Abstract
Klebsiella pneumoniae (Kp) is an opportunistic pathogen and the leading cause of healthcare-associated infections, mostly affecting subjects with compromised immune systems or suffering from concurrent bacterial infections. However, the dramatic increase in hypervirulent strains and the emergence of new multidrug-resistant clones resulted in Kp occurrence among previously healthy people and in increased morbidity and mortality, including neonatal sepsis and death across low- and middle-income countries. As a consequence, carbapenem-resistant and extended spectrum β-lactamase-producing Kp have been prioritized as a critical anti-microbial resistance threat by the World Health Organization and this has renewed the interest of the scientific community in developing a vaccine as well as treatments alternative to the now ineffective antibiotics. Capsule polysaccharide is the most important virulence factor of Kp and plays major roles in the pathogenesis but its high variability (more than 100 different types have been reported) makes the identification of a universal treatment or prevention strategy very challenging. However, less variable virulence factors such as the O-Antigen, outer membrane proteins as fimbriae and siderophores might also be key players in the fight against Kp infections. Here, we review elements of the current status of the epidemiology and the molecular pathogenesis of Kp and explore specific bacterial antigens as potential targets for both prophylactic and therapeutic solutions.
Keywords: Klebsiella pneumoniae, anti-microbial resistance, vaccines, monoclonal antibodies
1. Introduction
Multidrug-resistant (MDR) bacteria are defined as bacteria resistant to more than three antibiotic classes [1]. In February 2017, the World Health Organization (WHO) published a list of MDR pathogens to focus the research and development pipeline of new antibiotic treatments. Within this broad list, Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, Enterobacter species and Escherichia coli (ESKAPEE) pathogens were given the highest priority status [2]. Indeed, ESKAPEE pathogens have developed resistance mechanisms against most antibiotic treatments including those that are the last line of defense such as carbapenems and polymixins [3].
Klebsiella pneumoniae (Kp) was first identified as a causative agent of pneumonia in 1882 and has constantly gained significant public health attention. During the 1960s and 1970s, Kp became one of the most important causes of opportunistic healthcare-associated infections. In addition to pneumonia, Kp can also cause infections in the urinary tract and lower biliary tract, infections in surgical wound sites as well as bloodstream-associated infections [4]. The most vulnerable patients are neonates and the elderly, especially those that are immunocompromised.
Kp is naturally equipped with enzymes able to hydrolyze β-lactam antibiotics such as ampicillin and amoxicillin. However, the genome of Kp continued to accumulate antimicrobial resistance (AMR) genes through acquisition of plasmids and transferable genetic elements [5]. To date, over 400 AMR genes have been identified in different Kp genomes [6]. The introduction in the early 1980s of the third generation cephalosporins to treat ampicillin-resistant Enterobacteriaceae resulted in the emergence of extended-spectrum β-lactamase Kp (ESBL-Kp), which are currently prevalent worldwide and most frequently linked to high mortality rates [7,8]. Carbapenems were then the drug of choice for the treatment of ESBL-Kp in the 1990s, but this has resulted in the emergence of carbapenem-resistant Kp (CR-Kp), which have been alarmingly increasing in incidence since then [8,9]. It is noteworthy that a case–control study, conducted in two different hospitals in Greece, showed no statistical difference in hospital mortality between patients with CR-Kp (30.1%) and patients with carbapenem-susceptible Kp (CS-Kp) (33.9%). However, this may be due to the significant use of polymyxins for the treatment of severe multidrug-resistant infections in these hospitals, suggesting that the mortality of the CR-Kp could be possibly higher, if polymyxins were not used [10].
Different AMR mechanisms have been described in CR-Kp, including acquired carbapenemases (KPC), New Delhi metallo-β-lactamase (NDM) and carbapenemases of the oxacillinase-48 (OXA-48) type [11]. KPC-producing Kp has been most commonly occurring in the United States, after its first isolation in North Carolina [12,13]. NDM-producing Kp has been most commonly associated with the Indian subcontinent, since after its first detection in New Delhi [14], but also with specific countries in Europe, including Ireland [15], the Netherlands [16] and Italy [17]. The epicenter of OXA-48-like-producing Kp is in Turkey and surrounding countries [18], however health care-associated outbreaks have also been recently described in various European countries, including Greece [19], Ireland [20], France [21] and Germany [22], as well as in China [23] and Taiwan [24].
Beside the role of opportunistic pathogen in hospital settings, Kp can cause severe community-acquired infections in otherwise healthy individuals, such as pyrogenic liver abscess and meningitis often accompanied by bacteremia and metastatic spread [25]. Kp strains able to cause such infections are recognized as hypervirulent Kp (Hv-Kp) and were first detected in the 1980s in Taiwan [26], Hong Kong [27] and South Korea [28]. Hypervirulence-associated genes are often encoded on a large virulence plasmid but can also be encoded in the chromosome as part of integrative conjugative elements [29]. Such genes include rmpA/rmpA2 (associated to the hypermucoviscous phenotype), iroBCDN, iutA, iuacABCD and ybt genes (encoding multiple siderophores and their receptors) and the clb locus (encoding the colibactin genotoxin) [30,31,32].
Interestingly, MDR-Kp and Hv-Kp evolved from two different directions resulting in largely non-overlapping phenotypes [33]. Indeed, MDR-Kp clones maintained low virulence while Hv-Kp remained susceptible to carbapenems. The reasons for the apparent separation of phenotypes are still unclear, however growing reports of bi-directional convergence represent an alarming threat to public health [34,35,36].
The rising rates of AMR have a substantial economic impact globally, even though it remains difficult to be accurately calculated. This is related to extended duration of hospitalization and higher costs of new drugs to treat the infections. Low- and middle-income countries (LMIC) are already affected by AMR and are considered to be at higher risk of additional morbidity and mortality in the near future [37,38,39]. In particular, MDR bacterial infections are a leading cause of neonatal sepsis which accounts for over one-third of the global burden of child mortality [40]. Gram-negative rods are major pathogens of neonatal sepsis in LMIC and Kp is responsible for 16–28% of blood-culture-confirmed sepsis in different regions of the world. Assuming conservatively that Kp is responsible for 20% of the estimated 1.6 million neonatal sepsis-related deaths in the developing world, Kp-related neonatal deaths could amount to 320,000 deaths per year [40]. Child mortality remains disproportionately high in sub-Saharan Africa and South Asia [41]. A systematic review and meta-analysis of available data from the African continent, with a focus on the past decade (2008–2018), identified Klebsiella spp (mostly Kp) as accountable for 21% of all reported cases of neonatal bacteremia or sepsis [42]. The Child Health and Mortality Prevention Surveillance (CHAMPS) Network, established to collect data on under-5 mortality and stillbirths in sub-Saharan Africa and South Asia, also identified Kp as the most common contributory pathogen of neonatal deaths with at least one infectious condition in the causal chain, together with Acinetobacter baumannii and E. coli [43]. While more studies, including the Burden of Antibiotic Resistance in Neonates from Developing Societies (BARNARDS) study and the Global Neonatal Sepsis Observational Study (NeoOBS), are currently collecting more field data, the available estimates clearly highlight the need to acknowledge Kp as one of the most important neonatal pathogens in LMIC.
2. Klebsiella pneumoniae Diversity
The genus Klebsiella was named after the German microbiologist Edwin Klebs and is part of the Enterobacteriaceae family. However, it was termed Friedlander bacillus for many years in honor of the German pathologist Carl Friedländer, who first described this bacterium as the causative agent of pneumonia in immunocompromised patients. Members of this genus are Gram-negative rod-shaped bacteria that are found everywhere in nature, including water, soil, plant, insects and animals [44]. In humans, members of the genus Klebsiella are part of the normal flora in the nose, mouth and intestines. The species Klebsiella pneumoniae was first distinguished from other species of the genus Klebsiella on the basis of gyrA and parC genes and it was divided into three subspecies named K. pneumoniae subsp. pneumoniae, K. pneumoniae subsp. ozaenae and K. pneumoniae subsp. rhinoscleromatis [45]. However, closely related species of the genus Klebsiella might share 95–96% average nucleotide identity with Kp and only the implementation of whole-genome sequencing (WGS) facilitated the correct identification of clinical isolates [46].
Phenotypic methods developed for typing Kp isolates include phage typing, bacteriocin typing and serotyping. However, multilocus sequence typing (MLST) was first used by Diancourt et al., to accurately describe the genetic relationships among Kp isolates [47]. The MLST scheme developed was based on the sequences of seven housekeeping genes and allowed one to differentiate forty distinct allelic profiles, also known as sequence types (ST). A different MLST scheme, based on the sequences of 694 highly conserved core genes, was later developed by Bialek-Davene et al. and enabled the precise definition of globally distributed Kp clonal groups (CG) [48]. Most recently, WGS allowed one to show the existence of >150 deeply branching Kp lineages [31]. Whether defined as lineages, CG or ST, the resulting groups are typically referred to as clones. Kp infections are caused by different clones worldwide, but only a subset of these clones is responsible for the global disease burden. MDR-Kp clones are included in the largely described CG258, together with CG15, CG20, CG29, CG37, CG147, CG101 and CG307 [49,50]. In contrast, Hv-Kp clones are included in CG23, together with CG25, CG66 and CG380 [51,52]. While MDR-Kp CGs are each widely geographically distributed, Hv-Kp CGs are highly prevalent in Asia and the Pacific rim [53].
Historically, Kp has been classified into serotypes and tracked using typing antisera. Kp produces two types of polysaccharide on the cell surface: capsular polysaccharides (K-antigens) and O-antigens as part of the lipopolysaccharide (LPS) [54]. Both contribute to pathogenicity and form the basis for the identification of isolates, or serotyping. The most recently developed serotyping method relies on the genetic sequence analysis of the cps locus (K-antigen) and the rfb locus (O-antigen) [55]. To date, 141 distinct K-antigens have been identified, however, only 77 of these have been distinguished by traditional serological typing [56]. A substantial K-antigen diversity is observed in MDR-Kp clones, while K1 and K2 antigens are strongly associated to Hv-Kp clones [54]. In contrast, Kp has a surprisingly low number of reported O-antigens and only 11 distinct serotypes have been described. O1, O2, O3 and O5 are most commonly found in Kp clinical isolates, despite the fact that the prevalence of each serotype can vary based on the geographical regions and the MDR or Hv phenotypes [54,57,58].
3. Klebsiella pneumoniae Virulence Factors and Key Target Antigens
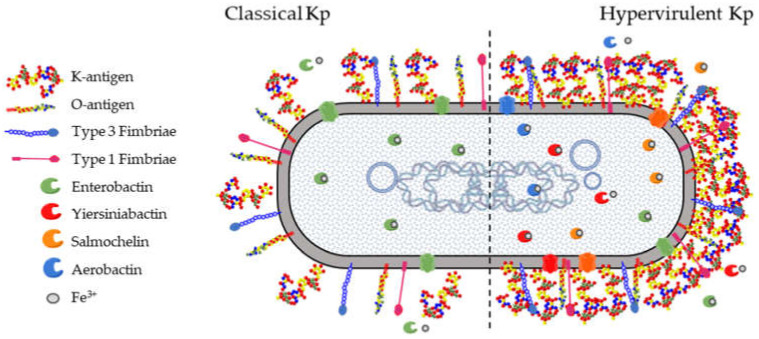
All Kp isolates carry a set of core genes responsible for their pathogenicity and required to establish opportunistic infections in humans and other hosts (Figure 1). These include the K-antigen locus (cps locus) and the O-antigen locus (rfb locus), collectively involved in immune evasion mechanisms. The core fim and mrk loci are responsible for the biosynthesis of type 1 and type 3 fimbriae which mediate processes in the earlier stages of infection, such as adhesion and colonization of host epithelia as well as biofilm formation on abiotic surfaces such as catheters. Finally, the ent locus encoding the siderophore enterobactin, as well as alternative acquired siderophores, is required for optimal growth in different niches.
Figure 1.
K. pneumoniae virulence factors. There are four well-characterized virulence factors for pathogenic Klebsiella pneumoniae (Kp). (1) The capsule is an extracellular polysaccharide matrix that envelops the bacteria and is overproduced in hypervirulent Kp (Hv-Kp) strains. (2) Lipopolysaccharide (LPS) is an integral part of the outer leaflet of the outer membrane and is produced by both classical and Hv-Kp strains. (3) Type 1 and type 3 fimbriae are membrane-bound adhesive structures. (4) Iron-scavenging siderophores are secreted small molecules recognized by specific membrane receptors mediating their uptake. Enterobactin is produced by virtually all Kp strains while other siderophores are typically secreted by Hv-Kp strains.
3.1. K-Antigens
The surface of Klebsiella species is shielded by a thick layer of capsular polysaccharide, historically known as K-antigen, that protects the bacteria from the environment as also observed in E. coli [59]. K-antigens play a crucial role in protecting Kp from innate immune response mechanisms, evading complement deposition and opsonization, reducing recognition and adhesion to epithelial cells and phagocytes, and abrogating lysis by antimicrobial peptides and the complement cascade [60].
Traditionally, 77 K-antigens have been identified among Klebsiella spp. based on the diversity in their sugar composition, type of glycosidic linkages, and the nature of enantiomeric and epimeric forms [61,62]. Recently, additional K-types have been reported based on the arrangement of the cps locus or K-locus (KL), known as the KL series [56].
The K-antigen biosynthesis occurs via the Wzx/Wzy-dependent pathway, similarly to group 1 capsules in E. coli [63,64]. The cps locus comprises two regions: the 5′ part of the locus contains four conserved genes (wzi, wza, wzb, and wzc), present in all group 1 capsule loci. The 3′ region of the locus is serotype-specific and encodes the enzymes for producing the repeating units and two integral inner membrane proteins (Wzy and Wzx) [65]. In particular, capsules biosynthesis takes place in the cytoplasmic leaflet of the inner membrane where the repeating units are assembled on the lipid carrier undecaprenol-pyrophosphate (Und-PP), with the contribution of sugar-specific glycosyltransferases. In particular, the specific initialization glycosyltransferase can either be WbaP (generating galactose-Und-PP) or WcaJ (generating glucose-Und-PP) [66]. Next, the Wzx flippase transfers the complete repeating units to the periplasmic leaflet of the inner membrane where the Wzy polymerase generates high molecular weight polysaccharides. Finally, Wza (an outer-membrane translocon), Wzc (a tyrosin autokinase) and Wzb (a phosphatase) synergistically transport the complete capsule onto the bacterial surface, where it remains associated to the outer-membrane protein Wzi (a lecto-aqua-porin) [62] (Figure 2A).
Figure 2.
(A) Model for biosynthesis and assembly of group 1 capsules. Undecaprenol-pyrophosphate (Und-PP)-linked repeating units are assembled on the cytoplasmic leaflet of the inner membrane. Wzx then flips the newly synthesized und-PP-linked repeats across the inner membrane. In the periplasmic leaflet of the inner membrane, Wzy polymerizes the repeating units. Continued polymerization requires transphosphorylation of the Wzc oligomer and dephosphorylation by the Wzb phosphatase. Finally, the polysaccharide is translocated by Wza in the extracellular milieu where it associates with the surface protein Wzi. PP = Undecaprenyl diphosphate, GT = glycosyltransferase. (B) Structures of K-antigens most commonly associated to Hv-Kp strains.
Among the different K-antigens, K1, K2, K5, K16, K23, K27, K28, K54, K62 and K64 are some of the most commonly isolated serotypes globally [54]. Interestingly, K1 and K2 serotypes are often found in Hv-Kp strains, indicating that this clonal lineage has a specific genetic background conferring hypervirulence [67,68] (Figure 2B). Moreover, Hv-Kp strains are known to produce a hypercapsule, resulting in a hypermucoviscous phenotype which further contribute to increased resistance to complement-mediated or phagocyte-mediated killing [69]. The rmpA gene was first identified as a regulator of the mucoid phenotype in 1989 [70], located either on the chromosome or on a large virulence plasmid. The correlation between rmpA (or the closely related rmpA2 gene) and hypermucoid phenotype is very high, and the presence of rmpA is among a set of genes proposed as biomarkers to identify potential Hv-Kp strains [29,71]. Other capsule types which are frequently found among Hv-Kp strains are K5, K20, K47, K54, K57, and K64 [29,72,73] (Figure 2B).
3.2. O-Antigens
The O-antigen moiety of the LPS has a limited range of structures, resulting from different sugar composition, glycosidic linkages, epimeric or enantiomeric forms of the sugars [54,74]. The nomenclature of O-serotypes has been revised multiple times over the years [75,76], however, the most recent classification includes 11 O-serotypes: O1, O2a, O2ac, O2afg, O2aeh (previously known as O9), O3 (divided in sub-serotypes O3, O3a and O3b), O4, O5, O7, O8 and O12 [56]. Moreover, additional O-types have been reported based on the arrangement of the rfb locus or O-locus (OL), known as the OL series.
Kp O-antigens are biosynthesized in the cytoplasm and transported to the bacterial surface through an adenosine triphosphate (ATP)-binding cassette (ABC) transporter dependent-pathway [77] (Figure 3A). In detail, the O-antigen is synthesized in the cytoplasmic leaflet of the inner membrane by serotype-specific glycosyltransferases and transferred to the periplasmic leaflet by the Wzm/Wzt complex. In parallel, the biosynthesis of the lipid A-core oligosaccharide also takes place in the cytoplasmic leaflet of the inner membrane and the sugars are then flipped to the periplasmic leaflet through the MsbA transporter. Finally, the polymerized O-antigen and lipid A-core oligosaccharide are linked by the WaaL ligase and the LptA-G complex transports the complete LPS to the bacterial surface [62]. In the whole process, three different gene clusters are involved: lpx, waa and rfb for the biosynthesis of lipid A, core-oligosaccharide and O-antigens, respectively [78,79,80].
Figure 3.
(A) Model of O-antigen biosynthesis via the adenosine triphosphate-binding cassette (ABC) transporter pathway. Und-PP-linked O-polyaccharides are processively assembled on the cytoplasmic leaflet of the inner membrane, through the sequential activity of different glycosyltransferases. The assembled O-antigen component is then transported via the Wzm/Wzt ABC transporter across the inner membrane. In the periplasmic leaflet of the inner membrane, the O-antigen is ligated to the lipid A-core oligosaccharide by WaaL and finally transported on the bacterial surface by the Lpt complex. (B) Structures of Kp O-antigens.
The O1, O2 and O8 serotypes are galactose-based polysaccharides and they share a common backbone named O2a (also known as Galactan-I) characterized by the [→3)-α-D-Galp-(1 → 3)-β-D-Galf(1→] repeating unit (Figure 3B). The biosynthesis of the O2a antigen is attributed to the rfb locus involving wzm, wzt, wbbM, glf, wbbN, and wbbO genes [81]. The O2 serotype includes different variants, represented by modified versions of the O2a backbone. The O2afg and O2aeh (O9) serotypes modify the O2a repeating unit by side-chain addition of (α-1 → 4)- or (α-1 → 2)-Galp residues catalyzed by a set of three enzymes encoded by genes gmlABC and gmlABD, respectively [82]. The O8 serotype modifies the O2a repeating unit by non-stoichiometric O-acetylation [83] (Figure 3B). The O1 serotype results from the covalent attachment of the O1 antigen (also known as Galactan-II) to the non-reducing terminus of O2a (as well as O2afg and O2aeh) and is characterized by the immunodominant [→3)-α-D-Galp-(1→3)-β-D-Galp-(1→] repeating unit, which has been associated to the wbbY-wbbZ genes [84] (Figure 3B). The O2c antigen can also be covalently attached to the non-reducing terminus of O2a (as well as O2afg and O2aeh), resulting in serotype O2ac: this is characterized by the immunodominant [→3)-β-D-GlcpNAc-(1 → 5)-β-D-Galf-(1→] repeating unit, which has been associated to the wbmVWX genes [84] (Figure 3B).
The O3 and O5 serotypes are mannose-based saccharides and their structures are identical to the E. coli O9 and O8 O-antigens, respectively [85,86] (Figure 3B). Biosynthetic enzymes of O3 and O5 are extremely similar but differ in the sequence of their mannosyltransferase (WbdA) and methyltransferase (WbdD). WbdD, in complex with WbdA, regulates the mannose chain length by capping the growing chain with a phosphate and methyl group in O3 and a methyl group only in O5 [87,88]. Therefore, the O3 and O5 repeating units differ from each other in the number of mannoses, anomeric configurations and/or their intra-mannose linkages [89]. In particular, the trimeric O5 repeating unit is composed of α-mannose (α-Man) and β-mannose (β-Man) with 1 → 2 and 1 → 3 linkage, whereas O3 uses pentameric 1 → 2- and 1 → 3-linked α-Man repeating units [89]. In addition, two Kp O3 serotype variants, O3a and O3b, have been identified with only four α-Man residues (O3a) or three α-Man residues (O3b), instead of five α-Man residues per repeating unit [90] (Figure 3B).
The O4, O7 and O12 serotypes are very different to the other O-antigens for their repeating units sugar composition (Figure 3B). The Kp O4 repeating unit is characterized by the disaccharide [→4)-α-D-Galp-(1 → 2)-β-D-Ribf-(1→], the O7 by a tetrasaccharide [→2)-α-L-Rhap-(1 → 2)-β-D-Ribf-(1 → 3)-α-L-Rhap-(1 → 3)-α-L-Rhap-(1→] while the O12 by the disaccharide [→4)α-L-Rhap-(1 → 3)-β-D-GlcpNAc-(1→] [89]. In addition, it was found that the O4 and O12 O-antigen chains terminate with Kdo residues.
3.3. Fimbriae
The majority of Gram-negative enterobacteria differentially express surface-associated fimbriae, organelles appointed to facilitate attachment and adherence to eukaryotic cells, but also involved in other functions, such as interaction with macrophages, biofilm formation, intestinal persistence, and bacterial aggregation [91,92,93]. Fimbriae are typically extracellular appendages with 0.5–10 μm length and 2–8 nm width. Most clinical Kp isolates express two types of fimbrial adhesins, type 1 fimbriae and type 3 fimbriae, that are assembled by the chaperone/usher-assembly pathway [94,95,96]. Besides type 1 and type 3 fimbriae, a third type named KPC fimbria was identified and the heterologous expression in E. coli demonstrated an active role of this protein in biofilm formation [97]. Since the late 1990s, another Kp fimbrial antigen named KPF-28 has been reported to be expressed by several Kp circulating strains and its role in colonization has been demonstrated by using KPF-28 antisera to inhibit bacterial adhesion to intestinal cells. However, besides a correlation of the expression of this adhesin with an antibiotic-resistance phenotype, no further information has been collected about KPF-28 over the last two decades [98,99].
Type 1 fimbriae are 7 nm wide and approximately 1 µm long surface polymers found on the majority of Enterobacteriaceae and encoded by the genes in the fimAICDFGHK operon. This organelle is a helical cylinder that belongs to the chaperon-usher pili family, made by a polymer of the major building element FimA. FimH, together with the minor subunits FimF and FimG, forms a flexible tip fibrillum that is connected to the distal end of the pilus rod and that is responsible of the adhesion to the host [100,101,102,103]. FimH recognizes mannosylated glycoproteins, including those present on the host urinary epithelium as demonstrated by using D-mannose or oligosaccharides containing terminal mannose residues in FimH-mediated adhesion inhibition experiments [104,105].
Type 1 fimbriae were described to be phase-variable with a different role in lungs and urinary or intestinal tract infections. Indeed, fimbrial expression was found to be highly upregulated in the bacterial population in urine and infected bladders and downregulated in the lungs. In this organ, expression of type 1 fimbriae may be a disadvantage for the bacteria because of their ability to adhere to phagocytic cells in the lungs and therefore to be rapidly eliminated [106,107,108]. Moreover, FimH was shown to be required for Kp invasion and biofilm formation in a murine model of UTI [109] but the role of type 1 fimbriae in colonization of the gastrointestinal tract still remains controversial. Indeed, while Jung et al. observed the decreased colonization ability of a Kp fimD mutant of antibiotic-treated mice intestine (FimD is appointed to facilitate assembly and translocation of the pilus), another recent work showed data supporting that type 1 fimbriae do not contribute to gastrointestinal colonization as no significant difference in adhesion was noticed between wild-type and fimH mutant strains [110,111].
The components of type 3 fimbriae are encoded by the genes in the mrkABCDF operon [112,113]. The mrk gene cluster, as other fimbrial operons of the chaperone-usher class, contains genes encoding the chaperone (mrkB), the usher (mrkC) and the protein-based filament composed by a major (mrkA) and a minor (mrkF) subunit. MrkD is the tip adhesion protein, which is appointed for the adhesive properties of the whole complex, whereas to MrkE has been attributed a regulatory activity [112,114].
Among different Klebsiella strains, it is possible to find a plasmid-borne determinant, mrkD1P, and a chromosomally borne gene, mrkD1C, which are not genetically related, and the proteins encoded have been shown to have differences in functionality. MrkD adhesin located within a chromosomally borne gene cluster mediates binding to collagen types IV and V, whereas only few strains presenting the plasmid-borne form showed collagen-binding activity [115].
The presence of mrk genes in multiple genomic locations, including conjugative plasmids or transposons, can explain why this operon is so widespread among Gram-negative bacteria [116,117].
Type 3 fimbriae have been extensively proven to mediate binding to extracellular matrix (ECM) proteins such as collagen molecules [115,118] and to strongly promote biofilm formation [119,120,121].
A recent study revealed that MrkA expression was downregulated after treatment of Kp with the phytosynthesized silver nanoparticles (AgNPs) fabricated from Mespilus germanica, as demonstrated by RT-PCR analysis. In correlation with MrkA expression, also a strong reduction in biofilm formation was observed following treatment with AgNPs, confirming once again the leading role played by MrkA in this process [122]. Interestingly, investigation of MrkA regulation by Wilksch et al. has led to the understanding of another key player in type 3 fimbriae regulation, named MrkH, which directly activates transcription of the mrkA promoter. MrkH strongly binds to the mrkA regulatory region only in the presence of c-di-GMP, a second messenger molecule known to regulate the expression of many bacterial factors involved in colonization and biofilm formation [123,124,125,126,127].
3.4. Siderophores
Siderophores are secreted small molecules that can bind the iron present externally and re-enter the bacterial cells through specific receptors [128,129]. Kp production of siderophores such as enterobactin, yersiniabactin, salmochelin, and aerobactin has been shown to strongly correlate with in vivo virulence and differentiate Hv-Kp from classical Kp strains. For this reason, siderophores have been suggested as potential biomarkers to be further implemented in screening laboratory tests [29,130,131,132]. Each of these molecules presents a specific receptor on the bacterial surface able to bind the siderophore and to mediate the iron uptake, which is essential for the pathogen survival.
4. Vaccines and Monoclonal Antibodies (mAb) Strategies
No vaccines are currently licensed against Kp, but several vaccine targets have been described in the past few decades [133]. Moreover, a therapeutic approach based on the use of monoclonal antibodies (mAbs) against AMR pathogens has been taking place over the last years [134].
Several research models have been implemented in the processes of vaccine development or mAbs identification, chiefly rats and mice. Vaccine antigens or mAbs have been tested in the mouse model to investigate two clinical manifestations of Klebsiella infections: pneumonia and sepsis. The mouse pneumonia model recapitulates key features of Klebsiella-induced pneumonia in humans, characterized by a massive inflammation with influx of polymorphonuclear neutrophils and oedema. However, the results obtained with this model have uncovered many aspects implicated in host defense against the pathogen, due to the differences between mice and humans in immune system development and response to infection [135,136]. More recently, other models are proving to be useful in assessing Kp infection biology, such as the non-mammalian models Galleria mellonella and Danio rerio (zebrafish) that are more easy to handle and still recapitulates interaction with the innate immune system [136]. Certainly, non-human primates, which are also naturally infected by Kp, would provide the best model for the investigation of vaccines/mAbs efficacy, especially squirrel monkeys that were described to better mimic human respiratory pneumonia compared to rodents, but so far have been only employed to test antibiotics [137,138].
4.1. K-Antigen Based Approaches
Bacterial capsule polysaccharides have been used historically as vaccine target antigens and several formulations have been developed against different pathogens and licensed worldwide in the last 40 years [139]. However, a K-antigen-based immunotherapeutic strategy against Kp is complicated because of the high variability and structural diversity.
Different preparations of K-antigen vaccines have been tested in preclinical and clinical studies and used to produce hyperimmune human sera as a therapeutic. One of the first tries dates back to 1982, when serum recovered from mice immunized with UV-irradiated Kp was added to the inoculum of bacteria used to infect the animals, producing 50% of mice protection. In the study, a relationship between protective ability and hemagglutinating activity of the serum was demonstrated, which indicates the presence of antibodies directed against the capsule [140].
Several published studies have shown the ability of anti-K antibodies to confer protection against Kp in animal models of infection. Cryz et al. demonstrated that rabbit anti-K antibodies were able to reduce mortality when administered to mice before challenge with homologous Kp [141]. The same authors later showed that anti-K1 IgG isolated from human volunteers are able to protect mice from Kp sepsis [142]. Additionally, a murine anti-K2 IgM monoclonal antibody was tested in rats against experimental Kp challenge and, even though the invasion was not prevented, mAb-treated animals showed a more rapid bacterial clearance resulting in an accelerated resolution of infection [143]. In 1985, human volunteers were vaccinated using a preparation of K1 polysaccharide detoxified from trace quantities of LPS by treatment with 95% ethanol-0.1 N NaOH solution. This treatment reduced local reactions compared to the control group vaccinated with the untreated K1 and all vaccinees responded with a fourfold or greater rise in IgG and IgM titers. These antibodies were further proven to prevent Kp sepsis when tested in mice [142].
Considering that the ideal capsule-based vaccine should be multivalent in order to cover the majority of all bacteraemic isolates, Cryz et al. subsequently tested a polyvalent Klebsiella vaccine, composed of six K-serotypes (K2, K3, K10, K21, K30, and K55), which resulted to be safe and immunogenic in humans [144,145]. However, a following epidemiological study raised the awareness that a Kp vaccine able to cover around 70% of clinically relevant Kp strains should include at least 25 capsular polysaccharides [146]. With this aim, the same group formulated a vaccine consisting of 24 unconjugated K-antigens, manufactured by the Swiss Serum and Vaccine Institute, together with a Pseudomonas aeruginosa vaccine consisting of eight O-antigen polysaccharides conjugated to Pseudomonas toxin A. This formulation showed safety and immunogenicity in a phase 1 human trial and allowed one to collect hyperimmune globulin for intravenous use (H-IVIG) from the plasma of vaccinees [147,148]. Although the administration of H-IVIG proved to be effective in reducing the severity of Kp infection, these results were not statistically significant, and many patients suffered from adverse reactions [149].
Other groups followed the conjugation approach to increase K-antigens immunogenicity. Already in 1985, an octasaccharide obtained from K11 was conjugated to either bovine serum albumin or to keyhole limpet hemocyanin and tested in wild type and nude mice. This work showed that the coupling of this oligosaccharide to proteins converted the antigen from T-independent to T-dependent [150]. More recently, another group identified a shared hexasaccharide repeating unit (hexasaccharide 1) in the K-antigens of CR-Kp isolates responsible for a deadly outbreak in 2011 and conjugated it to CRM197, obtaining a vaccine that resulted immunogenic in mice and rabbits [151].
An interesting study was performed in 2019 by Feldman et al. who tested for the first time a bioconjugate vaccine encompassing capsules from the K1 and K2 serotypes, causing around 70% of all Hv-Kp infections worldwide [152]. The bioconjugate vaccine, produced in a glycoengineered E. coli strain, was made by the transfer of K1/K2 polysaccharides from a lipid-liked precursor to a genetically deactivated exotoxin A from Pseudomonas aeruginosa. K1 and K2 bioconjugates elicited serotype-specific IgG responses in mice and were also able to protect animals from Hv-Kp infection, confirming this strategy to be very promising for the development of a Kp vaccine. For their relevance in protecting most of the Hv-Kp strains, K1 and K2 were also investigated as potential targets for mAbs.
Recently, Diago-Navarro et al. isolated two mAbs, 17H12 and 8F12, targeting the above-mentioned hexasaccharide 1 and able to agglutinate several CR-Kp strains, presenting both K1, K2 and other K-serotypes. The functionality of these mAbs was extensively proven in vitro (e.g., revealing the ability to promote opsonophagocytic killing and neutrophil extracellular trap (NET) release), and in in vivo studies, where protection against K1 strains in three distinct murine infection models was demonstrated. The authors claimed that these two mAbs can be considered promising candidates for an antibody-based approach to the treatment of CR-Kp infections and that the hexasaccharide 1 should be further explored for vaccine development [153].
4.2. O-Antigen Based Approaches
Vaccination strategies against Kp have largely focused on the capsule polysaccharides. However, Kp O-antigens are alternative targets for the development of a vaccine, due to their limited range of structures. Indeed, four serotypes (O1, O2, O3 and O5) were predicted to cover over 80% of clinically relevant Kp strains [54,76,154,155]. Nevertheless, the development of O-antigen-based vaccines against Kp has not yet progressed beyond the preclinical phase.
Clements et al. showed that immunization with purified O1 LPS vaccine could protect mice against K2:O1 challenges [155]. Additionally, Chhibber et al., reported that O1 LPS, incorporated either into liposomes or sodium alginate microparticles, could protect rodents against lobar pneumonia and resulted to be less toxic than free LPS in mice [156]. A year later, the same authors prepared a glycoconjugate vaccine using the O1 O-antigen covalently linked to tetanus toxoid: the conjugate was found to be non-pyrogenic in rabbits and immunoprotective, as verified from the decrease in the relative colonization of bacteria in lungs of immunized rats as compared to the control animals [157].
Recently, Hegerle et al. reported the development of a combined Kp and Pseudomonas aeruginosa glycoconjugate vaccine comprising O1, O2, O3, O5 Kp O-serotypes, chemically linked to the two Pseudomonas flagellin types (FlaA, FlaB) [158]. The quadrivalent conjugate vaccine generated antibody titers to the four Kp O-antigens and both Fla antigens in rabbits. Innovative strategies have been recently proposed for the generation of O2a glycoconjugate vaccines including a semi-synthetic approach and a bioconjugation approach [159,160].
Of note, anti-Kp O-antigen mAbs also proved to be able to reduce bacterial burden, enhance survival, and show synergy with current standard of care therapy [161]. Pennini et al. demonstrated that the serotype specific anti-O1 (KPE33) and anti-O2 (KPN42) human mAbs are able to protect mice against Kp infection via opsonophagocytic killing [58]. Guachalla et al., generated an anti-O3 mAb 2F8, able to cross-react with all three O3 sub-serogroups (O3, O3a and O3b) [162].
4.3. Protein Based Approaches
Protein virulence factors have also been proposed as targets for the development of vaccines and therapeutic mAb against Kp. These antigens are characterized by low variability, especially when compared to the K-antigens and O-antigens polysaccharides.
In particular, type 3 fimbriae structural protein MrkA has gained a strong interest as a putative target for a vaccine as, besides being expressed by most of Kp strains, including Hv-Kp, it shows a structural position on the pilus filament that may ensure easy access to antibodies. There are already data showing that immunization with purified fimbriae was able to protect mice against a lethal challenge in a model of acute pneumonia [163]. In 2016, Wang et al., immunized mice subcutaneously with both monomeric and oligomeric MrkA formulated with Freund’s adjuvant and the vaccinated mice showed a reduction in the organs’ bacterial burden after intranasal challenge with Kp [164]. In the same work, using high throughput opsonophagocytic killing (OPK) screens, the authors identified an anti-MrkA mAb, named KP3, able not only to determine OPK activity against different Kp serotypes, but also to reduce Kp adhesion to human pulmonary epithelial cells A549. Interestingly, these data also showed that OPK may be a reliable assay to measure in vitro antibodies’ ability to mediate in vivo protection [164].
Type 1 and 3 fimbriae were used as carrier proteins conjugated to E. coli core-oligosaccharide, proving to be immunogenic also in rabbits [165]. Furthermore, Cross and coauthors have recently submitted a patent on a multiple antigen presenting system (MAPS) vaccine encompassing MrkA conjugated to four Kp O-antigens (O1, O2, O3 and O5), providing experimental evidence of good immunogenicity in mice [133].
Recently, using bioinformatic prediction tools, Zargaran et al., found four B cell epitopes (each for one Fim antigen) that were proposed as suitable vaccine candidates for Kp as these epitopes may be recognized by the B cell receptor, triggering humoral responses. Indeed, in silico analysis suggested that these epitopes are immunogenic and antigenic, not similar to human peptides, not allergenic and not toxic. However, these results still need to be supported by in vitro and in vivo testing [166].
Dar et al., in 2019, applied a reverse vaccinology approach on a total of 222 available complete genomes of Kp and four outer membrane proteins were shortlisted for vaccine designing: the outer membrane protein OmpA, the copper/silver efflux RND transporter, the phosphoporin PhoE and the peptidoglycan-associated lipoprotein Pal. Of these antigens, a total of four epitopes were joined together using flexible linkers resulting in a potential multi-epitope vaccine against Kp [167].
Another virulence factor recently investigated as a potential vaccine target is the highly conserved YidR, an ATP/GTP-binding protein that mediates the hyperadherence phenotype and is involved in biofilm formation [168]. Recombinant YidR has been proven to protect mice from challenge with a low dose of Kp [168].
5. Conclusions
The use of antimicrobials has historically enabled the treatment of life-threatening diseases, however the global increase in AMR has compromised the ability to cure a wide range of infections and has been paralleled by a general slowdown in the development and commercialization of novel antibiotics [169].
Klebsiella pneumoniae is a genetically and phenotypically variable bacterium which has become a significant threat to public health over the past few decades. MDR-Kp lineages, such as CG258, have evolved resistances to carbapenems and are responsible for hospital-acquired infections worldwide. Hv-Kp lineages, such as CG23, are instead associated with community-acquired invasive infections.
In both cases, novel strategies for prophylaxis and treatment of infections are urgently needed, including vaccines and monoclonal antibodies. Indeed, the use of mAbs could be advantageous to protect against hospital-acquired infections in patients at high risk, compared to vaccination. A mAb-based approach would be potentially attractive particularly for immunocompromised or elderly subjects, who may respond sub-optimally to vaccination. On the other hand, a vaccination approach would be more suitable to prevent community-acquired or hospital-acquired infections, especially in LMIC, where maternal immunization could potentially protect newborns from hospital acquired infections leading to neonatal sepsis.
There are, however, some barriers in developing vaccines and mAbs against Kp infections. First of all, the great serotype variability requires either high-valency vaccines or mAbs cocktails to obtain acceptable coverage. K-antigens have been historically considered key protective antigens for both active and passive immunization. However, less diverse polysaccharides as Kp O-antigens or highly conserved surface proteins as fimbriae are recently gaining attention as alternative approaches. While there are growing evidences supporting the ability of anti-Kp O-antigens and anti-fimbriae mAbs to protect mice in therapeutic models, there are still concerns around the protective immunity conferred by antibodies to sub-capsular antigens, especially considering the hypercapsule production in Hv-Kp. A second limitation is represented by the available in vivo models. Indeed, differences have been highlighted in Kp pathogenicity between humans and animal models (e.g., the CR-Kp CG258 is responsible for serious infections in humans but is rapidly cleared in mice and rats [170]). Finally, in vitro models need to reflect the diverse mechanisms operating in vivo to prevent infection and should allow the understanding of how mAbs work in the context of the immune system.
6. Future Perspectives
Preventive vaccines as well as therapeutic mAbs represent complementary approaches to fight AMR. Innovative technologies are needed to tackle complex pathogens such as Klebsiella, by targeting multiple antigens at the same time.
Recently, outer membrane vesicles (OMVs) from Kp were proposed as a vaccine candidate and conferred protection in a preclinical animal model, with a mechanism dependent on both humoral and cellular immunity [171]. This result emphasizes the need of novel strategies combining polysaccharide and protein antigens in a single formulation [172]. Additionally, optimal delivery systems need to be identified according to the target population, including a preference for single dose vaccination for use in pregnant women.
In parallel, improvements in the field of mAbs (e.g., new technologies enabling the generation of bispecific mAb) could result in passive administration of few unique mAbs (e.g., 2–3 mAbs) targeting key virulence factors and providing protection against the vast majority of clinical isolates [173].
Funding
This research received no external funding.
Conflicts of Interest
This work was undertaken at the request of and sponsored by GlaxoSmithKline Biologicals SA. GSK Vaccines Institute for Global Health Srl is an affiliate of GlaxoSmithKline Biologicals SA. All authors are employees of the GSK group of companies.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Magiorakos A.P., Srinivasan A., Carey R.B., Carmeli Y., Falagas M.E., Giske C.G., Harbarth S., Hindler J.F., Kahlmeter G., Olsson-Liljequist B., et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery and Development of New Antibiotics. [(accessed on 13 April 2021)]; Available online: https://www.who.int/medicines/publications/global-priority-list-antibiotic-resistant-bacteria/en/
- 3.Rice L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J. Infect. Dis. 2008;197:1079–1081. doi: 10.1086/533452. [DOI] [PubMed] [Google Scholar]
- 4.Paczosa M.K., Mecsas J. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol. Mol. Biol. Rev. 2016;80:629–661. doi: 10.1128/MMBR.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Navon-Venezia S., Kondratyeva K., Carattoli A. Klebsiella pneumoniae: A major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 2017;41:252–275. doi: 10.1093/femsre/fux013. [DOI] [PubMed] [Google Scholar]
- 6.Wyres K.L., Holt K.E. Klebsiella pneumoniae as a key trafficker of drug resistance genes from environmental to clinically important bacteria. Curr. Opin. Microbiol. 2018;45:131–139. doi: 10.1016/j.mib.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 7.Lee C.C., Lee C.H., Hong M.Y., Hsieh C.C., Tang H.J., Ko W.C. Propensity-matched analysis of the impact of extended-spectrum beta-lactamase production on adults with community-onset Escherichia coli, Klebsiella species, and Proteus mirabilis bacteremia. J. Microbiol. Immunol. Infect. 2018;51:519–526. doi: 10.1016/j.jmii.2017.05.006. [DOI] [PubMed] [Google Scholar]
- 8.Shu L.B., Lu Q., Sun R.H., Lin L.Q., Sun Q.L., Hu J., Zhou H.W., Chan E.W., Chen S., Zhang R. Prevalence and phenotypic characterization of carbapenem-resistant Klebsiella pneumoniae strains recovered from sputum and fecal samples of ICU patients in Zhejiang Province, China. Infect. Drug Resist. 2019;12:11–18. doi: 10.2147/IDR.S175823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han J.H., Goldstein E.J., Wise J., Bilker W.B., Tolomeo P., Lautenbach E. Epidemiology of Carbapenem-Resistant Klebsiella pneumoniae in a Network of Long-Term Acute Care Hospitals. Clin. Infect. Dis. 2017;64:839–844. doi: 10.1093/cid/ciw856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falagas M.E., Rafailidis P.I., Kofteridis D., Virtzili S., Chelvatzoglou F.C., Papaioannou V., Maraki S., Samonis G., Michalopoulos A. Risk factors of carbapenem-resistant Klebsiella pneumoniae infections: A matched case control study. J. Antimicrob. Chemother. 2007;60:1124–1130. doi: 10.1093/jac/dkm356. [DOI] [PubMed] [Google Scholar]
- 11.Nordmann P., Cuzon G., Naas T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect. Dis. 2009;9:228–236. doi: 10.1016/S1473-3099(09)70054-4. [DOI] [PubMed] [Google Scholar]
- 12.Yigit H., Queenan A.M., Anderson G.J., Domenech-Sanchez A., Biddle J.W., Steward C.D., Alberti S., Bush K., Tenover F.C. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2001;45:1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Duin D., Perez F., Rudin S.D., Cober E., Hanrahan J., Ziegler J., Webber R., Fox J., Mason P., Richter S.S., et al. Surveillance of carbapenem-resistant Klebsiella pneumoniae: Tracking molecular epidemiology and outcomes through a regional network. Antimicrob. Agents Chemother. 2014;58:4035–4041. doi: 10.1128/AAC.02636-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yong D., Toleman M.A., Giske C.G., Cho H.S., Sundman K., Lee K., Walsh T.R. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 2009;53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Connor C., Cormican M., Boo T.W., McGrath E., Slevin B., O’Gorman A., Commane M., Mahony S., O’Donovan E., Powell J., et al. An Irish outbreak of New Delhi metallo-beta-lactamase (NDM)-1 carbapenemase-producing Enterobacteriaceae: Increasing but unrecognized prevalence. J. Hosp. Infect. 2016;94:351–357. doi: 10.1016/j.jhin.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 16.Bosch T., Lutgens S.P.M., Hermans M.H.A., Wever P.C., Schneeberger P.M., Renders N.H.M., Leenders A., Kluytmans J., Schoffelen A., Notermans D., et al. Outbreak of NDM-1-Producing Klebsiella pneumoniae in a Dutch Hospital, with Interspecies Transfer of the Resistance Plasmid and Unexpected Occurrence in Unrelated Health Care Centers. J. Clin. Microbiol. 2017;55:2380–2390. doi: 10.1128/JCM.00535-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tavoschi L., Forni S., Porretta A., Righi L., Pieralli F., Menichetti F., Falcone M., Gemignani G., Sani S., Vivani P., et al. Prolonged outbreak of New Delhi metallo-beta-lactamase-producing carbapenem-resistant Enterobacterales (NDM-CRE), Tuscany, Italy, 2018 to 2019. Eur. Surveill. 2020;25:85. doi: 10.2807/1560-7917.ES.2020.25.6.2000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carrer A., Poirel L., Eraksoy H., Cagatay A.A., Badur S., Nordmann P. Spread of OXA-48-positive carbapenem-resistant Klebsiella pneumoniae isolates in Istanbul, Turkey. Antimicrob. Agents Chemother. 2008;52:2950–2954. doi: 10.1128/AAC.01672-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Avgoulea K., Di Pilato V., Zarkotou O., Sennati S., Politi L., Cannatelli A., Themeli-Digalaki K., Giani T., Tsakris A., Rossolini G.M., et al. Characterization of Extensively Drug-Resistant or Pandrug-Resistant Sequence Type 147 and 101 OXA-48-Producing Klebsiella pneumoniae Causing Bloodstream Infections in Patients in an Intensive Care Unit. Antimicrob. Agents Chemother. 2018;62:457. doi: 10.1128/AAC.02457-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wrenn C., O’Brien D., Keating D., Roche C., Rose L., Ronayne A., Fenelon L., Fitzgerald S., Crowley B., Schaffer K. Investigation of the first outbreak of OXA-48-producing Klebsiella pneumoniae in Ireland. J. Hosp. Infect. 2014;87:41–46. doi: 10.1016/j.jhin.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 21.Semin-Pelletier B., Cazet L., Bourigault C., Juvin M.E., Boutoille D., Raffi F., Hourmant M., Blancho G., Agard C., Connault J., et al. Challenges of controlling a large outbreak of OXA-48 carbapenemase-producing Klebsiella pneumoniae in a French university hospital. J. Hosp. Infect. 2015;89:248–253. doi: 10.1016/j.jhin.2014.11.018. [DOI] [PubMed] [Google Scholar]
- 22.Borgmann S., Pfeifer Y., Becker L., Riess B., Siegmund R., Sagel U. Findings from an outbreak of carbapenem-resistant Klebsiella pneumoniae emphasize the role of antibiotic treatment for cross transmission. Infection. 2018;46:103–112. doi: 10.1007/s15010-017-1103-3. [DOI] [PubMed] [Google Scholar]
- 23.Guo L., An J., Ma Y., Ye L., Luo Y., Tao C., Yang J. Nosocomial Outbreak of OXA-48-Producing Klebsiella pneumoniae in a Chinese Hospital: Clonal Transmission of ST147 and ST383. PLoS ONE. 2016;11:e0160754. doi: 10.1371/journal.pone.0160754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen C.M., Guo M.K., Ke S.C., Lin Y.P., Li C.R., Vy Nguyen H.T., Wu L.T. Emergence and nosocomial spread of ST11 carbapenem-resistant Klebsiella pneumoniae co-producing OXA-48 and KPC-2 in a regional hospital in Taiwan. J. Med. Microbiol. 2018;67:957–964. doi: 10.1099/jmm.0.000771. [DOI] [PubMed] [Google Scholar]
- 25.Liu Y.C., Cheng D.L., Lin C.L. Klebsiella pneumoniae liver abscess associated with septic endophthalmitis. Arch. Intern. Med. 1986;146:1913–1916. doi: 10.1001/archinte.1986.00360220057011. [DOI] [PubMed] [Google Scholar]
- 26.Wang J.H., Liu Y.C., Lee S.S., Yen M.Y., Chen Y.S., Wang J.H., Wann S.R., Lin H.H. Primary liver abscess due to Klebsiella pneumoniae in Taiwan. Clin. Infect. Dis. 1998;26:1434–1438. doi: 10.1086/516369. [DOI] [PubMed] [Google Scholar]
- 27.Lok K.H., Li K.F., Li K.K., Szeto M.L. Pyogenic liver abscess: Clinical profile, microbiological characteristics, and management in a Hong Kong hospital. J. Microbiol. Immunol. Infect. 2008;41:483–490. [PubMed] [Google Scholar]
- 28.Chung D.R., Lee S.S., Lee H.R., Kim H.B., Choi H.J., Eom J.S., Kim J.S., Choi Y.H., Lee J.S., Chung M.H., et al. Emerging invasive liver abscess caused by K1 serotype Klebsiella pneumoniae in Korea. J. Infect. 2007;54:578–583. doi: 10.1016/j.jinf.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 29.Russo T.A., Olson R., Fang C.T., Stoesser N., Miller M., MacDonald U., Hutson A., Barker J.H., La Hoz R.M., Johnson J.R. Identification of Biomarkers for Differentiation of Hypervirulent Klebsiella pneumoniae from Classical K. pneumoniae. J. Clin. Microbiol. 2018;56:18. doi: 10.1128/JCM.00776-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsu C.R., Lin T.L., Chen Y.C., Chou H.C., Wang J.T. The role of Klebsiella pneumoniae rmpA in capsular polysaccharide synthesis and virulence revisited. Microbiology. 2011;157:3446–3457. doi: 10.1099/mic.0.050336-0. [DOI] [PubMed] [Google Scholar]
- 31.Holt K.E., Wertheim H., Zadoks R.N., Baker S., Whitehouse C.A., Dance D., Jenney A., Connor T.R., Hsu L.Y., Severin J., et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc. Natl. Acad. Sci. USA. 2015;112:E3574–E3581. doi: 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walker K.A., Miller V.L. The intersection of capsule gene expression, hypermucoviscosity and hypervirulence in Klebsiella pneumoniae. Curr. Opin. Microbiol. 2020;54:95–102. doi: 10.1016/j.mib.2020.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lai Y.C., Lu M.C., Hsueh P.R. Hypervirulence and carbapenem resistance: Two distinct evolutionary directions that led high-risk Klebsiella pneumoniae clones to epidemic success. Expert Rev. Mol. Diagn. 2019;19:825–837. doi: 10.1080/14737159.2019.1649145. [DOI] [PubMed] [Google Scholar]
- 34.Gu D., Dong N., Zheng Z., Lin D., Huang M., Wang L., Chan E.W., Shu L., Yu J., Zhang R., et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: A molecular epidemiological study. Lancet Infect. Dis. 2018;18:37–46. doi: 10.1016/S1473-3099(17)30489-9. [DOI] [PubMed] [Google Scholar]
- 35.Yao B., Xiao X., Wang F., Zhou L., Zhang X., Zhang J. Clinical and molecular characteristics of multi-clone carbapenem-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in a tertiary hospital in Beijing, China. Int. J. Infect. Dis. 2015;37:107–112. doi: 10.1016/j.ijid.2015.06.023. [DOI] [PubMed] [Google Scholar]
- 36.Karlsson M., Stanton R.A., Ansari U., McAllister G., Chan M.Y., Sula E., Grass J.E., Duffy N., Anacker M.L., Witwer M.L., et al. Identification of a Carbapenemase-Producing Hypervirulent Klebsiella pneumoniae Isolate in the United States. Antimicrob. Agents Chemother. 2019;63:519. doi: 10.1128/AAC.00519-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frost I., Van Boeckel T.P., Pires J., Craig J., Laxminarayan R. Global geographic trends in antimicrobial resistance: The role of international travel. J. Travel Med. 2019;26:36. doi: 10.1093/jtm/taz036. [DOI] [PubMed] [Google Scholar]
- 38.Khan M.S., Durrance-Bagale A., Legido-Quigley H., Mateus A., Hasan R., Spencer J., Hanefeld J. ‘LMICs as reservoirs of AMR’: A comparative analysis of policy discourse on antimicrobial resistance with reference to Pakistan. Health Policy Plan. 2019;34:178–187. doi: 10.1093/heapol/czz022. [DOI] [PubMed] [Google Scholar]
- 39.Founou R.C., Founou L.L., Essack S.Y. Clinical and economic impact of antibiotic resistance in developing countries: A systematic review and meta-analysis. PLoS ONE. 2017;12:e0189621. doi: 10.1371/journal.pone.0189621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zaidi A.K., Huskins W.C., Thaver D., Bhutta Z.A., Abbas Z., Goldmann D.A. Hospital-acquired neonatal infections in developing countries. Lancet. 2005;365:1175–1188. doi: 10.1016/S0140-6736(05)71881-X. [DOI] [PubMed] [Google Scholar]
- 41.Liu L., Oza S., Hogan D., Chu Y., Perin J., Zhu J., Lawn J.E., Cousens S., Mathers C., Black R.E. Global, regional, and national causes of under-5 mortality in 2000-15: An updated systematic analysis with implications for the Sustainable Development Goals. Lancet. 2016;388:3027–3035. doi: 10.1016/S0140-6736(16)31593-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okomo U., Akpalu E.N.K., Le Doare K., Roca A., Cousens S., Jarde A., Sharland M., Kampmann B., Lawn J.E. Aetiology of invasive bacterial infection and antimicrobial resistance in neonates in sub-Saharan Africa: A systematic review and meta-analysis in line with the STROBE-NI reporting guidelines. Lancet Infect. Dis. 2019;19:1219–1234. doi: 10.1016/S1473-3099(19)30414-1. [DOI] [PubMed] [Google Scholar]
- 43.Taylor A.W., Blau D.M., Bassat Q., Onyango D., Kotloff K.L., Arifeen S.E., Mandomando I., Chawana R., Baillie V.L., Akelo V., et al. Initial findings from a novel population-based child mortality surveillance approach: A descriptive study. Lancet Glob. Health. 2020;8:e909–e919. doi: 10.1016/S2214-109X(20)30205-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bagley S.T. Habitat association of Klebsiella species. Infect. Control. 1985;6:52–58. doi: 10.1017/S0195941700062603. [DOI] [PubMed] [Google Scholar]
- 45.Brisse S., Verhoef J. Phylogenetic diversity of Klebsiella pneumoniae and Klebsiella oxytoca clinical isolates revealed by randomly amplified polymorphic DNA, gyrA and parC genes sequencing and automated ribotyping. Int. J. Syst. Evol. Microbiol. 2001;51:915–924. doi: 10.1099/00207713-51-3-915. [DOI] [PubMed] [Google Scholar]
- 46.Long S.W., Linson S.E., Ojeda Saavedra M., Cantu C., Davis J.J., Brettin T., Olsen R.J. Whole-Genome Sequencing of Human Clinical Klebsiella pneumoniae Isolates Reveals Misidentification and Misunderstandings of Klebsiella pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae. mSphere. 2017;2:290. doi: 10.1128/mSphereDirect.00290-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Diancourt L., Passet V., Verhoef J., Grimont P.A., Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 2005;43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bialek-Davenet S., Criscuolo A., Ailloud F., Passet V., Jones L., Delannoy-Vieillard A.S., Garin B., Le Hello S., Arlet G., Nicolas-Chanoine M.H., et al. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg Infect. Dis. 2014;20:1812–1820. doi: 10.3201/eid2011.140206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wyres K.L., Holt K.E. Klebsiella pneumoniae Population Genomics and Antimicrobial-Resistant Clones. Trends Microbiol. 2016;24:944–956. doi: 10.1016/j.tim.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 50.Wyres K.L., Hawkey J., Hetland M.A.K., Fostervold A., Wick R.R., Judd L.M., Hamidian M., Howden B.P., Lohr I.H., Holt K.E. Emergence and rapid global dissemination of CTX-M-15-associated Klebsiella pneumoniae strain ST307. J. Antimicrob. Chemother. 2019;74:577–581. doi: 10.1093/jac/dky492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi Q., Lan P., Huang D., Hua X., Jiang Y., Zhou J., Yu Y. Diversity of virulence level phenotype of hypervirulent Klebsiella pneumoniae from different sequence type lineage. BMC Microbiol. 2018;18:94. doi: 10.1186/s12866-018-1236-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin J.C., Koh T.H., Lee N., Fung C.P., Chang F.Y., Tsai Y.K., Ip M., Siu L.K. Genotypes and virulence in serotype K2 Klebsiella pneumoniae from liver abscess and non-infectious carriers in Hong Kong, Singapore and Taiwan. Gut Pathog. 2014;6:21. doi: 10.1186/1757-4749-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wyres K.L., Lam M.M.C., Holt K.E. Population genomics of Klebsiella pneumoniae. Nat. Rev. Microbiol. 2020;18:344–359. doi: 10.1038/s41579-019-0315-1. [DOI] [PubMed] [Google Scholar]
- 54.Follador R., Heinz E., Wyres K.L., Ellington M.J., Kowarik M., Holt K.E., Thomson N.R. The diversity of Klebsiella pneumoniae surface polysaccharides. Microb. Genom. 2016;2:e000073. doi: 10.1099/mgen.0.000073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wyres K.L., Wick R.R., Gorrie C., Jenney A., Follador R., Thomson N.R., Holt K.E. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb. Genom. 2016;2:e000102. doi: 10.1099/mgen.0.000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Patro L.P.P., Sudhakar K.U., Rathinavelan T. K-PAM: A unified platform to distinguish Klebsiella species K- and O-antigen types, model antigen structures and identify hypervirulent strains. Sci. Rep. 2020;10:16732. doi: 10.1038/s41598-020-73360-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choi M., Hegerle N., Nkeze J., Sen S., Jamindar S., Nasrin S., Sen S., Permala-Booth J., Sinclair J., Tapia M.D., et al. The Diversity of Lipopolysaccharide (O) and Capsular Polysaccharide (K) Antigens of Invasive Klebsiella pneumoniae in a Multi-Country Collection. Front. Microbiol. 2020;11:1249. doi: 10.3389/fmicb.2020.01249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pennini M.E., De Marco A., Pelletier M., Bonnell J., Cvitkovic R., Beltramello M., Cameroni E., Bianchi S., Zatta F., Zhao W., et al. Immune stealth-driven O2 serotype prevalence and potential for therapeutic antibodies against multidrug resistant Klebsiella pneumoniae. Nat. Commun. 2017;8:1991. doi: 10.1038/s41467-017-02223-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Amako K., Meno Y., Takade A. Fine structures of the capsules of Klebsiella pneumoniae and Escherichia coli K1. J. Bacteriol. 1988;170:4960–4962. doi: 10.1128/JB.170.10.4960-4962.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martin R.M., Bachman M.A. Colonization, Infection, and the Accessory Genome of Klebsiella pneumoniae. Front. Cell Infect. Microbiol. 2018;8:4. doi: 10.3389/fcimb.2018.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pan Y.J., Lin T.L., Lin Y.T., Su P.A., Chen C.T., Hsieh P.F., Hsu C.R., Chen C.C., Hsieh Y.C., Wang J.T. Identification of capsular types in carbapenem-resistant Klebsiella pneumoniae strains by wzc sequencing and implications for capsule depolymerase treatment. Antimicrob. Agents Chemother. 2015;59:1038–1047. doi: 10.1128/AAC.03560-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Patro L.P.P., Rathinavelan T. Targeting the Sugary Armor of Klebsiella Species. Front. Cell Infect. Microbiol. 2019;9:367. doi: 10.3389/fcimb.2019.00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rahn A., Drummelsmith J., Whitfield C. Conserved organization in the cps gene clusters for expression of Escherichia coli group 1K antigens: Relationship to the colanic acid biosynthesis locus and the cps genes from Klebsiella pneumoniae. J. Bacteriol. 1999;181:2307–2313. doi: 10.1128/JB.181.7.2307-2313.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sachdeva S., Palur R.V., Sudhakar K.U., Rathinavelan T. E. coli Group 1 Capsular Polysaccharide Exportation Nanomachinary as a Plausible Antivirulence Target in the Perspective of Emerging Antimicrobial Resistance. Front. Microbiol. 2017;8:70. doi: 10.3389/fmicb.2017.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Whitfield C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 2006;75:39–68. doi: 10.1146/annurev.biochem.75.103004.142545. [DOI] [PubMed] [Google Scholar]
- 66.Shu H.Y., Fung C.P., Liu Y.M., Wu K.M., Chen Y.T., Li L.H., Liu T.T., Kirby R., Tsai S.F. Genetic diversity of capsular polysaccharide biosynthesis in Klebsiella pneumoniae clinical isolates. Microbiology. 2009;155:4170–4183. doi: 10.1099/mic.0.029017-0. [DOI] [PubMed] [Google Scholar]
- 67.Fung C.P., Chang F.Y., Lee S.C., Hu B.S., Kuo B.I., Liu C.Y., Ho M., Siu L.K. A global emerging disease of Klebsiella pneumoniae liver abscess: Is serotype K1 an important factor for complicated endophthalmitis? Gut. 2002;50:420–424. doi: 10.1136/gut.50.3.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Struve C., Roe C.C., Stegger M., Stahlhut S.G., Hansen D.S., Engelthaler D.M., Andersen P.S., Driebe E.M., Keim P., Krogfelt K.A. Mapping the Evolution of Hypervirulent Klebsiella pneumoniae. mBio. 2015;6:e00630. doi: 10.1128/mBio.00630-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pomakova D.K., Hsiao C.B., Beanan J.M., Olson R., MacDonald U., Keynan Y., Russo T.A. Clinical and phenotypic differences between classic and hypervirulent Klebsiella pneumonia: An emerging and under-recognized pathogenic variant. Eur. J. Clin. Microbiol. Infect. Dis. 2012;31:981–989. doi: 10.1007/s10096-011-1396-6. [DOI] [PubMed] [Google Scholar]
- 70.Nassif X., Fournier J.M., Arondel J., Sansonetti P.J. Mucoid phenotype of Klebsiella pneumoniae is a plasmid-encoded virulence factor. Infect. Immun. 1989;57:546–552. doi: 10.1128/IAI.57.2.546-552.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu F., Lv J., Niu S., Du H., Tang Y.W., Pitout J.D.D., Bonomo R.A., Kreiswirth B.N., Chen L. Multiplex PCR Analysis for Rapid Detection of Klebsiella pneumoniae Carbapenem-Resistant (Sequence Type 258 [ST258] and ST11) and Hypervirulent (ST23, ST65, ST86, and ST375) Strains. J. Clin. Microbiol. 2018;56:731. doi: 10.1128/JCM.00731-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu W.L., Ko W.C., Cheng K.C., Lee C.C., Lai C.C., Chuang Y.C. Comparison of prevalence of virulence factors for Klebsiella pneumoniae liver abscesses between isolates with capsular K1/K2 and non-K1/K2 serotypes. Diagn Microbiol. Infect. Dis. 2008;62:1–6. doi: 10.1016/j.diagmicrobio.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 73.Shon A.S., Bajwa R.P., Russo T.A. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: A new and dangerous breed. Virulence. 2013;4:107–118. doi: 10.4161/viru.22718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Clarke B.R., Ovchinnikova O.G., Kelly S.D., Williamson M.L., Butler J.E., Liu B., Wang L., Gou X., Follador R., Lowary T.L., et al. Molecular basis for the structural diversity in serogroup O2-antigen polysaccharides in Klebsiella pneumoniae. J. Biol. Chem. 2018;293:4666–4679. doi: 10.1074/jbc.RA117.000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Trautmann M., Ruhnke M., Rukavina T., Held T.K., Cross A.S., Marre R., Whitfield C. O-antigen seroepidemiology of Klebsiella clinical isolates and implications for immunoprophylaxis of Klebsiella infections. Clin. Diagn. Lab. Immunol. 1997;4:550–555. doi: 10.1128/CDLI.4.5.550-555.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hansen D.S., Mestre F., Alberti S., Hernandez-Alles S., Alvarez D., Domenech-Sanchez A., Gil J., Merino S., Tomas J.M., Benedi V.J. Klebsiella pneumoniae lipopolysaccharide O typing: Revision of prototype strains and O-group distribution among clinical isolates from different sources and countries. J. Clin. Microbiol. 1999;37:56–62. doi: 10.1128/JCM.37.1.56-62.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kalynych S., Morona R., Cygler M. Progress in understanding the assembly process of bacterial O-antigen. FEMS Microbiol. Rev. 2014;38:1048–1065. doi: 10.1111/1574-6976.12070. [DOI] [PubMed] [Google Scholar]
- 78.Regue M., Izquierdo L., Fresno S., Pique N., Corsaro M.M., Naldi T., De Castro C., Waidelich D., Merino S., Tomas J.M. A second outer-core region in Klebsiella pneumoniae lipopolysaccharide. J. Bacteriol. 2005;187:4198–4206. doi: 10.1128/JB.187.12.4198-4206.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fresno S., Jimenez N., Canals R., Merino S., Corsaro M.M., Lanzetta R., Parrilli M., Pieretti G., Regue M., Tomas J.M. A second galacturonic acid transferase is required for core lipopolysaccharide biosynthesis and complete capsule association with the cell surface in Klebsiella pneumoniae. J. Bacteriol. 2007;189:1128–1137. doi: 10.1128/JB.01489-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Okuda S., Sherman D.J., Silhavy T.J., Ruiz N., Kahne D. Lipopolysaccharide transport and assembly at the outer membrane: The PEZ model. Nat. Rev. Microbiol. 2016;14:337–345. doi: 10.1038/nrmicro.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Clarke B.R., Whitfield C. Molecular cloning of the rfb region of Klebsiella pneumoniae serotype O1:K20: The rfb gene cluster is responsible for synthesis of the D-galactan I O polysaccharide. J. Bacteriol. 1992;174:4614–4621. doi: 10.1128/JB.174.14.4614-4621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kelly R.F., Perry M.B., MacLean L.L., Whitfield C. Structures of the O-antigens of Klebsiella serotypes 02 (2a,2e), 02 (2a,2e,2h), and 02 (2a,2f,2g), members of a family of related D-galactan O-antigens in Klebsiella spp. Innate Immun. 1995;2:131–140. doi: 10.1177/096805199500200208. [DOI] [Google Scholar]
- 83.Kelly R.F., Severn W.B., Richards J.C., Perry M.B., MacLean L.L., Tomás J.M., Merino S., Whitfield C. Structural variation in the O-specific polysaccharides of Klebsiella pneumoniae serotype O1 and O8 lipopolysaccharide: Evidence for clonal diversity in rfb genes. Mol. Microbiol. 1993;10:615–625. doi: 10.1111/j.1365-2958.1993.tb00933.x. [DOI] [PubMed] [Google Scholar]
- 84.Hsieh P.F., Wu M.C., Yang F.L., Chen C.T., Lou T.C., Chen Y.Y., Wu S.H., Sheu J.C., Wang J.T. D-galactan II is an immunodominant antigen in O1 lipopolysaccharide and affects virulence in Klebsiella pneumoniae: Implication in vaccine design. Front. Microbiol. 2014;5:608. doi: 10.3389/fmicb.2014.00608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Prehm P., Jann B., Jann K. The O9 antigen of Escherichia coli. Structure of the polysaccharide chain. Eur. J. Biochem. 1976;67:53–56. doi: 10.1111/j.1432-1033.1976.tb10631.x. [DOI] [PubMed] [Google Scholar]
- 86.Jansson P.E.e.a. Structural studies of the O-antigen polysaccharides of Klebsiella O5 and Escherichia coli O8. Carbohydr. Res. 1985;145:59–66. doi: 10.1016/S0008-6215(00)90412-9. [DOI] [PubMed] [Google Scholar]
- 87.Greenfield L.K.E.A. Biosynthesis of the polymannose lipopolysaccharide O-antigens from Escherichia coli serotypes O8 and O9a requires a unique combination of single- and multiple-active site mannosyltransferases. J. Biol. Chem. 2012;287:35078–35091. doi: 10.1074/jbc.M112.401000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hagelueken G.E.A. Structure of WbdD: A bifunctional kinase and methyltransferase that regulates the chain length of the O antigen in Escherichia coli O9a. Mol. Microbiol. 2012;86:730–742. doi: 10.1111/mmi.12014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vinogradov E.E.A. Structures of lipopolysaccharides from Klebsiella pneumoniae. Eluicidation of the structure of the linkage region between core and polysaccharide O chain and identification of the residues at the non-reducing termini of the O chains. J. Biol. Chem. 2002;277:25070–25081. doi: 10.1074/jbc.M202683200. [DOI] [PubMed] [Google Scholar]
- 90.Rollenske T., Szijarto V., Lukasiewicz J., Guachalla L.M., Stojkovic K., Hartl K., Stulik L., Kocher S., Lasitschka F., Al-Saeedi M., et al. Cross-specificity of protective human antibodies against Klebsiella pneumoniae LPS O-antigen. Nat. Immunol. 2018;19:617–624. doi: 10.1038/s41590-018-0106-2. [DOI] [PubMed] [Google Scholar]
- 91.Duguid J.P., Smith I.W., Dempster G., Edmunds P.N. Non-flagellar filamentous appendages (fimbriae) and haemagglutinating activity in Bacterium coli. J. Pathol. Bacteriol. 1955;70:335–348. doi: 10.1002/path.1700700210. [DOI] [PubMed] [Google Scholar]
- 92.Althouse C., Patterson S., Fedorka-Cray P., Isaacson R.E. Type 1 fimbriae of Salmonella enterica serovar Typhimurium bind to enterocytes and contribute to colonization of swine in vivo. Infect. Immun. 2003;71:6446–6452. doi: 10.1128/IAI.71.11.6446-6452.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Johnson J.G., Murphy C.N., Sippy J., Johnson T.J., Clegg S. Type 3 fimbriae and biofilm formation are regulated by the transcriptional regulators MrkHI in Klebsiella pneumoniae. J. Bacteriol. 2011;193:3453–3460. doi: 10.1128/JB.00286-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Allen W.J., Phan G., Waksman G. Pilus biogenesis at the outer membrane of Gram-negative bacterial pathogens. Curr. Opin. Struct. Biol. 2012;22:500–506. doi: 10.1016/j.sbi.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 95.Clegg S., Wilson J., Johnson J. More than one way to control hair growth: Regulatory mechanisms in enterobacteria that affect fimbriae assembled by the chaperone/usher pathway. J. Bacteriol. 2011;193:2081–2088. doi: 10.1128/JB.00071-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Morrissey B., Leney A.C., Toste Rego A., Phan G., Allen W.J., Verger D., Waksman G., Ashcroft A.E., Radford S.E. The role of chaperone-subunit usher domain interactions in the mechanism of bacterial pilus biogenesis revealed by ESI-MS. Mol. Cell Proteom. 2012;11:15289. doi: 10.1074/mcp.M111.015289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wu C.C., Huang Y.J., Fung C.P., Peng H.L. Regulation of the Klebsiella pneumoniae Kpc fimbriae by the site-specific recombinase KpcI. Microbiology. 2010;156:1983–1992. doi: 10.1099/mic.0.038158-0. [DOI] [PubMed] [Google Scholar]
- 98.Di Martino P., Livrelli V., Sirot D., Joly B., Darfeuille-Michaud A. A new fimbrial antigen harbored by CAZ-5/SHV-4-producing Klebsiella pneumoniae strains involved in nosocomial infections. Infect. Immun. 1996;64:2266–2273. doi: 10.1128/IAI.64.6.2266-2273.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Di Martino P., Sirot D., Joly B., Rich C., Darfeuille-Michaud A. Relationship between adhesion to intestinal Caco-2 cells and multidrug resistance in Klebsiella pneumoniae clinical isolates. J. Clin. Microbiol. 1997;35:1499–1503. doi: 10.1128/JCM.35.6.1499-1503.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Brinton C.C., Jr. The structure, function, synthesis and genetic control of bacterial pili and a molecular model for DNA and RNA transport in gram negative bacteria. Trans. N. Y. Acad. Sci. 1965;27:1003–1054. doi: 10.1111/j.2164-0947.1965.tb02342.x. [DOI] [PubMed] [Google Scholar]
- 101.Zyla D.S., Prota A.E., Capitani G., Glockshuber R. Alternative folding to a monomer or homopolymer is a common feature of the type 1 pilus subunit FimA from enteroinvasive bacteria. J. Biol. Chem. 2019;294:10553–10563. doi: 10.1074/jbc.RA119.008610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Busch A., Phan G., Waksman G. Molecular mechanism of bacterial type 1 and P pili assembly. Philos. Trans. A Math. Phys. Eng. Sci. 2015;373:153. doi: 10.1098/rsta.2013.0153. [DOI] [PubMed] [Google Scholar]
- 103.Lillington J., Geibel S., Waksman G. Biogenesis and adhesion of type 1 and P pili. Biochim. Biophys. Acta. 2014;1840:2783–2793. doi: 10.1016/j.bbagen.2014.04.021. [DOI] [PubMed] [Google Scholar]
- 104.Firon N., Ashkenazi S., Mirelman D., Ofek I., Sharon N. Aromatic alpha-glycosides of mannose are powerful inhibitors of the adherence of type 1 fimbriated Escherichia coli to yeast and intestinal epithelial cells. Infect. Immun. 1987;55:472–476. doi: 10.1128/IAI.55.2.472-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Firon N., Ofek I., Sharon N. Carbohydrate-binding sites of the mannose-specific fimbrial lectins of enterobacteria. Infect. Immun. 1984;43:1088–1090. doi: 10.1128/IAI.43.3.1088-1090.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bernhard W., Gbarah A., Sharon N. Lectinophagocytosis of type 1 fimbriated (mannose-specific) Escherichia coli in the mouse peritoneum. J. Leukoc. Biol. 1992;52:343–348. doi: 10.1002/jlb.52.3.343. [DOI] [PubMed] [Google Scholar]
- 107.Sharon N. Bacterial lectins, cell-cell recognition and infectious disease. FEBS Lett. 1987;217:145–157. doi: 10.1016/0014-5793(87)80654-3. [DOI] [PubMed] [Google Scholar]
- 108.Struve C., Bojer M., Krogfelt K.A. Characterization of Klebsiella pneumoniae type 1 fimbriae by detection of phase variation during colonization and infection and impact on virulence. Infect. Immun. 2008;76:4055–4065. doi: 10.1128/IAI.00494-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rosen D.A., Pinkner J.S., Walker J.N., Elam J.S., Jones J.M., Hultgren S.J. Molecular variations in Klebsiella pneumoniae and Escherichia coli FimH affect function and pathogenesis in the urinary tract. Infect. Immun. 2008;76:3346–3356. doi: 10.1128/IAI.00340-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jung H.J., Littmann E.R., Seok R., Leiner I.M., Taur Y., Peled J., van den Brink M., Ling L., Chen L., Kreiswirth B.N., et al. Genome-Wide Screening for Enteric Colonization Factors in Carbapenem-Resistant ST258 Klebsiella pneumoniae. mBio. 2019;10:663. doi: 10.1128/mBio.02663-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Young T.M., Bray A.S., Nagpal R.K., Caudell D.L., Yadav H., Zafar M.A. Animal Model To Study Klebsiella pneumoniae Gastrointestinal Colonization and Host-to-Host Transmission. Infect. Immun. 2020;88:128. doi: 10.1128/IAI.00071-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Allen B.L., Gerlach G.F., Clegg S. Nucleotide sequence and functions of mrk determinants necessary for expression of type 3 fimbriae in Klebsiella pneumoniae. J. Bacteriol. 1991;173:916–920. doi: 10.1128/JB.173.2.916-920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Duguid J.P. Fimbriae and adhesive properties in Klebsiella strains. J. Gen. Microbiol. 1959;21:271–286. doi: 10.1099/00221287-21-1-271. [DOI] [PubMed] [Google Scholar]
- 114.Huang Y.J., Liao H.W., Wu C.C., Peng H.L. MrkF is a component of type 3 fimbriae in Klebsiella pneumoniae. Res. Microbiol. 2009;160:71–79. doi: 10.1016/j.resmic.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 115.Sebghati T.A., Korhonen T.K., Hornick D.B., Clegg S. Characterization of the type 3 fimbrial adhesins of Klebsiella strains. Infect. Immun. 1998;66:2887–2894. doi: 10.1128/IAI.66.6.2887-2894.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Burmolle M., Bahl M.I., Jensen L.B., Sorensen S.J., Hansen L.H. Type 3 fimbriae, encoded by the conjugative plasmid pOLA52, enhance biofilm formation and transfer frequencies in Enterobacteriaceae strains. Microbiology. 2008;154:187–195. doi: 10.1099/mic.0.2007/010454-0. [DOI] [PubMed] [Google Scholar]
- 117.Ong C.L., Beatson S.A., McEwan A.G., Schembri M.A. Conjugative plasmid transfer and adhesion dynamics in an Escherichia coli biofilm. Appl. Environ. Microbiol. 2009;75:6783–6791. doi: 10.1128/AEM.00974-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tarkkanen A.M., Allen B.L., Westerlund B., Holthofer H., Kuusela P., Risteli L., Clegg S., Korhonen T.K. Type V collagen as the target for type-3 fimbriae, enterobacterial adherence organelles. Mol. Microbiol. 1990;4:1353–1361. doi: 10.1111/j.1365-2958.1990.tb00714.x. [DOI] [PubMed] [Google Scholar]
- 119.Schroll C., Barken K.B., Krogfelt K.A., Struve C. Role of type 1 and type 3 fimbriae in Klebsiella pneumoniae biofilm formation. BMC Microbiol. 2010;10:179. doi: 10.1186/1471-2180-10-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Boddicker J.D., Anderson R.A., Jagnow J., Clegg S. Signature-tagged mutagenesis of Klebsiella pneumoniae to identify genes that influence biofilm formation on extracellular matrix material. Infect. Immun. 2006;74:4590–4597. doi: 10.1128/IAI.00129-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jagnow J., Clegg S. Klebsiella pneumoniae MrkD-mediated biofilm formation on extracellular matrix- and collagen-coated surfaces. Microbiology. 2003;149:2397–2405. doi: 10.1099/mic.0.26434-0. [DOI] [PubMed] [Google Scholar]
- 122.Foroohimanjili F., Mirzaie A., Hamdi S.M.M., Noorbazargan H., Hedayati Ch M., Dolatabadi A., Rezaie H., Bishak F.M. Antibacterial, antibiofilm, and antiquorum sensing activities of phytosynthesized silver nanoparticles fabricated from Mespilus germanica extract against multidrug resistance of Klebsiella pneumoniae clinical strains. J. Basic. Microbiol. 2020;60:216–230. doi: 10.1002/jobm.201900511. [DOI] [PubMed] [Google Scholar]
- 123.Wilksch J.J., Yang J., Clements A., Gabbe J.L., Short K.R., Cao H., Cavaliere R., James C.E., Whitchurch C.B., Schembri M.A., et al. MrkH, a novel c-di-GMP-dependent transcriptional activator, controls Klebsiella pneumoniae biofilm formation by regulating type 3 fimbriae expression. PLoS Pathog. 2011;7:e1002204. doi: 10.1371/journal.ppat.1002204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rosen D.A., Twentyman J., Hunstad D.A. High Levels of Cyclic Di-GMP in Klebsiella pneumoniae Attenuate Virulence in the Lung. Infect. Immun. 2018;86:647. doi: 10.1128/IAI.00647-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Arato V., Gasperini G., Giusti F., Ferlenghi I., Scarselli M., Leuzzi R. Dual role of the colonization factor CD2831 in Clostridium difficile pathogenesis. Sci. Rep. 2019;9:5554. doi: 10.1038/s41598-019-42000-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wu D.C., Zamorano-Sanchez D., Pagliai F.A., Park J.H., Floyd K.A., Lee C.K., Kitts G., Rose C.B., Bilotta E.M., Wong G.C.L., et al. Reciprocal c-di-GMP signaling: Incomplete flagellum biogenesis triggers c-di-GMP signaling pathways that promote biofilm formation. PLoS Genet. 2020;16:e1008703. doi: 10.1371/journal.pgen.1008703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zamorano-Sanchez D., Xian W., Lee C.K., Salinas M., Thongsomboon W., Cegelski L., Wong G.C.L., Yildiz F.H. Functional Specialization in Vibrio cholerae Diguanylate Cyclases: Distinct Modes of Motility Suppression and c-di-GMP Production. mBio. 2019;10:670. doi: 10.1128/mBio.00670-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Miethke M., Marahiel M.A. Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 2007;71:413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Neilands J.B. Iron absorption and transport in microorganisms. Annu Rev. Nutr. 1981;1:27–46. doi: 10.1146/annurev.nu.01.070181.000331. [DOI] [PubMed] [Google Scholar]
- 130.Russo T.A., Marr C.M. Hypervirulent Klebsiella pneumoniae. Clin. Microbiol. Rev. 2019;32:1128. doi: 10.1128/CMR.00001-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Russo T.A., Olson R., Macdonald U., Metzger D., Maltese L.M., Drake E.J., Gulick A.M. Aerobactin mediates virulence and accounts for increased siderophore production under iron-limiting conditions by hypervirulent (hypermucoviscous) Klebsiella pneumoniae. Infect. Immun. 2014;82:2356–2367. doi: 10.1128/IAI.01667-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hsieh P.F., Lin T.L., Lee C.Z., Tsai S.F., Wang J.T. Serum-induced iron-acquisition systems and TonB contribute to virulence in Klebsiella pneumoniae causing primary pyogenic liver abscess. J. Infect. Dis. 2008;197:1717–1727. doi: 10.1086/588383. [DOI] [PubMed] [Google Scholar]
- 133.Choi M., Tennant S.M., Simon R., Cross A.S. Progress towards the development of Klebsiella vaccines. Expert Rev. Vaccines. 2019;18:681–691. doi: 10.1080/14760584.2019.1635460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zurawski D.V., McLendon M.K. Monoclonal Antibodies as an Antibacterial Approach Against Bacterial Pathogens. Antibiotics. 2020;9:155. doi: 10.3390/antibiotics9040155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mizgerd J.P., Skerrett S.J. Animal models of human pneumonia. Am. J. Physiol. Lung Cell Mol. Physiol. 2008;294:L387–L398. doi: 10.1152/ajplung.00330.2007. [DOI] [PubMed] [Google Scholar]
- 136.Bengoechea J.A., Pessoa J.S. Klebsiella pneumoniae infection biology: Living to counteract host defences. FEMS Microbiol. Rev. 2019;43:123–144. doi: 10.1093/femsre/fuy043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Berendt R.F., Magruder R.D., Frola F.R. Treatment of Klebsiella pneumoniae respiratory tract infection of squirrel monkeys with aerosol administration of kanamycin. Am. J. Vet. Res. 1980;41:1492–1494. [PubMed] [Google Scholar]
- 138.Berendt R.F., Knutsen G.L., Powanda M.C. Nonhuman primate model for the study of respiratory Klebsiella pneumoniae infection. Infect. Immun. 1978;22:275–281. doi: 10.1128/IAI.22.1.275-281.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Micoli F., Costantino P., Adamo R. Potential targets for next generation antimicrobial glycoconjugate vaccines. FEMS Microbiol. Rev. 2018;42:388–423. doi: 10.1093/femsre/fuy011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Cooper J.M., Rowley D. Resistance to Klebsiella pneumoniae and the importance of two bacterial antigens. Aust. J. Exp. Biol. Med. Sci. 1982;60:629–641. doi: 10.1038/icb.1982.65. [DOI] [PubMed] [Google Scholar]
- 141.Cryz S.J.F.E., Germanier R. Protection against fatal Klebsiella pneumoniae burn wound sepsis by passive transfer of anticapsular polysaccharide. Infect. Immun. 1984;45:139–142. doi: 10.1128/IAI.45.1.139-142.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Cryz S.J., Jr., Furer E., Germanier R. Safety and immunogenicity of Klebsiella pneumoniae K1 capsular polysaccharide vaccine in humans. J. Infect. Dis. 1985;151:665–671. doi: 10.1093/infdis/151.4.665. [DOI] [PubMed] [Google Scholar]
- 143.Trautmann M., Cryz S.J., Jr., Sadoff J.C., Cross A.S. A murine monoclonal antibody against Klebsiella capsular polysaccharide is opsonic in vitro and protects against experimental Klebsiella pneumoniae infection. Microb. Pathog. 1988;5:177–187. doi: 10.1016/0882-4010(88)90020-4. [DOI] [PubMed] [Google Scholar]
- 144.Cryz S.J., Jr., Mortimer P., Cross A.S., Furer E., Germanier R. Safety and immunogenicity of a polyvalent Klebsiella capsular polysaccharide vaccine in humans. Vaccine. 1986;4:15–20. doi: 10.1016/0264-410X(86)90092-7. [DOI] [PubMed] [Google Scholar]
- 145.Granstrom M., Wretlind B., Markman B., Cryz S. Enzyme-linked immunosorbent assay to evaluate the immunogenicity of a polyvalent Klebsiella capsular polysaccharide vaccine in humans. J. Clin. Microbiol. 1988;26:2257–2261. doi: 10.1128/JCM.26.11.2257-2261.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Cryz S.J., Jr., Mortimer P.M., Mansfield V., Germanier R. Seroepidemiology of Klebsiella bacteremic isolates and implications for vaccine development. J. Clin. Microbiol. 1986;23:687–690. doi: 10.1128/JCM.23.4.687-690.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cryz S.J., Jr., Furer E., Sadoff J.C., Fredeking T., Que J.U., Cross A.S. Production and characterization of a human hyperimmune intravenous immunoglobulin against Pseudomonas aeruginosa and Klebsiella species. J. Infect. Dis. 1991;163:1055–1061. doi: 10.1093/infdis/163.5.1055. [DOI] [PubMed] [Google Scholar]
- 148.Edelman R., Taylor D.N., Wasserman S.S., McClain J.B., Cross A.S., Sadoff J.C., Que J.U., Cryz S.J. Phase 1 trial of a 24-valent Klebsiella capsular polysaccharide vaccine and an eight-valent Pseudomonas O-polysaccharide conjugate vaccine administered simultaneously. Vaccine. 1994;12:1288–1294. doi: 10.1016/S0264-410X(94)80054-4. [DOI] [PubMed] [Google Scholar]
- 149.Donta S.T., Peduzzi P., Cross A.S., Sadoff J., Haakenson C., Cryz S.J., Jr., Kauffman C., Bradley S., Gafford G., Elliston D., et al. Immunoprophylaxis against klebsiella and pseudomonas aeruginosa infections. The Federal Hyperimmune Immunoglobulin Trial Study Group. J. Infect. Dis. 1996;174:537–543. doi: 10.1093/infdis/174.3.537. [DOI] [PubMed] [Google Scholar]
- 150.Zigterman J.W., van Dam J.E., Snippe H., Rotteveel F.T., Jansze M., Willers J.M., Kamerling J.P., Vliegenthart J.F. Immunogenic properties of octasaccharide-protein conjugates derived from Klebsiella serotype 11 capsular polysaccharide. Infect. Immun. 1985;47:421–428. doi: 10.1128/IAI.47.2.421-428.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Seeberger P.H., Pereira C.L., Khan N., Xiao G., Diago-Navarro E., Reppe K., Opitz B., Fries B.C., Witzenrath M. A Semi-Synthetic Glycoconjugate Vaccine Candidate for Carbapenem-Resistant Klebsiella pneumoniae. Angew Chem. Int. Ed. Engl. 2017;56:13973–13978. doi: 10.1002/anie.201700964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Feldman M.F., Bridwell A.E.M., Scott N.E., Vinogradov E., McKee S.R., Chavez S.M., Twentyman J., Stallings C.L., Rosen D.A., Harding C.M. A promising bioconjugate vaccine against hypervirulent Klebsiella pneumoniae. Proc. Natl. Acad. Sci. USA. 2019;116:18655–18663. doi: 10.1073/pnas.1907833116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Diago-Navarro E., Motley M.P., Ruiz-Perez G., Yu W., Austin J., Seco B.M.S., Xiao G., Chikhalya A., Seeberger P.H., Fries B.C. Novel, Broadly Reactive Anticapsular Antibodies against Carbapenem-Resistant Klebsiella pneumoniae Protect from Infection. mBio. 2018;9:18. doi: 10.1128/mBio.01005-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Trautmann M., Held T.K., Cross A.S. O-antigen seroepidemiology of Klebsiella clinical isolates and implications for immunoprophylaxis of Klebsiella infections. Vaccine. 2004;22:818–821. doi: 10.1016/j.vaccine.2003.11.026. [DOI] [PubMed] [Google Scholar]
- 155.Clements A., Jenney A.W., Farn J.L., Brown L.E., Deliyannis G., Hartland E.L., Pearse M.J., Maloney M.B., Wesselingh S.L., Wijburg O.L., et al. Targeting subcapsular antigens for prevention of Klebsiella pneumoniae infections. Vaccine. 2008;26:5649–5653. doi: 10.1016/j.vaccine.2008.07.100. [DOI] [PubMed] [Google Scholar]
- 156.Chhibber S., Wadhwa S., Yadav V. Protective role of liposome incorporated lipopolysaccharide antigen of Klebsiella pneumoniae in a rat model of lobar pneumonia. Jpn J. Infect. Dis. 2004;57:150–155. [PubMed] [Google Scholar]
- 157.Chhibber S., Rani M., Vanashree Y. Immunoprotective potential of polysaccharide-tetanus toxoid conjugate in Klebsiella pneumoniae induced lobar pneumonia in rats. Indian. J. Exp. Biol. 2005;43:40–45. [PubMed] [Google Scholar]
- 158.Hegerle N., Choi M., Sinclair J., Amin M.N., Ollivault-Shiflett M., Curtis B., Laufer R.S., Shridhar S., Brammer J., Toapanta F.R., et al. Development of a broad spectrum glycoconjugate vaccine to prevent wound and disseminated infections with Klebsiella pneumoniae and Pseudomonas aeruginosa. PLoS ONE. 2018;13:e0203143. doi: 10.1371/journal.pone.0203143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Harduin-Lepers A. Glyco25, XXV International Symposium on Glycoconjugates. Glycoconj. J. 2019;36:267–397. doi: 10.1007/s10719-019-09880-4. [DOI] [PubMed] [Google Scholar]
- 160.Zhang L., Pan C., Feng E., Hua X., Yu Y., Wang H., Zhu L. Biosynthesis of polysaccharide conjugate vaccines against Klebsiella pneumoniae serotype O2 strains. Sheng Wu Gong Cheng Xue Bao. 2020;36:1899–1907. doi: 10.13345/j.cjb.200014. [DOI] [PubMed] [Google Scholar]
- 161.Cohen T.S., Pelletier M., Cheng L., Pennini M.E., Bonnell J., Cvitkovic R., Chang C.S., Xiao X., Cameroni E., Corti D., et al. Anti-LPS antibodies protect against Klebsiella pneumoniae by empowering neutrophil-mediated clearance without neutralizing TLR4. JCI Insight. 2017;2:774. doi: 10.1172/jci.insight.92774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Guachalla L.M., Stojkovic K., Hartl K., Kaszowska M., Kumar Y., Wahl B., Paprotka T., Nagy E., Lukasiewicz J., Nagy G., et al. Discovery of monoclonal antibodies cross-reactive to novel subserotypes of K. pneumoniae O3. Sci. Rep. 2017;7:6635. doi: 10.1038/s41598-017-06682-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lavender H., Jagnow J.J., Clegg S. Klebsiella pneumoniae type 3 fimbria-mediated immunity to infection in the murine model of respiratory disease. Int. J. Med. Microbiol. 2005;295:153–159. doi: 10.1016/j.ijmm.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 164.Wang Q., Chang C.S., Pennini M., Pelletier M., Rajan S., Zha J., Chen Y., Cvitkovic R., Sadowska A., Heidbrink T.J., et al. Target-Agnostic Identification of Functional Monoclonal Antibodies Against Klebsiella pneumoniae Multimeric MrkA Fimbrial Subunit. J. Infect. Dis. 2016;213:1800–1808. doi: 10.1093/infdis/jiw021. [DOI] [PubMed] [Google Scholar]
- 165.Witkowska D., Mieszała M., Gamian A., Staniszewska M., Czarny A., Przondo-Mordarska A., Jaquinod M., Forest E. Major structural proteins of type 1 and type 3 Klebsiella fimbriae are effective protein carriers and immunogens in conjugates as revealed from their immunochemical characterization. FEMS Immunol. Med. Microbiol. 2005;45:221–230. doi: 10.1016/j.femsim.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 166.Zargaran F.N., Akya A., Rezaeian S., Ghadiri K., Lorestani R.C., Madanchi H., Safaei S., Rostamian M. B Cell Epitopes of Four Fimbriae Antigens of Klebsiella pneumoniae: A Comprehensive In Silico Study for Vaccine Development. Int. J. Pept. Res. Ther. 2020;134:1–12. doi: 10.1007/s10989-020-10134-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Dar H.A., Zaheer T., Shehroz M., Ullah N., Naz K., Muhammad S.A., Zhang T., Ali A. Immunoinformatics-Aided Design and Evaluation of a Potential Multi-Epitope Vaccine against Klebsiella Pneumoniae. Vaccines. 2019;7:88. doi: 10.3390/vaccines7030088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Rodrigues M.X., Yang Y., de Souza M.E.B., Jr., do Carmo S.J., Bicalho R.C. Development and evaluation of a new recombinant protein vaccine (YidR) against Klebsiella pneumoniae infection. Vaccine. 2020;38:4640–4648. doi: 10.1016/j.vaccine.2020.03.057. [DOI] [PubMed] [Google Scholar]
- 169.Micoli F., Bagnoli F., Rappuoli R., Serruto D. The role of vaccines in combatting antimicrobial resistance. Nat. Rev. Microbiol. 2021;3:506. doi: 10.1038/s41579-020-00506-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Szijarto V., Guachalla L.M., Hartl K., Varga C., Badarau A., Mirkina I., Visram Z.C., Stulik L., Power C.A., Nagy E., et al. Endotoxin neutralization by an O-antigen specific monoclonal antibody: A potential novel therapeutic approach against Klebsiella pneumoniae ST258. Virulence. 2017;8:1203–1215. doi: 10.1080/21505594.2017.1279778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lee W.H., Choi H.I., Hong S.W., Kim K.S., Gho Y.S., Jeon S.G. Vaccination with Klebsiella pneumoniae-derived extracellular vesicles protects against bacteria-induced lethality via both humoral and cellular immunity. Exp. Mol. Med. 2015;47:e183. doi: 10.1038/emm.2015.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Micoli F., Alfini R., Di Benedetto R., Necchi F., Schiavo F., Mancini F., Carducci M., Palmieri E., Balocchi C., Gasperini G., et al. GMMA Is a Versatile Platform to Design Effective Multivalent Combination Vaccines. Vaccines. 2020;8:540. doi: 10.3390/vaccines8030540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Opoku-Temeng C., Kobayashi S.D., DeLeo F.R. Klebsiella pneumoniae capsule polysaccharide as a target for therapeutics and vaccines. Comput. Struct. Biotechnol. J. 2019;17:1360–1366. doi: 10.1016/j.csbj.2019.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]