Abstract
Background:
Idiopathic pulmonary fibrosis (IPF) is a rare form of immune-mediated interstitial lung disease characterized by progressive pulmonary fibrosis and scarring. The pathogenesis of IPF is still unclear. Gene fusion events exist universally during transcription and show alternated patterns in a variety of lung diseases. Therefore, the comprehension of the function of gene fusion in IPF might shed light on IPF pathogenesis research and facilitate treatment development.
Methods:
In this study, we included 91 transcriptome datasets from the National Center for Biotechnology Information (NCBI), including 52 IPF patients and 39 healthy controls. We detected fusion events in these datasets and probed gene fusion-associated differential gene expression and functional pathways. To obtain robust results, we corrected the batch bias across different projects.
Results:
We identified 1550 gene fusion events in all transcriptomes and studied the possible impacts of IL7 = AC083837.1 gene fusion. The two genes locate adjacently in chromosome 8 and share the same promoters. Their fusion is associated with differential expression of 282 genes enriched in six Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and 35 functional gene sets. Gene ontology (GO) enrichment analysis shows that IL7 = AC083837.1 gene fusion is associated with the enrichment of 187 gene sets. The co-expression network of interleukin-7 (IL7) indicates that decreased IL7 expression is associated with many pathways that regulate IPF progress.
Conclusion:
Based on the results, we conclude that IL7 = AC083837.1 gene fusion might exacerbate fibrosis in IPF via enhancing activities of natural killer cell-mediated cytotoxicity, skin cell apoptosis, and vessel angiogenesis, the interaction of which contributes to the development of fibrosis and the deterioration of respiratory function of IPF patients. Our work unveils the possible roles of gene fusion in regulating IPF and demonstrates that gene fusion investigation is a valid approach in probing immunologic mechanisms and searching potential therapeutic targets for treating IPF.
The reviews of this paper are available via the supplemental material section.
Keywords: angiogenesis, apoptosis, fibrosis, fusion gene, IL7, IPF
Introduction
Idiopathic pulmonary fibrosis (IPF) is a rare form of interstitial lung disease characterized by scar tissues and progressive fibrosis, continuously worsening of which leads to the death of 70% patients within 3–5 years, caused mostly by respiratory failure.1 Although its pathogenesis is still unclear, IPF is considered to be associated with the abnormality of the immune response under the presence of certain environmental factors, such as alveolar injury and inflammation.1–3 Studies have shown that the innate immunity system, macrophages, chemotactic cytokines, and interleukins have pivotal impacts on the scarring and fibrosis progress in IPF.4–6 As a component of the innate immunity, natural killer (NK) cells have been found depleted in lung tissue of IPF patients,7 and the increased bronchial NK cell count is associated with impairment of IPF patients’ respiratory function.8 The imbalanced immune response initiates endothelial apoptosis,9,10 which further exacerbates the irreversible process of fibrosis under the interaction with cell migration and angiogenesis.11–14
IPF is a family-aggregated disease15,16 associated with the differential expression of genes such as SFTPA2,17 CAV1,18 MUC5B,19,20 and MHC21,22; its possible relationship with more genes is still under investigation.23–26 Besides protein-coding genes, long non-coding RNA (lncRNA) is also involved in the regulation of IPF. Reverse transcription-polymerase chain reaction (RT-PCR) of human IPF fibroblasts identifies 14 lncRNAs that might be central regulators of the IL-1β induced inflammatory reaction in IPF.27 A knock-out experiment in cultured human fibroblast cells also demonstrates lncRNAs’ regulatory function in IPF.28
Studies have shown that gene fusion affects downstream signaling pathway and participates in the pathogenesis of a variety of diseases.29 Gene fusion studies have significantly deepened our understanding in diseases and have provided novel personalized therapeutic targets for some types of cancers, such as non-small cell lung cancer.30,31 Until now, however, studies have not been reported in probing possible functions and signaling pathways of fusion genes in IPF. In this study, we identify gene fusion events in transcriptome datasets of human IPF lung tissue from four projects and focus on the possible impacts of interleukin-7 (IL7) gene fusion in the progress of IPF. Our results show that the gene fusion between IL7 and lncRNA AC083837.1 might be associated with worsening symptoms of IPF caused by enhanced activity of NK cell-mediated cytotoxicity, apoptotic process, and vessel angiogenesis. Our study, as one of the first attempts in describing the regulatory function of gene fusion in IPF, might shed light on the research of pathogenesis and novel treatments to IPF.
Materials and methods
Downloading of transcriptome datasets of human IPF lung tissues
We searched the National Center for Biotechnology Information (NCBI)’s Gene Expression Omnibus (GEO) database32 and The Cancer Genome Atlas (TCGA) database (https://www.cancer.gov/tcga) for the transcriptomes datasets required for this study. The criteria for the expected transcriptomes for this study were: 1. they were transcriptomes of human lung tissues with IPF or matched healthy lung tissues; 2. The transcriptomes were pair-ended sequenced; 3. The transcriptomes had relatively higher sequencing depth.
As a result, we identified four projects in the GEO database that performed paired-end RNA sequencing for lung tissue from human IPF patients: GSE52463,33 GSE83717,34 GSE92592,35 and GSE99621.36 No projects from the TCGA database were included, as we did not have access to the raw datasets. The dataset contained 91 transcriptomes, including 52 IPF tissues and 39 healthy tissues. GSE52463 has eight IPF and seven control lung tissue samples, GSE83717 has six IPF and five control lung tissues, GSE92592 has 20 IPF and 19 control lung tissue samples, GSE99621 has 18 IPF and eight healthy control samples. The sequencing device used in GSE99621 was Illumina HiSeq 2500, as well as Illumina HiSeq 2000 in the other three projects. The average size of these datasets was 7269 ± 2575 Mb.
The transcriptome data sets were downloaded as sequence read archive (SRA) files using Aspera’s command line of ascp and the prefetch command from NCBI’s sratoolkit (http://ncbi.github.io/sra-tools/, version 2.9.6-1). The SRA files were then converted into fastq files by the fastq-dump command from sratoolkit.
Identification of fusion genes
We filtered out low-quality reads using “Trimmomatic”. After removing the adaptors, reads shorter than 50 bases were dropped, and bases with leading or trailing quality <3 were removed. The bases were scanned four-base wide, and all four bases were cut out if their average quality was lower than 15.
Using the software of STAR, RNA sequences were mapped to the reference genome Homo_sapiens.GRCh38, and the unmapped reads were kept for the detection of the fusion genes. The parameters used for the alignment were: number of threads: 8, outFilterMultimapNmax: 1, outFilterMismatchNmax: 3, chimSegmentMin: 10, chimOutType: WithinBAM SoftClip, -chimJunctionOverhangMin: 10, chimScoreMin: 1, -chimScoreDropMax: 30, chimScoreJunctionNonGTAG: 0, chimScoreSeparation: 1, alignSJstitchMismatchNmax: 5 -1 5 5, chimSegmentReadGapMax: 3.
We then detected gene fusion events in the unmapped reads in the last step, using Arriba (https://github.com/suhrig/arriba/), a piece of software based on STAR. The maximum of the expected number of fusions was set as 0.5. The information of gene fusion events was recorded in a separate file for each dataset and an illustration for each fusion event was plotted using an R function embedded in Arriba.
Fusion genes functionality analysis
We used the transcript counts of each gene to study the gene expression variance among disease status, the associations between fused genes and other IPF feature genes, and the impacted signaling pathways associated with the gene fusion of IL7:AC083837.1.
Transcript quantification
The read count of RNA transcripts of each gene were generated using featureCounts.37 The BAM files created by STAR were mapped against the annotated genome of Homo_sapiens.GRCh38.97.gtf. The numbers of matched fragments for each gene were summarized into a count table.
Analysis of differential expression
Firstly, to reduce the computation, genes with extremely low expression (no reads in any of the 91 samples) were removed from the expression matrix. The quality of the expression matrix was checked, then the R package DESEq2 was used to detect expression differences between three conditions following the standard protocol.38 The batch biases among different projects were controlled in the design function (design = ~ project + status). Differentially expressed (DE) genes between healthy tissues and IPF tissues with or without IL7:AC083837.1 gene fusion were identified for further investigation. For each gene, the statistics of base mean, log2 foldchange, p value, and adjusted p value were calculated.
Generation of gene expression matrix with batch variation removed
Read counts are required for gene set enrichment analysis (GSEA; Broad Institute) and Pearson correlation calculation. To generate robust results, we removed the variations from different projects in the gene expression matrix. The expression matrix was modeled using DESEq2::deseq, and then variance-stabilizing transformed by DESEq2::vst; afterwards the “removeBatch Effect” function in R package “limma” was used to remove batch variations in the matrix.39 Principal components analysis (PCA) plots were drawn to check the corrective effects of the datasets.
Expression correlation analysis
Using the gene expression matrix resulting from step (3), Pearson correlation coefficients between the expression of IL7, AC083837.1 and other genes were calculated using functions in R base package.40
We searched three databases, CTDbase,41 the Harmonizome database,42 and NCBI,32 to collect known IFP-associated genes. A unique gene list was created by removing duplicate records to study the possible correlations between the fused genes and IPF feature genes.
We then conducted the correlation analysis in a much larger database GeneFriends (http://genefriends.org/RNAseq/)43 which is an online co-expression analysis tool based on 46,475 human samples. The Pearson correlation coefficients between IL7 and known IPF-associated genes were collected.
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis
DE genes in the comparisons of IPF tissue with and without IL7 = AC083837.1 fusion were collected and submitted to KOBAS (KEGG Orthology Based Annotation System) (http://kobas.cbi.pku.edu.cn/kobas3/) for the online pathway enrichment analysis.44,45 The databases used for this pathways analysis included KEGG pathway, KEGG disease, and gene ontology (GO). The retrieval task ID in KOBAS is ccaec36e8b504309949bb98a91fd6f56.
GO enrichment analysis using GSEA
GSEA (Broad Institute) computes a normalized enrichment score for each gene set (GS) from the read counts following another mathematical method different from DEseq2.46,47 It provides robust results by weighting each gene in the GS, adjusting for the variation in GS size, and controlling the negative rate. The read counts of DE genes were extracted from the corrected gene expression matrix and input into GSEA for GO enrichment analysis. The parameters were set according to GSEA official instructions: GS size:15–500, permutation type: gene set, permutation number: 1000, reference: msigdb.v7.0.symbols.gmt, metric: signal to noise, normalization model: “meandiv”. The results were then visualized using the software Cytoscape.48
IL7 gene co-expression network
We also generated a co-expression network for IL7 based on the expression matrix of DE genes generated in the previous step. The parameters were set according to GSEA official instructions: GS size:15–500, permutation type: gene set, permutation number: 1000, reference: msigdb.v7.0.symbols.gmt, metric: Pearson, normalization model: “meandiv”. The results were then visualized using the software Cytoscape.
Results
Fused genes in IPF lung tissues
In the 91 transcriptome datasets, we detect 1550 gene fusion events. Eight gene fusions have significantly different incidence rates between two conditions (Chi-square test, p < 0.05) (Table 1); among them, five gene fusions have significantly higher incidence rate in IPF tissues. Particularly, the gene fusion of IL7 = AC083837.1 and MFAP4 = EPN2 are detected exclusively in IPF samples. The complete list of all gene fusion events and their Chi-square p values are available upon request.
Table 1.
Gene fusion events with significant different incident rate.
| Gene fusion event | IPF | Control | Chi-square pa |
|---|---|---|---|
| HSD17B4 = S100A3 | 0 | 13 | 2.74×10−5 |
| WSB1 = S100A3 | 0 | 6 | 0.012 |
| IL7 = AC083837.1 | 8 | 0 | 0.028 |
| RPS10P7 = NAV1 | 3 | 9 | 0.036 |
| CCDC120 = PIM2 | 14 | 3 | 0.040 |
| LGALS1 = MICALL1 | 12 | 2 | 0.040 |
| CAP1 = RLF | 12 | 2 | 0.040 |
| MFAP4 = EPN2 | 7 | 0 | 0.047 |
Eight gene fusion events with the highest occurrence rates are listed. The IPF and Control columns indicate the number of samples in which gene fusion events occur in each group.
p values when comparing the incidence rate of the gene fusion event in both groups using Chi-square test. The null hypothesis is that the incidence in both groups is equal, the type 1 error is set as 0.05.
IPF, idiopathic pulmonary fibrosis.
Based on the study of these gene fusion events in their gene loci, occurrence differences between groups, and known functions in biological processes, we determined to focus our research on the IL7 = AC083837.1 fusion, which is most possibly involved in IPF pathology. Firstly, IL7 = AC083837.1 and MFAP4 = EPN2 are the only two events found exclusively in IPF samples. Secondly, although many of the other fused genes, such as MFAP4 = EPN2 and CCDC120 = PIM2, also locate in the same chromosomes, IL7 = AC083837.1 are the only pair of genes that have common promotor zones. Thirdly, compared with other genes, IL7, as an immune promoter, has a higher possibility to be involved in the imbalanced immune responses in IPF. Notably, the level of IL7 in serum has been reported to be associated with an increased survival rate in IPF patients.49
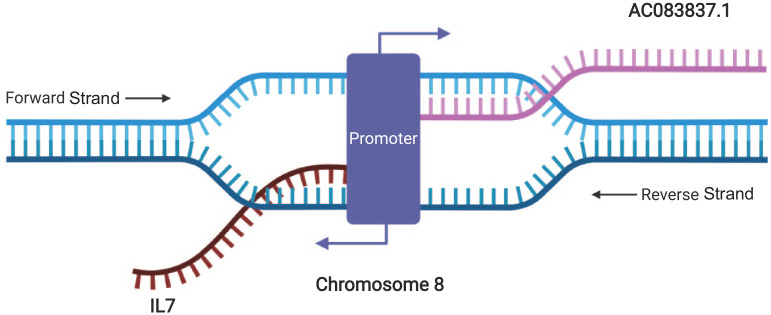
AC083837.1 and IL7 locate adjacently on chromosome 8 and have two shared promoters. IL7 locates between 78,675,743 and 78,805,523 in the reverse strand, while AC083837.1 locates between 78,805,293 and 78,956,082 of the forward strand.50 ENSR00001139954 (chr8:78,803,200–78,806,601) and ENSR00000226134 (chr8:78,807,000–78,807,401) are two shared promotors located in the overlapped zone of the two genes,51 indicating the possibility of the simultaneous initiation of both genes’ transcription and gene fusion under certain circumstances (Figure 1 and Supplemental file S1).
Figure 1.
Illustration of the locations of IL7 and AC083837.1 in chromosome 8 and their simultaneous transcription. Graphic created with Biorender.com.
IL7, interleukin-7.
IL7 = AC083837.1 is detected in eight IPF samples, and the illustrations of fused transcripts show that the translation of IL7 is arrested by this gene fusion. Three IL7 transcripts, IL7-201, IL7-204, and IL7-209 are involved in the fusion with AC083837.1. Not all these fused transcripts encode proteins due to antisense transcription (3'–3'). The plots of the fused transcripts in all tissues are provided in Supplemental file S2.
Differential expression analysis
The gene expression matrix is divided into three groups based on disease status and the occurrence of IL7 gene fusion: control, IPF with IL7 fusion, and IPF without IL7 fusion. Among these three groups, we studied the expression variance related to IL7 = AC083837.1 and identified significant DE genes.
Expression variance of IL7 and AC083837.1
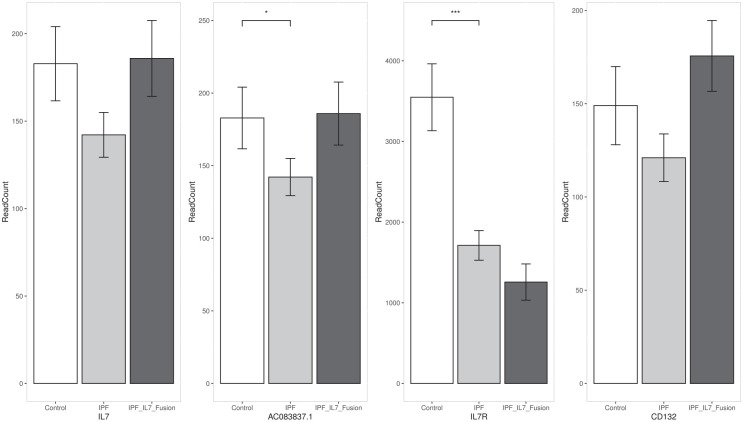
The expression of both fused genes, IL7 and AC083837.1, decreases in the IPF group, and the expression of AC083837.1 is lowered significantly (adjusted p < 0.05). In the samples with IL7 = AC083837.1 fusion, the expression of both genes has a non-significant increase (adjusted p ⩾ 0.05). We also study the expression variance of two component-genes of the IL7 receptor (IL7R, and CD132) to probe the possible impacts of the decreased IL7 translation. The expression of IL7R reduces significantly in IPF tissues and further decreases non-significantly in IPF tissues with IL7 gene fusion. The variance of CD132(IL-2RG) is similar to that of IL7: it decreases in IPF tissues but increases slightly in IPF tissues. The variance of CD132 expression does not have significant differences among these three groups. Figure 2 shows the bar plots of the read counts of these four genes [mean ± standard error of the mean (SEM)].
Figure 2.
Gene expression of the four genes directly associated with IL7 = AC083837.1 gene fusion. Bar values represent the mean ± SEM of the read counts of each gene, the p values were calculated by DESeq2.
IL7, interleukin-7; IPF, idiopathic pulmonary fibrosis; SEM, standard error of the mean.
Identification of DE genes in samples associated with gene fusion
We conduct pairwise comparisons of gene expressions among above-mentioned three groups to identify DE genes associated with IPF and IL7 = AC083837.1 gene fusion. When compared with healthy tissues, IPF tissues have 8924 DE genes (adjusted p < 0.01). When compared with IPF tissues without IL7 gene fusion, IPF tissues with IL7 gene fusion have 282 DE genes (p < 0.01). There are no known IPF feature genes included in these 282 DE genes. In this comparison, there are no genes that have a differential expression with adjusted p < 0.01, this might be because the difference between the two types of tissues is relatively small in the background of a multiple hypothesis testing for more than 58 000 genes. The DE analysis results are available upon request.
Pearson correlation analysis
Association of expression between IL7 and AC083837.1
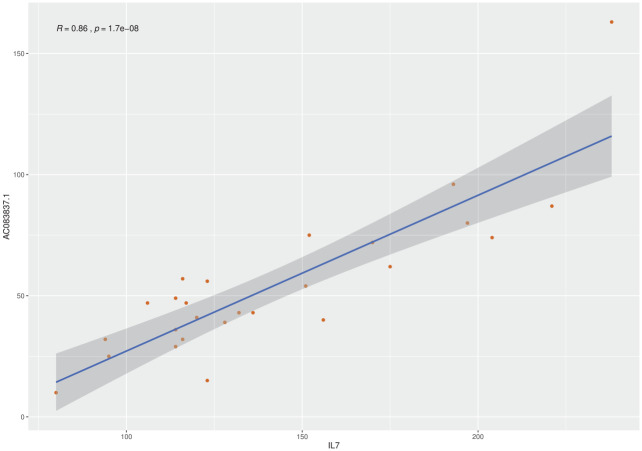
In the 91 IPF samples, the expression of AC083837.1 shows a moderate positive correlation with that of IL7, the Pearson correlation coefficient is 0.56 [p value = 5.9 × 10–9, confidence interval (CI): 0.40–0.69]. To diminish the impact of cross-experiment variance, we also study the Pearson correlation of these two genes in the 24 lung tissues of the project PRJNA388978, six IPF tissues that had IL7 gene fusion. In this project, the expressions of these two genes are strongly associated, with a coefficient of 0.86 (p value = 1.69 × 10–8, CI: 0.71–0.93) (Figure 3).
Figure 3.
Pearson correlation analysis of the expression of IL7 and AC083837.1.
IL7, interleukin-7.
Association between fused genes and IPF feature genes
We collect 94 IPF-associated genes in the databases of CTDbase,41 the Harmonizome,42 and NCBI.32 After the removal of duplicates, we obtained a list of 54 genes that have been experimentally verified to be associated with IPF. The Pearson correlation analysis based on the 91 samples shows that expression of two fused genes has only weak associations with these featured genes (r < 0.5).
The correlation analysis performed using the online tool on www.GeneFriends.org confirms the results about IL7: its expression has a weak association with all known IPF featured genes. As the GeneFriends.org database does not have RNA-seq data for AC083837.1, we are not able to obtain correlation coefficients for this gene. The list of IPF featured genes and all the expression correlation analysis results in this section are provided in Supplemental file S3.
KEGG pathway enrichment analysis by KOBAS
The 282 DE genes in IPF tissues with IL7- AC083837.1 gene fusion are enriched in six KEGG pathways and 35 functional GS (p < 0.01). There are no signaling pathways or GS that have an adjusted p value < 0.05, possibly because of the small number of submitted differentially expressed genes. In the enriched pathways and GS, we identify three pathways that are involved in the regulation of IPF: NK cell-mediated cytotoxicity (ranking #1, p = 0.0004), peroxisome (ranking #3, p = 0.004), and apoptosis (ranking #9, p = 0.019). There are eight genes enriched in NK cell-mediated cytotoxicity and four genes in both the peroxisome and apoptosis pathways.
Besides, these genes are also significantly enriched in the GS related to human immunodeficiency virus 1 (HIV-1) infection, non-small cell lung cancer (p = 0.0027), and cancers of the lung and pleura (p = 0.01), indicating that gene fused IPF tissues might demonstrate more characteristics of an immunological disease. No other GS in our investigation are related to immunology processes or other processes associated with IPF pathogenesis. The complete result of the KOBAS analysis is available upon request.
GSEA GO enrichment analysis
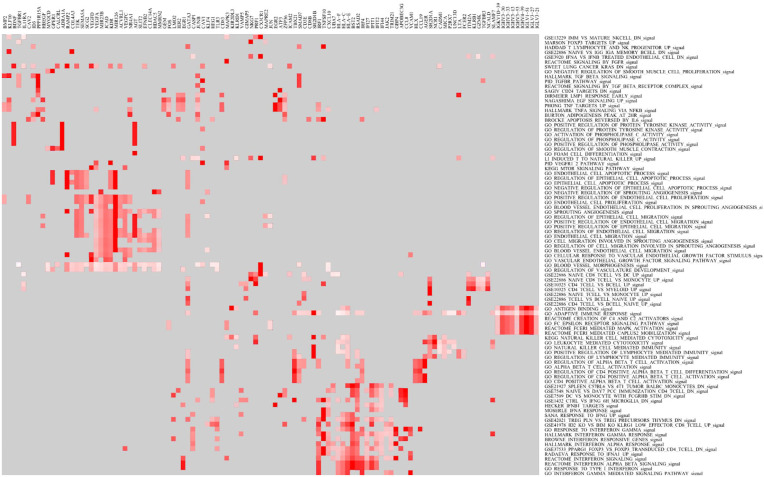
In the GSEA, 273 pathways and GS are enriched in the IPF tissues with the IL7 = AC083837.1 gene fusion (p < 0.01, FDR q < 0.25), while 187 GS are enriched significantly in IPF tissues without the gene fusion (p < 0.01, FDR q < 0.25). The enriched pathways are associated with various physiological and pathological processes, such as immunology, metabolism, and infection (Supplemental file S5). Heatmap shows the clustered genes in the leading-edge gene sets, such as IRF1, PRF1, IRF4, CXCL10, and GBP4 (Figure 4). A set-to-set leading-edge plot shows the overlap of the leading-edge gene sets, including the interferon signaling pathway, epithelial apoptotic process, and epithelial migration (Figure 5). Cytoscape visualization shows that the pathways are enriched in IPF tissues with and without IL7 = AC083837.1 gene fusion (Figure 6). The 187 enriched pathways in regular IPF tissues are clustered into four functional groups: immune responses and tumorigenesis, extracellular matrix activities, reproduction, and skin growth. However, the pathways enriched in IPF tissues with IL7 = AC083837.1 gene fusion do not have a significant clustering effect. Among them there are pathways associated with exacerbation of IPF progress, including immune cell development, apoptotic processes of endothelial and epithelial cells, angiogenesis and endothelial proliferation, and interferon-mediated responses. The full GO enrichment analysis reports are available upon request.
Figure 4.
Heatmap for the GO enrichment analysis. The heat map shows the clustered leading-edge genes in the gene sets. The x-axis represents the genes and y-axis represents gene sets. Red means positive correlation with IL7 = AC083837.1 gene fusion.
GO, gene ontology; IL7, interleukin-7.
Figure 5.
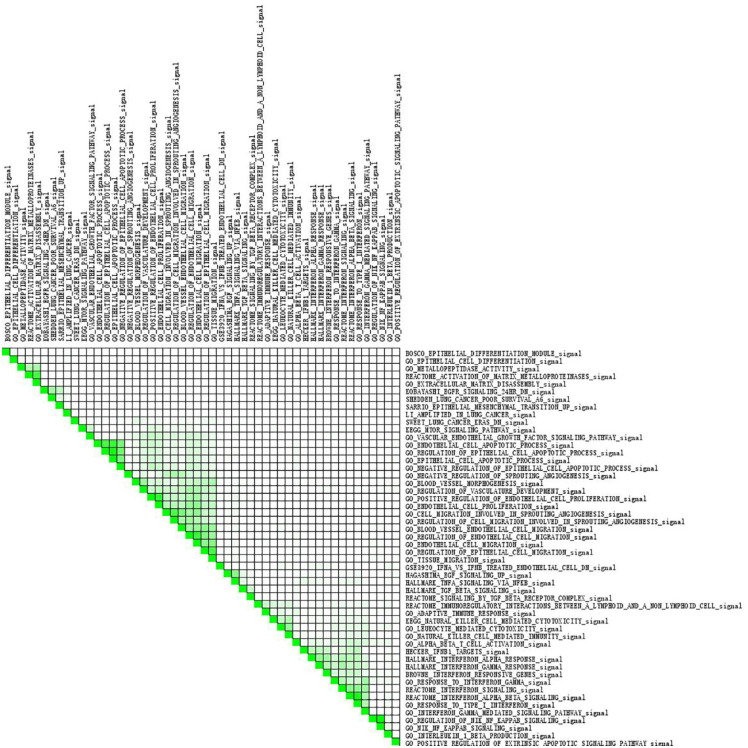
Subset analysis of all enriched gene sets. The range of color stands for the overlap of the two gene sets.
Figure 6.
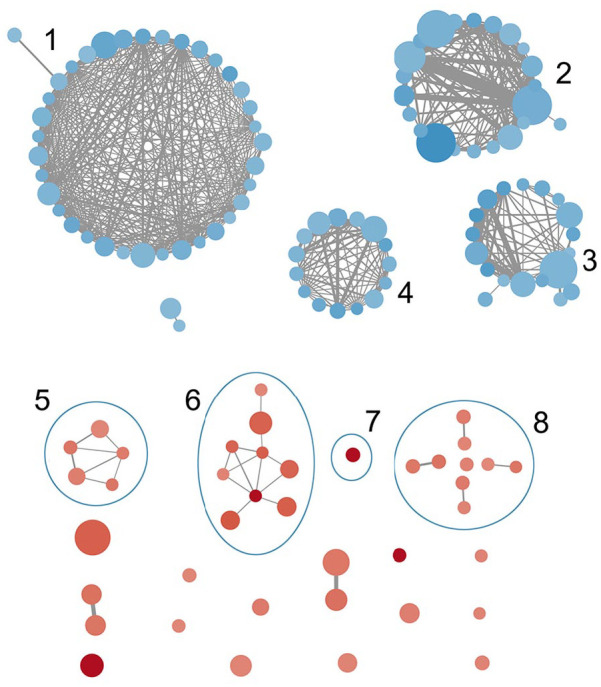
GO enrichment map associated with IL7 = AC083837.1 gene fusion. Node size stands for the GS size, edge width stands for the overlap size of the GS it connects, the color depth stands for the enrichment score (blue = negative, red = positive). Clusters: (1) GS associated with immune responses and tumor growth and regulation, (2) GS associated with reproduction, (3) GS associated with skin growth, (4) GS associated with extracellular matrix, (5) GS associated with IFN signaling pathways, (6) GS associated with immune cell growth, (7) GS of NK cell-mediated cytotoxicity, (8) GS associated with apoptosis and angiogenesis of endothelial and epithelial cells.
GO, gene ontology; GS, gene set; IFN, interferon; IL7, interleukin-7; NK, natural killer.
Co-expression network associated with IL7
The co-expression network of IL7 based on these 91 transcriptomes highlights that the expression of IL7 is associated strongly with the enrichment of 180 pathways and GS and the repression of 1545 pathways and GS (Figure 7, p < 0.01, FDR q < 0.25). The expression of IL7 is correlated positively with a cluster of reproduction-related pathways and inversely with a bunch of pathways involved in the processes of angiogenesis, metabolism, and extracellular matrix reproduction. The endothelial proliferation and extracellular matrix reconstruction now are generally considered as critical mechanisms that drive the fibrosis in IPF. This result validates the observed enrichment of these pathways in IPF tissue with IL7 = AC083837.1 gene fusion, which impairs IL7 protein coding. The full GO enrichment analysis reports are available upon request.
Figure 7.
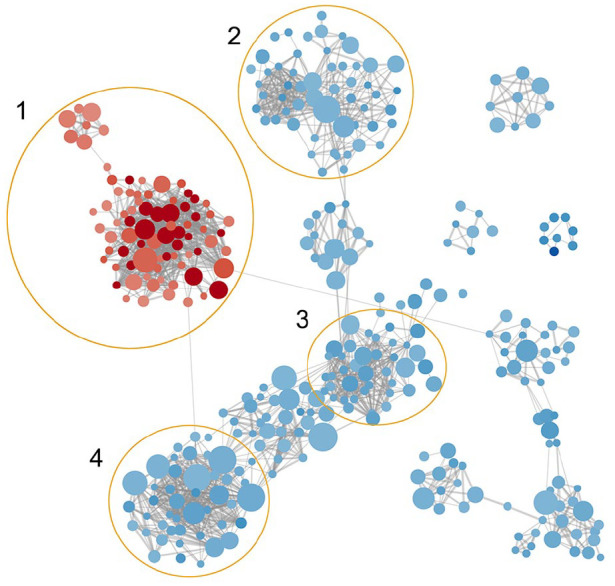
Co-expression network associated with expression of IL7. Node size stands for the GS size, edge width stands for the similarity coefficient of the GS it connects, the color depth stands for the enrichment score (blue = negative, red = positive). Clusters: (1) GS associated with reproduction, (2) GS associated with metabolism, (3) GS associated with extracellular matrix, (4) GS associated with vessel angiogenesis.
GS, gene set; IL7, interleukin-7.
To probe pathways that might be impacted by the gene fusion of IL7 = AC083837.1, we compare the results of KOBAS, GSEA GO enrichment analysis, and the co-expression network of IL7. As a result, we discover that the pathways of angiogenesis, apoptotic processes, and NK cell-mediated cytotoxicity most probably play significant roles in IPF pathogenesis. The NK cell-mediated cytotoxicity is the most significantly enriched signaling pathway expressed in IPF tissues with IL7 = AC083837.1 gene fusion. It is ranked first in the result of the KEGG analysis and is ranked third in the GSEA enrichment analysis. The activities of the pathways related to NK cell’s principal secretion, IFN, are also significantly enriched. Apoptotic process-related pathways are another cluster of recurring results from KEGG and GSEA analysis. The apoptosis signaling pathway is ranked ninth in the KEGG results. At the same time, there are five apoptotic process-related GS among the GSEA analysis results, such as the regulation of epithelial cell apoptotic process (36th), the endothelial cell apoptotic process (52nd), and the epithelial cell apoptotic process (83rd). Several GS associated with cell migration and sprouting angiogenesis appear in the GSEA enrichment analysis, including the cell migration involved in sprouting angiogenesis (32nd), the regulation of cell migration involved in sprouting angiogenesis (47th), and sprouting angiogenesis (92nd). Besides, two signaling pathways that regulate angiogenesis, transforming growth factor beta (TGF-β) and vascular endothelial growth factor (VEGF) pathways, are also significantly enriched in IPF tissues with gene fusion, ranked 55th and 109th, respectively. Additionally, the co-expression network shows that the expression of angiogenesis GS enhances with the decreased expression of IL7.
Discussion
Our knowledge of the immunological changes in IPF is still limited, and research on therapeutic targets has made little progress.1,2 Fusion genes and lncRNAs regulate biological and pathological processes, and novel therapeutic drugs have been developed based on the investigation of their functions. Thus, this study aimed to detect gene fusion events that occur in human lung IPF tissues and investigate their impact on disease progression.
To guarantee the validity and reliability of our research, we studied 91 high-quality transcriptomes collected from four IPF projects, with the cross-batch bias being corrected. The small sample size caused by rare incidence and relatively high-level variances of sequencing depth multiply the difficulties in the analysis. On the one hand, the small sample size of a single project leads to irrelevant results, which leads to a high false-positive rate. On the other hand, the variances in transcriptomes from different experiments cause nonsignificant results when investigating the combined matrix directly. Therefore, we controlled the cross-experiment variance in gene expression matrix before DE and GO enrichment analysis, which, as a result, yielded a moderate number of functional gene clusters that appear repeatedly in different analysis methods. Despite these efforts to increase sample size and reduce the batch effects, the authors still have to acknowledge that the results from our method could not be as robust as those from a well-designed large-scale transcriptome RNA-seq study.
We found 1550 gene fusion events that occur in 91 transcriptomes and, based on the research of the occurrence rate, gene loci, and known biological functions of fused genes, we determined to investigate the possible impacts of IL7 = AC083837.1 in IPF.
The expression of both genes decreases in normal IPF tissues and increases when their fusion occurred. These results are consistent with previous genome-wide expression analysis of IPF, where IL7 does not show significant expression differences.26 The expression of IL7R, although also decreased in normal IPF tissues, is further decreased in tissues with IL7 fusion. This might explain why the requirement of IL7R production is decreased by the depletion of encoded IL7 protein caused by this gene fusion. The expression of CD132 increases in tissues with gene fusion, which might be because it also encodes a common gamma chain for the receptors of IL2, IL4, IL7, IL9, IL15, and IL21.
Pearson correlation analysis based on the 91 transcriptome datasets and online transcriptome database suggests weak associations between the expression of IL7, AC083837.1 and all known featured genes of IPF. These correlation results, as well as non-significant expression variations, are consistent with previous genome-wide studies on IPF, indicating that this IL7 = AC083837.1 fusion might impact IPF through indirect pathways instead of direct regulation.
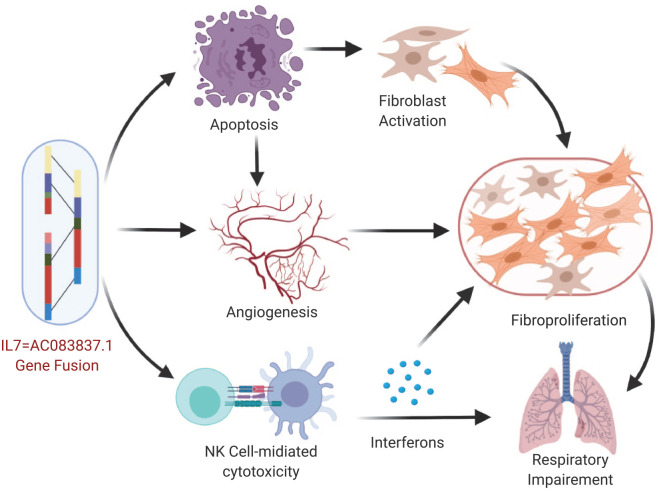
To explore the impacts of IL7 = AC083837.1 gene fusion on the pathogenesis of IPF, we study the significantly impacted signaling pathways identified in GSEA and KOBAS online analyses. We conclude that IL7 = AC083837.1 gene fusion might exacerbate IPF progress by enhancing the activities of angiogenesis, apoptotic processes, and NK cell-mediated cytotoxicity (Figure 8). The detailed mechanisms are described below.
Figure 8.
The impacting pathways of IL7 = AC083837.1 gene fusion on the progression of IPF. The gene fusion enhances the expression of GS associate with apoptosis, angiogenesis, and NK cell-mediated cytotoxicity. These strengthened signaling pathways then exacerbate IPF symptoms through initiating and promoting the fibrosis process. The graphic was created with Biorender.com.
GS, gene set; IL7, interleukin-7; IPF, idiopathic pulmonary fibrosis; NK, natural killer.
NK cell induces the programmed death of injured cells by releasing cytotoxic substances.52,53 This cell-mediated cytotoxicity has demonstrated effects on the deterioration of IPF in recent studies. A study in 2004 shows that, in a mouse IPF model, decreased NK cell recruitment leads to reduced production of IFN-γ, which further enhances pulmonary fibrosis.54 Although pre-clinical trials and small-scale clinical trials report therapeutic effects of exogenous IFN-γ in treating IPF,55–60double-blinded, multiple-centered clinical trials show that this IFN-γ treatment does not benefit IPF patients.61,62 Recent studies, on the contrary, demonstrate a possible deteriorating effect on IPF of NK cell-mediated cytotoxicity and its principle product, IFN. A 2017 study shows that NK cell-mediated cytotoxicity is particularly up-regulated in the IPF mouse model.63 A 2019 clinical trial shows that the percentage of NK cells in IPF patients’ bronchoalveolar lavage is associated inversely with their forced vital capacity and diffused lung carbon monoxide,8 and increased serum IFN-γ level is found to be associated with acute exacerbation of IPF patients.64 Similarly, monocytes primed by type-I IFNs (IFN-α, IFN-β) are considered to be a driving factor of aberrant injury repair and fibrosis. In our study, NK cell-mediated cytotoxicity, the signaling pathways and responses of IFN-α, -β, and -γ were significantly enriched in IPF tissues with IL7 = AC083837.1 gene fusion, indicating that they might be the pivotal pathways through which this gene fusion accelerates the exacerbation of IPF.
The impact of IL7 on NK cell-mediated cytotoxicity has not been well established. Although IL7 might enhance NK cell survival by inhibiting their apoptotic process,65,66 knockout experiments have shown that IL7 and IL7R were not required in the development and maturation of NK cells,67–70 neither do they impact NK cytotoxicity and their production of IFNs.66 Our study shows that the IL7 = AC083837.1 gene fusion is associated with significant enrichment of NK cell-mediated cytotoxicity, which, according to the latest studies, could further activate the fibrosis process and exacerbate patients’ respiratory impairment. We have few clues as to the mechanism of this impact of IL7; thus, more studies in this area are required.
Up-regulated epithelial cell apoptosis is a critical etiological change in IPF that is found commonly in bronchial and alveolar epithelial cells of IPF patients. 71–74 Epithelial apoptosis is involved in the initiation of pulmonary fibrosis, and enlarges the lesion area by inducing apoptosis of the adjacent epithelial cells.72 The loss of epithelial cells leads to impaired protection against the invasion of fibrosis in alveolar surfactant, and treatment blocking epithelial cell apoptosis could inhibit the development of fibrosis.75–78 The endothelial cell apoptotic process is also involved in the exacerbation of pulmonary fibrosis; it worsens the injury of epithelial cells, reduces wound closure,79 and enhances fibrotic responses in adjacent epithelial cells.12,80–82 Besides, endothelial apoptosis might be associated with the development of emphysema in lung tissues.83 Our analysis results indicate that the IL7 = AC083837.1 gene fusion is associated with the up-regulated apoptotic activities of epithelial cells and endothelial cells, leading to the initiation and proliferation of fibrosis in lung tissues. This gene fusion might activate these apoptotic processes via NK cell-mediated cytotoxicity,9,10 as described above, or some other signaling pathways that require further investigation.
Angiogenesis is a typical pathological change that exists universally in IPF lung tissues84; excessive angiogenesis enhances fibroproliferation and regulates fibrosis together with apoptotic processes.12,85,86 Studies show that productions of apoptotic endothelial cells initiate the angiogenesis process, which then activates and enhances the fibrosis process in lung tissues.13,14 Treatments inhibiting angiogenesis have shown to have therapeutic effects of fibrosis on the PF model in mice.87 Based on the results of our analysis, we conclude that the gene fusion of IL7 is associated with regulation of the initiation and progression of fibrosis via enhancing the activities of angiogenetic and apoptotic processes of epithelial and endothelial cells in IPF.
There are two possible explanations for the mechanism whereby IL7 gene fusion impacts these signaling pathways. One is that the IL7-mediated protective immune response is impaired by the reduction of IL7 protein production. IL7 has been revealed to play a protective role in IPF in clinical and in vitro studies. The concentration of IL7 in peripheral blood is associated with the survival chance of IPF patients.49 IL7 inhibits the promotive effect of TGF-β signaling pathways on angiogenesis and fibrosis,88 and relieves fibrosis in kidney cells.89 Another explanation is related to the regulatory effect of the lncRNA AC083837.1. Recent studies identify 14 lncRNAs regulating the inflammatory response in IPF,27,90 among which one antisense lncRNA of IL7 promotes the expression of several inflammatory genes.91 In our case, the increased expression of the lncRNA AC083837.1 might also regulate the progress of IPF via unrecognized pathways, the validation of which requires more studies. As the pathways and responders that are impacted directly by the gene fusion are unclear, we suggest that more evidence should be collected from bioinformatics research before a wet-lab validation experiment can be designed and conducted.
In summary, our study reveals the possible regulation effect of IL7 = AC083837.1 gene fusion, which occurs exclusively in IPF lung tissues with a significant incidence rate (8/52, 16%). The lncRNA AC083837.1 locates adjacent to IL7 in chromosome 8 and is possibly transcribed simultaneously with IL7 and fuses with its transcripts. This gene fusion disables the translation of IL7 transcripts and is associated with the exacerbation of IPF. Although the expression of fused genes neither leads to a significant change in IPF, nor is associated directly with known IPF feature genes, our study suggests that the gene fusion possibly exacerbates IPF symptoms, especially lung tissue fibrosis, by promoting the signaling pathways of NK cell-mediated cytotoxicity, angiogenesis, and the apoptotic process in IPF tissues. Despite the disadvantages we acknowledge in the Discussion, our study is a valuable attempt towards understanding the roles gene fusion plays in the pathogenesis of IPF. The results of this study will guide future research in the field of IPF mechanisms and therapeutic targets.
Supplemental Material
Supplemental material, sj-pdf-1-tar-10.1177_1753466621995045 for Gene fusion of IL7 involved in the regulation of idiopathic pulmonary fibrosis by Shixue Sun, Chen Huang, Dongliang Leng, Chang Chen, Teng Zhang, Kuan Cheok Lei and Xiaohua Douglas Zhang in Therapeutic Advances in Respiratory Disease
Supplemental material, sj-pdf-2-tar-10.1177_1753466621995045 for Gene fusion of IL7 involved in the regulation of idiopathic pulmonary fibrosis by Shixue Sun, Chen Huang, Dongliang Leng, Chang Chen, Teng Zhang, Kuan Cheok Lei and Xiaohua Douglas Zhang in Therapeutic Advances in Respiratory Disease
Supplemental material, sj-pdf-3-tar-10.1177_1753466621995045 for Gene fusion of IL7 involved in the regulation of idiopathic pulmonary fibrosis by Shixue Sun, Chen Huang, Dongliang Leng, Chang Chen, Teng Zhang, Kuan Cheok Lei and Xiaohua Douglas Zhang in Therapeutic Advances in Respiratory Disease
Supplemental material, sj-pdf-5-tar-10.1177_1753466621995045 for Gene fusion of IL7 involved in the regulation of idiopathic pulmonary fibrosis by Shixue Sun, Chen Huang, Dongliang Leng, Chang Chen, Teng Zhang, Kuan Cheok Lei and Xiaohua Douglas Zhang in Therapeutic Advances in Respiratory Disease
Supplemental material, sj-pdf-6-tar-10.1177_1753466621995045 for Gene fusion of IL7 involved in the regulation of idiopathic pulmonary fibrosis by Shixue Sun, Chen Huang, Dongliang Leng, Chang Chen, Teng Zhang, Kuan Cheok Lei and Xiaohua Douglas Zhang in Therapeutic Advances in Respiratory Disease
Supplemental material, sj-pdf-7-tar-10.1177_1753466621995045 for Gene fusion of IL7 involved in the regulation of idiopathic pulmonary fibrosis by Shixue Sun, Chen Huang, Dongliang Leng, Chang Chen, Teng Zhang, Kuan Cheok Lei and Xiaohua Douglas Zhang in Therapeutic Advances in Respiratory Disease
Supplemental material, sj-pdf-8-tar-10.1177_1753466621995045 for Gene fusion of IL7 involved in the regulation of idiopathic pulmonary fibrosis by Shixue Sun, Chen Huang, Dongliang Leng, Chang Chen, Teng Zhang, Kuan Cheok Lei and Xiaohua Douglas Zhang in Therapeutic Advances in Respiratory Disease
Supplemental material, sj-pdf-9-tar-10.1177_1753466621995045 for Gene fusion of IL7 involved in the regulation of idiopathic pulmonary fibrosis by Shixue Sun, Chen Huang, Dongliang Leng, Chang Chen, Teng Zhang, Kuan Cheok Lei and Xiaohua Douglas Zhang in Therapeutic Advances in Respiratory Disease
Supplemental material, sj-tif-4-tar-10.1177_1753466621995045 for Gene fusion of IL7 involved in the regulation of idiopathic pulmonary fibrosis by Shixue Sun, Chen Huang, Dongliang Leng, Chang Chen, Teng Zhang, Kuan Cheok Lei and Xiaohua Douglas Zhang in Therapeutic Advances in Respiratory Disease
Acknowledgments
We are grateful to NCBI for the provision of GSE52463, GSE83717, GSE92592, and GSE99621 Genome Expression Omnibus datasets.
Footnotes
Author contributions: All authors participated in the study design, the interpretation of the results, and the drafting and revision of the manuscript. CH conceived the idea, SS downloaded and analyzed the data and drafted the manuscript, XDZ supervised the research. All authors reviewed and commented on the manuscript and approved the final draft.
Conflict of interest statement: The authors declare that there is no conflict of interest.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was funded by the Science and Technology Development Fund, Macau SAR (File no. 0004/2019/AFJ and 0011/2019/AKP) and by the University of Macau (grant numbers: FHS-CRDA-029-002-2017, EF005/FHS-ZXH/2018/GSTIC and MYRG2018-00071-FHS).
ORCID iD: Xiaohua Douglas Zhang  https://orcid.org/0000-0002-2486-7931
https://orcid.org/0000-0002-2486-7931
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Shixue Sun, CRDA, Faculty of Health Sciences, University of Macau, Taipa, Macau SAR, China.
Chen Huang, CRDA, Faculty of Health Sciences, University of Macau, Taipa, Macau SAR, China.
Dongliang Leng, CRDA, Faculty of Health Sciences, University of Macau, Taipa, Macau SAR, China.
Chang Chen, CRDA, Faculty of Health Sciences, University of Macau, Taipa, Macau SAR, China.
Teng Zhang, CRDA, Faculty of Health Sciences, University of Macau, Taipa, Macau SAR, China.
Kuan Cheok Lei, CRDA, Faculty of Health Sciences, University of Macau, Taipa, Macau SAR, China.
Xiaohua Douglas Zhang, CRDA, Faculty of Health Sciences, University of Macau, Avenida de Universidade, Taipa, Macau SAR, China.
References
- 1. Bullers K. Merck manuals. J Med Libr Assoc 2016; 104: 369–371. [Google Scholar]
- 2. Xaubet A, Ancochea J, Molina-Molina M. Idiopathic pulmonary fibrosis. Med Clin (Barc) 2017; 148: 170–175. [DOI] [PubMed] [Google Scholar]
- 3. Wilson MS, Wynn TA. Pulmonary fibrosis: pathogenesis, etiology and regulation. Mucosal Immunol 2009; 2: 103–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Agostini C, Semenzato G. Immunology of idiopathic pulmonary fibrosis. Curr Opin Pulm Med 1996; 2: 364–369. [DOI] [PubMed] [Google Scholar]
- 5. Heukels P, Moor CC, von der Thusen JH, et al. Inflammation and immunity in IPF pathogenesis and treatment. Respir Med 2019; 147: 79–91. [DOI] [PubMed] [Google Scholar]
- 6. Butler MW, Keane MP. The Role of Immunity and Inflammation in IPF Pathogenesis. In: Meyer KC, Nathan SD. (eds) Idiopathic pulmonary fibrosis. Cham: Springer International Publishing, 2019, pp.97–131. [Google Scholar]
- 7. Galati D, De Martino M, Trotta A, et al. Peripheral depletion of NK cells and imbalance of the Treg/Th17 axis in idiopathic pulmonary fibrosis patients. Cytokine 2014; 66: 119–126. [DOI] [PubMed] [Google Scholar]
- 8. Bergantini L, Cameli P, d’Alessandro M, et al. NK and NKT-like cells in granulomatous and fibrotic lung diseases. Clin Exp Med 2019; 19: 487–494. [DOI] [PubMed] [Google Scholar]
- 9. Abel AM, Yang C, Thakar MS, et al. Natural killer cells: development, maturation, and clinical utilization. Front Immunol 2018; 9: 1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Khosravi-Far R, Esposti MD. Death receptor signals to mitochondria. Cancer Biol Ther 2004; 3: 1051–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pardo A, Selman M. Fibroblast senescence and apoptosis. “One-two punch” to slow down lung fibrosis? Am J Respir Cell Mol Biol 2018; 56: 145–146. [DOI] [PubMed] [Google Scholar]
- 12. Johnson A, DiPietro LA. Apoptosis and angiogenesis: an evolving mechanism for fibrosis. Faseb J 2013; 27: 3893–3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dimmeler S, Zeiher AM. Endothelial cell apoptosis in angiogenesis and vessel regression. Circ Res 2000; 87: 434–439. [DOI] [PubMed] [Google Scholar]
- 14. Cheresh DA, Stupack DG. Regulation of angiogenesis: apoptotic cues from the ECM. Oncogene 2008; 27: 6285–6298. [DOI] [PubMed] [Google Scholar]
- 15. Marshall RP, Puddicombe A, Cookson WO, et al. Adult familial cryptogenic fibrosing alveolitis in the United Kingdom. Thorax 2000; 55: 143–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Garcia-Sancho C, Buendia-Roldan I, Fernandez-Plata MR, et al. Familial pulmonary fibrosis is the strongest risk factor for idiopathic pulmonary fibrosis. Respir Med 2011; 105: 1902–1907. [DOI] [PubMed] [Google Scholar]
- 17. Wang Y, Kuan PJ, Xing C, et al. Genetic defects in surfactant protein A2 are associated with pulmonary fibrosis and lung cancer. Am J Hum Genet 2009; 84: 52–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang XM, Zhang Y, Kim HP, et al. Caveolin-1: a critical regulator of lung fibrosis in idiopathic pulmonary fibrosis. J Exp Med 2006; 203: 2895–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang Y, Noth I, Garcia JG, et al. A variant in the promoter of MUC5B and idiopathic pulmonary fibrosis. N Engl J Med 2011; 364: 1576–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Seibold MA, Wise AL, Speer MC, et al. A common MUC5B promoter polymorphism and pulmonary fibrosis. N Engl J Med 2011; 364: 1503–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Falfan-Valencia R, Camarena A, Juarez A, et al. Major histocompatibility complex and alveolar epithelial apoptosis in idiopathic pulmonary fibrosis. Hum Genet 2005; 118: 235–244. [DOI] [PubMed] [Google Scholar]
- 22. Aquino-Galvez A, Perez-Rodriguez M, Camarena A, et al. MICA polymorphisms and decreased expression of the MICA receptor NKG2D contribute to idiopathic pulmonary fibrosis susceptibility. Hum Genet 2009; 125: 639–648. [DOI] [PubMed] [Google Scholar]
- 23. Selman M, Lin HM, Montano M, et al. Surfactant protein A and B genetic variants predispose to idiopathic pulmonary fibrosis. Hum Genet 2003; 113: 542–550. [DOI] [PubMed] [Google Scholar]
- 24. Hodgson U, Pulkkinen V, Dixon M, et al. ELMOD2 is a candidate gene for familial idiopathic pulmonary fibrosis. Am J Hum Genet 2006; 79: 149–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kropski JA, Blackwell TS, Loyd JE. The genetic basis of idiopathic pulmonary fibrosis. Eur Respir J 2015; 45: 1717–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kaur A, Mathai SK, Schwartz DA. Genetics in idiopathic pulmonary fibrosis pathogenesis, prognosis, and treatment. Front Med (Lausanne) 2017; 4: 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hadjicharalambous MR, Roux BT, Feghali-Bostwick CA, et al. Long non-coding RNAs are central regulators of the IL-1β-induced inflammatory response in normal and idiopathic pulmonary lung fibroblasts. Front Immunol 2018; 9: 2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hadjicharalambous MR, Roux BT, Csomor E, et al. Long intergenic non-coding RNAs regulate human lung fibroblast function: implications for idiopathic pulmonary fibrosis. Sci Rep 2019; 9: 6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Parker BC, Zhang W. Fusion genes in solid tumors: an emerging target for cancer diagnosis and treatment. Chin J Cancer 2013; 32: 594–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007; 448: 561–566. [DOI] [PubMed] [Google Scholar]
- 31. Mascaux C, Tomasini P, Greillier L, et al. Personalised medicine for nonsmall cell lung cancer. Eur Respir Rev 2017; 26: 170066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res 2013; 41: D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nance T, Smith KS, Anaya V, et al. Transcriptome analysis reveals differential splicing events in IPF lung tissue. PLoS One 2014; 9: e92111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vukmirovic M, Herazo-Maya JD, Blackmon J, et al. Identification and validation of differentially expressed transcripts by RNA-sequencing of formalin-fixed, paraffin-embedded (FFPE) lung tissue from patients with idiopathic pulmonary fibrosis. BMC Pulm Med 2017; 17: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schafer MJ, White TA, Iijima K, et al. Cellular senescence mediates fibrotic pulmonary disease. Nat Commun 2017; 8: 14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Luzina IG, Salcedo MV, Rojas-Pena ML, et al. Transcriptomic evidence of immune activation in macroscopically normal-appearing and scarred lung tissues in idiopathic pulmonary fibrosis. Cell Immunol 2018; 325: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014; 30: 923–930. [DOI] [PubMed] [Google Scholar]
- 38. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015; 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Team RC. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing, 2017. [Google Scholar]
- 41. Davis AP, Grondin CJ, Johnson RJ, et al. The comparative toxicogenomics database: update 2019. Nucleic Acids Res 2019; 47: D948–D954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rouillard AD, Gundersen GW, Fernandez NF, et al. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database (Oxford) 2016; 2016: baw100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. van Dam S, Cordeiro R, Craig T, et al. GeneFriends: an online co-expression analysis tool to identify novel gene targets for aging and complex diseases. BMC Genomics 2012; 13: 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mao X, Cai T, Olyarchuk JG, et al. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005; 21: 3787–3793. [DOI] [PubMed] [Google Scholar]
- 45. Xie C, Mao X, Huang J, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 2011; 39: W316–W322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003; 34: 267–273. [DOI] [PubMed] [Google Scholar]
- 47. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005; 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003; 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tachibana K, Inoue Y, Nishiyama A, et al. Polymyxin-B hemoperfusion for acute exacerbation of idiopathic pulmonary fibrosis: serum Il-7 as a prognostic marker. Sarcoidosis Vasc Diffuse Lung Dis 2011; 28: 113–122. [PubMed] [Google Scholar]
- 50. Yates AD, Achuthan P, Akanni W, et al. Ensembl 2020. Nucleic Acids Res 2019; 48: D682–D688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Stelzer G, Rosen N, Plaschkes I, et al. The genecards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics 2016; 54: 1.30.1–1.30.33. [DOI] [PubMed] [Google Scholar]
- 52. Topham NJ, Hewitt EW. Natural killer cell cytotoxicity: how do they pull the trigger? Immunology 2009; 128: 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Packard BZ, Komoriya A, Ucker DS. NK cell-mediated target cell death. In: Lotze MT, Thomson AW. (eds) Natural killer cells. San Diego: Academic Press, 2010, pp.645–652. [Google Scholar]
- 54. Jiang D, Liang J, Hodge J, et al. Regulation of pulmonary fibrosis by chemokine receptor CXCR3. J Clin Invest 2004; 114: 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Datta A, Scotton CJ, Chambers RC. Novel therapeutic approaches for pulmonary fibrosis. Br J Pharmacol 2011; 163: 141–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gurujeyalakshmi G, Giri SN. Molecular mechanisms of antifibrotic effect of interferon gamma in bleomycin-mouse model of lung fibrosis: downregulation of TGF-beta and procollagen I and III gene expression. Exp Lung Res 1995; 21: 791–808. [DOI] [PubMed] [Google Scholar]
- 57. Strieter RM, Keane MP. Innate immunity dictates cytokine polarization relevant to the development of pulmonary fibrosis. J Clin Invest 2004; 114: 165–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Strieter RM, Starko KM, Enelow RI, et al. Effects of interferon-gamma 1b on biomarker expression in patients with idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 2004; 170: 133–140. [DOI] [PubMed] [Google Scholar]
- 59. Ziesche R, Hofbauer E, Wittmann K, et al. A preliminary study of long-term treatment with interferon gamma-1b and low-dose prednisolone in patients with idiopathic pulmonary fibrosis. N Engl J Med 1999; 341: 1264–1269. [DOI] [PubMed] [Google Scholar]
- 60. Raghu G, Brown KK, Bradford WZ, et al. A placebo-controlled trial of interferon gamma-1b in patients with idiopathic pulmonary fibrosis. N Engl J Med 2004; 350: 125–133. [DOI] [PubMed] [Google Scholar]
- 61. King TE, Jr, Albera C, Bradford WZ, et al. Effect of interferon gamma-1b on survival in patients with idiopathic pulmonary fibrosis (INSPIRE): a multicentre, randomised, placebo-controlled trial. Lancet 2009; 374: 222–228. [DOI] [PubMed] [Google Scholar]
- 62. Cottin V, Hansell DM, Sverzellati N, et al. Effect of emphysema extent on serial lung function in patients with idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 2017; 196: 1162–1171. [DOI] [PubMed] [Google Scholar]
- 63. Golec M, Wielscher M, Lemieszek MK, et al. Middle age enhances expression of innate immunity genes in a female mouse model of pulmonary fibrosis. Biogerontology 2017; 18: 253–262. [DOI] [PubMed] [Google Scholar]
- 64. Gui X, Qiu X, Tian Y, et al. Prognostic value of IFN-γ, sCD163, CCL2 and CXCL10 involved in acute exacerbation of idiopathic pulmonary fibrosis. Int Immunopharmacol 2019; 70: 208–215. [DOI] [PubMed] [Google Scholar]
- 65. Armant M, Delespesse G, Sarfati M. IL-2 and IL-7 but not IL-12 protect natural killer cells from death by apoptosis and up-regulate bcl-2 expression. Immunology 1995; 85: 331–337. [PMC free article] [PubMed] [Google Scholar]
- 66. Michaud A, Dardari R, Charrier E, et al. IL-7 enhances survival of human CD56bright NK cells. J Immunother 2010; 33: 382–390. [DOI] [PubMed] [Google Scholar]
- 67. Marcais A, Viel S, Grau M, et al. Regulation of mouse NK cell development and function by cytokines. Front Immunol 2013; 4: 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lin JX, Du N, Li P, et al. Critical functions for STAT5 tetramers in the maturation and survival of natural killer cells. Nat Commun 2017; 8: 1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. von Freeden-Jeffry U, Vieira P, Lucian LA, et al. Lymphopenia in interleukin (IL)-7 gene-deleted mice identifies IL-7 as a nonredundant cytokine. J Exp Med 1995; 181: 1519–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Maki K, Sunaga S, Komagata Y, et al. Interleukin 7 receptor-deficient mice lack gammadelta T cells. Proc Natl Acad Sci U S A 1996; 93: 7172–7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Uhal BD. The role of apoptosis in pulmonary fibrosis. Eur Respir Rev 2008; 17: 138–144. [Google Scholar]
- 72. Thannickal VJ, Horowitz JC. Evolving concepts of apoptosis in idiopathic pulmonary fibrosis. Proc Am Thorac Soc 2006; 3: 350–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Plataki M, Koutsopoulos AV, Darivianaki K, et al. Expression of apoptotic and antiapoptotic markers in epithelial cells in idiopathic pulmonary fibrosis. Chest 2005; 127: 266–274. [DOI] [PubMed] [Google Scholar]
- 74. Uhal BD, Joshi I, Hughes WF, et al. Alveolar epithelial cell death adjacent to underlying myofibroblasts in advanced fibrotic human lung. Am J Physiol 1998; 275: L1192–L1199. [DOI] [PubMed] [Google Scholar]
- 75. Hagimoto N, Kuwano K, Nomoto Y, et al. Apoptosis and expression of Fas/Fas ligand mRNA in bleomycin-induced pulmonary fibrosis in mice. Am J Respir Cell Mol Biol 1997; 16: 91–101. [DOI] [PubMed] [Google Scholar]
- 76. Wang R, Ibarra-Sunga O, Verlinski L, et al. Abrogation of bleomycin-induced epithelial apoptosis and lung fibrosis by captopril or by a caspase inhibitor. Am J Physiol Lung Cell Mol Physiol 2000; 279: L143–L151. [DOI] [PubMed] [Google Scholar]
- 77. Budinger GR, Mutlu GM, Eisenbart J, et al. Proapoptotic bid is required for pulmonary fibrosis. Proc Natl Acad Sci U S A 2006; 103: 4604–4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Xie T, Liang J, Geng Y, et al. MicroRNA-29c prevents pulmonary fibrosis by regulating epithelial cell renewal and apoptosis. Am J Respir Cell Mol Biol 2017; 57: 721–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Murray LA, Habiel DM, Hohmann M, et al. Antifibrotic role of vascular endothelial growth factor in pulmonary fibrosis. JCI Insight 2017; 2: e92192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Thannickal VJ, Toews GB, White ES, et al. Mechanisms of pulmonary fibrosis. Annu Rev Med 2004; 55: 395–417. [DOI] [PubMed] [Google Scholar]
- 81. Wang L, Scabilloni JF, Antonini JM, et al. Induction of secondary apoptosis, inflammation, and lung fibrosis after intratracheal instillation of apoptotic cells in rats. Am J Physiol Lung Cell Mol Physiol 2006; 290: L695–L702. [DOI] [PubMed] [Google Scholar]
- 82. Laplante P, Sirois I, Raymond MA, et al. Caspase-3-mediated secretion of connective tissue growth factor by apoptotic endothelial cells promotes fibrosis. Cell Death Differ 2010; 17: 291–303. [DOI] [PubMed] [Google Scholar]
- 83. Lu Q, Harrington EO, Rounds S. Apoptosis and lung injury. Keio J Med 2005; 54: 184–189. [DOI] [PubMed] [Google Scholar]
- 84. Turner-Warwick M. Precapillary systemic-pulmonary anastomoses. Thorax 1963; 18: 225–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Keane MP. Angiogenesis and pulmonary fibrosis: feast or famine? Am J Respir Crit Care Med 2004; 170: 207–209. [DOI] [PubMed] [Google Scholar]
- 86. Hanumegowda C, Farkas L, Kolb M. Angiogenesis in pulmonary fibrosis: too much or not enough? Chest 2012; 142: 200–207. [DOI] [PubMed] [Google Scholar]
- 87. Wang X, Zhu H, Yang X, et al. Vasohibin attenuates bleomycin induced pulmonary fibrosis via inhibition of angiogenesis in mice. Pathology 2010; 42: 457–462. [DOI] [PubMed] [Google Scholar]
- 88. Huang M, Sharma S, Zhu LX, et al. IL-7 inhibits fibroblast TGF-beta production and signaling in pulmonary fibrosis. J Clin Invest 2002; 109: 931–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hsieh PF, Liu SF, Lee TC, et al. The role of IL-7 in renal proximal tubule epithelial cells fibrosis. Mol Immunol 2012; 50: 74–82. [DOI] [PubMed] [Google Scholar]
- 90. Roux BT, Heward JA, Donnelly LE, et al. Catalog of differentially expressed long non-coding RNA following activation of human and mouse innate immune response. Front Immunol 2017; 8: 1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Liu X, Lu Y, Zhu J, et al. A long noncoding RNA, antisense IL-7, promotes inflammatory gene transcription through facilitating histone acetylation and switch/sucrose nonfermentable chromatin remodeling. J Immunol 2019; 203: 1548–1559. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-tar-10.1177_1753466621995045 for Gene fusion of IL7 involved in the regulation of idiopathic pulmonary fibrosis by Shixue Sun, Chen Huang, Dongliang Leng, Chang Chen, Teng Zhang, Kuan Cheok Lei and Xiaohua Douglas Zhang in Therapeutic Advances in Respiratory Disease
Supplemental material, sj-pdf-2-tar-10.1177_1753466621995045 for Gene fusion of IL7 involved in the regulation of idiopathic pulmonary fibrosis by Shixue Sun, Chen Huang, Dongliang Leng, Chang Chen, Teng Zhang, Kuan Cheok Lei and Xiaohua Douglas Zhang in Therapeutic Advances in Respiratory Disease
Supplemental material, sj-pdf-3-tar-10.1177_1753466621995045 for Gene fusion of IL7 involved in the regulation of idiopathic pulmonary fibrosis by Shixue Sun, Chen Huang, Dongliang Leng, Chang Chen, Teng Zhang, Kuan Cheok Lei and Xiaohua Douglas Zhang in Therapeutic Advances in Respiratory Disease
Supplemental material, sj-pdf-5-tar-10.1177_1753466621995045 for Gene fusion of IL7 involved in the regulation of idiopathic pulmonary fibrosis by Shixue Sun, Chen Huang, Dongliang Leng, Chang Chen, Teng Zhang, Kuan Cheok Lei and Xiaohua Douglas Zhang in Therapeutic Advances in Respiratory Disease
Supplemental material, sj-pdf-6-tar-10.1177_1753466621995045 for Gene fusion of IL7 involved in the regulation of idiopathic pulmonary fibrosis by Shixue Sun, Chen Huang, Dongliang Leng, Chang Chen, Teng Zhang, Kuan Cheok Lei and Xiaohua Douglas Zhang in Therapeutic Advances in Respiratory Disease
Supplemental material, sj-pdf-7-tar-10.1177_1753466621995045 for Gene fusion of IL7 involved in the regulation of idiopathic pulmonary fibrosis by Shixue Sun, Chen Huang, Dongliang Leng, Chang Chen, Teng Zhang, Kuan Cheok Lei and Xiaohua Douglas Zhang in Therapeutic Advances in Respiratory Disease
Supplemental material, sj-pdf-8-tar-10.1177_1753466621995045 for Gene fusion of IL7 involved in the regulation of idiopathic pulmonary fibrosis by Shixue Sun, Chen Huang, Dongliang Leng, Chang Chen, Teng Zhang, Kuan Cheok Lei and Xiaohua Douglas Zhang in Therapeutic Advances in Respiratory Disease
Supplemental material, sj-pdf-9-tar-10.1177_1753466621995045 for Gene fusion of IL7 involved in the regulation of idiopathic pulmonary fibrosis by Shixue Sun, Chen Huang, Dongliang Leng, Chang Chen, Teng Zhang, Kuan Cheok Lei and Xiaohua Douglas Zhang in Therapeutic Advances in Respiratory Disease
Supplemental material, sj-tif-4-tar-10.1177_1753466621995045 for Gene fusion of IL7 involved in the regulation of idiopathic pulmonary fibrosis by Shixue Sun, Chen Huang, Dongliang Leng, Chang Chen, Teng Zhang, Kuan Cheok Lei and Xiaohua Douglas Zhang in Therapeutic Advances in Respiratory Disease