Summary
Cellular reprogramming is driven by a defined set of transcription factors; however, the regulatory logic that underlies cell-type specification and diversification remains elusive. Single-cell RNA-seq provides unprecedented coverage to measure dynamic molecular changes at the single-cell resolution. Here, we multiplex and ectopically express 20 pro-neuronal transcription factors in human dermal fibroblasts and demonstrate a widespread diversification of neurons based on cell morphology and canonical neuronal marker expressions. Single-cell RNA-seq analysis reveals diverse and distinct neuronal subtypes, including reprogramming processes that strongly correlate with the developing brain. Gene mapping of 20 exogenous pro-neuronal transcription factors further unveiled key determinants responsible for neuronal lineage specification and a regulatory logic dictating neuronal diversification, including glutamatergic and cholinergic neurons. The multiplex scRNA-seq approach is a robust and scalable approach to elucidate lineage and cellular specification across various biological systems.
Keywords: single-cell multiplexing genomics, direct cell reprogramming, neuronal reprogramming
Highlights
-
•
Multiplexed scRNA-seq approach reveals combinations of genes to induce neuronal diversification
-
•
Neuronal diversification is deterministic early in the reprogramming process
-
•
PAX6 drives induced neurons away from fibroblasts
In this article Shin and colleagues combine single-cell RNA-seq with multiplexed induction of pro-neuronal genes to reveal novel gene combinations responsible to induce neuronal diversification. The study reveals that neuronal diversification is deterministic with a distinct set of transcription factors as early as 9 days.
Introduction
The brain consists of a wide range of neuronal subtypes. Understanding the mechanism that underlies neuronal diversification is critical to the study of brain development and neurodegenerative disease. The recent discoveries that fully differentiated somatic cells can be reprogrammed into alternative cell fates opened up exciting avenues to study cellular specification and regenerative medicine (Chanda et al., 2014; Liu et al., 2012; Smith et al., 2016; Takahashi et al., 2007; Vierbuchen et al., 2010; Shin et al., 2012). Despite the rapid development of cell reprogramming, the possibility to identify the gene regulatory logic that underlies neuronal reprogramming has remained elusive, which has limited our understanding of the cellular plasticity of stem cells and cellular diversification during development and reprogramming.
Several studies have aimed to identify neurogenic transcription factors (TFs) that allow the reprogramming of major neuronal subtypes, where distinct combinations of TFs have been shown to create dopaminergic neurons and cholinergic motor neurons, which are selective targets of degeneration in patients with Parkinson disease and amyotrophic lateral sclerosis, respectively (An et al., 2016; Caiazzo et al., 2011; Kim et al., 2011; Liu et al., 2013; Mazzoni et al., 2013; Pfisterer et al., 2011; Son et al., 2011). Moreover, in silico predictive models (Rackham et al., 2016) and combinatorial screens (Chen et al., 2015; Liu et al., 2018) provide high-throughput approaches to identify various combination of TFs involved in cell reprogramming. However, these approaches often require complex and costly experimental setups, which is further exacerbated by the time and labor-intensive retesting and validation of newly identified candidates.
Multiplexing offers a more efficient and scalable strategy, where a collection of genes can be perturbed either by small hairpin RNA or CRISPR in a single experiment. This is followed by a phenotypic selection and targeted DNA sequencing to reveal candidate genes that attribute to a pre-defined cellular phenotype (Chen et al., 2015; Hsu et al., 2014; Liu et al., 2018; Shalem et al., 2015). While the approach is highly scalable, it relies on a limited number of phenotypic readouts or requires specific markers to enrich for target cell populations, which are often not available or limited to a single cell type. The advancement in single-cell RNA sequencing (scRNA-seq) has opened up new opportunities to profile gene expression profiles of heterogeneous tissues at the single-cell resolution without the need for cell selection (Adamson et al., 2016; Dixit et al., 2016; Nowakowski et al., 2017; Picelli et al., 2014). Moreover, adaptation of scRNA-seq with CRISPR/sgRNA led to reconstruction of regulatory networks involved in immune activation (Shifrut et al., 2018), cholesterol biogenesis (Replogle et al., 2020), and zygotic genome activation (Alda-Catalinas et al., 2020). However, how TFs work in concert to drive neuronal diversification and profiling scRNA-seq with multiplexed TFs remain unexplored.
To address these challenges, we introduce a strategy that combines multiplexing ectopic expression of full-length TFs with scRNA-seq to retrospectively assign reprogrammed neurons with exogenous and endogenous expression profiles. We identify key determinants and regulatory processes governing the classification of neuronal subtypes in a single experiment. Single-cell RNA-seq of induced neurons generated by the multiplexed transduction of 20 pro-neuronal TFs reveals unique combinations of key TFs and genetic programs orchestrating the plasticity of human fibroblasts into various neuronal subtypes, including glutamatergic and cholinergic neurons. We demonstrate that the method efficiently gains deep insights into biological processes governing cell plasticity and cell fate decisions, and to systematically dissect regulatory processes controlling neuronal diversification during cell reprogramming.
Results
Inducing a Heterogeneous Population of Human Neuronal Subtypes
We established a system to identify TFs governing direct cell reprogramming toward diverse neuronal subtypes by infecting multiplexed TFs followed by scRNA-seq (Figure 1A). We first generated a pool of lentiviruses encoding TFs previously implicated in neuronal reprogramming, either as pioneering factors that can access closed chromatin (ASCL1, NEUROG2) (Smith et al., 2016; Wapinski et al., 2013), as factors that increase the efficiency of reprogramming (POU3F2, ZIC1, OLIG2, NEUROD1) (Pang et al., 2011; Vierbuchen et al., 2010) or as factors that convert human fibroblasts into GABAergic (DLX1, DLX2) (Victor et al., 2014), cholinergic (ISL1) (Son et al., 2011), serotonergic (FEV) (Xu et al., 2016), or dopaminergic (FOXA2, NR4A2, PITX3) (Liu et al., 2012; Pfisterer et al., 2011) neurons when co-expressed with other TFs (Figure S1A). These 13 TFs were also upregulated during human induced pluripotent stem cell (iPSC) differentiation into neuronal progenitor cells (NPCs) (Figure S1A). We also selected seven TFs that were upregulated during the differentiation but were not previously implicated in direct neuronal reprogramming. Altogether 20 TFs, hereafter termed the "TF-pool," were individually tuned for multiplicity of infection (MOI) to allow for ~85% of fibroblasts to express mixed combinations of 2–6 TFs after pooled infection (Figures S1B and S1C).
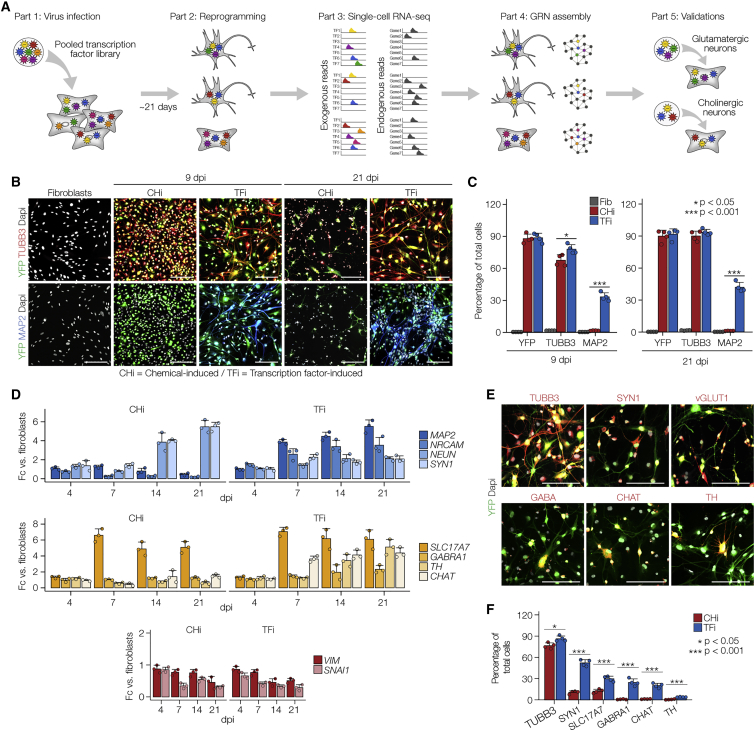
Figure 1.
Generation of a Heterogeneous Population of Human Induced Neurons
(A) Overview of the single-cell TF multiplex pipeline.
(B) Immunostaining for canonical neuronal marker genes of fibroblasts at day 0 and CHi and TFi at 9 and 21 dpi. Scale bars, 100 μm. YFP (green) marks infected cells and cell nuclei were visualized using DAPI nuclear stain (gray).
(C) Quantification of immunostainings in (B). n = 4 independent experiments, unpaired Student's t test. Error bars represent mean + SD.
(D) qPCR for pan-neuronal marker genes (MAP2, NRCAM, NEUN, SYN1; top), canonical neuronal subtype markers (SLC17A7, GABRA1, TH, CHAT; middle), and canonical fibroblast markers (VIM, SNAI1; bottom). n = 3 independent experiments, unpaired Student's t test. Error bars represent mean + SD.
(E) Immunostainings for canonical neuronal subtype markers (red) of TFi at 21 dpi. Scale bars, 100 μm. YFP (green) marks infected cells and cell nuclei were visualized using DAPI nuclear stain (gray).
(F) Quantification of immunostainings in (E). n = 4 independent experiments, unpaired Student's t test. Error bars represent mean + SD.
Transduction of the TF-pool in human dermal fibroblasts followed by a serial application of neuronal induction medium for 14 days and neuronal maturation medium for the subsequent 7 days led to heterogeneous neuron-like morphologies, exhibiting multiple elongated dendrites from the cell body and, in some cases, star shape-like features mimicking astrocytes (Figure 1B). At 9 days post-infection (dpi), 78.6% of TF-pool-induced (TFi) fibroblasts stained positive for the immature neuronal marker TUBB3 and 34.7% expressed MAP2, a microtubule-associated protein expressed specifically in neurons (Gelles et al., 1988) (Figures 1B and 1C). By 21 dpi, TUBB3+ and MAP2+ populations increased to 93.5% and 42.4%, respectively, indicating progressive transdifferentiation toward the neuronal lineage. Consistent with TUBB3 and MAP2 protein expressions, qPCR revealed upregulation of neuronal marker transcripts (MAP2, NRCAM, NEUN, SYN1) and neuronal subtype marker transcripts (SLC17A7 [glutamatergic neurons], GABRA1 [GABAergic neurons], TH [dopaminergic neurons], and CHAT [cholinergic neurons]) as well as significant downregulation of fibroblast marker expression (VIM, SNAI1) starting at 7 dpi (Figure 1D).
Previous studies have shown that mouse and human fibroblasts can be directly converted to induced neurons solely by chemical cocktails of small molecules (Hu et al., 2015; Li et al., 2015). We also found that application of neuronal induction medium to fibroblasts (hereafter termed chemical-induced [Chi] cells) generated TUBB3+ neurites, upregulated the RNA expression of NEUN, SYN1, SLC17A7, and TUBB3, and downregulated the expression of VIM and SNAI1 (Figures 1B–1D). However, CHi failed to express MAP2 at any time point analyzed, indicating that small molecules/growth factors, in the absence of key TFs, were insufficient to elicit neuronal maturation in human dermal fibroblasts. Consistently, neuronal complexity, measured as number of branch points and average neurite length, was significantly lower at 9 dpi (8.5- and 1.8-fold, respectively) and at 21 dpi (5.6- and 4.8-fold, respectively) when fibroblasts were treated with the neuronal induction medium alone compared with TF-pool induction (Figures S1E and S1F). Immunofluorescence at 21 dpi indicated that TFi exhibited a heterogeneous population of cells expressing glutamatergic (VGlut1+; ~29%), GABAergic (GABA+; ~23%), cholinergic (CHAT+; ~19%), and dopaminergic (TH+; ~4%) subtype-specific genes (Figures 1E and 1F). In contrast, only ~15% of CHi expressed VGlut1 at 21 dpi and none of the other subtype-specific genes were induced by small molecules alone. Together, pooled overexpression of pro-neuronal TFs generated a heterogeneous population of both immature and mature neuronal subtypes, while small molecules partially reprogrammed fibroblasts toward the glutamatergic fate at low efficiency.
Molecular Characterization of Induced Neurons Using scRNA-Seq
To gain further insights into the cellular heterogeneity of TFi and CHi fibroblasts, we used droplet-based massively parallel scRNA-seq (Zheng et al., 2017) to profile 2,092 CHi and 1,900 TFi at 14 dpi. Uniform Manifold Approximation and Projection (UMAP) (Mcinnes et al., 2018) distinctively separated CHi and TFi fibroblasts along the first dimension, UMAP1 (Figures 2A and S2A–S2D), and revealed activation of neuronal differentiation genes (NRCAM, STMN2, SST, DKK3) and synapse formation genes (SYT1, SERPINI1, SYNGR1, SYNPO2), which was accompanied by a decrease in fibroblast-specific genes (SNAI2, THY1) (Figure 2A). Other suppressed genes included extracellular matrix genes (COL3A1, COL15A1, EDIL3, HAPLN1, CPXM1, MFAP4), reflective of the morphological changes that occur during the cell transformation toward the neurons. UMAP analysis further partitioned TFi cells into several transcriptionally distinct clusters (clusters 4–9 [CL4–CL9]). Cells in CL4 dominantly expressed cell-cycle-related genes, including MIKI67 and TOP2A, and showed enrichment of gene ontology (GO) terms related to cell division, suggesting that CL4 cells did not successfully exit the cell cycle and failed to initiate the reprogramming process (Table S1). Interestingly, CL5–CL9 showed enrichment of genes and GO terms related to nervous system development and neurogenesis (Figures 2B, 2C, S2E, and S2F).
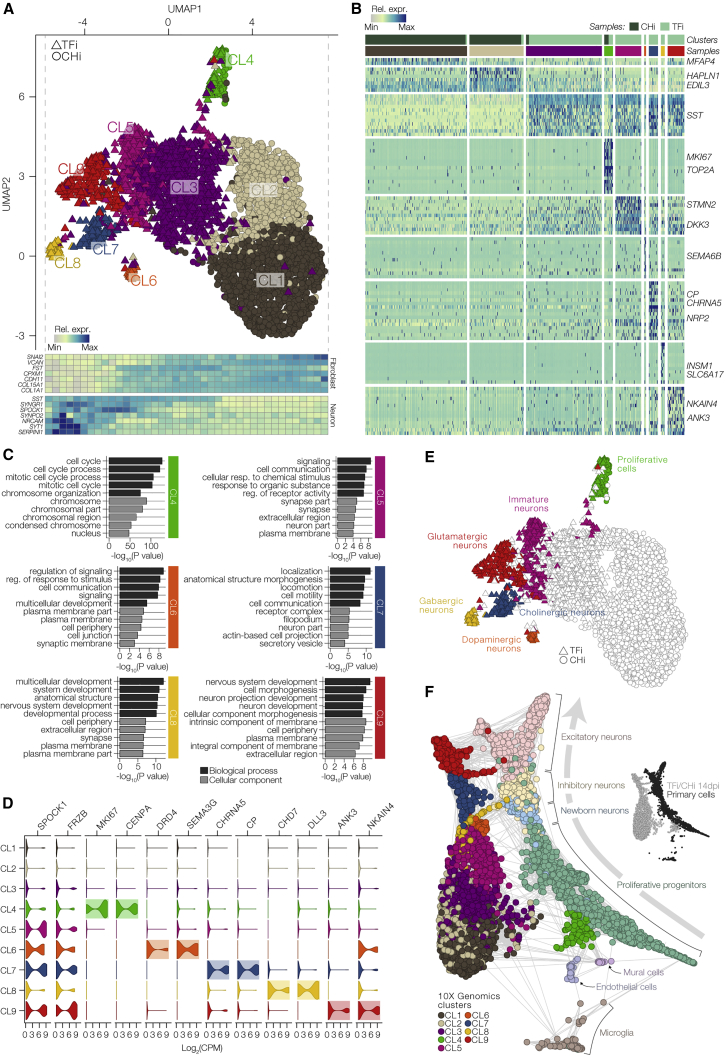
Figure 2.
Molecular Characterization of Induced Neurons using scRNA-Seq
(A) Top: visualization of droplet-based scRNA-seq data from CHi and TFi at 14 dpi using UMAP (n = 3,865 cells). The detected clusters are indicated by different colors. Bottom: heatmap of the relative expression of canonical fibroblast and neuron markers along UMAP1.
(B) Heatmap of the relative expression of top marker genes for each cluster in the UMAP plot in (A).
(C) GO analysis of cluster-specific marker genes in clusters CL4–CL9. Shown are the top 5 GO terms related to biological process (dark gray) and cellular component (light gray) for each cluster; colors as in (A).
(D) Violin plots of log2-transformed counts per million (CPM) values of marker genes in all clusters.
(E) Annotation of TFi clusters CL4–CL9 based on genes differentially expressed between each cluster.
(F) Relationship of CHi and TFi to primary human brain cells in a force-directed k-nearest neighbors graph created using SPRING. Primary cells are colored in light colors, induced cells are colored by cluster as in (A).
To further annotate the remaining clusters, from CL5 to CL9, we first interrogated top differentially expressed genes (Seurat [Satija et al., 2015]; p < 10−20) for known neuronal subtype marker genes. The analysis revealed CL5 to associate with immature neurons, CL6 with the dopaminergic neuron program (DRD4, SEMA3G), CL7 with the cholinergic neuron program (CHRNA5, CP), CL8 with the GABAergic neuron program (CHD7, DLL3), and CL9 with the glutamatergic neuron program (ANK3, NKAIN4) (Figures 2D, 2E, and S2E). To support these findings, we used a publicly available scRNA-seq dataset of human primary cortical and medial ganglionic eminence brain (Nowakowski et al., 2017) to infer cellular relationships among the cells in a force-directed k-nearest neighbors graph (Weinreb et al., 2018) (Figure 2F). The comparative analysis aligned the primary brain cells along a developmental progression from proliferative progenitor cells, immature neurons, and finally to mature inhibitory and excitatory neurons. Consistent with our previous results, CL4 cells expressing cell-cycle genes positioned near proliferative progenitors and non-neuronal cells, whereas all other clusters positively aligned with primary cells along the same developmental trajectory. Importantly, clusters CL6–CL9 exhibiting neuronal subtype markers aligned along the cellular trajectory with newborn neurons, and mature inhibitory and excitatory neurons. Cluster CL3, which represented almost half of the TF-induced data, showed lower expression of neuronal genes as compared with CL4 and lower expression of fibroblast genes as compared with CL1–CL2. Furthermore, the placement of CL3 in both UMAP and force-directed k-nearest neighbor graphs indicated that these cells were still in transition or partially reprogrammed toward neurons (Figures 2A and 2F). Notably, CHi cells, largely represented by CL1 and CL2, also showed neuronal GO terms, which is consistent with our notion that the chemical cocktail alone upregulated neuronal genes (Figure S2F). However, high expression levels of fibroblast-specific genes, including CHD11 and SNAI2, suggest a failure to suppress the fibroblastic network in the absence of exogenous TF expression. Together, these data revealed that transduction of the TF-pool converted fibroblasts into a heterogeneous population of cells exhibiting distinct neuronal subtype-specific molecular signatures and congruent progression of neuronal development.
Distinguished Detection of Exogenous and Endogenous Transcripts along Developmental Trajectories
Next, to identify which TF in the TF-pool was responsible for inducing distinct neuronal subtype specification, we performed single-cell Smart-seq2 to achieve high gene coverage across the transcripts. We relied on full-length reads to extract nucleotides at the 5′ and 3′ junctions of exogenous open reading frames (ORFs) where distinctive alignment of reads to the specific 5′ and 3′ junctions allows to discriminate between exogenous (ORFs with the attB Gateway cloning sequences) and endogenous (ORFs without attB) gene expression (Figure 3A).
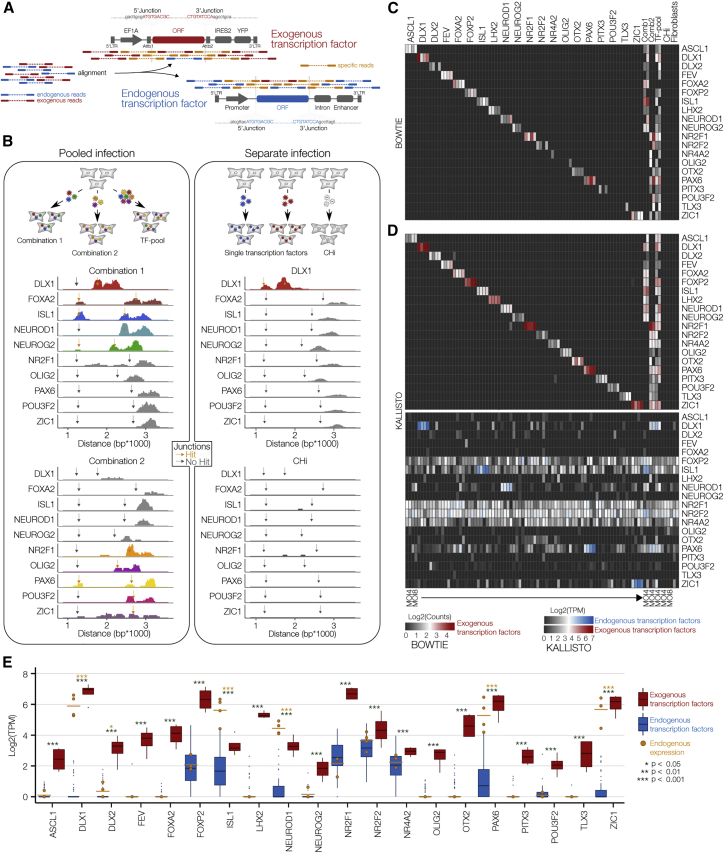
Figure 3.
Distinguished Detection of Exogenous and Endogenous Transcripts
(A) Schematic depicting the strategy to distinguish exogenous and endogenous sequencing reads.
(B) Bulk RNA-seq on pooled and separately infected fibroblasts. Horizontal dimension, distance from the 5ʹ end of the EF1A promoter; vertical dimension, number of aligned paired-end reads. Gray arrows (no overlap) and golden arrows (overlap) mark 5ʹ and 3′ junctions of exogenous ORFs.
(C and D) Heatmaps showing log2-transformed count values of exogenous TFs after alignment using Bowtie (C) and log2-transformed tags per million (TPM) values of exogenous and endogenous TF pairs after trimming junction sequences to ~100 base pairs and alignment using Kallisto (D). For individually infected fibroblasts and CHi, two replicates at an MOI of 4 and two replicates at an MOI of 8 were included. For pooled infected fibroblasts, two replicates at an MOI of 4 were included.
(E) Boxplots showing increased exogenous (red) versus endogenous (blue) expression of all TFs across all individually infected fibroblasts. Golden dots show endogenous expression in samples infected with the corresponding exogenous TFs. Unpaired Student's t test. Error bars represent mean + SD.
First, to benchmark the accuracy and sensitivity of detecting exogenous transcripts, we infected fibroblasts with single TFs, two combinations of 10 TFs, and the complete (20) TF-pool at two MOIs, low and high (Figures 3B and S3B). Sequence alignment to the exogenous transcript model using Bowtie (Langmead et al., 2009) resulted in 24.4% false-negative events, possibly due to the exclusion of multi-mapping reads (Figure 3C). To reduce the number of false-negative events, we implemented the pseudo-alignment tool Kallisto, providing the advantage of assigning multi-mapping reads to transcripts without pinpointing exactly how the sequences of the reads and transcripts align (Bray et al., 2016). This approach reduced the occurrence of false-negative events to 6.3% and yielded highly specific and sensitive detection efficiencies of 97.5% and 87.5% in individual infection and pooled infection samples, respectively (Figures 3D and S3C). Specific detection of lentivirus-mediated expression was further confirmed by revealing significantly higher mean expression of exogenous TFs compared with the mean expression of corresponding endogenous TFs (p < 0.05; Figure 3E).
Subsequently, we performed Smart-seq2 on TFi and CHi cells at an early (9 dpi) and late (21 dpi) time point (Figure 4A). Cells were sequenced at an average depth of ~2.6 million reads and a median of 10,143 genes per cell. We used unsupervised hierarchical clustering and UMAP (Figures S4A–S4D) to reveal that cells clustered largely by type rather than batch and that we could assign individual cells with exogenous TF expression (Figures 4B, 4C, S4E, and S4F). Most notably, 30%–40% of TFi 9 dpi and TFi 21 dpi cells clustered as CL5 (Figure 4B). CL5 showed the highest degree of neuronal reprogramming as indicated by GO terms associated to nervous system development, synaptic transmission, cholinergic regulation of neuron differentiation; CL3 and CL4 showed GO terms related to cytokine-mediated signaling pathway and blood vessel development, respectively (Figure S5A). This suggests that cells acquired a neuronal program fairly early in the reprogramming process and only a fixed number of cells undergo neuronal reprogramming. Based on the TF enrichment in CL5, it is plausible that only the cells that acquired the necessary TF combinations induced neuronal conversion. The exogenous genes detected in day 9 and 21 were similar, whereas the exogenous factors were significantly different between clusters CL3 versus CL5 or CL4 versus CL5. This suggests that cells that acquired a neuronal program, in this case CL5, harbor a distinct set of TF combinations. Indeed, we detected over-representation of PAX6, NEUROG2, POU3F2, ZIC1, and FEV in CL5 as compared with other clusters (Figure S5B). Overall, these data indicate that a distinct set of TF combinations was necessary, as opposed to the presence of pre-determined cells, to initiate cell conversion toward neurons as early as day 9.
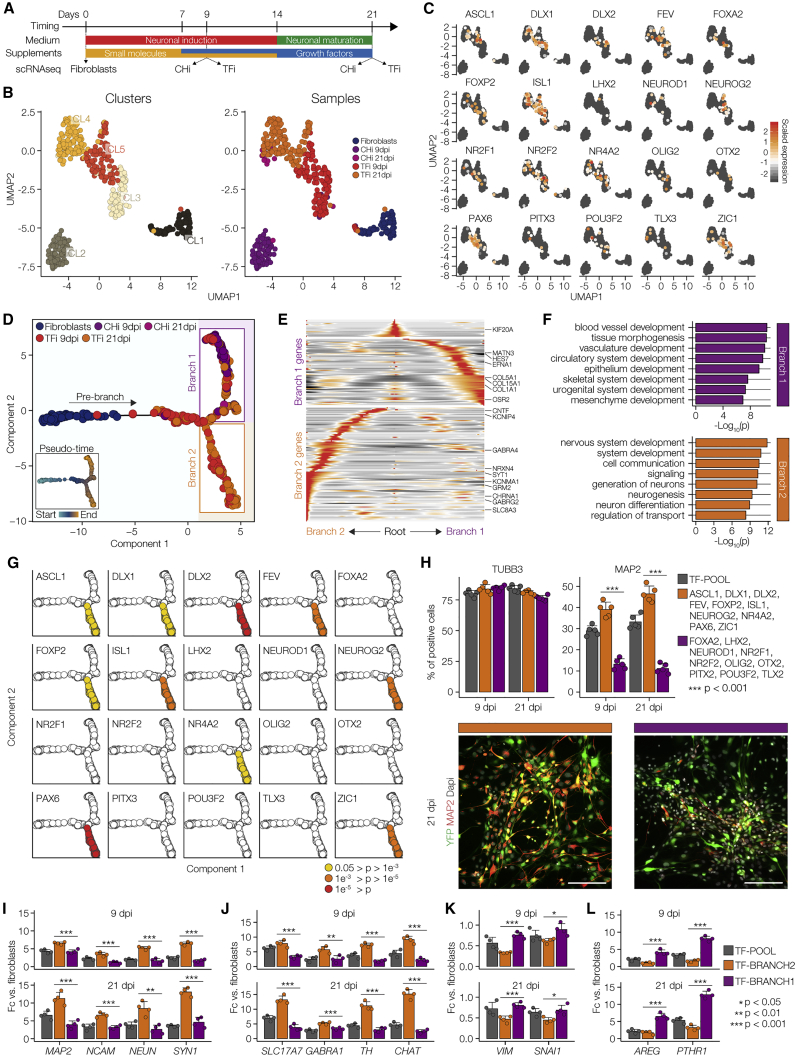
Figure 4.
Association of Developmental Trajectories with Exogenous Expression Profiles
(A) Diagram of the differentiation protocol of TFi and CHi, depicting the samples used for the time course Smart-seq experiment.
(B) UMAP 2D cell maps of the time course data. Left: cells were colored by cluster identity. Right: cells were colored by sample identity. Fibroblasts (n = 78 cells), CHi at 9 dpi (n = 87 cells), TFi at 9 dpi (n = 129), CHi at 21 dpi (n = 15 cells), and TFi at 21 dpi (n = 137 cells).
(C) Visualization of relative expression values of exogenous TFs on UMAP plots.
(D) Pseudo-temporal ordering of the Smart-seq time course based on the expression of 2,925 developmental genes (n = 446 cells). Small inset shows the same plot colored by pseudo-temporal values.
(E) Heatmap showing ~200 genes with branch-specific differential expression as determined by BEAM (R Package “Monocle”). In this heatmap, columns are points in pseudo-time, rows are genes, and the middle (Root) is the beginning of pseudo-time. Branch 1 goes from the middle of the heatmap to the right, while branch 2 goes to the left.
(F) GO analysis of genes differentially expressed in branch 1 (top panel) and branch 2 (bottom panel).
(G) Identification of exogenous TFs with branch-specific enrichment based on Fisher's exact tests.
(H) Top panels: quantification of TUBB3+ and MAP2+ cells in fibroblasts infected with the complete TF-pool (gray), branch 2-enriched TFs (orange), and unenriched TFs (purple) at 9 and 21 dpi. n = 5 independent experiments, unpaired Student's t test. Error bars represent mean + SD. Bottom panels: representative images of immunostainings for MAP2 (red) at 21 dpi; colors as in top panels. YFP (green) marks infected cells and cell nuclei were visualized using DAPI nuclear stain (gray). Scale bars, 100 μm.
(I–L) Neuronal differentiation, loss of fibroblast characteristics, and acquisition of alternative developmental fates as revealed by qPCR for pan-neuronal marker genes (MAP2, NRCAM, NEUN, SYN1) (I), canonical neuronal subtype markers (VGlut1, GABA, TH, CHAT) (J), canonical fibroblast markers (VIM, SNAI2) (K), and branch 1-enriched genes (AREG, PTHR1) (L) at 9 and 21 dpi; colors as in (H). n = 4 independent experiments, unpaired Student's t test. Error bars represent mean +SD.
To investigate the dynamic progression of neuronal reprogramming, we placed the cells in pseudo-temporal order (Trapnell et al., 2014) based on differentially expressed genes between 9 and 21 dpi (qval < 0.01) (Figure S4G). We found that genes along pseudo-time were enriched for GO terms related to several developmental lineages (Figures S6H and S6I; Table S2), including neurogenic genes (NRCAM, SFRP1, SNAP25, and SYT1) and genes regulating the development of mesodermal tissues: bone (BMP4), kidney (FAT4), and endothelial cells (PGF and VEGFA) (Figures S6J–S6L). Because genes involved in cell reprogramming generally overlap with developmental genes (Masserdotti et al., 2016), we reordered the cells only using genes implicated in the developmental processes (~3,000 genes, Figure 4D; Table S3). This secondary pseudo-temporal ordering revealed clear bifurcation of cells into two main trajectories: branch 1 associated with non-neuronal developmental fates (Figures 4E and 4F; Table S4) and branch 2 associated with the neuronal lineage based on gene program (fold-change > 4; p < 0.05).
Mapping the 20 exogenous genes to each branch, we found significant enrichment of 10 TFs in branch 2 (p < 10−2), while no exogenous TF was significantly enriched in branch 1 (Fisher's exact test; p > 0.05) (Figures 4G and S4M).
To confirm whether these 10 TFs derived from branch 2 can induce neuronal phenotype, we independently transduced fibroblasts with the 20 TF-pool, the 10 TFs enriched in branch 2 (neuron branch) and the remaining 10 TFs showing no enrichment in either branch (control). Profiling of neuronal markers revealed that TFs from the neuron branch markedly increased the efficiency of neuronal expression and morphological features as compared with both the complete 20 TF-pool and control TFs (Figures 4H–4K). In fact, infection with control TFs reduced the efficiency of neuronal conversion as compared with the complete TF-pool and induced upregulation of genes that were functionally relevant in breast and kidney, such as AREG and PTHR1, respectively (Figures 4L).
Identification of Novel TF Combinations Controlling Neuronal Subtype Specification
To further interrogate the 10 TFs enriched in the neuron branch, we leveraged the droplet-based scRNA-seq UMAP to annotate the cells derived from the Smart-seq2 time course experiment (Figure 5A). Consistent with our previous results (Figure 2), TFi cells mapped to mature neuronal subtypes, whereas chemically induced cells mapped to clusters containing partially reprogrammed cells and immature neurons (Figures S5A–S5C). The majority of TFi clusters were assigned to either the glutamatergic or cholinergic cluster, suggesting that these two neuronal subtypes prevailed in the Smart-seq2 time course experiment.
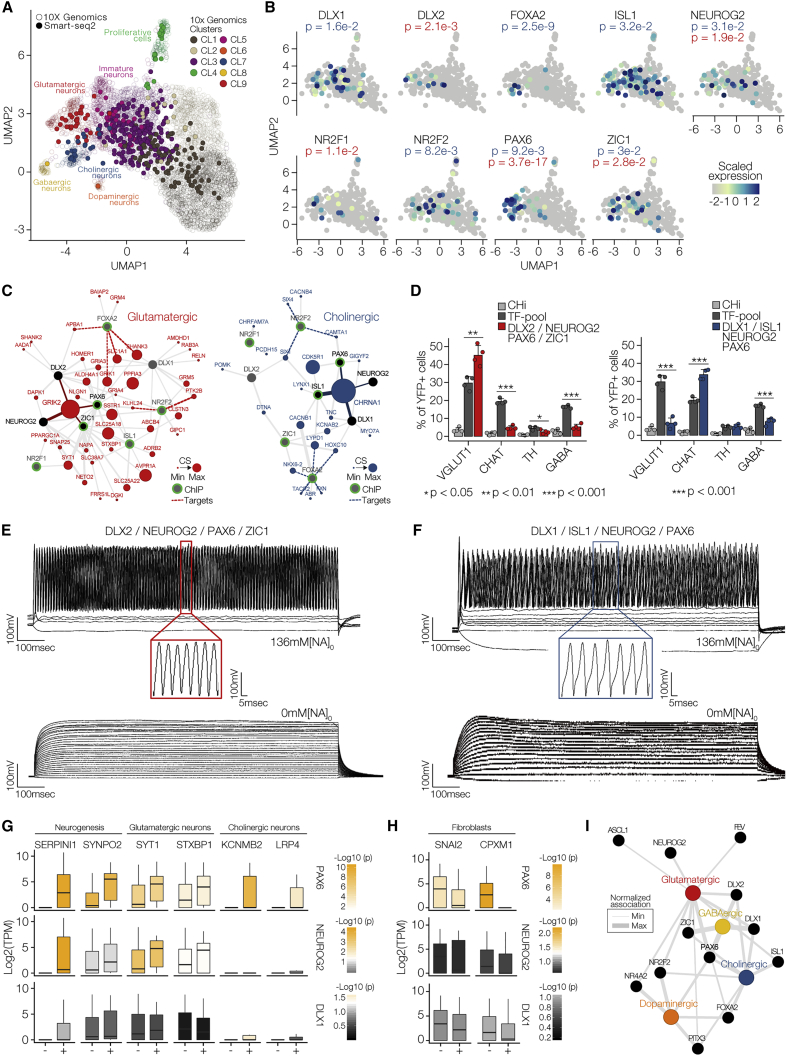
Figure 5.
PAX6 Acts as Master Regulator to Control Reprogramming of Glutamatergic and Cholinergic Neurons
(A) Computational mapping of the Smart-seq time course onto the 10X Genomics UMAP. Smart-seq cells are colored based on 10X Genomics cluster membership and positioned based on the five nearest neighbors.
(B) Visualization of scaled expression values of exogenous TFs that showed significant enrichment (Fisher's exact test, p < 0.05) in glutamatergic and/or cholinergic clusters on 2D UMAPs.
(C) Neuronal subtype-specific co-expression modules on the basis of significant associations (Fisher's exact test, p < 0.05) with exogenous TFs shown in (B). Exogenous TFs associated with genes showing highest CS in each module are shown in black, all other exogenous TFs are shown in gray. Neuronal subtype-specific genes are colored. Direct targets of exogenous TF are indicated with dashed lines based on TF with chromatin immunoprecipitation sequencing (ChIP-seq) evidence, highlighted with green borders.
(D) Validation of novel combinations of exogenous TFs by quantification of immunostainings for VGlut1, CHAT, TH, and GABA of CHi (light gray), fibroblasts infected with the complete TF-pool (dark gray) and fibroblasts infected with novel combinations (color) at 21 dpi. n = 4 independent experiments, unpaired Student's t test. Error bars represent mean + SD.
(E and F) The generation of repetitive action potentials in induced neurons infected with DLX2, NEUROG2, PAX6, ZIC1 (E) or DLX1, ISL1, NEUROG2, or PAX6 (F). Representative traces in the presence (upper panel) or absence (lower panel) of extracellular Na+ were recorded using the current-clamp protocol.
(G and H) Boxplots showing the log2-transformed TPM values of neurogenic and neuronal subtype-specific genes (G) and fibroblast-specific genes (H) in cells with (+) or without (−) exogenous PAX6 (top), NEUROG2 (middle), and DLX1 (bottom). Boxplots are colored based on −log10-transformed p values.
(I) Edge-normalized network summarizing the associations of exogenous TFs with glutamatergic, cholinergic, GABAergic, and dopaminergic modules.
To identify the major determinants controlling glutamatergic and cholinergic reprogramming, we performed co-expression module analysis (Fisher's exact tests; p < 0.05) and subsequently attributed genes in each subtype with combination scores (CSs) (Figure 5C). The CS represents the significance of enrichment of marker gene expression in cells containing at least three of the predicted exogenous TFs. Based on this approach, we identified DLX2, ZIC1, NEUROG2, and PAX6 controlling the reprogramming of glutamatergic-like neurons and DLX1, ISL1, NEUROG2, and PAX6 controlling the reprogramming of cholinergic-like neurons (Figure 5D). The analysis revealed that most gene modules found in the glutamatergic and cholinergic network are indirect targets of exogenous TFs, with few TFs directly linked to marker genes (Figure 5C, Table S5).
Vesicular glutamate transporter 1, VGlut1, mediates glutamate uptake into synaptic vesicles and is a hallmark of glutamatergic neurons (Zhou and Danbolt, 2014). To independently validate whether TFs derived from the gene module analysis can induce the expression of VGlut1, we infected fibroblasts with DLX2, ZIC1, NEUROG2, and PAX6 and revealed significant induction of VGlut1 when compared with fibroblasts infected with the complete TF-pool or CHi only (Figure 5D). Similarly, we observed a significant induction of choline acetyltransferase (CHAT), a transferase enzyme responsible for the synthesis of the neurotransmitter acetylcholine (Lin et al., 2005), upon infection of DLX1, ISL1, NEUROG2, and PAX6 in fibroblasts. We further characterized the electrophysiological properties and found that both glutamatergic-like and cholinergic-like neurons generated repetitive but distinct action potentials upon current injection at 21 dpi (Figures 5E, 5F, and S6F) and both action potentials were inhibited by withdrawal of extracellular Na+ ion in the medium (Figures 5E, 5F, and S6G–S6J).
Finally, NEUROG2 acts as a pioneering factor during direct fibroblast-to-neuron reprogramming (Smith et al., 2016). To identify whether other pioneering factors present in the TF-pool are involved in direct fibroblast-to-neuron reprogramming, we compared our gene module analyses for each subtype and revealed that, in addition to NEUROG2, PAX6 exhibited hallmarks of a common regulator of direct cell reprogramming toward the neurons based on its gene association analysis. During brain development, PAX6-expressing neuroectodermal cells can be readily patterned to region-specific neuro-progenitor cells that give rise to various neuronal subtypes, including cholinergic neurons (Li et al., 2005), dopaminergic neurons (Liu et al., 2012), and GABAergic neurons (Kallur et al., 2008); however, its role in neuronal direct reprogramming has not been described. Thus, we hypothesized that cells expressing exogenous PAX6 should exhibit increased neuronal molecular phenotype as compared with cells lacking exogenous PAX6. To test this, we grouped cells with and without exogenous PAX6 and assessed expression signatures of neurogenic, neuronal subtype-specific, and fibroblast genes (Figures 5G and 5H). Indeed, we found that cells expressing PAX6 consistently showed significantly increased expression of neurogenic and neuronal subtype-specific genes and decreased expression of fibroblast genes compared with cells lacking exogenous PAX6. Moreover, the same extraction analysis for other exogenous genes, such as NEUROG2, DLX1, DLX2, ISL1, and ZIC1, suggests that PAX6 is the most dominant factor in inducing neurons (Figures 5G, 5H, S6K, and S6L). In line with these findings, expanding our analysis to additional neuronal subtypes placed PAX6 at the core of the association network, possibly as a pioneering factor during direct neuronal reprogramming, while TFs that induce neuronal subtype specification were placed at its periphery for a defined specification (Figure 5I).
Discussion
This study adapts retrospective identification of vector-based gene expression in single cells for the scale of massively parallel scRNA-seq. Our results demonstrate reliable distinction of endogenous and exogenous gene expression by a deep full-length scRNA-seq approach and further identify the most influential pro-neuronal TFs and alternative combinations of TFs from a large pool of candidate genes that govern the reprogramming of multiple and distinct neuronal subtypes.
This approach is set apart from earlier studies testing various possible combinations of TFs in a one-by-one approach (An et al., 2016; Caiazzo et al., 2011; Kim et al., 2011; Liu et al., 2012; Pang et al., 2011; Vierbuchen et al., 2010). Unlike other perturbation single-cell approaches (Adamson et al., 2016; Dixit et al., 2016), the multiplex overexpression method combined with scRNA-seq is compatible with virtually any vector-based expression system without the need for barcoding, avoiding possible barcode swapping and reconstruction of existing vectors (Griffiths et al., 2018); however, profiling of integrated barcodes in a vector-based expression system may allow the inclusion of higher cell numbers using either the 5′ or 3′ droplet-based single-cell RNA-seq.
Based on single-cell expression profiles of perturbed cells, we identified distinct developmental trajectories emerging during neuronal lineage specification. Specifically, we could associate NEUROG2 with glutamatergic (Winpenny et al., 2011), DLX1/DLX2 with GABAergic (Pla et al., 2018), ISL1 with cholinergic (Cho et al., 2014), and NR4A2 with dopaminergic neuron development (Caiazzo et al., 2011). Collectively this study strongly advocates that: (1) the multiplex scRNA-seq efficiently identifies key determinants of transdifferentiation in a single experiment, (2) genes identified by this approach are not random but specifically enriched in a defined neuronal program and function, (3) neuronal induction medium alone could not induce reprogramming, possibly due to the epigenetic gridlock of the original cell type, (4) multiple factors work in concert to drive cell fate specifications and diversification, and finally (5) our findings support a hierarchical reprogramming model which predicts that replacement of only a few factors can alter the fate of generated cells (Wapinski et al., 2013). However, retrospective identification of exogenous gene expression necessitates continuous exogenous gene expression over the whole time period analyzed. Here, we did not address temporal aspects of exogenous gene expression, which is still largely unexplored. Future studies that aim to explore temporal aspects of exogenous gene expression during cell reprogramming will likely require temporally controlled activation and suppression of gene sets and sequential gene delivery methods. In addition, due to the massively increased complexity, a much larger number of single cells will need to be analyzed.
Our results also suggest that PAX6 acts as a key driver of direct neuronal reprogramming. PAX6 acts as a major regulator of mammalian nervous system development and is expressed in a region-specific manner in NPCs and uniformly in neuroectodermal cells differentiated from embryonic stem cells and iPSCs (Chapouton et al., 1999; Stoykova et al., 2000; Yun et al., 2001; Zhang et al., 2010). In line with this, combination of PAX6 with NEUROG2, DLX2, and ZIC1 generates mainly glutamatergic neurons, whereas DLX1 and ISL1 generate mainly cholinergic neurons, suggesting that combinations of TFs are involved in both activation and repression toward cell specification. Previous reports implicated that DLX1 and DLX2 act as repressors to glia lineage tilting the cell fate toward the neuronal lineage (Petryniak et al., 2007). In another study, ZIC1 represses dopaminergic specification (Tiveron et al., 2017) and PAX6/NEUROG2 direct the cells toward glutamatergic fate (Winpenny et al., 2011), while Isl1 further pushes the cells toward cholinergic development (Cho et al., 2014). While precise dynamics will require systematic perturbation analysis, our approach demonstrates that a defined set of TFs important for cell-type switching and lineage specification can be precisely extrapolated.
In conclusion, our data corroborate previous studies on neuronal reprogramming, predict novel combinations of pro-neuronal TFs that allow the generation of neuronal subtypes, and yield the regulatory logic of neuronal reprogramming. We envision that this approach is an effective strategy to identify transcriptional codes controlling cell fate conversions, speaking to its potential to become a standard strategy to unravel molecular mechanisms governing other cell reprogramming pathways.
Experimental procedures
Cell Culture and Generation of Induced Neurons
Human neonatal dermal fibroblasts were purchased from Lonza (C-2509; passages 4–8) and cultured in DMEM containing 10% FBS/L-glutamine/10% penicillin and streptomycin at 37°C 5% CO2 until virus infection. For TFi generation, a total of six MOI (~0.3 MOI per virus) were pooled into culture medium containing 8 μg mL−1 polybrene (Sigma) to increase infection efficiency. For CHi cell generation, dermal fibroblasts were infected with a virus containing multiple cloning sites (e.g., no TF) at an MOI of 6 expressing YFP. The virus pool was infected and incubated overnight at 37°C 5% CO2. On the following day, the virus-containing medium was removed and cells were incubated for an additional day in the fresh culture medium at 37°C 5% CO2. Henceforth, CHi and TFi were cultured under identical culture conditions: cells were split onto poly-D-lysine (100 μg mL−1; Sigma)/laminin (50 μg mL−1; Sigma)-coated culture plates and incubated overnight in culture medium at 37°C 5% CO2. On the following day, the medium was changed to neuronal induction medium containing DMEM/F12 and Neurobasal-A (Thermo Fisher Scientific) mixed at a 1:1 ratio, 2% (vol/vol) B27 supplement, and 0.5% N2 supplement (Gibco), 1× nonessential amino acids (Thermo Fisher Scientific), 1% GlutaMAX supplement, VPA (0.5 mM, Wako), L-ascorbic acid (200 nM, Sigma), Y-27632 (10 μM, Wako), dcAMP (0.5 mM, Sigma), 10% FBS, and 10% penicillin and streptomycin (Wako). The concentrations of small molecules used were as follows: CHIR99021 (2 μM, Abcam), SB-431542 (10 μM, Sigma), LDN-193189 (0.5 μM, Stemgent), and Noggin (100 ng mL−1, Sigma). Neuronal induction medium was changed every third day, during which the concentration of FBS was gradually reduced from 10% to 0%. After 2 weeks, neuronal induction medium was replaced with neuronal maturation medium without small molecules, but containing 10 ng mL−1 BDNF (Gibco), 10 ng mL−1 NT3 (R&D Systems), and 10 ng mL−1 GDNF (Thermo Fisher Scientific) and the medium was changed every third day until further analysis.
Electrophysiology
Whole-cell current-clamp recordings were performed as described (Ichikawa et al., 2012). All experiments were conducted at 25°C. Patch pipettes with resistances ranging from 3 to 7 MΩ were pulled from capillary tubes using a DMZ-Universal Puller (Zeitz Instruments, Martinsried, Germany) and then backfilled with intracellular solution. Action potentials were recorded using a patch-clamp amplifier (Axopatch 200B; Axon Instruments, Foster City, CA) with a series of current steps from 0 to 200 pA with a 2,000-ms duration. The action potentials were monitored and stored using pCLAMP software (Molecular Devices, CA) after digitizing the analog signals at 5 kHz (DigiData 1322A; Axon Instruments). For patch-clamp recordings, the extracellular solution (ECS) consisted of the following: 137 mM NaCl, 5 mM KCl, 0.44 mM KH2PO4, 0.33 mM Na2HPO4, 10 mM glucose, 12 mM NaHCO3, 0.5 mM MgCl2, and 10 mM HEPES, adjusted to pH 7.4 with tris(hydroxymethyl)aminomethane. To examine the Na+ selectivity, extracellular 136 mM NaCl was substituted with equimolar extracellular LiCl (Na+-free ECS). To record ionic currents under physiological conditions, intracellular solution containing 150 mM KCl, 10 mM HEPES, and 2 mM magnesium adenosine triphosphate (pH 7.2 by tris(hydroxymethyl)aminomethane) was used.
Computational Methods for scRNA-Seq Data
Artificial Transcript Model
To determine the exact nucleotide sequences flanking the ORF of each exogenous transcript, we sequenced recombined plasmids using a 3730/3730xl DNA Analyzer (Applied Biosystems) following the manufacturer's protocol. In brief, we first amplified templates by PCR using primers annealing to the EF1A promoter sequence near the 5′ end of each ORF to amplify the 5′ junction sequences, and primers annealing to the IRES2 sequence near the 3′ end of each ORF to amplify the 3′ junction sequences. After gel purification, we sequenced templates using BigDye Terminator v.3.1 Cycle Sequencing Kits (Applied Biosystems). We integrated results derived from three primers (three replicates each) at both the 5′ and 3′ junction of each ORF and combined the resulting 5′ and 3′ junction sequences with known sequences of the ORFs of each TF and the CSII-EF-RfA-IRES2-VENUS pENTR lentivirus vector to compile the artificial exogenous transcript model. Finally, we combined our artificial exogenous transcript model with the human transcriptome (version GRCh38.p5) to obtain the final artificial transcript model.
Mapping of the Smart-Seq Time Course Data onto the 10X Genomics UMAP
To integrate the Smart-seq time course data with the 10X Genomics data, we computed a pairwise correlation matrix (Spearman correlation) of all cells based on the expression of ~1,300 highly variable genes in cells of clusters CL4–CL9. Smart-seq cells were mapped onto the 10X Genomics UMAP by averaging x and y coordinates of the five 10X Genomics cells showing the highest correlation values and assigned to their most likely 10X Genomics cluster by determining to which cluster the majority of these five 10X Genomics cells belonged to. If two or more clusters tied for majority, clusters were assigned at random. To identify exogenous TFs with enrichment in any cluster, we performed Fisher's exact tests to calculate the significance of association of a given exogenous TF with each cluster. Exogenous TFs with p < 0.05 were considered significantly enriched.
Gene Co-expression Module Assembly
To construct neuronal subtype-specific gene co-expression modules, we calculated the significance of association of each exogenous TF with all other genes using Fisher's exact tests. Exogenous TFs were attributed to neuronal subtype-specific module based on the following criteria: (1) p < 0.05 in Fisher's exact test and (2) at least three edges to three neuronal subtype-specific genes. Gene co-expression modules were visualized with Cytoscape using the organic layout.
Combination Score
The CS represents the −log10-transformed p value of the significance (Mann-Whitney U test) of increased gene expression in cells containing at least two of the predicted exogenous TFs in a network compared with all other cells.
Data and Code Availability
All analysis code used in this study is available upon request. Custom code for the main analytical steps can be found in: https://github.com/JoachimLu/Decoding-neuronal-diversification-by-multiplexed-single-cell-RNA-seq. All sequence data are accessible with accession number GEO: GSE117075.
Author contributions
Formulation of research goals and aims: J.L. and J.W.S.; data curation: J.L., T.K., and C.P.; formal analysis: J.L., T.K., R.N., and J.W.S.; funding acquisition: J.L., P.C., and J.W.S.; performing experiments: J.L., T.K., R.N., T.E.C., D.M.S., M.K., and F.R.; project supervision: P.C. and J.W.S.; writing – original draft: J.L. and J.W.S.; writing – review & editing: J.L. and J.W.S.
Acknowledgments
This work was supported by a Research Grant for RIKEN Center for Life Science Technology, Division of Genomic Technologies (CLST DGT) and RIKEN Center for Integrative Medical Sciences (IMS) from the Japanese Ministry of Education, Culture, Sports, Science and Technology (MEXT) and a Postdoctoral Fellowship for Research in Japan by the Japan Society for the Promotion of Science (JSPS). The authors wish to acknowledge RIKEN GeNAS for the sequencing of the libraries.
Published: March 11, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.02.006.
Supplemental information
References
- Adamson B., Norman T.M., Jost M., Cho M.Y., Nunez J.K., Chen Y., Villalta J.E., Gilbert L.A., Horlbeck M.A., Hein M.Y. A multiplexed single-cell CRISPR screening platform enables systematic dissection of the unfolded protein response. Cell. 2016;167:1867–1882.e21. doi: 10.1016/j.cell.2016.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alda-Catalinas C., Bredikhin D., Hernando-Herraez I., Eckersley-Maslin A., Stegle O., Reik W. A single-cell transcriptomics CRISPR-activation screen identifies epigenetic regulators of the zygotic genome activation program. Cell. Syst. 2020;11:25–41. doi: 10.1016/j.cels.2020.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An N., Xu H., Gao W.Q., Yang H. Direct conversion of somatic cells into induced neurons. Mol. Neurobiol. 2016;55:642–651. doi: 10.1007/s12035-016-0350-0. [DOI] [PubMed] [Google Scholar]
- Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- Caiazzo M., Dell’Anno M.T., Dvoretskova E., Lazarevic D., Taverna S., Leo D., Sotnikova T.D., Menegon A., Roncaglia P., Colciago G. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476:224–227. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- Chanda S., Ang C.E., Davila J., Pak C., Mall M., Lee Q.Y., Ahlenius H., Jung S.W., Sudhof T.C., Wernig M. Generation of induced neuronal cells by the single reprogramming factor ASCL1. Stem Cell Reports. 2014;3:282–296. doi: 10.1016/j.stemcr.2014.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapouton P., Gartner A., Gotz M. The role of Pax6 in restricting cell migration between developing cortex and basal ganglia. Development. 1999;126:5569–5579. doi: 10.1242/dev.126.24.5569. [DOI] [PubMed] [Google Scholar]
- Chen S., Sanjana N.E., Zheng K., Shalem O., Lee K., Shi X., Scott D.A., Song J., Pan J.Q., Weissleder R. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell. 2015;160:1246–1260. doi: 10.1016/j.cell.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H., Cargnin F., Kim Y., Lee B., Kwon R., Nam H., Shen R., Barnes A.P., Lee J.W., Lee S. Isl1 directly controls a cholinergic neuronal identity in the developing forebrain and spinal cord by forming cell type-specific complexes. PLoS Genet. 2014;10:e1004280. doi: 10.1371/journal.pgen.1004280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit A., Parnas O., Li B., Chen J., Fulco C.P., Jerby-Arnon L., Marjanovic N.D., Dionne D., Burks T., Raychowdhury R. Perturb-seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell. 2016;167:1853–1866.e17. doi: 10.1016/j.cell.2016.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelles J., Schnapp B.J., Sheetz M.P. Tracking kinesin-driven movements with nanometer-scale precision. Nature. 1988;331:450–453. doi: 10.1038/331450a0. [DOI] [PubMed] [Google Scholar]
- Griffiths J.A., Richard A.C., Bach K., Lun A.T.L., Marioni J.C. Detection and removal of barcode swapping in single-cell RNA-seq data. Nat. Commun. 2018;9:1–6. doi: 10.1038/s41467-018-05083-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Qiu B., Guan W., Wang Q., Wang M., Li W., Gao L., Shen L., Huang Y., Xie G. Direct conversion of normal and Alzheimer’s disease human fibroblasts into neuronal cells by small molecules. Cell Stem Cell. 2015;17:204–212. doi: 10.1016/j.stem.2015.07.006. [DOI] [PubMed] [Google Scholar]
- Ichikawa H., Kim H., Shuprisha A., Shikano T., Tsumura M., Shibukawa Y., Tazaki M. Voltage-dependent sodium channels and calcium-activated potassium channels in human odontoblasts in vitro. J. Endod. 2012;38:1355–1362. doi: 10.1016/j.joen.2012.06.015. [DOI] [PubMed] [Google Scholar]
- Kallur T., Gisler R., Lindvall O., Kokaia Z. Pax6 promotes neurogenesis in human neural stem cells. Mol. Cell Neurosci. 2008;38:616–628. doi: 10.1016/j.mcn.2008.05.010. [DOI] [PubMed] [Google Scholar]
- Kim J., Su S.C., Wang H., Cheng A.W., Cassady J.P., Lodato M.A., Lengner C.J., Chung C.Y., Dawlaty M.M., Tsai L.H. Functional integration of dopaminergic neurons directly converted from mouse fibroblasts. Cell Stem Cell. 2011;9:413–419. doi: 10.1016/j.stem.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.J., Du Z.W., Zarnowska E.D., Pankratz M., Hansen L.O., Pearce R.A., Zhang S.C. Specification of motoneurons from human embryonic stem cells. Nat. Biotechnol. 2005;23:215–221. doi: 10.1038/nbt1063. [DOI] [PubMed] [Google Scholar]
- Li X., Zuo X., Jing J., Ma Y., Wang J., Liu D., Zhu J., Du X., Xiong L., Du Y. Small-molecule-driven direct reprogramming of mouse fibroblasts into functional neurons. Cell Stem Cell. 2015;17:195–203. doi: 10.1016/j.stem.2015.06.003. [DOI] [PubMed] [Google Scholar]
- Lin W., Dominguez B., Yang J., Aryal P., Brandon E.P., Gage F.H., Lee K., Jolla L. Neurotransmitter acetylcholine negatively regulates neuromuscular synapse formation by a Cdk5-dependent mechanism. 2005;46:569–579. doi: 10.1016/j.neuron.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Liu X., Li F., Stubblefield E.A., Blanchard B., Richards T.L., Larson G.A., He Y., Huang Q., Tan A.C., Zhang D. Direct reprogramming of human fibroblasts into dopaminergic neuron-like cells. Cell Res. 2012;22:321–332. doi: 10.1038/cr.2011.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M.L., Zang T., Zou Y., Chang J.C., Gibson J.R., Huber K.M., Zhang C.L. Small molecules enable neurogenin 2 to efficiently convert human fibroblasts into cholinergic neurons. Nat. Commun. 2013;4:2183. doi: 10.1038/ncomms3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Yu C., Daley T.P., Wong W.H., Wernig M., Qi L.S. CRISPR activation screens systematically identify factors that drive neuronal fate and reprogramming. Stem Cell. 2018;23:758–771.e8. doi: 10.1016/j.stem.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masserdotti G., Gascon S., Gotz M. Direct neuronal reprogramming: learning from and for development. Development. 2016;143:2494–2510. doi: 10.1242/dev.092163. [DOI] [PubMed] [Google Scholar]
- Mazzoni E.O., Mahony S., Closser M., Morrison C.A., Nedelec S., Williams D.J., An D., Gifford D.K., Wichterle H. Synergistic binding of transcription factors to cell-specific enhancers programs motor neuron identity. Nat. Neurosci. 2013;16:1219–1227. doi: 10.1038/nn.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcinnes L., Healy J., Melville J. UMAP: uniform manifold approximation and projection for dimension reduction. arXiv. 2018 1802.03426v2. [Google Scholar]
- Nowakowski T.J., Bhaduri A., Pollen A.A., Alvarado B., Mostajo-Radji M.A., Di Lullo E., Haeussler M., Sandoval-Espinosa C., Liu S.J., Velmeshev D. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science. 2017;358:1318–1323. doi: 10.1126/science.aap8809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Z.P., Yang N., Vierbuchen T., Ostermeier A., Fuentes D.R., Yang T.Q., Citri A., Sebastiano V., Marro S., Sudhof T.C. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryniak M.A., Potter G.B., Rowitch D.H., Rubenstein J.L.R. Dlx1 and Dlx2 control neuronal versus oligodendroglial cell fate acquisition in the developing forebrain. Neuron. 2007;55:417–433. doi: 10.1016/j.neuron.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfisterer U., Kirkeby A., Torper O., Wood J., Nelander J., Dufour A., Bjorklund A., Lindvall O., Jakobsson J., Parmar M. Direct conversion of human fibroblasts to dopaminergic neurons. Proc. Natl. Acad. Sci. U S A. 2011;108:10343–10348. doi: 10.1073/pnas.1105135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S., Faridani O.R., Bjorklund A.K., Winberg G., Sagasser S., Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- Pla R., Stanco A., Howard M.A., Rubin A.N., Vogt D., Mortimer N., Cobos I., Potter G.B., Lindtner S., Price J.D. Dlx1 and Dlx2 promote interneuron GABA synthesis, synaptogenesis, and dendritogenesis. Cereb. Cortex. 2018;28:3797–3815. doi: 10.1093/cercor/bhx241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rackham O.J., Firas J., Fang H., Oates M.E., Holmes M.L., Knaupp A.S., Consortium F., Suzuki H., Nefzger C.M., Daub C.O. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016;48:331–335. doi: 10.1038/ng.3487. [DOI] [PubMed] [Google Scholar]
- Replogle J.M., Norman T.M., Xu A., Hussmann J.A., Chen J., Cogan J.Z., Meer E.J., Terry J.M., Riordan D.P., Srinivas N. Combinatorial single-cell CRISPR screens by direct guide RNA capture and targeted sequencing. Nat. Biotechnol. 2020;38:954–961. doi: 10.1038/s41587-020-0470-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R., Farrell J.A., Gennert D., Schier A.F., Regev A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O., Sanjana N.E., Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat. Rev. Genet. 2015;16:299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shifrut E., Carnevale J., Tobin V., Diolaiti M.E., Ashworth A., Marson A., Shifrut E., Carnevale J., Tobin V., Roth T.L. Genome-wide CRISPR screens in primary human T cells reveal key regulators of immune function resource. Cell. 2018;175:1958–1971. doi: 10.1016/j.cell.2018.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J.W., Suzuki T., Ninomiya N., Kishima M., Hasegawa Y., Kubosaki A., Yabukami H., Hayashizaki Y., Suzuki H. Establishment of single-cell screening system for the rapid identification of transcriptional modulators involved in direct cell reprogramming. Nucleic Acids Res. 2012;40:e165. doi: 10.1093/nar/gks732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D.K., Yang J., Liu M.L., Zhang C.L. Small molecules modulate chromatin accessibility to promote NEUROG2-mediated fibroblast-to-neuron reprogramming. Stem Cell Reports. 2016;7:955–969. doi: 10.1016/j.stemcr.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son E.Y., Ichida J.K., Wainger B.J., Toma J.S., Rafuse V.F., Woolf C.J., Eggan K. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell. 2011;9:205–218. doi: 10.1016/j.stem.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoykova A., Treichel D., Hallonet M., Gruss P. Pax6 modulates the dorsoventral patterning of the mammalian telencephalon. J. Neurosci. 2000;20:8042–8050. doi: 10.1523/JNEUROSCI.20-21-08042.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Tiveron M., Beclin C., Murgan X.S., Wild S., Angelova A., Marc J., Core N., De Chevigny X.A., Herrera X.E., Bosio A. Zic-proteins are repressors of dopaminergic forebrain fate in mice and C. elegans. J. Neurosci. 2017;37:10611–10623. doi: 10.1523/JNEUROSCI.3888-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N.J., Livak K.J., Mikkelsen T.S., Rinn J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victor M.B., Richner M., Hermanstyne T.O., Ransdell J.L., Sobieski C., Deng P.Y., Klyachko V.A., Nerbonne J.M., Yoo A.S. Generation of human striatal neurons by microRNA-dependent direct conversion of fibroblasts. Neuron. 2014;84:311–323. doi: 10.1016/j.neuron.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T., Ostermeier A., Pang Z.P., Kokubu Y., Südhof T.C., Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski O.L., Vierbuchen T., Qu K., Lee Q.Y., Chanda S., Fuentes D.R., Giresi P.G., Ng Y.H., Marro S., Neff N.F. Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell. 2013;155:621–635. doi: 10.1016/j.cell.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreb C., Wolock S., Klein A.M. SPRING: a kinetic interface for visualizing high dimensional single-cell expression data. 2018;34:1246–1248. doi: 10.1093/bioinformatics/btx792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winpenny E., Lebel-Potter M., Fernandez M.E., Brill M.S., Götz M., Guillemot F., Raineteau O. Sequential generation of olfactory bulb glutamatergic neurons by Neurog2-expressing precursor cells. Neural Dev. 2011;6:1–18. doi: 10.1186/1749-8104-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Jiang H., Zhong P., Yan Z., Chen S., Feng J. Direct conversion of human fibroblasts to induced serotonergic neurons. Mol. Psychiatry. 2016;21:62–70. doi: 10.1038/mp.2015.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun K., Potter S., Rubenstein J.L. Gsh2 and Pax6 play complementary roles in dorsoventral patterning of the mammalian telencephalon. Development. 2001;128:193–205. doi: 10.1242/dev.128.2.193. [DOI] [PubMed] [Google Scholar]
- Zhang X., Huang C.T., Chen J., Pankratz M.T., Xi J., Li J., Yang Y., Lavaute T.M., Li X.J., Ayala M. Pax6 is a human neuroectoderm cell fate determinant. Cell Stem Cell. 2010;7:90–100. doi: 10.1016/j.stem.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G.X., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R., Ziraldo S.B., Wheeler T.D., McDermott G.P., Zhu J. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Danbolt N.C. Glutamate as a neurotransmitter in the healthy brain. J. Neural Transm. (Vienna) 2014;121:799–817. doi: 10.1007/s00702-014-1180-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All analysis code used in this study is available upon request. Custom code for the main analytical steps can be found in: https://github.com/JoachimLu/Decoding-neuronal-diversification-by-multiplexed-single-cell-RNA-seq. All sequence data are accessible with accession number GEO: GSE117075.