Abstract
DNA end resection has a key role in double-strand break repair and DNA replication. Defective DNA end resection can cause malfunctions in DNA repair and replication, leading to greater genomic instability. DNA end resection is initiated by MRN-CtIP generating short, 3′-single-stranded DNA (ssDNA). This newly generated ssDNA is further elongated by multiple nucleases and DNA helicases, such as EXO1, DNA2, and BLM. Effective DNA end resection is essential for error-free homologous recombination DNA repair, the degradation of incorrectly replicated DNA and double-strand break repair choice. Because of its importance in DNA repair, DNA end resection is strictly regulated. Numerous mechanisms have been reported to regulate the initiation, extension, and termination of DNA end resection. Here, we review the general process of DNA end resection and its role in DNA replication and repair pathway choice.
Subject terms: Double-strand DNA breaks, Cell signalling
DNA repair: tying up loose ends
Carefully regulated enzymatic processing of the ends of DNA strands is essential for efficient replication and damage repair while also minimizing the risk of genomic instability. Replication and repair depend on a mechanism known as DNA resection, in which enzymes trim back double-stranded DNA ends to leave single-stranded overhangs. Zhenkun Lou and colleagues at the Mayo Clinic in Rochester, USA, have reviewed the various steps involved in the initiation and control of DNA resection. There are multiple different DNA repair processes, and the manner in which resection occurs can determine which of these processes subsequently takes place. The authors note that cancer cells rely heavily on these repair pathways to survive radiotherapy and chemotherapy, and highlight research opportunities that might reveal therapeutically useful vulnerabilities in the resection mechanism.
Introduction
DNA double-strand breaks (DSBs) are seriously harmful genomic lesions that threaten genomic stability and cell survival. Defective DSB repair is associated with embryonic death, aging, immunodeficiency, neurological disorders, and cancer1–3. In response to DSBs, the kinases ATM, ATR, and DNA-PKcs become activated and phosphorylate multiple substrates to initiate the DNA-damage response (DDR), leading to the recruitment of DDR factors to DNA-damage sites, cell cycle arrest, and activation of DNA repair. The two major DSB repair pathways that have been extensively investigated are homologous recombination (HR) and non-homologous end joining (NHEJ). NHEJ functions throughout interphase. Error-free repair by HR requires a homologous sister chromatid as a recombination template and hence is favored in the S and G2 phases.
DNA end resection plays a key role in error-free repair because of its essential role in HR4. DNA end resection initially generates 3′ single-stranded DNA (ssDNA), which provides a platform for recruiting HR repair-related proteins and prevents DNA repair by NHEJ5. For the initiation of DNA end resection, the CtBP-interacting protein (CtIP) functions together with the MRE11–RAD50–NBS1 (MRN) complex to generate a short ssDNA at the DSB ends. After ssDNA is generated by the CtIP/MRN complex, downstream nucleases and helicases, such as exonuclease 1 (EXO1) or DNA replication ATP-dependent helicase/nuclease DNA replication helicase/nuclease 2 (DNA2) and Bloom syndrome protein (BLM), are recruited to extend the 3′-ssDNA for HR-mediated repair6,7. End resection is essential not only for HR repair but also for mediating accurate DNA replication by degrading faulty replication forks and activating the ATR-CHK1 pathway. Another important function for DNA end resection is the regulation of DSB repair pathway choice8. Sufficient end resection is important for RPA complex and RAD51 loading, which are essential for homologous recombination and error-free repair. In fact, blocking DNA end resection leads to NHEJ repair.
In this review, we summarize the general process of DNA end resection and highlight the function of DNA end resection in DNA replication and DSB repair pathway choice.
Initiation of DNA end resection
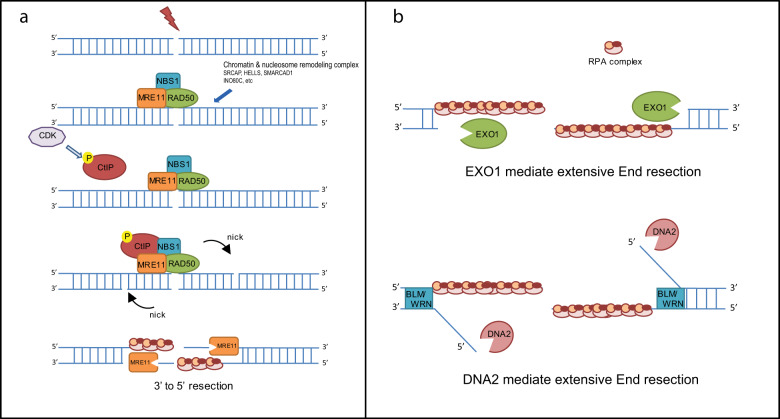
In mammalian cells, the MRN complex consists of three subunits, MRE11, RAD50, and Nibrin (NBS1). All of these components are established regulators of DNA damage signal transduction and DNA end resection initiation. Following DNA damage, the MRN complex is recruited to DSB sites and binds DNA through the RAD50 globular ABC ATPase head domain, which is a tetramer formed by the RAD50 walker A/B ATP-binding motif and MRE11. The extended coiled-coil tail of RAD50 is important for tethering the complex9. This tethering function of RAD50 allows the MRN complex to form bridges between free DNA ends and is an important step in the regulation of DSB signal transduction. ATM dimers and CtIP are then recruited to the DSB by the C terminus and Forkhead-associated (FHA) domain/BRCA1 C terminus (BRCT) domain of NBS1, respectively10,11. Last, MRE11 acts as the catalytic subunit for the initiation of end resection through its nuclease activity4.
MRE11 contains four N-terminal phosphodiesterase motifs that make up the catalytic domain and two DNA-binding domains in the C-terminal region12. MRE11 exhibits both 3′–5′ exonuclease and endonuclease activity in vitro. Both the endonuclease and exonuclease activity of MRE11 have been shown to be important for DNA end resection. During end resection, endonuclease activity generates a nick in double-stranded DNA (dsDNA). Then, the 3′–5′ exonuclease activity generates a 3′-ssDNA overhang from the nick. In vitro experiments have also revealed that the phosphodiesterase motifs of MRE11 are essential for its nuclease activities13.
RAD50 is a structural maintenance of chromosome family member that contains two ATP-binding motifs (Walker A and Walker B) that together exhibit ATPase activity14. The ATPase activity of RAD50 is important for the nuclease activity of MRE1115. A previous publication suggested that the 3′–5′ exonuclease activity of the MRN complex is very limited because RAD50 strongly inhibits the exonuclease activity of MRE11 in the presence of ATP16. ATP binding to RAD50 induces the conformation of RAD50-MRE11 to close, and MRE11 then exhibits mainly endonuclease activity. ATP hydrolysis changes the conformation of the RAD50-MRE11 complex to an open state, allowing MRE11 exonuclease activity17. Thus, RAD50 acts as a molecular switch for controlling MRE11 endonuclease/exonuclease activities.
NBS1 is an important regulator of the MRN complex18. NBS1 contains two adjacent BRCT domains and an FHA domain, which are established phosphorylation residue-binding domains. Thus, NBS1 is regarded as an important adaptor protein for MRN complex function19. NBS1 modulates both the DNA binding and nuclease activity of MRE1120. The FHA domain of NBS1 binds phosphorylated CtIP, which is important for CtIP recruitment to DSB sites21. In addition, the C-terminal domain is important for ATM recruitment10,22,23. These characteristics make NBS1 absolutely indispensable in the mammalian system, which differ from that in yeast where the Xrs2/NBS1 subunit is not required for end resection reaction in vitro24.
CtIP, a cofactor for the MRN complex, also has an essential role in DNA end resection initiation. Using an in vitro system containing purified proteins and DNA substrates with blunt ends, in the absence of CtIP, MRN was unable to stimulate DNA end resection4,25. The function of CtIP is dependent on CtIP phosphorylation in a CDK target motif at Thr-847, which is important for its association with the MRN complex26. Thus, MRE11 endonuclease associated with RAD50, NBS1, and phosphorylated CtIP preferentially generates a short 3′-ssDNA overhang, which is indispensable for extensive DNA end resection and RPA complex loading (Fig. 1a)18,27.
Fig. 1. Initiation and extension of DNA end resection.
a The model of MRN-CtIP and chromatin/nucleosome remodeling proteins in the regulation of DNA end resection initiation. b The model of EXO1, DNA2, BLM, WRN and the RPA complex in the regulation of DNA end resection extension.
Extension of DNA end resection
The MRN complex and CtIP consistently generate a short (~100 nt) 3′-ssDNA overhang28. MRE11 has limited exonuclease activity for producing long 3′-ssDNA overhangs for RPA complex binding. Instead, extensive end resection is performed by EXO14. EXO1 exhibits 5′-to-3′ dsDNA exonuclease and 5′-flap endonuclease activities in vitro29. Interestingly, EXO1 prefers dsDNA substrates with a 3′-ssDNA overhang end, which are produced by the MRN complex and CtIP30,31. Following MRN-CtIP-mediated end resection initiation, EXO1 is recruited to the DSB site by the MRN complex, and the MRN complex stimulates EXO1 nuclease activity in vitro24,26. In addition to EXO1, DNA2 is a key regulator for extensive end resection. DNA2 is a DNA helicase/ssDNA endonuclease that cleaves only free ssDNA ends, with no nuclease effect on dsDNA. It contains a PD-(D/E)XK superfamily nuclease motif and a helicase domain32. In vitro DNA2 exhibits 5ʹ- and 3ʹ-endonuclease activities and DNA helicase activity33,34. However, extensive end resection is dependent on the nuclease activity of DNA2 combined with the helicase activity of the RecQ helicase BLM35. In fact, the DNA helicase BLM is essential for DNA2-mediated extensive end resection. Several studies have shown that DNA2 cleaves ssDNA via BLM-mediated DNA unwinding during end resection36–38.
In addition to BLM, Werner syndrome ATP-dependent helicase (WRN), another RecQ family helicase, also functions in DNA end resection in parallel to BLM39. Loss-of-function mutations in BLM and WRN cause Bloom and Werner syndromes, respectively40. Patients with these syndromes show developmental problems, premature aging, increased genomic instability, and elevated tumorigenesis41. However, the distinct roles of BLM and WRN in DNA end resection are still not clearly understood.
Even though EXO1 and the DNA2/BLM/WRN complex show nuclease and DNA helicase activity, they are insufficient to initiate DNA end resection in the absence of the MRN complex37. Both WRN-DNA2 and BLM-DNA2 require a short 3′-ssDNA overhang to efficiently extend DNA end resection in vitro, which is in agreement with the current model showing that the initial 5′-end trimming is performed by the MRN complex and CtIP, while EXO1 and DNA2/BLM/WRN are critical for more-extensive resection25,36. The RPA complex is also important for promoting the helicase activity of BLM in part by coating unwound ssDNA strands and regulating DNA2 nuclease activity by blocking its 3′–5′ exonuclease activity34,42. RPA depletion eliminates long-range extensive DNA end resection and leads to the loss of the 3′-ssDNA overhang ends generated by MRN-CtIP25. Thus, EXO1, DNA2, BLM, WRN and the RPA complex constitute the minimal complex that can carry out long-range extensive DNA end resection (Fig. 1b)36.
In addition to the DNA end resection machineries (MRN-CtIP, EXO1/DNA2-BLM, and RPA), multiple chromatin remodeling proteins have been reported to regulate the initiation or extension of DNA end resection by relaxing chromatin and thus facilitating access of the core end resection regulators to the broken DNA ends. Dong et al.43 reported that the human SNF2-related CBP activator protein (SRCAP) chromatin remodeling complex, which consists of four subunits, SRCAP, ZNHIT1, Arp6, and YL-1, promotes DNA end resection by enhancing CtIP recruitment and chromatin decondensation in an ATPase-dependent manner. Similarly, lymphoid-specific helicase (also known as SMARCA6, LSH, or PASG)44, a SNF2-like chromatin remodeling ATPase, was also reported to promote DNA end resection by facilitating the accumulation of CtIP at IR-induced foci. The nucleosome remodeling enzyme, SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily A containing DEAD/H box 1 (SMARCAD1, Fun30 in yeast) decreases histone–DNA interactions in nucleosomes flanking DSBs and promotes ssDNA production by the EXO1/DNA2 resection machineries45,46. INO80 complex subunit C (INO80C), another nucleosome remodeler involved in the DSB response, shows two distinct functions during HR: promoting DNA end resection and forming the RAD51 presynaptic filament in both yeast and mammalian cells47,48.
Termination of DNA end resection
Proper initiation of end resection is essential for HR-mediated DNA repair. However, uncontrolled end resection also threatens genomic stability, as hyperresection can cause mutational recombination through microhomology-mediated end joining or single-strand annealing, leading to the loss of genetic information49. Moreover, unlimited end resection may also reduce RPA recycling efficiency in cells, leading to increased ssDNA exposure, replication fork collapse and genomic instability50. Therefore, mechanisms must terminate end resection when the length of ssDNA is sufficient for HR repair, because unlimited extensive end resection is toxic to cells.
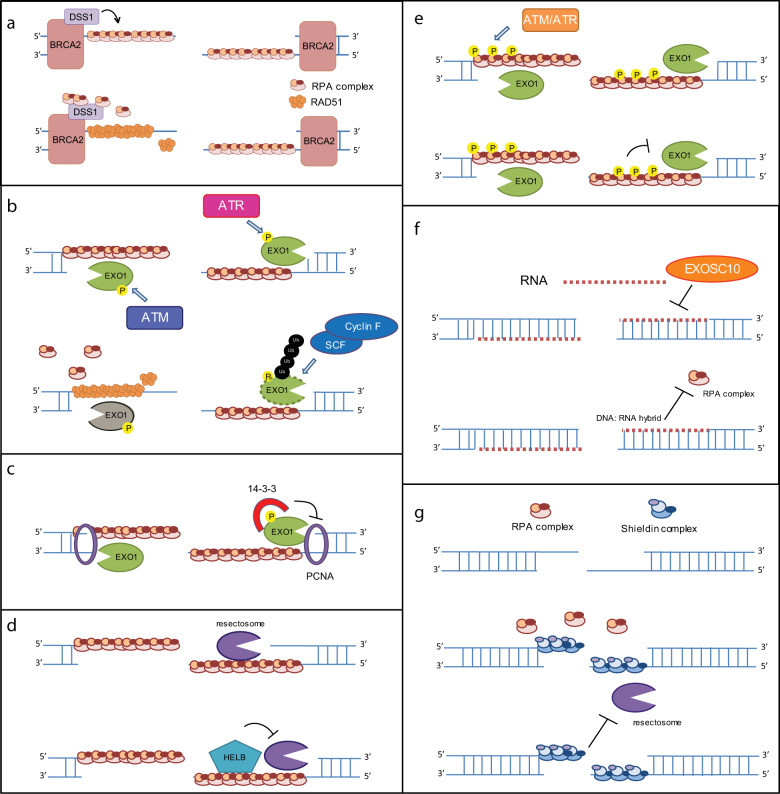
Although the regulation of DNA end resection termination is still not well studied, several mechanisms have been suggested. Under physiological conditions, end resection is terminated by RAD51-RPA switching. This progress is regulated by BRCA2-DSS1. DSS1 is a small (70 residues) and highly acidic protein that functions by ssDNA mimicry to remove RPA from real ssDNA. Then, RAD51 is recruited by BRCA2, to finish completing the switch (Fig. 2a)51.
Fig. 2. Current models of DNA end resection termination.
The regulatory mechanism of DNA end resection termination by BRCA2-DSS1-RAD51 (a), EXO1 phosphorylation/ubiquitination (b), 14-3-3 (c), HELB (d), RPA phosphorylation (e), EXOSC10 (f) and the shieldin complex (g).
Numerous publications show that end resection can be terminated by targeting end resection-regulating proteins. In mammals, EXO1 is rapidly degraded by the Skp1-Cullin1-F-box family of ubiquitin ligases in a proteasome-dependent manner soon after DSB induction52. ATR inhibition attenuates EXO1 degradation following DNA damage, suggesting that ATR-mediated EXO1 phosphorylation further promotes EXO1 degradation (Fig. 2b)52. Another study shows that ATM-mediated phosphorylation of EXO1 appears to regulate the activity of EXO1 following end resection, promoting RPA disassociation and the completion of HR repair (Fig. 2b)53,54. Together, these observations suggest that DNA damage-induced phosphorylation of EXO1 attenuates its function to terminate resection55. Several publications also report that 14-3-3 disrupts the EXO1–PCNA interaction following DNA damage, thus attenuating EXO1 exonuclease activity and terminating end resection Fig. 3. The well-known phosphorylation motif binding protein 14-3-3 may inhibit the EXO1–PCNA interaction in an ATM/ATR-dependent manner (Fig. 2c)56,57. Taken together, these studies suggest that ATM/ATR may be involved in end resection termination through multiple different pathways. In addition to EXO1-dependent end resection termination, other groups have also reported that DNA helicase B (HELB), a DNA helicase translocates ssDNA in the 5′–3′ direction, inhibits the action of the BLM-DNA2 and EXO1 nucleases58. The exact mechanism by which HELB functions remains to be determined (Fig. 2d).
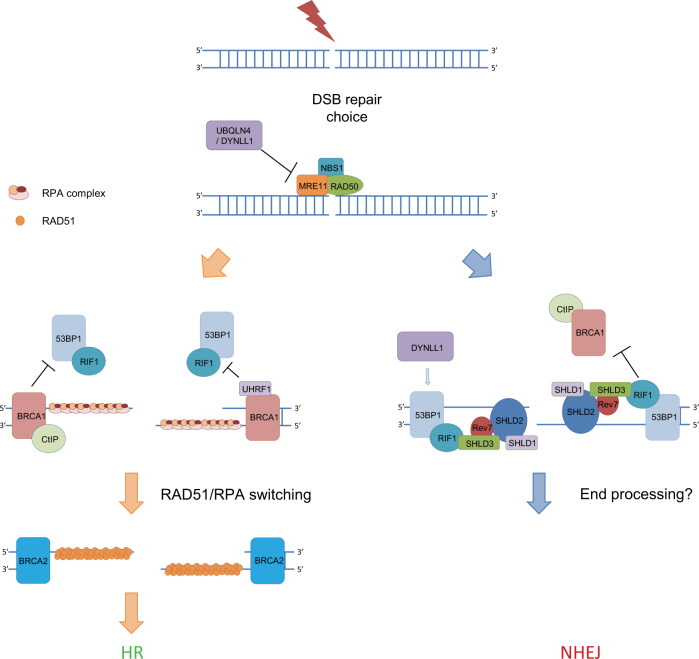
Fig. 3. DNA end resection and DSB repair choice.
The current view of regulators involved in DNA end resection and DSB repair choice.
Recently, Ilya Finkelstein’s group reported that the RPA complex has functions in end resection regulation59. The unphosphorylated RPA complex stimulates the initiation of the BLM-EXO1 and BLM-DNA2 resectosomes and promotes rapid, progressive DNA resection60. However, phosphorylated RPA32 (pRPA32) drastically slows both BLM-EXO1 and BLM-DNA2 resectosomes and stimulates BLM strand switching when the nuclease is omitted from the reaction60. Moreover, BLM-EXO1 and BLM-DNA2 can resect past nucleosomes in the presence of the RPA complex but are blocked when pRPA32 is added to the in vitro reaction system. These findings imply that the RPA complex may stimulate extensive resection early, whereas increased ATM/ATR-mediated RPA complex phosphorylation terminates end resection at a later time point. However, the authors use a heavily phosphorylated RPA32 protein in their in vitro system, (Fig. 2e). End resection termination mediated by phosphorylated RPA needs further evaluation in vivo.
Interestingly, some papers also show that RNAs play roles in end resection termination. DNA damage-induced long noncoding RNAs (dilncRNAs) may block RPA complex recruitment to ssDNA by forming DNA:RNA hybrids61. Exosome component 10, an exosome catalytic subunit, decreases dilncRNA and DNA–RNA hybrid levels, which may facilitate RPA complex recruitment62. However, it is unclear whether RNA-mediated end resection termination is a universal mechanism in cells or if whether it happens at specific regions, such as transcriptively active regions (Fig. 2f).
In addition to these resectosome proteins, some NHEJ proteins also have functions in end resection termination. Several publications show that the shieldin complex is important to inhibit end resection. The shieldin complex consists of four subunits: SHLD3, REV7, SHLD2, and SHLD163. Similar to RPA70, SHLD2 is also an oligonucleotide/oligosaccharide fold domain-containing protein that attenuates end resection by competing with the RPA complex for ssDNA binding and blocks end resection-related nuclease or helicase recruitment (Fig. 2g)64,65.
DNA end resection and DNA replication
DNA end resection is important for precise DNA replication. ssDNA produced by end resection at stalled replication forks activates the RPA-ATR-CHK1 checkpoint and arrests the cell cycle, which is important for fork remodeling and restart. Stalled replication forks are also sensitive to nucleases involved in end resection. Stabilization of stalled replication forks prevents them from collapsing into noxious DSBs, which increases their chance for recovery. Multiple HR factors were reported to regulate the stabilization of replication forks and prevent excess resection at stalled forks, suggesting a tight link between replication fork remodeling and degradation66. In particular, BRCA1 and BRCA2, as well as RAD51, are suggested to be the main regulators for protecting the replication fork from nuclease-mediated degradation.
During replication stress, ssDNA is covered by the RPA complex. The RPA complex promotes replication fork remodeling from three-way junctions to four-way junctions, called fork reversal. Fork reversal helps repair collapsed replication forks and restart stalled replication forks, increasing genomic stability. After a fork is remodeled, the junction of the reversed fork can be targeted by structure-dependent endonucleases67. Studies in yeast and human cells suggest that MRE11 has the key role in the processing and restarting of stalled replication forks68. However, end resection also has a “dark side” that is evident at stalled replication forks. Unrestricted end resection by MRE11 and EXO1 can initiate the degradation of stalled forks, leading to fork collapse69,70. Several mechanisms have been shown to prevent abnormal degradation of reversed strands. Saravanabhavan et al. reported that DNA2 and WRN function together to degrade reversed replication forks with 5′-to-3′ polarity and promote replication restart, thus preventing aberrant processing of unresolved replication intermediates. In addition, ATP-dependent DNA helicase Q1 limits DNA2 activity by preventing extensive nascent strand degradation71. RAD51 has a key role in fork stabilization. RAD51 requires mediator proteins such as BRCA1 and BRCA2 to access the RPA-ssDNA complex72. Recruited BRCA–RAD51 promotes fork reversal formation and protects reversed replication forks from MRE11-, EXO1-, and MUS81-mediated nucleolytic degradation66. However, excessive RAD51 activity slows replication and causes replication forks to stall. David Cortez’s group showed that RAD51 antagonist on the X chromosome (RADX) prevents MUS81-dependent replication fork collapse. Furthermore, RADX competes with RAD51 and prevents excessive replication fork remodeling by antagonizing RAD5173,74. Other factors, such as the biorientation of chromosomes in cell division protein 1-like 1 (BOD1L), protect reversed forks from DNA2-mediated degradation75. Loss of BOD1L confers exquisite cellular sensitivity to replication stress and uncontrolled resection of damaged replication forks because of a failure to stabilize RAD51 at the replication forks. However, no study has shown that BOD1L functions in HR repair. MRN complex-interacting protein (MRNIP), an MRE11 interaction protein, was also reported to function as a replication protector by directly binding with MRE11 and suppressing the exonuclease activity of MRE11 on replication forks76. In contrast to BOD1L, MRNIP reportedly affects the MRN complex-mediated DSB response77. However, it is still not clear whether MRNIP affects end resection in HR repair.
In addition to end resection regulators in HR, NHEJ regulators also show functions in DNA replication. In DSB repair, RIF1 functions together with PTIP and the shieldin complex to promote NHEJ repair by inhibiting DNA end resection. Chirantani et al. reported that RIF1-knockout cells show elevated replication degradation and defective fork restart, which further contribute to genome instability78. Moreover, Lu et al. found that the CST complex (CTC1-STN1-TEN1), a downstream effector of RIF1 in end resection suppression, localizes at stalled replication forks to protect the forks from MRE11-mediated degradation under DNA replication stress79. CST complex deficiency leads to nascent DNA strand degradation, ssDNA accumulation at stalled replication forks and decay during replication restart. In contrast to RIF1 and the CST complex, another end resection antagonist, PTIP, was suggested to play an opposite role in replication fork stability80. PTIP promotes the localization of MRE11 to stalled replication forks in BRCAness (BRCA1/2 deficient) cells, thus promoting MRE11-mediated nascent DNA strand degradation and replication fork collapse. Loss of PTIP rescues the lethality of fork-stalling compounds in BRCA-deficient cells by stabilizing and remodeling stalled replication forks80. This study strongly suggests that the stability of nascent DNA strands can confer drug resistance to chemotherapies in BRCA-deficient cancers, indicating their great importance for cancer therapeutics.
Overall, regulated end resection at the reversed replication fork is beneficial for replication restart by inducing recombination. However, resected ends of reversed replication forks can also form a platform for the recruitment of DSB proteins. These proteins can interfere with replication fork stability, and restoration leads to chromosomal instability through uncontrolled recombination events. Therefore, end resection factors at the reversed fork need to be tightly regulated for proper fork restart and genomic stability.
DNA end resection and DSB repair pathway choice
HR and NHEJ are the two main DSB repair pathways. At present, the pathway choice between HR and NHEJ is of particular interest in the DNA damage field. Part of this choice is dependent on the cell cycle. HR occurs in the S and G2 phases because it needs a template that is provided by the sister chromatin. Conversely, NHEJ can occur throughout interphase. In the current model, BRCA1 and 53BP1 play important roles in repair choice. BRCA1 interacts with CtIP following CDK-mediated S372 phosphorylation7,81. In fact, phosphorylation of CtIP is required for BRCA1 interaction but is dispensable for RPA and RAD51 foci formation in DT40 cells, implying that CtIP-dependent resection does not require interaction with BRCA182. However, cells with a mutated CtIP phosphorylation site still show hypersensitivity to camptothecin and etoposide83. Later, Pablo Huertas’s group proved that CtIP is able to initiate end resection in the absence of BRCA181. BRCA1-CtIP also suppresses the recruitment of RIF1, an essential NHEJ regulator, to DNA damage sites84,85. However, the mechanism for this is still unclear. In addition, our group reported that UHRF1, a ubiquitin E3 ligase, inhibits RIF1 recruitment in a BRCA1-dependent manner. UHRF1 is recruited to DSB sites through the BRCT domain of BRCA1. UHRF1-mediated RIF1 ubiquitination disrupts the 53BP1–RIF1 interaction, thus inhibiting NHEJ and promoting end resection86.
In addition to BRCA1/CtIP, some proteins were also reported to regulate DSB repair choice through the initiation of MRE11-mediated end resection initiation. Dynein light chain LC8 type 1 (DYNLL1) is one protein recently reported to promote NHEJ and inhibit HR87. DYNLL1 was shown to physically interact with MRE11 and inhibit end resection in the presence of MRN, EXO1, DNA2, and BLM in a cell-free system. However, other publications showed that DYNLL1 promotes NHEJ by promoting 53BP1 oligomerization and loading to DSB sites88,89. The proteasomal shuttle factor ubiquilin 4 (UBQLN4) was also reported to regulate DSB repair choice through MRE1190. Upon DNA damage, UBQLN4 is phosphorylated by ATM and interacts with ubiquitylated MRE11 to promote MRE11 degradation. UBQLN4 deficiency causes chromatin retention of MRE11, enhancing nonphysiological HR activity. In contrast, UBQLN4 overexpression represses HR and promotes NHEJ90. Moreover, UBQLN4 is overexpressed in aggressive tumors, leading to deficient HR and conferring sensitivity to PARP inhibitor treatment.
53BP1 works as an antagonist of BRCA1 and has a key role in DSB repair choice. 53BP1 promotes classical non-homologous end joining (c-NHEJ) by recruiting RIF1 to inhibit BRCA1 recruitment to break sites, thereby antagonizing BRCA1-CtIP-mediated end resection and blocking BRCA2/RAD51-mediated homologous recombination91. RIF1 has been shown to inhibit BRCA1 recruitment in the G1 phase, although the mechanism is unclear. This may be one reason that HR is restricted to the S and G2 phase85. In addition to its role in “pre-resection” regulation for repair choice, RIF1 has been reported in recent publications to function in a “post-resection” step by recruiting the shieldin complex to resected DNA ends92. Within the shieldin complex, SHLD3 directly interacts with RIF1 and facilitates the recruitment of REV7, which is an adaptor protein that recruits SHLD264. SHLD2 contains three tandem OB-fold domains that have ssDNA-binding activity. SHLD2 binds to a ssDNA end and protects the ssDNA end from extensive resection by EXO1/DNA2 and consequently inhibits HR63. The knockdown of REV7 or SHLD2 restores PARP inhibitor resistance in BRCA1-deficient cells93. The function of SHLD1 is still not clear. It has been shown that knocking down SHLD1 decreases SHLD2 foci, implying that SHLD1 may stabilize SHLD2 on ssDNA. In addition to protecting the ssDNA end, the shieldin complex was reported by Callen et al. to inhibit RNF168-mediated PALB2-RAD51 loading to DSB sites in BRCA1Δ1153BP1S25A cells (S25A abolishes the PTIP interaction), which suggests that the shieldin complex has dual functions in antagonizing HR94. Recently, one publication suggested that TRIP13, an ATPase, functions as an antagonist to the shieldin complex and promotes HR. TRIP13 physically interacts with REV7 and inhibits NHEJ by changing the closed conformation of REV7 to an open conformation, which disrupts the shieldin complex93. Similar to the shieldin complex, the CST complex is a novel end resection regulator that also functions in DSB repair choice. The mechanism involves CST recruiting DNA polymerase α (polα) to fill in the resected 3′ single-stranded DNA end95. Titia de Lange’s group concluded that CST/polα is a downstream effector of the shieldin complex because CST can be recruited by 53BP1 or SHLD296. CST mainly functions at telomeric regions97. However, Sven Rottenberg’s group thinks that CST/polα is an alternative pathway to shieldin complex activity and functions at nontelomeric regions98. CTC1 is also an OB-fold domain-containing protein, which is similar to RPA1 and SHLD2, and CTC1 is capable of binding to ssDNA independently of SHLD299. Thus, they presumed that the three complexes (RPA, shieldin, CST) likely compete at the 3′ single-stranded DNA end. However, some researchers have suggested that polα only has limited DNA polymerase activity, which might be insufficient to process ssDNA ends64. The detailed mechanism for this regulation still needs further study.
In addition to the shieldin and the CST complex, deletion of PTIP also shows increased DNA end resection, suggesting that PTIP is an antagonist to DNA end resection. However, the distinct role of PTIP and the shieldin/CST complex in DNA end resection attenuation is not very clear. Callen et al. suggested that PTIP preferentially inhibits DNA2-mediated end resection, whereas the shieldin complex prefers to inhibit EXO1-mediated end resection. In this model, the two function independently while also complementing each other94. The PTIP-associated DNA exo/endonuclease Artemis is also reported to function in DSB repair choice. Artemis shows both 5′–3′ exonuclease and endonuclease activity in vitro. Independent of RIF1, Artemis is recruited by 53BP1-PTIP following DNA damage and trims the resected ssDNA with its endonuclease activity, thus promoting NHEJ100.
Concluding remarks
As end resection has an essential role in DSB repair, it also shows significant clinical relevance in cancer therapy. Cancer cells with a mutation or deficiency of a core end resection-regulating gene are more sensitive to radiotherapy and chemotherapy. However, in BRCA1-deficient patients, loss of important end resection antagonists, such as 53BP1, RIF1, and the shieldin complex, leads to enhanced resistance to PARP inhibitor treatment due to the recovery of end resection. Thus, the status of end resection suggests its importance for guiding cancer therapy. Because of its critical role in DNA repair and cancer therapeutics, the mechanism for end resection has been thoroughly studied in the past decade. However, there are still many unanswered questions, several of which we highlight here:
Because of the toxicity induced by hyperresection in cells, how is the termination of end resection determined under physiological conditions?
What are the downstream effectors of 53BP1-RIF1-shieldin that contribute to the inhibition of end resection? As these proteins have no enzymatic activity, how is the resected ssDNA end processed?
What is the role of the shieldin complex, an important antagonist of end resection in DSB repair, in DNA replication?
The answers to these questions will further broaden our current view of end resection, DNA repair, and cancer therapeutics.
Acknowledgements
This work was supported by the National Institutes of Health [RO1CA203971 and RO1CA203561 to Z.L.]. J.A.K. was supported by T32GM65841.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Fei Zhao, Wootae Kim
References
- 1.White RR, Vijg J. Do DNA double-strand bbreaks drive Aging? Mol. Cell. 2016;63:729–738. doi: 10.1016/j.molcel.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gennery AR, Cant AJ, Jeggo PA. Immunodeficiency associated with DNA repair defects. Clin. Exp. Immunol. 2000;121:1–7. doi: 10.1046/j.1365-2249.2000.01257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McKinnon PJ. DNA repair deficiency and neurological disease. Nat. Rev. Neurosci. 2009;10:100–112. doi: 10.1038/nrn2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu T, Huang J. DNA end resection: facts and mechanisms. Genomics Proteom. Bioinformatics. 2016;14:126–130. doi: 10.1016/j.gpb.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huertas P. DNA resection in eukaryotes: deciding how to fix the break. Nat. Struct. Mol. Biol. 2010;17:11–16. doi: 10.1038/nsmb.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huertas P, Jackson SP. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J. Biol. Chem. 2009;284:9558–9565. doi: 10.1074/jbc.M808906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yun MH, Hiom K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009;459:460–463. doi: 10.1038/nature07955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- 9.Longhese MP, Bonetti D, Manfrini N, Clerici M. Mechanisms and regulation of DNA end resection. EMBO J. 2010;29:2864–2874. doi: 10.1038/emboj.2010.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 11.Wang H, et al. The interaction of CtIP and Nbs1 connects CDK and ATM to regulate HR-mediated double-strand break repair. PLoS Genet. 2013;9:e1003277. doi: 10.1371/journal.pgen.1003277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hopfner KP, et al. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell. 2001;105:473–485. doi: 10.1016/s0092-8674(01)00335-x. [DOI] [PubMed] [Google Scholar]
- 13.Tsubouchi H, Ogawa H. A novel mre11 mutation impairs processing of double-strand breaks of DNA during both mitosis and meiosis. Mol. Cell Biol. 1998;18:260–268. doi: 10.1128/mcb.18.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koroleva O, Makharashvili N, Courcelle CT, Courcelle J, Korolev S. Structural conservation of RecF and Rad50: implications for DNA recognition and RecF function. EMBO J. 2007;26:867–877. doi: 10.1038/sj.emboj.7601537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Westmoreland J, et al. RAD50 is required for efficient initiation of resection and recombinational repair at random, gamma-induced double-strand break ends. PLoS Genet. 2009;5:e1000656. doi: 10.1371/journal.pgen.1000656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee JH, et al. Ataxia telangiectasia-mutated (ATM) kinase activity is regulated by ATP-driven conformational changes in the Mre11/Rad50/Nbs1 (MRN) complex. J. Biol. Chem. 2013;288:12840–12851. doi: 10.1074/jbc.M113.460378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Majka J, Alford B, Ausio J, Finn RM, McMurray CT. ATP hydrolysis by RAD50 protein switches MRE11 enzyme from endonuclease to exonuclease. J. Biol. Chem. 2012;287:2328–2341. doi: 10.1074/jbc.M111.307041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Syed A, Tainer JA. The MRE11-RAD50-NBS1 complex conducts the orchestration of damage signaling and outcomes to stress in DNA replication and repair. Annu. Rev. Biochem. 2018;87:263–294. doi: 10.1146/annurev-biochem-062917-012415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams GJ, Lees-Miller SP, Tainer JA. Mre11-Rad50-Nbs1 conformations and the control of sensing, signaling, and effector responses at DNA double-strand breaks. DNA Repair. 2010;9:1299–1306. doi: 10.1016/j.dnarep.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson LH. Recognition, signaling, and repair of DNA double-strand breaks produced by ionizing radiation in mammalian cells: the molecular choreography. Mutat. Res. 2012;751:158–246. doi: 10.1016/j.mrrev.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 21.Chen L, Nievera CJ, Lee AY, Wu X. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J. Biol. Chem. 2008;283:7713–7720. doi: 10.1074/jbc.M710245200. [DOI] [PubMed] [Google Scholar]
- 22.You Z, Chahwan C, Bailis J, Hunter T, Russell P. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol. Cell Biol. 2005;25:5363–5379. doi: 10.1128/MCB.25.13.5363-5379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 24.Oh J, Al-Zain A, Cannavo E, Cejka P, Symington LS. Xrs2 dependent and independent functions of the Mre11-Rad50 complex. Mol. Cell. 2016;64:405–415. doi: 10.1016/j.molcel.2016.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Symington LS. Mechanism and regulation of DNA end resection in eukaryotes. Crit. Rev. Biochem. Mol. Biol. 2016;51:195–212. doi: 10.3109/10409238.2016.1172552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anand R, Ranjha L, Cannavo E, Cejka P. Phosphorylated CtIP functions as a co-factor of the MRE11-RAD50-NBS1 endonuclease in DNA end resection. Mol. Cell. 2016;64:940–950. doi: 10.1016/j.molcel.2016.10.017. [DOI] [PubMed] [Google Scholar]
- 27.Cejka P. DNA end resection: nucleases team up with the right partners to initiate homologous recombination. J. Biol. Chem. 2015;290:22931–22938. doi: 10.1074/jbc.R115.675942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamarche BJ, Orazio NI, Weitzman MD. The MRN complex in double-strand break repair and telomere maintenance. FEBS Lett. 2010;584:3682–3695. doi: 10.1016/j.febslet.2010.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson DM, 3rd, et al. Hex1: a new human Rad2 nuclease family member with homology to yeast exonuclease 1. Nucleic Acids Res. 1998;26:3762–3768. doi: 10.1093/nar/26.16.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shim EY, et al. Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J. 2010;29:3370–3380. doi: 10.1038/emboj.2010.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cannavo E, Cejka P, Kowalczykowski SC. Relationship of DNA degradation by Saccharomyces cerevisiae exonuclease 1 and its stimulation by RPA and Mre11-Rad50-Xrs2 to DNA end resection. Proc. Natl Acad. Sci. USA. 2013;110:E1661–E1668. doi: 10.1073/pnas.1305166110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steczkiewicz K, Muszewska A, Knizewski L, Rychlewski L, Ginalski K. Sequence, structure and functional diversity of PD-(D/E)XK phosphodiesterase superfamily. Nucleic Acids Res. 2012;40:7016–7045. doi: 10.1093/nar/gks382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levikova M, Pinto C, Cejka P. The motor activity of DNA2 functions as an ssDNA translocase to promote DNA end resection. Genes Dev. 2017;31:493–502. doi: 10.1101/gad.295196.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou, C., Pourmal, S. & Pavletich, N. P. Dna2 nuclease-helicase structure, mechanism and regulation by Rpa. Elife4, e09832 (2015). [DOI] [PMC free article] [PubMed]
- 35.Levikova M, Klaue D, Seidel R, Cejka P. Nuclease activity of Saccharomyces cerevisiae Dna2 inhibits its potent DNA helicase activity. Proc. Natl Acad. Sci. USA. 2013;110:E1992–E2001. doi: 10.1073/pnas.1300390110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nimonkar AV, et al. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011;25:350–362. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sturzenegger A, et al. DNA2 cooperates with the WRN and BLM RecQ helicases to mediate long-range DNA end resection in human cells. J. Biol. Chem. 2014;289:27314–27326. doi: 10.1074/jbc.M114.578823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pinto, C., Kasaciunaite, K., Seidel, R. & Cejka, P. Human DNA2 possesses a cryptic DNA unwinding activity that functionally integrates with BLM or WRN helicases. Elife5, e18574 (2016). [DOI] [PMC free article] [PubMed]
- 39.Bernstein KA, Gangloff S, Rothstein R. The RecQ DNA helicases in DNA repair. Annu. Rev. Genet. 2010;44:393–417. doi: 10.1146/annurev-genet-102209-163602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cunniff C, Bassetti JA, Ellis NA. Bloom’s syndrome: clinical spectrum, molecular pathogenesis, and cancer predisposition. Mol. Syndromol. 2017;8:4–23. doi: 10.1159/000452082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Renty C, Ellis NA. Bloom’s syndrome: why not premature aging?: a comparison of the BLM and WRN helicases. Ageing Res. Rev. 2017;33:36–51. doi: 10.1016/j.arr.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qin, Z. et al. Human RPA activates BLM’s bidirectional DNA unwinding from a nick. Elife9, e54098 (2020). [DOI] [PMC free article] [PubMed]
- 43.Dong S, et al. The human SRCAP chromatin remodeling complex promotes DNA-end resection. Curr. Biol. 2014;24:2097–2110. doi: 10.1016/j.cub.2014.07.081. [DOI] [PubMed] [Google Scholar]
- 44.Kollarovic G, Topping CE, Shaw EP, Chambers AL. The human HELLS chromatin remodelling protein promotes end resection to facilitate homologous recombination and contributes to DSB repair within heterochromatin. Nucleic Acids Res. 2020;48:1872–1885. doi: 10.1093/nar/gkz1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chakraborty S, et al. SMARCAD1 phosphorylation and ubiquitination are required for resection during DNA double-strand break repair. iScience. 2018;2:123–135. doi: 10.1016/j.isci.2018.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen X, et al. The Fun30 nucleosome remodeller promotes resection of DNA double-strand break ends. Nature. 2012;489:576–580. doi: 10.1038/nature11355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gospodinov A, et al. Mammalian Ino80 mediates double-strand break repair through its role in DNA end strand resection. Mol. Cell Biol. 2011;31:4735–4745. doi: 10.1128/MCB.06182-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lademann CA, Renkawitz J, Pfander B, Jentsch S. The INO80 complex removes H2A.Z to promote presynaptic filament formation during homologous recombination. Cell Rep. 2017;19:1294–1303. doi: 10.1016/j.celrep.2017.04.051. [DOI] [PubMed] [Google Scholar]
- 49.Truong LN, et al. Microhomology-mediated end joining and homologous recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc. Natl Acad. Sci. USA. 2013;110:7720–7725. doi: 10.1073/pnas.1213431110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen H, Lisby M, Symington LS. RPA coordinates DNA end resection and prevents formation of DNA hairpins. Mol. Cell. 2013;50:589–600. doi: 10.1016/j.molcel.2013.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao W, et al. Promotion of BRCA2-dependent homologous recombination by DSS1 via RPA targeting and DNA mimicry. Mol. Cell. 2015;59:176–187. doi: 10.1016/j.molcel.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tomimatsu N, et al. DNA-damage-induced degradation of EXO1 exonuclease limits DNA end resection to ensure accurate DNA repair. J. Biol. Chem. 2017;292:10779–10790. doi: 10.1074/jbc.M116.772475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bolderson E, et al. Phosphorylation of Exo1 modulates homologous recombination repair of DNA double-strand breaks. Nucleic Acids Res. 2010;38:1821–1831. doi: 10.1093/nar/gkp1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kijas AW, et al. ATM-dependent phosphorylation of MRE11 controls extent of resection during homology directed repair by signalling through exonuclease 1. Nucleic Acids Res. 2015;43:8352–8367. doi: 10.1093/nar/gkv754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tomimatsu N, et al. Phosphorylation of EXO1 by CDKs 1 and 2 regulates DNA end resection and repair pathway choice. Nat. Commun. 2014;5:3561. doi: 10.1038/ncomms4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Andersen SD, et al. 14-3-3 checkpoint regulatory proteins interact specifically with DNA repair protein human exonuclease 1 (hEXO1) via a semi-conserved motif. DNA Repair. 2012;11:267–277. doi: 10.1016/j.dnarep.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen X, et al. 14-3-3 proteins restrain the Exo1 nuclease to prevent overresection. J. Biol. Chem. 2015;290:12300–12312. doi: 10.1074/jbc.M115.644005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tkac J, et al. HELB is a feedback inhibitor of DNA end resection. Mol. Cell. 2016;61:405–418. doi: 10.1016/j.molcel.2015.12.013. [DOI] [PubMed] [Google Scholar]
- 59.Soniat MM, Myler LR, Kuo HC, Paull TT, Finkelstein IJ. RPA Phosphorylation Inhibits DNA Resection. Mol. Cell. 2019;75:145–153. e145. doi: 10.1016/j.molcel.2019.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Myler LR, Finkelstein IJ. Eukaryotic resectosomes: a single-molecule perspective. Prog. Biophys. Mol. Biol. 2017;127:119–129. doi: 10.1016/j.pbiomolbio.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thapar, R. Regulation of DNA double-strand break repair by non-coding RNAs. Molecules23, 2789 (2018). [DOI] [PMC free article] [PubMed]
- 62.Domingo-Prim J, et al. EXOSC10 is required for RPA assembly and controlled DNA end resection at DNA double-strand breaks. Nat. Commun. 2019;10:2135. doi: 10.1038/s41467-019-10153-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Setiaputra, D. & Durocher, D. Shieldin - the protector of DNA ends. EMBO Rep20, e47560 (2019). [DOI] [PMC free article] [PubMed]
- 64.Liang L, et al. Molecular basis for assembly of the shieldin complex and its implications for NHEJ. Nat. Commun. 2020;11:1972. doi: 10.1038/s41467-020-15879-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Scully R, Panday A, Elango R, Willis NA. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol. 2019;20:698–714. doi: 10.1038/s41580-019-0152-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Quinet A, Lemacon D, Vindigni A. Replication fork reversal: players and guardians. Mol. Cell. 2017;68:830–833. doi: 10.1016/j.molcel.2017.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Neelsen KJ, Lopes M. Replication fork reversal in eukaryotes: from dead end to dynamic response. Nat. Rev. Mol. Cell Biol. 2015;16:207–220. doi: 10.1038/nrm3935. [DOI] [PubMed] [Google Scholar]
- 68.Trenz K, Smith E, Smith S, Costanzo V. ATM and ATR promote Mre11 dependent restart of collapsed replication forks and prevent accumulation of DNA breaks. EMBO J. 2006;25:1764–1774. doi: 10.1038/sj.emboj.7601045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lemacon D, et al. MRE11 and EXO1 nucleases degrade reversed forks and elicit MUS81-dependent fork rescue in BRCA2-deficient cells. Nat. Commun. 2017;8:860. doi: 10.1038/s41467-017-01180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sidorova J. A game of substrates: replication fork remodeling and its roles in genome stability and chemo-resistance. Cell Stress. 2017;1:115–133. doi: 10.15698/cst2017.12.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thangavel S, et al. DNA2 drives processing and restart of reversed replication forks in human cells. J. Cell Biol. 2015;208:545–562. doi: 10.1083/jcb.201406100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bhat KP, Cortez D. RPA and RAD51: fork reversal, fork protection, and genome stability. Nat. Struct. Mol. Biol. 2018;25:446–453. doi: 10.1038/s41594-018-0075-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dungrawala H, et al. RADX promotes genome stability and modulates chemosensitivity by regulating RAD51 at replication forks. Mol. Cell. 2017;67:374–386. e375. doi: 10.1016/j.molcel.2017.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bhat KP, et al. RADX modulates RAD51 activity to control replication fork protection. Cell Rep. 2018;24:538–545. doi: 10.1016/j.celrep.2018.06.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Higgs MR, Stewart GS. Protection or resection: BOD1L as a novel replication fork protection factor. Nucleus. 2016;7:34–40. doi: 10.1080/19491034.2016.1143183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bennett LG, et al. MRNIP is a replication fork protection factor. Sci. Adv. 2020;6:eaba5974. doi: 10.1126/sciadv.aba5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Staples CJ, et al. MRNIP/C5orf45 interacts with the MRN complex and contributes to the DNA damage response. Cell Rep. 2016;16:2565–2575. doi: 10.1016/j.celrep.2016.07.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mukherjee C, et al. RIF1 promotes replication fork protection and efficient restart to maintain genome stability. Nat. Commun. 2019;10:3287. doi: 10.1038/s41467-019-11246-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lyu, X. et al. Human CST complex protects replication fork stability by directly blocking MRE11 degradation of nascent strand DNA. bioRxiv797647 (2019). [DOI] [PMC free article] [PubMed]
- 80.Ray Chaudhuri A, et al. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature. 2016;535:382–387. doi: 10.1038/nature18325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cruz-Garcia A, Lopez-Saavedra A, Huertas P. BRCA1 accelerates CtIP-mediated DNA-end resection. Cell Rep. 2014;9:451–459. doi: 10.1016/j.celrep.2014.08.076. [DOI] [PubMed] [Google Scholar]
- 82.Reczek CR, Szabolcs M, Stark JM, Ludwig T, Baer R. The interaction between CtIP and BRCA1 is not essential for resection-mediated DNA repair or tumor suppression. J. Cell Biol. 2013;201:693–707. doi: 10.1083/jcb.201302145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sartori AA, et al. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Escribano-Diaz C, et al. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol. Cell. 2013;49:872–883. doi: 10.1016/j.molcel.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 85.Daley JM, Sung P. RIF1 in DNA break repair pathway choice. Mol. Cell. 2013;49:840–841. doi: 10.1016/j.molcel.2013.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang H, et al. A cell cycle-dependent BRCA1-UHRF1 cascade regulates DNA double-strand break repair pathway choice. Nat. Commun. 2016;7:10201. doi: 10.1038/ncomms10201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.He YJ, et al. DYNLL1 binds to MRE11 to limit DNA end resection in BRCA1-deficient cells. Nature. 2018;563:522–526. doi: 10.1038/s41586-018-0670-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Becker JR, et al. The ASCIZ-DYNLL1 axis promotes 53BP1-dependent non-homologous end joining and PARP inhibitor sensitivity. Nat. Commun. 2018;9:5406. doi: 10.1038/s41467-018-07855-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.West KL, et al. LC8/DYNLL1 is a 53BP1 effector and regulates checkpoint activation. Nucleic Acids Res. 2019;47:6236–6249. doi: 10.1093/nar/gkz263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jachimowicz RD, et al. UBQLN4 represses homologous recombination and is overexpressed in aggressive tumors. Cell. 2019;176:505–519. doi: 10.1016/j.cell.2018.11.024. [DOI] [PubMed] [Google Scholar]
- 91.Zimmermann M, de Lange T. 53BP1: pro choice in DNA repair. Trends Cell Biol. 2014;24:108–117. doi: 10.1016/j.tcb.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Di Virgilio M, et al. Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science. 2013;339:711–715. doi: 10.1126/science.1230624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Clairmont CS, et al. TRIP13 regulates DNA repair pathway choice through REV7 conformational change. Nat. Cell Biol. 2020;22:87–96. doi: 10.1038/s41556-019-0442-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Callen E, et al. 53BP1 enforces distinct pre- and post-resection blocks on homologous recombination. Mol. Cell. 2020;77:26–38. e27. doi: 10.1016/j.molcel.2019.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jachimowicz RD, Goergens J, Reinhardt HC. DNA double-strand break repair pathway choice - from basic biology to clinical exploitation. Cell Cycle. 2019;18:1423–1434. doi: 10.1080/15384101.2019.1618542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mirman Z, et al. 53BP1-RIF1-shieldin counteracts DSB resection through CST- and Polalpha-dependent fill-in. Nature. 2018;560:112–116. doi: 10.1038/s41586-018-0324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rice C, Skordalakes E. Structure and function of the telomeric CST complex. Comput. Struct. Biotechnol. J. 2016;14:161–167. doi: 10.1016/j.csbj.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Barazas M, et al. The CST complex mediates end protection at double-strand breaks and promotes PARP inhibitor sensitivity in BRCA1-deficient cells. Cell Rep. 2018;23:2107–2118. doi: 10.1016/j.celrep.2018.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Flynn RL, Zou L. Oligonucleotide/oligosaccharide-binding fold proteins: a growing family of genome guardians. Crit. Rev. Biochem. Mol. Biol. 2010;45:266–275. doi: 10.3109/10409238.2010.488216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang J, et al. PTIP associates with Artemis to dictate DNA repair pathway choice. Genes Dev. 2014;28:2693–2698. doi: 10.1101/gad.252478.114. [DOI] [PMC free article] [PubMed] [Google Scholar]