Abstract
Background
Glioblastoma is an aggressive form of brain cancer. Approximately five in 100 people with glioblastoma survive for five years past diagnosis. Glioblastomas that have a particular modification to their DNA (called methylation) in a particular region (the O6‐methylguanine–DNA methyltransferase (MGMT) promoter) respond better to treatment with chemotherapy using a drug called temozolomide.
Objectives
To determine which method for assessing MGMT methylation status best predicts overall survival in people diagnosed with glioblastoma who are treated with temozolomide.
Search methods
We searched MEDLINE, Embase, BIOSIS, Web of Science Conference Proceedings Citation Index to December 2018, and examined reference lists. For economic evaluation studies, we additionally searched NHS Economic Evaluation Database (EED) up to December 2014.
Selection criteria
Eligible studies were longitudinal (cohort) studies of adults with diagnosed glioblastoma treated with temozolomide with/without radiotherapy/surgery. Studies had to have related MGMT status in tumour tissue (assessed by one or more method) with overall survival and presented results as hazard ratios or with sufficient information (e.g. Kaplan‐Meier curves) for us to estimate hazard ratios. We focused mainly on studies comparing two or more methods, and listed brief details of articles that examined a single method of measuring MGMT promoter methylation. We also sought economic evaluations conducted alongside trials, modelling studies and cost analysis.
Data collection and analysis
Two review authors independently undertook all steps of the identification and data extraction process for multiple‐method studies. We assessed risk of bias and applicability using our own modified and extended version of the QUality In Prognosis Studies (QUIPS) tool. We compared different techniques, exact promoter regions (5'‐cytosine‐phosphate‐guanine‐3' (CpG) sites) and thresholds for interpretation within studies by examining hazard ratios. We performed meta‐analyses for comparisons of the three most commonly examined methods (immunohistochemistry (IHC), methylation‐specific polymerase chain reaction (MSP) and pyrosequencing (PSQ)), with ratios of hazard ratios (RHR), using an imputed value of the correlation between results based on the same individuals.
Main results
We included 32 independent cohorts involving 3474 people that compared two or more methods. We found evidence that MSP (CpG sites 76 to 80 and 84 to 87) is more prognostic than IHC for MGMT protein at varying thresholds (RHR 1.31, 95% confidence interval (CI) 1.01 to 1.71). We also found evidence that PSQ is more prognostic than IHC for MGMT protein at various thresholds (RHR 1.36, 95% CI 1.01 to 1.84). The data suggest that PSQ (mainly at CpG sites 74 to 78, using various thresholds) is slightly more prognostic than MSP at sites 76 to 80 and 84 to 87 (RHR 1.14, 95% CI 0.87 to 1.48). Many variants of PSQ have been compared, although we did not see any strong and consistent messages from the results. Targeting multiple CpG sites is likely to be more prognostic than targeting just one. In addition, we identified and summarised 190 articles describing a single method for measuring MGMT promoter methylation status.
Authors' conclusions
PSQ and MSP appear more prognostic for overall survival than IHC. Strong evidence is not available to draw conclusions with confidence about the best CpG sites or thresholds for quantitative methods. MSP has been studied mainly for CpG sites 76 to 80 and 84 to 87 and PSQ at CpG sites ranging from 72 to 95. A threshold of 9% for CpG sites 74 to 78 performed better than higher thresholds of 28% or 29% in two of three good‐quality studies making such comparisons.
Keywords: Adult; Humans; Antineoplastic Agents, Alkylating; Antineoplastic Agents, Alkylating/therapeutic use; Bias; Brain Neoplasms; Brain Neoplasms/drug therapy; Brain Neoplasms/enzymology; Brain Neoplasms/mortality; Cohort Studies; CpG Islands; CpG Islands/genetics; DNA Methylation; DNA Modification Methylases; DNA Modification Methylases/metabolism; DNA Repair Enzymes; DNA Repair Enzymes/metabolism; Glioblastoma; Glioblastoma/drug therapy; Glioblastoma/enzymology; Glioblastoma/mortality; High-Throughput Nucleotide Sequencing; Immunohistochemistry; Polymerase Chain Reaction; Polymerase Chain Reaction/methods; Predictive Value of Tests; Prognosis; Promoter Regions, Genetic; Promoter Regions, Genetic/genetics; Temozolomide; Temozolomide/therapeutic use; Tumor Suppressor Proteins; Tumor Suppressor Proteins/metabolism
Plain language summary
Which method of determining MGMT promoter methylation best predicts survival in people with glioblastoma treated with temozolomide?
What was the aim of this review?
Glioblastoma is a very aggressive type of brain cancer. People with glioblastoma are usually treated with surgical removal of the tumour followed by radiotherapy, chemotherapy, or both. The standard chemotherapy is a medicine called temozolomide. Some glioblastoma tumours have a particular modification in their DNA (which contains the genetic code of organisms), and knowing whether a person has this modification is useful to predict how long the person may live after their diagnosis with cancer and how they may respond to temozolomide. The modification is known as 'methylation of the MGMT promoter region' and it can also affect MGMT protein expression (the way MGMT is made and modified). There are many ways to work out whether a tumour has this modification. In this review, we attempted to work out which method is best.
What we found
We identified 32 studies comparing different ways to measure whether the MGMT promoter region is methylated. The main three methods were called 'methylation‐specific polymerase chain reaction (PCR),' 'pyrosequencing' (both of which look directly at the MGMT promoter region) and 'immunohistochemistry' (which looks at MGMT protein expression). We found that methylation‐specific PCR and pyrosequencing are better at predicting overall survival than immunohistochemistry. Methylation‐specific PCR and pyrosequencing can be carried out by targeting different parts of the tumour DNA. Pyrosequencing can be performed using different cut‐off thresholds to determine whether a tumour is methylated or unmethylated. We did not identify very clear signals in terms of the best parts of the DNA to target or which are the best cut‐off thresholds.
How reliable are results of the studies in this review?
We rated our confidence in the evidence as 'moderate' for our conclusions about methylation‐specific PCR, but as 'low' for pyrosequencing. Although there were many studies, they all looked at different variants of the methods, so it is difficult to work out exactly which variant is best.
What are the implications of this review?
Our review indicates both methylation‐specific PCR and pyrosequencing provide better predictions of survival than immunohistochemistry. There is some evidence that pyrosequencing may be better than methylation‐specific PCR at predicting overall survival, depending on the DNA targets and cut‐off thresholds used. We documented the most frequent DNA targets used in methylation‐specific PCR and pyrosequencing. We described cut‐off thresholds used in pyrosequencing, although it is unclear which of these is best.
Summary of findings
Summary of findings 1. Methods for measuring MGMT promoter methylation status.
| Methods for measuring MGMT promoter methylation status | ||||
|
Patient or population: people with glioblastoma undergoing treatment with temozolomide Outcome being predicted: overall survival (time to death) | ||||
| Technique/method | Ratio of hazard ratios (95% CI) | No of participants (studies) | Certainty of the evidence (GRADE) | Comments |
| MSP compared with IHC | 1.31 (1.01 to 1.71) | 913 (7 studies) |
⊕⊕⊕⊝ Moderatea | — |
| PSQ compared with IHC | 1.36 (1.01 to 1.84) | 871 (5 studies) |
⊕⊕⊝⊝ Lowb | — |
| PSQ compared with MSP | 1.14 (0.87 to 1.48) | 1119 (9 studies) |
⊕⊕⊝⊝ Lowb | — |
| Variants of PSQ | Not estimated | 876 (11 studies) |
⊕⊝⊝⊝ Very lowc | — |
| qMSP (against MSP or PSQ) | Not estimated | 765 (7 studies) |
⊕⊝⊝⊝ Very lowc | — |
| Bead array (against MSP or PSQ) | Not estimated | 81 (2 studies) |
⊕⊝⊝⊝ Very lowd | — |
| PCR‐mRNA (against MSP or PSQ) | Not estimated | 148 (2 studies) |
⊕⊝⊝⊝ Very lowe | — |
| MS‐MLPA (against MSP or PSQ) | Not estimated | 48 (1 study) |
⊕⊝⊝⊝ Very lowf | — |
| PCR‐HRM (against MSP or PSQ) | Not estimated | 309 (3 studies) |
⊕⊝⊝⊝ Very lowg | — |
| Other techniques (against MSP or PSQ) | Not estimated | 1209 (7 studies) across various other techniques |
⊕⊝⊝⊝ Very lowd | — |
| Grades of evidence High certainty: further research is very unlikely to change our confidence in the conclusion. Moderate certainty: further research is likely to have an important impact on our confidence in the conclusion. Low certainty: further research is very likely to have an important impact on our confidence in the conclusion. Very low certainty: we are very uncertain about the conclusion. | ||||
| CpG: 5'‐cytosine‐phosphate‐guanine‐3'; HRM: high‐resolution melting; IHC: immunohistochemistry; MGMT: O6‐methylguanine–DNA methyltransferase; MS‐MLPA: methylation‐specific multiplex ligation‐dependent probe amplification; MSP: methylation‐specific polymerase chain reaction; PCR: polymerase chain reaction; PCR‐mRNA: polymerase chain reaction‐messenger ribonucleic acid; PSQ: pyrosequencing; qMSP: quantitative methylation‐specific polymerase chain reaction. | ||||
aDowngraded one level for imprecision. bDowngraded two levels for imprecision and indirectness (due to variability in CpG sites and thresholds used for PSQ). cDowngraded three levels for serious risk of bias, imprecision, inconsistency and indirectness. dDowngraded three levels for serious imprecision, inconsistency and indirectness. eDowngraded three levels for imprecision, inconsistency and indirectness. fDowngraded three levels for serious risk of bias, serious imprecision, inconsistency and indirectness. gDowngraded three levels for risk of bias, serious imprecision, inconsistency and indirectness.
Background
Description of the health condition and context
Glioblastoma is an aggressive form of brain cancer. Approximately five of every 100 people with glioblastoma survives for five years past diagnosis (Ostrom 2014). Glioblastomas that have a particular modification to their DNA (called methylation) in a particular region (the O6‐methylguanine–DNA methyltransferase (MGMT) promoter) respond better to treatment with chemotherapy using a drug called temozolomide. Although we know that modification of this DNA region is important (Butler 2020), we do not know the best way to measure it. In this Cochrane Review, we aimed to assess which way of measuring methylation of the MGMT promoter best predicts survival for people with glioblastoma who are treated with temozolomide.
Gliomas are a group of brain tumours that share some features with glial cells, which are the cells that support and insulate neurons and are thought to originate from a population of stem or progenitor cells in the brain. The World Health Organization (WHO) divides gliomas into astrocytic, oligodendroglial and ependymal tumours, and other rarer subtypes depending on the type of glial cell the tumour shares features with (Louis 2016). Glioblastoma is the most malignant (aggressive) type of astrocytic tumour (Louis 2016), and the most common primary brain tumour among adults. Age‐adjusted incidence of primary (isocitrate dehydrogenase (IDH)‐wild‐type) glioblastoma (ICD‐O‐3 morphology codes 9440 to 9442, WHO grade IV) ranges from 0.59 to 3.69 per 100,000 people (Ostrom 2014). IDH‐wild‐type glioblastomas are more common in older people, peaking in 74 to 84‐year olds (Ostrom 2014). These glioblastomas are associated with poor prognosis, with a five‐year relative survival of approximately 5% (Ostrom 2014). The median overall survival is 9.9 months for people treated with surgery plus radiotherapy, and 15 months for people treated with surgery plus radiotherapy plus chemotherapy (Louis 2016). For people with secondary (IDH‐mutant) glioblastomas, median overall survival is 24 months for people treated with surgery plus radiotherapy, and 31 months for people treated with surgery plus radiotherapy plus chemotherapy (Louis 2016).
Glioblastomas are commonly diagnosed by a neurosurgical multidisciplinary team following brain imaging with computerised tomography (CT) and magnetic resonance imaging (MRI). If appropriate, the person has a biopsy or resection (surgical removal) of the tumour to confirm the histopathological diagnosis. For newly diagnosed glioblastoma, the standard treatment is maximal surgical resection followed by radiotherapy with concomitant and adjuvant temozolomide (Stupp 2005). Temozolomide is an alkylating chemotherapeutic agent. It causes DNA damage, which inhibits DNA replication. However, not all people respond to temozolomide therapy to the same extent. There is evidence that people with newly diagnosed glioblastoma who start treatment with radiotherapy and temozolomide more than six weeks after neurosurgery have worse overall survival than people who start treatment within six weeks (Sun 2015).
In the UK, it is estimated that on average just over 20 years of life are lost per person with a brain tumour, the most of any form of cancer (Burnet 2005). Olesen 2012 estimated the total annual costs of brain tumours in Europe to be EUR 5.2 billion, based upon purchasing power parity rates for 2010.
Description of the prognostic factors
MGMT is a DNA repair enzyme in glioblastoma cells that can repair the damage caused by alkylating agents such as temozolomide. If the MGMT gene promoter is methylated, it is thought the glioblastoma cell is less able to repair this damage and is more likely to die, therefore making the tumour more sensitive to alkylating therapy (Brandner 2015). If the MGMT gene promoter in the glioblastoma cell is unmethylated, it is thought that the glioblastoma cell can repair the damage caused by temozolomide and, therefore, temozolomide is less effective. Consequently, epigenetic silencing of the MGMT gene by promoter methylation is associated with longer overall survival in people with glioblastoma receiving alkylating therapy in addition to radiotherapy (Alnahhas 2020; Esteller 2000; Hegi 2004; Hegi 2005). A key retrospective analysis of one randomised phase III trial found that treatment with temozolomide and radiotherapy conferred a significant survival benefit versus radiotherapy alone in people with MGMT promoter methylation (median survival: 21.7 months, 95% confidence interval (CI) 17.4 to 30.4 with temozolomide plus radiotherapy versus 15.3 months, 95% CI 13.0 to 20.9 with radiotherapy alone; P = 0.007), whereas there was a smaller difference in survival in people with unmethylated MGMT (median survival: 12.7 months, 95% CI 11.6 to 14.4 with temozolomide plus radiotherapy versus 11.8 months, 95% CI 9.7 to 14.1 with radiotherapy alone) (Hegi 2005).
There is clear evidence that MGMT promoter methylation status testing is important in older people. When older people with glioblastomas with an unmethylated MGMT promoter were treated with single‐agent temozolomide chemotherapy, they had worse outcomes than those treated with radiotherapy (Malmström 2012; Wick 2012). Professional bodies, such as the European Association for Neuro‐Oncology (EANO), recommend evaluation of MGMT promoter methylation status in older people (Weller 2017a). The National Institute for Health and Care Excellence (NICE) recommends that all high‐grade gliomas are tested for MGMT promoter methylation to inform prognosis and guide treatment (NICE 2018). Most non‐elderly (aged under 65 years) people are treated with temozolomide chemotherapy irrespective of MGMT promoter status, possibly due to the lack of alternative treatments (Hegi 2015). Despite this, MGMT promoter status is still a useful prognostic marker which may impact clinical management. It can also inform recruitment into clinical trials for novel therapies.
There are many ways of assessing methylation status. These include:
methylation‐specific polymerase chain reaction (MSP);
quantitative (real‐time) methylation‐specific polymerase chain reaction (qMSP), including MethyLight;
methylation‐specific sequencing, including pyrosequencing (PSQ);
bead array;
methylation‐specific multiplex ligation‐dependent probe amplification (MS‐MLPA);
polymerase chain reaction with high‐resolution melting (PCR‐HRM);
co‐amplification at lower denaturation temperature (COLD)‐PCR; and
digestion‐based assays.
We describe these techniques briefly in Table 2. In addition, protein expression or enzymatic activity may be used as a proxy for methylation status. Internationally accepted consensus about the most appropriate diagnostic method for MGMT promoter status is lacking (Brandner 2015). MSP was used to assess MGMT promoter status in the landmark study by Hegi 2005. In practice, the choice of technique used to assess MGMT promoter status may depend on the amount and quality of the DNA sample(s) (e.g. formalin‐fixed paraffin‐embedded (FFPE) versus frozen tissue‐derived DNA), the robustness and simplicity of the method, the availability of equipment and reagents, cost and experience. In the most recent UK National Quality Assessment (UK NEQAS) External Quality Assessment report, 10 of 18 UK laboratories used PSQ, five used MSP, two used HRM and one used MS‐MLPA.
1. Techniques of determining methylation status.
| Test | Brief description |
| Methylation‐specific polymerase chain reaction (MSP) | In MSP, DNA is extracted from tumour tissue and then treated with sodium bisulfite. Sodium bisulfite causes changes in the sequence of unmethylated DNA, as it changes the DNA base cytosine into uracil. Methylated DNA is protected and remains unchanged. Regions of DNA can then be amplified using PCR in a manner that is dependent on whether the changed (containing uracil) or original sequence (containing cytosine) is present. |
| Quantitative (or real time) methylation‐specific PCR (qMSP) | qMSP is very similar to MSP, but there is a measure of the amount of changed and original DNA sequence. |
| Methylation‐specific sequencing, including pyrosequencing (PSQ) | In methylation‐specific sequencing, DNA is extracted from tumour tissue and treated with sodium bisulfite, which changes unmethylated DNA. The DNA can then be sequenced to determine if it contains the changed or original sequence, i.e. whether it contains uracil in place of cytosine. There are many ways of sequencing DNA, but one commonly used method is called PSQ. |
| Bead array | In bean array, DNA is extracted from tumour tissue and treated with sodium bisulfite, which changes unmethylated DNA. The DNA is then hybridised to sequences that are either complementary to the original sequence or changed sequence. The hybridisation produces a signal that can be measured. |
| Methylation‐specific multiplex ligation‐dependent probe amplification (MS‐MLPA) | In MS‐MLPA, the DNA is treated with an enzyme that cleaves unmethylated DNA at specific sequences, but methylated DNA is protected. PCR to amplify regions of DNA is then performed. Amplification will only occur if the DNA was not cleaved. |
| PCR with methylation‐sensitive high‐resolution melting (PCR‐HRM) | This technique relies on the changes to DNA caused by sodium bisulfite (i.e. the replacement of cytosine by uracil) leading to it having a lower melting temperature, which is the temperature at which the 2 different DNA strands come apart. Methylated DNA will have a higher melting temperature. A dye that changes fluorescence depending on whether the DNA strands are together or apart can be added. |
| Co‐amplification at lower denaturation temperature PCR (COLD‐PCR) | COLD‐PCR relies on the same principle as PCR‐HRM. In this case only sequences with low melting temperatures will be amplified. This means that only unmethylated regions will be amplified. |
| Digestion‐based assays | This technique relies on enzymes that cleave unmethylated DNA at specific sequences, but methylated DNA is protected. |
COLD: co‐amplification at lower denaturation temperature; MS‐MLPA: methylation‐specific multiplex ligation‐dependent probe amplification; MSP: methylation‐specific polymerase chain reaction; PCR: polymerase chain reaction; PCR‐HRM: polymerase chain reaction with high‐resolution melting; PSQ: pyrosequencing; qMSP: quantitative methylation‐specific polymerase chain reaction.
Additionally, there is within‐technique heterogeneity arising from differences in the regions of the MGMT promoter tested to determine MGMT methylation status. The prognostic impact of these differences is not well understood. Similarly, there is variation in the cut‐offs used for categorising methylation status for techniques that quantify the amount of methylation present. The manufacturers do not recommend specific thresholds and there is no consensus on what are the most appropriate, with individual laboratories left to determine their own thresholds, for example by running tests on healthy control samples, or examining survival of people with glioblastoma.
The result of each method for measuring MGMT status can be considered a separate prognostic factor for predicting overall survival in people with glioblastoma treated with temozolomide.
Health outcomes
The health outcome of interest for this review was overall survival. We did not limit the period of follow‐up. Glioblastomas are associated with poor prognosis, so we anticipated that most studies would assess overall survival within five years of diagnosis.
Why it is important to do this review
It is important to reach a consensus regarding which is the best method for assessing MGMT methylation status based on the prognostic value of each method in predicting overall survival in people with glioblastoma treated with temozolomide, so that people living with glioblastoma can be confident that they are having the appropriate molecular analysis performed. The regions of the promoter that need to be analysed and the most relevant cut‐offs for quantitative tests need to be established. Systematic reviews and meta‐analyses have determined the prognostic value of MGMT promoter status assessed by a specific technique, for example by PSQ (Zhao 2016), MSP (Zhang 2013), or qMSP (Hegi 2019). However, we are aware of no systematic review that has determined which method is best correlated with prognosis. One narrative overview addressed the question, but provided no quantitative synthesis of the results (Dullea 2016).
In this Cochrane Review, we seek to determine which technique, assessing which regions and (if relevant) which cut‐off is best associated with overall survival in people with glioblastoma treated with temozolomide. We consider each MGMT test as a separate prognostic factor. We extract or calculate (where possible) hazard ratios (HRs) for those who tested positive compared with those who tested negative. A test that is not better than flipping a coin is expected to have an HR of one. The better the test's ability to discriminate between people with a good overall survival versus people with poor overall survival, the further the HR value will be from one.
The review aims to answer part of the question "Do molecular subtyping techniques improve treatment selection, prediction and prognostication in people with brain and spinal cord tumours," one of the top 10 topics identified by the James Lind Alliance Neuro‐Oncology Priority Setting Partnership (JLA PSP 2018), by addressing the predictive ability of one specific molecular modification (MGMT methylation status) in people with glioblastoma. The James Lind Alliance is an organisation that brings people, carers and clinicians together to set research priorities. The National Cancer Research Institute Brain Tumour Clinical Studies Group has also identified this as an area for future research.
It is also important to consider the cost effectiveness of alternative methods of assessing MGMT promoter methylation status. Each method of assessment will incur costs, such as laboratory costs, clinic costs and subsequent treatment costs. The benefits of targeting treatment may include greater survival and less exposure to potentially toxic treatments, as well as potential cost‐savings from the avoidance of waste from the use of ineffective drugs. This review aims to consider the costs alongside the consequences of the prognostic tests to understand the value that they provide to the healthcare system.
Objectives
Primary objective
To determine which method for assessing MGMT methylation status best predicts overall survival in people diagnosed with glioblastoma who are treated with temozolomide. We consider each MGMT method as a separate prognostic factor.
See Table 3 for the review question in population, index prognostic factor, comparator prognostic factor(s), outcome, timing and setting (PICOTS) format.
2. Review question in PICOTS format.
| Population | People with diagnosed glioblastoma (at any point after diagnosis) who go onto be treated with temozolomide |
| Index prognostic factors | Tests for MGMT promoter methylation. We considered each method as a separate prognostic factor. |
| Outcome | Overall survival |
| Timing | The outcome is to be predicted at any point after the start of treatment. |
| Setting | To give prognostic information before the start of treatment with temozolomide. |
MGMT: O6‐methylguanine–DNA methyltransferase; PICOTS: Population, Index prognostic factor, Comparator prognostic factor(s), Outcome, Timing, Setting.
Secondary objective
We undertake an integrated economic review to identify economic evaluations in relation to the different methods of assessing MGMT methylation status effect on overall survival, and undertake a simple economic analysis exploring the cost‐effectiveness of alternative approaches to assessing MGMT methylation status.
Investigation of sources of heterogeneity
We examine for each technique whether any of the following features was best associated with overall survival.
Promoter region/CpGs analysed (or the antibody used in the case of IHC).
Cut‐off used (where relevant).
Type of tumour sample (FFPE or frozen).
We planned to investigate the effect of population characteristics including the following if sufficient data allowed us to do this.
Age.
Extent of tumour resection.
Karnofsky performance status.
IDH status.
Recurrent tumour versus first diagnosis.
We are assuming constant HRs. To confirm the validity of this assumption, we hoped to investigate length of follow‐up as a source of heterogeneity, again if sufficient replications of the same methods had been available.
Methods
Criteria for considering studies in this review
Types of studies
We included longitudinal studies of adults with diagnosed glioblastoma treated with temozolomide with/without radiotherapy/surgery that had related MGMT status in tumour tissue (assessed by one or more method) with overall survival. This included the temozolomide‐treated arms of randomised controlled trials (RCT). We also sought nested case‐control studies. To be included, studies must have determined MGMT status from samples taken before the initiation of treatment. Studies could have had any length of follow‐up. We excluded cohort studies performed exclusively in people who had survived a particular amount of time, or case reports.
Studies were only eligible if they reported HRs, or if we could calculate HRs from the data reported.
Types of studies for the economic component
We sought economic evaluations conducted alongside trials, modelling studies and cost analyses to inform the identification of cost‐effectiveness outcomes.
Targeted participants
Eligible studies were of adults with diagnosed glioblastoma treated with temozolomide with or without radiotherapy/surgery. If studies included people with other forms of glioma (and we could not extract results for the population with glioblastoma), we included these if other forms of glioma made up less than 10% of the population. We included studies of participants with either first diagnosis or recurrent glioblastoma. Participants in eligible studies could receive concomitant and adjuvant therapies in addition to temozolomide (e.g. surgery or radiotherapy, or both, or additional chemotherapeutics). If not all participants received temozolomide (e.g. in the context of an RCT), we included data on people who did receive temozolomide if these were available. We excluded studies performed exclusively in children (under 18 years of age).
Types of prognostic factors
Eligible studies had to assess MGMT promoter methylation status in tumour tissue by at least one method. We treated each method as a separate prognostic factor. Eligible techniques included, but were not restricted to, MSP; quantitative MSP (real‐time PCR or MethyLight methylation‐specific quantitative PCR); methylation‐specific sequencing, including PSQ; bead array; MS‐MLPA; PCR‐HRM; COLD‐PCR and digestion‐based assays. We also included testing strategies that considered MGMT expression (e.g. IHC for protein expression, or tests measuring messenger ribonucleic acid (mRNA) levels) or MGMT enzymatic activity.
Eligible techniques had to be molecular techniques and performed directly on tumour tissue. We excluded studies that assessed MGMT promoter methylation status from blood samples because insufficient quantities of brain tumour DNA cross the blood–brain barrier for testing to be appropriate. In addition, we excluded studies that inferred MGMT methylation status due to macroscopic morphological changes that can be detected by, for example, imaging (i.e. MRI, CT, positron emission tomography (PET)).
We excluded studies that did not report the method of determining MGMT promoter methylation status, as this information is essential for this review.
Types of outcome to be predicted
Overall survival.
Outcomes of the economic component
Resources use, costs, cost effectiveness and cost‐utility of different methods of assessing MGMT promoter methylation status based on full economic review.
Relative efficiency of each method of testing for MGMT promoter methylation status based on a decision model using the outcomes from the review of effectiveness and from the full integrated economic review.
Search methods for identification of studies
Electronic searches
We searched the following databases in December 2018 (Appendix 1):
Ovid MEDLINE (1946 to 4 December 2018);
PubMed NOT MEDLINE (4 December 2018);
Ovid Embase (1980 to 2018, week 49);
BIOSIS (1969 to 3 December 2018) and
Web of Science Conference Proceedings Citation Index (CPCI‐S) (1900 to 3 December 2018).
We applied no restrictions on language or date of publication to the searches.
Searching other resources
The Society of Neuro‐Oncology (SNO), and its partner associations the EANO and the Japan Society of Neuro‐Oncology hold meetings where relevant research may be presented. We searched for abstracts from these meetings and other relevant conferences via the Web of Science Conference Proceedings Citation Index (CPCI‐S) (from 1990 to 3 December 2018), as listed above. We translated the BIOSIS search for CPCI‐S, since both databases are hosted on Web of Science.
Additional searches for the economic component
We searched the NHS Economic Evaluation Database (EED), with combinations of relevant keywords from the search strategy, up to the end of December 2014, when the last records were added to that database. The NHS EED was based on a comprehensive search of bibliographic databases including MEDLINE and Embase.
Data collection
We used EPPI‐Reviewer 4 and EPPI‐Reviewer Web for the screening and selection of studies, and for select data extraction tasks (EPPI‐Reviewer). Further data extraction was undertaken into a Microsoft Excel spreadsheet.
Selection of studies
Two review authors (of AM, KMK and AH) independently screened titles and abstracts of all identified search results. We retrieved the full text of any articles that either review author deemed relevant, or whose relevance could not be determined from the abstract. Two review authors (of AM, CK, FS, KMK, AH, SB and CLF) independently assessed the full‐text articles for eligibility. We resolved any disagreements about eligibility as multiple‐method studies by consensus, or by consulting a third review author where necessary. One review author made decisions about studies considered eligible as single‐method studies. We constructed a PRISMA flow diagram to depict the flow of information through the different phases of the review.
Two review authors (AK and TR) screened studies retrieved for full‐text screening for potentially relevant economic studies.
Data extraction and management
As planned in the protocol (McAleenan 2019), we performed full data extraction, risk of bias assessment and synthesis on studies that evaluated MGMT promoter methylation status of the same people using two or more methods (i.e. multiple techniques, CpG sites, cut‐offs or types of tissue sample) so that these tests could be compared on the same samples of people. We performed limited data extraction on studies that evaluated MGMT promoter methylation status using a single method.
Two review authors (of AM, CK, FS, LS, HC and JPTH) independently performed data extraction on each article describing two or more methods for MGMT promoter methylation status, using forms piloted on several articles. We resolved any disagreements by consensus and consulted a third review author where necessary. We extracted data on the following items relevant to prognostic factor studies, derived from the CHecklist for critical Appraisal and data extraction for systematic Reviews of prediction Modelling Studies (CHARMS) (Moons 2014). We did not contact primary investigators for information that was not available in the reports.
Study characteristics
Author.
Year.
Country and setting.
Length of follow‐up.
Study dates.
Study design.
Population characteristics
Number of participants.
Population source and setting.
Timing of MGMT promoter methylation assessment.
Inclusion/exclusion criteria.
Tumour type.
Age.
Gender.
Karnofsky performance status.
Extent of resection.
Treatment regimen.
Length of time between neurosurgery and start of treatment.
IDH mutation status.
First diagnosis or recurrent disease.
Deaths during follow‐up.
Prevalence of MGMT promoter methylation (by each technique).
Method(s) of MGMT promoter methylation assessment
Technique.
Tumour sample type (i.e. FFPE or frozen tissue).
Region/CpGs analysed (for PCR‐based tests); antibody used (for immunohistochemistry).
Cut‐off/threshold used to determine MGMT promoter methylation status (where relevant).
Method of determining threshold and whether it was prespecified.
Outcome assessment
Time point from which overall survival was measured.
Missing data
Number of participants with any missing data.
Association between MGMT methylation status and overall survival
Data sufficient to determine computer HRs and their CIs.
Adjusted HRs and their CIs (where reported), and factors for which the result was adjusted.
To obtain HRs, we followed strategies described by Tierney 2007 and Parmar 1998. We primarily sought unadjusted HRs, and used these if they were presented directly. We computed standard errors of log HRs from CIs or exact P values, assuming these were based on Wald tests. Where only a P value threshold was stated, we set the P value to be equal to this; this only occurred in cases where small thresholds had been used (P < 0.001, or < 0.000001). When we could not obtain unadjusted HRs directly using these approaches, we obtained HRs using (in order of preference): 1. individual participant data (IPD) from publications; 2. reported adjusted HRs or 3. published Kaplan‐Meier curves. From Kaplan‐Meier curves, we reconstructed approximate IPD following Guyot 2012. We derived plot co‐ordinates from the published curves using Engauge Digitizer 12.1 as input into Guyot's algorithm (Engauge Digitizer). Where possible, we followed Guyot and colleagues' suggestion of including information from risk tables and total numbers of events. Depending on the information provided in study reports, we reconstructed IPD using the best information (i.e. in preferential order 'all information,' 'no numbers at risk' then 'no total events' as referred to by Guyot 2012). However, for most study reports there was insufficient information, in which case we followed Guyot and colleagues' 'neither' case. We reconstructed the IPD using the R script from the supplement of Guyot 2012. These analyses were conducted using R (version 4.0.3) in RStudio (version 1.2.5042). Reconstructed data from these plots are available at the data.bris repository (data.bris.ac.uk/data/).
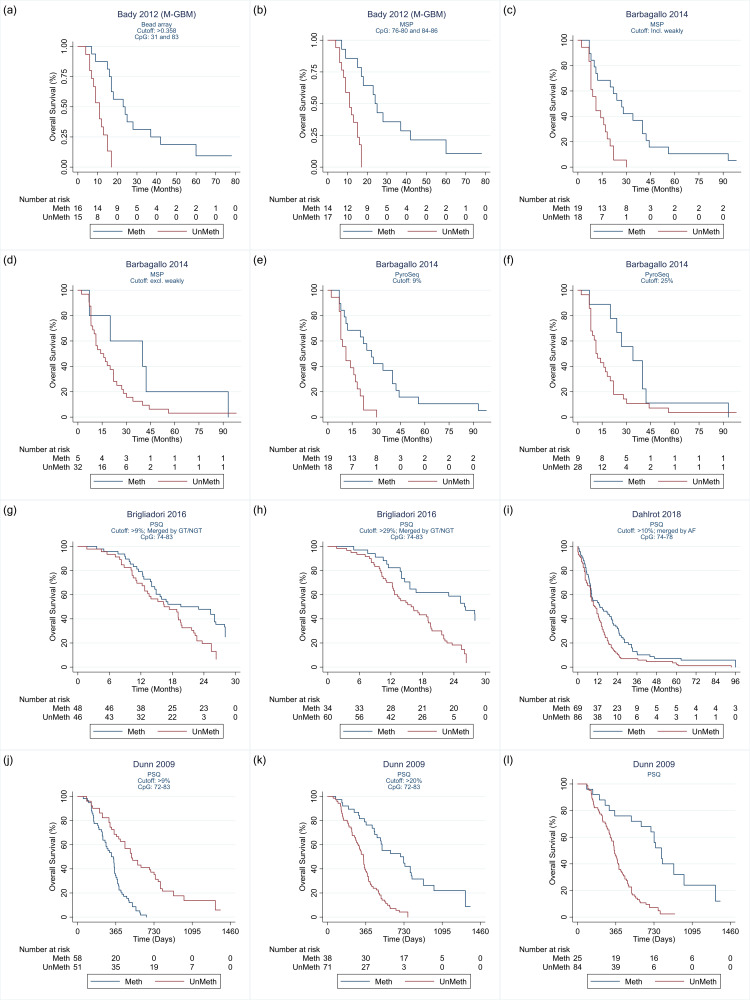
We analysed the IPD or reconstructed IPD (from Kaplan‐Meier curves) to estimate HRs using Cox proportional hazards regression, using the stcox command in Stata. Some study reports categorised participants by the extent of methylation, e.g. "unmethylated (0 to 9%)," "weakly methylated (10% to 29%)," and "methylated (30% or greater)." Where survival data for these groups were presented in Kaplan‐Meier curves, we combined the individuals across categories to dichotomise the data at each cut‐off. To illustrate, for the categories in the above example, we regrouped the data to estimate the HR for the comparison "unmethylated" and "weakly methylated" combined versus "methylated" (cut‐off at 29%), and the HR for the comparison "unmethylated" versus "weakly methylated" and "methylated (cut‐off 9%). These analyses, including plotting of reconstructed Kaplan‐Meier curves, were performed using Stata (version 16).
For studies that evaluated MGMT promoter status using a single method, we extracted details on author, year, country, length of follow‐up, number of participants, tumour type, IDH mutation status and technique used for MGMT promoter methylation assessment.
Economic studies
In addition to the data extracted from clinical studies, we planned to extract relevant data from economic evaluations (had any been identified). We aimed to collect the following data from the economic evaluation studies.
Type of evaluations.
Sources of effectiveness data.
Cost data.
Sources of cost data.
Sources of outcome valuations.
Analytical approach.
Assessment of risk of bias in included studies
We assessed risk of bias in studies that evaluated MGMT promoter methylation status of the same people using at least two methods.
The QUality In Prognosis Studies (QUIPS) tool is designed to assess risk of bias in prognostic factor studies (Hayden 2013). It assesses bias across six domains: study participation, study attrition, prognostic factor measurement, outcome measurement, study confounding, and statistical analysis and reporting. We assessed risk of bias across QUIPS domains, although we added a domain on subsequent treatment. We renamed the study confounding domain to 'adjustment for other potential prognostic factors'; and we limited the domain about statistical analysis and reporting to selective reporting alone because we sought only results of a standard proportional hazards regression analysis. We replaced the prompting items and considerations, which mainly assessed reporting, with signalling questions to help us reach domain‐level judgements. The domain modifications and signalling questions were informed by the CHARMS checklist (Moons 2014), a framework for assessing internal validity of articles dealing with prognosis described in Altman 2001, as well as ROBINS‐I (risk of bias in non‐randomised studies of interventions) (Sterne 2016) and QUADAS‐2 (Whiting 2011). In addition, for each domain apart from study attrition and selective reporting, we added questions assessing the applicability of the study as in QUADAS‐2 (Whiting 2011) and PROBAST (Wolff 2019). We assessed risk of bias in the first three domains (participant selection, subsequent treatment and outcome measurement) at the study level, and the other four domains (prognostic factor measurement, study attrition, adjustment for other potential prognostic factors and selective reporting) for each result within each study. We judged risk of bias and concerns regarding applicability as high, low or unclear. The tool is detailed in Appendix 2. Two review authors (of AM, CK, FS, LS, HC and JPTH) independently performed assessments using a form that had been piloted on several articles. These review authors sought to reach a consensus judgement and resolved any remaining disagreements by consulting a third review author. We did not contact primary investigators for information that was not available in the reports.
Assessment of risk of bias in studies included in the economic component
We planned to perform assessment of the quality of the economic evaluations captured in this review in two stages. The first stage was to assess the risk of bias in the clinical studies informing the evaluation. If the economic evaluation was carried out alongside a single study, then we planned to use our bespoke tool described in Appendix 2. Should any economic evaluations based on models have been identified, we planned to assess any summary effect sizes from systematic reviews used as data inputs in these model‐based economic evaluations using the ROBIS tool (Whiting 2016). The second stage for assessing any identified economic evaluations was to assess the overall methodological quality of the economic component of the evaluation. Based on the methods from the Cochrane Handbook chapter on economic methods (Aluko 2020), we planned to assess evaluations carried out alongside single empirical studies using the CHEERS checklist (Husereau 2013). In addition, we planned to assess any model‐based economic evaluations using the NICE methodology checklist (NICE 2012).
Assessment of reporting bias
For each meta‐analysis that contained 10 or more studies, we planned to examine the symmetry of funnel plots and test for asymmetry using Debray's funnel inverse variance test based on HRs (Debray 2018). Asymmetry may be an indicator of publication bias.
Data synthesis
Data synthesis and meta‐analysis approaches
To assess the relative prognostic ability of the different methods we focused on data from direct, within‐study comparisons, where the MGMT promoter methylation status of the same series of people was evaluated in multiple ways and the results correlated with overall survival. We undertook full data extraction, risk of bias assessment and synthesis on studies only for this subset of studies.
We harmonised the direction of the HRs from each study so that each represented hazard rate among people with an unmethylated MGMT promoter divided by the hazard rate among people with a methylated MGMT promoter. This means that a value greater than one indicates favourable outcomes in people with a methylated MGMT promoter. The greater the HR, the better the method was at predicting time to death. In the main analyses focusing on unadjusted HRs, we substituted an adjusted HR if an unadjusted HR was not available. We present each statistical result with a 95% CI.
Where at least five studies had compared the same pair of methods, we compared the HRs within studies to produce a ratio of hazard ratios (RHR). A complication here was how to account for the correlation between the log HRs to reflect that the different methods were applied to the same people. We computed the correlation between the original test results from studies for which we could extract IPD from the publications. We assumed this correlation would carry approximately through to the HRs comparing the two methods. We then computed log HRs and their variances (the latter as var(logHR1) + var(logHR2) – 2Cov(logHR1, logHR2), with covariances computed from the imputed correlation coefficient). We performed standard random‐effects meta‐analyses (with DerSimonian‐Laird estimator of between‐study variance) to estimate an overall RHR. We performed a sensitivity analysis using higher and lower values for the correlation coefficient. In these analyses, we quantified heterogeneity across results of the studies using an estimate of the between‐study variance in logRHRs and portrayed these using prediction intervals. We also reported between‐study variance (Tau2). In addition, we describe the extent of inconsistency in the findings using the I2 statistic, which describes the percentage of variation across studies that is due to heterogeneity rather than chance (Higgins 2002).
The prognostic value of each test may be dependent on other prognostic factors of overall survival, and these may have been adjusted for. In addition to analyses of unadjusted HRs, we planned to extract and meta‐analyse adjusted results, to confirm that the tests have added prognostic value in addition to (easier to measure) prognostic factors such as age, gender, disease stage at diagnosis and comorbidity. We present HRs adjusting for age and extent of resection, which were the most common factors adjusted for.
We also expected to identify studies that had evaluated MGMT promoter using only one method. We present only brief details of these studies. At a later date we may investigate these studies further to supplement inferences from the comparative studies. Specifically, there may be a possibility of comparing methods indirectly across studies. Such unadjusted indirect comparisons rely on the assumption that the studies assessing each test for MGMT promoter methylation are similar for all important characteristics (i.e. that they were conducted on similar populations that had been given similar treatments (or that these factors were adjusted for) and that the risk of bias was similar). This is a very strong assumption, and not one we were willing to make in this review.
We planned to present the results from the full economic review as a narrative analysis, describing the results of the economic evaluations identified by the search. In addition to the narrative summary of the economic evaluations, we planned to use both the clinical and economic outcomes to inform a decision model to estimate the cost effectiveness of assessing MGMT status in the management of glioma.
Subgroup analysis and investigations of heterogeneity
We aimed to investigate potential sources of heterogeneity in the results for each method using subgroup analyses or meta‐regression, depending on the number of studies identified and the nature of the source of heterogeneity.
We examined, for each technique, whether any of the following features was best associated with overall survival.
The promoter region/CpGs analysed (or the antibody used in the case of immunohistochemistry).
The cut‐off used (where relevant).
The type of tumour sample (FFPE or frozen).
We also planned to investigate the effect of population characteristics including:
age;
extent of tumour resection;
Karnofsky performance status;
IDH status;
recurrent versus first diagnosis.
We assumed constant HRs. To test the validity of this assumption, we planned to investigate length of follow‐up as a source of heterogeneity, and if studies had started follow‐up for overall survival from different time points, we aimed to investigate this as a source of heterogeneity.
Sensitivity analyses
We planned sensitivity analyses restricting the analysis to studies at low or unclear risk of bias. We also performed sensitivity analyses imputing different correlation coefficients between logHRs within studies, as described in the section on 'Data synthesis and meta‐analysis approaches.'
Decision model
We aimed to create an economic model using outcomes from both the clinical and economic evidence we identified. The aim was to use the extracted data to populate a decision analytic model, to assess the cost‐effectiveness of different methods of testing for MGMT promoter methylation status in people with glioblastoma. The effect of the different methods of assessing MGMT promoter methylation status (including not assessing for promoter methylation status at all) was to be compared in terms of probability of effectiveness and overall survival. The model was to be conducted from the UK National Health Service perspective for a target population aged 65 years or over. The time horizon of the model in terms of costs considered would have been six weeks (i.e. until the start of temozolomide treatment). Key uncertainties were to be explored using sensitivity analysis. However, due to the paucity of evidence with which to parameterise a cost‐effectiveness decision model, particularly in reliable estimates of costs, this was not possible.
As an alternative, we considered cost comparison ratios (CCRs) of the three main techniques (PSQ, MSP and IHC). The principle underpinning a CCR comes from the conditions required for an efficient allocation of resources. Economic theory determines that when resources are efficiently allocated, the ratio of marginal costs (MC) to marginal benefits (MB) for all treatments 'a' to 'n' must be equal (i.e. MCa/MBa = MCb/MBb = MCc/MBc = MCn/MBn). Rearranging this equation and simplifying shows that when allocation of resources is efficient, the ratio of marginal costs is equal to the ratio of marginal benefits of all care, such that MCa/MCb = MBa/MBb. To inform these analyses, we used costs for performing these tests in the UK.
Summary of findings
We present the prognostic value of each method on overall survival in a 'Summary of findings' table. We assessed confidence in each result using the GRADE approach (Guyatt 2008). Guidance on the use of GRADE for prognostic factor studies has not yet been published, although adaptations have been proposed (Huguet 2013). We rated the overall strength of evidence as 'high,' 'moderate,' 'low' or 'very low.' We considered risk of bias, indirectness, inconsistency, imprecision and publication bias, which may lead to downgrading of the strength of the evidence; and size of effect, which may lead to upgrading of the strength of the evidence (see Appendix 3).
Results
Results of the search
The search identified 5494 records and we included 223 of these in the review (see Figure 1).
1.
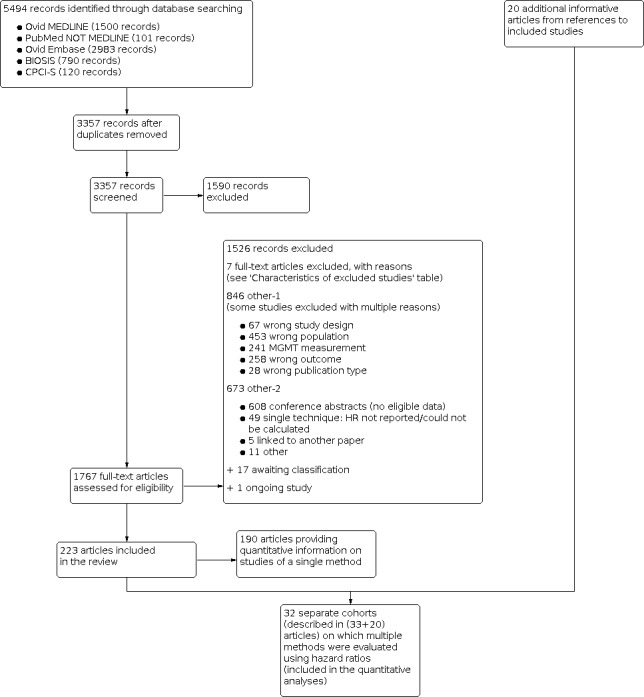
Study flow diagram. N, number of records.
We included 32 distinct cohorts of people (referred to hereafter as studies) in our main analysis of studies comparing two or more methods, including studies comparing different variants of the same technique. These were reported in 33 articles. We drew on an additional 20 publications that had been cited in these 33 articles; some cohorts were reported in multiple publications, and some publications reported on two studies. In addition, we included 190 articles describing single‐method studies. We summarise these in less detail than the main (multiple‐method) studies, as planned in the protocol (McAleenan 2019).
None of the papers examined in full text met the criteria for inclusion into the review of economic evidence. The search of the NHS EED database also identified no relevant economic evaluations. Thus, we identified no economic evaluations assessing the cost‐effectiveness of tests for MGMT promoter methylation status people with glioblastoma treated with temozolomide. This was true for evaluations based on a single study or using decision modelling to synthesise data from multiple studies. Thus, there is a lack of evidence for determining the most efficient strategies for assessing tests for MGMT promoter methylation status of people with glioblastoma treated with temozolomide.
Characteristics of the included studies
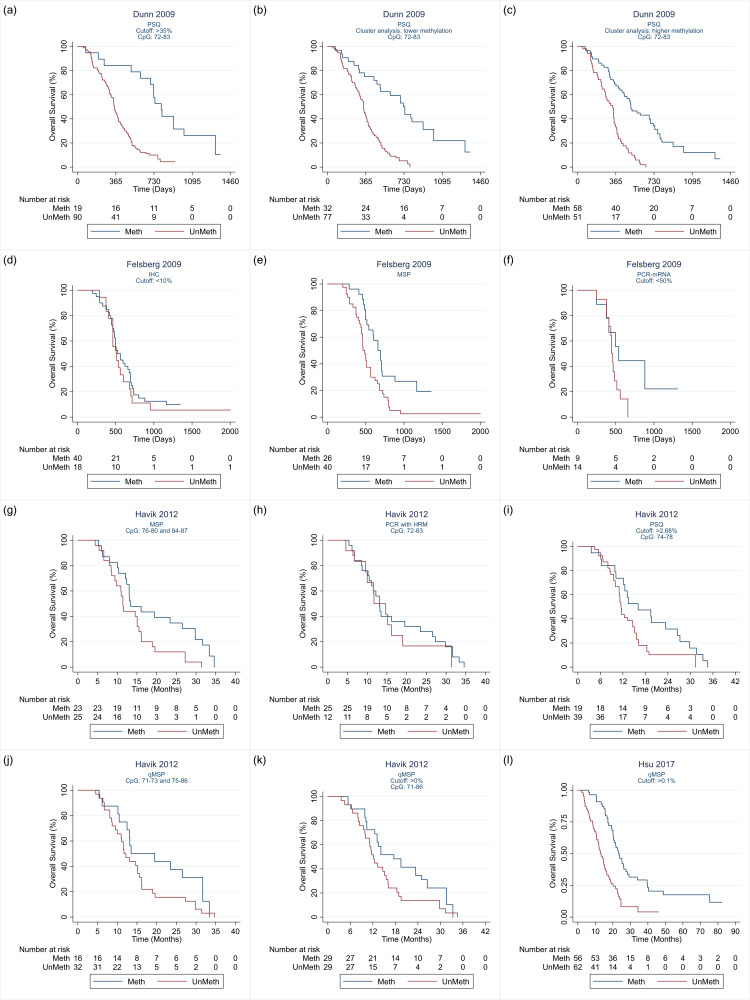
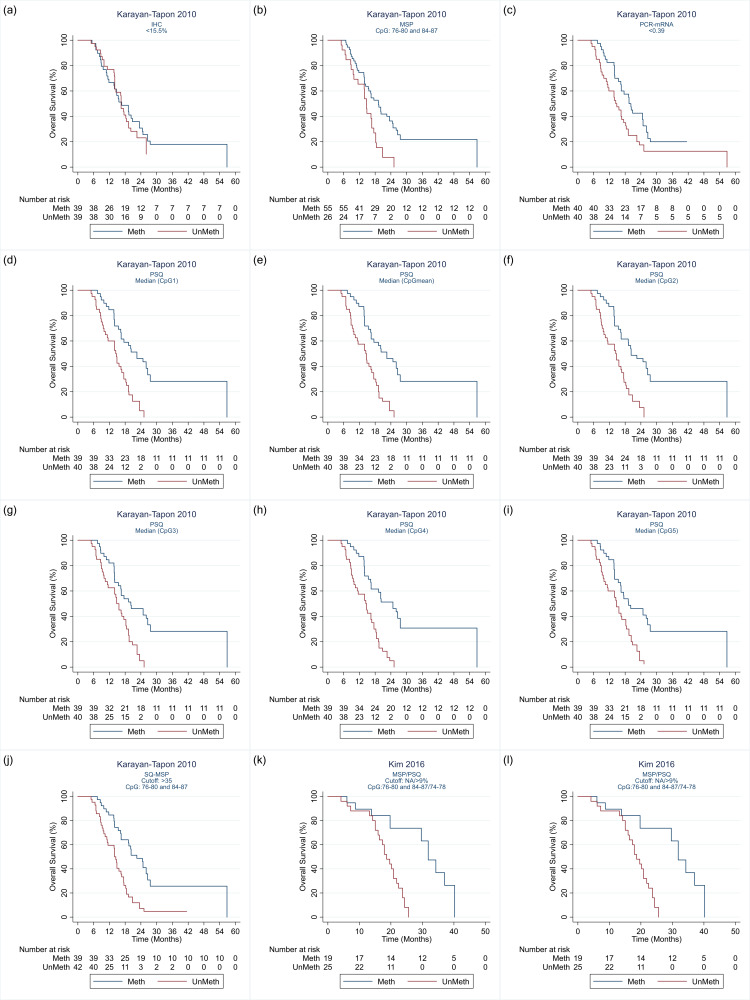
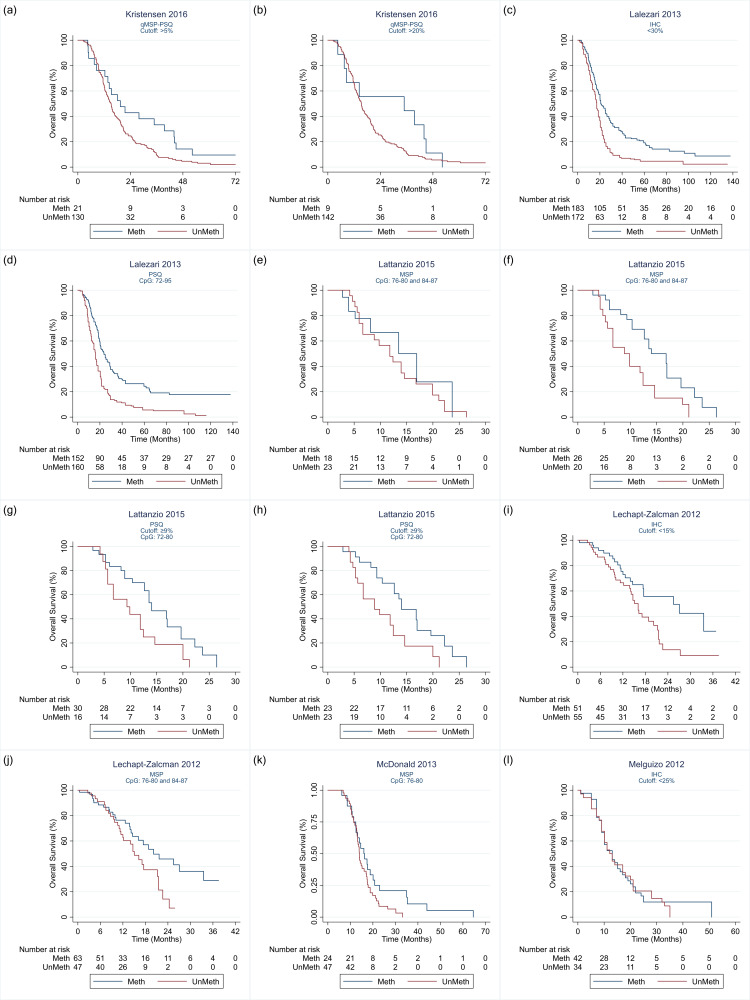
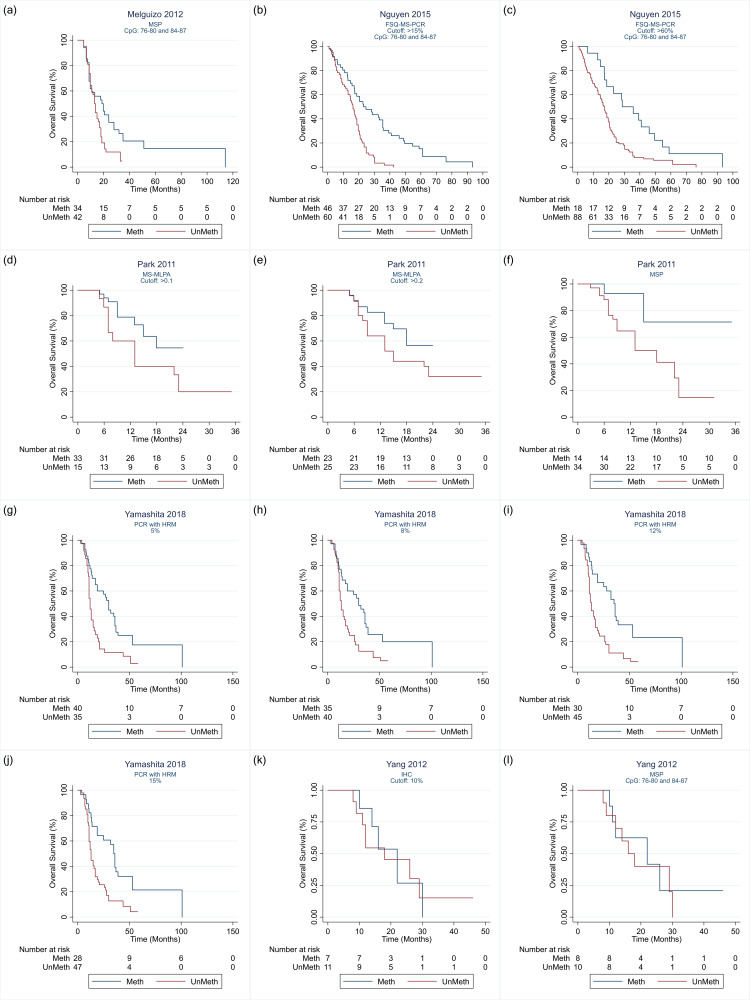
Details of the 32 studies, including 3474 participants, are presented in the Characteristics of included studies table. In descending order, the techniques investigated in the most studies were PSQ in 20 studies; MSP in 17 studies, IHC in nine studies, quantitative MSP (qMSP) in eight studies (including semi‐quantitative polymerase chain reaction (SQ‐PCR), fluorescent semi‐quantitative methylation‐specific polymerase chain reaction (FSQ‐MS‐PCR) and MethyLight‐MSP), PCR‐HRM in three studies (Havik 2012; Quillien 2014 (test); Yamashita 2018); bead array in two studies (Bady 2012 (M‐GBM); Bady 2012 (E‐GBM)); polymerase chain reaction‐targeting messenger ribonucleic acid (PCR‐mRNA) in two studies (Felsberg 2009; Karayan‐Tapon 2010); and MS‐MLPA in one study (Park 2011). Other techniques studied were methylation‐specific restriction enzyme quantitative polymerase chain reaction (MS‐RE‐qPCR; Almuqate 2018), methyl‐beaming (Barault 2015), quantitative fluorescence immunohistochemistry (QF‐IHC AQUA; Bell 2017), double immunofluorescence (DIF; Dahlrot 2018 (NS cohort); Dahlrot 2018 (RSD cohort)) qMSP combined with PSQ (qMSP‐PSQ; Kristensen 2016), and sequencing (Thon 2017). The largest study compared qMSP with QF‐IHC AQUA in 452 tumour samples (Bell 2017). The second largest compared MSP, PSQ and IHC in 418 samples (Lalezari 2013), and the third largest compared PSQ against DIF in 234 samples (Dahlrot 2018 (RSD cohort)). All other studies included fewer than 160 samples, with the smallest including 18.
All studies had a standard cohort design (with one being embedded in a randomised trial; Bell 2017). Nineteen studies were undertaken in Europe, two in North America, eight in East Asia, one in Australia and two across multiple countries. Mean ages ranged from 44 to 64 years. All studies had more men than women (overall, 60% were men where reported). Most studies were exclusively in people with glioblastoma at first diagnosis (where reported). In most studies, the majority of participants had undergone total resection. In 10 studies, it was explicitly stated that treatment followed the Stupp protocol (Stupp 2005), and in most of the others it was clear that temozolomide and radiotherapy were provided in a way that appeared consistent with the Stupp protocol.
We illustrate the comparisons made in the different studies in Table 4 and Appendix 4. Details of the specific methods implemented are provided in Appendix 5. We illustrate the CpG sites targeted in Figure 2.
3. Table of comparisons made.
| Study | IHC | MSP | PSQ | qMSP | Bead array | MS‐MLPA | PCR‐HRM | PCR‐mRNA | Other |
| Almuqate 2018 | — | — | — | — | — | — | — | — | 2 |
| Bady 2012 (E‐GBM) | — | — | 1 | — | 2 | — | — | — | — |
| Bady 2012 (M‐GBM) | — | 1 | — | — | 1 | — | — | — | — |
| Barault 2015 | — | — | 1 | — | — | — | — | — | 1 |
| Barbagallo 2014 | — | 2 | 2 | — | — | — | — | — | — |
| Bell 2017 | — | — | — | 1 | — | — | — | — | 1 |
| Brigliadori 2016 | — | — | 2 | — | — | — | — | — | — |
| Chai 2018 (7‐site cohort) | — | — | 3 | — | — | — | — | — | — |
| Chai 2018 (8‐site cohort) | — | — | 3 | — | — | — | — | — | — |
| Dahlrot 2018 (NS cohort) | — | — | 1 | — | — | — | — | — | 1 |
| Dahlrot 2018 (RSD cohort) | — | — | 1 | — | — | — | — | — | 1 |
| Dunn 2009 | — | — | 6 | — | — | — | — | — | — |
| Felsberg 2009 | 1 | 1 | — | — | — | — | — | 1 | — |
| Havik 2012 | — | 1 | 9 | 2 | — | — | 1 | — | — |
| Hsu 2015 | 1 | 1 | 1 | 2 | — | — | — | — | — |
| Karayan‐Tapon 2010 | 1 | 1 | 6 | 1 | — | — | — | 1 | — |
| Kim 2016 | — | 1 | 1 | — | — | — | — | — | — |
| Kristensen 2016 | 1 | — | 1 | — | — | — | — | — | 3 |
| Lalezari 2013 | 1 | 1 | 1 | — | — | — | — | — | — |
| Lattanzio 2015 | — | 2 | 2 | — | — | — | — | — | — |
| Lechapt‐Zalcman 2012 | 1 | 1 | — | — | — | — | — | — | — |
| McDonald 2013 | — | 1 | 1 | — | — | — | — | — | — |
| Melguizo 2012 | 1 | 1 | — | — | — | — | — | — | — |
| Nguyen 2015 | — | — | — | 2 | — | — | — | — | — |
| Park 2011 | — | 1 | — | — | — | 2 | — | — | — |
| Quillien 2016 | — | — | 12 | 5 | — | — | — | — | — |
| Quillien 2014 (test) | 1 | 1 | 32 | 1 | — | — | 1 | — | — |
| Quillien 2014 (validation) | — | — | 3 | — | — | — | — | — | — |
| Thon 2017 | — | 1 | — | — | — | — | — | — | 1 |
| Yamashita 2018 | — | 1 | — | — | — | — | 5 | — | — |
| Yang 2012 | 1 | 1 | — | — | — | — | — | — | — |
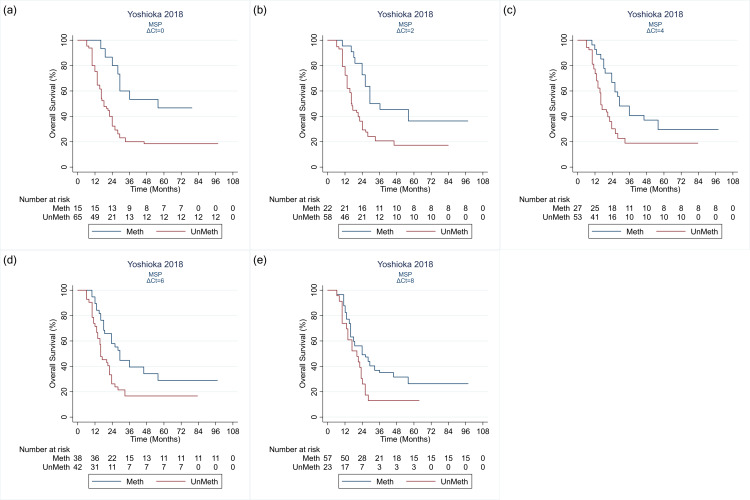
| Yoshioka 2018 | — | — | — | 5 | — | — | — | — | — |
Numbers in cells indicate the number of variants of that technique in the respective study for which we could extract hazard ratios.
IHC: immunohistochemistry; MSP: methylation‐specific polymerase chain reaction; PSQ: pyrosequencing; qMSP: methylation‐specific polymerase chain reaction; MS‐MLPA: methylation‐specific multiplex ligation‐dependent probe amplification; PCR‐HRM: polymerase chain reaction with high‐resolution melting; PCR‐mRNA: polymerase chain reaction‐messenger ribonucleic acid.
2.
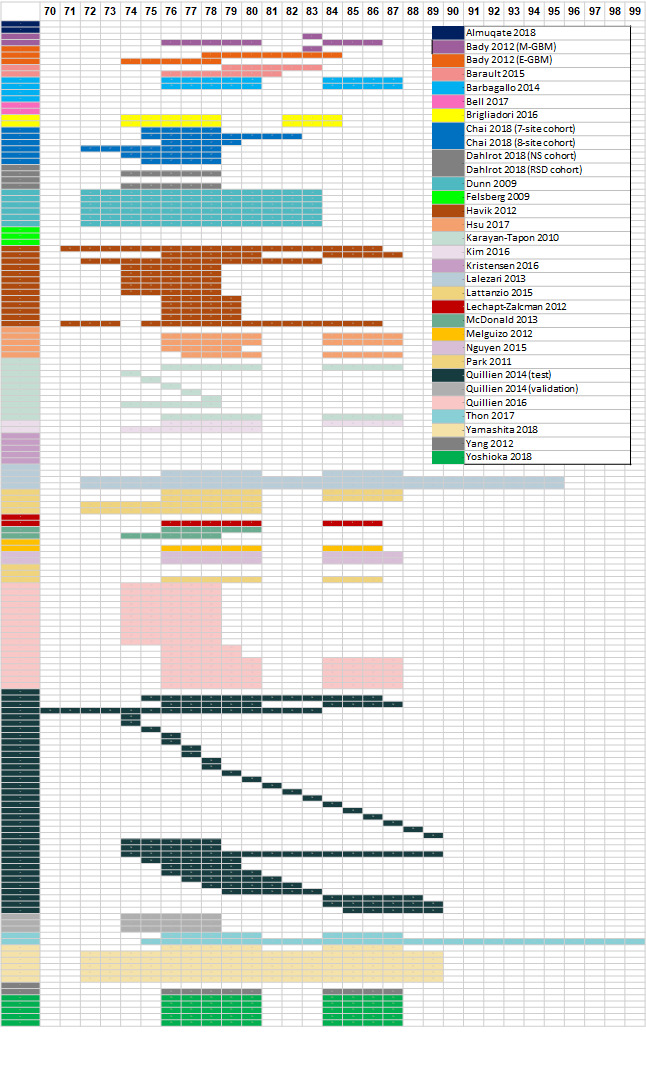
Graphical illustration of the CpG sites examined. The top row indicates the number of the CpG site. Each row is colour‐coded, corresponding to the enclosed legend indicating the study ID. Rows with blank cells (i.e. no colour‐coded CPG sites) indicate that a method was not PCR‐based test or that CpG information is not available. For studies using PCR primers as described by Esteller 1999, CpG sites location is based on Bienkowski 2015.
CpG: 5'‐cytosine‐phosphate‐guanine‐3'; GBM: glioblastoma; NS: Nordic Study; RSD: Region of Southern Denmark.
Risk of bias assessment of included studies
We present results of the risk of bias assessment for the three domains that apply to the whole studies in Figure 3. All studies were assessed at low or unclear risk of bias for participant selection (domain D1). All studies except one were assessed to be at low risk of bias arising from variation in subsequent treatment after collection of the tumour sample (D2). All studies were assessed to be at low risk of bias in measurement of the outcome (all‐cause mortality; D3). We present result‐level risk of bias judgements in forest plots of these results in subsequent sections. We were mostly free of concerns about risk of bias in the domains for study attrition (D5), problems with other prognostic factors adjusted for (D6) and selective reporting (D7). One PSQ result from Lalezari 2013 was deemed to be at high risk of bias due to attrition because more than 25% of the sample was missing from the analysis. Results for PCR‐HRM from Yamashita 2018 were considered to be at high risk of bias from selective reporting because two other primer sets were used; one was discarded because of its inferior predictive performance. Full details of the risk of bias assessments and their justifications are available in Appendix 6.
3.
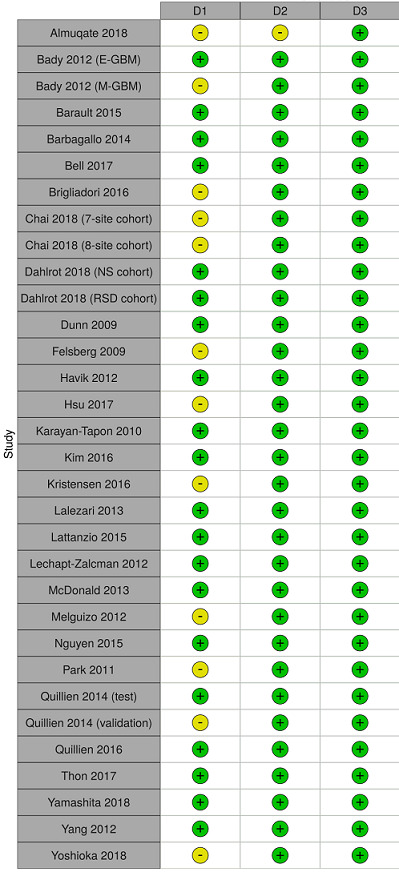
Study‐level risk of bias assessments for studies comparing two or more methods.
D1 = participant selection; D2 = subsequent treatment; D3 = outcome measurement. Green (+) = low risk of bias; yellow (–) = unclear risk of bias.
GBM: glioblastoma; NS: Nordic Study; RSD: Region of Southern Denmark.
Our assessment of the applicability of the studies to the typical clinical context found almost all the studies to be generally applicable. We had no concerns for any studies regarding applicability issues in domains of subsequent treatment (D2) or prognostic factor measurement (D4). All studies except one were free of applicability concerns in the domains of outcome measurement (D3) and other prognostic factors adjusted for (D6); in these exceptional studies we were unclear rather than concerned. In the final domain of assessment for applicability (participant selection), we had high concerns for just one study, which included only people who were treated with open resection and who received at least two cycles of chemotherapy, thus excluding people of poorer clinical condition and those who did not survive to complete two cycles of chemotherapy (Felsberg 2009). In all other studies, we had low concerns or were uncertain. Full details of the applicability assessments and their justifications are available in Appendix 7.
Findings: comparisons of different methods
We provide all of the 160 HRs we extracted in Appendix 8. In all cases, the estimated HR was above 1, indicating higher hazard of death in people with an unmethylated MGMT promoter (or MGMT protein expression on IHC). In the vast majority of cases, the lower limit of a 95% CI for the HR was above 1, confirming the prognostic value of MGMT promoter methylation status. When examining these results and the forest plots that follow, we emphasise that comparisons should only be made of different methods within studies. HRs should not be compared across studies because there were many (more substantial) differences between these results than the choice of technique, tumour sample, CpG islands or cut‐off thresholds.
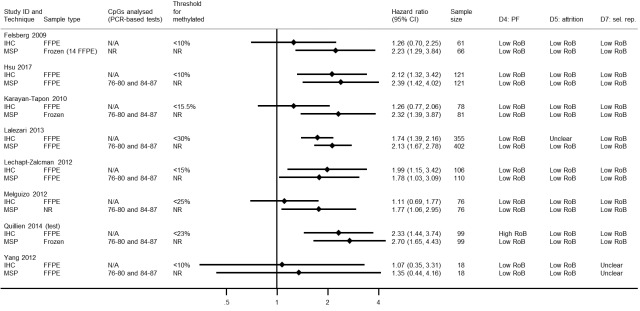
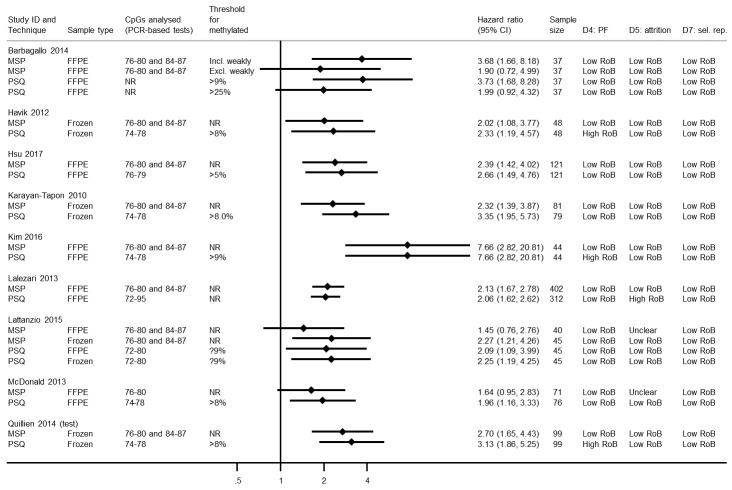
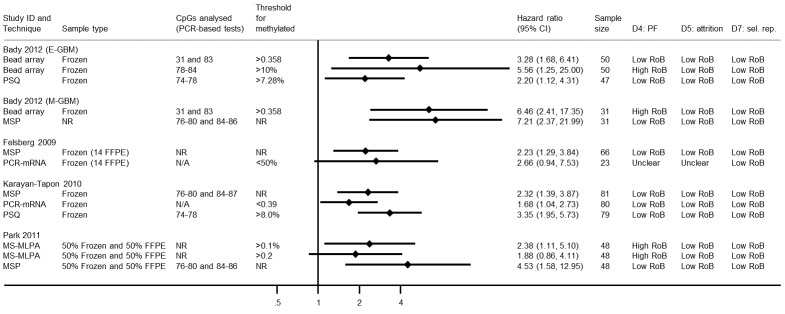
Comparison of methylation‐specific polymerase chain reaction versus immunohistochemistry
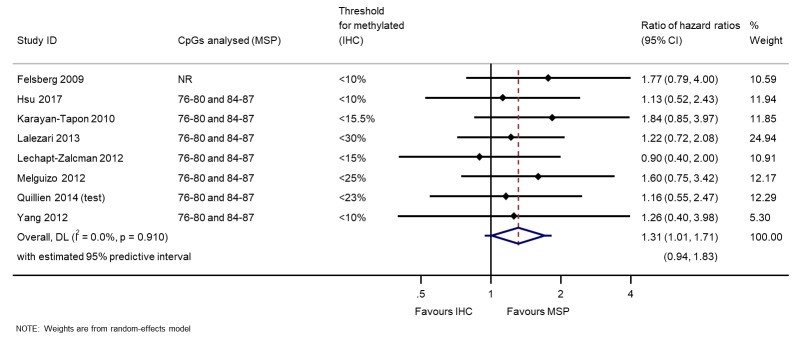
We illustrate results from eight studies that included both MSP and IHC in Figure 4. Seven reported they had targeted CpG sites 76 to 80 and 84 to 87 in MSP (the other did not report their CpG sites). Risk of bias was assessed to be low (or unclear) in all of these results except in one study where the cut‐off threshold for interpretation of IHC for the MGMT protein was based on its performance (Quillien 2014 (test)), which we assessed to be at high risk of bias. There was a tendency for MSP to produce larger HRs than IHC, suggesting that MSP provides a more discriminating predictor of time to death than IHC for MGMT protein. RHRs providing direct within‐study comparisons of MSP versus IHC for these studies are presented in Figure 5. A meta‐analysis of these gave a summary RHR of 1.31 (95% CI 1.01 to 1.71), providing some statistical support for the observation that MSP is more prognostic for overall survival than IHC for MGMT protein. There was no evidence of heterogeneity between the studies (estimated between‐study variance = 0; I2 = 0%), and a 95% prediction interval from 0.94 to 1.83 was therefore barely any wider than the CI.
4.
Hazard ratios from studies comparing MSP with IHC.
CI: confidence interval; CpG: 5'‐cytosine‐phosphate‐guanine‐3'; FFPE: formalin‐fixed paraffin embedded; HR: hazard ratio; IHC: immunohistochemistry; MSP: methylation‐specific polymerase chain reaction; N/A: not applicable; NR: not reported; PCR: polymerase chain reaction; PF: prognostic factor; RoB: risk of bias; sel. rep.: selective reporting.
5.
Meta‐analysis of ratios of hazard ratios (RHR) providing within‐study comparisons of MSP and IHC (an RHR greater than 1 indicates that MSP is more prognostic than IHC).
CpG: 5'‐cytosine‐phosphate‐guanine‐3'; CI: confidence interval; DL: DerSimonian and Laird estimation; IHC: immunohistochemistry; MSP: methylation‐specific polymerase chain reaction.
Technical notes: in the four data sets from the whole review for which we had IPD, the correlations between results on pairs of test results for the same people (categorised as methylated versus unmethylated) were 0.75 (MSP versus PSQ; McDonald 2013), 0.88 (MSP versus bead array; Bady 2012 (M‐GBM)), 0.65 (IHC versus qMSP; Hsu 2015) and –0.03 (MSP versus IHC; Yang 2012). Based on these observations, in our statistical comparison of HRs between methods within the same study, we used a value of 0.7 for the correlation, and repeated the analysis using correlations of 0.5 and 0.9 as our sensitivity analysis.
Sensitivity analysis: when we assumed a correlation of 0.5, the RHR was 1.31 (95% CI 0.93 to 1.85); when we assumed a correlation of 0.9, the RHR was 1.32 (95% CI 1.12 to 1.56). There was a small amount of heterogeneity in the latter analysis (between‐study variance = 0.008) due to the narrowing of CIs for the RHRs in individual studies.
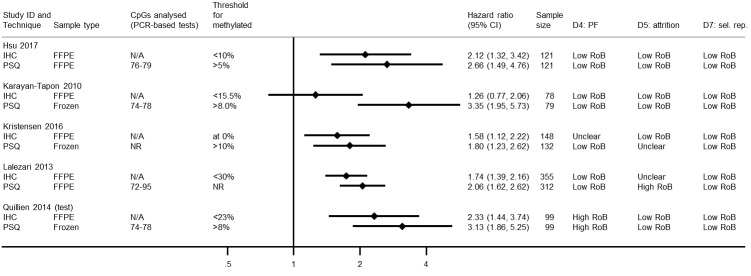
Comparison of pyrosequencing versus immunohistochemistry
We illustrate results from five studies that included both PSQ and IHC for MGMT protein in Figure 6. Risks of bias were variable, with seven of the 10 results being at low or unclear risk of bias in all domains. In one study (in which cut‐off thresholds were derived based on performance) both results were assessed to be at high risk of bias (Quillien 2014 (test)), and in another study we rated the PSQ result to be at high risk of bias in the attrition domain because it omitted more than 25% of the sample without explanation (Lalezari 2013). In all the studies, IHC had been performed using FFPE samples, but in three PSQ was performed using frozen samples.
6.
Hazard ratios from studies comparing PSQ with IHC.
CI: confidence interval; CpG: 5'‐cytosine‐phosphate‐guanine‐3'; FFPE: formalin‐fixed paraffin embedded; HR: hazard ratio; IHC: immunohistochemistry; N/A: not applicable; NR: not reported; PCR: polymerase chain reaction; PF: prognostic factor; PSQ: pyrosequencing; RoB: risk of bias; sel. rep.: selective reporting
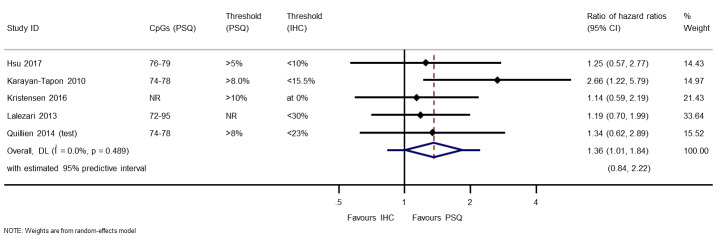
RHRs providing direct within‐study comparisons of PSQ versus IHC for these studies are presented in Figure 7. There was evidence of superiority of PSQ over IHC (RHR 1.36, 95% CI 1.01 to 1.84). There was no evidence of statistical heterogeneity (between‐study variance = 0; I2 = 0%; the wide 95% prediction interval from 0.84 to 2.22 represents uncertainty in estimation of the between‐study variance).
7.
Meta‐analysis of ratios of hazard ratios (RHR) providing within‐study comparisons of PSQ and IHC (an RHR greater than 1 indicates that PSQ is more prognostic than IHC).
CI: confidence interval; CpG: 5'‐cytosine‐phosphate‐guanine‐3'; DL: DerSimonian and Laird estimation; IHC: immunohistochemistry; NR: not reported; PSQ: pyrosequencing.
Technical notes: four studies had used more than three variants of PSQ, and all of these had examined CpG sites 74 to 78 with a cut‐off of 8%, applied to frozen tumour samples (Havik 2012; Karayan‐Tapon 2010; Quillien 2014 (test); Quillien 2016). This combination had also been used in two other studies comparing PSQ with MSP (Bady 2012 (E‐GBM); Kim 2016). Therefore, we included only this combination from these four studies in the forest plot. (Detailed comparison of PSQ variants are covered below).
Sensitivity analysis: when we assumed a correlation of 0.5, the RHR was 1.36 (95% CI 0.92 to 2.01); when we assumed a correlation of 0.9, the RHR was 1.41 (95% CI 1.06 to 1.87). Between‐study variance was estimated to be 0.06 in the latter analysis.
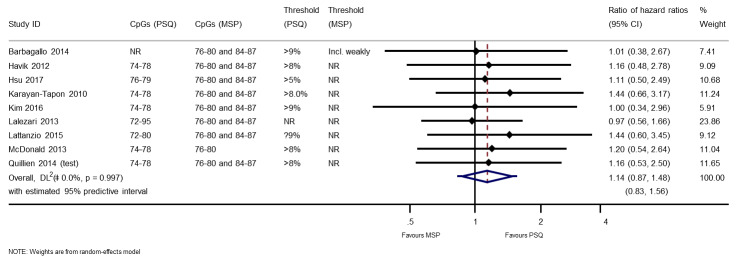
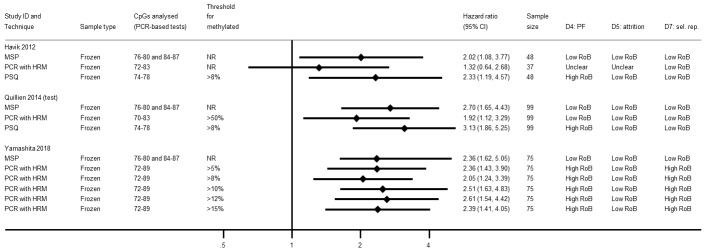
Comparison of methylation‐specific polymerase chain reaction versus pyrosequencing
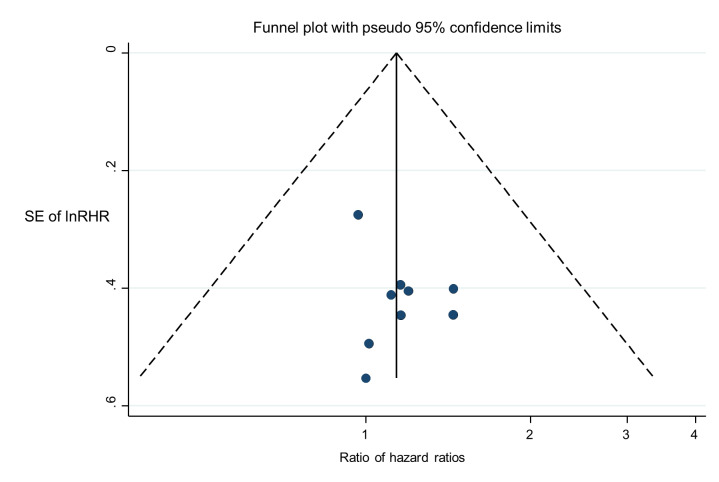
We illustrate results from nine studies that included both MSP and PSQ in Figure 8. Three of the 22 results in this plot were considered to be at high risk of bias: for Havik 2012 and Quillien 2014 (test) due to a data‐based selection of a threshold for PSQ, and for Lalezari 2013 due to missing data for PSQ. RHRs providing direct within‐study comparisons of MSP versus PSQ for these studies are presented in Figure 9. While there was a consistent pattern that PSQ seemed to be a slightly better predictor than MSP, there was not strong statistical evidence to confirm this (RHR 1.14, 95% CI 0.87 to 1.48). There was no evidence of statistical heterogeneity (between‐study variance = 0; I2 = 0%), with a 95% prediction interval for the HRH ranging from 0.83 to 1.56. No pattern could be discerned in a funnel plot of these results (Figure 10); tests for asymmetry were not undertaken.
8.
Hazard ratios from studies comparing PSQ with MSP.
CI: confidence interval; CpG: 5'‐cytosine‐phosphate‐guanine‐3'; FFPE: formalin‐fixed paraffin embedded; HR: hazard ratio; MSP: methylation‐specific polymerase chain reaction; N/A: not applicable; NR: not reported; PCR: polymerase chain reaction; PF: prognostic factor; PSQ: pyrosequencing; RoB: risk of bias; sel. rep.: selective reporting
9.
Meta‐analysis of ratios of hazard ratios (RHR) providing within‐study comparisons of PSQ and MSP (an RHR greater than 1 indicates that PSQ is more prognostic than MSP).
CI: confidence interval; CpG: 5'‐cytosine‐phosphate‐guanine‐3'; HR: hazard ratio; MSP: methylation‐specific polymerase chain reaction; NR: not reported; PSQ: pyrosequencing.
10.
Funnel plot of ratios of hazard ratios (RHR) providing within‐study comparisons of PSQ and MSP (an RHR greater than 1 indicates that PSQ is more prognostic than MSP).
MSP: methylation‐specific polymerase chain reaction; PSQ: pyrosequencing; SE: standard error.
Technical notes: we restricted these forest plots (and funnel plot) to PSQ results for CpG sites 74 to 78 with a cut‐off of 8% and frozen tumour samples for four studies (Havik 2012; Karayan‐Tapon 2010; Quillien 2014 (test); Quillien 2016). To select a single HR for each technique in each study to estimate RHRs, we applied two further rules: we selected FFPE over frozen (as it was used more often across studies), and a cut‐off for PSQ of less than 9% over less than 25% (for consistency with other studies).
Sensitivity analysis: when we assumed a correlation of 0.5, the RHR was 1.14 (95% CI 0.81 to 1.60); when we assumed a correlation of 0.9, the RHR was 1.14 (95% CI 0.98 to 1.33). Heterogeneity was estimated to be 0 in these analyses. Thus, assuming HRs for MSP and PSQ were highly correlated (0.9), the CI narrowed sufficiently to be suggestive of a difference in favour of PSQ.
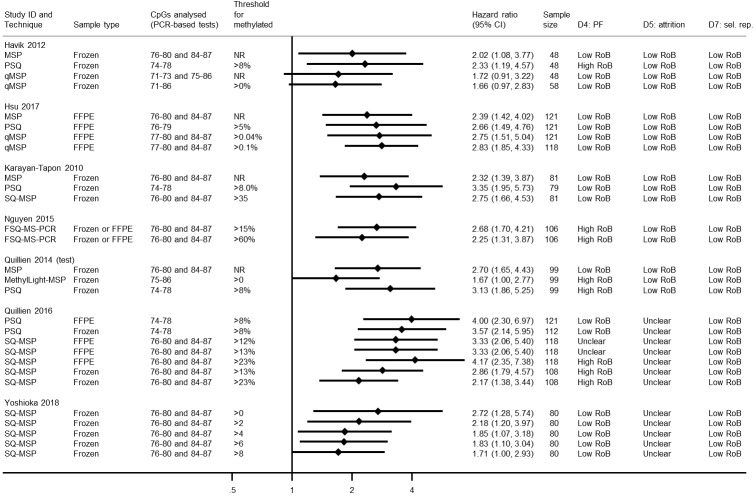
Quantitative methylation‐specific polymerase chain reaction
Figure 11 shows HRs for studies comparing different methods for qMSP, or comparing qMSP against either MSP or PSQ. The qMSP methods most commonly targeted CpG sites 76 to 80 and 84 to 87. Quillien 2016 and Yoshioka 2018 both compared SQ‐MSP, using frozen tissue samples, targeting CpGs 76 to 80 and 84 to 87 using different thresholds. Both observed that HRs were higher the lower the cut‐off point. Quillien 2016 observed the opposite when they looked also at FFPE samples. Across the studies, there was no indication that qMSP methods were superior to PSQ. The one study that looked at MethyLight‐MSP did not find it very prognostic (Quillien 2014 (test)).
11.
Hazard ratios from studies comparing different methods for qMSP, or comparing qMSP against either MSP or PSQ.
CI: confidence interval; CpG: 5'‐cytosine‐phosphate‐guanine‐3'; FFPE: formalin‐fixed paraffin embedded; FSQ‐MS‐PCR: fluorescent semi‐quantitative methylation‐specific polymerase chain reaction; HR: hazard ratio; MSP: methylation‐specific polymerase chain reaction; NR: not reported; PCR: polymerase chain reaction; PF: prognostic factor; PSQ: pyrosequencing; qMSP: quantitative methylation‐specific polymerase chain reaction; RoB: risk of bias; sel. rep.: selective reporting; SQ‐MSP: semi quantitative methylation‐specific polymerase chain reaction.
Comparing 5'‐cytosine‐phosphate‐guanine‐3' sites, thresholds and sample types
MSP was almost always studied in relation to CpG sites 76 to 80 and 84 to 87, so comparisons of CpG sites for MSP could not be made. One study compared MSP in FFPE versus frozen samples, and observed it to be more prognostic in frozen samples (Lattanzio 2015; see Figure 8). Many variants of PSQ were used across the studies. In particular, Dunn 2009 compared six threshold definitions for CpG sites 72 to 83; Havik 2012 compared five thresholds for CpGs 74 to 76 and four for CpGs 78 to 79; Karayan‐Tapon 2010 compared six CpG sites; Quillien 2014 (test) compared 32 combinations of CpG sites and thresholds; and Quillien 2016 compared two CpG sites with multiple thresholds and both FFPE and frozen tumour samples. Figure 12 illustrates the 80 HRs for PSQ variants that we extracted or computed from 11 studies.
12.
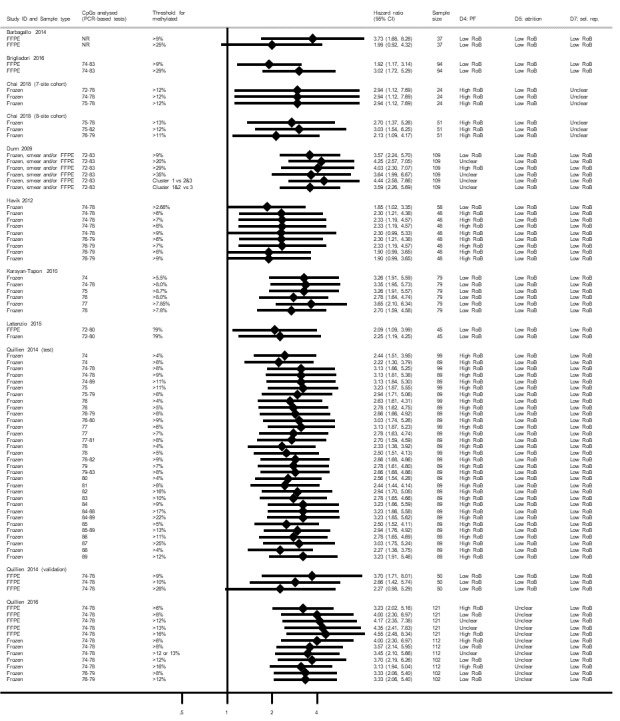
Hazard ratios from studies comparing different methods for PSQ.
CpG: 5'‐cytosine‐phosphate‐guanine‐3'; CI: confidence interval; FFPE: formalin‐fixed paraffin embedded; HR: hazard ratio; NR: not reported; PSQ: pyrosequencing; RoB: risk of bias; sel. rep.: selective reporting.
The range of CpG sites examined in studies comparing PSQ variants was from 72 to 89 (although in a comparison against IHC, Lalezari 2013 had examined sites 72 to 95). The most commonly examined CpG sites using PSQ were 74 to 78. Cut‐off thresholds used when a single CpG site was targeted ranged from 4% to 25%, and when multiple CpG sites were targeted thresholds ranged from 2.68% to 35%. In frozen tissue, Havik 2012 and Quillien 2014 (test) observed no clear dependency of HRs on threshold for CpGs 74 to 78, although Quillien 2016 observed a slight worsening as cut‐off thresholds increased from 6% to 8% to 12%. In FFPE, Quillien 2014 (validation) observed best prognostic value using a 9% threshold, followed by 10%, followed by 28% (a validation study with low risk of bias), although Brigliadori 2016 (a good‐quality study) observed the opposite when comparing 9% with 29%. In a third study comparing thresholds that was free of high risks of bias, findings echoed Quillien 2014 (validation), with a threshold of 9% more prognostic than a threshold of 25%, although it was unclear what CpG islands had been targeted (Barbagallo 2014).
Quillien 2014 (test) examined a large variety of combinations of CpG sites and thresholds. Most of their highest HRs were observed for scenarios in which multiple CpG sites were targeted. There was no clear difference apparent between using PSQ on FFPE versus frozen tissue (Lattanzio 2015; Quillien 2016).
Other techniques
Bead array, PCR‐mRNA and MS‐MLPA
Results for studies that had examined bead array (Bady 2012 (E‐GBM); Bady 2012 (M‐GBM)), PCR‐mRNA (Felsberg 2009; Karayan‐Tapon 2010) or MS‐MLPA (Park 2011) are illustrated in Figure 13. These had mostly been examined in frozen samples. One study compared two approaches to bead array with one approach to PSQ and found the highest HR for one of the bead array methods (CpG sites 76 to 84, using a cut‐off threshold of 10%) with an HR of 5.56 (compared with 2.20 for PSQ). However, this particular bead array result was assessed to be at high risk of bias due to selection of the threshold. Across the evidence base, there was no clear evidence that bead array or PCR for mRNA expression differed in any consistent direction from MSP or PSQ. The only study to report MS‐MLPA found it to be less prognostic than MSP (Park 2011).
13.
Hazard ratios from studies comparing bead array, mRNA or MS‐MLPA against either MSP or PSQ.
CI: confidence interval; CpG: 5'‐cytosine‐phosphate‐guanine‐3'; FFPE: formalin‐fixed paraffin embedded; HR: hazard ratio; mRNA: messenger ribonucleic acid; MS‐MLPA: methylation‐specific multiplex ligation‐dependent probe amplification; MSP: methylation‐specific polymerase chain reaction; NR: not reported; PCR: polymerase chain reaction; PF: prognostic factor; PSQ: pyrosequencing; qMSP: quantitative methylation‐specific polymerase chain reaction; RoB: risk of bias; sel. rep.: selective reporting; SQ‐MSP: semi quantitative methylation‐specific polymerase chain reaction.
PCR‐HRM
Results for three studies that had examined PCR‐HRM against either MSP or PSQ (mostly in frozen samples) are presented in Figure 14. Two studies observed it to be less prognostic than MSP (Havik 2012; Quillien 2014 (test)), although a third study obtained HRs for PCR‐HRM using different thresholds that fell both sides of a result for MSP, although MSP had targeted different CpG sites (Yamashita 2018).
14.
Hazard ratios from studies comparing PCR‐HRM against either MSP or PSQ.
PCR‐HRM: polymerase chain reaction with high‐resolution melting; MSP: methylation‐specific polymerase chain reaction; PSQ: pyrosequencing; FFPE: formalin‐fixed paraffin embedded; NR: not reported; N/A: not applicable; CpG: 5'‐cytosine‐phosphate‐guanine‐3'; CI: confidence interval. RoB: risk of bias; sel. rep.: selective reporting; HRs: hazard ratios.
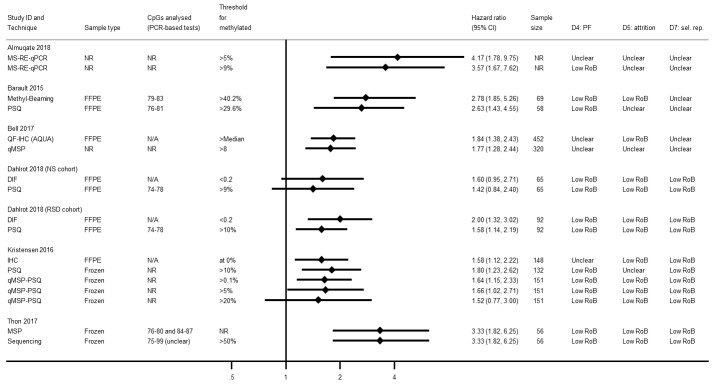
Further techniques
Figure 15 provides HRs for other techniques. Two linked studies (at low risk of bias) examined DIF and found it provided a better prognostic factor than PSQ targeting CpG sites 74 to 78 in FFPE (Dahlrot 2018 (NS cohort); Dahlrot 2018 (RSD cohort)).Thon 2017 observed no difference between PSQ and sequencing.
15.
Hazard ratios from of studies involving other techniques not included in previous forest plots.
CI: confidence interval; CpG: 5'‐cytosine‐phosphate‐guanine‐3'; DIF: double immunofluorescence; FFPE: formalin‐fixed paraffin embedded; HR: hazard ratio; IHC: immunohistochemistry; MS‐RE‐qPCR: methylation‐specific restriction enzyme quantitative polymerase chain reaction; MSP: methylation‐specific polymerase chain reaction; PSQ: pyrosequencing; QF‐IHC: quantitative fluorescence immunohistochemistry; qMSP‐PSQ: quantitative methylation‐specific polymerase chain reaction; NR: not reported; N/A: not applicable; RoB: risk of bias; sel. rep.: selective reporting.
Other prognostic variables
Seven studies provided both unadjusted and adjusted results for at least one method, either because these were both reported in the publication or because we could compute them from IPD. We only extracted results adjusted for age and extent of resection, or the closest to this that was available. A comparison of 15 instances of unadjusted and adjusted results for these studies is provided in Table 5. Most of these adjusted for age, but we were able to adjust additionally for resection in only one study. Two studies (six results) adjusted for Karnofsky performance status. Results were generally very similar across unadjusted and adjusted analyses, demonstrating that MGMT promoter methylation status remains prognostic after accounting for these other factors.
4. Impact of adjustment for other prognostic factors.
| Study ID | Technique | Sample type | CpGs analysed (PCR‐based tests) | Threshold for methylated | Unadjusted HR (95% CI) | Adjusted HR (95% CI) | Factors adjusted for (if an adjusted HR reported) | Source of results | Risk of bias (Domain 6) |
| Bady 2012 (M‐GBM) | Bead array | Frozen | 31 and 83 | > 0.358 | 6.46 (2.41 to 17.35) | 6.51 (2.42 to 17.54) | Age | IPD | Low |
| MSP | NR | 76–80 and 84–86 | NR | 7.21 (2.37 to 21.99) | 7.38 (2.41 to 22.60) | Age | IPD | Low | |
| Dahlrot 2018 (NS cohort) | DIF | FFPE | N/A | < 0.2 | 1.60 (0.95 to 2.71) | 1.50 (0.93 to 2.43) | Age, ECOG performance status and gender | Directly reported | Low |
| Hsu 2015 | IHC | FFPE | N/A | < 10% | 2.12 (1.32 to 3.42) | 1.80 (1.01 to 3.21) | Age, sex, KPS, extent of resection, bevacizumab treatment and IDH1 status | Directly reported | Low |
| MSP | FFPE | 76–80 and 84–87 | NR | 2.39 (1.42 to 4.02) | 2.62 (1.50 to 4.55) | Age, sex, KPS, extent of resection, bevacizumab treatment and IDH1 status | Directly reported | Low | |
| PSQ | FFPE | 76–79 | > 5% | 2.66 (1.49 to 4.76) | 2.51 (1.46 to 4.33) | Age, sex, KPS, extent of resection, bevacizumab treatment and IDH1 status | Directly reported | Low | |
| qMSP | FFPE | 77–80 and 84–87 | > 0.04% | 2.75 (1.51 to 5.04) | 2.65 (1.47 to 4.76) | Age, sex, KPS, extent of resection, bevacizumab treatment and IDH1 status | Directly reported | Low | |
| McDonald 2013 | MSP | FFPE | 76–80 | NR | 1.64 (0.95 to 2.83) | 1.63 (0.95 to 2.81) | Age | IPD | Low |
| PSQ | FFPE | 74–78 | > 8% | 1.96 (1.16 to 3.33) | 1.68 (0.99 to 2.84) | Age | Directly reported | Low | |
| Thon 2017 | MSP | Frozen | 76–80 and 84–87 | NR | 3.33 (1.82 to 6.25) | 3.23 (1.72 to 6.25) | Age, RTOG and KPS | Directly reported | Low |
| Sequencing | Frozen | 75–99 (unclear) | > 50% | 3.33 (1.82 to 6.25) | 3.23 (1.72 to 6.25) | Age, RTOG and KPS | Directly reported | Low | |
| Yamashita 2018 | MSP | Frozen | 76–80 and 84–87 | NR | 2.36 (1.62 to 5.05) | 1.63 (0.86 to 3.09) | Surgery (gross‐total resection vs other) and MGMT status by PCR‐HRM | Directly reported | High (model includes other MGMT status using alternative method) |
| PCR‐HRM | Frozen | 72–89 | > 10% | 2.51 (1.63 to 4.83) | 2.36 (1.20 to 4.65) | Surgery (gross‐total resection vs other) and MGMT status by MSP | Directly reported | High (model includes other MGMT status using alternative method) | |
| Yang 2012 | IHC | FFPE | N/A | < 10% | 1.07 (0.35 to 3.31) | 1.50 (0.37 to 6.04) | Age and extent of resection | IPD | Low |
| MSP | FFPE | 76–80 and 84–87 | NR | 1.35 (0.44 to 4.16) | 0.99 (0.29 to 3.45) | Age and extent of resection | IPD | Low |
CpG: 5'‐cytosine‐phosphate‐guanine‐3'; CI: confidence interval; DIF: double immunofluorescence; ECOG: Eastern Cooperative Oncology Group; FFPE: formalin‐fixed paraffin embedded; HR: hazard ratios; IDH: isocitrate dehydrogenase; IHC: immunohistochemistry; IPD: individual participant data; KPS: Karnofsky performance status; MGMT: O6‐methylguanine–DNA methyltransferase; MSP: methylation‐specific polymerase chain reaction; N/A: not applicable; NR: not reported; PCR: polymerase chain reaction; PCR‐HRM: polymerase chain reaction with high‐resolution melting; PSQ: pyrosequencing; qMSP: quantitative methylation‐specific polymerase chain reaction; RTOG: Radiation Therapy Oncology Group prognostic factor class.
We did not perform formal analyses to investigate whether heterogeneity in HRs may have been due to age, extent of tumour resection, Karnofsky performance status, IDH status, recurrent versus first diagnosis, length of follow‐up time of start of follow‐up, due to the very limited replication of specific methods and large amounts of missing data for many of these study characteristics.
Studies examining only a single method
Table 6 provides details of the 190 articles describing studies that presented HRs from survival analysis in people in which MGMT methylation was measured by one method, and studies that used more than one method but only MGMT methylation data from one method were used in the survival analysis.
5. Details of articles describing a single method.
| Author (year) | Country of population/cohort | Length of follow‐up (median in months (range)) | Number of participants | Tumour type | IDH mutation status (% of WT) | Technique used to assess MGMT methylation status |
| Abhinav 2013 | UK | NR | 19 | GBM | NR | MSP |
| Adeberg 2015 | Germany | NR | 32 | GBM | NR | MSP |
| Ahmed 2015 | US | 11.4 | 214 | GBM | NR | mRNA levels |
| Alonso 2017 | Spain | NR | 63 | GBM: 97.2%; gliosarcoma: 2.8% | 88.7 (data from 63/71 people) | MSP |
| Appin 2013 | US | NR | 236 | GBM: primary: 89% (GBM with oligodendroglioma: 13.3%); secondary: 11% | GBM: 91.4 (data from 116/208 people); GBM‐O: 65 (data from 20/28 people) | MSP |
| Ardon 2012 | Belgium | 25.0 (10.5–42.2) | 77 | GBM (primary) | NR | MSP |
| Arita 2016 | Japan | NDR | 193 | GBM | 100 | PSQ |
| Badruddoja 2017 | US | NR | 30 | GBM | NR | qMSP |
| Balana 2016 | Spain | NR | 93 | GBM | NR | MSP |
| Balana 2017 | Spain | 17.0 (10.7–24.5) | 256 | GBM | 94.5 (data from 162/256 people) | MSP |
| Blumenthal 2017 | US | NR | 1395 | GBM | NR | qMSP |
| Boots‐Sprenger 2013 | The Netherlands | NR | 333 | GBM | 84 (data from 226/333 people) | MS‐MLPA |
| Brandes 2008 | Italy | 18.93 (6.6–62) | 208 | GBM | NR | MSP |
| Brandes 2009 | Italy | NR | 37 | GBM | NR | MSP |
| Brandes 2010 | Italy | NR | 44 | GBM | NR | MSP |
| Brandes 2014 | Italy | NR | 116 | GBM | NR | MSP |
| Brandes 2017 | Italy | NR | 108 | GBM | NR | MSP |
| Brennan 2013 | US | NR | 332 | GBM | 93.4 (data from 423/543 people) | Bead array |
| Burford 2013 | UK | NR | NDR | GBM | NDR | MSP |
| Burger 2017 | Germany | NR | 32 | GBM | NR | MSP |
| Butowski 2011 | US | NR | 66 | GBM and gliosarcoma | NR | MSP |
| Capellades 2018 | Spain | NR | 292 | GBM | 96.6 | MSP |
| Chakhoyan 2018 | US | NR | 23 | GBM | NR | MSP |
| Chen 2015 | China | 12.8 (4.0–37.7) | 78 | GBM | NR | IHC |
| Chen 2016 | China | NR | 300 | GBM | 85.7 | Bead array |
| Cheng 2015 | China, US | NR | 285 (CGGA: 55; TCGA: 235) | GBM | NR | Bead array (TCGA); PSQ (CGGA) |
| Chinot 2007 | France | 6 (0.9–19) | 29 | GBM | NR | IHC |
| Choi 2016 | South Korea | NR | 112 (training cohort: 74; test cohort 38) | GBM | NR | MSP |
| Clarke 2009 | US | 18.8 | 85 | GBM | NR | MSP |
| Coburger 2017 | Germany | 40 (37–43) | 170 | GBM | NR | MSP |
| Colman 2010 | US | NR | 101 | GBM | NR | qMSP |
| Combs 2011 | Germany | NR | 160 | GBM (primary) | 97.1 (data from 140/160 people) | MSP |
| Cominelli 2015 | Italy | NR | 70 | GBM | 95.7 | MSP |
| Costa 2010 | Portugal | NR | 90 | GBM (primary) | NR | MSP |
| Criniere 2007 | France | 57.2 | 77 | GBM | NR | MSP |
| Dahlrot 2017 | Denmark | NR | 226 | GBM (primary) | NR | PSQ |
| Das 2011 | India | NR | 6 | GBM | NR | MSP |
| Etcheverry 2014 | France | 15.5 | 399 | GBM (primary) | 91 | PSQ |
| Felsberg 2011 | Germany | 48.6 | 64 | GBM | NR | MSP |
| Fiano 2014 | Italy | NR | 32 | GBM | NR | MSP |
| Fontana 2016 | Italy | NDR | 128 | GBM | NDR | PSQ |
| Franceschi 2016 | Italy | NR | 21 | GBM | NR | MSP |
| Franceschi 2018 | Italy | NR | 169 | GBM | NR | MSP |
| Galldiks 2015 | Germany | NR | 21 | GBM | NR | MSP |
| Gallego Perez‐Larraya 2011 | France | NR | 31 | GBM | NR | qMSP |
| Gilbert 2013 | Canada, European multicentres, US | NR | 762 | GBM | NR | qMSP |
| Gilbert 2014 | Canada, European multicentres, US | NR | 637 | GBM | NR | qMSP |
| Gittleman 2017 | US | NR | 799 | GBM | NR | qMSP |
| Glas 2009 | Germany | 41.5 | 23 | GBM | NR | MSP |
| Gorlia 2008 | Belgium, Canada, Italy, Germany, Switzerland, the Netherlands | NR | 287 | GBM | NR | MSP |
| Gramatzki 2016 | Switzerland | 9 | 108 | GBM | 100 | MSP |
| Gutenberg 2013a | Germany | NR | 17 | GBM (primary) | NR | MSP |
| Gutenberg 2013b | Germany | 16.2 (1.4–54.1) | 191 | GBM (primary) | NR | MSP |
| Ha 2013 | South Korea | NR | 10 | GBM | 75 | qMSP |
| Haemmig 2014 | Switzerland | NR | 60 | GBM | 85 | qMSP |
| Han 2014 | US | NR | 28 | GBM | NR | MSP |
| Han 2015a | China | 13.7 (1–43) | 152 | GBM | NR | MSP |
| Han 2015b | China | 13.7 (1–43) | 79 | GBM | 93.5 (data from 214 people) | MS‐MLPA |
| Happold 2018 | European multicentres | 29 (25–35; CENTRIC cohort) | 797 | GBM | NR | qMSP |
| Hayes 2015 | US | 14.1 | 219 | GBM | 94.5 (data from 475 people) | Bead array |
| Hegi 2004 | Switzerland | NR | 38 | GBM | NR | MSP |
| Hegi 2005 | Canada, European multicentres | NR | 106 | GBM | NR | MSP |
| Herrlinger 2006 | Germany | NR | 31 | GBM | NR | MSP |
| Herrlinger 2009 | Germany | 41.5 | 31 | GBM | NR | MSP |
| Hervouet 2009 | France | NR | 53 | GBM | NR | MSP |
| Hobbs 2012 | US | 19.1 (8.1–74.6; survivors only) | 312 | GBM | NR | qMSP |
| Huang 2017 | US | NR | 301 | GBM | NR | mRNA levels |
| Hudson 2018 | Australia | NR | 16 | GBM | NR | PSQ |
| Inoges 2017 | Spain | NR | 31 | GBM | NR | MSP |
| Ishida 2015 | Japan | NR | 46 | GBM | NR | IHC |
| Ishikawa 2014 | Japan | 19.6 (7.3–48.7) | 23 | GBM | 75 (data from the whole cohort) | IHC |
| Ius 2018 | Italy | NR | 116 | GBM | NR | PSQ |
| Iwadate 2017 | Japan | NR | 70 | GBM | 92 | IHC |
| Jan 2018 | Taiwan | NR | NDR | GBM | NDR | MSP |
| Karim 2012 | Egypt | NR | 34 | GBM | NR | MSP |
| Kessler 2018 | Germany, US | NR | 404 (Heidelberg cohort: 143; TCGA: 261) | GBM | 100 | Bead array |
| Kim 2012 | South Korea | 22 (3–88) | 93 | GBM | NR | MSP |
| Kim 2017 | South Korea | 16.3 (0.3–105.1) | 750 | GBM | NR | MSP |
| Kim 2018 | South Korea | NR | 93 | GBM | 86.8 (data from 91 people) | qMSP |
| Klitkou 2014a | Denmark | NR | 173 | GBM (primary) | NR | IHC |
| Klitkou 2014b | Denmark | NR | 173 | GBM (primary) | NR | IHC |
| Klitkou 2014c | Denmark | NR | 173 | GBM (primary) | NR | IHC |
| Kong 2011 | South Korea | 16.5 (6.2–48) | 90 | GBM | NR | MSP |
| Kreth 2013 | Germany | NR | 222 | GBM | NR | MSP |
| Lakomy 2011 | Czech Republic | NR | 38 | GBM | NR | PCR‐HRM |
| Laxton 2013 | UK | NR | 288 | GBM | 95.3 (data from 107/288 people) | MSP |
| Lee 2013 | South Korea | 15 | 36 | GBM | NR | qMSP |
| Lee 2017 | South Korea | NR | 65 | GBM | 80 | MSP |
| Li 2016a | China | NR | 145 | GBM | 83.4 | MSP |
| Li 2016b | China | NR | 50 | GBM (primary) | 85.9 (data from 50/78 people) | PSQ |
| Lombardi 2015 | Italy | NR | 151 | GBM | 94 (data from 100/237 people) | MSP and PSQ |
| Lombardi 2017 | Italy | NR | 128 | GBM | NR | PSQ |
| Ma 2016 | China | NR | 56 | GBM (primary) | NR | MSP |
| Majewska 2017 | United Kingdom | NR | 99 | GBM | NR | PSQ |
| Malmström 2012 | Austria, Denmark, France, Norway, Sweden, Switzerland, Turkey | NR | 72 | GBM | 99.7 (data from 291 people) | qMSP |
| Malmström 2017 | Denmark, Finland, Norway, Sweden | 20 | 78 | GBM | 96.10% | PSQ |
| Martini 2008 | Italy | NR | 46 | GBM | NR | MSP |
| McDonald 2015 | Australia | NR | 33 | GBM | 93.90% | PSQ |
| Metellus 2011 | France | 18.9 | 61 | GBM | NR | qMSP |
| Meyronet 2017 | Austria, Denmark, France, Norway, Sweden, Switzerland, Turkey | NR | 6 | GBM | 100 | PSQ |
| Michaelsen 2013 | Denmark | 60 (23–92) | 163 | GBM | NR | IHC |
| Michaelsen 2018 | Denmark | NR | 415 | GBM | 100 | mRNA levels |
| Minniti 2011a | Italy | NR | 36 | GBM (recurrent) | NR | MSP |
| Minniti 2011b | Italy | NR | 83 | GBM | NR | MSP |
| Minniti 2015 | Italy | 24.0 (standard RT + TMZ group); 22.5 (short course RT + TMZ group) | 243 | GBM | NR | MSP |
| Miyazaki 2014 | Japan | NR | 117 | GBM: 83.8%; GBM with oligodendroglioma: 16.2% | 93.4 | IHC |
| Montano 2011 | Italy | NR | 73 | GBM (primary) | NR | MSP |
| Morandi 2010 | Italy | NR | 159 | GBM | NR | qMSP |
| Motomura 2011 | Japan | 16.7 (3.4–46.7) | 68 | GBM (primary) | 94.1 | PSQ |
| Mur 2015 | Spain | NR | 68 | GBM | 70.6 (unknown in 25%) | Bead array |
| Nabors 2012 | US | NR | 69 | GBM | NR | qMSP |
| Nagane 2007 | Japan | 7.1 (2.4–16.7) | 19 | GBM (recurrent) | NR | Western blot analysis |
| Ohka 2011 | Japan | NR | 51 | GBM (primary) | 94 | PSQ |
| Ohno 2013 | Japan | NR | 85 | GBM | NR | PSQ |
| Ohno 2016 | Japan | NR | 112 | GBM | 92 | PSQ |
| Omuro 2014 | US | 42 | 40 | GBM | 100 | qMSP |
| Pallini 2008 | Italy | NR | 44 | GBM | NR | MSP |
| Pambuku 2016 | Italy | NR | 128 | GBM | NR | PSQ |
| Park 2013 | South Korea | NR | 75 | GBM | NR | MSP |
| Pei 2013 | China | NR | 54 | GBM | NR | MSP |
| Picart 2018 | France | NR | 14 | GBM (cerebellar) | 100 | PSQ |
| Poulsen 2017 | Denmark | 14 | 146 | GBM | 98 | IHC |
| Prados 2009 | US | 33.7 | 65 | GBM and gliosarcoma | NR | MSP |
| Purkait 2016 | India | NR | 114 | GBM | 93.3 | MSP |
| Qi 2012 | China | NR | 86 | GBM (secondary) | 26.6 (data from 79 people) | MSP |
| Rankeillor 2014 | UK | NR | 29 | GBM (primary) | NR | MS‐MLPA |
| Rapkins 2015 | Australia, US | NR | 319 (AGOG cohort: 160; UCLA cohort: 159) | GBM | NR | MSP (UCLA); PSQ (AGOG) |
| Rapp 2013 | Germany | NR | 85 | GBM (primary) | NR | MSP |
| Reifenberger 2012 | Germany | 29 | 104 | GBM | NR | MSP |
| Roh 2017 | South Korea | 20.8 | 252 | GBM: 84.5%; giant‐cells GBM: 4.0%; GBM with oligodendroglioma: 11.5% | 93.4 (data from 106/252 people) | MSP |
| Romano 2013 | Italy | NR | 47 | GBM | NR | MSP |
| Rosati 2013 | Italy | 11.5 (1.5–58) | 83 | GBM: 95.2%; gliosarcoma: 2.4%; GBM with oligodendroglia: 2.4% | 97.6 | MSP |
| Rosenschold 2019 | Denmark | NR | 412 | GBM (primary) | NR | IHC |
| Rubio Fernandez 2014 | Spain | NR | 65 | GBM | NR | MSP |
| Sadones 2009 | Belgium | NR | 32 | GBM (recurrent) | NR | qMSP |
| Saito 2017a | Japan | NR | 53 | GBM (supratentorial) | 49 (unknown in 49%) | IHC |
| Saito 2017b | Japan | NR | 36 | GBM (supratentorial) | NR | IHC |
| Saito 2018a | Japan | NR | 102 | GBM (supratentorial) | 75.5 | IHC |
| Saito 2018b | Japan | NR | 50 | GBM (supratentorial) | 100 | IHC |
| Saito 2018c | Japan | NR | 50 | GBM (supratentorial) | NR | IHC |
| Salvati 2012 | Italy | NR | 105 | GBM (primary supratentorial) | NR | MSP |
| Sana 2014 | Czech Republic | NR | 58 | GBM (primary) | NR | PCR‐HRM |
| Saraiva‐Esperon 2014 | Spain | NR | 25 | GBM | NR | MSP |
| Sasaki 2018 | Japan | NR | 101 | GBM: 99%; gliosarcoma: 1% | 99 | qMSP |
| Schaich 2009 | Germany | NR | 64 | GBM (supratentorial) | NR | MSP |
| Schiffgens 2016 | Germany, Italy | NR | 225 (Hannover cohort 1: 120); Bologna cohort: 105) | GBM (primary) | 91.4 (data from the Bologna cohort) | MSP |
| Schulze Heuling 2017 | Germany, US | NR | 275 | GBM | 100 | Bead array |
| Shu 2018 | China | NR | 265 | GBM (primary) | 100 | PSQ |
| Sijben 2008 | Canada | NR | 16 | GBM (supratentorial) | NR | MSP |
| Singh 2012 | India | 13.15 (1.5–46) | 16 | Gliosarcoma | NR | MSP |
| Soike 2018 | US | NR | 74 | GBM | 89.2 | IHC |
| Stetson 2016 | US | NR | 203 (training cohort: 102; validation cohort: 101) | GBM | Training cohort: 75 (unknown: 21%); validation cohort: 77 (unknown: 17%) | Bead array |
| Stummer 2012 | Germany | 24 | 143 | GBM | NR | MSP |
| Stupp 2009 | Canada, European multicentres | 61 (0.36–79) | 287 | GBM | NR | MSP |
| Stupp 2010 | Germany, Switzerland | 34 | 52 | GBM | NR | MSP |
| Suchorska 2015 | Germany | NR | 79 | GBM (supratentorial) | NR | MSP |
| Tanaka 2014 | Japan | NR | 45 | GBM | NR | mRNA levels |
| Thon 2011 | Germany | 11 (5–33) | 56 | GBM | NR | MSP |
| Tini 2015 | Italy | NR | 144 | GBM | NR | MSP |
| Tini 2016 | Italy | 12 (6–84) | 169 | GBM | 100 | MSP |
| Tini 2017 | Italy | NR | 222 | GBM | NR | MSP |
| Toms 2018 | Multicentre (North America, Europe, South Korea, Israel) | NR | 466 | GBM (supratentorial) | NR | MSP |
| Trabelsi 2016 | Tunisia | NR | 20 | GBM | NR | MS‐MLPA |
| Urbschat 2017 | Germany | NR | 72 | GBM | NR | MSP |
| van Dijken 2019 | China, Taiwan, the Netherland, UK | NR | 50 | GBM | 84 (missing in 8%) | PSQ |
| Villani 2015 | Italy | 12 (3–27) | 51 | GBM (primary) | NR | PSQ |
| Wang 2014 | China | 47 (6–65) | 78 | GBM | 52.6 | MSP |
| Wang 2015a | China | NR | 216 | GBM | 48.1 | MSP |
| Wang 2015b | Multicentre (North America and Europe) | 31.9 (0.2–53.3) | 831 | GBM | NR | qMSP |
| Wang 2016 | China, US | NR | 84 (CGGA: 21; TCGA: 63) | GBM | NR | Bead array (TCGA); PSQ (CGGA) |
| Watanabe 2011 | Japan | 15.4 | 41 | GBM | NR | IHC |
| Wee 2017 | South Korea | 20.5 | 340 | GBM | 93.8 | MSP |
| Wee 2018 | Japan, South Korea | 18.4 | 692 | GBM | 92.3 | MSP |
| Wei 2017 | Taiwan | NR | 25 | GBM | NR | PSQ |
| Weller 2015 | Austria, Germany, Switzerland | NR | 105 | GBM (recurrent) | NR | MSP |
| Weller 2017b | Multicentre (165 hospitals in 22 countries) | NR | 745 | GBM (EGFRvIII‐expressing) | NR | MSP |
| Weller 2009 | Germany | 29.4 (data from the whole cohort) | 189 | GBM: 96.7; giant cells GBM: 2.6%; gliosarcoma: 0.75 | 94.4 (data from the whole cohort) | MSP |
| Westphal 2015 | Germany | NR | 66 | GBM | NR | PSQ |
| Wu 2018 | US | NR | 285 | GBM | NR | Bead array |
| Yan 2017 | UK | NR | 31 | GBM | 90.3 | PSQ |
| Yang 2015 | China | NR | 229 | GBM | 76.3 (data from 274 people; unknown in 3.3%) | PSQ |
| Yin 2017 | France | NR | 106 | GBM | NR | Bead array |
| Yin 2018 | France | 53 (8–113; Rennes and Angers datasets) | 129 | GBM | NR | Bead array |
| You 2013 | Taiwan | NR | 32 | GBM | NR | qMSP |
| Younis 2016 | Egypt | 13 (9–27) | 73 | GBM | NR | IHC |
| Yuan 2017a | China | NR | 84 | GBM | NR | PSQ |
| Yuan 2017b | China | NR | 48 | GBM | NR | PSQ |
| Yue 2014 | China | 17.5 (3–77) | 62 | GBM | NR | IHC |
| Zhang 2014 | China | NR | 80 | GBM | NR | PCR‐HRM |
| Zunarelli 2011 | Italy | 10.9 | 46 | GBM (primary) | NR | MSP |
EGFRvIII: epidermal growth factor receptor variant 3; GBM: glioblastoma; IDH: isocitrate dehydrogenase; IHC: immunohistochemistry; MGMT: O6‐methylguanine–DNA methyltransferase; mRNA: messenger ribonucleic acid; MS‐MLPA: methylation‐specific multiplex ligation‐dependent probe amplification; MSP: methylation‐specific polymerase chain reaction; NDR: not directly reported; NR: not reported; PCR‐HRM: polymerase chain reaction with high‐resolution melting; PSQ: pyrosequencing; qMSP: quantitative methylation‐specific polymerase chain reaction; RT: radiotherapy; TMZ: temozolomide; WT: wild‐type.
Of the 190 articles identified, 29 described studies conducted in Italy, 21 in Germany; 20 in the US; 18 in Japan; 17 in China; 11 in South Korea; nine in France; eight each in Denmark and Spain; six in the UK; three each in India, Switzerland and Taiwan; two each in Australia, Belgium, Czech Republic and Egypt; and one each in Canada, Portugal, the Netherlands and Tunisia. In addition, 23 articles described studies performed in more than one country.
Fifty‐four articles reported information about follow‐up, as median in months, with a minimum median of six months and a maximum of 61 months. Of these 54 articles, 29 reported data on the range of follow‐up, with a minimum follow‐up of 0.2 months and a maximum of 113 months. The total number of participants among the 190 articles was 27,899, with the smallest study of six and the largest study of 1395 participants. For two articles, data on number of participants were not directly reported but IPD were included.
Sixty‐two articles reported data on IDH‐1 and IDH‐2 mutations: in 11 articles, people were 100% wild‐type (i.e. no mutation), while 47 articles reported presence of IDH mutations, with mutation proportions ranging between 0.3% and 73.4% (in one study of secondary glioblastoma). Three articles did not directly report data on IDH mutations. With regard to type of tumour, 183 articles described studies only on glioblastoma, six on mixed glioblastoma and gliosarcoma, and one on gliosarcoma only.
Ninety‐four articles used MSP to measure MGMT methylation, 27 used PSQ, 22 used qMSP (real‐time PCR or MethyLight), 10 used bead array, four used MS‐MLPA and three used PCR‐HRM. In addition, 21 articles measured MGMT protein by IHC and one article by Western blotting (WB), and in four articles measured mRNA levels. Four articles reporting data from two cohorts, used a different method for each cohort.
As anticipated in our protocol (McAleenan 2019), we did not examine the results of these studies, because comparisons of HRs across studies would not provide reliable indicators of differences between the methods.
Findings: economic issues
We identified cost estimates for the three main techniques identified in the review: PSQ, MSP and IHC. These costs are expressed in 2020 Pounds Sterling (GBP). Where necessary, costs were converted using the EPPI‐Centre Cost Converter (Shemilt 2019). Costs were inclusive of staff, consumables, equipment and overheads associated with preparing the sample, running the analysis and feeding back findings.
We estimated PSQ costs from personal correspondence with H Liu, Consultant Clinical Scientist, Molecular Malignancy Laboratory, Haematopathology and Oncology Diagnostic Service (HODS), Addenbrooke's Hospital, Cambridge University Hospitals NHS Foundation Trust, UK (29 October 2020). The costs were estimated to be GBP 325 per sample. Costs for IHC were estimated from a Health Technology Assessment testing for Lynch syndrome (Snowsill 2017). The costs were GBP 214.21 per sample. Finally, the costs for MSP were derived from published costing schedule from North East Thames Regional Genetics Service, the cost was GBP 198.91 (GOSH 2018).
We produced illustrative CCRs for these three tests. The comparison of PSQ to IHC would produce a CCR of 1.5. This means that to justify the increased cost, PSQ would need to have a 50% better performance. The results above indicate supremacy of PSQ over IHC (RHR 1.36, 95% CI 1.01 to 1.84), but it was unclear whether the magnitude of the superiority of the test was great enough. The CCR for PSQ and MSP was 1.6, meaning an additional 60% better performance was required. While the results presented above suggest that PSQ may have a higher RHR (1.14, 95% CI 0.87 to 1.48), the point estimate of the CCR is above the upper limit of the CI. Finally, when comparing MSP with IHC, the CCR was lower at 1.08, meaning that IHC would only require an additional 8% better performance to justify its additional cost. The results found that the lower costing intervention MSP has better ability to predict survival (RHR 1.31, 95% CI 1.01 to 1.71) and the CCR lay within the CIs for the RHR.
Discussion
Summary of the main results
We examined 32 independent cohorts (3474 people) that compared different techniques, regions, cut‐offs or tumour sample types for predicting overall survival in people with glioblastoma treated with temozolomide. We found evidence through meta‐analysis (of RHRs) that MSP (CpG sites 76 to 80 and 84 to 87) was more prognostic than IHC (varying thresholds). Since a large majority of MSP studies had examined CpG sites 76 to 80 and 84 to 87, we were unable to compare alternative CpG sites for MSP. We also found evidence that PSQ was more prognostic than IHC, although studies of PSQ feeding into this analysis had targeted different CpG sites. The CpG sites targeted by PSQ ranged between 72 and 95, and several studies had examined sites 74 to 78. There was a suggestion that PSQ (mainly at CpG sites 74 to 78, but with varying thresholds) was slightly more prognostic than MSP (sites 76 to 80 and 84 to 87). Many variants of PSQ have been compared, although we did not see any strong and consistent messages from the results. Cut‐off thresholds varied substantially (from 4% to 25% for single CpG sites; and from 2.68% to 35% for multiple CpG sites). Two of the three studies with low (or unclear) risk of bias that compared different thresholds found that a 9% cut‐off was more prognostic than higher cut‐offs (of 28% or 29%). It appears that targeting multiple CpG sites is likely to be preferable to targeting just one. We found no evidence of superiority of qMSP over MSP or PSQ, and found a suggestion that lower cut‐off thresholds for interpreting SQ‐MSP in frozen tissue (for sites 76 to 80 and 84 to 87) were more prognostic for survival. There was very little evidence about other techniques (such as bead array, MS‐MLPA PCR‐HRM) and few suggestions that any of these outperform MSP or PSQ.
Certainty of the evidence
We rated the evidence for the comparison between MSP and IHC at moderate certainty, and the evidence for comparisons of PSQ with MSP or IHC at low certainty. For all other comparisons, we rated certainty as very low. Although risk of bias and publication bias were not major concerns for us, we downgraded many of our assessments for indirectness because there was a wide variety of different CpG sites and thresholds investigated, without systematic replications of findings using the same methods across studies. The amount of evidence was small, with only 10s or at most the low 100s of participants contributing to evidence for many of the techniques.
Strengths and weaknesses of the review
We took a systematic approach to identifying, appraising and collecting information from the evidence, and assessed risk of bias and applicability concerns using a modification of QUIPS specific to the topic of the review. To ensure comparisons of methods were fair, we restricted our attention to comparisons made within studies (i.e. on the same people and tumours).
A large variety of methods have been examined, particularly use of different CpG sites and thresholds for PSQ, as well as a mixture of use of FFPE and frozen tumour samples. There was only a small amount of direct replicability across studies, meaning that firm conclusions were difficult to draw.
We limited eligibility for the review to studies that reported HRs or data sufficient for us to estimate them. In many instances, we reconstructed time‐to‐event data from Kaplan‐Meier curves, allowing us to include 14 studies that we would not have included otherwise. However, there was still a small number of studies that had sought to compare methods but not presented data compatible with computation of HRs, which therefore did not meet our eligibility criteria. To compare HRs statistically within studies, strong correlations between different results based on the same tumour samples need to be considered. We imputed such correlations using a rather ad hoc approach and accompanied these with sensitivity analyses. We could not see an obvious way to derive appropriate correlations between these results, a problem that a statistical simulation study might help to resolve.
We listed brief details of articles describing studies examining only one method. When writing the protocol we were unsure whether these would prove to be informative about comparisons between methods. Among the studies that compared multiple methods, we observed that HRs varied markedly across studies, and we were unwilling to make naive indirect comparisons of methods across different studies. Therefore, we do not present quantitative results for these single‐method studies, and did not undergo the process of establishing which articles related to the same underlying cohorts of people.
Economic issues
The results of the economic review demonstrated a paucity of economic evidence for different MGMT strategies in prediction of overall survival. Some illustrative examples showed that more costly tests might be worthwhile. However, a full economic evaluation would be required to investigate fully the costs and consequences of each of the methods for determining for MGMT promoter methylation status. The economic analyses we presented in this review were at best illustrative at this stage, since there is a lack of data to populate the decision model required to investigate the costs and consequences more robustly. A particular limitation is that cost estimates are available for only single sources and for only three of the techniques.
There are limitations with the use of overall survival as a primary outcome in terms of economic analysis. Although survival is important to consider from the perspective of patients and clinicians, other factors such as quality of life and psychological welfare associated with a patient's diagnosis and treatment are also important. This might manifest in a variety of ways, both positive and negative. For example, a false‐positive test result would not have an impact on a patient's survival but would potentially cause psychological distress associated with the finding until the diagnosis was clarified, whereas a true negative might result in a sense of relief and reassurance.
For a future model‐based economic evaluation, data would be needed on the costs of all the different techniques in sufficient detail to allow readers to judge applicability of the cost data, and available from more than one source to allow uncertainty to be explored. Further information would be needed to model the consequences of providing the test information: it is currently unclear how practitioners, participants and their families might use these data. Furthermore, it would be important to know whether the test results be used to stratify treatment or follow‐up, and if so, what are the consequences of these changes. For both changes in management and the consequences of those changes, the impact on the use of resources, survival and health‐related quality of life would be needed. As part of this, it would be important to understand fully the costs of consequences of events such as false positive and false negatives when stratifying patients, both from an overall survival and a quality‐of‐life perspective. This could be derived from further evidence synthesis or primary research. Alternatively, if the goal of measuring methylation status is to help the patient and their family plan for end‐of‐life care, then a valuation method such as willingness to pay approach, which can capture the health and non‐health impacts, could be used to place a value on tests. We are not aware of any such data and hence primary research would be needed.
Applicability of findings to clinical practice and policy
We found the evidence identified to be generally applicable to clinical practice. We included only studies in which at least 90% of people had glioma, and nearly all people were treated with temozolomide. The assessments of applicability we made in our appraisal of included studies were all either of low concerns or unclear due to limited information. The relevance of our findings will depend to an extent on the availability of methods at local sites. For example, although many centres do PSQ, it is not available universally. Finally, we focused on overall survival only as an outcome, so are unable to draw conclusions about using these methods to predict quality of life or progression‐free survival.
Agreements and disagreements with other studies or reviews
Previous systematic reviews and meta‐analyses have summarised the evidence across studies using specific techniques that MGMT promoter methylation is associated with prognosis of people with glioblastoma (Hegi 2019; Zhang 2013; Zhao 2016). This is the first systematic review to our knowledge that compares methods for categorising tumours as methylated in relation to their ability to predict survival in people with glioblastoma. The findings concur with one recent consensus review on management of glioblastoma in adults from the Society for Neuro‐Oncology and the European Society of Neuro‐Oncology (Wen 2020). That review noted that PSQ may provide the best stratification in terms of outcomes and argued that IHC should not be used because it is unreliable. One 2020 update to evidence‐based guidelines on management of newly diagnosed glioblastoma recommends the assessment of tumour MGMT promoter methylation status as a significant predictor of survival (Farrell 2020). They provided a descriptive review of the evidence about different methods but did not provide recommendations on which one(s) to use.
Authors' conclusions
Implications for practice
Among methods for categorising MGMT promoter methylation status in people with glioblastoma treated with temozolomide, pyrosequencing (PSQ) and methylation‐specific polymerase chain reaction (MSP) appear to be more prognostic for overall survival than immunohistochemistry (IHC). Strong evidence is not available to draw conclusions with confidence about the best CpG sites or cut‐off thresholds for quantitative methods. MSP has been studied mainly for CpG sites 76 to 80 and 84 to 87 and PSQ at CpG sites ranging from 72 to 95. A cut‐off threshold of 9% for CpG sites 74 to 78 was found to perform better than higher thresholds of 28% or 29% in two of three good‐quality studies making such comparisons. We are unable to draw strong conclusions about use of frozen tissue versus formalin‐fixed paraffin‐embedded (FFPE) in MSP and PSQ, although one study observed that MSP was more prognostic when based on frozen tissue.
Implications for research
Further large studies would be needed to establish which of the techniques has the best predictive value, and which CpG sites should be tested. Furthermore, remains important to identify a cut‐off value to call the MGMT promoter "methylated," or "unmethylated," or determine ranges that predict response to temozolomide in a more graded manner. Future research should focus on MSP and PSQ (rather than IHC) as well as other up‐and‐coming technologies in the methylation array analysis, such as bead chip arrays.
History
Protocol first published: Issue 4, 2019 Review first published: Issue 3, 2021
Acknowledgements
We thank Robin Grant (Co‐ordinating Editor) and Gail Quinn (Managing Editor) for the Cochrane Gynaecological, Neuro‐oncology and Orphan Cancer Group (GNOC) for their editorial guidance and patience. We also thank Joanne Platt (Information Specialist, GNOC) for aiding with the development of the search strategy.
We thank all our external peer reviewers, including Andrew Bryant, Helen Bulbeck and Catherine McBain.
This project was supported in part by the National Institute for Health Research (NIHR), via Cochrane Programme Grant funding (16/144) to the Cochrane Gynaecological, Neuro‐oncology and Orphan Cancers Group. AM, FS and JPTH were supported in part by Cancer Research UK (grant number C18281/A19169). The views and opinions expressed herein are those of the review authors and do not necessarily reflect those of the NIHR Systematic Reviews Programme, the NIHR, the National Health Service (NHS), the Department of Health or Cancer Research UK.
Appendices
Appendix 1. Database search strategies
Date of search: 3 December 2018
Ovid MEDLINE (1946 to 4 December 2018), 1500 records.
PubMed NOT MEDLINE (4 December 2018), 101 records.
Ovid Embase (1980 to 2018, week 49), 2983 records.
BIOSIS (1969 to 3 December 2018), 790 records.
Web of Science Conference Proceedings Citation Index (CPCI‐S) (1900 to 3 December 2018), 120 records.
Total: 5494 records
Duplicates removed: 2137 records
Records to screen: 3357
Ovid MEDLINE(R) and Epub Ahead of Print, In‐Process & Other Non‐Indexed Citations and Daily <1946 to 4 December 2018>
Search strategy:
‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐
1 glioma/ or astrocytoma/ or glioblastoma/ (65202)
2 (glioblastom* or GBM or astrocytom* or gliosarcom*).mp. (55603)
3 1 or 2 (81995)
4 "O(6)‐Methylguanine‐DNA Methyltransferase"/ (2192)
5 ((methylguanin* or methyl guanin* or alkylguanin* or alkyl guanin*) adj5 (methyltransferas* or methyl transferas* or alkyltransferas* or alkyl transferas* or transmethylas* or trans methylas*)).mp. (3857)
6 (methyl* DNA protein cystein* adj (methyltransferas* or methyl transferas*)).mp. (3)
7 (AGT or MGMT or AGAT).ti,ab,kf,ot. (6403)
8 or/4‐7 (7811)
9 exp Prognosis/ (1466623)
10 (prognos* or predict*).mp. (2042311)
11 exp mortality/ (350654)
12 survival/ (4532)
13 survival rate/ (158119)
14 exp survival analysis/ (264438)
15 (mortality or death* or surviv*).mp. (2412078)
16 Follow‐Up Studies/ (602886)
17 ((followup or follow‐up) adj (study or studies)).ti,ab,kf. (48020)
18 or/9‐17 (4920356)
19 3 and 8 and 18 (1515)
20 exp animals/ not humans.sh. (4519948)
21 19 not 20 (1500)
***************************
Ovid Embase <1980 to 2018 week 49>
Search strategy:
‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐‐
1 glioma/ or astrocytoma/ or glioblastoma/ (104447)
2 (glioblastom* or GBM or astrocytom* or gliosarcom*).mp. (88786)
3 1 or 2 (119931)
4 methylated DNA protein cysteine methyltransferase/ (5065)
5 (methyl* DNA protein cystein* adj (methyltransferas* or methyl transferas*)).ti,ab,kw,ot. (3)
6 ((methylguanin* or methyl guanin* or alkylguanin* or alkyl guanin*) adj5 (methyltransferas* or methyl transferas* or alkyltransferas* or alkyl transferas* or transmethylas* or trans methylas*)).mp. (4545)
7 (AGT or MGMT or AGAT).ti,ab,kw,ot. (10397)
8 or/4‐7 (13071)
9 prognosis/ or cancer prognosis/ (629552)
10 (prognos* or predict*).mp. (2614430)
11 exp mortality/ (931698)
12 exp survival/ (972881)
13 survival analysis/ (12408)
14 (mortality or death? or surviv*).ti,ab,kw,ot. (2769350)
15 or/9‐14 (4989016)
16 Methylation/ or DNA methylation/ (107678)
17 methylat*.ti,ab,kw,ot. (123232)
18 ((amount? or express* or level? or activ* or status) adj5 (protein? or AGT or MGMT or AGAT or ((methylguanin* or methyl guanin* or alkylguanin* or alkyl guanin*) adj5 (methyltransferas* or methyl transferas* or alkyltransferas* or alkyl transferas* or transmethylas* or trans methylas*)))).ti,ab,kw,ot. (962065)
19 or/16‐18 (1093057)
20 3 and 8 and 15 and 19 (3054)
21 ((animal or nonhuman) not human).de. (5171408)
22 (cell line or cell culture).hw. not ((human or adult).sh. or patient.hw.) (344719)
23 (20 not (21 or 22)) (2983)
***************************
BIOSIS Citation Index (3 December 2018)
[Search‐1: ((tumour ‘near’ enzyme) and prognosis)]
#1 ((TS=((glioblastoma* OR GBM* OR astrocytom*) NEAR (methylguanin* OR “methyl guanin*” OR alkylguanin* OR “alkyl guanin*” OR AGT OR MGMT OR AGAT)) OR TS=((gliosarcom*) AND (methylguanin* OR “methyl guanin*” OR alkylguanin* OR “alkyl guanin*” OR AGT OR MGMT OR AGAT))) AND (TS=(prognos* or predict* or mortalit* or death* or surviv*)))n=437
[Search‐2: ((prognosis ‘near’ enzyme) and tumour)]
#2 TS=((prognos* OR predict* OR mortalit* OR death* OR surviv*) NEAR (methylguanin* OR “methyl guanin*” OR alkylguanin* OR “alkyl guanin*” OR AGT OR MGMT OR AGAT)) AND TS=(glioblastom* OR GBM* OR astrocytom* OR gliosarcom*) n=425
[Search‐3: ((prognosis ‘near’ tumour) and enzyme)]
#3 ((TS=((prognos OR predict* OR mortalit* OR death* OR surviv*) NEAR (glioblastom* OR GBM* OR astrocytom*)) OR TS=((prognos OR predict* OR mortalit* OR death* OR surviv*) AND gliosarcom*)) AND (TS=("O(6)‐Methylguanine‐DNA Methyltransferase" OR "O‐6‐Methylguanine‐DNA Methyltransferase" or “methylated DNA protein cysteine methyltransferase” OR AGT or MGMT or AGAT) OR TS=((methylguanin* or “methyl guanin*” or alkylguanin* or “alkyl guanin*”) NEAR (methyltransferas* or “methyl transferas*” or alkyltransferas* or “alkyl transferas*” or transmethylas* or “trans methylas*”)))) n=413
[Search‐4: prognosis AND tumour AND enzyme AND methylation/expression]
#4 (TS=(glioblastom* or GBM or astrocytom* or gliosarcom*) AND (TS=("O(6)‐Methylguanine‐DNA Methyltransferase" or "O‐6‐Methylguanine‐DNA Methyltransferase" or “methylated DNA protein cysteine methyltransferase” or AGT or MGMT or AGAT) OR TS=((methylguanin* or “methyl guanin*” or alkylguanin* or “alkyl guanin*”) NEAR (methyltransferas* or “methyl transferas*” or alkyltransferas* or “alkyl transferas*” or transmethylas* or “trans methylas*”))) AND (TS=(prognos* or predict* or mortality or death or deaths or surviv*)) AND (TS= methylat* OR TS=((amount or amounts or express* or level or levels or activ* or status) NEAR (protein* or AGT or MGMT or AGAT or methylguanin* or “methyl guanin*” or alkylguanin* or “alkyl guanin*”)))) n=722
#5 (#4 or #3 or #2 or #1) n=790
[N.B. Gliosarcoma is a much rarer tumour, so the proximity operator was not used in this context, in search lines 1 and 3)]
***************************
Conference Proceedings Citation Index‐ Science (CPCI‐S) (1990 to 3 December 2018)
#1 TS=((glioblastom* or GBM or astrocytom* or gliosarcom*) and (prognos* or predict* or mortality or death or deaths or surviv*)) n=2200
#2 TS=("O(6)‐Methylguanine‐DNA Methyltransferase" or "O‐6‐Methylguanine‐DNA Methyltransferase" or “methylated DNA protein cysteine methyltransferase” or AGT or MGMT or AGAT) n=1042
#3 TS=((methylguanin* or “methyl guanin*” or alkylguanin* or “alkyl guanin*”) AND (methyltransferas* or “methyl transferas*” or alkyltransferas* or “alkyl transferas*” or transmethylas* or “trans methylas*”)) n=277
#4 (#3 OR #2) n=1135
#5 (#4 and #1) n=120
***************************
PubMed 4 December 2018
#12 Search (#10 AND #11) (101)
#11 Search pubmednotmedline[sb] (2531482)
#10 Search (#3 AND #9) (1855)
#9 Search (#4 OR #5 OR #6 OR #7 OR #8) (11388)
#8 Search (AGT[Title/Abstract] OR MGMT[Title/Abstract] OR AGAT[Title/Abstract]) (6400)
#7 ((“methylated DNA protein cysteine methyltransferas*” or “methylated DNA protein cysteine methyl transferas*” or “methylguanine deoxyribonucleic acid methyltransferas*” or “methyl guanine deoxyribonucleic acid methyltransferas*” or “methylguanine deoxyribonucleic acid methyl transferas*” or “methyl guanine deoxyribonucleic acid methyl transferas*” or “methylguanin* dna protein methyltransferas*” or “methyl guanin* dna protein methyltransferas*” or “methylguanin* dna protein methyl transferas*” or “methyl guanin* dna protein methyl transferas*”)) (3730)
#6 ((“methylguanin* DNA methyltransferas*” OR “methylguanin* DNA methyl transferas*” OR “methylguanin* DNA alkyltransferas*” OR “methylguanin* DNA alkyl transferas*” OR “methyl guanin* DNA methyltransferas*” OR “methyl guanin* DNA methyl transferas*” OR “methyl guanin* DNA alkyltransferas*” OR “methyl guanin* DNA alkyl transferas*” OR “alkylguanin* DNA methyltransferas*” OR “alkylguanin* DNA methyl transferas*” OR “alkylguanin* DNA alkyltransferas*” OR “alkylguanin* DNA alkyl transferas*” OR “alkyl guanin* DNA methyltransferas*” OR “alkyl guanin* DNA methyl transferas*” OR “alkyl guanin* DNA alkyltransferas*” OR “alkyl guanin* DNA alkyl transferas*” or “methylguanin* dna transmethylas*” or “methylguanin* dna trans methylas*” )) (5973)
#5 Search ((“methylguanin* methyltransferas*” OR “methylguanin* methyl transferas*” OR “methylguanin* alkyltransferas*” OR “methylguanin* alkyl transferas*” OR “methyl guanin* methyltransferas*” OR “methyl guanin* methyl transferas*” OR “methyl guanin* alkyltransferas*” OR “methyl guanin* alkyl transferas*” OR “alkylguanin* methyltransferas*” OR “alkylguanin* methyl transferas*” OR “alkylguanin* alkyltransferas*” OR “alkylguanin* alkyl transferas*” OR “alkyl guanin* methyltransferas*” OR “alkyl guanin* methyl transferas*” OR “alkyl guanin* alkyltransferas*” OR “alkyl guanin* alkyl transferas*”)) (7450)
#4 Search "O(6)‐Methylguanine‐DNA Methyltransferase"[Mesh:NoExp] (2192)
#3 Search (#1 OR #2) (81976)
#2 Search (glioblastom* OR GBM OR astrocytom* OR gliosarcom*) (55573)
#1 ("Glioma"[Mesh:NoExp] OR "Astrocytoma"[Mesh:NoExp] OR "Glioblastoma"[Mesh:NoExp]) (65225)
***************************
Appendix 2. Risk of bias and applicability assessment
Bespoke tool to assess risk of bias and applicability of prognostic factor studies. SQ: signalling question.
| Domain 1: participant selection | |
| Risk of bias | SQ1.1: was a consecutive or random sample of people enrolled? |
| SQ1.2: was a case‐control or cross‐sectional design avoided? | |
| SQ1.3: did the study avoid inappropriate exclusions? | |
| Applicability | Are there concerns that the included participants and setting do not match the review question? |
| Domain 2: subsequent treatment | |
| Risk of bias | SQ2.1: did treatment vary across participants? (or "Was treatment either standardised or randomised?") |
| Applicability | Are there concerns that treatments received do not match the review question? |
| Domain 3: outcome measurement | |
| Risk of bias | SQ3.1: was the method of outcome measurement used adequately valid and reliable? |
| SQ3.2: was the method and setting of outcome measurement the same for all study participants? | |
| SQ3.3: was the outcome objective or assessed without knowledge of the prognostic factor? | |
| SQ3.4: do the prognostic factors investigated form part of the outcome? | |
| Applicability | Are there concerns that outcome does not match the question or that follow‐up was not of sufficient duration, or both? |
| Domain 4: prognostic factor measurement | |
| Risk of bias | SQ4.1: was the method and setting of measurement of the prognostic factor the same for all participants? |
| SQ4.2: was the prognostic factor objective or measured without knowledge of the outcome or risk of the outcome? | |
| SQ4.3: if a threshold was used, was it prespecified? | |
| Applicability | Are there concerns that prognostic factor, the way that it was measured, or the way that it was interpreted, differ from the review question? |
| Domain 5: study attrition | |
| Risk of bias | SQ5.1: were all participants included in the analysis? |
| If no to SQ5.1: SQ5.2: were there important differences between participants who completed the study/were included in the analysis and those who were not? | |
| Domain 6: adjustment for other potential prognostic factors (where relevant) | |
| Risk of bias | SQ6.1: were other potential prognostic factors measured adequately and reliably and in a similar manner for all participants, and is the method of adding them to the model appropriate? |
| Applicability | Did the prognostic factors adjusted for match the review question? |
| Domain 7: selective reporting | |
| Risk of bias | SQ7.1: is the reported estimate likely to be selected on the basis of the results from: multiple outcome measurements, multiple analyses of the prognostic factor‐outcome relationship, from different subgroups, or a combination of these? |
Appendix 3. Domains to be considered when judging the strength of the body of evidence
We considered the following domains when we assessed the strength of the body of evidence, based on the GRADE approach (Guyatt 2008).
| Domain | Explanation |
| Risk of bias | Based on results of 'Risk of bias' assessments, we downgraded confidence in the evidence base if most evidence was from studies that we judged at high risk of bias. |
| Indirectness | We downgraded confidence in the evidence base if we had concerns that the study sample, the prognostic factor, the outcome, other factors in the models in the primary studies or a combination of these did not reflect the review question. |
| Inconsistency | We downgraded confidence in the evidence base if there was unexplained heterogeneity or variability in results across studies. |
| Imprecision | We downgraded confidence in the evidence base if the estimate of the effect size from a meta‐analysis was not precise or, if no meta‐analysis was performed, if the estimate of the size of effect from individual studies was not precise. |
| Publication bias | Studies showing no association are likely to be unpublished, unless part of a larger study that specifically aimed to compare tests. We downgraded our confidence in the evidence base if we had reason to suspect publication bias from our assessments of reporting bias. |
| Size of effect | We upgraded our confidence in the evidence base if the size of effect was moderate or large. If a meta‐analysis is not possible, we upgraded if the size of effect was moderate or large for most included studies. |
Appendix 4. Identification of studies making different comparisons among the techniques
Comparison numbers are for [row‐defining technique] versus [column‐defining technique], for example Kristensen 2016 (top right cell) compares one version of IHC versus three 'other' techniques.
| IHC | MSP | PSQ | qMSP | Bead array | MS‐MLPA | PCR‐HRM | PCR‐mRNA | Other | |
| IHC | None | Felsberg 2009 (1 vs 1) Hsu 2015 (1 vs 2) Karayan‐Tapon 2010 (1 vs 1) Lalezari 2013 (1 vs 1) Lechapt‐Zalcman 2012 (1 vs 1) Melguizo 2012 (1 vs 1) Quillien 2014 (test) (1 vs 1) Yang 2012 (1 vs 1) |
Hsu 2017 (1 vs 1) Karayan‐Tapon 2010 (1 vs 6) Kristensen 2016 (1 vs 1) Lalezari 2013 (1 vs 1) Quillien 2014 (test) (1 vs 32) |
Hsu 2017 (1 vs 2) Karayan‐Tapon 2010 (1 vs 1) Quillien 2014 (test) (1 vs 1) |
None | None | Quillien 2014 (test) (1 vs 1) | Felsberg 2009 (1 vs 1) Karayan‐Tapon 2010 (1 vs 1) |
Kristensen 2016 (1 vs 3) |
| MSP | — | Barbagallo 2014 (2) Hsu 2017 (2) Lattanzio 2015 (2) |
Barbagallo 2014 (2 vs 2) Havik 2012 (1 vs 9) Hsu 2017 (2 vs 1) Karayan‐Tapon 2010 (1 vs 6) Kim 2016 (1 vs 1) Lalezari 2013 (1 vs 1) Lattanzio 2015 (1 vs 2) McDonald 2013 (1 vs 1); Quillien 2014 (test) (1 vs 32) |
Havik 2012 (1 vs 2) Hsu 2017 (2 vs 2) Karayan‐Tapon 2010 (1 vs 1) Quillien 2014 (test) (1 vs 1) |
Bady 2012 (M‐GBM) (1 vs 1) | Park 2011 (1 vs 2) | Havik 2012 (1 vs 1) Quillien 2014 (test) (1 vs 1) Yamashita 2018 (1 vs 5) |
Felsberg 2009 (1 vs 1) Karayan‐Tapon 2010 (1 vs 1) |
Thon 2017 (1 vs 1) |
| PSQ | — | — | Barbagallo 2014 (2) Brigliadori 2016 (2) Chai 2018 (7‐site cohort) (3) Chai 2018 (8‐site cohort) (3) Dunn 2009 (6) Havik 2012 (9) Karayan‐Tapon 2010 (6) Lattanzio 2015 (2) Quillien 2014 (test) (32) Quillien 2014 (validation) (3) Quillien 2016 (12) |
Havik 2012 (9 vs 2) Hsu 2017 (1 vs 2) Karayan‐Tapon 2010 (6 vs 1) Quillien 2014 (test) (32 vs 1) Quillien 2016 (12 vs 5) |
Bady 2012 (E‐GBM) (1 vs 2) | None | Havik 2012 (9 vs 1) Quillien 2014 (test) (32 vs 1) |
Karayan‐Tapon 2010 (6 vs 1) | Barault 2015 (1 vs 1) Dahlrot 2018 (NS Cohort) (1 vs 1) Dahlrot 2018 (RSD Cohort) (1 vs 1) Kristensen 2016 (1 vs 3) |
| qMSP | — | — | — | Havik 2012 (2) Hsu 2017 (2) Nguyen 2015 (2) Quillien 2016 (5) Yoshioka 2018 (5) |
None | None | Havik 2012 (2 vs 1) Quillien 2014 (test) (1 vs 1) |
Karayan‐Tapon 2010 (1 vs 1) | Bell 2017 (1 vs 1) |
| Bead array | — | — | — | — | Bady 2012 (E‐GBM) (1) | None | None | None | None |
| MS‐MLPA | — | — | — | — | — | Park 2011 (2) | None | None | None |
| PCR‐HRM | — | — | — | — | — | — | Yamashita 2018 (5) | None | None |
| PCR‐mRNA | — | — | — | — | — | — | — | None | None |
| Other | — | — | — | — | — | — | — | — | Almuqate 2018 (2) Kristensen 2016 (3) |
| IHC: immunohistochemistry; MS‐MLPA: methylation‐specific multiplex ligation‐dependent probe amplification; MSP: methylation‐specific polymerase chain reaction; PCR‐HRM: polymerase chain reaction with high‐resolution melting; PCR‐mRNA: polymerase chain reaction‐messenger ribonucleic acid; PSQ: pyrosequencing; qMSP: quantitative methylation‐specific polymerase chain reaction. | |||||||||
Appendix 5. Details of methods implemented
| Study ID | Method | Technique description | Primers used | Antibody/mRNA measurement/enzymatic activity assay | How cut‐off threshold determined |
| Almuqate 2018 | Technique: MS‐RE‐qPCR Sample type: NR CpG sites: NR Threshold: > 5% |
MS‐RE‐qPCR | NR | — | Mention of "optimal cut‐off;" unclear whether the 5% cut‐off was prespecified, or whether multiple cut‐offs were investigated |
| Technique: MS‐RE‐qPCR Sample type: NR CpG sites: NR Threshold: > 9% |
Described as "current cut‐off" and "analytically validated" | ||||
| Bady 2012 (E‐GBM) | Technique: bead array Sample type: frozen CpG sites: 31 and 83 Threshold: > 0.358 |
Infinium HumanMethylation27 (HM‐27K) BeadChip | NR | — | From M‐GBM dataset |
| Technique: bead array Sample type: frozen CpG sites: 78–84 Threshold: > 10% |
Infinium HumanMethylation27 beadchip (Illumina Inc.) | NR | — | Selected the threshold that gave the best stratification value according to the log‐rank test | |
| Technique: PSQ Sample type: frozen CpG sites: 74–78 Threshold: > 7.28% |
Methylation‐specific PSQ performed with PyroMark Q96 CpG MGMT kit Qiagen | — | — | "The percentage of MGMT methylation was averaged over the 5 CpG‐sites interrogated… The data was dichotomized into unmethylated and methylated status using an iterative procedure based on segmented regression [5]. The optimal cut‐off obtained was 7.28%, defined as the point where the sum of squares of residuals is minimal." | |
| Bady 2012 (M‐GBM) | Technique: bead array Sample type: frozen CpG sites: 31 and 83 Threshold: > 0.358 |
Infinium HumanMethylation27 (HM‐27K) BeadChip | NR | — | Threshold derived empirically to maximise sensitivity and specificity: "For classification, we used a probability cut‐off of 0.358, which empirically maximized the sum of sensitivity and specificity." |
| Technique: MSP Sample type: NR CpG sites: 76–80 and 84–86 Threshold: NR |
"Performed basically as reported by Esteller et al." Esteller M et al. New England Journal of Medicine 2000;343:1350–4. | See Figure 1 of publication | — | NR | |
| Barault 2015 | Technique: Methyl‐beaming Sample type: FFPE CpG sites: 79–83 Threshold: > 40.2% |
Methyl‐beaming assay. "BEAMing analysis is a multistep digital PCR based technique published by Diehl and colleagues [7]. Its application for methylation is named Methyl‐BEAMing and has been previously described to detect methylation of the VIM gene [5]…The percentage of methylation was calculated dividing the methylated specific signal by the sum of methylated plus unmethylated specific signal." Workflow for methyl‐beaming: bisulfite treatment; locus enrichment; digital PCR; hybridisation flow cytometry | Methyl‐beaming 1st PCR: forward 5'‐TCCCGCGAAATTAATACGACGTTTAGGATATGTTGGGATAGT‐3', reverse 5'‐GCTGGAGCTCTGCAGCTAAACCACCCAAACACTCACCAA‐3'. Methyl‐beaming emulsion PCR: forward 5'‐TCCCGCGAAATTAATACGAC‐3', reverse 5'‐GCTGGAGCTCTGCAGCTA‐3' (Table S2 of publication). | — | "ROC analysis was carried out to evaluate the threshold best fitting the overall survival (OS) at 1 year" on a cohort of 98 participants with GBM diagnosed before TMZ was introduced as a component of standard treatment. The cut‐off was then validated in this cohort of participants. |
| Technique: PSQ Sample type: FFPE CpG sites: 76–81 Threshold: > 29.6% |
Bisulfite‐PSQ. "Pyrograms were analyzed using PyroMark Q24 Software, average of the 6 CpG sites methylation values was used for further analyses." | Forward 5'‐GTTTAGGATATGTTGGGATAGT‐3', reverse 5'‐GGACACCGCTGATCGTTTAAACCACCCAAACACTCACCAA‐3', universal 5'‐GGGACACCGCTGATCGTTTA‐3', sequencing 5'‐GTTTTTAGAAYGTTTTGYGTTT‐3' (Table S2 of publication) | — | ||
| Barbagallo 2014 | Technique: MSP Sample type: FFPE CpG sites: 76–80 and 84–87 Threshold: including weakly |
"MSP assay was performed using a 2‐step nested PCR approach as previously described. The MSP reactions were performed in 25 ml by 2720 Thermal Cycler Applied Biosystem PCR. Universal unmethylated and polymethylated DNA were included as controls in each set of reactions, in addition to a negative control sample without DNA." | Primers from Esteller 1999 | — | "Universal unmethylated and polymethylated DNA were included as controls in each set of reactions, in addition to a negative control sample without DNA. Individual tumors showing only very weak PCR products for the methylated MGMT sequence promoter but strong PCR products for the unmethylated MGMT sequence promoter were judged as "weakly methylated"." |
| Technique: MSP Sample type: FFPE CpG sites: 76–80 and 84–87 Threshold: excluding weakly | |||||
| Technique: PSQ Sample type: FFPE CpG sites: NR Threshold: > 9% |
"Templates for pyrosequencing were amplified with primers that were biotinylated for template strands (MGMT PyroMark CpG Assay kit, Qiagen). The biotinylated polymerase chain reaction (PCR) products were then immobilized on streptavidin‐coated Sepharose beads (GE Healthcare), and the single‐stranded DNA templates were analyzed by PyroMark Q24 (Qiagen)." | Primers from MGMT PyroMark CpG Assay kit, Qiagen | — | NR | |
| Technique: PSQ Sample type: FFPE CpG sites: NR Threshold: > 25% | |||||
| Bell 2017 | Technique: QF‐IHC (AQUA) Sample type: FFPE CpG sites: N/A Threshold: > median |
QF‐IHC (AQUA). Median cut‐off tumour mask. 4 tissue microarrays containing paraffin‐embedded tumour cores from the 452 RTOG 0525 people were cut at 5 µm and sections were placed on positively charged slides. As a surrogate for tumour colocalisation, proteins were colocalised with glial fibrillary acidic protein (DAKO;1:100) to stain the cytoplasmic compartments of glial cells. Deparaffinisation and retrieval were performed as previously described. Slides were scanned by HistoRx PM‐2000 and analysed by AQUAnalysis software. Each protein was scored in the tumour, cytoplasm, and nuclear components of each tissue microarray core using the HistoRxTM AQUA platform and fluorescent IHC. | N/A | Antibody: MGMT (MT3.1) (Santa Cruz; 1:100) | To determine the best cut‐off points for markers with continuous values significantly associated with survival for inclusion in the RPA model, the technique of using ROC curves was applied. Because the area under the ROC curve for all markers was ≤ 0.65, limiting the ability to determine optimal cut points, methods using quartiles, tertiles and medians were used. |
| Technique: qMSP Sample type: NR CpG sites: NR Threshold: > 8 |
qMSP assay (detail from the original NRG RTOG 0525 paper). Performed centrally by Oncomethylome Science – direct, real‐time MSP (RTOG 0525 Gilbert paper references Vlassenbroeck 2008, MSP method taken from Vlassenbroeck. "Validation of Real‐Time Methylation‐Specific PCR to Determine O6‐Methylguanine‐DNA Methyltransferase Gene Promoter Methylation in Glioma"). "Analyte (m_MGMT and β‐actin [ACTB]) quantification was performed by real‐time MSP assays. These consisted of parallel amplification/quantification processes using specific primer and primer/detector pairs for each analyte using the Amplifluor assay format on an ABI Prism 7900HT instrument (Applied Biosystems, Foster City, CA). The analyte defined in the direct, real‐time MSP was the MGMT promoter sequence and detects the fully methylated version. ACTB was used as a reference gene in the assay, using primers that are outside any CpG islands. The Amplifluor direct forward primers are preceded by the detection elements (underlined). The amplicon size is 136 bp for the m_MGMT analyte and 125 bp for the ACTB analyte, including the Amplifluor detection sequence." | See Vlassenbroeck. | — | From Vlassenbroeck paper: "These cutoffs had been defined previously in a smaller data set and are consistent with the present study suggesting the cutoff at ratio value 8." | |
| Brigliadori 2016 | Technique: PSQ Sample type: FFPE CpG sites: 74–83 Threshold: > 9% |
PSQ. 10 CpG sites of the MGMT promoter (74–83) located in a gene region recognised as critical for transcriptional control (DMR2) were analysed using a PyroMark 96 system (Diatech, Iesi, Italy), according to the manufacturer's protocol. All tumour and control samples were analysed in triplicate. Templates for PSQ were amplified using a Rotorgene 6000 with primers that had been biotinylated for template strands (MGMT plus kit, Diatech, Iesi, Italy). 20 μL of the biotinylated PCR products were then immobilised on streptavidin‐coated Sepharose beads (GE Healthcare, Uppsala, Sweden), and the single‐stranded DNA templates were analysed by PyroMark Q96 system (Diatech, Iesi, Italy). Subsequent quantification of the methylation density for the 10 investigated CpG sites was performed using the PyroMark Q96 software. Methylation percentages for each sample were obtained by calculating the mean of all 10 methylation sites. The median value of the 3 analyses was considered for each methylation level. | Primers that had been biotinylated for template strands (MGMT plus kit, Diatech, Iesi, Italy). | — | References literature |
| Technique: PSQ Sample type: FFPE CpG sites: 74–83 Threshold: > 29% | |||||
| Chai 2018 (7‐site cohort) | Technique: PSQ Sample type: frozen CpG sites: 72–78 Threshold: > 12% |
"Bisulfite‐treated DNA was preamplified with the primers (a) F‐primer 5'‐GTT TYG GAT ATG TTG GGA TAG TT‐3'; (b) biotinylated R‐primer 5'‐biotin‐ACR ACC CAA ACA CTC ACC AA‐3'. Different samples were analyzed with two independent assays, and the PSQ primers were (a) 5'‐GAT ATG TTG GGA TAG T‐3' (for CpGs 72–78)… PSQ testing was performed on a PyroMarker Q96 instrument, and the results were analyzed with PyroMarker Q96 software." (Qiagen) | Bisulfite‐treated DNA was preamplified with the primers (a) F‐primer 5'‐GTT TYG GAT ATG TTG GGA TAG TT‐3'; (b) biotinylated R‐primer 5'‐biotin‐ACR ACC CAA ACA CTC ACC AA‐3'. Different samples were analysed with 2 independent assays, and the PSQ primers were (a) 5'‐GAT ATG TTG GGA TAG T‐3' (for CpGs 72–78). | — | "We determined the cutoff in this study by similar strategy, comprehensively considering the ROC likelihood value, sensitivity, specificity, and cutoffs used in reported studies." |
| Technique: PSQ Sample type: frozen CpG sites: 74–78 Threshold: > 12% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 75–78 Threshold: > 12% | |||||
| Chai 2018 (8‐site cohort) | Technique: PSQ Sample type: frozen CpG sites: 75–78 Threshold: > 13% |
PSQ. CpGs 75–78. "Briefly, bisulfite‐treated DNA was preamplified with the primers (a) F‐primer 5'‐GTT TYG GAT ATG TTG GGA TAG TT‐3'; (b) biotinylated R‐primer 5'‐biotin‐ACR ACC CAA ACA CTC ACC AA‐3'. Different samples were analyzed with two independent assays, and the PSQ primers were…(b) 5'‐GTT TTT AGA AYG TTT TG‐3' (for CpGs 75–82)… PSQ testing was performed on a PyroMarker Q96 instrument, and the results were analyzed with PyroMarker Q96 software." (Qiagen) | Bisulfite‐treated DNA was preamplified with the primers (a) F‐primer 5'‐GTT TYG GAT ATG TTG GGA TAG TT‐3'; (b) biotinylated R‐primer 5'‐biotin‐ACR ACC CAA ACA CTC ACC AA‐3'. Different samples were analysed with 2 independent assays, and the PSQ primers were…(b) 5'‐GTT TTT AGA AYG TTT TG‐3' (for CpGs 75–82). | — | "We determined the cutoff in this study by similar strategy, comprehensively considering the ROC likelihood value, sensitivity, specificity, and cutoffs used in reported studies." |
| Technique: PSQ Sample type: frozen CpG sites: 75–82 Threshold: > 12% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 76–79 Threshold: > 11% | |||||
| Dahlrot 2018 (NS cohort) | Technique: DIF Sample type: FFPE CpG sites: N/A Threshold: < 0.2 |
DIF was performed on FFPE tissue on the Autostainer Plus platform (DAKO Denmark A/S, Glostrup, Denmark). Detection was performed using DAKO CSA II, Biotin‐Free Catalyzed Amplification System (DAKO ref. K1497). Positive controls consisting of tissue cores from different normal and cancer tissues, including 11 high‐grade gliomas, were included in each run. | N/A | Antibody: MT23.2; Invitrogen 1 + 100, CA, USA. | Median value. The AF‐all of all MGMT positive nuclei (defined as the area of all MGMT‐positive nuclei divided by the area of all nuclei), the AF‐t of MGMT‐positive tumour nuclei (defined as the area of MGMT‐positive tumour nuclei divided by the area of all tumour nuclei), and the AF‐nt of MGMT positive non‐tumour nuclei (defined as the area of MGMT positive non‐tumour nuclei divided by the area of all non‐tumour nuclei) were identified. Only the AF‐t of MGMT‐positive tumour nuclei (defined as the area of MGMT positive tumour nuclei divided by the area of all tumour nuclei) was evaluated. |
| Technique: PSQ Sample type: FFPE CpG sites: 74–78 Threshold: > 9% |
PyroMark Q96. MGMT promoter status was determined, measured, established using PSQ (MGMT Pyro kit; Qiagen, Hilden, Germany). A modified PSQ method published by Collins et al. [29] was used. After bisulfite conversion of 50–200 ng of DNA using EZ DNA Methylation Kit (Zymo Research, Irvine, CA, USA), nested PCR was carried out with HotStarTaq Master Mix (Qiagen). Obtained PCR product was used as a template in 4 PSQ assays. PSQ was performed on a PyroMark Q96 MD instrument (Qiagen) using PyroMark Gold Q96 CDT Reagents (Qiagen). as described by the manufacturer. | NR | — | NR | |
| Dahlrot 2018 (RSD cohort) | Technique: DIF Sample type: FFPE CpG sites: N/A Threshold: < 0.2 |
DIF was performed on formalin fixed paraffin embedded tissue on the Autostainer Plus platform (DAKO Denmark A/S, Glostrup, Denmark). Detection was performed using DAKO CSA II, Biotin‐Free Catalyzed Amplification System (DAKO ref. K1497). Positive controls consisting of tissue cores from different normal and cancer tissues, including 11 high‐grade gliomas, were included in each run. | N/A | Antibody: MT23.2; Invitrogen 1 + 100, CA, USA. | Median value. The AF‐all of all MGMT‐positive nuclei (defined as the area of all MGMT positive nuclei divided by the area of all nuclei), the AF‐t of MGMT‐positive tumour nuclei (defined as the area of MGMT positive tumour nuclei divided by the area of all tumour nuclei), and the AF‐nt of MGMT‐positive non‐tumour nuclei (defined as the area of MGMT‐positive non‐tumour nuclei divided by the area of all non‐tumour nuclei) were identified. Only the AF‐t of MGMT‐positive tumour nuclei (defined as the area of MGMT‐positive tumour nuclei divided by the area of all tumour nuclei) was evaluated. |
| Technique: PSQ Sample type: FFPE CpG sites: 74–78 Threshold: > 10% |
MGMT promoter status was determined, measured and established using PSQ (MGMT Pyro kit; Qiagen, Hilden, Germany) as described by the manufacturer. DNA was purified from 10 lm paraffin slides using QIAamp DNA FFPE Tissue kit (Qiagen), and MGMT PSQ was performed according to the kit instructions. | NR | — | NR | |
| Dunn 2009 | Technique: PSQ Sample type: frozen, smear, FFPE or a combination CpG sites: 72–83 Threshold: > 9% |
"The pyrosequencing assay was performed as described earlier (Shaw et al, 2006). The primers used for amplification of bisulphite‐treated DNA were forward: 5'‐gGGATAGTTGGGATAGTT‐3' (the first g avoids formation of hairpin loops) and reverse: 5'‐biotin‐ATTTGGTGAGTGTTTGGG‐3' giving a 99‐bp amplicon at genomic position 131 155 467–131 155 565…pyrosequencing on a PSQ96MA System (Biotage, Uppsala, Sweden) using the primer 5'‐GGATATGTTGGGATAGT‐3' and PyroGold reagents (Biotage). The Pyro Q‐CpG software 1.0.9 (Biotage) was used to analyse data…Pyrosequencing yields data for 12 CpG sites within the MGMT promoter. For data analysis, the percentage methylation obtained for each CpG was averaged across the 12 CpGs in duplicate PCR reactions (average methylation per sample)." | "The primers used for amplification of bisulphite‐treated DNA were forward: 5'‐gGGATAGTTGGGATAGTT‐3' (the first g avoids formation of hairpin loops) and reverse: 5'‐biotin‐ATTTGGTGAGTGTTTGGG‐3' giving a 99‐bp amplicon at genomic position 131 155 467–131 155 565…pyrosequencing on a PSQ96MA System (Biotage, Uppsala, Sweden) using the primer 5'‐GGATATGTTGGGATAGT‐3'." | — | ≥ mean ± 2 SD for non‐neoplastic brain |
| Technique: PSQ Sample type: frozen, smear, FFPE, or a combination CpG sites: 72–83 Threshold: > 20% |
"methylated cases were ranked according to methylation and divided into three groups." | ||||
| Technique: PSQ Sample type: frozen, smear, FFPE or a combination CpG sites: 72–83 Threshold: > 29% |
"Methylated cases were dichotomised using receiver operator characteristic (ROC) plots comparing average methylation per case with the Cox regression survival function for OS…Receiver operator characteristic analysis used to separate methylated cases into two prognostic groups yielded a cut‐off of 29.4%." | ||||
| Technique: PSQ Sample type: frozen, smear, FFPE or a combination CpG sites: 72–83 Threshold: > 35% |
"methylated cases were ranked according to methylation and divided into three groups." | ||||
| Technique: PSQ Sample type: frozen, smear, FFPE or a combination CpG sites: 72–83 Threshold: cluster 1 vs 2 and 3 |
"Unsupervised hierarchical cluster analysis of average methylation at each CPG site." | ||||
| Technique: PSQ Sample type: frozen, smear, FFPE or a combination CpG sites: 72–83 Threshold: cluster 1 and 2 vs 3 | |||||
| Felsberg 2009 | Technique: IHC Sample type: FFPE CpG sites: N/A Threshold: < 10% |
Negative controls were carried out by omission of the primary antibodies. Each IHC staining was scored blinded to clinical or molecular information. For the assessment of MGMT protein expression, only nuclear staining was considered. Staining of vascular endothelial cells served as an internal positive control. The DAKO catalysed signal amplification horseradish peroxidase system was used as the detection systems according to the manufacturer's protocol to show MGMT expression. | N/A | Antibody: mouse monoclonal antibody MT 3.1 (Dako). | The fraction of immunopositive tumour cells was evaluated semiquantitatively and categorised according to the following immunoreactivity scores: 0, no positive tumour cells; 1, weak expression < 10% positive tumour cells; 2, moderate expression 10–50% positive tumour cells; 3, strong expression > 50% positive tumour cells. |
| Technique: MSP Sample type: frozen (14 FFPE) CpG sites: NR Threshold: NR |
Methylation‐specific PCR | Methylated MGMT promoter: 5'‐gtttttagaacgttttgcgtttcgac‐3' and 5'‐ caccgtcccgaaaaaaaactccg‐3', amplify a 122‐bp fragment. Unmethylated MGMT promoter sequences were 5'‐tgtgtttttagaatgttttgtgttttgat‐3' and 5'‐ctaccaccatcccaaaaaaaaactcca‐3', amplify a 129‐bp fragment. | — | N/A | |
| Technique: PCR‐mRNA Sample type: frozen (14 FFPE) CpG sites: N/A Threshold: < 50% |
Expression of MGMT transcripts was determined by real‐time reverse transcription‐PCR using the ABI PRISM 5700 sequence detection system (Applied Biosystems). The transcript level of MGMT was normalised to the transcript level of ARF1 (ADP‐ribosylation factor 1, GenBank accession‐no. M36340). | MGMT‐RT‐F, 5'‐tgcacagcctggctgaatg‐3' and MGMT‐RT‐R, 5' ggtgaacgactcttgctggaaa‐3' resulting in a 102‐bp fragment. | mRNA: commercially available adult human brain RNA (BD Biosciences) was used as reference for the mRNA expression. | NR | |
| Havik 2012 | Technique: MSP Sample type: frozen CpG sites: 76–80 and 84–87 Threshold: NR |
"For MSP, melting curve analysis was used to detect PCR products in our samples (35)…Three replicates of each sample were used to ensure statistical representativity. Real‐time PCR followed by melting curve analysis was run on a CFX96 Touch™ Real‐ Time PCR Detection system (Bio‐Rad Laboratories)…Following the last cycle, PCR products were incubated for 10 s at 95°C before the melting curve was generated by heating from 65°C to 95°C in increments of 0.5°C/5 s while continuously measuring the fluorescence. The melting curves were analyzed using Bio‐Rad CFX Manager Software (Bio‐Rad Laboratories). Melting peaks determined for methylated and unmethylated controls, respectively, were used to identify methylated and unmethylated PCR products in the samples (EpiTect PCR Control DNA Set, cat. number 59695; Qiagen). Samples having only methylated PCR products and samples having both methylated and unmethylated PCR products were both scored as methylation‐positive." | Correspond to those used in Esteller 1999. MSP‐MGMT‐methylated forward 5'‐TTTCGACGTTCGTAGGTTTTCGC‐3', MSP‐MGMT‐methylated reverse 5'‐GCACTCTTCCGAAAACGAAACG‐3', MSP‐MGMT‐unmethylated forward 5'‐TTTGTGTTTTGATGTTTGTAGGTTTTTGT‐3', MSP‐MGMT‐unmethylated reverse 5'‐AACTCCACACTCTTCCAAAAACAAAACA‐3'. | — | "Samples having only methylated PCR products and samples having both methylated and unmethylated PCR products were both scored as methylation‐positive." |
| Technique: PCR‐HRM Sample type: frozen CpG sites: 72–83 Threshold: NR |
"Three replicates of each sample were used…Real‐time PCR followed by a melting curve step was run on a CFX96 Touch Real‐Time PCR Detection system (Bio‐Rad Laboratories)…The melting curve step was performed according to the company's recommendation (Bio‐Rad Laboratories)…The data files generated by the CFX96 system were imported using Precision Melt Analysis software (Bio‐Rad Laboratories) and further analyzed." | MGMT MS‐HRM2‐forward 5'‐GCGTTTCGGATATGTTGGGATA‐3', MGMT MS‐HRM2‐reverse 5'‐AACGACCCAAACACTCACCAAA‐3' | — | NR | |
| Technique: PSQ Sample type: frozen CpG sites: 74–78 Threshold: > 2.68% |
PSQ using the PyroMark MD System (Qiagen). Bisulfite‐treated DNA was amplified in a PCR reaction using primers from the PyroMark Q96 CpG MGMT kit (part number 972032, Qiagen). | NR | — | "The pyrosequencing threshold was determined from the mean methylation value in the five analyzed CpG sites and the mean standard deviation (X + 2SD) in the four meningiomas. Glioma samples were scored as methylation positive by pyrosequencing if all five CpG sites had methylation values higher than the resulting threshold of 2.68%." | |
| Technique: PSQ Sample type: frozen CpG sites: 74–78 Threshold: > 6% |
"PyroMark Q96 CpG MGMT kit (cat. number 972032; Qiagen) and the PyroMark MD system (Qiagen)." | PyroMark Q96 CpG MGMT kit (cat. number 972032; Qiagen). | — | "Receiver operating characteristic (ROC) curve analysis was used to estimate the optimal cut‐off value for the two PSQ assays, using the mean percentage MGMT methylation for the CpGs covered by the two assays. The area under the ROC curve (AUROC) was calculated after fitting ordinary logistic regressions with the dependent variable indicating if a patient lived at least 18 months after diagnosis or not. Methylation was included as an independent variable." | |
| Technique: PSQ Sample type: frozen CpG sites: 74–78 Threshold: > 7% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 74–78 Threshold: > 8% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 74–78 Threshold: > 9% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 76–79 Threshold: > 6% |
Thera 6%. PSQ. 6% cut‐off. "PyroMark therascreen MGMT kit (cat. number 971061; Qiagen) and the PyroMark Q24 system (Qiagen)" | PyroMark therascreen MGMT kit (catalogue number 971061; Qiagen) | — | "Receiver operating characteristic (ROC) curve analysis was used to estimate the optimal cut‐off value for the two PSQ assays, using the mean percentage MGMT methylation for the CpGs covered by the two assays. The area under the ROC curve (AUROC) was calculated after fitting ordinary logistic regressions with the dependent variable indicating if a patient lived at least 18 months after diagnosis or not. Methylation was included as an independent variable." | |
| Technique: PSQ Sample type: frozen CpG sites: 76–79 Threshold: > 7% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 76–79 Threshold: > 8% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 76–79 Threshold: > 9% | |||||
| Technique: qMSP Sample type: frozen CpG sites: 71–73 and 75–86 Threshold: NR |
"Quantitation of MGMT promoter methylation assessed by qMSP is described in (34)." | MGMT qMSP forward primer: 5'‐GCGTTTCGACGTTCGTAGGT‐3', reverse primer: 5'‐CACTCTTCCGAAAACGAAACG‐3'. MGMT_1 qMSP forward primer: 5'‐CGAATATACTAAAACAACCCGCG‐3', reverse primer: 5'‐TTTTTTCGGGAGCGAGGC‐3' (as in Havik 2012) | — | Percentage methylated reference 0 (from Havik 2012) (stated "None" in Johannessen 2018) "A threshold value for scoring methylation positive samples was defined based on the qMSP result of four meningiomas, which all had PMR values of zero in both qMSP assays." (from Havik 2012) | |
| Technique: qMSP Sample type: frozen CpG sites: 71–86 Threshold: > 0% |
MGMT promoter methylation was quantitatively assessed by 2 qMSP assays, each covering 11 CpG sites (CpGs). The 2 assays analysed CpGs in partially overlapping regions (Additional file 1: Figure S1), but detected methylation on opposite DNA strands. Primers (Medprobe) and 6‐FAM labelled minor groove binder (MGB) probes (Applied Biosystems, Life Technologies) were modified from 2 previously reported assays. | qMSP: forward primer GCGTTTCGACGTTCGTAGGT; reverse primer CACTCTTCCGAAAACGAAACG MGMT_1 qMSP: forward primer CGAATATACTAAAACAACCCGCG; reverse primer TTTTTTCGGGAGCGAGGC ALU qMSP: forward primer GGTTAGGTATAGTGGTTTATATTTGTAATTTTAGTA; reverse primer ATTAACTAAACTAATCTTAAACTCCTAACCTCA. | — | "Samples with a Ct‐value above 35 were censored (resulting in a quantity of 0). The percentage of methylated reference (PMR) was calculated for each sample from the median quantity value from the triplicates by dividing the MGMT/ALU quantity ratio in the target by the MGMT/ALU quantity ratio in the methylated control, and multiplying by 100. A threshold value for scoring methylation positive samples was defined based on the qMSP result of four meningiomas, which all had PMR values of zero in both qMSP assays. Only samples with a PMR value above zero in both assays were scored as methylation positive." | |
| Hsu 2017 (see Hsu 2015) | Technique: IHC Sample type: FFPE CpG sites: N/A Threshold: < 10% |
"Tissue sections were immunostained on BOND‐MAX immunostainer (Leica Microsystems). Normal brain was used as external positive control, a previously proven MGMT methylated GBM was used as negative control." | N/A | Antibody: clone MT3.1 (1:100; Thermo, Fremont, CA) | The staining intensity of endothelial cells was used as a reference for interpretation of positive or negative staining of tumour cells. Positive MGMT staining (IHC+) was defined as > 10% of tumour nuclei with the staining intensity similar to or slightly weaker than that of the adjacent endothelial cells (Fig. 1A). Negative MGMT staining was defined as staining that did not fulfil the positive criteria. |
| Technique: MSP Sample type: FFPE CpG sites: 76–80 and 84–87 Threshold: NR |
1‐step MSP was performed as previously described (Hsu 2013). | Unmethylated: USP‐F1 5' TTTGTGTTTTGATGTTTGTAGGTTTTTGT 3' and USP‐R1 5' AACTCCACACTCTTCCAAAAACAAAACA 3'; methylated: were MSP‐F1 50 TTTCGACGTTCGTAGGTTTTCGC 30 and MSP‐R1 5' GCACTCTTCCGAAAACGAAACG 3'. | — | Serial dilutions of the positive control were performed and the lowest concentration of methylated DNA to have a PCR product was 0.5%. MSP of each sample, including DNA extraction and bisulfite modification, was performed in duplicates in accompany with 100%, 0.5%, and 0% methylated DNA as positive, cut‐off, and negative controls in each run. Samples with PCR products of any intensity were regarded as a positive result, whereas those with no PCR products were negative for MSP. | |
| Technique: PSQ Sample type: FFPE CpG sites: 76–79 Threshold: > 5% |
"The methylation status of 4 CpG sites within MGMT promoter region (genomic sequence on chromosome 10 from 131,265,519 to 131,265,537: CGACGCCCGCAGGTCCT CG) was analyzed by therascreen MGMT Pyro Kit (Qiagen GmbH)." | Primers from the MGMT Pyro Kit. | — | According to the recommendation by the manufacturer. | |
| Technique: qMSP Sample type: FFPE CpG sites: 77–80 and 84–87 Threshold: > 0.04% |
"The qMSP was performed using QuantiTect SYBR Green PCR Kit (Qiagen GmbH, Hilden, Germany) as previously described (Hsu 2015)." | Methylated MGMT‐F1 5' TTTCGACGTTCGTAGGTTTTCGC 3', methylated MGMT‐R1 5' GCACTCTTCCGAAAACGAAACG 3'. | — | Median value based on results of the assay. | |
| Technique: qMSP Sample type: FFPE CpG sites: 77–80 and 84–87 Threshold: > 0.1% |
Based on authors previous data. | ||||
| Karayan‐Tapon 2010 | Technique: IHC Sample type: FFPE CpG sites: N/A Threshold: < 15.5% |
Percentage of positive cells was determined in the most highly stained areas of each tumour section by counting ≥ 200 contiguous cells. All tumoural cells with nuclear immuno‐staining (high or low intensity) were counted as positive. | N/A | Antibody: MT3.1 (Novus Biologicals) | Median value used as cut‐off. |
| Technique: MSP Sample type: frozen CpG sites: 76–80 and 84–87 Threshold: NR |
"The methylation status of the CpG island of MGMT promoter was determined using two‐stage PCR [32]." | Study references Palmisano et al. Cancer Research 60:5954–8 which in turn references Esteller et al. 1999. Primers for stage 1 (amplification) from Palmisano et al. 2000 MGMT‐forward, 5'‐GGATATGTTG GGATAGTT‐3'; and MGMT‐reverse, 5'‐CCAAAAACCCCAAACCC‐3'. Primers for stage 2 from Esteller et al. 1999: Primer sequences for the unmethylated reaction were 5'‐TTTGTGTTTTGATGTTTGTAGGTTTTTGT‐3' (forward primer) and 5'‐AACTCCACACTCTTCCAAAAACAAAACA‐3' (reverse primer), and for the methylated reaction they were 5'‐TTTCGACGTTCGTAGGTTTTCGC‐3' (forward primer) and 5'‐GCACTCTTCCGAAAACGAAACG‐3' (reverse primer). | — | NR | |
| Technique: PCR‐mRNA Sample type: frozen CpG sites: N/A Threshold: < 0.39 |
Quantitative real‐time PCR. "RNA (1 μg) was reversed‐transcribed using the Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA). The relative expression of MGMT was quantified using the Applied Biosystems TaqMan FAM‐labeled probes for MGMT and three housekeeping genes: 18S, RPLPO, and GAPDH. The expression of MGMT in tumors was compared with the expression of MGMT in PBMC (unmethylated DNA) by the 2^‐ΔΔCt method [34] using the average Ct of the three housekeeping genes for normalization." | N/A | mRNA: "RNA (1 μg) was reversed‐transcribed using the Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA). The relative expression of MGMT was quantified using the Applied Biosystems TaqMan FAM‐labeled probes for MGMT and three housekeeping genes: 18S, RPLPO, and GAPDH. The expression of MGMT in tumors was compared with the expression of MGMT in PBMC (unmethylated DNA) by the 2^‐ΔΔCt method [34] using the average Ct of the three housekeeping genes for normalization." | Median value used as cut‐off. | |
| Technique: PSQ Sample type: frozen CpG sites: 74 Threshold: > 5.5% |
PSQ. CpG 1. "The pyrosequencing methylation assay was performed with the PyroMarkTM MGMT kit (Biotage, Uppsala, Sweden) on a PSQTM96 MA system (Biotage, Uppsala, Sweden), according to the manufacturer's protocol. The PyroMarkTM MGMT kit detects the level of methylation of five CpG sites located in the first exon of the MGMT gene." | PyroMarkTM MGMT kit (Biotage, Uppsala, Sweden). | — | Median value used as cut‐off. | |
| Technique: PSQ Sample type: frozen CpG sites: 74–78 Threshold: > 8.0% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 75 Threshold: > 8.7% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 76 Threshold: > 8.0% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 77 Threshold: > 7.85% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 78 Threshold: > 7.8% | |||||
| Technique: SQ‐MSP Sample type: frozen CpG sites: 76–80 and 84–87 Threshold: > 35 |
SQ‐MSP. "Amplifications were carried out on an MX4000 instrument with the Brilliant SYBR Green Core kit (Stratagene, La Jolla, CA) or on an Applied Biosystems ABI‐PRISM 7900 with the Applera SYBR Green master mix (Applied Biosystems, Foster City, CA). Methylation index (MI) was calculated using a modification of the formula proposed by Fackler et al.: %M = 100 – [(CtM/ CtM + CtUM) × 100] [33]." | Study references Palmisano et al. Cancer Research 60:5954–8 which in turn references Esteller et al. 1999. Primers for stage 1 (amplification) from Palmisano et al. 2000 MGMT‐forward, 5'‐GGATATGTTG GGATAGTT‐3'; and MGMT‐reverse, 5'‐CCAAAAACCCCAAACCC‐3'. Primers for stage 2 from Esteller et al. 1999: Primer sequences for the unmethylated reaction were 5'‐TTTGTGTTTTGATGTTTGTAGGTTTTTGT‐3' (forward primer) and 5'‐AACTCCACACTCTTCCAAAAACAAAACA‐3' (reverse primer), and for the methylated reaction they were 5'‐TTTCGACGTTCGTAGGTTTTCGC‐3' (forward primer) and 5'‐GCACTCTTCCGAAAACGAAACG‐3' (reverse primer). | — | Median value used as cut‐off. | |
| Kim 2016 | Technique: MSP Sample type: FFPE CpG sites: 76–80 and 84–87 Threshold: NR |
SQ‐MSP. FFPE, 12% cut‐off. "sqMSPCR was performed with primers specific for either "methylated" or "unmethylated" DNA. Forward primers were labeled at their 5' end with a fluorescent reporter dye (FAM), as previously described [17]. The PCR products corresponding to the "methylated" sequences have a size of 82bp while the "unmethylated" sequences have 12 additional nucleotides (94bp). Both fragments were amplified in the same reaction and PCR products were analyzed by capillary electrophoresis. Estimation of the amount of methylated DNA was calculated with the following formula, abbreviations are as follows; MF‐ "methylated" fraction, UM‐"unmethylated" fraction: (peak height of the MF/peak height of the UM + MF) × 100." Reference 17: Nguyen et al. Current Cancer Drug Targets 2015;15:624–40. | Study references Palmisano WA et al. Cancer Research 60:5954–8 which in turn references Esteller et al. 1999. Primers for stage 1 (amplification) from Palmisano et al. 2000 MGMT‐forward, 5'‐GGATATGTTG GGATAGTT‐3'; and MGMT‐reverse, 5'‐CCAAAAACCCCAAACCC‐3'. Primers for stage 2 from Esteller et al. 1999: Primer sequences for the unmethylated reaction were 5'‐TTTGTGTTTTGATGTTTGTAGGTTTTTGT‐3' (forward primer) and 5'‐AACTCCACACTCTTCCAAAAACAAAACA‐3' (reverse primer), and for the methylated reaction they were 5'‐TTTCGACGTTCGTAGGTTTTCGC‐3' (forward primer) and 5'‐GCACTCTTCCGAAAACGAAACG‐3' (reverse primer). | — | NR |
| Technique: PSQ Sample type: FFPE CpG sites: 74–78 Threshold: > 9% |
PSQ. "The PyroMark Q96 CpG MGMT kit5,10) (Ensembl ID: OTTHUMT00000051009) (Qiagen, Hilden, Germany)…PyroGold reagents were used for the PSQ reaction, and the signal was analyzed using the PSQ 96MA System (Biotage, Uppsala, Sweden). Target CpGs were evaluated by PSQ96MA 2.1 instrument software (Biotage, Uppsala, Sweden)." | PyroMark Q96 CpG MGMT kit (Ensembl ID: OTTHUMT00000051009) (Qiagen, Hilden, Germany). | — | "Receiver operating characteristic (ROC) curve analysis was used to determine the cut‐off value of mean percentage of methylation at the five CpGs for predicting the longer survival3). The area under the ROC curve (AUC) was used to determine the optimal threshold of the mean percentage of the methylation at the five CpGs." | |
| Kristensen 2016 | Technique: IHC Sample type: FFPE CpG sites: N/A Threshold: at 0% |
"Formalin‐fixed, paraffin‐embedded sections were deparaffinized in xylene and rehydrated in decreasing concentrations of ethanol; Immunoreactivity was visualized with DAB + (DAKO K3468) as chromogen. The immunohistochemical reactions were semiquantitatively evaluated according to the number of tumor cells stained; For MGMT evaluation, positive endothelial cells, lymphocytes, and microglia served as positive internal controls." | N/A | Antibody: monoclonal mouse anti‐human antibody against MGMT (MAB16200, 1:200, Millipore) | NR |
| Technique: PSQ Sample type: frozen CpG sites: NR Threshold: > 10% |
Standard PSQ. "PCR and pyrosequencing were performed using the Therascreen (R) MGMT Pyro (R) kit according to the manufacturer's instructions with slight modifications." | Supp Fig 1 | — | NR | |
| Technique: qMSP‐PSQ Sample type: frozen CpG sites: NR Threshold: > 0.1% |
qMSP‐PSQ. Quantitative and allelic methylation analyses were performed using qMSP and melting analyses followed by PSQ of methylation‐positive samples being heterozygous for the rs16906252 SNP. Flowchart of method in Fig 1 of the publication. Sodium bisulfite conversion of the samples was performed using the EZ DNA Methylation kit (Zymo Research) according to the manufactures' instructions, with slight modifications; samples were incubated at 42 °C for 30 minutes instead of 37 °C for 15 minutes. For the bisulfite reaction the alternative incubation conditions described in the appendix were used. The LightCycler 480 (Roche Life Science) was used for real‐time PCR and melting analysis. The real‐time PCR cycling protocol started with one cycle of 95 °C for 10 minutes, followed by 40 cycles of 95 °C for 20 seconds, 70 °C for 20 seconds, 72 °C for 20 seconds. The melting step was performed from 65 °C to 95 °C after a denaturation step of 1 minute at 95 °C and a hybridization step of 40 °C for 1 minute. For the reaction mixtures the SYBR Green Master Mix (Roche) was used at a final concentration at 1×. Final primer concentrations were 200 nM of each primer, and 25 ng of DNA was used as template. The final reaction volume was 20 μL. Primer sequences have been published previously. The Alu assay used for normalisation was used without the TaqMan probe using an intercalating fluorescent dye instead as previously described. PSQ was performed on the PyroMark Q24 (Qiagen) using the PyroMark Gold Q24 reagents (Qiagen), according to the manufactures' instructions. | Supp Fig 1 | — | This technical cut‐off was defined following an evaluation of a serial dilution series of methylated DNA into unmethylated. | |
| Technique: qMSP‐PSQ Sample type: frozen CpG sites: NR Threshold: > 5% | |||||
| Technique: qMSP‐PSQ Sample type: frozen CpG sites: NR Threshold: > 20% | |||||
| Lalezari 2013 | Technique: IHC Sample type: FFPE CpG sites: N/A Threshold: < 30% |
"MGMT immunoreactivity was semi‐quantitatively assessed by counting the immunostained tumor nuclei as a percentage of the total tumor nuclei…All immunohistochemical analyses were performed blinded to methylation status and clinical information." | N/A | Antibody: MT3.1 (Millipore) | Median value used as cut‐off. |
| Technique: MSP Sample type: FFPE CpG sites: 76–80 and 84–87 Threshold: NR |
MGMT methylation analysis was performed by MSP according to a previously published protocol with slight modifications. Samples were subjected to a 2‐stage nested PCR strategy using 2 sets of primers. | First‐stage primers (5'‐GGATATGTTGGGATAGTT‐3' and 5'‐CCAAAAACCCCAAACCC‐3') and second‐stage primers (unmethylated reaction: 5'‐TTTGTGTTTTGATGTTTGTA‐GGTTTTTGT‐3' and 5'‐AACTCCACACTCTTCCAAAAACAAAACA‐3'; methylated reaction: 5_‐TTTCGACGTTCGTAGGTTTTCGC‐3' and 5'‐GCACTCTTCCGAAAACGAA‐ACG‐3'). | — | NR | |
| Technique: PSQ Sample type: FFPE CpG sites: 72–95 Threshold: NR |
Bisulfite‐modified DNA, generated by the method described above, was used to sequence a portion of the MGMT promoter contiguous with and inclusive of the MSP region. Samples were sequenced with a 2‐stage nested PCR using the same first‐stage primers as those that were used in MSP: 5'‐GGATATGTTGGGATAGTT‐3' and 5'‐CCAAAAACCCCAAACCC‐3', and second stage primers 5'‐GGATATGTTGGGATAGTT‐3' and 5'‐CACCTAAAAAACACTTAAAAC‐3'. The sequence of each sample was determined using Chromas Lite 2.33 (Technelysium Pty Ltd). There did not appear to be any significant difference in yield compared to MSP. | First‐stage primers as those that were used in MSP: 5'‐GGATATGTTGGGATAGTT‐3’ and 5'‐CCAAAAACCCCAAACCC‐3', and second stage primers 5'‐GGATATGTTGGGATAGTT‐3' and 5'‐CACCTAAAAAACACTTAAAAC‐3'. | — | Median number of methylated CpG sites used as the threshold defining hypomethylated (< 3 sites) and hypermethylated (≥ 3 sites) | |
| Lattanzio 2015 | Technique: MSP Sample type: FFPE CpG sites: 76–80 and 84–87 Threshold: NR |
MSP using "primers amplifying the exon 1 region of the MGMT gene including the CpG island and subsequently the specific primers for either methylated or unmethylated DNA established by Esteller et al (12). Primers used in the first PCR reaction were: 5'‐GGATATGTTGGGATAGTT‐3' (forward primer, GenBank accession number AL355531, nucleotides 46891 to 46908) and 5'‐CCAAAAACCCCAAACCC‐3' (reverse primer, GenBank accession number AL355531, nucleotides 47162 to 47179) amplifying a 289‐bp fragment…Frozen and FFPE tissue samples were analyzed in triplicate." DNA extracted from snap‐frozen samples. | "primers amplifying the exon 1 region of the MGMT gene including the CpG island and subsequently the specific primers for either methylated or unmethylated DNA established by Esteller et al (12). Primers used in the first PCR reaction were: 5'‐GGATATGTTGGGATAGTT‐3' (forward primer, GenBank accession number AL355531, nucleotides 46891 to 46908) and 5'‐CCAAAAACCCCAAACCC‐3' (reverse primer, GenBank accession number AL355531, nucleotides 47162 to 47179) amplifying a 289‐bp fragment." Primers from Esteller et al. 1999: primer sequences for the unmethylated reaction were 5'‐TTTGTGTTTTGATGTTTGTAGGTTTTTGT‐3' (forward primer) and 5'‐AACTCCACACTCTTCCAAAAACAAAACA‐3' (reverse primer), and for the methylated reaction they were 5'‐TTTCGACGTTCGTAGGTTTTCGC‐3' (forward primer) and 5'‐GCACTCTTCCGAAAACGAAACG‐3' (reverse primer). | — | "the results were qualitatively interpreted as follows: a visible band in the M primer set and absence of the U primer set product indicated a positive methylation status, whereas absence of a M primer set product and presence of a band in the U primer set was evaluated as a negative methylation status." |
| Technique: MSP Sample type: frozen CpG sites: 76–80 and 84–87 Threshold: NR |
MSP using "primers amplifying the exon 1 region of the MGMT gene including the CpG island and subsequently the specific primers for either methylated or unmethylated DNA established by Esteller et al (12). Primers used in the first PCR reaction were: 5'‐GGATATGTTGGGATAGTT‐3' (forward primer, GenBank accession number AL355531, nucleotides 46891 to 46908) and 5'‐CCAAAAACCCCAAACCC‐3' (reverse primer, GenBank accession number AL355531, nucleotides 47162 to 47179) amplifying a 289‐bp fragment…Frozen and FFPE tissue samples were analyzed in triplicate." DNA extracted from snap‐frozen samples. | "primers amplifying the exon 1 region of the MGMT gene including the CpG island and subsequently the specific primers for either methylated or unmethylated DNA established by Esteller et al (12). Primers used in the first PCR reaction were: 5'‐GGATATGTTGGGATAGTT‐3' (forward primer, GenBank accession number AL355531, nucleotides 46891 to 46908) and 5'‐CCAAAAACCCCAAACCC‐3' (reverse primer, GenBank accession number AL355531, nucleotides 47162 to 47179) amplifying a 289‐bp fragment." Primers from Esteller et al. 1999: primer sequences for the unmethylated reaction were 5'‐TTTGTGTTTTGATGTTTGTAGGTTTTTGT‐3' (forward primer) and 5'‐AACTCCACACTCTTCCAAAAACAAAACA‐3' (reverse primer), and for the methylated reaction they were 5'‐TTTCGACGTTCGTAGGTTTTCGC‐3' (forward primer) and 5'‐GCACTCTTCCGAAAACGAAACG‐3' (reverse primer). | — | "the results were qualitatively interpreted as follows: a visible band in the M primer set and absence of the U primer set product indicated a positive methylation status, whereas absence of a M primer set product and presence of a band in the U primer set was evaluated as a negative methylation status." | |
| Technique: PSQ Sample type: FFPE CpG sites: 72–80 Threshold: ≥ 9% |
DNA extracted from FFPE samples. "PSQ was performed using the PyroMark ID System (Biotage, Uppsala, Sweden). The PSQ primers used for amplification of bisulfite‐treated DNAs were designed to cover a region including 9 CpG sites of the MGMT promoter at the beginning of the first exon, adjacent to the region covered by MSP primers (specifically, CpGs 5–6–7–8–9 in the pyrograms corresponded to CpGs included in specific M/U MSP primers). The primers were 5'‐GGATATGTTGGGATAGTT‐3' (forward primer, GenBank accession number AL355531, nucleotides 46891 to 46908) and 5'‐biotin‐ ACCCAAACACTCACCAAA‐3' (reverse primer, GenBank accession number AL355531, nucleotides 46972 to 46990), which amplified a 99‐bp region…PSQ using the forward primer as sequencing primer…Resulting data were analyzed and quantified with PyroMark CpG Software (Biotage)…All samples were analyzed in duplicate." | 5'‐GGATATGTTGGGATAGTT‐3' (forward primer, GenBank accession number AL355531, nucleotides 46891 to 46908) and 5'‐biotin‐ ACCCAAACACTCACCAAA‐3' (reverse primer, GenBank accession number AL355531, nucleotides 46972 to 46990). Forward primer also used as the sequencing primer. | — | "To determine the methylation cutoff value for PSQ analysis, we extracted DNA from a pool of 5 normal brain tissues derived from autopsies; the average percentage of methylation of the 5 samples was 8%; thus we considered methylated any tumor sample carrying ≥9% methylation." | |
| Technique: PSQ Sample type: frozen CpG sites: 72–80 Threshold: ≥ 9% |
DNA extracted from snap‐frozen samples. PSQ performed as above. | ||||
| Lechapt‐Zalcman 2012 | Technique: IHC Sample type: FFPE CpG sites: N/A Threshold: < 15% |
Immunostaining was performed using heat‐induced epitope retrieval, pH 9.0, a labelled method (EnVision Kit; Dako SA, Trappes, France), and automate immunostainer (Dako SA) according to the manufacturer's protocol. Negative controls consisted of omitting the primary antibody and replacing it with an irrelevant antibody of similar isotype. Endothelial staining was used as an internal positive control. A pathologist who was blind to the people' clinical and MGMT methylation data independently evaluated MGMT staining using a light microscope at 400 magnification. Specimens without valid internal positive controls were excluded from the analysis. For each specimen, 5–10 images of representative fields were then acquired at 400 magnification. 360–1790 tumour cells were counted in specimen, and the percentage of positive tumour nuclei was calculated. Endothelial and inflammatory cells were excluded from the cell counts. | N/A | Antibody: a mouse primary antibody against MGMT (clone MT3.1; Chemicon International, Temecula, Calif) was used at 1:200 dilution. mRNA: NA | This cut‐off was the median value of reactivity in the GBM series. |
| Technique: MSP Sample type: FFPE CpG sites: 76–80 and 84–87 Threshold: NR |
MSP was performed using a 2‐step approach. Bisulfite modification of genomic DNA was undertaken by means of the Epitect Kit (Qiagen SA) according to the manufacturer's recommendation. PCR amplification was carried out as described by Esteller et al. PCR products were loaded onto 5% agarose gels, stained with GelRed (Interchim, Montlucon, France), and observed under ultraviolet illumination. | 5'‐TTTGTGTTTTGATGTTTGTAGGTTTTTGT‐3' (forward primer) and 5'‐AACTCCACACTCTTCCAAAAA CAAAACA‐3' (reverse primer) for the unmethylated product and 5'‐TTTCGACGTTCGTAGGTTTTCGC‐3' (forward primer) and 5'‐GCACTCTTCCGAAAACGAAA CG‐3' (reverse primer) for the methylated product. | — | N/A | |
| McDonald 2013 | Technique: MSP Sample type: FFPE CpG sites: 76–80 Threshold: NR |
(From Estellar et al., 1999) 1 μg of DNA was denatured by sodium hydroxide and modified by sodium bisulfite. DNA samples were then purified using Wizard DNA purification resin (Promega), again treated with sodium hydroxide, precipitated with ethanol, and resuspended in water. Controls without DNA were performed for each set of PCRs. Each PCR reaction (10 μL) was directly loaded onto nondenaturing 6% polyacrylamide gels, stained with ethidium bromide, and visualised under ultraviolet illumination. | Primer sequences of MGMT were for the unmethylated reaction 5'‐TTTGTGTTTTGATGTTTGTAGGTTTTTGT‐3' (upper primer) and 5'‐AACTCCACACTCTTCCAAAAACAAAACA‐3' (lower primer) and for the methylated reaction 5'‐TTTCGACGTTCGTAGGTTTTCGC‐3' (upper primer) and 5'‐GCACTCTTCCGAAAACGAAACG‐3' (lower primer) | — | NR |
| Technique: PSQ Sample type: FFPE CpG sites: 74–78 Threshold: > 8% |
PSQ. Tumour DNA (500 ng) was bisulphite modified using the EZ DNA methylation kit (Zymo Research, Orange CA) according to the manufacturer's recommendations. The CpG PSQ methylation assay was performed with the PyroMark MGMT kit (Qiagen) on a PSQe96 MA system (Qiagen) according to the manufacturer's protocol. Methylation was quantified using the Pyromark CpG software (Qiagen). | NR | — | Determined through a series of segmented regressions where the CpG PSQ values were regressed against their rank order. The model with the cut‐off of 8% CpG PSQ resulted in the minimum mean square error and was thus chosen (Supplementary Fig 1). | |
| Melguizo 2012 | Technique: IHC Sample type: FFPE CpG sites: N/A Threshold: < 25% |
Immunostaining was performed using the Bond Polymer Refine Detection system (Leica Microsistemas S.L.U, Barcelona, Spain). Readings were taken automatically with the ACIS III DAKO system for quantification IHC and were verified by 2 experienced pathologists. | N/A | Antibody: 1:50; Santa Cruz Biotechnology, incmRNA: NA | NR |
| Technique: MSP Sample type: NR CpG sites: 76–80 and 84–87 Threshold: NR |
Methylation patterns in the CpG island of MGMT were determined by chemical modification of unmethylated, but not methylated, cytosine to uracil. | 5'‐TTTGTGTTTTGATGTTTGTAGGTTTTTGT‐3' (forward primer) and 5'‐AACTCCACACTCTTCCAAAAA CAAAACA‐3' (reverse primer) for the unmethylated product and 5'‐TTTCGACGTTCGTAGGTTTTCGC‐3' (forward primer) and 5'‐GCACTCTTCCGAAAACGAAA CG‐3' (reverse primer) for the methylated product. | — | NR | |
| Nguyen 2015 | Technique: FSQ‐MS‐PCR Sample type: frozen or FFPE CpG sites: 76–80 and 84–87 Threshold: > 15% |
FSQ‐MS‐PCR was using specific primers in a semiquantitative multiplexed fluorescent MS‐PCR. | Unmethylated cytosines, were 5'‐TTTGTGTTTTGATGTTTGTAGGTTTTTGT‐3' (forward primer, UF) and 5'‐AACTCCACACTCTTCCAAAAAC AAAACA‐3' (reverse primer, UR), and the specific primers for methylated cytosines were 5'‐TTTCGACGTTCGTA GGTTTTCGC‐3' (forward primer, MF) and 5'‐GCACTCTT CCGAAAACGAAACG‐3' (reverse primer, MR). | — | Outcome‐based approach used. Assessed effect of multiple cut‐off points on survival and determined the cut‐off point with the best statistical significance (p value) and the ones associated with the shortest and longest survivals. |
| Technique: FSQ‐MS‐PCR Sample type: frozen or FFPE CpG sites: 76–80 and 84–87 Threshold: > 60% | |||||
| Park 2011 | Technique: MS‐MLPA Sample type: 50% frozen and 50% FFPE CpG sites: NR Threshold: > 0.1% |
Used MS‐MLPA probe mix prepared by MRC‐Holland (Salsa MS‐MLPA Kit ME011 MMR), which included 3 probes specific for the MGMT promoter region containing a HhaI recognition site. The procedure was performed according to the manufacturer's protocol. HhaI (R6441; Promega), a methylation‐sensitive restriction enzyme that cuts unmethylated GCGC sites was applied to each set of samples. The resultant PCR fragments were separated by capillary gel electrophoresis (ABI Prism 7000/7700, Applied Biosystems). The methylation status was quantified using GeneMarker software (version 1.5, Soft Genetics). To compensate for the differences in the efficiency of the PCR for the individual samples, the peak value of each probe was normalised by dividing it by the peak of the control probes. To evaluate the methylation status, the methylation ratio was calculated by the mean of dividing each normalised peak value of the digested sample by that of the corresponding undigested sample. This value corresponds to the percentage of methylated sequences. | NR | — | Outcome‐based approach: chose the best cut‐off to predict early‐response evalution (progression/pseudoprogression). |
| Technique: MS‐MLPA Sample type: 50% frozen and 50% FFPE CpG sites: NR Threshold: > 0.2 | |||||
| Technique: MSP Sample type: 50% frozen and 50% FFPE CpG sites: 76–80 and 84–86 Threshold: NR |
The obtained PCR products were electrophoresed in 2% agarose gels and visualised under ultraviolet illumination after staining with ethidium bromide. For the evaluation of the assay results, the products from the controls were examined first. The MGMT gene promoter fragments in the controls were observed at 80 and 92 bp for the methylated DNA–methylated primer and unmethylated DNA–unmethylated primer combinations, respectively. The methylated DNA–unmethylated primer and unmethylated DNA–methylated primer controls were not expected to show any bands. If the control results were acceptable, participant samples were evaluated for the presence of amplification with the methylated and unmethylated primers. | The primer sequences for the MGMT were as follows: methylated forward: 5'‐TTT CGA CGT TCG TAG GTT TTC GC‐3', methylated reverse: 5'‐GCA CTC TTC CGA AAA CGA AAC G‐3', unmethylated forward: 5'‐TTT GTG TTT TGA TGT TTG TAG GTT TTT GT‐3', unmethylated reverse: 5'‐AAC TCC ACA CTC TTC CAA AAA CAA AAC A‐3'. | — | NR | |
| Quillien 2014 (test) | Technique: IHC Sample type: FFPE CpG sites: N/A Threshold: < 23% |
References Chinot et al. Journal of Clinical Oncology 2007;25:1470–5. Percentage of positive tumour cells determined by a pathologist. | N/A | Antibody: MT‐1; Chemicon, Temecula, CA (From Chinot et al. Journal of Clinical Oncology 2007;25:1470–5, NR in Quillien 2012) | "Optimal risk cutoffs were therefore determined as the threshold values of the continuous distribution that best separated low‐ and high‐risk people according to their outcomes (outcome based method). More precisely, they were defined as the thresholds that optimized the area under the receiver operating characteristic (ROC) curve obtained with a Cox model 25 using overall survival (OS) adjusted for age and Karnofsky score (the proportional hazard assumption was checked)." |
| Technique: MSP Sample type: frozen CpG sites: 76–80 and 84–87 Threshold: NR |
2‐stage PCR | Study references Karayan‐Tapon et al. Journal of Neuro‐oncology 2010;97:311–22, which in turn references Palmisano et al. Cancer Research 20;60:5954–8 which in turn references Esteller et al. 1999. Primers for stage 1 (amplification) from Palmisano et al. 2000 MGMT‐forward, 5'‐GGATATGTTG GGATAGTT‐3'; and MGMT‐reverse, 5'‐CCAAAAACCCCAAACCC‐3'. Primers for stage 2 from Esteller et al. 1999: primer sequences for the unmethylated reaction were 5'‐TTTGTGTTTTGATGTTTGTAGGTTTTTGT‐3' (forward primer) and 5'‐AACTCCACACTCTTCCAAAAACAAAACA‐3' (reverse primer), and for the methylated reaction they were 5'‐TTTCGACGTTCGTAGGTTTTCGC‐3' (forward primer) and 5'‐GCACTCTTCCGAAAACGAAACG‐3' (reverse primer). | — | NR | |
| Technique: MethyLight‐MSP Sample type: frozen CpG sites: 75–86 Threshold: > 0 |
MethyLight. Paper cites Metellus P et al. Cancer 2009;115:4783–94. "real‐time, fluorescence‐based polymerase chain reaction (PCR) was performed using the Light Cycler 480 (Roche Diagnostics, Meylan, France). Bisulfite‐converted genomic DNA was amplified using a set of primers and a fluorescent dye‐labeled oligonucleotide probe, resulting in a semiquantitative methylation analysis." In Quillien 2012: "The differences in amounts of input genomic DNA were normalized by the collagen type II, alpha 1 gene (COL2A1). The percentage of methylated reference was calculated as follows: the methylated MGMT/COL2A1 ratio for each sample was divided by the same ratio obtained for a SssI‐treated genomic DNA used as standard, and values were multiplied by 100." | References Widschwendter et al. Cancer Research 2004;64:3807–13. Forward primer sequence 5'‐GCGTTTCGACGTTCGTAGGT‐3', reverse primer sequence 5'‐CACTCTTCCGAAAACGAAACG‐3' | — | "Optimal risk cutoffs were therefore determined as the threshold values of the continuous distribution that best separated low‐ and high‐risk people according to their outcomes (outcome based method). More precisely, they were defined as the thresholds that optimized the area under the receiver operating characteristic (ROC) curve obtained with a Cox model 25 using overall survival (OS) adjusted for age and Karnofsky score (the proportional hazard assumption was checked)." | |
| Technique: PCR‐HRM Sample type: frozen CpG sites: 70–83 Threshold: > 50% |
"PCR amplification and high‐resolution melting analysis were performed using a Mx3000P apparatus (Stratagene, La Jolla, Calif)…(forward: 5' GCGTTTCGGATATGTTGGGATAGT 3' and reverse: 5' AACGACCCAAACACTCACCAAA 3')…After amplification, a postamplification melting curve program was initiated by heating to 95°C for 1 minute, cooling to 70°C for 30 seconds, and increasing the temperature to 95°C (heating rate 0.01°C/s) while continuously measuring fluorescence. Control DNAs were extracted from blood. The methylated control was obtained by treatment with CpG Methylase M.sssI M0226S (New England Biolabs, Ipswich, Mass). Sample melting curves were compared with control melting curves obtained with unmethylated and methylated controls. When the peak corresponding to methylated DNA was >50% of the peak corresponding to unmethylated DNA, the sample was considered methylated. All reactions were performed in duplicate." | Forward: 5'‐GCGTTTCGGATATGTTGGGATAGT‐3', reverse: 5'‐AACGACCCAAACACTCACCAAA‐3'. | — | Melting‐curve method. "When the peak corresponding to methylated DNA was >50% of the peak corresponding to unmethylated DNA, the sample was considered methylated." | |
| Technique: PSQ Sample type: frozen CpG sites: 74 Threshold: > 4% |
"Pyrosequencing was performed with the PyroMark Q96 MGMT kit (Qiagen, Courtaboeuf, France) on a PSQTM96 MA system (Biotage, Uppsala, Sweden)." | PyroMark Q96 MGMT kit (Qiagen, Courtaboeuf, France) used. | — | "Optimal risk cutoffs were therefore determined as the threshold values of the continuous distribution that best separated low‐ and high‐risk people according to their outcomes (outcome based method). More precisely, they were defined as the thresholds that optimized the area under the receiver operating characteristic (ROC) curve obtained with a Cox model 25 using overall survival (OS) adjusted for age and Karnofsky score (the proportional hazard assumption was checked)." | |
| Technique: PSQ Sample type: frozen CpG sites: 74–78 Threshold: > 8% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 75 Threshold: > 11% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 76 Threshold: > 4% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 77 Threshold: > 6% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 78 Threshold: > 5% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 74 Threshold: > 8% |
Assay for CpG 74–83. Templates for PSQ were prepared by amplifying bisulfite modified DNA with a forward primer (GTTTYGGATATG TTGGGATAG) and a biotinylated reverse primer (AAAA CCACTCRAAACTACCAC). PSQ was performed using PyroGold Q96 SQA Reagents and the Pyro Q‐CpG software on a PyroMark ID pyrosequencer (Qiagen, Crawley, UK) as per manufacturer's recommendation. Full details for CpG location and PSQ can be found in Malley et al. [6] and Mullolland et al. [11]. | "forward primer (GTTTYGGATATGTTGGGATAG) and a biotinylated reverse primer (AAAACCACTCRAAACTACCAC). Two assays were designed and run on this template using two PSQ primers: GAT‐AGTTYGYGTTTTTAGAA (assay for CpGs 74–83) andGYGATTTGGTGAGTGTTTG (assay for CpGs 84–89)." | — | "For each of the 16 tested CpG, as well as for the mean of consecutive selected CpGs, optimal risk cut‐off was determined as the threshold value of the continuous distribution which best discriminates low‐ and high‐risk people according to their outcomes (outcome‐based method). these values were defined as the thresholds that optimized the area under the ROC curve obtained with a Cox model [12] using overall survival (OS) and progression‐free survival (PFS) adjusted for age and Karnofsky score (the proportional hazard assumption was checked)." | |
| Technique: PSQ Sample type: frozen CpG sites: 74–78 Threshold: > 9% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 74–89 Threshold: > 11% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 75–79 Threshold: > 8% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 76 Threshold: > 5% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 76–79 Threshold: > 8% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 76–80 Threshold: > 9% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 77 Threshold: > 7% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 77–81 Threshold: > 8% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 78 Threshold: > 4% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 78–82 Threshold: > 9% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 79 Threshold: > 7% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 79–83 Threshold: > 8% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 80 Threshold: > 4% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 81 Threshold: > 8% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 82 Threshold: > 16% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 83 Threshold: > 10% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 84 Threshold: > 9% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 84–88 Threshold: > 17% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 84–89 Threshold: > 22% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 85 Threshold: > 5% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 85–89 Threshold: > 13% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 86 Threshold: > 11% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 87 Threshold: > 25% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 88 Threshold: > 4% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 89 Threshold: > 12% | |||||
| Quillien 2014 (validation) | Technique: PSQ Sample type: FFPE CpG sites: 74–78 Threshold: > 9% |
Assay for CpG 74–83. PSQ performed using PyroGold Q96 SQA Reagents and the Pyro Q‐CpG software on a PyroMark ID pyrosequencer (Qiagen, Crawley, UK) as per manufacturer's recommendation. | Forward primer (GTTTYGGATATG TTGGGATAG) and a biotinylated reverse primer (AAAA CCACTCRAAACTACCAC). | — | This was the optimal risk cut‐off in the initial population of 89 participants with GBM. |
| Technique: PSQ Sample type: FFPE CpG sites: 74–78 Threshold: > 10% | |||||
| Technique: PSQ Sample type: FFPE CpG sites: 74–78 Threshold: > 28% | |||||
| Quillien 2016 | Technique: PSQ Sample type: FFPE CpG sites: 74–78 Threshold: > 6% |
FFPE, 6% cut‐off. "PSQ was performed as previously described [10, 12] using the PyroMark CpG MGMT kit (ref. 972032, Qiagen, France). All assays were performed in duplicate and each result was averaged together. The average percentage of the 5 CpGs tested was considered." | PyroMark CpG MGMT kit (ref. 972032, Qiagen, France). | — | Optimised cut‐off (current series/frozen samples). "Optimal risk cut‐offs were determined as previously described with age and performance status introduced as adjustment factors [10]." Reference 10: Quillien et al. Cancer 2012;118:4201–11. |
| Technique: PSQ Sample type: FFPE CpG sites: 74–78 Threshold: > 8% |
Optimised cut‐off (previous series/frozen samples). | ||||
| Technique: PSQ Sample type: FFPE CpG sites: 74–78 Threshold: > 12% |
Best level of concordance between frozen and FFPE samples. | ||||
| Technique: PSQ Sample type: FFPE CpG sites: 74–78 Threshold: > 13% |
Best level of concordance between frozen and FFPE samples. | ||||
| Technique: PSQ Sample type: FFPE CpG sites: 74–78 Threshold: > 16% |
Optimised cut‐off (current series/frozen samples). "Optimal risk cut‐offs were determined as previously described with age and performance status introduced as adjustment factors [10]." Reference 10: Quillien et al. Cancer 2012; 118:4201–11. | ||||
| Technique: PSQ Sample type: frozen CpG sites: 74–78 Threshold: > 6% | |||||
| Technique: PSQ Sample type: frozen CpG sites: 74–78 Threshold: > 8% |
Optimised cut‐off (previous series/frozen samples) | ||||
| Technique: PSQ Sample type: frozen CpG sites: 74–78 Threshold: > 12% or 13% |
Best level of concordance between frozen and FFPE samples | ||||
| Technique: PSQ Sample type: frozen CpG sites: 74–78 Threshold: > 12% |
PSQ cut‐off 12% (Qiagen kit) Hs_MGMT_01_PM PyroMark CpG assay (ref 970032 and 972032). | PyroMark CpG assay (ref 970032 and 972032). | — | The mean of the methylation at the 4 CpG sites as predefined in previous study (Quillien 2014) | |
| Technique: PSQ Sample type: frozen CpG sites: 74‐78 Threshold: > 16% |
PSQ. Frozen, 16% cut‐off. "PSQ was performed as previously described [10, 12] using the PyroMark CpG MGMT kit (ref. 972032, Qiagen, France). All assays were performed in duplicate and each result was averaged together. The average percentage of the 5 CpGs tested was considered." | PyroMark CpG MGMT kit (ref. 972032, Qiagen, France). | — | Optimised cut‐off (current series/frozen samples). "Optimal risk cut‐offs were determined as previously described with age and performance status introduced as adjustment factors [10]." Reference 10: Quillien et al. Cancer 2012; 118:4201–11. | |
| Technique: PSQ Sample type: frozen CpG sites: 76–79 Threshold: > 8% |
Therascreen MGMT Pyro Kit (ref. 971061, Qiagen, France) according to the manufacturer's instructions. | N/A | — | The mean of the methylation at the 4 CpG sites as predefined in previous study (Quillien 2014) | |
| Technique: PSQ Sample type: frozen CpG sites: 76–79 Threshold: > 12% | |||||
| Technique: SQ‐MSP Sample type: FFPE CpG sites: 76–80 and 84–87 Threshold: > 12% |
SQ‐MSP. FFPE, 12% cut‐off. "sqMSPCR was performed with primers specific for either "methylated" or "unmethylated" DNA. Forward primers were labeled at their 5' end with a fluorescent reporter dye (FAM), as previously described [17]. The PCR products corresponding to the "methylated" sequences have a size of 82bp while the "unmethylated" sequences have 12 additional nucleotides (94bp). Both fragments were amplified in the same reaction and PCR products were analyzed by capillary electrophoresis. Estimation of the amount of methylated DNA was calculated with the following formula, abbreviations are as follows; MF‐ "methylated" fraction, UM‐"unmethylated" fraction: (peak height of the MF/peak height of the UM + MF) × 100." Reference 17: Nguyen et al. Current Cancer Drug Targets 2015;15:624–40. | "The technique is using specific primers in a semi‐quantitative multiplexed fluorescent MS‐PCR. Primer sequences recognizing unmethylated cytosines were 5'‐TTTGTGTTTTGATGTTTGTAGGTTTTTGT‐3' (forward primer, UF) and 5'‐AACTCCACACTCTTCCAAAAACAAAACA‐3' (reverse primer, UR), and the specific primers for methylated cytosines were 5'‐TTTCGACGTTCGTAGGTTTTCGC‐3' (forward primer, MF) and 5'‐GCACTCTTCCGAAAACGAAACG‐3' (reverse primer, MR). The analyzed sequences were designed to target the distal part of the CpG island region of MGMT promoter, whose complete methylation has been correlated with MGMT promoter silencing in cancer cell lines and primary tumors [18]. Primers were labeled at their 5' end with a fluorescent reporter dye (FAM)." From: Nguyen et al. Current Cancer Drug Targets 2015;15:624–40. | — | Best level of concordance between frozen and FFPE samples. | |
| Technique: SQ‐MSP Sample type: FFPE CpG sites: 76–80 and 84–87 Threshold: > 13% |
Best level of concordance between frozen and FFPE samples. | ||||
| Technique: SQ‐MSP Sample type: FFPE CpG sites: 76–80 and 84–87 Threshold: > 23% |
Optimised cut‐off (current series/FFPE samples). "Optimal risk cut‐offs were determined as previously described with age and performance status introduced as adjustment factors [10]." Reference 10: Quillien V et al. Cancer 2012;118:4201–11. | ||||
| Technique: SQ‐MSP Sample type: frozen CpG sites: 76–80 and 84–87 Threshold: > 13% |
Optimised cut‐off (current series/frozen samples) and best level of concordance between frozen and FFPE samples. "Optimal risk cut‐offs were determined as previously described with age and performance status introduced as adjustment factors [10]." Reference 10: Quillien et al. Cancer 2012;118:4201–11. | ||||
| Technique: SQ‐MSP Sample type: frozen CpG sites: 76–80 and 84–87 Threshold: > 23% |
Optimised cut‐off (current series/FFPE samples). "Optimal risk cut‐offs were determined as previously described with age and performance status introduced as adjustment factors [10]." Reference 10: Quillien V et al. Cancer 2012;118:4201–11. | ||||
| Thon 2017 | Technique: MSP Sample type: frozen CpG sites: 76–80 and 84–87 Threshold: NR |
Bisulfite conversion of 200–400 ng DNA was performed with the EpiTect Bisulfite Kit (Qiagen) as described previously. | 2 pairs of primers, each specific for either the methylated or the unmethylated MGMT promoter region, were used as originally described by Esteller and colleagues. | — | NR |
| Technique: sequencing Sample type: frozen CpG sites: 75–99 (unclear) Threshold: > 50% |
Taken from Eigenbrod 2014: "The sequencing reaction covers a 316 bp region of the MGMT promoter with 25 CpG sites, including those detected by MSP (corresponding to CpG positions 2–14)." | — | — | "The MGMT promoter was considered "methylated" when more than half of the CpG sites (≥13 of the 25 CpG sites) were found to be "methylated" or "partially methylated." A "partially methylated" CpG site was defined as the cytosine peaks being 50 % or more of the corresponding thymine peak. Positions with cytosine peaks as small as 10–50 % of the thymine peak were considered weakly methylated. When 9 – 12 of 25 CpG sites were "methylated/partially methylated" the MGMT promoter was considered "partially" methylated. When more than 9 of the 25 CpG sites were "methylated/partially methylated" the MGMT promoter was considered not methylated." "Sequencing of bisulfite‐modified DNA indicated a methylated promoter when more than half of the CpG sites (13 of 25 CpG sites) were found to be methylated; partial methylation was defined as the cytosine and thymine peaks being equally sized or the cytosine peak being twice as high as the corresponding thymine peak." | |
| Yamashita 2018 | Technique: MSP Sample type: frozen CpG sites: 76–80 and 84–87 Threshold: NR |
"Converted DNA was subjected to MS‐PCR using 2 primer pairs designed for the amplification of methylated and unmethylated alleles of the MGMT promoter…Amplified products were loaded on 16% polyacrylamide gels and visualized under ultraviolet light using ethidium bromide staining." | "The primer sequences for unmethylated reactions was 5'‐TTTGTGTTTTGATGTTTGTAGGTTTTTGT‐3' (forward), 5'‐AACTCCACACTCTTCCAAAAACAAAACA‐3' (reverse); for methylated reactions it was 5'‐TTTCGACGTTCGTAGGTTTTCGC‐3' (forward), 5'‐GCACTCTTCCGAAAACGAAACG‐3' (reverse)." | — | Samples with 5% methylation featured a faint positive band, suggesting that the appropriate cut‐off value for MS‐PCR was 5%. |
| Technique: PCR‐HRM Sample type: frozen CpG sites: 72–89 Threshold: > 5% |
StepOne system (Thermo Fisher Scientific). "MS‐HRM data were analyzed using HRM software (version 3.0.1, Thermo Fisher Scientific). Output plots were produced as aligned melting curves…The area under the curve (AUC) was calculated from the aligned melting curves using ImageJ (NIH); linear regression was applied to interpolate unknown samples from the standards…All measurements were performed in triplicate." | "The primers sets were 5'‐GCGTTTCGGATATGTTGGGATAGT‐3' (forward), 5'‐CCTACAAAACCACTCGAAACTACCA‐3' (reverse) primer 1." | — | Validation of ROC analysis. | |
| Technique: PCR‐HRM Sample type: frozen CpG sites: 72–89 Threshold: > 8% |
Validation of ROC analysis. | ||||
| Technique: PCR‐HRM Sample type: frozen CpG sites: 72–89 Threshold: > 10% |
From ROC analysis. | ||||
| Technique: PCR‐HRM Sample type: frozen CpG sites: 72–89 Threshold: > 12% |
Validation of ROC analysis. | ||||
| Technique: PCR‐HRM Sample type: frozen CpG sites: 72–89 Threshold: > 15% |
Validation of ROC analysis. | ||||
| Yang 2012 | Technique: IHC Sample type: FFPE CpG sites: N/A Threshold: < 10% |
"Staining for MGMT protein on primary tumor samples was performed using anti‐MGMT antibody clone MT3.1 (Abcam, Cambridge, England)…To calculate the MGMT labeling index of MGMT‐positive cells, the number of immunoreactive tumor cells was determined for at least 1,000 cells in randomly selected fields." | N/A | Antibody: MT3.1 (Abcam, Cambridge, UK). | NR |
| Technique: MSP Sample type: FFPE CpG sites: 76–80 and 84–87 Threshold: NR |
"The converted DNA was subjected to methylation‐specific PCR using two primer sets designed for amplifying the methylated or unmethylated allele of the MGMT promoter…Amplified products were separated on a 3% agarose gel and visualized under UV illumination." | "The primer sequences of MGMT for the unmethylated and methylated reactions were as follows: 5'‐TTTGTGTTTTGATGTTTGTAGGTTTTTGT‐3' (forward) and 5'‐AACTCCACACTCTTCCAAAAACAAAACA‐3' (reverse); and 5'‐TTTCGACGTTCGTAGGTTTTCGC‐3' (forward) and 5'‐GCACTCTTCCGAAAACGAAACG‐3' (reverse), respectively." | — | NR | |
| Yoshioka 2018 | Technique: SQ‐MSP Sample type: frozen CpG sites: 76–80 and 84–87 Threshold: > 0 |
Brilliant II SYBR Green qPCR Master Mix and 2 types of primers were used for MSP. | Corresponded to those used in Esteller 1999. mMGMT forward5'‐TTTCGACGTTCGTAGGTTTTCGC‐3', mMGMTreverse 5'‐GCACTCTTCCGAAAACGAAACG‐3', uMGMT forward 5'‐TTTGTGTTTTGATGTTTGTAGGTTTTTGT‐3', and uMGMT reverse 5'‐AACTCCACACTCTTCCAAAAACAAAACA‐3'. | — | Based on Delta Ct values "The ΔCt values of the tumors having no peak at 81° C in dissociation curve were between 4 and 10;" "The smaller the ΔCt value is, the greater the proportion of methylated cells and the greater the extent of the methylated region in each cell. Therefore, we set five cutoffs." |
| Technique: SQ‐MSP Sample type: frozen CpG sites: 76–80 and 84–87 Threshold: > 2 | |||||
| Technique: SQ‐MSP Sample type: frozen CpG sites: 76–80 and 84–87 Threshold: > 4 | |||||
| Technique: SQ‐MSP Sample type: frozen CpG sites: 76–80 and 84–87 Threshold: > 6 | |||||
| Technique: SQ‐MSP Sample type: frozen CpG sites: 76–80 and 84–87 Threshold: > 8 | |||||
| AF: area fraction; bp: base pair; CpG: 5'‐cytosine‐phosphate‐guanine‐3'; DIF: double immunofluorescence; FFPE: formalin‐fixed paraffin‐embedded; GBM: glioblastoma; IHC: immunohistochemistry; MGMT: O6‐methylguanine–DNA methyltransferase; mRNA: messenger ribonucleic acid; MS‐MLPA: methylation‐specific multiplex ligation‐dependent probe amplification; MS‐RE‐qPCR: methylation‐specific restriction enzyme quantitative polymerase chain reaction; MSP: methylation‐specific polymerase chain reaction; N/A: not applicable; NR: not reported; PCR: polymerase chain reaction; PCR‐HRM: polymerase chain reaction with high‐resolution melting; PCR‐mRNA: polymerase chain reaction‐messenger ribonucleic acid; PSQ: pyrosequencing; qMSP: quantitative methylation‐specific polymerase chain reaction; qMSP‐PSQ: quantitative methylation‐specific polymerase chain reaction with pyrosequencing; RNA: ribonucleic acid; ROC: receiver operating characteristic; SD: standard deviation; TMZ: temozolomide. | |||||
Appendix 6. Detailed risk of bias assessments
| CpGs analysed (PCR‐based tests) | Threshold for methylated | D4: PF | D4 (justification) | D5: attrition | D5 (justification) | D6: other PFs | D6 (justification) | D7: sel. rep. | D7 (justification) | ||
| Almuqate 2018 | MS‐RE‐qPCR | NR | > 5% | Unclear | Cut‐off may have been based on performance. | Unclear | Insufficient information | Low RoB | No concerns | Unclear | Conference abstract – little information reported. |
| MS‐RE‐qPCR | NR | > 9% | Low RoB | No concerns | Unclear | Insufficient information | Low RoB | No concerns | Unclear | Conference abstract – little information reported. | |
| Bady 2012 (E‐GBM) | Bead array | 31 and 83 | > 0.358 | Low RoB | No concerns | Low RoB | No missing data | N/A | — | Low RoB | No concerns |
| Bead array | 78–84 | > 10% | High RoB | Threshold derived from outcome measurement. | Low RoB | No missing data | N/A | — | Low RoB | No concerns | |
| PSQ | 74–78 | > 7.28% | Low RoB | Cut‐off does not seem to be determined by performance. | Low RoB | Only 3/50 missing. | N/A | — | Low RoB | No concerns | |
| Bady 2012 (M‐GBM) | Bead array | 31 and 83 | > 0.358 | High RoB | Threshold derived from outcome measurement. | Low RoB | No missing data | Low RoB | No concerns | Low RoB | No concerns |
| MSP | 76–80 and 84–86 | NR | Low RoB | No concerns | Low RoB | No missing data | Low RoB | No concerns | Low RoB | No concerns | |
| Barault 2015 | Methyl‐beaming | 79–83 | > 40.2% | Low RoB | No concerns | Low RoB | No missing data | N/A | — | Unclear | Unclear why there was no result for MSP for this cohort of people (MSP investigated in other cohorts of people in this study). |
| PSQ | 76–81 | > 29.6% | Low RoB | No concerns | Unclear | Missing data for 11/69. Unclear whether there were important differences between those included in the study and those who were not. | N/A | — | Unclear | Unclear why there is no result for MSP for this cohort of people (MSP investigated in other cohorts of people in this study). | |
| Barbagallo 2014 | MSP | 76–80 and 84–87 | Including weakly | Low RoB | No concerns | Low RoB | No missing data | Low RoB | No concerns | Low RoB | No concerns |
| MSP | 76–80 and 84–87 | Excluding weakly | Low RoB | No concerns | Low RoB | No missing data | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | NR | > 9% | Low RoB | No concerns | Low RoB | No missing data | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | NR | > 25% | Low RoB | No concerns | Low RoB | No missing data | Low RoB | No concerns | Low RoB | No concerns | |
| Bell 2017 | QF‐IHC (AQUA) | N/A | > median | Unclear | — | Low RoB | No concerns | Low RoB | No concerns | Unclear | Multiple HRs are reported for different overlapping subgroups of this population. |
| qMSP | NR | > 8 | Unclear | MSP was performed centrally in original RCT. | Unclear | Of the 833 people from the original RCT, 452 had available tissue and underwent the microarray of 22 proteins. From these, 320 had data for MSP. The study authors did compare OS between the 452 microarray samples and the remainder of the 833 from the RCT cohort that were not included in this secondary analysis – no significant difference. However, there does not appear to be any further examination of missing data. | Low RoB | No concerns | Unclear | Multiple HRs are reported for different overlapping subgroups of this population. | |
| Brigliadori 2016 | PSQ | 74–83 | > 9% | Low RoB | No concerns | Low RoB | No concerns | N/A | — | Low RoB | No concerns |
| PSQ | 74–83 | > 29% | Low RoB | No concerns | Low RoB | No concerns | N/A | — | Low RoB | No concerns | |
| Chai 2018 (7‐site cohort) | PSQ | 72–78 | > 12% | High RoB | Cut‐offs appeared to have been selected based on performance (ROC curve analysis, sensitivity, specificity). | Low RoB | people had to have information on methylation status and OS to be included, and therefore there is no missing data. The bias that selecting on this basis has already been covered in domain 1. | N/A | — | Unclear | The whole set of CpGs analysed was correlated with OS. In addition a subset that is part of a Qiagen kit was tested, and a third combination. Unclear why results for other combinations not presented. |
| PSQ | 74–78 | > 12% | High RoB | Cut‐offs appeared to have been selected based on performance (ROC curve analysis, sensitivity, specificity). | Low RoB | People had to have information on methylation status and OS to be included, and therefore there were no missing data. The bias that selecting on this basis has already been covered in domain 1. | N/A | — | Unclear | The whole set of CpGs analysed was correlated with OS. In addition a subset that is part of a Qiagen kit was tested, and a third combination. Unclear why results for other combinations not presented. | |
| PSQ | 75–78 | > 12% | High RoB | Cut‐offs appeared to have been selected based on performance (ROC curve analysis, sensitivity, specificity). | Low RoB | Participants had to have information on methylation status and OS to be included, and therefore there is no missing data. The bias that selecting on this basis has already been covered in domain 1. | N/A | — | Unclear | The whole set of CpGs analysed was correlated with OS. In addition a subset that is part of a Qiagen kit was tested, and a third combination. Unclear why results for other combinations not presented. | |
| Chai 2018 (8‐site cohort) | PSQ | 75–78 | > 13% | High RoB | Cut‐offs appeared to have been selected based on performance (ROC curve analysis, sensitivity, specificity). | Low RoB | People had to have information on methylation status and OS to be included, and therefore there were no missing data. The bias that selecting on this basis has already been covered in domain 1. | N/A | — | Unclear | The whole set of CpGs analysed was correlated with OS. In addition a subset that is part of a Qiagen kit was tested, and a third combination. Unclear why results for other combinations not presented. |
| PSQ | 75–82 | > 12% | High RoB | Cut‐offs appeared to have been selected based on performance (ROC curve analysis, sensitivity, specificity). | Low RoB | People had to have information on methylation status and OS to be included, and therefore there were no missing data. The bias that selecting on this basis has already been covered in domain 1. | N/A | — | Unclear | The whole set of CpGs analysed was correlated with OS. In addition a subset that is part of a Qiagen kit was tested, and a third combination. Unclear why results for other combinations not presented. | |
| PSQ | 76–79 | > 11% | High RoB | Cut‐offs appeared to have been selected based on performance (ROC curve analysis, sensitivity, specificity). | Low RoB | People had to have information on methylation status and OS to be included, and therefore there were no missing data. The bias that selecting on this basis has already been covered in domain 1. | N/A | — | Unclear | The whole set of CpGs analysed was correlated with OS. In addition a subset that is part of a Qiagen kit was tested, and a third combination. Unclear why results for other combinations not presented. | |
| Dahlrot 2018 (NS cohort) | DIF | N/A | < 0.2 | Low RoB | Although no information was provided about blindness of analysis, all nuclear identification was performed automatically so there is reason to assume that the measurements ere objectives. | Low RoB | No concerns | Low RoB | No concerns | Low RoB | No concerns |
| PSQ | 74–78 | > 9% | Low RoB | No concerns | Low RoB | No concerns | N/A | — | Low RoB | No concerns | |
| Dahlrot 2018 (RSD cohort) | DIF | N/A | < 0.2 | Low RoB | Although no information was provided about blindness of analysis, all nuclear identification was performed automatically so there is reason to assume that the measurements ere objectives. | Low RoB | No concerns | Low RoB | No concerns | Low RoB | No concerns |
| PSQ | 74–78 | > 10% | Low RoB | No concerns | Low RoB | No concerns | N/A | — | Low RoB | No concerns | |
| Dunn 2009 | PSQ | 72–83 | > 9% | Low RoB | Cut‐off may or may not have been prespecified, but it was not data driven (i.e. not based on ROC curve analysis). | Low RoB | Missing data for 6/115 people treated with chemoradiation during the study period: four had histology elsewhere and two had inadequate tissue. Median OS for the complete cohort of 115 people was 12.8 months vs 12.4 months in the 109 included people. | Low RoB | No concerns | Low RoB | No concerns |
| PSQ | 72–83 | > 20% | Unclear | Methylated cases were ranked according to methylation and divided into 3 groups. This does not seem to have been a prespecified analysis, but not data driven. | Low RoB | Missing data for 6/115 people treated with chemoradiation during the study period: 4 had histology elsewhere and 2 had inadequate tissue. Median OS for the complete cohort of 115 people was 12.8 months vs 12.4 months in the 109 included people. | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 72–83 | > 29% | High RoB | ROC analysis used to separate cases into 2 prognostic groups. | Low RoB | Missing data for 6/115 people treated with chemoradiation during the study period: 4 had histology elsewhere and 2 had inadequate tissue. Median OS for the complete cohort of 115 people was 12.8 months vs 12.4 months in the 109 included people. | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 72–83 | > 35% | Unclear | Methylated cases were ranked according to methylation and divided into 3 groups. This does not seem to have been a prespecified analysis, but not data driven. | Low RoB | Missing data for 6/115 people treated with chemoradiation during the study period: 4 had histology elsewhere and 2 had inadequate tissue. Median OS for the complete cohort of 115 people was 12.8 months vs 12.4 months in the 109 included people. | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 72–83 | Cluster 1 vs 2 and 3 | Unclear | Methylated cases were ranked according to methylation and divided into 3 groups. This does not seem to have been a prespecified analysis, but not data driven. | Low RoB | Missing data for 6/115 people treated with chemoradiation during the study period: 4 had histology elsewhere and 2 had inadequate tissue. Median OS for the complete cohort of 115 people was 12.8 months vs 12.4 months in the 109 included people. | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 72–83 | Cluster 1 and 2 vs 3 | Unclear | Methylated cases were ranked according to methylation and divided into 3 groups. This does not seem to have been a prespecified analysis, but not data driven. | Low RoB | Missing data for 6/115 people treated with chemoradiation during the study period: 4 had histology elsewhere and 2 had inadequate tissue. Median OS for the complete cohort of 115 people was 12.8 months vs 12.4 months in the 109 included people. | Low RoB | No concerns | Low RoB | No concerns | |
| Felsberg 2009 | IHC | N/A | < 10% | Low RoB | No concerns | Low RoB | Missing data were due to issues with the IHC staining. | N/A | — | Low RoB | No concerns |
| MSP | NR | NR | Low RoB | No concerns | Low RoB | No concerns | N/A | — | Low RoB | No concerns | |
| PCR‐mRNA | N/A | < 50% | Unclear | Insufficient information | Unclear | mRNA data not available for 64% of study cohort. Large amount of missing data but unclear differences between missing and included participants. | N/A | — | Low RoB | No concerns | |
| Havik 2012 | MSP | 76–80 and 84–87 | NR | Low RoB | No concerns | Low RoB | Missing data vs the population studied in Havik 2012 covered in Domain 1. | N/A | — | Low RoB | No concerns |
| PCR‐HRM | 72–83 | NR | Unclear | States that there is "no" threshold, although there must have been one. | Unclear | Missing data for methylation status for 11/48 participants. | N/A | — | Low RoB | No concerns | |
| PSQ | 74–78 | > 2.68% | Low RoB | No concerns | Low RoB | Only participants treated with radiotherapy + TMZ included. | N/A | — | Low RoB | No concerns | |
| PSQ | 74–78 | > 6% | High RoB | Not prespecified, data driven. "In order to compare the prognostic ability of the different methods, the optimal cut‐off value for PSQ needed to be identified. ROC curve analysis is the method of choice for predicting optimal cut‐off values (37, 38). The mean percentage methylation of the CpGs analyzed in the two PSQ assays were used in our ROC curve analysis, where methylation cut‐off scores (1–15%) were plotted to identify the optimum cut‐off value for the prediction of OS of 18 months or more after surgery. The AUROC results, including HR values, are listed in Table II. The highest values for AUROC were at a cut‐off of 7% for PSQ Therascreen and 7 and 8% for PSQ 96." | Low RoB | Missing data vs the population studied in Havik 2012 covered in Domain 1. | N/A | — | Low RoB | No concerns | |
| PSQ | 74–78 | > 7% | High RoB | Not prespecified, data driven. "In order to compare the prognostic ability of the different methods, the optimal cut‐off value for PSQ needed to be identified. ROC curve analysis is the method of choice for predicting optimal cut‐off values (37, 38). The mean percentage methylation of the CpGs analyzed in the two PSQ assays were used in our ROC curve analysis, where methylation cut‐off scores (1–15%) were plotted to identify the optimum cut‐off value for the prediction of OS of 18 months or more after surgery. The AUROC results, including HR values, are listed in Table II. The highest values for AUROC were at a cut‐off of 7% for PSQ Therascreen and 7 and 8% for PSQ 96." | Low RoB | Missing data vs the population studied in Havik 2012 covered in Domain 1. | N/A | — | Low RoB | No concerns | |
| PSQ | 74–78 | > 8% | High RoB | Not prespecified, data driven. But this was not the optimal cut‐off. "In order to compare the prognostic ability of the different methods, the optimal cut‐off value for PSQ needed to be identified. ROC curve analysis is the method of choice for predicting optimal cut‐off values (37, 38). The mean percentage methylation of the CpGs analyzed in the two PSQ assays were used in our ROC curve analysis, where methylation cut‐off scores (1–15%) were plotted to identify the optimum cut‐off value for the prediction of OS of 18 months or more after surgery. The AUROC results, including HR values, are listed in Table II. The highest values for AUROC were at a cut‐off of 7% for PSQ Therascreen and 7 and 8% for PSQ 96." | Low RoB | Missing data vs the population studied in Havik 2012 covered in Domain 1. | N/A | — | Low RoB | No concerns | |
| PSQ | 74–78 | > 9% | High RoB | Not prespecified, data driven. But this was not the optimal cut‐off. "In order to compare the prognostic ability of the different methods, the optimal cut‐off value for PSQ needed to be identified. ROC curve analysis is the method of choice for predicting optimal cut‐off values (37, 38). The mean percentage methylation of the CpGs analyzed in the two PSQ assays were used in our ROC curve analysis, where methylation cut‐off scores (1–15%) were plotted to identify the optimum cut‐off value for the prediction of OS of 18 months or more after surgery. The AUROC results, including HR values, are listed in Table II. The highest values for AUROC were at a cut‐off of 7% for PSQ Therascreen and 7 and 8% for PSQ 96." | Low RoB | Missing data vs the population studied in Havik 2012 covered in Domain 1. | N/A | — | Low RoB | No concerns | |
| PSQ | 76–79 | > 6% | High RoB | Not prespecified, data driven. But this was not the optimal cut‐off. "In order to compare the prognostic ability of the different methods, the optimal cut‐off value for PSQ needed to be identified. ROC curve analysis is the method of choice for predicting optimal cut‐off values (37, 38). The mean percentage methylation of the CpGs analyzed in the two PSQ assays were used in our ROC curve analysis, where methylation cut‐off scores (1–15%) were plotted to identify the optimum cut‐off value for the prediction of OS of 18 months or more after surgery. The AUROC results, including HR values, are listed in Table II. The highest values for AUROC were at a cut‐off of 7% for PSQ Therascreen and 7 and 8% for PSQ 96." | Low RoB | Missing data vs the population studied in Havik 2012 covered in Domain 1. | N/A | — | Low RoB | No concerns | |
| PSQ | 76–79 | > 7% | High RoB | Not prespecified, data driven. "In order to compare the prognostic ability of the different methods, the optimal cut‐off value for PSQ needed to be identified. ROC curve analysis is the method of choice for predicting optimal cut‐off values (37, 38). The mean percentage methylation of the CpGs analyzed in the two PSQ assays were used in our ROC curve analysis, where methylation cut‐off scores (1–15%) were plotted to identify the optimum cut‐off value for the prediction of OS of 18 months or more after surgery. The AUROC results, including HR values, are listed in Table II. The highest values for AUROC were at a cut‐off of 7% for PSQ Therascreen and 7 and 8% for PSQ 96." | Low RoB | Missing data vs the population studied in Havik 2012 covered in Domain 1. | N/A | — | Low RoB | No concerns | |
| PSQ | 76–79 | > 8% | High RoB | Not prespecified, data driven. But this was not the optimal cut‐off. "In order to compare the prognostic ability of the different methods, the optimal cut‐off value for PSQ needed to be identified. ROC curve analysis is the method of choice for predicting optimal cut‐off values (37, 38). The mean percentage methylation of the CpGs analyzed in the two PSQ assays were used in our ROC curve analysis, where methylation cut‐off scores (1–15%) were plotted to identify the optimum cut‐off value for the prediction of OS of 18 months or more after surgery. The AUROC results, including HR values, are listed in Table II. The highest values for AUROC were at a cut‐off of 7% for PSQ Therascreen and 7 and 8% for PSQ 96." | Low RoB | Missing data vs the population studied in Havik 2012 covered in Domain 1. | N/A | — | Low RoB | No concerns | |
| PSQ | 76–79 | > 9% | High RoB | Not prespecified, data driven. But this was not the optimal cut‐off. "In order to compare the prognostic ability of the different methods, the optimal cut‐off value for PSQ needed to be identified. ROC curve analysis is the method of choice for predicting optimal cut‐off values (37, 38). The mean percentage methylation of the CpGs analyzed in the two PSQ assays were used in our ROC curve analysis, where methylation cut‐off scores (1–15%) were plotted to identify the optimum cut‐off value for the prediction of OS of 18 months or more after surgery. The AUROC results, including HR values, are listed in Table II. The highest values for AUROC were at a cut‐off of 7% for PSQ Therascreen and 7 and 8% for PSQ 96." | Low RoB | Missing data vs the population studied in Havik 2012 covered in Domain 1. | N/A | — | Low RoB | No concerns | |
| qMSP | 71–73 and 75–86 | NR | Low RoB | No concerns | Low RoB | Missing data vs the population studied in Havik 2012 covered in Domain 1. | N/A | — | Low RoB | No concerns | |
| qMSP | 71–86 | > 0% | Low RoB | No concerns | Low RoB | Only people treated with radiation therapy + TMZ included. | N/A | — | Low RoB | No concerns | |
| Hsu 2017 (see Hsu 2015) | IHC | N/A | < 10% | Low RoB | No concerns | Low RoB | No missing data | Low RoB | No concerns | Low RoB | No concerns |
| MSP | 76–80 and 84–87 | NR | Low RoB | No concerns | Low RoB | No missing data | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 76–79 | > 5% | Low RoB | No concerns | Low RoB | No missing data | Low RoB | No concerns | Low RoB | No concerns | |
| qMSP | 77–80 and 84–87 | > 0.04% | Low RoB | No concerns | Low RoB | No missing data | Low RoB | No concerns | Low RoB | No concerns | |
| qMSP | 77–80 and 84–87 | > 0.1% | Low RoB | No concerns | Low RoB | No missing data | Low RoB | No concerns | Low RoB | No concerns | |
| Karayan‐Tapon 2010 | IHC | N/A | < 15.5% | Low RoB | No concerns | Low RoB | Missing data for 3/81 people. | N/A | — | Low RoB | No concerns |
| MSP | 76–80 and 84–87 | NR | Low RoB | Likely to be based on presence/absence of bands on a gel but threshold not reported. | Low RoB | No missing data | N/A | — | Low RoB | No concerns | |
| PCR‐mRNA | N/A | < 0.39 | Low RoB | No concerns | Low RoB | Missing data for 1/81 people. | N/A | — | Low RoB | No concerns | |
| PSQ | 74 | > 5.5% | Low RoB | No concerns | Low RoB | Missing data for 2/81 people. | N/A | — | Low RoB | No concerns | |
| PSQ | 74–78 | > 8.0% | Low RoB | No concerns | Low RoB | Missing data for 2/81 people. | N/A | — | Low RoB | No concerns | |
| PSQ | 75 | > 8.7% | Low RoB | No concerns | Low RoB | Missing data for 2/81 people. | N/A | — | Low RoB | No concerns | |
| PSQ | 76 | > 8.0% | Low RoB | No concerns | Low RoB | Missing data for 2/81 people. | N/A | — | Low RoB | No concerns | |
| PSQ | 77 | > 7.85% | Low RoB | No concerns | Low RoB | Missing methylation data for 2/81 people. | N/A | — | Low RoB | No concerns | |
| PSQ | 78 | > 7.8% | Low RoB | No concerns | Low RoB | Missing data for 2/81 people. | N/A | — | Low RoB | No concerns | |
| SQ‐MSP | 76–80 and 84–87 | > 35 | Low RoB | No concerns | Low RoB | No missing data | N/A | — | Low RoB | No concerns | |
| Kim 2016 | MSP | 76–80 and 84–87 | NR | Low RoB | Likely to be based on presence/absence of bands on a gel but threshold not reported. | Low RoB | Data for all people treated with TMZ. | N/A | — | Low RoB | No concerns |
| PSQ | 74–78 | > 9% | High RoB | Data driven – based on the results of ROC analysis (although for the whole cohort, not just those treated with TMZ). | Low RoB | Data for all people treated with TMZ. | N/A | — | Low RoB | No concerns | |
| Kristensen 2016 | IHC | N/A | at 0% | Unclear | It is unclear whether the investigators analysing the results were blinded to clinical outcomes. | Low RoB | Low proportion with missing data. | N/A | — | Low RoB | No concerns |
| PSQ | NR | > 10% | Low RoB | No concerns | Unclear | 12% missing data. | N/A | — | Low RoB | No concerns | |
| qMSP‐PSQ | NR | > 0.1% | Low RoB | No concerns | Low RoB | No missing data | N/A | — | Low RoB | No concerns | |
| qMSP‐PSQ | NR | > 5% | Low RoB | No concerns | Low RoB | No missing data | N/A | — | Low RoB | No concerns | |
| qMSP‐PSQ | NR | > 20% | Low RoB | No concerns | Low RoB | No missing data | N/A | — | Low RoB | No concerns | |
| Lalezari 2013 | IHC | N/A | < 30% | Low RoB | No concerns | Unclear | Missing data on MGMT status for about 15% of the study population. Unclear if there are systematic differences between those with and without data. | Low RoB | No concerns | Low RoB | No concerns |
| MSP | 76–80 and 84–87 | NR | Low RoB | No concerns | Low RoB | MGMT methylation by MSP was measured in 402 people (missing < 5% of the population). No information about reason for exclusion from this analysis is given. | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 72–95 | NR | Low RoB | Cut‐off based on median number of methylated CpG sites as resulted from the analysis. | High RoB | MGMT methylation by BiSEQ was measured in 312 people (> 25% is missing). No information about reason for exclusion from this analysis is given. | Low RoB | No concerns | Low RoB | No concerns | |
| Lattanzio 2015 | MSP | 76–80 and 84–87 | NR | Low RoB | No concerns | Unclear | Missing data for 6/46 enrolled people. No information to judge whether there were important differences between participants who completed the study and those who did not. | N/A | — | Low RoB | No concerns |
| MSP | 76–80 and 84–87 | NR | Low RoB | No concerns | Low RoB | Missing data for just 1 participant, and not due to missing data on MGMT status. | N/A | — | Low RoB | No concerns | |
| PSQ | 72–80 | ≥ 9% | Low RoB | No concerns | Low RoB | Missing data for just 1 participant, and not due to missing data on MGMT status. | N/A | — | Low RoB | No concerns | |
| PSQ | 72–80 | ≥ 9% | Low RoB | No concerns | Low RoB | Missing data for just 1 participant, and not due to missing data on MGMT status. | N/A | — | Low RoB | No concerns | |
| Lechapt‐Zalcman 2012 | IHC | N/A | < 15% | Low RoB | Single centre for MGMT testing, analysis of tumour specimens blinded to MSP data and clinical outcomes. Cut‐off was defined by the median value of reactivity – as done previously by other authors (referenced). | Low RoB | Low proportion of missing data. No information on differences between missing and non‐missing people. | N/A | — | Low RoB | No concerns |
| MSP | 76–80 and 84–87 | NR | Low RoB | Single centre for MGMT testing, analysis of tumour specimens blinded to participant identity, threshold not reported but mentions a "detailed protocol" so I think can assume this included a prespecified cut‐off. | Low RoB | Low proportion of missing data in univariate analysis. No information on differences between missing and non‐missing people. | N/A | — | Low RoB | No concerns | |
| McDonald 2013 | MSP | 76–80 | NR | Low RoB | No concerns | Unclear | Insufficient information | Low RoB | No concerns | Low RoB | No concerns |
| PSQ | 74–78 | > 8% | Low RoB | Although cut‐off was determined post‐hoc, this was only to dichotomise the data and was not determined based on outcomes, which does not seem like it would increase risk of bias. | Low RoB | Only 2/78 missing data. | Low RoB | No concerns | Low RoB | No concerns | |
| Melguizo 2012 | IHC | N/A | < 25% | Low RoB | No concerns | Low RoB | Only 2.5% samples missing data. | N/A | — | Low RoB | No concerns |
| MSP | 76–80 and 84–87 | NR | Low RoB | Unclear as no information about threshold. | Low RoB | No concerns | N/A | — | Low RoB | No concerns | |
| Nguyen 2015 | FSQ‐MS‐PCR | 76–80 and 84–87 | > 15% | High RoB | Cut‐offs appeared to have been determined based on performance. | Low RoB | States that all clinical and molecular data were fully complete in all people. | Low RoB | — | Low RoB | No concerns |
| FSQ‐MS‐PCR | 76–80 and 84–87 | > 60% | High RoB | Cut‐offs appeared to have been determined based on performance. | Low RoB | States that all clinical and molecular data were fully complete in all people. | N/A | — | Low RoB | No concerns | |
| Park 2011 | MS‐MLPA | NR | > 0.1% | High RoB | Outcome‐based cut‐off chosen. | Low RoB | No missing data | N/A | — | Low RoB | No concerns |
| MS‐MLPA | NR | > 0.2 | High RoB | Outcome‐based cut‐off chosen. | Low RoB | No missing data | N/A | — | Low RoB | No concerns | |
| MSP | 76–80 and 84–86 | NR | Low RoB | No concerns | Low RoB | No concerns | N/A | — | Low RoB | No concerns | |
| Quillien 2014 (test) | IHC | N/A | < 23% | High RoB | Threshold was not prespecified. Chosen based on performance. | Low RoB | 1/100 people excluded because of missing results on IHC due to a technical problem during the staining process (this participant was also excluded from all other analyses). | Low RoB | No concerns | Low RoB | No concerns |
| MSP | 76–80 and 84–87 | NR | Low RoB | Likely to be based on presence/absence of bands on a gel but threshold not reported. | Low RoB | 1/100 people excluded because of missing results on IHC due to a technical problem during the staining process (this participant was also excluded from all other analyses). | Low RoB | No concerns | Low RoB | No concerns | |
| MethyLight‐MSP | 75–86 | > 0 | High RoB | Threshold was not prespecified. Chosen based on performance. | Low RoB | 1/100 people excluded because of missing results on IHC due to a technical problem during the staining process (this participant was also excluded from all other analyses). | Low RoB | No concerns | Low RoB | No concerns | |
| PCR‐HRM | 70–83 | > 50% | Low RoB | No concerns | Low RoB | 1/100 people excluded because of missing results on IHC due to a technical problem during the staining process (this participant was also excluded from all other analyses). | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 74 | > 4% | High RoB | Threshold was not prespecified. Chosen based on performance. | Low RoB | 1/100 people excluded because of missing results on IHC due to a technical problem during the staining process (this participant was also excluded from all other analyses). | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 74 | > 8% | High RoB | Cut‐off of methylation based on outcome. | Low RoB | No concerns | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 74–78 | > 8% | High RoB | Threshold was not prespecified. Chosen based on performance. | Low RoB | 1/100 people excluded because of missing results on IHC due to a technical problem during the staining process (this participant was also excluded from all other analyses). | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 74–78 | > 9% | High RoB | Cut‐off of methylation based on outcome. | Low RoB | No concerns | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 74‐89 | > 11% | High RoB | Cut‐off of methylation based on outcome. | Low RoB | No concerns | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 75 | > 11% | High RoB | Threshold was not prespecified. Chosen based on performance. | Low RoB | 1/100 people excluded because of missing results on IHC due to a technical problem during the staining process (this participant was also excluded from all other analyses). | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 75–79 | > 8% | High RoB | Cut‐off of methylation based on outcome. | Low RoB | No concerns | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 76 | > 4% | High RoB | Threshold was not prespecified. Chosen based on performance. | Low RoB | 1/100 people excluded because of missing results on IHC due to a technical problem during the staining process (this participant was also excluded from all other analyses). | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 76 | > 5% | High RoB | Cut‐off of methylation based on outcome. | Low RoB | No concerns | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 76–79 | > 8% | High RoB | Cut‐off of methylation based on outcome. | Low RoB | No concerns | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 76–80 | > 9% | High RoB | Cut‐off of methylation based on outcome. | Low RoB | No concerns | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 77 | > 6% | High RoB | Threshold was not prespecified. Chosen based on performance. | Low RoB | 1/100 people excluded because of missing results on IHC due to a technical problem during the staining process (this participant was also excluded from all other analyses). | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 77 | > 7% | High RoB | Cut‐off of methylation based on outcome. | Low RoB | No concerns | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 77–81 | > 8% | High RoB | Cut‐off of methylation based on outcome. | Low RoB | No concerns | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 78 | > 4% | High RoB | Cut‐off of methylation based on outcome. | Low RoB | No concerns | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 78 | > 5% | High RoB | Threshold was not prespecified. Chosen based on performance. | Low RoB | 1/100 people excluded because of missing results on IHC due to a technical problem during the staining process (this participant was also excluded from all other analyses). | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 78–82 | > 9% | High RoB | Cut‐off of methylation based on outcome. | Low RoB | No concerns | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 79 | > 7% | High RoB | Cut‐off of methylation based on outcome. | Low RoB | No concerns | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 79–83 | > 8% | High RoB | Cut‐off of methylation based on outcome. | Low RoB | No concerns | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 80 | > 4% | High RoB | Cut‐off of methylation based on outcome. | Low RoB | No concerns | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 81 | > 8% | High RoB | Cut‐off of methylation based on outcome. | Low RoB | No concerns | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 82 | > 16% | High RoB | Cut‐off of methylation based on outcome. | Low RoB | No concerns | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 83 | > 10% | High RoB | Cut‐off of methylation based on outcome. | Low RoB | No concerns | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 84 | > 9% | High RoB | Cut‐off of methylation based on outcome. | Low RoB | No concerns | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 84–88 | > 17% | High RoB | Cut‐off of methylation based on outcome. | Low RoB | No concerns | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 84–89 | > 22% | High RoB | Cut‐off of methylation based on outcome. | Low RoB | No concerns | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 85 | > 5% | High RoB | Cut‐off of methylation based on outcome. | Low RoB | No concerns | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 85–89 | > 13% | High RoB | Cut‐off of methylation based on outcome. | Low RoB | No concerns | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 86 | > 11% | High RoB | Cut‐off of methylation based on outcome. | Low RoB | No concerns | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 87 | > 25% | High RoB | Cut‐off of methylation based on outcome. | Low RoB | No concerns | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 88 | > 4% | High RoB | Cut‐off of methylation based on outcome. | Low RoB | No concerns | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 89 | > 12% | High RoB | Cut‐off of methylation based on outcome. | Low RoB | No concerns | Low RoB | No concerns | Low RoB | No concerns | |
| Quillien 2014 (validation) | PSQ | 74–78 | > 9% | Low RoB | Cut‐off of methylation based on outcome of testing cohort. | Low RoB | No concerns | Low RoB | No concerns | Low RoB | No concerns |
| PSQ | 74–78 | > 10% | Low RoB | Cut‐off of methylation based on outcome of testing cohort. | Low RoB | No concerns | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 74–78 | > 28% | Low RoB | Cut‐off of methylation based on outcome of testing cohort. | Low RoB | No concerns | Low RoB | No concerns | Low RoB | No concerns | |
| Quillien 2016 | PSQ | 74–78 | > 6% | High RoB | Data driven – based on the results of ROC analysis. | Unclear | Numerous missing data but unclear if those with missing data different to those without. | Low RoB | No concerns | Low RoB | No concerns |
| PSQ | 74–78 | > 8% | Low RoB | Threshold found in a previous publication. | Unclear | Numerous missing data but unclear if those with missing data different to those without. | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 74–78 | > 12% | Unclear | Unclear why this was chosen/no justification. | Unclear | Numerous missing data but unclear if those with missing data different to those without. | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 74–78 | > 13% | Unclear | Unclear why this was chosen/no justification. | Unclear | Numerous missing data but unclear if those with missing data different to those without. | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 74–78 | > 16% | High RoB | Data driven – based on the results of ROC analysis. | Unclear | Numerous missing data but unclear if those with missing data different to those without. | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 74–78 | > 6% | High RoB | Data driven – based on the results of ROC analysis. | Unclear | Numerous missing data but unclear if those with missing data different to those without. | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 74–78 | > 8% | Low RoB | Threshold found in a previous publication. | Unclear | Numerous missing data but unclear if those with missing data different to those without. | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 74–78 | > 12% or 13% | Unclear | Unclear why this was chosen/no justification. | Unclear | Numerous missing data but unclear if those with missing data different to those without. | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 74–78 | > 12% | Low RoB | From Quillien 2016: Standard Operating Procedure and 10 quality control samples for the determination of MGMT promoter methylation were sent to the different centres as way of standardisation of the process throughout the multiple centres. This approach reduced risk of bias due to different setting for prognostic factor measurement. Cut‐off of methylation based on outcome. | Unclear | From the original cohort, 10 people who had successful initial PSQ could not have the Thera PSQ. Therefore, the number of people included in this analysis was 102, but should have been 112. Authors commented in paper: "These data are almost identical to those obtained for the overall population (n=112 people, 49%, 44% and AUCROC values of 0.69 and 0.70), indicating the absence of bias in the selection of the 102 people for the present cohort." | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 74–78 | > 16% | High RoB | Data driven – based on the results of ROC analysis. | Unclear | Numerous missing data but unclear if those with missing data different to those without. | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 76–79 | > 8% | Low RoB | Analysis of intralaboratory reproducibility of Thera showed highly reproducibility of results from the different centres. | Unclear | From the original cohort, 10 people who had successful initial PSQ could not have the Thera PSQ. Therefore, the number of people included in this analysis was 102, but should have been 112. Authors commented in paper: "These data are almost identical to those obtained for the overall population (n=112 people, 49%, 44% and AUCROC values of 0.69 and 0.70), indicating the absence of bias in the selection of the 102 people for the present cohort." | Low RoB | No concerns | Low RoB | No concerns | |
| PSQ | 76–79 | > 12% | Low RoB | Analysis of intralaboratory reproducibility of Thera showed highly reproducibility of results from the different centres. | Unclear | From the original cohort, 10 people who had successful initial PSQ could not have the Thera PSQ. Therefore, the number of people included in this analysis was 102, but should have been 112. Authors commented in paper: "These data are almost identical to those obtained for the overall population (n=112 people, 49%, 44% and AUCROC values of 0.69 and 0.70), indicating the absence of bias in the selection of the 102 people for the present cohort." | Low RoB | No concerns | Low RoB | No concerns | |
| SQ‐MSP | 76–80 and 84–87 | > 12% | Unclear | Unclear why this was chosen/no justification. | Unclear | Numerous missing data but unclear if those with missing data different to those without. | Low RoB | No concerns | Low RoB | No concerns | |
| SQ‐MSP | 76–80 and 84–87 | > 13% | Unclear | Unclear why this was chosen/no justification. | Unclear | Numerous missing data but unclear if those with missing data different to those without. | Low RoB | No concerns | Low RoB | No concerns | |
| SQ‐MSP | 76–80 and 84–87 | > 23% | High RoB | Data driven – based on the results of ROC analysis. | Unclear | Numerous missing data but unclear if those with missing data different to those without. | Low RoB | No concerns | Low RoB | No concerns | |
| SQ‐MSP | 76–80 and 84–87 | > 13% | High RoB | Data driven – based on the results of ROC analysis. Although also the cut‐off that corresponds to best concordance which we rated as unclear elsewhere. | Unclear | Numerous missing data but unclear if those with missing data different to those without. | Low RoB | No concerns | Low RoB | No concerns | |
| SQ‐MSP | 76–80 and 84–87 | > 23% | High RoB | Data driven – based on the results of ROC analysis. | Unclear | Numerous missing data but unclear if those with missing data different to those without. | Low RoB | No concerns | Low RoB | No concerns | |
| Thon 2017 | MSP | 76–80 and 84–87 | NR | Low RoB | MSP is usually evaluated by visibility of a band indicating methylation, therefore, we judged as prespecified threshold in the absence of a clear description in the text. | Low RoB | 1 participant lost to follow‐up after 6 months. | Low RoB | No concerns | Low RoB | No concerns |
| Sequencing | 75–99 (unclear) | > 50% | Low RoB | No concerns | Low RoB | 1 participant lost to follow‐up after 6 months. | Low RoB | No concerns | Low RoB | No concerns | |
| Yamashita 2018 | MSP | 76–80 and 84–87 | NR | Low RoB | No cut‐off/presence or absence of band. | Low RoB | 1 participant lost to follow‐up after 6 months. | High RoB | Model included other MGMT status using alternative method. | Low RoB | No concerns |
| PCR‐HRM | 72–89 | > 5% | High RoB | Data driven – based on the results of ROC analysis. This was not the optimal cut‐off. | Low RoB | No missing data for MGMT status or OS. Multivariate analyses reportedly for all 75 participants, although 1 participant with missing IDH1 status. | N/A | — | High RoB | 2 other primer sets were used for PCR‐HRM. Only have ROC curve data for these (discarded as not as predictive as primer set 1). | |
| PCR‐HRM | 72–89 | > 8% | High RoB | Data driven – based on the results of ROC analysis. This was not the optimal cut‐off. | Low RoB | No missing data for MGMT status or OS. Multivariate analyses reportedly for all 75 participants, although 1 participant with missing IDH1 status. | N/A | — | High RoB | 2 other primer sets were used for PCR‐HRM. Only have ROC curve data for these (discarded as not as predictive as primer set 1). | |
| PCR‐HRM | 72–89 | > 10% | High RoB | Data driven – based on the results of ROC analysis. | Low RoB | No missing data for MGMT status or OS. Multivariate analyses reportedly for all 75 participants, although 1 participant with missing IDH1 status. | High RoB | Model includes other MGMT status using alternative method | High RoB | 2 sets were used for PCR‐HRM. Only have ROC curve data for these (discarded as not as predictive as primer set 1). | |
| PCR‐HRM | 72–89 | > 12% | High RoB | Data driven – based on the results of ROC analysis. This was not the optimal cut‐off. | Low RoB | No missing data for MGMT status or OS. Multivariate analyses reportedly for all 75 participants, although 1 participant with missing IDH1 status. | N/A | — | High RoB | 2 other primer sets were used for PCR‐HRM. Only have ROC curve data for these (discarded as not as predictive as primer set 1). | |
| PCR‐HRM | 72–89 | > 15% | High RoB | Data driven – based on the results of ROC analysis. This was not the optimal cut‐off. | Low RoB | No missing data for MGMT status or OS. Multivariate analyses reportedly for all 75 participants, although 1 participant with missing IDH1 status. | N/A | — | High RoB | 2 other primer sets were used for PCR‐HRM. Only have ROC curve data for these (discarded as not as predictive as primer set 1). | |
| Yang 2012 | IHC | N/A | < 10% | Low RoB | To a degree we can set the cut‐off in this study. | Low RoB | No missing data | Low RoB | No concerns | Unclear | HRM analyses also performed but extractable data not presented. |
| MSP | 76–80 and 84–87 | NR | Low RoB | To a degree we can set the cut‐off in this study. | Low RoB | No missing data | Low RoB | No concerns | Unclear | HRM analyses also performed but extractable data not presented. | |
| Yoshioka 2018 | SQ‐MSP | 76–80 and 84–87 | > 0 | Low RoB | No concerns | Unclear | No information regarding 4 missing samples. | Low RoB | No concerns | Low RoB | No concerns |
| SQ‐MSP | 76–80 and 84–87 | > 2 | Low RoB | No concerns | Unclear | No information regarding 4 missing samples. | Low RoB | No concerns | Low RoB | No concerns | |
| SQ‐MSP | 76–80 and 84–87 | > 4 | Low RoB | No concerns | Unclear | No information regarding 4 missing samples. | Low RoB | No concerns | Low RoB | No concerns | |
| SQ‐MSP | 76–80 and 84–87 | > 6 | Low RoB | No concerns | Unclear | No information regarding 4 missing samples. | Low RoB | No concerns | Low RoB | No concerns | |
| SQ‐MSP | 76–80 and 84–87 | > 8 | Low RoB | No concerns | Unclear | No information regarding 4 missing samples. | Low RoB | No concerns | Low RoB | No concerns | |
| AUROC: area under receiver operating characteristic; CpG: 5'‐cytosine‐phosphate‐guanine‐3'; DIF: double immunofluorescence; FSQ‐MS‐PCR: fluorescent semi‐quantitative methylation‐specific polymerase chain reaction; GBM: glioblastoma; HR: hazard ratio; IDH: isocitrate dehydrogenase; IHC: immunohistochemistry; MGMT: O6‐methylguanine–DNA methyltransferase; mRNA: messenger ribonucleic acid; MS‐MLPA: methylation‐specific multiplex ligation‐dependent probe amplification; MS‐RE‐qPCR: methylation‐specific restriction enzyme quantitative polymerase chain reaction; MSP: methylation‐specific polymerase chain reaction; N/A: not applicable; NR: not reported; OS: overall survival; PCR: polymerase chain reaction; PCR‐HRM: polymerase chain reaction with high‐resolution melting; PCR‐mRNA: polymerase chain reaction‐messenger ribonucleic acid; PF: prognostic factor; PSQ: pyrosequencing; QF‐IHC: quantitative fluorescence immunohistochemistry; qMSP: quantitative methylation‐specific polymerase chain reaction; qMSP‐PSQ: quantitative methylation‐specific polymerase chain reaction with pyrosequencing; RCT: randomised controlled trial; RoB: risk of bias; ROC: receiver operating characteristic; sel. rep.: selective reporting; SQ‐MSP: semi‐quantitative methylation‐specific polymerase chain reaction; TMZ: temozolomide. | |||||||||||
Appendix 7. Applicability assessments
| Study ID | Domain 1: participant selection | Domain 1 (justification) | Domain 2: subsequent treatment | Domain 2 (justification) | Domain 3: outcome measurement | Domain 3 (justification) | Domain 4: prognostic factor measurement | Domain 4 (justification) | Domain 6: adjustment for other prognostic factors | Domain 6 (justification) |
| Almuqate 2018 | Unclear concerns | Conference abstract: little information | Low concerns | All people received TMZ | Low concerns | Outcome was all‐cause mortality | Low concerns | No concerns | Low concerns | No concerns |
| Bady 2012 (E‐GBM) | Low concerns | No concerns | Low concerns | People received Stupp | Low concerns | Outcome was all‐cause mortality | Low concerns | No concerns | N/A | No concerns |
| Bady 2012 (M‐GBM) | Low concerns | No concerns | Low concerns | All people received TMZ | Low concerns | Outcome was all‐cause mortality | Low concerns | No concerns | N/A | No concerns |
| Barault 2015 | Low concerns | No concerns | Low concerns | All people received TMZ | Low concerns | Outcome was all‐cause mortality | Low concerns | No concerns | N/A | No concerns |
| Barbagallo 2014 | Low concerns | No concerns | Low concerns | All people received TMZ | Low concerns | Outcome was all‐cause mortality | Low concerns | No concerns | Low concerns | No concerns |
| Bell 2017 | Low concerns | No concerns | Low concerns | No concerns | Unclear concerns | Note that OS was measured from a later start point than other papers | Low concerns | No concerns | Low concerns | No concerns |
| Brigliadori 2016 | Unclear concerns | People undergoing biopsy were not included in our analysis due to the way data were presented (they are not included in the KM‐curve) | Low concerns | People received Stupp | Low concerns | Outcome was all‐cause mortality | Low concerns | No concerns | N/A | No concerns |
| Chai 2018 (7‐site cohort) | Low concerns | No concerns | Low concerns | All people received TMZ. | Low concerns | Outcome was all‐cause mortality | Low concerns | No concerns | N/A | No concerns |
| Chai 2018 (8‐site cohort) | Low concerns | No concerns | Low concerns | All people received TMZ | Low concerns | Outcome was all‐cause mortality | Low concerns | No concerns | N/A | No concerns |
| Dahlrot 2018 (NS cohort) | Low concerns | No concerns | Low concerns | No concerns | Low concerns | Outcome was all‐cause mortality | Low concerns | No concerns | Low concerns | No concerns |
| Dahlrot 2018 (RSD cohort) | Low concerns | No concerns | Low concerns | Not all people received TMZ but only people that received TMZ are included in the analysis | Low concerns | Outcome was all‐cause mortality | Low concerns | No concerns | N/A | No concerns |
| Dunn 2009 | Low concerns | No concerns | Low concerns | All people received TMZ | Low concerns | Outcome was all‐cause mortality | Low concerns | No concerns | Low concerns | No concerns |
| Felsberg 2009 | High concerns | Study cohort included only people who were treated with open resection and who received ≥ 2 cycles of chemotherapy, thus excluding people with tumour biopsy only or too poor clinical condition for chemotherapy, or both | Low concerns | No concerns | Low concerns | Outcome was all‐cause mortality | Low concerns | No concerns | N/A | No concerns |
| Havik 2012 | Low concerns | No concerns | Low concerns | Only people treated with radiotherapy + TMZ were included in the analysis | Low concerns | Outcome was all‐cause mortality | Low concerns | No concerns | N/A | No concerns |
| Hsu 2017 (see Hsu 2015) | Low concerns | No concerns | Low concerns | TMZ with concomitant radiotherapy for all people | Low concerns | Outcome was all‐cause mortality | Low concerns | No concerns | Low concerns | No concerns |
| Karayan‐Tapon 2010 | Low concerns | No concerns | Low concerns | All people received TMZ | Low concerns | Outcome was all‐cause mortality | Low concerns | No concerns | N/A | No concerns |
| Kim 2016 | Low concerns | No concerns | Low concerns | No concerns | Low concerns | Outcome was all‐cause mortality | Low concerns | No concerns | N/A | No concerns |
| Kristensen 2016 | Low concerns | All people have glioblastoma | Low concerns | People received Stupp | Low concerns | Outcome was all‐cause mortality | Low concerns | No concerns | N/A | No concerns |
| Lalezari 2013 | Low concerns | 1/418 participants did not receive TMZ; 2 died before TMZ | Low concerns | Nearly all people received TMZ, only 1 surviving participant did not | Low concerns | Outcome was all‐cause mortality | Low concerns | No concerns | Low concerns | No concerns |
| Lattanzio 2015 | Low concerns | No concerns | Low concerns | All people received TMZ | Low concerns | Outcome was all‐cause mortality | Low concerns | No concerns | N/A | No concerns |
| Lechapt‐Zalcman 2012 | Low concerns | People had TMZ but additionally got Gliadel | Low concerns | All people received TMZ | Low concerns | Outcome was all‐cause mortality | Low concerns | No concerns | N/A | No concerns |
| McDonald 2013 | Low concerns | No concerns | Low concerns | All people received TMZ. | Low concerns | Outcome was all‐cause mortality | Low concerns | No concerns | N/A | No concerns |
| Melguizo 2012 | Unclear concerns | People had to have KPS > 60 to be included | Low concerns | All people received concurrent TMZ | Low concerns | Outcome was all‐cause mortality | Low concerns | No concerns | N/A | No concerns |
| Nguyen 2015 | Low concerns | No concerns | Low concerns | No concerns | Low concerns | Outcome was all‐cause mortality | Low concerns | No concerns | Low concerns | No concerns |
| Park 2011 | Low concerns | No concerns | Low concerns | No concerns | Low concerns | Outcome was all‐cause mortality | Low concerns | No concerns | N/A | No concerns |
| Quillien 2014 (test) | Low concerns | No concerns | Low concerns | All people received TMZ | Low concerns | Outcome was all‐cause mortality | Low concerns | No concerns | Low concerns | No concerns |
| Quillien 2014 (validation) | Low concerns | No concerns | Low concerns | No concerns | Low concerns | Outcome was all‐cause mortality | Low concerns | No concerns | Low concerns | No concerns |
| Quillien 2016 | Low concerns | Frozen samples with a histologically estimated tumour cell content < 40% were excluded from the study | Low concerns | All people received TMZ | Low concerns | Outcome was all‐cause mortality | Low concerns | No concerns | Low concerns | No concerns |
| Thon 2017 | Unclear concerns | Special population: unsuitable for GTR | Low concerns | All people were assigned to receive radiotherapy + TMZ | Low concerns | Outcome was all‐cause mortality | Low concerns | No concerns | Low concerns | No concerns |
| Yamashita 2018 | Low concerns | No concerns | Low concerns | All people received TMZ | Low concerns | Outcome was all‐cause mortality | Low concerns | No concerns | Unclear concerns | Unclear whether putting multiple methods of determining MGMT status into the model together would this reflect real practice |
| Yang 2012 | Low concerns | No concerns | Low concerns | All people received TMZ | Low concerns | Outcome was all‐cause mortality | Low concerns | No concerns | N/A | No concerns |
| Yoshioka 2018 | Low concerns | No concerns | Low concerns | No concerns | Low concerns | Outcome was all‐cause mortality | Low concerns | No concerns | Low concerns | No concerns |
| GTR: gross total resection; KM: Kaplan‐Meier; KPS: Karnofsky performance status; N/A: not applicable; OS: overall survival; TMZ: temozolomide. | ||||||||||
Appendix 8. Hazard ratios for all methods examined in included studies
| Study | Technique | Sample type | CpGs analysed (PCR‐based tests) | Threshold for methylated | HR (95% confidence interval) | Data source |
| Almuqate 2018 | MS‐RE‐qPCR | NR | NR | > 5% | 4.17 (1.78 to 9.75) | Adjusted HR (for age, sex and surgery) |
| MS‐RE‐qPCR | NR | NR | > 9% | 3.57 (1.67 to 7.62) | Adjusted HR (for age, sex and surgery) | |
| Bady 2012 (E‐GBM) | Bead array | Frozen | 31 and 83 | > 0.358 | 3.28 (1.68 to 6.41) | Unadjusted HR |
| Bead array | Frozen | 78–84 | > 10% | 5.56 (1.25 to 25.0) | Unadjusted HR | |
| PSQ | Frozen | 74–78 | > 7.28% | 2.20 (1.12 to 4.31) | Unadjusted HR | |
| Bady 2012 (M‐GBM) | Bead array | Frozen | 31 and 83 | > 0.358 | 6.46 (2.41 to 17.3) | IPD |
| MSP | NR | 76–80 and 84–86 | NR | 7.21 (2.37 to 22.0) | IPD | |
| Barault 2015 | Methyl‐beaming | FFPE | 79–83 | > 40.2% | 2.78 (1.85 to 5.26) | Unadjusted HR |
| PSQ | FFPE | 76–81 | > 29.6% | 2.63 (1.43 to 4.55) | Unadjusted HR | |
| Barbagallo 2014 | MSP | FFPE | 76–80 and 84–87 | Including weakly | 3.68 (1.66 to 8.18) | KM curves (Fig 3A or 3C, Barbagallo 2014) |
| MSP | FFPE | 76–80 and 84–87 | Excluding weakly | 1.90 (0.72 to 4.99) | KM curves (Fig 3C, Barbagallo 2014) | |
| PSQ | FFPE | NR | > 9% | 3.73 (1.68 to 8.28) | KM curves (Fig 3A or 3B, Barbagallo 2014) | |
| PSQ | FFPE | NR | > 25% | 1.99 (0.92 to 4.32) | KM curves (Fig 3B, Barbagallo 2014) | |
| Bell 2017 | QF‐IHC (AQUA) | FFPE | N/A | > Median | 1.84 (1.38 to 2.43) | Adjusted HR (for age, KPS, resection status and treatment) |
| qMSP | NR | NR | > 8 | 1.77 (1.28 to 2.44) | Adjusted HR (for age, KPS, resection status and treatment) | |
| Brigliadori 2016 | PSQ | FFPE | 74–83 | > 9% | 1.92 (1.17 to 3.14) | KM curves (Fig 1; stratified by extent of resection, Brigliadori 2016) |
| PSQ | FFPE | 74–83 | > 29% | 3.02 (1.72 to 5.29) | KM curves (Fig 2; stratified by extent of resection, Brigliadori 2016) | |
| Chai 2018 (7‐site cohort) | PSQ | Frozen | 72–78 | > 12% | 2.94 (1.12 to 7.69) | Unadjusted HR |
| PSQ | Frozen | 74–78 | > 12% | 2.94 (1.12 to 7.69) | Unadjusted HR | |
| PSQ | Frozen | 75–78 | > 12% | 2.94 (1.12 to 7.69) | Unadjusted HR | |
| Chai 2018 (8‐site cohort) | PSQ | Frozen | 75–78 | > 13% | 2.70 (1.37 to 5.26) | Unadjusted HR |
| PSQ | Frozen | 75–82 | > 12% | 3.03 (1.54 to 6.25) | Unadjusted HR | |
| PSQ | Frozen | 76–79 | > 11% | 2.13 (1.09 to 4.17) | Unadjusted HR | |
| Dahlrot 2018 (NS cohort) | DIF | FFPE | N/A | < 0.2 | 1.60 (0.95 to 2.71) | Unadjusted HR |
| PSQ | FFPE | 74–78 | > 9% | 1.42 (0.84 to 2.40) | Unadjusted HR | |
| Dahlrot 2018 (RSD cohort) | DIF | FFPE | N/A | < 0.2 | 2.00 (1.32 to 3.02) | Unadjusted HR |
| PSQ | FFPE | 74–78 | > 10% | 1.58 (1.14 to 2.19) | KM curves (Fig 3A, Dahlrot 2018) | |
| Dunn 2009 | PSQ | Frozen, smear, FFPE or a combination | 72–83 | > 9% | 3.57 (2.24 to 5.70) | KM curves (Fig 2B, Dunn 2009) |
| PSQ | Frozen, smear, FFPE or a combination | 72–83 | > 20% | 4.25 (2.57 to 7.05) | KM curves (Fig 2D, Dunn 2009) | |
| PSQ | Frozen, smear, FFPE or a combination | 72–83 | > 29% | 4.03 (2.30 to 7.07) | KM curves (Fig 2F, Dunn 2009) | |
| PSQ | Frozen, smear, FFPE or a combination | 72–83 | > 35% | 3.64 (1.99 to 6.67) | KM curves (Fig 2D, Dunn 2009) | |
| PSQ | Frozen, smear, FFPE or a combination | 72–83 | Cluster 1 vs 2 and 3 | 4.44 (2.58 to 7.66) | KM curves (Fig Suppl 4C, Dunn 2009) | |
| PSQ | Frozen, smear, FFPE or a combination | 72–83 | Cluster 1 and 2 vs 3 | 3.59 (2.26 to 5.69) | KM curves (Fig Suppl 4C, Dunn 2009) | |
| Felsberg 2009 | IHC | FFPE | N/A | < 10% | 1.26 (0.70 to 2.25) | KM curves (Fig Suppl 1C, 2nd column, Felsberg 2009) |
| MSP | Frozen (14 FFPE) | NR | NR | 2.23 (1.29 to 3.84) | KM curves (Fig Suppl 1A, 2nd column, Felsberg 2009) | |
| PCR‐mRNA | Frozen (14 FFPE) | N/A | < 50% | 2.66 (0.94 to 7.53) | KM curves (Fig Suppl 1B, 2nd column, Felsberg 2009) | |
| Havik 2012 | MSP | Frozen | 76–80 and 84–87 | NR | 2.02 (1.08 to 3.77) | KM curves (Fig 2A, Johannessen 2018) |
| PCR‐HRM | Frozen | 72–83 | NR | 1.32 (0.64 to 2.68) | KM curves (Fig 2B, Johannessen 2018) | |
| PSQ | Frozen | 74–78 | > 2.68% | 1.85 (1.02 to 3.35) | KM curves (Fig 1B, Havik 2012) | |
| PSQ | Frozen | 74–78 | > 6% | 2.30 (1.21 to 4.38) | Unadjusted HR | |
| PSQ | Frozen | 74–78 | > 7% | 2.33 (1.19 to 4.57) | Unadjusted HR | |
| PSQ | Frozen | 74–78 | > 8% | 2.33 (1.19 to 4.57) | Unadjusted HR | |
| PSQ | Frozen | 74–78 | > 9% | 2.30 (0.99 to 5.33) | Unadjusted HR | |
| PSQ | Frozen | 76–79 | > 6% | 2.30 (1.21 to 4.38) | Unadjusted HR | |
| PSQ | Frozen | 76–79 | > 7% | 2.33 (1.19 to 4.57) | Unadjusted HR | |
| PSQ | Frozen | 76–79 | > 8% | 1.90 (0.99 to 3.65) | Unadjusted HR | |
| PSQ | Frozen | 76–79 | > 9% | 1.90 (0.99 to 3.65) | Unadjusted HR | |
| qMSP | Frozen | 71–73 and 75–86 | NR | 1.72 (0.91 to 3.22) | KM curves (Fig 2C, Johannessen 2018) | |
| qMSP | Frozen | 71–86 | > 0% | 1.66 (0.97 to 2.83) | KM curves (Fig 1A, Havik 2012) | |
| Hsu 2015 | IHC | FFPE | N/A | < 10% | 2.12 (1.32 to 3.42) | Unadjusted HR |
| MSP | FFPE | 76–80 and 84–87 | NR | 2.39 (1.42 to 4.02) | Unadjusted HR | |
| PSQ | FFPE | 76–79 | > 5% | 2.66 (1.49 to 4.76) | Unadjusted HR | |
| qMSP | FFPE | 77–80 and 84–87 | > 0.04% | 2.75 (1.51 to 5.04) | Unadjusted HR | |
| qMSP | FFPE | 77–80 and 84–87 | > 0.1% | 2.83 (1.85 to 4.33) | IPD | |
| Karayan‐Tapon 2010 | IHC | FFPE | N/A | < 15.5% | 1.26 (0.77 to 2.06) | KM curves (Fig 3B, Karayan‐Tapon 2010) |
| MSP | Frozen | 76–80 and 84–87 | NR | 2.32 (1.39 to 3.87) | KM curves (Fig 1A, Karayan‐Tapon 2010) | |
| PCR‐mRNA | Frozen | N/A | < 0.39 | 1.68 (1.04 to 2.73) | KM curves (Fig 3A, Karayan‐Tapon 2010) | |
| PSQ | Frozen | 74 | > 5.5% | 3.26 (1.91 to 5.59) | KM curves (Fig Elec Suppl 1B, Karayan‐Tapon 2010) | |
| PSQ | Frozen | 74–78 | > 8.0% | 3.35 (1.95 to 5.73) | KM curves (Fig 2B; cpg mean, Karayan‐Tapon 2010) | |
| PSQ | Frozen | 75 | > 8.7% | 3.26 (1.91 to 5.57) | KM curves (Fig Elec Suppl 1B, Karayan‐Tapon 2010) | |
| PSQ | Frozen | 76 | > 8.0% | 2.78 (1.64 to 4.74) | KM curves (Fig Elec Suppl 1B, Karayan‐Tapon 2010) | |
| PSQ | Frozen | 77 | > 7.85% | 3.65 (2.10 to 6.34) | KM curves (Fig 2B, Karayan‐Tapon 2010) | |
| PSQ | Frozen | 78 | > 7.8% | 2.70 (1.59 to 4.58) | KM curves (Fig Elec Suppl 1B, Karayan‐Tapon 2010) | |
| SQ‐MSP | Frozen | 76–80 and 84–87 | > 35 | 2.75 (1.66 to 4.53) | KM curves (Fig 1B, Karayan‐Tapon 2010) | |
| Kim 2016 | MSP | FFPE | 76–80 and 84–87 | NR | 7.66 (2.82 to 20.8) | KM curves (Fig 2G, Kim 2016) |
| PSQ | FFPE | 74–78 | > 9% | 7.66 (2.82 to 20.8) | KM curves (Fig 2G, Kim 2016) | |
| Kristensen 2016 | IHC | FFPE | N/A | At 0% | 1.58 (1.12 to 2.22) | Unadjusted HR |
| PSQ | Frozen | NR | > 10% | 1.80 (1.23 to 2.62) | Unadjusted HR | |
| qMSP‐PSQ | Frozen | NR | > 0.1% | 1.64 (1.15 to 2.33) | Unadjusted HR | |
| qMSP‐PSQ | Frozen | NR | > 5% | 1.66 (1.02 to 2.71) | KM curves (Fig 5a, Kristensen 2016) | |
| qMSP‐PSQ | Frozen | NR | > 20% | 1.52 (0.77 to 3.00) | KM curves (Fig 5a, Kristensen 2016) | |
| Lalezari 2013 | IHC | FFPE | N/A | < 30% | 1.74 (1.39 to 2.16) | KM curves (Fig 1A, Lalezari 2013) |
| MSP | FFPE | 76–80 and 84–87 | NR | 2.13 (1.67 to 2.78) | Adjusted HR (for age, sex, KPS, extent of resection, bevacizumab treatment at any time and IDH1R132 mutation status) | |
| PSQ | FFPE | 72–95 | NR | 2.06 (1.62 to 2.62) | KM curves (Fig 1E; unadjusted, Lalezari 2013) | |
| Lattanzio 2015 | MSP | FFPE | 76–80 and 84–87 | NR | 1.45 (0.76 to 2.76) | KM curves (Fig 3B, Lattanzio 2015) |
| MSP | Frozen | 76–80 and 84–87 | NR | 2.27 (1.21 to 4.26) | KM curves (Fig 3A, Lattanzio 2015) | |
| PSQ | FFPE | 72–80 | ≥ 9% | 2.09 (1.09 to 3.99) | KM curves (Fig 3D, Lattanzio 2015) | |
| PSQ | Frozen | 72–80 | ≥ 9% | 2.25 (1.19 to 4.25) | KM curves (Fig 3C, Lattanzio 2015) | |
| Lechapt‐Zalcman 2012 | IHC | FFPE | N/A | < 15% | 1.99 (1.15 to 3.42) | KM curves (Fig 4d; unadjusted, Lechapt‐Zalcman 2012) |
| MSP | FFPE | 76–80 and 84–87 | NR | 1.78 (1.03 to 3.09) | KM curves (Fig 4c; unadjusted, Lechapt‐Zalcman 2012) | |
| McDonald 2013 | MSP | FFPE | 76–80 | NR | 1.64 (0.95 to 2.83) | IPD |
| PSQ | FFPE | 74–78 | > 8% | 1.96 (1.16 to 3.33) | Unadjusted HR | |
| Melguizo 2012 | IHC | FFPE | N/A | < 25% | 1.11 (0.69 to 1.77) | KM curves (Fig 4B, Melguizo 2012) |
| MSP | NR | 76–80 and 84–87 | NR | 1.77 (1.06 to 2.95) | KM curves (Fig 4A, Melguizo 2012) | |
| Nguyen 2015 | FSQ‐MS‐PCR | Frozen or FFPE | 76–80 and 84–87 | > 15% | 2.68 (1.70 to 4.21) | KM curves (Fig 3D, Nguyen 2015) |
| FSQ‐MS‐PCR | Frozen or FFPE | 76–80 and 84–87 | > 60% | 2.25 (1.31 to 3.87) | KM curves (Fig 3D, Nguyen 2015) | |
| Park 2011 | MS‐MLPA | 50% frozen and 50% FFPE | NR | > 0.1% | 2.38 (1.11 to 5.10) | KM curves (Fig 2B, Park 2011) |
| MS‐MLPA | 50% frozen and 50% FFPE | NR | > 0.2 | 1.88 (0.86 to 4.11) | KM curves (Fig 2B, Park 2011) | |
| MSP | 50% frozen and 50% FFPE | 76–80 and 84–86 | NR | 4.53 (1.58 to 12.9) | KM curves (Fig 2A, Park 2011) | |
| Quillien 2014 (test) | IHC | FFPE | N/A | < 23% | 2.33 (1.44 to 3.74) | Adjusted HR (for age and KPS) |
| MSP | Frozen | 76–80 and 84–87 | NR | 2.70 (1.65 to 4.43) | Adjusted HR (for age and KPS) | |
| MethyLight‐MSP | Frozen | 75–86 | > 0 | 1.67 (1.00 to 2.77) | Adjusted HR (for age and KPS) | |
| PCR‐HRM | Frozen | 70–83 | > 50% | 1.92 (1.12 to 3.29) | Adjusted HR (for age and KPS) | |
| PSQ | Frozen | 74 | > 4% | 2.44 (1.51 to 3.95) | Adjusted HR (for age and KPS) | |
| PSQ | Frozen | 74 | > 8% | 2.22 (1.30 to 3.79) | Adjusted HR (for age and KPS) | |
| PSQ | Frozen | 74–78 | > 8% | 3.13 (1.86 to 5.25) | Adjusted HR (for age and KPS) | |
| PSQ | Frozen | 74–78 | > 9% | 3.13 (1.81 to 5.38) | Adjusted HR (for age and KPS) | |
| PSQ | Frozen | 74–89 | > 11% | 3.13 (1.84 to 5.30) | Adjusted HR (for age and KPS) | |
| PSQ | Frozen | 75 | > 11% | 3.23 (1.87 to 5.55) | Adjusted HR (for age and KPS) | |
| PSQ | Frozen | 75–79 | > 8% | 2.94 (1.71 to 5.06) | Adjusted HR (for age and KPS) | |
| PSQ | Frozen | 76 | > 4% | 2.63 (1.61 to 4.31) | Adjusted HR (for age and KPS) | |
| PSQ | Frozen | 76 | > 5% | 2.78 (1.62 to 4.75) | Adjusted HR (for age and KPS) | |
| PSQ | Frozen | 76–79 | > 8% | 2.86 (1.66 to 4.92) | Adjusted HR (for age and KPS) | |
| PSQ | Frozen | 76–80 | > 9% | 3.03 (1.74 to 5.26) | Adjusted HR (for age and KPS) | |
| PSQ | Frozen | 77 | > 6% | 3.13 (1.87 to 5.23) | Adjusted HR (for age and KPS) | |
| PSQ | Frozen | 77 | > 7% | 2.78 (1.63 to 4.74) | Adjusted HR (for age and KPS) | |
| PSQ | Frozen | 77–81 | > 8% | 2.70 (1.59 to 4.59) | Adjusted HR (for age and KPS) | |
| PSQ | Frozen | 78 | > 4% | 2.33 (1.38 to 3.92) | Adjusted HR (for age and KPS) | |
| PSQ | Frozen | 78 | > 5% | 2.50 (1.51 to 4.13) | Adjusted HR (for age and KPS) | |
| PSQ | Frozen | 78–82 | > 9% | 2.86 (1.68 to 4.86) | Adjusted HR (for age and KPS) | |
| PSQ | Frozen | 79 | > 7% | 2.78 (1.61 to 4.80) | Adjusted HR (for age and KPS) | |
| PSQ | Frozen | 79–83 | > 8% | 2.86 (1.68 to 4.86) | Adjusted HR (for age and KPS) | |
| PSQ | Frozen | 80 | > 4% | 2.56 (1.54 to 4.28) | Adjusted HR (for age and KPS) | |
| PSQ | Frozen | 81 | > 8% | 2.44 (1.44 to 4.14) | Adjusted HR (for age and KPS) | |
| PSQ | Frozen | 82 | > 16% | 2.94 (1.70 to 5.08) | Adjusted HR (for age and KPS) | |
| PSQ | Frozen | 83 | > 10% | 2.78 (1.65 to 4.66) | Adjusted HR (for age and KPS) | |
| PSQ | Frozen | 84 | > 9% | 3.23 (1.86 to 5.59) | Adjusted HR (for age and KPS) | |
| PSQ | Frozen | 84–88 | > 17% | 3.23 (1.86 to 5.58) | Adjusted HR (for age and KPS) | |
| PSQ | Frozen | 84–89 | > 22% | 3.23 (1.85 to 5.62) | Adjusted HR (for age and KPS) | |
| PSQ | Frozen | 85 | > 5% | 2.50 (1.52 to 4.11) | Adjusted HR (for age and KPS) | |
| PSQ | Frozen | 85–89 | > 13% | 2.94 (1.76 to 4.92) | Adjusted HR (for age and KPS) | |
| PSQ | Frozen | 86 | > 11% | 2.78 (1.65 to 4.69) | Adjusted HR (for age and KPS) | |
| PSQ | Frozen | 87 | > 25% | 3.03 (1.75 to 5.24) | Adjusted HR (for age and KPS) | |
| PSQ | Frozen | 88 | > 4% | 2.27 (1.38 to 3.75) | Adjusted HR (for age and KPS) | |
| PSQ | Frozen | 89 | > 12% | 3.23 (1.91 to 5.46) | Adjusted HR (for age and KPS) | |
| Quillien 2014 (validation) | PSQ | FFPE | 74–78 | > 9% | 3.70 (1.71 to 8.01) | Unadjusted HR |
| PSQ | FFPE | 74–78 | > 10% | 2.86 (1.42 to 5.74) | Unadjusted HR | |
| PSQ | FFPE | 74–78 | > 28% | 2.27 (0.98 to 5.29) | Unadjusted HR | |
| Quillien 2016 | PSQ | FFPE | 74–78 | > 6% | 3.23 (2.02 to 5.16) | Adjusted HR (for age and KPS) |
| PSQ | FFPE | 74–78 | > 8% | 4.00 (2.30 to 6.97) | Adjusted HR (for age and KPS) | |
| PSQ | FFPE | 74–78 | > 12% | 4.17 (2.35 to 7.38) | Adjusted HR (for age and KPS) | |
| PSQ | FFPE | 74–78 | > 13% | 4.35 (2.41 to 7.83) | Adjusted HR (for age and KPS) | |
| PSQ | FFPE | 74–78 | > 16% | 4.55 (2.48 to 8.34) | Adjusted HR (for age and KPS) | |
| PSQ | Frozen | 74–78 | > 6% | 4.00 (2.30 to 6.97) | Adjusted HR (for age and KPS) | |
| PSQ | Frozen | 74–78 | > 8% | 3.57 (2.14 to 5.95) | Adjusted HR (for age and KPS) | |
| PSQ | Frozen | 74–78 | > 12% or 13% | 3.45 (2.10 to 5.66) | Adjusted HR (for age and KPS) | |
| PSQ | Frozen | 74–78 | > 12% | 3.70 (2.19 to 6.26) | Adjusted HR (for age and KPS) | |
| PSQ | Frozen | 74–78 | > 16% | 3.13 (1.94 to 5.04) | Adjusted HR (for age and KPS) | |
| PSQ | Frozen | 76–79 | > 8% | 3.33 (2.06 to 5.40) | Adjusted HR (for age and KPS) | |
| PSQ | Frozen | 76–79 | > 12% | 3.33 (2.06 to 5.40) | Adjusted HR (for age and KPS) | |
| SQ‐MSP | FFPE | 76–80 and 84–87 | > 12% | 3.33 (2.06 to 5.40) | Adjusted HR (for age and KPS) | |
| SQ‐MSP | FFPE | 76–80 and 84–87 | > 13% | 3.33 (2.06 to 5.40) | Adjusted HR (for age and KPS) | |
| SQ‐MSP | FFPE | 76–80 and 84–87 | > 23% | 4.17 (2.35 to 7.38) | Adjusted HR (for age and KPS) | |
| SQ‐MSP | Frozen | 76–80 and 84–87 | > 13% | 2.86 (1.79 to 4.57) | Adjusted HR (for age and KPS) | |
| SQ‐MSP | Frozen | 76–80 and 84–87 | > 23% | 2.17 (1.38 to 3.44) | Adjusted HR (for age and KPS) | |
| Thon 2017 | MSP | Frozen | 76–80 and 84–87 | NR | 3.33 (1.82 to 6.25) | Unadjusted HR |
| Sequencing | Frozen | 75–99 (unclear) | > 50% | 3.33 (1.82 to 6.25) | Unadjusted HR | |
| Yamashita 2018 | MSP | Frozen | 76–80 and 84–87 | NR | 2.36 (1.62 to 5.05) | Unadjusted HR |
| PCR‐HRM | Frozen | 72–89 | > 5% | 2.36 (1.43 to 3.90) | KM curves (Fig Suppl 13, Yamashita 2018) | |
| PCR‐HRM | Frozen | 72–89 | > 8% | 2.05 (1.24 to 3.39) | KM curves (Fig Suppl 13, Yamashita 2018) | |
| PCR‐HRM | Frozen | 72–89 | > 10% | 2.51 (1.63 to 4.83) | Unadjusted HR | |
| PCR‐HRM | Frozen | 72–89 | > 12% | 2.61 (1.54 to 4.42) | KM curves (Fig Suppl 13, Yamashita 2018) | |
| PCR‐HRM | Frozen | 72–89 | > 15% | 2.39 (1.41 to 4.05) | KM curves (Fig Suppl 13, Yamashita 2018) | |
| Yang 2012 | IHC | FFPE | N/A | < 10% | 1.07 (0.35 to 3.31) | IPD |
| MSP | FFPE | 76–80 and 84–87 | NR | 1.35 (0.44 to 4.16) | IPD | |
| Yoshioka 2018 | SQ‐MSP | Frozen | 76–80 and 84–87 | > 0 | 2.72 (1.28 to 5.74) | KM curves (Fig 2A, Yoshioka 2018) |
| SQ‐MSP | Frozen | 76–80 and 84–87 | > 2 | 2.18 (1.20 to 3.97) | KM curves (Fig 2B, Yoshioka 2018) | |
| SQ‐MSP | Frozen | 76–80 and 84–87 | > 4 | 1.85 (1.07 to 3.18) | KM curves (Fig 2C, Yoshioka 2018) | |
| SQ‐MSP | Frozen | 76–80 and 84–87 | > 6 | 1.83 (1.10 to 3.04) | KM curves (Fig 2D, Yoshioka 2018) | |
| SQ‐MSP | Frozen | 76–80 and 84–87 | > 8 | 1.71 (1.00 to 2.93) | KM curves (Fig 2E, Yoshioka 2018) | |
| CpG: 5'‐cytosine‐phosphate‐guanine‐3'; DIF: double immunofluorescence; FFPE: formalin‐fixed paraffin‐embedded; FSQ‐MS‐PCR: fluorescent semi‐quantitative methylation‐specific polymerase chain reaction; HR: hazard ratio; IDH: isocitrate dehydrogenase; IHC: immunohistochemistry; IPD: individual participant data; KM: Kaplan‐Meier; KPS: Karnofsky performance status; MS‐MLPA: methylation‐specific multiplex ligation‐dependent probe amplification; MS‐RE‐qPCR: methylation‐specific restriction enzyme quantitative polymerase chain reaction; MSP: methylation‐specific polymerase chain reaction; N/A: not applicable; NR: not reported; PCR: polymerase chain reaction; PCR‐HRM: polymerase chain reaction with high‐resolution melting; PCR‐mRNA: polymerase chain reaction‐messenger ribonucleic acid; PSQ: pyrosequencing; QF‐IHC: quantitative fluorescence immunohistochemistry; qMSP: quantitative methylation‐specific polymerase chain reaction; qMSP‐PSQ: quantitative methylation‐specific polymerase chain reaction with pyrosequencing; SQ‐MSP: semi‐quantitative methylation‐specific polymerase chain reaction. | ||||||
Appendix 9. Reconstructed Kaplan‐Meier plots
Reconstructed Kaplan‐Meier plots based on reported IPD or published Kaplan‐Meier curves are presented in Figure 16, Figure 17, Figure 18, Figure 19, Figure 20, and Figure 21.
16.
Reconstructed Kaplan‐Meier curves (1/6).
AF: area fraction; CpG: 5'‐cytosine‐phosphate‐guanine‐3'; GT: gross total; Incl: including; M‐GBM: methylated glioblastoma; Meth: methylated; MSP: methylation‐specific polymerase chain reaction; NGT: non‐gross total; PSQ: pyrosequencing; PyroSeq: pyrosequencing; UnMeth: unmethylated.
17.
Reconstructed Kaplan‐Meier curves (2/6).
CpG: 5'‐cytosine‐phosphate‐guanine‐3'; IHC: immunohistochemistry; Meth: methylated; MSP: methylation‐specific polymerase chain reaction; NGT: ; PCR‐HRM: polymerase chain reaction with high‐resolution melting; PCR‐mRNA: polymerase chain reaction targeting messenger ribonucleic acid; PSQ: pyrosequencing; qMSP: quantitative methylation‐specific polymerase chain reaction; UnMeth: unmethylated.
18.
Reconstructed Kaplan‐Meier curves (3/6).
CpG: 5'‐cytosine‐phosphate‐guanine‐3'; IHC: immunohistochemistry; Meth: methylated; MSP: methylation‐specific polymerase chain reaction; NA: not available; PCR‐mRNA: polymerase chain reaction‐messenger ribonucleic acid; PSQ: pyrosequencing; SQ‐MSP: semi‐quantitative methylation‐specific polymerase chain reaction; UnMeth: unmethylated.
19.
Reconstructed Kaplan‐Meier curves (4/6).
CpG: 5'‐cytosine‐phosphate‐guanine‐3'; IHC: immunohistochemistry; Meth: methylated; MSP: methylation‐specific polymerase chain reaction; PSQ: pyrosequencing; qMSP‐PSQ: quantitative methylation‐specific polymerase chain reaction with pyrosequencing; UnMeth: unmethylated.
20.
Reconstructed Kaplan‐Meier curves (5/6).
CpG: 5'‐cytosine‐phosphate‐guanine‐3'; FSQ‐MS‐PCR: fluorescent semi‐quantitative methylation‐specific polymerase chain reaction; IHC: immunohistochemistry; Meth: methylated; MS‐MLPA: methylation‐specific multiplex ligation‐dependent probe amplification; MSP: methylation‐specific polymerase chain reaction; PCR with HRM: polymerase chain reaction with HRM: high‐resolution melting; UnMeth: unmethylated.
21.
Reconstructed Kaplan‐Meier curves (6/6).
Meth: methylated; MSP: methylation‐specific polymerase chain reaction; UnMeth: unmethylated.
Characteristics of studies
Characteristics of included studies [ordered by study ID]
Almuqate 2018.
| Study characteristics | ||
| Study design | Cohort | |
| Study setting | Setting: University of Calgary Country: Canada Dates: tumour samples were tested for MGMT methylation in 2015 and 2016. |
|
| Selection of participants | Cases were retrieved from the Molecular Diagnostic Laboratory database. | |
| Participant characteristics | Sample size: 158 (deaths: NR) Age: mean 61 years Sex: 53.8% men KPS: NR |
|
| Tumour characteristics | GBM: 100% First diagnosis: NR Biopsy: 8.9%; subtotal resection: 38%; total resection: 53.2% IDH1 wild‐type: NR; IDH2 wild‐type: NR |
|
| Treatment regimen | NR | |
| MGMT promoter methylation tests implemented | MS‐RE‐qPCR | |
| Dates and follow‐up | Timing of MGMT assessment: at diagnosis Start time for follow‐up: NR; follow‐up: median NR; range NR |
|
| Notes | ||
Bady 2012 (E‐GBM).
| Study characteristics | ||
| Study design | Cohort | |
| Study setting | Setting: Neurosurgery Departments of Rennes and Angers University Hospitals Country: France Dates: NR |
|
| Selection of participants | Cases from an external dataset (Etcheverry 2010). Prospectively collected samples from people with newly diagnosed GBM | |
| Participant characteristics | Sample size: 50 (deaths: NR) Age: median 57.5, SD NR; range 26–80 years Sex: 51% men KPS: median 78.6; range 40–100 |
|
| Tumour characteristics | GBM: 100% First diagnosis: 100% Biopsy: NR; subtotal resection: NR; total resection: NR IDH1 wild‐type: NR; IDH2 wild‐type: NR |
|
| Treatment regimen | Stupp protocol | |
| MGMT promoter methylation tests implemented | PSQ, bead array | |
| Dates and follow‐up | Timing of MGMT assessment: at diagnosis Start time for follow‐up: NR; follow‐up: median NR; range NR |
|
| Notes | ||
Bady 2012 (M‐GBM).
| Study characteristics | ||
| Study design | Cohort | |
| Study setting | Setting: Neurosurgery Departments of Rennes and Angers University Hospitals Country: France Dates: NR |
|
| Selection of participants | Cases from an external dataset (Etcheverry 2010). Prospectively collected samples from people with newly diagnosed GBM | |
| Participant characteristics | Sample size: 50 (deaths: NR) Age: median 57.5, SD NR; range 26–80 years Sex: 51% men KPS: median 78.6, range 40–100 |
|
| Tumour characteristics | GBM: 100% First diagnosis: 100% Biopsy: NR; subtotal resection: NR; total resection: NR IDH1 wild‐type: NR; IDH2 wild‐type: NR |
|
| Treatment regimen | Stupp protocol | |
| MGMT promoter methylation tests implemented | PSQ, bead array | |
| Dates and follow‐up | Timing of MGMT assessment: at diagnosis Start time for follow‐up: NR; follow‐up: median NR; range NR |
|
| Notes | ||
Barault 2015.
| Study characteristics | ||
| Study design | Cohort | |
| Study setting | Setting: VU University Medical Center, Amsterdam Country: the Netherlands Dates: diagnosis between 2005 and 2011 |
|
| Selection of participants | Eligible people had a histopathological diagnosis of supratentorial GBM. The GBM validation‐set consisted of tissue samples from people with newly diagnosed GBM, who had surgery and chemoradiation with follow‐up ≥ 2 years. Inclusion criteria: adults aged > 17 years; a new histopathological diagnosis of supratentorial GBM between 2005 and 2011, verified by an independent neuropathologist; no prior brain tumour treatment to exclude dedifferentiated glioma; pre‐ and postoperative MRI within 3 days of surgery; standard adjuvant therapy |
|
| Participant characteristics | Sample size: 66 (deaths: NR) Age: NR Sex: % men NR KPS: NR |
|
| Tumour characteristics | GBM: 100% First diagnosis: 100% Biopsy: 3%; subtotal resection: NR; total resection: 97% IDH1 wild‐type: NR; IDH2 wild‐type: NR |
|
| Treatment regimen | Standard adjuvant therapy consisting of radiotherapy and concomitant TMZ, followed by 6 monthly cycles of adjuvant TMZ | |
| MGMT promoter methylation tests implemented | PSQ, methyl‐beaming | |
| Dates and follow‐up | Timing of MGMT assessment: not explicitly reported, but presumably on material obtained during resective surgery, prior to adjuvant therapy with 30 × 2 Gy radiotherapy and concomitant TMZ, followed by 6 monthly cycles of adjuvant TMZ. Start time for follow‐up: NR; follow‐up: median NR; range NR |
|
| Notes | ||
Barbagallo 2014.
| Study characteristics | ||
| Study design | Cohort | |
| Study setting | Setting: Department of Neurosurgery, Policlinico "G. Rodolico" Policlinico "G. Rodolico" University Hospital, University of Catania Country: Italy Dates: surgery between 2004 and 2012 |
|
| Selection of participants | All people underwent surgery for primary GBM with the aid of neuronavigation, and all but 2 people received gross tumour resection. The study was aimed at comparing short‐term vs long‐term TMZ treatment (people who received > 6 cycles), therefore some data were given as group A vs group B based on duration of treatment. | |
| Participant characteristics | Sample size: 37 (deaths: NR) Age: mean 60.4, SD 11.8; range 30–82 years Sex: 51.4% men KPS: mean 67.1, SD 15.2 |
|
| Tumour characteristics | GBM: 100% First diagnosis: 100% Biopsy: 0%; subtotal resection: 5.4%; total resection: 94.6% IDH1 wild‐type: NR; IDH2 wild‐type: NR |
|
| Treatment regimen | Stupp protocol. For adjuvant TMZ therapy, people in Group A received TMZ every 28 days for > 6 cycles (up to 101), those in Group B were treated with the same adjuvant TMZ dose regimen for ≤ 6 cycles. | |
| MGMT promoter methylation tests implemented | MSP, PSQ | |
| Dates and follow‐up | Timing of MGMT assessment: NR Start time for follow‐up: date of surgery; follow‐up: median NR; range NR |
|
| Notes | ||
Bell 2017.
| Study characteristics | ||
| Study design | Cohort nested within RCT | |
| Study setting | Setting: NR Country: multiple (Northern America and European centres) Dates: recruitment between 2006 and 2008; follow‐up to 2011 |
|
| Selection of participants | Participants were a subset of the NRG Oncology 0525 cohort (Gilbert 2013) with available specimens. This phase 3 trial compared standard adjuvant TMZ with a dose‐dense schedule in people with newly diagnosed GBM and KPS ≥ 60. | |
| Participant characteristics | Sample size: 452 (deaths: NR) Age: NR Sex: % men NR KPS: NR |
|
| Tumour characteristics | GBM: 100% First diagnosis: 100% Biopsy: NR; subtotal resection: NR; total resection: NR IDH1 wild‐type: NR; IDH2 wild‐type: NR |
|
| Treatment regimen | Standard TMZ or dose‐dense TMZ | |
| MGMT promoter methylation tests implemented | qMSP, QF‐IHC | |
| Dates and follow‐up | Timing of MGMT assessment: at diagnosis Start time for follow‐up: date of randomisation, i.e. after the initial 6 weeks of chemoradiotherapy; follow‐up: median NR; range NR |
|
| Notes | ||
Brigliadori 2016.
| Study characteristics | ||
| Study design | Cohort | |
| Study setting | Setting: Oncology Network of Romagna Country: Italy Dates: tumour samples collected between 2008 and 2013 |
|
| Selection of participants | Participants had GBM treated with surgery and Stupp regimen. People undergoing biopsy were not included in our analysis. | |
| Participant characteristics | Sample size: 105 (deaths: 73) Age: median 61, SD NR; range 23–76 years Sex: 61.9% men KPS: median NR; KPS ≤ 70: 19.1%, KPS: 80–90: 43.8%, KPS 100: 37.1% |
|
| Tumour characteristics | GBM: 100% First diagnosis: NR Biopsy: 0%; subtotal resection: 51.1%; total resection: 49% IDH1 wild‐type: NR; IDH2 wild‐type: NR |
|
| Treatment regimen | Stupp protocol | |
| MGMT promoter methylation tests implemented | PSQ, Bead array | |
| Dates and follow‐up | Timing of MGMT assessment: at diagnosis Start time for follow‐up: date of diagnosis; follow‐up: median 55; range 5–79 months |
|
| Notes | ||
Chai 2018 (7‐site cohort).
| Study characteristics | ||
| Study design | Cohort | |
| Study setting | Setting: Chinese Glioma Genome Atlas (CGGA) database Country: China Dates: NR |
|
| Selection of participants | Inclusion criteria: diagnosed with WHO grade III or IV glioma; containing MGMT promoter methylation PSQ testing data in detail; including exact MGMT mRNA sequencing data; having received radiotherapy + TMZ treatment; containing overall survival information | |
| Participant characteristics | Sample size: 24 (deaths: NR) Age: median 55; range 29–79 years Sex: 58.3% men KPS: median NR; KPS < 80: 11/24 (45.8%); KPS ≥ 80: 9/24 (37.5%); KPS not available: 4/24 (16.7%) |
|
| Tumour characteristics | GBM: 100% First diagnosis: NR Biopsy: 0%; subtotal resection: 50%; total resection: 50% IDH mutant 5/24 (20.8%), IDH wild‐type 18/24 (75.0%), not available 1/24 (4.2%). IDH status combined results of IDH1 and IDH2 testing |
|
| Treatment regimen | Radiotherapy + TMZ | |
| MGMT promoter methylation tests implemented | PSQ | |
| Dates and follow‐up | Timing of MGMT assessment: not explicitly reported, but presumably on freshly frozen tumour samples obtained during resection/biopsy. Start time for follow‐up: NR; follow‐up: median NR; range NR |
|
| Notes | ||
Chai 2018 (8‐site cohort).
| Study characteristics | ||
| Study design | Cohort | |
| Study setting | Setting: Chinese Glioma Genome Atlas (CGGA) database Country: China Dates: NR |
|
| Selection of participants | Inclusion criteria: diagnosed with WHO grade III or IV glioma; containing MGMT promoter methylation PSQ testing data in detail; including exact MGMT mRNA sequencing data; having received radiotherapy + TMZ treatment; containing overall survival information | |
| Participant characteristics | Sample size: 24 (deaths: NR) Age: median 55; range 29–79 years Sex: 58.3% men KPS: median NR; KPS < 80: 11/24 (45.8%); KPS ≥ 80: 9/24 (37.5%); KPS not available: 4/24 (16.7%) |
|
| Tumour characteristics | GBM: 100% First diagnosis: NR Biopsy: 0%; subtotal resection: 50%; total resection: 50% IDH mutant 5/24 (20.8%), IDH wild‐type 18/24 (75.0%), not available 1/24 (4.2%). IDH status combined results of IDH1 and IDH2 testing. |
|
| Treatment regimen | Radiotherapy + TMZ | |
| MGMT promoter methylation tests implemented | PSQ | |
| Dates and follow‐up | Timing of MGMT assessment: not explicitly reported, but presumably on freshly frozen tumour samples obtained during resection/biopsy. Start time for follow‐up: NR; follow‐up: median NR; range NR |
|
| Notes | ||
Dahlrot 2018 (NS cohort).
| Study characteristics | ||
| Study design | Cohort | |
| Study setting | Setting: Nordic study (validation cohort) Country: Denmark Dates: diagnosis between January 2003 and May 2008. |
|
| Selection of participants | People included in a collaborative Nordic Study (NS) with WHO grade 3 and 4 gliomas treated with radiotherapy and different combinations of TMZ. Minimum 15 mm2 vital tumour tissue was required for inclusion. | |
| Participant characteristics | Sample size: 92 (deaths: 64) Age: NR Sex: 57% men KPS: median NR ECOG performance status: 0–1: 64 (94%); 2: 4 (6%) |
|
| Tumour characteristics | GBM: 100% First diagnosis: 100% Biopsy: NR; subtotal resection: NR; total resection: NR IDH1 wild‐type: 94%; IDH2 wild‐type: NR |
|
| Treatment regimen | Radiotherapy and different combinations of TMZ | |
| MGMT promoter methylation tests implemented | PSQ, DIF | |
| Dates and follow‐up | Timing of MGMT assessment: NR Start time for follow‐up: date of randomisation; follow‐up: median 17.5; range 0.5–129 months |
|
| Notes | ||
Dahlrot 2018 (RSD cohort).
| Study characteristics | ||
| Study design | Cohort | |
| Study setting | Setting: Region of Southern Denmark Country: Denmark Dates: diagnosis between 1 January 2005 to 31 December 2009 |
|
| Selection of participants | Inhabitants in the Region of Southern Denmark (RSD), and no treatment received prior to surgery. Minimum 15 mm2 vital tumour tissue required for inclusion. | |
| Participant characteristics | Sample size: 234 (deaths: 168) Age: median NR; aged < 65 years: 83 (49%); aged > 65 years: 88 (51%) Sex: 57% men KPS: median NR; ECOG performance status: 0–1: 106 (62%); 2–4: 65 (38%) |
|
| Tumour characteristics | GBM: 100% First diagnosis: NR Biopsy: NR; subtotal resection: NR; total resection: NR IDH1 wild‐type: 98%; IDH2 wild‐type: NR |
|
| Treatment regimen | Stupp regimen (57%); palliative (25%); none (18%) | |
| MGMT promoter methylation tests implemented | PSQ, DIF | |
| Dates and follow‐up | Timing of MGMT assessment: NR Start time for follow‐up: date of primary surgery; follow‐up: median 11; range 0.03–96 months |
|
| Notes | ||
Dunn 2009.
| Study characteristics | ||
| Study design | Cohort | |
| Study setting | Setting: surgery at Walton Centre for Neurology and Neurosurgery and treatment at Clatterbridge Centre for Oncology Country: UK Dates: diagnosis between June 2004 and October 2007 |
|
| Selection of participants | Newly diagnosed, previously untreated GBMs WHO grade IV. These people had cytoreductive surgery where possible or biopsy before radical treatment with radiotherapy and concurrent TMZ + radiotherapy followed 4 weeks later by adjuvant TMZ. | |
| Participant characteristics | Sample size: 109 (deaths: 94) Age: median 55; range 18–68 years Sex: 66.1% men KPS: median NR; WHO performance status 0: 37/109 (33.9%); WHO performance status 1: 54/109 (49.5%); WHO performance status 2: 16/109 (14.7%); WHO performance status 3: 2/109 (1.8%) |
|
| Tumour characteristics | GBM: 100% First diagnosis: 100% Biopsy: 23.9%; subtotal resection: 0%; total resection: 76.1%; dichotomised as biopsy or resection IDH1 wild‐type: NR; IDH2 wild‐type: NR |
|
| Treatment regimen | Radiotherapy and concurrent TMZ + radiotherapy followed 4 weeks later by adjuvant TMZ | |
| MGMT promoter methylation tests implemented | PSQ | |
| Dates and follow‐up | Timing of MGMT assessment: not explicitly reported, but presumably on freshly frozen tumour samples obtained during resection/biopsy Start time for follow‐up: date of diagnosis; follow‐up: median NR; range NR |
|
| Notes | ||
Felsberg 2009.
| Study characteristics | ||
| Study design | Cohort | |
| Study setting | Setting: Department of Neurosurgery, Heinrich‐Heine‐University Düsseldorf Country: Germany Dates: recruitment between 1998 and 2004; follow‐up to 2006 |
|
| Selection of participants | Participants had been treated with open resection and ≥ 2 cycles of chemotherapy with TMZ first‐line. They had sufficient tissue for molecular analysis, and available follow‐up data. | |
| Participant characteristics | Sample size: 67 (deaths: 58) Age: median 56; range 26–80 years Sex: 61.2% men KPS: median 80; range 20–90 |
|
| Tumour characteristics | GBM: 100% First diagnosis: 100% Biopsy: 0%; subtotal resection: NR; total resection: NR; NCR defined by a residual tumour volume of < 5 mL on early postoperative MRI IDH1 wild‐type: 100%; IDH2 wild‐type: NR |
|
| Treatment regimen | Radiotherapy followed by adjuvant TMZ according to the standard 5‐day schedule every 28 days | |
| MGMT promoter methylation tests implemented | MSP, IHC, PCR‐mRNA | |
| Dates and follow‐up | Timing of MGMT assessment: at diagnosis Start time for follow‐up: date of surgery for primary tumour; follow‐up: median 40.6; range 16.5–96.0 months |
|
| Notes | ||
Havik 2012.
| Study characteristics | ||
| Study design | Cohort | |
| Study setting | Setting: Department of Neurosurgery, Oslo University Hospital Country: Norway Dates: surgery between January 2005 and January 2009 |
|
| Selection of participants | Tumour samples from 134 people with glioma (diffuse astrocytoma WHO grade II (n = 10), oligodendroglioma WHO grade II (n = 6), oligoastrocytoma WHO grade II (n = 17), low‐grade neuroepithelial tumour not otherwise specified (n = 2), anaplastic astrocytoma WHO grade III (n = 4), anaplastic oligodendroglioma, WHO grade III (n = 6), anaplastic oligoastrocytoma WHO grade III (n = 3), GBM WHO grade IV (n = 86)) and 4 people with meningioma | |
| Participant characteristics | Sample size: 134 (deaths: NR) Age: mean 58.5, SD 9.1 years Sex: 53.5% men KPS: NR |
|
| Tumour characteristics | GBM: 64.2% First diagnosis: NR Biopsy: NR; subtotal resection: NR; total resection: NR IDH1 wild‐type: NR; IDH2 wild‐type: NR |
|
| Treatment regimen | Standard radiotherapy and concomitant TMZ, some also adjuvant TMZ | |
| MGMT promoter methylation tests implemented | MSP, PSQ, qMSP, PCR‐HRM | |
| Dates and follow‐up | Timing of MGMT assessment: not explicitly reported, but presumably on freshly frozen tumour samples obtained during resection/biopsy. Start time for follow‐up: date of first surgery; follow‐up: median NR; range NR |
|
| Notes | ||
Hsu 2015.
| Study characteristics | ||
| Study design | Cohort | |
| Study setting | Setting: Taipei Veterans General Hospital Country: Taiwan, Republic of China Dates: enrolment between October 2007 and January 2014 |
|
| Selection of participants | People with primary GBM, TMZ chemotherapy with concomitant radiotherapy and adequate follow‐up data | |
| Participant characteristics | Sample size: 121 (deaths: 119) Age: median 55; range 40–65 years Sex: 59.5% men KPS: median NR; KPS ≥ 80: 66 (54.6%) |
|
| Tumour characteristics | GBM: 100% First diagnosis: NR Biopsy: NR; subtotal resection: NR; total resection: 81% IDH1 wild‐type: 91.7%; IDH2 wild‐type: NR |
|
| Treatment regimen | TMZ chemotherapy with concomitant radiotherapy | |
| MGMT promoter methylation tests implemented | MSP, PSQ, IHC, qMSP | |
| Dates and follow‐up | Timing of MGMT assessment: NR, probably after surgery Start time for follow‐up: date of surgery; follow‐up: median NR; range NR |
|
| Notes | ||
Karayan‐Tapon 2010.
| Study characteristics | ||
| Study design | Cohort | |
| Study setting | Setting: Centres in Poitiers, Rennes and Nantes Country: France Dates: NR |
|
| Selection of participants | Participants had GBM treated with surgery and Stupp regimen | |
| Participant characteristics | Sample size: 81 (deaths: NR) Age: median 61; range 30–78 years Sex: 55.6% men KPS: median NR; WHO performance status: 0–2: 52/81 (64.2%); WHO performance status 3–4: 23/81 (28.4%); WHO performance status not available: 6/81 (7.4%) |
|
| Tumour characteristics | GBM: 100% First diagnosis: 100% Biopsy: NR; subtotal resection: NR; total resection: NR; extent of resection determined perioperatively by neurosurgeon IDH1 wild‐type: NR; IDH2 wild‐type: NR |
|
| Treatment regimen | Stupp protocol | |
| MGMT promoter methylation tests implemented | MSP, PSQ, IHC, SQ‐MSP, PCR‐mRNA | |
| Dates and follow‐up | Timing of MGMT assessment: at diagnosis Start time for follow‐up: date of surgery; follow‐up: median 16; range 5–57 months |
|
| Notes | ||
Kim 2016.
| Study characteristics | ||
| Study design | Cohort | |
| Study setting | Setting: University College of Medicine, Busan; Ungkyunkwan University School of Medicine, Changwon Country: South Korea Dates: tissue collected between 1997 and 2012 |
|
| Selection of participants | Study set included FFPE brain tumour tissue diagnosed as GBM. All people underwent surgical resection or biopsy sampling of their tumours | |
| Participant characteristics | Sample size: 104 (deaths: 79) Age: mean 51.4; range 26.4–87.2 years Sex: 55.8% men KPS: median NR; KPS ≥ 70: 71.2%; KPS < 70: 28.8% |
|
| Tumour characteristics | GBM: 100% First diagnosis: NR Biopsy: 4.8%; subtotal resection: 57.7%; total resection: 37.5% IDH1 wild‐type: NR; IDH2 wild‐type: NR |
|
| Treatment regimen | Concurrent TMZ chemoradiotherapy | |
| MGMT promoter methylation tests implemented | MSP, PSQ | |
| Dates and follow‐up | Timing of MGMT assessment: not explicitly reported but presumably on tissue harvested during biopsy/resection Start time for follow‐up: date of diagnosis; follow‐up: median NR; range 3.2–41.5 months |
|
| Notes | ||
Kristensen 2016.
| Study characteristics | ||
| Study design | Cohort | |
| Study setting | Setting: Rigshospitalet Country: Denmark Dates: cases diagnosed between 2005 and 2010. Study ended 2015 |
|
| Selection of participants | Participants had available samples and received Stupp regimen | |
| Participant characteristics | Sample size: 151 (deaths: 146) Age: median 59, SD NR; range 22–74 years Sex: 62.9% men KPS: NR |
|
| Tumour characteristics | GBM: 100% First diagnosis: NR Biopsy: 3.3%; subtotal resection: NR; total resection: 41.1%; NR IDH1 wild‐type: 96%; IDH2 wild‐type: NR |
|
| Treatment regimen | Stupp protocol | |
| MGMT promoter methylation tests implemented | IHC, qMSP‐PSQ | |
| Dates and follow‐up | Timing of MGMT assessment: at diagnosis Start time for follow‐up: NR; follow‐up: median 94; range 53–123 months |
|
| Notes | ||
Lalezari 2013.
| Study characteristics | ||
| Study design | Cohort | |
| Study setting | Setting: University of California Los Angeles and Kaiser Permanente Los Angeles Country: USA Dates: diagnosed between 2000 and 2010. |
|
| Selection of participants | People were retrospectively identified based on an electronic database query of adults with primary GBM receiving upfront TMZ and treated at the University of California Los Angeles or Kaiser Permanente Los Angeles. People whose samples were directed to the laboratory in an unselected manner were also included. | |
| Participant characteristics | Sample size: 418 (deaths: 356) Age: median 57.6; range 22.3–90.0 years Sex: 60.8% men KPS: median NR; KPS 100: 13.9%; KPS 90: 47.6%; KPS 80: 24.6%; KPS 70: 6.2%; KPS ≤ 60: 7.2%; KPS missing: 0.5% |
|
| Tumour characteristics | GBM: 100% First diagnosis: 100% Biopsy: 9.1%; subtotal resection: 47.8%; total resection: 41.9%; NR IDH1 wild‐type: NR; IDH2 wild‐type: NR |
|
| Treatment regimen | Combination of radiotherapy and TMZ: concurrent daily radiotherapy/TMZ followed by TMZ (Stupp, n = 235), maintenance dose TMZ overlapping with radiotherapy (modified Stupp, n = 127), TMZ after radiotherapy (pre‐Stupp, n = 48). | |
| MGMT promoter methylation tests implemented | MSP, PSQ, IHC | |
| Dates and follow‐up | Timing of MGMT assessment: FFPE samples from initial surgery prior to any treatment Start time for follow‐up: NR; follow‐up: median 70; range 2–137 years |
|
| Notes | ||
Lattanzio 2015.
| Study characteristics | ||
| Study design | Cohort | |
| Study setting | Setting: Department of Neurosurgery, Santa Croce University Hospital, Cuneo Country: Italy Dates: tissue collected between 2006 and 2013 |
|
| Selection of participants | People with newly diagnosed GBM and treated with standard TMZ‐containing chemoradiotherapy protocols | |
| Participant characteristics | Sample size: 46 (deaths: 29) Age: median 64.5; range 24–84 years Sex: 76.1% men KPS: NR |
|
| Tumour characteristics | GBM: 100% First diagnosis: 100% Biopsy: NR; subtotal resection: NR; total resection: NR IDH1 wild‐type: NR; IDH2 wild‐type: NR |
|
| Treatment regimen | Standard TMZ‐containing chemoradiotherapy protocols | |
| MGMT promoter methylation tests implemented | MSP, PSQ | |
| Dates and follow‐up | Timing of MGMT assessment: for each participant, 2 samples of the primary tumour obtained: 1 collected during surgery, immersed in RNA later (Life Technologies, Carlsbad, CA, USA) and immediately snap‐frozen in liquid nitrogen, and 1 assembled from biopsy in FFPE sections using standard procedures. Start time for follow‐up: date of first surgery; follow‐up: median 7.4 months; range NR |
|
| Notes | ||
Lechapt‐Zalcman 2012.
| Study characteristics | ||
| Study design | Cohort | |
| Study setting | Setting: network of 11 neurosurgical university departments Country: France Dates: tumour samples collected between 2005 and 2009 |
|
| Selection of participants | Tumour samples collected from 2 observational studies analysing the use of Gliadel implants in people with newly diagnosed GBM | |
| Participant characteristics | Sample size: 111 (deaths: 56) Age: median 58, SD NR; range 33–77 years Sex: 65.8% men KPS: mean 80.2, SD 13.5 |
|
| Tumour characteristics | GBM: 100% First diagnosis: 100% Biopsy: 0%; subtotal resection: 30%; total resection: 55.5%; total 100% disappearance of contrast enhancement, subtotal ≥ 90% disappearance of contrast enhancement, partial < 90% disappearance of contrast enhancement. IDH1 wild‐type: NR; IDH2 wild‐type: NR |
|
| Treatment regimen | Gliadel (carmustine) wafers followed by the Stupp protocol | |
| MGMT promoter methylation tests implemented | MSP, PSQ | |
| Dates and follow‐up | Timing of MGMT assessment: at diagnosis Start time for follow‐up: NR; follow‐up: median 13.6; range 0–37.6 months |
|
| Notes | ||
McDonald 2013.
| Study characteristics | ||
| Study design | Cohort | |
| Study setting | Setting: Royal North Shore Hospital and the North Shore Private Hospital, Sydney Country: Australia Dates: NR |
|
| Selection of participants | Retrospective cohort of people with primary GBM treated by gross total resection | |
| Participant characteristics | Sample size: 78 (deaths: 74) Age: mean 58.4, SD 12.4; range 22–83 years Sex: 75.6% men KPS: NR |
|
| Tumour characteristics | GBM: 100% First diagnosis: 100% Biopsy: 0%; subtotal resection: 0%; total resection: 100% IDH1 wild‐type: 97.4%; IDH2 wild‐type: NR |
|
| Treatment regimen | Concurrent radiotherapy and TMZ followed by adjuvant TMZ (61.5%) or TMZ as an adjuvant therapy after radiotherapy (38.5%) | |
| MGMT promoter methylation tests implemented | MSP, PSQ | |
| Dates and follow‐up | Timing of MGMT assessment: at diagnosis Start time for follow‐up: NR; follow‐up: median NR; range NR |
|
| Notes | ||
Melguizo 2012.
| Study characteristics | ||
| Study design | Cohort | |
| Study setting | Setting: Hospital Virgen de las Nieves from Granada (Spain) and the University Hospital of Sassari (Italy) Country: Spain and Italy Dates: tumour samples collected between 2001 and 2009 |
|
| Selection of participants | Participants aged ≥ 70 years with newly diagnosed GBM and postoperative KPS ≥ 60 | |
| Participant characteristics | Sample size: 78 (deaths: NR) Age: mean 56; range 24–81 years Sex: 53.8% men KPS: median NR. All participants had KPS ≥ 60 |
|
| Tumour characteristics | GBM: 100% First diagnosis: 100% Biopsy: NR; subtotal resection: NR; total resection: NR IDH1 wild‐type: NR; IDH2 wild‐type: NR |
|
| Treatment regimen | Concurrent chemoradiotherapy with TMZ followed by adjuvant TMZ | |
| MGMT promoter methylation tests implemented | MSP, IHC | |
| Dates and follow‐up | Timing of MGMT assessment: at diagnosis Start time for follow‐up: date of diagnosis; follow‐up: median NR; range NR |
|
| Notes | ||
Nguyen 2015.
| Study characteristics | ||
| Study design | Cohort | |
| Study setting | Setting: Oncological Centre, University Hospital of Strasbourg Country: France Dates: cases treated and followed up between 2006 and 2010 |
|
| Selection of participants | Participants aged > 18 years with treatment‐naive GBM | |
| Participant characteristics | Sample size: 106 (deaths: NR) Age: median NR, SD NR; aged ≥ 50 years: 78%, aged < 50 years: 22% Sex: 63% men KPS: NR |
|
| Tumour characteristics | GBM: 100% First diagnosis: 100% Biopsy: 12%; subtotal resection: 34%; total resection: 34%; gross‐total (no residual tumour on MRI), subtotal (residual tumour on MRI) IDH1 wild‐type: 98%; IDH2 wild‐type: NR |
|
| Treatment regimen | Stupp protocol | |
| MGMT promoter methylation tests implemented | FSQ‐MS‐PCR | |
| Dates and follow‐up | Timing of MGMT assessment: at diagnosis Start time for follow‐up: NR; follow‐up: median 17.4; range 2–92.8 months |
|
| Notes | ||
Park 2011.
| Study characteristics | ||
| Study design | Cohort | |
| Study setting | Setting: Seoul National University Hospital and Seoul National University Bundang Hospital Country: South Korea Dates: NR |
|
| Selection of participants | Participants with newly diagnosed supratentorial GBM treated with surgery and Stupp regimen | |
| Participant characteristics | Sample size: 48 (deaths: 22) Age: mean 53.4; range 28–74 years Sex: 62.5% men KPS: NR |
|
| Tumour characteristics | GBM: 100% First diagnosis: 100% Biopsy: 24%; subtotal resection: NR; total resection: NR IDH1 wild‐type: NR; IDH2 wild‐type: NR |
|
| Treatment regimen | Stupp protocol | |
| MGMT promoter methylation tests implemented | MSP, MS‐MLPA | |
| Dates and follow‐up | Timing of MGMT assessment: at diagnosis Start time for follow‐up: date of surgery; follow‐up: median 16 months; range NR |
|
| Notes | ||
Quillien 2014 (test).
| Study characteristics | ||
| Study design | Cohort | |
| Study setting | Setting: 4 centres (Marseille, Paris, Poitiers and Rennes) Country: France Dates: treatment between November 2003 and September 2007 |
|
| Selection of participants | People with newly diagnosed primary GBM, excluding giant‐cell GBM, were given standard care treatment and followed up for ≥ 18 months. For each participant, a frozen tumour sample and paraffin‐embedded tissue specimens had to be available. People treated between November 2003 and September 2007 |
|
| Participant characteristics | Sample size: 100 (deaths: 75) Age: median 57.5; range 21.0–73.0 years Sex: 64% men KPS: median NR; KPS 90–100: 28%; KPS 70–80: 56%; KPS < 70: 16% |
|
| Tumour characteristics | GBM: 100% First diagnosis: 100% Biopsy: 7%; subtotal resection: 22%; total resection: 71% IDH1 wild‐type: 100; IDH2 wild‐type: NR |
|
| Treatment regimen | TMZ chemotherapy with concomitant radiotherapy, followed by cycles of adjuvant TMZ. 57 people required second‐line treatment (nitrosourea chemotherapy (n = 30); surgery with carmustine wafers (n = 12); surgery + nitrosourea chemotherapy (n = 1); bevacizumab + irinotecan (n = 6); other chemotherapy (n = 8). 12 people required third‐line treatment (bevacizumab + irinotecan (n = 5); nitrosourea chemotherapy (n = 3); other chemotherapy (n = 4) | |
| MGMT promoter methylation tests implemented | MSP, PSQ, IHC, MethyLight‐MSP, PCR‐HRM | |
| Dates and follow‐up | Timing of MGMT assessment: tumour samples obtained during surgery Start time for follow‐up: NR; follow‐up: median 17.9 months; range NR |
|
| Notes | ||
Quillien 2014 (validation).
| Study characteristics | ||
| Study design | Cohort | |
| Study setting | Setting: NR Country: France Dates: NR |
|
| Selection of participants | Independent validation cohort comprised 50 people with newly diagnosed GBM treated with radiotherapy and concurrent/adjuvant TMZ | |
| Participant characteristics | Sample size: 50 (deaths: NR) Age: median 59; range 41–78 years Sex: % men NR KPS: median NR; KPS 90–100: 22 (44%); KPS 70–80: 23 (46%); KPS < 70: 5 (10%) |
|
| Tumour characteristics | GBM: 100% First diagnosis: 100% Biopsy: NR; subtotal resection: NR; total resection: NR IDH1 wild‐type: NR; IDH2 wild‐type: NR |
|
| Treatment regimen | Radiotherapy and concurrent/adjuvant TMZ | |
| MGMT promoter methylation tests implemented | PSQ | |
| Dates and follow‐up | Timing of MGMT assessment: NR Start time for follow‐up: NR; follow‐up: median NR; range NR |
|
| Notes | ||
Quillien 2016.
| Study characteristics | ||
| Study design | Cohort | |
| Study setting | Setting: 8 centres Country: France Dates: enrolled between March 2009 and June 2011 |
|
| Selection of participants | Inclusion criteria: histologically confirmed de novo‐GBM, aged 18–70 years, presented with no contraindications as dictated by the Stupp protocol and not included in experimental therapeutic protocols | |
| Participant characteristics | Sample size: 139 (deaths: 119) Age: median 55.9; range 23.0–71.0 years Sex: 70.5% men KPS: median NR; KPS 90–100: 41 (29.5%); KPS 70–80: 76 (54.7%); KPS < 70: 20 (14.4%); KPS missing: 2 (1.4%) |
|
| Tumour characteristics | GBM: 100% First diagnosis: 100% Biopsy: 14.4%; subtotal resection: 29.5%; total resection: 56.1% IDH1 wild‐type: NR; IDH2 wild‐type: NR |
|
| Treatment regimen | Stupp protocol | |
| MGMT promoter methylation tests implemented | PSQ, SQ‐MSP | |
| Dates and follow‐up | Timing of MGMT assessment: not explicitly reported but presumably on tissue harvested during biopsy/resection Start time for follow‐up: NR; follow‐up: median NR; range NR |
|
| Notes | ||
Thon 2017.
| Study characteristics | ||
| Study design | Cohort | |
| Study setting | Setting: University of Munich Country: Germany Dates: enrolment between March 2006 and August 2008; last follow‐up June 2016 |
|
| Selection of participants | Adults with supratentorial GBM not suitable for gross total tumour resection with histology being confirmed by stereotactic biopsy; no severe mass effect of the tumour demanding debulking surgery; no prior history of surgery, radiotherapy or chemotherapy (or both); KPS ≥ 60 and adequate haematological, renal and hepatic function (Thon 2011) | |
| Participant characteristics | Sample size: 56 (deaths: 53) Age: median 62.5; range 23–85 years Sex: 58.9% men KPS: median 70; inclusion criterion was KPS ≥ 60. 24 (42.9%) participants had KPS 70 and 13 had KPS 60 (23.2%) |
|
| Tumour characteristics | GBM: 100% First diagnosis: 100% Biopsy: 100%; subtotal resection: 0%; total resection: 0% IDH1 wild‐type: NR; IDH2 wild‐type: NR |
|
| Treatment regimen | Radiotherapy/TMZ followed by adjuvant TMZ (adjuvant TMZ was not initiated in 14 people because of clinical deterioration with disorientation and confusion). Salvage treatment for progressive disease was initiated in 22 people and best supportive care in 33 people. | |
| MGMT promoter methylation tests implemented | MSP, PSQ | |
| Dates and follow‐up | Timing of MGMT assessment: tissue samples collected during biopsy Start time for follow‐up: date of biopsy; follow‐up: median NR; range NR |
|
| Notes | ||
Yamashita 2018.
| Study characteristics | ||
| Study design | Cohort | |
| Study setting | Setting: Miyazaki University Hospital Country: Japan Dates: surgery between February 2008 and July 2015 |
|
| Selection of participants | People with newly diagnosed GBM who had undergone surgery | |
| Participant characteristics | Sample size: 75 (deaths: NR) Age: median 64; range 32–84 years Sex: 61.3% men KPS: median NR; KPS 90–100: 18 (24%); KPS 70–80: 34 (45.3%); KPS < 70: 23 (30.7%) |
|
| Tumour characteristics | GBM: 100% First diagnosis: 100% Biopsy: 2.7%; subtotal resection: 30.7%; total resection: 66.7% 92% (IDH1 mutated 6.7%; unknown 1.3%); IDH2 wild‐type: NR |
|
| Treatment regimen | Stupp protocol | |
| MGMT promoter methylation tests implemented | MSP, PCR‐HRM | |
| Dates and follow‐up | Timing of MGMT assessment: tissue specimens obtained at surgery Start time for follow‐up: NR; follow‐up: median 17 months; range NR |
|
| Notes | ||
Yang 2012.
| Study characteristics | ||
| Study design | Cohort | |
| Study setting | Setting: Catholic University of Korea, Suwon Country: South Korea Dates: surgery between 2000 and 2006 |
|
| Selection of participants | People undergoing surgery with new histological diagnosis of supratentorial GBMs classified according to the WHO 2007 criteria | |
| Participant characteristics | Sample size: 18 (deaths: 13) Age: mean 53.3, SD 14.1; range 23–71 years Sex: 50% men KPS: NR |
|
| Tumour characteristics | GBM: 100% First diagnosis: NR Biopsy: 5.6%; subtotal resection: 27.8%; total resection: 66.7% IDH1 wild‐type: NR; IDH2 wild‐type: NR |
|
| Treatment regimen | Radiotherapy + TMZ (66.7%); radiotherapy + PCV + TMZ (33.3%) | |
| MGMT promoter methylation tests implemented | MSP, IHC | |
| Dates and follow‐up | Timing of MGMT assessment: tumour samples obtained during surgery Start time for follow‐up: date of histological diagnosis; follow‐up: median NR; range NR |
|
| Notes | ||
Yoshioka 2018.
| Study characteristics | ||
| Study design | Cohort | |
| Study setting | Setting: Chiba University Hospital Country: Japan Dates: NR |
|
| Selection of participants | People under a protocol approved by the Ethics Committee of the Chiba University Graduate School of Medicine, with informed consent obtained from the people or their guardians | |
| Participant characteristics | Sample size: 84 (deaths: NR) Age: median NR; aged < 60 years: 36 (43%); aged ≥ 60 years: 48 (57%) Sex: 51% men KPS: median NR; KPS ≤ 70: 47 (56%); KPS > 70: 37 (44%) |
|
| Tumour characteristics | GBM: 100% First diagnosis: NR Biopsy: NR; subtotal resection: 0.57%; total resection: 0.43% IDH1 wild‐type: 94%; IDH2 wild‐type: NR |
|
| Treatment regimen | Stupp protocol | |
| MGMT promoter methylation tests implemented | MSP | |
| Dates and follow‐up | Timing of MGMT assessment: time of the first surgery Start time for follow‐up: date of initial surgery; follow‐up: median NR; range NR |
|
| Notes | ||
DIF: double immunofluorescence; ECOG: Eastern Cooperative Oncology Group; FFPE: formalin‐fixed paraffin‐embedded; FSQ‐MS‐PCR: fluorescent semi‐quantitative methylation‐specific polymerase chain reaction; GBM: glioblastoma; IDH: isocitrate dehydrogenase; IHC: immunohistochemistry; KPS: Karnofsky performance status; MGMT: O6‐methylguanine–DNA methyltransferase; mRNA: messenger ribonucleic acid; MRI: magnetic resonance imaging; MS‐MLPA: methylation‐specific multiplex ligation‐dependent probe amplification; MS‐RE‐qPCR: methylation‐specific restriction enzyme quantitative polymerase chain reaction; MSP: methylation‐specific polymerase chain reaction; n: number of participants; NR: not reported; PCR: polymerase chain reaction; PCR‐HRM: polymerase chain reaction with high‐resolution melting; PCR‐mRNA: polymerase chain reaction‐messenger ribonucleic acid; PCV: procarbazine plus lomustine plus vincristine; PSQ: pyrosequencing; QF‐IHC: quantitative fluorescence immunohistochemistry; qMSP: quantitative methylation‐specific polymerase chain reaction; qMSP‐PSQ: quantitative methylation‐specific polymerase chain reaction with pyrosequencing; RCT: randomised controlled trial; RNA: ribonucleic acid; SD: standard deviation; SQ‐MSP: semi‐quantitative methylation‐specific polymerase chain reaction; TMZ: temozolomide; WHO: World Health Organization.
Characteristics of excluded studies [ordered by study ID]
| Study | Reason for exclusion |
|---|---|
| Becker 2016 | HR not reported/could not be calculated. |
| Becker 2018 | Unclear if all people received TMZ. |
| Christians 2012 | HR not reported/could not be calculated. |
| Crosby 2013 | HR not reported/could not be calculated. |
| Gurrieri 2018 | HR not reported/could not be calculated. |
| Jung 2010 | People had to survive until recurrence to be included. |
| Kang 2011 | IPD were available for the 9 eligible people, but these were too few to estimate HRs with any reliability. |
HR: hazard ratio; IPD: individual participant data; TMZ: temozolomide.
Characteristics of studies awaiting classification [ordered by study ID]
Cao 2009.
| Notes | Unclear report, further information required to make a decision. |
Dreval 2009.
| Notes | Requires translation (Russian). |
Ellingson 2012.
| Notes | Unclear report, further information required to make a decision. |
Fosmark 2017.
| Notes | Unclear report, further information required to make a decision. |
Grabenbauer 2010.
| Notes | Unclear report, further information required to make a decision. |
Hou 2011.
| Notes | Requires translation (Chinese). |
Jarboe 2012.
| Notes | Unclear report, further information required to make a decision. |
Kalkan 2015.
| Notes | Unclear report, further information required to make a decision. |
Kamoshima 2012.
| Notes | Requires translation (Japanese). |
Lin 2008.
| Notes | Requires translation (Chinese). |
Liu 2018.
| Notes | Requires translation (Chinese). |
Lobanova 2016.
| Notes | Requires translation (Russian). |
Shen 2011.
| Notes | Requires translation (Chinese). |
Sun 2004.
| Notes | Requires translation (Chinese). |
Tang 2012.
| Notes | Unclear report, further information required to make a decision. |
Yan 2015.
| Notes | Requires translation (Chinese). |
Yang 2011.
| Notes | Requires translation (Chinese). |
Characteristics of ongoing studies [ordered by study ID]
Rapp 2018.
| Study name | GlioVax |
| Starting date | March 2018 |
| Contact information | Michael Sabel (Michael.Sabel@med.uni‐duesseldorf.de) |
| Notes | Trial identifier: EudraCT‐Number 2017–000304‐14 Country: Germany Inclusion criteria: monofocal GBM, IDH wild‐type; near‐complete resection (≤ 5 mL residual tumour volume); Karnofsky performance status ≥ 70% |
GBM: glioblastoma; IDH: isocitrate dehydrogenase.
Differences between protocol and review
We had planned to search Open Grey (www.opengrey.eu/) using the free text terms from our MEDLINE search and in dissertations and theses using ProQuest Dissertations & Theses Global (search.proquest.com/pqdtglobal/dissertations/) and Networked Digital Library of Theses and Dissertations (search.ndltd.org/index.php). We did not implement these.
We had planned meta‐analyses (if deemed appropriate), although had not clarified that these would target comparisons of techniques (or of variants of techniques) to address the prespecified objectives of the review. Our meta‐analyses all addressed such comparisons. We developed our specific approach for this while undertaking the analyses (specifically, our imputation of a correlation between log HRs based on correlations among tests results in some individual participant data that we found in publications of included studies).
We rearranged the domains of our risk of bias assessment, although it covers the same issues and uses the same signalling questions. This was to allow us to separate better the study‐level issues from the result‐level issues.
We had aimed to create an economic model using outcomes from both the clinical and economic evidence. This was to be a decision analytic model, assessing the cost‐effectiveness of different methods of testing for MGMT promoter methylation status in people with glioma. We decided against this, in consultation with the hosting Cochrane Review Group, on the basis that it would involve a comparison of different policies for managing people using the information about MGMT promoter methylation status, thus extending substantially beyond the scope of the review.
Contributions of authors
AM led development of the protocol and organised retrieval of papers. SD designed and undertook the searches. AM, KMK and AH screened search results. AM, CK, FS, KMK, AH, SB and CLF screened retrieved papers against eligibility criteria. AK and TR screened retrieved papers against economic eligibility criteria. AM, CK, FS, LS, HC and JPTH extracted data from papers. AM, CK, FS, LS, HC and JPTH assessed risk of bias and applicability of included studies. JPTH and HC undertook the statistical analyses. JPTH undertook GRADE assessments. KMK, JPTH, FS and CLF interpreted the results. JPTH and KMK co‐ordinated the review. KMK, CLF, SB, SJ and CW provided a clinical perspective. LV, AK and TR provided economics expertise. JPTH drafted the manuscript with contributions from KMK, FS and HC.
Sources of support
Internal sources
University of Bristol, UK
External sources
-
National Institute for Health Research (NIHR), UK
This work was supported in part by a NIHR Systematic Reviews Cochrane Programme Grant 16/144
-
Cancer Research UK, UK
AM, FS and JPTH were supported in part by Cancer Research UK (grant number C18281/A19169).
The views and opinions expressed are those of the review authors and do not necessarily reflect those of the NIHR, its Systematic Reviews Programme, the National Health Service (NHS), the Department of Health or Cancer Research UK.
Declarations of interest
AM: none known CK: none known FS: none known AK: none known HC: none known SD: none known LS: none known TR: none known SB: member of the NICE Primary Brain Tumours Guideline Committee CLF: none known CW: none known SJ: member of the National Institute for Health and Care Excellence (NICE) Primary Brain Tumours Guideline Committee AH: none known LV: none known JPTH: none known KMK: none known
New
References
References to studies included in this review
Almuqate 2018 {published data only}
- Almuqate AT, Sayeed W, Kornaga E, Nikolic A, Roldan-Urgoiti GB, Itani D. MGMT promoter methylation status in glioblastoma: a single institutional experience. Laboratory Investigation 2018;98(Suppl 1):655. [Google Scholar]
Bady 2012 (E‐GBM) {published data only}
- Bady P, Sciuscio D, Diserens AC, Bloch J, den Bent MJ, Marosi C, et al. MGMT methylation analysis of glioblastoma on the Infinium methylation BeadChip identifies two distinct CpG regions associated with gene silencing and outcome, yielding a prediction model for comparisons across datasets, tumor grades, and CIMP-status. [Erratum appears in Acta Neuropathologica 2013;126(1):159]. Acta Neuropathologica 2012;124(4):547-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etcheverry A, Aubry M, Tayrac M, Vauleon E, Boniface R, Guenot F, et al. DNA methylation in glioblastoma: impact on gene expression and clinical outcome. BMC Genomics 2010;11:701. [DOI] [PMC free article] [PubMed] [Google Scholar]
Bady 2012 (M‐GBM) {published data only}
- Bady P, Sciuscio D, Diserens AC, Bloch J, den Bent MJ, Marosi C, et al. MGMT methylation analysis of glioblastoma on the Infinium methylation BeadChip identifies two distinct CpG regions associated with gene silencing and outcome, yielding a prediction model for comparisons across datasets, tumor grades, and CIMP-status. [Erratum appears in Acta Neuropathologica 2013;126(1):159]. Acta Neuropathologica 2012;124(4):547-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bady P, Sciuscio D, Stupp R, Delorenzi M, Hegi ME. MGMT methylation based outcome prediction is associated with two CpG regions separated by a prediction minimum centred at the initiation start site. Cancer Research 2012;72(Suppl 8):4031. [Google Scholar]
- Stupp R, Dietrich PY, Ostermann Kraljevic S, Pica A, Maillard I, Maeder P, et al. Promising survival for patients with newly diagnosed glioblastoma multiforme treated with concomitant radiation plus temozolomide followed by adjuvant temozolomide. Journal of Clinical Oncology 2002;20(5):1375-82. [DOI] [PubMed] [Google Scholar]
- Stupp R, Mason WP, den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. New England Journal of Medicine 2005;352(10):987-96. [DOI] [PubMed] [Google Scholar]
Barault 2015 {published data only}
- Barault L, Amatu A, Bleeker FE, Moutinho C, Falcomata C, Fiano V, et al. Digital PCR quantification of MGMT methylation refines prediction of clinical benefit from alkylating agents in glioblastoma and metastatic colorectal cancer. Annals of Oncology 2015;26(9):1994-9. [DOI] [PubMed] [Google Scholar]
Barbagallo 2014 {published data only}
- Barbagallo GM, Paratore S, Caltabiano R, Palmucci S, Parra HS, Privitera G, et al. Long-term therapy with temozolomide is a feasible option for newly diagnosed glioblastoma: a single-institution experience with as many as 101 temozolomide cycles. Neurosurgical Focus 2014;37(6):E4. [DOI] [PubMed] [Google Scholar]
Bell 2017 {published data only}
- Bell EH, Pugh SL, McElroy JP, Gilbert MR, Mehta M, Klimowicz AC, et al. Molecular-based recursive partitioning analysis model for glioblastoma in the temozolomide era: a correlative analysis based on NRG Oncology RTOG 0525. JAMA Oncology 2017;3(6):784-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert MR, Wang M, Aldape KD, Stupp R, Hegi ME, Jaeckle KA, et al. Dose-dense temozolomide for newly diagnosed glioblastoma: a randomized phase III clinical trial. Journal of Clinical Oncology 2013;31(32):4085-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Brigliadori 2016 {published data only}
- Brigliadori G, Foca F, Dall'Agata M, Rengucci C, Melegari E, Cerasoli S, et al. Defining the cutoff value of MGMT gene promoter methylation and its predictive capacity in glioblastoma. Journal of Neuro-oncology 2016;128(2):333-9. [DOI] [PubMed] [Google Scholar]
- Rocca A, Brigliadori G, Calistri D, Foca F, Dall'Agata M, Rengucci C, et al. Defining the cutoff value of MGMT gene promoter methylation and its predictive capacity. Journal of Clinical Oncology 2015;33(15 Suppl 1):2017. [DOI: 10.1200/jco.2015.33.15_suppl.2017] [DOI] [PubMed] [Google Scholar]
Chai 2018 (7‐site cohort) {published data only}
- Chai RC, Zhang KN, Liu YQ, Wu F, Zhao Z, Wang KY, et al. Combinations of four or more CpGs methylation present equivalent predictive value for MGMT expression and temozolomide therapeutic prognosis in gliomas. CNS Neuroscience and Therapeutics 2018;25(3):314-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Chai 2018 (8‐site cohort) {published data only}
- Chai RC, Zhang KN, Liu YQ, Wu F, Zhao Z, Wang KY, et al. Combinations of four or more CpGs methylation present equivalent predictive value for MGMT expression and temozolomide therapeutic prognosis in gliomas. CNS Neuroscience and Therapeutics 2018;25(3):314-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Dahlrot 2018 (NS cohort) {published data only}
- Dahlrot RH, Dowsett J, Fosmark S, Malmstrom A, Henriksson R, Boldt H, et al. Prognostic value of O-6-methylguanine-DNA methyltransferase (MGMT) protein expression in glioblastoma excluding nontumour cells from the analysis. Neuropathology and Applied Neurobiology 2018;44(2):172-84. [DOI] [PubMed] [Google Scholar]
- Dahlrot RH, Hermansen SK, Hansen S, Kristensen BW. What is the clinical value of cancer stem cell markers in gliomas? International Journal of Clinical & Experimental Pathology 2013;6(3):334-48. [PMC free article] [PubMed] [Google Scholar]
Dahlrot 2018 (RSD cohort) {published data only}
- Dahlrot RH, Dowsett J, Fosmark S, Malmstrom A, Henriksson R, Boldt H, et al. Prognostic value of O-6-methylguanine-DNA methyltransferase (MGMT) protein expression in glioblastoma excluding nontumour cells from the analysis. Neuropathology and Applied Neurobiology 2018;44(2):172-84. [DOI] [PubMed] [Google Scholar]
Dunn 2009 {published data only}
- Dunn J, Baborie A, Alam F, Joyce K, Moxham M, Sibson R, et al. Extent of MGMT promoter methylation correlates with outcome in glioblastomas given temozolomide and radiotherapy. British Journal of Cancer 2009;101(1):124-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Felsberg 2009 {published data only}
- Felsberg J, Rapp M, Loeser S, Fimmers R, Stummer W, Goeppert M, et al. Prognostic significance of molecular markers and extent of resection in primary glioblastoma patients. Clinical Cancer Research 2009;15(21):6683-93. [DOI] [PubMed] [Google Scholar]
Havik 2012 {published data only}
- Havik AB, Brandal P, Honne H, Dahlback HS, Scheie D, Hektoen M, et al. MGMT promoter methylation in gliomas-assessment by pyrosequencing and quantitative methylation-specific PCR. Journal of Translational Medicine 2012;10:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen LE, Brandal P, Myklebust TA, Heim S, Micci F, Panagopoulos I. MGMT gene promoter methylation status – assessment of two pyrosequencing kits and three methylation-specific PCR methods for their predictive capacity in glioblastomas. Cancer Genomics & Proteomics 2018;15(6):437-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
Hsu 2015 {published data only}
- Hsu CY, Ho HL, Lin SC, Chang-Chien YC, Chen MH, Hsu SP, et al. Prognosis of glioblastoma with faint MGMT methylation-specific PCR product. Journal of Neuro-oncology 2015;122(1):179-88. [DOI] [PubMed] [Google Scholar]
- Hsu CY, Ho HL, Lin SC, Chen MH, Hsu SP, Yen YS, et al. Comparative assessment of 4 methods to analyze MGMT status in a series of 121 glioblastoma patients. Applied Immunohistochemistry & Molecular Morphology 2017;25(7):497-504. [DOI] [PubMed] [Google Scholar]
- Hsu CY, Ho HL, Lin SC, Ho TD, Ho DM. The MGMT promoter single-nucleotide polymorphism rs1625649 had prognostic impact on patients with MGMT methylated glioblastoma. PloS One 2017;12(10):e0186430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CY, Lin SC, Ho HL, Chang-Chien YC, Hsu SP, Yen YS, et al. Exclusion of histiocytes/endothelial cells and using endothelial cells as internal reference are crucial for interpretation of MGMT immunohistochemistry in glioblastoma. American Journal of Surgical Pathology 2013;37(2):264-71. [DOI] [PubMed] [Google Scholar]
- Yang CF, Ho HL, Lin SC, Hsu CY, Ho DMT. Detection of human cytomegalovirus in glioblastoma among Taiwanese subjects. PloS One 2017;12(6):e0179366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Karayan‐Tapon 2010 {published data only}
- Karayan-Tapon L, Quillien V, Guilhot J, Wager M, Fromont G, Saikali S, et al. Prognostic value of O6-methylguanine-DNA methyltransferase status in glioblastoma patients, assessed by five different methods. Journal of Neuro-oncology 2010;97(3):311-22. [DOI] [PubMed] [Google Scholar]
Kim 2016 {published data only}
- Kim D, Kim Y, Song Y, Kim K, Choi Y, Choi S. MGMT gene promoter methylation analysis by pyrosequencing of glial tumours. European Journal of Cancer 2011;47(Suppl 1):S175-S6. [Google Scholar]
- Kim DC, Kim KU, Kim YZ. Prognostic role of methylation status of the MGMT promoter determined quantitatively by pyrosequencing in glioblastoma patients. Journal of Korean Neurosurgical Society 2016;59(1):26-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DC, Song YJ, Lee EH, Kim KU, Kim YZ. Prognostic role of methylation status of MGMT promoter on glioblastoma patients estimated quantitatively by pyrosequencing. Neuro-oncology 2015;17(Suppl 5):141. [DOI: 10.1093/neuonc/nov222.14] [DOI] [Google Scholar]
Kristensen 2016 {published data only}
- Kristensen LS, Michaelsen SR, Dyrbye H, Aslan D, Grunnet K, Christensen IJ, et al. Assessment of quantitative and allelic MGMT methylation patterns as a prognostic marker in glioblastoma. Journal of Neuropathology and Experimental Neurology 2016;75(3):246-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
Lalezari 2013 {published data only}
- Lai A, Lalezari S, Chou AP, Tran A, Solis OE, Carrillo JA, et al. Prediction of GBM outcome using combined analysis of MGMT protein expression and promoter methylation. Journal of Clinical Oncology 2011;29(15 Suppl 1):2003. [DOI: 10.1200/jco.2011.29.15_suppl.2003] [DOI] [Google Scholar]
- Lalezari S, Chou AP, Tran A, Solis OE, Khanlou N, Chen W, et al. Combined analysis of O6-methylguanine-DNA methyltransferase protein expression and promoter methylation provides optimized prognostication of glioblastoma outcome. Neuro-oncology 2013;15(3):370-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Lattanzio 2015 {published data only}
- Lattanzio L, Borgognone M, Mocellini C, Giordano F, Favata E, Fasano G, et al. MGMT promoter methylation and glioblastoma: a comparison of analytical methods and of tumor specimens. International Journal of Biological Markers 2015;30(2):e208-16. [DOI] [PubMed] [Google Scholar]
Lechapt‐Zalcman 2012 {published data only}
- Lechapt-Zalcman E, Levallet G, Dugue AE, Vital A, Diebold MD, Menei P, et al. O(6)-methylguanine-DNA methyltransferase (MGMT) promoter methylation and low MGMT-encoded protein expression as prognostic markers in glioblastoma patients treated with biodegradable carmustine wafer implants after initial surgery followed by radiotherapy with concomitant and adjuvant temozolomide. Cancer 2012;118(18):4545-54. [DOI] [PubMed] [Google Scholar]
McDonald 2013 {published data only}
- McDonald KL, Rapkins R, Zhau L, Hitchins M. The t genotype of the MGMT C > T (rs16906252) enhancer SNP is associated with promoter methylation and longer survival in patients with glioblastoma. Neuro-oncology 2012;14(Suppl 3):iii8. [DOI] [PubMed] [Google Scholar]
- McDonald KL, Rapkins RW, Olivier J, Zhao L, Nozue K, Lu D, et al. The T genotype of the MGMT C>T (rs16906252) enhancer single-nucleotide polymorphism (SNP) is associated with promoter methylation and longer survival in glioblastoma patients. European Journal of Cancer 2013;49(2):360-8. [DOI] [PubMed] [Google Scholar]
Melguizo 2012 {published data only}
- Gonzalez-Astorga B, Luque R, Castellon V, Gonzalez E, Sanchez C, Soberino J, et al. MGMT promoter methylation status and MGMT and CD133 immunohistochemical expression as prognostic markers in glioblastoma patients treated with temozolomide plus radiotherapy. European Journal of Cancer 2013;49(Suppl 2):S794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melguizo C, Prados J, Gonzalez B, Ortiz R, Concha A, Alvarez PJ, et al. MGMT promoter methylation status and MGMT and CD133 immunohistochemical expression as prognostic markers in glioblastoma patients treated with temozolomide plus radiotherapy. Journal of Translational Medicine 2012;10:250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Nguyen 2015 {published data only}
- Nguyen A, Legrain M, Noel G, Coca A, Meyer E, Schott R, et al. An innovative fluorescent semi-quantitative methylation-specific PCR method for the determination of MGMT promoter methylation is reflecting intra-tumor heterogeneity. Current Cancer Drug Targets 2015;15(7):624-40. [DOI] [PubMed] [Google Scholar]
Park 2011 {published data only}
- Park CK, Kim J, Yim SY, Lee AR, Han JH, Kim CY, et al. Usefulness of MS-MLPA for detection of MGMT promoter methylation in the evaluation of pseudoprogression in glioblastoma patients. Neuro-oncology 2011;13(2):195-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Quillien 2014 (test) {published data only}
- Quillien V, Bellissant E, Sanson M, Karayan-Tapon L, Lesimple T, Chinot O, et al. Comparison of MS-PCR, MethyLight, pyrosequencing, MS-HRM, and immunohistochemistry for MGMT analysis. Neuro-oncology 2010;12(Suppl 3):iii4. [Google Scholar]
- Quillien V, Lavenu A, Karayan-Tapon L, Carpentier C, Labussiere M, Lesimple T, et al. Comparative assessment of 5 methods (methylation-specific polymerase chain reaction, MethyLight, pyrosequencing, methylation-sensitive high-resolution melting, and immunohistochemistry) to analyze O6-methylguanine-DNA-methyltranferase in a series of 100 glioblastoma patients. Cancer 2012;118(17):4201-11. [DOI] [PubMed] [Google Scholar]
- Quillien V, Lavenu A, Sanson M, Legrain M, Dubus P, Karayan-Tapon L, et al. Outcome-based determination of optimal pyrosequencing assay for MGMT methylation detection in glioblastoma patients. Journal of Neuro-oncology 2014;116(3):487-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
Quillien 2014 (validation) {published data only}
- Quillien V, Lavenu A, Sanson M, Legrain M, Dubus P, Karayan-Tapon L, et al. Outcome-based determination of optimal pyrosequencing assay for MGMT methylation detection in glioblastoma patients. Journal of Neuro-oncology 2014;116(3):487-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
Quillien 2016 {published data only}
- Quillien V, Lavenu A, Ducray F, Joly MO, Chinot O, Fina F, et al. Validation of the high-performance of pyrosequencing for clinical MGMT testing on a cohort of glioblastoma patients from a prospective dedicated multicentric trial. Oncotarget 2016;7(38):61916-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillien V, Lavenu A, Ducray F, Meyronet D, Chinot O, Fina F, et al. Clinical validation of the CE-IVD marked Therascreen MGMT kit in a cohort of glioblastoma patients. Cancer Biomarkers: Section A of Disease Markers 2017;20(4):435-41. [DOI] [PubMed] [Google Scholar]
Thon 2017 {published data only}
- Thon N, Eigenbrod S, Grasbon-Frodl EM, Lutz J, Kreth S, Popperl G, et al. Predominant influence of MGMT methylation in non-resectable glioblastoma after radiotherapy plus temozolomide. Journal of Neurology, Neurosurgery, and Psychiatry 2011;82(4):441-6. [DOI] [PubMed] [Google Scholar]
- Thon N, Eigenbrod S, Lutz J, Egensperger R, Kretzschmar H, Tonn J-C, et al. Prognostic relevance of MGMT promoter hypermethylation as demonstrated in serial stereotactic specimens from glioblastomas after primary concomitant radiochemotherapy followed by adjuvant temozolomide. Proceedings of the American Association for Cancer Research 2009;50:623. [Google Scholar]
- Thon N, Grasbon-Frodl EM, Eros C, Kretzschmar H, Tonn JC, Kreth FW. Prognostic relevance of MGMT promoter methylation as demonstrated in serial stereotactic specimens from anaplastic astrocytomas and glioblastomas after primary radio-/radiochemotherapy. Neuro-oncology 2008;10(5):865-6. [Google Scholar]
- Thon N, Thorsteinsdottir J, Eigenbrod S, Schuller U, Lutz J, Kreth S, et al. Outcome in unresectable glioblastoma: MGMT promoter methylation makes the difference. Journal of Neurology 2017;264(2):350-8. [DOI] [PubMed] [Google Scholar]
Yamashita 2018 {published data only}
- Yamashita S, Yokogami K, Matsumoto F, Saito K, Mizuguchi A, Ohta H, et al. MGMT promoter methylation in patients with glioblastoma: is methylation-sensitive high-resolution melting superior to methylation-sensitive polymerase chain reaction assay? Journal of Neurosurgery 2018;130(3):780-8. [DOI] [PubMed] [Google Scholar]
- Yamashita S, Yokogami K, Takeshima H. Detection of MGMT promoter methylation levels with MS-HRM in patients with glioblastoma. Neuro-oncology 2017;19(Suppl 6):vi99. [Google Scholar]
Yang 2012 {published data only}
- Yang SH, Lee KS, Yang HJ, Jeon BH, Lee YS, Nam SW, et al. O6-methylguanine-DNA-methyltransferase promoter methylation assessment by microdissection-assisted methylation-specific PCR and high resolution melting analysis in patients with glioblastomas. Journal of Neuro-oncology 2012;106(2):243-50. [DOI] [PubMed] [Google Scholar]
Yoshioka 2018 {published data only}
- Yoshioka M, Matsutani T, Hara A, Hirono S, Hiwasa T, Takiguchi M, et al. Real-time methylation-specific PCR for the evaluation of methylation status of MGMT gene in glioblastoma. Oncotarget 2018;9(45):27728-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
References to studies excluded from this review
Becker 2016 {published data only}
- Becker AP, Bell EH, McElroy JP, Cui T, Geurts M, Fleming J, et al. Comprehensive survival analysis of MGMT protein expression by traditional and quantitative fluorescence immunohistochemistry compared to MGMT promoter methylation in a large institutional glioblastoma cohort treated with the stupp protocol. Neuro-oncology 2016;18(Suppl 6):vi117. [Google Scholar]
Becker 2018 {published data only}
- Becker A, Bell EH, McElroy J, Cui T, Geurts M, Liu Z, et al. MGMT protein expression adds prognostic value beyond MGMT promoter methylation and stratifies survival prognoses of un-methylated glioblastoma patients. International Journal of Radiation Oncology, Biology, Physics 2018;102(3):S47. [Google Scholar]
Christians 2012 {published data only}
- Christians A, Hartmann C, Benner A, Meyer J, Deimling A, Weller M, et al. Prognostic value of three different methods of MGMT promoter methylation analysis in a prospective trial on newly diagnosed glioblastoma. PloS One 2012;7(3):e33449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Crosby 2013 {published data only}
- Crosby C, Faulkner C, Smye-Rumsby T, Kurian K, Williams M, Hopkins K. A retrospective review of the influence of quantitative MGMT methylation on survival after chemoradiotherapy for patients with glioblastoma. Neuro-oncology 2013;15(Suppl 3):iii142. [Google Scholar]
Gurrieri 2018 {published data only}
- Gurrieri L, De Carlo, Gerratana L, De Maglio, Macerelli M, Pisa FE, et al. MGMT pyrosequencing-based cut-off methylation level and clinical outcome in patients with glioblastoma multiforme. Future Oncology 2018;14(8):699-707. [DOI] [PubMed] [Google Scholar]
Jung 2010 {published data only}
- Jung TY, Jung S, Moon KS, Kim IY, Kang SS, Kim YH, et al. Changes of the O6-methylguanine-DNA methyltransferase promoter methylation and MGMT protein expression after adjuvant treatment in glioblastoma. Oncology Reports 2010;23(5):1269-76. [DOI] [PubMed] [Google Scholar]
Kang 2011 {published data only}
- Kang SH, Park KJ, Kim CY, Yu MO, Park CK, Park SH, et al. O6-methylguanine DNA methyltransferase status determined by promoter methylation and immunohistochemistry in gliosarcoma and their clinical implications. Journal of Neuro-oncology 2011;101(3):477-86. [DOI] [PubMed] [Google Scholar]
References to studies awaiting assessment
Cao 2009 {published data only}
- Cao VT, Jung TY, Jung S, Jin SG, Moon KS, Kim IY, et al. The correlation and prognostic significance of MGMT promoter methylation and MGMT protein in glioblastomas. Neurosurgery 2009;65(5):866-75. [DOI] [PubMed] [Google Scholar]
Dreval 2009 {published data only}
- Dreval ON, Belokhvostov AS, Feniksov VM. Clinical and cytogenetic factors of prognosis in glial tumors of the brain. Zhurnal Voprosy Neirokhirurgii Imeni N. N. Burdenko 2009;4:7-13. [PubMed] [Google Scholar]
Ellingson 2012 {published data only}
- Ellingson BM, Cloughesy TF, Pope WB, Zaw TM, Phillips H, Lalezari S, et al. Anatomic localization of O6-methylguanine DNA methyltransferase (MGMT) promoter methylated and unmethylated tumors: a radiographic study in 358 de novo human glioblastomas. Neuroimage 2012;59(2):908-16. [DOI] [PubMed] [Google Scholar]
Fosmark 2017 {published data only}
- Fosmark S, Hellwege S, Dahlrot RH, Jensen KL, Derand H, Lohse J, et al. APNG as a prognostic marker in patients with glioblastoma. PloS One 2017;12(6):e0178693. [DOI] [PMC free article] [PubMed] [Google Scholar]
Grabenbauer 2010 {published data only}
- Grabenbauer GG. Long-term survival of patients with glioblastoma multiforme treated with chemoradiation: correlation with MGMT promoter methylation status. Strahlentherapie und Onkologie 2010;186(3):185-7. [Google Scholar]
Hou 2011 {published data only}
- Hou X, Zhao Y, Zheng YR, Wang JJ, Wu ZC, Sun JH. Comparison of MGMT and ERCC2 expression in temozolomide for the treatment of malignant glioma drug resistance and their genetic relationship. Chung-Hua i Hsueh Tsa Chih [Chinese Medical Journal] 2011;91(1):56-8. [PubMed] [Google Scholar]
Jarboe 2012 {published data only}
- Jarboe JS, Anderson JC, Duarte CW, Mehta T, Nowsheen S, Hicks PH, et al. MARCKS regulates growth and radiation sensitivity and is a novel prognostic factor for glioma. Clinical Cancer Research 2012;18(11):3030-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Kalkan 2015 {published data only}
- Kalkan R, Atli EI, Ozdemir M, Ciftci E, Aydin HE, Artan S, et al. IDH1 mutations is prognostic marker for primary glioblastoma multiforme but MGMT hypermethylation is not prognostic for primary glioblastoma multiforme. Gene 2015;554(1):81-6. [DOI] [PubMed] [Google Scholar]
Kamoshima 2012 {published data only}
- Kamoshima Y, Motegi H, Terasaka S, Kobayashi H, Yamaguchi S, Murata J, et al. Analyses of IDH1 mutation and MGMT promoter methylation status for 5 cases of long-term survivors with glioblastoma. No Shinkei Geka [Neurological Surgery] 2012;40(2):129-35. [PubMed] [Google Scholar]
Lin 2008 {published data only}
- Lin ZX, Tan SL, Zhou AP, Mei WZ, He LS, Jiang CZ, et al. The impact of non-pathological factors on TMZ treatment of cerebral glioma. Chinese Journal of Contemporary Neurology and Neurosurgery 2008;8(5):437-41. [Google Scholar]
Liu 2018 {published data only}
- Liu J, Lou M, Ji P, Li C, Feng F, Li B, et al. Analysis of prognostic factors for survival in elderly patients with glioma. Zhong Nan da Xue Xue Bao. Yi Xue Ban [Journal of Central South University. Medical Sciences] 2018;43(4):403-9. [DOI] [PubMed] [Google Scholar]
Lobanova 2016 {published data only}
- Lobanova NV, Shishkina LV, Ryzhova MV, Kobyakov GL, Sycheva RV, Burov SA, et al. Clinical, immunohistochemical, and molecular genetic prognostic factors in adult patients with glioblastoma [Klinicheskie, immunogistokhimicheskie i molekulyarno-geneticheskie faktory prognoza u bol'nykh c glioblastomoi]. Arkhiv Patologii 2016;78(4):10-9. [DOI] [PubMed] [Google Scholar]
Shen 2011 {published data only}
- Shen D, Yang Q, Sai K, Mou Y, Zhang X, Jiang X, et al. Efficacy of salvage chemotherapy based on MGMT protein expression in patients with recurrent malignant gliomas: a report of 30 cases. Chinese Journal of Clinical Oncology 2011;38(13):781-3, 787. [Google Scholar]
Sun 2004 {published data only}
- Sun YH, Zhang YZ, Wang ZC, Sun MZ, Zhao D H. Relationship between the expression of O6-methylguanine-DNA methyltransferase in glioma and the survival time of patients. Chinese Journal of Cancer 2004;23(9):1052-5. [PubMed] [Google Scholar]
Tang 2012 {published data only}
- Tang K, Jin Q, Yan W, Zhang W, You G, Liu Y, et al. Clinical correlation of MGMT protein expression and promoter methylation in Chinese glioblastoma patients. Medical Oncology 2012;29(2):1292-6. [DOI] [PubMed] [Google Scholar]
Yan 2015 {published data only}
- Yan H, Han N, Ding Y, Zhang H, Hou Y, He Y. Prognostic value of CDKN2A mRNA level in glioblastoma. Cancer Research and Clinic 2015;27(11):766-70. [Google Scholar]
Yang 2011 {published data only}
- Yang QY, Shen D, Sai K, Mu YG, Jiang XB, Zhang XH, et al. Nimotuzumab in combination with chemotherapy for patients with malignant gliomas. Chung-Hua Chung Liu Tsa Chih [Chinese Journal of Oncology] 2011;33(3):232-5. [PubMed] [Google Scholar]
References to ongoing studies
Rapp 2018 {published data only}
- Rapp M, Grauer OM, Kamp M, Sevens N, Zotz N, Sabel M, et al. A randomized controlled phase II trial of vaccination with lysate-loaded, mature dendritic cells integrated into standard radiochemotherapy of newly diagnosed glioblastoma (GlioVax): study protocol for a randomized controlled trial. Trials 2018;19(1):293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Additional references
Abhinav 2013
- Abhinav K, Aquilina K, Gbejuade H, La M, Hopkins K, Iyer V. A pilot study of glioblastoma multiforme in elderly patients: treatments, O-6-methylguanine-DNA methyltransferase (MGMT) methylation status and survival. Clinical Neurology and Neurosurgery 2013;115(8):1375-8. [DOI] [PubMed] [Google Scholar]
Adeberg 2015
- Adeberg S, Bostel T, Harrabi S, Bernhardt D, Welzel T, Wick W, et al. Impact of delays in initiating postoperative chemoradiation while determining the MGMT promoter-methylation statuses of patients with primary glioblastoma. BMC Cancer 2015;15:558. [DOI] [PMC free article] [PubMed] [Google Scholar]
Ahmed 2015
- Ahmed KA, Chinnaiyan P, Fulp WJ, Eschrich S, Torres-Roca JF, Caudell JJ. The radiosensitivity index predicts for overall survival in glioblastoma. Oncotarget 2015;6(33):34414-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Alnahhas 2020
- Alnahhas I, Alsawas M, Rayi A, Palmer JD, Raval R, Ong S, et al. Characterizing benefit from temozolomide in MGMT promoter unmethylated and methylated glioblastoma: a systematic review and meta-analysis. Neuro-oncology Advances 2020;2(1):vdaa082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Alonso 2017
- Alonso D, Matallanas M, Riveros-Perez A, Perez-Payo M, Blanco S. Prognostic and predictive factors in high-grade gliomas. Experience at our institution [Factores pronosticos y predictivos en gliomas de alto grado. Experiencia en nuestro centro]. Neurocirugia (Asturias, Spain) 2017;28(6):276-83. [DOI] [PubMed] [Google Scholar]
Altman 2001
- Altman DG. Systematic reviews of evaluations of prognostic variables. BMJ 2001;323(7306):224-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Aluko 2020
- Aluko P, Graybill E, Craig D, Henderson C, Drummond M, Wilson ECF, et al, on behalf of the Campbell and Cochrane Economics Methods Group. Chapter 20: Economic evidence. In: Higgins JPT, Thomas J, Chandler J, Cumpston M, Li T, Page MJ, et al, editor(s). Cochrane Handbook for Systematic Reviews of Interventions Version 6.1 (updated September 2020). Cochrane, 2020. Available from training.cochrane.org/handbook.
Appin 2013
- Appin CL, Gao J, Chisolm C, Torian M, Alexis D, Vincentelli C, et al. Glioblastoma with oligodendroglioma component (GBM-O): molecular genetic and clinical characteristics. Brain Pathology 2013;23(4):454-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Ardon 2012
- Ardon H, Van Gool SW, Verschuere T, Maes W, Fieuws S, Sciot R, et al. Integration of autologous dendritic cell-based immunotherapy in the standard of care treatment for patients with newly diagnosed glioblastoma: results of the HGG-2006 phase I/II trial. Cancer Immunology, Immunotherapy 2012;61(11):2033-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
Arita 2016
- Arita H, Yamasaki K, Matsushita Y, Nakamura T, Shimokawa A, Takami H, et al. A combination of TERT promoter mutation and MGMT methylation status predicts clinically relevant subgroups of newly diagnosed glioblastomas. Acta Neuropathologica Communications 2016;4(1):79. [DOI] [PMC free article] [PubMed] [Google Scholar]
Badruddoja 2017
- Badruddoja MA, Pazzi M, Sanan A, Schroeder K, Kuzma K, Norton T, et al. Phase II study of bi-weekly temozolomide plus bevacizumab for adult patients with recurrent glioblastoma. Cancer Chemotherapy & Pharmacology 2017;80(4):715-21. [DOI] [PubMed] [Google Scholar]
Balana 2016
- Balana C, De Las Penas, R, Sepulveda JM, Gil-Gil MJ, Luque R, et al. Bevacizumab and temozolomide versus temozolomide alone as neoadjuvant treatment in unresected glioblastoma: the GENOM 009 randomized phase II trial. Journal of Neuro-Oncology 2016;127(3):569-79. [DOI] [PubMed] [Google Scholar]
Balana 2017
- Balana C, Capellades J, Pineda E, Estival A, Puig J, Domenech S, et al. Pseudoprogression as an adverse event of glioblastoma therapy. Cancer Medicine 2017;6(12):2858-66. [DOI] [PMC free article] [PubMed]
Bienkowski 2015
- Bienkowski M, Berghoff AS, Marosi C, Wöhrer A, Heinzl H, Hainfellner JA, et al. Clinical Neuropathology practice guide 5-2015: MGMT methylation pyrosequencing in glioblastoma: unresolved issues and open questions. Clinical Neuropathology 2015;34(5):250-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Blumenthal 2017
- Blumenthal DT, Won M, Mehta MP, Gilbert MR, Brown PD, Bokstein F, et al. Short delay in initiation of radiotherapy for patients with glioblastoma-effect of concurrent chemotherapy: a secondary analysis from the NRG Oncology/Radiation Therapy Oncology Group database. Neuro-oncology 2018;20(7):966-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Boots‐Sprenger 2013
- Boots-Sprenger SH, Sijben A, Rijntjes J, Tops BB, Idema AJ, Rivera AL, et al. Significance of complete 1p/19q co-deletion, IDH1 mutation and MGMT promoter methylation in gliomas: use with caution. Modern Pathology 2013;26(7):922-9. [DOI] [PubMed] [Google Scholar]
Brandes 2008
- Brandes AA, Franceschi E, Tosoni A, Blatt V, Pession A, Tallini G, et al. MGMT promoter methylation status can predict the incidence and outcome of pseudoprogression after concomitant radiochemotherapy in newly diagnosed glioblastoma patients. Journal of Clinical Oncology 2008;26(13):2192-7. [DOI] [PubMed]
Brandes 2009
Brandes 2010
- Brandes AA, Franceschi E, Tosoni A, Bartolini S, Bacci A, Agati R, et al. O(6)-methylguanine DNA-methyltransferase methylation status can change between first surgery for newly diagnosed glioblastoma and second surgery for recurrence: clinical implications. Neuro-oncology 2010;12(3):283-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Brandes 2014
- Brandes AA, Franceschi E, Ermani M, Tosoni A, Albani F, Depenni R, et al. Pattern of care and effectiveness of treatment for glioblastoma patients in the real world: results from a prospective population-based registry. Could survival differ in a high-volume center? Neuro-oncology Practice 2014;1(4):166-71. [DOI] [PMC free article] [PubMed]
Brandes 2017
- Brandes AA, Franceschi E, Paccapelo A, Tallini G, De B, Ghimenton C, et al. Role of MGMT methylation status at time of diagnosis and recurrence for patients with glioblastoma: clinical implications. Oncologist 2017;22(4):432-7. [DOI] [PMC free article] [PubMed]
Brandner 2015
- Brandner S, Deimling A. Diagnostic, prognostic and predictive relevance of molecular markers in gliomas. Neuropathology and Applied Neurobiology 2015;41(6):694-720. [DOI] [PubMed] [Google Scholar]
Brennan 2013
- Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The somatic genomic landscape of glioblastoma. [Erratum appears in Cell 2014;157(3):753]. Cell 2013;155(2):462-77. [DOI] [PMC free article] [PubMed]
Burford 2013
- Burford A, Little SE, Jury A, Popov S, Laxton R, Doey L, et al. Distinct phenotypic differences associated with differential amplification of receptor tyrosine kinase genes at 4q12 in glioblastoma. PloS One 2013;8(8):e71777. [DOI] [PMC free article] [PubMed]
Burger 2017
- Burger MC, Breuer S, Cieplik HC, Harter PN, Franz K, Bahr O, et al. Bevacizumab for patients with recurrent multifocal glioblastomas. International Journal of Molecular Sciences 2017;18(11):2469. [DOI] [PMC free article] [PubMed]
Burnet 2005
- Burnet NG, Jefferies SJ, Benson RJ, Hunt DP, Treasure FP. Years of life lost (YLL) from cancer is an important measure of population burden and should be considered when allocating research funds. British Journal of Cancer 2005;92(2):241-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Butler 2020
- Butler M, Pongor L, Su YT, Xi L, Raffeld M, Quezado M, et al. MGMT status as a clinical biomarker in glioblastoma. Trends in Cancer 2020;6(5):380-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Butowski 2011
- Butowski N, Chang SM, Lamborn KR, Polley MY, Pieper R, Costello JF, et al. Phase II and pharmacogenomics study of enzastaurin plus temozolomide during and following radiation therapy in patients with newly diagnosed glioblastoma multiforme and gliosarcoma. Neuro-oncology 2011;13(12):1331-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Capellades 2018
- Capellades J, Puig J, Domenech S, Pujol T, Oleaga L, Camins A, et al. Is a pretreatment radiological staging system feasible for suggesting the optimal extent of resection and predicting prognosis in glioblastoma? An observational study. Journal of Neuro-oncology 2018;137(2):367-77. [DOI] [PubMed] [Google Scholar]
Chakhoyan 2018
- Chakhoyan A, Woodworth DC, Harris RJ, Lai A, Nghiemphu PL, Liau LM, et al. Mono-exponential, diffusion kurtosis and stretched exponential diffusion MR imaging response to chemoradiation in newly diagnosed glioblastoma. Journal of Neuro-oncology 2018;139(3):651-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Chen 2015
- Chen L, Lin ZX, Lin GS, Zhou CF, Chen YP, Wang XF, et al. Classification of microvascular patterns via cluster analysis reveals their prognostic significance in glioblastoma. Human Pathology 2015;46(1):120-8. [DOI] [PubMed] [Google Scholar]
Chen 2016
- Chen W, Yu Q, Chen B, Lu X, Li Q. The prognostic value of a seven-microRNA classifier as a novel biomarker for the prediction and detection of recurrence in glioma patients. Oncotarget 2016;7(33):53392-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
Cheng 2015
- Cheng W, Li M, Cai J, Wang K, Zhang C, Bao Z, et al. HDAC4, a prognostic and chromosomal instability marker, refines the predictive value of MGMT promoter methylation. Journal of Neuro-oncology 2015;122(2):303-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Chinot 2007
- Chinot OL, Barrie M, Fuentes S, Eudes N, Lancelot S, Metellus P, et al. Correlation between O6-methylguanine-DNA methyltransferase and survival in inoperable newly diagnosed glioblastoma patients treated with neoadjuvant temozolomide. Journal of Clinical Oncology 2007;25(12):1470-5. [DOI] [PubMed] [Google Scholar]
Choi 2016
- Choi YS, Ahn SS, Kim DW, Chang JH, Kang SG, Kim EH, et al. Incremental prognostic value of ADC histogram analysis over MGMT promoter methylation status in patients with glioblastoma. Radiology 2016;281(1):175-84. [DOI] [PubMed] [Google Scholar]
Clarke 2009
- Clarke JL, Iwamoto FM, Sul J, Panageas K, Lassman AB, DeAngelis LM, et al. Randomized phase II trial of chemoradiotherapy followed by either dose-dense or metronomic temozolomide for newly diagnosed glioblastoma. Journal of Clinical Oncology 2009;27(23):3861-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Coburger 2017
- Coburger J, Wirtz CR, Konig RW. Impact of extent of resection and recurrent surgery on clinical outcome and overall survival in a consecutive series of 170 patients for glioblastoma in intraoperative high field magnetic resonance imaging. Journal of Neurosurgical Sciences 2017;61(3):233-44. [DOI] [PubMed] [Google Scholar]
Colman 2010
- Colman H, Zhang L, Sulman EP, McDonald JM, Shooshtari NL, Rivera A, et al. A multigene predictor of outcome in glioblastoma. Neuro-oncology 2010;12(1):49-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
Combs 2011
- Combs SE, Rieken S, Wick W, Abdollahi A, von Deimling, Debus J, et al. Prognostic significance of IDH-1 and MGMT in patients with glioblastoma: one step forward, and one step back? Radiation Oncology 2011;6:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Cominelli 2015
- Cominelli M, Grisanti S, Mazzoleni S, Branca C, Buttolo L, Furlan D, et al. EGFR amplified and overexpressing glioblastomas and association with better response to adjuvant metronomic temozolomide. Journal of the National Cancer Institute 2015;107(5):djv041. [DOI] [PubMed] [Google Scholar]
Costa 2010
- Costa BM, Caeiro C, Guimaraes I, Martinho O, Jaraquemada T, Augusto I, et al. Prognostic value of MGMT promoter methylation in glioblastoma patients treated with temozolomide-based chemoradiation: a Portuguese multicentre study. Oncology Reports 2010;23(6):1655-62. [DOI] [PubMed] [Google Scholar]
Criniere 2007
- Criniere E, Kaloshi G, Laigle-Donadey F, Lejeune J, Auger N, Benouaich-Amiel A, et al. MGMT prognostic impact on glioblastoma is dependent on therapeutic modalities. Journal of Neuro-oncology 2007;83(2):173-9. [DOI] [PubMed] [Google Scholar]
Dahlrot 2017
- Dahlrot RH, Larsen PV, Boldt H, Kreutzfeldt MS, Hjelmborg JV, Hansen S, et al. Time-varying effect of MGMT methylation level on survival of glioblastoma multiforme. Neuro-oncology 2017;19(Suppl 6):vi182. [Google Scholar]
Das 2011
- Das P, Puri T, Jha P, Pathak P, Joshi N, Suri V, et al. A clinicopathological and molecular analysis of glioblastoma multiforme with long-term survival. Journal of Clinical Neuroscience 2011;18(1):66-70. [DOI] [PubMed] [Google Scholar]
Debray 2018
- Debray TP, Moons KG, Riley RD. Detecting small-study effects and funnel plot asymmetry in meta-analysis of survival data: a comparison of new and existing tests. Research Synthesis Methods 2018;9(1):41-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Dullea 2016
- Dullea A, Marignol L. MGMT testing allows for personalised therapy in the temozolomide era. Tumor Biology 2016;37(1):87-96. [DOI] [PubMed] [Google Scholar]
Engauge Digitizer [Computer program]
- Engauge Digitizer Software. Mitchell M, Muftakhidinov B, Winchen T, Schaik B, Wilms A, Jędrzejewski-Szmek Z, et al, Version 12.2.1. Geneva: Zenodo, 2020. Available at markummitchell.github.io/engauge-digitizer. [DOI: 10.5281/zenodo/3941227] [DOI]
EPPI‐Reviewer [Computer program]
- EPPI-Reviewer 4: software for research synthesis. EPPI-Centre Software. Thomas J, Brunton J, Graziosi S. London: Social Science Research Unit, UCL Institute of Education, 2010.
Esteller 1999
- Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Research 1999;59(4):793-7. [PubMed] [Google Scholar]
Esteller 2000
- Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. New England Journal of Medicine 2000;343(19):1350-4. [DOI] [PubMed] [Google Scholar]
Etcheverry 2014
- Etcheverry A, Aubry M, Idbaih A, Vauleon E, Marie Y, Menei P, et al. DGKI methylation status modulates the prognostic value of MGMT in glioblastoma patients treated with combined radio-chemotherapy with temozolomide. PloS One 2014;9(9):e104455. [DOI] [PMC free article] [PubMed] [Google Scholar]
Farrell 2020
- Farrell C, Shi W, Bodman A, Olson JJ. Congress of neurological surgeons systematic review and evidence-based guidelines update on the role of emerging developments in the management of newly diagnosed glioblastoma. Journal of Neuro-oncology 2020;150:269-359. [DOI] [PubMed] [Google Scholar]
Felsberg 2011
- Felsberg J, Thon N, Eigenbrod S, Hentschel B, Sabel MC, Westphal M, et al. Promoter methylation and expression of MGMT and the DNA mismatch repair genes MLH1, MSH2, MSH6 and PMS2 in paired primary and recurrent glioblastomas. International Journal of Cancer 2011;129(3):659-70. [DOI] [PubMed] [Google Scholar]
Fiano 2014
- Fiano V, Trevisan M, Trevisan E, Senetta R, Castiglione A, Sacerdote C, et al. MGMT promoter methylation in plasma of glioma patients receiving temozolomide. Journal of Neuro-oncology 2014;117(2):347-57. [DOI] [PubMed] [Google Scholar]
Fontana 2016
- Fontana L, Tabano S, Bonaparte E, Marfia G, Pesenti C, Falcone R, et al. MGMT-methylated alleles are distributed heterogeneously within glioma samples irrespective of IDH status and chromosome 10q deletion. Journal of Neuropathology and Experimental Neurology 2016;75(8):791-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Franceschi 2016
- Franceschi E, Depenni R, Paccapelo A, Ermani M, Faedi M, Sturiale C, et al. Which elderly newly diagnosed glioblastoma patients can benefit from radiotherapy and temozolomide? A PERNO prospective study. Journal of Neuro-oncology 2016;128(1):157-62. [DOI] [PubMed] [Google Scholar]
Franceschi 2018
- Franceschi E, Tosoni A, Minichillo S, Depenni R, Paccapelo A, Bartolini S, et al. The prognostic roles of gender and O6-Methylguanine-DNA methyltransferase methylation status in glioblastoma patients: the female power. World Neurosurgery 2018;112:e342-7. [DOI] [PubMed] [Google Scholar]
Galldiks 2015
- Galldiks N, Dunkl V, Stoffels G, Hutterer M, Rapp M, Sabel M, et al. Diagnosis of pseudoprogression in patients with glioblastoma using O-(2-[18F]fluoroethyl)-L-tyrosine PET. European Journal of Nuclear Medicine and Molecular Imaging 2015;42(5):685-95. [DOI] [PubMed] [Google Scholar]
Gallego Perez‐Larraya 2011
- Gallego Perez-Larraya J, Ducray F, Chinot O, Catry-Thomas I, Taillandier L, Guillamo JS, et al. Temozolomide in elderly patients with newly diagnosed glioblastoma and poor performance status: an ANOCEF phase II trial. Journal of Clinical Oncology 2011;29(22):3050-5. [DOI] [PubMed] [Google Scholar]
Gilbert 2013
- Gilbert MR, Wang M, Aldape KD, Stupp R, Hegi ME, Jaeckle KA, et al. Dose-dense temozolomide for newly diagnosed glioblastoma: a randomized phase III clinical trial. Journal of Clinical Oncology 2013;31(32):4085-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Gilbert 2014
- Gilbert MR, Dignam JJ, Armstrong TS, Wefel JS, Blumenthal DT, Vogelbaum MA, et al. A randomized trial of bevacizumab for newly diagnosed glioblastoma. New England Journal of Medicine 2014;370(8):699-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
Gittleman 2017
- Gittleman H, Lim D, Kattan MW, Chakravarti A, Gilbert MR, Lassman AB, et al. An independently validated nomogram for individualized estimation of survival among patients with newly diagnosed glioblastoma: NRG Oncology RTOG 0525 and 0825. Neuro-oncology 2017;19(5):669-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
Glas 2009
- Glas M, Happold C, Rieger J, Wiewrodt D, Bahr O, Steinbach JP, et al. Long-term survival of patients with glioblastoma treated with radiotherapy and lomustine plus temozolomide. Journal of Clinical Oncology 2009;27(8):1257-61. [DOI] [PubMed] [Google Scholar]
Gorlia 2008
- Gorlia T, den Bent MJ, Hegi ME, Mirimanoff RO, Weller M, Cairncross JG, et al. Nomograms for predicting survival of patients with newly diagnosed glioblastoma: prognostic factor analysis of EORTC and NCIC trial 26981-22981/CE.3. Lancet Oncology 2008;9(1):29-38. [DOI] [PubMed] [Google Scholar]
GOSH 2018
- Great Ormand Street Hospital for Children NHS Foundation Trust. North East Thames Regional Genetics Service. Pricing 2018/9. www.labs.gosh.nhs.uk/media/1390838/nhs_joint_price_list_01.01.2019.pdf (accessed 30 November 2020).
Gramatzki 2016
- Gramatzki D, Dehler S, Rushing EJ, Zaugg K, Hofer S, Yonekawa Y, et al. Glioblastoma in the Canton of Zurich, Switzerland revisited: 2005 to 2009. Cancer 2016;122(14):2206-15. [DOI] [PubMed] [Google Scholar]
Gutenberg 2013a
- Gutenberg A, Bock HC, Bruck W, Doerner L, Mehdorn HM, Roggendorf W, et al. MGMT promoter methylation status and prognosis of patients with primary or recurrent glioblastoma treated with carmustine wafers. British Journal of Neurosurgery 2013;27(6):772-8. [DOI] [PubMed] [Google Scholar]
Gutenberg 2013b
- Gutenberg A, Bock HC, Reifenberger G, Bruck W, Giese A. Toxicity and survival in primary glioblastoma patients treated with concomitant plus adjuvant temozolomide versus adjuvant temozolomide: results of a single-institution, retrospective, matched-pair analysis. Acta Neurochirurgica 2013;155(3):429-35. [DOI] [PubMed] [Google Scholar]
Guyatt 2008
- Guyatt GH, Oxman AD, Vist GE, Kunz R, Falck-Ytter Y, Alonso-Coello P, et al. GRADE: an emerging consensus on rating quality of evidence and strength of recommendations. BMJ 2008;336(7650):924-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Guyot 2012
- Guyot P, Ades AE, Ouwens MJ, Welton NJ. Enhanced secondary analysis of survival data: reconstructing the data from published Kaplan-Meier survival curves. BMC Medical Research Methodology 2012;12:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Ha 2013
- Ha SY, Kang SY, Do IG, Suh YL. Glioblastoma with oligodendroglial component represents a subgroup of glioblastoma with high prevalence of IDH1 mutation and association with younger age. Journal of Neuro-oncology 2013;112(3):439-48. [DOI] [PubMed] [Google Scholar]
Haemmig 2014
- Haemmig S, Baumgartner U, Gluck A, Zbinden S, Tschan MP, Kappeler A, et al. miR-125b controls apoptosis and temozolomide resistance by targeting TNFAIP3 and NKIRAS2 in glioblastomas. Cell Death & Disease 2014;5:e1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
Han 2014
- Han SJ, Rolston JD, Molinaro AM, Clarke JL, Prados MD, Chang SM, et al. Phase II trial of 7 days on/7 days off temozolomide for recurrent high-grade glioma. Neuro-oncology 2014;16(9):1255-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
Han 2015a
- Han S, Liu Y, Li Q, Li Z, Hou H, Wu A. Pre-treatment neutrophil-to-lymphocyte ratio is associated with neutrophil and T-cell infiltration and predicts clinical outcome in patients with glioblastoma. BMC Cancer 2015;15:617. [DOI] [PMC free article] [PubMed] [Google Scholar]
Han 2015b
- Han S, Huang Y, Li Z, Hou H, Wu A. The prognostic role of preoperative serum albumin levels in glioblastoma patients. BMC Cancer 2015;15(1):108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Happold 2018
- Happold C, Gorlia T, Nabors LB, Erridge SC, Reardon DA, Hicking C, et al. Do statins, ACE inhibitors or sartans improve outcome in primary glioblastoma? Journal of Neuro-oncology 2018;138(1):163-71. [DOI] [PubMed] [Google Scholar]
Hayden 2013
- Hayden JA, Windt DA, Cartwright JL, Côté P, Bombardier C. Assessing bias in studies of prognostic factors. Annals of Internal Medicine 2013;158(4):280-6. [DOI] [PubMed] [Google Scholar]
Hayes 2015
- Hayes J, Thygesen H, Tumilson C, Droop A, Boissinot M, Hughes TA, et al. Prediction of clinical outcome in glioblastoma using a biologically relevant nine-microRNA signature. Molecular Oncology 2015;9(3):704-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Hegi 2004
- Hegi ME, Diserens AC, Godard S, Dietrich PY, Regli L, Ostermann S, et al. Clinical trial substantiates the predictive value of O-6-methylguanine-DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clinical Cancer Research 2004;10(6):1871-4. [DOI] [PubMed] [Google Scholar]
Hegi 2005
- Hegi ME, Diserens AC, Gorlia T, Hamou MF, De Tribolet N, Weller M, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. New England Journal of Medicine 2005;352(10):997-1003. [DOI] [PubMed] [Google Scholar]
Hegi 2015
- Hegi ME, Stupp R. Withholding temozolomide in glioblastoma patients with unmethylated MGMT promoter-still a dilemma? Neuro-oncology 2015;17(11):1425-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Hegi 2019
- Hegi ME, Genbrugge E, Gorlia T, Stupp R, Gilbert MR, Chinot OL, et al. Promoter methylation cutoff with safety margin for selecting glioblastoma patients into trials omitting temozolomide: a pooled analysis of four clinical trials. Clinical Cancer Research 2019;25(6):1809-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Herrlinger 2006
- Herrlinger U, Rieger J, Koch D, Loeser S, Blaschke B, Kortmann RD, et al. Phase II trial of lomustine plus temozolomide chemotherapy in addition to radiotherapy in newly diagnosed glioblastoma: UKT-03. Journal of Clinical Oncology 2006;24(27):4412-7. [DOI] [PubMed] [Google Scholar]
Herrlinger 2009
- Herrlinger U, Glas M, Happold C, Rieger J, Wiewrodt D, Bixhr O, et al. Long-term survival of patients with glioblastoma treated with radiotherapy and lomustine plus temozolomide. Journal of Clinical Oncology 2009;27(8):1257-61. [DOI] [PubMed] [Google Scholar]
Hervouet 2009
- Hervouet E, Debien E, Campion L, Charbord J, Menanteau J, Vallette FM, et al. Folate supplementation limits the aggressiveness of glioma via the remethylation of DNA repeats element and genes governing apoptosis and proliferation. Clinical Cancer Research 2009;15(10):3519-29. [DOI] [PubMed] [Google Scholar]
Higgins 2002
- Higgins JPT, Thompson SG. Quantifying heterogeneity in a meta-analysis. Statistics in Medicine 2002;21(11):1539-58. [DOI] [PubMed] [Google Scholar]
Hobbs 2012
- Hobbs J, Nikiforova MN, Fardo DW, Bortoluzzi S, Cieply K, Hamilton RL, et al. Paradoxical relationship between the degree of EGFR amplification and outcome in glioblastomas. American Journal of Surgical Pathology 2012;36(8):1186-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
Huang 2017
- Huang SP, Chang YC, Low QH, Wu AT, Chen CL, Lin YF, et al. BICD1 expression, as a potential biomarker for prognosis and predicting response to therapy in patients with glioblastomas. Oncotarget 2017;8(69):113766-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Hudson 2018
- Hudson AL, Parker NR, Khong P, Parkinson JF, Dwight T, Ikin RJ, et al. Glioblastoma recurrence correlates with increased APE1 and polarization toward an immuno-suppressive microenvironment. Frontiers in Oncology 2018;8:314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Huguet 2013
- Huguet A, Hayden JA, Stinson J, McGrath PJ, Chambers CT, Tougas ME, et al. Judging the quality of evidence in reviews of prognostic factor research: adapting the GRADE framework. Systematic Reviews 2013;2:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Husereau 2013
- Husereau D, Drummond M, Petrou S, Carswell C, Moher D, Greenberg D, et al. Consolidated Health Economic Evaluation Reporting Standards (CHEERS) statement. BMJ 2013;346:f1049. [DOI] [PubMed] [Google Scholar]
Inoges 2017
- Inoges S, Tejada S, Cerio AL, Gallego Perez-Larraya J, Espinos J, Idoate MA, et al. A phase II trial of autologous dendritic cell vaccination and radiochemotherapy following fluorescence-guided surgery in newly diagnosed glioblastoma patients. Journal of Translational Medicine 2017;15(1):104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Ishida 2015
- Ishida J, Kurozumi K, Ichikawa T, Otani Y, Onishi M, Fujii K, et al. Evaluation of extracellular matrix protein CCN1 as a prognostic factor for glioblastoma. Brain Tumor Pathology 2015;32(4):245-52. [DOI] [PubMed] [Google Scholar]
Ishikawa 2014
- Ishikawa E, Muragaki Y, Yamamoto T, Maruyama T, Tsuboi K, Ikuta S, et al. Phase I/IIa trial of fractionated radiotherapy, temozolomide, and autologous formalin-fixed tumor vaccine for newly diagnosed glioblastoma. Journal of Neurosurgery 2014;121(3):543-53. [DOI] [PubMed] [Google Scholar]
Ius 2018
- Ius T, Cesselli D, Isola M, Toniato G, Pauletto G, Sciacca G, et al. Combining clinical and molecular data to predict the benefits of carmustine wafers in newly diagnosed high-grade gliomas. Current Treatment Options in Neurology 2018;20(2):3. [DOI] [PubMed] [Google Scholar]
Iwadate 2017
- Iwadate Y, Suganami A, Tamura Y, Matsutani T, Hirono S, Shinozaki N, et al. The pluripotent stem-cell marker alkaline phosphatase is highly expressed in refractory glioblastoma with DNA hypomethylation. Neurosurgery 2017;80(2):248-56. [DOI] [PubMed] [Google Scholar]
Jan 2018
- Jan CI, Tsai WC, Harn HJ, Shyu WC, Liu MC, Lu HM, et al. Predictors of response to autologous dendritic cell therapy in glioblastoma multiforme. Frontiers in Immunology 2018;9:727. [DOI] [PMC free article] [PubMed] [Google Scholar]
JLA PSP 2018
- James Lind Alliance Priority Setting Partnerships. Neuro-oncology top 10. www.jla.nihr.ac.uk/priority-setting-partnerships/neuro-oncology/top-10-priorities/ (accessed 29 May 2018).
Karim 2012
- Karim KA, El Mahdy MM, Wahab MM, Ei Arab LR, El Shehaby A, Raouf SA. Temozolomide and radiotherapy in newly diagnosed glioblastoma patients: O6-methylguanine-DNA methyltransferase (MGMT) promotor methylation status and Ki-67 as biomarkers for survival and response to treatment. Chinese-German Journal of Clinical Oncology 2012;11(3):168-76. [Google Scholar]
Kessler 2018
- Kessler T, Sahm F, Sadik A, Stichel D, Hertenstein A, Reifenberger G, et al. Molecular differences in IDH wildtype glioblastoma according to MGMT promoter methylation. Neuro-oncology 2018;20(3):367-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
Kim 2012
- Kim YS, Kim SH, Cho J, Kim JW, Chang JH, Kim DS, et al. MGMT gene promoter methylation as a potent prognostic factor in glioblastoma treated with temozolomide-based chemoradiotherapy: a single-institution study. International Journal of Radiation Oncology, Biology, Physics 2012;84(3):661-7. [DOI] [PubMed] [Google Scholar]
Kim 2017
- Kim BS, Nam DH, Kim IH, Yoon SM, Kang SG, Suh CO, et al. Concurrent chemoradiotherapy with temozolomide followed by adjuvant temozolomide for newly diagnosed glioblastoma patients: a retrospective multicenter observation study in Korea. Cancer Research and Treatment 2017;49(1):193-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
Kim 2018
- Kim BS, Kim ST, Kim JH, Seol HJ, Nam DH, Shin HJ, et al. Apparent diffusion coefficient as a predictive biomarker for survival in patients with treatment-naive glioblastoma using quantitative multiparametric magnetic resonance profiling. World Neurosurgery 2018;122:e812-20. [DOI] [PubMed] [Google Scholar]
Klitkou 2014a
- Klitkou J, Dahlrot RH, Hansen S, Kristensen BW. The biomarker potential of MGMT protein in glioblastoma is improved by exclusion of non-tumor cells. Clinical Neuropathology 2014;33(3):205. [ePPI-R 38858612 ] [Google Scholar]
Klitkou 2014b
- Klitkou J, Dahlrot RH, Hansen S, Kristensen BW. The biomarker potential of MGMT protein in glioblastoma is improved by exclusion of non-tumor cells. Brain Pathology 2014;1:99. [ePPI-R 38858611] [Google Scholar]
Klitkou 2014c
- Klitkou J, Dahlrot RH, Hansen S, Kristensen BW. The biomarker potential of MGMT protein expression in glioblastoma is improved by exclusion of non-tumor cells. Neuro-oncology 2014;2:ii47. [ePPI-R 38858610] [Google Scholar]
Kong 2011
- Kong DS, Kim ST, Kim EH, Lim DH, Kim WS, Suh YL, et al. Diagnostic dilemma of pseudoprogression in the treatment of newly diagnosed glioblastomas: the role of assessing relative cerebral blood flow volume and oxygen-6-methylguanine-DNA methyltransferase promoter methylation status. American Journal of Neuroradiology 2011;32(2):382-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Kreth 2013
- Kreth FW, Thon N, Simon M, Westphal M, Schackert G, Nikkhah G, et al. Gross total but not incomplete resection of glioblastoma prolongs survival in the era of radiochemotherapy. Annals of Oncology 2013;24(12):3117-23. [DOI] [PubMed] [Google Scholar]
Lakomy 2011
- Lakomy R, Sana J, Hankeova S, Fadrus P, Kren L, Lzicarova E, et al. MiR-195, miR-196b, miR-181c, miR-21 expression levels and O-6-methylguanine-DNA methyltransferase methylation status are associated with clinical outcome in glioblastoma patients. Cancer Science 2011;102(12):2186-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
Laxton 2013
- Laxton RC, Popov S, Doey L, Jury A, Bhangoo R, Gullan R, et al. Primary glioblastoma with oligodendroglial differentiation has better clinical outcome but no difference in common biological markers compared with other types of glioblastoma. Neuro-oncology 2013;15(12):1635-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Lee 2013
- Lee D, Suh YL, Park TI, Do IG, Seol HJ, Nam DH, et al. Prognostic significance of tetraspanin CD151 in newly diagnosed glioblastomas. Journal of Surgical Oncology 2013;107(6):646-52. [DOI] [PubMed] [Google Scholar]
Lee 2017
- Lee Y, Koh J, Kim SI, Won JK, Park CK, Choi SH, et al. The frequency and prognostic effect of TERT promoter mutation in diffuse gliomas. Acta Neuropathologica Communications 2017;5(1):62. [DOI] [PMC free article] [PubMed] [Google Scholar]
Li 2016a
- Li H, Li J, Cheng G, Zhang J, Li X. IDH mutation and MGMT promoter methylation are associated with the pseudoprogression and improved prognosis of glioblastoma multiforme patients who have undergone concurrent and adjuvant temozolomide-based chemoradiotherapy. Clinical Neurology and Neurosurgery 2016;151:31-6. [DOI] [PubMed] [Google Scholar]
Li 2016b
- Li Q, Chen B, Cai J, Sun Y, Wang G, Li Y, et al. Comparative analysis of matrix metalloproteinase family members reveals that MMP9 predicts survival and response to temozolomide in patients with primary glioblastoma. PloS One 2016;11(3):e0151815. [DOI] [PMC free article] [PubMed] [Google Scholar]
Lombardi 2015
- Lombardi G, Pace A, Pasqualetti F, Rizzato S, Faedi M, Anghileri E, et al. Predictors of survival and effect of short (40 Gy) or standard-course (60 Gy) irradiation plus concomitant temozolomide in elderly patients with glioblastoma: a multicenter retrospective study of AINO (Italian Association of Neuro-Oncology). Journal of Neuro-oncology 2015;125(2):359-67. [DOI] [PubMed] [Google Scholar]
Lombardi 2017
- Lombardi G, Bellu L, Bertorelle R, Pambuku A, Gardiman M, Fiduccia P, et al. MGMT promoter methylation status in glioblastoma (GBM) patients: a quantitative pyrosequencing approach and its prognostic role. Neuro-oncology 2017;19(Suppl 3):iii76. [Google Scholar]
Louis 2016
- Louis DN, Perry A, Reifenberger G, Deimling A, Figarella-Branger D, Cavenee W, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathologica 2016;131(6):803-20. [DOI] [PubMed] [Google Scholar]
Ma 2016
- Ma C, Zhou W, Yan Z, Qu M, Bu X. beta-Elemene treatment of glioblastoma: a single-center retrospective study. OncoTargets and therapy 2016;9:7521-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Majewska 2017
- Majewska P, Ioannidis S, Raza MH, Tanna N, Bulbeck H, Williams M. Postprogression survival in patients with glioblastoma treated with concurrent chemoradiotherapy: a routine care cohort study. CNS Oncology 2017;6(4):307-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Malmström 2012
- Malmström A, Grønberg BH, Marosi C, Stupp R, Frappaz D, Schultz H, et al. Temozolomide versus standard 6-week radiotherapy versus hypofractionated radiotherapy in patients older than 60 years with glioblastoma: the Nordic randomised, phase 3 trial. Lancet Oncology 2012;13(9):916-26. [DOI] [PubMed] [Google Scholar]
Malmström 2017
- Malmström A, Poulsen HS, Gronberg BH, Stragliotto G, Hansen S, Asklund T, et al. Postoperative neoadjuvant temozolomide before radiotherapy versus standard radiotherapy in patients 60 years or younger with anaplastic astrocytoma or glioblastoma: a randomized trial. Acta Oncologica 2017;56(12):1776-85. [DOI] [PubMed] [Google Scholar]
Martini 2008
- Martini M, Pallini R, Luongo G, Cenci T, Lucantoni C, Larocca LM. Prognostic relevance of SOCS3 hypermethylation in patients with glioblastoma multiforme. International Journal of Cancer 2008;123(12):2955-60. [DOI] [PubMed] [Google Scholar]
McDonald 2015
- McDonald KL, Tabone T, Nowak AK, Erber WN. Somatic mutations in glioblastoma are associated with methylguanine-DNA methyltransferase methylation. Oncology Letters 2015;9(5):2063-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Metellus 2011
- Metellus P, Nanni-Metellus I, Delfino C, Colin C, Tchogandjian A, Coulibaly B, et al. Prognostic impact of CD133 mRNA expression in 48 glioblastoma patients treated with concomitant radiochemotherapy: a prospective patient cohort at a single institution. Annals of Surgical Oncology 2011;18(10):2937-45. [DOI] [PubMed] [Google Scholar]
Meyronet 2017
- Meyronet D, Esteban-Mader M, Bonnet C, Joly MO, Uro-Coste E, Amiel-Benouaich A, et al. Characteristics of H3 K27M-mutant gliomas in adults. Neuro-oncology 2017;19(8):1127-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Michaelsen 2013
- Michaelsen SR, Christensen IJ, Grunnet K, Stockhausen MT, Broholm H, Kosteljanetz M, et al. Clinical variables serve as prognostic factors in a model for survival from glioblastoma multiforme: an observational study of a cohort of consecutive non-selected patients from a single institution. BMC Cancer 2013;13:402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Michaelsen 2018
- Michaelsen SR, Urup T, Olsen LR, Broholm H, Lassen U, Poulsen HS. Molecular profiling of short-term and long-term surviving patients identifies CD34 mRNA level as prognostic for glioblastoma survival. Journal of Neuro-oncology 2018;137(3):533-42. [DOI] [PubMed] [Google Scholar]
Minniti 2011a
- Minniti G, Armosini V, Salvati M, Lanzetta G, Caporello P, Mei M, et al. Fractionated stereotactic reirradiation and concurrent temozolomide in patients with recurrent glioblastoma. Journal of Neuro-oncology 2011;103(3):683-91. [ePPI-R 38859039] [DOI] [PubMed] [Google Scholar]
Minniti 2011b
- Minniti G, Salvati M, Arcella A, Buttarelli F, D'Elia A, Lanzetta G, et al. Correlation between O6-methylguanine-DNA methyltransferase and survival in elderly patients with glioblastoma treated with radiotherapy plus concomitant and adjuvant temozolomide. Journal of Neuro-oncology 2011;102(2):311-6. [ePPI-R 38859042] [DOI] [PubMed] [Google Scholar]
Minniti 2015
- Minniti G, Scaringi C, Lanzetta G, Terrenato I, Esposito V, Arcella A, et al. Standard (60 Gy) or short-course (40 Gy) irradiation plus concomitant and adjuvant temozolomide for elderly patients with glioblastoma: a propensity-matched analysis. International Journal of Radiation Oncology, Biology, Physics 2015;91(1):109-15. [DOI] [PubMed] [Google Scholar]
Miyazaki 2014
- Miyazaki M, Nishihara H, Terasaka S, Kobayashi H, Yamaguchi S, Ito T, et al. Immunohistochemical evaluation of O6-methylguanine DNA methyltransferase (MGMT) expression in 117 cases of glioblastoma. Neuropathology 2014;34(3):268-76. [DOI] [PubMed] [Google Scholar]
Montano 2011
- Montano N, Cenci T, Martini M, D'Alessandris QG, Pelacchi F, Ricci-Vitiani L, et al. Expression of EGFRvIII in glioblastoma: prognostic significance revisited. Neoplasia (New York, N.Y.) 2011;13(12):1113-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Moons 2014
- Moons KG, Groot JA, Bouwmeester W, Vergouwe Y, Mallett S, Altman DG, et al. Critical appraisal and data extraction for systematic reviews of prediction modelling studies: the CHARMS checklist. PLoS Medicine 2014;11(10):e1001744. [DOI] [PMC free article] [PubMed] [Google Scholar]
Morandi 2010
- Morandi L, Franceschi E, de Biase, Marucci G, Tosoni A, Ermani M, et al. Promoter methylation analysis of O6-methylguanine-DNA methyltransferase in glioblastoma: detection by locked nucleic acid based quantitative PCR using an imprinted gene (SNURF) as a reference. BMC Cancer 2010;10:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Motomura 2011
- Motomura K, Natsume A, Kishida Y, Higashi H, Kondo Y, Nakasu Y, et al. Benefits of interferon-beta and temozolomide combination therapy for newly diagnosed primary glioblastoma with the unmethylated MGMT promoter. Cancer 2011;117(8):1721-30. [DOI] [PubMed] [Google Scholar]
Mur 2015
- Mur P, Rodriguez de Lope A, Diaz-Crespo FJ, Hernandez-Iglesias T, Ribalta T, Fiano C, et al. Impact on prognosis of the regional distribution of MGMT methylation with respect to the CpG island methylator phenotype and age in glioma patients. Journal of Neuro-oncology 2015;122(3):441-50. [DOI] [PubMed] [Google Scholar]
Nabors 2012
- Nabors LB, Mikkelsen T, Hegi ME, Ye X, Batchelor T, Lesser G, et al. A safety run-in and randomized phase 2 study of cilengitide combined with chemoradiation for newly diagnosed glioblastoma (NABTT 0306). Cancer 2012;118(22):5601-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Nagane 2007
- Nagane M, Kobayashi K, Ohnishi A, Shimizu S, Shiokawa Y. Prognostic significance of O6-methylguanine-DNA methyltransferase protein expression in patients with recurrent glioblastoma treated with temozolomide. Japanese Journal of Clinical Oncology 2007;37(12):897-906. [DOI] [PubMed] [Google Scholar]
NICE 2012
- National Institute for Health and Care Excellence. Appendices B–I. The guidelines manual. Process and methods (PMG6), 2012. www.nice.org.uk/process/pmg6/resources/the-guidelines-manual-appendices-bi-2549703709/ (accessed 30 November 2020).
NICE 2018
- National Institute for Health and Care Excellence. Brain tumours (primary) and brain metastases in adults. NICE guideline (NG99), 2018. www.nice.org.uk/guidance/ng99 (accessed 11 July 2018).
Ohka 2011
- Ohka F, Natsume A, Motomura K, Kishida Y, Kondo Y, Abe T, et al. The global DNA methylation surrogate LINE-1 methylation is correlated with MGMT promoter methylation and is a better prognostic factor for glioma. PloS One 2011;6(8):e23332. [DOI] [PMC free article] [PubMed] [Google Scholar]
Ohno 2013
- Ohno M, Narita Y, Miyakita Y, Arita H, Matsushita Y, Yoshida A, et al. Clinical and molecular characteristics of newly diagnosed glioblastomas with IDH1 mutation and correlation of IDH1 mutations with prognosis. Neuro-oncology 2013;3:iii124. [Google Scholar]
Ohno 2016
- Ohno M, Narita Y, Miyakita Y, Matsushita Y, Arita H, Yonezawa M, et al. Glioblastomas with IDH1/2 mutations have a short clinical history and have a favorable clinical outcome. Japanese Journal of Clinical Oncology 2016;46(1):31-9. [DOI] [PubMed] [Google Scholar]
Olesen 2012
- Olesen J, Gustavsson A, Svensson M, Wittchen HU, Jönsson B, CDBE2010 study group, European Brain Council. The economic cost of brain disorders in Europe. European Journal of Neurology 2012;19(1):155-62. [DOI] [PubMed] [Google Scholar]
Omuro 2014
- Omuro A, Beal K, Gutin P, Karimi S, Correa DD, Kaley TJ, et al. Phase II study of bevacizumab, temozolomide, and hypofractionated stereotactic radiotherapy for newly diagnosed glioblastoma. Clinical Cancer Research 2014;20(19):5023-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Ostrom 2014
- Ostrom QT, Bauchet L, Davis FG, Deltour I, Fisher JL, Langer CE, et al. The epidemiology of glioma in adults: a "state of the science" review. Neuro-oncology 2014;16(7):896-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
Pallini 2008
- Pallini R, Ricci-Vitiani L, Banna GL, Signore M, Lombardi D, Todaro M, et al. Cancer stem cell analysis and clinical outcome in patients with glioblastoma multiforme. Clinical Cancer Research 2008;14(24):8205-12. [DOI] [PubMed] [Google Scholar]
Pambuku 2016
- Pambuku A, Lombardi G, Bertorelle R, Bellu L, Fiduccia P, Gardiman M, et al. MGMT promoter methylation status in glioblastoma (GBM) patients: a quantitative pyrosequencing approach and its prognostic role. Annals of Oncology 2016;27(6):vi107. [Google Scholar]
Park 2013
- Park CK, Lee SH, Kim TM, Choi SH, Park SH, Heo DS, et al. The value of temozolomide in combination with radiotherapy during standard treatment for newly diagnosed glioblastoma. Journal of Neuro-oncology 2013;112(2):277-83. [DOI] [PubMed] [Google Scholar]
Parmar 1998
- Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Statistics in Medicine 1998;17(24):2815-34. [DOI] [PubMed] [Google Scholar]
Pei 2013
- Pei C, Chen H, Jia X, Yan L, Zou Y, Jiang C, et al. A high frequency of MSH6 G268A polymorphism and survival association in glioblastoma. International Journal of Neuroscience 2013;123(2):114-20. [DOI] [PubMed] [Google Scholar]
Picart 2018
- Picart T, Barritault M, Berthillier J, Meyronet D, Vasiljevic A, Frappaz D, et al. Characteristics of cerebellar glioblastomas in adults. Journal of Neuro-oncology 2018;136(3):555-63. [DOI] [PubMed] [Google Scholar]
Poulsen 2017
- Poulsen SH, Urup T, Grunnet K, Christensen IJ, Larsen VA, Jensen ML, et al. The prognostic value of FET PET at radiotherapy planning in newly diagnosed glioblastoma. European Journal of Nuclear Medicine and Molecular Imaging 2017;44(3):373-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Prados 2009
- Prados MD, Chang SM, Butowski N, DeBoer R, Parvataneni R, Carliner H, et al. Phase II study of erlotinib plus temozolomide during and after radiation therapy in patients with newly diagnosed glioblastoma multiforme or gliosarcoma. Journal of Clinical Oncology 2009;27(4):579-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
Purkait 2016
- Purkait S, Mallick S, Sharma V, Kumar A, Pathak P, Jha P, et al. Prognostic stratification of GBMs using combinatorial assessment of IDH1 mutation, MGMT promoter methylation, and TERT mutation status: experience from a tertiary care center in India. Translational Oncology 2016;9(4):371-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Qi 2012
- Qi ST, Yu L, Gui S, Ding YQ, Han HX, Zhang XL, et al. IDH mutations predict longer survival and response to temozolomide in secondary glioblastoma. Cancer Science 2012;103(2):269-73. [DOI] [PubMed] [Google Scholar]
Rankeillor 2014
- Rankeillor KL, Cairns DA, Loughrey C, Short SC, Chumas P, Ismail A, et al. Methylation-specific multiplex ligation-dependent probe amplification identifies promoter methylation events associated with survival in glioblastoma. Journal of Neuro-oncology 2014;117(2):243-51. [DOI] [PubMed] [Google Scholar]
Rapkins 2015
- Rapkins RW, Wang F, Nguyen HN, Cloughesy TF, Lai A, Ha W, et al. The MGMT promoter SNP rs16906252 is a risk factor for MGMT methylation in glioblastoma and is predictive of response to temozolomide. Neuro-oncology 2015;17(12):1589-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
Rapp 2013
- Rapp M, Goeppert M, Felsberg J, Steiger HJ, Sabel M. The impact of sequential vs. combined radiochemotherapy with temozolomide, resection and MGMT promoter hypermethylation on survival of patients with primary glioblastoma – a single centre retrospective study. British Journal of Neurosurgery 2013;27(4):430-5. [DOI] [PubMed] [Google Scholar]
Reifenberger 2012
- Reifenberger G, Hentschel B, Felsberg J, Schackert G, Simon M, Schnell O, et al. Predictive impact of MGMT promoter methylation in glioblastoma of the elderly. International Journal of Cancer 2012;131(6):1342-50. [DOI] [PubMed] [Google Scholar]
Roh 2017
- Roh TH, Park HH, Kang SG, Moon JH, Kim EH, Hong CK, et al. Long-term outcomes of concomitant chemoradiotherapy with temozolomide for newly diagnosed glioblastoma patients: a single-center analysis. Medicine 2017;96(27):e7422. [DOI] [PMC free article] [PubMed] [Google Scholar]
Romano 2013
- Romano A, Calabria LF, Tavanti F, Minniti G, Rossi-Espagnet MC, Coppola V, et al. Apparent diffusion coefficient obtained by magnetic resonance imaging as a prognostic marker in glioblastomas: correlation with MGMT promoter methylation status. European Radiology 2013;23(2):513-20. [DOI] [PubMed] [Google Scholar]
Rosati 2013
- Rosati A, Poliani PL, Todeschini A, Cominelli M, Medicina D, Cenzato M, et al. Glutamine synthetase expression as a valuable marker of epilepsy and longer survival in newly diagnosed glioblastoma multiforme. Neuro-oncology 2013;15(5):618-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Rosenschold 2019
- Rosenschold PM, Law I, Engelholm S, Engelholm SA, Muhic A, Lundemann MJ, et al. Influence of volumetric modulated arc therapy and FET-PET scanning on treatment outcomes for glioblastoma patients. Radiotherapy and Oncology 2019;130:149-55. [DOI] [PubMed] [Google Scholar]
Rubio Fernandez 2014
- Rubio Fernandez A, López Macias M, Toro Zambrano W, Campos de Orellana AM, Catalina Fernandez I, Díaz Delgado M, et al. Study of the methylation status of O-6-methylguanine-DNA methyltransferase in glioblastomas and related overall survival. Virchows Archiv 2014;Suppl 1:S250-1. [Google Scholar]
Sadones 2009
- Sadones J, Michotte A, Veld P, Chaskis C, Sciot R, Menten J, et al. MGMT promoter hypermethylation correlates with a survival benefit from temozolomide in patients with recurrent anaplastic astrocytoma but not glioblastoma. European Journal of Cancer 2009;45(1):146-53. [DOI] [PubMed] [Google Scholar]
Saito 2017a
- Saito T, Sugiyama K, Hama S, Yamasaki F, Takayasu T, Nosaka R, et al. High expression of glypican-1 predicts dissemination and poor prognosis in glioblastomas. World Neurosurgery 2017;105:282-8. [ePPI-R 38859633] [DOI] [PubMed] [Google Scholar]
Saito 2017b
- Saito T, Sugiyama K, Ikawa F, Yamasaki F, Ishifuro M, Takayasu T, et al. Permeability surface area product using perfusion computed tomography is a valuable prognostic factor in glioblastomas treated with radiotherapy plus concomitant and adjuvant temozolomide. World Neurosurgery 2017;97:21-6. [ePPI-R 38859634] [DOI] [PubMed] [Google Scholar]
Saito 2018a
- Saito T, Muragaki Y, Shioyama T, Komori T, Maruyama T, Nitta M, et al. Malignancy index using intraoperative flow cytometry is a valuable prognostic factor for glioblastoma treated with radiotherapy and concomitant temozolomide. Neurosurgery 2018;30:30. [ePPI-R 38859631] [DOI] [PubMed] [Google Scholar]
Saito 2018b
- Saito T, Sugiyama K, Hama S, Yamasaki F, Takayasu T, Nosaka R, et al. Prognostic importance of temozolomide-induced neutropenia in glioblastoma, IDH-wildtype patients. Neurosurgical Review 2018;41(2):621-8. [DOI] [PubMed] [Google Scholar]
Saito 2018c
- Saito T, Sugiyama K, Takeshima Y, Amatya VJ, Yamasaki F, Takayasu T, et al. Prognostic implications of the subcellular localization of survivin in glioblastomas treated with radiotherapy plus concomitant and adjuvant temozolomide. Journal of Neurosurgery 2018;128(3):679-84. [DOI] [PubMed] [Google Scholar]
Salvati 2012
- Salvati M, Pichierri A, Piccirilli M, Floriana Brunetto GM, D'Elia A, Artizzu S, et al. Extent of tumor removal and molecular markers in cerebral glioblastoma: a combined prognostic factors study in a surgical series of 105 patients. Journal of Neurosurgery 2012;117(2):204-11. [DOI] [PubMed] [Google Scholar]
Sana 2014
- Sana J, Radova L, Lakomy R, Kren L, Fadrus P, Smrcka M, et al. Risk score based on microRNA expression signature is independent prognostic classifier of glioblastoma patients. Carcinogenesis 2014;35(12):2756-62. [DOI] [PubMed] [Google Scholar]
Saraiva‐Esperon 2014
- Saraiva-Esperon U, Ruibal A, Herranz M. The contrasting epigenetic role of RUNX3 when compared with that of MGMT and TIMP3 in glioblastoma multiforme clinical outcomes. Journal of the Neurological Sciences 2014;347(1-2):325-31. [DOI] [PubMed] [Google Scholar]
Sasaki 2018
- Sasaki T, Fukai J, Kodama Y, Hirose T, Okita Y, Moriuchi S, et al. Characteristics and outcomes of elderly patients with diffuse gliomas: a multi-institutional cohort study by Kansai Molecular Diagnosis Network for CNS Tumors. Journal of Neuro-oncology 2018;140(2):329-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
Schaich 2009
- Schaich M, Kestel L, Pfirrmann M, Robel K, Illmer T, Kramer M, et al. A MDR1 (ABCB1) gene single nucleotide polymorphism predicts outcome of temozolomide treatment in glioblastoma patients. Annals of Oncology 2009;20(1):175-81. [DOI] [PubMed] [Google Scholar]
Schiffgens 2016
- Schiffgens S, Wilkens L, Brandes AA, Meier T, Franceschi E, Ermani M, et al. Sex-specific clinicopathological significance of novel (Frizzled-7) and established (MGMT, IDH1) biomarkers in glioblastoma. Oncotarget 2016;7(34):55169-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
Schulze Heuling 2017
- Schulze Heuling E, Knab F, Radke J, Eskilsson E, Martinez-Ledesma E, Koch A, et al. Prognostic relevance of tumor purity and interaction with MGMT methylation in glioblastoma. Molecular Cancer Research : MCR 2017;15(5):532-40. [DOI] [PubMed] [Google Scholar]
Shemilt 2019
- Shemilt I, The Campbell and Cochrane Economics Methods Group (CCEMG) and the Evidence for Policy and Practice Information and Coordinating Centre (EPPI-Centre). CCEMG-EPPI-Centre cost converter, version 1.6 (updated April 2019). eppi.ioe.ac.uk/costconversion/ (accessed 30 November 2020).
Shu 2018
- Shu C, Wang Q, Yan X, Wang J. The TERT promoter mutation status and MGMT promoter methylation status, combined with dichotomized MRI-derived and clinical features, predict adult primary glioblastoma survival. Cancer Medicine 2018;7(8):3704-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Sijben 2008
- Sijben AE, McIntyre JB, Roldan GB, Easaw JC, Yan E, Forsyth PA, et al. Toxicity from chemoradiotherapy in older patients with glioblastoma multiforme. Journal of Neuro-oncology 2008;89(1):97-103. [DOI] [PubMed] [Google Scholar]
Singh 2012
- Singh G, Mallick S, Sharma V, Joshi N, Purkait S, Jha P, et al. A study of clinico-pathological parameters and O6-methylguanine DNA methyltransferase (MGMT) promoter methylation status in the prognostication of gliosarcoma. Neuropathology 2012;32(5):534-42. [DOI] [PubMed] [Google Scholar]
Snowsill 2017
- Snowsill T, Coelho H, Huxley N, Jones-Hughes T, Briscoe S, Frayling IM, et al. Molecular testing for Lynch syndrome in people with colorectal cancer: systematic reviews and economic evaluation. Health Technology Assessment (Winchester, England) 2017;21(51):1-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Soike 2018
- Soike MH, McTyre ER, Shah N, Puchalski RB, Holmes JA, Paulsson AK, et al. Glioblastoma radiomics: can genomic and molecular characteristics correlate with imaging response patterns? Neuroradiology 2018;60(10):1043-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
Sterne 2016
- Sterne JAC, Hernán MA, Reeves BC, Savović J, Berkman ND, Viswanathan M, et al. ROBINS-I: a tool for assessing risk of bias in non-randomized studies of interventions. BMJ 2016;355:i4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
Stetson 2016
- Stetson LC, Dazard JE, Barnholtz-Sloan JS. Protein markers predict survival in glioma patients. Molecular & Cellular Proteomics : MCP 2016;15(7):2356-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
Stummer 2012
- Stummer W, Meinel T, Ewelt C, Martus P, Jakobs O, Felsberg J, et al. Prospective cohort study of radiotherapy with concomitant and adjuvant temozolomide chemotherapy for glioblastoma patients with no or minimal residual enhancing tumor load after surgery. Journal of Neuro-oncology 2012;108(1):89-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
Stupp 2005
- Stupp R, Mason WP, den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. New England Journal of Medicine 2005;352:987-96. [DOI] [PubMed] [Google Scholar]
Stupp 2009
- Stupp R, Hegi ME, Mason WP, den Bent MJ, Taphoorn MJ, Janzer RC, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncology 2009;10(5):459-66. [DOI] [PubMed] [Google Scholar]
Stupp 2010
- Stupp R, Hegi ME, Neyns B, Goldbrunner R, Schlegel U, Clement PM, et al. Phase I/IIa study of cilengitide and temozolomide with concomitant radiotherapy followed by cilengitide and temozolomide maintenance therapy in patients with newly diagnosed glioblastoma. Journal of Clinical Oncology 2010;28(16):2712-8. [DOI] [PubMed] [Google Scholar]
Suchorska 2015
- Suchorska B, Jansen NL, Linn J, Kretzschmar H, Janssen H, Eigenbrod S, et al. Biological tumor volume in 18FET-PET before radiochemotherapy correlates with survival in GBM. Neurology 2015;84(7):710-9. [DOI] [PubMed] [Google Scholar]
Sun 2015
- Sun MZ, Oh T, Ivan ME, Clark AJ, Safaee M, Sayegh ET, et al. Survival impact of time to initiation of chemoradiotherapy after resection of newly diagnosed glioblastoma. Journal of Neurosurgery 2015;122(5):1144-50. [DOI] [PubMed] [Google Scholar]
Tanaka 2014
- Tanaka S, Akimoto J, Narita Y, Oka H, Tashiro T. Is the absolute value of O-6-methylguanine-DNA methyltransferase gene messenger RNA a prognostic factor, and does it predict the results of treatment of glioblastoma with temozolomide? Journal of Neurosurgery 2014;121(4):818-26. [DOI] [PubMed] [Google Scholar]
Thon 2011
- Thon N, Eigenbrod S, Grasbon-Frodl EM, Lutz J, Kreth S, Popperl G, et al. Predominant influence of MGMT methylation in non-resectable glioblastoma after radiotherapy plus temozolomide. Journal of Neurology, Neurosurgery, and Psychiatry 2011;82(4):441-6. [DOI] [PubMed] [Google Scholar]
Tierney 2007
- Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials 2007;8:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Tini 2015
- Tini P, Cerase A, Cevenini G, Carbone SF, Miracco C, Pirtoli L. Epidermal growth factor receptor expression may correlate with survival through clinical and radiological features of aggressiveness in glioblastoma treated with radiochemotherapy. Anticancer Research 2015;35(7):4117-24. [PubMed] [Google Scholar]
Tini 2016
- Tini P, Pastina P, Nardone V, Sebaste L, Toscano M, Miracco C, et al. The combined EGFR protein expression analysis refines the prognostic value of the MGMT promoter methylation status in glioblastoma. Clinical Neurology and Neurosurgery 2016;149:15-21. [DOI] [PubMed] [Google Scholar]
Tini 2017
- Tini P, Nardone V, Pastina P, Battaglia G, Miracco C, Sebaste L, et al. Patients affected by unmethylated O(6)-methylguanine-DNA methyltransferase glioblastoma undergoing radiochemotherapy may benefit from moderately dose-escalated radiotherapy. BioMed Research International 2017;2017:9461402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Toms 2018
- Toms SA, Kim CY, Nicholas G, Ram Z. Increased compliance with tumor treating fields therapy is prognostic for improved survival in the treatment of glioblastoma: a subgroup analysis of the EF-14 phase III trial. Journal of Neuro-oncology 2018;141(2):467-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Trabelsi 2016
- Trabelsi S, Mama N, Ladib M, Karmeni N, Haddaji Mastouri M, Chourabi M, et al. MGMT methylation assessment in glioblastoma: MS-MLPA versus human methylation 450K beadchip array and immunohistochemistry. Clinical and Translational Oncology 2016;18(4):391-7. [DOI] [PubMed] [Google Scholar]
Urbschat 2017
- Urbschat S, Sippl C, Engelhardt J, Kammers K, Oertel J, Ketter R. Importance of biomarkers in glioblastomas patients receiving local BCNU wafer chemotherapy. Molecular Cytogenetics 2017;10(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
van Dijken 2019
- Dijken BR, Laar PJ, Li C, Yan JL, Boonzaier NR, Price SJ, et al. Ventricle contact is associated with lower survival and increased peritumoral perfusion in glioblastoma. Journal of Neurosurgery 2019;131:717-23. [DOI] [PubMed] [Google Scholar]
Villani 2015
- Villani V, Casini B, Pace A, Prosperini L, Carapella CM, Vidiri A, et al. The prognostic value of pyrosequencing-detected MGMT promoter hypermethylation in newly diagnosed patients with glioblastoma. Disease Markers 2015;2015:604719. [DOI] [PMC free article] [PubMed] [Google Scholar]
Wang 2014
- Wang K, Wang YY, Ma J, Wang JF, Li SW, Jiang T, et al. Prognostic value of MGMT promoter methylation and TP53 mutation in glioblastomas depends on IDH1 mutation. Asian Pacific Journal of Cancer Prevention 2014;15(24):10893-8. [DOI] [PubMed] [Google Scholar]
Wang 2015a
- Wang X, Zhang K, Chen X, Zhao C, Sun Z. Epilysin is overexpressed in glioblastoma and related to clinical outcome of patients. Medical Oncology 2015;32(1):363. [ePPI-R 38860220] [DOI] [PubMed] [Google Scholar]
Wang 2015b
- Wang M, Dignam JJ, Won M, Curran W, Mehta M, Gilbert MR. Variation over time and interdependence between disease progression and death among patients with glioblastoma on RTOG 0525. Neuro-oncology 2015;17(7):999-1006. [ePPI-R 38860205] [DOI] [PMC free article] [PubMed] [Google Scholar]
Wang 2016
- Wang W, Zhang L, Wang Z, Yang F, Wang H, Liang T, et al. A three-gene signature for prognosis in patients with MGMT promoter-methylated glioblastoma. Oncotarget 2016;7(43):69991-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Watanabe 2011
- Watanabe R, Nakasu Y, Tashiro H, Mitsuya K, Ito I, Nakasu S, et al. O6-methylguanine DNA methyltransferase expression in tumor cells predicts outcome of radiotherapy plus concomitant and adjuvant temozolomide therapy in patients with primary glioblastoma. Brain Tumor Pathology 2011;28(2):127-35. [DOI] [PubMed] [Google Scholar]
Wee 2017
- Wee CW, Kim E, Kim N, Kim IA, Kim TM, Kim YJ, et al. Novel recursive partitioning analysis classification for newly diagnosed glioblastoma: a multi-institutional study highlighting the MGMT promoter methylation and IDH1 gene mutation status. Radiotherapy and Oncology 2017;123(1):106-11. [DOI] [PubMed] [Google Scholar]
Wee 2018
- Wee CW, Kim IH, Park CK, Kim JW, Dho YS, Ohka F, et al. Validation of a novel molecular RPA classification in glioblastoma (GBM-molRPA) treated with chemoradiation: a multi-institutional collaborative study. Radiotherapy and Oncology 2018;129(2):347-51. [DOI] [PubMed] [Google Scholar]
Wei 2017
- Wei KC, Chen CY, Feng LY, Huang WT, Chen CH, Hsu PW, et al. The rs16906252:C>T SNP is not associated with increased overall survival or temozolomide response in a Han-Chinese glioma cohort. PloS One 2017;12(6):e0178842. [DOI] [PMC free article] [PubMed] [Google Scholar]
Weller 2009
- Weller M, Felsberg J, Hartmann C, Berger H, Steinbach JP, Schramm J, et al. Molecular predictors of progression-free and overall survival in patients with newly diagnosed glioblastoma: a prospective translational study of the German Glioma Network. Journal of Clinical Oncology 2009, 2009;27(34):5743-50. [DOI] [PubMed] [Google Scholar]
Weller 2015
- Weller M, Tabatabai G, Kastner B, Felsberg J, Steinbach JP, Wick A, et al. MGMT promoter methylation Is a strong prognostic biomarker for benefit from dose-intensified temozolomide rechallenge in progressive glioblastoma: the DIRECTOR Trial. Clinical Cancer Research 2015;21(9):2057-64. [DOI] [PubMed] [Google Scholar]
Weller 2017a
- Weller M, den Bent M, Tonn JC, Stupp R, Preusser M, Cohen-Jonathan-Moyal E, et al. European Association for Neuro-Oncology (EANO) guideline on the diagnosis and treatment of adult astrocytic and oligodendroglial gliomas. Lancet Oncology 2017;18(6):e315-29. [DOI] [PubMed] [Google Scholar]
Weller 2017b
- Weller M, Butowski N, Tran DD, Recht LD, Lim M, Hirte H, et al. Rindopepimut with temozolomide for patients with newly diagnosed, EGFRvIII-expressing glioblastoma (ACT IV): a randomised, double-blind, international phase 3 trial. Lancet Oncology 2017;18(10):1373-85. [ePPI-R 38860275] [DOI] [PubMed] [Google Scholar]
Wen 2020
- Wen PY, Weller M, Lee EQ, Alexander BM, Barnholtz-Sloan JS, Barthel FP, et al. Glioblastoma in adults: a Society for Neuro-Oncology (SNO) and European Society of Neuro-Oncology (EANO) consensus review on current management and future directions. Neuro-oncology 2020;22(8):1073-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Westphal 2015
- Westphal M, Heese O, Steinbach JP, Schnell O, Schackert G, Mehdorn M, et al. A randomised, open label phase III trial with nimotuzumab, an anti-epidermal growth factor receptor monoclonal antibody in the treatment of newly diagnosed adult glioblastoma. European Journal of Cancer 2015;51(4):522-32. [DOI] [PubMed] [Google Scholar]
Whiting 2011
- Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Annals of Internal Medicine 2011;155(8):529-36. [DOI] [PubMed] [Google Scholar]
Whiting 2016
- Whiting P, Savović J, Higgins JPT, Caldwell DM, Reeves BC, Shea B, et al. ROBIS: a new tool to assess risk of bias in systematic reviews was developed. Journal of Clinical Epidemiology 2016;69:225-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Wick 2012
- Wick W, Platten M, Meisner C, Felsberg J, Tabatabai G, Simon M, et al. Temozolomide chemotherapy alone versus radiotherapy alone for malignant astrocytoma in the elderly: the NOA-08 randomised, phase 3 trial. Lancet Oncology 2012;13(7):707-15. [DOI] [PubMed] [Google Scholar]
Wolff 2019
- Wolff RF, Moons KG, Riley RD, Whiting PF, Westwood M, Collins GS, et al. PROBAST: a tool to assess the risk of bias and applicability of prediction model studies. Annals of Internal Medicine 2019;170(1):51-8. [DOI] [PubMed] [Google Scholar]
Wu 2018
- Wu L, Bernal GM, Cahill KE, Pytel P, Fitzpatrick CA, Mashek H, et al. BCL3 expression promotes resistance to alkylating chemotherapy in gliomas. Science Translational Medicine 2018;10(448):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Yan 2017
- Yan JL, Hoorn A, Larkin TJ, Boonzaier NR, Matys T, Price SJ. Extent of resection of peritumoral diffusion tensor imaging-detected abnormality as a predictor of survival in adult glioblastoma patients. Journal of Neurosurgery 2017;126(1):234-41. [DOI] [PubMed] [Google Scholar]
Yang 2015
- Yang P, Zhang W, Wang Y, Peng X, Chen B, Qiu X, et al. IDH mutation and MGMT promoter methylation in glioblastoma: results of a prospective registry. Oncotarget 2015;6(38):40896-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
Yin 2017
- Yin A, Etcheverry A, He Y, Aubry M, Barnholtz-Sloan J, Zhang L, et al. Integrative analysis of novel hypomethylation and gene expression signatures in glioblastomas. Oncotarget 2017;8(52):89607-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Yin 2018
- Yin AA, Lu N, Etcheverry A, Aubry M, Barnholtz-Sloan J, Zhang LH, et al. A novel prognostic six-CpG signature in glioblastomas. CNS Neuroscience and Therapeutics 2018;24(3):167-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
You 2013
- You WC, Chiou SH, Huang CY, Chiang SF, Yang CL, Sudhakar JN, et al. Mitochondrial protein ATPase family, AAA domain containing 3A correlates with radioresistance in glioblastoma. Neuro-oncology 2013;15(10):1342-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
Younis 2016
- Younis SG, Khedr RA, El-Shorbagy SH. Immunohistochemical analysis of O6-methylguanine-DNA methyltransferase (MGMT) protein expression as prognostic marker in glioblastoma patients treated with radiation therapy with concomitant and adjuvant temozolomide. Journal of Egyptian National Cancer Institute 2016;28(1):23-30. [DOI] [PubMed] [Google Scholar]
Yuan 2017a
- Yuan G, Niu L, Zhang Y, Wang X, Ma K, Yin H, et al. Defining optimal cutoff value of MGMT promoter methylation by ROC analysis for clinical setting in glioblastoma patients. Journal of Neuro-oncology 2017;133(1):193-201. [ePPI-R 38860460] [DOI] [PubMed] [Google Scholar]
Yuan 2017b
- Yuan GQ, Wei NL, Mu LY, Wang XQ, Zhang YN, Zhou WN, et al. A 4-miRNAs signature predicts survival in glioblastoma multiforme patients. Cancer Biomarkers: Section A of Disease Markers 2017;20(4):443-52. [ePPRI-R 38860461] [DOI] [PubMed] [Google Scholar]
Yue 2014
- Yue Q, Zhang X, Ye HX, Wang Y, Du ZG, Yao Y, et al. The prognostic value of Foxp3+ tumor-infiltrating lymphocytes in patients with glioblastoma. Journal of Neuro-oncology 2014;116(2):251-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Zhang 2013
- Zhang K, Wang XQ, Zhou B, Zhang L. The prognostic value of MGMT promoter methylation in glioblastoma multiforme: a meta-analysis. Familial Cancer 2013;12(3):449-58. [DOI] [PubMed] [Google Scholar]
Zhang 2014
- Zhang L, Wang M, Wang W, Mo J. Incidence and prognostic value of multiple gene promoter methylations in gliomas. Journal of Neuro-oncology 2014;116(2):349-56. [DOI] [PubMed] [Google Scholar]
Zhao 2016
- Zhao H, Wang S, Song C, Zha Y, Li L. The prognostic value of MGMT promoter status by pyrosequencing assay for glioblastoma patients' survival: a meta-analysis. World Journal of Surgical Oncology 2016;14:261. [DOI] [PMC free article] [PubMed] [Google Scholar]
Zunarelli 2011
- Zunarelli E, Bigiani N, Sartori G, Migaldi M, Sgambato A, Maiorana A. INI1 immunohistochemical expression in glioblastoma: correlation with MGMT gene promoter methylation status and patient survival. Pathology 2011;43(1):17-23. [DOI] [PubMed] [Google Scholar]
References to other published versions of this review
McAleenan 2019
- McAleenan A, Howell A, Kernohan A, Faulkner CL, Dawson S, Wragg C, et al. Prognostic value of test(s) for O6‐methylguanine–DNA methyltransferase (MGMT) promoter methylation for predicting overall survival in people with glioblastoma treated with temozolomide. Cochrane Database of Systematic Reviews 2019, Issue 4. Art. No: CD013316. [DOI: 10.1002/14651858.CD013316] [DOI] [PMC free article] [PubMed] [Google Scholar]