Abstract
Heat shock proteins, in particular Hsp70, play a central role in proteostasis in eukaryotic cells. Due to its chaperone properties, Hsp70 is involved in various processes after stress and under normal physiological conditions. In contrast to mammals and many Diptera species, inducible members of the Hsp70 family in Drosophila are constitutively synthesized at a low level and undergo dramatic induction after temperature elevation or other forms of stress. In the courtship suppression paradigm used in this study, Drosophila males that have been repeatedly rejected by mated females during courtship are less likely than naive males to court other females. Although numerous genes with known function were identified to play important roles in long-term memory, there is, to the best of our knowledge, no direct evidence implicating Hsp70 in this process. To elucidate a possible role of Hsp70 in memory formation, we used D. melanogaster strains containing different hsp70 copy numbers, including strains carrying a deletion of all six hsp70 genes. Our investigations exploring the memory of courtship rejection paradigm demonstrated that a low constitutive level of Hsp70 is apparently required for learning and the formation of short and long-term memories in males. The performed transcriptomic studies demonstrate that males with different hsp70 copy numbers differ significantly in the expression of a few definite groups of genes involved in mating, reproduction, and immunity in response to rejection. Specifically, our analysis reveals several major pathways that depend on the presence of hsp70 in the genome and participate in memory formation and consolidation, including the cAMP signaling cascade.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12192-021-01203-7.
Keywords: Hsp70, Courtship suppression paradigm, Drosophila, Learning and memory, Transcriptome analysis
Introduction
Drosophila melanogaster has been employed as a model system to understand the genetic and molecular basis of learning and memory in eukaryotic organisms (Tully et al. 1990; Akalal et al. 2010; Mery et al. 2007; Zhuravlev et al. 2015). The use of different paradigms including a memory of courtship rejection approach in males (Kamyshev et al. 1999; Winbush et al. 2012; Gerstner and Yin 2010; Keleman et al. 2007) has provided insights into molecular mechanisms underlying the formation of long- and short-term memories (Tully et al. 1994; Beck et al. 2000; Davis 2005; Dubnau and Tully 1998; Griffith and Ejima 2009; Jones et al. 2018). Fruit flies have a memory system that permits sophisticated forms of learning and information processing necessary for survival and reproduction (Mery et al. 2007; Ishimoto et al. 2009). However, the numerous genes and signaling pathways discovered in flies to date (Akalal et al. 2011; Bozler et al. 2017; Kahsai and Zars 2011) apparently represent only a small number of the factors participating in learning and memory formation. In particular, various findings and observations suggest some interactions between memory formation and storage and very ancient stress–response systems including heat shock genes, innate immunity genes, and genes involved in the production of gaseous signaling molecules such as H2S (Bozler et al. 2017; Barajas-Azpeleta et al. 2018; Kuntz et al. 2017; Nagpure and Bian 2015; Snijder et al. 2016).
Heat shock proteins (Hsps) are at the heart of proteostasis in eukaryotic cells, given their essential roles in protein folding and degradation pathways (Hartl et al. 2011). Hsps participate in damaged protein degradation and clearing either by the UPS or autophagy (Carman et al. 2013; Ciechanover and Kwon 2017; Witt 2013; Wyatt et al. 2013). Under normal physiological conditions, Hsp70 binding to client proteins in the early stages of protein folding controls the formation of proper protein folding and transport of mature proteins, while inhibiting aggregate formation. Due to these chaperone properties, Hsp70 is involved in various processes including adaptation to stress (Evgen'ev et al. 2014), development and apoptosis (Kennedy et al. 2014; Kumar and Tiwari 2018), and probably learning and memory formation (Thekkuveettil and Lakhotia 1996; Gyurko et al. 2014). Heat shock treatment (HS) can restore memory in D. melanogaster mutant flies (“agnostic”) and mollusks, as well as several other eukaryotic organisms (Hooper et al. 2016; Porto et al. 2018; Savvateeva-Popova et al. 2017). There is abundant information concerning the involvement of individual Hsps induced by HS or administration of recombinant Hsps in memory formation and storage in various organisms (Kandel 2001; Foster et al. 2015; Sunada et al. 2016; Lukowiak et al. 2010). Furthermore, in our experiments, administration of recombinant Hsp70 by different means including intranasal injections has been found to ameliorate the cognitive impairments in mouse models of AD (Bobkova et al. 2014; Bobkova et al. 2015). Similarly, it has been shown that the expression of Hsp70 significantly improves memory in transgenic flies carrying Aβ42 oligomers (Martín-Peña et al. 2018). To this end, the heat shock transcription factor (HSF1), which induces the expression of hsp70 and other heat shock genes, plays the central role in synaptic fidelity and memory consolidation (Hooper et al. 2016; Murshid et al. 2010).
It is possible to speculate that during evolution, mechanisms underlying memory formation and the stress response were formed in parallel, and the observed simultaneous activation of Hsps synthesis and major neurotrophic factor (BDNF) production corroborates this speculation (Hooper et al. 2016; Zhao et al. 2015; Scaccianoce et al. 2003).
The cytosolic chaperone Hsp70 is evolutionarily conserved and represents one of the most abundant chaperones in mammalian cells. In contrast to mammals and many studied Diptera species, the major stress proteins in fruit flies belonging to the Hsp70 family under normal conditions are constitutively synthesized at very low levels and undergo dramatic induction immediately after heat shock and other stressful stimuli (Evgen'ev et al. 2014; Dahlgaard et al. 1998; Shilova et al. 2018). Using a “rejection” paradigm, we have previously demonstrated a severe memory deficit in the D. melanogaster strain (ts403) with abnormal function of the heat shock genes system (Evgen'ev et al. 1979). However, in this ts-mutant, the synthesis of all Hsps including Hsp70 was inhibited and we were not able to pin-point the observed effects to specific hsp genes.
Given these findings and observations, herein, we used D. melanogaster strains containing different hsp70 copy numbers, including strains with deletion of all six hsp70 genes. Our investigations exploring the mating rejection courtship paradigm demonstrated that a low constitutive level of Hsp70 present in the brain is required for the formation of short and long-term memories in male fruit flies. The transcriptomic studies revealed characteristic differences in the expression of a few definite groups of genes in response to rejection in the heads of males with different hsp70 copy numbers.
Materials and methods
Fly strains
All flies were reared on standard sugar–yeast–agar medium at 25°С, 60% humidity, and the light–dark cycle 12:12 h. The following strains were obtained from Bloomington Drosophila Stock Center: the original strain w1118 (Bloomington #6326) containing all six copies of hsp70; the derived strain w1118 (df(3R) Hsp70A, df(3R)Hsp70B), referred to as “hsp70-”, in which all the hsp70 genes were deleted by genetic manipulation (Bloomington #8841) (Gong and Golic 2004). We also used a derived w1118 strain which comprised four hsp70 copies located in the 87B region named “hsp70-4c” (Bloomington # 8842). We introduced into the “hsp70-” strain one hsp70 copy originating from the 87A locus via P-mediated transformation (Shilova et al. 2018) and designated this strain “hsp70-1c”. Since hsp70- flies were phenotypically white and it is known that the white mutation influences memory characteristics (Sitaraman et al. 2008; Anaka et al. 2008), the (w+/hsp70-) pCaSpeR5 vector plasmid carrying the mini-white gene was injected into preblastoderm embryos of 8841 strain to obtain an hsp70- strain with red eyes, as described previously (Astakhova et al. 2015). Therefore, we developed a strain lacking all hsp70 genes with normally pigmented eyes and used these red-eyed males in our studies as an additional control. The obtained strain was designated w+/hsp70-. We used the Canton-S strain, which carries all hsp70 genes, in the courtship experiments. This strain is routinely used as a control in all behavior tests in our laboratory.
Strain Elav GFP/FM7; Hsp70-/Hsp70- was constructed by several consequent crosses of # 108077 strain (Genotype P{GawB}elavC155, P{UAS-mCD8::GFP. L}Ptp4ELL4, P{hsFLP}1, w*/FM7c (short name Elav GFP/FM7)), and #8841 strain (genotype w[1118]; Df(3R)Hsp70A, Df(3R)Hsp70B (short name hsp70-)). In the resulted strain with deleted copies of hsp70, the GFP gene coding region is driven by the Elav gene promoter, expressed in neurons. Confocal microscopy was used to study the fluorescence of GFP in the brains isolated from third instar larvae and adult flies of this strain.
Transgenic strains containing various constructs were maintained under the same conditions as the abovementioned laboratory strains.
Heat shock treatment
Heat shock (HS) treatment was used to modulate the HS stress response. Behavioral patterns of flies were assessed after HS treatment was applied to 5-day-old adult flies 1 h before the test as described (Nikitina et al. 2003). The flies were subjected to heat shock in empty culture flasks immersed in a water bath for 30 min at 37°C.
Test for learning and memory of flies in a conditioned courtship suppression paradigm
To evaluate memory formation in Drosophila males, we used a conditioned courtship suppression paradigm (CCSP) (Kamyshev et al. 1999).
Males were collected and kept individually, and virgin wild-type Canton-S females were maintained in small groups (10 per vial) until 5 days of age. Before the day of the experiment, the females were allowed to mate overnight. The collection of naive flies was done at the same circadian time as the trained flies. The studies were performed on adult flies at the first half of the day by the temperature +25 ± 0.5°С.
For training, a naive male (with no previous courtship experience) was placed together with a 5-day-old fertilized Canton-S female for 30 min in the experimental chamber (test for learning and short-term memory) or 5 h in a beaker containing medium (volume of free space of approximately 3 cm3) (test for learning and long-term memory) according to an established protocol (McBride et al. 1999). In the retraining test, a trained male was placed in a fresh chamber together with another fertilized female.
To observe courtship, a male of a strain understudy and a fertilized 5-day-old wild-type female were introduced by shaking through a funnel into different halves of a Plexiglas experimental chamber (15 mm in diameter, 5 mm high) separated by a sliding opaque partition. After 45 s of adaptation, the partition was withdrawn, and the flies were left together. During 300 s of observation, the temporal sequence of behavioral elements was recorded using a specially designed program (Kamyshev et al. 1999). The elements related to courtship include orientation and pursuit, wing vibration, licking, and attempted copulation. The non-courtship elements recorded were activity (running), preening, and rest.
In the test for learning and short-term memory for each strain, three independent samples of experimental males were examined: naive males (with no previous courtship experience) and trained males (having previous experience of courtship), tested either immediately (for learning acquisition) or 3 h after training (for memory retention).
In the test for learning and long-term memory, four independent samples of experimental males were examined: naive males (with no previous courtship experience) and trained males (with previous courtship experience), tested either immediately (for learning acquisition) or 2 or 8 days after training (for memory retention). For each group (naive males, immediately following training and at given time intervals after training), 20 flies were tested.
The resulting courtship index [CI], the percentage of time spent in courtship over a 300-s period, was calculated for each male (Fig. S1). The CI was used to calculate the learning index (LI) as follows:
LI = ([CIna − Citr]/CIna) × 100 = (1 − CItr/CIna) × 100 (McBride et al. 1999), where CIna and CItr are the mean courtship indices for independent samples of naive and trained males, respectively.
Statistical comparisons of behavioral data were performed using a two-sided randomization test (Rohlf and Sokal 1981) by directly computing the probability of rejection of the null-hypothesis αR. The sampled randomization test with 10,000 permutations was used. The null hypothesis was rejected at αR<0.05. Besides, we compared all experimental groups with each other.
Incorporation of [35S]_methionine into D. melanogaster brain proteins
The brains were dissected from 5-day old naïve males kept at 25°C (control), from males subjected to HS for 30 min at 37°C (immediately and 4 h after HS), and from males after 5-h training with fertilized females. For each point, 15 brains were dissected and incubated for 1 h at 25°C in 20 μL of PBS supplemented with 1 μL (1 μCu) of [35S]_methionine (Amersham Biosciences, USA). Labeled brains were lysed in 20 μL Laemmli buffer. Protein extracts were separated by electrophoresis in 8% SDS_polyacrylamide gel. The incorporation of the radioactive label was evaluated by radioautography exposure.
Western blotting
Protein extracts were obtained from the heads of 5-day naïve males kept at 25°C, males subjected to HS 30 min 37°C (all tested strains) and males after 5-h training with fertilized females (Canton-S and strain hsp70-4c). After electrophoresis in 8% SDS PAGE, proteins were transferred to nitrocellulose membrane (Amersham, USA). Hsp70 was detected using monoclonal antibodies 7FB, specific for the inducible Drosophila Hsp70 (Kind gift of Dr. Suzanne Lindquist). Actin was detected with the anti-Actin antibody, clone 4C (Sigma-Aldrich). After incubation with secondary antibodies conjugated with horseradish peroxidase, immune complexes were detected on a ChemiDoc MP system (Bio-Rad, USA) using a reagent for chemiluminescent detection (Thermo Scientific SuperSignal West Pico Plus Chemiluminescent substrate) of Hsp70 after heat shock and actin as an internal control. For detection of Hsp70 under control conditions, we used the Thermo Scientific Super Signal West Femto Maximum Sensitivity substrate. The results were processed using Image J. The measured levels of Hsp70 in each sample were normalized to the amount of actin.
Collection of fly heads and RNA extraction
To obtain the libraries, naïve males were placed with fertilized females at 10 a.m. for 5 h and, hence, the isolation of RNA from the heads of the males was carried out at 3 p.m. For each behavioral experiment, four biological replicates of approximately 100 flies were collected and frozen. Naive or trained males snap-frozen in liquid nitrogen at the appropriate time point after training were manually decapitated. The heads were collected on a dry ice-cooled surface. Total RNA extraction was performed with RNAzol reagent (Molecular Research Center, USA) according to the manufacturer’s protocol. The concentration of RNA was measured with a Qubit Fluorometer (Invitrogen). The quality of RNA was determined with an Agilent BioAnalyzer 2100 using an RNA 6000 nano kit. The RNA Integrity Number (RIN) of all RNA samples taken for mRNA library preparation was not less than 8.
cDNA library preparation and data analysis
Libraries for RNA-seq were prepared using the NEB Next Ultra II Directional RNA Library Prep Kit for Illumina (NEB, USA) according to the manufacturer’s guidelines. The 75-bp single-end sequencing was conducted on an Illumina NextSeq 500 platform.
Deep sequencing provided ~15–20 million reads for each library. Processing of the raw sequence data was performed using PPLine script (Krasnov et al. 2015), which included mapping of the reads to the D. melanogaster genome (Dm6) with STAR (Dobin et al. 2013) following adapter, length, and quality trimming by Trimmomatic (Bolger et al. 2014). Differential gene expression analysis was performed with the edgeR package (Robinson et al. 2010). Gene Ontology and KEGG enrichment analyses were performed using the top GO (v.2.36.0) and cluster Profiler Bioconductor packages (Yu et al. 2012). Visualization of the gene set enrichment analysis (GSEA) was performed using custom scripts written in Python and R. Sequence data were deposited in the NCBI GEO database under the number - GSE152647.
Quantitative PCR
One microgram of total RNA was used for cDNA synthesis with an MMLV RT kit (Evrogen, Moscow, Russia). All qRT-PCR reactions were conducted using the SYBR Green fluorescent dye (Evrogen, Russia) in an ABI PRISM VR 7500 device (Applied Biosystems). The relative expression of the studied genes was calculated based on the ΔΔCt method (Schefe et al. 2006). Quantifications were normalized to the housekeeping gene rp49 (Ponton et al. 2011). Experiments were performed with three replicates and three experimental replicates. The primer sequences are listed in Table S1.
Results
Classical studies involving various learning paradigms and courtship rejection have identified major genes and learning and memory circuits in the Drosophila brain (Griffith and Ejima 2009; Kahsai and Zars 2011). However, to the best of our knowledge, there are no direct data concerning the role of the constitutive expression of the major Drosophila stress gene hsp70 in LTM and STM formation and consolidation. The data presented below help to define the role of hsp70 transcription in LTM and STM and reveal various gene systems that interact with Hsp70 in the processes of learning and memory formation and consolidation in male flies.
Learning and short-term memory
The conditioned courtship suppression paradigm (CCSP) is based on naturally occurring Drosophila mating behavior stimuli. In male courtship of a fertilized female, two unconditioned stimuli are combined: attracting (courtship-stimulating hormone, aphrodisiac) and aversive (courtship-repressing hormone, antiaphrodisiac). The antiaphrodisiac is characteristic only of fertilized females, which release it in response to male courtship. As a result of the combination, the attracting stimulus transforms into the aversive conditioned one, which reduces its attracting properties. After a 30-min stay of a naive male with a fertilized female, the intensity of its subsequent courtship of a tester female dramatically decreased. In the latter case, memory was retained for up to 8 h because retrieval of the memory trace is facilitated by the presentation of a fertilized female to the male (Kamyshev et al. 1999). The CCSP is used widely for evaluation of basic courtship behavior and learning ability and memory retention in Drosophila (Kuzin et al. 2014; Savvateeva-Popova et al. 2007; Savvateeva-Popova et al. 2008; Godenschwege et al. 2004; Redt-Clouet et al. 2012; Savvateeva et al. 2000; Fedotov et al. 2020).
At the first stage, the courtship index [CI] was calculated for each male (Fig. S1). A low index in males white1118 and hsp70- strains is apparently associated with a mutation in the gene white (Anaka et al. 2008). At the same time, in males hsp70- and w +/hsp70-, the courtship time of the female immediately and after training is comparable to that of naive males and does not depend on the eye color. Therefore, the males hsp70- and w+/hsp70- after different times of training maintain a level of courtship comparable to naïve which means that dramatic disturbance of learning, as well as STM and LTM take place in hsp70- and w +/hsp70- males (Fig. S1).
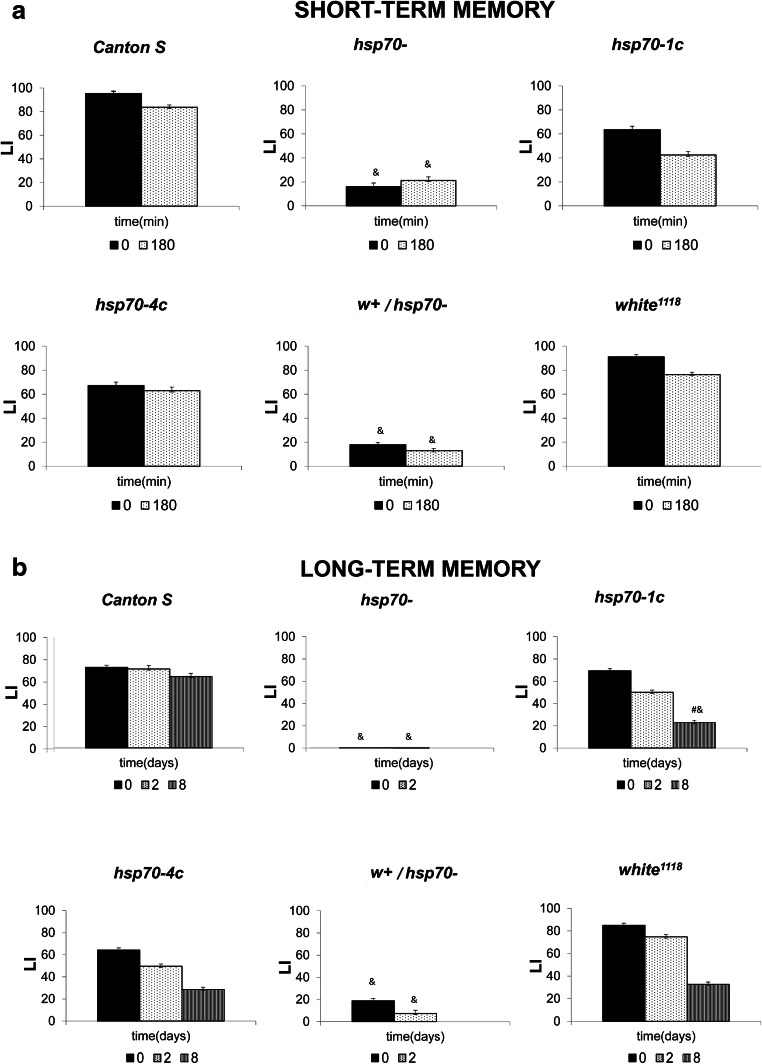
Figure 1 presents the results of the learning acquisition, short-term (A) analyses under normal conditions. Surprisingly, the mutant flies without hsp70 genes (hsp70- and w+/hsp70- ) showed a 4-fold lower 3-h memory and learning ability than control Canton-S flies. This result clearly demonstrated the involvement of Hsp70 in the learning process, specifically the formation of short-term memory (STM). Concurrently, hsp70-1c and hsp70-4c males demonstrated approximately the same learning and short-memory formation as Canton-S flies. Thus, the presence of at least one copy of the hsp70 gene (belonging to either A or B loci) was necessary and sufficient to restore learning ability and STM formation (Fig. 1a). During the next stage, we investigated learning acquisition and long-term memory retention (LTM) in the studied strains (Fig. 1b). The hsp70- males, both intact and treated with heat shock ( Fig. 2, exhibited an even more drastic, 70-fold reduction in 2-day memory and learning ability compared with Canton-S flies. Thus, the mutant hsp70- male flies demonstrated poor ability to learn and to form STM and inability to form LTM in CCSP. Additionally, an increase in the duration of training led to a catastrophic decline in learning of these males (Fig. 2b).
Fig. 1.
Dynamics of learning acquisition, short-term (a) and long-term memory retention (b) as revealed by conditioned courtship suppression in mutant males under normal conditions. Males of the Canton-S, hsp70-, hsp70-1c, hsp70-4c, w+/hsp70-, and white1118 strains were tested. Abscissa: time after training (min or days); ordinate: LI - learning index, standard units (see M&M). The sample size for each time point was 20 males. & - LI significantly lower than the wild type Canton-S strain under similar conditions (two-sided randomization test, αR<0.05); # - LI in the delayed test was significantly lower than in the test immediately following training (two-sided randomization test, αR<0.05)
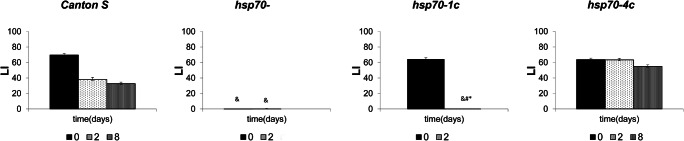
Fig. 2.
Dynamics of learning acquisition and long-term memory retention as revealed by conditioned courtship suppression in mutant males under heat shock (37°C 30 min). Males of the Canton-S, hsp70-, hsp70-1c and hsp70-4c strains were tested. Abscissa: time after training (days); ordinate: LI - learning index, standard units. The sample size for each time point was 20 males. * - LI after heat shock significantly differed from LI at 25°C (two-sided randomization test, αR<0.05); & - LI was significantly lower than the wild type Canton-S strain under similar conditions (two-sided randomization test, αR<0.05); # - LI in the delayed test was significantly lower than in the test immediately following training (two-sided randomization test, αR<0.05)
This dramatic effect observed in hsp70- males concerning learning acquisition and memory retention may be explained by the presence of the white mutation, which makes it difficult for a male to visually find a partner. To test this possibility, we conducted a series of similar experiments using two related strains lacking hsp70 but differing by eye color (i.e., hsp70- and w+/hsp70-strains). However, these experiments failed to demonstrate a significant influence of the white mutation on learning acquisition and memory retention in (Fig. 1a, b). This observation seems quite logical since CCSP is based predominantly on olfactory rather than visual stimuli. The results obtained in the analysis of learning acquisition and long-term memory retention in the mutant hsp70-4c containing four copies of group B were consistent with those obtained for short-term memory. Learning indices immediately and 2 and 8 days after training remained at a high level and did not significantly differ from the wild type (Fig. 1) validating the normal implementation of learning processes and formation of long-term memory in this strain. Thus, the absence of the hsp70 two copies of group A is not critical for the implementation of learning and memory processes and, hence, we used hsp70-4c as a control.
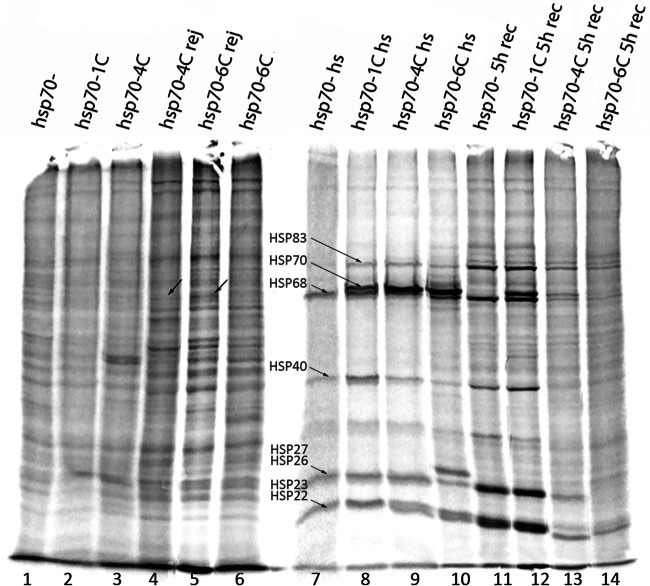
The most intriguing results were obtained in the analysis of the formation of LTM in the mutant strain hsp70-1c after HS treatment (Fig. 2). Learning acquisition was not impaired in this strain, regardless of the duration of training (30 min or 5 h) and HS treatment. At a normal temperature, LI remained at a high level for 2 days, but it decreased notably after 8 days, significantly differing from the wild type (Fig. 1B). Finally, in heat-treated males of this strain, LI was dramatically reduced after 2 days (60-fold compared with LI immediately after training and 70-fold compared with the wild type under similar conditions), indicating a disturbance of LTM formation in hsp70-1c flies after HS. Our experiments involving the analysis of protein synthesis under normal conditions and after HS treatment help to explain the obtained results. Specifically, it is evident that in hsp70- and hsp70-1c strains even 5 h after HS treatment, the synthesis of Hsp68 and other Hsps continues at a relatively high rate, in contrast to the strains hsp70-4c and hsp70-6c where only trace level of Hsps synthesis is observed. At the same time, the hsp70-6c strain demonstrates slightly better recovery than hsp70-4c one. Interestingly, the level of synthesis of Hsp83 and small Hsps after 5 h of recovery is even higher in hsp70- and hsp70-1c strains than immediately after HS. This indicates that mild HS (37 °C, 30 min) probably causes serious damage to the brain tissues in hsp70- and hsp70-1c strains. A more long-lasting synthesis of all Hsps is required to restore the normal functioning of cellular protein synthesis necessary for LTM formation in these strains in contrast to the strains comprising four or six hsp70 copies (Fig. 3).
Fig. 3.
Incorporation of S35 methionine in Drosophila melanogaster imago brains. Strains containing different hsp70 copy numbers hsp70-, hsp70-1c, hsp70-4c, and Canton-S (hsp70-6c) were used. Lanes 1–6: brains were dissected from male flies kept at 25 °C. Lanes 4–5: brains were dissected from males after 5-h training (rejection experiments) with a fertilized female, lanes 1–3, 6–10 from naïve males. Lanes 7–10: brains were dissected just after HS 37 °C 30min. Lanes 11–14: brains were dissected after 4 h of recovery at 25 °C after HS. The labeling period lasted 1 h in all cases. Arrows indicate Hsp70 induction after rejection
Thus, learning acquisition, regardless of the duration of training and effect of stress, was disturbed only in the absence of both groups of hsp70 genes (A and B). The presence of at least one hsp70 copy is sufficient to restore the ability to form STM memory and partially LTM under normal conditions (Fig. 1).
Transcriptome analysis
It is known that the behavioral phenotype and ability to form memory involve complex interactions of various gene networks (Sokolowski 2001). To determine the roles of major stress genes belonging to the hsp70 family in memory formation and storage, we analyzed the gene expression patterns in the heads of naive males and males after rejection. To achieve this goal, we used a rejection paradigm, which included 5 h of training with a mated female (5h) of several strains with different hsp70 gene copy numbers (see Methods). We compared the results with those obtained in the hsp70- strain lacking all inducible members of the hsp70 family. We also monitored the transcriptomic changes in the investigated strains (hsp70-4c; hsp70-1c; w+/hsp70 and hsp70-) that occurred due to the stress of rejection after 5h. The pairwise analysis of differentially expressed genes between all experimental groups and strains is depicted in Table S2.
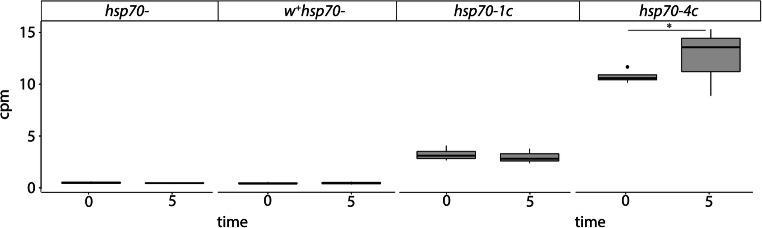
Analysis of transcriptomic data revealed a low basal level of hsp70 transcription in the hsp70-1c strain, which was not upregulated after 5 h of training. In the strain carrying 4 copies of hsp70, the basal level of transcription was higher and slightly increased after 5h of training ( RT-PCR analysis confirmed the upregulation of hsp70 expression after 1h of training in the male heads of hsp70-4c strain (Fig. S2A) and in the male heads of Canton-S strain, which contains 6 copies of the hsp70 gene (Fig. S2B). These results were corroborated by our Western analysis using 7FB antibodies that recognize inducible Hsp70 in the heads of Drosophila naïve males (Fig. S3). We analyzed the difference in the Hsp70 expression level in naïve males at normal temperature (Fig. S3A) and after 30 min of recovery following HS 370C (Fig. S3B) in the investigated strains. Our analysis confirmed that the basal and inducible level of Hsp70 in the studied strains exhibited a clear-cut correlation with the number of hsp70 copies present in the genome. Slight induction of Hsp70 after rejection was revealed in hsp70-4c and hsp70-6c strains (Fig. 3, Fig. S3).
The comparative transcriptome analysis demonstrated significantly fewer differences in terms of differentially expressed gene numbers after 5h of training in the two strains lacking hsp70 copies (Table 1). Tables 1 and 2). By contrast, the hsp70-4c strain carrying a maximal number of hsp70 copies exhibited the largest number of significantly changed genes (up or downregulation) at the level of transcription after rejection (5h). Characteristically, the introduction of a single copy of the hsp70 gene into the hsp70- strain increased the differentially expressed gene number (Tables 1 and 2).
Table 1.
The number of differentially expressed genes in the strains with different hsp70 copy numbers after 5-h training. p value <0.05
| Drosophila strain (after 5-h training) | Number of genes | |
|---|---|---|
| Upregulated | Downregulated | |
| hsp70-4c | 1111 | 1191 |
| hsp70-1c | 758 | 939 |
| w+hsp70 | 521 | 527 |
| hsp70- | 248 | 266 |
Table 2.
The number of up- and down-regulated genes in naïve males in the strains with different hsp70 copy numbers compared to the strain without hsp70 genes (hsp70-) in control conditions. p value <0.05
| Drosophila strain (naïve males) | Number of genes | |
|---|---|---|
| Upregulated | Downregulated | |
| hsp70-1c vs hsp70- | 1046 | 713 |
| hsp70-4c vs hsp70- | 1387 | 1192 |
| w+hsp70- vs hsp70- | 286 | 218 |
The pairwise analysis of differentially expressed genes using all studied strains exhibited in Table 2 is depicted in Table S2.
Genes involved in reproduction and memory processes
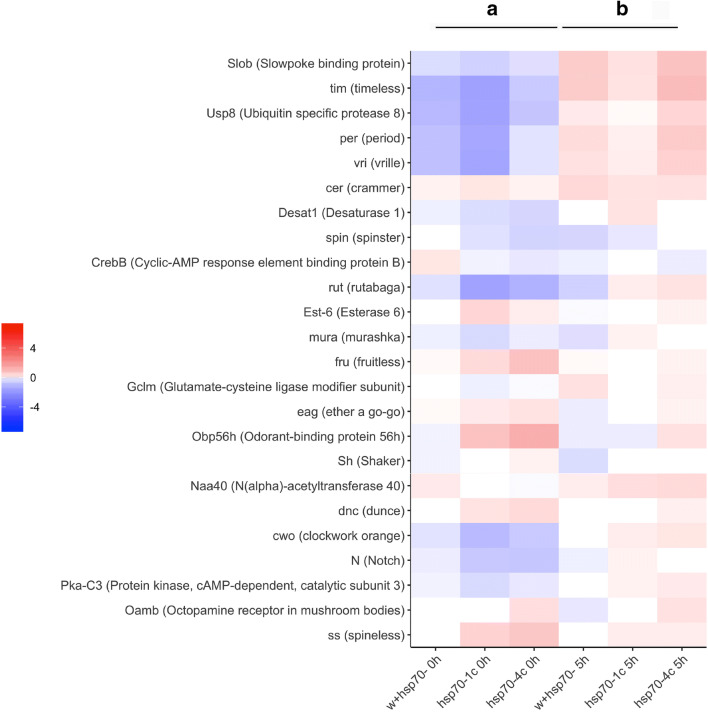
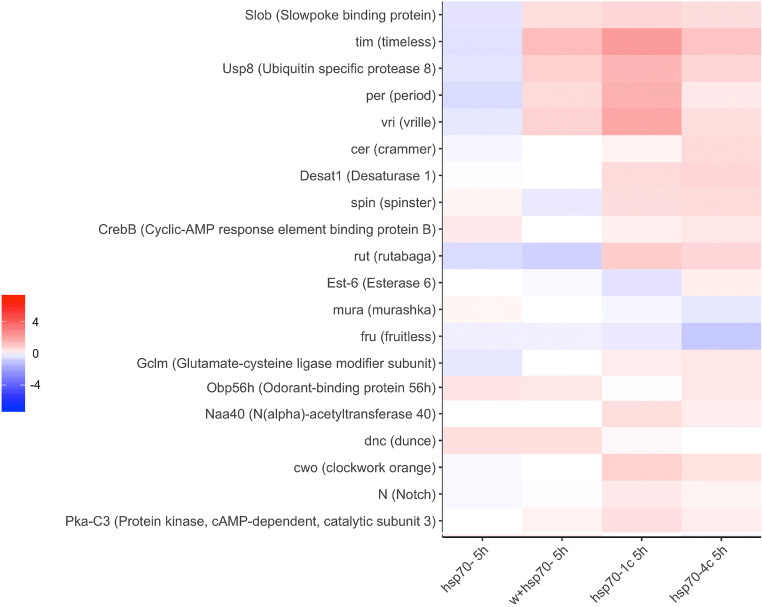
The performed analysis demonstrated characteristic differences in the expression of gene fruitless -fru that play important role in the courtship behavior of male fruit flies (Billeter et al. 2006). Thus, while the fru gene was expressed at higher levels in the heads of naive males containing hsp70 copies (hsp70-4c and hsp70-1c strains) compared with those lacking representatives of this family (i.e., w+/hsp70- and hsp70- strains) ( Fig. 5a), the rejection resulted in a significant drop in fru expression in all the studied strains ( Figs. 5b and 6).
Fig. 5.
Heatmap illustrating the comparative analysis of differentially expressed genes involved in reproduction and memory processes. Differentially expressed genes in the heads of naive males – 0 h (a) and the males after 5 h of training with fertilized females – 5 h (b) in strains hsp70-4с, hsp70-1с, w+/hsp70- in comparison to hsp70-. Genes with p < 0.05 (quasi-likelihood F-test) in at least one comparison were considered significant. (Statistic data for described genes are presented in Table S3 and Fig. 7)
Fig. 6.
Heatmap illustrating the comparative analysis of differentially expressed genes involved in reproduction and memory processes. Differentially expressed genes in the heads of males after 5 h of training (5h) with fertilized females in comparison to corresponding naive males of the hsp70-4с, hsp70-1с, w+/hsp70- and hsp70- strains. Genes with p < 0.05 (quasi-likelihood F-test) in at least one comparison were considered significant. (Statistic data for described genes are presented in Table S3 and Fig. 7)
Our transcriptome analysis revealed several genes that play an important role in mating and reproductive behavior (Obp56h and Est-6), as well as in memory formation (dunce and ss), that were expressed at higher levels in the heads of naive males of hsp70-1c and hsp70-4c strains than in the hsp70- and w+/hsp70- strains (Fig. 5a). It is noteworthy that the Oamb gene has higher expression in the heads of naive and trained males of the hsp70-4c strain carrying the maximal number of hsp70 copies (Fig. 5a Fig. S3). The neurotransmitter octopamine (OA) and its receptor OAMB are expressed in the neurons of mushroom bodies and are necessary for courtship conditioning (Zhou et al. 2012). It is assumed that the LTM formed during courtship is not under the influence of most clock genes (Sakai and Kitamoto 2006). Interestingly, a significant increase in the expression of per, tim, vri, and Usp8 after 5-h training was observed in all red-eyed strains carrying the normal allele of the white gene. By contrast, these genes were downregulated after 5-h training in the white-eyed hsp70- strain (Fig. 6).
Interestingly, the expression of the Clockwork orange gene (cwo), which activates transcription of the period and timeless genes (Gerstner and Yin 2010), was upregulated after 5-h training only in the strains containing hsp70 genes.
The historical genes rutabaga (rut) and dunce (dun) that regulate the level of cAMP and are expressed in mushroom bodies (MBs), one of the main structures in the adult insect brain, play a critical role in olfactory learning and memory (Guan et al. 2011; Livingstone et al. 1984; Levin et al. 1992). In the hsp70-1c and hsp70-4c strains, the basal level in naïve males of rut transcription was reduced in comparison to both strains lacking hsp70 (Fig. 5a). Characteristically, an increase in rut gene expression after 5-h training is observed (Fig. 6) only in the strains containing hsp70 copies (i.e., hsp70-1c and hsp70-4c strains), characterized by normal memory formation. Similarly, the dunce gene, which is involved in regulating the level of the cAMP-specific phosphodiesterase that degrades cAMP slightly increases its expression after 5-h training in the strains lacking hsp70 copies in the genome (Fig. 6).
The genes encoding the subunits of protein kinase A (Pka) represent another group of critical genes expressed in mushroom body neurons. In the strains containing copies of the hsp70 gene, the Pka-C3 catalytic subunit was expressed at a higher level in the heads of trained males in comparison to the hsp70- strain (Fig. 5b). It is known that activated Pka phosphorylates the transcriptional activator CREB. The level of CrebB isoform expression was increased after 5-h training in all strains used in the study with the prominent exception of w+/hsp70- flies (Fig. 6). Such changes in gene expression may play critical roles in long-term memory formation. It is known that activated Pka modulates the activity of K+ channels (Yao and Wu 2001). Our analysis demonstrated that the relative expression level of the eag gene, which exhibits voltage-gated potassium channel activity and is involved in learning (Cowan and Siegel 1986), was slightly higher in the heads of naïve males of the strains with hsp70 copies (Fig. 4a). A similar expression pattern was revealed for the Shaker (Sh) gene, which encodes the A potassium ion channel (Timpe et al. 1988).
Fig. 4.
The expression level of hsp70 genes in the heads of naïve males (point 0) and after 5-h training (point 5) in the following strains: hsp70- 4c, hsp70-1c, w+/hsp70-and hsp70-. * - p < 0.05 (Fisher exact test)
It is well-known that epigenetic post-translational modifications (PTMs) of histones that control gene expression patterns play important role in memory formation (Peixoto and Abel 2013; Xu et al. 2014). Our transcriptome analysis demonstrated an increase in the expression level of the Naa40 (N (alpha)-acetyltransferase 40) gene in the strains containing hsp70 after 5-h training (Fig. 6) This gene plays an important role in epigenetic protein modifications that may be involved in the processes of learning and memory formation (Gupta et al. 2010; Zovkic et al. 2013).
It was shown that LTM but not STM requires Notch (Presente et al. 2004) signaling in courtship conditioning, and Notch is also required for memory consolidation, a process believed to require remodeling of existing neurons in adults. In our experiments, the expression of Notch has a tendency for upregulation after 5-h training only in the strains containing hsp70 copies (Fig. 6). We also detected downregulation of expression of gene Murashka in the hsp70-4c strain after 5-h training. The selective translational repression of gene Murashka during LTM formation was earlier observed (Mastushita-Sakai et al. 2010).
Collectively, our experiments demonstrated that 5-h training upregulated the transcription of a certain set of genes involved in memory formation and consolidation, such as cwo, Gclm, Desat1, cer, spin, and ru, only in strains containing hsp70 copies (Fig. 6).
Immune system gene expression depends on the presence of hsp70 genes
In the last decade, several groups have demonstrated that putative antimicrobial peptides expressed in the brain of flies and other organisms influence LTM (Bozler et al. 2017; Barajas-Azpeleta et al. 2018). Mating appears to switch on the immune system in preparation for the effects of external agents, including various pathogens, as well as possible stress of rejection by females.
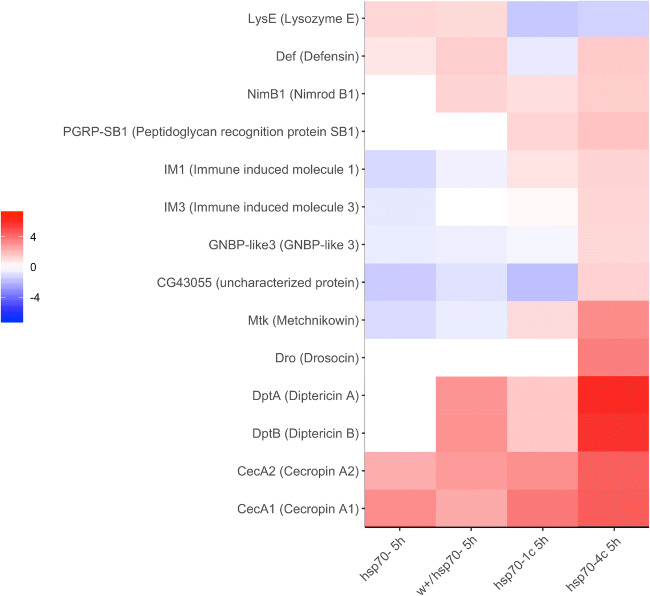
Comparison of immune response gene expression in the heads of naive males and males after 5-h training from the studied strains demonstrated similar patterns of expression in the strains lacking hsp70 genes (i.e., hsp70- and w+/hsp70-) suggesting a role of constitutive Hsp70 expression in this vital process (Fig. S6). Interestingly, in the heads of naive males of the strain with four copies of hsp70, the relative level of several immune genes (i.e., LyzE, Mtk, CecA2 CecA1, Def, DptA, and DptB) was lower than in strain hsp70-1c containing only one copy of hsp70 (Fig. S5). On the other hand, the relative expression level of genes such as CecA1, CecA2, Mtk, Im1, Im3, and GNBP-like3 after 5-h training was higher in the hsp70-4c strain (Fig. S6).
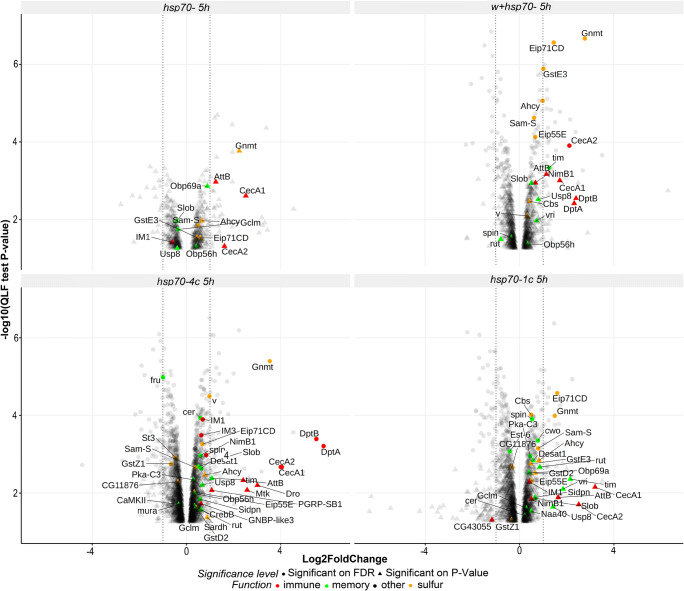
Only two genes belonging to the innate immune system (CecA1 and CecA2) were upregulated in all the studied strains after 5-h training. Figs. 7 and 8). Notably, in both strains containing hsp70 copies, the induction of Im1, Mtk, and Pgrp-SB1 after 5-h training was evident (Fig. 8). Moreover, the maximal upregulation of all differentially expressed genes was observed in strain hsp70-4c (Fig. 8), which was the only strain that showed a significant upregulation of the genes Dro, Cg43055, GNBP-like3, and IM3. These data suggest that the basal level of Hsp70 expression plays an important role in modulating the immune response and in the function of certain immune antibacterial peptides that participate in LTM formation.
Fig. 7.
Volcano plot. Differentially expressed genes in the heads of males after 5 h of training (5h) with fertilized females in comparison to corresponding naive males of the hsp70-4с, hsp70-1с, w+/hsp70-, and hsp70-strains. Only genes that exhibited statistically significant differences are included in the plot (p value <0.05 or FDR<0.05). LogFC values are displayed on the abscissa. Upregulated genes are depicted on the right side of the plot, and downregulated genes are shown on the left side of the plot
Fig. 8.
Heatmap illustrating the comparative analysis of differentially expressed genes involved in immune processes. Differentially expressed genes in the heads of males after 5 h of training with fertilized females in comparison to corresponding naive males of the hsp70-4с, hsp70-1с, w+/hsp70-, and hsp70-strains. Genes with p < 0.05 (quasi-likelihood F-test) in at least one comparison were considered significant. (Statistic data for the described genes are summarized in Table S3 and Fig. 7)
Interestingly, after 5-h training, the expression of the LysE gene, functioning in the negative regulation of the innate immune response, was decreased in strains carrying hsp70 genes and upregulated in hsp70- strains.
It is of note that differentially expressed genes Dro, PGRP-SB1, Mtk, CecA1, and CecA2 (Fig. 8) belong to a category of genes with a cyclic pattern of transcription in the heads of D. melanogaster (McDonald and Rosbash 2001), which corroborates their involvement in the function of the Drosophila central nervous system, including LTM formation and consolidation (Gerstner and Yin 2010).
Genes involved in methionine metabolism and glutathione metabolic processes showed altered expression in trained males
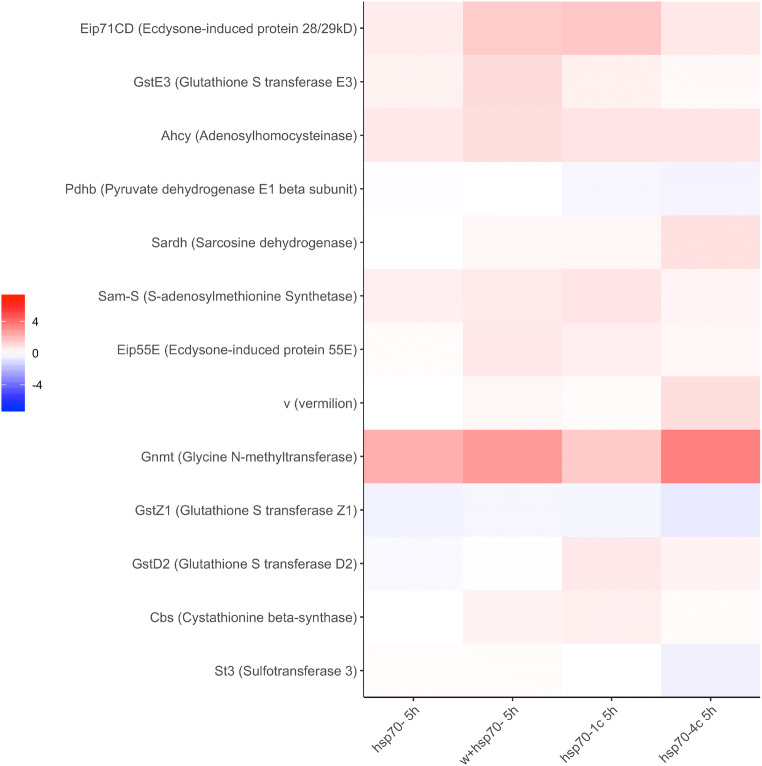
Genes involved in methionine metabolism (transsulfuration pathway) comprise another distinct group that exhibited characteristic changes after 5-h training in all strains used in the study ( Fig. 9). Interestingly, maximal induction of expression (especially in the hsp70-4c strain) was demonstrated for the glycine N-methyltransferase (Gnmt) gene, which controls the amount of the methyl donor S-adenosylmethionine (Obata et al. 2014) and plays a key role in life extension in Caenorhabditis elegans and fruit flies (Obata and Miura 2015). In the hsp70-4c strain, we also observed a strong induction of the genes Sardh and vermillion (v). Sardh plays an important role in the S-adenosylmethionine cycle. Vermillion gene participates in tryptophan metabolism and plays a role in memory performance, as revealed in a courtship-conditioning paradigm (Savvateeva et al. 1999).
Fig. 9.
Heatmap illustrating the comparative analysis of differentially expressed genes involved in the processes of sulfur metabolism. Differentially expressed genes in the heads of males after 5-h training with fertilized females in comparison to corresponding naive males of the hsp70-4с, hsp70-1с, w+/hsp70-, and hsp70- strains. Genes with p < 0.05 (quasi-likelihood F-test) in at least one comparison were considered significant. (Statistic data for described genes is represented in Table S3 and Fig. 7)
All strains with the prominent exception of hsp70- were also characterized by upregulation of the cbs gene encoding cystathionine beta-synthase and representing one of the three major genes responsible for H2S production in Drosophila and other organisms including humans (Kabil et al. 2011; Kimura 2014). Additionally, training of the males (5h) resulted in the induction of ecdysone-induced genes (i.e., Eip55E; Eip71CD), as well as several other methionine metabolism genes including Sam-S, Ahcy, and GstE3 (Fig. 8). Eip55E encodes cystathionine γ-lyase (cse) and together with cbs, is responsible for H2S production in various organisms (Kabil et al. 2011; Zars 2017).
Analysis of the transcriptome data revealed a significant effect of 5-h training on the expression of the gene system participating in sulfur metabolism and glutathione metabolic processes.
Glutathione transferases (GSTs), represented by 40 genes in Drosophila, are ubiquitous key enzymes that catalyze the conjugation of glutathione to xenobiotic compounds in the detoxification process and play a critical role in insect chemo detection by modifying odorant molecules (Gonzalez et al. 2018). When we compared the expression levels of “sulfur” and GST genes in naive males in the studied strains, it became clear that strains without hsp70 copies were characterized by comparatively lower expression levels of these genes (Figs. S6 and S7). By contrast, after 5-h training, the minimal relative expression level of these genes was detected in hsp70-4c flies and maximal level in the w+/hsp70- (Figs. S6, S7). Glutathione (GSH) is a downstream critical metabolite of the transsulfuration pathway (TSP), and hence, it is not surprising that genes making up these two olfactory-related systems exhibit a rather similar pattern of expression after 5-h training (Durand et al. 2018). In our experiments, the expression levels of the number of genes belonging to this category increased after 5-h training: GstE3, GstE7, GstE1, and Eip55E decreased in all the studied strains: GstZ1, gfzf. It should be noted that maximum induction of these genes was observed in the w+/hsp70- strain (Fig. S8). Only strains carrying hsp70 copies were characterized by an upregulation of GstD2 and Gclm gene expression (Fig. S8). GstD2 participates in the modulation of chemoperception in insects (Gonzalez et al. 2018) while glutamate–cysteine ligase modifier subunit (Gclm) is involved in glutathione biosynthesis, and its overexpression was shown to extend the mean life span (Orr et al. 2005). Additionally, only in the hsp70-4c strain, the GstD9 and GstZ2 genes were induced after 5-h training.
The observed differences in the learning and memory formation as well as in brain transcription patterns characteristic for strains lacking hsp70 genes in principle may be due to some developmental abnormalities in the brains of the strains with deletions accumulated in the process of hsp70 deletion generation. To check this possibility, we obtained a new hsp70- strain with GFP gene coding region controlled by the Elav gene promoter (see Materials and methods). We examined brain samples from third instar larvae and adult flies by confocal microscopy. We monitored the GFP fluorescence caused by the activity of the Elav gene promoter in the neurons of the mushroom body. In the course of this analysis, we failed to detect any visible gross deviations in the structure of the mushroom body in the hsp70- strain (Fig. S9).
Discussion
At present, a plethora of reports has described numerous genes that are somehow involved in the learning process and STM and LTM in fruit flies and other organisms including humans (Griffith and Ejima 2009; Winbush et al. 2012; Keleman et al. 2007; Akalal et al. 2011; Kahsai and Zars 2011; Sokolowski 2001; Guan et al. 2011). Many circuits and signaling pathways involved in memory formation are rather conservative because memory exists at all levels of organization from worms to humans. Learning and long-term memory formation in D. melanogaster represent important neuronal functions that are controlled by various interacting gene systems. Early stages of memory formation do not require de novo protein synthesis and are achieved by covalent modifications of pre-existing target proteins in the neurons responsible for cellular membrane conductivity and neuronal excitation (Bailey et al. 1996; Tully 1996; Kandel 2012). Long-term memory requires the synthesis of new proteins and the formation of new synapses (Kandel 2012). The regulation of gene expression during the formation of long-term memory partially is due to the phosphorylation of the transcription factor CREB (CRE-binding protein) and the induction of transcription of genes having CRE motifs (CREB targets) in the promoter region (Flexner et al. 1963; Kandel 2012). Notably, the level of CrebB isoform expression was increased after 5-h training in all strains used in our studies except for w+/hsp70- flies (Fig. 6).
Several groups studying LTM using conditioned courtship or courtship suppression in males that have been repeatedly rejected by mating females have used RNA sequencing and described a large number of differentially expressed genes with known functions, including those that function in cyclic AMP, neuronal development, chromatin biology, translation, and various aspects of behavior (Ge et al. 2004; Winbush et al. 2012; Keleman et al. 2007; Ishimoto et al. 2009). However, it is clear that LTM formation is an extremely complex process and includes contributions from additional mechanisms. Thus, here we used several D. melanogaster strains with different copy numbers of hsp70 genes to determine the role of this major stress protein in STM and LTM formation. Available data suggest a coupled evolution of stress response and memory formation (Hooper et al. 2016; Porto et al. 2018; Scaccianoce et al. 2003; Thekkuveettil and Lakhotia 1996).
This is also confirmed by the fact that psychophysiological stress can cause the induction of proteins belonging to the Hsp70 family. Thus, Hsp70 and Hsp60 levels were elevated in daphnia exposed to a predator (fishes) (Pijanowska and Kloc 2004). Similarly, stress caused by predators induces Hsp70 expression in goldfish and cats (Kagawa et al. 1999; Fleshner et al. 2004). Immersion of rats in water caused a significant increase in the level of Hsp70 and Hsc70 mRNA in the hippocampus (Fukudo et al. 1997; Fukudo et al. 1999).
Heat shock proteins and, in particular, Hsp70 is important in the processes of protein folding and degradation (Hartl et al. 2011), and, hence, they participate in the processes of protein synthesis and transport, that are necessary for the maintenance of existing synapses and the formation of new synapses during memory formation. In the promoter region of mammalian hsp70 genes, the CRE-motifs were identified. Furthermore, in the promoter region of Drosophila hsp70 genes, sites for binding of transcriptional factor FOXO were found (Donovan and Marr 2nd 2016). It was shown that FOXO-DAF-16 regulates learning and memory, regeneration, and stress resistance in C. elegans neurons (Kaletsky et al. 2016: Kim and Webb 2017). Collectively, these data suggest a possible role of Hsp70 in the memory processes.
Similar to our results (Fig. 4) in several rodent models, the induction of hsp70 was detected in postsynaptic neuronal structures, hippocampus, and cerebellum during learning, as well as memory formation and consolidation (Porto et al. 2018; Ambrosini et al. 2005; Pizarro et al. 2003; Suzuki et al. 1999; Igaz et al. 2004). It was also demonstrated that an optimal level of neuronal Hsp70 is required for normal memory formation (Ambrosini et al. 2005; Ammon-Treiber et al. 2008; Pizarro et al. 2003). These results explain the observed severe memory disturbances observed in strain hsp70- (Figs. 1 and 2).
Taking into account the observation that LTM formation and consolidation require the synthesis of new proteins and various modifications of existing proteins in the brain regions, the presence and induction of Hsp70 during memory formation are not surprising. However, most of the abovementioned studies are correlative, and, therefore, the investigators were able to only speculate regarding specific gene systems and signal pathways that require the presence of Hsp70 for memory formation.
Thus, the Drosophila strains used in this study either lacking or carrying different numbers of hsp70 copies represent a unique tool to pin-point gene systems that interact with Hsp70 during memory formation. Generally speaking, the dramatic memory deficit observed in hsp70- flies may be a consequence of the deletion of all copies of hsp70 per se because the deletion may have some adverse neurodevelopmental effects and indirectly affects learning and memory formation. However, when comparing the LI in different training schemes (30 min or 5 h), it is evident (Fig. 1) that a low level of learning is still observed in hsp70- and w+/hsp70- flies after 30 min of training. This does not allow us to explain the low ability of the hsp70- strain in learning by possible neuronal developmental disturbances inflicted by deletion per se. Besides, the introduction of a single hsp70 copy into the strain with the deletion significantly restored learning and memory parameters at normal temperatures. The performed analysis of GFP fluorescence in the neurons of third instar larvae and the brains of adult flies revealed no visible gross abnormalities in the development of the nervous system in the hsp70- strain. Therefore, we assume that the absence of memory formation in hsp70- flies most likely is not due to some defects in the development of the nervous system, but is associated with the deficit of Hsp70 necessary for the induction of several transcription cascades participating in memory formation. It is also clear that this problem requires more in-depth study. In our experiments, we also used red-eyed flies lacking all hsp70 genes but containing a normal allele of the white gene (“mini-white” construct). This strain developed by us from the white-eyed strain hsp70- was used as an additional control to eliminate the effect of the white gene, which has been previously shown to significantly affect synaptic transmission and memory in fruit flies (Anaka et al. 2008; Sitaraman et al. 2008).
Specifically, our transcriptomic analysis revealed several major pathways that depend on the presence of hsp70 copies in the genome and are involved in memory formation. Thus, it is well-known that the cAMP signaling cascade, which changes the excitability of mushroom body neurons, plays a critical role in learning, LTM, and STM formation (Levin et al. 1992; Livingstone et al. 1984). Adenylate cyclase (Ac) is encoded by rutabaga (rut), while cAMP-specific phosphodiesterase is encoded by dunce, which degrades cAMP. Additionally, cAMP signaling (rut-dnc) modulates synaptic morphology and physiology (Renger et al. 2000), a requirement for learning and memory formation (Zhong and Wu 1993). Characteristically, here we established that the expression of these two “historical” genes in response to 5-h training depends on the presence of hsp70 copies in the fly genome (Figs. 5 and 6) Thus, in both strains carrying hsp70 copies (i.e., hsp70-1c and hsp70-4c), 5-h training led to the upregulation of rut expression while the transcription of gene dnc was not changed. By contrast, in the strains lacking hsp70 copies (hsp70-and w+/hsp70-) the expression of the rut was downregulated in parallel with a slight increase in dnc transcription, which should result in a drop in the cAMP level and, hence, may interfere with memory formation.
In our studies, we also observed an upregulation of critical components of CREB-responsive transcription such as the Pka-C3 catalytic subunit, preferentially in the strains with hsp70 copies. Phosphorylated CREB binds to cAMP-responsive elements (CRE) in the regulatory regions of cAMP-inducible genes. This system has an essential and well-established role in long-term memory formation throughout a diverse set of organisms (Presente et al. 2004; Zhang et al. 2013). It was shown that exogenous Hsp70 stimulates CREB phosphorylation in mice hippocampus, and improves memory (Kwon et al. 2019).
The effect of hsp70 deletions herein is not surprising since LTM requires protein synthesis, and, hence, normal CREB activity and intact signaling through CaMKII (Yu et al. 2006; Akalal et al. 2010).
We observed differential expression of many immune response genes in the strains with different hsp70 copy numbers. Earlier, it was found that Diptericin B, an immune peptide with antimicrobial activity, and Gram-Negative Bacteria Binding Protein like 3 (GNBP-like3), are upregulated following behavioral training (Barajas-Azpeleta et al. 2018). Deletion and knockdown experiments revealed that Diptericin B, GNBP-like3, and IM18 (Barajas-Azpeleta et al. 2018; Bozler et al. 2017) regulate long-term but not short-term memory in Drosophila. In our results, most of the immune genes related to LTM formation exhibited significantly higher induction after 5-h training in the strains containing hsp70 copies, i.e., hsp70-1c and especially hsp70-4c, confirming the need for a certain basal level of Hsp70 synthesis (Figs. 7 and 8). This result confirms that Hsp70 modulates the immune response (Kim and Yenari 2013) and that hsp70 genes may be involved in the activation of immune peptides in the nervous system during LTM formation. These data help to understand how some immune peptides may have been repurposed to influence the function of the nervous system (Harris et al. 2015; Stevens et al. 2007).
Furthermore, there is growing evidence in favor of cross-talk between the stress gene system comprising hsp genes and another ancient and highly conserved H2S-producing system which also provides an antioxidant defense in all organisms (Kimura 2014; Paul and Snyder 2015; Stevens et al. 2007; Yurinskaya et al. 2020). It is known that H2S acts as a neuromodulator and promotes long-term potentiation and regulates intracellular calcium levels, necessary for learning and memory (Nagai et al. 2004; Yong et al. 2010). It has been proposed that H2S is essential for visual memory (Zars 2017). Also, it was shown that mutant flies containing an insertion in the first exon of cbs gene are characterized by memory deficit (Kuntz et al. 2017).
Our transcriptomic analysis demonstrated that naive males from the strains without hsp70 copies but differing by eye color were very similar in terms of sulfur and GSH metabolism (Figs. S6, S7). However, the presence of the wild-type allele of the white gene significantly modulated the response of genes participating in sulfur metabolism, courtship behavior, and the circadian rhythm in response to the 5-h training procedure (Figs. 6 and 9).
It is of note, that we observed comparatively higher induction of the most sulfur metabolism genes in the w+/hsp70- and hsp70-1c strains after 5-h training. These findings suggest that the presence of the hsp70 gene can modulate the activation pathways of sulfur metabolism genes.
We constructed the network for protein–protein interaction using genes differentially expressed in the process of memory formation (5-h training) described in the article (Fig. S10). It is seen that Hsp70 proteins (A and B) interact with a few definite protein groups and are located in the center of the interactome.
Our experiments exploring strains with different hsp70 copy numbers demonstrated a positive correlation between the number of hsp70 copies and LTM formation (Fig. 1). However, when we applied a heat shock treatment (37°C, 30 min) to the studied strains, we obtained seemingly paradoxical effects. Thus, in the strain with four hsp70 copies, HS treatment slightly improves memory formation, but in the strain containing only one copy of hsp70 (hsp70-1c) and characterized by comparatively lower LTM, HS treatment dramatically disturbed LTM formation (Fig. 2). We speculate that here we have some trade-off and in hsp70-4c strain, the induced Hsp70 amount is optimal and sufficient to perform chaperone properties eliminating stress consequences and participate in LTM formation. By contrast, in the case of the hsp70-1c strain, a low level of Hsp70 is insufficient to protect the brain tissues from the deleterious effect of HS which results in strong memory impairment in this strain after HS.
Our experiments exploring 35S-methionine labeling of proteins synthesized in the brain shed new light on this issue. It was shown that in hsp70- and hsp70-1c strains even 5 h after HS, the synthesis of all Hsps and especially small ones continues at a high rate. It is known that low molecular weight chaperones after HS associate with misfolded proteins and facilitate their folding or degradation by other chaperones (Sun and MacRae 2005). To this end, it was shown that Hsp23 and Hsp26 show high expression levels in CNS during fly ontogenesis, suggesting a role in neural system development (Santana et al. 2020). Probably in the case of strains hsp70- and hsp70-1c, small Hsps are required for recovery of neuronal structures after HS. Higher level of small Hsps observed in strains hsp70- and hsp70-1c probably represents compensation for the absence or low level of Hsp70 synthesis in the brains of strains hsp70- and hsp70-1c that are not able to recover 5 h after HS (Figs. 2 and 3).
Our experiments also demonstrated that the rejection accompanied by antiaphrodisiac excretion represents serious stress for Drosophila males and results in memory formation and modulation of multiple gene expression. The constitutive level of hsp70 gene expression in the brain and probably a modest induction of these genes by 5h training (Figs. 3 and 4, Figs. S2, S3) are necessary to cope with the stress consequences and provide conditions for memory formation.
Conclusions
This is the first report describing an important role of constitutive hsp70 expression in learning and memory formation in D. melanogaster males in CCSP. In this study, exploring the courtship rejection paradigm and D. melanogaster strains with different hsp70 copy numbers including strains carrying a deletion of all six hsp70 genes, we demonstrated that a low constitutive level of Hsp70 is required for learning and the formation of short and long-term memories in males. The courtship-dependent genes interacting with the hsp70 system that we identified contribute to many vital biological processes, including behavior and reproduction, stress, and immune responses as well as the production of gaseous signaling molecules such as H2S. Furthermore, our results revealed a positive correlation between the number of hsp70 copies present in Drosophila genome and the number of differentially expressed genes that responded to training with a fertilized female. Further studies are necessary to dissect in detail the molecular mechanisms underlying the demonstrated role of hsp70 expression in memory formation and consolidation in flies and other organisms.
Supplementary Information
(DOCX 2105 kb)
(XLSX 2422 kb)
(XLSX 22 kb)
Acknowledgements
The bioinformatics was performed using the computational facilities of Engelhardt Institute of Molecular Biology RAS Genome center (http://www.eimb.ru/rus/ckp/ccu_genome_c.php).
Availability of data and material
Sequence data were deposited in the NCBI GEO database under the number GSE152647.
Author contribution
Study conception and design were performed by (Evgen’ev M.B., Zatsepina O.G., Nikitina E.A.). Material preparation, data collection, and analysis were performed by (Shilova V.Y., Chuvakova L.N., Sorokina S., Tokmacheva E.V.). Libraries for RNA-seq were prepared by (Chuvakova L.N., Funikov S.Y.). Differential gene expression analysis was performed by (Rezvykh A.P.). Construction of Elav GFP/FM7; Hsp70-/Hsp70- strain for confocal analysis was done by Vorontsova J.E. The first draft of the manuscript was written by (Zatsepina O.G., Nikitina E.A., Evgen’ev M.B.). All authors read and approved the final manuscript.
Funding
All genetic and behavioral experiments were supported by the Russian Foundation for Basic Research grant no. 18-04-00865 (to O.Z.). Transcriptomic studies were funded by Russian Science Foundation Grant 17-74-30030 to M.E.
Declarations
\Consent to participate
Consent for participation was obtained from all authors of this paper.
Consent for publication
The Author grants the Publisher the sole and exclusive license of the full copyright in the Contribution, which license the Publisher hereby accepts.
Conflicts of interest/Competing interest
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zatsepina O.G. and Nikitina E.A. contributed equally to the work.
References
- Akalal D-BG, Yu D, Davis RL. A late-phase, long-term memory trace forms in the γ neurons of Drosophila mushroom bodies after olfactory classical conditioning. J Neurosci. 2010;30:16699–16708. doi: 10.1523/JNEUROSCI.1882-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akalal D-BG, Yu D, Davis RL. The long-term memory trace formed in the Drosophila α/β mushroom body neurons is abolished in long-term memory mutants. J Neurosci. 2011;31:5643–5647. doi: 10.1523/JNEUROSCI.3190-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosini MV, Mariucci G, Tantucci M, Van Hooijdonk L, Ammassari-Teule M. Hippocampal 72-kDa heat shock protein expression varies according to mice learning performance independently from chronic exposure to stress. Hippocampus. 2005;15:413–417. doi: 10.1002/hipo.20069. [DOI] [PubMed] [Google Scholar]
- Ammon-Treiber S, Grecksch G, Angelidis C, Vezyraki P, Höllt V, Becker A. Emotional and learning behaviour in mice overexpressing heat shock protein 70. Neurobiol Learn Mem. 2008;90:358–364. doi: 10.1016/j.nlm.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Anaka M, Macdonald CD, Barkova E, Simon K, Rostom R, Godoy RA, Haigh AJ, Meinertzhagen IA, Lloyd V. The white gene of Drosophila melanogaster encodes a protein with a role in courtship behavior. J Neurogenet. 2008;22:243–276. doi: 10.1080/01677060802309629. [DOI] [PubMed] [Google Scholar]
- Astakhova LN, Zatsepina OG, Funikov SY, Zelentsova ES, Schostak NG, Orishchenko KE, Evgen’ev MB, Garbuz DG. Activity of heat shock genes' promoters in thermally contrasting animal species. PLoS One. 2015;10:e0115536. doi: 10.1371/journal.pone.0115536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey CH, Bartsch D, Kandel ER. Toward a molecular definition of long-term memory storage. Proc Natl Acad Sci. 1996;93:13445–13452. doi: 10.1073/pnas.93.24.13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barajas-Azpeleta R, Wu J, Gill J, Welte R, Seidel C, McKinney S, Dissel S, Si K. Antimicrobial peptides modulate long-term memory. PLoS Genet. 2018;14:e1007440. doi: 10.1371/journal.pgen.1007440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck CD, Schroeder B, Davis RL. Learning performance of normal and mutant Drosophila after repeated conditioning trials with discrete stimuli. J Neurosci. 2000;20:2944–2953. doi: 10.1523/JNEUROSCI.20-08-02944.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billeter JC, Villella A, Allendorfer JB, Dornan AJ, Richardson M, Gailey DA, Goodwin SF. Isoform-specific control of male neuronal differentiation and behavior in Drosophila by the fruitless gene. Curr Biol. 2006;16:1063–1076. doi: 10.1016/j.cub.2006.04.039. [DOI] [PubMed] [Google Scholar]
- Bobkova NV, Evgen'ev M, Garbuz DG, Kulikov AM, Morozov A, et al. Exogenous Hsp70 delays senescence and improves cognitive function in aging mice. Proc Natl Acad Sci U S A. 2015;112:16006–16011. doi: 10.1073/pnas.1516131112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobkova NV, Garbuz DG, Nesterova I, Medvinskaya N, Samokhin A, Alexandrova I, Yashin V, Karpov V, Kukharsky MS, Ninkina NN, Smirnov AA, Nudler E, Evgen'ev M. Therapeutic effect of exogenous hsp70 in mouse models of Alzheimer's disease. J Alzheimers Dis. 2014;38:425–435. doi: 10.3233/JAD-130779. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozler J, Kacsoh BZ, Chen H, Theurkauf WE, Weng Z, Bosco G. A systems level approach to temporal expression dynamics in Drosophila reveals clusters of long term memory genes. PLoS Genet. 2017;13:e1007054. doi: 10.1371/journal.pgen.1007054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman A, Kishinevsky S, Koren J, Lou W, Chiosis G. Chaperone-dependent Neurodegeneration: a molecular perspective on therapeutic intervention. J Alzheimers Dis Parkinsonism. 2013;2013:s10. doi: 10.4172/2161-0460.S10-007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A, Kwon YT. Protein quality control by molecular chaperones in neurodegeneration. Front Neurosci. 2017;11:185. doi: 10.3389/fnins.2017.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan TM, Siegel RW. Drosophila mutations that alter ionic conduction disrupt acquisition and retention of a conditioned odor avoidance response. J Neurogenet. 1986;3:187–201. doi: 10.3109/01677068609106849. [DOI] [PubMed] [Google Scholar]
- Dahlgaard J, Loeschcke V, Michalak P, Justesen J. Induced thermotolerance and associated expression of the heat-shock protein Hsp70 in adult Drosophila melanogaster. Funct Ecol. 1998;12:786–793. doi: 10.1046/j.1365-2435.1998.00246.x. [DOI] [Google Scholar]
- Davis RL. Olfactory memory formation in Drosophila: from molecular to systems neuroscience. Annu Rev Neurosci. 2005;28:275–302. doi: 10.1146/annurev.neuro.28.061604.135651. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan MR, Marr MT., 2nd dFOXO activates large and small heat shock protein genes in response to oxidative stress to maintain proteostasis in drosophila. J Biol Chem. 2016;291(36):19042–19050. doi: 10.1074/jbc.M116.723049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau J, Tully T. Gene discovery in Drosophila: new insights for learning and memory. Annu Rev Neurosci. 1998;21:407–444. doi: 10.1146/annurev.neuro.21.1.407. [DOI] [PubMed] [Google Scholar]
- Durand N, Pottier MA, Siaussat D, Bozzolan F, Maïbèche M, Chertemps T. Glutathione-S-transferases in the olfactory organ of the noctuid moth. Front Physiol. 2018;9:1283. doi: 10.3389/fphys.2018.01283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evgen'ev M, Levin A, Lozovskaya E. The analysis of a temperature-sensitive (ts) mutation influencing the expression of heat shock-inducible genes in Drosophila melanogaster. Mol Gen Genet. 1979;176:275–280. doi: 10.1007/BF00273222. [DOI] [PubMed] [Google Scholar]
- Evgen'ev MB, Garbuz DG, Zatsepina OG (2014) Heat shock proteins and whole body adaptation to extreme environments. Springer, Berlin
- Fedotov SA, Besedina NG, Bragina JV, Danilenkova LV, Kamysheva EA, Kamyshev NG. The involvement of neurons expressing gene factor of interpulse interval (fipi) in regulation of courtship behavior of Drosophila melanogaster males. Integr Physiol. 2020;4:366–379. doi: 10.33910/2687-1270-2020-1-4-365-378. [DOI] [Google Scholar]
- Fleshner M, Campisi J, Amiri L, Diamond D. Cat exposure induces both intra- and extracellular Hsp72: the role of adrenal hormones. Psychoneuroendocrinology. 2004;29:1142–1152. doi: 10.1016/j.psyneuen.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Flexner JB, Flexner LB, Stellar E. Memory in mice as affected by intracerebral puromycin. Science. 1963;141:57–59. doi: 10.1126/science.141.3575.57. [DOI] [PubMed] [Google Scholar]
- Foster NL, Lukowiak K, Henry TB. Time-related expression profiles for heat shock protein gene transcripts (HSP40, HSP70) in the central nervous system of Lymnaea stagnalis exposed to thermal stress. Commun Integr Biol. 2015;8:e1040954. doi: 10.1080/19420889.2015.1040954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukudo S, Abe K, Hongo M, Utsumi A, Itoyama Y. Brain-gut induction of heat shock protein (HSP) 70 mRNA by psychophysiological stress in rats. Brain Res. 1997;757(1):146–148. doi: 10.1016/s0006-8993(97)00179-0. [DOI] [PubMed] [Google Scholar]
- Fukudo S, Abe K, Itoyama Y, Mochizuki S, Sawai T, Hongo M. Psychophysiological stress induces heat shock cognate protein 70 messenger RNA in the hippocampus of rats. Neuroscience. 1999;91(4):1205–1208. doi: 10.1016/s0306-4522(99)00069-x. [DOI] [PubMed] [Google Scholar]
- Ge X, Hannan F, Xie Z, Feng C, Tully T, Zhou H, Xie Z, Zhong Y. Notch signaling in Drosophila long-term memory formation. Proc Natl Acad Sci U S A. 2004;101:10172–10176. doi: 10.1073/pnas.0403497101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstner JR, Yin JC. Circadian rhythms and memory formation. Nat Rev Neurosci. 2010;11:577–588. doi: 10.1038/nrn2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godenschwege TA, Reisch D, Diegelmann S, Eberle K, Funk N, Heisenberg M, Hoppe V, Hoppe J, Klagges BRE, Martin JR, Nikitina EA, Putz G, Reifegerste R, Reisch N, Rister J, Schaupp M, Scholz H, Schwarzel M, Werner U, Zars TD, Buchner S, Buchner E. Flies lacking all synapsins are unexpectedly healthy but are impaired in complex behaviour. Eur J Neurosci. 2004;20(3):611–622. doi: 10.1111/j.1460-9568.2004.03527.x. [DOI] [PubMed] [Google Scholar]
- Gong WJ, Golic KG. Genomic deletions of the Drosophila melanogaster Hsp70 genes. Genetics. 2004;168:1467–1476. doi: 10.1534/genetics.104.030874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez D, Fraichard S, Grassein P, Delarue P, Senet P, Nicolaï A, Chavanne E, Mucher E, Artur Y, Ferveur JF, Heydel JM, Briand L, Neiers F. Characterization of a Drosophila glutathione transferase involved in isothiocyanate detoxification. Insect Biochem Mol Biol. 2018;95:33–43. doi: 10.1016/j.ibmb.2018.03.004. [DOI] [PubMed] [Google Scholar]
- Griffith LC, Ejima A. Courtship learning in Drosophila melanogaster: diverse plasticity of a reproductive behavior. Learn Mem. 2009;16:743–750. doi: 10.1101/lm.956309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Z, Buhl LK, Quinn WG, Littleton JT. Altered gene regulation and synaptic morphology in Drosophila learning and memory mutants. Learn Mem. 2011;18:191–206. doi: 10.1101/lm.2027111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Kim SY, Artis S, Molfese DL, Schumacher A, Sweatt JD, Paylor RE, Lubin FD. Histone methylation regulates memory formation. J Neurosci. 2010;30:3589–3599. doi: 10.1523/JNEUROSCI.3732-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyurko DM, Soti C, Stetak A, Csermely P. System level mechanisms of adaptation, learning, memory formation and evolvability: the role of chaperone and other networks. Curr Protein Pept Sci. 2014;15:171–188. doi: 10.2174/1389203715666140331110522. [DOI] [PubMed] [Google Scholar]
- Harris N, Braiser DJ, Dickman DK, Fetter RD, Tong A, Davis GW. The innate immune receptor PGRP-LC controls presynaptic homeostatic plasticity. Neuron. 2015;88:1157–1164. doi: 10.1016/j.neuron.2015.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Hooper PL, Durham HD, Török Z, Crul T, Vígh L. The central role of heat shock factor 1 in synaptic fidelity and memory consolidation. Cell Stress Chaperones. 2016;21:745–753. doi: 10.1007/s12192-016-0709-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igaz LM, Bekinschtein P, Izquierdo I, Medina JH. One-trial aversive learning induces late changes in hippocampal CaMKIIα, Homer 1a, Syntaxin 1a and ERK2 protein levels. Mol Brain Res. 2004;132:1–12. doi: 10.1016/j.molbrainres.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Ishimoto H, Sakai T, Kitamoto T. Ecdysone signaling regulates the formation of long-term courtship memory in adult Drosophila melanogaster. Proc Natl Acad Sci U S A. 2009;106:6381–6386. doi: 10.1073/pnas.0810213106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SG, Nixon KC, Chubak MC, Kramer JM. Mushroom body specific transcriptome analysis reveals dynamic regulation of learning and memory genes after acquisition of long-term courtship memory in Drosophila. G3: Genes. Genom Genet. 2018;8:3433–3446. doi: 10.1534/g3.118.200560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabil H, Kabil O, Banerjee R, Harshman LG, Pletcher SD. Increased transsulfuration mediates longevity and dietary restriction in Drosophila. Proc Natl Acad Sci U S A. 2011;108:16831–16836. doi: 10.1073/pnas.1102008108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa N, Ryo K, Mugiya Y. Enhanced expression of stress protein 70 in the brains of goldfish, Carassius auratus, reared with bluegills, Lepomis macrochirus. Fish Physiol Biochem. 1999;21:103–110. doi: 10.2108/zsj.17.1061. [DOI] [Google Scholar]
- Kahsai L, Zars T. Learning and memory in Drosophila: behavior, genetics, and neural systems. Int Rev Neurobiol. 2011;99:139–167. doi: 10.1016/B978-0-12-387003-2.00006-9. [DOI] [PubMed] [Google Scholar]
- Kaletsky R, Lakhina V, Arey R, Williams A, Landis J, Ashraf J, Murphy CT. The C. elegans adult neuronal IIS/FOXO transcriptome reveals adult phenotype regulators. Nature. 2016;529(7584):92–96. doi: 10.1038/nature16483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamyshev NG, Iliadi KG, Bragina JV. Drosophila conditioned courtship: two ways of testing memory. Learn Mem. 1999;6:1–20. [PMC free article] [PubMed] [Google Scholar]
- Kandel ER. The molecular biology of memory storage: a dialogue between genes and synapses. Science. 2001;294:1030–1038. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- Kandel ER. The molecular biology of memory: cAMP, PKA, CRE, CREB-1, CREB-2, and CPEB. Mol Brain. 2012;5:14. doi: 10.1186/1756-6606-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keleman K, Krüttner S, Alenius M, Dickson BJ. Function of the Drosophila CPEB protein Orb2 in long-term courtship memory. Nat Neurosci. 2007;10:1587–1593. doi: 10.1038/nn1996. [DOI] [PubMed] [Google Scholar]
- Kennedy D, Jäger R, Mosser DD, Samali A. Regulation of apoptosis by heat shock proteins. IUBMB Life. 2014;66:327–338. doi: 10.1002/iub.1274. [DOI] [PubMed] [Google Scholar]
- Kim JY, Yenari MA. The immune modulating properties of the heat shock proteins after brain injury. Anatol Cell Biol. 2013;46:1–7. doi: 10.5115/acb.2013.46.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Webb AE. Neuronal functions of FOXO/DAF-16. Nutr Healthy Aging. 2017;4(2):113–126. doi: 10.3233/NHA-160009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H. Production and physiological effects of hydrogen sulfide. Antioxid Redox Signal. 2014;20:783–793. doi: 10.1089/ars.2013.5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasnov GS, Dmitriev AA, Kudryavtseva AV, Shargunov AV, Karpov DS, Uroshlev LA, Melnikova NV, Blinov VM, Poverennaya EV, Archakov AI, Lisitsa AV, Ponomarenko EA. PPLine: An automated pipeline for SNP, SAP, and splice variant detection in the context of proteogenomics. J Proteome Res. 2015;14:3729–3737. doi: 10.1021/acs.jproteome.5b00490. [DOI] [PubMed] [Google Scholar]
- Kumar A, Tiwari AK. Molecular chaperone Hsp70 and its constitutively active form Hsc70 play an indispensable role during eye development of Drosophila melanogaster. Mol Neurobiol. 2018;55:4345–4361. doi: 10.1007/s12035-017-0650-z. [DOI] [PubMed] [Google Scholar]
- Kuntz S, Poeck B, Strauss R. Visual working memory requires permissive and instructive NO/cGMP signaling at presynapses in the Drosophila central brain. Curr Biol. 2017;27:613–623. doi: 10.1016/j.cub.2016.12.056. [DOI] [PubMed] [Google Scholar]
- Kuzin BA, Nikitina EA, Cherezov RO, Vorontsova JE, Slezinger MS, Zatsepina OG, Simonova OB, Enikolopov GN, Savvateeva-Popova EV. Combination of hypomorphic mutations of the Drosophila homologues of aryl hydrocarbon receptor and nucleosome assembly protein family genes disrupts morphogenesis, memory and detoxification. PLoS One. 2014;9:e94975. doi: 10.1371/journal.pone.0094975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon HJ, Kim W, Jung HY, Kang MS, Kim JW, Hahn KR, Yoo DY, Yoon YS, Hwang IK, Kim DW. Heat shock protein 70 increases cell proliferation, neuroblast differentiation, and the phosphorylation of CREB in the hippocampus. Lab Anim Res. 2019;35:21. doi: 10.1186/s42826-019-0020-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin LR, Han PL, Hwang PM, Feinstein PG, Davis RL, Reed RR. The Drosophila learning and memory gene rutabaga encodes a Ca2+/Calmodulin-responsive adenylyl cyclase. Cell. 1992;68:479–489. doi: 10.1016/0092-8674(92)90185-f. [DOI] [PubMed] [Google Scholar]
- Livingstone MS, Sziber PP, Quinn WG. Loss of calcium/calmodulin responsiveness in adenylate cyclase of rutabaga, a Drosophila learning mutant. Cell. 1984;37:205–215. doi: 10.1016/0092-8674(84)90316-7. [DOI] [PubMed] [Google Scholar]
- Lukowiak K, Orr M, de Caigny P, Lukowiak KS, Rosenegger D, Han JI, Dalesman S. Ecologically relevant stressors modify long-term memory formation in a model system. Behav Brain Res. 2010;214:18–24. doi: 10.1016/j.bbr.2010.05.011. [DOI] [PubMed] [Google Scholar]
- Martín-Peña A, Rincón-Limas DE, Fernandez-Fúnez P. Engineered Hsp70 chaperones prevent Aβ42-induced memory impairments in a Drosophila model of Alzheimer’s disease. Sci Rep. 2018;8:1–14. doi: 10.1038/s41598-018-28341-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastushita-Sakai T, White-Grindley E, Samuelson J, Seidel C, Si K. Drosophila Orb2 targets genes involved in neuronal growth, synapse formation, and protein turnover. Proc Natl Acad Sci U S A. 2010;107:11987–11992. doi: 10.1073/pnas.1004433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride SM, Giuliani G, Choi C, Krause P, Correale D, et al. Mushroom body ablation impairs short-term memory and long-term memory of courtship conditioning in Drosophila melanogaster. Neuron. 1999;24:967–977. doi: 10.1016/s0896-6273(00)81043-0. [DOI] [PubMed] [Google Scholar]
- McDonald MJ, Rosbash M. Microarray analysis and organization of circadian gene expression in Drosophila. Cell. 2001;107:567–578. doi: 10.1016/s0092-8674(01)00545-1. [DOI] [PubMed] [Google Scholar]
- Mery F, Belay AT, So AK-C, Sokolowski MB, Kawecki TJ. Natural polymorphism affecting learning and memory in Drosophila. Proc Natl Acad Sci. 2007;104:13051–13055. doi: 10.1073/pnas.0702923104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshid A, Chou SD, Prince T, Zhang Y, Bharti A, Calderwood SK. Protein kinase A binds and activates heat shock factor 1. PLoS One. 2010;5:e13830. doi: 10.1371/journal.pone.0013830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai Y, Tsugane M, Oka J, Kimura H. Hydrogen sulfide induces calcium waves in astrocytes. FASEB J. 2004;18:557–559. doi: 10.1096/fj.03-1052fje. [DOI] [PubMed] [Google Scholar]
- Nagpure BV, Bian JS. Brain, learning, and memory: role of H2S in neurodegenerative diseases. Handb Exp Pharmacol. 2015;230:193–215. doi: 10.1007/978-3-319-18144-8_10. [DOI] [PubMed] [Google Scholar]
- Nikitina E, Tokmatcheva E, Savateeva-Popova E. Heat shock during the development of central structures of the Drosophila brain: memory formation in the l (1) ts403 mutant of Drosophila melanogaster. Russ J Genet. 2003;39:25–31. doi: 10.1023/A:1022062609102. [DOI] [PubMed] [Google Scholar]
- Obata F, Kuranaga E, Tomioka K, Ming M, Takeishi A, Chen CH, Soga T, Miura M. Necrosis-driven systemic immune response alters SAM metabolism through the FOXO-GNMT axis. Cell Rep. 2014;7:821–833. doi: 10.1016/j.celrep.2014.03.046. [DOI] [PubMed] [Google Scholar]
- Obata F, Miura M. Enhancing S-adenosyl-methionine catabolism extends Drosophila lifespan. Nat Commun. 2015;6:8332. doi: 10.1038/ncomms9332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr WC, Radyuk SN, Prabhudesai L, Toroser D, Benes JJ, Luchak JM, Mockett RJ, Rebrin I, Hubbard JG, Sohal RS. Overexpression of glutamate-cysteine ligase extends life span in Drosophila melanogaster. J Biol Chem. 2005;280:37331–37338. doi: 10.1074/jbc.M508272200. [DOI] [PubMed] [Google Scholar]
- Paul BD, Snyder SH. H2S: A novel gasotransmitter that signals by sulfhydration. Trends Biochem Sci. 2015;40:687–700. doi: 10.1016/j.tibs.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peixoto L, Abel T. The role of histone acetylation in memory formation and cognitive impairments. Neuropsychopharmacology. 2013;38:62–76. doi: 10.1038/npp.2012.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijanowska J, Kloc M. Daphnia response to predation threat involves heat-shock proteins and the actin and tubulin cytoskeleton. Genesis. 2004;38(2):81–86. doi: 10.1002/gene.20000. [DOI] [PubMed] [Google Scholar]
- Pizarro JM, Haro LS, Barea-Rodriguez EJ. Learning associated increase in heat shock cognate 70 mRNA and protein expression. Neurobiol Learn Mem. 2003;79:142–151. doi: 10.1016/s1074-7427(02)00008-4. [DOI] [PubMed] [Google Scholar]
- Ponton F, Chapuis MP, Pernice M, Sword GA, Simpson SJ. Evaluation of potential reference genes for reverse transcription-qPCR studies of physiological responses in Drosophila melanogaster. J Insect Physiol. 2011;57:840–850. doi: 10.1016/j.jinsphys.2011.03.014. [DOI] [PubMed] [Google Scholar]
- Porto RR, Dutra FD, Crestani AP, Holsinger RMD, Quillfeldt JA, Homem de Bittencourt PI, Jr, de Oliveira Alvares L. HSP70 Facilitates memory consolidation of fear conditioning through MAPK pathway in the hippocampus. Neuroscience. 2018;375:108–118. doi: 10.1016/j.neuroscience.2018.01.028. [DOI] [PubMed] [Google Scholar]
- Presente A, Boyles RS, Serway CN, de Belle JS, Andres AJ. Notch is required for long-term memory in Drosophila. Proc Natl Acad Sci U S A. 2004;101:1764–1768. doi: 10.1073/pnas.0308259100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redt-Clouet C, Trannoy S, Boulanger A, Tokmatcheva E, Savvateeva-Popova E, Parmentier ML, Preat T, Dura JM. Mushroom body neuronal remodelling is necessary for short-term but not for long-term courtship memory in Drosophila. Eur J Neurosci. 2012;35(11):1684–1691. doi: 10.1111/j.1460-9568.2012.08103.x. [DOI] [PubMed] [Google Scholar]
- Renger JJ, Ueda A, Atwood HL, Govind CK, Wu CF. Role of cAMP cascade in synaptic stability and plasticity: ultrastructural and physiological analyses of individual synaptic boutons in Drosophila memory mutants. J Neurosci. 2000;20:3980–3992. doi: 10.1523/JNEUROSCI.20-11-03980.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlf FJ, Sokal RR (1981) Biometry: the principles and practice of statistics in biological research. Freeman New York
- Sakai T, Kitamoto T. Clock, love and memory: circadian and non-circadian regulation of Drosophila mating behavior by clock genes. Sleep Biol Rhythms. 2006;4:255–262. doi: 10.1111/j.1479-8425.2006.00224.x. [DOI] [Google Scholar]
- Santana E, de Los RT, Casas-Tintó S. Small heat shock proteins determine synapse number and neuronal activity during development. PLoS One. 2020;15(5):e0233231. doi: 10.1371/journal.pone.0233231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savvateeva EV, Popov AV, Kamyshev NG, Iliadi KG, Bragina JV, Heisenberg M, Kornhuber J, Riederer P. Age-dependent changes in memory and mushroom bodies in the Drosophila mutant vermilion deficient in the kynurenine pathway of tryptophan metabolism. Ross Fiziol Zh Im I M Sechenova. 1999;85:167–183. [PubMed] [Google Scholar]
- Savvateeva E, Popov A, Kamyshev N, Bragina J, Heisenberg M, Senitz D, Kornhuber J, Riederer P. Age-dependent memory loss, synaptic pathology and altered brain plasticity in the Drosophila mutant cardinal accumulating 3-hydroxykynurenine. J Neural Transm (Vienna) 2000;107(5):581–601. doi: 10.1007/s007020070080. [DOI] [PubMed] [Google Scholar]
- Savvateeva-Popova E, Popov A, Grossman A, Nikitina E, Medvedeva A, Molotkov D, Kamyshev N, Pyatkov K, Zatsepina O, Schostak N, Zelentsova E, Pavlova G, Panteleev D, Riederer P, Evgen`ev M. Non-coding RNA as a trigger of neuropathologic disorder phenotypes in transgenic Drosophila. J Neural Transm (Vienna) 2008;115:1629–1642. doi: 10.1007/s00702-008-0078-8. [DOI] [PubMed] [Google Scholar]
- Savvateeva-Popova E, Popov A, Grossman A, Nikitina E, Medvedeva A, Peresleni A, Korochkin L, Moe JG, Davidowitz E, Pyatkov K, Myasnyankina E, Zatsepina O, Schostak N, Zelentsova E, Evgen'ev M. Pathogenic chaperone-like RNA induces congophilic aggregates and facilitates neurodegeneration in Drosophila. Cell Stress Chaperones. 2007;12:9–19. doi: 10.1379/csc-222r.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savvateeva-Popova EV, Zhuravlev AV, Brázda V, Zakharov GA, Kaminskaya AN, Medvedeva AV, Nikitina EA, Tokmatcheva EV, Dolgaya JF, Kulikova DA, Zatsepina OG, Funikov SY, Ryazansky SS, Evgen‘ev MB. Model for the analysis of genesis of LIM-kinase 1-dependent Williams-Beuren syndrome cognitive phenotypes: INDELs, transposable elements of the Tc1/ Front Genet. 2017;8:123. doi: 10.3389/fgene.2017.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaccianoce S, Del Bianco P, Caricasole A, Nicoletti F, Catalani A. Relationship between learning, stress and hippocampal brain-derived neurotrophic factor. Neuroscience. 2003;121:825–828. doi: 10.1016/s0306-4522(03)00514-1. [DOI] [PubMed] [Google Scholar]
- Schefe JH, Lehmann KE, Buschmann IR, Unger T, Funke-Kaiser H. Quantitative real-time RT-PCR data analysis: current concepts and the novel "gene expression's CT difference" formula. J Mol Med (Berl) 2006;84:901–910. doi: 10.1007/s00109-006-0097-6. [DOI] [PubMed] [Google Scholar]
- Shilova VY, Zatsepina OG, Garbuz DG, Funikov SY, Zelentsova ES, Schostak NG, Kulikov AM, Evgen'ev MB. Heat shock protein 70 from a thermotolerant Diptera species provides higher thermoresistance to Drosophila larvae than correspondent endogenous gene. Insect Mol Biol. 2018;27:61–72. doi: 10.1111/imb.12339. [DOI] [PubMed] [Google Scholar]
- Sitaraman D, Zars M, Laferriere H, Chen YC, Sable-Smith A, et al. Serotonin is necessary for place memory in Drosophila. Proc Natl Acad Sci U S A. 2008;105:5579–5584. doi: 10.1073/pnas.0710168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder PM, Baratashvili M, Grzeschik NA, Leuvenink HGD, Kuijpers L, Huitema S, Schaap O, Giepmans BNG, Kuipers J, Miljkovic JL, Mitrovic A, Bos EM, Szabó C, Kampinga HH, Dijkers PF, Bos EM, Szabó C, Kampinga HH, Dijkers PF, Dunnen WFAD, Filipovic MR, Goor HV, Sibon OCM. Overexpression of Cystathionine γ-lyase suppresses detrimental effects of spinocerebellar ataxia type 3. Mol Med. 2016;21:758–768. doi: 10.2119/molmed.2015.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolowski MB. Drosophila: genetics meets behaviour. Nat Rev Genet. 2001;2:879–890. doi: 10.1038/35098592. [DOI] [PubMed] [Google Scholar]
- Stevens B, Allen NJ, Vazquez LE, Howell GR, Christopherson KS, Nouri N, Micheva KD, Mehalow AK, Huberman AD, Stafford B, Sher A, Litke AM, Lambris JD, Smith SJ, John SWM, Barres BA. The classical complement cascade mediates CNS synapse elimination. Cell. 2007;131:1164–1178. doi: 10.1016/j.cell.2007.10.036. [DOI] [PubMed] [Google Scholar]
- Sun Y, MacRae TH. Small heat shock proteins: molecular structure and chaperone function. Cell Mol Life Sci. 2005;62(21):2460–2476. doi: 10.1007/s00018-005-5190-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunada H, Riaz H, de Freitas E, Lukowiak K, Swinton C, Swinton E, Protheroe A, Shymansky T, Komatsuzaki Y, Lukowiak K. Heat stress enhances LTM formation in Lymnaea: role of HSPs and DNA methylation. J Exp Biol. 2016;219:1337–1345. doi: 10.1242/jeb.134296. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Usuda N, Murata S, Nakazawa A, Ohtsuka K, Takagi H. Presence of molecular chaperones, heat shock cognate (Hsc) 70 and heat shock proteins (Hsp) 40, in the postsynaptic structures of rat brain. Brain Res. 1999;816:99–110. doi: 10.1016/s0006-8993(98)01083-x. [DOI] [PubMed] [Google Scholar]
- Thekkuveettil A, Lakhotia SC. Regulation of HSP70 in excitatory neurons: Possible implications for neuronal functioning. J Biosci. 1996;21:631–639. doi: 10.1007/BF02703141. [DOI] [Google Scholar]
- Timpe LC, Schwarz TL, Tempel BL, Papazian DM, Jan YN, Jan LY. Expression of functional potassium channels from Shaker cDNA in Xenopus oocytes. Nature. 1988;331:143–145. doi: 10.1038/331143a0. [DOI] [PubMed] [Google Scholar]
- Tully T. Discovery of genes involved with learning and memory: an experimental synthesis of Hirschian and Benzerian perspectives. Proc Natl Acad Sci. 1996;93:13460–13467. doi: 10.1073/pnas.93.24.13460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tully T, Boynton S, Brandes C, Dura JM, Mihalek R, Preat T, Villella A. Genetic dissection of memory formation in Drosophila melanogaster. Cold Spring Harb Symp Quant Biol. 1990;55:203–211. doi: 10.1101/sqb.1990.055.01.022. [DOI] [PubMed] [Google Scholar]
- Tully T, Preat T, Boynton SC, Del Vecchio M. Genetic dissection of consolidated memory in Drosophila. Cell. 1994;79:35–47. doi: 10.1016/0092-8674(94)90398-0. [DOI] [PubMed] [Google Scholar]
- Winbush A, Reed D, Chang PL, Nuzhdin SV, Lyons LC, Arbeitman MN. Identification of gene expression changes associated with long-term memory of courtship rejection in Drosophila males. G3: Genes, Genomes, Genetics. 2012;2:1437–1445. doi: 10.1534/g3.112.004119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt SN. Molecular chaperones, α-synuclein, and neurodegeneration. Mol Neurobiol. 2013;47:552–560. doi: 10.1007/s12035-012-8325-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt AR, Yerbury JJ, Ecroyd H, Wilson MR. Extracellular chaperones and proteostasis. Annu Rev Biochem. 2013;82:295–322. doi: 10.1146/annurev-biochem-072711-163904. [DOI] [PubMed] [Google Scholar]
- Xu S, Wilf R, Menon T, Panikker P, Sarthi J, Elefant F. Epigenetic control of learning and memory in Drosophila by Tip60 HAT action. Genetics. 2014;198:1571–1586. doi: 10.1534/genetics.114.171660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao WD, Wu CF. Distinct roles of CaMKII and PKA in regulation of firing patterns and K(+) currents in Drosophila neurons. J Neurophysiol. 2001;85(4):1384–1394. doi: 10.1152/jn.2001.85.4.1384. [DOI] [PubMed] [Google Scholar]
- Yong QC, Choo CH, Tan BH, Low CM, Bian JS. Effect of hydrogen sulfide on intracellular calcium homeostasis in neuronal cells. Neurochem Int. 2010;56:508–515. doi: 10.1016/j.neuint.2009.12.011. [DOI] [PubMed] [Google Scholar]
- Yu D, Akalal D-BG, Davis RL. Drosophila α/β mushroom body neurons form a branch-specific, long-term cellular memory trace after spaced olfactory conditioning. Neuron. 2006;52:845–855. doi: 10.1016/j.neuron.2006.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurinskaya MM, Krasnov GS, Kulikova DA, Zatsepina OG, Vinokurov MG, Chuvakova LN, Rezvykh AP, Funikov SY, Morozov AV, Evgen’ev MB. H2S counteracts proinflammatory effects of LPS through modulation of multiple pathways in human cells. Inflamm Res. 2020;69:481–495. doi: 10.1007/s00011-020-01329-x. [DOI] [PubMed] [Google Scholar]
- Zars T. Working memory: It’sa gas, gas, gas. Curr Biol. 2017;27:R179–RR81. doi: 10.1016/j.cub.2017.01.005. [DOI] [PubMed] [Google Scholar]
- Zhang J, Little CJ, Tremmel DM, Yin JC, Wesley CS. Notch-inducible hyperphosphorylated CREB and its ultradian oscillation in long-term memory formation. J Neurosci. 2013;33:12825–12834. doi: 10.1523/JNEUROSCI.0783-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Sabirzhanov B, Wu J, Faden AI, Stoica BA. Voluntary exercise preconditioning activates multiple antiapoptotic mechanisms and improves neurological recovery after experimental traumatic brain injury. J Neurotrauma. 2015;32:1347–1360. doi: 10.1089/neu.2014.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Wu CF. Differential modulation of potassium currents by cAMP and its long-term and short-term effects: dunce and rutabaga mutants of Drosophila. J Neurogenet. 1993;9:15–27. doi: 10.3109/01677069309167273. [DOI] [PubMed] [Google Scholar]
- Zhou C, Huang H, Kim SM, Lin H, Meng X, Han KA, Chiang AS, Wang JW, Jiao R, Rao Y. Molecular genetic analysis of sexual rejection: roles of octopamine and its receptor OAMB in Drosophila courtship conditioning. J Neurosci. 2012;32:14281–14287. doi: 10.1523/JNEUROSCI.0517-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuravlev A, Nikitina E, Savvateeva-Popova E. Learning and memory in Drosophila: physiologic and genetic bases. Usp Fiziol Nauk. 2015;46:76–92. [PubMed] [Google Scholar]
- Zovkic IB, Guzman-Karlsson MC, Sweatt JD. Epigenetic regulation of memory formation and maintenance. Learn Mem. 2013;20:61–74. doi: 10.1101/lm.026575.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 2105 kb)
(XLSX 2422 kb)
(XLSX 22 kb)
Data Availability Statement
Sequence data were deposited in the NCBI GEO database under the number GSE152647.