Abstract
The extent to which the gut microbiota may play a role in latitudinal clines of body mass variation (i.e., Bergmann’s rule) remains largely unexplored. Here, we collected wild house mice from three latitudinal transects across North and South America and investigated the relationship between variation in the gut microbiota and host body mass by combining field observations and common garden experiments. First, we found that mice in the Americas follow Bergmann’s rule, with increasing body mass at higher latitudes. Second, we found that overall differences in the gut microbiota were associated with variation in body mass controlling for the effects of latitude. Then, we identified specific microbial measurements that show repeated associations with body mass in both wild-caught and laboratory-reared mice. Finally, we found that mice from colder environments tend to produce greater amounts of bacteria-driven energy sources (i.e., short-chain fatty acids) without an increase in food consumption. Our findings provide motivation for future faecal transplant experiments directly testing the intriguing possibility that the gut microbiota may contribute to Bergmann’s rule, a fundamental pattern in ecology.
Keywords: adaptation, body size, coevolution, latitude, microbiome, Mus musculus
1 |. INTRODUCTION
A major goal in ecology is to link individual processes to macro-ecological and evolutionary patterns. Accumulating evidence suggests that the gut microbiota affects a variety of host phenotypes and thus an organism’s fitness (McFall-Ngai et al., 2013; Suzuki, 2017). The known links between the gut microbiota and host phenotype raise the possibility that the gut microbiota might mediate geographical patterns of phenotypic variation in hosts. However, connecting variation in gut microbial communities to variation in host phenotype remains challenging. Many mammals, including humans, show latitudinal clines of increasing body mass in colder climates (Ashton, Tracy, & De Queiroz, 2000; Bergmann, 1847; Foster & Collard, 2013; Roberts, 1953). This pattern, termed Bergmann’s rule, is thought to reflect adaptation to cold environments either through heat conservation or greater fat storage (Ashton et al., 2000; Bergmann, 1847; Blackburn, Gaston, & Loder, 1999). Whether the gut microbiota is causally related to this body mass variation remains unclear.
Studies in obesity research suggest that the gut microbiota can play a causal role in host body mass variation at least under certain dietary conditions (Bäckhed et al., 2004; Ley et al., 2005; Turnbaugh et al., 2006). Increased energy extraction through bacteria-dependent digestion of plant polysaccharides, activation of fat storage and production of short-chain fatty acids (SCFAs) has been proposed to explain the link between microbiota and body mass (Bäckhed et al., 2004; Turnbaugh et al., 2006). For example, obese individuals of mice and humans have been classically characterized by a higher ratio of two dominant gut bacterial phyla, the Firmicutes and Bacteroidetes (F/B ratio) in some studies (Ley et al., 2005; Ley, Turnbaugh, Klein, & Gordon, 2006; Turnbaugh et al., 2006) but not in others (Arumugam et al., 2011; Finucane, Sharpton, Laurent, & Pollard, 2014). Germ-free mice that received a microbiota with a higher F/B ratio show significant increases in energy extraction and concentrations of SCFAs compared to controls (Turnbaugh et al., 2006), a pattern also seen in humans (Jumpertz et al., 2011). Because both body mass and the F/B ratio display latitudinal clines in humans (Suzuki & Worobey, 2014), the gut microbiota could be causally linked to host body mass variation. However, the F/B ratio is also associated with many other factors, including diet (De Filippo et al., 2010), age (Mariat et al., 2009) and inflammation (Hansen, Gulati, & Sartor, 2010). There are also many other microbial measurements that have been linked to host body mass variation in humans (Dao et al., 2016; Goodrich et al., 2014; Yun et al., 2017), but they have been less explored in natural populations of mammals.
House mice (Mus musculus) provide a unique opportunity to study the role of gut microbes in host phenotypic variation. House mice have a global distribution in association with humans, encompassing a wide range of latitudes and climates (Phifer-Rixey & Nachman, 2015). House mice colonized the Americas recently, and they show clinal variation in body mass in eastern North America consistent with Bergmann’s rule (Lynch, 1992; Phifer-Rixey et al., 2018). Population-specific differences in body mass persist in a common laboratory environment after multiple generations, suggesting that these differences have a genetic basis (Lynch, 1992; Phifer-Rixey et al., 2018). An experimental study demonstrates that wild house mice kept in cold conditions increase body weight and fat compared to controls in fewer than 10 generations (Barnett & Dickson, 1989). The gut microbiota of wild house mice is also known to differ across geography and over different genotypes (Linnenbrink et al., 2013; Suzuki, Martins, & Nachman, 2018; Suzuki et al., 2019), and the alpha-diversity of the gut microbiota has been associated with differences in body weight (Suzuki et al., 2019; Weldon et al., 2015). Finally, wild house mice can easily be kept in captivity to experimentally test hypotheses concerning their gut microbiota (Rosshart et al., 2017).
Here, we looked for associations between gut microbiota variation and body mass variation in wild house mice collected from three latitudinal transects across the Americas by combining field observations and laboratory experiments. We found that differences in body mass were significantly associated with differences in the gut microbiota of wild mice accounting for geography and other covariates. Mice from colder environments generally had larger body mass and greater faecal SCFAs compared to mice from warmer environments. Larger body size was repeatedly associated with greater microbial alpha-diversity and higher relative abundance of “Rikenellaceae_RC9_gut_group” and “Ruminiclostridium” in wild and laboratory mice. However, the strength of these correlations was heterogeneous among the three transects. This suggests that the relationship between microbial variation and host body mass is complex, and that teasing apart cause and effect will require carefully controlled experiments in which individual microbes are introduced onto a common host genetic background.
2 |. MATERIALS AND METHODS
2.1 |. Samples
A total of 166 wild house mice (Mus musculus) were collected from three latitudinal transects across the Americas (Figure 1a) and five individuals were excluded from all analyses due to low sequence reads (see below). We used Sherman live traps with a mix of oats and peanut butter as bait to collect the animals overnight. Microhabitats included farms, barns, haystacks, feed stores, houses and zoos. The animals were euthanized in the field, prepared as museum specimens, and have been deposited in the collections of the U.C. Berkeley Museum of Vertebrate Zoology (Table S1). The Eastern North American transect (East-NA; latitude range: 29.10–44.13°N, altitude range: 1–98 m) includes the same individuals as in Suzuki et al. (2019), collected in summer between May and August 2012 (Phifer-Rixey et al., 2018). The Western North American transect (West-NA; latitude range: 32.08–53.52°N, altitude range: 642–1,419 m) includes five populations collected in summer between May and August 2012 in the vicinity of the following locations: (a) Tucson, Arizona; (b) St. George, Utah;, (c) Provo, Utah; (d) Stevensville, Montana; and (e) Edmonton, Alberta. The South American transect (SA; latitude range: 8.76–54.80°N, altitude range: 3–1,264 m) includes seven populations collected mostly in summer between February and September 2013 in the vicinity of the following locations: (a) Porto Velho, Brazil; (b) Brasilia, Brazil; (c) Maringa, Brazil; (d) Uruguaiana, Brazil; (e) Tandil, Argentina; (f) Gaiman, Argentina; and (g) Ushuaia, Argentina. To minimize the effect of phenology, animals were collected during the warmest months of the year in 15 out of 17 populations. The two exceptions were Uruguaiana and Maringa, where mice were collected during May and June, respectively. All mice were collected at least 500 m apart from each other to avoid collecting relatives, except at two sites in Edmonton and two sites in Ushuaia. Detailed sample information, including exact latitude and longitude, is provided in Table S1. Body weight and body mass index (i.e., BMI = body weight/body length2) were recorded, and caecal samples were collected within 24 hr after capture (the majority of animals were dissected in the morning following overnight trapping). Caecal samples were flash-frozen in the field using liquid nitrogen or dry ice and stored at −80°C until DNA extraction. Following the protocol of Suzuki and Nachman (2016), carbon (δ13C) and nitrogen (δ15N) stable isotopes from mouse hair were analysed to infer diet. We used the WorldClim database (Hijmans, Cameron, Parra, Jones, & Jarvis, 2005) to download 19 climatic variables based on GPS localities using the r package “dismo.” Principal components (PCs) were calculated, and the first two PC axes, Climate PC1 (48.9%) and Climate PC2 (21.7%), were used for downstream analyses (Table S2). The top three eigenvectors of Climate PC1 were temperature-related variables (e.g., mean temperature of coldest quarter, minimum temperature of coldest month and annual mean temperature) and those of Climate PC2 were precipitation-related variables (e.g., precipitation of driest month, precipitation of driest quarter and precipitation of coldest quarter; Table S2). All procedures involving animals were approved by the Institutional Animal Care and Use Committee at the University of Arizona (07–004) and the Animal Care and Use Committee at the University of California, Berkeley (R361–0514).
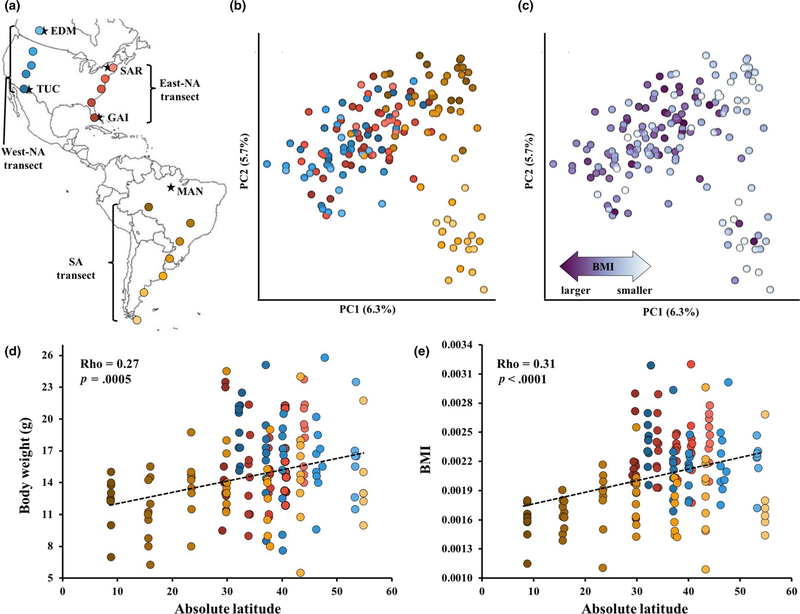
FIGURE 1.
The relationship between geography, body mass and the gut microbiota of wild house mice. (a) A map of 17 population samples of house mice across the Americas (n = 161). Populations from the East-NA transect (red), West-NA transect (blue) and SA transect (yellow) are shown in circles. Black stars indicate source locations of wild-derived inbred lines from MAN, GAI, TUC, SAR and EDM (see Methods for detailed locations). (b) PCoA plot showing the effect of geography on the Bray–Curtis dissimilarity (Mantel r = .41, p < .0001). Colours correspond to the map. (c) PCoA plot showing the effect of body mass index (BMI) on Bray–Curtis dissimilarity (Mantel r = .13, p = .002). Colour intensity reflects greater BMI. Significant correlations (d) between body weight and latitude (ρ = 0.27, p = .0005) and (e) between BMI and latitude (ρ = 0.31, p < .0001). Raw p-values are shown and they all remain significant after FDR correction (Table S6)
2.2 |. Laboratory-reared animals and wild-derived inbred lines
In addition to the collections described above, laboratory colonies of wild mice were established from animals collected at five locations: (a) Manaus, Amazonas, Brazil (MAN); (b) Gainesville, Florida, USA (GAI); (c) Tucson, Arizona, USA (TUC); (d) Saratoga Springs, New York, USA (SAR); and (e) Edmonton, Alberta, Canada (EDM; Figure 1a). Wild-caught animals and their descendants were maintained in a standard laboratory environment at 23°C with 10-hr dark and 14-hr light cycles. Teklad Global food (18% Protein Rodent Diet) was provided ad libitum. Wild-caught mice from each locality were paired to establish wild-derived inbred lines, propagated through sib–sib mating each generation. Up to 10 independent lines were established from each of these five populations. Within each population, the lines were established from unrelated pairs.
We used two sets of laboratory-reared individuals in this study, one from the first generation and another from later generations (up to generation 14). For the first-generation animals, we collected body weight and fresh faecal samples for 16S rRNA amplicon sequencing. A total of 120 individuals comprising 40 individuals each from three populations (MAN, GAI and SAR) were used (Table S3), although four individuals were excluded from all analyses due to low sequence reads (see below). Four adult individuals (i.e., two males and two females) from each of 10 independent inbred lines from each population were included. GAI (Florida) and SAR (New York) individuals are the same as in Suzuki et al. (2019).
For the later-generation animals, we collected body weight, fresh faecal samples for SCFA quantification, food intake and faecal weight. A total of 32 individuals representing five populations comprising five to seven age-matched (100–112 days) adult males per population were used (Table S4). Only one male per line was sampled, and thus each male represents a different inbred line. Food intake and faecal weight were measured every 24 hr for three consecutive days, and the measurements were averaged. The animals were housed in standard conditions as described above, but with a mesh floor to avoid coprophagy. Dry mass was used for food intake measurements and a mix of dry and fresh faeces were used for faecal weight measurements. Collection time and method were consistent across treatments. To account for body mass differences, average food intake and faecal weight were divided by body weight. Fresh faecal samples were immediately stored at −80°C and shipped to the West Coast Metabolomics Center at the University of California, Davis, for quantification of SCFAs, including acetate, propionate and butyrate. Mass spectrometry was conducted on an Agilent 6890A Gas Chromatograph with an Agilent 5977A Mass Selective Detector as in Moreau et al. (2003) and Richardson (1989). Although microbial communities were not characterized from the later-generation animals in this study, previous work has shown that the population differences in microbial communities among these lines are maintained for at least 11 generations in the laboratory (Moeller, Suzuki, Phifer-Rixey, & Nachman, 2018).
2.3 |. DNA extraction and 16S rRNA amplicon sequencing
To characterize the gut microbiota, frozen caecal samples were used for the wild-caught individuals and frozen faecal samples were used for laboratory-reared individuals. Although these sample types are different, previous studies have shown that caecal and faecal samples capture nearly identical patterns of interindividual variation in wild mice (Suzuki & Nachman, 2016). The protocols of DNA extraction and 16S rRNA gene sequencing are identical to those of Suzuki et al. (2019). Briefly, we used the QIAamp DNA stool Minikit (Qiagen) with a modified protocol adding a bead-beating step. The V4 region of the 16S rRNA gene was sequenced with 150-bp paired-end Illumina MiSeq at the Next Generation Sequencing Core Facility at Argonne National Laboratory. Negative controls were included in every set of amplifications. No amplification products were observed in gels, and thus no negative controls were sequenced. The polymerase chain reaction (PCR) primers (515F and 806R) and the barcodes are described in Caporaso et al. (2012).
2.4 |. Data analyses
All bioinformatic analyses on the 16S rRNA sequence data were conducted in QIIME2 version 2019.10.0 (Bolyen et al., 2019) and all statistical tests were conducted in r version 3.5.1. Sequence reads were quality-filtered and pair-end reads were merged using DADA2 (Callahan et al., 2016) resulting in an average sequence length of 226 bp (±6.0 SD), an average number of reads of 44,526 (±34,237 SD) per sample, and a total of 4,552 amplicon sequence variants (ASVs, 100% sequence identity). Separately, the feature table was also clustered at 94% sequence identity (resulting in a total of 1,420 operational taxonomic units [OTUs]) to test for robustness. The two clustering methods (100% and 94% sequence identities) produced nearly identical results, and thus we used ASVs for all analyses as recommended by Edgar (2018). All samples were rarefied to 5,500 reads. Five wild-caught individuals and four laboratory-reared individuals were excluded from all analyses due to the low number of reads (<3,000): MPR.137, FMM111, FMM112, FMM148, FMM274, BR07F1, BR08M1, FL08M1 and FL08M2. Taxonomy was assigned using the Naïve Bayes classifier trained on SILVA 132 99% (Quast et al., 2013), and phylogenetic trees were computed using FASTTREE (Price, Dehal, & Arkin, 2009).
Beta-diversity of the gut microbiota was calculated using Bray–Curtis dissimilarity (BCD) and principal coordinates analysis (PCoA) plots were generated. Mantel tests were used to test for correlations between BCD and seven predictor variables (geographical distance [km], climate PC1, climate PC2, body weight [g], BMI, δ13C diet and δ15N diet) using all individuals combined and using each transect separately. Similarly, partial Mantel tests were used to control for the effects of geographical distance and to test for a correlation between BCD and the other six predictor variables. ADONIS tests were also used to test for correlations between BCD and seven predictor variables (population, climate PC1, climate PC2, body weight [g], BMI, δ13C diet and δ15N diet) separately and also in a model incorporating all other variables as covariates. Alpha-diversity was calculated using phylogenetic diversity (Faith, 1992), 99% OTU counts and Shannon index.
To identify bacterial taxa whose relative abundance was correlated with body size measurements (i.e., body weight and BMI) or latitude, we selected phyla (taxonomic level 2) and species (taxonomic level 7) that had an average relative abundance >0.1% and were present in >50% of individuals in wild-caught and laboratory-reared mice separately. Correlations between microbial measurements (i.e., alpha-diversity and relative abundance of taxa) and metadata (e.g., body weight, BMI, latitude) were conducted for all wild-caught individuals, for each transect separately, and for all laboratory-reared individuals. Correlations were based on Spearman’s rho. To account for effects of latitude on the relationship between body size measurements and microbial measurements, residual values between body size measurements and latitude were used. To identify microbial measurements that show robust associations with body size measurements or latitude, we looked for repeated associations across the three latitudinal transects. When the three transects showed associations in the same direction, we used Fisher’s combined p-values to determine the significance (Fisher, 1932) as in Suzuki et al. (2018). Additionally, to test whether the collection site affected the relationship between body size measurements and latitude, a multiple regression model was used with population as a covariate.
Body weight, food intake, faecal weight and concentrations of SCFAs were compared among 32 wild-derived inbred lines in a common environment using Kruskal–Wallis tests, and pairwise comparisons were performed using the Wilcoxon test. We also conducted the same test using the residual values between body weight and SCFAs. To test whether the metadata show clinal patterns among the inbred lines, Spearman rho correlation between metadata and latitude of collection site was used. False discovery rate (FDR) corrections were used for all tests, and corrected p-values are indicated in each table and figure.
3 |. RESULTS
3.1 |. Geography and body mass are associated with compositional variation in the gut microbiota
To identify factors that correlate with overall differences in the gut microbiota in wild mice, we first looked for associations between BCD and seven predictor variables using all individuals from three latitudinal transects across the Americas (Figure 1a, Table 1). We found that geographical distance (Mantel r = .41, p < .0001) and BMI (Mantel r = .13, p = .002) were each significantly correlated with BCD. Principal coordinate plots of BCD show that microbial variation is associated with geography (Figure 1b) and with BMI (Figure 1c). In contrast, climate PCs, body weight and diet measured by stable isotopes (δ13C and δ15N) did not show significant correlations with BCD after FDR correction (Table 1). Similar results were found using ADONIS tests where population, climate PC1 and BMI were the top predictors of BCD by testing each variable separately or in a model accounting for other variables as covariates (Table S5). Calculating BCD using different sequencing identities (100% or 94% OTUs) provided similar results (Figure S1 and Table S5).
TABLE 1.
Correlations between predictor variables and Bray–Curtis dissimilarity
| All transects |
||||
|---|---|---|---|---|
| Predictor variables | n | Mantel r |
Raw p-value |
FDR p-value |
| Geographical distance | 161 | .406 | <.0001 | <.0007 |
| Climate PC1 | 161 | .077 | .03 | .07 |
| Climate PC2 | 161 | .001 | .97 | .97 |
| Body weight | 158 | .067 | .10 | .18 |
| BMI | 158 | .132 | .002 | .007 |
| Carbon | 158 | .057 | .13 | .18 |
| Nitrogen | 158 | .033 | .49 | .57 |
The association between BMI and microbial variation is interesting because body weight and BMI both show positive correlations with latitude when all individuals are analysed together, a pattern consistent with Bergmann’s rule (Figure 1d,e; Table S6). Latitude remained as a significant predictor of body size in multiple regression models adding collection site as a covariate (body weight; p = .01 and BMI; p = .05). However, individual transects revealed some differences from this overall pattern. These differences may be artefacts of sampling (i.e., low power) or they may reflect actual differences among mice from different broad geographical regions and climates (Figure S2). For example, positive correlations between body weight and absolute latitude were observed in both the East-NA (ρ = 0.34, p = .016) and SA transects (ρ = 0.26, p = .045), but not in the West-NA transect (ρ = −0.28, p = .06; Table S6). Similar correlations with body size were observed for Climate PC1 (mostly explained by temperature-related variables) but not for Climate PC2 (mostly explained by precipitation-related variables; Table S7).
To explore whether associations between BCD and body weight or BMI exist within each transect, we looked for correlations between BCD and predictor variables in each of the three transects separately (Table S8). We found differences among transects. For example, within the East-NA transect, BMI showed the strongest correlation with BCD (Mantel r = .20, p = .004). However, within West-NA transect and within SA transect, only geographical distance and climate PC1 remained significant after correcting for multiple tests (Table S8). To ask whether the correlations between body size measurements and BCD remain significant after controlling for the effect of geographical distance within each transect, we conducted Partial Mantel tests. Although climate PC1 remained the strongest predictor of BCD in the SA transect, BMI (Partial Mantel r = .19, p = .002) and body weight (Partial Mantel r = .18, p = .02) were the strongest predictors of BCD within the East-NA and West-NA transects after controlling for geographical distance, respectively (Table S9).
Together, these results suggest that geographical distance is significantly associated with differences in microbial composition. Differences in body size measurements (i.e., body weight and BMI) are also associated with overall differences in the gut microbial composition independent of geographical distance in the North American transects, but not in the South American transect.
3.2 |. Microbial measurements that show repeated associations with latitude and body weight in wild-caught mice
To identify specific microbial measurements that associate with latitude and host body size measurements (i.e., body weight and BMI), we evaluated a total of 58 microbial measurements. Microbial measurements included three alpha-diversity measurements (phylogenetic diversity, OTU counts and Shannon index) and relative abundances of seven bacterial phyla and 48 bacterial species that had an average relative abundance of >0.1% and were present in >50% of all the wild-caught individuals.
Among the 58 microbial measurements, 16 showed consistent associations with latitude across all three transects, and seven were significant after FDR correction (Table S10). For example, the relative abundances of four taxa in the family Ruminococcaceae including Ruminiclostridium and Oscillibacter were negatively correlated with latitude across all individuals (ρ = −0.30, p = .0001 and ρ = −0.32, p < .0001, respectively) and within each of the three transects (Fisher’s combined p < .000001). None of the seven phyla-level or alpha-diversity measurements showed significant associations with latitude based on Fisher’s combined p-value. However, although the trend is nonsignificant, the relative abundance of the phylum Bacteroidetes was negatively correlated with latitude (r = −.15, Fisher’s combined p = .09) and that of the phylum Cyanobacteria was positively correlated with latitude (r = −.26; Fisher’s combined p = .09) across all three transects (Table S10). There was also a nonsignificant trend towards a higher F/B ratio with increasing latitude among all wild-caught mice (ρ = .15, p = .051; Figure S3, Table S11), as observed in humans (Suzuki & Worobey, 2014). However, among the three transects, only the SA-transect showed a significant positive correlation between the F/B ratio and latitude (ρ = .25, p = .04; Figure S3, Table S11).
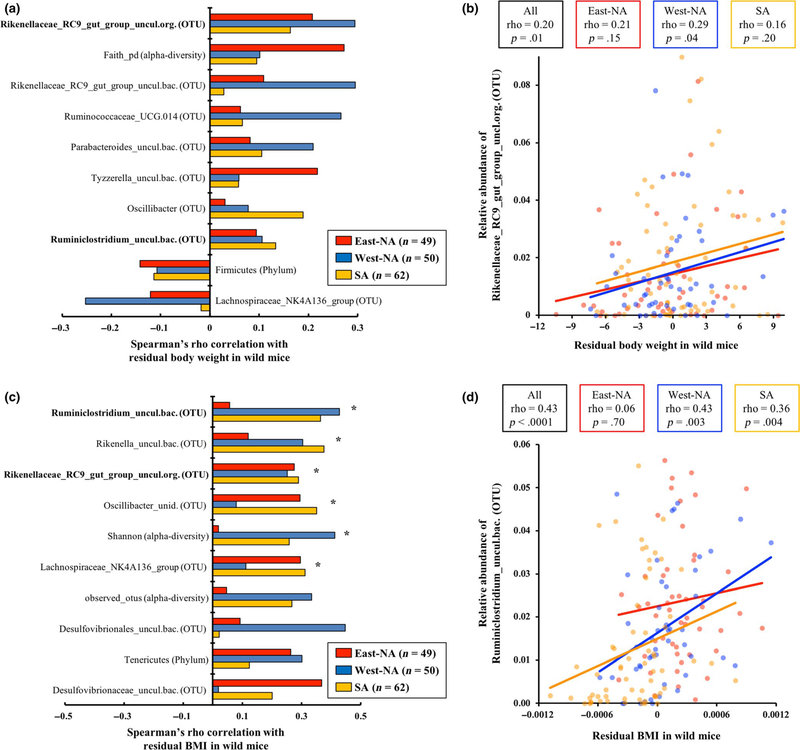
We next asked whether there are microbial measurements that show consistent associations with body weight and/or BMI after accounting for the effect of latitude using residual regression (Figure 2). Among the 58 microbial measurements tested, 15 showed consistent associations with body weight (Table S12) and 27 showed consistent associations with BMI (Table S13) across all three transects regardless of the significance. The relative abundances of uncultured taxa in “Rikenellaceae_RC9_gut_group” and an alpha-diversity measurement (i.e., phylogenetic diversity) were the top microbial measurements that positively correlated with body weight across all three transects based on raw Fisher’s combined p-values, although neither was significant after FDR correction (Figure 2a,b; Table S12). In contrast, six microbial measurements showed consistent correlations with BMI across all three transects and were significant after FDR correction (Figure 2c,d; Table S13). Ruminiclostridium and Rikenella were the top taxa that showed significant positive associations with BMI. Not surprisingly, microbial measurements that positively correlated with body weight also showed significant positive correlations with BMI, including “Rikenellaceae_RC9_gut_group” and alpha-diversity measurements (Figure 2c; Table S13). Associations between the F/B ratio and body mass were not significant in wild-caught mice with or without accounting for the effect of latitude (Table S11).
FIGURE 2.
Consistent correlations between microbial measurements and body size measurements in wild-caught individuals across all three transects. (a) The top 10 microbial measurements that are consistently associated with residual values between body weight and latitude across all transects based on Fisher’s combined p-value. Each bar represents the Spearman’s rho value from each of the three transects. Asterisks indicate significant Fisher’s combined p-value after FDR correction (p < .05). Taxa in bold show the same trend for both body weight and BMI. (b) Scatter plot of the top microbial taxon (Rikenellaceae_RC9_gut_group_uncl.org.) associated with residual body weight. Colours correspond to different transects. (c) The top 10 microbial measurements that are consistently associated with residual values between BMI and latitude across all transects based on Fisher’s combined p-value. Taxa in bold show the same trend for both body weight and BMI. (d) Scatter plot of the top microbial taxon (Ruminiclostridium_uncul.bac.) associated with residual BMI in wild mice. Species names of uncultured taxa are abbreviated (“uncul.bac.,” uncultured bacterium; and “uncul.org.,” uncultured organism). Full taxon names and Spearman p-values within and across transects are reported in Tables S12 and S13
3.3 |. Microbiota–body size links that persist in a common environment
The observed associations between body size measurements and microbial measurements (i.e., alpha-diversity and relative abundance of specific taxa) among wild-caught mice could be driven by unknown factors that differ among field sites such as environmental microbes, climate or aspects of diet that were not captured by the isotope analyses. To test whether the observed associations persist in a common environment, we collected live mice from three locations (New York, Florida and Brazil; Figure 1a) and generated 40 animals from each location by crossing wild-caught parents in a common laboratory environment. We measured body weight in laboratory-reared animals and found that population differences in body weight persisted in a common environment after one generation (Figure S4).
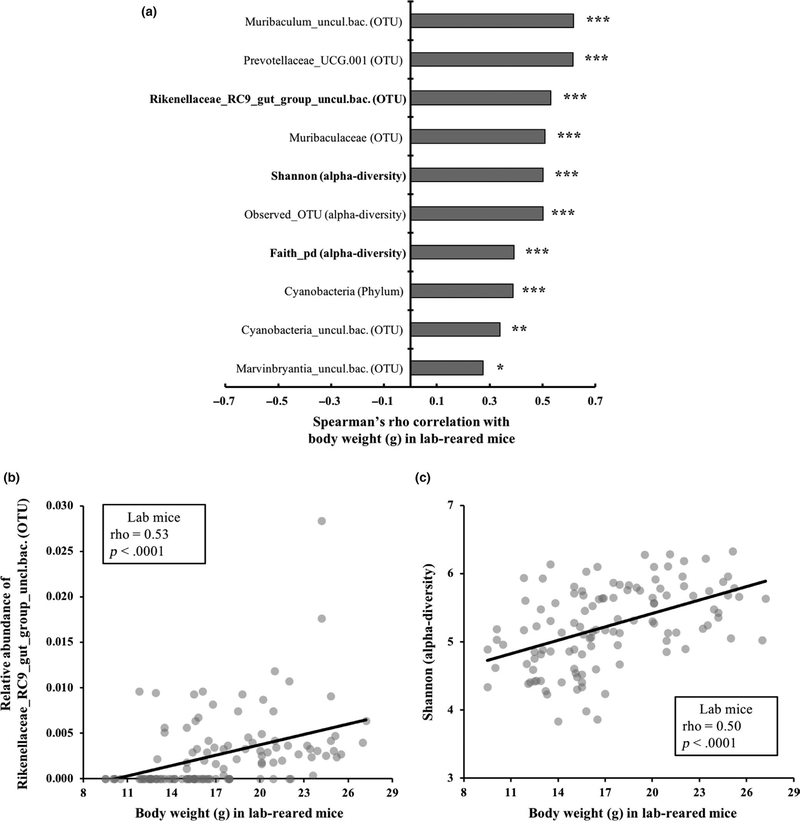
To test whether the links between microbial measurements and body weight also persisted in the common environment, we evaluated 59 microbial measurements. Microbial measurements included three alpha-diversity measurements (phylogenetic diversity, OTU counts and Shannon index) and seven bacterial phyla and 49 bacterial species that had an average relative abundance of >0.1% and were present in >50% of all the laboratory-reared individuals. We found a highly significant positive correlation between body weight and the relative abundance of “Rikenellaceae_RC9_gut_group” (ρ = 0.53, p = 9 × 10−10) in laboratory-reared individuals (Figure 3a,b), the same genus that positively correlated with body weight and BMI in the field (Figure 2a–c). Similarly, all three alpha-diversity measurements showed significant positive correlations with body weight in laboratory-reared individuals (Figure 3a,c), consistent with observations in the field (Figure 2a,c). The relative abundance of several microbial taxa that were not common in wild-caught individuals (i.e., <0.1% relative abundance and <50% prevalence) also showed significant correlations with body weight in laboratory-reared mice including Muribaculum (ρ = 0.62, p = 2 × 10−13) and Prevotellaceae_UCG.001 (ρ = 0.62, p = 2 × 10−13; Figure 3a; Table S14).
FIGURE 3.
Correlations between body weight and bacterial measurements in laboratory-reared individuals (n = 116). (a) The top 10 microbial measurements that showed significant correlations with body weight in laboratory-reared mice are shown based on Spearman’s rho p-value. Significance after FDR correction are indicated: *<.05, **<.01, ***<.001. Microbial measurements that show consistent correlations with body size measurements (body weight and/or BMI) in both wild-caught mice and laboratory-reared mice are in bold. (b) Scatter plot of the microbial taxon (Rikenellaceae_RC9_gut_group_uncl.bac.) in laboratory mice that showed consistent correlations with body size in wild mice. (c) Scatter plot of Shannon index in laboratory mice that showed consistent correlations with body size in wild mice. Full taxon names and the full list of microbial measurements that showed a significant association with body weight in laboratory-reared mice are reported in Table S14
3.4 |. Mice from colder environments produce greater amounts of faecal SCFAs
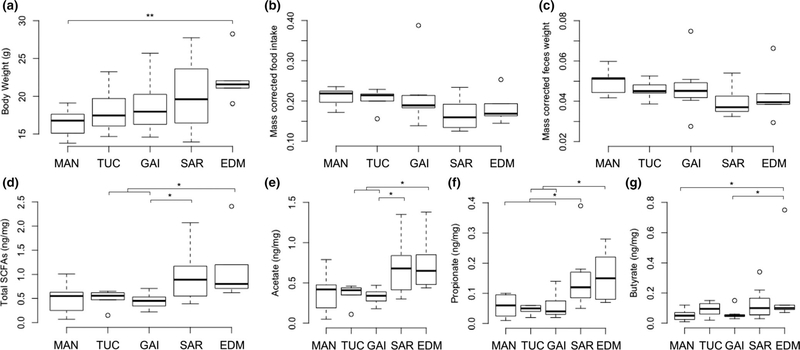
To test whether microbiome-derived energy sources are associated with differences in body mass, we measured amounts of faecal SCFAs in wild-derived inbred lines collected from five populations that vary in body mass: MAN, TUC, GAI, SAR and EDM (Figures 1a and 4a). We used five to seven males per population and measured body weight, food intake, faecal weight, and amount of SCFAs between 100 and 112 days of age in a common environment (Table S4). Laboratory-born mice showed significant clinal variation in body weight that increased from low- to high-latitude populations (ρ = 0.48, p = .005), consistent with our observations among wild-caught animals and consistent with Bergmann’s rule (Figure 4a). Food intake and faecal weight (corrected for body weight) did not differ among populations (Kruskal–Wallis test, p = .26 and p = .17, respectively; Figure 4b,c), but showed a trend towards a negative correlation with the latitude of the collection site (food intake; ρ = −0.33, p = .069 and faecal weight; ρ = −0.39, p = .026). The total amount of SCFAs per milligram of faeces differed among populations (Kruskal–Wallis test p = .033) and showed clinal variation with the latitude of the collection site (ρ = 0.50, p = .003) and with body weight (ρ = 0.41, p = .02; Figure 4d). Population differences in the total amount of SCFAs were not significant after accounting for differences in body weight by residual regression (Kruskal–Wallis test p = .18), suggesting that population differences in SCFAs are dependent on body weight differences despite the fact that there were no differences in food intake or faecal weight among populations (Figure 4b,c). In particular, the clinal variation in the total amount of SCFAs seems to be driven by the two northernmost populations (Figure 4d). This pattern was consistent among all three major SCFAs produced by bacteria: acetate (Figure 4e), propionate (Figure 4f) and butyrate (Figure 4g). The greater amount of total SCFAs, acetate and propionate observed in SAR mice compared to GAI mice is particularly interesting because the two populations are from the same East-NA transect (Figure 1a), and mice in this transect showed clinal variation in body mass (Table S6) and also showed correlations between body size measurements and beta-diversity of the gut microbiota (Tables S8 and S9).
FIGURE 4.
Differences in the amounts of faecal SCFAs among laboratory-reared populations. (a) Significant clinal variation of body weight persisted in laboratory-reared populations (ordered from MAN [Manaus, Brazil], TUC [Arizona, USA], GAI [Florida, USA], SAR [New York, USA] and EDM [Edmonton, Canada], ρ = 0.48, p = .005). (b) Average food intake (g per 24 hr) corrected for body weight (g) did not differ among populations (Kruskal–Wallis test, p = .26). (c) Average faeces weight (g per 24 hr) corrected for body weight (g) did not differ among populations (Kruskal–Wallis test, p = .17). (d) Total SCFAs (sum of acetate, propionate and butyrate, ng/mg) differed significantly among populations (Kruskal–Wallis test p = .033). The amount of (e) acetate (ng/mg) (f) propionate (ng/mg) varied among populations (Kruskal–Wallis test p < .05), but (g) butyrate (ng/mg) was not significant (Kruskal–Wallis test p = .086). Significant comparisons between populations are shown based on Wilcoxon tests: *p < .05, **p < .01
4 |. DISCUSSION
We studied variation in the gut microbiota of wild mice and laboratory-reared descendants of wild mice sampled across a large latitudinal range in North and South America. Several general patterns emerged. First, when all wild mice were analysed together, we observed a significant correlation between body size and latitude. This pattern was also seen among laboratory-born descendants of wild-caught mice. This example of Bergmann’s rule is striking because M. m. domesticus, which is native to Western Europe, has only been in the Americas for a short period of time (perhaps several hundred generations). Second, body size measurements (body weight and BMI) were significantly associated with overall differences in the microbiota (beta-diversity) when all wild mice were analysed together. However, the general patterns seen in the analysis of all mice together were not always recapitulated in the analyses of each of the transects separately. For example, patterns of body size variation in mice from Eastern North America and in South America conformed to Bergmann’s rule, but that was not the case in Western North America. Associations between body mass and variation in the microbiota were seen in both North American transects, but not in South America. Whether these differences among transects are biologically meaningful or simply reflect low power is unclear. Although we collected the majority of the animals in summer, differences in phenology and microclimates might contribute to some of the differences observed among transects. Longitudinal sampling and larger sample sizes from each location would help to clarify the causes of heterogeneity among transects.
We observed significant positive correlations between alpha-diversity and body size measurements across the three transects, and this pattern also persisted in the laboratory under a common environment. The common garden experiment suggests that the link between alpha-diversity and body size is not driven by the population differences in climate or diet. Previous studies have also demonstrated a positive correlation between body mass and alpha-diversity both within and between species of mammals (Godon, Arulazhagan, Steyer, & Hamelin, 2016; Nishida & Ochman, 2017; Suzuki et al., 2019; Weldon et al., 2015). The simplest explanation for this association is that larger animals possess a larger gut capacity, allowing for the colonization and persistence of a larger number of microbial taxa (Godon et al., 2016). However, in humans, obese individuals tend to have lower alpha-diversity compared to healthy controls (Le Chatelier et al., 2013; Turnbaugh et al., 2009; but see Finucane et al., 2014; Walters, Xu, & Knight, 2014), probably due to inflammation and the increase in abundance of a few pathogenic bacteria.
In contrast with previous work in humans (Suzuki & Worobey, 2014), we failed to find a significant positive correlation between the F/B ratio and either latitude or body mass (body weight or BMI) in mice (Table S11). The expectation that the F/B ratio might be associated with body mass was motivated by the observations in humans and also by studies in laboratory mice showing that a greater F/B ratio is associated with increased SCFAs and energy extraction (Turnbaugh et al., 2006). Intriguingly, the wild mice in this study showed a trend towards having a higher F/B ratio at higher latitudes, but this pattern was not significant and no correlation was seen between the F/B ratio and body size measurements. Thus, variation in the F/B ratio in these mice does not appear to be the cause of host body mass variation (Finucane et al., 2014; Walters et al., 2014).
We did identify bacterial taxa that showed associations with body size measurements in both wild-caught mice and laboratory-reared mice (Figures 2 and 3). Notably, taxa in the “Rikenellaceae_RC9_gut_group” were significantly associated with larger body size measurements in both wild and laboratory mice. Previous studies have shown an enrichment of the family Rikenellaceae in obese leptin-resistant mouse models compared to lean controls (Geurts et al., 2011) and the ability to produce SCFAs including propionate, acetate and/or succinate (Graf, 2014). Although little is known about the genus “Rikenellaceae_RC9_gut_group”, it has been suggested to play a role in lipid metabolism in mice (Zhou et al., 2018).
We also observed that a taxon in Ruminiclostridium was positively associated with BMI in the field and with body weight in the laboratory. All known members of the genus Ruminiclostridium can ferment cellulose and produce acetate as well as propionate and butyrate (Yutin & Galperin, 2013). The genus Ruminiclostridium, formally known as Clostridium thermocellum, has been positively associated with adiponectin and negatively associated with triglycerides, glucose and leptin in humans (Karlsson et al., 2013). Interestingly, the wild-derived inbred lines used in this study show population differences in the same metabolic markers where mice from colder climates tend to have higher levels of adiponectin and lower levels of triglycerides, glucose and leptin compared to mice from warmer climates (Phifer-Rixey et al., 2018). Although a link between Ruminiclostridium and BMI has not been observed in humans, the similar associations between Ruminiclostridium and host blood chemistry in both mice and humans are intriguing. However, whether Ruminiclostridium or any of the other taxa identified in this study play a causal role in energy extraction and host body mass variation remains unknown. Bacterial transplant experiments using germ-free models would help to identify the specific physiological effects of these taxa on host body mass variation.
Finally, we found greater concentrations of faecal SCFAs in wild-derived inbred mice collected from higher latitudes compared to mice collected from lower latitudes in a controlled laboratory environment. This observation cannot be explained by differences in food intake because there was no difference in food consumption or faecal weight in mice from higher latitudes relative to mice from lower latitudes. It is possible that larger mice produce more SCFAs per gram of faeces as a consequence of greater fermentation due to the longer retention time of food materials in the hindgut of larger animals compared to smaller animals (Hume, Morgan, & Kenagy, 1993). Obese and overweight individuals have been associated with increased amounts of SCFAs in mice (Murphy et al., 2010; Turnbaugh et al., 2006) and humans (Fernandes, Su, Rahat-Rozenbloom, Wolever, & Comelli, 2014; Rahat-Rozenbloom, Fernandes, Gloor, & Wolever, 2014; Schwiertz et al., 2010). Our results are consistent with the idea that the gut microbiota of mice living in colder environments produce greater amounts of SCFAs and may have an increased capacity to extract energy from a given diet. However, we cannot exclude the possibility that the observed differences in faecal SCFAs are due to differences in the ability of the host to absorb SCFAs in the large intestine. Quantifying the levels of SCFAs in the caecum and the physiological effects of the microbiome by conducting faecal transplant experiments using cold- and warm-adapted donors into common recipients will directly test the significance of population differences in SCFAs.
Together, our results show that wild house mice in the Americas (i) conform to Bergmann’s rule, (ii) show associations between microbial composition (beta-diversity) and body size measurements independent of geography, (iii) show associations between specific bacterial taxa and body size measurements in the laboratory and in the field, and (iv) harbour gut microbes producing more SFCAs in populations from colder environments. However, whether any of the observed differences in microbial taxa and SCFAs play a causal role in body size variation in wild house mice remains uncertain. Controlled transplant experiments of microbes in the laboratory will be needed to test the hypothesis that the gut microbiota plays a role in the pattern described by Bergmann’s rule, a fundamental pattern in ecology.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Nachman laboratory and two anonymous reviewers for valuable comments and discussions. This work was supported by NIH grants (R01 GM074245, R01 GM127468) to M.W.N. and an NSF grant (1501646) to T.A.S.
Funding information
NIH, Grant/Award Number: R01, GM074245 and R01 GM127468; NSF, Grant/Award Number: 1501646
Footnotes
DATA AVAILABILITY STATEMENT
Museum specimens (i.e., skins and skulls) and associated data (i.e., location, body size measurements) have been deposited in the Museum of Vertebrate Zoology at the University of California, Berkeley and uploaded to a public database, ARCTOS (accession nos. MVZ:Mamm:230441–MVZ:Mamm:231430). Individual stable isotope data and specimen IDs are available in Tables S1 and S2. 16S rRNA gene sequence data are available in the European Molecular Biology Laboratory, European Nucleotide Archive (ENA) database (accession nos. PRJEB38188 and PRJEB32701).
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section.
REFERENCES
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, … Bork P (2011). Enterotypes of the human gut microbiome. Nature, 473(7346), 174–180. 10.1038/nature09944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashton KG, Tracy MC, & De Queiroz A (2000). Is Bergmann’s rule valid for mammals? American Naturalist, 156(4), 390–415. 10.1086/303400 [DOI] [PubMed] [Google Scholar]
- Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, … Gordon JI (2004). The gut microbiota as an environmental factor that regulates fat storage. Proceedings of the National Academy of Sciences of the United States of America, 101(44), 15718–15723. 10.1073/pnas.0407076101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett SA, & Dickson RG (1989). Wild mice in the cold: Some findings on adaptation. Biological Reviews of the Cambridge Philosophical Society, 64(4), 317–340. 10.1111/j.1469-185X.1989.tb00679.x [DOI] [PubMed] [Google Scholar]
- Bergmann C (1847). Uber die verhaltnisse der warmeokonomie der thiere zu ihrer grosse. Göttinger Studien, 1, 595–708. [Google Scholar]
- Blackburn TM, Gaston KJ, & Loder N (1999). Geographic gradients in body size: A clarification of Bergmann’s rule. Diversity and Distributions, 5(4), 165–174. 10.1046/j.1472-4642.1999.00046.x [DOI] [Google Scholar]
- Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, … Caporaso JG (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature Biotechnology, 37(8), 852–857. 10.1038/s41587-019-0209-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, & Holmes SP (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nature Methods, 13(7), 581–583. 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, … Knight R (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. The ISME Journal, 6(8), 1621–1624. 10.1038/ismej.2012.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao MC, Everard A, Aron-Wisnewsky J, Sokolovska N, Prifti E, Verger EO, … Clément K (2016). Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: Relationship with gut microbiome richness and ecology. Gut, 65(3), 426–436. 10.1136/gutjnl-2014-308778 [DOI] [PubMed] [Google Scholar]
- De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, … Lionetti P (2010). Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proceedings of the National Academy of Sciences of the United States of America, 107(33), 14691–14696. 10.1073/pnas.1005963107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC (2018). Updating the 97% identity threshold for 16S ribosomal RNA OTUs. Bioinformatics, 34(14), 2371–2375. 10.1093/bioinformatics/bty113 [DOI] [PubMed] [Google Scholar]
- Faith DP (1992). Conservation evaluation and phylogenetic diversity. Biological Conservation, 61(1), 1–10. 10.1016/0006-3207(92)91201-3 [DOI] [Google Scholar]
- Fernandes J, Su W, Rahat-Rozenbloom S, Wolever TMS, & Comelli EM (2014). Adiposity, gut microbiota and faecal short chain fatty acids are linked in adult humans. Nutrition & Diabetes, 4(6), e121. 10.1038/nutd.2014.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finucane MM, Sharpton TJ, Laurent TJ, & Pollard KS (2014). A taxonomic signature of obesity in the microbiome? Getting to the guts of the matter. PLoS One, 9(1), 1–5. 10.1371/journal.pone.0084689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA (1932). Statistical methods for research workers, Edinburgh and London:. Oliver and Boyd. [Google Scholar]
- Foster F, & Collard M (2013). A reassessment of Bergmann’s rule in modern humans. PLoS One, 8(8), e72269. 10.1371/journal.pone.0072269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts L, Lazarevic V, Derrien M, Everard A, Van Roye M, Knauf C, … Cani PD (2011). Altered gut microbiota and endocannabinoid system tone in obese and diabetic leptin-resistant mice: Impact on apelin regulation in adipose tissue. Frontiers in Microbiology, 2, 149. 10.3389/fmicb.2011.00149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godon JJ, Arulazhagan P, Steyer JP, & Hamelin J (2016). Vertebrate bacterial gut diversity: Size also matters. BMC Ecology, 16(1), 1–9. 10.1186/s12898-016-0071-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, … Ley RE (2014). Human genetics shape the gut microbiome. Cell, 159(4), 789–799. 10.1016/j.cell.2014.09.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf J (2014). The family Rikenellaceae. In Eugene R, Stephen DEFL, Erko S, & Fabiano T (Eds.), The prokaryotes (pp. 857–859). Berlin Heidelberg: Springer-Verlag. [Google Scholar]
- Hansen J, Gulati A, & Sartor RB (2010). The role of mucosal immunity and host genetics in defining intestinal commensal bacteria. Current Opinion in Gastroenterology, 26(6), 564–571. 10.1097/MOG.0b013e32833f1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijmans RJ, Cameron SE, Parra JL, Jones G, & Jarvis A (2005). Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology, 25, 1965–1978. 10.1002/joc.1276 [DOI] [Google Scholar]
- Hume ID, Morgan KR, & Kenagy GJ (1993). Digesta retention and digestive performance in sciurid and microtine rodents: Effects of hindgut morphology and body size. Physiological Zoology, 66(3), 396–411. 10.1086/physzool.66.3.30163700 [DOI] [Google Scholar]
- Jumpertz R, Le DS, Turnbaugh PJ, Trinidad C, Bogardus C, Gordon JI, & Krakoff J (2011). Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. The American Journal of Clinical Nutrition, 94(1), 58–65. 10.3945/ajcn.110.010132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson FH, Tremaroli V, Nookaew I, Bergström G, Behre CJ, Fagerberg B, … Bäckhed F (2013). Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature, 498(7452), 99–103. 10.1038/nature12198 [DOI] [PubMed] [Google Scholar]
- Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, … Pedersen O (2013). Richness of human gut microbiome correlates with metabolic markers. Nature, 500(7464), 541–546. 10.1038/nature12506 [DOI] [PubMed] [Google Scholar]
- Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, & Gordon JI (2005). Obesity alters gut microbial ecology. Proceedings of the National Academy of Sciences of the United States of America, 102(31), 11070–11075. 10.1073/pnas.0504978102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, & Gordon JI (2006). Microbial ecology: Human gut microbes associated with obesity. Nature, 444(7122), 1022–1023. 10.1038/4441022a [DOI] [PubMed] [Google Scholar]
- Linnenbrink M, Wang J, Hardouin EA, Künzel S, Metzler D, & Baines JF (2013). The role of biogeography in shaping diversity of the intestinal microbiota in house mice. Molecular Ecology, 22, 1904–1916. 10.1111/mec.12206 [DOI] [PubMed] [Google Scholar]
- Lynch C (1992). Clinal variation in cold adaptation in Mus domesticus: Verification of predictions from laboratory populations. American Naturalist, 139(6), 1219–1236. 10.1086/285383 [DOI] [Google Scholar]
- Mariat D, Firmesse O, Levenez F, Guimarăes V, Sokol H, Doré J, … Furet J-P (2009). The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiology, 9, 123. 10.1186/1471-2180-9-123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, Domazet-Lošo T, Douglas AE, … Wernegreen JJ (2013). Animals in a bacterial world, a new imperative for the life sciences. Proceedings of the National Academy of Sciences, 110(9), 3229–3236. 10.1073/pnas.1218525110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller AH, Suzuki TA, Phifer-Rixey M, & Nachman MW (2018). Transmission modes of the mammalian gut microbiota. Science, 362(6413), 453–457. 10.1126/science.aat7164 [DOI] [PubMed] [Google Scholar]
- Moreau NM, Goupry SM, Antignac JP, Monteau FJ, Le Bizec BJ, Champ MM, … Dumon HJ (2003). Simultaneous measurement of plasma concentrations and 13C-enrichment of short-chain fatty acids, lactic acid and ketone bodies by gas chromatography coupled to mass spectrometry. Journal of Chromatography B: Analytical Technologies in the Biomedical and Life Sciences, 784(2), 395–403. 10.1016/S1570-0232(02)00827–9 [DOI] [PubMed] [Google Scholar]
- Murphy EF, Cotter PD, Healy S, Marques TM, O’Sullivan O, Fouhy F, … Shanahan F (2010). Composition and energy harvesting capacity of the gut microbiota: Relationship to diet, obesity and time in mouse models. Gut, 59(12), 1635–1642. 10.1136/gut.2010.215665 [DOI] [PubMed] [Google Scholar]
- Nishida AH, & Ochman H (2017). Rates of Gut Microbiome Divergence in Mammals. Molecular Ecology, 12(10), 3218–3221. 10.1111/mec.14473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phifer-Rixey M, Bi K, Ferris KG, Sheehan MJ, Lin D, Mack KL, … Nachman MW (2018). The genomic basis of environmental adaptation in house mice. PLoS Genetics, 14(9), e1007672. 10.1371/journal.pgen.1007672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phifer-Rixey M, & Nachman MW (2015). Insights into mammalian biology from the wild house mouse Mus musculus. eLife, 4, e05959. 10.7554/eLife.05959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, & Arkin AP (2009). Fasttree: Computing large minimum evolution trees with profiles instead of a distance matrix. Molecular Biology and Evolution, 26(7), 1641–1650. 10.1093/molbev/msp077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, … Glöckner FO (2013). The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Research, 41(D1), 590–596. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahat-Rozenbloom S, Fernandes J, Gloor GB, & Wolever TMS (2014). Evidence for greater production of colonic short-chain fatty acids in overweight than lean humans. International Journal of Obesity, 38(12), 1525–1531. 10.1038/ijo.2014.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson AJ (1989). Simultaneous determination of volatile and non-volatile acidic ferment. Letters in Applied Microbiology, 9, 5–8. [Google Scholar]
- Roberts DF (1953). Body weight, race and climate. American Journal of Physical Anthropology, 11(4), 533–558. 10.1002/ajpa.1330110404 [DOI] [PubMed] [Google Scholar]
- Rosshart SP, Vassallo BG, Angeletti D, Hutchinson DS, Morgan AP, Takeda K, … Rehermann B (2017). Wild mouse gut microbiota promotes host fitness and improves disease resistance. Cell, 171(5), 1015–1028.e13. 10.1016/j.cell.2017.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwiertz A, Taras D, Schäfer K, Beijer S, Bos NA, Donus C, & Hardt PD (2010). Microbiota and SCFA in lean and overweight healthy subjects. Obesity, 18(1), 190–195. 10.1038/oby.2009.167 [DOI] [PubMed] [Google Scholar]
- Suzuki TA (2017). Links between natural variation in the microbiome and host fitness in wild mammals. Integrative and Comparative Biology, 57(4), 756–769. 10.1093/icb/icx104 [DOI] [PubMed] [Google Scholar]
- Suzuki TA, Martins FM, & Nachman MW (2018). Altitudinal variation of the gut microbiota in wild house mice. Molecular Ecology, 28(9), 2378–2390. 10.1111/mec.14905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki TA, & Nachman MW (2016). Spatial heterogeneity of gut microbial composition along the gastrointestinal tract in natural populations of house mice. PLoS One, 11(9), 1–15. 10.1371/journal.pone.0163720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki TA, Phifer-Rixey M, Mack KL, Sheehan MJ, Lin D, Bi K, & Nachman MW (2019). Host genetic determinants of the gut microbiota of wild mice. Molecular Ecology, 28(13), 3197–3207. 10.1111/mec.15139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki TA, & Worobey M (2014). Geographical variation of human gut microbial composition. Biology Letters, 10(2), 20131037. 10.1098/rsbl.2013.1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, … Gordon JI (2009). A core gut microbiome in obese and lean twins. Nature, 457(7228), 480–484. 10.1038/nature07540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, & Gordon JI (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature, 444(7122), 1027–1031. 10.1038/nature05414 [DOI] [PubMed] [Google Scholar]
- Walters WA, Xu Z, & Knight R (2014). Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Letters, 588(22), 4223–4233. 10.1016/j.febslet.2014.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weldon L, Abolins S, Lenzi L, Bourne C, Riley EM, & Viney M (2015). The gut microbiota of wild mice. PLoS One, 10(8), 1–15. 10.1371/journal.pone.0134643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun Y, Kim H-N, Kim SE, Heo SG, Chang Y, Ryu S, … Kim H-L (2017). Comparative analysis of gut microbiota associated with body mass index in a large Korean cohort. BMC Microbiology, 17(1), 151. 10.1186/s12866-017-1052-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N, & Galperin MY (2013). A genomic update on clostridial phylogeny: Gram-negative spore formers and other misplaced clostridia. Environmental Microbiology, 15(10), 2631–2641. 10.1111/1462-2920.12173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Xiao X, Zhang Q, Zheng J, Li M, Yu M, … Li R (2018). Improved glucose and lipid metabolism in the early life of female offspring by maternal dietary genistein is associated with alterations in the gut microbiota. Frontiers in Endocrinology, 9, 516. 10.3389/fendo.2018.00516 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.