Abstract
Scavenger receptor class B type I (SR-BI), is a physiologically relevant HDL receptor that mediates selective uptake of lipoprotein (HDL)-derived cholesteryl ester (CE) in vitro and in vivo. Mammalian SR-BI is a 509-amino acid, ∼82 kDa glycoprotein, that contains N- and C-terminal cytoplasmic domains, two-transmembrane domains, as well as a large extracellular domain containing 5-6 cysteine residues and multiple sites for N-linked glycosylation. The size and structural characteristics of SR-BI, however, vary considerably among lower vertebrates and insects. Recently, significant progress has been made in understanding the molecular mechanisms involved in the posttranscriptional/posttranslational regulation of SR-BI in a tissue specific manner. The purpose of this review is to summarize the current body of knowledge about the events and molecules connected with the posttranscriptional/posttranslational regulation of SR-BI and to update the molecular and functional characteristics of the insect SR-BI orthologs.
Keywords: HDL receptor, selective uptake, steroid hormones
Introduction
Scavenger receptor class B type I (SR-BI), is a physiologically relevant HDL receptor that mediates selective uptake of lipoprotein (HDL)-derived cholesteryl ester (CE) in vitro and in vivo [1-3]. The SR-BI-mediated CE uptake/selective pathway is different from that of the LDL receptor (LDLR)/endocytic pathway in that it binds CE-rich lipoprotein on the cell surface and selectively delivers the CE from the hydrophobic core of the lipoprotein to the inside of the cells without internalization of the intact lipoprotein particle [1, 4]. Recent studies have expanded the lipids it facilitates across the plasma membrane to include lipid-soluble vitamins such as vitamin E and carotenoids [5, 6]. SR-BI facilitates the bidirectional flux of free cholesterol (FC) and phospholipids between HDL and cells, thus influencing plasma membrane cholesterol content [7, 8]. It has also been implicated in the entry into cells of the hepatitis C virus (HCV) [9, 10], phagocytosis of apoptotic cells [11], protection against female infertility [12], modulation of platelet reactivity [13, 14], and plays a regulatory role in HDL-induced signaling in the vasculature [15, 16]. Most recently, it has been shown that SR-BI expression in bone marrow-derived cells is protective against diet-induced atherosclerosis and myocardial infarction [17]. SR-BI is evolutionally conserved, but exhibits diverse molecular properties among the various species in which it is expressed. In this review, we aim to summarize the current knowledge about the events and molecules connected to the regulation of SR-BI and to update the molecular and functional characteristics of the insect SR-BI orthologs.
SR-BI/CD36 family: distribution across different species and different tissues
SR-BI is a member of the class B scavenger receptor family, which in mammals includes the cluster determinant 36 (CD36) family, lysosomal integral membrane protein II (LIMPII, a lysosomal protein) and SR-BII (an isoform of SR-BI with an alternate C-terminal cytoplasmic tail). Structurally, all these proteins contain N- and C-terminal cytoplasmic domains, two-transmembrane domains, as well as a large extracellular domain containing 5-6 cysteine residues and multiple sites for N-linked glycosylation [1, 18]. In murine SR-BI, 11 N-linked glycosylation sites have been shown to be glycosylated, of which two glycosylation sites (Asn-108 and Asn-173) are required for proper expression and function of SR-BI [18]. Both SR-BI and CD36 are physiologically regulated by palmitoylation [19-22].
As seen with selective CE uptake [23-26], prominent expression of SR-BI has been observed in the liver and steroidogenic cells of the adrenal and gonads [27-31]. In the liver, SR-BI is mainly expressed in parenchymal cells (hepatocytes), which account for ≥90% of liver mass. Thus, on an organ basis, the liver expresses the highest amount of SR-BI. Furthermore, hepatocytes express twice as much SR-BI as that of Kupffer cells [32]. Ovarian steroidogenic cells, such as luteinized ovarian granulosa cells [4], luteal cells [29], and theca cells [33], express high levels of SR-BI. High SR-BI expression is also detected in fetal and adult adrenals, adrenocortical tumor cells, adrenal glomerulosa cells and adrenal carcinoma cells [31, 34]. Rat SR-BI expression is induced to high levels in response to treatment of animals with gonadotropin [30]. Exceptionally high levels of SR-BI are also reported in R2C rat testicular Leydig tumor cells [35]. In contrast to rodents, high levels of SR-BI/CLA-1 (the human homolog) are detected in human placental trophoblasts and in trophoblast-like human choriocarcinoma cell lines [36, 37]. Interestingly, SR-BI is also expressed in testicular Sertoli cells, which are not generally classified as steroid producing cells [38]. Testicular germ cells, on the other hand, preferentially express SR-BII, an isoform of SR-BI [39]. Brain astrocytes, which synthesize neurosteroids, and microglia express SR-BI [40]. Finally, the expression of SR-BI, albeit at varying levels, is also reported in many non-steroidogenic cells, including adipocytes [1, 41], macrophages [42, 43], endothelial cells [44, 45], smooth muscle cells [46], intestinal cells [47, 48], retinal cells [49] and in skin keratinocytes [50].
SR-BI has also been identified in the livers of many non-mammalian species (turtle, goldfish, shark, chicken, frog, and skate), suggesting it emerged early in vertebrate evolutionary history. The expression of SB-RI in turtles is up-regulated during egg development, correlating with peak cholesterol efflux during developmental stages [51].
Multiple orthologs have been identified for the SR-BI/CD36 family in invertebrates. There are six CD36-like proteins identified in Caenorhabditis elegans; these proteins have been shown to be involved in pathogen recognition and the clearance of apoptotic cells [52, 53]. A genome-wide analysis identified thirteen SR-BI orthologs in the silk worm (Bombyx mori) [54], some of which are responsible for selective uptake of carotenoids into the silk gland [55, 56]. Multiple genes have been identified within the CD36 family in insects as well, eight in hymenopteran and twelve to fourteen in dipterans [57, 58]. Recent studies analyzing the expression pattern of the fourteen genes in Drosophila melanogaster during three different developmental larval stages prior to and during the peak of the insect steroid hormone ecdysone show highly regulated expression patterns of these genes, specifically three of the genes are up-regulated in steroidogenic tissues at the onset of pupariation when steroidogenesis is crucial [57]. These studies show the conserved function of SR-BI across species, revealing its role as an important regulator for cholesterol efflux and steroid hormone production.
In order to understand the mechanisms underlying the structure-function relation of the SR-BI/CD36 gene family, we aligned various orthologs of SR-BI/CD36 genes from different species and analyzed the structural features that are important for the function of SR-BI. As shown in the phylogenetic tree of some of the orthologs of SR-BI from the fruit fly to human (Fig. 1), the SR-BI homologs from mammals to other vertebrates are clustered together; similarly, those of nematodes and silk worms are clustered. The orthologs from the fruit fly, however, seem to have diverged into different groups. The 14 fruit fly SR-BI orthologs are clustered into four groups: 1) CG2736 seems to have diverged from most of the other SR-BI orthologs; 2) CG10345 and CG40006 of the fruit fly are more closely related to silkworm SR-BI homologs (SR-B11, SR-B12, SR-B13 and SR-B15); 3) CG3829, CG1887 and emp are more closely related to SR-BI homologs of nematodes (SCAV-4, SCAV-5 and SCAV-6); 4) crq, CG31741, CG7227, ninaD, peste, santa_maria, snmp1 and snmp2 are more closely related to those SR-BI from mammals and other vertebrates.
Figure 1.
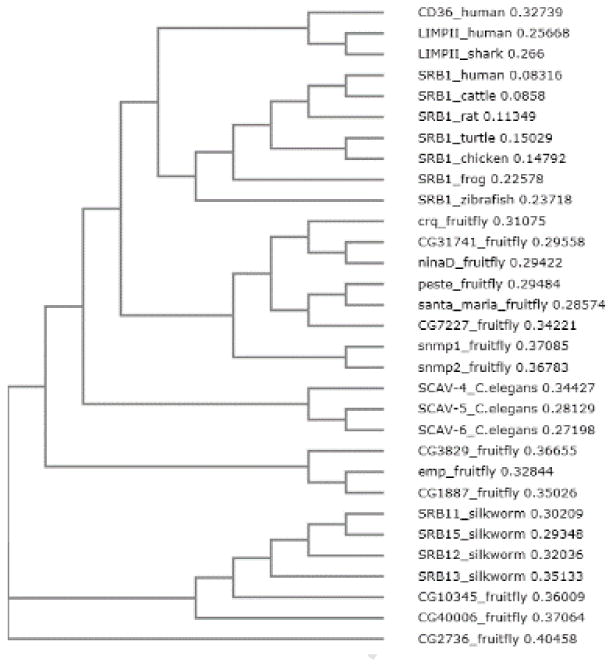
Phylogenetic tree of representative orthologs of SR-BI from different species. Amino acid sequences from the various SR-BI orthologs were analyzed using the multiple sequence alignment program Clustal Omega from EMBL-EBI (http://www.ebi.ac.uk/Tools/msa/clustalo).
Some of the structural features of SR-BI have been demonstrated to be important for its function. The N-terminal transmembrane glycine dimerization motif (G15_G18_G25) has been defined to be required for normal receptor oligomerization and lipid transport [59]. Recent studies of the crystal structure of LIMP-2 shed light on the structure of this group of proteins, and showed that the main ectodomain of the protein contains an antiparallel β-barrel core with many short α-helical segments [60]. Two disulfide bridges stabilize the fold. The disulfide bridge pattern for LIMP-2 (C274-C329 and C312-C318) is similar to that predicted for SR-BI (C321-C323, C274-C329) and that of CD36 (C313-C322, C272-C333), and is consistent with experimental data [61-64]. There are nine N-linked glycosylation sites that have been confirmed with well-defined electron density. When the sites Asn-68 and Asn-325 from LIMP-2 were mutated, the protein failed to be targeted to lysosomes and was retained in the endoplasmic reticulum. With regard to N-linked glycosylation, human SR-BI has 11 putative sites, and a mutational study of each of them shows that the protein failed to locate to the plasma membrane and has a marked reduction in the ability to transfer lipid from HDL to cells when Asn-108 or Asn-173 was mutated [18]. Most of the proteins in this family have lipid transport activity, and the crystal structure of the LIMP-2 showed that 8 amino acids work coordinately to form a tunnel that forms an interconnected cavity through the entire length of the ectodomain to facilitate lipid transfer. These 8 acidic and basic amino acids form a network of hydrogen and ionic bonds and contribute to the lining of the cavity, which is predominately hydrophobic to accommodate lipid moieties.
We have analyzed the above-mentioned structural features in the different SR-BI orthologs, and summarized them in Table 1. The alignment of the sequences highlights some of the structural features (Figure 2). As shown, the orthologs that are more closely related to the SR-BI of nematodes all have extended N-terminal and C-terminal extensions and stretches of internal insertions within their sequences (CG4006_fruitfly, CG3829_fruitfly, emp_fruitfly, CG1887_fruitfly, SRB11_silkworm, SRB13_silkworm, SRB15_silkworm, SCAV-4_c.elegans, SCAV-5_c.elegans, SCAV-6_c.elegans). Most of the proteins have at least one of the two disulfide bridges that stabilize the fold; specifically, the C274-C329 bridge of LIMP-2 is present in all the proteins except the fruit fly protein CG31741. The N-terminal transmembrane glycine dimerization motif is conserved in SR-BI from human to frog, and also in the fruit fly group that is more closely related to mammals and other vertebrates (crq_fruitfly, ninaD_fruitfly, santa maria_fruitfly).
Table 1.
Structural features of SR-BI orthologs from different species. % similarity was calculated using the T-coffee program from EMBL-EBI (http://tcoffee.org.cat/apps/tcoffee).
| Accessio n # |
Over all simil arity |
Seque nce Extensi ons |
N- termin al length |
TM len gth |
EC M len gth |
ECM simila rity |
TM len gth |
C- termin al length |
Glycine dimerization motif |
N- Glycosyla tion |
S-S bonds |
Tunnel cavity (E93, R95, K97, K115,D252, D254, K381, E413) |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G15 G18 G25 | NXS(T) | 2 pairs | |||||||||||
| CD-36-human | NP_00006 3.2 | 76% | 9 | 21 | 409 | 80% | 20 | 12 | G__G___G | 4 | 2 | 2/8 ()()()()D()() E | |
| LIMP-II_human | NP_00549 7.1 | 76% | 7 | 22 | 395 | 82% | 18 | 26 | 0__G___0 | 10 | 2 | 8/8 ERKKDDKE | |
| LIMPII shark | AFK11285. 1 | 81% | 10 | 22 | 397 | 76% | 21 | 25 | G__G___0 | 7 | 2 | 4/8 ER()()D()()E | |
| SR-BI Human | NP_00549 6.4 | 78% | 11 | 21 | 395 | 100% | 21 | 48 | G__G___G | 9 | 2 | 4/8ERK()()()()E | |
| SR-B1 cattle | NP_77702 2.1 | 81% | 11 | 21 | 395 | 80% | 21 | 48 | G__G___G | 9 | 2 | 4/8 ERK()()()()E | |
| SR-BI rat | NP_11372 9.1 | 78% | 11 | 21 | 386 | 82% | 21 | 48 | G__G___G | 10 | 2 | 3/8 ERK()()()() | |
| SR-B1 turtle | XP_00529 9633.1 | 78% | 10 | 21 | 387 | 80% | 21 | 47 | G__G___G | 8 | 2 | 4/8 ERK()()()()E | |
| SR-B1 Chicken | XP_41510 6.2 | 69% | 9 | 21 | 406 | 82% | 21 | 44 | G__G___G | 9 | 2 | 3/8 E()K()()()()E | |
| SRB1 frog | XP_00293 5333.1 | 74% | 5 | 21 | 406 | 80% | 21 | 39 | G__G___G | 8 | 2 | 3/8 E()K()()()()E | |
| SR-B1 Zebra-fish | NP_94460 3.1 | 78% | 8 | 0 | 358 | 82% | 21 | 57 | 0__0___0 | 6 | 2 | 3/8 E()K()()()()E | |
| SCAV-4_C_elegan s | NP_51015 7.2 | 74% | internal | 7 | 21 | 495 | 80% | 21 | 15 | 0__0___G | 2 | *1( no 321-323) | 4/8 E()KKD()()() |
| SCAV-5_C_elegan s | NP_50965 1.2 | 74% | internal | 4 | 21 | 488 | 80% | 21 | 16 | 0__0___G | 2 | *1( no 321-323) | 4/8E()KK()()K() |
| SCAV-6 C_elegans | NP_49208 1.1 | 74% | internal | 7 | 21 | 492 | 80% | 21 | 41 | 0__0___G | 2 | 2 | 3/8E()KK()()()() |
| SRB11_silk worm | NP_00126 6430.1 | 73% | N-terminal | 20 | 21 | 409 | 71% | 21 | 31 | 0__G___0 | 2 | *1( no 321-323) | 3/8 ER()K()()()() |
| SRB12_silk worm | NP_00126 6422.1 | 73% | 9 | 21 | 401 | 74% | 21 | 33 | 0__G___0 | 1 | 1( no 321-323) | 3/8 ER()K()()()() | |
| SRB13_silk worm | XP_00492 5280.1 | 73% | N,C-terminal | 33 | 21 | 371 | 74% | 21 | 77 | 0__G___0 | 3 | 2 | 3/8 ER()K()()()() |
| SRB15_silk worm | BAM6701 7.1 | 71% | N-terminal | 18 | 21 | 404 | 75% | 22 | 36 | 0__G___0 | 2 | *1( no 321-323) | 2/8 ER()()()()()() |
| crq_fruitfly | NP_78795 7.1 | 77% | 12 | 21 | 417 | 73% | 21 | 20 | G__G___G | 6 | 2 | 3/8 E()K()D()()() | |
| CG31741_fr uitfly | NP_72408 8.2 | 76% | 12 | 21 | 415 | 77% | 21 | 22 | 0__G___G | 3 | *1(no 274-329) | 2/8 ()()KK()()()() | |
| niniaD_fruitfl y | AAO11676 .1 | 76% | 12 | 21 | 428 | 77% | 21 | 31 | G__G___G | 4 | 2 | 5/8 ERK()DK() | |
| peste_fruitfly | NP_00113 7808.1 | 75% | C-terminal | 10 | 21 | 372 | 78% | 24 | 73 | 0__G___G | 4 | 2 | 4/8 E()KK()()()E |
| santa-maria_fruitfly | NP_72327 7.3 | 73% | C-terminal | 16 | 21 | 368 | 76% | 21 | 83 | G__G___G | 6 | 2 | 3/8 ERK()()()()() |
| CG7227_frui tfly | NP_60916 9.1 | 75% | C-terminal | 7 | 21 | 408 | 76% | 18 | 47 | 0__G___0 | 5 | 2 | 3/8 ERK()()()()() |
| snmp1_fruitfl y | NP_65095 3.1 | 74% | C-terminal | 7 | 19 | 413 | 77% | 21 | 73 | 0__0___0 | 2 | 2 | 4/8 E()K()D()()E |
| snmp2_fruitfl y | ABW7012 9.1 | 74% | 6 | 21 | 492 | 75% | 21 | 16 | 0__G___G | 3 | 2 | 3/8 ()()()K()D()()E | |
| CG10345_fr uitfly | NP_65056 3.2 | 75% | C-terminal | 7 | 21 | 443 | 76% | 21 | 19 | 0__G___G | 4 | 2 | 2/8 ()()()K()D()()() |
| CG2736_frui tfly | NP_61199 1.1 | 74% | 7 | 21 | 411 | 68% | 21 | 41 | 0__0___0 | 2 | 1( no 321-323) | 2/8 ()()()()D()() E | |
| CG40006_fr uitfly | NP_00103 6401.1 | 65% | N, C-terminal | 91 | 21 | 337 | 76% | 22 | 143 | 0__G___0 | 3 | 2 | 3/8 E()()()D()()E |
| CG3829_frui tfly | NP_61199 2.3 | 76% | N, C-terminal | 30 | 76 | 405 | 74% | 21 | 52 | 0__G___0 | 4 | 2 | 4/8 E()KK()()()E |
| emp_fruitfly | NP_72650 4.2 | 75% | N,C-terminal | 20 | 20 | 410 | 78% | 21 | 63 | 0__0___G | 4 | 2 | 5/8 E()KKD()()E |
| CG1887_frui tfly | NP_00109 7477.1 | 67% | N,C-terminal | 72 | 22 | 381 | 78% | 21 | 83 | 0__G___G | 5 | 1( no 321-323) | 4/8 E()KK()()()E |
Figure 2.
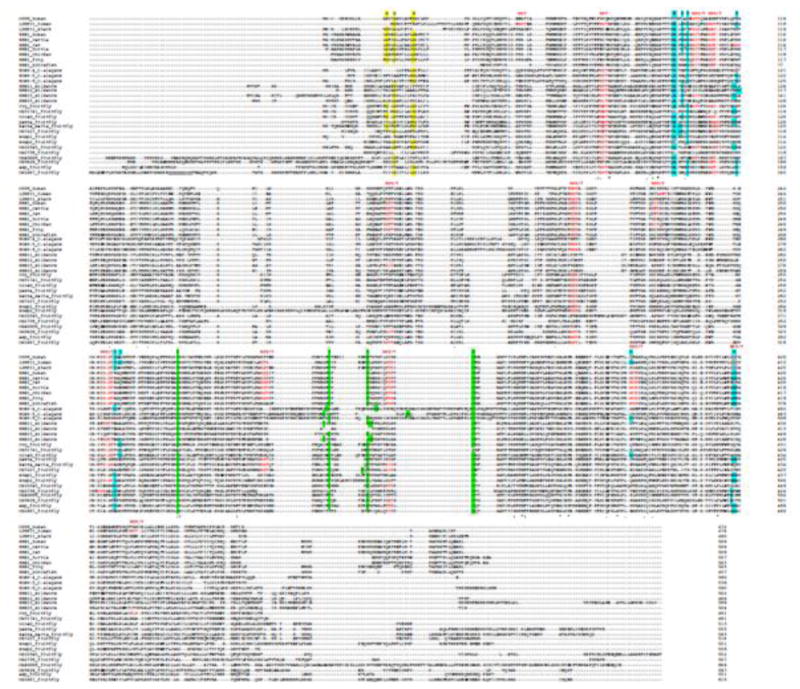
Sequence alignment of SR-BI orthologs from different species. Amino acid sequences from the various SR-BI orthologs were analyzed using the multiple sequence alignment program Clustal Omega from EMBL-EBI (http://www.ebi.ac.uk/Tools/msa/clustalo). Colored amino acids represent the structural features discussed in the text. G_G_G is the glycine dimerization motif, NXS(T) is the N-glycosylation site, C-C is the S-S bond, ERKKDDKE is the tunnel cavity.
Regulation of the expression of CD36/SR-BI gene family
As noted above, human and rodent SR-BI is expressed most abundantly in the liver parenchymal cells and steroidogenic cells of the adrenal gland and gonads, where the selective pathway exhibits its highest activity [27-31]. In steroidogenic cells, SR-BI is primarily localized on the cell surface, and its expression is regulated by trophic hormones (adrenocorticotropic hormone [ACTH] or gonadotropins luteinizing hormone [LH] or follicle-stimulating hormone [FSH]) in concert with the regulation of steroid hormone production [28, 65]. SR-BI expression in both lower vertebrates (i.e., turtle) and fruit fly correlates with their developmental stage when peak cholesterol flux is required. On the other hand, many dietary, hormonal and pharmacological agents can regulate hepatic SR-BI expression [66]. Dietary polyunsaturated fatty acids up-regulate, whereas myristic acid decreases, liver SR-BI expression [67, 68]. Fibrates down-regulate SR-BI levels in the liver, while they upregulate SR-BI levels in macrophages and have no effect on SR-BI levels in the adrenal gland [69]. Therefore, SR-BI is regulated in a cell type specific fashion and may have different modes of selective uptake depending on the cell type.
Recently, in the search for the cellular and molecular mechanisms involved in the regulation of SR-BI expression and function, particularly posttranscriptional/posttranslational regulation, we have shown that two microRNAs, miRNA-125a and miRNA-455, can bind to specific sites in the 3′ UTR of SR-BI mRNA and regulate the expression of SR-BI [70]. The expression of miRNA-125a and miRNA-455 is detected in adrenal, ovarian granulosa, and model Leydig cell lines, and their expression is down-regulated by trophic hormones or the second messenger, cAMP. When either miRNA-125a or miRNA-455 is overexpressed or inhibited, SR-BI-mediated selective HDL uptake and SR-BI-supported steroid hormone synthesis is inhibited or stimulated, respectively. Therefore, our findings suggest that miRNA-125a and miRNA-455 act as SR-BI attenuators to negatively regulate SR-BI-mediated selective delivery of lipoprotein cholesterol in steroidogenic cells and, consequently, inhibition of SR-BI-supported steroidogenesis. Furthermore, in a recent report, Wang et al provided evidence that the 3′-UTR of human SR-BI contains binding sites for miR-185, miR-96 and miR-223, and these three miRNAs were shown to inhibit selective HDL-CE transport into HepG2 cells via inhibition of SR-BI protein expression [71].
Mechanism of Actions
SR-BI facilitates selective HDL cholesteryl ester uptake in two separate independent steps: lipid-rich lipoprotein binding to the extracellular domain of SR-BI and the selective transfer of lipid to the plasma membrane [72, 73]. Studies with CD36/SR-BI chimeras and SR-BI mutants show that high affinity lipid rich-lipoprotein binding to SR-BI is important yet not sufficient for efficient lipid transfer [74]. Meanwhile, efficient lipid uptake via SR-BI or SR-BI/CD36 chimeric receptors is dependent on the extracellular domain of SR-BI [75]. In addition, compounds that can increase CE-rich lipoprotein binding to SR-BI can actually block lipid transfer [76]. Furthermore, protein-protein interaction involving SR-BI with many of its CE donors could be the predominant feature that drives productive SR-BI-lipoprotein complex formation. It has been shown that many of the CE donors (HDL, apoA-I/phospholipid bilayer disks, and lipid-free apoA-I) for SR-BI all share class A amphipathic helices that could be the structural feature to which SR-BI is binding [77, 78]. In addition, SR-BI exists as a multimeric complex with itself or other membrane proteins on the cell surface to facilitate lipid transfer, and the extracellular domain of SR-BI is essential for efficient CE transfer.
Many studies have also observed the formation of specialized cell surface structures, termed ‘microvillar channels’, whose expression is induced by SR-BI, that facilitate selective lipid transfer to the cell interior [79-82]. Immunolocalization studies at the electron microscopic level in rat ovarian luteal, testicular Leydig, and adrenocortical cells have demonstrated that SR-BI is preferentially localized on the microvillar membrane domains that form channels in which various lipoproteins, including HDL, get trapped [80, 81]. In addition, SR-BI was also shown to affect the flux of free cholesterol and the properties of the plasma membrane, and to facilitate the formation of specific lipid rafts that are necessary for the formation of the microvillar channels [82]. It is in these microvillar channels that HDL particles are trapped, apparently to boost the efficiency of the selective HDL-CE transport process.
Recently, we have also shown that the physical state of the SR-BI protein (i.e., monomeric versus dimeric and high-order oligomeric forms) and SR-BI-dependent architectural changes on the cell surface also play significant roles in SR-BI-mediated selective HDL-CE uptake [83]. Using a sulfhydryl-reactive reagent, such as cell-impermeant dithiothreitol (DTT), we showed that disulfide bond(s) exist in SR-BI and contribute to the oligomerization of SR-BI as well as to selective HDL-CE uptake. In addition, the reduction of higher complexes by DTT suggested that ectodomain (or ECD) dimer/oligomer formation may be mediated, at least in part, by disulfide bond formation via the six conserved cysteine residues (C251, C280, C321, C323, C334, and C384) found on the ECD of SR-BI.
Tissue Specific Mechanism of Actions
As summarized above, the regulation of steroidogenic SR-BI expression primarily occurs at the gene transcription level by trophic hormones. In contrast, the functional expression of hepatic SR-BI is primarily regulated at the post-transcriptional level. Many recent studies have shown that SR-BI interacts with other proteins, which, in turn influence the SR-BI-mediated HDL-CE transport into cells [84-89]. In the liver, the four PDZ (PSD-95/Discs-large/ZO-1) containing adaptor protein, PDZK1 (also called NHERF3), interacts with the C-terminal end of SR-BI (mouse sequence: EAKL), and is responsible for the post-transcriptional control of the expression, stability, localization, and function of SR-BI [89-94]. PDZK1 does not bind to the C-terminus of SR-BII, a minor splice variant of SR-BI (mouse sequence: SAMA) [87, 95]. Inactivation of the PDZK1 gene in mice (PDZK1-/-) leads to almost complete (∼95%) suppression of SR-BI protein expression in hepatocytes, ∼50% reduction of SR-BI protein expression in the small intestine, but no significant reduction in SR-BI protein levels in macrophages, steroidogenic tissues, adrenal, ovary and testis and endothelial cells [91, 96, 97]. In addition, overexpression of small PDZK1-associated protein (SPAP) downregulated PDZK1 in a liver-specific fashion, causing the subsequent down-regulation of hepatic SR-BI, but again having no effect on the levels of SR-BI in the adrenal gland or peritoneal macrophages [98]. The significance of these functional interactions has been shown in PDZK1-/- mice, SPAP transgenic mice and mice treated with fibrates [69, 91, 98], where plasma HDL levels are markedly increased due to a deficiency of hepatic SR-BI. PDZK1-/- mice differed from SR-BI-/- mice in that the ratio of unesterified to total plasma cholesterol was normal, females were fertile and CE stores in steroidogenic organs were unaffected in PDZK1-/- mice in contrast to the abnormalities observed in SR-BI-/- mice [91, 99] . These differences are presumably due to normal expression of SR-BI in extrahepatic tissues.
In other recent studies to search for factors involved in regulating SR-BI expression and function in steroidogenic tissue, we examined the expression pattern of PDZ domain containing proteins related to PDZK1 [100]. In fact, PDZK1, also known as Na+/H+ exchanger regulator factor-3 (NHERF3), belongs to a family of scaffolding proteins that also includes NHERF1 (EBP50), NHERF2 (E3KARP), and NHERF4 (IKEPP) [84, 101, 102]. All of these family members possess tandem PDZ domains; NHERF1 and NHERF2 have two and PDZK1/NHERF3 and NHERF4 have four tandem PDZ domains [84, 102]. In addition to PDZ domains, NHERF1 and NHERF2 possess C-terminal MERM (merlin-ezrin-radixin-moesin) binding domains, which indirectly tether these proteins to the actin cytoskeleton [103]. We have shown that NHERF1 and NHERF2 mRNA and protein are expressed at varying levels in model steroidogenic cell lines and the adrenal, with only low expression of PDZK1 (NHERF3) and NHERF4. Dibutyryl cyclic AMP decreased NHERF1 and NHERF2 and increased SR-BI mRNA expression in primary rat granulosa cells and MLTC-1 cells, whereas ACTH had no effect on NHERF1 and NHERF2 mRNA levels but decreased their protein levels in rat adrenals [100]. Co-immunoprecipitation, colocalization, bimolecular fluorescence complementation, and mutational analysis indicated that SR-BI associates with NHERF1 and NHERF2. NHERF1 and NHERF2 down-regulated SR-BI protein expression by inhibiting its de novo protein synthesis. NHERF1 and NHERF2 also inhibited SR-BI-mediated selective CE transport and steroidogenesis, which were markedly attenuated by partial deletions of the PDZ1 or PDZ2 domain of NHERF1, the PDZ2 domain of NHERF2, or the MERM domains of NHERF1/2 or by gene silencing of NHERF1/2. Moreover, an intact COOH-terminal PDZ recognition motif (EAKL) in SR-BI is needed. Transient transfection of hepatic cell lines with NHERF1 or NHERF2 caused a significant reduction in endogenous protein levels of SR-BI. Additional studies demonstrated that another member of the NHERF family, NHERF4, had no effect on either SR-BI protein levels or its selective transport function when CHO cells were transiently co-transfected with SR-BI plus NHERF4 as compared with SR-BI alone. In contrast, co-transfection of CHO cells with SR-BI plus PDZK1/NHERF3 up-regulated the expression of SR-BI protein as compared with SR-BI protein levels seen in cells transfected with the SR-BI construct alone. These results are in agreement with the earlier reports showing that PDZK1/NHERF3 is essential for normal expression of SR-BI in mouse hepatocytes. Our results further demonstrate that the presence of PDZK1/NHERF3 can stimulate SR-BI protein expression even in non-hepatic cells expressing SR-BI [100]. Collectively, these data establish NHERF1 and NHERF2 as SR-BI protein binding partners that play a negative role in the regulation of SR-BI expression, selective CE transport, and steroidogenesis.
SR-BI and Clinical Relevance
In addition to mediating selective cholesteryl ester transport from HDL to cells [1, 2, 34], bi-directional flux of unesterified cholesterol between HDL and cells [104, 105], phospholipid uptake by cells [106], and hepatic reverse cholesterol transport [107], it is becoming increasingly clear that SR-BI performs many other functions including its involvement in atherosclerosis [108, 109], apoptosis [110-113], platelet function [109, 114], immune responses [115], hepatitis C virus (HCV) [116-118] and dengue virus [119] entry, malaria parasite infection [120], and human HDL homeostasis [121, 122]. Evidence for the atheroprotective role of SR-BI is shown in genetically modified mouse models [108]. Both SR-BI transgenic [123, 124] and adenovirus-mediated [125] hepatic overexpression of SR-BI has been shown to markedly suppress atherosclerosis in LDL-receptor deficient mice that were chronically fed a high-fat diet. Conversely, complete deficiency of the SR-BI gene in chow-fed apoE-deficient (SR-BI/apoE DKO) mice [126] or high-fat diet-fed LDL receptor-deficient (SR-BI/LDLR DKO) mice [127] accelerated the development of atherosclerosis. Interestingly, SR-BI/apoE DKO mice exhibited severe dyslipidemia, developed early occlusive atherosclerotic coronary artery disease, spontaneous myocardial infarctions, severe cardiac dysfunction and died prematurely around 6 to 8 weeks of age [128, 129]. Other studies demonstrated that attenuated expression of SR-BI in LDL-receptor KO mice [130] or feeding a high-fat/high cholesterol diet to SR-BI-deficient mice [131, 132] also led to development of atherosclerosis. In addition, evidence is accumulating that SR-BI influences the functions of a number of vascular cells relevant to atherosclerosis, including macrophages, lymphocytes, endothelial cells and endothelial progenitor cells [109].
SR-BI is also implicated in modulating platelet function and mediating platelet-HDL interactions [109, 114]. Imachi et al [133] first provided evidence that human (h) SR-BI/CLA-1 is expressed in platelets and megakaryocytes, the platelet precursors within the bone marrow. Furthermore, it was reported that hSR-BI/CLA-1 abundance was significantly reduced on the surface of platelets from patients with atherosclerotic disease and that such attenuated levels showed an inverse correlation with increased cholesteryl ester content and platelet aggregation. Subsequent work of Dole et al [134] demonstrated that a high plasma UC:TC ratio in SR-BI deficient mice was associated with platelet abnormalities, including cholesterol content, abnormal morphology, high clearance rates, and thrombocytopenia. It was further demonstrated that abnormal circulating lipoproteins containing a high UC:TC ratio in SR-BI KO mice, rather than the SR-BI deficiency itself, variably impacted platelet structure, function and clearance. A similar functional phenotype has been observed in human carriers of an SR-BI genetic variant in which a proline was substituted for a serine at amino acid position 297 (P297S) [121]. Recently, it was reported that increased platelet activation in response to SR-BI deficiency in mice is caused by dyslipidemia and platelet cholesterol overload [13, 135].
While various metabolic functions of SR-BI have been well studied in rodents, the understanding of the role of hSR-BI/CLA-I in human lipid metabolism, particularly HDL metabolism and atherosclerotic disease, is still in its infancy stage. Human genetic studies of hSR-BI/CLA-I function have primarily focused on polymorphisms in hSR-BI/CLA-I for potential association with HDL-cholesterol and other metabolites [107]. As summarized by Kent and Stylianou, most of the early studies focused primarily on three SNPs located in exon-1, intron-5, and exon-8 [107]. However, these studies reported only a weak association between hSR-BI/CLA-I and HDL-cholesterol levels. A recent, large, genome-wide association study (GWAS) involving over 10000 individuals reported that genetic polymorphism in SCARBI was associated with a small, but significant elevation in plasma HDL-cholesterol [136]. More recently, Vergeer et al [121] identified a single loss-of-function mutation (P297S) in SR-BI that was associated with increased HDL cholesterol levels, reduced cholesterol efflux from macrophages, and reduced adrenal steroid secretion, but had no effect on atherosclerosis. Another study identified two novel missense mutations, S112F (nucleotide C588T) and T175A (nucleotide A776T), in the hSR-BI/CLA-I gene that were also associated with elevated HDL-cholesterol in heterozygous careers [122]. Currently, however, there is no evidence that any of these mutations in anyway impact atherosclerosis in humans.
Concluding Remarks
SR-BI is a member of the class B family of receptors, is evolutionarily conserved but exhibits variable molecular properties among the various species in which it is found. It serves as an HDL receptor that preferentially mediates the selective uptake of HDL-CE in the liver for biliary cholesterol secretion and bile acid formation and also mediates a bulk delivery of cholesterol to the adrenal, ovary and testis for steroidogenesis. In addition to the central role of SR-BI in facilitating selective CE uptake, it is becoming increasingly clear that this receptor protein performs many other important functions; e.g., SR-BI appears to regulate processes involved in cellular cholesterol homeostasis, bi-directional cholesterol flow, membrane lipid expression, female fertility (oocyte maturation), apoptosis, platelet function and, in addition, SR-BI may act as an athero-protective agent. SR-BI, along with CD81, has been implicated in the entry of the hepatitis C virus into liver cells. Recent findings of mammalian SR-BI provide new insights into the posttranscriptional/posttranslational regulation of this receptor, but also raise many questions. Understanding the molecular mechanisms and relative importance of various miRNAs and how they regulate and contribute to the function of SR-BI in a tissue specific manner will be very informative. Furthermore, investigations into NHERF1 and NHERF2 mediated negative regulation of SR-BI in steroidogenic cells, the liver and in other SR-BI expressing cells should clearly establish their critical role in SR-BI-dependent selective HDL-CE transport and potentially other SR-BI linked functions. This information will be especially useful for liver, where, until now, another member of the NHERF family, PDZK1/NHERF3, is considered to be a major regulator of the post-transcriptional/posttranslational control of the expression, stability, localization, and function of SR-BI. Finally, very little information is currently available about the expression, structural organization and function of SR-BI from lower vertebrates as well as insects. Further exploration of their properties may lead to identification of a ‘super SR-BI’ that can be exploited to increase the efficacy of selective HDL-CE function, especially in the liver for the removal (excretion) of excessive cholesterol.
Acknowledgments
We would like to acknowledge Yuan Cortez for her technical assistance.
Funding: This work was supported by the Office of Research and Development, Medical Service, Department of Veterans Affairs (SA and FBK) and NIH Public Health Services Grant, R01HL33881.
Abbreviations
- CD36
cluster determinant
- CE
cholesteryl ester
- CHO
Chinese hamster ovary cells
- CLA-I
the human CD36 and LIMPII A-nalogous-1
- FSH
follicle stimulating hormone
- HCV
Hepatitis C virus
- HDL
high-density lipoprotein
- LH
luteinizing hormone
- LIMPII
lysosomal integral membrane protein II
- NHERF1
Na+/H+ exchange regulatory factor 1
- NHERF2
Na+/H+ exchange regulatory factor 2
- NHERF3
Na+/H+ exchange regulatory factor 3
- PDZ
PSD-95/Discs-large/ZO-1
- SR-BI
scavenger receptor class B type I
Footnotes
Disclosure: The authors have nothing to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Acton S, Rigotti A, Landschultz K, et al. Identification of scavenger receptor sr-bi as a high density lipoprotein receptor. Science. 1996;271:518–20. doi: 10.1126/science.271.5248.518. [DOI] [PubMed] [Google Scholar]
- 2.Azhar S, Leers-Sucheta S, Reaven E. Cholesterol uptake is adrenal and gonadal tissues: The sr-bi and “selective” pathway connection. Front Biosci. 2003;8:s998–s1029. doi: 10.2741/1165. [DOI] [PubMed] [Google Scholar]
- 3.Connelly M, Klein S, Azhar S, et al. Comparison of class b scavenger receptors, cd36, and scavenger receptor bi (sr-bi), shows that both receptors mediate high-density lipoprotein-cholesteryl ester selective uptake bu sr-bi exhibits a unique enhancement of cholesteryl ester uptake. J Biol Chem. 1999;274:41–7. doi: 10.1074/jbc.274.1.41. [DOI] [PubMed] [Google Scholar]
- 4.Azhar S, Nomoto A, Leers-Sucheta S, et al. Simultaneous induction of an hdl receptor protein (sr-bi) and the selective uptake of hdl-cholesteryl esters in a physiologically relevant steroiodgenic cell model. J Lipid Res. 1998;39:1616–28. [PubMed] [Google Scholar]
- 5.During A, Dawson H, Harrison E. Carotenoid transport is decreased and expression of the lipid transporters sr-bi, npc1l1, and abca1 is downregulated in caco-2 cells treated with ezetimibe. J Nutr. 2005;135:2305–12. doi: 10.1093/jn/135.10.2305. [DOI] [PubMed] [Google Scholar]
- 6.Reboul E, Klein A, Bietrix F, et al. Scavenger receptor class b type i (sr-bi) is involved in vitamin e transport across the enterocyte. J Biol Chem. 2006;281:4739–45. doi: 10.1074/jbc.M509042200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rigotti A, Acton S, Krieger M. The class b scavenger receptors sr-bi and cd36 are receptors for anionic phospholipids. J Biol Chem. 1995;270(27):16221–4. doi: 10.1074/jbc.270.27.16221. [DOI] [PubMed] [Google Scholar]
- 8.Saddar S, Carriere V, Lee W, et al. Scavenger receptor class b type i (sr-bi) is a plasma membrane cholesterol sensor. Circ Res. 2013;112(1):140–51. doi: 10.1161/CIRCRESAHA.112.280081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burlone M, Budkowska A. Hepatitis c virus cell entery: Role of lipoproteins and cellular receptors. J Gen Virol. 2009;90:1055–70. doi: 10.1099/vir.0.008300-0. [DOI] [PubMed] [Google Scholar]
- 10.Barth H, Schnober E, Neumann-Haefelin C, et al. Scavenger receptor class b is required for hepatitis c virus uptake and cross-presentation by human dendritic cells. J Virol. 2008;82(7):3466–79. doi: 10.1128/JVI.02478-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Osada Y, Sunatani T, Kim I, et al. Signalling pathway involving gulp, mapk and rac1 for sr-bi-induced phagocytosis of apoptotic cells. J Biochem. 2009;145(3):387–94. doi: 10.1093/jb/mvn176. [DOI] [PubMed] [Google Scholar]
- 12.Miettinen H, Rayburn H, Krieger M. Abnormal lipoprotein metabolism and reversible female infertility in hdl receptor (sr-bi)-deficient mice. J Clin Invest. 2001;108(11):1717–22. doi: 10.1172/JCI13288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma Y, Ashraf M, Podrez E. Scavenger receptor bi modulates platelet reactivity and thrombosis in dyslipidemia. Blood. 2010;116(11):1932–41. doi: 10.1182/blood-2010-02-268508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zimman A, Podrez E. Regulation of platelet function by class b scavenger receptors in hyperlipidemia. Arterioscler Thromb Vasc Biol. 2010;30:2350–6. doi: 10.1161/ATVBAHA.110.207498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al-Jarallah A, Trigatti B. A role for the scavenger receptor, class b type i in high density lipoprotein dependent activation of cellular signaling pathways. Biochim Biophys Acta. 2010;1801:1239–49. doi: 10.1016/j.bbalip.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 16.Saddar SMC, Shaul PW. Signaling by the high-affinity hdl receptor b type i. Arterioscler Thromb Vasc Biol. 2010;30:144–50. doi: 10.1161/ATVBAHA.109.196170. [DOI] [PubMed] [Google Scholar]
- 17.Pei Y, Chen X, Aboutouk D, et al. Sr-bi in bone marrow derived cells protects mice from diet induced coronary artery atherosclerosis and myocardial infarction. PLoS One. 2013;8(8):e72492. doi: 10.1371/journal.pone.0072492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Viñals M, Xu S, Vasile E, et al. Identification of the n-linked glycosylation sites on the high density lipoprotein (hdl) receptor sr-bi and assessment of the effects on hdl binding and selective lipid uptake. J Biol Chem. 2003;278:5325–32. doi: 10.1074/jbc.M211073200. [DOI] [PubMed] [Google Scholar]
- 19.Tao N, Wagner S, Lublin D. Cd36 is palmitoylated on both n- and c-terminal cytoplasmic tails. J Biol Chem. 1996;271:22315–20. doi: 10.1074/jbc.271.37.22315. [DOI] [PubMed] [Google Scholar]
- 20.Babitt J, Trigatti B, Rigotti A, et al. Murine sr-bi, a high density lipoprotein receptor that mediates selective lipid uptake, is n-glycosylated and aftty acylated and colocalizes with plasma membrane caveolae. J Biol Chem. 1997;272:13242–9. doi: 10.1074/jbc.272.20.13242. [DOI] [PubMed] [Google Scholar]
- 21.Thorne R, Ralston K, deBock C, et al. Palmitoylation of cd36/fat regulates the rate of its post-transcriptional processing in the endoplasmic reticulum. Biochim Biophys Acta. 2010;1803:1298–307. doi: 10.1016/j.bbamcr.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 22.Wei X, Song H, Semenkovitch CF. Insulin-regulated protein palmitoylation impactsendothelial cell function. Arterioscler Thromb Vasc Biol. 2014;34:346–54. doi: 10.1161/ATVBAHA.113.302848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glass C, Pittman R, Weinstein D, et al. Dissociation of tissue uptake of cholesterol ester from that of apoprotein a-1 of rat plasma high density lipoprotein: Selective delivery of cholesterol ester to liver, adrenal, and gonad. Proc Natl Acad Sci, USA. 1983;80:5435–9. doi: 10.1073/pnas.80.17.5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Azhar S, Stewart D, Reaven E. Utilization of cholesterol-rich lipoproteins by perfused rat adrenals. J Lipid Res. 1989;30:1799–810. [PubMed] [Google Scholar]
- 25.Khoo J, Pittman R, Rubin E. Selective uptake of hdl cholesteryl esters is active in transgenic mice expressing human apolipoprotein a-i. J Lipid Res. 1995;(36):593–600. [PubMed] [Google Scholar]
- 26.Reaven E, Tsai L, Azhar S. Intracellular events in the selective transport of lipoprotein-derived cholesteryl esters. J Biol Chem. 1996;271:16208–17. doi: 10.1074/jbc.271.27.16208. [DOI] [PubMed] [Google Scholar]
- 27.Landschulz K, Pathak R, Rigotti A, et al. Regulation of scavenger receptor, class b, type i, a high density lipoprotein receptor, in liver and steroidogenic tissues of the rat. J Clin Invest. 1996;98:984–5. doi: 10.1172/JCI118883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rigotti A, Edelmann E, Seifert P, et al. Regulation by adrenocorticotropic hormone of the in vivo expression of scavenger receptor class b type i (sr-bi), a high density lipoprotein receptor, in steroidogenic cells of the murine adrenal gland. J Biol Chem. 1996;271:33545–9. doi: 10.1074/jbc.271.52.33545. [DOI] [PubMed] [Google Scholar]
- 29.Reaven E, Nomoto A, Leers-Sucheta S, et al. Expression and microvillar localization of scavenger receptor, class b, type i (a high density lipoprotein receptor) in luteinized and hormone-desensitized rat ovarian models. Endocrinology. 1998;139:2847–56. doi: 10.1210/endo.139.6.6056. [DOI] [PubMed] [Google Scholar]
- 30.Reaven E, Zhan L, Nomoto A, et al. Expression and microvillar localization of scavenger receptor class b, type i (sr-bi) and selective cholesteryl ester uptake in leydig cells from rat testis. J Lipid Res. 2000;41:343–56. [PubMed] [Google Scholar]
- 31.Azhar S, Nomoto A, Reaven E. Hormonal regulation of adrenal microvillar channel formation. J Lipid Res. 2002;43:861–71. [PubMed] [Google Scholar]
- 32.Fluiter K, van der Westhuijzen D, van Berkel T. In vivo regulation of scavenger receptor bi and the selective uptake of high density lipoprotein cholesteryl esters in rat liver parenchymal and kupffer cells. J Biol Chem. 1998;273:8434–8. doi: 10.1074/jbc.273.14.8434. [DOI] [PubMed] [Google Scholar]
- 33.Wu Q, Sucheta S, Azhar S, et al. Lipoprotein enhancement of ovarian theca-interstitial cell steroiodgensis: Relative contribution of scavenger receptor class b (type i) and adenosine 5′-triphosphate-binding cassette (type a1) transporter in high-density lipoprotein-cholesterol transport and androgen synthesis. Endocrinology. 144:2437–45. doi: 10.1210/en.2002-221110. [DOI] [PubMed] [Google Scholar]
- 34.Azhar S, Reaven E. Scavenger receptor class bi and selective cholesteryl ester uptake: Partners in the regulation of steroidogenesis. Mol Cell Endocrinol. 2002;195:1–26. doi: 10.1016/s0303-7207(02)00222-8. [DOI] [PubMed] [Google Scholar]
- 35.Rao R, Leers-Sucheta S, Bose H, et al. Differential regulation od f steroid hormone biosynthesis in r2c and ma-10 leydig tumor cells: Role of sr-bi-mediated selective choleryl ester transport. Biol Reprod. 2003;68:114–21. doi: 10.1095/biolreprod.102.007518. [DOI] [PubMed] [Google Scholar]
- 36.Wadsack C, Hammer A, Levak-Frank S, et al. Selective cholesteryl ester uptake from high density lipoprotein by human first trimester and term villous trophoblast cells. Placenta. 2003;24:131–43. doi: 10.1053/plac.2002.0912. [DOI] [PubMed] [Google Scholar]
- 37.Wadsack C, Hrzenjak A, Hammer A, et al. Traophoblast –like human chriocarcinoma cells serve as a suitable in vitro model for selective cholesteryl ester uptake from high density lipoproteins. Eur J Biochem. 2003;270:451–62. doi: 10.1046/j.1432-1033.2003.03394.x. [DOI] [PubMed] [Google Scholar]
- 38.Nakagawa A, Nagaosa K, Hirose T, et al. Expression and function of class b scavenger receptor type i on both apical and basolateral sides of the plasma membrane of polarized testicular sertoli cells of the rat. Dev Growth Differ. 2004;46:283–98. doi: 10.1111/j.1440-169x.2004.00746.x. [DOI] [PubMed] [Google Scholar]
- 39.Webb N, de Villiers W, de Beer F, et al. Alternate forms of the scavenger receptor bi (sr-bi) J Lipid Res. 1997;38:1490–5. [PubMed] [Google Scholar]
- 40.Husamann J, Loike J, Anankov R, et al. Scavenger receptors in neurobiology and neuropathology: Their role on microglia and other cells of the nervous system. Glia. 2002;40:195–205. doi: 10.1002/glia.10148. [DOI] [PubMed] [Google Scholar]
- 41.Tondu A, Robichon C, Yvan-Charvet L, et al. Insulin and angiotensin induce the translocation of scavenger receptor class b, type i from intracellular sites to the plasma membrane of adipocytes. J Biol Chem. 2005;280:33536–40. doi: 10.1074/jbc.M502392200. [DOI] [PubMed] [Google Scholar]
- 42.Yu L, Cao G, Repa J, et al. Sterol regulation of scavenger receptor class b type i in macrophages. J Lipid Res. 2004;45:889–99. doi: 10.1194/jlr.M300461-JLR200. [DOI] [PubMed] [Google Scholar]
- 43.Svensson P, Englund M, Snäckestrand M, et al. Regulation and splicing of scavenger receptor class b type i in human macrophages and atherosclerotic plaques. BMC Cardiovasc Disord. 2005;5:25. doi: 10.1186/1471-2261-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu X, Murao K, Imachi H, et al. Regulation of scavenger receptor class bi gene expression by angiotensin ii in vascular endothelial cells. Hypertension. 2007;49:1378–84. doi: 10.1161/HYPERTENSIONAHA.106.082479. [DOI] [PubMed] [Google Scholar]
- 45.Kimura T, Mogi C, Tomura H, et al. Induction of scavenger receptor class b type i is critical for simvastatin enhancement of high-density lipoprotein-induced anti-inflammatory actions in endothelial cells. J Immunol. 2008;181:7332–40. doi: 10.4049/jimmunol.181.10.7332. [DOI] [PubMed] [Google Scholar]
- 46.Yeh Y, Hwang G, Liu I, et al. Identification and expression of scavenger receptor sr-bi in endothelial cells and smooth muscle cells of rat aorta in vitro and in vivo. Atherosclerosis. 2002;161:95–103. doi: 10.1016/s0021-9150(01)00642-6. [DOI] [PubMed] [Google Scholar]
- 47.Cai S, Kirby R, Howles P, et al. Differentiation-dependent expression and localization of the class b type i scavenger receptor in intestine. J Lipid Res. 2001;42:902–9. [PubMed] [Google Scholar]
- 48.Voshol P, Schwarz M, Rigotti A, et al. Down-regulation of intestinal scavenger receptor class b, type i (sr-bi) expression in rodents under conditions of deficient bile delivery to the intestine. Biochem J. 2001;356:317–25. doi: 10.1042/0264-6021:3560317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duncan K, Bailey K, Kane J, et al. Human retinal pigment epithelial cells express scavenger receptors bi and bii. Biochem Biophys Res Commun. 2002;292:1017–22. doi: 10.1006/bbrc.2002.6756. [DOI] [PubMed] [Google Scholar]
- 50.Tsuruoka H, Khovidhunkit W, Brown B, et al. Scavenger receptor class b type i is expressed in cultured keratinocytes and epidermis. Regulation in response to changes in cholesterol homeostasis and barrier requirements. J Biol Chem. 2002;277:2916–22. doi: 10.1074/jbc.M106445200. 277. [DOI] [PubMed] [Google Scholar]
- 51.Duggan A, Marie R, Callard I. Expression of sr-bi (scavenger receptor class b type i) in turtle (chrysemys picta) tissues and other nonmammalian vertebrates. J Exp Zool. 2002;292(5):430–4. doi: 10.1002/jez.10067. [DOI] [PubMed] [Google Scholar]
- 52.Means T, Mylonakis E, Tampakakis E, et al. Evolutionarily conserved recognition and innate immunity to fungal pathogens by the scavenger receptors scarf1 and cd36. J Exp Med. 2009;206(3):637–53. doi: 10.1084/jem.20082109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Means T. Fungal pathogen recognition by scavenger receptors in nematodes and mammals. Virulence. 2010;1(1):37–41. doi: 10.4161/viru.1.1.10228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tanaka H, Ishibashi J, Fujita K, et al. A genome-wide analysis of genes and gene families involved in innate immunity of bombyx mori. Insect Biochem Mol Biol. 2008;38(12):1087–110. doi: 10.1016/j.ibmb.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 55.Sakudoh T, Kuwazaki S, Iizuka T, et al. Cd36 homolog divergence is responsible for the selectivity of carotenoid species migration to the silk gland of the silkworm bombyx mori. J Lipid Res. 2013;54(2):482–95. doi: 10.1194/jlr.M032771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sakudoh T, Iizuka T, Narukawa J, et al. A cd36-related transmembrane protein is coordinated with an intracellular lipid-binding protein in selective carotenoid transport for cocoon coloration. J Biol Chem. 2010;285(10):7739–51. doi: 10.1074/jbc.M109.074435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Herboso L, Talamillo A, Pérez C, et al. Expression of the scavenger receptor class b type i (sr-bi) family in drosophila melanogaster. Int J Dev Biol. 2011;55(6):603–11. doi: 10.1387/ijdb.103254lh. [DOI] [PubMed] [Google Scholar]
- 58.Nichols Z, Vogt R. The snmp/cd36 gene family in diptera, hymenoptera and coleoptera: Drosophila melanogaster, d. Pseudoobscura, anopheles gambiae, aedes aegypti, apis mellifera, and tribolium castaneum. Insect Biochem Mol Biol. 2008;38(4):398–415. doi: 10.1016/j.ibmb.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 59.Gaidukov L, Nager A, Xu S, et al. Glycine dimerization motif in the n-terminal transmembrane domain of the high density lipoprotein receptor sr-bi required for normal receptor oligomerization and lipid transport. J Biol Chem. 2011;286(21):18452–64. doi: 10.1074/jbc.M111.229872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Neculai D, Schwake M, Ravichandran M, et al. Structure of limp-2 provides functional insights with implications for sr-bi and cd36. Nature. 2013;504:172–6. doi: 10.1038/nature12684. [DOI] [PubMed] [Google Scholar]
- 61.Rasmussen J, Berglund L, Rasmussen M, et al. Assignment of disulfide bridges in bovine cd36. Eur J Biochem. 1998;257:488–94. doi: 10.1046/j.1432-1327.1998.2570488.x. [DOI] [PubMed] [Google Scholar]
- 62.Miao Y, Romera K, Nieland T, et al. Exoplasmic cysteine cys384 of the hdl receptor sr-bi is critical for its sensitivity to a small-molecule inhibitor and normal lipid transport activity. Proc Natl Acad Sci U S A. 2011;108:12243–8. doi: 10.1073/pnas.1109078108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Papale G, Hanson P, Sahoo D. Extracellular disulfide bonds support scavenger receptor class b type i-mediated cholesterol transport. Biochemistry. 2011;50(28):6245–54. doi: 10.1021/bi2005625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu M, Lau T, Carr S, et al. Contributions of a disulfide bond and a reduced cysteine side chain to the intrinsic activity of the high-density lipoprotein receptor sr-bi. Biochemistry. 2012;51(50):10044–55. doi: 10.1021/bi301203x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cao G, Zhao L, Stangl H, et al. Developmental and hormonal regulation of murine scavenger receptor, class b, type 1. Mol Endocrinol. 1999;13(9):1460–73. doi: 10.1210/mend.13.9.0346. [DOI] [PubMed] [Google Scholar]
- 66.Rigotti A, Miettinen H, Krieger M. The role of the high density lipoprotein receptor sr-bi in the lipid metabolism of endocrine and other tissues. Endocr Rev. 2003;(24):357–87. doi: 10.1210/er.2001-0037. [DOI] [PubMed] [Google Scholar]
- 67.Spady D, Kearney D, Hobbs H. Polyunsaturated fatty acids up-regulate hepatic scavenger receptor b1 (sr-bi) expression and hdl cholesteryl ester uptake in the hamster. JLR. 1999;40:1384–94. [PubMed] [Google Scholar]
- 68.Loison C, Mendy F, Sérougne C, et al. Dietary myristic acid modifies the hdl-cholesterol concentration and liver scavenger receptor bi expression in the hamster. Br J Nutr. 2002;87:199–210. doi: 10.1079/BJNBJN2002521. [DOI] [PubMed] [Google Scholar]
- 69.Mardones P, Pilon A, Bouly M, et al. Fibrates down-regulate hepatic scavenger receptor class b type i protein expression in mice. J Biol Chem. 2003;278(10):7884–90. doi: 10.1074/jbc.M211627200. [DOI] [PubMed] [Google Scholar]
- 70.Hu Z, Shen W, Kraemer F, et al. Micrornas 125a and 455 repress lipoprotein-supported steroidogenesis by targeting scavenger receptor class b type i in steroidogenic cells. Mol Cell Biol. 2012;32(24):5035–45. doi: 10.1128/MCB.01002-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang L, Jia XJ, Jiang HJ, et al. Micrornas 185, 96, and 223 repress selective high-density lipoprotein cholesterol uptake through posttranscriptional inhibition. Mol Cell Biol. 2013;33:1956–64. doi: 10.1128/MCB.01580-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rigotti A, Miettinen H, Krieger M. The role of the high-density lipoprotein receptor sr-bi in the lipid metabolism of endocrine and other tissues. Endocr Rev. 2003;24(3):357–87. doi: 10.1210/er.2001-0037. [DOI] [PubMed] [Google Scholar]
- 73.Gu X, Trigatti B, Xu S, et al. The efficient cellular uptake of high density lipoprotein lipids via scavenger receptor class b type i requires not only receptor-mediated surface binding but also receptor-specific lipid transfer mediated by its extracellular domain. J Biol Chem. 1998;273(41):26338–48. doi: 10.1074/jbc.273.41.26338. [DOI] [PubMed] [Google Scholar]
- 74.Sun B, Boyanovsky B, Connelly M, et al. Distinct mechanisms for oxldl uptake and cellular trafficking by class b scavenger receptors cd36 and sr-bi. J Lipid Res. 2007;48(12):2560–70. doi: 10.1194/jlr.M700163-JLR200. [DOI] [PubMed] [Google Scholar]
- 75.Connelly M, Klein S, Azhar S, et al. Comparison of class b scavenger receptors, cd36 and scavenger receptor bi (sr-bi), shows that both receptors mediate high density lipoprotein-cholesteryl ester selective uptake but sr-bi exhibits a unique enhancement of cholesteryl ester uptake. J Biol Chem. 1999;274(1):41–7. doi: 10.1074/jbc.274.1.41. [DOI] [PubMed] [Google Scholar]
- 76.Yu M, Romer K, Nieland T, et al. Exoplasmic cysteine cys384 of the hdl receptor sr-bi is critical for its sensitivity to a small-molecule inhibitor and normal lipid transport activity. Proc Natl Acad Sci U S A. 2011;108(30):12243–8. doi: 10.1073/pnas.1109078108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bocharov A, Baranova I, Vishnyakova T, et al. Targeting of scavenger receptor class b type i by synthetic amphipathic alpha-helical-containing peptides blocks lipopolysaccharide (lps) uptake and lps-induced pro-inflammatory cytokine responses in thp-1 monocyte cells. J Biol Chem. 2004;279(34):36072–82. doi: 10.1074/jbc.M314264200. [DOI] [PubMed] [Google Scholar]
- 78.Thuahnai S, Lund-Katz S, Anantharamaiah G, et al. A quantitative analysis of apolipoprotein binding to sr-bi: Multiple binding sites for lipid-free and lipid-associated apolipoproteins. J Lipid Res. 2003;44(6):1132–42. doi: 10.1194/jlr.M200429-JLR200. [DOI] [PubMed] [Google Scholar]
- 79.Reaven E, Zhan L, Nomoto A, et al. Expression and microvillar localization of scavenger receptor class b, type i (sr-bi) and selective cholesteryl ester uptake in leydig cells from rat testis. J Lipid Res. 2000;41:343. [PubMed] [Google Scholar]
- 80.Reaven E, Leers-Sucheta S, Nomoto A, et al. Expression of scavenger receptor class b type 1 (sr-bi) promotes microvillar channel formation and selective cholesteryl ester transport in a heterologous reconstituted system. Proc Natl Acad Sci U S A. 2001;98:1613. doi: 10.1073/pnas.98.4.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Williams D, Wong J, Hamilton R. Sr-bi is required for microvillar channel formation and the localization of hdl particles to the surface of adrenocortical cells in vivo. J Lipid Res. 2002;43:544. [PubMed] [Google Scholar]
- 82.Peng Y, Akmentin W, Connelly M, et al. Scavenger receptor bi (sr-bi) clustered on microvillar extensions suggests that this plasma membrane domain is a way station for cholesterol trafficking between cells and high-density lipoprotein. Mol Biol Cell. 2004;15:384–96. doi: 10.1091/mbc.E03-06-0445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hu J, Zhang Z, Shen W, et al. Differential roles of cysteine residues in the cellular trafficking, dimerization, and function of the high-density lipoprotein receptor, sr-bi. Biochemistry. 2011;50(50):10860–75. doi: 10.1021/bi201264y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Donowitz M, Cha B, Zachos N, et al. Nherf family and nhe3 regulation. J Physiol. 2005;567:3–11. doi: 10.1113/jphysiol.2005.090399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Claperon A, Mergey M, Fouassier L. Roles of the scaffolding proteins nherf in liver biology. Clin Res Hepatol Gastroenterol. 2011;35(3):176–81. doi: 10.1016/j.clinre.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 86.Silver DL. Sr-bi and protein-protein interactions in hepatic high density lipoprotein metabolism. Rev Endocr Metab Disord. 2004;5(4):327–33. doi: 10.1023/B:REMD.0000045104.38104.8e. [DOI] [PubMed] [Google Scholar]
- 87.Kocher O, Krieger M. Role of the adaptor protein pdzk1 in controlling the hdl receptor sr-bi. Curr Opin Lipidol. 2009;20(3):236–41. doi: 10.1097/MOL.0b013e32832aee82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cunningham R, Biswas R, Steplock D, et al. Role of nherf and scaffolding proteins in proximal tubule transport. Urol Res. 2010;38(4):257–62. doi: 10.1007/s00240-010-0294-1. [DOI] [PubMed] [Google Scholar]
- 89.Tsukamoto K, Wales T, Daniels K, et al. Noncanonical role of the pdz4 domain of the adaptor protein pdzk1 in the regulation of the hepatic high density lipoprotein receptor scavenger receptor class b, type i (sr-bi) J Biol Chem. 2013;288:19845–60. doi: 10.1074/jbc.M113.460170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ikemoto M, Arai H, Feng D, et al. Identification of a pdz-domain-containing protein that interacts with the scavenger receptor class b type i. Proc Natl Acad Sci USA. 2000;97:6538–43. doi: 10.1073/pnas.100114397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kocher O, Yesilaltay A, Cirovic C, et al. Targeted disruption of the pdzk1 gene in mice causes tissue-specific depletion of the high density lipoprotein receptor scavenger receptor class b type i and altered lipoprotein metabolism. J Biol Chem. 2003;278:52820–5. doi: 10.1074/jbc.M310482200. [DOI] [PubMed] [Google Scholar]
- 92.Fenske S, Yesilaltay A, Pal R, et al. Overexpression of the pdz1 domain of pdzk1 blocks the activity of hepatic scavenger receptor, class b, type i by altering its abundance and cellular localization. J Biol Chem. 2008;283:22097–104. doi: 10.1074/jbc.M800029200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fenske S, Yesilaltay A, Pal R, et al. Normal hepatic cell surface localization of the high density lipoprotein receptor, scavenger receptor class b, type i, depends on all four pdz domains of pdzk1. J Biol Chem. 2009;284:5797–806. doi: 10.1074/jbc.M808211200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kocher O, Birrane G, Yesilaltay A, et al. Identification of the pdz3 domain of the adaptor protein pdzk1 as a second, physiologically functional binding site for the c terminus of high density lipoprotein receptor scavenger receptor class b type i. J Biol Chem. 2011;286:25171–86. doi: 10.1074/jbc.M111.242362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Webb N, de Villiers W, de Beer F, et al. Alternate forms of the scavenger receptor bi (sr-bi) J Lipid Res. 1997;38:1490–5. [PubMed] [Google Scholar]
- 96.Kocher O, Yesilalttay A, Shen CH, et al. Influence of pdzk1 on lipoprotein metabolism and atherosclerosis. Biochim Biophys Acta. 2008;1782:310–6. doi: 10.1016/j.bbadis.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhu W, Saddar S, Seetharam D, et al. The scavenger receptor class b type i adaptor protein pdzk1 maintains endothelial monolayer integrity. Circ Res. 2008;102:480–7. doi: 10.1161/CIRCRESAHA.107.159079. [DOI] [PubMed] [Google Scholar]
- 98.Silver D, Wang N, Vogel S. Identification of small pdzk1-associated protein, dd96/map17, as a regulator of pdzk1 and plasma high density lipoprotein levels. J Bio Chem. 2003;278:28528–32. doi: 10.1074/jbc.M304109200. [DOI] [PubMed] [Google Scholar]
- 99.Rigotti A, Trigatti B, Penman M, et al. A targeted mutation in the murine gene encoding the high density lipoprotein (hdl) receptor scavenger receptor class b type i reveals its key role in hdl metabolism. Proc Natl Acad Sci U S A. 1997;94:12610–5. doi: 10.1073/pnas.94.23.12610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hu Z, Hu J, Zhang Z, et al. Regulation of expression and function of scavenger receptor class b, type i (sr-bi) by na+/h+ exchanger regulatory factors (nherfs) J Biol Chem. 2013;288(16):11416–35. doi: 10.1074/jbc.M112.437368. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Seidler U, Singh A, Cinar A, et al. The role of the nherf family of pdz scaffolding proteins in the regulation of salt and water transport. Ann N Y Acad Sci. 2009;1165:249–60. doi: 10.1111/j.1749-6632.2009.04046.x. [DOI] [PubMed] [Google Scholar]
- 102.Ardura J, Friedman P. Regulation of g protein-coupled receptor function by na+/h+ exchange regulatory factors. Pharmacol Rev. 2011;63(4):882–900. doi: 10.1124/pr.110.004176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bretscher A, Edwards K, Fehon R. Erm proteins and merlin: Integrators at the cell cortex. Nat Rev Mol Cell Biol. 2002;3(8):586–99. doi: 10.1038/nrm882. [DOI] [PubMed] [Google Scholar]
- 104.de La Llera-Moya M, Rothblat G, Conelly M, et al. Scavenger receptor bi (sr-bi) mediates free cholesterol flux independently of hdl tethering to the cell surface. J Lipid Res. 1999;40:575–80. [PubMed] [Google Scholar]
- 105.de La Llera-Moya M, Connelly M, Drazul D, et al. Scavenger receptor class b type i affects cholesterol homeostasis by magnifying cholesterol flux between cells and hdl. J Lipid Res. 2001;42:1969–078. [PubMed] [Google Scholar]
- 106.Engelman B, Weidmann M. Cellular phospholipid uptake: Flexible paths to coregulate the functions of intracellular lipids. Biochim Biophys Acta. 2010;1801:609–16. doi: 10.1016/j.bbalip.2010.02.013. [DOI] [PubMed] [Google Scholar]
- 107.Kent A, Stylianou I. Scavenger receptor class b member i protein: Hepatic regulation and its effects on lipids, reverse cholesterol transport, and atherosclerosis. Hepat Med. 2011;3:29–44. doi: 10.2147/HMER.S7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Trigatti B, Krieger M, Rigotti A. Influence of the hdl receptor sr-bi on lipoprotein metabolism and atherosclerosis. Arterioscler Thromb Vasc Biol. 2003;23:1732–8. doi: 10.1161/01.ATV.0000091363.28501.84. [DOI] [PubMed] [Google Scholar]
- 109.Mineo C, Shaul P. Functions of scavenger receptor class b, type i in atherosclerosis. Curr Opin Lipidol. 2012;23:487–93. doi: 10.1097/MOL.0b013e328357ba61. [DOI] [PubMed] [Google Scholar]
- 110.Murao K, Terpdtra V, Green S, et al. Characterization of cla-1, a human homologue of rodent scavenger receptor bi, as a receptor for high density lipoprotein and apoptotic thymocytes. J Biol Chem. 1997;272:17551–7. doi: 10.1074/jbc.272.28.17551. [DOI] [PubMed] [Google Scholar]
- 111.Shiratsuchi A, Kawasaki Y, Ikemoto M, et al. Role of class b scavenger receptor type i in phagocytosis of apoptotic rat spermatogenic cells by sertoli cells. J Biol Chem. 1999;274:5901–8. doi: 10.1074/jbc.274.9.5901. [DOI] [PubMed] [Google Scholar]
- 112.Fuhrman B, Gantman A, Aviram M. Paraoxonase 1 (pon1) deficiency in mice is associated with esduced expression of macrophage sr-bi and consequently the loss of hdl cytoprotection against apoptosis. Atherosclerosis. 2010;211:61–8. doi: 10.1016/j.atherosclerosis.2010.01.025. [DOI] [PubMed] [Google Scholar]
- 113.Nishiuchi T, Murao K, Imachi H, et al. Scavenger receptor class bi mediates the anti-apoptotic effect of erythropoietin. Ann Med. 2010;42:151–60. doi: 10.3109/07853891003601556. [DOI] [PubMed] [Google Scholar]
- 114.Nofer JR, van Eck M. Hdl scavenger receptor class b type i and platelet function. Curr Opin Lipidol. 2011;22:277–82. doi: 10.1097/MOL.0b013e32834701de. [DOI] [PubMed] [Google Scholar]
- 115.Zheng Z, Ai J, Li XA. Scavenger receptor class b type i and immune dysfunctions. Curr Opin Endocrinol Diabetes Obes. 2014;21(2):121–8. doi: 10.1097/MED.0000000000000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Catanese M, Graziani R, von Hahn T, et al. High-avidity monoclonal antibodies against the human scavenger class b type i receptor efficiently block hepatitis c virus infection in the presence of high-density lipoprotein. J Virol. 2007;81:8063–71. doi: 10.1128/JVI.00193-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Grove J, Huby T, Stamataki Z, et al. Scavenger receptor bi and bii expression levels modulate hepatitis c virus infectivity. J Virol. 2007;81:3162–9. doi: 10.1128/JVI.02356-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Syder A, Lee H, Zeisel M, et al. Small molecule scavenger receptor bi antagonists are potent hcv entry inhibitors. J Hepatol. 2011;54:48–55. doi: 10.1016/j.jhep.2010.06.024. [DOI] [PubMed] [Google Scholar]
- 119.Li Y, Kakinami C, Li Q, et al. Human apolipoprotein a-i is associated with dengue virus and enhances virus infection through sr-bi. PLoS ONE. 2013;8:e70390. doi: 10.1371/journal.pone.0070390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rodrigues CD, Hannus M, Prudencio M, et al. Host scavenger receptor sr-bi plays a dual role in the establishment of malaria parasite liver infection. Cell Host Microbe. 2008;4(3):271–82. doi: 10.1016/j.chom.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 121.Vergeer M, Korporaal S, Franssen R, et al. Genetic variant of the scavenger receptor bi in humans. N Engl J Med. 2011;364:136–45. doi: 10.1056/NEJMoa0907687. [DOI] [PubMed] [Google Scholar]
- 122.Brunham L, Tietjen I, Bochem A, et al. Novel mutations in scavenger receptor bi associated with high hdl cholesterol in humans. Clin Genet. 2011;79:575–81. doi: 10.1111/j.1399-0004.2011.01682.x. [DOI] [PubMed] [Google Scholar]
- 123.Arai T, Wang N, Benzouevski M, et al. Decreased atherosclerosis in heterozygous low density lipoprotein-deficient mice expressing the scavenger receptor bi transgene. J Biol Chem. 1999;274:2366–271. doi: 10.1074/jbc.274.4.2366. [DOI] [PubMed] [Google Scholar]
- 124.Ueda Y, Gong E, Royer L, et al. Relationship between expression levels and atherogenesis in scavenger receptor class b, type i transgenics. J Biol Chem. 2000;275:20368–73. doi: 10.1074/jbc.M000730200. [DOI] [PubMed] [Google Scholar]
- 125.Kozarsky K, Donahee M, Glick J, et al. Gene transfer and hepatic overexpression of the hdl receptor sr-bi reduces atherosclerosis in the cholesterol-fed ldl receptor-deficient mouse. Arterioscler Thromb Vasc Biol. 2000;20:721–7. doi: 10.1161/01.atv.20.3.721. [DOI] [PubMed] [Google Scholar]
- 126.Trigatti B, Rayburn H, Viñals M, et al. Influence of the hdl receptor sr-bi on reproductive and cardiovascular pathophysiology. Proc Natl Acad Sci USA. 1999;96:9322–7. doi: 10.1073/pnas.96.16.9322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Covey SD, Krieger M, Wang W, et al. Scavenger receptor class b type i-mediated protection against atherosclerosis in ldl receptor-negative mice involves its expression in bone marrow-derived cells. Arterioscler Thromb Vasc Biol. 2003;23(9):1589–94. doi: 10.1161/01.ATV.0000083343.19940.A0. [DOI] [PubMed] [Google Scholar]
- 128.Braun A, Trigatti BL, Post MJ, et al. Loss of sr-bi expression leads to the early onset of occlusive atherosclerotic coronary artery disease, spontaneous myocardial infarctions, severe cardiac dysfunction, and premature death in apolipoprotein e-deficient mice. Circ Res. 2002;90(3):270–6. doi: 10.1161/hh0302.104462. [DOI] [PubMed] [Google Scholar]
- 129.Yu H, Zhang W, Yancey P, et al. Macrophage apolipoprotein e reduces atherosclerosis and prevents premature death in apolipoprotein e and scavenger receptor-class bi double-knockout mice. Arterioscler Thromb Vasc Biol. 2006;26(1):150–6. doi: 10.1161/01.ATV.0000194096.89476.73. [DOI] [PubMed] [Google Scholar]
- 130.Huszar D, Varban M, Rinninger F, et al. Increased ldl cholesterol and atherosclerosis in ldl receptor-deficient mice with attenuated expression scavenger receptor bi. Arterioscler Thromb Vasc Biol. 2000;20:1068–73. doi: 10.1161/01.atv.20.4.1068. [DOI] [PubMed] [Google Scholar]
- 131.Van Eck M, Twisk J, Hoekstra M, et al. Differential effects of scavenger receptor bi deficiency on lipid metabolism in cells of the arterial wall and in the liver. J Biol Chem. 2003;278(26):23699–705. doi: 10.1074/jbc.M211233200. [DOI] [PubMed] [Google Scholar]
- 132.Huby T, Doucet C, Dachet C, et al. Knockdown expression and hepatic deficiency reveal an atheroprotective role for sr-bi in liver and peripheral tissues. J Clin Invest. 2006;116:2787–76. doi: 10.1172/JCI26893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Imachi H, Murao K, Cao W, et al. Expression of human scavenger receptor b1 on and in human platelets. Arterioscler Thromb Vasc Biol. 2003;23(5):898–904. doi: 10.1161/01.ATV.0000067429.46333.7B. [DOI] [PubMed] [Google Scholar]
- 134.Dole VS, Matuskova J, Vasile E, et al. Thrombocytopenia and platelet abnormalities in high-density lipoprotein receptor-deficient mice. Arterioscler Thromb Vasc Biol. 2008;28(6):1111–6. doi: 10.1161/ATVBAHA.108.162347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Korporaal S, Meurs I, Hauer A, et al. Deletion of the high-density lipoprotein receptor scavenger receptor bi in mice modulates thrombosis susceptibility and indirectly affects platelet function by elevation of plasma free cholesterol. Arterioscler Thromb Vasc Biol. 2011;31:34–42. doi: 10.1161/ATVBAHA.110.210252. [DOI] [PubMed] [Google Scholar]
- 136.Teslovich TM, Musunuru K, Smith AV, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466(7307):707–13. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]


