Significance
Heterosis, referring to the better performance of an F1 hybrid compared with its parents, has been greatly exploited in agriculture to improve yield. Nevertheless, our understanding of its genetic and molecular mechanism has remained limited. We investigated the temporally dynamic phenotypes and transcriptomes during early seedling development in two Arabidopsis ecotypes together with their hybrid and unveiled differences in the transcriptional regulatory network between the hybrid and parents, orchestrating the hybrid growth vigor. Moreover, we found complementary high-parent expression in the hybrid between the network hub genes involved in photosynthesis and cell cycle. Our findings provide molecular-level evidence that dominant expression complementation between genes involved in two fundamental biological pathways contributes to heterosis.
Keywords: biomass heterosis, WGCNA, dominance, cell cycle, photosynthesis
Abstract
The mechanisms underlying heterosis have long remained a matter of debate, despite its agricultural importance. How changes in transcriptional networks during plant development are relevant to the continuous manifestation of growth vigor in hybrids is intriguing and unexplored. Here, we present an integrated high-resolution analysis of the daily dynamic growth phenotypes and transcriptome atlases of young Arabidopsis seedlings (parental ecotypes [Col-0 and Per-1] and their F1 hybrid). Weighted gene coexpression network analysis uncovered divergent expression patterns between parents of the network hub genes, in which genes related to the cell cycle were more highly expressed in one parent (Col-0), whereas those involved in photosynthesis were more highly expressed in the other parent (Per-1). Notably, the hybrid exhibited spatiotemporal high-parent–dominant expression complementation of network hub genes in the two pathways during seedling growth. This suggests that the integrated capacities of cell division and photosynthesis contribute to hybrid growth vigor, which could be enhanced by temporal advances in the progression of leaf development in the hybrid relative to its parents. Altogether, this study provides evidence of expression complementation between fundamental biological pathways in hybrids and highlights the contribution of expression dominance in heterosis.
Heterosis refers to the superior performance of an F1 hybrid compared with its parental inbred lines in terms of growth, yield, and adaption to the environment. Despite its successful utilization in agriculture, the genetic basis underlying this phenomenon remains elusive (1). Because each chromosome in the F1 genome possesses an allele from each parent, dominance is one of the proposed hypotheses explaining heterosis. This hypothesis attributes hybrid vigor to genome-wide dominant complementation between beneficial alleles from either parent (2–4). For example, function complementation between dominant superior loci in hybrids has been reported to trigger heterosis in crops (5, 6). In addition, single-parent expression has been found to be a general mechanism driving expression complementation in some hybrids (7–9). However, despite these findings, concurrent complementation of gene expression and biological function in a hybrid has rarely been reported, and it has remained a challenge to associate the genetic complementation in a hybrid with its heterotic phenotypes.
Biomass heterosis, which has been interpreted as a complicated quantitative trait (10), was recently found to be associated with multiple loci from the perspective of hybrid genomic architecture (11, 12), suggesting that genome-wide determinants contribute to hybrid vigor. In addition, global gene expression profiles have uncovered prevalent nonadditive transcriptional changes in various tissues of Arabidopsis and other plant species hybrids relative to their parents (13–22). However, most of these studies did not link gene expression differences to the heterotic phenotype and did not find a uniform pattern of differential gene expression between hybrids and parents (1, 10), likely due to the limitations of tissue sampling at individual or discrete time points during plant development. Therefore, understanding of the mechanism of heterosis may progress by comparing time-series transcriptional architectures of a target tissue between a hybrid and its parents and by studying the dynamic manifestation of heterosis during the entire developmental process.
In Arabidopsis, obvious biomass heterosis at the vegetative growth stage has been shown to be common in crosses between different ecotypes (11, 23–25), making it an excellent model for studying the genetic and molecular basis of heterosis. Nonadditive expression of genes involved in cell cycle and photosynthesis in hybrids has been reported in previous studies (14, 15, 23, 26), and some genes in these two pathways were revealed to be associated with heterosis (11). However, how these genes and biological processes contribute to hybrid vigor is largely uncharacterized. Arabidopsis cotyledons and true leaves comprise most of the above-ground seedling biomass during vegetative growth, and their development is precisely controlled by dynamically regulated gene expression in multiple pathways, along with programmed cellular and physiological behaviors (27–29). Therefore, analyzing the dynamic transcriptome landscapes of early shoot or true leaf development in a hybrid and its parents can provide a better understanding of the regulatory networks underlying the enhanced growth and development of hybrids.
Here, we generated integrated high temporal-resolution phenotypes and transcriptome atlases of the early seedling shoots (3–8 d after sowing [DAS]) and first true leaves (7–21 DAS) of Arabidopsis ecotypes Col-0, Per-1, and their F1 hybrid (Col-0 × Per-1). Using weighted gene coexpression network analysis (WGCNA), we defined the gene coexpression networks underlying two developmental processes and uncovered a conserved coexpression network among the hybrid and parents for each organ. We discovered that the hybrid’s coexpression modules had gene expression profiles and interaction patterns different from those of the parents. In addition, network hub genes exhibiting opposite differential expression directions for the two parents were involved in the cell cycle and photosynthesis, reflecting a natural transcriptional divergence in Arabidopsis ecotypes at key periods of shoot apex cell division and building photosynthesis capacity. Remarkably, these two sets of hub genes exhibited a complementary spatiotemporal high-parent–dominant expression pattern in the hybrid, suggesting that integrated cell division and photosynthesis abilities from the parents contributed to the hybrid’s growth vigor. Thus, through transcriptional complementation of these two fundamental biological pathways, we uncovered evidence supporting the dominance model of heterosis, and our high temporal-resolution transcriptome data provide a valuable resource for further functional genomics studies of seedling development.
Results
Dynamic Growth Heterosis Established during Early Stage of Seedling Development.
In a previous study, we revealed a high positive correlation between heterosis for shoot biomass and the first true leaf area in hundreds of Arabidopsis hybrids, and the Col-0 × Per-1 F1 hybrid was recognized as exhibiting the most significant heterosis for both shoot biomass and the first true leaf growth at 14 DAS (11). However, when and how the strong growth vigor of the Col-0 × Per-1 hybrid was established during seedling growth and development was unclear. To obtain a detailed understanding of the underlying attributes, we investigated the time-series manifestation of seedling growth heterosis in Col-0 × Per-1 by determining the daily growth dynamics of the cotyledon during 3–12 DAS and the first true leaf during 3–21 DAS. The cotyledon size was significantly larger for the hybrid than the parents during 3–12 DAS, but we found a dynamically changing pattern in the level of heterosis (SI Appendix, Figs. S1A and S2A and Dataset S1). Best-parent heterosis (BPH) of cotyledon growth was established (47.61%) as early as 3 DAS and reached its peak (71.85–74.69%) during 4–6 DAS. For the first true leaf, the F1 hybrid exhibited significant growth vigor compared with at least one parent during 3–21 DAS (SI Appendix, Figs. S1B and S2B and Dataset S1). Notably, leaf area heterosis underwent dramatic changes during the early stage of first true leaf development, with BPH increasing from 70.37% at 5 DAS to 289.50% at 6 DAS, significantly peaking at 7 DAS (346.95%), holding at 188.91% at 8 DAS, and decreasing sharply to 50.02% at 9 DAS. From the cellular perspective, 6–7 DAS is approximately the cell proliferation phase and the proliferation-to-expansion transition phase for the first true leaf (27). Therefore, these results indicate that, compared with its parents, the F1 hybrid undergoes accelerated cell cycle or cell division during early leaf growth, implying that the growth heterosis of the mature leaf may result largely from a cumulative effect of increased cell number in the hybrid at an early leaf development stage. In addition, as the cotyledon’s growth heterosis was established earlier than that of the first true leaf, it highlights the embryonic tissue’s contribution to hybrid vigor (30).
Highly Preserved Gene Regulatory Networks in the Hybrid and Parents during Early Shoot Development.
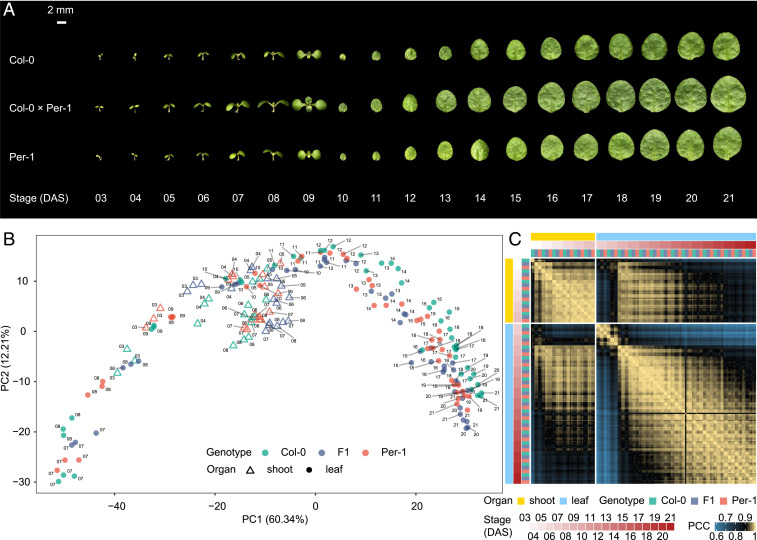
As our phenotypic observations showed that hybrid seedling growth vigor is continuously manifested and highly dynamic, we wondered how that process is affected by transcriptomic reprogramming in the hybrid during seedling development. We performed RNA-seq on early shoots (3–8 DAS) and first true leaves (7–21 DAS) from Col-0, Per-1, and the Col-0 × Per-1 F1 hybrid, generating daily temporal dynamic transcriptome atlases based on ∼1.2 Tb of RNA-seq data from 189 samples in three biological replicates (Fig. 1A and SI Appendix, Table S1 and Dataset S2). As expected, principal component analysis (PCA) clearly showed transcriptome profile clustering (Fig. 1B) that correlated with continuous developmental states and dynamic changes in gene expression throughout early shoot and first true leaf development in all three genotypes. In general, the shoot expression profiles were not always tightly related to the true leaf profiles, which reflected both conservation and divergence of the developmental regulatory circuitry between the two organs. Nevertheless, for both organs, expression profiles at the earlier developmental stages (3–5 DAS for the shoot, and before 12 DAS for the true leaf) were mostly different from the profiles in the later stages (6–8 DAS for the shoot, and after 12 DAS for the true leaf), suggesting the relatively large transcriptomic changes during developmental progression. Furthermore, the global gene expression in both parents and the hybrid remained closely correlated, indicating highly conserved transcriptional features in the parental and hybrid lines during early shoot and true leaf development. The relationship among gene expression profiles in different organs and genotypes was supported by the pairwise Pearson correlation coefficients (Fig. 1C).
Fig. 1.
Dynamic transcriptome landscapes of shoots at 3–8 d after sowing (DAS) and the first true leaves at 7–21 DAS in Arabidopsis Col-0, Per-1, and Col-0 × Per-1. (A) Representative photographs of harvested organs for RNA-seq. (B) PCA of 189 transcriptome profiles of different organs and genotypes, including three biological replicates. (C) Heat map illustrating pairwise Pearson correlation coefficients (PCCs) between each pair of 189 transcriptome profiles.
Because previous studies have not explored whether and how transcriptional regulatory network differences between a hybrid and its parents function in heterosis, we took advantage of our high-resolution time-series transcriptome atlases to gain insights into growth heterosis in a hybrid. WGCNA can be used to construct coexpressed gene modules based on the correlation of global gene transcription across experiments or samples (31). The genes with similar expression behaviors cluster together and are involved in closely connected biological processes (32). Thus, a coexpression network can represent the complex transcriptional architecture of the whole genome throughout dynamic developmental progression. Here, we performed WGCNA to construct gene coexpression networks underlying early shoot development in Col-0, Per-1, and their F1 hybrid, which summarized the genotypes’ global gene expression status across this process. Gene sets containing 14,088 (Col-0), 13,781 (Per-1), or 13,834 (F1) genes with expression level changes in shoots during 3–8 DAS were used to construct a signed, scale-free network for each genotype according to their topological overlap matrix of gene expression correlations produced by introducing the corresponding power (β) (SI Appendix, Fig. S3 A–C and E and Table S2). Overall, the gene coexpression network comprised 8 modules of 524–4,664 genes, 10 modules of 111–5,877 genes, and 9 modules of 374–2,878 genes in Col-0, Per-1, and their hybrid, respectively (SI Appendix, Fig. S4 and Table S2). In view of the close relatedness revealed by the PCA, we assessed whether the gene coexpression networks were well conserved between the hybrid and its parents by conducting permutation tests. These tests summarize the evidence that network properties are preserved between two independent datasets (33). By projecting the Per-1 transcriptome dataset onto the Col-0 network, we found that seven out of eight modules were strongly preserved (Zsummary > 10), and one module exhibited weak to moderate preservation (10 > Zsummary > 2) (SI Appendix, Fig. S4). Furthermore, by considering modules identified in the hybrid as a reference network, we found that eight out of nine modules and all nine modules were at least weakly (Zsummary > 2) preserved in Col-0 and Per-1 compared with the hybrid, respectively (SI Appendix, Fig. S4). These results confirmed an overall highly conserved transcriptional architecture underlying the dynamic regulation of early shoot development in Col-0, Per-1, and their hybrid.
Divergent Core Gene Regulatory Networks between the Hybrid and Parents during Early Shoot Development.
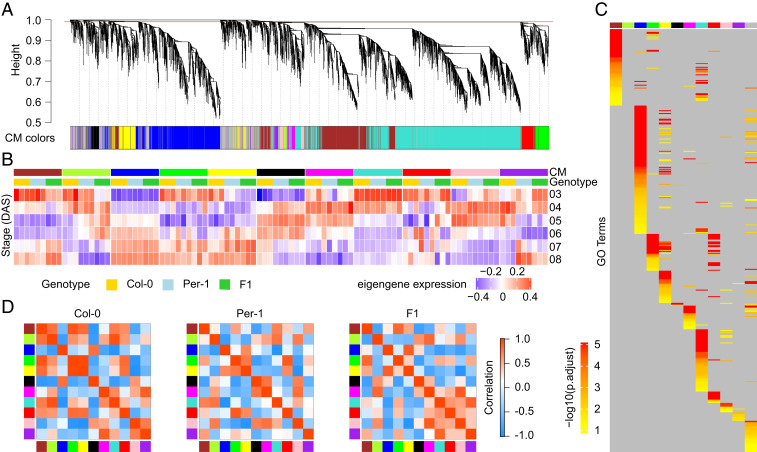
As the overall transcriptional networks were highly conserved in both the hybrid and the parents, we investigated whether and how transcriptional differences in the core gene regulatory network are associated with the significantly greater growth vigor of the hybrid compared with its parents. First, based on the network modules above, we identified a conserved gene coexpression network underlying early shoot development among the hybrid and its parents. This network was represented by consensus modules (CMs), clusters of genes presenting coexpression in all three genotypes (SI Appendix, Fig. S3 D and F). Genes in these modules were regulated consistently in different genotypes, comprising the core regulatory mechanisms commonly involved in dynamic early shoot growth and development in both the parents and hybrid, which affected their shoot growth vigor and biomass. The 9,670 genes assigned to the conserved network were clustered in 11 CMs consisting of 78–3,346 coexpressed genes (Fig. 2A and SI Appendix, Table S2). We used each module eigengene (ME), defined as the first principal component of the gene expression profiles of a given module, to describe the global gene expression behavior within each module during shoot development (32). In the conserved network, each CM presented a specific eigengene expression pattern for early shoot development during 3–8 DAS (Fig. 2B). We also observed differing eigengene expression patterns between the hybrid and its parents for some of the CMs, indicating a genotype-specific perturbation of gene expression levels in these modules, a phenomenon that could potentially play a role in the heterosis regulatory mechanism. For example, brown CM eigengene expression levels decreased over 3–5 DAS but increased over 6–8 DAS in all three genotypes (Fig. 2B). However, compared with the parents, brown CM eigengene expression in the hybrid was more down-regulated over 3–5 DAS and more up-regulated over 6–8 DAS. Gene Ontology (GO) enrichment analysis indicated that genes of the brown CM were significantly overrepresented in cell cycle-related processes (Fig. 2C and Dataset S3). In contrast, the black CM indicated similar eigengene expression patterns in the hybrid and Per-1, which were different from that in Col-0 (Fig. 2B), and GO analysis showed that the genes within that CM were significantly enriched in the photosynthetic process (Fig. 2C and Dataset S3). The hybrid and its parents also exhibited different eigengene expression patterns in some of the other CMs (Fig. 2B), and the genes in those CMs were associated with distinct biological functions (Fig. 2C and Dataset S3).
Fig. 2.
Preservation and divergence of gene coexpression networks between the hybrid and its parents underlying early shoot development (3–8 DAS). (A) Hierarchical cluster dendrogram showing consensus gene coexpression modules (CMs) among Arabidopsis ecotypes (Col-0, Per-1) and their F1 hybrid. In the dendrogram, each leaf represents one gene, and each module below the dendrogram is labeled with one color. The red line above is the cut height (0.99) for CM identification. CM colors are labeled independent of the separately identified modules in each genotype. Genes not coexpressed in all three genotypes are marked in gray. (B) Differential eigengene expression patterns of CMs in the three genotypes across 3–8 DAS. Blue and red represent lesser and greater expression, respectively. (C) Heat map summarizing GO enrichment of genes in each CM. Color intensity represents the significance (−log10[adjusted P value]) of GO term enrichment. (D) Heat maps of eigengene networks showing the relationships among CMs in each genotype.
The gene coexpression network structure describes the gene regulatory relationships between genes in functionally divergent modules. To test for network structure differences between the hybrid and its parents, we analyzed network intermodule connections among the 11 CMs in each genotype by generating an eigengene network represented by pairwise correlation coefficients among their CM eigengenes (34). The hybrid’s CM eigengene network had an obviously different pattern from the network of either parent (Fig. 2D). Due to the unique expression pattern and the involvement in specific biological processes of each CM in the network, the above result indicates changed genetic interactions and strengths in various biological processes associated with early shoot growth or development in the hybrid compared with its parents. Therefore, despite its overall conservation, the hybrid’s core transcriptional regulatory network during early shoot development diverged greatly from that of its parents.
Complementary High-Parent–Dominant Expression of Network Hub Genes Involved in the Cell Cycle and Photosynthesis in the Hybrid.
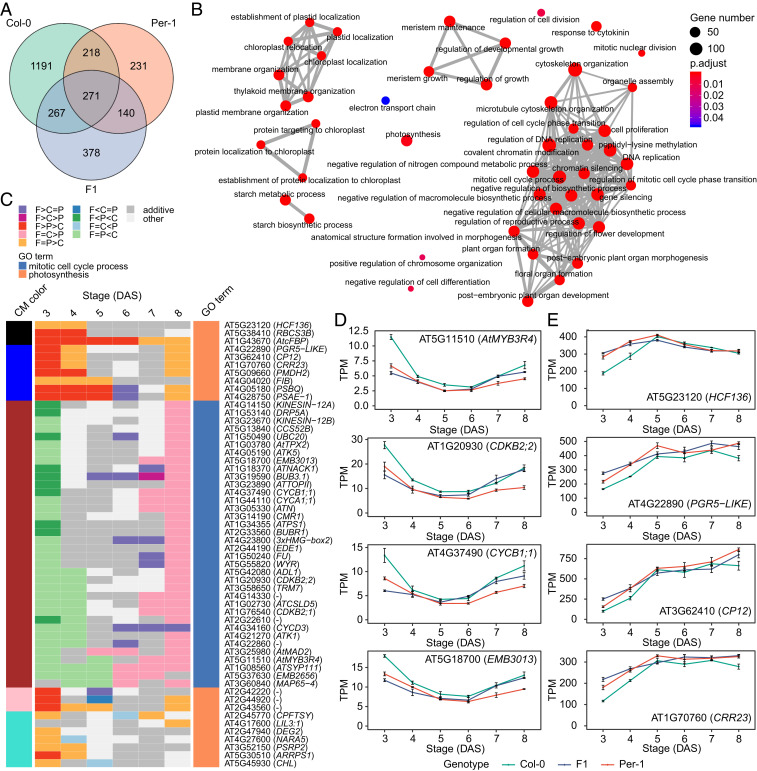
To further narrow our transcriptomic network analysis and identify key regulators that may participate in the generation of network differences underlying the dynamic regulation of heterosis in the hybrid during early shoot growth and development, we detected hub genes that were highly connected to other genes in each genotype’s conserved gene coexpression network. First, we determined the correlation between each gene’s expression profile and its corresponding ME to find the module membership value for each gene (kME) (32, 35). We subsequently identified 1,947, 860, and 1,056 hub genes (kME > 0.90, P < 10−6) in Col-0, Per-1, and their F1 hybrid, respectively (Fig. 3A and Dataset S4). Among these hub genes, 489 were shared between the two parents, and 271 were shared by all three genotypes (Fig. 3A), indicating that these genes have preserved functional features across genotypes. GO enrichment analysis revealed that these hub genes were significantly overrepresented in growth-related functional categories, such as cell cycle, photosynthesis, development, and other cellular metabolic processes (Fig. 3B and Dataset S5). These results supported the key roles of hub genes in early shoot growth and development.
Fig. 3.
Hub genes in the core gene coexpression network underlying the dynamic regulation of early shoot development. (A) Venn diagram showing the shared hub genes between Col-0, Per-1, and the F1 hybrid. (B) GO enrichment analysis of the hub genes in A. Dot color intensity represents the significance (adjusted P value) of the GO term enrichment, and dot size indicates the number of genes in each GO term. (C) Heat map showing the hybrid’s complementary differential expression patterns (adjusted P < 0.05) for hub genes involved in the mitotic cell cycle and photosynthesis. The cell colors represent differential expression patterns, and the colors on the Left and Right represent the corresponding consensus module (CM) and GO term to which each gene belongs, respectively. C, Col-0; P, Per-1; F, F1; DAS, days after sowing. (D and E) Expression levels of representative hub genes involved in the mitotic cell cycle process (D) and photosynthesis (E) across 3–8 DAS. Data are mean TPM ± SE in each genotype at each stage (n = 3).
The mitotic cell cycle continuously produces new cells from the shoot apical meristem (SAM), enabling leaf growth and development (36), whereas photosynthesis provides fundamental energy for seedling metabolic processes and biomass accumulation (37, 38). Genes in these two pathways were previously revealed to be associated with growth heterosis in Arabidopsis (14, 23). Coincidentally, evidence has been found of interfaces between cell cycle-related and photosynthesis-related energy metabolism (39). Accordingly, we compared the hybrid’s and its parents’ mitotic cell cycle and photosynthesis hub gene expression profiles across early shoot development. We found that many cell cycle regulator genes were expressed at higher levels in Col-0 than in Per-1, whereas genes involved in photosynthesis were expressed at higher levels in Per-1 than in Col-0 (Fig. 3C and Dataset S6). Intriguingly, in the F1 hybrid, a large set of hub genes involved in the cell cycle were expressed in high-parent or above high-parent patterns at 6–8 DAS, which is the key period for true leaf initiation from the SAM in Arabidopsis. These genes include those encoding the cyclin/CDK family of proteins and transcription factors regulating cell division, as well as other genes encoding essential proteins involved in cell cycle progression (Fig. 3C and Dataset S6). Among these genes, CDKB2;1, CDKB2;2, and CYCB1;1 are involved in regulation of the G2/M transition of the mitotic cell cycle (40); MYB3R4 acts as a transcriptional factor that positively regulates cytokinesis (41); and EMB3013 encodes a microtubule-associated kinase-like protein that colocalizes with the mitotic preprophase band, spindle, and phragmoplast and is required for cell plate expansion in cytokinesis (42) (Fig. 3D). All of this suggests that Col-0 and the F1 hybrid have greater cell division activity than Per-1. In temporal complementation, most of the hub genes involved in photosynthesis (e.g., genes encoding photosystem-associated proteins and the rubisco subunit, as well as genes involved in photosynthetic electron transport, thylakoid membrane organization, and that positively regulate photosynthesis efficiency) were also expressed in high-parent or above high-parent patterns in the F1 hybrid during 3–5 DAS, the key period for seedling photomorphogenesis and photosynthesis capacity establishment (Fig. 3C and Dataset S6). Among these genes, HCF136 encodes a photosystem II stability/assembly factor (43), PGR5-LIKE A encodes a thylakoid transmembrane protein involved in photosynthetic electron transport in photosystem I (44), and CP12 is involved in the formation of a Calvin cycle complex and important in Calvin cycle functioning, as well as the photosynthetic capacity (45, 46). CRR23, a subunit of the chloroplast NAD(P)H dehydrogenase complex, is also involved in photosynthetic electron transport in photosystem I (47) (Fig. 3E). These findings suggest that Per-1 and the F1 hybrid had better photosynthesis capacity than Col-0. In summary, the hybrid’s expression of network hub genes involved in both the cell cycle and photosynthesis shows prominent, complementary high-parent–dominant patterns in different developmental periods. Therefore, we propose that the growth vigor or biomass heterosis of early seedling shoots in the Col-0 × Per-1 hybrid is likely achieved by integrating the higher cell division capability of the Col-0 accession and the higher photosynthesis capability of the Per-1 accession.
Gene Coexpression Networks Underlying True Leaf Development Emphasize the Fundamental Contribution of Early Shoot Vigor to Heterosis.
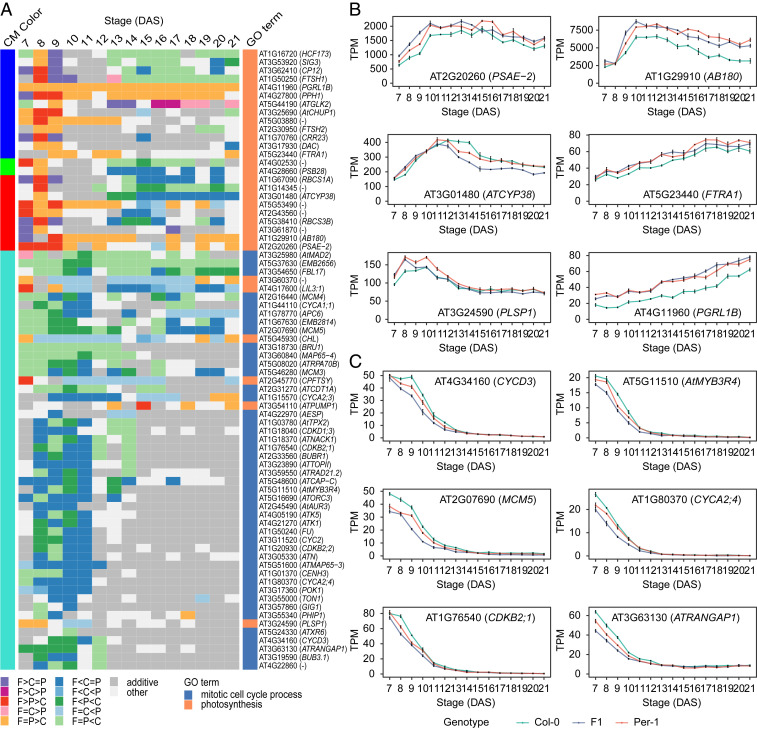
Next, we investigated whether true leaves exhibit transcriptional differences in the core gene regulatory network that are associated with the hybrid seedling’s growth heterosis. Again, we constructed gene coexpression networks for Col-0, Per-1, and the F1 hybrid, but these networks were underlying the dynamic control of first true leaf growth and development during 7–21 DAS (SI Appendix, Figs. S5 A–C and E and S6A and Table S2). Based on module preservation analysis (SI Appendix, Fig. S6A), we compiled a conserved network among the three genotypes (SI Appendix, Figs. S5 D and F and S6B and Table S2). The six CMs in the network showed both distinct gene expression patterns during 7–21 DAS and different expression levels between the hybrid and its parents (SI Appendix, Fig. S6C), whereas genes in these CMs were involved in divergent biological processes (SI Appendix, Fig. S6D and Dataset S7). Interestingly, GO analysis revealed that the network hub genes (kME > 0.90, P < 10−6) were also significantly enriched in the pathways directly related to plant growth and development, such as the mitotic cell cycle, photosynthesis, organ development, and primary metabolic processes (Datasets S8 and S9). Remembering that the hybrid’s cell cycle and photosynthesis pathways had both contributed to its growth vigor in the early shoot stage, we realized that these processes would go on to influence the subsequent growth and development of the true leaf. Therefore, we focused our next analyses on the hub genes involved in these two pathways. We found hub genes related to photosynthesis were in several CMs (including blue, green, red, and turquoise CMs), and they showed different expression patterns across leaf development (Fig. 4 A and B). Consistent with our observations of early shoot development during 3–5 DAS, a majority of the photosynthesis-related hub genes exhibited higher expression during true leaf development in Per-1 than in Col-0 (Fig. 4A and Dataset S10). They also exhibited high-parent or above high-parent expression patterns in the hybrid through the leaf’s entire development (e.g., AB180, FTRA1, and PGRL1B) or at earlier stages in its development (e.g., PSAE-2, ATCYP38, and PLSP1) (Fig. 4B). This suggests that key genes in the photosynthesis pathway maintained consistent expression patterns and functions in all above-ground organs throughout seedling development, contributing continuously to hybrid growth vigor.
Fig. 4.
Differential expression patterns of the cell cycle and photosynthesis associated hub genes in the core regulatory network of true leaf development between the hybrid and its parents. (A) Heat map showing the differential expression patterns of hub genes involved in the mitotic cell cycle and photosynthesis in the first true leaf of the F1 hybrid (F) and its parents (C, Col-0; P, Per-1) (adjusted P < 0.05) across 7–21 d after sowing (DAS). The cell colors represent the differential expression patterns, and the colors on the Left and Right of the heat map represent the corresponding consensus module (CM) and GO term to which each gene belongs, respectively. (B and C) Expression levels of representative hub genes involved in photosynthesis (B) and the mitotic cell cycle process (C) across 7–21 DAS. Data are mean TPM ± SE in each genotype at each stage (n = 3).
In contrast, the hub genes involved in the cell cycle exhibited decreasing expression levels from 7 to 13 DAS, and then maintained low expression levels until 21 DAS (Fig. 4C). This corresponds to the dynamic cellular behaviors within the first true leaf in Arabidopsis during development of 7–21 DAS, which include successive cell proliferation, division-to-expansion transition, and cell expansion until leaf maturation (27, 48). Cell cycle-related hub genes once again exhibited higher expression in Col-0 than in Per-1 (Fig. 4 A and C and Dataset S10), indicating a stable, superior cell division capability in proliferating tissues during seedling development in the Col-0 accession. However, these cell cycle genes [i.e., cell cycle regulator genes including CYCD3, which is involved in the switch from cell proliferation to the final stages of differentiation (49)] were down-regulated in the hybrid compared with its parents during leaf development 7–13 DAS and later kept the same expression levels as its parents (Fig. 4 A and C and Dataset S10). Due to the gradual decrease in cell cycle gene expression level and cell division activity during leaf growth and development (50), our results suggested an earlier transition from cell division to cell expansion in true leaves in hybrid than the parents. However, cell cycle genes showed high-parent expression in hybrid shoot as early as 6–8 DAS. Thus, the enhancement of this pathway in hybrid had already happened at the true leaf initiation and proliferation stages and contributed to heterosis before its down-regulation over the leaf development. Although these genes were down-regulated in hybrid later, the adverse effect of this expression pattern on heterosis was very limited due to their very low expression levels at later stages as well as the already increased cell number and more cells undergoing expansion in hybrid. This both explains the leaf growth heterosis peak at 6–7 DAS and further highlights the effects of the hybrid’s early shoot stage growth vigor on heterosis at later developmental periods.
Discussion
Since agriculture’s earliest beginnings, farmers have exploited heterosis to improve both biomass and yield. However, even with its advances, modern agriculture still does not adequately understand the genetic and molecular mechanisms of heterosis. Although genetic models, such as dominance, overdominance, and epistasis, have been proposed to explain heterosis, only a few studies have provided molecular-level support for these hypotheses, mostly because of the hybrids’ complex genomic architecture and limited understanding of the regulatory mechanisms of quantitative traits. Transcriptome analysis is an effective tool for genomic studies in plant biology and has been applied widely to characterize the molecular basis of heterosis (10, 20, 51). However, how genome-wide changes in gene expression in hybrids cause hybrid vigor phenotypes is largely unknown due to the complicated dynamic gene regulatory networks in different tissues and at various developmental stages. Here, we developed temporally dynamic transcriptome atlases during early seedling shoot and true leaf development in an Arabidopsis hybrid and its parents, allowing us to identify gene coexpression network differences between the hybrid and its parents that underlie the dynamic regulation of growth heterosis. Our study uncovered highly preserved gene regulatory networks with distinct levels of module gene expression and patterns of network structures shared by the hybrid and its parents. Unlike traditional transcriptome studies that examine differences between a hybrid and its parents, gene regulatory network analysis, with time-series dynamic transcriptome mapping, alleviates transcriptional noise due to single-stage tissue sampling. In addition, hub gene determination helps identify the hybrid’s differentially expressed genes that most effectively contribute to the seedling growth heterosis phenotype. Furthermore, traditional transcriptome analyses usually focus on genes that exhibit large changes in expression in the hybrid. Here, we found that the key genes associated with plant growth vigor were not greatly differentially expressed in the hybrid, but maintained small, stable, and functional up-regulation for a certain duration during plant development, a phenomenon that may explain the difficulties in discovering a uniform global differential gene expression pattern associated with heterosis manifestations in hybrids (1, 10, 51). The gene coexpression information we provide will aid future research into plant seedling growth regulatory mechanisms, as well as leaf development control.
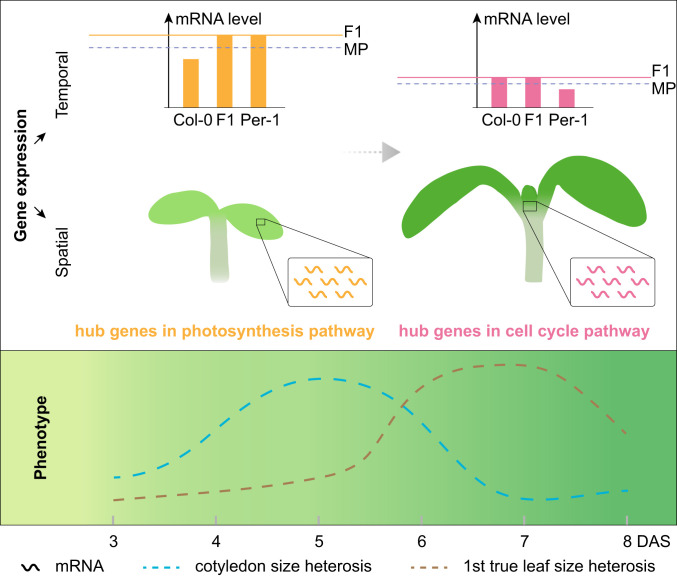
Previous studies reported that enhanced photosynthesis or increased leaf cell number is associated with vegetative growth vigor in different hybrids (14, 23), suggesting critical roles of these biological processes in heterosis. Knowing the mechanisms by which genes in these pathways function in hybrids can promote more efficient utilization of heterosis in plants. Recent studies have proposed dominance as the most likely mechanism for hybrid vigor, but it has been challenging to uncover relevance between alleles in the dominance model and the heterotic phenotypes (3). Here, we revealed a hybrid’s (Col-0 × Per-1) complementary dominant gene expression pattern between transcriptional regulatory network hub genes involved in the photosynthesis and cell division pathways at different stages of seedling development. Mostly during the photosynthesis capacity building stages of photosynthetic organs, the hybrid’s photosynthesis genes exhibited Per-1–like higher expression, whereas cell cycle genes exhibited Col-0-like higher expression at the true leaf initiation stages in the shoot apex (Fig. 5). This implies that coordinated gene expression and function complementation during plant development contributes to heterosis, which is consistent with the F1 hybrid’s genomic features and provides biological pathway-level evidence supporting the dominance hypothesis of heterosis.
Fig. 5.
A model for seedling growth heterosis triggered by complementary high-parent–dominant expression between growth-related key genes involved in the cell cycle and photosynthesis. At the early stages of seedling growth (3–8 d after sowing [DAS]), regulatory network hub genes involved in photosynthesis had higher expression levels in parent Per-1 than in parent Col-0, and had expression levels similar to Per-1 in the F1 hybrid mainly during 3–5 DAS (the key period for establishing seedling cotyledon photosynthesis capability). In contrast, the regulatory network hub genes involved in the cell cycle had higher expression levels in Col-0 than in Per-1, and in temporal complementarity had expression patterns similar to Col-0 in the hybrid mainly during 6–8 DAS (the key period for cell division-driven true leaf initiation in the shoot apex). Thus, the hybrid spatiotemporally integrates one parent’s higher expression of cell cycle pathway genes and the other parent’s higher expression of photosynthesis pathway genes to realize enhanced growth vigor triggered by the complementary capacities of these two fundamental biological pathways (MP: midparent, the average level of both parents).
We also found that a few photosynthesis hub genes continued high-parent expression in the hybrid through true leaf development (Fig. 4 A and B and Dataset S10). These genes are involved in light harvesting, photosynthetic electron transport, and light reaction (e.g., AB180, FTRA1, and PGRL1B), which are all processes that continually function along with photosynthesis in the true leaf throughout its development. In contrast, the genes participating in photosystem assembly or thylakoid organization (e.g., PSAE-2, ATCYP38, and PLSP1) were expressed in a high-parent pattern in the hybrid only at the earlier stage of leaf development. These two patterns demonstrate the functional divergence of genes in the photosynthesis pathway, and this is also reflected in their distinct expression patterns across leaf development (Fig. 4 A and B). Moreover, in the true leaf, some genes (e.g., HCF173, CP12, PSB28, RBCS1A, RBCS3B, ATCYP38, and LIL3:1) were down-regulated in the hybrid after 12 DAS (Fig. 4 A and B), and most of these genes’ functions were associated with biosynthesis or formation of the photosynthetic machinery (Dataset S10). These energy-consuming processes (52) worked mostly during the early stage of leaf development to build photosynthetic capacity, so their down-regulation at later stages could be a resource-saving strategy to avoid excessive energy waste in the hybrid. Besides, the expression of hub genes in the cell cycle pathway was significantly down-regulated in the shoot during 3–5 DAS, before the most active true leaf initiation in the SAM (Fig. 3 C and D and Dataset S6). This may have occurred because of the higher cell division activity in the parents’ cotyledons; it was no longer observed after cell division ceased in the cotyledons.
In addition to the prominent expression complementation of genes involved in cell cycle and photosynthesis, the above high-parent expression pattern of a few hub genes in these two pathways was also observed in hybrid (Figs. 3C and 4A). This type of nonadditive gene expression may be a result of the joint function of both cis and trans regulation in the hybrid genome (53). It cannot be excluded that the overdominance effect of the above high-parent gene expression may lead to the improvement of one of the two biological pathways in hybrid and contribute to heterosis. However, the complementation of two different pathways in hybrid could have much stronger effects on the level of heterosis.
In summary, the early establishment of dynamic shoot and leaf growth heterosis that we revealed using high–temporal-resolution phenotype analysis, together with the transcriptional complementation in early hybrid seedlings, illustrates the significance of understanding and manipulating heterosis in the early plant development stage in future research. Moreover, given the similarity in the leaf growth regulation mechanism between dicots and monocots (54), our study also benefits future studies of heterosis in crops. Taken together, our findings provide insight and valuable guidance for future applications of the dominance model in hybrid breeding.
Materials and Methods
Arabidopsis thaliana accessions (Col-0 and Per-1) were obtained from the Arabidopsis Biological Resource Center. The F1 hybrid Col-0 × Per-1 was obtained using Col-0 as the maternal line through hand-pollination. Details on the plant materials and sampling and phenotyping are described in SI Appendix, SI Materials and Methods.
For RNA-seq, shoots at 3–8 DAS and the first or second true leaves at 7–21 DAS for both the parents and the hybrid were sampled to extract total RNA with three biological replicates. Normalized gene expression value (transcripts per million [TPM]) was quantified using Salmon and tximport softwares. Differential gene expression analysis was conducted using the voom function in limma software with an adjusted P value < 0.05. The transcriptional network analyses were performed with WGCNA. The details and procedures for RNA-seq and the data analysis are provided in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (31871221 and 31621001), the State Key Laboratory of Protein and Plant Gene Research, and the Peking–Tsinghua Center for Life Sciences (to X.W.D.).
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2023278118/-/DCSupplemental.
Data Availability
RNA-seq data sets have been deposited in the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo) under accession number GSE157957. All other study data are included in the article and/or supporting information.
References
- 1.Hochholdinger F., Hoecker N., Towards the molecular basis of heterosis. Trends Plant Sci. 12, 427–432 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Chen Z. J., Genomic and epigenetic insights into the molecular bases of heterosis. Nat. Rev. Genet. 14, 471–482 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Hochholdinger F., Baldauf J. A., Heterosis in plants. Curr. Biol. 28, R1089–R1092 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Birchler J. A., Plant science: Hybrid vigour characterized. Nature 537, 620–621 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Huang X., et al., Genomic analysis of hybrid rice varieties reveals numerous superior alleles that contribute to heterosis. Nat. Commun. 6, 6258 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li X., Li X., Fridman E., Tesso T. T., Yu J., Dissecting repulsion linkage in the dwarfing gene Dw3 region for sorghum plant height provides insights into heterosis. Proc. Natl. Acad. Sci. U.S.A. 112, 11823–11828 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baldauf J. A., et al., Single-pparent expression is a general mechanism driving extensive complementation of non-syntenic genes in maize hybrids. Curr. Biol. 28, 431–437.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Shao L., et al., Patterns of genome-wide allele-specific expression in hybrid rice and the implications on the genetic basis of heterosis. Proc. Natl. Acad. Sci. U.S.A. 116, 5653–5658 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z., et al., Single-parent expression drives dynamic gene expression complementation in maize hybrids. Plant J 105, 93–107 (2021). [DOI] [PubMed] [Google Scholar]
- 10.Lippman Z. B., Zamir D., Heterosis: Revisiting the magic. Trends Genet. 23, 60–66 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Yang M., et al., Genomic architecture of biomass heterosis in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 114, 8101–8106 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang X., et al., Genomic architecture of heterosis for yield traits in rice. Nature 537, 629–633 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Meyer R. C., et al., Heterosis manifestation during early Arabidopsis seedling development is characterized by intermediate gene expression and enhanced metabolic activity in the hybrids. Plant J 71, 669–683 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Fujimoto R., Taylor J. M., Shirasawa S., Peacock W. J., Dennis E. S., Heterosis of Arabidopsis hybrids between C24 and Col is associated with increased photosynthesis capacity. Proc. Natl. Acad. Sci. U.S.A. 109, 7109–7114 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu A., et al., Early changes of gene activity in developing seedlings of Arabidopsis hybrids relative to parents may contribute to hybrid vigour. Plant J 88, 597–607 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Alonso-Peral M. M., et al., Patterns of gene expression in developing embryos of Arabidopsis hybrids. Plant J 89, 927–939 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Zhou P., Hirsch C. N., Briggs S. P., Springer N. M., Dynamic patterns of gene expression additivity and regulatory variation throughout maize development. Mol. Plant 12, 410–425 (2019). [DOI] [PubMed] [Google Scholar]
- 18.He G., et al., Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell 22, 17–33 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He G., et al., Conservation and divergence of transcriptomic and epigenomic variation in maize hybrids. Genome Biol. 14, R57 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schnable P. S., Springer N. M., Progress toward understanding heterosis in crop plants. Annu. Rev. Plant Biol. 64, 71–88 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Saeki N., et al., Molecular and cellular characteristics of hybrid vigour in a commercial hybrid of Chinese cabbage. BMC Plant Biol. 16, 45 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen Y., et al., Analysis of transcriptional and epigenetic changes in hybrid vigor of allopolyploid Brassica napus uncovers key roles for small RNAs. Plant J. 91, 874–893 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Groszmann M., et al., Intraspecific Arabidopsis hybrids show different patterns of heterosis despite the close relatedness of the parental genomes. Plant Physiol. 166, 265–280 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer R. C., Törjék O., Becher M., Altmann T., Heterosis of biomass production in Arabidopsis. Establishment during early development. Plant Physiol. 134, 1813–1823 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barth S., Busimi A. K., Friedrich Utz H., Melchinger A. E., Heterosis for biomass yield and related traits in five hybrids of Arabidopsis thaliana L. Heynh. Heredity 91, 36–42 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Offermann S., Peterhansel C., Can we learn from heterosis and epigenetics to improve photosynthesis? Curr. Opin. Plant Biol. 19, 105–110 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Gonzalez N., Vanhaeren H., Inzé D., Leaf size control: Complex coordination of cell division and expansion. Trends Plant Sci. 17, 332–340 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Vercruysse J., Baekelandt A., Gonzalez N., Inzé D., Molecular networks regulating cell division during Arabidopsis leaf growth. J. Exp. Bot. 71, 2365–2378 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chandler J. W., Cotyledon organogenesis. J. Exp. Bot. 59, 2917–2931 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Wang L., et al., Cotyledons contribute to plant growth and hybrid vigor in Arabidopsis. Planta 249, 1107–1118 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Zhang B., Horvath S., A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 4, Article17 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Langfelder P., Horvath S., WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langfelder P., Luo R., Oldham M. C., Horvath S., Is my network module preserved and reproducible? PLOS Comput. Biol. 7, e1001057 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langfelder P., Horvath S., Eigengene networks for studying the relationships between co-expression modules. BMC Syst. Biol. 1, 54 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langfelder P., Mischel P. S., Horvath S., When is hub gene selection better than standard meta-analysis? PLoS One 8, e61505 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Polyn S., Willems A., De Veylder L., Cell cycle entry, maintenance, and exit during plant development. Curr. Opin. Plant Biol. 23, 1–7 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Leister D., Schneider A., From genes to photosynthesis in Arabidopsis thaliana. Int. Rev. Cytol. 228, 31–83 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Wang P., Hendron R. W., Kelly S., Transcriptional control of photosynthetic capacity: Conservation and divergence from Arabidopsis to rice. New Phytol. 216, 32–45 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Siqueira J. A., Hardoim P., Ferreira P. C. G., Nunes-Nesi A., Hemerly A. S., Unraveling interfaces between energy metabolism and cell cycle in plants. Trends Plant Sci. 23, 731–747 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Inzé D., De Veylder L., Cell cycle regulation in plant development. Annu. Rev. Genet. 40, 77–105 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Haga N., et al., R1R2R3-Myb proteins positively regulate cytokinesis through activation of KNOLLE transcription in Arabidopsis thaliana. Development 134, 1101–1110 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Krupnova T., et al., Microtubule-associated kinase-like protein RUNKEL needed [corrected] for cell plate expansion in Arabidopsis cytokinesis. Curr. Biol. 19, 518–523 (2009). [DOI] [PubMed] [Google Scholar]
- 43.Plöchinger M., Schwenkert S., von Sydow L., Schröder W. P., Meurer J., Functional update of the auxiliary proteins PsbW, PsbY, HCF136, PsbN, TerC and ALB3 in maintenance and assembly of PSII. Front Plant Sci 7, 423 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DalCorso G., et al., A complex containing PGRL1 and PGR5 is involved in the switch between linear and cyclic electron flow in Arabidopsis. Cell 132, 273–285 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Gontero B., Maberly S. C., An intrinsically disordered protein, CP12: Jack of all trades and master of the Calvin cycle. Biochem. Soc. Trans. 40, 995–999 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Elena López-Calcagno P., Omar Abuzaid A., Lawson T., Anne Raines C., Arabidopsis CP12 mutants have reduced levels of phosphoribulokinase and impaired function of the Calvin-Benson cycle. J. Exp. Bot. 68, 2285–2298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shimizu H., et al., CRR23/NdhL is a subunit of the chloroplast NAD(P)H dehydrogenase complex in Arabidopsis. Plant Cell Physiol. 49, 835–842 (2008). [DOI] [PubMed] [Google Scholar]
- 48.Kalve S., De Vos D., Beemster G. T., Leaf development: A cellular perspective. Front Plant Sci 5, 362 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dewitte W., et al., Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc. Natl. Acad. Sci. U.S.A. 104, 14537–14542 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Powell A. E., Lenhard M., Control of organ size in plants. Curr. Biol. 22, R360–R367 (2012). [DOI] [PubMed] [Google Scholar]
- 51.Birchler J. A., Yao H., Chudalayandi S., Vaiman D., Veitia R. A., Heterosis. Plant Cell 22, 2105–2112 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paul M. J., Pellny T. K., Carbon metabolite feedback regulation of leaf photosynthesis and development. J. Exp. Bot. 54, 539–547 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Zhang H. Y., et al., A genome-wide transcription analysis reveals a close correlation of promoter INDEL polymorphism and heterotic gene expression in rice hybrids. Mol. Plant 1, 720–731 (2008). [DOI] [PubMed] [Google Scholar]
- 54.Nelissen H., Gonzalez N., Inzé D., Leaf growth in dicots and monocots: So different yet so alike. Curr. Opin. Plant Biol. 33, 72–76 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data sets have been deposited in the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo) under accession number GSE157957. All other study data are included in the article and/or supporting information.