Abstract
Rubella virus (RuV) is the causative agent of rubella (“German measles”) and remains a global health concern. Until recently, RuV was the only known member of the genus Rubivirus and the only virus species classified within the Matonaviridae family of positive-sense RNA viruses. Recently, two new rubella-like matonaviruses, Rustrela virus and Ruhugu virus, have been identified in several mammalian species, along with more divergent viruses in fish and reptiles. To screen for the presence of additional novel rubella-like viruses, we mined published transcriptome data using genome sequences from Rubella, Rustrela, and Ruhugu viruses as baits. From this, we identified a novel rubella-like virus in a transcriptome of Tetronarce californica—order Torpediniformes (Pacific electric ray)—that is more closely related to mammalian Rustrela virus than to the divergent fish matonavirus and indicative of a complex pattern of cross-species virus transmission. Analysis of host reads confirmed that the sample analysed was indeed from a Pacific electric ray, and two other viruses identified in this animal, from the Arenaviridae and Reoviridae, grouped with other fish viruses. These findings indicate that the evolutionary history of the Matonaviridae is more complex than previously thought and highlights the vast number of viruses that remain undiscovered.
Keywords: metatranscriptomics, virus discovery, rubella, ruhugu, rustrela, Matonaviridae, Arenaviridae, Reoviridae, fish, virus evolution, phylogeny
1. Introduction
Rubella virus (RuV) (Matonaviridae: Rubivirus) is a single-stranded positive-sense RNA virus [1]. It is best known as the causative agent of rubella, sometimes known as “German measles”—a relatively mild measles-like illness [2]. On occasion, however, RuV can result in more severe disease, including complications such as miscarriage or congenital rubella syndrome (CRS) if contracted during pregnancy [3,4]. Despite the availability of effective vaccines, including the combination measles, mumps, and rubella (MMR) vaccine [5], they are deployed in only around half of the world’s countries [6]. RuV, therefore, remains a significant global health concern, particularly in developing countries where childhood infection rates are high and vaccination efforts are low or non-existent [7].
The disease rubella was first described in 1814 [8], and until recently, RuV was the only known species within the genus Rubivirus, the only genus in the family Matonaviridae [9]. This picture has changed with the discovery of other rubella-like viruses through the use of metagenomic technologies. The first of these, Guangdong Chinese water snake rubivirus, was discovered via a large virological survey of vertebrates in China and is highly divergent from RuV, only sharing around 34% amino acid identity [10]. Similarly, Tiger flathead matonavirus was identified through a metatranscriptomic exploration of Australian marine fish [11], and, along with Guangdong Chinese water snake rubivirus, forms a sister clade to RuV. Even more recently, two novel rubella-like virus species were described: Rustrela virus, identified in several mammalian species in, and in close proximity to, a zoo in Germany, and Ruhugu virus from bats in Uganda [12]. These viruses represent the first non-human mammalian viruses within this family. Such findings begin to shed light on the deeper evolutionary history of this previously elusive viral family.
The popularisation of metagenomic methods to identify novel viruses within tissue or environmental samples has enabled their rapid discovery and provided important insights into viral diversity and evolution [13,14]. Here, we performed data mining of metagenomic data to determine if related matonaviruses were present in other vertebrate taxa, screening the National Center for Biotechnology Information (NCBI) Transcriptome Shotgun Assembly (TSA) database against the genome sequences of Rubella, Ruhugu, and Rustrela viruses.
2. Materials and Methods
2.1. TSA Mining for Novel Matonaviruses
To identify the presence of novel rubella-like viruses in vertebrates, translated genome sequences of Rustrela virus (MN552442, MT274274, and MT274275), Ruhugu virus (MN547623), and RuV strain 1A (KU958641) were screened against transcriptome assemblies available in NCBI’s TSA database (https://www.ncbi.nlm.nih.gov/genbank/tsa/, accessed on 1 February 2021) using the online translated Basic Local Alignment Search Tool (tBLASTn) [15]. Searches were restricted to vertebrates (taxonomic identifier: 7742), excluding Homo sapiens (taxonomic identifier: 9606), and utilised the BLOSUM45 matrix to increase the chance of finding highly divergent viruses. Putative viral sequences were then queried using the DIAMOND BLASTx (v.2.02.2) [16] algorithm against the non-redundant (nr) protein database for confirmation using the “more-sensitive” flag.
2.2. Pacific Electric Ray Transcriptome Assembly and Annotation
To recover further fragments of the novel matonavirus genome and screen for other viruses in the Pacific electric ray transcriptome (see Results), raw sequencing reads were downloaded from the NCBI Short Read Archive (SRA) (Bioproject: PRJNA322346). These reads were first quality trimmed and then assembled de novo using Trinity RNA-Seq (v2.11) with the “trimmomatic” flag option [17]. Assembled contigs were annotated based on similarity searches against the NCBI nucleotide (nt) database using the BLASTn algorithm and the non-redundant protein (nr) database using DIAMOND BLASTx (v.2.02.2) [16]. Novel viral sequences were collated by searching the annotated contigs, and these potential viruses were confirmed with additional BLASTx searches against nr and nt databases.
2.3. Abundance Estimations
Virus abundance, expressed as the standardised total number of raw viral sequencing reads that comprise a given contig, was calculated using the “align and estimate” module available for Trinity RNA-seq with the “prep_reference” flag enabled, RNA-seq by Expectation-Maximization (RSEM) [18] as the method for abundance estimation, and Bowtie 2 [19] as the alignment method.
2.4. Phylogenetic Analysis
A range of virus genomes representative of each virus family and sub-family under consideration, and for a range of host species, were retrieved from GenBank for phylogenetic analysis. The amino acid sequences of previously described viruses were aligned with those generated here with Multiple Alignment using Fast Fourier Transform (MAFFT) (v.7.4) [20] employing the E-INS-i algorithm. All sequence alignments were visualised in Geneious Prime (v2020.2.4) [21]. Ambiguously aligned regions were removed using trimAl (v.1.2) [22] with the ”gappyout” flag. The maximum likelihood approach available in IQ-TREE (v.1.6.12) [23] was used to generate phylogenetic trees for each family. ModelFinder (MFP) [24] was used to determine the best-fit model of amino acid substitution for each tree, and 1000 ultrafast bootstrap replicates [25] were performed to assess nodal support. Phylogenetic trees were annotated with FigTree (v.1.4.4) [26].
2.5. Virus Nomenclature
The new T. californica viruses described were named with “Tetronarce” preceding their respective family name to signify their electric ray host.
2.6. Data Availability
The Rustrela and Ruhugu virus sequences used for data mining in the project are available under Bioproject PRJNA576343. The raw T. californica sequence reads are available at Bioproject PRJNA322346. All other publicly available sequences used in analyses can be accessed via their corresponding accession codes (see relevant figures). Consensus sequences of the new viruses identified in this study are available on GenBank under accession numbers BK014632, BK014633, BK014634 and BK014635.
3. Results
3.1. Identification of a Novel Matonaviridae in Pacific Electric Ray
We identified a complete structural polyprotein and several fragments of the RNA-dependent RNA polymerase (RdRp) from a divergent rubi-like virus that we tentatively named Tetronarce matonavirus. Screening of Rustrela, Ruhugu, and Rubella 1A virus sequences against the TSA database revealed matches to a contig of 3006 nucleotides (1001 amino acids) from a transcriptome of the electric organ of a female Pacific electric ray (Tetronarce californica) [27], ranging in sequence identity from 37.6 to 44.9% at the protein level. Querying this contig against the non-redundant protein database using the BLASTx algorithm confirmed that it had ~38% sequence identity to the structural protein of Rubella virus (accession: AAY34244.1; query coverage: 87%; e-value 0.0; see Table 1).
Table 1.
Novel Tetronarce viruses and Matonaviridae referenced in this study.
| Virus | Family (Sub-Family) | NCBI Accession Number | Host Species | Location | Sequence Length (Amino Acids) | Closest Amino Acid Match (GenBank Accession) |
|---|---|---|---|---|---|---|
| Rubella virus (1A) | Matonaviridae | KU958641 | Homo sapiens | Worldwide | NS: 2116 S: 1063 | - |
| Rustrela virus | Matonaviridae | MN552442, MT274274-5 | Equus asinus, Hydrochoerus hydrochaeris Linnaeu, Apodemus flavicollis | Germany | NS: 1921 S: 1017 | - |
| Ruhugu virus | Matonaviridae | MN547623 | Hipposideros cyclops | Africa | NS: 2048 S: 1098 | - |
| Tiger flathead matonavirus | Matonaviridae | - | Neoplatycephalus richardsoni | Australia | NS: 1590 | NS: Guangdong chinese water snake rubivirus 34.9% (AVM87614) |
| Gaungdong chinese water snake rubivirus | Matonaviridae | MG600129 | Enhydris chinensis | China | NS: 1710 * S: 687 * | - |
| Tetronarce matonavirus † | Matonaviridae | - | Tetronarce californica | USA | NS: 399 * S: 1001 * | NS: Rubella virus 64.9%(YP_004617078.1) S: Rubella virus 38.1% (AAY34244.1) |
| Tetronarce arenavirus † | Arenaviridae | - | Tetronarce californica | USA | NS: 753 * | Salmon pescarenavirus 1 53.0% (QEG08233.1) Wenling frogfish arenavirus 2 51.9% (YP_009551605.1) |
| Tetronarce reovirus † | Reoviridae (Sedoreovirinae) | - | Tetronarce californica | USA | NS: 615 * | Wenling scaldfish reovirus 54.4% (AVM87459.1) |
NS = non-structural proteins. S = structural proteins. † Viruses identified in this study. * Derived from contigs and may not represent complete (poly)protein length.
To recover further the genomic regions of Tetronarce matonavirus, we de novo assembled the raw sequencing reads from the Pacific electric ray sample. Accordingly, transcripts of a non-structural polyprotein, the p90 (RdRp) protein (815 amino acids in length in RuV), were recovered. By mapping contigs to the ruhugu virus non-structural polyprotein reference sequence, a continuous 399 amino acid section of the RdRp was identified and used for additional phylogenetic analysis. The RdRp fragment matched most closely to the RdRp of RuV (accession: YP_004617078.1; identity: 64.9%; query coverage: 99%; e-value: 0.0; see Table 1).
3.2. Identification of Novel Arenaviridae and Reoviridae in Pacific Electric Ray
To help confirm that the host sequence reads analysed were indeed from the Pacific electric ray, we mined for additional viruses in the relevant transcriptome data: the discovery of additional mammalian associated viruses would cast doubt on the provenance of these samples. Accordingly, BLASTx analyses revealed contigs containing divergent sequences most closely related to viruses within Arenaviridae and Reoviridae. Specifically, we obtained a 753 amino acid fragment of the L segment of a novel arenavirus (tentatively termed Tetronarce arenavirus) and a 615 amino acid segment of the segment 1 protein of a novel reovirus (Tetronarce reovirus). BLASTx searches indicated that the 753 amino acid Tetronarce arenavirus fragment was most closely related to salmon pescarenavirus 1 (53.0% sequence identity, Table 1), and the 615 Tetronarce reovirus segment was most closely related to Wenling scaldfish reovirus (54.4% sequence identity; Table 1).
3.3. Screening for Host Genes
To further address whether the host reads analysed were from the Pacific electric ray, rather than inadvertent mammalian contamination, the assembled Pacific electric ray contigs were screened for non-electric ray vertebrate annotations. We found that the closest genetic matches to all contigs with sequence homology to eukaryote genomes were either from the Pacific electric ray, related species within the Batoidea superorder, or genes that are highly conserved across vertebra, including cartilaginous and bony fish. Hence, these results strongly suggest that the Tetronarce matonavirus was indeed present in the Pacific electric ray transcriptome.
3.4. Virus Abundance
We estimated the standardised abundances of the three novel viruses in comparison to the stably expressed host gene, 40S ribosomal protein S13 (RPS13) (Figure 1a). RPS13 was chosen as the reference host gene for comparison due to evidence that it shows limited inter- and intra-tissue variation in expression levels [28]. Of the three viruses present, Tetronarce matonavirus was the most abundant (standardised abundance = 4.4. × 10−6), followed by Tetronarce arenavirus (8.0 × 10−7), and then Tetronarce reovirus (2.7 × 10−7). The abundance of reads from the host gene, RPS13, was 2.4 × 10−4.
Figure 1.
(a) Standardised abundance of viral reads (log) from the novel viruses (matonavirus, reovirus, and arenavirus) and a host gene (RPS13). (b,c) Phylogenetic trees of the non-structural and structural polyproteins of Tetronarce matonavirus within the Matonaviridae. (d) Phylogenetic tree of the L segment (RdRp) of Tetronarce arenavirus within the Arenaviridae. (e) Phylogenetic tree of segment 1 (RdRp) of Tetronarce reovirus within the Reoviridae. All phylogenetic trees are rooted at the midpoint. Viruses from fish species are highlighted within each phylogeny. Names of novel viruses identified in this study are in bold and highlighted. Nodes with bootstrap support values of >70% are indicated with asterisks (*).
3.5. Phylogenetic Analysis of the Novel Viruses
Maximum likelihood phylogenetic trees were estimated using amino acid sequences of the non-structural (Figure 1b) and structural (Figure 1c) polyproteins of Matonaviridae, as well as the RdRp (L segment) of Arenaviridae (Figure 1d) and the RdRp (segment 1) of Reoviridae (Figure 1e) to determine the evolutionary relationships of the new Pacific electric ray viruses in the context of previously described virus species. As there is no available structural polyprotein sequence for Tiger flathead matonavirus, this virus could not be included in the structural protein tree (Figure 1c).
For phylogenetic analysis of Tetronarce matonavirus, separate sequence alignments were made for the non-structural and structural polyprotein, and individual trees were generated for each genomic region. However, the phylogenetic analysis of both the non-structural and structural polyproteins revealed similar topologies, with the novel Tetronarce matonavirus being most closely related to Rustrela virus (found in donkeys, mice, and capybara) and forming an ingroup to Ruhugu virus (found in bats). Strikingly, therefore, Tetronarce matonavirus does not group with the only other fish (Neoplatycephalus richardsoni—scorpaeniformes) virus in this family, Tiger flathead matonavirus, and the evolutionary history of this virus family does not follow strict virus–host co-divergence.
In contrast, Tetronarce arenavirus falls within a group of viruses that have been identified in other fish host species, including Big eyed perch arenavirus and Wenling frogfish arenavirus-2. Unlike the Reoviridae and Matonaviridae phylogenetic trees, all fish arenaviruses identified to date form a distinct fish arenavirus clade.
The Reoviridae comprises two sub-families (Sedoreovirinae and Spinareovirinae). Reoviruses have previously been identified in fish (Figure 1e), although these viruses are primarily Orthoreoviruses and Aquareoviruses (sub-family: Spinareovirinae). The Tetronarce reovirus and its closest match in the NCBI database (Wenling scaldfish virus, Table 1) are the only two fish viruses that fall within the Sedoreovirinae sub-family and appear to be more closely related to vector-borne reoviruses (orbiviruses) known to infect various mammalian hosts [29], including African horse sickness virus (AHSV) and Equine encephalosis virus (EEV), and the more recently discovered snake-associated Letea virus [30].
3.6. Matonaviridae Amino Acid Conservation
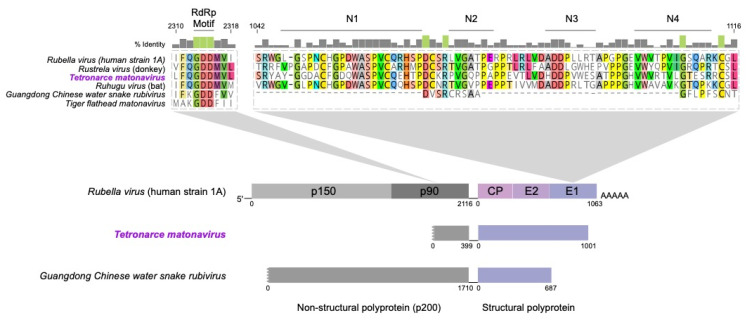
We screened for evidence of conserved sequences or motifs between the novel matonavirus and previously described viruses. An immune-reactive region within the E1 structural protein of RuV [31] containing four neutralising B cell epitopes N1–N4 [32] revealed significant levels of conservation with corresponding regions in the other mammalian matonaviruses [12]. These regions were also well conserved in Tetronarce matonavirus (Figure 2), compatible with our phylogenetic results. In contrast, the Guangdong Chinese water snake rubivirus, which has a very short structural protein sequence (687 amino acids compared to ~1098 amino acids), exhibited little conservation of the E1 protein and other structural proteins overall. There is clear conservation in regions corresponding to epitopes N2–N4 between the mammalian viruses and the electric ray virus, including a glycine residue in N4 conserved across all five viruses in the alignment (Figure 2). In addition, a highly conserved glycine-aspartic acid-aspartic acid (GDD) motif present in the RdRp [33] was also conserved in Tetronarce matonavirus as well as all other matonaviruses referenced here (Figure 2).
Figure 2.
Conserved sequence motifs between human Rubella virus (strain 1A), Rustrela virus (donkey), Ruhugu virus (bat), Tetronarce matonavirus, Tiger flathead matonavirus, and Guangdong Chinese water snake rubivirus shown. The non-structural (p200) polyprotein contains a conserved GDD amino acid motif in the p90 (RdRp) protein at positions 2313–2315. Regions corresponding to RuV B cell epitopes N1–N4 within E1 in the structural protein alignment at positions 1042–1116 are also illustrated. Bar graphs of percentage identity (%) of residues shown above the alignments with identities of 100% highlighted green. Genomic organisation of Matonaviridae (Rubella virus strain 1A) is shown below with segments able to be recovered from Tetronarce matonavirus and Guangdong Chinese water snake rubivirus indicated underneath for comparison. These comprise the complete structural polyprotein and partial non-structural polyproteins.
4. Discussion
We identified a complete structural polyprotein and a partial non-structural polyprotein (comprising a section of the RdRp) from a divergent matonavirus in a transcriptome of a Pacific electric ray. Additionally, partial RdRp transcripts of other novel viruses within the Reoviridae and Arenaviridae were identified in this same electric ray transcriptome.
The novel Tetronarce matonavirus identified here exhibited clear sequence identity to RuV, as well as to the Rustrela and Ruhugu viruses recently discovered in other mammals. High levels of sequence conservation within the E1 structural protein have been previously shown among RuV, Rustrela virus, and Ruhugu virus [12], including regions corresponding to various immune-reactive B and T cell epitopes in RuV [31,32]. Tetronarce matonavirus shares comparable levels of amino acid conservation to mammalian matonaviruses in this region of E1, whereas the other matonavirus from a non-mammalian host (Guangdong Chinese water snake virus) does not. However, we were unable to recover a structural polyprotein from Tiger flathead matonavirus to incorporate it into our analysis. The non-structural fragment of the novel virus also contains a highly conserved GDD motif within the RdRp, which is highly conserved across many RNA viruses and is essential for efficient replication [33].
Of particular note was that phylogenetic analysis suggested that Tetronarce matonavirus is more closely related to Rustrela virus than to the other, more divergent, fish matonavirus, and hence is indicative of a complex history of cross-species transmission events. Such a conclusion is not dependent on the position of the tree root. Given that this phylogenetic position is somewhat paradoxical, it is important to exclude sample contamination, or that the Tetronarce matonavirus was in fact obtained from a component of the diet of T. californica. Importantly, no significant non-fish matches were identified in the T. californica transcriptome library, with most host reads producing matches to T. californica or related fish species. Similarly, that the novel arenavirus and reovirus identified were most closely related to previously documented fish-associated viruses, rather than those from mammals, again suggests that the unusual phylogenetic position of Tetronarce matonavirus is bona fide. Hence, these data are indicative of a complex history of cross-species transmission of viruses in the Matonaviridae, potentially involving cross-species transmission events among different vertebrate classes. Evidence of such expansive host-jumping events has been observed in the evolution of other virus families, such as hepatitis B viruses in the Hepadnaviridae, in which a number of fish viruses group closely with those identified in mammals [34]. Indeed, cross-species transmission is commonplace in RNA viruses as a whole [35], and the expansion of the Matonaviridae with viruses from such a broad host range suggests this process has also played an important role in the evolutionary history of these viruses. Further screening of transcriptomes will also be paramount in solidifying Tetronarce matonavirus’ position in the Matonaviridae tree by revealing additional evolutionary links.
A potentially new reovirus—Tetronarce reovirus—was also identified in our analysis. The majority of reoviruses found in fish fall within the sub-family Spinareovirinae. In contrast, Tetronarce reovirus appears to be one of two viruses currently known that fall within the Sedoreovirinae sub-family that include vector-borne orbiviruses, such as AHSV and EEV, that cause fatal diseases in equines [36,37]. However, the host range of this sub-family is continually expanding and now contains a recently identified snake-associated Letea virus [30]. It is also notable that Tetronarce reovirus is most clearly related to another fish-associated reovirus—Wenling scaldfish reovirus. Similarly, the Arenaviridae has also been recently expanded, with a host range that was previously thought to be limited to rodents and humans now expanded to include other warm- and cold-blooded vertebrates [38]. The novel arenavirus identified here—Tetronarce arenavirus—clusters with a group of fish and amphibian viruses.
The recovery of a complete structural polyprotein and a partial RdRp, both with conserved amino acid sequences and motifs present across Matonaviridae, as well as a relatively high viral abundance compared with a stably expressed host gene, is highly suggestive of the existence of a novel rubi-like virus in T. californica. While screening of annotated contigs and the existence of two other potentially novel viruses sharing sequence identity to other fish viruses in the assembled transcriptome suggest the Pacific electric ray to be the true host of these viruses, further surveying of this species for viral genomes would be beneficial to further understanding the origin and emergence of matonaviruses like RuV.
Overall, these findings expand the currently established host range of Matonaviridae and suggest a more complex evolutionary history than previously suspected. More broadly, this study demonstrates the value of performing extensive transcriptome sequencing and screening of a wide range of potential hosts to identify animal relatives of viruses of significant health concern and to characterise more of the global virosphere.
Author Contributions
Conceptualisation, R.M.G. and J.L.G.; formal analysis, R.M.G.; resources, J.L.G.; writing—original draft preparation, R.M.G.; writing—review and editing, R.M.G., E.C.H. and J.L.G.; funding acquisition, E.C.H. and J.L.G. All authors have read and agreed to the published version of the manuscript.
Funding
This work was partly funded by ARC Discovery grant DP200102351 awarded to E.C.H. and J.L.G. E.C.H. is funded by an ARC Australian Laureate Fellowship (FL170100022), and J.L.G. is funded by a New Zealand Royal Society Rutherford Discovery Fellowship (RDF-20-UOO-007). R.M.G. was funded by a summer scholarship from GlycoSyn.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The raw T. californica sequence reads are available at Bioproject PRJNA322346. Consensus sequences of the new viruses identified in this study are available on GenBank under accession numbers BK014632, BK014633, BK014634 and BK014635.
Conflicts of Interest
The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Frey T.K. Molecular biology of rubella virus. Adv. Virus Res. 1994;44:69–160. doi: 10.1016/s0065248352760328-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lambert N., Strebel P., Orenstein W., Icenogle J., Poland G.A. Rubella. Lancet. 2015;385:2297–2307. doi: 10.1016/S0140-6736(14)60539-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gregg N.M. Congenital cataract following German measles in the mother. Epidemiol. Infect. 1991;107:iii–xiv. doi: 10.1017/S0950268800048627. discussion xiii–xiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alford C.A., Jr., Neva F.A., Weller T.H. Virologic and Serologic Studies on Human Products of Conception after Maternal Rubella. N. Engl. J. Med. 1964;271:1275–1281. doi: 10.1056/NEJM196412172712501. [DOI] [PubMed] [Google Scholar]
- 5.Schwarz A.J., Jackson J.E., Ehrenkranz N.J., Ventura A., Schiff G.M., Walters V.W. Clinical evaluation of a new measles-mumps-rubella trivalent vaccine. Am. J. Dis. Child. 1975;129:1408–1412. doi: 10.1001/archpedi.1975.02120490026008. [DOI] [PubMed] [Google Scholar]
- 6.Olivé J.M. Report of a Meeting on Preventing Congenital Rubella Syndrome: Immunization Strategies, Surveillance Needs. WHO; Geneva, Italy: 2000. Historical review and summary of meeting objectives; pp. 5–9. [Google Scholar]
- 7.Cutts F.T., Robertson S.E., Diaz-Ortega J.L., Samuel R. Control of rubella and congenital rubella syndrome (CRS) in developing countries, Part 1: Burden of disease from CRS. Bull. World Health Organ. 1997;75:55–68. [PMC free article] [PubMed] [Google Scholar]
- 8.Maton W.G. Some Account of a Rash Liable to be Mistaken for Scarlatina. Med. Trans. Coll. Physicians. 1815;5:149–165. [Google Scholar]
- 9.Ryman K.D., Klimstra S.C., Weaver S.C. Togaviruses: Molecular Biology. Encycl. Virol. 2008;3:116–124. doi: 10.1016/B978-012374410-4.00628-2. [DOI] [Google Scholar]
- 10.Shi M., Lin X.D., Chen X., Tian J.H., Chen L.J., Li K., Wang W., Eden J.S., Shen J.J., Liu L., et al. The evolutionary history of vertebrate RNA viruses. Nature. 2018;556:197–202. doi: 10.1038/s41586-018-0012-7. [DOI] [PubMed] [Google Scholar]
- 11.Geoghegan J.L., Di Giallonardo F., Wille M., Ortiz-Baez A.S., Costa V.A., Ghaly T., Mifsud J.C.O., Turnbull O.M.H., Bellwood D.R., Williamson J.E., et al. Host evolutionary history and ecology shape virome composition in fishes. Virus Evol. 2021;7:veab005. doi: 10.1093/ve/veab005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bennett A.J., Paskey A.C., Ebinger A., Pfaff F., Priemer G., Hoper D., Breithaupt A., Heuser E., Ulrich R.G., Kuhn J.H., et al. Relatives of rubella virus in diverse mammals. Nature. 2020;586:424–428. doi: 10.1038/s41586-020-2812-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi M., Zhang Y.Z., Holmes E.C. Meta-transcriptomics and the evolutionary biology of RNA viruses. Virus Res. 2018;243:83–90. doi: 10.1016/j.virusres.2017.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turnbull O.M.H., Ortiz-Baez A.S., Eden J.S., Shi M., Williamson J.E., Gaston T.F., Zhang Y.Z., Holmes E.C., Geoghegan J.L. Meta-Transcriptomic Identification of Divergent Amnoonviridae in Fish. Viruses. 2020;12:1254. doi: 10.3390/v12111254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 16.Buchfink B., Xie C., Huson D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 17.Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li B., Dewey C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katoh K., Misawa K., Kuma K., Miyata T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geneious Prime. [(accessed on 4 February 2020)]; Available online: https://www.geneious.com.
- 22.Capella-Gutierrez S., Silla-Martinez J.M., Gabaldon T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen L.T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., von Haeseler A., Jermiin L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoang D.T., Chernomor O., von Haeseler A., Minh B.Q., Vinh L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rambaut A. FigTree. [(accessed on 1 February 2021)]; Available online: http://tree.bio.ed.ac.uk/software/figtree/
- 27.Stavrianakou M., Perez R., Wu C., Sachs M.S., Aramayo R., Harlow M. Draft de novo transcriptome assembly and proteome characterization of the electric lobe of Tetronarce californica: A molecular tool for the study of cholinergic neurotransmission in the electric organ. BMC Genom. 2017;18:611. doi: 10.1186/s12864-017-3890-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robichaux S., Lacour N., Bagby G.J., Amedee A.M. Validation of RPS13 as a reference gene for absolute quantification of SIV RNA in tissue of rhesus macaques. J. Virol. Methods. 2016;236:245–251. doi: 10.1016/j.jviromet.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roy P. Orbivirus structure and assembly. Virology. 1996;216:1–11. doi: 10.1006/viro.1996.0028. [DOI] [PubMed] [Google Scholar]
- 30.Tomazatos A., Marschang R.E., Maranda I., Baum H., Bialonski A., Spinu M., Luhken R., Schmidt-Chanasit J., Cadar D. Letea Virus: Comparative Genomics and Phylogenetic Analysis of a Novel Reassortant Orbivirus Discovered in Grass Snakes (Natrix natrix) Viruses. 2020;12:243. doi: 10.3390/v12020243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DuBois R.M., Vaney M.C., Tortorici M.A., Kurdi R.A., Barba-Spaeth G., Krey T., Rey F.A. Functional and evolutionary insight from the crystal structure of rubella virus protein E1. Nature. 2013;493:552–556. doi: 10.1038/nature11741. [DOI] [PubMed] [Google Scholar]
- 32.McCarthy M., Lovett A., Kerman R.H., Overstreet A., Wolinsky J.S. Immunodominant T-cell epitopes of rubella virus structural proteins defined by synthetic peptides. J. Virol. 1993;67:673–681. doi: 10.1128/JVI.67.2.673-681.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X., Gillam S. Mutations in the GDD motif of rubella virus putative RNA-dependent RNA polymerase affect virus replication. Virology. 2001;285:322–331. doi: 10.1006/viro.2001.0939. [DOI] [PubMed] [Google Scholar]
- 34.Dill J.A., Camus A.C., Leary J.H., Di Giallonardo F., Holmes E.C., Ng T.F. Distinct Viral Lineages from Fish and Amphibians Reveal the Complex Evolutionary History of Hepadnaviruses. J. Virol. 2016;90:7920–7933. doi: 10.1128/JVI.00832-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geoghegan J.L., Duchene S., Holmes E.C. Comparative analysis estimates the relative frequencies of co-divergence and cross-species transmission within viral families. PLoS Pathog. 2017;13:e1006215. doi: 10.1371/journal.ppat.1006215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zientara S., Weyer C.T., Lecollinet S. African horse sickness. Rev. Sci. Technol. 2015;34:315–327. doi: 10.20506/rst.34.2.2359. [DOI] [PubMed] [Google Scholar]
- 37.Viljoen G.J., Huismans H. The characterization of equine encephalosis virus and the development of genomic probes. J. Gen. Virol. 1989;70:2007–2015. doi: 10.1099/0022-1317-70-8-2007. [DOI] [PubMed] [Google Scholar]
- 38.Pontremoli C., Forni D., Sironi M. Arenavirus genomics: Novel insights into viral diversity, origin, and evolution. Curr. Opin. Virol. 2019;34:18–28. doi: 10.1016/j.coviro.2018.11.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw T. californica sequence reads are available at Bioproject PRJNA322346. Consensus sequences of the new viruses identified in this study are available on GenBank under accession numbers BK014632, BK014633, BK014634 and BK014635.