Abstract
Computed Tomography (CT) plays an important role in lung malignancy diagnostics, therapy assessment, and facilitating precision medicine delivery. However, the use of personalized imaging protocols poses a challenge in large-scale cross-center CT image radiomic studies. We present an end-to-end solution called STAN-CT for CT image standardization and normalization, which effectively reduces discrepancies in image features caused by using different imaging protocols or using different CT scanners with the same imaging protocol. STAN-CT consists oftwo components: 1)a Generative Adversarial Networks (GAN) model where a latent-feature-based loss function is adopted to learn the data distribution of standard images within a few rounds of generator training, and 2) an automatic DICOM reconstruction pipeline with systematic image quality control that ensures the generation ofhigh-quality standard DICOM images. Experimental results indicate that the training efficiency and model performance of STAN-CT have been significantly improved compared to the state-of-the-art CT image standardization and normalization algorithms.
1. Introduction
Computed Tomography (CT), one of the widely used modalities for cancer diagnostics1,2, provides a flexible image acquisition and reconstruction protocol that allows adjusting kernel function, amount of radiation, slice thickness, etc. to meet clinical requirements3. The non-standard protocol setup broadens the scope of CT uses effectively, but at the same time, it creates a data discrepancy problem among the acquired images4. For example, the same clinical observation with two different CT acquisition protocols may result in images with significantly different radiomic features, esp. intensity and texture5,6. As a result, this hinders the effectiveness of inter-clinic data sharing and the performance of large-scale radiomics studies5.
The CT data discrepancy problem could be potentially addressed by defining and using a standard image acquisition protocol. However, it is impractical to use the same image acquisition protocol in all the clinical practices, not only because there are already multiple CT scanner manufactures in the market7, but also the limitations of using a fixed protocol for all patients under all situations in diagnosis, staging, therapy selection, and therapy assessment of tumor malignancies8. We propose to develop an image standardization and normalization tool to "translate" any CT images acquired using non-standard protocols into the standard one while preserving most of the anatomic details4. Mathematically, let target image x be an image acquired using a standard protocol, given any non-standard source image x', the image standardization and normalization tool aims to compose a synthetic image X from x' such that X is significantly more similar to x than to x' regarding radiomic features.
In recent years, deep-learning-based algorithms have been developed for image or data synthesis9,10. U-Net is a special kind of fully connected U-shaped neural network for image synthesis11. Built upon U-Net or a similar neural network structure, Generative Adversarial Network (GAN) is a class of deep learning models, in which two neural networks contest with each other9. Being one of the mostly-used deep learning architectures for image synthesis, GAN has been utilized on CT image standardization10. In GANai, a customized GAN model is trained using an alternative training strategy to effectively learn the data distribution, thus achieving significantly better performance than the classic GAN model and the traditional image processing algorithm called Histogram matching10,12,13. However, GANai focuses on the relatively easier image patch synthesis rather than the whole DICOM image synthesis problem10.
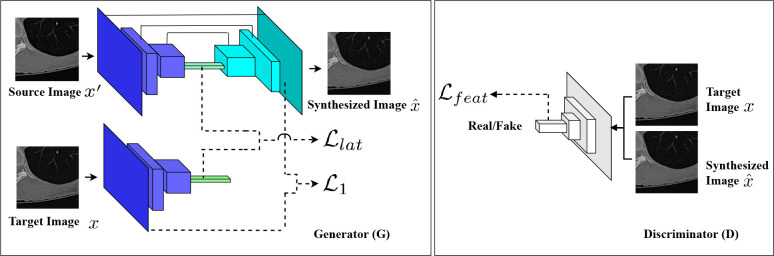
To address the CT image standardization and normalization problem, tools are needed to reconstruct synthesized data that have the common feature space as the target data10. This poses two fundamental computational challenges: 1) to effectively map between target images and synthesized images with great pixel-level details, and 2) to maintain the texture consistency among the synthesized images. In this paper, we present an end-to-end solution called STAN-CT. In STAN-CT, we introduce two new constrains in GAN loss shown in Fig. 1. Specifically, we adopt a latent-space-based loss for the generator to establish a one-to-one mapping from target images to synthesized images. Also, a feature-based loss is adopted for the discriminator to critic the texture features of the standard and the synthesized images. Furthermore, to synthesize CT images in the Digital Imaging and Communications in Medicine (DICOM) format14, STAN-CT introduces a DICOM reconstruction framework that can integrate all the synthesized image patches to generate a DICOM file for clinical use. The framework ensures the quality of the synthesized DICOM by systematically identifying and pruning low-quality image patches. In our experiment, by comparing the synthesized images with the ground truth, we demonstrate that STAN-CT significantly outperforms the current state-of-the-art models. In summary, STAN-CT has the following advantages:
STAN-CT provides an end-to-end solution for CT image standardization and normalization. The outcome of STAN-CT is DICOM image files that are ready to be loaded into clinical systems directly.
STAN-CT adopts a novel one-to-one mapping loss function on the latent space. It enforces the generator to draw samples from the same distribution where the standard image belongs to.
STAN-CT uses a novel feature-based loss to improve the performance of the discriminator.
STAN-CT is effective in model training. It can quickly converge within a few rounds of training processes.
2. Background
CT images are one of the key modalities in tumor malignancy studies15. The CT image discrepancy problem due to the common use of non-standard imaging protocols poses a gap between CT imaging and radiomics studies. To fill the gap, clinical image synthesis tools need to be developed to translate images acquired using non-standard protocols into standard ones.
Image or data synthesis
Image or data synthesis is an active research area in computer vision, computer graphics, and natural language pro-cessing9. By definition, image synthesis is a process of generating synthetic images using limited information16. The given information includes text description, random noise, or any other types of information. With the recent break through in deep learning, image synthesis algorithms have been applied in the areas of text-to-image generation17, detecting lost frame in a video18, image-to-image transformation19, and medical imaging20 successfully.
U-net
U-Net is a special fully connected neural network originally proposed for medical image segmentation11. Precise localization and relatively small training data requirements are the major advantages of using U-Net11. A U-Net usually has three parts, down-sampling, bottleneck, and up-sampling, where the up-sampling and down-sampling are symmetric. There are also connections from down-sampling layers to the corresponding up-sampling layers to recover lost information during down-sampling. However, while U-net is effective on generating structural information, it suffers from learning and keeping texture details 21. This issue can be overcome by adopting U-net in a more sophisticated generative model called Generative Adversarial Networks (GANs)9.
Generative Adversarial Network
Generative Adversarial Networks (GAN), which are often used for data and image synthesis9, normally consist of a generator G and a discriminator D. The generator that could be a U-Net is responsible for generating fake data from noise, and the discriminator tries to identify whether its input is drawn from the real or fake data. Among all the GAN models, cGAN is capable of synthesizing new images based on a prior distribution22. However, since the image features of the synthesized data and that of the target data may not fall into the same distribution, cGAN may not be directly applicable for the CT image standardization problem. GANai is a customized cGAN model, in which the generator and the discriminator are trained alternatively to learn the data distribution, thus achieving significantly better performance than the vanilla cGAN model. However, GANai focuses on the relatively easier image patch synthesis problem rather than the whole DICOM image synthesis problem.
Disentanglement
In a generative model, the latent space often plays a vital role in target domain mapping. Appropriate latent space learning is crucial for generating high quality data. Disentanglement is an effective metric that provides a deep understanding of a particular layer in a neural network23. Network disentanglement can assist to uncover the important factors that contribute to the data generation process24.
Alternative Training Strategy
Model training is one of the most crucial parts of GAN because of the special network architecture (i.e. the generator needs to fool the discriminator while the discriminator tries to detect true data distribution from the false one). In the alternative training mechanism, when one component is in training, the other one remains unchanged. Also, each component has a fixed number of training iterations. A variant of alternative training was proposed in Liang et al.10 named fully-optimized alternative training, where the model training is divided into two phases called G-phase and D-phase. In the G-phase, D is fixed, and G needs to achieve a certain accuracy 6G before reaching the maximum training step tmax. In the D-phase, G is fixed, and D needs to achieve a pre-defined performance 6D or it stops when reaching a maximum training step tmax. When one training phase is completed, the other phase will begin. The GANai training will continue until an optimal result is achieved or the maximum number of epochs is reached. Furthermore, instead of performance competing between a single copy of D and G, multiple copies of Gs and Ds compete with each other. For example, a G needs to fool multiple Ds before its phase is over. Also, a rollback mechanism is implemented in GANai so that if a component is not able to fool its counterpart within limited steps, it rolls back to the beginning status of the current phase and starts again. This alternative training mechanism has been successfully applied to address the CT image standardization problem.
3. Method
With STAN-CT, we attempt to address the long-standing CT image standardization problem. STAN-CT consists of a novel GAN model and a dedicated DICOM synthesis framework to meet the clinical requirements.
Standardizing CT image patches
Similar to the conventional GAN models, the STAN-CT GAN model has two components, the generator G and the discriminator D. G is a U-shaped network11 consisting of an encoder and a decoder. Both the encoder and the decoder consist of seven hidden layers. There is a skip connection from each layer of the encoder to the corresponding layer of the decoder to address the information loss problem during the down-sampling. D consists of five fully connected convolutional layers. Fig. 1 illustrates the GAN architecture of STAN-CT. The detailed architecture and the hyperparameters of STAN-CT used in our experiments are specified in section 4 sub-section titled STAN-CT architecture and hyperparameters. Mathematically, let x be a standard image and x' be its corresponding non-standard image. The aim of the generator is to create a new image X that has the same data distribution as x. Meanwhile, the discriminator determines whether x and X are from the standard image distribution.
Discriminator Loss. In a GAN model, the performance of D and G increases accordingly. We propose to adopt two losses for the discriminator training, i.e. the WGAN25 adversarial loss function to critic the standard and non-standard images and the fetcher-based loss. WGAN is a stable GAN training mechanism that provides a learning balance between G and D26. STAN-CT adopts the WGAN-based adversarial loss of the discriminator defined as:
<math/>
where w is the hyper-parameters of D, m is the batch size, x'(i) is the input (non-standard image), and x(i) is the corresponding standard image. In addition to the WGAN-based adversarial loss, STAN-CT introduces a new feature-based loss function L feat. To improve generator diversity, a similar feature-based loss function has been used in Yang et al.27. Here, we use the feature space of D instead of a secondary pre-trained network to maintain a balanced network (i.e. D and G are not too strong or too weak compared with other). The feature-based loss is described in Eq 2:
<math/>
where </> is the feature extractor and V is the volume of the feature space, and G(x') is an image generated by G and x is the target image. Finally, let A1 be a wight factor (A1 e [0,1]), the total loss of D that combines the WGAN-based loss Ladv(D) and the feature-based loss Lfeat is defined as:
<math/>
Generator Loss. The generator loss consists of three components, i.e. the WGAN-based loss, the latent loss, and the L1 regularization. The WGAN-based loss, which is used to improve network convergence, is defined as:
<math/>
where 6 represents all the hyper-parameters of G, x'(i) is a source image, and f is 1-Lipschitz function, which returns the Earth-Mover (EM) distance from one metric space to another.
It is the latent space that connects the distribution of an input domain and an output domain in the same generative model, which allows a smooth domain translation28. Inspired by Mao et al.28, we propose a new latent-feature-based loss function to enforce one-to-one mapping between the synthesized image and the standard image. Specifically, the latent loss Llat aims to minimize the distance between the latent distribution of the synthesized images and their corresponding standard images.
<math/>
where z stands for the latent vector, G(x'), and x is its corresponding standard image. Finally, the total loss of G L(G) is defined as:
<math/>
where A2 e [0,1] and A3 e [0,1] are wight factors. m J2m=1 |x — G(x')| is the L1 regularization function.
DICOM Reconstruction Framework
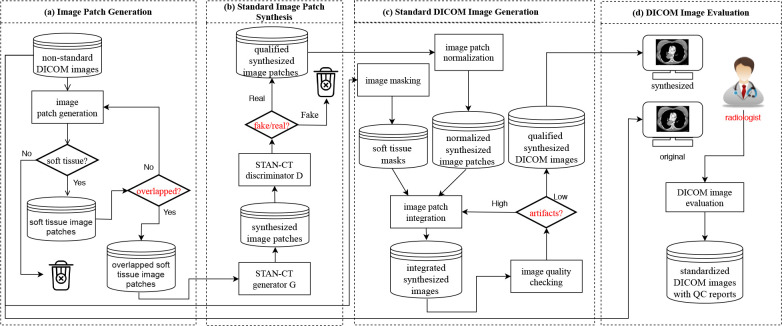
STAN-CT presents a DICOM-to-DICOM reconstruction framework for systematic DICOM reconstruction. The DICOM-to-DICOM reconstruction framework includes four additional components to facilitate processes such as image patch generation and fusion (see Fig. 2). Each component has a unique quality control unit (red diamond box) that ensures the outputs are free from defects.
Step 1. soft tissue image patch generation: The first step of STAN-CT DICOM-to-DICOM image standardization is soft tissue image patch generation. Image patches with size between 100 and 256 are randomly generated using the input DICOM image. An image patch is a soft tissue image patch if at least 70% of the pixels are in the soft tissue range (Hounsfield unit value ranging from -1000 to 900). The process will continue until each soft-tissue image patch contains at least 50% overlapped pixels.
Step 2. standard image patch synthesis: With a trained STAN-CT generator, a soft-tissue image patch obtained in the previous step will be standardized (see Section 3 for details).
Then, the synthesized image patches will be examined by STAN-CT discriminator. If a synthesized image patch can fool the discriminator, it is considered as a qualified synthesized image patch. Otherwise, the synthesized image patch will be discarded. This step ensures the quality of the synthesized image patches.
Step 3. standard DICOM image generation: Given all the qualified synthesized image patches, we first normalize the pixel intensity from gray-scale to the Hounsfield unit using:
<math/>
where PHU and Pg is the pixel value in Hounsfield unit and gray-scale unit respectively, Xg is a qualified synthesized image patch, and MAX and MIN are the maximum and minimum CT number of a source DICOM.
Meanwhile, with a soft tissue image mask created from the original DICOM images with Hounsfield unit ranging from — 1000 to 900, the non-soft tissue parts of the synthesized and normalized image patches will be discarded. Finally, we integrate all the valid soft tissue patches to generate the integrated synthesized images.
The quality of the integrated synthesized images will be checked using a quality control unit, which inspects whether there is any box artifacts or missing values. If some artifacts are identified, the corresponding image patches will be re-integrated by cropping boundary pixels.
Step 4. DICOM image evaluation: In the DICOM image evaluate step, both the synthesized and the original nonstandard DICOM image files will be viewed side-by-side by radiologists using a PACS reading workstation. Radiologists will be asked to evaluate image quality, estimate the acquisition protocol, and extract tumor properties. The radiologists' reports will be used to evaluate the quality of the standardized CT images. Meanwhile, with all the synthesized DICOM files generated in the previous step, image texture features will be automatically extracted and compared for automatic performance evaluation.
4. Experimental result
Data
For the training data, we used total of 14,688 CT image slices captured using three different kernels (BL57, BL64, and BR40) and four different slice thicknesses (0.5, 1, 1.5, 3mm) using Siemens CT Somatom Force scanner at the University of Kentucky Medical Center. STAN-CT adopted BL64 kernel and 1mm slice thickness as the standard protocol since it has been widely used in clinical practice. Random cropping was used for the image patch extraction and resized into 256 x 256 pixel patches. Data augmentation was done by rotating and shifting image patches. Finally, a total of 49,000 soft-tissue image patches were generated from the CT slices and were used as the training data of STAN-CT. Two testing data sets were prepared for STAN-CT performance evaluation. Both data sets were captured using Siemens CT Somatom Force scanner at the University of Kentucky Medical Center hospital. The first testing data were captured using the non-standard protocol BR40 and 1mm slice thickness. The second testing data were captured using the non-standard protocol BL57 and 1mm slice thickness. The image patch generation step was the same as that of the training data. Each test data set contains 3,810 image patches.
STAN-CT architecture and hyperparameters
STAN-CT GAN model consists of a U-Net with fifteen hidden layers and an FCN with five hidden layers. The 4 x 4 kernel is used in the convolutional layer. LeakyRelu29 is adopted as the activation function in all the hidden layers. Softmax is used in the last layer of FCN. Random weight is used during the network initialization phase. The prediction thresholds for determining fake or real images is 0.01 and 0.99 respectively. Maximum training epochs were set to 100 with a learning rate of 0.0001 with momentum 0.5. A fully optimized alternative training mechanism (the same as GANai) was used for the network training. STAN-CT was implemented in TensorFlow30 on a Linux computer server with eight Nvidia GTX 1080 GPU cards. The model took about 36 hours to train from scratch. Once the model was trained, it took about 0.1 seconds to synthesize and normalize every image patch.
Evaluation Metric
For performance evaluation, we computed five radiomic texture features (i.e. dissimilarity, contrast, homogeneity, energy, and correlation) using Gray Level Co-occurrence Matrix (GLCM). The absolute error E of each radiomic texture feature was computed using:
<math/>
where p is the GLCM feature extractor Isyn and Itarget is the synthesized image from STAN-CT and the target image respectively. J is the corresponding feature space.
Performance of image patch synthesis
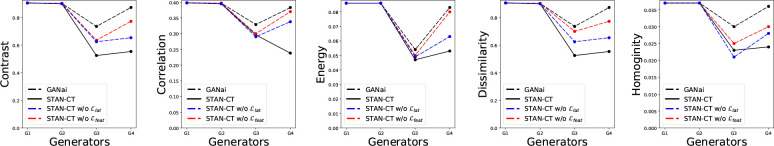
Table 1 shows the absolute error of five GLCM-based texture features of STAN-CT, GANai (the current state-of-the-art model), and two disentangled representation of STAN-CT on the soft tissues. In the model named "STAN-CT w/o Llat", we discarded from STAN-CT the latent loss function Llat of G. In the second one named "STAN-CT w/o L feat", we discarded the feature-based loss L feat of D from STAN-CT. All the models were tested using kernel BL57 and BR40 with the same slice thickness (1mm). For kernel BL57, STAN-CT and its variants outperformed GANai in all the texture features. For kernel BR40, STAN-CT was significantly better than GANai in four out of five features. Also, Table 3 shows the feature comparison on the same tumor tissue. On all the five GLCM-based texture features extracted from the images scanned using BL57 and BR40 kernels, STAN-CT clearly outperforms GANai.
Table 1: Texture feature comparison between GANai, STAN-CT and its two variants. Five texture features (dis- similarity, contrast, homogeneity, energy and correlation) were extracted from DICOM image patches. The absolute error was reported for each feature.
| Kernel | Features | GANai | STAN-CT | STAN-CT w/o LLat |
STAN-CT w/o Lfeat |
| dissimilarity | 0.313 | 0.228 | 0.234 | 0.245 | |
| contrast | 0.313 | 0.228 | 0.234 | 0.245 | |
| BL57 | homogeneity | 0.012 | 0.009 | 0.009 | 0.012 |
| energy | 0.032 | 0.035 | 0.038 | 0.022 | |
| correlation | 0.085 | 0.058 | 0.057 | 0.120 | |
| dissimilarity | 0.683 | 0.407 | 0.441 | 0.545 | |
| contrast | 0.683 | 0.407 | 0.441 | 0.545 | |
| BR40 | homogeneity | 0.028 | 0.018 | 0.019 | 0.024 |
| energy | 0.040 | 0.041 | 0.057 | 0.045 | |
| correlation | 0.315 | 0.203 | 0.273 | 0.303 |
Table 3: Performance on tumor-specific tissues. Five texture features, i.e. dissimilarity, contrast, homogeneity, energy, and corre- lation, were extracted from all the image patches of a tumor. The mean absolute error of each feature was reported.
| Kernel | Model | dissimilarity | contrast | homogeneity | energy | correlation |
| BL57 | GANai | 0.124 ± 0.098 | 0.124 ± 0.098 | 0.018 ± 0.012 | 0.098 ± 0.054 | 0.877 ± 0.368 |
| STAN-CT | 0.114 ± 0.026 | 0.114 ± 0.026 | 0.017 ± 0.005 | 0.041 ± 0.019 | 0.847 ± 0.408 | |
| BR40 | GANai | 0.616 ± 0.104 | 0.616 ± 0.104 | 0.091 ± 0.018 | 0.222 ± 0.061 | 1.184 ± 0.646 |
| STAN-CT | 0.602 ± 0.081 | 0.602 ± 0.081 | 0.076 ± 0.011 | 0.182 ± 0.061 | 1.184 ± 0.646 |
The first four generators of each GAN models were selected for further analysis. Fig. 3 illustrates the change of the absolute errors of the five GLCM-based texture features using the generators produced in the first four iterations of alternative training of STAN-CT or GANai. The result indicates that STAN-CT can quickly reduce the errors in the first a few iteration of the alternative training, while no clear trend was observed in the results of GANai.
Performance of DICOM reconstruction
A straightforward patch-based image reconstruction approach has three steps: 1) splitting a DICOM slice into overlapped or non-overlapped image patches; 2) standardizing each image patch; and 3) merging the standardized image patches into one DICOM slice. A common problem in such a patch-based image reconstruction process is image artifacts, such as boundary artifact or inconsistent texture. As shown in Table 2, the straightforward approach has the highest absolute error on all the tested image features.
Table 2: Texture feature comparison. Five texture fea- tures were extracted from DICOM images constructed from the same image patches using four different DICOM reconstruc- tion methods. The averaged absolute error is reported for each feature.
| Kernel | Features | straightforward | w/ overlapped check | w/ real/fake check | STAN-CT |
| dissimilarity | 0.727 | 0.485 | 0.334 | 0.201 | |
| contrast | 0.727 | 0.485 | 0.334 | 0.201 | |
| BL57 | homogeneity | 0.031 | 0.019 | 0.012 | 0.009 |
| energy | 0.072 | 0.063 | 0.046 | 0.032 | |
| correlation | 0.319 | 0.149 | 0.075 | 0.054 | |
| dissimilarity | 0.849 | 0.653 | 0.496 | 0.405 | |
| contrast | 0.849 | 0.653 | 0.496 | 0.405 | |
| BR40 | homogeneity | 0.035 | 0.027 | 0.022 | 0.016 |
| energy | 0.048 | 0.045 | 0.051 | 0.040 | |
| correlation | 0.386 | 0.345 | 0.289 | 0.201 |
In STAN-CT, three quality control units were inserted into the framework, each being adopted to address a specific image quality problem. Table 2 shows that STAN-CT achieved significantly better performance than the straightforward method regarding the absolute errors on five selected texture features. Fig. 4 visualized the reconstructed DICOM images using the two methods. The red (green) circle highlights the boundary effect where two image patches were merged (texture inconsistency within a DICOM slice) using the straightforward method. In the same DICOM reconstructed using STAN-CT, no visual artifacts were found according to the radiologist's report.
We further compared STAN-CT with its two variants. The method named "w/ overlapped check" used only the first quality control unit to check whether there were enough overlapped soft-tissue image patches. The method named "w/ real/fake check" used the first two quality control units, which not only checked if there were enough image patches, but also examined whether the image patches were successfully standardized. Table 2 shows that both approaches achieve better results than the straightforward method, but none of is better than STAN-CT, indicating all the three quality control units are critical regarding artifact detection and removal. The standardized DICOM images, along with the corresponding standard images, were reviewed by radiologists at the Department of radiology, University of Kentucky using the picture archiving and communication system (PACS) viewer (Barco, GA, USA). The radiologists, who were blinded to the image reconstruction algorithms, reported that no obvious difference was observed in lung regions between the two kinds of images.
5. Discussion
First, by systematically removing every single component in the GAN model and in the DICOM reconstruction pipeline using the the leave-one-out approach, we analyzed the impact of every component of STAN-CT. In STAN-CT, both the latent loss CLat and the feature loss C feat are key components. To evaluate the impact of the loss functions, two versions of STAN-CT GAN, where the latent loss or the feature loss has been removed respectively, were created. Table 1 shows that none of them can achieve the same performance as that of STAN-CT regarding the GLCM-based texture features. Also, Fig. 5 shows that the latent loss of STAN-CT CLat decreases during G-training, indicating that the generator can reduce the gap between the distributions of the target image and the synthetic image effectively, while maintaining flat during the D-training phases.
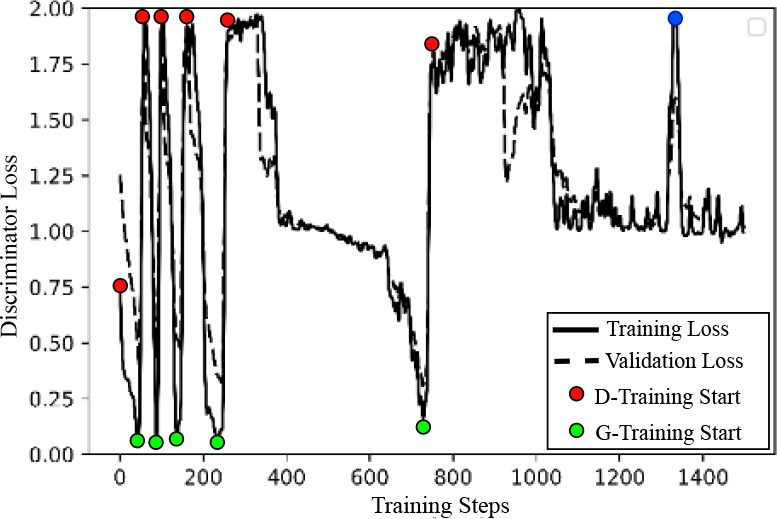
Second, we analyzed the STAN-CT training phase switches. During the STAN-CT training, the discriminator loss (shown in Fig. 6) bounced between 0 and 2 rapidly in the early phase switches, indicating an efficient discriminator and generator training. In the later phases, however, the training time increased significantly, indicating that both the generator and the discriminator were converging. At the blue colored point, the discriminator failed to distinguish real from fake images after certain iterations of discriminator training. In this situation, the new discriminator was ruled out and the D-training was restarted. If the D-training continued to fail, the STAN-CT alternative training can stop10.
Finally, the DICOM reconstruction pipeline includes four quality control units, each contributing to the improvements of the quality of the resulting DICOM images. Table 2 shows that the contrast error of the straightforward DICOM reconstruction (without using any of the quality control units) is 0.727, which can be reduced to 0.485 by adding the overlapped soft tissue quality control, which provides consistent texture throughout the DICOM. It can be further reduced to 0.334 (54% improvement) by adding the discriminator checker that ensures the success of image synthesis. Eventually, if all the four quality control units were used, the contract error was reduced to 0.201 (72% improvement).
6. Conclusion
Data discrepancy in CT images due to the use of non-standard image acquisition protocols adds extra burden to radiologists and also creates a gap in large-scale cross-center radiomic studies. We propose STAN-CT, a novel tool for CT DICOM image standardization and normalization. In STAN-CT, new loss functions are introduced for efficient GAN training, and a dedicated DICOM-to-DICOM image reconstruction framework has been developed to automate the DICOM standardization and normalization process. The experimental results show that STAN-CT is significantly better than the existing tools on CT image standardization. Our experiments demonstrate that inconsistency in CT image acquisition can be effectively harmonized using STAN-CT. This work fits well with large-scale radiomic studies in cancer researches where radiomic features can be extracted from standardized images rather than the original ones.
Acknowledgements
This research is supported by NIH NCI (grant no. 1R21CA231911) and Kentucky Lung Cancer Research (grant no. KLCR-3048113817).
Figures & Table
Figure 1: GAN architecture of STAN-CT. The generator G is a U-Net with a new latent loss for synthesizing image patches. The discriminator D is an Fully Convolutional Network classifier for determining whether a synthesized image patch is fake or real.
Figure 2: STAN-CT DICOM-to-DICOM image standardization framework. (a) Soft tissue image patches are generated from the input DICOM files, ready for image standardization and normalization. (b) For all the soft tissue image patches, new image patches are synthesized using STAN-CT generator. The quality of the new image patches is checked using STAN-CT discriminator. (c) All the synthesized soft-tissue image patches are integrated and are filtered by a soft tissue image mask generated using the input DICOM image. DICOM image quality is checked by examining box artifacts and empty pixels. (d) The synthesized and the original non-standard DICOM image files will be viewed side-by-side by radiologists using a PACS reading workstation. The radiologists’ reports will be used to further evaluate the quality of the standardized CT images. Meanwhile, image texture features will be extracted for automatic performance evaluation.
Figure 3: Performance evaluation of STAN-CT generator. The first four generators of STAN-CT and GANai were compared using the GLCM-based features. The x-axis denotes the training phase, and the y-axis denotes the absolute error of each selected texture feature. The result indicates STAN-CT archived overall the best performance.
Figure 4: DICOM Reconstruction Comparison.
Figure 5: Latent loss during GAN training. Latent loss is de- creasing effectively in the G-training phase, while it keeps stable in the D-training phase.
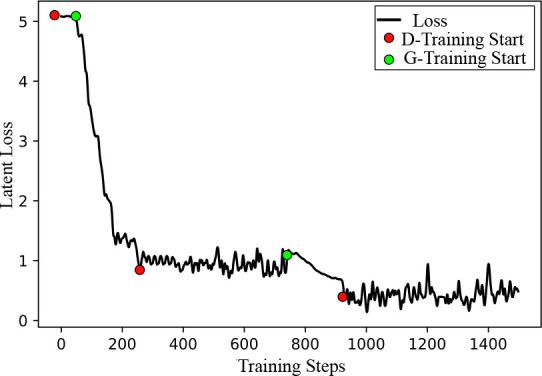
Figure 6: Performance of the discriminator of STAN-CT in different training phases. The discriminator loss decreases in the D-training phase and increases in the G-training phase. The blue-colored point indicates a failed-then-restart D-training.
References
- 1.Jerry L Prince, Jonathan M Links. Medical imaging signals and systems. Pearson Prentice Hall Upper Saddle River: 2006. [Google Scholar]
- 2.Mahesh Mahadevappa. Fundamentals of medical imaging. Medical Physics. 2011;38(3):1735–1735. [Google Scholar]
- 3.Midya Abhishek, Chakraborty Jayasree, Gonen Mithat, et al. Influence of ct acquisition and reconstruction parameters on radiomic feature reproducibility. Journal of Medical Imaging. 2018;5(1):011020. doi: 10.1117/1.JMI.5.1.011020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liang G, Zhang J, Brooks M, et al. radiomic features of lung cancer and their dependency on ct image acquisition parameters. Medical Physics. 2017;44(6):3024. [Google Scholar]
- 5.Berenguer Roberto, Pastor-Juan Maria del Rosario, Canales-Vazquez Jesus, et al. Radiomics of ct features may be nonrepro-ducible and redundant: Influence of ct acquisition parameters. Radiology. 2018. pp. 172–361. [DOI] [PubMed]
- 6.Hunter Luke A, Krafft Shane, Stingo Francesco, et al. High quality machine-robust image features: Identification in nonsmall cell lung cancer computed tomography images. Medical physics. 2013;40(12) doi: 10.1118/1.4829514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jijo Paul, Krauss B, Banckwitz R, et al. Relationships of clinical protocols and reconstruction kernels with image quality and radiation dose in a 128-slice ct scanner: study with an anthropomorphic and water phantom. European journal of radiology. 2012;81(5):e699–e703. doi: 10.1016/j.ejrad.2011.01.078. [DOI] [PubMed] [Google Scholar]
- 8.Gierada David S, Bierhals Andrew J, Choong Cliff K, et al. Effects of ct section thickness and reconstruction kernel on emphysema quantification: relationship to the magnitude of the ct emphysema index. Academic radiology. 2010;17(2):146–156. doi: 10.1016/j.acra.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He Huang, Yu Philip S, Changhu Wang. An introduction to image synthesis with generative adversarial nets. arXivpreprint arXiv:1803.04469. 2018.
- 10.Liang Gongbo, Fouladvand Sajjad, Zhang Jie, et al. Ganai: Standardizing ct images using generative adversarial network with alternative improvement. bioRxiv. 2018. p. 460188.
- 11.Ronneberger Olaf, Fischer Philipp, Brox Thomas. In Medical Image Computing and Computer-Assisted Intervention. Springer; 2015. U-net: Convolutional networks for biomedical image segmentation; pp. 234–241. [Google Scholar]
- 12.Robert Weeks Arthur, Lloyd J Sartor, Harley R Myler . Histogram specification of 24-bit color images in the color difference (cy) color space. Journal of electronic imaging. 1999;8(3):290–301. [Google Scholar]
- 13.Anil K Jain. Fundamentals of digital image processing. Englewood Cliffs, NJ: Prentice Hall; 1989. [Google Scholar]
- 14.Mildenberger Peter, Eichelberg Marco, Martin Eric. Introduction to the dicom standard. European radiology. 2002;12(4):920–927. doi: 10.1007/s003300101100. [DOI] [PubMed] [Google Scholar]
- 15.Rizzo Stefania, Botta Francesca, Raimondi Sara, et al. Radiomics: the facts and the challenges of image analysis. European radiology experimental. 2018;2(1):36. doi: 10.1186/s41747-018-0068-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roy A Hall, Donald P Greenberg. A testbed for realistic image synthesis. IEEE Computer Graphics and Applications. 1983;3(8):10–20. [Google Scholar]
- 17.Reed Scott, Akata Zeynep, Yan Xinchen, et al. Generative adversarial text to image synthesis. In Proceedings of the 33rd International Conference on Machine Learning. 2016;48:1060–1069. [Google Scholar]
- 18.Yong-Hoon Kwon, Min-Gyu Park. Predicting future frames using retrospective cycle gan. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2019. pp. 1811–1820.
- 19.Karras Tero, Laine Samuli, Aila Timo. A style-based generator architecture for generative adversarial networks. In Proceedings ofthe IEEE Conference on Computer Vision and Pattern Recognition. 2019. pp. 4401–4410. [DOI] [PubMed]
- 20.Yu Zekuan, Xiang Qing, Jiahao, et al. Meng. Retinal image synthesis from multiple-landmarks input with generative adversarial networks. Biomedical engineering online. 2019;18(1):62. doi: 10.1186/s12938-019-0682-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ravishankar Hariharan, Venkataramani Rahul, Thiruvenkadam Sheshadri, et al. Learning and incorporating shape models for semantic segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer; 2017. pp. 203–211. [Google Scholar]
- 22.Mirza Mehdi, Osindero Simon. Conditional generative adversarial nets. arXiv:1411.1784v1. 2014.
- 23.Quan-shi Zhang, Song-Chun Zhu. Visual interpretability for deep learning: a survey. Frontiers of Information Technology & Electronic Engineering. 2018;19(1):27–39. [Google Scholar]
- 24.Higgins Irina, David, et al. Amos. Towards a definition of disentangled representations. arXiv preprint arXiv:1812.02230. 2018.
- 25.Arjovsky Martin, Chintala Soumith, Bottou Leon. Wasserstein gan. arXiv preprint arXiv:1701.07875. 2017.
- 26.Gulrajani Ishaan, Ahmed Faruk, Arjovsky Martin, et al. Improved training of wasserstein gans. In Advances in neural information processing systems. 2017:5767–5777. [Google Scholar]
- 27.Yang Qingsong, Yan Pingkun, Zhang Yanbo, et al. Low-dose ct image denoising using a generative adversarial network with wasserstein distance and perceptual loss. IEEE transactions on medical imaging. 2018;37(6):1348–1357. doi: 10.1109/TMI.2018.2827462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mao Qi, Lee Hsin-Ying, Tseng Hung-Yu, et al. Mode seeking generative adversarial networks for diverse image synthesis. In Proceedings ofthe IEEE Conference on Computer Vision and Pattern Recognition. 2019. pp. 1429–1437.
- 29.Xu Bing, Wang Naiyan, Chen Tianqi, et al. Empirical evaluation of rectified activations in convolutional network. arXiv preprint arXiv:1505.00853. 2015.
- 30.Abadi Martin, Agarwal Ashish, Barham Paul, et al. TensorFlow: Large-scale machine learning on heterogeneous systems. 2015. Software available from tensorflow.org .