1. Introduction
On March 11, 2020, the World Health Organization declared Coronavirus Disease 2019 (COVID-19) a global pandemic [1]. As more information is collected regarding the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), it has become clear that patients with certain comorbidities, such as hypertension and diabetes mellitus, are more likely to develop severe COVID-19 complications [2]. In particular, patients with hematologic malignancies (HM) are uniquely immunocompromised and considered at high risk for COVID-19. It has been reported that patients with HM had mortality rates as high as 37 %, and when compared across all age groups, patients with HM demonstrated worse overall survival than the general population of their corresponding age group [3]. However, data regarding the diagnosis of COVID-19 in these patients is sparse, and in particular, the ability to successfully detect SARS-CoV-2 in patients with HM remains unknown. We have previously reported two cases of allogeneic (SCT) diagnosed with COVID-19 using clustered regularly interspaced short palindromic repeats (CRISPR) technique, following multiple negative nasopharyngeal RT-PCR testing [4]. Here, we examine 53 patients with a variety of HM with high suspicion for COVID-19 based on clinical presentation, lab results, and imaging, who were tested with CRISPR and/or RT-PCR based techniques.
2. Results
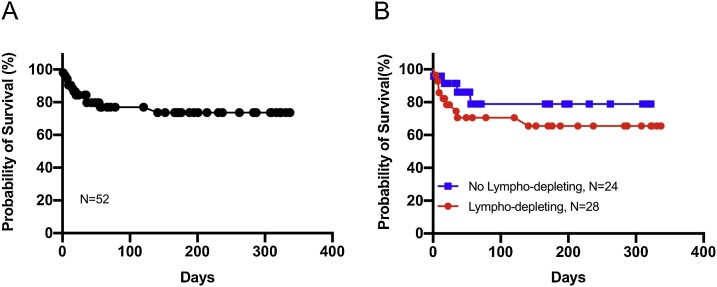
From 3/31/20 to 1/18/21, 53 patients (age 24–86) with a variety of HM (35 lymphoid, 18 myeloid; Table 1 ), were evaluated for COVID-19. Twenty-four patients were undergoing active chemotherapy, 12 had received an autologous SCT, six had received an allogeneic SCT, and 11 were on surveillance. Forty-seven patients presented symptomatic with an undiagnosed respiratory illness, six presented asymptomatic for testing prior to scheduled treatment. After initial testing, 40 patients tested positive for COVID-19 with guideline-directed nasopharyngeal RT-PCR testing (including the six asymptomatic patients), while 13 patients tested negative with the same technique. However, based on their clinical history, imaging, and disease course, concern for COVID-19 infection remained high in these 13 patients. After repeat nasopharyngeal RT-PCR tests, CRISPR technology [5] available at our institution, as previously described [4], was used to test eight patients who had initially tested negative by RT-PCR. Surprisingly, seven of the eight patients tested positive for COVID-19 with either a blood/serum, nasopharyngeal swab, urine, or saliva, for the SARS-CoV-2 specific N gene and/or ORF1ab gene (Supplemental Table 1). Excluding the patients who were negative by RT-PCR and not tested by CRISPR, the rate of false negativity with RT-PCR testing was found to be significantly elevated at 15 % (7/48) in our cohort. In addition, a high mortality rate was observed with 12 out of 53 patients (23 %) ultimately expiring, and an overall survival (OS) rate of 75 % (Fig. 1 A). Interestingly, the majority of deaths occurred within the first 25 days of diagnosis. By comparing survival of patients who did receive lympho-depleting chemotherapy within 30 days of their COVID-19 diagnosis to that of patients who did not, we noted a trend toward worse OS in the former group (Fig. 1B), although this difference was not found to be statistically significant (P = 0.268).
Table 1.
Demographics, Hematologic malignancy, chemotherapy regimen prior to diagnosis, stem cell transplant status, and absolute lymphocyte count at diagnosis.
| Patient | Demographics | Hematologic Malignancy | Chemotherapy Regimen (within 30 days of diagnosis) | Transplant status | Absolute Lymphocyte count at diagnosis |
|---|---|---|---|---|---|
| Patient-1 | 75 M | Multiple Myeloma | melphalan | Autologous SCT | 870 |
| Patient-2 | 67F | AML | None | Allogeneic SCT | N/A |
| Patient-3 | 65M | Atypical CML | hydroxyurea & ruxolitinib | None | N/A |
| Patient-4 | 30M | AML | busulfan & fludarabine | Allogeneic SCT | 380 |
| Patient-5 | 29M | Classical Hodgkin's Lymphoma | None | None | N/A |
| Patient-6 | 49M | CML | ponatinib | None | 160 |
| Patient-7 | 49F | B-Cell ALL | vincristine & dasatinib | None | 1050 |
| Patient-8 | 53F | B-Cell ALL | rituximab | Allogeneic SCT | 110 |
| Patient-9 | 65M | CLL | acalabrutinib | None | N/A |
| Patient-10 | 30F | AML | None | None | 800 |
| Patient-11 | 26F | CML | Negative for COVID-19 | None | 0 |
| Patient-12 | 42F | Classical Hodgkin's Lymphoma | None | None | N/A |
| Patient-13 | 86M | AML | imatinib | None | N/A |
| Patient-14 | 53F | Amyloidosis | bortezomib | Autologous SCT | N/A |
| Patient-15 | 67M | AML | ibrutinib | Allogeneic SCT | 330 |
| Patient-16 | 57F | CLL | ibrutinib | None | 2560 |
| Patient-17 | 38M | DLBCL | rituximab | Allogeneic SCT | N/A |
| Patient-18 | 35F | Classical Hodgkin's Lymphoma | None | None | 540 |
| Patient-19 | 59F | B-Cell ALL | rituximab, cytarabine, methotrexate, imatinib, | None | 530 |
| Patient-20 | 84F | CML | imatinib | None | 510 |
| Patient-21 | 71M | Multiple Myeloma | lenalidomide | Autologous SCT | N/A |
| Patient-22 | 64M | T-Cell ALL | venetoclax, cyclophosphamide, vincristine | None | 260 |
| Patient-23 | 24M | AML | None | None | 590 |
| Patient-24 | 61M | Multiple Myeloma | daratumumab | None | N/A |
| Patient-25 | 65M | CMML | decitabine | None | N/A |
| Patient-26 | 58F | Multiple Myeloma | bortezomib | Autologous SCT | N/A |
| Patient-27 | 45M | Follicular Lymphoma | rituximab & acalabrutinib | None | N/A |
| Patient-28 | 72F | Multiple Myeloma | None | Autologous SCT | 640 |
| Patient-29 | 52M | Classical Hodgkin's Lymphoma | None | None | N/A |
| Patient-30 | 55F | APML | None | None | 1820 |
| Patient-31 | 65F | MGUS | None | None | 980 |
| Patient-32 | 82M | Multiple Myeloma | daratumumab & lenalidomide | None | N/A |
| Patient-33 | 74M | Multiple Myeloma | lenalidomide | Autologous SCT | 400 |
| Patient-34 | 72M | Multiple Myeloma | lenalidomide | Autologous SCT | N/A |
| Patient-35 | 49F | CML | None | Allogeneic SCT | 2640 |
| Patient-36 | 24F | AML | None | None | N/A |
| Patient-37 | 65M | B-Cell ALL | rituximab, cytarabine, methotrexate, imatinib | None | N/A |
| Patient-38 | 54M | Multiple Myeloma | daratumumab & carfilzomib | None | 2430 |
| Patient-39 | 60M | Multiple Myeloma | bortezomib & lenalidomide | Autologous SCT | N/A |
| Patient-40 | 73M | Multiple Myeloma | lenalidomide | Autologous SCT | N/A |
| Patient-41 | 62M | APML | arsenic trioxide & all-trans retinoic acid | None | N/A |
| Patient-42 | 62F | T-Cell Lymphoma | None | Autologous SCT | N/A |
| Patient-43 | 55M | Amyloidosis | cyclophosphamide, bortezomib | None | 780 |
| Patient-44 | 59M | Follicular Lymphoma | bendamustine & obinatuzumab | None | 50 |
| Patient-45 | 78F | CML | dasatinib | None | 1090 |
| Patient-46 | 76F | MDS | lenalidomide | None | N/A |
| Patient-47 | 78M | T-Cell PLL | None | Autologous SCT | 70 |
| Patient-48 | 55F | Smoldering Multiple Myeloma | None | None | N/A |
| Patient-49 | 58F | CLL | None | None | N/A |
| Patient-50 | 56M | DLBCL | None | None | N/A |
| Patient-51 | 70F | DLBCL | rituximab, methotrexate, procarbazine, vincristine | None | 630 |
| Patient-52 | 81M | AML | decitabine & venetoclax | None | 270 |
| Patient-53 | 60M | Multiple Myeloma | daratumumab | Autologous SCT | N/A |
SCT, stem cell transplant; ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; CLL, chronic lymphocytic leukemia; CML, chronic myelogenous leukemia; DLBCL, diffuse large B-Cell lymphoma; MDS, myelodysplastic syndrome; PLL, prolymphocytic leukemia; NS, nodular sclerosing hodgkin's lymphoma; APML, acute promyelocytic leukemia; MGUS, monoclonal gammopathy of undetermined significance.
Fig. 1.
Kaplan Meier Survival Curve. (A) Kaplan Meier survival curve evaluating 52 patients with COVID-19. Overall survival was calculated to be 73 %. (B) Kaplan Meier survival curve evaluating patients who did not receive lympho-depleting chemotherapy within 30 days of their diagnosis (in blue), versus patients who did receive lympho-depleting chemotherapy within 30 days of their diagnosis (in red). P-value calculated to be 0.268.
Of the 47 COVID-19 positive patients (by RT-PCR or CRISPR), 12 patients received COVID-19-directed therapy with either hydroxychloroquine/azithromycin, remdesivir, and/or Covid-19 convalescent plasma (CCP) depending on their clinical status. Of the 12 treated patients, 11 initially improved while one patient expired. In the following months after their discharge, two additional patients who were treated also unfortunately died. For the five patients who were negative for RT-PCR with no CRISPR analyzed, one patient received hydroxychloroquine/azithromycin proactively due to clinical course and recovered, three patients expired at outside facilities due to unknown causes, and one patient was lost to follow up. Breakdown of testing and treatment is shown in Supplemental Fig. 1.
3. Discussion
Patient’s with HM are uniquely immunocompromised and at high-risk of developing severe complications from COVID-19. The majority of our patients had undergone SCT or were actively on chemotherapy, notably lymphodepleting chemotherapy. Associated with the fact that COVID-19 has been reported to cause a lymphopenia, specifically a B-cell lymphopenia [6], our patient’s symptoms and immune response to COVID-19 are likely to differ from that of immunocompetent hosts. This translated into an overall worse outcome as seen by the high mortality with our patients. In addition, our patients possess an altered immune system due to their underlying disease, as well as the treatments they have received, as many chemotherapy agents cause a lymphopenia which has been correlated with higher risk of in-hospital death and severity of COVID-19 infection [7]. Our study suggests that the ability to generate an adequate immune response is vital for the disease course of COVID-19, as our data revealed that while not statistically significant, there did appear to be a slight trend toward worse outcomes for patients who received lympho-depleting chemotherapy within 30 days of their diagnosis versus patients who did not receive lympho-depleting chemotherapy within 30 days of their diagnosis. Furthermore, we have previously reported that patients who received COVID-19 convalescent plasma, in essence a form of passive immunity, substantially improving [8]. This demonstrates that when patients have adept lymphocytes, they may be able to respond appropriately to the virus and improve their disease course, and if they have ineffective/absent lymphocytes, such as many of our patients, they may have worse outcomes.
It is thus imperative to establish the diagnosis of COVID-19 quickly, as faster initiation of treatment has been postulated to have better outcomes [9,10]. Eleven of the 12 patients who were subsequently diagnosed after their initial negative test substantially improved after treatment. However, as seen by our dataset, a strikingly high false negative rate was observed. Our study showed a 15 % false negative rate in our patient population (possibly higher as there was concern for COVID-19 in the five patients who were negative with RT-PCR but not tested with CRISPR). This compares slightly worse than the expected false negative rates of RT-PCR techniques [11]. The high false negative rate can be in part attributed to decreased viral shedding, as studies have shown patients who initially present with milder disease will have less viral shedding, and thus, a higher likelihood of a false negative test [12]. Therefore, our patients may have presented in an earlier disease state. Additionally, our patient’s altered immune systems may also be a big factor in the high false negative rate. Our patients may be severely immunocompromised that a small amount of virus may cause severe clinical effects but is not enough to be detected via nasopharyngeal RT-PCR and can only be detected by more sensitive assays such as the CRISPR test.
In conclusion, a high clinical suspicion must guide further workup and therapy in patients with HM who present with an undiagnosed respiratory illness consistent with COVID-19. Patients with HM can have a wide variety of presentations when infected with COVID-19. For this select patient population, we must establish an algorithm to diagnose COVID-19 efficiently as we reported a high number of initial false negative tests before the more sensitive CRISPR revealed a positive test. In addition, the patients who were CRISPR positive for Sars-CoV-2 tested positive in samples other than a nasopharyngeal source. This poses the question of whether patients with objective findings consistent with COVID-19 and a negative nasopharyngeal RT-PCR assay would benefit from testing of other tissue types, as COVID-19 has been isolated in other sources [13]. With a severe, undiagnosed respiratory illness, it may be appropriate to preemptively treat patients with HM for COVID-19 if there is high clinical suspicion, but guideline directed testing remains negative. Treatment pathways need to be instituted to not only treat COVID-19 infection, but also provide the best treatment for these patient’s underlying HM.
Authors contibutions
AN and NSS collected and analyzed data and wrote the manuscript, TH and BN performed the RT-PCR/CRISPR assay, all other authors contributed to treatment, reviewing, editing, writing of the manuscript.
Financial support
There was no funding for this project
Ethical approval
De-identified samples were obtained from patients following written informed consent in accordance with the Declaration of Helsinki overseen by the Institutional Review Board at Tulane University (IRB#M0600 and #2020−549). Leftover serum samples initially used for clinical care were tested from one patient following death and obtaining next of kin’s verbal consent.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgements
We thank our patients for participating and donating the tissue samples to make this research possible. We also thank the Louisiana Cancer Research Center (LCRC) biospecimen core for their help in consenting and sample collection process.
Footnotes
Presented in abstract form at the 62nd annual meeting of the American Society of Hematology, 6, December 2020, under abstract number 138611.
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.leukres.2021.106582.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Helmy Y.A., Fawzy M., Elaswad A., Sobieh A., Kenney S.P., Shehata A.A. The COVID-19 pandemic: a comprehensive review of taxonomy, genetics, epidemiology, diagnosis, treatment, and control. J. Clin. Med. 2020;9(4) doi: 10.3390/jcm9041225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanyaolu A., Okorie C., Marinkovic A., Patidar R., Younis K., Desai P., et al. Comorbidity and its impact on patients with COVID-19. SN Comprehens. Clin. Med. 2020;2(8):1069–1076. doi: 10.1007/s42399-020-00363-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Passamonti F., Cattaneo C., Arcaini L., Bruna R., Cavo M., Merli F., et al. Clinical characteristics and risk factors associated with COVID-19 severity in patients with haematological malignancies in Italy: a retrospective, multicentre, cohort study. Lancet Haematol. 2020;7(10) doi: 10.1016/S2352-3026(20)30251-9. e737-e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niu A., McDougal A., Ning B., Safa F., Luk A., Mushatt D.M., et al. COVID-19 in allogeneic stem cell transplant: high false-negative probability and role of CRISPR and convalescent plasma. Bone Marrow Transplant. 2020;55(12):2354–2356. doi: 10.1038/s41409-020-0972-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang Z., Tian D., Liu Y., Lin Z., Lyon C.J., Lai W., et al. Ultra-sensitive and high-throughput CRISPR-p owered COVID-19 diagnosis. Biosens. Bioelectron. 2020;164 doi: 10.1016/j.bios.2020.112316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin L., Lu L., Cao W., Li T. Hypothesis for potential pathogenesis of SARS-CoV-2 infection-a review of immune changes in patients with viral pneumonia. Emerg. Microbes Infect. 2020;9(1):727–732. doi: 10.1080/22221751.2020.1746199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu B., Fan C.-Y., Wang A.-L., Zou Y.-L., Yu Y-H He C., et al. Suppressed T cell-mediated immunity in patients with COVID-19: a clinical retrospective study in Wuhan, China. J. Infect. 2020 doi: 10.1016/j.jinf.2020.04.012. S0163-4453(20)30223-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pal P., Ibrahim M., Niu A., Zwezdaryk K.J., Tatje E., Robinson IV WR, et al. Safety and efficacy of COVID-19 convalescent plasma in severe pulmonary disease: A report of 17 patients. Transfusion Medicine.n/a(n/a). [DOI] [PubMed]
- 9.Grein J., Ohmagari N., Shin D., Diaz G., Asperges E., Castagna A., et al. Compassionate use of remdesivir for patients with severe Covid-19. N. Engl. J. Med. 2020;382(24):2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeng Q.L., Yu Z.J., Gou J.J., Li G.M., Ma S.H., Zhang G.F., et al. Effect of convalescent plasma therapy on viral shedding and survival in patients with Coronavirus disease 2019. J. Infect. Dis. 2020;222(1):38–43. doi: 10.1093/infdis/jiaa228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y., Yao L., Li J., Chen L., Song Y., Cai Z., et al. Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically diagnosed with COVID-19. J. Med. Virol. 2020;92(7):903–908. doi: 10.1002/jmv.25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng S., Fan J., Yu F., Feng B., Lou B., Zou Q., et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;369:m1443. doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323(18):1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.