Abstract
Epigenetic modifications are emerging as major players in the pathogenesis of neurodegenerative disorders and susceptibility to acute brain injury. DNA and histone modifications act together with noncoding RNAs to form a complex gene expression machinery that adapts the brain to environmental stressors and injury response. These modifications influence cell-level operations like neurogenesis and DNA repair to large, intricate processes such as brain patterning, memory formation, motor function and cognition. Thus, epigenetic imbalance has been shown to influence the progression of many neurological disorders independent of aberrations in the genetic code. This review aims to highlight ways in which epigenetics applies to several commonly researched neurodegenerative diseases and forms of acute brain injury as well as shed light on the benefits of epigenetics-based treatments.
Keywords: Alzheimer’s disease, Parkinson’s disease, Huntington’s disease, stroke, cerebral ischemia, traumatic brain injury
INTRODUCTION
Epigenetics are regulatory mechanisms that modulate gene expression without changing the genetic code. Epigenetics represent interactions between genes and the environment providing a connection between nutrition, toxins, medications, stress and cellular physiology.1–3 The concept of epigenetics was first described by Conrad Waddington in 1940’s with DNA methylation and has since been shown to encompass transcriptional regulation by histone modifications and long noncoding RNAs (lncRNAs) as well as post-transcriptional regulation by microRNAs (miRNAs).4–8 Epigenetic alterations are involved in neurodevelopmental processes such as brain patterning, neural stem cell maintenance and neurogenesis and has been implicated in many diseases of the brain.9 Epigenetics can significantly increase our understanding of the molecular mechanisms that contribute to brain damage as well as identify targets for efficient therapeutic targeting to promote neuronal survival. This review discusses the epigenetic targets for both chronic and acute conditions that lead to significant neuronal death and neurological dysfunction including Alzheimer’s Disease (AD), Parkinson’s Disease (PD), Huntington’s Disease (HD), epilepsy, stroke and traumatic brain injury (TBI).
EPIGENETIC MECHANISMS
DNA Methylation:
DNA methylation (addition of a methyl group to cytosine to form 5-methylcytosine; 5-mC) is the most studied epigenetic mechanism implicated in gene regulation.10, 11 This usually occurs in stretches of dense CG dinucleotide repeats known as CpG islands that when methylated often lead to gene silencing by interfering with the ability of transcription factors to bind (Fig. 1).3, 12, 13 DNA methylation is mediated by a family of DNA methyltransferases (DNMTs). Methyl groups donated by S-adenosyl-methionine (SAM) are added to CpG islands by DNMT3a and DNMT3b and maintained throughout successive cell generations by DNMT1.6, 14–16 DNMTs are highly expressed in the embryonic nervous system as well as post-mitotic neurons and glia where they facilitate synaptic plasticity, long-term potentiation and DNA repair.9, 10, 17, 18 In addition, methyl-CpG binding domain proteins (MBD) like MeCP2 can be recruited to 5-mC and play a role in gene regulation by mediating histone modifications and gene silencing.10, 19–21 The CNS shows the highest prevalence of DNA methylation of all organs which is thought to be involved in neurodevelopment, cognitive processes and aging.6, 22 In recent years, it has been shown that 5-mC patterning is strongly associated with aging and mortality.23 Thus, DNA methylation age (DNAm age) may be used as an estimation of biological age, a measure of an individual’s physiological health.24, 25 DNAm age has been proposed as a biomarker for predicting aging-associated brain disorders such as cognitive decline, dementia and AD.23, 26
Fig. 1: DNA methylation and hydroxymethylation.
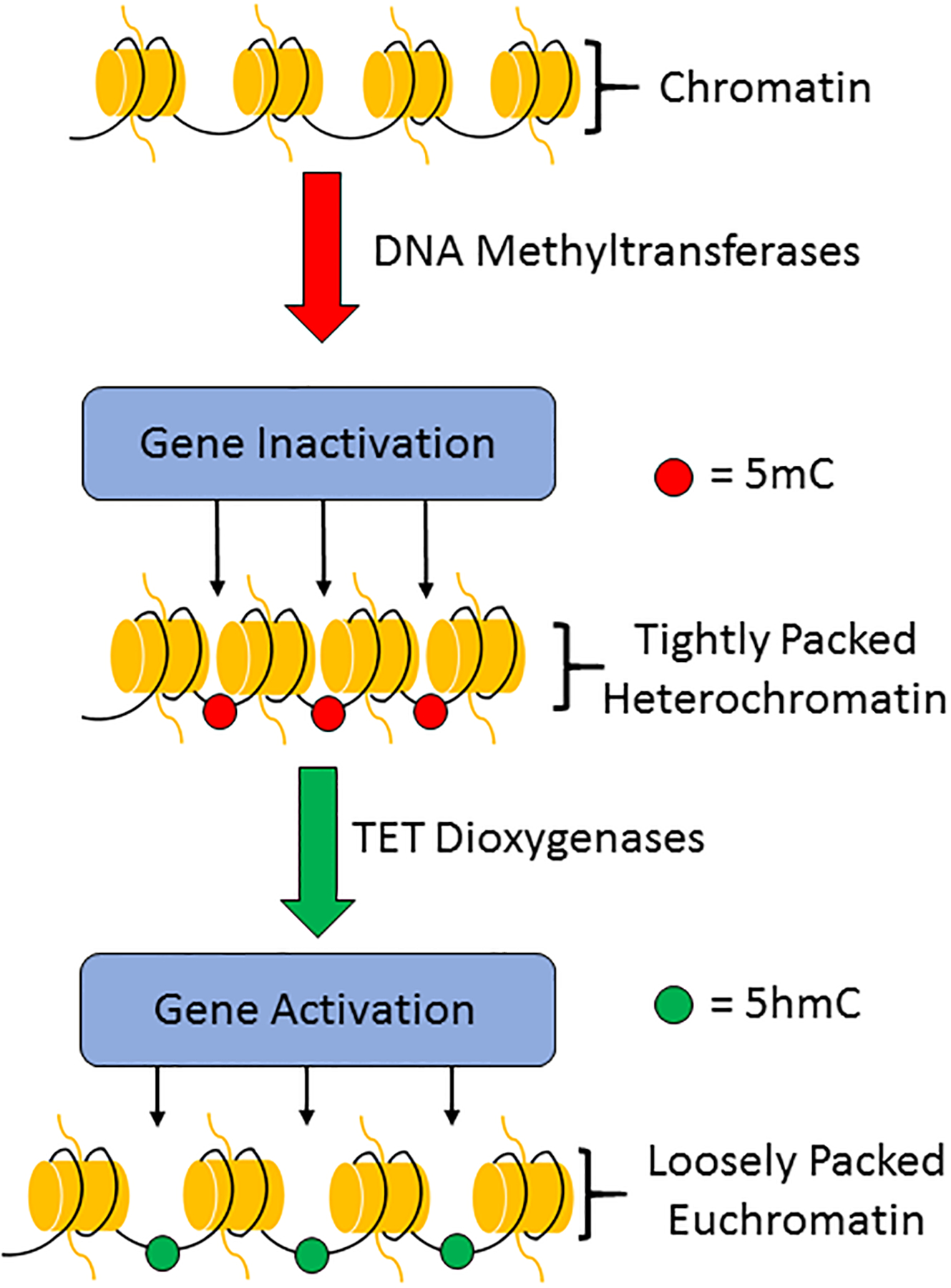
DNA methylation occurs through the addition of a methyl (CH3) group to the cytosine of DNA by DNA methyltransferases (DNMTs) to produce 5-methylcytosine (5-mC). DNA methylation leads to densely packed heterochromatin that is consistent with gene inactivation. 5-mC can subsequently be converted to 5-hydroxymethylcytosine (5-hmC) by the ten-eleven translocation (TET) dioxygenases. Hydroxymethylation loosens chromatin to promote gene activation.
DNA hydroxymethylation:
The family of ten-eleven translocase dioxygenases (TETs) oxidize 5-mC to 5-hydroxymethylcytosine (5-hmC).27–29 Similar to 5-mC, 5-hmC is also highly enriched in the brain where it is predominantly found in neurons.30, 31 Contrary to 5-mC, 5-hmC is often concentrated at euchromatin and is associated with transcriptional activation.32–34 The TET enzymes are recruited to methylated CpGs where they have been shown to inhibit methyltransferase interaction, hinder MeCP2, and promote demethylation by further oxidizing 5-hmC to yield an unmethylated DNA through base excision repair.35–38 In the CNS, 5-hmC plays a role in DNA repair, synaptic plasticity and neuronal aging.39
Histone modifications:
Nucleosomes, the basic units of chromatin, consist of DNA wrapped around histones that are essential for DNA structure. Histone organization can influence gene expression by affecting the architecture of promoters and the availability of DNA to transcription factors.10, 40, 41 Histones can undergo various epigenetic modifications including methylation, acetylation, ubiquitination, SUMOylation, citrullination and ADP-ribosylation.12 Of these, the roles of histone methylation and acetylation in CNS disorders have been studied in detail. Histone methyltransferases (HMTs) catalyze methylation using SAM as a donor on the amino acid side-chains of histones 3 and 4 (H3 and H4).6, 42 The degree, symmetry, and location of histone methylation dictates whether a gene is going to be expressed or suppressed (Fig. 2).43,44 Methylated histones do not change the overall shape of chromatin, but facilitate the recruitment of additional proteins that regulate gene expression.6, 42 Dysregulation of histone methylation has been linked to brain aging and neurological diseases.45 Histones are acetylated by histone acetyltransferases (HATs) which results in gene activation due to electrostatic reduction between histones and DNA leading to a relaxed state euchromatin (Fig. 3).6, 46–49 Histone deacetylation mediated by histone deacetylases (HDACs) and sirtuin deacetylases (SIRTs) lead to tightly wound chromatin and suppression of gene expression.6, 47, 48 HDACs are implicated in axon growth, oxidative stress, synaptic plasticity and cognition.50–57 Overall, epigenetic mechanisms that include DNA methylation and histone modifications, together with other proteins like MBDs, regulate gene expression in a highly dynamic manner in normal physiologic states and pathologic conditions.9, 16
Fig. 2: Histone methylation.
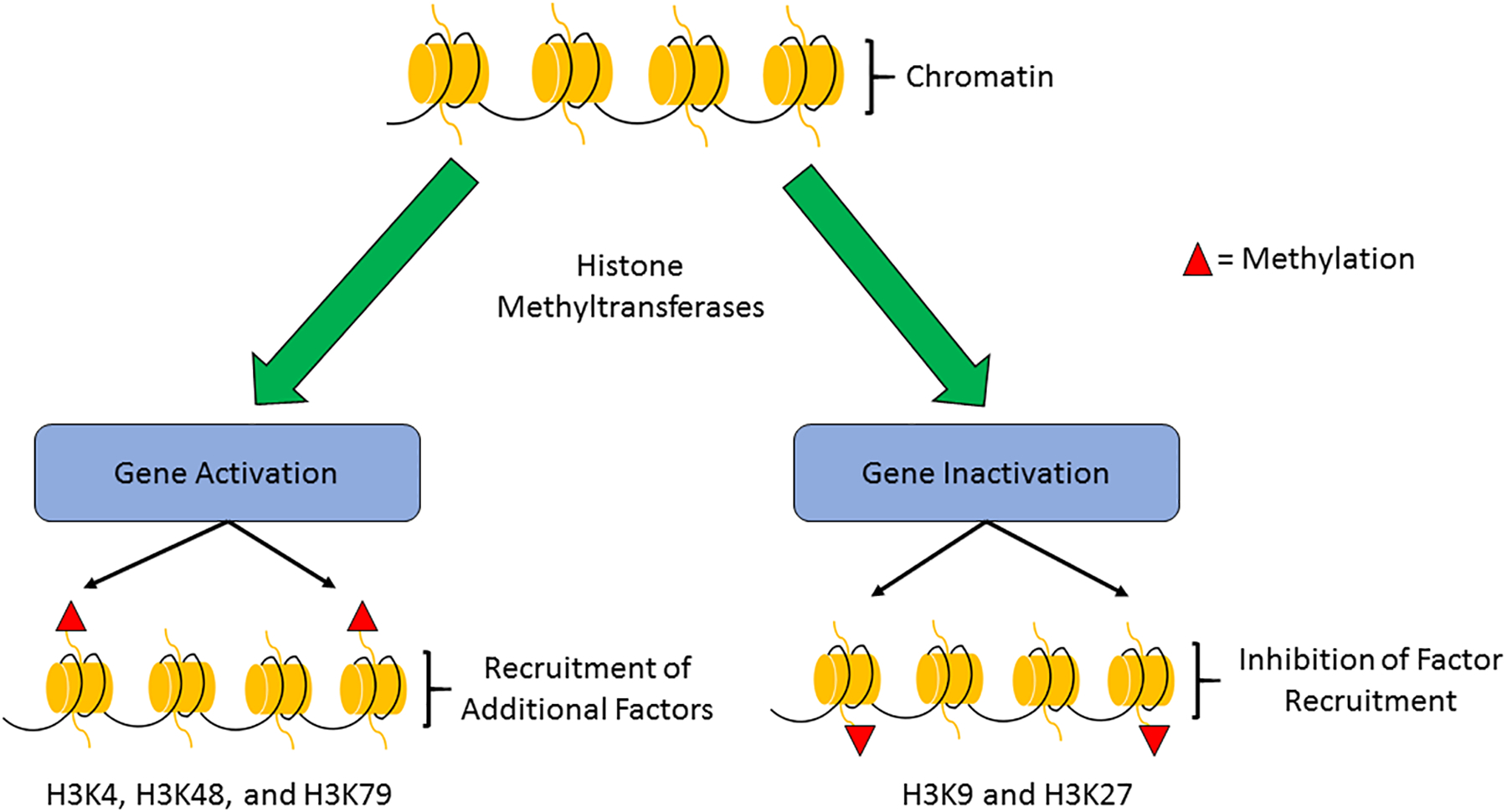
Histone methylation is the addition of methyl (CH3) groups to arginine and lysine side chains by histone methyltransferases (HMTs). The effect of histone methylation on gene transcription is dependent on the location and degree of methylation. For example, methylation of histone 3, lysine residue 4 (H3K4) is an activating mark whereas H3K9 is a deactivating mark.
Fig.3: Histone acetylation.
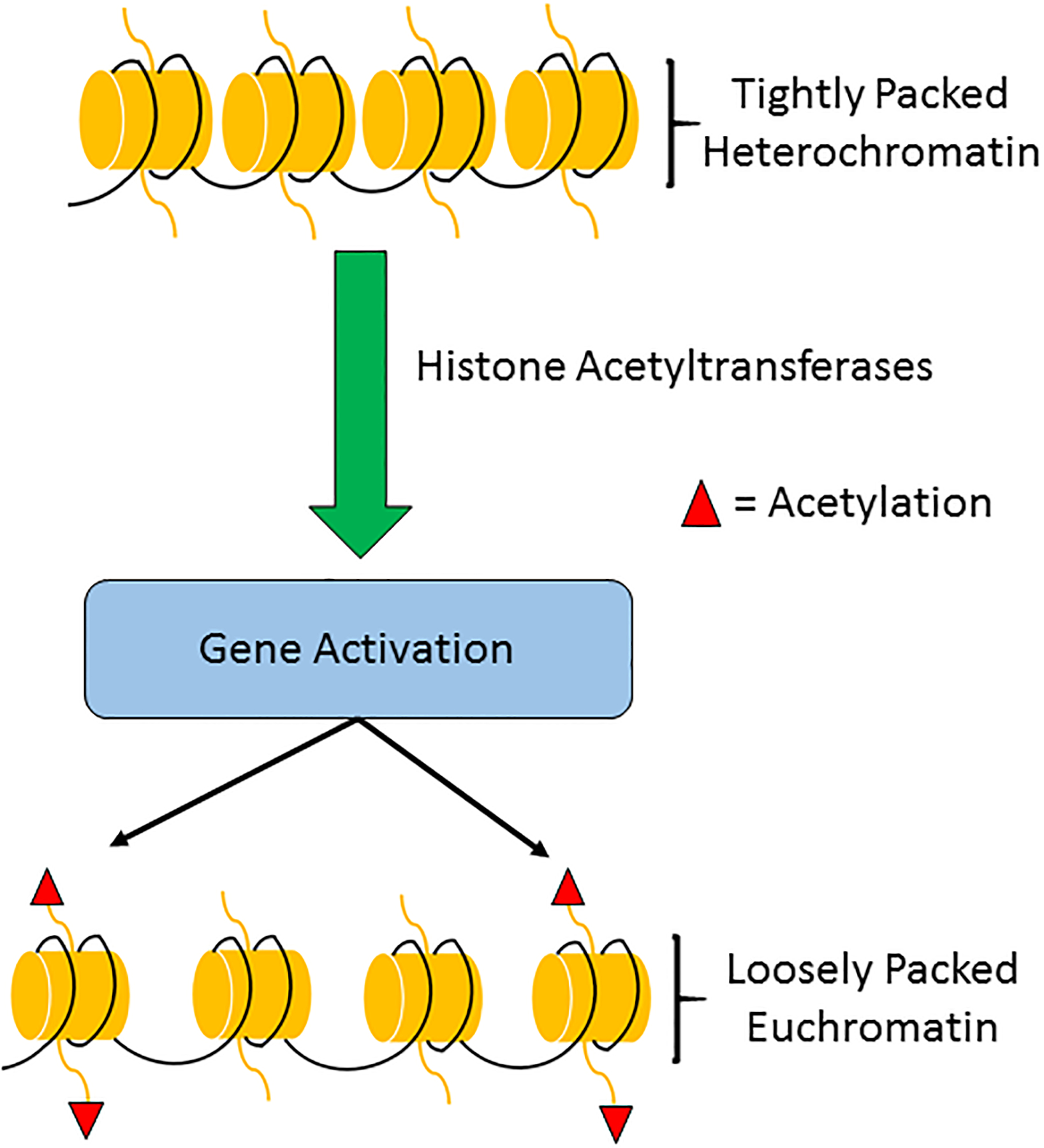
Histone acetylation occurs with the addition of acetyl (CH3CO) groups to lysine side chains by histone acetyltransferases (HATs). Acetylation reduces the steric clash between histones and DNA, opening up chromatin for gene transcription. Histone deacetylases (HDACs) reverse this modification and repress gene transcription.
Non-Coding RNA:
Non-coding RNAs (ncRNAs) are functional RNA molecules that are not translated into proteins, and instead regulate the expression of genes at the transcriptional or post-transcriptional level. The epigenetic-related ncRNAs include micro, circular, short-interfering, PIWI-interacting, and long non-coding RNA, among others.51 Of these ncRNAs, microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) are the most studied in relation to their epigenetic roles in diseases of the brain.
MicroRNA (miRNA) act post-transcriptionally on messenger RNA (mRNA), binding to their 3’ untranslated region (3’ UTR) and regulating gene expression by degradation or silencing of transcripts.7, 52–55 These 20–25 nucleotide long species, of which over 2000 have been classified, undergo splicing by Drosha and Dicer56 before individually acting on as many as 1000 target genes.9, 53 The ability of a single miRNA to act upon multiple genes produces an incredibly complex epigenetic environment, providing many therapeutic opportunities for human disease. In the CNS, miRNAs are important for neuronal signaling, synaptic plasticity and neurorepair mechanisms.57–59
Long non-coding RNA (lncRNA), RNA transcripts containing more than 200 nucleotides, are highly expressed in the human brain.60 While the biological function of most lncRNAs remains to be elucidated, lncRNAs have been shown to play a role within chromatin remodeling, often guiding other modifying proteins to specific histones or gene sites and thereby influencing gene expression.8, 45, 61, 62 In addition, lncRNA may come in the form of antisense transcripts, functionally masking genes and preventing degradation of their sequences by miRNA.61, 63 LncRNAs are important in normal brain development and function, while aberrant lncRNA expression has been implicated in neurological disorders.64–69
EPIGENETICS AND NEURODEGENERATIVE DISEASES
Alzheimer’s disease
Alzheimer’s disease (AD), the most common cause of dementia is a progressive neurodegenerative disease marked by the aggregation of proteins amyloid-beta (Aβ42) and phosphorylated tau leading to extracellular plaques and intracellular tangles in the brain.56, 70, 71 These plaques and tangles are accompanied by neuronal loss, dysregulation of microtubule assembly, apoptosis, and brain atrophy.56, 72, 73 Interestingly, <5% of AD cases are early onset/familial and can be accounted for by common variants.56, 70, 74 This indicates the possible mediation of epigenetics in the pathogenesis of AD.
The amyloid-β plaques fundamental to AD pathogenesis are caused by the dysregulation of the amyloid precursor protein gene (APP).75–77 Post-mortem studies with brain tissue from humans that died of natural causes showed that 13 cytosine residues in the promoter region of APP are differentially methylated with age.78 Additionally, those patients older than 70 showed ~50% reduction in methylation at these cytosines.78 Increased methylation of APP in AD subjects has been shown to increase APP expression leading to aggregation of the neurotoxic Aβ42 indicating the importance of differential methylation patterning in AD pathogenesis.79
Presilin1 (PS1) and β-secretase (BACE) are integral in the processing of APP and their dysregulation leads to the aberrant Aβ42 plaques observed in AD.80 PS1 and BACE methylation is highly dependent on SAM as methyl donor and severe decrease in SAM levels correlates with AD.81 In neuroblastoma cell lines, vitamin B12 and folate deprivation induces PS1 and BACE that are reversed by SAM.82, 83 In transgenic mice that overexpress human APP and display Aβ plaque deposition, SAM supplementation reduced the activity of β- and γ-secretase, decreased Aβ production and plaques, restored normal levels of tau phosphorylation and improved spatial memory.84, 85 These studies are supported by a clinical trial where increased plasma levels of SAM were correlated with decreased Aβ−40 and PS1 mRNA levels in newly diagnosed AD patients.86
Apolipoprotein E (APOE) regulates Aβ42 clearance and hence it is considered as an important risk factor for late-onset Alzheimer’s disease (LOAD).87 APOE has a non-classical (CpG-poor) promoter and hence its regulation is complex.88 The APOE haplotypes ε2–4 has been shown to be differentially correlated with LOAD risk.88 For example, APOE4 confers more risk than APOE3, but not all APOE4 carriers develop LOAD and many LOAD patients are not carriers of APOE4.87, 89–91 Furthermore, significant hypomethylation of the 2 APOE CpG sub-regions in the frontal lobe of Lewy body dementia and AD patients have been identified in post-mortem brain studies.92, 93 These studies indicate a role for methylation of the APOE promoter as an epigenetic regulator of LOAD.
Histone acetylation levels were reported to be markedly decreased in both AD transgenic mice and AD human brains.94, 95 In AD transgenic mice, HDAC2 was shown to be induced and treatment with HDAC2-specific inhibitor suberoylanilide hydroxamic acid (SAHA) improved learning and memory.96, 97 Treatment with another HDAC inhibitor sodium 4-phenylbutyrate (4-PBA) also decreased the number of phosphorylated tau tangles and enhanced cognitive function in transgenic AD mice.98 Furthermore, HDAC6 knockout mice showed improved learning and memory and protection against Aβ42-induced disruption of mitochondrial trafficking, which is related to amyloid pathology.99, 100
The majority of identified AD-related miRNAs are involved in regulation of APP. Bioinformatics analyses have revealed several putative APP 3’UTR miRNA binding sites that have been validated in vitro. MiR-16 and miR-101 were shown to target APP and reduce Aβ-induced cytotoxicity in both PC12 cells and hippocampal neurons.101–103 In human cell lines, several miRNAs including miR-106, miR-520c, miR-20a, miR-17–5p, miR-106b, miR-17, miR-153, miR-147, miR-644, miR-655, miR-323–3p and miR-20a have been shown to bind to APP and repress APP expression.104–106 MiR-195, miR-339–5p and miR-107 are reduced in the brain tissue of AD disease patients and have also been shown to directly target and reduce the APP processor BACE1 in human cell lines and mouse cell culture studies.107–110 Furthermore, overexpression of miR-195 was shown to reduce Aβ toxicity in neuroblastoma cells.107 BACE1 is also targeted by miR-485–5p and miR-485–5p overexpression returned BACE1 to its non-pathological levels in HEK293T cells.61
Several studies have also identified the role of miRNAs in APP regulation both in vitro and in vivo. For example, the miR-29 family (miR29a, −29b, −29c) is downregulated in AD brains and has been shown to target the 3’-UTR of the APP processor BACE1 in human and mouse cell lines.111–113 In vitro, suppression of miR-29a and miR-29b in human cells increases production of Aβ.111 Hippocampal injection of miR-29c mimic in SAMP8 mice decreased Aβ and improved learning and memory compared to untreated control mice.112 In APP/PS1 mice, reductions in miR-135, miR-200b and miR-429 were observed in the hippocampus. MiR-200b and miR-429 were shown to target APP and reduce APP expression while miR-135 targeted and decreased BACE1.114 MiR-124 is also a negative regulator of BACE1 and lentiviral overexpression of miR-124 in the dentate gyrus of APP/PS1 mice reduced of apoptotic and autophagy markers and ameliorated cognitive deficits.115
The miR-132/212 cluster has been shown to be downregulated in tauopathy-related diseases including AD.116, 117 Knockout of miR-132/212 in mice led to an increase in tau expression, phosphorylation and aggregation.118 Another study showed that miR-132/212 knockout mice display significant deficits in cognitive function.119 MiR-132 was shown to directly target and decrease tau mRNA and treatment with a miR-132 mimic restored tau and improved memory function in 3xTg-AD mice.118 Downregulation of miR-132 was most significantly observed in neurons showing hyper-phosphorylation of tau in the brains of late stage AD patients.120, 121 Furthermore miR-132 downregulation has been shown to correlate with cognitive decline in patients with AD.118 MiR-219 also targets and represses tau synthesis and decreased levels of miR-219 were also observed in the human AD brain.122 In a Drosophila model that produces human tau, reductions in miR-219 were associated with exacerbation of tau toxicity, linking this miRNA to AD-related pathology.122
The dysregulation of miRNAs has not only been shown to occur within the brain, but also within bio fluids such as serum, plasma, and CSF. A large push to develop AD detection methods has led to miRNA profiling within serum123–127, plasma125, 128–130, CSF125, 130–135, exosomes132, 134–136, extracellular fluid137, and PBMCs138 of humans and AD animal models. For example, one study showed that miR-135 was reduced in the blood of patients with mild cognitive impairment and miR-135 and miR-200b levels were decreased in CSF of individuals with dementia of Alzheimer’s type (DAT) group.114 In blood samples obtained from AD patients, both miR-29c and BACE1 expression were increased.112 Recently, Kumar et al. has developed a biomarker technique allowing AD patients to be distinguished from control patients with 95% specificity by hsa-miR-191–5p and hsa-miR-15b-5p signatures within plasma.139 Another study developed a 16 miRNA exosomal serum panel that predicts AD with 87% sensitivity and 77% specificity.137 Table 1 outlines the therapeutic potential of miRNAs and the other epigenetic regulators discussed in AD.
Table 1.
Therapeutic Potential of Epigenetic Modulators in Alzheimer’s Disease
| Model | Therapeutic Potential | Modification | Citation |
|---|---|---|---|
| Neuroblastoma Cells | SAM reduces vitamin B12/folate deficiency-induced overexpression of PS1 and BACE | DNA methylation | Fuso et al., 2005 |
| Neuroblastoma Cells | SAM reduces PS1 expression and Aβ plaques | DNA methylation | Scarpa et al., 2003 |
| TgCRND8 mice | SAM+SOD reduced vitamin B deficiency-induced AD features | DNA methylation | Cavallaro et al., 2017 |
| AD Humans | SAM reduces inflammation in AD patients and improves MMSE | DNA methylation | Chen et al., 2016 |
| CK-p25 mice | shRNA knockdown of HDAC2 reduces memory impairment | Histone acetylation | Graff et al., 2012 |
| Tg2576 mice | 4-PBA reverses brain hypoacetylation and reduces phosphorylated tau | Histone acetylation | Ricobaraza et al., 2009 |
| Tg2576 mice | Caloric restriction and NAD+ treatment is neuroprotective via α-secretase | Histone acetylation | Qin et al., 2006 |
| 3xTg-AD mice | Nicotinamide reduces phosphor-species of tau (Thr231) and improves memory | Histone acetylation | Green et al., 2008 |
| rTg4510 mice | AK1 is non-toxic and potentially neuroprotective | Histone acetylation | Spires-Jones et al., 2012 |
| APPPS-21 mice | MiR-34c seed inhibitors reverse memory deficit | MiRNA | Zovoilis et al., 2011 |
| Hippocampal Neurons | MiR-34c transfection reduces dendritic length and density | MiRNA | Kao et al., 2018 |
| Neuroblastoma | MiR-195 overexpression reduces Aβ-induced cytotoxicity | MiRNA | Zhu et al., 2012 |
| Hippocampal Neurons | Lentiviral overexpression of miR-101 decreases APP and Aβ | MiRNA | Vilardo et al., 2010 |
| AD PC12 cells and hippocampal neurons | miR-16 mimic targets APP and reduces Aβ-induced cytotoxicity | MiRNA | Zhang et al., 2015 |
| APP/PS1 mice | Lentiviral overexpression of miR-124 improves behavior through BACE1 | MiRNA | Du et al., 2017 |
| HEK293T | LNA inhibition of miR-485–5p or overexpression of BACE1-AS increase BACE1 levels | MiRNA, lncRNA | Faghihi et al., 2010 |
| Human plasma | Biomarker technique of 2 miRNAs identify AD with 95% specificity | MiRNA | Kumar et al., 2013 |
| Human serum exosomes | Biomarker panel of 16 miRNAs identify AD with 87% sensitivity | MiRNA | Cheng et al., 2015 |
| Neuroblastoma Cells | NEAT1 knockdown reduces AB-induced apoptosis and p-Tau levels | LncRNA | Ke et al., 2019 |
Bioinformatics studies have begun to elucidate the roles of lncRNAs in AD. In postmortem brain samples, the expression of hundreds of lncRNAs are significantly changed in AD patients versus age-matched controls in AD-related regions of the brain such as the hippocampus, middle temporal gyrus, entorhinal cortex and cortex.140–142 Gene ontological analysis identified significantly altered lncRNAs associated with mRNAS involved in protein ubiquitination, amyloid-β clearance, neural communication, electron transport chain, metabolic processes and cholesterol homeostasis.140–142 When neurofibrillary tangles were sampled, lncRNAs associated with development and morphogenesis of the neural tube and neural crest were significantly changed.143 Microarray analyses performed in rodent models of AD have similarly shown extensive changes in lncRNA expression.144, 145 While these bioinformatics studies revealed that several lncRNAs play roles in AD pathology, recent reports have identified Nuclear Paraspeckle Assembly Transcript 1 (NEAT1) as a key lncRNA in AD progression.141 Downregulation of NEAT1 in hippocampal tissue of APPswe/PS1dE9 mice was observed in the early stage of disease progression.146 Reduced levels of NEAT1 prevented clearance of Aβ by inhibiting expression of endocytosis-related genes.146 Alternatively, in vitro models of AD performed with mouse brain tissue or neuroblastoma cells have shown upregulation of NEAT1.147, 148 Knockdown of NEAT1 reduced Aβ-induced toxicity, apoptosis and promotion of p-Tau. Furthermore, NEAT1 was shown to reduce miR-107 and miR-124, thereby increasing BACE1.148 Another lncRNA that modulates miRNA efficacy is BACE1-AS, an anti-sense transcript that is upregulated in AD and expressed alongside BACE1. BACE-AS competes with the miR-485–5p binding site and prevents miR-485–5p from degrading the BACE1 transcript.61, 149 Dysregulated expression of both BACE-AS and miR-485–5p have been observed in RNA samples from the brains of AD patients.61
Parkinson’s disease
Parkinson’s Disease (PD) is the second most common neurodegenerative disease, affecting over 600,000 Americans, a number expected to double by 2040.150 PD is caused by degeneration of dopaminergic nigrostriatal pathways from the substantia nigra pars compacta (SNpc) to the striatum.6, 151, 152 The neurodegeneration of PD is marked by aggregates of α-synuclein (α-syn), a synaptic protein leading to dopaminergic neuron failure and resulting in tremors, rigidity, and non-motor symptoms like dementia and depression.6, 151, 153 Levodopa (L-Dopa), an amino acid precursor to neurotransmitters, provides relief to the motor-based symptoms of PD; however, L-Dopa can lead to the involuntary movement disorder, tardive dyskinesia, and other drugs like it fail to treat the non-motor symptoms of the disease.6, 154 Therefore, potential PD treatments in the realm of epigenetics are currently being explored (Table 2).
Table 2.
Therapeutic Potential of Epigenetic Modulators in Parkinson’s Disease
| Model | Therapeutic Potential | Modification | Citation |
|---|---|---|---|
| Tg-α-syn mice | Lentiviral administration of DNMT1 restores nuclear localization dysregulated by α-syn | DNA methylation | Desplats et al., 2011 |
| Tg-α-syn Drosophila | SB and SAHA decrease apoptosis in dorsomedial neurons | Histone acetylation | Kontopoulos et al., 2006 |
| N27 cells | Anarchic acid reverses Dieldrin-induced H3/H4 acetylation and apoptosis | Histone acetylation | Song et al., 2010 |
| N27 cells | Anarchic acid reverses Paraquat-induced H3/H4 acetylation and apoptosis | Histone acetylation | Song et al., 2011 |
| Tg-α-syn Drosophila | AKG2 reduces α-syn-induced neurotoxicity in dorsomedial neurons | Histone acetylation | Outeiro et al., 2007 |
| Dopaminergic neurons | SB and TSA increase GDNF and BDNF expression | Histone acetylation | Wu et al., 2008 |
| Dopaminergic neurons | SAHA is protective against neurotoxin-induced apoptosis | Histone acetylation | Chen et al., 2012 |
| MN9D cells, MPP+ | miR-124 mimic calpain 1, p25, and cdk5 to prevent cytotoxicity | MiRNA | Kanagaraj et al., 2014 |
| Tg-LRRK2 Cortical neurons | Overexpression of miR-205 reduces LRRK2 protein concentration and rescues neurite outgrowths | MiRNA | Cho et a., 2013 |
| Cortical neurons | Overexpression of miR-7 and miR-153 downregulates SNCA mRNA expression | MiRNA | Doxakis, 2010 |
| Tg-α-syn Neuroblastoma cells | Overexpression of miR-7 prevents α-syn-induced sensitivity to H2O2 cytotoxicity | MiRNA | Junn et al., 2009 |
| Neuroblastoma cells, MPP+ | Overexpression of miR-7 or VDAC1 reduces MPP+-induced oxidative stress and cell death | MiRNA | Chaudhuri et al., 2016 |
| MiR-155−/− mice | miR-155 knockout reduces inflammatory response to α-syn | MiRNA | Thome et al., 2016 |
| Human plasma | A miRNA biomarker strategy predicts PD wit 91% sensitivity and 100% specificity | MiRNA | Khoo et al., 2012 |
| Cortical neurons, MPP+ | MiR-7 and miR-153 is protective via mTOR and SAPK/JNK pathways | MiRNA | Fragkouli & Doxakis, 2014 |
| Neuroblastoma cells, MPP+ | MiR-7 targets RelA to provide neuroprotection | MiRNA | Choi et al., 2014 |
| Tg-α-Syn mice, MPTP | MiR-7 targets NLRP3 to provide neuroprotection | MiRNA | Zhou et al., 2016 |
| C57 mice, MPTP and neuroblastoma cells, MPP+ | Overexpression of miR-124 downregulates apoptotic and autophagic pathways | MiRNA | Wang et al., 2016 |
| PC12 cells, 6-OHDA | MiR-221 prevents cytotoxicity by targeting PTEN | MiRNA | Li et al., 2018 |
| C57 mice, MPTP | H2S treatment downregulates miR-135a-5p to increase ROCK2 expression | MiRNA | Liu et al., 2016 |
| Neuroblastoma cells, MPTP | MiR-185 overexpression prevents apoptosis | MiRNA | Wen et al., 2018 |
| PC12 cells, MPP+ | MiR-181c overexpression prevents apoptosis | MiRNA | Wei et al., 2017 |
| Neuroblastoma cells, rotenone | Inhibition of miR-384–5p reverses ER stress | MiRNA | Jiang et al., 2016 |
| Neuroblastoma cells, MPP+ | Knockout of HOTAIR attenuates neurotoxicity | LncRNA | Wang et al., 2017 |
| Tg-LRRK2 Drosophila | Overexpression of miR-7 or miR-184* in dopaminergic neurons targets DP and E2F1 as well as rescues flies from mLRRK2 | MiRNA | Gehrke et al., 2010 |
| C57 mice, MPTP and neuroblastoma cells, MPP+ | Inhibition of HAGLROS is protective by regulation of miR-100 and the mTOR pathway | LncRNA | Peng et al., 2019 |
| Neuroblastoma cells, MPP+ | P21 knockdown reduces ROS, neuroinflammation, and apoptosis | LncRNA | Ding et al., 2019 |
| Neuroblastoma cells, MPP+ | Knockdown of SNGH1 or overexpression of miR-15b-5p is neuroprotective | LncRNA, miRNA | Xie et al., 2019 |
| Neuroblastoma cells, MPP+ | Knockdown of SNGH1 decreases α-Syn aggregation through miR-15b-5p upregulation | LncRNA, miRNA | Chen et al., 2018 |
| MN9D cells, MPP+ and C57 mice, MPTP | Silencing of SNGH1 acts through miRNA and the mTOR pathway to provide neuroprotection | LncRNA | Qian et al., 2019 |
| C57 mice, MPTP | Downregulation of SNGH1 acts through upregulation of miR-7 suppress inflammation | LncRNA, miRNA | Cao et al., 2018 |
| MN9D cells, MPP+ | Knockdown of MALAT1 deceases α-Syn-mediated cytotoxicity | LncRNA | Chen et al., 2018 |
| Neuroblastoma cells, MPP+ and C57 mice, MPTP | β-Asarone downregulates MALAT1 to prevent α-Syn accumulation | LncRNA | Zhang et al., 2016 |
| Neuroblastoma cells and HEK293T cells, paraquat | NEAT1 is upregulated and mediates fenofibrate and simvastatin-induced neuroprotection | LncRNA | Simchovitz et al., 2019 |
| Neuroblastoma cells, MPP+ | Silencing NEAT1 derepresses miR-124, reduces inflammatory markers, and decreases apoptosis | LncRNA, miRNA | Xie et al., 2019 |
| Neuroblastoma cells, MPP+ | NORAD overexpression decreases apoptosis, ROS, and LDH | LncRNA | Song et al., 2019 |
| Neuroblastoma cells, MPP+ | UAC1 knockdown reduces caspase activity and apoptosis | LncRNA | Lu et al., 2018 |
| Wistar rats, 6-OHDA | UAC1 downregulation is neuroprotective by way of oxidative stress and inflammation reduction | LncRNA | Cai et al., 2019 |
Oligomerization and aggregation of α-synuclein (α-syn) is thought to be responsible for the onset of PD in humans.152 While phosphorylation and ubiquitination are known to promote α-syn aggregation, the precise mechanisms that control α-syn are still not known. Hypomethylation of the CpG2 (near intron 1) in the α-syn gene was shown in the blood, substantia nigra and putamen tissue samples of PD patients.155 This hypomethylation of the α-syn gene is thought to be responsible for increased levels of α-syn and correlate with age of onset of PD.156, 157 Furthermore, DNMT1 levels were shown to be decreased by ~50% that might be responsible for reduction in α-syn promoter methylation in PD brain samples.158 There has been evidence showing that α-syn mediates the sequestration of DNMT1 in neuronal cells causing its own hypomethylation in a feed-forward mechanism.158 Thus, epigenetic control by methylation of the α-syn promoter might be a factor responsible for PD onset as well as progression.
Many studies showed that histone acetylation also plays a significant role in PD pathogenesis. In α-syn overexpressing cells, α-syn binds to H3 leading to hypoacetylation of H3.159 Treatment with HDAC inhibitors sodium butyrate and SAHA reversed this and rescued cells from α-syn toxicity.159 Furthermore, H3 deacetylation inhibitors valproic acid (VPA), sodium butyrate, Trichostatin A (TSA) and SAHA protected the neuronal cells following MPTP treatment, a drug that produces a neurotoxin up taken by dopaminergic neurons and causes parkinsonism features.160, 161 In an MPTP mouse model of PD, H3 acetylation was observed to be upregulated and treatment with Levodopa (L-DOPA) reversed this effect.162 However, in primate PD models H3 acetylation was observed to be downregulated upon L-DOPA administration.162 Hence, HDAC inhibitors are promising to understand PD pathology, but the mechanistic role of histone acetylation in PD is still not completely clear. Histone acetylation has also been explored in the pesticide exposure-induced model of PD. In rat mesencephalic dopaminergic neurons, pesticides dieldrin and paraquat that are known to cause PD-like symptoms promoted H3 and/or H4 hyperacetylation.163 Furthermore, CREB-binding protein mediated histone hyperacetylation led to dopaminergic neuronal apoptosis that was rescued by the HAT inhibitor anacardic acid.163, 164
Familial and sporadic PD can be caused by gain-of-function mutations in leucine-rich repeat kinase 2 (LRRK2).165 It was previously shown that mutant LRRK2 leads to miRNA-induced transcriptional repression by negatively regulating Argonaute-2 of the RISC complex and by antagonizing let-7 and miR-184 in Drosophila.166 In mammalian PD models, LRKK2 has been shown to cause dysregulation of GTPase activity167, autophagy168, and actin stabilization.169 Cho et al. found that despite increased expression of LRRK2 protein in PD patients, LRRK2 mRNA levels are not significantly altered, leading them to investigate post-transcriptional modifications.170 Screening for miRNAs with target sites near the LRRK2 3’UTR, revealed that miR-205 is disproportionately downregulated in PD frontal cortex. Overexpression of miR-205 suppressed LRRK2 protein expression, and inhibition of miR-205 increased LRRK2 in primary neurons and dopaminergic MN9D cell lines.170, 171 Increased miR-205 levels were shown to inhibit defects in neurite outgrowth in hippocampal neurons of mice expressing mutant forms of LRRK2.170
SNCA is also post-transcriptionally regulated by several miRNAs. MiR-7 has been shown to reduce levels of α-syn by 30% while miR-153 reduced levels of α-syn by 19% in primary neurons.172 A synergistic effect was observed with overexpression of both miR-7 and miR-153 which reduced α-syn levels by 46% in primary neurons.172 In another study, miR-7 and miR-153 overexpression was protective via mTOR and SAPK/JNK pathway preservation in cortical neurons exposed to MPP+.173 Furthermore, inhibition of miR-7 upregulated α-syn and miR-7 was shown to protect against cellular α-syn-mediated susceptibility to oxidative stress, proteasome impairment, and prevent cell death by targeting RelA in dopaminergic neuroblastoma cells.174, 175 MiR-155 was shown to play a key role in α-syn-induced inflammation and knockout of miR-155 reduced α-syn neurotoxicity in mice.176 Finally, eight miRNA, including hsa-miR-21 and hsa-miR-301b were shown to deregulate the chaperone mediated autophagy proteins (CMA), lysosome-associated membrane protein type 2a (LAMP-2A) and heat shock protein 70 (hsc70), each degraders of α-syn, resulting in increased aggregates in neuroblastoma cells.177
MiR-34b/c and the previously reported miR-7 have been implicated in mitochondrial function in PD and PD-related models. MiR-34b and miR-34c were downregulated in human brain tissue from PD patients in Braak stages 4 and 5.178 Correspondingly, decreased levels of miR-34b and miR-34c correlated with mitochondrial dysfunction, reactive oxygen species (ROS) generation and increased α-syn aggregation in human dopaminergic neuroblastoma cells.178, 179 It has been proposed that miR-34b loss in PD patients leads to the characteristic upregulation of A2AR in the putamen observed in the disease.179 MiR-7 has also been shown to regulate mitochondrial protein expression, prevent ROS and mitochondrial permeability transition pore opening following MPP+ exposure in neuroblastoma cells.180 Finally, miR-7 has been shown to target the NLRP3 inflammasome in microglia, and a miR-7 mimic provided neuroprotection in Transgenic-α-syn mice subject to MPTP.181
Several other miRNAs involved in modulating PD pathology have been identified. For example, treatment with a miR-221 mimetic was shown to be protective against a 6-OHDA treatment model of PD in PC12 cells.182 Overexpression of miR-185 or miR-181c prevented MPTP-induced apoptosis in neuroblastoma or PC12 cells, respectively.183, 184 In a rotenone PD model of neuroblastoma cells, miR-384–5p inhibition reversed ER stress and attenuated apoptosis.185 Overexpression of miR-124 downregulated apoptotic and autophagic pathways to provide neuroprotection in both MPTP-treated mice models and MPP+-treated neuroblastoma cells.186 Loss of miR-133b has been observed in the midbrain of PD patients and miR-133 was shown to regulate maturation and function of dopaminergic neurons through a feedback loop with Pitx3.187 MiR-16–1-mediated downregulation of hsp70 was shown to worsen aggregation of α-syn in transgenic neuroblastoma cells.188 Finally, hydrogen sulfide treatment was shown to protect MPTP-treated mice by increasing miR-135a- 5p, which represses rho-associated protein kinase 2, an enzyme that promotes neurodegeneration.189
Profiles of miRNAs expressed in the prefrontal cortex tissue190, SNpc191, exosomes of the CSF192, and serum193 of PD patients have revealed extensive dysregulation of miRNAs. CSF miRNA profiling was shown to distinguish PD patients from controls and also correlated with different stages of PD pathology.194, 195 Overlapping miRNAs identified in serum studies have implicated several miRNAs that may be key to PD pathogenesis including miR-29c, miR-221, and miR-214.196–199 The role of miR-29c has not been elucidated, but miR-221 has been shown to promote survival in human dopaminergic neuronal cells and loss of miR-214 increased alpha-synuclein expression in human neuroblastoma cells.200, 201 In plasma, a strategy combining k-Top Scoring Pairs algorithm of differentially expressed miRNA can predict PD with 91% sensitivity and 100% specificity.202
Several profiling studies have shown aberrations in lncRNA expression in the human PD brain and mouse models of PD providing a basis for biomarker research.203–208 The majority of the lncRNAs investigated in PD have been shown to modulate miRNAs. For example, HAGLROS was upregulated in MPTP-treated mice and MPP+-treated neuroblastoma cells.208 HAGLROS was shown to sponge miR-100 and subsequent inhibition decreased apoptosis and autophagy both in vitro and in vivo through PI3K/Akt/mTOR regulation.209 P21 was upregulated in neuroblastoma cells subject to MPP+ and subsequent knockdown decreased ROS generation, neuroinflammation, and apoptosis.210 It was found that p21 exerts protective function through de-repression of miR-625 and thus upregulation of Transient receptor potential melastatin 2 (TRPM2).210 In addition, p21 promoted apoptosis in neuroblastoma cells by sponging miR-1277–5p and thereby increasing expression of α-syn.211 MALAT1 is increased in midbrains of MPTP-treated mice and was shown to suppress miR-205–5p, leading to a subsequent increase of LRRK2.171 Correspondingly, MALAT1 knockdown prevented apoptosis after MPP+ treatment in MN9D cells.171 In addition, β-asarone treatment has been shown to be protective in both in vitro and in vivo models of PD by downregulating MALAT1 expression.212
The small nucleolar host gene 1 (SNGH1) lncRNA was upregulated in MPP+-treated neuroblastoma cells and exacerbated toxicity by sponging miR-15b-5p.213 Knockdown of SNGH1 or overexpression of miR-15b-5p abrogated ROS production and cell death in the same model.213 The protective role of miR-15b-5p was again shown through SNGH1 knockdown leading to less α-syn aggregation in neuroblastoma cells.214 SNGH1 silencing has been shown to act through other axes like miR-221/222 and CDKN1B/p27/mTOR to enhance autophagy and prevent cell death in MPTP-treated mice and MPP+-treated MN9D cells.215 Importantly, SNGH1 was also elevated in brains of PD patients and was shown to promote neuroinflammation by suppressing miR-7 and enhancing microglia and inflammasome activation in MPTP-treated mice.216
MPTP-treated mice and MPP+-treated neuroblastoma cells were both shown to induce NEAT1 expression.217 Subsequent knockdown of NEAT1 suppressed autophagy in mice by stabilizing PINK 1.217 Importantly, NEAT1 was also increased in the substantia nigra of patients with PD and the neuroprotective drugs fenofibrate and simvastatin have been shown to require NEAT1 to prevent paraquat-induced cell death in neuroblastoma cells.218 Alternatively, NEAT1 silencing was also shown to be protective by reducing apoptosis and inflammatory signaling in MPP+-treated neuroblastoma cells by derepressing miR-124.219
The Urothelial Cancer Associated 1 (UCA1) lncRNA was highly expressed in MPTP-treated mice and MPP+-treated neuroblastoma cells, leading to increased SNCA expression.220 Knockdown of UCA1 decreased caspase-3 activity and apoptosis in the same cell model.220 Similarly, downregulation of UCA1 prevented inflammation and oxidative stress in a PD rat model induced by 6-hydroxydopamine injection (6-OHDA).221 MPTP treatment of neuroblastoma cells downregulated NORAD and subsequent overexpression protected against MPP+-mediated apoptosis, decreased ROS, and decreased lactose dehydrogenase activity.222 Finally, expression of the lncRNA HOTAIR has been shown to increase in both MPTP models of mice and MPP+ models of neuroblastoma cells along with a corresponding increase in LRRK2.66 Knockout of HOTAIR in the cellular model attenuated induced neurotoxicity.66
Huntington’s disease
Huntington’s Disease (HD) is a dominant, late-onset genetic disorder affecting 5–10 people of 100,000 globally.223, 224 In HD, CAG repeats form at exon 1 of the Huntingtin gene (HTT), producing the neurotoxic Huntingtin protein (mHTT) and resulting in neurodegeneration of GABAergic spiny striatal neurons.224–226 HD manifests in motor impairment and chorea, schizophrenia-like behavior and suicidal tendency, as well as changes in mood and judgement.224 HD patients undergo differing treatment depending on the course of the disease, and it is hopeful that altering gene expression through combinatorial approaches such as tacrine, moclobemide, and creatine will improve treatment options.227, 228 Several studies have revealed a role for epigenetic mechanisms in HD pathology, many of which display therapeutic potential (Table 3).
Table 3.
Therapeutic Potential of Epigenetic Modulators in Huntington’s Disease
| Model | Therapeutic Potential | Modification | Citation |
|---|---|---|---|
| mHTT Drosophila | Selisistat improves survival and decreases cytotoxic inclusions | Histone acetylation | Smith et al., 2014 |
| mHTT Drosophila | AK1 and AGK2 are neuroprotective by way of sterol biosynthesis downregulation | Histone acetylation | Luthi-Carter et al., 2010 |
| Htn-Q150 C. elegans | Knockdown of HDA3 reduces polyglutamine toxicity | Histone acetylation | Bates et al., 2006 |
| Tg-R6/2 mice | HDACi 4b reverses H3 hypoacetylation and improves functional recovery | Histone acetylation | Thomas et al., 2008 |
| HTT-Q73 PC12 cells | miR-10b-5p mimic increases cell survival | MiRNA | Hoss et al., 2014 |
| HTT-Q84 Neuroblastoma cells | Overexpression of miR-196a increases neurite outgrowth | MiRNA | Fu et al., 2015 |
| R6/2 mice | MiR-132 overexpression provided neuroprotection and delayed disease progression | MiRNA | Fukuoka et al., 2018 |
| Tg-R6/2 mice | Overexpression of miR-196a enhances neurite outgrowths and improves learning and memory | MiRNA | Her et al., 2017 |
| HD neural progenitor cells | Overexpression of miR-196a normalized mitochondrial activity and decreased apoptosis | MiRNA | Kunkanjanawan, et al., 2016 |
| STHdh(Q111)/Hdh(Q111) cells | Exogenous expression of miR-146a, miR-432, and miR-19a reversed cell cycle defects and apoptosis | MiRNA | Das et al., 2015 |
| STHdhQ7/HdhQ7 cells | Overexpression of HYPK and Hsp70 reverses miR-125b, miR-146a, and miR-150 expressions while reducing mHTT aggregates | MiRNA | Ghose et al., 2011 |
| Neuronal stem cells derived from R6/2 mice | MiR-27a overexpression decreased mHTT aggregates | MiRNA | Ban et al., 2017 |
| mHTT sheep | HTT targeting by way of artificial miRNA safely reduces mHTT mRNA and protein | MiRNA | Pfister et al., 2018 |
| Primary Striatal cell and cortical neurons, mHTT and 3-NP | MiR-22 overexpression is neuroprotective | MiRNA | Jovicic et al., 2013 |
| R6/2 neurons | MiR-27a overexpression decreases mHTT aggregates by way of MDR-1 | MiRNA | Ban et al., 2017 |
| Neuroblastoma cells, H2O2 | Neat1 transfection increases tolerance to oxidative stress | LncRNA | Sunwoo et al., 2017 |
| HEK293 cells and neuroblastoma cells | Overexpression of HTTAS_v1 decreases HTT levels in a gene-specific manner | LncRNA | Chung et al., 2011 |
In post-mortem tissue from the frontal and parietal cortex of HD patients, a higher level of global methylation was observed compared to control patients.229 Cultured mouse striatal cells from knock-in embryos expressing full-length huntingtin (HTT) show methylation of promoters and thus down-regulation of several genes that control developmental processes, neuronal migration and cell signaling genes.230 A correlation between global cortex hypermethylation and age of disease onset in the cerebral cortex has been observed, although differential methylation at probed sites around HTT was not identified.231 Genome-wide reduction of 5-hmC has also been seen in a HD mouse model, particularly in the cerebral cortex and striatum.232 Differentially hydroxymethylated promoter regions in these animals correspond to Wnt/β-catenin/Sox pathway, axon guidance, GABA signaling and dopamine feedback, which are all implicated in HD pathology.232 Stimulation of the adenosine A2A receptor (A2AR) has been shown to ameliorate neurodegeneration and several major HD symptoms in rodents.233, 234 However, A2AR levels are significantly reduced in the HD putamen of human brains potentially due to hypermethylation and decreased hydroxymethylation of the adenosine a2a receptor (ADORA2A) gene.235
HDAC inhibition is a major mechanism of neuroprotection in HD. Various chemical HDAC inhibitors such as SAHA, sodium butyrate, 4-PBA and TSA have been shown to ameliorate motor dysfunction, cognitive deficits or the neurodegenerative phenotype in transgenic C. elegans, Drosophila and mouse models of HD.236–242 Treatment with the HDAC inhibitor 4b was shown to reduce hypoacetylation of H3 and H4, and diminish transcriptional abnormalities caused by mutant HTT in the striatum, cortex and cerebellum of HD transgenic mice.243 The 4b treatment was additionally shown to improve motor function and decrease brain atrophy in HD mice.243 While chemical HDAC inhibition has proven to be effective in ameliorating HD deficits, studies employing genetic knockdown of various HDACs in HD are not clear. For example, knockout of HDAC4, HDAC6, or HDAC7 were not effective in ameliorating neurodegeneration in transgenic HD mouse models.244–246 Similarly, while HDAC3 chemical inhibition has been shown to have a therapeutic effect in HD, HDAC3 knockout was not effective in reducing transcriptional dysregulation or HTT aggregation in transgenic HD mice and increased nuclear HTT aggregates in HeLA and 293T cells expressing mutant HTT.241, 247, 248 Alternatively, RNA interference-mediated HDAC3 knockdown suppressed polyglutamine toxicity in a C. elegans model of HD using neuronal expression of mutant HTT with expanded polyglutamine repeats.239 These opposing results highlight the complexity of HDACs in HD as chemical inhibitors may target multiple HDACs, HDAC isoforms differ in target specificity and efficacy, and chemical and genetic inhibition have been shown to have differential compensatory mechanisms.249
Levels of brain-derived neurotrophic factor (BDNF) that is essential for striatal neuronal survival is severely reduced in the brain of HD patients.250–253 HTT is known to interact and recruit the repressor element-1 transcription factor (REST) complex (REST/coREST/Sin3A/HDAC1/HDAC2) to the cytosol which consequently allows BDNF gene expression.252, 253 A study assessing REST levels in brain tissues of HD patients found that cytoplasmic REST levels are reduced in neurons of the cortex and caudate of HD patients.254 Studies in mice have shown that the HTT mutation in HD causes the REST complex to accumulate in the nucleus, thereby silencing BDNF and contributing to neuronal death.253 In addition to BDNF, loss of several REST-controlled genes involved in neuronal maintenance have been observed in both the mouse and human HD brain.253 Since REST silencing occurs via HDAC-dependent chromatin remodeling, HDAC inhibitors that may also target REST activity may by therapeutically beneficial.
Differential expression of miRNAs has been observed in the brain and blood of HD patients and animal models of HD.255–260 MiR-9 was decreased in the cortices of HD patients and was shown to target REST and coREST in neuronal precursor cells.261 In a transgenic non-human primate model, miR-128 was downregulated from the time of birth and was shown to interact with HTT and huntingtin interacting protein 1 (HIP1).262 Other studies have implicated the miR-10b family which is upregulated in the serum and brain of HD patients.255, 263–265 In silico analysis revealed that miR-10b-5p targets BDNF, which displays reduced levels in HD leading to neuronal dysfunction.264 MiR-10b-5p expression in the prefrontal cortex has been correlated with HD age of onset in HD patients.263 This indicates that miR-10b-5p could serve as an important biomarker in HD treatment; however, it has not been examined in peripheral fluid.
Evidence of other miRNAs with potential involvement in HD pathology include miR-137, miR-148a and miR-214 which have been shown to directly target HTT and reduce HTT protein levels in HEK293T cells.266 In STHdh(Q111)/Hdh(Q111) cells, miR-214 was also responsible for the downregulation of β-catenin often seen in HD.267 MiR-196a has been identified as significantly upregulated in HD265 and bioinformatics analyses indicated it may target inflammation and apoptosis-related pathway.263, 268, 269 MiR-196a overexpression was shown to increase neurite outgrowth in neuroblastoma cells269 and suppress Ran-binding protein 10 (RANBP10), a protein which is elevated in HD mice.270 MiR-196a also suppressed apoptosis in neural progenitor cells and differentiated neural cells in HD non-human primates.271 In STHdh(Q111)/Hdh(Q111) cells, increased levels of p53 led to downregulation of miR-146a.272 Subsequent overexpression of miR-146a attenuated cell cycle abnormalities and decreased apoptosis in the same cell model.273 Similarly, in the R6/2 mouse model, miR-34a-5p levels have been shown to decrease with increased p53 expression, although the relationship between p53 and miRNA expression has not been elucidated.274
Downregulation of miR-22 was observed in the brains of both YAC128 and R6/2 transgenic mice.257 In vitro, miR-22 has been shown to provide neuroprotection in multiple primary striatal and cortical cell models of HD including mHTT and 3-NP exposure by reducing caspase activation and apoptosis.275 MiR-132 overexpression in the striatum of R6/2 mice was neuroprotective and delayed disease progression despite having no effect on mutant HTT.276 MiR-27a overexpression in R6/2-derived neuronal stem cells decreased mHTT aggregates, potentially by upregulating multidrug resistance protein 1 (MDR-1), a transporter of mHTT.277 Lastly, overexpression of an artificial miRNA targeting mHTT in sheep expressing human HD CAG repeat decreased HTT levels by 50–80% at 1 and 6 months following treatment.278 This study supports the therapeutic potential of miRNA modulation in the large animal brain.
One study identified the existence of a natural antisense HTT transcript (HTTAS), which manifests in two splice variants HTTAS_v1, containing exons 1 and 3, and HTTAS_v2, containing exons 2 and 3.279 Up to 50% loss of HTTAS_v1 has been observed in the human HD frontal cortex and depending upon levels of overexpression and HTTAS_v1 repeat length, HTT can be decreased by 20–90% in HEK293 and SH-SY5Y cells.279 Multiple mouse models of HD have shown decreased levels of the lncRNA Abhd11os.280 Lentiviral-mediated overexpression of Abhd11os was neuroprotective, while knockdown of Abhd11os exacerbated mHTT toxicity.280 Other differentially expressed lncRNAs identified in cell and animal models are maternally expressed 3 (MEG3) and NEAT1.281–283 NEAT1 levels were increased in the brains of HD patients and R6/2 mice and NEAT1 overexpression in neuroblastoma cells was shown to protect against H2O2-induced oxidative stress.282 Similarly, NEAT1 overexpression protected neuroblastoma cells cotransfected with HTT expression plasmids from cytotoxicity.284 However, knockdown of NEAT1 or MEG3 was also shown to decrease mHTT aggregation and p53 expression in neuroblastoma cells281, indicating the roles of these lncRNAs in HD needs to be studied further.
EPIGENETICS AND ACUTE BRAIN INJURY
Ischemic Stroke
Stroke is a major cause of death and disability worldwide.10, 285 Ischemic stroke (cerebral ischemia) is caused by the blockage of blood flow to the brain and results in energy depletion, mitochondrial dysfunction, excitotoxicity and ultimately cell death. During the reperfusion phase, return of the blood supply introduces inflammatory factors and induces oxidative stress which then cause secondary brain damage.286–288 Following ischemia/reperfusion injury, the cells within the penumbra (the area surrounding the infarcted core) are destined to die; however, some if not all, have the ability to recover if given therapeutic intervention.289, 290 At this time, recombinant tissue plasminogen activator (TPA) is the only stroke medication approved for treatment.291, 292 TPA works to digest clots through the degradation of fibrin293, 294; however, no significant decrease in mortality has been shown after treatment and increased incidence of intracerebral hemorrhage is linked to TPA.295 Recent studies showed that epigenetic changes play a significant role in modulating secondary brain damage and neurological dysfunction following stroke and hence may represent much-needed potential stroke therapeutic targets (Table 4).
Table 4.
Therapeutic Potential of Epigenetic Modulators in Ischemic Stroke
| Model | Therapeutic Potential | Modification | Citation |
|---|---|---|---|
| 129/SV mice, MCAO | 5’-aza-dC as well as TSA reduced infarct size | DNA methylation | Endres et al., 2000 |
| SD rats, HI | 5’-aza-dC protects against nicotine-induced susceptibility | DNA methylation | Li et al., 2013 |
| Wistar rats, photothrombotic stroke | TST or TST+5’-aza-dC upregulates BDNF and improves neurological score | DNA methylation | Choi et al., 2018 |
| HT22 cells | 5’aza-dC downregulates DNMT1, induces S phase arrest, and inhibits early apoptosis while promoting late apoptosis | DNA methylation | Yang et al., 2017 |
| ICR mice, MCAO | SC1 reverses mitochondrial 5hmC upregulation by inhibiting TET2 leading to increased levels of ATP | DNA hydroxymethylation | Ji et al., 2018 |
| C57 mice, MCAO | Overexpression of Polycomb proteins SCMH1 and BMI1 is neuroprotective | Polycomb protein | Stapels et al., 2010 |
| Cortical neurons, CA and 3-NP | Overexpression of BMI1 acts by way of antioxidant genes to provide protection | Polycomb protein | Abdouh et al., 2012 |
| Wistar rats, 4-VO | VPA confers inflammatory protection and improves functional recovery | Histone acetylation | Xuan et al., 2012 |
| SD rats, MCAO | VPA, SB, or TSA prevent hypoacetylation of H3, prevent inflammation, and reduce infarct volume | Histone acetylation | Kim et al., 2007 |
| C57 mice, MCAO | SAHA reverses H3 hypoacetylation, promotes Hsp70 and BCL-2, and reduces infarct volume | Histone acetylation | Faraco et al., 2006 |
| C57 mice, HI | 4-PBA improves functional recovery via protection from apoptotic mechanisms | Histone acetylation | Qi et al., 2004 |
| Optic nerves, OGD | SAHA and MS-275 rescue white matter | Histone acetylation | Baltan et al., 2011 |
| Cortical neurons | Despite reducing ischemic infarct, HDAC inhibitors are cytotoxic to cells they do not protect | Histone acetylation | Langley et al., 2008 |
| SD rats, forebrain ischemia | MiR-181a antagomir decreases CA1 neuron death | MiRNA | Moon et al., 2013 |
| Primary astrocytes, GD | MiR-181a inhibition decreases apoptosis via upregulation of BCL-2 and MCL-1 | MiRNA | Ouyang et al., 2012 |
| C57 mice, MCAO | Overexpression of miR-124 is neuroprotective by downregulation of REST and Usp-14 | MiRNA | Doeppner et al., 2013 |
| C57 mice, MCAO | MiR-124 agomir reduces infarct volume | MiRNA | Sun et al., 2013 |
| C57 mice, MCAO | Liposomated miR-124 reduces inflammatory markers and infarct volume | MiRNA | Hamzei et al., 2016 |
| PC12 cells, OGD | MiR-124 mimic activates the PI3K/AKT pathway and reduces apoptosis | MiRNA | Wang et al., 2017 |
| SD rats, MCAO | MiR-124 knockdown or miR-124 antagomir is neuroprotective | MiRNA | Zhu et al., 2014 |
| SD rats, MCAO | MiR-155 inhibition reduces infarct volume | MiRNA | Xing et al., 2016 |
| C57 mice, MCAO | MiR-155 inhibition reduces infarct volume | MiRNA | Caballero-Garrido et al., 2015 |
| Oligodendrocyte precursors | MiR-146a overexpression increases myelination | MiRNA | Liu et al., 2017 |
| Neuroblastoma cells, OGD | MiR-146a inhibition prevents apoptosis via Fblx10 upregulation | MiRNA | Li et al., 2017 |
| C57 mice, MCAO | Lentiviral overexpression of miR-424 is neuroprotective via cell cycle arrest and inflammatory suppression | MiRNA | Zhao et al., 2013 |
| C57 mice, MCAO | MiR-424 antagomir is neuroprotective by way of oxidative stress prevention | MiRNA | Liu et al., 2015 |
| C57 mice, MCAO | The inhibitor TAT-p53-DBD270–281 uncouples MEG3 from p53 to provide neuroprotection | LncRNA | Yan et al., 2016 |
| SHR rats, MCAO | Knockdown of FosDT derepresses REST genes and improves functional recovery | LncRNA | Mehta et al., 2015 |
| C57 mice, MCAO | MiR-181a antagomir reduces inflammation and infarct size while improving behavioral recovery | MiRNA | Xu et al., 2015 |
| PC12 cells, OGD | MiR-210 overexpression protects against apoptotic events | MiRNA | Qiu et al., 2013 |
| C57 mice, MCAO | MiR-210 overexpression increases BDNF and improves neurological scores | MiRNA | Zeng et al., 2016 |
| SD rats, MCAO | MiR-210 promotes vagus nerve stimulation-mediated recovery | MiRNA | Jiang et al., 2015 |
| C57 mice, MCAO | Inhibition of miR-210 attenuates inflammatory response | MiRNA | Huang et al., 2018 |
| C57 mice, MCAO | Inhibition of MALAT1 is neuroprotective by way of decreasing autophagy | LncRNA | Guo et al., 2017 |
| SD rats, MCAO | Sh-MEG3 administration improves functional recovery and promotes angiogenesis | LncRNA | Liu et al., 2017 |
| Human serum | Peng et al. used let-7e to predict acute stroke with 73.4% sensitivity and 82.8% specificity | MiRNA | Peng et al., 2015 |
| C57 mice, MCAO | MEG3 knockdown prevents apoptosis | LncRNA | Yan et al., 2017 |
| BV2 cells, OGD | H19 knockdown reduces inflammation to provide neuroprotection | LncRNA | Wang et al., 2017 |
| Neuroblastoma cells, OGD | Overexpression of N1LR prevents apoptosis | LncRNA | Wu et al., 2017 |
| C57 mice, MCAO and cortical neurons, OGD | Knockdown of GAS5 is neuroprotective via decreased competition with miR-137 | LncRNA, miRNA | Chen et al., 2018 |
The contribution of DNA hypermethylation to poor outcomes after cerebral ischemia has been well characterized. After middle cerebral artery occlusion (MCAO) induced focal ischemia in mice, levels of 5-mC were shown to be elevated in the striatum and cortex of mice.296 Heterozygous DNMT knockout mice or mice heterozygous for a mutant DNMT allele showed smaller infarcts after mild ischemic damage.296, 297 Furthermore, treatment with the DNMT inhibitor 5-aza-dC or other demethylating agents protected wild-type rodents after focal ischemia.296, 298, 299 However, in a severe ischemic mouse model, DNMT expression was not increased nor were mice protected by DNMT gene deletion.296 Interestingly, genomic methylation has been shown to be better predictor of biological age and stroke outcome than chronological age.300, 301
Recent studies also evaluated the role of 5-hmC in post-stroke brain damage. Following MCAO in mice, 5-hmC increased quickly after reperfusion (by 5 min) and remained elevated up to 2 days of reperfusion following focal ischemia.32, 302 It was observed that the post-stroke induction of 5-hmC was mediated by TET3 in the peri-infarct region or TET2 in the whole brain.32, 302 Pharmacological or genetic inhibition of 5-hmC exacerbated ischemia/reperfusion injury, while TET activation via ascorbate enhanced the expression of protective genes, prevented degeneration, and improved motor function recovery after focal ischemia.302 A recent study in mice reported that 5-hmC is increased in the mitochondrial genome after focal ischemia where it may influence mitochondrial gene expression and ATP levels.303
Inhibition of histone deacetylation was shown to protect the brain after stroke. VPA administration prevented deacetylation of H3 and H4 and ameliorated hippocampal CA1 neuronal death after global ischemia in adult rats.304 Treatment with VPA or sodium butyrate or TSA was shown to inhibit microglial activation, downregulate nitric oxide synthase and upregulated heat shock proteins leading to improved behavioral outcomes and reduced infarct volume in a rats following permanent MCAO.305 Importantly, VPA and sodium butyrate were shown to be most beneficial when given at 3 to 6 h of reperfusion after focal ischemia in rodents indicating the translational potential of these drugs in stroke therapy.305 Administration of SAHA following transient MCAO in mice was shown to suppress ischemia-induced H3 deacetylation which led to decreased proinflammatory levels of cytokine IL1β and increased levels of the chaperone HSC70.306 SAHA treatment also decreased size of the infarction after transient MCAO.306 Pre- or post-stroke treatment with the HDAC inhibitor 4-PBA was also shown to reduce infarct volume significantly in a mouse model of hypoxia-ischemia.307 An in vitro study using white matter cells isolated from the mouse optic nerve showed that HDAC inhibitor treatment (SAHA or MS-275) before or after oxygen glucose deprivation (OGD) preserved white matter architecture and reduced excitotoxicity.308
Several rodent studies have shown that miRNAs are significantly altered within the brain after cerebral ischemia.309–311 Additional studies have identified a number of miRNAs involved in various aspects of stroke pathophysiology including excitotoxicity (miR-223, miR-107, miR-125b), oxidative stress (miR-23, miR-99), apoptosis (miR-21, miR-25, miR-15, miR-497, miR-29), edema (miR-29, miR-9, miR-375, miR-150, miR-130, miR-320) inflammation (miR-22, miR-203, miR-9, miR-132), neurogenesis (miR-17) and angiogenesis (miR-107, miR-376, miR-140).312 Several key miRNAs studied within the field of stroke are discussed in more detail below.
Both focal and permanent ischemia have been shown to reduce miR-424 expression in the plasma of stroke patients and within the blood and brain of rodents.313, 314 Overexpression of miR-424 was shown to reduce focal ischemia-induced oxidative stress and infarct in the mouse brain by increasing Nrf2 and MnSOD.314 In a mouse model of permanent focal ischemia, miR-424 overexpression reduced edema and inflammatory processes by inhibiting microglial activation.313 MiR-424 expression was increased in human endothelial cells subjected to hypoxia and promoted angiogenesis by stabilizing HIF-1α. In the plasma of stroke patients, miR-424 levels were upregulated in lymphocytes and neutrophils, which was negatively correlated with TNF-α, IL-10, and IGF-1 expression as well as infarct size.315 Several studies have shown that miR-124 is increased after stroke and delivery of a miR-124 mimetic confers neuroprotection in both transient MCAO and OGD models.316–318 MiR-124 was shown to promote angiogenesis and inhibit excitotoxicity, apoptosis, BBB damage, and inflammation through modulation of glutamate receptors319, 320, Notch signaling321, DNA repair protein Ku70322, REST inhibition, calpain reduction316, PI3K/AKT activation323, upregulation of antiapoptotic proteins317, and regulation of and M2-like microglia/macrophage activation318 in several in vitro and in vivo models of rodent cerebral ischemia. MiR-155 was shown to upregulate inflammatory cytokines like IL-10, IL-4, and IL-6 in mice324 and increase apoptosis through the Rheb/mTOR pathway in rats325 following focal ischemic injury. Inhibition of miR-155 significantly decreased infarction in mice following transient MCAO by increasing nitric oxide (NO) production and the expression of Notch1 and endothelial NO synthase.326, 327
While the miRNAs discussed above have shown consistent roles in animal models of cerebral ischemia, the impact of other key miRNAs is more complex. For example, following mouse transient focal ischemia, miR-181a increased within the infarcted region where it was shown to negatively regulate binding immunoglobulin protein (GRP78/HSPA5), a protein involved in ER function and inhibition of apoptosis.328, 329 However, within the penumbra, miR-181a expression decreased and positively regulated GRP78, implicating miR-181a in ischemic pathogenesis as well as ischemic recovery.328 The evidence thus far indicates that miR-181 inhibition is protective as it has been shown to reduce infarct and inflammation by decreasing glutamate transporter 1 (GLT-1), apoptosis and mitochondrial dysfunction in rodent models of cerebral ischemia.330–332 Similarly, miR-210 has been implicated in both protection and injury following cerebral ischemia. For example, miR-210 overexpression has been shown to upregulate BDNF levels, reduce neuronal apoptosis and improve neurological severity scores following transient focal ischemia in rats and mice.333, 334 However, both pre- and post-MCAO inhibition of miR-210 improved neurological outcomes by reducing inflammation following mouse transient MCAO.335
As with neurodegenerative disorders, a significant effort has been made to develop biomarker procedures that can identify stroke with minimal invasiveness. MiRNA profiles have been established in serum336–341, plasma342–346, whole blood347–349, exosomes350, and CSF.341 MiR-145339, 340, 348 and let-7e341, 347 were identified as key miRNAs in multiple profiling studies. Based off the miRNA let-7e in serum, Peng et al. was also able to predict acute stroke in patients with 73.4% sensitivity and 82.8% specificity.341
Several studies have also been carried out to profile changes in lncRNA expression after stroke.351, 352 These studies implicate lncRNAs associated with genes involved in lipoprotein production, ABO blood type, prostaglandin synthesis, hematopoietic cell lineage, and glycolysis/gluconeogenesis.351, 352 Several lncRNAs have been shown to play significant roles in cerebral ischemia pathology. For example, MEG3 was increased following cerebral ischemia in mice where it was shown to bind p53 and promote post-ischemic neuronal death.353 MEG3 silencing reduced infarct size, improved neurological scoring, and promoted angiogenesis in rats and mice subjected to MCAO.354, 355 MEG3 has been implicated in apoptosis by targeting the miR-21/programmed cell death 4 pathway.355 ANRIL (CDKN2BAS) overexpression in diabetes mellitus rats upregulated VEGF, NF-κB, p-IκB/IκB and stimulated angiogenesis following MCAO.356 FosDT has been shown to increase following focal ischemia and associates with the chromatin modifying proteins sin3a and coREST to induce REST.357 Knockdown of FosDT and REST have been shown to decrease infarct size and improve functional recovery up to 7 days of reperfusion following focal ischemic injury.357, 358 Other lncRNAs implicated in ischemic stroke include H19, which provided BV2 cells neuroprotection following OGD when silenced68, N1LR, which prevented apoptosis in neuroblastoma cells when overexpressed359, and GAS5, which provided neuroprotection via decreased competition with miR-137 following MCAO in mice or OGD in cortical neurons when overexpressed.360
A key lncRNA identified in cerebral ischemia is MALAT1 (a.k.a NEAT2) which has been shown to be upregulated after OGD and MCAO.67, 361 Knockout of MALAT1 increased pro-apoptotic and pro-inflammatory factors and infarct size, and has been shown to directly associate with Bim and E-selectin following MCAO in mice.67 MALAT1 was shown to mediate autophagy and protect brain microvascular endothelial cells after OGD treatment.362 Conversely, MALAT1 inhibition downregulated autophagy, which induced neuroprotection following MCAO in mice363 indicating MALAT1 involvement in cerebral ischemia is complex and warrants additional research.
Hemorrhagic Stroke
Subarachnoid hemorrhage (SAH) and intracerebral hemorrhage (ICH) are two cerebrovascular events resulting in bleeding of the tissue around the brain and bleeding within the brain tissue, respectively. Hemorrhagic strokes account for only a small portion of strokes, but lead to high rates of disability364, 365, appear earlier in life than ischemic events365–367, and are responsible for over 25% of potential years of life lost to stroke.368, 369 Not only do hemorrhagic strokes often lead to secondary cerebrovascular events like vasospasms, ischemia, and hydrocephalus365, 370–373, but they also cause non-neurological complications like heart, lung, kidney, and liver injury or failure.365, 374 Risk factors for SAH and ICH include inflammation375, malformations and tumors376, anticoagulation medication377, and heavy alcohol consumption.378 Lack of screening technology and the reliance on surgical approaches for treatment are major contributors to the devastating effects of these brain bleeds.379–381 Epigenetics alterations represent potential mechanisms through which hemorrhagic stroke may not only be detected, but also treated (Table 5).
Table 5.
Therapeutic Potential of Epigenetic Modulators in Hemorrhagic Stroke
| Model | Therapeutic Potential | Modification | Citation |
|---|---|---|---|
| Intracranial aneurysm humans | RIC, a method used to protect against post-SAH events, dysregulates cell cycle and inflammatory genes | DNA methylation | Nikkola et al., 2015 |
| CD-1 mice, ICH | SAHA reverses H3 and H4 hypoacetylation to provide neuroprotection | Histone acetylation | Sukumari-Ramesh et al., 2016 |
| SD rats, ICH | VPA prevents inflammation via activation of H3 genes | Histone acetylation | Sinn et al., 2007 |
| Microglia, erythrocyte lysate ICH | miR-124 mimics reduce M1 markers and increase M2 markers | MiRNA | Yu et al., 2017 |
| C57 mice, endovascular perforation | Melatonin is neuroprotective by way of H19-let-7a-NGF interaction | MiRNA, lncRNA | Yang et al., 2018 |
| SD rats, ICH | Let-7c antagomir reduces reactive microglia and neutrophils as well as improves functional outcome | MiRNA | Kim et al., 2014 |
| SD rats, ICH | miR-126 mimics reduces reactive microglia and neutrophils as well as apoptosis | MiRNA | Xi et al., 2017 |
| C57 mice, ICH | Augmentation of miR-132 reduces permeability of the BBB | MiRNA | Zhang et al., 2017 |
| BALB/c mice, ICH | Inhibition of miR-144 downregulates autophagy, reduces inflammation, and leads to improved functional recovery | MiRNA | Yu et al., 2017 |
| SD rats, ICH | Restoration of miR-27a-3p is neuroprotective by way of aquaporin-11 | MiRNA | Xi et al., 2018 |
Research on the role of DNA methylation in hemorrhagic stroke is still in its infancy, but a few studies indicate this epigenetic modification may play a role in hemorrhagic stroke pathology. For example, ITPR3, a gene involved in vasospasms caused by SAH382, was significantly hypermethylated in patients that experience delayed cerebral ischemia following SAH.383 In addition, SAH patients with delayed cerebral ischemia had higher levels of DNMT1 as well as lower levels of ITPR3 mRNA and TET1.383 In an autologous blood injection model of mouse ICH, TET1, TET2, TET3 and 5hmC were downregulated from 24–72 hours following hemorrhage.384 AKT2, PDPK1, and VEGF genes displayed decreased hydroxymethylation and increased methylation, resulting in a downregulation of AKT2, PDPK1 and VEGF expression.384
Histone modifications relating to brain bleeds like SAH and ICH have not been thoroughly explored; however, as with each disorder and injury discussed in this review, HDACi may be a promising therapeutic strategy. SAHA administration after spontaneously induced ICH in mice was shown to not only reverse H3 hypoacetylation but also decrease apoptosis, hemin-induced cytotoxicity, behavior deficits, and microglial and astrocytic activation.385 Similar to the neuroprotective effects of SAHA, VPA administration in a rat model of ICH inhibited inflammation and caspase activity, upregulated BCL-2 and BCL-XL while downregulating BAX.386
Despite the lack of classical epigenetic marker studies in SAH and ICH, several studies have been performed assessing the role of noncoding RNAs. Extensive research into circulating biomarkers for hemorrhagic stroke has been conducted. A serum study evaluating miRNA levels has shown that miR-502–5p, miR-1297 and miR-4320 levels are higher in SAH patients when compared to control groups.387 Furthermore, miR-502–5p and miR-1297 were even significantly higher in those with severe SAH when compared to non-severe SAH.387 Another serum study identified 86 differently expressed miRNA, 69 upregulated and 17 downregulated, between three severities of intracranial aneurysms and the healthy control group.388 Important gene pathways found in this study included smooth muscle cell proliferation, apoptosis, myosin generation, and actin cytoskeleton organization.388 Plasma studies have identified miR-16 and miR-25 as dysregulated in intracranial aneurysm patients389 while inflammatory miRNAs are most pronounced in ICH patients.390 Studies in intracranial aneurysm tissue showed aberrations in several miRNA in common including miR-23b, miR-24–1, and isoforms of miR-143 and miR-145.391, 392 These studies both indicate gene networks involved in smooth muscle cell proliferation and movement may be involved.391, 392 Finally, potential biomarkers in cerebrospinal fluid393–395 and animal models396, 397 have also been proposed.
After ICH, plasma and brain tissue levels of miR-124 were increased followed by a slow decrease with patient recovery.398 This is mirrored in an induced ICH rat model, with miR-124 levels returning to normal by day 60.398 In an erythrocyte lysate model of ICH, miR-124 was significantly downregulated in microglia.399 However, when microglia were transduced with miR-124, M1 markers decreased, M2 markers increased, and microglia-induced cytotoxicity of neurons decreased.399 In vivo, mimics of miR-124 also reduced water content of the mouse brain.399 This study further showed that miR-124 acts by way of BCL-2, BCL-XL, and C/EBP-α to produce these neuroprotective effects.399
In ICH and SAH, let-7a and let-7c also play important roles and have shown to have neuroprotective efficacy. In both thrombin-induced cytotoxicity models and induced ICH rat models, let-7c was significantly upregulated.400 Administration of a let-7c antagomir reduced numbers of MPO+ neutrophils and OX42+ microglia in the basal ganglia and cortex.400 In addition, let-7c antagomir treatment improved functional outcome and decreased cell death by restoring IGF1 and p-AKT.400 In the endovascular perforation SAH mouse model, melatonin improved neurological scores and reduced brain water content through the H19 lncRNA, which associates with let-7a and the let-7a target NGF.401
A number of other miRNAs that have shown therapeutic potential by modulating inflammatory responses have also been identified. Administration of miR-126–3p mimic reduced MPO+ neutrophils, OX42+ microglia, and apoptosis in ICH rats.402 Restoration of miR-126 in ICH rats showed neuroprotection, by VEGF upregulation and caspase-3 inhibition.403 MiR-144 upregulation in ICH mice was shown to downregulate the mTOR pathway.404 Inhibition of miR-144 with an antagomir reduced autophagy and inflammation as well as improved function in ICH mice.404 Augmentation of miR-132 in ICH mice reduced brain edema and restored integrity to the BBB.405 MiR-233 was shown to directly bind to and downregulate NLRP3 of the inflammasome after ICH in mice406, providing another mechanism through which inflammation can be prevented. Finally, restoration of miR-27a-3p levels after collagenase-induced ICH in rats improved functional recovery by inhibiting aquaporin-11, increasing BBB integrity, and decreasing edema.407
Dysregulation of lncRNA expression has been observed in several models of hemorrhagic stroke. A study in rats found 64 upregulated and 144 downregulated lncRNA in early brain injury after SAH.408 In mice, SAH led to upregulation of 103 lncRNAs and downregulation of 514 lncRNAs.409 A preliminary study on ICH in rats found 625 differentially expressed lncRNA corresponding to 826 mRNA.410 Within human studies, 2926 differentially expressed lncRNA were identified in intracranial aneurysm tissue and superficial temporal arteries.411 However, further research is needed to identify the specific roles of lncRNAs in hemorrhagic stroke pathogenesis.
Traumatic Brain Injury (TBI)
Traumatic brain injury (TBI) affects nearly 4 million Americans each year and accounts for a large percentage of injury-related deaths.412 Diffuse brain injuries, affecting the entire brain, and focal brain injuries, affecting a specific area of the brain, are most commonly caused in the field of battle or motor vehicle accidents.412–415 TBI presents with a host of neurological symptoms from contusions and hemorrhage to mood changes and memory loss.412, 413, 416 At the molecular level, homeostasis of excitatory neurotransmitter release is disrupted, axonal stretching and shearing occurs, and neurodegenerative pathology like amyloid plaques may appear.412, 417–421 Although the epigenetic study of TBI is in its infancy, results pointing towards therapeutic intervention are promising (Table 6).
Table 6.
Therapeutic Potential of Epigenetic Modulators in Traumatic Brain Injury
| Model | Therapeutic Advantage | Modification | Citation |
|---|---|---|---|
| Sabra mice, CHI | ITF2357 reverses downregulation of Hsp70 and H3 hypoacetylation to provide neuroprotection | Histone acetylation | Shein et al., 2009 |
| C57 mice, CCI | SB in combination with water maze training improves memory | Histone acetylation | Dash et al., 2009 |
| SD rats, CCI | VPA reduces inflammation and apoptosis | Histone acetylation | Tai et al., 2014 |
| C57 mice, CCI | VPA in combination with lithium prevents H3 hypoacetylation and reduces lesion volume | Histone acetylation | Yu et al., 2013 |
| SD rats, CCI | VPA prevents the hypoacetylation of H3 and H4 while decreasing the permeability of the BBB | Histone acetylation | Dash et al., 2010 |
| C57 mice, CCI | Fluoxetine prevents H3 and H4 hypoacetylation while inducing hippocampal neurogenesis | Histone acetylation | Wang et al., 2011 |
| Wistar rats, HCb | RvD1 administration is neuroprotective via ALX/FPR2 | MiRNA | Bisicchia et al., 2018 |
| SD rats, FPI | MiR-21 agomir upregulates (Ang-1)/Tie-2 to decrease BBB leakage | MiRNA | Ge et al., 2015 |
| SD rats, CCI | H2 gas increases miR-21 and improves BBB permeability | MiRNA | Wang et al., 2018 |
| Wistar rats, weight drop | Formononetin reverses miR-155 downregulation and improves functional recovery | MiRNA | Li et al., 2017 |
| C57 mice, CCI | MiR-155 antagomir prevents against inflammation and reduces lesion volume | MiRNA | Henry et al., 2019 |
| C57 mice, CCI | MiR-23a and miR-27a mimics provide neuroprotection via decreased apoptotic markers | MiRNA | Sabirzhanov et al., 2014 |
| Cortical neurons, scratch | MiR-21 overexpression is neuroprotective | MiRNA | Han et al., 2014 |
| HT-22 neurons, scratch | MiR-21–5p overexpressing cells or exosomes aid in neuroprotection of other neurons | MiRNA | Li et al., 2019 |
| SD rats, fluid percussion | MiR-21 agomir prevents apoptosis and promotes angiogenesis | MiRNA | Ge et al., 2014 |
| SD rats, weight drop | MiR-23a overexpression downregulates ATG12 to suppress autophagy | MiRNA | Sun et al., 2018 |
| SD rats, weight drop | MiR-27a overexpression downregulates FoxO3a to suppress autophagy | MiRNA | Sun et al., 2017 |
| BV2 microglia, rTBI and C57 mice, rTBI | Exosomes derived from miR-124–3p overexpressing microglia are neuroprotective | MiRNA | Huang et al., 2018 |
| C57 mice, CCI | Let-7c-5p mimic decreases microglial activation to provide functional recovery | MiRNA | Lv et al., 2018 |
| C57 mice, CCI | Neat1 knockdown is neuroprotective | LncRNA | Zhong et al., 2017 |
| SD rats, weight drop | MiR-144 antagomir improves functional recovery and long-term potentiation | MiRNA | Sun et al., 2017 |
| C57 mice, CCI | Voluntary running wheel improves functional recovery and dysregulates miR-34a and miR-21 | MiRNA | Bao et al., 2014 |
| C57 mice, CCI | Voluntary running wheel improves functional recovery and dysregulates several miRNA | MiRNA | Miao et al., 2015 |
| SD rats, fluid percussion | Therapeutic hypothermia improves functional recovery and dysregulates miR-874 and miR-451 | MiRNA | Truettner et al., 2011 |
In a weight drop model of TBI in adult rats, global cellular DNA hypomethylation was observed within 24 hours and up to 48 hour post-injury.422 Activated microglia/macrophages were identified as the major source of the reduced 5-mC in the peri-lesion area.422 Within repeated blast-injury model of TBI in rats, significant differential methylation between neurons and glia has been shown in the transforming growth factor β (TGF-β) pathway, which includes genes like MAP2K6, RUNX3, NODAL, and SMAD1 which regulates cell survival.423 Hypermethylation and decreased expression of the aralkylamine N-acetyltransferase AANAT gene that mediates serotonin-to-melatonin conversion was also observed.423 In blast-induced TBI in rats, there was a negative correlation between blast severity and global DNA methylation levels in the hippocampus, indicating that the degree of injury may influence methylation imbalance.424 In a rat blast model of TBI, hippocampal DNMT1, DNMT3b, TET2, TET3 and TDG expression increased while prefrontal cortical DNMT3b expression was decreased and TET2 expression was increased two weeks following injury.425 In addition, controlled cortical impact (CCI) induced TBI in rats upregulates insulin-like growth factor 1B (IGF-1B) hippocampal and cortical mRNA levels.426 Three days after injury, when IGF-1B mRNA levels are highest, the P1 promoter region and sites upstream/within exon 5 are hypermethylated while the P2 promotor region is unchanged and sites downstream of exon 5 are hypomethylated with respect to sham animals.426 IGF-1B has been implicated in neural plasticity and regeneration and is considered as a potential therapeutic target after TBI.427
Several epigenetic marks of histone acetylation that activate gene expression including H3k9ac and H3K4me3 at the P1 promoter region, H3K9ac, H3K14ac, H3K36me3, and H3K4me3 at the P2 promoter region, and H3K9ac, H3K36me3, and H3K4me3 at exon 5 of IGF-1B are also increased after CCI injury in rats three days after injury.426 In the hippocampal CA3 region, both acetylation and methylation were decreased 6–72 hours after CCI injury in another rat study.428 Following closed head injury TBI in mice, administration of the HDAC inhibitor ITF2357 was shown to be neuroprotective by preventing H3 deacetylation.429 Several other HDAC inhibitors were also tested after TBI. VPA decreases lesion volume and improves motor function in rats subjected to CCI TBI.430, 431 Combined administration of sub-effective VPA and lithium doses had the same influence on mice subject to CCI injury.432 Fluoxetine induces neurogenesis in CCI injury mice; however, no improvement in gait or spatial learning and memory is seen.433 Furthermore, sodium butyrate administration to mice subject to CCI injury increases histone acetylation, but does not significantly improve function unless combined with behavioral training.434
Several studies have shown that dysregulation of hundreds of miRNAs occurs in the rodent hippocampus and cortex following TBI as early as 1 hour, and up to 7 days post injury.435–440 Distinctive miRNA expression profiles have been observed in a temporal manner following TBI, indicating the association of various miRNAs with differing stages of TBI progression. Altered miRNAs have been associated with processes that regulate transcriptional regulation, oxidative stress, metabolism, synaptic signaling and signal transduction, inflammation, neurogenesis, angiogenesis and apoptosis.435, 437, 439 Interestingly, a mouse study using a weight drop model of TBI, showed that differential expression of miRNAs may also be observed depending on the severity of TBI.441
A study performed in the rat hippocampus found 10 miRNAs that were consistently altered from 1 hour to 7 days following moderate TBI. Of these, miR-144, miR-153 and miR-340–5p were elevated and their predicted targets calcium/calmodulin-dependent serine protein kinase, nuclear factor erythroid 2-related factor (Nrf2) and alpha-synuclein were downregulated, respectively.437 Although the roles of miR-144, miR-152 and miR-340–5p in TBI have not been evaluated, their targets have been shown to play important roles in neuroprotection and are implicated in learning and memory following TBI in rats.437, 442 Other miRNAs with important roles in TBI include miR-23a and miR-27a which have been shown to reduce apoptosis and modulate autophagy in a neuroprotective manner following overexpression or mimetic treatment in rodent models of TBI.443–445 MiR-124–3p was shown to increase in microglia and microglial exosomes from mouse brain extracts after treatment with repetitive CCI injury.446 Administration of exosomes derived from microglia overexpressing miR-124–3p improved mice neurological recovery following repetitive CCI by downregulating mTOR and reducing inflammation.446 Let-7c-5p overexpression was also shown to reduce inflammatory processes by attenuating microglial activation, which led to reduced brain edema improved neurological scores in mice subjected to CCI.447 Interestingly, miR-155 has been shown to be both neuroprotective448, 449 and damaging to the brain.450 The differences in experimental design may have led to these results as the first study tested miR-155 knockout mice with CCI448, the second used the formononetin drug to modulate miR-155 in rats using weight drop injury449 and the third tested RNA silencing of miR-155 in mice with CCI.450
Other studies have shown that miR-21 expression is consistently upregulated from 6h to 72h in the cortex and from 24h to 72h in the hippocampus following CCI in rodents436, 440, 451, indicating miR-21 may play a pivotal role in pathology of TBI progression. In a study evaluating age differences, miR-21 was upregulated in adult mice subject to brain injury; however, miR-21 was not altered following injury in elderly mice.452 Despite increased expression of miR-21 upon TBI, miR-21 mimetic administration has been shown to be neuroprotective against scratch-cell injury in cortical neurons453 as well as rats subjected to fluid percussion.454 Co-culture of scratch-injured neurons with miR-21–5p-overexpressing neurons or administration of miR-21–5p overexpressing exosomes was also neuroprotective.455 Finally, MiR-21 mimic administration in rats with or without hydrogen gas treatment improved functional outcome following CCI or fluid percussion injury, respectively.456, 457 In addition to hippocampal tissue, miR-21 expression increased in extracellular vesicles after TBI, suggesting its promise in future biomarker screens.458
There have been several attempts to establish miRNA profiles to serve biomarker development. Differential expression has been surveyed in serum459–462, plasma463–467, CSF462, 468–470, saliva468, and extracellular vesicles471 of humans and animals. Several therapeutic interventions such as exercise hypothermia have been shown to alter the miRNA profiles and improve cognitive function and recovery following TBI in rodents.472–474 The let-7 family has been involved in multiple studies467, 469 and may prove to be a promising pathway in TBI.
Emerging evidence indicates lncRNAs may be implicated in TBI. Microarray studies have revealed significant alterations in lncRNA expression in the brain following rodent models of TBI.475–477 Administration of MALAT1 deficient exosomes derived from adipose stem cells caused rats to develop larger lesions following CCI.478 Bioinformatics analysis revealed that MALAT1 might modulate pathways related to inflammation and cell regeneration, indicating potential therapeutic effects of MALAT1 in TBI.478 In mice subjected to CCI, NEAT1 overexpression inhibited apoptosis and inflammation, while knockdown of NEAT1 downregulated hundreds of genes, many of which are involved in synaptic and axonal health.479
Epilepsy
Epilepsy, a group of neurological disorders characterized by recurrent seizures, affects 50 million people globally.480, 481 Epilepsy encompasses both focal and generalized seizures; however, most common is temporal lobe epilepsy (TLE) which often presents with hippocampal sclerosis.481–484 Epilepsy can be brought about by trauma, an infection, or improper neurodevelopment leading to neuronal hyperexcitability that can lie latent for years.481, 485 The disorders are commonly marked by anxiety, cognitive defects, and decreased social interaction.481, 486, 487 Although antiepileptic drugs are given to reverse excitability, they do not target the root cause of the disorder and over 30% of patients become drug resistant.483, 485, 488, 489 In addition, surgical intervention has a nearly 50% failure rate.483, 490 Target genes in epileptogenesis that lead to several pathways of dysfunction have been established, including synaptic plasticity, ion transport, and inflammation, suggesting epigenetic therapies may be possible (Table 7).483, 491–497
Table 7.
Therapeutic Potential of Epigenetic Modulators in Epilepsy
| Model | Therapeutic Potential | Modification | Citation |
|---|---|---|---|
| Wistar rats, pilocarpine | Ketogenic diet reduces global methylation and seizure severity | DNA methylation | Kobow et al., 2013 |
| C57 mice, kainic acid | RG108 prevents RASgrf1 methylation and reduces seizures | DNA methylation | Chen et al., 2017 |
| SD rats, kainic acid | Methionine restores BDNF methylation to improve memory | DNA methylation | Parrish et al., 2015 |
| SD rats, pilocarpine | TSA reverses deacetylation of H4 at the GRIA2 promoter | Histone acetylation | Huang et al., 2002 |
| WAG/Rij rats | Pretreatment of SB, VPA, or both reduces seizure severity | Histone acetylation | Citraro et al., 2019 |
| C57 mice, hippocampus kindling | SB improves functional recovery and reduces epileptic morphology | Histone acetylation | Reddy et al., 2018 |
| Human serum | Epilepsy can be predicted with 81% specificity by miRNA biomarkers | MiRNA | Wang et al., 2015 |
| C57 mice, pilocarpine | miR-134 antagomir reduced number of mice that developed seizures and reduced severity in those that did | MiRNA | 24874920 |
| C57 mice, pentylenetetrazol | miR-134 antagomir reduces seizure severity and epileptic behavior | MiRNA | 28325299 |
| C57 mice, kainic acid | miR-132 antagomir reduces neurotoxicity after seizures | MiRNA | 21945804 |
| SD rats, kainic acid | Upregulating miR-124 has both anti-epileptic and pro-epileptic effects | MiRNA | 26947066 |
| SD rats, kainic acid | Inhibition of H19 prevents neurotoxicity after seizures | LncRNA | 29795132 |
There have been several attempts to characterize DNA methylation within epilepsy. Global hypermethylation has been observed in epileptic human and mouse hippocampal specimens affecting pathways such as neuron remodeling and maturation.498, 499 Levels of DNMT1 and DNMT3a were increased in human TLE neocortices specifically in NeuN-positive neurons, but not GFAP-positive astrocytes.500 Comparisons of brain tissue between drug-refractory epileptic patients and control patients showed differential methylation of 224 genes.501 A ketogenic, high fat, low carbohydrate diet was shown to reverse global hypermethylation and ameliorate seizure progression in pilocarpine-induced epileptic rats499, indicating that aberrant methylation promotes pathology. Interestingly, induction of epilepsy in rats through focal amygdala stimulation, systemic pilocarpine injection, or lateral fluid-percussion induced traumatic brain injury did not result in a similar spatiality or degree of methylation despite global hypermethylation across all three models.502 This suggests that although global hypermethylation is a general feature of epilepsy, distinct DNA methylation patterns exist depending on etiology.502
In humans with focal epilepsy and febrile seizures, the promoter of carboxypeptidase 6 (CPA6), a gene involved in familial and sporadic cases of epilepsy, is highly methylated compared to controls.503 Patients with TLE were shown to have increased reelin promoter methylation, a gene important to hippocampal development.504 Subjects with juvenile myoclonic epilepsy have varied methylation of cation-chloride transporters, with lower sodium-potassium-chloride cotransporter 1 methylation and higher potassium-chloride transporter member 5 methylation in the epilepsy group.505 Increased methylation of ionotropic glutamate receptor 2 (GRIA2) has been observed in hippocampal slices of mice and rats subjected to kainic acid-induced epilepsy, which was reversed with administration of the DNMT inhibitor RG108.506 TLE patients and pilocarpine-induced epileptic rats display decreased levels of Ras-guanine nucleotide-releasing factor 1 (RASgrf1) in the neocortex and hippocampus, respectively.507 In mice subject to acute epileptic seizures using kainic acid, the RASgrf1 promoter was methylated, suppressing levels of RASgrf mRNA; however, treatment with the DNMT inhibitor RG108 reversed this trend as well as reduced seizures.508 During memory consolidation in kainic acid-induced TLE rats, methylation of BDNF was significantly decreased leading to an upregulation of BDNF mRNA and increased memory deficit; however, following administration of methionine, BDNF methylation was increased, BDNF mRNA was decreased, and memory deficits were reversed.509
The methylation pattern of noncoding gene promoters has also been studied. In human hippocampus specimens, 12 differentially methylated miRNA were discovered, with half hypomethylated and upregulated and half hypermethylated and downregulated.498 This same study found hypermethylation of several lncRNAs including UCA1, ADARB2-AS1, LINC324, and MAP3K14-AS1.498 In a study characterizing epileptic whole blood samples, 87% and 85% of differentially methylated lncRNA and miRNA promoters were hypermethylated, respectively.510 Differentially methylated lncRNA were related mRNAs involved in ion/gated channel activity, GABA receptor activity, and synaptic transmission while differentially methylated miRNA were related to neuronal projection and differentiation, protein kinase activity, and axonal guidance.510
Several studies have attempted to characterize the landscape of histone modifications within epilepsy. Expression of HDAC5 and HDAC9 was shown to increase significantly in mice subject to kainic acid, while expression of HDAC5 and HDAC9 decreased in pilocarpine-induced epileptic mice.511 In addition, both epileptic groups of mice showed a sharp decrease in HDAC7 expression during the acute seizure period.511 Kainic acid-induced epileptic mice displayed decreased HDAC1, 2, and 11 expression in the acute phase (2–6h after kainic acid treatment) followed by a subsequent increase of all class I HDACs from 12–48 hours.512 Pilocarpine-treated mice model showed similar acute results, but HDAC2, 3, and 8 were decreased during the chronic phase (14 and 28 days following pilocarpine treatment).512 In rats, pilocarpine exposures led to decreased hippocampal acetylation of H4 at the GRIA2 promoter, downregulating GRIA2 mRNA expression.513 However, administration of the HDACi TSA reversed deacetylation and prevented GRIA2 mRNA downregulation.513 Interestingly, this study also found that H4 acetylation increased at the BDNF promoter P2.513 The findings of this study complement the previously discussed methylation studies and point to both DNA methylation and histone acetylation playing a role in GRIA2 and BDNF levels in epilepsy.
Rodent studies have shown promise for HDACi treatment in epilepsy. Administration of SB or VPA in WAG/Rij rats (a model for absent epilepsy that displays H3 and H4 hypoacetylation) increased brain histone acetylation, decreased HDAC1 and HDAC3 expression, and reduced seizures.514 Furthermore, the protective effects were intensified by co-administration of SB and VPA.514 SB was also used in a mouse kindling model of TLE which showed decreased HDAC expression, reduced seizures, and decreased mossy fiber sprouting.515
The field of noncoding RNA within the pathogenesis, prediction, and prevention of epilepsy has been extensively explored. MiR-134 has been found to be an important factor in dendritic spine density and morphology in pilocarpine- or kainic acid-induced mouse models of epilepsy.516, 517 MiR-134 inhibition reduced seizures in multiple rodent models of epilepsy.516–518 In the pilocarpine mouse model of status epilepticus, dendritic spine volume increased upon administration of a cholesterol-tagged locked nucleotide acid (LNA) miR-134 antagomir in CA3 pyramidal neurons.516 Pre-treatment with the LNA antagomir and subsequent induction of status epilepticus resulted in increased survival and decreased seizures.516 Another study showed that miR-134 LNA antagomir reduced dendritic spine density in CA3 pyramidal neurons, but still prevented seizures in kainic acid-treated mice.517 In the mouse pentylenetetrazol-induced model of epilepsy, LNA miR-134 antagomir treatment reduced the number of spontaneous seizures and convulsive behavior.518 Similarly, rats subjected to the perforant pathway stimulation model showed reduced spontaneous seizures with LNA miR-134 antagomir treatment.518
Status epilepticus induced by kainic acid in mice was shown to increase levels of miR-132 as well as its binding to Argonaute-2.519 Subsequent inhibition of miR-123 with an LNA antagomir reduced hippocampal neuronal death.519 MiR-124 was significantly reduced after kainic acid induction in rats and subsequent supplementation with synthetic miR-124 inhibited NSRF, effectively contributing to neuroprotection against epilepsy.520 However, miR-124 supplementation also promoted inflammation, by enhancing microglia activation, effectively contributing to the epileptic state.520 Another study showed that intrahippocampal administration of miR-124 reduced the severity and occurrence of seizures in both the pentylenetetrazole- and pilocarpine-induced rat models of epilepsy by repressing of CREB, a key protein in epileptogenesis.520, 521 Importantly, it has been shown that miR-124 was decreased in the hippocampus of adult patients with TLE520, while miR-124 was upregulated in the hippocampus of children with mesial TLE522, revealing dynamic differences of this miRNA with age and etiology.
Pilocarpine-induced temporal lobe epilepsy in mice resulted in the upregulation of 22 lncRNA and downregulation of 83 lncRNA in the hippocampus.523 Another mouse study identified the dysregulation of 384 lncRNA in the pilocarpine model and 279 in the kainic acid model when analyzing whole brain, olfactory bulb, and cerebellum tissue.524 The lncRNA UCA1 was shown to increase in tandem with NF-κB after pilocarpine-induced epilepsy in the brain of rats525 and UCA1 overexpression inhibited apoptosis and suppressed seizures.526 Finally, the lncRNA H19 is significantly upregulated in the latent period of pilocarpine-induced and kainic acid-induced epilepsy in rats where it was shown to regulate apoptosis through sponging of let-7b.527 Microarray analysis following knockdown or overexpression of H19 in kainic-acid induced epileptic rats revealed involvement of H19 in a number of epileptogenic processes including demyelination, immune response, and inflammation.528 Inhibition of H19 was shown to protect against hippocampal neuronal death in kainic acid-treated rats527, indicating a potential role for H19 as a therapeutic target in epilepsy.
Several studies have been conducted to identify miRNA biomarkers of epilepsy in serum, of which the most accurate predict the disorder with 81.2% sensitivity.529, 530 Other studies have observed expression profiles that distinctly identify epilepsy in human cerebrospinal fluid531, rat hippocampus and peripheral blood532, rat hippocampal granule cells and plasma533, and rat synaptosomes.534, 535 Profiling studies have also characterized miRNA from the hippocampus of epilepsy patients with TLE, showed overall failure of mature miRNA processing due to Dicer loss in TLE and identified dysregulation of a number of miRNAs involved in immune response.536, 537 Mooney et al. has provided a comprehensive database known as EpimiRBase providing a plethora of miRNA-epilepsy associations in both human and animal studies538 facilitating future work in identifying key miRNAs involved pathophysiology or as biomarkers.
CONCLUSION & FUTURE DIRECTIONS
Advancements in epigenetics have revolutionized our understanding into the mechanisms involved in brain diseases. The evidence thus far from various disease models from cell lines to the post-mortem human brain reveals a critical role for epigenetic dysfunction in neurodegenerative diseases and acute brain injury. These studies demonstrate that epigenetic imbalances in DNA methylation and histone modifications by molecular readers, writers and erasers predispose the brain to disease as well as influence neurological recovery. While the role of epigenetics in diseases of the brain is still emerging, it is clear that restoration of epigenetic imbalances may provide potential treatments that lead to recovery. HDAC inhibitors have emerged as an overlapping therapy among all of the brain diseases discussed. However, despite the protective effects shown in animal studies, some studies have shown certain downsides to HDAC inhibitor therapy including inflammation, oxidative stress and apoptosis.539–541 Therefore, additional research is needed to further delineate the molecular mechanisms by which HDAC inhibitors function in various CNS disorders. Profiling of blood biomarkers is another promising clinical epigenetic approach that may help diagnosis and therapeutic development for CNS disorders. Biomarker profiling of DNA methylation and miRNA has already been used to develop prognostic and diagnostic indicators in brain disorders, and epigenetic profiles in neurological diseases continue to be developed. More recently, lncRNA profiling has emerged as an additional potential biomarker tool. Modulation of various epigenetic regulatory mechanisms has been shown to provide therapeutic potential against several pathophysiological mechanisms. However, the current challenge lies in further elucidating the interplay between epigenetic changes and the downstream effects on gene expression and the complex cellular environment of the diseased brain.
AKNOWLEDGEMENT
This work was supported by NIH grants RO1 NS101960, RO1 NS099531, RO1 NS109459 grants and R21 NS095192.
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflict of interest.
References
- 1.Barouki R, Melen E, Herceg Z, Beckers J, Chen J, Karagas M, et al. Epigenetics as a mechanism linking developmental exposures to long-term toxicity. Environ Int. 2018;114:77–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haggarty P Genetic and metabolic determinants of human epigenetic variation. Curr Opin Clin Nutr Metab Care. 2015;18:334–338 [DOI] [PubMed] [Google Scholar]
- 3.Feinberg AP. Epigenetics at the epicenter of modern medicine. Jama. 2008;299:1345–1350 [DOI] [PubMed] [Google Scholar]
- 4.Waddington CH. The epigenotype. 1942. Int J Epidemiol. 2012;41:10–13 [DOI] [PubMed] [Google Scholar]
- 5.Szulwach KE, Jin P. Integrating dna methylation dynamics into a framework for understanding epigenetic codes. Bioessays. 2014;36:107–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harrison IF, Dexter DT. Epigenetic targeting of histone deacetylase: Therapeutic potential in parkinson’s disease? Pharmacol Ther. 2013;140:34–52 [DOI] [PubMed] [Google Scholar]
- 7.Hobert O Gene regulation by transcription factors and micrornas. Science. 2008;319:1785–1786 [DOI] [PubMed] [Google Scholar]
- 8.Roberts TC, Morris KV, Weinberg MS. Perspectives on the mechanism of transcriptional regulation by long non-coding rnas. Epigenetics. 2014;9:13–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mehler MF. Epigenetic principles and mechanisms underlying nervous system functions in health and disease. Prog Neurobiol. 2008;86:305–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Felling RJ, Song H. Epigenetic mechanisms of neuroplasticity and the implications for stroke recovery. Exp Neurol. 2015;268:37–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holliday R, Pugh JE. Dna modification mechanisms and gene activity during development. Science. 1975;187:226–232 [PubMed] [Google Scholar]
- 12.Baccarelli A, Rienstra M, Benjamin EJ. Cardiovascular epigenetics: Basic concepts and results from animal and human studies. Circ Cardiovasc Genet. 2010;3:567–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deaton AM, Bird A. Cpg islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miranda TB, Jones PA. Dna methylation: The nuts and bolts of repression. J Cell Physiol. 2007;213:384–390 [DOI] [PubMed] [Google Scholar]
- 15.Rottach A, Leonhardt H, Spada F. Dna methylation-mediated epigenetic control. J Cell Biochem. 2009;108:43–51 [DOI] [PubMed] [Google Scholar]
- 16.Klose RJ, Bird AP. Genomic dna methylation: The mark and its mediators. Trends Biochem Sci. 2006;31:89–97 [DOI] [PubMed] [Google Scholar]
- 17.Levenson JM, Sweatt JD. Epigenetic mechanisms in memory formation. Nat Rev Neurosci. 2005;6:108–118 [DOI] [PubMed] [Google Scholar]
- 18.Feng J, Fouse S, Fan G. Epigenetic regulation of neural gene expression and neuronal function. Pediatr Res. 2007;61:58r–63r [DOI] [PubMed] [Google Scholar]
- 19.Zhao H, Han Z, Ji X, Luo Y. Epigenetic regulation of oxidative stress in ischemic stroke. Aging Dis. 2016;7:295–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-cpg binding proteins. Mol Cell Biol. 1998;18:6538–6547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du Q, Luu PL, Stirzaker C, Clark SJ. Methyl-cpg-binding domain proteins: Readers of the epigenome. Epigenomics. 2015;7:1051–1073 [DOI] [PubMed] [Google Scholar]
- 22.Ehrlich M, Gama-Sosa MA, Huang LH, Midgett RM, Kuo KC, McCune RA, et al. Amount and distribution of 5-methylcytosine in human dna from different types of tissues of cells. Nucleic Acids Res. 1982;10:2709–2721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCartney DL, Stevenson AJ, Walker RM, Gibson J, Morris SW, Campbell A, et al. Investigating the relationship between dna methylation age acceleration and risk factors for alzheimer’s disease. Alzheimers Dement (Amst). 2018;10:429–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horvath S Dna methylation age of human tissues and cell types. Genome Biol. 2013;14:R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49:359–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levine ME, Lu AT, Bennett DA, Horvath S. Epigenetic age of the pre-frontal cortex is associated with neuritic plaques, amyloid load, and alzheimer’s disease related cognitive functioning. Aging (Albany NY). 2015;7:1198–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of tet proteins in 5mc to 5hmc conversion, es-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian dna by mll partner tet1. Science. 2009;324:930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minor EA, Court BL, Young JI, Wang G. Ascorbate induces ten-eleven translocation (tet) methylcytosine dioxygenase-mediated generation of 5-hydroxymethylcytosine. J Biol Chem. 2013;288:13669–13674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin IH, Chen YF, Hsu MT. Correlated 5-hydroxymethylcytosine (5hmc) and gene expression profiles underpin gene and organ-specific epigenetic regulation in adult mouse brain and liver. PLoS One. 2017;12:e0170779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szulwach KE, Li X, Li Y, Song CX, Wu H, Dai Q, et al. 5-hmc-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci. 2011;14:1607–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miao Z, He Y, Xin N, Sun M, Chen L, Lin L, et al. Altering 5-hydroxymethylcytosine modification impacts ischemic brain injury. Hum Mol Genet. 2015;24:5855–5866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wen L, Li X, Yan L, Tan Y, Li R, Zhao Y, et al. Whole-genome analysis of 5-hydroxymethylcytosine and 5-methylcytosine at base resolution in the human brain. Genome Biol. 2014;15:R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mellen M, Ayata P, Heintz N. 5-hydroxymethylcytosine accumulation in postmitotic neurons results in functional demethylation of expressed genes. Proc Natl Acad Sci U S A. 2017;114:E7812–e7821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, et al. Genome-wide regulation of 5hmc, 5mc, and gene expression by tet1 hydroxylase in mouse embryonic stem cells. Mol Cell. 2011;42:451–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bassi S, Tripathi T, Monziani A, Di Leva F, Biagioli M. Epigenetics of huntington’s disease. Adv Exp Med Biol. 2017;978:277–299 [DOI] [PubMed] [Google Scholar]
- 38.Maiti A, Drohat AC. Thymine dna glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: Potential implications for active demethylation of cpg sites. J Biol Chem. 2011;286:35334–35338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen H, Dzitoyeva S, Manev H. Effect of aging on 5-hydroxymethylcytosine in the mouse hippocampus. Restor Neurol Neurosci. 2012;30:237–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qureshi IA, Mehler MF. Emerging role of epigenetics in stroke: Part 1: Dna methylation and chromatin modifications. Arch Neurol. 2010;67:1316–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cairns BR. The logic of chromatin architecture and remodelling at promoters. Nature. 2009;461:193–198 [DOI] [PubMed] [Google Scholar]
- 42.Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: A new frontier for drug discovery. Nat Rev Drug Discov. 2012;11:384–400 [DOI] [PubMed] [Google Scholar]
- 43.Ammal Kaidery N, Tarannum S, Thomas B. Epigenetic landscape of parkinson’s disease: Emerging role in disease mechanisms and therapeutic modalities. Neurotherapeutics. 2013;10:698–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Habibi E, Masoudi-Nejad A, Abdolmaleky HM, Haggarty SJ. Emerging roles of epigenetic mechanisms in parkinson’s disease. Funct Integr Genomics. 2011;11:523–537 [DOI] [PubMed] [Google Scholar]
- 45.Cacabelos R, Torrellas C. Epigenetics of aging and alzheimer’s disease: Implications for pharmacogenomics and drug response. Int J Mol Sci. 2015;16:30483–30543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wagner FF, Wesmall yi UM, Lewis MC, Holson EB. Small molecule inhibitors of zinc-dependent histone deacetylases. Neurotherapeutics. 2013;10:589–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chakravarty S, Pathak SS, Maitra S, Khandelwal N, Chandra KB, Kumar A. Epigenetic regulatory mechanisms in stress-induced behavior. Int Rev Neurobiol. 2014;115:117–154 [DOI] [PubMed] [Google Scholar]
- 48.Grayson DR, Kundakovic M, Sharma RP. Is there a future for histone deacetylase inhibitors in the pharmacotherapy of psychiatric disorders? Mol Pharmacol. 2010;77:126–135 [DOI] [PubMed] [Google Scholar]
- 49.Ratan RR. Epigenetics and the nervous system: Epiphenomenon or missing piece of the neurotherapeutic puzzle? Lancet neurol. England; 2009:975–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Finelli MJ, Wong JK, Zou H. Epigenetic regulation of sensory axon regeneration after spinal cord injury. J Neurosci. 2013;33:19664–19676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qureshi IA, Mehler MF. Epigenetics and therapeutic targets mediating neuroprotection. Brain Res. 2015;1628:265–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carthew RW, Sontheimer EJ. Origins and mechanisms of mirnas and sirnas. Cell. 2009;136:642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: Micrornas can up-regulate translation. Science. 2007;318:1931–1934 [DOI] [PubMed] [Google Scholar]
- 54.Trujillo RD, Yue SB, Tang Y, O’Gorman WE, Chen CZ. The potential functions of primary micrornas in target recognition and repression. Embo j. 2010;29:3272–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gruber AJ, Zavolan M. Modulation of epigenetic regulators and cell fate decisions by mirnas. Epigenomics. 2013;5:671–683 [DOI] [PubMed] [Google Scholar]
- 56.Millan MJ. The epigenetic dimension of alzheimer’s disease: Causal, consequence, or curiosity? Dialogues Clin Neurosci. 2014;16:373–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang Y, Shu X, Liu D, Shang Y, Wu Y, Pei L, et al. Epac null mutation impairs learning and social interactions via aberrant regulation of mir-124 and zif268 translation. Neuron. 2012;73:774–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gao R, Wang L, Sun J, Nie K, Jian H, Gao L, et al. Mir-204 promotes apoptosis in oxidative stress-induced rat schwann cells by suppressing neuritin expression. FEBS Lett. 2014;588:3225–3232 [DOI] [PubMed] [Google Scholar]
- 59.Davis TH, Cuellar TL, Koch SM, Barker AJ, Harfe BD, McManus MT, et al. Conditional loss of dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J Neurosci. 2008;28:4322–4330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The gencode v7 catalog of human long noncoding rnas: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Faghihi MA, Zhang M, Huang J, Modarresi F, Van der Brug MP, Nalls MA, et al. Evidence for natural antisense transcript-mediated inhibition of microrna function. Genome Biol. 2010;11:R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li LC. Chromatin remodeling by the small rna machinery in mammalian cells. Epigenetics. 2014;9:45–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, et al. Expression of a noncoding rna is elevated in alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ng SY, Bogu GK, Soh BS, Stanton LW. The long noncoding rna rmst interacts with sox2 to regulate neurogenesis. Mol Cell. 2013;51:349–359 [DOI] [PubMed] [Google Scholar]
- 65.Deng B, Cheng X, Li H, Qin J, Tian M, Jin G. Microarray expression profiling in the denervated hippocampus identifies long noncoding rnas functionally involved in neurogenesis. BMC Mol Biol. 2017;18:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang S, Zhang X, Guo Y, Rong H, Liu T. The long noncoding rna hotair promotes parkinson’s disease by upregulating lrrk2 expression. Oncotarget. 2017;8:24449–24456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang X, Tang X, Liu K, Hamblin MH, Yin KJ. Long noncoding rna malat1 regulates cerebrovascular pathologies in ischemic stroke. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2017;37:1797–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang J, Zhao H, Fan Z, Li G, Ma Q, Tao Z, et al. Long noncoding rna h19 promotes neuroinflammation in ischemic stroke by driving histone deacetylase 1-dependent m1 microglial polarization. Stroke. 2017;48:2211–2221 [DOI] [PubMed] [Google Scholar]
- 69.Chandran R, Mehta SL, Vemuganti R. Non-coding rnas and neuroprotection after acute cns injuries. Neurochem Int. 2017;111:12–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reitz C, Mayeux R. Alzheimer disease: Epidemiology, diagnostic criteria, risk factors and biomarkers. Biochem Pharmacol. 2014;88:640–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Benilova I, Karran E, De Strooper B. The toxic abeta oligomer and alzheimer’s disease: An emperor in need of clothes. Nat Neurosci. 2012;15:349–357 [DOI] [PubMed] [Google Scholar]
- 72.Jacobs HI, Radua J, Luckmann HC, Sack AT. Meta-analysis of functional network alterations in alzheimer’s disease: Toward a network biomarker. Neurosci Biobehav Rev. 2013;37:753–765 [DOI] [PubMed] [Google Scholar]
- 73.Alonso AD, Grundke-Iqbal I, Barra HS, Iqbal K. Abnormal phosphorylation of tau and the mechanism of alzheimer neurofibrillary degeneration: Sequestration of microtubule-associated proteins 1 and 2 and the disassembly of microtubules by the abnormal tau. Proc Natl Acad Sci U S A. 1997;94:298–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ertekin-Taner N Genetics of alzheimer disease in the pre- and post-gwas era. Alzheimers Res Ther. 2010;2:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Adwan L, Zawia NH. Epigenetics: A novel therapeutic approach for the treatment of alzheimer’s disease. Pharmacol Ther. 2013;139:41–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Citron M, Teplow DB, Selkoe DJ. Generation of amyloid beta protein from its precursor is sequence specific. Neuron. 1995;14:661–670 [DOI] [PubMed] [Google Scholar]
- 77.Shoji M, Golde TE, Ghiso J, Cheung TT, Estus S, Shaffer LM, et al. Production of the alzheimer amyloid beta protein by normal proteolytic processing. Science. 1992;258:126–129 [DOI] [PubMed] [Google Scholar]
- 78.Tohgi H, Utsugisawa K, Nagane Y, Yoshimura M, Genda Y, Ukitsu M. Reduction with age in methylcytosine in the promoter region −224 approximately −101 of the amyloid precursor protein gene in autopsy human cortex. Brain Res Mol Brain Res. 1999;70:288–292 [DOI] [PubMed] [Google Scholar]
- 79.Iwata A, Nagata K, Hatsuta H, Takuma H, Bundo M, Iwamoto K, et al. Altered cpg methylation in sporadic alzheimer’s disease is associated with app and mapt dysregulation. Hum Mol Genet. 2014;23:648–656 [DOI] [PubMed] [Google Scholar]
- 80.Nunan J, Small DH. Regulation of app cleavage by alpha-, beta- and gamma-secretases. FEBS Lett. 2000;483:6–10 [DOI] [PubMed] [Google Scholar]
- 81.Morrison LD, Smith DD, Kish SJ. Brain s-adenosylmethionine levels are severely decreased in alzheimer’s disease. J Neurochem. 1996;67:1328–1331 [DOI] [PubMed] [Google Scholar]
- 82.Fuso A, Seminara L, Cavallaro RA, D’Anselmi F, Scarpa S. S-adenosylmethionine/homocysteine cycle alterations modify dna methylation status with consequent deregulation of ps1 and bace and beta-amyloid production. Mol Cell Neurosci. 2005;28:195–204 [DOI] [PubMed] [Google Scholar]
- 83.Scarpa S, Fuso A, D’Anselmi F, Cavallaro RA. Presenilin 1 gene silencing by s-adenosylmethionine: A treatment for alzheimer disease? FEBS Lett. 2003;541:145–148 [DOI] [PubMed] [Google Scholar]
- 84.Fuso A, Nicolia V, Ricceri L, Cavallaro RA, Isopi E, Mangia F, et al. S-adenosylmethionine reduces the progress of the alzheimer-like features induced by b-vitamin deficiency in mice. Neurobiol Aging. 2012;33:1482.e1481–1416 [DOI] [PubMed] [Google Scholar]
- 85.Fuso A, Nicolia V, Cavallaro RA, Scarpa S. Dna methylase and demethylase activities are modulated by one-carbon metabolism in alzheimer’s disease models. J Nutr Biochem. 2011;22:242–251 [DOI] [PubMed] [Google Scholar]
- 86.Chen H, Liu S, Ji L, Wu T, Ji Y, Zhou Y, et al. Folic acid supplementation mitigates alzheimer’s disease by reducing inflammation: A randomized controlled trial. Mediators Inflamm. 2016;2016:5912146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Raber J, Huang Y, Ashford JW. Apoe genotype accounts for the vast majority of ad risk and ad pathology. Neurobiol Aging. 2004;25:641–650 [DOI] [PubMed] [Google Scholar]
- 88.Wang SC, Oelze B, Schumacher A. Age-specific epigenetic drift in late-onset alzheimer’s disease. PLoS One. 2008;3:e2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Caesar I, Gandy S. Evidence that an apoe epsilon4 ‘double whammy’ increases risk for alzheimer’s disease. BMC Med. 2012;10:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Miyata M, Smith JD. Apolipoprotein e allele-specific antioxidant activity and effects on cytotoxicity by oxidative insults and beta-amyloid peptides. Nat Genet. 1996;14:55–61 [DOI] [PubMed] [Google Scholar]
- 91.Alagiakrishnan K, Gill SS, Fagarasanu A. Genetics and epigenetics of alzheimer’s disease. Postgrad Med J. 2012;88:522–529 [DOI] [PubMed] [Google Scholar]
- 92.Tulloch J, Leong L, Chen S, Keene CD, Millard SP, Shutes-David A, et al. Apoe dna methylation is altered in lewy body dementia. Alzheimers Dement. 2018;14:889–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tulloch J, Leong L, Thomson Z, Chen S, Lee EG, Keene CD, et al. Glia-specific apoe epigenetic changes in the alzheimer’s disease brain. Brain Res. 2018;1698:179–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang K, Schrag M, Crofton A, Trivedi R, Vinters H, Kirsch W. Targeted proteomics for quantification of histone acetylation in alzheimer’s disease. Proteomics. 2012;12:1261–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Francis YI, Fa M, Ashraf H, Zhang H, Staniszewski A, Latchman DS, et al. Dysregulation of histone acetylation in the app/ps1 mouse model of alzheimer’s disease. J Alzheimers Dis. 2009;18:131–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Graff J, Rei D, Guan JS, Wang WY, Seo J, Hennig KM, et al. An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature. 2012;483:222–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, et al. Hdac2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ricobaraza A, Cuadrado-Tejedor M, Perez-Mediavilla A, Frechilla D, Del Rio J, Garcia-Osta A. Phenylbutyrate ameliorates cognitive deficit and reduces tau pathology in an alzheimer’s disease mouse model. Neuropsychopharmacology. 2009;34:1721–1732 [DOI] [PubMed] [Google Scholar]
- 99.Govindarajan N, Rao P, Burkhardt S, Sananbenesi F, Schluter OM, Bradke F, et al. Reducing hdac6 ameliorates cognitive deficits in a mouse model for alzheimer’s disease. EMBO Mol Med. 2013;5:52–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang X, Perry G, Smith MA, Zhu X. Amyloid-beta-derived diffusible ligands cause impaired axonal transport of mitochondria in neurons. Neurodegener Dis. 2010;7:56–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang B, Chen CF, Wang AH, Lin QF. Mir-16 regulates cell death in alzheimer’s disease by targeting amyloid precursor protein. Eur Rev Med Pharmacol Sci. 2015;19:4020–4027 [PubMed] [Google Scholar]
- 102.Barbato C, Pezzola S, Caggiano C, Antonelli M, Frisone P, Ciotti MT, et al. A lentiviral sponge for mir-101 regulates ranbp9 expression and amyloid precursor protein metabolism in hippocampal neurons. Front Cell Neurosci. 2014;8:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vilardo E, Barbato C, Ciotti M, Cogoni C, Ruberti F. Microrna-101 regulates amyloid precursor protein expression in hippocampal neurons. J Biol Chem. 2010;285:18344–18351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Patel N, Hoang D, Miller N, Ansaloni S, Huang Q, Rogers JT, et al. Micrornas can regulate human app levels. Mol Neurodegener. 2008;3:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hebert SS, Horre K, Nicolai L, Bergmans B, Papadopoulou AS, Delacourte A, et al. Microrna regulation of alzheimer’s amyloid precursor protein expression. Neurobiol Dis. 2009;33:422–428 [DOI] [PubMed] [Google Scholar]
- 106.Delay C, Calon F, Mathews P, Hebert SS. Alzheimer-specific variants in the 3’utr of amyloid precursor protein affect microrna function. Mol Neurodegener. 2011;6:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhu HC, Wang LM, Wang M, Song B, Tan S, Teng JF, et al. Microrna-195 downregulates alzheimer’s disease amyloid-beta production by targeting bace1. Brain Res Bull. 2012;88:596–601 [DOI] [PubMed] [Google Scholar]
- 108.Long JM, Ray B, Lahiri DK. Microrna-339–5p down-regulates protein expression of beta-site amyloid precursor protein-cleaving enzyme 1 (bace1) in human primary brain cultures and is reduced in brain tissue specimens of alzheimer disease subjects. J Biol Chem. 2014;289:5184–5198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nelson PT, Wang WX. Mir-107 is reduced in alzheimer’s disease brain neocortex: Validation study. J Alzheimers Dis. 2010;21:75–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang WX, Rajeev BW, Stromberg AJ, Ren N, Tang G, Huang Q, et al. The expression of microrna mir-107 decreases early in alzheimer’s disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci. 2008;28:1213–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hebert SS, Horre K, Nicolai L, Papadopoulou AS, Mandemakers W, Silahtaroglu AN, et al. Loss of microrna cluster mir-29a/b-1 in sporadic alzheimer’s disease correlates with increased bace1/beta-secretase expression. Proc Natl Acad Sci U S A. 2008;105:6415–6420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yang G, Song Y, Zhou X, Deng Y, Liu T, Weng G, et al. Microrna-29c targets beta-site amyloid precursor protein-cleaving enzyme 1 and has a neuroprotective role in vitro and in vivo. Mol Med Rep. 2015;12:3081–3088 [DOI] [PubMed] [Google Scholar]
- 113.Zong Y, Wang H, Dong W, Quan X, Zhu H, Xu Y, et al. Mir-29c regulates bace1 protein expression. Brain Res. 2011;1395:108–115 [DOI] [PubMed] [Google Scholar]
- 114.Liu CG, Wang JL, Li L, Xue LX, Zhang YQ, Wang PC. Microrna-135a and −200b, potential biomarkers for alzheimers disease, regulate beta secretase and amyloid precursor protein. Brain Res. 2014;1583:55–64 [DOI] [PubMed] [Google Scholar]
- 115.Du X, Huo X, Yang Y, Hu Z, Botchway BOA, Jiang Y, et al. Mir-124 downregulates bace 1 and alters autophagy in app/ps1 transgenic mice. Toxicol Lett. 2017;280:195–205 [DOI] [PubMed] [Google Scholar]
- 116.Wong HK, Veremeyko T, Patel N, Lemere CA, Walsh DM, Esau C, et al. De-repression of foxo3a death axis by microrna-132 and −212 causes neuronal apoptosis in alzheimer’s disease. Hum Mol Genet. 2013;22:3077–3092 [DOI] [PubMed] [Google Scholar]
- 117.Chen-Plotkin AS, Unger TL, Gallagher MD, Bill E, Kwong LK, Volpicelli-Daley L, et al. Tmem106b, the risk gene for frontotemporal dementia, is regulated by the microrna-132/212 cluster and affects progranulin pathways. J Neurosci. 2012;32:11213–11227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Smith PY, Hernandez-Rapp J, Jolivette F, Lecours C, Bisht K, Goupil C, et al. Mir-132/212 deficiency impairs tau metabolism and promotes pathological aggregation in vivo. Hum Mol Genet. 2015;24:6721–6735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hansen KF, Sakamoto K, Aten S, Snider KH, Loeser J, Hesse AM, et al. Targeted deletion of mir-132/−212 impairs memory and alters the hippocampal transcriptome. Learn Mem. 2016;23:61–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lau P, Bossers K, Janky R, Salta E, Frigerio CS, Barbash S, et al. Alteration of the microrna network during the progression of alzheimer’s disease. EMBO Mol Med. 2013;5:1613–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhu QB, Unmehopa U, Bossers K, Hu YT, Verwer R, Balesar R, et al. Microrna-132 and early growth response-1 in nucleus basalis of meynert during the course of alzheimer’s disease. Brain. 2016;139:908–921 [DOI] [PubMed] [Google Scholar]
- 122.Santa-Maria I, Alaniz ME, Renwick N, Cela C, Fulga TA, Van Vactor D, et al. Dysregulation of microrna-219 promotes neurodegeneration through post-transcriptional regulation of tau. J Clin Invest. 2015;125:681–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kumar S, Vijayan M, Reddy PH. Microrna-455–3p as a potential peripheral biomarker for alzheimer’s disease. Hum Mol Genet. 2017;26:3808–3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dong H, Li J, Huang L, Chen X, Li D, Wang T, et al. Serum microrna profiles serve as novel biomarkers for the diagnosis of alzheimer’s disease. Dis Markers. 2015;2015:625659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Galimberti D, Villa C, Fenoglio C, Serpente M, Ghezzi L, Cioffi SM, et al. Circulating mirnas as potential biomarkers in alzheimer’s disease. J Alzheimers Dis. 2014;42:1261–1267 [DOI] [PubMed] [Google Scholar]
- 126.Geekiyanage H, Jicha GA, Nelson PT, Chan C. Blood serum mirna: Non-invasive biomarkers for alzheimer’s disease. Exp Neurol. 2012;235:491–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tan L, Yu JT, Tan MS, Liu QY, Wang HF, Zhang W, et al. Genome-wide serum microrna expression profiling identifies serum biomarkers for alzheimer’s disease. J Alzheimers Dis. 2014;40:1017–1027 [DOI] [PubMed] [Google Scholar]
- 128.Bekris LM, Lutz F, Montine TJ, Yu CE, Tsuang D, Peskind ER, et al. Microrna in alzheimer’s disease: An exploratory study in brain, cerebrospinal fluid and plasma. Biomarkers. 2013;18:455–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Garza-Manero S, Arias C, Bermudez-Rattoni F, Vaca L, Zepeda A. Identification of age- and disease-related alterations in circulating mirnas in a mouse model of alzheimer’s disease. Front Cell Neurosci. 2015;9:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kiko T, Nakagawa K, Tsuduki T, Furukawa K, Arai H, Miyazawa T. Micrornas in plasma and cerebrospinal fluid as potential markers for alzheimer’s disease. J Alzheimers Dis. 2014;39:253–259 [DOI] [PubMed] [Google Scholar]
- 131.Riancho J, Vazquez-Higuera JL, Pozueta A, Lage C, Kazimierczak M, Bravo M, et al. Microrna profile in patients with alzheimer’s disease: Analysis of mir-9–5p and mir-598 in raw and exosome enriched cerebrospinal fluid samples. J Alzheimers Dis. 2017;57:483–491 [DOI] [PubMed] [Google Scholar]
- 132.Alexandrov PN, Dua P, Hill JM, Bhattacharjee S, Zhao Y, Lukiw WJ. Microrna (mirna) speciation in alzheimer’s disease (ad) cerebrospinal fluid (csf) and extracellular fluid (ecf). Int J Biochem Mol Biol. 2012;3:365–373 [PMC free article] [PubMed] [Google Scholar]
- 133.Denk J, Boelmans K, Siegismund C, Lassner D, Arlt S, Jahn H. Microrna profiling of csf reveals potential biomarkers to detect alzheimer`s disease. PLoS One. 2015;10:e0126423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.McKeever PM, Schneider R, Taghdiri F, Weichert A, Multani N, Brown RA, et al. Microrna expression levels are altered in the cerebrospinal fluid of patients with young-onset alzheimer’s disease. Mol Neurobiol. 2018;55:8826–8841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Muller M, Kuiperij HB, Claassen JA, Kusters B, Verbeek MM. Micrornas in alzheimer’s disease: Differential expression in hippocampus and cell-free cerebrospinal fluid. Neurobiol Aging. 2014;35:152–158 [DOI] [PubMed] [Google Scholar]
- 136.Lugli G, Cohen AM, Bennett DA, Shah RC, Fields CJ, Hernandez AG, et al. Plasma exosomal mirnas in persons with and without alzheimer disease: Altered expression and prospects for biomarkers. PLoS One. 2015;10:e0139233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Cheng L, Doecke JD, Sharples RA, Villemagne VL, Fowler CJ, Rembach A, et al. Prognostic serum mirna biomarkers associated with alzheimer’s disease shows concordance with neuropsychological and neuroimaging assessment. Mol Psychiatry. 2015;20:1188–1196 [DOI] [PubMed] [Google Scholar]
- 138.Ren RJ, Zhang YF, Dammer EB, Zhou Y, Wang LL, Liu XH, et al. Peripheral blood microrna expression profiles in alzheimer’s disease: Screening, validation, association with clinical phenotype and implications for molecular mechanism. Mol Neurobiol. 2016;53:5772–5781 [DOI] [PubMed] [Google Scholar]
- 139.Kumar P, Dezso Z, MacKenzie C, Oestreicher J, Agoulnik S, Byrne M, et al. Circulating mirna biomarkers for alzheimer’s disease. PLoS One. 2013;8:e69807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhou X, Xu J. Identification of alzheimer’s disease-associated long noncoding rnas. Neurobiol Aging. 2015;36:2925–2931 [DOI] [PubMed] [Google Scholar]
- 141.Wu J, Chen L, Zheng C, Xu S, Gao Y, Wang J. Co-expression network analysis revealing the potential regulatory roles of lncrnas in alzheimer’s disease. Interdiscip Sci. 2019 [DOI] [PubMed] [Google Scholar]
- 142.Magistri M, Velmeshev D, Makhmutova M, Faghihi MA. Transcriptomics profiling of alzheimer’s disease reveal neurovascular defects, altered amyloid-beta homeostasis, and deregulated expression of long noncoding rnas. J Alzheimers Dis. 2015;48:647–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wang LK, Chen XF, He DD, Li Y, Fu J. Dissection of functional lncrnas in alzheimer’s disease by construction and analysis of lncrna-mrna networks based on competitive endogenous rnas. Biochem Biophys Res Commun. 2017;485:569–576 [DOI] [PubMed] [Google Scholar]
- 144.Yang B, Xia ZA, Zhong B, Xiong X, Sheng C, Wang Y, et al. Distinct hippocampal expression profiles of long non-coding rnas in an alzheimer’s disease model. Mol Neurobiol. 2017;54:4833–4846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lee DY, Moon J, Lee ST, Jung KH, Park DK, Yoo JS, et al. Distinct expression of long non-coding rnas in an alzheimer’s disease model. J Alzheimers Dis. 2015;45:837–849 [DOI] [PubMed] [Google Scholar]
- 146.Wang Z, Zhao Y, Xu N, Zhang S, Wang S, Mao Y, et al. Neat1 regulates neuroglial cell mediating abeta clearance via the epigenetic regulation of endocytosis-related genes expression. Cell Mol Life Sci. 2019;76:3005–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhao MY, Wang GQ, Wang NN, Yu QY, Liu RL, Shi WQ. The long-non-coding rna neat1 is a novel target for alzheimer’s disease progression via mir-124/bace1 axis. Neurol Res. 2019;41:489–497 [DOI] [PubMed] [Google Scholar]
- 148.Ke S, Yang Z, Yang F, Wang X, Tan J, Liao B. Long noncoding rna neat1 aggravates abeta-induced neuronal damage by targeting mir-107 in alzheimer’s disease. Yonsei Med J. 2019;60:640–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang J, Yu JT, Tan MS, Jiang T, Tan L. Epigenetic mechanisms in alzheimer’s disease: Implications for pathogenesis and therapy. Ageing Res Rev. 2013;12:1024–1041 [DOI] [PubMed] [Google Scholar]
- 150.Kowal SL, Dall TM, Chakrabarti R, Storm MV, Jain A. The current and projected economic burden of parkinson’s disease in the united states. Mov Disord. 2013;28:311–318 [DOI] [PubMed] [Google Scholar]
- 151.Chaudhuri KR, Schapira AH. Non-motor symptoms of parkinson’s disease: Dopaminergic pathophysiology and treatment. Lancet Neurol. 2009;8:464–474 [DOI] [PubMed] [Google Scholar]
- 152.Moore DJ, West AB, Dawson VL, Dawson TM. Molecular pathophysiology of parkinson’s disease. Annu Rev Neurosci. 2005;28:57–87 [DOI] [PubMed] [Google Scholar]
- 153.Wu Y, Le W, Jankovic J. Preclinical biomarkers of parkinson disease. Arch Neurol. 2011;68:22–30 [DOI] [PubMed] [Google Scholar]
- 154.Buck K, Ferger B. L-dopa-induced dyskinesia in parkinson’s disease: A drug discovery perspective. Drug Discov Today. 2010;15:867–875 [DOI] [PubMed] [Google Scholar]
- 155.Ai SX, Xu Q, Hu YC, Song CY, Guo JF, Shen L, et al. Hypomethylation of snca in blood of patients with sporadic parkinson’s disease. J Neurol Sci. 2014;337:123–128 [DOI] [PubMed] [Google Scholar]
- 156.Tan YY, Wu L, Zhao ZB, Wang Y, Xiao Q, Liu J, et al. Methylation of alpha-synuclein and leucine-rich repeat kinase 2 in leukocyte dna of parkinson’s disease patients. Parkinsonism Relat Disord. 2014;20:308–313 [DOI] [PubMed] [Google Scholar]
- 157.Matsumoto L, Takuma H, Tamaoka A, Kurisaki H, Date H, Tsuji S, et al. Cpg demethylation enhances alpha-synuclein expression and affects the pathogenesis of parkinson’s disease. PLoS One. 2010;5:e15522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Desplats P, Spencer B, Coffee E, Patel P, Michael S, Patrick C, et al. Alpha-synuclein sequesters dnmt1 from the nucleus: A novel mechanism for epigenetic alterations in lewy body diseases. J Biol Chem. 2011;286:9031–9037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kontopoulos E, Parvin JD, Feany MB. Alpha-synuclein acts in the nucleus to inhibit histone acetylation and promote neurotoxicity. Hum Mol Genet. 2006;15:3012–3023 [DOI] [PubMed] [Google Scholar]
- 160.Wu X, Chen PS, Dallas S, Wilson B, Block ML, Wang CC, et al. Histone deacetylase inhibitors up-regulate astrocyte gdnf and bdnf gene transcription and protect dopaminergic neurons. Int J Neuropsychopharmacol. 2008;11:1123–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Chen SH, Wu HM, Ossola B, Schendzielorz N, Wilson BC, Chu CH, et al. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, protects dopaminergic neurons from neurotoxin-induced damage. Br J Pharmacol. 2012;165:494–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Nicholas AP, Lubin FD, Hallett PJ, Vattem P, Ravenscroft P, Bezard E, et al. Striatal histone modifications in models of levodopa-induced dyskinesia. J Neurochem. 2008;106:486–494 [DOI] [PubMed] [Google Scholar]
- 163.Song C, Kanthasamy A, Anantharam V, Sun F, Kanthasamy AG. Environmental neurotoxic pesticide increases histone acetylation to promote apoptosis in dopaminergic neuronal cells: Relevance to epigenetic mechanisms of neurodegeneration. Mol Pharmacol. 2010;77:621–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Song C, Kanthasamy A, Jin H, Anantharam V, Kanthasamy AG. Paraquat induces epigenetic changes by promoting histone acetylation in cell culture models of dopaminergic degeneration. Neurotoxicology. 2011;32:586–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Deng H, Le W, Guo Y, Hunter CB, Xie W, Jankovic J. Genetic and clinical identification of parkinson’s disease patients with lrrk2 g2019s mutation. Ann Neurol. 2005;57:933–934 [DOI] [PubMed] [Google Scholar]
- 166.Gehrke S, Imai Y, Sokol N, Lu B. Pathogenic lrrk2 negatively regulates microrna-mediated translational repression. Nature. 2010;466:637–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Tsika E, Moore DJ. Contribution of gtpase activity to lrrk2-associated parkinson disease. Small GTPases. 2013;4:164–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Orenstein SJ, Kuo SH, Tasset I, Arias E, Koga H, Fernandez-Carasa I, et al. Interplay of lrrk2 with chaperone-mediated autophagy. Nat Neurosci. 2013;16:394–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Bardai FH, Ordonez DG, Bailey RM, Hamm M, Lewis J, Feany MB. Lrrk promotes tau neurotoxicity through dysregulation of actin and mitochondrial dynamics. PLoS Biol. 2018;16:e2006265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Cho HJ, Liu G, Jin SM, Parisiadou L, Xie C, Yu J, et al. Microrna-205 regulates the expression of parkinson’s disease-related leucine-rich repeat kinase 2 protein. Hum Mol Genet. 2013;22:608–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Chen Q, Huang X, Li R. Lncrna malat1/mir-205–5p axis regulates mpp(+)-induced cell apoptosis in mn9d cells by directly targeting lrrk2. Am J Transl Res. 2018;10:563–572 [PMC free article] [PubMed] [Google Scholar]
- 172.Doxakis E Post-transcriptional regulation of alpha-synuclein expression by mir-7 and mir-153. J Biol Chem. 2010;285:12726–12734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Fragkouli A, Doxakis E. Mir-7 and mir-153 protect neurons against mpp(+)-induced cell death via upregulation of mtor pathway. Front Cell Neurosci. 2014;8:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Junn E, Lee KW, Jeong BS, Chan TW, Im JY, Mouradian MM. Repression of alpha-synuclein expression and toxicity by microrna-7. Proc Natl Acad Sci U S A. 2009;106:13052–13057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Choi DC, Chae YJ, Kabaria S, Chaudhuri AD, Jain MR, Li H, et al. Microrna-7 protects against 1-methyl-4-phenylpyridinium-induced cell death by targeting rela. J Neurosci. 2014;34:12725–12737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Thome AD, Harms AS, Volpicelli-Daley LA, Standaert DG. Microrna-155 regulates alpha-synuclein-induced inflammatory responses in models of parkinson disease. J Neurosci. 2016;36:2383–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Alvarez-Erviti L, Seow Y, Schapira AH, Rodriguez-Oroz MC, Obeso JA, Cooper JM. Influence of microrna deregulation on chaperone-mediated autophagy and alpha-synuclein pathology in parkinson’s disease. Cell Death Dis. 2013;4:e545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Minones-Moyano E, Porta S, Escaramis G, Rabionet R, Iraola S, Kagerbauer B, et al. Microrna profiling of parkinson’s disease brains identifies early downregulation of mir-34b/c which modulate mitochondrial function. Hum Mol Genet. 2011;20:3067–3078 [DOI] [PubMed] [Google Scholar]
- 179.Kabaria S, Choi DC, Chaudhuri AD, Mouradian MM, Junn E. Inhibition of mir-34b and mir-34c enhances alpha-synuclein expression in parkinson’s disease. FEBS Lett. 2015;589:319–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Chaudhuri AD, Choi DC, Kabaria S, Tran A, Junn E. Microrna-7 regulates the function of mitochondrial permeability transition pore by targeting vdac1 expression. J Biol Chem. 2016;291:6483–6493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zhou Y, Lu M, Du RH, Qiao C, Jiang CY, Zhang KZ, et al. Microrna-7 targets nod-like receptor protein 3 inflammasome to modulate neuroinflammation in the pathogenesis of parkinson’s disease. Mol Neurodegener. 2016;11:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Li L, Xu J, Wu M, Hu JM. Protective role of microrna-221 in parkinson’s disease. Bratisl Lek Listy. 2018;119:22–27 [DOI] [PubMed] [Google Scholar]
- 183.Wen Z, Zhang J, Tang P, Tu N, Wang K, Wu G. Overexpression of mir185 inhibits autophagy and apoptosis of dopaminergic neurons by regulating the ampk/mtor signaling pathway in parkinson’s disease. Mol Med Rep. 2018;17:131–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Wei M, Cao LJ, Zheng JL, Xue LJ, Chen B, Xiao F, et al. Microrna-181c functions as a protective factor in a 1-methyl-4-phenylpyridinium iodide-induced cellular parkinson’s disease model via bcl2l11. Eur Rev Med Pharmacol Sci. 2017;21:3296–3304 [PubMed] [Google Scholar]
- 185.Jiang M, Yun Q, Shi F, Niu G, Gao Y, Xie S, et al. Downregulation of mir-384–5p attenuates rotenone-induced neurotoxicity in dopaminergic sh-sy5y cells through inhibiting endoplasmic reticulum stress. Am J Physiol Cell Physiol. 2016;310:C755–763 [DOI] [PubMed] [Google Scholar]
- 186.Wang H, Ye Y, Zhu Z, Mo L, Lin C, Wang Q, et al. Mir-124 regulates apoptosis and autophagy process in mptp model of parkinson’s disease by targeting to bim. Brain Pathol. 2016;26:167–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, et al. A microrna feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zhang Z, Cheng Y. Mir-16–1 promotes the aberrant alpha-synuclein accumulation in parkinson disease via targeting heat shock protein 70. ScientificWorldJournal. 2014;2014:938348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Liu Y, Liao S, Quan H, Lin Y, Li J, Yang Q. Involvement of microrna-135a-5p in the protective effects of hydrogen sulfide against parkinson’s disease. Cell Physiol Biochem. 2016;40:18–26 [DOI] [PubMed] [Google Scholar]
- 190.Hoss AG, Labadorf A, Beach TG, Latourelle JC, Myers RH. Microrna profiles in parkinson’s disease prefrontal cortex. Front Aging Neurosci. 2016;8:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Cardo LF, Coto E, Ribacoba R, Menendez M, Moris G, Suarez E, et al. Mirna profile in the substantia nigra of parkinson’s disease and healthy subjects. J Mol Neurosci. 2014;54:830–836 [DOI] [PubMed] [Google Scholar]
- 192.Cao XY, Lu JM, Zhao ZQ, Li MC, Lu T, An XS, et al. Microrna biomarkers of parkinson’s disease in serum exosome-like microvesicles. Neurosci Lett. 2017;644:94–99 [DOI] [PubMed] [Google Scholar]
- 193.Gui Y, Liu H, Zhang L, Lv W, Hu X. Altered microrna profiles in cerebrospinal fluid exosome in parkinson disease and alzheimer disease. Oncotarget. 2015;6:37043–37053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Burgos K, Malenica I, Metpally R, Courtright A, Rakela B, Beach T, et al. Profiles of extracellular mirna in cerebrospinal fluid and serum from patients with alzheimer’s and parkinson’s diseases correlate with disease status and features of pathology. PLoS One. 2014;9:e94839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Marques TM, Kuiperij HB, Bruinsma IB, van Rumund A, Aerts MB, Esselink RAJ, et al. Micrornas in cerebrospinal fluid as potential biomarkers for parkinson’s disease and multiple system atrophy. Mol Neurobiol. 2017;54:7736–7745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Ma W, Li Y, Wang C, Xu F, Wang M, Liu Y. Serum mir-221 serves as a biomarker for parkinson’s disease. Cell Biochem Funct. 2016;34:511–515 [DOI] [PubMed] [Google Scholar]
- 197.Botta-Orfila T, Morato X, Compta Y, Lozano JJ, Falgas N, Valldeoriola F, et al. Identification of blood serum micro-rnas associated with idiopathic and lrrk2 parkinson’s disease. J Neurosci Res. 2014;92:1071–1077 [DOI] [PubMed] [Google Scholar]
- 198.Ding H, Huang Z, Chen M, Wang C, Chen X, Chen J, et al. Identification of a panel of five serum mirnas as a biomarker for parkinson’s disease. Parkinsonism Relat Disord. 2016;22:68–73 [DOI] [PubMed] [Google Scholar]
- 199.Dong H, Wang C, Lu S, Yu C, Huang L, Feng W, et al. A panel of four decreased serum micrornas as a novel biomarker for early parkinson’s disease. Biomarkers. 2016;21:129–137 [DOI] [PubMed] [Google Scholar]
- 200.Oh SE, Park HJ, He L, Skibiel C, Junn E, Mouradian MM. The parkinson’s disease gene product dj-1 modulates mir-221 to promote neuronal survival against oxidative stress. Redox Biol. 2018;19:62–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Wang ZH, Zhang JL, Duan YL, Zhang QS, Li GF, Zheng DL. Microrna-214 participates in the neuroprotective effect of resveratrol via inhibiting alpha-synuclein expression in mptp-induced parkinson’s disease mouse. Biomed Pharmacother. 2015;74:252–256 [DOI] [PubMed] [Google Scholar]
- 202.Khoo SK, Petillo D, Kang UJ, Resau JH, Berryhill B, Linder J, et al. Plasma-based circulating microrna biomarkers for parkinson’s disease. J Parkinsons Dis. 2012;2:321–331 [DOI] [PubMed] [Google Scholar]
- 203.Ni Y, Huang H, Chen Y, Cao M, Zhou H, Zhang Y. Investigation of long non-coding rna expression profiles in the substantia nigra of parkinson’s disease. Cell Mol Neurobiol. 2017;37:329–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Kraus TFJ, Haider M, Spanner J, Steinmaurer M, Dietinger V, Kretzschmar HA. Altered long noncoding rna expression precedes the course of parkinson’s disease-a preliminary report. Mol Neurobiol. 2017;54:2869–2877 [DOI] [PubMed] [Google Scholar]
- 205.Elkouris M, Kouroupi G, Vourvoukelis A, Papagiannakis N, Kaltezioti V, Matsas R, et al. Long non-coding rnas associated with neurodegeneration-linked genes are reduced in parkinson’s disease patients. Front Cell Neurosci. 2019;13:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Zhou Y, Gu C, Li J, Zhu L, Huang G, Dai J, et al. Aberrantly expressed long noncoding rnas and genes in parkinson’s disease. Neuropsychiatr Dis Treat. 2018;14:3219–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Jiao F, Wang Q, Zhang P, Bu L, Yan J, Tian B. Expression signatures of long non-coding rna in the substantia nigra of pre-symptomatic mouse model of parkinson’s disease. Behav Brain Res. 2017;331:123–130 [DOI] [PubMed] [Google Scholar]
- 208.Hossein-Nezhad A, Fatemi RP, Ahmad R, Peskind ER, Zabetian CP, Hu SC, et al. Transcriptomic profiling of extracellular rnas present in cerebrospinal fluid identifies differentially expressed transcripts in parkinson’s disease. J Parkinsons Dis. 2016;6:109–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Peng T, Liu X, Wang J, Liu Y, Fu Z, Ma X, et al. Long noncoding rna haglros regulates apoptosis and autophagy in parkinson’s disease via regulating mir-100/atg10 axis and pi3k/akt/mtor pathway activation. Artif Cells Nanomed Biotechnol. 2019;47:2764–2774 [DOI] [PubMed] [Google Scholar]
- 210.Ding XM, Zhao LJ, Qiao HY, Wu SL, Wang XH. Long non-coding rna-p21 regulates mpp(+)-induced neuronal injury by targeting mir-625 and derepressing trpm2 in sh-sy5y cells. Chem Biol Interact. 2019;307:73–81 [DOI] [PubMed] [Google Scholar]
- 211.Xu X, Zhuang C, Wu Z, Qiu H, Feng H, Wu J. Lincrna-p21 inhibits cell viability and promotes cell apoptosis in parkinson’s disease through activating alpha-synuclein expression. Biomed Res Int. 2018;2018:8181374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Zhang QS, Wang ZH, Zhang JL, Duan YL, Li GF, Zheng DL. Beta-asarone protects against mptp-induced parkinson’s disease via regulating long non-coding rna malat1 and inhibiting alpha-synuclein protein expression. Biomed Pharmacother. 2016;83:153–159 [DOI] [PubMed] [Google Scholar]
- 213.Xie N, Qi J, Li S, Deng J, Chen Y, Lian Y. Upregulated lncrna small nucleolar rna host gene 1 promotes 1-methyl-4-phenylpyridinium ion-induced cytotoxicity and reactive oxygen species production through mir-15b-5p/gsk3beta axis in human dopaminergic sh-sy5y cells. J Cell Biochem. 2019;120:5790–5801 [DOI] [PubMed] [Google Scholar]
- 214.Chen Y, Lian YJ, Ma YQ, Wu CJ, Zheng YK, Xie NC. Lncrna snhg1 promotes alpha-synuclein aggregation and toxicity by targeting mir-15b-5p to activate siah1 in human neuroblastoma sh-sy5y cells. Neurotoxicology. 2018;68:212–221 [DOI] [PubMed] [Google Scholar]
- 215.Qian C, Ye Y, Mao H, Yao L, Sun X, Wang B, et al. Downregulated lncrna-snhg1 enhances autophagy and prevents cell death through the mir-221/222 /p27/mtor pathway in parkinson’s disease. Exp Cell Res. 2019;384:111614. [DOI] [PubMed] [Google Scholar]
- 216.Cao B, Wang T, Qu Q, Kang T, Yang Q. Long noncoding rna snhg1 promotes neuroinflammation in parkinson’s disease via regulating mir-7/nlrp3 pathway. Neuroscience. 2018;388:118–127 [DOI] [PubMed] [Google Scholar]
- 217.Yan W, Chen ZY, Chen JQ, Chen HM. Lncrna neat1 promotes autophagy in mptp-induced parkinson’s disease through stabilizing pink1 protein. Biochem Biophys Res Commun. 2018;496:1019–1024 [DOI] [PubMed] [Google Scholar]
- 218.Simchovitz A, Hanan M, Niederhoffer N, Madrer N, Yayon N, Bennett ER, et al. Neat1 is overexpressed in parkinson’s disease substantia nigra and confers drug-inducible neuroprotection from oxidative stress. Faseb j. 2019;33:11223–11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Xie SP, Zhou F, Li J, Duan SJ. Neat1 regulates mpp(+)-induced neuronal injury by targeting mir-124 in neuroblastoma cells. Neurosci Lett. 2019;708:134340. [DOI] [PubMed] [Google Scholar]
- 220.Lu M, Sun WL, Shen J, Wei M, Chen B, Qi YJ, et al. Lncrna-uca1 promotes pd development by upregulating snca. Eur Rev Med Pharmacol Sci. 2018;22:7908–7915 [DOI] [PubMed] [Google Scholar]
- 221.Cai L, Tu L, Li T, Yang X, Ren Y, Gu R, et al. Downregulation of lncrna uca1 ameliorates the damage of dopaminergic neurons, reduces oxidative stress and inflammation in parkinson’s disease through the inhibition of the pi3k/akt signaling pathway. Int Immunopharmacol. 2019;75:105734. [DOI] [PubMed] [Google Scholar]
- 222.Song Q, Geng Y, Li Y, Wang L, Qin J. Long noncoding rna norad regulates mpp+-induced parkinson’s disease model cells. J Chem Neuroanat. 2019;101:101668. [DOI] [PubMed] [Google Scholar]
- 223.Cohen-Carmon D, Meshorer E. Polyglutamine (polyq) disorders: The chromatin connection. Nucleus. 2012;3:433–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Valor LM, Guiretti D. What’s wrong with epigenetics in huntington’s disease? Neuropharmacology. 2014;80:103–114 [DOI] [PubMed] [Google Scholar]
- 225.A novel gene containing a trinucleotide repeat that is expanded and unstable on huntington’s disease chromosomes. The huntington’s disease collaborative research group. Cell. 1993;72:971–983 [DOI] [PubMed] [Google Scholar]
- 226.Valor LM. Epigenetic-based therapies in the preclinical and clinical treatment of huntington’s disease. Int J Biochem Cell Biol. 2015;67:45–48 [DOI] [PubMed] [Google Scholar]
- 227.Morton AJ, Hunt MJ, Hodges AK, Lewis PD, Redfern AJ, Dunnett SB, et al. A combination drug therapy improves cognition and reverses gene expression changes in a mouse model of huntington’s disease. Eur J Neurosci. 2005;21:855–870 [DOI] [PubMed] [Google Scholar]
- 228.Valor LM. Transcription, epigenetics and ameliorative strategies in huntington’s disease: A genome-wide perspective. Mol Neurobiol. 2015;51:406–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Horvath S, Langfelder P, Kwak S, Aaronson J, Rosinski J, Vogt TF, et al. Huntington’s disease accelerates epigenetic aging of human brain and disrupts dna methylation levels. Aging (Albany NY). 2016;8:1485–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Ng CW, Yildirim F, Yap YS, Dalin S, Matthews BJ, Velez PJ, et al. Extensive changes in dna methylation are associated with expression of mutant huntingtin. Proc Natl Acad Sci U S A. 2013;110:2354–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.De Souza RA, Islam SA, McEwen LM, Mathelier A, Hill A, Mah SM, et al. Dna methylation profiling in human huntington’s disease brain. Hum Mol Genet. 2016;25:2013–2030 [DOI] [PubMed] [Google Scholar]
- 232.Wang F, Yang Y, Lin X, Wang JQ, Wu YS, Xie W, et al. Genome-wide loss of 5-hmc is a novel epigenetic feature of huntington’s disease. Hum Mol Genet. 2013;22:3641–3653 [DOI] [PubMed] [Google Scholar]
- 233.Chiu FL, Lin JT, Chuang CY, Chien T, Chen CM, Chen KH, et al. Elucidating the role of the a2a adenosine receptor in neurodegeneration using neurons derived from huntington’s disease ipscs. Hum Mol Genet. 2015;24:6066–6079 [DOI] [PubMed] [Google Scholar]
- 234.Chou SY, Lee YC, Chen HM, Chiang MC, Lai HL, Chang HH, et al. Cgs21680 attenuates symptoms of huntington’s disease in a transgenic mouse model. J Neurochem. 2005;93:310–320 [DOI] [PubMed] [Google Scholar]
- 235.Villar-Menendez I, Blanch M, Tyebji S, Pereira-Veiga T, Albasanz JL, Martin M, et al. Increased 5-methylcytosine and decreased 5-hydroxymethylcytosine levels are associated with reduced striatal a2ar levels in huntington’s disease. Neuromolecular Med. 2013;15:295–309 [DOI] [PubMed] [Google Scholar]
- 236.Gardian G, Browne SE, Choi DK, Klivenyi P, Gregorio J, Kubilus JK, et al. Neuroprotective effects of phenylbutyrate in the n171–82q transgenic mouse model of huntington’s disease. J Biol Chem. 2005;280:556–563 [DOI] [PubMed] [Google Scholar]
- 237.Hockly E, Richon VM, Woodman B, Smith DL, Zhou X, Rosa E, et al. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of huntington’s disease. Proc Natl Acad Sci U S A. 2003;100:2041–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Ferrante RJ, Kubilus JK, Lee J, Ryu H, Beesen A, Zucker B, et al. Histone deacetylase inhibition by sodium butyrate chemotherapy ameliorates the neurodegenerative phenotype in huntington’s disease mice. J Neurosci. 2003;23:9418–9427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Bates EA, Victor M, Jones AK, Shi Y, Hart AC. Differential contributions of caenorhabditis elegans histone deacetylases to huntingtin polyglutamine toxicity. J Neurosci. 2006;26:2830–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Steffan JS, Bodai L, Pallos J, Poelman M, McCampbell A, Apostol BL, et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in drosophila. Nature. 2001;413:739–743 [DOI] [PubMed] [Google Scholar]
- 241.Jia H, Pallos J, Jacques V, Lau A, Tang B, Cooper A, et al. Histone deacetylase (hdac) inhibitors targeting hdac3 and hdac1 ameliorate polyglutamine-elicited phenotypes in model systems of huntington’s disease. Neurobiol Dis. 2012;46:351–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Suelves N, Kirkham-McCarthy L, Lahue RS, Gines S. A selective inhibitor of histone deacetylase 3 prevents cognitive deficits and suppresses striatal cag repeat expansions in huntington’s disease mice. Sci Rep. 2017;7:6082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Thomas EA, Coppola G, Desplats PA, Tang B, Soragni E, Burnett R, et al. The hdac inhibitor 4b ameliorates the disease phenotype and transcriptional abnormalities in huntington’s disease transgenic mice. Proc Natl Acad Sci U S A. 2008;105:15564–15569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Benn CL, Butler R, Mariner L, Nixon J, Moffitt H, Mielcarek M, et al. Genetic knock-down of hdac7 does not ameliorate disease pathogenesis in the r6/2 mouse model of huntington’s disease. PLoS One. 2009;4:e5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Bobrowska A, Paganetti P, Matthias P, Bates GP. Hdac6 knock-out increases tubulin acetylation but does not modify disease progression in the r6/2 mouse model of huntington’s disease. PLoS One. 2011;6:e20696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Mielcarek M, Landles C, Weiss A, Bradaia A, Seredenina T, Inuabasi L, et al. Hdac4 reduction: A novel therapeutic strategy to target cytoplasmic huntingtin and ameliorate neurodegeneration. PLoS Biol. 2013;11:e1001717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Moumne L, Campbell K, Howland D, Ouyang Y, Bates GP. Genetic knock-down of hdac3 does not modify disease-related phenotypes in a mouse model of huntington’s disease. PLoS One. 2012;7:e31080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Mano T, Suzuki T, Tsuji S, Iwata A. Differential effect of hdac3 on cytoplasmic and nuclear huntingtin aggregates. PLoS One. 2014;9:e111277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Ajamian F, Salminen A, Reeben M. Selective regulation of class i and class ii histone deacetylases expression by inhibitors of histone deacetylases in cultured mouse neural cells. Neurosci Lett. 2004;365:64–68 [DOI] [PubMed] [Google Scholar]
- 250.Zuccato C, Ciammola A, Rigamonti D, Leavitt BR, Goffredo D, Conti L, et al. Loss of huntingtin-mediated bdnf gene transcription in huntington’s disease. Science. 2001;293:493–498 [DOI] [PubMed] [Google Scholar]
- 251.Zuccato C, Cattaneo E. Role of brain-derived neurotrophic factor in huntington’s disease. Prog Neurobiol. 2007;81:294–330 [DOI] [PubMed] [Google Scholar]
- 252.Cattaneo E, Zuccato C, Tartari M. Normal huntingtin function: An alternative approach to huntington’s disease. Nat Rev Neurosci. 2005;6:919–930 [DOI] [PubMed] [Google Scholar]
- 253.Zuccato C, Tartari M, Crotti A, Goffredo D, Valenza M, Conti L, et al. Huntingtin interacts with rest/nrsf to modulate the transcription of nrse-controlled neuronal genes. Nat Genet. 2003;35:76–83 [DOI] [PubMed] [Google Scholar]
- 254.Schiffer D, Caldera V, Mellai M, Conforti P, Cattaneo E, Zuccato C. Repressor element-1 silencing transcription factor (rest) is present in human control and huntington’s disease neurones. Neuropathol Appl Neurobiol. 2014;40:899–910 [DOI] [PubMed] [Google Scholar]
- 255.Hoss AG, Lagomarsino VN, Frank S, Hadzi TC, Myers RH, Latourelle JC. Study of plasma-derived mirnas mimic differences in huntington’s disease brain. Mov Disord. 2015;30:1961–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Mastrokolias A, Ariyurek Y, Goeman JJ, van Duijn E, Roos RA, van der Mast RC, et al. Huntington’s disease biomarker progression profile identified by transcriptome sequencing in peripheral blood. Eur J Hum Genet. 2015;23:1349–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Lee ST, Chu K, Im WS, Yoon HJ, Im JY, Park JE, et al. Altered microrna regulation in huntington’s disease models. Exp Neurol. 2011;227:172–179 [DOI] [PubMed] [Google Scholar]
- 258.Sinha M, Mukhopadhyay S, Bhattacharyya NP. Mechanism(s) of alteration of micro rna expressions in huntington’s disease and their possible contributions to the observed cellular and molecular dysfunctions in the disease. Neuromolecular Med. 2012;14:221–243 [DOI] [PubMed] [Google Scholar]
- 259.Marti E, Pantano L, Banez-Coronel M, Llorens F, Minones-Moyano E, Porta S, et al. A myriad of mirna variants in control and huntington’s disease brain regions detected by massively parallel sequencing. Nucleic Acids Res. 2010;38:7219–7235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Diez-Planelles C, Sanchez-Lozano P, Crespo MC, Gil-Zamorano J, Ribacoba R, Gonzalez N, et al. Circulating micrornas in huntington’s disease: Emerging mediators in metabolic impairment. Pharmacol Res. 2016;108:102–110 [DOI] [PubMed] [Google Scholar]
- 261.Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microrna mir-9/mir-9* regulates rest and corest and is downregulated in huntington’s disease. J Neurosci. 2008;28:14341–14346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Kocerha J, Xu Y, Prucha MS, Zhao D, Chan AW. Microrna-128a dysregulation in transgenic huntington’s disease monkeys. Mol Brain. 2014;7:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Hoss AG, Labadorf A, Latourelle JC, Kartha VK, Hadzi TC, Gusella JF, et al. Mir-10b-5p expression in huntington’s disease brain relates to age of onset and the extent of striatal involvement. BMC Med Genomics. 2015;8:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Muller S In silico analysis of regulatory networks underlines the role of mir-10b-5p and its target bdnf in huntington’s disease. Transl Neurodegener. 2014;3:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Hoss AG, Kartha VK, Dong X, Latourelle JC, Dumitriu A, Hadzi TC, et al. Micrornas located in the hox gene clusters are implicated in huntington’s disease pathogenesis. PLoS Genet. 2014;10:e1004188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Kozlowska E, Krzyzosiak WJ, Koscianska E. Regulation of huntingtin gene expression by mirna-137, −214, −148a, and their respective isomirs. Int J Mol Sci. 2013;14:16999–17016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Ghatak S, Raha S. Micro rna-214 contributes to proteasome independent downregulation of beta catenin in huntington’s disease knock-in striatal cell model sthdhq111/q111. Biochem Biophys Res Commun. 2015;459:509–514 [DOI] [PubMed] [Google Scholar]
- 268.Dong X, Cong S. Bioinformatic analysis of microrna expression in huntington’s disease. Mol Med Rep. 2018;18:2857–2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Fu MH, Li CL, Lin HL, Tsai SJ, Lai YY, Chang YF, et al. The potential regulatory mechanisms of mir-196a in huntington’s disease through bioinformatic analyses. PLoS One. 2015;10:e0137637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Her LS, Mao SH, Chang CY, Cheng PH, Chang YF, Yang HI, et al. Mir-196a enhances neuronal morphology through suppressing ranbp10 to provide neuroprotection in huntington’s disease. Theranostics. 2017;7:2452–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Kunkanjanawan T, Carter RL, Prucha MS, Yang J, Parnpai R, Chan AW. Mir-196a ameliorates cytotoxicity and cellular phenotype in transgenic huntington’s disease monkey neural cells. PLoS One. 2016;11:e0162788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Ghose J, Sinha M, Das E, Jana NR, Bhattacharyya NP. Regulation of mir-146a by rela/nfkb and p53 in sthdh(q111)/hdh(q111) cells, a cell model of huntington’s disease. PLoS One. 2011;6:e23837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Das E, Jana NR, Bhattacharyya NP. Delayed cell cycle progression in sthdh(q111)/hdh(q111) cells, a cell model for huntington’s disease mediated by microrna-19a, microrna-146a and microrna-432. Microrna. 2015;4:86–100 [DOI] [PubMed] [Google Scholar]
- 274.Reynolds RH, Petersen MH, Willert CW, Heinrich M, Nymann N, Dall M, et al. Perturbations in the p53/mir-34a/sirt1 pathway in the r6/2 huntington’s disease model. Mol Cell Neurosci. 2018;88:118–129 [DOI] [PubMed] [Google Scholar]
- 275.Jovicic A, Zaldivar Jolissaint JF, Moser R, Silva Santos Mde F, Luthi-Carter R. Microrna-22 (mir-22) overexpression is neuroprotective via general anti-apoptotic effects and may also target specific huntington’s disease-related mechanisms. PLoS One. 2013;8:e54222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Fukuoka M, Takahashi M, Fujita H, Chiyo T, Popiel HA, Watanabe S, et al. Supplemental treatment for huntington’s disease with mir-132 that is deficient in huntington’s disease brain. Mol Ther Nucleic Acids. 2018;11:79–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Ban JJ, Chung JY, Lee M, Im W, Kim M. Microrna-27a reduces mutant hutingtin aggregation in an in vitro model of huntington’s disease. Biochem Biophys Res Commun. 2017;488:316–321 [DOI] [PubMed] [Google Scholar]
- 278.Pfister EL, DiNardo N, Mondo E, Borel F, Conroy F, Fraser C, et al. Artificial mirnas reduce human mutant huntingtin throughout the striatum in a transgenic sheep model of huntington’s disease. Hum Gene Ther. 2018;29:663–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Chung DW, Rudnicki DD, Yu L, Margolis RL. A natural antisense transcript at the huntington’s disease repeat locus regulates htt expression. Hum Mol Genet. 2011;20:3467–3477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Francelle L, Galvan L, Gaillard MC, Petit F, Bernay B, Guillermier M, et al. Striatal long noncoding rna abhd11os is neuroprotective against an n-terminal fragment of mutant huntingtin in vivo. Neurobiol Aging. 2015;36:1601.e1607–1616 [DOI] [PubMed] [Google Scholar]
- 281.Chanda K, Das S, Chakraborty J, Bucha S, Maitra A, Chatterjee R, et al. Altered levels of long ncrnas meg3 and neat1 in cell and animal models of huntington’s disease. RNA Biol. 2018;15:1348–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Sunwoo JS, Lee ST, Im W, Lee M, Byun JI, Jung KH, et al. Altered expression of the long noncoding rna neat1 in huntington’s disease. Mol Neurobiol. 2017;54:1577–1586 [DOI] [PubMed] [Google Scholar]
- 283.Park H, Miyazaki H, Yamanaka T, Nukina N. Non-coding rna neat1 and abhd11os expressions are dysregulated in medium spiny neurons of huntington disease model mice. Neurosci Res. 2019;147:58–63 [DOI] [PubMed] [Google Scholar]
- 284.Cheng C, Spengler RM, Keiser MS, Monteys AM, Rieders JM, Ramachandran S, et al. The long non-coding rna neat1 is elevated in polyglutamine repeat expansion diseases and protects from disease gene-dependent toxicities. Hum Mol Genet. 2018;27:4303–4314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Blaha MJ, et al. Heart disease and stroke statistics--2014 update: A report from the american heart association. Circulation. 2014;129:e28–e292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Jhelum P, Karisetty BC, Kumar A, Chakravarty S. Implications of epigenetic mechanisms and their targets in cerebral ischemia models. Curr Neuropharmacol. 2017;15:815–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Smith WS. Pathophysiology of focal cerebral ischemia: A therapeutic perspective. J Vasc Interv Radiol. 2004;15:S3–12 [DOI] [PubMed] [Google Scholar]
- 288.Bramlett HM, Dietrich WD. Pathophysiology of cerebral ischemia and brain trauma: Similarities and differences. J Cereb Blood Flow Metab. 2004;24:133–150 [DOI] [PubMed] [Google Scholar]
- 289.Cifu DX, Stewart DG. Factors affecting functional outcome after stroke: A critical review of rehabilitation interventions. Arch Phys Med Rehabil. 1999;80:S35–39 [DOI] [PubMed] [Google Scholar]
- 290.Nahmani M, Turrigiano GG. Adult cortical plasticity following injury: Recapitulation of critical period mechanisms? Neuroscience. 2014;283:4–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Saver JL. Targeting the brain: Neuroprotection and neurorestoration in ischemic stroke. Pharmacotherapy. 2010;30:62s–69s [DOI] [PubMed] [Google Scholar]
- 292.Del Zoppo GJ, Saver JL, Jauch EC, Adams HP Jr. Expansion of the time window for treatment of acute ischemic stroke with intravenous tissue plasminogen activator: A science advisory from the american heart association/american stroke association. Stroke. 2009;40:2945–2948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Collen D, Lijnen HR. Tissue-type plasminogen activator: A historical perspective and personal account. J Thromb Haemost. 2004;2:541–546 [DOI] [PubMed] [Google Scholar]
- 294.Collen D, Lijnen HR. Basic and clinical aspects of fibrinolysis and thrombolysis. Blood. 1991;78:3114–3124 [PubMed] [Google Scholar]
- 295.Tissue plasminogen activator for acute ischemic stroke. N Engl J Med. 1995;333:1581–1587 [DOI] [PubMed] [Google Scholar]
- 296.Endres M, Meisel A, Biniszkiewicz D, Namura S, Prass K, Ruscher K, et al. Dna methyltransferase contributes to delayed ischemic brain injury. J Neurosci. 2000;20:3175–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Endres M, Fan G, Meisel A, Dirnagl U, Jaenisch R. Effects of cerebral ischemia in mice lacking dna methyltransferase 1 in post-mitotic neurons. Neuroreport. 2001;12:3763–3766 [DOI] [PubMed] [Google Scholar]
- 298.Li Y, Xiao D, Yang S, Zhang L. Promoter methylation represses at2r gene and increases brain hypoxic-ischemic injury in neonatal rats. Neurobiol Dis. 2013;60:32–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Choi IA, Lee CS, Kim HY, Choi DH, Lee J. Effect of inhibition of dna methylation combined with task-specific training on chronic stroke recovery. Int J Mol Sci. 2018;19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Soriano-Tarraga C, Giralt-Steinhauer E, Mola-Caminal M, Vivanco-Hidalgo RM, Ois A, Rodriguez-Campello A, et al. Ischemic stroke patients are biologically older than their chronological age. Aging (Albany NY). 2016;8:2655–2666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Soriano-Tarraga C, Mola-Caminal M, Giralt-Steinhauer E, Ois A, Rodriguez-Campello A, Cuadrado-Godia E, et al. Biological age is better than chronological as predictor of 3-month outcome in ischemic stroke. Neurology. 2017;89:830–836 [DOI] [PubMed] [Google Scholar]
- 302.Morris-Blanco KC, Kim T, Lopez MS, Bertogliat MJ, Chelluboina B, Vemuganti R. Induction of dna hydroxymethylation protects the brain after stroke. 2019;Accepted for publication [DOI] [PMC free article] [PubMed]
- 303.Ji F, Zhao C, Wang B, Tang Y, Miao Z, Wang Y. The role of 5-hydroxymethylcytosine in mitochondria after ischemic stroke. J Neurosci Res. 2018;96:1717–1726 [DOI] [PubMed] [Google Scholar]
- 304.Xuan A, Long D, Li J, Ji W, Hong L, Zhang M, et al. Neuroprotective effects of valproic acid following transient global ischemia in rats. Life Sci. 2012;90:463–468 [DOI] [PubMed] [Google Scholar]
- 305.Kim HJ, Rowe M, Ren M, Hong JS, Chen PS, Chuang DM. Histone deacetylase inhibitors exhibit anti-inflammatory and neuroprotective effects in a rat permanent ischemic model of stroke: Multiple mechanisms of action. J Pharmacol Exp Ther. 2007;321:892–901 [DOI] [PubMed] [Google Scholar]
- 306.Faraco G, Pancani T, Formentini L, Mascagni P, Fossati G, Leoni F, et al. Pharmacological inhibition of histone deacetylases by suberoylanilide hydroxamic acid specifically alters gene expression and reduces ischemic injury in the mouse brain. Mol Pharmacol. 2006;70:1876–1884 [DOI] [PubMed] [Google Scholar]
- 307.Qi X, Hosoi T, Okuma Y, Kaneko M, Nomura Y. Sodium 4-phenylbutyrate protects against cerebral ischemic injury. Mol Pharmacol. 2004;66:899–908 [DOI] [PubMed] [Google Scholar]
- 308.Baltan S, Murphy SP, Danilov CA, Bachleda A, Morrison RS. Histone deacetylase inhibitors preserve white matter structure and function during ischemia by conserving atp and reducing excitotoxicity. J Neurosci. 2011;31:3990–3999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Dharap A, Bowen K, Place R, Li LC, Vemuganti R. Transient focal ischemia induces extensive temporal changes in rat cerebral micrornaome. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2009;29:675–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Jeyaseelan K, Lim KY, Armugam A. Microrna expression in the blood and brain of rats subjected to transient focal ischemia by middle cerebral artery occlusion. Stroke. 2008;39:959–966 [DOI] [PubMed] [Google Scholar]
- 311.Yuan Y, Wang JY, Xu LY, Cai R, Chen Z, Luo BY. Microrna expression changes in the hippocampi of rats subjected to global ischemia. Journal of clinical neuroscience : official journal of the Neurosurgical Society of Australasia. 2010;17:774–778 [DOI] [PubMed] [Google Scholar]
- 312.Li G, Morris-Blanco KC, Lopez MS, Yang T, Zhao H, Vemuganti R, et al. Impact of micrornas on ischemic stroke: From pre- to post-disease. Prog Neurobiol. 2018;163–164:59–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Zhao H, Wang J, Gao L, Wang R, Liu X, Gao Z, et al. Mirna-424 protects against permanent focal cerebral ischemia injury in mice involving suppressing microglia activation. Stroke. 2013;44:1706–1713 [DOI] [PubMed] [Google Scholar]
- 314.Liu P, Zhao H, Wang R, Wang P, Tao Z, Gao L, et al. Microrna-424 protects against focal cerebral ischemia and reperfusion injury in mice by suppressing oxidative stress. Stroke. 2015;46:513–519 [DOI] [PubMed] [Google Scholar]
- 315.Li G, Ma Q, Wang R, Fan Z, Tao Z, Liu P, et al. Diagnostic and immunosuppressive potential of elevated mir-424 levels in circulating immune cells of ischemic stroke patients. Aging and disease. 2018;9:172–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Doeppner TR, Doehring M, Bretschneider E, Zechariah A, Kaltwasser B, Muller B, et al. Microrna-124 protects against focal cerebral ischemia via mechanisms involving usp14-dependent rest degradation. Acta Neuropathol. 2013;126:251–265 [DOI] [PubMed] [Google Scholar]
- 317.Sun Y, Gui H, Li Q, Luo ZM, Zheng MJ, Duan JL, et al. Microrna-124 protects neurons against apoptosis in cerebral ischemic stroke. CNS Neurosci Ther. 2013;19:813–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Hamzei Taj S, Kho W, Riou A, Wiedermann D, Hoehn M. Mirna-124 induces neuroprotection and functional improvement after focal cerebral ischemia. Biomaterials. 2016;91:151–165 [DOI] [PubMed] [Google Scholar]
- 319.Dutta R, Chomyk AM, Chang A, Ribaudo MV, Deckard SA, Doud MK, et al. Hippocampal demyelination and memory dysfunction are associated with increased levels of the neuronal microrna mir-124 and reduced ampa receptors. Ann Neurol. 2013;73:637–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Morel L, Regan M, Higashimori H, Ng SK, Esau C, Vidensky S, et al. Neuronal exosomal mirna-dependent translational regulation of astroglial glutamate transporter glt1. J Biol Chem. 2013;288:7105–7116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Liu XS, Chopp M, Zhang RL, Tao T, Wang XL, Kassis H, et al. Microrna profiling in subventricular zone after stroke: Mir-124a regulates proliferation of neural progenitor cells through notch signaling pathway. PloS one. 2011;6:e23461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Zhu F, Liu JL, Li JP, Xiao F, Zhang ZX, Zhang L. Microrna-124 (mir-124) regulates ku70 expression and is correlated with neuronal death induced by ischemia/reperfusion. Journal of molecular neuroscience : MN. 2014;52:148–155 [DOI] [PubMed] [Google Scholar]
- 323.Zhu H, Wang J, Shao Y, Wan D. Catalpol may improve axonal growth via regulating mir-124 regulated pi3k/akt/mtor pathway in neurons after ischemia. Ann Transl Med. 2019;7:306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Pena-Philippides JC, Caballero-Garrido E, Lordkipanidze T, Roitbak T. In vivo inhibition of mir-155 significantly alters post-stroke inflammatory response. Journal of neuroinflammation. 2016;13:287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Xing G, Luo Z, Zhong C, Pan X, Xu X. Influence of mir-155 on cell apoptosis in rats with ischemic stroke: Role of the ras homolog enriched in brain (rheb)/mtor pathway. Medical science monitor : international medical journal of experimental and clinical research. 2016;22:5141–5153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Caballero-Garrido E, Pena-Philippides JC, Lordkipanidze T, Bragin D, Yang Y, Erhardt EB, et al. In vivo inhibition of mir-155 promotes recovery after experimental mouse stroke. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2015;35:12446–12464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Jiang T, Zhou S, Li X, Song J, An T, Huang X, et al. Microrna-155 induces protection against cerebral ischemia/reperfusion injury through regulation of the notch pathway in vivo. Experimental and therapeutic medicine. 2019;18:605–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Ouyang YB, Lu Y, Yue S, Xu LJ, Xiong XX, White RE, et al. Mir-181 regulates grp78 and influences outcome from cerebral ischemia in vitro and in vivo. Neurobiol Dis. 2012;45:555–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Ni M, Zhang Y, Lee AS. Beyond the endoplasmic reticulum: Atypical grp78 in cell viability, signalling and therapeutic targeting. Biochem J. 2011;434:181–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Moon JM, Xu L, Giffard RG. Inhibition of microrna-181 reduces forebrain ischemia-induced neuronal loss. J Cereb Blood Flow Metab. 2013;33:1976–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Ouyang YB, Lu Y, Yue S, Giffard RG. Mir-181 targets multiple bcl-2 family members and influences apoptosis and mitochondrial function in astrocytes. Mitochondrion. 2012;12:213–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Xu LJ, Ouyang YB, Xiong X, Stary CM, Giffard RG. Post-stroke treatment with mir-181 antagomir reduces injury and improves long-term behavioral recovery in mice after focal cerebral ischemia. Experimental neurology. 2015;264:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Zeng LL, He XS, Liu JR, Zheng CB, Wang YT, Yang GY. Lentivirus-mediated overexpression of microrna-210 improves long-term outcomes after focal cerebral ischemia in mice. CNS neuroscience & therapeutics. 2016;22:961–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Jiang Y, Li L, Tan X, Liu B, Zhang Y, Li C. Mir-210 mediates vagus nerve stimulation-induced antioxidant stress and anti-apoptosis reactions following cerebral ischemia/reperfusion injury in rats. Journal of neurochemistry. 2015;134:173–181 [DOI] [PubMed] [Google Scholar]
- 335.Huang L, Ma Q, Li Y, Li B, Zhang L. Inhibition of microrna-210 suppresses pro-inflammatory response and reduces acute brain injury of ischemic stroke in mice. Experimental neurology. 2018;300:41–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Li P, Teng F, Gao F, Zhang M, Wu J, Zhang C. Identification of circulating micrornas as potential biomarkers for detecting acute ischemic stroke. Cellular and molecular neurobiology. 2015;35:433–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Wu J, Fan CL, Ma LJ, Liu T, Wang C, Song JX, et al. Distinctive expression signatures of serum micrornas in ischaemic stroke and transient ischaemic attack patients. Thrombosis and haemostasis. 2017;117:992–1001 [DOI] [PubMed] [Google Scholar]
- 338.Wang Y, Ma Z, Kan P, Zhang B. The diagnostic value of serum mirna-221–3p, mirna-382–5p, and mirna-4271 in ischemic stroke. Journal of stroke and cerebrovascular diseases : the official journal of National Stroke Association. 2017;26:1055–1060 [DOI] [PubMed] [Google Scholar]
- 339.He W, Chen S, Chen X, Li S, Chen W. Bioinformatic analysis of potential micrornas in ischemic stroke. Journal of stroke and cerebrovascular diseases : the official journal of National Stroke Association. 2016;25:1753–1759 [DOI] [PubMed] [Google Scholar]
- 340.Jia L, Hao F, Wang W, Qu Y. Circulating mir-145 is associated with plasma high-sensitivity c-reactive protein in acute ischemic stroke patients. Cell biochemistry and function. 2015;33:314–319 [DOI] [PubMed] [Google Scholar]
- 341.Peng G, Yuan Y, Wu S, He F, Hu Y, Luo B. Microrna let-7e is a potential circulating biomarker of acute stage ischemic stroke. Translational stroke research. 2015;6:437–445 [DOI] [PubMed] [Google Scholar]
- 342.Wang W, Sun G, Zhang L, Shi L, Zeng Y. Circulating micrornas as novel potential biomarkers for early diagnosis of acute stroke in humans. Journal of stroke and cerebrovascular diseases : the official journal of National Stroke Association. 2014;23:2607–2613 [DOI] [PubMed] [Google Scholar]
- 343.Tiedt S, Prestel M, Malik R, Schieferdecker N, Duering M, Kautzky V, et al. Rna-seq identifies circulating mir-125a-5p, mir-125b-5p, and mir-143–3p as potential biomarkers for acute ischemic stroke. Circulation research. 2017;121:970–980 [DOI] [PubMed] [Google Scholar]
- 344.Long G, Wang F, Li H, Yin Z, Sandip C, Lou Y, et al. Circulating mir-30a, mir-126 and let-7b as biomarker for ischemic stroke in humans. BMC neurology. 2013;13:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Wang Y, Zhang Y, Huang J, Chen X, Gu X, Wang Y, et al. Increase of circulating mir-223 and insulin-like growth factor-1 is associated with the pathogenesis of acute ischemic stroke in patients. BMC neurology. 2014;14:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Zhou J, Zhang J. Identification of mirna-21 and mirna-24 in plasma as potential early stage markers of acute cerebral infarction. Molecular medicine reports. 2014;10:971–976 [DOI] [PubMed] [Google Scholar]
- 347.Huang S, Lv Z, Guo Y, Li L, Zhang Y, Zhou L, et al. Identification of blood let-7e-5p as a biomarker for ischemic stroke. PloS one. 2016;11:e0163951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Gan CS, Wang CW, Tan KS. Circulatory microrna-145 expression is increased in cerebral ischemia. Genetics and molecular research : GMR. 2012;11:147–152 [DOI] [PubMed] [Google Scholar]
- 349.Tan KS, Armugam A, Sepramaniam S, Lim KY, Setyowati KD, Wang CW, et al. Expression profile of micrornas in young stroke patients. PloS one. 2009;4:e7689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Chen Y, Song Y, Huang J, Qu M, Zhang Y, Geng J, et al. Increased circulating exosomal mirna-223 is associated with acute ischemic stroke. Frontiers in neurology. 2017;8:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Dykstra-Aiello C, Jickling GC, Ander BP, Shroff N, Zhan X, Liu D, et al. Altered expression of long noncoding rnas in blood after ischemic stroke and proximity to putative stroke risk loci. Stroke. 2016;47:2896–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.He W, Wei D, Cai, Chen S, Li S, Chen W. Altered long non-coding rna transcriptomic profiles in ischemic stroke. Human gene therapy. 2018;29:719–732 [DOI] [PubMed] [Google Scholar]
- 353.Yan H, Yuan J, Gao L, Rao J, Hu J. Long noncoding rna meg3 activation of p53 mediates ischemic neuronal death in stroke. Neuroscience. 2016;337:191–199 [DOI] [PubMed] [Google Scholar]
- 354.Liu J, Li Q, Zhang KS, Hu B, Niu X, Zhou SM, et al. Downregulation of the long non-coding rna meg3 promotes angiogenesis after ischemic brain injury by activating notch signaling. Molecular neurobiology. 2017;54:8179–8190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Yan H, Rao J, Yuan J, Gao L, Huang W, Zhao L, et al. Long non-coding rna meg3 functions as a competing endogenous rna to regulate ischemic neuronal death by targeting mir-21/pdcd4 signaling pathway. Cell death & disease. 2017;8:3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Zhang B, Wang D, Ji TF, Shi L, Yu JL. Overexpression of lncrna anril up-regulates vegf expression and promotes angiogenesis of diabetes mellitus combined with cerebral infarction by activating nf-kappab signaling pathway in a rat model. Oncotarget. 2017;8:17347–17359 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 357.Mehta SL, Kim T, Vemuganti R. Long noncoding rna fosdt promotes ischemic brain injury by interacting with rest-associated chromatin-modifying proteins. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2015;35:16443–16449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Morris-Blanco KC, Kim T, Bertogliat MJ, Mehta SL, Chokkalla AK, Vemuganti R. Inhibition of the epigenetic regulator rest ameliorates ischemic brain injury. Molecular neurobiology. 2019;56:2542–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Wu Z, Wu P, Zuo X, Yu N, Qin Y, Xu Q, et al. Lncrna-n1lr enhances neuroprotection against ischemic stroke probably by inhibiting p53 phosphorylation. Molecular neurobiology. 2017;54:7670–7685 [DOI] [PubMed] [Google Scholar]
- 360.Chen F, Zhang L, Wang E, Zhang C, Li X. Lncrna gas5 regulates ischemic stroke as a competing endogenous rna for mir-137 to regulate the notch1 signaling pathway. Biochemical and biophysical research communications. 2018;496:184–190 [DOI] [PubMed] [Google Scholar]
- 361.Zhang J, Yuan L, Zhang X, Hamblin MH, Zhu T, Meng F, et al. Altered long non-coding rna transcriptomic profiles in brain microvascular endothelium after cerebral ischemia. Experimental neurology. 2016;277:162–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Li Z, Li J, Tang N. Long noncoding rna malat1 is a potent autophagy inducer protecting brain microvascular endothelial cells against oxygen-glucose deprivation/reoxygenation-induced injury by sponging mir-26b and upregulating ulk2 expression. Neuroscience. 2017;354:1–10 [DOI] [PubMed] [Google Scholar]
- 363.Guo D, Ma J, Yan L, Li T, Li Z, Han X, et al. Down-regulation of lncrna malat1 attenuates neuronal cell death through suppressing beclin1-dependent autophagy by regulating mir-30a in cerebral ischemic stroke. Cellular physiology and biochemistry : international journal of experimental cellular physiology, biochemistry, and pharmacology. 2017;43:182–194 [DOI] [PubMed] [Google Scholar]
- 364.Gangloff A, Nadeau L, Perry JJ, Baril P, Emond M. Ruptured aneurysmal subarachnoid hemorrhage in the emergency department: Clinical outcome of patients having a lumbar puncture for red blood cell count, visual and spectrophotometric xanthochromia after a negative computed tomography. Clinical biochemistry. 2015;48:634–639 [DOI] [PubMed] [Google Scholar]
- 365.Huang F, Yi J, Zhou T, Gong X, Jiang H, Yao X. Toward understanding non-coding rna roles in intracranial aneurysms and subarachnoid hemorrhage. Translational neuroscience. 2017;8:54–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Chen S, Feng H, Sherchan P, Klebe D, Zhao G, Sun X, et al. Controversies and evolving new mechanisms in subarachnoid hemorrhage. Progress in neurobiology. 2014;115:64–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Sacco RL, Kasner SE, Broderick JP, Caplan LR, Connors JJ, Culebras A, et al. An updated definition of stroke for the 21st century: A statement for healthcare professionals from the american heart association/american stroke association. Stroke. 2013;44:2064–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Taylor TN, Davis PH, Torner JC, Holmes J, Meyer JW, Jacobson MF. Lifetime cost of stroke in the united states. Stroke. 1996;27:1459–1466 [DOI] [PubMed] [Google Scholar]
- 369.D’Abbondanza JA, Ai J, Lass E, Wan H, Brathwaite S, Tso MK, et al. Robust effects of genetic background on responses to subarachnoid hemorrhage in mice. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2016;36:1942–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Connolly ES Jr., Rabinstein AA, Carhuapoma JR, Derdeyn CP, Dion J, Higashida RT, et al. Guidelines for the management of aneurysmal subarachnoid hemorrhage: A guideline for healthcare professionals from the american heart association/american stroke association. Stroke. 2012;43:1711–1737 [DOI] [PubMed] [Google Scholar]
- 371.Shimamura N, Ohkuma H. Phenotypic transformation of smooth muscle in vasospasm after aneurysmal subarachnoid hemorrhage. Translational stroke research. 2014;5:357–364 [DOI] [PubMed] [Google Scholar]
- 372.Fujii M, Yan J, Rolland WB, Soejima Y, Caner B, Zhang JH. Early brain injury, an evolving frontier in subarachnoid hemorrhage research. Translational stroke research. 2013;4:432–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Dupont S, Rabinstein AA. Extent of acute hydrocephalus after subarachnoid hemorrhage as a risk factor for poor functional outcome. Neurological research. 2013;35:107–110 [DOI] [PubMed] [Google Scholar]
- 374.Chen S, Li Q, Wu H, Krafft PR, Wang Z, Zhang JH. The harmful effects of subarachnoid hemorrhage on extracerebral organs. BioMed research international. 2014;2014:858496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Miller BA, Turan N, Chau M, Pradilla G. Inflammation, vasospasm, and brain injury after subarachnoid hemorrhage. BioMed research international. 2014;2014:384342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Veltkamp R, Purrucker J. Management of spontaneous intracerebral hemorrhage. Current neurology and neuroscience reports. 2017;17:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.de Schipper LJ, Baharoglu MI, Roos Y, de Beer F. Medical treatment for spontaneous anticoagulation-related intracerebral hemorrhage in the netherlands. Journal of stroke and cerebrovascular diseases : the official journal of National Stroke Association. 2017;26:1427–1432 [DOI] [PubMed] [Google Scholar]
- 378.Chen CJ, Brown WM, Moomaw CJ, Langefeld CD, Osborne J, Worrall BB, et al. Alcohol use and risk of intracerebral hemorrhage. Neurology. 2017;88:2043–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Unruptured intracranial aneurysms--risk of rupture and risks of surgical intervention. The New England journal of medicine. 1998;339:1725–1733 [DOI] [PubMed] [Google Scholar]
- 380.Wiebers DO, Whisnant JP, Huston J 3rd, Meissner I, Brown RD Jr., Piepgras DG, et al. Unruptured intracranial aneurysms: Natural history, clinical outcome, and risks of surgical and endovascular treatment. Lancet (London, England). 2003;362:103–110 [DOI] [PubMed] [Google Scholar]
- 381.Starke RM, Chalouhi N, Ding D, Hasan DM. Potential role of aspirin in the prevention of aneurysmal subarachnoid hemorrhage. Cerebrovascular diseases (Basel, Switzerland). 2015;39:332–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Abou-Saleh H, Pathan AR, Daalis A, Hubrack S, Abou-Jassoum H, Al-Naeimi H, et al. Inositol 1,4,5-trisphosphate (ip3) receptor up-regulation in hypertension is associated with sensitization of ca2+ release and vascular smooth muscle contractility. The Journal of biological chemistry. 2013;288:32941–32951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Kim BJ, Kim Y, Hong EP, Jeon JP, Yang JS, Choi HJ, et al. Correlation between altered dna methylation of intergenic regions of itpr3 and development of delayed cerebral ischemia in patients with subarachnoid hemorrhage. World neurosurgery. 2019;130:e449–e456 [DOI] [PubMed] [Google Scholar]
- 384.Tang Y, Han S, Asakawa T, Luo Y, Han X, Xiao B, et al. Effects of intracerebral hemorrhage on 5-hydroxymethylcytosine modification in mouse brains. Neuropsychiatric disease and treatment. 2016;12:617–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Sukumari-Ramesh S, Alleyne CH Jr., Dhandapani KM. The histone deacetylase inhibitor suberoylanilide hydroxamic acid (saha) confers acute neuroprotection after intracerebral hemorrhage in mice. Translational stroke research. 2016;7:141–148 [DOI] [PubMed] [Google Scholar]
- 386.Sinn DI, Kim SJ, Chu K, Jung KH, Lee ST, Song EC, et al. Valproic acid-mediated neuroprotection in intracerebral hemorrhage via histone deacetylase inhibition and transcriptional activation. Neurobiology of disease. 2007;26:464–472 [DOI] [PubMed] [Google Scholar]
- 387.Lai NS, Zhang JQ, Qin FY, Sheng B, Fang XG, Li ZB. Serum micrornas are non-invasive biomarkers for the presence and progression of subarachnoid haemorrhage. Bioscience reports. 2017;37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Jin H, Li C, Ge H, Jiang Y, Li Y. Circulating microrna: A novel potential biomarker for early diagnosis of intracranial aneurysm rupture a case control study. Journal of translational medicine. 2013;11:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Li P, Zhang Q, Wu X, Yang X, Zhang Y, Li Y, et al. Circulating micrornas serve as novel biological markers for intracranial aneurysms. Journal of the American Heart Association. 2014;3:e000972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Guo D, Liu J, Wang W, Hao F, Sun X, Wu X, et al. Alteration in abundance and compartmentalization of inflammation-related mirnas in plasma after intracerebral hemorrhage. Stroke. 2013;44:1739–1742 [DOI] [PubMed] [Google Scholar]
- 391.Jiang Y, Zhang M, He H, Chen J, Zeng H, Li J, et al. Microrna/mrna profiling and regulatory network of intracranial aneurysm. BMC medical genomics. 2013;6:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Liu D, Han L, Wu X, Yang X, Zhang Q, Jiang F. Genome-wide microrna changes in human intracranial aneurysms. BMC neurology. 2014;14:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Bache S, Rasmussen R, Rossing M, Laigaard FP, Nielsen FC, Moller K. Microrna changes in cerebrospinal fluid after subarachnoid hemorrhage. Stroke. 2017;48:2391–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Powers CJ, Dickerson R, Zhang SW, Rink C, Roy S, Sen CK. Human cerebrospinal fluid microrna: Temporal changes following subarachnoid hemorrhage. Physiological genomics. 2016;48:361–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Stylli SS, Adamides AA, Koldej RM, Luwor RB, Ritchie DS, Ziogas J, et al. Mirna expression profiling of cerebrospinal fluid in patients with aneurysmal subarachnoid hemorrhage. Journal of neurosurgery. 2017;126:1131–1139 [DOI] [PubMed] [Google Scholar]
- 396.Lee HJ, Yi JS, Lee HJ, Lee IW, Park KC, Yang JH. Dysregulated expression profiles of micrornas of experimentally induced cerebral aneurysms in rats. Journal of Korean Neurosurgical Society. 2013;53:72–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Holcomb M, Ding YH, Dai D, McDonald RJ, McDonald JS, Kallmes DF, et al. Rna-sequencing analysis of messenger rna/microrna in a rabbit aneurysm model identifies pathways and genes of interest. AJNR. American journal of neuroradiology. 2015;36:1710–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Wang Z, Lu G, Sze J, Liu Y, Lin S, Yao H, et al. Plasma mir-124 is a promising candidate biomarker for human intracerebral hemorrhage stroke. Molecular neurobiology. 2018;55:5879–5888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Yu A, Zhang T, Duan H, Pan Y, Zhang X, Yang G, et al. Mir-124 contributes to m2 polarization of microglia and confers brain inflammatory protection via the c/ebp-alpha pathway in intracerebral hemorrhage. Immunology letters. 2017;182:1–11 [DOI] [PubMed] [Google Scholar]
- 400.Kim JM, Lee ST, Chu K, Jung KH, Kim JH, Yu JS, et al. Inhibition of let7c microrna is neuroprotective in a rat intracerebral hemorrhage model. PloS one. 2014;9:e97946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Yang S, Tang W, He Y, Wen L, Sun B, Li S. Long non-coding rna and microrna-675/let-7a mediates the protective effect of melatonin against early brain injury after subarachnoid hemorrhage via targeting tp53 and neural growth factor. Cell death & disease. 2018;9:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Xi T, Jin F, Zhu Y, Wang J, Tang L, Wang Y, et al. Microrna-126–3p attenuates blood-brain barrier disruption, cerebral edema and neuronal injury following intracerebral hemorrhage by regulating pik3r2 and akt. Biochemical and biophysical research communications. 2017;494:144–151 [DOI] [PubMed] [Google Scholar]
- 403.Kong F, Zhou J, Zhou W, Guo Y, Li G, Yang L. Protective role of microrna-126 in intracerebral hemorrhage. Molecular medicine reports. 2017;15:1419–1425 [DOI] [PubMed] [Google Scholar]
- 404.Yu A, Zhang T, Zhong W, Duan H, Wang S, Ye P, et al. Mirna-144 induces microglial autophagy and inflammation following intracerebral hemorrhage. Immunology letters. 2017;182:18–23 [DOI] [PubMed] [Google Scholar]
- 405.Zhang Y, Han B, He Y, Li D, Ma X, Liu Q, et al. Microrna-132 attenuates neurobehavioral and neuropathological changes associated with intracerebral hemorrhage in mice. Neurochemistry international. 2017;107:182–190 [DOI] [PubMed] [Google Scholar]
- 406.Yang Z, Zhong L, Xian R, Yuan B. Microrna-223 regulates inflammation and brain injury via feedback to nlrp3 inflammasome after intracerebral hemorrhage. Molecular immunology. 2015;65:267–276 [DOI] [PubMed] [Google Scholar]
- 407.Xi T, Jin F, Zhu Y, Wang J, Tang L, Wang Y, et al. Mir-27a-3p protects against blood-brain barrier disruption and brain injury after intracerebral hemorrhage by targeting endothelial aquaporin-11. The Journal of biological chemistry. 2018;293:20041–20050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Zheng B, Liu H, Wang R, Xu S, Liu Y, Wang K, et al. Expression signatures of long non-coding rnas in early brain injury following experimental subarachnoid hemorrhage. Molecular medicine reports. 2015;12:967–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Peng J, Wu Y, Tian X, Pang J, Kuai L, Cao F, et al. High-throughput sequencing and co-expression network analysis of lncrnas and mrnas in early brain injury following experimental subarachnoid haemorrhage. Scientific reports. 2017;7:46577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Hanjin C, Tao L, Pengfei L, Ali Y, Huajun Z, Jiekun L, et al. Altered long noncoding rna and messenger rna expression in experimental intracerebral hemorrhage - a preliminary study. Cellular physiology and biochemistry : international journal of experimental cellular physiology, biochemistry, and pharmacology. 2018;45:1284–1301 [DOI] [PubMed] [Google Scholar]
- 411.Wang W, Li H, Yu L, Zhao Z, Wang H, Zhang D, et al. Aberrant expression of lncrnas and mrnas in patients with intracranial aneurysm. Oncotarget. 2017;8:2477–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Wong VS, Langley B. Epigenetic changes following traumatic brain injury and their implications for outcome, recovery and therapy. Neurosci Lett. 2016;625:26–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Namjoshi DR, Good C, Cheng WH, Panenka W, Richards D, Cripton PA, et al. Towards clinical management of traumatic brain injury: A review of models and mechanisms from a biomechanical perspective. Dis Model Mech. 2013;6:1325–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Elder GA, Cristian A. Blast-related mild traumatic brain injury: Mechanisms of injury and impact on clinical care. Mt Sinai J Med. 2009;76:111–118 [DOI] [PubMed] [Google Scholar]
- 415.Coronado VG, Xu L, Basavaraju SV, McGuire LC, Wald MM, Faul MD, et al. Surveillance for traumatic brain injury-related deaths--united states, 1997–2007. MMWR Surveill Summ. 2011;60:1–32 [PubMed] [Google Scholar]
- 416.Hicks RR, Fertig SJ, Desrocher RE, Koroshetz WJ, Pancrazio JJ. Neurological effects of blast injury. J Trauma. 2010;68:1257–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Smith DH, Wolf JA, Lusardi TA, Lee VM, Meaney DF. High tolerance and delayed elastic response of cultured axons to dynamic stretch injury. J Neurosci. 1999;19:4263–4269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Eucker SA, Smith C, Ralston J, Friess SH, Margulies SS. Physiological and histopathological responses following closed rotational head injury depend on direction of head motion. Exp Neurol. 2011;227:79–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Bullock R, Zauner A, Myseros JS, Marmarou A, Woodward JJ, Young HF. Evidence for prolonged release of excitatory amino acids in severe human head trauma. Relationship to clinical events. Ann N Y Acad Sci. 1995;765:290–297; discussion 298 [DOI] [PubMed] [Google Scholar]
- 420.Bullock R, Zauner A, Woodward JJ, Myseros J, Choi SC, Ward JD, et al. Factors affecting excitatory amino acid release following severe human head injury. J Neurosurg. 1998;89:507–518 [DOI] [PubMed] [Google Scholar]
- 421.Johnson VE, Stewart W, Smith DH. Widespread tau and amyloid-beta pathology many years after a single traumatic brain injury in humans. Brain Pathol. 2012;22:142–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Zhang ZY, Zhang Z, Fauser U, Schluesener HJ. Global hypomethylation defines a sub-population of reactive microglia/macrophages in experimental traumatic brain injury. Neurosci Lett. 2007;429:1–6 [DOI] [PubMed] [Google Scholar]
- 423.Haghighi F, Ge Y, Chen S, Xin Y, Umali MU, De Gasperi R, et al. Neuronal dna methylation profiling of blast-related traumatic brain injury. J Neurotrauma. 2015;32:1200–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Bailey ZS, Grinter MB, De La Torre Campos D, VandeVord PJ. Blast induced neurotrauma causes overpressure dependent changes to the dna methylation equilibrium. Neuroscience Letters. 2015;604:119–123 [DOI] [PubMed] [Google Scholar]
- 425.Bailey ZS, Grinter MB, De La Torre Campos D, VandeVord PJ. Blast induced neurotrauma causes overpressure dependent changes to the dna methylation equilibrium. Neurosci Lett. 2015;604:119–123 [DOI] [PubMed] [Google Scholar]
- 426.Schober ME, Ke X, Xing B, Block BP, Requena DF, McKnight R, et al. Traumatic brain injury increased igf-1b mrna and altered igf-1 exon 5 and promoter region epigenetic characteristics in the rat pup hippocampus. Journal of neurotrauma. 2012;29:2075–2085 [DOI] [PubMed] [Google Scholar]
- 427.Aberg D Role of the growth hormone/insulin-like growth factor 1 axis in neurogenesis. Endocr Dev. 2010;17:63–76 [DOI] [PubMed] [Google Scholar]
- 428.Gao WM, Chadha MS, Kline AE, Clark RS, Kochanek PM, Dixon CE, et al. Immunohistochemical analysis of histone h3 acetylation and methylation--evidence for altered epigenetic signaling following traumatic brain injury in immature rats. Brain Res. 2006;1070:31–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Shein NA, Grigoriadis N, Alexandrovich AG, Simeonidou C, Lourbopoulos A, Polyzoidou E, et al. Histone deacetylase inhibitor itf2357 is neuroprotective, improves functional recovery, and induces glial apoptosis following experimental traumatic brain injury. Faseb j. 2009;23:4266–4275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Tai YT, Lee WY, Lee FP, Lin TJ, Shih CL, Wang JY, et al. Low dose of valproate improves motor function after traumatic brain injury. BioMed research international. 2014;2014:980657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Dash PK, Orsi SA, Zhang M, Grill RJ, Pati S, Zhao J, et al. Valproate administered after traumatic brain injury provides neuroprotection and improves cognitive function in rats. PloS one. 2010;5:e11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Yu F, Wang Z, Tanaka M, Chiu CT, Leeds P, Zhang Y, et al. Posttrauma cotreatment with lithium and valproate: Reduction of lesion volume, attenuation of blood-brain barrier disruption, and improvement in motor coordination in mice with traumatic brain injury. Journal of neurosurgery. 2013;119:766–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Wang Y, Neumann M, Hansen K, Hong SM, Kim S, Noble-Haeusslein LJ, et al. Fluoxetine increases hippocampal neurogenesis and induces epigenetic factors but does not improve functional recovery after traumatic brain injury. Journal of neurotrauma. 2011;28:259–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Dash PK, Orsi SA, Moore AN. Histone deactylase inhibition combined with behavioral therapy enhances learning and memory following traumatic brain injury. Neuroscience. 2009;163:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Hu Z, Yu D, Almeida-Suhett C, Tu K, Marini AM, Eiden L, et al. Expression of mirnas and their cooperative regulation of the pathophysiology in traumatic brain injury. PloS one. 2012;7:e39357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Lei P, Li Y, Chen X, Yang S, Zhang J. Microarray based analysis of microrna expression in rat cerebral cortex after traumatic brain injury. Brain research. 2009;1284:191–201 [DOI] [PubMed] [Google Scholar]
- 437.Liu L, Sun T, Liu Z, Chen X, Zhao L, Qu G, et al. Traumatic brain injury dysregulates micrornas to modulate cell signaling in rat hippocampus. PloS one. 2014;9:e103948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Sun TY, Chen XR, Liu ZL, Zhao LL, Jiang YX, Qu GQ, et al. Expression profiling of micrornas in hippocampus of rats following traumatic brain injury. Journal of Huazhong University of Science and Technology. Medical sciences = Hua zhong ke ji da xue xue bao. Yi xue Ying De wen ban = Huazhong keji daxue xuebao. Yixue Yingdewen ban. 2014;34:548–553 [DOI] [PubMed] [Google Scholar]
- 439.Redell JB, Liu Y, Dash PK. Traumatic brain injury alters expression of hippocampal micrornas: Potential regulators of multiple pathophysiological processes. Journal of neuroscience research. 2009;87:1435–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Meissner L, Gallozzi M, Balbi M, Schwarzmaier S, Tiedt S, Terpolilli NA, et al. Temporal profile of microrna expression in contused cortex after traumatic brain injury in mice. Journal of neurotrauma. 2016;33:713–720 [DOI] [PubMed] [Google Scholar]
- 441.Chandran R, Sharma A, Bhomia M, Balakathiresan NS, Knollmann-Ritschel BE, Maheshwari RK. Differential expression of micrornas in the brains of mice subjected to increasing grade of mild traumatic brain injury. Brain injury. 2017;31:106–119 [DOI] [PubMed] [Google Scholar]
- 442.Sun L, Zhao M, Zhang J, Liu A, Ji W, Li Y, et al. Mir-144 promotes beta-amyloid accumulation-induced cognitive impairments by targeting adam10 following traumatic brain injury. Oncotarget. 2017;8:59181–59203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Sun L, Liu A, Zhang J, Ji W, Li Y, Yang X, et al. Mir-23b improves cognitive impairments in traumatic brain injury by targeting atg12-mediated neuronal autophagy. Behavioural brain research. 2018;340:126–136 [DOI] [PubMed] [Google Scholar]
- 444.Sun L, Zhao M, Wang Y, Liu A, Lv M, Li Y, et al. Neuroprotective effects of mir-27a against traumatic brain injury via suppressing foxo3a-mediated neuronal autophagy. Biochemical and biophysical research communications. 2017;482:1141–1147 [DOI] [PubMed] [Google Scholar]
- 445.Sabirzhanov B, Zhao Z, Stoica BA, Loane DJ, Wu J, Borroto C, et al. Downregulation of mir-23a and mir-27a following experimental traumatic brain injury induces neuronal cell death through activation of proapoptotic bcl-2 proteins. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2014;34:10055–10071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Huang S, Ge X, Yu J, Han Z, Yin Z, Li Y, et al. Increased mir-124–3p in microglial exosomes following traumatic brain injury inhibits neuronal inflammation and contributes to neurite outgrowth via their transfer into neurons. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2018;32:512–528 [DOI] [PubMed] [Google Scholar]
- 447.Lv J, Zeng Y, Qian Y, Dong J, Zhang Z, Zhang J. Microrna let-7c-5p improves neurological outcomes in a murine model of traumatic brain injury by suppressing neuroinflammation and regulating microglial activation. Brain research. 2018;1685:91–104 [DOI] [PubMed] [Google Scholar]
- 448.Harrison EB, Emanuel K, Lamberty BG, Morsey BM, Li M, Kelso ML, et al. Induction of mir-155 after brain injury promotes type 1 interferon and has a neuroprotective effect. Frontiers in molecular neuroscience. 2017;10:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Li Z, Wang Y, Zeng G, Zheng X, Wang W, Ling Y, et al. Increased mir-155 and heme oxygenase-1 expression is involved in the protective effects of formononetin in traumatic brain injury in rats. American journal of translational research. 2017;9:5653–5661 [PMC free article] [PubMed] [Google Scholar]
- 450.Henry RJ, Doran SJ, Barrett JP, Meadows VE, Sabirzhanov B, Stoica BA, et al. Inhibition of mir-155 limits neuroinflammation and improves functional recovery after experimental traumatic brain injury in mice. Neurotherapeutics : the journal of the American Society for Experimental NeuroTherapeutics. 2019;16:216–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Redell JB, Zhao J, Dash PK. Altered expression of mirna-21 and its targets in the hippocampus after traumatic brain injury. Journal of neuroscience research. 2011;89:212–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Sandhir R, Gregory E, Berman NE. Differential response of mirna-21 and its targets after traumatic brain injury in aging mice. Neurochemistry international. 2014;78:117–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Han Z, Chen F, Ge X, Tan J, Lei P, Zhang J. Mir-21 alleviated apoptosis of cortical neurons through promoting pten-akt signaling pathway in vitro after experimental traumatic brain injury. Brain research. 2014;1582:12–20 [DOI] [PubMed] [Google Scholar]
- 454.Ge XT, Lei P, Wang HC, Zhang AL, Han ZL, Chen X, et al. Mir-21 improves the neurological outcome after traumatic brain injury in rats. Scientific reports. 2014;4:6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Li D, Huang S, Zhu J, Hu T, Han Z, Zhang S, et al. Exosomes from mir-21–5p-increased neurons play a role in neuroprotection by suppressing rab11a-mediated neuronal autophagy in vitro after traumatic brain injury. Medical science monitor : international medical journal of experimental and clinical research. 2019;25:1871–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Ge X, Han Z, Chen F, Wang H, Zhang B, Jiang R, et al. Mir-21 alleviates secondary blood-brain barrier damage after traumatic brain injury in rats. Brain research. 2015;1603:150–157 [DOI] [PubMed] [Google Scholar]
- 457.Wang L, Zhao C, Wu S, Xiao G, Zhuge X, Lei P, et al. Hydrogen gas treatment improves the neurological outcome after traumatic brain injury via increasing mir-21 expression. Shock (Augusta, Ga.). 2018;50:308–315 [DOI] [PubMed] [Google Scholar]
- 458.Harrison EB, Hochfelder CG, Lamberty BG, Meays BM, Morsey BM, Kelso ML, et al. Traumatic brain injury increases levels of mir-21 in extracellular vesicles: Implications for neuroinflammation. FEBS open bio. 2016;6:835–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Di Pietro V, Ragusa M, Davies D, Su Z, Hazeldine J, Lazzarino G, et al. Micrornas as novel biomarkers for the diagnosis and prognosis of mild and severe traumatic brain injury. Journal of neurotrauma. 2017;34:1948–1956 [DOI] [PubMed] [Google Scholar]
- 460.Sharma A, Chandran R, Barry ES, Bhomia M, Hutchison MA, Balakathiresan NS, et al. Identification of serum microrna signatures for diagnosis of mild traumatic brain injury in a closed head injury model. PloS one. 2014;9:e112019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Yang T, Song J, Bu X, Wang C, Wu J, Cai J, et al. Elevated serum mir-93, mir-191, and mir-499 are noninvasive biomarkers for the presence and progression of traumatic brain injury. Journal of neurochemistry. 2016;137:122–129 [DOI] [PubMed] [Google Scholar]
- 462.Bhomia M, Balakathiresan NS, Wang KK, Papa L, Maheshwari RK. A panel of serum mirna biomarkers for the diagnosis of severe to mild traumatic brain injury in humans. Scientific reports. 2016;6:28148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Mitra B, Rau TF, Surendran N, Brennan JH, Thaveenthiran P, Sorich E, et al. Plasma micro-rna biomarkers for diagnosis and prognosis after traumatic brain injury: A pilot study. Journal of clinical neuroscience : official journal of the Neurosurgical Society of Australasia. 2017;38:37–42 [DOI] [PubMed] [Google Scholar]
- 464.Osei J, Kelly W, Toffolo K, Donahue K, Levy B, Bard J, et al. Thymosin beta 4 induces significant changes in the plasma mirna profile following severe traumatic brain injury in the rat lateral fluid percussion injury model. Expert opinion on biological therapy. 2018;18:159–164 [DOI] [PubMed] [Google Scholar]
- 465.Qin X, Li L, Lv Q, Shu Q, Zhang Y, Wang Y. Expression profile of plasma micrornas and their roles in diagnosis of mild to severe traumatic brain injury. PloS one. 2018;13:e0204051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Redell JB, Moore AN, Ward NH 3rd, Hergenroeder GW, Dash PK. Human traumatic brain injury alters plasma microrna levels. Journal of neurotrauma. 2010;27:2147–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Sajja V, Jablonska A, Haughey N, Bulte JWM, Stevens RD, Long JB, et al. Sphingolipids and microrna changes in blood following blast traumatic brain injury: An exploratory study. Journal of neurotrauma. 2018;35:353–361 [DOI] [PubMed] [Google Scholar]
- 468.Hicks SD, Johnson J, Carney MC, Bramley H, Olympia RP, Loeffert AC, et al. Overlapping microrna expression in saliva and cerebrospinal fluid accurately identifies pediatric traumatic brain injury. Journal of neurotrauma. 2018;35:64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.Balakathiresan N, Bhomia M, Chandran R, Chavko M, McCarron RM, Maheshwari RK. Microrna let-7i is a promising serum biomarker for blast-induced traumatic brain injury. Journal of neurotrauma. 2012;29:1379–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.You WD, Tang QL, Wang L, Lei J, Feng JF, Mao Q, et al. Alteration of microrna expression in cerebrospinal fluid of unconscious patients after traumatic brain injury and a bioinformatic analysis of related single nucleotide polymorphisms. Chinese journal of traumatology = Zhonghua chuang shang za zhi. 2016;19:11–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Ko J, Hemphill M, Yang Z, Sewell E, Na YJ, Sandsmark DK, et al. Diagnosis of traumatic brain injury using mirna signatures in nanomagnetically isolated brain-derived extracellular vesicles. Lab on a chip. 2018;18:3617–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Bao TH, Miao W, Han JH, Yin M, Yan Y, Wang WW, et al. Spontaneous running wheel improves cognitive functions of mouse associated with mirna expressional alteration in hippocampus following traumatic brain injury. Journal of molecular neuroscience : MN. 2014;54:622–629 [DOI] [PubMed] [Google Scholar]
- 473.Miao W, Bao TH, Han JH, Yin M, Yan Y, Wang WW, et al. Voluntary exercise prior to traumatic brain injury alters mirna expression in the injured mouse cerebral cortex. Brazilian journal of medical and biological research = Revista brasileira de pesquisas medicas e biologicas. 2015;48:433–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Truettner JS, Alonso OF, Bramlett HM, Dietrich WD. Therapeutic hypothermia alters microrna responses to traumatic brain injury in rats. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2011;31:1897–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Wang CF, Zhao CC, Weng WJ, Lei J, Lin Y, Mao Q, et al. Alteration in long non-coding rna expression after traumatic brain injury in rats. Journal of neurotrauma. 2017;34:2100–2108 [DOI] [PubMed] [Google Scholar]
- 476.Zhong J, Jiang L, Cheng C, Huang Z, Zhang H, Liu H, et al. Altered expression of long non-coding rna and mrna in mouse cortex after traumatic brain injury. Brain research. 2016;1646:589–600 [DOI] [PubMed] [Google Scholar]
- 477.Lyu Q, Zhang ZB, Fu SJ, Xiong LL, Liu J, Wang TH. Microarray expression profile of lncrnas and mrnas in rats with traumatic brain injury after a2b5+ cell transplantation. Cell transplantation. 2017;26:1622–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Patel NA, Moss LD, Lee JY, Tajiri N, Acosta S, Hudson C, et al. Long noncoding rna malat1 in exosomes drives regenerative function and modulates inflammation-linked networks following traumatic brain injury. Journal of neuroinflammation. 2018;15:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 479.Zhong J, Jiang L, Huang Z, Zhang H, Cheng C, Liu H, et al. The long non-coding rna neat1 is an important mediator of the therapeutic effect of bexarotene on traumatic brain injury in mice. Brain, behavior, and immunity. 2017;65:183–194 [DOI] [PubMed] [Google Scholar]
- 480.Ngugi AK, Bottomley C, Kleinschmidt I, Sander JW, Newton CR. Estimation of the burden of active and life-time epilepsy: A meta-analytic approach. Epilepsia. 2010;51:883–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Hauser RM, Henshall DC, Lubin FD. The epigenetics of epilepsy and its progression. The Neuroscientist : a review journal bringing neurobiology, neurology and psychiatry. 2018;24:186–200 [DOI] [PubMed] [Google Scholar]
- 482.Blumcke I, Thom M, Aronica E, Armstrong DD, Bartolomei F, Bernasconi A, et al. International consensus classification of hippocampal sclerosis in temporal lobe epilepsy: A task force report from the ilae commission on diagnostic methods. Epilepsia. 2013;54:1315–1329 [DOI] [PubMed] [Google Scholar]
- 483.Kobow K, Blumcke I. Epigenetics in epilepsy. Neuroscience letters. 2018;667:40–46 [DOI] [PubMed] [Google Scholar]
- 484.Falconer MA. Mesial temporal (ammon’s horn) sclerosis as a common cause of epilepsy. Aetiology, treatment, and prevention. Lancet (London, England). 1974;2:767–770 [DOI] [PubMed] [Google Scholar]
- 485.Younus I, Reddy DS. Epigenetic interventions for epileptogenesis: A new frontier for curing epilepsy. Pharmacology & therapeutics. 2017;177:108–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 486.Perrine K, Hermann BP, Meador KJ, Vickrey BG, Cramer JA, Hays RD, et al. The relationship of neuropsychological functioning to quality of life in epilepsy. Archives of neurology. 1995;52:997–1003 [DOI] [PubMed] [Google Scholar]
- 487.Henshall DC, Hamer HM, Pasterkamp RJ, Goldstein DB, Kjems J, Prehn JHM, et al. Micrornas in epilepsy: Pathophysiology and clinical utility. The Lancet. Neurology. 2016;15:1368–1376 [DOI] [PubMed] [Google Scholar]
- 488.Kwan P, Brodie MJ. Early identification of refractory epilepsy. The New England journal of medicine. 2000;342:314–319 [DOI] [PubMed] [Google Scholar]
- 489.Hesdorffer DC, Beck V, Begley CE, Bishop ML, Cushner-Weinstein S, Holmes GL, et al. Research implications of the institute of medicine report, epilepsy across the spectrum: Promoting health and understanding. Epilepsia. 2013;54:207–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490.Najm I, Jehi L, Palmini A, Gonzalez-Martinez J, Paglioli E, Bingaman W. Temporal patterns and mechanisms of epilepsy surgery failure. Epilepsia. 2013;54:772–782 [DOI] [PubMed] [Google Scholar]
- 491.Becker AJ, Chen J, Zien A, Sochivko D, Normann S, Schramm J, et al. Correlated stage- and subfield-associated hippocampal gene expression patterns in experimental and human temporal lobe epilepsy. The European journal of neuroscience. 2003;18:2792–2802 [DOI] [PubMed] [Google Scholar]
- 492.Elliott RC, Miles MF, Lowenstein DH. Overlapping microarray profiles of dentate gyrus gene expression during development- and epilepsy-associated neurogenesis and axon outgrowth. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2003;23:2218–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493.Jamali S, Bartolomei F, Robaglia-Schlupp A, Massacrier A, Peragut JC, Regis J, et al. Large-scale expression study of human mesial temporal lobe epilepsy: Evidence for dysregulation of the neurotransmission and complement systems in the entorhinal cortex. Brain : a journal of neurology. 2006;129:625–641 [DOI] [PubMed] [Google Scholar]
- 494.Majores M, Eils J, Wiestler OD, Becker AJ. Molecular profiling of temporal lobe epilepsy: Comparison of data from human tissue samples and animal models. Epilepsy research. 2004;60:173–178 [DOI] [PubMed] [Google Scholar]
- 495.Pitkanen A, Lukasiuk K. Mechanisms of epileptogenesis and potential treatment targets. The Lancet. Neurology. 2011;10:173–186 [DOI] [PubMed] [Google Scholar]
- 496.Sharma A Genome-wide expression analysis in epilepsy: A synthetic review. Current topics in medicinal chemistry. 2012;12:1008–1032 [DOI] [PubMed] [Google Scholar]
- 497.Henshall DC, Kobow K. Epigenetics and epilepsy. Cold Spring Harbor perspectives in medicine. 2015;5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.Miller-Delaney SF, Bryan K, Das S, McKiernan RC, Bray IM, Reynolds JP, et al. Differential dna methylation profiles of coding and non-coding genes define hippocampal sclerosis in human temporal lobe epilepsy. Brain : a journal of neurology. 2015;138:616–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 499.Kobow K, Kaspi A, Harikrishnan KN, Kiese K, Ziemann M, Khurana I, et al. Deep sequencing reveals increased dna methylation in chronic rat epilepsy. Acta neuropathologica. 2013;126:741–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 500.Zhu Q, Wang L, Zhang Y, Zhao FH, Luo J, Xiao Z, et al. Increased expression of dna methyltransferase 1 and 3a in human temporal lobe epilepsy. Journal of molecular neuroscience : MN. 2012;46:420–426 [DOI] [PubMed] [Google Scholar]
- 501.Wang L, Fu X, Peng X, Xiao Z, Li Z, Chen G, et al. Dna methylation profiling reveals correlation of differential methylation patterns with gene expression in human epilepsy. Journal of molecular neuroscience : MN. 2016;59:68–77 [DOI] [PubMed] [Google Scholar]
- 502.Debski KJ, Pitkanen A, Puhakka N, Bot AM, Khurana I, Harikrishnan KN, et al. Etiology matters - genomic dna methylation patterns in three rat models of acquired epilepsy. Scientific reports. 2016;6:25668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 503.Belhedi N, Perroud N, Karege F, Vessaz M, Malafosse A, Salzmann A. Increased cpa6 promoter methylation in focal epilepsy and in febrile seizures. Epilepsy research. 2014;108:144–148 [DOI] [PubMed] [Google Scholar]
- 504.Kobow K, Jeske I, Hildebrandt M, Hauke J, Hahnen E, Buslei R, et al. Increased reelin promoter methylation is associated with granule cell dispersion in human temporal lobe epilepsy. Journal of neuropathology and experimental neurology. 2009;68:356–364 [DOI] [PubMed] [Google Scholar]
- 505.Genc F, Kara M, Unal Y, Uygur Kucukseymen E, Bicer Gomceli Y, Kaynar T, et al. Methylation of cation-chloride cotransporters nkcc1 and kcc2 in patients with juvenile myoclonic epilepsy. Neurological sciences : official journal of the Italian Neurological Society and of the Italian Society of Clinical Neurophysiology. 2019;40:1007–1013 [DOI] [PubMed] [Google Scholar]
- 506.Machnes ZM, Huang TC, Chang PK, Gill R, Reist N, Dezsi G, et al. Dna methylation mediates persistent epileptiform activity in vitro and in vivo. PloS one. 2013;8:e76299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Zhu Q, Wang L, Xiao Z, Xiao F, Luo J, Zhang X, et al. Decreased expression of ras-grf1 in the brain tissue of the intractable epilepsy patients and experimental rats. Brain research. 2013;1493:99–109 [DOI] [PubMed] [Google Scholar]
- 508.Chen X, Peng X, Wang L, Fu X, Zhou JX, Zhu B, et al. Association of rasgrf1 methylation with epileptic seizures. Oncotarget. 2017;8:46286–46297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509.Parrish RR, Buckingham SC, Mascia KL, Johnson JJ, Matyjasik MM, Lockhart RM, et al. Methionine increases bdnf dna methylation and improves memory in epilepsy. Annals of clinical and translational neurology. 2015;2:401–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Xiao W, Cao Y, Long H, Luo Z, Li S, Deng N, et al. Genome-wide dna methylation patterns analysis of noncoding rnas in temporal lobe epilepsy patients. Molecular neurobiology. 2018;55:793–803 [DOI] [PubMed] [Google Scholar]
- 511.Jagirdar R, Drexel M, Bukovac A, Tasan RO, Sperk G. Expression of class ii histone deacetylases in two mouse models of temporal lobe epilepsy. Journal of neurochemistry. 2016;136:717–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Jagirdar R, Drexel M, Kirchmair E, Tasan RO, Sperk G. Rapid changes in expression of class i and iv histone deacetylases during epileptogenesis in mouse models of temporal lobe epilepsy. Experimental neurology. 2015;273:92–104 [DOI] [PubMed] [Google Scholar]
- 513.Huang Y, Doherty JJ, Dingledine R. Altered histone acetylation at glutamate receptor 2 and brain-derived neurotrophic factor genes is an early event triggered by status epilepticus. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2002;22:8422–8428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.Citraro R, Leo A, De Caro C, Nesci V, Gallo Cantafio ME, Amodio N, et al. Effects of histone deacetylase inhibitors on the development of epilepsy and psychiatric comorbidity in wag/rij rats. Molecular neurobiology. 2019 [DOI] [PubMed] [Google Scholar]
- 515.Reddy SD, Clossen BL, Reddy DS. Epigenetic histone deacetylation inhibition prevents the development and persistence of temporal lobe epilepsy. The Journal of pharmacology and experimental therapeutics. 2018;364:97–109 [DOI] [PubMed] [Google Scholar]
- 516.Jimenez-Mateos EM, Engel T, Merino-Serrais P, Fernaud-Espinosa I, Rodriguez-Alvarez N, Reynolds J, et al. Antagomirs targeting microrna-134 increase hippocampal pyramidal neuron spine volume in vivo and protect against pilocarpine-induced status epilepticus. Brain structure & function. 2015;220:2387–2399 [DOI] [PubMed] [Google Scholar]
- 517.Jimenez-Mateos EM, Engel T, Merino-Serrais P, McKiernan RC, Tanaka K, Mouri G, et al. Silencing microrna-134 produces neuroprotective and prolonged seizure-suppressive effects. Nature medicine. 2012;18:1087–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 518.Reschke CR, Silva LF, Norwood BA, Senthilkumar K, Morris G, Sanz-Rodriguez A, et al. Potent anti-seizure effects of locked nucleic acid antagomirs targeting mir-134 in multiple mouse and rat models of epilepsy. Molecular therapy. Nucleic acids. 2017;6:45–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 519.Jimenez-Mateos EM, Bray I, Sanz-Rodriguez A, Engel T, McKiernan RC, Mouri G, et al. Mirna expression profile after status epilepticus and hippocampal neuroprotection by targeting mir-132. The American journal of pathology. 2011;179:2519–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520.Brennan GP, Dey D, Chen Y, Patterson KP, Magnetta EJ, Hall AM, et al. Dual and opposing roles of microrna-124 in epilepsy are mediated through inflammatory and nrsf-dependent gene networks. Cell reports. 2016;14:2402–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521.Zhu X, Han X, Blendy JA, Porter BE. Decreased creb levels suppress epilepsy. Neurobiology of disease. 2012;45:253–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 522.Peng J, Omran A, Ashhab MU, Kong H, Gan N, He F, et al. Expression patterns of mir-124, mir-134, mir-132, and mir-21 in an immature rat model and children with mesial temporal lobe epilepsy. Journal of molecular neuroscience : MN. 2013;50:291–297 [DOI] [PubMed] [Google Scholar]
- 523.Jang Y, Moon J, Lee ST, Jun JS, Kim TJ, Lim JA, et al. Dysregulated long non-coding rnas in the temporal lobe epilepsy mouse model. Seizure. 2018;58:110–119 [DOI] [PubMed] [Google Scholar]
- 524.Lee DY, Moon J, Lee ST, Jung KH, Park DK, Yoo JS, et al. Dysregulation of long non-coding rnas in mouse models of localization-related epilepsy. Biochemical and biophysical research communications. 2015;462:433–440 [DOI] [PubMed] [Google Scholar]
- 525.Wang HK, Yan H, Wang K, Wang J. Dynamic regulation effect of long non-coding rna-uca1 on nf-kb in hippocampus of epilepsy rats. European review for medical and pharmacological sciences. 2017;21:3113–3119 [PubMed] [Google Scholar]
- 526.Geng JF, Liu X, Zhao HB, Fan WF, Geng JJ, Liu XZ. Lncrna uca1 inhibits epilepsy and seizure-induced brain injury by regulating mir-495/nrf2-are signal pathway. The international journal of biochemistry & cell biology. 2018;99:133–139 [DOI] [PubMed] [Google Scholar]
- 527.Han CL, Ge M, Liu YP, Zhao XM, Wang KL, Chen N, et al. Long non-coding rna h19 contributes to apoptosis of hippocampal neurons by inhibiting let-7b in a rat model of temporal lobe epilepsy. Cell death & disease. 2018;9:617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 528.Han CL, Liu YP, Zhao XM, Wang KL, Chen N, Hu W, et al. Whole-transcriptome screening reveals the regulatory targets and functions of long non-coding rna h19 in epileptic rats. Biochemical and biophysical research communications. 2017;489:262–269 [DOI] [PubMed] [Google Scholar]
- 529.Wang J, Yu JT, Tan L, Tian Y, Ma J, Tan CC, et al. Genome-wide circulating microrna expression profiling indicates biomarkers for epilepsy. Scientific reports. 2015;5:9522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 530.Wang J, Tan L, Tan L, Tian Y, Ma J, Tan CC, et al. Circulating micrornas are promising novel biomarkers for drug-resistant epilepsy. Scientific reports. 2015;5:10201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 531.Raoof R, Jimenez-Mateos EM, Bauer S, Tackenberg B, Rosenow F, Lang J, et al. Cerebrospinal fluid micrornas are potential biomarkers of temporal lobe epilepsy and status epilepticus. Scientific reports. 2017;7:3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 532.Hu K, Zhang C, Long L, Long X, Feng L, Li Y, et al. Expression profile of micrornas in rat hippocampus following lithium-pilocarpine-induced status epilepticus. Neuroscience letters. 2011;488:252–257 [DOI] [PubMed] [Google Scholar]
- 533.Roncon P, Soukupova M, Binaschi A, Falcicchia C, Zucchini S, Ferracin M, et al. Microrna profiles in hippocampal granule cells and plasma of rats with pilocarpine-induced epilepsy--comparison with human epileptic samples. Scientific reports. 2015;5:14143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 534.Pichardo-Casas I, Goff LA, Swerdel MR, Athie A, Davila J, Ramos-Brossier M, et al. Expression profiling of synaptic micrornas from the adult rat brain identifies regional differences and seizure-induced dynamic modulation. Brain research. 2012;1436:20–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 535.Risbud RM, Porter BE. Changes in microrna expression in the whole hippocampus and hippocampal synaptoneurosome fraction following pilocarpine induced status epilepticus. PloS one. 2013;8:e53464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 536.Kan AA, van Erp S, Derijck AA, de Wit M, Hessel EV, O’Duibhir E, et al. Genome-wide microrna profiling of human temporal lobe epilepsy identifies modulators of the immune response. Cellular and molecular life sciences : CMLS. 2012;69:3127–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 537.McKiernan RC, Jimenez-Mateos EM, Bray I, Engel T, Brennan GP, Sano T, et al. Reduced mature microrna levels in association with dicer loss in human temporal lobe epilepsy with hippocampal sclerosis. PloS one. 2012;7:e35921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 538.Mooney C, Becker BA, Raoof R, Henshall DC. Epimirbase: A comprehensive database of microrna-epilepsy associations. Bioinformatics (Oxford, England). 2016;32:1436–1438 [DOI] [PubMed] [Google Scholar]
- 539.Suuronen T, Huuskonen J, Pihlaja R, Kyrylenko S, Salminen A. Regulation of microglial inflammatory response by histone deacetylase inhibitors. Journal of neurochemistry. 2003;87:407–416 [DOI] [PubMed] [Google Scholar]
- 540.Boutillier AL, Trinh E, Loeffler JP. Selective e2f-dependent gene transcription is controlled by histone deacetylase activity during neuronal apoptosis. Journal of neurochemistry. 2003;84:814–828 [DOI] [PubMed] [Google Scholar]
- 541.Jin H, Kanthasamy A, Harischandra DS, Kondru N, Ghosh A, Panicker N, et al. Histone hyperacetylation up-regulates protein kinase cdelta in dopaminergic neurons to induce cell death: Relevance to epigenetic mechanisms of neurodegeneration in parkinson disease. The Journal of biological chemistry. 2014;289:34743–34767 [DOI] [PMC free article] [PubMed] [Google Scholar]


