The authors wish to make the following correction to this paper [1].
After the publication of the manuscript, the authors recognized a discrepancy due to a nucleotide numbering difference between the prototype Tn1546 (M97297) and the Tn1546 elements sequenced in this study. Therefore, a point mutation within the vanX gene of six Tn1546 elements described in this study was mistakenly reported.
We changed Table 1 and Figure 1 and Figure 2 and present the correct versions here.
Table 1.
Phenotypic and genotypic features of vancomycin-resistant Enterococcus spp. isolated from cattle, pigs, and poultry. Changed information is highlighted in grey.
| Host/Species | No. of Strains | Resistance Phenotype | Resistance Genotype | MLST | ||
|---|---|---|---|---|---|---|
| MIC [µg/mL] Vancomycin |
Additional Resistances | Resistance Genes | Tn1546 Type | |||
| Cattle | ||||||
| E. faecalis | 1 | ≥128 | – | dfrE, emeA, efrA, efrB, lsaA, vanA | I | 29 |
| Pigs | ||||||
| E. faecium | 1 | ≥256 | PEN, ERY, TE | aac(6′)-Ii, eat(A)v, cadA, cadC, copZ, czrA, merA, merR, tetW/N/W, vanA, zosA | I | 133 |
| E. faecium | 5 | ≥256 | PEN, TE | aac(6′)-Ii, eat(A)v, cadA, cadC, copZ, czrA, merA, merR, tetW/N/W, vanA, zosA | I | 133 |
| Poultry | ||||||
| E. faecium | 1 | ≥256 | ERY | aac(6′)-Ii, aadK, eat(A)v, vanA | I | 13 |
| E. faecium | 1 | ≥256 | PEN | aac(6′)-Ii, aadK, eat(A)v, vanA | I | 157 |
| E. faecium | 1 | ≥256 | – | aac(6′)-Ii, aadK, eat(A)v, vanA | I | 157 |
| E. faecium | 3 | ≥256 | ERY | aac(6′)-Ii, aadK, eat(A)v, vanA | I | 310 |
| E. durans | 1 | ≥256 | TE | aac(6′)-Iid, tetW/N/W, vanA | I | – |
| E. durans | 2 | ≥256 | ERY, TE | aac(6′)-Iid, ermB, vanA | I | – |
| E. durans | 1 | 256 | ERY, TE | aac(6′)-Iid, ermB tetW/N/W, vanA | I | – |
| E. durans | 1 | ≥256 | TE | aac(6′)-Iid, ermB, tetW/N/W, vanA | I | – |
| E. durans | 1 | ≥256 | ERY, TE | aac(6′)-Iid, ermB tetW/N/W, vanA | I | – |
| E. durans | 3 | ≥256 | ERY, TE | aac(6′)-Iid, ermB tetW/N/W, vanA | II | – |
| E. durans | 1 | ≥256 | TE | aac(6′)-Iid, tetW/N/W, vanA | II | – |
Abbreviations: aac(6′)-Ii and aac(6′)-Iid: genes for aminoglycoside N-acetyltransferases; aadK, aminoglycoside 6-adenylyl-transferase; cadA, cadC, cadmium resistance genes; cop, copper resistance gene; czrA, metal transport repressor gene; dfrE, dihydrofolate reductase gene; eat(A)v, allelic variant of eat(A) gene for resistance to lincosamides, streptogramins A, and pleuromutilins (LSAP); emeA, enterococcal multidrug resistance efflux gene; efrA, efrB, ABC multidrug efflux pump genes; ermB, gene for 23S ribosomal RNA methyl-transferase; ERY: erythromycin; lsaA, active efflux ABC transporter gene for intrinsic LSAP resistance; merA, merR, mercury resistance genes; MIC, minimal inhibitory concentration; MLST, multilocus sequence type; PEN: penicillin; TET: tetracyclin; tetW/N/W, mosaic tetracycline resistance gene and ribosomal protection protein; vanA, vancomycin resistance gene; zosA, zinc transporter gene.
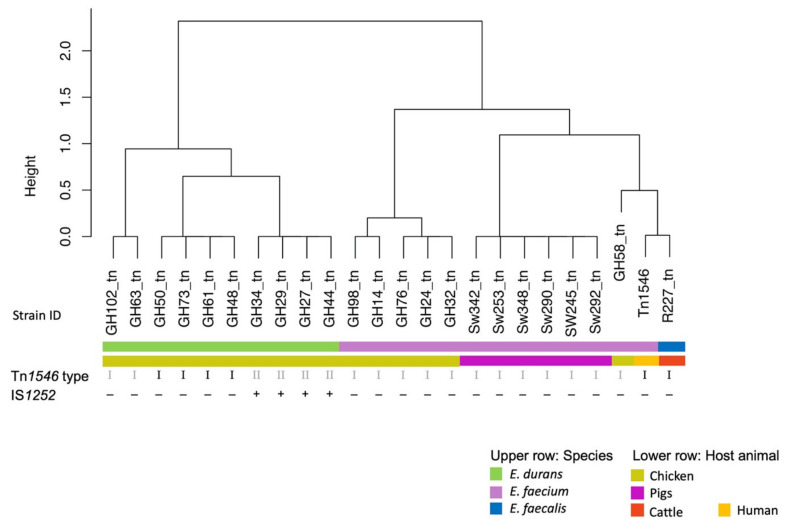
Figure 1.
Average nucleotide identity (ANI) based cluster dendrogram showing three types of Tn1546-like elements carrying vanA operons identified in 23 vanA- type vancomycin-resistant enterococci from healthy food-producing animals. Type I corresponds to the prototype Tn1546 (GenBank M97297) from human E. faecium B4147 [35]. Type II additionally carries an IS1252 in the orf2-vanR intergenic region. Changed information is highlighted in grey.
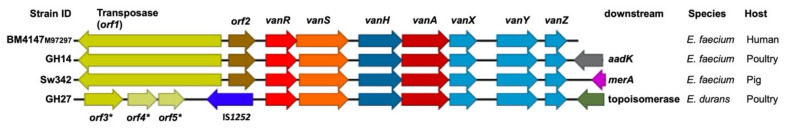
Figure 2.
Linear maps of vanA encoding regions of the prototype Tn1546 (GenBank M97297) from human E. faecium B4147 [35], and of vancomycin-resistant enterococci from healthy food-producing animals. aadK, aminoglycoside 6-adenylyltransferase; merA, mercury resistance gene; *, putative open reading frames.
The authors wish to point out the following:
The abstract should read: “two different types of Tn1546-like elements carrying the vanA operon were identified”.
The results Section 3.4. should read: “Analysis of the Tn1546 structures distinguished two different Tn1546-like types I and II, respectively (Table 1).
The structure of the van operon in type I was identical to the van operon prototype described previously (GenBank M97297) and included six E. durans isolates from poultry, the E. faecalis isolate from cattle and E. faecium from pigs (Table 1). The Tn1546-like type I elements detected in E. durans contained a topoisomerase gene downstream of vanZ, placing them in a highly similar but distinct cluster to the E. faecalis Tn1546 (Figure 1).
The Tn1546-like type I elements identified in E. faecium from poultry contained an aadK gene downstream of vanZ, whereas those found in pigs carried merA. Examples of the Tn1546-like elements are shown in Figure 2.
Finally, type II Tn1546 was identical to the type I structure, but disrupted by IS1252 in the orf2-vanR intergenic region. Type II elements were detected in four E. durans isolates from poultry and carried a topoisomerase gene located downstream of vanZ (Figure 2)”.
The discussion should read: “The majority corresponded to the prototype Tn1546, which has been found in enterococcal isolates from healthy and hospitalised humans, in pig isolates, in food isolates, and in environmental enterococci [53–56]”.
The authors would like to apologize for any inconvenience caused to the readers by these changes.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Reference
- 1.Wist V., Morach M., Schneeberger M., Cernela N., Stevens M.J.A., Zurfluh K., Stephan R., Nüesch-Inderbinen M. Phenotypic and genotypic traits of vancomycin-resistant enterococci from healthy food-producing animals. Microorganisms. 2020;8:261. doi: 10.3390/microorganisms8020261. [DOI] [PMC free article] [PubMed] [Google Scholar]