Abstract
Shadow enhancers are seemingly redundant transcriptional cis-regulatory elements that regulate the same gene and drive overlapping expression patterns. Recent studies have shown that shadow enhancers are remarkably abundant and control the majority of developmental gene expression in both invertebrates and vertebrates, including mammals. Shadow enhancers might provide an important mechanism for buffering against mutations in non-coding regulatory regions of genes implicated in human disease. Technological advances in genome editing and live imaging have shed light on how shadow enhancers establish precise gene expression patterns and confer phenotypic robustness. Shadow enhancers can interact in complex ways and may also help drive the formation of transcriptional hubs within the nucleus. Despite their apparent redundancy, the prevalence and evolutionary conservation of shadow enhancers underscore their key role in emerging metazoan gene regulatory networks.
Introduction
Transcriptional enhancers are non-coding DNA elements that are typically 200–2,000 bp in length and drive gene expression patterns in space and time. Enhancers contain numerous binding sites for sequence-specific transcription factors (TFs), which, upon binding to the enhancer, recruit cofactors to activate transcription from a target core promoter. A typical metazoan gene contains multiple cell-type-specific enhancers spread across large genomic distances, which collectively produce a complex gene expression pattern (for general reviews on enhancers see REFS.1–7). The classic textbook view is that, within a gene locus, different enhancers drive distinct spatiotemporal aspects of gene expression1,7. However, this model is an oversimplification because enhancers regulating the same gene often display overlapping or partially overlapping spatiotemporal activity8–12. Examples of such redundant enhancers were often overlooked until 2008 when Mike Levine and colleagues introduced the term “shadow enhancer”13. In that study, redundant enhancers were designated either as ‘primary’ (the enhancers closest to the core promoter) or ‘shadow’ (the enhancers located at a greater distance from the core promoter)13. This distinction was later revised owing to a lack of functional differences between primary and shadow enhancers14,15. In this Review, we define ‘shadow’ enhancers as sets of enhancers that regulate a common target gene and drive expression patterns that partially or completely overlap in space and time. This definition has become increasingly accepted in the gene regulation community14–24. The degree of overlap required for enhancers to qualify as shadow enhancers depends on what is functionally meaningful in a given biological context. In the early Drosophila melanogaster embryo, characterized shadow enhancers typically overlap in more than 50% of their expression domains at a given time point (Table 1). However, in other contexts, such as the nematode nervous system, even an overlap in a single neuron cell can be biologically significant25.
Table 1 |.
Examples of shadow enhancers in different organisms
| Tissue or cell type | Gene(s) with reported shadow enhancers | Gene class | Maximum distance between shadow enhancers | Refs |
|---|---|---|---|---|
| Plant | ||||
| Anthers, pollen | LAT | Signalling | ~ 3 kb | 50 |
| Leaf cells | rbcS-8B | Signalling | ~ 1 kb | 51 |
| Worm | ||||
| Nervous system | cog-1, ric-4, ric-19, snb-1, unc-10, unc-11, unc-31, unc-64, unc-108 | TF, pan-neuronal genes | ~ 10 kb | 25,147 |
| Fruit fly | ||||
| Neurogenic ectoderm | vnd, brk, sog, dan, SoxN | TF, signalling | ~ 40 kb | 10,13,43 |
| Dorsal ectoderm | tup | TF | ~ 20 kb | 43 |
| A–P blastoderm | slp1, wg, hb, Kr, kni, gt, oc/otd, ems, hkb, fkh, Abd-B, prd | TF, signalling | ~ 30 kb | 28,148,149 |
| Mesoderm | sna, miR-1, ade5, Traf1, rols, CG42788, CadN | Various | ~ 10 kb | 10,16,27 |
| Salivary glands | sens | TF | ~ 2 kb | 43 |
| Epidermis | Ser, svb, y | Various | ~ 40 kb | 26,53,150 |
| Wing imaginal disc | brk | Signalling | ~10 kb | 56 |
| Nervous system | Ddc | Signalling | ~ 1 kb | 151 |
| Eye | Dve, dac | TF, signalling | ~ 15 kb | 55,87 |
| Zebrafish | ||||
| Brain | krox20 | TF | ~ 100 kb | 152 |
| Fin | Shh | Signalling | ~ 2 kb | 19 |
| Neural tube | Shh | Signalling | ~ 2 kb | 57 |
| Mouse | ||||
| Brain, neural tube | Otx2, Pomc, Shh, Arx, Ngn1 | TF, signalling | ~ 800 kb | 11,12,58,78,153,154 |
| Neural crest | Pax3 | TF | ~ 30 kb | 155 |
| Eye | Pax6, Cryaa | TF, structural | ~ 150 kb | 18,156 |
| Blood | α-globin and β-globin genes, igk, igH | Haemoglobin subunits, immune response | ~ 25 kb | 68,91,114,130 |
| Limb | Gli3, Sox9, Shox2, Ihh, HoxD, HoxA, Tbx4 | TF, signalling | ~ 1.2 Mb | 29,59,104,157,158 |
| Tooth | Shh | Signalling | ~ 100 kb | 30 |
| Gut | Cdx2 | TF | ~ 7 kb | 23 |
| Human | ||||
| Liver | APOE | Metabolism | ~ 10 kb | 63 |
| Blood | β-globin genes | Haemoglobin subunits | ~ 15 kb | 60,61 |
| Eye | ATOH7 | TF | ~ 20 kb | 22,159 |
| Kidney | REN | Signalling | ~ 6 kb | 62 |
Only non-adjacent enhancers were included in the table. TF, transcription factor.
The existence of shadow enhancers has raised fundamental questions about the purpose and evolutionary origins of this apparent redundancy. In development, multiple mechanisms of regulatory redundancy ensure accurate patterning. Examples include redundant genetic interactions and multiple binding sites for the same TF within an enhancer. Shadow enhancers are increasingly appreciated as another mechanism of redundancy that safeguards against genetic and environmental perturbations. Seminal studies in D. melanogaster demonstrated that shadow enhancers improve the precision of gene expression and phenotypic robustness during animal development, especially under conditions of physiological or genetic stress26–28. Later work in mammals confirmed that shadow enhancers similarly confer robustness to mammalian development18,29,30. Together, these studies suggest that shadow enhancers may be a common mechanism of developmental robustness in animals. Understanding the mechanism of shadow enhancer function will therefore illuminate how multi-enhancer architecture can determine the robustness or fragility of a developmental process to perturbation.
Recent advances in enhancer mapping and novel genetic and imaging tools for enhancer analysis have deepened our understanding of shadow enhancer function and their crucial role in development and human disease. Chromatin and 3D genome profiling by large consortia such as ENCODE, FANTOM, Roadmap Epigenomics and the 4D Nucleome Project have produced genome-wide maps of putative enhancers across many cell types, tissues and time points, both in mice and humans31–36. Large-scale transgenic reporter assays have enabled characterization of in vivo activity for thousands of bona fide enhancers37–42, revealing an ever growing number of putative shadow enhancers25,43. Efficient genome and epigenome editing of enhancers within their native genomic context has enabled analysis of enhancer requirements for organismal function44–47. And finally, quantitative live imaging methods have allowed the assessment of shadow enhancer functions in whole embryos48,49. These advances have enabled scientists to address key questions in shadow enhancer biology: how common are shadow enhancers in the genomes of different animals? Is there a function of shadow enhancers beyond conferring robustness? How do shadow enhancers work together in the context of the 3D genome? What is the role of shadow enhancers in human disease?
In this Review, we discuss key points that have emerged from these technological advances. We discuss how these advances have provided insight into the prevalence of shadow enhancers in bilaterian genomes and their crucial role in ensuring normal development under conditions of stress. Several lines of evidence suggest that shadow enhancers control the majority of developmentally regulated genes and are a remarkably widespread feature of bilaterian genomes. Examples in vertebrates show that shadow enhancers are required for normal development under stressful conditions, confirming earlier observations from the Drosophila model. We illustrate how the action of multiple shadow enhancers on a single promoter can fine-tune gene expression. We also discuss a potential role of shadow enhancers in organizing ‘hubs’ of transcriptional activity in the nucleus. We review the evidence that shadow enhancers frequently regulate genes implicated in human disease and buffer against mutations in non-coding regulatory DNA. Lastly, we discuss theories for the origin of shadow enhancers and their unexpectedly high evolutionary constraint. We synthesize mechanistic studies of shadow enhancers in D. melanogaster with emerging genetic manipulations of enhancers in mice to provide a cohesive picture of the role of regulatory redundancy in animal systems.
Genomic prevalence of shadow enhancers
Enhancers with redundant activity have been described for more than 30 years, with examples from plants50,51, flies8–10,52–56, zebrafish57, mice11,12,58,59 and humans60–63 (Table 1). These individual gene locus studies showed that shadow enhancers are found in a broad set of multicellular organisms, but within a single genome the prevalence of shadow enhancers was unknown. Since these studies were often focused on enhancers that control important developmental regulators, it was also not clear whether shadow enhancers are associated with other classes of genes. Substantial increases in the throughput of enhancer identification and characterization (reviewed in REFS.1,7,64) have allowed researchers to determine the prevalence of shadow enhancers genome-wide.
Genome-wide enhancer predictions based on chromatin features, such as chromatin accessibility, histone modifications, and TF binding, have suggested that shadow enhancers might be common in animal genomes. Using a combination of mesodermal TF chromatin immunoprecipitation (ChIP) data and computational models, Cannavo et al. generated an exhaustive catalogue of muscle development enhancers in D. melanogaster16. They found that nearly two-thirds of examined muscle developmental genes were controlled by shadow enhancers, and the majority of these genes had three or more predicted shadow enhancers16. A genome-wide analysis combining ENCODE transcriptomic and epigenomic data from multiple mouse tissues showed ample enhancer redundancy among developmentally regulated genes29,35. Whereas housekeeping genes are typically controlled by one enhancer, developmentally regulated genes can have 10 or more shadow enhancers (Table 2).
Table 2 |.
Examples of genes controlled by more than two shadow enhancers
| Tissue or cell type | Enhancer identification method | Method of assigning enhancers to genes | Genes | Number of shadow enhancers per gene | Refs. |
|---|---|---|---|---|---|
| Fruit fly | |||||
| S2 cells (macrophage-like) | MPRA | Genomic proximity | Various (203 genes) | 5 or more | 74 |
| Embryonic mesoderm | Mesoderm TFs ChIP | Genomic proximity and correlation with gene expression | Various (150 genes) | 3 or more | 16 |
| Mouse | |||||
| Embryonic limb | H3K27ac ChIP | Genomic proximity within a TAD and correlation with gene expression | Limb TFs (41 genes) | Median of 8 | 29 |
| Embryonic heart | H3K27ac ChIP | Genomic proximity within a TAD and correlation with gene expression | Heart TFs (27 genes) | Median of 10 | 29 |
| Embryonic forebrain | H3K27ac ChIP | Genomic proximity within a TAD and correlation with gene expression | Forebrain TFs (21 genes) | Median of 4 | 29 |
ChIP, chromatin immunoprecipitation; H3K27ac, histone 3 lysine 27 acetylation; MPRA, massively parallel reporter assay, TAD, topologically associating domains.
In human cells, ChIP-based profiling of TFs, cofactors, chromatin regulators and enhancer-associated histone modifications revealed that hundreds of key cell identity genes are regulated by large clusters of putative transcriptional enhancers (super-enhancers and stretch enhancers), which could be clusters of shadow enhancers65–70. Many mammalian enhancers including human enhancers are actively transcribed, and the presence of enhancer-derived RNAs (eRNAs) was suggested to be predictive of enhancer activity31,71,72. Profiling of eRNAs using cap analysis of gene expression (CAGE) across hundreds of human cell lines and tissues revealed that ~80% of 2,206 examined genes were associated with two or more co-transcribed enhancers31, suggesting that enhancer redundancy is common in the human genome. Computational approaches have also found widespread evidence for shadow enhancers in the human genome, particularly in association with developmental and disease-causing genes73.
Most chromatin and TF profiling methods are based on indirect measures of enhancer activity, which is why they have to be followed by functional testing. Large-scale transgenic enhancer-reporter screens have verified that bona fide redundant enhancers are common in the D. melanogaster and Caenorhabditis elegans genomes. An analysis of nearly 8,000 enhancer fragments during D. melanogaster embryogenesis revealed that many developmentally regulated genes are controlled by two or more enhancers with overlapping activities (Table 1)43. Single-neuron imaging data from hundreds of enhancer-reporter constructs in C. elegans demonstrated that shadow enhancers control nearly all 23 studied pan-neuronal genes25. Even within a single cell type, massively parallel reporter assays (MPRAs) have shown that hundreds of genes in D. melanogaster cell lines are potentially controlled by two or more redundant enhancers74. Taken together, these studies indicate that enhancers driving overlapping expression patterns are common in organisms from worms to insects to mammals and are preferentially, albeit not exclusively, associated with the control of developmental genes.
Shadow enhancers confer robustness
Several early studies in D. melanogaster demonstrated that shadow enhancers are required to drive normal development under conditions of stress, but they may be dispensable in ‘ideal’ conditions. For example, the TF Snail is required for normal gastrulation, and its expression in mesoderm is controlled by two shadow enhancers. Deletion of either of snail’s shadow enhancers caused no apparent gastrulation defect27. However, individual shadow enhancer deletion led to abnormal gastrulation under elevated temperatures or in a sensitized genetic background where the dosage of an upstream regulator, Dorsal, was reduced27. Similarly, a deletion of three of six epidermal shadow enhancers of shavenbaby had no phenotype under normal conditions but caused a decrease of trichome numbers under temperature or genetic stress conditions26.
Advances in genome editing have enabled the efficient introduction of multiple mutations in mice75–77, enabling experiments to test whether shadow enhancers similarly provide developmental robustness in vertebrates. Whereas single shadow enhancer deletions in mice typically show either mild or no observable phenotypes, double enhancer deletions show severe phenotypes, often comparable to complete gene loss-of-function in relevant tissues18,29,30,68,78. Together, these observations indicate that both enhancers regulate the gene, and at least one shadow enhancer is required for normal development in ideal conditions (FIG. 1). Despite driving similar expression patterns, the individual shadow enhancers are not strictly redundant. In a sensitized genetic background with a reduced dosage of the target gene, single enhancer deletions show abnormal phenotypes, indicating that shadow enhancers can confer robustness to genetic perturbations (FIG. 1). This pattern has been demonstrated for Pax6, a gene required for early eye lens morphogenesis18, Shh in developing teeth30 and several limb development loci29.
Fig. 1 |. Shadow enhancers confer phenotypic robustness in mammals.
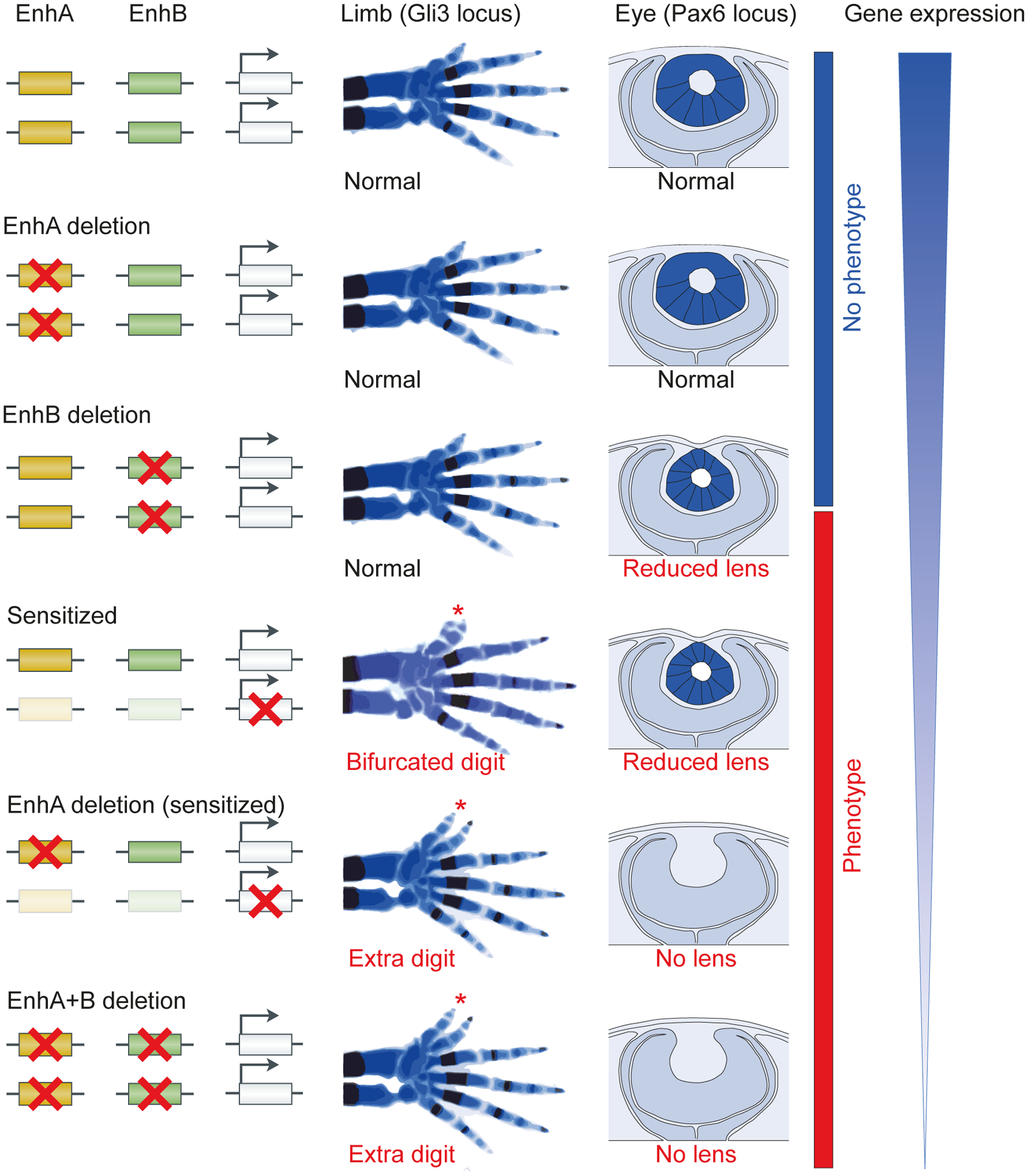
In mice, many individual shadow enhancer deletions yield no observable phenotypes. However, either the deletion of individual shadow enhancers in a sensitized background or the deletion of pairs of shadow enhancers leads to observable phenotypes. Schematics of perturbations (left) and resulting phenotypes in mice (right) are shown for two gene loci: Gli329 and Pax618,160. GLI3 is critical for proper limb development, and its knockout causes the formation of extra digits (among other phenotypes)161. Skeletal phenotypes in the absence of individual Gli3 shadow enhancers, pairs of shadow enhancers, or an individual shadow enhancer in a sensitized background are shown (centre). Red asterisks indicate the presence of extra digits. Pax6-deficient mice have arrested eye development and no lens formation162,163. A schematic diagram of an eye section showing a developing lens in the absence of individual PAX6 shadow enhancers, pairs of shadow enhancers, or an individual shadow enhancer in a sensitized background is shown (center-right). A schematic of gene dosage in the mutants is shown on the right. Figure adapted with permission from REF 29.
Taken together, both fruit fly and mouse studies emphasize that, while ostensibly redundant in the expression patterns they drive, the necessity of shadow enhancers is revealed when enhancer-deficient organisms are placed in stressful conditions. How shadow enhancers provide this robustness remains an area of open investigation and more than one mechanism may be at play. One potential scenario is that each enhancer alone can drive sufficient levels of gene expression for normal development, similar to the haplosufficiency of many developmental genes. By having multiple enhancers the probability that at least one is active increases, improving the chance for normal development14,24. A second potential mechanism was suggested by the observation that a pair of D. melanogaster shadow enhancers controlling the gene Krüppel are regulated by different combinations of TFs17. By responding to different sets of TFs, but converging on a single output, shadow enhancers could provide a mechanism to buffer against not only mutations in their sequences but, more importantly, perturbations in one of their upstream TFs (FIG. 2). Experimental measurements show that Krüppel’s independently controlled shadow enhancers drive lower expression noise than single or duplicated enhancer configurations, suggesting that simple enhancer duplications may not be sufficient to provide phenotypic robustness79. Independent regulation of shadow enhancers may be a widespread mechanism to confer robustness, as many mesodermal shadow enhancers are bound by different combinations of upstream TFs16.
Fig. 2 |. Independent TF inputs to shadow enhancers lead to more robust transcriptional output.
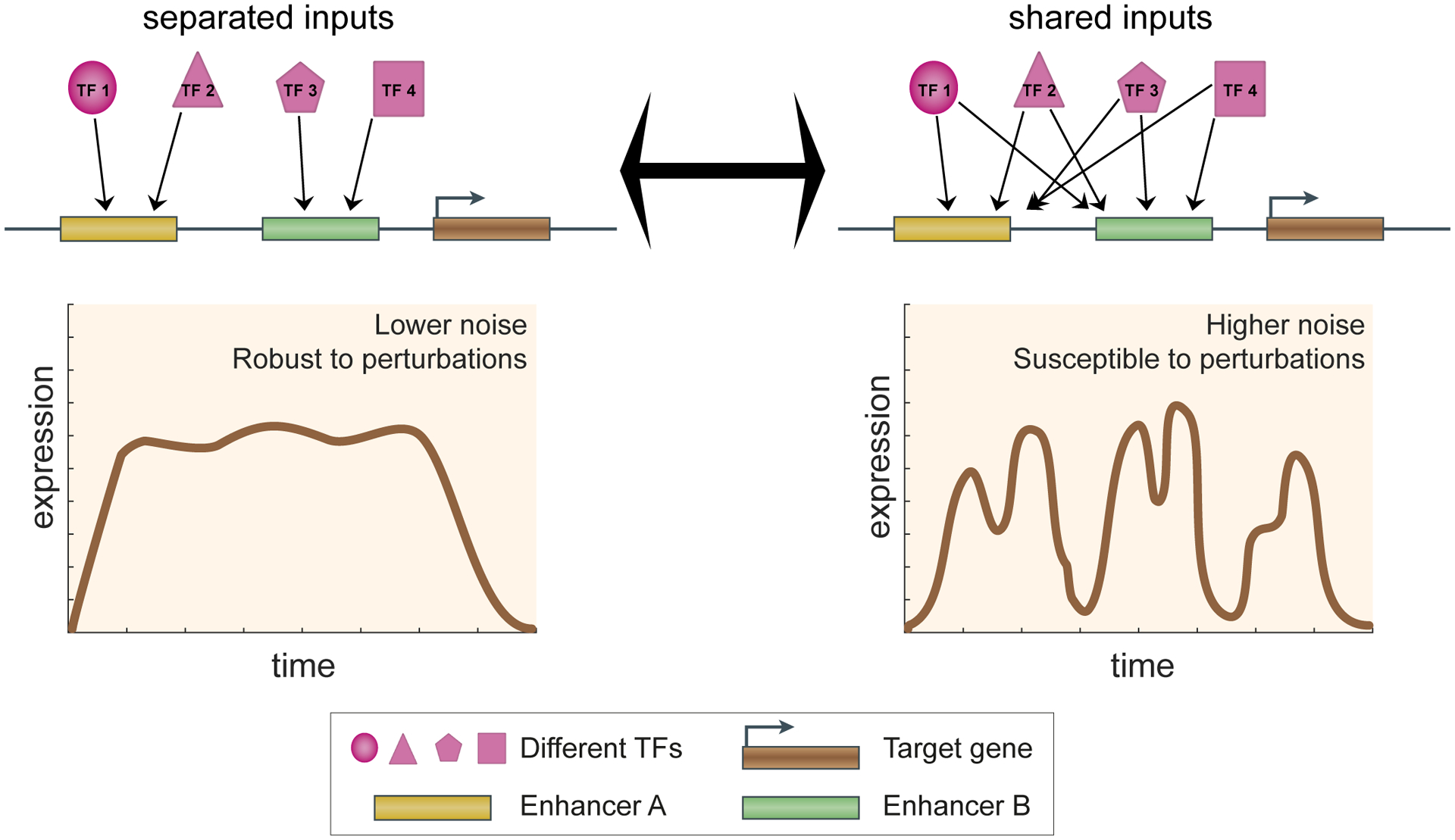
Shared and separated TF inputs to the individual shadow enhancers can have different effects on gene expression noise. In case of separated inputs, shadow enhancers regulating the same target gene do not share any of the same TF regulators (top left), while in case of shared inputs, shadow enhancers are regulated by the same set of TFs (top right). Below these two different models, we show the corresponding target gene expression dynamics in single cells as a function of time. Lower expression noise is seen with shadow enhancers with separated TF inputs than with shadow enhancers using shared TF inputs. See REF 79 for more details.
Modes of shadow enhancer interactions
The interactions between shadow enhancers can fine-tune the expression pattern of their target gene. Within an individual cell, shadow enhancers can interact in one of four ways: additively, super-additively (driving more expression than the sum of the individual enhancer activities), sub-additively (driving less expression than the sum of the individual enhancer activities), or repressively (FIG. 3). The classic view of enhancers is implicitly additive, as each enhancer functions independently to build up a gene’s total expression pattern80. Several studies in the fruit fly embryo used live mRNA tracking of reporter constructs to measure shadow enhancer interaction. Shadow enhancers can act additively, with a pair of shadow enhancers driving expression roughly equal to the sum of the individual enhancers’ expression output. For example, such additive behaviour is seen for the shadow enhancers controlling the genes knirps and hunchback81. However, this behaviour can change depending on the cell type or time point because of the varying levels and identities of TFs bound to each enhancer. For example, the knirps shadow enhancers act additively at some time points, and super-additively at others, indicating the presence of synergistic interactions between shadow enhancers81. The shadow enhancers controlling the mouse Pomc gene also show super-additivity at some time points and additivity at others82. In the case of the D. melanogaster hunchback gene, the behaviour depends on the concentration of its primary activating TF, Bicoid (Bcd). In cells where Bcd is low, the two enhancers combine additively, but in cells where Bcd is high, the enhancers combine sub-additively81. Such sub-additive behaviour could indicate the presence of competition between shadow enhancers for promoter occupancy. Sub-additivity has also been observed in the case of strong enhancers in the Krüppel locus83.
Fig. 3 |. Shadow enhancers can combine in complex and varied ways.
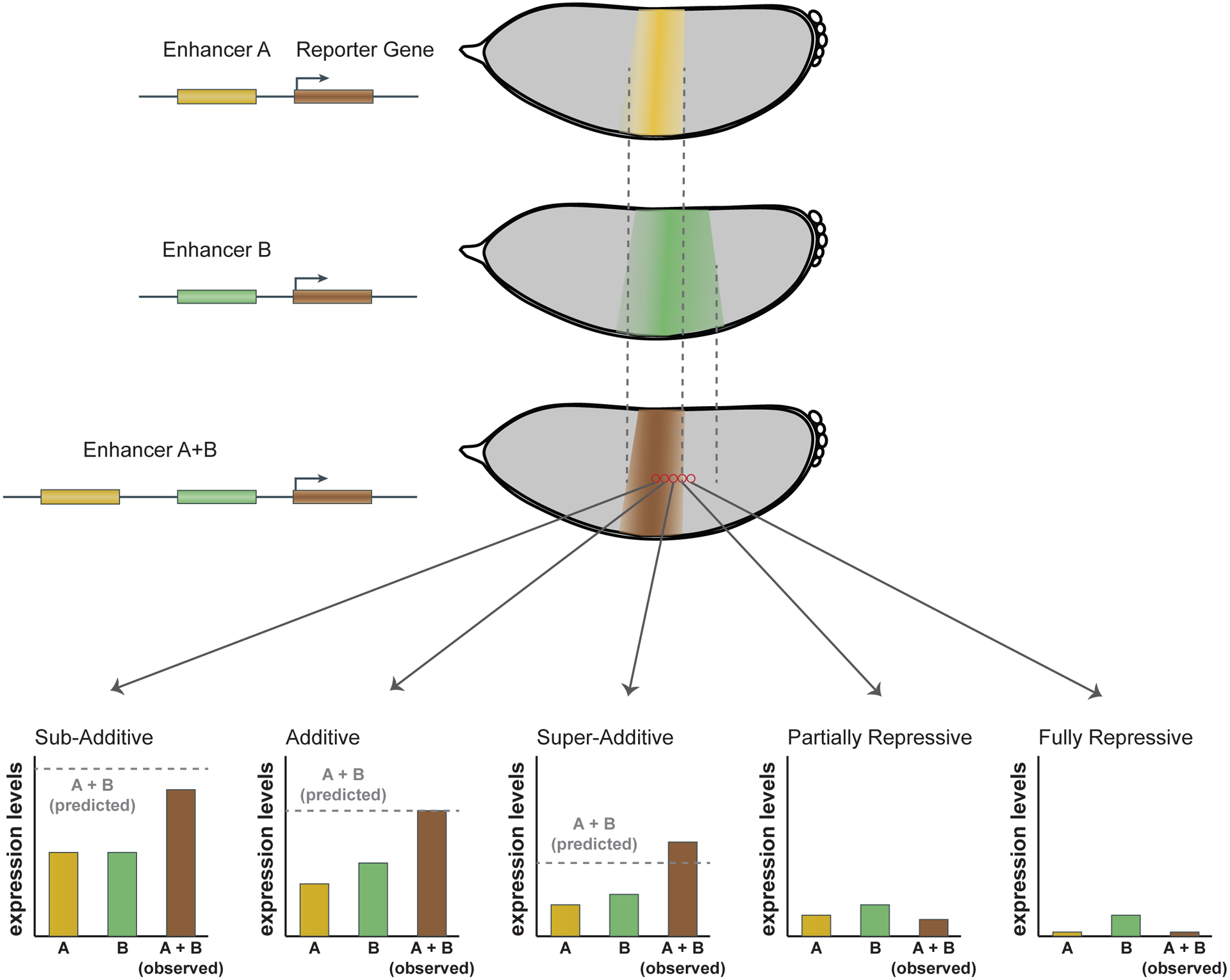
In an individual cell (red circles), enhancers can interact additively, sub-additively, super-additively, partially repressively, or fully repressively, as shown in the bar graphs. Cartoon embryos depicting individual (yellow and green) and combined (brown) enhancer activities, measured in transgenic D. melanogaster, are shown. Notably, the mechanism of interaction can vary from cell to cell, highlighting the importance of performing experiments in whole embryos. For example, two shadow enhancers can show each of these behaviours in different parts of their expression stripe, allowing them to combine to produce a sharper and stronger stripe than that produced by either individual enhancer.
In addition to the above interactions, one shadow enhancer can partially or completely repress the other, decreasing or shutting off expression entirely. In D. melanogaster embryonic cells at the boundary of the knirps and Krüppel expression domains, shadow enhancers can repress each other’s activity, yielding sharper expression patterns than either enhancer alone84. Examples from the sog, snail, and defective proventriculus loci show some shadow enhancer deletions can lead to higher expression levels, suggesting that one shadow enhancer represses the other81,85–87. The mechanisms that explain this repression are still unclear and could include quenching, interference of enhancer–promoter looping, or the spread of repressive chromatin marks.
On the tissue or organismal level, shadow enhancers can interact in nuanced ways to fine-tune both the levels and patterns of gene expression. The way that multiple shadow enhancers interact can vary from cell to cell, depending on the trans-regulatory environment. Multiple potential mechanisms may explain the variety of behaviours observed. For example, sub-additive behaviours between two strong shadow enhancers might occur because their target promoter has reached its maximum expression rate or because the enhancers are competing with each other for promoter access3,81,83. Super-additive behaviours might arise if there is synergy between the TFs bound at each shadow enhancer3,88. Additional experiments that manipulate number of shadow enhancers in a locus or their TF binding content, combined with experiments that probe the molecular details of shadow enhancer function (described below) may further illuminate the mechanisms at play.
Shadow enhancers and nuclear organization
The experiments described in the previous section measured the gene expression driven by shadow enhancers across the entire organism. How do shadow enhancers operate on a molecular level? Enhancers can regulate their target core promoters over long distances, sometimes up to several megabases (Table 1), a process mediated by TFs, co-activators and RNA polymerase II. Many studies observe the establishment of enhancer–promoter interactions coordinately with gene transcription. Various mechanisms and models of enhancer–promoter communication have been proposed, including tracking, linking, looping, and combinations thereof (for general reviews on enhancer–promoter interactions see REFS.2,4,89,90). The prevalence of shadow enhancers raises an intriguing question about how multiple enhancers interact with a single core promoter. Do shadow enhancers loop to the target promoter in a coordinated fashion, or is it a dynamic process with multiple enhancers competing for the same promoter (FIG. 4a)? Distinguishing between these possibilities may help illuminate how multiple shadow enhancers combine their activities to specify patterns and levels of gene expression.
Fig. 4 |. Interactions of shadow enhancers with target promoters.
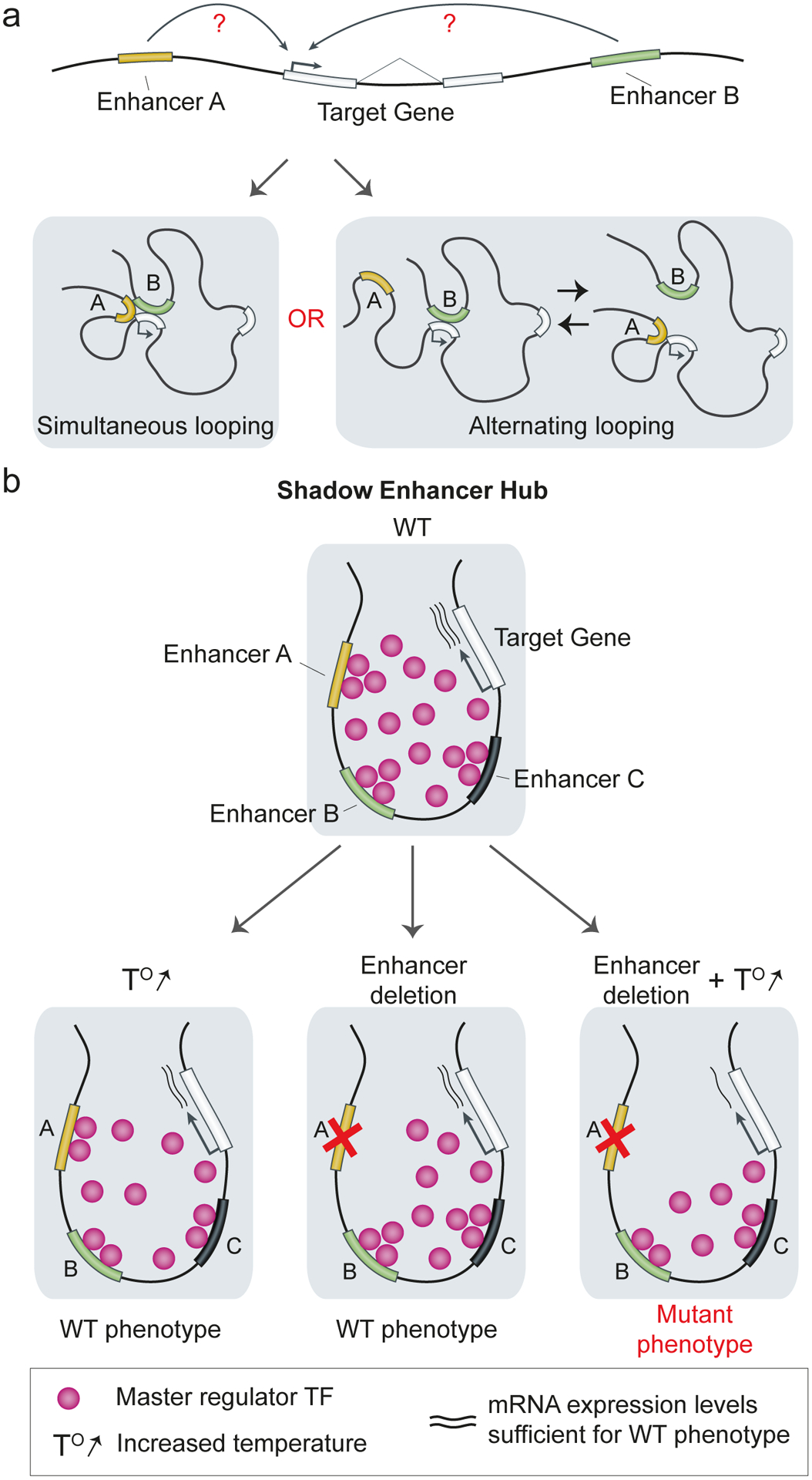
a | There are two possible models of enhancer–promoter looping. In the simultaneous looping model, enhancer A and B coordinately loop to the target core promoter to initiate transcription. In the alternating looping model, enhancer A competes with enhancer B, so, at a given time point, only one of the enhancers contacts the promoter. b | Multiple shadow enhancers may aid in the formation of transcriptional hubs by recruiting a high local amount of a master regulator TF (pink). Such transcription hubs can buffer against environmental stress and genetic perturbations110.
Experiments based on chromosome conformation capture provide indirect support for simultaneous promoter activation, as individual shadow enhancers often form contacts between each other and the target gene in the same cell68,91–94. These capture experiments were performed in populations of fixed cells and do not reflect the dynamics of enhancer–promoter interactions and transcription from the target promoter. Live imaging of transcription in D. melanogaster embryos suggests that a single enhancer can simultaneously activate two different promoters, even those located on different chromosomes, leading to synchronized transcription bursts95,96. Together, these studies suggest that enhancer–promoter loops can include more than two DNA elements. Therefore, it seems plausible that several shadow enhancers could simultaneously coordinate the expression of a single target promoter (FIG. 4a). A direct demonstration of such coordinated expression is challenging as it requires simultaneous labelling of several shadow enhancers and transcription from a target promoter. With the development of new live imaging tools, it may soon be possible to visualize how shadow enhancers activate target promoters in live nuclei97–100.
The concept of dynamic ‘transcriptional hubs’ (or the related concepts of ‘nuclear microenvironments’ or less dynamic ‘transcriptional condensates’) challenges the simple enhancer–promoter looping model and provides a plausible model for promoter regulation by multiple shadow enhancers90,101–106. These large hubs (>300 nm) are formed by TFs, components of the core transcriptional machinery102,103 and RNA polymerase II105,107 and may explain why some enhancers activate promoters even in the absence of close enhancer–promoter proximity108,109. The hub model suggests that shadow enhancers and their target promoter can simultaneously participate in the same microenvironment, forming a multi-enhancer hub. The observation of transcriptional coactivator condensates on super-enhancer-associated genes provides support for this model65,102,103 Recent work on the D. melanogaster shavenbaby locus showed that deleting one of the shadow enhancers results in decreased local density of the key activating TFs, suggesting that shadow enhancers are critical for maintaining high concentrations of TFs within the transcriptional hub (FIG. 4b)110. Through the formation of multi-enhancer transcriptional hubs with high concentrations of TFs, transcriptional coactivators and RNA polymerase II, shadow enhancers may improve phenotypic resilience to stress by buffering against environmental and genetic perturbations.
Shadow enhancers and human disease
Many human genetic disorders are caused by mutations in developmental genes. A strong association of shadow enhancers with developmental genes suggests that enhancer redundancy provides an important safeguard against deactivating non-coding mutations in cis-regulatory regions of disease-causing genes (FIG. 5). Indeed, evidence from human genetics studies and experiments in mice suggests that disease-associated genes contain shadow enhancers that likely buffer against the effect of loss-of-function (LoF) non-coding mutations.
Fig. 5 |. Shadow enhancers buffer against non-coding mutations in disease-causing genes.
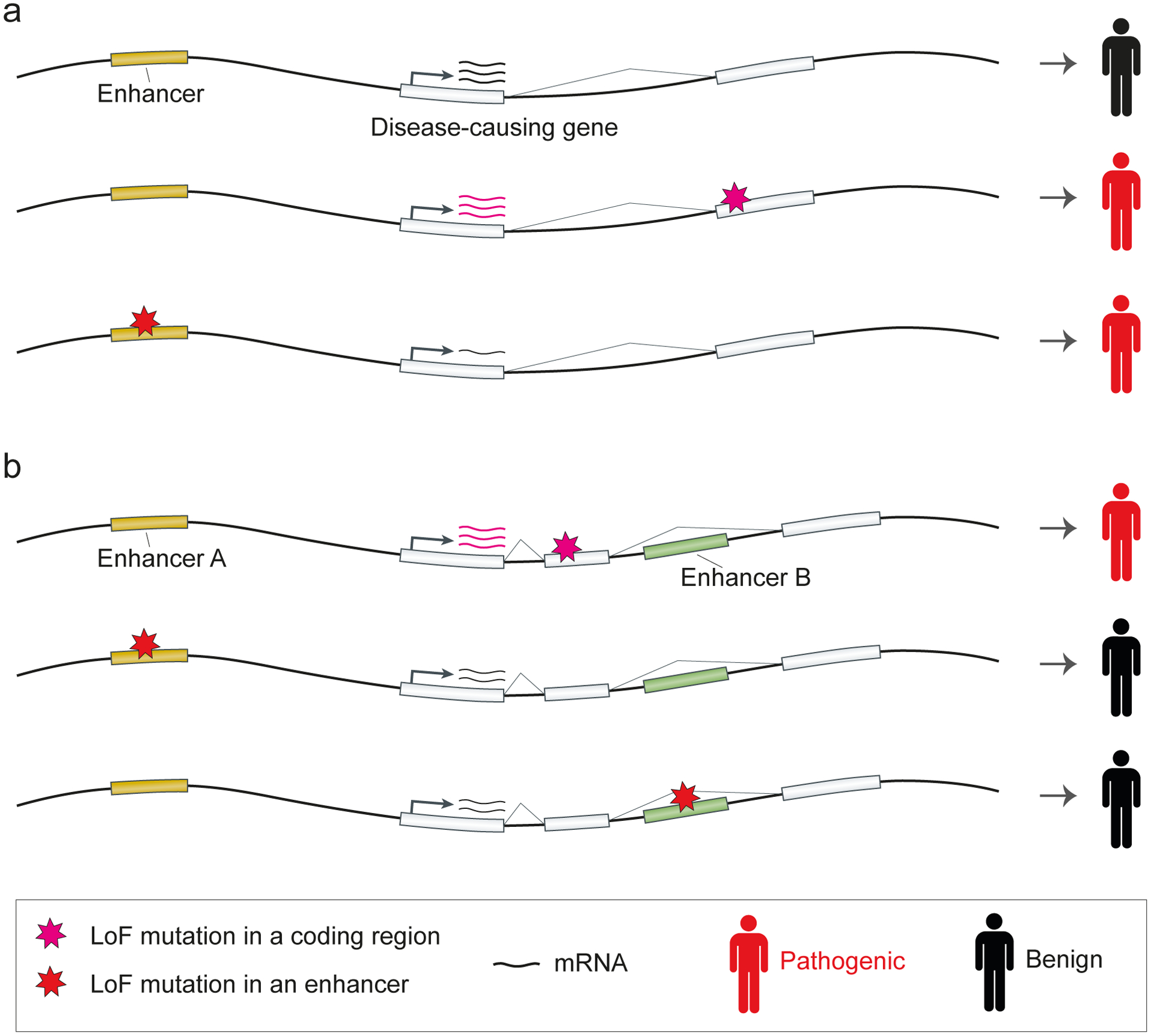
a | Many genetic disorders are caused by loss-of-function (LoF) mutations in coding regions of disease-causing genes. If the gene is controlled by a single enhancer, a LoF mutation in the enhancer will mimic the loss of gene function in the tissue and the time point of enhancer activity. b | If a disease-causing gene is controlled by shadow enhancers, non-coding mutations that deactivate one of the shadow enhancers will be buffered by another shadow enhancer.
A recent study used chromatin profiling data across 127 human tissues from the Roadmap Epigenomic Consortium to calculate an ‘enhancer-domain score’ for each human gene33,73. Enhancer-domain scores indicate the amount of redundant regulatory DNA for each gene, based on the total number of predicted enhancers and the redundancy of TF motifs within them. High enhancer-domain scores are predictive of gene pathogenicity, suggesting that the number of shadow enhancers is closely related to the gene’s importance in human disease73. This analysis is consistent with previous observations in fruit flies and mice, where important developmental genes tend to have larger regulatory domains111 and contain more enhancers per tissue29.
The strong association between shadow enhancers and developmental and disease-associated genes explains why many targeted deletions of enhancers of these genes cause fairly mild phenotypes or no observable phenotypes in mice12,112–116. Moreover, deletions of ultraconserved enhancers, which retain almost perfect sequence conservation across vertebrates and are located next to important developmental genes, have also led to viable mice with subtle phenotypes78,117,118. With the availability of highly efficient CRISPR–Cas9 genome editing, the number of enhancer knockout mice that lack observable phenotypes has grown18,29,68,78,119. These studies further suggest that a significant fraction of LoF mutations in human shadow enhancers will cause relatively subtle phenotypes in patients.
Shadow enhancer buffering predicts that LoF genetic variants in human shadow enhancers would have less severe effects on gene expression and phenotypes than variants in non-redundant enhancers. Indeed, genes with redundant enhancer domains are depleted for common and rare non-coding variants associated with gene expression changes73. This pattern indicates that genes with shadow enhancers are more resilient against naturally occurring non-coding mutations in the human population73.
It remains to be seen whether shadow enhancers also buffer against gain-of-function mutations in enhancers that cause misexpression of disease-associated genes. Studies in D. melanogaster showed that one shadow enhancer can repress another shadow enhancer in a dominant fashion84, suggesting that enhancer mutations causing gene misexpression could, in principle, be buffered by repression from another shadow enhancer. By contrast, both rare and common gain-of-function enhancer variants are associated with congenital malformations120, heart disease121, intellectual disabilities122 and cancer123–125, potentially through misexpression or upregulation of important developmental genes. In these examples, it is not always clear whether an additional shadow enhancer was also present. Systematic mutagenesis of human enhancers using MPRAs followed by in vivo validation in mice will help identify how frequently such gain-of-function mutations affect enhancers40,126–129.
Evolution of shadow enhancers
Evolutionary origin of shadow enhancers.
Despite their importance, the evolutionary origins of the majority of shadow enhancers are unclear. Like non-redundant enhancers (reviewed in REF.3), shadow enhancers may arise by one of several mechanisms: de novo from existing non-coding DNA, duplication of existing enhancers, or co-option of transposable elements or unrelated enhancers. Another potential mechanism is splitting an enhancer with redundancy across its length into two parts through the insertion of non-functional DNA (FIG. 6). The redundancy of shadow enhancers suggests that they may emerge as a result of duplication events, an idea proposed for some Drosophila shadow enhancers13. However, there are only few documented examples of such origins63,130, and many shadow enhancers seem to have little sequence similarity79,131. Some shadow enhancers can arise from transposon co-option events. For example, MER41 endogenous retroviruses (ERVs) have been co-opted to redundantly regulate genes involved in the interferon response132. Mammalian-apparent long terminal repeat (LTR) and short interspersed element (SINE) retrotransposons were independently co-opted to redundantly regulate the brain expression of the Pomc gene, which is important for the control of food intake133 (Table 1). A recent study used enhancer predictions based on eRNA profiling across hundreds of human and mouse cell lines31 to estimate that 31% of all redundant enhancer pairs in human and 17% of those in mouse have evolved by transposon co-option. Interestingly, for most transposon-derived redundant enhancer pairs, both enhancers have evolved through independent transposons co-option events, suggesting that duplication may not be a dominant route of shadow enhancer acquisition131. Most shadow enhancers have only partially overlapping activity patterns (Table 1), suggesting that one of the main mechanisms of shadow enhancer birth could be through co-option of enhancers with initially non-overlapping activities. Selection may favour the recruitment of shadow enhancers to genes whose robust expression is required for a newly emerging key developmental process (for example, pectoral fins in jawed fish19).
Fig. 6 |. Many evolutionary routes potentially lead to shadow enhancer birth.
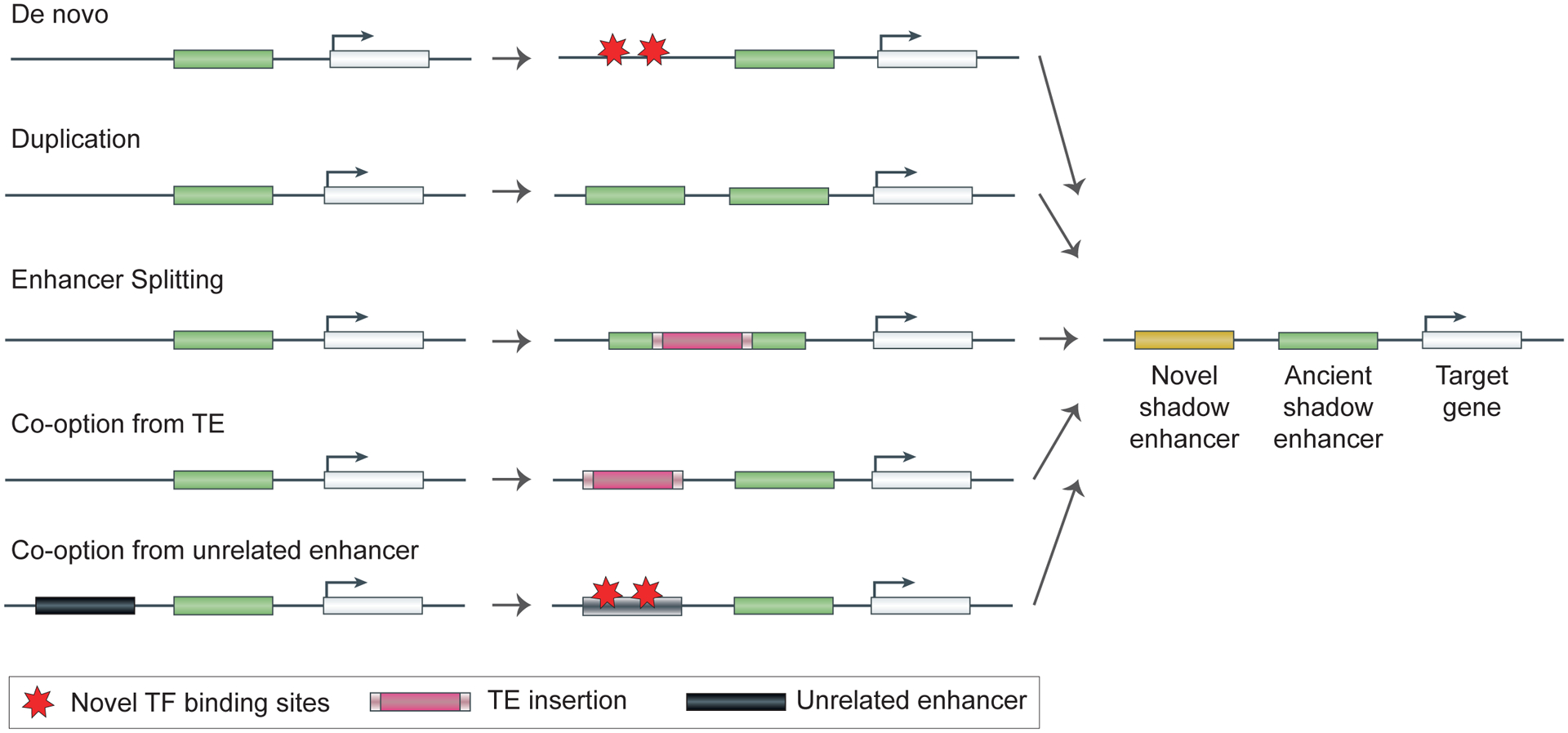
Proposed mechanisms include mutations in non-coding regions that generate a novel enhancer, duplication of an existing enhancer, splitting of a large enhancer into two by TE insertion, and co-option of either a transposable element (TE) or an unrelated enhancer to become a shadow enhancer.
Evolutionary conservation of shadow enhancers.
Given the redundancy of shadow enhancers, it was initially hypothesized that they would be subject to relaxed evolutionary constraint, allowing them to evolve novel regulatory functions13. If true, this hypothesis would predict a greater rate of mutations in shadow enhancers than in non-redundant enhancers. In a large group of D. melanogaster mesoderm-specific enhancers, shadow enhancers have higher sequence conservation than non-redundant enhancers, and there is no evidence of relaxed constraint on shadow enhancers16. Among ultraconserved enhancers, many have activity that is redundant with another ultraconserved enhancer in the locus29,78,117,118. This observation is again in contrast to the prediction that shadow enhancers are subject to weak evolutionary constraint. Growing evidence also suggests that evolution acts on groups of shadow enhancers as regulatory units, instead of on each enhancer individually. Similar to the stabilizing selection that maintains a single enhancer’s function134, mutations that cause a reduction in the activity of one shadow enhancer could be compensated by other mutations that increase the activity of another shadow enhancer and vice versa. Indeed, stabilizing selection has been shown to act on shadow enhancers to maintain conserved expression levels across different species17,135,136.
A full understanding of the evolutionary patterns of shadow enhancers remains to emerge, but the data collected so far suggest that shadow enhancers may not be an evolutionarily special and distinct class of enhancers per se. The conservation of shadow enhancers and the growing evidence that individual shadow enhancers can have distinct functions suggest that shadow enhancers can be fine-tuned for multiple purposes79,84,86.
Perspectives
Work over the last 10 years has shown that enhancer redundancy is a common feature of animal genomes, with shadow enhancers potentially controlling the majority of developmental genes. The primary purpose of this redundancy seems to be providing a mechanism to drive robust developmental patterning, irrespective of genetic and environmental stress. Shadow enhancers can also interact in complex ways to drive finely-tuned expression patterns, similar to the intricate interactions between TF binding sites within an enhancer. Shadow enhancers may also drive the formation of transcriptional condensates or hubs via increased TF recruitment, which may increase the fidelity of transcription. Evidence from human genetics studies indicates that shadow enhancers are key to regulating many disease-associated genes. The importance of shadow enhancers is also underscored by their surprising evolutionary conservation.
There remain a number of open questions in the shadow enhancer field. One of the most persistent questions about shadow enhancer prevalence is whether multiple enhancers are intrinsically capable of regulation that is unachievable by a single enhancer. Many of the ways that shadow enhancers interact, that is, synergistically or repressively, are reminiscent of interactions observed between TF binding sites within a single enhancer. So why have more than one enhancer? It may be possible that there is a limit on the stretch of DNA that can serve as an enhancer, so multiple enhancers allow for more room to encode complex biological functions. Or perhaps the formation of stable transcriptional hubs requires multiple clusters of TF binding sites spread throughout a locus to recruit the necessary transcriptional machinery. Alternatively, the flexibility of 3D genome organization may allow regulatory information to be either encoded in a single enhancer or multiple shadow enhancers located within the same topologically associating domain (TAD). If true, this suggests that shadow enhancers exist in the genome because there is no selective pressure to consolidate them into a single enhancer. A comprehensive answer to these questions will require several types of experiments. Measuring the activity of large numbers of individual enhancers and shadow enhancer sets may identify the behaviours that are possible with multiple enhancers, but not a single enhancer. Experiments that visualize the dynamic 3D conformation of loci with multiple enhancers would improve our ability to predict how multiple enhancers interact to control a single target gene.
Despite the prevalence of shadow enhancers in animal genomes, their evolutionary origins are largely a mystery. Once present in a genome, shadow enhancers are typically more conserved than other enhancers16. Some shadow enhancers are even among the most conserved sequences in the genome, that is, ultraconserved enhancers38,118. Since many shadow enhancers seem to be dispensable for organismal function and display superficial redundancy, their high degree of evolutionary conservation is puzzling. Most shadow enhancer knockout studies have been performed in lab conditions, which do not recapitulate native environments. Therefore, it may be hard to observe the potentially small reductions in fitness that can result in strong purifying selection. Future studies of shadow enhancer mutants in more natural environments may generate a fuller picture of the contributions of enhancer redundancy to organismal fitness.
Finally, our ability to predict the effect of enhancer sequence variation on human phenotypes is still limited. Most trait- and disease-causing variants discovered in genome-wide association studies (GWAS) fall outside coding sequences and are hypothesized to affect enhancer sequences137,138. Similarly, whole-genome sequencing (WGS) of patients has identified a growing number of rare non-coding variants that affect developmental genes and are linked to disease139–142. In contrast to findings from GWAS and WGS studies, disease-associated genes with large redundant regulatory domains show a relative depletion of functional non-coding variants73. How can we synthesize the fact that shadow enhancers can buffer sequence variation with the prevalence of disease-associated enhancer mutations? It is possible that disease-causing non-coding variants primarily affect genes lacking shadow enhancers or cause a gain of enhancer activity, which may not be buffered by the presence of shadow enhancers. Alternatively, variants in shadow enhancers may have a fairly small effect on target gene expression, which can be amplified by the presence of other mutations or the environment, leading to disease. The rapid increase in WGS of individuals143–145 combined with large-scale functional assays of human enhancer variant activity127,146 will shed more light on the role shadow enhancers play in human disease.
Acknowledgements
This work was supported by National Institutes of Health grants R01HD095246 (to Z.W.) and R00HG009682 (to E.Z.K). R.W. was supported by an ARCS Foundation award. We thank Axel Visel, Diane Dickel, Alexander Stark, Daria Shlyueva, and the anonymous reviewers for helpful comments on the manuscript.
Glossary
- Shadow enhancers
Sets of enhancers that drive overlapping expression patterns and regulate the same gene.
- Expression domains
The specific tissues or cell types where an enhancer drives expression of its target gene.
- Phenotypic robustness
The ability of a system to reliably produce a wild-type phenotype in the presence of environmental (e.g., temperature) or genetic (e.g., decreased expression levels of an upstream TF) stress.
- Super-enhancers (or the closely-related stretch enhancers)
Clusters of enhancers that are strongly bound by TFs, co-activators, or modified histones (as measured by ChIP-seq) and that control key cell identity genes.
- Enhancer RNAs (eRNAs)
Short, non-coding RNAs that are transcribed from the DNA of enhancer sequences and whose transcription correlates with enhancer activity.
- Genes expression noise
Variability in gene expression across either time or space, owing to the stochastic nature of the molecular interactions underlying gene expression.
- Quenching
A form of repression whereby the binding of repressive TFs within an enhancer sequence blocks the binding of activating TFs.
- Topologically Associating Domains (TADs)
Large genomic domains (~1 megabase) that display more frequent physical contacts between sequences within the same domain than between sequences from different domains.
- Evolutionary constraint
Factors that serve to limit the divergence of a particular phenotype; conserved DNA sequences are interpreted as evidence of evolutionary constraint.
- Haplosufficiency
A property of an allele whereby a single copy of that allele in a diploid organism is sufficient to drive a wild-type phenotype.
- Transcriptional bursting
Periods of rapid transcription interspersed with periods of transcriptional silence.
- Ultraconserved enhancers
Enhancers overlapping “ultraconserved” sequences, which are stretches of DNA that share perfect sequence conservation between human, mouse, and rat.
- Transposon co-option
The process by which a transposon changes its function (e.g., becomes a new gene or enhancer) through the introduction of sequence mutations.
- Transcriptional hubs
Three-dimensional nuclear compartments (>300 nm) formed around actively transcribed genes with a high local concentration of TFs, co-activators, RNA polymerase II and other components of the core transcriptional machinery.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Shlyueva D, Stampfel G & Stark A Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet 15, 272–286 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Furlong EEM & Levine M Developmental enhancers and chromosome topology. Science 361, 1341–1345 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Long HK, Prescott SL & Wysocka J Ever-Changing Landscapes: Transcriptional Enhancers in Development and Evolution. Cell 167, 1170–1187 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schoenfelder S & Fraser P Long-range enhancer-promoter contacts in gene expression control. Nat. Rev. Genet 20, 437–455 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Andersson R & Sandelin A Determinants of enhancer and promoter activities of regulatory elements. Nat. Rev. Genet 21, 71–87 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Field A & Adelman K Evaluating Enhancer Function and Transcription. Annu. Rev. Biochem 89, 213–234 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Visel A, Rubin EM & Pennacchio LA Genomic views of distant-acting enhancers. Nature 461, 199–205 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoch M, Schröder C, Seifert E & Jäckle H cis-acting control elements for Krüppel expression in the Drosophila embryo. The EMBO Journal vol. 9 2587–2595 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kassis JA Spatial and temporal control elements of the Drosophila engrailed gene. Genes Dev. 4, 433–443 (1990). [DOI] [PubMed] [Google Scholar]
- 10.Zeitlinger J et al. Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev. 21, 385–390 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeong Y, El-Jaick K, Roessler E, Muenke M & Epstein DJ A functional screen for sonic hedgehog regulatory elements across a 1 Mb interval identifies long-range ventral forebrain enhancers. Development 133, 761–772 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Kurokawa D et al. Regulation of Otx2 expression and its functions in mouse forebrain and midbrain. Development 131, 3319–3331 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Hong J-W, Hendrix DA & Levine MS Shadow enhancers as a source of evolutionary novelty. Science 321, 1314 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper describes the first use of the term “shadow enhancer” to describe several enhancers important for dorsal-ventral patterning in the fly embryo and speculate on the evolutionary role of shadow enhancers.
- 14.Barolo S Shadow enhancers: frequently asked questions about distributed cis-regulatory information and enhancer redundancy. Bioessays 34, 135–141 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hobert O Gene regulation: enhancers stepping out of the shadow. Curr. Biol 20, R697–9 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Cannavò E et al. Shadow Enhancers Are Pervasive Features of Developmental Regulatory Networks. Curr. Biol 26, 38–51 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study, the authors identify more than 1000 shadow enhancers that pattern D. melanogaster mesoderm; sequence conservation suggests that these shadow enhancers play a key role in development.
- 17.Wunderlich Z et al. Krüppel Expression Levels Are Maintained through Compensatory Evolution of Shadow Enhancers. Cell Rep. 12, 1740–1747 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Antosova B et al. The Gene Regulatory Network of Lens Induction Is Wired through Meis-Dependent Shadow Enhancers of Pax6. PLoS Genet. 12, e1006441 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]; This is one of the first studies to examine phenotypic impacts of double shadow enhancer deletions and shadow enhancer deletions on a sensitized background in mice.
- 19.Letelier J et al. A conserved Shh cis-regulatory module highlights a common developmental origin of unpaired and paired fins. Nat. Genet 50, 504–509 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nolte C, Jinks T, Wang X, Martinez Pastor MT & Krumlauf R Shadow enhancers flanking the HoxB cluster direct dynamic Hox expression in early heart and endoderm development. Dev. Biol 383, 158–173 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Swami M Transcription: Shadow enhancers confer robustness. Nature reviews. Genetics vol. 11 454 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Ghiasvand NM et al. Deletion of a remote enhancer near ATOH7 disrupts retinal neurogenesis, causing NCRNA disease. Nat. Neurosci 14, 578–586 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watts JA et al. Study of FoxA pioneer factor at silent genes reveals Rfx-repressed enhancer at Cdx2 and a potential indicator of esophageal adenocarcinoma development. PLoS Genet. 7, e1002277 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lagha M, Bothma JP & Levine M Mechanisms of transcriptional precision in animal development. Trends Genet. 28, 409–416 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stefanakis N, Carrera I & Hobert O Regulatory Logic of Pan-Neuronal Gene Expression in C. elegans. Neuron 87, 733–750 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frankel N et al. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature 466, 490–493 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper describes the phenotypic impact of shadow enhancer deletions on bristle formation in fly larvae and, along with Perry, et al. was among the first to show that shadow enhancers are required for proper development under stressful conditions.
- 27.Perry MW, Boettiger AN, Bothma JP & Levine M Shadow enhancers foster robustness of Drosophila gastrulation. Curr. Biol 20, 1562–1567 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper describes the phenotypic impact of shadow enhancer deletions on gastrulation in fly embryos and, along with Frankel, et al. was among the first to show that shadow enhancers are required for proper development under stressful conditions.
- 28.Perry MW, Boettiger AN & Levine M Multiple enhancers ensure precision of gap gene-expression patterns in the Drosophila embryo. Proc. Natl. Acad. Sci. U. S. A 108, 13570–13575 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Osterwalder M et al. Enhancer redundancy provides phenotypic robustness in mammalian development. Nature 554, 239–243 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study used mouse ENCODE data and systematic deletions (both individual and pairs) of 10 different mouse limb enhancers to show widespread redundancy in mammalian genomes.
- 30.Sagai T et al. SHH signaling directed by two oral epithelium-specific enhancers controls tooth and oral development. Sci. Rep 7, 13004 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andersson R et al. An atlas of active enhancers across human cell types and tissues. Nature 507, 455–461 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yue F et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature 515, 355–364 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abascal F et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583, 699–710 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gorkin DU et al. An atlas of dynamic chromatin landscapes in mouse fetal development. Nature 583, 744–751 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dekker J et al. The 4D nucleome project. Nature 549, 219–226 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Visel A, Minovitsky S, Dubchak I & Pennacchio LA VISTA Enhancer Browser--a database of tissue-specific human enhancers. Nucleic Acids Res. 35, D88–92 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pennacchio LA et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature 444, 499–502 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Manning L et al. A resource for manipulating gene expression and analyzing cis-regulatory modules in the Drosophila CNS. Cell Rep. 2, 1002–1013 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kvon EZ Using transgenic reporter assays to functionally characterize enhancers in animals. Genomics 106, 185–192 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Gallo SM et al. REDfly v3.0: toward a comprehensive database of transcriptional regulatory elements in Drosophila. Nucleic Acids Res. 39, D118–23 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bonn S et al. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat. Genet 44, 148–156 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Kvon EZ et al. Genome-scale functional characterization of Drosophila developmental enhancers in vivo. Nature 512, 91–95 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Lopes R, Korkmaz G & Agami R Applying CRISPR-Cas9 tools to identify and characterize transcriptional enhancers. Nat. Rev. Mol. Cell Biol 17, 597–604 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Catarino RR & Stark A Assessing sufficiency and necessity of enhancer activities for gene expression and the mechanisms of transcription activation. Genes Dev. 32, 202–223 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klein JC, Chen W, Gasperini M & Shendure J Identifying Novel Enhancer Elements with CRISPR-Based Screens. ACS Chem. Biol 13, 326–332 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wright JB & Sanjana NE CRISPR Screens to Discover Functional Noncoding Elements. Trends Genet. 32, 526–529 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garcia HG, Tikhonov M, Lin A & Gregor T Quantitative imaging of transcription in living Drosophila embryos links polymerase activity to patterning. Curr. Biol 23, 2140–2145 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lucas T et al. Live imaging of bicoid-dependent transcription in Drosophila embryos. Curr. Biol 23, 2135–2139 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Twell D, Yamaguchi J, Wing RA, Ushiba J & McCormick S Promoter analysis of genes that are coordinately expressed during pollen development reveals pollen-specific enhancer sequences and shared regulatory elements. Genes Dev. 5, 496–507 (1991). [DOI] [PubMed] [Google Scholar]
- 51.Poulsen C & Chua NH Dissection of 5’ upstream sequences for selective expression of the Nicotiana plumbaginifolia rbcS-8B gene. Mol. Gen. Genet 214, 16–23 (1988). [DOI] [PubMed] [Google Scholar]
- 52.Camprodón FJ & Castelli-Gair JE Ultrabithorax protein expression in breakpoint mutants: localization of single, co-operative and redundant cis regulatory elements. Rouxs. Arch. Dev. Biol 203, 411–421 (1994). [DOI] [PubMed] [Google Scholar]
- 53.Bachmann A & Knust E Dissection of cis-regulatory elements of the Drosophila gene Serrate. Dev. Genes Evol 208, 346–351 (1998). [DOI] [PubMed] [Google Scholar]
- 54.Schroeder MD et al. Transcriptional control in the segmentation gene network of Drosophila. PLoS Biol. 2, E271 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pappu KS et al. Dual regulation and redundant function of two eye-specific enhancers of the Drosophila retinal determination gene dachshund. Development 132, 2895–2905 (2005). [DOI] [PubMed] [Google Scholar]
- 56.Yao LC et al. Multiple modular promoter elements drive graded brinker expression in response to the Dpp morphogen gradient. Development 135, 2183–2192 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ertzer R et al. Cooperation of sonic hedgehog enhancers in midline expression. Dev. Biol 301, 578–589 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Nakada Y, Parab P, Simmons A, Omer-Abdalla A & Johnson JE Separable enhancer sequences regulate the expression of the neural bHLH transcription factor neurogenin 1. Dev. Biol 271, 479–487 (2004). [DOI] [PubMed] [Google Scholar]
- 59.Menke DB, Guenther C & Kingsley DM Dual hindlimb control elements in the Tbx4 gene and region-specific control of bone size in vertebrate limbs. Development 135, 2543–2553 (2008). [DOI] [PubMed] [Google Scholar]
- 60.Li Q, Peterson KR, Fang X & Stamatoyannopoulos G Locus control regions. Blood 100, 3077–3086 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grosveld F, van Assendelft GB, Greaves DR & Kollias G Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell 51, 975–985 (1987). [DOI] [PubMed] [Google Scholar]
- 62.Zhou X & Sigmund CD Chorionic enhancer is dispensable for regulated expression of the human renin gene. American Journal of Physiology - Regulatory Integrative and Comparative Physiology 294, R279 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Allan CM, Walker D & Taylor JM Evolutionary duplication of a hepatic control region in the human apolipoprotein E gene locus. Identification of a second region that confers high level and liver-specific expression of the human apolipoprotein E gene in transgenic mice. J. Biol. Chem 270, 26278–26281 (1995). [DOI] [PubMed] [Google Scholar]
- 64.Babbitt CC, Markstein M & Gray JM Recent advances in functional assays of transcriptional enhancers. Genomics 106, 137–139 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hnisz D et al. Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Whyte WA et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Parker SCJ et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc. Natl. Acad. Sci. U. S. A 110, 17921–17926 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hay D et al. Genetic dissection of the α-globin super-enhancer in vivo. Nat. Genet 48, 895–903 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pott S & Lieb JD What are super-enhancers? Nat. Genet 47, 8–12 (2015). [DOI] [PubMed] [Google Scholar]
- 70.Dukler N, Gulko B, Huang Y-F & Siepel A Is a super-enhancer greater than the sum of its parts? Nature genetics vol. 49 2–3 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li W, Notani D & Rosenfeld MG Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat. Rev. Genet 17, 207–223 (2016). [DOI] [PubMed] [Google Scholar]
- 72.Arner E et al. Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science 347, 1010–1014 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang X & Goldstein DB Enhancer Domains Predict Gene Pathogenicity and Inform Gene Discovery in Complex Disease. Am. J. Hum. Genet 106, 215–233 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study used human chromatin data from the Roadmap Epigenomics Consortia to show that human genes with large redundant domains are depleted of cis-acting genetic variants that disrupt gene expression, and they are buffered against the effects of disruptive non-coding mutations
- 74.Arnold CD et al. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science 339, 1074–1077 (2013). [DOI] [PubMed] [Google Scholar]
- 75.Wang H et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Joung JK & Sander JD TALENs: a widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol 14, 49–55 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Doudna JA & Charpentier E Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096 (2014). [DOI] [PubMed] [Google Scholar]
- 78.Dickel DE et al. Ultraconserved Enhancers Are Required for Normal Development. Cell 172, 491–499.e15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Waymack R, Fletcher A, Enciso G & Wunderlich Z Shadow enhancers can suppress input transcription factor noise through distinct regulatory logic. Elife 9, e59351 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study in fly embryos shows that a pair of shadow enhancers, each regulated by different TFs, drives lower expression noise than do single or duplicated enhancers.
- 80.Visel A et al. Functional autonomy of distant-acting human enhancers. Genomics 93, 509–513 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bothma JP et al. Enhancer additivity and non-additivity are determined by enhancer strength in the Drosophila embryo. Elife 4, e07956 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper investigates how the activities of individual shadow enhancers interact to establish their combined activity in fly embryos and find a wide range of possible interactions (super-additive, additive, sub-additive, and repressive) that may depend on the strength of the individual enhancers.
- 82.Lam DD et al. Partially redundant enhancers cooperatively maintain Mammalian pomc expression above a critical functional threshold. PLoS Genet. 11, e1004935 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Scholes C, Biette KM, Harden TT & DePace AH Signal Integration by Shadow Enhancers and Enhancer Duplications Varies across the Drosophila Embryo. Cell Rep. 26, 2407–2418.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.El-Sherif E & Levine M Shadow Enhancers Mediate Dynamic Shifts of Gap Gene Expression in the Drosophila Embryo. Curr. Biol 26, 1164–1169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dunipace L, Ozdemir A & Stathopoulos A Complex interactions between cis-regulatory modules in native conformation are critical for Drosophila snail expression. Development 138, 4075–4084 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dunipace L, Ákos Z & Stathopoulos A Coacting enhancers can have complementary functions within gene regulatory networks and promote canalization. PLoS Genet. 15, e1008525 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yan J et al. Regulatory logic driving stable levels of defective proventriculus expression during terminal photoreceptor specification in flies. Development 144, 844–855 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bentovim L, Harden TT & DePace AH Transcriptional precision and accuracy in development: from measurements to models and mechanisms. Development 144, 3855–3866 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Robson MI, Ringel AR & Mundlos S Regulatory Landscaping: How Enhancer-Promoter Communication Is Sculpted in 3D. Mol. Cell 74, 1110–1122 (2019). [DOI] [PubMed] [Google Scholar]
- 90.Zabidi MA & Stark A Regulatory Enhancer-Core-Promoter Communication via Transcription Factors and Cofactors. Trends Genet. 32, 801–814 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Proudhon C et al. Active and Inactive Enhancers Cooperate to Exert Localized and Long-Range Control of Gene Regulation. Cell Rep. 15, 2159–2169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jiang T et al. Identification of multi-loci hubs from 4C-seq demonstrates the functional importance of simultaneous interactions. Nucleic Acids Res. 44, 8714–8725 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hughes JR et al. Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nat. Genet 46, 205–212 (2014). [DOI] [PubMed] [Google Scholar]
- 94.Allahyar A et al. Enhancer hubs and loop collisions identified from single-allele topologies. Nat. Genet 50, 1151–1160 (2018). [DOI] [PubMed] [Google Scholar]
- 95.Fukaya T, Lim B & Levine M Enhancer Control of Transcriptional Bursting. Cell 166, 358–368 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lim B, Heist T, Levine M & Fukaya T Visualization of Transvection in Living Drosophila Embryos. Mol. Cell 70, 287–296.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Godin AG, Lounis B & Cognet L Super-resolution microscopy approaches for live cell imaging. Biophys. J 107, 1777–1784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schermelleh L et al. Super-resolution microscopy demystified. Nat. Cell Biol 21, 72–84 (2019). [DOI] [PubMed] [Google Scholar]
- 99.Chen H et al. Dynamic interplay between enhancer-promoter topology and gene activity. Nat. Genet 50, 1296–1303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen H & Gregor T Using RNA Tags for Multicolor Live Imaging of Chromatin Loci and Transcription in Drosophila Embryos. in RNA Tagging: Methods and Protocols (ed. Heinlein M) 373–384 (Springer; US, 2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Reiter F, Wienerroither S & Stark A Combinatorial function of transcription factors and cofactors. Curr. Opin. Genet. Dev 43, 73–81 (2017). [DOI] [PubMed] [Google Scholar]
- 102.Sabari BR et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hnisz D, Shrinivas K, Young RA, Chakraborty AK & Sharp PA A Phase Separation Model for Transcriptional Control. Cell 169, 13–23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Montavon T et al. A regulatory archipelago controls Hox genes transcription in digits. Cell 147, 1132–1145 (2011). [DOI] [PubMed] [Google Scholar]
- 105.Cho W-K et al. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361, 412–415 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jackson DA, Iborra FJ, Manders EM & Cook PR Numbers and organization of RNA polymerases, nascent transcripts, and transcription units in HeLa nuclei. Mol. Biol. Cell 9, 1523–1536 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cisse II et al. Real-Time Dynamics of RNA Polymerase II Clustering in Live Human Cells. Science 341, 664–667 (2013). [DOI] [PubMed] [Google Scholar]
- 108.Benabdallah NS et al. Decreased Enhancer-Promoter Proximity Accompanying Enhancer Activation. Mol. Cell 76, 473–484.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Alexander JM et al. Live-cell imaging reveals enhancer-dependent Sox2 transcription in the absence of enhancer proximity. Elife 8, e41769 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tsai A, Alves MR & Crocker J Multi-enhancer transcriptional hubs confer phenotypic robustness. Elife 8, e45325 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study describes how shadow enhancers may be critical for establishing or maintaining local regions of high TF concentrations required for achieving a certain threshold of gene expression.
- 111.Zeitlinger J & Stark A Developmental gene regulation in the era of genomics. Dev. Biol 339, 230–239 (2010). [DOI] [PubMed] [Google Scholar]
- 112.Cretekos CJ et al. Regulatory divergence modifies limb length between mammals. Genes Dev. 22, 141–151 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Manis JP et al. Class Switching in B Cells Lacking 3′ Immunoglobulin Heavy Chain Enhancers. J. Exp. Med 188, 1421–1431 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bender MA et al. Description and targeted deletion of 5’ hypersensitive site 5 and 6 of the mouse beta-globin locus control region. Blood 92, 4394–4403 (1998). [PubMed] [Google Scholar]
- 115.Fiering S et al. Targeted deletion of 5’HS2 of the murine beta-globin LCR reveals that it is not essential for proper regulation of the beta-globin locus. Genes Dev. 9, 2203–2213 (1995). [DOI] [PubMed] [Google Scholar]
- 116.Attanasio C et al. Fine tuning of craniofacial morphology by distant-acting enhancers. Science 342, 1241006 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nolte MJ et al. Functional analysis of limb transcriptional enhancers in the mouse. Evol. Dev 16, 207–223 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ahituv N et al. Deletion of ultraconserved elements yields viable mice. PLoS Biol. 5, e234 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cunningham TJ, Lancman JJ, Berenguer M, Dong PDS & Duester G Genomic Knockout of Two Presumed Forelimb Tbx5 Enhancers Reveals They Are Nonessential for Limb Development. Cell Rep. 23, 3146–3151 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hill RE & Lettice LA Alterations to the remote control of Shh gene expression cause congenital abnormalities. Philos. Trans. R. Soc. Lond. B Biol. Sci 368, 20120357 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gupta RM et al. A Genetic Variant Associated with Five Vascular Diseases Is a Distal Regulator of Endothelin-1 Gene Expression. Cell 170, 522–533.e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Huang L et al. A noncoding, regulatory mutation implicates HCFC1 in nonsyndromic intellectual disability. Am. J. Hum. Genet 91, 694–702 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wright JB, Brown SJ & Cole MD Upregulation of c-MYC in cis through a large chromatin loop linked to a cancer risk-associated single-nucleotide polymorphism in colorectal cancer cells. Mol. Cell. Biol 30, 1411–1420 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sur IK et al. Mice lacking a Myc enhancer that includes human SNP rs6983267 are resistant to intestinal tumors. Science 338, 1360–1363 (2012). [DOI] [PubMed] [Google Scholar]
- 125.Oktay Y et al. IDH-mutant glioma specific association of rs55705857 located at 8q24.21 involves MYC deregulation. Sci. Rep 6, 27569 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Inoue F & Ahituv N Decoding enhancers using massively parallel reporter assays. Genomics 106, 159–164 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kvon EZ et al. Comprehensive In Vivo Interrogation Reveals Phenotypic Impact of Human Enhancer Variants. Cell 180, 1262–1271.e15 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kircher M et al. Saturation mutagenesis of twenty disease-associated regulatory elements at single base-pair resolution. Nat. Commun 10, 3583 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Canver MC et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature 527, 192–197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Saleque S et al. Dyad symmetry within the mouse 3’ IgH regulatory region includes two virtually identical enhancers (C alpha3’E and hs3). J. Immunol 158, 4780–4787 (1997). [PubMed] [Google Scholar]
- 131.Barth NKH, Li L & Taher L Independent Transposon Exaptation Is a Widespread Mechanism of Redundant Enhancer Evolution in the Mammalian Genome. Genome Biol. Evol 12, 1–17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chuong EB, Elde NC & Feschotte C Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 351, 1083–1087 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Franchini LF et al. Convergent evolution of two mammalian neuronal enhancers by sequential exaptation of unrelated retroposons. Proc. Natl. Acad. Sci. U. S. A 108, 15270–15275 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ludwig MZ, Bergman C, Patel NH & Kreitman M Evidence for stabilizing selection in a eukaryotic enhancer element. Nature 403, 564–567 (2000). [DOI] [PubMed] [Google Scholar]
- 135.Arnold CD et al. Quantitative genome-wide enhancer activity maps for five Drosophila species show functional enhancer conservation and turnover during cis-regulatory evolution. Nat. Genet 46, 685–692 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Berthelot C, Villar D, Horvath JE, Odom DT & Flicek P Complexity and conservation of regulatory landscapes underlie evolutionary resilience of mammalian gene expression. Nat Ecol Evol 2, 152–163 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Maurano MT et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.McGuire AL et al. The road ahead in genetics and genomics. Nat. Rev. Genet (2020) doi: 10.1038/s41576-020-0272-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Short PJ et al. De novo mutations in regulatory elements in neurodevelopmental disorders. Nature 555, 611–616 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Turro E et al. Whole-genome sequencing of patients with rare diseases in a national health system. Nature 583, 96–102 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Richter F et al. Genomic analyses implicate noncoding de novo variants in congenital heart disease. Nat. Genet (2020) doi: 10.1038/s41588-020-0652-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Turner TN et al. Genomic Patterns of De Novo Mutation in Simplex Autism. Cell 171, 710–722.e12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Karczewski KJ et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.1000 Genomes Project Consortium et al. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Taliun D et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. bioRxiv 563866 (2019) doi: 10.1101/563866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.van Arensbergen J et al. High-throughput identification of human SNPs affecting regulatory element activity. Nat. Genet 51, 1160–1169 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.O’Meara MM et al. Cis-regulatory mutations in the Caenorhabditis elegans homeobox gene locus cog-1 affect neuronal development. Genetics 181, 1679–1686 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Fujioka M & Jaynes JB Regulation of a duplicated locus: Drosophila sloppy paired is replete with functionally overlapping enhancers. Dev. Biol 362, 309–319 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bell K, Skier K, Chen KH & Gergen JP Two pair-rule responsive enhancers regulate wingless transcription in the Drosophila blastoderm embryo. Dev. Dyn 249, 556–572 (2020). [DOI] [PubMed] [Google Scholar]
- 150.Kalay G, Lachowiec J, Rosas U, Dome MR & Wittkopp P Redundant and Cryptic Enhancer Activities of the Drosophila yellow Gene. Genetics 212, 343–360 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Johnson WA, McCormick CA, Bray SJ & Hirsh J A neuron-specific enhancer of the Drosophila dopa decarboxylase gene. Genes Dev. 3, 676–686 (1989). [DOI] [PubMed] [Google Scholar]
- 152.Torbey P et al. Cooperation, cis-interactions, versatility and evolutionary plasticity of multiple cis-acting elements underlie krox20 hindbrain regulation. PLoS Genet. 14, e1007581 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yao Y et al. Cis-regulatory architecture of a brain signaling center predates the origin of chordates. Nat. Genet 48, 575–580 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.de Souza FSJ et al. Identification of neuronal enhancers of the proopiomelanocortin gene by transgenic mouse analysis and phylogenetic footprinting. Mol. Cell. Biol 25, 3076–3086 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Degenhardt KR et al. Distinct enhancers at the Pax3 locus can function redundantly to regulate neural tube and neural crest expressions. Dev. Biol 339, 519–527 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.McGreal-Estrada RS, Wolf LV & Cvekl A Promoter-enhancer looping and shadow enhancers of the mouse αA-crystallin locus. Biol. Open 7, bio036897 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Berlivet S et al. Clustering of tissue-specific sub-TADs accompanies the regulation of HoxA genes in developing limbs. PLoS Genet. 9, e1004018 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Will AJ et al. Composition and dosage of a multipartite enhancer cluster control developmental expression of Ihh (Indian hedgehog). Nat. Genet 49, 1539–1545 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Miesfeld JB et al. The Atoh7 remote enhancer provides transcriptional robustness during retinal ganglion cell development. Proc. Natl. Acad. Sci. U. S. A (2020) doi: 10.1073/pnas.2006888117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Dimanlig PV, Faber SC, Auerbach W, Makarenkova HP & Lang RA The upstream ectoderm enhancer in Pax6 has an important role in lens induction. Development 128, 4415–4424 (2001). [DOI] [PubMed] [Google Scholar]
- 161.Hui CC & Joyner AL A mouse model of greig cephalopolysyndactyly syndrome: the extra-toesJ mutation contains an intragenic deletion of the Gli3 gene. Nat. Genet 3, 241–246 (1993). [DOI] [PubMed] [Google Scholar]
- 162.Hogan BL et al. Small eyes (Sey): a homozygous lethal mutation on chromosome 2 which affects the differentiation of both lens and nasal placodes in the mouse. J. Embryol. Exp. Morphol 97, 95–110 (1986). [PubMed] [Google Scholar]
- 163.Hill RE et al. Mouse small eye results from mutations in a paired-like homeobox-containing gene. Nature 354, 522–525 (1991). [DOI] [PubMed] [Google Scholar]


