubiquitin ligase deletion strains. The advantage of using this variant is that the restoration of levels phenotype would not require deleting MSH6 in the ligase mutant strains as would need to be done for wild-type Msh2.
Figure 6.
Different E3 Ubiquitin ligases control Msh2 variant levels. (A) Deletion of the San1 ubiquitin ligase stabilizes all the low level Msh2 variants except for Msh2R542P. The isogenic WT (+) and san1Δ (–) expressing Msh2 or variants were grown to exponential phase and processed for immunoblotting as described in the Materials and Methods. Band intensities were quantified using ImageJ and shown below the immunoblot images as the fold increase detected in the absence of San1. (B) San1 interacts with the unstable Msh2L521P variant, but not with wild-type Msh2 or Msh2R542P. Yeast 2-hybrid was used as described in Figure 2 to determine the interaction between San1 and wild-type Msh2 or Msh2 low level variants, Msh2L521P or Msh2R542P. Growth on medium lacking histidine (–HIS) indicates a positive interaction with San1C279S, a variant of San1 that allows for better detection of the interaction with substrates. (C) Genetic inhibition of the proteasome stabilizes Msh2R542P. Turnover experiments were conducted using the Msh2R542P variant in wild-type (PRE2 PRE1) or in a proteasome mutant strain (pre2-2 pre1-1) as described in in the Materials and Methods. Cells were processes for immunoblotting at 0, 2 and 4 h after shutting off synthesis of Msh2R542P as indicated. The immunoblot was probed to detect the Msh2R542P variant as well as PGK, the loading control. Band intensities of Msh2 were normalized to the loading controls using ImageJ and are shown below the immunoblot image as the percentage Msh2 expressed at 0 h for the strains with a wild-type proteasome or a defective proteasome over the 0, 2, and 4 h time points. (D) Deletion of NOT4 increases steady-state levels of Msh2R542P. Deletion strains not3Δ, not4Δ, not5Δ or the wild-type strain (WT) expressing MSH2 (Msh2), msh2-R542P (R452P), or the empty vector (VEC) were grown to exponential phase and processed for immunoblotting as described above. Band intensities of Msh2 were normalized to the loading controls using ImageJ and are shown below the immunoblot image as the percentage WT Msh2.
We verified that Msh2R542P is turned over by a mechanism similar to wild-type Msh2 using turnover assays in proteasome defective and wild-type strains (Figure 6C). The data confirmed that Msh2R542P is also regulated by the ubiquitin-proteasome pathway. To identify the ubiquitin ligase targeting Msh2R542P (and potentially wild-type Msh2) we screened a collection of strains with deletions in the ubiquitin ligase genes. We transformed the ligase deletion strains with a construct expressing the msh2-R542P allele on a low-copy, centromere-based plasmid and examined the steady-state levels of the Msh2R542P variant compared to the levels in the wild-type control. We found that in the absence of NOT4 (not4Δ), levels of Msh2R542P were stabilized from ∼14% to ∼50% of wild-type Msh2 levels (Figure 6D). Not4 has been shown to work in concert with Not3 and Not5 (Collart et al. 2013). Because Msh2R542P was not stabilized in strains deleted for NOT3 (not3Δ) or NOT5 (not5Δ) (Figure 6D), we concluded that Not4 is acting independently of Not3/Not5 to target Msh2R542P for degradation.
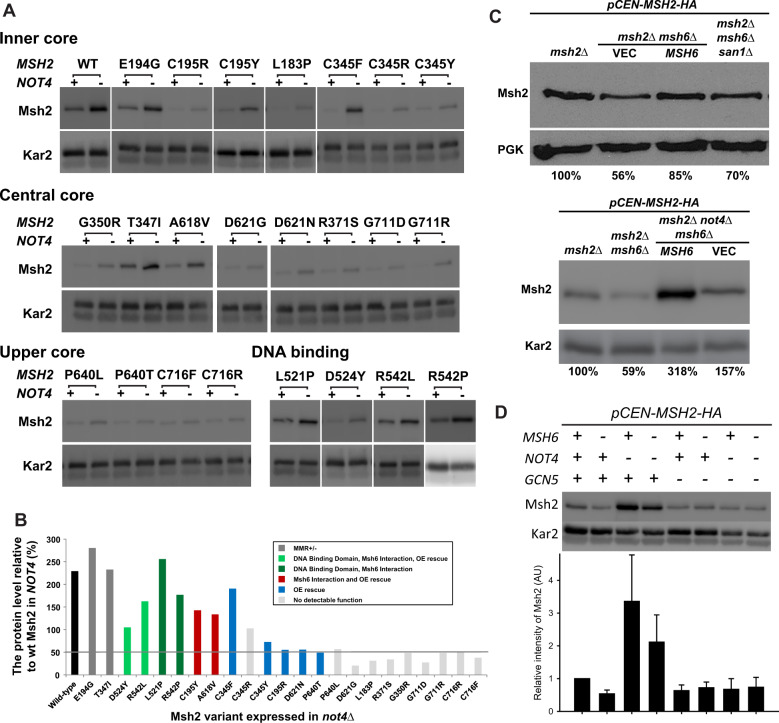
To determine whether Not4 acts exclusively on the Msh2R542P variant, we tested wild-type Msh2 and the entire panel of low-level variants (Figure 7A). We find that wild-type Msh2 and most of the variants are slightly stabilized (∼2-fold) by deleting NOT4; however, certain low-level variants are stabilized to near wild-type levels (∼2- to 13-fold). Although these stabilized variants do not fall into a single category, most have either (1) some residual mismatch repair function at low copy, (2) the ability to interact with Msh6, or (3) display an overexpression rescue phenotype (Figure 7B). These data suggest that deleting Not4 from the cell allows for stabilization of Msh2 and Msh2 low-level variants with partial activity.
Figure 7.
Low-level Msh2 variants with partial function are targeted by the Not4 ubiquitin ligase. (A) Deletion of NOT4 increases steady-state levels of Msh2 and Msh2 variants. The isogenic wild type (+) and not4Δ (–) expressing MSH2 or variants from a centromere-based plasmid (pCEN-MSH2-HA) were grown to exponential phase and processed for immunoblotting as described in Figure 1A. (B) Deleting NOT4 stabilizes Msh2 and Msh2 variants with partial function. Immunoblots were quantified and the levels of Msh2 or variants in not4Δ were presented as the relative level (%) to wild-type Msh2 level in NOT4. Data bars for Msh2 missense variants are colored according to the partial functioning including, partial mismatch repair function(MMR±) when expressed from a centromere-based plasmid, the ability to interact with Msh6 based on a yeast 2-hybrid assay, and the ability to function in mismatch repair function when overexpressed (OE rescue). The horizontal gray bar highlights the variants whose levels are increased to at least 50% of wild-type Msh2 levels. (C) Lower Wild-type Msh2 levels in strains lacking Msh6 is not a function of San1 or Not4 activity. Indicated cells were grown to exponential phase and processed for immunoblotting as described in Figure 1A. MSH6 was endogenously expressed from a centromere-based plasmid. PGK and Kar2 were used as a loading control. Band intensities of Msh2 were normalized to the loading controls using ImageJ and are shown below the immunoblot image as the percentage of the wild-type control (msh2Δ expressing MSH2 on a centromere-based plasmid). (D) Not4 specific turnover of Msh2 is not observed in the absence of Gcn5. Msh2 steady-state levels in the presence (+) or absence (–) of MSH6, NOT4 or GCN5. Cells were grown to exponential phase and processed for immunoblotting as described previously. The levels from duplicate experiments were quantified, normalized to Kar2 and graphed as the relative intensity of Msh2 (AU, arbitrary units). Error bars are standard error of the mean.
We next determined whether Not4 acted on the wild-type monomer in the absence of Msh6. The controls show that wild-type Msh2 is turned over in the absence of Msh6 and the levels are restored if MSH6 is supplied on a centromere-based plasmid (Figure 7C); however, as expected from our previous work (Arlow et al. 2013), this effect is not reversed if the San1 is deleted from the cells (Figure 7C). Likewise, the decrease in Msh2 levels in the absence of Msh6 is not alleviated by deleting NOT4 (Figure 7C). These data confirm that the characteristic decrease in levels of wild-type Msh2 in the absence of Msh6 is not eliminated by deleting either SAN1 or NOT4; however, deleting Not4 does cause an overall increase in Msh2 levels. Interestingly, when GCN5 is deleted the stabilizing effect of Msh6 and the NOT4 deletion is lost (Figure 7D). Taken together, the data suggest that Msh2 and certain Msh2 variants with partial functioning are targeted for degradation by Not4 and that the turnover is dependent upon acetylation by Gcn5. The most straightforward interpretation of the data is that acetylation of Msh6 by Gcn5 enhances dimer formation, as is the case in other systems (Yuan et al. 2005; Zhang et al. 2020) and that dimer formation is a prerequisite for serving as a target for Not4.
Discussion
Conservation of mismatch repair includes regulatory mechanisms
In this work, we provide evidence for a dimer stabilization mechanism to control the levels of the MutSα subunits, but not MutSβ, and show that the process is mediated by acetylation and ubiquitination. Interestingly, in human cells where MutSα is the major complex, the levels of hMsh6 are not detectable in the absence of hMsh2 and this relationship is used to assess mismatch repair status of tumor samples (Hall et al. 2010). Furthermore, human MutSα is also regulated by ubiquitination and acetylation (Hernandez-Pigeon et al. 2004; Hernandez-Pigeon et al. 2005; Zhang et al. 2014; Piekna-Przybylska et al. 2016). These findings underscore the conservation of function, structure, and regulation of mismatch repair proteins from yeast to humans.
Yeast MutSα, but not MutSβ, is reciprocally regulated during normal cell growth
The components of the mismatch recognition complex must be appropriately regulated and translocated to the nucleus to meet the needs of the cell during replication, recombination and in response to DNA damage. The reciprocal subunit regulation of MutSα, but not of MutSβ, could reflect the needs of the cell to maintain genome fidelity under a variety of conditions. During replication, the frequencies of the different types of mismatches are likely to occur at a relatively constant level, determined by the intrinsic error rate of the DNA polymerases (Lujan et al. 2012). However, MutSα also recognizes certain mispairings caused by lesions formed during oxidative stress (Bridge et al. 2014). We speculate that MutSα levels need to be responsive to environmental changes contributing to the generation of reactive oxygen species, whereas, MutSβ requirements would be more constant. Along these lines, it is of interest that MSH6 is induced over 10-fold upon re-oxygenation after anaerobic growth (Lai et al. 2006). Thus, the observed reciprocal regulation of MutSα, but not MutSβ, is potentially understandable in the context of being responsive to oxidative stress.
The regulation of the MutSα/β subunits in this work was examined during normal cell growth conditions where detecting mismatches during replication is the primary role for these proteins. It would be of interest to see if the regulation changes under DNA damaging conditions or during meiosis when recombination is occurring at a higher rate.
Dimer stabilization could contribute to fitness and ensure essential repair proteins are in balance
Several hypotheses could account for the dimer stabilization regulatory mechanism. For example, it is possible that free monomers are cleared from the cell because it is energetically favorable. In this model, excess monomers are cleared when cells are not generating mismatches to recycle resources. In support of this, we found that the stabilizing effect of Msh6 on Msh2 was particularly important as cells exited exponential phase and reached saturation. Moreover, the ubiquitin pathway is up-regulated upon entering stationary phase (Fujimuro et al. 1998) and it has been suggested that the regulation of the ubiquitin-proteasome pathway in early and late stationary phase is important for fitness (discussed in De Virgilio 2012). Additionally, the regulatory mechanisms controlling Msh6 and Msh2 levels may reflect the need to keep the stoichiometry of repair proteins in balance. For example, excess Msh2 or Msh6 might titrate other proteins important for maintaining genome integrity. Using genetic and biochemical approaches, the stoichiometry of mismatch repair proteins was previously shown to be important for the fidelity of the genome. Specifically, both haplo-insufficiency and increased dosage, of certain mismatch repair genes result in defects in mismatch repair (Drotschmann et al. 1999; Shcherbakova and Kunkel 1999; Drotschmann et al. 2000; Shcherbakova et al. 2001).
A proposed novel role for Not4 in targeting mismatch repair proteins after surveillance and repair
In this work we show that Msh2 and Msh2 variants with full or partial function are targeted by the Not4 ubiquitin ligase, representing a novel role for this protein; however, it has been shown previously, that Ubc4 and Not4 regulate steady-state levels of DNA polymerase-α to promote efficient and accurate DNA replication (Haworth et al. 2010). Although the precise role that Not4 plays in regulating mismatch repair is not known, we hypothesize that Not4 acts to clear mismatch repair proteins from DNA, potentially after surveillance and repair is completed.
The role of Not4 was discovered by looking for ubiquitin ligase deletion alleles that stabilize the Msh2R542P variant. This arginine to proline substitution variant, Msh2R542P, is unstable and causes a severe mismatch repair defect (Gammie et al. 2007). The amino acid substitution (R524P in humans) is in domain 4, known as the connector domain in the human MutSα heterodimer (Warren et al. 2007). In the wild-type protein, the positively charged R524 forms a hydrogen bond with the negatively charged mismatched DNA phosphodiester backbone (Warren et al. 2007). The proline substitution changes the charge and is likely to alter the interaction with the mismatched DNA. Because Msh2R542P is not stabilized in the absence of San1, we conclude that the variant does not display significant misfolding. Our previous work showed that the Msh2R542P variant is able to complex with Msh6 (Gammie et al. 2007) and that it is found in the nucleus (data not shown). We hypothesize that the variant may stabilize one of the conformations that MutS has been shown to form when associated with DNA (Qiu et al. 2012) and that this form is targeted for degradation by Not4. In support of this conclusion, the deletion of Not4 stabilized wild-type Msh2 and unstable Msh2 variants that had partial function, whereas Msh2 variants that had no detectable function were not stabilized in the absence of Not4. If the model that acetylation is a regulatory signal for MutS heterodimer formation is correct, then it would explain why Gcn5 acetylation is required to see the effects of deleting Not4, as Not4 appears to act on functional Msh2 and Msh2 variants. Future studies are required to reveal the role of Not4 in mismatch repair regulation.
Acknowledgement
This research was conducted solely at the Department of Molecular Biology, Princeton University, Princeton, New Jersey, 08544.
Funding
This research was supported by a New Jersey Commission on Cancer Research (NJCCR) Grant 10-1064-CCR-E0 and the Cancer Institute of New Jersey NCI Cancer Center Support Grant [P30CA072720] awarded to A. E. Gammie, and the Center for Quantitative Biology National Institutes of Health Grant P50 [GM071508].
Conflicts of interest: None declared.
Literature cited
- Acharya S, Wilson T, Gradia S, Kane MF, Guerrette S, et al. 1996. hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc Natl Acad Sci USA. 93:13629–13634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg DC, Burke DJ, Strathern JN.. 2005. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Plainview, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- American Cancer Society 2020. Cancer Facts & Figures 2020. Atlanta, GA: American Cancer Society. [Google Scholar]
- Arlow T, Scott K, Wagenseller A, Gammie A.. 2013. Proteasome inhibition rescues clinically significant unstable variants of the mismatch repair protein Msh2. Proc Natl Acad Sci USA. 110:246–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, et al. 1989. Short Protocols in Molecular Biology. New York: John Wiley & Sons Inc. [Google Scholar]
- Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C.. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucl Acids Res. 21:3329–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li JC, et al. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 14:115–132. [DOI] [PubMed] [Google Scholar]
- Bridge G, Rashid S, Martin SA.. 2014. DNA mismatch repair and oxidative DNA damage: implications for cancer biology and treatment. Cancers (Basel). 6:1597–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart MA, Panasenko OO, Nikolaev SI.. 2013. The Not3/5 subunit of the Ccr4-Not complex: a central regulator of gene expression that integrates signals between the cytoplasm and the nucleus in eukaryotic cells. Cell Signal. 25:743–751. [DOI] [PubMed] [Google Scholar]
- De Virgilio C. 2012. The essence of yeast quiescence. FEMS Microbiol Rev. 36:306–339. [DOI] [PubMed] [Google Scholar]
- Downey M, Johnson JR, Davey NE, Newton BW, Johnson TL, et al. 2015. Acetylome profiling reveals overlap in the regulation of diverse processes by sirtuins, gcn5, and esa1. Mol Cell Proteomics. 14:162–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drotschmann K, Clark AB, Tran HT, Resnick MA, Gordenin DA, et al. 1999. Mutator phenotypes of yeast strains heterozygous for mutations in the MSH2 gene. Proc Natl Acad Sci USA. 96:2970–2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drotschmann K, Shcherbakova PV, Kunkel TA.. 2000. Mutator phenotype due to loss of heterozygosity in diploid yeast strains with mutations in MSH2 and MLH1. Toxicol Lett. 112-113:239–244. [DOI] [PubMed] [Google Scholar]
- Fujimuro M, Takada H, Saeki Y, Toh A, Tanaka K, et al. 1998. Growth-dependent change of the 26S proteasome in budding yeast. Biochem Biophys Res Commun. 251:818–823. [DOI] [PubMed] [Google Scholar]
- Gammie AE, Erdeniz N, Beaver J, Devlin B, Nanji A, et al. 2007. Functional characterization of pathogenic human MSH2 missense mutations in Saccharomyces cerevisiae. Genetics. 177:707–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AL, McCusker JH.. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 15:1541–1553. [DOI] [PubMed] [Google Scholar]
- Guex N, Diemand A, Peitsch MC.. 1999. Protein modelling for all. Trends Biochem Sci. 24:364–367. [DOI] [PubMed] [Google Scholar]
- Gupta D, Heinen CD.. 2019. The mismatch repair-dependent DNA damage response: Mechanisms and implications. DNA Repair (Amst). 78:60–69. [DOI] [PubMed] [Google Scholar]
- Habraken Y, Sung P, Prakash L, Prakash S.. 1996. Binding of insertion/deletion DNA mismatches by the heterodimer of yeast mismatch repair proteins MSH2 and MSH3. Curr Biol. 6:1185–1187. [DOI] [PubMed] [Google Scholar]
- Hall G, Clarkson A, Shi A, Langford E, Leung H, et al. 2010. Immunohistochemistry for PMS2 and MSH6 alone can replace a four antibody panel for mismatch repair deficiency screening in colorectal adenocarcinoma. Pathology. 42:409–413. [DOI] [PubMed] [Google Scholar]
- Haworth J, Alver RC, Anderson M, Bielinsky AK.. 2010. Ubc4 and Not4 regulate steady-state levels of DNA polymerase-alpha to promote efficient and accurate DNA replication. MBoC. 21:3205–3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haye JE, Gammie AE.. 2015. The eukaryotic mismatch recognition complexes track with the replisome during DNA synthesis. PLoS Genet. 11:e1005719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes AP, Sevi LA, Feldt MC, Rose MD, Gammie AE.. 2009. Reciprocal regulation of nuclear import of the yeast MutSalpha DNA mismatch repair proteins Msh2 and Msh6. DNA Repair (Amst). 8:739–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemeyer W, Gruhler A, Mohrle V, Mahe Y, Wolf DH.. 1993. PRE2, highly homologous to the human major histocompatibility complex-linked RING10 gene, codes for a yeast proteasome subunit necessary for chrymotryptic activity and degradation of ubiquitinated proteins. J Biol Chem. 268:5115–5120. [PubMed] [Google Scholar]
- Hernandez-Pigeon H, Laurent G, Humbert O, Salles B, Lautier D.. 2004. Degadration of mismatch repair hMutSalpha heterodimer by the ubiquitin-proteasome pathway. FEBS Lett. 562:40–44. [DOI] [PubMed] [Google Scholar]
- Hernandez-Pigeon H, Quillet-Mary A, Louat T, Schambourg A, Humbert O, et al. 2005. hMutS alpha is protected from ubiquitin-proteasome-dependent degradation by atypical protein kinase C zeta phosphorylation. J Mol Biol. 348:63–74. [DOI] [PubMed] [Google Scholar]
- Hsieh P, Yamane K.. 2008. DNA mismatch repair: Molecular mechanism, cancer, and ageing. Mech Ageing Dev. 129:391–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iaccarino I, Palombo F, Drummond J, Totty NF, Hsuan JJ, et al. 1996. MSH6, a Saccharomyces cerevisiae protein that binds to mismatches as a heterodimer with MSH2. Curr Biol. 6:484–486. [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA.. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 144:1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jariel-Encontre I, Bossis G, Piechaczyk M.. 2008. Ubiquitin-independent degradation of proteins by the proteasome. Biochim Biophys Acta Rev Cancer. 1786:153–177. [DOI] [PubMed] [Google Scholar]
- Keppler BR, Archer TK.. 2010. Ubiquitin-dependent and ubiquitin-independent control of subunit stoichiometry in the SWI/SNF complex. J Biol Chem. 285:35665–35674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA, Erie DA.. 2005. DNA mismatch repair. Annu Rev Biochem. 74:681–710. [DOI] [PubMed] [Google Scholar]
- Lai LC, Kosorukoff AL, Burke PV, Kwast KE.. 2006. Metabolic-state-dependent remodeling of the transcriptome in response to anoxia and subsequent reoxygenation in Saccharomyces cerevisiae. Eukaryot Cell. 5:1468–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laney JD, Hochstrasser M.. 2002. Assaying protein ubiquitination in Saccharomyces cerevisiae. In: Gerald Fink and Christine Guthrie, editors. Guide to Yeast Genetics and Molecular and Cell Biology, Pt C. San Diego: Academic Press Inc. p. 248–257. [DOI] [PubMed] [Google Scholar]
- Lorenz MC, Muir RS, Lim E, McElver J, Weber SC, et al. 1995. Gene disruption with PCR products in Saccharomyces cerevisiae. Gene. 158:113–117. [DOI] [PubMed] [Google Scholar]
- Lujan SA, Williams JS, Pursell ZF, Abdulovic-Cui AA, Clark AB, et al. 2012. Mismatch repair balances leading and lagging strand DNA replication fidelity. PLoS Genet. 8:e1003016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch HT, Lanspa S, Shaw T, Casey MJ, Rendell M, et al. 2018. Phenotypic and genotypic heterogeneity of Lynch syndrome: a complex diagnostic challenge. Fam Cancer. 17:403–414. [DOI] [PubMed] [Google Scholar]
- Mao X, Gluck N, Li D, Maine GN, Li H, et al. 2009. GCN5 is a required cofactor for a ubiquitin ligase that targets NF-kappaB/RelA. Genes Dev. 23:849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsischky GT, Filosi N, Kane MF, Kolodner R.. 1996. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 10:407–420. [DOI] [PubMed] [Google Scholar]
- Obmolova G, Ban C, Hsieh P, Yang W.. 2000. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature. 407:703–710. [DOI] [PubMed] [Google Scholar]
- Ohashi A, Gibson J, Gregor I, Schatz G.. 1982. Import of proteins into mitochondria. The precursor of cytochrome c1 is processed in two steps, one of them heme-dependent. J Biol Chem. 257:13042–13047. [PubMed] [Google Scholar]
- Palombo F, Iaccarino I, Nakajima E, Ikejima M, Shimada T, et al. 1996. hMutSbeta, a heterodimer of hMSH2 and hMSH3, binds to insertion/deletion loops in DNA. Curr Biol. 6:1181–1184. [DOI] [PubMed] [Google Scholar]
- Piekna-Przybylska D, Bambara RA, Balakrishnan L.. 2016. Acetylation regulates DNA repair mechanisms in human cells. Cell Cycle. 15:1506–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu R, DeRocco VC, Harris C, Sharma A, Hingorani MM, et al. 2012. Large conformational changes in MutS during DNA scanning, mismatch recognition and repair signalling. EMBO J. 31:2528–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes GX, Schmidt TT, Kolodner RD, Hombauer H.. 2015. New insights into the mechanism of DNA mismatch repair. Chromosoma. 124:443–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan NAJ, Glaire MA, Blake D, Cabrera-Dandy M, Evans DG, et al. 2019. The proportion of endometrial cancers associated with Lynch syndrome: a systematic review of the literature and meta-analysis. Genet Med. 21:2167–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW.. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbakova PV, Hall MC, Lewis MS, Bennett SE, Martin KJ, et al. 2001. Inactivation of DNA mismatch repair by increased expression of yeast MLH1. Mol Cell Biol. 21:940–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbakova PV, Kunkel TA.. 1999. Mutator phenotypes conferred by MLH1 overexpression and by heterozygosity for mlh1 mutations. Mol Cell Biol. 19:3177–3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P.. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennen RI, Haye JE, Wijayatilake HD, Arlow T, Ponzio D, et al. 2013. Cell-cycle and DNA damage regulation of the DNA mismatch repair protein Msh2 occurs at the transcriptional and post-transcriptional level. DNA Repair (Amst). 12:97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren JJ, Pohlhaus TJ, Changela A, Iyer RR, Modrich PL, et al. 2007. Structure of the human MutSalpha DNA lesion recognition complex. Mol Cell. 26:579–592. [DOI] [PubMed] [Google Scholar]
- Yuan ZL, Guan YJ, Chatterjee D, Chin YE.. 2005. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 307:269–273. [DOI] [PubMed] [Google Scholar]
- Zhang M, Xiang S, Joo HY, Wang L, Williams KA, et al. 2014. HDAC6 deacetylates and ubiquitinates MSH2 to maintain proper levels of MutSalpha. Mol Cell. 55:31–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Feng Y, Guo Q, Guo W, Xu H, et al. 2020. SIRT1 modulates cell cycle progression by regulating CHK2 acetylation-phosphorylation. Cell Death Differ. 27:482–496. [DOI] [PMC free article] [PubMed] [Google Scholar]