Critically endangered sifaka lemurs exhibit high genetic diversity and genomic evidence for dietary adaptation.
Abstract
Sifakas (genus Propithecus) are critically endangered, large-bodied diurnal lemurs that eat leaf-based diets and show corresponding anatomical and microbial adaptations to folivory. We report on the genome assembly of Coquerel’s sifaka (P. coquereli) and the resequenced genomes of Verreaux’s (P. verreauxi), the golden-crowned (P. tattersalli), and the diademed (P. diadema) sifakas. We find high heterozygosity in all sifakas compared with other primates and endangered mammals. Demographic reconstructions nevertheless suggest declines in effective population size beginning before human arrival on Madagascar. Comparative genomic analyses indicate pervasive accelerated evolution in the ancestral sifaka lineage affecting genes in several complementary pathways relevant to folivory, including nutrient absorption and xenobiotic and fatty acid metabolism. Sifakas show convergent evolution at the level of the pathway, gene family, gene, and amino acid substitution with other folivores. Although sifakas have relatively generalized diets, the physiological challenges of habitual folivory likely led to strong selection.
INTRODUCTION
Sifakas (genus Propithecus) are lemurs of the Indriid family and represent a radiation of nine species that occupy various forested regions across Madagascar (1). Lemurs are a primate clade endemic to Madagascar that exhibit exceptional taxonomic and ecological diversity (1, 2). Lemurs’ lengthy geographic isolation (>50 million years) and their arrival earlier than the handful of other mammalian orders on Madagascar have resulted in a remarkable array of adaptations to diverse niches (2, 3). Madagascar is also characterized by unpredictable and hypervariable rainfall environments, marked seasonality, low primary productivity, and nitrogen-poor fruits, which are likely to have been important selective pressures shaping lemur evolution (4–6). Genomic analyses of lemurs are of interest for understanding the molecular bases for lemur traits; however, to date, genomic studies in primates have tended to focus on haplorrhines (7–9). Moreover, lemurs are among the mammalian groups most vulnerable to extinction, with >90% of species currently under threat (10). With one exception, all species within the genus Propithecus are classified as Critically Endangered (11), making it among the most threatened lemur genera. Hence, conservation efforts are urgently needed and genomic data can shed light on factors like inbreeding, genetic connectivity, historical population dynamics, and species delineation/discovery (12) that will be important in future assessments and planning.
Sifakas are diurnal, gregarious, and relatively large-bodied lemurs. They are considered anatomical folivores, showing phenotypic adaptations to folivory that include aspects of dental morphology, an elongated gastrointestinal (GI) tract, and a sacculated cecum and gut microbial community that facilitate hindgut fermentation (13, 14). However, behaviorally, sifakas are less strict folivores compared with the members of the two other indriid genera, Indri and Avahi (15, 16), incorporating a sizeable portion of fruit in the diet when available (16–22) (fig. S1). Nevertheless, leaves comprise a considerable and crucial part of the diet in all populations studied, and anatomical adaptations and the ability to tolerate plant secondary compounds may enable sifakas to exploit foliage of many species as fallback foods (16, 17, 20, 21). Sifaka diets vary considerably by season, habitat, and species, and generally are highly diverse, including greater numbers of different plant species than sympatric diurnal lemurs (18–21, 23).
Here, we report on a whole-genome reference assembly (Pcoq_1.0; GenBank accession GCA_000956105.1) that we generated for Coquerel’s sifaka (Propithecus coquereli), a species with a large but fragmented distribution in the dry, deciduous forests of northwestern Madagascar (24). We also generated whole-genome sequence data for three other sifaka species: Verreaux’s sifaka (Propithecus verreauxi) of the arid deciduous forests of southwestern Madagascar (25), the golden-crowned sifaka (Propithecus tattersalli) from the dry deciduous forests of the northern tip of Madagascar (26), and the diademed sifaka (Propithecus diadema) of the eastern rainforest (27) (Fig. 1). These species share a common ancestor estimated to have lived sometime between 6 and 16 million years ago (28).
Fig. 1. Sifaka samples and demographic history.
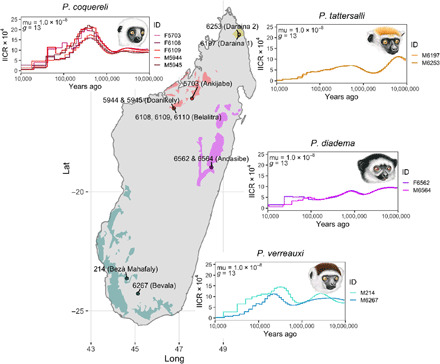
The current known geographic distributions of the four sifaka species included in this study with sample origins. (Illustrations copyright 2020 Stephen D. Nash/IUCN SSC Primate Specialist Group. Used with permission.)
Our goals in this analysis are twofold: (i) to use genomic data to estimate key parameters that are relevant to conservation efforts and (ii) to assess patterns of selection on protein-coding genes in this genus. First, to better understand the conservation status and historical population dynamics of these species, we assessed genomic diversity and reconstructed demographic change in each species. An important outstanding question in Madagascar conservation biology is the degree to which loss of forested habitats is recent and anthropogenic (29) or more ancient and driven by nonanthropogenic climate change (10). Analysis of multilocus microsatellite data of the golden-crowned sifaka, one of the species included here, indicated that population declines far predate human settlement of Madagascar and instead are more likely attributable to habitat change caused by Holocene droughts before human arrival (24). However, many lemurs, including some sifaka species, show evidence of more recent bottlenecks that likely coincide with anthropogenic disturbance (30–34). Different forest habitats and their resident lemurs are likely to have been influenced differently by climate and human activities across the island. Additional genomic analyses from sifaka species inhabiting ecologically distinct regions of Madagascar will expand our knowledge of historical population dynamics in lemurs and help determine whether population size changes occurred well before, during, or well after human settlement of the island.
Second, to gain insights into sifaka physiology, we assessed genome-wide accelerated protein-coding change in the ancestral sifaka lineage. The genomes of other foliage specialists, including other primates (colobine monkeys), giant pandas, red pandas, and koalas (fig. S1), have revealed positive selection and gene family expansion in pathways related to xenobiotic and nutrient metabolism, which could aid in detoxification of plant toxins and maximization of nutritional gain from leaves, respectively (35–37). Compared to other primate clades, folivores are overrepresented among lemurs (2, 6). However, molecular adaptation to folivory has not been extensively studied in lemurs [though see (38)]. The other lemurs represented by genomic data in our comparative analysis were the gray mouse lemur (Microcebus murinus) and the closely related black (Eulemur macaco) and blue-eyed black lemurs (Eulemur flavifrons), all of which are frugivorous/omnivorous (39–41). We also include several colobine species in our analysis and specifically compare our results to findings in this clade. The results presented here expand our knowledge of the genetic basis of mammalian folivore digestive physiology to an ecologically unique group of mammals, sifaka lemurs.
RESULTS
We sequenced and assembled the P. coquereli genome using DNA from a female sifaka that was born in Madagascar (Belalitra, Mahajanga Province) but housed at the Duke Lemur Center (DLC; see Materials and Methods; Fig. 1). We generated 104.7× whole-genome sequence coverage using 100–base pair (bp) paired-end sequence libraries and Illumina Hi-Seq 2000 and 2500 instruments. The assembly process, which used ALLPATHS-LG, Atlas-Link, and AtlasGapFill software (see Materials and Methods), produced a draft assembly (Pcoq_1.0; GenBank accession GCA_000956105.1) of total length 2.80 Gb, with contig N50 of 28.1 kb and scaffold N50 of 5.60 Mb (Table 1).
Table 1. Propithecus coquereli genome assembly description.
| GC content | 43.2% |
| Total size (Mbp) | 2,798.15 |
| Number of contigs | 299,070 |
| Number of scaffolds | 22,539 |
| Contig N50 | 28,129 |
| Contig L50 | 18,523 |
| Scaffold N50 | 5.60 Mb |
| Scaffold L50 | 149 |
The annotation of Pcoq_1.0 was performed by the National Center for Biotechnology Information (NCBI) annotation team using their Eukaryotic Annotation Pipeline (www.ncbi.nlm.nih.gov/genome/annotation_euk/process). This draft assembly provides the sequences for 19,484 protein-coding genes, 408 noncoding genes, and 2867 pseudogenes. We assessed the completeness of this annotation by analyzing the BUSCO (benchmarking universal single-copy orthologs) euarchontoglires gene set of 6192 highly conserved protein-coding genes (42) and found that 91.25% of those genes are complete, while another 3.92% are fragmented. We additionally sequenced five other, unrelated, wild-born P. coquereli individuals originating from Ankijabe, Belalitra, and Doanikely (Mahajanga Province) and housed at the DLC. We also sequenced two wild-born diademed sifakas (P. diadema) from near Andasibe (Toamasina), two wild-born golden-crowned sifakas (P. tattersalli) originating from two sites near Daraina (Antsiranana Province), all also housed at the DLC, as well as a single male Verreaux’s sifaka (P. verreauxi) born to wild-caught parents from Bevala (Toliara Province) at the DLC (Fig. 1). Last, we sequenced an additional wild Verreaux’s sifaka from Bezà Mahafaly Special Reserve (Toliara Province; Fig. 1). All additional genomes were sequenced to an average of >23× coverage (Table 2). The sequencing data demonstrated a high mapping rate (>94%), low polymerase chain reaction (PCR) duplication rate (<15%), and high average base quality (Phred score > 35; Table 2).
Table 2. Additional Propithecus spp. genome sequence descriptions. SNVs, single nucleotide variants.
| Measure | P. diadema | P. diadema | P. tattersalli | P. tattersalli | P. verreauxi | P. verreauxi |
| Reads | 765,447,178 | 749,239,612 | 717,945,044 | 716,893,458 | 954,251,816 | 790,776,474 |
| Mapping rate | 94.3% | 94.5% | 94.6% | 95.1% | 94.5% | 94.6% |
| PCR duplication rate | 4.0% | 3.4% | 3.2% | 2.5% | 15.0% | 3.4% |
| Average phred quality |
36.1 | 36.1 | 36.3 | 36.8 | 38.9 | 35.8 |
| Average depth coverage |
23× | 23× | 23× | 23× | 39× | 33× |
| Variants called (after filtering) |
40,822,967 | 40,851,734 | 22,519,595 | 22,701,370 | 26,512,635 | 24,814,200 |
| SNVs | 37,479,654 | 37,516,145 | 20,714,023 | 20,865,986 | 24,376,639 | 22,875,767 |
| Indels | 3,343,313 | 3,335,589 | 1,805,572 | 1,835,384 | 2,281,800 | 1,938,433 |
All sifakas exhibit high genome-wide heterozygosity (Fig. 2). We estimated genome-wide individual heterozygosity as the number of high confidence heterozygous sites divided by the number of bases in the genome for which variants could be reliably identified based on coverage and mapping quality. Heterozygosity ranged from 0.00569 to 0.00587 for the Coquerel’s sifakas, 0.00314 to 0.00468 for Verreaux’s sifaka, 0.00239 to 0.00249 for golden-crowned sifakas, and 0.00396 to 0.00397 for diademed sifakas (Fig. 2).
Fig. 2. Genome-wide heterozygosity compared across primates and other mammals.
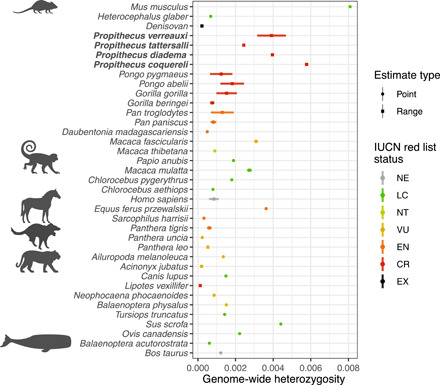
IUCN “Red List” status: NE, not evaluated; LC, least concern; NT, near threatened; VU, vulnerable; EN, endangered; CR, critically endangered; EX, extinct. Mammalian families included represented by clip art. Tasmanian devil silhouette by Sarah Werning from PhyloPic.org. Data from (54, 114–117).
We used the pairwise sequentially Markovian coalescent [PSMC; (43)] to infer the inverse instantaneous coalescence rate (IICR) (44), which can reflect changes in effective population size (Ne) under panmixia where coalescent rate is directly inversely proportional to Ne, and/or changes in population connectivity under a scenario of population structure. We used several different generation time estimates, spanning 9 to 17.5 years, which represent estimates of the average age of female first reproduction (45) and the time required for the population to grow by the net reproductive rate (46), respectively, based on demographic analysis of a population of P. verreauxi studied long term (see section S2). The genomes of all individual sifakas analyzed indicated cycles of increasing and declining IICR over the past million years, with pronounced decreases in the past 100,000 years (Fig. 1). The two western species (P. coquereli and P. verreauxi) show higher peaks and sharper declines in IICR, whereas the two eastern species (P. diadema and P. tattersalli) showed more gradual declines. Different individuals of the same species generally show highly similar demographic reconstructions (Fig. 1); P. verreauxi showed the greatest differences between individuals. Estimates of IICR were highly sensitive to the mutation rate used (fig. S2), but marked declines in IICR, possibly reflecting Ne, before human arrival on Madagascar were detected regardless of the assumed mutation rate.
Genome-wide comparative genomic analyses revealed that genes showing accelerated evolution were enriched for Gene Ontology (GO) terms related to small-molecule transport, processing, and metabolism (table S3). To detect potential positive selection, we compared sequence data for the sifaka species with orthologous coding region alignments for 30 other mammals, including 27 primates. We identified rapidly evolving genes (REGs) that show a significantly elevated branch dN/dS ratio on the ancestral sifaka branch relative to the rest of the phylogenetic tree or contain positively selected sites on the ancestral sifaka branch.
Several pathways potentially relevant to diet were enriched for REGs, including digestion, intestinal absorption, lipid metabolism, and xenobiotic metabolism (Table 3 and table S3). REGs involved in digestion included a number of lipase and protease enzymes, and REGs involved in intestinal absorption included genes encoding two proteins associated with intestinal microvilli morphology, which could influence intestinal surface area (Table 4). REGs involved in drug metabolism and transport included several loci encoding proteins involved in detoxification. In particular, among these proteins were a number of cytochrome P450 monooxygenases, enzymes important for xenobiotic detoxification via oxidative metabolism (47); members of solute carrier families involved in toxin elimination (48); members of adenosine 5′-triphosphate (ATP)–binding cassette families, which extrude dietary phytotoxins (49); and members of the uridine 5′-diphospho (UDP)–glucuronosyltransferase family, which are known to play roles in the excretion of plant secondary compounds in bile or urine (50) (Table 4).
Table 3. Enriched pathways related to folivory.
| Pathway name | Pathway GO |
| Digestion | 0007586 |
| Intestinal absorption | 0050892 |
| Lipid metabolic process | 0006629 |
| Fatty acid derivative metabolic process |
1901568 |
| Bile acid biosynthetic process | 0006699 |
| Long-chain fatty acid metabolic process |
0001676 |
| Drug metabolic process | 0017144 |
| Drug transport | 0015893 |
Table 4. REGs involved in digestion and xenobiotic metabolism.
| Functional category | Gene symbol | Gene name | Notes |
| Digestion | PNLIP | Pancreatic lipase | The main digestive enzyme in triglyceride hydrolysis |
| Digestion | CLPS | Pancreatic colipase | Necessary pancreatic lipase cofactor |
| Digestion | UCN3 | Urocortin 3 | Hormone regulating gut passage and energy homeostasis |
| Digestion | CAPN9 | Calpain 9 | Protease |
| Digestion | CTRB1 | Chymotrypsinogen b1 | Protease |
| Digestion | CTRB2 | Chymotrypsinogen b2 | Protease |
| Digestion | PGA3 | Pepsinogen 3 | Protease |
| Digestion | VIL1 | Villin-1 | Regulatory of microvilli morphology |
| Digestion | PLS1 | Plastin-1 | Regulatory of microvilli morphology |
| Xenobiotic metabolism | CYP2B6 | Cytochrome P450 family 2 subfamily B member 6 |
Cytochrome P450 monooxygenase |
| Xenobiotic metabolism | CYP2A13 | Cytochrome P450 family 2 subfamily A member 13 |
Cytochrome P450 monooxygenase |
| Xenobiotic metabolism | SLC22A2 | Solute carrier family 22 member 2 | Cation transporter |
| Xenobiotic metabolism | ABCG2 | ATP binding cassette subfamily G member 2 |
Transmembrane transporter |
| Xenobiotic metabolism | UGT1A8 | UDP- glucuronosyltransferase 1 family, polypeptide a8 |
UDP-glucuronosyltransferase |
We also identified several bitter taste receptors that showed provisional evidence of positive selection, although none of the analyses reached significance after correcting for multiple testing (q < 0.05). TAS2R40 had dN/dS > 1 (sifaka dN/dS = 1.330; background dN/dS = 0.494; P = 0.022). Positively selected sites were identified within TAS2R7 (C60V, P = 0.029, Bayes empirical Bayes score = 0.816) and TAS2R16 (S70R, P = 0.007, Bayes empirical Bayes score = 0.95).
In comparing our data to previous studies, we found overlap between our results and those of the analysis of the snub-nosed monkey (genus Rhinopithecus) genomes (35). Snub-nosed monkeys are folivorous colobine monkeys whose ancestors diverged from the ancestors of Propithecus approximately 70 million years ago (28). Parallel changes were observed at the levels of the genetic pathway, the gene, and particular amino acid substitution. Loci that show rapid evolution in both sifakas and snub-nosed monkeys included genes involved in both lipid metabolism and xenobiotic metabolism (table S4). One gene, ACE (angiotensin-converting enzyme), showed the same two convergent amino acid substitutions in both lineages (M109L and I869V).
We also found positive selection on the pancreatic ribonuclease (RNase) gene, RNASE1, which is thought to be important for breaking down bacterial RNA in herbivores (51). In primates, there have been independent duplications of RNASE1 in African and Asian colobine monkeys, including snub-nosed monkeys that, like bovine ruminants, have evolved foregut fermentation (51, 52).
RNASE1 exhibited an elevated dN/dS (sifaka dN/dS = 5.574, background dN/dS = 0.291) and 11 positively selected sites on the ancestral sifaka branch. Sifakas showed amino acid substitutions at five of the same sites predicted to change the catalytic activity of the duplicated RNASE1B gene found in douc langurs (Pygathrix nemaeus) (Fig. 3) (53). All mutations were confirmed with Sanger sequencing (section S6). Site 83, which is one of the two sites predicted to have the greatest influence on the enzymatic activity of RNASE1 based on site-directed mutagenesis experiments (53), shows the same amino acid substitution (D > E) in sifakas and duplicated colobine sequences. Sifakas also show the same substituted amino acid residue (S) at position 39 as two Asian colobine species (Semnopithecus entellus and Trachypithecus francoisi). As in colobines, substitutions in sifakas include many losses of arginines, although sifakas have also gained one arginine. The loss of positively charged arginine residues contributes to the change in enzymatic activity. The loss of arginines might also enhance the stability of the enzyme in the gut as arginines are especially vulnerable to hydrolysis by trypsin, one of the primary digestive enzymes active in the upper GI tract, and to alteration during bacterial fermentation (52). P. diadema and P. tattersalli additionally show substitutions at position 1, another site predicted to change enzymatic activity in colobines. These two sifaka species show different substitutions: K1N and K1T, respectively.
Fig. 3. Sequence alignment of RNASE1 sites predicted to affect enzymatic activity including duplicated colobine sequences.

Positions of positively selected sites in sifakas highlighted in yellow. Duplicated colobine RNASE1 genes and sifaka species bolded.
In total, we found 10 gene families to have undergone rapid expansion and 33 gene families to have undergone rapid contraction on the ancestral sifaka branch (fig. S4). Gene families of ribosomal proteins, oxidases, and transcription factors showed both expansion and contraction. Families showing expansion were involved in response to oxidative damage, apoptosis, and DNA damage. Families of immune receptors, heat shock proteins, olfactory receptors, and proteins involved in translation and protein folding were among those showing contraction.
DISCUSSION
Detailed genomic and genetic analysis of sifakas and other strepsirrhines can provide critical insight into their phylogenetic, demographic, and adaptive histories. Our genome assembly (Pcoq_1.0) is comparable to other strepsirrhine assemblies, with global assembly statistics (Table 1) similar to those of the small-eared greater galago (Otolemur garnetti) draft assembly (OtoGar3; Contig N50: 27,100; Contig L50: 21,634; Scaffold N50: 13,852,661; Scaffold L50: 47). Although not a chromosome-level assembly and not as complete as the most recent gray mouse lemur (M. murinus) reference assembly (Mmur_3.0; Contig N50: 210,702; Contig L50: 2987; Scaffold N50: 108,171,978; Scaffold L50: 10), the annotation pipeline resulted in the identification of 19,484 protein-coding genes, 408 noncoding genes, and 2867 pseudogenes. For comparison, Mmur_3.0 contains 20,671 protein-coding and 7142 noncoding genes, and OtoGar3 contains 19,558 protein-coding and 4827 noncoding genes. Thus, our assembly is of sufficient quality to assess protein evolution. This new sifaka reference assembly extends our knowledge of lemur genomics to an ecologically unique branch of strepsirrhine evolution.
Although ecological and population pressures currently face sifaka species, we found that standing genetic variation remains high. Three of the four sifaka species analyzed showed higher genome-wide heterozygosity than any of the other primates for which comparable data are available (Fig. 2). Our findings are consistent with the results of several previous genetic diversity assessments of various sifaka species, including P. coquereli and P. tattersalli, studied here (30, 33, 54–56). However, despite evidence for relatively high genetic diversity in sifakas, several studies have found evidence of population bottlenecks or declines in various species (30, 33, 34). There is some evidence that heterozygosity can temporarily increase following a population bottleneck (57), as may be the case here. Our finding of long-term declines in IICR in all sifaka species would seem to be at odds with the high estimates of genome-wide heterozygosity. A slow and steady population decline need not be accompanied by a reduction in heterozygosity, however, as numerous demographic and genetic factors (mutation rate, dispersal distance, overlapping generations, subpopulation connectivity, and admixture) can blur the relationship between past demographic events and genetic diversity (58). This trend of steady declines in IICR during the late Pleistocene is consistent with some demographic reconstructions of other lemur species using whole-genome sequence data (59–61). These declines have been attributed to a drying climate and resulting loss of prime forest habitat (59, 60, 62). Lake drill cores offer evidence of severe drying in East Africa from ~145,000 to 60,000 years ago (63), a time range overlapping the period when a marked decline in the four species’ effective population sizes seems to have occurred. The demographic history inferred here indicates that these sifaka populations had already declined markedly before the arrival of people in Madagascar, even when considering recent, earlier estimates of human arrivals (64). This result, however, does not rule out more recent declines driven by anthropogenic disturbance and/or Holocene droughts (34, 65), as detected in other studies of sifakas (30, 33, 34) and other lemurs (31, 32). A paucity of recombination events in the last several thousand years precludes resolution at this time scale using the IICR approach (43).
We also note that there are other limitations to PSMC reconstruction. Parameter choice can have a large influence on PSMC results (43). Generation time estimates used in the model have a sizable influence on the estimated timing of demographic events, while mutation rate influences the inferred magnitude of change (Fig. 1 and fig. S2). Empirical estimates of per generation mutation rate for sifaka species would be desirable given the large difference in estimates of IICR depending on whether the human mutation rate or a faster mutation rate similar to that estimated for mouse lemurs is used (Fig. 1 and fig. S2) (66). Population structure may also confound interpretation of the demographic analyses. The PSMC assumes panmixia, which is often violated in real-world cases. Changes in population structure may produce the same signal in an individual’s genome as would historical change in population size (44), which has led to criticism of simplistic interpretations about species’ past population sizes. Thus, it has been argued that Ne estimates from PSMC analyses are better termed IICR (44). We sought to obtain more robust estimates of IICR by including multiple individuals per species and particularly individuals from more than one geographic locale when possible (Fig. 1). Within a species, we find that individuals’ genomes produce highly similar past IICR trajectories. A greater intraspecific difference is apparent for P. verreauxi, providing possible evidence for population structure (Fig. 1). Nevertheless, we are unable to distinguish between changes in population size, changes in population structure, or the combined effects of both. Future work sequencing more genomes of each species to determine the pattern of rare versus intermediate frequency variants combined with improved paleoecological reconstructions of changes in forest cover will aid in modeling sifaka demographic history.
The maintenance of genetic diversity by these sifaka populations has several possible explanations. It may be that sifakas are relatively resilient to habitat fragmentation and able to disperse across deforested areas. This is consistent with evidence for continued gene flow in crowned sifakas (P. coronatus) (55) and P. verreauxi (67) despite uneven forest cover, as well as patterns of isolation-by-distance in genetic diversity observed across localities in P. tattersalli (56), Perrier’s sifaka (P. perrieri) (33), and P. verreauxi (67). This would seem to be in contrast to some species, like black-and-white ruffed lemurs (Varecia variegata), for which habitat cover is a major predictor of genetic structure (68). Specifically, the capacity for seasonally flexible folivory may buffer sifakas from energetic deficits as leaves remain relatively abundant even as habitat quality diminishes, aiding them in comparison to more specialized, dedicated folivores or frugivores (69–71). P. tattersalli appear to exhibit a high level of dietary flexibility that may allow it to tolerate disturbed and/or degraded habitat (23), and sifakas generally have diverse diets (18), including when compared with other sympatric diurnal lemurs (19, 21). Our findings of positive selection on pathways that may be involved in nutrient absorption and xenobiotic metabolism further lend support to this proposition. This finding is consistent with observations that other folivorous primate taxa, like howler monkeys (genus Alouatta), which also show extensive dietary flexibility, are relatively resistant to anthropogenic disturbance compared to sympatric frugivores (72, 73). In addition, the degree of disturbance, rather than disturbance alone, may be crucial to understand the impact on population dynamics. For example, there is evidence that some lemurs may benefit nutritionally from “edge effects” in landscapes where discontinuous tree cover allows greater sunlight exposure and leaves with higher protein content as a result (74). Thus, other lemurs in these same and other fragmented and/or degraded habitats in Madagascar may be more affected.
Moreover, it is unclear to what degree any ability of sifakas to cope with fragmentation is long term. There may be lag times between habitat disturbance and/or fragmentation and population declines (75–77). In particular, the long generation times observed in sifakas (45, 46) may obscure the impact of relatively recent connectivity loss. Further, there is evidence that habitat fragmentation leads to declines in diet quality, energy intake, physiological health measures that may reflect immune function, and body condition in P. diadema (78–80), indicating that habitat fragmentation is not without harm to these lemurs.
There is also evidence that regional fadys, or taboos, against hunting and consuming sifakas have offered a degree of protection from human hunting pressure (81–83). Nevertheless, hunting of P. verreauxi has been observed in the southwest (82), some surveys report relatively high rates of consumption of both P. diadema and P. verreauxi (12 to 29% of respondents had eaten sifaka in the last year) (84), and there is some indication that the strength of traditional fadys may be diminishing in some regions (85).
A relatively large geographic distribution, even in a fragmented habitat where populations are still connected by dispersal and gene flow, may contribute to high heterozygosity in P. coquereli, P. verreauxi, and P. diadema (24, 25, 27). The congener P. coronatus also shows a large geographic distribution and high genetic diversity (55). Although census population size estimates are lacking for P. verreauxi and P. diadema and population size is thought to be decreasing in all four species, the P. coquereli population size was nevertheless recently estimated to be around 47,000 in Ankarafantsika National Park alone (25–27, 86), which is consistent with the especially high estimates of genome-wide heterozygosity in this species. Last, social structure may maintain heterozygosity (87). Sifakas exhibit male-biased dispersal, although some females may also leave their natal group to establish a new group (88). Males may disperse multiple times over the course of their lives, usually among neighboring groups (88), and social group boundaries tend to become porous during the very short breeding season, during which time males may sire offspring in neighboring groups (89). These dynamics lead to a “neighborhood”-like intermediate level of hierarchical social organization (88). P. tattersalli and P. verreauxi social groups have been found to exhibit excess heterozygosity (89, 90), indicating that relatively small-scale social structure can generate and maintain genetic diversity, although the relationship between heterozygosity maintained at the social group level and at a larger spatial scale, such as a metapopulation, is not straightforward (90).
Our results do offer a glimmer of optimism. They confirm that considerable genetic diversity still persists in these endangered sifaka species despite habitat fragmentation, likely due to sifakas’ dietary and behavioral flexibility, including the ability to disperse across open landscapes (33, 55, 56, 67). Although there may be a lag time between anthropogenic disturbance and population declines, our results, along with those of others (30, 33, 54–56), indicate that genetically viable populations of these species may be maintained if further habitat loss or fragmentation is prevented.
The results of our comparative genomic analyses indicate accelerated genetic change in sifakas in several pathways that may play a role in folivory. Evolutionary changes in genes involved in digestion and intestinal absorption (Table 4) likely contribute to maximizing nutrient acquisition. Similarly, some of the genes in the enriched pathways of small-molecule and biosynthetic processes may also aid in nutrient absorption and utilization. Enhanced nutrient uptake capacity would be advantageous under conditions of the unpredictable food resource availability and low primary productivity that characterize Madagascar (2, 4). This nutritional edge might be particularly beneficial when transitioning to a leaf-based diet. For example, genes involved in fatty acid, amino acid, and vitamin processing have been found to show adaptive convergence in giant and red pandas, two carnivorans specialized for a leaf-based diet. In sifakas, SLC44A4 may contribute to absorption of vitamin B1 (thiamine) produced by microbiota (91), while SLC52A1 may aid in efficient vitamin B2 (riboflavin) transport (92), SLC23A1 in vitamin C uptake (92), and SLC6A20 in amino acid uptake (93). We identified rapid evolution in many genes involved in lipid metabolism (tables S3 and S5), some of which may enable sifakas to maximize the capture of lipids from a lipid-poor diet. All but one of the genes that display overlapping rapid evolution in sifakas and snub-nosed monkeys also show accelerated change in cattle (35), further implicating these genes in adaptation to herbivory or folivory (table S4).
Enhanced detoxification capacity, indicated by enriched GO terms related to drug metabolism, could confer tolerance of toxic plant secondary compounds like alkaloids (94, 95), which are particularly abundant in the diets of sifakas compared to other folivorous indriids (16). These xenobiotic molecules are small enough to be absorbed by the host GI tract, after which they may have harmful effects or be energetically expensive to neutralize (94). In particular, two genes in the cytochrome P450 family 2 (CYP2B6 and CYP2A13) showed accelerated change on the ancestral sifaka branch. Gonzalez and Nebert (47) argued that this gene family has expanded in particular species to aid in detoxification of plant secondary compounds as part of an animal-plant arms race. In koalas, one subfamily (cytochrome P450 family 2 subfamily C) in this group expanded and some genes show evidence of selection, which has been proposed can help detoxify eucalyptus leaves (36). Convergent amino acid substitutions in genes in the cytochrome P450 family 2 have also been identified in giant and red pandas (37), suggesting a common role for these genes in adaptation to a foliage-based diet in mammals.
Overall, these findings also raise the interesting question of how host and microbial genes and metabolism may operate synergistically to mediate folivory across evolutionary and ecological time scales. Gut microbiota are fundamental to folivore and herbivore nutrition, contributing to plant fiber and secondary compound degradation, essential amino acid, vitamin, and nutrient biosynthesis (96). Previous work has shown the gut microbiota of sifakas to comprise taxa and metabolic processes tuned to their hosts’ specialized diets (14) and that respond, in real time, to changing diets (97). The ecological and metabolic success of folivory in sifakas suggests that these lemurs could be prime candidates in which to investigate the coevolution of the genomes of the host organism and its associated microbiota to better understand the shift to novel, digestively challenging feeding strategies.
We also found evidence for positive selection on several bitter taste receptors on the ancestral sifaka branch. This finding is interesting given that Verreaux’s sifakas demonstrate a higher threshold for avoiding bitter-tasting tannins than do sympatric Lemur catta (95). Genetic variation in the binding domains of these genes is thought to be shaped by environmental fine-tuning involving a tradeoff between the conflicting needs to avoid toxins versus preventing the unwarranted exclusion of valuable food resources (98). One of the genes identified, TAS2R16, is one of the best-studied human bitter taste receptor genes. It primarily binds β-glucopyranosides, compounds produced by many plants and insects as deterrents, including several common crops like cruciferous vegetables (e.g., Brussels sprouts and broccoli) and manioc. In the latter, β-glucopyranosides contribute to the production of cyanide when the root is consumed (99). There is potential evidence for functional and possibly adaptive variation in this gene in humans [discussed in (98)]. Sifakas showed amino acid substitutions at three sites within this protein that may be important for signal transduction (V45L, L185V, and L244V) (99).
It is possible that greater tolerance for bitter compounds could coevolve with greater xenobiotic neutralization capacity. Analyses of the koala genome identified, for example, concomitant evolution of bitter taste receptors and cytochrome P450 genes (36). Verreaux’s sifakas consume more alkaloids than do sympatric Lemur catta at Berenty Reserve and Bezà Mahafaly Special Reserve (21, 95). Some bitter taste receptor ligands are also targets of cytochrome P450 enzymes and/or solute carriers, including one, SLC22A2, identified specifically in the present study (100). Moreover, bitter taste receptors are expressed throughout the GI tract, and detection in the gut may actually stimulate the expression of other xenobiotic metabolizers, like ATP-binding cassette transporters (101) and UDP-glucuronosyltransferases (50), many of which were identified in this study.
It is worth noting that we observe in sifakas positive selection on RNase 1 and substitutions at five amino acid sites within it that were experimentally shown to alter enzymatic activity. Selection on duplicated RNASE1 genes in colobine and bovine foregut fermenters is thought to be related to the breakdown of bacterial RNA from large bacterial communities living in the foregut. This breakdown facilitates absorption of important nutrients, like nitrogen and phosphorus (52). Independent amino acid substitutions in colobines and bovine ruminants both result in a lowered optimal pH for RNase 1 enzymatic activity, presumably to boost enzymatic efficiency in the acidic environment of the foregut (51).
A recent study reported that the duplication of the RNASE1 gene in howler monkeys (Alouatta spp.) was followed by amino acid changes paralleling those in colobines (102). Howler monkeys, like sifakas, rely on hindgut fermentation to obtain energy from foliage (94). Although colobines have ruminant capacity (94), they also have an enlarged cecum and colon, which enable an additional round of fermentation and energy capture in the hindgut (94). These collective findings suggest a role for RNASE1 in hindgut and foregut fermentation. Nevertheless, the lack of evidence for RNASE1 duplications in sifakas is notable and warrants further study given the mode of RNASE1 evolution observed in these other lineages. Last, because the RNase1 enzymes play nondigestive roles in other tissues, in particular in the immune system (22), we cannot rule out alternative explanations for selection on RNASE1; however, that alternate pressures could lead to parallel gene changes between sifakas and other folivores seems less likely than parallelism due to dietary adaptation.
Although a smaller proportion of the sifaka diet is composed of leaves compared with colobines and other indriids (16, 94), our findings indicate extensive selection for the capacity for folivory in sifakas. These signatures in the genome may reflect a more folivorous indriid ancestor and/or a scenario in which leaves represent an essential fallback food. Because we currently lack data for other indriid genera, we cannot rule out the possibility that many of the protein-coding changes we assign to the sifaka branch actually lie deeper within indriids. Future genomic analyses of the indri (I. indri) and woolly lemurs (Avahi spp.) are warranted to answer this question. Data for members of the Lepilemuridae family, in which folivory evolved independently (28, 103), could also potentially shed greater light on common physiological mechanisms involved in adaptation to a leaf-based diet in lemurs.
The rapid evolution of ribosomal proteins, as well as other gene families involved in translation and proteome maintenance, is of potential interest as these families have been found to show rapid evolution in desert- and/or heat-adapted animal species (104, 105). High temperatures can disrupt transcription and translation and cause the denaturation and degradation of proteins (106). Western sifakas, including P. coquereli, which provides the reference genome reported here, live in seasonally very hot and dry tropical desert/savannah/dry forest biomes (1).
The lemurs of Madagascar represent an adaptive radiation of primates exploiting a diverse array of habitats and ecological niches. Sifakas evolved to exploit a variety of forest types across Madagascar through a series of genetic, morphological, and microbial adaptations to folivory in combination with locomotor adaptations to life in the trees. Although folivory presents many digestive challenges (94), the capacity to meet nutritional needs with a leaf-based diet might have been especially beneficial to sifakas, given the erratic fruit availability in Madagascar caused by unpredictable seasonality and/or the low nitrogen content of Malagasy fruit (5, 6). Our analysis of sifaka genomes reveals a number of genetic characteristics that likely played a role in sifakas’ successful movement into and exploitation of forest niches where leaves were an abundant resource. Moreover, a flexibly folivorous dietary repertoire may today contribute to an ability to tolerate a degree of habitat disturbance and partly account for the high levels of genetic diversity that sifakas currently appear to retain. Conservation efforts to prevent further habitat loss and/or fragmentation while these populations appear to largely remain viable are critical. Further study of other lineages within the lemuriform clade is likely to be equally revealing of aspects of primate evolutionary adaptation and their underlying molecular mechanisms.
MATERIALS AND METHODS
DNA extraction and sequencing
For the sifakas housed at the DLC, we extracted DNA using the QIAGEN Blood and Cell Culture Kit. For the wild Verreaux’s sifaka individual, we extracted DNA as described in (107) from banked ear tissue. All samples then underwent quality control via PicoGreen quantification and gel imaging for concentration and purity. We used the manufacturer’s recommended procedures to generate Illumina 100-bp paired-end whole-genome sequencing libraries. We sequenced those libraries to 104.7× whole-genome coverage via runs of 12 lanes on an Illumina Hi-Seq 2000 instrument and 1 lane on an Illumina Hi-Seq 2500 instrument for the individual used for the assembly. The additional individuals were sequenced on two lanes of an Illumina Hi-Seq 2000 instrument each. For the Verreaux’s sifaka from Bezà Mahafaly Special Reserve, 150-bp paired-end libraries were generated and sequenced on a single Illumina Hi-Seq X lane.
Genome assembly and annotation
We assembled the resulting quality-filtered sequence reads using standard approaches and methods (BioProject PRJNA260602). Briefly, we performed the initial assembly steps using ALLPATHS-LG version R43839 (108). We then conducted additional scaffolding using AtlasLink v1.0 (www.hgsc.bcm.edu/software/atlas-link). Where possible, we filled remaining gaps within scaffolds using the original Illumina read data and AtlasGapFill v2.2 (www.hgsc.bcm.edu/software/atlas-gapfill). The NCBI annotation team performed the annotation of Pcoq_1.0 using their Eukaryotic Annotation Pipeline (www.ncbi.nlm.nih.gov/genome/annotation_euk/process).
P. diadema, P. tattersalli, and P. verreauxi sequences
We aligned diademed, golden-crowned, and Verreaux’s sifaka reads to the Pcoq_1.0 assembly and called variants in regions of sufficient coverage. To obtain sequences for the comparative genomic and gene family evolution analyses, we applied variants in the various sifakas’ sequences to the Coquerel’s sifaka reference sequence using GATK (Genome Analysis Toolkit) FastaAlternateReferenceMaker (109).
Diversity analysis
We divided the number of heterozygous sites by the number of callable sites to estimate genome-wide heterozygosity for each sifaka individual. We used vcftools (110) to count heterozygous sites.
Demographic reconstruction
We used PSMC (43) to infer past effective population size (Ne). We used several different ages for the generation time parameter and three mutation rate estimates to scale coalescent time to historic time (see section S2). To obtain confidence intervals, we performed 100 bootstrap replicates.
Comparative genomics
We used branch and branch site models in codeml (111) within the python wrapper Environment for Tree Exploration (ETE) Toolkit [v3; (111)] using orthologous gene alignments for 30 mammals, including 27 primates for 8862 genes and a species tree (28, 112) to model protein-coding evolution along the ancestral sifaka branch. We used likelihood ratio tests to determine whether a model of accelerated evolution on the branch of interest relative to the rest of the tree was better than a null model. To identify genes potentially involved in adaptation to folivory, we compared REGs in the sifaka lineage with genes previously identified as REGs in the genomes of several snub-nosed monkey species (Rhinopithecus roxellana, R. bieti, R. brelichi, and R. strykeri), which also showed rapid evolution in bovine ruminants (35).
Gene family evolution
We assessed gene family expansion and contraction on the ancestral sifaka branch using CAFE (Computational Analysis of gene Family Evolution) (113) and a reduced dataset of 12 species, including 11 primates and the Northern treeshrew (Tupaia belangeri; section S5 and fig. S4).
Acknowledgments
We would like to thank Madagascar National Parks for allowing us to conduct research at Bezà Mahafaly Special Reserve; the DLC and the Bezà Mahafaly Special Reserve monitoring team for samples; K. Glander, D. Haring, and K. Gerard for helpful information; A. Richard, E. Sargis, and D. Watts for helpful feedback on earlier drafts of this work; P. Cingolani, the creator of SnpSift; S. Langdon of the Duke DNA Analysis Facility for support; and E. Tapanes and G. Tiley for helpful discussion. We thank the Yale Center for Research Computing and Duke Compute Cluster team for guidance and use of the research computing infrastructure. We also thank the library and sequence production teams and the project managers’ group within the Human Genome Sequencing Center, Baylor College of Medicine for essential assistance in generating the data reported here. This is DLC publication #1474. Funding: We thank the Center for the Advanced Study of Human Paleobiology at The George Washington University, Duke University, and the Wenner-Gren Foundation for funding. Genome sequencing and assembly were funded by National Human Genome Research Institute grant U54 HG003273 to R. Gibbs (HGSC, Baylor College of Medicine). The research reported here complied with governmental and institutional regulations and ethical guidelines. Author contributions: Designed the study and supervised the analysis: J.Rog., K.C.W., and A.D.Y.; managed or supervised the sequence production: M.R. and D.M.M.; managed or supervised the preparation of sequencing libraries: M.R. and D.M.M.; produced the assembly: K.C.W., Y.L., S.M., R.A.H., and D.S.T.H.; project and data management: D.M.M., M.R., and K.C.W.; coding region assembly from resequencing data: E.E.G. and T.H.W.; demographic reconstruction, selection analysis, and gene family analysis: E.E.G.; RNASE1 analysis: E.E.G. and L.K.G.; provided essential biomaterials: R.R.L., B.J.B., L.K.G., J.Ran., J.Rat., and A.D.Y.; wrote the paper: E.E.G., J.Rog., T.H.W., R.R.L., B.J.B., L.K.G., and A.D.Y. All authors read and approved the final manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All the raw sequence data, as well as sample metadata and additional information for this genome assembly and analysis project, are available under BioProject PRJNA251429 at www.ncbi.nlm.nih.gov. This includes the Pcoq_1.0 genome assembly (RefSeq accession GCF_000956105.1) and the supporting read data (SRA accessions SRX701282, SRX701294 to SRX701300, SRX763489 to SRX763493, and SRX1025581 to SRX1025582). The data used for the diversity analyses across several Propithecus species can be accessed under umbrella project PRJNA260602, which covers the data for P. diadema (PRJNA260543), P. tattersalli (PRJNA260541), and P. verreauxi (PRJNA260540). Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/17/eabd2274/DC1
REFERENCES AND NOTES
- 1.Mittermeier R. A., Ganzhorn J. U., Konstant W. R., Glander K., Tattersall I., Groves C. P., Rylands A. B., Hapke A., Ratsimbazafy J., Mayor M. I., Louis E. E., Rumpler Y., Schwitzer C., Rasoloarison R. M., Lemur diversity in Madagascar. Int. J. Primatol. 29, 1607–1656 (2008). [Google Scholar]
- 2.Richard A. F., Dewar R. E., Lemur ecology. Annu. Rev. Ecol. Syst. 22, 145–175 (1991). [Google Scholar]
- 3.Dewar R. E., Richard A. F., Madagascar: A history of arrivals, what happened, and will happen next. Annu. Rev. Anthropol. 41, 495–517 (2012). [Google Scholar]
- 4.Wright P. C., Lemur traits and Madagascar ecology: Coping with an island environment. Yearb. Phys. Anthropol. 42, 42 (1999). [DOI] [PubMed] [Google Scholar]
- 5.Dewar R. E., Richard A. F., Evolution in the hypervariable environment of Madagascar. Proc. Natl. Acad. Sci. U.S.A. 104, 13723–13727 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donati G., Santini L., Eppley T. M., Arrigo-Nelson S. J., Balestri M., Boinski S., Bollen A., Bridgeman L. L., Campera M., Carrai V., Chalise M. K., Lewis A. D., Hohmann G., Kinnaird M. F., Koenig A., Kowalewski M., Lahann P., McLennan M. R., Nekaris A. K. I., Nijman V., Norscia I., Ostner J., Polowinsky S. Y., Schülke O., Schwitzer C., Stevenson P. R., Talebi M. G., Tan C., Tomaschewski I., Vogel E. R., Wright P. C., Ganzhorn J. U., Low levels of fruit nitrogen as drivers for the evolution of Madagascar’s primate communities. Sci. Rep. 7, 14406 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kronenberg Z. N., Fiddes I. T., Gordon D., Murali S., Cantsilieris S., Meyerson O. S., Underwood J. G., Nelson B. J., Chaisson M. J. P., Dougherty M. L., Munson K. M., Hastie A. R., Diekhans M., Hormozdiari F., Lorusso N., Hoekzema K., Qiu R., Clark K., Raja A., Welch A. E., Sorensen M., Baker C., Fulton R. S., Armstrong J., Graves-Lindsay T. A., Denli A. M., Hoppe E. R., Hsieh P., Hill C. M., Pang A. W. C., Lee J., Lam E. T., Dutcher S. K., Gage F. H., Warren W. C., Shendure J., Haussler D., Schneider V. A., Cao H., Ventura M., Wilson R. K., Paten B., Pollen A., Eichler E. E., High-resolution comparative analysis of great ape genomes. Science 360, eaar6343 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Svardal H., Jasinska A. J., Apetrei C., Coppola G., Huang Y., Schmitt C. A., Jacquelin B., Ramensky V., Müller-Trutwin M., Antonio M., Weinstock G., Grobler J. P., Dewar K., Wilson R. K., Turner T. R., Warren W. C., Freimer N. B., Nordborg M., Ancient hybridization and strong adaptation to viruses across African vervet monkey populations. Nat. Genet. 49, 1705–1713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rogers J., Raveendran M., Harris R. A., Mailund T., Leppälä K., Athanasiadis G., Schierup M. H., Cheng J., Munch K., Walker J. A., Konkel M. K., Jordan V., Steely C. J., Beckstrom T. O., Bergey C., Burrell A., Schrempf D., Noll A., Kothe M., Kopp G. H., Liu Y., Murali S., Billis K., Martin F. J., Muffato M., Cox L., Else J., Disotell T., Muzny D. M., Phillips-Conroy J., Aken B., Eichler E. E., Marques-Bonet T., Kosiol C., Batzer M. A., Hahn M. W., Tung J., Zinner D., Roos C., Jolly C. J., Gibbs R. A., Worley K. C., Baboon Genome Analysis Consortium, The comparative genomics and complex population history of Papio baboons. Sci. Adv. 5, eaau6947 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwitzer C., Mittermeier R. A., Johnson S. E., Donati G., Irwin M., Peacock H., Ratsimbazafy J., Razafindramanana J., Louis E. E., Chikhi L., Averting lemur extinctions amid Madagascar’s political crisis. Science 343, 842–843 (2014). [DOI] [PubMed] [Google Scholar]
- 11.IUCN, The IUCN Red List of Threatened Species Version 2020–2 (IUCN, 2020); www.iucnredlist.org.
- 12.Lawler R. R., Emerging and enduring issues in primate conservation genetics. Annu. Rev. Anthropol. 47, 395–415 (2018). [Google Scholar]
- 13.Campbell J. L., Eisemann J. H., Williams C. V., Glenn K. M., Description of the gastrointestinal tract of five lemur species: Propithecus tattersalli, Propithecus verreauxi coquereli, Varecia variegata, Hapalemur griseus, and Lemur catta. Am. J. Primatol. 52, 133–142 (2000). [DOI] [PubMed] [Google Scholar]
- 14.Greene L. K., Williams C. V., Junge R. E., Mahefarisoa K. L., Rajaonarivelo T., Rakotondrainibe H., O’Connell T. M., Drea C. M., A role for gut microbiota in host niche differentiation. ISME J. 14, 1675–1687 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ganzhorn J. U., Abraham J. P., Razanahoera-Rakotomalala M., Some aspects of the natural history and food selection of Avahi laniger. Primates 26, 452–463 (1985). [Google Scholar]
- 16.Powzyk J. A., Mowry C. B., Dietary and feeding differences between sympatric Propithecus diadema diadema and Indri indri. Int. J. Primatol. 24, 1143–1162 (2003). [Google Scholar]
- 17.M. T. Irwin, Developments in primatology: Progress and prospect, in Lemurs, L. Gould, M. L. Sauther, Eds. (Springer US, 2006), pp. 305–326. [Google Scholar]
- 18.Norscia I., Carrai V., Borgognini-Tarli S. M., Influence of dry season and food quality and quantity on behavior and feeding strategy of Propithecus verreauxi in Kirindy, Madagascar. Int. J. Primatol. 27, 1001–1022 (2006). [Google Scholar]
- 19.Simmen B., Hladik A., Ramasiarisoa P., Food intake and dietary overlap in native Lemur catta and Propithecus verreauxi and introduced Eulemur fulvus at Berenty, Southern Madagascar. Int. J. Primatol. 24, 949–968 (2003). [Google Scholar]
- 20.Richard A., Intra-specific variation in the social organization and ecology of Propithecus verreaux. Folia Primatol. 22, 178–207 (1974). [DOI] [PubMed] [Google Scholar]
- 21.Yamashita N., Chemical properties of the diets of two lemur species in southwestern Madagascar. Int. J. Primatol. 29, 339–364 (2008). [Google Scholar]
- 22.Sato H., Santini L., Patel E. R., Campera M., Yamashita N., Colquhoun I. C., Donati G., Dietary flexibility and feeding strategies of Eulemur: A comparison with Propithecus. Int. J. Primatol. 37, 109–129 (2016). [Google Scholar]
- 23.Quéméré E., Hibert F., Miquel C., Lhuillier E., Rasolondraibe E., Champeau J., Rabarivola C., Nusbaumer L., Chatelain C., Gautier L., Ranirison P., Crouau-Roy B., Taberlet P., Chikhi L., A DNA metabarcoding study of a primate dietary diversity and plasticity across its entire fragmented range. PLOS ONE 8, e58971 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salmona J., Jan F., Rasolondraibe E., Besolo A., Ousseni D. S., Beck A., Zaranaina R., Rakotoarisoa H., Rabarivola C. J., Chikhi L., Extensive survey of the endangered Coquerel’s sifaka Propithecus coquereli. Endanger. Species Res. 25, 175–183 (2014). [Google Scholar]
- 25.E. Louis, T. Sefczek, C. Bailey, B. Raharivololona, R. Lewis, E. Rakotomalala, Propithecus verreauxi, IUCN Red List of Threatened Species 2020: e.T18354A115572044. IUCN Red List Threatened Species (2020); 10.2305/IUCN.UK.2020-2.RLTS.T18354A115572044.en. [DOI]
- 26.B. Semel, M. Semel, J. Salmona, R. Heriniaina, Propithecus tattersalli, The IUCN Red List of Threatened Species 2020: e.T18352A115571806. IUCN Red List Threatened Species (2020); 10.2305/IUCN.UK.2020-2.RLTS.T18352A115571806.en. [DOI]
- 27.M. Irwin, Propithecus diadema, The IUCN Red List of Threatened Species 2020: e.T18358A115572884. IUCN Red List Threatened Species (2020); 10.2305/IUCN.UK.2020-2.RLTS.T18358A115572884.en. [DOI]
- 28.dos Reis M., Gunnell G. F., Barba-Montoya J., Wilkins A., Yang Z., Yoder A. D., Using phylogenomic data to explore the effects of relaxed clocks and calibration strategies on divergence time estimation: Primates as a test case. Syst. Biol. 67, 594–615 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harper G. J., Steininger M. K., Tucker C. J., Juhn D., Hawkins F., Fifty years of deforestation and forest fragmentation in Madagascar. Environ. Conserv. 34, 325–333 (2007). [Google Scholar]
- 30.Bailey C. A., Mc Lain A. T., Paquette S., McGuire S. M., Shore G. D., Lei R., Randriamanana J. C., Rabekinaja J. D., Rakotoarisoa G., Razafindrakoto A., Brenneman R., Hawkins M. T. R., Louis E., Evaluating the genetic diversity of three endangered lemur species (Genus: Propithecus) from northern Madagascar. J. Primatol. 5, 132 (2015). [Google Scholar]
- 31.Brenneman R. A., Johnson S. E., Bailey C. A., Ingraldi C., Delmore K. E., Wyman T. M., Andriamaharoa H. E., Ralainasolo F. B., Ratsimbazafy J. H., Louis E. E., Population genetics and abundance of the Endangered grey-headed lemur Eulemur cinereicepsin south-east Madagascar: Assessing risks for fragmented and continuous populations. Oryx 46, 298–307 (2012). [Google Scholar]
- 32.Razakamaharavo V. R., McGuire S. M., Vasey N., Louis E. E. Jr., Brenneman R. A., Genetic architecture of two red ruffed lemur (Varecia rubra) populations of Masoala National Park. Primates 51, 53–61 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Salmona J., Teixeira H., Rasolondraibe E., Aleixo-Pais I., Kun-Rodrigues C., Rakotonanahary A. N., Jan F., Rabarivola C. J., Zaonarivelo J. R., Andriaholinirina N. V., Chikhi L., Genetic diversity, population size, and conservation of the critically endangered Perrier’s sifaka (Propithecus perrieri). Int. J. Primatol. 36, 1132–1153 (2015). [Google Scholar]
- 34.Salmona J., Heller R., Quéméré E., Chikhi L., Climate change and human colonization triggered habitat loss and fragmentation in Madagascar. Mol. Ecol. 26, 5203–5222 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Zhou X., Wang B., Pan Q., Zhang J., Kumar S., Sun X., Liu Z., Pan H., Lin Y., Liu G., Zhan W., Li M., Ren B., Ma X., Ruan H., Cheng C., Wang D., Shi F., Hui Y., Tao Y., Zhang C., Zhu P., Xiang Z., Jiang W., Chang J., Wang H., Cao Z., Jiang Z., Li B., Yang G., Roos C., Garber P. A., Bruford M. W., Li R., Li M., Whole-genome sequencing of the snub-nosed monkey provides insights into folivory and evolutionary history. Nat. Genet. 46, 1303–1310 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Johnson R. N., O’Meally D., Chen Z., Etherington G. J., Ho S. Y. W., Nash W. J., Grueber C. E., Cheng Y., Whittington C. M., Dennison S., Peel E., Haerty W., O’Neill R. J., Colgan D., Russell T. L., Alquezar-Planas D. E., Attenbrow V., Bragg J. G., Brandies P. A., Chong A. Y.-Y., Deakin J. E., Palma F. D., Duda Z., Eldridge M. D. B., Ewart K. M., Hogg C. J., Frankham G. J., Georges A., Gillett A. K., Govendir M., Greenwood A. D., Hayakawa T., Helgen K. M., Hobbs M., Holleley C. E., Heider T. N., Jones E. A., King A., Madden D., Graves J. A. M., Morris K. M., Neaves L. E., Patel H. R., Polkinghorne A., Renfree M. B., Robin C., Salinas R., Tsangaras K., Waters P. D., Waters S. A., Wright B., Wilkins M. R., Timms P., Belov K., Adaptation and conservation insights from the koala genome. Nat. Genet. 50, 1102–1111 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu Y., Wu Q., Ma S., Ma T., Shan L., Wang X., Nie Y., Ning Z., Yan L., Xiu Y., Wei F., Comparative genomics reveals convergent evolution between the bamboo-eating giant and red pandas. Proc. Natl. Acad. Sci. U.S.A. 114, 1081–1086 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.E. Lauterbur, thesis, Stony Brook University, Stony Brook, NY (2019). [Google Scholar]
- 39.N. Rowe, M. Myers, All the World’s Primates (Pogonias Press, 2016). [Google Scholar]
- 40.Simmen B., Bayart F., Marez A., Hladik A., Diet, nutritional ecology, and birth season of Eulemur macaco in an anthropogenic forest in Madagascar. Int. J. Primatol. 28, 1253–1266 (2007). [Google Scholar]
- 41.Dammhahn M., Kappeler P. M., Comparative feeding ecology of sympatric Microcebus berthae and M. murinus. Int. J. Primatol. 29, 1567–1589 (2008). [Google Scholar]
- 42.Waterhouse R. M., Seppey M., Simão F. A., Manni M., Ioannidis P., Klioutchnikov G., Kriventseva E. V., Zdobnov E. M., BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 35, 543–548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H., Durbin R., Inference of human population history from individual whole-genome sequences. Nature 475, 493–496 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mazet O., Rodríguez W., Grusea S., Boitard S., Chikhi L., On the importance of being structured: Instantaneous coalescence rates and human evolution—Lessons for ancestral population size inference? Heredity 116, 362–371 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lawler R. R., Testing for a historical population bottleneck in wild Verreaux’s sifaka (Propithecus verreauxi verreauxi) using microsatellite data. Am. J. Primatol. 70, 990–994 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Lawler R. R., Caswell H., Richard A. F., Ratsirarson J., Dewar R. E., Schwartz M., Demography of Verreaux’s sifaka in a stochastic rainfall environment. Oecologia 161, 491–504 (2009). [DOI] [PubMed] [Google Scholar]
- 47.Gonzalez F. J., Nebert D. W., Evolution of the P450 gene superfamily. Trends Genet. 6, 182–186 (1990). [DOI] [PubMed] [Google Scholar]
- 48.Koepsell H., Lips K., Volk C., Polyspecific organic cation transporters: Structure, function, physiological roles, and biopharmaceutical implications. Pharm. Res. 24, 1227–1251 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Jonker J. W., Buitelaar M., Wagenaar E., van der Valk M. A., Scheffer G. L., Scheper R. J., Plösch T., Kuipers F., Oude Elferink R. P. J., Rosing H., Beijnen J. H., Schinkel A. H., The breast cancer resistance protein protects against a major chlorophyll-derived dietary phototoxin and protoporphyria. Proc. Natl. Acad. Sci. U.S.A. 99, 15649–15654 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guillemette C., Pharmacogenomics of human UDP-glucuronosyltransferase enzymes. Pharm. J. 3, 136–158 (2003). [DOI] [PubMed] [Google Scholar]
- 51.Zhang J., Parallel adaptive origins of digestive RNases in Asian and African leaf monkeys. Nat. Genet. 38, 819–823 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Schienman J. E., Holt R. A., Auerbach M. R., Stewart C.-B., Duplication and divergence of 2 distinct pancreatic ribonuclease genes in leaf-eating African and Asian colobine monkeys. Mol. Biol. Evol. 23, 1465–1479 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Zhang J., Zhang Y., Rosenberg H. F., Adaptive evolution of a duplicated pancreatic ribonuclease gene in a leaf-eating monkey. Nat. Genet. 30, 411–415 (2002). [DOI] [PubMed] [Google Scholar]
- 54.Perry G. H., Reeves D., Melsted P., Ratan A., Miller W., Michelini K., Louis E. E. Jr., Pritchard J. K., Mason C. E., Gilad Y., A genome sequence resource for the aye-aye (Daubentonia madagascariensis), a nocturnal lemur from Madagascar. Genome Biol. Evol. 4, 126–135 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sgarlata G. M., Salmona J., Razanaparany T. P., Joseph C., Rabarivola F. J., Rasolondraibe E., Andriaholinirina N. V., Lafosse S., Chikhi L., Manni F., Mitochondrial genetic diversity in the crowned sifaka (Propithecus coronatus) in a fragmented landscape. Primate Conserv. 30, 39–57 (2016). [Google Scholar]
- 56.Quéméré E., Crouau-Roy B., Rabarivola C., Louis E. E. Jr., Chikhi L., Landscape genetics of an endangered lemur (Propithecus tattersalli) within its entire fragmented range. Mol. Ecol. 19, 1606–1621 (2010). [DOI] [PubMed] [Google Scholar]
- 57.Watterson G. A., The homozygosity test after a change in population size. Genetics 112, 899–907 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Epps C. W., Keyghobadi N., Landscape genetics in a changing world: Disentangling historical and contemporary influences and inferring change. Mol. Ecol. 24, 6021–6040 (2015). [DOI] [PubMed] [Google Scholar]
- 59.Meyer W. K., Venkat A., Kermany A. R., van de Geijn B., Zhang S., Przeworski M., Evolutionary history inferred from the de novo assembly of a nonmodel organism, the blue-eyed black lemur. Mol. Ecol. 24, 4392–4405 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Williams R. C., Blanco M. B., Poelstra J. W., Hunnicutt K. E., Comeault A. A., Yoder A. D., Conservation genomic analysis reveals ancient introgression and declining levels of genetic diversity in Madagascar’s hibernating dwarf lemurs. Heredity 124, 236–251 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hawkins M. T. R., Culligan R. R., Frasier C. L., Dikow R. B., Hagenson R., Lei R., Louis E. E., Genome sequence and population declines in the critically endangered greater bamboo lemur (Prolemur simus) and implications for conservation. BMC Genomics 19, 445 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Quéméré E., Amelot X., Pierson J., Crouau-Roy B., Chikhi L., Genetic data suggest a natural prehuman origin of open habitats in northern Madagascar and question the deforestation narrative in this region. Proc. Natl. Acad. Sci. 109, 13028–13033 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scholz C. A., Cohen A. S., Johnson T. C., King J., Talbot M. R., Brown E. T., Scientific drilling in the Great Rift Valley: The 2005 Lake Malawi Scientific Drilling Project—An overview of the past 145,000 years of climate variability in Southern Hemisphere East Africa. Palaeogeogr. Palaeoclimatol. Palaeoecol. 303, 3–19 (2011). [Google Scholar]
- 64.Hansford J., Wright P. C., Rasoamiaramanana A., Pérez V. R., Godfrey L. R., Errickson D., Thompson T., Turvey S. T., Early Holocene human presence in Madagascar evidenced by exploitation of avian megafauna. Sci. Adv. 4, eaat6925 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Virah-Sawmy M., Willis K. J., Gillson L., Evidence for drought and forest declines during the recent megafaunal extinctions in Madagascar. J. Biogeogr. 37, 506–519 (2010). [Google Scholar]
- 66.C. R. Campbell, G. P. Tiley, J. W. Poelstra, K. E. Hunnicutt, P. A. Larsen, M. dos Reis, A. D. Yoder, Pedigree-based measurement of the de novo mutation rate in the gray mouse lemur reveals a high mutation rate, few mutations in CpG sites, and a weak sex bias. bioRxiv 724880 [Preprint]. 5 August 2019. 10.1101/724880. [DOI]
- 67.C. Chen-Kraus, thesis, Yale University (2020). [Google Scholar]
- 68.Baden A. L., Mancini A. N., Federman S., Holmes S. M., Johnson S. E., Kamilar J., Louis E. E., Bradley B. J., Anthropogenic pressures drive population genetic structuring across a Critically Endangered lemur species range. Sci. Rep. 9, 16276 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Herrera J. P., Wright P. C., Lauterbur E., Ratovonjanahary L., Taylor L. L., The effects of habitat disturbance on lemurs at Ranomafana National Park, Madagascar. Int. J. Primatol. 32, 1091–1108 (2011). [Google Scholar]
- 70.Eppley T. M., Santini L., Tinsman J. C., Donati G., Do functional traits offset the effects of fragmentation? The case of large-bodied diurnal lemur species. Am. J. Primatol. 82, e23104 (2020). [DOI] [PubMed] [Google Scholar]
- 71.Craul M., Chikhi L., Sousa V., Olivieri G. L., Rabesandratana A., Zimmermann E., Radespiel U., Influence of forest fragmentation on an endangered large-bodied lemur in northwestern Madagascar. Biol. Conserv. 142, 2862–2871 (2009). [Google Scholar]
- 72.J. C. Bicca-Marques, in Primates in Fragments (Springer, 2003), pp. 283–303. [Google Scholar]
- 73.Rimbach R., Link A., Heistermann M., Gómez-Posada C., Galvis N., Heymann E. W., Effects of logging, hunting, and forest fragment size on physiological stress levels of two sympatric ateline primates in Colombia. Conserv. Physiol. 1, cot031 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ganzhorn J. U., Low-level forest disturbance effects on primary production, leaf chemistry, and lemur populations. Ecology 76, 2084–2096 (1995). [Google Scholar]
- 75.Brooks T. M., Pimm S. L., Oyugi J. O., Time lag between deforestation and bird extinction in tropical forest fragments. Conserv. Biol. 13, 1140–1150 (1999). [Google Scholar]
- 76.Worman C. O., Chapman C. A., Densities of two frugivorous primates with respect to forest and fragment tree species composition and fruit availability. Int. J. Primatol. 27, 203–225 (2006). [Google Scholar]
- 77.Chapman C., Balcomb S. B., Gillespie T. R., Skorupa J., Struhsaker T. T., Long-term effects of logging on African primate communities: A 28-year comparison from Kibale National Park, Uganda. Conserv. Biol. 14, 207–217 (2000). [Google Scholar]
- 78.Irwin M. T., Feeding ecology of Propithecus diadema in forest fragments and continuous forest. Int. J. Primatol. 29, 95–115 (2008). [Google Scholar]
- 79.Irwin M. T., Junge R. E., Raharison J.-L., Samonds K. E., Variation in physiological health of diademed sifakas across intact and fragmented forest at Tsinjoarivo, eastern Madagascar. Am. J. Primatol. 72, 1013–1025 (2010). [DOI] [PubMed] [Google Scholar]
- 80.Irwin M. T., Samonds K. E., Raharison J.-L., Junge R. E., Mahefarisoa K. L., Rasambainarivo F., Godfrey L. R., Glander K. E., Morphometric signals of population decline in diademed sifakas occupying degraded rainforest habitat in Madagascar. Sci. Rep. 9, 8776 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Anania A., Salmona J., Rasolondraibe E., Jan F., Chikhi L., Fichtel C., Kappeler P. M., Rasoloarison R., Taboo adherence and presence of Perrier’s sifaka (Propithecus perrieri) in Andrafiamena forest. Madag. Conserv. Dev. 13, 6–14 (2018). [Google Scholar]
- 82.Randrianandrianina F. H., Racey P. A., Jenkins R. K. B., Hunting and consumption of mammals and birds by people in urban areas of western Madagascar. Oryx 44, 411–415 (2010). [Google Scholar]
- 83.R. A. Mittermeier, E. E. Louis Jr, M. J. Richardson, C. Schwitzer, O. Langrand, A. B. Rylands, F. Hawkins, S. Rajaobelina, J. Ratsimbazafy, R. Rasoloarison, Lemurs of Madagascar (Conservation International, 2010). [Google Scholar]
- 84.Razafimanahaka J. H., Jenkins R. K., Andriafidison D., Randrianandrianina F., Rakotomboavonjy V., Keane A., Jones J. P., Novel approach for quantifying illegal bushmeat consumption reveals high consumption of protected species in Madagascar. Oryx 46, 584–592 (2012). [Google Scholar]
- 85.Jenkins R. K., Keane A., Rakotoarivelo A. R., Rakotomboavonjy V., Randrianandrianina F. H., Razafimanahaka H. J., Ralaiarimalala S. R., Jones J. P., Analysis of patterns of bushmeat consumption reveals extensive exploitation of protected species in eastern Madagascar. PLOS ONE 6, e27570 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kun-Rodrigues C., Salmona J., Besolo A., Rasolondraibe E., Rabarivola C., Marques T. A., Chikhi L., New density estimates of a threatened sifaka species (Propithecus coquereli) in Ankarafantsika National Park. Am. J. Primatol. 76, 515–528 (2014). [DOI] [PubMed] [Google Scholar]
- 87.Parreira B. R., Chikhi L., On some genetic consequences of social structure, mating systems, dispersal, and sampling. Proc. Natl. Acad. Sci. U.S.A. 112, E3318–E3326 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Richard A. F., Rakotomanga P., Schwartz M., Dispersal by Propithecus verreauxi at Beza Mahafaly, Madagascar: 1984-1991. Am. J. Primatol. 30, 1–20 (1993). [DOI] [PubMed] [Google Scholar]
- 89.Lawler R. R., Richard A. F., Riley M. A., Genetic population structure of the white sifaka (Propithecus verreauxi verreauxi) at Beza Mahafaly Special Reserve, southwest Madagascar (1992–2001). Mol. Ecol. 12, 2307–2317 (2003). [DOI] [PubMed] [Google Scholar]
- 90.Parreira B., Quéméré E., Vanpé C., Carvalho I., Chikhi L., Genetic consequences of social structure in the golden-crowned sifaka. Heredity 125, 328–339 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nabokina S. M., Inoue K., Subramanian V. S., Valle J. E., Yuasa H., Said H. M., Molecular identification and functional characterization of the human colonic thiamine pyrophosphate transporter. J. Biol. Chem. 289, 4405–4416 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hediger M. A., Clémençon B., Burrier R. E., Bruford E. A., The ABCs of membrane transporters in health and disease (SLC series): Introduction. Mol. Asp. Med. 34, 95–107 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Takanaga H., Mackenzie B., Suzuki Y., Hediger M. A., Identification of mammalian proline transporter SIT1 (SLC6A20) with characteristics of classical system imino. J. Biol. Chem. 280, 8974–8984 (2005). [DOI] [PubMed] [Google Scholar]
- 94.Milton K., Physiological ecology of howlers (Alouatta): Energetic and digestive considerations and comparison with the Colobinae. Int. J. Primatol. 19, 513–548 (1998). [Google Scholar]
- 95.B. Simmen, A. Hladik, P. L. Ramasiarisoa, S. Iaconelli, C. M. Hladik, in New Directions in Lemur Studies, B. Rakotosamimanana, H. Rasamimanana, J. U. Ganzhorn, S. M. Goodman, Eds. (Springer US, 1999), pp. 201–219. [Google Scholar]
- 96.Dearing M. D., Kohl K. D., Beyond fermentation: Other important services provided to endothermic herbivores by their gut microbiota. Integr. Comp. Biol. 57, 723–731 (2017). [DOI] [PubMed] [Google Scholar]
- 97.Greene L. K., McKenney E. A., O’Connell T. M., Drea C. M., The critical role of dietary foliage in maintaining the gut microbiome and metabolome of folivorous sifakas. Sci. Rep. 8, 14482 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Behrens M., Meyerhof W., Vertebrate bitter taste receptors: Keys for survival in changing environments. J. Agric. Food Chem. 66, 2204–2213 (2018). [DOI] [PubMed] [Google Scholar]
- 99.Thomas A., Sulli C., Davidson E., Berdougo E., Phillips M., Puffer B. A., Paes C., Doranz B. J., Rucker J. B., The bitter taste receptor TAS2R16 achieves high specificity and accommodates diverse glycoside ligands by using a two-faced binding pocket. Sci. Rep. 7, 7753 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Di Pizio A., Ben Shoshan-Galeczki Y., Hayes J. E., Niv M. Y., Bitter and sweet tasting molecules: It’s complicated. Neurosci. Lett. 700, 56–63 (2019). [DOI] [PubMed] [Google Scholar]
- 101.Jeon T.-I., Seo Y.-K., Osborne T. F., Gut bitter taste receptor signalling induces ABCB1 through a mechanism involving CCK. Biochem. J. 438, 33–37 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Janiak M. C., Burrell A. S., Orkin J. D., Disotell T. R., Duplication and parallel evolution of the pancreatic ribonuclease gene (RNASE1) in folivorous non-colobine primates, the howler monkeys (Alouatta spp.). Sci. Rep. 9, 20366 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.C. M. Hladik, P. Charles-Dominique, in Prosimian Biology, R. Martin, G. Doyle, A. Walker, Eds. (Duckworth, 1974), pp. 25–37. [Google Scholar]
- 104.Porcelli D., Butlin R. K., Gaston K. J., Joly D., Snook R. R., The environmental genomics of metazoan thermal adaptation. Heredity 114, 502–514 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tigano A., Colella J. P., MacManes M. D., Comparative and population genomics approaches reveal the basis of adaptation to deserts in a small rodent. Mol. Ecol. 29, 1300–1314 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sonna L. A., Fujita J., Gaffin S. L., Lilly C. M., Invited Review: Effects of heat and cold stress on mammalian gene expression. J. Appl. Physiol. 92, 1725–1742 (2002). [DOI] [PubMed] [Google Scholar]
- 107.Lawler R. R., Richard A. F., Riley M. A., Characterization and screening of microsatellite loci in a wild lemur population (Propithecus verreauxi verreauxi). Am. J. Primatol. 55, 253–259 (2001). [DOI] [PubMed] [Google Scholar]
- 108.Gnerre S., Callum I. M., Przybylski D., Ribeiro F. J., Burton J. N., Walker B. J., Sharpe T., Hall G., Shea T. P., Sykes S., Berlin A. M., Aird D., Costello M., Daza R., Williams L., Nicol R., Gnirke A., Nusbaum C., Lander E. S., Jaffe D. B., High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc. Natl. Acad. Sci. U.S.A. 108, 1513–1518 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M. A., The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Danecek P., Auton A., Abecasis G., Albers C. A., Banks E., DePristo M. A., Handsaker R. E., Lunter G., Marth G. T., Sherry S. T., McVean G., Durbin R.; 1000 Genomes Project Analysis Group , The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Huerta-Cepas J., Dopazo J., Gabaldón T., ETE: A python Environment for Tree Exploration. BMC Bioinformatics. 11, 24 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Arnold C., Matthews L. J., Nunn C. L., The 10kTrees website: A new online resource for primate phylogeny. Evol. Anthropol. Issues News Rev. 19, 114–118 (2010). [Google Scholar]
- 113.De Bie T., Cristianini N., Demuth J. P., Hahn M. W., CAFE: A computational tool for the study of gene family evolution. Bioinformatics 22, 1269–1271 (2006). [DOI] [PubMed] [Google Scholar]
- 114.Prado-Martinez J., Sudmant P. H., Kidd J. M., Li H., Kelley J. L., Lorente-Galdos B., Veeramah K. R., Woerner A. E., O’Connor T. D., Santpere G., Cagan A., Theunert C., Casals F., Laayouni H., Munch K., Hobolth A., Halager A. E., Malig M., Hernandez-Rodriguez J., Hernando-Herraez I., Prüfer K., Pybus M., Johnstone L., Lachmann M., Alkan C., Twigg D., Petit N., Baker C., Hormozdiari F., Fernandez-Callejo M., Dabad M., Wilson M. L., Stevison L., Camprubí C., Carvalho T., Ruiz-Herrera A., Vives L., Mele M., Abello T., Kondova I., Bontrop R. E., Pusey A., Lankester F., Kiyang J. A., Bergl R. A., Lonsdorf E., Myers S., Ventura M., Gagneux P., Comas D., Siegismund H., Blanc J., Agueda-Calpena L., Gut M., Fulton L., Tishkoff S. A., Mullikin J. C., Wilson R. K., Gut I. G., Gonder M. K., Ryder O. A., Hahn B. H., Navarro A., Akey J. M., Bertranpetit J., Reich D., Mailund T., Schierup M. H., Hvilsom C., Andrés A. M., Wall J. D., Bustamante C. D., Hammer M. F., Eichler E. E., Marques-Bonet T., Great ape genetic diversity and population history. Nature 499, 471–475 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fan Z., Zhao G., Li P., Osada N., Xing J., Yi Y., Du L., Silva P., Wang H., Sakate R., Zhang X., Xu H., Yue B., Li J., Whole-genome sequencing of Tibetan macaque (Macaca thibetana) provides new insight into the macaque evolutionary history. Mol. Biol. Evol. 31, 1475–1489 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Robinson J. A., Ortega-Del Vecchyo D., Fan Z., Kim B. Y., vonHoldt B. M., Marsden C. D., Lohmueller K. E., Wayne R. K., Genomic flatlining in the endangered island fox. Curr. Biol. 26, 1183–1189 (2016). [DOI] [PubMed] [Google Scholar]
- 117.Prüfer K., Racimo F., Patterson N., Jay F., Sankararaman S., Sawyer S., Heinze A., Renaud G., Sudmant P. H., de Filippo C., Li H., Mallick S., Dannemann M., Fu Q., Kircher M., Kuhlwilm M., Lachmann M., Meyer M., Ongyerth M., Siebauer M., Theunert C., Tandon A., Moorjani P., Pickrell J., Mullikin J. C., Vohr S. H., Green R. E., Hellmann I., Johnson P. L. F., Blanche H., Cann H., Kitzman J. O., Shendure J., Eichler E. E., Lein E. S., Bakken T. E., Golovanova L. V., Doronichev V. B., Shunkov M. V., Derevianko A. P., Viola B., Slatkin M., Reich D., Kelso J., Pääbo S., The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505, 43–49 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.H. Li, Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997 [q-bio.GN] (16 March 2013).
- 119.Faust G. G., Hall I. M., SAMBLASTER: Fast duplicate marking and structural variant read extraction. Bioinformatics 30, 2503–2505 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cingolani P., Patel V. M., Coon M., Nguyen T., Land S. J., Ruden D. M., Lu X., A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly 6, 1–12 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yang Z., PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- 124.A. Dabney, J. D. Storey, G. R. Warnes, qvalue: Q-value estimation for false discovery rate control. R Package Version (2010).
- 125.Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z., GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10, 48 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Supek F., Bošnjak M., Škunca N., Šmuc T., REVIGO summarizes and visualizes long lists of gene ontology terms. PLOS ONE 6, e21800 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Smedley D., Haider S., Ballester B., Holland R., London D., Thorisson G., Kasprzyk A., BioMart—Biological queries made easy. BMC Genomics 10, 22 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Enright A. J., Van Dongen S., Ouzounis C. A., An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30, 1575–1584 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kumar S., Stecher G., Suleski M., Hedges S. B., TimeTree: A resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 34, 1812–1819 (2017). [DOI] [PubMed] [Google Scholar]
- 130.Hansen R. L., Carr M. M., Apanavicius C. J., Jiang P., Bissell H. A., Gocinski B. L., Maury F., Himmelreich M., Beard S., Ouellette J. R., Kouba A. J., Seasonal shifts in giant panda feeding behavior: Relationships to bamboo plant part consumption. Zoo Biol. 29, 470–483 (2009). [DOI] [PubMed] [Google Scholar]
- 131.Wei F., Feng Z., Wang Z., Zhou A., Hu J., Use of the nutrients in bamboo by the red panda (Ailurus fulgens). J. Zool. 248, 535–541 (1999). [Google Scholar]
- 132.Melzer A., Baudry C., Kadiri M., Ellis W., Tree use, feeding activity and diet of koalas on St Bees Island, Queensland. Aust. Zool. 35, 870–875 (2011). [Google Scholar]
- 133.Wang R. J., Thomas G. W., Raveendran M., Harris A. R., Doddapaneni H., Muzny D. M., Capitanio J. P., Radivojac P., Rogers J., Hahn M. W., Paternal age in rhesus macaques is positively associated with germline mutation accumulation but not with measures of offspring sociability. Genome Res. 30, 826–834 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/17/eabd2274/DC1


