Abstract
Objective:
The objective of this study was to identify key genes and shed light on the underlying molecular mechanisms of vulvar squamous cell carcinoma (VSCC).
Methods:
Bioinformatic software was utilized for the identification and characterization of key differentially expressed genes (DEGs) from microarrays GSE63678 and GSE38228, which contain VSCC and normal vulvar tissue data. These microarrays were obtained from Gene Expression Omnibus (GEO). Immunohistochemical assays (55 VSCC and 50 normal vulvar tissues) were utilized to validate the expression of VEGF, IGF1, BIRC5, and MMP1 screened from the identified DEGs. SPSS 18.0 software was used for statistical analyses of the relationships between IGF1, BIRC5, VEGF, MMP1 expression levels and patient clinicopathological characteristics.
Results:
A total of 141 DEGs were identified, among which 18 genes were closely correlated with the biological characteristics of VSCC. Four of the 18 genes (VEGF, IGF1, BIRC5, and MMP1) screened from the GEO database were markedly enriched in pathways in cancer (P < 0.05), and could be considered key genes in VSCC based on KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis in DAVID (Database for Annotation, Visualization and Integrated Discovery).The expression levels of these 4 hub genes, determined by immunohistochemical assays, were consistent with the bioinformatics results. Higher expression of IGF1 showed significant association with well-differentiated carcinomas (P = 0.017).BIRC5 expression levels showed a positive correlation with clinical stage (P = 0.039); compared with those in menopause for over 10 years, patients in menopause for less than 10 years at the time of diagnosis tended to have significantly higher expression of BIRC5 (P = 0.003). VEGF and MMP1 expression levels were not correlated with any of the tested clinicopathological characteristics.
Conclusion:
VEGF, IGF1, BIRC5, and MMP1 were identified as being associated with VSCC using integrated bioinformatic methods, which may provide important insights into the pathogenesis of this disease and help to identify new biomarkers.
Keywords: vulvar squamous cell carcinoma, gene chip, VEGF, IGF1, BIRC5, MMP1
Introduction
Vulvar cancer is an uncommon disease that comprises 5% of all gynecological tumors.1 The vast majority of vulvar cancers are of the squamous epithelial carcinoma type, which accounts for approximately 95% of vulvar malignant tumors.2 The average age of onset is 50-60 years old. It is reported that the 5-year specific survival rates were 100% and 100% in patients with stage I and Ⅱ vulvar cancer, which were higher than that in late-stage vulvar cancer (Stage Ⅲ 86%, Stage Ⅳ 29%).3 It seems that the prognosis of patients with early-stage vulvar cancer is better, which may be due to a slowly progression and a mature treatment strategy for patients. However, the driving factors in tumor progression and the underlying molecular mechanism are still unclear.
As a reliable and high-throughput research technique, gene chips based on microarray technology have been commonly used for more than ten years.4They enable the simultaneous detection of expression levels of numerous genes, and have become increasingly useful in understanding the genetic basis, prognosis and drug development of tumors. Only a few studies using this technique have focused on the identification of molecular targets in VSCC. One study,5 which for the first time investigated the expression profile of VSCC using microarray technology, aimed to delineate the pathways involved in the pathogenesis of VSCC. Another study,6 focused on genomic aberration patterns and expression profiles of VSCC, illustrated a concordant relationship between aCGH results and the level of gene expression found by microarray. Genes located in gained regions were generally overexpressed while those located in lost regions were downregulated. One fascinating study,7 revealed common specific pathogenetic patterns and gene markers for cervical (CC), endometrial (EC) and vulvar cancer (VC). Altogether, previous studies5-7 have laid the foundations for the present study to integrate, reanalyze and experimentally validate clinical samples. Herein, we performed data remining based on the GSE38228 and GSE63678 datasets.6,7 Furthermore, 4 hub genes (IGF1, BIRC5, VEGF and MMP1) selected in the present research were verified with clinical samples, and the relationships between their expression levels and patient clinicopathological characteristics were analyzed.
In this research, the VSCC-associated gene expression datasets GSE63678 and GSE38228 were downloaded from Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/). Then, differentially expressed genes between VSCC and normal vulvar tissues (DEGs) in the 2 datasets were identified using the GEO2R online tool and Venn diagram software. Subsequently, a protein-protein interaction (PPI) network was established, and Cytotype MCODE (Molecular Complex Detection) was used for additional analysis of the DEGs to identify the hub genes. In addition, these core DEGs were imported into the Kaplan-Meier Plotter online database to determine whether the core DEGs provided significant prognostic information for cervical squamous cell carcinoma. Finally, an immunohistochemical assay was conducted, and the relationships between the expression levels and patient clinicopathological characteristics were analyzed. In conclusion, the bioinformatic study and experimental verification of our research provide a number of key biomarkers that could be of vital consequence in the progression of VSCC.
Methods
Microarray Data Information
We searched NCBI-GEO, a public database8 of microarray/gene maps, and acquired the gene expression profiles of GSE63678 and GSE38228 in VSCC and normal vulvar tissues. Microarray data of GSE63678 comprise 6 VSCC tumor and 13 normal vulvar tissue samples, while the GSE38228 profile consists of 7 VSCC tumor and 5 normal vulvar tissues samples.
Identification of DEGs
The relevant samples included in GSE63678 and GSE38228 were categorized into 2 groups: -VSCC and normal vulvar tissues. DEGs between the 2 groups were identified by using the GEO2R online tool8 with statistical significance reached at |logFC| > 1 and adjusted P value < 0.05. GEO2R is an interactive web tool especially developed for the identification of differentially expressed genes among selected groups (https://www.ncbi.nlm.nih.gov/geo/geo2r/). Then, we input the raw data in TXT format in Venn software to determine the common DEGs among the 2 datasets (http://bioinformatics.psb.ugent.be/webtools/Venn/). The DEGs with logFC<-1 were considered downregulated genes, while the DEGs with logFC>1 were considered upregulated genes.
PPI Network and Module Analysis
STRING (Search Tool for the Retrieval of Interacting Genes), an online tool, helps us to evaluate PPI information (https://string-db.org/).9 Next, Cytoscape 10 was applied to examine the underlying correlation between these DEGs (maximum number of interactions = 0 and confidence score≥0.4). Furthermore, the MCODE plug-in in Cytoscape was used to check modules of the PPI network to identify the hub genes with parameters as follows: degree cutoff = 2, maximum depth = 100, k-core = 2, and node score cutoff = 0.2.
KEGG Pathway Enrichment Analysis
KEGG is a collection of databases concerning genomes, diseases, biological pathways, drugs, and chemical materials.11,12 As an online bioinformatic tool (https://david.ncifcrf.gov/), DAVID bioinformatics resources consist of an integrated biological knowledge base and analytic tools aimed at systematically extracting biological meaning from large gene/protein lists.13 In this study, a gene list containing 18 gene identifiers was analyzed with the Kyoto Encyclopedia of Gene and Genome (KEGG) pathway using one or more text and pathway-mining tools, such as gene functional classification, functional annotation chart or clustering and functional annotation table. In addition, DAVID provided help to visualize the DEG pathway enrichment (P < 0.05).
Immunohistochemical Assay
In this study, 55 VSCC tumor and 50 normal vulvar tissue samples from June 2013 to October 2019 were collected from the institutional database of Women’s Hospital, School of Medicine, Zhejiang University, China. Among these 55 VSCC tumor samples, 37 were well differentiated, 15 were moderately differentiated and 3 were poorly differentiated; 45 were at clinical stage I, 5 were at clinical stage II and 5 were at clinical stag III; 42 patients were in menopause for over ten years while the other 13 patients were in menopause for less ten years. The average age of all cases was 66.84 years. Each enrolled patient signed an informed consent form to allow her samples and records to be used for scientific research.
Immunohistochemical assays were performed on the VSCC sections. Antibodies against VEGF (1:250 dilution; Abcam cat# ab32152), IGF1 (1:2000 dilution; Abcam cat# ab40657), BIRC5 (1:100 dilution; Abcam cat# ab76424), and MMP1 (1:200 dilution; Abcam cat# ab52631) were incubated overnight at 4°C after deparaffinization, hydration and antigen retrieval. After a washing procedure, a broad-spectrum secondary antibody was incubated. The immunostaining was individually interpreted by the researchers (ZT and LQ).
Positive cells were indicated by the presence of yellow to brown DAB staining in the nucleus or cytoplasm. The number of positively stained cells out of 100 in 10 random fields (400× objective) was counted and represented as the percentage of positive cells. The semiquantitative immunoreactive score was based on the percentage of positive cells and the staining intensity. 0, <5% positive cells; 1, 5%-25% positive cells; 2, 26%-50% positive cells; 3, 51%-75%; 4, more than 76% positive cells. According to the DAB staining intensity, the cells were scored as follows: 0, no staining; 1, faint yellow; 2, brown yellow; and 3, dark brown. The expression categories were finally divided into a high expression group and a low expression group according to expression scores. Because the expression of IGF1 is at a low level, 4 was set as the cutoff value, for which ≥4 is highly expressed and <4 is expressed at low levels. In the expression analysis of VEGF, BIRC5, and MMP1, cutoff value was set at 6: ≥6 indicates high expression; <6 indicates low expression. Five random fields (400× objective) of each slip were observed, and the average of the five fields obtained was the score of the cover flip immunostaining.
Statistical Analysis
The SPSS 18.0 software was used for statistical analyses. Fisher’s Exact tests were used to evaluate protein expression.
Results
Identification of DEGs in VSCC
In total, there were 13 VSCC and 18 normal vulvar tissues samples in GSE63678 and GSE38228. Analysis with the GEO2R online tool, we identified 850 and 1012 DEGs in the 2 datasets, respectively. By screening with |logFC|>1 and adjusted P value < 0.05, we identified 195 upregulated genes and 655 downregulated genes in GSE63678, while 398 upregulated genes and 614 downregulated genes were identified from GSE38228. Next, Venn diagram software was used to identify the common DEGs between the 2 datasets. The results showed that a total of 141 common DEGs were filtered out, of which 39 were upregulated (logFC>1) and 102 were downregulated (logFC<-1) in the VSCC tissues (Table 1, Figure 1).
Table 1.
Entire 141 Common DEGs Were Identified From 2 Profile Datasets, Including 39 Up-Regulated Genes and 102 Down-Regulated Genes in the Vulvar Cancer Tissues Compared to Normal Vulvar Tissues.
| DEGs | Genes name |
|---|---|
| Up-regulated | CDH3, TNFAIP6, MYO1B, COL4A1, PLAU, SLC28A3, HOMER3, IL1A, MMP3, LPAR3, LAMC2, SUGCT, SCD, BASP1, POSTN, BIRC5, KIF20A, MYO5A, AIM2, GALNT6, UBE2C, NQO1, ITGA6, CCNB2, TDO2, PCDH7, IL36G, LIPG, VEGFA, TOP2A, CCNA2, NCAPG, PC, PRNP, MMP1, TYMP, MMP12, AURKA |
| Down-regulated | CHPT1, NR3C2, ZSCAN18, IGF1, VEZF1, ADD3, SLC27A6, SLC11A2, SASH1, MAOB, PTGDS, DHRS11, DIAPH2, SPAG16, ID4, ISL1, DACH1, TACC1, NFIB, ALDH3A2, ACKR1, NBEA, SLC25A4, NAAA, DDAH1, MEIS2, BCL2, SECISBP2L, PAMR1, CXCL12, EFHD1, GLUL, APOD, LDB2, CBX7, ARMCX1, ERMP1, NCOA1, ZBTB20, TLE2, ARMCX6, METTL7A, ITM2A, ZFP36L2, CBX6, MPC1, PLAGL1, FAM189A2, MSRA, ABCA8, DEPTOR, ISOC1, CYP3A5, JUND, FBLN1, NDRG2, SVEP1, PLPP3, PLLP, MEIS1, COBL, TGFBR3, EDN3, PLPP1, KLF2, P4HTM, SOSTDC1, NYNRIN, PIK3R1, CDKN2C, ACACB, CLDN8, FAM107A, GPX3, DDAH2, RRAGD, IL20RA, ABI3BP, DCN, PER2, SLIT2, CX3CR1, LONRF1, LDOC1, ATP6V0E2, MLPH, AR, GATM, KLHL3, GPD1L, OCA2, GULP1, CFD, SYNGR1, ALDH5A1, RNASE4, IGFBP5, EPHX2, ARMCX2, CYP4B1, NR2F2, NDN |
Abbreviation: DEGs, differentially expressed genes.
Figure 1.
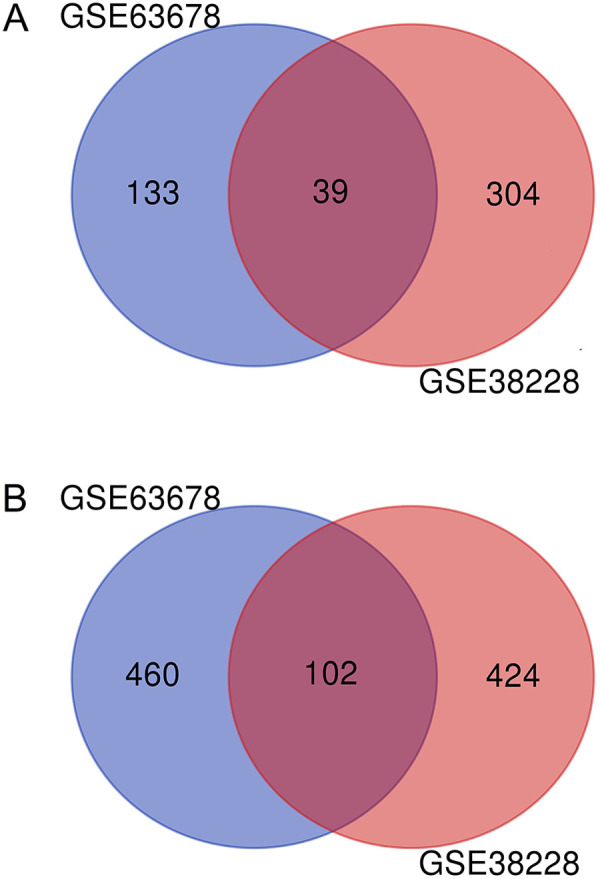
Identification of 141 common DEGs in the 2 datasets. A, 39 common up-regulated genes were searched by GSE63678 and GSE38228. B, 102 common down-regulated genes were searched by GSE63678 and GSE38228.
Protein–Protein Interaction (PPI) Network and Modular Analysis
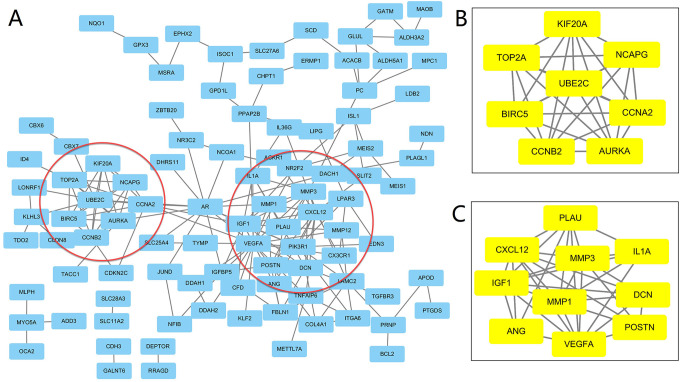
A total of 141 DEGs were imported into the Search Tool for the Retrieval of Interacting Genes (STRING) database (https://string-db.org/) which then obtained 141 nodes and 196 edges, including 102 downregulated and 39 upregulated genes. Then, Cytoscape MCODE was applied to further analysis and 18 central nodes are highlighted in yellow: NCAPG, BIRC5, AURKA, CCNA2, UBE2C, KIF20A, CCNB2, TOP2A, IL1A, PLAU, POSTN, IGF1, MMP1, CXCL12, VEGF, MMP3, DCN, and ANG (Figure 2).
Figure 2.
Common DEGs PPI network constructed by STRING online database and Module analysis. PPI network complex. The nodes meant proteins; the edges meant the interaction of proteins. The yellow part is the 19 hub nodes which was obtained by Module analysis via Cytoscape software (degree cutoff = 2, node score cutoff = 0.2, k-core = 2, and max. Depth = 100).
Analysis of 18 Core Genes for KEGG Pathway Enrichment
Ultimately, 18 genes via DAVID were analyzed via DAVID for KEGG pathway enrichment, which indicated that 4 genes (VEGF, IGF1, BIRC5, MMP1) were markedly enriched in pathways in cancer (P < 0.05), and could be considered key genes in VSCC (Table 2).
Table 2.
KEGG Pathway Analysis of 18 Core Genes in VSCC.
| Pathway ID | Term | Number of genes | % | P-value | Genes |
|---|---|---|---|---|---|
| hsa05200 | Pathways in cancer | 4 | 17.6 | 0.030 | VEGFA,IGF1, BIRC5,MMP1 |
| hsa04114 | Oocyte meiosis | 3 | 13.2 | 0.022 | CCNB2,IGF1, AURKA |
| hsa04914 | Progesterone-mediated oocyte maturation | 3 | 13.2 | 0.014 | CCNB2,IGF1, CCNA2 |
Abbreviation: KEGG, Kyoto Encyclopaedia of Genes and Genomes.
Validation of VEGF, IGF1, BIRC5, and MMP1 Expression With Immunohistochemical Assay in Normal Vulvar Tissues Versus Vulvar Cancer Samples
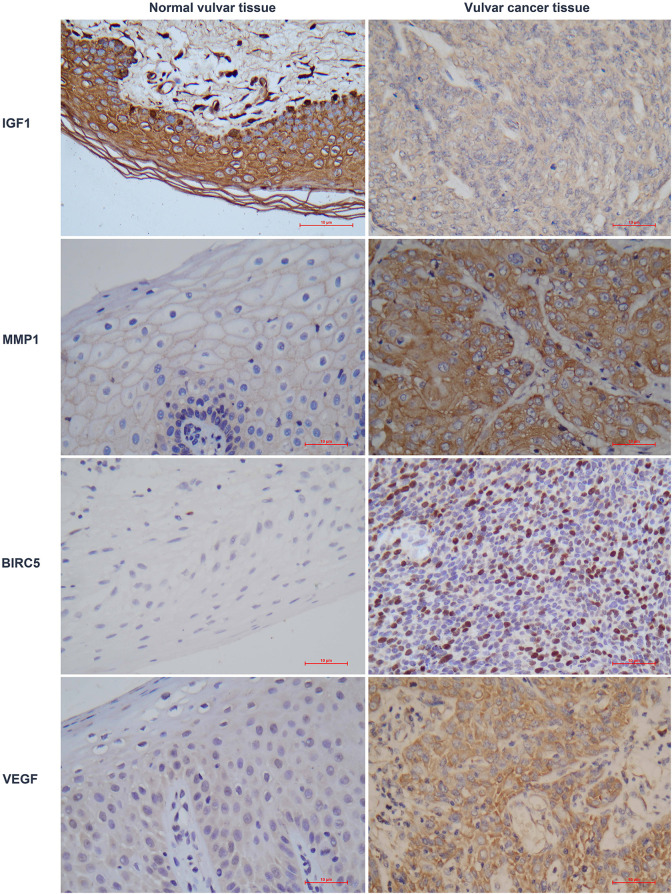
IHC assays of 55 VSCC and 50 normal vulvar tissue sections were conducted to validate the expression levels of VEGF, IGF1, BIRC5, and MMP1 (Table 3). The VEGF, IGF1 and MMP1 proteins were mainly located in the cytoplasm, while BIRC5 was mainly located in the nucleus. The IHC analysis showed consistent results with the bioinformatic analysis (Figure 3).
Table 3.
Protein Expression in Different Vulvar Tissue.a
| Genes | Sample types | Number of samples | Number of samples | P value | |
|---|---|---|---|---|---|
| Low expression | High expression | ||||
| IGF1 | vulvar cancer | 55 | 16 | 39 | 0.000 |
| normal vulvar | 50 | 0 | 50 | ||
| MMP1 | vulvar cancer | 55 | 6 | 49 | 0.000 |
| normal vulvar | 50 | 50 | 0 | ||
| BIRC5 | vulvar cancer | 55 | 20 | 35 | 0.000 |
| normal vulvar | 50 | 50 | 0 | ||
| VEGF | vulvar cancer | 55 | 7 | 48 | 0.000 |
| normal vulvar | 50 | 50 | 0 | ||
aFisher Exact test, normal vulvar versus vulvar cancer. A P value less than 0.05 was considered significant.
Figure 3.
Validation of VEGF, IGF1, BIRC5, and MMP1 with immunohistochemical assay of normal vulvar tissues versus vulvar cancer tissues.
Relationships Between IGF1, BIRC5, VEGF, MMP1 Expression Levels and Patients’ Clinicopathological Characteristics
Clinicopathological data of 55 VSCC patients were collected including age, tumor dimension, tumor differentiation, clinical stage and menopausal duration. The 55 VSCC patients were divided into a high expression group and a low expression group according to the median value of the corresponding protein expression level (Table 4). The IGF1 expression status was not correlated with age (P = 0.662), tumor dimension (P = 0.575), clinical stage (P = 0.86) or menopausal duration (P = 0.141). However, tumor differentiation (P = 0.017) was significantly associated with IGF1 expression, and well-differentiated carcinomas frequently presented higher expression of IGF1. BIRC5 expression was not related to age (P = 0.147), tumor dimension (P = 0.182) or tumor differentiation (P = 0.309). However, BIRC5 expression levels showed a positive correlation with clinical stage (P = 0.039) and menopause duration (P = 0.003). VEGF and MMP1 expression levels were not correlated with any of the tested clinicopathological characteristics.
Table 4.
Association Between the Expression of IGF1, BIRC5, VEGF, and MMP1 and Clinicopathological Characteristics.a
| Patients characteristics | No. of patients | Samples of IGF1 expression | Samples of BIRC5 expression | Samples of VEGF expression | Samples of MMP1 expression | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| High level | Low level | P | High level | Low level | P | High level | Low level | P | High level | Low level | P | ||
| Age group (years) | 0.662 | 0.147 | 0.508 | 0.452 | |||||||||
| <50 | 5 | 3 | 2 | 5 | 0 | 4 | 1 | 4 | 1 | ||||
| ≥50 | 50 | 36 | 14 | 30 | 20 | 44 | 6 | 45 | 5 | ||||
| Tumor dimension (cm) | 1.60 ± 1.23 | 1.91 ± 1.45 | 0.575 | 1.91 ± 1.38 | 1.30 ± 1.04 | 0.182 | 1.76 ± 1.27 | 1.19 ± 1.38 | 0.376 | 1.70 ± 1.30 | 1.62 ± 1.35 | 0.771 | |
| Tumor differentiation | 0.017* | 0.309 | 0.774 | 0.101 | |||||||||
| Well-differentiated | 37 | 29 | 8 | 21 | 16 | 31 | 6 | 35 | 2 | ||||
| Moderately- differentiated | 15 | 10 | 5 | 11 | 4 | 14 | 1 | 12 | 3 | ||||
| Poorly-differentiated | 3 | 0 | 3 | 3 | 0 | 3 | 0 | 2 | 1 | ||||
| Clinical stage | 0.86 | 0.039* | 0.776 | 1 | |||||||||
| Ⅰ | 45 | 32 | 13 | 26 | 19 | 39 | 6 | 39 | 6 | ||||
| Ⅱ | 5 | 4 | 1 | 5 | 0 | 4 | 1 | 5 | 0 | ||||
| Ⅲ | 5 | 3 | 2 | 5 | 0 | 5 | 0 | 5 | 0 | ||||
| Menopause duration (years) | 0.141 | 0.003* | 0.265 | 0.069 | |||||||||
| 0 | 4 | 1 | 3 | 4 | 0 | 4 | 0 | 2 | 2 | ||||
| <10 | 9 | 7 | 2 | 9 | 0 | 8 | 1 | 9 | 0 | ||||
| ≥10 | 42 | 31 | 11 | 22 | 20 | 36 | 6 | 38 | 4 | ||||
aFisher Exact test. A P value less than 0.05 was considered significant.
Discussion
In this study, we first utilized bioinformatic methods to identify differentially expressed genes (DEGs) in VSCC patients. Next, we investigated the clinical significance and prognostic value of IGF1, BIRC5, VEGF, and MMP1 in VSCC. The results showed a significant upregulation of BIRC5, VEGF, and MMP1 expression and a distinct downregulation of IGF1 expression in VSCC compared to normal vulvar tissues.
Kaplan Meier Plotter was utilized to determine the prognostic values of IGF1, BIRC5, VEGF, and MMP1, which showed that increased IGF1 or BIRC5 expression correlated with better survival, while increased VEGF or MMP1 expression correlated with poorer survival in cervical carcinoma patients (Supplementary Material). Previous studies have consistently reported14-18 that VEGF, MMP1 and BIRC5 gene expression is up-regulated and IGF1 expression is down-regulated in both cervical and vulvar tumors. We also performed follow-up and survival analyses, which showed that the 4 genes had statistically insignificant prognostic value in VSCC, which may be due to the insufficient sample size and the short follow-up time of some cases. These results are presented in the Supplementary Material.
In addition, the IGF1 expression status displayed in clinical samples was significantly related to tumor differentiation; well-differentiated carcinomas frequently presented a higher expression of IGF1. On the other hand, lower BIRC5 expression was often shown in clinical stage I patients.
Studies investigating the carcinogenesis of VSCC have been conducted for decades. However, there are few studies about biomarkers that could help tailor conventional treatment and follow-up. Kalliopi5 systematically investigated the expression profile of VSCC for the first time using the microarray technology and delineated the molecular parameters to reveal the cellular pathways involved in the pathogenesis of VSCC. Recently, second harmonic generation (SHG) microscopy has been used for the analysis of collagen fibers in VSCC and preneoplastic lesions and an evident decrease in the values of collagen fiber parameters in the VSCC was discovered by Leuridan et al. 19
Previous research,6 which mainly focused on genomic aberration patterns and expression profiles of VSCC, illustrated a concordant relationship between the imbalances scored by aCGH and the level of gene expression found by microarray. Genes located in gained regions were generally overexpressed while those located in lost regions were downregulated. In addition, the GSE63678 dataset contributor7 revealed common specific pathogenetic patterns and gene markers for cervical (CC), endometrial (EC) and vulvar cancer (VC). We further investigated the possible pathogenesis of vulvar cancer and verified the results with clinical samples based on the aforementioned meaningful explorations. The relationship between IGF1, BIRC5, VEGF, and MMP1 expression levels and patient clinicopathological characteristics were analyzed in our study, which would be informative for mining new biomarkers involved in the carcinogenesis of VSCC.
Some studies have shown that VEGF plays a substantial role in driving the expansion of the tumor vascular bed.20 One study reported that the median serum VEGF concentration in 41 patients affected by VSCC was higher than that in 130 controls,21 which is consistent with the present study. Other studies reported a significant correlation between microvessel density (MVD), VEGF IHC staining, and poorer OS in vulvar cancer patients.22 Subsequently, Hantschmann23 described a high MVD in 29% of VSCCs and a correlation with TGF-α expression, outlining its role in promoting angiogenesis; tumors with both features tended toward having worse DFS, although this was not statistically significant. Not only a high vessel number but also increased vessel size and other vessel characteristics (shape and staining intensity) appeared to be related to prognosis.24
Insulin-like growth factor-1 (IGF1) is a pivotal regulator of normal tissue growth and development and is involved in the occurrence and progression of various cancers, including breast cancer.25-27 Numerous studies confirmed that IGF1 receptor (IGF1R) is overexpressed in approximately 90% of breast cancer cases and that IGF1R levels are higher in breast cancer cells than in normal breast tissues28; thus, the IGF1 system appears to be a promising therapeutic target.29 In addition, the IGF1 signaling pathway has been implicated in HNSCC development and progression.30 Furthermore, Mark31 demonstrated the presence of an IGF1 regulated VEGF autocrine loop in HNSCC. In oral squamous cell carcinoma (OSCC), Eik Schiegnitz32 reported that serum levels of IGF1 in OSCC patients were significantly lower than those in healthy subjects (P < 0.001), which is consistent this study’s findings for VSCC. Additionally, they demonstrated that OSCC patients with a lower IGF1 serum levels showed a significantly worse survival rate than the high expression group (P = 0.049). Little is known about IGF1 tissue expression in VSCC.
BIRC5 is located at the crossroads of a number of cancer cell signaling networks and its functions are controlled and regulated by many upstream cellular signaling molecules33 that make up the upstream signaling pathways of BIRC5. The upstream molecules34,35 include binding proteins, protein regulators, various enzymes (protease, kinase, phosphatase), transcription factors, miRNAs, transporter and channel proteins, and receptors with or without kinase activity. By using BIRC5 as a target, future translational research related to drug discovery and cancer therapeutics is flourishing.36,37 It has been reported38 that BIRC5 expression increases significantly from normal squamous vulvar epithelia to high-grade classic vulvar intraepithelial neoplasia, and vulvar invasive keratinizing squamous cell carcinoma. In this study, the results that BIRC5 is overexpressed in VSCC confirmed its involvement as an early event in vulvar carcinogenesis.
MMP1 is an interstitial collagenase that pertains to matrix metalloproteinases (MMPs), a family of zinc-dependent proteases that aim at the degradation and proteolytic process of components of the extracellular matrix.39,40 Increasing evidence suggests that abnormal expression of MMP1 is related to the progression of malignant tumors. It was reported that high expression levels of MMP1 have significant prognostic value in bladder cancer,41 prostate carcinoma42 and gastric cancer.43 Furthermore, MMP1 has been shown to function as an oncogene.44-46 High MMP1 expression levels were significantly associated with lymph node metastasis, microvessel density and advanced TNM stage in esophageal cancer and head and neck squamous cell carcinoma (HNSCC). HNSCC of patients in the MMP1 high expression group proved to have worse disease-free survival and overall survival than those in the MMP1 low expression group.47 However, there have been no reports about MMP1 expression in VSCC.
On the other hand, vulvar cancer is an uncommon gynecological malignancy primarily affecting postmenopausal women. Few data are available on hormonal receptor expression in VCSS tissues, but hormonal therapy is considered a valuable field of investigation.48 It is worth noting in the present study, 2 of 55 VSCC patients were menopausal at an early age of 28; moreover, patients in menopause for less than 10 years at the time of diagnosis tended to have higher expression of BIRC5. This suggests that hormone levels may affect VSCC genesis and is worth further study.
Few studies have been reported about these 4 genes in VSCC thus far. Therefore, the current study could provide helpful information and directions for future studies in VSCC. The results demonstrated that these 4 genes could be involved in vulvar carcinogenesis. Subsequent experiments validated that the 4 hub DEGs (VEGF, IGF1, BIRC5, MMP1) participated in the molecular mechanisms of VSCC. The results may provide useful information and direction regarding the potential biomarkers and biological mechanisms of VSCC.
Supplementary Material
Supplemental Material, sj-jpg-1-tct-10.1177_15330338211004922 for Expression Profiles Reveal Involvement of VEGF, IGF1, BIRC5, and MMP1 in Vulvar Carcinogenesis by Tao Zhang, Qin Liu, Minghua Yu, Yibing Lan and Jianghong Zhou in Technology in Cancer Research & Treatment
Supplemental Material, sj-pdf-1-tct-10.1177_15330338211004922 for Expression Profiles Reveal Involvement of VEGF, IGF1, BIRC5, and MMP1 in Vulvar Carcinogenesis by Tao Zhang, Qin Liu, Minghua Yu, Yibing Lan and Jianghong Zhou in Technology in Cancer Research & Treatment
Supplemental Material, sj-tif-1-tct-10.1177_15330338211004922 for Expression Profiles Reveal Involvement of VEGF, IGF1, BIRC5, and MMP1 in Vulvar Carcinogenesis by Tao Zhang, Qin Liu, Minghua Yu, Yibing Lan and Jianghong Zhou in Technology in Cancer Research & Treatment
Abbreviations
DEGs, differentially expressed genes; GO, Gene Ontology; HR, hazard ratio; KEGG, Kyoto Encyclopedia of Genes and Genomes; MCODE, molecular complex detection; MVD, microvessel density; PPI, protein-protein interaction; STRING, Search Tool for the Retrieval of Interacting Genes; VSCC, vulvar squamous cell carcinoma.
Acknowledgments
The authors thank Caiyun Zhou and Wenwen Wang for their immunohistochemical technique and interpretation support.
Authors' Note: Approval was obtained from the Human Ethics Committee of Women’s Hospital, Zhejiang University School of Medicine (No. IRB-20200158-R). Each enrolled patient permitted and signed up informed consent to use her samples and records for scientific research.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by the National Natural Science Foundation of China (No. 81801405) and the Zhejiang Provincial Natural Science Foundation of China (No.LY18H040003).
ORCID iD: Qin Liu, MD  https://orcid.org/0000-0003-1891-2078
https://orcid.org/0000-0003-1891-2078
Supplementary Material: Supplemental material is available online for this article.
References
- 1. Barlow EL, Kang YJ, Hacker NF, Canfell K. Changing trends in vulvar cancer incidence and mortality rates in Australia Since 1982. Int J Gynecol Cancer. 2015;25(9):1683-1689. [DOI] [PubMed] [Google Scholar]
- 2. Kang YJ, Smith M, Barlow E, Coffey K, Hacker N, Canfell K. Vulvar cancer in high-income countries: Increasing burden of disease. Int J Cancer. 2017;141(11):21742186. [DOI] [PubMed] [Google Scholar]
- 3. Chan JK, Sugiyama V, Pham H, et al. Margin distance and other clinico-pathologic prognostic factors in vulvar carcinoma: a multivariate analysis. Gynecol Oncol. 2007;104(3):636-641. [DOI] [PubMed] [Google Scholar]
- 4. Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kalliopi IP, Jasmine JH, George DV, et al. Expression profiling of vulvar carcinoma: clues for deranged extracellular matrix remodeling and effects on multiple signaling pathways combined with discrete patient. Transl Oncol. 2011;4(5):301-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Micci F, Panagopoulos I, Haugom L, et al. Genomic aberration patterns and expression profiles of squamous cell carcinomas of the vulva. Genes Chromosomes Cancer. 2013;52(6):551-563. [DOI] [PubMed] [Google Scholar]
- 7. Pappa KI, Polyzos A, Jacob-Hirsch J, et al. Profiling of discrete gynecological cancers reveals novel transcriptional modules and common features shared by other cancer types and embryonic stem cells. Plos One. 2015;10(11):e142229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Davis S, Meltzer PS. GEO query, a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23(14):1846-1847. [DOI] [PubMed] [Google Scholar]
- 9. Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(D1):447-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498-2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25(1):25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44-57. [DOI] [PubMed] [Google Scholar]
- 14. Zhao Q, He Y, Wang XL, Zhang YX, Wu YM. Differentially expressed proteins among normal cervix, cervical intraepithelial neoplasia and cervical squamous cell carcinoma. Clin Transl Oncol. 2015;17(8):620-631. [DOI] [PubMed] [Google Scholar]
- 15. Liu Y, Yi Y, Wu W, Wu K, Zhang W. Bioinformatics prediction and analysis of hub genes and pathways of three types of gynecological cancer. Oncol Lett. 2019;18(1):617-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu X, Peng L, Zhang Y, et al. Identification of key genes and pathways in cervical cancer by bioinformatics analysis. Int J Med Sci. 2019;16(6):800-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhao M, Huang W, Zou S, Shen Q, Zhu X. A five-genes-based prognostic signature for cervical cancer overall survival prediction. Int J Genomics. 2020;2020:8347639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang J, Zheng H, Han Y, Wang G, Li Y. A novel four-gene prognostic signature as a risk biomarker in cervical cancer. Int J Genomics. 2020;2020:4535820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Maria G, Leuridan CT, José V, Natal RA, Vassallo J. Study on collagen parameters in vulvar cancer and preneoplastic lesions by second harmonic generation microscopy. Sci Rep. 2020;10(1):5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Claesson-Welsh L, Welsh M. VEGF and tumour angiogenesis. J Intern Med. 2013;273(2):114-127. [DOI] [PubMed] [Google Scholar]
- 21. Hefler L, Tempfer C, Obermair A, et al. Serum concentrations of vascular endothelial growth factor in vulvar cancer. Clin Cancer Res. 1999;5(10):2806-2809. [PubMed] [Google Scholar]
- 22. Obermair A, Kohlberger P, Bancher-Todesca D, et al. Influence of microvessel density and vascular permeability factor/vascular endothelial growth factor expression on prognosis in vulvar cancer. Gynecol Oncol. 1996;63(2):204-209. [DOI] [PubMed] [Google Scholar]
- 23. Hantschmann P, Jeschke U, Friese K. TGF-alpha, c-erbB-2 expression and neoangiogenesis in vulvar squamous cell carcinoma. Anticancer Res. 2005;25(3A):1731-1737. [PubMed] [Google Scholar]
- 24. Näyhä VV, Stenbäck FG. Increased angiogenesis is associated with poor prognosis of squamous cell carcinoma of the vulva. Acta Obstet Gynecol Scand 2007;86(11):1392-1397. [DOI] [PubMed] [Google Scholar]
- 25. Weroha SJ, Haluska P. The insulin-like growth factor system in cancer. Endocrinol Metab Clin N Am. 2012;41(2):335-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Karamouzis MV, Papavassiliou AG. Targeting insulin-like growth factor in breast cancer therapeutics. Crit Rev Oncol Hematol. 2012;84(1):8-17. [DOI] [PubMed] [Google Scholar]
- 27. Hormones E, Key TJ, Appleby PN, Reeves GK, Roddam AW. Insulin-like growth factor 1 (IGF1), IGF binding protein 3 (IGFBP3), and breast cancer risk: pooled individual data analysis of 17 prospective studies. Lancet Oncol. 2010;11(6):530-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nielsen TO, Andrews HN, Cheang M, et al. Expression of the insulin-like growth factor I receptor and urokinase plasminogen activator in breast cancer is associated with poor survival: potential for intervention with 17-allylamino geldanamycin. Cancer Res. 2004;64(1):286-291. [DOI] [PubMed] [Google Scholar]
- 29. Mauro DS, Annibalini G, Barbieri E, et al. Human IGF1 pro-forms induce breast cancer cell proliferation via the IGF1 receptor. Cell Oncol. 2016;39(2):149-159. [DOI] [PubMed] [Google Scholar]
- 30. Valentina F, Claudia F, Elena G, et al. ΔNp63 promotes IGF1 signalling through IRS1 in squamous cell carcinoma. Aging (Albany NY). 2018;10(12):4224-4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mark GS, Leigh AB, Megan MK, Day TA, Rosenzweig SA. IGF-1 induced vascular endothelial growth factor secretion in head and neck squamous cell carcinoma. Biochem Biophys Res Commun. 2006;342(3):851-858. [DOI] [PubMed] [Google Scholar]
- 32. Schiegnitz E, Kämmerer PW, Schön H, et al. The matrix metalloproteinase and insulin-like growth factor system in oral cancer—a prospective clinical study. Onco Targets Ther. 2017;10:5099-5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li F, Aljahdali I, Ling X. Cancer therapeutics using BIRC5 as a target: what can we do after over two decades of study? J Exp Clin Canc Res. 2019;38(1):368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wheatley SP, Altieri DC. Survivin at a glance. J Cell Sci. 2019;132(7):jcs223826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li F. Discovery of survivin inhibitors and beyond: FL118 as a proof of concept. Int Rev Cell Mol Biol. 2013;305:217-252. [DOI] [PubMed] [Google Scholar]
- 36. Suzuki S, Yamamoto M, Sanomachi T, et al. Brexpiprazole, a serotonin-dopamine activity modulator, can sensitize glioma stem cells to osimertinib, a third-generation EGFR-TKI, via BIRC5 reduction. Cancers (Basel). 2019;11(7):947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Plescia J, Salz W, Xia F, et al. Rational design of shepherdin, a novel anticancer agent. Cancer Cell. 2005;7(5):457-468. [DOI] [PubMed] [Google Scholar]
- 38. Hermann B, Susanne H, Andreas B. Immunohistochemical expression of survivin and γ-H2AX in vulvar intraepithelial neoplasia and low-stage squamous cell carcinoma. Int J Gynecol Pathol. 2011;30(6):583-590. [DOI] [PubMed] [Google Scholar]
- 39. Liu M, Hu Y, Zhang MF. MMP1 promotes tumor growth and metastasis in esophageal squamous cell carcinoma. Cancer Lett. 2016;377(1):97-104. [DOI] [PubMed] [Google Scholar]
- 40. Roy R, Yang J, Moses MA. Matrix metalloproteinases as novel biomarkers and potential therapeutic targets in human cancer. J Clin Oncol. 2009;27(31):5287-5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shin DH, Dier U, Melendez JA, Hempel N. Regulation of MMP-1 expression in response to hypoxia is dependent on the intracellular redox status of metastatic bladder cancer cells. Biochim Biophys Acta. 2015;1852(12):2593-2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ozden F, Saygin C, Uzunaslan D. Expression of MMP-1, MMP-9 and TIMP-2 in prostate carcinoma and their influence on prognosis and survival. J Cancer Res Clin Oncol. 2013;139(8):1373-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cai QW, Li J, Li XQ, Wang JQ, Huang Y. Expression of STAT3, MMP-1 and TIMP-1 in gastric cancer and correlation with pathological features. Mol Med Rep. 2012;5(6):1438-1442. [DOI] [PubMed] [Google Scholar]
- 44. Juncker-Jensen A, Deryugina EI, Rimann I, et al. Tumor MMP-1 activates endothelial PAR1 to facilitate vascular intravasation and metastatic dissemination. Cancer Res. 2013;73(14):4196-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hu F, Zhang Y, Li M, et al. BMP-6 inhibits the metastasis of MDA-MB-231 breast cancer cells by regulating MMP-1 expression. Oncol Rep. 2016;35(3):1823-1830. [DOI] [PubMed] [Google Scholar]
- 46. Anand M, Van Meter TE, Fillmore HL. Epidermal growth factor induces matrix metalloproteinase-1 (MMP-1) expression and invasion in glioma cell lines via the MAPK pathway. J Neurooncol. 2011;104(3):679-687. [DOI] [PubMed] [Google Scholar]
- 47. Liu LQ, Chen ZW, Yi SJ. Association between the expression of MMP1 gene and prognosis in head and neck squamous cell carcinoma. Lin Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2018;32(4):287-291. [DOI] [PubMed] [Google Scholar]
- 48. Giulia M, Simona MF, Frediano I, et al. Molecular pathways in vulvar squamous cell carcinoma: implications for target therapeutic strategies. J Cancer Res Clin Oncol. 2020; 146(7):1647-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, sj-jpg-1-tct-10.1177_15330338211004922 for Expression Profiles Reveal Involvement of VEGF, IGF1, BIRC5, and MMP1 in Vulvar Carcinogenesis by Tao Zhang, Qin Liu, Minghua Yu, Yibing Lan and Jianghong Zhou in Technology in Cancer Research & Treatment
Supplemental Material, sj-pdf-1-tct-10.1177_15330338211004922 for Expression Profiles Reveal Involvement of VEGF, IGF1, BIRC5, and MMP1 in Vulvar Carcinogenesis by Tao Zhang, Qin Liu, Minghua Yu, Yibing Lan and Jianghong Zhou in Technology in Cancer Research & Treatment
Supplemental Material, sj-tif-1-tct-10.1177_15330338211004922 for Expression Profiles Reveal Involvement of VEGF, IGF1, BIRC5, and MMP1 in Vulvar Carcinogenesis by Tao Zhang, Qin Liu, Minghua Yu, Yibing Lan and Jianghong Zhou in Technology in Cancer Research & Treatment