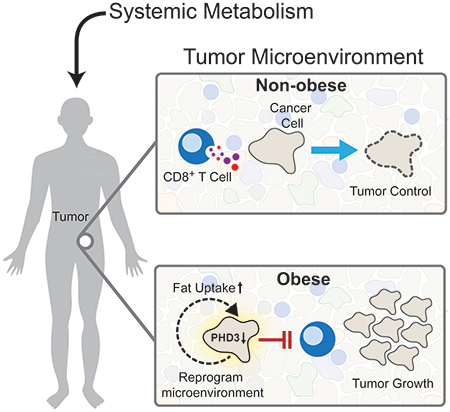
Summary
Obesity is a major cancer risk factor, but how differences in systemic metabolism change the tumor microenvironment (TME) and impact anti-tumor immunity is not understood. Here, we demonstrate that high-fat diet (HFD)-induced obesity impairs CD8+ T cell function in the murine TME, accelerating tumor growth. We generate a single-cell resolution atlas of cellular metabolism in the TME, detailing how it changes with diet-induced obesity. We find that tumor and CD8+ T cells display distinct metabolic adaptations to obesity. Tumor cells increase fat uptake with HFD, whereas tumor-infiltrating CD8+ T cells do not. These differential adaptations lead to altered fatty acid partitioning in HFD tumors, impairing CD8+ T cell infiltration and function. Blocking metabolic reprogramming by tumor cells in obese mice improves anti-tumor immunity. Analysis of human cancers reveals similar transcriptional changes in CD8+ T cell markers, suggesting interventions that exploit metabolism to improve cancer immunotherapy.
Graphical Abstract
Introduction
Obesity is an epidemic in the Western world and a risk factor for at least 13 types of cancer, including colorectal carcinoma (Lauby-Secretan et al., 2016). Of cancers in patients >30 years of age in the United States, ~5% and 10% are attributable to excess body weight in men and women, respectively (Islami et al., 2018). Moreover, while overall cancer incidence has decreased over the past 10 years in the United States, rates are rising for several obesity-related cancers, such as liver, pancreatic, thyroid and uterine cancer, as well as for colorectal cancer in patients under 55 (Lauby-Secretan et al., 2016; Siegel et al., 2019; Sung et al., 2019). Thus, it is crucial to elucidate mechanisms by which obesity increases tumor burden.
Obesity induces systemic perturbations to organismal metabolism, leading to dyslipidemia, hypercholesterolemia, insulin resistance, altered hormone levels and baseline changes in inflammation (Deng et al., 2016). Cellular changes associated with obesity include transcriptional and epigenetic alterations in the intestinal epithelium favoring colorectal tumor initiation (Beyaz et al., 2016; Li et al., 2014, 2018) and progression (Li et al., 2014). Previous studies on obesity and cancer have focused on tumor-intrinsic effects or on the endocrine-tumor cell regulatory axis. It has not yet been reported how changes in systemic metabolism induced by obesity affect immune cells in the local tumor microenvironment (TME).
The TME is a unique metabolic niche, containing cellular components (tumor cells, immune cells, and stromal cells) as well as the contents of the tumor interstitial space. A hallmark feature of tumor cell metabolism is increased nutrient consumption to meet energetic, anabolic, and pro-survival demands (Pavlova and Thompson, 2016; Spinelli and Haigis, 2018). Activated T cells are highly proliferative and rely on specific metabolic pathways to sustain T cell effector functions (Buck et al., 2015). Given the limited blood supply of solid tumors, high nutrient consumption by tumor cells may pose a barrier to the metabolic requirements of intratumoral T cells (Chang et al., 2015; Ho et al., 2015). Tumor cell metabolism is known to alter many aspects of the local metabolic landscape in ways that inhibit anti-tumor immunity (Sugiura and Rathmell, 2018). How systemic metabolism affects local metabolism within the TME, however, is not well understood. It is also not known how obesity impacts the interplay between tumor and immune cells.
Here we investigate how obesity shifts the metabolic landscape of the TME to inhibit T cell function and promote tumor growth. Using metabolic, proteomic, and genomic approaches, as well as multiplexed tissue imaging, we systematically dissect how diet-induced obesity reshapes metabolism in the TME using syngeneic mouse tumor models. We use single-cell RNA-sequencing to characterize metabolic responses to obesity by immune subsets in the TME. These multi-omic data reveal a detailed map of the dynamic landscape of tumor-immune cell interactions with HFD-induced obesity at single-cell resolution. Tumor cells, but not CD8+ T cells, dynamically respond to HFD by upregulating pathways that mobilize free fatty acids (FFAs). This shift results in altered fuel partitioning among cell populations, contributing to a TME that is paradoxically depleted for major fatty acid fuel sources during obesity. Genetic intervention to block metabolic reprogramming in tumor cells increases the availability of major FFA fuel sources and promotes tumor control by the immune system. Thus, adaptive metabolic plasticity in tumors with obesity instigates a tug of war in the TME between tumor cells and CD8+ T cells for beneficial fatty acids.
Results
HFD accelerates MC38 tumor growth in a CD8+ T cell-dependent manner
To model human obesity in mice, we randomized C57BL/6J animals at five weeks of age to control diet (CD) or HFD groups fed ad libitum (Figure 1A). While CD chow contains 13.2% kilocalories from fat, 60% of the kilocalories in the HFD come from saturated and unsaturated fats (Figure S1A). After 8-10 weeks of feeding, HFD mice gained significantly more weight (Figure S1B) and exhibited systemic obesity-associated metabolic changes, such as hypercholesterolemia and mild hyperglycemia, as well as changes in circulating levels of leptin, resistin, adiponectin, and IL-6, but without changes in fed or fasting insulin levels (Figures S1C–S1I).
Figure 1. MC38 tumor growth is accelerated by HFD feeding in a CD8+ T cell-dependent manner.
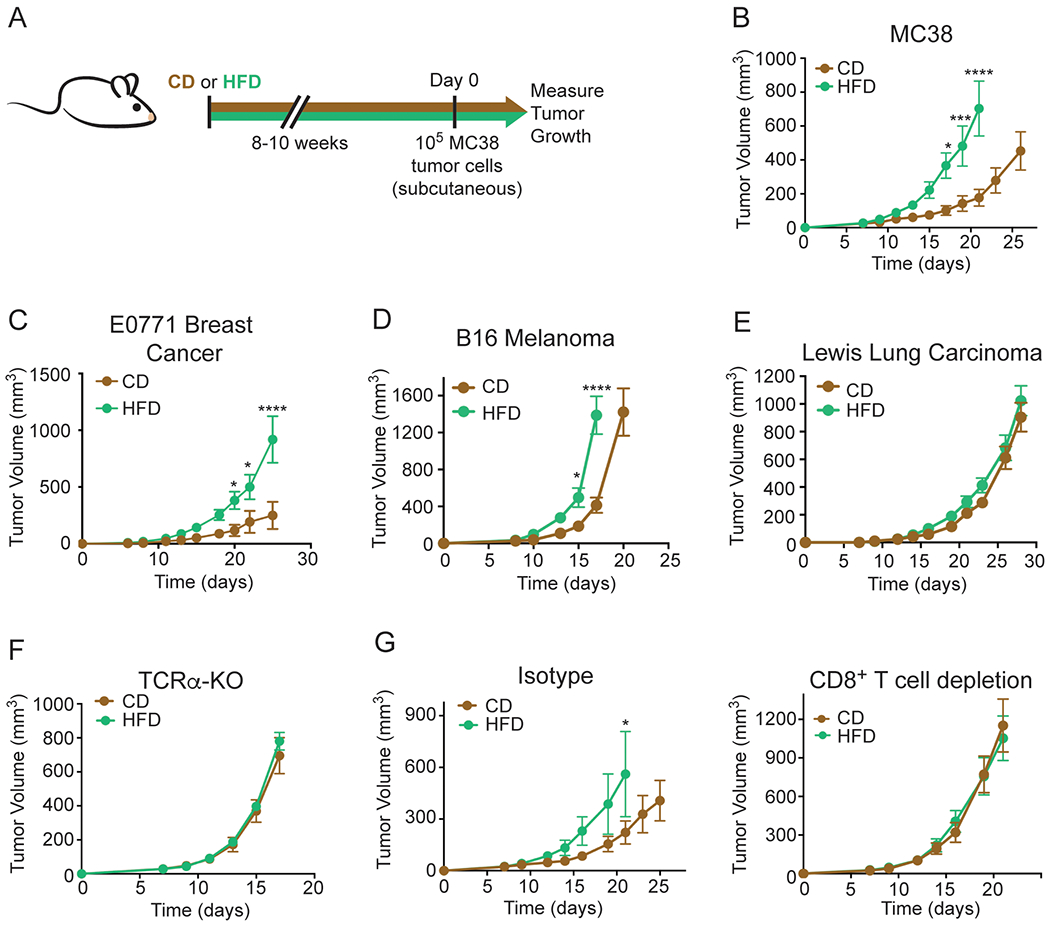
(A) Schematic depicting experimental setup.
(B-E) Tumor growth curves of WT C57BL/6J mice inoculated with 105 MC38 (B), 2x105 E0771 (C), 105 B16 melanoma (D), or 105 Lewis Lung Carcinoma (E) tumor cells.
(F) Tumor growth curves of TCRα-KO mice fed CD or HFD inoculated with 105 MC38 tumor cells.
(G) Tumor growth curves of WT C57BL/6J mice inoculated with 105 MC38 tumor cells and treated with isotype control (left) or depleting anti-CD8 (right) antibodies after CD or HFD feeding for 8-10 weeks.
Data represent ≥ two independent experiments with ≥ 5 mice per group. (*p≤0.05, **p<≤.01, ***p≤0.001, ****p≤0.0001). Graphs display mean +/− SEM (B-G).
See also Figure S1.
After adaptation to CD or HFD, mice were injected with syngeneic MC38 colorectal adenocarcinoma cells, which establish highly immunogenic tumors. As observed previously, MC38 tumors grew more rapidly in mice fed HFD compared to CD (Figure 1B) (Algire et al., 2011; Nimri et al., 2015). We also studied the growth kinetics of three other C57BL/6J syngeneic tumor models of varying immunogenicity: E0771 breast adenocarcinoma, B16 melanoma and Lewis Lung Carcinoma (LLC) (Crosby et al., 2018; Lechner et al., 2013; Mosely et al., 2017). Highly immunogenic orthotopic E0771 breast tumors grew faster in HFD animals (Figure 1C), while moderately immunogenic B16 melanoma tumors demonstrated a modest increase in growth rate with HFD (Figure 1D), and the growth rate of poorly immunogenic LLC tumors did not change with diet (Figure 1E). To further investigate how immunogenicity affects growth kinetics, we monitored growth of B16 tumors expressing the immunogenic model antigen ovalbumin (B16-OVA-RFP) for 11 days, and found that even at this early time point, HFD tumors were substantially larger than CD tumors (Figure S1J). We also measured the effect of HFD on the growth of CT26 (colon carcinoma) and RENCA (renal adenocarcinoma) tumors, which are syngeneic in BALB/cJ mice. CT26 tumors grew faster in HFD- than CD-fed mice, but RENCA tumors did not (Figures S1K and S1L).
To interrogate whether the reduced growth rates of tumors in CD animals were due to control by T cells, we implanted MC38 tumors in T cell receptor α chain knock-out (TCRα-KO) mice, which lack conventional αβ T cells. Although TCRα-KO mice exhibited similar weight gain on HFD as wildtype (WT) mice (Figure S1B), there was no difference in the growth rate of MC38 tumors in TCRα-KO mice fed CD compared to HFD (Figure 1F). Similarly, there were no diet-dependent changes in tumor growth rate in mice depleted of CD8+ T cells (Figure 1G, Figures S1M and S1N). While CD8+ T cell depletion expectedly accelerated tumor growth in both conditions, the difference between tumors in mice with and without CD8+ T cell depletion was smaller in the HFD context (Figures S1O and S1P). Taken together, these data provide evidence that HFD-induced changes in metabolism increase MC38 tumor growth by limiting anti-tumor CD8+ T cell responses.
HFD feeding reduces the number and functionality of intratumoral CD8+ T cells
To understand how HFD feeding alters the immune landscape of MC38 tumors, we used flow cytometry to profile tumor-infiltrating immune cell populations in tumors 10-14 days after implantation, when tumors were similar in volume (Figures 1B and 2A). In HFD tumors we observed large changes in the lymphocyte compartment, including cytotoxic T cell populations capable of performing antigen-specific cell killing. Specifically, HFD MC38 tumors contained fewer CD8+ T cells as a fraction of the CD45+ leukocyte infiltrate (Figure 2B, Figure S2A). The reduction in CD8+ T cells was specific to the tumor, since we did not observe this in the spleen or draining lymph node (dLN) (Figure 2B). To determine whether the reduced CD8+ T cell fraction corresponded to an overall reduction in number, we used GFP-expressing MC38 cells and counted the number of CD45+ leukocytes and CD8+ T cells relative to tumor cells by flow cytometry. This showed that HFD mice had a decreased leukocyte to tumor cell ratio (Figure 2C, Figure S2B), as well as a lower CD8+ T cell to tumor cell ratio (Figure 2D, Figure S2B). The ratio of CD4+ T cells to tumor cells was not significantly changed (Figures S2B and S2C), and the proportion of regulatory FOXP3+ CD4+ T cells (Tregs) of all CD4+ T cells was comparable in CD and HFD tumors (Figure S2A, Figure S2D). As a result, the ratio of CD8+ T cells to Tregs was lower in HFD tumors (Figure S2E).
Figure 2. HFD reduces intratumoral CD8+ T cell numbers and functionality.
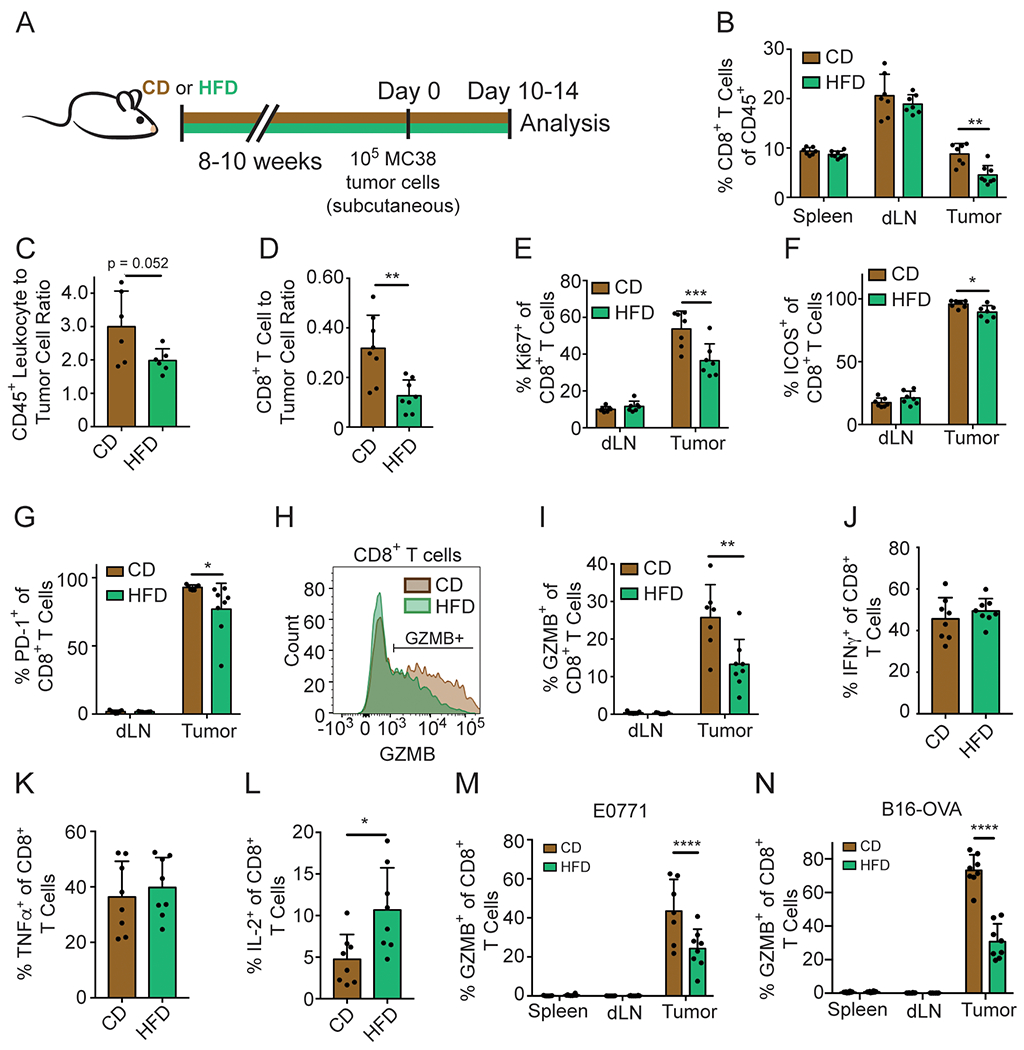
(A) Schematic depicting experimental setup.
(B-L) Flow cytometry analysis of MC38 (B, E-L), MC38-GFP (C-D), E0771 (M) or B16-OVA-RFP (N) tumors on day 10-14 after inoculation.
(B) Quantification of the percentage of CD8+ T cells among intratumoral CD45+ cells.
(C-D) The ratio of CD45+ cells (C) or CD8+ T cells (D) to MC38-GFP tumor cells.
(E-G) Quantification of Ki67 (E), ICOS (F), and PD-1 (G) expression among CD8+ TILs.
(H-I) Representative flow plot (H) and quantification (I) of GZMB expression among CD8+ TILs.
(J-L) Quantification of IFNγ (J), TNFα (K) and IL-2 (L) expression among CD8+ TILs after ex vivo phorbol myristate acetate (PMA)/ionomycin stimulation.
(M-N) Quantification of GZMB expression among CD8+ TILs in E0771 (M) and B16-OVA-RFP (N) tumors.
Data represent ≥ two independent experiments with ≥ 6 mice per group. (ns p>0.05, *p≤0.05, **p≤0.01). Graphs display mean +/− SD (B-G, I-N).
See also Figure S2.
In addition to CD8+ T cells, we also evaluated the effects of HFD on other immune cell populations in MC38 tumors. Natural killer (NK) cell numbers were comparable across conditions (Figure S2F). However, the percentage of CD11b+ myeloid cells increased with HFD (Figures S2A and S2G), corresponding to an expansion of both GR1+ CD11b+ myeloid-derived suppressor cell numbers (Figures S2A and S2H) and F4/80+ GR1− CD11b+ tumor-associated macrophages (TAMs) (Figures S2A and S2I), two populations known to promote tumor growth. The ratio of CD11b+ myeloid cells to tumor cells did not increase, since there were fewer infiltrating immune cells in HFD tumors (Figure S2J). We also looked at CD11c+ dendritic cells, which stimulate T cells by presenting antigen. The percentage of CD11c+ dendritic cells in tumors was similar in CD and HFD, as were the expression levels of MHC-I, MHC-II, and CD40 on CD11c+ cells (Figures S2K–S2N). Diet did not alter MHC-I and PD-L1 levels on MC38 tumor cells (Figures S2O and S2P). These findings suggest that CD8+ T cells are the immune cell type in the MC38 TME most dramatically impacted by HFD.
To study the effect of HFD on the activity and function of CD8+ T cells in tumors we assayed markers indicative of T cell function. CD8+ tumor-infiltrating lymphocytes (TILs) from HFD mice were less proliferative, based on Ki67 levels (Figure 2E, Figure S2Q). A smaller proportion of HFD CD8+ TILs expressed the costimulatory receptor ICOS (Figure 2F, Figure S2R). We also examined PD-1 expression, which is induced during activation, and observed fewer CD8+ TILs expressing PD-1 in HFD animals (Figure 2G). Thus, CD8+ TILs express lower levels of both costimulatory and co-inhibitory receptors, consistent with decreased activation. Accordingly, we found that fewer CD8+ TILs expressed the cytolytic molecule Granzyme B (GZMB) with HFD compared to CD (Figures 2H and 2I), suggesting reduced functionality. HFD did not alter the ability of CD8+ T cells to produce the inflammatory cytokines IFNγ and TNFα, but increased IL-2 production (Figures 2J–2L). We also analyzed the expression of key T cell functional markers from the diet-sensitive E0771 and B16-RFP-OVA tumor models, finding similar functional deficits in intratumoral T cells. GZMB and PD-1 expression were significantly lower in CD8+ T cells from HFD tumors, whereas CD8+ T cell infiltration was reduced in B16-OVA, but not E0771, tumors (Figures 2M and 2N, Figures S2S–S2V). In contrast, CD8+ T cell infiltration and functionality did not change with HFD in CT26 tumors in BALB/cJ mice (Figures S2W and S2X). Thus, HFD reduces intratumoral T cell function across many, but not all, diet-sensitive tumor models.
We then asked whether obesity altered the activation of CD8+ T cells ex vivo after antibody stimulation. HFD-derived naïve splenic CD8+ T cells performed similarly or better than their CD-derived counterparts in terms of proliferation, GZMB expression and Ki67 expression when stimulated with anti-CD3/anti-CD28 antibodies in vitro (Figures S2Y–S2AA). Thus, changes in CD8+ T cell proliferation and state were specific to the HFD environment in vivo and did not appear to involve an intrinsic defect in T cell activation.
Single-cell RNA-sequencing shows diet-induced alterations in tumor-infiltrating immune populations
Next, we used single-cell RNA-sequencing of tumor-infiltrating CD45+ leukocytes from CD and HFD MC38 tumors to map the tumor-immune transcriptional landscape in an unbiased and comprehensive manner (Figure 3A). To define major cell populations, we performed unsupervised clustering analysis on integrated single-cell datasets from CD and HFD tumors. This identified 16 distinct clusters (Figure 3B), where each of the 16 clusters contained cells from both diet conditions (Figure S3A). We annotated clusters based on the expression of known genetic markers (Figure S3B) and categorized those cell populations into groups with common pro- or anti-tumorigenic properties. Lymphocytes were significantly reduced in animals on HFD, whereas the relative proportions of immunosuppressive myeloid cell populations did not change (Figure 3C).
Figure 3. Single-cell analysis reveals global metabolic remodeling of tumor-immune infiltrate.
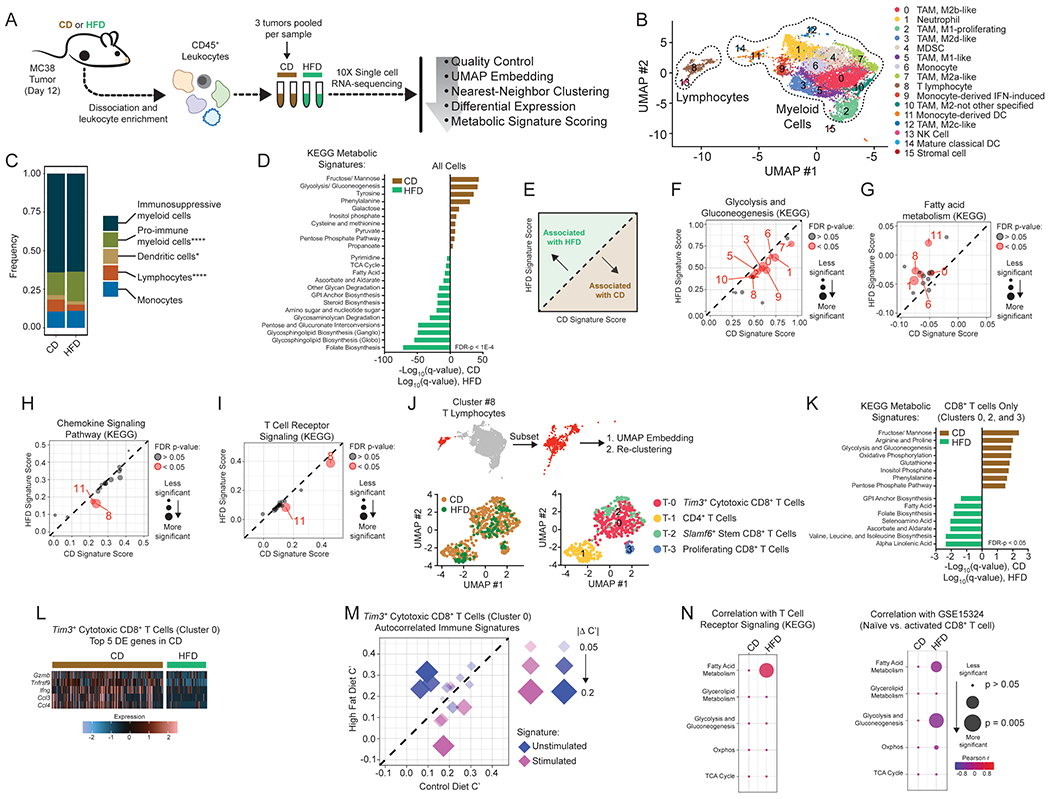
(A) Schematic depicting single-cell RNA-seq experiment and analysis.
(B) Identification of tumor-infiltrating immune cell populations. Uniform Manifold Approximation and Projection (UMAP) embeddings of single-cell RNA-seq profiles from 9,104 CD45+ leukocyte cells showing 16 clusters identified by integrated analysis, colored by cluster. Representative of one experiment, n = 6 pooled CD mice and n = 3 pooled HFD mice.
(C) Barplot depicting proportional differences in leukocyte infiltrate from HFD versus CD tumors. Each class contains the following clusters from 3B: immunosuppressive (all M2 macrophage clusters #0, #3, #7, #10, #12; neutrophils #1, and MDSCs #4), pro-immune (all M1 macrophage clusters #2 and #5), dendritic cells (clusters #11 and #14), monocytes (clusters #6 and #9), and lymphocytes (T lymphocytes #8 and natural killer cells #13).
(D) Enrichment of KEGG metabolic signature scores in all single-cell transcriptomes for HFD versus CD tumors.
(E) Schematic depicting the interpretation of panels F-I.
(F-I) Scatterplots showing average signature score, calculated in VISION, for curated KEGG pathways on a cluster-by-cluster basis in HFD versus CD for glycolysis and gluconeogenesis (F), fatty acid metabolism (G), chemokine signaling pathway (H), and T cell receptor signaling (I).
(J) Subset and re-clustering of T lymphocytes from cluster #8 (top), colored by diet (lower left) or cluster (lower right).
(K) Enrichment of KEGG metabolic signature scores that are altered by diet in single-cell transcriptomes from re-clustered CD8+ T cells. CD and HFD q-values are depicted in positive and negative directions, respectively.
(L) Heatmap of the top 5 differentially expressed genes enriched in Tim3+ cytotoxic CD8+ tumor-infiltrating lymphocytes from CD animals (cluster #T-0).
(M) Scatterplot comparing autocorrelation scores computed in Vision for curated immune gene signatures in Tim3+ cytotoxic CD8+ tumor-infiltrating lymphocytes (cluster #T-0). Plot depicts immune signatures that are significantly autocorrelated in at least one diet condition, and the point size reflects the magnitude of the difference in autocorrelation between HFD and CD.
(N) Correlation between KEGG metabolic pathway signatures involved in major carbon-handling pathways and KEGG T Cell Receptor Signaling (left) or Naïve vs. activated CD8+ T cell (GSE15324) signature (right) in Tim3+ cytotoxic CD8+ T cells (cluster #T-0).
Statistical significance was assessed by two-sided binomial test (C), Wilcoxon rank sum with FDR correction using the method of Benjamini and Hochberg (D, F-I, K), empirical p-value calculation with FDR-correction within Vision (M), and by asymptotic t approximation (N). (ns p>0.05, *p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001).
See also Figure S3.
To map metabolic alterations with obesity at a single-cell resolution, we projected a curated set of 61 KEGG metabolic gene signatures onto all cells (DeTomaso et al., 2019). KEGG signatures enriched in leukocytes from CD tumors included sugar metabolism (fructose/mannose metabolism, glycolysis/gluconeogenesis, galactose metabolism, and inositol phosphate metabolism) as well as redox pathways (cysteine and methionine metabolism and the pentose phosphate pathway) (Figure 3D). By contrast, leukocytes from HFD tumors were enriched for multiple pathways involved in fat and cholesterol metabolism (glycosphingolipid biosynthesis, steroid biosynthesis, fatty acid metabolism, and TCA cycle), folate biosynthesis, and pentose and glucuronate interconversion (Figure 3D). Next, we calculated KEGG metabolic signature scores for each cluster and compared average scores in CD versus HFD (Figures 3E–3G, Figures S3C–S3H). These comparisons largely corroborated the KEGG signatures for the complete dataset. For example, glycolysis and gluconeogenesis were significantly enriched with CD for 9 out of 16 clusters (Figures 3D and 3F) and fatty acid metabolism was significantly enriched with HFD across multiple clusters (Figures 3D and 3G).
We also identified cluster-specific metabolic adaptations that were not reflected in the overall cell analysis. For example, oxidative phosphorylation was enriched with CD in four clusters, including T lymphocytes, and enriched in HFD in cluster #10, corresponding to M2 macrophages (Figure S3F). Interestingly, some pathways related to fat synthesis, including glycerolipid and sphingolipid metabolism, were not altered by HFD (Figures S3G and S3H). Clusters #6, #8, and #10, corresponding to monocytes, T cells, and M2 TAMs, were particularly sensitive to HFD, as each of these populations showed significant differences for three out of four of the major metabolic signatures that direct carbon into glycolysis and the TCA cycle (Figures 3E–3G, Figures S3E and S3F).
We next profiled KEGG signaling signatures involved in metabolic regulation and immune activity. As expected, transcriptional signatures of insulin signaling were enriched with HFD, but only in certain myeloid clusters (Figure S3I). Likewise, mTOR signaling was more associated with HFD, but only altered in myeloid populations (Figure S3J). On the other hand, phosphatidyl inositol signaling was enriched in CD for T lymphocytes (S3K). We saw other signatures consistent with impaired T cell function with HFD. In T lymphocytes (cluster #8), we observed significant reductions in chemokine signaling and T cell receptor signaling with HFD (Figures 3H and 3I). Collectively, these data reveal the existence of both common and cell type-specific modes of metabolic adaptations to HFD.
Since our initial clustering did not resolve different T lymphocyte populations, we re-clustered T lymphocytes (cluster #8) into subsets, which identified four sub-clusters (Figure 3J) that all express Cd3g (Figure S3L). Lymphocyte sub-clusters were identified as CD4+ or CD8+ T cells by the expression of Ikzf2 and Cd8a, respectively (Figures S3M and S3N). CD8+ T cells were further characterized as proliferating, Tim3+ cytotoxic, or Slamf6+ stem cell-progenitor subsets based on the expression of the corresponding cell signature (Figures S3O–S3R) (Kowalczyk et al., 2015; Miller et al., 2019). We then scored KEGG metabolic signatures within all CD8+ T cells, which revealed metabolic pathways enriched with CD versus HFD (Figure 3K). We observed metabolic signatures enriched with CD that have been correlated with T cell activation, including sugar and amino acid metabolism (Figure 3K) (Geiger et al., 2016; MacIver et al., 2013; Sinclair et al., 2013; Wei et al., 2017). Due to the small number of cells in clusters T-2 and T-3, we focused subsequent analysis on the T-0 Tim3+ cytotoxic CD8+ T cell population.
When we performed differential expression analysis on Tim3+ cytotoxic CD8+ T cells, the top five genes enriched in CD CD8+ T cells were involved in T cell effector function and included Gzmb, Tnfrsf9, Ifng, Ccl3, and Ccl4 (Figure 3L). To examine changes associated with diet, we scored CD8+ T cells in cluster T-0 against the C7 immunological signature database for MSigDB. We manually curated gene sets to focus on genes involved in CD8+ T cell stimulation and filtered for signatures that were significantly autocorrelated in ≥ 1 condition (Figures S3S and S3T), which represent features that are associated with the cell cluster. Overall, signatures associated with naïve or unstimulated T cells tended to be enriched with HFD, whereas those corresponding to stimulated T cells were enriched with CD (Figure 3M, Figure S3T). To determine if cells scoring higher for T cell stimulation signatures also scored higher for specific metabolic signatures (or vice versa), we calculated the correlation between T cell stimulation signatures and a core set of KEGG metabolic pathways for CD and HFD CD8+ T cells. Indeed, metabolic pathways were more significantly correlated with T cell activation in the HFD TME (Figure 3N). In sum, single-cell profiling revealed that immune cells in the TME undergo unique metabolic adaptations in response to HFD and the differences are distinctive in the T cells, which display altered expression of major central carbon metabolic pathways.
HFD remodels the tumor-immune landscape in the TME
For tumor cell killing, CD8+ T cells require direct cell-cell contact and sufficient metabolic resources. Thus, we sought to understand if obesity affects the location of TILs within the TME as well as whether the positions of T cells in a tumor relate to changes in the intratumoral metabolic niche. We mapped the locations and states of cells in the TME using cyclic immunofluorescence (CyCIF), which enables multiplexed tissue imaging. The resulting 23-plex, sub-cellular resolution images of formalin fixed paraffin embedded (FFPE) tissue sections made it possible to generate an atlas of immune, tumor, and stromal cell populations, map their locations within the TME, and identify key features of their metabolic state (Lin et al., 2018).
Cell clusters were defined computationally, based on patterns of marker expression and the score for their metabolic signature (see materials and methods for details; Figures 4A and 4B, FiguresS4A and S4B). We assigned identities to individual clusters based on the expression of lineage markers (Figure 4C). Overall, nine distinct cell types could be identified and mapped (Figure 4C). Similar to our earlier findings, CyCIF analysis independently showed that HFD tumors contained fewer CD8+ T cells, and further revealed that T cells were not concentrated at the tumor margin, a hallmark of T cell exclusion (Figure 2D, Figure S4C).
Figure 4. Multiplexed imaging reveals metabolic remodeling in tumors with HFD.
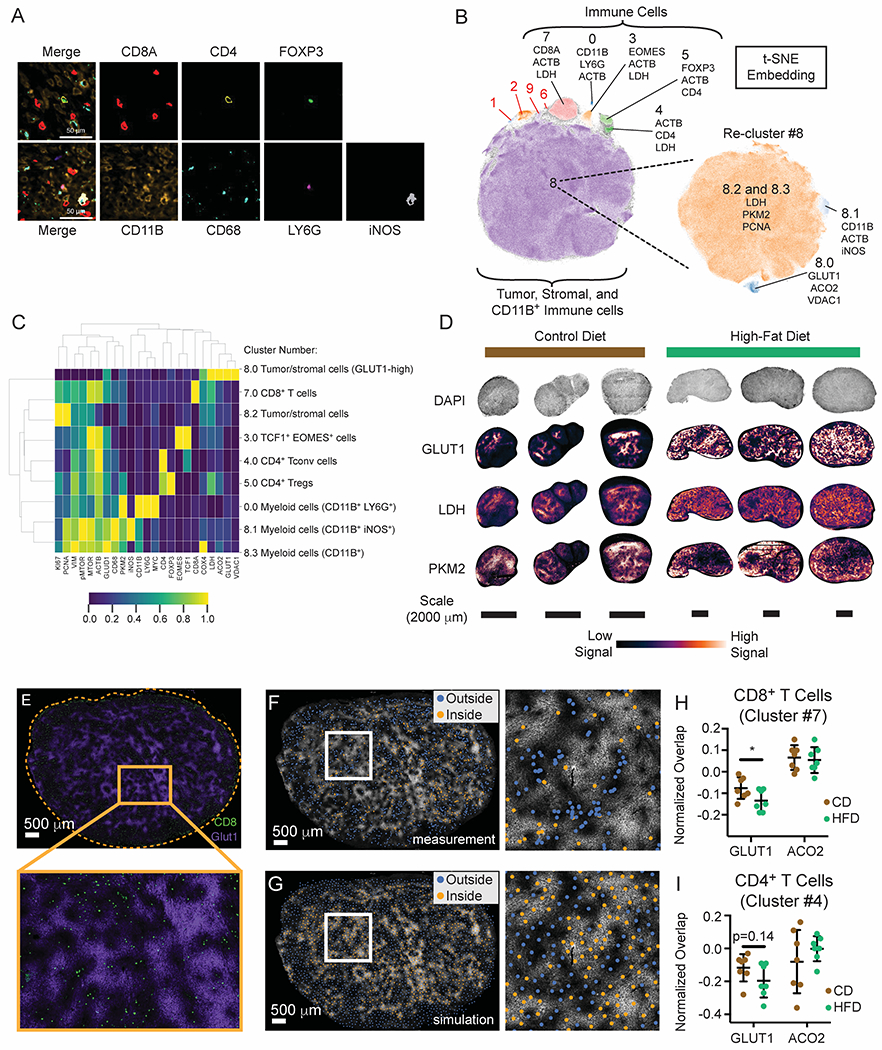
(A-E) CyCIF analysis of MC38 HFD versus CD tumors.
(A) Representative image of CD tumors depicting segregation of immune lineage markers. Scale bars are 50 μm.
(B) Cell populations identified by t-SNE embedding and density-based clustering, showing the top three markers expressed per population.
(C) Heatmap depicting cell populations identified by HDBSCAN from Figure 4B.
(D) Expression pattern of glycolytic genes in CD and HFD tumors.
(E) Representative t-CyCIF image showing GLUT1 (purple) and CD8α (green) expression in the MC38 TME (HFD tumor shown). Scale bar is 500 μm.
(F-G) Representative images depicting real and simulated data used for spatial analysis. GLUT1 expression in the HFD TME superimposed with scatter points representing the x, y coordinates of cells classified as CD8+ T cells (F) or a similar number of uniformly-distributed data points across the same tissue area as generated by Poisson-Disc sampling (G). Data points are colored according to their inclusion (orange) or exclusion (blue) from areas of high GLUT1 expression. Scale bars are 500 μm.
(H-I) Normalized fraction of CD8+ (H) and CD4+ (I) T cells overlapping areas of high GLUT1 or ACO2 expression in the MC38 tumor microenvironment.
Statistical significance was assessed by student’s t-test (H-I). (ns p>0.05, *p≤0.05, **p≤0.01, ***p≤0.001).
See also Figure S4.
We observed substantial variation in the expression of metabolic and cell state markers with cell type and across the TME. Of note, the expression of glycolytic markers (GLUT1, PKM2, and LDH) was non-uniform across tumors with regions of both bright and dim staining (Figure 4D). By contrast, the spatial distributions of other metabolic markers such as GLUD1, ACO2, COX4, and VDAC1 were more uniform across tissue sections (Figure S4D). To determine whether variation in metabolic state across tumors correlated spatially with the locations of immune cells, we measured the overlap between immune cell populations and regions of the tumor with high expression of GLUT1 or ACO2. GLUT1- and ACO2-high regions were defined by protein expression and were not associated with any particular cell type. For this analysis, the proportion of intratumoral CD8+ T cells located in GLUT1-high or ACO2-high regions (Figures 4E and 4F) was compared to a simulated null distribution involving the same number of CD8+ T cells (Figure 4G, Figure S4E). This analysis showed that both CD4+ and CD8+ T cells were significantly less abundant in GLUT1-high regions than expected by chance (Figures S4F and S4G). Exclusion was not observed either for CD8+ T cells in ACO2-high regions (Figure S4F) or for other immune cell types, such as CD11b+ LY6G+ myeloid cells (Figure S4H). To determine if diet changes the propensity for T cells to avoid GLUT1-high regions, we controlled for differences in cell density for each tissue by normalizing the percent overlap between CD8+ T cells and GLUT1-high regions to the corresponding simulated distribution. The normalized overlap between CD8+ and CD4+ T cells and GLUT1 was reduced in HFD tumors (Figures 4H and 4I), with CD8+ T cells significantly more excluded from GLUT1-high areas in HFD than in CD. Thus, while CD8+ T cells are found within HFD tumors, our data suggest that HFD feeding changes metabolic niche interactions within tumors and impacts local T cell infiltration patterns.
HFD causes opposing metabolic changes in CD8+ T cells versus tumor cells
CD8+ T cells rely on many of the same fuel sources and metabolic pathways as tumor cells to support proliferation, survival, and effector functions. To study how diet impacts metabolic reprogramming in different types of cells within tumors, we used bulk RNA-sequencing (RNA-seq) on sorted populations of GFP+ MC38 tumor cells, CD8+ TILs, and CD8+ T cells residing in the dLN from day 12 tumors in CD- or HFD-fed mice. By comparing CD8+ T cells from the tumor and dLN, we identified patterns of gene expression specific to the TME. Principal component (PC) analysis revealed robust separation along PC1 of T cells derived from tumor versus dLN, involving genes with well-described roles in T cell activation (Figure 5A, Figures S5A and S5B). PC2 distinguished TILs isolated from HFD and CD animals (Figure 5A). Thus, PC analysis revealed a larger effect of diet on the transcriptional profiles of CD8+ T cells in tumors than in dLNs, suggesting that the differences in CD8+ T cells with obesity are specific to the TME.
Figure 5. HFD induces distinct metabolic adaptations in MC38 tumor cells and CD8+ TILs.
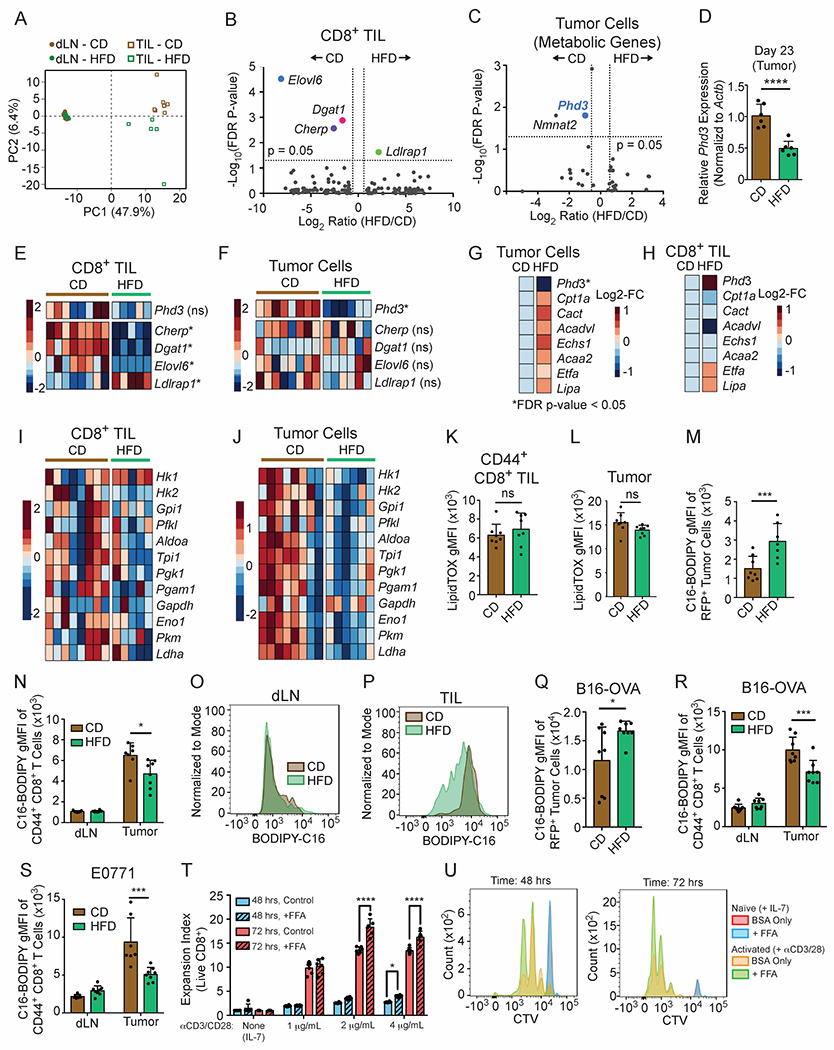
(A-C, E-J) Analysis of RNA-sequencing data performed on cells sorted from day 12 MC38 tumors from CD-fed and HFD-fed animals.
(A) Principal component analysis of the top 400 genes with the largest variance from CD8+ TILs versus CD8+ T cells from the dLN in animals fed HFD or CD.
(B) Volcano plot comparing gene expression levels in CD8+ TILs from CD and HFD tumors. Genes with FDR-corrected p-value < 0.05 are highlighted. Dotted lines indicate 1.5-fold change.
(C) Volcano plot depicting differentially expressed metabolic genes in MC38-GFP tumor cells. Metabolic genes were defined as the union of the following GO gene subsets: GO:0006520 Cellular Amino Acid Metabolic Process, GO:0005975 Carbohydrate Metabolic Process, and GO0006629 Lipid Metabolic Process, or GO:0006099 Tricarboxylic acid cycle, excluding transcription factors. Dotted lines indicate 1.5-fold change.
(D) Phd3 expression in day 23 MC38 tumors measured by qPCR.
(E-F) Heatmaps showing relative expression in CD8+ TILs (E) and MC38 tumor cells (F) of genes that are significantly differently expressed between CD and HFD tumor cells (Phd3) or CD8+ TILs (Cherp, Dgat1, Elovl6, Ldlrap1).
(G-H) Average expression for genes involved in FAO from tumor cells (G) and CD8+ TILs (H).
(I-J) Heatmaps depicting glycolytic genes in CD8+ TILs (I) versus tumor cells (J).
(K-L) Ex vivo LipidTox neutral lipid staining in CD8+ TILs (K) and GFP+ MC38 cells (L) in day 10-14 tumors.
(M) Quantification of C16-BODIPY uptake ex vivo in MC38-RFP tumor cells.
(N-P) Quantification of C16-BODIPY uptake in ex vivo CD8+ T cells (N). Representative histograms for ex vivo C16-BODIPY uptake in CD8+ T cells isolated from dLN (O) or tumor (P) from day 10-14 MC38 tumors.
(Q-S) Quantification of C16-BODIPY uptake ex vivo from dissociated tumors: B16-OVA-RFP tumor cells (Q) or CD8+ TILs isolated from B16-OVA-RFP (R) and E0771 tumors (S) or dLN.
(T-U) Expansion index (T) and representative flow plots (U) measuring proliferation of CTV-labeled CD- and HFD-derived naïve CD8+ T cells after 48 and 72 hours on 1, 2, or 4 μg/mL each of plate-bound anti-CD3 and anti-CD28, with or without supplementation of BSA-conjugated free fatty acids (FFAs).
Data represent ≥ two independent experiments with ≥ 6 mice per group. Graphs display mean +/− SD (K-N, Q-T). (ns p>0.05, *p≤0.05, **≤<0.01).
See also Figure S5.
To understand TME-specific adaptive responses in CD8+ T cells, we studied transcriptional changes in CD8+ TILs. Only four genes exhibited FDR-corrected statistically significant differential expression with HFD (Figure 5B), and three of these genes were involved in fat synthesis or cholesterol metabolism: ELOVL6, DGAT1, and LDLRAP1. To determine whether tumor cells displayed similar adaptations to HFD, we profiled gene expression in sorted MC38 tumor cells. 32 genes were differentially expressed between CD and HFD in MC38 tumor cells (GEO subseries GSE157994) and gene set enrichment analysis (GSEA) revealed hallmarks of hypoxia (FDR q-value = 0.022) and inflammation (TNFα signaling via NFκB, FDR q-value = 0.027) enriched in CD tumors (Figure S5C). Transcriptional changes in CD8+ TILs were non-overlapping with tumor cells (Figures 5C–5F), providing evidence for distinct metabolic adaptations to HFD. Using a less stringent cut-off for significance (p-value < 0.01, non-FDR corrected), only 12 out of 703 differentially expressed genes were common between tumor and CD8+ T cells (Figure S5D). Diet did not alter the expression of genes involved in antigen presentation and immunogenicity in MC38 tumor cells (Figure S5E). These data indicate that HFD has different effects on T cells and tumor cells and that changes in T cells are specific to the TME.
To probe for metabolic adaptations with HFD, we profiled changes in >1800 metabolic genes and known metabolic regulators belonging to four GO categories (GO:0006520, GO:0005975, GO:0006629, and GO:0006099). We identified a significant reduction in Phd3 (Prolyl Hydroxylase-3, alternatively known as Egln3) and Nmnat2 expression in tumor cells as the top significantly changed genes with diet-induced obesity (Figure 5C). We confirmed by qPCR that Phd3 mRNA expression was reduced with HFD in later stage (day 23) tumor lysates (Figure 5D). By contrast, Phd3 expression did not change in the CD8+ TILs (Figure 5E).
PHD3 is a member of the prolyl hydroxylase family, best known for regulating the response to hypoxia by hydroxylating the transcription factor HIF1α (Kaelin and Ratcliffe, 2008). PHD3 has been associated with a number of signaling pathways important in inflammation and immune control (Fu and Taubman, 2010; Garvalov et al., 2014; Xue et al., 2010; Yano et al., 2018). Another study in tumor cells found that PHD3 regulates fatty acid oxidation (FAO) by repressing the import of long-chain fatty acids into mitochondria (German et al., 2016). Thus, we hypothesized that HFD might reprogram tumor cell metabolism at the expense of local CD8+ T cells. We therefore compared average expression levels of a panel of FAO genes within the two RNA-seq datasets. We found that while HFD tumor cells displayed overall changes in gene expression that would promote FAO (Figure 5G), these changes were absent in CD8+ TILs (Figure 5H). In addition, transcript levels for glycolytic genes tended to decrease with HFD in MC38 tumor cells to a greater extent than in CD8+ TILs (Figures 5I and 5J). These data reveal that the metabolic adaptations to the systemic stress of HFD, including changes in fat metabolism, differ between tumor cells and CD8+ TILs.
HFD reprograms fat utilization in the TME
To monitor lipid storage profiles in CD8+ TILs and tumor cells, we measured neutral lipid accumulation using LipidTOX staining. We found that both CD8+ TILs and MC38 tumor cells contained similar levels of neutral lipids in both diets (Figures 5K and 5L, Figure S5F). To test whether diet alters fatty acid uptake, we measured palmitate influx ex vivo using BODIPY-labeled palmitate (C16-BODIPY). Tumor cells derived from HFD mice took up more fatty acid than CD tumor cells (Figure 5M). We reasoned that alterations in tumor-intrinsic fat utilization might impact fat uptake by CD8+ T cells in the same microenvironment. Whereas diet did not alter baseline palmitate uptake by CD8+ T cells in the dLN (Figures 5N and 5O), CD44+ CD8+ TILs from HFD-fed mice acquired less palmitate from media than their CD counterparts (Figures 5N and 5P). This was also true in B16-OVA-RFP and E0771 tumors (Figures 5Q–5S). Data on CD44+ CD8+ TILs contrast with other tumor-infiltrating leukocyte populations from HFD tumors, such as CD11b+ myeloid cells, which did not reduce fatty acid uptake (Figures S5G–S5I). Thus, tumor and CD8+ T cells appear to rewire their metabolism differently: tumor cells adapt and increase fatty acid utilization, whereas CD8+ T cells do not.
Enhanced fatty acid uptake by tumor cells may leave T cells deprived of fatty acids in the TME. Consistent with this hypothesis, naïve CD8+ T cells activated in vitro in charcoal-stripped serum-containing media, which contains few fatty acids, proliferated better with fatty acid supplementation (Figures 5T and 5U). In contrast, FFA supplementation did not affect MC38 tumor cell proliferation (Figure S5J), demonstrating a cell-type specific vulnerability to fat availability. In total, these findings provide functional evidence that individual cell populations in the TME mount distinct responses to systemic metabolic perturbations (obesity), resulting in differences in fatty acid utilization between immune and tumor cells.
Proteomic analysis reveals fatty acid uptake and oxidation signatures by HFD tumor cells
To gain a deeper molecular understanding of tumor cell adaptations to HFD, we compared the proteome of GFP+ tumor cells sorted from CD or HFD tumors using tandem mass tag (TMT)-based quantitative proteomics (Figure 6A). Principal component analysis and hierarchical clustering of 7,178 proteins showed that CD- and HFD-derived MC38 cells have distinct proteomes (Figures S6A and S6B). Using fast pre-ranked gene set enrichment analysis, we found that fatty acid metabolism and oxidative phosphorylation were among the pathways most enriched in HFD tumor cells (Figure 6B). IFNγ response in HFD was reduced relative to CD, which could be explained by reduced CD8+ T cell infiltration (Figure 6B).
Figure 6. Protein-level analysis confirms enhanced fatty acid uptake and oxidation by HFD tumor cells.
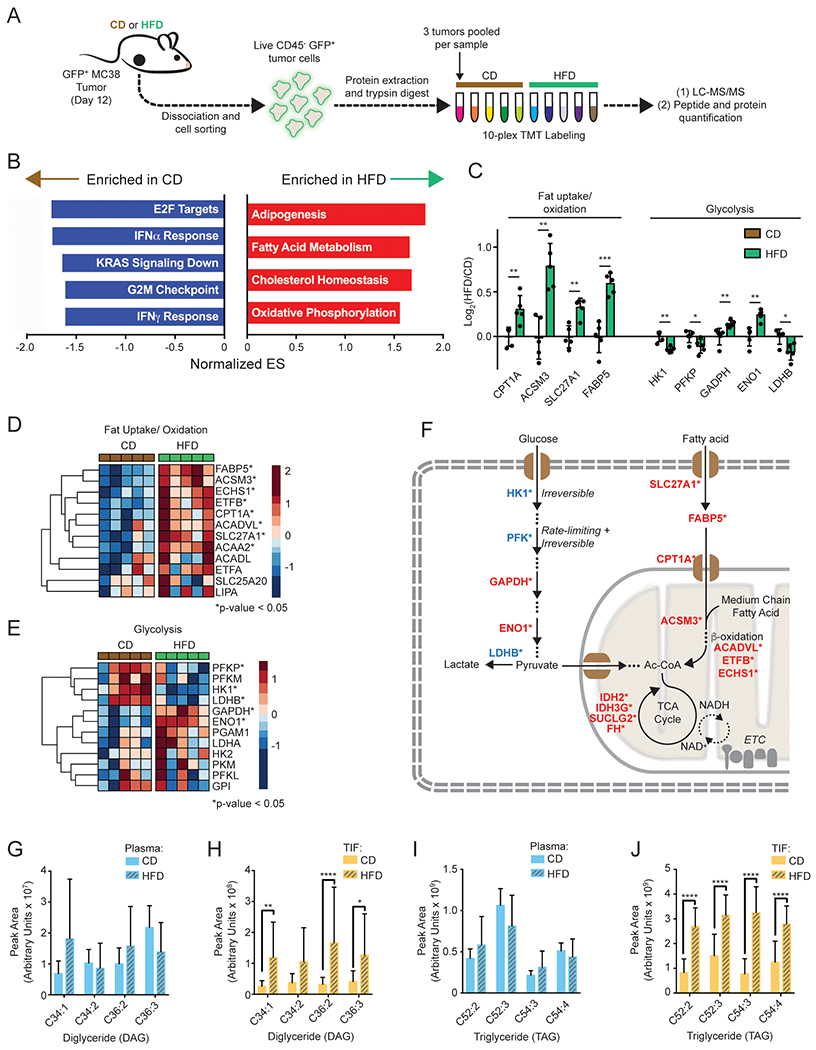
(A) Schematic depicting TMT-proteomics experiment.
(B) Enrichment analysis using Hallmark gene sets from MSigDB.
(C) Bar graph showing relative expression of key proteins involved in fat oxidation or glycolysis.
(D-E) Heatmaps depicting relative expression levels of proteins involved in fat uptake and oxidation (D) or glycolysis (E).
(F) Schematic depicting key upregulated (red) or downregulated (blue) proteins in fat uptake and oxidation, glycolysis, and TCA cycle.
(G-J) Relative abundance of indicated DAG (G-H) and TAG (I-J) lipid species in CD and HFD plasma (G,I) and TIF (H,J).
Key abbreviations: DAG, diglyceride. ES, Enrichment Score, TAG, triglyceride. TIF, tumor interstitial fluid. Graphs display mean +/− SD. Statistical significance was assessed by Student’s t-test (C-J). (ns p>0.05, *p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001).
See also Figure S6.
Proteomic analysis revealed additional mechanisms whereby HFD supports fat utilization in tumors, via induction of transporters (SLC27A1), fatty acid binding proteins (FABP5), and proteins involved in mitochondrial beta-oxidation (CPT1A, ACSM3, ACADVL, ETFB, and ECHS1) (Figures 6C, 6D, and 6F). By contrast, glycolytic enzymes catalyzing irreversible and/or rate-limiting steps were downregulated with HFD (Figures 6C–6F). There was no clear change in proteins involved in fat synthesis, while the expression of several TCA cycle proteins increased with HFD, in line with the gene set enrichment analysis (Figure 6F, Figures S6C and S6D). The expression of proteins mediating antigen processing and presentation was not significantly changed with diet, suggesting that reduced anti-tumor immunity with HFD was not caused by altered antigen presentation, in line with RNA-seq data (Figures S5E and S6E). Overall, these proteomic data support that HFD MC38 tumor cells rewire metabolism to increase fatty acid uptake and oxidation.
HFD alters the neutral lipid composition of the TME
Since fat oxidation signatures were highly enriched in tumor cells with HFD, we performed targeted lipidomics to measure the effect of HFD on lipid levels in the circulation and in the TME. We analyzed the lipid composition of plasma and two intratumoral compartments: sorted GFP+ MC38 tumor cells and tumor interstitial fluid (TIF). Whereas few lipid species were significantly altered in tumor cells (Figure S6F), HFD had a larger impact on lipid availability in both plasma and TIF (Figures S6G and S6H). Because circulating nutrients are a main source of metabolites in TIF, we anticipated many metabolites would scale uniformly between TIF and plasma (Sullivan et al., 2019). Thus, we plotted the TIF-to-plasma ratio for each lipid with either diet to identify the lipid classes specific to the TME that were most altered by HFD. As expected, there was a strong positive correlation between the TIF-to-plasma ratios in CD and HFD for all lipids analyzed (Pearson r = 0.8071, p<0.0001), showing that TIF composition primarily reflects lipid levels in circulation (Figure S6I). However, we noticed that triglyceride (TAG) and, to a lesser extent, diglyceride (DAG) species tended to occupy off-diagonal positions (Figures S6I–S6L). Removing these lipid classes increased the correlation between HFD and CD (Pearson r = 0.9543, p<0.0001), and improved the goodness-of-fit to a linear scaling model (Figures S6L and S6M), indicating that DAG and TAG levels are major differences in the TME in mice fed HFD. Indeed, the top four TAG and DAG species (by peak intensity) revealed significant enrichment in TIF with HFD, but not in plasma (Figures 6G–6J). To check if local lipoprotein lipase (LPL) activity could account for the lipid enrichment in TIF with HFD, we measured LPL activity in homogenized tumor lysates and mouse heart for comparison. We observed similar LPL activity in CD and HFD tumors (Figure S6N), suggesting that HFD tumors contain a lipid-rich microenvironment with local lipase activity that facilitates cellular uptake.
Tumor cell PHD3 expression controls fatty acid availability in the HFD TME
Since we found that HFD reprograms the TME to enhance fat uptake in tumors, we hypothesized that these HFD-induced changes in tumor cell fat metabolism may impact FFA availability and CD8+ T cell function in the TME. We postulated that preventing HFD-induced metabolic rewiring may restore CD8+ T cell responses and prevent increased tumor growth on HFD. To test this idea, we overexpressed PHD3 in MC38 cells (MC38 PHD3-OE), which is one of the main metabolic regulators changed in MC38 cells with HFD (Figure S7A). PHD3-OE cells had higher levels of hydroxyl ACC2, which is a PHD3 substrate that regulates fat oxidation and read-out of PHD3 activity (Figure S7B). MC38 PHD3-OE cell lines grew at the same rate as control cells in vitro (Figure S7C). PHD3 overexpression was maintained when cells were injected into CD and HFD animals (Figures S7D and S7E). Moreover, PHD3 overexpression did not alter the expression of MHC-I or PD-L1, but did significantly reduce fatty acid uptake by the tumor cells (Figures S7F–S7H).
To test the hypothesis that PHD3 expression in tumor cells modulates local availability of FFAs, we used targeted metabolomics to measure FFAs in plasma and TIF from MC38 tumors (Figure 7A). To verify the purity of our TIF isolation, we compared normalized NAD+ levels between TIF and whole tumor tissue lysate, which indicated little contamination of our TIF preparation with intracellular contents (Figure S7I). HFD increased circulating levels of many FFAs, including palmitic and oleic acid (C16:0 and C18:1) (Figures 7B and 7C, Figure S7J). By contrast, local FFA levels in TIF were reduced in animals on HFD as compared to CD (Figure 7D). Notably, most metabolites in TIF were unchanged by diet (data not shown), indicating a unique role for FFAs in the HFD TME. Depletion of fatty acids in the TME is paradoxical given the high level of dietary fatty acids in HFD and suggests that local metabolic reprogramming may affect FFA availability.
Figure 7. Modulating Phd3 expression in tumor cells increases CD8+ T cell infiltration and reduces tumor growth kinetics during HFD.
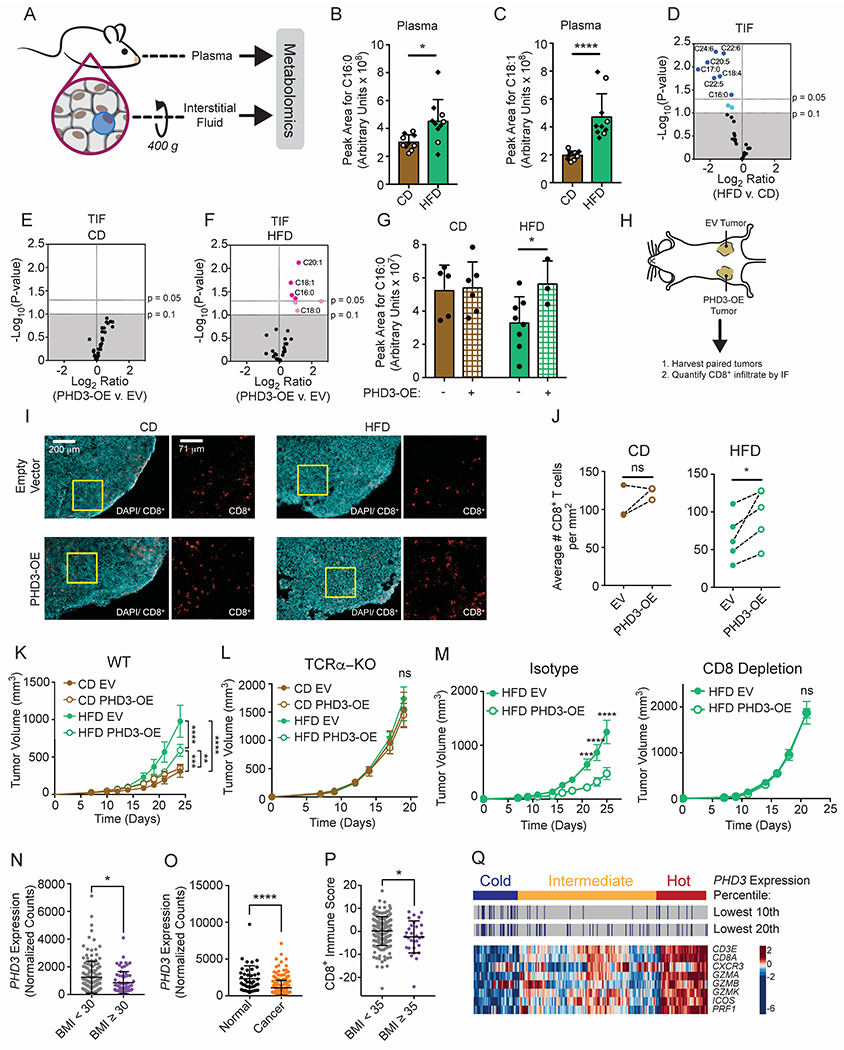
(A-G) Metabolomic analysis for FFA content of TIF and plasma from CD- or HFD-fed day 14 MC38 tumors.
(A) Experimental schematic for fractionation of interstitial fluid.
(B-C) Comparison of palmitate (B) and oleate (C) levels in plasma from tumor-bearing mice fed HFD versus CD. Open circles correspond to mice bearing PHD-OE tumors and diamonds correspond to mice bearing empty vector-transduced tumors.
(D-F) Volcano plots comparing FFA abundance in TIF that change with diet (D), or PHD3-OE versus empty vector (EV)-transduced MC38 tumors from CD-fed (E) and HFD-fed (F) animals. Blue circles represent FFAs that decrease across the tested conditions, where light blue corresponds to 0.05<p-value<0.1 and dark blue corresponds to FFAs with p-value<0.05. Pink circles represent FFAs that increase across the tested conditions, where light pink corresponds to 0.05<p-value<0.1 and dark pink corresponds to FFAs with p-value<0.05.
(G) Comparison of palmitate (C16:0) levels in TIF among HFD versus CD and PHD3-OE versus EV-transduced MC38 tumors.
(H-J) Histological analysis of PHD3-OE versus EV-transduced day 12 MC38 tumors.
(H) Schematic depicting experimental setup with paired tumors in HFD or CD-fed mice.
(I) Images showing tissue architecture as well as numbers and localization of CD8+ T cells.
(J) Blinded quantification of CD8+ T cell numbers in tissue sections.
(K-L) Tumor growth curves of CD-fed and HFD-fed WT C57BL/6J (K) or TCRα-KO (L) mice inoculated with 105 EV-transduced or PHD3-OE MC38 tumor cells.
(M) Tumor growth curves of HFD-fed WT C57BL/6J mice inoculated with 105 EV-transduced or PHD3-OE MC38 tumor cells and treated with isotype control (left) or depleting anti-CD8 (right) antibodies.
(N-Q) Bioinformatics analysis of colon adenocarcinoma (COAD) RNA-seq TCGA data.
(N) PHD3 expression in obese and non-obese COAD patients.
(O) PHD3 expression in cancer versus normal tissue in COAD patients.
(P) CD8+ T cell Immune Score from severely obese and non-obese COAD patients, calculated as the genewise z-score sum of CD8+ T cell marker genes shown in panel (Q).
(Q) COAD samples clustered by CD8+ T cell expression signature. PHD3 expression was stratified based on a percentile cut-off and combined with the clustering results.
Data represent one independent experiment with > 6 mice per group (K-M). Graphs display mean +/− SD (B, C, G, N-O) or mean +/− SEM (K-M). (ns p>0.05, *p≤0.05, **p≤0.01, ***p≤0.001, ***p≤0.0001).
See also Figure S7.
Next, we compared relative FFA levels in TIF from MC38 PHD3-OE and empty vector (EV) control tumors. Whereas PHD3 overexpression had no significant impact on FFA levels in CD TIF (Figure 7E), several FFAs increased in PHD3-OE TIF with HFD (Figure 7F). FFAs constituting major lipid carbon sources in circulation, including palmitate and oleate (C16:0 and 18:1) were among the significantly changed fatty acids (Figure 7F). Of note, tumor cell PHD3 overexpression was sufficient to restore palmitate availability in the TME (Figure 7G). Thus, restoring PHD3 expression in tumor cells is sufficient to alter nutrient availability in the TME.
Tumor cell PHD3 overexpression promotes CD8+ T cell tumor control
If metabolic reprogramming and depletion of FFAs within the TME diminish local anti-tumor immunity, then counteracting this local metabolic rewiring may improve tumor control by infiltrating CD8+ T cells. We therefore tested whether altering PHD3 expression in MC38 cells would affect tumor control by CD8+ T cells. As numbers of CD8+ TILs might be reduced in a TME depleted for critical fuels, we asked whether raising PHD3 levels in tumor cells would increase CD8+ infiltration in HFD tumors. We injected EV-transduced and PHD3-OE MC38 tumor cells in opposing flanks of mice fed CD or HFD and measured intratumoral CD8+ T cell numbers and localization (Figure 7H). MC38 tumor sections were stained with DAPI to reveal overall architecture, and then for CD8 to reveal infiltration of CD8+ T cells (Figure 7I). Quantification of images revealed depletion of CD8+ T cells in HFD, without major changes to CD8+ T cell localization around or within the tumors (Figure 7J). PHD3 overexpression in tumor cells significantly increased CD8+ T cell infiltration in HFD animals (Figure 7J), providing evidence that changing a metabolic pathway in tumor cells can impact tumor control via a non-cell autonomous mechanism.
We investigated the effect of PHD3 overexpression on tumor growth in vivo. Ectopic PHD3 expression did not alter tumor growth kinetics in mice fed a CD (Figure 7K). However, MC38 PHD3-OE tumor growth was reduced compared to EV control in HFD animals (Figure 7K). To test the hypothesis that this finding was a result of better tumor control by CD8+ T cells, we examined tumor growth in TCRα-KO mice (Figure 7L) and in CD8+ T cell-depleted mice (Figure 7M). PHD3-OE had no effect on tumor growth in TCRα-KO mice on HFD (Figure 7L), which shows that PHD3-OE MC38 cells do not have an intrinsic reduction in growth over control MC38 cells. Moreover, while PHD3-OE reduced tumor growth rates in HFD mice treated with isotype control antibody, PHD3 expression status did not affect tumor growth rate in CD8+ T cell-depleted mice (Figure 7M). These data show that maintaining high PHD3 expression in MC38 tumor cells improves the anti-tumor T cell response in HFD mice and mitigates the effects of HFD on anti-tumor immunity. Overall, multiple lines of evidence reveal that HFD-induced local metabolic rewiring in the tumor alters fuel partitioning and reduces anti-tumor immunity in the TME.
PHD3 loss correlates with reduced anti-tumor CD8+ T cell function across multiple human cancers
To explore whether obesity alters the tumor metabolic landscape in human patients, we analyzed the colon adenocarcinoma (COAD) RNA-seq dataset available on the public domain of The Cancer Genome Atlas (TCGA) with corresponding BMI data. Expression of PHD3, but not of PHD1 or PHD2, was significantly lower in tumors from obese patients with BMI ≥ 30 kg/m2 (Figure 7N, Figure S7K). We also observed a reduction in PHD3 expression in cancer compared to normal tissue from COAD patients (Figure 7O) (Radhakrishnan et al., 2016; Rawluszko et al., 2013).
Based on these findings, we hypothesized that obesity might reduce T cell infiltration or function in human tumors. We scored COAD tumors based on the expression of a CD8+ gene signature. Using this metric, CD8+ T cell infiltration was reduced in tumors from severely obese patients (BMI ≥ 35 kg/m2) (Figure 7P), consistent with a recent study that found fewer CD8+ TILs in colorectal tumors from obese patients (Wang et al., 2018). We also analyzed the association between PHD3 transcript levels and markers of CD8+ T cell-mediated immunity and inflammation in human patient samples. There were significant positive correlations between MHC-I and GZMB with PHD3 expression across multiple cancer types (Figures S7L and S7M), suggesting that PHD3-low tumors are less inflamed.
Next, we asked whether patient samples with low PHD3 expression might be enriched among immunologically “cold” tumors. We tested this hypothesis in five TCGA datasets composed of the most significantly correlated cancers from the Spearman analysis (COAD, PRAD, KIRC, LUAD, and THCA) (Figures S7L and S7M) as well as melanoma (SKCM) for comparison. Patient samples were stratified as PHD3-high or PHD3-low based on percentile cut-offs for PHD3 expression of 10% or 20%. We then clustered patient samples into immunologically “hot”, “intermediate”, or “cold” categories based on CD8+ gene signature score (Figure 7Q). Not only was PHD3 expression lower in cold COAD tumors (Figure S7N), we also found that PHD3-low samples were significantly enriched among cold tumors for five out of six cancer types, and for all cancer types tested with a strong correlation between PHD3 and MHC-I or GZMB expression (Figures S7L, S7M and S7O). As a control, GLUD1 expression, an unrelated metabolic gene involved in glutaminolysis, was not enriched in immunologically cold tumors (Figure S7O). These data show that PHD3 downregulation occurs in human cancers and correlates with reduced immunity.
Discussion
In this paper, we identify systemic metabolism, as altered by diet, to be a critical determinant of metabolic programs in the TME. Tumors develop in metabolically diverse contexts, since patients have a broad spectrum of dietary habits, blood chemistry parameters, adiposity, and basal metabolic properties. Previous studies have focused primarily on tumor cell-intrinsic metabolic rewiring associated with these variables. Our work demonstrates that cellular components of the TME, for example immune and tumor cells, can behave differently. Adaptations by tumor cells to HFD cause T cell dysfunction due to altered fatty acid partitioning and local depletion of essential metabolites, revealing that the metabolic states of cells within tumors can be coupled. These findings highlight the complexity of local metabolism within tumors and demonstrate that it is critical to consider metabolic reciprocity between intratumoral cell populations as a factor regulating immune control over tumor growth.
Several recent studies have found that dietary perturbations altering systemic metabolic state can also impact anti-tumor immunity (Di Biase et al., 2016; Pietrocola et al., 2016; Rubio-Patiño et al., 2018). For example, low-protein diet enhances cytokine production by tumor cells, which promotes the anti-tumor immune response (Rubio-Patiño et al., 2018). Another study showed that obesity diminished CD8+ T cell function in the PyMT tumor model, which was associated with increased FAO and reduced glycolysis in T cells (Zhang et al., 2020). One key difference with our study is that PyMT tumors occur in the lipid-rich environment of the mammary fat pad. Also, we do not see changes in glycolytic gene expression with diet in CD8+ T cells (Figure 5I). Finally, a recent study found that diet-induced obesity enhances responses to PD-1/PD-L1 therapy in the B16 mouse melanoma model (Wang et al., 2018), which matches clinical data showing better outcomes for obese melanoma patients treated with immune checkpoint blockade (Cortellini et al., 2019; McQuade et al., 2018; Murphy and Longo, 2019; Woodall et al., 2020). In agreement with these studies, we also find a defective baseline anti-tumor T cell response with HFD. Melanoma is not among the cancer types in which we find a correlation between low PHD3 expression and reduced CD8+ T cell signatures (Figure S7O), suggesting that metabolic properties unique to specific cancer types may impact intratumoral changes in response to obesity. Clearly, metabolic state may have profound effects on therapeutic outcomes, and these effects are under-explored.
The current study provides insight into the immunometabolic landscape within tumors at single-cell resolution. Our studies reveal that tumor metabolism may significantly differ in a lean versus an obese setting, and that dietary stress may amplify the metabolic tug of war in tumors with a direct effect on the local function of CD8+ T cells. We show that obesity potentiates transcriptional and metabolic reprogramming events leading to altered nutrient availability in the TME and immune dysfunction. Thus, the full impact of metabolic reprogramming in tumors cannot be captured by studying tumor cells in culture without pressures imposed by the microenvironment. Rather, to fully harness the power of targeting metabolism in patients, we must understand the interplay between systemic metabolism, changes in the TME, and the potential for different effects on the metabolism of tumor and immune cells. An improved understanding of how systemic metabolism affects nutrient partitioning and immune function in the TME may have implications for therapeutic interventions targeting cancer metabolism and/or anti-tumor immunity with impacts on precision medicine and future patient care.
Limitations of Study
The study presented here has several important considerations. First, our data are suggestive of a T cell activation defect in tumors, rather than terminal, irreversible exhaustion, and it would be interesting for future studies to dissect this further. Second, we show that HFD alters the nutrient content within the TME, which can be reversed by PHD3 overexpression in MC38 tumor cells to improve anti-tumor CD8+ T cell responses. While these data provide proof of concept that the transcriptional changes with HFD in tumor cells dampen anti-tumor immunity, we did not formally demonstrate this is caused by enhancing fat oxidation in MC38 cells. As diet-induced obesity leads to systemic perturbations and PHD3 overexpression only partially rescues CD8+ T cell infiltration and tumor growth kinetics, other mechanisms likely contribute to reduced anti-tumor CD8+ T cell responses. For instance, our studies do not rule out the role of other immune cell types in these phenotypes. Alternatively, HFD may induce the production of a metabolite downstream of altered metabolism within the TME that is inhibitory to immune cells. Finally, as tumors display an array of metabolic reprogramming, the full spectrum of cancer adaptation to systemic stress will depend upon tumor types, as well as the systemic physiology of the individual. Our data determine conclusively that an individual’s systemic metabolism can transmit signals to the TME. It will be exciting for future investigation to study other systemic perturbations in this context, such as other models of metabolic dysfunction and aging.
STAR Methods
Resource Availability
Lead Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Marcia C. Haigis (Marcia_Haigis@hms.harvard.edu).
Materials Availability
Plasmids generated in this study will be made available by request to the lead contact.
Data Code and Availability
RNA-sequencing datasets generated during this study are available at the GEO repository within the GSE157999 reference series. TMT-proteomics data generated during this study have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD019495. All other relevant data are available from the corresponding author on request.
Experimental Model and Subject Details
Cell Lines
MC38, LLC, B16, Phoenix, and 293T cells were cultured in normal DMEM without pyruvate supplemented with 10% FBS and 1% penicillin/streptomycin. E0771 and CT26 cells were cultured in RPMI 1640 supplemented with 10% FBS, 10 mM HEPES and 1% penicillin/streptomycin. RENCA cells were cultured in RPMI 1640 supplemented with 10% FBS, 1% penicillin/streptomycin, 0.1 mM non-essential amino acids, 1 mM sodium pyruvate, and 2 mM extra L-glutamine. Cells derived from dissociated tumors were cultured in R10 or R2, which consists of RPMI 1640 medium supplemented with either 10% or 2% FBS, 1% penicillin/streptomycin, 10 mM HEPES, and 0.05 mM 2-mercaptoethanol. MC38, Phoenix, 293T and E0771 cells are female. B16 and RENCA cells are male. The sex of LLC and CT26 cell lines is not published. All cells were cultured at 37°C in a humidified 5% CO2 incubator. All FBS was heat-inactivated prior to use.
Mice
4-week old C57BL/6, TCRα knock-out (B6.129S2-Tcrαtm1Mom/J), and C57BL/6-Tg(TcraTcrb)1100Mjb/J (OT-1) female mice were purchased from Jackson Laboratories. OT-1 CD8+ TCR-transgenic mice have been previously described (Hogquist et al. 1994). For all experiments, 5-week old mice were assigned to CD (PicoLab Rodent Diet 20 5053; Lab Diet) or HFD (#12492; Research Diets, Inc.) for 8-10 weeks. All mouse colonies and experimental animals were maintained in the same animal facility at Harvard Medical School and housed in specific pathogen-free conditions. All animals were used in accordance with animal care guidelines from the Harvard Medical School Standing Committee on Animals and the National Institutes of Health. All mouse protocols were approved by the Harvard Medical Area Standing Committee on Animals.
Method Details
Cloning
PHD3-overexpression vectors were constructed by PCR-amplifying mouse PHD3 (Egln3) using CloneAmp HiFi PCR Premix (Clontech) from a plasmid containing full-length PHD3 (MmCD00320451) using primers containing BamHI and Sal I restriction sites. The digested insert was ligated into pLenti CMV GFP Blast (659-1) using Quick Ligase (NEB BioLabs) according to the manufacturer’s instructions. Resulting ligation reactions were transformed in Stbl3 E. coli for vector propagation and validation. RFP-overexpression vectors were constructed in the same way, but using primers containing Xho I and EcoR V restriction sites. The digested insert was ligated into MSCV-PIG (Puro-IRES-GFP, Addgene #18751), which was digested with the same enzymes, to replace GFP with RFP. For the generation of B16-OVA-RFP cells from B16 melanoma cells, plasmids were generated by replacing the puromycin resistance cassette with OVA and replacing GFP with RFP in the MSCV-PIG plasmid.
Ectopic Gene Overexpression
Phoenix-ECO packaging cells were transfected with MSCV-PIG, which constitutively expresses GFP, or MSCV-PIR (generated in this paper, which constitutively expresses RFP) or MSCV OVA-IRES-RFP to produce retrovirus. Fluorescent MC38 cell lines were generated by infecting MC38 cells with viral supernatants for 24 hours, and then sorting for the brightest 20% of GFP+ or RFP+ cells after 48 hours of resting in normal DMEM. Cells were sorted a second time 7-14 days later and the brightest 20% of GFP+ or RFP+ cells were collected and maintained in vitro under standard culture conditions. B16-OVA-RFP cells were created following the same protocol, infecting B16 melanoma cells with OVA-IRES-RFP retrovirus. Cell stocks were frozen and stored in liquid nitrogen. Fluorescent cell lines were routinely selected for 24-48 hours in puromycin after recovery to ensure retention of the fluorescent marker.
Lentivirus containing PHD3-OE plasmid or empty vector control were produced by co-transfecting HEK 293T cells with the target plasmid plus four helper plasmids (5 μg target, 2 μg pHRΔ8.2, 2 μg CMV-VSVG, 0.5 μg pMD.G, and 0.5 μg CMVΔR8.2) using a 1:3 ratio of Fugene 6 to DNA. Virus was harvested 48 hours post-infection and filtered through a 0.45 μm filter. Lentivirus was frozen at −80°C for long-term storage or used immediately. For MC38 infection, MC38 cells were trypsinized and resuspended in media containing 10 μg/mL Polybrene (Santa Cruz). Trypsinized cells were diluted 1:1 with fresh or thawed lentivirus and then plated for 24 hours before changing the media. 48 hours post-infection, MC38 cells were selected with 7 μg/mL blasticidin (Sigma-Aldrich) for 48-72 hours before using for assays. Frozen MC38 PHD3-OE cell lines were cultured for 24 hours in the presence of 7 μg/mL blasticidin before expanding for tumor injections.
Mouse Tumor Models
After 8-10 weeks of CD or HFD feeding, mice were anesthetized with 2.5% Avertin diluted in 1X DPBS, shaved at the injection site, and then injected subcutaneously in the abdominal flank with 105 (MC38, LLC, and B16) or 2.5x105 (B16-OVA-RFP, CT26, and RENCA) cells, or in the mammary gland with 2x105 E0771 cells. Once palpable tumors were present, tumors measurements were performed using a caliper every 2-3 days. Tumor volumes were calculated using the following formula for ellipsoid volume: 0.5 x D x d2, where D is the long and d is the short diameter. Mice were sacrificed at humane endpoints or day 10-14 for tissue harvest.
Metabolic Phenotyping
Plasma glucose concentrations were measured in blood collected from the tail vein using a Contour blood glucose meter. To measure plasma insulin, leptin, IL-6, adiponectin, resistin, and cholesterol concentrations, mice were separated into fed or fasted groups and fasted overnight for 16 hours before blood was collected via cardiac puncture into EDTA-coated tubes. Whole blood was spun at 1500 g for 15 minutes at 4°C and the upper plasma layer was moved to a new tube. Plasma samples were sent for analysis at the Vanderbilt University Medical Center (VUMC) Lipid Core or VUMC Hormone Assay & Analytical Services Core.
Antibody-Mediated T Cell Depletions
Mice were treated with six doses of depleting antibodies or isotype control delivered by intraperitoneal injection on days-1 (300 vg), 1 (200 μg), 4 (200μg), 8 (200μg), 12 (200 μg), and 16 (200 μg) relative to tumor injection (day 0). Depletion efficiency was checked by flow cytometry on cheek bleeds on days 3, 10, and 18 using antibodies targeting non-competing CD8 epitopes. For depletion experiments with MC38 cell lines, the following antibodies were used: rat IgG2b isotype control (BioXCell, Clone #LTF-2) or anti-CD8α (BioXCell, Clone #2.43). For depletion experiments with PHD3-OE cell lines, the following antibodies were used: rat IgG1 isotype control (BioXCell, Clone #TNP6A7) or anti-CD8β (BioXCell, Clone #53-5.8)
Protein Extraction and Western Blotting
MC38 cells were washed once with ice-cold 1X DPBS and then lysed directly in ice-cold RIPA lysis buffer (150 mM NaCl, 5 mM EDTA, 50 mM Tris pH 8.0, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) supplemented with 1 mM DTT, EDTA-free protease inhibitor (Sigma), phosphatase inhibitor cocktail 2 (Sigma), and phosphatase inhibitor cocktail 3 (Sigma). Cells were scraped into Eppendorf tubes and incubated on ice for 15 minutes with occasional vortexing before clarification by centrifugation at >16,000 g in a microcentrifuge at 4°C for 10 minutes. Protein concentrations were determined by BCA assay (ThermoFisher Scientific). Equal concentrations of protein were diluted into 1X SDS-PAGE loading buffer, boiled at 95°C for 10 minutes, and then loaded onto Criterion TGX 4-20% gels (Biorad). Proteins were transferred onto nitrocellulose membranes (Biorad) and transfer quality was assessed by Ponceau staining. All Western blotting solutions were prepared in 1X TBST. The membrane was blocked for 1 hour at room temperature in 5% nonfat milk and then incubated overnight at 4°C with the indicated antibodies diluted in 3% BSA: αPHD3 (ThermoFisher Scientific, Cat# PA1-20196, 1:1000), αFLAG (Cell Signaling, Cat# 2368, 1:1000), αBeta-Actin (Sigma, Cat# A2066, 1:10,000). The membrane was washed three times for 5 minutes with 1X TBST and then incubated for one hour at room temperature with the corresponding secondary HRP-conjugate antibody diluted in 5% nonfat milk. The membrane was washed three times with 1X TBST and then developed using ECL solution (Perkin Elmer).
Protein Immunoprecipitation (IP)
ACC2 immunoprecipitation from MC38 cells and Western blotting for hydroxy-proline post-translational modifications was performed as previously described (German et al., 2016).
Tumor Dissociation and Tumor-Infiltrating Leukocyte Isolation
Tumors were harvested on day 10-14 post-injection and digested in 1X DPBS containing calcium, magnesium, and 250 units/mL of Type 1 Collagenase (Worthington Biochemical Corporation). For samples that were not run through Percoll gradients, this buffer also contained 20 units/mL DNase I. Samples were dissociated by GentleMACS (MiltenyI), incubated for 20 minutes at 37°C with gentle rocking, dissociated by GentleMACS again, and then filtered through a 70 μm filter. In some experiments, TILs were enriched by centrifuging samples through a Percoll gradient. Briefly, dissociated tumor cells were resuspended in 5 mL 40% salt-adjusted Percoll (GE Healthcare Lifesciences), which was layered over 2 mL 70% salt-adjusted Percoll. Samples were spun at room temperature for 20 minutes at 800 g with the acceleration and brake off. Leukocytes were recovered from the interface of the 40% and 70% Percoll layers.
Flow Cytometry and Staining
Primary mouse cells isolated from spleen, draining lymph node, and tumor were stained with fluorescent antibodies and analyzed by flow cytometry. For experiments with live/dead criteria, cells were first stained with LIVE/DEAD Fixable Near-IR stain (ThermoFisher Scientific) in 1X DPBS according to the manufacturer’s instructions. Subsequent surface marker staining was performed in MACS buffer containing 1X DPBS supplemented with 1% FBS and 2 mM EDTA. Intracellular staining for flow panels containing nuclear proteins was performed using the eBioscience FoxP3/ Transcription Factor Staining Buffer Set (ThermoFisher Scientific). For intracellular staining of cytoplasmic proteins, such as cytokines, the Fixation/Permeabilization Solution Kit (BD Biosciences) was used. Intracellular cytokine staining was performed after a 4 hour stimulation with PMA (100 ng/mL) and ionomycin (500 ng/mL) in the presence of GolgiStop at 37°C. Please see Key Resources Table for the fluorescently labeled antibodies used for staining.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| DYKDDDDK Tag Antibody (FLAG) | Cell Signaling | Cat#2368S; RRID: AB_2217020 |
| PHD3 Polyclonal Antibody | ThermoFisher Scientific | Cat#PA1-20196; RRID: AB_2096876 |
| Anti-Actin antibody produced in rabbit | Sigma | Cat#A2066; RRID: AB_476693 |
| Acetyl-CoA Carboxylase (C83B10) Rabbit mAb #3676 | Cell Signaling | Cat#C83B10; RRID: AB_2219397 |
| Acetyl-CoA Carboxylase 2 (D5B9) Rabbit mAb #8578 | Cell Signaling | Cat#D5B9; RRID: AB_10949898 |
| Anti-Hydroxyproline antibody (ab37067) | Abcam | Cat#ab37067; RRID: AB_873885 |
| αTubulin Antibody (B-7) | Santa Cruz Biotechnology | Cat#sc-5286; RRID: AB_628411 |
| Rabbit IgG HRP Linked Whole Ab | GE Healthcare/Sigma | Cat#NA934-1ML; RRID: AB_2722659 |
| Mouse IgG HRP Linked Whole Ab | GE Healthcare/Sigma | Cat#NA931-1ML; RRID: AB_772210 |
| InVivoMAb anti-mouse CD3ε, Clone #145-2C11 | BioXCell | Cat#BE0001-1; RRID: AB_1107634 |
| InVivoMAb anti-mouse CD28, Clone #37.51 | BioXCell | Cat#BE0015-1-A050MG; RRID: AB_1107624 |
| InVivoMAb rat IgG2b isotype control, anti-keyhole limpet hemocyanin | BioXCell | Clone: LTF-2; Cat#BE0090; RRID AB_1107780 |
| InVivoMAb anti-mouse CD8α | BioXCell | Clone: 2.43; Cat#BE0061; RRID: AB_1125541 |
| InVivoMAb rat IgG1 Isotype control, anti-trinitrophenol | BioXCell | Clone: TNP6A7; Cat#BE0290; RRID: AB_2687813 |
| InVivoMAb anti-mouse CD8β (Lyt 3.2) | BioXCell | Clone: 53-5.8; Cat#BE0223; RRID: AB_2687706 |
| TruStain FcX™ (anti-mouse CD16/32) Antibody | BioLegend | Clone: 93; RRID: AB_1574973 |
| PE anti-mouse CD45.1 Antibody | BioLegend | Clone: A20; RRID: AB_313496 |
| Alexa Fluor® 647 anti-mouse CD45.2 Antibody | BioLegend | Clone: 104; RRID: AB_492870 |
| APC anti-mouse CD45.2 Antibody | BioLegend | Clone: 104; RRID: AB_389210 |
| Brilliant Violet 421™ anti-mouse CD45.2 Antibody | BioLegend | Clone: 104; RRID: AB_10900256 |
| BUV395 Mouse Anti-Mouse CD45.2 | BD Biosciences | Clone: 104; RRID: RRID: AB_2738867 |
| APC anti-mouse CD3ε Antibody | BioLegend | Clone: 145-2C11; RRID: AB_312676 |
| PE anti-mouse CD3ε Antibody | BioLegend | Clone: 145-2C11; RRID: AB_312672 |
| FITC anti-mouse CD3ε Antibody | BioLegend | Clone: 145-2C11; RRID: AB_312670 |
| Alexa Fluor® 700 anti-mouse CD4 Antibody | BioLegend | Clone: RM4-5; RRID: AB_493701 |
| APC/Cy7 anti-mouse CD4 Antibody | BioLegend | Clone: RM4-5; RRID: AB_312726 |
| BUV737 Rat Anti-Mouse CD4 | BD Biosciences | Clone: RM4-5; RRID: AB_2732918 |
| Pacific Blue™ anti-mouse CD4 | BioLegend | Clone: RM4-5; RRID: AB_493375 |
| Brilliant Violet 421™ anti-mouse CD8α Antibody | BioLegend | Clone: 53-6.7; RRID: AB_10897101 |
| Brilliant Violet 510™ anti-mouse CD8α Antibody | BioLegend | Clone: 53-6.7; RRID: AB_2561389 |
| FITC anti-mouse CD8b Antibody | BioLegend | Clone: YTS156.7.7; RRID: AB_961293 |
| V500 Rat anti-Mouse CD8a | BD Biosciences | Clone: 53-6.7; RRID: AB_1937317 |
| Pacific Blue™ anti-mouse CD8b.2 Antibody | BioLegend | Clone: 53-5.8; RRID: AB_10641278 |
| Alexa Fluor® 700 anti-mouse CD8b Antibody | BioLegend | Clone: YTS156.7.7; RRID: AB_2563948 |
| APC/Cy7 anti-mouse CD8b Antibody | BioLegend | Clone: YTS156.7.7; RRID: AB_2563950 |
| PE/Cy7 anti-mouse/human CD11b Antibody | BioLegend | Clone: M1/70; RRID: AB_312798 |
| V500 Rat anti-CD11b | BD Biosciences | Clone: M1/70; RRID: AB_10893815 |
| Brilliant Violet 510™ anti-mouse/human CD11b Antibody | BioLegend | Clone: M1/70; RRID: AB_2561390 |
| Brilliant Violet 605™ anti-mouse/human CD11b Antibody | BioLegend | Clone: M1/70; RRID: AB_11126744 |
| PerCP/Cy5.5 anti-mouse/human CD44 Antibody | BioLegend | Clone: IM7; RRID: AB_2076206 |
| FITC anti-mouse/human CD44 Antibody | BioLegend | Clone: IM7; RRID: AB_312956 |
| PE anti-mouse/human CD44 Antibody | BioLegend | Clone: IM7; RRID: AB_312958 |
| PE/Cy7 anti-mouse CD62L Antibody | BioLegend | Clone: MEL-14; RRID: AB_313102 |
| FOXP3 Monoclonal Antibody (FJK-16s), eFluor 450; eBioscience™ | ThermoFisher Scientific | Clone: FJK-16s; RRID: AB_1518812 |
| PerCP-Cy™5.5 Mouse anti-Ki-67 | BD Biosciences | Clone: B56; RRID: AB_10611574 |
| FITC anti-human/mouse Granzyme B Antibody | BioLegend | Clone: GB11; AB_2114575 |
| Pacific Blue™ anti-human/mouse Granzyme B Antibody | BioLegend | Clone: GB11; RRID: AB_2562195 |
| PE/Cy7 anti-mouse CD279 (PD-1) Antibody | BioLegend | Clone: RMP1-30; RRID: AB_572016 |
| Brilliant Violet 605™ anti-mouse CD279 (PD-1) Antibody | BioLegend | Clone: 29F.1A12; RRID: AB_11125371 |
| Brilliant Violet 605™ anti-mouse CD19 Antibody | BioLegend | Clone: 6D5; RRID: AB_11203538 |
| PerCP/Cy5.5 anti-mouse CD11c Antibody | BioLegend | Clone: N418; RRID: AB_2129642 |
| APC/Cy7 anti-mouse NK-1.1 Antibody | BioLegend | Clone: PK136; RRID: AB_830870 |
| Ly-6G/Ly-6C Monoclonal Antibody (RB6-8C5), FITC, eBioscience™ | ThermoFisher Scientific | Clone: RB6-8C5; RRID: AB_465314 |
| Pacific Blue™ anti-mouse F4/80 Antibody | BioLegend | Clone: BM8; RRID: AB_893487 |
| Brilliant Violet 421™ anti-mouse CD11c antibody | BioLegend | Clone: N418; RRID: AB_10897814 |
| Alexa fluor® 647 anti-mouse H-2Kb/H-2Db antibody | BioLegend | Clone: 28-8-6; RRID: AB_492931 |
| FITC anti-mouse I-Ab antibody | BioLegend | Clone: AF6-120.1; RRID: AB_313724 |
| PE/Cy7 anti-mouse CD274 (PD-L1) antibody | BioLegend | Clone: 10F.9G2; RRID: AB_10639934 |
| PE anti-mouse CD273 (PD-L2) antibody | BioLegend | Clone: TY25; RRID: AB_2299418 |
| Brilliant Violet® 711 anti-mouse CD40 antibody | BD Biosciences | Clone: 3/23; RRID: AB_2740384 |
| APC anti-mouse IFN-γ antibody | BioLegend | Clone: XMG1.2; RRID: AB_315403 |
| PerCP/Cy5.5 anti-mouse TNF-α antibody | BioLegend | Clone: MP6-XT22; RRID: AB_961435 |
| PE anti-mouse IL-2 | BioLegend | Clone: JES6-5H4; RRID: AB_315301 |
| CD8a Monoclonal Antibody (4SM15) | eBioscience | Cat#14-0808-82; Clone: 4SM15; RRID: AB_2572861 |
| Anti-CD68 antibody | Abcam | Clone: ab125212; RRID: AB_10975465 |
| Recombinant Anti-Lactate Dehydrogenase antibody-Alexa Fluor® 488 | Abcam | Cat#ab202652; Clone: EP1566Y |
| CD4 Monoclonal Antibody (4SM95), eFluor 570 | eBioscience | Cat#41-9766-82; Clone: 4SM95; RRID: AB_2573637 |
| FOXP3 Monoclonal Antibody (FJK-16s), Alexa Fluor 488, eBioscience | eBioscience | Cat#53-5773-82; Clone: FJK-16s; RRID: AB_763537 |
| EOMES Monoclonal Antibody (Dan11mag), PE, eBioscience | ThermoFisher Scientific | Cat#12-4875-82; Clone: Dan11mag; RRID: AB_1603275 |
| Alexa Fluor® 647 anti-mouse Ly-6G Antibody | BioLegend | Clone: 1A8; RRID: AB_1134159 |
| Ki-67 (D3B5) Rabbit mAb (Alexa Fluor® 488 Conjugate) | Cell Signaling | Cat#11882S; Clone: D3B5; RRID: AB_2687824 |
| Anti-CD11b antibody [EPR1344] (Alexa Fluor® 647) | Abcam | Cat#ab204471; Clone: EPR1344 |
| Recombinant Anti-GLUD1 antibody [EPR11370] (Alexa Fluor® 488) | Abcam | Cat#ab204001; Clone: EPR11370 |
| Vimentin (D21H3) XP® Rabbit mAb (Alexa Fluor® 555 Conjugate) #9855 | Cell Signaling | Cat#9855; Clone: D21H3; RRID: AB_10859896 |
| Recombinant Anti-Glucose Transporter GLUT1 antibody [EPR3915] (Alexa Fluor® 647) | Abcam | Cat#ab195020; Clone: EPR3915; RRID: AB_2783877 |
| PCNA (PC10) Mouse mAb (Alexa Fluor® 488 Conjugate) | Cell Signaling | Cat#8580; Clone: PC10; RRID: AB_11178664 |
| Recombinant Anti-COX IV Antibody [EPR9442(ABC)] - Mitochondrial Loading Control (Alexa Fluor® 555) | Abcam | Cat#ab210675; Clone: EPR9442; RRID: AB_2857975 |
| Phospho-mTOR (Ser2448) Monoclonal Antibody, eFluor 660 | eBioscience | Cat#50-9718-41; Clone: MRRBY; RRID: AB_2574351 |
| Recombinant Anti-iNOS antibody [EPR16635] (Alexa Fluor® 555) | Abcam | Cat#ab209594; Clone: EPR16635 |
| Anti-Aconitase 2 antibody [6F12BD9] (Alexa Fluor® 647) | Abcam | Cat#ab198050; Clone: 6F12BD9; RRID: AB_2857971 |
| TCF1/TCF7 (C63D9) Rabbit mAb (Alexa Fluor® 488 Conjugate) | Cell Signaling | Cat#6444S; Clone: C63D9; RRID: AB_2797627 |
| PKM2 (D78A4) XP® Rabbit mAb (PE Conjugate) | Cell Signaling | Cat#89367; Clone: D78A4; RRID: AB_2800137 |
| mTOR (7C10) Rabbit mAb (Alexa Fluor® 647 Conjugate) | Cell Signaling | Cat#5048; Clone: 7C10; RRID: AB_10828101 |
| Recombinant Anti-c-Myc antibody [Y69] (Alexa Fluor® 555) | Abcam | Cat#ab201780; Clone: Y69; RRID: AB_2728791 |
| Anti-VDAC1 / Porin antibody [20B12AF2] (Alexa Fluor® 647) | Abcam | Cat#ab179840; Clone: 20B12AF2 |
| Bacterial and Virus Strains | ||
| Stbl3™ Chemically Competent E. coli | ThermoFisher Scientific | Cat#C737303 |
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| DMEM (high glucose, glutamine, no pyruvate) | ThermoFisher Scientific | Cat#11965118 |
| RPMI 1640 Medium | ThermoFisher Scientific | Cat #11875093 |
| 1X DPBS | ThermoFisher Scientific | Cat#14190250 |
| 1X DPBS (calcium, magnesium) | ThermoFisher Scientific | Cat#14040133 |
| Penicillin-Streptomycin | ThermoFisher Scientific | Cat#15140122 |
| Fetal Bovine Serum (FBS) | Sigma | Cat#F2442 Lot#17L189 |
| Charcoal-Stripped Fetal Bovine Serum (FBS) | ThermoFisher Scientific | Cat#A3382101 |
| 2-mercaptoethanol | ThermoFisher Scientific | Cat#21985023 |
| EDTA (0.5 M) | ThermoFisher Scientific | Cat#15575020 |
| HEPES | ThermoFisher Scientific | Cat#15630080 |
| Fugene 6 Transfection Reagent | Promega | Cat#E2691 |
| Hexadimethrine bromide (Polybrene) | Santa Cruz | Cat#sc-255611 |
| Collagenase, Type I | Worthington Biochemical Corporation | Cat#LS004194 |
| Collagenase P | Roche | Cat#11249002001 |
| Percoll density gradient media | GE Healthcare LifeSciences | Cat#17089101 |
| Complete Mini Protease Inhibitor | Sigma | Cat#11836170001 |
| Phosphatase Inhibitor Cocktail 2 | Sigma | Cat#P5726-5ML |
| Phosphatase Inhibitor Cocktail 3 | Sigma | Cat#P0044 |
| Blasticidin | Sigma-Aldrich | Cat#15205 |
| BamH I-HF Restriction Endonuclease | NEB BioLabs | Cat#R3136S |
| Sal I-HF Restriction Endonuclease | NEB BioLabs | Cat#R3138S |
| Xho I Restriction Endonuclease | NEB BioLabs | Cat#R0146S |
| EcoR V-HF Restriction Endonuclease | NEB BioLabs | Cat#R3195S |
| CloneAmp™ HiFi PCR Pre-Mix | Clontech | Cat#639298 |
| Quick Ligation™ Kit | NEB BioLabs | Cat#M2200S |
| TRIzol™ Reagent | ThermoFisher Scientific | Cat#15596018 |
| iScript cDNA Synthesis Kit | BioRad | Cat#1708891 |
| PerfeCTa SYBR® Green FastMix | Quantabio | Cat#101414-270 |
| Buffer RLT | Qiagen | Cat#79216 |
| LB Broth LB | Sigma-Aldrich | Cat#L7275 |
| HCS LipidTOX™ Deep Red Neutral Lipid Stain, for cellular imaging | ThermoFisher Scientific | Cat#H34477 |
| BODIPY™ FL C16 (4,4-Difluoro-5,7-Dimethyl-4-Bora-3a,4a-Diaza-s-Indacene-3-Hexadecanoic Acid) | ThermoFisher Scientific | Cat#D3821 |
| Prolong Glass Antifade Mountant | ThermoFisher Scientific | Cat#P36982 |
| Recombinant Mo-se IL-7 (carrier-free)-10 ug | BioLegend | Cat#577802 |
| Ionomycin from Streptomyces conglobatus | Sigma Aldrich | Cat#I9657-1MG |
| GolgiStop™ Protein transport inhibitor | BD Biosciences | Cat#554724 |
| Bovine Serum Albumin (BSA), ≥98%, Fatty Acid-free | MP Biomedicals | Cat#IC15240110 |
| Sodium palmitate | Sigma | Cat#P9767 |
| Sodium oleate | Sigma | Cat#O7501 |
| Critical Commercial Assays | ||
| LIVE/DEAD Fixable Near-IR stain | ThermoFisher Scientific | Cat#L10119 |
| Naive CD8a+ T Cell Isolation Kit, mouse | Miltenyi Biotec | Cat#130-096-543 |
| CD45 MicroBeads, mouse | Miltenyi Biotec | Cat#130-052-301 |
| eBioscience™ Foxp3 / Transcription Factor Staining Buffer Set | ThermoFisher Scientific | Cat#00-5523-00 |
| Fixation/Permeabilization Solution Kit | BD Biosciences | Cat#554714 |
| Direct-zol RNA Miniprep Kit | Zymo Research | Cat#R2050 |
| RNeasy Micro Kit | Qiagen | Cat#74004 |
| Pierce™ BCA Protein Assay Kit | ThermoFisher Scientific | Cat#23227 |
| Western Lightning ECL Pro | Perkin Elmer | Cat#NEL120001EA |
| Cell Trace™ Violet Cell Proliferation Kit, for flow cytometry | ThermoFisher Scientific | Cat#C34557 |
| 10’ Chromium Single Cell 3′ v2 | 10X Genomics | Cat#PN-120267 |
| Chromium Single Cell A Chip Kit | 10X Genomics | Cat#PN-1000009 |
| Chromium i7 Multiplex Kit, 96 rxns | 10X Genomics | Cat#PN-120262 |
| Lipoprotein Lipase Assay Kit (Fluorometric) | Abcam | Cat#ab204721 |
| Deposited Data | ||
| TMT-proteomics of sorted MC38 tumor cells with high-fat diet | This manuscript (ProteomeXchange) | Accession#PXD019495 |
| Raw and analyzed RNA-sequencing data | This paper (GEO repository) | GSE157999 |
| Experimental Models: Cell Lines | ||
| MC38 colorectal adenocarcinoma | Laboratory of D. Vignali, University of Pittsburgh School of Medicine, Pittsburgh, PA | RRID: CVCL_B288 |
| Lewis Lung Carcinoma | N/A | RRID: CVCL_4358 |
| B16.F10 Melanoma | Gift from G. Dranoff (Novartis Institutes for Biomedical Research) | RRID: CVCL_0159 |
| E0771 breast cancer cell line | Corporate Cell Line Sales (CH3 Biosystems) | Cat#94A001; RRID: CVCL_GR23 |
| RENCA kidney renal adenocarcinoma cell line | ATCC | Cat#CRL-2947; RRID: CVCL_2174 |
| CT26 colon carcinoma cell line | N/A | RRID: CVCL_7256 |
| HEK293T | N/A | RRID: CVCL_0063 |
| Phoenix-ECO | ATCC | Cat#CRL-3214; RRID: CVCL_H717 |
| Experimental Models: Organisms/Strains | ||
| C57BL6/J | The Jackson Laboratory | #000664; RRID: IMSR_JAX:000664 |
| TCRa knock-out mice: B6.129S2-Tcratm1Mom/J | The Jackson Laboratory | #002116; RRID: IMSR_JAX:002116 |
| OT-1 mice: C57BL/6-Tg(TcraTcrb)1100Mjb/J | The Jackson Laboratory | #003831; RRID: IMSR_JAX:003831 |
| Oligonucleotides | ||
| Cloning mouse PHD3-OE vector with C-terminal
FLAG: Fwd - CGTAGAGGATCCATGCCTCTGGGACACAT Rev -GGACGCGTCGACCTACTTGTCGTCGTCGTCCTTGTAGTCGATGTCGTGGTCCTTGTAGTCACCGTCGTGGTCCTTGTAGTCGTCTTTAGCAAGAGCA |
This manuscript | N/A |
| Cloning RFP to generate MSCV-PIR: Fwd - cttccgctcgagATGGCCTCCTCCGAGGACG Rev - gattcggatatcTTAGGCGCCGGTGGAGTG |
This manuscript | N/A |
| PHD3 qPCR Primers (mouse): Fwd - CAGACCGCAGGAATCCACAT Rev - TTCAGCATCGAAGTACCAGACAGT |
German et al., 2016 | N/A |
| β-Actin qPCR Primers
(mouse): Fwd - AGCCATGTACGTAGCCATCC Rev - CTCTCAGCTGTGGTGGTGAA |
German et al., 2016 | N/A |
| Recombinant DNA | ||
| pCMV-SPORT6 Egln3 (Phd3) | Harvard PlasmID Database | MmCD00320451 |
| MSCV-PIG (Retroviral vector containing puromycin-IRES-GFP) | Addgene | Addgene#18751 |
| MSCV-PIR (Retroviral vector containing puromycin-IRES-RFP) | This manuscript | N/A |
| pLenti CMV GFP Blast (659-1) | Addgene | Addgene#17445 |
| pLenti CMV Phd3 Blast (C-terminal FLAG-tag) | This manuscript | N/A |
| Software and Algorithms | ||
| GraphPad Prism V7 | GraphPad Software | https://www.graphpad.com |
| FlowJo 10.4.1 | FlowJo LLC | https://www.flowjo.com |
| CLC Genomics Workbench Version 8.0.1 | Qiagen | https://www.qiagenbioinformatics.com/products/clc-genomics-workbench/ |
| GEPIA (Gene Expression Profiling Interactive Analysis) | Laboratory of Z. Zhang, Peking University, Beijing Shi, China | http://gepia.cancer-pku.cn/ |
| GenePattern | Broad Institute | https://www.genepattern.org/ |
| Other | ||
| PicoLab® Rodent Diet 20 | LabDiet | Cat#5053 |
| Rodent Diet With 60 kcal% Fat | Research Diets | Cat#D12492 |
| GentleMACS C Tubes | Miltenyi | Cat#130-093-237 |
| Criterion TGX gel 4-20% | BioRad | Cat#5671095 |
| Nitrocellulose Membrane 0.2uM | BioRad | Cat#162-0112 |
| Lithium Heparin Tubes (2 mL) | VWR | Cat#454237 |
| MICROVETTE CB300 EDTA/PK100 | Sarstedt Inc | Cat#NC9141704 |
| Nylon Net Filters | EMD Millipore | Cat#NY2004700 |
To stain for neutral lipids, TILs or dissociated GFP+ tumors were incubated with HCS LipidTOX™ Deep Red (ThermoFisher Scientific) diluted 1:200 in MACS buffer for 1 hour at 37°C. After LipidTox staining, TIL samples were stained with the flow cytometry antibodies. Ex vivo palmitate uptake was measured in purified TIL samples or dissociated RFP+ tumors after overnight incubation at 37°C in R2 with 1 μM BODIPY™ FL C16 added. The following morning, TIL samples were stained with flow cytometry antibodies.
Data collection was performed on a BD FACSymphony or LSR II flow cytometer and analyzed using FlowJo v10.4.1.
T Cell Isolation and Activation
Naïve CD8+ T cells were isolated from the spleens of 13-15 week old female wildtype or OT-1 mice using negative magnetic selection (Miltenyi Biotec) and stained with Cell Trace Violet (CTV, ThermoFisher Scientific) as per the manufacturer’s instructions. 50,000 CD8+ T cells were plated per well on 96-well plates pre-coated with αCD3/αCD28 antibodies at concentrations ranging from 0 to 8 μg/mL and incubated at 37°C in a humidified 5% CO2 incubator. For free fatty acid supplementation, palmitate and oleate were individually conjugated to fatty acid-free BSA (MP Biomedicals) in 150 mM NaCl at a 6:1 molar ratio to make a 4 mM, pH-adjusted FA stock solution. For fatty acid supplementation, 100 μM each of BSA-conjugated palmitate and oleate was added to the culture medium containing charcoal-stripped serum, or an equivalent concentration of BSA alone as a control. After 48-72 hours of stimulation, cell numbers, viability, and proliferation were measured by flow cytometry. A naïve control was maintained with 10 ng/mL IL-7 (BioLegend).
Lipoprotein Lipase Activity Assays
LPL activity assays were performed using a commercially available fluorometric assay from Abcam. Briefly, dissected tissues were weighed on an analytical balance, rinsed in ice-cold PBS, and then homogenized using 10-15 strokes in a dounce homogenizer. The LPL assay was performed in accordance with the manufacturer’s instructions.
RNA Extraction and qPCR
RNA was extracted from flash-frozen, powderized tumor tissue using TRIzol™ reagent (ThermoFisher Scientific) and purified using the Direct-zol RNA miniprep kit (Zymo Research) according to the manufacturer’s instructions. cDNA was synthesized using the iScript cDNA synthesis kit (BioRad). Quantitative real-time PCR was performed with PerfeCTa SYBR® Green FastMix (Quantabio) on a Roche Lightcycler 480 and analyzed using ΔΔCt calculations. β-actin was used as a reference gene for normalization.
RNA-Sequencing
For RNA-sequencing experiments, CD8+ TILs and GFP+ tumor cells were sorted from dissociated MC38-PIG tumors, as were CD8+ T cells from dLNs, after staining with the following antibodies for mouse antigens: CD45.2-AF647 (BioLegend, Clone #104), CD3ε-PE (BioLegend, Clone#145-2C11), CD8α-BC510 (BioLegend, Clone #53-6.7), CD4-PacBlue (BioLegend, Clone #RM4-5), and CD11b-PE/Cy7 (BioLegend Clone #M1/70). 20-100K CD8+ T cells were sorted by gating away from debris and then gating for singlets that were CD45+, CD3+/CD11b−, CD8+/CD4−. 200K GFP+ tumor cells were sorted from each sample by gating away from debris and then gating for singlets that were CD45−/GFP+. Samples were sorted into tubes containing R10 supplemented with an extra 10% FBS, stored on ice, and then spun at 750 g for 10 minutes at 4°C. Cell pellets were resuspended in 30 μL RLT buffer (Qiagen) with 1% (v/v) 2-mercaptoethanol added and frozen at −80°C. RNA-seq library preparations were performed as previously described (Sage et al., 2016).
Single-Cell RNA-Sequencing
CD45+ tumor-infiltrating leukocytes were enriched by positive selection from dissociated day 12 MC38 tumors for single-cell analysis. Prior to magnetic enrichment, MC38 tumors were minced in RPMI containing 2% FBS, 2 mg/mL collagenase P (Roche), and 50 mcg/mL DNase I (Sigma) and then incubated at 37°C with gentle rocking for 10 minutes. CD45+ leukocytes were magnetically labeled and enriched by positive selection (Miltenyi Biotec). Cells were diluted with Trypan Blue and counted using a hemocytometer. Tumors from three different mice were pooled per sample, and two samples were prepared per diet condition. For a target recovery of 5,000 single cells, ~8-9,000 live cells were loaded onto the Chromium Controller (10X genomics) and processed according to the manufacturer’s instructions. One sample failed at this step, leaving two CD and one HFD sample, which were sequenced on an Illumina NextSeq500 sequencer using a 75-bp kit with paired-end reads.
Metabolite Extractions
12-14 days after tumor injections, plasma, tumor interstitial fluid, and tumor tissue were harvested for metabolomics analysis. Blood was collected by heart puncture into heparinized tubes (VWR) and separated into plasma by centrifugation at 1500 g for 20 minutes at 4°C. Tumor interstitial fluid was collected as previously described (Wiig et al., 2003). Briefly, dissected tumors were wrapped in nylon net filters (EMD Millipore) and placed over Eppendorf tubes on ice. Tumors were spun at 400 g for 15 minutes at 4°C. Tumor tissue was moved to a new tube so that both the tumor interstitial fluid and tumor tissue could be snap-frozen in liquid nitrogen and stored at −80°C. Tumor tissue was manually powderized by mortar and pestle and accurately weighed prior to metabolite isolation.
To extract metabolites from serum and tumor interstitial samples, 50 μL of cold extraction solvent (40:40:20 methanol:acetonitrile:water stored at −20°C) was added to 1.5 μL of sample. After vortexing, the samples were incubated on ice for 20 minutes, and then centrifuged at 16,000 g for 20 minutes at 4°C. Finally, the supernatants were transferred to LC vials. For tissue samples, the volume of the extraction solution (μL) was 40 x the weight of tissue (mg) to make an extract of 25 mg tissue per mL solvent. Metabolite extraction from tissues was performed by adding the extraction solution to the powderized tissues followed by vortexing. Samples were incubated at −20°C for 2 hours, then centrifuged at 16,000 g for 20 minutes at 4°C and the clean supernatants transferred to LC vials (Jang et al., 2018).
For lipidomics analysis, 2 μL TIF or plasma was extracted with water : methanol : chloroform (1:1:1) containing 4 μM TAG (19:0/19:0/19:0) as internal standard. Samples were vortexed and sonicated for 5 minutes. After centrifugation at 3000g for 20 minutes, the chloroform phase was dried under nitrogen gas and then reconstituted in 50 μL methanol : chloroform (9:1).
LC-MS Analysis
For untargeted metabolomics of polar metabolites, extracts were analyzed using a quadrupole-orbitrap mass spectrometer (Q Exactive, Thermo Fisher Scientific, San Jose, CA) coupled to hydrophilic interaction chromatography via electrospray ionization. lC separation was on a XBridge BEH Amide column (2.1 mm x 150 mm, 2.5 μm particle size; Waters, Milford, MA) using a gradient of solvent A (20 mM ammonium acetate, 20 mM ammonium hydroxide in 95:5 water: acetonitrile, pH 9.45) and solvent B (acetonitrile). Flow rate was 150 μL/minute, column temperature was 25 °C, autosampler temperature was 5°C, and injection volume was 10 μL. The LC gradient was: 0 min, 90% B; 2 min, 85% B; 3 min, 75% B; 7 min, 75% B; 8 min, 70% B; 9 min, 70% B; 10 min, 50% B; 12 min, 50% B; 13 min, 25% B; 14 min, 25% B; 16 min, 0% B; 21 min, 0% B; 22 min, 90% B; 25 min, 90% B. The mass spectrometer was operated in negative ion or positive ionizations mode to scan from m/z 70 to 1000 at 1Hz and a resolving power of 140,000 (Jang et al., 2018).
For the analysis of free fatty acids, extracts were analyzed using an orbitrap mass spectrometer (Exactive, Thermo Fisher Scientific, San Jose, CA) coupled to ion-pairing reverse phase chromatography via electrospray ionization. LC separation was on a Luna C8 reversed-phase column (2.0 x 150 mm, 3μm particle size, 100 Å poresize, Phenomenex, Torrance, CA) using a gradient of solvent A (97/3 water/methanol with 10 mM tributylamine and 15 mM acetic acid, pH 4.5) and solvent B (methanol). Flow rate was 250 μL/minute, column temperature was 25 °C, autosampler temperature was 5°C, and injection volume was 10 μL. The LC gradient was: 0 min, 80% B; 10 min, 90% B; 11 min, 99% B; 25 min, 99% B; 26 min, 80% B; 30 min, 80% B. The mass spectrometer was operated in negative ion mode to scan from m/z 120 to 400 at 1Hz and a resolving power of 100,000 (Kamphorst et al., 2011).
For lipidomics analysis, the lipid extract was separated by using a Kinetex evo C18 column (2.6 um, 150 mm × 2.0 mm I.D., Phenomenex) coupled to a Thermo Scientific SII UPLC system. The C18 column was used with the following buffers and linear gradient: A = water with 0.1% formic acid, B = 90% isopropanol, 10% methanol; 5% to 100% from 0-20min, 100% for additional 5min; flow rate 200 μL/min. Mass spectrometry detection was carried out on a Q Extractive HF-X orbitrap mass spectrometer with an HESI source operated in positive mode. Metabolite quantification was done using TraceFinder software (ThermoFisher). The identity of the metabolite was confirmed by matching accurate mass or MS/MS fragmentation pattern to databases.
Histology
Empty vector-transduced and PHD3-OE tumor cells were implanted into mice on opposing abdominal flanks as described above. On day 12 post-implantation, both left and right flank tumors were excised and immediately frozen in Optimal Cutting Temperature compound (OCT) on dry ice. Tumors sections were cut at 7 μm, fixed in ice-cold acetone, and stored at −20°C until staining. Before staining, slides were re-fixed in ice-cold acetone for 10 minutes. Tissues were then re-hydrated in 1X PBS and blocked at room temperature with 5% BSA/1X PBS solution for one hour. The slides were incubated with CD8-PE antibody (BioLegend, Clone#YTS156.7.7) diluted in 1X PBS for one hour in a humidified chamber at room temperature. Slides were then washed 3 times with 1X PBS and stained with DAPI for an additional 10 minutes. Finally, slides were washed once in 1X PBS and mounted with Prolong Glass Antifade Mountant (Thermofisher Scientific). Slides were imaged with a confocal Olympus FV3000 microscope and the resulting images (each 1024 x 1024 pixels) were stitched together using microscope software to generate the final reconstructed tissue sections with a scale of 1 μm = 1.6091 pixels. CD8+ T cell numbers were counted by two independent blinded observers and obtained by counting the number of positive signals in randomly drawn 1000x1000 pixel boxes throughout the tissue image.
TMT-Proteomics
Cell pellets were processed using the streamlined TMT labelling protocol (Navarrete-Perea et al., 2018). Samples were lysed in 8M urea in 200 mM EPPS pH 8.5 with protease (Pierce A32953) and phosphatase (Pierce A32957) inhibitors, and passed through a 21-guage needle 10x. Samples were reduced with 5 mM TCEP, alkylated with 10 mM iodoacetamide, and quenched with 5 mM DTT, followed by methanol/chloroform precipitation of protein. Pellets were reconstituted in 200 mM EPPS pH 8.5, digested overnight with LysC protease (Wako 129-02541) at 1:100 while shaking at room temperature, followed by digestion with trypsin (Pierce 90305) at 1:100 while shaking at 37°C. Anhydrous acetonitrile (Honeywell AS017-0100) was added to ~30%, followed by labelling with TMT10 (Thermo 90110) reagent. 1% of each labeled sample was combined and analyzed unfractionated to ensure labeling efficiency was >97%. After confirmation, the reaction was quenched by adding ~0.3% hydroxylamine, and incubating at RT for 15min. The samples were then mixed at a 1:1 (total amount) ratio across all conditions. After mixing, labelled peptide samples were de-salted using a 200 mg Sep-Pak cartridge (Waters WAT054925), followed by drying in a rotary evaporator. Samples were then reconstituted in 5% ACN 10 mM ammonium bicarbonate for basic reverse phase fractionation on an Agilent 300extend-C18 column (3.5μm, 4.6x250mm) using an Agilent Infinity 1260 HPLC. Peptides were subjected to a 75 min linear gradient from 13% to 42% of Buffer B (10 mM ammonium bicarbonate, 90% ACN, pH 8) at a flow rate of 0.6 mL/min, resulting in a total of 96 fractions which were consolidated into 24 by combining (in a chessboard pattern) four alternating wells down columns of the 96-well plate. Assuming adjacent fractions contain overlapping peaks, only 12 non-adjacent samples were analyzed by the mass spectrometer. The pooling scheme has been illustrated previously (Navarrete-Perea et al., 2018; Paulo et al., 2016a). Each eluted fraction was desalted via StageTip for SPS-MS3 analysis.
Mass spectra were collected on an Orbitrap Lumos mass spectrometer equipped with a Field Asymmetric-waveform Ion-Mobility Spectrometry (FAIMS) device coupled to a Proxeon EASY-nLC 1200 LC pump (ThermoFisher Scientific). Peptides were separated on a 35 cm column (i.d. 100 μm, Accucore, 2.6 μm, 150 Å) packed in-house using a 90 min gradient (from 5% -30% acetonitrile with 0.1% formic acid) at 500 nl/min. A multi-notch FAIMS method was used to additionally separate peptides at 40, 60 and 80 CV (Schweppe et al., 2019, 2020). Each analysis used an SPS-MS3-based TMT method (McAlister et al., 2014; Ting et al., 2011), which has been shown to reduce ion interference compared to MS2-based quantification (Paulo et al., 2016b). MS1 data were collected using the Orbitrap (120,000 resolution; maximum injection time 50 ms; AGC 4e5, 400-1400 m/z). Determined charge states between 2 and 5 were required for sequencing and a 90 s dynamic exclusion window was used. MS2 scans consisted of collision-induced dissociation (CID), quadrupole ion trap analysis, automatic gain control (AGC) 2E4, NCE (normalized collision energy) 45, q-value 0.25, maximum injection time 35 ms, and isolation window of 0.7 Da using a Top10 method. MS3 scans were collected in the Orbitrap at a resolution of 50,000, NCE of 45%, maximum injection time of 100 ms, and AGC of 1.5e5. Data from all 12 fractions were combined to achieve a <1% false discovery rate (FDR) at the protein level.
Mass spectra were processed using a SEQUEST-based software pipeline. Data were searched against the UniProt Mouse database (December 2018), using a 20-ppm precursor ion tolerance for total protein-level analysis and 0.9 Da product ion tolerance. TMT tags on lysine residues and peptide N termini (+229.163 Da) and carbamidomethylation of cysteine residues (+57.021 Da) were set as static modifications, while oxidation of methionine residues (+15.995 Da) was set as a variable modification. Peptide-spectrum matches (PSMs) were identified, quantified, and filtered to a 1% peptide false discovery rate (FDR) and then collapsed further to a final protein-level FDR of 1%. Proteins were quantified by summing reporter ion counts across all matching PSMs. Briefly, a 0.003 Da (3 millidalton) window around the theoretical m/z of each reporter ion was scanned and the maximum intensity nearest the theoretical m/z was used. Reporter ion intensities were adjusted to correct for the isotopic impurities of the different TMT reagents according to manufacturer specifications and adjusted to normalize ratios across labelling channels. Lastly, for each protein, signal-to-noise (S:N) measurements of the peptides were summed and then normalized to 100.
Cyclic Immunofluorescence
Cyclic immunofluorescence (CyCIF) (Lin et al., 2015, 2018) was performed on formalin-fixed paraffin embedded (FFPE) tissue sections of mouse MC38 colorectal tumors engrafted into syngeneic mice. Resected tissues were fixed overnight in 10% neutral buffered formalin at room temperature and then moved to 70% ethanol at 4°C. Fourteen tissue sections were taken from a cohort of 7 mice fed CD and 7 mice fed HFD. Each tissue section was probed with antibodies against the following immune or metabolism targets: CD8A, CD4, FOXP3, CD11b, CD68, LY6G, NOS2, EOMES, TCF1, MTOR, phospho-MTOR, MYC, GLUT1, PKM2, LDH, COX4, ACO2, GLUD1, VDAC1, PCNA, KI67, ACTB and VIM. Tissues were imaged at 20x (2x2 binning) using a CyteFinder slide-scanning fluorescence microscope (RareCyte inc.) equipped with an automated stage controller. Raw imaging data from each of 4 CyCIF cycles were flat-field corrected using a previously described method for background and shading correction (Peng et al., 2017), then stitched and aligned using ASHLAR: a program for seamless mosaic image processing across imaging cycles (manuscript in preparation). The resulting mosaic images were segmented by first training a convolutional neural network to recognize cell boundaries using training data, then using the model to predict cell boundaries in each of the 14 tissue sections used in the experiment (Ronneberger et al., 2015).
Single-cell data were computationally analyzed using a set of Python-based data analysis libraries (pandas, numpy, scipy, matplotlib, etc.). Several data pre-processing steps were taken; first, cells associated with adipose tissue surrounding many of the tissue sections were filtered from the analysis by using a Gaussian kernel density estimator to identify and isolate areas of high cell density (i.e. tumor tissue) (Figure S4B). Second, under- and over-segmented cells were removed by setting lower and upper bounds on cell area (Figure S4B). Third, DNA signal intensity (Hoechst dye) was correlated across imaging cycles to isolate cells present across all imaging cycles (Figure S4B). Finally, raw signal intensities were log10-transformed, filtered to remove extreme outliers, and rescaled between the values of 0 and 1.
Overall, the cleaned dataset consisted of 14 tissues. To identify major cell populations, dimensional reduction using t-Distributed Stochastic Neighbor Embedding (t-SNE) was performed on the cleaned dataset followed by density-based clustering by HDBSCAN (Figure 4B) (Campello et al., 2013; van der Maaten, L. J. P., 2008). We excluded cell clusters that were comprised of cells from fewer than two samples (#1, #2, #6, and #9). This approach identified six major clusters (#0, #3, #4, #5, #7, and #8). We next re-clustered the largest cell population (#8), which revealed three sub-clusters (#8.0, #8.1, and #8.2). We noticed that cluster #8.2 contained a mixture of CD11b+ myeloid cells and tumor/stromal cells, the latter of which do not express immune lineage markers but are dimly positive for CD11b expression. Thus, we thresholded cluster #8.2 by CD11b expression to separate the tumor/stromal fraction (#8.2) and CD11b+ myeloid cells (#8.3) (Figures 4B and 4C). We used the expression of lineage markers to identify intratumoral cell populations including CD8+ T cells (#7), CD4+ Tconv cells (#4), CD4+ Treg cells (#5), double negative cells expressing T cell transcription factors (#3), several myeloid populations (#0, #8.1 and #8.3), and tumor/stromal cells (#8.0 and #8.2) (Figures 4B and 4C).
Spatial relationships between immune cells and areas of high metabolic gene expression within the MC38 TME were quantified by calculating the percentage of immune cells overlapping regions of strong metabolic immunomarker signal intensity (referred to as % overlap). Poisson-Disc sampling (Bridson, 2007) was used to calculate the % overlap resulting from uniformly distributing a similar number of cells across the same 2D tissue space to achieve expectation values which assume the absence of spatial patterning with respect to metabolic gene expression. Independent, two-tailed, Student’s t-tests were computed on % overlap values from experiment and simulation to test for the presence of biological patterning within the CD and HFD groups independently. In cases where biological patterning was detected under both treatment conditions, experimental % overlap values were subtracted from their respective simulated values and the differences were used to test for differences in the magnitude and direction of biological pattern between CD and HFD tumors.
Quantification and Statistical Analysis
Statistical Analyses
Statistics were computed with GraphPad Prism 7 software using unpaired Student’s t-test for comparisons between two groups and two-way ANOVA for tumor growth curves with multiple variables, followed by the Bonferroni posthoc test for comparison of tumor sizes at multiple individual time points. Graphs containing tumor growth curves display mean values with error bars corresponding to standard error of the mean (SEM). All other data are represented as mean +/− standard deviation. P-values are denoted in figures as: *p<0.05, **p<0.01, ***p<0.001, ****<0.0001.
Metabolomics Analysis
Fold-changes between experimental groups were computed based on blanked, integrated peak areas for each metabolite. Significance was determined using an unpaired Student’s t-test.
Bulk RNA-Seq Analysis
RNA-seq data was analyzed using the CLC Genomics Workbench version 8.0.1 RNA-seq analysis software package (Qiagen). Read alignment was performed to the mouse genome using the following parameters: (mismatch cost=2, insertion cost=3, deletion cost=3, length fraction=0.8, similarity fraction=0.8). Samples were normalized to reads per million before performing differential expression analysis (total count filter cutoff=5.0). Heatmaps were generated using the pheatmap package from bioconductor in R (version 3.4.4). Raw and processed RNA-seq data files were uploaded to the GEO repository within the GSE157999 reference series.
Single-Cell RNA-Seq Analysis
Sample demultiplexing, barcode processing, alignment, filtering, and UMI counting were performed using the Cell Ranger analysis pipeline (v.1.2). Ambient RNA contamination estimation and removal was performed in R using the SoupX package (v1.2.2) with three lists of nonexpressed genes: (i) Cd3g, Cd3e, Cd8a, Thy1, Lat, Lck, and Ptprcap, (ii) Cd74, Lyz2, C1qa, C1qb, H2-Ab1, H2-Eb1, Tyrobp, Tpt1, Fth1, and (iii) Col3a1, Col6a1, Serpinh1, and Sparc (v.1.2.2) (Young and Behjati, 2018). Strained counts from SoupX were written to a directory in CellRanger format using the R package DropletUtils (v.1.6.1) and used for downstream analysis in R using Seurat (v.3.1.5) (Stuart et al., 2019). Raw and processed RNA-seq data files were uploaded to the GEO repository within the GSE157999 reference series.
Data pre-processing, normalization, integration, and clustering was performed using the R package Seurat. After merging all samples into a single Seurat object, single-cell transcriptomes were initially filtered using four metrics for quality control. First, cells in which fewer than 400 genes or more than 2,500 genes were detected were removed from the analysis. Second, cells where mitochondrial encoded transcripts represented greater than 12% or less than 0.25% of the total library were excluded. Third, cells with more than 12,000 RNA molecules detected per cell were discarded. Finally, genes detected in fewer than three cells across the dataset were removed. The resulting expression matrix contained 9,104 cells by 13,484 genes, with 5,721 cells from CD tumors and 3,383 from HFD tumors. Normalization and variance stabilization was performed using the R package sctransform, which interfaces directly with Seurat, on each diet condition separately (v0.2.1) (Hafemeister and Satija, 2019). CD and HFD datasets were integrated using canonical correlation analysis to harmonize the datasets by finding shared sources of variation (Butler et al., 2018). 2,000 variable gene features were chosen using the SelectIntegrationFeatures function in Seurat, followed by the PrepSCTIntegration function to make sure that all Pearson residuals required for downstream analysis were calculated. Anchors between datasets were identified using the FindIntegrationAnchors function and passed to the IntegrateData function using normalization.method = ‘SCT’ to produce an integrated dataset. Dimensional reduction was accomplished by performing principal component analysis (PCA) on the integrated dataset and then using the first 48 principal components for Uniform Manifold Approximation and Projection (UMAP) using default parameters associated with the RunUMAP function. Unsupervised clustering was done by constructing a shared nearest neighbor (SNN) graph using the FindNeighbors function and then performing graph-based clustering using the “Louvain” algorithm with resolution = 0.5 by the FindClusters function. Differential expression analysis between clusters and comparing diet conditions was performed using a Wilcoxon rank sum test. Cluster markers were identified using the FindConservedMarkers function. Comparisons between population sizes among diet conditions were performed using a two-sided exact binomial test.
Metabolic and signaling pathway gene signatures were curated from the KEGG subset of canonical pathways from the C2 collection within MSigDB. Immune signatures were curated from the C7 collection of immunologic signatures within MSigDB. Single-cell signature scores were calculated using the Vision package v.2.1.0 (DeTomaso et al., 2019). Signatures that were highly autocorrelated within clusters were evaluated by Geary’s C, using C′=1− C for autocorrelation effect size and computation of an empirical p-value with FDR-correction within Vision for significance. For each cluster, signatures that were significantly altered by diet were assessed by Wilcoxon rank sum test with false discovery rate correction using the method of Benjamini and Hochberg. Pearson correlations and p-values between signatures were calculated in R using the rcorr function from the Hmisc package v.4.4.0.
TCGA Data Analysis
Spearman correlation analysis was performed on TCGA data using the GEPIA web portal (http://gepia.cancer-pku.cn/). For each cancer type represented in TCGA, Spearman correlation coefficients and p-values were calculated between PHD3 (Egln3) and MHC-I or GZMB expression using normalized transcripts per million values (TPM).
To evaluate the impact of body mass index (BMI) on PHD3 expression and how PHD3 expression levels stratify across immunologically “hot” versus “cold” tumors, normalized counts (RNA-seq) were obtained using the TCGAbiolinks package (version 3.8) from bioconductor in R for the COAD, PRAD, KIRC, LUAD, THCA, and SKCM datasets, along with clinical annotations. Normal tissue samples and metastases were removed from the analysis, unless otherwise noted. Each sample was ranked within the corresponding dataset for PHD3 expression and classified as PHD3-high or PHD3-Low using percentile cut-offs of 10% or 20%. Next, patient samples were clustered based on a list of CD8+ signature genes comprised of CD8+ T cell lineage markers (CD3D and CD8A) as well as genes involved in CD8+ T cell trafficking, activation, and cytotoxicity (CXCR3, GZMA, GZMB, GZMK, ICOS, and PRF1) using the Pheatmap package from bioconductor in R, generating sample clusters that were “hot”, “intermediate”, or “cold” for T cell infiltration and functionality. CD8+ immune scores were calculated as the genewise sum of z-scores for the CD8+ signature genes. For subsequent enrichment analysis the “intermediate” and “hot” groups were combined. PHD3 status was layered over the clustering and the number of PHD3-Low samples was counted in “cold” tumors versus all others. Fisher’s Exact Test was used to quantify enrichment of PHD3-low samples in “cold” tumors for each TCGA dataset.
Proteomics Analysis
Enrichment analysis was performed using the fgsea package implemented in R (v1.14.0) (Sergushichev, 2016). Pre-ranked lists were generated by weighting fold-change by the negative logarithm of the p-value comparing CD and HFD. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD019495 (Deutsch et al., 2020; Perez-Riverol et al., 2019).
Supplementary Material
Figure S1. Related to Figure 1
(A) Comparison of percentages of kilocalories (kcal) derived from fat, protein, and carbohydrate in CD and HFD.
(B) Body weights of WT C57BL/6J and TCRα-KO mice after 8-10 weeks of CD or HFD feeding.
(C-I) Systemic metabolic and inflammatory parameters from WT C57BL/6J mice after 8-10 weeks of CD or HFD feeding in plasma: cholesterol (C), glucose (D), insulin (E), leptin (F), IL-6 (G), resistin (H) and adiponectin (I).
(J) Tumor volumes 11 days after subcutaneous inoculation with 2.5 x 105 B16-OVA-RFP tumor cells in WT C57BL/6J mice after CD or HFD feeding for 8-10 weeks.
(K-L) Tumor growth curves of WT BALB/cJ mice inoculated with 2.5x105 CT26 (K) or 2.5x105 RENCA (L) tumor cells.
(M-N) Representative flow cytometry plots, pre-gated on live T cells, showing relative abundance of CD4+ and CD8+ T cells on day 10 after tumor implantation with isotype control (M) and depleting anti-CD8 (N) antibody treatment.
(O-P) Tumor growth curves of WT C57BL/6J mice inoculated with 105 MC38 tumor cells and treated with isotype control or depleting anti-CD8 antibodies after CD (O) or HFD (P) feeding for 8-10 weeks.
Graphs display mean +/− SD (B-J) or mean +/− SEM (K-L, O-P). (ns p>0.05, *p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001).
Figure S2. Related to Figure 2
(A) Gating strategies for flow cytometry analysis in Fig. 2B, 2E–2N, S2D-S2I, and S2Q-S2X.
(B) Gating strategies for flow cytometry analysis in Fig. 2C–2D, S2C, S2J, S2O, and S2P.
(C) The ratio of CD4+ T cells to MC38-GFP tumor cells, as measured by flow cytometry.
(D-I) Flow cytometry analysis of CD45+ leukocytes isolated from MC38 tumors on day 10-14 after inoculation with 105 tumor cells. Quantification of FoxP3+ regulatory T cells (Tregs) (D), Treg to CD8+ T cell ratio (E), NK1.1+ cells (F), CD11b+ myeloid cells (G), GR1+ CD11b+ cells (H) and F4/80+ Gr1− CD11b+ cells (I).
(J) Leukocyte census enumerating CD8+ T cells, CD4+ T cells, CD11b+ myeloid cells, and all other CD45+ cells per GFP+ tumor cell from HFD- or CD-fed mice.
(K-N) Flow cytometric analysis of CD11c+ cells, showing % CD11c+ cells of CD45+ cells (K), % MHC-I+ of CD11c+ cells (L), % MHC-II+ of CD11c+ cells (M), and % CD40+ of CD11c+ cells (N).
(O-P) Flow cytometric analysis of GFP+ MC38 tumor cells, showing % PD-L1+ (O) and % MHC-I+ (P).
(Q-R) Representative histograms for Ki67 (Q) or ICOS (R) staining.
(S-V) Flow cytometry analysis of CD8+ T cells from indicated tissues in CD or HFD mice bearing E0771 (S-T) or B16-OVA-RFP (U-V) tumors, quantifying % CD8+ T cells of CD45+ cells (S,U) and % PD-1+ of CD8+ T cells (T,V).
(W-X) Flow cytometry analysis of CD8+ T cells from indicated tissues in CD or HFD BALB/cJ mice bearing CT26 tumors, quantifying % CD8+ T cells of CD45+ cells (W) and % GZMB+ of CD8+ T cells (X).
(Y-AA) Flow cytometry analysis of ex vivo stimulated naïve CD8+ T cells from CD and HFD mice on 2, 4, or 8 μg/mL each of plate-bound anti-CD3 and anti-CD28 antibodies, quantifying live cell numbers (Y), GZMB expression (Z), and Ki67 expression (AA).
Data represent ≥ two independent experiments. Graphs display mean +/− SD (C-P, S-AA). (ns p>0.05, *p≤0.05, **p≤0.01, ***p≤0.001).
Figure S3. Related to Figure 3
(A) UMAP embeddings of CD45+ leukocyte infiltrate colored by diet.
(B) Overview of cell populations and key differentially expressed transcripts that define each cell cluster. Quantification of the proportion of each cell cluster among all CD45+ leukocyte infiltrate (left) and table showing cluster identifications with key differentially transcripts (right). Statistical significance was assessed by two-sided binomial test.
(C-K) Scatterplots showing average signature score for curated KEGG pathways on a cluster-by-cluster basis in HFD versus CD for fructose and mannose metabolism (C), folate biosynthesis (D), TCA cycle (E), oxidative phosphorylation (F), glycerolipid metabolism (G), sphingolipid metabolism (H), insulin signaling pathway (I), mTOR signaling pathway (J), and phosphatidyl inositol signaling pathway (K).
(L-N) Expression of key marker genes within lymphocyte clusters: Cd3g (L), Ikzf2 (M), and Cd8a (N).
(O-Q) Projection of gene signatures onto lymphocyte clusters using AddModuleScore in Seurat: G2M-S signature (O), Slamf6+ stem cell progenitor signature (P), and Tim3+ cytotoxic signature (Q).
(R) Table showing sub-cluster identifications within T lymphocytes with key differentially transcripts and associated signatures.
(S) Schematic for scoring in VISION against Immunological Signatures (C7) from ImmuneSigDB.
(T) Table of filtered/curated immunological signatures and whether they correspond to the activated/stimulated or naïve/unstimulated state.
Statistical significance was assessed by Wilcoxon rank sum with FDR correction using the method of Benjamini and Hochberg (C-K). (ns p>0.05, * p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001)
Figure S4. Related to Figure 4
(A) Representative CyCIF image of CD tumors showing staining for non-lineage antibody targets. Scale bar is 2000 μm (full tissue section) and 100 μm (regions of interest).
(B) Schematic showing CyCIF analysis pipeline.
(C) Representative CyCIF images of CD and HFD tumors showing infiltration and general localization of cells bearing lymphocyte markers. Scale bars are 1000 μm.
(D) Expression pattern of mitochondrial metabolic genes in CD or HD tumors. Scale bars are 2000 μm.
(E) This panel is a control for subsequent spatial overlap analysis (S4F-H), showing a comparison between the number of simulated CD8+ T cells and the actual number of CD8+ T cells per CyCIF tissue sample. For each tissue, the number of measured CD8+ T cells (y-axis) is plotted versus the number of simulated Poisson-Disc sample points (x-axis), the latter of which represent randomly distributed CD8+ T cells within the tissue section. There is a strong correlation between the number of events used to compute fractional overlap under actual and simulated conditions.
(F-H) Spatial overlap between GLUT1-high or ACO2-high regions of tumors and CD8+ T cells (F), CD4+ T cells (G), and CD11b+ LY6G+ myeloid cells (H). Graphs depict measured and simulated data.
Graphs display mean +/− SD (F-H). (ns p>0.05, *p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001).
Figure S5. Related to Figure 5
(A-E) Analysis of RNA-sequencing data performed on cells sorted out of day 12 MC38 tumors from CD-fed and HFD-fed animals.
(A) Screen plot depicting variation explained by the first ten principal components.
(B) Heatmap of genes with the highest loadings that contribute to PC#1.
(C) Significantly enriched gene sets from GSEA analysis against the Hallmarks MSigDB collection for tumor cell RNA-seq data.
(D) Venn diagram depicting overlap between significantly altered genes with HFD in CD8+ TILs and MC38 tumor cells using a p-value cut-off = 0.01.
(E) Heatmap depicting expression of genes related to T cell stimulation and suppression in MC38 tumor cells.
(F) Representative flow plot of ex vivo LipidTox neutral lipid staining in MC38-GFP tumor cells in day 10-14 tumors.
(G-I) Quantification of C16-BODIPY uptake ex vivo in CD11b+ myeloid cells in MC38 (G), E0771 (H) and B16-OVA-RFP (I) tumors.
(J) MC38 tumor cell number after 48 hours of growth in the presence of 200 μM each of BSA-conjugated palmitate plus BSA-conjugated oleate versus BSA control.
Bar graphs display mean +/− SD (G-J). (ns p>0.05, *p≤0.05, **p≤0.01, ***p≤0.001).
Figure S6. Related to Figure 6
(A) Principal component analysis of proteomics data.
(B) Hierarchical clustering of proteomics samples.
(C-E) Heatmaps showing relative expression of proteins involved in fat synthesis (C), TCA cycle (D), or T cell stimulation and suppression (E).
(F-M) Lipidomics analysis of plasma, TIF and FACS-sorted GFP+ tumor cells from day 13 MC38-GFP tumor-bearing mice fed CD or HFD for 8-10 weeks.
(F-H) Volcano plots of lipid species in GFP+ tumor cells (F), plasma (G) and TIF (H).
(I-L) Correlation between the TIF to plasma ratio of lipid abundance in the CD and HFD context, displaying all lipids (I), all lipids except DAG (J), all lipids without TAG (K), and all lipids without DAG and TAG (L).
(M) Plot displaying the strength of correlation in panels I-L.
(N) Lipoprotein lipase (LPL) activity in homogenized mouse hearts and MC38 tumors.
Key abbreviations: DAG, diglyceride. TAG, triglyceride. TIF, tumor interstitial fluid.
Figure S7. Related to Figure 7
(A-C) Characterization of PHD3-OE MC38 cell lines.
(A) Western blot of C-terminally FLAG-tagged PHD3-OE cell lines.
(B) IP-Western blot for hydroxylated ACC2 from PHD3-OE versus empty vector-transduced MC38 cell lines.
(C) In vitro growth curve of PHD3-OE and EV-transduced MC38 cells in DMEM media.
(D-E) Phd3 expression in MC38 EV-transduced versus PHD3-OE tumors measured by qPCR in mice fed CD (D) and HFD (E) and sacrificed at humane endpoints.
(F-G) Flow cytometric analysis of MHC-I (F) and PD-L1 (G) expression on RFP+ tumor cells from day 14 MC38-PHD3-OE and control tumors.
(H) Quantification of C16-BODIPY uptake ex vivo in RFP-expressing MC38-PHD3-OE and control tumor cells from dissociated tumors isolated from mice fed HFD.
(I) NAD+ levels in TIF versus tumor tissue from targeted metabolomics analysis of day 14 tumors. NAD+ signal is internally normalized to the alanine signal, which represents an abundant metabolite in both sample types that is not altered by diet (data not shown).
(J) Volcano plot depicting metabolomics analysis of circulating FFAs from the plasma of tumor-bearing HFD and CD mice. Pink corresponds to FFAs with p-value<0.05 that increase with HFD relative to CD and blue corresponds to FFAs with p-value<0.05 that decrease with HFD relative to CD.
(K-O) Bioinformatics analysis of TCGA RNA-seq data across multiple cancer types.
(K) Expression of PHD1, PHD2, and PHD3 in colorectal cancer in obese and non-obese patients.
(L-M) Graph depicting Spearman correlation coefficients and p-values comparing PHD3 versus MHC-I (L) or GZMB (M) expression across patient samples in all TCGA datasets available from the GEPIA web portal.
(N) Quantification of PHD3 expression in cold tumors versus all others (Int+Hot).
(O) Chart detailing quantification for the enrichment of PHD3-low samples in immunologically cold tumors using Fisher’s Exact Test across multiple cancer types, as well as GLUD1 as a control.
Graphs display mean +/− SD (C-I, K, N). (ns p>0.05, *p≤0.05, **p≤0.01, ***p≤0.001).
Highlights:
Defined a metabolic single cell atlas of the tumor immune landscape with obesity
Obesity induces a metabolic tug of war between tumor and T cells for lipids
Tumor cells induce fat metabolism and change their microenvironment during obesity
Blocking metabolic adaptations to obesity in cancers restores anti-tumor immunity
Acknowledgments
We are grateful for resources provided by the “MicRoN core at Harvard Medical School” and “HMS Immunology Flow Cytometry Core Facility”. This study was supported by grants from the Ludwig Center at Harvard Medical School (M.C.H. A.H.S. and P.K.S), NCI grant U54-CA225088 (M.C.H., A.H.S. and P.K.S.), the Evergrande Center for Immunologic Disease (A.H.S.), the Glenn Foundation for Medical Research (M.C.H.), NCI Grant R01CA213062 (M.C.H.), and NIH grant from the National Institute of Diabetes and Digestive and Kidney Diseases R01DK103295 (M.C.H.) and P01 AI56299 (to A.H.S). A.E.R was supported by postdoctoral fellowships from the American Cancer Society (130373-PF-17-132-01-CCG) and Cell Biology Education and Fellowship Fund at Harvard Medical School. J.M.D. was supported by a predoctoral F31 NIH fellowship (5F31CA224601). M.W.L. was supported by a predoctoral T32 NIH fellowship (T32CA207021). The VUMC Hormone Assay & Analytical Services and Lipid Cores are supported by NIH grants DK059637 (MMPC) and DK020593 (DRTC).
Declaration of Interests
A.H.S. has patents/pending royalties on intellectual property on the PD-1 pathway from Roche and Novartis. A.H.S. is on advisory boards for Surface Oncology, Elstar, SQZ Biotechnologies, Selecta, Elpiscience, and Monopteros and has research funding from Novartis, Roche, Ipsen, Quark, and Merck. M.C.H. has patents pending on the PHD3 pathway and is on the scientific advisory board for Pori Therapeutics and has research funding from Roche. J.M.D. has consulted for ElevateBio and Third Rock Ventures. The remaining authors declare no competing interests.
References
- Algire C, Amrein L, Bazile M, David S, Zakikhani M, and Pollak M (2011). Diet and tumor LKB1 expression interact to determine sensitivity to anti-neoplastic effects of metformin in vivo. Oncogene 30, 1174–1182. [DOI] [PubMed] [Google Scholar]
- Beyaz S, Mana MD, Roper J, Kedrin D, Saadatpour A, Hong S-J, Bauer-Rowe KE, Xifaras ME, Akkad A, Arias E, et al. (2016). High-fat diet enhances stemness and tumorigenicity of intestinal progenitors. Nature 531, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Biase S, Lee C, Brandhorst S, Manes B, Buono R, Cheng C-W, Cacciottolo M, Martin-Montalvo A, de Cabo R, Wei M, et al. (2016). Fasting-Mimicking Diet Reduces HO-1 to Promote T Cell-Mediated Tumor Cytotoxicity. Cancer Cell 30, 136–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridson R (2007). Fast Poisson Disk Sampling in Arbitrary Dimensions. In ACM SIGGRAPH 2007 Sketches, (New York, NY, USA: Association for Computing Machinery; ), pp. 22–es. [Google Scholar]
- Buck MD, O’Sullivan D, and Pearce EL (2015). T cell metabolism drives immunity. J. Exp. Med 212, 1345 LP–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campello RJGB, Moulavi D, and Sander J (2013). Density-Based Clustering Based on Hierarchical Density Estimates BT-Advances in Knowledge Discovery and Data Mining. Pei J, Tseng VS, Cao L, Motoda H, and Xu G, eds. (Berlin, Heidelberg: Springer Berlin Heidelberg; ), pp. 160–172. [Google Scholar]
- Chang CH, Qiu J, O’Sullivan D, Buck MD, Noguchi T, Curtis JD, Chen Q, Gindin M, Gubin MM, Van Der Windt GJW, et al. (2015). Metabolic Competition in the Tumor Microenvironment Is a Driver of Cancer Progression. Cell 162, 1229–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortellini A, Bersanelli M, Buti S, Cannita K, Santini D, Perrone F, Giusti R, Tiseo M, Michiara M, Di Marino P, et al. (2019). A multicenter study of body mass index in cancer patients treated with anti-PD-1/PD-L1 immune checkpoint inhibitors: when overweight becomes favorable. J. Immunother. Cancer 7, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby EJ, Wei J, Yang XY, Lei G, Wang T, Liu CX, Agarwal P, Korman AJ, Morse MA, Gouin K, et al. (2018). Complimentary mechanisms of dual checkpoint blockade expand unique T-cell repertoires and activate adaptive anti-tumor immunity in triple-negative breast tumors. Oncoimmunology 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng T, Lyon CJ, Bergin S, Caligiuri MA, and Hsueh WA (2016). Obesity, Inflammation, and Cancer. Annu. Rev. Pathol 11, 421–449. [DOI] [PubMed] [Google Scholar]
- DeTomaso D, Jones MG, Subramaniam M, Ashuach T, Ye CJ, and Yosef N (2019). Functional interpretation of single cell similarity maps. Nat. Commun 10, 4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch EW, Bandeira N, Sharma V, Perez-Riverol Y, Carver JJ, Kundu DJ, García-Seisdedos D, Jarnuczak AF, Hewapathirana S, Pullman BS, et al. (2020). The ProteomeXchange consortium in 2020: enabling “big data” approaches in proteomics. Nucleic Acids Res. 48, D1145–D1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, and Taubman MB (2010). Prolyl hydroxylase EGLN3 regulates skeletal myoblast differentiation through an NF-kappaB-dependent pathway. J. Biol. Chem 285, 8927–8935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvalov BK, Foss F, Henze A-T, Bethani I, Gräf-Höchst S, Singh D, Filatova A, Dopeso H, Seidel S, Damm M, et al. (2014). PHD3 regulates EGFR internalization and signalling in tumours. Nat. Commun 5, 5577. [DOI] [PubMed] [Google Scholar]
- Geiger R, Rieckmann JC, Wolf T, Basso C, Feng Y, Fuhrer T, Kogadeeva M, Picotti P, Meissner F, Mann M, et al. (2016). L-Arginine Modulates T Cell Metabolism and Enhances Survival and Anti-tumor Activity. Cell 167, 829–842.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German NJ, Yoon H, Yusuf RZ, Murphy JP, Finley LWS, Laurent G, Haas W, Satterstrom FK, Guarnerio J, Zaganjor E, et al. (2016). PHD3 Loss in Cancer Enables Metabolic Reliance on Fatty Acid Oxidation via Deactivation of ACC2. Mol. Cell 63, 1006–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafemeister C, and Satija R (2019). Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho PC, Bihuniak JD, MacIntyre AN, Staron M, Liu X, Amezquita R, Tsui YC, Cui G, Micevic G, Perales JC, et al. (2015). Phosphoenolpyruvate Is a Metabolic Checkpoint of Anti-tumor T Cell Responses. Cell 162, 1217–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islami F, Goding Sauer A, Gapstur SM, and Jemal A (2018). Proportion of Cancer Cases Attributable to Excess Body Weight by US State, 2011-2015. JAMA Oncol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang C, Hui S, Lu W, Cowan AJ, Morscher RJ, Lee G, Liu W, Tesz GJ, Birnbaum MJ, and Rabinowitz JD (2018). The Small Intestine Converts Dietary Fructose into Glucose and Organic Acids. Cell Metab. 27, 351–361.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG, and Ratcliffe PJ (2008). Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell 30. [DOI] [PubMed] [Google Scholar]
- Kamphorst JJ, Fan J, Lu W, White E, and Rabinowitz JD (2011). Liquid chromatography-high resolution mass spectrometry analysis of fatty acid metabolism. Anal. Chem 83, 9114–9122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk MS, Tirosh I, Heckl D, Rao TN, Dixit A, Haas BJ, Schneider RK, Wagers AJ, Ebert BL, and Regev A (2015). Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 25, 1860–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauby-Secretan B, Scoccianti C, Loomis D, Grosse Y, Bianchini F, Straif K, and International Agency for Research on Cancer Handbook Working Group (2016). Body Fatness and Cancer--Viewpoint of the IARC Working Group. N. Engl. J. Med 375, 794–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner MG, Karimi SS, Barry-Holson K, Angell TE, Murphy KA, Church CH, Ohlfest JR, Hu P, and Epstein AL (2013). Immunogenicity of murine solid tumor models as a defining feature of in vivo behavior and response to immunotherapy. J. Immunother 36, 477–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Grimm SA, Chrysovergis K, Kosak J, Wang X, Du Y, Burkholder A, Janardhan K, Mav D, Shah R, et al. (2014). Obesity, Rather Than Diet, Drives Epigenomic Alterations in Colonic Epithelium Resembling Cancer Progression. Cell Metab. 19, 702–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Grimm SA, Mav D, Gu H, Djukovic D, Shah R, Merrick BA, Raftery D, and Wade PA (2018). Transcriptome and DNA Methylome Analysis in a Mouse Model of Diet-Induced Obesity Predicts Increased Risk of Colorectal Cancer. Cell Rep. 22, 624–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J-R, Fallahi-Sichani M, and Sorger PK (2015). Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method. Nat. Commun 6, 8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J-R, Izar B, Wang S, Yapp C, Mei S, Shah PM, Santagata S, and Sorger PK (2018). Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maaten LJP, H.G.E. (2008). Visualizing Data using t-SNE. JournalofMachineLearningResearch 2579–2605. [Google Scholar]
- MacIver NJ, Michalek RD, and Rathmell JC (2013). Metabolic Regulation of T Lymphocytes. Annu. Rev. Immunol 31, 259–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlister GC, Nusinow DP, Jedrychowski MP, Wühr M, Huttlin EL, Erickson BK, Rad R, Haas W, and Gygi SP (2014). MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal. Chem 86, 7150–7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuade JL, Daniel CR, Hess KR, Mak C, Wang DY, Rai RR, Park JJ, Haydu LE, Spencer C, Wongchenko M, et al. (2018). Association of body-mass index and outcomes in patients with metastatic melanoma treated with targeted therapy, immunotherapy, or chemotherapy: a retrospective, multicohort analysis. Lancet. Oncol 19, 310–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BC, Sen DR, Al Abosy R, Bi K, Virkud YV, LaFleur MW, Yates KB, Lako A, Felt K, Naik GS, et al. (2019). Subsets of exhausted CD8(+) T cells differentially mediate tumor control and respond to checkpoint blockade. Nat. Immunol 20, 326–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosely SIS, Prime JE, Sainson RCA, Koopmann J-O, Wang DYQ, Greenawalt DM, Ahdesmaki MJ, Leyland R, Mullins S, Pacelli L, et al. (2017). Rational Selection of Syngeneic Preclinical Tumor Models for Immunotherapeutic Drug Discovery. Cancer Immunol. Res 5, 29–41. [DOI] [PubMed] [Google Scholar]
- Murphy WJ, and Longo DL (2019). The Surprisingly Positive Association Between Obesity and Cancer Immunotherapy Efficacy. JAMA 321, 1247–1248. [DOI] [PubMed] [Google Scholar]
- Navarrete-Perea J, Yu Q, Gygi SP, and Paulo JA (2018). Streamlined Tandem Mass Tag (SL-TMT) Protocol: An Efficient Strategy for Quantitative (Phospho)proteome Profiling Using Tandem Mass Tag-Synchronous Precursor Selection-MS3. J. Proteome Res 17, 2226–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimri L, Saadi J, Peri I, Yehuda-Shnaidman E, and Schwartz B (2015). Mechanisms linking obesity to altered metabolism in mice colon carcinogenesis. Oncotarget 6, 38195–38209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulo JA, O’Connell JD, Everley RA, O’Brien J, Gygi MA, and Gygi SP (2016a). Quantitative mass spectrometry-based multiplexing compares the abundance of 5000 S. cerevisiae proteins across 10 carbon sources. J. Proteomics 148, 85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulo JA, O’Connell JD, and Gygi SP (2016b). A Triple Knockout (TKO) Proteomics Standard for Diagnosing Ion Interference in Isobaric Labeling Experiments. J. Am. Soc. Mass Spectrom 27, 1620–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlova NN, and Thompson CB (2016). The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 23, 27–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng T, Thorn K, Schroeder T, Wang L, Theis FJ, Marr C, and Navab N (2017). A BaSiC tool for background and shading correction of optical microscopy images. Nat. Commun 8, 14836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, et al. (2019). The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrocola F, Pol J, Vacchelli E, Rao S, Enot DP, Baracco EE, Levesque S, Castoldi F, Jacquelot N, Yamazaki T, et al. (2016). Caloric Restriction Mimetics Enhance Anticancer Immunosurveillance. Cancer Cell 30, 147–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan P, Ruh N, Harnoss JM, Kiss J, Mollenhauer M, Scherr A-L, Platzer LK, Schmidt T, Podar K, Opferman JT, et al. (2016). Prolyl Hydroxylase 3 Attenuates MCL-1–Mediated ATP Production to Suppress the Metastatic Potential of Colorectal Cancer Cells. Cancer Res. 76, 2219 LP–2230. [DOI] [PubMed] [Google Scholar]
- Rawluszko AA, Bujnicka KE, Horbacka K, Krokowicz P, and Jagodziński PP (2013). Expression and DNA methylation levels of prolyl hydroxylases PHD1, PHD2, PHD3 and asparaginyl hydroxylase FIH in colorectal cancer. BMC Cancer 13, 526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronneberger O, Fischer P, and Brox T (2015). U-Net: Convolutional Networks for Biomedical Image Segmentation BT - Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015 . Navab N, Hornegger J, Wells WM, and Frangi AF, eds. (Cham: Springer International Publishing; ), pp. 234–241. [Google Scholar]
- Rubio-Patiño C, Bossowski JP, De Donatis GM, Mondragón L, Villa E, Aira LE, Chiche J, Mhaidly R, Lebeaupin C, Marchetti S, et al. (2018). Low-Protein Diet Induces IRE1α-Dependent Anticancer Immunosurveillance. Cell Metab. 27, 828–842.e7. [DOI] [PubMed] [Google Scholar]
- Sage PT, Ron-Harel N, Juneja VR, Sen DR, Maleri S, Sungnak W, Kuchroo VK, Haining WN, Chevrier N, Haigis M, et al. (2016). Suppression by TFR cells leads to durable and selective inhibition of B cell effector function. Nat. Immunol 17, 1436–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweppe DK, Prasad S, Belford MW, Navarrete-Perea J, Bailey DJ, Huguet R, Jedrychowski MP, Rad R, McAlister G, Abbatiello SE, et al. (2019). Characterization and Optimization of Multiplexed Quantitative Analyses Using High-Field Asymmetric-Waveform Ion Mobility Mass Spectrometry. Anal. Chem 91, 4010–4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweppe DK, Rusin SF, Gygi SP, and Paulo JA (2020). Optimized Workflow for Multiplexed Phosphorylation Analysis of TMT-Labeled Peptides Using High-Field Asymmetric Waveform Ion Mobility Spectrometry. J. Proteome Res 19, 554–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergushichev AA (2016). An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. BioRxiv 60012. [Google Scholar]
- Siegel RL, Miller KD, and Jemal A (2019). Cancer statistics, 2019. CA. Cancer J. Clin [DOI] [PubMed] [Google Scholar]
- Sinclair LV, Rolf J, Emslie E, Shi Y-B, Taylor PM, and Cantrell DA (2013). Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nat. Immunol 14, 500–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinelli JB, and Haigis MC (2018). The multifaceted contributions of mitochondria to cellular metabolism. Nat. Cell Biol 20, 745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura A, and Rathmell JC (2018). Metabolic Barriers to T Cell Function in Tumors. J. Immunol 200, 400 LP–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan MR, Danai LV, Lewis CA, Chan SH, Gui DY, Kunchok T, Dennstedt EA, Vander Heiden MG, and Muir A (2019). Quantification of microenvironmental metabolites in murine cancers reveals determinants of tumor nutrient availability. Elife 8, e44235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H, Siegel RL, Torre LA, Pearson-Stuttard J, Islami F, Fedewa SA, Goding Sauer A, Shuval K, Gapstur SM, Jacobs EJ, et al. (2019). Global patterns in excess body weight and the associated cancer burden. CA. Cancer J. Clin 69, 88–112. [DOI] [PubMed] [Google Scholar]
- Ting L, Rad R, Gygi SP, and Haas W (2011). MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat. Methods 8, 937–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Aguilar EG, Luna JI, Dunai C, Khuat LT, Le CT, Mirsoian A, Minnar CM, Stoffel KM, Sturgill IR, et al. (2018). Paradoxical effects of obesity on T cell function during tumor progression and PD-1 checkpoint blockade. Nat. Med [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Raynor J, Nguyen T-LM, and Chi H (2017). Nutrient and Metabolic Sensing in T Cell Responses. Front. Immunol 8, 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiig H, Aukland K, and Tenstad O (2003). Isolation of interstitial fluid from rat mammary tumors by a centrifugation method. Am. J. Physiol. Heart Circ. Physiol 284, H416–24. [DOI] [PubMed] [Google Scholar]
- Woodall MJ, Neumann S, Campbell K, Pattison ST, and Young SL (2020). The Effects of Obesity on Anti-Cancer Immunity and Cancer Immunotherapy. Cancers (Basel). 12, 1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J, Li X, Jiao S, Wei Y, Wu G, and Fang J (2010). Prolyl hydroxylase-3 is downregulated in colorectal cancer cells and inhibits IKKbeta independent of hydroxylase activity. Gastroenterology 138, 606–615. [DOI] [PubMed] [Google Scholar]
- Yano H, Sakai M, Matsukawa T, Yagi T, Naganuma T, Mitsushima M, Iida S, Inaba Y, Inoue H, Unoki-Kubota H, et al. (2018). PHD3 regulates glucose metabolism by suppressing stress-induced signalling and optimising gluconeogenesis and insulin signalling in hepatocytes. Sci. Rep 8, 14290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MD, and Behjati S (2018). SoupX removes ambient RNA contamination from droplet based single cell RNA sequencing data. BioRxiv 303727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Yue C, Herrmann A, Song J, Egelston C, Wang T, Zhang Z, Li W, Lee H, Aftabizadeh M, et al. (2020). STAT3 Activation-Induced Fatty Acid Oxidation in CD8+ T Effector Cells Is Critical for Obesity-Promoted Breast Tumor Growth. Cell Metab. 31, 148–161.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Related to Figure 1
(A) Comparison of percentages of kilocalories (kcal) derived from fat, protein, and carbohydrate in CD and HFD.
(B) Body weights of WT C57BL/6J and TCRα-KO mice after 8-10 weeks of CD or HFD feeding.
(C-I) Systemic metabolic and inflammatory parameters from WT C57BL/6J mice after 8-10 weeks of CD or HFD feeding in plasma: cholesterol (C), glucose (D), insulin (E), leptin (F), IL-6 (G), resistin (H) and adiponectin (I).
(J) Tumor volumes 11 days after subcutaneous inoculation with 2.5 x 105 B16-OVA-RFP tumor cells in WT C57BL/6J mice after CD or HFD feeding for 8-10 weeks.
(K-L) Tumor growth curves of WT BALB/cJ mice inoculated with 2.5x105 CT26 (K) or 2.5x105 RENCA (L) tumor cells.
(M-N) Representative flow cytometry plots, pre-gated on live T cells, showing relative abundance of CD4+ and CD8+ T cells on day 10 after tumor implantation with isotype control (M) and depleting anti-CD8 (N) antibody treatment.
(O-P) Tumor growth curves of WT C57BL/6J mice inoculated with 105 MC38 tumor cells and treated with isotype control or depleting anti-CD8 antibodies after CD (O) or HFD (P) feeding for 8-10 weeks.
Graphs display mean +/− SD (B-J) or mean +/− SEM (K-L, O-P). (ns p>0.05, *p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001).
Figure S2. Related to Figure 2
(A) Gating strategies for flow cytometry analysis in Fig. 2B, 2E–2N, S2D-S2I, and S2Q-S2X.
(B) Gating strategies for flow cytometry analysis in Fig. 2C–2D, S2C, S2J, S2O, and S2P.
(C) The ratio of CD4+ T cells to MC38-GFP tumor cells, as measured by flow cytometry.
(D-I) Flow cytometry analysis of CD45+ leukocytes isolated from MC38 tumors on day 10-14 after inoculation with 105 tumor cells. Quantification of FoxP3+ regulatory T cells (Tregs) (D), Treg to CD8+ T cell ratio (E), NK1.1+ cells (F), CD11b+ myeloid cells (G), GR1+ CD11b+ cells (H) and F4/80+ Gr1− CD11b+ cells (I).
(J) Leukocyte census enumerating CD8+ T cells, CD4+ T cells, CD11b+ myeloid cells, and all other CD45+ cells per GFP+ tumor cell from HFD- or CD-fed mice.
(K-N) Flow cytometric analysis of CD11c+ cells, showing % CD11c+ cells of CD45+ cells (K), % MHC-I+ of CD11c+ cells (L), % MHC-II+ of CD11c+ cells (M), and % CD40+ of CD11c+ cells (N).
(O-P) Flow cytometric analysis of GFP+ MC38 tumor cells, showing % PD-L1+ (O) and % MHC-I+ (P).
(Q-R) Representative histograms for Ki67 (Q) or ICOS (R) staining.
(S-V) Flow cytometry analysis of CD8+ T cells from indicated tissues in CD or HFD mice bearing E0771 (S-T) or B16-OVA-RFP (U-V) tumors, quantifying % CD8+ T cells of CD45+ cells (S,U) and % PD-1+ of CD8+ T cells (T,V).
(W-X) Flow cytometry analysis of CD8+ T cells from indicated tissues in CD or HFD BALB/cJ mice bearing CT26 tumors, quantifying % CD8+ T cells of CD45+ cells (W) and % GZMB+ of CD8+ T cells (X).
(Y-AA) Flow cytometry analysis of ex vivo stimulated naïve CD8+ T cells from CD and HFD mice on 2, 4, or 8 μg/mL each of plate-bound anti-CD3 and anti-CD28 antibodies, quantifying live cell numbers (Y), GZMB expression (Z), and Ki67 expression (AA).
Data represent ≥ two independent experiments. Graphs display mean +/− SD (C-P, S-AA). (ns p>0.05, *p≤0.05, **p≤0.01, ***p≤0.001).
Figure S3. Related to Figure 3
(A) UMAP embeddings of CD45+ leukocyte infiltrate colored by diet.
(B) Overview of cell populations and key differentially expressed transcripts that define each cell cluster. Quantification of the proportion of each cell cluster among all CD45+ leukocyte infiltrate (left) and table showing cluster identifications with key differentially transcripts (right). Statistical significance was assessed by two-sided binomial test.
(C-K) Scatterplots showing average signature score for curated KEGG pathways on a cluster-by-cluster basis in HFD versus CD for fructose and mannose metabolism (C), folate biosynthesis (D), TCA cycle (E), oxidative phosphorylation (F), glycerolipid metabolism (G), sphingolipid metabolism (H), insulin signaling pathway (I), mTOR signaling pathway (J), and phosphatidyl inositol signaling pathway (K).
(L-N) Expression of key marker genes within lymphocyte clusters: Cd3g (L), Ikzf2 (M), and Cd8a (N).
(O-Q) Projection of gene signatures onto lymphocyte clusters using AddModuleScore in Seurat: G2M-S signature (O), Slamf6+ stem cell progenitor signature (P), and Tim3+ cytotoxic signature (Q).
(R) Table showing sub-cluster identifications within T lymphocytes with key differentially transcripts and associated signatures.
(S) Schematic for scoring in VISION against Immunological Signatures (C7) from ImmuneSigDB.
(T) Table of filtered/curated immunological signatures and whether they correspond to the activated/stimulated or naïve/unstimulated state.
Statistical significance was assessed by Wilcoxon rank sum with FDR correction using the method of Benjamini and Hochberg (C-K). (ns p>0.05, * p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001)
Figure S4. Related to Figure 4
(A) Representative CyCIF image of CD tumors showing staining for non-lineage antibody targets. Scale bar is 2000 μm (full tissue section) and 100 μm (regions of interest).
(B) Schematic showing CyCIF analysis pipeline.
(C) Representative CyCIF images of CD and HFD tumors showing infiltration and general localization of cells bearing lymphocyte markers. Scale bars are 1000 μm.
(D) Expression pattern of mitochondrial metabolic genes in CD or HD tumors. Scale bars are 2000 μm.
(E) This panel is a control for subsequent spatial overlap analysis (S4F-H), showing a comparison between the number of simulated CD8+ T cells and the actual number of CD8+ T cells per CyCIF tissue sample. For each tissue, the number of measured CD8+ T cells (y-axis) is plotted versus the number of simulated Poisson-Disc sample points (x-axis), the latter of which represent randomly distributed CD8+ T cells within the tissue section. There is a strong correlation between the number of events used to compute fractional overlap under actual and simulated conditions.
(F-H) Spatial overlap between GLUT1-high or ACO2-high regions of tumors and CD8+ T cells (F), CD4+ T cells (G), and CD11b+ LY6G+ myeloid cells (H). Graphs depict measured and simulated data.
Graphs display mean +/− SD (F-H). (ns p>0.05, *p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001).
Figure S5. Related to Figure 5
(A-E) Analysis of RNA-sequencing data performed on cells sorted out of day 12 MC38 tumors from CD-fed and HFD-fed animals.
(A) Screen plot depicting variation explained by the first ten principal components.
(B) Heatmap of genes with the highest loadings that contribute to PC#1.
(C) Significantly enriched gene sets from GSEA analysis against the Hallmarks MSigDB collection for tumor cell RNA-seq data.
(D) Venn diagram depicting overlap between significantly altered genes with HFD in CD8+ TILs and MC38 tumor cells using a p-value cut-off = 0.01.
(E) Heatmap depicting expression of genes related to T cell stimulation and suppression in MC38 tumor cells.
(F) Representative flow plot of ex vivo LipidTox neutral lipid staining in MC38-GFP tumor cells in day 10-14 tumors.
(G-I) Quantification of C16-BODIPY uptake ex vivo in CD11b+ myeloid cells in MC38 (G), E0771 (H) and B16-OVA-RFP (I) tumors.
(J) MC38 tumor cell number after 48 hours of growth in the presence of 200 μM each of BSA-conjugated palmitate plus BSA-conjugated oleate versus BSA control.
Bar graphs display mean +/− SD (G-J). (ns p>0.05, *p≤0.05, **p≤0.01, ***p≤0.001).
Figure S6. Related to Figure 6
(A) Principal component analysis of proteomics data.
(B) Hierarchical clustering of proteomics samples.
(C-E) Heatmaps showing relative expression of proteins involved in fat synthesis (C), TCA cycle (D), or T cell stimulation and suppression (E).
(F-M) Lipidomics analysis of plasma, TIF and FACS-sorted GFP+ tumor cells from day 13 MC38-GFP tumor-bearing mice fed CD or HFD for 8-10 weeks.
(F-H) Volcano plots of lipid species in GFP+ tumor cells (F), plasma (G) and TIF (H).
(I-L) Correlation between the TIF to plasma ratio of lipid abundance in the CD and HFD context, displaying all lipids (I), all lipids except DAG (J), all lipids without TAG (K), and all lipids without DAG and TAG (L).
(M) Plot displaying the strength of correlation in panels I-L.
(N) Lipoprotein lipase (LPL) activity in homogenized mouse hearts and MC38 tumors.
Key abbreviations: DAG, diglyceride. TAG, triglyceride. TIF, tumor interstitial fluid.
Figure S7. Related to Figure 7
(A-C) Characterization of PHD3-OE MC38 cell lines.
(A) Western blot of C-terminally FLAG-tagged PHD3-OE cell lines.
(B) IP-Western blot for hydroxylated ACC2 from PHD3-OE versus empty vector-transduced MC38 cell lines.
(C) In vitro growth curve of PHD3-OE and EV-transduced MC38 cells in DMEM media.
(D-E) Phd3 expression in MC38 EV-transduced versus PHD3-OE tumors measured by qPCR in mice fed CD (D) and HFD (E) and sacrificed at humane endpoints.
(F-G) Flow cytometric analysis of MHC-I (F) and PD-L1 (G) expression on RFP+ tumor cells from day 14 MC38-PHD3-OE and control tumors.
(H) Quantification of C16-BODIPY uptake ex vivo in RFP-expressing MC38-PHD3-OE and control tumor cells from dissociated tumors isolated from mice fed HFD.
(I) NAD+ levels in TIF versus tumor tissue from targeted metabolomics analysis of day 14 tumors. NAD+ signal is internally normalized to the alanine signal, which represents an abundant metabolite in both sample types that is not altered by diet (data not shown).
(J) Volcano plot depicting metabolomics analysis of circulating FFAs from the plasma of tumor-bearing HFD and CD mice. Pink corresponds to FFAs with p-value<0.05 that increase with HFD relative to CD and blue corresponds to FFAs with p-value<0.05 that decrease with HFD relative to CD.
(K-O) Bioinformatics analysis of TCGA RNA-seq data across multiple cancer types.
(K) Expression of PHD1, PHD2, and PHD3 in colorectal cancer in obese and non-obese patients.
(L-M) Graph depicting Spearman correlation coefficients and p-values comparing PHD3 versus MHC-I (L) or GZMB (M) expression across patient samples in all TCGA datasets available from the GEPIA web portal.
(N) Quantification of PHD3 expression in cold tumors versus all others (Int+Hot).
(O) Chart detailing quantification for the enrichment of PHD3-low samples in immunologically cold tumors using Fisher’s Exact Test across multiple cancer types, as well as GLUD1 as a control.
Graphs display mean +/− SD (C-I, K, N). (ns p>0.05, *p≤0.05, **p≤0.01, ***p≤0.001).


